Abstract
Human respiratory syncytial virus (HRSV) is the leading viral cause of serious pediatric respiratory disease, and lifelong reinfections are common. Its 2 major subgroups, A and B, exhibit some antigenic variability, enabling HRSV to circulate annually. Globally, research has increased the number of HRSV genomic sequences available. To ensure accurate molecular epidemiology analyses, we propose a uniform nomenclature for HRSV-positive samples and isolates, and HRSV sequences, namely: HRSV/subgroup identifier/geographic identifier/unique sequence identifier/year of sampling. We also propose a template for submitting associated metadata. Universal nomenclature would help researchers retrieve and analyze sequence data to better understand the evolution of this virus.
Keywords: HRSV, Human orthopneumovirus, human respiratory syncytial virus, isolates, nomenclature, respiratory infections, specimens, strains, subspecies nomenclature, viruses
Human respiratory syncytial virus (HRSV) is the leading cause of severe respiratory illness in children <5 years of age and is associated with substantial illness from lower respiratory tract infections in industrialized countries and substantial illness and death in low- and middle-income countries (1–5). HRSV also causes severe disease among elderly and high-risk adults (6).
In 2016, HRSV was reclassified by the International Committee on Virus Taxonomy (ICTV) into a new family, Pneumoviridae, genus, Orthopneumovirus, and species, Human orthopneumovirus. (7). The wider availability of viral sequencing technologies has increased submissions of HRSV sequences to databases (Figure 1), a trend we anticipate will continue. Although ICTV provides nomenclature standards for virus taxa, there is currently no standardized format for HRSV nomenclature below the species level. Given the current interest in both HRSV and database submissions, a standard nomenclature is needed to simplify studies of the genomic diversity of HRSV strains and variants below the species level. ICTV’s taxonomic reassignment provides us a timely opportunity to propose a universal naming convention for HRSV strains, sequences, and isolates, including a framework for database submissions that are rich in contextual information and associated metadata.
Figure 1.
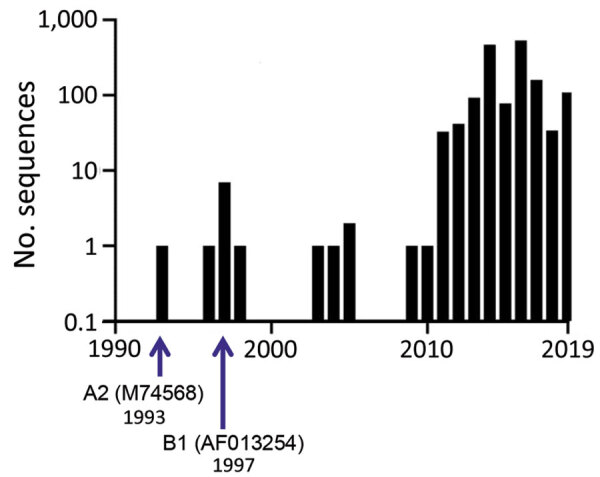
Annual numbers of HRSV whole-genome sequences released in GenBank since publication of the whole-genome sequence of HRSV A2, M74568, in 1993. HRSV, human respiratory syncytial virus.
Several large laboratory HRSV surveillance and epidemiology studies are currently in progress. These studies include the World Health Organization’s Global Respiratory Syncytial Virus (WHO RSV) Surveillance Project (https://www.who.int/influenza/rsv), which conducts large-scale testing for HRSV and extensive sequencing of HRSV-positive clinical specimens from >20 countries worldwide. Focused molecular analyses have helped elucidate HRSV household (8) and local (9) transmission dynamics and may guide development of strategies for the control of HRSV transmission. For example, molecular analysis showed that HRSV in healthcare facilities can be acquired from sources within the facility or introduced from the community (10,11).
In temperate climates, annual HRSV epidemics usually occur in winter months; it remains to be seen how social distancing measures and nonpharmaceutical interventions due to the current coronavirus disease (COVID-19) pandemic will affect global HRSV circulation patterns. One of the 2 major genetic and antigenic HRSV subgroups, A or B, usually predominates in alternating years, but both subgroups can also co-circulate in the same season. Early research has shown that subgroup A HRSV is associated with slightly greater clinical severity than subgroup B (12). Disease severity has been correlated with specific strains, genotypes, or clades, but to date, no consistent association has been established between strains (13–15), genotypes, or clades (16–19) and virulence. Thus, a possible role of different HRSV strains in disease severity remains to be elucidated. The lack of standard nomenclature and the scarcity of rich metadata in databases currently limit and complicate such studies.
Reliable and concise nomenclature systems below the species level are available for measles virus, influenza virus, rotavirus, filovirus isolates (20–23), and many other human viral pathogens. A similar nomenclature system tailored to HRSV and its pathology would support the requirements of researchers and the public health community by minimizing information errors when handling, storing, and shipping HRSV samples and when submitting, searching, and displaying sequencing data and associated metadata. Moreover, consistent nomenclature would improve the ability of researchers to pool and analyze data and associated information from different sources. To fill this need, an international group of researchers, in conjunction with the WHO RSV Global Surveillance Project, proposes a concise nomenclature system for HRSV below the species level.
HRSV Genome Organization
HRSV has a single-stranded nonsegmented negative-sense RNA genome ≈15,191–15,277 nt long (Figure 2, panel A) (7). The HRSV genome contains 10 genes, each encoding a separate mRNA with a single open reading frame (ORF) (Figure 2, panel A; Table), except for the M2 mRNA, which contains 2 overlapping ORFs. The 11 HRSV proteins are 2 nonstructural proteins (NS1 and NS2), nucleoprotein (N), phosphoprotein (P), matrix protein (M), small hydrophobic envelope protein (SH), attachment glycoprotein (G), fusion glycoprotein (F), the transcription processivity factor (M2–1), RNA regulatory factor (M2–2), and large RNA polymerase protein (L) (Table) (24,25). The F glycoprotein is the major viral neutralization and protective antigen, followed by the G glycoprotein (26).
Figure 2.
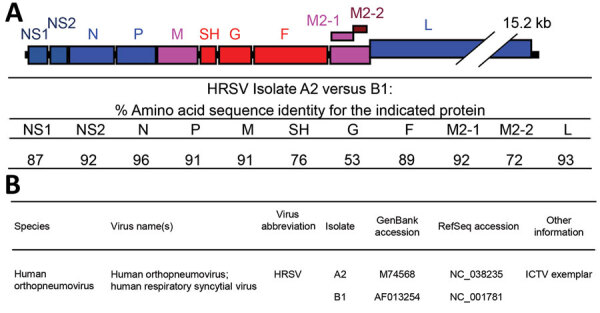
A) Schematic overview of the HRSV gene order and comparison of the amino acid identities of the reference strains of subgroups A (HRSV A2, GenBank accession no. M74568/NC_038235) and B (HRSV B1, GenBank accession no. AF013254/NC_001781). B) ICTV-proposed species designation, virus name, and associated GenBank reference sequences. HRSV, human respiratory syncytial virus; ICTV, International Committee on Virus Taxonomy; RefSeq, reference sequence.
Table. Widely accepted nomenclature for HRSV genes and proteins and gene annotation of strains HRSV A2 and HRSV B1*.
| Gene or genome region |
Proteins |
Open reading frame genome position, nt |
||||||
|---|---|---|---|---|---|---|---|---|
| Annotation† | Genome position, nt† |
|||||||
| Strain A2‡ | Strain B1¶ | Annotation | Abbreviation | Strain A2 | Strain B1 | |||
| Leader region |
1–44 |
1–44 |
|
|
|
|
|
|
| NS1 | 45–576 | 45–577 | Nonstructural protein 1 | NS1 | 99–518 | 99–518 | ||
| NS2 |
596–1098 |
594–1098 |
|
Nonstructural protein 2 |
NS2 |
|
628–1002 |
626–1000 |
| N | 1126–2328 | 1125–2327 | Nucleoprotein | N | 1141–2316 | 1140–2315 | ||
| P | 2330–3243 | 2331–3244 | Phosphoprotein | P | 2347–3072 | 2348–3073 | ||
| M | 3253–4210 | 3254–4208 | Matrix protein | M | 3262–4032 | 3263–4033 | ||
| SH | 4220–4629 | 4218–4630 | Small hydrophobic protein | SH | 4304–4498 | 4303–4500 | ||
| G | 4674–5596 | 4675–5600 | Attachment glycoprotein | G | 4689–5585 | 4690–5589 | ||
| F |
5649–7551 |
5653–7552 |
|
Fusion glycoprotein |
F |
|
5662–7386 |
7666–7390 |
| M2 |
7598–8558 |
7609–8568 |
|
Matrix protein 2 | ||||
| Matrix protein M2–1 | M2–1 | 7607–8191 | 7618–8205 | |||||
| Matrix protein M2–2 |
M2–2 |
|
8160–8432 |
8171–8443 |
||||
| L |
8491–15068 |
8501–15080 |
|
Polymerase protein |
L |
|
8499–14996 |
8509–15009 |
| Trailer region | 15069–15223 | 15081–15225 | ||||||
*HRSV, human respiratory syncytial virus. †Nucleotide annotations for leader and trailer region, and for indicated HRSV genes from the first nucleotide of the gene start signal [GGGGCAAAT(A/G); GGAGCAAAAT in case of the L gene] through the last adenosine residue of the gene end signal [AGT(T/A)A(T/A/G)(A/T)(A/T)(A/T)An]. ‡HRSV/A/USA/A2/2015 (45); GenBank accession number KT992094. ¶HRSV/B/USA/B1/1985/B1 (46); GenBank accession number AF013254.1.
HRSV Subgroups and Genotype Designations: Status and Outlook
HRSV subgroups A and B exhibit genomewide nucleotide and amino acid divergence (Figure 2, panel A) (25,27). The reference sequences for the 2 subgroups are derived from strains HRSV A2 (28–31; GenBank accession number M74568.1; RefSeq accession number NC_038235) and HRSV B1 (32; GenBank accession number AF013254.1; RefSeq accession number NC_001781.1; Figure 2, panel B). F glycoprotein sequences between the 2 subgroups are well conserved (89% aa identity), whereas the G glycoproteins are the most divergent (53% aa identity between the subgroups) among the HRSV proteins (Figure 2, panel A) and undergo continuous molecular evolution. The ectodomain of the G glycoproteins of both subgroups contains a conserved central domain, representing an important antigenic site, flanked by 2 hypervariable domains (33). Except for the central conserved region, the antigenic cross-reactivity between G glycoproteins of the 2 subgroups is low (26).
Because the G ORF exhibits the greatest degree of genetic variability between isolates, it is most commonly used for studies on the molecular evolution of HRSV. The genetic variability of HRSV strains over time has been commonly determined by sequencing the distal C-terminal third of the G ORF, which includes the second hypervariable domain. The variability in the G ORF is characterized by a high rate of nonsynonymous nucleotide changes, suggesting that evolution may be driven by immune pressure, even though this factor may be partially antibody independent (34). It is likely that variability in the G protein contributes to the capacity of HRSV to cause yearly outbreaks in the community (35–37). The nomenclature proposal outlined herein will be useful for the sequence analyses required to follow the molecular evolution of HRSV.
In a parallel effort, several research groups are working together on a genotyping proposal to provide a consensus on uniform genotype designations (38,39). As virus evolution continues, we expect new genotypes to emerge and older genotypes to become extinct. HRSV genotyping designations will need to capture present molecular evolutionary status and be adaptable to changes and will need to be reevaluated periodically by a global consortium.
Nomenclature Proposal for HRSV Strains and Isolates
For molecular epidemiology studies, a concise standard for short identifiers of specific HRSV sequences, suitable for the short definition lines that give context to a sample and its derived sequence, would be useful. Ideally, concise standardized identifiers should convey key information about each individual sequence in an alignment or phylogram, including source, date, and type, if known. Here, we aim to define this type of common naming convention for HRSV samples and isolates. We also propose using standard names and appropriate annotations for HRSV genes, provide examples to guide the annotation of sequence data during the sequence submission process, and suggest how to submit metadata associated with the source materials of HRSV sequences.
Definition Lines for HRSV Sequence Submissions
GenBank records available through the National Center for Biotechnology Information (NCBI) are identified by 2 elements: a unique alphanumeric accession number and a definition line. The definition line is the portion of the identifier commonly associated with GenBank records shown in BLAST results and other searches. Definition lines are generated by the submitter during the sequence submission process and include the species and isolate name (e.g., Human orthopneumovirus isolate HRSV/A/USA/001/2011, complete genome [proposed]). (40). We propose a standardized format to capture 5 sample-specific parameters of HRSV-positive clinical samples or isolates to be included in sequence definition lines compatible with database naming requirements (Figure 3), in this specific order: [virus name abbreviation]/[HRSV subgroup]/[geographic identifier]/[unique sequence identifier]/[year of sampling].
Figure 3.
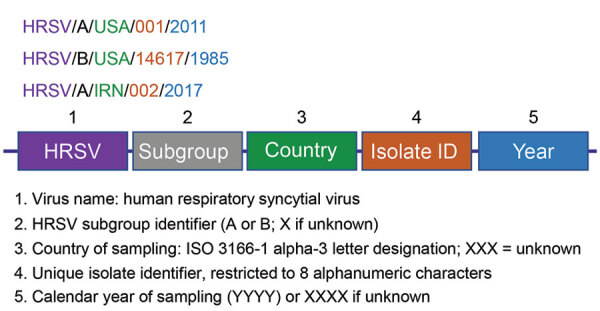
Schematic representation of the 5 consensus nomenclature elements of HRSV strains and isolates, with examples (top) and an explanation of each element (bottom). HRSV, human respiratory syncytial virus; ID, identification number; ISO, International Organization for Standardization.
Elements of Sequence Definition
I. Organism name; virus name abbreviation: HRSV
ICTV’s species name, Human orthopneumovirus (7), will be reflected as HRSV in the NCBI definition line. During submission to databases, the organism name can be entered as either Human orthopneumovirus or human respiratory syncytial virus. The abbreviation HRSV should be used in the definition line regardless of which organism name is provided.
II. HRSV subgroup: A or B; X, if unknown
III. Geographic identifier for the location of sampling. Because individual HRSV research networks have predefined requirements, we suggest some flexibility for this field:
If not specified by a research network, the ISO 3166–1 α-3 letter country code (https://www.iso.org/iso-3166-country-codes.html) should be used to indicate the country of sampling (XXX, if unknown). We strongly suggest that submitters provide any more specific geographic information on the sampling location (e.g., city or state) in metadata fields rather than in the definition line.
The WHO Global RSV Surveillance Project plans to use just the simple English-language name for the country.
Individual national studies may require a state, province, or city designation, in addition to the mandatory country name. If required, a period should be used to set off the country name ([state/province/city].[country name]).
IV. Unique isolate identifier
This field must be restricted to 8 alphanumeric characters. Underscores are permitted, but neither other special characters (e.g., /, %, $, @) nor spaces can be used. Controversial names or phrases, names of prominent people, and trademarked names or phrases cannot be used. To prevent duplication of sample or isolate identifiers by different groups, we recommend inserting a lettercode identifying a study or institute before the sample number. For example, unique isolate identifiers for samples from the INFORM-RSV study might use “INF” followed by a number (e.g., INF001 in HRSV/A/COUNTRY/INF001/2019).
V. Year of sampling; YYYY or XXXX, if unknown.
Examples of Sequence Definition Lines Using Proposed Nomenclature
HRSV/A/USA/001/2011
HRSV/B/Denver.USA/14617/1985
HRSV/A/IRN/001/2017
HRSV/A/Iran/001/2017
HRSV/X/IRN/001/2017 (subgroup unknown)
HRSV/B/New_Zealand/FR123/2020
Our nomenclature proposal prioritizes a short, concise definition line that will be easy to use in the laboratory, easily readable, and be a uniform system for HRSV in public databases. Additional host, virus, location or temporal information if desired could be submitted in metadata fields, which would allow researchers, epidemiologists, and database users to apply specific metadata filters, as needed for data retrieval and specific applications, analyses, or for displaying designations, such as in dendrograms.
Terminology for Annotations
To support efficient data analysis, uniform designations must be used at the database submission stage. Commonly accepted names for HRSV genes and proteins are shown in the table. An HRSV gene comprises a gene start signal GGGGCAAAT(A/G), an ORF with adjacent noncoding regions, if present, and the gene end signal through the last adenosine residue [AGT(T/A)A(T/A/G)(A/T)(A/T)(A/T)An] (Figure 4; 41). Each HRSV gene contains a single ORF, except for the M2 gene, which has 2 overlapping ORFs, M2–1 and M2–2. Nucleotide annotations of genes and ORFs for the HRSV A2 (Figure 4) and HRSV B1 reference sequences are shown in the table.
Figure 4.
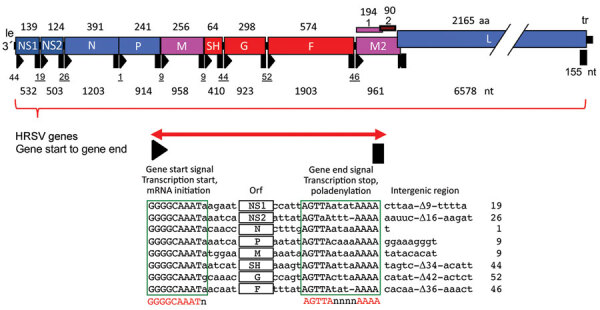
Schematic aid to gene annotations of HRSV whole-genome sequences. An HRSV gene comprises the sequence from the first nucleotide of the conserved HRSV gene start signal (GGGGCAAATa) to the last adenosine residue of the HRSV gene end signal (AGTTAnnnnAAAA) (25,41). Gene start signals are represented by black triangles, and gene end signals are shown as black rectangles, separated by intergenic regions (underlined). Note the M2/L gene overlap (annotations derived from HRSV A2; GenBank accession no. M74568/NC_038235). le, leader region; HRSV, human respiratory syncytial virus; tr, trailer region.
Metadata for HRSV Sequence Submissions
What is the most pertinent host data will depend on the interests and objectives of individual study groups. For example, when studying HRSV in a pediatric setting, prematurity may be of interest, but when studying HRSV in an adult setting, researchers may be more interested in whether participants are immunocompromised. We suggest information that could be included in metadata fields for HRSV:
1. Isolation source: sample type (upper or lower airways)
-
Viral RNA can be extracted directly from a clinical sample, from an isolate grown in cell culture, or possibly from a cDNA-derived recombinant virus. The sources of sequences from isolated viruses can be identified by the following designations:
wt: wild-type; sequences derived from RNA extracted directly from clinical specimens
tc: tissue culture; sequences derived from RNA extracted from HRSV isolates propagated in tissue culture
rec: recombinant; sequences of cDNA-derived recombinant virus (including vaccine strain)
2. Host:
Homo sapiens; subject age. Indicate years and months if <5 years of age, years only if ≥5 years of age. Sex should be spelled out if known.
3. Country, state, and (nearest) city of sampling.
Metadata information must include the full country name (not the 3-letter abbreviation) from the NCBI list of accepted country designations (https://www.ncbi.nlm.nih.gov/GenBank/collab/country). City, state, or province can also be included. Names should be written based on the standard ASCII letters including spaces if required (https://www.nist.gov/system/files/documents/2021/03/23/ansi-nist_2010_traditional_encoding.pdf). Geolocation coordinates of the location where sampling took place should be included if known.
4. Collection date
We highly recommend that the exact date of specimen collection (DD-Mon-YYYY format; e.g., 17-Feb-2002) be used; if exact date is not known, at least the month and year should be indicated (Mon-YYYY format).
5. Genotype according to the consensus in genotype classification by an HRSV working group [in progress (38,39)].
Associated with the International RSV Society, a special interest group of the International Society for Influenza and other respiratory viruses https://www.isirv.org/site/index.php/special-interest-groups/international-respiratory-syncytial-virus-society.
6. Metadata on the patient-host and the clinical disease should be included in the notes field in a structured format. Protected personally identifiable health information will be excluded from metadata submissions.
If >6 months of age, birthweight and gestational age at birth.
Significant pediatric co-morbidities, including prematurity, congenital cardiac disease, and broncho-pulmonary dysplasia (BPD).
Twin? (Y/N)
Exposed to specific HRSV therapeutic, vaccine, antibody, or antiviral? (Y/N)
Viral or bacterial co-infections, if known; pathogen species should be spelled out.
Adult underlying conditions, such as chronic obstructive pulmonary disease (COPD) or asthma, or altered immune status (e.g., immunocompromised, bone marrow transplant recipient).
-
Disease outcomes. Five grades are distinguished:
No medical care.
Outpatient or emergency room.
Hospital admission.
ICU admission.
Death.
For NCBI submissions, data can be entered through the web interface, or uploaded as tab-delimited text files. Sequences can be uploaded in FASTA format (https://blast.ncbi.nlm.nih.gov; Appendix), with associated metadata provided in a plain text, tab-delimited, source modifier table (Appendix Table 1) and gene or protein annotations provided in a plain text, tab-delimited, 5-column feature table (Appendix Table 2).
Outlook
Molecular surveillance has revealed that multiple HRSV genotypes circulate simultaneously in communities. Circulating genotypes often vary between communities, and circulation patterns within a community can change from year to year. Extended monitoring of circulating viruses is necessary to better understand transmission and molecular evolution (42). As HRSV vaccine candidates and antivirals are being developed, molecular epidemiology studies may reveal potential effects of prevention strategies on viral evolution and possible antibody-escape variants. Timely sharing of HRSV data worldwide through the use of public databases is essential. We propose that sequence data be uploaded to publicly accessible databases, such as NCBI (31). Although NCBI is the most complete repository for HRSV sequence information, studies may require that sequences first be submitted to other databases, such as GISAID (https://www.gisaid.org).
Public access will provide useful availability for investigators to submit, query, and analyze HRSV sequence data, enabling the evolutionary analysis of sequence diversity within or between HRSV genotypes. The utility of public access has been clearly demonstrated with the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the critical role that genetic sequence analysis has played. Notably, a nomenclature model for SARS-CoV-2 similar to what we have proposed for HRSV has been adopted, although some differences remain between databases (e.g., NCBI, SARS-CoV-2/human/USA/COVID20–1096/2020; GISAID, hCoV-19/Australia/VIC12/2020). When an HRSV vaccine becomes available, high-quality, geographically representative, country-specific data on circulating strains and rich datasets of well-curated, standardized, and parsable data will be required to monitor and trace possible evolutionary changes in response to vaccine-induced selective pressure (43,44). This proposal will profit from strong support by members of the International RSV Society, a special interest group of the International Society for Influenza and other Respiratory Virus Diseases (https://isirv.org), members of the WHO Global RSV Surveillance Project, and the HRSV research community.
Background genomic sequencing information on human respiratory syncytial virus.
Acknowledgments
This manuscript is dedicated to the memory of Jose A. Melero, a superb scientist and leader in the HRSV field and a wonderful and generous friend to us all.
Support for this work was provided by the Intramural Research Programs of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. M.Z. was supported by Public Health England (PHE), and I.N. was supported by PHE and by the National Institute of Health Research (NIHR) Respiratory Health Protection Unit. The work of J.R.B. and E.L.H. was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. T.C.W. was the recipient of a Wellcome Trust Award [204802/Z/16/Z]. The WHO’s Global RSV Surveillance is supported by the Bill and Melinda Gates Foundation grant award no. OPP1127419.
S.H., S.J., and W.Z. work with the World Health Organization. The authors are responsible for the views expressed in this publication and they do not necessarily represent the decisions, policy or views of the World Health Organization. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the World Health Organization. L.A. has done paid consultancies on RSV vaccines for Bavarian Nordic, Novavax, ClearPath Vaccines Company, and Pfizer; his laboratory is currently receiving funding through Emory University from Pfizer and Advac for laboratory studies for HRSV surveillance studies in adults, and he holds a subcontract on an NIH SBIR award to Sciogen on G protein HRSV vaccines. L.A. is a co-inventor on several CDC patents on the HRSV G protein and its CX3C chemokine motif relative to immune therapy and vaccine development, and on a patent filing for use of HRSV platform VLPs with the F and G proteins for vaccines. U.B. reports CRADA support to NIH from Sanofi Pasteur, outside the submitted work; in addition, she has patents on live-attenuated HRSV with royalties paid to NIH by Sanofi Pasteur. There are no additional conflicts of interest by any of the authors.
Biography
Dr. Salimi is a virologist, associate professor, and head of the virology department at the School of Public Health, Tehran University of Medical Sciences. His primary research interests include genetic characterization, immunopathogenesis, and vaccine design of respiratory viruses.
Footnotes
Suggested citation for this article: Salimi V, Viegas M, Trento A, Agoti CN, Anderson LJ, Avadhanula V, et al. Proposal for human respiratory syncytial virus nomenclature below the species level. Emerg Infect Dis. 2021 Jun [date cited]. https://doi.org/10.3201/eid2706.204608
These authors contributed equally to this article.
These last authors contributed equally to this article.
References
- 1.Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. ; RSV Global Epidemiology Network. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–58. 10.1016/S0140-6736(17)30938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–55. 10.1016/S0140-6736(10)60206-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salimi V, Tavakoli-Yaraki M, Yavarian J, Bont L, Mokhtari-Azad T. Prevalence of human respiratory syncytial virus circulating in Iran. J Infect Public Health. 2016;9:125–35. 10.1016/j.jiph.2015.05.005 [DOI] [PubMed] [Google Scholar]
- 4.Stein RT, Bont LJ, Zar H, Polack FP, Park C, Claxton A, et al. Respiratory syncytial virus hospitalization and mortality: Systematic review and meta-analysis. Pediatr Pulmonol. 2017;52:556–69. 10.1002/ppul.23570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Brien KL, Baggett HC, Brooks WA, Feikin DR, Hammitt LL, Higdon MM, et al. ; Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet. 2019;394:757–79. 10.1016/S0140-6736(19)30721-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–59. 10.1056/NEJMoa043951 [DOI] [PubMed] [Google Scholar]
- 7.Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, et al. ; Ictv Report Consortium. ICTV virus taxonomy profile: Pneumoviridae. J Gen Virol. 2017;98:2912–3. 10.1099/jgv.0.000959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agoti CN, Phan MVT, Munywoki PK, Githinji G, Medley GF, Cane PA, et al. Genomic analysis of respiratory syncytial virus infections in households and utility in inferring who infects the infant. Sci Rep. 2019;9:10076. 10.1038/s41598-019-46509-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trovao NS, Khuri-Bulos N, Tan Y, Puri V, Shilts MH, Halpin RA, et al. Molecular characterization of respiratory syncytial viruses circulating in a paediatric cohort in Amman, Jordan. Microb Genom. 2019. Sep 18 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 10.Geis S, Prifert C, Weissbrich B, Lehners N, Egerer G, Eisenbach C, et al. Molecular characterization of a respiratory syncytial virus outbreak in a hematology unit in Heidelberg, Germany. J Clin Microbiol. 2013;51:155–62. 10.1128/JCM.02151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabeya D, Kinjo T, Parrott GL, Uehara A, Motooka D, Nakamura S, et al. The clinical and phylogenetic investigation for a nosocomial outbreak of respiratory syncytial virus infection in an adult hemato-oncology unit. J Med Virol. 2017;89:1364–72. 10.1002/jmv.24800 [DOI] [PubMed] [Google Scholar]
- 12.Hall CB, Walsh EE, Schnabel KC, Long CE, McConnochie KM, Hildreth SW, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–90. 10.1093/infdis/162.6.1283 [DOI] [PubMed] [Google Scholar]
- 13.Malekshahi SS, Razaghipour S, Samieipoor Y, Hashemi FB, Manesh AAR, Izadi A, et al. Molecular characterization of the glycoprotein and fusion protein in human respiratory syncytial virus subgroup A: Emergence of ON-1 genotype in Iran. Infect Genet Evol. 2019;71:166–78. 10.1016/j.meegid.2019.03.026 [DOI] [PubMed] [Google Scholar]
- 14.Vos LM, Oosterheert JJ, Kuil SD, Viveen M, Bont LJ, Hoepelman AIM, et al. High epidemic burden of RSV disease coinciding with genetic alterations causing amino acid substitutions in the RSV G-protein during the 2016/2017 season in The Netherlands. J Clin Virol. 2019;112:20–6. 10.1016/j.jcv.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 15.Pierangeli A, Viscido A, Bitossi C, Frasca F, Gentile M, Oliveto G, et al. Differential interferon gene expression in bronchiolitis caused by respiratory syncytial virus-A genotype ON1. Med Microbiol Immunol (Berl). 2020;209:23–8. 10.1007/s00430-019-00633-6 [DOI] [PubMed] [Google Scholar]
- 16.Vandini S, Biagi C, Lanari M. Respiratory syncytial virus: the influence of serotype and genotype variability on clinical course of infection. Int J Mol Sci. 2017;18:1717. 10.3390/ijms18081717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laham FR, Mansbach JM, Piedra PA, Hasegawa K, Sullivan AF, Espinola JA, et al. Clinical profiles of respiratory syncytial virus subtypes A and B among children hospitalized with bronchiolitis. Pediatr Infect Dis J. 2017;36:808–10. 10.1097/INF.0000000000001596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilca R, De Serres G, Tremblay M, Vachon ML, Leblanc E, Bergeron MG, et al. Distribution and clinical impact of human respiratory syncytial virus genotypes in hospitalized children over 2 winter seasons. J Infect Dis. 2006;193:54–8. 10.1086/498526 [DOI] [PubMed] [Google Scholar]
- 19.Martinello RA, Chen MD, Weibel C, Kahn JS. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis. 2002;186:839–42. 10.1086/342414 [DOI] [PubMed] [Google Scholar]
- 20.Measles virus nomenclature update: 2012. Wkly Epidemiol Rec. 2012;87:73–81. [PubMed] [Google Scholar]
- 21.Kuhn JH, Bao Y, Bavari S, Becker S, Bradfute S, Brister JR, et al. Virus nomenclature below the species level: a standardized nomenclature for laboratory animal-adapted strains and variants of viruses assigned to the family Filoviridae. Arch Virol. 2013;158:1425–32. 10.1007/s00705-012-1594-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol. 2011;156:1397–413. 10.1007/s00705-011-1006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reconsideration of influenza A virus nomenclature: a WHO memorandum. Bull World Health Organ. 1979;57:227–33. [PMC free article] [PubMed] [Google Scholar]
- 24.Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol. 2013;372:3–38. 10.1007/978-3-642-38919-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins PL, Karron RA. Respiratory syncytial virus and metapneumovirus. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields Virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 1086–123. [Google Scholar]
- 26.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162:80–99. 10.1016/j.virusres.2011.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mufson MA, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–24. 10.1099/0022-1317-66-10-2111 [DOI] [PubMed] [Google Scholar]
- 28.Collins PL, Olmsted RA, Spriggs MK, Johnson PR, Buckler-White AJ. Gene overlap and site-specific attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus. Proc Natl Acad Sci U S A. 1987;84:5134–8. 10.1073/pnas.84.15.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mink MA, Stec DS, Collins PL. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology. 1991;185:615–24. 10.1016/0042-6822(91)90532-G [DOI] [PubMed] [Google Scholar]
- 30.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020;48(D1):D9–16. 10.1093/nar/gkz899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94:13961–6. 10.1073/pnas.94.25.13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez I, Dopazo J, Melero JA. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol. 1997;78:2419–29. 10.1099/0022-1317-78-10-2419 [DOI] [PubMed] [Google Scholar]
- 34.Trento A, Ábrego L, Rodriguez-Fernandez R, González-Sánchez MI, González-Martínez F, Delfraro A, et al. Conservation of G-protein epitopes in respiratory syncytial virus (group A) despite broad genetic diversity: is antibody selection involved in virus evolution? J Virol. 2015;89:7776–85. 10.1128/JVI.00467-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melero JA, García-Barreno B, Martínez I, Pringle CR, Cane PA. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78:2411–8. 10.1099/0022-1317-78-10-2411 [DOI] [PubMed] [Google Scholar]
- 36.Sullender WM, Mufson MA, Anderson LJ, Wertz GW. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J Virol. 1991;65:5425–34. 10.1128/JVI.65.10.5425-5434.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cane PA, Matthews DA, Pringle CR. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J Clin Microbiol. 1994;32:1–4. 10.1128/JCM.32.1.1-4.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goya S, Galiano M, Nauwelaers I, Trento A, Openshaw PJ, Mistchenko AS, et al. Toward unified molecular surveillance of RSV: A proposal for genotype definition. Influenza Other Respir Viruses. 2020;14:274–85. 10.1111/irv.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramaekers K, Rector A, Cuypers L, Lemey P, Keyaerts E, Van Ranst M. Towards a unified classification for human respiratory syncytial virus genotypes. Virus Evol. 2020;6:veaa052. [DOI] [PMC free article] [PubMed]
- 40.The GenBank Submissions Handbook. Bethesda (MD): National Center for Biotechnology Information; 2011. [Google Scholar]
- 41.Collins PL, Dickens LE, Buckler-White A, Olmsted RA, Spriggs MK, Camargo E, et al. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci U S A. 1986;83:4594–8. 10.1073/pnas.83.13.4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trento A, Casas I, Calderón A, Garcia-Garcia ML, Calvo C, Perez-Breña P, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84:7500–12. 10.1128/JVI.00345-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham BS. Vaccine development for respiratory syncytial virus. Curr Opin Virol. 2017;23:107–12. 10.1016/j.coviro.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazur NI, Higgins D, Nunes MC, Melero JA, Langedijk AC, Horsley N, et al. ; Respiratory Syncytial Virus Network (ReSViNET) Foundation. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect Dis. 2018;18:e295–311. 10.1016/S1473-3099(18)30292-5 [DOI] [PubMed] [Google Scholar]
- 45.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci U S A. 1995;92:11563–7. 10.1073/pnas.92.25.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crowe JE Jr, Bui PT, Firestone CY, Connors M, Elkins WR, Chanock RM, et al. Live subgroup B respiratory syncytial virus vaccines that are attenuated, genetically stable, and immunogenic in rodents and nonhuman primates. J Infect Dis. 1996;173:829–39. 10.1093/infdis/173.4.829 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Background genomic sequencing information on human respiratory syncytial virus.


