Summary
DNA double-strand break (DSB) repair is mediated by multiple pathways. It is thought that the local chromatin context affects the pathway choice, but the underlying principles are poorly understood. Using a multiplexed reporter assay in combination with Cas9 cutting, we systematically measure the relative activities of three DSB repair pathways as a function of chromatin context in >1,000 genomic locations. This reveals that non-homologous end-joining (NHEJ) is broadly biased toward euchromatin, while the contribution of microhomology-mediated end-joining (MMEJ) is higher in specific heterochromatin contexts. In H3K27me3-marked heterochromatin, inhibition of the H3K27 methyltransferase EZH2 reverts the balance toward NHEJ. Single-stranded template repair (SSTR), often used for precise CRISPR editing, competes with MMEJ and is moderately linked to chromatin context. These results provide insight into the impact of chromatin on DSB repair pathway balance and guidance for the design of Cas9-mediated genome editing experiments.
Keywords: CRISPR, Chromatin, DNA repair, double strand break, MMEJ, NHEJ, SSTR, reporter assay, nuclear lamina, heterochromatin
Graphical abstract
Highlights
-
•
Sequencing-based reporter detects activities of multiple DSB repair pathways
-
•
Multiplexing approach to integrate reporter in >1,000 genomic locations
-
•
Overlay with epigenome data reveals effects of chromatin context on pathway balance
-
•
Pathway balance differs between heterochromatin and euchromatin
Schep et al. designed a reporter to probe the activities of three DNA double-strand break repair pathways. Integration of this reporter in >1,000 genomic locations in a human cell line provided a detailed view of the impact of local chromatin context on the balance between the repair pathways.
Introduction
The repair of DNA double-strand breaks (DSBs) is crucial for genetic stability. In addition, it is a key step in CRISPR-Cas9-mediated genome editing (Jasin and Haber, 2016; Yeh et al., 2019). Several pathways can repair DSBs, including classical non-homologous end-joining (NHEJ), homologous recombination (HR), and microhomology-mediated end-joining (MMEJ) (Chang et al., 2017; Iliakis et al., 2015; McVey and Lee, 2008; Scully et al., 2019; Yeh et al., 2019). NHEJ directly re-joins blunt-ended DSBs, while HR typically uses the intact sister chromatid in G2 phase as a template to mend the break. In contrast, MMEJ recombines short homologous sequences that are close to either end of the DSB and consequently results in a small deletion. An additional variant is single-stranded template repair (SSTR), a type of homology-directed repair that requires a single-stranded oligodeoxynucleotide (ssODN) donor sequence (Lin et al., 2014; Richardson et al., 2016). SSTR is highly relevant because it is leveraged in CRISPR-Cas9-mediated genome editing to generate precisely designed small mutations, such as point mutations or small insertions or deletions (indels) (DeWitt et al., 2016; Okamoto et al., 2019; Riesenberg et al., 2019).
Which pathway repairs a particular DSB depends in part on the local DNA sequence (Allen et al., 2018; Chakrabarti et al., 2019; Chen et al., 2019; Shen et al., 2018) and on the stage of the cell cycle (reviewed in Chapman et al., 2012; Hustedt and Durocher, 2016; Mladenov et al., 2016). In addition, local chromatin packaging can affect the choice of repair pathway (Clouaire and Legube, 2015; Jeggo and Downs, 2014; Kalousi and Soutoglou, 2016; Scully et al., 2019). Most studies of chromatin effects so far have focused on the balance between HR and NHEJ. For example, the histone modification H3K36me3, which is present along active transcription units, is thought to promote HR (Aymard et al., 2014; Carvalho et al., 2014; Clouaire et al., 2018; Daugaard et al., 2012; Pfister et al., 2014). Paradoxically, H3K9 di- or trimethylated (H3K9me2/3) heterochromatin, which packages transcriptionally inactive regions of the genome, has also been implicated in promoting HR (Alagoz et al., 2015; Baldeyron et al., 2011; Lee et al., 2013; Soria and Almouzni, 2013; Sun et al., 2009), although some single-locus studies in mouse and fruit fly found no major change in the balance between NHEJ and HR when a sequence was shifted between heterochromatin and euchromatin states (Janssen et al., 2016; Kallimasioti-Pazi et al., 2018). Furthermore, reduced binding of HR proteins was observed at a locus that was artificially tethered to the nuclear lamina (Lemaître et al., 2014), suggesting that spatial positioning of the DSB inside the nucleus may also play a role.
Much less is known about the impact of chromatin on MMEJ and SSTR. Like HR, these pathways require resection of the DNA ends to produce single-stranded DNA overhangs, but downstream of this step, the mechanisms and responsible proteins diverge (Chang et al., 2017; Scully et al., 2019; Yeh et al., 2019). It is thus possible that the local chromatin environment also modulates MMEJ and SSTR in unique ways, but this has remained largely unexplored (Clouaire and Legube, 2019; Mitrentsi et al., 2020).
One strategy to investigate the impact of local chromatin context on repair pathway balance is to generate DSBs at various genomic locations with known chromatin states and compare pathway use across these locations (Chakrabarti et al., 2019; Clouaire et al., 2018; van Overbeek et al., 2016). However, with such an approach it is difficult to separate the effects of chromatin context from the effects of sequence context because both vary simultaneously along the genome. Ideally, different chromatin contexts are compared while the sequence context is kept fixed.
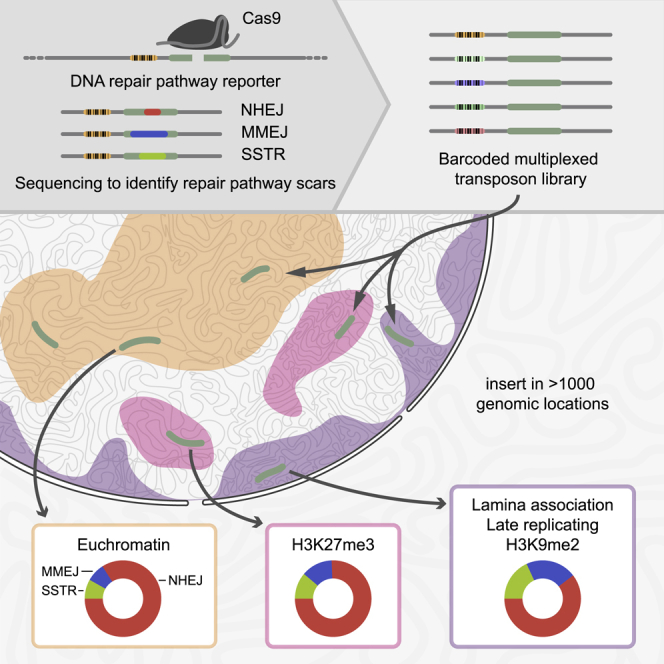
Here, we report a strategy that effectively tackles these challenges in human cells. The strategy consists of two parts. First, we used a reporter that, when cut with Cas9, produces distinct “scars” when repaired by either NHEJ, MMEJ, or SSTR; high-throughput sequencing of these scars provides highly accurate measurements of the relative contributions of the three pathways. Second, we used a modification of our TRIP (thousands of reporters integrated in parallel) method (Akhtar et al., 2013) to insert this reporter into >1,000 random genomic locations, tracking each individual reporter in parallel by molecular barcoding. We thus systematically measured the relative contributions of NHEJ, MMEJ, and SSTR as a function of chromatin context in >1,000 genomic locations. This yielded datasets that (1) comprehensively sample the broad diversity of chromatin “flavors” across the entire genome, (2) bypass the confounding effects of varying sequence context, and (3) probe the three pathways with high accuracy and sensitivity. The results provide a detailed view of the impact of chromatin context on the relative activities of the three repair pathways.
Results
Multiplexed DSB repair pathway assay: Principle
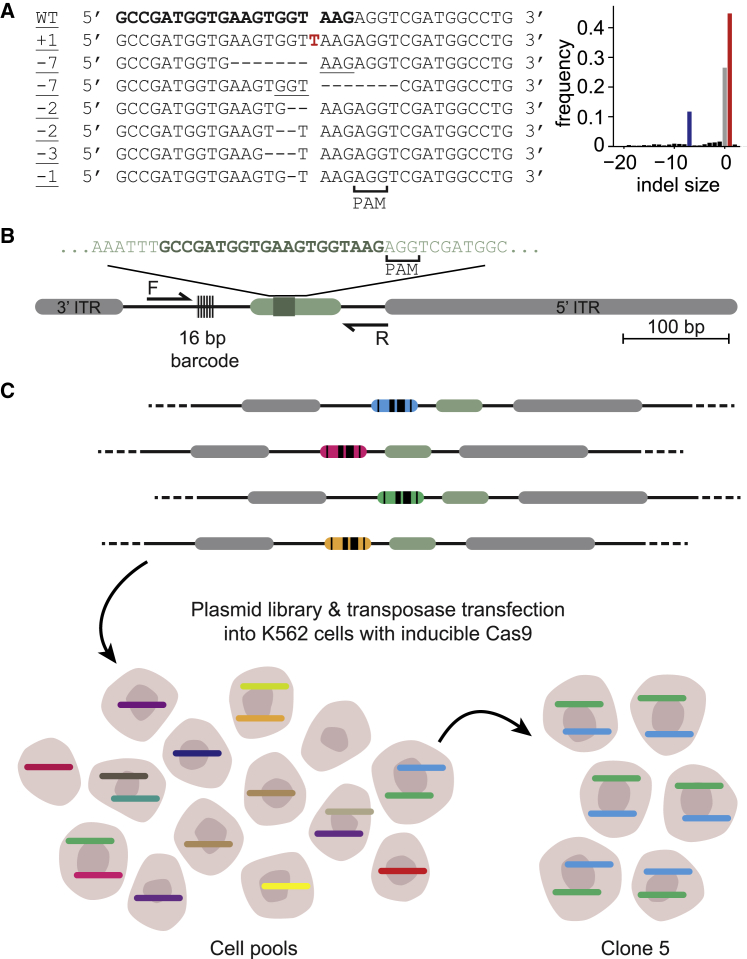
We developed a strategy to measure the relative contribution of several DSB repair pathways in more than 1,000 genomic locations that include all known common chromatin states. The strategy involves a pathway-specific reporter construct that contains a short DNA sequence (derived from the human LBR gene) that predominantly produces a +1 insertion or a −7 deletion when cut at a specific base pair position by Cas9 (Figure 1A). We previously found that these two indels are primarily the result of NHEJ (+1) and MMEJ (−7), respectively (Brinkman et al., 2018), and we provide additional support below. The relative abundance of these signature indels can therefore be interpreted as a measure of the relative contribution of these two pathways. Furthermore, this readout can be extended to include SSTR and translocations (see below). We note that HR cannot be detected with this assay, because HR generally repairs DSBs perfectly, and perfectly repaired DNA cannot be distinguished from uncut DNA. However, we previously estimated that perfect repair of this reporter sequence is rare and by inference that the contribution of HR is likely to be very minor (Brinkman et al., 2018). Further evidence supporting this inference is presented below.
Figure 1.
Principle of multiplexed DSB repair pathway reporter assay
(A) Left panel: sequences of the most common insertions and deletions produced after break and repair induced with sgRNA LBR2. Inserted nucleotide in red, microhomologies are underlined. Right panel: indel frequency distribution after cutting the LBR gene. Negative indel sizes refer to deletions, positive sizes refer to insertions. The +1 and −7 indels are marked in red and blue, respectively.
(B) Schematic of the TRIP construct. ITR, inverted terminal repeat of PiggyBac transposable element; the LBR gene-derived sequence is shown in light green, with the sgRNA target sequence in dark green. PCR primers are indicated by the arrows (F and R).
(C) Schematic of the TRIP experimental setup. See main text.
With this reporter, we implemented a variant of the TRIP technology (Akhtar et al., 2013) to systematically probe the effects of many chromatin environments on the repair pathway use. We inserted the reporter sequence into a PiggyBac transposon vector, together with a 16 bp random barcode sequence that was located 56 bp from the DSB site (Figure 1B). We then randomly integrated this construct into the genomes of pools of K562 cells (Figure 1C). We chose K562 cells because the chromatin landscape has been extensively characterized. From one of these pools, we also generated several clones for smaller scale experiments. Each copy of the integrated reporter carried a different barcode. We mapped the genomic locations of these integrated pathway reporters (IPRs) together with their barcode sequences by inverse PCR (Akhtar et al., 2013). Next, after Cas9-mediated DSB induction and the ensuing repair, we determined the accumulated spectrum of indels of each individual IPR in a multiplexed fashion, by PCR amplification (see primer locations in Figure 1C) followed by high-throughput sequencing. Because each barcode is linked to its genomic location, the sequence information enabled us to infer the relative DSB repair pathway use at each location. Comparison of the resulting data with the local chromatin state of the IPRs then provides insight into the impact of chromatin context on DSB repair pathway use.
Implementation and validation of the multiplexed reporter assay
For these experiments we used a human K562 cell line that expresses Cas9 protein in an inducible manner (Brinkman et al., 2018). We generated two cell pools with 979 and 1,099 (total 2,078) uniquely mapped IPRs (Figures 2A and S1A). In addition, we established 14 clonal cell lines, for which the barcodes and locations were also mapped by inverse PCR and tagmentation (Stern, 2017). On average these clones carried 6.8 integrations, which we take as an estimate of the numbers of integrations per cell in the cell pools. For some additional analyses described below, we used one clone (clone 5) with 19 mapped IPRs that are located across most major chromatin types (Figures S1A and S1B).
Figure 2.
Multiplexed detection of DSB repair pathway use
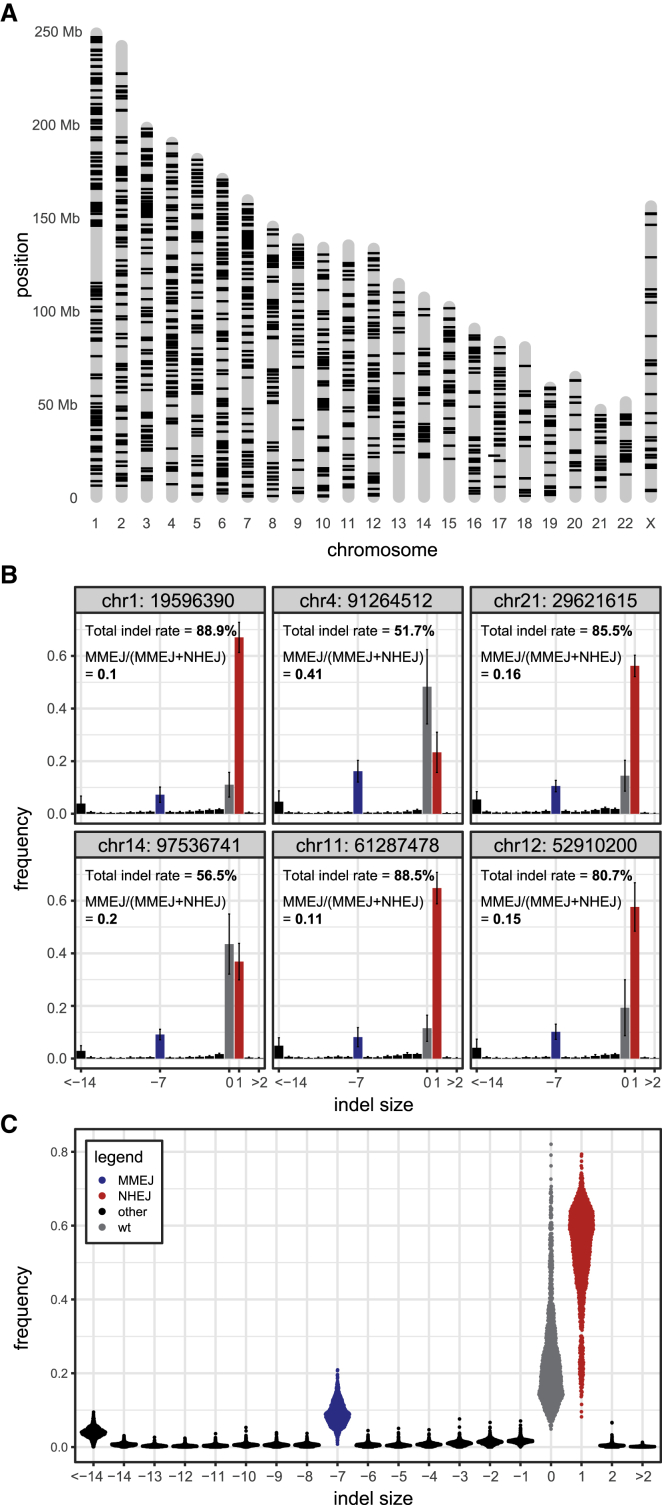
(A) Genomic integration coordinates of 1,229 uniquely mapped IPRs (both cell pools combined) that passed filtering as described in STAR Methods and are used in this work.
(B) Indel frequency distributions of six randomly selected IPRs, 64 h after Cas9 induction. Data are average of six independent replicates. Error bars are ±SD. Gray, wild-type sequence; red, +1 insertion, diagnostic of NHEJ; blue, −7 deletion, diagnostic of MMEJ; black, other indels.
(C) Indel frequencies of all IPRs shown in (A), 64 h after Cas9 induction. Data are average of two to six independent replicates.
See also Figures S1 and S2.
Next, we induced Cas9 in the cell pools by ligand-dependent stabilization (Banaszynski et al., 2006) and transfection with the single guide RNA (sgRNA) (named sgRNA-LBR2). We collected genomic DNA after 64 h and determined the indel spectra of all IPRs. At this time point, indel accumulation in the LBR gene has reached near saturation (Brinkman et al., 2018). After applying stringent quality criteria (see STAR Methods), we obtained robust indel spectra of 1,229 IPRs (Figure S1A); the other IPRs were mostly discarded because they were insufficiently represented in the cell pools. Overall, the IPRs of both pools showed a similar pattern of indels as the endogenous LBR sequence in the same cells, dominated by +1 and −7 indels (Figures 2B, 2C, S1C, and S1D). This supports previous findings that the sequence determines the overall indel pattern (Allen et al., 2018; Chakrabarti et al., 2019; Chen et al., 2019; Shen et al., 2018; van Overbeek et al., 2016), but we also observe clear variations in indel frequencies.
As noted before (Brinkman et al., 2018), the −7 deletions come in two variants that both involve 3 nt microhomologies (Figure 1A), consistent with MMEJ. To further verify that the +1 and −7 indels indeed represent NHEJ and MMEJ, respectively, we depleted or inhibited several pathway-specific proteins (Chang et al., 2017; Scully et al., 2019) in either the pools or clone 5 (Figure S2). The +1 insertion was strongly reduced, and the −7 deletion was increased by inhibition of DNA-PKcs by the compounds NU7441 (Figures S2A and S2B) or M3814 (Figure S2C). DNA-PKcs is a key component of NHEJ (Gottlieb and Jackson, 1993). In contrast, the −7 deletions but not the +1 insertion were selectively reduced upon depletion of DNA polymerase theta (POLQ) and CTIP (also known as RBBP8), which are proteins of the MMEJ pathway (Chan et al., 2010; Mateos-Gomez et al., 2015; Sartori et al., 2007) (Figures S2D–S2F). Knockdown of HR factors BRCA1 and BRCA2 did not affect the −7 deletion, and BRCA1 knockdown caused a slight decrease of the wild-type sequence and an increase of the +1 insertion (Figures S2G–S2I). The latter observation could point to a small role of HR in repairing DSBs in an error-free manner. Depletion of Rad51 resulted in a reduction of the −7 deletion (Figures S2D–S2H); however, even without DSB induction, this caused a reduction in cell viability to 78% (95% confidence interval [CI] = 65.3%–91.1%, p = 1.98e-05, one-sample t test, n = 6), making it difficult to interpret these results. Aside from this latter experiment, all evidence indicates that the +1 and −7 indels are primarily the result of NHEJ and MMEJ, respectively, and that the contribution of HR is minor at Cas9-induced DSBs.
Detection of large genomic rearrangements
The sequencing of the repair “scars” indicated that large indels are very rare (Figure S1C). However, because of the length of the sequence reads, we could not capture any deletions >119 bp, nor translocations and other rearrangements that might occur. We therefore modified a tagmentation-based approach (Giannoukos et al., 2018; Stern, 2017) to identify distal genomic sequences that became ligated to the IPRs after Cas9 activation (Figure S2J). We applied this assay to clone 5. The results indicate that large indels, inversions, and translocations occur with a relative frequency of approximately 4% (Figure S2K). As may be expected (Frock et al., 2015), these rearrangements preferentially occur in cis, with a bias for shorter distances (Figures S2L and S2M). The LBR gene, which is also cut by sgRNA-LBR2, is involved in about 22.2% and 16.4% of the long-range rearrangements with IPRs in cis and trans respectively (Figures S2L and S2M). This is particularly frequent for an IPR located ~0.5 Mb away on the same chromosome. Unfortunately, because of a technical limitation (see STAR Methods) we were unable to reliably detect junctions between two IPRs. However, considering that there are four alleles of the LBR gene in K562 cells (Brinkman et al., 2018) and an estimated 6.8 IPRs/cell in our cell pools, it seems unlikely that IPR-IPR junctions occur with a total frequency of more than 1% (see STAR Methods). Although the detection of rearrangements by this approach may not be fully quantitative because of possible technical biases, we conclude that IPRs are involved in inversions, large deletions, and translocations. Yet the frequency of these rearrangements is generally too low to substantially skew our estimates of MMEJ and NHEJ.
Effects of chromatin context on overall indel frequencies
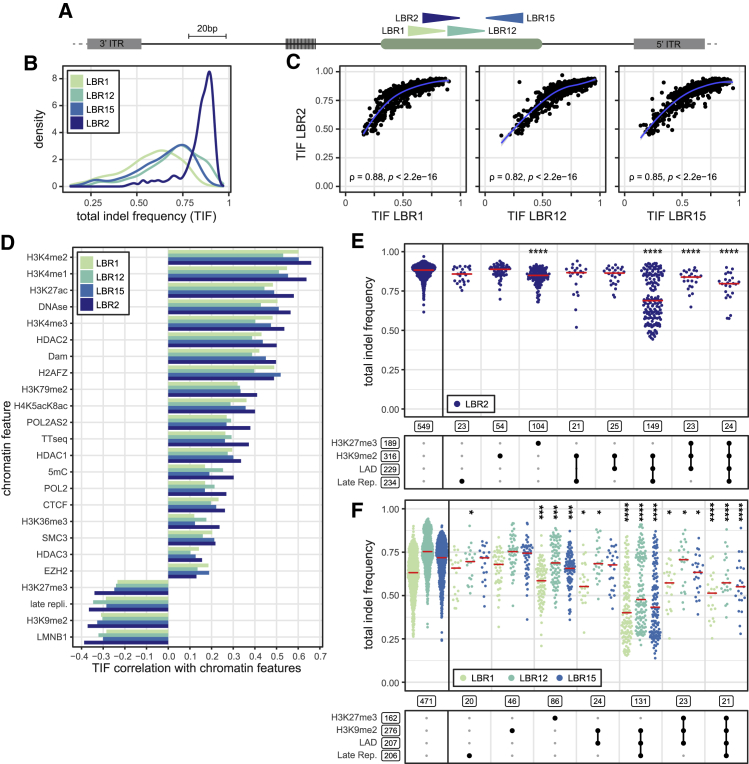
Using the indel spectra from the two cell pools, we first investigated the impact of chromatin context on total indel frequencies (TIFs; i.e., the proportion of reporter sequences carrying any type of indel). Across the IPRs these frequencies varied from ~25% to ~100% (Figures 3A and 3B). This variation most likely reflects differences in either the cutting efficiency by Cas9 or the DSB repair rate, or both. The fact that in some IPRs the TIFs approached 100% indicates that virtually all cells received the sgRNA in our transfection protocol. We repeated these experiments with three different sgRNAs targeting different sequences in the same reporter (Figures 3A, 3B, and S3A). This yielded overall indel frequencies that strongly correlated with sgRNA-LBR2 and with each other, although with sgRNA-LBR2 we may have approximated saturation of indels more than with the other sgRNAs (Figures 3B, 3C, and S3B).
Figure 3.
IPR total indel frequency varies as a function of chromatin context
(A) Schematic of the IPR with four different gRNA target sites indicated by arrowheads, oriented toward the PAM.
(B) Total indel frequency distributions of IPRs after cutting with each sgRNA, shown as density plots, which can be interpreted as smoothed histograms. For each sgRNA, IPRs were included only if they yielded reliable data in at least two independent experiments (LBR2: 1,010 IPRs [n = 2–8]; LBR1: 956 IPRs [n = 2]; LBR12: 942 IPRs [n = 2]; LBR15: 932 IPRs [n = 2]). Indel frequencies were not corrected for transfection efficiency, which we infer to be close to 100% because some IPRs have indel frequencies near 100%.
(C) Scatterplots of total indel frequencies obtained with LBR2 versus the three other sgRNAs. ρ is Spearman’s rank correlation coefficient.
(D) Spearman’s correlations between total indel frequencies in IPRs and the local intensities of 24 chromatin features at the IPR integration sites, for each sgRNA. p < 0.01 for all correlations. Chromatin features are ordered by the LBR2 correlation coefficients.
(E) Total indel frequency at each IPR obtained with LBR2 sgRNA, split into different combinations of heterochromatin features present, as indicated by black dots in the scheme below the graph. Red lines show median values. Boxed numbers indicate the number of IPRs in each group; only groups with >20 IPRs are shown. Asterisks mark p values according to Wilcoxon test, compared with euchromatin IPRs (leftmost column): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(F) Same as (E) but for the other three sgRNAs.
See also Figure S3.
Because the sequences of the IPRs are identical (except for the short barcodes located 56 bp from the cut site), the differences in indel frequencies across integration sites are presumably due to variation in the local chromatin environment. To investigate this, we correlated the indel data for each sgRNA with a curated set of 24 genome-wide maps of chromatin features that represent most of the known main chromatin types, including a multitude of markers of transcription and active regulatory elements; chromatin accessibility; DNA methylation; and heterochromatic features such as the histone modifications H3K27me3, H3K9me2, late replication, and nuclear lamina interactions (Tables S1 and S2) (Chen et al., 2018; Dekker et al., 2017; ENCODE Project Consortium, 2012; Leemans et al., 2019; Ott et al., 2018; Salzberg et al., 2017; Schmidl et al., 2015; Schwalb et al., 2016; Shah et al., 2018). The IPRs lack gene regulatory elements and are only 640 bp long and may thus be expected to adopt the local chromatin state. Chromatin immunoprecipitation (ChIP) experiments for three histone modifications generally confirmed this (Figure S3C). This is consistent with previous studies showing that integrated reporters adopt and strongly respond to the local chromatin state (Akhtar et al., 2013; Corrales et al., 2017; Leemans et al., 2019). We therefore assume that the chromatin state of the integration positions is a reasonable approximation of the chromatin state of the IPRs themselves.
Indel frequencies at the IPRs generally correlated positively with various markers of euchromatin and negatively with markers of heterochromatin. These correlations were highly consistent between the four sgRNAs that we tested (Figure 3D); minor differences may reflect differences in statistical power or subtle modulating effects of the DNA sequence at the broken ends. On average, IPRs integrated in heterochromatin regions showed lower indel frequencies than in euchromatic regions. However, within heterochromatin, the magnitude of this effect varied depending on the specific combination of features (Figures 3E and 3F). The most pronounced effect was observed in regions marked by the combination of H3K9me2, lamina-associated domains (LADs), and late replication (further referred to as triple heterochromatin). In these regions the distribution of indel frequencies is bimodal. This is similar to TRIP reporters of promoter activity, which also tend to show a bimodal distribution of activities across LADs, correlating with local chromatin features (Leemans et al., 2019). Differences in lamina interactions and replication time could explain part of the bimodal distribution of indel frequencies (Figure S3D). Remarkably, when H3K27me3 is additionally present, the reduction of indel frequencies is less pronounced, and regions marked by H3K27me3 alone only slightly affect indel frequencies compared with euchromatin. Thus, H3K27me3 only mildly impedes Cas9 editing and may even counteract the effects of other heterochromatin features. Regions marked by H3K9me2 together with either late replication or lamina interactions (but not both) show only marginally reduced indel frequencies, compared with the triple-marked regions (Figures 3E and 3F). For euchromatic regions, we did not survey combinatorial effects, because there are too many possible combinations and hence statistical power is insufficient.
Together, these results indicate that the overall indel frequency depends on the local chromatin context and that heterochromatin features are correlated with the efficiency of indel accumulation in a combinatorial manner. These effects may be through modulation of Cas9 cutting efficiency, modulation of indel-forming repair rates, or both.
Impact of chromatin context on MMEJ:NHEJ balance
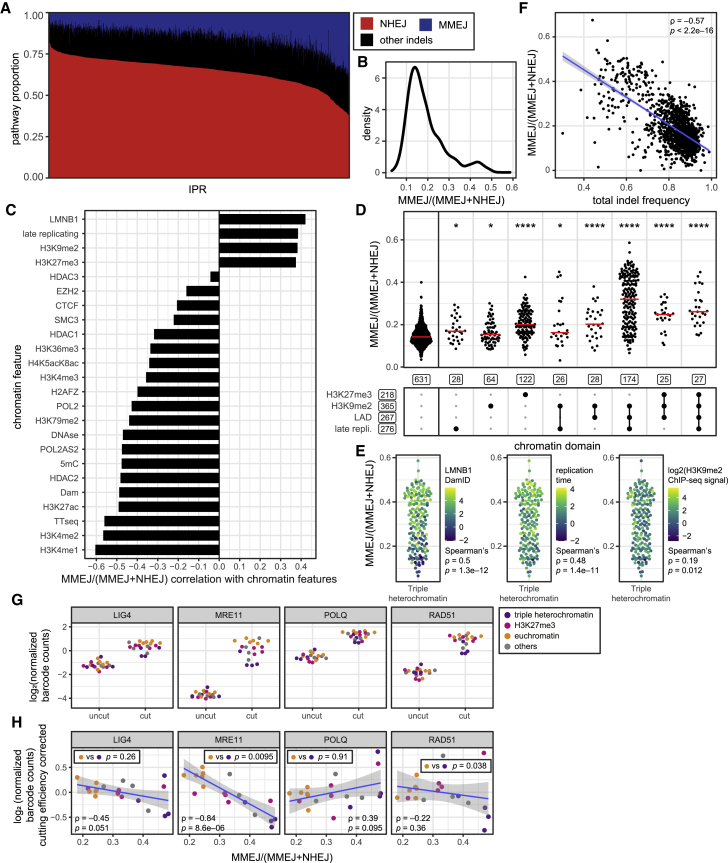
Next, we analyzed the variation in the balance between MMEJ and NHEJ, throughout this paper referred to as “MMEJ:NHEJ balance” and defined for each IPR as the number of −7 reads over the sum of −7 and +1 reads. Importantly, this metric intrinsically corrects for any differences in cutting efficiencies, because it scores only sequences that were broken and repaired. The MMEJ:NHEJ balance varies profoundly depending on the integration site (Figures 4A, 4B, and S4A). Globally, it correlates negatively with markers of euchromatin and positively with markers of heterochromatin (Figures 4C and S4B). The strongest negative correlations were observed for H3K4me1, H3K4me2, and H3K27ac, which are histone modifications that primarily mark enhancers and to a lesser extent promoters (Gasperini et al., 2020), and TT-seq, which measures transcription activity (Schwalb et al., 2016). Positive correlations occur with multiple markers of heterochromatin such as H3K27me3, H3K9me2, and LADs. Thus, in heterochromatin the MMEJ:NHEJ balance is generally higher than in euchromatin.
Figure 4.
Chromatin context effects on MMEJ:NHEJ balance and protein binding
(A) Variation in indel composition across IPRs. Red, +1 (NHEJ); blue, −7 (MMEJ); black, other indels (unknown pathway). IPRs are ordered by +1 insertion frequency (1,171 IPRs, two to eight independent experiments).
(B) MMEJ:NHEJ balance distribution across all IPRs.
(C) Spearman’s correlation coefficients of MMEJ:NHEJ balance versus the local intensities of 24 chromatin features. p < 0.001 for all correlations.
(D) MMEJ:NHEJ balance per IPR, split into different combinations of heterochromatin features similar to Figure 3E; see Figure S4D for all groups. Asterisks mark p values according to the Wilcoxon test, compared with euchromatin IPRs (leftmost column): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(E) MMEJ:NHEJ balance of IPRs in triple heterochromatin, colored by levels of H3K9me2, LMNB1, or replication timing.
(F) Correlation between total indel frequency and MMEJ:NHEJ balance across all IPRs.
(G) Barcode counts (normalized log2 values) of IPRs in clone 5 after ChIP of indicated proteins, 16 h after Cas9 induction with (cut) or without (uncut) sgRNA LBR2.
(H) Same ChIP data as (G) under cut conditions, but now corrected for estimated cutting efficiencies at t = 72 h and plotted against MMEJ:NHEJ balance. Bottom p values refer to the Pearson’s correlation with MMEJ:NHEJ balance; top p values refer to the difference between IPRs in euchromatin and triple heterochromatin (Wilcoxon test); blue line and gray shading show linear regression fit with 95% confidence interval. Triple heterochromatin is the combination of H3K9me2, lamina association, and late replication; euchromatin is here defined as the absence of any heterochromatin features.
See also Figure S4.
Cutting of the IPRs by Cas9 in combination with each of the other three sgRNAs yields rather complex indel spectra (Figure S3A). When we provisionally assigned each of these indels to either MMEJ or NHEJ (on the basis of their sensitivity to M3814 treatment; see STAR Methods), we observed consistently that the MMEJ:NHEJ balance is higher in heterochromatin than in euchromatin (Figure S4C). Thus, the results are consistent among all sgRNAs tested, although sgRNA LBR2 is more suited to measure the two pathways.
Within heterochromatin, we further explored whether certain combinations of chromatin features are more predictive than others. The strongest effect on the balance was observed in IPRs located in regions marked by triple heterochromatin (Figures 4D and S4D). As for the TIF, the bimodal distribution of the triple heterochromatin group can be partly explained by local differences in levels of lamina interactions and replication timing (Figure 4E). Regions marked by two of these three features showed less pronounced but significant increases in MMEJ:NHEJ balance compared with euchromatin regions, as did regions marked by H3K27me3. Altogether, these data show that the balance between MMEJ and NHEJ is broadly linked to the global heterochromatin/euchromatin dichotomy, and within heterochromatin depends on the local combination of heterochromatin features.
Overall, the patterns of MMEJ:NHEJ balance (Figure 4C) and indel frequencies (Figure 3D) appeared to mirror each other. Indeed, across all IPRs the two variables correlate, although imperfectly (Spearman’s correlation coefficient = −0.57) (Figure 4F). This correlation does not necessarily imply a causal relationship. But it seems improbable that the cutting rate itself determines the pathway balance.
Repair protein binding is linked to pathway balance and chromatin context
To test whether the variation in pathway balances across the IPRs may be explained by differences in presence of pathway-specific repair proteins, we conducted ChIP experiments on clone 5 cells. We probed the occupancy of several proteins at the IPR barcodes (i.e., close to the break sites) within the resolution of ChIP. We focused on time point 16 h after Cas9 induction, at which we previously found the largest amount of broken DNA (Brinkman et al., 2018). The results show that MRE11 (involved in early DSB detection and processing), LIG4 (specific to NHEJ), POLQ (specific to MMEJ), and RAD51 (specific to HR) can all be detected at each IPR when Cas9 cutting is induced (Figure 4G). Thus, the binding of these proteins appears to be universal across different chromatin contexts. However, correction of the ChIP signals for the approximate cutting frequency (see STAR Methods) revealed quantitative trends. As expected, the levels of LIG4 and POLQ at the IPRs are negatively and positively correlated with the MMEJ:NHEJ balance, respectively, although these trends are of borderline statistical significance (Figure 4H). Surprisingly, binding of MRE11 shows a strong negative correlation with the MMEJ:NHEJ balance and a significant preference for euchromatin compared with triple heterochromatin. Finally, binding of RAD51 shows no significant correlation with the MMEJ:NHEJ balance, although is shows an overall preference for euchromatin over triple heterochromatin. We conclude that proteins of all major pathways are present at each DSB, with quantitative differences that may in part explain the observed pathway preferences.
Different kinetics of MMEJ engagement between chromatin types
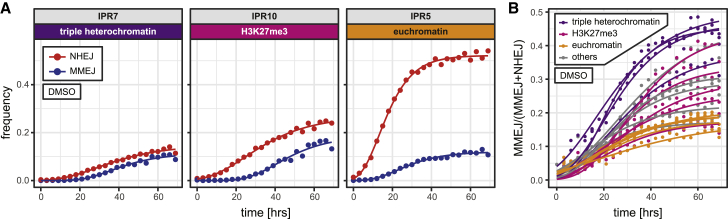
To explore how the difference in pathway balance between heterochromatin and euchromatin develops over time after DSB induction, we conducted time-series experiments. We used a robotics setup to collect DNA samples every 3 h over a period of 3 days following Cas9 activation. For these experiments we focused on clone 5; because all 19 IPRs in this clone are in the same cell, their repair kinetics can be directly compared.
As expected, Cas9 activation resulted in a gradual accumulation of +1 and −7 indels in all IPRs, concomitant with a loss of wild-type sequence (Figures 5A and S5). These kinetics were generally slower in regions in triple heterochromatin (e.g., IPRs 7, 14, and 16; Figure S5). Remarkably, the MMEJ:NHEJ balance was not constant over time but was strongly skewed toward NHEJ at the early time points and gradually shifted toward MMEJ for all IPRs, culminating in a plateau approximately 50 h after Cas9 activation (Figure 5B). This points to a delayed use of the MMEJ pathway, as we had observed previously for a single locus (Brinkman et al., 2018). This buildup of MMEJ use may occur eventually at most DSBs. However, over time the pathway balance diverged between IPRs in different chromatin environments, with nearly all heterochromatic IPRs developing a higher MMEJ:NHEJ balance than the euchromatin IPRs (Figure 5B).
Figure 5.
Accumulation of indels over time
(A) Time curves of the +1 insertion (red) and −7 deletion (blue) in single IPRs located in three different types of chromatin. See Figure S5 for plots of all 19 IPRs in clone 5. Dots are measured values; lines are fitted sigmoid curves.
(B) Shifting MMEJ:NHEJ balance over time in 19 IPRs of clone 5, colored by chromatin type.
Data in (A) and (B) are averages of two independent experiments. See also Figure S5.
Overall robustness of pathway balance in heterochromatin
We then investigated the role of several heterochromatin features in pathway balance by perturbation experiments. To distinguish direct from indirect effects, we compared the MMEJ:NHEJ balance of IPRs in regions with the to-be-perturbed feature to that of IPRs in regions already lacking the feature prior to treatment. Direct effects should primarily alter the MMEJ:NHEJ balance in regions originally marked by the feature.
We first reduced the levels of H3K9me2 by treatment with the G9a inhibitor BIX01294 (Figures S6A–S6C). This did not alter the MMEJ:NHEJ balance in H3K9me2 domains, except when in combination with LADs and late replication, where the balance increased slightly (p = 0.02; Figure S6A).
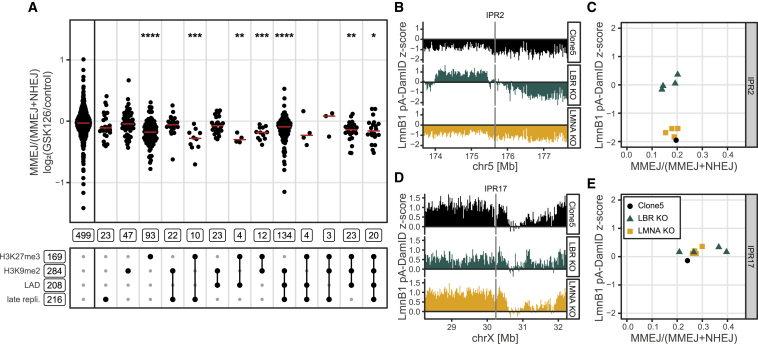
We then tested the effect of GSK126, a compound that inhibits the H3K27me3 methyltransferase EZH2 and causes a global loss of H3K27me3 (Figures S6D and S6E). This inhibitor caused a significant reduction of the MMEJ:NHEJ balance in H3K27me3-only domains compared with euchromatin regions (p = 2e-11), as well as in virtually all domain combinations that include H3K27me3 (Figure 6A). Because the IPRs in the H3K27me3 domains are in the same cell pools and receive exactly the same drug treatment as the euchromatic IPRs, the effect of GSK126 must be local in the genome; as GSK126 reduces H3K27me3 levels, we conclude that its effect in the H3K27me3 domains is direct. Unexpectedly, GSK126 treatment also reduced levels of H3K9me2 (Figures S6B and S6C) and also slightly lowered the MMEJ:NHEJ balance in the triple heterochromatin domains (p = 5.6e-05), but not in the single H3K9me2 domains. The most prominent shift in balance was, however, in H3K27me3 domains, pointing to a local effect of this histone modification on MMEJ:NHEJ balance.
Figure 6.
Effects of heterochromatin perturbations on pathway balance
(A) Log2 fold change of MMEJ:NHEJ balance in GSK126 treated cells compared with control cells, for 917 IPRs divided by heterochromatin type. Data are average of two independent biological replicates. Wilcoxon test compared with euchromatin IPRs (leftmost column): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(B and D) Nuclear lamina interaction tracks around IPR2 (B) and IPR17 (D). The tracks for the KO clones are the average of four separate clones (individual tracks are shown in Figures S7K and S7L). All data are average of two independent biological replicates.
(C and E) Comparison of MMEJ:NHEJ balance (n = 3) and average lamina interaction score in a 20 kb window centered on the IPR (n = 2), for IPR2 (C) and IPR17 (E).
See also Figure S6.
Finally, because IPRs in LADs often show a high MMEJ:NHEJ balance, we used CRISPR-Cas9 editing to derive cell lines from clone 5 that lacked Lamin A/C (LMNA) or Lamin B receptor (LBR) (Figures S6F–S6I). These two lamina proteins are important for the peripheral positioning of heterochromatin (Clowney et al., 2012; Solovei et al., 2013), and LMNA has been implicated in the control of NHEJ by sequestering 53BP1 (Redwood et al., 2011). Using the pA-DamID method, we mapped genome-wide changes in lamina interactions in four knockout (KO) clones each of LMNA and LBR. LMNA-KO cells showed very few changes in lamina interactions, while the LBR-KO clones showed many regions with either gains or losses in lamina interactions. A detailed analysis of these changes will be reported elsewhere. Here, we investigated whether changes in lamina interactions of the IPRs coincide with changes MMEJ:NHEJ balance.
The majority of the IPRs did not undergo substantial changes in lamina interactions in either the LMNA- or LBR-KO clones compared with the parental clone 5, and they also did not show significant changes in MMEJ:NHEJ balance (Figure S6J). An exception was IPR 2, in which the lamina interactions became stronger in all four LBR-KO clones (Figures 6B and 6C). However, the MMEJ:NHEJ balance in IPR 2 was not detectably altered in these clones (Figures 6C and S6K). This suggests that lamina contacts do not modulate this balance, but we cannot rule out that effects on this balance emerge only when lamina contacts are stronger than those of IPR 2 in the LBR-KO clones (note that the lamina interaction Z score in these clones is about 0, which corresponds to a moderate level of lamina interactions). Interestingly, IPR 17 showed a marked increase in the MMEJ:NHEJ balance in two of the four LBR-KO clones (Figures 6D, 6E, and S6J). However, for this IPR the lamina interactions did not change (Figure S6L), and we do not understand why only two of the four clones show this behavior. Nevertheless, this result underscores that it is possible to shift the MMEJ:NHEJ balance in an IPR markedly without any change in its sequence. Presumably, an unknown change in the local chromatin state in the two clones is responsible for this.
Together, these data indicate that the MMEJ:NHEJ balance in specific heterochromatin types is not easily shifted by targeting individual key markers of the respective heterochromatin types. Pathway balance may be redundantly controlled by multiple factors in each heterochromatin type. Nevertheless, depletion of H3K27me3 did cause a detectable reduction in MMEJ:NHEJ balance in heterochromatin domains that normally carry this mark.
Impact of chromatin context on SSTR
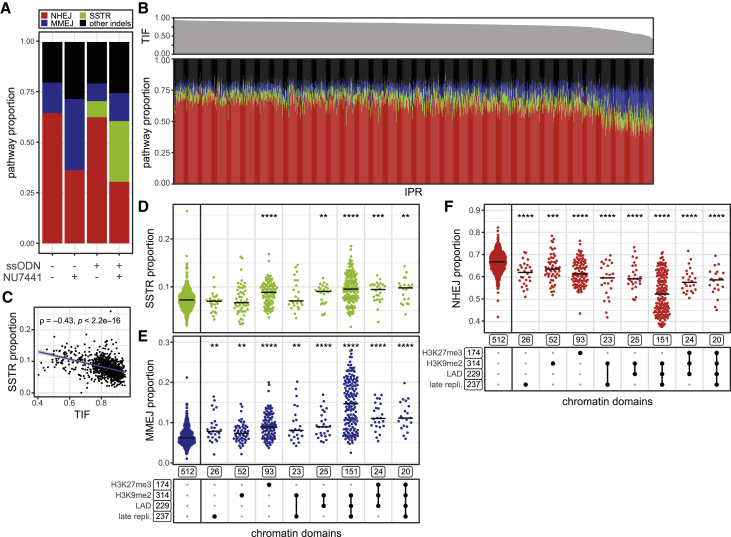
Finally, we investigated a third repair pathway, SSTR, which is commonly used to create specific mutations by CRISPR-Cas9 editing. We hypothesized that this pathway may also be modulated by the local chromatin environment. To test this, we triggered DSB formation in our reporter sequence in the presence of a template ssODN containing a specific +2 insertion (ssODN insertion) (Figure S7A). We designed this insertion within the PAM site, so that a successful editing event destroyed the PAM site and prevented further cutting by Cas9. We then transfected the IPR cell pools with this ssODN (together with the sgRNA) to probe the impact of chromatin context on the relative contribution of SSTR, NHEJ, and MMEJ in parallel. In these experiments, we found that a median 6% of the indels consisted of the SSTR insertion (Figure S7B). Accumulation of this insertion was mostly at the expense of the −7 deletion but not the +1 insertion (Figures 7A, S7C, and S7D), suggesting competition between SSTR and MMEJ. Indeed, depletion of POLQ (a key protein of MMEJ) caused an increase in ssODN insertions (Figures S2D and S2E). Consistent with an earlier study (Richardson et al., 2018), we find that knockdown of RBBP8, a factor that promotes end-resection (Sartori et al., 2007), strongly reduces SSTR and MMEJ usage (Figures S2D and S2E). This suggests that DSB end-resection is an early step in SSTR. In agreement with previous work (Richardson et al., 2018), we conclude that SSTR is distinct from NHEJ and MMEJ.
Figure 7.
Balance between NHEJ, MMEJ, and SSTR in euchromatin and heterochromatin
(A) Average pathway contribution across all IPRs in the cell pools, in the absence or presence of an ssODN donor, and with or without NU7441 treatment (DMSO: 1,219 IPRs [n = 2–8, same data as in Figures 3 and 4]; DMSO + ssODN: 1,032 IPRs [n = 2 or 3]; NU7441: 1,186 IPRs [n = 2–4, same data as in Figures S2A and S2B]; NU7441 + ssODN: 1,047 IPRs [n = 2 or 3]). Red, +1 insertion (NHEJ); blue, −7 deletion (MMEJ); green, +2 insertion due to SSTR; black, other indels.
(B) Pathway contributions in all individual IPRs in the presence of ssODN donor but without NU7441 (bottom panel), sorted by overall indel frequency (top panel).
(C) Correlation between total indel frequency and SSTR proportion across all IPRs. Blue line and gray shading show linear regression fit with 95% confidence interval. ρ is Pearson’s rank correlation coefficient.
(D–F) Proportions of total indels generated by SSTR (D), MMEJ (E), and NHEJ (F) in the presence of the ssODN donor, split according to heterochromatin features as indicated in the bottom panel, similar to Figure 3E. Asterisks mark p values according to the Wilcoxon test, compared with euchromatin IPRs (most left column): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
See also Figure S7.
We then investigated the balance of the three simultaneously probed pathways as a function of chromatin type. In the presence of ssODN, the proportion of indels created by SSTR is inversely correlated with the overall indel frequency (Figures 7B and 7C) and is higher in various types of heterochromatin compared with euchromatin (Figure 7D). Under this condition, the proportions of indels produced by MMEJ (Figure 7E) and NHEJ (Figure 7F) show similar effects of various types of heterochromatin as we observed in the absence of ssODN (cf. Figure 4D). We conclude that the proportion of SSTR is higher in several types of heterochromatin compared with euchromatin.
Discussion
Genome-wide survey of DSB repair
Here, we present a powerful reporter system to query effects of chromatin on DSB pathway use. It consists of (1) a simple short DNA sequence that, when cut with Cas9, produces a signature indel for three repair pathways and (2) an adaptation of the TRIP multiplexed reporter assay (Akhtar et al., 2013). In combination, these tools offer precise measurements of the relative contribution of NHEJ, MMEJ, and SSTR, combined with the throughput that is needed to query the impact of a wide diversity of chromatin contexts. The sequencing-based “scar-counting” readout renders the assay highly quantitative. Furthermore, because the same reporter sequence is integrated throughout the genome, differences in cutting and repair must be due to the local chromatin context of the integration sites.
The PiggyBac transposable element is not known to use host DNA repair factors (Mitra et al., 2008) that could bias its integration across the genome, but it shows a ~3-fold preference for transcriptionally active regions (Akhtar et al., 2013). Otherwise, integration of this element is thought to be largely random, and the large number of IPRs provides enough statistical power to compare all major chromatin types.
A key aspect of our analysis is the comparison of the relative pathway activities across chromatin types. For this we used pathway balance metrics that are intrinsically corrected for cutting efficiencies. Because these metrics are based on specific, well-characterized indels (−7, +1, and +2) and disregard the (relatively low abundant) other indels with uncertain pathway origin, these metrics should not be interpreted as definitive measures of relative pathway activities; however, they can be used to compare pathway balances between different chromatin contexts in the same cell pools.
Effects of chromatin context on DNA repair pathway preference
We found that the balance between MMEJ and NHEJ varies >5-fold across chromatin contexts. Generally, in heterochromatin we observe a higher MMEJ:NHEJ balance than in euchromatin, but this shift depends on the precise heterochromatin features that are present. A previous study of smaller scale (Kallimasioti-Pazi et al., 2018) found that the indel spectrum after a Cas9-induced cut was not affected by imprinted heterochromatin, a type of heterochromatin that we did not probe. This underscores the notion that different types of heterochromatin may have distinct effects on repair pathway balance.
Two classes of models may explain the chromatin effects that we observed. These models are not mutually exclusive. First, it is possible that euchromatin carries one or more features that activate or bind the NHEJ machinery, or conversely, that certain heterochromatin features promote MMEJ. If the latter is true, then it should be considered that multiple heterochromatin features can play such a role, as H3K27me3 modulates the MMEJ:NHEJ balance in H3K27me3-marked heterochromatin, but this does not explain the high MMEJ:NHEJ balance in triple heterochromatin. By ChIP we found that MRE11 is enriched at euchromatic DSBs relative to heterochromatic DSBs. MRE11 has been implicated in both MMEJ and NHEJ (reviewed in Reginato and Cejka, 2020), but perhaps its role is quantitatively more important for NHEJ than for MMEJ, which would cause a reduced MMEJ:NHEJ balance at euchromatic sites where it is more abundant.
A second class of models involves the differential accessibility of heterochromatin and euchromatin. By default (particularly in G1 phase), NHEJ may be globally more active than MMEJ. Hence, at a DSB in “open” euchromatin, NHEJ may mostly outcompete MMEJ. In contrast, in heterochromatin a DSB may be inaccessible to either pathway until the heterochromatin is de-compacted. This remodeling of heterochromatin may be a slow process, which would allow time for MMEJ to be activated and give both MMEJ and NHEJ a more similar chance to repair the DSB. Indeed, DSB-induced unfolding of heterochromatin has been reported (Chiolo et al., 2011; Goodarzi et al., 2008; Jakob et al., 2011; Janssen et al., 2016; Ryu et al., 2015; Tsouroula et al., 2016). However, as seen by microscopy, this remodeling process occurs within ~20 min, while we found that the MMEJ indels accumulate only after several hours. It is possible that additional biochemical or structural changes in heterochromatin are involved that take place over a timescale of hours. We also considered that one early cutting event in euchromatin may trigger slow upregulation of MMEJ activity globally throughout the nucleus, which would then increase the probability of MMEJ repairing a DSB that is formed later in heterochromatin (which may be cut more slowly). However, this explanation seems unlikely, because early breaks caused by ionizing radiation do not boost MMEJ repair at a Cas9 cut ~16 h later (Brinkman et al., 2018). This result suggests that the slow MMEJ activation does not occur globally throughout the nucleus but rather locally at the DSB. Further studies are needed to understand the different kinetics of MMEJ and NHEJ.
DSB repair pathways are known to vary in activity depending on the cell cycle stage (Chapman et al., 2012; Hustedt and Durocher, 2016; Mladenov et al., 2016). Our cell cultures were unsynchronized, and we assumed that all IPRs in the pools were subject to the same cell cycle stage distribution; thus, cell cycle effects should be averaged out, and the differences in indel spectra must be due to effects of chromatin context. In the future it will be interesting to explore whether any cross-talk exists between cell cycle and chromatin effects on pathway balance.
Comparison with previous studies
As summarized in the Introduction, previous studies have addressed the impact of specific chromatin contexts or proteins on NHEJ and HR but generally did not monitor MMEJ and SSTR. Although the latter two pathways share components with HR, they are mechanistically distinct from HR, and thus the effects of chromatin context may differ. For example, several previous studies have indicated that H3K36me3, which is generally present along active transcription units, promotes HR (Aymard et al., 2014; Carvalho et al., 2014; Clouaire and Legube, 2015; Pfister et al., 2014). We find H3K36me3 to correlate negatively with MMEJ. Possibly, HR and MMEJ respond differently to H3K36me3, or the repair of Cas9-induced breaks differs from breaks induced by other means.
The activities of MMEJ and SSTR that we detect indicate that end-resection is generally not impeded by various types of heterochromatin. Previous work has pointed to a role of HP1α and HP1β in tethering proteins involved in end-resection (Soria and Almouzni, 2013), but not all types of heterochromatin are marked by these proteins. Our data indicate that multiple heterochromatin types can create an environment that is more prone to be repaired by MMEJ. This includes LADs, which is in agreement with a previous study that implicated various MMEJ-specific proteins in the repair of DSBs near the nuclear lamina (Lemaître et al., 2014).
Practical implications for genome editing
The results obtained in this study have practical implications for genome editing by means of Cas9. First, the efficiency of Cas9 editing is generally lower in most types of heterochromatin compared with euchromatin. This has been noted before but on the basis of data that covered only a small number of loci that did not compare all heterochromatin types (Chen et al., 2016; Daer et al., 2017; Jensen et al., 2017; Kallimasioti-Pazi et al., 2018). Our data indicate that Cas9 editing is primarily suppressed in triple heterochromatin. Most likely the relatively low accessibility of the DNA in these loci is preventing efficient cutting by Cas9. Regions that carry only one of these marks, or H3K27me3, show only a modestly reduced editing efficiency.
From a genome editing perspective, the skew toward the MMEJ and SSTR pathways in heterochromatin is a convenient partial compensation for the lower overall editing efficiency, because MMEJ and SSTR are generally more useful than NHEJ to generate specific types of mutations. MMEJ is better suited to generate frameshifts and deletions that can result in functional KO of genes, while SSTR is particularly useful to generate specifically designed mutations. Maps of heterochromatin features are thus useful resources to choose the optimal target loci for CRISPR-Cas9 editing, particularly when combined with algorithms that predict editing outcomes on the basis of sequence (Allen et al., 2018; Chakrabarti et al., 2019; Chen et al., 2019; Shen et al., 2018; van Overbeek et al., 2016).
Multiplexed DSB repair reporters: outlook
This work complements a recent study that used a multiplexed reporter for DNA mismatch repair, which did not reveal significant effects of chromatin context on the repair outcome (Pokusaeva et al., 2019). Another multiplexed integrated reporter study also found evidence that genomic location can affect Cas9 editing efficiency (Gisler et al., 2019), but these results were more difficult to interpret because the reporter sequence itself was not transcriptionally inert. Importantly, neither of these studies addressed the impact of chromatin context on the balance between specific DSB repair pathways. Multiplexed reporter assays provide new opportunities to systematically investigate the role of local chromatin context in DSB repair by multiple pathways. Moreover, our time-series experiments demonstrate that the assay can be performed in 96-well format, making it scalable for applications such as drug screens and CRISPR screens. In the future, the assay may also be modified to include the detection of DSB resection or other intermediates, computational modeling of time series to infer perfect repair (Brinkman et al., 2018), and perhaps measurements of HR activity.
Limitations of study
By using the repair scar as a readout, the method is limited to detecting mutagenic repair. Perfect repair (e.g., by HR or NHEJ) is therefore untraceable. HR might never be measurable with the current setup. However, perfect repair by NHEJ might be if the time-series approach quantifiable by mathematical modeling of time-series data (Figure 5; Brinkman et al., 2018) is improved. Related to this, our degron control of Cas9 activity had a slow response. Given the repeated cutting and repair, it is very difficult to know what the actual rates are in our TRIP setting. Emerging Cas9 inhibitors (Kundert et al., 2019; Maji et al., 2019) and photocleavable (Carlson-Stevermer et al., 2020; Zou et al., 2021) and photoactivatable (Liu et al., 2020) guide RNAs (gRNAs) might provide help overcome this limitation by having a tighter control of Cas9 activity.
Although our data contain much more information in the various other indels, we limited our analysis to the main mutations produced by our gRNA for clarity. A more thorough analysis is feasible to better dissect the smaller nuances in DNA repair. We have also only looked at spacing for the indel size and ignored more complex mutations. The Tn5 approach (Figures S2J–S2M) can be optimized and used for better understanding of the balance between more complex mutations and their local chromatin environment. Finally, it is possible that the repair of Cas9-induced DSBs is different from the repair of DSBs generated by other agents. It will therefore be interesting to apply our approach to other sequence-specific endonucleases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-H3K4me1 (ChIP) | Abcam | Cat# ab8895; RRID: AB_306847 |
| Anti-H3K27me3 (ChIP) | Active Motif | Cat# 39155; RRID: AB_2561020 |
| Anti-H3K27ac (ChIP) | Active Motif | Cat# 39133; RRID: AB_2561016 |
| Anti-POLQ (ChIP) | Sigma-Aldrich | Cat# SAB1402530.; RRID: AB_10639636 |
| Anti-LIG4 (ChIP) | GeneTex | Cat# GTX55592; RRID: AB_2887931 |
| Anti-RAD51 (ChIP) | Santa Cruz Biotechnology | Cat# sc-8349; RRID: AB_2253533 |
| Anti-MRE11 (ChIP) | Novus | Cat# NB100-142; RRID: AB_10077796 |
| Anti-LMNB2 (pA-DamID) | Abcam | Cat# ab8983; RRID: AB_306912 |
| Anti-Mouse (pA-DamID) | Abcam | Cat# ab6709; RRID: AB_956006 |
| Anti-H3K27me3 (Western Blot) | Cell Signaling | Cat# C36B11; RRID: AB_11220433 |
| Anti-H3K9me2 (Western Blot) | Millipore | Cat# 07-441; RRID: AB_11212297 |
| Anti-LBR (Western Blot) | Abcam | Cat# ab122919; RRID: AB_10902156 |
| Anti-LMNA (Western Blot) | Santa Cruz Biotechnology | Cat# sc-376248; RRID: AB_10991536 |
| Bacterial and virus strains | ||
| CloneCatcher DH5α electrocompetent E. coli | Genlantis | Cat# C810111 |
| JM109 Competent Cells | Promega | Cat# L2001 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 | GIBCO | Cat#: 21875034 |
| Fetal Bovine Serum | Sigma | Cat#: F7524 |
| Penicillin-Streptomycin (##10,000 U/mL) | GIBCO | Cat#: 15070063 |
| Lipofectamine 2000 | Invitrogen | Cat#: 11668019 |
| Tn5 enzyme | Luca Braccioli & this study | N/A |
| PEG 8000x | Sigma | Cat#: P1458 |
| TAPS-NAOH | Sigma | Cat#: T5130 |
| dimethylformamide | Sigma | Cat#: D4551 |
| Shield-1 | Aeobius | Cat#: AOB1848 |
| Phusion® Hot Start Flex DNA Polymerase | New England BioLabs | Cat#: M0535L |
| Phusion® HF DNA Polymerase | New England BioLabs | Cat#: M0530L |
| MyTac Red Mix 2x | Bioline | Cat#: BIO-25044 |
| CleanPCR | CleanNA | Cat#: CPCR-0500 |
| Shrimp Alkaline Phosphatase (1U/μl) | New England Biolabs | Cat#: M0371S |
| Exonuclease I | New England Biolabs | Cat#: M0293S |
| RNase-Free DNase Set | QIAGEN | Cat#: 79254 |
| SensiFast no-ROX | Bioline | Cat#: BIO-86050 |
| ATP Solution (100 mM) | Thermo Scientific | Cat#: R0441 |
| DMSO | Sigma | Cat#: D4540 |
| NU7441 | Cayman | Cat#: 14881 |
| M3814 | MCE | Cat#: HY-101570 |
| GSK126 | Cayman | Cat#: 15415 |
| BIX01294 | Sigma | Cat#: B9311 |
| Lipofectamine RNAiMAX Transfection Reagent | Invitrogen | Cat#: 13778150 |
| Opti-MEM | GIBCO | Cat#: 31985047 |
| DirectPCR® Lysis | Viagen | Cat#: 302-C |
| Proteinase K | Bioline | Cat#: BIO-37084 |
| MyTaq HS Red mix | Bioline | Cat#: BIO-25048 |
| CellTiter-Blue® Cell Viability Assay | Promega | Cat#: G8080 |
| spermidine | Sigma | Cat#: S0266 |
| digitonin | Millipore | Cat#: 300410 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#: 11873580001 |
| SAM | New England BioLabs | Cat#: B9003S |
| Dam | New England BioLabs | Cat#: #M0222L |
| Critical commercial assays | ||
| ISOLATE II Genomic DNA kit | Bioline | Cat#: BIO-52067 |
| PCR Isolate II PCR and Gel Kit | Bioline | Cat#: BIO-52060 |
| Tetro Reverse Transcriptase | Bioline | Cat#: BIO-65050 |
| PureLink HiPure Plasmid Midiprep Kit | Invritrogen | Cat#: K210004 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat#: Q32854 |
| RNeasy Mini Kit | QIAGEN | Cat#: 74104 |
| Deposited data | ||
| Raw data | This study | SRA: PRJNA686952 |
| Processed data | This study | https://osf.io/cywxd/ |
| Code | This study | GitHub: https://github.com/vansteensellab/DSB_repair_TRIP |
| Unprocessed images | This study | https://osf.io/cywxd/ |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/human |
| Experimental models: cell lines | ||
| K562#17 ddCas9 | Brinkman et al., 2018 | N/A |
| Oligonucleotides | ||
| Tables S3 and S4 | This study | N/A |
| Rad51 siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-003530-00-0005 |
| PolQ siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-015180-01-0005 |
| BRCA1 siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-003461-00-0005 |
| BRCA2 siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-003462-00-0005 |
| Lig4 siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-004254-00-0005 |
| RBBP8 siRNA ON-TARGETplus Smart Pool | Dharmacon/Horizon | Cat#: L-011376-00-0005 |
| Oligo(dT)20 | Invitrogen | Cat#: 18418020 |
| Recombinant DNA | ||
| pPTK-Gal4-tet-Off-Puro-IRES-eGFP-sNRP-pA-trim1 | Akhtar et al., 2014 | GenBank: KC710229 |
| pPTK-P.CMV.584-eGFP-trim1-PI04 | Alexey Pindyurin and Waseem Akhtar | N/A |
| pBlue-sgRNA | Brinkman et al., 2018 | N/A |
| pPTK-BC-IPR | This study | GenBank: MW408732 |
| mPB-L3-ERT2-mCherry | Akhtar et al., 2014 | N/A |
| Software and algorithms | ||
| Bowtie2 v2.3.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools v1.5 | Li et al., 2009 | RRID:SCR_002105; https://www.htslib.org/ |
| Cutadapt v1.9.1 | Martin, 2011 | RRID:SCR_011841; https://cutadapt.readthedocs.io/en/stable/ |
| Starcode v1.1 | Zorita et al., 2015 | https://github.com/gui11aume/starcode |
| TagMeppr | This paper | https://github.com/robinweide/tagmeppr |
| Sambamba v0.6.6 | Tarasov et al., 2015 | https://lomereiter.github.io/sambamba/ |
| deeptools v3.3.1 | Ramírez et al., 2016 | RRID:SCR_016366; https://deeptools.readthedocs.io/en/develop/ |
| HMMt | Guillaume Filion | https://github.com/gui11aume/HMMt |
| BBTools v38.86 | Bushnell et al., 2017 | RRID:SCR_016968; https://sourceforge.net/projects/bbmap/ |
| FASTX-Toolkit v0.0.14 | Hannon Lab | RRID:SCR_005534; http://hannonlab.cshl.edu/fastx_toolkit/ |
| Bedtools v2.26.0 | Quinlan and Hall, 2010 | RRID:SCR_006646; https://github.com/arq5x/bedtools2 |
| BWA MEM | Heng Li arXiv:1303.3997v2 [q-bio.GN] |
https://github.com/lh3/bwa |
| GreyListChIP | Brown, 2020 | https://doi.org/doi:10.18129/B9.bioc.GreyListChIP |
| chipseq-greylist | Rory Kirchner | https://github.com/roryk/chipseq-greylist |
| Custom code for this study | This paper | https://github.com/vansteensellab/DSB_repair_TRIP |
| R code for this study | This paper | https://github.com/vansteensellab/DSB_repair_TRIP |
| RStudio Server Version 1.3.1073 | RStudio Team, 2020 | https://rstudio.com/ |
| R version 3.6.3 (2020-02-29) | R Core Team, 2020 | https://www.r-project.org/ |
| ggplot2 | Wickham, 2016 | https://ggplot2.tidyverse.org |
| CHOPCHOP | Labun et al., 2019 | https://chopchop.cbu.uib.no/ |
| inDelphi | Shen et al., 2018 | https://indelphi.giffordlab.mit.edu/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Bas van Steensel (b.v.steensel@nki.nl).
Material availability
Plasmids and cell lines generated in this study are available without restriction upon request from the Lead Contact.
Data and code availability
Processed data, script outputs, and original images are available at https://osf.io/cywxd/. Raw sequencing data are available at the Sequence Read Archive: PRJNA686952. Code: https://github.com/vansteensellab/DSB_repair_TRIP; https://github.com/robinweide/tagmeppr
Experimental model and subject details
Cell culture
We used clonal cell line K562#17, which is a female human K562 cell line (ATCC) stably expressing DD-Cas9 (Brinkman et al., 2018). K562#17 cells were cultured in RPMI 1640 (GIBCO) supplemented with 10% fetal bovine serum (FBS, Sigma), 1% penicillin/streptomycin. Cells were free of mycoplasma according to tests performed every 1-2 months.
Method details
Constructs
The pPTK-BC-IPR (PiggyBac) construct was derived from the pPTK-Gal4-tet-Off-Puro-IRES-eGFP-sNRP-pA-trim1 plasmid (GenBank: KC710229). The enhanced green fluorescent protein (eGFP) expression transcription unit including promoter, the puromycin resistance cassette (PuroR) and the internal ribosome entry site (IRES) were replaced with the sgRNA-LBR2 target sequence and its flanking region. This plasmid did contain a point mutation which was removed by restriction cloning using a derivative of plasmid with a shortened 3′ITR of 67 bp (pPTK-P.CMV.584-eGFP-trim1-PI04 – kindly provided by Alexey Pindyurin and Waseem Akhtar). The target sequence was obtained by annealing ODS001 and ODS002 (400 pM each) (for primer sequences see Table S4) in 50 μl MyTaq Red mix followed by 5 cycles of PCR. This PCR product was then further amplified with TAC0001 and TAC0002 (50 pM each). This sequence was then inserted in the PB backbone by restriction cloning with NheI and KpnI. This construct (IPR-PB) was then used to make the barcoded plasmid libraries, the 3′-ITR of PiggyBac was amplified with primers TAC0003 (containing a 16 nucleotide random barcode) and TAC0004. The PCR product was digested with KpnI and BssHII, ligated into the KpnI and MluI sites of the IPR-PB plasmid and transformed into CloneCatcher DH5α electrocompetent E. coli (Genlantis). A pool of ~500,000 transformed bacterial cells were grown, and plasmids were purified, resulting in the pPTK-BC-IPR (GenBank: MW408732) plasmid library. The PB transposase expression vector (mPB-L3-ERT2-mCherry) is described in (Akhtar et al., 2014). The sgRNAs were designed using CHOPCHOP (Labun et al., 2019; Montague et al., 2014) and cloned into expression vector pBlue-sgRNA (Brinkman et al., 2018) (see Table S3). All the plasmids were extracted and purified using the PureLink HiPure Plasmid Midiprep Kit (Invitrogen). The vectors for the sgRNA sequences are listed in Table S3.
Generation of IPR cell pools
Cell pools carrying IPRs were produced as described (Akhtar et al., 2014). Briefly, K562#17 cells were transfected with 32 μg of barcoded pPTK-BC-IPR plasmid library and 6 μg of PB transposase plasmid using Lipofectamine 2000 (ThermoFisher). Mock-transfected (without PB transposase) and GFP plasmid controls were included. After 24 h, the cells were sorted by fluorescence-activated cell sorting (FACS) based on mCherry signals. We discarded cells without any detectable mCherry signal because they most likely failed to take up any plasmid. 0.5 μM of 4-hydroxytamoxifen (4-OHT) was added to the samples to activate the transposase. Sixteen hours later the cells were washed to remove 4-OHT. After sorting, the population was grown for 8 days to clear the cells from free plasmid. Then, the mCherry negative cells were FACS sorted in aliquots of ~2000 cells, which were expanded to establish two cell pools, each with a different collection of IPRs. We also isolated single cells to make clonal TRIP lines originating from pool B, including clone 5.
Cloning, expression and purification of Tn5
The gene encoding Tn5 was cloned into the pETNKI-his-SUMO3-LIC vector, containing a N-terminal 6xHis-SUMO3-tag, using Ligation Independent Cloning (LIC) (Luna-Vargas et al., 2011). The recombinant Tn5 protein was expressed in Rosetta2(DE3) cells in 3 l of LB medium, supplemented with 30 μg/ml kanamycin and 40 μg/ml chloramphenicol. Cells were grown at 37°C to OD600 = 0.6-0.8. Cells were cooled to 18°C before 0.4 mM IPTG was added and protein was expressed overnight. Cells were harvested by centrifugation (3,000 g, 15 min, 20°C) and pellet was stored at −20°C until further use.
For protein purification, the pellet was resuspended in lysis buffer (20 mM HEPES pH 7.5, 800 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM TCEP) containing 0.2% Triton X-100. Cells were lysed by sonication and the lysate was clarified by centrifugation (50,000 g, 30 min, 4°C). Polyethylenimine (0.1% w/v) was added dropwise to the supernatant, incubated for 30 min and the precipitate was removed by centrifugation (50,000 g, 30 min, 4°C). The soluble fraction was used for affinity purification (1 mL nickel Sepharose Excel, GE healthcare). Beads were washed with lysis buffer, containing 20 mM imidazole, and protein was eluted by 200 mM imidazole in lysis buffer. Fractions were analyzed by SDS-PAGE. To remove the 6xHis-SUMO3-tag, pooled elution fractions were incubated with his-Senp2 protease, followed by overnight dialysis at 4°C against 20 mM HEPES pH 7.5, 800 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM TCEP. After reverse affinity purification using 1 mL nickel Sepharose excel, the 6xHis-SUMO3-tag was found to be efficiently cleaved off from Tn5. The flow-through and wash fractions were pooled and concentrated to 1 ml. The protein was further purified by size exclusion chromatography on a SEC 650 10/300 column (Bio-Rad), equilibrated with 20 mM HEPES pH 7.5, 800 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM TCEP. Fractions containing dimeric Tn5 were pooled, concentrated and flash frozen in liquid nitrogen before storing at −80°C.
Tagmentation-based mapping of IPRs in the clones
Not all the IPRs were mapped in the clones by iPCR. They were additionally mapped using a Tn5 transposon based IPR mapping technique based on Stern (2017). In brief, PCR-products covering both the up- and downstream junctions between the genome and the inverted terminal inverted repeats (ITRs) were obtained by designing divergent internal primers. The library preparation is carried out as follows. 45 μl of 100 μM of TAC0101 & TAC0102 each were mixed with 10 μl 10x TE and annealed using the following PCR reaction; 10 min at 95°C, 1 min at 90°C, followed by a slow ramp down (0.1°C/sec) until 4°C. The transposome is obtained by combining the adapters (1 μl of 1:2 diluted adapters) and the Tn5 transposon (1.5 μl of 2.7 mg/mL stock) in 18.7 μl Tn5 dilution buffer (20 mM HEPES, 500 mM NaCl, 25% Glycerol) and incubating the mix for 1 hour at 37°C. The DNA tagmentation was performed by mixing 100 ng of gDNA with 1 μl of transposome, 4 μl 5x TAPS-PEG buffer (50mM TAPS-NAOH, 25mM MgCl2, 8% vol/vol PEG8000) in a final volume of 20 μl. This was incubated at 55°C for 10 minutes and quenched afterward with 4ul of 0.2% SDS. Library preparation was as follows. Both sides of the PiggyBac transposon were processed for the best mapping results by generating 3′ ITR and 5′ITR libraries. First, we enriched our target region with a linear enrichment PCR amplification using TAC0006 (3′ ITR) and TAC0099 (5′ ITR). The PCR mix was 3 μl of tagmented DNA, 1 μl of 1 μM primer, 2 μl dNTPs (10mM), 4 μl 5x Phusion® HF Buffer (Promega), 0.25 Phusion® HS Flex polymerase (2 U/μl - Promega), in a final volume of 20 μl and amplified as follows; 30 s at 98°C, 45 cycles of 10 s at 98°C, 20 s at 62°C and 30 s at 72°C. PCR1 of the library preparation was done with TAC0161 (3′ITR) and TAC0110 (5′ ITR) in combination with N5xx (Nextera Index Kit – Illumina). The PCR mix was 5 μl of enrichment PCR, 1 μl of 10 μM primers, 2 μl dNTPs (10mM), 4 μl 5x Phusion® HF Buffer and 0.25 Phusion® HS Flex polymerase, in a final volume of 25 μl and amplified as follows; 30 s at 98°C, 3 cycles of 10 s at 98°C, 20 s at 62°C and 30 s at 72°C, 8 cycles of 10 s at 98°C, 50 s at 72°C. PCR2 of the library preparation was done with TAC0103 (both ITRs) and N7xx (Nextera Index Kit – Illumina). The PCR mix was 2 μl of PCR1, 1 μl of 10 μM primers, 2 μl dNTPs (10mM), 4 μl 5x Phusion® HF Buffer and 0.25 Phusion® polymerase (Promega), in a final volume of 22 μl and amplified as follows; 30 s at 98°C, 10 cycles of 10 s at 98°C, 20 s at 63°C and 30 s at 72°C. The library was then checked and purified with bead purification as for the indel libraries, quantitated with Qubit and sequenced on a Miseq (150 bp, paired-end).
Tagmentation-based indel and rearrangement detection
To assess potential rearrangements and very large deletions in our setup, we have adapted the protocol to map IPRs (above) to read out the IPR barcode and mutations. As input, we used three control replicates (clone 5) from the LBR & LMNA knock out experiments (Figure 6B) that were transfected either with the LBR2 sgRNA or an empty vector. The protocol follows the same steps as the IPR mapping except that we used an improved tagmentation buffer and the PCR primers are different. Instead of 5x TAPS-PEG buffer we used 10 ul of 2x TD buffer (20 mM Tris, 10mM MgCl2, adjusted to pH 7.6 with 100% acetic acid before the addition of 20% (vol/vol) dimethylformamide). For the library PCR we first enriched the IPR with TAC0078. PCR1 was carried out with TAC0238 (primer F on Figure S2J) & Nextera N5xx. PCR2 was unchanged.
Transfection of sgRNA plasmids and ssODN
For transient transfection of the sgRNAs, 1 to 6 x106 cells (lower limit for clonal experiments, higher limit for pooled experiments) were resuspended in transfection buffer (100 mM KH2PO4, 15 mM NaHCO3, 12 mM MgCl2, 8 mM ATP, 2 mM glucose (pH 7.4)) (Hendel et al., 2014). After addition of 3.0-9.0 μg plasmid, the cells were electroporated in an Amaxa 2D Nucleofector using program T-016. DD-Cas9 was induced directly for ~16 hours after transfection with a final concentration of 500 nM Shield-1 (Aobious). For uncut controls we transfected either a GFP containing plasmid or pBlue-sgRNA vector without a sgRNA sequence. To probe SSTR, 3-9 μg sgRNA was co-transfected with 1.5-4.5 μg ssODN (5′ TAGAATGCTAGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTAATTTCTACTTCATAATAAAGTGAACTCCCAGGCCATCGACATCTCTTACCACTTCACCATCGGCAAATTTCCTACTTGGCATT 3′, Ultramer grade, IDT). The specific mutation that disrupts the PAM is underlined.
LMNA & LBR knock out generation
The guides for LBR and LMNA knock outs were selected using a combination of inDelphi (Shen et al., 2018) and CHOPCHOP (Labun et al., 2019; Montague et al., 2014) of optimal frameshift chances and efficiency. One million clone 5 cells were transfected with 3 μg of plasmid expressing the following sgRNAs per clone: LMNA_KO1 & LMNA_KO2 (each 1.5 μg) for LMNA KO 1 and 2; LMNA_KO4 for LMNA KO 3 and 4; LBR_KO1 for LBR KO 1-4 (Table S3), and cultured in complete RPMI medium with 500 nM Shield-1 (to activate Cas9) for 3 days. To obtain individual clones, cells were plated in two 96-well plates by limiting dilution (2 cells per ml; 100 μl per well). Each clone was then tested by TIDE (Brinkman et al., 2014) for frameshifts in all alleles (primers in Table S4). For each sgRNA we selected two clones with complete frameshifts for further experiments.
TIDE method
Prior to high throughput sequencing, all the samples were checked for general transfection and cutting efficiency by TIDE. For this we used the primers TAC0017 and TAC0018, that cover the endogenous LBR locus and the TIDE method was performed as described in Brinkman et al. (2014). Briefly, PCR reactions were carried out with ~100 ng genomic DNA in MyTaq Red mix (Bioline) and purified using the PCR Isolate II PCR and Gel Kit (Bioline) or by ExoSAP (for primers see Table S4). ExoSAP was done by adding 0.125 μl Shrimp Alkaline Phosphatase (1U/μl; New England Biolabs), 0.0125 μl Exonuclease I (20 U/μl; New England Biolabs) and 2.3625 μl H2O per 10 μl PCR reaction. Samples were incubated 30 minutes at 37°C and inactivated for 10 minutes at 95°C. About 2 μl (50-100 ng) of purified PCR product was then subjected to Sanger Sequencing by Eurofins Genomics. The sequence traces were analyzed using the TIDE analysis tool (https://tide.nki.nl).
siRNA lipofection
All siRNAs were obtained from Dharmacon as ON-TARGETplus Smartpool and transfected on 1 million cells with the RNAiMAX Transfection kit (Invitrogen) at a final concentration of 25 nM, 24h prior to sgRNA electroporation according to the manufacturer’s protocol for 6 well plates. Samples were partially collected 24h after electroporation for subsequent RNA isolation, reverse transcription and qPCR analysis. The rest was left to grow for 48 more hours CRISPR/Cas9 editing analysis.
RT-qPCR
Cells were collected in RLT buffer with 1% 2-mercaptoethanol and stored at −80C until processed. Total RNA was extracted using RNeasy mini kit (QIAGEN) with on-column DNase treatment (QIAGEN) according to the manufacturer’s instructions. The RNA was eluted in 30 ul RNase-free H2O and quantitated by Nanodrop. RNA was reverse-transcribed using Tetro Reverse Transcriptase (Bioline) with random hexamers and Oligo(dT)20 primers (Invitrogen) according to the manufacturer’s instructions. The RT reaction was diluted 10x and 4ul RT was used for the qPCR reaction. qPCR was performed using SensiFast no-ROX mix (Bioline) in a 10 μL reaction. Melt curves after each PCR and all samples yielded a single peak. Gene-specific primers were obtained from PrimerBank (Wang et al., 2012), see Table S4. Data were normalized to the levels of TBP.
Western blots
Whole-cell extracts of ~0.5x106 cells were prepared by washing cultures in PBS and lysing with 50 μL lysis buffer (Tris pH 7.6, 10% SDS, Roche cOmplete Protease Inhibitor Cocktail). Western blotting was performed according to standard procedures using the following antibodies and dilutions: H3K27me3 (1:1000 Cell Signaling C36B11, rabbit), H3K9me2 (1:1000 Millipore 07-441, rabbit), LMNA (1:800 Santa Cruz Biotechnology sc-376248, mouse), LBR (1:1000, Abcam, ab122919, rabbit).
Cell viability
Cells transfected with Rad51 siRNA were tested for cell viability using the Cell Titer Blue 48 hours after lipofection. After mixing, 100 ul cells were plated in a 96-well opaque-walled tissue culture plates plus 20μl/well of CellTiter-Blue® Reagent (Promega), in 3 technical replicates per sample. The cells were briefly shaken and incubated for 3 hours in at cell culturing conditions. The fluorescence was measured by a Perkin Elmer EnVision plate reader. The three technical replicates were averaged, then the biological replicates were averaged, and the result was normalized of the non-targeting siRNA control. One sample t test was used for the statistics.
Inhibitor treatments
DNA-PKcs inhibitor NU7441 (Cayman; diluted 1:1000 from 1 mM stock in dimethylsulfoxide [DMSO]), M3814 (MedChemExpress; diluted 1:1000 from 1 mM stock in DMSO), GSK126 (Selleckchem; diluted 1:2000 from 1 mM stock in DMSO), BIX01294 (Sigma; diluted 1:1000 from 1 mM stock in H2O), or respective solvent-only controls at equal volumes, was added to the cells at the same time when the cells were supplemented with Shield-1 to induce DD-Cas9 or 24 hours prior to nucleofection for GSK126 and BIX01294. DMSO was also present in the experiments in Figures 3, 4, 5, 6A, 7, S2A–S2F, S3A, S3B, S4, S5, and S7.
pA-DamID
pA-DamID maps were generated and processed as described (van Schaik et al., 2020). Briefly, 1 million cells were collected by centrifugation (3 minutes, 500 g) and washed in ice-cold PBS and subsequently in ice-cold digitonin wash buffer (DigWash) (20 mM HEPES-KOH pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.02% digitonin, cOmplete Protease Inhibitor Cocktail). Cells were resuspended in 200 μL DigWash with 1:100 mouse Lamin B2 antibody (Abcam, ab8983) and rotated for 2 hours at 4°C, followed by a wash step with 0.5 mL DigWash buffer. This was repeated with a 1:100 mouse anti-rabbit antibody (Abcam, ab6709) and 1 hour of rotation, and afterward with 1:100 pA-Dam (~60 NEB units). After two washes with DigWash, cells were resuspended in 100 μL DigWash supplemented with 80 μM SAM to activate Dam and incubated for 30 minutes at 37°C. Genomic DNA was extracted using the ISOLATE II Genomic DNA kit (Bioline cat. no. BIO-52067) and DNA was processed for high-throughput sequencing similar to conventual DamID (Leemans et al., 2019; Vogel et al., 2007), except that the DpnII digestion was omitted. To control for DNA accessibility and amplification bias, 1 million permeabilized cells (without any antibodies bound) were incubated with 4 units of Dam enzyme (NEB, M0222L) during the activation step. This sample functions as “dam-control” over which a log2-ratio is determined. Log2-ratios were converted to z-scores to account for small differences in dynamic range between experiments.
Generation of indel sequencing libraries
After 64 or 72 (for clone 5) hours incubation, the cells were collected, and genomic DNA was extracted using the ISOLATE II Genomic DNA kit (Bioline cat. no. BIO-52067). PCR was performed in two steps and pooled experiments were performed in triplicates for a higher coverage. IndelPCR1 was performed with 200 ng genomic DNA each using primers TAC0007 (indexed) and TAC0012 that amplify 1 bp upstream of the barcode 46 bp downstream of the cut-site (see Figure 1A; Table S4). indelPCR2 used 2 μl of each indelPCR1 product with TAC0009 and either TAC0011 (non-indexed) or TAC0159 (indexed). Each sample was generated with a unique combination of one or two indexes. Both PCR reactions were carried out with 25 μl MyTaq Red mix (Bioline cat. no. BIO-25044), 0.5 μM of each primer and 50 μl final volume. PCR conditions for both steps were 1 min at 95°C, followed by 15 s at 95°C, 15 s at 58°C and 1 min at 72°C (5x), followed by 15 s at 95°C, 15 s at 65°C and 1 min at 72°C (10x). The indelPCR2 was pooled per experiment after quantification on a 1% agarose gel and cleaned up using CleanPCR (CleanNA) beads at 0.8:1 beads:sample ratio. 5 μl PCR product was run on a 1% agarose gel to check for remaining primer dimers and if required reloaded on a 1% agarose gel and cut out to remove remaining primer dimers and cleaned with PCR Isolate II PCR and Gel Kit (Bioline). The libraries were quantitated using the Qubit DNA dsHS Assay Kit. The purified libraries were sequenced on an Illumina HiSeq2500 or MiSeq (150bp, singe-end) depending on the expected complexity of the library.
Time series
Sample collection in the time series experiments was done automatically using a Hamilton Microlab® STAR equipped with a Cytomat 2 C450 incubator. One million cells were transfected as described above with the sgLBR2. After 16 hours, 40,000 cells in 100 μl medium were seeded per well in 96-well plates. The automated system then added 100 μl RPMI medium (with 1 μM Shield-1) to each well, for a final 500 nM Shield-1 concentration. The first time point was directly collected and required a brief centrifugation step (10 s at 500 g) to precipitate the cells, before returning the cell culture plate to the robot. Then for each time point 170 μl medium was removed from the well and discarded, the left-over was mixed and transferred to a new 96-well PCR plate at 8°C. Each newly collected well was then filled with 50 μl of DirectPCR® Lysis (Viagen) buffer with 1 mg/ml Proteinase K (Bioline) to pre-lyse the cells. The cell culture plate was returned to the incubator and every 3 hours a new time point was collected as described above. One 96-well plate included 4 timeseries of each 24 time points. After 69 hours the collection of samples was finished, and the cell lysates were sealed and incubated for 3 hours at 55°C and heat-inactivated for 10 minutes at 95°C.
Library preparation for the timeseries was very similar to the pool experiments except for indelPCR1. 20 μl of crude lysate was used in a total PCR volume of 80 μl, with 40 μl MyTaq HS Red mix (Bioline) and 0.5 μM of each primer. PCR cycles were as described above.
Chromatin immunoprecipitation analysis of IPRs
Chromatin immunoprecipitation was performed as described (Schmidt et al., 2009). Main steps and modifications are described here. 50 μL protein A Dynabeads were precleared with 0.5% BSA, 5 μL of specific antibody (H3K4me1[Abcam]; H3K27me3 [Active Motif]; H3K27ac [Active Motif], MRE11 [Novus]; DNA ligase IV [Genetex]; Rad51 [Santa Cruz Biotechnology];DNA polymerase theta [Sigma]) and beads incubated at 4°C overnight. 10 million clone 5 cells (uncalibrated experiments) and 3 million cells (calibrated experiments) were fixed at a final concentration of 1% formaldehyde for 10 minutes. Fixation was quenched with 125nM glycine for 5min and a PBS wash. Equal amounts of clone 9 cells were fixed and quenched for calibrated experiments. Spike-in cells (clone 9) were mixed in a 1:1 ratio with clone 5 cells after fixation. After nuclear extraction, chromatin was sonicated (~10 cycles 30 s on/ 30 s off in BioRuptor Pico), Triton X-100 added to a final concentration of 1% and centrifuged to remove cell debris. Antibody coupled beads were washed with 0.5% BSA in PBS, chromatin was added (5% was kept as input) and rotated overnight at 4°C. Beads were washed 10 times with RIPA buffer and once with TBS. After last wash, 200 μL of elution buffer was added and samples eluted and de-crosslinked at 65°C for 6 hours or overnight. 200 μL of TE buffer and 0.9 μL of 10mg/ml RNase A was added the samples and were incubated at 37°C for 1 hour and with 4 μL of 20mg/ml Proteinase K at 55°C for 2 hours. DNA was extracted by phenol:chloroform extraction and resuspended in 50 μL of 10mM Tris-HCl. IPR barcodes were collectively amplified using two step PCR. For indelPCR1, 100ng DNA was taken from input samples and same input volume added from pull-downs. PCR1 was performed in a final volume of 50 μL with 25 μL of MyTaq HS Red mix and 0.5 μM of each primer (indelPCR: TAC00012 and TAC0007 or bcPCR: TAC0162 and TAC0007). 5 μL of PCR1 was taken for PCR2 with 25 μL of MyTaq Red Mix and 0.5 μM of each primer (TAC0009 and TAC0159) for 12 PCR cycles (3 cycles with 58°C annealing followed by 9 cycles with 65°C annealing). PCR products were pooled, purified and quantitated as described above, and sequenced on an Illumina MiSeq.
For calibrated ChIP, clone 5 IPR barcode counts were normalized by clone 9 IPRs (spike-in clone). Each sample was normalized for library size (total clone 9 barcode counts) and input counts. Then, clone 9 read counts were used to normalize every sample to a reference sample. Normalized barcode counts were used for further plotting and analysis.
Quantification and statistical analysis
The data generated by the indel scoring was further processed and analyzed in R (R Core Team, 2020) with Rstudio (RStudio Team, 2020), the figures were generated using ggplot2 (Wickham, 2016). The main packages and software are listed in the key resource table and an extensive bibliography is available in all the scripts (github & OSF for markdowns). Statistical details for individual experiments have been provided in the main text, figure legends, and Method Details, as well as in the available markdown documents on OSF.
Mapping of IPR integration sites by inverse PCR
Mapping of IPR integration sites was performed in two replicates by inverse PCR (iPCR) followed by 2 × 150 bp paired end sequencing on an Illumina HiSeq2500 as previously described (Akhtar et al., 2014). Linking of IPR barcodes to the integration sites was adapted from Akhtar et al. (2013). Reads of both replicates were pooled. The first read in each read pair was used to extract the barcode. This was done using the ‘GTCACAAGGGCCGGCCACAAC’ constant sequence followed by a regular expression ‘TCGAG[ACGT]{16}TGATC’. From the sequence matching this regular expression, the 16 bp barcode was extracted. To identify barcodes arising from mutations during PCR and sequencing, starcode v1.1 (Zorita et al., 2015) was used with the sphere clustering setting and a maximum Levenshtein distance of 2. The second read of each pair was used to locate the site of integration after removing the ‘GTACGTCACAATATGATTATCTTTCTAGGGTTAA’ sequence matching the transposon arm. The flanking sequence was aligned to GRCh38 using bowtie2 (Langmead and Salzberg, 2012) using the very-sensitive-local option (20 seed extension attempts, up to 3 re-seed attempts for repetitive seeds, 0 mismatches per seed, with a seed-length of 20 and using a multi-seed function: . Locations of integration sites were required to be supported by at least 5 reads with an average mapping quality larger than 10 at the primary location, having at least 95% of the reads located at this locus, with not more than 2.5% of the reads at a secondary location.
TagMeppR
To infer the integration sites, we created a software package (TagMeppR) specially designed to map, identify and visualize tagmentation mapping reads. This R-package is available at https://github.com/robinweide/tagmeppr. TagMeppR enables the creation of a hybrid reference genome made up of a full genome with additional two pseudo-chromosomes, consisting of the two PiggyBac ITRs. Next, the align-tool enables the mapping of read-pairs from both indexes separately with BWA MEM (arXiv:1303.3997v2 [q-bio.GN] - https://github.com/lh3/bwa). After mapping, suspected PCR-duplicate reads are removed with SAMtools rmdup (Li et al., 2009). For each putative IPR, TagMeppR will check if (1) reads from both indexes are enriched and (2) whether the read-densities are on opposite sides of the IPR. To assess the latter, we perform a binomial test on reads up- and downstream of the IPR. Next, we filter out IPRs in which both sets are biased on the same side of the IPR. We compute an IPR-specific p value with the conservative Edgington’s sum-p method and compute the family-wise error rate. TagMeppR also enables the visualization of single insertions and genome-wide insertion maps.
The two IPRs that were mapped only with Tagmeppr were confirmed by PCR (with TAC0065 & TAC0128 for IPR5 [chr20:18569153]; TAC0065 & TAC0126 (Table S4) for IPR8 [chr7:13259711] followed by Sanger sequencing (see TIDE method) to identify the barcodes associated to them.
Indel scoring
Indel reads after induction and repair of DSBs consist of single-end reads of 150 bp that span both the DSB site and the barcode. Indel scoring was adapted from Brinkman et al. (2018). Barcodes were extracted from the reads with an in-house script using functions of cutadapt 1.11 (Martin, 2011). The 16 bp barcode was located using the 20 bp constant ‘GTCACAAGGGCCGGCCACAA’ sequence preceding the barcode and ‘TGATCGGT’ expected immediately after the barcode. For the 20 bp constant sequence, 2 mismatches were allowed.
To determine the indel size in each read, we used the distance (number of nucleotides) between two fixed sequences at the start and at the end of the read. The indel size was calculated as the difference between the measured distance and the expected distance based on the wild-type sequence. We used the following anchor sequences: before the break site, ‘TGATCGGT’ and after the break site, ‘TGGCCTT’, ‘GGAGTT’, ‘CACTTT’, ‘ATTATG’, ‘GAAGTA’, ‘ATTAGA’ and ‘GGAAGA’. The most proximal match found with the selection of these sequences found after the break site of the specific guide was used to calculate the indel size by subtracting the expected location from the observed location. Insertions and deletions have indel sizes > 0 and < 0, respectively. Wild-type sequence is defined as indel size 0. Point mutations were not analyzed. Per replicate experiment we observed a mean 17.3% (95% CI: 16.7%–17.8%) sequence reads in which we could not find a match with the constant parts; we discarded these reads in subsequent analyses. Potentially these represent large deletions, complex mutations, sequencing errors or a combination thereof.
Per barcode, the reads of all technical replicates were pooled if applicable. Mutated barcodes were included or discarded as described above for the mapping of IPR integrations. Because in the cell pools not all IPRs are equally represented (the cell pools consist of a mix of clones that each carry different IPRs, and some cell clones grow faster than others), we then discarded IPRs that were too underrepresented to provide reliable data. Specifically, we required that each IPR is represented by at least 50 cells among the ~100,000 cells that were used in each experiment. We assumed an average of 6 IPRs per cell. Accordingly, the number of total reads per IPR was divided by the library size and multiplied by 6 ∗ 100,000 to obtain the estimated number of cells for each IPR. IPRs for which this score was > 50 were used for subsequent analyses. Then each replicate was normalized over library size and biological replicates were averaged. The frequency of each indel type as proportion of total reads was calculated on that average. Pathway frequency per IPR was calculated as a proportion of the specific mutation over all indels (excluding wild-type sequences).
For LBR2, 7bp deletions were classified as MMEJ, and 1bp insertions were classified as NHEJ. For the other guides, a slightly more complicated approach was used. For these guides all specific deletions with a microhomology at the site of deletion of 2 or more nucleotides were classified as MMEJ. Indels with an indel ratio of at least 0.01 in the DMSO setting and a significant decrease (adjusted p value < 0.05) in the M3814 inhibitor setting (one-sided Wilcoxon test) were classified as NHEJ.
Tagmentation to identify rearrangements
Similar methods were used to identify the barcode of the IPR for each read pair, similar to the iPCR mapping. After the barcode was identified, the sequence before the targeted break-site was removed using fastx_trimmer (http://hannonlab.cshl.edu/fastx_toolkit/). Only read pairs containing a barcode found in the pool were kept. The mate pair starting from the Tn5 part was scanned for the sequence of the Tn5 constant part and this sequence was removed whenever there was a match. The mate pair starting at the barcode was also scanned for the reverse complement of the Tn5 constant part, reads containing that fragment were discarded. The mate pairs were then tidied up using BBTools (Bushnell et al., 2017). The parts of both mates that were left were aligned to an in silico engineered genome in which transposon sequence was added at the previously identified sites of integration. The resulting bam file was converted to a bed file with the start and end of the complete fragment using bedtools (Quinlan and Hall, 2010) and awk (Aho et al., 1992). This bed file was used to identify overlap with each in silico engineered transposon site. The fragments with overlap of the transposon were used to quantify the small indels using the same method as for the general indel scoring, except with additional anchor sequences after the break site (TCTGA, CTAGC, GTTGA, TCTAT, AAGTT, AGAAC, TCGTA, AAGTC, TGACT, AGTGA, ACGCC, AGCTC, TGCAC, GAAAG, TGCAT, ACGCA, GGGTT). Fragments without any overlap with any of the transposon sites were used to identify putative transposition and big deletion events (> 480bp) (Figure S2J, 3ii). For the unique event counts of these fragments, PCR duplicates were removed by grouping all fragments with the exact same start (break site) and end (tagmentation adaptor) position, or 1 nucleotide difference on each side. The most abundant barcode found at these duplicates was assigned and the start of the mate pair beginning from the IPR was used as the putative side of translocation/big deletion event. To identify putative rearrangements/big deletions with the native LBR locus, we looked at the overlap of the putative rearrangement side and the gene annotation of the LBR gene. We were not able to identify rearrangements between IPRs for two reasons. First, only a fraction of the fragments was long enough to span outside of the IPR (517 bp from TAC0238 (Table S4) to the end of the transposon, see Figure S2J). Second, we detected a lot of PCR template switching between IPRs most likely due to the repetitive nature of the transposon ITR. This means that read pair 1 would pick up a specific barcode (starting from TAC0238) and read pair 2 from the Tn5 adaptor end would pick up genomic sites that is linked with another IPR. These events were detected in similar quantities in both cut and uncut setting, indicating that this is most likely a PCR artifact, and not a biological rearrangement after a DSB.
We estimated the chance of having rearrangements between IPRs to be < 1% by looking at the rearrangements with the cut-site in the native LBR locus. Simulations showed us that the chance of IPRs landing within 1Mb from each other are very small. In this simulation we assumed equal probability of landing in every 1Mb region of a fully triploid human genome. This resulted in an average 1.2 barcodes, meaning between 0 or one pair of barcodes landing within 1Mb of each other in our pools of 2000 cells. Considering this contribution to be minor we assumed rearrangements in trans between IPRs and the 4 LBR alleles could be used to calculate an estimate of rearrangements between IPRs. The ratio calculated for all rearrangements outside of the IPRs was multiplied by the ratio of LBR trans rearrangements. This was divided by the number of endogenous LBR alleles in K562 (4) and multiplied by the average of 6.8 IPRs per cell. This resulted in an estimate of 0.39% rearrangements between IPRs in trans. We therefore concluded that, together with very rare occurrence of IPRs being located in cis and within 1Mb of each other, the total percentage of rearrangements between IPRs is probably below 1%.
Preprocessing of previously published chromatin data
Published ChIP-seq data from various sources (Table S1) were re-processed for consistency. Raw sequencing data were obtained from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/). Reads were aligned to the human genome GRCh38 using bowtie2 with default options. Replicate datasets were processed separately, while the sequences from the input were combined. After alignment, reads were filtered on a minimum mapping quality of 30 (Tarasov et al., 2015). Duplicate reads were removed except for the reads coming from experiments using tagmentation, in which duplicates were kept. After this, genomic regions were masked based on blacklist regions identified by the ENCODE project (ENCFF419RSJ) (ENCODE Project Consortium, 2012) and putative artifact regions were identified based on the input reads using chipseq-greylist (https://github.com/roryk/chipseq-greylist) (Brown, 2020), a python implementation of GreyListChIPs. We considered ChIP-seq datasets to be of sufficient quality for our analyses if there was well annotated input and sample data available and consistent read lengths were used. Mean ChIP-seq signals for IPR integration sites were calculated by taking the sum of the reads in a region of 2 kb around the IPR using deeptools2 (Ramírez et al., 2016), scaling input and sample counts by the smallest library size, adding a pseudo count of 1 and subsequently dividing sample over input normalized counts. After this, replicate experiments were averaged. For domain calling, ChIP-seq signals were calculated in similar fashion for bins of 5kb. HMMt (https://github.com/gui11aume/HMMt), an R package implementing a Hidden Markov model with t emission, was used to subsequently call domains.
DamID data of Lamin B1 are from Leemans et al. (2019). The DamID score was calculated by scaling counts to the smallest library size, adding a pseudocount of 1 and dividing over Dam-only. The normalized dam-only score was log2-transformed before averaging between replicates to calculate the dam accessibility score. Replication timing data was obtained from the 4DN data portal in the form of read coverage for late and early fraction separately. Counts were processed in the same way as for the ChIP data. For TTseq coverage from forward and reverse tracks were summed and the lowest coverage score above zero was used as pseudo count before log2-transforming and averaging between replicates. For DNase hypersensitivity data of both paired-end and single-end sequencing reactions were used from encode. Coverage tracks were used and for the single ended reaction a small pseudo count of half the minimum value above 0 was used before log2 transforming. Paired-end and single-end coverage was log2 transformed before averaging. Whole genome bisulfite sequencing tracks from encode were used and coverage was calculated and log2 transformed without the need for a pseudo count. Replicates were subsequently averaged. Data sources are available in Table S2.
Z-scores of above chromatin information for the clonal line was calculated by using the mean and standard deviation of the signals in the TRIP pool. For pA-DamID on the knock-out clones and clone 5, the scores were calculated in a window 10kb up and downstream of the IPR. Except for the different window size, pA-DamID scores were calculated similar to the overall DamID scores. Z-scores for pA-DamID were calculated using the mean and standard deviation of the pA-DamID score for 20kb binned tracks over the whole genome using the same formula as for the individual IPR’s.
Correction of ChIP signals at IPRs for differences in cutting frequencies
Across IPRs, differences in ChIP signals of repair proteins may be confounded by differences in DSB frequencies. We assume that in a population of cells, an IPR that is cut twice as often as another IPR also yields a ChIP signal that is twice as strong. On top of this simple relationship there may be quantitative effects of chromatin context on the binding of these proteins. Because we are primarily interested in the latter chromatin effects, we computationally corrected the ChIP signals for differences in cutting frequency as follows. 1. As an approximation of the cutting rates, we used the total indel frequency of each IPR after 72 hours, i.e., after essentially all breaks were repaired. 2. Because sgRNA-LBR exhibits a mild saturation of indel frequencies, we corrected for this. For this we correction we used the indel frequencies of sgRNAs LBR1, 12 and 15 (which do not show this saturation), using the loess fits of the pairwise scatterplots as shown in Figure 3C; we averaged the results of these three independent corrections, yielding a value TIFcor for each IPR. 3. We then fitted for each protein a model log2(ChIP) ~1∗log2(TIFcor) + b using the nls() function of R. This fit forces a slope of 1, modeling the above assumption. 4. The residuals of this fit were taken as the variation in ChIP signals due to chromatin context effects (now corrected for cutting efficiency) and are shown in Figure 4G.
Time series analyses
Indels in clone 5 were identified and counted as described above. Indel frequencies and MMEJ:NHEJ balance was calculated before averaging across replicates. Only IPRs mapped to a single genomic location were used (19 total). Sigmoid curves were fitted to time series data using the following formula:
Where t is time and a, b and c are parameters that determine the shape and plateau of the curve. For the decay of wild-type sequence over time, the ratio was fitted as 1-y. Fitting was done using the nls package in R. Starting values 20, 10 and 0.1 were used for fitting of the parameters a, b and c, respectively.
Acknowledgments
We thank the NKI Genomics, Flow Cytometry, and Research High Performance Computing core facilities for excellent support and members from our laboratories for inspiring and helpful discussions. We thank Luca Braccioli and the NKI Protein core facility for Tn5 protein. The NKI Protein Facility is an Instruct-ERIC center. This work was supported by ZonMW TOP grant 91215067 (to R.H.M. and B.v.S.), European Research Council (ERC) Advanced Grant 694466 (to B.v.S), and NIH Common Fund “4D Nucleome” Program grant U54DK107965 (B.v.S.). S.G.M. is funded by Marie Curie/AIRC iCARE2.0 fellowship 800924. The Oncode Institute is partly supported by KWF Dutch Cancer Society.
Author contributions
E.K.B., R.S., R.H.M., and B.v.S. conceived and designed the study. R.S., E.K.B., X.V., T.v.S., S.G.M., D.P.-H., B.M., and J.v.d.B. performed experiments. C.L., R.S., E.K.B., R.H.v.d.W., X.V., T.v.S., and S.G.M. performed data processing, bioinformatics, and figure preparation. B.v.S., R.H.M., and R.L.B. performed supervision and project management. R.S., E.K.B., and B.v.S. wrote the manuscript with input from all authors.
Declaration of interests
B.v.S. is a member of the Advisory Board of Molecular Cell.
Published: April 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2021.03.032.
Supplemental information
References
- Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Akhtar W., Pindyurin A.V., de Jong J., Pagie L., Ten Hoeve J., Berns A., Wessels L.F., van Steensel B., van Lohuizen M. Using TRIP for genome-wide position effect analysis in cultured cells. Nat. Protoc. 2014;9:1255–1281. doi: 10.1038/nprot.2014.072. [DOI] [PubMed] [Google Scholar]
- Alagoz M., Katsuki Y., Ogiwara H., Ogi T., Shibata A., Kakarougkas A., Jeggo P. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015;43:7931–7944. doi: 10.1093/nar/gkv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho A.V., Kernighan B.W., Weinberger P.J. The awk programming language. Comput. Hum. 1992;26:293–297. [Google Scholar]
- Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Palenikova P., Khodak A., Kiselev V., Kosicki M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4317. Published online November 27, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeyron C., Soria G., Roche D., Cook A.J., Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Chen L.C., Maynard-Smith L.A., Ooi A.G., Wandless T.J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell. 2018;70:801–813.e6. doi: 10.1016/j.molcel.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. R package version; 2020. GreyListChIP: Grey Lists – Mask Artefact Regions Based on ChIP Inputs. [Google Scholar]
- Bushnell B., Rood J., Singer E. BBMerge—accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson-Stevermer J., Kelso R., Kadina A., Joshi S., Rossi N., Walker J., Stoner R., Maures T. CRISPRoff enables spatio-temporal control of CRISPR editing. Nat. Commun. 2020;11:5041. doi: 10.1038/s41467-020-18853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S., Vítor A.C., Sridhara S.C., Martins F.B., Raposo A.C., Desterro J.M., Ferreira J., de Almeida S.F. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P. Target-specific precision of CRISPR-mediated genome editing. Mol. Cell. 2019;73:699–713.e6. doi: 10.1016/j.molcel.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.H., Yu A.M., McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen X., Rinsma M., Janssen J.M., Liu J., Maggio I., Gonçalves M.A. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44:6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang Y., Wang Y., Zhang L., Brinkman E.K., Adam S.A., Goldman R., van Steensel B., Ma J., Belmont A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018;217:4025–4048. doi: 10.1083/jcb.201807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., Noble W.S., Shendure J. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019;47:7989–8003. doi: 10.1093/nar/gkz487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T., Legube G. DNA double strand break repair pathway choice: a chromatin based decision? Nucleus. 2015;6:107–113. doi: 10.1080/19491034.2015.1010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T., Legube G. A snapshot on the cis chromatin response to DNA double-strand breaks. Trends Genet. 2019;35:330–345. doi: 10.1016/j.tig.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Clouaire T., Rocher V., Lashgari A., Arnould C., Aguirrebengoa M., Biernacka A., Skrzypczak M., Aymard F., Fongang B., Dojer N. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol. Cell. 2018;72:250–262.e6. doi: 10.1016/j.molcel.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney E.J., LeGros M.A., Mosley C.P., Clowney F.G., Markenskoff-Papadimitriou E.C., Myllys M., Barnea G., Larabell C.A., Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales M., Rosado A., Cortini R., van Arensbergen J., van Steensel B., Filion G.J. Clustering of Drosophila housekeeping promoters facilitates their expression. Genome Res. 2017;27:1153–1161. doi: 10.1101/gr.211433.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daer R.M., Cutts J.P., Brafman D.A., Haynes K.A. The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth. Biol. 2017;6:428–438. doi: 10.1021/acssynbio.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M., Baude A., Fugger K., Povlsen L.K., Beck H., Sørensen C.S., Petersen N.H., Sorensen P.H., Lukas C., Bartek J. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O’Shea C.C., Park P.J., Ren B., 4D Nucleome Network The 4D nucleome project. Nature. 2017;549:219–226. doi: 10.1038/nature23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M., Tome J.M., Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020;21:292–310. doi: 10.1038/s41576-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G., Ciulla D.M., Marco E., Abdulkerim H.S., Barrera L.A., Bothmer A., Dhanapal V., Gloskowski S.W., Jayaram H., Maeder M.L. UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics. 2018;19:212. doi: 10.1186/s12864-018-4561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler S., Gonçalves J.P., Akhtar W., de Jong J., Pindyurin A.V., Wessels L.F.A., van Lohuizen M. Multiplexed Cas9 targeting reveals genomic location effects and gRNA-based staggered breaks influencing mutation efficiency. Nat. Commun. 2019;10:1598. doi: 10.1038/s41467-019-09551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Gottlieb T.M., Jackson S.P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Hendel A., Kildebeck E.J., Fine E.J., Clark J., Punjya N., Sebastiano V., Bao G., Porteus M.H. Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep. 2014;7:293–305. doi: 10.1016/j.celrep.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N., Durocher D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- Iliakis G., Murmann T., Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;793:166–175. doi: 10.1016/j.mrgentox.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Jakob B., Splinter J., Conrad S., Voss K.O., Zink D., Durante M., Löbrich M., Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Breuer G.A., Brinkman E.K., van der Meulen A.I., Borden S.V., van Steensel B., Bindra R.S., LaRocque J.R., Karpen G.H. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 2016;30:1645–1657. doi: 10.1101/gad.283028.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Haber J.E. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst.) 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P.A., Downs J.A. Roles of chromatin remodellers in DNA double strand break repair. Exp. Cell Res. 2014;329:69–77. doi: 10.1016/j.yexcr.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Jensen K.T., Fløe L., Petersen T.S., Huang J., Xu F., Bolund L., Luo Y., Lin L. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591:1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
- Kallimasioti-Pazi E.M., Thelakkad Chathoth K., Taylor G.C., Meynert A., Ballinger T., Kelder M.J.E., Lalevée S., Sanli I., Feil R., Wood A.J. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 2018;16:e2005595. doi: 10.1371/journal.pbio.2005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousi A., Soutoglou E. Nuclear compartmentalization of DNA repair. Curr. Opin. Genet. Dev. 2016;37:148–157. doi: 10.1016/j.gde.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Kundert K., Lucas J.E., Watters K.E., Fellmann C., Ng A.H., Heineike B.M., Fitzsimmons C.M., Oakes B.L., Qu J., Prasad N. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 2019;10:2127. doi: 10.1038/s41467-019-09985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.H., Kuo C.Y., Stark J.M., Shih H.M., Ann D.K. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013;41:5784–5798. doi: 10.1093/nar/gkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans C., van der Zwalm M.C.H., Brueckner L., Comoglio F., van Schaik T., Pagie L., van Arensbergen J., van Steensel B. Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell. 2019;177:852–864.e14. doi: 10.1016/j.cell.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., Heyer V., Rogier M., Attwood K.M., Kessler P. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–2463. doi: 10.1101/gad.248369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zou R.S., He S., Nihongaki Y., Li X., Razavi S., Wu B., Ha T. Very fast CRISPR on demand. Science. 2020;368:1265–1269. doi: 10.1126/science.aay8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas M.P., Christodoulou E., Alfieri A., van Dijk W.J., Stadnik M., Hibbert R.G., Sahtoe D.D., Clerici M., Marco V.D., Littler D. Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J. Struct. Biol. 2011;175:113–119. doi: 10.1016/j.jsb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019;177:1067–1079.e19. doi: 10.1016/j.cell.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S.E. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R., Fain-Thornton J., Craig N.L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrentsi I., Yilmaz D., Soutoglou E. How to maintain the genome in nuclear space. Curr. Opin. Cell Biol. 2020;64:58–66. doi: 10.1016/j.ceb.2020.02.014. [DOI] [PubMed] [Google Scholar]
- Mladenov E., Magin S., Soni A., Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin. Cancer Biol. 2016;37–38:51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Amaishi Y., Maki I., Enoki T., Mineno J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019;9:4811. doi: 10.1038/s41598-019-41121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C.J., Federation A.J., Schwartz L.S., Kasar S., Klitgaard J.L., Lenci R., Li Q., Lawlor M., Fernandes S.M., Souza A. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018;34:982–995.e7. doi: 10.1016/j.ccell.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S.X., Ahrabi S., Zalmas L.P., Sarkar S., Aymard F., Bachrati C.Z., Helleday T., Legube G., La Thangue N.B., Porter A.C., Humphrey T.C. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva V.O., Diez A.R., Espinar L., Filion G.J. Strand asymmetry influences mismatch repair during single-strand annealing. bioRxiv. 2019 doi: 10.1101/847160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. https://www.R-project.org/ [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011;10:2549–2560. doi: 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato G., Cejka P. The MRE11 complex: a versatile toolkit for the repair of broken DNA. DNA Repair (Amst.) 2020;91–92:102869. doi: 10.1016/j.dnarep.2020.102869. [DOI] [PubMed] [Google Scholar]
- Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Richardson C.D., Kazane K.R., Feng S.J., Zelin E., Bray N.L., Schäfer A.J., Floor S.N., Corn J.E. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 2018;50:1132–1139. doi: 10.1038/s41588-018-0174-0. [DOI] [PubMed] [Google Scholar]
- Riesenberg S., Chintalapati M., Macak D., Kanis P., Maricic T., Pääbo S. Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res. 2019;47:e116. doi: 10.1093/nar/gkz669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . PBC; Boston, MA: 2020. RStudio: Integrated Development for R. RStudio.http://www.rstudio.com/ [Google Scholar]
- Ryu T., Spatola B., Delabaere L., Bowlin K., Hopp H., Kunitake R., Karpen G.H., Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg A.C., Harris-Becker A., Popova E.Y., Keasey N., Loughran T.P., Claxton D.F., Grigoryev S.A. Genome-wide mapping of histone H3K9me2 in acute myeloid leukemia reveals large chromosomal domains associated with massive gene silencing and sites of genome instability. PLoS ONE. 2017;12:e0173723. doi: 10.1371/journal.pone.0173723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Wilson M.D., Spyrou C., Brown G.D., Hadfield J., Odom D.T. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B., Michel M., Zacher B., Frühauf K., Demel C., Tresch A., Gagneur J., Cramer P. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- Scully R., Panday A., Elango R., Willis N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R.N., Grzybowski A.T., Cornett E.M., Johnstone A.L., Dickson B.M., Boone B.A., Cheek M.A., Cowles M.W., Maryanski D., Meiners M.J. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol. Cell. 2018;72:162–177.e7. doi: 10.1016/j.molcel.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Soria G., Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle. 2013;12:422–429. doi: 10.4161/cc.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.L. Tagmentation-based mapping (TagMap) of mobile DNA genomic insertion sites. bioRxiv. 2017 doi: 10.1101/037762. [DOI] [Google Scholar]
- Sun Y., Jiang X., Xu Y., Ayrapetov M.K., Moreau L.A., Whetstine J.R., Price B.D. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K., Furst A., Rogier M., Heyer V., Maglott-Roth A., Ferrand A., Reina-San-Martin B., Soutoglou E. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell. 2016;63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- van Schaik T., Vos M., Peric-Hupkes D., Hn Celie P., van Steensel B. Cell cycle dynamics of lamina-associated DNA. EMBO Rep. 2020;21:e50636. doi: 10.15252/embr.202050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Wang X., Spandidos A., Wang H., Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Yeh C.D., Richardson C.D., Corn J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019;21:1468–1478. doi: 10.1038/s41556-019-0425-z. [DOI] [PubMed] [Google Scholar]
- Zorita E., Cuscó P., Filion G.J. Starcode: sequence clustering based on all-pairs search. Bioinformatics. 2015;31:1913–1919. doi: 10.1093/bioinformatics/btv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou R.S., Liu Y., Wu B., Ha T. Cas9 deactivation with photocleavable guide RNAs. Mol. Cell. 2021 doi: 10.1016/j.molcel.2021.02.007. Published online February 23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed data, script outputs, and original images are available at https://osf.io/cywxd/. Raw sequencing data are available at the Sequence Read Archive: PRJNA686952. Code: https://github.com/vansteensellab/DSB_repair_TRIP; https://github.com/robinweide/tagmeppr