Abstract

Streptomyces scabiei is a key causative agent of common scab disease, which causes significant economic losses to potato growers worldwide. This organism produces several phytotoxins that are known or suspected to contribute to host–pathogen interactions and disease development; however, the full metabolic potential of S. scabiei has not been previously investigated. In this study, we used a combined metabolomic and genomic approach to investigate the metabolites that are produced by S. scabiei. The genome sequence was analyzed using antiSMASH and DeepBGC to identify specialized metabolite biosynthetic gene clusters. Using untargeted liquid chromatography-coupled tandem mass spectrometry (LC-MS2), the metabolic profile of S. scabiei was compared after cultivation on three different growth media. MS2 data were analyzed using Feature-Based Molecular Networking and hierarchical clustering in BioDendro. Metabolites were annotated by performing a Global Natural Products Social Molecular Networking (GNPS) spectral library search or using Network Annotation Propagation, SIRIUS, MetWork, or Competitive Fragmentation Modeling for Metabolite Identification. Using this approach, we were able to putatively identify new analogues of known metabolites as well as molecules that were not previously known to be produced by S. scabiei. To our knowledge, this study represents the first global analysis of specialized metabolites that are produced by this important plant pathogen.
Introduction
The ability to infect living plant tissues and to cause disease is a rare trait among bacteria belong to the genus Streptomyces, with approximatively a dozen pathogenic species out of the hundreds described having this capability. The first reported and best-characterized pathogenic species is Streptomyces scabiei (syn. S. scabies), which has a worldwide distribution and is one of the main causative agents of the economically important crop disease called potato common scab. This disease is characterized by the formation of superficial, raised, or deep-pitted scablike lesions on the tuber surface, and these lesions negatively impact the quality and market value of table stock, processing, and seed potatoes.1 There is also some evidence that common scab can reduce the yield of the potato crop2 as well as the overall size of affected tubers.3S. scabiei is neither tissue nor host-specific; it can cause scab disease on taproot crops such as carrot, radish, beet, and turnip, and it has been associated with “pod wart” disease of peanuts.1 In addition, seedlings of both monocot and dicot plants can be infected by S. scabiei, resulting in root stunting, swelling, necrosis, and seeding death.4 Control strategies for common scab disease are largely inadequate and inconsistent,5 and thus a thorough understanding of the molecular mechanisms of S. scabiei plant pathogenicity is critical to the development of strategies that can effectively manage the disease.
S. scabiei can produce at least five different types of bioactive specialized metabolites, of which three are known or suspected to play a role in mediating host–pathogen interactions. The thaxtomins are a family of nitrated 2,5-diketopiperazines that exhibit potent phytotoxic activity and are the key pathogenicity determinants produced by S. scabiei and other scab-causing Streptomyces species.6 Several thaxtomin analogues have been described, of which thaxtomin A is the predominant analogue produced by S. scabiei.7 Thaxtomin A functions as a cellulose biosynthesis inhibitor in higher plants, and it has been proposed to facilitate the penetration of expanding plant tissues by the pathogen during host colonization and infection.8S. scabiei has also been reported to produce N-coronafacoyl-l-isoleucine (CFA-Ile), which is a member of the coronafacoyl family of phytotoxins.9 Coronafacoyl phytotoxins are non-host-specific toxins that are produced by several different plant pathogenic bacteria and which function as molecular mimics of the bioactive plant hormone jasmonoyl-l-isoleucine (JA-Ile). It is thought that the production of these molecules enables the pathogen to manipulate jasmonate signaling in the plant host to overcome host defenses during infection.9 Disruption of CFA-Ile production in S. scabiei results in reduced disease symptom development on tobacco seedlings,10 while elevated phytotoxin production has been associated with increased necrosis and pitting of potato tuber tissue,11 suggesting that CFA-Ile enhances the virulence phenotype of S. scabiei, though it is not required for pathogenicity. Concanamycins are a family of specialized metabolites that are characterized by an 18-membered macrolide ring and a β-hydroxyhemiacetal side chain.12 They function as vacuolar-type adenosinetriphosphatase (ATPase) inhibitors and are biologically active against fungi, plants, and cancer cells.13,14S. scabiei produces two members of the concanamycin family, concanamycin A and B, both of which exhibit root growth inhibitory activity against different plant species.15−18 Natsume and colleagues have suggested that concanamycin A and thaxtomin A might act synergistically to induce necrosis of potato tuber tissue, and that concanamycin A may contribute to the development of deep-pitted lesions by strains of S. scabiei.19 Other specialized metabolites that can be produced by S. scabiei include the antibacterial bottromycins and the siderophores desferrioxamine E, scabichelin, and pyochelin.20−22 The production of bottromycins may allow S. scabiei to compete for limited nutrients in the soil environment by killing or inhibiting the growth of other microorganisms, while siderophore-mediated iron uptake may play an important role in pathogen survival within the plant host.23,24
In this study, we used a combined metabolomic and genomic approach to further investigate the chemical potential of S. scabiei. The genome sequence of S. scabiei was analyzed using antiSMASH25 and DeepBGC26 to identify specialized metabolite biosynthetic gene clusters (BGCs), and untargeted liquid chromatography-coupled tandem mass spectrometry (LC-MS2) was used to characterize the metabolome of S. scabiei under different culturing conditions. MS2 data were analyzed using the Feature-Based Molecular Networking (FBMN) workflow within the Global Natural Products Social Molecular Networking (GNPS) web platform27 and using hierarchical clustering in BioDendro.28 FBMN allows for the discrimination of isomers and quantitative interpretation of the molecular network, while BioDendro uses dynamic binning and hierarchical clustering of MS2 spectra and can function as a complementary analysis to molecular networking.27,28 Metabolites were annotated by matching the observed MS2 spectra with reference spectra in the GNPS libraries29 or using Network Annotation Propagation (NAP),30 SIRIUS,31 MetWork,32 or Competitive Fragmentation Modeling for Metabolite Identification 3.0 (CFM-ID 3.0).33
Results and Discussion
Genomic Analysis of Specialized Metabolite Biosynthetic Gene Clusters in S. scabiei
To examine the S. scabiei genome for specialized metabolite BGCs, we employed two different genome mining tools: antiSMASH 5.0 and DeepBGC. antiSMASH uses Hidden Markov Models (HMM) and a human-defined rules-based approach to identify BGCs for specialized metabolites,34 while DeepBGC excels at detecting novel BGC classes by employing Recurrent Neural Networks (RNNs) and Pfam domains to detect BGCs.26 As shown in Table 1, antiSMASH predicted 34 putative specialized metabolite BGCs in the S. scabiei genome, including eight terpenes, six polyketides (PKs), six nonribosomal peptides (NRPs), five ribosomally synthesized and post-translationally modified peptides (RiPPs), and one hybrid PK-NRP BGC. Ten of the predicted BGCs displayed high levels of similarity (≥70%) to BGCs in the MIBiG database, while five displayed moderate similarity (30–70%) and the remaining 19 showed low similarity (<30%) to known BGCs. The combined length of the predicted BGCs is ca. 1167 kb, accounting for ∼11.5% of the S. scabiei genome. In contrast, DeepBGC identified 146 putative specialized metabolite BGCs for S. scabiei, of which 112 were not detected by antiSMASH. The predicted BGCs include 15 PKs, 11 RiPPs, 5 NRPs, 4 PKs-Terpenes, 3 terpenes, and 1 hybrid PK-NRP BGC, as well as 99 unclassified BGCs (Table S1). In addition, the predicted products of the BGCs include metabolites with antibacterial, cytotoxic, antibacterial–cytotoxic activities. The total length of these predicted BGCs is ca. 2540 kb, making up ∼25.03% of the genome.
Table 1. Specialized Metabolite Biosynthetic Gene Clusters Predicted in the Genome of S. scabiei Using antiSMASH 5.0.
| BGC | BGC type | start | stop | length (bp) | most similar known clustera (% similarity)b | MIBiG ID |
|---|---|---|---|---|---|---|
| 1 | NRPS | 122 258 | 207 140 | 84 883 | pyochelin (73%) | BGC0001801 |
| 2 | betalactone | 210 499 | 245 699 | 35 201 | esmeraldin (8%) | BGC0000935 |
| 3 | NRPS | 331 948 | 400 458 | 68 511 | cadaside A/B (28%) | BGC0001968 |
| 4 | lanthipeptide | 402 211 | 422 773 | 20 563 | labyrinthopeptin A1/A2/A3 (60%) | BGC0000519 |
| 5 | terpene | 561 366 | 581 352 | 19 987 | ebelactone (5%) | BGC0001580 |
| 6 | terpene | 602 733 | 628 185 | 25 453 | isorenieratene (100%) | BGC0001456 |
| 7 | lanthipeptide, bacteriocin | 958 523 | 983 885 | 25 363 | informatipeptin (100%) | BGC0000518 |
| 8 | butyrolactone | 1 354 553 | 1 365 035 | 10 483 | lactonamycin (5%) | BGC0000238 |
| 9 | terpene | 1 445 117 | 1 469 780 | 24 664 | hopene (92%) | BGC0000663 |
| 10 | siderophore | 2 067 195 | 2 079 101 | 11 907 | grincamycin (8%) | BGC0000229 |
| 11 | NRPS-like | 2 214 965 | 2 255 782 | 40 818 | s56-p1 (11%) | BGC0001764 |
| 12 | terpene | 2 273 305 | 2 294 341 | 21 037 | geosmin (100%) | BGC0001181 |
| 13 | bacteriocin | 2 343 879 | 2 354 292 | 10 414 | unknown | |
| 14 | terpene | 2 615 998 | 2 636 826 | 20 829 | FD-594 (8%) | BGC0000222 |
| 15 | siderophore | 2 789 747 | 2 799 722 | 9976 | unknown | |
| 16 | NRPS | 3 581 513 | 3 645 548 | 64 036 | thaxtomin A (50%) | BGC0000444 |
| 17 | T2PKS | 4 831 093 | 4 903 608 | 72 516 | spore pigment (83%) | BGC0000271 |
| 18 | T1PKS, NRPS | 4 907 595 | 4 958 875 | 51 281 | herboxidiene (2%) | BGC0001065 |
| 19 | lanthipeptide | 5 327 434 | 5 350 019 | 22 586 | unknown | |
| 20 | bottromycin, bacteriocin | 6 301 273 | 6 323 906 | 22 634 | bottromycin A2 (54%) | BGC0000469 |
| 21 | siderophore | 6 440 983 | 6 452 205 | 11 223 | desferrioxamine B/E (66%) | BGC0000940 |
| 22 | melanin | 6 584 191 | 6 592 550 | 8360 | melanin (80%) | BGC0000909 |
| 23 | T1PKS, butyrolactone | 6 949 013 | 7 031 953 | 82 941 | 4-hexadecanoyl-3-hydroxy-2-(hydroxymethyl)-2H-furan-5-one (54%) | BGC0000140 |
| 24 | ectoine | 7 828 037 | 7 838 435 | 10 399 | ectoine (100%) | BGC0000853 |
| 25 | NRPS-like | 8 050 515 | 8 093 153 | 42 639 | granaticin (5%) | BGC0000227 |
| 26 | terpene | 8 147 349 | 8 166 320 | 18 972 | unknown | |
| 27 | T1PKS, indole | 8 700 909 | 8 763 617 | 62 709 | 5-isoprenylindole-3-carboxylate/β-D-glycosyl ester (28%) | BGC0001483 |
| 28 | T1PKS | 8 793 628 | 8 848 502 | 54 875 | lasalocid (22%) | BGC0001648 |
| 29 | T3PKS | 8 870 608 | 8 911 792 | 41 185 | daptomycin (10%) | BGC0000336 |
| 30 | terpene | 9 109 516 | 9 130 190 | 20 675 | unknown | |
| 31 | T1PKS | 9 288 913 | 9 413 896 | 124 984 | concanamycin A (89%) | BGC0000040 |
| 32 | siderophore | 9 430 360 | 9 443 981 | 13 622 | unknown | |
| 33 | NRPS | 9 526 250 | 9 591 840 | 65 591 | scabichelin (100%) | BGC0000423 |
| 34 | terpene | 9 618 596 | 9 639 636 | 21 041 | formicamycins A-M (4%) | BGC0001590 |
BGCs for which the associated specialized metabolite(s) were annotated in the metabolomic experiments are indicated in bold font.
% Similarity represents the percentage of genes in the query cluster which are present in the hit BGC from MIBiG.
Metabolomic Profiling of S. scabiei Using Untargeted LC-MS2
To characterize the specialized metabolites produced by S. scabiei, the bacterium was cultured on three different agar growth media: modified yeast extract-malt extract-starch agar (YMSm), modified maltose-yeast extract-malt extract agar (MYMm), and oat bran agar (OBA). OBA is a plant-based medium that is known to support the production of phytotoxins such as thaxtomins,35 CFA-Ile,10 and concanamycins,36 while MYMm is a modified version of MYM, a rich medium used for assessing the production of specialized metabolites and other natural products by Streptomyces species.37−39 Agar cores prepared from MYMm and YMSm plate cultures of S. scabiei are able to cause necrosis and pitting of potato tuber tissue to varying degrees (Figure 1), suggesting that secreted phytotoxic compounds are produced during growth on the two media. The plate cultures were extracted with ethyl acetate, and the recovered metabolites were analyzed by untargeted LC-MS2 in both positive and negative ionization mode to detect as many compounds as possible. The resulting spectral data were then analyzed using the FBMN workflow within the GNPS platform to generate molecular networks along with quantitative results for statistical analysis within the networks.27 In addition, we employed the recently described BioDendro workflow, which enables hierarchical clustering of MS2 spectra and presents the results as a tree.28 After molecular networking, background and media subtraction, a total of 6260 compounds were detected (Figure 2A,B). The majority of these molecules were detected under all three culturing conditions, though distinct sets of metabolites were also found in each extract, with the OBA extract containing the highest number of unique metabolites (Figure 2B). The 15 most intense ions (by peak area) detected in the extracts (Figure 2C and Table 2) were annotated by performing a GNPS spectral library search or using NAP, SIRIUS, MetWork, or CFM-ID. Of these compounds, CFA-Ile, pyochelin, concanamycin A, and thaxtomin A are known metabolites produced by S. scabiei,1 while the other compounds have not been previously reported to be produced by this organism.
Figure 1.

Biological activity S. scabiei agar cores on potato tuber tissue. S. scabiei was cultured on MYMm and YMSm media, after which agar cores of each culture (87.22/MYMm, 87.22/YMSm) were placed onto tuber tissue slices along with agar cores from uninoculated media (MYMm, YMSm). The slices were photographed after 10 days incubation. The assay was conducted three times in total, and a representative image is shown.
Figure 2.
Metabolomic analysis of S. scabiei. (A) FBMN of S. scabiei metabolites extracted from MYMm, YMSm, and OBA and analyzed by untargeted LC-MS2 in both positive and negative ionization modes. Each node represents one fragmentation spectrum from a detected compound, and node size represents the summed intensity (peak area) of the ion from all samples. Edge thickness indicates the relative similarity of MS2 data between nodes. The pie charts indicate the relative abundance of each compound in the different extracts: MYM—red, YMS—blue, and OBA—yellow. The networks containing annotated compounds are circled in thick black: 1, thaxtomin A in negative ionization mode; 2, thaxtomin A in positive ionization mode; 3, CFA-Ile in negative ionization mode; 4, CFA-Ile in positive ionization mode; 5, concanamycin A in positive ionization mode; 6, bottromycin A2 in positive ionization mode; 7, bottromycin A2 in negative ionization mode; 8, desferrioxamine E in positive and negative ionization modes; 9, pyochelin in positive ionization mode; 10, indole acetic acid in positive ionization mode; 11, ectoine in positive ionization mode; 12, cyclo(l-Val-l-Pro) in positive ionization mode; 13, 211A decahydroquinoline and mairine B in positive ionization mode; 14, andrachcinidine in positive ionization mode; and 15, aerugine in positive ionization mode. The dots at the bottom of the figure indicate that other networks were detected but are not shown. (B) Venn diagram displaying node counts according to distribution among the S. scabiei culture extracts: MYM—red, YMS—blue, and OBA—yellow. (C) Base peak chromatogram from LC-MS2 analysis of S. scabiei culture extracts in positive (upper) and negative (lower) modes: MYM—red: YMS—blue, and OBA—green.
Table 2. Fifteen Most Intense Ions (by Peak Area) Detected in the S. scabiei Culture Extracts.
| m/z[M + H]+ | m/z [M – H]− | retention time (min) | formulaa | compound name | annotated using |
|---|---|---|---|---|---|
| 212.2006 | 210.1860 | 9.7489 | C13H25NO | (2S,4aS,5S,6R,8aR)-5-methyl-2-propyl-decahydroquinolin-6-ol (211A decahydroquinoline) | NAP |
| 322.2004 | 320.1865 | 15.6427 | C18H27NO4 | CFA-Ile | CFM-ID |
| 224.0365 | 222.0219 | 11.72 | C10H9NO3S | 2-(1,3-benzoxazol-2-ylthio)propanoic acid | NAP |
| 197.1153 | 195.1007 | 9.9636 | C10H16N2O2 | cyclo(l-Val-l-Pro) | GNPS libraries |
| 228.1957 | 226.1811 | 8.8787 | C13H25NO2 | 1-[(2S,6R)-6-[(2S)-2-hydroxypentyl]piperidin-2-yl]propan-2-one (andrachcinidine) | NAP |
| 228.196 | 226.1814 | 10.079 | C13H25NO2 | 1-[(2S,6R)-6-[(2S)-2-hydroxypentyl]piperidin-2-yl]propan-2-one (andrachcinidine) | NAP |
| 325.0676 | 323.0530 | 10.9854 | C14H16N2O3S2 | pyochelin | GNPS libraries |
| 308.1888 | 306.1706 | 10.3184 | C18H29NO3 | methyl-substituted CFA-Ile | CFM-ID |
| 184.1675 | 182.1529 | 9.2423 | C11H21NO | (4R,4aR,7R,7aR)-2,4,7-trimethyl-hexahydro-1H-cyclopenta[c]pyridin-7a-ol (mairine B) | NAP |
| 210.0596 | 208.0450 | 9.8113 | C10H11NO2S | 2-[(4R)-4-(hydroxymethyl)-4,5-dihydro-1,3-thiazol-2-yl]phenol (aerugine) | NAP |
| 421.1512 | 419.1366 | 9.6884 | C22H20N4O5 | thaxtomin A derivative | MetWork |
| 213.2039 | 211.1893 | 9.8261 | b | b | |
| 699.4439 | 697.4293 | 13.6265 | C40H60NO9 | steroids and steroid derivatives | NAP |
| 888.5071 | c | 12.6819 | C46H75NO14 | concanamycin A | GNPS libraries |
| 439.1598 | 437.1452 | 9.4529 | C22H22N4O6 | thaxtomin A | GNPS libraries |
Predicted using SIRIUS.
No annotation/structure prediction.
Not detected.
Of the annotation tools used, the GNPS spectral library search is the most reliable method, as it compares the experimental MS2 spectra obtained to a catalogue of more than 221 000 MS2 reference spectra for known metabolites.29 However, for a large number of microbial specialized metabolites, there are no matching MS2 spectra in the reference libraries, and thus web-based in silico prediction tools can be used to obtain putative identifications for such molecules. For example, CFM-ID is a widely used web server that employs a probabilistic generative model to predict possible candidate structures for a target MS2 spectrum.33 NAP uses a combination of spectral similarity molecular networks and in silico fragmentation to improve annotation rates and quality through automated propagation.30 MetWork is an annotation propagation tool that incorporates in silico metabolization of known metabolites,32 and SIRIUS is mainly used for determining the sum formula of a metabolite.31 Overall, these annotations tools are complementary to each other as they each have unique advantages for the annotation of MS2 data.
The following is a description of the known metabolites annotated in the S. scabiei metabolome along with molecules that were not previously known to be produced by this organism.
Thaxtomins
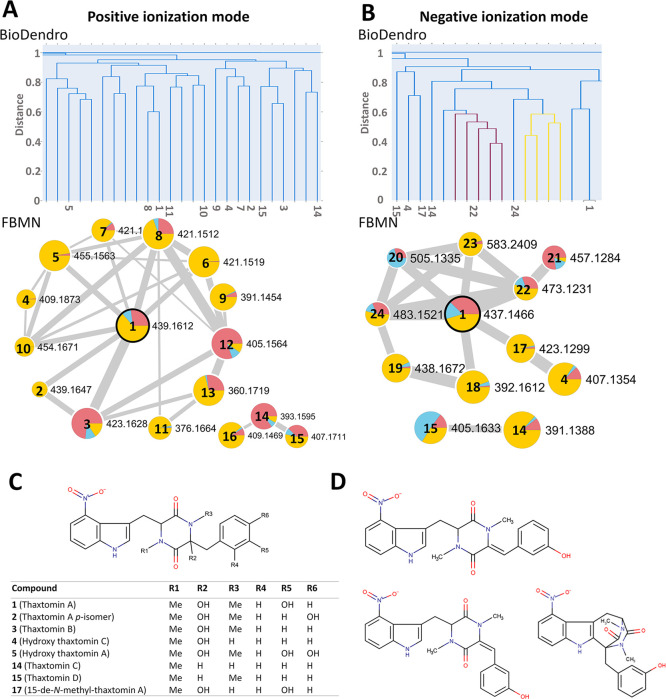
The BGC for production of thaxtomins (Figure S1) has been well-characterized and was detected by both antiSMASH and DeepBGC (Tables 1 and S1).6,8,40 Eleven thaxtomin analogues have been previously identified from different pathogenic Streptomyces species, and these vary in the presence or absence of hydroxyl and N-methyl groups on the thaxtomin backbone.7 Using FBMN, two thaxtomin networks were annotated from the metabolome of S. scabiei, with one in positive ionization mode (15 compounds) and the other in negative ionization mode (9 compounds) (Figures 2A, 3A,B, and Table S2). Results indicated that OBA is the best medium for supporting the production of most compounds in the networks, consistent with the presence of cellobiose and other cello-oligosaccharides in oat-based media, which are known inducers of thaxtomin production in S. scabiei and other Streptomyces species.35,41 In contrast, the production of thaxtomins in MYMm and YMSm has not been described before, and neither contain cello-oligosaccharides or the other known thaxtomin inducer, suberin.42
Figure 3.
Visualization of fragmentation spectra for thaxtomin metabolites from S. scabiei. Similarity between MS2 spectra was explored using BioDendro (top) and FBMN (bottom) in positive ionization mode (A) and in negative ionization mode (B). In FBMN, edges are created if the cosine score is >0.7 and there are at least four matched fragment ions. In the BioDendro trees, all links connecting nodes with Jaccard distances ≥0.6 are indicated in blue, and those with distances <0.6 are indicated in other colors. The numbers indicated in the trees correspond to the node numbers shown in the FBMN networks. Features in the FBMN networks are as described in the legend for Figure 2. Nodes that have MS2 matches in the GNPS spectral libraries are outlined in black. (C) Chemical structures of the metabolites annotated in the thaxtomin networks. (D) Chemical structures of the dehydrated analogues of thaxtomin A that have been described before.47
As expected, thaxtomin A (1; m/z 439.1612, [M + H]+; m/z 437.1466, [M – H]−) was the predominant thaxtomin analogue and was annotated using the GNPS spectral libraries (Figure 3). The annotation was supported by the comparison of key MS2 fragments with those reported for thaxtomin A43 (Table S2). Based on the precursor ion mass, MS2 fragments, and retention time (Table S2), 2 was predicted to be an isomer of thaxtomin A. Two thaxtomin A isomers, p-isomer and o-isomer, have been reported to be produced in minor amounts by S. scabiei,44 and using CFM-ID 3.0, we predicted that the p-isomer is the best candidate match for 2. Compound 3 (m/z 423.1628, [M + H]+) was predicted to be thaxtomin B based on its precursor ion mass and MS2 fragmentation pattern (Table S2).45 Based on the mass, comparative chromatographic data and CFM-ID prediction, 4 (m/z 409.1873, [M + H]+) was predicted to be a monooxygenated analogue of thaxtomin C, 5 (m/z 455.1563, [M + H]+) was predicted to be a monooxygenated derivative of thaxtomin A, and 17 (m/z 423.1299, [M – H]−) was predicted to be a 15-de-N-methyl analogue of thaxtomin A. The three thaxtomin analogues mentioned above have all been previously reported to be isolated from S. scabiei.7,46 Using MetWork, 6 (m/z 421.1519, [M + H]+), 7 (m/z 421.1477, [M + H]+), and 8 (m/z 421.1512, [M + H]+) were putatively identified as C-14 dehydrated analogues of thaxtomin A, of which 8 was among the 15 most intense ions detected in the culture extracts in the current study (Table 2). Dehydrated analogues of thaxtomin A have only been reported as biotransformation products of Aspergillus niger(47) and have not been detected in S. scabiei before. Notably, thaxtomin C (14; m/z 393.1595, [M + H]+; m/z 391.1388, [M – H]−) and thaxtomin D (15; m/z 407.1711, [M + H]+; m/z 405.1633, [M – H]−) were not readily annotated using molecular networking, whereas they could be annotated in the hierarchical MS2 spectral trees generated by BioDendro (Figure 3A,B). When crosschecked with the FBMN network, thaxtomin C and D were found to form a separate network from the main thaxtomin network in both the positive and negative ionization mode (Figure 3A,B). On the other hand, other related ions from the network were not clustered in the BioDendro tree, which illustrates how different similarity and visualization methods are complementary to explore spectral similarity in metabolomic experiments. Thaxtomin C is the major form of thaxtomin produced by the sweet potato pathogen Streptomyces ipomoeae,48 and trace amount of thaxtomin C and thaxtomin D have also been reported from S. scabiei,7,49 but their production has never been reported from S. scabiei cultured on MYMm and YMSm. Other compounds in these networks have mass fragmentation pattern similar to thaxtomins (Table S2), but the predicted structures based on NAP, SIRIUS, MetWork, or CFM-ID are unrelated to the thaxtomins, and thus they might be new derivatives. Therefore, further studies will need to be conducted to characterize these compounds.
Coronafacoyl Phytotoxins
The 31 kb PK BGC responsible for the production of coronafacoyl phytotoxins in S. scabiei has been described before (Figure S2).10 antiSMASH detected a 54 kb BGC (#28), while DeepBGC identified a 116 kb BGC (#120), both containing the 31 kb coronafacoyl phytotoxin BGC (Tables 1 and S1). CFA-Ile has been shown to be the main coronafacoyl phytotoxin produced by S. scabiei, though other minor compounds that are likely coronafacoyl derivatives have also been detected in culture extracts.9,50 In this study, the metabolomic analysis of S. scabiei using FBMN allowed us to observe and annotate two putative coronafacoyl phytotoxin networks. One was observed in positive ionization mode and one in negative ionization mode (Figure 4A,B), and they contained 10 and 14 compounds, respectively (Table S3). Most of the compounds from both networks could be detected in all three of the culture extracts, although they were generally more abundant in the MYMm and YMSm extracts. While production of coronafacoyl phytotoxins in oat-based media has been described before,51 the current study is the first to detect the production of these molecules in YMSm and MYMm.
Figure 4.
Visualization of fragmentation spectra for the putative coronafacoyl phytotoxins from S. scabiei. Similarity between MS2 spectra was explored using BioDendro (top) and FBMN (bottom) in positive ionization mode (A) and in negative ionization mode (B). The features of the BioDendro trees and the FBMN networks are as described in the legends for Figures 2 and 3. (C) Chemical structures of metabolites putatively annotated in the coronafacoyl phytotoxin networks.
Coronafacoyl phytotoxins possess a readily recognized MS2 fragmentation pattern, which includes peaks at m/z 191 and 163.52 The mass spectra for all of the compounds in the two networks displayed the characteristic fragments m/z 191 and 163 except for 27, 29, 42, and 43, which had prominent fragments corresponding to m/z 177 and 149 (Figure S3 and Table S3). Based on the precursor ion mass, MS2 fragments, and CFM-ID prediction, 25 was annotated as CFA-Ile (m/z 322.2004, [M + H]+; m/z 320.1863, [M – H]−) (Figures 4C and S3) and was the main coronafacoyl analogue as expected.9 We annotated 35 (m/z 320.1865, [M – H]−) as an isomer of CFA-Ile on the basis of the precursor ion mass, retention time, and mass fragmentation pattern (Table S3), and the only other reported isomer for CFA-Ile is N-coronafacoyl-l-allo-isoleucine (CFA-aIle) produced by Pseudomonas savastanoi.9 The mass fragmentation pattern of 27, 29, 42, and 43 suggested that these metabolites differ from the other coronafacoyl derivatives in that they contain a methyl group at position C-7 of the bicyclic hydrindane ring instead of an ethyl group (Figure S3). The production of methyl-substituted coronafacoyl derivatives was previously proposed based on our studies of the biosynthesis of CFA-Ile in S. scabiei,50 but this is the first time that such molecules have been detected in culture extracts of wild-type S. scabiei. Based on the mass fragmentation pattern and CFM-ID prediction, 27 was annotated as the methyl-substituted derivative of CFA-Ile (Figures 4C and S3) and was among the most intense ions detected in the extracts (Table 2). It is notable that the production of methyl-substituted coronafacoyl derivatives has not been observed in other coronafacoyl phytotoxin-producing bacteria and would presumably result from the incorporation of methylmalonyl-CoA instead of ethylmalonyl-CoA during the synthesis of the coronafacic acid polyketide moiety.50
28 (m/z 308.1856, [M + H]+) and 29 (m/z 308.1856, [M + H]+) both have the same mass as 27, but their fragmentation patterns matched that of CFA-Ile. As N-coronafacoyl-valine (CFA-Val) is a known coronafacoyl phytotoxin,9 and we previously showed that S. scabiei likely produces this compound,51 we annotated 28 and 29 as isomers of CFA-Val. Using MetWork, 37 and 38 are predicted to be decarboxylated derivatives of CFA-Ile and CFA-Val, respectively. Using BioDendro, we detected four additional coronafacoyl-related compounds, 46 (m/z 322.1978, [M + H]+), 47 (m/z 308.1895, [M + H]+), 48 (m/z 645.107, [M – H]−), and 49 (m/z 322.2010, [M – H]−), with mass fragmentation patterns similar to that of CFA-Ile. Based on their precursor ion mass and fragmentation pattern, we annotated 46 and 47 as isomers of CFA-Ile and CFA-Val, respectively. The remaining compounds in these two networks have mass fragmentation pattern similar to coronafacoyl phytotoxins, but the predicted structures based on NAP, SIRIUS, MetWork, or CFM-ID are completely unrelated to the coronafacoyl phytotoxins, and thus further characterization of these compounds is required.
Concanamycins
The BGC for the PK macrolide concanamycin was previously identified in S. scabiei (Figure S4)53 and was also detected by both antiSMASH and DeepBGC in the current study (Tables 1 and S1). It has been reported that oat-based media can support the production of concanamycin A and B in S. scabiei and other Streptomyces species,15−17,36 whereas production in MYMm and YMSm has not been previously investigated. Using FBMN, a concanamycin MS2 network consisting of 21 compounds was annotated in the metabolome of S. scabiei in positive ionization mode (Figure 5A and Table S4), whereas no concanamycin derivatives were detected in negative ionization mode. OBA was the best medium for supporting production of most of the concanamycins, though 50 and 52 are evenly distributed across all three media tested, and 54, 63, and 64 were found to be most abundant in the MYMm extract.
Figure 5.
Visualization of fragmentation spectra for the concanamycin metabolites from S. scabiei. (A) Similarity between MS2 spectra was explored using BioDendro (top) and FBMN (bottom) in positive ionization mode. The features of the BioDendro tree and the FBMN network are as described in the legends for Figures 2 and 3. (B) Chemical structures of metabolites annotated in the concanamycin network.
50 (m/z 874.4908, [M + Na]+) was annotated as concanamycin B by spectral matching with the GNPS libraries (Figure 5B), and we annotated 51 as an isomer of concanamycin B based on the precursor ion mass, retention time, and MS2 fragments (Table S4). 52 (m/z 888.5071, [M + Na]+) was annotated as concanamycin A using the GNPS spectral libraries and by inspection of its MS2 fragmentation pattern,12 and 53 and 54 were annotated as isomers of concanamycin A based on the precursor ion mass, retention time, and MS2 fragments (Table S4). The only previously reported isomer for concanamycin A was O-methyl-concanamycin B,13 which could be either 53 or 54 (Figure 5B). Using MetWork, 55 (m/z 902.5233, [M + Na]+) was predicted to be O-methyl-concanamycin A, and 56 (m/z 886.4888, [M + Na]+) and 57 (m/z 886.4922, [M + Na]+) were predicted to be oxidized analogues of concanamycin A based on the expected molecular formula differences (Figure 5B). Seven additional previously unreported concanamycin-related compounds (71–76) were discovered using BioDendro (Figure 5A and Table S4), suggesting that they may be novel derivatives, though further investigations are required.
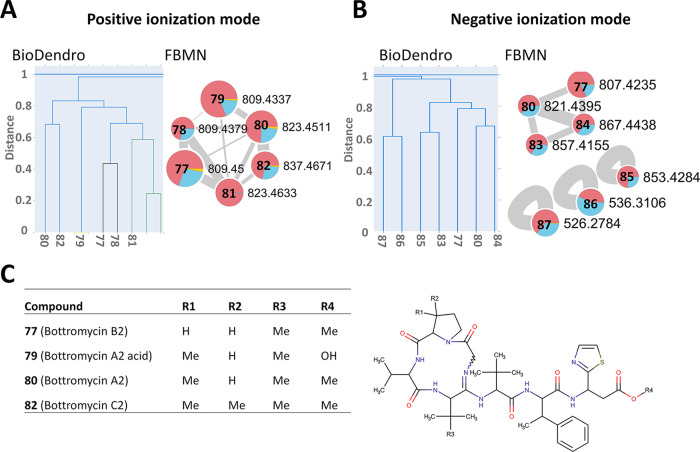
Bottromycins
The bottromycin BGC has been characterized in S. scabiei,54,55 where the production of the metabolite was detected on glucose-yeast extract-malt extract (GYM) medium, which is similar in composition to MYMm. The same RiPP BGC (Figure S5) was identified in the genome of S. scabiei by antiSMASH and DeepBGC (Tables 1 and S1). Using FBMN, we detected two putative bottromycin networks, one each in positive and negative ionization mode, containing six and four molecules, respectively. Their masses and fragmentation patterns of the metabolites are in good agreement with those of reported bottromycin molecules55 (Figure 6A,B and Table S5). MYMm was found to be the best medium for bottromycin production followed by YMSm, whereas very little production was detected when S. scabiei was cultured on OBA.
Figure 6.
Visualization of fragmentation spectra for the putative bottromycins from S. scabiei. Similarity between MS2 spectra was explored using BioDendro (left) and FBMN (right) in positive ionization mode (A) and in negative ionization mode (B). The features of the BioDendro trees and the FBMN networks are as described in the legends for Figures 2 and 3. (C) Chemical structures of metabolites annotated in the bottromycin networks.
Based on their precursor ion masses, MS2 fragmentation patterns, and retention times, 77 (m/z 809.4500, [M + H]+; m/z 807.4235, [M – H]−) was annotated as bottromycin B2, and 78 was annotated as bottromycin B1 (Figure 6A,B and Table S5). In addition, using NAP, it was predicted that 80 (m/z 823.4511, [M + H]+; m/z 821.4395, [M – H]−) is bottromycin A2, 79 (m/z 809.4377, [M + H]+) is bottromycin A2 acid based on shared MS2 fragments (141.1, 169.1, 268.2, 301.1, 363.2, 476.3, 639.4), 81 is an isomer of bottromycin A2, and 82 (m/z 837.4670, [M + H]+) is bottromycin C2. Analysis of the hierarchical MS2 tree created by BioDendro revealed the presence of three additional putative bottromycin-related compounds: 85 (m/z 853.4284, [M + H]+), 86 (m/z 526.2784, [M + H]+), and 87 (m/z 536.3106, [M + H]+), where the mass and fragmentation pattern of 85 matched that of carboxylated O-desmethyl bottromycins A2 (Table S5).55 Other compounds in these networks have mass fragmentation pattern similar to bottromycins, but the predicted structures based on NAP, SIRIUS, MetWork, or CFM-ID are completely unrelated to the bottromycins. Therefore, further experiments need to be done to characterize them.
Siderophores
Analysis of the S. scabiei genome revealed the presence of four siderophore BGCs: desferrioxamine, pyochelin, scabichelin, and one for an unknown product (Tables 1 and S1, Figures S6 and S7). The production of desferrioxamine, pyochelin, and scabichelin has previously been confirmed in this organism.20,22 The metabolomic analysis conducted here allowed us to annotate one pyochelin network in positive ionization mode and two desferrioxamine networks in positive and negative ionization modes (Figure 7A). However, we did not detect the presence of scabichelin in any of the samples in either positive or negative ionization mode. MYMm and OBA supported desferrioxamine and pyochelin production, while only low levels of the metabolites were detected in YMSm extracts. Spectral matching with the GNPS libraries enabled the annotation of 88 (m/z 601.3532, [M + H]+; m/z 599.3431, [M – H]−), 92 (m/z 325.0607, [M + H]+), and 94 (m/z 583.3459, [M – H]−) as desferrioxamine E, pyochelin, and dehydroxynocardamine (a derivative of desferrioxamine), respectively, which was further strengthened by comparison with published spectra.20,56 The use of NAP allowed us to putatively annotate 89 (m/z 401.2392, [M + H]+) as bisucaberin,30 which is part of a family of dihydroxamate siderophores originally isolated from the marine bacterium Alteromonas haloplanktis.56,57 Bisucaberin production has been reported in some Streptomyces species,58 though it has not been previously described for S. scabiei. The identical building blocks are used for the biosynthesis of bisucaberins and desferrioxamines,58,59 suggesting that the two metabolites are likely synthesized by gene products from the same BGC.
Figure 7.
Visualization of the similarity of fragmentation spectra for the siderophore metabolites from S. scabiei. Spectral dendrogram and network of desferrioxamine E in positive ionization mode (A) and in negative ionization mode (B), and the dendrogram and network of pyochelin in positive ionization mode (C). The dendrograms were created by BioDendro and the networks by FBMN, and the features of each are as described in the legends for Figures 2 and 3. (D) Putative structures of metabolites annotated in the siderophore networks.
Other Compounds
In the current study, a compound corresponding to indole-3-acetic acid (IAA) (m/z 176.0800, [M + H]+) (Figure 8) was detected in the S. scabiei OBA extract but not in the other extracts (Figure 2A). IAA is the major active form of auxins, which are plant hormones responsible for cell division, differentiation, root architecture formation, apical dominance, and senescence.60 Production of IAA has been reported in many plant pathogenic and plant growth-promoting microorganisms, including S. scabiei, and homologues of IAA biosynthetic genes were previously reported in the genome of S. scabiei 87.22.53 Ectoine (m/z 143.0700, [M + H]+) (Figure 8) was detected in MYMm, YMSm, and OBA extracts, with levels being highest in MYMm (Figure 2A). Ectoine is a water-soluble organic osmolyte that is produced by various Streptomyces spp., helping them to cope with extreme osmotic stress.61 The BGC for ectoine was also identified in S. scabiei by antiSMASH with 100% similarity (Table 1).
Figure 8.
Chemical structures of select metabolites identified/predicted from the metabolome of S. scabiei: (i) indole-3-acetic acid (IAA); (ii) ectoine; (iii) cyclo(l-Val-l-Pro); (iv) aerugine; (v) 211A decahydroquinoline; (vi) andrachcinidine; and (vii) mairine B.
Besides the known metabolites, several putative compounds not previously known to be produced by S. scabiei were also identified from the list of the 15 most intense ions detected. These included cyclo(l-Val-l-Pro), aerugine, decahydroquinoline, andrachcinidine, and mairine B (Figure 8 and Table 2). The cyclodipeptide cyclo(l-Val-l-Pro) and related metabolites are mainly formed by NRPSs or cyclodipeptide synthases, and they exhibit a variety of biological activities including plant growth promotion,62 cell-to-cell communication,63 antimicrobial, and anticancer.64 The production of cyclodipeptides has been demonstrated in other Streptomyces spp.;65 however, their corresponding BGCs have not been identified. Genomic analysis of S. scabiei revealed the presence of many short or incomplete NRPS BGCs, though none are predicted to utilize Val or Pro as substrates. In addition, cyclodipeptide synthase-encoding genes were not identified in the S. scabiei genome based on homology searches. Thus, it is currently unclear which BGC is involved in the biosynthesis of cyclo(l-Val-l-Pro) in S. scabiei. Aerugine is a siderophore that has been reported to be produced by Pseudomonas and Streptomyces species66 and is proposed to be derived from the hydrolytic cleavage and subsequent reduction of pyochelin.66 Therefore, aerugine and pyochelin, which were detected in our study, are likely synthesized using the same BGC. The decahydroquinolines are lipophilic alkaloids that have important pharmacological activities and have been reported in extracts from the skin of neotropical poison frogs.67 Andrachcinidine is a 2,6-disubstituted piperidine alkaloid that has been isolated from a small perennial plant Andrachne aspera Spreng and may function as a chemical defense agent.68 Mairine B is a new skytanthine-type monoterpenoid alkaloid that has been isolated from the plant Incarvillea mairei.69 The production of decahydroquinoline, andrachcinidine, and mairine B by Streptomyces species has not been previously reported. Therefore, further studies will be required to characterize these metabolites and to identify the BGCs responsible for their production in S. scabiei.
Conclusions
In this study, we used a combined genomic and metabolomic approach to explore the metabolic potential of S. scabiei. We showed that the genome of S. scabiei contains a large number of putative specialized metabolite BGCs that are predicted to produce a variety of molecules with diverse bioactivities. Using untargeted LC-MS2 along with FBMN and hierarchical clustering of MS2 spectra, we annotated known S. scabiei metabolites as well as putative new analogues, and these annotations were supported by the BGC annotations in several instances. Furthermore, we were able to detect and annotate new molecules that were not previously known to be produced by S. scabiei. We showed that the metabolic profile of S. scabiei varies among the three culture media tested, with most metabolites being observed in the plant-based medium OBA. This may reflect the plant pathogenic lifestyle of S. scabiei and the importance of specialized metabolism in mediating interactions between S. scabiei and plants, a notion supported by the fact that several of the known S. scabiei specialized metabolites exhibit phytotoxic activity. Overall, our study represents the first detailed analysis of the specialized metabolite potential of S. scabiei, an economically important plant pathogen that has a worldwide distribution. Further research is required to characterize the predicted novel metabolites identified and the associated BGCs to determine what role, if any, they may play in plant–microbe or microbe–microbe interactions.
Experimental Section
General Experimental Procedures
All media/reagents used in this study were obtained from Fisher Scientific or VWR International (Canada) unless otherwise specified. LC-MS2 analysis was carried out using a Thermo Fisher Scientific Vanquish UHPLC System coupled to a Thermo Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer. Separation was conducted on a Scherzo SM-C18 column (2 × 250 mm2, 3 μm, 130 Å; Imtakt) maintained at 40 °C using a water/acetonitrile gradient with 0.1% formic acid following the program from AbuSara et al.73 Mass spectra were recorded in mixed mode as described by AbuSara et al.73 The LC-MS2 data are available through the Mass Spectrometry Interactive Virtual Environment (MassIVE) data repository under the accession number MSV000085858.
Bacterial Strains and Culture Conditions
The pathogenic strain S. scabiei 87.22 was originally isolated by R. Loria from a scab lesion on a Russet Burbank potato tuber from Wisconsin.70 The strain was routinely cultured at 28 °C on potato mash agar (PMA) or in Trypticase Soy Broth (TSB) with shaking (200 rpm).51 For metabolite analysis, 100 μL of a spore stock of S. scabiei 87.22 was inoculated in TSB and was incubated for 24 h. Then, 100 μL of the TSB culture was spread onto OBA,35 YMSm, and MYMm plates, and the plates were incubated at 28 °C for 14 days. YMSm and MYMm are the same as YMS71 and MYM,72 respectively, except that they contain Bacto Malt Extract Broth (BD Biosciences) in place of malt extract.
Annotation of Specialized Metabolite Biosynthetic Gene Clusters
The complete genome sequence of S. scabiei 87.22 (NC_013929.1) was previously obtained by the Wellcome Trust Sanger Institute in collaboration with R. Loria (Cornell University) (http://www.sanger.ac.uk/Projects/S_scabies/). The sequence was uploaded to antiSMASH 5.0 (https://antismash.secondarymetabolites.org/#!/start) and DeepBGC (https://github.com/Merck/deepbgc) to identify specialized metabolite BGCs using the default parameters.26,34
Extraction of S. scabiei Specialized Metabolites
Metabolites were extracted from whole plate cultures of S. scabiei grown on YMSm, MYMm, and OBA, as well as from uninoculated control plates for each medium. Briefly, each agar plate was cut into small pieces using a sterile pipette tip and was then transferred to a clean 250 mL flask. Ethyl acetate (20 mL) was added to each flask, and the suspension was incubated overnight at room temperature with periodic shaking. The ethyl acetate extracts were each transferred to a clean evaporation flask, and the agar was rinsed with another 10 mL of solvent, which was then added to the corresponding evaporation flask. The solvent was removed by rotary evaporation, and the dried extracts were each redissolved in 1 mL of 100% v/v LC-MS-grade methanol. An aliquot of each extract (10 μL) was then used for LC-MS2 analysis.
Feature-Based Molecular Networking
Raw LC-MS2 data files were converted into mzXML format using MSConvert,74 and the data in positive and negative ionization mode were analyzed with MZmine2 (v2.53) for Feature-Based Molecular Networking analysis to generate three MS2 MGF files and three quantification CSV files: one in positive ionization mode, one in negative ionization mode, and another in mixed ionization mode.27 The MZmine2 parameter settings are outlined in Table S7. The peak areas of the control (uninoculated medium) samples were manually subtracted from the corresponding test sample data before uploading to the GNPS web platform for Feature-Based Molecular Networking.27,29,75 Cytoscape 3.7.276 was used to visualize the resulting molecular networks, and known metabolites were annotated by comparing the mass and mass fragmentation pattern with the GNPS spectral libraries29 and by cross-checking with the published result. Molecules that exhibited the same mass and mass fragmentation patterns but differed in their retention times were designated as isomers. Network Annotation Propagation (NAP)30 and the CFM-ID web server33 were used to putatively annotate known compounds. SIRIUS (version 4.0.1)31 was used for molecular formula prediction, and MetWork32 was used to predict the structures of unknown metabolites that were detected by spectral similarity analysis. Parameter settings used within SIRIUS, MetWork, and CFM-ID are outlined in Table S8, and web links to the FBMN, NAP, and MetWork jobs are provided in Table S9.
Spectral Hierarchical Clustering Using BioDendro
MS2 MGF files and the quantification table CSV files were exported from MZmine2 (v2.53) following the FBMN method. The MGF were manually edited to meet the requirements for BioDendro, and the quantification table file was then converted to a TXT file containing the feature list. The edited MGF file and the converted TXT file were then submitted to BioDendro using a distance threshold of 0.6. The detailed parameter settings are outlined in Table S10. The resulting trees were visualized using Plotly (Plotly Technologies Inc., Dendrograms in Python).
Phytotoxic Activity Assay
The potato tuber slice bioassay was performed as described before.70S. scabiei was cultured on YMSm and MYMm agar for 7 days until well sporulated, after which agar plugs from the plates were inverted onto the tuber slices along with control agar plugs from uninoculated media plates. The tuber slices were incubated in a moist chamber at 22–25 °C in the dark and were photographed after 10 days. The assay was conducted three times in total.
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada Grant No. RGPIN-2018-05155 to D.R.D.B., a Natural Sciences and Engineering Research Council of Canada Grant No. RGPIN-2018-05949 to K.T., and by the U.S. National Institutes of Health (NIH) R01 GM107550 to P.C.D. L-F.N. was supported by the NIH (R01 GM107550), the European Union’s Horizon 2020 program (MSCA-GF, 704786), and the Centre for Microbiome Innovation from the UC San Diego (Support Program award). J.L. was supported by a China Scholarship Council Grant No. 201603250060. The authors would like to thank Arshad Ali Shaikh (Memorial University of Newfoundland) for technical assistance with the FBMN, and Xin Lin (Verafin) for assistance with the DeepBGC and BioDendro analyses.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00526.
Supplementary bioinformatic and metabolomic data and parameters used for metabolomic data analysis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Li Y.; Liu J.; Díaz-Cruz G.; Cheng Z.; Bignell D. R. D. Virulence mechanisms of plant-pathogenic Streptomyces species: an updated review. Microbiology 2019, 165, 1025–1040. 10.1099/mic.0.000818. [DOI] [PubMed] [Google Scholar]
- Hiltunen L. H.; Ojanperä T.; Kortemaa H.; Richter E.; Lehtonen M. J.; Valkonen J. P. T. Interactions and biocontrol of pathogenic Streptomyces strains co-occurring in potato scab lesions. J. Appl. Microbiol. 2009, 106, 199–212. 10.1111/j.1365-2672.2008.03992.x. [DOI] [PubMed] [Google Scholar]
- Wanner L. A.; Kirk W. W. Streptomyces – from basic microbiology to role as a plant pathogen. Am. J. Potato Res. 2015, 92, 236–242. 10.1007/s12230-015-9449-5. [DOI] [Google Scholar]
- Loria R.; Bukhalid R. A.; Fry B. A.; King R. R. Plant pathogenicity in the genus Streptomyces. Plant Dis. 1997, 81, 836–846. 10.1094/PDIS.1997.81.8.836. [DOI] [PubMed] [Google Scholar]
- Dees M. W.; Wanner L. A. In search of better management of potato common scab. Potato Res. 2012, 55, 249–268. 10.1007/s11540-012-9206-9. [DOI] [Google Scholar]
- Li Y.; Liu J.; Adekunle D.; Bown L.; Tahlan K.; Bignell D. R. D. TxtH is a key component of the thaxtomin biosynthetic machinery in the potato common scab pathogen Streptomyces scabies. Mol. Plant Pathol. 2019, 20, 1379–1393. 10.1111/mpp.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. R.; Calhoun L. A. The thaxtomin phytotoxins: Sources, synthesis, biosynthesis, biotransformation and biological activity. Phytochemistry 2009, 70, 833–841. 10.1016/j.phytochem.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Loria R.; Bignell D. R. D.; Moll S.; Huguet-Tapia J. C.; Joshi M. V.; Johnson E. G.; Seipke R. F.; Gibson D. M. Thaxtomin biosynthesis: The path to plant pathogenicity in the genus Streptomyces. Antonie van Leeuwenhoek 2008, 94, 3–10. 10.1007/s10482-008-9240-4. [DOI] [PubMed] [Google Scholar]
- Bignell D. R. D.; Cheng Z.; Bown L. The coronafacoyl phytotoxins: structure, biosynthesis, regulation and biological activities. Antonie van Leeuwenhoek 2018, 111, 649–666. 10.1007/s10482-017-1009-1. [DOI] [PubMed] [Google Scholar]
- Bignell D. R. D.; Seipke R. F.; Huguet-Tapia J. C.; Chambers A. H.; Parry R. J.; Loria R. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant–microbe interactions. Mol. Plant-Microbe Interact. 2010, 23, 161–175. 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- Cheng Z.; Bown L.; Piercey B.; Bignell D. R. D. Positive and negative regulation of the virulence-associated coronafacoyl phytotoxin in the potato common scab pathogen Streptomyces scabies. Mol. Plant-Microbe Interact. 2019, 32, 1348–1359. 10.1094/MPMI-03-19-0070-R. [DOI] [PubMed] [Google Scholar]
- Haydock S. F.; Appleyard A. N.; Mironenko T.; Lester J.; Scott N.; Leadlay P. F. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology 2005, 151, 3161–3169. 10.1099/mic.0.28194-0. [DOI] [PubMed] [Google Scholar]
- Kinashi H.; Someno K.; Sakaguchi K. Isolation and characterization of concanamycins A, B and C. J. Antibiot. 1984, 37, 1333–1343. 10.7164/antibiotics.37.1333. [DOI] [PubMed] [Google Scholar]
- Seki-Asano M.; Okazaki T.; Yamagishi M.; Sakai N.; Hanada K.; Mizoue K. Isolation and characterization of new 18-membered macrolides FD-891 and FD-892. J. Antibiot. 1994, 47, 1226–1233. 10.7164/antibiotics.47.1226. [DOI] [PubMed] [Google Scholar]
- Natsume M.; Ryu R.; Abe H. Production of phytotoxins, concanamycins A and B by Streptomyces spp. causing potato scab. Jpn. J. Phytopathol. 1996, 62, 411–413. 10.3186/jjphytopath.62.411. [DOI] [Google Scholar]
- Natsume M.; Yamada A.; Tashiro N.; Abe H. Differential production of the phytotoxins thaxtomin A and concanamycins A and B by potato common scab-causing Streptomyces spp.. Jpn. J. Phytopathol. 1998, 64, 202–204. 10.3186/jjphytopath.64.202. [DOI] [Google Scholar]
- Natsume M.; Taki M.; Tashiro N.; Abe H. Phytotoxin production and aerial mycelium formation by Streptomyces scabies and S. acidiscabies in vitro. J. Gen. Plant Pathol. 2001, 67, 299–302. 10.1007/PL00013035. [DOI] [Google Scholar]
- Natsume M.; Komiya M.; Koyanagi F.; Tashiro N.; Kawaide H.; Abe H. Phytotoxin produced by Streptomyces sp. causing potato russet scab in Japan. J. Gen. Plant Pathol. 2005, 71, 364–369. 10.1007/s10327-005-0211-6. [DOI] [Google Scholar]
- Natsume M.; Tashiro N.; Doi A.; Nishi Y.; Kawaide H. Effects of concanamycins produced by Streptomyces scabies on lesion type of common scab of potato. J. Gen. Plant Pathol. 2017, 83, 78–82. 10.1007/s10327-017-0696-9. [DOI] [Google Scholar]
- Seipke R. F.; Song L.; Bicz J.; Laskaris P.; Yaxley A. M.; Challis G. L.; Loria R. The plant pathogen Streptomyces scabies 87-22 has a functional pyochelin biosynthetic pathway that is regulated by TetR- and AfsR-family proteins. Microbiology 2011, 157, 2681–2693. 10.1099/mic.0.047977-0. [DOI] [PubMed] [Google Scholar]
- Bicz J.Investigations of Siderophore and Tetronic Acid Biosynthesis in Streptomyces scabies 87.22. Ph.D. Thesis, University of Warwick, 2013. [Google Scholar]
- Kodani S.; Bicz J.; Song L.; Deeth R. J.; Ohnishi-Kameyama M.; Yoshida M.; Ochi K.; Challis G. L. Structure and biosynthesis of scabichelin, a novel tris-hydroxamate siderophore produced by the plant pathogen Streptomyces scabies 87.22. Org. Biomol. Chem. 2013, 11, 4686–4694. 10.1039/c3ob40536b. [DOI] [PubMed] [Google Scholar]
- Luo M.; Fadeev E. A.; Groves J. T. Mycobactin-mediated iron acquisition within macrophages. Nat. Chem. Biol. 2005, 1, 149–153. 10.1038/nchembio717. [DOI] [PubMed] [Google Scholar]
- Miethke M.; Marahiel M. A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. N.; Yen M. R.; Chiang C. Y.; Lin H. C.; Chen P. Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 2019, 103, 3277–3287. 10.1007/s00253-019-09708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G. D.; Prihoda D.; Palicka A.; Soukup J.; Klempir O.; Rampula L.; Durcak J.; Wurst M.; Kotowski J.; Chang D.; et al. A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 2019, 47, e110 10.1093/nar/gkz654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias L. F.; Petras D.; Schmid R.; Dührkop K.; Rainer J.; Sarvepalli A.; Protsyuk I.; Ernst M.; Tsugawa H.; Fleischauer M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson C.; Jones D.; Rakshit S.; Meka S.; Moffat C. S.; Moolhuijzen P. Hierarchical clustering of MS/MS spectra from the firefly metabolome identifies new lucibufagin compounds. Sci. Rep. 2020, 10, 6043 10.1038/s41598-020-63036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva R. R.; Wang M.; Nothias L.-F.; van der Hooft J. J. J.; Caraballo-Rodríguez A. M.; Fox E.; Balunas M. J.; Klassen J. L.; Lopes N. P.; Dorrestein P. C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089 10.1371/journal.pcbi.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcker S.; Letzel M. C.; Lipták Z.; Pervukhin A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics 2009, 25, 218–224. 10.1093/bioinformatics/btn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauxis Y.; Genta-Jouve G. MetWork: A web server for natural products anticipation. Bioinformatics 2019, 35, 1795–1796. 10.1093/bioinformatics/bty864. [DOI] [PubMed] [Google Scholar]
- Allen F.; Pon A.; Wilson M.; Greiner R.; Wishart D. CFM-ID: A web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014, 42, W94–W99. 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Steinke K.; Villebro R.; Ziemert N.; Lee S. Y.; Medema M. H.; Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. G.; Joshi M. V.; Gibson D. M.; Loria R. Cello-oligosaccharides released from host plants induce pathogenicity in scab-causing Streptomyces species. Physiol. Mol. Plant Pathol. 2007, 71, 18–25. 10.1016/j.pmpp.2007.09.003. [DOI] [Google Scholar]
- Fyans J. K.; Bown L.; Bignell D. R. D. Isolation and characterization of plant-pathogenic Streptomyces species associated with common scab-infected potato tubers in Newfoundland. Phytopathology 2016, 106, 123–131. 10.1094/PHYTO-05-15-0125-R. [DOI] [PubMed] [Google Scholar]
- Daniel-Ivad M.; Hameed N.; Tan S.; Dhanjal R.; Socko D.; Pak P.; Gverzdys T.; Elliot M. A.; Nodwell J. R. An engineered allele of afsQ1 facilitates the discovery and investigation of cryptic natural products. ACS Chem. Biol. 2017, 12, 628–634. 10.1021/acschembio.6b01002. [DOI] [PubMed] [Google Scholar]
- Hegemann J. D.; Zimmermann M.; Xie X.; Marahiel M. A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015, 48, 1909–1919. 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- Gehrke E. J.; Zhang X.; Pimentel-Elardo S. M.; Johnson A. R.; Rees C. A.; Jones S. E.; Hindra; Gehrke S. S.; Turvey S.; Boursalie S.; et al. Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated protein Lsr2. eLife 2019, 8, e47691 10.7554/eLife.47691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell D. R. D.; Francis I. M.; Fyans J. K.; Loria R. Thaxtomin A production and virulence are controlled by several bld gene global regulators in Streptomyces scabies. Mol. Plant-Microbe Interact. 2014, 27, 875–885. 10.1094/MPMI-02-14-0037-R. [DOI] [PubMed] [Google Scholar]
- Wach M. J.; Krasnoff S. B.; Loria R.; Gibson D. M. Effect of carbohydrates on the production of thaxtomin A by Streptomyces acidiscabies. Arch. Microbiol. 2007, 188, 81–88. 10.1007/s00203-007-0225-x. [DOI] [PubMed] [Google Scholar]
- Lerat S.; Simao-Beaunoir A.-M.; Wu R.; Beaudoin N.; Beaulieu C. Involvement of the plant polymer suberin and the disaccharide cellobiose in triggering thaxtomin A biosynthesis, a phytotoxin produced by the pathogenic agent Streptomyces scabies. Phytopathology 2010, 100, 91–96. 10.1094/PHYTO-100-1-0091. [DOI] [PubMed] [Google Scholar]
- Winn M.; Francis D.; Micklefield J. De novo biosynthesis of “non-natural” thaxtomin phytotoxins. Angew. Chem., Int. Ed. 2018, 57, 6830–6833. 10.1002/anie.201801525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. R.; Lawrence C. H.; Gray J. A. Herbicidal properties of the thaxtomin group of phytotoxins. J. Agric. Food Chem. 2001, 49, 2298–2301. 10.1021/jf0012998. [DOI] [PubMed] [Google Scholar]
- Jiang G.; Zuo R.; Zhang Y.; Powell M. M.; Zhang P.; Hylton S. M.; Loria R.; Ding Y. One-pot biocombinatorial synthesis of herbicidal thaxtomins. ACS Catal. 2018, 8, 10761–10768. 10.1021/acscatal.8b03317. [DOI] [Google Scholar]
- King R. R.; Lawrence C. H. Characterization of new thaxtomin A analogues generated in vitro by Streptomyces scabies. J. Agric. Food Chem. 1996, 44, 1108–1110. 10.1021/jf950243o. [DOI] [Google Scholar]
- Lazarovits G.; Hill J.; King R. R.; Calhoun L. A. Biotransformation of the Streptomyces scabies phytotoxin thaxtomin A by the fungus Aspergillus niger. Can. J. Microbiol. 2004, 50, 121–126. 10.1139/w03-110. [DOI] [PubMed] [Google Scholar]
- King R. R.; Lawrence C. H.; Calhoun L. A.; Ristaino J. B. Isolation and characterization of thaxtomin-type phytotoxins associated with Streptomyces ipomoeae. J. Agric. Food Chem. 1994, 42, 1791–1794. 10.1021/jf00044a042. [DOI] [Google Scholar]
- Jiang G.; Zhang Y.; Powell M. M.; Zhang P.; Zuo R.; Zhang Y.; Kallifidas D.; Tieu A. M.; Luesch H.; Loria R.; et al. High-yield production of herbicidal thaxtomins and thaxtomin analogs in a nonpathogenic Streptomyces strain. Appl. Environ. Microbiol. 2018, 84, e00164-18 10.1128/AEM.00164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown L.; Li Y.; Berrué F.; Verhoeven J. T. P.; Dufour S. C.; Bignell D. R. D. Coronafacoyl phytotoxin biosynthesis and evolution in the common scab pathogen Streptomyces scabiei. Appl. Environ. Microbiol. 2017, 83, e01169-17 10.1128/AEM.01169-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyans J. K.; Altowairish M. S.; Li Y.; Bignell D. R. D. Characterization of the coronatine-like phytotoxins produced by the common scab pathogen Streptomyces scabies. Mol. Plant-Microbe Interact. 2015, 28, 443–454. 10.1094/MPMI-09-14-0255-R. [DOI] [PubMed] [Google Scholar]
- Mitchell R. E. A naturally-occurring structural analogue of the phytotoxin coronatine. Phytochemistry 1984, 23, 791–793. 10.1016/S0031-9422(00)85028-1. [DOI] [Google Scholar]
- Bignell D. R. D.; Huguet-Tapia J. C.; Joshi M. V.; Pettis G. S.; Loria R. What does it take to be a plant pathogen: genomic insights from Streptomyces species. Antonie van Leeuwenhoek 2010, 98, 179–194. 10.1007/s10482-010-9429-1. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P.; Song L.; Bibb M. J.; Challis G. L. Posttranslational β-methylation and macrolactamidination in the biosynthesis of the bottromycin complex of ribosomal peptide antibiotics. Chem. Sci. 2012, 3, 3522–3525. 10.1039/c2sc21183a. [DOI] [Google Scholar]
- Crone W. J. K.; Vior N. M.; Santos-Aberturas J.; Schmitz L. G.; Leeper F. J.; Truman A. W. Dissecting bottromycin biosynthesis using comparative untargeted metabolomics. Angew. Chem., Int. Ed. 2016, 55, 9639–9643. 10.1002/anie.201604304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senges C. H. R.; Al-Dilaimi A.; Marchbank D. H.; Wibberg D.; Winkler A.; Haltli B.; Nowrousian M.; Kalinowski J.; Kerr R. G.; Bandow J. E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 2490–2495. 10.1073/pnas.1715713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. J.; Kimura N.; Yokose H.; Otsuka M. Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol. Biosyst. 2012, 8, 482–485. 10.1039/C1MB05431G. [DOI] [PubMed] [Google Scholar]
- Barona-Gómez F.; Wong U.; Giannakopulos A. E.; Derrick P. J.; Challis G. L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- Kadi N.; Song L.; Challis G. L. Bisucaberin biosynthesis: An adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem. Commun. 2008, 5119–5121. 10.1039/b813029a. [DOI] [PubMed] [Google Scholar]
- Morel M. A.; Castro-sowinski S.. The Complex Molecular Signaling Network in Microbe–Plant Interaction. In Plant Microbe Symbiosis: Fundamentals and Advances; Arora N. K., Ed.; Springer, New Delhi, 2013; pp 169–199. [Google Scholar]
- Sadeghi A.; Soltani B. M.; Nekouei M. K.; Jouzani G. S.; Mirzaei H. H.; Sadeghizadeh M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. 10.1016/j.micres.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Ortiz-Castro R.; Díaz-Pérez C.; Martínez-Trujillo M.; del Río R. E.; Campos-García J.; López-Bucio J. Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 7253–7258. 10.1073/pnas.1006740108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin P.; Moutiez M.; Lautru S.; Seguin J.; Pernodet J. L.; Gondry M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 2012, 29, 961–979. 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- Mishra A. K.; Choi J.; Choi S. J.; Baek K. H. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 2017, 22, 1796 10.3390/molecules22101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosse J. T.; Ghosh S.; Sproule A.; Overy D.; Cheeptham N.; Boddy C. N. Whole genome sequencing and metabolomic study of cave Streptomyces isolates ICC1 and ICC4. Front. Microbiol. 2019, 10, 1020 10.3389/fmicb.2019.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inahashi Y.; Zhou S.; Bibb M. J.; Song L.; Al-Bassam M. M.; Bibb M. J.; Challis G. L. Watasemycin biosynthesis in Streptomyces venezuelae: thiazoline C-methylation by a type B radical-SAM methylase homologue. Chem. Sci. 2017, 8, 2823–2831. 10.1039/C6SC03533G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama T.; Nishimori N.; Shimada A.; Edwards M. W.; Daly J. W. New classes of amidine, indolizidine and quinolizidine alkaloids from a poison-frog, dendrobates pumilio (dendrobatidae). Tetrahedron 1987, 43, 643–652. 10.1016/S0040-4020(01)89998-1. [DOI] [Google Scholar]
- Apichaisataienchote B.; Korpraditskul V.; Fotso S.; Laatsch H. Aerugine, an antibiotic from Streptomyces fradiae strain SU-1. Kasetsart J.: Nat. Sci. 2006, 40, 335–340. [Google Scholar]
- Xing A.; Tian J.; Liu C.; Li H.; Zhang W.; Shan L. Three new monoterpene alkaloids and a new caffeic acid ester from Incarvillea mairei var. multifoliolata. Helv. Chim. Acta 2010, 93, 718–723. 10.1002/hlca.200900284. [DOI] [Google Scholar]
- Loria R.; Bukhalid R. A.; Creath R. A.; Leiner R. H.; Olivier M.; Steffens J. C. Differential production of thaxtomins by pathogenic Streptomyces species in vitro. Phytopathology 1995, 85, 537–541. 10.1094/Phyto-85-537. [DOI] [Google Scholar]
- Ikeda H.; Kotaki H.; Omura S. Genetic studies of avermectin biosythesis in Streptomyces avermitilis. J. Bacteriol. 1987, 169, 5615–5621. 10.1128/JB.169.12.5615-5621.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuttard C. Temperate phages of Streptomyces venezuelae: Lysogeny and host specificity shown by phages SV1 and SV2. Microbiology 1982, 128, 115–121. 10.1099/00221287-128-1-115. [DOI] [Google Scholar]
- AbuSara N. F.; Piercey B. M.; Moore M. A.; Shaikh A. A.; Nothias L.-F.; Srivastava S. K.; Cruz-Morales P.; Dorrestein P. C.; Barona-Gómez F.; Tahlan K. Comparative genomics and metabolomics analyses of clavulanic acid-producing Streptomyces species provides insight into specialized metabolism. Front. Microbiol. 2019, 10, 2550 10.3389/fmicb.2019.02550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T.; Castillo S.; Villar-Briones A.; Orešič M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010, 11, 395 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.