A proposed machine learning method capable of mining feature interactions reveals the mechanisms of bio-nano interactions.
Abstract
The development of machine learning provides solutions for predicting the complicated immune responses and pharmacokinetics of nanoparticles (NPs) in vivo. However, highly heterogeneous data in NP studies remain challenging because of the low interpretability of machine learning. Here, we propose a tree-based random forest feature importance and feature interaction network analysis framework (TBRFA) and accurately predict the pulmonary immune responses and lung burden of NPs, with the correlation coefficient of all training sets >0.9 and half of the test sets >0.75. This framework overcomes the feature importance bias brought by small datasets through a multiway importance analysis. TBRFA also builds feature interaction networks, boosts model interpretability, and reveals hidden interactional factors (e.g., various NP properties and exposure conditions). TBRFA provides guidance for the design and application of ideal NPs and discovers the feature interaction networks that contribute to complex systems with small-size data in various fields.
INTRODUCTION
Nanoparticles (NPs) have attracted increasing attention in health care, biosensor, and immunotherapy research because of their excellent physicochemical properties (1, 2). NPs induce an immune system response once they enter or contact the body of humans or other organisms, and their pharmacokinetics also play critical roles in nanomedicine or other biologically related applications (3–5). For conventional biological experiments performed to assess the immune responses and organ burden of NPs, the cost is high, the reproducibility is low, the experimental time is long, and many animals are sacrificed (4, 6, 7). Quickly and accurately predicting the immune response and organ burden of NPs based on their physical and chemical properties is urgent for NP design and applications and pharmacokinetic assessments (8, 9). Given the complexity of the immune system and complicated properties of NPs, conventional methods [e.g., quantitative structure-activity relationships (QSARs) and molecular dynamics simulations] cannot precisely predict the immune response and organ burden of NPs.
The molecular structure must be known for QSARs and molecular dynamics simulations (10). However, the molecular structure of the immune system is difficult to depict. In addition, the complicated molecular calculations required for QSAR and molecular dynamics simulations require a long time to complete (11), which limits the ability to manage thousands of NPs with many properties (e.g., size, shape, and surface modifications) and the capacity to address data heterogeneity (12). As a type of robust nonparametric model, machine learning approaches, such as random forest (RF), artificial neural network (ANN), and support vector machine (SVM), have good potential to construct models that simulate complex relationships (12, 13). The anti-interference capability of machine learning may overcome data heterogeneity and provide a solution to predicting complicated biological responses to NPs (14, 15). However, a great challenge for machine learning methods is their poor interpretability, thus affecting the trustworthiness of models (16). Although high-precision predictions or classification tasks can be achieved by configuring the appropriate parameters (17, 18), the internal operations of the model are obscure, and poor interpretability also obscures causality (19). Current interpretable studies in the field of machine learning, such as prototype networks, local interpretable model-agnostic explanations, and Shapley additive explanations (20–22), are devoted to revealing how machine learning works to achieve classification tasks to judge the rationality of decision-making, but they have not paid attention to the interaction among multiple features. Understanding the interaction among multiple features is useful to design NPs with ideal features and explore the mechanisms of bio-nano interactions.
To improve the interpretability of machine learning, it is urgent to understand how features affect labels and interact with each other (23), which widely occur in various machine learning models, such as RF and ANN (12, 19, 24). The present work proposed a feature interaction network concept along with a tree-based RF feature importance and feature interaction network analysis framework (TBRFA) as a proof-of-principle demonstration. TBRFA disassembles the trees implied by RF and then improves the interpretability of the RF model. The scheme is shown in Fig. 1. Inspired by meta-analysis workflow concepts and comparisons of RF, ANN, and SVM models, this study predicted the immune response and organ burden of various NPs with complicated properties. High correlation coefficients between the observations and predictions are achieved and further verified by validation set and animal experiments, thus ensuring the reliability of the model for the TBRFA framework. TBRFA includes two parts: importance analysis and feature interaction network analysis. TBRFA used a multiple-indicator importance analysis approach for RF based on existing methods to comprehensively screen the important features for the immune response and organ burden of NPs, which resolved the problems caused by the unbalanced data structure and the routine importance analysis method. Moreover, TBRFA proposes an interaction coefficient, uses the working mechanism of models to explore the interaction relationships among multiple features, and builds feature interaction networks to provide guidance for the design and application of ideal NPs.
Fig. 1. Overview of the machine learning workflow and TBRFA.
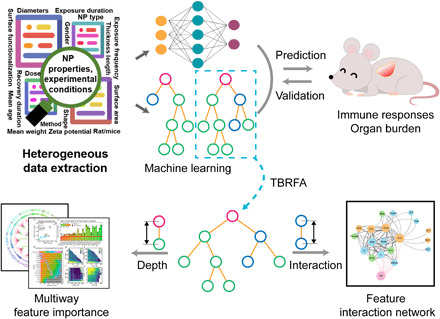
Data extracted from publications are regressed by the RF and ANN methods. The random forest mechanism is explored in depth by TBRFA to screen important features and build feature interaction networks to provide guidance for the design and application of ideal NPs. Com.2, main element/component 2; D, diameter; Dim, dimension; E.D., exposure duration; IH., inhalation; IN.N., intranasal inoculation; IN.T., intranasal instillation, L, length; M.C., macromolecular compound; M.W., mean weight; Mφ, macrophages; R.D., recovery duration; SSA, specific surface area.
RESULTS
Description and pretreatment of heterogeneous data
The data were extracted following the process in Materials and Methods. To comprehensively correlate immune responses and lung burden with the physical and chemical properties of NPs, data and literature on pulmonary immune responses and NP burden caused by lung exposure were assembled and integrated. On the basis of a comprehensive and rigorous extraction criterion of published data, a total of 1620 samples containing 16 features (including three parts: NP properties, animal properties, and experimental conditions) and 12 toxicity labels [biomarkers, e.g., total protein, total cells, and interleukin-1β (IL-1β)] were mined for immune response datasets. A total of 301 samples containing the same 16 features and 3 burden labels [i.e., lung, liver, and bronchoalveolar lavage fluid (BALF)] were mined for burden datasets. The references for the above samples are given in supplementary note S1. The datasets are described in Fig. 2 and tables S1 and S2. Six characteristic variables (NP type, shape, surface functionalization, rat/mouse, sex, and method) were described by the reported frequency (percentage). Inhalation as an exposure method was not recorded because of the difficulty in normalizing the inhaled doses. Ten numeric variables [diameter, thickness/length, zeta potential, specific surface area (SSA), mean age, mean weight, exposure duration, exposure frequency, recovery duration, and dose] were described by mathematical statistics (mean, SD, median, and distribution range). The abovementioned 6 characteristic variables and 10 numeric variables covered the main factors in the immune response analysis (25, 26). The distribution of the samples was visualized and arranged in descending order of NP length (Fig. 2). Heterogeneity of the immune response datasets was mainly caused by NP diversity (number of NP types, 57). The literature on biological responses usually focuses on widely used NPs [for example, multiwalled carbon nanotube (MWCNT) and TiO2 accounted for 22.54 and 16.6% in the immune response dataset, respectively], leading to a small proportion of some uncommon or previously unknown NPs. Encoding the discrete NP types into several continuous features reduced biases caused by the imbalance of NP types. The inconsistent NP characterization standards and differences in NP properties and the exposure protocols among laboratories also led to challenges for precise prediction. Systematic evaluations of the immune response to 57 NPs by routine experiments are costly and time-consuming, and it is also difficult to finish such complex model construction for QSAR analysis.
Fig. 2. Visualization of the data distribution of the raw datasets was performed using the “tabplot” package in R software.
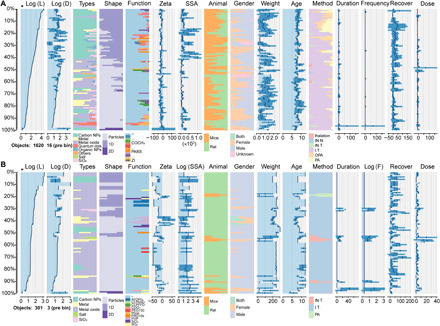
(A) Immune response dataset (data file S1). (B) NP burden dataset (data file S2). Samples are arranged in descending order of material length. The figure divided the raw data (data files S1 and S2) into 100 parts based on the material length and displayed the data distribution of the sample features. Logarithmic processing is performed for features with large distribution ranges. Characteristic variables (e.g., type and shape) are simply classified because they contain too many elements. The immunotoxicity dataset contains a variety of NPs, while metal oxides have a high proportion in the burden dataset. The distribution of exposure duration shows that most studies only focus on the acute toxicity of NPs. E.F, exposure frequency; IN.N., intranasal inoculation; I.T., intratracheal instillation; OPA, oropharyngeal aspiration; PA, pharyngeal aspiration.
Many immune response biomarkers have been reported, although the choice of immune response index varies from laboratory to laboratory. Given the high reported frequency and the strong connections with immune responses (27, 28), the following 12 biomarkers were individually used for machine learning model construction: total protein, lactate dehydrogenase (LDH), total cells, macrophages, neutrophils), IL-1β, tumor necrosis factor–α (TNF-α), IL-6, IL-4, IL-10, monocyte chemoattractant protein-1 (MCP-1), and macrophage inflammatory protein–1α (MIP-1α). The burden datasets contained fewer NP types (n = 17) and had lower heterogeneity than the immune datasets. The NP burdens in the lung, liver, and BALF were chosen as labels. The details of the subsets are listed in table S3. General regression methods, such as multiple linear regressions, presented poor performance on heterogeneous data in complex systems because of the missing values (16). As shown in fig. S1, most of the multiple linear regressions obtained low correlation coefficients [coefficient of determination (R2), minimum value: 0.324] and high root mean square errors (RMSEs; maximum value: 0.902), especially in the large subsets, indicating that traditional regression methods were not suitable for heterogeneous data. In contrast, machine learning can achieve accurate predictions of these data, and the internal relationships of complex systems can be mined by TBRFA-based machine learning, as indicated in the following sections.
Model performance and feature selection
Nonparametric and nonlinear machine learning methods have the ability to resist noise and are expected to build accurate prediction models using aggregated data (24). To eliminate the dimensional effects and balance the weights of features, z-score normalization and encoding of the character variables were applied before the model training (details are provided in Materials and Methods). The label values need to be normalized to improve the accuracy of the models. However, the distribution range of the label data was too wide (e.g., total proteins ranged from 1.5 to 2752.9%), and a considerable number of outliers occurred. The rough use of the z score can lead to serious collapse of the model accuracy. For the immune response dataset, we used the percentage change of the experimental group relative to the control group as the label and compressed the values between −1 and 1 (Materials and Methods, formula 2). For the organ burden dataset, we directly used the ratio of the detection concentration to the total exposure dose as the label. We initially compared the performance of ANN, SVM, and RF using all features for regression. To ensure that credible results are obtained from machine learning with a meta-analysis workflow, high R2 values are necessary (16). The R2 of the regression (Fig. 3, A to C) showed that in terms of the test set, the performance of RF (average of all models, 0.75 ± 0.12) was better than that of ANN (average of all models, 0.67 ± 0.11) and SVM (average of all models, 0.64 ± 0.10). Moreover, RF spent less time during the training process than ANN and had simpler adjustable parameters than ANN and SVM.
Fig. 3. Performance of the machine learning models.
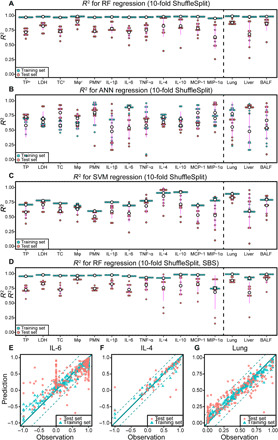
(A) R2 distribution of the RF regression (10-fold ShuffleSplit cross-validation). (B) R2 distribution of the ANN regression (10-fold ShuffleSplit cross-validation). (C) R2 distribution of the SVM regression (10-fold ShuffleSplit cross-validation). (D) R2 distribution of the RF regression with sequential backward selection (SBS) feature selection (10-fold ShuffleSplit cross-validation). (E to G) Prediction performance of IL-6, IL-4, and lung burden using RF models. The slope of the solid lines is 1, and the dotted lines represent the intercepts of ±RMSE. TP, total proteins; TC, total cells; PMN, polymorphonuclear neutrophil.
Figure S2 (A and B) shows that most of the features have low linear correlations, indicating that the features obtained via the literature and generated through coding will not cause overfitting due to multicollinearity. Figure S2 (A and B) also shows that there are low linear correlations between features and labels. The immune responses and accumulation burden of NPs are complicated because a single feature contributes little information to the label. Although the tested features may all affect the performance of models (29), a suitable feature selection procedure is still necessary to determine whether there is undetectable redundant information in the features. A greedy algorithm, sequential backward selection (SBS; see Materials and Methods), was used here to eliminate redundant information. Figure 3D shows that the SBS algorithm hardly improved the performance of the models, and we found that SBS abandoned some important NPs properties. Therefore, the RF models constructed on the basis of all features were selected for the subsequent analysis. The R2 values of the test set of most models were greater than 0.7, where R2 values for macrophages, lung burden, and BALF burden were >0.85, and the maximum value reached 0.896 (Fig. 3A and table S3). For some biological indicators, the testing R2 values less than 0.7 were probably due to the biases of data from interlaboratory studies. Moreover, we performed permutation tests, and the intercepts of the cross-validation coefficients (Q2) on the y axis were all less than 0.05, indicating that the models did not overfit (fig. S3) (30). Figure 3 (E to G) lists three examples (IL-6, IL-4, and lung burden) of regression results, and the others are given in fig. S4. To ensure that the features contributed valid information to the models, we performed feature value shuffling, and the predictive performance was abrogated after feature value shuffling (fig. S5). Because the published immune literature contained few negative samples (experimental group > control group), the scatters were mostly distributed in the top right of the scatter plots (Fig. 3, E and F), and the predicted results of the negative samples were distributed outside the RMSE interval. This flaw is inherently unavoidable because of the published datasets, although the prediction accuracy of the positive samples is not influenced. Multilabel prediction was performed by ANN, but the prediction accuracy rate was low, with most of the R2 values less than 0.7 (fig. S6), because the labels were independent of each other and represented different biological meanings.
NPs with a wide range of responses were used to verify the models. Five samples of each subset were randomly sampled as a validation set before building the model. The validation set did not participate in the construction of the model at all to ensure that the model did not learn them during the cross-validation. Figure 4A shows the prediction errors of the models on the validation set, where 76% of validation errors were less than 0.2. To verify the model further, animal experiments were performed in our laboratory. MWCNTs functionalized with triethoxycaprylylsilane and three MWCNTs of different sizes (small, medium, and large MWCNT, named S-MWCNT, M-MWCNT, and L-MWCNT, respectively) were chosen to conduct animal experiments. The above materials were all outside the scope of the datasets. Figure S7 shows the characterization of the NPs, and Fig. 4 (B to F) shows the immunofluorescence results of IL-1β. The expressions of IL-1β induced by MWCNTs distributed within the error bounds of the RF model (Fig. 4G). Figure 4G also contains the observation prediction of the validation set. The validation set of IL-1β contained five additional samples, which were randomly sampled from the main NPs in the IL-1 subset. The quantitative comparison between the observed and predicted results of the validation set indicated that the model was reliable (Fig. 4G). Compared with SVM and ANN, RF exhibited high accuracy and reliability for dealing with heterogeneous data in a complex system.
Fig. 4. Validation of models and immunofluorescence imaging of lung tissue.
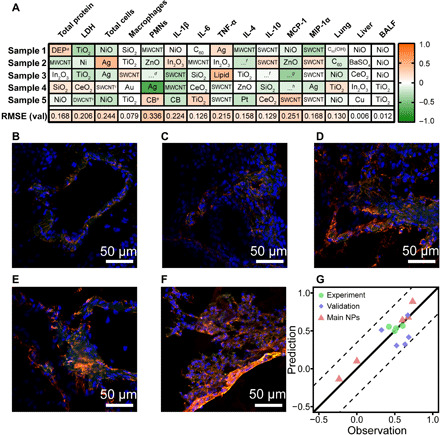
(A) Prediction errors of the validation sets. DEPa, diesel engine particles; DWCNTb, double-wall carbon nanotubes; SWCNTb, single-wall carbon nanotubes; …d, SWGe-imogolite; CBe, carbon black; …f, cellulose nanocrystals; …g, QD-CdSe-ZnS; …h, Rosette nanotubes. (B) Immunofluorescence imaging of control. (C) F-MWCNTs. (D) S-MWCNTs. (E) M-MWCNTs. (F) L-MWCNTs. (G) Validation of models using IL-1β fluorescence intensity. Red channel, nuclear factor κB (NF-κB) p65; green channel, IL-1β; and blue channel, 4′,6-diamidino-2-phenylindole (DAPI).
Discovering important and unbiased features by TBRFA
The RF model was used to measure the importance of features by calculating the change in the error (increase in MSE) on the out-of-bag (OOB) data based on permutations of each feature (19). Figure 5 visualizes the feature importance measured by the MSE increase. Each model was normalized by its most important features. Figure 5A shows that the exposure dose (Fig. 5B) and recovery duration (the time from last exposure to animal euthanasia; Fig. 5C) have a great impact on the immune responses and NP burden. However, as a mathematical statistic, increases in the MSE are limited by the quality of the dataset. Thus, using increased MSE values as the only criterion for analyzing feature importance may lead to bias. For example, increased MSE values indicated that sex (male and female) contributed significantly to the IL-4 model (Fig. 5A). However, sex was not supposed to be the main factor affecting the immune response, and we confirmed this bias in the follow-up analysis. Therefore, absolute dominant features may be incorrectly identified when using a single importance evaluation index.
Fig. 5. Conventional feature importance analysis.
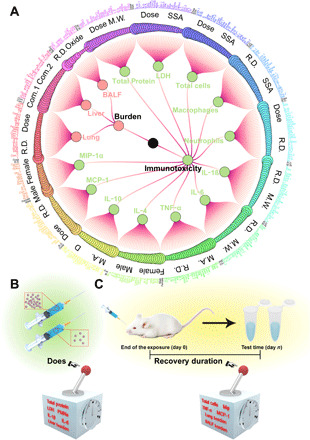
(A) Features importance measured by the increase in MSE. The dot size represents the importance of the features, and the connecting lines indicate the hierarchical relationship. The black words represent the two most important features of each model. Dose (B) and recovery duration (C) were identified as important features in most of the models.
TBRFA uses multiple indicators to perform a multiway feature importance analysis and can evaluate the importance of features from different perspectives to balance the absolute dominant features achieved by a single indicator. Three other indicators, i.e., node purity increase, mean minimal depth, and P value, were combined with increased MSE values to comprehensively screen important features (Fig. 6, A and B). Although gender led to a higher increase in MSE and node purity than the other features, the P value indicated that its importance was not statistical significant (Fig. 6A). The distribution of the features’ mean minimal depth also suggested that the zeta potential, dose, length/weight, and SSA appeared more frequently near the roots than the other features (Fig. 6B). The TBRFA results of all models indicated that recovery duration and exposure dose were the main factors affecting the immune response and organ burden (Fig. 6, A and B, and figs. S8 to S21). TBRFA successfully overcame the importance bias caused by a single indicator with small datasets.
Fig. 6. TBRFA importance analysis of IL-4 RF model.
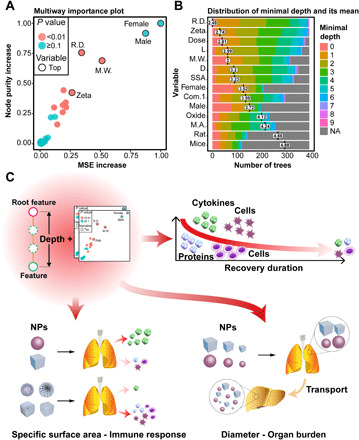
(A) Multiway feature importance analysis of the IL-4 model combining the MSE increase, node purity increase, and P values of the features. The importance of gender is not significant. (B) Distribution of the features’ mean minimal depth, recovery duration, zeta potential, dose, and length/weight are closer to the root of the trees than the other features. (C) Summary diagram of TBRFA importance analysis. Immune responses decreased as recovery duration increased. NPs with a large SSA cause low levels of cytokine release and high levels of total protein and cell numbers. NPs with a small diameter (<100 nm) could easily penetrate biological membranes and achieve cross-organ transport. NA, not available (missing value).
Figure 6C summarizes the conclusions of the TBRFA importance analysis. Exposure dose is well known as one of the most important features, while recovery duration, which represents the acute or chronic responses induced by NPs, has been ignored (9, 10). A partial dependence analysis (fig. S22A) showed that immune responses decreased as recovery duration increased and NPs tended to induce short-term acute immune responses. In the initial stage of exposure, the immune indicators increased rapidly; and within 50 days after the end of exposure, all immune indicators were decreased to less than 166.7% (corresponding to the normalized value 0.4 in fig. S22A) of the control group. The mechanisms of NP clearance by the immune system are complicated and include immune cell phagocytosis and extracellular traps (31). The clearance rate of NPs is also highly correlated with their structure and physical and chemical properties (32, 33). Figure S22B also confirmed that NPs could be gradually cleared by the immune system, with the accumulation of NPs in the lungs rapidly declining to approximately 20% of the total exposure dose within 100 days; hence, recovery duration became an important feature affecting the final observed immune responses. However, the immune responses did not decrease to the level of the control groups even after a long recovery duration. The optimized models proposed that the immune responses induced by NPs were persistent during the postexposure period; thus, the chronic and long-term toxicity of NPs deserve attention in future studies.
Although NP properties determine their immune response and organ burden, key properties of NPs remain inconclusive because of the limited number of animal experiments (34). The models proposed that SSA was important for immune responses, while diameter was important for organ burden compared with other properties (fig. S22, C and D). The SSA of NPs will allow the particles to adsorb proteins and form NP-protein corona complexes, which mediate immune responses (35). NPs with a large SSA are more likely to be internalized by the cell, thus causing a decrease in the number of molecules released by the cell (36). As the diameter decreased, fewer NPs stayed in the alveoli, and more NPs were transported to the lung and liver, which indicated that NPs with a small diameter (<100 nm) could easily penetrate biological membranes and achieve cross-organ transport (Fig. 6C and fig. S22D). A small size confers enhanced permeability to NPs, which is attributable to the binding to integrins on the surface of epithelial cells (37). Moreover, studies have shown that smaller NPs are less likely to be taken up by cells than larger NPs; thus, large NPs may preferentially translocate into the reticuloendothelial system, while small NPs may not be capable of being distributed among organs (38).
Animal studies, especially toxicological tests, are time-consuming, costly, and often impractical (10, 39). Because of these constraints, animal experiments can only screen the importance of specific or few properties (e.g., type, size, and surface functionalization) for one or a few NPs. In contrast, machine learning can quickly screen and sort various important features for various NPs at the same time. Moreover, the TBRFA approach overcame the conventional important feature analysis bias caused by the unbalanced data structure, identified the critical features (recovery duration, dose, SSA, and diameter), and quantified the importance of features and the response degree of biological biomarkers to features, thus providing a reference for the design of ideal NPs.
Feature interaction networks created by TBRFA
The mechanism underlying the ability of NPs to induce immune toxicity is complicated. Understanding how the different properties of materials interact with each other (feature interaction networks) and influence their immune response and organ burden is critical to the design and application of NPs, although the related information remains largely unknown (39). Because most machine learning methods are black-box models, identifying the interactions among features represents a difficult challenge. The conditional minimal depth (Fig. 7A) in RF represents the strength of interactions between two features (40). As demonstrated in Fig. 7B and figs. S8 to S21C, the conditional minimal depth was calculated to obtain the order of the interaction strength between features, and then the strongest four feature interaction relationships were displayed through a double-variable partial dependence analysis (Fig. 7, C to F, and figs. S8 to S21, D to G). However, strong interactions between features were not found for most of the partial dependence analyses, and they tended to show a simple additive effect. A comparison with the unconditional minimal depth of the feature showed that the conditional minimal depth was greatly affected by the importance of the feature itself. If the two features are both important, then they will have small and similar depths; therefore, even if they are close in the tree, they may not interact with each other.
Fig. 7. Feature interaction analysis using the conditional depth.
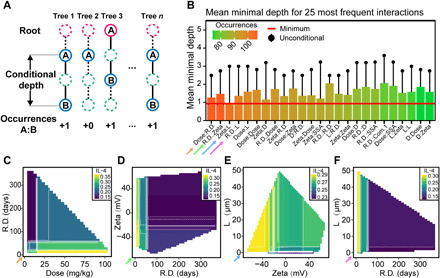
(A) Schematic diagram of conditional minimum depth in the RF model. (B) Mean minimal (conditional and unconditional) depth for the 25 most frequent interactions. (C to F) Double-variable partial dependence on the four strongest feature interactions, which corresponds to the four arrows in (B).
To weaken the influence of feature importance on the interaction represented by the conditional minimal depth, the RF based on the decision tree was explored again, which was called feature interaction network analysis. We redesigned an interaction coefficient on the basis of the conditional minimal depth (see Materials and Methods and Fig. 8A). To calculate the interaction coefficient between features, we explored the working mechanism of RF in depth, traversed each decision tree constituting the RF, and obtained the properties of the tree and its features. Subsequently, we integrated the interaction coefficients and built interaction networks for each RF model. The interactions between NP properties were mainly analyzed to provide guidance for NP design. Interaction networks of total proteins, total cells, IL-6, IL-4, lung burden, and liver burden are provided as examples in Fig. 8 (B to G), and the other networks are given in figs. S22 to S30. As shown in Fig. 8 (B to G), the network analysis for the immune response dataset indicated that SSA had a strong interaction with the zeta potential and length/width, while the network analysis of the burden dataset indicated that diameter had strong interactions with length. The above feature interaction network explained the critical roles of SSA in immune responses (fig. S22C). The feature interaction network clearly showed the connections between NP properties and immune responses or organ burden (Fig. 9, A to F). For example, NPs with negative charges (<0 mV) and small SSAs (0 to 200 m2/g) induced low levels of total protein and total cells increase and high levels of IL-4 and IL-6 release (Fig. 9, A to D). The total protein value reflects the trend of inflammation as a whole, and the complicated trend between different cytokines and NP zeta potentials may be related to the electrostatic interaction between the particle surface and cytokines (41, 42). There was an important influence of both zeta potential and SSA on the formation of protein coronas and the uptake of nanomaterials (43, 44). The interactions excavated by TBRFA indicated that both factors were mutually restrictive and affected the biocompatibility and toxicity of NPs. Long NPs caused more severe immune responses than short NPs, while a suitable SSA reduced different immune responses (Fig. 9, E to F). For the burden dataset, the interactions between diameter and length caused the opposite degree of lung and liver burden. One-dimensional NPs had the lowest accumulation in the lung and the highest accumulation in the liver, suggesting that length determined the translocation capacity of NPs, which was consistent with the conclusion drawn by Huang et al. (45) that long NPs were cleared from the lung quickly. The above results also agreed that gold nanorods accumulated significantly less than gold spheres with similar size and surface chemistry (46). NPs with nonspherical shapes exhibited the properties of targeted accumulation because of the deviating hydrodynamic behavior (e.g., roll and rotate) and lateral drifting speed in the vessel (47). In addition, NPs with a length and width in the range of 100 and 500 nm showed greater transport across organs (Fig. 9, G to I). Figure 9J visualizes the conclusions of the above partial dependence analysis. This result further supplements the conclusion on the NP size effect on organ burden obtained from the TBRFA importance analysis, indicating that feature interaction network analysis was a powerful method of boosting the interpretability of machine learning. The models are consistent with the experiments, ensuring the reliability of TBRFA. TBRFA discovers hidden feature interactions that are difficult to explore through few experiments and other machine learning methods [e.g., ANN and deep neural network (DNN)]. TBRFA extracts the complex interaction network among NP properties, immune response, and organ burden, thereby providing guidance for the design and discovery of ideal NPs.
Fig. 8. TBRFA feature interaction network analysis.
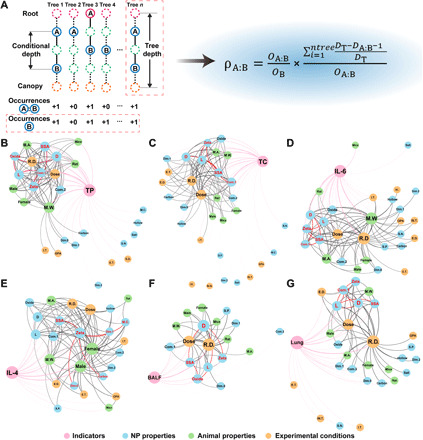
(A) TBRFA interaction coefficient calculation method. (B to G) Feature interaction networks for total proteins (B), total cells (C), IL-6 (D), IL-4 (E), BALF (F), and lung (G). Different colors are used to represent different types of features and indicators. The thickness of the lines represents the strength of the interaction. The size of the circle represents the number of times the feature interacts with other features. The five strongest interaction NP properties in the networks are highlighted in red.
Fig. 9. Double-variable partial dependence for strong feature interactions.

(A) Total proteins and SSA-Zeta. (B) IL-6 and SSA-Zeta. (C) IL-4 and SSA-Zeta. (D) Total cells and Zeta-SSA. (E) Total cells and L-SSA. (F) IL-6 and L-SSA. (G) Lung burden and L-D. (H) Liver burden and L-D. (I) BALF burden and L-D. (J) Summary diagram of TBRFA feature interaction network analysis. NPs with different properties are sorted according to the level of immune response. One-dimensional NPs tend to be transported to the live.
DISCUSSION
Given the high costs and limitations associated with animal protection, comprehensive biological response evaluation experiments for various NPs are not practicable. Although a number of studies have applied machine learning approaches to solve the above problems, the interpretability of the models is poor, thus hindering the application of machine learning in the design and discovery of ideal NPs. Here, a rigorous TBRFA approach that boosts the interpretability of machine learning successfully predicted the pulmonary immune response and organ burden of NPs. The optimized prediction accuracy was achieved, with R2 values of all training sets >0.9 and half of the test sets >0.75. To overcome the shortcomings of traditional importance analyses, TBRFA used multiway importance analysis and reduced the biases caused by the unbalanced structure of small sample datasets. The TBRFA framework also established feature interaction networks and boosted the interpretability of machine learning. It is difficult for researchers to fully explore the joint effects of multiple features through experiments. In most machine learning studies, the exploration of interpretability usually stops at revealing the importance of features and ignores the relationships under the joint action (e.g., antagonism and synergy) of multiple features (12, 48). It is well known that the interactions between NPs and biology are determined by multiple features or properties of NPs (49), but the key features that regulate biological responses remain controversial. For the design of NPs, studies usually focus on one or two features (50), but how the other features influence the global properties of NPs is unclear. The feature interaction network analysis in Fig. 8 provides insights into the above challenging question.
The TBRFA approach revealed and explained the critical roles of SSA and diameter in the NP immune response and organ burden, respectively. The improved interpretability of machine learning is useful for models that explore causation and the design of excellent NPs in the medicine, biosensor, drug delivery, and other health care fields, and it also provides accurate predictions. Some animal experiments conducted by different laboratories are not completely consistent due to the biological response affected by multiple factors (e.g., the sources of animals, the environments of animal growth, and the operation skills of researchers) (51, 52). The above biases of data, especially for small-size data, would lead to a rather large variation in test predictive performance across labels. As the above explanation, the biases of data from interlaboratory studies may contribute to the misclassified (very off-diagonal) samples in Fig. 3E. Drafting standardized nanomaterial characterization protocols and animal exposure protocols and improving the quality of the literature will ensure the authenticity of the label value and greatly improve the accuracy and applicability of TBRFA (49). Further improving the degree of automation of interaction coefficient calculations will also make TBRFA more suitable for big data. Overall, the models established in this work are suitable for predicting the lung immune response induced by inorganic NPs and their lung burden. TBRFA also deserves application in other fields, and the discovered feature interaction networks could contribute to human disease or anticancer drug research.
MATERIALS AND METHODS
Extracting data to establish datasets
The immune response data were obtained from published articles in the ISI Web of Knowledge database (the datasets before 31 December 2020 were collected). A total of 2548 studies were initially searched using the following search formula: TS = nano* AND TS = immun* NOT TS = Immunosensor AND (TS = mice OR TS = rat OR TS = mouse) AND (TS = pulmonary OR TS = lung). Given the heterogeneity of the biological data, the acquired publications were then filtered by the following conditions: (i) full text was available; (ii) the topic was the pulmonary immune responses of mice or rats induced by NPs; (iii) at least one of the following immune metrics contained: total protein, LDH, alkaline phosphatase, cell count [total cells, macrophages, and neutrophils (polymorphonuclear neutrophils)], cytokines, and chemokines; (iv) exposure methods were instillation, oropharyngeal aspiration, or similar methods; and (v) basic material characterization data and experimental conditions were provided. Last, 1620 samples were identified to establish datasets. To precisely and comprehensively establish the relationships between NP/exposure features and pulmonary immune responses, 16 features were included in the dataset, and they consisted of seven material properties, four animal properties, and five experimental conditions. Two biochemical indicators, three types of cell counts, and six different cytokines were selected as labels for the immune responses. The details are shown in supplementary Excel files.
Acquisition of lung burden data followed the above workflow. A total of 3525 studies (search date, June 2020) were initially searched using the following search formula: TS = nano* AND (TS = mice OR TS = rat OR TS = mouse) AND (TS = pulmonary OR TS = lung) AND TS = (accumulat* or burden* or clear*). Taking the exposure method and the determined features from the immune response dataset as the main filtering basis, 302 samples were lastly used to establish datasets. Liver and BALF burden data were also included in this burden dataset (supplementary Excel files). The data of biological responses to NPs are very complex and distributed in the texts, tables, and figures of publications. It is difficult to extract the required data by machine. The data were extracted from the publications by hand. The data directly given in texts and tables were copied by hand. For the data in the figures, the “Digitizer” tool provided by OriginLab was used to read each point three times and then the averages were calculated. To reduce the errors, the extracted data were checked thoroughly by another person again.
Preprocessing data
To calculate characteristic variables, the six characteristic variables in the dataset (NP type, shape, surface functionalization, animal, gender, and method) need to be coded. “One-hot” is a general encoding method that converts disordered and discrete variables into binary vectors to overcome the problem that such variables cannot be recognized by machine learning algorithms. For the three discrete features with few unique types (i.e., animal, gender, and method), the “one-hot” coding was adopted. For one-hot encoding features, there were no obvious correlations through correlation coefficient analysis (fig. S2). However, the one-hot method was not suitable for the descriptor “NP types” because the diversity of NP types (n = 57) caused a rapid increase in the data dimension. New descriptors were created on the basis of chemical properties for the other three characteristic variables to distinguish different properties. The new descriptors are as follows:
1) NP types: carbide (0/1), macromolecular compound (M.C., 0/1), oxide (0/1), salt (0/1), component 1 (Com.1, relative atomic mass), and component 2 (Com.2, relative atomic mass).
2) Shape: hollow (0/1), granular NPs (Dim.0, 0/1), one-dimensional NPs (Dim.1, 0/1), and two-dimensional NPs (Dim.2, 0/1).
3) Surface functionalization: positive charge (S.P., 0/1) and negative charge (S.N., 0/1).
To prevent certain features from contributing excessively to the models, the z-score (Eq. 1) normalization method was applied to the features. The formula is listed as follows
| (1) |
where x is the feature value, μ is the mean of each feature, and σ is the SD of each feature. The raw data in literatures are difficult to use for direct comparison. The difference in experimental design and used animals may produce biases in models. To intuitively reflect the relative degrees of the immune response caused by different NPs in different literatures, the label data were converted to values between −1 and 1 on the basis of the treated and control groups in each literature individually, rather than using the data in all literatures together, to reduce the biases.
The formula is listed as follows
| (2) |
where y and c are the values of the experimental group and control group, respectively.
Machine learning regression
RF models were trained using scikit-learn in Python 3.7. According to different labels, the datasets were split into 15 subsets (12 immune response subsets and 3 burden subsets) that corresponded to 15 regression models. The RF models used 500 random decision trees and selected 5 random features at each node, which were determined by the grid search method (53). RF is based on bootstrapping to avoid inherent overfitting (54, 55). A 10-fold cross-validation (ShuffleSplit) method was used for each machine learning algorithm to prevent overfitting, with 90% of the samples in each subset chosen as training sets and 10% chosen as test sets. Approximately 36.8% of the samples in the training set, which were called OOB data, were used as the validation sets and not used during the training process (56). The percentage of OOB was calculated by the following formula
| (3) |
where m is the frequency of sampling and e is the natural constant with a value of approximately 2.71828. Each model was trained 10 times, and the average of the R2 and RMSE values between the predictions and observations were calculated to measure the model performance.
Two-layer fully connected ANN models were trained using Keras in Python 3.7. The workflow was similar to that of the RF regression except that 25% of the samples in the training set were split randomly as validation data to monitor overfitting. Multilayer feedforward neural networks with different layers and different units in the hidden layer were trained to find the optimal configuration, and 57 and 55 units in the hidden layers were adopted in the ANN immunotoxicity and burden models, respectively. Stochastic gradient descent was used as the optimizer, and a small learning rate decay (less than 0.0001) was set to prevent overfitting during the training process. SVM models were trained using scikit-learn in Python 3.7. Tenfold ShuffleSplit method was also applied. “Rbf” was chosen as the kernel function. The regularization parameter was set to 1.
Overfitting test
To judge whether the RF models used were overfitted, we adopted a permutation test method. In each process of 10-fold cross-validation, 20, 40, 60, 80, and 100% of the label values in the training set were randomly replaced by random values within the original label range, and the corresponding cross-validation coefficients (Q2) were calculated. The calculation formula of Q2 is as follows
| (4) |
where yi is the observed label value, is the predicted label value, and is the average of the label. The permutation of each ratio (20, 40, 60, 80, and 100%) was performed 10 times, resulting in 500 permutation Q2 values for each model (5 ratio × 10 times/ratio × 10 folds). Subsequently, linear regressions were performed on the Q2 values and the correlation coefficients between the original labels and the permutation labels. The intercept of the regression result on the y axis less than 0.05 proves that the model is not overfitting (30).
NP preparation and characterization for model verification
Three types of MWCNTs with axial lengths ranging from approximately 0.5 to 2 μm were obtained from XFNANO (China): S-MWCNTs (production number XFM10; diameter, 8 to 15 nm; purity > 95%), M-MWCNTs (production number XFM16; diameter, 10 to 20 nm; purity > 95%), and L-MWCNTs (production number XFM28; diameter, 30 to 50 nm; purity > 95%). All the MWCNTs were produced by the chemical vapor deposition method. To investigate the effects of surface modification on immune responses, nano-TiO2 (production number XFI02; diameter, 20 to 40 nm; purity > 99%) and hydroxylated MWCNTs (production number XFM16; diameter, 10 to 20 nm; purity > 95%) functionalized with triethoxycaprylylsilane (F-MWCNT) were synthesized. Functionalization was carried out by stirring a mixture of 100 mg of hydroxylated MWCNTs with 10 mg of triethoxycaprylylsilane (DB, Shanghai) in 100 ml of 90% ethanol solution at 70°C for 8 hours. The mixture was then centrifuged at 1699g for 15 min, washed with Milli-Q water (18.2 megaohm cm−1), and then vacuum freeze-dried. To calculate the sizes of the NPs, transmission electron microscopy (TEM) images were obtained on a TEM instrument (JEM-2010 FEF, JEOL, Japan). Fourier transform infrared (FTIR) spectroscopy (Bruker Tensor 27, Germany) with a resolution of 2 cm−1 from 4000 to 400 cm−1 was used to confirm the synthesis of F-MWCNTs. Zeta potentials were determined by a ZetaSizer Nano instrument (BI-200SM, Brookhaven, USA). BET surface areas were measured with a surface area and porosity analyzer (ASAP 2460, Macrometrics, USA). The results of characterization were given in the supplementary note S2 and tables S4 and S5.
Animal experiment
Male Institute of Cancer Research (ICR) mice with a body weight range of 24 to 26 g were purchased from Vital River Laboratories (Beijing, China) and acclimated for 2 weeks in animal facilities supplied with normal food and water. All animal studies were performed in accordance with the guidelines and regulations of the Human and Animal Experiments Ethical Committee of the Nankai University. The four types of NPs were individually suspended in sterile phosphate-buffered saline (PBS) at 1 mg/ml. Suspensions were processed with ice-bath ultrasonication before instillation to ensure even dispersion. ICR mice (n = 6 per group) were treated individually with a single intratracheal instillation of 25 μl of (1 mg/kg) NPs. Equal volumes of PBS were instilled as a control.
Immune response analysis
Frozen sections (thickness, 5 μm) of lung lobes were made by a clinical cryostat (CM1850, Leica, Germany) for immunofluorescence. Anti–nuclear factor κB (NF-κB) p65 with Alexa Fluor 555 (bs-0465R-AF555, Bioss, China) and anti–IL-1β with fluorescein isothiocyanate (FITC) (bs-0812R-FITC, Bioss, China) were used to label NF-κB p65 and IL-1β, respectively. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei throughout the lung section. Immunofluorescence images of NF-κB (red), IL-1β (green), and DAPI (blue) were obtained with a confocal laser scanning microscope (LSM880 with Airyscan, Zeiss, Germany) at 543, 488, and 405 nm, respectively.
Model validation
Model validation was composed of two parts: animal experimental and validation sets. In terms of animal experiments, the characterization data and experimental conditions of NPs were input into the trained RF models to obtain predicted values. To validate the accuracy of the model, the immunofluorescence intensity of the IL-1β protein was statistically analyzed and compared with the predicted IL-1β results. The interval formed by ±RMSE was considered to be the allowable error bound of the model (11). As the validation set, random seeds were set to randomly sample the validation set (n = 5) for each subset before modeling. The five validation sets covered the NPs with a wide range of responses. The validation set did not participate in the construction of the model at all to ensure that the model did not learn them during the cross-validation. During the 10-fold cross-validation process, the average predicted values of 10 times were obtained for comparison with the observed values. According to the data source, the animal experiment validated the three critical NP properties (diameters, SSA, and zeta; see table S4), and the validation set validated all features.
Feature selection
Models were optimized using a SBS procedure. This method started from the full set of features to eliminate redundant features one by one to find the optimal feature subset. The R2 of each selection process was calculated and recoded to compare the performances of different feature combinations. To avoid the models losing too much information, we set the minimum number of features selected by SBS to not be less than one-half of the total number of features.
Feature importance analysis of TBRFA
Feature importance analysis was based on the “randomForest” and “randomForestExplainer” packages in R 4.0.2. To avoid the bias produced by a single indicator (i.e., MSE) used in comment RF, a total of four indicators (MSE increase, node purity increase, P value, and mean minimal depth) were selected to represent different perspectives and to comprehensively evaluate the importance of features. MSE increase is based on the decrease in predictive accuracy of the forest after perturbation of the variable; node purity increase is based on changes in node purity after splits on the variable; and P value is based on the one-sided binomial test to evaluate the significance of feature importance (40, 53). The mean minimal depth is based on the structure of the forest. The significant and important features were screened by calculating the MSE and node purity (measured by residual sum of squares) increase and the P value. During the training process, feature importance was also reflected by the mean minimal depth of the feature among trees. In this study, five random features were set to be randomly sampled as candidates at each node and one of the five that contributed the most to the overall split was retained at the node. Therefore, features near the root were more important than others.
Feature interaction network analysis of TBRFA
The conditional minimal depth of features represents the strength of the interaction between two features. The original conditional minimal depth of features was initially calculated using the randomForestExplainer package. Four groups of strong interaction features were selected for the double-variable partial dependence analysis. However, there was actually no specific interaction between some features. To improve the present method and weaken the influence of the feature importance (minimal depth) on the interaction among features, we explored the conditional minimal depth of each feature in each tree of an RF model and used the occurrences of interaction among the feature and its corresponding root feature to normalize its conditional minimal depth. The interaction coefficient is defined as follows
| (5) |
where A and B are two of the features, A:B means the interaction of B and the maximal A-subtree [see the description by Ishwaran et al. (40)], ρA:B is the directed interaction coefficient, OA:B is the occurrence of the interaction of B and the maximal A-subtree among the trees (ntree = 500), OB is the occurrence of B among the trees (ntree = 500), DT is the tree depth, and DA:B is the minimal depth of B in the maximal A-subtree, that is, the conditional minimal depth. All interaction coefficients in each RF model were calculated, and the interaction coefficients that had two directions (using average values) were merged. Then, the importance of features was combined to build feature interaction networks, and new strong interaction features for double-variable partial dependence analysis were screened out.
Statistical analysis
A two-sided Kolmogorov-Smirnov test was implemented in R software to test whether the distribution of the data was normal (P > 0.05).
Acknowledgments
We acknowledge the technical support provided by H. Zhang, C. Zou, J. Xu, W. Li, L. Zhang, and R. Cai from the Nankai University. Funding: This work was financially supported by the National Natural Science Foundation of China (grant nos. 21722703 and 42077366), the 111 program (grant no. T2017002), and the National Key Research and Development Project (grant no. 2019YFC1804603). Author contributions: X.H. designed the project; X.H. and F.Y. contributed to the writing and revision of the manuscript; F.Y. ran the models; F.Y. and P.D. collected the data; C.W., F.Y., and T.P. performed the animal experiments. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Datasets containing all relevant literature data are provided as supplementary Excel files, and the datasets are also available on Zenodo at https://doi.org/10.5281/zenodo.4661099. Performance of models, TEM, FTIR, and other characterization of materials are also provided in the Supplementary Materials. The custom codes written to develop the machine learning models, perform TBRFA and other analysis, and generate figures are all available on Zenodo at https://doi.org/10.5281/zenodo.4660115.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabf4130/DC1
REFERENCES AND NOTES
- 1.Sun Q., Barz M., De Geest B. G., Diken M., Hennink W. E., Kiessling F., Lammers T., Shi Y., Nanomedicine and macroscale materials in immuno-oncology. Chem. Soc. Rev. 48, 351–381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioannidis J. P. A., Kim B. Y. S., Trounson A., How to design preclinical studies in nanomedicine and cell therapy to maximize the prospects of clinical translation. Nat. Biomed. Eng. 2, 797–809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musetti S., Huang L., Nanoparticle-mediated remodeling of the tumor microenvironment to enhance immunotherapy. ACS Nano 12, 11740–11755 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Fadeel B., Farcal L., Hardy B., Vazquez-Campos S., Hristozov D., Marcomini A., Lynch I., Valsami-Jones E., Alenius H., Savolainen K., Advanced tools for the safety assessment of nanomaterials. Nat. Nanotechnol. 13, 537–543 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Mu Q., Jiang G., Chen L., Zhou H., Fourches D., Tropsha A., Yan B., Chemical basis of interactions between engineered nanoparticles and biological systems. Chem. Rev. 114, 7740–7781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reina G., Gonzalez-Dominguez J. M., Criado A., Vazquez E., Bianco A., Prato M., Promises, facts and challenges for graphene in biomedical applications. Chem. Soc. Rev. 46, 4400–4416 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Fadeel B., Bussy C., Merino S., Vázquez E., Flahaut E., Mouchet F., Evariste L., Gauthier L., Koivisto A. J., Vogel U., Martín C., Delogu L. G., Buerki-Thurnherr T., Wick P., Beloin-Saint-Pierre D., Hischier R., Pelin M., Carniel F. C., Tretiach M., Cesca F., Benfenati F., Scaini D., Ballerini L., Kostarelos K., Prato M., Bianco A., Safety assessment of graphene-based materials: Focus on human health and the environment. ACS Nano 12, 10582–10620 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Baalousha M., Cornelis G., Kuhlbusch T. A. J., Lynch I., Nickel C., Peijnenburg W., van den Brink N. W., Modeling nanomaterial fate and uptake in the environment: Current knowledge and future trends. Environ. Sci. Nano 3, 323–345 (2016). [Google Scholar]
- 9.Bai X., Wang S., Yan X., Zhou H., Zhan J., Liu S., Sharma V. K., Jiang G., Zhu H., Yan B., Regulation of cell uptake and cytotoxicity by nanoparticle core under the controlled shape, size, and surface chemistries. ACS Nano 14, 289–302 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Sedykh A., Sun H. N., Zhao L., Russo D. P., Zhou H., Yan B., Zhu H., Predicting nano-bio interactions by integrating nanoparticle libraries and quantitative nanostructure activity relationship modeling. ACS Nano 11, 12641–12649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Huang Y., Cheng T., Goddard W. A. III, Identifying active sites for CO2 reduction on dealloyed gold surfaces by combining machine learning with multiscale simulations. J. Am. Chem. Soc. 141, 11651–11657 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Oh E., Liu R., Nel A., Gemill K. B., Bilal M., Cohen Y., Medintz I. L., Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 11, 479–486 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Pun G. P. P., Batra R., Ramprasad R., Mishin Y., Physically informed artificial neural networks for atomistic modeling of materials. Nat. Commun. 10, 2339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Zheng M., Jagota A., Learning to predict single-wall carbon nanotube-recognition DNA sequences. NPJ Comput. Mater. 5, 3 (2019). [Google Scholar]
- 15.Brown K. A., Brittman S., Maccaferri N., Jariwala D., Ceano U., Machine learning in nanoscience: Big data at small scales. Nano Lett. 20, 2–10 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Ban Z., Zhou Q., Sun A., Mu L., Hu X., Screening priority factors determining and predicting the reproductive toxicity of various nanoparticles. Environ. Sci. Technol. 52, 9666–9676 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ekins S., Puhl A. C., Zorn K. M., Lane T. R., Russo D. P., Klein J. J., Hickey A. J., Clark A. M., Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deringer V. L., Caro M. A., Csanyi G., Machine learning interatomic potentials as emerging tools for materials science. Adv. Mater. 31, 1902765 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Hu X., Belle J. H., Meng X., Wildani A., Waller L. A., Strickland M. J., Liu Y., Estimating PM2.5 concentrations in the conterminous united states using the random forest approach. Environ. Sci. Technol. 51, 6936–6944 (2017). [DOI] [PubMed] [Google Scholar]
- 20.C. Chen, O. Li, C. Tao, A. J. Barnett, J. Su, C. Rudin, This looks like that: Deep learning for interpretable image recognition (2018); https://ui.adsabs.harvard.edu/abs/2018arXiv180610574C.
- 21.M. Tulio Ribeiro, S. Singh, C. Guestrin, "Why should I trust you?": Explaining the predictions of any classifier (2016); https://ui.adsabs.harvard.edu/abs/2016arXiv160204938T.
- 22.S. Lundberg, S.-I. Lee, A unified approach to interpreting model predictions (2017); https://ui.adsabs.harvard.edu/abs/2017arXiv170507874L.
- 23.Walker S. W. C., Anwar A., Psutka J. M., Crouse J., Liu C., Le Blanc J. C. Y., Montgomery J., Goetz G. H., Janiszewski J. S., Campbell J. L., Hopkins W. S., Determining molecular properties with differential mobility spectrometry and machine learning. Nat. Commun. 9, 5096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gernand J. M., Casman E. A., A meta-analysis of carbon nanotube pulmonary toxicity studies–How physical dimensions and impurities affect the toxicity of carbon nanotubes. Risk Anal. 34, 583–597 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Dobrovolskaia M. A., Mcneil S. E., Immunological properties of engineered nanomaterials. Nat. Nanotechnol. 2, 469–478 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Kim S., Seo J., Park H. H., Kim N., Oh J.-W., Nam J.-M., Plasmonic nanoparticle-interfaced lipid bilayer membranes. Acc. Chem. Res. 52, 2793–2805 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Herda L. M., Hristov D. R., Lo Giudice M. C., Polo E., Dawson K. A., Mapping of molecular structure of the nanoscale surface in bionanoparticles. J. Am. Chem. Soc. 139, 111–114 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Lo Giudice M. C., Herda L. M., Polo E., Dawson K. A., In situ characterization of nanoparticle biomolecular interactions in complex biological media by flow cytometry. Nat. Commun. 7, 13475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S., Zhou Q., Ouyang Y., Guo Y., Li Q., Wang J., Accelerated discovery of stable lead-free hybrid organic-inorganic perovskites via machine learning. Nat. Commun. 9, 3405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. Eriksson, T. Byrne, E. Johansson, J. Trygg, C. Vikström, Multi-and Megavariate Data Analysis Basic Principles and Applications (Umetrics Academy, 2013), vol. 1. [Google Scholar]
- 31.Bartneck M., Keul H. A., Zwadlo-Klarwasser G., Groll J., Phagocytosis independent extracellular nanoparticle clearance by human immune cells. Nano Lett. 10, 59–63 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Han S. G., Lee J. S., Ahn K., Kim Y. S., Kim J. K., Lee J. H., Shin J. H., Jeon K. S., Cho W. S., Song N. W., Gulumian M., Shin B. S., Yu I. J., Size-dependent clearance of gold nanoparticles from lungs of Sprague-Dawley rats after short-term inhalation exposure. Arch. Toxicol. 89, 1083–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Cartwright A. N. R., Griggs J., Davis D. M., The immune synapse clears and excludes molecules above a size threshold. Nat. Commun. 5, 5479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannon G., Lysaght J., Liptrott N. J., Prina-Mello A., Immunotoxicity considerations for next generation cancer nanomedicines. Adv. Sci. 6, 1900133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baimanov D., Wu J., Chu R. X., Cai R., Wang B., Cao M., Tao Y., Liu J., Guo M., Wang J., Yuan X., Ji C., Zhao Y., Feng W., Wang L., Chen C., Immunological responses induced by blood protein coronas on two-dimensional MoS2 nanosheets. ACS Nano 14, 5529–5542 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Maurer-Jones M. A., Lin Y. S., Haynes C. L., Functional assessment of metal oxide nanoparticle toxicity in immune cells. ACS Nano 4, 3363–3373 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Lamson N. G., Berger A., Fein K. C., Whitehead K. A., Anionic nanoparticles enable the oral delivery of proteins by enhancing intestinal permeability. Nat. Biomed. Eng. 4, 84–96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu Q., Su G., Li L., Gilbertson B. O., Yu L. H., Zhang Q., Sun Y.-P., Yan B., Size-dependent cell uptake of protein-coated graphene oxide nanosheets. ACS Appl. Mater. Interfaces 4, 2259–2266 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Fourches D., Pu D., Tassa C., Weissleder R., Shaw S. Y., Mumper R. J., Tropsha A., Quantitative nanostructure-activity relationship modeling. ACS Nano 4, 5703–5712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishwaran H., Kogalur U. B., Gorodeski E. Z., Minn A. J., Lauer M. S., High-dimensional variable selection for survival data. J. Am. Stat. Assoc. 105, 205–217 (2010). [Google Scholar]
- 41.Orr G., Panther D. J., Phillips J. L., Tarasevich B. J., Dohnalkova A., Hu D. H., Teeguarden J. G., Poundsl J. G., Submicrometer and nanoscale inorganic particles exploit the actin machinery to be propelled along microvilli-like structures into alveolar cells. ACS Nano 1, 463–475 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Shi C., Wang X., Wang L., Meng Q., Guo D., Chen L., Dai M., Wang G., Cooney R., Luo J., A nanotrap improves survival in severe sepsis by attenuating hyperinflammation. Nat. Commun. 11, 3384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilar Calatayud M., Sanz B., Raffa V., Riggio C., Ricardo Ibarra M., Goya G. F., The effect of surface charge of functionalized Fe3O4 nanoparticles on protein adsorption and cell uptake. Biomaterials 35, 6389–6399 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Shannahan J. H., Brown J. M., Chen R., Ke P. C., Lai X., Mitra S., Witzmann F. A., Comparison of nanotube-protein corona composition in cell culture media. Small 9, 2171–2181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X., Li L., Liu T., Hao N., Liu H., Chen D., Tang F., The shape effect of mesoporous silica nanoparticles on biodistribution, clearance, and biocompatibility in vivo. ACS Nano 5, 5390–5399 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Arnida, Janat-Amsbury M. M., Ray A., Peterson C. M., Ghandehari H., Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages. Eur. J. Pharm. Biopharm. 77, 417–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Z., Ukidve A., Krishnan V., Mitragotri S., Effect of physicochemical and surface properties on in vivo fate of drug nanocarriers. Adv. Drug Deliv. Rev. 143, 3–21 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Cordier T., Esling P., Lejzerowicz F., Visco J., Ouadahi A., Martins C., Cedhagen T., Pawlowski J., Predicting the ecological quality status of marine environments from eDNA metabarcoding data using supervised machine learning. Environ. Sci. Technol. 51, 9118–9126 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Faria M., Bjornmalm M., Thurecht K. J., Kent S. J., Parton R. G., Kavallaris M., Johnston A. P. R., Gooding J. J., Corrie S. R., Boyd B. J., Thordarson P., Whittaker A. K., Stevens M. M., Prestidge C. A., Porter C. J. H., Parak W. J., Davis T. P., Crampin E. J., Caruso F., Minimum information reporting in bio-nano experimental literature. Nat. Nanotechnol. 13, 777–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang G., Liu H., Qu J., Li J., Two-dimensional layered MoS2: Rational design, properties and electrochemical applications. Energy Environ. Sci. 9, 1190–1209 (2016). [Google Scholar]
- 51.Martin J.-C., Maillot M., Mazerolles G., Verdu A., Lyan B., Migné C., Defoort C., Canlet C., Junot C., Guillou C., Manach C., Jabob D., Bouveresse D. J.-R., Paris E., Pujos-Guillot E., Jourdan F., Giacomoni F., Courant F., Favé G., Gall G. L., Chassaigne H., Tabet J.-C., Martin J.-F., Antignac J.-P., Shintu L., Defernez M., Philo M., Alexandre-Gouaubau M.-C., Amiot-Carlin M.-J., Bossis M., Triba M. N., Stojilkovic N., Banzet N., Molinié R., Bott R., Goulitquer S., Caldarelli S., Rutledge D. N., Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics 11, 807–821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feswick A., Ankley G. T., Denslow N., Ellestad L. E., Fuzzen M., Jensen K. M., Kroll K., Lister A., MacLatchy D. L., McMaster M. E., Orlando E. F., Servos M. R., Tetreault G. R., Van Den Heuvel M. R., Munkittrick K. R., An inter-laboratory study on the variability in measured concentrations of 17β-estradiol, testosterone, and 11-ketotestosterone in white sucker: Implications and recommendations. Environ. Toxicol. Chem. 33, 847–857 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 54.Basu S., Kumbier K., Brown J. B., Yu B., Iterative random forests to discover predictive and stable high-order interactions. Proc. Natl. Acad. Sci. U.S.A. 115, 1943–1948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B., Fan Z. T., Zhang X. L., Huang D. S., Robust dimensionality reduction via feature space to feature space distance metric learning. Neural Netw. 112, 1–14 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Meng X., Hand J. L., Schichtel B. A., Liu Y., Space-time trends of PM2.5 constituents in the conterminous United States estimated by a machine learning approach, 2005-2015. Environ. Int. 121, 1137–1147 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabf4130/DC1


