Abstract
We assessed antimicrobial resistance (AMR) in Neisseria gonorrhoeae in Nunavut, Canada, using remnant gonorrhea nucleic acid amplification test–positive urine specimens. This study confirms the feasibility of conducting N. gonorrhoeae AMR surveillance and highlights the diversity of gonococcal sequence types and geographic variation of AMR patterns in the territory.
Keywords: antimicrobial resistance, Arctic, bacteria, Canada, DNA, gonorrhea, Inuit Nunangat, molecular characterization, Neisseria gonorrhoeae, sequence analysis, sexually transmitted infections
In 2015, the prevalence of Neisseria gonorrhoeae reported in the territory of Nunavut, Canada, 837.6 cases/100,000 residents, was 15 times the national rate in Canada (55.4 cases/100,000 residents) (1). Gonorrhea is a notifiable disease in Nunavut, and all positive cases are reported to the department of health. Globally, reports are increasing of N. gonorrhoeae with resistance to currently recommended first-line antimicrobial treatment agents (2). These resistance rates in some global regions approach or exceed World Health Organization thresholds of 5% to warrant changes to the prescribed gonorrhea treatments (3).
The Gonococcal Antimicrobial Surveillance Program of Canada is a culture-based laboratory surveillance system monitoring antimicrobial resistance (AMR) trends and gonococcal sequence types (STs) in N. gonorrhoeae (4). Traditionally, N. gonorrhoeae AMR surveillance has required obtaining cultures from patients for phenotypic testing. In the Canadian Arctic regions, including Nunavut, no information has been available on the prevalence and distribution of N. gonorrhoeae AMR patterns because long transport times and conditions preclude transporting cultures; assessment is exclusively done by using nucleic acid amplification tests (NAATs). The National Microbiology Laboratory (Winnipeg, Manitoba, Canada) has addressed this issue by developing molecular assays to predict AMR and molecular STs from remnant NAAT specimens. We sought to assess the prevalence and distribution of AMR N. gonorrhoeae STs in Nunavut using NAAT-tested N. gonorrhoeae–positive specimens.
The Study
Nunavut, at 1,877,787 km2 the largest territory in Canada, is divided into 3 regions: Qikiqtaaluk (also called Qikiqtani) in the east, Kivalliq in the center, and Kitikmeot in the west. Nunavut’s reported 2016 population was 35,994 (5), ≈85% of whom identify as Inuit (6). Nunavut is one region in Inuit Nunangat, a Canadian Inuktitut term inclusive of the land, water, and ice of the Inuit homeland (7).
For this study we used remnant N. gonorrhoeae NAAT–positive specimens collected in Nunavut for routine gonococcal diagnostics during January 1, 2018–December 31, 2019. We submitted specimens from the Kitikmeot region to DynaLife Laboratories (https://www.dynalife.ca) and from the Qikiqtaaluk and Kivalliq regions to the Qikiqtani General Hospital Laboratory (Iqaluit, Nunavut, Canada) to the National Microbiology Laboratory for molecular antimicrobial susceptibility prediction using NAATs and N. gonorrhoeae multiantigen sequence typing (NG-MAST). Urine specimens submitted to DynaLife laboratories were tested using the Gen-Probe Aptima Combo 2 test (Hologic, https://www.hologic.com). Testing at the Qikiqtani General Hospital Laboratory was done using the Roche-Cobas test (Roche Molecular Diagnostics, https://diagnostics.roche.com). We did not submit repeat samples or test-of-cure specimens. Ethics approval was obtained from the University of Alberta’s Health Research Ethics Board and the Nunavut Research Institute.
We performed NG-MAST as described elsewhere (8,9). We then tested specimens successfully typed with NG-MAST using single-nucleotide polymorphism (SNP) assays targeting cephalosporin-DS mutations (ponA, mtrR delA, porB, penA A311V, penA A501, penA N513Y, and penA G545S), ciprofloxacin-resistance mutations (gyrA and parC), and azithromycin-resistance mutations (23S rRNA A2059G, C2611T, and mtrR) to predict antimicrobial susceptibility. We performed DNA extraction, preparation, real-time PCR, and results analysis as described elsewhere (9,10). For specimens identified with the same ST, we performed SNP assay testing for a subset of samples and for the remainder of samples within identical ST groups. We inferred AMR profiles based on STs with 4–70 samples if ≥50% of those samples had identical AMR predictions based on SNP results and STs with >70 samples if ≥30 samples were tested with identical AMR predictions. These results are molecular based and have been validated against MIC phenotypes in a previous study (9).
All NAAT-tested N. gonorrhoeae–positive specimens (n = 1,128) from Nunavut collected between January 1, 2018–December 31, 2019, were included in the study (Table 1). Of these, 106 (9.4%) samples were nontypeable for NG-MAST because of low concentrations of DNA and therefore excluded from further testing. We identified a total of 75 different STs among the remaining 1,022 NAAT-identified isolates from samples submitted from Nunavut (Figure 1). The most prevalent was ST16840 (16.5%), followed by ST5985 (15.3%).
Table 1. Sex and geographic distribution of gonorrhea nucleic acid amplification test positive specimens from Nunavut, Canada, 2018–2019*.
| Year |
|
Region |
Total by sex | Total | ||
|---|---|---|---|---|---|---|
| Sex | Kivalliq | Qikiqtaaluk | Kitikmeot | |||
| 2018 |
M | 62 | 112 | 8 | 182 | 432 |
| F |
98 |
135 |
17 |
250 |
||
| 2019 |
M | 94 | 166 | 14 | 274 | 696 |
| F | 142 | 256 | 22 | 420 | ||
| Unk |
0 |
0 |
2 |
2 |
||
| Total |
396 |
669 |
63 |
1,128 |
||
| *Unk, unknown | ||||||
Figure 1.

Distribution of prevalent Neisseria gonorrhoeae multiantigen sequence typing sequence types of gonorrhea-positive nucleic acid amplification specimens (n = 1,022) from a study of antimicrobial resistance in N. gonorrhoeae in the Nunavut region of Inuit Nunangat, Canada, 2018–2019.
SNP assay results for predicting AMR were successfully determined for cephalosporin (687 samples), ciprofloxacin (541 samples), and azithromycin (435 samples). We predicted AMR for an additional 289 cephalosporin results, 436 ciprofloxacin results, and 533 azithromycin results on the basis of expected results of prevalent STs tested (Table 2). Within Nunavut, the Qikiqtaaluk region had the highest prevalence of intermediate cephalosporin MICs (51.3%) and resistance to azithromycin (11.3%). The Kivalliq region had the only sample (0.3%) with predicted decreased susceptibility to cephalosporins. We inferred the genetic relationships between the NG-MAST STs by using the maximum-likelihood method and Tamura-Nei model (Figure 2) (11,12). We predicted most samples in clusters A, B, and D would have elevated MICs to cephalosporins but that most samples in cluster C would have azithromycin resistance. We also found that STs were clustered based on geography: in Qikiqtaaluk, we primarily identified STs from clusters A, B and C, but we identified STs from cluster D in all 3 jurisdictions.
Table 2. Predicted AMR of gonorrhea positive nucleic acid amplification specimens from Nunavut, 2018–2019*.
| Resistance | MICs, mg/L | No. specimens with AMR/no. predicted (%) |
|||
|---|---|---|---|---|---|
| Qikiqtaaluk | Kivalliq | Kitikmeot | Total | ||
| Decreased susceptibility to cephalosporins |
≥0.125 |
0/591 (0) |
1/350 (0.3) |
0/35 (0) |
1/976 (0.1) |
| Intermediate cephalosporin MICs |
0.032–0.063 |
303/591 (51.3) |
49/350 (14.0) |
8/35 (22.9) |
360/976 (36.9) |
| Ciprofloxacin resistant |
≥1 |
298/591 (50.4) |
50/351 (14.2) |
7/35 (20.0) |
355/977 (36.3) |
| Azithromycin resistant |
≥2 |
66/583 (11.3) |
2/350 (0.6) |
0/35 (0) |
68/968 (7.0) |
| *Denominators indicate predicted number of samples with susceptibility for the given antimicrobial. Not all samples were tested for antimicrobial susceptibilities and not all the samples that were tested gave conclusive results. AMR, antimicrobial resistance. | |||||
Figure 2.
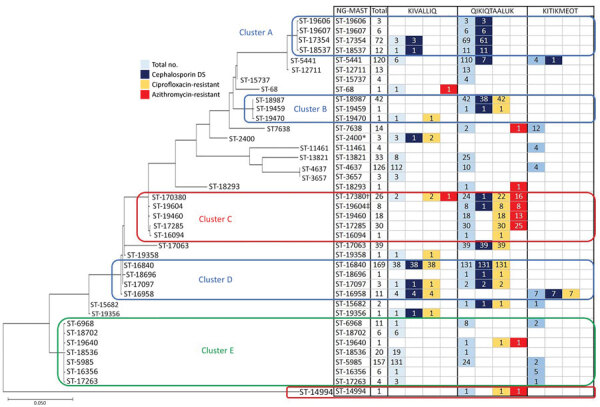
Genetic relationship of Neisseria gonorrhoeae multiantigen sequence typing sequence types (STs) of gonorrhea-positive nucleic acid amplification specimens with prevalent and predicted nonsusceptible SNP assay results (n = 975) from a study of antimicrobial resistance in N. gonorrhoeae in the Nunavut region of Inuit Nunangat, Canada, 2018–2019. Only prevalent STs or STs whose samples predicted decreased susceptibility to cephalosporins or resistance to ciprofloxacin or azithromycin were included. The evolutionary history was inferred by using the maximum-likelihood method and Tamura-Nei model (13). The tree with the highest log likelihood (–4321.41) is shown. Initial trees for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 39 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding; a total of 917 positions were in the final dataset. Evolutionary analyses were conducted in MEGA X (14). Clusters were identified as STs with ≤5 base pair differences between them. *One of the ST2400 samples was predicted to be cephalosporin decreased susceptibility (not cephalosporin/DS). †The sample that was predicted to be cephalosporin/DS was not azithromycin resistant. ‡The sample that was cephalosporin/DS was also azithromycin resistant. Cephalosporin/DS, cephalosporin intermediate/decreased susceptibility.
Our analysis of routinely collected diagnostic specimens for gonorrhea from Nunavut highlights the usefulness of SNP assays and NG-MAST typing in identifying population-level N. gonorrhoeae AMR and delineating transmission patterns (9). Current Nunavut treatment guidelines for gonorrhea recommend dual ceftriaxone/azithromycin therapy (13). Although decreased susceptibility to cephalosporins was identified in only 1 sample, the rate of predicted intermediate cephalosporin MICs in Nunavut at 36.9% is >2 times as high as the national rate of 16% in 2018, potentially limiting the long-term use of this agent (4). In addition, the 7.0% rate of predicted azithromycin resistance exceeds the World Health Organization threshold of 5% above which a drug is not routinely recommended for treatment (3). We also noted substantial geographic variation; the highest prevalence of predicted intermediate cephalosporin MICs (51.3%) and predicted resistance to azithromycin (11.3%) were in the Qikiqtaaluk region. In the adjacent province of Quebec, the reported rate of intermediate cephalosporin MICs was 22% and azithromycin resistance was 11.8% (4). NG-MAST typing detected a large number of STs, reflecting the genetic diversity of N. gonorrhoeae and suggesting the introduction of multiple strains into the territory.
The first limitation of our study was that no gonococcal cultures were available and cases were compared to national gonococcal cultures, which may not be representative of gonorrhea in all regions. In addition, only urine specimens were collected for gonorrhea in Nunavut. Previous research has highlighted the variability in ST from extragenital sites (14,15). There are limitations to using culture-independent techniques to predict AMR because specimens often contain low concentrations of DNA, which can limit SNP detection. Gonococcus is also known to mutate easily, which may lead to false-negative results in the SNP assays because of sequence variations in DNA (9). A molecular-based approach may overestimate AMR because mutations associated with resistance do not always correlate phenotypically. Finally, we acknowledge the biomedical and technical laboratory approach of this work and the limited partnerships with the Inuit people.
Conclusions
Because research across Inuit Nunangat has historically often been done in an exploitative way that has not respected Inuit self-determination, meaningful partnerships are required to prioritize research activities and develop approaches that add value for and are accepted by communities. Our findings highlight the feasibility of conducting molecular surveillance for N. gonorrhoeae AMR using remnant N. gonorrhoeae–positive NAAT specimens Data based on these findings can be used to provide Nunavut with up-to-date information about the best choices to treat gonorrhea.
Acknowledgments
We thank Pam Sawatzky, Norman Barairo, and Shelley Peterson for technical assistance.
This project was supported by internal funding from the Public Health Agency of Canada and Indigenous Services Canada.
Biography
Dr. Singh is a clinical professor with the Division of Infectious Diseases at the University of Alberta in Edmonton. Her primary research interests are clinical and epidemiologic aspects of bacterial STIs, HIV biomedical prevention, and the use of rapid diagnostic tests for diagnosing syphilis and HIV.
Footnotes
Suggested citation for this article: Singh AE, Pawa J, Kulleperuma K, Prasad E, Marchand S, Dionne K, et al. Molecular characterization and antimicrobial resistance in Neisseria gonorrhoeae, Nunavut region of Inuit Nunangat, Canada, 2018–2019. Emerg Infect Dis. 2021 Jun [date cited]. https://doi.org/10.3201/eid2706.204407
References
- 1.Choudhri Y, Miller J, Sandhu J, Leon A, Aho J. Gonorrhea in Canada, 2010-2015. Can Commun Dis Rep. 2018;44:37–42. 10.14745/ccdr.v44i02a01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unemo M, Seifert HS, Hook EW III, Hawkes S, Ndowa F, Dillon JR. Gonorrhoea. Nat Rev Dis Primers. 2019;5:79. 10.1038/s41572-019-0128-6 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization, Department of Reproductive Health and Research. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. 2012. [cited 2021 Apr 21]. https://www.who.int/reproductivehealth/publications/rtis/9789241503501/en
- 4.Public Health Agency of Canada, National Microbiology Laboratory. 2020. National surveillance of antimicrobial susceptibilities of Neisseria gonorrhoeae annual summary 2018 [cited 2021 Apr 21]. https://www.canada.ca/en/public-health/services/publications/drugs-health-products/national-surveillance-antimicrobial-susceptibilities-neisseria-gonorrhoeae-annual-summary-2018.html
- 5.Statistics Canada. Population and dwelling count highlight tables, 2016. census [cited 2021 Apr 21]. https://www12.statcan.gc.ca/census-recensement/2016/dp-pd/hlt-fst/pd-pl/Table.cfm?Lang=Eng&T=101&SR=1&S=3&O=D#tPopDwell
- 6.Nunavut Tunngavik Incorporated. Annual report on the state of Inuit culture and society, 2007–8 [cited 2021 Apr 21]. https://www.tunngavik.com/publications/annual-report-on-the-state-of-inuit-culture-and-society-2007-2008
- 7.Kanatami IT. National Inuit strategy on research, 2018. [cited 2021 Apr 21]. https://www.itk.ca/wp-content/uploads/2018/04/ITK_NISR-Report_English_low_res.pdf
- 8.Martin IM, Ison CA, Aanensen DM, Fenton KA, Spratt BG. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis. 2004;189:1497–505. 10.1086/383047 [DOI] [PubMed] [Google Scholar]
- 9.Peterson S, Martin I, Demczuk W, Barairo N, Naidu P, Lefebvre B, et al. Multiplex real-time polymerase chain reaction assays for the prediction of cephalosporin, ciprofloxacin, and azithromycin antimicrobial susceptibility of positive Neisseria gonorrhoeae nucleic acid amplification test samples. J Antimicrob Chemother. 2020;75:3485–90. 10.1093/jac/dkaa360 [DOI] [PubMed] [Google Scholar]
- 10.Trembizki E, Buckley C, Donovan B, Chen M, Guy R, Kaldor J, et al. Direct real-time PCR-based detection of Neisseria gonorrhoeae 23S rRNA mutations associated with azithromycin resistance. J Antimicrob Chemother. 2015;70:3244–9. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonorrhea public health protocol. Nunavut communicable disease and surveillance manual. Iqaluit (Canada): Nunavut Department of Health. 2018. [Google Scholar]
- 14.Didelot X, Dordel J, Whittles LK, Collins C, Bilek N, Bishop CJ, et al. Genomic analysis and comparison of two gonorrhea outbreaks. MBio. 2016;7:e00525–16. 10.1128/mBio.00525-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ota KV, Fisman DN, Tamari IE, Smieja M, Ng L-K, Jones KE, et al. Incidence and treatment outcomes of pharyngeal Neisseria gonorrhoeae and Chlamydia trachomatis infections in men who have sex with men: a 13-year retrospective cohort study. Clin Infect Dis. 2009;48:1237–43. 10.1086/597586 [DOI] [PubMed] [Google Scholar]


