Abstract

Kaixin Powder (KXP) is a classic formula for treating morbid forgetfulness in ancient China. To guarantee the efficacy and safety of KXP, a simple and accurate HPLC–DAD method has been established and validated for the quantitative analysis of seven bioactive compounds in KXP. Dehydrotumulosic acid (DTU) and dehydrotrametenolic acid (DTR) were quantified in KXP for the first time. Good chromatographic separation was conducted on a Kromasil 100-5 C18 column (250 mm × 4.6 mm, 5 μm) by gradient elution using mobile phases containing acetonitrile and 0.1% formic acid aqueous solution at different detection wavelengths. The calibration curves of each compound showed good linearity (r ≥ 0.9990), and the LOD and LOQ were in the ranges of 0.01–0.10 and 0.03–0.40 μg/mL, respectively. The relative standard deviations (RSDs) of intra-day and inter-day precisions were in the ranges of 0.45–1.74% and 0.56–2.32%, respectively. All recoveries were in the range of 93.6–105.5% with an RSD no more than 2.77%. These quantification results of seven compounds determined in the samples were further confirmed by HPLC–QTOF-MS/MS. This study provides a useful and simple method for analyzing the major bioactive compounds and improves the quality assessment research of KXP.
1. Introduction
Kaixin Powder (KXP) is a well-known formula for treating morbid forgetfulness in ancient China. Modern pharmacological research shows that KXP could significantly ameliorate the cognitive deficit symptoms and decrease the Aβ level in the hippocampus.1−3 To guarantee the efficacy of KXP, the quality evaluation of bioactive compounds contained in KXP is indispensable and significant.
KXP is comprised of Ginseng Radix et Rhizoma (the roots and rhizomes of Panax ginseng C. A. Mey.), Polygalae Radix (the roots of Polygala tenuifolia Willd.), Poria (the sclerotium of Poria cocos (Schw.) Wolf), and Acori Tatarinowii Rhizoma (the rhizomes of Acorus tatarinowii Schott) at a weight ratio of 1.3%:1.3%:64.9%:32.5%.4 Triterpene acids, volatile oils, xanthones, oligosaccharide esters, and saponins5,6 are representative bioactive compounds and the material basis in KXP and all the above compounds are also the key ones in our experiment.
Literature research showed that several quantitative analytical methods about KXP have already been established.7−9 Nevertheless, these methods mainly focused on quantifying the compounds from Ginseng Radix et Rhizoma and Polygalae Radix in KXP and neglected the bioactive ones in the other two herbs Poria and Acori Tatarinowii Rhizoma. Even though one method has detected the compounds from Poria, only one triterpene acid (pachymic acid) was selected as a quality marker of KXP.4 In our opinion, due to the high ratio of Poria and Acori Tatarinowii Rhizoma in KXP, their compounds are highly essential and representative in reflecting the quality of KXP. More triterpene acids and volatile oils should be detected to realize the comprehensive quality of KXP. Herein, a new and straightforward high-performance liquid chromatography with a diode array detector (HPLC–DAD) method has been established and validated for simultaneous quantitative analysis of seven bioactive compounds in KXP including three triterpene acids (dehydrotumulosic acid (DTU), pachymic acid (PAC), and dehydrotrametenolic acid (DTR)), two volatile oils (α-asarone (AAS) and β-asarone (BAS)), one xanthone (polygalaxanthone III (POL)), and one oligosaccharide ester (3,6′-disinapoyl sucrose (DISS)). Among them, DTU and DTR were quantified in KXP for the first time. In addition, due to the complicated matrix of KXP, it is insufficient to identify the peaks of target analytes in the sample chromatogram only by comparing to the retention time of reference standards. Therefore, for obtaining more reliable quantitative results, the samples were also analyzed using a high-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (HPLC–QTOF-MS/MS) method for further confirmation. This study provides a useful method for analyzing the major pharmacodynamic compounds and improves the quality assessment method of KXP.
2. Results and Discussion
2.1. Selection of Quality Markers of KXP
For the comprehensive quality evaluation of KXP, seven compounds, including DTU, PAC, DTR, AAS, BAS, POL, and DISS, were selected as quality markers. The chemical structures of these reference standards are shown in Figure 1. DTU, PAC, and DTR are three major bioactive triterpene acids in Poria, and they were usually selected as quality markers in the quality evaluation of Poria(10,11) and formulation containing Poria.12 Especially, PAC has been reported to induce autophagy concerning the IGF-1 signaling pathway in the aged cells13 and autophagy is closely related to the pathogenesis of AD.14 Similarly, the major bioactive compounds in Acori Tatarinowii Rhizoma are volatile oils, among which AAS and BAS are two representative and indispensable compounds in the quality evaluation of Acori Tatarinowii Rhizoma.15 Both AAS and BAS could ameliorate learning and memory abilities in AD rats.16 POL and DISS, the main xanthone and the main oligosaccharide ester in Polygalae Radix, respectively, are specified as the content determination compounds of Polygalae Radix in the Pharmacopoeia of the People’s Republic of China (2020 edition17). The previous reports indicated that POL and DISS exerted antidepressant-like effects18,19 and neuroprotective effects.20 In addition, saponins, main bioactive compounds of Polygalae Radix, were also chosen as target compounds of KXP at the beginning of our experiment; however, they could not be detected by HPLC–DAD due to their quite low ratio in the formulation and weak ultraviolet absorption. Thus, saponins were not selected as quality markers of KXP, and they could be further analyzed by the UPLC–MS/MS method with higher resolution and lower detection limit.21
Figure 1.
Chemical structures of seven bioactive compounds in KXP.
2.2. Optimization of HPLC Conditions
The HPLC conditions, involving the chromatographic column, mobile phase system, elution procedure, and detection wavelength, were improved to realize the good separation and high sensitivity of the seven target compounds in KXP. By comparing different columns, a Kromasil 100-5 C18 column was selected because peak shapes and the separation efficiency of the seven compounds were more satisfactory. Methanol, acetonitrile, and a variety of modifiers (including formic acid and phosphoric acid) were investigated for realizing better elution, and acetonitrile and formic acid aqueous solution were selected as mobile phases. Then, different concentrations of formic acid were compared, and 0.1% was more appropriate with a better peak shape. In addition, given that the fourth analytical peak (i.e., AAS) was hard to separate from the peak on the left, the flow rate was reduced from 1.00 mL/min to 0.80 mL/min, and the elution time of the low proportion of acetonitrile was prolonged to promote the separation of AAS. Then, to reduce the running time and improve the detection efficiency, the proportion of acetonitrile in the mobile phase was increased rapidly to shorten the elution time of low-polarity compounds. Moreover, according to the absorption characteristics of these seven compounds in the UV spectra, four UV wavelengths were chosen for detection, including 203 nm (PAC), 242 nm (DTU and DTR), 257 nm (AAS and BAS), and 320 nm (POL and DISS).
2.3. Optimization of Extraction Conditions
Ethanol is commonly used as an extraction solvent of TCM, and the ethanol extract of KXP was proved to exert neuroprotective and antidepressant effects on animal models.22,23 Therefore, ethanol was selected to extract the samples of KXP. To extract the compounds more efficiently, several concentrations of ethanol solutions (from 50 to 100%) were compared, and the optimal proportion of ethanol was found to be 70%, which produced more peaks with higher responses. In addition, for the HPLC analysis, the concentrations of methanol were also optimized, and the optimized preparation condition was to extract the dry extract with 80% methanol by ultrasonication at room temperature for 30 min.
2.4. Method Validation
The typical chromatograms of reference standards and KXP samples are presented in Figure 2, and the magnified HPLC spectra of Figure 2b for other six compounds are shown in Figure S1 of the Supporting Information. Each peak of target compounds in the KXP sample chromatogram did not interfere with other peaks. The resolutions and tailing factors of chromatographic peaks are shown in Table S1 of the Supporting Information, and the values met the requirements of FDA guidelines for validation of chromatographic methods, indicating that the developed HPLC–DAD method had good selectivity.
Figure 2.

HPLC–DAD chromatograms of KXP. (a) HPLC–UV chromatograms of seven major mixed standards; (b) HPLC–UV chromatogram of KXP samples: POL (1), DISS (2), BAS (3), AAS (4), DTU (5), PAC (6), and DTR (7).
The calibration curves with the regression equation, linear range, correlation coefficients (r), LOD, and LOQ of seven target compounds are listed in Table 1, and calibration curves are shown in Figure S2 of the Supporting Information. The calibration curves of seven reference standards showed good linearity with r no less than 0.9990 within their respective concentration ranges. The results of LOD and LOQ were in the ranges of 0.01–0.10 and 0.03–0.40 μg/mL, respectively.
Table 1. Linearity, LOD, and LOQ of Seven Bioactive Compounds in KXP.
| compound | regression equation | r | linear range (μg/mL) | LOD (μg/mL) | LOQ (μg/mL) |
|---|---|---|---|---|---|
| POL | y = 28.35x – 0.477 | 0.9998 | 0.20–10.0 | 0.05 | 0.10 |
| DISS | y = 33.44x + 16.31 | 0.9990 | 0.60–60.0 | 0.01 | 0.03 |
| AAS | y = 129.6x + 19.71 | 0.9999 | 1.00–50.0 | 0.10 | 0.20 |
| BAS | y = 89.86x – 92.17 | 0.9999 | 4.00–200 | 0.08 | 0.20 |
| DTU | y = 26.97x + 31.52 | 0.9999 | 1.00–100 | 0.10 | 0.40 |
| PAC | y = 16.53x + 13.46 | 0.9999 | 1.00–200 | 0.10 | 0.40 |
| DTR | y = 50.32x – 6.340 | 0.9997 | 0.20–10.0 | 0.05 | 0.10 |
As shown in Table 2, the RSDs of the intra-day and inter-day precisions were no more than 1.74 and 2.32%, respectively. The RSD of repeatability was no more than 1.84%. In the stability evaluation, RSD was no more than 1.73%, indicating the good stability of the established method. As shown in Table 3, the recovery of sample preparation was acceptable in the range of 93.6–105.5%, and RSD was no more than 2.77%.
Table 2. Precision, Repeatability, Stability, and Accuracy of Seven Bioactive Compounds in KXP (n = 6).
| precision |
||||
|---|---|---|---|---|
| compound | intra-day RSD (%) | inter-day RSD (%) | repeatability RSD (%) | stability RSD (%) |
| POL | 1.74 | 1.00 | 1.25 | 1.18 |
| DISS | 1.20 | 2.32 | 1.84 | 1.62 |
| AAS | 0.98 | 1.36 | 1.00 | 1.23 |
| BAS | 0.45 | 1.98 | 0.80 | 0.36 |
| DTU | 1.61 | 0.56 | 0.65 | 1.73 |
| PAC | 1.00 | 1.30 | 0.78 | 1.16 |
| DTR | 0.92 | 1.32 | 0.84 | 1.55 |
Table 3. Recovery of Seven Bioactive Compounds in KXP (n = 6).
| compound | initial amount (μg) | spiked amount (μg) | detected amount (μg, mean ± SD) | recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| POL | 0.75 | 0.75 | 1.47 ± 0.01 | 96.8 | 2.03 |
| DISS | 8.54 | 8.50 | 17.5 ± 0.19 | 105.5 | 1.94 |
| AAS | 7.97 | 8.00 | 16.1 ± 0.20 | 100.8 | 2.34 |
| BAS | 83.9 | 80.0 | 166 ± 1.61 | 102.5 | 2.77 |
| DTU | 24.4 | 24.0 | 49.5 ± 0.17 | 104.5 | 1.62 |
| PAC | 34.1 | 34.0 | 65.9 ± 0.50 | 93.6 | 1.57 |
| DTR | 0.96 | 0.96 | 1.91 ± 0.01 | 99.2 | 0.86 |
2.5. HPLC–QTOF-MS/MS Confirmation
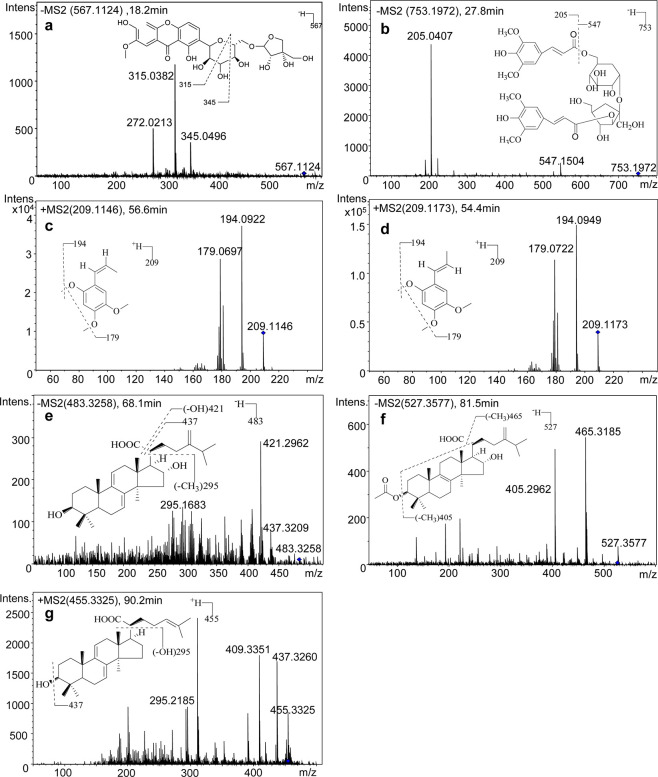
For further confirming the peaks of seven target compounds in the sample chromatograms, the KXP samples were also analyzed with the HPLC–QTOF-MS/MS method. The accurate masses of quasi-molecular ions and major fragment ions are shown in Table 4, and the MS/MS spectra are shown in Figure 3a–g. The results showed that the calculated error values between measured and theoretical values of accurate masses were all in the range of ±5 ppm. In addition, the fragmentation characteristics of seven identified compounds were the same as described in the previous reports.24−27,28
Table 4. Mass Data and Major Fragments of the Seven Bioactive Compounds in KXP Acquired by HPLC–QTOF-MS.
| compound | RT (min, mean ± SD) | formula | theoretical mass (m/z) | measured mass (m/z) | error (ppm) | major fragments (m/z) |
|---|---|---|---|---|---|---|
| POL | 18.2 ± 0.10 | C25H28O15 | 567.1350 | 567.1124 | –2.47 | 345.0496, 315.0382 |
| DISS | 27.8 ± 0.05 | C34H42O19 | 753.2242 | 753.1972 | –3.72 | 547.1504, 205.0407 |
| AAS | 56.6 ± 0.05 | C12H16O3 | 209.1178 | 209.1146 | 1.91 | 194.0922, 179.0697 |
| BAS | 54.4 ± 0.05 | C12H16O3 | 209.1178 | 209.1173 | 0.48 | 194.0949, 179.0722 |
| DTU | 68.1 ± 0.05 | C31H48O4 | 483.3474 | 483.3258 | –1.45 | 421.2962, 295.1683 |
| PAC | 81.5 ± 0.10 | C33H52O5 | 527.3737 | 527.3577 | 2.28 | 465.3185, 405.2962 |
| DTR | 90.2 ± 0.20 | C30H46O3 | 455.3525 | 455.3325 | 1.10 | 437.3260, 295.2185 |
Figure 3.
MS/MS spectra of seven bioactive compounds in KXP: POL (a), DISS (b), AAS (c), BAS (d), DTU (e), PAC (f), and DTR (g).
POL (peak 1, 18.2 min) showed a major molecular ion at m/z 567.1124 [M – H]−, indicating the molecular formula of C25H28O15. In Figure 3a, fragment ions at m/z 345.0496 and 315.0382 were generated by the cleavage of the cross ring, which were consistent with the data in the previous literature.24,25
DISS (peak 2, 27.8 min) showed an [M – H]− ion at m/z 753.1972, indicating the molecular formula of C34H41O19. In the MS/MS spectrum (Figure 3b), fragment ions at m/z 547.1507 and 205.0508 were assigned as [M – H – sinapic acid]− and [sinapic acid – H – H2O]−, respectively. Therefore, peak 2 was identified as DISS.24,25
The isomers BAS (peak 3, 54.4 min) and AAS (peak 4, 56.6 min) had the same molecular formula of C12H16O3, and BAS was eluted earlier than AAS due to higher polarity. AAS and BAS produced [M + H]+ ions at m/z 209.1146 and 209.1173 in the positive mode, respectively. As shown in Figure 3c,d, the m/z 194 [M + H – CH3]+ ion and the m/z 179 [M + H – 2CH3]+ ion were due to the loss of the methyl group. The fragmentation characteristics were the same as previously reported.26
DTU (peak 5, 68.1 min) presented a precursor ion at m/z 483.3258 [M – H]−. In Figure 3e, the m/z 437.3209 [M – H – HCOOH]− ion and the m/z 421.2962 [M – H – COOH – OH]− ion resulted from the loss of COOH or OH. In addition, a fragment ion at m/z 295.1683 [M – H – 2CH3 – C9H15O2]− was derived from the loss of side chain (C9H15O2) on the D ring and two methyl substituents. The fragmentation characteristics were the same as previously reported.27
For PAC (peak 6, 81.5 min), a precursor ion was observed at m/z 527.3577 [M – H]−. In Figure 3f, the fragment ion with an m/z value of 465.3185 was derived from the loss of the hydroxyl group (−OH) and the carboxyl group (−COOH), and the fragment ion at m/z 405.2962 was generated from the consequent loss of CH3COOH, COOH, and a single equivalent of H2O. The fragmentation characteristics were the same as previously reported.27
DTR (peak 7, 90.2 min) showed a precursor ion at m/z 455.3325 [M + H]+, indicating a molecular formula of C30H46O3. In Figure 3g, product ions with m/z values of 437.3260 [M + H – OH]+ and 295.2185 [M + H – H2O – C8H13O2]+ were respectively designated to the loss of the hydroxyl group and the side chain (C8H13O2) in the D ring, which were similar to a previous report.28
In summary, the seven target compounds in KXP were further identified according to the extract mass information and fragmentation characteristics obtained by HPLC–QTOF-MS analysis.
2.6. Sample Determination
The established and validated HPLC–DAD method was applied to determine seven bioactive compounds in three batches of KXP, including POL, DISS, AAS, BAS, DTU, PAC, and DTR. Each sample was triple-analyzed, and mean contents of each compound are presented in Table 5. The results of sample determination indicated that the contents of DISS, AAS, BAS, DTU, and PAC in KXP samples were higher, while the contents of POL and DTR were quite low. The contents of these compounds were affected not only by the content of each compound contained in the original medicinal material but also by the compatibility ratio of each medicinal material in KXP. Although POL and DTR were low in content, they may also contribute to the material basis for the efficacy of KXP. Additionally, the contents of seven bioactive compounds in three batches of KXP samples were compared with GraphPad Prism v6.0 software by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test, and the results showed that there were no significant differences (p <0.05) among different batches and the quality of KXP was stable.
Table 5. Contents of Seven Bioactive Compounds in KXP Determined by HPLC–DAD (Mean ± SD, μg/g, n = 3).
| content (μg/g) |
|||
|---|---|---|---|
| compound | batch 1 | batch 2 | batch 3 |
| POL | 9.28 ± 0.17 | 8.87 ± 0.10 | 9.02 ± 0.06 |
| DISS | 102 ± 1.80 | 107 ± 0.68 | 108 ± 1.07 |
| AAS | 97.1 ± 0.58 | 99.2 ± 1.43 | 98.8 ± 0.52 |
| BAS | 1033 ± 13.94 | 1044 ± 11.25 | 1037 ± 1.99 |
| DTU | 298 ± 1.71 | 292 ± 2.80 | 291 ± 0.62 |
| PAC | 410 ± 5.43 | 422 ± 1.14 | 419 ± 4.22 |
| DTR | 11.7 ± 0.17 | 11.8 ± 0.19 | 11.6 ± 0.17 |
3. Conclusions
In this study, a reliable and accurate HPLC–DAD method has been established and validated to simultaneously quantify seven bioactive compounds in KXP for the first time, including POL, DISS, AAS, BAS, DTU, PAC, and DTR. Moreover, the samples were further analyzed using the HPLC–QTOF-MS/MS method to further confirm the identification results of each target compound. However, saponins had not been detected in samples by HPLC–DAD because of low content and poor ultraviolet absorption. Therefore, in upcoming research, we intend to utilize a sensitive and fast LC–MS/MS method to analyze more compounds in KXP for the establishment of a more effective quality evaluation method. In a word, this study improved the comprehensive quality evaluation standard of KXP to some extent, which will be helpful to guarantee the effectiveness and facilitate a further study of the pharmacokinetics and pharmacological mechanism of KXP.
4. Materials and Methods
4.1. Chemicals and Reagents
Chinese herbs Ginseng Radix et Rhizoma (lot number: 20120523), Poria (lot number: 17111504), and Acorus Tatarinowii Rhizome (lot number: 17110103) were obtained from Beijing Nengji Chinese Medicine Pieces Co. Ltd. (Beijing, China); Polygalae Radix (lot number: 16080803) was purchased from Beijing Minghui Hengtong Pharmaceutical Co. Ltd. (Beijing, China). The above four herbs were authenticated by Associate Professor Jia Li (School of Traditional Chinese Medicine, Capital Medical University).
Reference standards of DISS (purity, 98%), DTU (purity, 98%), PAC (purity, 97%), and DTR (purity, 98%) were obtained from Shanghai Yuanye Biotechnology Co. Ltd. (Shanghai, China), POL (purity, 97.8%) and BAS (purity, 98%) were acquired from China National Institutes for Food and Drug Control (Beijing, China), and AAS (purity, 98%) was obtained from Chengdu Push Bio-technology Co. Ltd. (Chengdu, Sichuan, China).
HPLC-grade acetonitrile was provided by Thermo Fisher Scientific Co. Ltd. (Waltham, Massachusetts, USA). Pure water was obtained from Hangzhou Wahaha Group Co. Ltd. (Hangzhou, Zhejiang, China). Ethanol, methanol, and formic acid were all of analytical grade and were acquired from Beijing Chemical Works (Beijing, China).
4.2. HPLC–DAD Analysis Conditions
Chromatographic analysis was developed on an Agilent 1260 Infinity LC system (Agilent Technologies, USA) with a diode array detector, an autosampler, a quaternary pump, and a thermostatic column oven. All separations were achieved on a Kromasil 100-5 C18 column (250 mm × 4.6 mm, 5 μm). Acetonitrile (A) and 0.1% formic acid aqueous solution (B) were used as mobile phases, and the linear gradient elution was applied as follows: 0–15 min, 15% A; 15–30 min, 15–35% A; 30–60 min, 35–60% A; 60–67 min, 60–80% A; 67–85 min, 80% A; 85–90 min, 80–95% A; 90–100 min, 95%A; 100–100.1 min, 95–15% A; 100.1–110 min, 15% A. The other parameters were as follows: flow rate, 0.8 mL/min; column temperature, 35 °C; injection volume, 20 μL. Also, the detection wavelengths were 203 nm (PAC), 242 nm (DTU and DTR), 257 nm (AAS and BAS), and 320 nm (POL and DISS) where the analyzed compounds had their maximum response of ultraviolet spectrum, respectively.
4.3. HPLC–QTOF-MS/MS Analysis Conditions
For further confirming the peaks of seven target compounds in the sample chromatograms, the KXP samples were also analyzed using the HPLC–QTOF-MS/MS method. The HPLC apparatus was equipped with a micrOTOF-Q MS (Bruker, Germany) using an electrospray ionization (ESI) source. The HPLC conditions were implemented as above, and mass spectrometry ran in both positive and negative ESI modes. The parameters of mass spectrometry were set as follows: mass range, 50–2000 Da; drying gas flow rate, 8.0 L/min; drying gas temperature, 180 °C; nebulizer, 0.8 bar; capillary, 4500 V; fragmentation voltage, 130 V; collision energy, 15–50 V. Also, the data acquisition and analysis were performed on Bruker Compass DataAnalysis 4.0 (Bruker, Germany).
4.4. Preparation of Standard Solutions
Accurately weighed amounts of seven standards were prepared to stock standard solutions by dissolving in a certain volume of methanol separately. Then, the stock standard solutions were combined into one standard solution and the concentration of each standard was as follows: 0.4 mg/mL for BAS, 0.2 mg/mL for DTU and PAC, 0.05 mg/mL for DISS and AAS, and 0.01 mg/mL for POL and DTR.
4.5. Preparation of Sample Solutions
The four herbs of KXP were crushed and sifted through a 24-mesh sieve. Then, the powder was mixed together with 1 g of Ginseng Radix et Rhizoma, 1 g of Polygalae Radix, 50 g of Poria, and 25 g of Acori Tatarinowii Rhizoma and refluxed two times in 10-fold 70% ethanol (1:10, w/v) at 150 °C for 2 h. Next, the extracts were filtered and evaporated to dryness under vacuum. The dry extracts were stored at 4 °C. For the HPLC analysis, 0.5 g of KXP extract (equal to 3.28 g of crude drugs) was weighed and extracted in 20 mL of 80% methanol by ultrasonication (power: 500 W; ultrasonic frequency: 40 kHz) for 30 min. Then, the extraction solution was filtered with 0.22 μm membranes.
4.6. Method Validation
4.6.1. Calibration Curves, LOD, and LOQ
The calibration curves were carried out by analyzing a series of standard solutions, which were obtained by diluting the stock standard solutions into an appropriate concentration with methanol. The linear calibration curves for seven compounds were calculated with at least six different concentrations of standard solutions by plotting peak areas (y) versus the corresponding concentrations (x), and the linear regression equation and correlation coefficient (r) of each compound were obtained. The limit of detection (LOD) and limit of quantification (LOQ) of seven compounds were separately determined by the concentrations of diluted standard solutions at the signal-to-noise (S/N) ratios of 3:1 and 10:1.
4.6.2. Precision, Repeatability, and Stability
Intra-day and inter-day precisions were determined by analyzing one standard solution mixture for six times in 1 day and in 3 consecutive days, respectively. The peak area of each compound in six samples was recorded, and the results of RSD were calculated as the intra-day and inter-day precisions.
The repeatability was determined by the analysis of six samples of KXP prepared according to the preparation procedure of sample solutions. The peak area of each compound in six samples was recorded and the results of RSD were calculated.
The stability was evaluated by determining one sample solution at 0, 2, 4, 8, 12 and 24 h after initial storage at room temperature. The peak area of each compound in the sample at six time points was recorded and the results of RSD were calculated.
4.6.3. Accuracy
To ensure the accuracy of the quantitative analysis method, the recovery test was carried out by adding accurate amounts of seven reference standards (100% of the initial amount of each compound in the KXP sample) to KXP samples of known concentration with six replicates. Then, the mixture was prepared and analyzed using the established method. The spike recoveries were calculated by the formula as follows: spiked recovery (%) = (detected amount – initial amount)/spiked amount × 100%. Then, the average of recoveries and the RSD of six replicates were calculated.
4.7. HPLC–QTOF-MS/MS Confirmation
Due to the complexity system of KXP, all samples were also analyzed by HPLC–QTOF-MS/MS to further confirm that the identified peaks in the sample chromatogram indeed represented the determined compounds. Specifically, the reliability of the results was verified by comparing the exact molecular weight of precursor ions and the characteristics of fragment ions of the reference standards and the samples.
Acknowledgments
This work was financially supported by the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (no. CIT&TCD201504098) and Beijing Natural Science Foundation (no. 7182019).
Glossary
Abbreviations
- AD
Alzheimer’s disease
- TCM
traditional Chinese medicine
- KXP
Kaixin Powder
- HPLC–DAD
high-performance liquid chromatography with a diode array detector
- HPLC–QTOF-MS/MS
high-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry
- AAS
α-asarone
- BAS
β-asarone
- DISS
3,6′-disinapoyl sucrose
- DTR
dehydrotrametenolic acid
- DTU
dehydrotumulosic acid
- PAC
pachymic acid
- POL
polygalaxanthone III
- LOD
limit of detection
- LOQ
limit of quantification
- RSD
relative standard deviation
- ESI
electrospray ionization
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00289.
The authors declare no competing financial interest.
Supplementary Material
References
- Wang N.; Jia Y.; Zhang B.; Li Y.; Murtaza G.; Huang S.; Liu X. Kai-Xin-San, a Chinese Herbal Decoction, Accelerates the Degradation of β-Amyloid by Enhancing the Expression of Neprilysin in Rats. J. Evidence-Based Complementary Altern. Med. 2020, 2020, 3862342. 10.1155/2020/3862342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L.; Li H.; Tian J.; Jin N.; Zhang J.; Yang F.; Zhou L.; Wang Q.; Huang Z. The traditional formula Kai-Xin-San alleviates polyglutamine-mediated neurotoxicity by modulating proteostasis network in caenorhabditis elegans. Rejuvenation Res. 2020, 23, 207–216. 10.1089/rej.2018.2149. [DOI] [PubMed] [Google Scholar]
- Xu Y. M.; Wang X. C.; Xu T. T.; Li H. Y.; Hei S. Y.; Luo N. C.; Wang H.; Zhao W.; Fang S. H.; Chen Y. B.; Guan L.; Fang Y. Q.; Zhang S. J.; Wang Q.; Liang W. X. Kai Xin San ameliorates scopolamine-induced cognitive dysfunction. Neural Regener. Res. 2019, 14, 794–804. 10.4103/1673-5374.249227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K. Y.; Fu Q.; Xie H. Q.; Xu S. L.; Cheung A. W. H.; Zheng K. Y. Z.; Luk W. K. W.; Choi R. C. Y.; Lau D. T. W.; Dong T. T. X.; Jiang Z. Y.; Chen J. J.; Tsim K. W. K. Quality assessment of a formulated Chinese herbal decoction, Kaixinsan, by using rapid resolution liquid chromatography coupled with mass spectrometry: A chemical evaluation of different historical formulae. J. Sep. Sci. 2010, 33, 3666–3674. 10.1002/jssc.201000498. [DOI] [PubMed] [Google Scholar]
- Sun H.; Liu C.; Zhang A. H.; Han Y.; Yan G. L.; Wang P.; Wang X. J. Rapid discovery and global characterization of multiple constituents from Kai-Xin-San using an integrated MSE data acquisition mode strategy based on ultra-performance liquid chromatography coupled to electrospray ionization/quadrupole-time-of-flight mass spectrometry. Anal. Methods 2015, 7, 279–286. 10.1039/C4AY01954G. [DOI] [Google Scholar]
- Zhang X.; Li Q.; Lv C.; Xu H.; Liu X.; Sui Z.; Bi K. Characterization of multiple constituents in Kai-Xin-San prescription and rat plasma after oral administration by liquid chromatography with quadrupole time-of-flight tandem mass spectrometry. J. Sep. Sci. 2015, 38, 2068–2075. 10.1002/jssc.201500123. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Mu L. H.; Zhao R. Q.; Li M. H.; Wang X. D.; Liu P. Determination of three major components in Kaixin San by HPLC. Chin. J. Exp. Tradit. Med. Formulae 2016, 4, 65–68. 10.13422/j.cnki.syfjx.2016040065. [DOI] [Google Scholar]
- Dai Y.; Jiang Y.-Y.; Chen X.-H.; Shi R.-B. Quality control of Kaixinsan based on drug-like components,3,6’-disinapoylsucrose and α-asarone. J. Beijing Univ. Tradit. Chin. Med. 2011, 34, 254–257. [Google Scholar]
- Li H.-J.; Zhang L.; Quan J.-Y.; Yang S.-J.; Yang Y.; Chen W.; Sun D.-H.; Feng D.; Jiang Y.-Y.; Shi R.-B. Quality representation and correlation analysis of medicinal preparation of Kaixin San based on drug system. J. Beijing Univ. Tradit. Chin. Med. 2015, 38, 539–545. 10.3969/j.issn.1006-2157.2015.08.008. [DOI] [Google Scholar]
- Tian S. S.; Liu X. Q.; Feng W. H.; Zhang Q. W.; Yan L. H.; Wang Z. M.; Gao L. Quality evaluation of Poria based on specific chromatogram and quantitative analysis of multicomponents. China J. Chin. Mater. Med. 2019, 44, 1371. 10.19540/j.cnki.cjcmm.20181220.005. [DOI] [PubMed] [Google Scholar]
- Zhang G.; Wang H.; Xie W.; Wang Q.; Wang X.; Wang C.; Du Y.; Huo C.; Wang Q. Comparison of triterpene compounds of four botanical parts from Poria cocos (Schw.) wolf using simultaneous qualitative and quantitative method and metabolomics approach. Food Res. Int. 2019, 121, 666–677. 10.1016/j.foodres.2018.12.036. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Gao X.; Yan G.; Zhang A.; Sun H.; Han Y.; Li W.; Liu L.; Wang X. Chinmedomics facilitated quality-marker discovery of Sijunzi decoction to treat spleen qi deficiency syndrome. Front. Med. 2020, 14, 335. 10.1007/s11684-019-0705-9. [DOI] [PubMed] [Google Scholar]
- Lee S. G.; Kim M. M. Pachymic acid promotes induction of autophagy related to IGF-1 signaling pathway in WI-38 cells. Phytomedicine 2017, 36, 82–87. 10.1016/j.phymed.2017.09.020. [DOI] [PubMed] [Google Scholar]
- Park H.; Kang J. H.; Lee S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. 10.3390/ijms21093369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Zhang B.; Xin Z.; Ren D.; Yi L. GC-MS Fingerprinting Combined with Chemometric Methods Reveals Key Bioactive Components in Acori Tatarinowii Rhizoma. Int. J. Mol. Sci. 2017, 18, 1342. 10.3390/ijms18071342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Gao X.; Liu Q.; Zeng L.; Zhang K.; Mu K.; Zhang D.; Zou H.; Wu N.; Ou J.; Wang Q.; Mao S. Alpha-asarone improves cognitive function of aged rats by alleviating neuronal excitotoxicity via GABAA receptors. Neuropharmacology 2020, 162, 107843. 10.1016/j.neuropharm.2019.107843. [DOI] [PubMed] [Google Scholar]
- Pharmacopoeia of the People’s Republic of China; National Medical Products Administration, 2020. [Google Scholar]
- Hu Y.; Liu M.; Liu P.; Guo D. H.; Wei R. B.; Rahman K. Possible mechanism of the antidepressant effect of 3,6′-disinapoyl sucrose from Polygala tenuifolia Willd. J. Pharm. Pharmacol. 2011, 63, 869–874. 10.1111/j.2042-7158.2011.01281.x. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Liao H.-B.; Dai-Hong G.; Liu P.; Wang Y.-Y.; Rahman K. Antidepressant-like effects of 3,6′-disinapoyl sucrose on hippocampal neuronal plasticity and neurotrophic signal pathway in chronically mild stressed rats. Neurochem. Int. 2010, 56, 461–465. 10.1016/j.neuint.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Li J.; Liu P.; Chen X.; Guo D. H.; Li Q. S.; Rahman K. Protection of SH-SY5Y neuronal cells from glutamate-induced apoptosis by 3,6′-disinapoyl sucrose, a bioactive compound isolated from Radix Polygala. J. Biomed. Biotechnol. 2012, 2012, 1–5. 10.1155/2012/728342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C.; Li Q.; Zhang X.; He B.; Xu H.; Yin Y.; Liu R.; Liu J.; Chen X.; Bi K. Simultaneous quantitation of polygalaxanthone III and four ginsenosides by ultra-fast liquid chromatography with tandem mass spectrometry in rat and beagle dog plasma after oral administration of Kai-Xin-San: application to a comparative pharmacokinetic study. J. Sep. Sci. 2014, 37, 1103–1110. 10.1002/jssc.201400058. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Chao C.; Duan X.; Cheng X.; Liu P.; Su S.; Duan J.; Dong T. T.; Tsim K. W.-K. Kai-Xin-San series formulae alleviate depressive-like behaviors on chronic mild stressed mice via regulating neurotrophic factor system on hippocampus. Sci. Rep. 2017, 7, 1467. 10.1038/s41598-017-01561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Jia Y. M.; Zhang B.; Xue D.; Reeju M.; Li Y.; Huang S. M.; Liu X. W. Neuroprotective mechanism of Kai Xin San: upregulation of hippocampal insulin-degrading enzyme protein expression and acceleration of amyloid-beta degradation. Neural Regen. Res. 2017, 12, 654–659. 10.4103/1673-5374.205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q.; Chen J.; Zhou Q.; Lei H.; Luan L.; Liu X.; Wu Y. Indirect identification of antioxidants in Polygalae Radix through their reaction with 2,2-diphenyl-1-picrylhydrazyl and subsequent HPLC-ESI-Q-TOF-MS/MS. Talanta 2015, 144, 830–835. 10.1016/j.talanta.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Yang C.; Yin X.; Dong X.; Zhang X.; You L.; Wang W.; Wang J.; Chen Q.; Ni J. Determination of the phytochemical composition of Jingning Fang and the in vivo pharmacokinetics of its metabolites in rat plasma by UPLC-MS/MS. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2017, 1067, 71–88. 10.1016/j.jchromb.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Deng Z.; Zhang F.; Sun P.; Li J.; Zheng W. Development of an LC-MS/MS method for quantification of two isomeric phenylpropenes and the application to pharmacokinetic studies in rats. Biomed. Chromatogr. 2018, 32, e4115 10.1002/bmc.4115. [DOI] [PubMed] [Google Scholar]
- Wu L. F.; Wang K. F.; Mao X.; Liang W. Y.; Chen W. J.; Li S.; Qi Q.; Cui Y. P.; Zhang L. Z. Screening and Analysis of the Potential Bioactive Components of Poria cocos (Schw.) Wolf by HPLC and HPLC-MS(n) with the Aid of Chemometrics. Molecules 2016, 21, 227. 10.3390/molecules21020227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Xu J.; Zhang S.; Wang R.; Huang Q.; Chen H.; Dong X.; Zhao Z. Qualitatively and quantitatively comparing secondary metabolites in three medicinal parts derived from Poria cocos (Schw.) Wolf using UHPLC-QTOF-MS/MS-based chemical profiling. J. Pharm. Biomed. Anal. 2018, 150, 278–286. 10.1016/j.jpba.2017.11.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.