Abstract
Site-specifically modified protein bioconjugates have important applications in biology, chemistry and medicine. Functionalizing specific protein side chains with enzymes using mild reaction conditions is of significant interest, but remains challenging. Recently, the lysine-isopeptide bond forming activity of the sortase enzyme that builds surface pili in Corynebacterium diphtheriae (CdSrtA) has been reconstituted in vitro. A mutationally activated form of CdSrtA was shown to be a promising bioconjugating enzyme that can attach Leu-Pro-Leu-Thr-Gly peptide fluorophores to a specific lysine residue within the N-terminal domain of the SpaA protein (NSpaA), enabling the labeling of target proteins that are fused to NSpaA. Here we present a detailed analysis of the CdSrtA catalyzed protein labeling reaction. We show that the first step in catalysis is rate limiting, which is the formation of the CdSrtA-peptide thioacyl intermediate that subsequently reacts with a lysine ε-amine in NSpaA. This intermediate is surprisingly stable, limiting spurious proteolysis of the peptide substrate. We report the discovery of a new enzyme variant (CdSrtAΔ) that has significantly improved transpeptidation activity because it completely lacks an inhibitory polypeptide appendage (“lid”) that normally masks the active site. We show that the presence of the lid primarily impairs formation of the thioacyl intermediate and not recognition of the NSpaA substrate. Quantitative measurements reveal that CdSrtAΔ generates its crosslinked product with a catalytic turnover number of 1.4 ± 0.004 hrs−1 and that it has apparent KM values of 0.16 ± 0.04 and 1.6 ± 0.3 mM for its NSpaA and peptide substrates, respectively. CdSrtAΔ is 7-fold more active than previously studied variants, labeling >90% of NSpaA with peptide within 6 hours. The results of this study further improve the utility of CdSrtA as a protein labeling tool and provide insight into the enzyme catalyzed reaction that underpins protein labeling and pilus biogenesis.
Keywords: Bioconjugation, lysine-isopeptide bond, sortase, pili, Gram-positive bacteria
Introduction
New methods are needed to create protein bioconjugates that can be used as therapeutics, imaging tools, diagnostic reagents and materials.1–5 Labeling specific sites on the protein is often preferred as it enables the construction of well-defined antibody-drug conjugates, small molecule- and fluorophore-labeled proteins for biophysical experiments, orientation-specific protein immobilization and the preparation of ordered, multifunctional protein complexes.6–8 A variety of protein modification strategies have been developed that exhibit varying degrees of site-selectivity, efficiencies and ease of use. They range from chemical approaches that leverage the reactivity of amino acid specific functional groups (e.g., cysteine and lysine modifications), to highly selective, but less facile methods that require the incorporation of non-natural amino acids to facilitate bio-orthogonal conjugation chemistries (e.g., azide or alkyne-containing residues for click-chemistry).9,10 Bioconjugating enzymes (e.g., ligase, transferases, etc.) are particularly attractive for site-specific protein labeling because they can be employed using mild reaction conditions, and in principle can be highly selective.11,12 The sortase A enzyme from Staphylococcus aureus (SaSrtA) is one of the most widely used bioconjugating enzymes.12–14 It has been successfully deployed to catalyze protein-protein ligations, backbone cyclization, and to modify proteins with peptides, lipids, sugars and small molecules.15–20 However, SaSrtA bioconjugations do not modify protein side chains and are almost exclusively restricted to altering only the N- or C-terminus of a protein.
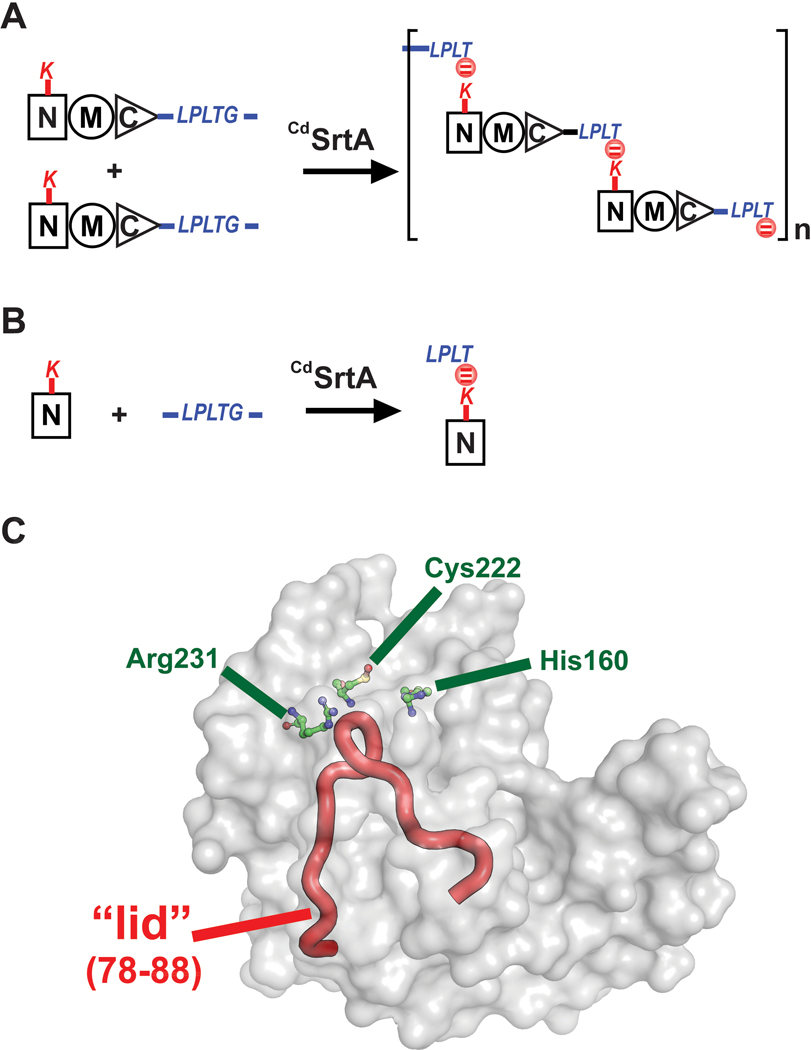
Recently, we demonstrated that the pilus-specific sortase from Corynebacterium diphtheriae (CdSrtA) can be used to attach a peptide fluorophore via an isopeptide bond to a specific lysine residue within a protein.21 Although CdSrtA and SaSrtA are both members of the sortase-superfamily of cysteine transpeptidases, they have distinct substrate specificities.22 SaSrtA is a class A sortase that catalyzes formation of backbone-backbone peptide-bonds, whereas CdSrtA is a class C sortase that joins molecules together via lysine-isopeptide bonds.23,24 CdSrtA assembles the SpaA pilus in C. diphtheriae by crosslinking SpaA “pilin” subunits via a lysine-isopeptide bond.25,26 In this process, a lysine residue (K190) on one SpaA pilin is joined to the C-terminal LPLTG sorting signal located in a second SpaA pilin.26 Repetition of this reaction forms the SpaA pilus, which is approximately 1–2 μm in length and further elaborated with unique tip and basal pilin proteins (Fig. 1A).26, 27 In vitro, the native CdSrtA enzyme is enzymatically inactive because it contains a polypeptide appendage that occludes its active site, called a “lid” (Fig. 1C). However, CdSrtA variants containing destabilizing mutations in the lid exhibit in vitro activity.27 The most active form of the enzyme thus far discovered is CdSrtA3M, which contains residues N37-Q257 in CdSrtA and D81G/W83G/N85A mutations in the lid.21 CdSrtA3M is a promising bioconjugation tool as it can be used to selectively modify proteins harboring the N-terminal domain of SpaA (NSpaA) with peptide fluorophores. Modification via lysine-isopeptide bonds is attractive, as these linkages may be less susceptible to proteolysis and enzymatic reversibility.
Figure 1.
The C. diphtheriae CdSrtA pilin sortase catalyzes lysine isopeptide bond formation. (A) Schematic showing the pilin polymerization reaction catalyzed by CdSrtA. The enzyme creates the SpaA pilus by polymerizing SpaA pilin proteins. In the reaction it recognizes lysine (K190) side chain nucleophile within the N-terminal domain of SpaA (NSpaA) and joins it the backbone threonine carbonyl carbon atom located in the C-terminal LPLTG sorting signal located within another SpaA protein. This reaction is repeated to construct the SpaA pilus that mediates bacterial adhesion. (B) Schematic of the reaction used to monitor lysine isopeptide bond formation. In this assay the CdSrtA enzyme ligates the isolated NSpaA domain to the peptide containing the LPLTG sorting signal (FELPLTGGSG). (C) The structure of CdSrtA showing H160, C222 and R231 active site residues. The “Lid” is highlighted in red (residues P77 to S89).
In this study we developed an HPLC-based assay to measure for the first time the kinetics of catalysis and we have used the assay to identify a new CdSrtA variant that has improved bioconjugation activity. In particular, we show that: (i) the bioconjugation reaction rate is limited by formation of an enzyme-acyl intermediate with the LPLTG sorting signal, (ii) the enzyme preferentially recognizes non-polar amino acids at the X position within the sorting signal, (iii) unlike SaSrtA, CdSrtA exhibits minimal proteolytic activity, (iv) mutations in the lid accelerate catalysis by facilitating enzyme-acyl intermediate formation, and (v) complete removal of the lid further activates the enzyme. These results increase the in vitro utility of CdSrtA as a bioconjugation tool to modify proteins and provides new insight into the enzymatic reaction that underpins the construction of surface pili in Gram-positive bacteria.
Results and Discussion
Kinetics of lysine-isopeptide bond formation.
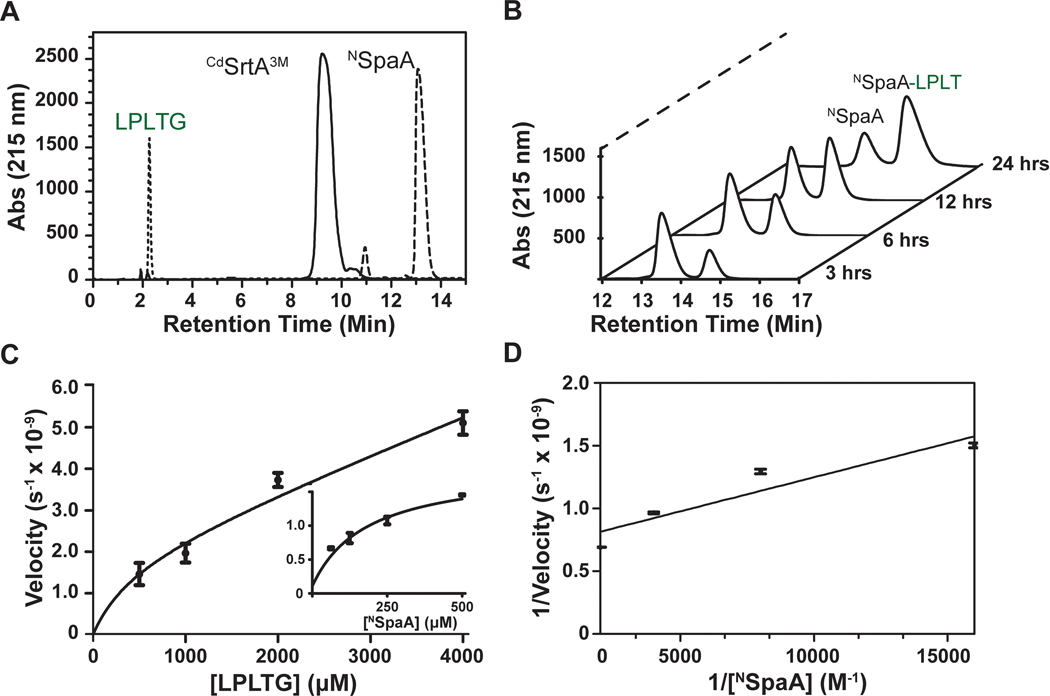
Previously, we monitored the lysine-isopeptide bond forming activity of CdSrtA3M using SDS-PAGE.21 However, the kinetics of this process could not be accurately determined because the reactants and products were difficult to separate and quantify. To overcome this problem, we developed an HPLC-based transpeptidation assay that monitors the ability of CdSrtA3M to join the N-terminal domain from SpaA (NSpaA, residues E30-S195 of SpaA) to a peptide containing its C-terminal sorting signal (FELPLTGGSG, hereafter called LPLTG peptide). This reaction represents an isolated ligation event in the polymerization reaction by producing a NSpaA-LPLT product in which the K190 side chain in NSpaA is joined via an isopeptide bond to the threonine carbonyl group in the FELPLT peptide (Fig. 1B). The reactants and products are readily separated by reverse phase HPLC (Fig. 2A). Moreover, this procedure enables facile monitoring of the time-dependent conversion of the protein substrate (NSpaA) into the crosslinked protein-peptide product (NSpaA-LPLT) (Fig. 2B). The identity of the product and the location of its isopeptide linkage were previously confirmed by LC-MS/MS.21,27 Initially, for each substrate (NSpaA and the LPLTG peptide) the dependence of the reaction velocity on substrate concentration was determined (Fig. 2C). This analysis reveals that NSpaA and peptide substrates do not saturate the enzyme even when they are present at concentrations of 500 μM (Fig. 2C, insert) and 4 mM (Fig. 2C, main), respectively. Because using higher concentrations of each substrate is not practical, we determined apparent steady state parameters using sub-saturating amounts of each substrate (representative data is shown in Fig. 2D). Two sets of Michaelis-Menten parameters were obtained. Either the concentration of NSpaA was varied from 62.5 to 500 μM with the amount of LPLTG peptide held constant at 1 mM, or the concentration of the LPLTG peptide was varied from 0.5 to 4 mM, while the concentration of NSpaA was held fixed at 500 μM. These measurements revealed that CdSrtA3M catalyzes isopeptide bond formation with apparent KM values for NSpaA (NKM) and the LPLTG peptide (SKM) of 70 ± 10 μM and 2 ± 1 mM, respectively (Table 1). Similar turnover numbers were measured in each experiment, with a maximal value of 26 ± 10 (x10−5 sec) (obtained when NSpaA is held constant at 500 μM and the LPLTG peptide is varied).
Figure 2.
CdSrtA transpeptidation assay (A) Three separate reversed phase HPLC traces on a Waters C4 column separated the sorting signal peptide (LPLTG), CdSrtA3M, and NSpaA. (B) Several representative HPLC traces showing the decrease in NSpaA and the increase of Peptide-NSpaA. The reaction (100 μM of enzyme, 200 μM of NSpaA, and 1 mM of LPLT-10 Peptide) was sampled at 3,6,12, and 24 h. (C) A velocity vs substrate graph for both LPLTG (the sorting signal) and NSpaA from a range of concentrations of 500 μM to 4 mM and 62.5 μM to 500 μM, respectively. (n=3) Reactions were frozen with liquid N2 after 3 hours then subsequently. Initial velocities were calculated from the linear portion of each assay and peak assay was converted to concentration described in the experimental methods to obtain the initial velocity. (D) A Lineweaver-Burk graph graphing the initial velocities of CdSrtA3M versus the increasing concentration of NSpaA. Kcat and KM were estimated from a linear trend line fit.
Table 1:
Kinetics of CdSrtA catalyzed lysine-isopeptide formation
| kcat × 10−5 (s−1)a | NKM × 10−4 (M) | SKM × 10−4 (M) | kcat/ NKM (s−1 M−1) | |
|---|---|---|---|---|
| CdSrtA | n.d.b | n.d. | n.d. | n.d. |
| CdSrtA3M | 5.6 ± 0.8 | 0.7 ± 0.1 | 20 ± 10 | 0.7 ± 0.1 |
| H160A | 3.1 ± 0.4 | 0.43 ± 0.05 | ----- | 0.72 ± 0.08 |
| C222A | n.d. | n.d. | ----- | n.d. |
| R231A | n.d. | n.d. | ----- | n.d. |
| CdSrtAΔ | 40 ± 0.1 | 1.6 ± 0.4 | 16 ± 3 | 2.5 ± 0.6 |
| H160A | 2.5 ± 0.6 | 0.70 ± 0.02 | ----- | 0.36 ± 0.09 |
| C222A | n.d. | n.d. | ----- | n.d. |
| R231A | n.d. | n.d. | ----- | n.d. |
| SaSrtAc | 1600 ± 100 | 1.8 ± 0.1 | 73.3 ± 10.1d | 86 ± 5 |
All kinetics are approximations as saturating concentrations were not able to be measured
Transpeptidation steady-state kinetic parameters for CdSrtA were determine by the monitoring rate at which the enzyme ligated an FELPLTGGSG peptide to the NSpaA domain via a lysine-isopeptide bond..
n.d., not determined because insufficient amount of product was detectable.
Transpeptidation steady-state kinetic parameters for SaSrtA were determine by monitoring the rate at which the enzyme ligated GGG and FELPLTGGSG peptides via a backbone peptide bond. Reported values are the average from three measurements, and the error is the standard deviation.
Values are reported from Frankel et al (2005) and measures reactions between a Abz-LPETG-Dap(Dnp)-NH2 and pentaglycine.28 NRefers to transpeptidation kinetics measure when NSpaA is varied and FELPLTGGSG concentration is held constant. SRefers to when FELPLTGGSG peptide is varied and NSpaA is held constant.
Formation of the CdSrtA-LPLT thioacyl intermediate is a rate limiting step in catalysis and can be accelerated by completely removing the lid.
Armed with an assay to measure the rate of lysine-isopeptide bond formation we further mutated CdSrtA to try to improve its activity. CdSrtA3M is the most active form of CdSrtA thus far discovered and contains three mutations in the inhibitory lid structure (D81G, W83G, N85A).21 We reasoned that an enzyme mutant with the entire lid deleted might exhibit even higher transpeptidation activity by completely unmasking the active site. Inspection of the crystal structure CdSrtA suggests that lid residues I78 to A88 can be deleted without disrupting its tertiary structure, as the remaining P77 and S89 amino acids are positioned adjacent to one another in three-dimensional space.27 Indeed, the measured steady state kinetic parameters for a lid-deletion mutant CdSrtAΔ (residues N37-Q257 of CdSrtA in which the amino acids I78 to A88 are deleted) reveal that it is more active than CdSrtA3M; there is a 7-fold improvement in the apparent kcat, while the NKM and SKM values are only modestly affected (Table 1).
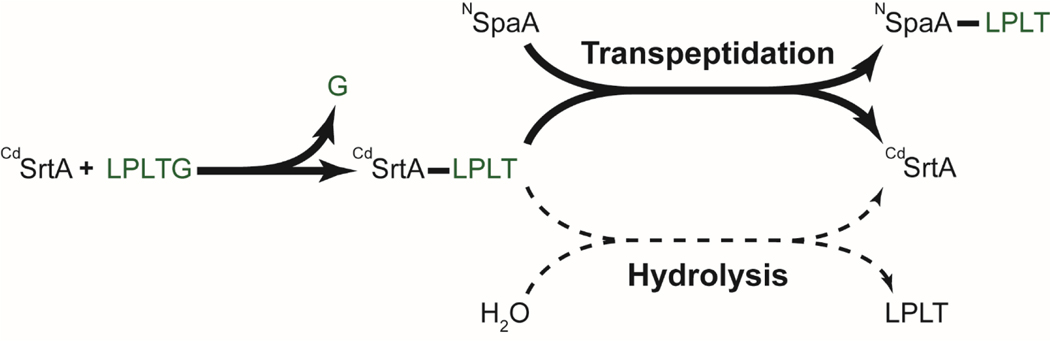
To address why CdSrtAΔ is catalytically more active than CdSrtA3M, we investigated how lid-removal affected catalysis. By analogy to the prototypical SaSrtA enzyme, CdSrtA presumably catalyzes lysine-isopeptide bond formation via a two-step process (Scheme 1).22, 28–30 In the transpeptidation mechanism CdSrtA’s C222 thiol presumably functions as a nucleophile to attack the threonine carbonyl carbon in the LPLTG sorting signal to form a CdSrtA-LPLT thioacyl intermediate. Then the enzyme recognizes the K190 sidechain amine group located within NSpaA, which resolves the thioacyl intermediate to form the lysine-isopeptide linked NSpaA-LPLT product. In SaSrtA, a side reaction also occurs in which a water molecule attacks the thioacyl intermediate instead of a primary amine, resulting in the hydrolysis of the intermediate to release the LPLT peptide.28 In this side reaction the enzyme effectively functions as a protease, cleaving the LPLTG peptide substrate at the peptide bond that joins the threonine and glycine residues. CdSrtA has also been shown to proteolyze its LPLTG peptide substrate, but the kinetics and extent of proteolysis have not been rigorously measured.21,27
Scheme 1.
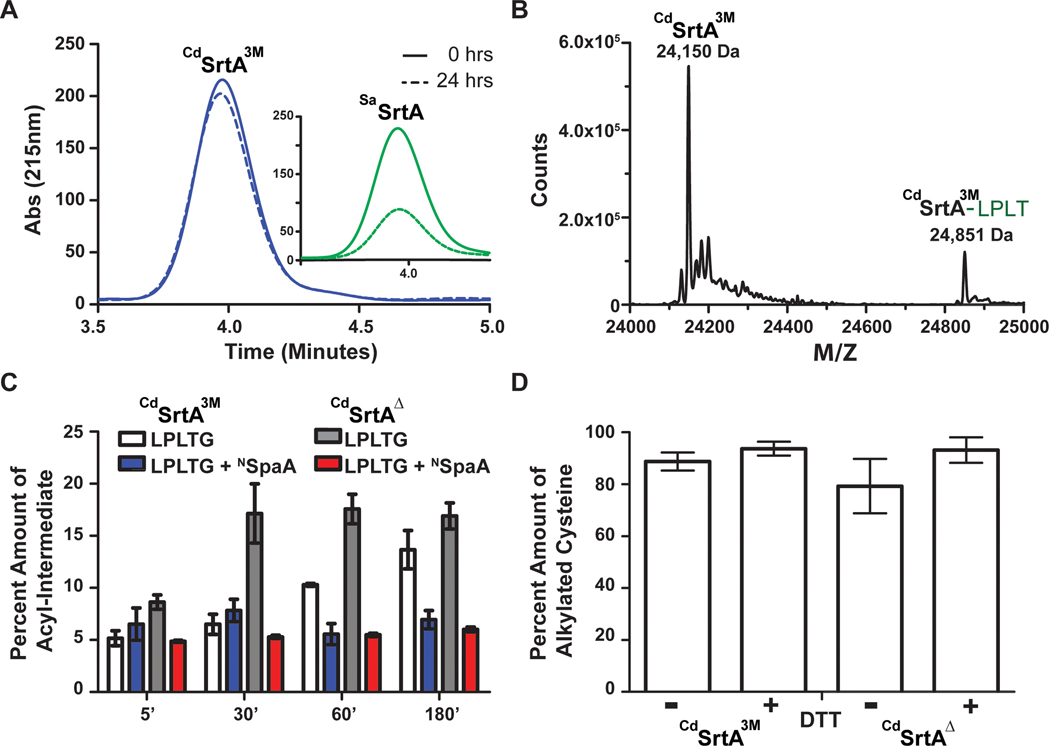
To determine if differences in the rate of the hydrolytic side reaction causes the CdSrtAΔ and CdSrtA3M enzymes to produce differing amounts of transpeptidation product, we measured the ability of each enzyme to proteolyze the LPLTG peptide substrate using reversed-phase HPLC. In these reactions only the enzyme and LPLTG peptide are present. Interestingly, even though CdSrtAΔ and CdSrtA3M catalyze transpeptidation at 25 °C, the rate at which the hydrolytic side reaction occurs at this temperature is very slow for both enzymes, with less than 5% of the LPLTG peptide substrate consumed by the enzyme (Fig. 3A, left). In fact, because this assay employed excess amounts of peptide relative to enzyme (50 μM enzyme and 500 μM peptide), much of the observed peptide consumption can be attributed to formation of the thioacyl intermediate and not to repeated rounds of proteolysis. This finding is in marked contrast to the archetypal SaSrtA enzyme, which proteolyzes more than 60% of its sorting signal peptide substrate when identical reaction conditions are used (Fig. 3A, right). Thus, we conclude that for both the CdSrtAΔ and CdSrtA3M enzymes the transpeptidation pathway is dominant and the hydrolytic side reaction occurs only to a minor extent.
Figure 3.
The characterization of CdSrtA3M and CdSrtAΔ (A) A representative hydrolysis HPLC trace showing the hydrolysis by CdSrtA3M and SaSrtAWT at 25°C for 0 hours (solid line) and 24 hours (Dashed line). (B) A mass deconvolution of LC-MS data of CdSrtA3M. The acyl-intermediate at 24,851 Da is approximately 700 Da higher than where the enzyme is at 24,150 Da. Masses are 88 Da higher than expected due to the presence of a formic acid, acetonitrile and proton mass adduct (C) A comparison of the amounts of acyl-intermediate with and without the presence of NSpaA over a period of three hours. CdSrtA3M without NSpaA (white) in comparison to CdSrtAΔ without NSpaA (gray) shows a must faster formation of acyl-intermediate within the first 30 minutes of mixing the reaction. Upon the introduction of NSpaA to CdSrtA3M (blue) and CdSrtAΔ (red), there are similar amounts of acyl-intermediate formed over a period of three hours. (D) Fresh samples of protein were expressed and either treated with excess amount of DTT or additional buffer for one hour. The proteins were then digested with trypsin and then iodoacetamide was added to alkylate the reduced cysteines. The digests were then run on an on-line EASY-Spray HPLC (Pepmap C18 column, 25 cm x75 μm) to a Q-Exactive Orbitrap mass spectrometer (Thermo). The data was analyzed with MASCOT and the percent amount of alkylated cysteine was compared to total amount of cysteine peptides present. (n=3)
We wondered whether the superior transpeptidation activity of CdSrtAΔ originated from its ability to form the enzyme-LPLT thioacyl intermediate more rapidly than CdSrtA3M (Scheme 1). For both the CdSrtAΔ and CdSrtA3M enzymes, this intermediate is readily observable in LC-MS mass spectra when they are incubated with the LPLTG peptide (Fig. 3B). This finding is consistent with the low proteolytic activity of each enzyme and suggests that the two steps of catalysis are independent of one another – each enzyme can form and maintain the enzyme-acyl intermediate in the absence of the NSpaA nucleophile. Moreover, it is compatible with our previously published cellular studies of class C sortases in which long-lived and stable acyl-enzyme intermediates were established to be important for catalysis.31 To determine if CdSrtAΔ and CdSrtA3M differed in their ability to perform the first step of catalysis, we tracked thioacyl intermediate formation at various times after mixing the enzymes with the LPLTG peptide (25 μM and 1 mM of enzyme and LPLTG peptide, respectively). The intermediate formation was followed over a 3 hour period, the same duration used to measure transpeptidation activity. This analysis revealed that CdSrtAΔ forms the thioacyl intermediate more rapidly than CdSrtA3M, with ~17± 2% of CdSrtAΔ joined to the peptide after 30 minutes, while it takes upwards to 3 hours for CdSrtA3M to form similar levels of this reaction intermediate (Fig. 3C). Interestingly, when similar time course experiments are performed in the presence of both NSpaA and LPLTG substrates, significantly lower amounts of thioacyl intermediate are observed for each type of enzyme (~5% for both CdSrtAΔ and CdSrtA3M). As the reaction conditions are identical to those used to measure transpeptidation activity, this data suggests that the formation of the acyl-intermediate is rate-limiting. Thus, we conclude that improved transpeptidation activity of CdSrtAΔ results from its ability to form the acyl-intermediate at a faster rate than CdSrtA3M.
The identity of the ‘X’ residue within the LPXTG sorting signal affects the rate of transpeptidation.
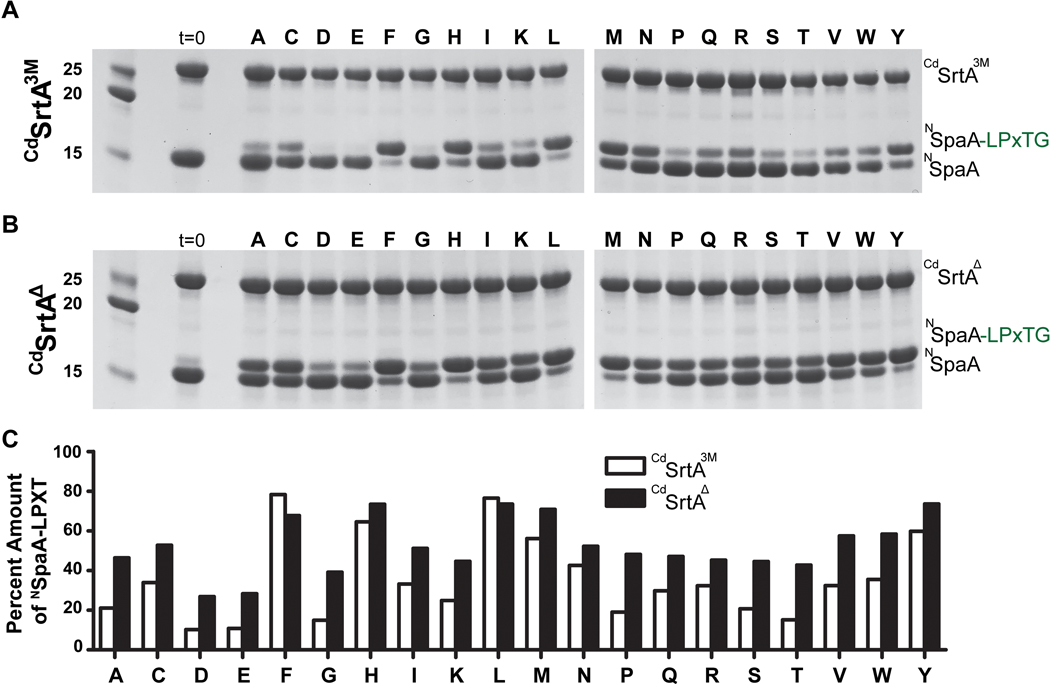
Structural and computational studies of class A and B sortases bound to their respective sorting signals have revealed that they do not recognize the side chain of the ‘X’ residue within their respective LPXTG-type sorting signal substrates because it projects away from the enzyme into the solvent.22,32,33 The structures are consistent with detailed substrate specificity analyses of the class A SaSrtA enzyme, which revealed that LPXTG sorting signals containing any amino acid at the ‘X’ position can be used as substrates.34 However, CdSrtA and other class C sortases are unique because they contain a lid structure whose proximity to the active site could affect recognition of the LPXTG sorting signal (Fig. 1C).32 Indeed, our prior studies of CdSrtA3M revealed that a leucine to alanine substitution at the X position of the LPLTG sorting signal slowed transpeptidation.21 To investigate this issue in greater detail, the CdSrtAΔ and CdSrtA3M enzymes were tested for their ability to utilize as transpeptidation substrates twenty distinct LPXTG peptides in which the ‘X’ position was varied. For these studies, the CdSrtAΔ and CdSrtA3M enzymes were incubated with NSpaA and each member of the peptide library, and the amount of cross-linked product was then determined by SDS-PAGE (Fig. 4A,B). Significant variation in reactivity is observed for the different library members. However, in nearly all cases, CdSrtAΔ is more active than CdSrtA3M, consistent with the steady state kinetic measurements that employed the LPLTG peptide (Table 1). Interestingly, both enzymes exhibit similar sequence preferences. In particular, they preferentially use sorting signals containing phenylalanine, histidine, methionine, tyrosine and leucine at the ‘X’ position, while their least reactive substrates contain negatively charged side chains at this site. In all cases, peptides containing leucine at the ‘X’ position are very reactive, explaining why this amino acid is present in the native LPLTG substrate present in SpaA. The ‘X’ position-dependent activity of CdSrtA is distinct from what has been observed for SaSrtA, as similar peptide library studies have shown that after 24 hours exposure all peptides in the library are processed to a similar extent by SaSrtA.35 The molecular basis underlying the observed variation in peptide reactivity is not known, but it is not caused by the presence of the lid as similar trends in activity are observed for CdSrtA3M and CdSrtAΔ.
Figure 4.
A) SDS-PAGE gel of the reactions of CdSrtA3M where the X position varied. Reactions (200 μM enzyme, 200 μM NSpaA, 5 mM DTT, and 1 mM Peptide) were measured after 24 hours. The majority of the more active sorting signals were non-polar while the polar residues tended to decrease the activity relative to the original sorting signal (LPLTG). B) SDS-PAGE gel of the reactions of CdSrtAΔ where the X position varied in the LPLTG Peptide. Reactions (200 μM enzyme, 200 μM NSpaA, 5 mM DTT, and 1 mM Peptide) were measured after 24 hours. C) Images were analyzed with Image J which estimated the amount of product produced for each peptide in the library.
Cysteine and arginine active site residues are required for catalysis.
Based on sequence homology with the well-studied SaSrtA sortase, three conserved residues in CdSrtA are presumably required for catalysis: H160, C222 and R231 (Fig. 1B).36,37 However, their role in catalyzing lysine-isopeptide bond transpeptidation in vitro has not been determined. We therefore used the HPLC-assay to measure the transpeptidation activities of CdSrtA3M and CdSrtAΔ enzymes containing alanine substitutions at these sites. For both enzyme variants, C222A and R231A substitutions completely abrogate transpeptidation activity, demonstrating that they have critical functions in catalysis (Table 1). However, CdSrtA3M and CdSrtAΔ enzymes harboring H160A substitutions retain some enzyme activity, exhibiting similar turnover numbers that are reduced by 44% and 94% as compared to their native forms, respectively. In the well-studied SaSrtA transpeptidation reaction the analogous histidine, cysteine and arginine residues are essential for catalysis in vitro (R197, C184 and H120 in the SaSrtA sequence).35 The cysteine thiol functions as a nucleophile, while the arginine guanidinium group (R197 in SaSrtA) has been proposed to facilitate catalysis by stabilizing oxyanion tetrahedral intermediates.22 Our finding that the C222A and R231A mutants are completely inactive is consistent with these residues having similar functions.27 It is also compatible with pH dependence of the transpeptidation reaction, which occurs most rapidly at pH values between 7.5 and 8. We were surprised that H160A mutants retain some activity, since histidine mutations in SaSrtA disrupt transpeptidation in vitro.36 In SaSrtA the histidine side chain has been proposed to function as a general acid and base, protonating the amine of the leaving group glycine residue as the threonine-glycine peptide bond in the LPLTG sorting signal is broken, and facilitating the last step of catalysis by deprotonating incoming amine nucleophile that resolves the thioacyl intermediate to produce the final transpeptidation product (Scheme 1).28,38 Whole cell studies have shown that H160A mutants of CdSrtA are incapable of assembling surface pili.27 The residual activity observed in the H160A mutants suggests residues in addition to H160 in CdSrtA may facilitate these steps in vitro, albeit less efficiently.
We wondered whether the superior activity of CdSrtAΔ relative to CdSrtA3M could be attributed to differences in the oxidation state of the C222 sulfhydryl group that is influenced by the presence of the lid. This is because the sulfhydryl group can in principle become oxidized to unreactive sulfenic, sulfinic and sulfonic forms that are non-nucleophilic.39 To investigate this issue, freshly purified CdSrtAΔ and CdSrtA3M enzymes were incubated for one hour in buffer A (50 mM Tris-HCl, 300 mM NaCl, at a pH of 8.0) with, and without, a reducing agent (5 mM DTT). The amount of C222 (the protein’s only cysteine) present in the active thiol form was then determined by adding iodoacetamide, digesting with trypsin, and analyzing by mass spectrometry. The percent of reduced thiol was calculated by measuring the amount of C222 in each enzyme that was derivatized with carboxyamidomethyl as compared to the total amount of C222 in its various oxidation states. In the absence of reducing agent, 89 ± 4 and 79 ± 10 % of CdSrtA3M and CdSrtAΔ contain a reactive thiol, respectively. Moreover, only small increases in the amount of reactive thiol are observed when the DTT reducing agent is present; under these conditions 94 ± 3 and 93 ± 5 % CdSrtA3M and CdSrtAΔ are reactive, respectively (Fig. 3D). Thus, the C222 sulfhydryl in freshly purified CdSrtA3M and CdSrtAΔ primarily exists in a reduced, transpeptidation competent state. A caveat is that ionization efficiencies may differ for alkylated and non-alkylated forms of a cysteine-containing peptide. Nevertheless, the similarity between the +/− DTT values implies that C222 is primarily reduced. It should be noted that we have found that C222 can become oxidized and unreactive if the enzymes are stored for more than several weeks and that their activity can be restored by incubating them with DTT. Therefore, including DTT in the labeling reactions is recommended as a precaution.
Improved protein lysine labeling using CdSrtAΔ.
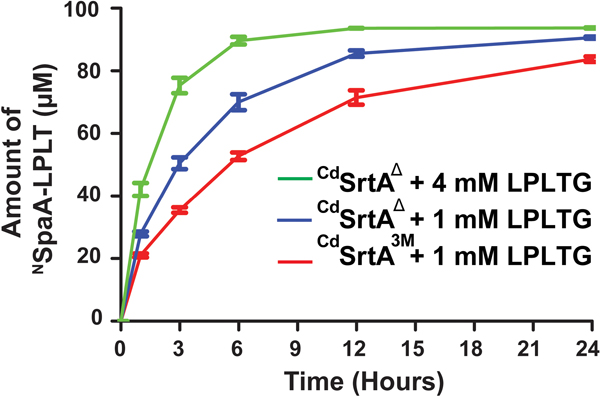
Having defined substrate and reaction conditions that are optimal for activity, we directly compared the peptide labeling efficiencies of CdSrtA3M with the newer CdSrtAΔ enzyme. Consistent with our steady-state kinetic analyses, a temporal analysis using identical conditions clearly shows that CdSrtAΔ (blue trace) produces more cross-linked product than CdSrtA3M (red trace) (Fig. 5) (100 μM enzyme, 100 μM NSpaA and 1 mM LPLTG peptide at 25 °C). In particular, when CdSrtAΔ is used, ~90% of NSpaA is modified with peptide within 12 hours, a higher amount than is achieved with CdSrtA3M even after 24 hours. Notably, even faster labeling can be achieved using CdSrtAΔ by increasing the concentration of LPLTG peptide in the reaction to 4 mM, which enables 90 ± 2 % of NSpaA to be modified within 6 hours (Fig. 5, green trace). This is finding is compatible with the relatively high SKM (2 ± 1 mM), which makes it challenging to saturate the enzyme with sorting signal substrate (Table 1). Similar comparisons were performed at 37 °C instead of 25 °C. CdSrtAΔ becomes less active at elevated temperatures because it is less thermostable than CdSrtA3M. However, both the rate and yield of product formed by CdSrtAΔ at 25 °C is superior to what CdSrtA3M produces at 37 °C. Thus, CdSrtAΔ should be used for labeling reactions that are performed at room temperature.
Figure 5.
A comparative study between CdSrtA3M and CdSrtAΔ at identical conditions of 1 mM LPLTG peptide and CdSrtAΔ at its ideal conditions of 4 mM of LPLTG peptide with 100 μM of enzyme and 100 μM of NSpaA. (n=3) After 6 hours, CdSrtAΔ with 4 mM peptide ligated 90% of NSpaA compared to 50% ligation by CdSrtA3M.
Comparison with the prototypical SaSrtA sortase.
SaSrtA is the best-studied member of the sortase superfamily and is routinely used to modify the N- and C-termini of proteins.13,22 CdSrtA and SaSrtA catalyze mechanistically related transpeptidation reactions, but differ in the type of nucleophile that they use, i.e., a lysine ε-amine within NSpaA and the N-terminal amine group within an oligoglycine peptide, respectively. In order to directly compare their activities, an established HPLC-assay was used to measure the rate at which SaSrtA joins the LPLTG peptide previously used in studies of CdSrtA to its triglycine substrate (Gly3), a reaction that creates a FELPTGGGG peptide product (Table 1).28,34 The steady state kinetic values measured from this analysis are generally consistent with published values that used a much shorter fluorogenic labeled peptide.28 Our direct comparison reveals that SaSrtA forms backbone peptide bonds ~40-times faster than CdSrtAΔ creates lysine-isopeptide bonds (Table 1). Interestingly, both enzymes exhibit generally similar KM values for their substrates, with each exhibiting millimolar KM values for the sorting signal substrate (CdSrtAΔ: 1.6 ± 0.3 mM SaSrtA: 7.33 ± 1.01 mM ), and approximately ~10-fold lower KM values for their respective amine nucleophiles (CdSrtAΔ: 160 ± 30 μM SaSrtA: 180 ± 10 μM ).36 In addition, based on prior studies by the McCafferty group and the studies presented here (Fig. 3), for both SaSrtA and CdSrtA the first step of catalysis is rate limiting – formation of the thioacyl intermediate (Scheme 1).28,34 It is unclear why this step is so slow in both enzymes. However, it has been shown that SaSrtA primarily exists in an inactive form in which only a small fraction (ca. 0.06%) of enzyme contains catalytically capable Cys184 thiolate and His120 imidazolium forms that are capable of reacting with the sorting signal to form the thioacyl intermediate.29,40,41 Whether the active site of CdSrtA also primarily exists in a catalytically dormant state remains to be determined. Startlingly, we found that CdSrtA is quite inefficient at proteolyzing the LPLTG substrate, which is in stark contrast to previously reported studies of SaSrtA that have shown that it catalyzes this reaction with a kcat of 0.28 ± 0.02 s−1 (Fig. 3A).28 Presumably this difference originates from distinct active site features that enable SaSrtA to use water as a nucleophile much more efficiently than CdSrtA (Scheme 1). This idea is substantiated by our observation that the thioacyl intermediate in the CdSrtA reaction forms to an appreciable extent in the absence of the nucleophile (Fig. 3C). The reduced proteolytic activity of CdSrtA is presumably advantageous, limiting the release of partially assembled pili from the bacterial cell surface. Similar to CdSrtA, under certain conditions SaSrtA can catalyze formation of a lysine isopeptide bond.42–44 A thorough study by Dasgupta and colleagues demonstrated that SaSrtA can join sorting signal and lysine-containing peptides together via an isopeptide linkage.43 However, the reaction was inefficient and exhibited poor substrate specificity; even after 12 hours, the ligation reaction was incomplete and a range of distinct isopeptide-linked products were generated. Moreover, during these reactions a significant amount of the sorting signal was proteolyzed, suggesting that SaSrtA does not discriminate between lysine and water nucleophiles. This is in contrast to CdSrtAΔ and CdSrtA3M, which produce lysine isopeptide-linked products at high yields with only limited proteolysis of the sorting signal substrate.
In conclusion, we have characterized the in vitro kinetics and mechanism of lysine-isopeptide bond forming activity of CdSrtA, and discovered CdSrtAΔ which is ~7-fold more active than previously reported enzyme variants. This bioconjugation activity is beginning to rival that of microbial transglutaminases from Streptomyces mobaraensis, which can be used to join biomolecules isopeptide linkages between glutamine and lysine side chains.45 Although promising, these enzymes have not gained wide usage in site specific protein labeling, presumably because of their penchant to catalyze spurious ligations.45–48 In contrast, CdSrtA exhibits a high level of specificity for its substrates, ligating peptides containing a LPXTG sequence to a specific lysine residue within the NSpaA domain. The molecular basis of specificity for the K190 side chain remains unknown, but presumably originates from protein-protein interactions between the NSpaA and CdSrtA-LPLT thioacyl intermediate that function to properly position the nucleophile for catalysis. It may also arise from NSpaA structural features that provide an environment for K190 that lower its pKa. Our mechanistic analysis also provides insight into the function of the lid, which is widely conserved in sortase enzymes that assemble pili. We show that in vitro its presence primarily affects the rate of thioacyl intermediate formation, and that it does not have a significant role in recognizing NSpaA or the ‘X’ residue within the LP(X)TG sorting signal. Further improvements in CdSrtAΔ-mediated labeling activity may also be possible, as we estimate that current versions of the enzyme catalyze in vitro transpeptidations ~102−3 times slower than the native enzyme when it is located on the cell surface. Thus, to further improve this protein labeling system future research should be focused on obtaining a greater understanding of the process of substrate recognition and catalysis.
Methods
Protein reagents.
Purified CdSrtA3M pilin sortase (residues N37-Q257 of SrtA from C. diphtheriae) and mutants were expressed and purified as described previously.21 Briefly, proteins were expressed from a pE-SUMO (Lifesensors) plasmid in E. coli BL21 (DE3) cells. The cells were grown up in LB supplemented with 500 μg/mL of Kanamycin at 37 °C until they reached an OD600 of ~ 0.6. The cells were induced with 1M IPTG and then left to express at 17 °C for 8 to 12 hours. After, the cells were removed from the incubator and pelleted at 8670 x g for 15 minutes. The pellets were then dissolved in a buffer of 50 mM Tris-HCl, 300 mM NaCl, at a pH of 8.0 (lysis buffer). Subsequently the cells were lysed using high-pressure emulsification and then fractionated via centrifugation at 22,720 x g for 50 minutes. Afterwards, the cell lysate was purified via IMAC-Co2+ purification. Proteins were eluted from the resin using a lysis buffer supplemented with 200 mM Imidazole. The His6x-SUMO tags were removed via treatment by His6x-Ulp1 protease at 1 mg/mL and subsequent purification by IMAC-Co2+. Afterwards, the protein was purified by size exclusion chromatography via the AKTA Pure (GE) and with Superdex 75pg resin. Protein purity was confirmed by SDS-PAGE. pE-SUMO expression plasmids encoding CdSrtA3M mutants were created using standard molecular biology methods and confirmed by nucleotide sequencing. NSpaA (residues E30 to S195) and SaSrtA (S. aureus Sortase A, residues Q60-K206) were purified as described previously.27,49 All purified enzymes were stored at −20°C in buffer A (50 mM Tris-HCl, 300 mM NaCl, at a pH of 8.0) supplemented with 40% glycerol. The FELPLTGGSG peptide (LPLTG peptide) used in the transpeptidation and hydrolysis assays was synthesized by Peptide 2.0.
Transpeptidation assays.
An HPLC-based assay was developed to quantify the kinetics of CdSrtA catalyzed lysine-isopeptide bond formation. In this assay, the NSpaA protein containing the reactive lysine is ligated to a FELPLTGGSG peptide (LPLTG, where the underlined residues correspond to the sorting signal) by the pilin sortase, followed by quantification using a HPLC C4 column. Reactions were performed in 100 μL volumes and contained 25 μM of pilin sortase (either CdSrtA3M or CdSrtAΔ), DTT (5 mM), either constant or variable amounts of LPLTG peptide (1 mM or 0.5 to 4 mM), and either constant or variable amounts of NSpaA (500 μM, 62.5 to 500 μM). All components were dissolved in buffer A. At these substrate concentrations, an estimated KM for NSpaA and LPLTG was determined. Reactions were initiated by adding the pilin sortase from a 2 mM stock solution, incubated for 3 hours at 25°C and then flash frozen with liquid N2 and stored at −20 °C. The reactions were analyzed using a Phenomenex C4 column (5 μm, 4.6 × 150 mm) and with an initial dwell time of 3 minutes at 36% CH3CN/0.1% TFA followed by a linear gradient from 36% to 46% CH3CN/0.1% TFA for 10 minutes at 1 mL/min was applied. The column was subsequently flushed with high concentrations of CH3CN/0.1% at 1 mL/min. NSpaA containing peaks were detected at 215 nm and the amount of substrate converted to product was calculated by integrating the area under the HPLC traces. The identity of each peak in the HPLC chromatogram was confirmed via MALDI-TOF MS. The activity of CdSrtA was compared to SaSrtA, which catalyzes a transpeptidation reaction that forms a backbone-backbone peptide bond between LPXTG and oligoglycine peptides. SaSrtA transpeptidation activity was measured as described previously.35 These reactions were performed in an identical manner to the CdSrtA reaction described above, except that they contained 25 μM SaSrtA instead of CdSrtA and were supplemented with 10 mM calcium in Buffer A, and triglycine (Gly3 peptide) (62.5 μM to 1 mM) instead of NSpaA. Reactions were quenched 15 minutes after mixing by adding an equal amount of 1 N Hydrochloric Acid (HCl). The reaction products were separated by HPLC using a Phenomenex C18 column (10 μm, 4.6 × 150 mm) and a linear gradient from 26% to 30% CH3CN/0.1% TFA over 8 minutes (1 mL/min). All HPLC experiments were performed on an Agilent 1100 HPLC. For both the SaSrtA and CdSrtA reactions kinetic parameters were obtained by fitting the data with Sigmaplot 12.0.
Two types of transpeptidation assays were used to investigate sorting signal specificity of the CdSrtA3M and CdSrtAΔ enzymes for the ‘X’ residue within the LPXTG sorting signal. A total of 20 peptides were tested in which the ‘X’ residues in the FELPXTGGSG was varied (Peptide 2.0). Reactions were performed in buffer A with a total volume of 35 μL. The reactions contained: either CdSrtA3M or CdSrtAΔ (200 μM), DTT (5 mM), NSpaA (200 μM), and one of the FELPXTGGSG peptides (1 mM, Peptide 2.0). Transpeptidation reactions performed at 25°C for 24 hours, and then quenched by flash freezing in liquid N2 and stored at −20°C. 5 μL of each reaction was diluted 4 times in SDS loading buffer and separated using a 12% SDS-PAGE gel, then visualized by Coomassie staining. The resulting bands were analyzed with ImageJ with the zero hour time point used as a control for activity.
Hydrolysis and cysteine oxidation measurements.
The hydrolytic activity of SaSrtA and CdSrtA variants was determined using an HPLC-based assay that monitors the ability of each enzyme to cleave the LPLTG peptide between the threonine and glycine residues. Reactions were performed in buffer A supplemented with 10 mM calcium and contained a total volume of 100 μL: sortase (50 μM), LPLTG (500 μM), and DTT (5 mM). Reactions were incubated at 25°C for 24 hours, and then quenched by adding an equal volume of 1N HCl. Reaction products were separated on a Waters C18 Column (10 μm, 4.6×150 mm) using a linear gradient from 26% to 30% CH3CN/0.1% TFA over 8 minutes at 1 mL/min. The reaction was monitored at 215 nm and the identity of each peak in the HPLC chromatogram was confirmed via LC-MS.
The oxidation status of the active site cysteine by monitoring susceptibility to iodoacetamide alkylation. CdSrtA3M and CdSrtAΔ were purified and stored in buffer A. Enzymes were then either treated to a final concentration of 5 mM of DTT or a buffer control for 1 hour before being frozen at −20°C. Subsequently, the proteins were defrosted and alkylated in 25 mM iodoacetamide and exchanged four times into 100 mM ammonium bicarbonate buffer using Amicon® 3kDa centrifugal filters.50 Trypsin digestion was performed overnight at 37°C. C18 Stage Tips were used for desalting prior to tandem mass spectrometry. Peptides were separated and measured on an EASY-Spray HPLC column (25 cm × 75 μm ID packed with PepMap RSLC C18, 2 μm particles, Thermo Scientific) with an on-line Easy-nLC 100 chromatography system to a Orbitrap mass spectrometer (Q-Exactive Orbitrap, Thermo Scientific). Precursor ions were selected using data dependent acquisition (top 10) and fragmented using collision induced dissociation (CID) at a normalized collision energy of 27. Raw MS/MS files were converted to mgf format (Thermo Proteome Discoverer, Thermo Scientific v1.4) and were searched against a sequence database using MASCOT (Matrix Science). Searches employed variable cysteine carbamidomethylation and methionine oxidation. The precursor mass accuracy was set to 10 ppm, while that for product ions was set to 0.02 da. Once identified, the fully digested peptides’ intensities were quantified from the area under the curves. Intensities of carbamidomethylated peptides were normalized by dividing against the summed intensity of the 3 most abundant peptides identified.
Acyl-intermediate detection with LC-MS.
LC-MS reactions to compare acyl formation with and without the presence of NSpaA were performed in buffer A and contained a total volume of 100 μL: sortase (25 μM), DTT (5 mM), LPLTG (1 mM), and NSpaA (250 μM) or buffer. All reactions were incubated at 25°C and 10 μL time points were removed and frozen with liquid N2 before being stored at −20°C. Before being run on the LC-MS system, samples were then diluted with 90 μL of 200 mM L-tryptophan (internal standard) and then measured on a Zorbax 300SB-C3 (3.5 μm, 3.0 × 150 mm) with an Agilent 6530 Q-TOF and Agilent 1260 Infinity HPLC with a gradient of 30 to 99 % over 6 minutes at 0.8 mL/min. The data was analyzed with Agilent MassHunter Qualitative Analysis. The amount of enzyme and acyl-intermediate was calculating by integrating the area under the curve for each peak.
ACKNOWLEDGMENT
This work was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research program under Award Number DE-FC02–02ER63421 and National Institutes of Health Grants AI52217 (R.T.C. and H.T-T.), DE025015 (H. T-T) and GM103479 (J.A.L.). C.K.S and S.A.M. were supported by a Cellular and Molecular Biology Training Grant (Ruth L. Kirschstein National Research Service Award GM007185). J.M. was supported by a UCLA Molecular Biology Institute Whitcome Fellowship (S10OD016336) NMR equipment used in this research was purchased using funds from shared equipment grant NIH S10OD016336.
References
- 1.Agarwal P; Bertozzi CR, Site-Specific Antibody Drug Conjugates: The Nexus of Bioorthogonal Chemistry, Protein Engineering, and Drug Development. Bioconjugate Chemistry 2014, 26, (2), 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chudasama V; Maruani A; Caddick S, Recent advances in the construction of antibody drug conjugates. Nature Chemistry 2015, 8, (2), 114–119. [DOI] [PubMed] [Google Scholar]
- 3.Hoyt EA; Cal PMSD; Oliveira BL; Bernardes G. a. J. L., Contemporary approaches to site-selective protein modification. Nature Reviews Chemistry 2019, 3, (3), 147–171. [Google Scholar]
- 4.Lagasse HAD; Alexaki A; Simhadri VL; Katagiri NH; Jankowski W; Sauna ZE; Kimchi-Sarfaty C, Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elizabeth AS; Esther B; Amy EP, A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annual Review of Physiology 2016, 79, (1), 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sochaj AM; Åšwiderska KW; Otlewski J, Current methods for the synthesis of homogeneous antibody–drug conjugates. Biotechnology Advances 2015, 33, (6, Part 1), 775–784. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto T; Tanaka T; Kondo A, Enzyme-mediated methodologies for protein modification and bioconjugate synthesis. Biotechnology Journal 2012, 7, (9), 1137–1146. [DOI] [PubMed] [Google Scholar]
- 8.Mohamad NR; Marzuki NHC; Buang NA; Huyop F; Wahab RA, An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnology & Biotechnological Equipment 2015, 29, (2), 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krall N; da Cruz FP; Boutureira O; Bernardes G. a. J. L., Site-selective protein-modification chemistry for basic biology and drug development. Nature Chemistry 2015, 8, (2), 103–113. [DOI] [PubMed] [Google Scholar]
- 10.Spicer CD; Davis BG, Selective chemical protein modification. Nature Communications 2014, 5, (1), 4740. [DOI] [PubMed] [Google Scholar]
- 11.Li X; Fang T; Boons G-J, Preparation of Well-Defined Antibody Drug Conjugates through Glycan Remodeling and Strain-Promoted Azide-Alkyne Cycloadditions. Angewandte Chemie International Edition 2014, 53, (28), 7179–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y; Park K-Y; Suazo KF; Distefano MD, Recent progress in enzymatic protein labelling techniques and their applications. Chemical Society Reviews 2018, 47, (24), 9106–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antos JM; Truttmann MC; Ploegh HL, Recent advances in sortase-catalyzed ligation methodology. Current Opinion in Structural Biology 2016, 38, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmohl L; Schwarzer D, Sortase-mediated ligations for the site-specific modification of proteins. Current Opinion in Chemical Biology 2014, 22, 122–128. [DOI] [PubMed] [Google Scholar]
- 15.Popp MW-L; Ploegh HL, Making and Breaking Peptide Bonds: Protein Engineering Using Sortase. Angewandte Chemie International Edition 2011, 50, (22), 5024–5032. [DOI] [PubMed] [Google Scholar]
- 16.Samantaray S; Marathe U; Dasgupta S; Nandicoori VK; Roy RP, Peptide−Sugar Ligation Catalyzed by Transpeptidase Sortase:  A Facile Approach to Neoglycoconjugate Synthesis. Journal of the American Chemical Society 2008, 130, (7), 2132–2133. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian SK; Liu G; Ton-That H; Schneewind O, Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 1999, 285, (5428), 760–3. [DOI] [PubMed] [Google Scholar]
- 18.Antos JM; Chew G-L; Guimaraes CP; Yoder NC; Grotenbreg GM; Popp MW-L; Ploegh HL, Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. Journal of the American Chemical Society 2009, 131, (31), 10800–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popp MW; Dougan SK; Chuang TY; Spooner E; Ploegh HL, Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci U S A 2011, 108, (8), 3169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukiji S; Nagamune T, Sortase-Mediated Ligation: A Gift from Gram-Positive Bacteria to Protein Engineering. ChemBioChem 2009, 10, (5), 787–798. [DOI] [PubMed] [Google Scholar]
- 21.McConnell SA; Amer BR; Muroski J; Fu J; Chang C; Ogorzalek Loo RR; Loo JA; Osipiuk J; Ton-That H; Clubb RT, Protein Labeling via a Specific Lysine-Isopeptide Bond Using the Pilin Polymerizing Sortase from Corynebacterium diphtheriae. Journal of the American Chemical Society 2018, 140, (27), 8420–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobitz AW; Kattke MD; Wereszczynski J; Clubb RT, Sortase Transpeptidases: Structural Biology and Catalytic Mechanism. Advances in protein chemistry and structural biology 2017, 109, 223–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HJ; Baker EN, Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem 2009, 284, (31), 20729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comfort D; Clubb RT, A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun 2004, 72, (5), 2710–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Echelman DJ; Alegre-Cebollada J; Badilla CL; Chang C; Ton-That H; Fernandez JM, CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc Natl Acad Sci U S A 2016, 113, (9), 2490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ton-That H; Schneewind O, Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 2003, 50, (4), 1429–38. [DOI] [PubMed] [Google Scholar]
- 27.Chang C; Amer BR; Osipiuk J; McConnell SA; Huang IH; Hsieh V; Fu J; Nguyen HH; Muroski J; Flores E; Ogorzalek Loo RR; Loo JA; Putkey JA; Joachimiak A; Das A; Clubb RT; Ton-That H, In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking. Proceedings of the National Academy of Sciences 2018, 115, (24), E5477–E5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frankel BA; Kruger RG; Robinson DE; Kelleher NL; McCafferty DG, Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry 2005, 44, (33), 11188–200. [DOI] [PubMed] [Google Scholar]
- 29.Clancy KW; Melvin JA; McCafferty DG, Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers 2010, 94, (4), 385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradshaw WJ; Davies AH; Chambers CJ; Roberts AK; Shone CC; Acharya KR, Molecular features of the sortase enzyme family. FEBS J 2015, 282, (11), 2097–114. [DOI] [PubMed] [Google Scholar]
- 31.Guttilla IK; Gaspar AH; Swierczynski A; Swaminathan A; Dwivedi P; Das A; Ton-That H, Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J Bacteriol 2009, 191, (18), 5603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobitz AW; Naziga EB; Yi SW; McConnell SA; Peterson R; Jung ME; Clubb RT; Wereszczynski J, The “Lid” in the Streptococcus pneumoniae SrtC1 Sortase Adopts a Rigid Structure that Regulates Substrate Access to the Active Site. J Phys Chem B 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan AH; Yi SW; Terwilliger AL; Maresso AW; Jung ME; Clubb RT, Structure of the Bacillus anthracis Sortase A Enzyme Bound to Its Sorting Signal: A Flexible Amino-Terminal Appenage Modulates Substrate Access. J Biol Chem 2015, 290, (42), 25461–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruger RG; Dostal P; McCafferty DG, Development of a high-performance liquid chromatography assay and revision of kinetic parameters for the Staphylococcus aureus sortase transpeptidase SrtA. Anal Biochem 2004, 326, (1), 42–8. [DOI] [PubMed] [Google Scholar]
- 35.Kruger RG; Otvos B; Frankel BA; Bentley M; Dostal P; McCafferty DG, Analysis of the Substrate Specificity of the Staphylococcus aureus Sortase Transpeptidase SrtA. Biochemistry 2004, 43, (6), 1541–1551. [DOI] [PubMed] [Google Scholar]
- 36.Frankel BA; Tong Y; Bentley ML; Fitzgerald MC; McCafferty DG, Mutational analysis of active site residues in the Staphylococcus aureus transpeptidase SrtA. Biochemistry 2007, 46, (24), 7269–78. [DOI] [PubMed] [Google Scholar]
- 37.Zong Y; Bice TW; Ton-That H; Schneewind O; Narayana SV, Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem 2004, 279, (30), 31383–9. [DOI] [PubMed] [Google Scholar]
- 38.Huang X; Aulabaugh A; Ding W; Kapoor B; Alksne L; Tabei K; Ellestad G, Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry 2003, 42, (38), 11307–15. [DOI] [PubMed] [Google Scholar]
- 39.Alcock LJ; Perkins MV; Chalker JM, Chemical methods for mapping cysteine oxidation. Chemical Society Reviews 2018, 47, (1), 231–268. [DOI] [PubMed] [Google Scholar]
- 40.Weiner EM; Robson S; Marohn M; Clubb RT, The Sortase A enzyme that attaches proteins to the cell wall of Bacillus anthracis contains an unusual active site architecture. J Biol Chem 2010, 285, (30), 23433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly KM; Smith BT; Pilpa R; Ilangovan U; Jung ME; Clubb RT, Sortase from Staphylococcus aureus does not contain a thiolate-imidazolium ion pair in its active site. J Biol Chem 2003, 278, (36), 34061–5. [DOI] [PubMed] [Google Scholar]
- 42.Bellucci JJ; Bhattacharyya J; Chilkoti A, A Noncanonical Function of Sortase Enables Site-Specific Conjugation of Small Molecules to Lysine Residues in Proteins. Angewandte Chemie International Edition 2014, 54, (2), 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dasgupta S; Samantaray S; Sahal D; Roy RP, Isopeptide ligation catalyzed by quintessential sortase A: mechanistic cues from cyclic and branched oligomers of indolicidin. The Journal of biological chemistry 2011, 286, (27), 23996–24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohlmann S; Mahlert C; Greven S; Scholz P; Harrenga A, In vitro Sortagging of an Antibody Fab Fragment: Overcoming Unproductive Reactions of Sortase with Water and Lysine Side Chains. ChemBioChem 2011, 12, (11), 1774–1780. [DOI] [PubMed] [Google Scholar]
- 45.Deweid L; Avrutina O; Kolmar H, Microbial transglutaminase for biotechnological and biomedical engineering. In Biological Chemistry, 2018; Vol. 400, p 257. [DOI] [PubMed] [Google Scholar]
- 46.Malešević M; Migge A; Hertel TC; Pietzsch M, A Fluorescence-Based Array Screen for Transglutaminase Substrates. ChemBioChem 2015, 16, (8), 1169–1174. [DOI] [PubMed] [Google Scholar]
- 47.Maullu C; Raimondo D; Caboi F; Giorgetti A; Sergi M; Valentini M; Tonon G; Tramontano A, Site-directed enzymatic PEGylation of the human granulocyte colony-stimulating factor. The FEBS Journal 2009, 276, (22), 6741–6750. [DOI] [PubMed] [Google Scholar]
- 48.Jeger S; Zimmermann K; Blanc A; Grünberg J; Honer M; Hunziker P; Struthers H; Schibli R, Site-Specific and Stoichiometric Modification of Antibodies by Bacterial Transglutaminase. Angewandte Chemie International Edition 2010, 49, (51), 9995–9997. [DOI] [PubMed] [Google Scholar]
- 49.Ilangovan U; Ton-That H; Iwahara J; Schneewind O; Clubb RT, Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A 2001, 98, (11), 6056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melvin JA; Murphy CF; Dubois LG; Thompson JW; Moseley MA; McCafferty DG, Staphylococcus aureus Sortase A Contributes to the Trojan Horse Mechanism of Immune Defense Evasion with Its Intrinsic Resistance to Cys184 Oxidation. Biochemistry 2011, 50, (35), 7591–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]