Abstract
Protein cysteine residues are susceptible to oxidative modifications that can affect protein functions. Proteomic techniques that comprehensively profile the cysteine redoxome, the repertoire of oxidized cysteine residues, are pivotal towards a better understanding of the protein redox signaling. Recent technical advances in chemical tools and redox proteomic strategies have greatly improved selectivity, in vivo applicability, and quantification of the cysteine redoxome. Despite this substantial progress, still many challenges remain. Here, we provide an update on the recent advances in proteomic strategies for cysteine redoxome profiling, compare the advantages and disadvantages of current methods and discuss the outstanding challenges and future perspectives for plant redoxome research.
Current cysteine redoxome profiling can characterize systematically diverse oxidative posttranslational modifications
Advances
The chemical toolbox for Cys redoxome profiling has extensively expanded.
Advanced chemoproteomic platforms have been applied to target specific Cys oxidative posttranslational modifications (OxiPTMs).
Various reductomic workflows have been widely implemented for reversible Cys OxiPTMs quantification.
Workflows have been integrated to measure the occupancy of multiple OxiPTMs simultaneously.
Disulfide-based traps enable the in situ profiling for –SOH sites.
Introduction
Numerous biochemical reactions involve electron transfers that depend on cellular reduction–oxidation (redox) potential. Incomplete reduction yields reactive electrophilic species, such as reactive oxygen/nitrogen/sulfur species (ROS/RNS/RSS; see Box 1), that can react with other biomolecules, including nucleic acids, lipids, and proteins, thus altering cellular homeostasis. Although ROS had traditionally been regarded as toxic side products, their role as signaling molecules in plants is now widely acknowledged (Mignolet-Spruyt et al., 2016; Mittler, 2017). Relatively stable ROS, such as hydrogen peroxide (H2O2), can modify protein function by inducing oxidative posttranslational modifications (OxiPTMs). Protein Cys thiols (–SH) are very susceptible to H2O2, and their reactivity depends on the Cys pKa and local microenvironment (Poole, 2015; Trost et al., 2017). The oxidation state of the thiol sulfur spans a wide range from −2 to +6 with the occurrence of diverse OxiPTMs (Paulsen and Carroll, 2013). The initial reaction of H2O2 with –SH (−2) forms sulfenic acid (–SOH, 0) that is intrinsically unstable and an intermediary en route to other OxiPTMs, such as the relatively more stable sulfinic (–SO2H, +2) and sulfonic (–SO3H, +4) acids (Figure 1). The –SO3H formation is irreversible, whereas –SO2H can be recycled in an ATP-dependent manner by sulfiredoxin (Biteau et al., 2003). Alternatively, –SOH can react with proximal –SHs, either within or from other proteins, and with the tripeptide glutathione (GSH), forming disulfides (–SSR, −1) and S-GSH (–SSG, −1), respectively (Figure 1). Disulfides can be enzymatically reduced to –SH by thiol reductases, such as thioredoxins (Trxs) and glutaredoxins (Grxs; Meyer et al., 2012). Besides ROS, RNS triggers the formation of S-nitrosothiols (–SNO, 0) via diverse mechanisms (see Box 1). Additionally, the RSS hydrogen sulfide (H2S) reacts with –SOH and forms persulfide (–SSH, −1) through S-sulfhydration (Figure 1; Aroca et al., 2018).
Box 1.
Reactive electrophilic species causing Cys OxiPTMs
Three major categories of reactive electrophilic species cause OxiPTMs on protein Cys thiols with a signaling function in plants.
Reactive oxygen species
Among diverse ROS, the relatively stable H2O2 is a well-established signaling molecule. Besides metabolic ROS leakage, as in chloroplastic and mitochondrial electron transfer chains, ROS production can be enzymatically orchestrated by, for instance, respiratory burst oxidase homologs (RBOHs) that generate a superoxide radical (), which, in turn, is converted to H2O2 by superoxide dismutases (SODs). In addition, many other detoxifying and producing enzymes occur in plant compartments (for details, see Mignolet-Spruyt et al., 2016).
Reactive nitrogen species
The major RNS refers to nitric oxide (NO) and its derived molecules. Whereas the origin of NO mainly depends on NO synthase (NOS) in mammals, nonenzymatic reduction of nitrite () is the best recognized NO source in plants. NO can react with O2 to form dinitrogen trioxide (N2O3) and nitrogen dioxide (NO2), or with to yield peroxynitrite (ONOO), which plays an important role in protein nitration. NO can also react with GSH in the presence of O2 to form S-nitrosoglutathione (GSNO), a major endogenous NO donor. NO serves as a signaling radical molecule that, through protein Cys S-nitrosylation, is involved in many well-described physiological processes in plants. Several mechanisms have been proposed for protein –SNO formation (Alcock et al., 2018). For instance, NO could directly react with thiyl radicals, or N2O3 with thiols. In addition, transnitrosylation between thiols and GSNO can occur, and more mechanisms have been suggested (for more details, see Alcock et al., 2018).
Reactive sulfur species
Cys persulfidation provoked by H2S is a major mechanism in sulfide (S2−) signaling (Aroca et al., 2018). H2S is, rather, a reductant and, upon reaction with –SOH, can generate persulfide (–SSH) and protect against overoxidation (Zivanovic et al., 2019). An important H2S source is provided by sulfate () assimilation in the plastids, in which sulfite reductase reduces sulfite () to sulfide. In addition, H2S is formed as a byproduct during cyanide detoxification in mitochondria and l-cysteine desulfhydrase1 (DES1) in the cytosol. For a more complete description and alternative production mechanisms, we refer to Aroca et al. (2018).
Figure 1.
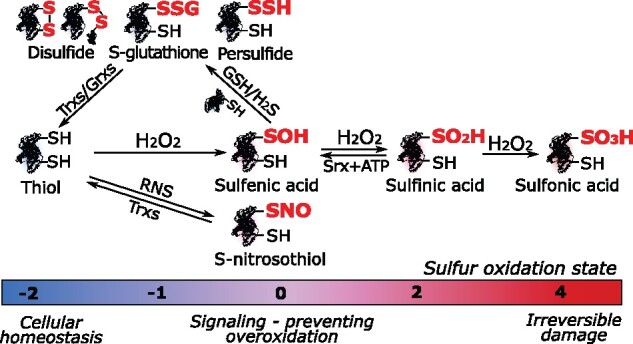
Simplified overview of Cys redox posttranslational modifications. The various Cys OxiPTMs are highlighted using red bold font and their respective sulfur oxidation states are indicated by a color code. Srx, sulfiredoxin.
Importantly, OxiPTMs provide a rapid posttranslational regulation mechanism that can fine-tune protein functions. A systematic characterization of the repertoire of cellular Cys OxiPTMs, or “redoxome” (Chiappetta et al., 2010), is essential to guide downstream functional research and to uncover redox-regulatory processes (Yang et al., 2015; Huang et al., 2018a, 2019; Shen et al., 2020). Here, we outline contemporary technologies for redoxome profiling in three groups: (1) chemoproteomic methods that use chemical probes to target specific OxiPTMs, (2) indirect reductomic methods that rely on general or selective reduction of OxiPTMs, and (3) recombinantly produced or genetically encoded proteinaceous disulfide traps. Besides highlighting plant redoxome studies, we describe several cutting edge profiling methods, mostly established in mammals. Lastly, we discuss current challenges and future perspectives for (plant) redox PTM research.
Chemoproteomics for detection of specific Cys OxiPTMs
Chemoproteomics is an interdisciplinary field in which protein interactors of small chemical molecules are identified by mass spectrometry (MS). Various chemical probes with high affinity toward –SH or specific Cys OxiPTMs have been developed and used for redoxome profiling (Figure 2). Below, chemoproteomic workflows are discussed per Cys OxiPTM together with several representative studies.
Figure 2.
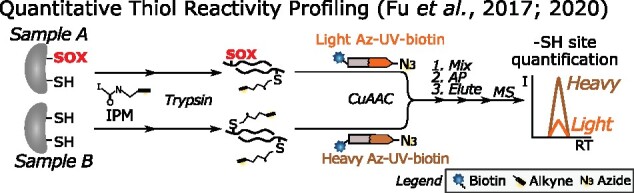
QTRP for pairwise quantitation of –SH levels (Fu et al., 2017, 2020). Cys –SH in protein lysates are labeled by IPM, unlike oxidized Cys thiols (SOX, bold red font). After trypsin digestion, IPM-tagged Cys are differentially conjugated with light (orange) or heavy (brown) isotope-labeled Az-UV-biotin depending on the conditions (for instance, H2O2-treated versus nontreated cells). After mixing light- and heavy-tagged samples, biotinylated peptides are captured on streptavidin and modified peptides are photoreleased by 365-nm UV light. Eluted peptides are identified and quantified by MS analysis. Comparison of peptide precursor intensities allows deduction of relative –SH levels between conditions. I, intensity; RT, retention time; SOX, oxidized thiol.
Quantification of native –SH levels
In redoxome workflows, free –SHs are typically blocked by thiol alkylating agents, such as iodoacetamide (IAM) and N-ethylmaleimide (NEM). Instead of mere blocking, they can be used to monitor –SH levels. Such chemicals are typically equipped with an alkyne handle to enable conjugation with azide-containing reporter groups via Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) or “click chemistry” (Speers et al., 2003), thus facilitating protein or peptide enrichment via affinity purification (AP). For instance, an IAM-alkyne (IA) probe was designed and used to determine –SH reactivity in mouse cell cultures (Weerapana et al., 2010) and later in human mitochondrial and endoplasmic reticulum subcellular fractions (Bak et al., 2017; Bechtel et al., 2020). By performing low and high IA probe labeling, hyperreactive –SHs are identified as those that saturate labeling at low probe concentration. Similarly, another alkyne-functionalized IAM probe, 2-iodo-N-(prop-2-yn-1-yl)acetamide (IPM) was applied for quantitative thiol reactivity profiling (QTRP) in human (Fu et al., 2017, 2020; Figure 2). IPM-labeled Cys from control and H2O2-treated cells were differentially conjugated with light or heavy isotope-labeled, UV-cleavable azido biotin (Az-UV-biotin). Subsequently, biotin-tagged peptides were captured by streptavidin and photoreleased for quantitative MS analysis of –SH levels.
Other reactive warheads have been developed and used for in situ –SH labeling in HeLa cells. For instance, alkyne or desthiobiotin-conjugated heteroaromatic sulfone probes, reacting with –SHs via nucleophilic aromatic substitutions, had an increased labeling rate when compared with IAM-based probes (Motiwala et al., 2020). Alternatively, trimethylsilyl-ethynylbenziodoxolone compounds can introduce acetylene groups to –SHs through ethynylation, resulting in thioalkynes that can be directly used for CuAAC (Tessier et al., 2020). Thus far, no in situ labeling of –SH has been done for quantitative analysis in plants.
In situ –SOH labeling with carbon nucleophiles
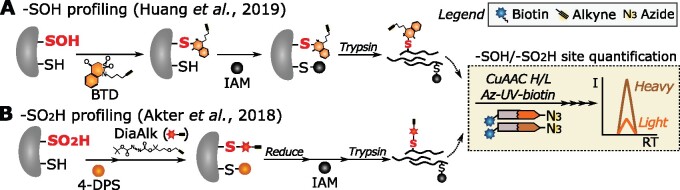
Almost 50 years ago, the cyclic diketone dimedone had been described for selective –SOH targeting (Benitez and Allison, 1974). The carbon-based nucleophilic chemistry of dimedone instigated the design of several optimized –SOH probes (Huang et al., 2018b; Shi and Carroll, 2020). A dimedone-based probe with alkyne handle, designated 4-(pent-4-yn-1-yl)cyclohexane-1,3-dione (DYn-2), was initially used to identify S-sulfenylated proteins in human cells (Paulsen et al., 2012), and later in Arabidopsis (Arabidopsis thaliana) cells (Akter et al., 2015). Meanwhile, isotope-labeled DYn-2 probes were used for –SOH site identification and quantification after enrichment of DYn-2-modified peptides (Yang et al., 2014). More recently, various carbon nucleophiles with different reactivities toward –SOH have been developed and tested in human cells (Gupta et al., 2017). Among these probes, benzo[c][1,2]thiazine-based (BTD) shows superior reactivity to –SOH and has been used for in situ identification and quantification of –SOH sites by conjugating Az-UV-biotin via CuAAC in human (Akter et al., 2018; Fu et al., 2019a) and Arabidopsis cell cultures (Huang et al., 2019; Figure 3). Compared with the previous Arabidopsis study identifying hundreds of S-sulfenylated proteins (Akter et al., 2015), the use of the BTD-based chemoproteomic workflow detected and quantified more than a thousand –SOH sites (Huang et al., 2019). The site-specific quantification under conditions of interest is a tremendous advance in plant –SOH profiling.
Figure 3.
Chemoproteomic strategies for –SOH and –SO2H redoxome profiling. Samples are purified and quantified by conjugation of isotopically labeled Az-UV-biotin tags similarly as for the QTRP method (tan box, more details see Figure 2). A, Schematic workflow for –SOH profiling (Huang et al., 2019). –SOH sites (red bold font) in mock- and H2O2-treated cells are labeled with BTD in Arabidopsis cell culture. The extracted proteomes then undergo a reduction-alkylation step and trypsin digestion. The peptides from mock- and H2O2-treated samples are differentially conjugated to light (orange) or heavy (brown) isotope-labeled Az-UV-biotin for quantification. B, Schematic workflow for –SO2H profiling (Akter et al., 2018). After proteome extraction, free thiols are blocked with 4-DPS and subsequently –SO2H sites (red bold font) are labeled by DiaAlk probe. After a reduction–alkylation step and trypsin digestion, DiaAlk-modified peptides are differentially conjugated with light (orange) or heavy (brown) isotope-labeled Az-UV-biotin for relative quantification between two conditions. I, intensity; RT, retention time; –SOH, sulfenic acid; –SO2H, sulfinic acid.
In addition to carbon nucleophilic probes, –SOH can also react with electrophilic probes containing strained alkene or alkyne groups. Initially, bicyclo[6.1.0]nonyne-based probes were used in human cells (Poole et al., 2014; McGarry et al., 2016) and, more recently, trans-cycloocten-5-ol (Scinto et al., 2019) and norbornene (Alcock et al., 2019, 2020). Whereas this alternative class of electrophilic probes offers a complementary view, some concerns have been raised about their possible cross-reactivity with –SH or –SSH (Shi and Carroll, 2020).
Electrophilic nitrogen species for –SO2H profiling
Thus far, the chemical probes used to selectively study –SO2H are mainly based on electrophilic nitrogen compounds that can form stable sulfonamide adducts with –SO2H. First, an aryl-nitroso probe coupled to biotin (NO-Bio) was utilized for –SO2H protein labeling in human cell lysates (Lo Conte and Carroll, 2012; Lo Conte et al., 2015) and, subsequently, biotin–S-nitrosoglutathione (GSNO; Majmudar et al., 2016). Lately, an electrophilic diazene alkyne (DiaAlk) probe was applied with superior –SO2H sensitivity (Akter et al., 2018; Figure 3). Here, after blocking –SH with 4,4 ’-dipyridyldisulfide (4-DPS), DiaAlk was used to label –SO2H in protein lysates, followed by conjugation of Az-UV-biotin via CuAAC. DiaAlk-modified peptides from samples treated with low and high doses of H2O2 were subsequently enriched and quantified (Figure 3). Using BTD probes in parallel for –SOH profiling, both the –SOH and –SO2H levels were measured, providing insight into the dynamic exchange between S-sulfenylation and S-sulfinylation. Furthermore, measurement of the turnover of the “S-sulfinome” in sulfiredoxin-deficient mutants hinted at more than 55 new sulfiredoxin substrates in mouse cells (Akter et al., 2018). Two of these substrates, PTPN12 and DJ-1, have been further confirmed biochemically to be reduced by sulfiredoxin. Thus far, no –SO2H profiling has been conducted in plants, but adoption of –SO2H chemical probes could help discover biological functions of S-sulfinylation and possibly expand the sulfiredoxin substrate repertoire in plants, currently restricted to 2-Cys peroxiredoxins and peroxiredoxin IIF (Iglesias-Baena et al., 2010, 2011).
Diverse strategies for –SSH detection
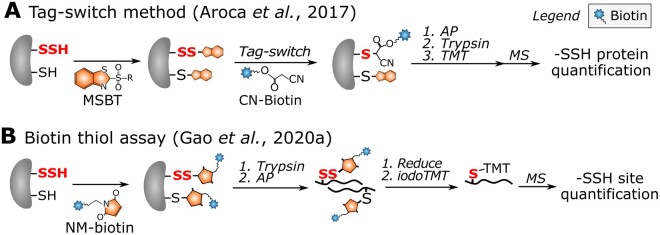
In the past decade, various chemoproteomic strategies have emerged to study protein S-sulfhydration. Discriminative labeling of –SSH and –SH is extremely challenging due to their similar reactivity toward –SH reagents (Pan and Carroll, 2013). Therefore, selective labeling of –SSH is typically indirect. Methyl methanethiosulfonate (MMTS) had been thought to specifically block –SH, whereafter –SSH would be targeted and enriched with pyridylthiol-biotin (biotin-HPDP; Mustafa et al., 2009). This method has been applied in Arabidopsis leaf extracts, and identified 106 S-sulfhydrated proteins (Aroca et al., 2015). However, –SH and –SSH were found to have a similar reactivity toward MMTS, questioning the aforementioned method (Pan and Carroll, 2013). In a tag-switch method, both –SH and –SSH were blocked by methylsulfonyl benzothiazole (MSBT), whereafter cyanoacetate biotin (CN-biotin) selectively reacted with MSBT disulfide adducts (Zhang et al., 2014). Given the shortcomings of using MMTS (Aroca et al., 2015), the tag-switch method was adopted and identified 2,015 S-sulfyhydrated proteins in Arabidopsis, which is a substantial increase compared with the 106 proteins identified previously (Aroca et al., 2017; Figure 4). In an adapted tag-switch method, –SSH, –SH, and –SOH first react with 4-chloro-7-nitrobenzofurazan (NBF-Cl) and dimedone-based probes, then selectively switch with –SS–NBF disulfides to label –SSH (Zivanovic et al., 2019). Besides tag-switch methods, –SH labeling reagents have been used to detect –SSH. For instance, IPM probes were used in human cells for –SSH level quantification after NaHS treatment by a low pH QTRP method (Fu et al., 2019b). The pKa of –SSH is 4.3, which is ∼4 units lower than that of –SH (pKa = 8.29); thus, –SSH maintains a relatively higher reactivity at a pH of 5 than the protonated –SH. Alternatively, isotope-coded affinity tag (ICAT) reagents (see Box 2) have been used to label both –SH and –SSH, in which the ICAT-tagged –SSH sites (+32 Da compared with tagged –SH) can be further distinguished and quantified (Wu et al., 2019). Additionally, in a biotin thiol assay (BTA) method, –SSH and –SH were first labeled with maleimide-polyethylenegylcol (PEG)2-biotin (NM-biotin) and trapped on the avidin column, whereafter only tagged –SSHs could be reduced by dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP). This approach was initially used for the identification of S-sulfhydrated proteins in mouse cells (Gao et al., 2015), and has recently been improved to quantify –SSH sites in human cells using iodoacetyl isobaric tandem mass tag (iodoTMT) reagents (see Box 2 and Figure 1 Box 2; Gao et al., 2020a; Figure 4). Similarly, –SSHs in human and yeast cells were detected with IAM-PEG2-biotin (Doka et al., 2016; Longen et al., 2016). However, the specificity of IAM-PEG2-biotin toward –SSHs has recently been questioned (Fan et al., 2020). Taken together, although plant –SSH studies are currently restricted to protein identification, given the availability of diverse methods established in mammals, we believe that sensitive quantification of –SSH sites will soon be achieved in plants.
Figure 4.
Redoxome workflows for –SSH profiling. A, Tag-switch method for –SSH protein quantitation by means of TMT isobaric reagents in Arabidopsis (Aroca et al., 2017). After protein extraction, both –SHs and –SSHs are blocked by MSBT. The –SSHs (red bold font) are further selectively trapped by incubating with CN-biotin, because solely MSBT disulfide adducts react with CN-biotin. Proteomes are digested with trypsin and CN-biotin-tagged peptides are enriched for identification of –SSH proteins. B, BTA method for quantification of –SSH with iodoTMT-labeling agents (Gao et al., 2020a). After protein extraction, both –SHs and –SSHs are blocked with NM-biotin. Proteomes are digested with trypsin and NM-biotin-tagged peptides are captured on an avidin column. The NM-biotin-tagged –SSHs are specifically eluted by DTT or TCEP through reduction. The newly generated –SHs are modified by 6-plex iodoTMT reagents for –SSH site quantification in mock- or NaHS-treated cells. –SSH, persulfide.
Box 2.
Reagents used in quantitative redox proteomics
Isobaric reagents for general protein quantification: TMT and iTRAQ
Isobaric labeling is a mass spectrometric strategy widely adopted for quantitative proteomics. Most commonly used are the amine-reactive (peptide N-terminus, Lys side chain) iTRAQ and TMT reagents (Pappireddi et al., 2019). These reagents have an identical mass, but their structures consist of differently distributed heavy and light isotopes. Upon tandem MS fragmentation, a linker is cleaved at a specific location, yielding characteristic reporter ion masses whose intensities can be used for multiplexed quantification (Pappireddi et al., 2019).
iodoTMT and ICAT quantitative thiol reagents
Of particular interest to redoxome profiling are the isobaric reagents that had been coupled with IAM for quantification of –SH peptides. iodoTMT reagents are IAM-functionalized isobaric mass tags that allow 6-plex quantification (see Figure 1 Box 2). Alternatively, ICAT reagents couple IAM to biotin through a cleavable light or heavy isotope linker for pairwise quantification of –SH levels (see Figure 1 Box 2).
In vivo metabolic labeling: SILAC
Besides quantitative labeling, heavy nonradioactive isotopic amino acids can be incorporated in vivo by SILAC. Heavy amino acids are incubated with cell cultures to enable quantitative peptide comparison. Typically, heavy Arg and Lys are used because they precede trypsin cleavage sites and constitute the C-terminal amino acid of tryptic peptides. SILAC labeling approaches are not widely used in plants, because plants are autotrophic organisms that synthesize all amino acids and, hence, challenge the efficient incorporation of heavy amino acids. In addition, the growth of green algae has been shown to be affected by the application of high concentrations of heavy amino acids (Andriukonis and Gorokhova, 2017).
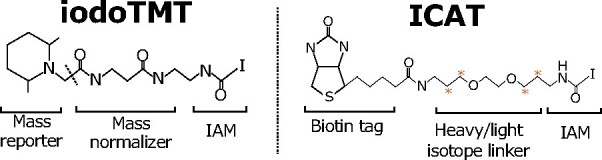
Figure 1 Box 2 Chemical structures of iodoTMT and ICAT. Orange asterisks indicate atoms carrying isotopic labels.
Phosphine-based probes for –SNO capture
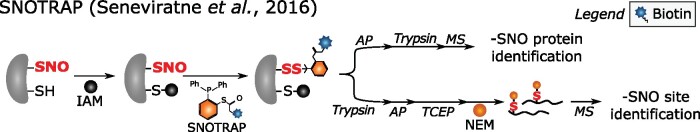
Initially, direct reaction of –SNO with mercury (Hg; Doulias et al., 2010, 2013) and gold (Au) centers (Faccenda et al., 2010) had been used for –SNO detection. Alternatively, based on the cross-reactivity of –SNO with –SO2H, a biotin-SO2H probe was applied to capture and identify –SNO proteins and sites in human cells (Majmudar et al., 2016). However, the most recently developed chemical probes are based on the reaction of –SNO with phosphine compounds to generate reducible disulfide adducts (Zhang et al., 2010). For instance, a phosphine-biotin probe, designated SNOTRAP, has been used to profile –SNO proteins and sites in mouse cell lysates (Seneviratne et al., 2016; Figure 5). Here, bulky biotin moieties were replaced with NEM after TCEP reduction to facilitate –SNO site identification. More recently, another phosphine-based probe, termed PBZyn, containing an o-phosphino-benzoyl warhead and an alkyne handle, has been developed and applied in human cells for in situ –SNO trapping (Clements et al., 2020). As reporter moieties are attached via disulfides that can be reduced or exchanged, additional care has to be taken to preserve these disulfides during processing. Despite the extended possibilities in direct –SNO profiling, –SNO is still detected exclusively in an indirect manner thus far (see below).
Figure 5.
Schematic representation of –SNO capture with the SNOTRAP probe (Seneviratne et al., 2016). Free thiols are alkylated and –SNO sites (red bold font) are selectively tagged with SNOTRAP. Two enrichment strategies aiming at the identification of –SNO proteins or sites are performed. For –SNO protein identification, modified proteins are captured by streptavidin beads, the enriched –SNO proteins are digested by trypsin, and peptides are subjected to MS analysis. For –SNO site identification, streptavidin capture is done after trypsin digestion at the peptide level, and the disulfide-linked SNOTRAP moiety is exchanged with NEM after TCEP reduction. Identification of NEM-modified peptides then indirectly informs on –SNO sites. –SNO, S-nitrosothiol.
Incorporation of functional GSH analogs for –SSG capture
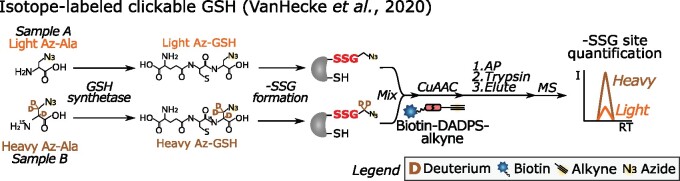
S-Glutathionylated proteins have been identified in Arabidopsis cell suspension cultures by incubating with either a biotinylated GSH homodimer (Dixon et al., 2005) or a GSH ethyl ester (Ito et al., 2003), but such biotinylated GSH analogs probably interfere with enzymatic (de)S-glutathionylation and alter the endogenous redox state (Samarasinghe et al., 2014). This drawback was overcome in a mammalian study in which the GSH synthetase was mutagenized for efficient incorporation of azido-alanine (Az-Ala) into GSH, thereby allowing a click reaction and AP of –SSG (Samarasinghe et al., 2014, 2016). More recently, a heavy isotope-labeled Az-Ala was used for a –SSG quantitative comparison (VanHecke et al., 2020; Figure 6). For similar systematic –SSG site detection and quantification in plants, a first prerequisite would be the efficient incorporation of Az-Ala by plant GSH synthetases.
Figure 6.
Metabolic incorporation of isotopically labeled Az-Ala in GSH for site-specific quantification of –SSG (VanHecke et al., 2020). Light (orange) or heavy (brown) Az-Ala is administered to a GSH synthetase mutant cell line to metabolically produce Az-GSH. After protein extraction of mock- or H2O2-treated cells, the isotopically labeled Az –SSG sites (red bold font) are conjugated with biotin-dialkoxydiphenylsilane (DADPS)-alkyne, where the DADPS is acid sensitive. After streptavidin capture and on-bead trypsin digestion, isotope-labeled –SSG peptides are released by acidic cleavage of DADPS. MS analysis not only identifies the –SSG sites but also determines the relative –SSG site levels between samples by comparison of light- and heavy-labeled –SSG peptide intensities. I, intensity; –SSG, S-glutathione; RT, retention time.
Reductomic approaches for reversible Cys OxiPTM profiling
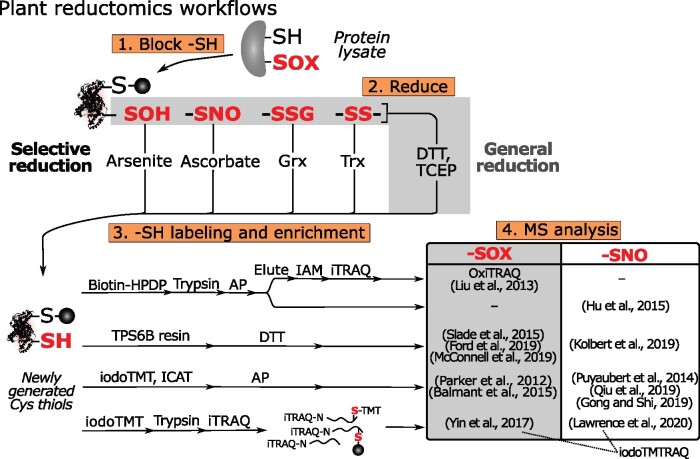
Reductomic workflows have been widely used to identify and quantify reversible Cys OxiPTMs (–SOXs), including –SOH, –SNO, and –SSH/SSG/SSR. In contrast to labeling by chemical probes, reductomic approaches are indirect, because they reduce –SOXs to –SHs. Typically, these workflows consist of four main steps: (1) blocking native –SHs via alkylation; (2) reduction of –SOXs to new –SHs; (3) labeling and enrichment of the newly generated –SHs; and (4) quantitative MS analysis (Figure 7). The reduction step can proceed either indiscriminately to target all –SOXs, typically by TCEP or DTT, or selectively to target a specific OxiPTM. The quantification of newly generated –SHs provides an indirect read-out of the initial OxiPTM(s). To this end, quantitative reagents (see Box 2) are often used for multiplexed –SH quantification (Pappireddi et al., 2019). Below, we review the reductomic approaches used in plants and highlight recently developed enrichment or quantification methods.
Figure 7.
Simplified scheme for reversible Cys OxiPTMs profiling approaches. Four main steps are outlined (orange boxes). After protein extraction, free –SHs are blocked and –SOXs (red bold font) are reduced indiscriminately (gray box) or specifically. Newly generated –SHs are then labeled, enriched, and quantified by various approaches. Plant studies are organized in a table, distinguishing studies analyzing –SOX (DTT/TCEP reduction, gray box) and –SNO (ascorbate reduction, white box). iodoTMTRAQ, combined iodoTMT and iTRAQ labeling; –SNO, S-nitrosothiol; –SOH, sulfenic acid; –SOX, reversible Cys OxiPTM; –SS–, disulfide.
Quantification of reversible Cys OxiPTMs in plants
Many plant studies have quantified –SOXs after indiscriminate reduction (Figure 7). OxiTRAQ is among the first site-specific quantitative plant redoxome methods (Liu et al., 2014, 2015). After initial Cys blocking, –SOXs are reduced by DTT and the newly generated –SHs are enriched with biotin-HPDP, followed by quantification across samples by means of isobaric tags for relative and absolute quantitation (iTRAQ) reagents (Figure 7; see Box 2). The more recent workflows typically require fewer processing steps. For instance, newly generated –SHs are captured directly via a disulfide exchange reaction on a thiol-affinity resin, thiopropyl sepharose 6B (TPS6B), enriching –SOX peptides after on-bead proteolysis and DTT/TCEP elution (Figure 7; Guo et al., 2014). This workflow has been applied in Arabidopsis and the green algae Chlamydomonas (Chlamydomonas reinhardtii), followed by label-free quantification of –SOXs (Slade et al., 2015; Ford et al., 2019; McConnell et al., 2019). Alternatively, iodoTMT reagents have been used for labeling and multiplexed quantification of –SOXs during Pseudomonas syringae infection in tomato (Solanum lycopersicum; Parker et al., 2012; Balmant et al., 2015).
Toward the absolute –SOX Cys occupancy
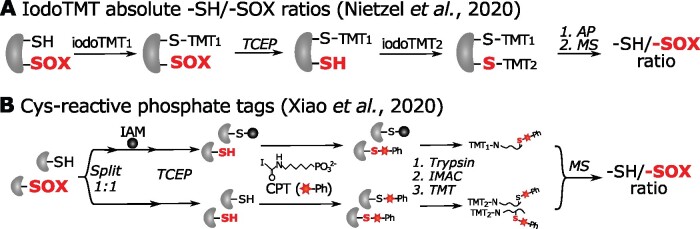
Whereas the abovementioned workflows provide relative –SOX levels across samples, other methods aim at the quantification of absolute –SOX levels. Determination of absolute occupancy is particularly interesting when conditions are anticipated to alter the cellular protein levels, such as prolonged stress treatments. For instance, both iTRAQ and stable isotope labeling with amino acids in cell culture (SILAC, see Box 2) have been combined with iodoTMT labeling of –SOXs (Parker et al., 2015; Vajrychova et al., 2019) to quantify both –SOX and protein levels in mammals. While SILAC has not been applied widely in plants and has been found to affect the growth of green algae (Andriukonis and Gorokhova, 2017), the consecutive iTRAQ and iodoTMT labeling, designated iodoTMTRAQ, has been applied in Arabidopsis to study –SOX (Yin et al., 2017) and –SNO (Lawrence et al., 2020; Figure 7). Alternatively, absolute –SOX and –SH ratios for Cys residues can be obtained by distinct –SH quantitative labeling before and after the reducing step. This rationale had initially been described in the OxICAT method (Leichert et al., 2008) that uses ICAT reagents (see Box 2). Similarly, absolute –SH/–SOX ratios were estimated by means of iodoTMT labels for the study of mitochondrial proteins during Arabidopsis seed germination (Nietzel et al., 2020; Figure 8). In mammals, isotope-labeled IA probes have been used as well for this purpose (Abo et al., 2018; Bechtel et al., 2020). Recently, phosphate-coupled IAM probes, designated cysteine-reactive phosphate tags (CPTs), have been utilized to label and quantify –SH and –SOX with TMT isobaric reagents (see Box 2) in mouse tissue lysates (Xiao et al., 2020; Figure 8). Of interest, a more advanced method, designated OxSWATH, quantifies –SH, –SOX, and protein abundance simultaneously by data-independent acquisition MS (Anjo et al., 2019). In contrast to routine selection of abundant peptide precursors, in data-independent acquisition, all peptide precursors are fragmented and recorded in consecutive m/z windows, thereby greatly increasing reproducibility and detection of low-abundant peptides (Bruderer et al., 2015).
Figure 8.
Determination of absolute –SH/–SOX ratios. A, IodoTMT labeling method for measurement of absolute –SH/–SOX ratios in Arabidopsis mitochondria (Nietzel et al., 2020). Free –SH and –SOX (red bold font) are distinctly tagged by different iodoTMT reagents by labeling –SHs prior to and after TCEP reduction, respectively. Six-plex-TMT quantification was used to measure absolute –SH/–SOX ratios. B, Cys-reactive phosphate tags (CPTs) method for absolute –SH/–SOX ratio quantification (Xiao et al., 2020). Protein extracts are divided into two fractions for labeling both –SHs and –SOXs (without IAM blocking before TCEP) and solely –SOXs (with IAM blocking before TCEP). Subsequently, immobilized metal affinity chromatography (IMAC) is used to capture phosphate tags, and phosphorylation is removed enzymatically. CPT-modified peptides in both fractions are differentially labeled with TMT. After the fractions have been mixed, a quantitative TMT analysis can determine the absolute –SH/–SOX ratios by MS. –SOX, reversible Cys OxiPTM.
Selective reduction of different Cys OxiPTMs
Chemicals and enzymes, such as arsenite, ascorbate, Trxs, and Grxs, have been used to selectively reduce –SOH, –SNO, –SSR, and –SSG, respectively (Figure 7). Thus far, arsenite and Grx-based reduction have been scarcely utilized for the systematic detection of –SOH (Saurin et al., 2004; Wojdyla et al., 2015; Li et al., 2016) and –SSG (Duan et al., 2016), respectively. Below, we describe in more detail methods used for –SNO and –SSR detection.
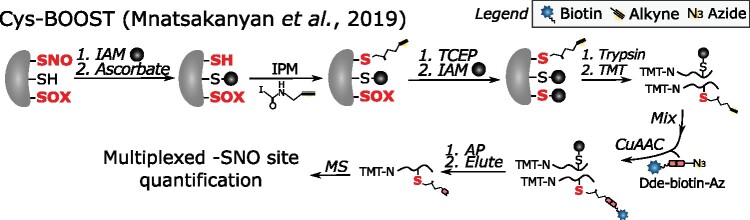
In plants, –SNO has been extensively studied by means of selective ascorbate reduction. Nearly two decades ago, this method was introduced as the biotin switch method (BSM) and involved three steps: (1) blocking of native –SHs by MMTS; (2) reduction of –SNOs by ascorbate; and (3) labeling of the newly generated –SHs with biotin-HPDP for enrichment of the labeled proteins (Jaffrey et al., 2001). This “classical” BSM has been used in various plant species, such as Arabidopsis (Lindermayr et al., 2005; Chaki et al., 2015), gray poplar (Populus × canescens; Vanzo et al., 2016), cucumber (Cucumis sativus; Niu et al., 2019), and sunflower (Helianthus annuus; Jain et al., 2018). Meanwhile, modified BSM workflows have been applied in plants. For instance, biotin-based enrichment was done at the peptide level to identify –SNO sites in wild-type Arabidopsis plants and mutants lacking S-nitrosoglutathione reductase (Hu et al., 2015). Alternatively, S-nitrosylated proteins were captured in Arabidopsis by means of the thiol-affinity resin TPS6B (Kolbert et al., 2019). Furthermore, –SNO sites were quantified in Arabidopsis (Puyaubert et al., 2014), tomato (Gong and Shi, 2019), and tea (Camellia sinensis) plants (Qiu et al., 2019) by ICAT or iodoTMT reagents. Of interest, a more advanced workflow, designated Cys-BOOST (Figure 9), has been applied for –SNO quantification in HeLa cells (Mnatsakanyan et al., 2019). Here, clickable IPM was conjugated to cleavable biotin-azide reagents for AP, and TMT isobaric reagents were used for multiplexed quantification. This approach nearly tripled the number of identified –SNO sites with a superior quantification reproducibility compared with an iodoTMT quantification workflow (Mnatsakanyan et al., 2019).
Figure 9.
Cys-BOOST method for –SNO site quantification with TMT reagents and bioorthogonal cleavable linker-based enrichment (Mnatsakanyan et al., 2019). After free thiol blocking, –SNO sites (red bold font) are selectively reduced by ascorbate and tagged with IPM. Afterward, other reversible Cys OxiPTMs (–SOXs, red bold font) are blocked with a TCEP reduction step. After trypsin digestion, samples are labeled with distinct isobaric TMT reagents (see Box 2) and pooled. Subsequently, biotin azide containing a hydrazine-cleavable Dde bond (Dde-biotin-Az) linkers are conjugated to IPM-modified Cys for streptavidin capture and elution of –SNO peptides by breaking the Dde bond with hydrazine. After a pH fractionation step, multiplexed quantitative –SNO sites are analyzed by MS. –SNO, S-nitrosothiol; –SOX, reversible Cys OxiPTM.
Besides reducing chemicals, recombinant Trxs have been applied in reductomic approaches to study –SSR. The newly generated Trx-reduced –SHs can be quantified to reveal putative Trx substrates. For instance, recombinant Trx-h was added to protein extracts of rapeseed (Brassica napus) and newly generated –SHs were quantified with cysTMT reagents (Zhang et al., 2016). In Chlamydomonas, extracted proteomes were incubated with a Trx reduction system consisting of NADPH, Arabidopsis NAPDH-dependent Trx reductase-B (NTRB), and cytosolic Chlamydomonas Trx-h1 (Pérez-Pérez et al., 2017), and quantified by means of ICAT reagents. Although these indirect methods can hint at putative Trx substrates, substrates can also be identified with monocysteinic Trx/Grx disulfide traps (see below).
Disulfide-based proteinaceous traps for –SOH, –SSR, and –SSG profiling
In addition to chemoproteomic and reductomic approaches, disulfide-based traps are an alternative strategy to capture –SOH, –SSR, and –SSG. They are based on the formation of intermolecular (or mixed) disulfide bonds between a target Cys OxiPTM and a proteinaceous thiol trap. Such thiol traps can either be recombinantly produced for substrate trapping in protein lysates or be genetically encoded to trap OxiPTMs in situ.
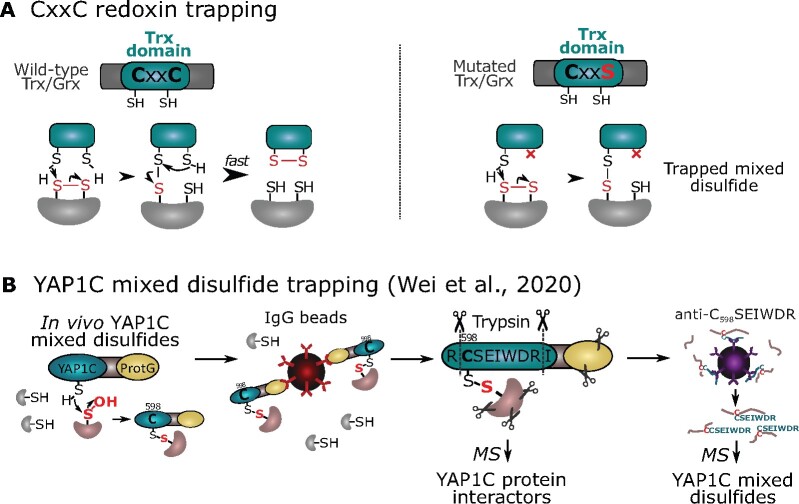
Trapping by monocysteinic redoxin mutants
Trxs and Grxs contain an active CxxC motif in which the first nucleophilic Cys forms transiently mixed disulfides with oxidized substrates (–SSR/–SSG from other proteins) that are quickly resolved by the second Cys to release reduced substrates (Figure 10). In turn, the oxidized Trxs are reduced by NAPDH-dependent Trx reductase (NTR) or ferredoxin-thioredoxin reductase, whereas oxidized Grxs are reduced by GSH. To trap substrates of CxxC redoxins, the second, resolving Cys is typically mutagenized to Ala/Ser to stabilize the mixed disulfides with oxidized substrates (Figure 10). Recombinant monocysteinic redoxins can be loaded on affinity columns to trap putative interactors in proteome extracts. Such a strategy has been used for Arabidopsis NUCLEOREDOXIN1 (Kneeshaw et al., 2017), Trx-f1 (Yoshida and Hisabori, 2016), Trx-m (Da et al., 2017, 2018), the atypical TrxL2 (YoshiDa et al., 2018), and Chlamydomonas Trx-h1 (Pérez-Pérez et al., 2017). For the Chlamydomonas Trx-h1, a reductomic approach has been used in parallel with incubation of the Trx-reducing system (Trx-h1, NTR, and NAPDH) and identified 394 out of 980 proteins (40.2%) that were trapped by the Trx-h1 monocysteinic mutant (Pérez-Pérez et al., 2017).
Figure 10.
Mixed disulfide-based traps. A, Disulfide exchange for wild-type (CxxC) redoxins (left) or trapped mixed disulfides in mutated (typically CxxS or CxxA) redoxins (right). B, Two-step enrichment of Yap1-based proteinaceous –SOH probe (YAP1C) Cys598 mixed disulfides (Wei et al., 2020). After IgG-based enrichment for YAP1C interacting proteins, the C598SEIWDR disulfide-linked peptides are enriched by a specific antibody and identified by MS. ProtG, protein G.
Besides Trxs, other redoxins have been studied in plants. For instance, substrates of the Arabidopsis NADPH-Trx reductase C (NTRC), an enzyme containing both a Trx domain and an NTR domain and of which the resolving Cys of both domains had been mutated for separate trapping, identified shared targets, such as Trx-z (Yoshida and Hisabori, 2016). In another NTRC trapping study, transgenic Arabidopsis plants expressing functional NTRC (nonmutagenized) coupled to a tandem AP (TAP) tag were used (González et al., 2019). The TAP-MS revealed 50 putative, including known, substrates, suggesting that general protein interaction methods with nonmutagenized redoxins could be used for substrate detection.
YAP1C disulfide trapping for in vivo –SOH detection
A genetically encoded yeast AP-1-like (Yap1) Cys598-based probe used to systematically trap –SOH in Escherichia coli and yeast (Takanishi et al., 2007; Takanishi and Wood, 2011) had later been introduced into Medicago truncatula (Oger et al., 2012). In Arabidopsis, a Yap1-based probe fused to a TAP tag, designated YAP1C, allowed the identification of hundreds of S-sulfenylated proteins in cell cultures (Waszczak et al., 2014). More recently, YAP1C trapping was directed to chloroplasts by the incorporation of a N-terminal transit peptide, entrapping 132 S-sulfenylated plastidial proteins (De Smet et al., 2019). A more advanced proteomic workflow now enables the identification of YAP1C-resolved Cys sites (Wei et al., 2020; Figure 10). Here, YAP1C interactors are first purified from the YAP1C-TAP-overexpressing Arabidopsis cells. Trypsin digestion results in disulfide-linked peptides between the YAP1C-derived C598SEIWDR peptide, and the resolved –SOH peptides are enriched by a generated C598SEIWDR antibody. The MS analysis after the purification steps of both the protein and peptide levels provides complementary information on the YAP1C protein interactors and their cross-linked sites. Taken together, YAP1C site-specific –SOH detection by enriching and identifying disulfides paves the way for future applications of disulfide traps for in situ discovery of OxiPTMs.
Current challenges and future perspectives
With the fast development of redox proteomic techniques, plant redoxome profiling has advanced substantially. Instead of reiterating these achievements here, it is worth mentioning the current limitations and challenges in redoxome profiling (see the “Outstanding questions”).
Underexplored Cys OxiPTM sites in plants
Several advanced techniques established in mammalian research still await introduction into plants for the thorough profiling of OxiPTMs. For instance, thus far, –SSH and –SSG proteins have been identified in Arabidopsis, but not at a site-specific level. Whereas –SNO has been extensively studied in plants by indirect ascorbate reduction, direct in situ labeling by chemical probes has not yet been implemented. Furthermore, other OxiPTMs, such as –SO2H, have never been studied on a global scale in plants. Adoption of contemporary profiling strategies for still underexplored OxiPTMs in plants will prove a promising future endeavor.
Selection of an optimal redoxome workflow
Diverse methodologies are available for Cys redoxome profiling, each of them with advantages and disadvantages (Table 1). Suitable workflows should be chosen to confidently address the biological question at hand. Ideally, OxiPTM sites are studied in situ within their physiological cellular environment. By means of chemoproteomics strategies, cell-permeable probes enable the direct detection and quantification of OxiPTMs sites in situ. Meanwhile, chemical probes containing an extra moiety for subcellular targeting are being developed, such as mitochondria-directed –SOH probes (Li et al., 2019; Gao et al., 2020b). However, chemical probes are mostly used in cell cultures and their application on plant seedlings under physiological conditions is extremely challenging. Besides the limited permeability within rigid plant tissues, possible side-reactions to undesired biomolecules or instability of chemical probes can additionally restrict their usage in plants. Thus, development of more suitable chemical probes for plant research would be greatly beneficial.
Table 1.
Comparison of current techniques for Cys OxiPTM profiling
| OxiPTM approaches | Advantages | Disadvantages |
|---|---|---|
| Chemoproteomics |
|
|
| Reductomics |
|
|
| Protein disulfide traps |
|
|
Alternatively, OxiPTMs can be labeled in situ by means of genetically encoded disulfide traps that benefit from the flexibility of the genetic construct. For example, specific subcellular targeting can be achieved by incorporation of signal peptides (De Smet et al., 2019). Although subcellular redoxome studies have also been carried out in plant nuclei (Chaki et al., 2015) and mitochondria (Nietzel et al., 2020) fractionation, endogenously expressed disulfide traps are a better option in terms of physiological relevance, because cell lysis will immediately change the redox environment of proteins. Nevertheless, generation of transgenic plants expressing disulfide traps is time-consuming and it is important to ensure that their expression does not affect cellular homeostasis and plant development. Although site-specific identification of YAP1C-trapped Cys was achieved recently (Wei et al., 2020), quantification of these sites remains a future challenge.
Redoxome trapping in protein lysates might lead to a different result than targeting inside the cell, especially when labile OxiPTMs (–SOH, –SNO, and –SSH/G/R) are studied that are susceptible to overoxidation, reduction, or shuffling. However, some approaches, such as reductomic strategies, are restricted to protein lysates. To minimalize artifactual oxidation events during sample preparations, the post-lysis processing time should be minimized, and the protein extraction buffer should be optimized by means of oxygen depletion and inclusion of ROS scavengers (such as catalase) and –SH blocking reagents. This is particularly challenging in plant studies, because rigid plant tissues typically necessitate extensive homogenization procedures. Nevertherless, reductomics approaches enable the direct usage of isobaric reagents for multiplexed sample quantification. Whereas until now chemoproteomic approaches have been mostly restricted to pairwise comparisons with heavy/light-labeled reagents, a possible combination with peptide isobaric (iTRAQ or TMT) labeling could be considered for multiplexed quantification. Overall, the diverse pros and cons of each approach have to be weighed to select an appropriate redoxome workflow (Table 1).
Specificity of thiol reagents and Cys OxiPTM reductants
Thiol blocking reagents that are essential for redoxome research have long been thought to be –SH selective. However, evidence has shown that several of the utilized thiol blocking reagents are not specific for –SH. For example, IAM, MMTS, and NEM have been reported to cross-react with –SOH (Reisz et al., 2013), and IAM and NEM with –SO2H (Lo Conte et al., 2015). Thus, caution needs to be taken when choosing –SH reagents, and more specific blocking agents should be considered. For instance, MSBT does not cross-react with –SOH, –SO2H, and –SNO (Shi and Carroll, 2020). Furthermore, development of real –SH-selective reagents would be greatly valuable. Similarly, the specificity of certain reductants relied on by selective reductomics has been questioned as well. For example, reduction by ascorbate and arsenite has been associated with OxiPTMs other than –SNO and –SOH, respectively (Wang and Xian, 2011; Gupta and Carroll, 2014; Anschau et al., 2020). Thus, each redoxome study has to be carefully scrutinized and interpreted in regard to the function of the respective Cys-labeling/reducing reactions.
Cys OxiPTMs (co-)occupancy
Advances in proteomic techniques and MS sensitivity result in increasing numbers of OxiPTM identifications. For instance, more than 1,500 –SOH sites (Huang et al., 2019) and 2,000 S-sulfhydrated proteins (Aroca et al., 2017) have been detected in Arabidopsis. In such extensive datasets, quantification of relative or absolute OxiPTM levels becomes increasingly important to discern the physiologically relevant OxiPTMs. Identified OxiPTMs modifying low percentages of a given Cys are probably less functionally relevant than Cys with an enhanced OxiPTM occupancy (Duan et al., 2017). For instance, analysis of the OxiPTM occupancy of 34,000 Cys in mouse (Xiao et al., 2020) allowed researchers to distinguish a minority of highly oxidized Cys sites. However, such Cys OxiPTM occupancy analysis is very limited in plants and remains a future challenge.
A complicating factor in OxiPTM quantification is that a Cys residue can be targeted by several OxiPTMs, as highlighted by the comparison of different OxiPTM redoxomes in plants (Aroca et al., 2017; Ford et al., 2019; Wei et al., 2020). Knowledge of the OxiPTM co-occupancy can help understand the biological function of different Cys OxiPTMs and their interplay in diverse signaling pathways. The simultaneous assessment of diverse OxiPTMs is a technical challenge, and redoxome workflows are often tailored for specific OxiPTM types. Nevertheless, in an extensive reductomic approach, –SH, –SNO, and –SOH have been quantified simultaneously through reduction by TCEP, ascorbate, and arsenite, respectively (Wojdyla et al., 2015). Additionally, benzothiazine (pBTD) and isotope-labeled halogenated benzothiazine (ipBTD) have been used for tagging –SOH and –SH/–SOX within samples, respectively (Albertolle et al., 2019). Combining the usage of different OxiPTM chemical probes could enable the study of OxiPTM co-occupancy systematically in the future.
The use of proteomic approaches to map Cys OxiPTMs is critical for fundamental redox biology. In plants, research on still underexplored OxiPTMs, profiling of OxiPTMs within cellular contexts, and quantification of absolute OxiPTM levels remains major challenges and will provide unique opportunities for the plant redox biology community. Given the ongoing advances in the Cys redoxome profiling, fascinating times await plant redox research.
Outstanding questions
Which Cys sites undergo –SSH, –SSG, or –SO2H modifications in plants?
Given the diverse redoxome profiling techniques, which is the optimal strategy to address the biological questions at hand?
Can OxiPTMs be monitored in real-time within plant cells under physiological conditions?
How can the physiological relevance of OxiPTMs on a given Cys be assessed in extensive datasets?
At which OxiPTM occupancy are protein functions affected?
To what extent are Cys modified by multiple OxiPTMs, and how can they be analyzed simultaneously?
How do Cys OxiPTMs interplay with each other and other PTMs?
Acknowledgments
The authors thank Martine De Cock for her kind support on the manuscript preparation.
Funding
This work was supported by the Research Foundation-Flanders (FWO) [Projects No. G006916N and G055416N to F.V.B.], FWO-Fonds de la Recherche Scientifique [Excellence of Science Project No. 30829584], and by the FWO Senior Postdoctoral fellowship [no. 1227020N to J.H.].
Conflict of interest statement. The authors declare no conflict of interest.
P.W., F.V.B., and J.H. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Jingjing Huang (Jingjing.Huang@psb.vib-ugent.be).
References
- Abo M, Li C, Weerapana E (2018) Isotopically-labeled iodoacetamide-alkyne probes for quantitative cysteine-reactivity profiling. Mol Pharm 15:743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, Carroll KS (2018) Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat Chem Biol 14:995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F. , et al. (2015) DYn-2 based identification of Arabidopsis sulfenomes. Mol Cell Proteomics 14:1183–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertolle ME, Glass SM, Trefts E, Guengerich FP (2019) Isotopic tagging of oxidized and reduced cysteines (iTORC) for detecting and quantifying sulfenic acids, disulfides, and free thiols in cells. J Biol Chem 294:6522–6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock LJ, Langini M, Stühler K, Remke M, Perkins MV, Bernardes GJL, Chalker JM (2020) Proteome-wide survey of cysteine oxidation by using a norbornene probe. ChemBioChem 21:1329–1334 [DOI] [PubMed] [Google Scholar]
- Alcock LJ, Oliveira BL, Deery MJ, Pukala TL, Perkins MV, Bernardes GJL, Chalker JM (2019) Norbornene probes for the detection of cysteine sulfenic acid in cells. ACS Chem Biol 14:594–598 [DOI] [PubMed] [Google Scholar]
- Alcock LJ, , Perkins MV, , Chalker JM ( 2018) Chemical methods for mapping cysteine oxidation. Chem Soc Rev 47: 231–268 [DOI] [PubMed] [Google Scholar]
- Andriukonis E, Gorokhova E (2017) Kinetic 15N-isotope effects on algal growth. Sci Rep 7: 44181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjo SI, Melo MN, Loureiro LR, Sabala L, Castanheira P, Grãos M, Manadas B (2019) oxSWATH: an integrative method for a comprehensive redox-centered analysis combined with a generic differential proteomics screening. Redox Biol 22:101130. [Corrigendum. Redox Biol 26: 101257] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anschau V, Ferrer-Sueta G, Aleixo-Silva RL, Bannitz Fernandes R, Tairum CA, Tonoli CCC, Murakami MT, de Oliveira MA, Netto LES (2020) Reduction of sulfenic acids by ascorbate in proteins, connecting thiol-dependent to alternative redox pathways. Free Radic Biol Med 156:207–216 [DOI] [PubMed] [Google Scholar]
- Aroca Á, Benito JM, Gotor C, Romero LC (2017) Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J Exp Bot 68:4915–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca Á, Gotor C, Romero LC (2018) Hydrogen sulfide signaling in plants: emerging roles of protein persulfidation. Front Plant Sci 9:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca Á, Serna A, Gotor C, Romero LC (2015) S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol 168:334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmant KM, Parker J, Yoo M-J, Zhu N, Dufresne C, Chen S (2015) Redox proteomics of tomato in response to Pseudomonas syringae infection. Hortic Res 2:15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak DW, Pizzagalli MD,, Weerapana E (2017) Identifying functional cysteine residues in the mitochondria. ACS Chem Biol 12:947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel TJ, Li C, Kisty EA, Maurais AJ, Weerapana E (2020) Profiling cysteine reactivity and oxidation in the endoplasmic reticulum. ACS Chem Biol 15:543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez LV, Allison WS (1974) The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. J Biol Chem 249:6234–6243 [PubMed] [Google Scholar]
- Biteau B, Labarre J, Toledano MB (2003) ATP-dependent reduction of cysteine–sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425:980–984 [DOI] [PubMed] [Google Scholar]
- Bruderer R, Bernhardt OM, Gandhi T, Miladinović SM, Cheng L-Y, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C. et al. (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics 14:1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Shekariesfahlan A, Ageeva A, Mengel A, von Toerne C, Durner J, Lindermayr C (2015) Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci 238:115–126 [DOI] [PubMed] [Google Scholar]
- Chiappetta G, Ndiaye S, Igbaria A, Kumar C, Vinh J, Toledano MB (2010) Proteome screens for Cys residues oxidation: the redoxome. In Cadenas E, Packer L, eds., Thiol Redox Transitions in Cell Signaling, Part A: Chemistry and Biochemistry of Low Molecular Weight and Protein Thiols, Vol. 473 (Methods in Enzymology). Academic Press, San Diego, pp. 199–216 [DOI] [PubMed] [Google Scholar]
- Clements JL, Pohl F, Muthupandi P, Rogers SC, Mao J, Doctor A, Birman VB, Held JM (2020) A clickable probe for versatile characterization of S-nitrosothiols. Redox Biol 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Q, Sun T, Wang M, Jin H, Li M, Feng D, Wang J, Wang H-B, Liu B (2018) M-type thioredoxins are involved in the xanthophyll cycle and proton motive force to alter NPQ under low-light conditions in Arabidopsis. Plant Cell Rep 37:279–291 [DOI] [PubMed] [Google Scholar]
- Da Q, Wang P, Wang M, Sun T, Jin H, Liu B, Wang J, Grimm B, Wang H-B (2017) Thioredoxin and NADPH-dependent thioredoxin reductase C regulation of tetrapyrrole biosynthesis. Plant Physiol 175:652–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet B, Willems P, Fernandez-Fernandez AD, Alseekh S, Fernie AR, Messens J, Van Breusegem F (2019) In vivo detection of protein cysteine sulfenylation in plastids. Plant J 97:765–778 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R (2005) Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol 138:2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, et al. (2016) A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci Adv 2:e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H (2010) Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A 107:16958–16963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias P-T, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H (2013) Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal 6:rs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gaffrey MJ, Qian W-J (2017) Quantitative proteomic characterization of redox-dependent post-translational modifications on protein cysteines. Mol Biosyst 13:816–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Kodali VK, Gaffrey MJ, Guo J, Chu RK, Camp DG, Smith RD, Thrall BD, Qian W-J (2016) Quantitative profiling of protein S-glutathionylation reveals redox-dependent regulation of macrophage function during nanoparticle-induced oxidative stress. ACS Nano 10:524–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccenda A, Bonham CA, Vacratsis PO, Zhang X, Mutus B (2010) Gold nanoparticle enrichment method for identifying S-nitrosylation and S-glutathionylation sites in proteins. J Am Chem Soc 132:11392–11394 [DOI] [PubMed] [Google Scholar]
- Fan K, Chen Z, Liu H (2020) Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity. Sci Adv 6:eabb6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MM, Smythers AL,, McConnell EW, Lowery SC, Kolling DRJ, Hicks LM (2019) Inhibition of TOR in Chlamydomonas reinhardtii leads to rapid cysteine oxidation reflecting sustained physiological changes. Cells 8:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Li Z, Liu K, Tian C, He J, He J, He F, Xu P, Yang J (2020) A quantitative thiol reactivity profiling platform to analyze redox and electrophile reactive cysteine proteomes. Nat Protoc 15: 2891–2919 [DOI] [PubMed] [Google Scholar]
- Fu L, Liu K, Ferreira RB, Carroll KS, Yang J (2019a) Proteome-wide analysis of cysteine S-sulfenylation using a benzothiazine-based probe. Curr Protoc Protein Sci 95:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Liu K, He J, Tian C, Yu X, Yang J (2019b) Direct proteomic mapping of cysteine persulfidation. Antioxid Redox Signal 33:1061–1076 [DOI] [PubMed] [Google Scholar]
- Fu L, Liu K, Sun M, Tian C, Sun R, Morales Betanzos C, Tallman KA, Porter NA, Yang Y, Guo D, et al. (2017) Systematic and quantitative assessment of hydrogen peroxide reactivity with cysteines across human proteomes. Mol Cell Proteomics 16:1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X-H, Krokowski D, Guan B-J, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, et al. (2015) Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife 4:e10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X-H, Li L, Parisien M, Wu J, Bederman I, Gao Z, Krokowski D, Chirieleison SM, Abbott D, Wang B, et al. (2020a) Discovery of a redox thiol switch: implications for cellular energy metabolism. Mol Cell Proteomics 19:852–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sun R, Zhao M, Ding J, Wang A, Ye S, Zhang Y, Mao Q, Xie W, Ma G, et al. (2020b) Sulfenic acid-mediated on-site-specific immobilization of mitochondrial-targeted NIR fluorescent probe for prolonged tumor imaging. Anal Chem 92:6977–6983 [DOI] [PubMed] [Google Scholar]
- Gong B, Shi Q (2019) Identifying S-nitrosylated proteins and unraveling S-nitrosoglutathione reductase-modulated sodic alkaline stress tolerance in Solanum lycopersicum L. Plant Physiol Biochem 142:84–93 [DOI] [PubMed] [Google Scholar]
- González M, Delgado-Requerey V, Ferrández J, Serna A, Cejudo FJ (2019) Insights into the function of NADPH thioredoxin reductase C (NTRC) based on identification of NTRC-interacting proteins in vivo. J Exp Bot 70:5787–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaffrey MJ, Su D, Liu T, Camp DG 2nd, Smith RD, Qian W-J (2014) Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat Protoc 9:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Carroll KS (2014) Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta 1840:847–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Yang J, Liebler DC, Carroll KS (2017) Diverse redoxome reactivity profiles of carbon nucleophiles. J Am Chem Soc 139:5588–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167:1731–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Niazi AK, Young D, Rosado LA, Vertommen D, Bodra N, Abdelgawwad MR, Vignols F, Wei B, Wahni K, et al. (2018a) Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis. J Exp Bot 69:3491–3505 [DOI] [PubMed] [Google Scholar]
- Huang J, Willems P, Van Breusegem F, Messens J (2018b) Pathways crossing mammalian and plant sulfenomic landscapes. Free Radic Biol Med 122:193–201 [DOI] [PubMed] [Google Scholar]
- Huang J, Willems P, Wei B, Tian C, Ferreira RB, Bodra N, Martínez Gache SA, Wahni K, Liu K, Vertommen D, et al. (2019) Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc Natl Acad Sci U S A 116:21256–21261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Baena I, Barranco-Medina S, Lazaro-Payo A, Lopez-Jaramillo FJ, Sevilla F, Lazaro JJ (2010) Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin. J Exp Bot 61: 1509–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Baena I, Barranco-Medina S, Sevilla F, Lazaro JJ (2011) The dual-targeted plant sulfiredoxin retroreduces the sulfinic form of atypical mitochondrial peroxiredoxin. Plant Physiol 155:944–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Iwabuchi M, Ogawa K (2003) The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol 44:655–660 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3:193–197 [DOI] [PubMed] [Google Scholar]
- Jain P, von Toerne C, Lindermayr C, Bhatla SC (2018) S-Nitrosylation/denitrosylation as a regulatory mechanism of salt stress sensing in sunflower seedlings. Physiol Plant 162:49–72 [DOI] [PubMed] [Google Scholar]
- Kneeshaw S, Keyani R, Delorme-Hinoux V, Imrie L, Loake GJ, Le Bihan T, Reichheld J-P, Spoel SH (2017) Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc Natl Acad Sci U S A 114:8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Molnár Á, Oláh D, Feigl G, Horváth E, Erdei L, Ördög A, Rudolf E, Barth T, Lindermayr C (2019) S-Nitrosothiol signaling is involved in regulating hydrogen peroxide metabolism of zinc-stressed Arabidopsis. Plant Cell Physiol 60:2449–2463 [DOI] [PubMed] [Google Scholar]
- Lawrence SR 2nd, Gaitens M, Guan Q, Dufresne C, Chen S (2020) S-Nitroso-proteome revealed in stomatal guard cell response to flg22. Int J Mol Sci 21:1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U (2008) Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105:8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Klockenbusch C, Lin L, Jiang H, Lin S, Kast J (2016) Quantitative protein sulfenic acid analysis identifies platelet releasate-induced activation of integrin β2 on monocytes via NADPH oxidase. J Proteome Res 15:4221–4233 [DOI] [PubMed] [Google Scholar]
- Li Z, Forshaw TE, Holmila RJ, Vance SA, Wu H, Poole LB, Furdui CM, King SB (2019) Triphenylphosphonium-derived protein sulfenic acid trapping agents: synthesis, reactivity, and effect on mitochondrial function. Chem Res Toxicol 32:526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhang H, Wang H, Xia Y (2014) Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics 14:750–762 [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang H, Yu B, Xiong L, Xia Y (2015) Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci Rep 5:8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte M, Carroll KS (2012) Chemoselective ligation of sulfinic acids with aryl-nitroso compounds. Angew Chem Int Ed Engl 51:6502–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte M, Lin J, Wilson MA, Carroll KS (2015) A chemical approach for the detection of protein sulfinylation. ACS Chem Biol 10:1825–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S, Richter F, Köhler Y, Wittig I, Beck K-F, Pfeilschifter J (2016) Quantitative persulfide site identification (qPerS-SID) reveals protein targets of H2S releasing donors in mammalian cells. Sci Rep 6:29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmudar JD, Konopko AM, Labby KJ, Tom CTMB, Crellin JE, Prakash A,, Martin BR (2016) Harnessing redox cross-reactivity to profile distinct cysteine modifications. J Am Chem Soc 138:1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell EW, Berg P, Westlake TJ, Wilson KM, Popescu GV, Hicks LM, Popescu SC (2019) Proteome-wide analysis of cysteine reactivity during effector-triggered immunity. Plant Physiol 179:1248–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry DJ, Shchepinova MM, Lilla S, Hartley RC, Olson MF (2016) A cell-permeable biscyclooctyne as a novel probe for the identification of protein sulfenic acids. ACS Chem Biol 11:3300–3304 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld J-P, Riondet C (2012) Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17:1124–1160 [DOI] [PubMed] [Google Scholar]
- Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J (2016) Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J Exp Bot 67:3831–3844 [DOI] [PubMed] [Google Scholar]
- Mittler R (2017) ROS are good. Trends Plant Sci 22:11–19 [DOI] [PubMed] [Google Scholar]
- Mnatsakanyan R, Markoutsa S, Walbrunn K, Roos A, Verhelst SHL, Zahedi RP (2019) Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique. Nat Commun 10:2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala HF, Kuo YH, Stinger BL, Palfey BA, Martin BR (2020) Tunable heteroaromatic sulfones enhance in-cell cysteine profiling. J Am Chem Soc 142:1801–1810 [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH (2009) H2S signals through protein S-sulfhydration. Sci Signal 2:ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzel T, Mostertz J, Ruberti C, Née G, Fuchs P, Wagner S, Moseler A, Müller-Schüssele SJ, Benamar A, Poschet G, et al. (2020) Redox-mediated kick-start of mitochondrial energy metabolism drives resource-efficient seed germination. Proc Natl Acad Sci U S A 117:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Yu J, Liao W, Xie J, Yu J, Lv J, Xiao X, Hu L, Wu Y (2019) Proteomic investigation of S-nitrosylated proteins during NO-induced adventitious rooting of cucumber. Int J Mol Sci 20:5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oger E, Marino D, Guigonis J-M, Pauly N, Puppo A (2012) Sulfenylated proteins in the Medicago truncatula–Sinorhizobium meliloti symbiosis. J Proteomics 75:4102–4113 [DOI] [PubMed] [Google Scholar]
- Pan J, Carroll KS (2013) Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem Biol 8:1110–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappireddi N, Martin L, Wühr M (2019) A review on quantitative multiplexed proteomics. ChemBioChem 20:1210–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Balmant K, Zhu F, Zhu N, Chen S (2015) cysTMTRAQ–an integrative method for unbiased thiol-based redox proteomics. Mol Cell Proteomics 14:237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Zhu N, Zhu M, Chen S (2012) Profiling thiol redox proteome using isotope tagging mass spectrometry. J Vis Exp 61:e3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS (2013) Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev 113:4633–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS (2012) Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol 8:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Mauriès A, Maes A, Tourasse NJ, Hamon M, Lemaire SD, Marchand CH (2017) The deep thioredoxome in Chlamydomonas reinhardtii: new insights into redox regulation. Mol Plant 10:1107–1125 [DOI] [PubMed] [Google Scholar]
- Poole LB (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 80:148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, King SB (2014) Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc 136:6167–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyaubert J, Fares A, Rézé N, Peltier J-B, Baudouin E (2014) Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci 215–216:150–156 [DOI] [PubMed] [Google Scholar]
- Qiu C, Sun J, Wang Y, Sun L, Xie H, Ding Y, Qian W, Ding Z (2019) First nitrosoproteomic profiling deciphers the cysteine S-nitrosylation involved in multiple metabolic pathways of tea leaves. Sci Rep 9:17525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz JA, Bechtold E, King SB, Poole LB, Furdui CM (2013) Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J 280:6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe KTG, Munkanatta Godage DNP, VanHecke GC, Ahn Y-H (2014) Metabolic synthesis of clickable glutathione for chemoselective detection of glutathionylation. J Am Chem Soc 136:11566–11569 [DOI] [PubMed] [Google Scholar]
- Samarasinghe KTG, Munkanatta Godage DNP, Zhou Y, Ndombera FT, Weerapana E, Ahn Y-H (2016) A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol Biosyst 12:2471–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AT, Neubert H, Brennan JP, Eaton P (2004) Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A 101:17982–17987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scinto SL, Ekanayake O, Seneviratne U, Pigga JE, Boyd SJ, Taylor MT, Liu J, am Ende CW, Rozovsky S, Fox JM (2019) Dual-reactivity trans-cyclooctenol probes for sulfenylation in live cells enable temporal control via bioorthogonal quenching. J Am Chem Soc 141:10932–10937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai LH, Tannenbaum SR (2016) S-Nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration. Proc Natl Acad Sci U S A 113:4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang J, Zhou M, Zhou H, Cui B, Gotor C, Romero LC, Fu L, Yang J, Foyer CH, et al. (2020) Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 32:1000–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Carroll KS (2020) Activity-based sensing for site-specific proteomic analysis of cysteine oxidation. Acc Chem Res 53:20–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade WO, Werth EG, McConnell EW, Alvarez S, Hicks LM (2015) Quantifying reversible oxidation of protein thiols in photosynthetic organisms. J Am Soc Mass Spectrom 26:631–640 [DOI] [PubMed] [Google Scholar]
- Speers AE, Adam GC, Cravatt BF (2003) Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc 125:4686–4687 [DOI] [PubMed] [Google Scholar]
- Takanishi CL, Ma L-H, Wood MJ (2007) A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry 46:14725–14732 [DOI] [PubMed] [Google Scholar]
- Takanishi CL, Wood MJ (2011) A genetically encoded probe for the identification of proteins that form sulfenic acid in response to H2O2 in Saccharomyces cerevisiae. J Proteome Res 10:2715–2724 [DOI] [PubMed] [Google Scholar]
- Tessier R, Nandi RK, Dwyer BG, Abegg D, Sornay C, Ceballos J, Erb S, Cianférani S, Wagner A, Chaubet G, et al. (2020) Ethynylation of cysteine residues: from peptides to proteins in vitro and in living cells. Angew Chem Int Ed Engl 59:10961–10970 [DOI] [PubMed] [Google Scholar]
- Trost P, Fermani S, Calvaresi M, Zaffagnini M (2017) Biochemical basis of sulphenomics: how protein sulphenic acids may be stabilized by the protein microenvironment. Plant Cell Environ 40:483–490 [DOI] [PubMed] [Google Scholar]
- Vajrychova M, Salovska B, Pimkova K, Fabrik I, Tambor V, Kondelova A, Bartek J, Hodny Z (2019) Quantification of cellular protein and redox imbalance using SILAC-iodoTMT methodology. Redox Biol 24:101227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHecke GC, Yapa Abeywardana M, Huang B, Ahn Y-H (2020) Isotopically labeled clickable glutathione to quantify protein S-glutathionylation. ChemBioChem 21:853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo E, Merl-Pham J, Velikova V, Ghirardo A, Lindermayr C, Hauck SM, Bernhardt J, Riedel K, Durner J, Schnitzler J-P (2016) Modulation of protein S-nitrosylation by isoprene emission in poplar. Plant Physiol 170:1945–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xian M (2011) Chemical methods to detect S-nitrosation. Curr Opin Chem Biol 15:32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Van Molle I, De Smet B, Vertommen D, Gevaert K, et al. (2014) Sulfenome mining in Arabidopsis thaliana. Proc Natl Acad Sci U S A 111:11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468:790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Willems P, Huang J, Tian C, Yang J, Messens J, Van Breusegem F (2020) Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. Front Plant Sci 11:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla K, Williamson J, Roepstorff P, Rogowska-Wrzesinska A (2015) The SNO/SOH TMT strategy for combinatorial analysis of reversible cysteine oxidations. J Proteomics 113:415–434 [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhao B, Weng Y, Shan Y, Li X, Hu Y, Liang Z, Yuan H, Zhang L, Zhang Y (2019) Site-specific quantification of persulfidome by combining an isotope-coded affinity tag with strong cation-exchange-based fractionation. Anal Chem 91:14860–14864 [DOI] [PubMed] [Google Scholar]
- Xiao H, Jedrychowski MP, Schweppe DK, Huttlin EL, Yu Q, Heppner DE,, Li J, Long J, Mills EL, Szpyt J, et al. (2020) A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 180:968–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou J-M, Zuo J (2015) S-Nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol 167:1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gupta V, Carroll KS, Liebler DC (2014) Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun 5:4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Balmant K, Geng S, Zhu N, Zhang T, Dufresne C, Dai S, Chen S (2017) Bicarbonate induced redox proteome changes in Arabidopsis suspension cells. Front Plant Sci 8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hara A, Sugiura K, Fukaya Y, Hisabori T (2018) Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc Natl Acad Sci U S A 115:E8296–E8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hisabori T (2016) Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc Natl Acad Sci U S A 113:E3967–E3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park C-M, Carroll KS, Filipovic MR, Xian M (2014) Detection of protein S-sulfhydration by a tag-switch technique. Angew Chem Int Ed Engl 53:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M (2010) Reductive ligation mediated one-step disulfide formation of S-nitrosothiols. Org Lett 12:4208–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhu M, Zhu N, Strul JM, Dufresne CP, Schneider JD, Harmon AC, Chen S (2016) Identification of thioredoxin targets in guard cell enriched epidermal peels using cysTMT proteomics. J Proteomics 133:48–53 [DOI] [PubMed] [Google Scholar]
- Zivanovic J, Kouroussis E, Kohl JB, Adhikari B, Bursac B, Schott-Roux S, Petrovic D, Miljkovic JL, Thomas-Lopez D, Jung Y, et al. (2019) Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metab 30:1152–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]