Abstract
People living with HIV (PLWH) continue to develop HIV-associated neurocognitive disorders despite combination anti-retroviral therapy. Lipocalin-2 (LCN2) is an acute phase protein that has been implicated in neurodegeneration and is upregulated in a transgenic mouse model of HIV-associated brain injury. Here we show that LCN2 is significantly upregulated in neocortex of a subset of HIV-infected individuals with brain pathology and correlates with viral load in CSF and pro-viral DNA in neocortex. However, the question if LCN2 contributes to HIV-associated neurotoxicity or is part of a protective host response required further investigation. We found that the knockout of LCN2 in transgenic mice expressing HIVgp120 in the brain (HIVgp120tg) abrogates behavioral impairment, ameliorates neuronal damage, and reduces microglial activation in association with an increase of the neuroprotective CCR5 ligand CCL4. In vitro experiments show that LCN2 neurotoxicity also depends on microglia and p38 MAPK activity. Genetic ablation of CCR5 in LCN2-deficient HIVgp120tg mice restores neuropathology, suggesting that LCN2 overrides neuroprotection mediated by CCR5 and its chemokine ligands. RNA expression of 168 genes involved in neurotransmission reveals that neuronal injury and protection are each associated with genotype- and sex-specific patterns affecting common neural gene networks. In conclusion, our study identifies LCN2 as a novel factor in HIV-associated brain injury involving CCR5, p38 MAPK and microglia. Furthermore, the mechanistic interaction between LCN2 and CCR5 may serve as a diagnostic and therapeutic target in HIV patients at risk of developing brain pathology and neurocognitive impairment.
Keywords: Lipocalin-2, CCR5, knockout, p38 MAPK, HIV gp120-transgenic, HIV-associated neurocognitive disorders, neurodegeneration, behavior, HIV neuropathology, sexual dimorphism
1. Introduction
About 50% of people living with human immunodeficiency virus-1 (HIV)-1 exhibit associated neurocognitive disorders (HAND) (Antinori et al., 2007; Saylor et al., 2016). Introduction of combination antiretroviral therapy (cART) decreased the number of deaths from acquired immunodeficiency syndrome (AIDS) and caused a shift from HIV-associated dementia (HAD) towards less severe forms of HAND. Nonetheless, the overall incidence of HAND remained unchanged (Maung et al., 2012; McArthur et al., 2010; Sanchez and Kaul, 2017; Saylor et al., 2016; Stevenson, 2003). The pathological, cellular and molecular mechanisms leading to HAND remain incompletely understood and treatments or reliable biomarkers to assess risk of HAND or track disease progression are lacking.
HIV-infected (HIV+) patients develop neuropathological changes despite cART (Ellis et al., 2007; Gelman et al., 2013; Levine et al., 2016; Saylor et al., 2016). The most severe neuropathology is referred to as HIV encephalitis (HIVE) with activated resident microglia, multinucleated giant cells, infiltration of monocytoid cells, and decreased synaptic and dendritic density, combined with selective neuronal loss, widespread reactive astrocytosis, and myelin pallor (Ellis et al., 2007; Gelman et al., 2012; Gelman et al., 2013; Levine et al., 2016). A transgenic (tg) mouse model expressing the viral envelope protein gp120 of the CXCR4-utilizing HIV-1 isolate lymphadenopathy associated virus (LAV) in the brain (HIVgp120tg) displays key neuropathological features observed in HIVE/AIDS brains, including microgliosis, astrocytosis, decreased synaptic terminals and dendritic processes (Maung et al., 2014; Thaney et al., 2018; Toggas et al., 1994). HIVgp120tg animals express gp120 under the control of a modified glial fibrillary acidic protein (GFAP) in astrocytes (Thaney et al., 2018; Toggas et al., 1994) and develop behavioral impairment, such as declining spatial learning and memory by 9 to 12 months of age (D’Hooge et al., 1999; Kaul et al., 2005; Maung et al., 2014; Thaney et al., 2018). Furthermore, a microarray analysis and comparison of central nervous system (CNS) gene expression between human HIV+ and HIVE patients and HIVgp120tg animals showed a significant overlap for differentially regulated genes (Gelman et al., 2012; Maung et al., 2014). The microarray identified LCN2 as one of the most upregulated genes in HIVgp120tg mouse brains and increased expression was confirmed at protein level (Maung et al., 2014). Furthermore, in both, human and mouse brains the expression of Lipocalin-2 (LCN2) and astrocytic GFAP were highly correlated (Maung et al., 2014).
LCN2 is a component of the innate immune system that counteracts bacterial infection by sequestering iron, but also regulates a range of biological process, such as cellular energy metabolism and apoptosis (Bachman et al., 2009; Bao et al., 2010; Ferreira et al., 2015; Flo et al., 2004; Yang et al., 2002). In the CNS, LCN2 has been implicated in the regulation of behaviors, such as anxiety, emotional and contextual discrimination, and cognition (Ferreira et al., 2015; Ferreira et al., 2013; Ferreira et al., 2018; Jha et al., 2015; Mucha et al., 2011). In addition, LCN2 has been shown to act as a regulator of inflammation (Jha et al., 2015; Jin et al., 2014; Kang et al., 2018) and to modulate microglial activation (Lee et al., 2007; Xing et al., 2014) and possibly neurodegeneration (Bi et al., 2013; Maung et al., 2014).
Here we show that LCN2 is significantly upregulated in neocortex of a subset of HIV+ patients with brain pathology (microglial nodules or HIVE) when compared to non-infected controls. LCN2 is also highly upregulated in the HIVgp120tg murine CNS. We find that the genetic knockout (KO) of LCN2 abrogates behavioral impairment, ameliorates neuronal damage, and reduces microglial activation induced by HIV gp120 protein. The protection is associated with an increase in the CCR5 ligand CCL4 and reduced activation of p38 mitogen activated protein kinase (p38 MAPK). In vitro experiments show LCN2 neurotoxicity requires microglia and p38 MAPK. Genetic ablation of CCR5 in LCN2-deficient HIVgp120tg mice restores neuronal damage and the increase in microglial cell numbers, suggesting that LCN2 overrides neuroprotection mediated by CCR5, at least in part, by limiting the expression of the neuroprotective β-chemokine CCL4. Furthermore, RNA expression analysis of 168 genes involved in neurotransmission reveals that gp120 expression, with and without genetic ablation of LCN2, or CCR5, or both together leads to alterations of the GABAergic, glutamatergic, dopaminergic and serotonergic systems. Altogether, our findings identify LCN2 as a novel factor in HIV-associated brain injury requiring p38 MAPK and microglia. Furthermore, we demonstrate a mechanistic interaction between LCN2 and CCR5 that may serve as a diagnostic and therapeutic target in HIV patients at risk of developing brain pathology and neurocognitive impairment.
2. Materials and Methods
2.1. Quantitative RT-PCR of human samples
RNA samples derived from the middle frontal gyrus matter (neocortex) of HIV+ patients and non-infected age matched individuals were prepared by Dr. Benjamin Gelman’s laboratory (UTMB Galveston, TX) for the National NeuroAIDS Tissue Consortium (NNTC). Demographic and virological characteristics of the cohort from which the samples were derived have previously been described (Gill et al., 2014; Kovacsics et al., 2017; Nguyen et al., 2010) and are summarized in Table S1 (see Supplementary File 1). RNA quality was assessed using the Bioanalyzer System (RNA Bioanalyzer 6000 Nano Chip (Cat. No. 5067–1511), Agilent Technologies, USA) and included as RNA Integrity Numbers (RIN) in Table S1. All samples were coded, and the investigators were blind to the HIV status and HIV brain damage diagnostic of the patients during the qRT-PCR experiments. QRT-PCR was performed as previously described (Hoefer et al., 2015; Maung et al., 2014; Thaney et al., 2017). Briefly, reverse transcription was performed using SuperScript II reverse transcriptase (Invitrogen, USA) following the manufacturer’s instructions. QRT-PCR was performed using Power PCR SYBR Green mastermix (Applied Biosystems, USA) on an Mx3005P system (Stratagene, Agilent Technologies, USA). For primers see Table S2. The results obtained were analyzed using MxPro QPCR software v4.10 (Stratagene, Agilent Technologies). Levels of mRNA of LCN2 in the brain were expressed as relative levels of the internal control, β-actin (Actb).
2.2. Immunoblotting of human samples
Frontal cortex tissues from a subset of human brains for LCN2 mRNA were homogenized using a buffer that facilitates separation of the membrane and cytosolic fractions (1.0 mmol/L HEPES, 5.0 mmol/L benzamidine, 2.0 mmol/L 2-mercaptoethanol, 3.0 mmol/L EDTA, 0.5 mmol/L magnesium sulfate, 0.05% sodium azide; final pH 8.8) as previously described (Fields et al., 2013). In brief, tissue from human brain samples (0.1 g) was homogenized in fractionation buffer containing a complete protease inhibitor cocktail (Roche). Samples were precleared by centrifugation and supernatant was retained as the whole lysate. Samples were denatured in Laemmli Sample buffer (Bio Rad) and 15 μg total protein/lane were loaded along with a recombinant human LCN2 control (3.75 ng) onto a 4–15 % Criterion TGX stain free gel (Bio Rad) and electrophoresed in Tris/Glycine/SDS running buffer (Bio Rad). Proteins were transferred onto a LF PVDF membrane with Bio Rad transfer stacks and transfer buffer (Bio Rad) using Bio Rad Trans Blot Turbo transfer system. After the transfer, total protein was imaged using a Bio Rad ChemiDoc imager under the stain free blot setting for normalization purposes. The membrane(s) were then blocked before being incubated overnight at 4°C with the primary antibody, rat anti-LCN2 (1:400; R&D Systems). Anti-rat IgG, HRP linked antibody was used as secondary antibody (1:2000; Cell signaling technologies) visualized with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific). Images were obtained, and semi-quantitative analysis was performed with the ChemiDoc gel imaging system and Quantity One software (Bio-Rad).
2.3. Mouse models
HIVgp120tg mice (CCR5WT and KO) expressing envelope glycoprotein gp120 of the HIV-1 strain LAV under the control of the GFAP promoter have previously been described (Maung et al., 2014; Toggas et al., 1994). Mice deficient in LCN2 (LCN2KO) were kindly provided by Drs. Shizuo Akira (Osaka University, Osaka, Japan) and Alan Aderem (Institute for Systems Biology, Seattle, WA) (Flo et al., 2004). LCN2KO mice were in a C57BL/6 genetic background and were crossed once with SJL mice. The F2 generation was subsequently crossbred with HIVgp120tg mice (CCR5WT and KO) which are mixed C57BL/6.129/SJL. The F3 generation of HIVgp120tghet-LCN2het-CCR5het mice was used to generate eight genotypes: 1) LCN2KO-gp120, 2) HIVgp120tg, 3) LCN2KO (control), 4) WT (control), 5) LCN2/CCR5 double knockout-gp120 (DKO-gp120), 6) CCR5KO-gp120, 7) LCN2/CCR5DKO (control) and 8) CCR5KO (control). Subsequently, LCN2KO-gp120, DKO-gp120, CCR5KO-gp120 and LCN2/CCR5 WT HIVgp120tg mice and their non-tg littermates were generated in separate breeding programs with all genotypes maintained in a mixed C57BL/6.129/SJL genetic background. During all subsequent experiments, mice were coded and the investigators were blinded to genotype. All procedures involving animals were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committees of the Sanford-Burnham-Prebys Medical Discovery Institute, The Scripps Research Institute and the University of California, Riverside.
2.4. Behavioral testing
Wild-type (male: 12, female: 14), HIVgp120tg (male: 9, female: 9), LCN2KO (male: 10, female: 13), and LCN2KO-gp120 (male: 8, female: 8) mice were tested at 7–8 months of age by The Scripps Research Institute’s (TSRI) Animal Models Core Facility. The behavioral test battery was designed to examine cognitive functionality as well as general activity, anxiety-like behavior and visual ability. The tests were performed as previously published (Hoefer et al., 2015; Maung et al., 2014) with minor modifications. Briefly, the locomotor activity test used two levels of photocell beams to record both horizontal (ambulation and center activity) and vertical (rearing) behavior. Mice were tested for 120 minutes and data were collected in 5 min intervals. The novel object recognition test assays recognition memory while leaving the spatial location of the objects intact and is presumably involves the hippocampus, perirhinal cortex, and raphe nuclei (Lieben et al., 2006; Mumby et al., 2005; Winters et al., 2004). Mice were tested with two identical objects placed in the field followed by replacing one of the familiar objects for a novel object. Habituation to the objects (decreased contact time) was an initial measure of learning and renewed interest (increased contact time) in the new object indicated successful object memory. The Barnes maze test assesses spatial learning and memory (Barnes, 1979) and is sensitive to impaired hippocampal function (Paylor et al., 2001). The maze consists of an opaque disc with twenty holes, and an escape box placed under one of the holes. Distinct spatial cues are located all around the maze and are kept constant throughout the study. An initial training session was performed, by placing the mouse in the escape box for one minute. Subsequently, the first session was started whereby the mouse was placed in the middle of the maze in an opaque start chamber. After 10 seconds the start chamber was removed, a buzzer and a light were turned on, and the mouse was free to explore the maze. Sessions ended when the mouse entered the escape tunnel or after 3 min elapsed. The tunnel was always located underneath the same hole (stable within the spatial environment), which was randomly determined for each mouse. Mice were tested once a day for 6 days for the acquisition portion of the study. A probe test was performed on the day following the final acquisition trial to assess use of the spatial cues. For this test, the escape tunnel was removed, and the mouse was allowed to freely explore the maze. The time spent in each quadrant was determined and the percent time spent in the target quadrant (the one originally containing the escape box) was compared with the average percent time in the other three quadrants. Each acquisition session was videotaped and scored by an experimenter blind to the genotype of the mouse. The probe data was collected using Noldus Ethovision software to determine time spent in each quadrant of the maze as well as to assess activity measures. A detailed description of behavioral testing is provided in Supplementary Information (see Supplementary File 2).
2.5. Isolation of mRNA and RT-PCR for mouse genes
RNA of murine cerebral cortex was isolated using the Qiagen RNeasy Lipid Tissue Midi Kit (Qiagen) and hippocampal RNA was isolated using Qiagen Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA quality was tested and qRT-PCR was performed as previously published (Hoefer et al., 2015; Maung et al., 2014; Thaney et al., 2017). Of the 50 samples total, a randomly selected subset of 12 samples was in addition tested for RNA quality using the RT2 RNA QC PCR Array (PAMM-999ZE-1; Cat. No. 330291) and all samples passed the quality control check. QRT-PCR results obtained were analyzed using MxPro QPCR software v4.10 (Stratagene, Agilent Technologies) and the relative amount of mRNA of every gene versus the internal controls (GAPDH) was calculated following the 2−ΔΔCt method.
2.6. GABA & Glutamate and Dopamine & Serotonin RT2 Profiler™ PCR Array
Neurotransmission related gene expression analysis was performed using the GABA & Glutamate (PAMM-152Z) and Dopamine & Serotonin (PAMM-158Z) RT2 Profiler™ Arrays following supplier’s instructions (Qiagen). A QuantStudio™ 6 Flex System (Applied Biosystems by Thermo Fisher Scientific) was used to run the arrays. The RT2 Profiler™ Array Data Analysis software package (version 3.5) used the 2-(ΔΔCT)-based method (Livak and Schmittgen, 2001) to calculate fold change and a modified Student’s t-test to compute two-tail, equal variance P-values. This method calculates the difference between the gene of interest and the average of selected housekeeping genes (Heat Shock Protein 90 Alpha Family Class B Member 1 (Hsp90ab1), Actin Beta (Actb), Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) and Glucuronidase Beta (Gusb)).
2.7. Bioinformatics analysis
A list including all 168 genes measured in the RT2 Profiler™ PCR array was further analyzed for identification of functional gene networks using Ingenuity Pathway Analysis (IPA; Ingenuity® Systems, www.ingenuity.com; build version: 486617M; content version: 46901286; release date: 2018–11-21). The complete list of settings used for analysis shown in Supplementary Information (see Supplementary File 2).
2.8. Immunoblotting of mouse brain samples
Preparation of brain tissue lysates and Western blotting using lysates from cerebral cortex of 9–10 months-old mice was conducted as previously published with minor modifications (Maung et al., 2014; Medders et al., 2010). Briefly, 25 μg of protein were added to 4 X LDS sample buffer and 10 X reducing agent (Invitrogen) and boiled for 5 min. Samples were loaded in a 4–12 % SDS-PAGE gel (Invitrogen) for electrophoretic separation and followed by electro-transference to PVDF membranes. Membranes were blocked with bovine serum albumin solution and subsequently incubated overnight at 4˚C with primary antibodies as follows: Active p38 MAPK (1:1000); total p38 MAPK (1:2000; both Cell Signaling Technology); and α-Tubulin (1:4000; Sigma). Following each primary antibody, membranes were incubated with goat anti-rabbit (1:5000; Jackson ImmunoResearch) or goat anti-mouse (1:20000; Pierce) secondary antibodies conjugated with horseradish-peroxidase. SuperSignal Dura chemiluminescent detection kit (Pierce) was used to detect the signal. Membranes were imaged using the ChemiDoc™ XRS+ imager. Densitometry analysis was performed using ImageJ 1.52a software (http://rsb.info.nih.gov/nih-image/) and normalized against α-Tubulin signal.
2.9. Multiplex assays
Levels of soluble cytokines in lysates of cerebral cortex of 9–10 months-old animals were determined using a Milliplex Mouse Cytokine/Chemokine Magnetic Bead Premixed kit (Millipore Sigma) following manufacturer’s instructions. Bead-bound protein concentrations were measured with Luminex MAGPIX system (Bio-Rad) and analyzed using Milliplex Analyst (Vigene Tech; version 5.1).
2.10. Immunohistology, quantitative fluorescence and deconvolution microscopy
Brain tissue harvest, immunofluorescence staining, deconvolution and quantitative fluorescence microscopy and cell counting to assess neuronal injury, astrocytosis and microglial activation were performed as previously described (Hoefer et al., 2015; Maung et al., 2014; Thaney et al., 2017). In brief, 9 to 10 months-old mice were terminally anesthetized with isoflurane, placed on an ice bed and immediately transcardially perfused with 0.9 % saline. The brain was divided into two hemispheres. One was fixed in 4 % paraformaldehyde for histological analysis and the second hemisphere was dissected to isolate the hippocampus and cortex and subsequently snap frozen in liquid nitrogen. For neuropathological assessment, 30 or 40 μm thick sagittal brain sections were immunostained using antibodies against Syp (1:50; Dako) and MAP-2 (1:200; Sigma) as neuronal markers, or GFAP (1:250; Dako) and Iba1 (1:125; Wako) as astrocytic and microglial markers, respectively. Images (Z-stacks and 2D) were acquired using an Axiovert 200M fluorescence microscope (Zeiss) equipped with a computer-controlled 3D stage and filters for DAPI, FITC, CY3 and CY5. Slidebook software (Intelligent Imaging Innovations, version 6) was used for image acquisition and analysis as reported previously (Maung et al., 2014; Thaney et al., 2017). A detailed description of immunohistology is provided in Supplementary Information (see Supplementary File 2).
2.11. RNAscope® in situ hybridization and immunofluorescent staining
Nine to 10 months-old mice were terminally anesthetized and immediately transcardially perfused with 0.9 % saline. The brain was then embedded in Optimal Cutting Temperature Compound (OCT; Sakura, Torrance, CA) and frozen in dry ice for 5 min and then stored at −80˚C until sectioning. A Leica cryostat (Leica CM 1950, Leica Microsystems, Buffalo Grove, IL) was used to cut 15 μm thick coronal sections. Slices were directly mounted onto charged microscope slides (Superfrost Plus, Cat. No. 22–037-246, Fisher Scientific, Pittsburg, PA) and stored at −80˚C until use. RNAscope® Multiplex Fluorescent Assay (Cat. No. 323100; ACD) was performed following supplier’s instructions. Briefly, slides were removed from −80˚C and brought to room temperature (RT). Tissue slices were fixed using chilled 4 % paraformaldehyde for 15 min at RT. Slices were then dehydrated using an increasing ethanol gradient (50 %, 70 %, 100 %, 100 %) for 5 min each. Next, slices were treated with hydrogen peroxide for 10 min at RT and followed with protease III treatment for 30 min at RT. RNAscope® mouse Lcn2 probe (Cat. No. 313971) and the RNAscope® 3-plex Negative Control Probe (Cat. No. 320871) which targets bacterial dapB gene were incubated each with separate tissue sections for 2 hrs at 40˚C. Amplification steps were performed according to manufacturer’s instructions. Hybridized probe was visualized using Opal 520 dye (Cat. NEL810001KT; Akoya Biosciences, 1:750).
Following in situ hybridization, the slices were blocked using 5 % goat serum diluted in 1X PBS with 0.2 % Tween (PBST). For immunofluorescent staining, the slices were incubated overnight at 4˚C with GFAP antibody (Dako, Cat. No. Z0334, 1:250). Alexa Fluor 488 (Thermo Fisher Scientific, Cat. No. A11034, 1:200) was used as secondary antibody. Nuclear DNA was stained with Hoechst (H) 33342 (Sigma, Cat. No. B2261) at a final concentration of 12 μM. Immunolabelled brain sections were placed on glass slides in fluorescence-protecting mounting medium (Vector Laboratories, Burlingame, CA 94010, USA, cat# H1000), overlaid by coverslips and sealed with nail polish. Image Z-stacks were acquired using an Axiovert 200M fluorescence microscope (Zeiss) with filters for DAPI, FITC, and CY3 and analyzed as described above employing Slidebook software (Intelligent Imaging Innovations, Denver, CO). To assess and visualize co-localization, Z-stacks were deconvolved using the Nearest Neighbor algorithm and 3D Surface View was used for visualization (Kaul et al., 2007).
2.12. Preparation of rat cerebrocortical cell cultures (RCC)
Mixed neuronal-glial cerebrocortical cell cultures from rat (RCC) were prepared as previously described by our group (Kaul and Lipton, 1999; Kaul et al., 2007; Medders et al., 2010; Sanchez et al., 2016a; Sanchez et al., 2016b; Thaney et al., 2017). Briefly, cells were isolated from embryos of Sprague-Dawley rats (Harlan) at day 15 to 17 of gestation and cultured poly-L-lysine-coated clear bottom 96 well plates for imaging (Falcon). Cultures were used after 17 days in vitro when the majority of neurons were considered to be fully differentiated and susceptible to NMDA toxicity (Kaul et al., 2007). For experiments in which microglia were depleted, cultures were pre-treated with 7.5 mM L-leucine methyl ester (LME) for 16 hrs prior to the neurotoxicity assay in conditioned media without LME (Kaul and Lipton, 1999).
2.13. In vitro neurotoxicity assay
Recombinant rat LCN2 and the specific p38 MAPK inhibitor, SB203580, were purchased from R&D systems and Calbiochem, respectively. LCN2 was reconstituted in 0.1 % bovine serum albumin (BSA; Gibco). SB203580 was dissolved in DMSO (ATCC) at 10 mM (stock solution) and added to the cerebrocortical cell cultures (RCC) at 10 μM final concentration 15 min prior to treatment with 4 nM LCN2 for 72 hrs. Controls received BSA vehicle alone (0.001 % final concentration) and DMSO (0.1 %). After treatment, RCC were washed with phosphate-buffered saline (PBS; Hyclone), and fixed with 4 % PFA (Electron Microscopy Sciences) at 4˚C for 25 min. The cells were permeabilized for 5 min with 0.2 % Triton X-100 at room temperature and then non-specific binding sites blocked with heat-inactivated goat serum. Primary mouse anti-MAP-2 IgG1 (1:250; clone HM2, Sigma) was used in combination with goat anti-mouse-Rhodamine Red (1:100; Jackson ImmunoResearch) as a secondary antibody. Non-specific murine IgG1 served as primary antibody control. Hoechst H33342 was used to label nuclear DNA. Image acquisition and analysis was performed as described previously by quantifying neuronal injury and survival using MAP-2 immunofluorescence (Sanchez et al., 2016b) and counting MAP2+ neurons and total cell numbers. Control samples were defined as 100% neuronal survival (Kaul and Lipton, 1999; Kaul et al., 2007; Medders et al., 2010; Sanchez et al., 2016a; Thaney et al., 2017).
2.14. Statistical analysis
Sample size in the animal experiments was determined based on previous studies (Hoefer et al., 2015; Maung et al., 2014). Since the experimental animal groups were defined by genotype, no randomization was employed but the groups were matched for sex and age. Animals were coded and investigators were blinded to the genotype during experiments and analysis. Since any gp120+ genotype is associated with pronounced astrocytosis, blinding was not possible for the viral transgene in the analysis of astrocytic GFAP although blinding remained for the genotype with regard to Lcn2 and Ccr5. Data collection utilized instrumentation–specific softwares and Excel. Analysis of histopathological data, mRNA expression, Western blotting data, and correlation analysis were performed using Prism software (GraphPad Software, Inc., CA, USA), behavioral data were evaluated using StatView (SAS Institute, Cary, NC). Comparisons of two groups were made by unpaired Student’s t-test, whereas multiple groups were compared by analysis of variance (ANOVA) followed by Fisher’s PLSD post hoc test. P-values < 0.05 were considered statistically significant. Besides qRT-PCR, CT values were measured using RT2 Profiler™ PCR Arrays. QIAGEN data analysis center was used for 2–(ΔΔCT)-based fold change calculations and a modified Student’s t-test to compute two tails, equal variance P-values (http://www.qiagen.com/geneglobe). No statistical methods were employed to predetermine the numbers of human autopsy samples. After relative gene expression levels for the genes of interest were assessed in a blinded fashion, the samples were separated for further analysis based on HIV infection status (HIV+ versus un-infected) and subsequently the HIV+ group was divided into two groups based on presence and absence of HIV brain pathology. The 1.5X Interquartile Range (IQR) rule was used to identify outliers in all three groups (Schwertman et al., 2004). Samples determined to be outliers were excluded from the correlation analysis. Pearson coefficient was used to determine significant correlations.
3. Results
3.1. LCN2 expression levels are increased in brains of HIV patients with pathology
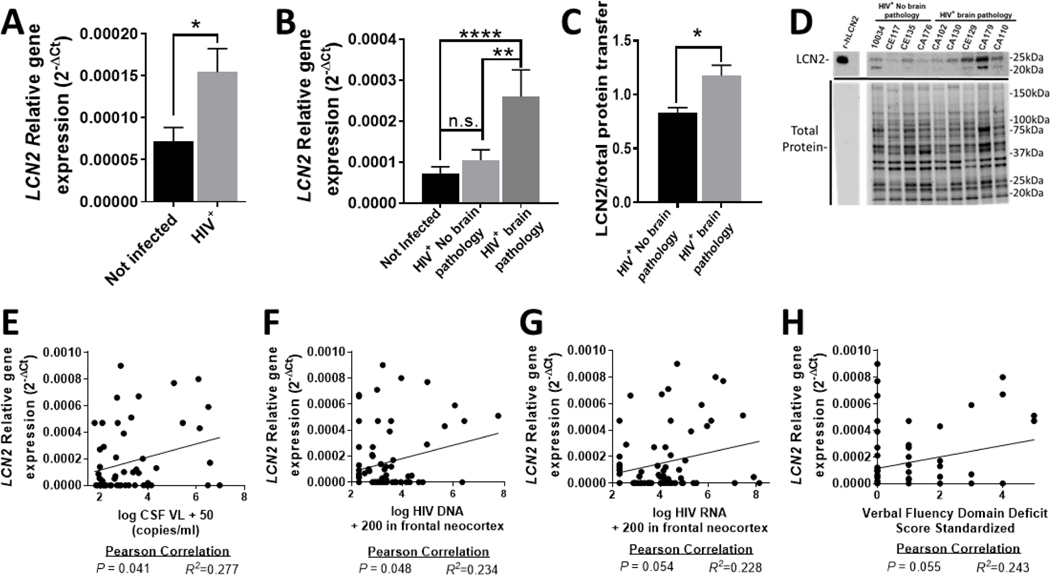
In order to assess relative gene expression of LCN2 in the brains of HIV-infected individuals, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using RNA extracted from the middle frontal gyrus matter (neocortex) of 72 HIV+ individuals and 56 non-infected age-matched controls. All human samples were provided by the National NeuroAIDS Tissue Consortium (NNTC). The demographic and virological information and neuropathological evaluation of the patients in this particular autopsy cohort have been established in multiple published reports (Gelman et al., 2013; Gill et al., 2014; Kovacsics et al., 2017; Nguyen et al., 2010) and are summarized in Table S1 (see Supplementary File 1). We found that the HIV-infected patients expressed on average significantly higher levels of LCN2 than the non-infected individuals (Fig. 1A). HIV+ samples were then separated based on the presence and absence of HIV brain pathology, defined as patients diagnosed with either HIVE or microglial nodule encephalitis (Gelman et al., 2013). HIV+ patients that displayed HIV-associated brain pathology had on average significantly higher levels of LCN2 than non-infected controls and infected patients without neuropathology (Fig. 1B). Protein extracts from a subset of the human brain specimens in this autopsy cohort were assayed for LCN2 protein levels (Gelman et al., 2013). Western blotting showed that LCN2 was significantly higher in brain specimens from the patients with HIV brain pathology (Fig. 1C, 1D and Fig. S1, see Supplementary File 3). Next, we correlated the LCN2 mRNA levels to previously published neurovirological data pertaining to the brain specimens (Gelman et al., 2013; Gill et al., 2014; Kovacsics et al., 2017; Nguyen et al., 2010). Pearson correlation analysis showed significance for LCN2 expression levels with HIV RNA loads in the CSF (Fig. 1E; R2 = 0.277; P = 0.041) and for LCN2 expression levels with HIV DNA levels in frontal neocortex (Fig. 1F; R2 = 0.2324; P = 0.048). We observed slightly weaker correlations between LCN2 levels and HIV RNA levels in frontal neocortex (Fig. 1G; R2 = 0.228; P = 0.054), and between LCN2 expression and the deficit score for verbal fluency domain (Fig. 1H; R2 = 0.243; P = 0.055). No significant correlation was observed between LCN2 levels and age at death (R2 = −0.138; P = 0.249), hours post-mortem (R2 = −0.093; P = 0.440), RNA integrity number (R2 = 0.137; P = 0.134) or history of any substance abuse (R2 = 0.178; P = 0.327; Table S3). Altogether, our findings are supportive of a potential role of LCN2 in HIV-associated brain injury.
Fig. 1. LCN2 levels are increased in brains of individuals with HIV neuropathology.
(A-B) Relative gene expression levels of LCN2 were assessed using mRNA from the middle frontal gyrus (neocortex) of 72 HIV+ and 56 non-infected patients. (B) Patients were then separated based on the presence or absence of histological signs of HIV brain pathology (Gelman et al., 2013) and levels of LCN2 mRNA were compared (HIV+ No brain pathology, n = 49; HIV+ brain pathology, n = 23). (C-D) Frontal gyrus protein of a subset of infected patients was analyzed using Western blotting (HIV+ No brain pathology, n = 4, and HIV+ with brain pathology, n = 5). (E-H) Pearson correlation was calculated for the patients’ virological parameters (Gelman et al., 2013) and their corresponding LCN2 levels. Values in graphs are mean ± s.e.m.; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, ANOVA followed by Fisher’s PLSD post hoc test (B) or Student’s t test (A, C).
3.2. LCN2 mediates learning and memory impairment caused by HIV gp120
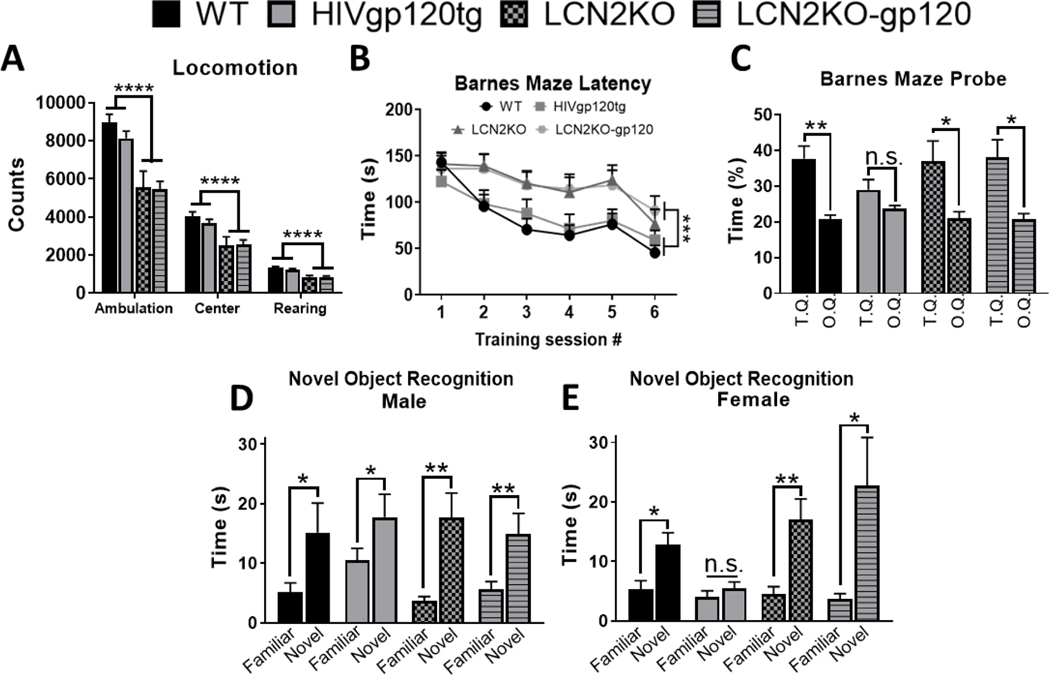
In order to further investigate the potential role of LCN2, we employed a model system of HIV-associated brain injury. HIVgp120tg mice develop behavioral impairment, including deficits in spatial learning and memory (Toggas et al., 1994). To test whether LCN2 affected behavioral deficits associated with gp120 expression, we crossbred HIVgp120tg and LCN2 knockout (LCN2KO) mice as described in the materials and methods section. We subjected the resulting LCN2+/+ gp120+ (HIVgp120tg) and LCN2−/− gp120+ (LCN2KO-gp120) animals between the ages of 7 to 8 months to a battery of behavioral tests using non-tg, LCN2+/+ gp120- (WT) and LCN2−/− gp120- knockouts (LCN2KO) as controls. The behavioral tests were designed to examine cognitive function as well as general activity, anxiety-like behavior and vision. Results of the optomotor test of vision and the light/dark transfer test of anxiety-like behavior showed no significant differences between the genotypes, suggesting intact vision and no difference in anxiety-like behavior (Fig. S2, see Supplementary File 3). Locomotor activity testing showed significantly less ambulation (P < 0.0001; F = 26.265) as well as center (P < 0.0001; F = 17.547) and rearing activity (P < 0.0001; F = 48.381) in LCN2KO animals regardless of the expression of the gp120 transgene (Fig. 2A). LCN2KO animals showed normal levels of exploration in the light/dark transfer test, suggesting that, while they may be less active overall, they don’t have motor impairments and respond similarly to novel environments.
Fig. 2. LCN2 affects locomotor activity and mediates learning and memory impairment caused by HIV gp120.
(A) Locomotor test showing difference for ambulation, center activity and rearing between Lcn2KO and Lcn2WT independently of viral gp120 expression. (B-C) Barnes maze test for spatial learning and memory; (B) Latency to escape showing time to enter the escape tunnel during six subsequent training sessions, (C) Probe test showing percent time spent in the target quadrant (T.Q) versus the average of the other three quadrants (O.Q). (D-E) Novel object recognition test for recognition memory showing time animals spent in contact with each object. Note sex-dependence is only observed in the HIVgp120tg group. Mice at 7–8 months of age were used for these experiments; n = 26 WT (male: 12, female: 14), n = 18 HIVgp120tg (male: 9, female: 9), n = 23 LCN2KO (male: 10, female: 13), and n = 16 LCN2KO-gp120 (male: 8, female: 8). Values in graphs are mean ± s.e.m.; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, ANOVA followed by Fisher’s PLSD post hoc test.
To assess the effects of LCN2 on spatial learning and memory, animals were assessed using the Barnes maze test (Barnes, 1979). Latency to escape was measured during six training sessions and showed that animals deficient in LCN2 took significantly longer to escape (P < 0.043; F = 8.678; Fig. 2B). This is consistent with our observation in the locomotor test in which LCN2KO animals, regardless of gp120, showed decreased activity (Fig. 2A). The probe test in which the escape chamber was removed allowed for the direct examination of spatial cognition, independent of overall activity levels. In this test WT animals spent more time in the target quadrant than the average of the other quadrants (P < 0.0014; F = 12.9; Fig. 2C). In contrast, HIVgp120tg animals did not spend significantly more time in the target than other quadrants, which indicates a deficit in spatial learning and memory (P < 0.1813; F = 1.943; Fig. 2C). Animals lacking LCN2 with and without gp120 spent equally and significantly more time in the target than other quadrants (P < 0.0454; F = 4.556 and P < 0.0178; F = 6.977; respectively, Fig. 2C), suggesting that genetic ablation of LCN2 abrogated the deficit in spatial learning and memory associated with gp120 expression. To assess whether gp120 leads to deficits in recognition memory, animals underwent a novel object recognition test. Here we observed a sex-dependent difference but only within the HIVgp120tg genotype group. Males and Females of WT (P < 0.0325; F = 6.378 and P < 0.0218; F = 6.547), LCN2KO (P < 0.0067; F = 12.782 and P < 0.0013; F = 17.538), and LCN2KO-gp120 (P < 0.0072; F = 12.782 and P < 0.038; F = 6.509) spent significantly more time with the novel than the familiar object, suggesting intact recognition memory. In contrast, only male HIVgp120tg distinguished between new and familiar object ((P < 0.0297; F = 6.416) while female animals failed to do so and spent equal time with both objects (P < 0.3757; F = 0.915; respectively, Fig.2D, 2E).
Together, these data suggest that HIVgp120tg animals have impaired learning and spatial memory but only female HIVgp120tg mice display a deficit in recognition memory. Moreover, the behavioral impairments associated with gp120 expression are prevented by genetic ablation of LCN2 suggesting a prominent role of LCN2 in mediating the detrimental effects of the viral envelope protein.
3.3. LCN2 facilitates neuronal damage in HIVgp120tg mice
Previous studies using brains from HIV+ individuals, have shown that MAP-2 and synaptophysin significantly correlate with neurocognitive functioning and severity of HIV-associated neurocognitive disorders (Levine et al., 2016). HIVgp120tg animals recapitulate the loss of presynaptic terminals and neuronal dendrites observed in the brains of AIDS/HIVE patients (Toggas et al., 1994). They also share other characteristic neuropathological features, such as pronounced astrocytosis, and an increased number of microglia (Maung et al., 2014; Thaney et al., 2018; Toggas et al., 1994). To elucidate the role of LCN2 in brain injury initiated in vivo by the transgenic viral gp120, brains of 9 to 10 months-old animals were collected for histopathological analysis. Neuronal injury in layer III of the mid-frontal cortex and CA1 region of the hippocampus was assessed using two methods; 1) quantification of fluorescent intensity of neuronal dendrite marker MAP-2, and 2) deconvolution microscopy to estimate the percentage of neuropil-positive for the presynaptic terminal marker synaptophysin (Syp). The HIVgp120tg animals displayed a significant reduction of both markers, MAP-2 fluorescence (P < 0.0001) and Syp+ neuropil (P < 0.0001) in layer III of the cortex as well as the stratum radiatum in CA1 of the hippocampus when compared WT controls (Fig. 3A-C, F-H, Fig. S3A-D, see Supplementary File 3). In contrast, LCN2KO-gp120 animals showed no change in MAP-2 immunoreactivity compared to WT or LCN2KO controls. Still, LCN2KO-gp120 animals showed a modest but significant decrease of Syp+ neuropil compared to WT mice in both cerebral cortex (P < 0.002) and hippocampus (P < 0.0185; Fig. S3A-D, see Supplementary File 3); however, levels of Syp+ neuropil were significantly higher in LCN2KO-gp120 animals than HIVgp120tg in both brain regions (P < 0.0001; Fig. S3A-B, see Supplementary File 3). Altogether the findings suggest that LCN2 mediates a significant portion of gp120-induced neurotoxicity, in particular the loss of MAP-2+ dendrites and to a slightly lesser extent the reduction of SYP+ presynaptic terminals.
Fig. 3. LCN2 mediates neuronal damage and microglial activation in HIVgp120tg mice.
(A & F) Representative images of frontal cortex layer III and CA1 of the hippocampus immunolabeled for neuronal synaptophysin (Syp; deconvolution microscopy, scale bar = 20 μm) and MAP-2, astrocytic GFAP and microglial Iba1 (fluorescence microscopy, scale bar = 100 μm). (B & G) Quantification of neuropil positive for Synaptophysin, (C & H) the fluorescence signal for neuronal MAP-2 and (D & I) astrocytic GFAP and (E & J) microglial Iba1+ cell numbers. Analysis used deconvolution microscopy, quantitative fluorescence microscopy (sum of fluorescence intensity, SFLI) and counting, respectively, in the frontal cortex and hippocampal CA1 region in sagittal brain sections of 9–10-month-old mice. Values in graph are mean ± s.e.m.; n = 6 (n = 3 males and 3 females) per genotype; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, student’s t-test.
3.4. LCN2 contributes to microglial activation but not astrocytosis in the presence of viral gp120
We next immunostained brain sections for astrocytic GFAP, and the microglial marker, ionized calcium-binding adaptor molecule 1 (Iba1) in order to assess astrocytosis and microglial activation using quantitative fluorescence microscopy and cell counting, respectively. HIVgp120tg mice showed a significant increase in GFAP (P < 0.0001) in both cerebral cortex and CA1 of the hippocampus when compared to WT controls (Fig. 3A, D, 3F, I, Fig. S3E-F, see Supplementary File 3). This increase was independent of LCN2 since LCN2KO-gp120 animals displayed significantly higher levels of GFAP than the LCN2KO and WT controls (P < 0.0001; Fig. 3D, 3I, Fig. S3E-F, see Supplementary File 3). Interestingly, RNAscope analysis shows that Lcn2 mRNA co-localizes with GFAP in HIVgp120tg animals suggesting astrocytes as the local source of LCN2 (Fig. S4, see Supplementary File 3). Furthermore, the presence of gp120 was associated with significantly increased numbers of microglia in the cerebral cortex in LCN2-expressing (P < 0.0001) and -deficient animals (P < 0.0011; Fig. 3E, Fig. S3G, see Supplementary File 3), but microglial numbers were significantly lower in LCN2KO-gp120 when compared to HIVgp120tg animals (P < 0.0008; Fig. S3G, see Supplementary File 3). In contrast, in hippocampus, HIVgp120tg animals had a significantly higher number of microglia cells than WT (P < 0.0001; Fig. 3J, Fig. S3H, see Supplementary File 3), while LCN2KO-gp120 animals showed no difference to the LCN2KO group (Fig. 3J, Fig. S3H, see Supplementary File 3). Thus, microglial activation in the presence of viral gp120 appears to depend more on LCN2 in hippocampus than cerebral cortex.
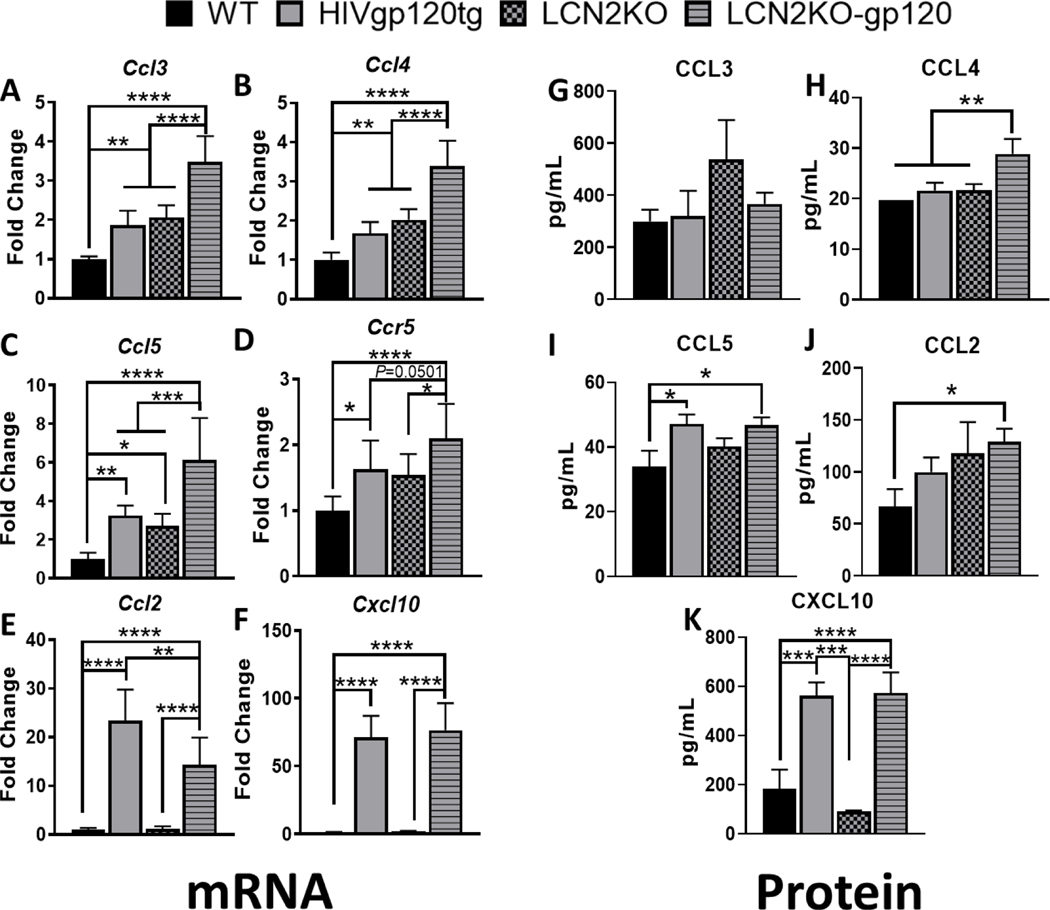
3.5. LCN2 deficiency is associated with higher expression of CCL4
LCN2 was been implicated in the regulation of brain inflammation as it can dampen the levels of pro-inflammatory cytokines and chemokines (Kang et al., 2018). To test whether LCN2 affects the induction of pro-inflammatory cytokines in our model, we compared their relative gene expression levels using qRT-PCR as described in Methods (for primers see Table S2, see Supplementary File 1). CCR5 and its natural ligands (CCL4 and CCL5) were of particular interest because we have previously observed in vitro that these ligands are protective against gp120 neurotoxicity (Kaul et al., 2007; Medders et al., 2010; Thaney et al., 2017). LCN2KO-gp120 animals showed a significant upregulation of CCR5 natural ligands (Ccl3, Ccl4, and Ccl5) when compared to all other genotypes including HIVgp120tg animals (Fig. 4A-C). We also observed a significant increase in Ccr5 expression in HIVgp120tg, LCN2KO, and LCN2KO-gp120 animals when compared to WT animals (Fig. 4D). LCN2 deficiency also resulted in increased expression of gp120 and Cxcr4 in gp120tg brains (Fig. S5). In contrast, LCN2KO-gp120 animals downregulated Ccl2 mRNA compared to HIVgp120tg animals expressing LCN2, although this chemokine remained significantly upregulated when compared to LCN2KO and WT controls (Fig. 4E). LCN2KO-gp120 animals showed a significant increase in both pro- and anti-inflammatory microglial associated genes when compared to WT and HIVgp120tg animals (Fig. S6, see Supplementary File 3). However, LCN2KO-gp120 and HIVgp120tg groups equally showed a significant increase in Cxcl10 expression independent of LCN2 (Fig. 4F). At protein level we found a significant increase in CCL4, but not CCL3, in LCN2KO-gp120 animals when compared to all genotypes (4H, G), and an increase in CCL5 when compared to WT animals (4I). LCN2KO-gp120 brains also had elevated CCL2 levels compared to WT controls but no difference to HIVgp120tg samples (4J). Consistent with mRNA data, LCN2KO-gp120 and HIVgp120tg animals equally showed a significant increase in CXCL10. Hence, our data indicate that LCN2 limits the expression of the CCR5 ligand CCL4, which has been implicated in the protection from of HIV-induced neuronal damage (Kaul and Lipton, 1999; Thaney et al., 2017).
Fig. 4. Differential expression of chemokines in LCN2 deficient mice.
(A-F) Expression of mRNA in cerebral cortex of 9 months-old mice was analyzed by qRT-PCR as described in Materials and Methods. Relative expression was calculated using the 2-ΔΔCT method. (G-K) Protein was extracted from cerebral cortices of 9–10 months old animals and 60 μg per sample were used to determine levels of soluble chemokines using a multiplex assay as described in Materials and Methods. Note that Milliplex Analyst 5.1 returned CCL4 values for all WT as 19.74 pg/ml which is the lower detection limit for this factor. Values are mean ± s.e.m.; n = 6 (female n = 3; male n = 3) per genotype; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, ANOVA followed by Fisher’s PLSD post hoc test.
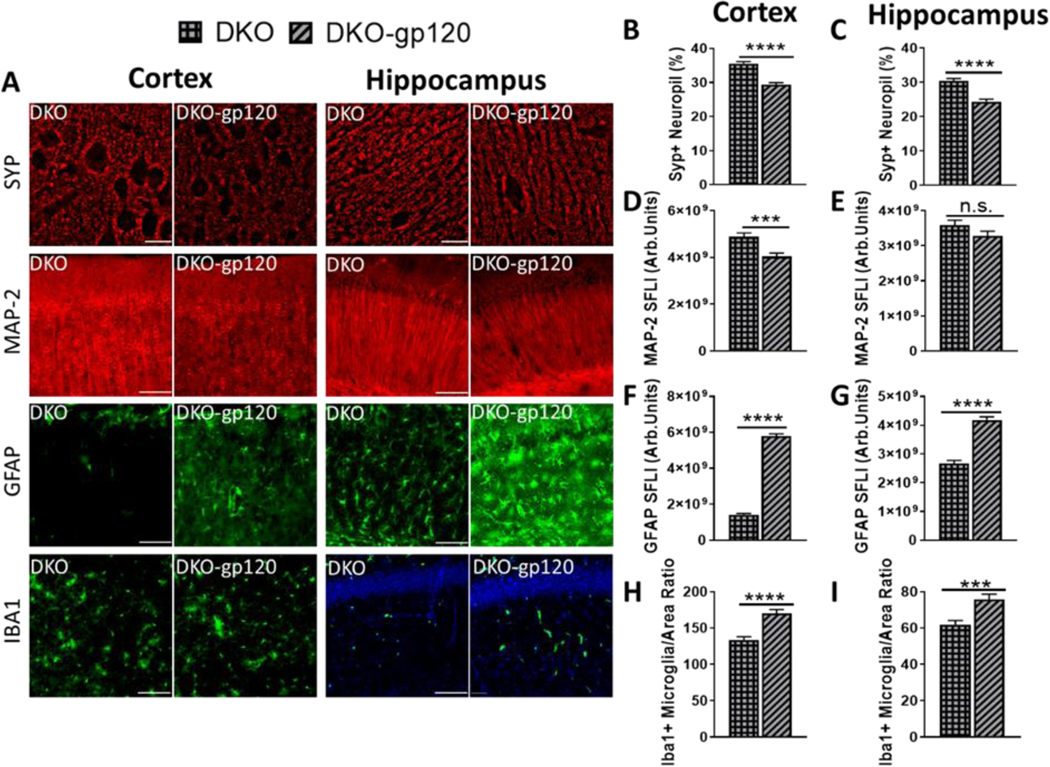
3.6. CCR5 is required for neuroprotection in LCN2-deficient HIVgp120tg mice
We hypothesized that besides the absence of LCN2, higher levels of CCR5 ligands enabled the protection observed in LCN2KO-gp120 animals. To test our hypothesis in vivo, we analyzed LCN2KO-gp120 animals also deficient in CCR5 (Huffnagle et al., 1999). The double knockout animals, CCR5KO-LCN2KO-gp120 mice are hereafter referred to as double-knockout (DKO). Brains from animals between 9 to 10 months old were harvested for histopathological analysis of neuronal injury in layer III of the mid-frontal cortex and CA1 of the hippocampus as described above. The DKO-gp120 animals displayed a significant reduction in both MAP-2 fluorescence intensity and Syp+ neuropil, in the cortex when compared to DKO control animals without gp120 expression (P < 0.0001 and P < 0.001 respectively; Fig. 5B, D, Fig. S3A, C, see Supplementary File 3). In the hippocampus, Syp+ neuropil was significantly lower in DKO-gp120 animals (P < 0.0001), but there was only a trend toward reduction in MAP-2 marker when compared to the DKO animals (P < 0.12; Fig. 5C, E, Fig. S3B, D, see Supplementary File 3). Quantitative fluorescence microscopy showed an increase in GFAP as a result of gp120 expression in both genetic backgrounds in the cortex as well as in the hippocampus (P < 0.0001; Fig. 5F-G, Fig. S3E-F, see Supplementary File 3). Microglial numbers were significantly increased in DKO-gp120 animals when compared to DKO without gp120 in both cortex and hippocampus (P < 0.0001 and P < 0.0002 respectively; Fig. 5H-I, Fig. S3G-H, see Supplementary File 3). QRT-PCR analysis shows that damage observed in DKO-gp120 animals is not associated with increased expression of the gp120 transgene whereas its co-receptor Cxcr4 is upregulated (fig. S5). Yet, DKO-gp120 animals display neuronal damage and microgliosis very similar to that observed in HIVgp120tg animals (Fig. S3, see Supplementary File 3).
Fig. 5. CCR5KO permits neuronal damage and increased gliosis in LCN2-deficient gp120tg mice.
(A) Histopathological analysis was performed in frontal cortex layer III and hippocampal CA1 region of sagittal brain sections of 9–10-month-old mice as described in Material and Methods using immunofluorescence staining for Synaptophysin (40X images of Layer III, deconvolution microscopy, scale bar = 20 μm), neuronal MAP-2, astrocytic GFAP and microglial Iba1 (10X images, fluorescence microscopy, scale bar = 100 μm). (B & C) Percentage of neuropil positive for Synaptophysin, and (D & E) the fluorescence signal for neuronal MAP-2 and (F & G) astrocytic GFAP and (H & I) microglial Iba1+ cell numbers were determined using deconvolution microscopy, quantitative fluorescence microscopy and counting, respectively. Values in graphs are mean ± s.e.m.; n = 6 (3 males and 3 females) for each condition; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001, student’s t-test.
Furthermore, qRT-PCR revealed that DKO-gp120 animals have significantly increased TNFα mRNA and a trend towards increase in CD68 (P < 0.081), two pro-inflammatory microglial factors, when compared to WT controls (Fig. S6A, C, see Supplementary File 3). There was no change in anti-inflammatory microglial associated genes (fig. S6D-F). Altogether, the results show that CCR5 is required for protection against neurotoxicity triggered by viral gp120 in the absence of LCN2.
3.7. LCN2, CCR5 and viral gp120 affect expression of genes involved in neurotransmission
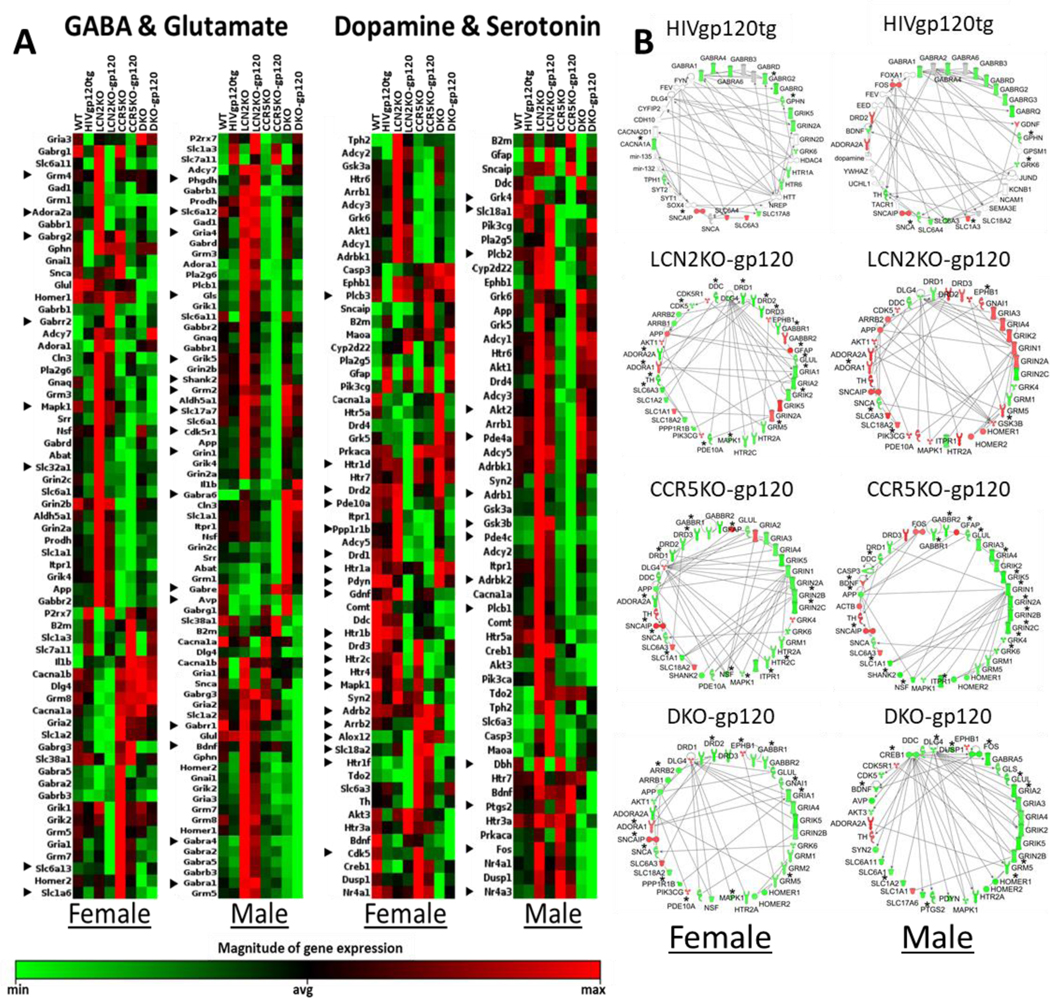
Decreased synaptic and dendritic density in hippocampus and cerebral cortex is one of the best correlates of ante mortem signs of cognitive impairment in HIV/AIDS patients (Ellis et al., 2007; Kaul et al., 2001). The same pathological features are also associated with behavioral impairment in HIVgp120tg mice (D’Hooge et al., 1999; Maung et al., 2014; Toggas et al., 1994), which reportedly display abnormal glutamatergic and GABAergic neurotransmission (Hoefer et al., 2015; Krucker et al., 1998). Therefore, we decided to investigate in hippocampus and cortex if expression of genes involved in neurotransmission was altered by genetic ablation of Lcn2 and Ccr5 in HIVgp120tg brains using RT2 Profiler™ PCR Arrays. Since we had previously found that CCR5 deficiency alone resulted in neuroprotection of HIVgp120tg mice if LCN2 is present (Maung et al., 2014), we also included samples of CCR5KO LCN2WT HIVgp120tg and non-tg controls in this analysis. Two qRT-PCR arrays were employed to analyze components of the GABAergic and glutaminergic systems (GG array), and the dopaminergic and serotonergic systems (DS array), respectively. The results revealed sex-dependent differences within the various genotypes in the expression of genes involved in cerebrocortical and hippocampal neurotransmission (Fig. 6A, Fig. S7, see Supplementary File 3, and Tables S4, S5, see Supplementary Files 4 and 5). Moreover, gp120, LCN2 and CCR5, all affect multiple components of the four neurotransmission systems ranging from receptors to transporters to signaling and regulatory factors (Tables S4, S5, see Supplementary Files 4 and 5). The pattern of gene expression and thus the configuration of neurotransmission components differs not only between the WT controls and genotypes that show neuronal injury (HIVgp120tg and DKO-gp120) but also between genotypes associated with neuroprotection (LCN2KO-gp120 and CCR5KO-gp120). Thus, protection from neuronal damage is not associated with restoration of the normal (control) configuration of neurotransmission-related gene expression.
Fig. 6. Differential gene expression signatures associated with neurodegeneration and protection are genotype- and sex-dependent.
(A) RNA isolation and RT2 RNA profiler PCR arrays were performed as described in Materials and Methods. The clustergram heat maps show gene expression differences in components of GABA- & glutamate- and dopamine- & serotonergic neurotransmission systems in the cerebral cortex of eight genotypes: HIVgp120tg, LCN2KO, LCN2KO-gp120, CCR5KO, CCR5KO-gp120, DKO, and DKO-gp120 animals compared to WT control. The QIAGEN data analysis center was used for 2–(ΔΔCT)-based fold change calculations and a modified student’s test to compute two tails, equal variance P-values. Heat maps for significantly differentially regulated genes (P ≤ 0.05) are row normalized; n = 3–6 males and n = 3–6 females per genotype. ► indicates gene only differentially expressed in male or female regardless of genotype. (B) Male and female neurotransmission-related gene networks were generated for cerebral cortex of HIVgp120tg, LCN2KO-gp120, CCR5KO-gp120, and DKO-gp120 mice employing Ingenuity Pathway Analysis software (IPA) by interrogating RNA expression data obtained with RT2 Profiler™ PCR Arrays. Networks display experimentally analyzed genes (up-regulated (red); down-regulated (green) in gp120-expressing genotypes compared to WT and indicate direct interactions. Genes included by IPA based on published data are shown without color. * Indicates genes significantly differentially regulated (P ≤ 0.05) as determined by the RT2 Profiler™ PCR Arrays.
All of the genotypes, whether displaying neuronal damage or protection express different patterns of neurotransmission-related genes. Ingenuity pathway analysis (IPA) enables the identification of gene networks and downstream targets shared between sexes and/or genotypes in association with either neuroprotection or neuronal damage using the 168 genes analyzed in the qRT-PCR arrays. The resulting top gene networks are shown in Fig. 6B, and Fig. S7B (representation of subcellular localization for hippocampus in Fig. S8, see Supplementary File 3) and indicate that male and female brains of the same genotype share several downstream genes. HIVgp120tg-associated gene networks feature huntingtin (HTT) as a central network component as well as the downstream interactions with PER1, CLOCK, and CDK5. LCN2KO-gp120 male and female animals share downstream engagement of FOS and FOXA1. The CCR5KO-gp120 animals share involvement of CREB1. In contrast, male DKO-gp120, but not females, show alteration in CDK5 and CDK5R, similarly to their HIVgp120tg counterparts. Thus, the bioinformatic analysis indicates shared and separate signaling pathways of neurotransmission affected by LCN2, CCR5 and viral gp120 in both sexes.
The top-scoring gene networks predicted by IPA link HTT, CDK5, FOS, DUSP1, and CREB1 to neuronal injury and dysfunction triggered by HIVgp120 via the p38 MAPK pathway (Fig. 7A) (Chang et al., 2010; Kalra and Kumar, 2004; Tan et al., 1996; Taylor et al., 2013). We have previously demonstrated that p38 MAPK is a critical mediator of neuronal injury and death caused by gp120-induced macrophage toxicity (Medders et al., 2010). A Western blot analysis of cerebral cortex of 9 months-old animals showed that HIVgp120tg animals had significantly higher levels of active p38 MAPK when compared to WT controls (Fig. 7B, C). DKO-gp120 animals displayed no significant increase in active p38 MAPK due to gp120 (Fig. 7B, C) but LCN2KO and DKO genotypes that lack gp120 and neuronal injury, also displayed in comparison to WT elevated levels of phosphorylated p38 MAPK which reached significance only in LCN2KO. Genotypes associated with protection (LCN2KO-gp120 and CCR5KO-gp120) showed no significant change in active p38 MAPK compared to WT (Fig. 7B, C) whereas DKO-gp120 samples showed a trend to higher kinase activity. No significant differences were detected in the levels of total p38 MAPK protein between the different genotypes, and no differences between males and females. Altogether, the data suggests that while elevated activity of p38 MAPK in the absence of HIVgp120 is not necessarily indicative of neuronal damage, LCN2 and CCR5 modulate neurotoxicity of HIVgp120 and activity of p38 MAPK and expression of neurotransmission-related genes and in the presence and absence of the viral envelope protein.
Fig. 7. LCN2 neurotoxicity depends on p38 MAPK and microglia.
(A) Predicted interactions between components of the gene networks shown in (Fig. 6B) and p38 MAPK. IPA software identified direct (solid lines) and indirect interactions (dashed lines). (B) Protein was extracted from cerebral cortices of 9–10 months old animals and 25 μg per sample were used for Western blotting as described in Materials and Methods. ImageJ software was used for densitometry analysis of proteins. (C) Quantification of phosphorylated p38 MAPK (pp38)/Total p38. Data was normalized to their corresponding tubulin signal. Values in graphs are mean ± s.e.m.; n = 6 (3 males and 3 females) per genotype; * P ≤ 0.05 for difference to WT control, ANOVA followed by Fisher’s PLSD post hoc test. (D-H) Rat mixed neuronal-glial cerebrocortical cell cultures (RCC) were pre-incubated with or without a p38 MAPK inhibitor, SB203580 (SB; 10 μM), for 15 min prior to treatment with LCN2 (4 nM) for 72 hrs. (D, F, and H) RCC were treated with LME (7.5 mM) to deplete microglia for 16 hrs and then pre-incubated with or without a p38 MAPK inhibitor and treated with LCN2 as described above. (E-F) Fluorescence intensity of immuno-labeled MAP2 in fixed cells was used to analyze neuronal injury and loss as described in Materials and Methods. (H-G) % Neuronal Survival was calculated by counting MAP2+ neurons and total cells, and calculating the percentage of neurons relative to control. Control samples were defined as 100% survival. Values in graphs are mean ± s.e.m.; n = 3 independent experiments; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, P ≤ 0.0001, ANOVA followed by Fisher’s PLSD post hoc test; scale bar = 100 μm.
3.8. LCN2 neurotoxicity involves p38 MAPK and microglia
LCN2 reportedly affects microglial activation and activity of p38 MAPK (Jang et al., 2013; Jeon et al., 2013). Therefore, we used rat cerebrocortical cell cultures (RCC) comprising neurons, astrocytes and microglia (Kaul and Lipton, 1999; Medders et al., 2010; Sanchez et al., 2016b) to determine, if the neurotoxic effect of LCN2 is dependent on microglia or p38 MAPK activity. RCC were treated with LCN2 in the presence and absence of SB203580, a specific inhibitor of p38 MAPK. Neuronal injury and loss were scored in fixed and immunofluorescence-labeled RCC by cell counting and fluorescent quantification of MAP-2+ neurons (Medders et al., 2010; Sanchez et al., 2016b). Treatment with LCN2 resulted in significant neuronal loss and injury but blockade of p38 MAPK diminished neurotoxicity (Fig. 7E & G). Inhibition of p38 MAPK prevented neuronal cell loss triggered by LCN2 exposure (Fig, 7G) while the reduction of MAP2 fluorescence intensity suggested residual neuronal injury in comparison to control (Fig. 7E).
Next, we pre-treated RCC with L-leucine-methyl ester (LME) for 16 hrs to deplete microglia (Kaul and Lipton, 1999; Medders et al., 2010). Microglia-depleted cultures exposed to LCN2 displayed a significant reduction of neurons and MAP2 fluorescence intensity similar to RCC with microglia (Fig. 7F & H). However, the combination of LME pre-treatment and SB203580 abrogated a significant loss of MAP2 intensity and MAP2+ neurons due to LCN2 (Fig. 7F & H). SB203580 showed no significant effect in RCC with or without microglia. Taken together this data shows that LCN2 toxicity depends on p38 MAPK activity and to a lesser extent on the presence of microglia.
4. Discussion
In this study, we show that the level of LCN2, an acute-phase protein of the innate immune response, is elevated in the brain of a subset of HIV-infected patients with neuropathology and neurocognitive impairment as well as in a transgenic model of HIV-induced brain injury. In the human samples, we observed significant correlations between LCN2 expression levels and HIV RNA loads in the CSF and HIV DNA levels in frontal neocortex. Correlations between LCN2 and HIV RNA levels in frontal neocortex and the deficit score for verbal fluency domain were only slightly weaker. Our observations, and a recent report of increased plasma and serum levels of LCN2 in HIV+ individuals in correlation with worse processing speed and motor function domain (Williams et al., 2019) lend support to a potential role of LCN2 in HIV associated brain injury.
Our study provides further support for a pathological role of LCN2 by showing in vivo that it contributes to neuronal damage caused by HIV envelope protein gp120. Genetic ablation of LCN2 prevents behavioral deficits and ameliorates neuronal damage associated with CNS expression of the viral gp120. LCN2 deficiency also results in higher expression levels of the neuroprotective CCR5 ligand Ccl4 and of Ccr5 itself. This β-chemokine ligand-receptor interaction contributes to antiviral and neuronal protection since the CCR5 ligands are major suppressors of HIV infection in CD8+ T lymphocytes (Cocchi et al., 1995), and microglia (Kitai et al., 2000a).In humans, elevated levels of β-chemokines in blood and cerebrospinal fluid (CSF) are associated with a slower progression to AIDS (Garzino-Demo et al., 1999) and less neurocognitive impairment, respectively (Letendre et al., 2011). We have shown that CCL4 and −5 reduce p38 MAPK activation and neurotoxicity in vitro in cerebrocortical cell cultures (Medders et al., 2010). Our finding that LCN2 limits the expression of the β-chemokine CCL4 may potentially explain why LCN2 levels correlate with higher viral loads in brain specimen of HIV infected individuals, although this possibility requires further investigation. In any case, disruption of the CCR5 ligand-receptor system by knocking out the receptor in LCN2KO-gp120tg mice largely restores neuronal injury and reveals that CCR5 contributes to neuroprotection in LCN2-deficient gp120tg brains. Thus, LCN2 presumably can override CCR5-mediated neuroprotection, at least in part, by suppressing expression of protective CCR5 ligands and their receptor. However, we cannot exclude that LCN2 affects CCR5-mediated protective signaling in other indirect ways, and this possibility remains to be explored. Paradoxically, stimulation of neuronal CCR5 can apparently also impair learning and memory as well as recovery after stroke and traumatic brain injury (Joy et al., 2019; Zhou et al., 2016). Of note, in HIV+ individuals low detectable levels of β-chemokines, in particular CCL3, in CSF were associated with worse neurocognitive performance than higher levels (Letendre et al., 1999). Seemingly, the role of CCR5 and its natural ligands in the brain depends on ligand concentration and context, such as the presence or absence of LCN2, and warrants further investigation.
Our findings raise also interesting questions about the effect of maraviroc (MVC), a clinically approved inhibitor of infection by CCR5-preferring viruses (Dorr et al., 2005). Similar to the physiological CCR5 ligands CCL3, CCL4, and CCL5, MVC appears to act as inhibitor of CCR5-preferring viruses, which are often referred to as macrophage-tropic, but can also infect CD8+ T lymphocytes (Cocchi et al., 1995; Kitai et al., 2000b). However, the question arises, if MVC can also interfere with the neuroprotective function of the physiological CCR5 ligands. This possibility remains to be investigated, but one consideration is that if MVC can reduce CCR5-mediated HIV-1 infection of macrophages, neurotoxicity may also be reduced and thus diminish the need for neuroprotection by physiological CCR5 ligands. We did not investigate MVC in this study but we previously observed in mixed neuronal-glial cerebrocortical cell cultures, that the combination of MVC with LCN2 in the presence of a CXCR4-preferring gp120 resulted in a significant loss of microglia and completely abrogated neurotoxicity (Maung et al., 2014). This is presumably due to the fact, that paradoxically, CCR5-CCL5 interactions can provide an essential pro-survival and activation signal to macrophages/microglia when exposed to viral components (Tyner et al., 2005). MVC likely disrupted that interaction leading to depletion of microglia activated by LCN2 and gp120, and depletion of microglia has been shown to prevent neurotoxicity of gp120 (Maung et al., 2014; Medders et al., 2010). Thus, it appears that MVC can disrupt CCR5-ligand interactions which, however, may not result in neuronal damage because it also limits activation and survival of neurotoxin-producing macrophages/microglia.
Our behavioral testing confirmed that HIVgp120tg animals exhibit impaired spatial memory (Maung et al., 2014; Toggas et al., 1994). Interestingly, testing recognition memory and expression of genes related to neurotransmission, both revealed sex-dependent differences in HIVgp120tg animals. The exact reason why recognition memory is only compromised in females, but not males, remains to be elucidated. The alterations in expression of genes involved in neurotransmission will guide those future experiments. Of note, protection from neuronal damage does not restore the normal (control) configuration of neurotransmission-related gene expression but possibly a compensatory pattern that permits normal function in terms of behavioral performance in the continued presence of viral gp120. In HIV+ patients, sex-dependent differences are observed in the progression towards AIDS, with female patients progressing faster than their male counterparts displaying similar levels of viremia (Addo and Altfeld, 2014). Our findings are also in line with recent reports of sex-dependent behavioral changes in other models of HIV brain injury (McLaurin et al., 2017; Putatunda et al., 2019). In contrast, our data suggest that LCN2 contributes in a sex-independent fashion to HIV-1’s detrimental effects on spatial learning and recognition memory, similar to its effects on other neurological and cognition-related capabilities, such as verbal fluency and disturbance of processing speed and motor function (Williams et al., 2019). The influence of LCN2 on the latter two functions may also be reflected by our observation that LCN2KO animals display overall diminished locomotor activity compared to LCN2 WT, independently of viral gp120.
Genetic ablation of LCN2 in HIVgp120tg animals prevented loss of neuronal dendrites, ameliorated the loss of presynaptic terminals, and limited the increase in microglial numbers, all prominent pathological features of HIV brain injury (Gelman et al., 2012). Interestingly, a modest, yet significant decrease in Syp+ neuropil in the cortex and hippocampus persisted in LCN2 deficient animals, which indicates additional mechanisms are contributing to neuronal damage caused by gp120. The elevated expression of the viral gp120 and host CXCR4 in the brain of LCN2KO-gp120 could counteract the overall protective effect of LCN2-deficiency and warrants further investigation.
However, LCN2 contributes to the increase in microglia numbers associated with gp120 expression. LCN2KO-gp120 mice showed no increase in microglial numbers in the hippocampus and a modest but significant increase in the cortex that remained significantly lower than in LCN2-expressing HIVgp120tg animals. This finding is in line with previous reports for Parkinson’s disease (Kim et al., 2016) and a model of neuropathic pain (Jeon et al., 2013), where LCN2 deficiency was associated with reduced numbers of microglia. Furthermore, LCN2KO-gp120 animals showed an increased expression of Arg1, Cd163, and Mrc1, which limit inflammation and promote tissue repair (Tang and Le, 2016). At the same time, LCN2KO-gp120 brains show an increase in Cd68, iNos, and Tnfα, all of which are associated with an inflammatory response (Tang and Le, 2016). The upregulation of both pro- and anti-inflammatory microglial genes indicates a role for LCN2 in balancing these two opposing biological functions. LCN2 has been shown to modulate inflammation in a context-dependent manner (Jeon et al., 2013; Jha et al., 2015; Kang et al., 2018; Sickinger et al., 2013). Limiting the inflammatory response can be beneficial or damaging depending on the context of the injury. During bacterial lipopolysaccharide (LPS) challenge, LCN2 protects the brain by preventing an increase in inflammatory factors (Kang et al., 2018). However, in neuropathic pain, LCN2 is detrimental as it helps maintain elevated levels of CCL2 which activate microglia and promote pain (Jeon et al., 2013). In the context of gp120 neurotoxicity the concentration of LCN2 seems to reach toxic levels in conjunction with the reduction of neuroprotective chemokines, such as CCL4. However, increased levels of CCL2 and CXCL10 may counteract the protective effect of the LCN2KO since both chemokines have been implicated in HIV neuropathogenesis (Kaul and Lipton, 1999; Letendre et al., 2011; Sanders et al., 1998). Consequently, LCN2 may well contribute to neurotoxicity also by mitigating the expression of the protective β-chemokine CCL4 and despite limiting expression of the potentially injury-promoting CCL2. However, the exact mechanisms by which LCN2 modulates expression of certain cytokines and chemokines remain to be elucidated. Moreover, our findings in the HIVgp120tg mouse model need to be evaluated in the human system. For example, LCN2 can modulate levels of intracellular iron (Dekens et al., 2018) and can contribute to activation of p38 MAPK (Jeon et al., 2013), and both of these factors are known modulators of cytokine production (Wang et al., 2009; Yu and Richardson, 2011). While we found an influence of LCN2 on the activity of p38 MAPK, we did not investigate the effect on cellular iron in this study. Thus, more research is necessary to determine LCN2’s mechanism of action.
Additionally, we and others have recently shown that LCN2 itself can cause neurotoxicity, and the RNAscope analysis supports the suggestion that neurotoxic LCN2 originates in astrocytes (Bi et al., 2013; Maung et al., 2014). However, upregulation of GFAP in astrocytosis does seemingly not depend on LCN2 in the presence or absence of CCR5 (Maung et al., 2014). Thus, our observations differ from an earlier report proposing LCN2 as driver of astrocytosis (Lee et al., 2009).
Of note, DKO mice show a significant increase in microglia numbers, although the number of microglia in cortex remains significantly lower than in LCN2- and CCR5-expressing HIVgp120tg animals. The elevated microglial number is accompanied by a significant increase of Tnfα expression, but not Arg1, Cd163 or Mrc1, suggesting a pro-inflammatory shift in the DKO-gp120 compared to LCN2KO-gp120 animals. In any case, while the knockout of LCN2 abrogated neurotoxicity, concomitant ablation of CCR5 restored neuronal damage and microglial activation in the presence of viral gp120, showing that in vivo LCN2 is not required for HIV neurotoxicity but can override CCR5-mediated neuroprotection.
Gene expression analysis showed that LCN2KO, CCR5KO, and DKO with and without gp120 displayed alterations in different components of the glutaminergic, dopaminergic, serotonergic and GABAergic neurotransmission systems compared to WT controls. Notably, all genotypes showed sex-dependent differences in the expression of neurotransmission-related genes but not in histopathology, p38 MAPK activity or spatial learning and memory. Only recognition memory was subject to sexual dimorphism in one genotype, HIVgp120tg mice expressing LCN2 and CCR5. Ingenuity Pathway Analysis (IPA) identified top gene networks affected by viral gp120 and deficiency of LCN2 and CCR5. The results showed that despite different expression patterns for neurotransmission-related genes, similar downstream factors are affected in males and females of the same genotype. In particular, the networks suggested a potential role of p38 MAPK since all networks showed molecules linked to this kinase, such as Cdk5, Creb1, and Dusp1. CREB1 can be activated by p38 MAPK (Tan et al., 1996) and DUSP1 and CDK5 play important roles in controlling p38 MAPK activation (Chang et al., 2010; Taylor et al., 2013). These reports and our findings are in line with a regulatory role of p38 MAPK in neurotransmission in both the presence and absence of HIV-induced neurotoxicity. Of note, LCN2-deficiency in the absence of gp120 clearly indicates that increased activity of p38 MAPK is not always linked to neuronal damage or behavioral impairment.
Furthermore, genotypes in which neurons are protected, LCN2KO-gp120 and CCR5KO-gp120, lack a significant increase of active p38 MAPK due to gp120 expression. Since p38 MAPK is a critical mediator of HIV-induced toxicity in neurons (Medders et al., 2010), the limitation of its activity points to a mechanism as to how protection is accomplished in the presence of the viral neurotoxin.
However, we find in vitro that LCN2 itself directly exerts neurotoxicity via p38 MAPK activation. In mixed neuronal-glial cell cultures LCN2 causes loss of MAP-2+ neurons which is largely, but not entirely, prevented by pharmacological inhibition of p38 MAPK and not diminished by depletion of microglia alone. Thus, in contrast to HIVgp120, complete protection against LCN2 neurotoxicity is only achieved when p38 MAPK is inhibited in conjunction with microglial depletion.
Conclusions
Our findings provide evidence that LCN2 contributes to neurotoxicity during HIV infection and suggest the overriding of CCR5-mediated neuroprotection as a mechanistic explanation (See Fig. 8 for a schematic summary). Therefore, our observation of higher levels of LCN2 in post mortem tissues from patients with HIV brain injury and our experimental findings have two potential implications. First, our results lend support to the recent proposal of LCN2 as a biomarker for the vulnerability to HIV-induced brain pathology and development of HAND (Williams et al., 2019). Second, neutralizing or depleting LCN2 in HIV+ patients with virus-induced, elevated CCR5 ligand expression may diminish the risk of developing neuronal injury and HAND.
Fig. 8. Schematic summary of the proposed roles of LCN2 and CCR5 in promotion and prevention of HIV-1 neurotoxicity triggered by the viral envelope protein gp120.
Activation of p38 MAPK in microglia initiated by HIV gp120 viral protein is necessary for neuronal damage. Microglia can produce pro-inflammatory factors and viral proteins that lead to activation of astrocytes, which release LCN2. LCN2 alone contributes to neurotoxicity by interacting with receptors in the neurons and presumably, at least in part, by limiting the expression of CCR5 natural ligands (CCL3, −4, −5). If LCN2 is absent, an increased expression of CCR5 natural ligands mitigates activation of p38 MAPK, and in turn abrogates neuronal damage and behavioral alterations. The CCR5 β-chemokine receptor is necessary for this protection to occur. In the absence of both CCR5 and LCN2, β-chemokines, such as CCL4 and −5, no longer provide protection. Hence, HIVgp120tg mice that are double knockout for LCN2 and CCR5 display an increase in active p38 MAPK as well as neuronal damage similar to gp120tg animals that express both components of the innate immune system.
Supplementary Material
Table S1 Demographic information for human brain samples.
Table S2 Primer sequences.
Table S3 Pearson correlations between LCN2 mRNA and cohort characteristics.
Detailed descriptions of experimental and analytical procedures for behavioral testing, immunohistology, quantitative fluorescence and deconvolution microscopy, and bioinformatics.
Table S4 Neurotransmission-related genes in mouse cerebral cortex, differential expression in HIVgp120tg with and without LCN2 and/or CCR5 knockout compared to wild-type controls
Table S5 Neurotransmission-related genes in mouse hippocampus, differential expression in HIVgp120tg with and without LCN2 and/or CCR5 knockout compared to wild-type controls
Fig. S1. LCN2 levels are increased in patients with HIV brain pathology.
Fig. S2. HIVgp120tg animals and/or LCN2 deficiency do not result in anxiety-like behavior or impaired vision.
Fig. S3. Combined analysis of immunofluorescence and deconvolution microscopy of HIVgp120tg, LCN2KO-gp120tg and CCR5KO-LCN2KO-gp120tg brains and the respective non-tg, gp120- controls.
Fig. S4. Lcn2 mRNA is expressed in astrocytes.
Fig. S5. Expression of HIV gp120 transgene and its receptor are not reduced in LCN2KO gp120 nor DKO animals.
Fig. S6. Expression of pro- and anti-inflammatory microglial genes.
Fig. S7. Differential gene expression signatures in genotypes associated with neurodegeneration and protection are sex-dependent in hippocampus.
Fig. S8. Top functional gene networks predicted based on gene expression changes.
Acknowledgments:
We thank Drs. Shizuo Akira (Osaka University, Osaka, Japan) and Alan Aderem (Institute for Systems Biology, Seattle, WA) for providing Lcn2KO mice. We are grateful for the kind assistance of Joshua G. Lisinicchia, UTMB, Galveston, TX, with the handling of the human brain specimen, Dr. Michael Walker for advice with statistical analysis, Samantha Bruinsma and Dr. Nancy Lainez for their help with RNAscope and Milliplex experiments respectively, the Genomic Core Facility, Institute for Integrative Genome Biology, at UC Riverside for running the bioanalyzer tests, and the teams of the animal facilities of the Sanford-Burnham-Prebys Medical Discovery Institute, The Scripps Research Institute and the University of California, Riverside, for expert assistance with the various mouse lines.
Funding
This work was supported by funds form the National Institute of Health NIH, R01 MH087332, MH104131, MH105330 and P50 DA026306 (P5) to M.K. Human brain specimens were obtained from the National NeuroAIDS Tissue Consortium supported by U24 MH100930 (BBG). The sponsor(s) did not play any role in study design, data collection or decision to submit the article for publication.
Footnotes
Declarations of interests
The authors have no conflict or competing interests.
Ethics approval and consent to participate
All human specimen were provided by the National NeuroAIDS Tissue Consortium (NNTC), which is an NIH funded tissue repository. All samples received for this study were de-identified. The NNTC requires making results publicly available.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author upon request. The newly generated mouse lines will be shared through material transfer agreements (MTA).
References:
- Addo MM, Altfeld M, 2014. Sex-based differences in HIV type 1 pathogenesis. J Infect Dis 209 Suppl 3, S86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, 2007. Updated research nosology for HIV-associated neurocognitive disorders. Neurology 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Miller VL, Weiser JN, 2009. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 5, e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Clifton M, Hoette TM, Mori K, Deng SX, Qiu A, Viltard M, Williams D, Paragas N, Leete T, Kulkarni R, Li X, Lee B, Kalandadze A, Ratner AJ, Pizarro JC, Schmidt-Ott KM, Landry DW, Raymond KN, Strong RK, Barasch J, 2010. Iron traffics in circulation bound to a siderocalin (Ngal)-catechol complex. Nat Chem Biol 6, 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CA, 1979. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J Comp Physiol Psychol 93, 74–104. [DOI] [PubMed] [Google Scholar]
- Bi F, Huang C, Tong J, Qiu G, Huang B, Wu Q, Li F, Xu Z, Bowser R, Xia XG, Zhou H, 2013. Reactive astrocytes secrete lcn2 to promote neuron death. Proc Natl Acad Sci U S A 110, 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, de Pablo Y, Lee HP, Lee HG, Smith MA, Shah K, 2010. Cdk5 is a major regulator of p38 cascade: relevance to neurotoxicity in Alzheimer’s disease. J Neurochem 113, 1221–1229. [DOI] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P, 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270, 1811–1815. [DOI] [PubMed] [Google Scholar]
- D’Hooge R, Franck F, Mucke L, De Deyn PP, 1999. Age-related behavioural deficits in transgenic mice expressing the HIV-1 coat protein gp120. Eur J Neurosci 11, 4398–4402. [DOI] [PubMed] [Google Scholar]
- Dekens DW, Naudé PJW, Keijser JN, Boerema AS, De Deyn PP, Eisel ULM, 2018. Lipocalin 2 contributes to brain iron dysregulation but does not affect cognition, plaque load, and glial activation in the J20 Alzheimer mouse model. Journal of Neuroinflammation 15, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M, 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49, 4721–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R, Langford D, Masliah E, 2007. HIV and antiretroviral therapy in the brain: neuronal injury and repair. Nat Rev Neurosci 8, 33–44. [DOI] [PubMed] [Google Scholar]
- Ferreira AC, Da Mesquita S, Sousa JC, Correia-Neves M, Sousa N, Palha JA, Marques F, 2015. From the periphery to the brain: Lipocalin-2, a friend or foe? Prog Neurobiol 131, 120–136. [DOI] [PubMed] [Google Scholar]
- Ferreira AC, Pinto V, Da Mesquita S, Novais A, Sousa JC, Correia-Neves M, Sousa N, Palha JA, Marques F, 2013. Lipocalin-2 is involved in emotional behaviors and cognitive function. Front Cell Neurosci 7, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AC, Santos T, Sampaio-Marques B, Novais A, Mesquita SD, Ludovico P, Bernardino L, Correia-Neves M, Sousa N, Palha JA, Sousa JC, Marques F, 2018. Lipocalin-2 regulates adult neurogenesis and contextual discriminative behaviours. Mol Psychiatry 23, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Fields J, Dumaop W, Rockenstein E, Mante M, Spencer B, Grant I, Ellis R, Letendre S, Patrick C, Adame A, Masliah E, 2013. Age-dependent molecular alterations in the autophagy pathway in HIVE patients and in a gp120 tg mouse model: reversal with beclin-1 gene transfer. J Neurovirol 19, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A, 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. [DOI] [PubMed] [Google Scholar]
- Garzino-Demo A, Moss RB, Margolick JB, Cleghorn F, Sill A, Blattner WA, Cocchi F, Carlo DJ, DeVico AL, Gallo RC, 1999. Spontaneous and antigen-induced production of HIV-inhibitory β-chemokines are associated with AIDS-free status. Proceedings of the National Academy of Sciences 96, 11986–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman BB, Chen T, Lisinicchia JG, Soukup VM, Carmical JR, Starkey JM, Masliah E, Commins DL, Brandt D, Grant I, Singer EJ, Levine AJ, Miller J, Winkler JM, Fox HS, Luxon BA, Morgello S, National Neuro ATC, 2012. The National NeuroAIDS Tissue Consortium brain gene array: two types of HIV-associated neurocognitive impairment. PLoS One 7, e46178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman BB, Lisinicchia JG, Morgello S, Masliah E, Commins D, Achim CL, Fox HS, Kolson DL, Grant I, Singer E, Yiannoutsos CT, Sherman S, Gensler G, Moore DJ, Chen T, Soukup VM, 2013. Neurovirological correlation with HIV-associated neurocognitive disorders and encephalitis in a HAART-era cohort. J Acquir Immune Defic Syndr 62, 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill AJ, Kovacsics CE, Cross SA, Vance PJ, Kolson LL, Jordan-Sciutto KL, Gelman BB, Kolson DL, 2014. Heme oxygenase-1 deficiency accompanies neuropathogenesis of HIV-associated neurocognitive disorders. J Clin Invest 124, 4459–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefer MM, Sanchez AB, Maung R, de Rozieres CM, Catalan IC, Dowling CC, Thaney VE, Pina-Crespo J, Zhang D, Roberts AJ, Kaul M, 2015. Combination of methamphetamine and HIV-1 gp120 causes distinct long-term alterations of behavior, gene expression, and injury in the central nervous system. Exp Neurol 263, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, McNeil LK, McDonald RA, Murphy JW, Toews GB, Maeda N, Kuziel WA, 1999. Cutting Edge: Role of C-C Chemokine Receptor 5 in Organ-Specific and Innate Immunity to Cryptococcus neoformans. The Journal of Immunology 163, 4642–4646. [PubMed] [Google Scholar]
- Jang E, Lee S, Kim J-H, Kim J-H, Seo J-W, Lee W-H, Mori K, Nakao K, Suk K, 2013. Secreted protein lipocalin-2 promotes microglial M1 polarization. The FASEB Journal 27, 1176–1190. [DOI] [PubMed] [Google Scholar]
- Jeon S, Jha MK, Ock J, Seo J, Jin M, Cho H, Lee WH, Suk K, 2013. Role of lipocalin-2-chemokine axis in the development of neuropathic pain following peripheral nerve injury. J Biol Chem 288, 24116–24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Lee S, Park DH, Kook H, Park K-G, Lee I-K, Suk K, 2015. Diverse functional roles of lipocalin-2 in the central nervous system. Neuroscience & Biobehavioral Reviews 49, 135–156. [DOI] [PubMed] [Google Scholar]
- Jin M, Kim J-H, Jang E, Lee YM, Soo Han H, Woo DK, Park DH, Kook H, Suk K, 2014. Lipocalin-2 deficiency attenuates neuroinflammation and brain injury after transient middle cerebral artery occlusion in mice. Journal of Cerebral Blood Flow & Metabolism 34, 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy MT, Ben Assayag E, Shabashov-Stone D, Liraz-Zaltsman S, Mazzitelli J, Arenas M, Abduljawad N, Kliper E, Korczyn AD, Thareja NS, Kesner EL, Zhou M, Huang S, Silva TK, Katz N, Bornstein NM, Silva AJ, Shohami E, Carmichael ST, 2019. CCR5 Is a Therapeutic Target for Recovery after Stroke and Traumatic Brain Injury. Cell 176, 1143–1157.e1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra N, Kumar V, 2004. c-Fos is a mediator of the c-myc-induced apoptotic signaling in serum-deprived hepatoma cells via the p38 mitogen-activated protein kinase pathway. J Biol Chem 279, 25313–25319. [DOI] [PubMed] [Google Scholar]
- Kang SS, Ren Y, Liu CC, Kurti A, Baker KE, Bu G, Asmann Y, Fryer JD, 2018. Lipocalin-2 protects the brain during inflammatory conditions. Mol Psychiatry 23, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA, 2001. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410, 988–994. [DOI] [PubMed] [Google Scholar]
- Kaul M, Lipton SA, 1999. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci U S A 96, 8212–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Ma Q, Medders KE, Desai MK, Lipton SA, 2007. HIV-1 coreceptors CCR5 and CXCR4 both mediate neuronal cell death but CCR5 paradoxically can also contribute to protection. Cell Death Differ 14, 296–305. [DOI] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA, 2005. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ 12, 878–892. [DOI] [PubMed] [Google Scholar]
- Kim BW, Jeong KH, Kim JH, Jin M, Kim JH, Lee MG, Choi DK, Won SY, McLean C, Jeon MT, Lee HW, Kim SR, Suk K, 2016. Pathogenic Upregulation of Glial Lipocalin-2 in the Parkinsonian Dopaminergic System. J Neurosci 36, 5608–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai R, Zhao M-L, Zhang N, Hua LL, Lee SC, 2000a. Role of MIP-1β and RANTES in HIV-1 infection of microglia: inhibition of infection and induction by IFNβ. Journal of neuroimmunology 110, 230–239. [DOI] [PubMed] [Google Scholar]
- Kitai R, Zhao ML, Zhang N, Hua LL, Lee SC, 2000b. Role of MIP-1beta and RANTES in HIV-1 infection of microglia: inhibition of infection and induction by IFNbeta. J Neuroimmunol 110, 230–239. [DOI] [PubMed] [Google Scholar]
- Kovacsics CE, Gill AJ, Ambegaokar SS, Gelman BB, Kolson DL, 2017. Degradation of heme oxygenase-1 by the immunoproteasome in astrocytes: A potential interferon-gamma-dependent mechanism contributing to HIV neuropathogenesis. Glia 65, 1264–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucker T, Toggas SM, Mucke L, Siggins GR, 1998. Transgenic mice with cerebral expression of human immunodeficiency virus type-1 coat protein gp120 show divergent changes in short- and long-term potentiation in CA1 hippocampus. Neuroscience 83, 691–700. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee J, Kim S, Park J-Y, Lee W-H, Mori K, Kim S-H, Kim IK, Suk K, 2007. A Dual Role of Lipocalin 2 in the Apoptosis and Deramification of Activated Microglia. The Journal of Immunology 179, 3231–3241. [DOI] [PubMed] [Google Scholar]
- Lee S, Park JY, Lee WH, Kim H, Park HC, Mori K, Suk K, 2009. Lipocalin-2 is an autocrine mediator of reactive astrocytosis. J Neurosci 29, 234–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letendre SL, Lanier ER, McCutchan JA, 1999. Cerebrospinal fluid beta chemokine concentrations in neurocognitively impaired individuals infected with human immunodeficiency virus type 1. J Infect Dis 180, 310–319. [DOI] [PubMed] [Google Scholar]
- Letendre SL, Zheng JC, Kaul M, Yiannoutsos CT, Ellis RJ, Taylor MJ, Marquie-Beck J, Navia B, Consortium HIVN, 2011. Chemokines in cerebrospinal fluid correlate with cerebral metabolite patterns in HIV-infected individuals. J Neurovirol 17, 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Soontornniyomkij V, Achim CL, Masliah E, Gelman BB, Sinsheimer JS, Singer EJ, Moore DJ, 2016. Multilevel analysis of neuropathogenesis of neurocognitive impairment in HIV. J Neurovirol 22, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieben CK, Steinbusch HW, Blokland A, 2006. 5,7-DHT lesion of the dorsal raphe nuclei impairs object recognition but not affective behavior and corticosterone response to stressor in the rat. Behav Brain Res 168, 197–207. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Maung R, Hoefer MM, Sanchez AB, Sejbuk NE, Medders KE, Desai MK, Catalan IC, Dowling CC, de Rozieres CM, Garden GA, Russo R, Roberts AJ, Williams R, Kaul M, 2014. CCR5 knockout prevents neuronal injury and behavioral impairment induced in a transgenic mouse model by a CXCR4-using HIV-1 glycoprotein 120. J Immunol 193, 1895–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maung R, Medders KE, Sejbuk NE, Desai MK, Russo R, Kaul M, 2012. Genetic knockouts suggest a critical role for HIV co-receptors in models of HIV gp120-induced brain injury. J Neuroimmune Pharmacol 7, 306–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JC, Steiner J, Sacktor N, Nath A, 2010. Human immunodeficiency virus-associated neurocognitive disorders: Mind the gap. Ann Neurol 67, 699–714. [DOI] [PubMed] [Google Scholar]
- McLaurin KA, Booze RM, Mactutus CF, Fairchild AJ, 2017. Sex Matters: Robust Sex Differences in Signal Detection in the HIV-1 Transgenic Rat. Front Behav Neurosci 11, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medders KE, Sejbuk NE, Maung R, Desai MK, Kaul M, 2010. Activation of p38 MAPK is required in monocytic and neuronal cells for HIV glycoprotein 120-induced neurotoxicity. J Immunol 185, 4883–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha M, Skrzypiec AE, Schiavon E, Attwood BK, Kucerova E, Pawlak R, 2011. Lipocalin-2 controls neuronal excitability and anxiety by regulating dendritic spine formation and maturation. Proc Natl Acad Sci U S A 108, 18436–18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby DG, Tremblay A, Lecluse V, Lehmann H, 2005. Hippocampal damage and anterograde object-recognition in rats after long retention intervals. Hippocampus 15, 1050–1056. [DOI] [PubMed] [Google Scholar]
- Nguyen TP, Soukup VM, Gelman BB, 2010. Persistent hijacking of brain proteasomes in HIV-associated dementia. Am J Pathol 176, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R, Zhao Y, Libbey M, Westphal H, Crawley JN, 2001. Learning impairments and motor dysfunctions in adult Lhx5-deficient mice displaying hippocampal disorganization. Physiol Behav 73, 781–792. [DOI] [PubMed] [Google Scholar]
- Putatunda R, Zhang Y, Li F, Fagan PR, Zhao H, Ramirez SH, Pratico D, Barbe MF, Hu W, 2019. Sex-specific neurogenic deficits and neurocognitive disorders in middle-aged HIV-1 Tg26 transgenic mice. Brain Behav Immun 80, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AB, Kaul M, 2017. Neuronal Stress and Injury Caused by HIV-1, cART and Drug Abuse: Converging Contributions to HAND. Brain Sci 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AB, Medders KE, Maung R, Sanchez-Pavon P, Ojeda-Juarez D, Kaul M, 2016a. CXCL12-induced neurotoxicity critically depends on NMDA receptor-gated and L-type Ca(2+) channels upstream of p38 MAPK. J Neuroinflammation 13, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AB, Varano GP, de Rozieres CM, Maung R, Catalan IC, Dowling CC, Sejbuk NE, Hoefer MM, Kaul M, 2016b. Antiretrovirals, Methamphetamine, and HIV-1 Envelope Protein gp120 Compromise Neuronal Energy Homeostasis in Association with Various Degrees of Synaptic and Neuritic Damage. Antimicrob Agents Chemother 60, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders VJ, Pittman CA, White MG, Wang G, Wiley CA, Achim CL, 1998. Chemokines and receptors in HIV encephalitis. AIDS 12, 1021–1026. [PubMed] [Google Scholar]
- Saylor D, Dickens AM, Sacktor N, Haughey N, Slusher B, Pletnikov M, Mankowski JL, Brown A, Volsky DJ, McArthur JC, 2016. HIV-associated neurocognitive disorder - pathogenesis and prospects for treatment. Nat Rev Neurol 12, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman NC, Owens MA, Adnan R, 2004. A simple more general boxplot method for identifying outliers. Computational Statistics & Data Analysis 47, 165–174. [Google Scholar]
- Sickinger S, Maier H, Konig S, Vallant N, Kofler M, Schumpp P, Schwelberger H, Hermann M, Obrist P, Schneeberger S, Margreiter R, Troppmair J, Pratschke J, Aigner F, 2013. Lipocalin-2 as mediator of chemokine expression and granulocyte infiltration during ischemia and reperfusion. Transpl Int 26, 761–769. [DOI] [PubMed] [Google Scholar]
- Stevenson M, 2003. HIV-1 pathogenesis. Nat Med 9, 853–860. [DOI] [PubMed] [Google Scholar]
- Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb MJ, 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. The EMBO Journal 15, 4629–4642. [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Le W, 2016. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol Neurobiol 53, 1181–1194. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Moser R, Regulier E, Breuillaud L, Dixon M, Beesen AA, Elliston L, Silva Santos Mde F, Kim J, Jones L, Goldstein DR, Ferrante RJ, Luthi-Carter R, 2013. MAP kinase phosphatase 1 (MKP-1/DUSP1) is neuroprotective in Huntington’s disease via additive effects of JNK and p38 inhibition. J Neurosci 33, 2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaney VE, O’Neill AM, Hoefer MM, Maung R, Sanchez AB, Kaul M, 2017. IFNβ Protects Neurons from Damage in a Murine Model of HIV-1 Associated Brain Injury. Sci Rep 7, 46514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaney VE, Sanchez AB, Fields JA, Minassian A, Young JW, Maung R, Kaul M, 2018. Transgenic mice expressing HIV-1 envelope protein gp120 in the brain as an animal model in neuroAIDS research. J Neurovirol 24, 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toggas SM, Masliah E, Rockenstein EM, Rall GF, Abraham CR, Mucke L, 1994. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature 367, 188–193. [DOI] [PubMed] [Google Scholar]
- Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O’Sullivan MP, Walter MJ, Schwendener RA, Cook DN, Danoff TM, Holtzman MJ, 2005. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nat Med 11, 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Harrington L, Trebicka E, Shi HN, Kagan JC, Hong CC, Lin HY, Babitt JL, Cherayil BJ, 2009. Selective modulation of TLR4-activated inflammatory responses by altered iron homeostasis in mice. J Clin Invest 119, 3322–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Ipser JC, Stein DJ, Joska JA, Naude PJW, 2019. The Association of Immune Markers with Cognitive Performance in South African HIV-Positive Patients. J Neuroimmune Pharmacol 14, 679–687. [DOI] [PubMed] [Google Scholar]
- Winters BD, Forwood SE, Cowell RA, Saksida LM, Bussey TJ, 2004. Double dissociation between the effects of peri-postrhinal cortex and hippocampal lesions on tests of object recognition and spatial memory: heterogeneity of function within the temporal lobe. J Neurosci 24, 5901–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C, Wang X, Cheng C, Montaner J, Mandeville E, Leung W, van Leyen K, Lok J, Wang X, Lo EH, 2014. Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke 45, 2085–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Goetz D, Li JY, Wang W, Mori K, Setlik D, Du T, Erdjument-Bromage H, Tempst P, Strong R, Barasch J, 2002. An iron delivery pathway mediated by a lipocalin. Mol Cell 10, 1045–1056. [DOI] [PubMed] [Google Scholar]
- Yu Y, Richardson DR, 2011. Cellular iron depletion stimulates the JNK and p38 MAPK signaling transduction pathways, dissociation of ASK1-thioredoxin, and activation of ASK1. J Biol Chem 286, 15413–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Greenhill S, Huang S, Silva TK, Sano Y, Wu S, Cai Y, Nagaoka Y, Sehgal M, Cai DJ, Lee YS, Fox K, Silva AJ, 2016. CCR5 is a suppressor for cortical plasticity and hippocampal learning and memory. Elife 5, e20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Demographic information for human brain samples.
Table S2 Primer sequences.
Table S3 Pearson correlations between LCN2 mRNA and cohort characteristics.
Detailed descriptions of experimental and analytical procedures for behavioral testing, immunohistology, quantitative fluorescence and deconvolution microscopy, and bioinformatics.
Table S4 Neurotransmission-related genes in mouse cerebral cortex, differential expression in HIVgp120tg with and without LCN2 and/or CCR5 knockout compared to wild-type controls
Table S5 Neurotransmission-related genes in mouse hippocampus, differential expression in HIVgp120tg with and without LCN2 and/or CCR5 knockout compared to wild-type controls
Fig. S1. LCN2 levels are increased in patients with HIV brain pathology.
Fig. S2. HIVgp120tg animals and/or LCN2 deficiency do not result in anxiety-like behavior or impaired vision.
Fig. S3. Combined analysis of immunofluorescence and deconvolution microscopy of HIVgp120tg, LCN2KO-gp120tg and CCR5KO-LCN2KO-gp120tg brains and the respective non-tg, gp120- controls.
Fig. S4. Lcn2 mRNA is expressed in astrocytes.
Fig. S5. Expression of HIV gp120 transgene and its receptor are not reduced in LCN2KO gp120 nor DKO animals.
Fig. S6. Expression of pro- and anti-inflammatory microglial genes.
Fig. S7. Differential gene expression signatures in genotypes associated with neurodegeneration and protection are sex-dependent in hippocampus.
Fig. S8. Top functional gene networks predicted based on gene expression changes.