Aerosol delivery of an ultrapotent nanobody efficiently prevents and treats SARS-CoV-2 infection.
Abstract
Globally, there is an urgency to develop effective, low-cost therapeutic interventions for coronavirus disease 2019 (COVID-19). We previously generated the stable and ultrapotent homotrimeric Pittsburgh inhalable Nanobody 21 (PiN-21). Using Syrian hamsters that model moderate to severe COVID-19 disease, we demonstrate the high efficacy of PiN-21 to prevent and treat SARS-CoV-2 infection. Intranasal delivery of PiN-21 at 0.6 mg/kg protects infected animals from weight loss and substantially reduces viral burdens in both lower and upper airways compared to control. Aerosol delivery of PiN-21 facilitates deposition throughout the respiratory tract and dose minimization to 0.2 mg/kg. Inhalation treatment quickly reverses animals’ weight loss after infection, decreases lung viral titers by 6 logs leading to drastically mitigated lung pathology, and prevents viral pneumonia. Combined with the marked stability and low production cost, this innovative therapy may provide a convenient and cost-effective option to mitigate the ongoing pandemic.
INTRODUCTION
By January 2021, a year after the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported (1), close to 100 million people have been infected by this highly transmissible virus, resulting in substantial morbidity and mortality worldwide. In addition to vaccines, there is an unparalleled quest to develop innovative and cost-effective therapeutics to combat the coronavirus disease 2019 (COVID-19) pandemic (2, 3). Early treatment using high-titer convalescent plasma (CP) may reduce the risk of severe disease in seniors (4), although CP is limited by supply. Potent neutralizing monoclonal antibodies (mAbs), predominantly isolated from COVID-19 patients for recombinant productions, have been developed for passive immunotherapy (5–14). In vivo evaluations of mAbs in animal models of COVID-19 disease such as murine, hamster, and nonhuman primates (NHPs) have provided critical insights into efficacy and the mechanisms by which they alter the course of infection (15–24). While mAb therapy lifts hopes to treat mild symptom onset in patients, they nevertheless require exceedingly high administration doses—typically several grams for intravenous injection (25, 26). The requirement of high doses for efficient neutralization may reflect SARS-CoV-2 virulence, pathogenesis, and the notoriously low efficiency of intravenously delivering these relatively large biomolecules across the plasma-lung barrier to treat pulmonary infections (27). Moreover, the associated high costs and challenges in bulk manufacturing may further limit the broad clinical use of mAbs worldwide (2).
In parallel efforts, we and others have recently developed camelid single-domain antibody fragments or nanobodies (Nbs) that primarily target the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) glycoprotein for virus neutralization (14, 28–31). Highly selected Nbs and the multivalent forms obtain high neutralization potency comparable to, or even better (per mass) than, some of the most successful SARS-CoV-2 neutralizing mAbs. In particular, an ultrapotent homotrimeric construct, Pittsburgh inhalable Nanobody 21 (PiN-21), efficiently blocked SARS-CoV-2 infectivity at below 0.1 ng/ml in vitro (28). Compared to mAbs, Nbs are substantially cheaper to produce. Moreover, affinity-matured, ultrapotent Nbs are characterized by high solubility and stability (32) that facilitate drug scaling, storage, and transportation, all of which are critical in response to pandemics. The excellent physicochemical properties and small sizes of Nbs raise an exciting possibility of efficient pulmonary delivery by aerosolization with characteristics of rapid onset of action, high local drug concentration/bioavailability, and improved patient compliance (needle-free) that may benefit a large population of SARS-CoV-2–infected patients (27–29, 33). However, despite the promise, no successful in vivo studies have been reported to date. The inferior pharmacokinetics of monomeric Nbs due to their small size (~15 kDa) and a lack of Fc-mediated immune effectors’ function, which is often required to augment the in vivo neutralizing activities of mAbs (34–36), drive potential concerns for Nb-based therapy. It remains unknown if the high in vitro neutralization potency of SARS-CoV-2 Nbs can be translated into in vivo therapeutic benefits.
In this study, we systematically evaluated the efficacy of PiN-21 for prophylaxis and treatment of SARS-CoV-2–infected Syrian hamsters that model moderate to severe COVID-19 disease. We provided direct evidence that ultra-low administration of PiN-21 efficiently treats the virus infection. Notably, PiN-21 aerosols can be inhaled to target respiratory infection that drastically reduces viral loads and prevents lung damage and viral pneumonia. This novel Nb-based therapy shows high potentials for the treatment of early infection and may provide a robust and affordable solution to address the current health crisis.
RESULTS
PiN-21 efficiently protects and treats SARS-CoV-2 infection in Syrian hamsters
To assess the in vivo efficacy of PiN-21, 12 hamsters were divided into two groups and infected with 9 × 104 plaque-forming units (PFU) of SARS-CoV-2 via the intratracheal route. Shortly after infection, Nb was delivered intranasally at an average dose of 0.6 mg/kg (Fig. 1A). Animals were monitored daily for weight change and clinical signs of disease. Half of the animals were euthanized 5 days post-infection (d.p.i.), and the remaining were euthanized 10 d.p.i. Virus titers in lung samples from the euthanized animals were measured by plaque assay. Nasal washes and throat swabs were collected at 2 and 4 d.p.i. to determine viral loads in the upper respiratory tract (URT). Consistent with published studies (37, 38), intratracheal inoculation of hamsters with SARS-CoV-2 resulted in a robust infection, rapid weight loss in all animals up to 16% at 7 d.p.i., and resulting recovery and reversal of weight loss by 10 d.p.i. before recovery. However, concurrent intranasal delivery of PiN-21 eliminated any significant weight loss in the infected animals (Fig. 1B). This marked protection was accompanied by a reduction of viral titer in the lungs, with an average decrease of four orders of magnitude in the lung tissue, respectively, compared to control on 5 d.p.i. (Fig. 1C). Infectious virus was essentially cleared by 10 d.p.i. (Fig. 1C). Consistently, a 3-log reduction of the viral genomic RNA (gRNA) by reverse transcription quantitative polymerase chain reaction (RT-qPCR) was evident on 5 and 10 d.p.i. (fig. S1, A and B).
Fig. 1. PiN-21 protects Syrian hamsters from SARS-CoV-2 infection.
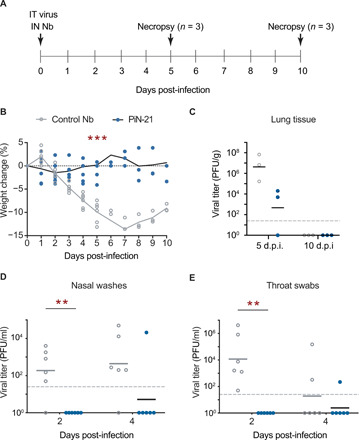
(A) Overview of the experimental design. SARS-CoV-2 (9 × 104 PFU) was intratracheally (IT) inoculated followed by intranasal (IN) delivery of 100 μg of PiN-21 (shown in blue dots) or a control Nb (shown in gray circles). Animal weight changes were monitored daily. Nasal washes and throat swabs were collected on 2 and 4 d.p.i. Animals were euthanized for necropsy on 5 (n = 3) and 10 d.p.i (n = 3), with viral titers and gRNAs of lung tissues measured. (B) Protection of weight loss of infected hamsters treated with PiN-21. ***P < 0.001. (C to E) Measurement of viral titers by the plaque assay. **P < 0.01. The dashed line indicates the detection limit of the assay. The color scheme is consistent across all the panels.
Notably, the virus was undetectable in the URT including both nasal washes and throat swabs of all PiN-21–treated animals on 2 d.p.i. This is significantly different from the control group, where varying levels of infectious virus were present (Fig. 1, D and E). Furthermore, five of six PiN-21–treated animals remained protected from detectable infection 4 d.p.i. The results were further supported by a drastic decrease of gRNA in the URT (fig. S1, C and D). Together, this demonstrates that the high in vitro neutralization potency of PiN-21 can be translated into therapeutic benefits in vivo independent of Fc-mediated immune responses. PiN-21 can efficiently protect SARS-CoV-2 infection in hamsters by rapidly and drastically suppressing viral replication in both the URT and lower respiratory tract (LRT).
Previous studies reveal that clinical mAbs are less effective for COVID-19 treatment (after infection) than for prophylaxis (before infection) in animal models, possibly reflecting the virulence of SARS-CoV-2, speed of virus replication, and rapid symptom onset (15, 22, 39). Therefore, we evaluated the therapeutic potential of PiN-21 because it was highly effective when coadministered. To explore the second route of infection, hamsters were inoculated intranasally with 3 × 104 PFU of SARS-CoV-2. PiN-21 or a control Nb (0.6 mg/kg) was intranasally delivered to animals 6 hours post-infection (h.p.i.). Animal weights were monitored daily, and throat swabs and nasal washes were collected, before they were euthanized 6 d.p.i. (fig. S2A). Similar to the intratracheal route, intranasal infection of hamsters with SARS-CoV-2 resulted in precipitous weight losses in the control animals. Encouragingly, intranasal treatment using PiN-21 significantly reduced weight loss throughout the assessment period (fig. S2B), paralleling the results of clinical mAbs in the same model, albeit using substantially higher doses. Less than 100-fold reduction in virus titers was found in nasal washes and throat swabs on 2 and 4 d.p.i. (fig. S2, C and D). Moreover, infectivity was undetectable in lung tissues 6 d.p.i. (fig. S2E), indicating that the virus has been predominantly cleared. Analysis of early time points will be needed to better understand virus suppression by Nb treatment.
PiN-21 aerosolization effectively treats SARS-CoV-2–infected hamsters at an ultra-low dose
The marked physicochemical properties of PiN-21 prompted us to evaluate pulmonary delivery by inhalation. To evaluate the impact of construct size and pharmacokinetics on lung uptake, we fused monomeric Nb21 and PiN-21 to an Nb that binds serum albumin (Alb) of both human and rodents with high affinity to generate two serum-stable constructs (Nb-21Alb and PiN-21Alb) (40). Using a portable mesh nebulizer, we aerosolized Nb21Alb, PiN-21, and PiN-21Alb and evaluated their post-aerosolization neutralization activities by pseudovirus neutralization assay. All constructs retained high neutralization potency in vitro (fig. S3B). The amount of Nbs recovered after aerosolization was inversely correlated with the size of constructs (fig. S3A). Moreover, while Nb-21Alb had the highest recovery, the post-aerosolization in vitro neutralization activity was substantially lower than for other constructs. Nb-21Alb was therefore excluded from downstream therapeutic analyses.
Next, we compared two ultrapotent constructs PiN-21 and PiN-21Alb for targeted aerosolization delivery into hamsters. Nbs were aerosolized using a nebulizer (Aerogen, Solo) that produces small aerosol particles with a mass median aerodynamic diameter of ~3 μm (table S1). Animals were sacrificed for 8 and 24 hours after administration to assess Nb distribution and activities when recovered from various respiratory compartments and sera (Fig. 2A). Consistent with the result using the portable nebulizer, we found that the inhalation dose of PiN-21 was approximately two times PiN-21Alb (at 8 hours, 41.0 μg or 0.24 mg/kg for PiN-21 versus 23.7 μg or 0.13 mg/kg for PiN-21Alb) (table S1), while neutralization activity as assessed by plaque reduction neutralization test (PRNT50) of SARS-CoV-2 remained essentially unchanged after aerosolization of both Nb constructs (Fig. 2B).
Fig. 2. Assessment of Nb delivery in the hamster respiratory system.
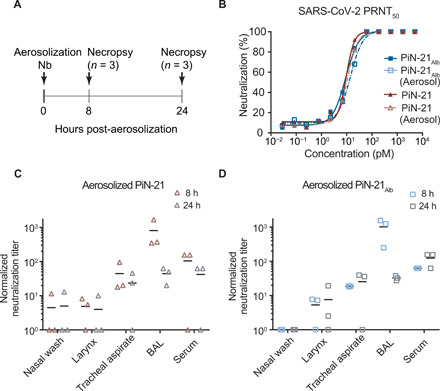
(A) Schematic design of PiN-21 (shown in red triangles) and PiN-21Alb (shown in blue squares) aerosolization in hamster models. (B) Nb neutralization potency before and after aerosolization measured by PRNT50 assay. (C and D) Normalized overall neutralization activity by plaque assay of PiN-21 and PiN-21Alb of different time points after aerosolization.
The neutralization activities of both Nb constructs were detected throughout the respiratory tract and in sera. As expected, within the airways, the neutralizing activities were predominantly associated with bronchoalveolar lavage (BAL) fluid, followed by tracheal aspirate, larynx wash, and nasal wash samples (Fig. 2, C and D). Compared to 8 hours after inhalation, we found that the amounts and activities of Nbs in BAL, but not in sera, were substantially lower 24 hours after inhalation, possibly indicating more rapid clearance. In addition, Nb conjugation to serum albumin did not seem to affect the activities in the airways, while stability was enhanced in the serum. These data underscore the requirement of an ultra-low dose of the ultrapotent PiN-21 construct to neutralize SARS-CoV-2 infectivity in vivo efficiently. Last, PiN-21 was preferentially selected for further evaluation owing to the high stability and resistance to aerosolization, which are likely critical for clinical applications.
To assess the therapeutic efficacy of PiN-21 by inhalation, 12 hamsters were intranasally inoculated with SARS-CoV-2 (3 × 104 PFU) followed by single-dose aerosolization treatment (~0.2 mg/kg) of either PiN-21 or the control Nb at 6 h.p.i. Animals were monitored for weight loss, and throat swabs and nasal washes were collected daily. Animals were euthanized (3 d.p.i.), and lungs and trachea were collected for virological, histopathology, and immunohistochemical analysis (Fig. 3A). Notably, pulmonary delivery of PiN-21 aerosols, despite only a minute amount, led to a remarkable reverse of weight loss in the treated animals. The average weight gain was 2% in PiN-21 versus 5% loss in the control on 3 d.p.i. (Fig. 3B). The weight loss in the control group was highly reproducible when compared with the above experiments. Critically, aerosolization treatment diminished infectious viruses in lung tissue by six orders of magnitude (Fig. 3C). The treatment also substantially decreased virus gRNA in the lungs (fig. S4C). Moreover, we observed a substantial reduction of viral titers in nasal washes and throat swabs (fig. S4, A and B). This indicates that Nb administration by aerosolization may limit human-to-human transmission of SARS-CoV-2.
Fig. 3. Treatment efficacy of aerosolized PiN-21 in the hamster model of SARS-CoV-2.
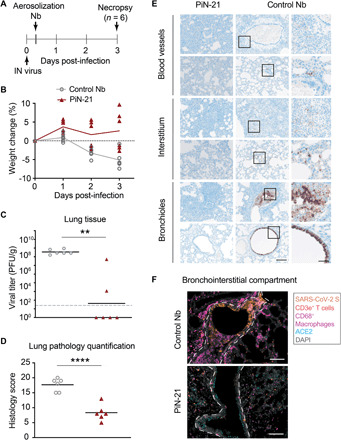
(A) Overview of the experiment design. SARS-CoV-2 (3 × 104 PFU) was intranasally inoculated. PiN-21 (shown in red triangles) or a control Nb (shown in gray circles) was aerosolized to hamsters in the cage 6 h.p.i. Animal weight changes were monitored, and nasal washes and throat swabs were taken daily. Animals were euthanized 3 d.p.i. for necropsy, with viral titers and gRNA of lung tissues measured. (B) Percentage of body weight change of PiN-21 aerosol–treated animals compared to the control (n = 6). (C) Reduction of viral titers in hamster lungs (3 d.p.i.). Significant differences were observed between treated and control groups. **P < 0.01; *P < 0.05. The dashed line indicates the detection limit of the assay. (D) Lung pathology scores of treated and control groups. Significant difference was denoted by ****P < 0.0001. (E) Hematoxylin and eosin staining of necrotizing bronchointerstitial pneumonia affiliate with abundant SARS-CoV-2 S antigen in bronchiole epithelium and alveolar type 1 and 2 pneumocytes in the control group. All images were acquired at 20×; scale bar, 100 μm. Areas marked by boxes are shown at higher magnification in the rightmost panel (scale bar, 25 μm). (F) Immunostainings of bronchointerstitial compartments (3 d.p.i.). Orange, SARS-CoV-2S; magenta, CD68/macrophages; red, CD3e+ T cells; teal, ACE2; gray, DAPI. The bronchiole is outlined by white hash. Total magnification, ×200; scale bar, 100 μm.
Effective control of SARS-CoV-2 infection in the LRT of PiN-21 aerosolized animals
To understand the mechanisms by which Nb aerosols prevent and/or ameliorate lower respiratory disease caused by SARS-CoV-2 infection better, we performed whole lung semiquantitative ordinal histologic analysis of control (n = 6) and PiN-21 (n = 6)–treated animals euthanized 3 d.p.i (tables S2 and S3). Cumulative scores encompassed pathologic features of airways, blood vessels, and alveoli/pulmonary interstitium. PiN-21 aerosolization protected most animals (5 of 6) from severe COVID-related histopathologic disease reflected by decreased ordinal scores (P < 0.0001) when compared to Nb-treated controls (Fig. 3D and table S4). Histopathologic findings in the control group resembled previous reports of SARS-CoV-2 inoculation in Syrian hamsters (41, 42). Pulmonary disease observed in the PiN-21–treated animals was very mild (Fig. 3E and fig. S5), being characterized by the absence of severe necrotizing bronchiolitis in most of the animals (five of six), a pathological finding ubiquitously observed in all control animals. Furthermore, the single PiN-21–treated animal with necrotizing bronchiolitis had localized disease compared to the multifocal and bilateral distribution observed in most Nb controls (four of six). Bronchiolitis was also affiliated with less severe bronchial hyperplasia and hypertrophy and the absence of syncytial cells when compared to Nb controls. The predominant histologic finding in PiN-21–treated animals was minimal-to-mild perivascular and peribronchial mononuclear inflammation consisting of macrophages and lymphocytes. Furthermore, aside from the one animal already mentioned, the PiN-21 group had considerably less interstitial inflammation with decreased vascular permeability, as indicated by the absence of perivascular and intra-alveolar edema, hemorrhage, and fibrin exudation (fig. S5).
In control animals, S antigen was abundant in the cytoplasm of the bronchiolar epithelium, with less common detection in alveolar type 1 and 2 pneumocytes. Interstitial and peribronchiolar infiltrates were composed of large numbers of CD3e+ T cells and CD68+ macrophages, with a complete absence of angiotensin-converting enzyme 2 (ACE2) in the apical cytoplasm of bronchiole epithelium in areas with abundant viral S (Fig. 3F, top). Consistent with a striking 6-log virus reduction after aerosolization, S antigen was extremely sparse (<1% of permissive cells) in all PiN-21–treated animals, with decreased T cell and macrophage immune cell infiltrate, and retention of native apical bronchiole ACE2 expression (Fig. 3F, bottom).
To determine the impact of PiN-21 on the upper airways of the LRT, we also examined the trachea histologically. In PiN-21–treated animals, tracheas for all animals were within normal limits, while mild to moderate neutrophilic and lymphohistiocytic tracheitis, with variable degrees of degeneration and necrosis, and segmental hyperplasia and hypertrophy were observed in all the control animals (Fig. 3F). In summary, our data demonstrate that PiN-21 aerosolization given during early disease course is highly effective in decreasing SARS-CoV-2 entry and subsequent replication in permissive epithelial cells of the LRT; this, in turn, has a major impact on viral shedding. The result is the prevention of disease, including decreased cytopathic effect on permissive epithelial cells, retention of ACE2 expression on permissive bronchioles, and decreased recruitment of inflammatory cells to sites of replication.
DISCUSSION
In this work, we demonstrate the high therapeutic efficacy of a trimeric Nb against SARS-CoV-2 infection in Syrian hamsters. Our investigations leverage both intranasal and aerosol delivery of PiN-21 and demonstrate that Nb treatment effectively targets the deep and local pulmonary structures such as terminal alveoli, which are lined with alveolar cells rich in ACE2 receptor to block viral entry and replication efficiently. Moreover, infection-induced weight loss correlates with pulmonary virus titer (Pearson r = −0.7) (fig. S6) and this clinical sign may be used to indicate the onset of infections (13). Notably, the ability of PiN-21 to eradicate viral replication and lung pathology almost completely in both the URT and LRT in hamsters contrasts the effects recently shown by clinical antibodies, which, despite administrating at high doses (e.g., from 10 to 50 mg/kg), remain particularly challenging to treat SARS-CoV-2 infection in the same model (15).
Significantly improved delivery upon aerosolization may be anticipated in NHPs and humans because airway anatomical structures differ considerably from small rodents in which a high degree of inertial impaction is seen using liquid droplets. Several inhalation therapeutics with excellent safety profiles are commercially available, and many are under clinical trials (27, 43). A combination of extremely low deposit doses will minimize potential adverse effects. Nevertheless, further preclinical analysis including an extensive toxicopathologic investigation, preferentially in an NHP model, will be needed before moving this technology into human trials. We envision that PiN-21 aerosolization treatment could provide both a convenient and cost-effective solution to alleviate disease onset and reduce virus transmission, especially for mild COVID-19 patients who constitute major populations of infections. It may also benefit high-risk groups, such as seniors, immunocompromised individuals, and infants, in both inpatient and outpatient settings. Last, as prevalent circulating variants of SARS-CoV-2 have emerged to evade clinical antibodies and wane vaccine-elicited serologic responses (44–48), this proof-of-concept study will shed light on the use of stable, multi-epitope, and multivalent Nb constructs, in combination with PiN-21, as a novel aerosol cocktail that can be rapidly generated to block virus mutational escape (28).
MATERIALS AND METHODS
Ethics
The animal work performed adhered to the highest level of humane animal care standards. The University of Pittsburgh is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animal work was performed under the standards of the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH) and according to the Animal Welfare Act guidelines. All animal studies adhered to the principles stated in the Public Health Services Policy on Humane Care and Use of Laboratory Animals. The University of Pittsburgh Institutional Animal Care and Use Committee approved and oversaw the animal protocols for these studies (no. 20067405).
Biological safety
All work with SARS-CoV-2 was conducted under biosafety level-3 (BSL-3) conditions in the University of Pittsburgh Center for Vaccine Research (CVR) and the Regional Biocontainment Laboratory (RBL). Respiratory protection for all personnel when handling infectious samples or working with animals was provided by powered air-purifying respirators (PAPRs; Versaflo TR-300; 3M, St. Paul, MN). Liquid and surface disinfection was performed using Peroxigard disinfectant (1:16 dilution), while solid wastes, caging, and animal wastes were steam-sterilized in an autoclave.
Nb production
The PiN-21 gene (ANTE-CoV2-Nab21TGS) was synthesized from Synbio Biotechnologies and cloned into pET-21b vector as previously described (28, 32). Nb21Alb and PiN-21Alb were generated by subcloning a human serum albumin binding Nb (40) into the N terminus of Nb21 and PiN-21 constructs. The plasmid was transformed into BL21(DE3) cells and plated on LB agar with ampicillin (50 μg/ml) at 37°C overnight. Single bacterial colonies were picked and cultured in LB broth to reach an optical density of ~0.5 to 0.6 before isopropyl-β-d-thiogalactopyranoside induction (0.5 mM) at 16°C overnight. Cells were then harvested, sonicated, and lysed on ice with a lysis buffer [1× phosphate-buffered saline (PBS), 150 mM NaCl, and 0.2% Triton X-100 with protease inhibitor]. After cell lysis, his-tagged Nbs were purified by Cobalt resin and natively eluted using the imidazole buffer. Eluted Nbs were subsequently dialyzed into 1× Dulbecco’s PBS (DPBS) (pH 7.4). For animal studies, endotoxin was removed with the ToxinEraser Endotoxin Removal Kit (GenScript), and the endotoxin level was measured using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (GenScript) to make sure that it is <1 EU/ml. The proteins were sterile-filtered using 0.22-μm centrifuge filters (Costar) before use.
Virology
SARS-CoV-2/München-1.1/2020/929 (Munich) (multiplicity of infection of 0.03) was added to confluent monolayers of Vero E6 cells in 25 T175. After 1 hour of incubation at 37°C, 5% (v/v) CO2, virus growth medium (20 ml/flask) [Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS; Life Technologies), 1% (v/v) l-glutamine (Gibco), and 1% (v/v) penicillin/streptomycin (Life Technologies)] was added and incubation was continued for 66 to 72 hours until cytopathic effect was observed. Virus-containing supernatant was collected and clarified by centrifugation at 3500 rpm for 30 min at 4°C. The cleared virus supernatant was aliquoted and stored at −80°C.
Plaque assay
Samples were prepared in Opti-MEM (Gibco) and were added, in duplicate, to confluent Vero E6 monolayers in six-well plates (200 μl per well; Thermo Fisher Scientific). After 1 hour of incubation at 37°C, 5% CO2, virus growth medium (2 ml per well) containing 0.1% (w/v) immunodiffusion agarose (MP Biomedicals) was added and incubation was continued for 72 hours. Plates were fixed with formaldehyde (2 ml per well) [37% (w/v) formaldehyde stabilized with 10 to 15% (v/v) methanol; Thermo Fisher Scientific] for 15 min at room temperature. Agarose and fixative were discarded, and 1% (w/v) crystal violet (1 ml per well) in 10% (v/v) methanol (both Thermo Fisher Scientific) was added. Plates were incubated at room temperature for 20 min and then rinsed thoroughly with water. Plaques were then enumerated.
General animal procedures
Syrian hamsters (aged 3 to 6 months old, both male and female) were obtained from Charles River, MA. For procedures (virus infection, throat swab, and nasal wash collection), each animal was sedated with 3 to 5% isoflurane. Baseline body weights were measured for all animals before infection. The animals were monitored twice daily for signs of COVID-19 disease (ruffled fur, hunched posture, labored breathing, anorexia, lethargy) after challenge with SARS-CoV-2. Body weight was measured once daily during the study period. At necropsy, small pieces of the lung were collected for viral load determination. Throat swabs were collected using ultrathin swabs (Puritan PurFlock Ultra Sterile Flocked Swabs) that were placed in Opti-MEM (Invitrogen) containing double-strength antibiotic-antimycotic (Life Technologies). Nasal washes were collected using 500 μl of PBS with antibiotic-antimycotic. All samples were stored at −80°C until viral load determination. The whole trachea and lungs were collected in Opti-MEM, TRIzol, or 4% paraformaldehyde (PFA), respectively, for virus titrations, RT-qPCR, and histopathological examinations.
Under isoflurane anesthesia, hamsters were infected (300 μl) with 9 × 104 PFU (300 μl) of SARS-CoV-2 via intratracheal administration, immediately followed by intranasal administration of 100 μg (50 μl per nare) of PiN-21 or a control Nb. Under isoflurane anesthesia, hamsters were infected (50 μl per nare) with 3 × 104 PFU of SARS-CoV-2. At 6 h.p.i., animals were administered with 100 μg (50 μl per nare) of PiN-21 (n = 6) or a control Nb (n = 6). Hamsters were administered with PiN-21 and PiN-21Alb via aerosol route and sacrificed at 8 (n = 3) and 24 hours (n = 3) after administration. Under isoflurane anesthesia, hamsters were infected intranasally (50 μl per nare) with 3 × 104 PFU of SARS-CoV-2. At 6 h.p.i., animals were administered with PiN-21 (n = 6) or a control Nb (n = 6) via the aerosol route.
BAL collection
Lungs with trachea were harvested from euthanized animals. A Sovereign feeding tube (Covetrus) was cut to the optimal length and connected to a 5-ml syringe (BD Biosciences) containing 3 ml of PBS with antibiotic-antimycotic before placement into the trachea. The PBS was gently pushed into the lungs until they were fully inflated, after which the liquid (BAL) was drawn back into the syringe.
Sample extraction and processing
For tissues, 100 to 200 mg of tissue were harvested, suspended in 1 ml of Opti-MEM supplemented with 2× antibiotic-antimycotic, and homogenized using a D2400 homogenizer (Benchmark Scientific). The eluate from swabs and nasal washes was analyzed directly. Virus isolations were performed by inoculation of tissue homogenates (100 μl) onto Vero E6 cells (22). For the preparation of RNA, tissue homogenate, swab eluate, or nasal wash (100 μl) was added to 400 μl of TRIzol LS (Ambion) and thoroughly mixed by vortexing. To ensure virus inactivation, the samples were incubated for 10 min at room temperature and stored overnight at −80°C before removal from the BSL-3 facility. Subsequent storage at −80°C or RNA isolation and one-step RT-qPCR analyses were performed at BSL-2. RNA was extracted from these samples using Direct-zol RNA purification kits (Zymo Research) according to the manufacturer’s instructions. Viral RNA was detected by RT-qPCR targeting the SARS-CoV-2 nucleocapsid (N) segment, as previously described (49). The primers used are forward primer: 2019-nCoV_N2F (TTACAAACATTGGCCGCAAA), reverse primer: 2019-nCoV_N2R (GCGCGACATTCCGAAGAA), and probe: 2019-nCoV_N2probe (FAM- ACAATTTGCCCCCAGCGCTTCAG-BHQ1). The PCR conditions and the standard curve generation were carried out as described previously (49). Data were normalized by tissue weight and are reported as copies of RNA determined by comparing the cycle threshold (CT) values from the unknown samples to CT values from a positive-sense SARS-CoV-2 vRNA standard curve, as previously described (49). Graphs were generated using GraphPad Prism version 9.
Neutralization assay
Nbs or hamster serum dilution (100 μl) was mixed with 100 μl of SARS-CoV-2 (Munich: P3 virus) containing 75 PFU of the virus in Opti-MEM. The serum-virus mixes (200 μl total) were incubated at 37°C for 1 hour, after which they were added dropwise onto confluent Vero E6 cell monolayers in six-well plates. After incubation at 37°C, 5% (v/v) CO2 for 1 hour, 2 ml of 0.1% (w/v) immunodiffusion agarose in DMEM supplemented with 10% (v/v) FBS and 2× antibiotic-antimycotic was added to each well. After incubation at 37°C, 5% (v/v) CO2 for 72 hours, the agarose overlay was removed and the cell monolayer was fixed with formaldehyde (1 ml per well) [37% (w/v) formaldehyde stabilized with 10 to 15% (v/v) methanol] for 20 min at room temperature. Fixative was discarded and 1% (w/v) crystal violet (1 ml per well) in 10% (v/v) methanol was added. Plates were incubated at room temperature for 20 min and rinsed thoroughly with water. Plaques were then enumerated, and the 80% and/or 50% plaque reduction neutralization titer (PRNT80 or PRNT50) was calculated (28, 49). A SARS-CoV-2–positive convalescent patient serum and naive human serum were used as positive and negative controls, respectively, for Nb neutralization assay.
Nb aerosolization
Aerosol exposures of hamsters to Nbs were performed under the control of the Aero3G aerosol management platform (Biaera Technologies, Hagerstown, MD), as previously described for rodents (50). Hamsters were loaded into metal exposure cages and transported via mobile transfer cart to the Aerobiology suite in the RBL. There, they were transferred into a class III biological safety cabinet and placed inside a rodent whole-body exposure chamber. Hamsters were exposed for 12 to 15 min to small particle aerosols containing Nbs generated with an Aerogen Solo vibrating mesh nebulizer (Aerogen, Chicago, IL) (19). The system was set in a push/pull configuration with an equal volume of input air [19.5 liters per minute (lpm) total: 7.5 lpm generator, 12 lpm dilution air] and exhaust (19.5 lpm total: 6 lpm sampler, 5 lpm particle sizer, 8.5 additional vacuum) equal to 0.5 air changes per minute in the exposure chamber. To determine inhaled dose, an all-glass impinger (AGI; catalog no. 7541-10, Ace Glass, Vineland, NJ) containing 10 ml of PBS + 0.001% antifoam was attached to the chamber and operated at 6 lpm, −6 to −15 psi. Particle size was measured once during each exposure at 5 min using the Aerodynamic Particle Sizer (TSI, Shoreview, MN) operating at 5 lpm. A 5-min air wash followed each aerosol, after which animals were returned to their cage. AGI samples were evaluated to determine the concentration of Nbs recovered from the aerosol. The inhaled dose was determined as the product of the Nb aerosol concentration, duration of exposure, and the minute volume of the individual hamster (51). Minute volume was determined using Guyton’s formula (52).
Histologic processing and analysis
Tissue samples were fixed for a minimum of 24 hours in 4% PFA before being removed from BSL-3 and subsequently processed in a Tissue-Tek VIP-6 automated vacuum infiltration processor (Sakura Finetek) and embedded in paraffin using a HistoCore Arcadia paraffin embedding machine (Leica). Tissue sections (5 μm) were generated using an RM2255 rotary microtome (Leica) and transferred to positively charged slides, deparaffinized in xylene, and dehydrated in graded ethanol. Tissue sections were stained with hematoxylin and eosin for histologic examination, with additional serial sections used for immunohistochemistry. A Ventana Discovery Ultra (Roche) tissue autostainer was used for immunohistochemistry. Specific protocol details are outlined in tables S5 and S6. The histomorphological analysis was performed by a single board-certified veterinary pathologist (N.A.C.), who developed an ordinal grading score encompassing the diversity and severity of histologic findings using isotype control administered animals as a baseline. Histologic criteria were broken down into three compartments: airways, blood vessels, and interstitium, with results used to generate a cumulative lung injury score. This score also incorporated the overall degree of immunoreactivity to the SARS-CoV-2 S antigen. A summary of individual animal scores and specific criteria used to score lungs is included in tables S2 and S3.
Multispectral whole imaging
Brightfield and fluorescent images were acquired using the Mantra 2.0 Quantitative Pathology Imaging System (Akoya Biosciences). To maximize signal-to-noise ratios, fluorescent images were spectrally unmixed using a synthetic library specific for the Opal fluorophores used for each assay and for 4′,6-diamidino-2-phenylindole (DAPI). An unstained Syrian hamster lung section was used to create an autofluorescence signature that was subsequently removed from images using InForm software version 2.4.8 (Akoya Biosciences).
Nb aerosolization using the mesh nebulizer
Nb (Nb21Alb, PiN-21, and PiN-21Alb) was concentrated to 1 ml (1.5 mg/ml) in 1× DPBS. 0.5 ml (50% of the Nb) was saved as a control for enzyme-linked immunosorbent assay and pseudovirus assay. The other 0.5 ml was aerosolized by using a portable mesh atomizer nebulizer (MayLuck). No obvious dead volume was observed. The aerosolized droplets were collected in a microcentrifuge tube. The concentration was measured to calculate the recovery of the proteins.
Pseudotyped SARS-CoV-2 neutralization assay
Pseudotype neutralization assay was carried out, and IC50 (median inhibitory concentration) was calculated as previously described (28).
Acknowledgments
We would like to thank H. P. Gertje (histology), K. O’Malley (aerosol), M. Midgett (aerosol), M. D. Dunn (animal studies), T. Gilliland (animal studies), E. L. Cottle (animal studies), R. C. Walker (animal studies), S. R. Barrick (animal studies), and K. A. Thakkar (laboratory) for excellent technical assistance. Funding: This work was funded by NIH 1R35GM137905-01 (Y.S.), a CTSI pilot grant (Y.S.), the University of Pittsburgh (W.P.D.), the Center for Vaccine Research (W.P.D.), the Commonwealth of Pennsylvania Department of Community and Economic Development (W.P.D.), the Richard King Mellon Foundation (W.P.D.), and the Henry L. Hillman Foundation (W.P.D.). Author contributions: Y.S. and W.P.D. conceived and supervised the study. W.B.K. oversaw the study. S.N. performed animal studies and virology experiments. Y.X. and N.L.T.-L. helped S.N. in animal studies. Y.X. performed Nb purification, molecular biology, and biochemistry. Y.X. and N.L.T.-L. performed viral RNA analysis. L.J.R. performed tissue culture and helped in viral RNA analysis. D.S.R. carried out the aerosolization of Nbs. Z.S. helped with the biochemistry analysis. N.A.C. designed and optimized immunohistochemical assays and carried out the histopathologic analysis. Y.S., W.P.D., S.N., and Y.X. designed the experiments and interpreted the results. Y.X., Y.S., S.N., and N.A.C. prepared the figures. Y.S. drafted the manuscript with edits from Y.X., S.N., N.A.C., and W.P.D. Competing interests: Y.S. and Y.X. are inventors on a pending patent related to this work filed by University of Pittsburgh (no. 63067567, filed on 28 August 2020). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. PiN-21 plasmid can be provided by Y.S. pending scientific review and a completed material transfer agreement. Requests for the PiN-21 plasmid should be submitted to Y.S.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabh0319/DC1
REFERENCES AND NOTES
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFrancesco L., COVID-19 antibodies on trial. Nat. Biotechnol. 38, 1242–1252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Libster R., Marc G. P., Wappner D., Coviello S., Bianchi A., Braem V., Esteban I., Caballero M. T., Wood C., Berrueta M., Rondan A., Lescano G., Cruz P., Ritou Y., Viña V. F., Paggi D. Á., Esperante S., Ferreti A., Ofman G., Ciganda Á., Rodriguez R., Lantos J., Valentini R., Itcovici N., Hintze A., Oyarvide M. L., Etchegaray C., Neira A., Name I., Alfonso J., Castelo R. L., Caruso G., Rapelius S., Alvez F., Etchenique F., Dimase F., Alvarez D., Aranda S. S., Yanotti C. S., De Luca J., Baglivo S. J., Laudanno S., Nowogrodzki F., Larrea R., Silveyra M., Leberzstein G., Debonis A., Molinos J., González M., Perez E., Kreplak N., Argüello S. P., Gibbons L., Althabe F., Bergel E., Polack F. P.; Fundación INFANT–COVID-19 Group , Early high-titer plasma therapy to prevent severe Covid-19 in older adults. N. Engl. J. Med. 384, 610–618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen J., Baum A., Pascal K. E., Russo V., Giordano S., Wloga E., Fulton B. O., Yan Y., Koon K., Patel K., Chung K. M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.-Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D. C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Zost S. J., Gilchuk P., Case J. B., Binshtein E., Chen R. E., Nkolola J. P., Schäfer A., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Martinez D. R., Williamson L. E., Chen E. C., Jones T., Day S., Myers L., Hassan A. O., Kafai N. M., Winkler E. S., Fox J. M., Shrihari S., Mueller B. K., Meiler J., Chandrashekar A., Mercado N. B., Steinhardt J. J., Ren K., Loo Y.-M., Kallewaard N. L., McCune B. T., Keeler S. P., Holtzman M. J., Barouch D. H., Gralinski L. E., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Wang P., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F.-W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.-Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z., Huang Y., Shapiro L., Ho D. D., Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brouwer P. J. M., Caniels T. G., van der Straten K., Snitselaar J. L., Aldon Y., Bangaru S., Torres J. L., Okba N. M. A., Claireaux M., Kerster G., Bentlage A. E. H., van Haaren M. M., Guerra D., Burger J. A., Schermer E. E., Verheul K. D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M. J., Bijl T. P. L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N. A., Wiersinga W. J., Vidarsson G., Haagmans B. L., Ward A. B., de Bree G. J., Sanders R. W., van Gils M. J., Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortorici M. A., Beltramello M., Lempp F. A., Pinto D., Dang H. V., Rosen L. E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., de Marco A., Guarino B., Bianchi S., Lauron E. J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J. A., Case J. B., Chen R. E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.-Y. C., Schmid M. A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M. S., Neyts J., Diamond M. S., Virgin H. W., Snell G., Corti D., Fink K., Veesler D., Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S., Hillyer C., Du L., Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses: (Trends in Immunology 41, 355-359; 2020). Trends Immunol. 41, 545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., Atwal G. S., Oyejide A., Goez-Gazi Y., Dutton J., Clemmons E., Staples H. M., Bartley C., Klaffke B., Alfson K., Gazi M., Gonzalez O., Dick E. Jr., Carrion R. Jr., Pessaint L., Porto M., Cook A., Brown R., Ali V., Greenhouse J., Taylor T., Andersen H., Lewis M. G., Stahl N., Murphy A. J., Yancopoulos G. D., Kyratsous C. A., REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 370, 1110–1115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinnon K. H. III, Leist S. R., Schäfer A., Edwards C. E., Martinez D. R., Montgomery S. A., West A., Yount B. L. Jr., Hou Y. J., Adams L. E., Gully K. L., Brown A. J., Huang E., Bryant M. D., Choong I. C., Glenn J. S., Gralinski L. E., Sheahan T. P., Baric R. S., A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J. F., Zhang A.-J., Yuan S., Poon V.-K., Chan C.-C., Lee A.-C., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.-P., Kok K.-H., Chu H., Chan K.-H., Sridhar S., Chen Z., Chen H., To K.-K., Yuen K.-Y., Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 71, 2428–2446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munster V. J., Feldmann F., Williamson B. N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V. A., Rosenke R., Hanley P. W., Saturday G., Scott D., Fischer E. R., de Wit E., Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585, 268–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J., Tostanoski L. H., Peter L., Mercado N. B., McMahan K., Mahrokhian S. H., Nkolola J. P., Liu J., Li Z., Chandrashekar A., Martinez D. R., Loos C., Atyeo C., Fischinger S., Burke J. S., Slein M. D., Chen Y., Zuiani A., Lelis F. J. N., Travers M., Habibi S., Pessaint L., van Ry A., Blade K., Brown R., Cook A., Finneyfrock B., Dodson A., Teow E., Velasco J., Zahn R., Wegmann F., Bondzie E. A., Dagotto G., Gebre M. S., He X., Jacob-Dolan C., Kirilova M., Kordana N., Lin Z., Maxfield L. F., Nampanya F., Nityanandam R., Ventura J. D., Wan H., Cai Y., Chen B., Schmidt A. G., Wesemann D. R., Baric R. S., Alter G., Andersen H., Lewis M. G., Barouch D. H., DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., Xu Y., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Liu Y., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C., Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 369, 818–823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Fontela C., Dowling W. E., Funnell S. G. P., Gsell P.-S., Riveros-Balta A. X., Albrecht R. A., Andersen H., Baric R. S., Carroll M. W., Cavaleri M., Qin C., Crozier I., Dallmeier K., de Waal L., de Wit E., Delang L., Dohm E., Duprex W. P., Falzarano D., Finch C. L., Frieman M. B., Graham B. S., Gralinski L. E., Guilfoyle K., Haagmans B. L., Hamilton G. A., Hartman A. L., Herfst S., Kaptein S. J. F., Klimstra W. B., Knezevic I., Krause P. R., Kuhn J. H., Grand R. L., Lewis M. G., Liu W.-C., Maisonnasse P., McElroy A. K., Munster V., Oreshkova N., Rasmussen A. L., Rocha-Pereira J., Rockx B., Rodríguez E., Rogers T. F., Salguero F. J., Schotsaert M., Stittelaar K. J., Thibaut H. J., Tseng C.-T., Vergara-Alert J., Beer M., Brasel T., Chan J. F. W., García-Sastre A., Neyts J., Perlman S., Reed D. S., Richt J. A., Roy C. J., Segalés J., Vasan S. S., Henao-Restrepo A. M., Barouch D. H., Animal models for COVID-19. Nature 586, 509–515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartman A. L., Nambulli S., McMillen C. M., White A. G., Tilston-Lunel N. L., Albe J. R., Cottle E., Dunn M. D., Frye L. J., Gilliland T. H., Olsen E. L., O’Malley K. J., Schwarz M. M., Tomko J. A., Walker R. C., Xia M., Hartman M. S., Klein E., Scanga C. A., Flynn J. A. L., Klimstra W. B., McElroy A. K., Reed D. S., Duprex W. P., SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLOS Pathog. 16, e1008903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., Guo Y., Deng Y.-Q., Huang W.-J., Liu Q., Liu Q.-M., Shen Y.-L., Zhou Y., Yang X., Zhao T.-Y., Fan C.-F., Zhou Y.-S., Qin C.-F., Wang Y.-C., A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar A., Liu J., Martinot A. J., McMahan K., Mercado N. B., Peter L., Tostanoski L. H., Yu J., Maliga Z., Nekorchuk M., Busman-Sahay K., Terry M., Wrijil L. M., Ducat S., Martinez D. R., Atyeo C., Fischinger S., Burke J. S., Slein M. D., Pessaint L., van Ry A., Greenhouse J., Taylor T., Blade K., Cook A., Finneyfrock B., Brown R., Teow E., Velasco J., Zahn R., Wegmann F., Abbink P., Bondzie E. A., Dagotto G., Gebre M. S., He X., Jacob-Dolan C., Kordana N., Li Z., Lifton M. A., Mahrokhian S. H., Maxfield L. F., Nityanandam R., Nkolola J. P., Schmidt A. G., Miller A. D., Baric R. S., Alter G., Sorger P. K., Estes J. D., Andersen H., Lewis M. G., Barouch D. H., SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369, 812–817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P., Nirula A., Heller B., Gottlieb R. L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Adams A. C., Van Naarden J., Custer K. L., Shen L., Durante M., Oakley G., Schade A. E., Sabo J., Patel D. R., Klekotka P., Skovronsky D. M.; BLAZE-1 Investigators , SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 384, 229–237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinreich D. M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B. J., Soo Y., Rofail D., Im J., Perry C., Pan C., Hosain R., Mahmood A., Davis J. D., Turner K. C., Hooper A. T., Hamilton J. D., Baum A., Kyratsous C. A., Kim Y., Cook A., Kampman W., Kohli A., Sachdeva Y., Graber X., Kowal B., Cioccio T. D., Stahl N., Lipsich L., Braunstein N., Herman G., Yancopoulos G. D.; Trial investigators , REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 384, 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton J. S., Byron P. R., Inhaling medicines: Delivering drugs to the body through the lungs. Nat. Rev. Drug Discov. 6, 67–74 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W. P., Schneidman-Duhovny D., Zhang C., Shi Y., Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370, 1479–1484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoof M., Faust B., Saunders R. A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C. B., Puchades C., Azumaya C. M., Kratochvil H. T., Zimanyi M., Deshpande I., Liang J., Dickinson S., Nguyen H.-C., Chio C.-M., Merz G. E., Thompson M. C., Diwanji D., Schaefer K., Anand A. A., Dobzinski N., Zha B.-S., Simoneau C. R., Leon K., White K. M., Chio U.-S., Gupta M., Jin M., Li F., Liu Y., Zhang K., Bulkley D., Sun M., Smith A. M., Rizo A. N., Moss F., Brilot A. F., Pourmal S., Trenker R., Pospiech T., Gupta S., Barsi-Rhyne B., Belyy V., Barile-Hill A. W., Nock S., Liu Y., Krogan N. J., Ralston C. Y., Swaney D. L., García-Sastre A., Ott M., Vignuzzi M.; QCRG Structural Biology Consortium, Walter P., Manglik A., An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konwarh R., Nanobodies: Prospects of expanding the gamut of neutralizing antibodies against the novel Coronavirus, SARS-CoV-2. Front. Immunol. 11, 1531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrapp D., de Vlieger D., Corbett K. S., Torres G. M., Wang N., van Breedam W., Roose K., van Schie L.; VIB-CMB COVID-19 Response Team, Hoffmann M., Pöhlmann S., Graham B. S., Callewaert N., Schepens B., Saelens X., McLellan J., Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1436–1441 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang Y., Sang Z., Bitton L., Xu J., Liu Y., Schneidman-Duhovny D., Shi Y., Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst. 12, 220–234.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang W., Pan H. W., Vllasaliu D., Lam J. K. W., Pulmonary delivery of biological drugs. Pharmaceutics 12, 1025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimmerjahn F., Ravetch J. V., Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 310, 1510–1512 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Schäfer A., Muecksch F., Lorenzi J. C. C., Leist S. R., Cipolla M., Bournazos S., Schmidt F., Maison R. M., Gazumyan A., Martinez D. R., Baric R. S., Robbiani D. F., Hatziioannou T., Ravetch J. V., Bieniasz P. D., Bowen R. A., Nussenzweig M. C., Sheahan T. P., Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 218, e20201993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bournazos S., Ravetch J. V., Anti-retroviral antibody FcγR-mediated effector functions. Immunol. Rev. 275, 285–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Custódio T. F., Das H., Sheward D. J., Hanke L., Pazicky S., Pieprzyk J., Sorgenfrei M., Schroer M. A., Gruzinov A. Y., Jeffries C. M., Graewert M. A., Svergun D. I., Dobrev N., Remans K., Seeger M. A., McInerney G. M., Murrell B., Hällberg B. M., Löw C., Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 11, 5588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanke L., Vidakovics Perez L., Sheward D. J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G. B., Hällberg B. M., Murrell B., McInerney G. M., An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 11, 4420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan W., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.-S. C., du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Z. Shen, Y. Xiang, S. Vegara, A. Chen, Z. Xiao, U. Santiago, C. Jin, Z. Sang, J. Luo, K. Chen, D. Schneidman-Duhovny, C. Camacho, G. Calero, B. Hu, Y. Shi, A robust and versatile nanobody platform for drug delivery. bioRxiv 2020.08.19.257725 [Preprint]. 20 August 2020. 10.1101/2020.08.19.257725. [DOI]
- 41.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P. J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., Yamada S., Fan S., Chiba S., Kuroda M., Guan L., Takada K., Armbrust T., Balogh A., Furusawa Y., Okuda M., Ueki H., Yasuhara A., Sakai-Tagawa Y., Lopes T. J. S., Kiso M., Yamayoshi S., Kinoshita N., Ohmagari N., Hattori S.-i., Takeda M., Mitsuya H., Krammer F., Suzuki T., Kawaoka Y., Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U.S.A. 117, 16587–16595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sia S. F., Yan L.-M., Chin A. W. H., Fung K., Choy K.-T., Wong A. Y. L., Kaewpreedee P., Perera R. A. P. M., Poon L. L. M., Nicholls J. M., Peiris M., Yen H.-L., Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laube B. L., Aerosolized medications for gene and peptide therapy. Respir. Care 60, 806–821; discussion 821-804 (2015). [DOI] [PubMed] [Google Scholar]
- 44.N. G. Davies, S. Abbott, R. C. Barnard, C. I. Jarvis, A. J. Kucharski, J. Munday, C. A. B. Pearson, T. W. Russell, D. C. Tully, A. D. Washburne, T. Wenseleers, A. Gimma, W. Waites, K. L. M. Wong, K. van Zandvoort, J. D. Silverman, CMMID COVID-19 Working Group, K. Diaz-Ordaz, R. Keogh, R. M. Eggo, S. Funk, M. Jit, K. E. Atkins, W. J. Edmunds, Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv 2020.2012.2024.20248822 (2020).
- 45.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C. O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J. A., Yang Z., Abernathy M. E., Huey-Tubman K. E., Hurley A., Turroja M., West K. A., Gordon K., Millard K. G., Ramos V., Silva J. D., Xu J., Colbert R. A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P. J., Casellas R., Hatziioannou T., Bieniasz P. D., Nussenzweig M. C., mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592, 616–622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Tegally, E. Wilkinson, M. Giovanetti, A. Iranzadeh, V. Fonseca, J. Giandhari, D. Doolabh, S. Pillay, E. J. San, N. Msomi, K. Mlisana, A. von Gottberg, S. Walaza, M. Allam, A. Ismail, T. Mohale, A. J. Glass, S. Engelbrecht, G. Van Zyl, W. Preiser, F. Petruccione, A. Sigal, D. Hardie, G. Marais, M. Hsiao, S. Korsman, M.-A. Davies, L. Tyers, I. Mudau, D. York, C. Maslo, D. Goedhals, S. Abrahams, O. Laguda-Akingba, A. Alisoltani-Dehkordi, A. Godzik, C. K. Wibmer, B. Trevor Sewell, J. Lourenço, L. C. J. Alcantara, S. L. K. Pond, S. Weaver, D. Martin, R. J. Lessells, J. N. Bhiman, C. Williamson, T. de Oliveira, Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.2012.2021.20248640 (2020).
- 47.Greaney A. J., Loes A. N., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., Bloom J. D., Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.E6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson E. C., Rosen L. E., Shepherd J. G., Spreafico R., da Silva Filipe A., Wojcechowskyj J. A., Davis C., Piccoli L., Pascall D. J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R. M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V. B., Nix J., Jarrett R. F., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Johnson N., Wickenhagen A., Ceschi A., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Semple M. G., Openshaw P. J. M., Baillie J. K.; The ISARICC Investigators; COVID- Genomics UK (COG-UK) consortium, Rihn S. J., Lycett S. J., Virgin H. W., Telenti A., Corti D., Robertson D. L., Snell G., Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.E20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klimstra W. B., Tilston-Lunel N. L., Nambulli S., Boslett J., McMillen C. M., Gilliland T., Dunn M. D., Sun C., Wheeler S. E., Wells A., Hartman A. L., McElroy A. K., Reed D. S., Rennick L. J., Duprex W. P., SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 101, 1156–1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faith S. A., Smith L. P., Swatland A. S., Reed D. S., Growth conditions and environmental factors impact aerosolization but not virulence of Francisella tularensis infection in mice. Front. Cell. Infect. Microbiol. 2, 126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowling J. D., O’Malley K. J., Klimstra W. B., Hartman A. L., Reed D. S., A vibrating mesh nebulizer as an alternative to the collison three-jet nebulizer for infectious disease aerobiology. Appl. Environ. Microbiol. 85, e00747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guyton A. C., Measurement of the respiratory volumes of laboratory animals. Am. J. Phys. 150, 70–77 (1947). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabh0319/DC1


