Abstract
The emergence of CRISPR-Cas9 as a powerful genome editing tool has led to several studies exploring its potential to treat neurological disorders. Cas9 and its sgRNA can be readily engineered to target any gene and can be multiplexed to target several genes at once. Furthermore, the use of adeno-associated virus (AAV) to deliver with Cas9 and its sgRNA is a promising therapeutic combination with strong potential to reach the clinic. Here we discuss how Cas9 editing has been utilized for gene insertion, knockout, and deletion in vivo for applications in the central nervous system (CNS). Furthermore, we highlight major challenges that remain for AAV-Cas9-sgRNA clinical translation.
Introduction
Neurological disorders are a set of diseases that affect the brain, spinal cord, retina, and peripheral nervous system. In the United States alone, nearly 100 million people are affected by a neurological disease, ranging from epilepsy to schizophrenia to stroke, and their prevalence increases with the aging population.1–3 Patient symptoms can vary widely depending on the neurological disease. For example, Alzheimer’s disease causes memory loss and disrupts mental function in patients, while Parkinson’s disease and amyotrophic lateral sclerosis (ALS) impair motor function.4 Alzheimer’s, Parkinson’s, and ALS in particular are part of a sub-class of neurological disorders termed neurodegenerative diseases that lead to atrophy and eventual loss of neurons. In general, the neurodegenerative disease prognosis is poor, and current therapies target the symptoms of the disease rather than slowing or halting disease progression. To improve patient outcomes, therapies targeting the underlying cause of the disease are needed.
Genome editing is a promising approach that can address the root cause of a disorder. With the advent of site-specific nuclease technologies, precise editing of a patient’s genome is possible, allowing for targeted genetic mutations to mitigate disease. Site-specific nucleases induce a double-stranded DNA break (or in some variations a nick) that is resolved by one of two primary repair mechanisms: non-homologous end joining (NHEJ) or homology-directed repair (HDR). With NHEJ, small nucleotide insertions or deletions (referred to as “indels”) can be introduced, which may disrupt a protein’s reading frame and thereby knock out a gene, whereas HDR introduces exogenous sequence at the target site. Zinc finger nucleases (ZFNs)- heterodimers of zinc finger repeats fused to a nuclease domain5,6 - were the first breakthrough gene editing tools and are being harnessed in ongoing clinical trials for hemophilia B, HIV, and other targets.7–15 Transcription activator-like effector nucleases (TALENs), another class of engineered site-specific nucleases containing 33–35 repeat domains that each recognize a single base pair, offer more modular design than ZFNs. However, TALENs are much larger in size than ZFNs and are highly repetitive, rendering delivery with viral vectors challenging.16–19
The recent emergence of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 has largely displaced ZFN and TALEN as editing tools in academic research.20 Compared to both initial technologies, CRISPR-Cas9 is readily targeted and has multiplexing capabilities, allowing for simultaneous editing at multiple genomic locations.21 Engineered CRISPR-Cas9 is a two-component system comprised of an endonuclease Cas9 protein and a short RNA scaffold termed a single guide RNA (sgRNA). The sgRNA contains a modular 20-nucleotide targeting sequence that directs the Cas protein to a genomic target site, which must be adjacent to a specific protospacer adjacent motif (PAM) that varies depending on the Cas9 variant used. Early landmark studies demonstrated the efficacy of Cas9 editing in human cells in vitro,21–24 and ongoing studies are demonstrating efficacy in vivo. Furthermore, Cas9 has been engineered and fused to other proteins in order to enable base editing,25 transcriptional interference or repression,26 and transcriptional activation.27
While CRISPR-Cas9 offers broad potential for therapeutic genome editing, it must be delivered to the nuclei of target cells. Among several delivery methods that have been recently explored, vectors based on adeno-associated virus (AAV) have been extensively researched for the nervous system. AAV has already demonstrated broad clinical potential and became the basis for an FDA approved gene therapy in 2017 for the treatment of the retinal disorder Leber’s congenital amaurosis type 2 (LCA2).28–34 AAV has several desirable features, including the lack of pathogenicity of the parent virus, its generally low immunogenicity, and moderate transduction efficiency on a broad range of cell types including non-dividing cells. Additionally, recombinant AAV can deliver up to ~5 kb of DNA that becomes predominantly maintained episomally after delivery, thus reducing genotoxicity risks. The biodistribution and cellular tropism of AAV is conferred by its capsid. For example, AAV serotype 9 (AAV9) is often harnessed for central nervous system (CNS) applications because of its capacity cross the blood brain barrier (BBB) and transduce neurons and astrocytes in the neonatal CNS (or predominantly astrocytes in the adult CNS).
In addition to natural serotypes, engineered AAVs can offer the potential for highly efficient and targeted gene delivery,35 and directed evolution of AAV has been increasingly implemented to improve AAV tropism. In the retina, the AAV variant Shh10 transduces Müller glia with high specificity, while 7m8 transduces all retinal cells, including photoreceptor neurons which are challenging to target from intravitreal administration.36,37 Several variants have been optimized for high transduction in other parts of the CNS, of which Sun et al. offer an extensive description.38–44
This review discusses recent studies using AAV and CRISPR-Cas9 for in vivo editing to treat neurodegenerative disorders. CRISPR-Cas9 genome editing techniques are summarized in Figure 1, and recent proof of concept experiments for AAV-Cas9-sgRNA including major hurdles to clinical translation are discussed.
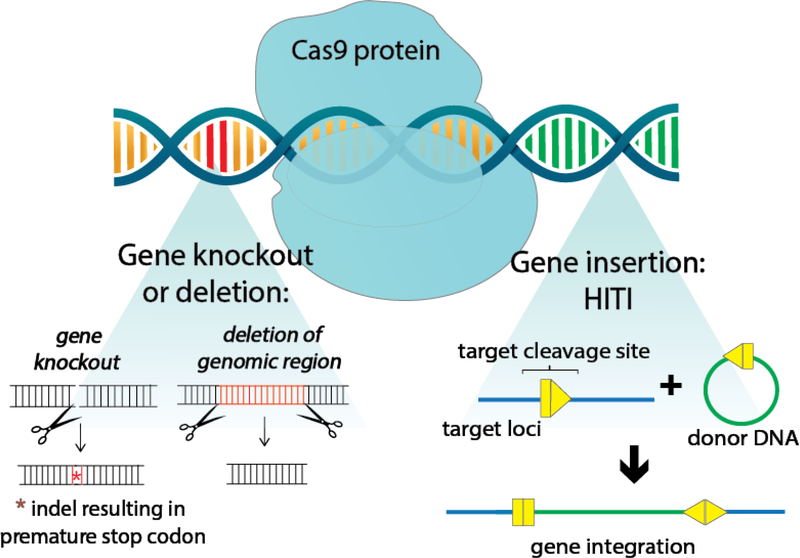
Figure 1: CRISPR-Cas9 genome editing.
After Cas9 introduces a break at the target locus, insertions, NHEJ-mediated knock out, or DNA deletion can be affected. In gene knockout, an indel at the target site disrupts gene expression. In addition, deletion of genomic regions is possible by using a pair of sgRNA to induce a double stranded break at two locations on the same gene, excising out a region of DNA. Finally, in homology independent targeted integration (HITI), a donor template is co-delivered to insert DNA at the target site. The introduction of exogenous gene sequences is also possible by homology directed repair (not shown), but its use is limited in post-mitotic cells since this repair mechanism is highly suppressed in G1 phase.45
In vivo AAV-mediated delivery of Cas9 and its sgRNA in the Central Nervous System
Gene Disruption by Indel Formation
AAV delivery of Cas9 and its sgRNA has potential as a powerful therapeutic modality to treat neurodegenerative diseases. The first proof-of-principle experiments demonstrating in vivo Cas9 editing in the brain targeted the mecp2 gene, which is broadly expressed in neurons and plays a role in learning.46 The Cas9 protein and the mecp2-targeted sgRNA were delivered using two AAV1 vectors, which were stereotaxically injected into the murine brain. In one vector, the ~4.2 kb S. pyogenes Cas9 (SpCas9) was packaged with a minimal promoter, a truncated version of the mecp2 promoter, and polyadenylation sequence to fit the size limitations of AAV. In the second vector, the sgRNA was delivered along with a neuronal promoter driving GFP expression to mark and enable sorting of cells transduced with the sgRNA. Based on tissue staining, approximately 80% of infected cells were co-transduced with Cas9 and sgRNA. Indels determined by next-generation sequencing occurred in approximately 68% of transduced cells, whereas off-target indel frequencies at top predicted off-target sites ranged from 0–1.6%. Behavioral changes were characterized by contextual fear-conditioning paradigm as well as V1 region response to visual stimuli of specific orientations. Additionally, sgRNA’s against the dnmt1, dmnt3a, and dmnt3b genes were co-delivered within the dentate gyrus to achieve multiplexing, and approximately 35% transduced cells were edited at these three loci. Another recent study used an engineered AAV vector to deliver Cas9 and its sgRNA to projection neurons and thereby disrupt tdTomato expression in vivo.39 Approximately 88% of Cas9-expressing cells demonstrated suppressed tdTomato expression. In another study, Cas9 editing enabled in vivo disruption of YFP expression in the retina after AAV delivery.47
Deletion of Genomic Regions
Another powerful gene editing technique is the deletion of large genomic regions to alleviate disease phenotype (Figure 1). As a proof-of-principle, a pair of sgRNA targeting both ends of a target region – the mir137 allele containing single nucleotide polymorphisms (SNPs) correlated with schizophrenia - were delivered by AAV2g9 in a Cas9-expressing transgenic mouse. 41. Using droplet digital PCR, targeted deletion was found in ~5% of the target sites in the brain. Future work may involve co-delivery of both the sgRNA’s and Cas9. In another example, a mutated intronic region was targeted to ablate a cryptic splice donor site that leads to a premature stop codon in Leber congenital amaurosis 10 (LCA10).48 Two AAV5 vectors were used to deliver the SpCas9 and the sgRNA pair. Next generation sequencing analysis of four retinas revealed a range of genomic deletion rates from 7.5–25%, with lower editing rates attributed to poor transduction. These two examples highlight genomic excisions are possible by designing a sgRNA pair against both ends of a target region.
Allele Specific Editing
Allele specific Cas9 editing would enable treatment of autosomal dominant neurodegenerative diseases by specifically targeting the mutated allele while maintaining expression from the wildtype allele. Most allele specificity efforts involve designing the sgRNA and PAM sequence to overlap with the SNPs (i.e. genetic variations),49–52 such that a mismatch relative to the wildtype allele, reduces Cas9 cleavage of the wildtype sequence. Allele specific editing was achieved in the brain of a transgenic mouse line expressing a mutant human Huntington (HTT) gene by including a SNP in the PAM sequence.49 Following AAV1 vector delivery of Cas9 with a sgRNA targeting mutant HTT, HTT mRNA levels at injected sites were reduced approximately 50% relative to uninjected brain regions, though it remains unknown if this approach can lead to improvements in disease symptoms.
One challenge for this strategy is the relative rarity of autosomal dominant diseases where unique SNP containing PAM sites are present in the disease-causing allele. To extend the applicability of allele specific editing, other studies have designed the unique SNP into the sgRNA sequence and proximal to the PAM. Allele specific editing was thereby achieved in the retina of a mouse model for an autosomal dominant form of retinitis pigmentosa.52 Two AAV PHP.B vectors, a variant with greater transduction efficiencies in the CNS than AAV9, were injected intravitreally. One vector encoded SpCas9 and the second vector contained the sgRNA along with a rhodopsin promoter driving GFP expression to enrich for sgRNA transduced photoreceptors. Within cells sorted for high expression of GFP, the indel rate was ~18%. In another study, single nucleotide discrimination was achieved against the rhodopsin allele in mice using engineered SpCas9 variants evolved for altered PAM specificities and truncated sgRNAs.51
Homology-Independent Targeted Integration
Introduction of new sequence, such as deleted gene regions, to alleviate disease phenotype is another strategy. Until recently, HDR involving donor sequence flanked by homology regions to the target site of interest was the primary strategy for introducing exogenous sequence into mammalian cells. HDR is most active during DNA replication, however, rendering this process problematic for non-dividing cells. As an alternative, Suzuki et al. developed a homology-independent targeted integration (HITI) method that relies on the NHEJ repair pathway.53 HITI uses Cas9 to target an integration site and induce a double-stranded break, and a DNA template co-delivered with the Cas9 system is then integrated into the genome. Cas9 cutting continues until the transgene is inserted in the forward orientation or indels are produced, a process that also removes the sgRNA recognition sequences (Figure 1). As a proof-of-principle, gene replacement was shown in a rat model for retinitis pigmentosa where a portion of the mertk gene is deleted. Replacement of this deleted region was mediated via subretinal injection of one AAV8 vector encoding SpCas9 and a sgRNA, and a second vector containing the donor template. Improved retinal morphology and ERG response were verified by immunohistochemistry and electroretinography.
Examples of In Vivo AAV-Cas9-sgRNA Improving Phenotypic Outcome in Neurodegenerative Models
The first example illustrating the therapeutic benefit of AAV delivered SpCas9 and sgRNA in a neurodegenerative disease model was for retinitis pigmentosa where disease progression was substantially delayed via disruption of the rod photoreceptor specific nrl gene, which encodes a transcription factor critical to photoreceptor development and function.54 Retinitis pigmentosa is a set of monogenic disorders involving initial loss of rod photoreceptors, which in turn leads to subsequent cone photoreceptor degeneration. Disruption of the nrl gene partially reprogrammed cells to a cone-like photoreceptor, thereby improving survival of cells with rod-specific gene mutations and preserving the function of surviving cone photoreceptors. Of importance, treatment was most beneficial when delivered before the onset of degeneration. In another study relevant to wet age-related macular degeneration, AAV delivered SpCas9 and sgRNA mediated disruption of the vascular endothelial growth factor receptor 2 (VEGFR2) expression was shown to abrogate retinal angiogenesis.55 Furthermore, in a study targeting the brain, AAV-SpCas9 and sgRNA delivery and editing alleviated motor deficits in a mouse model of Huntington’s disease.56
AAV titers drop when the total genome size exceeds ~5.0 kb.57 Because of these size limitations, packaging SpCas9 into AAV requires the use of a minimal promoter and/or minimal polyadenylation sequence, which are not as effective as their full length counterparts. Additionally, the sgRNA must be delivered using a second vector, such that dual vector delivery to every cell is necessary for editing to occur. Fortunately, smaller Cas9 variants have since emerged and include S. aureus Cas9 (SaCas9), approximately ~1 kb shorter than SpCas9, making delivery of the Cas9 and sgRNA in a single AAV vector possible.58,59 The first demonstration of AAV-SaCas9 to treat in vivo neurodegeneration used a single vector to target the SOD1 gene in a model of ALS, resulting in improved motor function and survival.60 Interestingly, while motor function and survival improved, a complete rescue was not observed, an outcome that was attributed to insufficient gene editing in astrocytes and microglia and highlighted the need for improved AAV vectors for translation to humans.
Challenges to Address before Proceeding to the Clinic
AAV gene delivery has succeeded in human trials for LCA2, hemophilia B, spinal muscular atrophy, and other disease targets,28–33 such that AAV-mediated genome editing in murine models of retinitis pigmentosa,54 ALS,60 and other neurological targets has future clinical potential. While ongoing work is establishing Cas9 efficacy in other neurodegenerative disease models, there are three critical areas that should be further explored and optimized: targeted delivery and potency, minimizing non-targeted delivery and off-target editing, and immunogenicity (Figure 2).
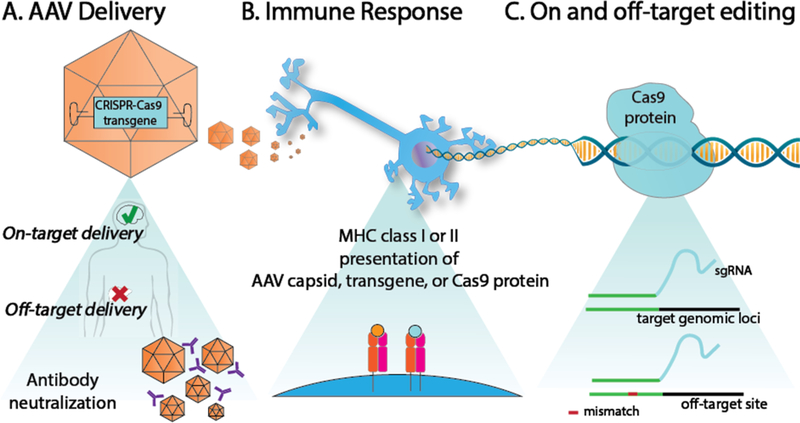
Figure 2: Challenges with in-vivo translation of AAV-Cas9-sgRNA.
A) Off-target transduction and antibody neutralization of the AAV capsid limit targeted tissue delivery. B) After AAV transduces a cell, parts of the capsid protein or vector-encoded protein, such as Cas9, can be presented on major histocompatability complex (MHC) cell surface protein to elicit an immune response. C) Efficient Cas9 editing at target loci is determined by the sgRNA design. Genomic regions with partial homology to the sgRNA can be prone to Cas9 editing.
Efficient targeted delivery and subsequent on-target editing of AAV-Cas9-sgRNA is necessary for clinical success. AAV tropism is conferred by the capsid and affected by the route of administration. To target the CNS, AAV can be injected locally, systemically, or intrathecally. Direct AAV injection enables high dosage, localized delivery but can be an invasive approach, particularly if more than one injection is needed to achieve the desired spread. Systemic administration is less invasive but requires an AAV variant capable of crossing the BBB and significantly exposes the vector to non-target tissues and to the immune system. Intrathecal injections deposit AAV into the cerebrospinal fluid, thereby bypassing the need to pass the BBB, and this route of administration is being translated to the clinic.61,62 To improve delivery efficiency and specificity, novel AAV vectors are being engineered in mouse and more importantly in non-human primates, as tropism will vary between species.63 Table 1 summarizes several AAV vectors tested in non-human primates, along with the route of administration (Table 1).
Table 1:
Several natural and engineered AAV vectors tested in non-human primates for applications in the CNS
| Variant | Parental serotype | Design | Tropism | Administration | Study |
|---|---|---|---|---|---|
| 7m8 | AAV2 | 7-mer peptide insertion at AA588 (LGETTRP) | Pan-retinal | subretinal and intravitreal | 37 |
| AAV1 and AAV2 | - | - | broad transduction of medium spiny neurons and cortico-striatal neurons | direct injection in caudate nucleus and putanem | 64 |
| AAV9 | - | - | broad transduction, including retrograde and anterograde transport under local administration; <3% transduction of neurons and astrocytes by intravenous injection; ~2% transduction of brain and spinal cord after intrathecal injection | direct injection into parenchyma; intravenous; intrathecal | 65,66 |
| PHP.B | AAV9 | 7-mer peptide insertion at AA588 (TLAVPFK) | <3% transduction of neurons and astrocytes | intravenous | 66 |
| AAV5 | - | - | neurons and astrocytes | direct injection into parenchyma | 67 |
Non-target transduction of AAV, particularly vectors based on natural serotypes with non-specific tropism, poses challenges. Higher doses can be required to compensate for non-target biodistribution, which can both lead to risks of genome editing in non-target tissues and raise the risk of immune responses to AAV capsids or transgene products, as discussed in more detail below. Tissue specific promoters can limit off-target transgene expression but are limited by size constraints of AAV and typically have lower expression levels than strong constitutively active promoters.
Following AAV delivery of Cas9 and its sgRNA, the vector genome becomes diluted in mitotic cells but persists as a stable episome in non-dividing cells.68 Since the majority of cells within the CNS are post-mitotic, the resulting indefinite persistence of Cas9 expression can pose a major issue, especially considering off-target indel formation is possible at sites with partial matches to the sgRNA.69 Additionally, more work is needed to characterize the potential undesired effects of Cas9 editing. Cas9 has induced large genomic deletions and rearrangements at two different targeted genomic loci in vitro.77 However more studies are needed to determine whether this observation occurs for other genomic target sites, and when doses of Cas9 are lowered to the levels mediated by AAV delivery.
Off-target editing for particular sgRNA(s) can be predicted using computational algorithms and measured with analytical tools such as next generation sequencing, aided for example by techniques such as Guide-seq.70,71 Several approaches have been pursued to minimize off-target editing. One is to modify Cas9 into a nickase that induces a single-stranded break,72,73 and co-delivery of two sgRNAs that are complementary to opposite strands of the target site and offset relative to on another stimulates HDR in the region flanked by the nick. Furthermore, the use of two offset nicks on complementary strands reduces the probability of a double-stranded break and subsequent indel formation at an off-target site. Another approach to reducing off-target editing is to shorten the sgRNA.74 A truncated sgRNA is believed to be more sensitive to mismatches between the sgRNA and DNA. As a result, the binding energy of the sgRNA to DNA is lowered, and a perfectly matched sgRNA and DNA sequence is more strongly favored over off-target sites containing mismatches relative to the shortened sgRNA. Cas9 nickases and truncated sgRNA, however, decrease Cas9’s editing efficiency. A third approach is a self-excising system, where DNA sequence sites matching the genomic target site is incorporated into the Cas9 transgene, or a second sgRNA targeting Cas9 is co-delivered with a sgRNA against a target genomic loci. 48,75,76 After delivery, the Cas9 sequence is ablated at the same time as the target genomic site is edited. Self-inactivation of the Cas9 prevents persistent Cas9 protein production and can potentially reduce off-target editing or an immune response against the bacterial protein. Future work will explore the tradeoff between on-site editing efficacy and off-target cleavage reduction.
A third major obstacle is potential immune responses against the AAV capsid or delivered Cas9. Immune responses to AAV have been discussed elsewhere.80 An immune response against the Cas9 itself is also possible,81,82 a concern in general for non-self proteins in both small animal models and in non-human primates.82 A screen of human subjects also showed that many individuals have pre-existing antibodies against Cas9,83 which could render delivery of recombinant Cas9 protein challenging, but may not especially impact Cas9 gene delivery since de novo immune responses against Cas9 protein are likely regardless. At any rate, methods to induce tolerance or to limit Cas9 expression may be needed to avoid these issues.
Due to these three primary obstacles, delivery still remains a significant challenge in the gene editing field. Alternative delivery approaches to AAV have been studied to potentially avoid concerns with off-target editing and immune responses against persistent Cas9 expression. For example, delivery of Cas9 protein and sgRNA as a ribonucleoprotein (RNP) complex could facilitate delivery and transient expression of Cas9 in vivo.84 RNPs can also be used with AAV genomic donor DNA for applications such as targeted gene knock-in by HDR.85 Other non-viral, synthetic delivery methods have been engineered for transient delivery of Cas9 as well.86,87 However, non-viral systems are not as efficient at delivery and cell transduction. As an alternative, engineered self-excising AAV-Cas9-sgRNA systems have the potential advantage of combining efficient delivery with transient Cas9 activity.
Finally, in addition to Cas9 nucleases, base editing Cas9 systems that mediate single base substitutions are an intriguing alternative approach that does not rely on induction of double stranded breaks in order to modify the genome, though they are challenged by the finding that hundreds of distinct mutations can result in a recessive mutation, or in some cases even a dominant allele (e.g. SOD1 for ALS). In addition, trans-splicing and other analogous approaches must be improved for delivery of Cas9 base editing systems, which are above the carrying capacity of AAV.88
Conclusion
AAV delivery of Cas9 and its sgRNA is a powerful combination for gene editing based therapies for neurodegenerative diseases. While extensive characterization of Cas9 editing and AAV delivery in vivo are needed for clinical translation, numerous key studies already highlight the promise of this approach. In particular proof-of-principle experiments have shown that Cas9-based gene insertion, knockout, and deletion of genomic regions is possible in vivo in the CNS of murine models. Furthermore, continued research and engineering of Cas9 and AAV will broaden the applicability to more disease targets and increase our understanding of the long term safety of this system.
Acknowledgements
CMF is supported by a National Science Foundation Graduate Research Fellowship, and DS is supported by NIH R01EY022975 and R21EB021572. We also thank Thomas Gaj for a critical read of the review, Rajan Kumar for edits, and Olivia Scheideler for feedback on illustrations.
Biography
Literature annotations
Komor et al, base editing – This study introduces a Cas9-cytidine deaminase fusion that retains the targeting capabilities of CRISPR-Cas9 and enables a C → T or G → A substitution.
Qi et al, interference – A nucleolytically inactive Cas9 is directed to a target loci and represses gene expression by interfering with transcriptional elongation, RNA polymerase binding, or transcription factor binding
Konermann et al, Cas9 activation – Cas9 is modified to be nucleolytically inactive (dCas9). Transactivation proteins are fused to dCas9 and the sgRNA backbone is modified to recruit additional transactivation proteins, enabling gene activation
Swiech et al, POC – This is the first studying showing in vivo Cas9 editing in the CNS. This study illustrates the use of Cas9 editing for gene knockdown and its multiplexing capabilities.
Suzuki et al, hiti paper – this study introduces a homology independent integration technique that enables transgene integration in post-mitotic cells
Gaj et al, ALS paper – This is the first study to demonstrate phenotypic changes in a neurodegenerative disease model using a single AAV vector to deliver SaCas9 and the corresponding sgRNA
Tsai, S et al, guideseq – In this study, a useful tool for determining Cas9 off target editing rates and location is introduced. This tool is used to study off target effects at the genome-wide level.
Samaranch L., et al, non-self proteins – Here, AAV mediated delivery of non-self proteins are studied in the CNS of non-human primates. This study highlights the need to consider the potential immune responses that are triggered by non-self proteins delivered by AAV, even in immune privileged regions.
Footnotes
Conflict of Interest Statement
DS is an inventor on patents related to AAV vector engineering and co-founder of a company focused on AAV gene therapy.
References
- 1.Gitler AD, Dhillon P & Shorter J Neurodegenerative disease: models, mechanisms, and a new hope. Dis. Model. Mech. 10, 499–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pritchard C, Rosenorn-Lanng E, Silk A & Hansen L Controlled population-based comparative study of USA and international adult [55–74] neurological deaths 1989–2014. Acta Neurol. Scand. 136, 698–707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooch CL, Pracht E & Borenstein AR The burden of neurological disease in the United States: A summary report and call to action. Ann. Neurol. 81, 479–484 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Yacoubian TA Neurodegenerative Disorders : Why Do We Need New Therapies? Drug Discovery Approaches for the Treatment of Neurodegenerative Disorders (Elsevier Inc., 2017). doi: 10.1016/B978-0-12-802810-0.00001-5 [DOI] [Google Scholar]
- 5.Gersbach CA, Gaj T & Barbas CF Synthetic zinc finger proteins: The advent of targeted gene regulation and genome modification technologies. Acc. Chem. Res 47, 2309–2318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urnov FD, Rebar EJ, Holmes MC, Zhang HS & Gregory PD Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet 11, 636–646 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Study of Molecular-targeted Therapy Using Zinc Finger Nuclease in Cervical Precancerous Lesions. at <https://clinicaltrials.gov/show/NCT02800369>
- 8.A Phase I Study of T-Cells Genetically Modified at the CCR5 Gene by Zinc Finger Nucleases SB-728mR in HIV-Infected Patients. at <https://clinicaltrials.gov/show/NCT02388594>
- 9.Ascending Dose Study of Genome Editing by the Zinc Finger Nuclease (ZFN) Therapeutic SB-318 in Subjects With MPS I. at <https://clinicaltrials.gov/show/NCT02702115>
- 10.Study of Autologous T-cells Genetically Modified at the CCR5 Gene by Zinc Finger Nucleases in HIV-Infected Subjects. at <https://clinicaltrials.gov/show/NCT01252641%0A>
- 11.Phase 1 Dose Escalation Study of Autologous T-cells Genetically Modified at the CCR5 Gene by Zinc Finger Nucleases in HIV-Infected Patients. at <https://clinicaltrials.gov/show/NCT01044654%0A>
- 12.Safety Study of Zinc Finger Nuclease CCR5-modified Hematopoietic Stem/Progenitor Cells in HIV-1 Infected Patients. at <https://clinicaltrials.gov/show/NCT02500849>
- 13.Ascending Dose Study of Genome Editing by the Zinc Finger Nuclease (ZFN) Therapeutic SB-913 in Subjects With MPS II. at <https://clinicaltrials.gov/show/NCT03041324>
- 14.Repeat Doses of SB-728mR-T After Cyclophosphamide Conditioning in HIV-Infected Subjects on HAART. at <https://clinicaltrials.gov/show/NCT02225665%0A>
- 15.A Study to Assess the Safety, Tolerability, and Efficacy of ST-400 for Treatment of Transfusion-Dependent Beta-thalassemia (TDT). at <https://clinicaltrials.gov/show/NCT03432364%0A>
- 16.Miller JC et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol 29, 143–8 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Christian M et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mussolino C et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol 29, 149–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M et al. RNA-programmed genome editing in human cells. Elife 2013, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho SW, Kim S, Kim JM & Kim J-S Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2–11 (2014). doi: 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathwani AC et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nathwani AC et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med 365, 2357–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett J et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci. Transl. Med. 4, 120ra15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bainbridge JWB et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med 358, 2231–9 (2008). [DOI] [PubMed] [Google Scholar]
- 32.MacLaren RE et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet (London, England) 383, 1129–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar SR, Markusic DM, Biswas M, High KA & Herzog RW Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev 3, 16034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smalley E First AAV gene therapy poised for landmark approval. Nat. Biotechnol. 35, 998–999 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Kotterman MA & Schaffer DV Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet 15, 445–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klimczak RR, Koerber JT, Dalkara D, Flannery JG & Schaffer DV A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS One 4, e7467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalkara D et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 5, 189ra76 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol 34, 204–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tervo DGR et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojala DS et al. In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the SVZ. Mol. Ther 26, 304–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murlidharan G et al. CNS-restricted Transduction and CRISPR/Cas9-mediated Gene Deletion with an Engineered AAV Vector. Mol. Ther. Nucleic Acids 5, e338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iida A, Takino N, Miyauchi H, Shimazaki K & Muramatsu S Systemic delivery of tyrosine-mutant AAV vectors results in robust transduction of neurons in adult mice. Biomed Res. Int 2013, 974819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhury SR et al. Widespread Central Nervous System Gene Transfer and Silencing After Systemic Delivery of Novel AAV-AS Vector. Mol. Ther 24, 726–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun S & Schaffer DV Engineered viral vectors for functional interrogation, deconvolution, and manipulation of neural circuits. Curr. Opin. Neurobiol 50, 163–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orthwein A et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Swiech L et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung SSC et al. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest. Ophthalmol. Vis. Sci 57, 3470–6 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Ruan G et al. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol. Ther 25, 331–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monteys AM, Ebanks SA, Keiser MS & Davidson BL CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol. Ther 25, 12–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gyorgy B et al. CRISPR/Cas9 mediated disruption of Swedish APP allele as a therapeutic approach for early onset Alzheimer’s disease. Mol. Ther. - Nucleic Acids 11, 429–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li P et al. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin- Associated Dominant Retinitis Pigmentosa. Cris. J 1, 55–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giannelli SG et al. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 27, 761–779 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Suzuki K et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu W et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 8, 14716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X et al. Genome editing abrogates angiogenesis in vivo. Nat. Commun 8, 112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest 127, 2719–2724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Z, Yang H & Colosi P Effect of genome size on AAV vector packaging. Mol. Ther 18, 80–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedland AE et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16, 257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaj T et al. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv 3, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bey K et al. Efficient CNS targeting in adult mice by intrathecal infusion of single-stranded AAV9-GFP for gene therapy of neurological disorders. Gene Ther. 24, 325–332 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Gray SJ, Nagabhushan Kalburgi S, McCown TJ & Jude Samulski R Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 20, 450–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hordeaux J et al. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther. 26, 664–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hadaczek P et al. Widespread AAV1- and AAV2-mediated transgene expression in the nonhuman primate brain: implications for Huntington’s disease. Mol. Ther. Methods Clin. Dev 3, 16037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Green F et al. Axonal transport of AAV9 in nonhuman primate brain. Gene Ther. 23, 520–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuzaki Y et al. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci. Lett 665, 182–188 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Samaranch L et al. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. 24, 253–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartel MA, Weinstein JR & Schaffer DV Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther. 19, 694–700 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Fu Y et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol 31, 822–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akcakaya P et al. In vivo CRISPR-Cas gene editing with no detectable genome-wide off-target mutations. (2018). doi: 10.1101/272724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mali P et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 31, 833–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ran FA et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu Y, Sander JD, Reyon D, Cascio VM & Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. 32, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Merienne N et al. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 20, 2980–2991 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Li F et al. Efficacy and dynamics of self-targeting CRISPR/Cas constructs for gene editing in the retina. (2018). doi: 10.1101/243683 [DOI] [Google Scholar]
- 77.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol (2018). doi: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haapaniemi E, Botla S, Persson J, Schmierer B & Taipale J CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med 24, 927–930 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Ihry RJ et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Bartel M, Schaffer D & Büning H Enhancing the Clinical Potential of AAV Vectors by Capsid Engineering to Evade Pre-Existing Immunity. Front. Microbiol. 2, 204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ciesielska A et al. Cerebral infusion of AAV9 vector-encoding non-self proteins can elicit cell-mediated immune responses. Mol. Ther 21, 158–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Samaranch L et al. AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol. Ther 22, 329–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charlesworth CT et al. Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans Carsten. (2018). doi: 10.1101/243345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeWitt MA, Corn JE & Carroll D Genome editing via delivery of Cas9 ribonucleoprotein. Methods 121–122, 9–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaj T et al. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. 45, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li L, Hu S & Chen X Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 171, 207–218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Montagna C et al. VSV-G-Enveloped Vesicles for Traceless Delivery of CRISPR-Cas9. Mol. Ther. - Nucleic Acids 12, 453–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ryu S et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol 1–7 (2018). doi: 10.1038/nbt.4148 [DOI] [PubMed] [Google Scholar]