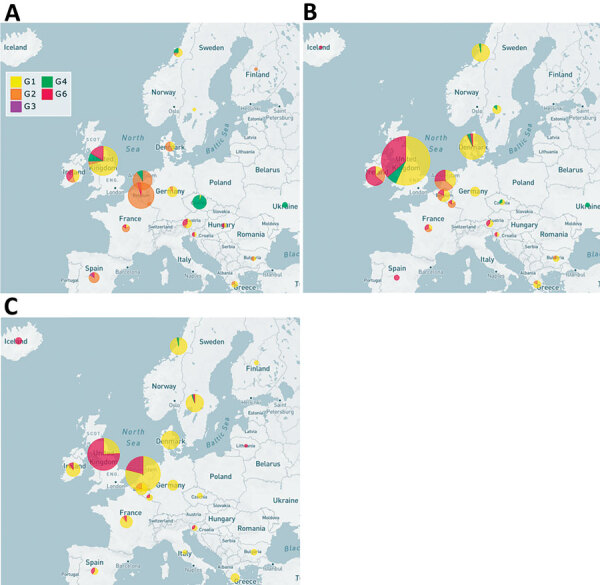
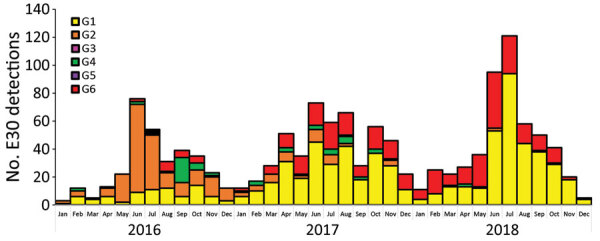
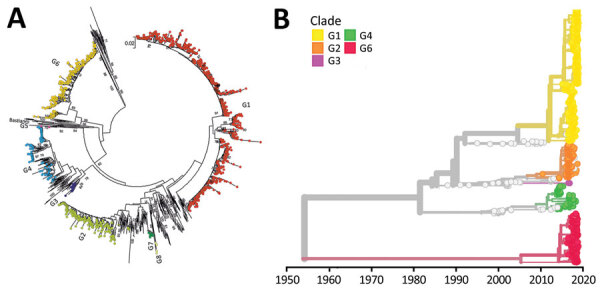
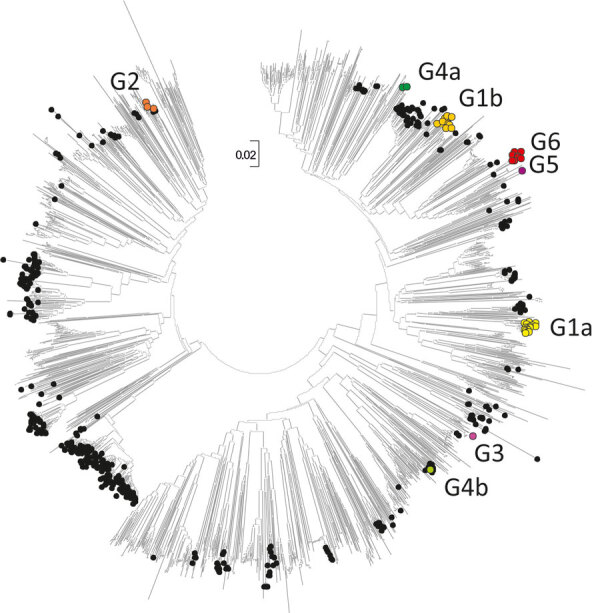
Kimberley SM Benschop
Kimberley SM Benschop
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,✉,
Eeva K Broberg
Eeva K Broberg
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Emma Hodcroft
Emma Hodcroft
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Dennis Schmitz
Dennis Schmitz
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jan Albert
Jan Albert
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Anda Baicus
Anda Baicus
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jean-Luc Bailly
Jean-Luc Bailly
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Gudrun Baldvinsdottir
Gudrun Baldvinsdottir
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Natasa Berginc
Natasa Berginc
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Soile Blomqvist
Soile Blomqvist
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Sindy Böttcher
Sindy Böttcher
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Mia Brytting
Mia Brytting
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Erika Bujaki
Erika Bujaki
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Maria Cabrerizo
Maria Cabrerizo
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Cristina Celma
Cristina Celma
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Ondrej Cinek
Ondrej Cinek
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Eric CJ Claas
Eric CJ Claas
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jeroen Cremer
Jeroen Cremer
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jonathan Dean
Jonathan Dean
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jennifer L Dembinski
Jennifer L Dembinski
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Iryna Demchyshyna
Iryna Demchyshyna
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Sabine Diedrich
Sabine Diedrich
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Susanne Dudman
Susanne Dudman
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jake Dunning
Jake Dunning
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Robert Dyrdak
Robert Dyrdak
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Mary Emmanouil
Mary Emmanouil
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Agnes Farkas
Agnes Farkas
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Cillian De Gascun
Cillian De Gascun
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Guillaume Fournier
Guillaume Fournier
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Irina Georgieva
Irina Georgieva
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Ruben Gonzalez-Sanz
Ruben Gonzalez-Sanz
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jolanda van Hooydonk-Elving
Jolanda van Hooydonk-Elving
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Anne J Jääskeläinen
Anne J Jääskeläinen
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Ruta Jancauskaite
Ruta Jancauskaite
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Kathrin Keeren
Kathrin Keeren
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Thea K Fischer
Thea K Fischer
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Sidsel Krokstad
Sidsel Krokstad
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Lubomira Nikolaeva–Glomb
Lubomira Nikolaeva–Glomb
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Ludmila Novakova
Ludmila Novakova
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Sofie E Midgley
Sofie E Midgley
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Audrey Mirand
Audrey Mirand
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Richard Molenkamp
Richard Molenkamp
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Ursula Morley
Ursula Morley
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Joël Mossong
Joël Mossong
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Svajune Muralyte
Svajune Muralyte
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jean-Luc Murk
Jean-Luc Murk
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Trung Nguyen
Trung Nguyen
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Svein A Nordbø
Svein A Nordbø
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Riikka Österback
Riikka Österback
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Suzan Pas
Suzan Pas
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Laura Pellegrinelli
Laura Pellegrinelli
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Vassiliki Pogka
Vassiliki Pogka
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Birgit Prochazka
Birgit Prochazka
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Petra Rainetova
Petra Rainetova
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Marc Van Ranst
Marc Van Ranst
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Lieuwe Roorda
Lieuwe Roorda
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Isabelle Schuffenecker
Isabelle Schuffenecker
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Rob Schuurman
Rob Schuurman
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Asya Stoyanova
Asya Stoyanova
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Kate Templeton
Kate Templeton
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Jaco J Verweij
Jaco J Verweij
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Androniki Voulgari-Kokota
Androniki Voulgari-Kokota
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Tytti Vuorinen
Tytti Vuorinen
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Elke Wollants
Elke Wollants
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Katja C Wolthers
Katja C Wolthers
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Katherina Zakikhany
Katherina Zakikhany
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Richard Neher
Richard Neher
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Heli Harvala
Heli Harvala
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,
Peter Simmonds
Peter Simmonds
1National Institute for Public Health and the Environment, Bilthoven, the Netherlands (K.S.M. Benschop, D. Schmitz, J. Cremer);
2European Centre for Disease Prevention and Control, Stockholm, Sweden (E.K. Broberg);
3Biozentrum, University of Basel and Swiss Institute of Bioinformatics, Basel, Switzerland (E. Hodcroft, R. Neher);
4Karolinska University Hospital and Karolinska Institute, Stockholm (J. Albert, R. Dyrdak);
5Cantacuzino, Bucharest, Romania (A. Baicus);
6CHU Clermont-Ferrand, National Reference Centre for Enteroviruses and Parechoviruses, Clermont-Ferrand, France (J. Bailly, A. Mirand);
7Landspitali-National University Hospital, Reykjavik, Iceland (G. Baldvinsdottir);
8National laboratory of Health, Environment and Food, Ljubljana, Slovenia (N. Berginc);
9National Institute for Health and Welfare, Helsinki, Finland (S. Blomqvist);
10Robert-Koch-Institue, Berlin, Germany (S. Böttcher S. Diedrich, K. Keeren);
11The Public Health Agency of Sweden, Solna, Sweden (M. Brytting, K. Zakikhany);
12National Public Health Center, Budapest, Hungary (E. Bujaki, A. Farkas);
13Instituto de Salud Carlos III, Madrid, Spain (M. Cabrerizo, R. Gonzalez-Sanz);
14Public Health England, Colindale, UK (C. Celma, J. Dunning);
15University of Oslo and Oslo University Hospital, Oslo, Norway (S. Dudman);
16Charles University, Prague, Czech Republic (O. Cinek);
17Leiden University Medical Center, Leiden, the Netherlands (E.C.J. Claas);
18University College Dublin, Dublin, Ireland, UK (J. Dean, C. De Gascun, U. Morley);
19World Health Organization National Polio Entero Reference Laboratory, Norwegian Institute of Public Health, Oslo (J.L. Dembinski, S. Dudman);
20Public Health Center of the Ministry of Health of Ukraine, Kiev, Ukraine (I. Demchyshyna);
21Hellenic Pasteur Institute, Athens, Greece (M. Emmanouil, V. Pogka, A. Voulgari-Kokota);
22Laboratoire National de Santé, Dudelange, Luxembourg (G. Fournier, T. Nguyen, J. Mossong);
23National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria (I. Georgieva, L. Nikolaeva-Glomb, A. Stoyanova);
24Microvida, Breda, the Netherlands (J. van Hooydonk-Elving, S. Pas);
25University of Helsinki and Helsinki University Hospital, Helsinki (A.J. Jääskeläinen);
26National Public Health Surveillance Laboratory, Vilnius, Lithuania (R. Jancauskaite, S. Muralyte);
27Nordsjaellands University Hospital, Hilleroed, Denmark (T.K. Fischer);
28Statens Serum Institute and University of Copenhagen, Copenhagen, Denmark (T.K. Fischer);
29University Hospital of Trondheim, Norway (S. Krokstad, S.A. Nordbø);
30National Institute of Public Health, Prague (L. Novakova, P. Rainetova);
31Danish WHO National Reference Laboratory for Poliovirus, Statens Serum Institut, Copenhagen (S.E. Midgley);
32Erasmus Medical Center, Rotterdam, the Netherlands (R. Molenkamp);
33Elisabeth Tweesteden Hospital, Tilburg, the Netherlands (J.-L. Murk, J.J. Verweij);
34Norwegian University of Science and Technology, Trondheim (S.A. Nordbø);
35Turku University Hospital, Turku, Finland (R. Österback, T. Vuorinen);
36University of Milan, Milan, Italy (L. Pellegrinelli);
37Austrian Agency for Health and Food Safety, Vienna, Austria (B. Prochazka); Rega Institute KU Leuven, Leuven, Belgium (M. Van Ranst, E. Wollants);
38Maasstad Ziekenhuis, Rotterdam (L. Roorda);
39Centre de Biologie Est des Hospices Civils de Lyon, Lyon, France (I. Schuffenecker);
40University Medical Center Utrecht, Utrecht, the Netherlands (R. Schuurman); N
41ational Health Services Scotland, Edinburgh, Scotland, UK (K. Templeton);
42University of Turku, Turku (T. Vuorinen) Amsterdam University Medical Center, Amsterdam, the Netherlands (K.C, Wolthers);
43University College London, London, UK (H. Harvala);
44National Health Service, Colindale (H. Harvala); University of Oxford, Oxford, UK (P. Simmonds)
1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44