Abstract
Aflatoxins seriously threaten the health of humans and animals due to their potential carcinogenic properties. Enzymatic degradation approach is an effective and environmentally friendly alternative that involves changing the structure of aflatoxins. In this study, Trametes versicolor aflatoxin B1-degrading enzyme gene (TV-AFB1D) was integrated into the genome of Pichia pastoris GS115 by homologous recombination approach. The recombinant TV-AFB1D was expressed in engineering P. pastoris with a size of approximately 77 kDa under the induction of methanol. The maximum activity of TV-AFB1D reached 17.5 U/mL after the induction of 0.8% ethanol (v/v) for 84 h at 28 °C. The AFB1 proportion of 75.9% was degraded using AFB1 standard sample after catalysis for 12 h. In addition, the AFB1 proportion was 48.5% using AFB1-contaminated peanuts after the catalysis for 18 h at 34 °C. The recombinant TV-AFB1D would have good practical application value in AFB1 degradation in food crops. This study provides an alternative degrading enzyme for the degradation of AFB1 in aflatoxin-contaminated grain and feed via enzymatic degradation approach.
Keywords: aflatoxin B1, Pichia pastoris GS115, peanuts, Trametes versicolor, degradation enzyme
1. Introduction
Aflatoxins are pathogenic fungal toxins produced by Aspergillus flavus and A. parasiticus [1]. Aflatoxins and their producing bacteria are usually present in food products, feedstuffs, and agricultural raw materials [2]. Aflatoxins are a real threat to human and animal health due to their carcinogenicity and teratogenicity [3]. Among more than 20 kinds of aflatoxin derivatives, AFB1 possesses the highest toxicity of liver cancer and immune system damage. AFB1 causes carcinogenesis in the presence of the double bond of AFB1 furan ring by inhibiting RNA synthesis [4]. The lactone ring of AFB1 is the main toxicity site. The cleavage of lactone ring reduces the biological activity of AFB1 with the formation of non-fluorescent compound [5]. In addition, a highly reactive AFB1-8,9-epoxide capable of reacting with DNA is produced under the activation of cytochrome P450 system [6,7].
Aflatoxins mainly pollute cereals, oils, and their products [8]. As an important agricultural raw material, peanut is directly in touch with A. flavus and A. parasiticus in the soil [9]. Peanut is highly susceptible to the contamination of aflatoxins. Aflatoxin-contaminated peanuts pollute the environment and cause the serious economic damage [10]. Despite remarkable improvements in the technologies of harvesting and storage processing, the contamination of aflatoxins is still a problem to be solved in the peanut industry [11]. Various physical, chemical, and biological methods have been developed to solve the problem of aflatoxin pollution [12]. The physical approach mainly includes heating treatment [13], UV radiation [14], high temperature [15], and adsorption [16]. The main disadvantages are long treatment time and low degradation rate [17,18]. The chemical methods include oxidant treatment and acid–base method by destroying the structure of aflatoxins [19,20]. Further, both the physical and chemical methods could reduce the quality of nutrition by changing the characteristics of raw materials [21].
Currently, the enzymatic degradation approach has been developed for its high specificity and efficiency of catalysis [22]. Laccase [23], peroxidase [24], and reductase [25] have been proved to be capable of degrading aflatoxins. The edible vegetable oils are vulnerable to aflatoxin pollution in the production process due to the raw materials and processing properties. Aflatoxin B1 is one of the most common toxic substances in edible vegetable oil. Recently, the quality and safety of edible vegetable oils has attracted great attention of China State Council and the State Food and Drug Administration. The government is concentrating R&D efforts to solve the problem of aflatoxin pollution.
Trametes versicolor is a common polypore mushroom found throughout the world and is also a well-known traditional medicinal mushroom, one that is found growing on tree trunks. We cloned AFB1-degrading enzyme gene from Trametes versicolor (TV-AFB1D) according to Genbank accession number txid717944 in NCBI. TV-AFB1D was expressed in engineering E. coli BL21(DE3) with TV-AFB1D activity of 9.3 U/mL (not reported). To further investigate the effectiveness of expression, we integrated TV-AFB1D into the genome of P. pastoris GS115 in this study. In addition, the application of TV-AFB1D for degradation was also investigated in AFB1-contaminated peanuts. This study provides a new AFB1-degrading enzyme for application in enzymatic degradation in the food and feed industry.
2. Results
2.1. TV-AFB1D Cloning and Evolutionary Tree Analysis
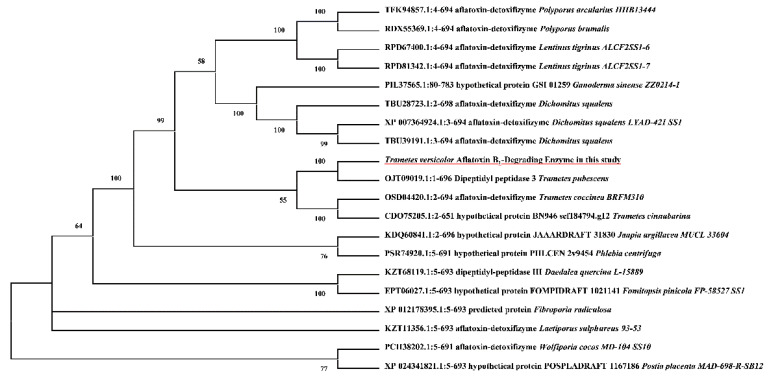
Aflatoxin B1-degrading enzyme TV-AFB1D from T. versicolor was cloned by using the designed primers with RT-PCR technique (Figure 1). The size of TV-AFB1D was 2100 bp according to electrophoresis analysis and sequencing confirmation. The evolutionary tree analysis of TV-AFB1D with AFB1 degradation enzymes from other species is shown in Figure 2. TV-AFB1D had closer genetic distance with Trametes coccinea than other aflatoxin-degrading enzymes from Dichomitus squalens, Polyporus arcularius, Lentinus tigrinus, and Laetiporus sulpbureus. All the aflatoxin-degrading enzymes from Trametes genus belonging to family Polyporaceae speculated that aflatoxin-degrading enzymes would have common traits according to the result of evolutionary tree analysis.
Figure 1.
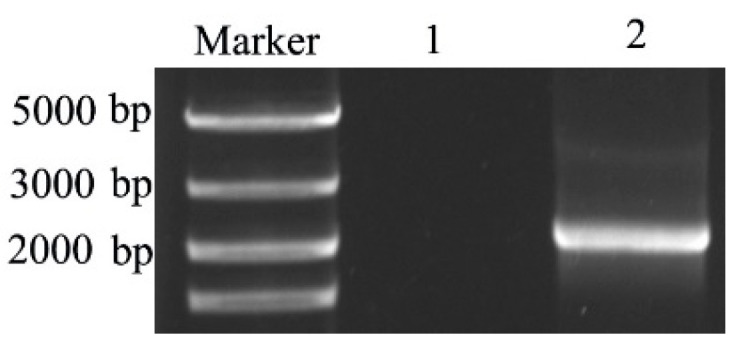
RT-PCR amplification of TV-AFB1D. Note: lane 1: control; lane 2: TV-AFB1D.
Figure 2.
Evolutionary tree analysis of TV-AFB1D with other AFB1-degrading enzymes.
2.2. Expression Vector Construction of TV-AFB1D
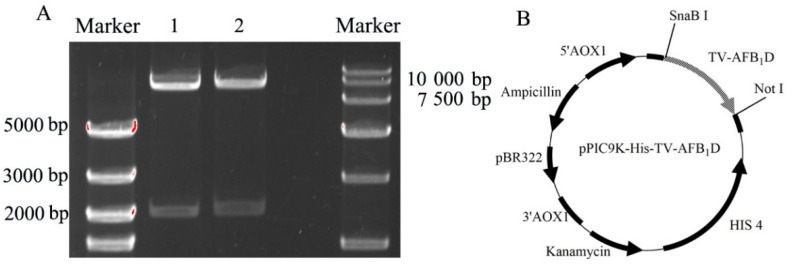
TV-AFB1D was integrated into pPIC9K-His vector by double digestion techniques. The constructed recombinant plasmid was identified by the double digestion of Sna B I and Not I. Two bands after digestion possessed the size of approximately 9300 and 2100 bp (Figure 3A). The confirmed recombinant vector was named pPIC9K-His-AFB1D after confirmation by gene sequencing (Figure 3B).
Figure 3.
Vector pPIC9K-His-TV-AFB1D construction. (A) Double-restriction digestion of vector pPIC9K-His-TV-AFB1D by SnaB I and Not I. Note: lanes 1, 2: pPIC9k-TV-AFB1D. (B) Schematic diagram of constructed pPIC9k-TV-AFB1D.
2.3. Transformation of TV-AFB1D into P. pastoris GS115
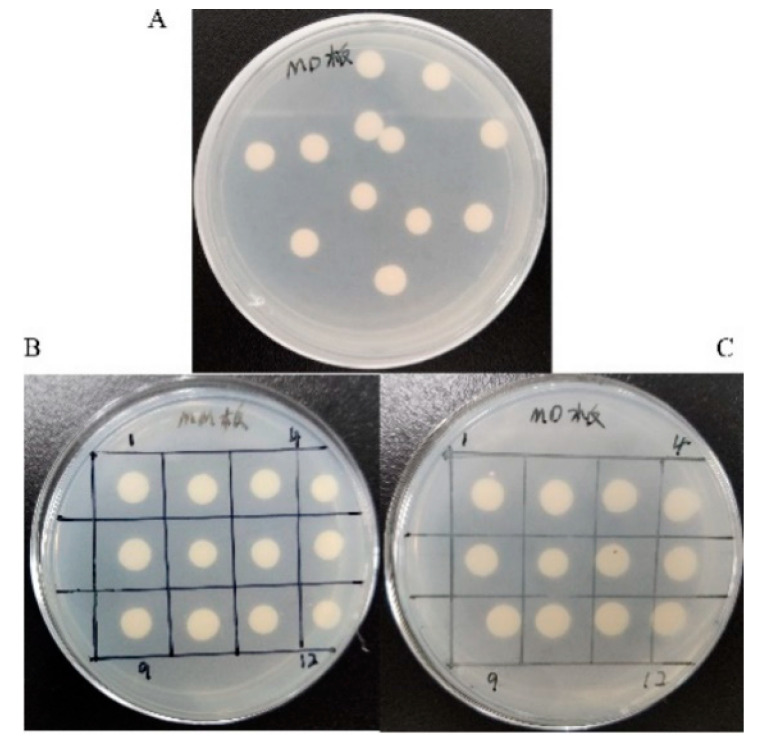
The P. pastoris transformants were cultured on the plates with MD media after transformation. The transformants were transferred to the plates equipped with MM media (Figure 4). The transformants were identified with the designed primers for TV-AFB1D amplification. P. pastoris GS115 transformants with His+Mut+ type for methanol utilization were integrated by pPIC9K-His-AFB1D according to the growth on the plates containing MD and MM media.
Figure 4.
Phenotype screening of recombinant P. pastoris GS115. (A) Screening with MD media; (B,C) recombinant P. pastoris screening with His+Mut+ phenotype using MM (B) and MD (C) plates.
2.4. Recombinant TV-AFB1D Expression in P. pastoris GS115
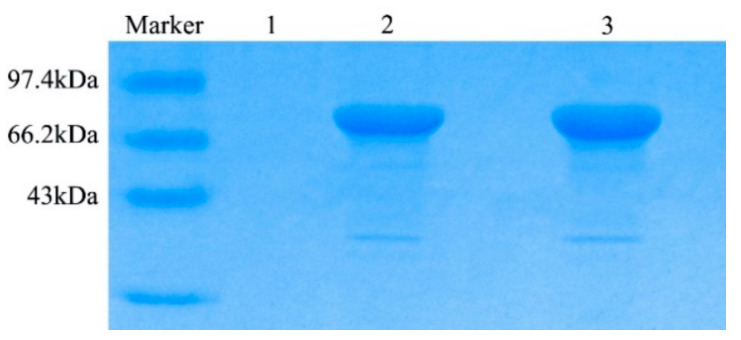
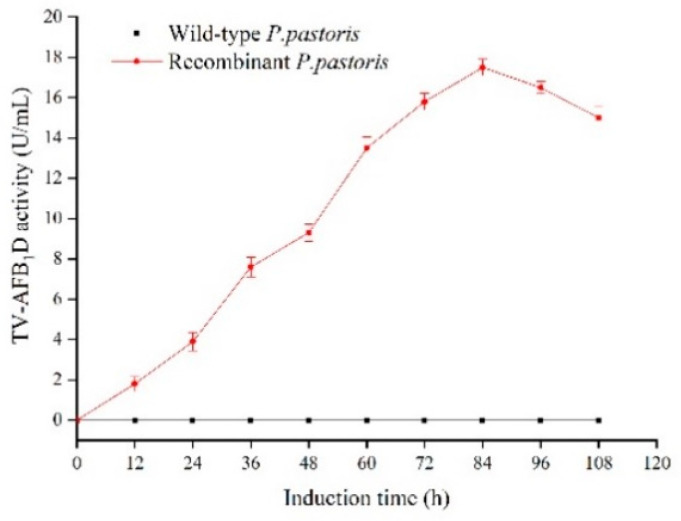
The recombinant TV-AFB1D was expressed by engineering P. pastoris GS115 with the size of approximately 77 kDa under the induction of 0.8% methanol. No TV-AFB1D was detected by the wild-type P. pastoris GS115 according to SDS-PAGE analysis (Figure 5). The highest activity of TV-AFB1D was 17.5 U/mL after the induction of 0.8% methanol (v/v) for 84 h at 30 °C. No TV-AFB1D activity of the fermentation broth from the wild-type P. pastoris GS115 was detected under the same treatment conditions (Figure 6). Therefore, the recombinant TV-AFB1D could express in engineering P. pastoris GS115.
Figure 5.
SDS-PAGE analysis of recombinant TV-AFB1D. Lane 1: wild-type P. pastoris expression; lanes 2 and 3: expression of the recombinant TV-AFB1D.
Figure 6.
Expression of TV-AFB1D from the transformants and wild-type P. pastoris.
2.5. Effect of Methanol Concentration and Time on the TV-AFB1D Expression
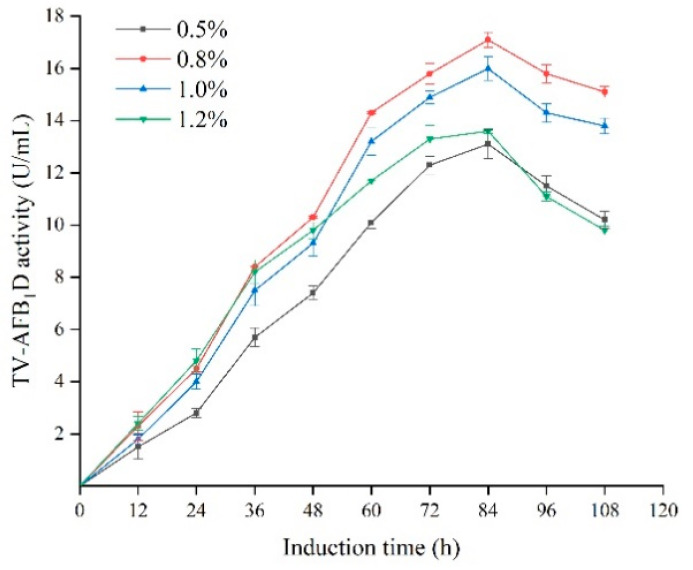
The effects of induction time and methanol concentration on TV-AFB1D activity were investigated (Figure 7). The highest activities of TV-AFB1D expressed by the recombinant P. pastoris GS115 were achieved after induction for 84 h and in the presence of 0.8% methanol (v/v). In particular, the activities of TV-AFB1D were substantially decreased with the addition of 1.2% and 0.5% of methanol. Therefore, the optimum methanol concentration and treatment time were 0.8% and 84 h for the inducible expression of TV-AFB1D, respectively.
Figure 7.
Effect of time and methanol concentration on the activity of TV-AFB1D.
2.6. Effect of Temperatures and Treatment Time on the Expression of TV-AFB1D
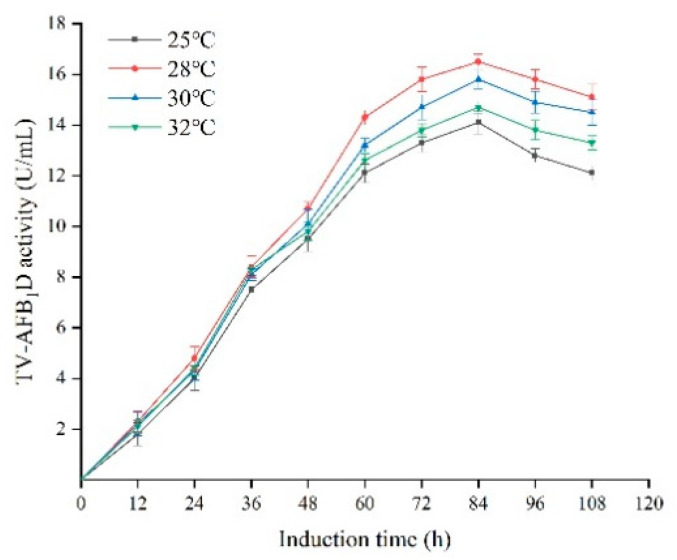
The effects of temperatures and treatment time on TV-AFB1D expression are shown in Figure 8. The TV-AFB1D activities reached the highest after induction for 84 h. The highest activity of TV-AFB1D was 16.5 U/mL at 28 °C among all the set temperatures of 25–32 °C. Therefore, the optimum temperature and time were 28 °C and 84 h for the inducible expression of TV-AFB1D in engineering P. pastoris GS115, respectively.
Figure 8.
Effect of temperature on the activity of TV-AFB1D.
2.7. Product Formation of AFB1-Contaminated Peanuts Catalyzed by TV-AFB1D
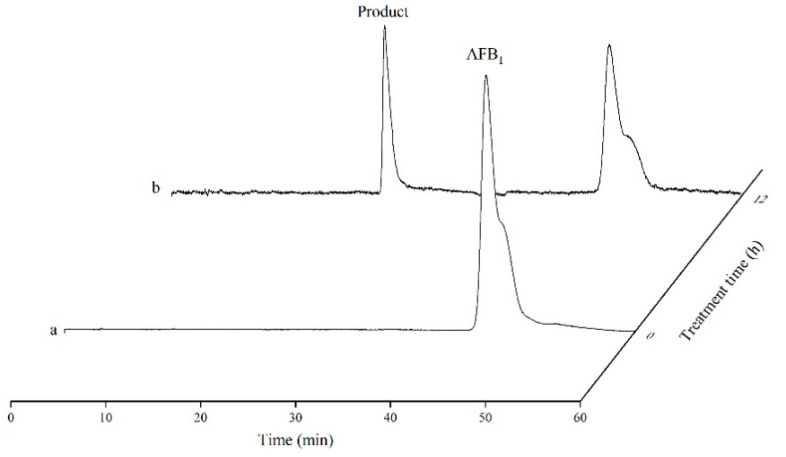
HPLC method was used to determine the product formation of AFB1 catalyzed by the recombinant TV-AFB1D. As the control group, the AFB1 standard possessed a special peak after running for 45 min at a wavelength of 365 nm (Figure 9). The profiles of catalytic products were determined by HPLC after the catalysis of AFB1-contaminated peanuts by the recombinant TV-AFB1D. With the exception of the AFB1 standard, another peak was detected under a wavelength of 210 nm after running for 22 min. The result indicated that new product was formed under the catalysis of TV-AFB1D with AFB1 as substrate.
Figure 9.
Determination of catalytic products of AFB1 catalyzed by TV-AFB1D using HPLC approach. Note: a and b represent HPLC profiles before and after catalysis, respectively.
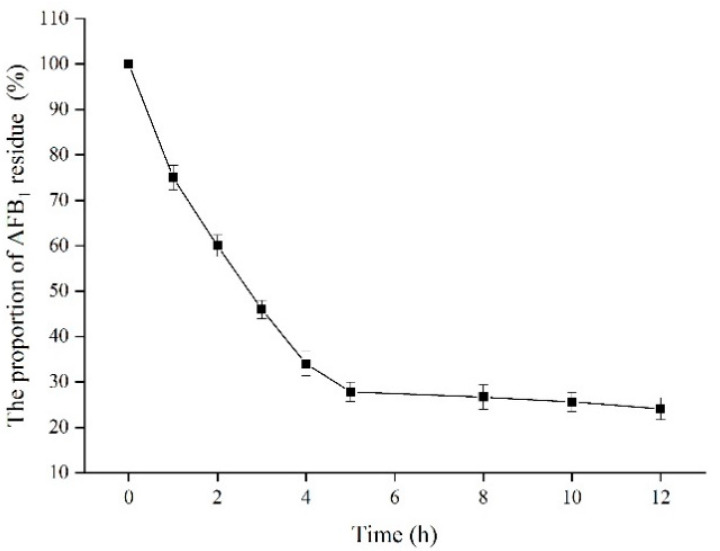
2.8. Effect of Time on the Residue of AFB1 Standard Sample
The effect of catalysis time on the residual proportion of the AFB1 standard sample was investigated at 32 °C (Figure 10). The proportion of AFB1 residue substantially decreased from 100% to 27.8% for 5 h of catalysis with the degradation rate of 72.2%. During the next catalysis for 5–12 h, the proportion of residual AFB1 decreased slowly. The proportion of residual AFB1 standard sample was 24.1% after catalysis for 12 h with the degradation rate of 75.9%.
Figure 10.
Effect of time on the concentration of AFB1 standard sample by TV-AFB1D.
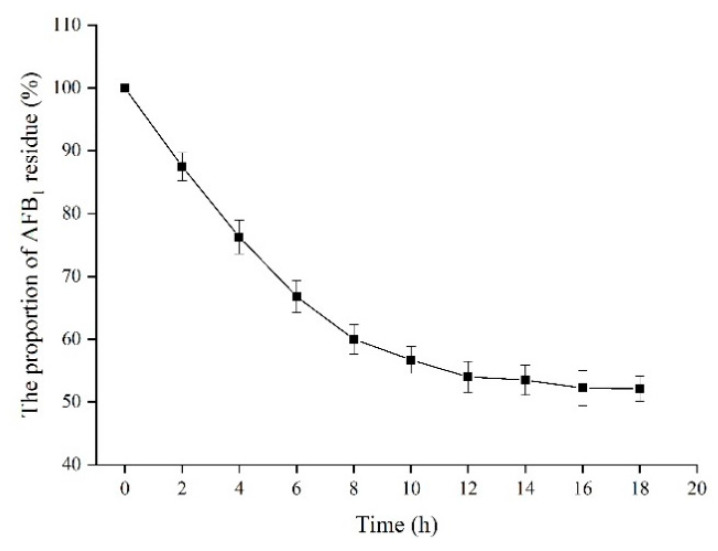
2.9. Effect of Time on the Proportion of AFB1 Residue in Peanuts
The effect of treatment time on the proportion of AFB1 residue in AFB1-contaminated peanuts was investigated at 32 °C (Figure 11). Within the initial 10 h, the proportion of AFB1 residue decreased rapidly from 100% to 55.7%. After treatment for 18 h, the proportion of AFB1 residue in AFB1-contaminated peanuts gradually decreased to 51.5% with the degradation rate of 48.5%. Therefore, the recombinant TV-AFB1D could effectively catalyze AFB1 in AFB1-contaminated peanuts.
Figure 11.
Effect of time on the concentration of AFB1 in AFB1-contaminated peanuts by TV-AFB1D.
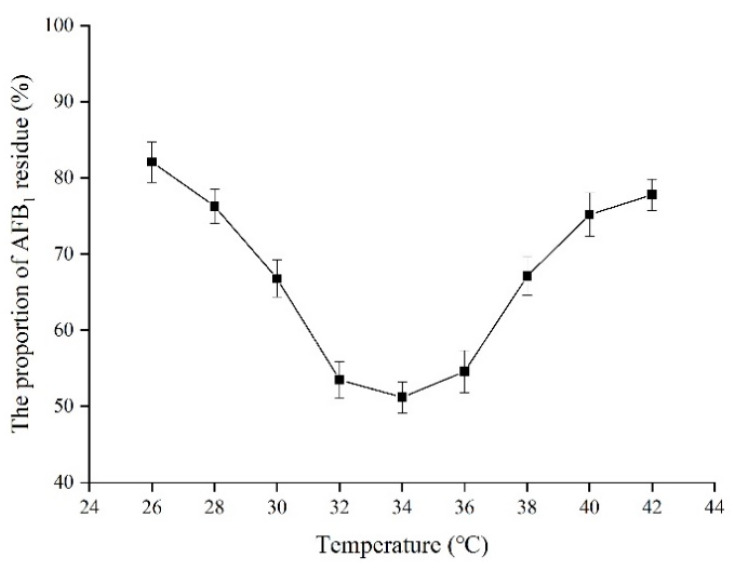
2.10. Effect of Temperature on the Proportion of AFB1 Residue in Peanuts
The effect of temperature on the proportion of AFB1 residue in AFB1-contaminated peanuts was investigated after catalysis for 5 h (Figure 12). In the range of 26 to 42 °C, the proportion curve of AFB1 residue was in the shape of a “V”. The proportion of AFB1 residue gradually decreased during the temperatures of 26 to 34 °C and increased during the temperatures of 34 to 42 °C. The lowest concentration of AFB1 residue in AFB1-contaminated peanuts was 52.1% at 34 °C with a degradation rate of 47.9%. Too low and too high temperatures were not conducive to the degradation of AFB1 by TV-AFB1D in AFB1-contaminated peanuts.
Figure 12.
Effect of temperature on the concentration of AFB1 in AFB1-contaminated peanuts.
3. Discussion
Although the physical and chemical methods have been applied in the detoxification of aflatoxins, these approaches are difficult to meet the requirement of clean, safe, and environmentally friendly production [26]. Currently, the enzymatic degradation has been applied in the degradation of aflatoxins for its safety and specificity [27]. A large number of studies have been reported on the enzymes related to aflatoxin degradation (Table 1). Enzymes related to aflatoxin degradation include various catalytic properties of F420H2-dependent reductase [25], laccase [23], and peroxidase [24]. In addition, aflatoxin-detoxifizyme (ADTZ) [28], AFB1-horse radish peroxidase (HRP) [29], Myxobacteria aflatoxin degradation enzyme (MADE) [30], Bacillus aflatoxin-degrading enzyme (BADE) [31], and Pantoea aflatoxin degradation enzyme (PADE) [32] also possess the capability of aflatoxin degradation. In the previous report, the catalytic performances of enzymes were identified by using AFB1 standard sample as catalytic substrate. These enzymes were rarely used for degradation of aflatoxins in food and feed. In this study, T. versicolor aflatoxin B1-degrading enzyme gene was expressed in engineering P. pastoris. AFB1 standard sample and AFB1-contaminated peanuts were used to detect the degradation effect of AFB1. The AFB1 proportions of 75.9% and 48.5% were degraded using AFB1 standard sample and AFB1-contaminated peanuts as substrates, respectively.
Table 1.
Aflatoxin-degrading enzymes and catalysis performances.
| Microbial Sources and Enzymes | Conditions | Catalytic Efficiency and Time |
|---|---|---|
| Mycobacterium smegmatis, F420H2-dependent reductase [25] | 20 mM Tris-HCl, pH 7.5 | 63%, 4 h |
| Myxococcus fulvus, MADE [30] | pH 6 | 96% |
| Armillariella tabescens, ADTZ [28] | 0.02 M PBS, pH 6 | 80% |
| Phanerochaete sordida, peroxidase [33] | 50 mM PBS, pH 6 | 86%, 48 h |
| Pleurotus ostreatus, peroxidase [34] | pH 4.5 | 90%, 48 h |
| Bacillus subtilis, BsCotA laccase [35] | 50 mM Tris–HCl, pH 7.0 | 98%, 10 h |
| Pleurotus pulmonarius, laccase [36] | 1 mM sodium acetate, pH 5 | 90%, 72 h |
| Aspergillus flavus, HRP [29] | 1mM sodium acetate, pH 5 | 42%, 1 h |
| Bacillus shackletonii, BADE [31] | 50 mM PBS, pH 7.0 | 47.5%, 72 h |
| Pseudomonas aeruginosa, PADE [32] | 0.02 M PBS, pH 7 | 72.5%, 12h |
| T. versicolor, TV-AFB1D, in this study | AFB1 standard sample | 75.9%, 12 h |
| T. versicolor, TV-AFB1D, in this study | AFB1-contaminated peanuts | 48.5%, 18 h |
4. Conclusions
In this study, the recombinant TV-AFB1D with a size of 77 kDa was effectively expressed by engineering P. pastoris. The optimal temperature and methanol concentration were 28 °C and 0.8% for the inducible expression of TV-AFB1D, respectively. The proportion of residual AFB1 standard sample was 24.1% after catalysis for 12 h with a degradation rate of 75.9%. In addition, the proportion of AFB1 residue in AFB1-contaminated peanuts decreased to 51.5% with a degradation rate of 48.5% after treatment for 18 h at 34 °C. This study provided an alternative TV-AFB1D degradation enzyme for the removal of AFB1 in food products and agricultural raw materials.
5. Materials and Methods
5.1. Materials and Reagents
Vector pPIC9K-His, TV-AFB1D gene, P. pastoris GS115, and T. versicolor were stored in the Experimental Center of Hefei University of Technology. Gene sequencing and primer synthesis were performed by Sangon Biotech. Taq polymerase, Stu I, Not I, and SnaB I were from NEB Company. Gel imaging systems, sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), polymerase chain reaction (PCR) amplification, and electrophoresis devices were manufactured by Bio-RAD Company. ProteinIsoNi-IDA resin and AFB1 immunoaffinity column were from Transgen Biotech and Welch Company, respectively.
5.2. Construction of pPIC9K-His-TV-AFB1D Vector
TV-AFB1D encoding aflatoxin degradation enzyme was cloned by reverse transcription PCR (RT-PCR) technique according to Genbank accession number txid717944 in the National Center for Biotechnology Information (NCBI). The sequences of restriction endonuclease SnaB I and Not I were added in the corresponding primers for the amplification of TV-AFB1D. The corresponding primers were designed as follows: F: 5′-GGGAAATACGTAATGGCTCGCGCGAAGTACTC-3′ (SnaB I); R: 5′-GGGAAAGCGGCCGCTTAAAGCTTCCGCTCTATGA3′ (Not I). Both the product of amplification and pPIC9K-His vector were double digested by SnaB I and Not I. The recovered gene was ligated by T4 ligase to produce vector pPIC9K-His-AFB1D. pPIC9K-His-AFB1D was confirmed by PCR amplification, double-enzyme digestion identification, and gene sequencing.
5.3. Transformation of pPIC9K-His-TV-AFB1D into P. pastoris GS115
The linearized pPIC9K-His-AFB1D was digested by Stu I and integrated into P. pastoris GS115 by electroporation transformation method [37]. P. pastoris GS115 had a mutation in histidine dehydrogenase (his4) and could not synthesize histidine. pPIC-9K-His contained his4 gene that was complementary to the host. The transformants were screened on the plate equipped with histidine-free medium. The recombinant P. pastoris with His+Mut+ phenotype grew on minimal methanol medium (MM) plates in the presence of methanol. The transformants were screened out using the solid plates containing minimal dextrose medium (MD) and MM. The true transformants were further confirmed by PCR identification and gene sequencing.
5.4. Inducible Expression and Condition Optimizations of TV-AFB1D
The His+Mut+ phenotype-positive clones were incubated with the shaking speed of 200 rpm at 30 °C. The collected cells of recombinant P. pastoris were mixed with methanol-complex buffer medium. The transformants were incubated at 30 °C with the shaking speed of 200 rpm. The fermentation broth was added with 0.8% methanol (v/v) every 24 h for the induction of recombinant TV-AFB1D. The parameters of temperatures, methanol concentrations, and time were optimized to enhance the inducible expression of TV-AFB1D.
5.5. Purification of Recombinant TV-AFB1D and Definition of Activity
The recombinant TV-AFB1D was purified by pre-equilibration with Ni2+-chelate affinity chromatography column and elution with imidazole eluent with ProteinIso Ni-IDA resin. The profile of TV-AFB1D protein was analyzed by SDS-PAGE. The unit of TV-AFB1D activity was defined as the amount of enzyme that catalyzed the consumption of 1 nmoL AFB1 per minute. The activity of TV-AFB1D was calculated by using the formula of (M0 − M1)/(312 × t × V), where M0, M1, t, and V represent the AFB1 weight in the control and experimental groups (µg), reaction time (min), and enzyme amount (mL), respectively; 312 indicates the molecular weight of AFB1.
5.6. Measurement of AFB1 Content and Degradation Products
The content of AFB1 was detected by HPLC using the following process parameters: methanol-to-water ratio of 40/60, isocratic elution, ultraviolet (UV) detector, C18 Shim-pack VP-ODS (5 µm, 250 mm × 4.6 mm), C18 reverse column, 0.7 mL/min of flow rate, 190–400 nm of detection wavelength, and 30 °C of column temperature. The signal of AFB1 was detected by the UV detector at a wavelength of 365 nm. The amount of AFB1 was calculated on the basis of the peak areas with the AFB1 standard sample as the control.
5.7. Application in the AFB1 Degradation of AFB1-Contaminated Peanuts
The AFB1-contaminated peanuts were immersed in the solution containing the recombinant TV-AFB1D from engineering P. pastoris GS115. After incubation for 12 h, the recovered peanuts were soaked with methanol solution containing 40% of water (v/v). Then, the supernatant was purified through the AFB1 immunoaffinity column with the flow rate of 2 drops per second. The collected eluate was further purified by the filtration with 0.22 μm size of filter membrane. The effects of treatment time and temperature on the degradation efficiency of AFB1-contaminated peanuts by TV-AFB1D were investigated.
5.8. Data Analysis
The data were statistically given with three replicates. Origin and SPSS software were used to analyze the data with mean ± standard deviation.
Author Contributions
Conceptualization, P.Y; formal analysis, W.X.; investigation, S.L. and M.Z.; resources, S.J. (Suwei Jiang); data curation, Z.Z.; writing—original draft preparation, W.X. and S.J. (Shuying Jiang); writing—review and editing, P.Y.; visualization, D.Z.; supervision, S.J. (Shaotong Jiang); project administration and funding acquisition, P.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation of Anhui Province (CN) (grant number 1908085MC80), Major Science and Technology Projects of Anhui Province (grant number 202003c08020001), Fundamental Research Funds for the Central Universities of China (grant number PA2020GDSK0058), The APC was funded by Natural Science Foundation of Anhui Province (CN)”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data in this study are available in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The recombinant TV-AFB1D expressed in Pichia pastoris GS115 could effectively degrade AFB1 from AFB1-contaminated peanuts.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hanvi M.D., Lawson-Evi P., Bouka E.C., Eklu-Gadegbeku K. Aflatoxins in maize dough and dietary exposure in rural populations of Togo. Food Control. 2021;121:107673. doi: 10.1016/j.foodcont.2020.107673. [DOI] [Google Scholar]
- 2.Fouche T., Claassens S., Maboeta M. Aflatoxins in the soil ecosystem: An overview of its occurrence, fate, effects and future perspectives. Mycotoxin Res. 2020;36:303–309. doi: 10.1007/s12550-020-00393-w. [DOI] [PubMed] [Google Scholar]
- 3.Pecorelli I., Guarducci N., von Holst C., Bibi R., Pascale M., Ciasca B., Logrieco A.F., Lattanzio V.M.T. Critical comparison of analytical performances of two immunoassay methods for rapid detection of aflatoxin M-1 in milk. Toxins. 2020;12:270. doi: 10.3390/toxins12040270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han C., Yu T., Qin W., Liao X., Huang J., Liu Z., Yu L., Liu X., Chen Z., Yang C., et al. Genome-wide association study of the TP53 R249S mutation in hepatocellular carcinoma with aflatoxin B1 exposure and infection with hepatitis B virus. J. Gastrointest. Oncol. 2020;11:1333–1339. doi: 10.21037/jgo-20-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee L.S., Dunn J.J., DeLucca A.J., Ciegler A. Role of lactone ring of aflatoxin B1 in toxicity and mutagenicity. Experientia. 1981;37:16–17. doi: 10.1007/BF01965543. [DOI] [PubMed] [Google Scholar]
- 6.Vanhoutte I., Audenaert K., De Gelder L. Biodegradation of Mycotoxins: Tales from Known and Unexplored Worlds. Front. Microbiol. 2016;7:20. doi: 10.3389/fmicb.2016.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guengerich F.P., Johnson W.W., Ueng Y.F., Yamazaki H., Shimada T. Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ. Health Perspect. 1996;104(Suppl. S3):557–562. doi: 10.1289/ehp.96104s3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrenk D., Bignami M., Bodin L., Chipman J.K., del Mazo J., Grasl-Kraupp B., Hogstrand C., Hoogenboom L., Leblanc J.C., Nebbia C.S., et al. Risk assessment of aflatoxins in food. EFSA J. 2020;18:1–112. doi: 10.2903/j.efsa.2020.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji J.M., Xie W.L. Removal of aflatoxin B-1 from contaminated peanut oils using magnetic attapulgite. Food Chem. 2021;339:128072. doi: 10.1016/j.foodchem.2020.128072. [DOI] [PubMed] [Google Scholar]
- 10.Javanmardi F., Khodaei D., Sheidaei Z., Bashiry M., Nayebzadeh K., Vasseghian Y., Khaneghah A.M. Decontamination of aflatoxins in edible oils: A comprehensive review. Food Rev. Int. 2020;17:1–17. doi: 10.1080/87559129.2020.1812635. [DOI] [Google Scholar]
- 11.Magagnoli S., Lanzoni A., Masetti A., Depalo L., Albertini M., Ferrari R., Spadola G., Degola F., Restivo F.M., Burgio G. Sustainability of strategies for Ostrinia nubilalis management in Northern Italy: Potential impact on beneficial arthropods and aflatoxin contamination in years with different meteorological conditions. Crop Prot. 2021;142:105529. doi: 10.1016/j.cropro.2020.105529. [DOI] [Google Scholar]
- 12.Adeyeye S.A.O. Aflatoxigenic fungi and mycotoxins in food: A review. Crit. Rev. Food Sci. Nutr. 2020;60:709–721. doi: 10.1080/10408398.2018.1548429. [DOI] [PubMed] [Google Scholar]
- 13.Kabak B. The fate of mycotoxins during thermal food processing. J. Sci. Food Agric. 2009;89:549–554. doi: 10.1002/jsfa.3491. [DOI] [Google Scholar]
- 14.Mao J., He B., Zhang L.X., Li P.W., Zhang Q., Ding X.X., Zhang W. A Structure Identification and Toxicity Assessment of the Degradation Products of Aflatoxin B-1 in Peanut Oil under UV Irradiation. Toxins. 2016;8:332. doi: 10.3390/toxins8110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samarajeewa U., Sen A.C., Cohen M.D., Wei C.I. Detoxification of Aflatoxins in Foods and Feeds by Physical and Chemical Methods 1. J. Food Prot. 1990;53:489–501. doi: 10.4315/0362-028X-53.6.489. [DOI] [PubMed] [Google Scholar]
- 16.Desheng Q., Fan L., Yanhu Y., Niya Z. Adsorption of aflatoxin B-1 on montmorillonite. Poult. Sci. 2005;84:959–961. doi: 10.1093/ps/84.6.959. [DOI] [PubMed] [Google Scholar]
- 17.Pankaj S.K., Shi H., Keener K.M. A review of novel physical and chemical decontamination technologies for aflatoxin in food. Trends Food Sci. Technol. 2018;71:73–83. doi: 10.1016/j.tifs.2017.11.007. [DOI] [Google Scholar]
- 18.Guo Y., Zhao L., Ma Q., Ji C. Novel strategies for degradation of aflatoxins in food and feed: A review. Food Res. Int. 2021;140:109878. doi: 10.1016/j.foodres.2020.109878. [DOI] [PubMed] [Google Scholar]
- 19.Mendez-Albores A., Martinez-Bustos F., Gaytan-Martinez M., Moreno-Martinez E. Effect of lactic and citric acid on the stability of B-aflatoxins in extrusion-cooked sorghum. Lett. Appl. Microbiol. 2008;47:1–7. doi: 10.1111/j.1472-765X.2008.02376.x. [DOI] [PubMed] [Google Scholar]
- 20.Ismail A., Goncalves B.L., de Neeff D.V., Ponzilacqua B., Coppa C., Hintzsche H., Sajid M., Cruz A.G., Corassin C.H., Oliveira C.A.F. Aflatoxin in foodstuffs: Occurrence and recent advances in decontamination. Food Res. Int. 2018;113:74–85. doi: 10.1016/j.foodres.2018.06.067. [DOI] [PubMed] [Google Scholar]
- 21.Rushing B.R., Selim M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019;124:81–100. doi: 10.1016/j.fct.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 22.Adebo O.A., Njobeh P.B., Gbashi S., Nwinyi O.C., Mavumengwana V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017;57:3208–3217. doi: 10.1080/10408398.2015.1106440. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y.P., Qin X.J., Tang Y., Ma Q.G., Zhang J.Y., Zhao L.H. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B-1 to aflatoxin Q(1) and epi-aflatoxin Q(1) Food Chem. 2020;325:126877. doi: 10.1016/j.foodchem.2020.126877. [DOI] [PubMed] [Google Scholar]
- 24.Loi M., Renaud J.B., Rosini E., Pollegioni L., Vignali E., Haidukowski M., Sumarah M.W., Logrieco A.F., Mule G. Enzymatic transformation of aflatoxin B-1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere. 2020;250:126296. doi: 10.1016/j.chemosphere.2020.126296. [DOI] [PubMed] [Google Scholar]
- 25.Li C.H., Li W.Y., Hsu I.N., Liao Y.Y., Yang C.Y., Taylor M.C., Liu Y.F., Huang W.H., Chang H.H., Huang H.L., et al. Recombinant aflatoxin-degrading F420H2-dependent reductase from Mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins. 2019;11:259. doi: 10.3390/toxins11050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H.W., Wang L.Z., Sun J.D., Wang L.P., Guo H.Y., Ye Y.L., Sun X.L. Microbial detoxification of mycotoxins in food and feed. Crit. Rev. Food Sci. Nutr. 2021;1:1–19. doi: 10.1080/10408398.2021.1879730. [DOI] [PubMed] [Google Scholar]
- 27.Brana M.T., Sergio L., Haidukowski M., Logrieco A.F., Altomare C. Degradation of aflatoxin B-1 by a sustainable enzymatic extract from spent mushroom substrate of Pleurotus eryngii. Toxins. 2020;12:49. doi: 10.3390/toxins12010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu Y.X., Wu X.Y., Xie C.F., Hu Y.D., Liu D.L., Ma Y., Yao D.S. A rational design for improving the trypsin resistance of aflatoxin-detoxifizyme (ADTZ) based on molecular structure evaluation. Enzym. Microb. Technol. 2016;86:84–92. doi: 10.1016/j.enzmictec.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Huang B., Xiao H.L., Zhang J., Zhang L.F., Yang H.L., Zhang Y., Jin J. Dual-label time-resolved fluoroimmunoassay for simultaneous detection of aflatoxin B1 and ochratoxin A. Arch. Toxicol. 2009;83:619–624. doi: 10.1007/s00204-009-0410-6. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L.H., Guan S., Gao X., Ma Q.G., Lei Y.P., Bai X.M., Ji C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011;110:147–155. doi: 10.1111/j.1365-2672.2010.04867.x. [DOI] [PubMed] [Google Scholar]
- 31.Xu L., Ahmed M.F.E., Sangare L., Zhao Y.J., Selvaraj J.N., Xing F.G., Wang Y., Yang H.P., Liu Y. Novel aflatoxin-degrading enzyme from Bacillus shackletonii L7. Toxins. 2017;9:36. doi: 10.3390/toxins9010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Y.L., Wang W., Zhang S.J. Purification and identification of an aflatoxin B-1 degradation enzyme from Pantoea sp. T6. Toxicon. 2019;157:35–42. doi: 10.1016/j.toxicon.2018.11.290. [DOI] [PubMed] [Google Scholar]
- 33.Wang X.L., Qin X., Hao Z.Z., Luo H.Y., Yao B., Su X.Y. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins. 2019;11:566. doi: 10.3390/toxins11100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yehia R.S. Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 2014;45:127–133. doi: 10.1590/S1517-83822014005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X.L., Bai Y.G., Huang H.Q., Tu T., Wang Y., Wang Y.R., Luo H.Y., Yao B., Su X.Y. Degradation of aflatoxin B-1 and zearalenone by Bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins. 2019;11:609. doi: 10.3390/toxins11100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loi M., Fanelli F., Zucca P., Liuzzi V.C., Quintieri L., Cimmarusti M.T., Monaci L., Haidukowski M., Logrieco A.F., Sanjust E., et al. Aflatoxin B₁ and M₁ degradation by lac2 from Pleurotus pulmonarius and redox mediators. Toxins. 2016;8:245. doi: 10.3390/toxins8090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu P., Yang J., Gu H. Expression of HpaI in Pichia pastoris and optimization of conditions for the heparinase I production. Carbohydr. Polym. 2014;106:223–229. doi: 10.1016/j.carbpol.2014.01.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in this study are available in the manuscript.