Abstract
Saponins are the group of plant specialized metabolites which are widely distributed in angiosperm plants and have various biological activities. The present study focused on α-tomatine, a major saponin present in tissues of tomato (Solanum lycopersicum) plants. α-Tomatine is responsible for defense against plant pathogens and herbivores, but its biological function in the rhizosphere remains unknown. Secretion of tomatine was higher at the early growth than the green-fruit stage in hydroponically grown plants, and the concentration of tomatine in the rhizosphere of field-grown plants was higher than that of the bulk soil at all growth stages. The effects of tomatine and its aglycone tomatidine on the bacterial communities in the soil were evaluated in vitro, revealing that both compounds influenced the microbiome in a concentration-dependent manner. Numerous bacterial families were influenced in tomatine/tomatidine-treated soil as well as in the tomato rhizosphere. Sphingomonadaceae species, which are commonly observed and enriched in tomato rhizospheres in the fields, were also enriched in tomatine- and tomatidine-treated soils. Moreover, a jasmonate-responsive ETHYLENE RESPONSE FACTOR 4 mutant associated with low tomatine production caused the root-associated bacterial communities to change with a reduced abundance of Sphingomonadaceae. Taken together, our results highlight the role of tomatine in shaping the bacterial communities of the rhizosphere and suggest additional functions of tomatine in belowground biological communication.
α-Tomatine is the major toxic saponin secreted from tomato roots at high levels during early growth stages and plays an important role in the formation of bacterial communities in the rhizosphere.
Introduction
Plant specialized metabolites (PSMs) are the family of at least 1,000,000 structurally and functionally diverse compounds (Afendi et al., 2012). These compounds have various biological activities and play important roles in plants’ adaptation to environments and defense against pathogens and herbivores (Pichersky and Lewinsohn, 2011). Plants secrete PSMs into the rhizosphere, which is the zone of soil in close proximity to the roots (Hiltner, 1904; Hartmann et al., 2008; van Dam and Bouwmeester, 2016). Secreted PSMs contribute to the plant’s response to nutrient deficiency and the symbiosis, attraction, and repelling of soil-borne organisms, with concomitant influence on the microbiota (Massalha et al., 2017; Guerrieri et al., 2019).
Saponins are the group of PSMs composed of an aglycone hydrophobic backbone bound to hydrophilic saccharides such as glycosides. The amphiphilicity of these molecules results in soap-like and surface-active properties. Saponins are classified into three types according to aglycone structure: triterpenoid saponins, steroidal saponins, and steroidal glycoalkaloids (SGAs). They are biosynthesized from a common precursor, 2,3-oxidosqualene, via cyclization, oxidation, and glycosylation; these biosynthetic pathways have been intensely studied (Moses et al., 2014; Cardenas et al., 2019). Saponins are widely distributed in angiosperm plants and have a diverse range of biological properties including antibacterial, antifungal, insecticidal, and molluscicidal activities (Sparg et al., 2004; Augustin et al., 2011). They have also been found to have hemolytic activity, sweet or bitter tastes, foaming and emulsifying properties, and various pharmaceutical effects (Vincken et al., 2007; Cheok et al., 2014; Upadhyay et al., 2018). In contrast to the biosynthesis and functions of saponins in planta, their secretion from plant roots and biological functions in the rhizosphere are not well characterized, although secreted saponins are thought to have protective properties against pathogens and herbivores due to their anti-microbial properties (Francis et al., 2002). For instance, avenacin A-1—an oat (Avena strigosa) root triterpenoid saponin—is secreted from the roots and has been suggested to inhibit fungal growth and protect against various soil-borne fungi (Carter et al., 1999; Field et al., 2006). Through their studies on hydroponically grown plants, Tsuno et al. (2018) demonstrated that leguminous plants, including soybean (Glycine max) and Medicago truncatula, secrete soyasaponins, which are triterpenoid saponins commonly accumulated in legumes. It was recently shown that soyasaponin Bb enrich Novosphingobium, suggesting a potential role of saponins in modulating the rhizosphere microbial communities (Fujimatsu et al., 2020).
Among the saponin family, SGAs are commonly found in Solanaceae plants and exhibit a variety of bioactivities including toxicity toward diverse organisms such as bacteria, fungi, insects, worms, animals, and humans (Milner et al., 2011). The well-known SGA α-tomatine is a major component of all tomato (Solanum lycopersicum) plant tissues, with particularly high levels existing in the leaves and green fruits (Friedman, 2002). This compound is composed of aglycone tomatidine with an oligosaccharide group, lycotetraose. To date there have been four oxidases, four glycosyltransferases, a dehydrogenase, and two reductases identified as enzymes involved in the biosynthesis of α-tomatine, which proceeds from 2,3-oxidosqualene via cholesterol as an intermediate (Itkin et al., 2011, 2013; Sawai et al., 2014; Umemoto et al., 2016; Nakayasu et al., 2017; Sonawane et al., 2018; Akiyama et al., 2019; Lee et al., 2019). In addition, the expression of these biosynthetic genes has been found to be coordinately regulated by a jasmonate-responsive transcription factor of the APETALA2/ETHYLENE RESPONSE FACTOR family, JRE4/GAME9 (Cardenas et al., 2016; Thagun et al., 2016). Although tomatine has been shown to have antifungal effects, tomato pathogens such as Fusarium oxysporum f. sp. lycopersici degrade tomatine to the less toxic compound tomatidine using tomatinase, an extracellular hydrolase (Lairini et al., 1996; Sandrock and VanEtten, 1998). This detoxification of tomatine has been reported to be essential for the full virulence of pathogens toward tomato plants to be realized (Pareja-Jaime et al., 2008; Okmen et al., 2013). A low tomatine-producing JRE4 mutant (jre4-1) shows decreased resistance to the generalist herbivore Spodoptera litura larvae in the above-ground parts (Nakayasu et al., 2018). Tomatine is also phytotoxic toward lettuce (Lactuca sativa) and two weeds (Echinochloa crusgalli and Lolium perenne), and inhibits the germination of a tomato parasitic plant, Phelipanche ramosa (Rial et al., 2018). These findings indicate that tomatine is involved in the defense against pathogens and herbivores, as well as suppression of the growth of surrounding plants. Although the secretion of tomatine from plant roots has been demonstrated (Kirwa et al., 2018; Rial et al., 2018; Korenblum et al., 2020), the function of tomatine in the rhizosphere remains unclear. In the present study, we discovered that tomatine is highly secreted during the early growth stages of hydroponically grown plants. By comparing the bacterial communities of tomatine- and tomatidine-treated soils with tomato rhizosphere (Figure 1), we elucidated the role of these PSMs in terms of their modulation of bacterial communities to form the tomato rhizosphere via enrichment of Sphingomonadaceae.
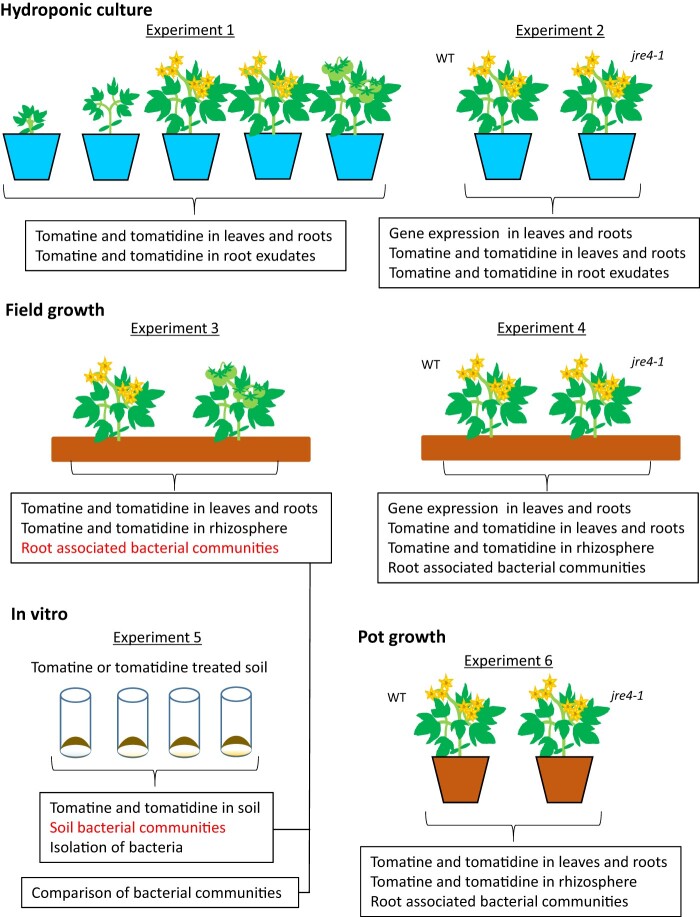
Figure 1.
Experimental design to study tomato growth in hydroponic system, field, and pot, and in vitro experiment to analyze the effect of tomatine or tomatidine on soil bacterial communities. Details for these experiments are described in the “Materials and methods” section.
Results
Accumulation and secretion of tomatine and tomatidine in hydroponically grown tomato plants
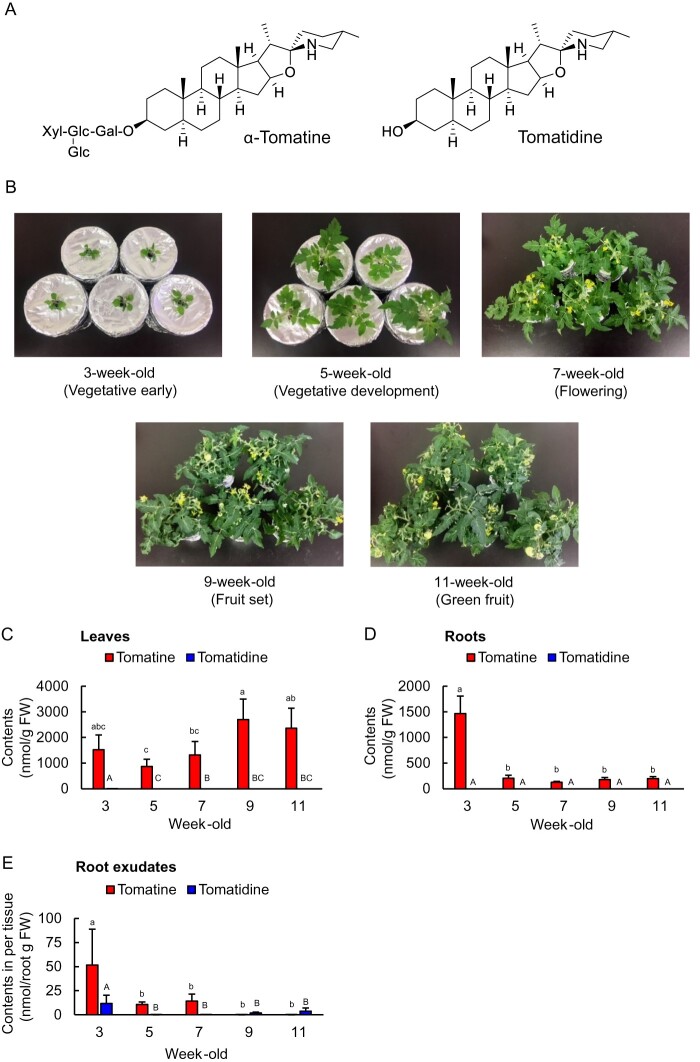
The results of quantification of the saponin contents of leaves, roots, and root exudates of 3-, 5-, 7-, 9-, and 11-week-old tomato plants are shown in Figure 2. The tomatine content in the leaves was found to be 1.3–2.7 µmol (g fresh weight [FW])−1, and it had a trend to increase at later growth stages. In contrast, the tomatine concentration in the roots was highest at 3 weeks old (1.5 µmol g FW−1), after which it decreased (Figure 2, C and D). The tomatine contents of both leaves and roots were substantially higher than those of its aglycone, tomatidine (Figure 2, C and D). In root exudates, the tomatine content per gram of root tissue (FW) was the highest at 3 weeks old (52 nmol g FW−1 day−1), after which it decreased (Figure 2, E). The tomatine concentrations of root exudates were higher than those of tomatidine at 3, 5, and 7 weeks (Figure 2, E).
Figure 2.
Levels of tomatine and tomatidine in tissue samples and root exudates of hydroponically grown tomatoes during different growth stages. A, The chemical structures of tomatine and tomatidine. B, Tomato plants at the different growth stages. The contents of tomatine and tomatidine in (C) leaves, (D) roots, and (E) root exudates; n = 5. Different letters (a, b, and c for tomatine; A, B, and C for tomatidine) indicate statistically significant differences (P < 0.05) for the contents of tomatine and tomatidine, respectively, by Tukey’s test. Error bars indicate standard deviation.
Accumulation and secretion of tomatine and tomatidine from field-grown tomato plants
Quantification of the saponin contents of leaves, roots, and rhizosphere soils of field-grown tomato plants at both the flowering and green-fruit stages (Figure 3, A and Supplemental Figure S1, A) revealed the tomatine contents of the leaves to be 1.3 and 1.5 µmol g FW−1 during the flowering and green-fruit stages, respectively, and 0.93 and 0.73 µmol g FW−1 during the flowering and green-fruit stages, respectively, in the roots (Figure 3, B and Supplemental Figure S1, B). The tomatine contents were substantially higher than those of tomatidine. The tomatine contents of leaves of hydroponically and field-grown tomato plants were comparable, whereas the roots of field-grown tomato plants contained substantially higher levels of tomatine than hydroponically grown plants (Figures 2, C and D, 3, B and Supplemental Figure S1, B). The tomatine contents of rhizosphere soil were 0.32 and 0.96 µmol g soil−1 during the flowering and green-fruit stages, respectively, and tomatine contents were substantially higher than those of tomatidine (Figure 3, C and Supplemental Figure S1, C). Tomatine levels in the rhizosphere during flowering and green-fruit stages of field-grown plants were comparable (Figure 3, C and Supplemental Figure S1, C). In the bulk soil, the levels of tomatine and tomatidine were below the quantification limit. Overall, these results suggest that tomatine is secreted from field-grown tomato roots and accumulates in the rhizosphere throughout all stages of growth.
Figure 3.
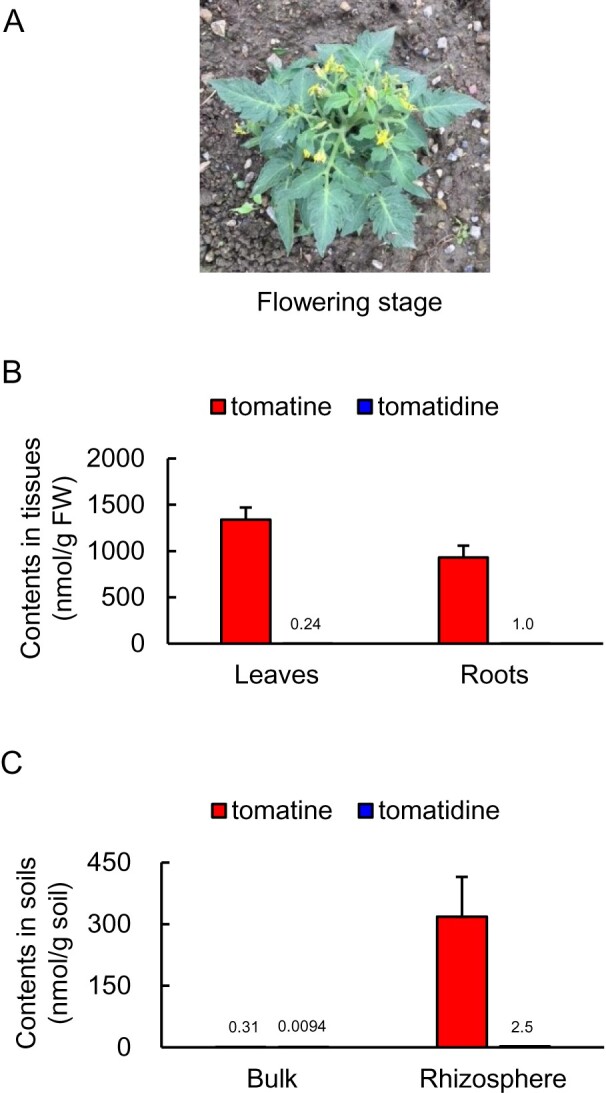
Analysis of tomatine and tomatidine concentrations in tissues of field-grown tomatoes and in soil samples. A, A photograph of a tomato plant at the flowering stage. The concentration of tomatine and tomatidine in (B) leaves and roots and (C) bulk and rhizosphere soil; n = 3. Error bars indicate standard deviation.
Effects of tomatine and tomatidine on bacterial communities in the soil
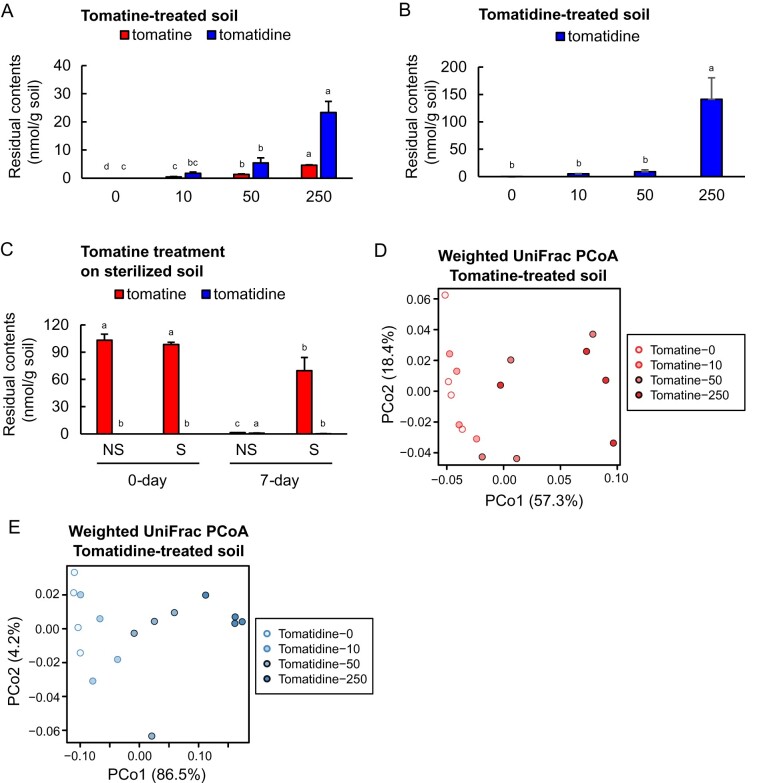
Although the effects of PSMs on the bacterial communities of the rhizosphere have been reported in several studies (Hu et al., 2018; Stringlis et al., 2018; Chen et al., 2019; Huang et al., 2019; Okutani et al., 2020; Voges et al., 2019), the effects of SGAs on the rhizosphere microbiota are not yet known. To analyze the effects of tomatine on the bacterial communities of the rhizosphere, we added 10, 50, and 250 µg g soil−1 of the pure compounds, that are, 10, 48, and 242 nmol g soil−1 for tomatine, and 22, 111, and 553 nmol g soil−1 for tomatidine, to field soil every 3 d for 15 d. After incubation, the tomatine contents of the soils were found to be 0.45–4.6 nmol g soil−1, depending on the concentration applied, and tomatidine was present in the range of 1.7–23 nmol g soil−1 (Figure 4, A). The contents of tomatidine in the field soils after incubation with authentic tomatidine ranged from 5.2 to 1.4 × 102 nmol g soil−1 (Figure 4, B). After 7 d of incubation, the tomatine content of nonsterile soils decreased to 3.7%, while >70% of the initial tomatine remained in γ-ray-sterilized soils after 7 d (Figure 4, C), indicating that biotic agents in the soil contribute to the degradation of tomatine.
Figure 4.
Analysis of the metabolites and bacterial communities of soils treated with tomatine and tomatidine. A, The residual contents of tomatine and its degradant tomatidine in soils treated with 10, 50, and 250 µg tomatine g soil−1, corresponding to 10, 48, and 242 nmol g soil−1 of tomatine (low, middle, or high concentrations) once every 3 d during the 15 d of incubation. B, The residual contents of tomatidine in soils treated with 10, 50, and 250 µg tomatidine g soil−1, corresponding to 22, 111, and 553 nmol g soil−1 of tomatidine once every 3 d during the 15 d of incubation. C, The residual contents of tomatine and tomatidine after 0 and 7 d of incubation in γ-ray-sterilized (S) and nonsterile (NS) soils treated with 100 μg (97 nmol) g soil−1 of tomatine. Error bars represent standard deviation in A and B (n = 4); C (n = 3). Different letters (a, b, and c for tomatine; A, B, and C for tomatidine) indicate statistically significant differences (P < 0.05) for the contents of tomatine and tomatidine, respectively, by Tukey’s test. D and E, Weighted UniFrac-based PCoA of tomatine- and tomatidine-treated soils as described in (A and B), respectively.
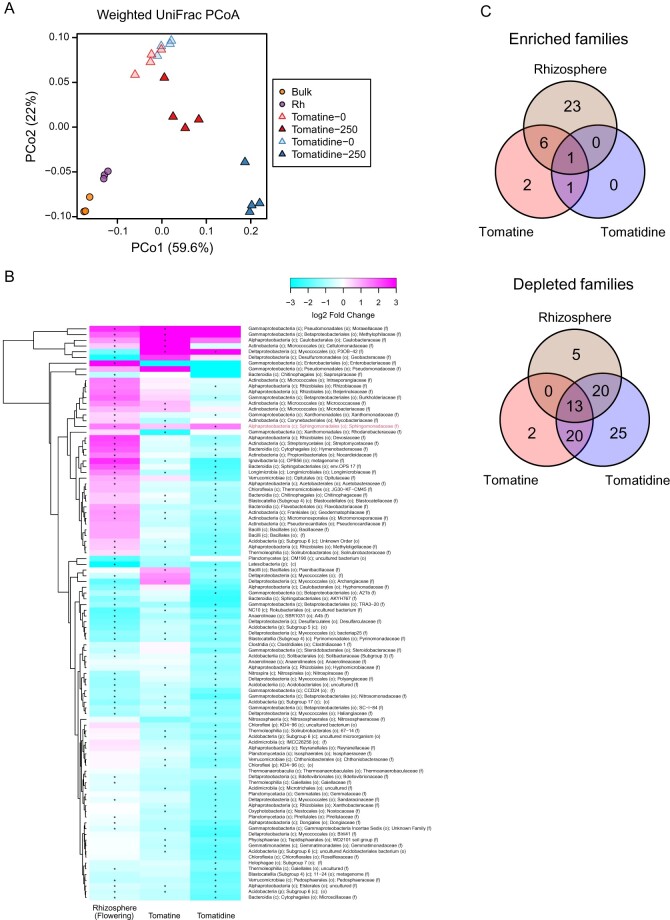
Analysis of the 16S rRNA amplicon sequence of tomatine- and tomatidine-treated soils through evaluation of the bacterial diversity in each sample (α-diversity), estimated as Shannon diversity, showed that both tomatine and tomatidine caused the α-diversity of soils to decrease when applied at the middle or high concentrations (Supplemental Figure S2). Distinct rhizosphere bacterial communities from bulk soil were observed both in flowering and green-fruit stages permutational multivariate analysis of variance (PERMANOVA, P = 0.001; Supplemental Figure S3 and Supplemental Table S1). The α-diversity of the rhizosphere soils was low compared with the bulk soil (Supplemental Figure S2). In the principal coordinate analysis (PCoA), the microbial diversity of untreated soil was clearly distinct from that of tomatine-treated soil (PERMANOVA, P = 0.003, Figure 4, D and Supplemental Table S1) and from that of tomatidine-treated (PERMANOVA, P = 0.001, Figure 4, E and Supplemental Table S1), indicating that both tomatine and tomatidine had a significant effect on bacterial assemblage. Comparison of the bacterial communities of tomatine- and tomatidine-treated soils with the bulk soil and rhizosphere soils of tomato fields in flowering and green-fruit stages revealed the distinction of bacterial communities between field and in vitro samples ( Figure 5, A and Supplemental Figure S4, A), but the weighted UniFrac distance between tomatine-treated soils and the rhizosphere was smaller than that between tomatine-treated soils and bulk soil (Supplemental Figure S5, A). The weighted UniFrac distance between tomatidine-treated soils and the rhizosphere was also smaller than that between tomatidine-treated soils and the bulk soil (Supplemental Figure S5, B). Tomatine and tomatidine treatments caused the enrichment of 10 and 2 families, respectively, including families that are commonly enriched in the tomato rhizosphere at the flowering and green-fruit stages, respectively (Figure 5, B and C, Supplemental Figure S4, B and C, and Supplemental Table S2). However, tomatine and tomatidine also caused the depletion of 35 and 78 families, respectively ( Figure 5, B and C, Supplemental Figure S4, B and C, and Supplemental Table S2). These results suggest that the two compounds influence the rhizosphere by depleting, rather than enriching bacterial taxa. Among the families that were enriched, only Sphingomonadaceae was enriched in both tomatine-treated, tomatidine-treated, and rhizosphere soils at both growth stages ( Figure 5, B and C, Supplemental Figure S4, B and C, and Supplemental Table S2). Among Sphingomonadaceae, the relative abundance of bacteria from genus Sphingobium was particularly enriched both in tomatine- or tomatidine-treated soil and rhizosphere soil (Supplemental Figure S6, A). Fifteen amplicon sequence variants (ASVs) were found in Sphingobium, but one ASV (ASV 6) predominantly accumulated in these soils (Supplemental Figure S6, B and Supplemental Table S3).
Figure 5.
Comparison of the bacterial communities in tomatine- and tomatidine-treated soil with those in the soil from the fields with tomato plants at the flowering stage. A, Results of the weighted UniFrac-based PCoA. B, Clustered heatmaps of the relative abundances of bacterial families (the mean of all samples >0.1%). The color of the heatmaps indicate log2 fold changes; teal denotes −3 while dark pink denotes 3. Asterisks indicate statistically significant differences in tomatine- and tomatidine-treated soils and tomato rhizosphere soil using LEfSe (an adjusted P-value < 0.05). Red letter indicates consistently enriched taxa. C, Venn diagrams to illustrate the overlap of enriched or depleted bacterial families in tomatine- and tomatidine-treated soil and rhizosphere soil. The numbers of bacterial families are shown in the circles. c, class; f, family; o, order; p, phylum; Rh, rhizosphere.
Effect of the jre4 mutation on the bacterial community of the rhizosphere
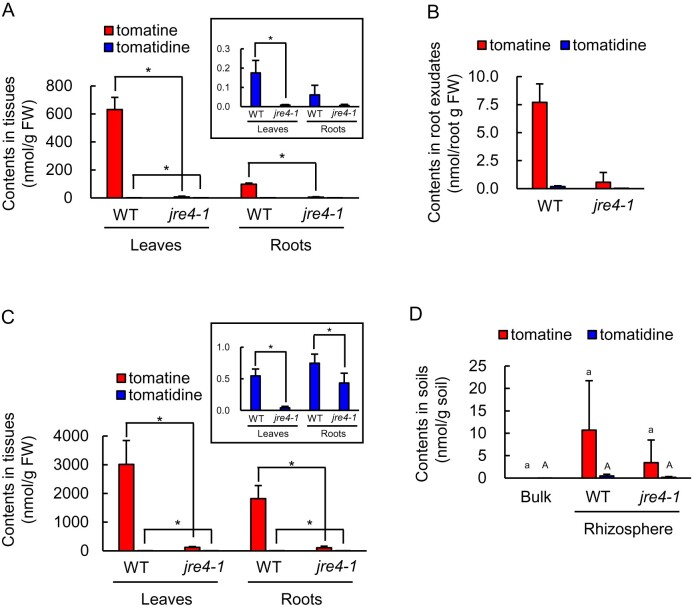
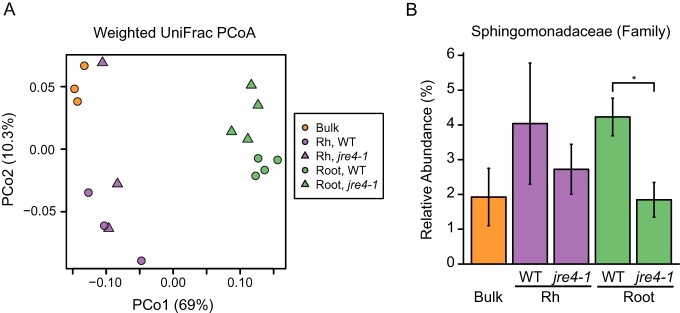
Previous studies have indicated that jre4-1 mutant accumulates only a 9% of tomatine content compared with wild type (WT) in the roots (Nakayasu et al., 2018). We found that the tomatine contents of leaves, roots, and root exudates of hydroponically grown jre4-1 tomato plants were >10-fold lower than those of WT plants (Figure 6, A and B). The effect of the jre4 mutation on the bacterial communities was analyzed both in field-grown and pot-grown plants. In field-grown conditions, the tomatine content in the leaves and roots of jre4-1 plants was >16-fold lower than those of WT plants (Figure 6, C). The tomatine content in the rhizosphere of jre4-1 plants was lower than the WT; however, the difference was not statistically significant (Figure 6, D). Bacterial communities of both rhizosphere and root endosphere were analyzed in jre4-1 and WT plants to reveal the effect of reduced tomatine biosynthesis on the bacterial communities. The α-diversity of the rhizosphere and root bacterial communities in jre4-1 plants was similar to those in the WT in field-grown condition (Supplemental Figure S7). PCoA showed a clear distinction between the root microbiome in mutant and WT plants (PERMANOVA, P = 0.027; Supplemental Table S1) but not between rhizosphere microbiome (PERMANOVA, P = 0.4; Supplemental Table S1; Figure 7, A). After removing the low-abundance taxa (mean < 0.1%), the differences between 123 families in the WT and jre4-1 mutant were statistically tested at an adjusted P value of < 0.05 (Supplemental Table S4). Nine families including Sphingomonadaceae were reduced in the root of the jre4-1 mutant (Figure 7, B and Supplemental Table S4). Among Sphingomonadaceae, the relative abundance of genus Sphingobium was notably lower both in the rhizosphere and root bacterial communities of the jre4-1 mutant than the WT (Supplemental Figure S8, A). Among ASVs closely related to Sphingobium; in particular, the same ASV (ASV 6) that was enriched in the tomatine- or tomatidine-treated soil and tomato rhizospheres was reduced in the jre4-1 mutant (Supplemental Figure S8, B and Supplemental Table S3).
Figure 6.
Metabolite concentrations in tissue and soil samples of WT and jre4-1 tomato plants grown in hydroponic and field conditions. The content of tomatine and tomatidine in (A) leaves and roots and (B) root exudates of hydroponically grown WT and jre4-1 plants at the flowering stage. The concentration of tomatine and tomatidine in (C) leaves and roots and (D) bulk and rhizosphere soil of field-grown WT and jre4-1 plants at the flowering stage. Error bars indicate standard deviation in A, B, C (roots of jre4-1), and D, n = 3; C (leaves of WT and jre4-1 and roots of WT), n = 4. Asterisks indicate statistically significant differences (P < 0.05) by Student’s t test. Different letters (a–c for tomatine; A–C for tomatidine) indicate statistically significant differences (P < 0.05) for the contents of tomatine and tomatidine, respectively, by Tukey’s test. Rh, Rhizosphere.
Figure 7.
Comparison of the bacterial communities in bulk and rhizosphere soil and root endosphere between field-grown WT and jre4-1 tomato plants. A, The weighted UniFrac-based PCoA. B, The relative abundances of Sphingomonadaceae in each compartment of WT and jre4-1 plants. Bulk and Rh, n = 3; Root, n = 4. Asterisk indicates statistically significant differences using LEfSe (an adjusted P-value < 0.05). Rh, rhizosphere. Error bars indicate standard deviation.
In contrast to field condition, light and temperature were controlled for pot cultivation, and the effect of jre4 mutation could be different from those in the field. The tomatine contents in the roots and rhizosphere soils of mutant plants were >5-fold lower than those of WT plants (Supplemental Figure S9, A and B). The α-diversity of the rhizosphere bacterial communities of 8-week-old plants was not significantly different between jre4-1 and WT plants (Supplemental Figure S9, C). PCoA showed a tendency of distinction between jre4-1 and WT plants in the rhizosphere and endosphere (PERMANOVA, P = 0.051; Supplemental Figure S9, D and Supplemental Table S1). Five families including Sphingomonadaceae were depleted in the roots of jre4-1 mutants in both pot and field cultivation (Supplemental Table S5). The relative abundance of Sphingomonadaceae was not reduced in the rhizosphere of jre4-1 plants (Supplemental Figure S9, E). The relative abundance of ASV 6 (Sphingobium) enriched in tomatine- or tomatidine-treated soil was higher in the rhizosphere than that in bulk soil but was not reduced in jre4-1 mutant compared with WT, which is inconsistent with the jre4-1 mutant grown in the field (Supplemental Figure S9, F and Supplemental Table S3). Collectively, these results demonstrate that JRE4 is involved in the modulation of root-associated bacterial communities both in field and pot cultivation, although the effect of JRE4 on bacterial communities was different between these conditions.
Discussion
Of all the carbon that is fixed during photosynthesis, up to 40% is secreted into the rhizosphere as root exudates (Badri and Vivanco, 2009). Rhizosphere microbial communities are key for maintaining plant growth and health (Berendsen et al., 2012), and recent evidence supports multiple roles of PSMs in shaping the composition and function of rhizosphere and root microbiome (Jacoby et al., 2021; Pascale et al., 2020). The involvement of triterpenoids, sesterterpenes, coumarins of Arabidopsis (Arabidopsis thaliana), and benzoxazinoids of maize (Zea mays) in the development of root-associated microbial communities has been demonstrated using mutants with disrupted PSM biosynthesis (Hu et al., 2018; Stringlis et al., 2018; Huang et al., 2019; Chen et al., 2019; Voges et al., 2019). We have previously reported that daidzein, a soybean isoflavone responsible for the initiation of symbiosis with rhizobia (Kosslak et al., 1987), shapes the bacterial communities of the rhizosphere (Okutani et al., 2020). The present study focused on the effects of tomatine, a major tomato saponin, on the bacterial communities of the rhizosphere, with the aim of deepening our understanding of the functions of PSMs within the rhizosphere. Our results show that the roots of hydroponically grown tomato plants secrete tomatine at high levels during the early growth stage, which is consistent with the secretion of PSMs from soybean roots. Soyasaponin and isoflavone secretion from soybean roots has been reported to occur during the early growth stage (Sugiyama et al., 2016; Tsuno et al., 2018). In field conditions, we found the tomatine content of the rhizosphere soil to be higher than the bulk soil, as has been observed for isoflavones secreted from field-grown soybean plants (Sugiyama et al., 2017). Several PSMs are metabolically activated by rhizosphere microbes and soil microbiota, leading to protection against biotic and abiotic stresses (Chialva et al., 2018; Korenblum and Aharoni, 2019). The increased tomatine contents of field-grown tomato roots compared with those of hydroponically grown tomato roots at the same growth stage suggest that soil-borne microbes may enhance tomatine biosynthesis in field-grown tomato roots by activating JRE4/GAME9 (Cardenas et al., 2016; Thagun et al., 2016). Indeed, the expression of JRE4/GAME9 in the roots of field-grown tomato was higher than that of hydroponically grown plant, while the expression of JRE4/GAME9 in leaves was comparable in both conditions (Supplemental Figure S10).
Tomatine secreted from tomato roots is degraded to tomatidine by soil microbes; thus, it is important to simultaneously analyze the effects of tomatidine as well as tomatine on soil microbes. The present study demonstrates that both tomatine and tomatidine modulate bacterial communities in the soil. Both compounds were found to increase the abundance of Sphingomonadaceae, a family commonly observed in the rhizosphere of tomato plants (Pii et al., 2016; Kwak et al., 2018; Lee et al., 2019). In addition, it should also be noted that Sphingobium isolated from tomatine-treated soil degrade tomatine and use it as a sole carbon source (Supplemental Figure S11), suggesting a possible role of tomatine as a nutrient source. It has been reported that several species of the Sphingomonadaceae family promote plant growth by producing phytohormones such as auxin and gibberellin, alleviating heavy metal and drought stress, and protecting against pathogens (Innerebner et al., 2011; Vogel et al., 2012; Khan et al., 2014; Pan et al., 2016; Asaf et al., 2017; Santiago et al., 2017; Luo et al., 2019). Based on these findings and those of the present study, we propose that tomato plant roots secrete tomatine in order to attract and/or stimulate the growth of Sphingomonadaceae in the rhizosphere, leading to the promotion of plant growth and protection against pathogens.
The relative abundance of Sphingomonadaceae was lower in the roots of the jre4-1 plants than in the WT in both field and pot conditions (Figure 7, B and Supplemental Tables S4, S5). At the ASV level, ASV 6 belonging to Sphingobium was predominantly enriched in the tomatine- or tomatidine-treated soil and tomato rhizosphere (Supplemental Figure S6, B and Supplemental Table S3). The relative abundance of ASV 6 was reduced both in the rhizosphere and root of the jre4-1 plants compared with the WT in the field-grown condition; however, the rhizosphere enrichment of this ASV still occurred in jre4-1 plants compared with bulk soil (Supplemental Figure S8, B and Supplemental Table S3). The increase of Sphingomonadaceae, particularly for ASV 6, in the rhizosphere of jre4-1 plants both in field and pot conditions may be attributed to the following nonexclusive reasons. First, tomatine content of the rhizosphere soil may be high enough to enrich Sphingomonadaceae; the tomatine content of hydroponically grown jre4-1 plant roots was 6% of the content of WT plant roots, whereas the tomatine content of the mutant increased 18-fold and 7.6-fold when grown in the field and in a pot, respectively. This suggests that the microbe-induced stimulation of tomatine biosynthesis is due to unknown factors other than JRE4. The induction of tomatine biosynthesis in the roots of the jre4-1 plants resulted in the accumulation of tomatine in the rhizosphere soils possibly high enough for the enrichment of Sphingomonadaceae. Second, other metabolites in tomato may contribute to the enrichment of Sphingomonadaceae. Reportedly, Sphingomonadaceae is enriched in the endosphere and rhizosphere of nontomatine-producing species such as lettuce and sugar beet (Beta vulgaris) (Cardinale et al., 2015; Chapelle et al., 2016). Although daidzein causes enrichment of Comamonadaceae, a family that is commonly enriched in the soybean rhizosphere, silencing of isoflavone synthase does not reduce the abundance of this family (White et al., 2017; Okutani et al., 2020), suggesting that multiple metabolites contribute to the enrichment of bacterial families in the rhizosphere.
In conclusion, this study demonstrates that tomatine, an SGA of tomato plants, is a key PSM which contributes to the modulation of the rhizosphere microbiota. Tomatine is a well-known chemical defense against pathogens and herbivores. In the present study, we provide new insight into the function of tomatine in the soil in terms of the enrichment of Sphingomonadaceae. Our results reveal potential avenues for the utilization of tomatine and related PSMs for modulating the microbiota of the rhizosphere to enhance the health and production of field-grown tomato plants.
Materials and methods
Chemicals and plant materials
Authentic compounds of tomatine and tetracycline hydrochloride were purchased from the Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan) and FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan), respectively. Tomatidine (hydrochloride), resazurin (sodium salt), and natamycin were purchased from Cayman Chemical (Ann Arbor, MI, USA). Seeds of WT tomato (S. lycopersicum) cv Micro-Tom were purchased from Inplanta Innovations Inc. (Kanagawa, Japan). Tomato plants with the jre4-1 mutant allele were constructed using Micro-Tom as genetic background during a previous study (Nakayasu et al., 2018).
Culture and sampling of hydroponically grown tomato plants
Experiments 1 and 2
Tomato seeds were sterilized by soaking in 70% (v/v) ethanol for 1 min then 5% bleach for 15 min, after which they were rinsed with sterile water four times, followed by soaking in fresh sterile water overnight. Seeds were sown on Murashige–Skoog medium supplemented with 3% sucrose and 0.3% gellan gum and grown for 1 week at 25°C in the dark. One-week-old seedlings were transferred to hydroponic culture medium: 470 mL of autoclave-sterilized tap water containing 0.2% (w/v) Otsuka House No. 1 and No. 2 standard nutrients (Otsuka Chemical Co., Osaka, Japan) in 500-mL plastic pots. Tomato plants were grown at 25°C under a 16-h-light/8-h-dark cycle, and culture medium was exchanged with fresh medium once per week. Three samples of upper-expanded leaves, roots, and root exudates (secreted into the culture medium over 24 h) were sampled from WT tomato plants at 3, 5, 7, 9, and 11 weeks old representing the early growth, vegetative development, flowering, fruit-set, and green-fruit stages, respectively. Samples from jre4-1 plants were taken at the flowering stage.
Cultivation and sampling of field-grown tomato plants
Experiments 3 and 4
Field cultivation of the tomato cultivar Micro-Tom (WT) was carried out at the Kyoto University of Advanced Science (KUAS), Kameoka, Kyoto, Japan (34°99ʹ38”N, 135°55ʹ14”E). Tomato seeds were sown in pots filled with a 1:1 mixture of vermiculite and Tsuchitaro (Sumitomo Forestry Landscaping, Tokyo, Japan), a culture soil. Seedlings were grown at 25°C under a 16-h-light/8-h-dark cycle in the laboratory for about 1 month, then planted in the field on May 31, 2019. Three samples of upper fully expanded leaves, roots, bulk soil, and rhizosphere soil were taken from each Micro-Tom plant at the flowering (July 3) and green-fruit (July 22) stages. Samples were immediately frozen in dry ice and transferred to the laboratory for storage at −80°C. Bulk soils were collected at least 20 cm from the plants and rhizosphere soil samples (adhering to the roots) were collected from five plants for one sample, using brushes as previously described (Sugiyama et al., 2014). In addition, WT and jre4-1 tomato seedlings were planted in the field on June 25, 2020. Leaves, roots, and rhizosphere and bulk soils were collected at the flowering stage on July 30 with the method described above, with a modification to collect root samples for microbiome analysis. After the rhizosphere soil was brush away, the roots were placed in a PBS buffer containing 130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4 (pH 7.0), and 0.02% Silwet L-77, washed, and sonicated for 5 min (Bulgarelli et al., 2012). After rinsing with tap water, the roots were stored at −80°C until DNA extraction.
Extraction of metabolites from tomato tissues, hydroponic culture medium, and field soils
Freeze-dried tissue samples of Micro-Tom plants were ground using a mortar and pestle with liquid nitrogen. Metabolites were extracted from the powdered samples using 300 µL of methanol, followed by vortex, sonication for 15 min, and centrifugation at 10,000 x g for 5 min. The supernatants were collected and the extraction repeated. Hydroponic culture medium containing root exudates was filtered through paper filter, then loaded onto a Sep-Pak C18 Plus Short cartridge (Waters, Milford, MA, USA). The cartridge was washed with 500 mL of water and metabolites eluted with 6 mL of methanol. Eluted fractions were dried in vacuo and the resulting solid dissolved in 1 mL of methanol. Metabolites in soil samples (300 mg) from Micro-Tom were extracted using 300 µL of 50% (v/v) acetonitrile containing 0.1% (v/v) formic acid three times. The combined supernatants were dried and the resulting solid dissolved in 1 mL of methanol. All prepared samples were filtered through a 0.45-μm Minisart RC4 filter (Sartorius, Göttingen, Germany) and analyzed using liquid chromatography–mass spectrometry (LC–MS).
Liquid chromatograph–mass spectrometry
Tomatine and tomatidine were analyzed on an Acquity ultra-high-performance liquid chromatography (UPLC) H-Class/Xevo TQD (Waters). Samples were injected onto an Acquity UPLC HSS T3 column (1.7 µm, 2.1 × 100 mm2; Waters) with a UPLC HSS T3 VanGuard Precolumn (1.7 µm, 2.1 × 5 mm2) at 40°C. The mobile phases were water containing 0.1% (v/v) formic acid (solvent A) and acetonitrile (solvent B), and elution programs were 10%–55% B from 0 to 15 min (linear gradient), 55% B from 15 to 17.5 min, 100% B from 17.5 to 22.5 min, and 10% B from 22.5 to 28.5 min. The flow rate was 0.2 mL min−1. Mass spectra were acquired using positive electrospray ionization mode with the following settings: cone voltage, 60 V; capillary voltage, 3.15 kV; source temperature, 150°C; desolvation gas temperature, 400°C; nebulizer and desolvation N2 gas flow rates, 50 and 800 L h−1, respectively. Tomatine and tomatidine were detected using selected ion recording modes with m/z 1034.7 and m/z 416.5, respectively. Data analysis was conducted using MassLynx v. 4.1 software (Waters). The contents of tomatine and tomatidine were estimated from peak areas in comparison with calibration curves constructed using known concentrations of the authentic compounds.
Characterization of degradation of tomatine in field soil
Tomatine (100 µg) was dissolved in methanol and dried in a 5-mL plastic tube. We added 1 g of KUAS field soil and 500 µL of distilled water, vortexed the mixture, and incubation at 28°C in the dark. The residual compounds after 7 d of incubation were extracted using 3 mL of 50% (v/v) acetonitrile containing 0.1% (v/v) formic acid, prepared as described above, and analyzed using LC–MS. Sterilization of field soil was performed by irradiating with gamma radiation at 30 kGy (Japan Irradiation Service, Tokai, Japan).
Treatment of field soil with tomatine and tomatidine
Experiment 5
All procedures in this section were performed as described in Okutani et al. (2020). Briefly, three different amounts (20, 100, or 500 µg) of tomatine and tomatidine hydrochloride were dissolved in methanol and dried in 5-mL tubes. We added 2 g of KUAS field soil and 600 µL of distilled water to each tube, vortexed the mixture, and incubated at 28°C in the dark. The addition of tomatine and tomatidine was carried out by transferring soil samples to a new tube containing the relevant compound once every 3 d during the 15 d of incubation. The residual contents of tomatine and tomatidine in the samples were quantified as described above.
Growth of WT and jre4-1 plants grown in pot
Experiment 6
Seeds of WT and jre4-1 plants were sown in pots filled with KUAS field soils. Tomato plants were grown at 25°C under a 16-h light/8-h dark cycle in the laboratory. Bulk and rhizosphere soil and root samples were taken during the flowering stage. Outer soil samples at the most distant point from the plants were taken as bulk soil samples. Rhizosphere soil samples were collected from three plant roots using brushes for one sample, and the roots were washed with running tap water (Kawasaki et al., 2016). Quantification of tomatine and tomatidine in the bulk and rhizosphere soil and root samples was conducted as described above.
Analysis of bacterial communities
DNA was extracted from tomatine- and tomatidine-treated, field-grown, and pot-grown tomato rhizosphere soils samples, bulk soil samples, and tomato roots using the DNeasy PowerSoil Kit (QIAGEN K.K., Tokyo, Japan). The DNA concentration was measured using a Qubit dsDNA HS Assay Kit and Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The V4 region of 16S rRNA genes was amplified using PCR with the following forward and reverse primers: 515F (5′‐ACACTCTTTCCCTACACGACGCTCTTCCGATCT‐GTGCCAGCMGCCGCGGTAA‐3′) and 806R (5′‐GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT‐GGACTACHVGGGTWTCTAAT‐3′), respectively, using a KOD FX Neo (TOYOBO, Osaka, Japan) in technical triplicate. The following PCR program was used: 94°C for 2 min and 20 cycles of 98°C for 10 s, 50°C for 30 s, and 68°C for 30 s. Mitochondrial- and chloroplast-specific peptide nucleic acids (mPNA [N-term‐GGCAAGTGTTCTTCGGA‐C-term] and pPNA [N-term‐GGCTCAACCCTGGACAG‐C-term], respectively [PANAGENE Inc, Daejeon, Republic of Korea]), were added to the PCR reaction mixture and PNA annealing at 78°C for 10 s was inserted before 50°C for 30 s of PCR cycle described above for amplification of 16S rRNA from root samples in order to prevent amplification of tomato mitochondrial and plastid sequences (Lundberg et al., 2013). The PCR products were purified using AMPure XP (Beckman Coulter, Danvers, MA, USA) then used as templates for PCR amplification for Miseq (Illumina, San Diego, CA, USA) adapter attachment. The PCR reaction was performed using primers provided by FASMAC Co., Ltd. (Kanagawa, Japan) using KOD FX Neo in technical duplicate. The following PCR program was used: 94°C for 2 min and 9–10 cycles of 98°C for 10 s, 59°C for 30 s, and 68°C for 30 s. The PCR products were purified and quantified as described above. Sequencing of the prepared 16S amplicon mixture was conducted to acquire 2 × 250-bp paired-end sequences on Miseq (Illumina) in FASMAC Co., Ltd. The accession number of sequence data has been registered in the DNA Data Bank of Japan (Sequence Read Archive is DRA009882 and DRA011415.
Sequence data analysis was performed using the QIIME2 platform version 2018.11 (Bolyen et al., 2019). The q2-dada2 plugin in QIIME2 was used to trim the first 20 bases and the bases after the 200th bases of all paired reads, and for quality filtering. The DADA2 algorithm was used to construct error-corrected ASVs from the trimmed reads (Callahan et al., 2016). Each ASV was assigned to a taxonomic group using the Naïve Bayes classifier from the SILVA release 132 (Quast et al., 2013; Bokulich et al., 2018). Reads for chloroplasts or mitochondria were removed, then MAFFT was used to align the obtained ASVs. FastTree2 was used to construct phylogenetic trees (Price et al., 2010; Katoh and Standley, 2013). After filtering chloroplastic and mitochondrial sequences, 50,494–196,608 reads per sample were remained for tomatine- and tomatidine-treated soils and field samples of 2019, 19,004–277,548 reads for field samples of 2020, and 11,586–59,495 reads for pot soil and roots (Supplemental Table S6). The α- and β-diversity indices of tomatine- and tomatidine-treated soils and field samples of 2019 were calculated using the core-metrics-phylogenetic pipeline in the q2-diversity plugin within QIIME2 from a subsampled ASV dataset with 45,000 sequences per sample (Supplemental Figure S12). These indices were calculated from a subsampled ASV dataset with 19,000 and 11,000 sequences per sample for field samples of 2020 and for pot soil and roots, respectively (Supplemental Figure S12).
Isolation of Sphingobium from tomatine-treated soils
The bacterial strains were isolated from the soil treated with tomatine at 250 nmol g soil−1. The tomatine-treated soil was suspended in sterile water at 1.0 g mL−1. Soil suspensions were diluted and distributed onto isolation media containing mineral salt buffer (Devers et al., 2004) with 20 µg mL−1 of tomatine or tomatidine as the sole carbon source. The plates of isolation media were incubated for up to 7 d at 28°C. Genomic DNA was extracted from the isolates using the hot-alkaline DNA extraction method with an extraction buffer comprising 25 mM NaOH, 0.2 mM EDTA, and 40 mM Tris–HCl, pH 6.8. 16S rRNA genes were amplified using the 10F (5′-GTTTGATCCTGGCTCA-3′) and 800R (5′-TACCAGGGTATCTAATCC-3′) primers set. PCR products were purified using Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) and sequenced with the 10F primer.
Tomatine and tomatidine degradation assay was performed using resting cells of three Sphingobium isolates (Sp1, Sp2, and Sp3). Each isolate was cultivated in 3 mL of Tryptone Yeast extract Glucose Medium (Bai et al., 2015) for 2 d and then harvested by centrifugation at 4,000 x g for 5 min. Cell pellets were washed twice and then resuspended, each with 1 mL of mineral salt buffer. The cells were inoculated at OD600 = 1.0, added to 100 µL of mineral salt buffer with 50 µM tomatine or tomatidine and incubated at 28°C for 1 d. The resting cell reaction was stopped with 100 µL of methanol. The reaction product was filtered; then it was analyzed using LC–MS as described above. The utilization of tomatine as a carbon source by Sphingobium was evaluated with Sp2 growth assay. The harvested cells were inoculated at OD600 = 0.1 and added to 150 µL of the mineral salt buffer with 300 µg mL−1 resazurin, an indicator for bacterial growth (Sarker et al, 2007), and 100 µM tomatine or d-glucose as the carbon source. The cells were incubated at 28°C for 10 d; then, Sp2 proliferation was quantified by observing the cellular color change from blueish purple to pink.
Reverse transcription quantitative PCR
Total RNA was prepared from leaves and roots with RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. DNA was eliminated by DNase I. The RNA was then reverse transcribed to cDNA using ReverTra Ace -α- (Toyobo) according to the manufacturer’s instructions. Reverse transcription quantitative PCR was performed with FX96 Touch DeepWell system (Bio Rad), using Thunderbird SYBR qPCR Mix (Toyobo) according to the manufacturers’ instructions. The cycling program consisted of initial denaturation at 95°C for 2 min; 45 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s for amplification; and ramping up from 65°C to 95°C to perform a melting curve analysis. The gene expression levels were normalized against the ubiquitin gene (UBI3), the internal reference. UBI3’s amplification used the forward primer 5′-CACCAAGCCAAAGAAGATCAAGC-3′ and reverse primer 5′-TCAGCATTAGGGCATCCCTTACG-3′ (Nakayasu et al., 2017). JRE4’s amplification used the forward primer 5′-CGATTTTTTCGAAACTCTTTCC-3′ and reverse primer 5′- TGTTTCCTCCGGTGTTACGG-3′ (Nakayasu et al., 2018).
Statistical analysis
T test and Wilcoxon rank-sum test were carried out using t.test function and wilcox.test function of R package “stats,” respectively. Tukey’s test was performed using Tukey’s HSD function of R package “multcomp.” Hierarchical clustering was performed using the complete-linkage method with hclust function in the R package “stats.” Linear discriminant analysis (LDA) effect size (LEfSe; Segata et al., 2011) was performed to find differentially abundant taxa at the family level. Significant difference was decided using default parameters at an adjusted P value of < 0.05 for Kruskal–Wallis and pairwise Wilcoxon test and an LDA effect size > 2. PERMANOVA was performed with adonis function in the R package “vegan” (Oksanen et al., 2019).
Accession numbers
The Genbank ID number of JRE4 is XP_004229751.1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of tomatine and tomatidine concentrations in tissues of field-grown tomatoes and in soil samples.
Supplemental Figure S2. Shannon diversity indices of bacterial communities in soils treated with 10, 50, and 250 μg (low, middle, and high concentrations) of tomatine and tomatidine g soil−1 and tomato field soils at the flowering and green-fruit stages.
Supplemental Figure S3. The weighted UniFrac-based PCoA of the bacterial communities in bulk and rhizosphere soil of field-grown tomato plants at the flowering and green-fruit stages.
Supplemental Figure S4. Comparison of the bacterial communities in tomatine- and tomatidine-treated soil with those in the soil from the fields with tomato plants at the green fruit stage.
Supplemental Figure S5. Comparison of the bacterial communities in tomatine- and tomatidine-treated soil with those in the soil from the fields with tomato plants.
Supplemental Figure S6. The presence of Sphingomonadaceae in different types of soils.
Supplemental Figure S7. Shannon diversity indices of bacterial communities in bulk and rhizosphere soil and root endosphere of field-grown wildtype and jre4-1 tomato plants at the flowering stage.
Supplemental Figure S8. The presence of Sphingomonadaceae in bulk and rhizosphere soil (n = 3) and root endosphere (n = 4) of field-grown WT and jre4-1 tomato plants at the flowering stage.
Supplemental Figure S9. Comparing metabolite concentrations and the bacterial communities in bulk and rhizosphere soil and roots between pot-grown WT and jre4-1 tomato plants at the flowering stage.
Supplemental Figure S10. RT-qPCR analysis of the expression patterns of the JRE4/GAME9 gene in tissues of tomato plants grown in hydroponic and field conditions.
Supplemental Figure S11. Tomatine degradation and utilization by Sphingobium isolated from tomatine-treated soil.
Supplemental Figure S12. Rarefaction curves for observed ASVs richness of tomatine- and tomatidine-treated soil, field samples of 2019 and 2020, and pot soil and roots.
Supplemental Table S1. Results of PERMANOVA.
Supplemental Table S2. Significantly enriched or depleted bacterial families in tomatine/tomatidine-treated soil compared with control soil and in the tomato rhizosphere compared with bulk soil in field 2019.
Supplemental Table S3. The relative abundances and sequences of ASVs found in genus Sphingobium detected in samples analyzed for bacterial communities.
Supplemental Table S4. Significantly enriched or depleted bacterial families in rhizosphere and root endosphere of jre4-1 compared with those of WT in the field in 2020.
Supplemental Table S5. Significantly enriched or depleted bacterial families in bulk, rhizosphere and roots of jre4-1 compared with those of WT in pots.
Supplemental Table S6. Sequencing depth per sample analyzed for bacterial communities.
Supplementary Material
Acknowledgments
The authors thank Keiko Kanai for technical assistance. They also thank DASH/FBAS, the Research Institute for Sustainable Humanosphere, Kyoto University, for supporting institutional setting.
Funding
This study was supported in part by grants from Core Research for Evolutional Science and Technology, Japan Science and Technology Agency (CREST, JST; JPMJCR17O2 to A.S. and Y.A.), and Japan Society for the Promotion of Science (JSPS; 18H02313 to A.S.); from the Research Institute for Sustainable Humanosphere (Mission 1), and Research Unit for Development of Global Sustainability, Kyoto University.
Conflict of interest statement. The authors have no conflict of interest.
M.N. and A.S. conceived and designed the research. K.Y. and A.S. supervised the experiments. T.S. supervised the analyses using jre4-1 tomato mutant. M.N. performed the experiments on microbial communities and M.N., K.O., K.T., H.T., and T.S. performed the experiments of tomatine analysis. Y.A. and S.Y. analyzed the data. M.N., Y.A., S.Y., and A.S. wrote the article with contributions of all the authors. A.S. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Akifumi Sugiyama (akifumi_sugiyama@rish.kyoto-u.ac.jp).
References
- Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, et al. (2012) KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol 53: 12. [DOI] [PubMed] [Google Scholar]
- Akiyama R, Lee HJ, Nakayasu M, Osakabe K, Osakabe Y, Umemoto N, Saito K, Muranaka T, Sugimoto Y, Mizutani M (2019) Characterization of steroid 5 alpha-reductase involved in alpha-tomatine biosynthesis in tomatoes. Plant Biotechnol 36: 253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaf S, Khan AL, Khan MA, Imran QM, Yun BW, Lee IJ (2017) Osmoprotective functions conferred to soybean plants via inoculation with Sphingomonas sp. LK11 and exogenous trehalose. Microbiol Res 205: 135–145 [DOI] [PubMed] [Google Scholar]
- Augustin JM, Kuzina V, Andersen SB, Bak S (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72: 435–457 [DOI] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32: 666–681 [DOI] [PubMed] [Google Scholar]
- Bai Y, Muller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Munch PC, Spaepen S, Remus-Emsermann M, et al. (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364–369 [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker P (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG (2018) Optimizing taxonomic classification of marker–gene amplicon sequences with QIIME 2' s q2-feature-classifier plugin. Microbiome 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich N, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95 [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas PD, Almeida A, Bak S (2019) Evolution of structural diversity of triterpenoids. Front Plant Sci 10: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, Tal L, Meir S, Rogachev I, Malitsky S, et al. (2016) GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun 7: 10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale M, Grube M, Erlacher A, Quehenberger J, Berg G (2015) Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ Microbiol 17: 239–252 [DOI] [PubMed] [Google Scholar]
- Carter JP, Spink J, Cannon PF, Daniels MJ, Osbourn AE (1999) Isolation, characterization, and avenacin sensitivity of a diverse collection of cereal-root-colonizing fungi. Appl Environ Microbiol 65: 3364–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle E, Mendes R, Bakker P, Raaijmakers JM (2016) Fungal invasion of the rhizosphere microbiome. Isme J 10: 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QW, Jiang T, Liu YX, Liu HL, Zhao T, Liu ZX, Gan XC, Hallab A, Wang XM, He J, et al. (2019) Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci China Life Sci 62: 947–958 [DOI] [PubMed] [Google Scholar]
- Cheok CY, Salman HAK, Sulaiman R (2014) Extraction and quantification of saponins: a review. Food Res Int 59: 16–40 [Google Scholar]
- Chialva M, Salvioli Di Fossalunga A, Daghino S, Ghignone S, Bagnaresi P, Chiapello M, Novero M, Spadaro D, Perotto S, Bonfante P (2018) Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol 220: 1296–1308 [DOI] [PubMed] [Google Scholar]
- Devers M, Soulas G, Martin-Laurent F (2004) Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Methods 56: 3–15 [DOI] [PubMed] [Google Scholar]
- Field B, Jordan F, Osbourn A (2006) First encounters—deployment of defence-related natural products by plants. New Phytol 172: 193–207 [DOI] [PubMed] [Google Scholar]
- Francis G, Kerem Z, Makkar HPS, Becker K (2002) The biological action of saponins in animal systems: a review. Br J Nutr 88: 587–605 [DOI] [PubMed] [Google Scholar]
- Friedman M (2002) Tomato glycoalkaloids: role in the plant and in the diet. J Agric Food Chem 50: 5751–5780 [DOI] [PubMed] [Google Scholar]
- Fujimatsu T, Endo K, Yazaki K, Sugiyama A. (2020) Secretion dynamics of soyasaponins in soybean roots and effects to modify the bacterial composition. Plant Direct 4: e00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrieri A, Dong LM, Bouwmeester HJ (2019) Role and exploitation of underground chemical signaling in plants. Pest Manag Sci 75: 2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M (2008) Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312: 7–14 [Google Scholar]
- Hiltner L (1904) Uber nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Grundungung und Broche. Arbeit Deut Landw Ges Berlin 98: 59–78 [Google Scholar]
- Hu LF, Robert CAM, Cadot S, Zhang X, Ye M, Li BB, Manzo D, Chervet N, Steinger T, van der Heijden MGA, et al. (2018) Root exudate metabolites drive plant–soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9: 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ACC, Jiang T, Liu YX, Bai YC, Reed J, Qu BY, Goossens A, Nutzmann HW, Bai Y, Osbourn A (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364: eaau6389. [DOI] [PubMed] [Google Scholar]
- Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77: 3202–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. (2011) Glycoalkaloid metabolism1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby RP, Koprivova A, Kopriva S (2021) Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J Exp Botany 72: 57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Donn S, Ryan PR, Mathesius U, Devilla R, Jones A, Watt M (2016) Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS ONE 11: e0164533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AL, Waqas M, Kang SM, Al-Harrasi A, Hussain J, Al-Rawahi A, Al-Khiziri S, Ullah I, Ali L, Jung HY, et al. (2014) Bacterial endophyte Sphingomonas sp. lk11 produces gibberellins and IAA and promotes tomato plant growth. J Microbiol 52: 689–695 [DOI] [PubMed] [Google Scholar]
- Kirwa HK, Murungi LK, Beck JJ, Torto B (2018) Elicitation of differential responses in the root-knot nematode meloidogyne incognita to tomato root exudate cytokinin, flavonoids, and alkaloids. J Agric Food Chem 66: 11291–11300 [DOI] [PubMed] [Google Scholar]
- Korenblum E, Aharoni A (2019) Phytobiome metabolism: beneficial soil microbes steer crop plants’ secondary metabolism. Pest Manag Sci 75: 2378–2384 [DOI] [PubMed] [Google Scholar]
- Korenblum E, Dong Y, Szymanski J, Panda S, Jozwiak A, Massalha H, Meir S, Rogachev I, Aharoni A (2020) Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc Natl Acad Sci U S A 117: 3874–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosslak RM, Bookland R, Barkei J, Paaren HE, Appelbaum ER (1987) Induction of bradyrhizobium–japonicum common nod genes by isoflavones isolated from glycine-max. Proc Natl Acad Sci U S A 84: 7428–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, et al. (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato (vol 36, pg 1100, 2018). Nat Biotechnol 36: 1117. [DOI] [PubMed] [Google Scholar]
- Lairini K, PerezEspinosa A, Pineda M, RuizRubio M (1996) Purification and characterization of tomatinase from Fusarium oxysporum f sp. lycopersici. Appl Environ Microbiol 62: 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Nakayasu M, Akiyama R, Kobayashi M, Miyachi H, Sugimoto Y, Umemoto N, Saito K, Muranaka T, Mizutani M (2019) Identification of a 3 beta-hydroxysteroid dehydrogenase/3-ketosteroid reductase involved in alpha-tomatine biosynthesis in tomato. Plant Cell Physiol 60: 1304–1315 [DOI] [PubMed] [Google Scholar]
- Lee SA, Kim Y, Kim JM, Chu B, Joa JH, Sang MK, Song J, Weon HY (2019) A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci Rep 9: 9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Wang F, Huang YL, Zhou M, Gao JL, Yan TZ, Sheng HM, An LZ (2019) Sphingomonas sp. Cra20 increases plant growth rate and alters rhizosphere microbial community structure of Arabidopsis thaliana under drought stress. Front Microbiol 10: 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL (2013) Practical innovations for high-throughput amplicon sequencing. Nat Methods 10: 999–1002 [DOI] [PubMed] [Google Scholar]
- Massalha H, Korenblum E, Tholl D, Aharoni A (2017) Small molecules below-ground: the role of specialized metabolites in the rhizosphere. Plant J 90: 788–807 [DOI] [PubMed] [Google Scholar]
- Milner SE, Brunton NP, Jones PW, O’Brien NM, Collins SG, Maguire AR (2011) Bioactivities of glycoalkaloids and their aglycones from Solanum species. J Agric Food Chem 59: 3454–3484 [DOI] [PubMed] [Google Scholar]
- Moses T, Papadopoulou KK, Osbourn A (2014) Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit Rev Biochem Mol Biol 49: 439–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu M, Shioya N, Shikata M, Thagun C, Abdelkareem A, Okabe Y, Ariizumi T, Arimura G, Mizutani M, Ezura H, et al. (2018) JRE4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. Plant J 94: 975–990 [DOI] [PubMed] [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M (2017) A dioxygenase catalyzes steroid 16 alpha-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol 175: 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okmen B, Etalo DW, Joosten M, Bouwmeester HJ, de Vos RCH, Collemare J, de Wit P (2013) Detoxification of -tomatine by Cladosporium fulvum is required for full virulence on tomato. New Phytol 198: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, et al. (2019) vegan: Community ecology package. R Package Version 2.5-6. https://CRAN.R-project.org/package=vegan.
- Okutani F, Hamamoto S, Aoki Y, Nakayasu M, Nihei N, Nishimura T, Yazaki K, Sugiyama A (2020) Rhizosphere modeling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ 43: 1036–1046 [DOI] [PubMed] [Google Scholar]
- Pan FS, Meng Q, Wang Q, Luo S, Chen B, Khan KY, Yang XE, Feng Y (2016) Endophytic bacterium Sphingomonas SaMR12 promotes cadmium accumulation by increasing glutathione biosynthesis in Sedum alfredii Hance. Chemosphere 154: 358–366 [DOI] [PubMed] [Google Scholar]
- Pareja-Jaime Y, Roncero MIG, Ruiz-Roldan MC (2008) Tomatinase from Fusarium oxysporum f. sp. lycopersici is required for full virulence on tomato plants. Mol Plant Microbe Interact 21: 728–736 [DOI] [PubMed] [Google Scholar]
- Pascale A, Proietti S, Pantelides IS, Stringlis IA (2020) Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci 10: 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. In Merchant SS, Briggs WR, Ort D, eds, Annual Review of Plant Biology, Vol. 62. Annual Reviews, Palo Alto, pp. 549–566 [DOI] [PubMed] [Google Scholar]
- Pii Y, Borruso L, Brusetti L, Crecchio C, Cesco S, Mimmo T (2016) The interaction between iron nutrition, plant species and soil type shapes the rhizosphere microbiome. Plant Physiol Biochem 99: 39–48 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial C, Gomez E, Varela RM, Molinillo JMG, Macias FA (2018) Ecological relevance of the major allelochemicals in Lycopersicon esculentum roots and exudates. J Agric Food Chem 66: 4638–4644 [DOI] [PubMed] [Google Scholar]
- Sandrock RW, VanEtten HD (1998) Fungal sensitivity to and enzymatic degradation of the phytoanticipin alpha-tomatine. Phytopathology 88: 137–143 [DOI] [PubMed] [Google Scholar]
- Santiago CD, Yagi S, Ijima M, Nashimoto T, Sawada M, Ikeda S, Asano K, Orikasa Y, Ohwada T (2017) Bacterial compatibility in combined inoculations enhances the growth of potato seedlings. Microbes Environ 32: 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker SD, Nahar L, Kumarasamy Y (2007) Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 42: 321–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane PD, Heinig U, Panda S, Gilboa NS, Yona M, Kumar SP, Alkan N, Unger T, Bocobza S,, Pliner M, et al. (2018) Short-chain dehydrogenase/reductase governs steroidal specialized metabolites structural diversity and toxicity in the genus Solanum. Proc Natl Acad Sci U S A 115: E5419–E5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparg SG, Light ME, van Staden J (2004) Biological activities and distribution of plant saponins. J Ethnopharmacol 94: 219–243 [DOI] [PubMed] [Google Scholar]
- Stringlis IA, Yu K, Feussner K, de Jonge R, van Bentum S, Van Verk MC, Berendsen RL, Bakker P, Feussner I, Pieterse CMJ (2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci U S A 115: E5213–E5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Ueda Y, Zushi T, Takase H, Yazaki K (2014) Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 9: e100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Yamazaki Y, Hamamoto S, Takase H, Yazaki K (2017) Synthesis and secretion of isoflavones by field-grown soybean. Plant Cell Physiol 58: 1594–1600 [DOI] [PubMed] [Google Scholar]
- Sugiyama A, Yamazaki Y, Yamashita K, Takahashi S, Nakayama T, Yazaki K (2016) Developmental and nutritional regulation of isoflavone secretion from soybean roots. Biosci Biotechnol Biochem 80: 89–94 [DOI] [PubMed] [Google Scholar]
- Thagun C, Imanishi S, Kudo T, Nakabayashi R, Ohyama K, Mori T, Kawamoto K, Nakamura Y, Katayama M, Nonaka S, et al. (2016) Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol 57: 961–975 [DOI] [PubMed] [Google Scholar]
- Tsuno Y, Fujimatsu T, Endo K, Sugiyama A, Yazaki K (2018) Soyasaponins: a new class of root exudates in soybean (Glycine max). Plant Cell Physiol 59: 366–375 [DOI] [PubMed] [Google Scholar]
- Umemoto N, Nakayasu M, Ohyama K, Yotsu-Yamashita M, Mizutani M, Seki H, Saito K, Muranaka T (2016) Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol 171: 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S, Jeena GSShikhaShukla RK (2018) Recent advances in steroidal saponins biosynthesis and in vitro production. Planta 248: 519–544 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Bouwmeester HJ (2016) Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci 21: 256–265 [DOI] [PubMed] [Google Scholar]
- Vincken JP, Heng L, de Groot A, Gruppen H (2007) Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68: 275–297 [DOI] [PubMed] [Google Scholar]
- Vogel C, Innerebner G, Zingg J, Guder J, Vorholt JA (2012) Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp. strain fr1 against Pseudomonas syringae DC3000. Appl Environ Microbiol 78: 5529–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges M, Bai Y, Schulze-Lefert P, Sattely ES (2019) Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci U S A 116: 12558–12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LJ, Ge XJ, Brozel VS, Subramanian S (2017) Root isoflavonoids and hairy root transformation influence key bacterial taxa in the soybean rhizosphere. Environ Microbiol 19: 1391–1406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.