Figure 8.
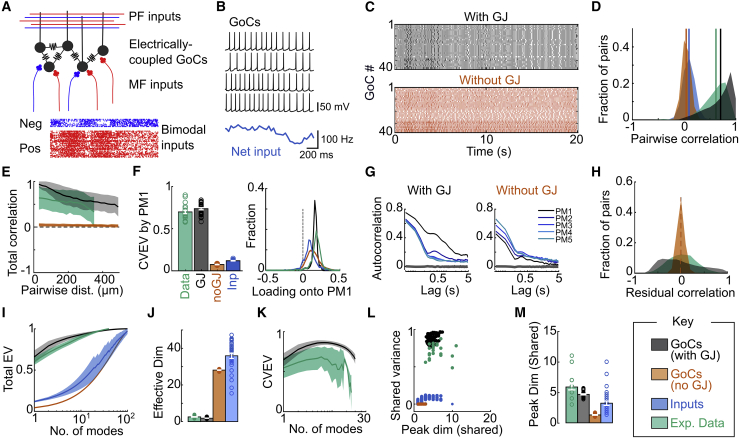
Biologically detailed GoC circuit model requires electrical coupling to reproduce multidimensional network activity with a common mode
(A) Schematic of GoC circuit model, with gap junctions (GJs, resistor symbols) between cells and positively (Pos, red) and negatively (Neg, blue) modulated input spike trains targeted to excitatory synapses on apical dendrites (PF, parallel fiber inputs) and soma (MF, mossy fiber inputs) of model GoCs.
(B) Example of somatic membrane potentials from model GoCs and net input rate.
(C) Raster plots of spiking activity from 40 GoCs with GJs (top, black) and without GJs (orange, bottom).
(D) Distribution of pairwise correlations between GoCs within local circuits with electrical coupling (black) and without electrical coupling (orange) together with correlations between inputs (blue) across varying input levels. The distribution of correlations observed experimentally with in vivo calcium imaging is shown in green.
(E) Dependence of total pairwise correlation on distance between model GoC somata and in experimental data.
(F) CVEV by PM1 (left) and (right) distribution of loadings for PM1, as measured experimentally (data), and for model circuits with (GJ) and without (noGJ) electrical coupling, together with PM1 of the simulated inputs (Inp).
(G) Autocorrelation of PM1–5 for modeled GoCs with (left) and without (right) GJs, as average profile across simulations with different input levels. The gray shaded area indicates autocorrelation of shuffled activity.
(H) Distribution of residual correlations (after projecting out PM1) across all input levels.
(I) Relationship between total explained variance (EV) and number of modes.
(J) Effective dimensionality (based on I) of population activity of GoCs and inputs.
(K) Relationship between CVEV and number of modes, for experimental data (green) and modeled GoCs with GJs (black). Bold lines are means across sessions and shaded regions SDs.
(L) Maximal CVEV (shared variance) versus number of modes at peak CVEV (shared dimensionality).
(M) Dimensionality of shared subspace: number of modes corresponding to maximal CVEV for each simulation or experimental session.
(D–M) Key for color labels. For all relevant panels, scatter denotes different network simulations (with a given input level) or experimental sessions.
See also Figures S9 and S10.