Abstract
Owing to the astounding biological properties, dietary plant flavonoids have received considerable attention toward developing unique supplementary food sources to prevent various ailments. Chemokines are chemotactic proteins involved in leukocyte trafficking through their interactions with G-protein-coupled receptors and cell surface glycosaminoglycans (GAGs). CCL2 chemokine, a foremost member of CC chemokines, is associated with the pathogenesis of various inflammatory infirmities, thus making the CCL2-Receptor (CCR2)/GAG axis a potential pharmacological target. The current study is designed to unravel the structural details of CCL2–flavonol interactions. Molecular interactions between flavonols (kaempferol, quercetin, and myricetin) with human/murine CCL2 orthologs and their monomeric/dimeric variants were systematically investigated using a combination of biophysical approaches. Fluorescence studies have unveiled that flavonols interact with CCL2 orthologs specifically but with differential affinities. The dissociation constants (Kd) were in the range of 10–5–10–7 μM. The NMR- and computational docking-based outcomes have strongly suggested that the flavonols interact with CCL2, comprising the N-terminal and β1- and β3-sheets. It has also been observed that the number of hydroxyl groups on the annular ring-B imposed a significant cumulative effect on the binding affinities of flavonols for CCL2 chemokine. Further, the binding surface of these flavonols to CCL2 orthologs was observed to be extensively overlapped with that of the receptor/GAG-binding surface, thus suggesting attenuation of CCL2-CCR2/GAG interactions in their presence. Considering the pivotal role of CCL2 during monocyte/macrophage trafficking and the immunomodulatory features of these flavonols, their direct interactions highlight the promising role of flavonols as nutraceuticals.
1. Introduction
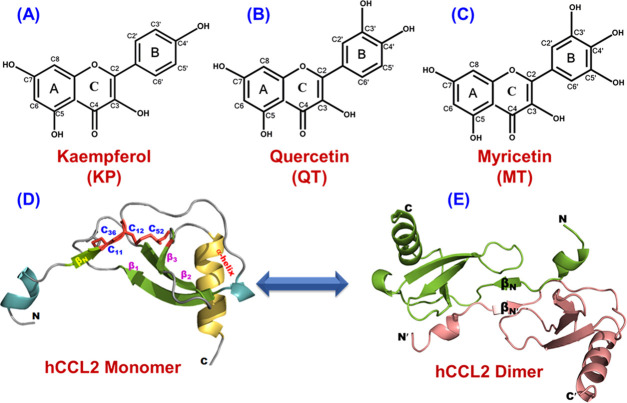
Natural products are in existence since time immemorial to furnish mankind with new panacea and effective remedies and also ensconce the foundation of sophisticated systems under the parasol of traditional medicine.1,2 Among various natural products, phytochemicals such as flavonoids have been consumed extensively to prevent various pernicious infirmities.3,4 Nearly 65–70% of people worldwide are solely dependent on herbal medicines for their health care, and more than 50% of the clinical drugs are based on natural products.5 Hence, the concept of consumption of flavonoids as nutraceuticals has been receiving significant interest. Flavonoids are categorized into seven subgroups: flavonols, flavones, flavanones, flavanols, chalcones, anthocyanidins, and isoflavones.4,6 Flavonol, also known as the 3-hydroxy derivatives of flavanones, is arguably the most widespread subgroup of flavonoids and is an eminent part of the human diet.4,7,8 Flavonols play a significant role in minimizing the effects of several noxious and inflamed conditions and exhibit innumerable beneficial therapeutic properties, including antiallergic,9,10 anticancer,11,12 antithrombogenic,13,14 antidiabetic,15,16 antioxidant,4,17 antifungal and antibacterial,18,19 and antiviral activities against the novel coronavirus (CoV).20,21 Apart from these biological effects, flavonols have also been reported to impart their anti-inflammatory activity to the biosynthesis of cytokines and chemokines, thereby restraining the attachment and migration of circulating leukocytes to the foci of inflammation.4,22,23 The most studied members of this family are kaempferol, quercetin, and myricetin. Structurally, these three flavonols differ in the number of hydroxyl groups (−OH) present on their annular ring-B (Figure 1A–C). The therapeutic properties of these flavonols make them provocative and astonishing candidates to be used as functional foods, nutraceuticals, preventive medicines, and pharma foods.24,25
Figure 1.
Characterization of the structural hallmarks of flavonols and the CCL2 protein. Chemical structure of flavonols kaempferol (KP) (A), quercetin (QT) (B), and myricetin (MT) (C). (D) Representative three-dimensional structure of the monomeric subunit of human CCL2 protein (PDB ID: 1dok) illustrating all of the crucial structural features. The two intra-disulfide bonds are represented in red. (E) Illustrative dimeric structure of human CCL2.
Chemokines, widely known as chemotactic cytokines, are crucial players in the leucocyte migration process that regulates the activation and migratory patterns of a subset of immune cells during infectious conditions.26,27 According to the positioning of N-terminal Cys residues, chemokines are categorized into four major subfamilies: C, CC, CXC, and CX3C.26,28 Chemokines’ biological functions are majorly governed by their compulsive interactions with cognate G-protein-coupled receptors (GPCRs) embedded on the leucocytes and glycosaminoglycans (GAGs) located on the cell surface.29,30 The monocyte chemoattractant protein (MCP) family is a crucial subgroup of the CC chemokine family, and the members of this subfamily are associated extensively with several inflammatory pathways.31,32 The MCP family consists of four pre-eminent members, including MCP-1, MCP-2, MCP-3, and MCP-4, and among these members, MCP-1 (CCL2) is the premier member.32 CCL2 has been extensively documented in numerous inflammatory conditions, such as multiple sclerosis,33 tumor conditions,34,35 HIV infection,31,36 diabetes,37 tuberculosis,38 cardiovascular disorders, and atherosclerosis.39,40 CCL2 exhibits monomer–dimer equilibrium with the dissociation constant (Kd) ∼50 μM.41,42 The structural elements of the CCL2 monomer constitute an N-terminal end, four consecutive antiparallel β-strands, and a C-terminal α-helix, and the dimer is formed through β0−β0′ interactions (Figure 1D,E).41,43,44 CCL2 engages its N-terminal segment and positive surface patches to interact with its cognate GPCR receptor CCR2 and GAGs.45,46 Thus, the CCL2-CCR2/GAG pivot has long been a thriving target for various pharmaceutical industries, tradipractitioners, and herbalists.47,48 On similar lines, baicalin (BA), a flavone-glycoside, was reported to modulate the chemotactic activity of CCL2 and other chemokines by interfering with the chemokine–receptor/GAG axis.49,50
Although flavonols have been widely studied as anti-inflammatory mediators, no molecular/structural information is available concerning their interaction with immune regulatory proteins such as chemokines. Hence, in the current study, all of the three potential members of the flavonol subgroup (kaempferol, quercetin, and myricetin) have been screened to decipher their binding characteristics with CCL2 chemokine. To gain comprehensive insights into the binding features, the flavonols were studied against monomeric and dimeric conformations of CCL2 for both human and murine CCL2 orthologs. Molecular interactions of CCL2–flavonol complexes were studied using fluorescence and NMR spectroscopies and molecular docking approaches. Our fluorescence quenching experiments have evidently suggested that the binding affinities significantly differ among the chosen flavonols. Further, NMR experiments and molecular docking studies have deciphered a pervasive overlap between the flavonol binding surface and the GAG/receptor-binding domain of the CCL2 protein. The intricate molecular details of flavonol–CCL2 interactions obtained in this study have firmly substantiated the anti-inflammatory properties of flavonols, thus proposing them as phytotherapeutic nutraceutical agents that fulfill the fundamental premise of “integrative medicine.”
2. Results
2.1. Assessing the Binding Affinity of Flavonols to CCL2 Orthologs by Fluorescence Spectroscopy
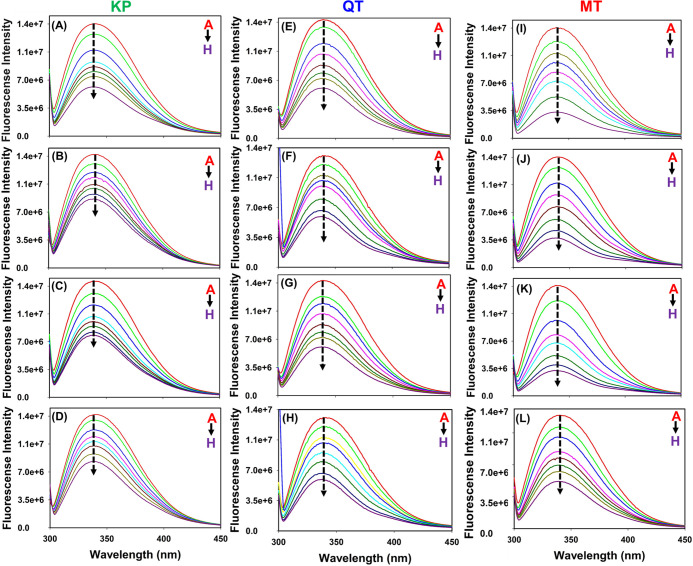
Flavonols kaempferol (KP), quercetin (QT), and myricetin (MT) are reported to have immunomodulatory effects as they can potentially target various immune regulatory proteins. Indeed, the involvement of chemokines such as CCL2 in various immune-related diseases has been extensively documented.31,32 Therefore, to decipher the direct molecular interactions of flavonols with chemokine proteins, fluorescence-based binding studies were performed between CCL2 orthologs (monomers and dimers) and KP, QT, and MT. Fluorescence titrations were carried out with increasing concentrations (10–100 μM) of flavonols (quenchers/titrants). A significant decrease in the Trp fluorescence intensities of CCL2 proteins was noticed upon increasing the concentration of flavonols, thus evidencing a noticeable interaction between them (Figure 2). Further, to confirm that the observed intensity changes are exclusively due to flavonol binding, control experiments were performed by adding a buffer without flavonols to CCL2 orthologs. No noticeable changes were observed in the spectra, indicating that the fluorescence intensity changes were due to formation of CCL2–flavonol complexes (Figure S1). Although there do exist apparent intensity changes in the fluorescence spectra of murine and human CCL2 (both monomers and wild type) orthologs upon the addition of flavonols, no noticeable spectral shifts were observed. Quenching parameters were assessed using the Stern–Volmer equation (Figure S2). For all of the flavonol–CCL2 complexes, the estimated quenching constant (Kq) values were observed to be ∼10–12 L mol–1 s–1 (Table 1). The observed Kq values for all three flavonol–CCL2 complexes are greater than the theoretical value of dynamic quenching (10–10 L mol–1 s–1), thus suggesting a static quenching mechanism.51,52
Figure 2.
Illustration of flavonol-mediated fluorescence quenching patterns of CCL2 orthologs. (A–D) Fluorescence quenching patterns of mCCL2-WT, mCCL2-P8A, hCCL2-WT, and hCCL2-P8A complexed with kaempferol (KP) (A–D), quercetin (QT) (E–H), and myricetin (MT) (I–L), respectively.
Table 1. Summarized Binding Parameters for the Interaction of Flavonols (Kaempferol, Quercetin, and Myricetin) with CCL2 Orthologsa.
| name of protein | quenching constant (Kq) (m–1 s–1) | dissociation constant (Kd) (μM) | number of binding sites | R2 |
|---|---|---|---|---|
| Kaempferol (KP) | ||||
| mCCL2-WT | 5.0 × 1012 | 24 ± 6 | 1.1 ± 0.2 | 0.96 |
| hCCL2-WT | 5.2 × 1012 | 26 ± 5 | 1.1 ± 0.1 | 0.97 |
| mCCL2-M | 5.1 × 1012 | 26 ± 5 | 1.2 ± 0. 1 | 0.97 |
| hCCL2-M | 5.3 × 1012 | 25 ± 4 | 1.1 ± 0.1 | 0.98 |
| Quercetin (QT) | ||||
| mCCL2-WT | 4.3 × 1012 | 3.9 ± 0.5 | 1.2 ± 0.2 | 0.98 |
| hCCL2-WT | 4.2 × 1012 | 3.5 ± 0.7 | 1.1 ± 0.1 | 0.99 |
| mCCL2-M | 5.7 × 1012 | 3.7 ± 0.4 | 1.1 ± 0.2 | 0.98 |
| hCCL2-M | 5.4 × 1012 | 3.6 ± 0.5 | 1.2 ± 0.1 | 0.99 |
| Myricetin (MT) | ||||
| mCCL2-WT | 6.1 × 1012 | 0.42 ± 0.03 | 1.1 ± 0.2 | 0.99 |
| hCCL2-WT | 5.8 × 1012 | 0.38 ± 0.04 | 1.3 ± 0.2 | 0.98 |
| mCCL2-M | 6.3 × 1012 | 0.40 ± 0.03 | 1.2 ± 0.1 | 0.99 |
| hCCL2-M | 6.2 × 1012 | 0.43 ± 0.02 | 1.2 ± 0.2 | 0.98 |
M represents monomer and WT represents wild type. For all CCL2 proteins, the concentrations were obtained in terms of the monomer.
Further, the dissociation constants (Kd) and the binding sites (n) were estimated using the double-logarithmic equation (Figure S3). Interestingly, significant variations in the Kd values for all of the three flavonol–CCL2 complexes were noticed (Table 1). The estimated Kd values for MT–CCL2, CCL2–KP, and CCL2–QT complexes were observed to be 0.4 ± 0.02, 3.5 ± 0.5, and 25 ± 5 μM, respectively (Table 1). A stoichiometry of approximately n = 1 for all of the complexes suggests that one CCL2 monomer binds to one flavonol molecule, as all of the protein concentrations were considered in monomeric fractions (Table 1). Nonlinear regression and Scatchard plot analysis also yielded similar stoichiometry and dissociation constant values, thus echoing the results obtained from double-logarithmic plots. A representative data analysis of MT binding to mCCL2-P8A using nonlinear regression and Scatchard plot analysis is presented in Figure S4. Fluorescence studies have clearly suggested that the oligomeric variation such as monomer/dimer and the orthologous nature of CCL2 does not influence the binding affinity for a chosen flavonol, as the observed binding/dissociation constants for a chosen flavonol (KP/QT/MT) are almost similar for both the monomeric and dimeric variants of hCCL2 and mCCL2 proteins (Table 1). Furthermore, the data establishes that CCL2 protein orthologs (monomer/dimers) differentially bind to the three chosen flavonols, and the efficacies of flavonols for CCL2 are as follows: kaempferol < quercetin < myricetin.
2.2. Delineating the Flavonol Binding Sites on CCL2 Chemokine Using NMR Spectroscopy
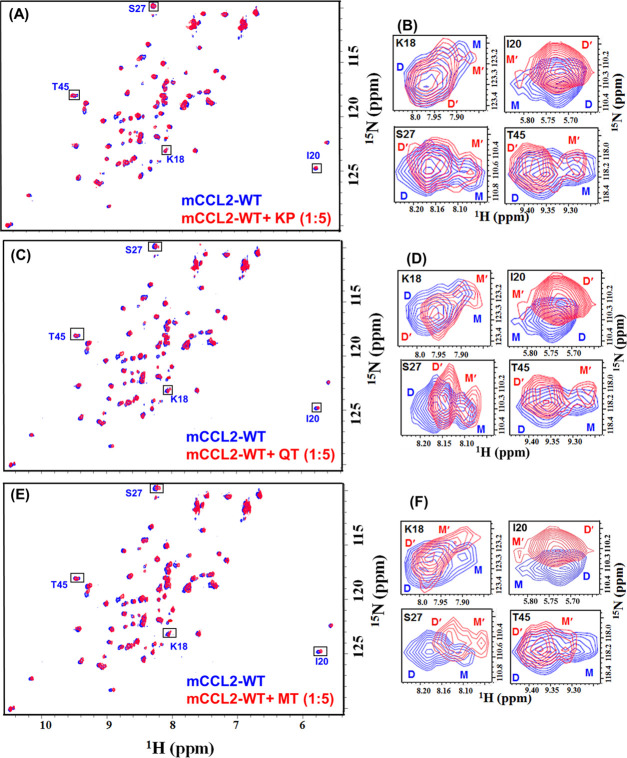
Taking the fluorescence quenching studies in the background, NMR-based studies were performed to unravel the residue-level information for flavonol interactions with mCCL2. Since it has been reported previously that CCL2 subsists in a monomer–dimer equilibrium at low concentration,42 all of the NMR titrations were performed at 100 μM protein concentration to assess the binding nature of both monomeric and dimeric species simultaneously. The titrants (KP, QT, and MT) were added to mCCL2-WT up to a ratio of 1:5 (protein/flavonol). A considerable extent of perturbation in the NH resonances of specific residues was detected. The change in the chemical shift position of the protein NH resonances upon addition of ligand indicates the binding interactions between the protein and the chosen ligand. Further, in the case of specific interactions, a subset of NH resonances of the protein that are involved in binding undergo exclusive spectral shifts. For all of the flavonol–mCCL2 complexes, the HSQC spectra overlay showing titrant-induced perturbations and the zoom-in section of monomer–dimer peaks (100 μM) are shown in Figure 3A–F. As the chemical shift perturbation (CSP) is the most crucial approach for evaluating the ligand-binding affinity,53 the nature of such perturbations induced by titrants was assessed exclusively by the CSP method. Residue-specific CSP plots were generated for the mCCL2 dimer and 16 separately identified monomeric resonances42 at a molar ratio of 1:5. Interestingly, it was noticed that a specific set of residues was considerably perturbed in both the dimeric and monomeric resonances of mCCL2-WT, thus signifying a prominent concurrency between their binding surfaces (Figure 4A,C,E).
Figure 3.
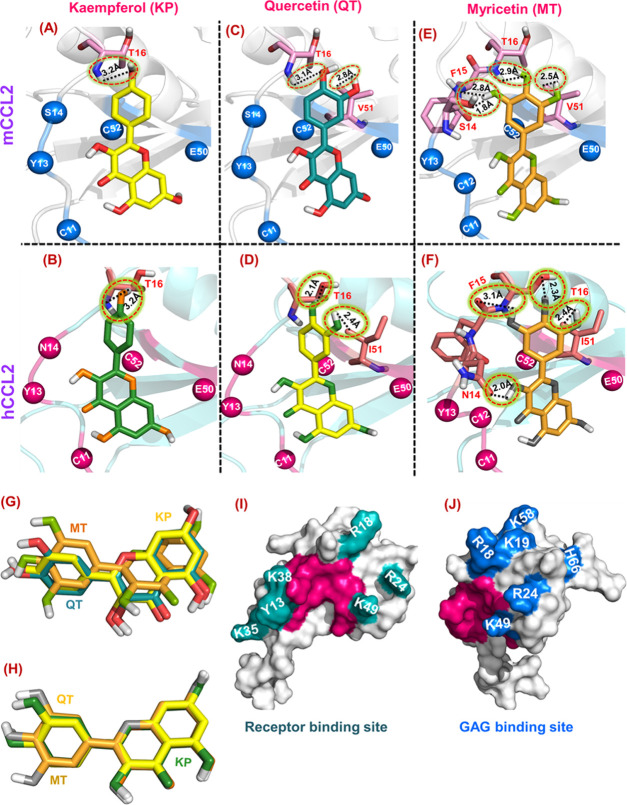
Deciphering the flavonols’ binding to the mCCL2 protein by NMR spectroscopy. HSQC overlay of (A) mCCL2-WT (red) and KP–mCCL2 complex (blue), (C) mCCL2 (red) and QT–mCCL2 complex (blue), and (E) mCCL2 (red) and MT–mCCL2 complex (blue). Residues exhibiting monomer–dimer equilibrium are signposted by rectangular boxes. The focalized HSQC sections of the boxed residues in (A), (C), and (E) are presented in (B), (D), and (F), respectively.
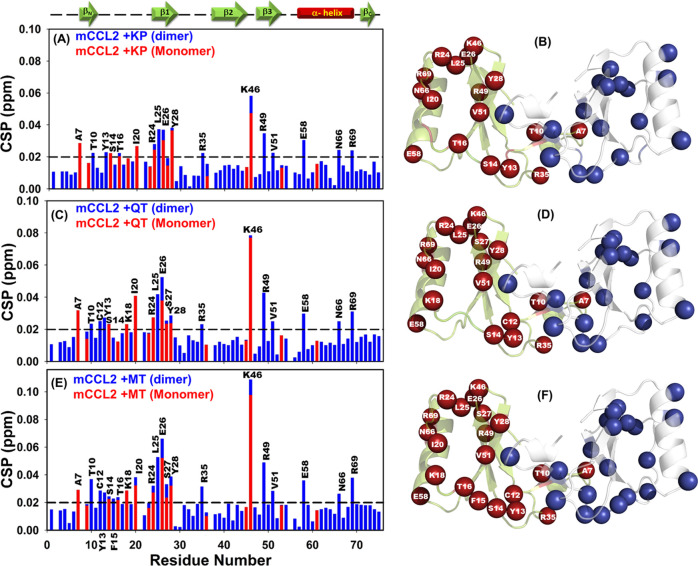
Figure 4.
1H–15N HSQC chemical shift perturbation (CSP) analysis of mCCL2–flavonol interactions. CSP histograms of (A) mCCL2–KP, (C) mCCL2–QT, and (E) mCCL2–MT complexes. The representative secondary structural features are presented on the top of CSP plot(s), and the horizontally dashed black line depicts the cutoff value. Residues exhibiting significant KP-induced (B), QT-induced (D), and MT-induced (F) perturbations are presented by spheres on the mCCL2 dimer and are marked only on one subunit.
A comparative analysis of CSP histograms of the three flavonols suggests that they exhibited a similar set of perturbed residues with minimal variations. For the KP–mCCL2 complex, the perturbed residues comprise A7, T10, Y13, S14, T16, and I20 from the N-terminal; R24, L25, E26, and Y28 from β1-sheet; R35 from the second loop; K46 from the third loop; R49 and V51 from β3-sheet; and E58, N66, and R69 from the C-terminal α-helix (Figure 4A). All of these perturbed residues are marked on one of the monomeric subunits of mCCL2 and are highlighted in both subunits by spheres (Figure 4B). For the QT–mCCL2 complex, the perturbed residues detected in the CSP map include A7, T10, C12, Y13, S14, T16, K18, and I20 from the N-terminal; R24, L25, E26, S27, and Y28 from β1-sheet; R35 from the second loop; K46 from the third loop; R49 and V51 from β3-sheet; and E58, N66, and R69 from the C-terminal α-helix (Figure 4C,D). Similarly, for the MT–mCCL2 interaction, the set of perturbed residues consists of A7, T10, C12, Y13, S14, F25, T16, K18, and I20 from the N-terminal; R24, L25, E26, S27, and Y28 from β1-sheet; R35 from the second loop; K46 from the third loop; R49 and V51 from β3-sheet; and E58, N66, and R69 from the C-terminal α-helix (Figure 4E,F).
NMR-based results have suggested that all of the three flavonols (KP, QT, and MT) specifically interact with the mCCL2-WT protein, and the binding surface for these ligands includes the N-terminal, β1- and β3-sheets, the third loop, and the C-terminal α-helix of the protein. The CSP results suggest that all of the three chosen flavonols bind to the CCL2 protein on the same surface/pocket, as the structural elements involving the perturbed residues are noted to be the same. Moreover, it is worth noting that there exist considerable variations in the chemical shift perturbation values of CCL2 upon binding of these three flavonols, which can be attributed to their differential affinities of these flavonols due to the increase of −OH groups on its annular ring-B, as evidenced by the binding constants derived from fluorescence quenching experiments. Moreover, no significant intensity ratio changes (dimer/monomer) were noticed (data not shown) in the mCCL2 HSQC spectrum upon addition of flavonols, thus indicating that these molecules do not alter/interfere with its monomer–dimer equilibrium. This suggests that the CCL2–flavonol interactions and their dimerization contacts might be mutually exclusive. Such a mutual exclusiveness of the dimerization and flavonol binding contacts of CCL2 explains the observed trend of similar Kd values for both monomeric/dimeric variants of CCL2 upon binding of a chosen flavonol (KP/QT/MT), as obtained from fluorescence quenching experiments (Table 1).
2.3. Molecular Docking Analysis of CCL2–Flavonol Binding Interactions
Considering the residues obtained from the NMR-based CSP analysis, the molecular docking analysis was performed to unveil the atomic-level contacts between mCCL2 monomer and flavonols. As it was observed that the human and murine CCL2 partners consist of similar dissociation constant (Kd) values for CCL2–flavonol complexes, the docking experiments were also executed for the hCCL2 monomer using a similar binding surface or grid dimension, as observed for the murine counterpart. Further, all of the docked CCL2–flavonol complexes were subjected to energy minimization and analyzed for potential energy and Coulomb short-range (Coul-SR) electrostatics to assess the stability of the complexes (Table 2 and Figure S5). It is evident from the potential and average values of Coul-SR electrostatics that the obtained complexes were stable. At the residue level, for the KP–mCCL2/hCCL2 complexes, the binding residues include C11, Y13, S14/N14, and T16 from the N-terminal and E50 and C52 from β3-sheet (Figure 5A,B), and the observed binding energies were around −5.1 kcal/mol. Further, for the QT–mCCL2/hCCL2 complexes, the stretch of interacting residues comprises C11, Y13, S14/N14, and T16 from the N-terminal and E50, V51/I51, and C52 from β3-sheet (Figure 5C,D), the estimated binding energies were observed to be −5.3 kcal/mol. Similarly, for the MT–mCCL2/hCCL2 complexes, the residues involved in the interaction consist of C11, C12, Y13, S14/N14, F15, and T16 from the N-terminal and E50, V51/I51, and C52 from β3-sheet (Figure 5E,F), and the binding energies were observed to be −6.4 kcal/mol. All of the crucial hydrogen bond and hydrophobic interactions for KP/QT/MT–mCCL2/hCCL2 complexes have been summarized in Tables S1–S3. It has been observed that for all three flavonols (MT, QT, and KP), the binding residues majorly belong to the N-terminal and β1- and β3-sheets of CCL2 orthologs.
Table 2. Summary of the Residues Involved in the Flavonol–mCCL2/hCCL2 Monomer Complexes’ Interactions along with Their Kd Values and Binding Energy Parameters.
| protein | ligand | residue involved in docking | Kd values (μM) | binding energy (kcal/mol) | potential (kJ/mol) | avg. Coul-SR electrostatics (kJ/mol) |
|---|---|---|---|---|---|---|
| mCCL2 | KP | C11, Y13, S14, T16, E50, C52 | 26 ± 5 | –5.08 | –5.682 × 105 | –6.250 × 105 |
| hCCL2 | KP | C11, Y13, N14, T16, E50, C52 | 25 ± 5 | –5.10 | –5.285 × 105 | –5.817 × 105 |
| mCCL2 | QT | C11, Y13, S14, T16, E50, V51, C52 | 3.7 ± 0.5 | –5.33 | –5.609 × 105 | –6.170 × 105 |
| hCCL2 | QT | C11, Y13, N14, T16, E50, I51, C52 | 3.6 ± 0.5 | –5.28 | –5.362 × 105 | –5.901 × 105 |
| mCCL2 | MT | C11, C12, Y13, S14, F15, T16, E50, V51, C52 | 0.4 ± 0.02 | –6.41 | –5.612 × 105 | –6.174 × 105 |
| hCCL2 | MT | C11, C12, Y13, N14, F15, T16, E50, I51, C52 | 0.43 ± 0.02 | –6.39 | –5.326 × 105 | –5.858 × 105 |
Figure 5.
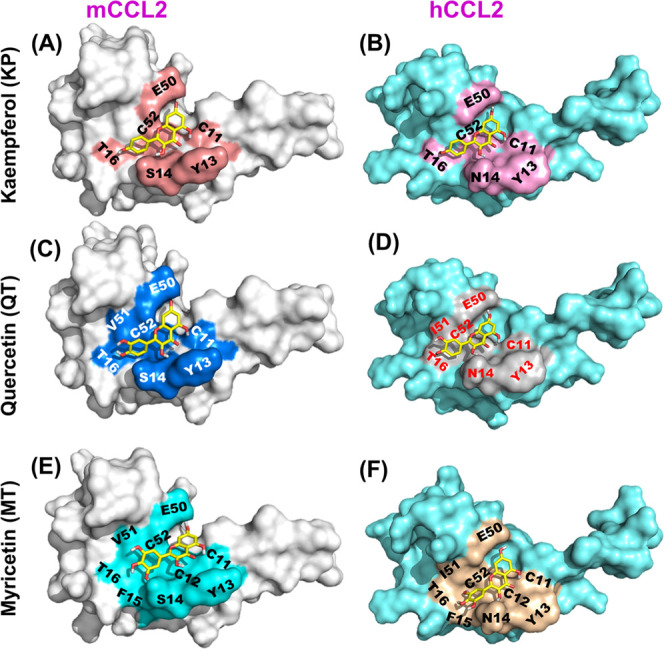
Docking profiles of flavonols onto the mCCL2/hCCL2 monomeric proteins. (A, B) mCCL2/hCCL2-monomeric surface structures depicting the KP binding sites are highlighted in salmon red and pink, respectively. (C, D) mCCL2/hCCL2-monomeric surface structures representing the QT binding sites are highlighted in marine blue and gray, respectively. (E, F) mCCL2/hCCL2-monomeric surface structures depicting the MT binding sites are highlighted in cyan and light olive, respectively.
3. Discussion
3.1. Hydroxyl Groups on Annular Ring-B Regulate the Binding Affinities of Flavonol–CCL2 Chemokine Interactions
The resilient appearance and consumption of plant-derived functional foods, dietary supplements, and nutraceuticals have blurred the discrepancy between nutrition and pharma and open the doors for seeking new therapeutic alternatives to prevent pernicious diseases.54 Flavonols are the most abundant and the largest subgroup of flavonoids with overwhelming therapeutic effects/benefits, hence gaining attention toward being essential components of pharmaceutical, cosmetic, medicinal, and nutraceutical applications.55 The intake of flavonols, especially kaempferol, quercetin, and myricetin, is broadly associated with various deleterious ailments like atherosclerosis,56 Alzheimer’s disease (AD),57 cancer,58 and cardiovascular59 and neurodegenerative disorders,60 where they mediate a wide range of health-promoting effects, including antioxidant and anti-inflammatory actions.61,62 Binding of flavonols with plasma proteins, DNA, RNA, and lipids has provided new insights into the mode of interactions and embolden the development of new therapeutic nutraceuticals.63,64
Indeed, flavonols exhibit different functional characteristics and structure–activity relationships (SARs) due to the presence of phenolic −OH groups at ring-B.65 In this regard, a myriad of epidemiological/biophysical studies has unveiled a strong correlation between chemical structures and biological activities of flavonols.66 Indeed, researchers established that the extent of hydroxylation and its positioning on ring-B influences the binding affinity of flavonols/flavonoids to proteins.67,68 For example, interaction of the canine distemper virus with flavonols unraveled that quercetin showed higher inhibitory activity than morin.67 The only structural difference between these two flavonols is the positioning of the hydroxyl groups on ring-B. On a similar note, the current study made an ardent attempt to unravel the effect of hydroxylation on the binding affinity of chosen flavonols for CCL2 orthologs in both dimeric and monomeric forms. The results indicated that all of the three flavonols (KP, QT, and MT) bound to the CCL2 chemokine (dimer/monomer) orthologs on a similar binding pocket. Although particular flavonols bind to the CCL2 chemokine variants with similar affinity, interestingly, a great degree of variations in their binding affinities was observed among the chosen flavonols toward CCL2 protein (Table 1). The results established that the number of hydroxyl groups on ring-B directly influenced the binding pattern. For the chosen compounds, kaempferol (KP) with only one OH group is present at the C13 position, quercetin (QT) comprises two successive OH groups that are present at C12 and C13 positions, and myricetin (MT) possess three consecutive OH groups that are situated at C12, C13, and C14 positions on ring-B (Figure 1A–C). The enhanced binding constants of the CCL2–flavonol complexes upon an increase in the number of −OH groups on annular ring-B of the flavonols are also evident from the enhanced number of molecular contacts upon complex formation in their energy-minimized docked complexes and their elevated CSP values. In line with the current results, previous studies on four flavonols’ (kaempferol, galangin, quercetin, and myricetin) interactions with bovine serum albumin protein have indicated that the hydroxylation on ring-B considerably upsurges their binding affinities.69 Further, the study has also demonstrated that the increase in the number of OH groups on ring-B consequently increases the binding affinity of flavonols. The observed binding constants (Ka) for BSA–flavonol interactions are as follows: myricetin (4.90 × 108 L/mol) > quercetin (3.65 × 107 L/mol) > kaempferol (2.57 × 106 L/mol) > galangin (6.43 × 105 L/mol). A significant difference of around 5–10-fold in the binding constants was observed with an increase in the single −OH group on ring-B of the flavonols.69 Consistent with these studies, the dissociation constants obtained in the current study for flavonol–CCL2 complexes were proportional to the number of OH groups on ring-B and are as follows: myricetin (Kd 0.4 μM) > quercetin (Kd 3.5 μM) > kaempferol (Kd 25 μM).
The differential affinities of these ligands to the CCL2 chemokine are also evident from the NMR-based CSP maps, where MT showed the largest perturbation of resonances and KP the least among the three flavonols (Figure 4A–F). The enhancement of the affinities was achieved by engaging the extra hydroxyl groups on the ring-B with the CCL2 protein. For the KP–CCL2 complex, the docking-based binding surface comprises only six residues, for the quercetin–CCL2 complex, the binding surface consists of seven residues, and for the myricetin–CCL2 complex, the involved residues are nine (Table 2). Remarkably, a closer look into the docking pose of flavonol–CCL2 complexes suggested that the increase in the number of OH groups on ring-B consequently enhanced the binding of flavonol to CCL2 residues on the binding surface, i.e., each OH group contributes toward the increase in the binding surface. For example, in the case of KP–mCCL2/hCCL2 complexes, the OH group at C4′ position on ring-B interacts with T16 of CCL2 (Figure 6A,B), while in QT, the OH groups at C3′ and C4′ positions engage with V51/I51 and T16 residues of CCL2 (Figure 6C,D). For MT, the OH groups at C3′, C4′, and C5′ positions interacted with V51/I51, T16, F15, and S14/N14 residues of mCCL2/hCCL2 (Figure 6E,F). Apart from these essential hydrogen-bonding contacts, several hydrophobic interactions were also observed to be involved in the interactions between flavonols (KP, QT, and MT) and CCL2 partners (Tables S1–S3). Similar to the observed flavonol–CCL2 complexes, the binding of flavones/flavonols with bovine lactoferrin protein (BLF) suggested that the binding affinities of these ligands were enhanced with an increase in the number of −OH groups present on ring-B, and the interactions were primarily governed by hydrogen bond and hydrophobic interactions.70 Hence, for flavonol–CCL2 complexes, a significant reduction in Kd values and a decrease in the number of binding site residues further corroborate the fact that hydroxylation on the annular ring-B directs the binding affinities of flavonols for CCL2 orthologs. Further, an overlay of these three flavonols (KP, QT, MT) on the binding surface of mCCL2/hCCL2 evidenced that, the flavonol family uses the single bond rotation between the rings (A, C) and B to orient the annular ring-B in a way so as to maximize the interactions between the −OH groups and the residues of CCL2 to achieve strong affinities upon an increase of the hydroxyl groups (Figure 6G,H). Essentially, the residues on the N-terminal (A7, T10, Y13, S14, T16, and I20) and β3-sheet (R49 and V51) of the CCL2 protein clamps the A,C rings of flavonols so that the rotation of annular ring-B across the single bond could become a preferable choice to maximize its binding contacts.
Figure 6.
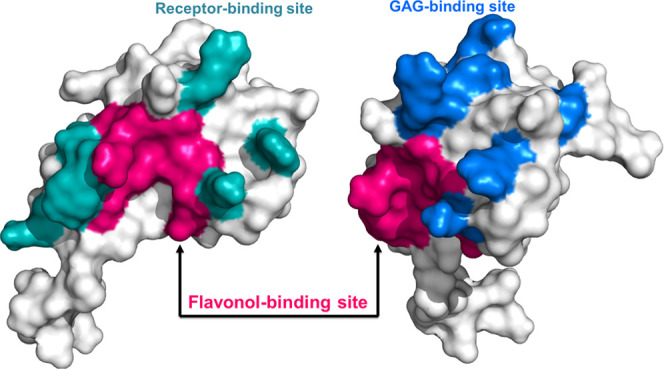
Unraveling the cumulative effect of −OH groups of the ring-B on the binding affinity of the CCL2 protein. (A–F) Representative hydrogen bond patterns (H-bonds) between mCCL2 and hCCL2 complexed with kaempferol (KP), quercetin (QT), and myricetin (MT), respectively. All H-bonds are encircled by a dotted black line, and the proximal bond length is shown in Å. (G, H) Overlay of three ligands, namely, kaempferol (yellow/green), quercetin (deep teal/lime), and myricetin (light orange/olive), docked onto mCCL2/hCCL2. The representative monomeric surface structures of hCCL2 showing receptor-binding (I) and GAG-binding (J) sites. The crucial binding site for flavonol (MT) is highlighted in dark pink.
The binding of all of the chosen flavonols with monomers and dimers of CCL2 orthologs was observed to be specific, as evinced by the perturbation of a subset of NH resonances (Figure 3A–F), where the binding domain comprises a set of residues from the N-terminal and β1- and β3-sheets of CCL2. It is extensively reported that the N-terminal end along with the β1- and β3-sheets of the CCL2 protein are the crucial sites for its receptor CCR2 interaction (Figure 6I).71 Indeed, several of these residues were also involved in the GAG binding to CCL2 (Figure 6J).72 Interestingly, significant concurrency in the binding pockets was observed between the flavonol binding surfaces and that of receptor/GAG-binding surfaces (Figure 6I,J). In vitro and structural studies on CCL2 family chemokines using the flavonoid glycoside baicalin evidenced that it uses the binding surface comprising the N-terminal and β1- and β3-sheets and significantly attenuated the in vitro binding features of chemokine–receptor interactions.49,50 As all of the three flavonols bind in the same pocket, it can be presumed that they do potentially interfere with the receptor/GAG interactions of CCL2 to modulate their activities. On similar lines, several small inhibitor molecules have been identified to block the chemokine–receptor/GAG (CCL2-CCR2/GAG) pivot, and this has become the most promising approach to develop them as potential therapeutic agents.73−75 For example, CCL2 binding mirror-image aptamers NOX-E36 (human) and mNOX-E36 (murine), also known as Spiegelmers, contain L-ribonucleotides. Both the NOX-E36 aptamers interact with CCL2 through the GAG and the receptor-binding surface and hinder the chemotactic activity of CCL2 chemokine.76,77 Similarly, interaction studies of CCR2 with various structurally different antagonists from cyclohexane and piperidine families have unveiled that these ligands interact with the CCR2 receptor with nanomolar affinity and abrogated the CCR2-mediated functioning/signaling cascade.78 As flavonoids/flavonols are nutraceutical targets for immune modulation, the current study unraveled a new class of natural molecules to target the chemokine–receptor/GAG axis for the regulation of leukocyte recruitment and also provided comprehensive structural insights into how flavonols interact with CCL2 chemokine.
4. Concluding Remarks
The present study offered a comparative and comprehensive piece of information on the molecular interactions of flavonols with CCL2 chemokine orthologs. The binding studies revealed considerable differences in apparent dissociation constants of flavonol members KP, QT, and MT toward CCL2 variants, thus suggesting their differential binding affinity. The differences in the binding interactions/affinities can be directly correlated to the number of hydroxyl groups on the annular ring-B of the flavonols. An increase in the number of −OH groups from 1 to 3 enhanced their binding affinities toward CCL2 chemokine by ∼50 times. A significant overlap between the binding surfaces of flavonols and the receptor/GAG-binding surface has been observed, suggesting plausible attenuation of chemokine-mediated leukocyte trafficking in their presence. Indeed, comprehensive cell-based studies and animal models need to be further explored to elucidate their cellular response. The structural insights obtained here on CCL2–flavonol interactions provide an inherent advantage of using flavonoids as nutraceuticals, which can be amalgamated in the field of pharmacokinetics and food sciences. In a nutshell, the inferences drawn from the current study can content the cradle of the search for new therapeutic nutraceutical agents that can act as immunomodulatory agents to regulate chemokine-mediated leukocyte trafficking.
5. Material and Methods
5.1. Protein Expression and Purification
The wild-type (dimer) and monomeric proteins of murine and human CCL2 orthologs were transformed, overexpressed, and purified in E. coli BL21(DE3) cells, as described elsewhere.42,50
5.2. Stock Solutions of Flavonols
Flavonols kaempferol (PubChem SID 57651711), quercetin (PubChem SID 329823865), and myricetin (PubChem SID 57652239) of purity 99% were purchased from Sigma-Aldrich. A stock solution of 20 mM concentration was prepared by suspending them in a 100% DMSO solution.
5.3. Flavonol Quenching Experiments
All of the fluorescence titration experiments were performed using a Fluorolog spectrophotometer at 25 °C and a slit width of 5 nm for excitation and emission. Titration experiments were acquired at a steady concentration (50 μM) of CCL2 protein(s) with increasing concentrations of flavonols (KP, QT, and MT). Trp (W59) of CCL2 proteins was excited at 295 nm and the emission spectra were acquired from 300 to 450 nm. All of the titration experiments were performed in triplicate to authenticate the data. The quenching parameters for flavonol–CCL2 complexes were determined using the following standard Stern–Volmer equation.
| 1 |
F0 and F represent the fluorescence intensities before and after the addition of the quencher, respectively. [Q] and Ksv indicate the quencher concentration and the Stern–Volmer quenching constant, respectively.52,79Kq and τ0 are the rate constant and the fluorophore lifetime without the quencher, respectively.79 The association between the apparent binding constant (Ka) and the number of binding sites (n) was deciphered by a double-logarithmic plot using eq 2 and the nonlinear regression and Scatchard plot analysis as described elsewhere.80
| 2 |
5.4. Nuclear Magnetic Resonance Spectroscopy
5.4.1. Sample Preparation and NMR Titrations
For NMR-based titrations, the 15N-labeled sample of mCCL2-WT protein was prepared in 50 mM Na2HPO4 and 50 mM NaCl buffer at pH 6.0. All of the flavonol–mCCL2 HSQC titrations were acquired at a protein concentration of 100 μM on a Bruker spectrometer (500 MHz) at 25 °C. The titrant (flavonol) was added with an increasing concentration up to a molar ratio (P/L) of 1:5. For all of the three flavonol–mCCL2 complexes, the variation in the chemical shift values was estimated using the following chemical shift perturbation (CSP) equation
| 3 |
ΔδH represents the variation in the chemical shift values of 1H, while ΔδN represents the variation in the chemical shift values of 15N.
5.5. Molecular Docking
Molecular docking interactions between CCL2–flavonol complexes were obtained using the Auto dock 4.2 tool. All of the three flavonols, kaempferol (IUPAC name: 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chromen-4-one), quercetin (IUPAC name: 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxychromen-4-one), and myricetin (IUPAC name: 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromen-4-one) (Figure 1A–C), were docked onto monomers of the human and murine CCL2 orthologs. For human CCL2, 1dok PDB id was used, while for mCCL2, a previously determined NMR structure was used.42 For docking, the hybrid genetic algorithm (Lamarckian) program was used, and the Gasteiger and Kollman charges were detected and distributed to CCL2 protein(s). The grid box was outlined onto the mCCL2/hCCL2 protein surface according to the CSP outcomes and was chosen at 70 × 66 × 60 dimension with a 0.37 Å spacing parameter. The stability of docked complexes was analyzed by performing energy minimization using CHARMM36 forcefield in Gromacs 2020.1.81,82 Ligand topologies were generated using the CGENFF server.83 The protein–ligand complex was solvated in a cubic box using the TIP3P water model, and the system was neutralized using chloride ions. Energy minimization was performed with the steepest descent algorithm with the cutoff set to 5000 steps. The gmx energy module was further used to analyze potential energy and Coulomb short-range (Coul-SR) electrostatics of energy-minimized CCL2–flavonoid complexes. Docking results were analyzed using LigPlot+ and PyMol.
Acknowledgments
K.M.P. acknowledges the receipt of Grants CRG/2018/001329 and SERB-SB/YS/LS-380/2013 from SERB-DST and DBT-IYBA fellowship, BT/07/IYBA/2013-19. N.J. acknowledges the receipt of JRF and SRF fellowships from UGC. The authors acknowledge the support of biophysical and 500 MHz NMR instrumentation facilities at Institute Instrumentation Centre, IIT-Roorkee, India.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00655.
Detailed summary of the all possible bonds for flavonol–mCCL2/hCCL2 complexes (Tables S1–S3); fluorescence quenching patterns of mCCL2-WT, mCCL2-P8A, hCCL2-WT, and hCCL2-P8A proteins in the presence of buffer devoid of flavonols (Figure S1); description of the fluorescence quenching patterns and double-log plots of flavonol–mCCL2/hCCL2 complexes (Figures S2 and S3); nonlinear regression and Scatchard plot analysis of mCCL2-P8A interaction with flavonol myricetin (MT) (Figure S4); and graphical description of Coulomb short-range (Coul-SR) electrostatics of flavonol–mCCL2/hCCL2 monomer complexes (Figure S5) (PDF)
Author Contributions
K.M.P. designed the project. N.J., D.K.T., and N.N. performed the experiments. N.J., D.K.T., N.N., and K.M.P. analyzed the data. N.J. and K.M.P. wrote the manuscript. All authors reviewed and accepted the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hosseinzadeh S.; Jafarikukhdan A.; Hosseini A.; Armand R. The application of medicinal plants in traditional and modern medicine: a review of Thymus vulgaris. Int. J. Clin. Med. 2015, 6, 635. 10.4236/ijcm.2015.69084. [DOI] [Google Scholar]
- Yuan H.; Ma Q.; Ye L.; Piao G. The traditional medicine and modern medicine from natural products. Molecules 2016, 21, 559. 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell M.; Rahman P. Medicinal plants: their use in anticancer treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Pandey A. K. Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013, 2013, 1–16. 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. 10.3389/fphar.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S.; Pal A.; Si S. C. Flavonoid as Nutraceuticals: A Therapeutic approach to Rheumatoid Arthritis. Res. J. Pharm. Technol. 2020, 13, 991–998. 10.5958/0974-360X.2020.00184.5. [DOI] [Google Scholar]
- Panche A.; Diwan A.; Chandra S. Flavonoids: an overview. J. Nutr. Sci. 2016, 5, e47 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Zhao X.; Qiu H. Flavones and flavonols: phytochemistry and biochemistry. Nat. Prod. 2013, 1821–1847. [Google Scholar]
- Cheong H.; Ryu S.-Y.; Oak M.-H.; Cheon S.-H.; Yoo G.-S.; Kim K.-M. Studies of structure activity relationship of flavonoids for the anti-allergic actions. Arch. Pharm. Res. 1998, 21, 478–480. 10.1007/BF02974647. [DOI] [PubMed] [Google Scholar]
- Kawai M.; Hirano T.; Higa S.; Arimitsu J.; Maruta M.; Kuwahara Y.; Ohkawara T.; Hagihara K.; Yamadori T.; Shima Y.; et al. Flavonoids and related compounds as anti-allergic substances. Allergol. Int. 2007, 56, 113–123. 10.2332/allergolint.R-06-135. [DOI] [PubMed] [Google Scholar]
- Kanadaswami C.; Lee L.-T.; Lee P.-P. H.; Hwang J.-J.; Ke F.-C.; Huang Y.-T.; Lee M.-T. The antitumor activities of flavonoids. In Vivo 2005, 19, 895–909. [PubMed] [Google Scholar]
- Kopustinskiene D. M.; Jakstas V.; Savickas A.; Bernatoniene J. Flavonoids as anticancer agents. Nutrients 2020, 12, 457. 10.3390/nu12020457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankari S. L.; Babu N. A.; Rani V.; Priyadharsini C.; Masthan K. Flavonoids–Clinical effects and applications in dentistry: A review. J. Pharm. BioAllied Sci. 2014, 6, S26. 10.4103/0975-7406.137256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irina I.; Mohamed G. Biological activities and effects of food processing on flavonoids as phenolic antioxidants. Adv. Appl. Microbiol. 2012, 101–124. [Google Scholar]
- Wang H.; Du Y.-J.; Song H.-C. α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 2010, 123, 6–13. 10.1016/j.foodchem.2010.03.088. [DOI] [Google Scholar]
- Li Y.; Ding Y. Minireview: Therapeutic potential of myricetin in diabetes mellitus. Food Sci. Hum. Wellness 2012, 1, 19–25. 10.1016/j.fshw.2012.08.002. [DOI] [Google Scholar]
- Chu Y. H.; Chang C. L.; Hsu H. F. Flavonoid content of several vegetables and their antioxidant activity. J. Sci. Food Agric. 2000, 80, 561–566. . [DOI] [Google Scholar]
- Agrawal A. Pharmacological activities of flavonoids: a review. Int. J. Pharm. Sci. Nanotechnol. 2011, 4, 1394–1398. 10.37285/ijpsn.2011.4.2.3. [DOI] [Google Scholar]
- Farhadi F.; Khameneh B.; Iranshahi M.; Iranshahy M. Antibacterial activity of flavonoids and their structure–activity relationship: An update review. Phytother. Res. 2019, 33, 13–40. 10.1002/ptr.6208. [DOI] [PubMed] [Google Scholar]
- Muchtaridi M.; Fauzi M.; Khairul Ikram N. K.; Mohd Gazzali A.; Wahab H. A. Natural Flavonoids as Potential Angiotensin-Converting Enzyme 2 Inhibitors for Anti-SARS-CoV-2. Molecules 2020, 25, 3980. 10.3390/molecules25173980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Islam M. S.; Wang J.; Li Y.; Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int. J. Biol. Sci. 2020, 16, 1708. 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey J. A. Biological properties of flavonoids pertaining to inflammation. Microcirculation 2000, 7, S29–S34. 10.1111/j.1549-8719.2000.tb00148.x. [DOI] [PubMed] [Google Scholar]
- Leyva-López N.; Gutierrez-Grijalva E. P.; Ambriz-Perez D. L.; Heredia J. B. Flavonoids as cytokine modulators: a possible therapy for inflammation-related diseases. Int. J. Mol. Sci. 2016, 17, 921. 10.3390/ijms17060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adefegha S. A. Functional foods and nutraceuticals as dietary intervention in chronic diseases; novel perspectives for health promotion and disease prevention. J. Diet. Suppl. 2018, 15, 977–1009. 10.1080/19390211.2017.1401573. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Kumar S.; Kumar V.; Thakur A. Comprehensive review on nutraceutical significance of phytochemicals as functional food ingredients for human health management. Int. J. Pharmacogn. Phytochem. 2019, 8, 385–395. 10.22271/phyto.2019.v8.i5h.9589. [DOI] [Google Scholar]
- Poluri K. Chemokines: the holy messengers of leukocyte trafficking. Austin J. Biotechnol. Bioeng. 2014, 1, 1–3. [Google Scholar]
- Sokol C. L.; Luster A. D. The chemokine system in innate immunity. Cold Spring Harbor Perspect. Biol. 2015, 7, a016303 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins B. J. Chemokines. Blood 1997, 90, 909–928. 10.1182/blood.V90.3.909. [DOI] [PubMed] [Google Scholar]
- Tripathi D. K.; Poluri K. M. Molecular insights into kinase mediated signaling pathways of chemokines and their cognate G protein coupled receptors. Front. Biosci. 2020, 25, 1361–1385. 10.2741/4860. [DOI] [PubMed] [Google Scholar]
- Gulati K.; Poluri K. M. Mechanistic and therapeutic overview of glycosaminoglycans: the unsung heroes of biomolecular signaling. Glycoconjugate J. 2016, 33, 1–17. 10.1007/s10719-015-9642-2. [DOI] [PubMed] [Google Scholar]
- Deshmane S. L.; Kremlev S.; Amini S.; Sawaya B. E. Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interferon Cytokine Res. 2009, 29, 313–326. 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Coillie E.; Van Damme J.; Opdenakker G. The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev. 1999, 10, 61–86. 10.1016/S1359-6101(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Sørensen T. L.; Ransohoff R.; Strieter R.; Sellebjerg F. Chemokine CCL2 and chemokine receptor CCR2 in early active multiple sclerosis. Eur. J. Neurol. 2004, 11, 445–449. 10.1111/j.1468-1331.2004.00796.x. [DOI] [PubMed] [Google Scholar]
- Trauzold A.; Siegmund D.; Schniewind B.; Sipos B.; Egberts J.; Zorenkov D.; Emme D.; Röder C.; Kalthoff H.; Wajant H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene 2006, 25, 7434–7439. 10.1038/sj.onc.1209719. [DOI] [PubMed] [Google Scholar]
- Wang J.; Ou Z.; Hou Y.; Luo J.; Shen Z.; Ding J.; Shao Z. Enhanced expression of Duffy antigen receptor for chemokines by breast cancer cells attenuates growth and metastasis potential. Oncogene 2006, 25, 7201–7211. 10.1038/sj.onc.1209703. [DOI] [PubMed] [Google Scholar]
- Cinque P.; Vago L.; Mengozzi M.; Torri V.; Ceresa D.; Vicenzi E.; Transidico P.; Vagani A.; Sozzani S.; Mantovani A.; et al. Elevated cerebrospinal fluid levels of monocyte chemotactic protein-1 correlate with HIV-1 encephalitis and local viral replication. AIDS 1998, 12, 1327–1332. 10.1097/00002030-199811000-00014. [DOI] [PubMed] [Google Scholar]
- Sartipy P.; Loskutoff D. J. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 7265–7270. 10.1073/pnas.1133870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Villanueva P. O.; Ruiz-Morales J. A.; Song C.-H.; Flores L. M.; Jo E.-K.; Montaño M.; Barnes P. F.; Selman M.; Granados J. A functional promoter polymorphism in monocyte chemoattractant protein–1 is associated with increased susceptibility to pulmonary tuberculosis. J. Exp. Med. 2005, 202, 1649–1658. 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boring L.; Gosling J.; Cleary M.; Charo I. F. Decreased lesion formation in CCR2–/– mice reveals a role for chemokines in the initiation of atherosclerosis. Nature 1998, 394, 894–897. 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- Cipollone F.; Marini M.; Fazia M.; Pini B.; Iezzi A.; Reale M.; Paloscia L.; Materazzo G.; D’Annunzio E.; Conti P.; et al. Elevated circulating levels of monocyte chemoattractant protein-1 in patients with restenosis after coronary angioplasty. Arterioscler., Thromb., Vasc. Biol. 2001, 21, 327–334. 10.1161/01.ATV.21.3.327. [DOI] [PubMed] [Google Scholar]
- Handel T. M.; Domaille P. J. Heteronuclear (1H, 13C, 15N) NMR assignments and solution structure of the monocyte chemoattractant protein-1 (MCP-1) dimer. Biochemistry 1996, 35, 6569–6584. 10.1021/bi9602270. [DOI] [PubMed] [Google Scholar]
- Joshi N.; Nagar N.; Gulati K.; Gangele K.; Mishra A.; Kumar D.; Poluri K. M. Dissecting the differential structural and dynamics features of CCL2 chemokine orthologs. Int. J. Biol. Macromol. 2020, 156, 239–251. 10.1016/j.ijbiomac.2020.04.067. [DOI] [PubMed] [Google Scholar]
- Lubkowski J.; Bujacz G.; Boqué L.; Alexander W.; et al. The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nat. Struct. Mol. Biol. 1997, 4, 64–69. 10.1038/nsb0197-64. [DOI] [PubMed] [Google Scholar]
- Paavola C. D.; Hemmerich S.; Grunberger D.; Polsky I.; Bloom A.; Freedman R.; Mulkins M.; Bhakta S.; McCarley D.; Wiesent L.; et al. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 1998, 273, 33157–33165. 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- Bartoli C.; Civatte M.; Pellissier J.; Figarella-Branger D. CCR2A and CCR2B, the two isoforms of the monocyte chemoattractant protein-1 receptor are up-regulated and expressed by different cell subsets in idiopathic inflammatory myopathies. Acta Neuropathol. 2001, 102, 385–392. 10.1007/s004010100394. [DOI] [PubMed] [Google Scholar]
- Crown S. E.; Yu Y.; Sweeney M. D.; Leary J. A.; Handel T. M. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J. Biol. Chem. 2006, 281, 25438–25446. 10.1074/jbc.M601518200. [DOI] [PubMed] [Google Scholar]
- Jacquelot N.; Duong C. P.; Belz G. T.; Zitvogel L. Targeting chemokines and chemokine receptors in melanoma and other cancers. Front. Immunol. 2018, 9, 2480. 10.3389/fimmu.2018.02480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crijns H.; Vanheule V.; Proost P. Targeting Chemokine—Glycosaminoglycan Interactions to Inhibit Inflammation. Front. Immunol. 2020, 11, 483 10.3389/fimmu.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. Q.; Fu T.; Gong W.-H.; Dunlop N.; Kung H.-f.; Yan Y.; Kang J.; Wang J. M. The flavonoid baicalin exhibits anti-inflammatory activity by binding to chemokines. Immunopharmacology 2000, 49, 295–306. 10.1016/S0162-3109(00)00244-7. [DOI] [PubMed] [Google Scholar]
- Joshi N.; Kumar D.; Poluri K. M. Elucidating the Molecular Interactions of Chemokine CCL2 Orthologs with Flavonoid Baicalin. ACS Omega 2020, 5, 22637–22651. 10.1021/acsomega.0c03428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.; Dan-Dan D.; Zhi-Juan C.; Qiong X.; LIANG J.-Y.; Jian-Zhong L. Interaction between serum albumin and four flavones by fluorescence spectroscopy and molecular docking. Chin. J. Anal. Chem. 2010, 38, 483–487. 10.1016/S1872-2040(09)60034-7. [DOI] [Google Scholar]
- Xiao J. B.; Chen J. W.; Cao H.; Ren F. L.; Yang C. S.; Chen Y.; Xu M. Study of the interaction between baicalin and bovine serum albumin by multi-spectroscopic method. J. Photochem. Photobiol., A 2007, 191, 222–227. 10.1016/j.jphotochem.2007.04.027. [DOI] [Google Scholar]
- Mureddu L.; Vuister G. W. Simple high-resolution NMR spectroscopy as a tool in molecular biology. FEBS J. 2019, 286, 2035–2042. 10.1111/febs.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul K.; Singh A.; Jabeen R. Nutraceuticals and functional foods: The foods for the future world. Crit. Rev. Food Sci. Nutr. 2016, 56, 2617–2627. 10.1080/10408398.2014.903384. [DOI] [PubMed] [Google Scholar]
- Sultana B.; Anwar F. Flavonols (kaempeferol, quercetin, myricetin) contents of selected fruits, vegetables and medicinal plants. Food Chem. 2008, 108, 879–884. 10.1016/j.foodchem.2007.11.053. [DOI] [PubMed] [Google Scholar]
- Phie J.; Krishna S. M.; Moxon J. V.; Omer S. M.; Kinobe R.; Golledge J. Flavonols reduce aortic atherosclerosis lesion area in apolipoprotein E deficient mice: A systematic review and meta-analysis. PLoS One 2017, 12, e0181832 10.1371/journal.pone.0181832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K.; Yoshiike Y.; Takashima A.; Hasegawa K.; Naiki H.; Yamada M. Potent anti-amyloidogenic and fibril-destabilizing effects of polyphenols in vitro: implications for the prevention and therapeutics of Alzheimer’s disease. J. Neurochem. 2003, 87, 172–181. 10.1046/j.1471-4159.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Majumder D.; Das A.; Saha C. Catalase inhibition an anti cancer property of flavonoids: A kinetic and structural evaluation. Int. J. Biol. Macromol. 2017, 104, 929–935. 10.1016/j.ijbiomac.2017.06.100. [DOI] [PubMed] [Google Scholar]
- Vinson J. A.; Dabbagh Y. A.; Serry M. M.; Jang J. Plant flavonoids, especially tea flavonols, are powerful antioxidants using an in vitro oxidation model for heart disease. J. Agric. Food Chem. 1995, 43, 2800–2802. 10.1021/jf00059a005. [DOI] [Google Scholar]
- Naoi M.; Shamoto-Nagai M.; Maruyama W. Neuroprotection of multifunctional phytochemicals as novel therapeutic strategy for neurodegenerative disorders: Antiapoptotic and antiamyloidogenic activities by modulation of cellular signal pathways. Future Neurol. 2019, 14, FNL9. 10.2217/fnl-2018-0028. [DOI] [Google Scholar]
- Wang L.; Tu Y.-C.; Lian T.-W.; Hung J.-T.; Yen J.-H.; Wu M.-J. Distinctive antioxidant and antiinflammatory effects of flavonols. J. Agric. Food Chem. 2006, 54, 9798–9804. 10.1021/jf0620719. [DOI] [PubMed] [Google Scholar]
- Pan M.-H.; Lai C.-S.; Ho C.-T. Anti-inflammatory activity of natural dietary flavonoids. Food Funct. 2010, 1, 15–31. 10.1039/c0fo00103a. [DOI] [PubMed] [Google Scholar]
- Xiao J.; Kai G. A review of dietary polyphenol-plasma protein interactions: characterization, influence on the bioactivity, and structure-affinity relationship. Crit. Rev. Food Sci. Nutr. 2012, 52, 85–101. 10.1080/10408398.2010.499017. [DOI] [PubMed] [Google Scholar]
- Jakobek L. Interactions of polyphenols with carbohydrates, lipids and proteins. Food Chem. 2015, 175, 556–567. 10.1016/j.foodchem.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S.; Chakraborty S.; Chorell E.; Sengupta P. K.; Bhowmik S. Importance of the hydroxyl substituents in the B–ring of plant flavonols on their preferential binding interactions with VEGF G–quadruplex DNA: Multi-spectroscopic and molecular modeling studies. Int. J. Biol. Macromol. 2018, 118, 629–639. 10.1016/j.ijbiomac.2018.06.115. [DOI] [PubMed] [Google Scholar]
- Wang T.-y.; Li Q.; Bi K.-s. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. 10.1016/j.ajps.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho O.; Botelho C.; Ferreira C.; Ferreira H.; Santos M.; Diaz M.; Oliveira T.; Soares-Martins J.; Almeida M.; Junior A. S. In vitro inhibition of canine distemper virus by flavonoids and phenolic acids: implications of structural differences for antiviral design. Res. Vet. Sci. 2013, 95, 717–724. 10.1016/j.rvsc.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Kim Y. S.; Ryu Y. B.; Curtis-Long M. J.; Yuk H. J.; Cho J. K.; Kim J. Y.; Kim K. D.; Lee W. S.; Park K. H. Flavanones and rotenoids from the roots of Amorpha fruticosa L. that inhibit bacterial neuraminidase. Food Chem. Toxicol. 2011, 49, 1849–1856. 10.1016/j.fct.2011.04.038. [DOI] [PubMed] [Google Scholar]
- Xiao J.; Suzuki M.; Jiang X.; Chen X.; Yamamoto K.; Ren F.; Xu M. Influence of B-ring hydroxylation on interactions of flavonols with bovine serum albumin. J. Agric. Food Chem. 2008, 56, 2350–2356. 10.1021/jf7037295. [DOI] [PubMed] [Google Scholar]
- Huang J.; Liu Z.; Ma Q.; He Z.; Niu Z.; Zhang M.; Pan L.; Qu X.; Yu J.; Niu B. Studies on the Interaction between Three Small Flavonoid Molecules and Bovine Lactoferrin. BioMed Res. Int. 2018, 2018, 1–10. 10.1155/2018/7523165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich S.; Paavola C.; Bloom A.; Bhakta S.; Freedman R.; Grunberger D.; Krstenansky J.; Lee S.; McCarley D.; Mulkins M.; et al. Identification of residues in the monocyte chemotactic protein-1 that contact the MCP-1 receptor, CCR2. Biochemistry 1999, 38, 13013–13025. 10.1021/bi991029m. [DOI] [PubMed] [Google Scholar]
- Lau E. K.; Paavola C. D.; Johnson Z.; Gaudry J.-P.; Geretti E.; Borlat F.; Kungl A. J.; Proudfoot A. E.; Handel T. M. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1 implications for structure and function in vivo. J. Biol. Chem. 2004, 279, 22294–22305. 10.1074/jbc.M311224200. [DOI] [PubMed] [Google Scholar]
- Zweemer A. J.; Nederpelt I.; Vrieling H.; Hafith S.; Doornbos M. L.; de Vries H.; Abt J.; Gross R.; Stamos D.; Saunders J.; et al. Multiple binding sites for small-molecule antagonists at the CC chemokine receptor 2. Mol. Pharmacol. 2013, 84, 551–561. 10.1124/mol.113.086850. [DOI] [PubMed] [Google Scholar]
- Gulati K.; Gangele K.; Kumar D.; Poluri K. M. An inter-switch between hydrophobic and charged amino acids generated druggable small molecule binding pocket in chemokine paralog CXCL3. Arch. Biochem. Biophys. 2019, 662, 121–128. 10.1016/j.abb.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Zweemer A. J.; Bunnik J.; Veenhuizen M.; Miraglia F.; Lenselink E. B.; Vilums M.; de Vries H.; Gibert A.; Thiele S.; Rosenkilde M. M.; et al. Discovery and mapping of an intracellular antagonist binding site at the chemokine receptor CCR2. Mol. Pharmacol. 2014, 86, 358–368. 10.1124/mol.114.093328. [DOI] [PubMed] [Google Scholar]
- Oberthür D.; Achenbach J.; Gabdulkhakov A.; Buchner K.; Maasch C.; Falke S.; Rehders D.; Klussmann S.; Betzel C. Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2. Nat. Commun. 2015, 6, 6923 10.1038/ncomms7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninichuk V.; Clauss S.; Kulkarni O.; Schmid H.; Segerer S.; Radomska E.; Eulberg D.; Buchner K.; Selve N.; Klussmann S.; et al. Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36–3′ PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am. J. Pathol. 2008, 172, 628–637. 10.2353/ajpath.2008.070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherney R. J.; Brogan J. B.; Mo R.; Lo Y. C.; Yang G.; Miller P. B.; Scherle P. A.; Molino B. F.; Carter P. H.; Decicco C. P. Discovery of trisubstituted cyclohexanes as potent CC chemokine receptor 2 (CCR2) antagonists. Bioorg. Med. Chem. Lett. 2009, 19, 597–601. 10.1016/j.bmcl.2008.12.062. [DOI] [PubMed] [Google Scholar]
- Qian X.; Dan-Dan D.; Zhi-Juan C.; Qiong X.; LIANG J.-Y.; Jian-Zhong L. Interaction between serum albumin and four flavones by fluorescence spectroscopy and molecular docking. Chin. J. Anal. Chem. 2010, 38, 483–487. 10.1016/S1872-2040(09)60034-7. [DOI] [Google Scholar]
- Montero M. T.; Hernández J.; Estelrich J. Fluorescence quenching of albumin. A spectrofluorimetric experiment. Biochem. Educ. 1990, 18, 99–101. 10.1016/0307-4412(90)90188-T. [DOI] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; De Groot B. L.; Grubmüller H.; MacKerell A. D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.