Abstract
Deciphering the mechanism of malate accumulation in apple (Malus domestica Borkh.) fruits can help to improve their flavor quality and enhance their benefits for human health. Here, we analyzed malate content as a quantitative trait that is determined mainly by genetic effects. In a previous study, we identified an R2R3−MYB transcription factor named MdMYB44 that was a candidate gene in qtl08.1 (quantitative trait locus mapped to chromosome 8) of fruit malate content. In the present study, we established that MdMYB44 negatively regulates fruit malate accumulation by repressing the promoter activity of the malate-associated genes Ma1 (Al-Activated Malate Transporter 9), Ma10 (P-type ATPase 10), MdVHA-A3 (V-type ATPase A3), and MdVHA-D2 (V-type ATPase D2). Two single-nucleotide polymorphisms (SNPs) in the MdMYB44 promoter, SNP A/G and SNP T/−, were experimentally shown to associate with fruit malate content. The TATA-box in the MdMYB44 promoter in the presence of SNP A enhances the basal activity of the MdMYB44 promoter. The binding of a basic-helix–loop–helix transcription factor MdbHLH49 to the MdMYB44 promoter was enhanced by the presence of SNP T, leading to increased MdMYB44 transcript levels and reduced malate accumulation. Furthermore, MdbHLH49 interacts with MdMYB44 and enhances MdMYB44 activity. The two SNPs could be used in combination to select for sour or non-sour apples, providing a valuable tool for the selection of fruit acidity by the apple breeding industry. This research is important for understanding the complex molecular mechanisms of fruit malate accumulation and accelerating the development of germplasm innovation in apple species and cultivars.
MdMYB44 modulates fruit malate accumulation and vacuolar acidification, and the genetic variation in the promoter of MdMYB44 determines fruit malate content in apple.
Introduction
Organic acids play essential roles in key fruit quality characteristics, such as flavor and palatability. Malate, which is one of the main organic acids, has various biological activities and pharmacological effects including the promotion of ATP production, antioxidant and anti-inflammatory activity, and the ability to reverse oxidative stress (Ding et al., 2016). Apple (Malus domestica Borkh.) has high nutritional value and is an important fruit crop worldwide. In apple, malate is the predominant organic acid, accounting for up to 90% of total organic acid content (Zhang et al., 2010; Ma et al., 2015a). The relatively high acidity plays a key role in the quality of apple juice (Huang et al., 2018).
Malate is involved in several pathways, including the tricarboxylic acid cycle, glyoxylate cycle, and gluconeogenesis (Sweetman et al., 2009; Etienne et al., 2013; Li et al., 2016a). Malate accumulation is influenced by various metabolic enzymes and malate-related transporters. Previous studies have revealed that vacuolar proton pumps and malate transporters play key roles in the determination of fruit acidity, such as tonoplast dicarboxylate transporter (AttDT; Emmerlichet al., 2003; Hurth et al., 2005), vacuolar-type H+-ATPase (V-ATPase; Schumacher and Krebs, 2010; Mauvezin et al., 2015), vacuolar H+ pumping pyrophosphatase (V-PPase; Suzuki et al., 2000; Hu et al., 2016), P-ATPase proton pumps (Verweij et al., 2008; Faraco et al., 2014; Shi et al., 2015; Li et al., 2016b; Ma et al., 2018; Strazzer et al., 2019), and aluminum-activated malate transporter (ALMT) family proteins (Kovermann et al., 2007; Barbier-Brygooet al., 2011; Meyer et al. 2011; Xu et al., 2012; Ma et al., 2015b; Sasaki et al., 2016; Ye et al., 2017; Li et al., 2020), all of which transport malate into vacuoles of the plant cell.
Recently, marker-assisted selection (MAS) has been widely adopted to accelerate the plant breeding process. MAS has been especially applied in woody plants, such as apple that feature high heterozygosity and a long juvenile period. The molecular markers used for MAS are primarily developed via quantitative trait locus (QTL) mapping (Xu et al., 2012; Cao et al., 2016; Ma et al., 2016; Ye et al., 2017). Fruit acidity, a quantitative trait, is controlled via one or several major genes along with the involvement of numerous minor genes (Chen et al., 2015; Cao et al., 2016; Ma et al., 2016; Jia et al., 2018). Ligaba (2009) reported that the phosphorylation level of the S384 residue of the TaALMT1 malate transporter regulated protein activity and affected malate transport. A 4-aa duplication in the CmPH coding region was found to cause differential acidity in Cucumis melo (Cohen et al., 2014). A 3-bp indel in the Sl-ALMT9 promoter region was linked to the higher malate content in tomato (Solanum lycopersicum) fruit (Ye et al., 2017). In apple fruit, the major QTLs associated with malate content have been mapped (Liebhard et al., 2003; Bai et al., 2012; Xu et al., 2012; Zhang et al., 2012; Sun et al., 2015; Ma et al., 2016; Jia et al., 2018). A single-nucleotide polymorphism (SNP) of A/G (252-bp upstream of the stop codon in the coding sequence) in Ma1 (MdALMT9) at the Ma (malic acid) locus on linkage group (LG) 16 was reportedly associated with fruit acidity in apple (Bai et al., 2012; Ma et al., 2015b). A non-synonymous A/G SNP of MdPP2CH was associated with malate content in apple fruit, and a 36-bp insertion in the MdSAUR37 promoter was linked to the higher malate content of apple fruit (Jia et al., 2018). These findings indicate that the mechanism controlling fruit acidity is complicated and requires further research.
MYB transcription factors have many different roles in plants, such as in development, hormone signaling, and metabolite biosynthesis (Allan and Espley, 2018). Previous studies have reported that MdMYB1 and MdMYB73 enhance vacuolar acidification and activate malate accumulation in apple fruit (Hu et al., 2016, 2017). The transcription factor MYB44 plays multifaceted roles in various species (Jung et al., 2008, 2010; Zhou et al., 2017). For example, with respect to fruit quality, FaMYB44.2 suppresses soluble sugar accumulation in strawberry (Fragaria × ananassa) fruits (Wei et al., 2018), and StMYB44 limits anthocyanin biosynthesis in potato (Solanum tuberosum) tubers (Liu et al., 2019). However, the contribution of MYB44 to fruit acidity remains unclear and merits further investigation.
In a previous study, we found that MdMYB44 was a candidate gene in the major qtl08.1 region for apple fruit acidity (Jia et al., 2018). Here, we analyzed the molecular and genetic mechanism by which natural variants in the MdMYB44 promoter might regulate apple fruit acidity. We provide evidence for two allelic variants in the MdMYB44 promoter, SNP A/G and SNP T/−, that can affect malate accumulation in apple fruit. Furthermore, our findings suggest that the transcription factor MdbHLH49, which binds to the MdMYB44 promoter via the presence of SNP T, enhances the MdMYB44 transcript levels and reduces malate accumulation. This research advances our ability to elucidate the complex molecular mechanisms regulating fruit acidity and should prove promising for use in the apple breeding industry.
Results
Phenotypic variation of malate content in apple fruit
To analyze the heritable trait of fruit malate content in apple fruit, phenotypic data of F1 hybrids from “Jonathan” × “Golden Delicious” and “Zisai Pearl” × “Red Fuji” crosses were detected in the years 2014–2016 (Table 1). The frequency distribution of malate content was abnormal and its coefficient of variation was 54.02%–62.51%, thus indicating that malate content is quantitatively inherited (Supplemental Figure S1 and Table 1). Broad sense heritability was 88.6%–89.8% (Table 1), indicating that the variation of malate content in apple fruits was primarily determined by genetics.
Table 1.
Phenotypic analysis of malate content in F1 progenies from “Jonathan” × “Golden Delicious” and “Zisai Pearl” × “Red Fuji” crosses in the years 2014 − 2016
| Hybrid crosses | Broad sense Heritability | Coefficient of variance (%) |
Shapiro−Wilk test |
||||
|---|---|---|---|---|---|---|---|
| 2014 | 2015 | 2016 | 2014 | 2015 | 2016 | ||
| “Jonathan” × “Golden Delicious” | 89.8% | 54.02 | 72.99 | 58.77 | Non | Non | Non |
| “Zisai Pearl” × “Red Fuji” | 88.6% | 56.46 | 57.71 | 62.51 | Non | Non | Normal |
“Non” represents non-normally distributed, and “Normal” represents normally distributed for fruit malate content.
Allelic variants in the MdMYB44 promoter and their effects on fruit malate content
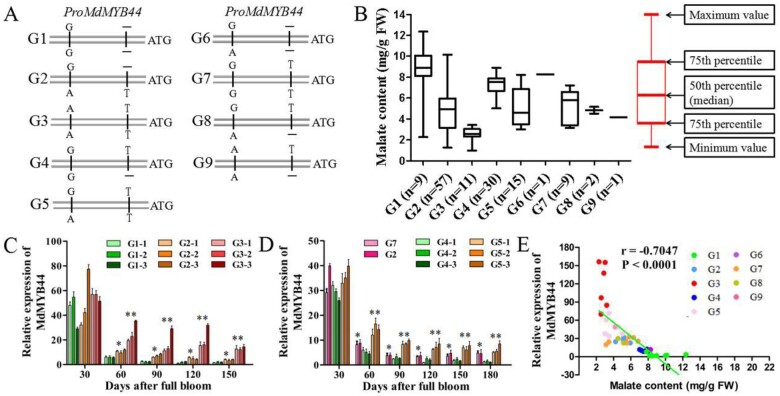
Our previous study indicated that MdMYB44 was a candidate gene in the major qtl08.1 region for apple fruit acidity (Jia et al., 2018). In the MdMYB44 promoter, we previously identified two SNPs: SNP T/− (−536 bp upstream of the ATG codon, where the − represents a T deletion) and SNP A/G (−1,010 bp upstream of the ATG codon; Jia et al., 2018). SNP A and SNP T, as well as SNP G and SNP−, were validated as complete linkages and associated with malate content (P = 6.3894 × 10−36) in 246 F1 hybrids of “Jonathan” (G2, AT/G−) × “Golden Delicious” (G2, AT/G−; Figure 1, A and Supplemental Table S1). SNP A and SNP T, SNP G and SNP−, and SNP G and SNP T were validated as complete linkages and associated with malate content (P = 5.5797 × 10−8) in 123 F1 hybrids of “Zisai Pearl” (G7, GT/GT) × “Red Fuji” (G2, AT/G−; Figure 1, A and Supplemental Table S2). Among the 135 Malus accessions, the genotype (AA, AG, and GG) of SNP A/G was associated with malate content and displayed this pattern: GG > AG > AA (P = 4.6119 × 10−14); the genotype (TT, T−, —) of SNP T/− was associated with malate content and showed the following pattern: —> T− > TT (P = 1.4193 × 10−7; Supplemental Table S3). Nine genotypes (combinations of SNP A/G and SNP T/−)—G1, G2, G3, G4, G5, G6, G7, G8, and G9—were found to exist in the 135 Malus accessions (Figure 1, A and Supplemental Table S3). The average malate content differed among these nine genotypes and the malate content was associated with the nine genotypes (P = 2.3328 × 10−14; Figure 1, B). Collectively, these results indicate that both SNP G and SNP− are associated with a higher malate content, whereas SNP A and SNP T are associated with a lower malate content.
Figure 1.
Allelic variants in the MdMYB44 promoter are associated with fruit malate content. A Nine genotypes formed by the SNP T/− and SNP A/G combinations in the Malus accessions. B, Boxplots showing the analysis between the genotypes (G1, G2, G3, G4, G5, G6, G7, G8, and G9) in the MdMYB44 promoter and fruit malate content in 135 Malus accessions. The instructions of boxplots were provided, and “n” represents number of individuals in each genotype (G1−G9). C, Transcription profiles of MdMYB44 in the F1 progenies from “Jonathan” × “Golden Delicious” with three genotypes (G1, G2, and G3) during apple fruit development. D, Transcription profiles of MdMYB44 in the F1 progenies from “Zisai Pearl” × “Red Fuji” with two genotypes (G4 and G5), parent “Zisai Pearl” with the genotype G7, and parent “Red Fuji” with the genotype G2, during fruit development. E, Correlation analysis between MdMYB44 expression levels and fruit malate content. Values represent the mean ± SE in C, D, and E. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01).
In the “Jonathan” × “Golden Delicious” F1 hybrids, the fruit malate content was greater in the three F1 hybrids carrying the G1 genotype (52-049, 52-120, and 52-201) than those with the G2 genotype (52-041, 52-157, and 53-064), and it was lowest in those harboring the G3 genotype (52-146, 54-047, and 55-008) at 60–150 d after full bloom (DAFB; Supplemental Figure S2, A and Supplemental Table S1). However, the average expression levels of MdMYB44 differed among the three genotypes: G1 < G2 < G3 (Figure 1, C). In the “Zisai Pearl” (G7) × “Red Fuji” (G2) F1 hybrids, MdMYB44 expression in the higher malate G4 genotype (02-123, 04-099, 06-140) was significantly lower than that in the lower malate G5 genotype (02-061, 04-050, 05-071; Figure 1, D; Supplemental Figure S2, B; and Supplemental Table S2). Furthermore, MdMYB44 expression exhibited a close correlation (r = –0.7047, P < 0.0001) with malate content in ripened fruit from nine genotypes (G1–G9) of 40 Malus accessions (Figure 1, E and Supplemental Table S4). Together, these results indicate that the expression level of MdMYB44 is negatively associated with the fruit malate content.
Genetic linkages between the SNP T/− and SNP A/G in the MdMYB44 promoter
The linkage between SNP T/− and SNP A/G was analyzed in two cross populations and 135 Malus germplasm accessions. In 246 F1 individuals from “Jonathan” × “Golden Delicious,” SNP T was linked to SNP A, and SNP − was linked to SNP G (Supplemental Table S5). For genotype segregation, the observed ratio almost met the expected ratio (χ2 = 7.10, P > 0.025; Supplemental Table S6). In 123 F1 individuals from “Zisai Pearl” × “Red Fuji,” SNP G was linked to SNP T except when it was linked to SNP − (Supplemental Table S5). For genotype segregation, the observed ratio agreed with the expected ratio (χ2 = 1.17, P > 0.05; Supplemental Table S6). In 135 Malus germplasm accessions, SNP T was linked to both SNP A and SNP G, and SNP − was linked to both SNP G and SNP A (Supplemental Table S5). Regarding the P-value for the association between genotype and malate content, each P was lower in the genotype combinations (G1–G9; 2.3328 × 10−14) than in either SNP T/− (1.4193 × 10−7) or SNP A/G (P = 4.6119 × 10−14). For example, the genotype is homozygous GG/TT in “Zisai Pearl,” which has a moderate malate content (5.55 mg/g FW), suggesting the combination of the SNP G related with high malate content and the SNP T related with low malate content leads to moderate malate content. The other genotype combinations exhibited similar trends (Supplemental Table S3). These results indicated the combinations of the two SNPs could be more effectively used for the selection of sour or non-sour apples.
MdMYB44 negatively regulates malate accumulation in transgenic lines
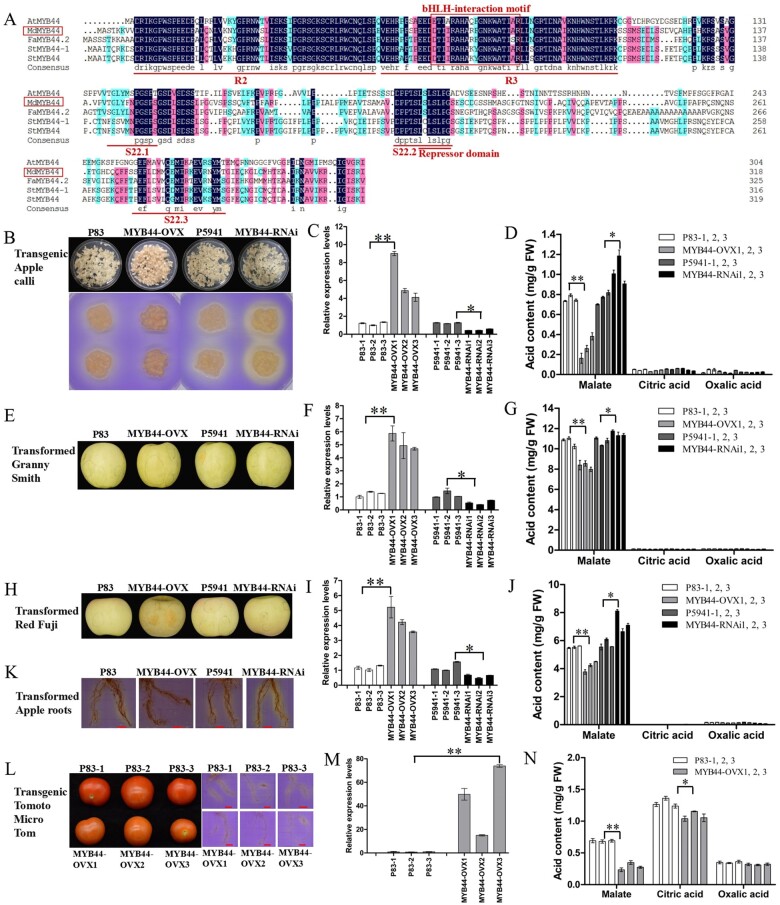
MdMYB44 is a typical R2R3 MYB transcription factor that harbors R2 and R3 MYB-repeat domains, a bHLH interaction motif, a transcription repressor domain LxLxL, and S22-subgroup motifs (Figure 2, A; Hiratsu et al., 2003; Kagale et al., 2010; Wei et al., 2018). In Arabidopsis, AtMYB44 contains the conserved motifs 22.1 (TGLYMSPxSP), 22.2 (D/EPP/MTxLSLP), and 22.3 (GxFMxVVQEMIxxEVRSYM; Zhou et al., 2017). MdMYB44 and FaMYB44.2 are highly similar in their 22.1 and 22.3 motifs, yet they differ distinctly from those of AtMYB44, allowing the prediction of specific physiological roles in apple fruit compared with its activity in Arabidopsis (Figure 2, A).
Figure 2.
MdMYB44 negatively regulates malate content in apple calli and fruit. A, Multiple sequence alignment was performed using the protein sequences of MdMYB44 (MDP0000463846 for M. domestica), AtMYB44 (AT5G67300 for Arabidopsis thaliana), FaMYB44.2 (XP_004291065.1 for Fragaia ananassa), StMYB44 (XP_006367421.1 for S. tuberosum), and StMYB44-1 (MK410941.1 for S. tuberosum). MdMYB44 has the R2 and R3 domains, bHLH-interaction motifs, subgroup S22 motifs, and the negative repressor motif LxLxL. B, E, and H, MdMYB44 was over-expressed (MYB44-OVX) and silenced (MYB44-RNAi) in apple calli, and the acidification of MdMYB44 transgenic apple calli was visualized (B), in apple fruit of “Granny Smith” (E) and “Red Fuji” (H). C and D, The transcripts of MdMYB44 (C) and the acid content (D) in transgenic apple calli. F and G, The transcripts of MdMYB44 (F) and the acid content (G) in transient transgenic “Granny Smith” fruit. I and J, The transcripts of MdMYB44 (I) and the acid content (J) in transient transgenic “Red Fuji” fruit. K, Visualization of rhizosphere acidification in the MdMYB44 transient transgenic apple roots. Scale bar = 10 mm. L, MYB44-OVX transgenic tomato were obtained and rhizosphere acidification of the MdMYB44 transgenic tomato roots was visualized. Scale bar = 10 mm. M and N, The transcripts of MdMYB44 in transgenic tomato (M) and its acid content (N). Values are the mean ± SE in C, D, F, G, I, J, M, and N. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01).
Constructs, 35S:MdMYB44 and RNAi:MdMYB44, were transformed into apple calli to investigate the roles of MdMYB44 in malate accumulation. The empty vectors PMDC83 (P83) and pFGC5941 (P5941) were transformed into apple calli to serve as controls. Using a pH imaging technique, the MdMYB44-OVX and MdMYB44-RNAi transgenic calli exhibited lower and higher acidification than P83 and P5941, respectively (Figure 2, B). The MdMYB44 expression levels were significantly higher in overexpressing transgenic calli (MYB44-OVX1, −2, and −3) yet much lower in the RNAi transgenic calli (MYB44-RNAi1, −2, and −3; Figure 2, C). The MdMYB44-GFP protein was detected in MdMYB44 transgenic calli (Supplemental Figure S3). The malate content was significantly lower in MYB44-OVX calli than in P83 calli, whereas it was greater in MYB44-RNAi calli than in P5941 calli (Figure 2, D). The contents of other organic acids, including oxalic acid and citric acid, were generally very low in apple calli, and were similar in the MYB44-OVX and MYB44-RNAi calli (Figure 2, D). These results suggest that MdMYB44 reduces the malate content in apple calli.
An Agrobacterium tumefaciens infiltration-based transient expression assay was used to overexpress MdMYB44 in the fruit of “Granny Smith” and “Red Fuji.” The MYB44-OVX and MYB44-RNAi apple fruit were harvested 8 d after the transient transformation of “Granny Smith” (Figure 2, E) and “Red Fuji” (Figure 2, H). The expression level of MdMYB44 was significantly higher in MYB44-OVX fruit but much lower in MYB44-RNAi fruit (Figure 2, F and I), whereas the malate content was significantly lower in MYB44-OVX fruit and higher in MYB44-RNAi fruit (Figure 2, G and J). The contents of other organic acids were very low and were comparable between MYB44-OVX and MYB44-RNAi fruit (Figure 2, G and J). Furthermore, MdMYB44-OVX transgenic apple roots were characterized by a lesser extent of rhizosphere acidification than those of MdMYB44-RNAi (Figure 2, K). These findings suggest MdMYB44 negatively regulates malate accumulation in both the fruits and roots of apple trees.
To further confirm the association between MdMYB44 and acid accumulation, we transformed MdMYB44 into tomato for heterologous expression. Transgenic lines were generated, and MdMYB44-OVX transgenic roots exhibited much lower rhizosphere acidification compared with P83 (Figure 2, L). MdMYB44 was highly expressed in tomato fruits of MdMYB44-OVX transgenic lines (Figure 2, M). Malate content was significantly lower in the MdMYB44-OVX fruits, and citric acid content was also slightly lower (Figure 2, N). These results indicate that MdMYB44 can reduce malate content and negatively regulates malate accumulation.
RNA-seq analysis of MdMYB44-OVX apple calli
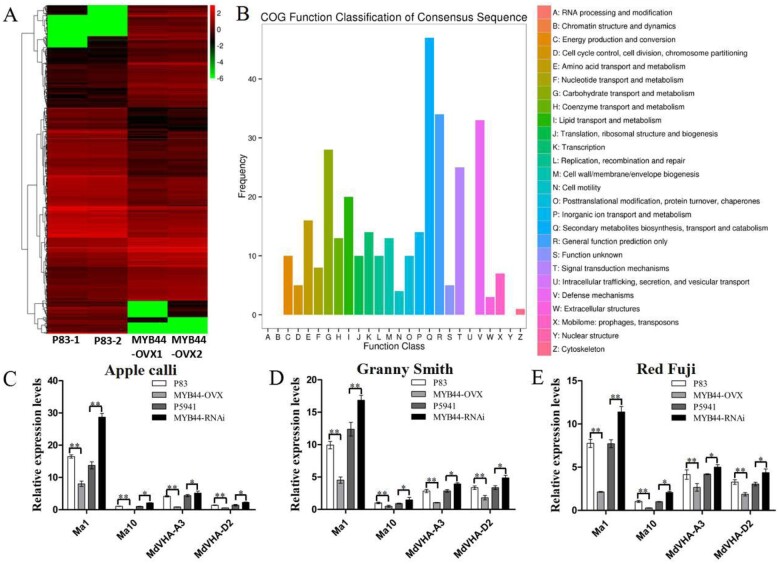
An RNA-seq analysis of P83 and MdMYB44-OVX transgenic apple calli was used to investigate the potential MdMYB44 regulation network (Figure 3, A and B; Supplemental Figure S4; and Supplemental Table S7). Among the 1, 436 differentially expressed genes (DEGs), several key genes involved in malate metabolism were identified (Supplemental Table S7). The gene Ma1 (MdALMT9 and MD16G1045200), encoding an aluminum-activated malate transporter, and the genes Ma10 (P-type ATPase and MD17G1046600), MdVHA-A3 (V-type ATPase, MD08G1021400), and MdVHA-D2 (V-type ATPase and MD09G1048000), encoding proton pumps, were identified, each of which were previously verified to influence fruit acidity (Etienne et al., 2013; Hu et al., 2016, 2017; Jia et al., 2018; Ma et al., 2018; Li et al., 2020). The expression levels of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 were all significantly lower in the MYB44-OVX lines compared with P83, whereas they were higher in the MYB44-RNAi lines compared with P5941 (Figure 3, C–E), indicating a negative correlation between MdMYB44 expression levels and that of the genes Ma1, Ma10, MdVHA-A3, and MdVHA-D2.
Figure 3.
RNA-seq analysis of MdMYB44-OVX transgenic apple calli. A, Hierarchical clustering of transcript accumulation between the MdMYB44-OVX transgenic apple calli lines (MdMYB44-OVX1 and MdMYB44-OVX2) and the P83 controls (P83-1and P83-2). B, Gene function was annotated based on the COG (clusters of orthologous groups of proteins) analysis of the DEGs, as determined by RNA-seq, in the MdMYB44-OVX transgenic apple calli lines compared with the control P83. C–E, Relative expression levels of the genes Ma1, Ma10, MdVHA-A3, and MdVHA-D2 were determined using RT-qPCR in transgenic calli (C), and the transient transgenic fruit of “Granny Smith” (D) and “Red Fuji” (E). Values are the mean ± SE in C, D, and E. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01).
MdMYB44 represses the transcription of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 by directly binding to their promoters
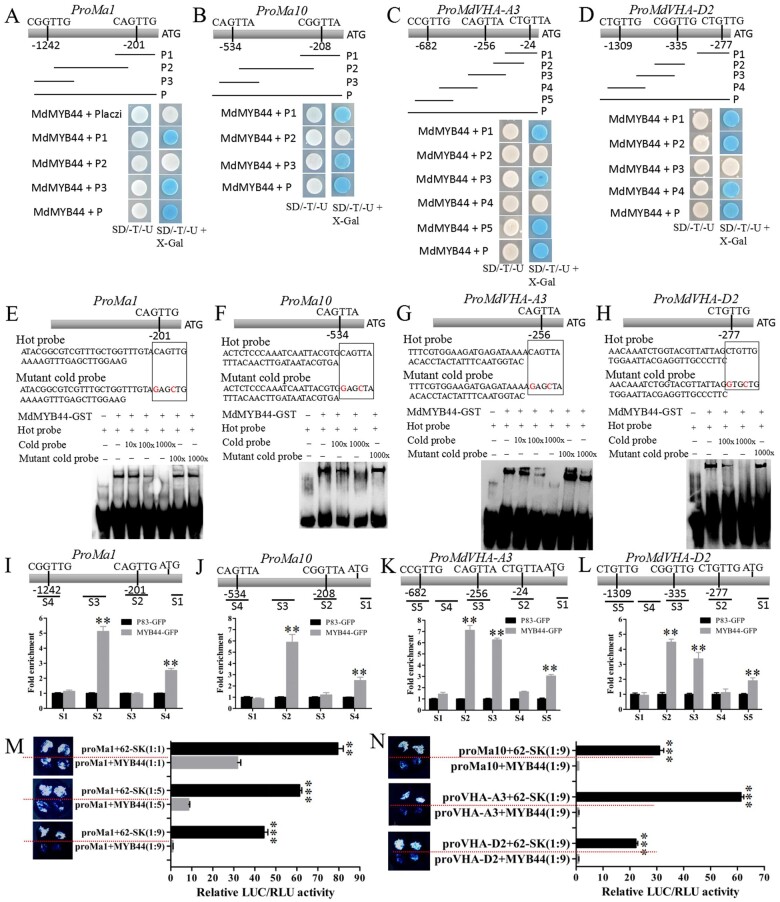
The core binding sequence of MYB44, 5′-CNGTTR-3′, was previously identified in Arabidopsis and potato (Shim et al., 2013; Zhou et al., 2017). Using a yeast one-hybrid (Y1H) assay, we found that MdMYB44 was able to bind to the fragments of the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoters containing the MYB-core CNGTTR motif (Figure 4, A–D). To further confirm this binding interaction, an electrophoretic mobility shift assay (EMSA) was performed. MdMYB44 was able to bind to the Ma1 promoter fragment containing the CAGTTG motif, the MdVHA-A3 promoter fragment containing the CAGTTA motif, the Ma10 promoter fragment containing the CAGTTA motif, and the MdVHA-D2 promoter fragment containing the CTGTTG motif (Figure 4, E–H).
Figure 4.
MdMYB44 represses the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 transcription. A–D, The Y1H showing that MdMYB44 binds to each Ma1 (ProMa1), Ma10 (ProMa10), MdVHA-A3 (ProMdVHA-A3), and MdVHA-D2 (ProVHA-D2) promoter fragment containing the MYB-core motif (CNGTTR). The MdMYB44 protein interacts with each Ma1 promoter’s fragment P1 (CAGTTG) and P3 (CGGTTG) (A), each Ma10 promoter’s fragment P1(CGGTTA) and P3 (CAGTTA) (B), each MdVHA-A3 promoter’s fragment P1(CTGTTA), P3 (CAGTTA), and P5 (CCGTTG) (C), and each MdVHA-D2 promoter’s fragment P1(CTGTTG), P3 (CGGTTG), and P5 (CTGTTG) (D). The Ma1 promoter (P2, 1,001 bp), Ma10 promoter (P2, 311 bp), MdVHA-A3 promoter (P2, 221 bp; P4, 413 bp), and MdVHA-D2 promoter (P3, 832 bp) served as negative controls. E−H, EMSA analysis showing that MdMYB44 binds to the CAGTTG motif of the Ma1 promoter (E), the CAGTTA motif of the Ma10 promoter (F), the CAGTTA motif of the MdVHA-A3 promoter (G), and the CTGTTG motif of the MdVHA-D2 promoter (H). I–L, ChIP-PCR showing that MdMYB44 binds to the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoters in vivo. Four regions (S1–S4) in Ma1 (I), four regions (S1–S4) in Ma10 (J), five regions (S1–S5) in MdVHA-A3 (K), and five regions (S1–-S5) in MdVHA-D2 (L) were investigated. M, LUC activity assay showing that MdMYB44 represses the Ma1 promoter. Three concentration combinations of ProMa1:LUC (ProMa1) and Pro35S:MdMYB44 (MYB44) were set up, for which the ratio of ProMa1 to MYB44 was 1:1, 1:5, and 1:9. N, LUC activity assays showing that MdMYB44 represses activity of Ma10, MdVHA-A3, and MdVHA-D2 promoters. Values are the mean ± SE in I–N. The ** indicates significantly different values (P < 0.01); *** indicates significantly different values (P < 0.001).
To confirm the binding of MdMYB44 to the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoters in vivo, a chromatin immunoprecipitation (ChIP)-PCR (Polymerase Chain Reaction) assay was conducted. The MdMYB44-GFP fusion was overexpressed in transgenic apple calli. The presence of MdMYB44 substantially enhanced the PCR-based detection of the promoters of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 (Figure 4, I–L), indicating that MdMYB44 can also bind to the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoters in vivo. Furthermore, to test how MdMYB44 may influence the activity of the Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoters, a luciferase (LUC) transactivation assay was performed. Three combinations of ProMa1:LUC (ProMa1) and Pro35S:MdMYB44 (MYB44) were used, consisting of ProMa1-to-MYB44 ratios of 1:1, 1:5, and 1:9, respectively. When MYB44 was co-transformed with ProMa1, Ma1 promoter activity was significantly decreased. As the ProMa1-to-MYB44 ratio increased from 1:1 to 1:9, Ma1 promoter activity markedly decreased (Figure 4, M). The activity of the Ma10, MdVHA-A3, and MdVHA-D2 promoters also decreased significantly when MYB44 was co-transformed (Figure 4, N). Together, these results support MdMYB44 repression of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 transcription.
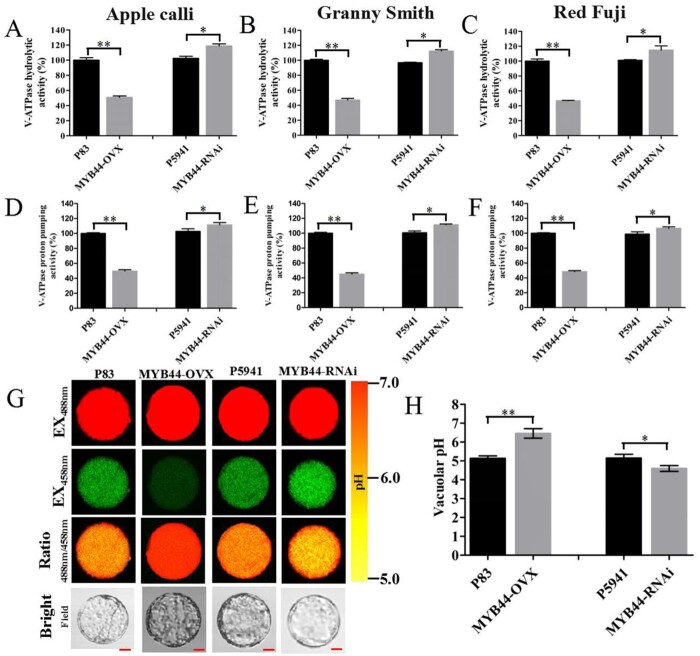
The activity of V-ATPase and proton-pumping decreased approximately two-fold in MYB44-OVX calli lines (Figure 5, A and D) and the fruits of “Granny Smith” (Figure 5, B and E) and “Red Fuji” (Figure 5, C and F) when compared with that in P83. By contrast, V-ATPase and proton-pumping activities were slightly augmented in the MYB44-RNAi lines. These results suggest that MdMYB44 negatively regulates tonoplast V-ATPase activity, an effect that is partially dependent on Ma10, MdVHA-A3, and MdVHA-D2 in apple calli.
Figure 5.
MdMYB44 negatively modulates malate accumulation and cell acidification. A−F, ATP hydrolysis activity (A–C) and proton-pumping activity (D–F) of V-ATPase in transgenic calli lines (A, D), and transient transgenic apple fruit of “Granny Smith” (B, E) and “Red Fuji” (C, F). G, Protoplast vacuoles of transgenic calli containing BCECF were imaged at 488 and 458 nm by confocal microscopy. Scale bar = 10 um. H, The pH level in the protoplast vacuoles of transgenic calli lines was determined. Values are the mean ± SE in A–H. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01).
The transport of H+ into vacuoles determines the extent of their acidification (Swanson and Jones, 1996; Gaxiola et al., 2007; Hu et al., 2016). We tested the vacuolar pH by using the ratiometric fluorescent pH indicator, 2′,,7′-bis-(2-carboxyethyl)-5-(6)-carboxyfluorescein (BCECF). The fluorescence ratios obtained from confocal images were used to establish the calibration curve and calculate the pH values (Supplemental Figure S5, A and B). The average vacuolar pH for MdMYB44-OVX apple calli was 6.46, a level much higher than that for P83 (5.13). By contrast, the average vacuolar pH for MdMYB44-RNAi apple calli was 4.59, which was much lower than that for P5941 (5.15; Figure 5, G and H). A lower H+ concentration in MdMYB44-OVX lines, as indicated by higher pH values, further supports the view that MdMYB44 negatively regulates vacuolar acidification and decreases malate content.
Involvement of the MdMYB44 allelic variant SNP A/G in the control of malate content
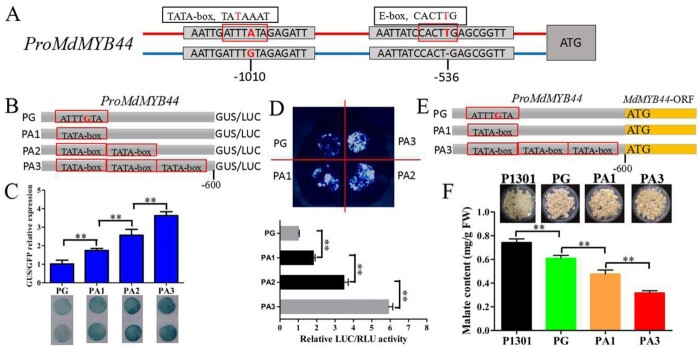
The SNP A/G variant in the MdMYB44 promoter is present at A in the TATA-box motif (Figure 6, A and Supplemental Figure S6). The TATA-box is involved in the activation of eukaryotic genes (Zhang et al., 2017; Huang et al., 2018). Here, we produced a series of synthetic constructs to test the effects of TATA-box units on the transcriptional activity of MdMYB44. These constructs contained the native promoter sequences with the addition of SNP G or up to three TATA-boxes fused to the GUS or LUC reporter (Figure 6, B). Compared with the SNP G lacking the TATA-box, the GUS assay revealed a strong positive correlation between the number of repeat TATA-box insertions and promoter activity (Figure 6, C). In the LUC assay, we found that the TATA-box significantly increased LUC activity (Figure 6, D). To further investigate the effects of the TATA-box on malate accumulation, we introduced the MdMYB44 ORF containing each promoter of PG (no TATA box), PA1 (one TATA box), and PA3 (three TATA boxes) into apple calli (Figure 6, E), and the empty vector pCAMBIA1301 (P1301) served as a control. The malate content of transgenic calli differed as follows: PA3 < PA1 < PG (Figure 6, F). Taken together, these results show that the presence of SNP A in the TATA-box in the MdMYB44 promoter enhances the basal activity of the MdMYB44 promoter and negatively influences malate content.
Figure 6.
Identification of the SNP A/G in the MdMYB44 promoter that influences its activity and malate accumulation. A, Schematic of the MdMYB44 promoter with SNP A/G and SNP T/−. The MdMYB44 promoter harbors the motif TATA-box when the SNP A/G is A, and the motif E-box when the SNP T/− is T. B, Synthetic constructs containing the native promoter sequences with the addition of SNP G or up to three TATA-boxes fused to the GUS reporter or LUC reporter. C, GUS reporter analysis showing that the TATA-box enhances the basal activity of the MdMYB44 promoter. D, LUC activity assays showing that the TATA-box enhances the basal activity of the MdMYB44 promoter. E, Synthetic constructs containing the native promoter sequences with the addition of SNP G or up to three TATA-boxes fused to the MdMYB44 open-reading frame. F, Malate content in transgenic apple calli after transforming each promoter PG, PA1, and PA3 driving the MdMYB44 ORF. Values are the mean ± SE in C, D, and F. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01).
MdbHLH49 enhances MdMYB44 expression and contributes to a decline in malate content by binding to the allelic variant, SNP T
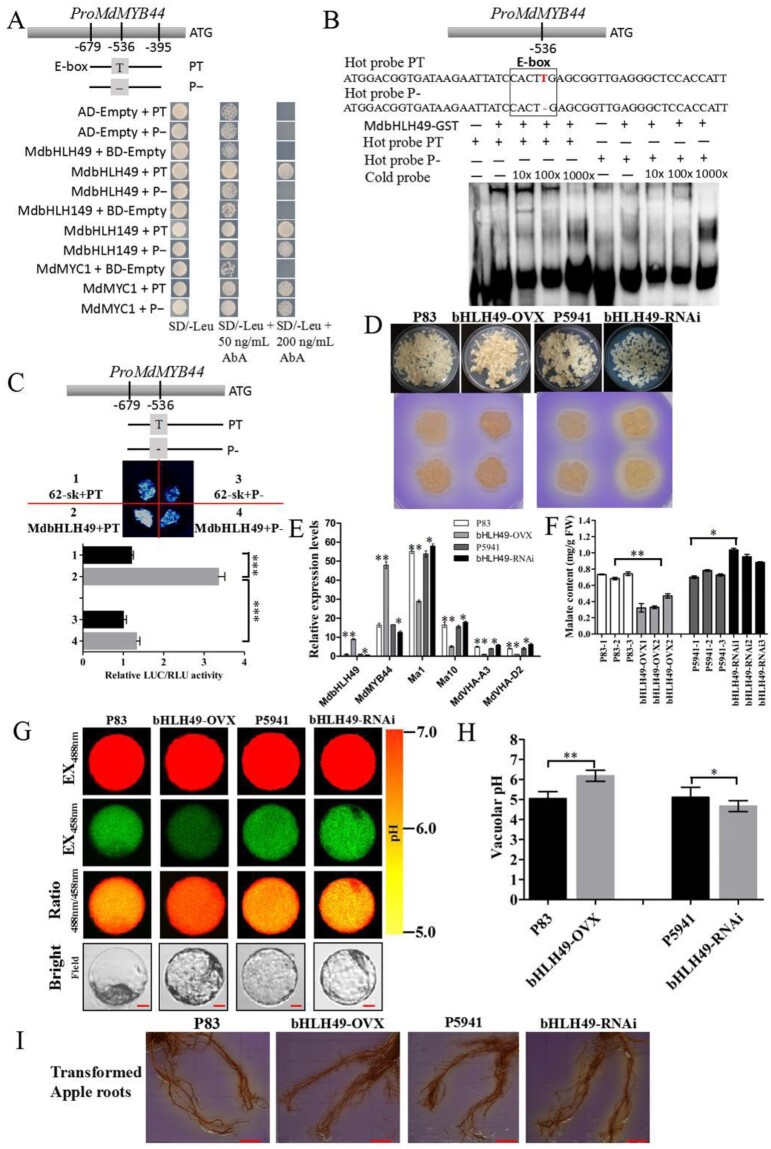
The SNP T/− variant in the MdMYB44 promoter is present at T in the E-box motif (Figure 6, A and Supplemental Figure S6). It has been reported that the E-box motif of 5′-CANNTG-3′, also referred to as the MYC cis-element, is recognized by bHLH proteins (Xie et al., 2012; Li et al., 2017). Using a Y1H screen, three bHLH-type proteins including MdbHLH49 protein (MD16G1053600), MdbHLH149 protein (MD12G1204600), and MdMYC1 protein (MD16G1055100) were identified that bind to the PT promoter of MdMYB44. The proteins MdbHLH149 and MdMYC1 showed no difference in their binding activity between the promoter PT and P−, since they were found bound to both, yet the MdbHLH49 protein was able to bind to the PT but not the P− promoter (Figure 7, A). The expression level of MdbHLH49 was negatively associated with fruit malate content (r = –0.6996, P < 0.001); however, the expression levels of MdbHLH149 and MdMYC1 showed no significant association with fruit malate content (Supplemental Table S4 and Supplemental Figure S7, A–C). In addition, the expression of MdbHLH49 was positively associated with that of MdMYB44 (r = 0.8914, P < 0.0001; Supplemental Figure S7, D).
Figure 7.
Transcription factor MdbHLH49 binds to PT (with SNP T), but not P− (with T deletion), in the MdMYB44 promoter to repress malate accumulation. A, Y1H assay showing the binding of MdbHLH49 to PT. B, EMSA showing MdbHLH49 binding to PT. C, LUC assay showing that MdbHLH49 significantly enhances the PT activity. D, MdbHLH49 was over-expressed (bHLH49-OVX) and silenced (bHLH49-RNAi) in apple calli, and the acidification of MdbHLH49 transgenic apple calli was visualized. E, The transcripts of MdbHLH49, MdMYB44, Ma1, Ma10, MdVHA-A3, and MdVHA-D2 in transgenic apple calli. F, Malate content in transgenic apple calli. G, Protoplast vacuoles of MdbHLH49 transgenic calli lines that contained BCECF were imaged at 488and 458 nm by confocal microscopy. Scale bar = 10 um. H, The pH level in the protoplast vacuoles of MdbHLH49 transgenic calli lines was determined. I, Visualization of rhizosphere acidification of MdbHLH49 transient transgenic apple roots. Scale bar = 10 mm. Values are the mean ± SE in C, E, F, and H. The * indicates significantly different values (P < 0.05); ** indicates significantly different values (P < 0.01); *** indicates significantly different values (P < 0.001).
To further confirm that the MdbHLH49 transcription factor binds to the PT promoter, an EMSA assay was conducted, which demonstrated that MdbHLH49 binds to PT but not P− (Figure 7, B). The LUC activity level was significantly higher when 35S:bHLH49 was co-transformed with PT:LUC than with P−:LUC (Figure 7, C). These results indicate that MdbHLH49 directly binds the PT promoter with an E-box to enhance MdMYB44 expression.
To investigate the effects of MdbHLH49 on malate accumulation, the constructs 35S:MdbHLH49 and RNAi:MdbHLH49 were separately transformed into apple calli. The MdbHLH49-OVX transgenic calli exhibited a lower degree of acidification, whereas MdbHLH49-RNAi exhibited a higher degree of acidification (Figure 7, D). The MdbHLH49-GFP protein was detected in MdbHLH49 transgenic calli (Supplemental Figure S8). The expression levels of MdbHLH49 and MdMYB44 were significantly higher in MdbHLH49-OVX calli compared with P83, whereas they were lower in MdbHLH49-RNAi calli compared with P5941 (Figure 7, E). The expression levels of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 were all significantly lower in MdbHLH49-OVX calli than in P83, and they were higher in MdbHLH49-RNAi calli than in P5941 (Figure 7, E). A positive correlation was found between the transcript levels of MdMYB44 and MdbHLH49, whereas the transcript levels of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 were negatively correlated with the transcript level of MdbHLH49.
Because the malate content was significantly lower in MdbHLH49-OVX calli compared with P83, whereas it was higher in MdbHLH49-RNAi calli (Figure 7, F), it is likely that MdbHLH49 can negatively regulate malate accumulation. The average vacuolar pH was 6.19 in MdbHLH49-OVX calli, which was much higher than that in P83 (5.05), whereas it was 4.67 in MdbHLH49-RNAi calli, which was lower than that in P5941 (5.11; Figure 7, G and H). MdbHLH49-OVX transgenic apple roots exhibited lower rhizosphere acidification compared with P83, whereas MdbHLH49-RNAi exhibited higher rhizosphere acidification compared with P5941 (Figure 7, I). This set of results further support a role of MdbHLH49 in the negative regulation of vacuolar acidification and malate accumulation.
MdbHLH49 interacts with MdMYB44 and enhances its effects on downstream target genes
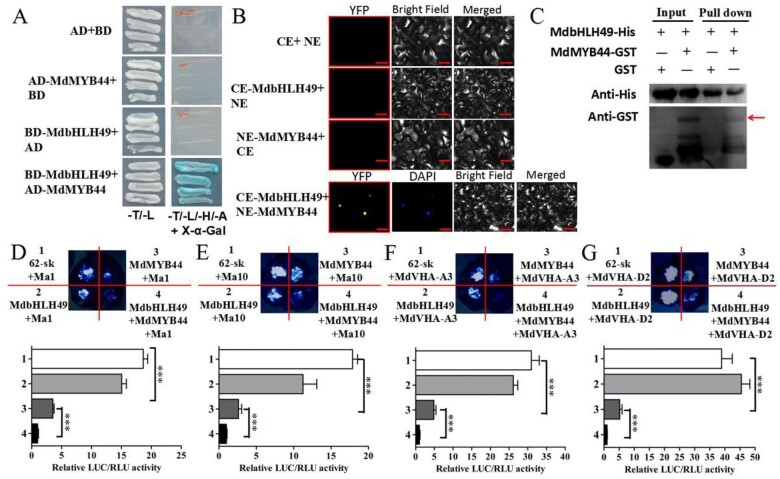
MdMYB44 harbors the bHLH interaction motif (Figure 2, A). Using a yeast two-hybrid (Y2H) assay to test whether MdbHLH49 could interact with MdMYB44, we found that MdbHLH49 interacts with MdMYB44 (Figure 8, A). A subsequent bimolecular fluorescence complementation (BiFC) assay confirmed the interaction between MdbHLH49 and MdMYB44, for which a yellow florescence signal was observed in the nucleus (Figure 8, B). Furthermore, a GST pull-down assay indicated the His-tagged MdbHLH49 protein interacted with the GST-tagged MdMYB44 protein in vitro (Figure 8, C). To determine how this MdbHLH49–MdMYB44 interaction affects the promoter activities of MdMYB44 downstream genes, a LUC assay was performed. LUC activity was significantly lower when both 35S:bHLH49 and 35S:MdMYB44 were co-transformed with each of ProMa1:LUC, ProMa10:LUC, ProMdVHA-A3:LUC, and ProMdVHA-D2:LUC compared with when only 35S:MdMYB44 was co-transformed with each of ProMa1:LUC, ProMa10:LUC, ProMdVHA-A3:LUC, and ProMdVHA-D2:LUC (Figure 8, D–G). These findings indicate that MdbHLH49 interacts with MdMYB44 and enhances MdMYB44 activity.
Figure 8.
MdbHLH49 interacts with MdMYB44 and enhances MdMYB44 activity. A, Y2H assay showing that MdbHLH49 interacts with MdMYB44. B, BiFC assay showing that MdbHLH49 interacts with MdMYB44 in the nucleus. The fluorescent images were obtained using confocal microscopy. Scale bar = 50 µm. C, GST pull-down showing that MdbHLH49 interacts with MdMYB44. D−G, LUC assay showing that MdbHLH49 significantly enhances MdMYB44 activity by affecting the MdMYB44 downstream gene promoters of Ma1 (D), Ma10 (E), MdVHA-A3 (F), and MdVHA-D2 (G). Values are the mean ± SE in D–G. The *** indicates significantly different values (P < 0.001).
The allelic variants SNP A/G and SNP T/− can jointly alter MdMYB44 expression levels to regulate malate accumulation
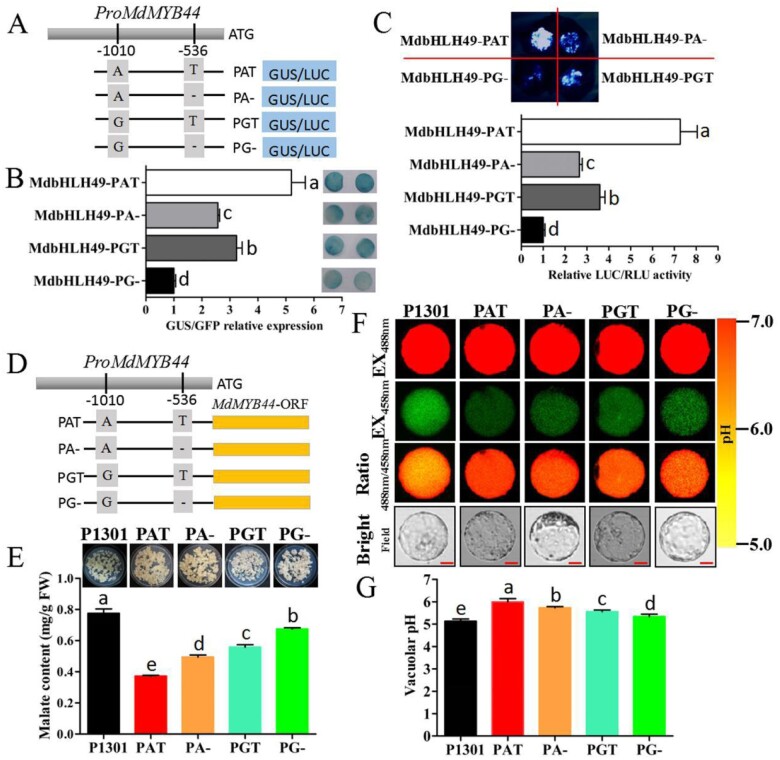
To test how different combinations of SNP A/G and SNP T/− could influence MdMYB44 transcript levels and malate accumulation, four types of the MdMYB44 promoter (PAT, PA−, PGT, and PG−) were synthesized and used (Figure 9, A). These constructs were all co-transformed with 35S:bHLH49, and the expression of the GUS reporter and LUC activity differed among the four combinations: PAT > PGT > PA− > PG− (Figure 9, B and C).
Figure 9.
The combinations (PAT, PA−, PGT, and PG−) of SNP A/G and SNP T/− in MdMYB44 promoter influence MdMYB44 transcription and malate accumulation. A, Synthetic constructs containing the combinations (PAT, PA−, PGT and PG−) fused to the GUS reporter or LUC reporter. B, The GUS reporter expressions exhibited significant differences among the four combinations: PAT > PGT > PA− > PG−. C, The LUC activity exhibited significant differences among the four combinations: PAT > PGT > PA− > PG−. D, Synthetic constructs containing the combinations (PAT, PA−, PGT, and PG−) fused to the MdMYB44 ORF. E, Malate content in transgenic apple calli by transforming each promoter PAT, PA−, PGT, and PG− driving the MdMYB44 ORF. F, Protoplast vacuoles of transgenic calli lines in (E) containing BCECF were imaged at 488 and 458 nm by confocal microscopy. Scale bar = 10 μm. G, The pH level in the protoplast vacuoles of transgenic calli lines was determined. Values are the mean ± SE in B, C, E, and G. The a–e letters denote differences at P < 0.01 by Dunnett’s multiple comparison.
To further investigate how the four combinations might affect malate accumulation, we introduced the MdMYB44 ORF driven by the PAT, PA−, PGT, or PG− promoter into apple calli (Figure 9, D). The malate content of transgenic calli differed significantly among the four combinations: PAT < PA− < PGT < PG− (Figure 9, E), as did their average vacuolar pH: PAT > PA− > PGT > PG− (Figure 9, F and G). The lower vacuolar H+ concentration in the combination AT lines confirmed that the AT combination was associated with a lower malate content, the A− and GT combinations were associated with an intermediate malate content, and the G− combination was associated with a higher malate content. These findings are consistent with our previous observations on the nine SNP genotypes and with MdMYB44 expression levels in apple fruit (Figure 1).
Discussion
In this study, we confirmed that fruit malate content is both a quantitative trait and its variation is primarily determined by genetic effects (Table 1 and Supplemental Figure S1). Previous studies have revealed the major QTLs controlling apple fruit acidity (Ma et al., 2016; Jia et al., 2018). For example, a premature termination mutation in the Ma1 gene in the Ma locus on LG16 leads to low fruit acidity in apple (Bai et al., 2012; Ma et al., 2015b; Li et al., 2020). The T268A variant of MdPP2CH that mapped to LG8 is strongly associated with fruit malate content and a 36-bp insertion in the MdSAUR37 promoter caused high fruit acidity in apple (Jia et al., 2018). Here, we further investigated the MdMYB44 transcription factor, which had emerged as a candidate gene in the major qtl08.1 region for apple fruit acidity in our prior study (Jia et al., 2018). We identified the SNPs A/G and T/− in the MdMYB44 promoter. SNP G and SNP − were associated with higher malate content, whereas SNP A and SNP T were associated with lower malate content (Figure 1 and Supplemental Table S6). SNP A/G and SNP T/− could be used as markers to distinguish between sour and non-sour apple cultivars.
Fruit acidity is a quantitative trait that is controlled by one or several major genes (Chen et al., 2015; Cao et al., 2016; Jia et al., 2018). The predicted phenotype values of malate content could be inferred by the genotype values of the major malate-related genes MdSAUR37, MdPP2CH, Ma1, and MdMYB44 (Jia et al., 2018). The present study indicated that MdMYB44 negatively regulates Ma1 transcripts, and we deduced that the existence of a hierarchical epistatic effect and an additive effect between the major genes MdMYB44 and Ma1. Ma1 accounted for only 7.46% of the variation in malate content (Ma et al., 2015b). We found that the three genotypes (AA, AG, and GG) of Ma1 were associated with malate content in 135 Malus germplasm accessions (P = 0.001505), whereas the two SNPs of MdMYB44 were highly associated with malate content (SNP T/−: P = 1.4193 × 10−7; SNP A/G: P = 4.6119 × 10−14; combination of two SNPs: P = 2.3328 × 10−14; Supplemental Table S3). Therefore, both SNPs in the MdMYB44 promoter and Ma1 are essential for the selection of fruit acidity, and the two SNPs in the MdMYB44 promoter are likely more important than Ma1. Previous studies showed that QTLs on LG8 explained greater phenotypic variances than Ma1 on LG16 in natural and some pedigree-based populations, and QTLs on LG8 could also be important (Liebhard et al., 2003; Kumar et al., 2012; Zhang et al., 2012; Ma et al., 2016; Jia et al., 2018). For example, major QTLs for apple fruit acidity detected on LG8 accounted for even higher phenotype variance (46%) than that on LG16 (42%; Liebhard et al., 2003). Similar results were found in the two major QTLs for malic acid content on LG8 (14.4%) and LG16 (12.4%; Ma et al., 2016). A major-effect QTL on LG8 and a moderate-effect QTL on LG16 for titratable acid of apple fruit were previously identified (Kumar et al., 2012). Furthermore, there is a complementary relationship between MdMYB44 and Ma1. For example, Ma1 plays a major role in the malate accumulation of the Malus accessions such as “Ruby,” “Xinjiang 8,” and “Holly,” whereas MdMYB44 figures prominently in “Qingxiang,” “Qiuxiang,” “Xinshijie,” “Qianxue,” “Meiba,” “Balenghaitang,” and “Xinjiang 3” (Supplemental Table S3).
In plant cells, malate accumulation is affected by malate metabolism and vacuolar storage (Sweetman et al., 2009; Etienne et al., 2013). Vacuolar transporters can transport malate into or out of the vacuole; therefore, their participation is crucial for controlling the extent of vacuolar acidification (Etienne et al., 2013). Recent studies demonstrated that Ma1 (MdALMT9) can transport malate from the cytoplasm into the vacuole and is responsible for much of the variation in apple fruit acidity (Bai et al., 2012; Ma et al., 2015b; Li et al., 2020). In addition, proton pumps, including those of V-ATPase and Ma10 (plasma H+-ATPas, P-ATPase), provide the driving force to move the malate into the vacuole (Hu et al., 2016; Veshaguri et al., 2016; Wilson et al., 2016; Ma et al., 2018). In our study, the malate transporters and proton pumps of Ma1, Ma10, MdVHA-A3, and MdVHA-D2 were identified in the MdMYB44-OVX transgenic apple calli, and their expression levels were significantly decreased in overexpressed MdMYB44 calli (Figure 3 and Supplemental Table S7). Hence, vacuolar acidification and malate accumulation of apple fruit appear to be genetically complex and are likely controlled by a complex regulatory network of genes.
MYB transcription factors play an essential role in regulating vacuolar acidification. For instance, PH4, a MYB transcription factor, enhances vacuolar acidification in petunia (Petunia hybrida) petal cells and grapevine (Vitis vinifera L.; Quattrocchio et al., 2006; Cavallini et al., 2014; Amato et al., 2019). Both MdMYB1 and MdMYB73 increase vacuolar acidification in apple (Hu et al., 2016, 2017). However, MdMYB1 is barely expressed in the flesh of apple fruit lacking the accumulation of anthocyanin; therefore, MdMYB1 is ineffective at influencing malate accumulation in the flesh of apples (Jia et al., 2020). Our previous experiments indicated that MdMYB73 is primarily expressed at 30 and 60 DAFB, but it is minimally expressed in mature fruit. In our study, the expression of MdMYB44 was closely correlated with fruit malate content in mature fruit (Figure 1, E). Interestingly, our findings suggest a mechanism by which MdMYB44 negatively regulates malate accumulation and vacuolar acidification via the direct repression of malate transporters and proton pumps, including Ma1, Ma10, MdVHA-A3, and MdVHA-D2 (Figure 4). Additionally, MdMYB44 interacts with MdbHLH49 to further decrease malate content and repress vacuolar acidification (Figure 8). This study reports the function of MdMYB44 in the regulation of malate accumulation. Furthermore, a suite of genes, such as ethylene-responsive transcription factors, sugar-related genes, PIN-like genes, ABC transporters, and flavonol-related genes, were significantly up- or down-regulated in MdMYB44 overexpressing transgenic apple calli when compared with the control P83 (Supplemental Table S7), indicating that MdMYB44 has other potential functions. Therefore, further research is warranted.
The two variants, SNP A/G and SNP T/−, co-regulate malate content through their influence on MdMYB44 transcript levels (Figure 9). Similar variants have been described in MdSAUR37 and Sl-ALMT9, which regulate malate accumulation in apples and tomatoes, respectively (Ye et al., 2017; Jia et al., 2018). The TATA-box plays a pivotal role in the activation of different types of genes (Zhang et al., 2017; Huang et al., 2018), including the MdMYB44 promoter, which harbors an intact TATA-box motif when the variant of SNP A/G is A. Our study suggests that the TATA-box can enhance the activity of the MdMYB44 promoter, leading to reduced malate content in transgenic apple calli (Figure 6). The E-box motif is recognizable by the bHLH proteins (Xie et al., 2012; Li et al., 2017). In our study, MdbHLH49 binds to the E-box when the variant of SNP T/− is T in the MdMYB44 promoter, thereby enhancing MdMYB44 expression and leading to a decline in the malate content of transgenic apple calli and roots (Figure 7). Furthermore, MdbHLH49 interacts with MdMYB44 to enhance MdMYB44 activity. Similarly, previous work has indicated that MdCIbHLH1 enhances MdMYB73 activity to modulate malate accumulation in apple fruit (Hu et al., 2017). MdBT2 controls malate accumulation via the MdCIbHLH1–MdMYB73 regulatory module (Zhang et al., 2020). MdbHLH3 plays a central role in the accumulation of both malate and anthocyanins by interacting with MdMYB1 and binding to the promoter of MdMYB1 (Hu et al., 2016). Additionally, previous study found that Noemi, encoding a bHLH transcription factor, controls fruit acidity and anthocyanin accumulation in Citrus spp. (Butelli et al., 2019).
Malate is a key factor in the regulation of apple fruit flavor and provides human health benefits. Maintaining fruit acidity is essential for ensuring the quality of apples both for immediate consumption and cider processing. Apple preferences differ drastically among countries and regions. For example, people in Asia prefer sweet, low-acid apples, whereas Europeans prefer sour, high-acid apples (Ma et al., 2015a). For the purpose of cider production, relatively high levels of malate in apples are critical and indispensable (Huang et al., 2018). Therefore, more detailed knowledge of the mechanism of malate accumulation in various apple cultivars is needed. This study developed two markers, SNP T/− and SNP A/G, which co-influence malate accumulation in apple fruit and could be used for the selection of appropriate fruit acidity for different kinds of utilization. The research is valuable for gaining insight into the evolution of Malus and accelerating the development of apple germplasm innovation.
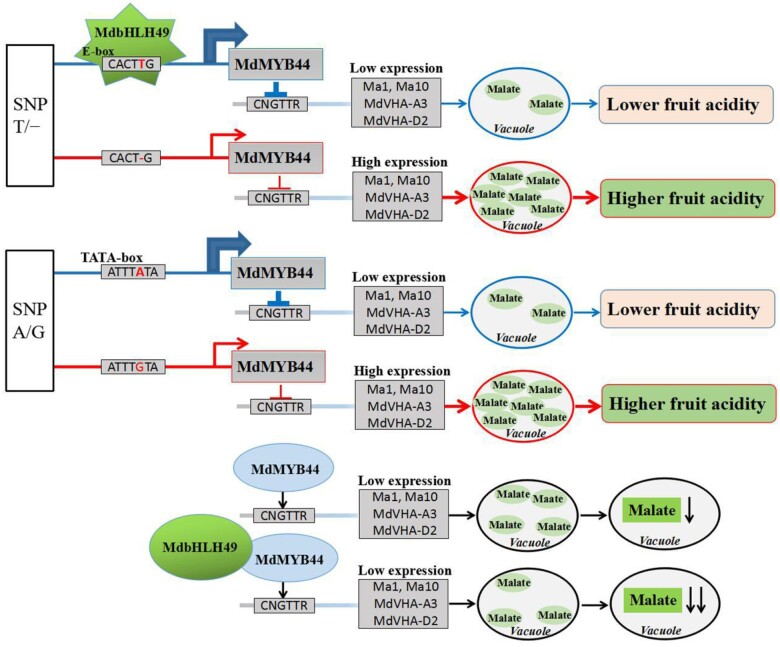
Herein, we elucidated a model that could explain the molecular basis for different levels of malate in different apple varieties (Figure 10). MdMYB44 negatively regulates fruit malate accumulation by repressing the promoter activity of the malate-associated genes Ma1, Ma10, MdVHA-A3, and MdVHA-D2. Two allelic variants in the MdMYB44 promoter, SNP T/− and SNP A/G, were experimentally shown to co-influence malate accumulation in fruit; therefore, they may prove useful in MAS. SNP G and SNP − are associated with a higher malate content, whereas SNP A and SNP T are associated with a lower malate content. The TATA-box in the MdMYB44 promoter caused by the presence of SNP A enhances the basal activity of the MdMYB44 promoter. The transcription factor MdbHLH49 can bind the MdMYB44 promoter via the presence of SNP T to enhance the MdMYB44 transcript levels and repress malate accumulation. Furthermore, MdbHLH49 interacts with MdMYB44 and promotes the activity of MdMYB44.
Figure 10.
Proposed model for the role of two natural genetic variants in the MdMYB44 promoter for regulating malate accumulation in apple fruit. In apple cultivars with lower fruit acidity, the transcription factor MdbHLH49 recognizes the E-box in the promoter of MdMYB44, resulting in augmented MdMYB44 transcript levels. TATA-box in the MdMYB44 promoter enhances MdMYB44 transcript levels. Then MdMYB44 silences their target malate-associated genes Ma1, Ma10, MdVHA-A3, and MdVHA-D2, resulting in decreases to the malate content. By contrast, in apple cultivars with higher fruit acidity, MdbHLH49 cannot bind to the promoter of MdMYB44 due to the replacement of a T residue with a deletion. Lacking the TATA-box in the MdMYB44 promoter, the latter can no longer enhance MdMYB44 transcript levels due to the replacement of an A residue with a G. Accordingly, very low amounts of MdMYB44 are produced, leading to the expression of the target genes Ma1, Ma10, MdVHA-A3, and MdVHA-D2, thereby increasing the overall malate content. Furthermore, MdbHLH49 interacts with MdMYB44 and enhances MdMYB44 activity.
Materials and methods
Plant materials
The apple (M. domestica Borkh.) trees used for genotyping consisted of 246 F1 progenies from M. domestica “Jonathan” × “Golden Delicious,” 123 F1 progenies from M. asiatica “Zisai Pearl” × M. domestica “Red Fuji,” and 135 Malus germplasm accessions (Jia et al., 2018). The trees were planted with a spacing density of 2.5 m × 0.5 m and were grown under identical management practices. All F1 progenies and Malus accessions were used to detect any association between molecular markers and malate content.
Apple fruits were collected and treated at 30, 60, 90, 120, 150, and 180 DAFB. Each sample included six fruits. Young leaves used for the DNA extractions were collected in late spring. The fruit flesh and leaves were frozen in liquid nitrogen and stored at –76°C until their use.
Apple calli used for the transformation and tonoplast extraction were induced from the flesh of the apple “Orin” (with a genotype AT/GT; Jia et al., 2018). The calli were cultured on Murashige and Skoog (MS) medium supplemented with 1.0 mg L−1 6-benzylaminopurine and 0.5 mg L−1 indole-3-butyric acid and were subcultured every 3 weeks at 25°C under dark conditions.
Organic acid extraction and determination
Fruit organic acid was isolated and measured as previously described (Jia et al., 2018). The malate content of fruit was measured and quantified using high-performance liquid chromatography (Waters 600 Chromatograph, USA) with a dual λ ultraviolet detector (Waters 2487). The statistical analysis of malate content in G1–G9 Malus accessions was conducted using the data collected on malate content in the years 2015, 2016, and 2017. The P-value was calculated for analyzing the significance of molecular markers based on the false discovery rate of malate content. The criterion of marker–trait association was set at P < 0.005 (Ma et al., 2015b). Significant difference was determined using GraphPad Prism 5 software (*P < 0.05; **P < 0.01; ***P < 0.001).
Genomic DNA extraction, RNA extraction, and RT-qPCR assay
Genomic DNA and total RNA were each isolated, and a reverse transcription quantitative PCR (RT-qPCR) was conducted as previously described (Jia et al., 2018). EF1α served as the reference gene. The RT-qPCR was performed on three biological replicates containing three technical replicates each. All the primers used for the RT-qPCR are listed in Supplemental Table S8.
Plasmid construction and genetic transformation
The plasmids were constructed as described by Jia et al. (2018). Briefly, each full-length coding sequence of MdMYB44 or MdbHLH49 was inserted into the overexpression vector pMDC83. For the RNAi vectors, a 383-bp fragment (located between +547 bp and +929 bp in the coding sequence) of MdMYB44 and a 332-bp fragment (located between +1,008 bp and +1,339 bp in the coding sequence) of MdbHLH49 were separately inserted into the RNAi vector pFGC5941. Each ensuing recombinant plasmid (35S:MdMYB44, RNAi:MdMYB44, 35S:MdbHLH49, and RNAi:MdbHLH49) was then transformed into “Orin” apple calli (Jia et al., 2018), apple fruits (Li et al., 2017), and apple roots (Sun et al., 2020), as previously described.
Different promoter fragments of MdMYB44, MdMa1, MdMa10, MdVHA-A3, and MdVHA-D2 were inserted into the vector pCAMBIA1301, as a Pro:GUS fusion vector. The recombinant plasmids were transformed into the Agrobacterium strain GV3101, and then transformed into 4-week-old Nicotiana benthamiana leaf epidermal cells. β-Galactosidase Reporter Gene Staining Kit (Solarbio, China) was used for GUS staining (Jia et al., 2018). All the primers used are listed in Supplemental Table S8.
RNA-seq analysis
Total RNA was isolated from MdMYB44 over-expression transgenic calli lines, by using an RNAprep Pure Plant Plus Kit (Polysaccharides and Polyphenolics-rich; Tiangen, China). Three biological replicates were performed for each transgenic calli line. The RNA-seq analysis was conducted by Biomarker Co. (Beijing, China). Two of the three transgenic calli lines were used to identify DEGs as the RNA-seq data of one line were not robust. RNA-seq analysis was performed as previously described (Ye et al., 2017; Ma et al., 2018). Clean reads were mapped to the genome sequences (Daccord et al., 2017) which was downloaded from Genome for Rosaceae (https://www.rosaceae.org/species/malus/malus_x_domestica/genome_GDDH13_v1.1). The number of unambiguous clean reads for each gene was calculated and normalized using Reads Per Kilobase of gene per Million reads value. The software edgeR was used to conduct differential expression analysis (Robinson et al., 2010) and analyze the difference in expression between the MdMYB44-OVX calli and the control P83 calli to calculate the fold change (Log2 ratio). P-value < 0.01 and log2-fold change of one were set as the threshold for significantly differential gene expression. DEGs were annotated based on NCBI RefSeq nucleotide database, UniPro protein and Swiss-Prot databases, followed by the pathway databases including KEGG (http://www.genome.jp/kegg/), COG (http://www.ncbi.nlm.nih.gov/COG/), and GO (http://www.geneontology.org/).
Protoplast isolation and transformation
Both the isolation and transformation of apple calli protoplasts were carried out as previously described (Yoo et al., 2007; Jia et al., 2018). The recombinant constructs were transformed into apple calli protoplasts, by using a PEG-mediated process. The transformed protoplasts were harvested after 16 h and stored at –76°C. Subsequently, the transformed protoplasts were used for RNA extraction, GUS expression analysis, and pH determination. Three independent biological replicates with three technical replicates each were tested.
V-ATPase activity assay
The tonoplast vacuoles of apple calli were isolated following the methodology described by Terrier et al. (2001). The V-ATPase hydrolase activity and proton transport activity were determined using a fluorescence spectrophotometer, as previously described (Jia et al., 2018). The data were tested for three biological replicates and three technical replicates.
Measuring the vacuolar pH
The pH of apple calli protoplast vacuoles was measured using a pH-sensitive fluorescent dye BCECF-AM, as previously described (Tang et al., 2012; Hu et al., 2016). The calibration curve was generated by plotting the fluorescence ratios (488/458 nm) against the pH value (Supplemental Figure S5, A and B). The data were tested for six biological replicates and three technical replicates. The images were generated using confocal microscopy.
Measurement of proton efflux capacity
The transient transgenic apple roots and transformed apple calli were separately transferred onto a medium containing 0.006% (w/v, g/L) bromocresol purple (the pH indicator) and 0.8% w/v agarose, at pH = 5.9, and incubated for 1–3 h (Zandonadi et al., 2010; Sun et al., 2020). The pH indicator bromocresol purple was used for staining, with acidification indicated by the yellow around the apple roots or calli. More yellowing around the apple roots or calli was indicative of a higher degree of acidification in the tissues.
Protein extraction and western blotting
Protein extraction and western blotting analysis were conducted as previously described (Jia et al., 2018). The protein concentration was analyzed using the BCA reagent (ComWin Biotech, China). The anti-GFP antibody and anti-ACTIN antibody were used, both obtained from the ComWin Biotech Company.
Y1H assay
The Y1H assays were performed as previously described (Tian et al., 2015; Li et al., 2017). The full-length coding sequence of MdMYB44 was inserted into the effector vector pJG4-5 (Clontech, USA). Each Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoter fragment was inserted into the reporter vector pLacZi. The full-length coding sequence of MdbHLH49 was inserted into the effector vector pGADT7 (Clontech). Each MdMYB44 promoter fragment was inserted into the reporter pAbAi vector (Clontech). All primers used are listed in Supplemental Table S8.
Y1H screening of apple fruit cDNA libraries
Y1H screening was implemented as described by Tran et al. (2004). The AD–cDNA (AD represents PGADT7-Rec) library from “Golden Delicious” apple fruit at various stages of development (30, 60, 90, 120, and 150 DAFB) was assembled by Takara Bio Inc. using the Clontech Matchmaker one-hybrid system. The reporter plasmid pAbAi-ProMdMYB44-T was constructed using a 197-bp fragment with T present in the E-box in the MdMYB44 promoter, then the pAbAi-ProMdMYB44-T plasmid was transformed into Y1H susceptible yeast. The AD-cDNA library was transfected into susceptible yeast containing pAbAi-ProMdMYB44-T to identify the binding specific transcription factors. All primers used are listed in Supplemental Table S8.
EMSA assays
The EMSA assays were performed as previously described (Li et al., 2017; Zhang et al., 2018). Each full-length coding sequence of MdMYB44 or MdbHLH49 was inserted into the pGEX-4T-1 vector (GST-tag), and then transformed into BL21 (DE3) Escherichia coli cells for protein expression. The 5ʹ biotin end-labeled double-stranded DNA probes were synthesized by Sangon Biotech. The EMSA assays were conducted using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, USA). The biotin-labeled Ma1 promoter, MdVHA-A3 promoter, and MdMYB44 promoter sequences are shown in Supplemental Table S8.
ChIP-PCR analysis
The ChIP-PCR assay was conducted following Hu et al. (2016) and Li et al. (2017). The full-length coding sequence (without the stop codon) of MdMYB44 was inserted into pMDC83-GFP vector and the fusion MdMYB44-GFP was generated. The recombinant pMDC83-MdMYB44-GFP was then transformed into apple calli. The ChIP-PCR assays were performed using the EpiQuik Plant ChIP Kit (Epigentek, USA) and anti-GFP antibody (ab290, Abcam, UK). Four Ma1 promoter regions, four Ma10 promoter regions, five MdVHA-A3 promoter regions, and five MdVHA-D2 promoter regions were analyzed to evaluate their enrichment. Each ChIP assay was repeated three times. The primers used are listed in Supplemental Table S8.
LUC assay
The LUC assay was performed as previously described (Ye et al., 2017). Each full-length coding sequence of MdMYB44 or MdbHLH49 was inserted into the effector vector pGreenII 62-SK under the control of the CaMV35S promoter. Each MdMYB44, Ma1, Ma10, MdVHA-A3, and MdVHA-D2 promoter fragment was inserted into the reporter vector pGreenII 0800-LUC. The combinations of effector and reporter vectors were co-transformed into 4-week-old N. benthamiana leaf epidermal cells. Firefly and Renilla LUC signals were observed with the treatment of dual LUC assay reagents (Promega, USA) using an Infinite M200 (Tecan, Switzerland). Each LUC assay was repeated three times. The primers used are given in Supplemental Table S8.
Y2H assay
The Y2H assay was conducted as previously described (Jia et al., 2018). The full-length coding sequence of MdMYB44 was cloned into the pGADT7 vector, and the full-length coding sequence of MdbHLH49 was cloned into the pGBKT7 vector. The primers used are given in Supplemental Table S8.
BiFC assay
The BiFC assay was implemented as previously described (Schütze et al., 2009; Jia et al., 2018). The full-length coding sequence (without the stop codon) of MdMYB44 was cloned into pSPYNE-35S, and the full-length coding sequence (without the stop codon) of MdbHLH49 was cloned into pSPYCE-35S. Leaf epidermal cells were observed under a confocal microscope. All the primers used are listed in Supplemental Table S8.
GST pull-down assay
The GST pull-down assay was conducted as previously described (Li et al., 2017; Huang et al., 2018). The full-length coding sequence of MdMYB44 was inserted into the pGEX-4T-1 vector, and the full-length coding sequence of MdbHLH49 was inserted into the pET-28a vector. For this assay, the primers used are listed in Supplemental Table S8.
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Frequency distribution for the fruit malate content of F1 progenies from “Jonathan” × “Golden Delicious” and “Zisai Pearl” × “Red Fuji” crosses in the years 2014–2016.
Supplemental Figure S2. Malate content in different apple varieties during fruit development.
Supplemental Figure S3. Western blotting analysis of MdMYB44-GFP protein in the overexpressing MdMYB44 transgenic calli, P83 calli, and wild-type calli.
Supplemental Figure S4. Gene Ontology (GO) term analysis of the DEGs determined by RNA-seq in MdMYB44-OVX transgenic apple calli lines compared with the control P83.
Supplemental Figure S5. Calibration curve of BCECF and pH in protoplast vacuoles of apple calli.
Supplemental Figure S6. The MdMYB44 promoter sequences cloned from “Jonathan” and “Golden Delicious’. J, “Jonathan”. G, “Golden Delicious.”
Supplemental Figure S7. Correlations between MdbHLH49, MdbHLH149, and MdMYC1 expression levels and the fruit malate content, and between MdbHLH49 and MdMYB44 expression levels.
Supplemental Figure S8. Western blotting analysis of MdbHLH49-GFP protein in the overexpressing MdbHLH49 transgenic calli, P83 calli, and wild-type calli.
Supplemental Table S1. The T/− genotype of MdMYB44 in 246 F1 progenies of “Jonathan (J)” × “Golden Delicious (G)”.
Supplemental Table S2. The T/− genotype of MdMYB44 in 123 F1 progenies of “Zisai Pearl (Z)” × “Red Fuji (F)”.
Supplemental Table S3. The SNP diversity and genotypes of MdMYB44 and Ma1, and fruit malate content, in 135 Malus accessions.
Supplemental Table S4. The nine genotypes of MdMYB44 variants and their fruit malate content in 40 Malus germplasm accessions.
Supplemental Table S5. The linkage analysis between SNP T/− and SNP A/G of the MdMYB44 promoter in 246 progenies from “Jonathan” × “Golden Delicious,” in 123 progenies from “Zisai Pearl” × “Red Fuji,” and in 135 Malus accessions.
Supplemental Table S6. Segregation of SNP T/− and SNP A/G genotypes in the F1 progenies from two crossed populations.
Supplemental Table S7. Profiling of DEGs in transgenic apple calli when compared between the MYB44-OVX and control P83.
Supplemental Table S8. The primers used in this study.
Supplementary Material
Acknowledgments
The authors thank Zhenyu Huang (China Agricultural University) for assistance with the sampling, and Yuanming Zhou (Qingdao Agricultural University) for measuring the fruit malate content in the experiments.
D. J., Y.Y., X.Z.Z., and Z.H. designed the experiments. D.J., P.W., and F.S. carried out the experiments. Y.Y., W.L., Y.W., X.D.Z., and X.Z.Z. contributed the plant materials, were responsible for the malate content determination, and performed the statistical analysis. D.J. wrote the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Zhenhai Han (rschan@cau.edu.cn), Xinzhong Zhang (zhangxinzhong999@126.com) and Yongbing Yuan (yyb@qau.edu.cn).
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 32001993 and 31972362), the Project of the Shandong Natural Science Foundation (grant no. ZR2020QC143), the Breeding Plan of the Shandong Provincial Qingchuang Research Team (grant no. 2019), and the High-Level Scientific Research Foundation of Qingdao Agricultural University (grant no. 663/1119045).
Conflict of interest statement. The authors declare no competing financial interests.
References
- Amato A, Cavallini E, Walker AR, Pezzotti M, Bliek M, Quattrocchio F, Koes R, Ruperti B, Bertini E, Zenoni S et al. (2019) The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J 99: 1220–1241 [DOI] [PubMed] [Google Scholar]
- Bai Y, Dougherty L, Li MJ, Fazio G, Cheng LL, Xu KN (2012) A natural mutation-led truncation in one of the two aluminum-activated malate transporter-like genes at the Ma locus is associated with low fruit acidity in apple. Mol Genet Genomics 287: 663–678 [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse JM, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Ramadugu C, Durand-Hulak M, Celant A, Reforgiato Recupero G, Froelicher Y, Martin C (2019) Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr Biol 29: 158–164.e2 [DOI] [PubMed] [Google Scholar]
- Cao K, Zhou ZK, Wang Q, Guo J, Zhao P, Zhu GR, Fang WC, Chen CW, Wang XW, Wang XL et al. (2016) Genome-wide association study of 12 agronomic traits in peach. Nat Commun 7: 13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini E, Zenoni S, Finezzo L, Guzzo F, Zamboni A, Avesani L, Tornielli GB (2014) Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol 55: 517–534 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang N, Fang LC, Liang ZC, Li SH, Wu BH (2015) Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Itkin M, Yeselson Y, Tzuri G, Portnoy V, Harel-Baja R, Lev S, Burger Y, Schafferet AA, et al. (2014) The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat Commun 5: 4026. [DOI] [PubMed] [Google Scholar]
- Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Ding S, Yang Y, Mei J (2016) Protective effects of L-malate against myocardial ischemia/reperfusion injury in rats. Evid Based Compl Alt 3803657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerlich V, Linka N, Reinhold T, Hurth MA, Traub M, Martinoia E, Neuhaus HE (2003) The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc Natl Acad Sci U S A 100: 11122–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne A, Génard M, Lobit P, Mbeguié-A-Mbéguié D, Bugaud C (2013) What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J Exp Bot 64: 1451–1469 [DOI] [PubMed] [Google Scholar]
- Faraco M, Spelt C, Bliek M, Verweij W, Hoshino A, Espen L, Prinsi B, Jaarsma R, Tarhan E, de Boer AH, et al. (2014) Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep 6: 32–43 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K (. 2007) Plant proton pumps. FEBS Lett 581: 2204–2214 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hu DG, Li YY, Zhang QY, Li M, Sun CH, Yu JQ, Hao YJ (2017) R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J 91: 443–454 [DOI] [PubMed] [Google Scholar]
- Hu DG, Sun CH, Ma QJ, You CX, Cheng L, Hao YJ (2016) MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol 170: 1315–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wang X, Tang ZZ, Yuan Y, Xu YT, He JX, Jiang XL, Peng SA, Li L, Butelli E, Deng XX, Xu Q (2018) Subfunctionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nat Plant 4: 930–941 [DOI] [PubMed] [Google Scholar]
- Huang ZY, Hu HY, Shen F, Wu B, Wang XX, Zhang BG, Wang WQ, Liu L, Liu J, Chen CJ, et al. (2018) Relatively high acidity is an important breeding objective for fresh juice-specific apple cultivars. Scientia Horticult 233: 29–37 [Google Scholar]
- Hurth MA, Suh SJ, Kretzschmar T, Geis T, Bregante M, Gambale F, Martinoia E, Neuhaus HE (2005) Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol 137: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia DJ, Li ZH, Dang QY, Shang LJ, Shen JL, Leng XP, Wang YZ, Yuan YB (2020) Anthocyanin biosynthesis and methylation of the MdMYB10 promoter are associated with the red blushed-skin mutant in the red striped-skin ‘Changfu 2’ apple. J Agr Food Chem 68: 4292–4304 [DOI] [PubMed] [Google Scholar]
- Jia DJ, Shen F, Wang Y, Wu T, Xu XF, Zhang XZ, Han ZH (2018) Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes MdSAUR37, MdPP2CH and MdALMTII. Plant J 95: 427–443 [DOI] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Shim JS, Seo JS, Lee HY, Kim CH, Choi YD, Cheong JJ (2010) Non-specific phytohormonal induction of AtMYB44 and suppression of jasmonate-responsive gene activation in Arabidopsis thaliana. Mol Cell 29: 71–76 [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann P, Meyer S, Hortensteiner S, Picco C, Scholz-Starke J, Ravera S, Lee Y, Martinoia E (2007) The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Kumar S, Chagné D, Bink MC, Volz RK, Whitworth C, Carlisle C (2010) Genomic selection for fruit quality traits in apple (Malus× domestica Borkh. PLoS One 7: e36674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Dougherty L, Coluccio AE, Meng D, El-Sharkawy I, Borejsza-Wysocka E, Liang D, Pineros MA, Xu KN, Cheng LL (2020) Apple ALMT9 requires a conserved C-terminal domain for malate transport underlying fruit acidity. Plant Physiol 182: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Li DG, Feng FJ, Zhang S, Ma FW, Cheng LL (2016a) Proteomic analysis reveals dynamic regulation of fruit development and sugar and acid accumulation in apple. J Exp Bot 67: 5145–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xu YX, Zhang LC, Ji YL, Tan DM, Yuan H, Wang AD (2017) The jasmonate-activated transcription factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YB, Provenzano S, Bliek M, Spelt C, Appelhagen I, Machado de Faria L, Verweij W, Schubert A, Sagasser M, Seidel T, et al. (2016b) Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification. New Phytol 211: 1092–1107 [DOI] [PubMed] [Google Scholar]
- Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C (. 2003) Mapping quantitative physiological traits in apple (Malus× domestica Borkh.). Plant Mol Biol 52: 511–526 [DOI] [PubMed] [Google Scholar]
- Ligaba A, Kochian L, Pineros M (2009) Phosphorylation at S384 regulates the activity of the TaALMT1 malate transporter that underlies aluminum resistance in wheat. Plant J 60: 411–423 [DOI] [PubMed] [Google Scholar]
- Liu YH, Lin-Wang K, Espley RV, Wang L, Li YM, Liu Z, Zhou P, Zeng LH, Zhang J, Zhang JL, et al. (2019) StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J Exp Bot 70: 3809–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BQ, Chen J, Zheng HY, Fang T, Ogutu C, Li SH, Han YP, Wu BH (2015a) Comparative assessment of sugar and malic acid composition in cultivated and wild apples. Food Chem 172: 86–91 [DOI] [PubMed] [Google Scholar]
- Ma BQ, Liao L, Fang T, Peng Q, Ogutu C, Zhou H, Ma FW, Han YP (2018) A Ma10 gene encoding P-type ATPase is involved in fruit organic acid accumulation in apple. Plant Biotechnol J 17: 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BQ, Liao L, Zheng HY, Chen J, Wu BH, Ogutu C, Li SH, Korban SS, Han YP (2015b) Genes encoding aluminum-activated malate trans-porter II and their association with fruit acidity in apple. Plant Gen 8: eplantgenome2015.03.0016 [DOI] [PubMed] [Google Scholar]
- Ma BQ, Zhao S, Wu BH, Wang DM, Peng Q, Owiti A, Fang T, Liao L, Ogutu C, Korban SS, et al. (2016) Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet Genomes 12: 1 [Google Scholar]
- Mauvezin C, Nagy P, Juhász G, Neufeld TP (2015) Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 6: 7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Scholz-Starke J, DeAngeli A, Kovermann P, Burla B, Gambale F, Martinoia E (2011) Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J 67: 247–257 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Tsuchiya Y, Ariyoshi M, Nakano R, Ushijima K, Kubo Y, Mori IC, Higashiizummi E, Galis I, Yamamoto Y (2016) Two members of the aluminum-activated malate transporter family, SlALMT4 and SlALMT5, are expressed during fruit development and the overexpression of SlALMT5 alters organic acid contents in seeds in tomato (Solanum lycopersicum). Plant Cell Physiol 57: 2367–2379 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Krebs M (2010) The V-ATPase: small cargo, large effects. Curr Opin Plant Biol 13: 724–730 [DOI] [PubMed] [Google Scholar]
- Shi CY, Song RQ, Hu XM, Liu X, Jin LF, Liu YZ (2015) Citrus PH5-like H+-ATPase genes: identification and transcript analysis to investigate their possible relationship with citrate accumulation in fruits. Front Plant Sci 6: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73: 483–495 [DOI] [PubMed] [Google Scholar]
- Strazzer P, Spelt CE, Li SJ, Bliek M, Federici CT, Roose ML, Koes R, Quattrocchio FM (2019) Hyperacidification of Citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nat Commun 10: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Chang YS, Yang FQ, Wang Y, Li H, Zhao YB, Chen DM, Wu T, Zhang XZ, Han ZH (2015) A dense SNP genetic map constructed using restriction site-associated DNA sequencing enables detection of QTLs controlling apple fruit quality. BMC Genomics 16: 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun RQ, Hao PB, Lv XM, Tian J, Wang Y, Zhang XZ, Xu XF, Han ZH, Wu T (2020) A long non-coding apple RNA, MSTRG.85814.11, acts as a transcriptional enhancer of SAUR32 and contributes to the Fe-deficiency response. Plant J 103: 53–67 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Shiratake K, Yamaki S (2000) Seasonal changes in the activities of vacuolar H+-pumps and their gene expression in the developing Japanese pear fruit. J Jpn Soc Hortic Sci 69: 15–21 [Google Scholar]
- Swanson SJ, Jones RL (1996) Gibberellic acid induces vacuolar acidification in barley aleurone. Plant Cell 8: 2211–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL (2009) Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70: 1329–1344 [DOI] [PubMed] [Google Scholar]
- Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, Luan S, Zhang HX (2012) Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res 22: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Sauvage FX, Ageorges A, Romieu C (2001) Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta 213: 20–28 [DOI] [PubMed] [Google Scholar]
- Tian J, Peng Z, Zhang J, Song TT, Wan HH, Zhang ML, Yao YC (2015) McMYB10 regulates coloration via activating McF’H and later structural genes in ever-red leaf crabapple. Plant Biotechnol J 13: 948–961 [DOI] [PubMed] [Google Scholar]
- Tran LP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 Promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 10: 1456–1462 [DOI] [PubMed] [Google Scholar]
- Veshaguri S, Christensen SM, Kemmer GC, Ghale1 G, Møller MP, Lohr C, Christensen AL, Justesen BH, Jørgensen IL, et al. (2016) Direct observation of proton pumping by a eukaryotic P-type ATPase. Science 351: 1469–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LZ, Mao WW, Jia MR, Xing SN, Ali U, Zhao YY, Chen YT, Cao ML, Dai ZR, Zhang K, et al. (2018) FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot 69: 4805–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, Chavda1 BJ, Pierre-Louis G, Quinn A, Tan-Wilson A (2016) Role of vacuolar membrane proton pumps in the acidification of protein storage vacuoles following germination. Plant Physiol Biochem 104: 242–249 [DOI] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Xu K, Wang A, Brown S (2012) Genetic characterization of the Ma locus with pH and titratable acidity in apple. Mol Breeding 30: 899–912 [Google Scholar]
- Ye J, Wang X, Hu TX, Zhang FX, Wang B, Li CX, Yang TX, Li HX, Lu YE, Giovannoni JJ, et al. (2017) An InDel in the promoter of Al-activated malate transporter 9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29: 2249–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Cho Y, Sheen J (. 2007) Arabidopsis mesophyll protoplasts: a versatile cell 782 system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zandonadi DB, Santos MP, Dobbss LB, Olivares FL, Canellas LP, Binzel ML, Okorokova-Façanha AL, Façanha AR (2010) Nitric oxide mediates humic acids-induced root development and plasma membrane H+-ATPase activation. Planta 231: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Zhang ML, Lv YD, Wang Y, Rose JKC, Shen F, Han ZY, Zhang XZ, Xu XF, Wu T, Han ZH (2017) TATA box insertion provides a selection mechanism underpinning adaptations to Fe deficiency. Plant Physiol 173: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ma BQ, Li H, Chang YS, Han YY, Li J, Wei GC, Zhao S, Khan MN, Zhou Y et al. (2012) Identification, characterization, and utilization of genome-wide simple sequence repeats to identify a QTL for acidity in apple. BMC Genomics 13: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QL, Ma C, Zhang Y, Gu ZY, Li W, Duan XW, Wang SN, Hao L, Wang YH, Wang SY, et al. (2018) A single-nucleotide polymorphism in the promoter of a hairpin RNA contributes to Alternaria alternata leaf spot resistance in apple (Malus × domestica). Plant Cell 30:1924–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Gu KD, Cheng LL, Wang JH, Yu JQ, Wang XF, You CX, Hu DG, Hao YJ (2020) BTB-TAZ domain protein MdBT2 modulates malate accumulation and vacuolar acidification in response to nitrate. Plant Physiol 183: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Li PM, Cheng LL (2010) Developmental changes of carbohydrates, organic acids, amino acids, and phenolic com-pounds in ‘Honeycrisp’ apple flesh. Food Chem 123: 1013–1018 [Google Scholar]
- Zhou X, Zha M, Huang J, Li L, Imran M, Zhang C (2017) StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. J Exp Bot 68: 265–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.