Cyanobacterial thioredoxin reductases diverged early in the evolution into flavo- or metallo-enzymes to adapt the regulation of their metabolism to the prevailing environment.
Abstract
Thioredoxin reductases control the redox state of thioredoxins (Trxs)—ubiquitous proteins that regulate a spectrum of enzymes by dithiol–disulfide exchange reactions. In most organisms, Trx is reduced by NADPH via a thioredoxin reductase flavoenzyme (NTR), but in oxygenic photosynthetic organisms, this function can also be performed by an iron-sulfur ferredoxin (Fdx)-dependent thioredoxin reductase (FTR) that links light to metabolic regulation. We have recently found that some cyanobacteria, such as the thylakoid-less Gloeobacter and the ocean-dwelling green oxyphotobacterium Prochlorococcus, lack NTR and FTR but contain a thioredoxin reductase flavoenzyme (formerly tentatively called deeply-rooted thioredoxin reductase or DTR), whose electron donor remained undefined. Here, we demonstrate that Fdx functions in this capacity and report the crystallographic structure of the transient complex between the plant-type Fdx1 and the thioredoxin reductase flavoenzyme from Gloeobacter violaceus. Thereby, our data demonstrate that this cyanobacterial enzyme belongs to the Fdx flavin-thioredoxin reductase (FFTR) family, originally described in the anaerobic bacterium Clostridium pasteurianum. Accordingly, the enzyme hitherto termed DTR is renamed FFTR. Our experiments further show that the redox-sensitive peptide CP12 is modulated in vitro by the FFTR/Trx system, demonstrating that FFTR functionally substitutes for FTR in light-linked enzyme regulation in Gloeobacter. Altogether, we demonstrate the FFTR is spread within the cyanobacteria phylum and propose that, by substituting for FTR, it connects the reduction of target proteins to photosynthesis. Besides, the results indicate that FFTR acquisition constitutes a mechanism of evolutionary adaptation in marine phytoplankton such as Prochlorococcus that live in low-iron environments.
Introduction
Thioredoxins (Trxs) are small redox proteins characterized by a redox-active disulfide in the form of a conserved WCGPC motif (Holmgren, 1985). The proteins relay redox information to multiple partner proteins via dithiol–disulfide exchange reactions, thereby regulating fundamental cellular processes, including photosynthesis (Balsera et al., 2014), general metabolism (Laurent et al., 1964; Porqué et al., 1970), removal of reactive oxygen species (ROS; Puerto-Galán et al., 2013; Perkins et al., 2015), and cell signaling (Shenton and Grant, 2003; Leichert et al., 2008; Balsera et al., 2014). Thioredoxin reductases—enzymes that reduce Trxs—can be divided into two phylogenetically unrelated groups based on their active sites: (i) ferredoxin–thioredoxin reductase (FTR), a well-characterized Fe-S enzyme in oxygenic photosynthetic organisms, that catalyzes the transfer of reducing equivalents from photochemically reduced ferredoxin (Fdx) to Trx (Dai et al., 2000); (ii) flavin thioredoxin reductase that comprises a diverse group of enzymes. The best-known member of this group—NADPH-thioredoxin reductase or NTR—receives reducing equivalents from NADPH (Arscott et al., 1997; Williams et al., 2000). Other members include Fdx flavin-thioredoxin reductase (FFTR), originally described in fermentative bacteria (Hammel et al., 1983), and deazaflavin-dependent thioredoxin reductase found in methanogenic archaea (Susanti et al., 2016). NTRs from prokaryotes, FFTR, and deazaflavin thioredoxin reductase form a homologous group of enzymes that connect metabolism, fermentation, and methanogenesis, respectively, to the reduction of Trx targets according to the needs of the cell.
We have recently found an enzyme, deeply rooted thioredoxin reductase or DTR, that is phylogenetically related to prokaryotic NTR but functions independently of pyridine nucleotides. The enzyme is present in photosynthetic organisms―including cyanobacteria, marine planktonic cyanobacteria, marine diazotrophic cyanobacteria, algae―and other bacteria (Buey et al., 2017b). In certain cyanobacteria, DTR is the only enzyme with thioredoxin reductase activity (9,11). GvDTR shows a conserved overall fold and a mode of subunit assembly similar to archetypal prokaryotic NTRs, while differing in amino acid motifs that form the NADPH-binding cavity (Buey et al., 2017b). GvDTR is a homodimer with each monomer composed of two conserved Rossmann-type modules that form the FAD-binding and redox-active disulfide domains. Of particular interest is the pi-stacking interaction between the side chain of the conserved tryptophan at the C-terminal tail of a monomer and the isoalloxazine ring of the FAD of an adjacent monomer at its re-face. This interaction seems to protect the flavin from the solvent—a distinctive feature of the GvDTR enzyme not found in other flavin thioredoxin reductases (Lennon et al., 2000; Buey et al., 2018). Nonetheless, until now, the electron donor of DTR has remained unknown.
Here, we have identified Fdx as the electron donor for the DTR enzyme from the cyanobacterium Gloeobacter violaceus (GvDTR). We also report the high-resolution crystallographic structure of the transient complex between Fdx and GvDTR. The biochemical data suggest that electron transfer proceeds sequentially from the [2Fe-2S] cluster in Fdx, to the flavin cofactor and the disulfide of the cyanobacterial thioredoxin reductase, and then to Trx. Our results relate the DTR enzyme to FFTR both structurally and functionally. The enzyme is, therefore, ascribed to the FFTR protein family and renamed accordingly. Interestingly, these observations indicate that FFTR represents an unidentified link between anaerobic fermentation and photosynthesis.
Finally, we demonstrate that the FFTR/Trx system of Gloeobacter is able to reduce CP12, a small protein functional in the regulation of two enzymes of the Calvin–Benson cycle in oxygenic photosynthetic organisms—phosphoribulokinase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The results suggest that FFTR can substitute for FTR in light-linked redox regulation in Gloeobacter. Further, our results indicate that organisms, such as Prochlorococcus, living in low-Fe environments have replaced the metalloenzyme FTR with the flavoenzyme FFTR as a mechanism of evolutionary adaptation. A similar adaptative response was earlier observed with Fdx and flavodoxin (Bottin and Lagoutte, 1992; Lodeyro et al., 2012). The results reported here pave the way to obtaining a complete picture of the complex evolutionarily relationships of thioredoxin reductases in relation to the metabolic adaptations of different organisms.
Results
Fdx is the redox partner of GvDTR
A comparison of the previously reported structure of GvDTR (PDB code 5J60; Buey et al., 2017b) with entries available in the protein data bank (PDB) using the DALI server (Holm and Laakso, 2016) enabled us to identify FFTR from Clostridium acetobutylicum (CaFFTR2, PDB code 6GNC) as the closest structural homolog. Since clostridial FFTR catalyzes the transfer of redox equivalents from Fdx to the disulfide bridge of Trxs (Hammel et al., 1983; Buey et al., 2018), the structural similarities found between GvDTR and CaFFTR2 (Supplemental Figure S1) prompted us to investigate whether Fdx could act also as substrate for the cyanobacterial enzyme.
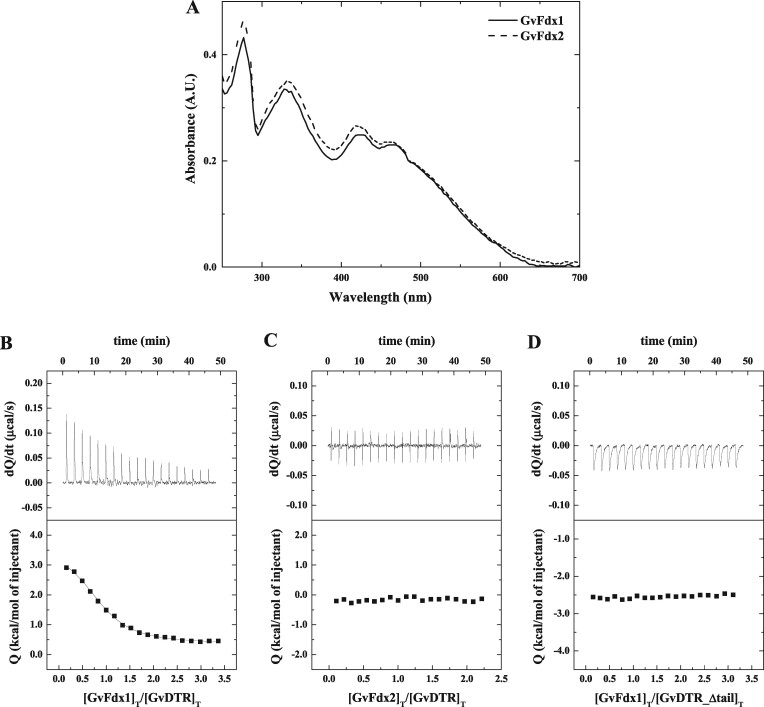
An analysis of the occurrence of Fdx-encoding genes in cyanobacteria revealed that organisms having GvDTR-like proteins commonly contain plant-type [2Fe-2S] Fdxs (Cassier-Chauvat and Chauvat, 2014). We, therefore, recombinantly produced and purified to homogeneity the two plant-type [2Fe-2S] Fdxs from Gloeobacter (GvFdx1 and GvFdx2). The UV–visible spectra obtained for both proteins (Figure 1A) showed the expected absorbance maxima at about 330, 420, and 460 nm (29). The interaction of the Fdxs with GvDTR was evaluated by isothermal titration calorimetry (ITC) experiments. We found that GvDTR and GvFdx1 interact with a dissociation constant (Kd) of ∼3.9 µM, with binding stoichiometry close to 1 (Figure 1B). By contrast, the binding of GvFdx2 was negligible according to the ITC data (Figure 1C).
Figure 1.
Binding affinity measurements for the interaction between Gloeobacter Fdxs and GvDTR by ITC. A, Absorption spectra of purified GvFdx1 (continuous line) and GvFdx2 (dashed line) in buffer 20-mM Tris-HCl, pH 7.6, 150-mM NaCl at 25°C. In the 300–600 nm region, the UV–visible spectra of the proteins are dominated by bands centered at about 330, 420, and 463 nm. B–D, Calorimetric assays for the binding of GvFdx1 to GvDTR (B), GvFdx2 to GvDTR (C), and GvFdx1 to GvDTR_Δtail (D). The upper plots show the thermograms corresponding to raw data of heat power associated with the sequential addition of the Fdxs solutions to the GvDTR or GvDTR_Δtail protein solutions. The lower parts are the binding isotherms (Fdx-normalized integrated heats per injection as a function of the molar ratio [(Fdx)Total/(GvDTR)Total]. Nonlinear least-squares regression employing a model with a single ligand-binding site per GvDTR monomer provided a dissociation constant, Kd, of 3.9 µM for the interaction of Fdx1 with GvDTR. No interaction was observed in the case of Fdx2 titrated into GvDTR or for Fdx1 titrated into GvDTR_Δtail.
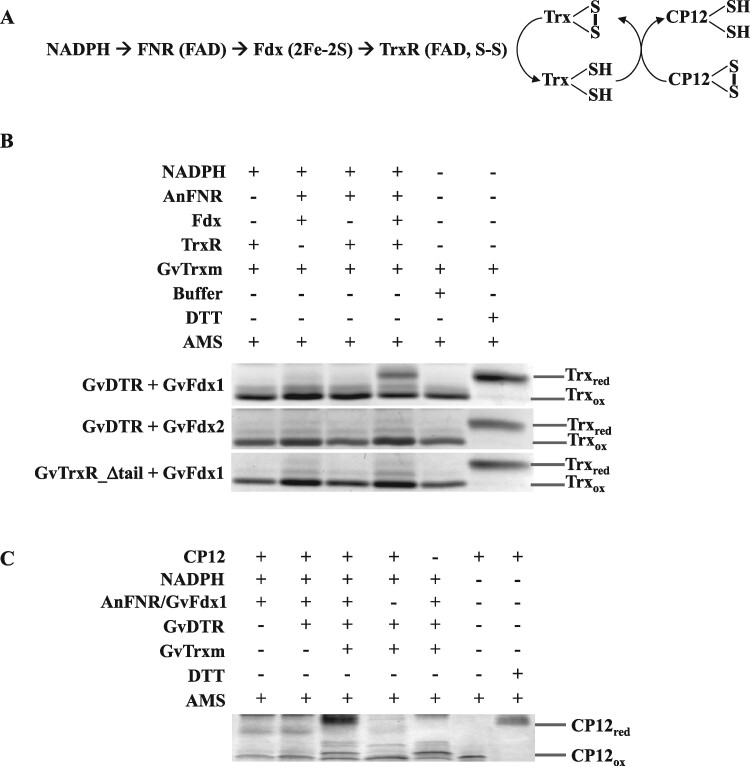
In a parallel experiment, we investigated GvDTR and GvFdx1 or GvFdx2 as functional redox partners by analyzing the reduction of photosynthetic m-type thioredoxin (Trx-m) in a mixture containing NADPH, the redox pair Anabaena ferredoxin-NADP+ reductase (AnFNR)/GvFdx1 or AnFNR/GvFdx2, GvDTR, and Gloeobacter Trx-m (GvTrx-m; Figure 2A). The redox state of GvTrx-m was examined with the thiol-specific reagent 4-acetamido-4′-maleimidyldistilbene-2,2′-disulfonic acid (AMS), separating the reduced and oxidized proteins with nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Balsera et al., 2009). As shown in Figure 2B and Supplemental Figure S2, only GvFdx1 with the complete reactants significantly increased the reduced/oxidized ratio of GvTrx-m, prompting the conclusion that Fdx, and particularly GvFdx1, is a functional electron-delivering partner for GvDTR.
Figure 2.
Fdx-dependent reduction of GvTrx-m and GvCP12 via GvDTR. A, Schematic illustration of the in vitro reaction performed in this study for the reduction of Trx (B), and further GvCP12 (C), by GvDTR with electrons derived from [2Fe-2S] GvFdx, itself reduced by Anabaena’s FNR in the presence of NADPH. B, The reaction mixtures contained equal amounts of Trx, NADPH, and some or all of the components included in the scheme depicted in (A), as indicated. After reaction, free sulfhydryl groups were labeled with AMS, which increases the mass of the protein by about 0.5 KDa per free thiol, allowing the two different redox states of GvTrx-m (reduced or oxidized, Trxred or Trxox respectively) to be separated by SDS-PAGE under nonreducing conditions. GvTrx-m in the oxidized state (buffer) and in the reduced state (after DTT incubation) are included for comparison. C, In a similar kind of reaction as above, the reduced/oxidized ratio of GvCP12 was determined by non-reducing SDS-PAGE after AMS treatment in the presence of components according to the complete scheme indicated in (A). The images of the complete gels can be found in Supplemental Figures S2–S3.
Then, the activity of the Trx system of Gloeobacter was assessed by analyzing its capacity to reduce CP12, a well-known Trx-linked regulatory protein in oxyphotosynthetic organisms, in vitro (Gontero and Maberly, 2012). The results showed that Gloeobacter CP12 (GvCP12) contains redox-sensitive Cys thiols that respond to Trx (Figure 2C; Supplemental Figure S3) and reinforce the idea that GvDTR receives electrons from GvFdx. These experiments further demonstrated that CP12 is a Trx target in Gloeobacter.
A unique feature of GvDTR is the presence of a C-terminal tail with a conserved aromatic amino acid that stacks onto the isoalloxazine ring of the FAD of the adjacent monomer. According to our SAXS analysis, deletion of the C-terminal tail does not significantly affect structure and both GvDTR and GvDTR_Δtail proteins are properly folded (Supplemental Figure S4). In agreement with these results, the mutant enzyme is able to reduce Trx when a non-physiological electron donor (dithionite) is used, as previously reported (Buey et al., 2017b). To further understand the contribution of the C-terminal tail to FFTR activity, we determined by ITC the affinity of GvDTR and the C-terminal deletion mutant (GvDTR_Δtail) enzymes for GvFdx1. The results showed that GvFdx1 binding to GvDTR was strictly dependent on the presence of the enzyme’s C-terminal tail (Figure 1D). We then examined the functional impact of the C-terminal deletion in GvDTR by assessing its effect on the reduction of Trx in the presence of NADPH and the redox pair AnFNR/GvFdx1. As expected from the ITC experiments, the deletion of the C-terminal tail in GvDTR abolished enzyme activity (Figure 2B; Supplemental Figure S2).
The C-terminal extension stabilizes the semiquinone state of the flavin
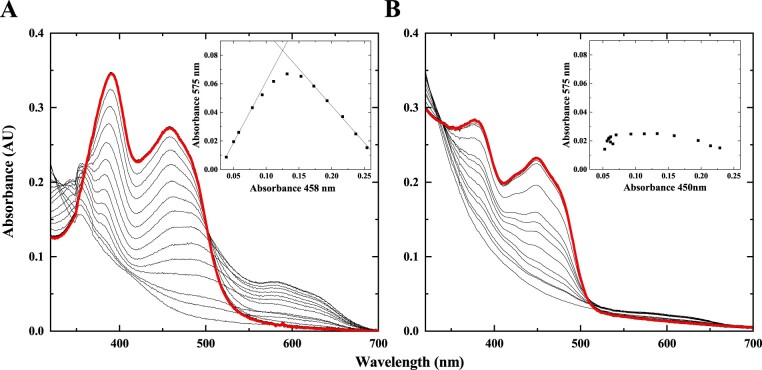
To obtain information on the properties of the cofactor of the flavoenzyme, we performed photoreduction experiments. Absorption spectra of oxidized FAD showed two peaks in the UV–visible region with maxima at 391 and 458 nm for GvDTR, and 380 and 450 nm for GvDTR_Δtail (Figure 3). Stepwise photoreduction of the flavin coincided with differences in the visible spectrum between GvDTR and GvDTR_Δtail. In the case of GvDTR (Figure 3A), the transient appearance of a long-wavelength absorbance band in the 550–700 nm region (with a maximum at 575 nm and a shoulder at 631 nm) suggests that the flavin is not being fully reduced to the two-electron state and that a large amount (nearly 80%) of the one-electron-reduced neutral semiquinone is stabilized. By contrast, GvDTR_Δtail marginally stabilized the semiquinone state (<10%) and full reduction of the cofactor was apparent with the loss in flavin absorption band I (Figure 3B). Upon admission of air in the dark, reoxidation occurred with trends similar to those observed for photoreduction, demonstrating reversibility (Figure 3A and B, red bold lines). These results indicate that the C-terminal tail of GvDTR stabilizes the flavin into the one-electron-reduced neutral semiquinone intermediate before being fully reduced, and highlights its essential role in flavin chemistry.
Figure 3.
Flavin spectral evolution for the photoreduction of GvDTR and GvDTR_Δtail. A, Absorption spectrum of oxidized flavin in GvDTR shows maxima of bands II and I at 391 and 458 nm, respectively. Flavin reduction occurs with neutral semiquinone stabilization as shown by the appearance of isosbestic points at 361 and 502 nm (for oxidized/semiquinone transition), and 324 nm (for semiquinone/hydroquinone transition). B, Reduction of oxidized flavin in GvDTR_Δtail (with maxima at 380 and 450 nm) occurs without semiquinone stabilization as shown by the isosbestic points at 340 and 509 nm for the oxidized/hydroquinone transition. The red bold lines show the spectra after one cycle of full protein photoreduction followed by molecular oxygen reoxidation, which is identical to the initial spectra. The insets show the absorption at the neutral semiquinone band maximum (575 nm) relative to absorption at the flavin band I maximum (450 nm) along with photoreduction. Experiments were performed under anaerobic conditions in the presence of 5-deazariboflavin and EDTA.
High-resolution crystal structure of GvDTR bound to GvFdx1
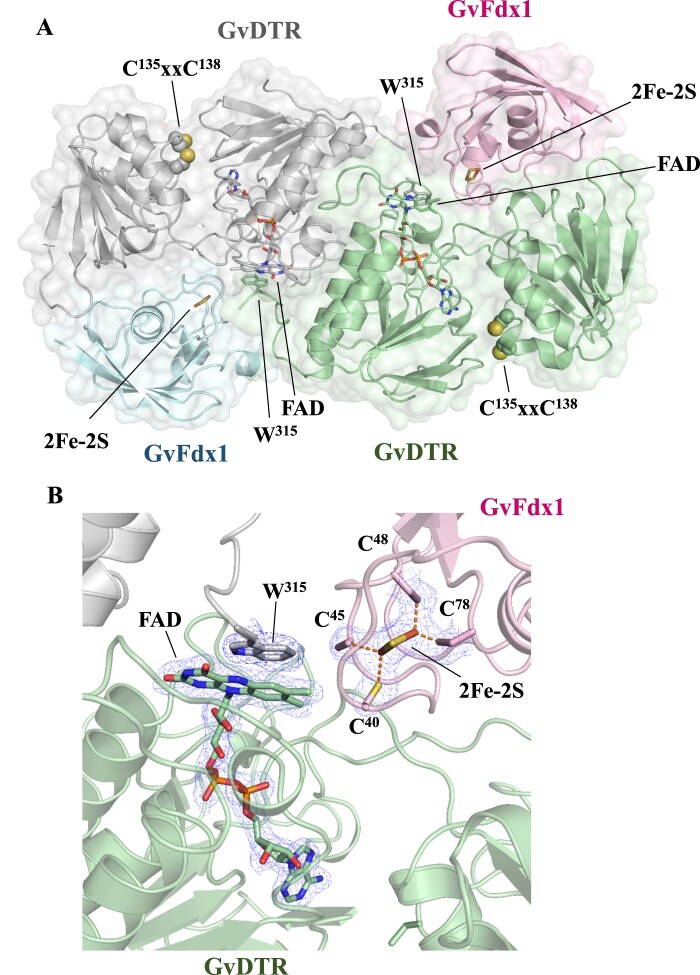
Finally, we sought to obtain structural information on the interaction of GvDTR with GvFdx1. To this end, we attempted first to isolate the GvDTR-GvFdx1 complex. This was not possible using gel filtration (Supplemental Figure S5), reflecting the transient nature of the interaction. We, therefore, employed a co-crystallization strategy with a molar excess of GvFdx1 to GvDTR (1.5:1) that produced single crystals amenable for X-ray diffraction experiments. The crystals belonged to the space group C2221, with two monomers in the asymmetric unit, allowing structure solution and refinement up to 2.25 Å resolution (Supplemental Table S1).
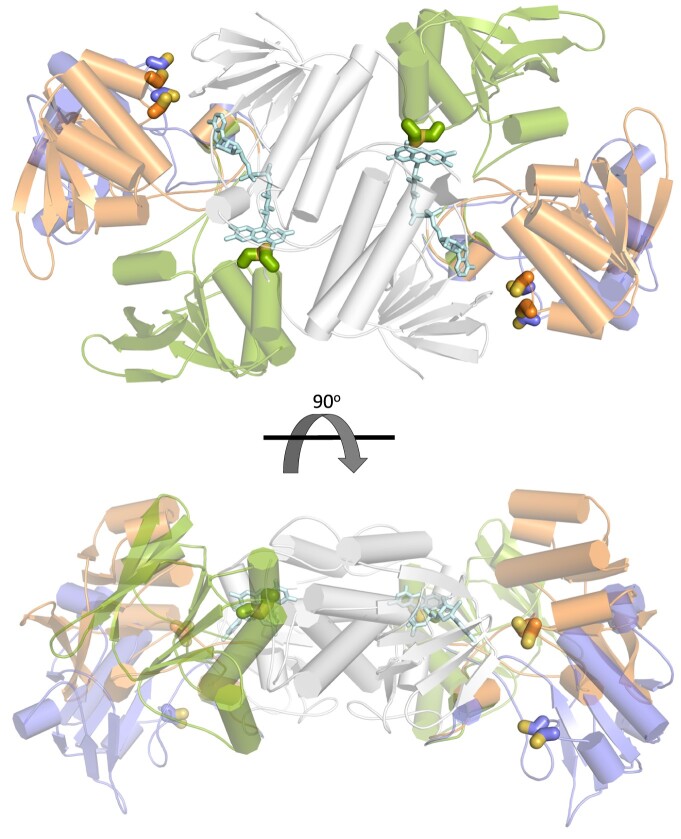
The structure showed that each GvDTR monomer binds one GvFdx1 (Figure 4A; PDB code 6XTF). Each monomer in the complex adopted an extended conformation with the two redox centers (the isoalloxazine ring of the flavin cofactor and the Cys amino acids of the CxxC motif) separated by more than 20 Å. The two structural domains formed a groove where GvFdx1 was bound (Figure 4B). Compared to the enzyme alone, the conformation of GvDTR is not substantially altered upon GvFdx1 binding (root-mean-square deviation or RMSDCα is 0.56 Å), though the domains rotate slightly as rigid bodies (Supplemental Figure S6). The structure of GvFdx1 superimposed well on Fdx1 from Synechocystis (PDB code 1OFF; RMSDCα is 0.47 Å), as expected from the close similarity between the two proteins (73% sequence identity). The binding of GvFdx1 to GvDTR is mostly driven by electrostatic interactions, as shown by the electrostatic potential surface (Supplemental Figure S7A), with the isoalloxazine ring of the flavin of a monomer conserving the stacking interaction with the tryptophan of the opposite monomer (W315; Figure 4A). In GvDTR, most of the Fdx-interacting residues are located in loops of the redox-active disulfide domain of one monomer or are provided by the C-terminal tail of the second monomer (Supplemental Figure S7B). The electron density is sufficiently clear to show the position of the [2Fe-2S] cluster with respect to FAD (Figure 4B). The structure shows that the C-7 and C-8 methyl groups of the isoalloxazine ring of FAD are oriented to the Fe-S cluster to accept electrons. The closest distance between the [2Fe-2S] cluster and the isoalloxazine ring of the flavin is about 6 Å, a distance that could facilitate rapid electron transfer from reduced Fdx to the flavin of GvDTR.
Figure 4.
Crystal structure of the GvDTR–GvFdx1 complex. A, Ribbon diagram of the GvDTR homodimer with the subunits in green and grey, each bound to an Fdx colored in magenta and blue, respectively. GvFdx1 has a plant-type Fdx fold and contains a [2Fe-2S] cluster. B, Electron density maps around FAD and Fe-S. The [2Fe-2S] cluster, coordinated by four Cys, is placed at the surface of the protein and close to the interface between GvDTR and GvFdx1 (A). The FAD cofactor, the [2Fe-2S] cluster and the side chain of amino acids Trp315, and Cys of the CxxC motif and participating in cluster coordination are shown as sticks.
Discussion
Oxygenic photosynthetic organisms utilize the Fdx–Trx system as an efficient strategy to connect metabolism to light, and to respond to changes in the environment. The basal system, regarded to be composed of the metalloenzyme Fdx, FTR, and Trx-m, was earlier developed in cyanobacteria and later acquired by eukaryotic phototrophs that greatly expanded during evolution. Remarkably, though FTR was long considered a hallmark of light regulation in oxygenic photosynthesis, this enzyme is not present in all cyanobacteria (Florencio et al., 2006; Balsera et al., 2013).
A few years ago, we reported a novel thioredoxin reductase flavoenzyme, denoted as DTR, present in a reduced group of cyanobacteria, including Gloeobacter and Prochlorococcus, a few algae and other bacteria. The enzyme was not functional with pyridine nucleotides but was shown to reduce Trx-m (Buey et al., 2017b). Nonetheless, essential questions related to its reaction mechanism, including who its electron donor is, remained open. Using the enzyme from Gloeobacter violaceus as a model, we have filled this gap and demonstrated that Fdx provides electrons to the enzyme for reducing Trx according to the following sequence: reduced Fdx ([2Fe-2S] cluster) → DTR (FAD to S-S) → Trx (S-S). Therefore, we assign DTR to the FFTR protein family (Hammel et al., 1983; Buey et al., 2018), which should be renamed accordingly.
These results indicate that the FFTR protein family comprised two classes of homolog enzymes with characteristic structural and functional properties: (i) clostridial FFTR in fermentative bacteria (Hammel et al., 1983; Buey et al., 2018), and (ii) cyanobacterial FFTR in cyanobacteria, a few marine algae and in certain members of bacteria that include representatives of the Aquificace, Chloroflexi, Bacillus, Firmicutes, Chlorobi, and Nitrospirae groups (Buey et al., 2017b). The most pronounced structural difference between these two types of enzymes in the presence of a C-terminal tail in cyanobacterial FFTR that contains a conserved aromatic residue that stacks over the flavin at its re-face. The structural distinctiveness of the FFTRs may account for the different substrate specificities, because clostridial FFTRs are functional with bacterial-type Fdx with two [4Fe-4S] clusters, whereas cyanobacterial FFTRs are functional with plant-type Fdx with one [2Fe-2S] cluster. It is, therefore, reasonable to assume that cyanobacterial FFTRs evolved the C-terminal tail to interact with plant-type Fdxs.
Our crystal structure analysis has provided details on the interaction of GvFdx1 with GvDTR, henceforth called GvFFTR. The FAD and Fe-S redox cofactors are located at a distance and orientation optimal for the electron transfer reactions. Since [2Fe-2S] Fdxs provide one electron at a time, a complete reduction of FAD would require the sequential binding of two Fdxs. Our results show that the C-terminal tail in cyanobacterial FFTR is an important structural element not only for interaction with Fdx, but also for enzyme activity. The absolutely conserved tryptophan in cyanobacteria participates in stabilizing the semiquinone form of the flavin and in setting its midpoint reduction potential during the reaction. This molecular mechanism closely resembles that proposed for thioredoxin reductase-like FNR, which houses a similar C-terminal extension (Seo et al., 2014; Buey et al., 2017b). Taken together, these results prompt the view that the redox systems rely on a limited number of structural modules and regulatory elements that have been combined during evolution to form enzymes specifically adapted to the metabolic and environmental requirements of each particular organism.
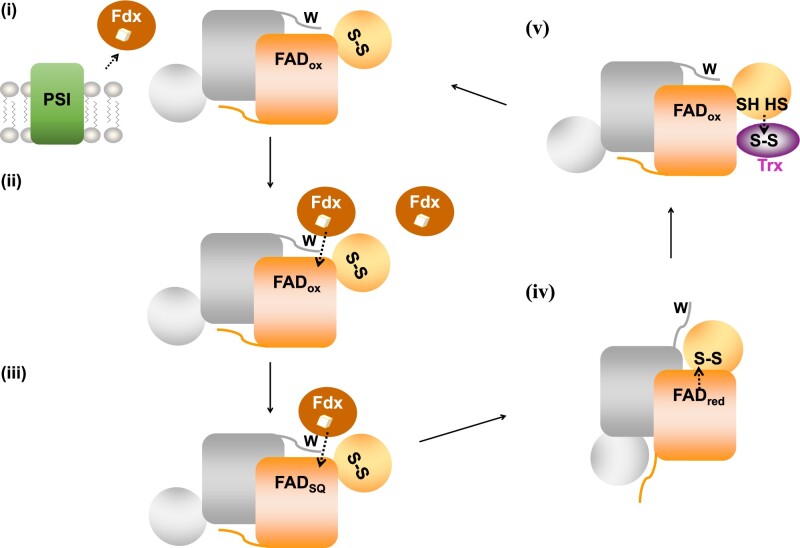
Based on our structural and biochemical data, we propose a functional working model for cyanobacterial FFTR (Figure 5): (i) The [2Fe-2S] cluster of Fdx1 is first reduced by photosystem I under light; (ii) reduced Fdx1 binds FFTR, resulting in electron transfer to the FAD cofactor forming the semiquinone of FAD, which is stabilized by the C-terminal tail; (iii) a second reduced Fdx1 binds to FFTR to reduce the FAD to the fully reduced quinone state; (iv) complete reduction of FAD triggers the release of the C-terminal tail, thereby opening the flavin active site and allowing the redox-active disulfide domain to swing into a position that facilities the intramolecular electron transfer from the isoalloxazine ring of FAD to the S-S; (v) subsequently, a domain rearrangement exposes the newly formed sulfhydryls for interaction with Trx.
Figure 5.
Working model for GvFFTR. The enzyme is homodimeric (monomers colored in orange and grey). For the sake of clarity, the redox cofactors (FAD and S-S) of a monomer and the tryptophan (W) residue at the C-terminal tail of the second monomer are shown only for one functional unit. (i) Transfer of reducing equivalents from photosystem I (PSI) to Fdx; (ii) Reduced Fdx transfers one electron to the flavin cofactor of a monomer; (iii) A second Fdx protein reduces FAD to the hydroquinone state; (iv) A conformational rearrangement allows the intramolecular transfer of electrons from reduced FAD to the redox-active disulfide; (v) Conformational rearrangement for dithiol–disulfide exchange reaction between GvFFTR and oxidized Trx. Upon Trx reduction, the enzyme recovers the resting state (i). Protein structural elements are not represented in scale. The three-dimensional structures that GvFFTR adopts in the different steps are shown in Figure 6.
In this model, cyanobacterial FFTR should adopt at least three different conformations during the catalytic cycle (Figure 6). In steps i–iii, the enzyme would adopt a conformation that corresponds to the resting state of the enzyme, ready for interaction with Fdx (Figure 6, orange). Indeed, there are no significant differences between the structures of GvFFTR alone and in complex with Fdx. During step iv, the enzyme must adopt a second conformation, in which the C-terminal tail flips out from the FAD, allowing the disulfide to move over the re-face of the FAD isoalloxazine ring for reduction (Figure 6, green). We speculate this conformation to be similar to the flavin-oxidizing (FO) conformation of archetypical NTRs (Lennon et al., 2000). Finally, in step v, the two domains of FFTR rearrange to adopt a third conformation (Figure 6, blue), in which the reduced Cys is exposed for interaction with Trx, as previously reported in the structure of the clostridial FFTR-Trx complex (Buey et al., 2018). Future experiments will be directed to decipher the pathways FFTR follows to adopt the redox-dependent conformational changes subsequent to the reduction of the flavin cofactor.
Figure 6.
Conformations adopted by GvDTR during its catalytic cycle. The structures are displayed in ribbons representation, with alpha-helices shown as cylinders and beta-strands as arrows. Cysteine residues of the CxxC motif (in CPK coloring) and the FAD cofactor (light blue) are represented as sticks. The three conformations are superimposed based on the structural alignment of the FAD-binding domain (grey). The structure shown in orange color represents the resting state of the enzyme (PDB codes: 6XTF, this work, and 5J60 (Buey et al., 2017b); states ii–iii in Figure 5; in green, the S-S of the CxxC motif in the redox-active disulfide domain is interacting with the flavin cofactor (state iv in Figure 5). This structure has been obtained by homology modeling using the flavin-oxidizing conformation of NTR from Escherichia coli as template (PDB code 1TRB; Kuriyan et al., 1991); the structure in blue represents the conformation of the enzyme for Trx binding (state v in Figure 5). This conformation has been modeled using the structure of the CaFFTR2:CaTrx2 complex as template (PDB code 6GND; Buey et al., 2018).
According to our ITC experiments, GvFFTR interacts with GvFdx1 but not with GvFdx2, which has a unique C-terminal extension (Cassier-Chauvat and Chauvat, 2014). It is therefore plausible that this C-terminal extension prevents the interaction of GvFdx2 with GvFFTR. GvFdx2 has been reported to play physiological roles unrelated to photosynthesis, in tolerance to environmental stresses, whose direct targets remain unknown (Cassier-Chauvat and Chauvat, 2014). By contrast, Fdx1 function has been associated with photosynthetic electron transport (Cassier-Chauvat and Chauvat, 2014), thus linking the cyanobacterial FFTR enzyme to photosynthesis. Indeed, our in vitro experiments have shown that CP12, a well-known Trx target in cyanobacteria and plants, is reduced by the FFTR-Trx system from Gloeoabacter.
It should be noted that cyanobacteria such as Gloeobacter and Prochlorococuss lack FTR but instead contain FFTR as the only enzyme with thioredoxin reductase activity (Buey et al., 2017a, 2017b). Alternatively, other organisms, such as the marine Trichodesmium erythraeum IMS101 and Leptolyngbya sp. PCC 7376, have the two Fdx-dependent thioredoxin reductase genes (FTR and FFTR). The functional substitution, redundancy, or crosstalk between the FTR and FFTR enzymes remains worthy of further investigation in the future.
The data indicate that cyanobacterial Fdx-dependent thioredoxin reductases might have diverged early in the evolution into flavo- or metalloenzymes. A plausible explanation of the existence of the two thioredoxin reductase enzymes in cyanobacteria is the adaptation of the thioredoxin-dependent redox mechanism to the prevailing environment. This hypothesis is supported by the fact that marine phytoplankton organisms such as Prochlorococcus appear to have sacrificed the iron-sulfur FTR gene in favor of FFTR, likely owing to Fe deficiency in their natural habitats (Biller et al., 2015). In a similar way, the isofunctional proteins Fdx and flavodoxin are differentially selected in cyanobacteria for the distribution of photosynthetically derived electrons from photosystem I to redox partners according to environmental conditions, such as Fe deficiency (Bottin and Lagoutte, 1992; Hurley et al., 2006; Medina, 2009).
Taken together, we have demonstrated that the FFTR family of enzymes has been evolutionarily disseminated, with representatives in clostridia, cyanobacteria, and other bacteria. The finding that FFTR is the only thioredoxin reductase present in certain cyanobacteria prompts the need to further investigate the metabolic pathways functional in these organisms. The results reported here pave the way to obtaining a complete picture of the complex evolutionary relationships of thioredoxin reductases in relation to the metabolic adaptations of different organisms.
Methods
Protein expression and purification
Production and purification of GvDTR and GvTrx-m were carried out as described previously (Buey et al., 2017b). Open-reading frames encoding GvFdx1 (petF, gvip492), GvFdx2 (gvip440), and GvCP12 (gsr3698) were cloned into a bacterial expression vector developed in house (de Pereda et al., 2009), which harbors an N-terminal 8xHis tag and a Tobacco Etch Virus (TEV) cleavage site preceding the Fdx or CP12 sequence. For protein expression, competent Escherichia coli Rosetta (DE3) pLys cells were transformed with the respective plasmids. The cells were grown at 37°C in Luria Bertani medium supplemented with ampicillin and chloramphenicol, and ferrous sulfate in the case of the Fdxs. Protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (0.1 mM) at a cell density of about 0.6. After growth overnight at 20°C, the cells were harvested by centrifugation, and the pellets were suspended in 20 mM Tris-HCl, pH 8, 300 mM NaCl, 10% (v/v) glycerol, and lysed by sonication. The insoluble fraction was removed by centrifugation. The supernatant fraction was subjected to purification by immobilized metal ion affinity chromatography (GE Healthcare HisTrap HP). Protein was eluted with buffer containing 20 mM Tris-HCl, pH 8, 300 mM NaCl, and 500 mM imidazole. After overnight incubation with TEV protease in buffer containing 20 mM Tris-HCl, pH 8, 300 mM NaCl, and 0.5 mM DTT, the protein solution was subjected to a second immobilized metal ion affinity chromatography (GE Healthcare HisTrap HP) to remove the protease (His-tagged). The uncleaved protein was subjected to size exclusion chromatography (GE Healthcare Sephacryl S-300 HR) in buffer containing 20 mM Tris-HCl, pH 8, 150 mM NaCl, and 2 mM β-mercaptoethanol. Fractions containing protein were pooled and concentrated using an Amicon centrifugation device (Millipore). AnFNR was prepared as described previously (Medina et al., 1998). Analytical gel filtration was performed with a Superdex 200 Increase 10/300 GL column equilibrated in buffer containing 20 mM Tris-HCl, pH 8, and 150 mM NaCl.
ITC
Standard ITC experiments were performed using an Auto-iTC200 system (MicroCal, Malvern-Panalytical). Enzyme (GvDTR or GvDTR_Δtail) 20 µM in 100-mM potassium phosphate, 2 mM EDTA, pH 7.0 was titrated at 25°C with 300 µM Fdx protein (GvFdx1 or GvFdx2). The resulting heats were integrated and normalized to the amount of protein injected. A control experiment performed by injecting Fdx into buffer showed that the heats of dilution were negligible. The binding isotherm was analyzed employing a model considering a single ligand-binding site per GvDTR subunit in the homodimer. Although GvDTR is a homodimer, no evidence for ligand binding cooperativity was observed when employing a general binding model for two binding sites per dimer (Freire et al., 2009; Vega et al., 2015). Thermodynamic binding parameters were estimated by non-linear least-squares regression and standard thermodynamic relationships.
Redox state of thioredoxin and CP12
Thioredoxin reductase activity was assessed by estimating the ratio of reduced/oxidized state of Trx in a reaction mixture that contained 500 µM NADPH, 1 µM FNR, 10 µM Fdx, 1 µM thioredoxin reductase, and 5 µM Trx in buffer, 50 mM Tris-HCl, pH 7.6, and 100 mM NaCl. After 1 h incubation at room temperature, protein was precipitated with trichloroacetic acid and resuspended in a solution containing 1% (w/v) SDS and 10 mM AMS. Samples were subjected to 12% (w/v) or 4–20% (w/v) nonreducing SDS-PAGE for Trx and CP12, respectively, and visualized with BlueSafe.
Small-angle X-ray scattering
SAXS experiments were performed at beamline B21 of the Diamond Light Source. SEC-SAXS data were collected for GvDTR and GvDTR_Δtail using a Superdex S200 PC 3.2/30 (GE Healthcare) column connected to the measurement cell. The SEC-SAXS data were analyzed using the Chromixs program (Panjkovich and Svergun, 2018) within the software package ATSAS (Franke et al., 2017). Crysol (Svergun et al., 1995) was used to compute the theoretical SAXS profile of the crystallographic structure and compare it to the experimental one.
Protein photoreduction
UV–visible absorption spectra were recorded with a UV–Vis Cary 100 spectrophotometer (Agilent Technologies) at 25°C. The molar absorption coefficient for GvDTR (ε458nm=10.1 ± 1.3 mM−1cm−1) and GvDTR_Δtail (ε450nm = 13.9 ± 1.1 mM−1cm−1) were spectrophotometrically determined by thermal denaturation of the protein for 10 min at 90°C, followed by centrifugation, separation of the precipitated apoprotein, and spectroscopic quantification of the FAD released in the supernatant as previously described (Macheroux, 1999). Spectral development of the stepwise photoreduction of GvDTR (27 µM) and GvDTR_Δtail (17 µM) was achieved by illumination of protein samples in the presence of 5-diazariboflavin (4 µM) and 3 mM EDTA under anaerobic conditions (Frago et al., 2010). Spectra were recorded in 20 mM Tris-HCl, pH 7.6, 100 mM NaCl.
Protein crystallization and data collection
GvDTR and GvFdx1 proteins were co-crystallized at a 1.5 molar excess of GvFdx1 over GvDTR. Crystals were grown in sitting drops at 293 K using the vapor diffusion method by mixing the protein in buffer 10 mM Tris-HCl, pH 8.0, with an equal volume of mother liquor consisting of 40% (v/v) PEG-300, 100 mM sodium cacodylate-HCl, pH 6.5, and 200 mM sodium acetate. Diffraction data were collected at 100 K with monochromatic X-rays of 0.916 Å wavelength using synchrotron radiation at the Diamond Light Source beamline I04-1. Diffraction intensities were indexed and integrated using the autoPROC software (Vonrhein et al., 2011) that makes use of the STARANISO program (Tickle et al., 2018). Experimental data were truncated according to an anisotropic ellipsoidal resolution of 2.23 (a), 2.90 (b), and 2.39 (c) Å, using as threshold I/sigI ≥ 1.2. The anisotropically truncated data were phased by molecular replacement using the Phaser program (McCoy et al., 2007) and the crystal structures of GvDTR (PDB code 5J60) and Fdx1 from Synechocystis (PDB code 1OFF) as templates. The structure was iteratively refined by alternating visual inspection and manual modeling using Coot (Emsley et al., 2010) with automated refinement with the Phenix crystallographic software suite (Adams et al., 2010). Rigid body, gradient-driven positional, simulated annealing, restrained individual isotropic B-factor, and TLS (Winn et al., 2001) were used for structure refinement. The crystallographic and refinement statistics are summarized in Supplemental Table S1. Molecular representations were generated using PyMOL (Schrödinger). Electrostatic potential surfaces were computed using the APBS (Baker et al., 2001) plug-in for PyMOL (Schrödinger, 2015). Homology models were built with SwissModel (Schwede et al., 2003).
Accession numbers
Sequence data from this article can be found in the UniProt data library under the accession number Q7NMP6. X-ray crystallographic data from this article can be found in the PDB database under accession number 6XTF.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structural comparison of GvDTR with CaFFTR2.
Supplemental Figure S2. Fdx-dependent reduction of GvTrx-m via GvDTR.
Supplemental Figure S3. Fdx-dependent reduction of GvCP12 via GvDTR.
Supplemental Figure S4. SAXS analysis of GvDTR-WT and GvDTR-ΔTail.
Supplemental Figure S5. GvDTR–GvFdx1 interaction analysis monitored by analytical gel filtration.
Supplemental Figure S6. Conformation of GvDTR in the presence and absence of Fdx.
Supplemental Figure S7. Interaction of GvDTR with GvFdx.
Supplemental Table S1. X-ray crystallography data collection and refinement statistics.
Supplementary Material
Acknowledgments
We thank the beamline staff at the Diamond Light Source and ALBA Synchrotron for assistance in data collection and Jorge Herrero-Vicente for help with initial cloning experiments. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Funding
This work was supported by the Spanish Ministry of Science and Innovation—State Research Agency (MICINN) Grants [PID2019-110900GB-I00 to M.B., PID2019-109671GB-I00 to R.M.B., BFU-2016-78232-P to A.V.-C., and PID2019-103901GB-I00 to M.M.], and the Government of Aragón-FEDER [Grupo de Referencia Biología Estructural (E35_20R to M.M., and E45_17R to A.V.-C.)]. D.F.J. was supported by a predoctoral contract from the Junta de Castilla y León. A.G.-L. acknowledges support from CSIC Jae-Intro program.
Conflict of interest statement. The authors declare there is no conflict of interest.
R.M.B., B.B.B., and M.B. conceived and designed the research; R.M.B., G.G.-H., D.F.-J., M.M.-J., A.G.-L., A.V.-C., M.M., and M.B. conducted experiments; R.M.B., M.M.-J, A.V.-C., M.M., B.B.B., and M.B. analyzed data; R.M.B, B.B.B., and M.B. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Rubén M. Buey (ruben.martinez@usal.es) and Monica Balsera (monica.balsera@irnasa.csic.es).
References
- Adams PD, Afonine PV,, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arscott LD, Gromer S, Schirmer RH, Becker K, Williams CH (1997) The mechanism of thioredoxin reductase from human placenta is similar to the mechanisms of lipoamide dehydrogenase and glutathione reductase and is distinct from the mechanism of thioredoxin reductase from Escherichia coli. Proc Natl Acad Sci USA 94: 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M, Goetze T, Kovacs Bogdan E, Schürmann P, Wagner R, Buchanan BB, Soll J, Bolter B (2009) Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca(2+) and a stromal regulatory disulfide bridge. J Biol Chem 284: 2603–2616 [DOI] [PubMed] [Google Scholar]
- Balsera M, Uberegui E, Schürmann P, Buchanan BB (2014) Evolutionary development of redox regulation in chloroplasts. Antioxid Redox Signal 21: 1327–1355 [DOI] [PubMed] [Google Scholar]
- Balsera M, Uberegui E, Susanti D, Schmitz R, Mukhopadhyay B, Schürmann P, Buchanan B (2013) Ferredoxin:thioredoxin reductase (FTR) links the regulation of oxygenic photosynthesis to deeply rooted bacteria. Planta 237: 619–635 [DOI] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Lindell D, Chisholm SW (2015) Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13: 13–27 [DOI] [PubMed] [Google Scholar]
- Bottin H, Lagoutte B (1992) Ferrodoxin and flavodoxin from the cyanobacterium Synechocystis sp PCC 6803. Biochim Biophys Acta 1101: 48–56 [DOI] [PubMed] [Google Scholar]
- Buey RM, Arellano JB, López-Maury L, Galindo-Trigo S, Velázquez-Campoy A, Revuelta JL, de Pereda JM, Florencio FJ, Schürmann P, Buchanan BB, et al. M (2017a) Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase. Proc Natl Acad Sci USA 114: 12725–12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Fernández-Justel D, de Pereda JM, Revuelta JL, Schürmann P, Buchanan BB, Balsera M (2018) Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases. Proc Natl Acad Sci USA 115: 12967–12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Galindo-Trigo S, López-Maury L, Velázquez-Campoy A, Revuelta JL, Florencio FJ, de Pereda JM, Schürmann P, Buchanan BB, Balsera M (2017b) A new member of the thioredoxin reductase family from early oxygenic photosynthetic organisms. Mol Plant 10: 212–215 [DOI] [PubMed] [Google Scholar]
- Cassier-Chauvat C, Chauvat F (2014) Function and regulation of ferredoxins in the cyanobacterium, Synechocystis PCC6803: Recent advances. Life 4: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Schwendtmayer C, Schürmann P, Ramaswamy S, Eklund H (2000) Redox signaling in chloroplasts: cleavage of disulfides by an iron-sulfur cluster. Science 287: 655–658 [DOI] [PubMed] [Google Scholar]
- de Pereda JM, Lillo MP, Sonnenberg A (2009) Structural basis of the interaction between integrin α6β4 and plectin at the hemidesmosomes. EMBO J 28: 1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florencio F, Pérez-Pérez M, López-Maury L, Mata-Cabana A, Lindahl M (2006) The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynthesis Res 89: 157–171 [DOI] [PubMed] [Google Scholar]
- Frago S, Lans I, Navarro JA, Hervás M, Edmondson DE, De la Rosa MA, Gómez-Moreno C, Mayhew SG, Medina M (2010) Dual role of FMN in flavodoxin function: Electron transfer cofactor and modulation of the protein–protein interaction surface. Biochim Biophys Acta 1797: 262–271 [DOI] [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, et al. (2017) ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Cryst 50: 1212–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire E, Schon A, Velazquez-Campoy A (2009) Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol 455: 127–155 [DOI] [PubMed] [Google Scholar]
- Gontero B, Maberly SC (2012) An intrinsically disordered protein, CP12: jack of all trades and master of the Calvin cycle. Biochem Soc Trans 40: 995–999 [DOI] [PubMed] [Google Scholar]
- Hammel KE, Cornwell KL, Buchanan BB (1983) Ferredoxin/flavoprotein-linked pathway for the reduction of thioredoxin. Proc Natl Acad Sci USA 80: 3681–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Laakso LM (2016) Dali server update. Nucleic Acids Res 44: W351–W355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A (1985) Thioredoxin. Annu Rev Biochem 54: 237–271 [DOI] [PubMed] [Google Scholar]
- Hurley JK, Tollin G, Medina M, Gómez-Moreno C (2006) Electron transfer from ferredoxin and flavodoxin to ferredoxin: NADP+ reductase. In JH Golbeck, ed, Photosystem I: The Light-Driven Plastocyanin: Ferredoxin Oxidoreductase. Springer Netherlands, Dordrecht, pp 455–476 [Google Scholar]
- Kuriyan J, Krishna TSR, Wong L, Guenther B, Pahler A, Williams CH, Model P (1991) Convergent evolution of similar function in two structurally divergent enzymes. Nature 352: 172–174 [DOI] [PubMed] [Google Scholar]
- Laurent TC, Moore EC, Reichard P (1964) Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli b. J Biol Chem 239: 3436–3444 [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U (2008) Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA 105: 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon BW, Williams CH Jr, Ludwig ML (2000) Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science 289: 1190–1194 [DOI] [PubMed] [Google Scholar]
- Lodeyro AF, Ceccoli RD, Pierella Karlusich JJ, Carrillo N (2012) The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett 586: 2917–2924 [DOI] [PubMed] [Google Scholar]
- Macheroux P (1999) UV-Visible spectroscopy as a tool to study flavoproteins. In Chapman SK, Reid GA, eds, Flavoprotein Protocols. Humana Press, Totowa, NJ, pp 1–7 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M (2009) Structural and mechanistic aspects of flavoproteins: photosynthetic electron transfer from photosystem I to NADP+. FEBS J 276: 3942–3958 [DOI] [PubMed] [Google Scholar]
- Medina M, Martínez-Júlvez M, Hurley JK, Tollin G, Gómez-Moreno C (1998) Involvement of glutamic acid 301 in the catalytic mechanism of Ferredoxin-NADP+ Reductase from Anabaena PCC 7119. Biochemistry 37: 2715–2728 [DOI] [PubMed] [Google Scholar]
- Panjkovich A, Svergun DI (2018) CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics 34: 1944–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA (2015) Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 40: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porqué PG, Baldesten A, Reichard P (1970) The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulfate. J Biol Chem 245: 2371–2374 [PubMed] [Google Scholar]
- Puerto-Galán L, Pérez-Ruiz J, Ferrández J, Cano B, Naranjo B, Nájera V, González M, Lindahl AM, Cejudo F (2013) Overoxidation of chloroplast 2-Cys peroxiredoxins: balancing toxic and signaling activities of hydrogen peroxide. Front Plant Sci 4: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Inc . (2015) The PyMOL Molecular Graphics System. Version 2.0 Schrödinger LLC
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Asano T, Komori H, Sakurai T (2014) Role of the C-terminal extension stacked on the re-face of the isoalloxazine ring moiety of the flavin adenine dinucleotide prosthetic group in ferredoxin-NADP+ oxidoreductase from Bacillus subtilis. Plant Physiol Biochem 81: 143–148 [DOI] [PubMed] [Google Scholar]
- Shenton D, Grant CM (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susanti D, Loganathan U, Mukhopadhyay B (2016) A novel F420-dependent thioredoxin reductase gated by low potential FAD: a tool for redox regulation in an anaerobe. J Biol Chem 291: 23084–23100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ (1995) CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst 28: 768–773 [Google Scholar]
- Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G (2018) STARANISO. Global Phasing Ltd, Cambridge [Google Scholar]
- Vega S, Abian O, Velazquez-Campoy A (2015) A unified framework based on the binding polynomial for characterizing biological systems by isothermal titration calorimetry. Methods 76: 99–115 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G (2011) Data processing and analysis with the autoPROC toolbox. Acta Cryst D 67: 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CH, Arscott LD,, Müller S, Lennon BW, Ludwig ML, Wang P-F, Veine DM, Becker K, Schirmer RH (2000) Thioredoxin reductase. Eur J Biochem 267: 6110–6117 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.