Abstract
LC-HRMS-based nontarget screening (NTS) has become the method of choice to monitor organic micropollutants (OMPs) in drinking water and its sources. OMPs are identified by matching experimental fragmentation (MS2) spectra with library or in silico-predicted spectra. This requires informative experimental spectra and prioritization to reduce feature numbers, currently performed post data acquisition. Here, we propose a different prioritization strategy to ensure high-quality MS2 spectra for OMPs that pose an environmental or human health risk. This online prioritization triggers MS2 events based on detection of suspect list entries or isotopic patterns in the full scan or an additional MS2 event based on fragment ion(s)/patterns detected in a first MS2 spectrum. Triggers were determined using cheminformatics; potentially toxic compounds were selected based on the presence of structural alerts, in silico-fragmented, and recurring fragments and mass shifts characteristic for a given structural alert identified. After MS acquisition parameter optimization, performance of the online prioritization was experimentally examined. Triggered methods led to increased percentages of MS2 spectra and additional MS2 spectra for compounds with a structural alert. Application to surface water samples resulted in additional MS2 spectra of potentially toxic compounds, facilitating more confident identification and emphasizing the method’s potential to improve monitoring studies.
Introduction
Organic Micropollutants in Water
Issues with water quality occur worldwide due to the large spread of the human population and their extensive use of chemicals, which leads to chemical pollution in a large number of water systems.1 These systems cause distribution of the pollution with long-range effects, ultimately posing a threat to drinking water sources.2−4 Various types of organic micropollutants (OMPs), that is, anthropogenic chemicals that are present at trace levels (μg/L), have been detected in ground and surface waters used for drinking water production. These include OMPs such as pesticides, pharmaceuticals, and industrial and consumer products. Despite their low concentrations, OMPs can pose a risk to human and environmental health as they can be toxic, persistent or easily degraded into more toxic (bio)transformation products.5 Compounds that pose a potential health risk need to be monitored to be able to assess the actual risks. Monitoring is typically performed using quantitative target analyses. As target analyses are limited to a set of known compounds, nontarget screening (NTS) based on liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) is often applied to monitor chemical water quality more comprehensively and broaden contaminant discovery.6,7
NTS-Based Micropollutant Identification
The structural identification of unknown compounds from NTS data remains challenging due to the large number of signals detected per experiment—typically referred to as features (accurate mass and retention time pairs associated with a signal intensity), and the need for high quality fragmentation spectra.8,9 The latter facilitates identification through spectral matching, where experimental spectra are compared to library spectra or in silico-predicted spectra. Software tools can connect the experimentally obtained mass spectrum with candidate structures from various sources.10−14
Prioritization
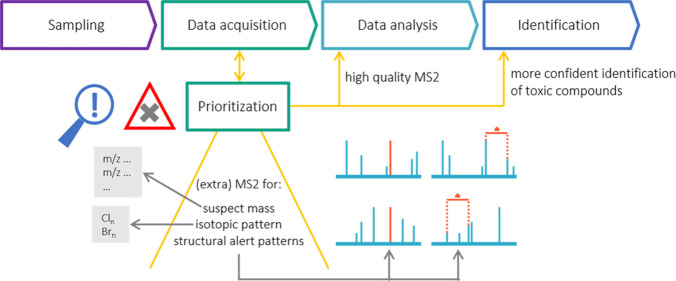
To limit the features that need to be identified, prioritization can be applied by selecting peaks of interest based on intensity, occurrence, persistence, or potential toxicity.9,15 Prioritization is currently performed offline during data analysis (Figure S1a). This entails that the structure of prioritized features without fragmentation spectrum or with uninformative, low-quality fragmentation spectra cannot be identified in a sufficiently confident manner. Instead, the sample has to be reanalyzed to obtain high-quality fragmentation spectra requiring more measurement time and resulting in delayed identification. Here, we hypothesize that the high costs and laboriousness of NTS offline prioritization could be remedied by using online prioritization for potentially toxic compounds in the mass spectrometer during data acquisition (Figure S1b).
Structural Alerts
Toxic compounds often comprise one or more structural alerts, that is, molecular (sub)structures related to the toxicity of a chemical. Several databases and software programs have been developed to derive and screen molecules for the presence of a structural alert, such as ToxAlerts,16 DEREK,17 and MultiCASE.18 Structural alerts can be specific for a toxic end point, that is, a measured biological effect in a toxicity test.19 Most are derived from the end points carcinogenicity and mutagenicity, with several lists published,20−23 including a revised list by Benigni and Bossa24 of 33 structural alerts included in the ToxAlerts database. Other water relevant toxic end points are examined less extensively, but some structural alerts were available in ToxAlerts for genotoxicity, endocrine disruption, and developmental toxicity.
Intelligent Acquisition
Structural alerts could be used for the “rough” selection of potentially toxic compounds that need to be identified in NTS methods. To this end, fragment ion masses and/or patterns indicating the presence of one or more structural alerts could be used as an MS trigger for further fragmentation events. In addition, suspect lists of toxic compounds and isotopic patterns suggesting anthropogenic origin of a compound were used to prompt a fragmentation event. This novel combination leads to an intelligent acquisition method, which would thereby prioritize (potentially) toxic compounds in contrast to the currently used data-dependent acquisition (DDA) that selects features using only the intensity in MS1 scans as selection criteria for fragmentation.
Overview
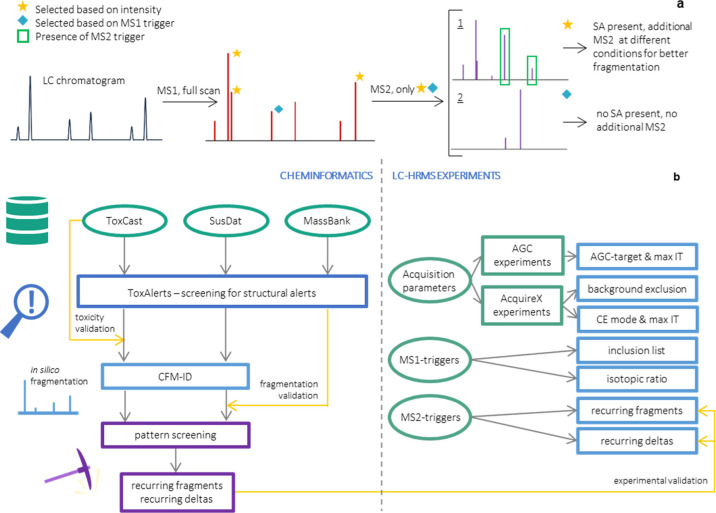
Here, we developed an intelligent acquisition method that utilizes online prioritization of potentially toxic compounds circumventing reanalysis of the sample due to lacking (high-quality) fragmentation spectra of features that are prioritized post-analysis (Figure 1). Cheminformatics were applied to determine triggers for (additional) MS2 events to be used in the LC-HRMS method. MS1-triggers exploited accurate mass and isotopic ratios detected in the full scan MS1 spectrum that suggested potential toxicity. MS2-triggers were based on fragment ion masses and/or patterns detected in the MS2 spectrum and linked to the presence of a structural alert. To this end, in silico fragmentation predictors were used to predict fragmentation of molecules with a structural alert and screen these spectra for patterns. The derived triggers were experimentally evaluated with LC-HRMS experiments. Finally, the developed method including MS1- and MS2-triggers was compared to a regular NTS method to evaluate whether the prioritization was successful.
Figure 1.
(A) Schematic representation of the proposed LC-HRMS/MS workflow using intelligent acquisition based on structural alerts (SAs). A full MS1 scan is taken after chromatographic separation, and the peaks are screened for their intensity and the presence of MS1-triggers (blue diamond marker). The most intense peaks (based on DDA-approach, yellow star marker) and those that contain a MS1-trigger are selected for a MS2 scan. The MS2 scans are screened for MS2-triggers, indicating the presence of a SA, resulting in two possible scenarios: 1. SA is present, so an additional MS2 scan at different conditions is taken. 2. No SA is present, so structure identification is not necessary, and no additional MS2 is taken. B) Schematic overview of the strategy that was used to develop the intelligent acquisition method, both cheminformatics (left) and LC-HRMS experiments (right) were applied.
Methods and Materials
Screening of Compounds for Structural Alerts
The detailed workflow for the screening and fragmentation of the ToxCast13 data set is given in S2.1. First, the CAS registry numbers of the 9224 compounds registered in the ToxCast data file Chemical_Summary_190708.csv25 were converted into MS-ready SMILES using the CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard).12 MS-ready SMILES are defined as structural representations that are observed in HRMS.26 Not all CAS registry numbers could be converted, and some lead to the same MS-ready SMILES, resulting in 7571 unique MS-ready SMILES. In addition to ToxCast entries (n = 7571), the MS-ready SMILES of the two databases NORMAN MassBank11 (n = 2304), and NORMAN SusDat14 (n = 65,697) were screened for structural alerts. NORMAN MassBank is a subset of MassBank Europe (https://massbank.eu) containing the majority environmental contributors. The compounds in the NORMAN MassBank are also included in NORMAN SusDat; however, MassBank contains fragmentation data and this was used for validation purposes. In the case of MassBank, only the 1903 compounds having available positive ionization HCD data were screened as this ionization mode was later used in the LC-HRMS experiments. Regarding SusDat, compounds were filtered for those with an EPISuite predicted log Kow value between −2.5 and +3.5 (provided in SusDat), resulting in 46,688 compounds. This filtering step was applied to eliminate compounds that are not detectable by RPLC.
Four toxic endpoints were selected for screening with ToxAlerts: “endocrine disruption” (EDC), “nongenotoxic carcinogenicity” (NGC), “genotoxic carcinogenicity, mutagenicity” (GCM), and “developmental and mitochondrial toxicity” (DMT). These end points and their corresponding 187 structural alerts were chosen based on their relevance for drinking water and potential human health risk. The endocrine disruption alerts belonged to both estrogenic and androgenic endocrine disruptors.27 This selection was made based on in vitro and in vivo (mammalian) data.
The output of ToxAlerts was formatted in R28 (version 3.6.1 (2019-07-05)) for fragmentation with CFM-ID. A text file was generated per structural alert containing the InChIKey and SMILES code as input for CFM-ID 2.0.
Validation
ToxCast assays relevant for the end points that were linked to the structural alerts were selected based on literature.9 These assays are listed in Table S1. The AC50 values of the ToxCast compounds with an alert were obtained from “ac50_Matrix190708.csv” (downloaded at 04 December 2019).29 In this file, inactive compounds are given an AC50 value of 1 × 106. Lower values indicate that the compound is active. Per toxic end point, that is, EDC, DMT, NGC, and GCM, the percentage of molecules with both a structural alert and activity in one of the specified assays was calculated. This percentage was compared to the percentage of active compounds for the total ToxCast data set, irrespective of the presence of a structural alert.
In ToxCast, MS-ready SMILES can occur multiple times but with a different DSSTox Substance identifier and in some cases, varying toxicity information. The toxicity validation was based on the DSSTox Substance ID to include all bioassay results for the same MS-ready SMILES and prevent information loss.
In silico Fragmentation
Compounds with a structural alert were in silico-fragmented with the combinatorial fragmentation predictor CFM-ID 2.0 using single energy competitive fragmentation modeling (SE-CFM) in the command line. The main reason for using CFM-ID is that it can be accessed in batch mode. CFM-ID includes assumptions of the fragmentation process such as that the molecule needs to carry a single positive charge, removal or addition of sigma bonds during a break is not allowed, and the valence and even electron rules must be satisfied in all fragments.30 Note that here, in silico fragmentation was not used for subsequent fragment matching but to predict spectra and screen these for patterns.
The command-line utility cfm-predict.exe31 was used to generate fragments with CFM-ID 2.0; the standard trained CFM model and its standard configuration parameters were used (S2.1). The postprocessing option was not included, and the probability threshold was set to 0.001 (default setting). The program output consisted of three lists containing m/z values and corresponding intensities for low energy CID (10 V), medium energy CID (20 V), and high energy CID (40 V). These energies reflect the type of spectra the model is based on. CFM-ID is based on CID QTOF data, which are comparable to HCD data from an Orbitrap instrument. The output was processed in R.
Validation
The in silico-predicted fragmentation results of SusDat generated with CFM-ID were validated with experimental data obtained from NORMAN MassBank.11 MassBank data was available for 2.25% of the 26,081 fragmented molecules with an alert from SusDat. The overlap in percentage of MassBank and CFM-ID fragments was calculated using eqs 1 and 2 to account for the differing total number of MassBank and CFM-ID fragments per spectrum. Since experimental data are also prone to errors, the output of these calculations must be considered as approximations.
| 1 |
| 2 |
Pattern Screening
The in silico-predicted fragmentation spectra of compounds with a structural alert were screened for characteristic patterns, that is, recurring fragment masses and recurring mass shifts (deltas). All structural alerts which were found in more than four molecules were included in the analysis. The CFM-ID data set was screened, with the control set being the in silico-predicted MS2 spectra of all molecules for each fragmentation method. To be able to compare the effect of the three CFM-ID energy levels on the recurring fragments and deltas, an intensity threshold was set at a minimum of 5% of the maximum peak intensity (100). The energy levels had an effect on the signal intensity only and not on the type of predicted fragments. Setting this threshold led to elimination of low-intensity fragments, resulting in different fragmentation spectra for the energy levels.
The frequencies of each m/z value and delta recurring within the MS2 spectra of the molecules of one structural alert were calculated and compared to the frequencies in the total fragmented data set. An extra control step for the frequencies was performed to show the difference in frequencies between a random sample and alerts. A random set of compounds (n = 3953) from NORMAN SusDat (ntotal = 65,697) that had not been screened for structural alerts was fragmented with CFM-ID. The frequencies of recurring fragment masses and recurring deltas within this random sample were then compared to the frequencies within MS2 spectra of compounds with structural alerts derived from ToxCast.
HRMS Method Development
Sample Preparation
The chemicals used in this study are listed in Tables S2–S8. An internal standard mixture of atrazine-d5 (CDN isotopes, Pointe-Claire, Canada) and benzotriazole-d4 (LGC Standards, Wesen, Germany) was added to each sample to a final concentration of 1 μg/L. Surface water (SW) (Lekkanaal, the Netherlands) and wastewater treatment plant (WWTP) influent samples, with and without spike-in (see Tables S2–S8) were filtered using 0.2 μm Phenex-RC 15 mm Syringe Filters (Phenomenex, Torrance, USA) prior to analysis. The WWTP-influent samples were 10 times diluted after spike-in and prior to filtration. The blanks used for these analyses were filtered as well. The spiking solution with water-relevant contaminants (see Table S2) was added to the samples to final concentrations of 10 μg/L, 1 μg/L, 100 ng/L, 10 ng/L, and 1 ng/L.
MS1-Trigger Experiments
Inclusion lists for MS1-trigger experiments (SusDat,14 SusDat + tR,14 UBAPMT,32 Sjerps,33 and Spike) were retrieved from the NORMAN Suspect List Exchange (https://www.norman-network.com/?q=suspect-list-exchange) and an in-house database and filtered for organic compounds within the full scan mass range (80 to 1000 Da) and polarity amenable to RP-HPLC, that is, log KOW between −2.5 and +3.5 (see the calculation method described in S2.3).
Based on the distribution of the number of chlorine and bromine atoms in the compounds registered in the CompTox Chemicals dashboard (n = 869,027),34 the isotopic ratios covering ≥99% of the chlorinated compounds (n = 128,650) and brominated compounds (n = 53,258) were used for the MS1-triggers. The isotopic ratios of Cl up to Cl6 and Br up to Br5 were calculated with the software Xcalibur (Thermo Fisher Scientific, San Jose, USA) and are shown in Table S9. The inclusion lists and the isotopic ratio trigger were tested separately and combined. The design of the resulting acquisition decision trees is shown in Figure S2. The methods were evaluated using surface water and WWTP-influent samples spiked with water-relevant contaminants; see Table S2.
MS2-Trigger Experiments
The performance of four different MS2-triggers, that is, two recurring deltas and two recurring fragments, was evaluated using ultrapure water samples spiked with compounds (Tables S3–S8) predicted to exhibit these fragments or deltas in their MS2 spectra based on the in silico experiments. Due to in-house availability of chemicals, only four different MS2-triggers were tested. The spike-in compounds were also added to surface water at concentrations ranging from 1 ng/L to 10 μg/L to determine sensitivity of the triggers. The MS2-trigger experiments were performed separately, together, and combined with the MS1-triggers using isotopic ratios and the Sjerps inclusion list. Detection of an MS2-trigger led to an additional MS2 event using alternative collision energies (CEs), that is, stepped CE (10, 75, 90) or assisted CE (20, 35, 50, 75, 90), or longer ITs, that is, stepped CE (20, 35, 50) with 200 ms IT instead of the regular 50 ms. These alternative fragmentation events were hypothesized to result in spectra with complementary fragments in the case of alternative energies, and higher-quality spectra in the case of longer ITs. The 11 different methods are described in Table S10 and the design of their decision trees in Figure S3. The experimental data obtained with the MS2-trigger experiments were used to validate the in silico-predicted fragmentation spectra and the pattern screening.
Data Analysis
The details of the data analysis are reported in S2.4 and S2.5. Data preprocessing and compound annotation were performed using Compound Discoverer 3.1 (Thermo Fisher Scientific, San Jose, USA). Further processing was done in R. Spectrum similarity scores were calculated using the function SpectrumSimilarity() from the R-package OrgMassSpecR (version 0.5–3).35 Fragment annotation was performed with the R-package metfRag (version 2.4.2)36 using the function frag.generateMatchingFragments() on the centroided MS2-spectra, using default settings. The spectrum similarity scores and number and percentage of annotated fragments and percentage of the annotated peak area were used to gain insights into the quality of the fragmentation spectra acquired with different acquisition settings.
Results and Discussion
Screening of Compounds for Structural Alerts
Three databases were screened with ToxAlerts for compounds with structural alerts (Figure S4). Screening of the ToxCast database revealed the presence of 139 unique structural alerts in one or more molecules (Figure S4). A total of 109 of these exceeded the pattern detection cutoff of a minimum of five molecules. Screening for structural alerts of SusDat compounds was performed accordingly, resulting in the detection of 152 unique alerts and 133 after the cutoff (Figure S4). The compounds in the NORMAN MassBank data set contained 103 unique structural alerts, of which 59 alerts were present in at least 5 compounds (Figure S4).
Validation of Toxicity
To validate the ToxAlerts approach for structural alert detection, we investigated whether compounds with a given structural alert were active in a bioassay linked to the toxic endpoint which was related to that alert. For all four end points, the compounds with structural alerts showed higher percentages of active chemicals in bioassays related to that alert (S3.1) than ToxCast compounds, regardless of the respective structural alert. Based on these results, structural alerts could indeed indicate toxicity, but the alerts used for screening did not cover all chemicals active in these toxic end points. Moreover, many chemicals have not been tested on all included ToxCast assays,37 causing a data gap.
In silico Fragmentation
To be able to determine patterns in the MS2 spectra characteristic for a structural alert, fragmentation spectra were generated in silico using the fragmentation software CFM-ID 2.0. CFM-ID provided intensity values to filter for the most likely fragments.
Validation with NORMAN MassBank Data
The in silico fragmentation results generated by CFM-ID were validated with experimental HCD data retrieved from NORMAN MassBank.11 Positive ionization HCD data were available for 1903 compounds, 587 of which were NORMAN SusDat compounds with a structural alert. To account for the experimental error in the MassBank data, a 10 ppm mass tolerance was used to find overlapping fragments between the CFM-ID predicted and experimental MassBank fragments. Depending on the CFM-ID energy, for 144 up to 398 of the 587 compounds ≥50% of the CFM-ID fragments were matched with a MassBank fragment (S3.2, Table S3.2, Figure S3.2). As no CFM-ID fragmentation energy setting outperformed the others, all energies were included in the further analyses.
Pattern Screening
After in silico generation of predicted fragmentation spectra, these predicted spectra of compounds with structural alerts were screened for patterns characteristic for each structural alert for subsequent use as MS2-triggers. These patterns included recurring fragment masses and recurring mass differences between two fragments referred to as deltas. All three CFM-ID fragmentation energies were included in the pattern screening, and patterns were filtered for occurrence in the spectra of at least two fragmentation energies to remove less relevant fragments and/or deltas. To further increase specificity, only fragments and deltas with a frequency higher than 0.5 in both the ToxCast and SusDat data sets were taken into consideration. These strict requirements led to a relatively low number of alerts: 6 recurring fragments and 11 deltas exceeded this frequency cut-off (Table 1). m/z 62.99960 was a recurring fragment in mustard-like structural alerts, which could correspond to C2ClH+, a fragment that is likely to form from these alerts. The recurring fragments m/z 55.01784 and m/z 109.01632 could correspond to C3H3O+ and C2H6ClON2+, respectively. Five structural alerts corresponded to the same recurring fragment, that is, m/z 62.99960 (Table 1), and four structural alerts to two recurring deltas, that is, Δ m/z 27.99491 and Δ m/z 42.01056 (Table 1) due to the similarity in their structures.
Table 1. Structural Alerts with a Recurring Fragment (Top) and Deltas (Bottom) and Their Frequencies in Each Data Set.
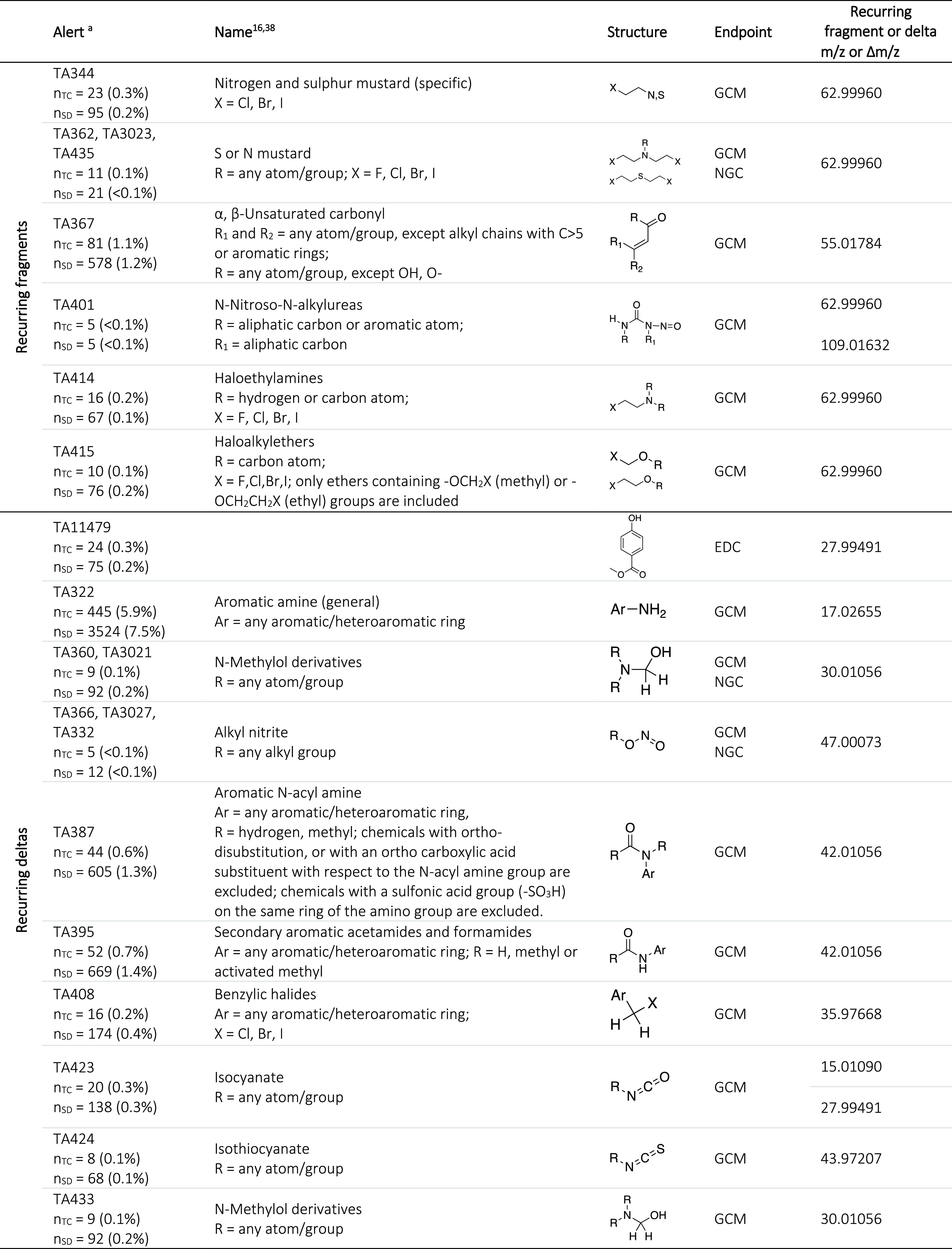
nTC and nSD represent the number of compounds in the ToxCast and SusDat data set, respectively. A description of the structural alert is given in the second column.38
For both the recurring fragments and deltas, their frequencies within an alert were significantly higher than the highest frequency observed in the three different control data sets, that is, in all fragmented molecules with an alert from ToxCast, a random sample from SusDat, regardless of the presence of an alert, and all fragmented molecules with an alert from SusDat (Tables S11–S13). This confirmed that the recurring fragments and deltas were characteristic for their structural alerts. Two deltas detected with high frequency were 2.01565 and 18.01056 Da. These were not considered as relevant deltas because there was no significant difference between their frequencies in the compounds with alerts compared to the total data set. These deltas are expected to correspond to a loss of 2H and H2O, respectively.
In order to increase the “yield” of alerts that could be used as trigger, other data mining approaches could be applied such as hierarchical clustering, random forest or multiple linear regression to find patterns characteristic for a specific structural alert. However, one has to take into account that the output of more advanced pattern recognition needs to be in a format that is suitable for implementation in acquisition software used to operate mass spectrometers. Moreover, even more reliable results could be generated when experimental fragmentation data is used instead of in silico-predicted fragments.
Based on in-house availability of chemicals, the recurring fragments m/z 62.99960 of ToxAlert alert TA344/TA362 (Table 1) and m/z 55.01784 of alert TA367 and the recurring deltas m/z 17.02655 of alert TA322 and m/z 42.01056 of alert TA387/TA395 were selected for use in the MS2-trigger experiments.
LC-HRMS Experiments
MS1-Trigger Experiments
Prior to implementing MS triggers, background exclusion and selected MS acquisition parameters were optimized to maximize available cycle time for (additional) MS2 scans and MS2 spectral quality during online prioritization (S3.3 Acquisition parameter optimization). Subsequently, the potential of MS1-triggers for the prioritization of toxic compounds was assessed experimentally. The MS1-triggers consisted of five different inclusion lists and isotopic ratios for chlorinated and brominated compounds.
Based on the Cl/Br pattern, which is a parameter in Compound Discoverer stating whether a chlorine- or bromine-specific isotopic pattern is present in the MS1, there was a significant increase in the percentage of MS2 scans for the surface water (μNTS = 94.2 ± 0.4%, μMS1-trig = 100 ± 0%, p-value of 0.001292, Figure S7) but not the WWTP-influent samples (μNTS = 82.7 ± 5.2%, μMS1-trig = 84.5 ± 1.3%, Figure S7). The lesser performance in the WWTP-influent samples could be due to the more complex MS1 spectra confounding isotopic ratios, in particular when low error tolerances are set. This is also supported by the pattern matches determined during the Compound Discoverer analysis. The peaks of Cl- and/or Br-containing features should contain a characteristic isotopic pattern due to the natural abundance of chlorine and bromine isotopes. For some brominated and/or chlorinated compounds, no additional MS2 was triggered because the isotopic ratio deviated more than the allowed 10% ratio tolerance. Additional experiments with increased mass tolerance (10 ppm instead of 3 ppm, which was chosen to test the extreme effect) and ratio tolerance (15% instead of 10%) did not improve this. Setting priority of the decision tree to the branch with the targeted isotopic ratio node, however, led to a significant increase in percentage of Cl and/or Br containing features with an MS2 spectrum (p-value of 0.04225, one-sided t-test). Further experiments could focus on optimizing the isotopic ratio and mass tolerance of the MS1-trigger to balance a more tolerant threshold and the subsequent increase in false-positive triggers.
Based on these results, the isotopic ratio was implemented (with the narrow tolerances) in the intelligent acquisition method as MS1-trigger as it increased MS2 spectral availability for Cl-/Br- containing features which are mostly anthropogenic and often toxic, and the risk of triggering fragmentation of irrelevant features was low.
Regarding the use of inclusion lists as MS1-triggers, there was a significant increase in percentage of MS2 scans for m/z values present in the inclusion lists SusDat, SusDat + tR, and Sjerps in the WWTP-influent and SusDat + tR in the SW samples (Table 2). The lesser effect observed in SW samples can be explained by the fact that the standard NTS method without an inclusion list was able to separate and identify the features present in the SW but not WWTP influent samples. Due to the large number of compounds in SusDat (+tR), including non water-relevant ones, the Sjerps list was used for subsequent MS2-trigger experiments.
Table 2. Comparison of Percentage MS2 Scans of the Inclusion List m/z Values between Methodsa.
| inclusion list type | sample type | method with inclusion list μ% features with MS2 | standard NTS method μ% features with MS2 | p-value | test type |
|---|---|---|---|---|---|
| SusDat | WWTP-influent | 95.86 | 91.76 | 0.01576 | t-test |
| SW | 97.68 | 97.31 | 0.1039 | t-test | |
| UBAPMT | WWTP-influent | 100.0 | 100.0 | - | |
| SW | 96.97 | 96.97 | - | ||
| Sjerps | WWTP-influent | 98.36 | 93.32 | 0.01485 | t-test |
| SW | 95.76 | 96.58 | 0.8779 | t-test | |
| Spike | WWTP-influent | 96.41 | 95.60 | 0.3425 | t-test |
| SW | 98.58 | 97.65 | 0.1250 | Sign test | |
| SusDat + tR | WWTP-influent | 97.74 | 92.53 | 0.004934 | t-test |
| SW | 98.80 | 97.87 | 0.0005877 | t-test |
In one case (Spike SW), a Sign test is applied since the data was not normally distributed.
Overall, less complex matrices such as SW samples seemed to benefit more from the isotopic ratio MS1-trigger, demonstrated by the significant increase in the percentage of MS2 scans for these samples. The analysis of more complex matrices such as WWTP influent improved through the use of inclusion lists that ensured that water relevant compounds were fragmented. The inclusion list MS1-trigger showed promising results for the inclusion lists SusDat, with and without retention time estimate, and Sjerps. As the Sjerps list consisted of water-relevant compounds, this list was used in subsequent experiments in combination with the MS2-triggers.
MS2-Trigger Experiments
Next to MS1-triggers that trigger an MS2 scan, MS2-triggers were developed that trigger an additional MS2 scan in the presence of a structural alert, indicating a potentially toxic compound. Four specific fragment masses and deltas were used as MS2-triggers: the recurring fragments m/z 62.99960 of alert TA344/TA362 and m/z 55.01784 of alert TA367 and the recurring deltas m/z 17.02655 of alert TA322 and m/z 42.01056 of alert TA387/TA395. These alerts correspond to the toxic end points genotoxic carcinogenicity and mutagenicity. A total of 12 reference compounds were selected, which were hypothesized to contain an alert and MS2-trigger based on pattern screening (Tables S3–S7).
The recurring fragments were present in the MS2 spectra of all 12 detected compounds, thereby confirming the usefulness of the in silico-predicted spectra generated with CFM-ID. MS2 scans were triggered in all cases, except ifosfamide and diacetone acrylamide. For these compounds, the ppm mass error tolerance was too narrow. Increasing the tolerance to 20 ppm lead to triggering of additional MS2 scans. Therefore, a higher error tolerance or potentially a combination of a low relative tolerance and an absolute tolerance of m/z 0.001 would be advantageous. Alternatively, the calibration range of the instrument could be expanded to lower m/z values.
In addition to the recurring fragments, the use of recurring deltas as MS2-triggers was investigated. The recurring delta m/z 17.02650 corresponding to alert TA322 was detected in the MS2 spectra of all reference compounds that contained this alert, thereby validating the approach of using CFM-ID to in silico predict spectra. Additional MS2 scans were triggered for all compounds with this recurring delta.
Examples of spectra where an additional MS2 was successfully MS2-triggered are shown in Figure 2. The recurring delta m/z 42.01060 corresponding to the alerts TA387 and TA395 was detected in all spectra except those of diatrizoic acid and one of the three triplicates of n-acetylsulfamethoxazole. The delta m/z 42.01060 triggered additional MS2 scans in all other compounds, where the recurring delta was detected. The measured MS2 spectra of diatrizoic acid did not match the in silico-predicted spectrum (see Figure S8), and the peaks that were expected to form the recurring delta (m/z 614.7769272 and m/z 572.7663625 or m/z 596.7663625 and m/z 554.7557979) were not present.
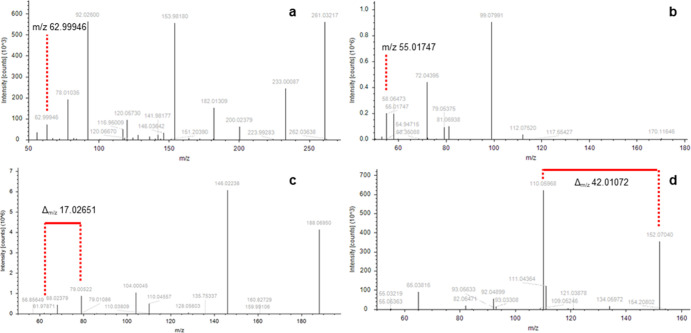
Figure 2.
Four experimentally obtained MS2 scans with expected MS2-trigger marked in red, of which an additional MS2 was triggered. (a) MS2 spectrum of ifosfamide and its recurring fragment, (b) MS2 spectrum of diacetone acrylamide and its recurring fragment, (c) MS2 spectrum of desethylatrazine and its recurring delta, and (d) MS2 spectrum of paracetamol and its recurring delta.
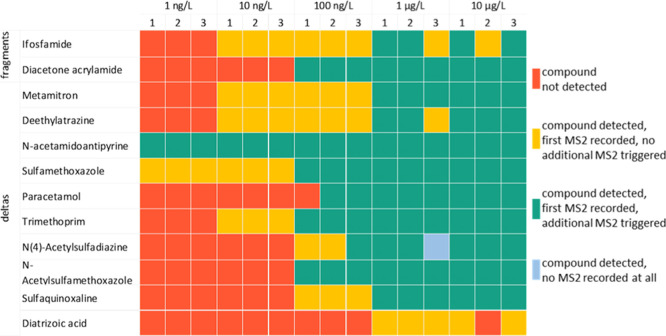
Next, the effect of compound concentration levels on the MS2-triggers was investigated (Figure 3). To this end, a concentration range from the 10 μg/L used in the proof-of-principle experiments down to 1 ng/L was used. At first, the precursor ion of the compound containing a structural alert has to be selected for a MS2 scan, in which the MS2-trigger can be detected. Thereafter, this trigger can prompt the consecutive MS2 scan. Generally, once a compound was detected and a MS2 scan recorded, an additional MS2 scan was triggered as well, indicating the sensitivity of the MS2-trigger. However, some exceptions were observed (marked in yellow in Figure 3). In these cases, the compound was detected, but no additional MS2 scans were triggered due to the absence of the trigger in the MS2 scan (in case of metamitron, desethylatrazine up to 100 ng/L, sulfamethoxazole, trimethoprim, and sulfaquinoxaline) or the selected error tolerance (5 ppm, in case of ifosfamide and desethylatrazine in the third measurement at 1 μg/L). In one case, no MS2 scan was recorded. Consequently, no additional MS2 scan could be triggered. This was the case for a single measurement of N(4)-acetylsulfadiazine at 1 μg/L.
Figure 3.
Schematic overview of the detection of the spike-in compounds, and whether an additional MS2 was triggered or not.
MS2-triggers were applied to prompt an additional MS2 scan that would ensure more informative fragmentation spectra, that is, higher spectral quality or complementary fragments to the first MS2 scan, of features with a structural alert. Different acquisition parameters were used for this additional MS scan: stepped CE (10, 75, 90 instead of the regular 20, 35, 50), assisted CE (20, 35, 50, 75) and longer ITs (200 ms IT instead of the regular 50 ms). The effect of the acquisition parameter to increase the information content of the spectra was assessed based on the mzCloud scores assigned to the identified features because these could be easily extracted from the Compound Discoverer results. The mzCloud scores tended to increase slightly (approximately 0.1–1%) with the additional MS2 scan using assisted CE and longer IT. As mzCloud scores are based on experimental spectra that might have not been generated with the optimal acquisition parameters, as an alternative performance evaluation MetFrag annotation was examined. This showed that generally, the additional MS2 scans using assisted CE had a higher percentage of annotated intensity (Figure S9) but no higher percentage of annotated fragments (Figure S10). However, to reach the maximum advantage of the additional MS2, higher spectral quality that facilitates identification, spectral quality metrics need to be developed and implemented online, that is, during the measurement.
Application of Triggered Methods to SW Samples
To compare the online prioritization methods to the standard NTS method, a SW sample spiked with water-relevant contaminants was analyzed. Three versions of the intelligent acquisition method combining the MS1- (isotopic ratio and Sjerps inclusion list) and MS2-triggers (fragment m/z 62.99960, fragment m/z 55.01784, delta m/z 42.01060, and delta m/z 17.02650) were used: with the additional MS2 with either stepped CE, ACE or longer IT. Ten of the spiked compounds contained an alert related to these MS2-triggers, and for eight of them, an additional MS2 was triggered. The spectra of 2-aminobenzothiazole and 2,4-dichloroaniline did not exhibit the expected delta m/z 17.02650. Consequently, no additional MS2 was triggered. Using the regular NTS method (see Tables S14–S15), the mzCloud best match and mzVault best match scores (S1.2) ranged from 97.1 to 99.8 out of 100 and from 89.6 to 99.8, respectively. This indicates that these scores are already high. Despite these high scores, for the compounds desisopropylatrazin and desethylatrazin, the mzCloud scores increased with all three tested intelligent acquisition methods (Table S15).
Conclusions
Overall, the intelligent acquisition method, using the Sjerps inclusion list and additional MS2’s with ACE or longer IT, directed prioritization toward potentially toxic compounds. The isotopic ratio MS1-trigger significantly improved the percentage of Cl-/Br-containing compounds with a MS2 spectrum if priority was assigned in the method. The use of an inclusion list increased the percentage of MS2 spectra of features with m/z values present in the inclusion list. The MS2-trigger method successfully triggered additional MS2 scans of molecules with a structural alert for the four alerts that were tested. Therefore, the method could prioritize these potentially toxic compounds online, and further developments will improve the added value. Once fully developed, it could be far more efficient than many current strategies involving post-acquisition processing.
Future work could expand the developed method with more structural alerts targeting different toxic endpoints, implementing the method in our laboratory, and making it available for other laboratories to use. Ultimately, application of intelligent acquisition methods in routine monitoring studies is necessary to expose the benefits in practice for safety monitoring of drinking water sources. While a clear benefit was demonstrated for MS1- and MS2-triggers, the automatic triggering of an additional MS2 scan will reach its maximum benefit once more knowledge is available on how spectral quality can be optimized in a directed manner through selection of appropriate acquisition parameters.
Acknowledgments
The authors acknowledge Astrid Reus, Tessa Pronk, and Margo van der Kooi from the KWR Water Research Institute for advice about relevant toxic end points, advice in programming in R, and preparation of the samples. Caroline Ding, Lena Becciolini, and Seema Sharma from Thermo Fisher Scientific are acknowledged for their help with the data acquisition and data processing software. Christian Panse from ETH Zürich is acknowledged for the development of the centroid function in R. Eelco Pieke from Het Waterlaboratorium and Jan van der Kooi from WLN for critical reading of the manuscript and Igor Tetko from VCCLAB for helping with ToxAlerts. This work was funded by the Joint Research Program of the Dutch and Belgian drinking water companies.
Glossary
Abbreviations
- AC50
concentration at 50% of maximum activity
- CE
collision energy
- CFM
competitive fragmentation modeling
- CID
collision-induced dissociation
- DDA
data-dependent acquisition
- DMT
developmental and mitochondrial toxicity
- EDC
endocrine disruption
- GCM
genotoxic carcinogenicity, mutagenicity
- HCD
higher-energy collisional dissociation
- HPLC
high-performance liquid chromatography
- HRMS
high-resolution mass spectrometry
- InChI
International Chemical Identifier
- IT
ion injection time
- KOW
octanol–water partition coefficient
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry, fragmentation spectrum
- MS2
tandem mass spectrometry, fragmentation spectrum
- NGC
nongenotoxic carcinogenicity
- NTS
nontarget screening
- OMP
organic micropollutants
- QTOF
quadrupole-time-of-flight mass spectrometer
- RPLC
reversed-phase liquid chromatography
- tR
retention time
- SA
structural alert
- SMILES
simplified molecular-input line-entry specification
- SusDat
NORMAN Substance Database
- SW
surface water
- WWTP
wastewater treatment plant
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c04473.
Acquisition parameters, spectral libraries and chemical databases, workflow screening with ToxAlerts and fragmentation, instrument settings for LC-HRMS experiments, inclusion lists, data analysis, compound discoverer workflow parameters, toxicity validation, validation of in silico fragmentation, acquisition parameter optimization, NTS workflow, design of acquisition decision trees, ToxAlerts screening results, frequency distributions of recurring fragments and deltas, number of detected features per MS1-trigger method, number of detected chlorinated and brominated features per MS1-trigger method, in silico predicted and experimental MS2 of diatrizoic acid, comparison of annotated intensity of regular MS2 scan and triggered MS2 scan, and comparison of annotated fragments of regular MS2 scan and triggered MS2 scan (PDF)
ToxCast assays used in toxicity validation, lists of spiked compounds and sample compositions, isotopic ratios, methods of MS2-trigger experiments, frequencies of recurring fragments and deltas within compounds with an alert, frequencies of recurring fragments and deltas in control data sets, mzVault and mzCloud best match scores from the total performance analysis (XLSX)
Screening results ToxAlerts (ZIP)
The authors declare no competing financial interest.
This article was initially published with an incorrect copyright statement and was corrected on or around May 5, 2021.
Supplementary Material
References
- Stamm C.; Räsänen K.; Burdon F. J.; Altermatt F.; Jokela J.; Joss A.; Ackermann M.; Eggen R. I. L. In Large-Scale Ecology: Model Systems to Global Perspectives; Dumbrell A. J., Kordas R. L., Woodward G., Eds.; Academic Press, 2016, pp 183–223. [Google Scholar]
- Ruff M.; Mueller M. S.; Loos M.; Singer H. P. Quantitative target and systematic non-target analysis of polar organic micro-pollutants along the river Rhine using high-resolution mass-spectrometry - Identification of unknown sources and compounds. Water Res. 2015, 87, 145–154. 10.1016/j.watres.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Bernhardt E. S.; Rosi E. J.; Gessner M. O. Synthetic chemicals as agents of global change. Front. Ecol. Environ. 2017, 15, 84–90. 10.1002/fee.1450. [DOI] [Google Scholar]
- Brack W.; Dulio V.; Ågerstrand M.; Allan I.; Altenburger R.; Brinkmann M.; Bunke D.; Burgess R. M.; Cousins I.; Escher B. I.; Hernández F. J.; Hewitt L. M.; Hilscherová K.; Hollender J.; Hollert H.; Kase R.; Klauer B.; Lindim C.; Herráez D. L.; Miège C.; et al. Towards the review of the European Union Water Framework Directive: Recommendations for more efficient assessment and management of chemical contamination in European surface water resources. Sci. Total Environ. 2017, 576, 720–737. 10.1016/j.scitotenv.2016.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach R. P.; Escher B. I.; Fenner K.; Hofstetter T. B.; Johnson C. A.; von Gunten U.; Wehrli B. Science 2006, 313, 1072. 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Bletsou A. A.; Jeon J.; Hollender J.; Archontaki E.; Thomaidis N. S. Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment. TrAC, Trends Anal. Chem. 2015, 66, 32–44. 10.1016/j.trac.2014.11.009. [DOI] [Google Scholar]
- Samanipour S.; Martin J. W.; Lamoree M. H.; Reid M. J.; Thomas K. V. Letter to the Editor: Optimism for Nontarget Analysis in Environmental Chemistry. Environ. Sci. Technol. 2019, 53, 5529–5530. 10.1021/acs.est.9b01476. [DOI] [PubMed] [Google Scholar]
- Hollender J.; Schymanski E. L.; Singer H. P.; Ferguson P. L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go?. Environ. Sci. Technol. 2017, 51, 11505–11512. 10.1021/acs.est.7b02184. [DOI] [PubMed] [Google Scholar]
- Brunner A. M.; Dingemans M. M. L.; Baken K. A.; van Wezel A. P. Prioritizing anthropogenic chemicals in drinking water and sources through combined use of mass spectrometry and ToxCast toxicity data. J. Hazard. Mater. 2019, 364, 332–338. 10.1016/j.jhazmat.2018.10.044. [DOI] [PubMed] [Google Scholar]
- HighChem LLC . mzCloud Features. https://www.mzcloud.org/Features (accessed Nov 25, 2019).
- Schymanski E. L.; Schulze T.; Alygizakis N.; Meier R.. S1|MASSBANK|NORMAN Compounds in MassBank, version NORMAN-SLE-S1.0.1.1. Zenodo.
- Williams A. J.; Grulke C. M.; Edwards J.; McEachran A. D.; Mansouri K.; Baker N. C.; Patlewicz G.; Shah I.; Wambaugh J. F.; Judson R. S.; Richard A. M. J. Cheminf. 2017, 9, 61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. M.; Judson R. S.; Houck K. A.; Grulke C. M.; Volarath P.; Thillainadarajah I.; Yang C.; Rathman J.; Martin M. T.; Wambaugh J. F.; Knudsen T. B.; Kancherla J.; Mansouri K.; Patlewicz G.; Williams A. J.; Little S. B.; Crofton K. M.; Thomas R. S. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 2016, 29, 1225–1251. 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- NORMAN Network . Aalizadeh R.; Alygizakis N.; Schymanski E.; Slobodnik J., S0|SUSDAT|Merged NORMAN Suspect List: SusDat, version NORMAN-SLE-S0.0.2.1. Zenodo.
- Brunner A. M.; Bertelkamp C.; Dingemans M. M. L.; Kolkman A.; Wols B.; Harmsen D.; Siegers W.; Martijn B. J.; Oorthuizen W. A.; Ter Laak T. L. Integration of target analyses, non-target screening and effect-based monitoring to assess OMP related water quality changes in drinking water treatment. Sci. Total Environ. 2020, 705, 135779. 10.1016/j.scitotenv.2019.135779. [DOI] [PubMed] [Google Scholar]
- Sushko I.; Salmina E.; Potemkin V. A.; Poda G.; Tetko I. V. ToxAlerts: A Web Server of Structural Alerts for Toxic Chemicals and Compounds with Potential Adverse Reactions. J. Chem. Inf. Model. 2012, 52, 2310–2316. 10.1021/ci300245q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridings J. E.; Barratt M. D.; Cary R.; Earnshaw C. G.; Eggington C. E.; Ellis M. K.; Judson P. N.; Langowski J. J.; Marchant C. A.; Payne M. P.; Watson W. P.; Yih T. D. Computer prediction of possible toxic action from chemical structure: an update on the DEREK system. Toxicology 1996, 106, 267–279. 10.1016/0300-483x(95)03190-q. [DOI] [PubMed] [Google Scholar]
- Saiakhov R. D.; Klopman G. MultiCASE Expert Systems and the REACH Initiative. Toxicol. Mech. Methods 2008, 18, 159–175. 10.1080/15376510701857460. [DOI] [PubMed] [Google Scholar]
- Organisation for Economic Co-operation and Development (OECD) . Appendix I - Collection of working definitions. http://www.oecd.org/chemicalsafety/testing/49963576.pdf (accessed Nov 18, 2019).
- Ashby J.; Tennant R. W. Chemical structure, Salmonella mutagenicity and extent of carcinogenicity as indicators of genotoxic carcinogenesis among 222 chemicals tested in rodents by the U.S. NCI/NTP. Mutat. Res. 1988, 204, 17–115. 10.1016/0165-1218(88)90114-0. [DOI] [PubMed] [Google Scholar]
- Bailey A. B.; Chanderbhan R.; Collazo-Braier N.; Cheeseman M. A.; Twaroski M. L. The use of structure-activity relationship analysis in the food contact notification program. Regul. Toxicol. Pharmacol. 2005, 42, 225–235. 10.1016/j.yrtph.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Kazius J.; McGuire R.; Bursi R. Derivation and Validation of Toxicophores for Mutagenicity Prediction. J. Med. Chem. 2005, 48, 312–320. 10.1021/jm040835a. [DOI] [PubMed] [Google Scholar]
- Kazius J.; Nijssen S.; Kok J.; Bäck T.; IJzerman A. P. Substructure Mining Using Elaborate Chemical Representation. J. Chem. Inf. Model. 2006, 46, 597–605. 10.1021/ci0503715. [DOI] [PubMed] [Google Scholar]
- Benigni R.; Bossa C. Structure alerts for carcinogenicity, and the Salmonella assay system: A novel insight through the chemical relational databases technology. Mutat. Res. 2008, 659, 248–261. 10.1016/j.mrrev.2008.05.003. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency . Chemical_Summary_190708 from invitrodb_v3.2. https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data (accessed Dec 4, 2019).
- McEachran A. D.; Mansouri K.; Grulke C.; Schymanski E. L.; Ruttkies C.; Williams A. J. Cheminf. 2018, 10, 45. 10.1186/s13321-018-0299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nendza M.; Wenzel A.; Muller M.; Lewin G.; Simetska N.; Stock F.; Arning J. Environ. Sci. Eur. 2016, 28, 26. 10.1186/s12302-016-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing, 3.6.1; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- United States Environmental Protection Agency . ac50_Matrix_190708 from invitrodb_v3.2. https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data (accessed Dec 4, 2019).
- Allen F.; Greiner R.; Wishart D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, 98–110. 10.1007/s11306-014-0676-4. [DOI] [Google Scholar]
- Allen F.CFM-ID: Competitive Fragmentation Modeling for Metabolite Identification. https://sourceforge.net/p/cfm-id/wiki/Home/ (accessed Dec 20, 2019).
- Von der Ohe P.; Fischer S.. S36|UBAPMT|Potential Persistent, Mobile and Toxic (PMT) Substances, version NORMAN-SLE-S36.0.1.0. Zenodo.
- Sjerps R. M. A.S27|KWRSJERPS2|Extended Suspect List from Sjerps et al (KWRSJERPS), version NORMAN-SLE-S27.0.1.1. Zenodo.
- United States Environmental Protection Agency . DSSTox MS Ready Mapping File. https://comptox.epa.gov/dashboard/downloads (accessed Feb 12, 2020).
- Dodder N.; Mullen K.. Organic Mass Spectrometry, version 0.5-3; 2017.
- Ruttkies C.; Schymanski E. L.; Wolf S.; Hollender J.; Neumann S. J. Cheminf. 2016, 8, 3. 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louisse J.; Dingemans M. M. L.; Baken K. A.; van Wezel A. P.; Schriks M. Exploration of ToxCast/Tox21 bioassays as candidate bioanalytical tools for measuring groups of chemicals in water. Chemosphere 2018, 209, 373–380. 10.1016/j.chemosphere.2018.06.056. [DOI] [PubMed] [Google Scholar]
- Sushko I.; Novotarskyi S.; Körner R.; Pandey A. K.; Rupp M.; Teetz W.; Brandmaier S.; Abdelaziz A.; Prokopenko V. V.; Tanchuk V. Y.; Todeschini R.; Varnek A.; Marcou G.; Ertl P.; Potemkin V.; Grishina M.; Gasteiger J.; Schwab C.; Palyulin V. A.; et al. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J. Comput.-Aided Mol. Des. 2011, 25, 533–554. 10.1007/s10822-011-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.