Abstract
During pandemics (e.g., COVID-19) physicians have to focus on diagnosing and treating patients, which often results in that only a limited amount of labeled CT images is available. Although recent semi-supervised learning algorithms may alleviate the problem of annotation scarcity, limited real-world CT images still cause those algorithms producing inaccurate detection results, especially in real-world COVID-19 cases. Existing models often cannot detect the small infected regions in COVID-19 CT images, such a challenge implicitly causes that many patients with minor symptoms are misdiagnosed and develop more severe symptoms, causing a higher mortality. In this paper, we propose a new method to address this challenge. Not only can we detect severe cases, but also detect minor symptoms using real-world COVID-19 CT images in which the source domain only includes limited labeled CT images but the target domain has a lot of unlabeled CT images. Specifically, we adopt Network-in-Network and Instance Normalization to build a new module (we term it NI module) and extract discriminative representations from CT images from both source and target domains. A domain classifier is utilized to implement infected region adaptation from source domain to target domain in an Adversarial Learning manner, and learns domain-invariant region proposal network (RPN) in the Faster R-CNN model. We call our model NIA-Network (Network-in-Network, Instance Normalization and Adversarial Learning), and conduct extensive experiments on two COVID-19 datasets to validate our approach. The experimental results show that our model can effectively detect infected regions with different sizes and achieve the highest diagnostic accuracy compared with existing SOTA methods.
Keywords: COVID-19 diagnosis, Semi-supervised learning, Adversarial learning, Network-in-Network, Instance normalization
1. Introduction
1.1. Background
The outbreak of COVID-19 has spread quickly [1] over the world, resulting in a severe public health crisis. Currently, diagnosing COVID-19 disease relies on reverse-transcription polymerase chain reaction (RT-PCR), IgM-IgG antibody test and Computed Tomography (CT) [2], [3], [4], [5]. Considering the limited medical resources and the high false negative rates of RT-PCR [6], physicians usually rely on the later two methods to diagnose COVID-19 patients and adopt different strategies to treat infected individuals. People with positive IgM-IgG but CT holding no infected regions are suggested to be isolated in their house. Only people with positive IgM-IgG and infected regions within CT need to be hospitalized for treatment. Moreover, inpatients have to check their lungs regularly [2]. Obviously, CT image is not only an important diagnosis tool, but also useful to determine treatment for COVID-19 patients.
In general, minor infections in COVID-19 lung CT images show the interstitial changing and patchy shadows [2]. People with middle-level symptoms show bilateral multilobar ground-glass opacification and infiltrating shadow [7], while patients with severe symptoms have bilateral with diffuse infiltration of all segments of their lungs [8], [9]. These characteristics of COVID-19 implicitly indicate that we can employ a deep learning model (e.g., Faster R-CNN [10]) to assess the infection degree, and prescribe drugs according to different infections. Accurate detecting on the COVID-19 CT images can significantly lighten the burden of physicians, given that physicians overwork heavily during outbreak of pandemics.
However, the COVID-19 is an outbreak with large scale and spreads by droplet transmission and fomite transmission due to the characteristics of COVID-19 with high infection [11]. Under such scenarios, physicians give their attention to diagnose and treat patients, and have no extra time to label a large number of CT images which is a requirement of successfully training a deep learning diagnosis model with supervised learning strategy [12], [13], [14], [15], [16], [17]. The semi-supervised learning strategy [18] may help train a powerful machine learning model with a small amount of labeled CT images and a large amount of unlabeled CT images, and the state-of-the-art semi-supervised object detection models (e.g., Domain adaptive Faster R-CNN [19] and Few-Shot Adaptive Faster R-CNN [20]) have been validated on the public scenery datasets. However, they have challenges to deal with real-world COVID-19 CT case. Those algorithms cannot detect the small infected regions in COVID-19 CT images. This is because those algorithms focus on the overall distribution of data, ignoring the details of a single instance. Moreover, CT images are represented by different gray levels, which reflects the absorption of X-ray by organs and tissues. For example, bright regions within CT represents high density regions, while shadow regions refers to low density regions. If the infections of small size are similar to the healthy tissues, traditional algorithms may mis-classify infections as healthy tissues.
Detecting small infected regions in real-world COVID-19 CT images is very important. This is because the infections with small regions often correspond to minor symptoms, and may develop to severe symptoms. The aggravation occurs usually within 4 to 9 days [21], [22] and the death rate for COVID-19 patients with severe symptom is over 67% [23], [24]. Since CT images are usually from different hospitals (or CT machines) in practice, and deep learning researchers always assume images from different hospitals (or CT machines) as different domains [25], [26], and detecting the small infection regions in different domains becomes more difficult under such a scenario. Considering the significance of challenges of aforementioned models and diagnosis burden of physicians as well as rapid aggravation from minor symptoms to severe symptoms, it is critical to develop a new approach to effectively detect COVID-19 infected regions, especially for minor symptoms.
1.2. NIA-Network description
In this paper, towards this crucial real-world problem, we construct a new module (we call it NI-Module) with multiple Network-in-Network (NIN) [27], [28] and Instance Normalization (IN) [29] layers to capture non-linear concepts and to normalize representations of CT, and utilize a Conv-based domain classifier to implement infected region adaption from source domain to target domain in an Adversarial Learning manner and to learn domain-invariant region proposal network (RPN) in a Faster R-CNN model [10], given that only limited CT images within source domain are labeled while a lot of CT images within target domain are unlabeled and they are taken by different CT machines. Although our proposed approach is built on existing work (i.e., Network-in-Network, Instance Normalization, GRL and Faster R-CNN), the synergic integration of these components into an effective architecture holds novelty. First, the goal of this paper is to detect infected symptoms in CT images, especially for minor symptoms. Hence, each pixel cannot be ignored, because a minor symptom may be encoded by several pixels in CT images. The study “Domain adaptive Faster R-CNN” focuses on the overall distribution of data samples, ignoring details of each CT image. Our proposed approach adopts Instance Normalization strategy, which can help a model attend to pixel-level information in each CT image. Second, feature extractor in our NIA-Network achieves better extracting performance than “Domain adaptive Faster R-CNN”, because we adopt Network-in-Network module which captures non-linear property of data representations, given that representations of data samples are usually lie on a non-linear manifold [27]. Finally, our approach does not require images within the target domain to hold labels. Other studies (e.g., Domain adaptive Faster R-CNN) requires images within target domain holding labels, otherwise they will result in low detection quality. This improvement has significant impact in practice as high-qualify labeled data is often scarce especially in medical domain. More details are shown in experiment section.
Specifically, first we randomly select two images from source and target domains separately, and feed them into an extractor to extract representations. The goal of extractor is to capture shift-invariant of objects within CT images, because CT images may be from different CT machines and different CT machines may have different imaging protocols [25], [30]. In this way, the extractor is usually a neural network (ResNet [31] in our paper). Then, we construct a 9-layer network to implement the NI-Module and feed the representations into this module to extract non-linear concepts with NIN and capture unique details of each instance to obtain discriminative features with IN. After that, a classifier is employed to implement infected regions adaptation from source domain to target domain in an Adversarial Learning manner, with Gradient Reversal Layer (GRL) [32]. Such an operation can transfer labels from source domain to target domain if representations of CT images from source domain are similar to those from target domain. Finally, Faster R-CNN learns domain-invariant region proposal network, outputting the framed result. We call our approach NIA-Network. In this way, we can effectively detect COVID-19 infected regions in the target images without labels even if an infected area is very small.
In summary, our contributions are as follows.
-
•
This paper proposes a new approach, named NIA-Network, to significantly improve detection performance in unlabeled target domain, providing new insights into the success of semi-supervised learning strategy in real-world COVID-19 case.
-
•
This paper demonstrates how to combine Network-in-Network model with Instance Normalization strategy to construct a new NI-Module, and how to implement infected region adaptation with a domain classifier in an Adversarial Learning manner.
-
•
Extensive experiments using two COVID-19 datasets demonstrate that our approach consistently improves detection performance, outperforming existing SOTA methods.
The rest of this paper is organized as follows. In Section 2 we discuss existing work. We then review ResNet, NIN, Faster R-CNN, Instance Normalization and adversarial learning in Section 3. In Section 4, we present our NIA-Network. In Section 5, we show our experimental results. Section 6 is our conclusion.
2. Related work
Object detection method has been widely used in many real-world applications. Here, we group them into two categories according to the level of automacy.
Supervised learning object detection. You Only Look Once (YOLO) [33] views object detection as a regression problem to spatially separate bounding boxes and to associate class probabilities. A single neural network directly predicts bounding boxes and class probabilities from full images in one evaluation. The unified architecture can help accelerate detection process in an end-to-end manner. However, YOLO makes more localization errors but is less likely to predict false positives on background [33]. Single Shot MultiBox Detector (SSD) [34] also uses a single deep neural network to detect objects. Different from YOLO, SSD discretizes output space of bounding boxes into a set of default boxes over different aspect ratios and scales per feature map location. The model predicts multiple feature maps with different resolutions to naturally handle objects of various sizes. However, SSD strategy often fails to detect small objects, because we need a large of feature maps to provide more fine features and do more intensive sampling. Also, it needs a powerful semantic tool to segment front object from background, and this is a non-trivial task in practice.
To improve detection performance, Faster R-CNN [10] model with Region Proposal Network (RPN) that shares full-image convolutional features with the detection network has been introduced, thus enabling nearly cost-free region proposals. Moreover, Faster R-CNN utilizes softmax loss function to perform classification and adopts smooth L1 loss function to perform bounding box regression. RPN predicts whether current anchors belong to foreground or background, and bounding box regression can help RPN revise anchors to get more accurate proposals. Since Faster R-CNN employs two networks to detect objects, detection performance of Faster R-CNN is better than YOLO and SSD models. However, training a Faster R-CNN model requires a large amount of labeled images, and collecting enough labeled images is hard in real-world applications. This significantly hurts its performance.
Semi-supervised learning object detection. Since collecting limited labeled images is often the case in practice, semi-supervised learning algorithms have been developed. It utilizes a small amount of labeled images to train a model but detect objects in unlabeled images. Traditional semi-supervised learning algorithms (e.g., Pseudo-Label [35] and ladderNet [36]) focus on object detection on the same dataset, and suffer challenge if objects are from different datasets. Domain adaptive Faster R-CNN [19] is the first work, to the best of our knowledge, to deal with this problem. It designs two domain adaptation components, on image and instance levels, to reduce domain discrepancy. The two domain adaptation components are based on H-divergence theory which is utilized to measure the divergence between data distribution of source domain and that of target domain, and to be implemented by learning a domain classifier in an adversarial training manner. Domain classifiers on different levels are further reinforced with a consistency regularization to learn a domain-invariant region proposal network (RPN) in the Faster R-CNN model. However, Domain adaptive Faster R-CNN focuses on matching overall distribution of source domain to that of target domain, ignoring details of one single instance. Moreover, it still requires images within target domain to hold labels, otherwise resulting in low quality detection performance. Therefore, applying this model to COVID-19 case results in low quality detection.
Khodabandeh, Mehran et al. [37] proposed to utilize noise to label images within a target domain. Based on a set of noisy object bounding boxes obtained via a detection model trained in source domain only, a final detection model is trained. Although this study improves robust learning, detection performance is not satisfactory in our case. This is because this study adopts the KL divergence to measure correlation between images within source domain and within target domain, and KL could be or when two feature maps have no correlation. This may hurt the model performance. A recent model is Few-Shot Adaptive Faster R-CNN proposed by [20], and they termed their model FAFRCNN. The architecture of FAFRCNN is similar to Domain adaptive Faster R-CNN. The differences between two models are that FAFRCNN couples with a feature pairing mechanism and a strong regularization for stable adaptation. Although FAFRCNN can detect objects with limited data, they have challenges. FAFRCNN adopts two ROI Pooling modules and utilizes the share-parameters strategy to train model. However, share-parameters strategy may lead to networks which receive feature maps from two ROI Pooling modules outputting the same bounding boxes even if they have different objects, causing detection failure. More details are shown the experiment section.
Note that some researchers segment CT images to figure out infected regions. However, CT segmentation just shows the infections with bright regions and masks the rest of lungs with shadow regions [38], given that CT images are represented by different gray levels. Physicians cannot guarantee the output produced by segmentation containing the COVID-19 and do not know the degree of infection if there is no healthy regions as reference. Hence, object detection is a better choice than segmentation in COVID-19 case.
3. Preliminaries
In our approach, five modules are employed, which are ResNet Faster R-CNN, Adversarial Learning, Network-in-Network and Instance Normalization. We introduce them one by one as follows.
3.1. ResNet
Traditional Conv-based neural network or full connection network suffer from information loss when performing information transmission. Moreover, vanishing/exploding gradients is also a serious issue during training. Hence, it is a non-trivial task to train a deeper neural network. ResNet adopts deep residual learning framework to address this problem [31]. Specifically, ResNet utilizes skip connection to add the outputs from previous layer to the outputs of current stacked layer with where indicates a mapping from current stacked layer, and denotes identity, preserving information to the most extent. Such a combination of is termed as a residual module. There are two kinds of residual modules used in ResNet, one is that two Conv-based layers with kernel size 3 3 are connected in a sequential manner to form a residual module, and the other is that three Conv-based layers with kernel size 1 1 and 3 3 as well as 1 1 are connected in a cascade manner to form a residual module.
In our case, a minor symptom may only contain several pixels, which requires that we keep information of CT images to the most extent. Deeper neural network can extract more rich information in practice. Hence, we employ ResNet to extract valuable information from COVID-19 CT for infected region detection.
3.2. Faster R-CNN
Although Faster R-CNN has been briefly introduced in previous sections, we formally describe it below to establish continuity. A vanilla Faster R-CNN [10] consists of three main components: a vanilla VGG16 network, a region proposal network (RPN) and an ROI classifier. The first component focuses on extracting features of images, the second one proposes regions of interests (ROI) for object detection, while the last one predicts object labels for the proposed bounding boxes. The last two components share first convolutional layers, and the shared layers extract feature maps when images are input. RPN calculates the probability of a set of pre-defined anchor boxes to predict whether current anchor box is an object or background. Anchor boxes are a fixed pre-defined set of boxes with varying positions, sizes and aspect ratios across an image. Similar to RPN, region classifier predicts object labels for ROI proposed by RPN as well as refinements for location and size of the boxes. Features passed to the classifier are obtained with a ROI-pooling layer. Both networks are trained jointly by minimizing the loss function:
| (1) |
where and refer to losses which are used for updating the parameters of whole network. The losses consist of a cross-entropy cost measuring mis-classification error and a regression loss quantifying localization error.
3.3. Adversarial learning
Adversarial learning is a technique employed in the field of machine learning which attempts to fool models through malicious input [39], [40], [41]. It can improve robustness of network training. Here, we take Generative Adversarial Net (GAN) [42] as an example to demonstrate how an adversarial learning method works. In general, the GAN consists of two components: a discriminator () and a generator (). Both of them play a minimax game. produces simulation data to fool into accepting it as the real one by maximizing its score where indicates noise code. strives to distinguish simulation data from real data by minimizing and to maximize the score it assigns to real data by maximising . Hence the combined loss for GAN can be written as:
| (2) |
In this work, and are alternatively optimized, and Jensen-Shannon (JS) divergence is utilized to measure the difference between original data distribution and generated data distribution. JS divergence reaches its optimal value as both components reach the Nash equilibrium where = = 0.5. Under such a scenario, GAN gets converged.
For our case, CT images from target domain can be viewed as generated data while CT images from source domain can be regarded as original data. If a domain classifier cannot distinguish interest regions within target domain from that within source domain, we can label interest regions in target domain with the labels belonging to source domain.
3.4. Network-in-Network
In general, a DNN model assumes latent representation of data is linearly separable, i.e., variants of a concept all live on one side of a separation plane defined by a generalized linear model (GLM) [27]. However, data for the same concept often live on a non-linear manifold, therefore representations that capture these concepts are generally highly non-linear function of the input data. Network-in-Network (NIN) [27] replaces GLM with a micro network (e.g., multilayer perceptron (MLP)) which is a general non-linear function approximator. The advantages of adopting MLP is: (1) parameters of MLP can be updated by back-propagation, such that MLP can be integrated with a neural network to be trained. (2) We can enhance capabilities of MLP by adding hidden layers, which satisfies the idea of feature reuse. (3) Feature maps from linear combination of multiple channels are changed to non-linear combination, which improves feature abstraction capability. These boil down to employ multiple kernel operations in convolutional layers, because multiple convolutional concatenations bring non-linear combination of feature maps from multiple channels.
3.5. Instance Normalization
In general, Instance Normalization (IN) [29] is used to transfer a style from an image into another, which is applied to the generator architecture. The stylized image matches simultaneously selected statistics of a style image and of a content image, and a normalization process allows to remove instance-specific contrast information from the content image, which simplifies generation during training [29]. Although this method just replaces batch normalization with Instance Normalization, its performance is significantly improved in real-time image generation. One advantage of IN in our case is that it enforces a model to attend to details of a single instance rather than overall distribution of batch data. This can help detect minor symptoms in COVID-19 CT images.
4. NIA-Network
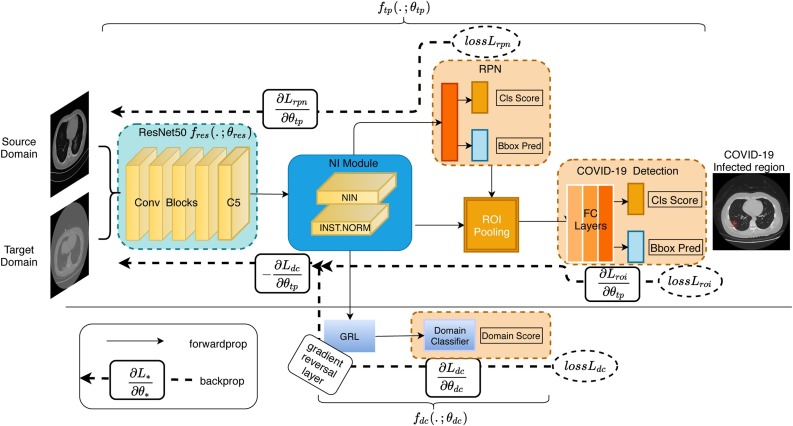
Inspired by component adaptation algorithm, NIA-Network transfers labels from a source domain to a target domain by minimizing the difference between infected regions which are from both source domain and target domain via an adversarial learning loss in latent space. As illustrated in Fig. 1 , NIA-Network comprises the following four major components.
-
•
A vanilla CNN serves as feature extractor, extracting representations from source and target domains. In this paper, we adopt the widely used vanilla ResNet50 as the extractor. One can choose other models according to different tasks. ResNet can keep shift-invariant of objects [43], and residual structure can help extract features of different levels of an image. These features commonly represent an image. Specifically, each time we randomly select one single CT image from each domain, and feed them into ResNet50. The outputs of ResNet50 are considered as a positive pair representations.
-
•
We design a new NI-Module to extract non-linear concepts and to normalize the representations. Our NI-Module is constituted by Network-in-Network (NIN) [27] and Instance Normalization (IN) [29]. The former component can capture non-linear concepts of data representations, while the latter one can keep unique details of each representation. Because NIN can be achieved by a Conv-based layer with kernel and IN is also a layer, we combine them to form a new module.
-
•
Adversarial loss is utilized to implement infected regions adaptation from source domain to target domain with Gradient Reversal Layer (GRL) [44], based on a classifier. GRL belongs to adversarial learning and its loss can be replaced with loss of GAN, while its implementation is easier than GAN if it is integrated with other components [32]. In that case “adversarial” loss is easily obtained by swapping domain labels [32]. In other words, if two representations are similar to each other which is evaluated by a classifier, we can transfer labels from source domain to target domain.
-
•
A vanilla Faster R-CNN [10] is employed to learn domain-invariant region proposal network (RPN), outputting the framed result.
Fig. 1.
Illustration of proposed NIA-Network architecture. For easier understanding, we utilize two functions ( and ) to illustrate our NIA-Network. The function contains an extractor (a vanilla ResNet50), NI-Module and a vanilla Faster R-CNN, and it is a feed-forward Conv-based architecture. The function indicates the domain adaptation architecture, and it contains a domain classifier and one gradient reversal layer (GRL). The function ensures that representations over the two domains are similar which enables to transfer labels from source domain to target domain. Our NI-Module is shown in Section 4.1 and domain adaptation architecture is introduced in Section 4.2.
Next, we discuss the backbone of NI-Module and our proposed NIA-Network, given that other two components are vanilla ResNet50 and Faster R-CNN.
4.1. NI-Module backbone
NI-Module backbone is shown in Table 1 , which holds 9 layers. Our NI-Module consists of three components: NIN, IN and ReLU. In the rest of this subsection, we introduce them one by one. NIN consists of MLP with kernel size, which is shown in Eq. (3).
| (3) |
where is the number of layers. From cross feature map perspective, Eq. (3) is a cascaded cross channel parametric layer. The cross channel-pooled feature maps are cross channel-pooled again and again in the next layers. This cascaded cross channel parametric pooling structure allows complex and learnable interactions of cross channel information [27]. Here, representations produced by are where refers to the quantity of representations and is the channel index, and indicate spatial location of representations. Since a conventional network implicitly assumes that latent concepts are linearly separable, and data for the same concept often live on a non-linear manifold, NIN is utilized to capture non-linear concepts of data representations in our case [27].
Table 1.
NI-Module backbone in which NIN indicates the Network-in-Network and IN refers to the Instance Normalization.
| Representation size | Layer | |
|---|---|---|
| Input | 32 32 | |
| NIN 1 | 32 32 | 1 1 kernel, 1024, stride 1, pad 0 |
| IN 1 | 32 32 | 1024 channels |
| ReLU 1 | 32 32 | |
| NIN 2 | 32 32 | 1 1 kernel, 1024, stride 1, pad 0 |
| IN 2 | 32 32 | 1024 channels |
| ReLU 2 | 32 32 | |
| NIN 3 | 32 32 | 1 1 kernel, 1024, stride 1, pad 0 |
| IN 3 | 32 32 | 1024 channels |
| ReLU 3 | 32 32 | |
| Output | 32 32 |
IN is immediately follows NIN. For COVID-19 case, each pixel within CT image cannot be ignored, given that a minor symptom just holds several pixels. Traditional normalization strategies (i.e., batch normalization) focus on overall distribution of data, ignoring unique details of each instance. However, IN considers all elements in each pass of each instance. Hence, it is necessary to employ IN to keep data details during training. IN is formally described in Eq. (4).
| (4) |
where is instance normalized response (also known as “instance normalization”). The input to the classifier is rather than the raw image . In this study, we view each component as a layer, and the NI-Module is stacked by 9 layers, which is shown in Table 1. Note that the number 1024 in IN indicates the channel size. Here, we assume that the size of the representations from ResNet is . From Table 1, we can see that the size is unchanged but the concept of data and the unique details are obtained by our NI-Module.
4.2. The algorithm
In our case, images within source domain hold labels while images within target domain have no labels. The goal of our algorithm is to measure similarity between symptoms from both source and target domains. If two symptoms are similar, we can transfer labels from CT in source domain to those CT images in target domain. Faster R-CNN then outputs detected results in target domain.
With NI-Module, representations are fed into a classifier. Note that the purpose of a classifier is to accurately predict labels of objects, and this is achieved by minimizing cross entropy loss. In our case, we need to maximize this loss to make the two representations as similar as possible while simultaneously minimize the loss of this classifier to predict which CT image is from source domain or target domain, resulting in that a classifier cannot distinguish target objects from source objects. Since transferring labels only happens on two similar representations [19]. The architecture of domain classifier is shown in Table 2 .
Table 2.
Architecture of the domain classifier.
| Representation size | Layer | |
|---|---|---|
| Input | 32 32 | |
| Conv2D 1 | 32 32 | 3 3 kernel, 1024, stride 1, pad 1 |
| ReLU 2 | 32 32 | |
| Conv2D 2 | 32 32 | 3 3 kernel, 1, stride 1, pad 1 |
| Sigmoid | 32 32 | |
| Output | 32 32 |
To achieve this, Gradient Reversal Layer (GRL) [32] is employed. During the forward propagation, GRL acts as an identity transform. On the other hand, GRL takes gradient from subsequent level, multiplies it by , and passes it to the preceding layer during backpropagation [32]. Here, refers to a coefficient and it controls the trade-off between two objects that shape representations during learning, and it is defined as where is empirically set to 10 while is the training progress linearly changing from 0 to 1 [32], which formally is shown as follows.
| (5) |
where indicates the vector of parameters of all layers within domain classifier in an representation and representations are mapped to label (label predictor) and to domain . The parameters of such a mapping in the former case are denoted as and in the latter case. Hence, is the loss for label prediction and is the loss for domain classification in Eq. (5).
Algorithm 1
Training and testing procedure of our proposed NIA-Network.
After that, representations are fed into RPN and ROI components, which are from Faster R-CNN. RPN is used to generate region proposals, and ROI collects representations and proposals to align anchors. After that, these anchors are fed into the fully convolution layers (FC layers) to classify those proposals and to make bounding box regression. In this way, our NIA-Network can be formulated as follows.
| (6) |
where denotes probability of anchor being an object. The ground-truth label is 1 if an anchor is positive, and 0 if an anchor is negative. is a vector representing the 4 parameterized coordinates of predicted bounding box and is that of ground-truth box associated with a positive anchor [10]. refers to loss. In this way, our NIA-Network is shown in Algorithm 1. Note that our algorithm adopts gradient descent to optimize our DNN model, and gradient descent makes training converged [45], [46]. In our study, fluctuation of detection loss is getting small as training converges. We illustrate training convergence with detection loss in Section 5. After training NIA-Network, only is applied to testing data samples for COVID-19 infected region detection. Our NIA-Network finally outputs the detected infection regions.
4.3. The difference between GRL and GAN
In our NIA-Network, we adopt Gradient Reversal Layer (GRL) to conduct adversarial training to transfer labels from source domain to target domain. GRL is different from GAN. For GAN, the minimax game is played by both discriminator and generator. It pursues Nash Equilibrium in which the discriminator achieves its maximum in [0, 1] at . Moreover, GAN training is unstable, because gradients could be vanishing during training. For GRL, it transforms the minimax game into a loss function . In forward-propagation, GRL can be viewed as an identity mapping function, because it does not change any values. However, GRL multiplies a coefficient in back-propagation step. In this way, GRL minimizes loss function to conduct adversarial training, which is more robust than GAN, because gradients are less likely to vanish during training. It can improve performance of domain classifier and capability of feature extractor that tries to confuse a domain classifier [44]. Therefore, GRL achieves a better performance than GAN.
5. Experiments
For experiments, three SOTA methods are chosen for comparison, which are Domain adaptive Faster R-CNN [19], Faster R-CNN [10] and Few-Shot Adaptive Faster R-CNN [20]. Moreover, to validate the impact of each component within our NIA-Network, we conduct an extensive series of ablation experiments.
To make a fair comparison, we deploy all models on the same machine with Intel Core E5 2.80 GHz, 32 GB RAM and 2080Ti GPU and set the same experimental settings for all models. Specifically, all models are trained with stochastic gradient descent (SGD) for 80 K iterations, with the initial learning rate . The learning rate is reduced by a factor of 10 at iteration 60 K and 80 K, respectively. Weight decay and momentum are set to 0.0001 and 0.9, respectively. We initialize ResNet50 with the weights pre-trained on ImageNet [47]. Note that parameters of the ResNet-50 backbone would be updated with the training process. On the one hand, extracted features from natural images focus on low-level features such as edge and texture. Those low-level features between natural images and CT images are similar to each other. For example, ImageNet images contain gray color and coarse textures, and COVID-19 CT images also have the gray color and ground-glass opacification and infiltrating shadow textures. The pre-trained parameters would help the backbone get better performance [48]. On the other hand, we also validate the performance of our NIA-Network with random initialization on the same dataset, and the results are 94% for sensitivity, 90.2% for specificity and 92.1% for accuracy, respectively. Those results are less than 94.2% for sensitivity, 99.5% for specificity and 96.82% for accuracy in which ResNet-50 is initialized by pre-trained model. Hence, utilizing the pre-trained parameters to initialize the model is plausible. As to other components like NI-Module, domain classifier, RPN and ROI, we adopt random initialization. Moreover, the hyper-parameter in Gradient Reversal Layer is initialized to 0.1. In addition, we set RPN anchor size as [20, 32, 43, 58, 92] and the aspect ratio size as [0.5, 1.0, 2.0] after capturing the distribution of COVID-19 infected region from training dataset.
5.1. Dataset description
The COVID-19 CT images in this paper are from Zhongnan Hospital, Wuhan University (ZNWU) and Italian Society of Medical and Interventional Radiology (ISMIR). The size of all CT images is . Here are details of the two datasets.
ZNWU dataset: This dataset is collected from Zhongnan Hospital, Wuhan University, and this hospital is a designated site to receive patients with COVID-19 in the early pandemic outbreak. We collected data of 78 patients and healthy chest CT data from 60 people. The definition of healthy status is that the lung CT image has not any infected symptoms. The patient group contains 12 patients with minor symptoms and others have severe symptoms, the median age is about 50 years, and 68% is male. All CT images are checked by physicians. Note that there are 8 patients from severe case which are aggravated from minor symptoms in 4 days [21]. We collect 300 slices from the group of patients and these slices are carefully annotated by physicians. Moreover, we randomly select 1000 slices from the health group to validate our approach. The original CT volume data of each patient is format with bits which holds a dynamic range from 32,768 to 32,767. We load the format data and use Hounsfield Units (HU) normalization technique to normalize those CT images into a range from 0 to 255, and then convert to with 512 512 size.
ISMIR dataset[49]: Italian Society of Medical and Interventional Radiology releases their COVID-19 data to the public. We used 2000 CT images of 69 patients from this dataset, of which 1000 are labeled images and the other 1000 are unlabeled images. The original format of this dataset is grey scaled and compiled into several NIFTI files. ISMIR dataset is reversely intensity-normalized by taking RGB values from JPG images from areas of air (either externally from the patient or in the trachea) and of fat (subcutaneous fat from the chest wall or pericardial fat). The reversely intensity-normalized output is used to establish unified Houndsfield Unit-scale (the air was normalized to 1000, fat to 100). We load all NIFTI data and convert it to (512 512) format images in our experiments. Since this dataset includes segment infected regions, we extract bounding box information from segmented volumetric CT mask for COVID-19 infected region detection.
Since our study focuses on label transferring from one domain (i.e., ZNWU dataset) to another domain (i.e., ISMIR dataset), 300 annotated samples from ZNWU dataset and 1000 samples without labels from ISMIR dataset are used as training data for Few-Shot Adaptive Faster R-CNN, Domain Adaptive Faster R-CNN, and our NIA-Network. More details are shown in Table 3 . Given that Faster R-CNN model belongs to supervised learning, only 300 annotated samples are used as the training data to train Faster R-CNN. Moreover, 1000 healthy samples from ZNWU dataset are integrated with the 1000 COVID-19 samples from ISMIR dataset as the testing data (See Table 4 ). We assess the performance of object detection on the testing data for all models.
Table 3.
Training data. The symbol indicates images with label while the symbol denotes images without label.
| Source domain () | Target domain () | |
|---|---|---|
| Number of images | 300 | 1000 |
Table 4.
Testing data. The quantity is totally 2000, half of them belongs to COVID-19 case from ISMIR dataset and another half belongs to healthy group from ZNWU dataset.
| COVID-19 (ISMIR) | Healthy (ZNWU) | |
|---|---|---|
| Number of images | 1000 | 1000 |
5.2. Evaluation metric
To quantify the performance of object detection for all models, three metrics including accuracy, sensitivity, specificity are employed as follows:
| (7) |
where True Positives (TP) refers to correctly predicted COVID-19, False Negatives (FN) indicates that COVID-19 patients are classified as healthy group. True Negatives (TN) indicates that healthy cases are correctly classified as non-COVID-19 group, and False Positives (FP) refers to that healthy cases are classified as COVID-19 group. In this work, we use object detection to detect COVID-19 infected regions. Specifically, the overlap between predicted infection region and ground-truth object is greater than 0.5 Intersection over Union (IoU) to be considered “COVID-19”, and other scenarios are considered as healthy. A sample is classified as a COVID-19 positive case if at least one infection region is detected, otherwise the image is judged as a healthy case if no infected regions are detected.
5.3. Experimental results
The baselines and our NIA-Network are trained on training data (Table 3) and validated on testing data (Table 4). Note that one convention in medical domain is to utilize data augmentation to improve model performance [50], [51], [52]. Therefore, we augment the COVID-19 CT images with method, given that the quantity of medical data is limited. Here, we randomly rotate each CT image to left or right direction. In this way, we obtain 2600 training samples. We compare detection performance on both original training data and augmented training data, and the results are shown in Tables 5 and 6 respectively. From both tables, we can observe that our NIA-Network significantly outperforms the baselines on both cases. We also show the training cost and model parameters in Table 7 . From Table 7, we can clearly observe that although the quantity of parameters of our NIA-Network is twice as big as Faster R-CNN, the training time is similar (9.7 h for Faster R-CNN vs. 11.2 h for NIA-Network). The running time indicates the detection time per slice. It further proves the effectiveness of our NIA-Network.
Table 5.
Detection results of all models which are trained on the original training samples but tested on the COVID-19 testing samples.
Table 6.
Detection results of all models which are trained on the augmented training samples but tested on the COVID-19 testing samples.
Table 7.
Training cost and model parameters on all models.
| Training time | Running time | RAM memory | Parameters quantity | |
|---|---|---|---|---|
| Faster R-CNN | 9.7 h | 0.218 s | 8.3% | 32,990,485 |
| Few-Shot Adaptive Faster R-CNN | 10.7 h | 0.227 s | 8.33% | 56,876,118 |
| Domain Adaptive Faster R-CNN | 9.8 h | 0.223 s | 8.32% | 42,439,958 |
| NIA-Network | 11.2 h | 0.228 s | 8.4% | 61,316,374 |
To further validate the effectiveness of our approach, we replace ResNet-50 with other backbones (i.e., VGG16). The detection results are shown in Table 8 . Obviously, our NIA-Network still outperforms the baselines.
Table 8.
Detection results of all models with VGG16 backbone.
| Sensitivity (%) | Specificity (%) | Accuracy (%) | |
|---|---|---|---|
| Faster R-CNN | 66.7 | 73.6 | 70.15 |
| Few-Shot Adaptive Faster R-CNN | 74.6 | 74.9 | 74.75 |
| Domain Adaptive Faster R-CNN | 77.3 | 77.5 | 77.4 |
| NIA-Network | 84.3 | 79.8 | 82.05 |
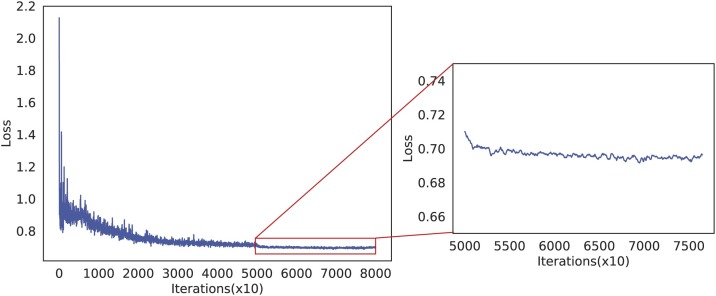
In addition, we illustrate the training convergence with loss optimization process for our NIA-Network, which is shown in Fig. 3. Obviously, we can observe that the loss gets converged from = 50,000, because the loss values have very small fluctuation from this point. Moreover, to validate the importance of each component in our NIA-Network, we conduct a series of ablation experiments, and the results are shown in Table 9 . Form Table 9, we can observe that each component in our NIA-Network is necessary, because removing any one decreases the detection accuracy. On the other hand, the ablation experiments demonstrate importance of NIN and IN. The former module captures non-linear property of data, which benefits feature extractor. The latter one considers all elements in each channel of each instance to better keep unique details of each sample. Hence, data details are kept to the most extent during training. The two components significantly improve detection performance.
Fig. 3.
The training convergence illustration of our proposed NIA-Network.
Table 9.
Ablation study of our NIA-Network on the COVID-19 testing dataset. The mark indicates that our NIA-Network holds the corresponding component. Note that ResNet + GRL corresponds to Domain Adaptive Faster R-CNN model and Faster R-CNN model utilizes ResNet module, so the two ablation experiments could be referred to Table 6. The last row in this table indicates the full NIA-Network.
| ResNet | NIN | IN | GRL | Sensitivity (%) | Specificity (%) | Accuracy (%) | |
|---|---|---|---|---|---|---|---|
| NIA-Network | 76.8 | 98.5 | 87.65 | ||||
| 70.9 | 98.0 | 84.45 | |||||
| 75.2 | 98.8 | 87.0 | |||||
| 91.0 | 98.1 | 94.55 | |||||
| 93.8 | 95.8 | 94.8 | |||||
| 63.0 | 94.4 | 50.35 | |||||
| 94.2 | 99.5 | 96.85 |
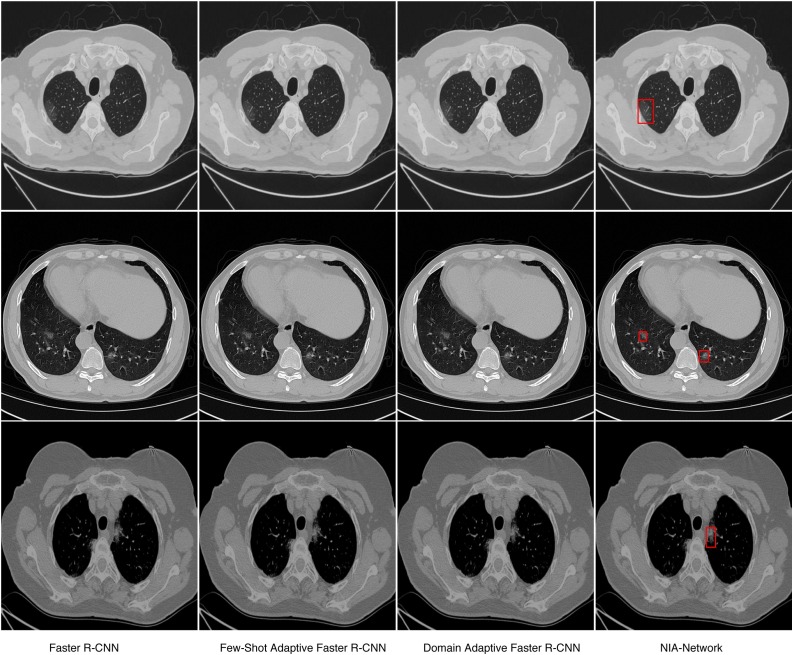
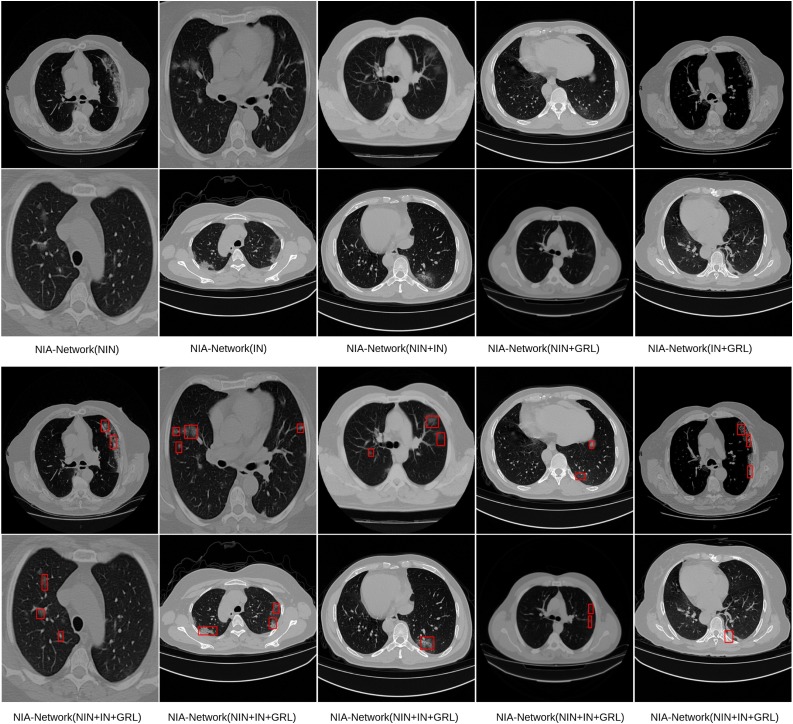
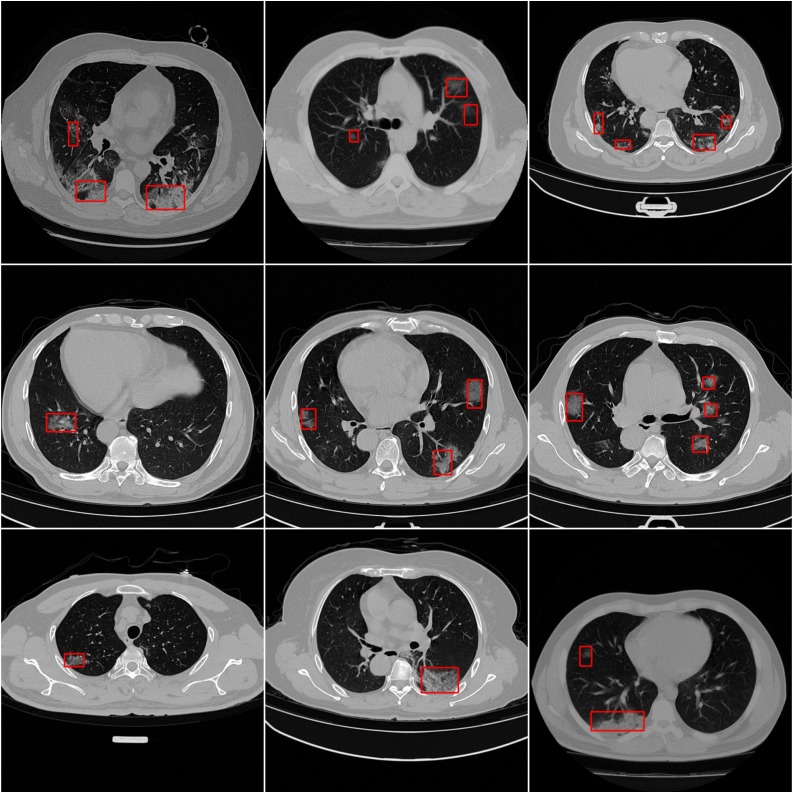
We also illustrate detection performance on slices with minor symptoms, and results are shown in Fig. 2 . In Fig. 2, we can see that only our NIA-Network successfully detects infected regions with small size, the baselines fail to detect such minor symptoms. Fig. 4 shows importance of each component within our NIA-Network. The first two rows (row 1 and row 2) of Fig. 4 indicates the failed detection CT images with oblation architecture. NIA-Network(NIN) is the model with NIN component and removing other components, which corresponds to the first row of Table 9. The last two rows (row 3 and row 4) of Fig. 4 indicates the successfully detected results of full NIA-Network on the same CT images. The oblation experiments demonstrate significant importance of each component. Fig. 5 shows that we apply our NIA-Network to different slices with different infected regions, which are taken by different CT machines. From Fig. 5, we can observe that infected regions of both large size and small size are successfully detected by our proposed NIA-Network.
Fig. 2.
The detection results of baselines and our NIA-Network on the same CT images. These examples only include minor symptoms. By comparison, we can observe that minor symptoms are successfully detected by our NIA-Network but fail to be detected by baselines. The red rectangle marks the infected regions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Illustration of ablation experiment results. The first two rows (row 1 and row 2) indicate failed detection with ablation networks, e.g.,: NIA-Network(NIN) indicates that NIA-Network just holds NIN component without other components. The last two rows (row 3 and row 4) indicate the detected results of NIA-Network (with NIN, IN, and GRL) on the same CT images. These detection results demonstrate that removing any component within NIA-Network hurts detection performance.
Fig. 5.
Illustration of detection results of our NIA-Network on different CT images within target domain. The infected regions are marked by red rectangles. Obviously, infected regions of both large size and small size are successfully captured by our NIA-Network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Here, we give an explanation about COVID-19 detection with detection model, given that non-experts may feel difficult to distinguish infected regions from healthy tissues. In general, the tissues (e.g., alveoli, bronchial tube and blood capillary) have inflammatory infiltration after lungs suffer from COVID-19. Hence, the color of infected regions is similar to ground-glass opacification and infiltrating shadow. However, there is no inflammatory infiltration in the healthy tissues, such that the color of those tissues is clearer than infected regions. Note that these characteristics of different colors can be reflected by pixels with different gray values in CT images, and deep-learning-based model can capture such characteristics. Hence, the infected regions are accurately detected by our detection model.
5.4. Discussion
From patients’ perspective, the higher diagnostic accuracy a model outputs, the better. Higher diagnostic accuracy indicates that a machine learning tool can significantly alleviate the burden of physicians. However, obtaining good diagnostic accuracy is a non-trivial task when a pandemic breaks out suddenly, because physicians have to focus on patients diagnosis and have no extra time to label CT images. Although semi-supervised deep learning algorithms (e.g., Domain adaptive Faster R-CNN and Few-shot adaptive Faster R-CNN) can help train a diagnosis model with limited labeled images, these algorithms have challenges. They cannot detect infected regions of small size in real-world COVID-19 case, such that patients with minor symptoms may be undetected, aggravating to the severe case. Our NIA-Network can detect not only small infected regions within CT images but also severe cases (See Figs. 2, 4, 5, Tables 5, 6, 8 and 9). This is very important to both patients and physicians. On the one hand, detecting infected regions of small size can significantly prevent patients with minor symptoms developing to the severe case, because latter scenario takes up a lot of medical resources with high death rate. On the other hand, the physicians may mis-detect minor symptoms, especially in a pandemic outbreak [53], [54]. Our model can fill in such gaps, improving the performance of COVID-19 detection.
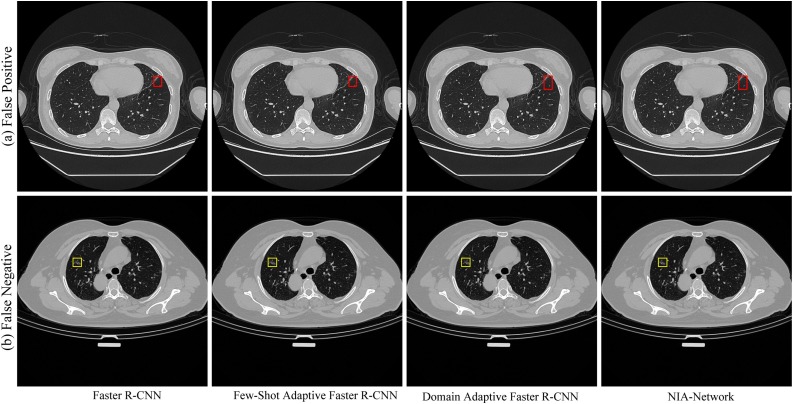
Although experimental results from our proposed NIA-Network are better than baselines, the accuracy is not 100% (See Table 6). Consider that our goal is to help physicians diagnose COVID-19 by identifying the infected regions in each CT image, higher metric scores can improve the diagnosis efficiency, especially in the early pandemic outbreak. Since our model is to reduce the diagnosis burden of physicians rather than replacing them to diagnose patients, diagnosis decision still relies on the physicians, our work can help physicians pay more attention to treatments. On the one hand, we can observe that the advantages of our NIA-Network are to successfully detect the infections of different sizes, especially for minor symptoms (See Fig. 2). On the other hand, there still exist failure cases where healthy regions are wrongly detected as infected regions (red rectangle in sub-figure (a)) and infected regions (yellow rectangle in sub-figure (b)) are not detected by models, which are shown in Fig. 6 . The characteristics of healthy region might be very similar to infected symptom's characteristics in the former case, while infected symptom is hard to be perceived even by physicians in the latter case. To analyze whether the performance improvement of our method is statistically significant, we conduct paired t-test for our method with baselines, given that the goal of our work is to improve the performance of detecting infected regions within COVID-19 CT images. We utilize accuracy as the evaluation measurement and set the significance level as 0.05. To get accuracy of all models, we divide test data into 5 different subsets and apply all trained models to those subsets. The accuracy results are shown in Table 10 . The paired t-test results are shown in Table 11 . All paired t-test results show -values smaller than 0.05, demonstrating that our improvements compared with these baseline approaches are statistically significant. We would like to keep reducing the false-positive and false-negative rates in our future work.
Fig. 6.
Failure cases. (a) Illustrates that all models wrongly detect healthy region as infected symptom, which are framed by red rectangles. As to sub-figure (b), all models cannot detect infected symptoms, and the ground truth is marked by yellow rectangles. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 10.
Accuracy results on all models.
| Acc (%) | Acc (%) | Acc (%) | Acc (%) | Acc (%) | |
|---|---|---|---|---|---|
| Faster R-CNN | 90.0 | 77.0 | 91.25 | 82.0 | 90.25 |
| Few-Shot Adaptive Faster R-CNN | 89.5 | 76.0 | 90.25 | 81.75 | 89.75 |
| Domain Adaptive Faster R-CNN | 92.0 | 80.75 | 90.75 | 87.25 | 90.25 |
| NIA-Network | 98.0 | 93.25 | 98.0 | 96.25 | 98.75 |
Table 11.
Paired t-test for our method with baselines via accuracy results.
| -Value | |
|---|---|
| Faster R-CNN | 0.0157 |
| Few-Shot Adaptive Faster R-CNN | 0.0129 |
| Domain Adaptive Faster R-CNN | 0.0090 |
6. Conclusion
In this paper, to improve the performance of lung infection detection, we propose a new model, called NIA-Network. The NIA-Network utilizes Network-in-Network and Instance Normalization to preserve key features of data samples from source and target domains. Moreover, to train our network, we employ adversarial loss and combine it with loss of both RPN and ROI to jointly update NIA-Network parameters. Comprehensive experiments on two COVID-19 CT image datasets demonstrate the following capabilities of our proposed NIA-Network. (i). Achieving the state-of-the-art accuracy, sensitivity, and specificity scores; (ii). Successfully detecting not only dramatic symptoms but also small infected regions; (iii). Strong generalization. NIA-Network diagnostic model can be successfully applied to CT images taken by different CT machines in different hospitals.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors would like to acknowledge the support provided by the Fundamental Research Funds for the Central Universities (No. JUSRP121073). The authors also appreciate the physicians who work tirelessly to diagnose and treat the patients during the outbreak of COVID-19.
References
- 1.Novel C.P.E.R.E., et al. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (covid-19) in China. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi. 2020;41:145. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 2.CNHC . 2020. Diagnostic and treatment protocol for novel coronavirus pneumonia: trial version 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyu Q., You C., Shan H., Zhang Y., Wang G. vol. 11113. International Society for Optics and Photonics; 2019. Super-resolution mri and ct through gan-circle; p. 111130X. (Developments in X-ray tomography XII). [Google Scholar]
- 4.You C., Li G., Zhang Y., Zhang X., Shan H., Li M., et al. Ct super-resolution gan constrained by the identical, residual, and cycle learning ensemble (gan-circle) IEEE Trans Med Imaging. 2019;39:188–203. doi: 10.1109/TMI.2019.2922960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.You C., Yang Q., Gjesteby L., Li G., Ju S., Zhang Z., et al. Structurally-sensitive multi-scale deep neural network for low-dose ct denoising. IEEE Access. 2018;6:41839–41855. doi: 10.1109/ACCESS.2018.2858196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Liu H., Liu W., Liu J., Liu K., Shang J., et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chin J Tuberc Respir Dis. 2020;43:E005. doi: 10.3760/cma.j.issn.1001-0939.2020.0005. [DOI] [PubMed] [Google Scholar]
- 8.Lei J., Li J., Li X., Qi X. Ct imaging of the 2019 novel coronavirus (2019-ncov) pneumonia. Radiology. 2020:200236. doi: 10.1148/radiol.2020200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (covid-19): a systematic review of imaging findings in 919 patients. Am J Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 10.Ren S., He K., Girshick R., Sun J. Advances in neural information processing systems. 2015. Faster r-cnn: towards real-time object detection with region proposal networks; pp. 91–99. [DOI] [PubMed] [Google Scholar]
- 11.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of sars-cov-2 as compared with sars-cov-1. N Engl J Med. 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., et al. 2020. Deep learning system to screen coronavirus disease 2019 pneumonia.arXiv:2002.09334 (arXiv preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., et al. 2020. Lung infection quantification of covid-19 in ct images with deep learning.arXiv:2003.04655 (arXiv preprint) [Google Scholar]
- 14.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H., et al. 2020. Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification.arXiv:2003.09860 (arXiv preprint) [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., et al. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) medRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv. 2020 doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Liu X., Liu J., Chen P., Wan S., Cui X. On improving the accuracy with auto-encoder on conjunctivitis. Appl Soft Comput. 2019;81:105489. [Google Scholar]
- 18.Zhu X., Goldberg A.B. Introduction to semi-supervised learning. Synth Lect Artif Intell Mach Learn. 2009;3:1–130. [Google Scholar]
- 19.Chen Y., Li W., Sakaridis C., Dai D., Van Gool L. Domain adaptive faster r-cnn for object detection in the wild. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018:3339–3348. [Google Scholar]
- 20.Wang T., Zhang X., Yuan L., Feng J. Few-shot adaptive faster r-cnn. Proceedings of the IEEE conference on computer vision and pattern recognition. 2019:7173–7182. [Google Scholar]
- 21.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., et al. Time course of lung changes on chest ct during recovery from 2019 novel coronavirus (covid-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., et al. Characteristics and outcomes of 21 critically ill patients with covid-19 in Washington state. JAMA. 2020 doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei X., Xiao Y.-T., Wang J., Chen R., Zhang W., Yang Y., et al. 2020. Sex differences in severity and mortality among patients with covid-19: evidence from pooled literature analysis and insights from integrated bioinformatic analysis.arXiv:2003.13547 (arXiv preprint) [Google Scholar]
- 25.Liu Q., Dou Q., Yu L., Heng P.A. Ms-net: multi-site network for improving prostate segmentation with heterogeneous mri data. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2974574. [DOI] [PubMed] [Google Scholar]
- 26.Sankaranarayanan S., Balaji Y., Jain A., Nam Lim S., Chellappa R. Learning from synthetic data: addressing domain shift for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018:3752–3761. [Google Scholar]
- 27.Lin M., Chen Q., Yan S. 2013. Network in network.arXiv:1312.4400 (arXiv preprint) [Google Scholar]
- 28.You C., Yang L., Zhang Y., Wang G. Low-dose ct via deep cnn with skip connection and network-in-network. Developments in X-Ray tomography XII, volume 11113; International Society for Optics and Photonics; 2019. p. 111131W. [Google Scholar]
- 29.Ulyanov D., Vedaldi A., Lempitsky V. 2016. Instance normalization: the missing ingredient for fast stylization.arXiv:1607.08022 (arXiv preprint) [Google Scholar]
- 30.Wang L., Li B., Tian L.-F. Eggdd: an explicit dependency model for multi-modal medical image fusion in shift-invariant shearlet transform domain. Inf Fusion. 2014;19:29–37. [Google Scholar]
- 31.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. Thirty-first AAAI conference on artificial intelligence. 2017 [Google Scholar]
- 32.Ganin Y., Lempitsky V. 2014. Unsupervised domain adaptation by backpropagation.arXiv:1409.7495 (arXiv preprint) [Google Scholar]
- 33.Redmon J., Divvala S., Girshick R., Farhadi A. You only look once: unified, real-time object detection. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016:779–788. [Google Scholar]
- 34.Liu W., Anguelov D., Erhan D., Szegedy C., Reed S., Fu C.-Y., et al. Ssd: single shot multibox detector. European conference on computer vision. 2016:21–37. [Google Scholar]
- 35.Lee D.-H. Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. Workshop on challenges in representation learning, ICML, vol. 3. 2013:2. [Google Scholar]
- 36.Rasmus A., Berglund M., Honkala M., Valpola H., Raiko T. Semi-supervised learning with ladder networks. Advances in neural information processing systems. 2015:3546–3554. [Google Scholar]
- 37.Khodabandeh M., Vahdat A., Ranjbar M., Macready W.G. A robust learning approach to domain adaptive object detection. Proceedings of the IEEE international conference on computer vision. 2019:480–490. [Google Scholar]
- 38.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., et al. 2020. Inf-net: automatic covid-19 lung infection segmentation from ct scans. [DOI] [PubMed] [Google Scholar]
- 39.Kurakin A., Goodfellow I., Bengio S. 2016. Adversarial machine learning at scale.arXiv:1611.01236 (arXiv preprint) [Google Scholar]
- 40.Li W., Ding W., Sadasivam R., Cui X., Chen P. His-gan: a histogram-based gan model to improve data generation quality. Neural Netw. 2019;119:31–45. doi: 10.1016/j.neunet.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Li W., Fan L., Wang Z., Ma C., Cui X. Tackling mode collapse in multi-generator gans with orthogonal vectors. Pattern Recognit. 2021;110:107646. [Google Scholar]
- 42.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial nets. Advances in neural information processing systems. 2014:2672–2680. [Google Scholar]
- 43.Girshick R., Donahue J., Darrell T., Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition. 2014:580–587. [Google Scholar]
- 44.Ganin Y., Lempitsky V. Unsupervised domain adaptation by backpropagation. International conference on machine learning, PMLR. 2015:1180–1189. [Google Scholar]
- 45.Ruder S. 2016. An overview of gradient descent optimization algorithms.arXiv:1609.04747 (arXiv preprint) [Google Scholar]
- 46.Nacson M.S., Srebro N., Soudry D. Stochastic gradient descent on separable data: exact convergence with a fixed learning rate. The 22nd international conference on artificial intelligence and statistics, PMLR. 2019:3051–3059. [Google Scholar]
- 47.Deng J., Socher R., Fei-Fei L., Dong W., Li K., Li L.-J. Imagenet: a large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition (CVPR) 2009:248–255. doi: 10.1109/CVPR.2009.5206848. https://ieeexplore.ieee.org/abstract/document/5206848/ [DOI] [Google Scholar]
- 48.He K., Girshick R., Dollár P. Rethinking imagenet pre-training. Proceedings of the IEEE international conference on computer vision. 2019:4918–4927. [Google Scholar]
- 49.I. S. of Medical, I. Radiology . 2020. Covid-19 ct segmentation dataset. [Google Scholar]
- 50.Shin H.-C., Tenenholtz N.A., Rogers J.K., Schwarz C.G., Senjem M.L., Gunter J.L., et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. International workshop on simulation and synthesis in medical imaging. 2018:1–11. [Google Scholar]
- 51.Zhao A., Balakrishnan G., Durand F., Guttag J.V., Dalca A.V. Data augmentation using learned transformations for one-shot medical image segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition. 2019:8543–8553. [Google Scholar]
- 52.Andreeva O., Li W., Ding W., Kuijjer M., Quackenbush J., Chen P. Catalysis clustering with gan by incorporating domain knowledge. Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining. 2020:1344–1352. [Google Scholar]
- 53.Zhu J., Ji P., Pang J., Zhong Z., Li H., He C., et al. Clinical characteristics of 3062 covid-19 patients: a meta-analysis. J Med Virol. 2020 doi: 10.1002/jmv.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]