Abstract
Monoclonal antibodies (mAbs) have revolutionized the treatment of several human diseases, including cancer and autoimmunity and inflammatory conditions, and represent a new frontier for the treatment of infectious diseases. In the last 20 years, innovative methods have allowed the rapid isolation of mAbs from convalescent subjects, humanized mice, or libraries assembled in vitro and have proven that mAbs can be effective countermeasures against emerging pathogens. During the past year, an unprecedentedly large number of mAbs have been developed to fight coronavirus disease 2019 (COVID-19). Lessons learned from this pandemic will pave the way for the development of more mAb-based therapeutics for other infectious diseases. Here, we provide an overview of SARS-CoV-2-neutralizing mAbs, including their origin, specificity, structure, antiviral and immunological mechanisms of action, and resistance to circulating variants, as well as a snapshot of the clinical trials of approved or late-stage mAb therapeutics.
Keywords: monoclonal antibody, COVID-19, neutralization, SARS-CoV-2, therapeutics
Intense efforts have been made to develop or identify drugs to treat people with COVID-19. Monoclonal antibodies are one of the few types of drugs that have shown efficacy in the clinic.
Monoclonal antibodies in infectious disease
Serotherapy of infectious diseases
Serotherapy refers to the therapeutic transfusion of blood serum from either previous human survivors of a disease or animals that have been immunized against specific organisms in order to transfer passive immunity. The use of serotherapy in infectious disease medicine was pioneered in 1890 by von Behring and Kitasato to combat tetanus and diphtheria (Behring and Kitasato, 2013) and led to the development of diphtheria antitoxin in 1894. This product is still in use and contributed to the dramatic reduction of mortality due to diphtheria many years before the advent of the diphtheria vaccine in the late 1940s. Twenty years after the first use of antitoxin, convalescent serum from survivors of the 1918 Spanish flu was used to treat pneumonia, showing a modest level of efficacy when administered early after symptom onset (Redden, 1919). In the following years until the present day, the efficacy of convalescent plasma for the treatment of viral respiratory infections was probed in several studies that have shown some evidence for a reduction in mortality, especially when given early after symptom onset (Mair-Jenkins et al., 2015). However, these studies did not formally show efficacy due to the moderate risk of bias and lack of control groups. The largest trial assessing the efficacy of convalescent plasma was recently conducted in the United Kingdom (Randomized Evaluation of COVID-19 Therapy [RECOVERY] trial) in which a range of potential treatments for coronavirus disease 2019 (COVID-19) are being tested side by side in hospitalized patients. In mid-January 2021, recruitment for the convalescent plasma arm of the trial was closed due to futility. The data monitoring committee reviewed all trial data, including 1,873 reported deaths among 10,406 randomized patients, and concluded no significant difference in the primary endpoint of mortality. The analysis of the complete results will be important to understand whether convalescent plasma has any therapeutic role in particular patient subgroups. Indeed, in two other recent studies, convalescent plasma was shown to be a valuable treatment option for hospitalized patients if provided early in the course of disease before patients require ventilation or are admitted to the intensive care unit (ICU) (Ma et al., 2021; Tworek et al., 2021). One important limitation of convalescent plasma therapy is the difficulty in standardizing the neutralizing potency of plasma doses and the overall modest-to-low titers of neutralizing antibodies that are administered, suggesting that monoclonal antibody (mAb)-based approaches are better suited for patient treatment.
Introduction to mAbs
A mAb is defined as an antibody derived from a single B cell clone and recognizes a single and unique epitope. Monoclonal antibodies were generated for the first time in 1975 in mice using the hybridoma technology, leading to the first licensed mAb in 1986 (muronomab against human CD3) used primarily to prevent kidney transplant rejection. Upon binding to their cognate epitope on target antigens, mAbs can mediate multiple effects such as disruption of the function of the targeted antigen and elimination of cells or pathogens. In many cases, the function of mAbs is mediated not only by the binding to the target antigen by the antigen-binding fragment (Fab) but also by the recruitment of immune cells or serum complement through the fragment crystallizable (Fc) portion, known as effector functions.
These general concepts hold true for antiviral neutralizing mAbs that act through multiple mechanisms. Neutralizing antibodies can be effective in vivo by targeting free virus and virally infected cells through a range of mechanisms, including direct blocking of viral entry (neutralization), mAb-mediated effector functions, or indirect blocking of viral entry by cross-linking virions, inactivating the viral entry glycoprotein (e.g., receptor mimicry) (Walls et al., 2019), preventing egress of virus from infected cells, or blocking cell-to-cell spread of virus (e.g., blocking the formation of cellular syncytia).
The advent of neutralizing mAbs targeting RSV in infants
The first attempt to develop a prophylactic approach to prevent respiratory syncytial virus (RSV)-induced disease in children was in 1966 and relied on the use of a formalin-inactivated vaccine. This approach failed and was even detrimental to patients, resulting in vaccine-associated enhanced respiratory disease (Kim et al., 1969), possibly associated with the elicitation of high levels of non-neutralizing antibodies. Despite this vaccine failure, in 1998, an RSV-neutralizing mAb (palivizumab) was shown to protect at-risk infants from RSV-induced severe disease (IMpact-RSV Study Group, 1998), demonstrating a beneficial, rather than detrimental, effect by neutralizing antibodies, even in a vulnerable population like premature newborns. Palivizumab became the first mAb approved to combat an infectious disease. The prophylactic use of palivizumab in at-risk adults was not explored due to the prohibitive cost of this mAb, as pricing was based on the body weight in infants (Georgescu and Chemaly, 2009). Palivizumab was also tested in therapeutic settings in at-risk adults, showing only modest efficacy in some studies, in agreement with the concept that antibodies are relatively ineffective when administered late after establishment of infection. Palivizumab was developed as monotherapy and has been proven to work for more than 20 years despite the fact that viral escape is readily observed in vitro (Zhao et al., 2004; Zhu et al., 2011). However, use in a larger population could potentially play a role in driving the selection of viral resistance, as has been seen with oseltamivir for influenza A virus (IAV).
The seasonality of viral infections such as RSV and influenza virus heralded mAb engineering strategies to extend half-life. This allows decreasing the frequency of dosing and potentially retaining effective antibody concentrations in patients throughout a season along with decreasing the cost of treatments. In the following years after palivizumab was approved, two additional RSV mAbs (motavizumab and suptavumab) were evaluated in phase 3 clinical trials but safety and resistance concerns halted further development in 2010 and 2018, respectively (Carbonell-Estrany et al., 2010; Simões et al., 2020). Another highly potent RSV mAb (nirsevimab) harboring an Fc fragment engineered for half-life extension recently showed a 78% reduction in hospitalization in pre-term infants in a phase 2b study (Griffin et al., 2020; Zhu et al., 2017). Nirsevimab is now being tested as a single-injection regimen in a phase 3 study expanded to all-term infants. Its success may revolutionize the prophylactic use of mAbs to a very large population, possibly reducing RSV-associated mortality and hospitalization globally. The development of mAbs against RSV is a paradigm for the field of anti-infective mAbs, with failures and successes highlighting the complexity of developing first-generation and next-generation mAbs against a given pathogen. In addition, this example illustrates how a successful product can pave the way for developing better mAbs in terms of efficacy, cost, and an expanded target population.
The development of neutralizing mAbs against Ebola, IAV, and HIV-1
Ebola and IAV are responsible for acute infections, for which mAbs have been developed clinically, with successes and failures. Two mAb-based products to treat Ebola virus disease have been approved in 2020, and their efficacy in reducing mortality in infected individuals represents a key medical milestone. These mAbs, along with a recently approved vaccine, will contribute to controlling the recurrent outbreaks of Ebola virus disease in Central and West Africa. These two products are based on a monotherapy (ansuvimab) and a cocktail of three mAbs (atoltivimab, maftivimab, and odesivimab). Both have shown similar efficacy in a randomized control trial whereas two other products tested in parallel failed, including one product based on a cocktail of three mAbs (ZMapp) (Mulangu et al., 2019). These results provide important insights in the development of anti-infective mAbs, including that (1) mAbs can be used to treat severe disease and not just in a prevention setting, (2) monotherapy can be successful (i.e., the use of cocktails is not the only suitable approach), and (3) not all mAb-based products are effective, even when cocktails are used.
In the case of influenza, all mAbs that have been tested in clinical trials so far are directed to the stem region of IAV hemagglutinin (HA), except for one directed to matrix protein 2 (M2). Modest efficacy was observed in clinical studies where participants were intentionally infected with IAV (the so-called human challenge model), while no signs of efficacy were observed when trials were run in outpatient settings with uncomplicated influenza A infection or in hospitalized patients (Ali et al., 2018; Corti et al., 2017; Hershberger et al., 2019; Lim et al., 2020). Peak viral load and symptoms onset typically occur 2–5 days after influenza virus infection. In these studies, enrollment included individuals having experienced symptoms for ≤5 days, which would imply that individuals may have already been infected for ≥7 days before the mAbs were administered. Indeed, the only indication of modest efficacy was observed in outpatients when a cocktail of two mAbs (CT-P27) was administered early after symptom onset (≤48 h), similarly to the recommended use of the approved antiviral oseltamivir (H. Yang et al., 2019, 29th ECCMID, abstract). There is not yet evidence of efficacy of HA stem mAbs in a prophylactic setting. In this regard, the most advanced program is represented by VIR-2482, a half-life-extended version of the previously identified MEDI8852 mAb, which recognizes a conserved epitope on the HA stem shared by all 18 IAV subtypes (Kallewaard et al., 2016). This study will assess VIR-2482 for immune prophylaxis in outpatients and could be pivotal for the field to investigate if stem-directed HA mAbs can be effective in prophylaxis, as indirectly suggested by a serological study in humans (Ng et al., 2019). The breadth of reactivity of HA stem mAbs toward all IAVs represents an opportunity to develop an off-the-shelf approach to prepare for future influenza A pandemics.
Finally, several mAbs are under investigation to prevent or treat human immunodeficiency virus 1 (HIV-1) infection. A comprehensive and updated overview is provided by Nussenzweig and Barouch to reflect recent progress in the field (Caskey et al., 2019; Julg and Barouch, 2021). These mAbs are broadly neutralizing and target a subset of vulnerability sites on the HIV-1 envelope (i.e., CD4-binding site, base of the V3 loop, membrane proximal external region, and V1/V2 region). Results in non-human primates suggest that broadly neutralizing mAbs can be effective in prophylaxis against highly diverse circulating HIV-1 strains. These data have prompted the development of some of these mAbs, such as VRC01, for prophylaxis in humans. Corey and co-authors have recently shown in two trials that administration of VRC01 did not prevent HIV-1 infection (Corey et al., 2021). However, 75% protection was observed in subjects exposed to HIV-1 isolates that were potently neutralized in vitro by VRC01, suggesting that prophylaxis of HIV-1 could be achieved by using more potent mAbs with broader coverage of field isolates. In a therapeutic setting, preclinical work showed that in contrast to monotherapy, a combination of mAbs provided prolonged control of infection (using simian-HIV models in macaques), with no evidence of escape. It was hypothesized that this control was mediated by T cell immunity (Barouch et al., 2013; Schoofs et al., 2016) and boosted by viral antigen presentation through immune complexes formed between HIV-1 envelope glycoprotein and the infused mAbs, through the so-called antibody vaccinal effect. Human trials showed that monotherapy only transiently controlled virus replication, with the rapid emergence of escape variants mirroring the results obtained with small-molecule drugs. Combination therapy was far more effective than monotherapy in viremic individuals, but it did not completely suppress viremia in participants with high baseline viral loads. The combination of two broadly neutralizing mAbs was shown to be effective in maintaining prolonged suppression after antiretroviral therapy interruption (Mendoza et al., 2018). Engineered mAbs with dual or triple specificities, as well as engineered Fc to modulate the adaptive immune response (i.e., vaccinal effect), are under development, and results in the coming years will inform on the potential of HIV-1 broadly neutralizing mAbs to provide sustained remission or represent an ultimate cure for HIV-1-infected patients. The lessons from HIV-1 trials may be relevant for the development of COVID-19 mAbs in that coverage of highly divergent co-circulating viruses is key in the long-term and that in therapeutic settings, a combination of mAbs, even if broadly reactive, might be required to prevent viral escape during treatment. However, this may not necessarily apply to acute viral infections and be a consideration in the treatment of prolonged infections in immunocompromised individuals, such as has been observed with COVID-19 (Avanzato et al., 2020; Choi et al., 2020; Kemp et al., 2021; McCarthy et al., 2021). Reciprocally, the HIV-1 field might benefit from the knowledge that will be gained from deployment of COVID-19 mAbs, particularly from use of an antibody (VIR-7832, described below) that was Fc engineered to leverage the so-called vaccinal effect (i.e., the boost of T cell responses during infection), as shown for anti-IAV mAbs in transgenic human Fcγ receptor mice (Bournazos et al., 2020a).
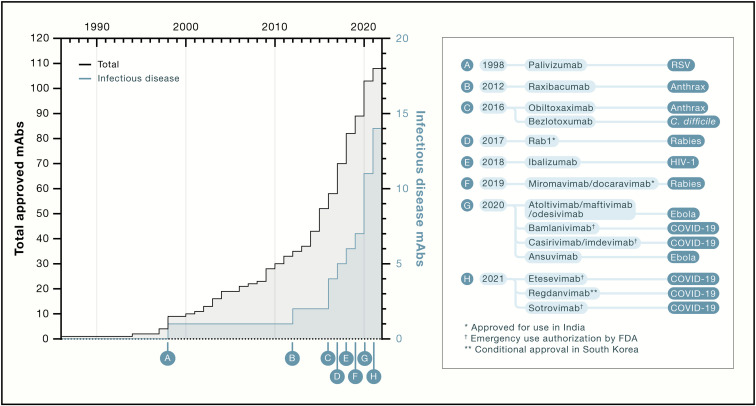
A total of 14 mAbs are now available for use against infectious diseases, and 7 of them were approved or authorized for emergency use in 2020 and 2021 (Figure 1 ). This might be just the beginning of an even larger wave of approved mAbs, and it is likely that the lessons learned with COVID-19 mAbs will cross-fertilize the field, driving an acceleration in the development of mAbs against various pathogens, including bacteria, viruses, fungi, and parasites, in both acute and chronic disease settings, along with exploration of alternative delivery platforms like viral-vectored mAbs.
Figure 1.
Timeline of approval of mAbs for all indications (black) and for infectious disease (blue)
Sotrovimab received EUA at the time of publication.
Developing mAbs to fight COVID-19
As of May 2021, the COVID-19 pandemic counts more than 160 million cases and over 3 million deaths worldwide. The rapid spread of the disease has prompted the development of several effective vaccines at an unprecedented pace (Jackson et al., 2020; Polack et al., 2020), as well as the intense research for potential novel treatments, including the reuse of existing drugs approved for other indications.
The recognition of the urgent need for therapies available on a global scale has prompted the rapid development of a large number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-neutralizing mAbs. In only 16 months, six mAbs have been developed and received an Emergency Use Authorization (EUA) by the United States or South Korea regulatory agencies, and several additional ones are being evaluated in phase 3 clinical trials or currently seeking an EUA. Importantly, these mAbs are being tested in different clinical settings and target populations with different endpoints, in prophylaxis as well as early- or late-disease therapeutic settings. In addition, mAbs in development display a variety of mutations in the constant Fc region aimed to enhance or eliminate effector functions or improve mAb half-life and are being utilized as monotherapies or cocktails. The successes and failures of these trials will be key for the development of additional anti-infective mAbs, at least for respiratory viral pathogens. Although sterilizing immunity may be required for viruses establishing chronic infection, for acute viral infections such as COVID-19, it might be sufficient to blunt viral replication such that the passively administered mAb can act in concert with the host immune response to avoid the development of severe complications and limit onward transmission.
Finally, the recent emergence of several SARS-CoV-2 variants of concern (VOCs) has shown limitation in the coverage by some of these mAbs that target epitopes mutating rapidly (Chen et al., 2021c; Collier et al., 2021; Davies et al., 2021a; McCallum et al., 2021a; Tegally et al., 2021; Wibmer et al., 2021). This review will provide an up-to-date overview on a selection of 14 clinical mAbs developed to tackle COVID-19.
Molecular targets of clinical COVID-19 mAbs
The SARS-CoV-2 spike (S) glycoprotein is a transmembrane homotrimer and the target of neutralizing antibodies, including all mAbs currently authorized or in early or late development. The S glycoprotein has two functional subunits that respectively mediate host cell attachment (S1 subunit, formed by four domains, the most relevant being the N-terminal domain [NTD] and receptor-binding domain [RBD]) and fusion of the viral and cellular membranes (S2 subunit) (Tortorici and Veesler, 2019; Walls et al., 2016, 2020b; Wrapp et al., 2020).
Monoclonal antibodies in development for COVID-19 are all fully human and were discovered from SARS-CoV-2-immune donors (the majority), SARS-CoV-immune donors (VIR-7831 and ADG2), immunized humanized immunoglobulin mice (REGN10933 and ABBV-47D11), or wild-type mice (ABBV-2B04). Only the ADG2 mAb has been affinity matured in vitro (Rappazzo et al., 2021), while all others were developed in their native or semi-native forms.
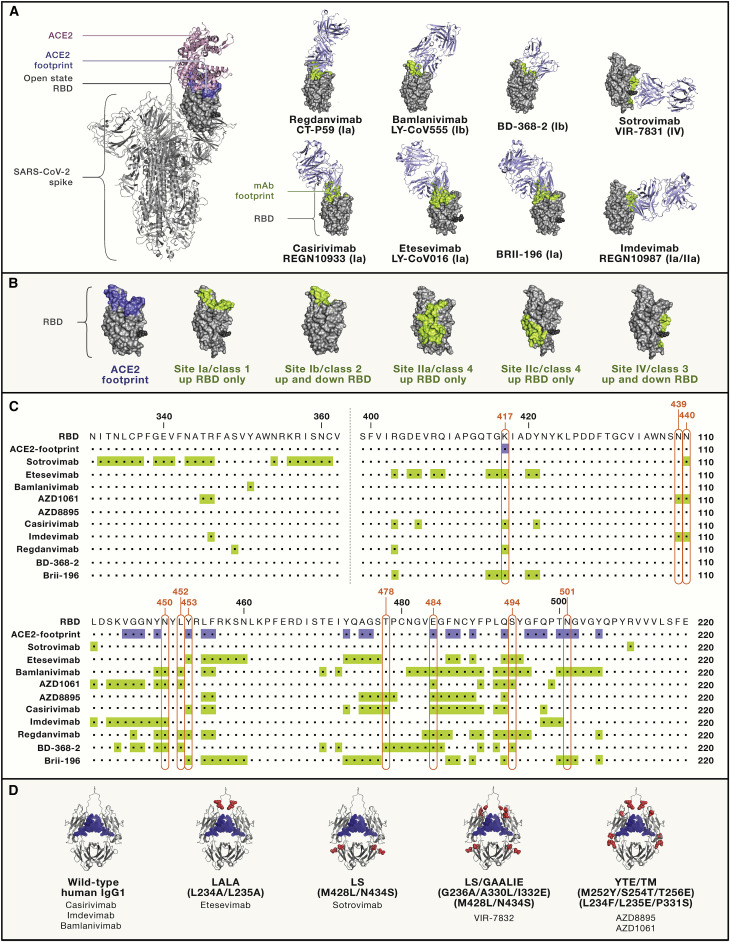
All mAbs authorized or in development are directed to the RBD, which interacts with the target receptor angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Letko et al., 2020; Walls et al., 2020b; Zhou et al., 2020). The RBD, and also the NTD, have been proposed to also interact with other receptors or co-receptors, including neuropilin-1 (NRP-1), dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN or CD209), and liver-/lymph-node-specific intracellular adhesion molecules-3 grabbing non-integrin (L-SIGN or CD209L) (Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Soh et al., 2020). We recently showed that transmembrane lectins DC-SIGN, L-SIGN, and SIGLEC1 act as attachment receptors rather than entry receptors, thus facilitating SARS-CoV-2 infection via the canonical ACE2 pathway (Lempp et al., 2021). The majority of mAbs (11 out of the 14) presented in this review recognize different epitopes fully or partially overlapping with the receptor-binding motif (RBM) on the RBD. In the context of the S trimer, the RBDs exist in two conformational states, described as open and closed, the former conformation exposing the RBM for ACE2 engagement (Barnes et al., 2020a; Piccoli et al., 2020; Walls et al., 2020b). Some mAbs directed to the RBM can bind their cognate epitope only in the open RBD state, and all recognize epitopes in the same RBD site (site Ia or class 1: REGN10933, LY-CoV016, AZD8895, Brii-196, ADG2, CT-P59, and likely ABBV-2B04). The site recognized by the ACE2-blocker cross-reactive mAb ADG2 is also only accessible in the open RBD state and appears to be located between site Ia/class 1 and the cryptic site II/class 4 site. The epitope recognized by REGN10987 mAb (located between sites Ia/class 1 and the cryptic site II/class 4) is mostly exposed at the surface of the RBD in the closed state but is not accessible to the mAb due to steric hindrance with a neighboring protomer. Therefore, REGN10987 requires RBD opening for mAb binding to occur. Conversely, other RBM mAbs can recognize both open and closed states of the RBD (site Ib or class 2; LY-CoV555, AZD1061, and BGB-DXP593) (Figures 2A and 2B). There are two exceptions of non-ACE2 blocker mAbs in early and late stages of development, VIR-7831 (alias for an engineered form of S309) and 47D11. The epitope of VIR-7831 is located in a site distal to the RBM and comprises a glycan bound to N343. Importantly, N343 is highly conserved within the sarbecovirus subgenus (known also as lineage B) of betacoronaviruses that comprises SARS-CoV-2, SARS-CoV, and other animal viruses, predominantly of bat origin. The epitope of mAb 47D11 resides in a conserved hydrophobic pocket nearby glycan N343 (structural data not publicly available; Fedry et al., 2020). No mAbs directed to the cryptic site IIc/class 3 represented by the CR3022 or S304 mAbs (poorly or not neutralizing) are currently under consideration for development (Piccoli et al., 2020; Yuan et al., 2020).
Figure 2.
Fab-RBD complexes, epitopes, and Fc mutations of clinically relevant mAbs
(A) Full spike (S) of SARS-CoV-2 (PDB: 6ZGG) is shown on the left, where ACE2 (pink cartoon) is modeled on the open-state RBD (gray space-filling model) (ACE-2 PDB: 6M0J); the structures of eight Fab-RBD complexes were determined by a combination of X-ray crystallography and/or cryoelectron microscopy (cryo-EM) analysis (Fabs shown as light-blue cartoons and RBD orientation fixed in the upward state shown on S trimer [left image]; see Table 1 for PDB accession numbers). ACE2 and mAb footprints are shown in blue and light green, respectively. Footprints on RBDs were defined according to 5 Å distance from ACE2- or mAb-contacting residues. The stem of N343 glycan is shown as a black sphere. mAbs are labeled using both their original and generic (nonproprietary) names.
(B) Antigenic sites nomenclature (Ia–IV versus classes 1–4) according to Barnes et al., 2020a, Cohen et al. (2021), and Piccoli et al. (2020) and colored as in (A).
(C) Sequences of the full RBD of SARS-CoV-2 (Wuhan-1 strain), where ACE2 and mAb footprints are highlighted in blue and light green, respectively, as in (A) and (B). The key RBD mutations found in VOCs/VOIs are boxed in red.
(D) Structural representation of human IgG1 Fc where amino acids changed in COVID-19 mAbs in late development are shown as red spheres. N297-bound glycans are shown as blue spheres. List of Fc abbreviations: LALA, L234A/L235A (Hezareh et al., 2001; Xu et al., 2000); GAALIE, G236A/A330L/I332E (Weitzenfeld et al., 2019); YTE, M252Y/S254T/T256E (Dall’Acqua et al., 2002); LS: M428L/N434S (Zalevsky et al., 2010); TM, triple mutant in Fc, L234F/L235E/P331S (Oganesyan et al., 2008).
The NTD is also a target of neutralizing mAbs (Cerutti et al., 2021; Chi et al., 2020; McCallum et al., 2021b; Suryadevara et al., 2021; Voss et al., 2020; Wu et al., 2021) that were shown to be potent in vitro and were all directed toward the same antigenic site of vulnerability (designated site i). However, NTD-specific neutralizing mAbs were shown to have a low barrier to resistance through the selection of a plethora of escape mutants in vitro and suffer from reduced activity against most of the currently circulating VOCs (Chen et al., 2021c; Collier et al., 2021; McCallum et al., 2021a; Wang et al., 2021b). To the best of our knowledge, there are no mAbs directed to this domain in clinical development.
Another important characteristic of mAbs in development is the ability to broadly neutralize multiple coronaviruses of the same lineage (e.g., sarbecoviruses). The only clinical mAb with neutralization breadth is VIR-7831, which just received EUA in the US , while ADG2 and 47D11 are in early phases of development. The recognition of epitopes that are conserved across different viruses of the same lineage may be important to reduce the risk of viral escape during treatment (i.e., under drug selection) as well as provide long-term coverage of circulating and emerging variants and strains.
Mechanisms of action of COVID-19 clinical mAbs
All mAbs in development for COVID-19 are defined as neutralizing antibodies. However, as described above, the concept of viral neutralization in vivo is complex and widespread. This section describes in more detail what is currently known about the multiple mechanisms of action of COVID-19 clinical-stage mAbs.
Blocking of infection of target cells
Twelve out of the 14 mAbs presented in this review neutralize SARS-CoV-2 entry by blocking engagement of ACE2 by targeting epitopes overlapping with the RBM. In some cases, mAbs binding to RBM can also prematurely trigger fusogenic S conformational changes that could result in the inactivation of the protein (Lempp et al., 2021), as has also been previously shown for the SARS-CoV-specific neutralizing mAb S230 (Walls et al., 2019). It is therefore possible that RBM mAbs may act not only by blocking binding to the ACE2 entry receptor but also by inactivating S by acting as receptor mimics before viral encounter of a host cell. As described below, this mechanism may also alter the ability of these mAbs to promote effector functions. RBM mAbs can also lock the RBD conformation in the closed state, thus preventing the engagement of ACE2 on target cells. This class of mAbs is exemplified by S2M11 and C144 mAbs, which lock RBDs in a closed state through the recognition of a quaternary epitope on neighboring RBDs (Barnes et al., 2020a; Tortorici et al., 2020). S2M11 was not further developed, while C144 mAb is in early clinical development by Bristol Myers Squibb (phase 1, NCT04700163) in combination with another RBM mAb named C135 (both developed with LS mutations in Fc for half-life extension) (Robbiani et al., 2020; Weisblum et al., 2020).
An important concept is that virus neutralization assays produce variable results depending on the cells used for virus propagation and infection. Determining the potency of neutralizing mAbs using target cells overexpressing ACE2 (to increase infectivity) was shown to alter the apparent potency and maximal neutralization achieved by non-RBM mAbs, such as S309 (parent of VIR-7831) and NTD-specific mAbs, without necessarily reflecting the bona fide potency and in vivo efficacy of these mAbs (Lempp et al., 2021; McCallum et al., 2021a; Pinto et al., 2020; Rappazzo et al., 2021; Suryadevara et al., 2021). Therefore, cell-line selection and the level of ACE2 expression are important variables in assessing the potency of SARS-CoV-2-neutralizing mAbs (Chen et al., 2021c; Lempp et al., 2021). Overexpression of ACE2 was shown by in situ cryoelectron tomographic analysis to promote S structural changes to the post-fusion conformation when virions were released from cells overexpressing ACE2 (Klein et al., 2020).
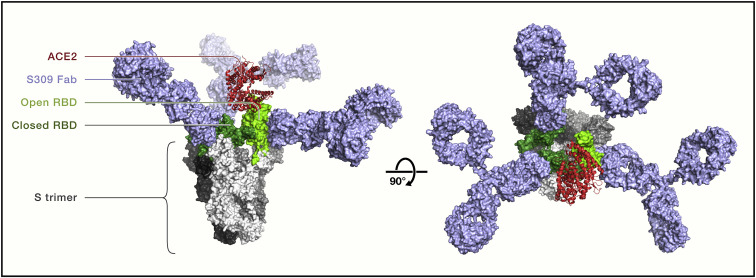
The different mAbs in development are expected to be able to achieve various levels of occupancy of binding to S. mAbs binding to site Ia/class 1 or site II/class 3 have more constrained access to their cognate epitope, as ∼50% of S trimers have all three RBDs closed, whereas the remaining half possess a single open RBD (Ke et al., 2020; Walls et al., 2020b), and the proximity of the epitopes on neighboring RBDs might not be compatible with multiple immunoglobulin G (IgG) molecules binding to each trimer. These classes of mAbs was found to trigger S rearrangement to the post-fusion state, likely due to conformational selection for open RBDs, and promote membrane fusion (Lempp et al., 2021). The access to site Ib/class 2 is not dependent on the opening state of the RBD, although the angle of mAb binding to the RBD may sterically prevent binding of other IgGs to neighboring RBDs in the same S trimer, thus reducing overall occupancy. Conversely, non-ACE2-blocking mAbs target highly exposed and distal sites on the RBD and are expected to have access to all three epitopes on the S trimer, allowing full occupancy (i.e., three IgG molecules bound per S trimer) not only on virions but also on S displayed at the surface of infected cells. High-density IgG binding on the surface of virions may sterically shield the engagement of ACE2 on the surface of target cells (Figure 3 ), thus possibly explaining the blocking of infection by S309 and NTD-specific mAbs. A recent study showed that S309 effectively blocked the ACE2-dependent entry of SARS-CoV-2 in the presence of membrane lectins and inhibited the fusogenic rearrangement induced by receptor-mimicking RBM antibodies that leads to cell-to-cell fusion. Thus, non-RBM mAbs might block viral infection by sterically interfering with ACE2 engagement and by preventing structural rearrangement of the S glycoprotein that is required for viral fusion (Lempp et al., 2021).
Figure 3.
Full occupancy of binding to S trimer by the non-RBM mAb S309
Side (left) and top (right) views of a structural model of SARS-CoV-2 S trimer (gray) with two RBDs in the closed state (dark green) and one in the open state (light green monomer) based on PDB: 6WPT. S309 Fab bound to all three RBDs is modeled as full IgG1 (light blue), and ACE2 monomer (red) bound to the open RBD is shown as a ribbon diagram. This model illustrates how the non-RBM mAb S309 may shield S trimers to sterically prevent ACE2 binding on target cells. S309 is the precursor of VIR-7831, recently renamed sotrovimab.
Whether occupancy is relevant to viral neutralization is unclear, but it might affect the ability of mAbs to trigger effector functions, since the density of mAbs on virions or infected cells can influence the ability to engage and activate low-affinity Fcγ-receptors (Ortiz et al., 2016). Additional studies are needed to address the role of bivalent IgG binding within an S trimer, as well as the cross-linking of neighboring S proteins on virions or between virions (i.e., virus aggregation) as relevant mechanisms of SARS-CoV-2 neutralization.
Finally, another important mechanistic aspect is whether combination of mAbs may benefit from synergistic effects in vitro or in vivo. Most of the available data have indicated thus far a lack of clear synergy for cocktails of mAbs targeting noncompeting sites, with the exceptions of S309 and S304 (Pinto et al., 2020) or COV2-2195 and COV2-2130 mAbs (Zost et al., 2020), with the latter combination showing enhanced neutralization potency in vitro, but not in vivo.
Role of Fc-dependent effector functions of SARS-CoV-2-neutralizing mAbs
The relevance of effector functions for the efficacy of SARS-CoV-2-neutralizing mAbs in humans is not well established. These could even be putatively detrimental, enhancing inflammation and therefore disease, at least under certain circumstances. mAbs can recruit immune cells and complement, facilitate the elimination of infected cells that display S at their surface, and promote phagocytosis and elimination of opsonized virions. In addition, the formation of immune complexes with mAbs can contribute to enhanced presentation of viral antigens to CD4+ and CD8+ T cells that may contribute to controlling viral infection. The relevance for antibody-dependent enhancement of disease (ADE) in the context of a range of different viral infections, including SARS-CoV and SARS-CoV-2, is reviewed elsewhere (Arvin et al., 2020; Bournazos et al., 2020b; Lee et al., 2020). Safety analysis in multiple mAb-based ongoing clinical trials for COVID-19 will reveal if ADE is an issue in any of the treatment modalities under investigation.
Several factors may play a role in determining the ability of anti-RBD mAbs to exert Fc-dependent effector function: (1) epitope specificities of the mAbs, as previously shown for mAbs against IAV HA and NA (DiLillo et al., 2014, 2016); (2) orientation and distance of the Fc fragment relative to the plasma membrane (Dilillo et al., 2016; Pinto et al., 2020; Tang et al., 2019); (3) steric hindrance of effector cells; (4) high-density binding of mAbs for efficient Fcγ receptor (FcγR) cross-linking and engagement of the hexameric C1q; and (5) the ability of some RBD-specific mAbs to trigger shedding of the S1 subunit and S conformational change, which limit their ability to trigger effector functions (Lempp et al., 2021; Tortorici et al., 2021). Indeed, mAbs that do not promote Fc-mediated effector functions seem to be those selecting for open RBD states and all are likely triggering S, leading to S1 (and mAb) shedding.
The study of antibody effector functions in vitro does not necessarily correlate with in vivo activity, due to the high complexity of the complement and FcγR biology. Protection by antiviral neutralizing antibodies in animal models of influenza A and HIV-1 was shown to require the contribution of effector functions mediated by Fc-FcγR interactions for optimal efficacy (DiLillo et al., 2014; Hessell et al., 2007). Similarly, in a mouse model of COVID-19, the elimination of effector functions through the introduction of GRLR mutations (G236R/L328R) in the Fc fragment that abrogates binding to FcγRs and complement resulted in a reduced antiviral efficacy in terms of viral load in lungs of infected mice (Schäfer et al., 2021). In the same study, the authors also showed a lack of correlation between in vitro neutralization and in vivo potency, arguing that neutralization of viral entry in vitro may not be the sole determinant of antiviral activity in vivo. Another study by Diamond and collaborators showed that in huACE2 mice (K18) and hamsters, Fc-dependent effector functions are dispensable in prophylactic settings but required for optimal protection under therapeutic settings. In particular, mAbs with wild-type Fc were shown to reduce SARS-CoV-2 burden and lung disease in animals more efficiently than mAbs with Fc mutated to abrogate binding to FcγRs and required monocyte intervention for therapeutic efficacy (Winkler et al., 2021). Of note, studies testing SARS-CoV-2-neutralizing mAbs at limiting doses to reach sub-neutralizing levels did not provide evidence for enhancement of disease, even when the Fc was modified to match the target host Fc receptors (e.g., hamsterized in hamsters) (Baum et al., 2020a; Cathcart et al., 2021; Lempp et al., 2021; Rogers et al., 2020). Collectively, these data indicate that at least in the animal models and therapeutic modalities tested, the antibody Fc fragment can contribute to in vivo efficacy.
Due to the initial uncertainty around the beneficial or detrimental role of effector functions, different groups have adopted opposite strategies in the choice of Fc formats for the clinical development of human-IgG1-antibody-based products. In particular, 5 of the 14 mAbs analyzed here have been engineered to remove (e.g., LALA or TM [triple mutant] Fc mutations) or reduce (e.g., YTE) mAb effector functions, while the remaining were developed as wild-type IgG1 (REGN-COV2) or half-life-extended Fc versions (VIR-7831 and ADG2) (Figure 2C). VIR-7832, which is a derivative of VIR-7831, is an exception, as it was further Fc engineered to carry the GAALIE mutation that was previously shown to mediate enhanced dendritic cell maturation and the induction of protective CD8+ T cell responses against IAV (Bournazos et al., 2020a). VIR-7832 is currently being tested in a phase 2 study in parallel with its parent, VIR-7831 (sotrovimab), to assess the potential beneficial effect of the so-called antibody vaccinal effect in humans. The use of half-life-extending mutations such as LS or YTE may also contribute to an improved biodistribution in mucosal tissues, such as lungs, as previously observed in non-human primates (Ko et al., 2014).
In conclusion, this section illustrates how different classes of S-specific mAbs can provide in vivo efficacy through multiple and different mechanisms of action beyond direct blocking of ACE2 binding and that the relevance of these mechanisms may differ in prophylactic or therapeutic settings.
Virus evolution and escape from mAbs
RNA viruses, such as SARS-CoV-2, are evolving biological entities. During the first few months of the COVID-19 pandemic, a modest rate of sequence divergence was observed, likely due to the coronavirus exonuclease “proofreading” activity enhancing replication fidelity (Denison et al., 2011). This initial low level of mutations led the field to underestimate the risk for the virus to escape the immunity elicited by infection, vaccination, or mAb administration. It was also assumed that targeting the RBM with mAbs could offer the potential advantage of functional constrains for mutations in this region, possibly increasing the barrier to resistance for RBM-directed mAbs. However, the in vitro selection of resistant viral variants from RBM mAbs provided evidence for a high degree of plasticity in this region, leading to the clinical development of several cocktails of noncompeting RBM mAbs (Liu et al., 2021b; Weisblum et al., 2020). Deep mutational scanning of all possible RBD mutations (Starr et al., 2020) highlighted the considerable mutational tolerance of this domain, although some substitutions are deleterious for RBD expression or ACE2 binding. These analyses were also extended to map the escape landscape of polyclonal antibodies and several mAbs, including some that are in the clinic (imdevimab, casirivimab, sotrovimab, bamlanivimab, etesevimab, AZD8895, and AZD1061 mAbs; see Figure 2A or Table 1 for correspondence between initial and generic names of mAbs) (Dong et al., 2021; Greaney et al., 2021a, 2021b; Starr et al., 2021a, 2021b, 2021c). These in vitro experiments provided evidence that the RBM can accommodate a large set of mutations while retaining or even increasing ACE2 binding. The higher frequency of mutations in the RBM over the rest of the RBD (Starr et al., 2021a; Thomson et al., 2021) may result from immune pressure, consistent with the observation that antibody responses to the RBD are dominated by anti-RBM antibodies (Piccoli et al., 2020). A similar phenomenon is observed with the highly immunogenic globular head domain of the influenza virus HA that evolves faster than the stem region, although it comprises the sialic-acid-receptor-binding site. This process is the basis for antigenic drift and the continuous requirement to update influenza vaccines (Doud et al., 2018; Kirkpatrick et al., 2018).
Table 1.
mAbs under investigation for prophylaxis or therapy of SARS-CoV-2/COVID-19
| Name | Alternate name(s) | Company | Phase | Clinical trial identifier (disease indication) | Target population | Route | Dose (g) | Study name | Fc | PDB ID | Source and VH germline (% identity) | RBD site | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIR-7831 VIR-7832 | S309 GSK4182136 sotrovimab | Vir Biotechnology GSK | 2/3 EUA | NCT04545060 (early treatment) | at-risk adults | IV | 0.5 | COMET-ICE | LS | 7JX3 | SARS-CoV-immune donor VH3–23 (96.5%) | IV/class 3 | Cathcart et al., 2021; Gupta et al., 2021; Lempp et al., 2021; Pinto et al., 2020; Tortorici et al., 2021 |
| 2 | NCT04634409, VIR-7831+ bamlanivimab (early treatment) | adults | IV | 0.5 + 0.7 | BLAZE-4 | LS | |||||||
| 3 | NCT04501978 (late treatment) | hospitalized | IV | 0.5 | ACTIV-3 | LS | |||||||
| 1b/2a | (UK) (1b–2a) (early treatment) (VIR-7831 versus VIR-7832) | at-risk adults | IV | 0.5 | AGILE | LS-GAALIE versus LS | |||||||
| 2 | NCT04779879 (safety and pharmacokinetics) | adults | IV/IM | 0.5 | COMET-PEAK | LS | |||||||
| REGN-COV2 (REGN10933, REGN10987 not co-formulated) | casirivimab imdevimab | Regeneron | 1/2/3 | NCT04426695 (late treatment) | hospitalized | IV | 2.4 | Study 2066 | WT | 6XDG | SARS-CoV-2-immunized huIg mice (REGN10987) and SARS-CoV-2 donor (REGN10987) VH3–11 (98.6%) VH3–30 (98.6%) | Ia/class 1 | Baum et al., 2020a, 2020b; Copin et al., 2021; Hansen et al., 2020; Weinreich et al., 2021 |
| 8 | |||||||||||||
| 3 | NCT04452318 (household contact prevention) | adults/pediatrics | SC/IM | 1.2 | Study 2069 | ||||||||
| 3 | NCT04381936 (late treatment) | 12 years and older (hospitalized) | IV | 8 | RECOVERY | ||||||||
| 1/2/3 EUA | NCT04425629 (early treatment) | adult/pediatrics and pregnant | IV | 2.4 | Study 2067 | ||||||||
| 8 | |||||||||||||
| 2 | NCT04666441 (early treatment dose ranging study) | adults | IV/SC | ||||||||||
| 1 | NCT04519437 (safety repeat dosing) | adults | SC | Study 2093 | |||||||||
| LY-CoV016 | CB6, JS016, LY3832479, etesevimab | AbCellera Eli Lilly | N/A | N/A | N/A | N/A | N/A | N/A | LALA | 7C01 | SARS-CoV-2-immune donor VH3–66 (99.7%) | Ia/class 1 | Shi et al., 2020 |
| LY-CoV555 | Ab169 LY3819253 bamlanivimab | 1 | NCT04411628 (healthy volunteer) | adults | IV | BLAZE-5 | WT | 7KMG | SARS-CoV-2-immune donor VH1–69 (99.7%) | Ib/class 2 | Chen et al., 2021a; Gottlieb et al., 2021; ACTIV-3/TICO LY-CoV555 Study Group et al., 2021; Jones et al., 2021 | ||
| 2 | NCT04701658 (early treatment) | 12 years and older | IV | BLAZE-5 | |||||||||
| 2/3 | NCT04518410 (early treatment) | adults | IV | ACTIV-2 | |||||||||
| 4 | NCT04656691 (early treatment, at-home infusion) | older adults | IV | UNITED | |||||||||
| NCT04603651 (expanded access) | 12 years and older | IV | ACTIV-2 | ||||||||||
| LY-CoV555 and LY-CoV016 | CB6, JS016, LY3832479, etesevimab and Ab169 LY3819253 bamlanivimab | 3 | NCT04497987 prevention in nursing home residents and staff (post-exposure prophylaxis?) | adults | IV | BLAZE-2 | |||||||
| EUA | NCT04427501 (early treatment) | 12 years and older | IV | BLAZE-1 | |||||||||
| AZD7442 (cocktail of AZD8895 and AZD1061) | COV2-2196 | AstraZeneca | 3 | NCT04625725 (pre-exposure prophylaxis) | adults | IM | 0.15+0.15 | PROVENT | TM/YTE | N/A | SARS-CoV-2-immune donor VH1–58, VH3–15 | Ia/class 1 Ib/class 2 | Dong et al., 2021; Suryadevara et al., 2021; Zost et al., 2020 |
| COV2-2130 | 3 | NCT04625972 (post-exposure prophylaxis) | adults | IM | 0.15+0.15 | STORM CHASER | |||||||
| 3 | NCT04723394 (early treatment) | adults | IM | 0.6 | TACKLE | ||||||||
| 3 | NCT04501978 (late treatment) | hospitalized | IM | ACTIV-3 | |||||||||
| BRII-196 | 1F11 | Brii Biosciences | 1 | NCT04479631 (safety) | healthy volunteers | IV | 1 | ? | 7CDI | SARS-CoV-2-immune donor VH3–53 (?) | Ia/class 1 | Ju et al., 2020 | |
| BRII-198 | 1 | NCT04479644 (safety) | healthy volunteers | IV | 1 | ? | N/A | ? | |||||
| BRII-196 and BRII-198 combination | 2 | NCT04770467 (early treatment) | adults | IV | |||||||||
| 3 | NCT04501978 (late treatment) | hospitalized | IV | 1+1 | ACTIV-3 | ||||||||
| CT-P59 | regdanvimab | Celltrion | 2/3 EUA in South Korea | NCT04602000 (early treatment) | at-risk adults | IV | 40 mg/kg | ? | 7CM4 | SARS-CoV-2-immune donor VH-70 (?) | Ia/class 1 | Du et al., 2020; Kim et al., 2021; Ryu et al., 2021 | |
| ADG20 | ADG-2 parent ADI-55688 | Adagio | 1/2/3 | NCT04805671 (early treatment) | adults | IM/IV | 1 dose | WT/half-life ext. (?) | N/A | SARS-CoV-immune donor VH1–69? | Ia/class 1 | Dejnirattisai et al., 2021; Rappazzo et al., 2021; Wec et al., 2020 | |
| IIa/class 4 | |||||||||||||
| BGB-DXP593 | BD-368-2 | BeiGene Singlomics | 2 | NCT04551898 (early treatment) | adults | IV | 3 doses | ? | 7CHH | SARS-CoV-2-immune donor VH3–23 (?) | Ib/class 2 | Cao et al., 2020 | |
| ABBV-47D11 | 47D11 | AbbVie | 1 | NCT04644120 (safety and late treatment) | hospitalized | IV | 3 doses | ? | N/A | SARS-CoV-immunized huIg mice, N/A | ? | Wang et al., 2020 | |
| ABBV-2B04 | 2B04 | ? | N/A | RBD-immunized mice (B6), humanized? | Ia/class 1 | Alsoussi et al., 2020; Chen et al., 2021b; Liu et al., 2021b |
huIg, humanized immunoglobulin; IV, intravenous; IM, intramuscular; N/A, not available; SC, subcutaneous; WT, wild type.
Overall, the emergence of resistant variants has at least three constraints. (1) The low rate of mutations of SARS-CoV-2 limits the frequency of random mutations to 1 nt per codon, restricting the spectrum of possible amino acid substitutions at each position. This is further limited by the fact that positive-sense single-stranded RNA viruses undergo more transitions in their genomes than transversions. Indeed, a three-times-higher frequency of transitions over transversions was observed in the SARS-CoV-2 genome (Roy et al., 2020; Sarkar et al., 2021). (2) High-affinity binding to ACE2 and possibly other co-receptors or attachment receptors needs to be retained for viral fitness. In a recent study, the mutation N501Y, found in multiple SARS-CoV-2 VOCs, was shown to increase by 6-fold the affinity for human ACE2 (Collier et al., 2021) and was also associated to an increased interaction with murine ACE2 (Gu et al., 2020), raising the possibility that mutations in S may also emerge as a result of inter-species exchange of SARS-CoV-2, like the two-way transmission of SARS-CoV-2 on mink farms observed in Denmark in late 2020 (Oude Munnink et al., 2021). (3) Mutations shall not affect RBD folding and expression. As an example, the RBD N343 glycan, which is part of the S309 mAb epitope (Pinto et al., 2020), is highly conserved in all sarbecoviruses as well in 100% of the more than 1 million sequences of SARS-CoV-2 available to date. The removal of this glycosylation site by mutagenesis resulted in impaired RBD folding, decreased expression, and reduced infectivity (Li et al., 2020; Starr et al., 2020, 2021a), most likely explaining its strict conservation.
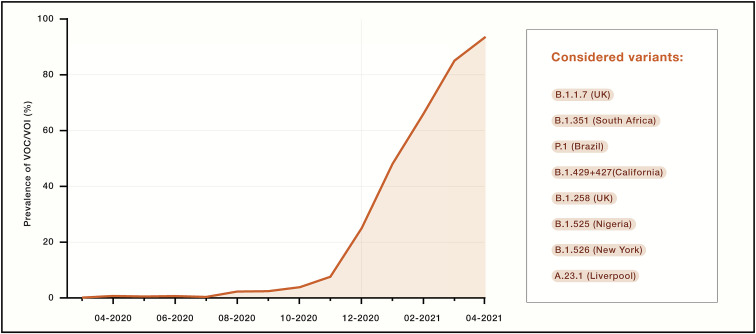
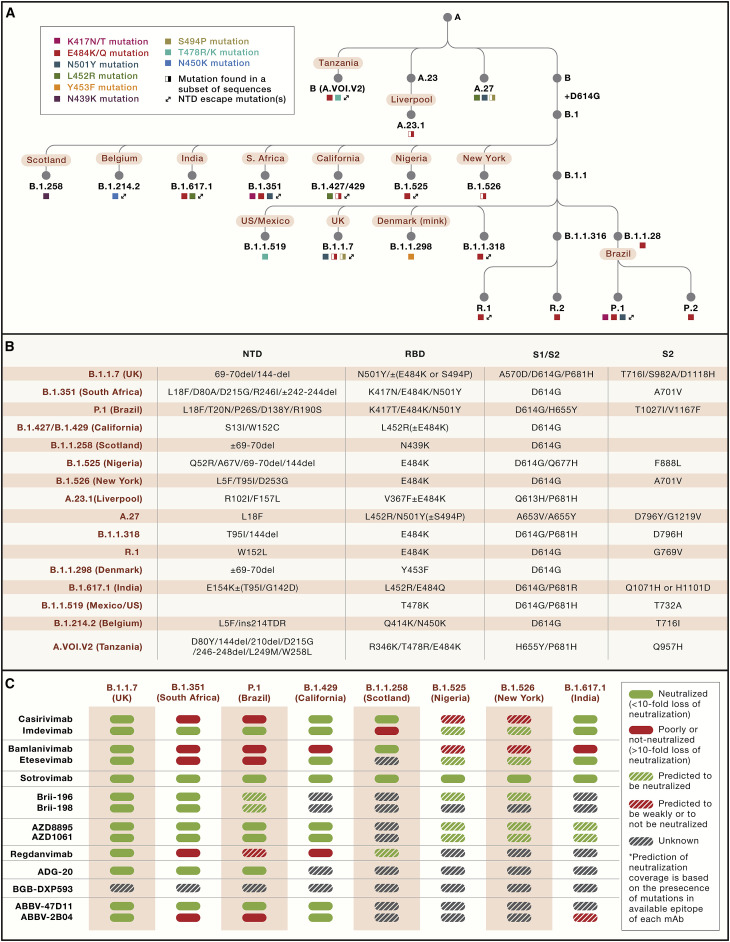
It is possible that the colossal number of infected patients, with very large estimated local seroprevalence at some locations, has imposed an immune pressure on the virus. In the early phase of the pandemic, the only mutation in S that became prevalent was D614G and was associated with higher viral loads and younger patient age (Volz et al., 2021). Since November 2020, SARS-CoV-2 has started to mutate more drastically, with the accumulation of several mutations and deletions in the RBD, NTD, and S2 subunit. This rapid evolution led to the simultaneous appearance of a plethora of SARS-CoV-2 VOCs or variants of interest (VOIs), such as B.1.1.7 (United Kingdom), B.1.351 (South Africa), B.1.525 (Nigeria), B.1.526 (New York), P.1 (Brazil), B.1.427/B.1.429 (California), B.1.258 (Scotland), and A.23.1 (Liverpool), which collectively now account for more than 90% of sequenced viruses (Figure 4 ).
Figure 4.
Prevalence of VOCs over time
Prevalence was calculated based on the cumulative counts of sequences retrieved from GISAID belonging to any of the listed VOCs over the total number of sequences deposited by month. GISAID sequences were filtered based on the quality of the sequences (<10% Xs) and the coverage of most of the S sequence (>80% full length). The Phylogenetic Assignment of Named Global Outbreak (PANGO) lineage designation (Rambaut et al., 2020) of each variant, along with the location where they were first identified, is indicated on the right.
Several mutations found in these VOCs/VOIs are found to reduce or abolish the neutralizing activity of several mAbs, including those already approved or in late stages of development (Figure 5 ). Some of these VOCs/VOIs have also been shown to reduce neutralizing titers of vaccine-elicited antibodies, possibly reducing vaccine effectiveness and the duration of protection (Cele et al., 2021; Chen et al., 2021c; Collier et al., 2021; Kustin et al., 2021; Liu et al., 2021a; McCallum et al., 2021a; Muik et al., 2021; Wang et al., 2021b, 2021c; Wu et al., 2021). In this context, the resurgence of COVID-19 in Manaus, Brazil is associated with the emergence of the P.1 SARS-CoV-2 VOC despite an estimated seroprevalence of ∼75% in this region (Buss et al., 2021; Sabino et al., 2021). Similarly, the recent surge of COVID-19 cases in India may be in part associated with a new VOI, named B.1.617.1, that carries a combination of two key RBD escape mutations (E484Q and L452R), found in other variants, in addition to several other mutations. Importantly, most of the RBD mutations found in current VOCs/VOIs were already previously identified as resistant variants in vitro by using mAbs and convalescent or vaccine-elicited serum antibodies (Andreano et al., 2020; Baum et al., 2020b; Liu et al., 2021b; Weisblum et al., 2020), suggesting that the emergence of these VOCs/VOIs may result from immune selection. It was hypothesized that antibody-resistant variants may exhibit pathogenesis and transmission deficits, but all recent findings have argued for the opposite. Indeed, the B.1.1.7 variant was found to be more transmissible and cause more severe infections compared to the parental virus (Davies et al., 2021a, 2021b; Munitz et al., 2021). However, two recent studies did not confirm that the B.1.1.7 variant is associated with more severe disease (Frampton et al., 2021; Graham et al., 2021). Preliminary data have suggested increased transmissibility for the B.1.427/B.1.429 VOCs (Deng et al., 2021). Whether a similar trend would be observed for B.1.351, P.1, and other VOCs that showed a more marked effect on antibody escape remains to be established.
Figure 5.
Mutations on the SARS-CoV-2 S in VOCs and resistance profile of clinical mAbs
(A) Ancestry tree of SARS-CoV-2 VOCs/VOIs.
(B) Schematic of SARS-CoV-2 S and the mutation landscape in each VOC/VOI. Del, deletion; ins, insertion.
(C) Neutralization of a selection of VOCs/VOIs by clinical-stage mAbs as reported previously (Baum et al., 2020b; Chen et al., 2021b, 2021c; Copin et al., 2021; Dejnirattisai et al., 2021; Dong et al., 2021; FDA (US Food and Drug Administration), 2020a, 2020b; Hoffmann et al., 2021; Liu et al., 2021b; McCallum et al., 2021a; Ryu et al., 2021; Starr et al., 2021b, 2021c; Thomson et al., 2021; Wang et al., 2021a, 2021b). Prediction of neutralization coverage is based on the presence of mutations in the available epitope of each mAb. mAbs developed clinically as cocktails are grouped.
Overall, immune-evading SARS-CoV-2 variants, which may mark the beginning of antigenic drift of SARS-CoV-2, may potentially continue to emerge and co-evolve when herd immunity is reached, with implications for reinfection, vaccines, and both mAb and polyclonal antibody therapeutics. Monitoring resistance of mAbs to circulating new variants will be key to define whether some of the developed mAbs should be discontinued or if different combinations of clinical-stage mAbs should be investigated, as recommended in a recent guideline developed by the US Food and Drug Administration (FDA) (FDA (US Food and Drug Administration), 2021). As a striking example of the effect of the evolution of SARS-CoV-2, in mid-April 2021, Eli Lilly requested revocation of the EUA for bamlanivimab, citing an increase in variants resistant to the mAb monotherapy. However, some clinical mAbs are not sensitive in vitro and in vivo to the mutations present in the current VOCs/VOIs due to targeting of highly conserved epitopes, as is the case for VIR-7831, COV2-2196, and other mAbs (Chen et al., 2021b; Dong et al., 2021; McCallum et al., 2021a; Pinto et al., 2020; Starr et al., 2021a; Tortorici et al., 2021). Monotherapy approaches are not necessarily inferior to mAb cocktails if they are based on mAbs with high barrier to resistance and optimal coverage of circulating variants. Monitoring the emergence of resistance may be important if single mAbs are used in the clinic, but it becomes equally important when one of the mAbs of a cocktail is found to be sensitive to the circulating variants. This is already the case for the REGN-COV2, bamlanivimab/etesevimab, and ABBV-2B04/ABBV-47D11 cocktails, which are sensitive to the E484K and/or K417N/T mutations found in the B.1.351, B.1.525, B.1.526, A.23.1, and P.1 VOCs (Figure 5C). Moreover, regdanvimab (CT-P59) and, to a smaller extent, etesevimab, showed a reduction in neutralization potency against the B.1.427/B.1.429 VOCs, whereas bamlanivimab (LY-CoV555) entirely lost its neutralizing activity due to the central location of L452R in the epitopes recognized by these mAbs (McCallum et al., 2021a). If one mAb in a cocktail is inactive against a VOC, then the clinical dosing of the administered mAbs is reduced by half, and the cocktail becomes a monotherapy.
Overview of COVID-19 mAb clinical trials
mAbs approved for early therapy of COVID-19
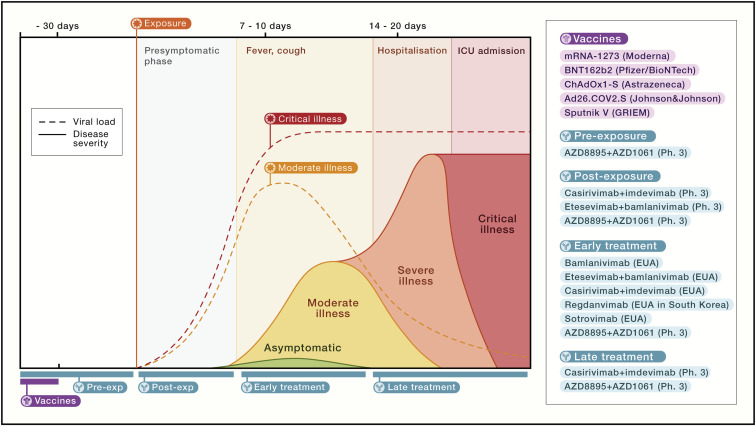
The pandemic has revolutionized the design and speed of clinical trials, including phase 1/2/3 adaptive trials that have led to approvals of therapeutic mAbs for COVID-19 in record time. To date, four mAb products have been approved under an EUA or conditional marketing authorization based on phase 2 and/or phase 3 interim data for COVID-19 for the same indication (i.e., early therapy in at-risk individuals) (Figure 6 ). An overview of the performed and ongoing clinical trials for all 14 mAbs described in this review is provided in Table 1 .
Figure 6.
Prophylactic and therapeutic approaches to COVID-19
Vaccines are listed in purple, and mAb-based prophylactic or therapeutic modalities completed successfully or in progress are shown in blue. Other therapeutic modalities are not shown.
Bamlanivimab was the first COVID-19-authorized mAb (November 9, 2020) and received from the FDA an EUA for the treatment of mild-to-moderate COVID-19 in adults and pediatric patients 12 years (over 40 kg) and older with a positive COVID-19 test result who are at high risk for progressing to severe COVID-19 and/or hospitalization (Chen et al., 2021a). The EUA was based on interim data from the BLAZE-1 phase 3 study in patients with recently diagnosed mild-to-moderate COVID-19 in the outpatient setting. The approved dose was 0.7 g to be administered by a single intravenous infusion within 10 days of symptom onset. Two weeks later, the REGN-COV2 cocktail (casirivimab and imdevimab) also received an EUA for use in the same patient population for the treatment of mild-to-moderate COVID-19 in high-risk patients at a dose of 2.4 g (1.2 g casirivimab and 1.2 g imdevimab) administered as a single intravenous infusion (Weinreich et al., 2021). In March 2021, interim analysis showed a 70% reduction in hospitalization or death but no dose-dependent effect (1.2, 2.4, or 8 g). In February 2021, the FDA also granted an EUA for the combined use of bamlanivimab (0.7 mg) and etesevimab (1.4 g) for the same indication for which bamlanivimab was approved (i.e., treatment of mild-to-moderate COVID-19 in at-risk patients) (Gottlieb et al., 2021). In addition, the EUA for this combination allowed for a much-reduced infusion time of 21 min compared to that authorized for bamlanivimab alone (60 min). The use of these mAbs is now authorized in multiple countries. A fourth approval for the same indication (i.e., early therapy in adults) occurred in early February 2021 for regdanvimab by Celltrion. Regdanvimab received a conditional marketing authorization by South Korea’s drug safety agency. This approval was based on the first part of a global phase 2/3 trial showing that progression rates to severe COVID-19 were reduced by 54% for patients with mild-to-moderate symptoms and 68% for patients aged 50 years and older. Currently, Celltrion is seeking conditional approvals by the European Medicines Agency (EMA) and FDA. Finally, the FDA granted an EUA also for the early-therapy use of VIR-7831 (sotrovimab) in late May 2021. VIR-7831 is a derivative of the cross-reactive S309 mAb that was engineered for half-life extension and potentially improved biodistribution in the lungs by the introduction of the LS mutation in Fc. This mAb was investigated in a phase 3 study in patients with early-stage COVID-19 infection who are at high risk for hospitalization (i.e., patients ≥55 years with preexisting lung or cardiovascular disease) and administered as a 0.5-g dose via intravenous infusion. This phase 3 study has been stopped early due to outstanding efficacy (i.e., 85% reduction in hospitalization or death) (Gupta et al., 2021).
The clinical studies that led to the approval of these mAbs have provided important insights influencing multiple aspects of the further development of these and other mAbs. A first important point is that mAbs were likely overdosed, since no dose response was observed in any of the reported studies (REGN-COV2 dosed at 1.2, 2.4, and 8 g; bamlanivimab dosed at 0.7, 2.8, and 7 g; and regdanvimab dosed at 40 and 80 mg/kg). The broad range of doses tested forced the initial use of intravenous administration due to the high volume that would be required for the highest doses that would not be compatible with intramuscular or subcutaneous routes. Dose sparing is also key to increase the overall number of available doses for each lot of produced drug product. So far, intravenous administration has limited the use of these approved mAbs, since this route requires a hospital setting or access to infusion centers. The shift to different routes, such as intramuscular or subcutaneous (Table 1), is underway and will possibly contribute to a facilitated and larger access to these mAbs.
Another important finding relates to primary endpoints used to measure the success of these clinical trials. Virological endpoints did not always reflect clinical endpoints, and the latter were ultimately the key drivers for approval. For instance, combination of bamlanivimab with etesevimab or bamlanivimab monotherapy showed similar efficacy on clinical endpoints, while combination therapy appeared to be more efficacious on virological endpoints. It is unclear whether measuring viral replication in the upper airways with molecular-based methods (i.e., RT-PCR) is an accurate measure of viral neutralization, since viral RNA may persist even in the absence of replication-competent virus. In addition, viral load in the lower respiratory tract would better reflect the injury response than the viral load in nasopharyngeal secretions, but this type of sampling is highly invasive and overall unpractical. Two factors were shown to influence the level of benefit of the mAb in the early therapy setting, the immune status at baseline (i.e., seropositive versus seronegative) and the viral titer at baseline (i.e., high versus low). The greatest benefit from mAbs (particularly for REGN-COV2 mAbs) was observed for seronegative patients with a high viral load at baseline, suggesting that the early development of endogenous antibodies may contribute to reduced disease severity and that high viral replication is associated with higher probability of severe disease outcome.
It is remarkable that approval was obtained in only 9–10 months from the initial discovery of these mAbs. This rapid process was made possible thanks to several factors, including a rapid isolation of the mAbs, an accelerated manufacturing process to generate clinical grade mAbs in only 3–4 months (Kelley, 2020), all-in-one clinical studies assessing both safety and efficacy, and a rapid analysis and approval by regulatory agencies. An inherent limitation of this rapid development process was that these mAbs could not be characterized for their resistance profile against variants that did not exist at that time. Indeed, all these four mAbs (etesevimab, bamlanivimab, casirivimab, and regdanvimab) are sensitive to one or more of the mutations found in RBD residues at positions 417, 452, or 484 that are carried by multiple circulating VOCs, including the recently emerging B.1.617 (Hoffmann et al., 2021; McCallum et al., 2021a; Starr et al., 2021b; Wang et al., 2021b) (Figure 5C).
Large-scale manufacturing for these approved mAbs is underway, and hundreds of thousands of doses have already been deployed in multiple countries. It is worth noting that each batch of production derived from the largest bioreactors (15,000 L) delivers ∼100–200,000 doses. Historically, the typical yearly supply of doses required for mAbs in oncology and inflammation is generally lower and fully covered by existing production plants, while the need for millions of doses that might be needed for COVID-19 is unprecedented in the field. Thus, if the global demand of mAbs exceeds the current production capacity of individual companies in the near future, then a cooperation between multiple private and public sectors will be needed for the simultaneous access to multiple very-large-scale bioreactors (Kelley, 2020; Kelley et al., 2021).
Importantly, these are the first antiviral mAbs authorized for use in a therapeutic setting for respiratory pathogens, proving the concept that established respiratory infections can be cured with mAbs, at least when mAbs are administered at an early stage of the disease. These same approved mAbs are also under investigation in multiple clinical trials (see below) to assess their efficacy in prophylaxis, post-exposure prophylaxis in residents and staff of long-term care facilities, and late therapy, where mAbs are tested in hospitalized patients with different degrees of disease severity (Table 1).
mAbs tested for early therapy of COVID-19 in advanced clinical stages
The success of early therapy of mild-to-moderate COVID-19 in high-risk patients by the mAbs presented above represents an important proof of concept that similar mAbs or mAbs that may provide an optimal coverage of evolving VOCs arewarranted.
VIR-7831 is also tested in a phase 2 early therapy study in adults in combination with bamlanivimab (BLAZE-4). A derivative of VIR-7831, VIR-7832, in which the GAALIE mutation was added to provide a potential vaccinal effect is currently being tested in the United Kingdom in parallel with VIR-7831 in an early-therapy phase 2 study (Agile study) to assess in humans the potential effect of the so-called vaccinal effect introduced by the Fc GAALIE mutation.
Three other studies are testing the efficacy of early therapy by mAbs: (1) a phase 3 study testing the cocktail of AZD7442 mAbs (YTE-TM) administered intramuscularly at a dose of 0.6 g (Zost et al., 2020), (2) a phase 2 study of the single RBM mAb BGB-DXP593 tested at three undisclosed doses administered intravenously (Cao et al., 2020), and (3) a phase 1/2/3 study of the single mAb ADG20 recognizing a conserved site on RBD tested as a single undisclosed dose administered either intravenously or intramuscularly (Rappazzo et al., 2021).
mAbs tested for late therapy of COVID-19 in advanced clinical stages
Late therapy with antiviral mAbs have historically shown limited success, but this is changing with the discovery of antibodies with remarkable neutralization breadth and potency, as exemplified by the successful use of mAbs to treat Ebola virus disease. mAbs might not be effective late in the course of COVID-19 disease, when viral replication is already decreasing and the disease course is mainly driven by host responses, leading to immune dysregulation, immunopathology, and exacerbation of underlying comorbid conditions. On the other hand, the successful use of convalescent plasma in hospitalized patients when provided before patients require ventilation (Ma et al., 2021; Tworek et al., 2021) argues for a careful assessment of the potential for mAb therapy in hospitalized patients.
An early study testing bamlanivimab in hospitalized patients (ACTIV-3 study organized by the NIH) was halted due to the lack of efficacy and unspecified safety concerns, suggesting that bamlanivimab is unlikely to help hospitalized COVID-19 patients recover from this advanced stage of disease (ACTIV-3/TICO LY-CoV555 Study Group et al., 2021). In the same trial (i.e., ACTIV-3), three more mAb products, in combination with standard of care, are under investigation (VIR-7831, Brii-196 + Brii-198, and AZD8895 + AZD1061). Both the VIR-7831 and Brii mAbs sub-studies were recently halted by the trial sponsor, the National Institute of Allergy and Infectious Diseases (NIAID), following an interim review and recommendation from the independent Data and Safety Monitoring Board (DSMB). There were no safety concerns with VIR-7831 or Brii-196 + Brii-198, and it was noted that VIR-7831, upon initial analysis, did meet the prespecified conditions for continuation (NIH (National Institutes of Health, 2021). Data continue to be collected, and publication is expected shortly. AbbVie is also testing ABBV-47D11 and ABBV-2B04 mAbs (Alsoussi et al., 2020; Wang et al., 2020) alone and in combination for treating adults hospitalized with COVID-19 in a phase 1 study. The treatment with REGN-COV2 mAbs is part of another comparative trial, the RECOVERY trial, in which multiple therapeutic modalities are being tested (hydroxychloroquine, aspirin, baricitinib, azithromycin, lopinavir-ritonavir convalescent plasma, dexamethasone, colchicine ,and tocilizumab). This study has so far shown a lack of efficacy for azithromycin, convalescent plasma, and lopinavir-ritonavir. The testing of REGN-COV2 in hospitalized patients receiving mechanical ventilation or high-flow oxygen was halted after the observation of a potential safety signal and an unfavorable risk/benefit profile. However, the testing of REGN-COV2 continued for hospitalized patients with less serious cases. The overall lack of efficacy of antiviral mAbs observed in hospitalized patients in late stages of disease is reminiscent of what was observed with the influenza HA stem mAb MHAA4549A that did not improve clinical outcomes over the standard of care (i.e., oseltamivir) alone when tested as late therapy in hospitalized patients (Lim et al., 2020).
The efficacy of mAbs in hospitalized patients, and the understanding of which disease stage would be compatible with the beneficial effect of mAbs, are key to add more therapeutic options to the current armamentarium of drugs to reduce severity and mortality in the advanced stages of COVID-19. Importantly, the use of anti-inflammatory drugs, which can limit the local damage triggered by SARS-CoV-2 infection, has also involved the use of mAbs. Two approved mAbs, sarilumab and tolicizumab, directed to the receptor of the pro-inflammatory cytokine interleukin-6 (IL-6) and approved for the treatment of rheumatoid arthritis, have also been tested in COVID-19. Over 350 studies have been initiated to study the effect of inhibition of IL-6 receptor (IL-6R) or IL-6 for COVID-19. Three major studies (REMAC-CAP, COVACTA, and RECOVERY) have published the validity of this approach in COVID-19 hospitalized patients, with contrasting results (Horby et al., 2021; REMAP-CAP Investigators et al., 2021; Rosas et al., 2021). In two of these studies (RECOVERY and REMAC-CAP), a reduced risk of death and an overall improvement of medical conditions was observed, while the third study did not observe a beneficial effect over placebo. Several variables may have influenced the disparate outcomes of these trials, including the definition of the patient population eligible for enrollment. The most notable is that in the two successful studies most of the patients received glucocorticoid therapy as standard of care, while only a minority of patients of the COVACTA study did. These results may suggest a possible additive or synergistic effect in using anti-inflammatory drugs (i.e., IL-6 and glucocorticoids) acting with different modalities. These results may call for combination therapies utilizing antiviral mAbs (or small-molecule drugs) with anti-inflammatories designed to address immune dysregulation (e.g., tolicizumab, sarilumab, and glucocorticoids), with the goal to improve efficacy in hospitalized patients and possibly expand the therapeutic window of intervention.
mAbs tested for prophylaxis of COVID-19 in advanced clinical development
In the setting of prophylaxis, mAbs could potentially be used alone or as a complement to vaccines. mAbs could also be used to provide an additional layer of protection for individuals at high risk of developing severe complications of COVID-19, such as the elderly and individuals with comorbidities or in those who are immunocompromised, and also to complement vaccine efficacy in case of significant SARS-CoV-2 antigenic drift.
Two mAbs have so far shown efficacy in interim analyses of phase 3 data in a prophylactic setting, bamlanivimab (BLAZE-2) and the REGN-COV2 cocktail (Study 2069), although publication of data from either trial is still pending, and data from these trials were only available via press releases. Bamlanivimab was reported to prevent COVID-19 in nursing homes where both residents and staff who tested negative for SARS-CoV-2 were enrolled. The subjects were followed for 8 weeks, and treatment with mAb was reported to significantly lower the frequency of symptomatic COVID-19, suggesting that subjects had up to an 80% lower risk of COVID-19. Results for the REGN-COV2 cocktail was reported from ∼400 subjects who were seronegative and did not have COVID-19 at baseline but who had a household member who had COVID-19. Here, prophylaxis with the cocktail was reported to prevent 100% of the symptomatic infections and had 50% lower rates of infection. Although viral loads were not reported for the bamlanivimab study, the peak viral loads in the placebo group of the REGN-COV2 household contact study were reported to be more than 100-fold higher than those treated with mAb, and treatment with REGN-COV2 was shown to reduce the duration of viral shedding. Both bamlanivimab and REGN-COV2 are IgG1 mAbs without Fc modifications, and data reported for these trials only looked at subjects soon after mAb administration (i.e., ≤2 months). Taken together, these data indicate a high potential for mAb-based immune prophylaxis of COVID-19 and support the hypothesis that protection from infection and disease can be mediated by neutralizing mAbs, furthermore indicating that vaccine-elicited antibodies are likely the major correlate of protection, as observed in non-human primates (Arunachalam et al., 2021; McMahan et al., 2021; Walls et al., 2020a).
These preliminary findings represent an important proof of concept for the development of mAbs with half-life extension to enable safe and reliable protection for up to 6–12 months that could be used as a vaccine alternative for individuals who respond poorly to or are not eligible for vaccination. The combination of RBM mAbs AZD8895 and AZD1061 was engineered for half-life extension with the YTE mutation and to abrogate effector functions with the addition of the TM mutation in the Fc. This mAb cocktail is being tested in an early-therapy study and in a large pre-exposure prophylactic study enrolling up to 5,000 participants administered with a low dose of the AZD8895/AZD1061 cocktail (0.15 + 0.15 g of each mAb) that is compatible with intramuscular delivery. A primary completion date of the prophylactic trial is expected in August 2021 and will assess the incidence of SARS-CoV-2 PCR-positive symptomatic illness against placebo for a period up to 6 months. The lack of effector functions of the AZD8895/AZD1061 cocktail and the results of its efficacy may be important to determine the contribution of effector functions to efficacy in prophylactic or therapeutic settings. The use of low doses such as the one used for the AZD8895/AZD1061 cocktail is key for a low-volume intramuscular or subcutaneous injection of a high-concentration product. As mentioned above, the shift from intravenous to these routes of administration is required for a larger access to these mAbs.
Concluding remarks and future perspectives
This review provides a comprehensive and up-to-date report on a selection of neutralizing mAbs against SARS-CoV-2 that have been or are being developed clinically. These mAbs target multiple sites on the SARS-CoV-2 S RDB and have shown different degrees of vulnerability to circulating VOCs/VOIs. Collectively, the data published to date suggest that their efficacy in preventing or controlling SARS-CoV-2 infection in animal models relies not only on viral neutralization but also on other indirect mechanisms, such as effector functions. The rapid successes obtained with these mAbs in both prophylactic and therapeutic clinical trials has been made possible thanks to the extensive experience accumulated in the past several years on the swift isolation of human mAbs, the availability of multiple Fc-engineering approaches aimed to fine-tune effector functions and improve half-life, and the great advances made in the manufacturing process. In a single year, the field of anti-infective mAbs has gained more knowledge regarding the clinical use of these molecules than what has been accumulated in the past 20 years. Several factors still limit the successful contribution of approved mAbs to the control of the COVID-19 pandemic, including the need for very-large-scale manufacturing (which could benefit from cooperation among public and private sectors), the awareness that these are now available treatments among both patients and providers, and the need to rapidly shift to routes of administration not requiring hospital settings. The partial or complete success in filling these gaps may ultimately foster the global access to mAbs to reduce the overall COVID-19 morbidity and mortality.
Acknowledgments
We would like to thank Siro Bianchi for the help in the preparation of the figures and Julia Di Julio for the analysis of prevalence of VOCs. Thanks to Christy Hebner, Bolyn Hubby, and Herbert W. Virgin for their insightful comments. This study was supported by the National Institute of General Medical Sciences (R01GM120553 to D.V.), a Pew Biomedical Scholars Award (to D.V.), and an Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (to D.V.).
Declaration of interests
D.C., L.A.P., and G.S. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. L.A.P. is a former employee and may hold shares in Regeneron Pharmaceuticals. D.V. is a consultant for Vir Biotechnology. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology, Inc.
References
- ACTIV-3/TICO LY-CoV555 Study Group. Lundgren J.D., Grund B., Barkauskas C.E., Holland T.L., Gottlieb R.L., Sandkovsky U., Brown S.M., Knowlton K.U., Self W.H., et al. A Neutralizing Monoclonal Antibody for Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384:905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S.O., Takas T., Nyborg A., Shoemaker K., Kallewaard N.L., Chiong R., Dubovsky F., Mallory R.M. Evaluation of MEDI8852, an Anti-Influenza A Monoclonal Antibody, in Treating Acute Uncomplicated Influenza. Antimicrob. Agents Chemother. 2018;62:1–47. doi: 10.1128/AAC.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021 doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020;183:1901–1912.e1909. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S., et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behring E.v., Kitasato S. Philipps-Universität Marburg; 2013. Ueber das Zustandekommen der Diphtherie-Immunität und der Tetanus-Immunität bei Thieren. [Google Scholar]
- Bournazos S., Corti D., Virgin H.W., Ravetch J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss L.F., Prete C.A., Jr., Abrahim C.M.M., Mendrone A., Jr., Salomon T., de Almeida-Neto C., França R.F.O., Belotti M.C., Carvalho M.P.S.S., Costa A.G., et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Estrany X., Simões E.A.F., Dagan R., Hall C.B., Harris B., Hultquist M., Connor E.M., Losonsky G.A., Motavizumab Study Group Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: a noninferiority trial. Pediatrics. 2010;125:e35–e51. doi: 10.1542/peds.2008-1036. [DOI] [PubMed] [Google Scholar]
- Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart A.L., Havenar-Daughton C., Lempp F.A., Ma D., Schmid M., Agostini M.L., Guarino B., Di Iulio J., Rosen L., Tucker H., et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Winkler E., Case J., Aziati I., Bricker T., Joshi A., Darling T., Ying B., Errico J., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Research Square. 2021 doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., et al. In vitro and in vivo preclinical studies predict REGEN-COV protection against emergence of viral escape in humans. bioRxiv. 2021 doi: 10.1101/2021.03.10.434834. [DOI] [Google Scholar]
- Corey L., Gilbert P.B., Juraska M., Montefiori D.C., Morris L., Karuna S.T., Edupuganti S., Mgodi N.M., deCamp A.C., Rudnicki E., et al. HVTN 704/HPTN 085 and HVTN 703/HPTN 081 Study Teams Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Cameroni E., Guarino B., Kallewaard N.L., Zhu Q., Lanzavecchia A. Tackling influenza with broadly neutralizing antibodies. Curr. Opin. Virol. 2017;24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Acqua W.F., Woods R.M., Ward E.S., Palaszynski S.R., Patel N.K., Brewah Y.A., Wu H., Kiener P.A., Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J. Immunol. 2002;169:5171–5180. doi: 10.4049/jimmunol.169.9.5171. [DOI] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Antón-Plágaro C., Kavanagh Williamson M., Shoemark D.K., Simón-Gracia L., Klein K., Bauer M., Hollandi R., Greber U.F., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. CMMID COVID-19 Working Group. COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:372. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021 doi: 10.1016/j.cell.2021.03.055. Published online March 30, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021 doi: 10.1016/j.cell.2021.04.025. Published online April 20, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zost S., Greaney A., Starr T.N., Dingens A.S., Chen E.C., Chen R., Case B., Sutton R., Gilchuk P., et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 doi: 10.1101/2021.01.27.428529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud M.B., Lee J.M., Bloom J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (US Food and Drug Administration) 2020. Emergency Use Authorization (EUA) for bamlanivimab. Center for Drug Evaluation and Research (CDER)https://www.fda.gov/media/144118/download [Google Scholar]
- FDA (US Food and Drug Administration) 2020. FACT SHEET FOR HEALTH CARE PROVIDERS EMERGENCY USE AUTHORIZATION (EUA) OF CASIRIVIMAB AND IMDEVIMAB. Emergency Use Authorization.https://www.fda.gov/media/143892/download [Google Scholar]
- FDA (US Food and Drug Administration) 2021. Development of Monoclonal Antibody Products Targeting SARS-CoV-2, Including Addressing the Impact of Emerging Variants, During the COVID 19 Public Health Emergency.https://www.fda.gov/regulatory-information/search-fda-guidance-documents/development-monoclonal-antibody-products-targeting-sars-cov-2-including-addressing-impact-emerging [Google Scholar]
- Fedry J., Hurdiss D.L., Wang C., Li W., Obal G., Drulyte I., Howes S.C., van Kuppeveld F.J.M., Förster F., Bosch B.-J. A human monoclonal antibody targeting a conserved pocket in the SARS-CoV-2 receptor-binding domain core. bioRxiv. 2020 doi: 10.1101/2020.09.30.318261. [DOI] [Google Scholar]
- Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00170-5. Published online April 12, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu G., Chemaly R.F. Palivizumab: where to from here? Expert Opin. Biol. Ther. 2009;9:139–147. doi: 10.1517/14712590802610692. [DOI] [PubMed] [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M.S., Sudre C.H., May A., Antonelli M., Murray B., Varsavsky T., Kläser K., Canas L.S., Molteni E., Modat M., et al. COVID-19 Genomics UK (COG-UK) Consortium Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021;6:e335–e345. doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin M.P., Yuan Y., Takas T., Domachowske J.B., Madhi S.A., Manzoni P., Simões E.A.F., Esser M.T., Khan A.A., Dubovsky F., et al. Nirsevimab Study Group Single-Dose Nirsevimab for Prevention of RSV in Preterm Infants. N. Engl. J. Med. 2020;383:415–425. doi: 10.1056/NEJMoa1913556. [DOI] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Gonzales-Rojas Y., Juarez E., Casal M.C., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early Covid-19 Treatment With SARS-CoV-2 Neutralizing Antibody Sotrovimab. medRxiv. 2021 doi: 10.1101/2021.05.27.21257096. [DOI] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger E., Sloan S., Narayan K., Hay C.A., Smith P., Engler F., Jeeninga R., Smits S., Trevejo J., Shriver Z., Oldach D. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: Results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine. 2019;40:574–582. doi: 10.1016/j.ebiom.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W., et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Hezareh M., Hessell A.J., Jensen R.C., van de Winkel J.G., Parren P.W. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 2001;75:12161–12168. doi: 10.1128/JVI.75.24.12161-12168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Sidarovich A., Moldenhauer A., Winkler M., S., Schulz S., et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv. 2021 doi: 10.1101/2021.05.04.442663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.A.P., Gross R., Seidel A., Hornich B., Hahn A., Kruger N., Graichen L., Pohlmann S. SARS-CoV-2 variants B.1.351 and B.1.1.248: Escape from therapeutic antibodies and antibodies induced by infection and vaccination. bioRxiv. 2021 doi: 10.1101/2021.02.11.430787. [DOI] [Google Scholar]
- Horby P.W., Pessoa-Amorim G., Peto L., Brightling C.E., Sarkar R., Thomas K., Jeebun V., Ashish A., Tully R., Chadwick D., et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): preliminary results of a randomised, controlled, open-label, platform trial. medRxiv. 2021 doi: 10.1101/2021.02.11.21249258. [DOI] [Google Scholar]
- IMpact-RSV Study Group Palivizumab, a Humanized Respiratory Syncytial Virus Monoclonal Antibody, Reduces Hospitalization From Respiratory Syncytial Virus Infection in High-risk Infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Julg B., Barouch D. Broadly neutralizing antibodies for HIV-1 prevention and therapy. Semin. Immunol. 2021 doi: 10.1016/j.smim.2021.101475. Published online April 12, 2021. [DOI] [PubMed] [Google Scholar]
- Kallewaard N.L., Corti D., Collins P.J., Neu U., McAuliffe J.M., Benjamin E., Wachter-Rosati L., Palmer-Hill F.J., Yuan A.Q., Walker P.A., et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell. 2016;166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley B. Developing therapeutic monoclonal antibodies at pandemic pace. Nat. Biotechnol. 2020;38:540–545. doi: 10.1038/s41587-020-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley B., Renshaw T., Kamarck M. Process and operations strategies to enable global access to antibody therapies. Biotechnol. Prog. 2021 doi: 10.1002/btpr.3139. Published online March 8, 2021. [DOI] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick E., Qiu X., Wilson P.C., Bahl J., Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018;8:10432. doi: 10.1038/s41598-018-28706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situcryo-electron tomography. Nat. Commun. 2020;3:237–238. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S.-Y., Pegu A., Rudicell R.S., Yang Z.-Y., Joyce M.G., Chen X., Wang K., Bao S., Kraemer T.D., Rath T., et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leschinsky M., Dror S.K., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2 mRNA vaccinated individuals. medRxiv. 2021 doi: 10.1101/2021.04.06.21254882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempp F.A., Soriaga L., Montiel-Ruiz M., Benigni F., Noack J., Park Y.-J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., et al. Membrane lectins enhance SARS-CoV-2 infection and influence the neutralizing activity of different classes of antibodies. bioRxiv. 2021 doi: 10.1101/2021.04.03.438258. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.J., Nilsson A.C., Silverman M., Assy N., Kulkarni P., McBride J.M., Deng R., Li C., Yang X., Nguyen A., et al. A Phase 2 Randomized, Double-Blind, Placebo-Controlled Trial of MHAA4549A, a Monoclonal Antibody, plus Oseltamivir in Patients Hospitalized with Severe Influenza A Virus Infection. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00352-20. 1285–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Wiggins C.C., Kornatowski B.M., Hailat R.S., Clayburn A.C., Guo W., Johnson P.W., Senefeld J.W., Klassen S.A., Baker S.E., et al. The Role of Disease Severity and Demographics in the Clinical Course of COVID-19 Patients Treated with Convalescent Plasma. medRxiv. 2021 doi: 10.3389/fmed.2021.707895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.-M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R., Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Bassi J., Marco A., Chen A., Walls A.C., Iulio J.D., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. 2021 doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza P., Gruell H., Nogueira L., Pai J.A., Butler A.L., Millard K., Lehmann C., Suárez I., Oliveira T.Y., Lorenzi J.C.C., et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature. 2018;561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., et al. PALM Writing Group. PALM Consortium Study Team A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munitz A., Yechezkel M., Dickstein Y., Yamin D., Gerlic M. BNT162b2 Vaccination Effectively Prevents the Rapid Rise of SARS-CoV-2 Variant B.1.1.7 in high risk populations in Israel. Cell Rep. Med. 2021;2:100264. doi: 10.1016/j.xcrm.2021.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Nachbagauer R., Balmaseda A., Stadlbauer D., Ojeda S., Patel M., Rajabhathor A., Lopez R., Guglia A.F., Sanchez N., et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med. 2019;25:962–967. doi: 10.1038/s41591-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH (National Institutes of Health . 2021. Statement—NIH-Sponsored ACTIV-3 Clinical Trial Closes Enrollment into Two Sub-Studies.https://www.nih.gov/news-events/news-releases/nih-sponsored-activ-3-clinical-trial-closes-enrollment-into-two-sub-studies [Google Scholar]
- Oganesyan V., Gao C., Shirinian L., Wu H., Dall’Acqua W.F. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr. D Biol. Crystallogr. 2008;64:700–704. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D.F., Lansing J.C., Rutitzky L., Kurtagic E., Prod’homme T., Choudhury A., Washburn N., Bhatnagar N., Beneduce C., Holte K., et al. Elucidating the interplay between IgG-Fc valency and FcγR activation for the design of immune complex inhibitors. Sci. Transl. Med. 2016;8:365ra158. doi: 10.1126/scitranslmed.aaf9418. [DOI] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redden W.R. Treatment of Influenza-Pneumonia by Use of Convalescent Human Serum. Boston Med. Surg. J. 1919;181:688–691. [Google Scholar]
- REMAP-CAP Investigators. Gordon A.C., Mouncey P.R., Al-Beidh F., Rowan K.M., Nichol A.D., Arabi Y.M., Annane D., Beane A., van Bentum-Puijk W., et al. Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N. Engl. J. Med. 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas I.O., Bräu N., Waters M., Go R.C., Hunter B.D., Bhagani S., Skiest D., Aziz M.S., Cooper N., Douglas I.S., et al. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N. Engl. J. Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C., Mandal S.M., Mondal S.K., Mukherjee S., Mapder T., Ghosh W., Chakraborty R. Trends of mutation accumulation across global SARS-CoV-2 genomes: Implications for the evolution of the novel coronavirus. Genomics. 2020;112:5331–5342. doi: 10.1016/j.ygeno.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D.-K., Song R., Kim M., Kim Y.-I., Kim C., Kim J.-I., Kwon K.-S., Tijsma A.S.L., Nuijten P.M., van Baalen C.A., et al. Therapeutic effect of CT-P59 against SARS-CoV-2 South African variant. bioRxiv. 2021 doi: 10.1101/2021.04.27.441707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R., Mitra S., Chandra P., Saha P., Banerjee A., Dutta S., Chawla-Sarkar M. Comprehensive analysis of genomic diversity of SARS-CoV-2 in different geographic regions of India: an endeavour to classify Indian SARS-CoV-2 strains on the basis of co-existing mutations. Arch. Virol. 2021;166:801–812. doi: 10.1007/s00705-020-04911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:218. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T., Klein F., Braunschweig M., Kreider E.F., Feldmann A., Nogueira L., Oliveira T., Lorenzi J.C.C., Parrish E.H., Learn G.H., et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Simões E.A.F., Forleo-Neto E., Geba G.P., Kamal M., Yang F., Cicirello H., Houghton M.R., Rideman R., Zhao Q., Benvin S.L., et al. Suptavumab for the Prevention of Medically Attended Respiratory Syncytial Virus Infection in Preterm Infants. Clin. Infect. Dis. 2020;390:946–949. doi: 10.1093/cid/ciaa951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh W.T., Liu Y., Nakayama E.E., Ono C., Torii S., Nakagami H., Matsuura Y., Shioda T., Arase H. The N-terminal domain of spike glycoprotein mediates SARS-CoV-2 infection by associating with L-SIGN and DC-SIGN. bioRxiv. 2020 [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Zatta F., Park Y.J., Liu Z., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape. bioRxiv. 2021 doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Chen Z., Cox K.S., Su H.-P., Callahan C., Fridman A., Zhang L., Patel S.B., Cejas P.J., Swoyer R., et al. A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein. Nat. Commun. 2019;10:4153. doi: 10.1038/s41467-019-12137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Iulio J.D., Wang Z., et al. Structural basis for broad sarbecovirus neutralization by a human monoclonal antibody. bioRxiv. 2021 doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworek A., Jaroń K., Uszyńska-Kałuża B., Rydzewski A., Gil R., Deptała A., Franek E., Wójtowicz R., Życińska K., Walecka I., et al. Convalescent plasma treatment is associated with lower mortality and better outcomes in high-risk COVID-19 patients: propensity-score matched case-control study. Int. J. Infect. Dis. 2021;105:209–215. doi: 10.1016/j.ijid.2021.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. COG-UK Consortium Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Kim J.E., Delidakis G., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., Abbasi S.A., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv. 2021 doi: 10.1101/2021.03.01.433466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzenfeld P., Bournazos S., Ravetch J.V. Antibodies targeting sialyl Lewis A mediate tumor clearance through distinct effector pathways. J. Clin. Invest. 2019;129:3952–3962. doi: 10.1172/JCI128437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e1816. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Alegre M.L., Varga S.S., Rothermel A.L., Collins A.M., Pulito V.L., Hanna L.S., Dolan K.P., Parren P.W., Bluestone J.A., et al. In vitro characterization of five humanized OKT3 effector function variant antibodies. Cell. Immunol. 2000;200:16–26. doi: 10.1006/cimm.2000.1617. [DOI] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., Desjarlais J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010;28:157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen F.P., Sullender W.M. Respiratory syncytial virus escape mutant derived in vitro resists palivizumab prophylaxis in cotton rats. Virology. 2004;318:608–612. doi: 10.1016/j.virol.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., McAuliffe J.M., Patel N.K., Palmer-Hill F.J., Yang C.-F., Liang B., Su L., Zhu W., Wachter L., Wilson S., et al. Analysis of respiratory syncytial virus preclinical and clinical variants resistant to neutralization by monoclonal antibodies palivizumab and/or motavizumab. J. Infect. Dis. 2011;203:674–682. doi: 10.1093/infdis/jiq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., McLellan J.S., Kallewaard N.L., Ulbrandt N.D., Palaszynski S., Zhang J., Moldt B., Khan A., Svabek C., McAuliffe J.M., et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci. Transl. Med. 2017;9:eaaj1928. doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]