Abstract
Ethiopian mustard (Brassica carinata) in the Brassicaceae family possesses many excellent agronomic traits. Here, the high-quality genome sequence of B. carinata is reported. Characterization revealed a genome anchored to 17 chromosomes with a total length of 1.087 Gb and an N50 scaffold length of 60 Mb. Repetitive sequences account for approximately 634 Mb or 58.34% of the B. carinata genome. Notably, 51.91% of 97,149 genes are confined to the terminal 20% of chromosomes as a result of the expansion of repeats in pericentromeric regions. Brassica carinata shares one whole-genome triplication event with the five other species in U’s triangle, a classic model of evolution and polyploidy in Brassica. Brassica carinata was deduced to have formed ∼0.047 Mya, which is slightly earlier than B. napus but later than B. juncea. Our analysis indicated that the relationship between the two subgenomes (BcaB and BcaC) is greater than that between other two tetraploid subgenomes (BjuB and BnaC) and their respective diploid parents. RNA-seq datasets and comparative genomic analysis were used to identify several key genes in pathways regulating disease resistance and glucosinolate metabolism. Further analyses revealed that genome triplication and tandem duplication played important roles in the expansion of those genes in Brassica species. With the genome sequencing of B. carinata completed, the genomes of all six Brassica species in U’s triangle are now resolved. The data obtained from genome sequencing, transcriptome analysis, and comparative genomic efforts in this study provide valuable insights into the genome evolution of the six Brassica species in U’s triangle.
With the release of this high-quality genome for Brassica carinata, complete genomes are now available for all six species in the Brassica U's triangle, providing rich resources for comparative and functional genomics analyses of Brassicaceae species.
Introduction
Brassica carinata (Ethiopian mustard) is in the family of Brassicaceae. The genus Brassica contains a diverse group of important vegetables, oilseed, and feed crops (Cheng et al., 2014; Yang et al., 2016). Crops of particular agricultural importance include three diploid species, namely, Brassica rapa (AA, 2n = 2x = 20), Brassica nigra (BB, 2n = 2x = 16), and Brassica oleracea (CC, 2n = 2x = 18), and three tetraploid species, namely, Brassica napus (AACC, 2n = 4x = 38), Brassica juncea (AABB, 2n = 4x = 36), and B. carinata (BBCC, 2n = 4x = 34). Their evolutionary relationships are described in U’s triangle model of Brassica, which proposes how the genomes of the three ancestral Brassica species, B. rapa, B. nigra, and B. oleracea, combined to give rise to the three allopolyploid species (Nagaharu, 1935). The common names of the three allopolyploid species are African rapeseed or Ethiopian mustard (B. carinata), European rapeseed (B. napus), and oilseed mustard (B. juncea; Young et al., 2012; Wang et al., 2014).
Brassica carinata originated in Sudan and Ethiopia in northeastern Africa and has a long history of cultivation that can be traced back to 4000 BC (Kumar et al., 1984). It has a very important role in agricultural production, producing seeds that can be used as condiments (Cardone et al., 2002). Brassica carinata possesses many desirable agronomic characteristics, such as heat, drought, and lodging tolerance (Ojiewo et al., 2014). Especially, it has strong disease resistance, including against white rust and downy mildew, and even has cancer-preventing potential (Tonguç and Griffiths, 2004; Sharma et al., 2016; Odongo et al., 2017; Raman et al., 2017). With these characteristics, B. carinata can adapt to a much wider range of environmental conditions than other Brassica species (Ojiewo et al., 2014). Recently, B. carinata has attracted increasing attention as an energy crop, and its cultivation for biofuel production has increased significantly over the past decade in Canada and the United States (Taylor et al., 2010; Ban et al., 2017). Of particular importance, B. carinata can grow in extremely harsh environments, such as hot, arid, or semiarid areas, which are not suitable for B. napus (Ban et al., 2017).
To date, genome sequencing has been completed for five of the six species in U’s triangle model. The genomes of B. rapa, B. oleracea, B. napus, B. nigra, and B. juncea have been published (Wang et al., 2011; Chalhoub et al., 2014; Liu et al., 2014; Parkin et al., 2014; Yang et al., 2016; Cai et al., 2017; Zhang et al., 2018; Paritosh et al., 2020). Each of the five sequenced Brassica species underwent a lineage-specific, whole-genome triplication (WGT; Cheng et al., 2013; Woodhouse et al., 2014). These genomes provide valuable resources for research on Brassica (Cheng et al., 2016; Su et al., 2018; Lu et al., 2019; Zou et al., 2019; Song et al., 2020).
Brassica carinata genome’s characterization will successfully complete the sequencing of all six species in U’s triangle model of Brassica, as well as provide a rich resource for comparative and functional genomics analysis of Brassicaceae species. Therefore, the completion of B. carinata genome sequencing will be a milestone in the study of Brassica. In this study, the genome of allopolyploid B. carinata was de novo assembled using the latest sequencing technologies, including Nanopore, PacBio, Illumina, and Hi-C sequencing. Brassica carinata offers a distinctive model to study the underlying genomic basis for selection in breeding for improvement. In addition to improving breeding in Brassica, this genome resource can contribute to comparative and functional genomics analysis in the broader context, with insights gained that could even be extended to other polyploid crops.
Results
Genome sequencing and assembly
To distinguish genomes and subgenomes of Brassica species, they were defined as the following: B. rapa as BraA; B. nigra as BniB; B. oleracea as BolC; B. juncea A subgenome as BjuA; and B subgenome as BjuB; B. napus A subgenome as BnaA and C subgenome as BnaC; and B. carinata B subgenome as BcaB and C subgenome as BcaC.
An advanced generation inbred line of B. carinata (“zd-1”) was selected for whole-genome sequencing. The estimated size of the B. carinata genome was 1,150.76 Mb by Kmer (Supplemental Figure S1, Table S1, and Note S1). In total, 327.86 Gb of data were obtained, including 64.24 Gb (59.11X) of Illumina shotgun reads, 83.09 Gb (76.45X) of Nanopore single-molecule long reads, and 180.53 Gb (166.11X) of Hi-C sequencing reads (Table 1; Supplemental Tables S2–S7 and Notes S1–S3). The assembled genome was 1,086.8 Mb, accounting for 94.44% of the estimated genome (Table 2). The assembled B. carinata genome was larger than that of the other two tetraploid species B. juncea (∼955 Mb) and B. napus “Darmor-bzh” (849.7 Mb) in addition to eight other latest reported B. napus genomes (1,001–1,033 Mb; Chalhoub et al., 2014; Yang et al., 2016; Song et al., 2020). In B. carinata, the contig N50 was 1.44 Mb, and the scaffold N50 was 60.00 Mb. The contig N50 was slightly smaller than that in the latest reported eight B. napus genomes (2.1–3.1 Mb); whereas the scaffold N50 was larger than that previously reported (46.68–57.88 Mb; Song et al., 2020).
Table 1.
Summary of the genome sequencing data of B. carinata
| Paired-end libraries | Insert size (bp) | Total data (Gb) | Read length (bp) | Coverage (×) |
|---|---|---|---|---|
| Illumina reads | 400 | 64.24 | 150 | 59.11 |
| Nanopore reads | – | 83.09 | – | 76.45 |
| HiC reads | – | 180.53 | 150 | 166.11 |
Table 2.
Statistics of the assembly quality of the B. carinata genome
| Terms | Contig length (bp) | Contig number | Scaffold length (bp) | Scaffold number |
|---|---|---|---|---|
| N50 | 1,437,473 | 172 | 60,001,024 | 8 |
| N60 | 941,266 | 268 | 58,209,656 | 10 |
| N70 | 565,390 | 415 | 56,612,299 | 12 |
| N80 | 344,764 | 662 | 55,501,767 | 13 |
| N90 | 169,672 | 1,102 | 53,286,246 | 15 |
| Longest | 10,952,811 | 1 | 72,379,882 | 1 |
| Total | 1,086,791,901 | 3,593 | 1,019,073,312 | 17 |
| Length≥1 Kb | 1,086,791,901 | 3,593 | 1,019,073,312 | 17 |
| Length≥2 Kb | 1,086,791,901 | 3,593 | 1,019,073,312 | 17 |
| Length≥5 Kb | 1,086,776,935 | 3,590 | 1,019,073,312 | 17 |
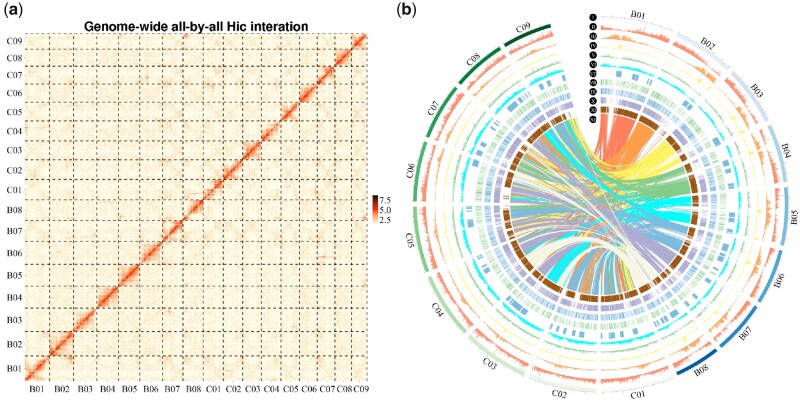
Sequences totaling 1,019.1 Mb were anchored to 17 chromosomes, accounting for 93.77% of the assembled genome (Figure 1A and Table 2; Supplemental Note S4). Of these sequences, 530.6 and 488.5 Mb were anchored onto the eight (BcaB) and nine (BcaC) pseudo-chromosomes, respectively (Supplemental Table S8). The assembled size of BcaB was smaller than that of the BjuB subgenome (547.5 Mb) but larger than that of the diploid BniB genome (396.9 Mb; Supplemental Figure S2). The assembled size of the BcaC subgenome was smaller than that of both the BnaC subgenome (525.8 Mb) and the diploid BolC genome (539.9 Mb). Brassica carinata was also compared with the eight B. napus genomes released recently.
Figure 1.
Hi-C map, chromosomal features, and functional annotation of the Brassica carinata genome. A, Hi-C map showing genome-wide all-by-all interactions between 17 chromosomes (B01–B08, C01–C09). B, I. 17 chromosomes of B. carinata. B01–B08 were from the BcaB subgenome, and C01–C09 were from the BcaC subgenome. II. Gene density. III–VII. Repeat sequences distribution on each chromosome: III. Gypsy; IV. Copia; V. Long interspersed nuclear elements (LINEs); VI. DNA repeat; VII. Simple sequence repeats (SSR). VIII. The gene expression was normalized as Fragments Per Kilobase of transcript sequence per Million base pairs. Gene expression levels (Log2FPKM) under drought treatment. IX. Gene expression levels (Log2FPKM) of control. X. Tandem genes distribution on each chromosome. XI. Orthologous genes distribution on each chromosome. XII. Lines connecting colinear blocks between BcaB and BcaC subgenomes, and the colors assigned according to each BcaB chromosome.
Genome annotation and assessment
In the B. carinata genome, 633.99 Mb of repetitive sequences was identified, which accounted for 58.34% of the assembled genome (Figure 1B; Supplemental Table S9 and Note S4). Long terminal repeats (LTRs) were the predominant transposable element (TE) family, accounting for 35.79% of the assembled genome. A similar trend in LTRs was also found in the other Brassica genomes using the same detection method. Among all LTRs, Copia- and Gypsy-type represented the two most abundant TE subfamilies, which accounted for 8.44 and 15.59% of the B. carinata genome, respectively (Supplemental Figure S3 and Supplemental Table S10). Moreover, the percentage of the Gypsy-type in the genome of B. carinata was higher than that in the other five species in U’s triangle. The analyses showed that LTR expansion occurred ∼0.86 million years ago (Mya) in B. carinata, which was slightly later than that in the other five Brassica species and Arabidopsis thaliana (Supplemental Figure S4). Based on the comparative analysis of 3′- and 5′-LTR terminal sequences, the expansion of LTRs in the six Brassica species in U’s triangle and A. thaliana occurred after their split. Notably, LTR expansion occurred at a similar time in the Brassica species, indicating their genome sizes increased in parallel (Supplemental Figures S5–S7).
Based on the assembled genome and full-length transcriptome, 97,149 genes were identified in B. carinata, which was fewer than that of B. napus (101,040) but more than that of B. juncea (79,593; Supplemental Figure S8, Supplemental Tables S11–S15 and Note S5). Notably, 13.99%, 27.54%, and 51.91% of the genes were located in the 5%, 10%, and 20% terminal regions, respectively, of each B. carinata chromosome, as a result of the expansion of repeats in pericentromeric regions (Supplemental Figure S9 and Supplemental Tables S16, S17). Approximately 97.14% (94,374) of B. carinata genes was annotated using six databases (Supplemental Table S18). In addition, 3.68 Mb of noncoding RNAs was detected, accounting for 0.25% of the B. carinata genome (Supplemental Table S19 and Note S4).
To validate genome assembly and annotation, the genome of B. carinata was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO; v4.1.4) and compared with the embryophyta_odb10 database. According to the results, 1,264 (91.64%) complete BUSCO genes were detected in the assembled B. carinata genome (Supplemental Table S20 and Note S4). Although this value was slightly smaller than that of the other five species in U’s triangle, it was larger than the most recently released plant genomes. Particularly, the scaffold N50 and genome integrity of B. carinata were much larger than those of the other five species in U’s triangle (Supplemental Table S12). These results indicated that a high-quality genome was obtained in this study, which could therefore be used for further comparative genomic analyses.
Comparative analysis of gene families in three tetraploid species
To investigate the evolution of gene families in B. carinata and the other two tetraploid species, a comparative analysis of the orthologous and paralogous gene families was conducted. A total of 88,537 genes were classified into gene families in B. carinata, 75,800 in B. juncea, and 90,486 in B. napus (Supplemental Figure S10a and Supplemental Table S21). Brassica carinata had 1,752 unique gene families, which was more than B. juncea (1,322) but fewer than B. napus (2,803). The three tetraploid species shared 26,708 gene families (Supplemental Figure S10b).
Furthermore, the gene families in B. carinata, B. juncea, and the pan-genome of B. napus were compared. The number of unique gene families was similar in B. carinata and B. juncea (Supplemental Figure S10, c and d and Supplemental Table S22). However, 4,931 unique gene families were detected in the pan-genome of B. napus, which was more than that in single B. napus.
Genome duplication and divergence time estimation
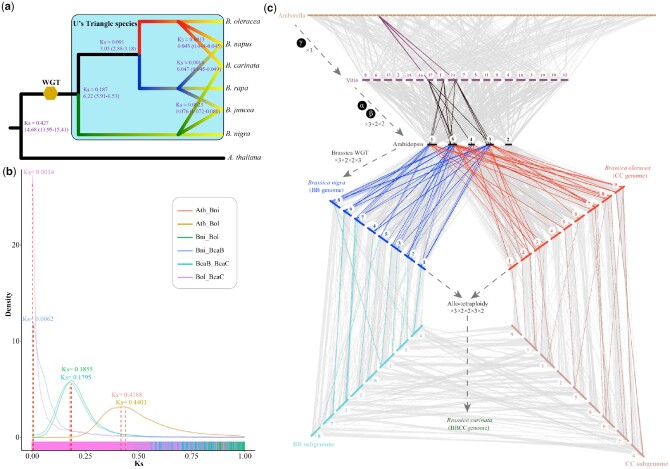
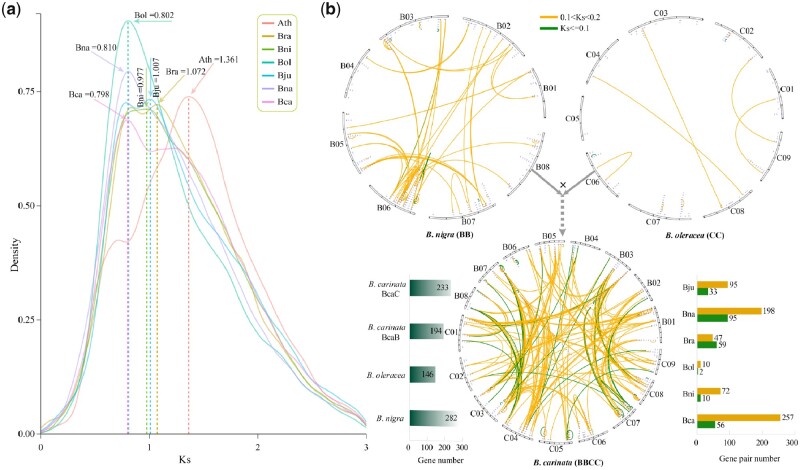
According to the transversion of four-fold degenerate site and synonymous mutation rate (Ks) distribution, B. carinata shared one WGT event with the other five species in U’s triangle (Supplemental Figure S11). Based on the single-copy gene families, we inferred that A. thaliana and Brassica species separated ∼29.50 Mya, and B. carinata and B. oleracea separated ∼6.08 (3.35–8.65) Mya (Supplemental Figure S12). The divergence time estimated by single-copy genes was accurate for most species. However, it might be not suitable to estimate the formation time of the three tetraploid species in U’s triangle according to the previously reports in B. juncea and B. napus genomes (Chalhoub et al., 2014; Yang et al., 2016).
Therefore, the times that BcaB and BcaC, respectively, diverged from the genomes of their two diploid species B. nigra and B. oleracea were calculated using colinear genes, as previously reported (Yang et al., 2016). Brassica carinata formed ∼0.047 (0.045–0.049) Mya, which is slightly earlier than B. napus but later than B. juncea (Figure 2, A and B; Supplemental Figure S13 and Supplemental Table S23). Brassica napus was deduced to form ∼0.043 (0.041–0.045) Mya, which is similar to the previous estimate of 0.038–0.051 Mya (Yang et al., 2016). The formation of B. juncea occurred ∼0.076 (0.072–0.080) Mya, which is slightly earlier than the previous estimate of 0.039–0.055 Mya (Yang et al., 2016).
Figure 2.
Divergence time estimation and recurrent genome duplications in B. carinata. A, Divergence time estimation among the six species in U’s triangle model of Brassica and Arabidopsis. The numbers on the nodes represent the Ks values and divergence time of the species (million years ago, Mya). The 95% confidence intervals of divergence time are in parentheses at each node. WGT indicates whole-genome triplication. B, Ks density plot of colinear genes for the two subgenomes (BcaB and BcaC) of B. carinata, B. nigra (Bni), B. oleracea (Bol), and Arabidopsis (Ath). C, Recurrent genome duplications in B. carinata. Genomic alignments are shown between the basal angiosperm Amborella trichopoda, the basal eudicot Vitis vinifera, the model Brassicaceae A. thaliana, and B. oleracea, B. nigra, and B. carinata.
Genome colinearity analysis of B. carinata and other selected species
The BcaB and BcaC subgenomes of B. carinata had high colinearity with the corresponding diploid BniB and BolC genomes. A total of 33,154 and 31,751 colinear gene pairs were detected between the BcaB and BcaC subgenomes, respectively, and their respective progenitor genomes (Supplemental Tables S24, S25; Supplemental Figure S14; and Note S6). To trace the genome evolution of B. carinata, a colinear analysis of Amborella trichopoda (basal angiosperm species), Vitis vinifera (basal eudicot species), A. thaliana, B. nigra, and B. oleracea was conducted (Figure 2C). The colinearity with Arabidopsis indicated the triplicated mesoploid structure of BcaB and BcaC subgenomes. With the recent allopolyploidy event, B. carinata became an aggregate 72× (3 × 2 × 2 × 3 × 2) genome multi-duplication following the origin of angiosperms. The evolutionary trajectory was similar with B. juncea and B. napus.
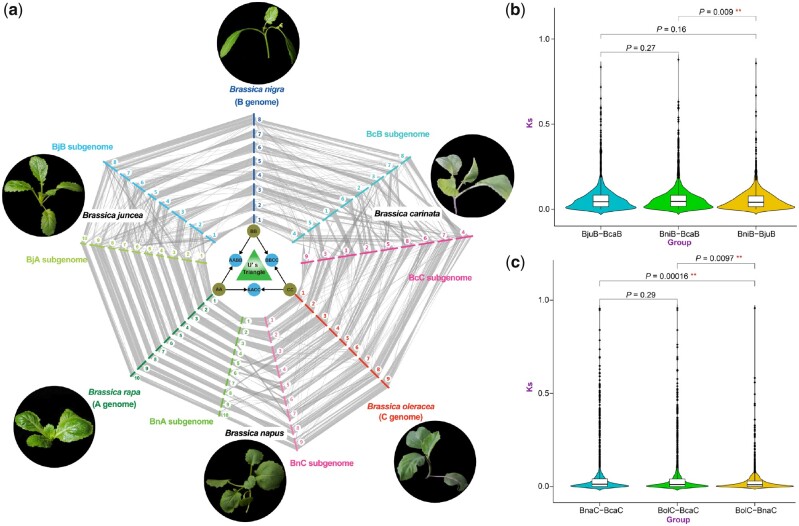
Furthermore, genome colinearity was determined among the six Brassica species in U’s triangle (Figure 3A; Supplemental Table S26). The BnaA and BnaC subgenomes of B. napus showed stronger colinearity than the two subgenomes of B. carinata or B. juncea. Many chromosomal rearrangements occurred between BcaB and BcaC of B. carinata, and a similar phenomenon was found between BjuA and BraA of B. juncea (Figure 3A). Colinear analysis between the two A subgenomes (BjuA and BnaA) and their corresponding diploid genome (BraA) showed higher genome colinearity than that between the two B subgenomes (BjuB and BcaB) and their diploid genome (BniB) or that between the two C subgenomes (BcaC and BnaC) and their diploid genome (BolC). These results could be explained by more chromosomal rearrangements or conversions occurring among the B subgenomes or the C subgenomes.
Figure 3.
Genome colinear analyses of Brassica carinata and five other Brassica species in U’s triangle. A, Global colinearity of the genomes of the six species in U’s triangle, including three diploid species, B. rapa (A genome), B. nigra (B genome), and B. oleracea (C genome) and three tetraploid species, B. napus (AACC, BnaA, and BnaC subgenomes); B. juncea (AABB, BjuA and BjuB subgenomes); and B. carinata (BBCC, BcaB and BcaC subgenomes). B, Boxplots of the relationships between two B subgenomes (BcaB and BjuB) and their corresponding diploid genomes (BniB) by calculating the Ks of colinear genes. A significance test was conducted using the R program. C, Boxplots of the relationships between two C subgenomes (BnaC and BcaC) and their corresponding diploid genomes (BolC) by calculating the Ks of colinear genes.
Furthermore, the relationships between two B subgenomes or two C subgenomes and their corresponding diploid genomes were estimated by calculating the Ks of colinear genes. The average Ks of colinear genes between BjuB and BniB was 0.0568, which was significantly smaller than that between BcaB and BniB (average Ks = 0.0675, P = 0.009; Figure 3B). This result indicates that the relationship between BjuB and BniB is closer than that between BcaB and BniB. Similarly, the average Ks value of colinear genes between BnaC and BolC was 0.0254, which was significantly smaller than that between BcaC and BolC (average Ks = 0.0527, P = 0.0097; Figure 3C). This result indicates that the relationship between BnaC and BolC is closer than that between BcaC and BolC. Finally, the nonet list was constructed, which contained nine genes (three genes from three diploid species, and six genes from six subgenomes of three tetraploid species), according to the colinearity of the six Brassica species in U’s triangle (Supplemental Table S26). In total, 1,461 nonets were identified, which provide a very important resource for the comparative genomics of Brassica.
Tandem gene analysis of two subgenomes in B. carinata
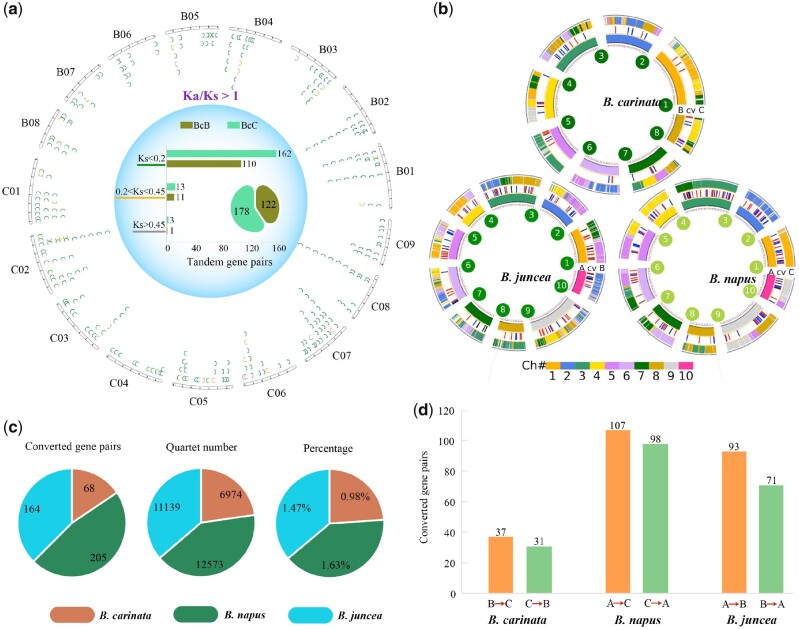
A total of 2,251 and 3,042 tandem gene pairs were identified in the BcaB and BcaC subgenomes of B. carinata, respectively (Supplemental Figures S15–S18). The Ks analyses showed a similar peak for BcaB and BcaC subgenomes. The average peak value was 0.025, and the corresponding tandem gene formation time was ∼0.83 Mya (Supplemental Figure S19 and Supplemental Table S27). Furthermore, positive selection analyses indicated that most tandem genes underwent purifying selection, whereas only 122 and 178 gene pairs were detected that underwent positive selection in BcaB and BcaC, respectively (Figure 4A; Supplemental Figures S20 and S21). Notably, there were 224 and 380 gene pairs with Ks = 0 in BcaB and BcaC, respectively, indicating they might have been produced recently, so no synonymous substitutions of bases have occurred (Supplemental Figure S22).
Figure 4.
Tandem genes and gene conversion analyses in the two subgenomes of B. carinata. A, Chromosomal distribution of tandem genes with different Ks values in the two subgenomes of B. carinata. The diagram shows tandem gene pairs with Ka/Ks >1. The green line represents Ks <0.2, the orange line 0.2 <Ks <0.45, and the gray line Ks >0.45. B, Distribution of gene conversions between the two subgenomes of B. carinata, B. juncea, and B. napus. The connected lines of different colors between the two circles of each species indicate a converted gene. The red lines indicate the inner circle subgenome is the donor; the blue lines indicate the outer circle subgenome is the donor; the black lines indicate the gene conversion was bidirectional between two subgenomes; and the gray lines indicate the direction of the gene conversion is unknown. C, Statistics of converted gene pairs, quartet gene number, and the percentage of converted genes pairs accounting for quartet number in the three tetraploid species B. carinata, B. napus, and B. juncea. D, The bar chart showed the number of converted gene pairs between two subgenomes in three tetraploid species.
Gene conversion between two subgenomes of three tetraploid Brassica species
Gene conversion is found in many plants during multiple rounds of genome polyploidizations (Wang and Paterson, 2011). It is a major driver of genome evolution and involves the exchange of DNA sequences from donor to acceptor (Wijnker et al., 2013; Gardiner et al., 2019).
On the basis of one tetraploid species and its related two diploid ancestral parents, the six Brassica species in U’s triangle were divided into three groups. In the first group, 6,974 quartets were constructed using BcaB–BcaC–BniB–BolC to study gene conversion between the two subgenomes of B. carinata (Figure 4, B–D; Supplemental Table S28). Similarly, 11,139 and 12,573 quartets were constructed between the two subgenomes of B. juncea and B. napus, respectively (Figure 4, C and D, Supplemental Tables S29, S30). Thus, the number of quartets in B. carinata was fewer than that in B. juncea and B. napus.
There were 68 converted gene pairs in B. carinata, which was fewer than that in B. juncea (164) and B. napus (205; Figure 4D; Supplemental Figures S22 and S23; Supplemental Tables S31 and S32). In B. carinata, the number of converted gene pairs from BcaB to BcaC was 37, which was greater than that from BcaC to BcaB (31). Notably, most converted genes were distributed in the two ends of Brassica chromosomes.
All 136 converted genes in B. carinata were colinear genes (Supplemental Figure S24). Furthermore, 14 and 18 converted genes belonged to tandem genes in the BcaB and BcaC subgenomes, respectively. Notably, four segmental gene conversions were detected, which contained 20 genes in B. carinata (Supplemental Table S33). Two tandem gene pairs were in the segmental gene conversion region, and the Ks of one pair (BcaB05g21732 vs. BcaB05g21733) was zero, indicating that it was a young tandem gene pair. These results will be a rich resource in studies to increase understanding of the mechanisms of gene conversion in Brassica species.
Colinear homoeologous gene expression dominance in the two subgenomes in B. carinata
To explore the expression patterns of the allopolyploid subgenomes, RNA-seq was used to analyze the expression of colinear homoeologous genes from BcaB and BcaC. The datasets of six tissues and from a drought treatment study were analyzed (Supplemental Figures S25, S26 and Supplemental Tables S34–S36). A total of 16,531 homoeologous gene pairs were detected between the two subgenomes (Supplemental Tables S37 and S38).
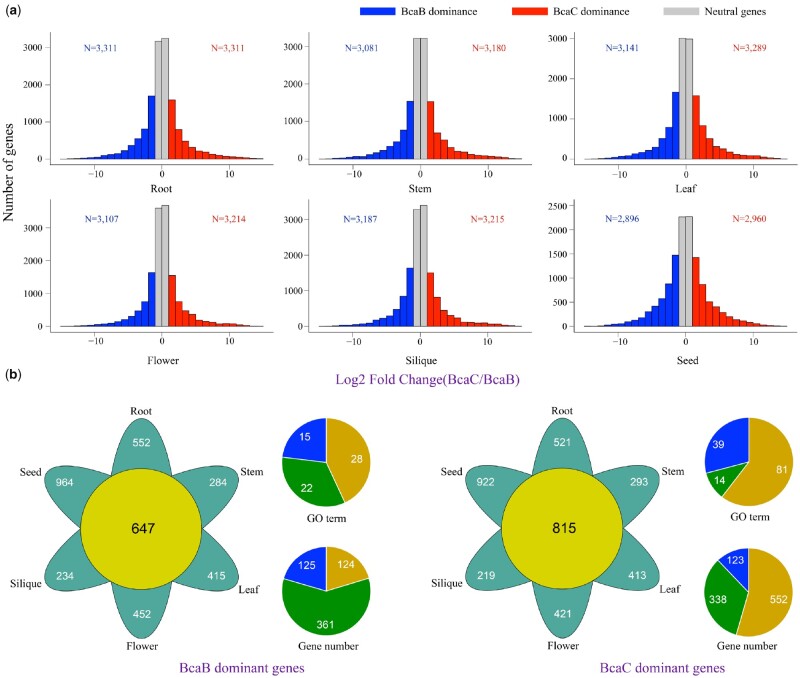
In the different tissues, 2,896–3,311 colinear gene pairs showed expression dominance between BcaB and BcaC, accounting for only ∼6.50% of whole-genome genes (Figure 5A; Supplemental Table S39). There was no significant dominance between the two subgenomes based on a double-sided binomial test with the P = 0.06–1.00 (Supplemental Table S39), which is a result similar to that with the other two polyploids in U’s triangle, B. napus and B. juncea (Chalhoub et al., 2014; Yang et al., 2016). On average, only 1.50% of genes displayed homoeolog expression dominance in all examined tissues, of which 647 (0.66%) and 815 (0.84%) genes were dominantly expressed toward BcaB and BcaC, respectively (Figure 5B; Supplemental Figure S27 and Supplemental Table S40).
Figure 5.
Colinear homologous gene expression dominance analyses between the two subgenomes of B. carinata. A, Homoeolog expression dominance analyses using the RNA-seq datasets of six tissues (top: root [left], stem, leaf [right]; bottom: flower [left], silique, seed [right]) of B. carinata. Blue and red represent the number of dominant genes in the BcaB and BcaC subgenomes, respectively; gray represents the neutral genes in the two subgenomes. B, Venn diagrams of the tissue-specific and common dominant genes in the two subgenomes (left, BcaB; right, BcaC). The pie diagrams show the enriched GO terms and the related gene numbers of the dominant genes in the BcaB and BcaC subgenomes. Orange indicates the number of terms and related genes in biological process; blue indicates those in molecular function; and green indicates those in cellular component.
The Gene Ontology (GO) database was used to perform functional enrichment analysis of the dominantly expressed genes shared by all six tissues (Figure 5B; Supplemental Figures S28–S30). The BcaB dominant genes were primarily enriched in cellular component biogenesis, mRNA cleavage, and ribonucleoprotein complex binding (Supplemental Table S41). The genes showing expression dominance in the BcaC subgenome were primarily enriched in organelle organization, cellular macromolecule metabolic process, and GTPase activator (Supplemental Table S42).
Identification of disease-related genes in the six Brassica species in U’s triangle
Generally, polyploid species possess stronger stress tolerances than their corresponding diploid species (te Beest et al., 2012). In this study, the disease-related gene family of nucleotide-binding site (NBS) was selected, and a comparative analysis was conducted with the five other Brassica species in U’s triangle (Supplemental Table S43). A total of 449 NBS family genes were identified in B. carinata, which was fewer than that in B. napus (463) but more than that in B. juncea (289; Supplemental Figures S31–S34; Supplemental Table S44). The BcaC subgenome contained 233 NBS genes, which was more than the corresponding diploid species B. oleracea (146). However, the BcaB subgenome had 194 NBS genes, which was fewer than the corresponding diploid species B. nigra (282; Supplemental Figures S31–S34; Supplemental Table S44).
The NBS family genes of B. carinata were further divided into three subtypes, including 281 Toll-interleukin-1 receptor-like NBS leucine-rich repeat (TNL), 146 coiled coil (CC)-NBS-LRR, and 22 resistance to powdery mildew8 (RPW8)-NBS-LRR subtypes (Supplemental Table S45). Sequence divergence (Ks) estimation showed the NBS gene family expansion in B. carinata occurred at ∼26.6 Mya (Ks = 0.798), which was later than that in the other five Brassica species and A. thaliana (Figure 6A). There were more NBS gene duplications in B. napus (190) and B. carinata (130) than in the other four Brassica species (Supplemental Figure S35). For gene loss, there were 31 NBS genes lost in B. carinata, which were fewer than that in B. juncea and the three diploid species (Supplemental Figure S35). In the latest ∼3.33 Mya (Ks ≤ 0.1), B. napus produced the most NBS genes (95) followed by B. rapa (59) and B. carinata (56; Figure 6B; Supplemental Figure S36). Notably, there were more B. carinata NBS genes (257) produced between 3.33 and 6.67 Mya (0.1 < Ks < 0.2) than that in the five other species in U’s triangle.
Figure 6.
Analyses of NBS (nucleotide-binding site) family genes in Brassica carinata. A, Ks density plot of NBS family genes in Bca, Bra, Bni, Bol, Bju, Bna, and Arabidopsis (Ath). B, Connection lines of homologous NBS family gene pairs in the two diploid species B. nigra (BB) and B. oleracea (CC) and the tetraploid species B. carinata (BBCC). The orange lines represent the gene pairs with 0.1 < Ks < 0.2 and the green lines those with Ks ≤ 0.1. The left bar chart shows the numbers of NBS family genes in B. nigra, B. oleracea, and B. carinata; the right bar chart shows the numbers of pairs of NBS family genes with Ks < 0.2 in the six species in U’s triangle.
Glucosinolate metabolic pathways in the six Brassica species in U’s triangle
Glucosinolates (GSLs) and their breakdown products have attracted much interest because of their important roles in plant defense and their anticancer properties (Wang et al., 2011; Yang et al., 2020). Arabidopsis and Brassica species share similar metabolic pathways for GSL biosynthesis and breakdown, and therefore, GSL metabolic pathway genes between Arabidopsis and the six Brassica species in U’s triangle were compared in order to explore their evolution.
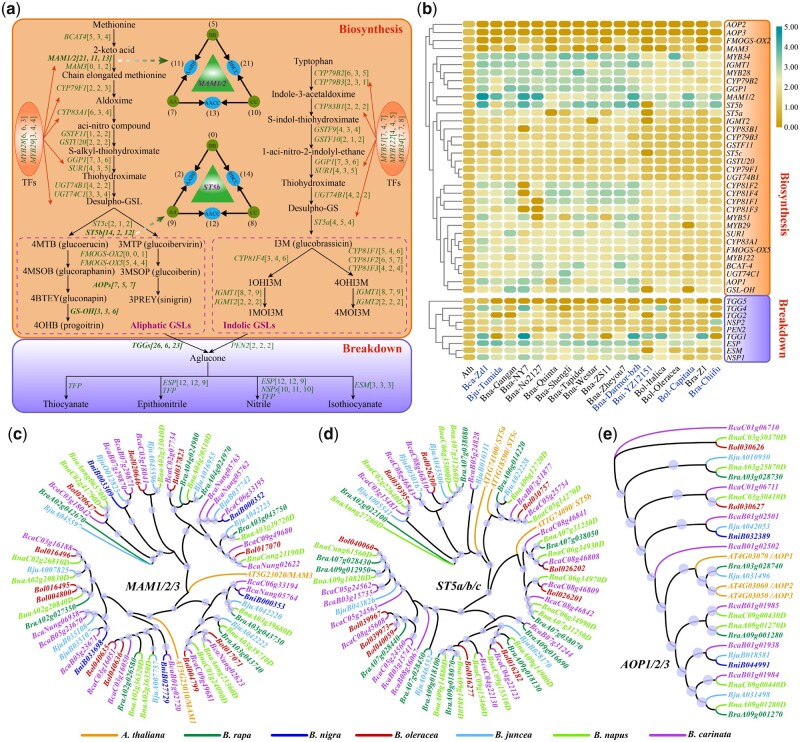
Arabidopsis GSL genes were used as seed, and 911 GSL biosynthesis and 276 GSL catabolism genes were detected in the six Brassica species in U’s triangle using Blastp (Figure 7A; Supplemental Table S46). Furthermore, 2,211 GSL biosynthesis and 575 GSL catabolism genes were also identified in other species in U’s triangle that had their genomes sequenced, including nine B. napus, two B. oleracea, and one B. rapa. The heat map showed that more methylthioalkylmalate (MAM1/2), ST5b, and TGG1 genes were detected in most species (Figure 7B). However, there were some exceptions, such as fewer ST5b genes in B. nigra (YZ12151) and fewer TGG1 genes in B. juncea (Tumida), B. napus (ZS11), and B. rapa (Chiifu) than in other species. For 2-oxoglutarate-dependent dioxygenase (AOP) genes, the AOP1 copy number was greater than that of the other two AOP genes in almost all Brassica species.
Figure 7.
Whole-genome comparison of genes involved in GSL metabolic pathways in B. carinata and five other Brassica species in U’s triangle. A, Aliphatic and indolic GSL biosynthesis and catabolism pathways in B. carinata and the two other tetraploid species (B. napus, B. juncea). The copy numbers of GSL biosynthetic genes in B. carinata, B. juncea, and B. napus are listed in square brackets. Two important amino acid chain elongation loci MAM1/2 and ST5b with significantly different numbers are highlighted in U’s triangle models. B, Heat map of the log2-transformed number of GSL metabolic pathway-related genes in Arabidopsis, the six species in U’s triangle (blue font), and other Brassica species with genomes released. The range from yellow to blue represents a gradual increase in the number of genes. C–E, Maximum-likelihood trees of MAM, ST5, and AOP genes that were generated on the basis of amino acid sequences with 1,000 bootstrap repeats in Arabidopsis, B. carinata, and the five other species in U’s triangle.
Among the three tetraploid species, the significantly different copies were primarily found in several genes, such as MAM1/2, ST5b, and TGGs, among others (Figure 7, A, C, and D; Supplemental Table S46). Although AOP genes play important roles in the biosynthesis of gluconapin and sinigrin in GSLs (Zhang et al., 2015), their copies were similar in the three tetraploid species (Figure 7, A and E). In Arabidopsis, the MAM family contains three tandemly duplicated genes, and MAM1 and MAM2 catalyze the condensation reaction of the elongation cycles of aliphatic GSL biosynthesis, whereas MAM3 contributes to the production of all GSL chain lengths (Textor et al., 2007; Benderoth et al., 2008). In B. carinata, MAM1/2 genes experienced independent tandem duplication to produce 21 copies, which was more than that in the other two tetraploid species B. juncea (11) and B. napus (13; Figure 7, A and C). For ST5b, there were only two genes in B. juncea, which was much fewer than that in B. carinata (14) and B. napus (12) and even fewer than that in its ancient diploid parent species B. rapa (9; Figure 7, A and D). This phenomenon revealed that the ST5b gene has been significantly lost during evolution. Similarly, the copies of TGGs genes in B. juncea (6) were also much fewer than those in B. carinata (26) and B. napus (23; Figure 7A). All these variations in gene copy number could lead to differences in the amounts of GSL biosynthesized and its breakdown products in Brassica species.
In B. carinata, 210 biosynthesis and 72 breakdown GSL genes were identified, of which 60 (21.28%) were tandem genes and 116 (41.13%) were colinear genes between BcaB and BcaC subgenomes (Supplemental Figure S37). Therefore, tandem genes and genome triplication might have contributed to the generation of multiple copies of GSL genes in Brassica species, and this phenomenon is consistent with previous reports (Liu et al., 2014; Yi et al., 2015). Duplicated GSL genes might increase the quantitative variation in glucosinolates in Brassica species. Furthermore, the GSL gene expression map in six tissues was constructed, which could be used to explore the specific functions of GSL genes, especially of the duplicated genes in B. carinata (Supplemental Figures S38, S39 and Supplemental Table S47).
Discussion
A high-quality genome of allopolyploid B. carinata was constructed using the latest sequencing technologies. The B. carinata genome and transcriptome, together with genomic data for A. thaliana and the five other Brassica species in U’s triangle, provide abundant resources for both fundamental and applied research in the Brassicaceae. The Brassica U’s triangle consists of three diploid and three tetraploid species and is a good and widely used analytical polyploid model. With the completion of the B. carinata genome, the genome sequences of all six species of U’s triangle have been obtained. The rich data resources provided by this study will greatly promote comparative genomics and functional genomics among Brassicaceae species and even other polyploid species.
Duplicated genes produced by polyploidization are in ancestral chromosomal regions and might interact directly through DNA sequence recombination (Wang and Paterson, 2011). All six Brassica species in U’s triangle underwent a recent WGT event, which provided a high number of duplicated genes. Sequence exchange through gene conversion is highly conserved among plants, leading to genome stability and genetic diversity (Gardiner et al., 2019). Gene conversion occurs when paralogous sequences are aligned during recombination, especially in young tandem genes (Innan and Kondrashov, 2010; Harpak et al., 2017). Gene conversion can maintain the similarity of paralogous genes but accelerate their divergence from their related orthologous genes (Wang et al., 2007). The recombination and gene duplication might facilitate the evolution of novel genes that confer functional innovations to improve the biological fitness of plants (Wang and Paterson, 2011). In this study, the converted genes were identified between the two subgenomes of three tetraploid species. Further analysis indicated that all converted genes were tandem genes or colinear genes between the two subgenomes of B. carinata. Gene conversion may have a critical role in plants, but the understanding of gene conversion is far from comprehensive. Therefore, future efforts are needed to uncover the mechanisms of gene conversion, such as sequencing the four products of meiotic tetrads or additional populations of Brassica species.
Interspecific hybridization can rapidly alter genomes and possibly introduce beneficial phenotypic variation, which can be useful in crop germplasm improvement and innovation (Xiong et al., 2011; Soltis et al., 2016; Zou et al., 2018). The Brassica species in U’s triangle are an ideal model system to study the interspecific hybridization process. Thus, the completion of genome sequencing in those species will enable better understanding of the hybridization mechanism at the gene level.
There is an abundant variation in the Brassica species in U’s triangle due to speciation, geographic differentiation, hybridization, domestication, and artificial selection (Wang et al., 2011; Chalhoub et al., 2014; Liu et al., 2014; Yang et al., 2016). This rich variation provides strong potential for crop improvement through interspecific hybridization. Allopolyploid species can benefit from the genomic plasticity obtained by frequent homologous gene exchanges between their two subgenomes (Chalhoub et al., 2014; Yang et al., 2016; Zou et al., 2018). On the basis of the genomes of six Brassica species in U’s triangle, a systematic analysis of whole-genome genes was conducted, including the detection of gene conversions, tandem genes, colinear genes, and species-specific genes within and between the genomes or subgenomes of those species. This information will be a very useful resource in applied research to explore the variation in the Brassica species in U’s triangle.
Brassica carinata has rich genetic resources, with strong tolerance to various stresses and disease resistance, thus representing a potentially very good germplasm resource (Tonguç and Griffiths, 2004; Taylor et al., 2010; Fredua-Agyeman et al., 2014). However, B. napus has undergone approximately 400 years of cultivation and domestication history, resulting in a narrow genetic base and high linkage disequilibrium, especially in the C subgenome (Chalhoub et al., 2014; Hu et al., 2019). Therefore, introducing novel genetic diversity into the A or C subgenome is highly desirable to broaden the genetic base of B. napus. Several targeted gene transfer and genome introgressions into B. napus have been conducted through interspecific crosses, which broadened the B. napus genetic base and promoted improvement of agronomic traits (Chatterjee et al., 2016; Zou et al., 2018). Therefore, how to further broaden the genetic basis of B. napus and expand its germplasm resources has been the focus of breeders. The advantageous traits of B. carinata can meet the multiple objectives needed to improve rapeseed varieties. In this study, a comprehensive analysis of disease-related family genes in B. carinata and other Brassica species was conducted by combining data from genome sequences and transcriptomes. This valuable genetic resource can be used to better direct the breeding of disease tolerance in B. napus and other Brassica crops.
GSLs are a group of amino acid-derived secondary metabolites that are diverse in the Brassicaceae. GSLs and their degradation products have important roles in insect and pathogen interactions, as well as having anticarcinogenic properties beneficial to human health (Wang et al., 2011). In this study, with the integration of genomes and transcriptomes, glucosinolate biosynthesis-related genes were identified in the six Brassica species in U’s triangle, followed by further exploration of their expansion, positive selection, and expression pattern. Hopefully, the results of this study will provide a solid foundation from which to dissect the genetic mechanisms regulating the biosynthesis of glucosinolates in Brassica and ultimately information that can be applied to crop breeding.
Conclusions
The high-quality B. carinata genome sequence described in this study, in combination with comparative transcriptome analysis, was used to identify related genes in B. carinata and explore their expression, providing a solid foundation from which to dissect the genetic mechanisms regulating disease resistance and glucosinolate biosynthesis. The information obtained can be applied to crop breeding. More importantly, the release of the B. carinata genome will provide a rich gene resource for comparative and functional genomics among Brassicaceae species and even other polyploid species. Combined with the rich germplasm resources of B. carinata, the results of this study will benefit Brassicaceae research and guide Brassica breeders to efficiently use any valuable genes.
Materials and methods
Genome sequencing
Leaf samples were collected from B. carinata “zd-1” and processed for genomic DNA isolation and library construction. A total of 327.86 Gb of genome sequencing data was obtained. Three sequencing strategies were used (Supplemental Notes S1–S3). (1) Paired-end Illumina libraries (Illumina Inc., CA, USA) were constructed with 400-bp insertion fragments (PE150). In total, 64.24 Gb of data was obtained, which covered the genome at ∼59.11×. (2) Nanopore libraries were constructed according to the protocol and then sequenced using a Nanopore sequencer. In total, 83.09 Gb of data was obtained, with ∼76.45× coverage of the genome, and the mean and max length of reads were 18.28 and 169.45 kb, respectively. (3) A Hi-C library was constructed and sequenced using Illumina HiSeq with the sequencing strategy PE150. In total, 180.53 Gb of data was obtained, with ∼166.11× coverage of the genome.
Estimation of genome size
A K-mer method was used to estimate genome size before genome assembly. The K-mer was used to give discrete probability distributions of many possible K-mer combinations (Marcais and Kingsford, 2011). If the read length is L, and the K-mer length is K, then L − K + 1 K-mers can be obtained. The copy number of K-mer (17-mer) in clean reads was counted and divided by the total length of each sequence read. Then, the distribution of copy numbers was plotted, and the K-mer distribution was used to estimate the genome size. To avoid the influence of sequencing error, K-mers with low frequency (<3) were discarded. The algorithm was (N × (L – K + 1))/D = G, where N is the total reads number, L is the average length of reads, K is the K-mer length, D is the overall depth estimated from the K-mer distribution, and G is the estimated B. carinata genome size. According to the algorithm, the B. carinata genome size was approximately 1,150.0 Mb.
De novo genome assembly
All reads that passed quality control were used in B. carinata genome assembly. The software Canu v1.3 was selected for the error correction (Koren et al., 2017). The software wtdbg v1.2.8 was used to assemble the genome, with the parameters set as follows: (wtdbg-1.2.8 -tidy-reads 5,000 -k 0 -p 16 -S 3; wtdbg-cns -c 0 -k 16; kbm-1.2.8 -k 0 -p 15 -S 3 -O 0; Jain et al., 2018a, b; Ghurye et al., 2019). Next-generation sequencing data were mapped to the assembled genome using bwa v0.7.12 with default settings (Giannoulatou et al., 2014) and then iterative corrected twice by the Pilon program v1.22 to obtain the B. carinata genome data (Walker et al., 2014).
Hi-C technology improved genome assembly
Hi-C-assisted assembly technology is used to explore the relationship of the spatial position of whole chromatin DNA in the genome. Hi-C technology can map genomes to chromatin with high precision and constructs sequence position structure information for intact chromatin. The Hi-C analysis primarily included the following five steps (Supplemental Note S3): (1) library construction and sequencing; (2) data filtering and quality control; (3) data comparison, which was primarily performed using bowtie2 (v2.3.2; alignment mode: -end-to-end; parameter: -very-sensitive -L 30; Langmead and Salzberg, 2012); (4) effective data evaluation; and (5) assisted assembly using interactions, including clustering scaffold by hierarchical clustering, scaffold orientation using interactions, and heat map evaluation. Finally, genome assembly quality and completeness were assessed using BUSCO v4.1.4 analysis pipelines using embryophyta_odb10 as the database (Seppey et al., 2019).
Gene prediction
Gene prediction and annotation in the B. carinata genome were conducted by combining de novo prediction, homology prediction, and RNA-seq. For de novo prediction, model parameters were established to c using Augustus v3.0.2 (Stanke and Morgenstern, 2005), Genscan v1.0 (Burge and Karlin, 1997), and semi-HMM-based nucleic acid parser software (Korf, 2004). For homology-based prediction, Uniprot protein sequences from the eight sequenced plants, including B. juncea, B. napus, B. nigra, B. oleracea, B. rapa, A. thaliana, V. vinifera, and Oryza sativa, were initially mapped onto the B. carinata genome using TBLASTN with an E-value cutoff of 1e-5 (Altschul et al., 1990). The homologous genome sequences were aligned against the matching proteins using GeneWise v2.4.1 (Birney and Durbin, 2000) and Gemoma (Keilwagen et al., 2019) for spliced alignments. For RNA-seq, the reads were aligned to the reference genome by TopHat v2.0.10 to identify exon regions, acceptor sites, and splicing donor (Kim et al., 2013). Then, the alignments were assembled into transcripts by Cufflinks v2.2.1 (Trapnell et al., 2010), and the unigenes were aligned to the reference genome by the Program to Assemble Spliced Alignments to annotate protein-coding genes (Haas et al., 2008). Furthermore, full-length transcriptome sequencing (PacBio Iso-Seq) was used to identify the new gene and its iso-form and to achieve an accurate analysis of variable splicing and fusion genes (Zhou et al., 2019). Finally, a consensus gene set was produced by combining all predictions using the three methods with EvidenceModeler (Haas et al., 2008). To remove transposons, TransposonPSI using default parameters (https://sourceforge.net/projects/transposonpsi/) was used to align the consensus gene set to the transposon database.
Genome annotation
Genome annotation in this study included three primary steps: repeated sequence annotation, gene function annotation, and noncoding RNA annotation. In repeated sequence annotation, two strategies were used, homologous sequence alignment and de novo prediction. Homologous sequence alignment was mainly based on the Repbase and Mips-REdat database and used Repeatmasker and repeatproteinmask programs to identify repeat sequences (Tarailo-Graovac and Chen, 2009; Bao et al., 2015; Spannagl et al., 2017). Tandem repeats were predicted using the tandem repeats finder program (Benson, 1999). In de novo prediction, first, the repeat sequence database was constructed using RepeatModeler v1.0.11, and then repeat sequences of B. carinata was predicted using the Repeatmasker program (http://www.repeatmasker.org/RepeatModeler/). The LTRs were detected with the LTR_FINDER program (Xu and Wang, 2007). The simple sequence repeats were identified using MIcroSAtellite (MISA) software (Beier et al., 2017). Gene function annotation was conducted by comparing with known databases, including SwissProt, TrEMBL, [A]KEGG, COG, GO, and InterProscan. In noncoding RNA annotation, rRNA, snRNA, and miRNA were annotated by aligning ncRNA to the known noncoding RNA database using Rfam (Kalvari et al., 2018). The tRNA sequences in the genome were predicted using tRNAscan-SE (Chan and Lowe, 2019). The rRNA and its various subunits were predicted by building models using RNAmmer (Lagesen et al., 2007). Additional specific information about the genome annotation is in Note S4.
Transcriptome sequencing
PacBio Iso-Seq: Full-length transcriptome sequencing was conducted to assist the gene annotation of B. carinata (Supplemental Note S5). The PacBio single-molecule real-time sequencing technology provides high-quality full-length transcript information because of its long read-length advantage. The long-read length easily spans the complete sequence of a transcript from 5′-end to 3′-polyA tail, enabling accurate identification of the full length of the gene. In this study, six tissues (root, stem, leaf, flower, silique, and seed) of B. carinata were mixed and then full-length transcriptome sequencing was performed using PacBio technology.
Illumina RNA-seq: RNA-seq of B. carinata was also conducted. Six tissues and drought-treatment leaf samples were sequenced using Illumina Hiseq sequencing (Supplemental Note S5). The six tissues were root, stem, leaf, flower, silique, and seed. The drought treatment was for 24 h, and each sample was treated three times. RNA was isolated from the samples using a kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. There were three primary steps in the RNA-seq: (1) RNA sample detection; (2) library construction and inspection; and (3) sequencing and bioinformatics. Clean reads were aligned to the B. carinata genome by HISAT2 software (Kim et al., 2019). The Fragments Per Kilobase of transcript sequence per Million base pairs were used to calculate gene expression values using StringTie software (Trapnell et al., 2010; Pertea et al., 2015). DESeq2 software was used to conduct analyses of differentially expressed genes (DEGs; Love et al., 2014). The P-value was corrected by multiple hypothesis tests, and the range of the P-value was determined by controlling the false discovery rate (Korthauer et al., 2019). In this study, the |log2(fold-change)| ≥ 1 and P-adj <0.05 were used as the threshold for screening DEGs. The gene annotation and enrichment analyses were conducted using the GO with Q ≤0.05 (Mi et al., 2019).
Gene family identification, expansion, and contraction
Protein sequences from the whole genomes of B. carinata and eight species (A. thaliana, B. nigra, B. oleracea, B. rapa, Medicago truncatula, O. sativa, Populus trichocarpa, and V. vinifera) were selected for gene family analysis. Only the longest transcript was retained when a gene had multiple alternative splicing transcripts. Pairwise sequence similarities between all sequences were calculated using Blastp (E-value <1e-5). Gene family clusters in all species were identified using OrthoFinder (inflation value: 1.5) to obtain single- and multi-copy gene families (Emms and Kelly, 2019). The NBS gene family was identified using the pfam program (Punta et al., 2012). The glucosinolate-related genes were identified using the Blastp program (E-value <1e-5; identify >50%; score >200). The MEGA X and FastTree (v2.1) software were used to perform phylogenetic analysis using the maximum likelihood method with 1,000 bootstrap replications (Price et al., 2009; Kumar et al., 2018).
Divergence time estimation
First, the protein sequences of genes from single-copy gene families were aligned using Mafft v7.427 (Nakamura et al., 2018) and then converted into coding sequences (CDS) alignment using ParaAT v2.0 (Zhang et al., 2012). A better alignment of the CDS was created after filtering regions with poor alignment with the Gblocks program (Castresana, 2000). Then, the sequences were concatenated, and the four-fold degenerate sites were extracted using a Perl program. Last, the phylogenetic trees were constructed using concatenated sequences of fourfold-degenerate sites by RaxML v8.0.19 under the GTRGAMMA model with 1,000 as the bootstrap value (Stamatakis, 2014). Based on the phylogenetic tree and concatenated sequences alignment, the divergence time was estimated using MCMCtree of the PAML v4.9 package (Yang, 2007). The following time points obtained from the Timetree database (http://www.timetree.org/) were used for the time estimate correction: O. sativa and V. vinifera (115–308 Mya), A. thaliana and V. vinifera (107–135 Mya), M. truncatula and A. thaliana (98–117 Mya), A. thaliana and B. rapa (23.4–33.5 Mya), and B. nigra and B. rapa (4.6–21.9 Mya). The operating parameters of MCMCtree were set as burn-in = 5,000,000; sample number = 1,000,000; and sample frequency = 50. Divergence time was estimated for each node of a phylogenetic tree with 95% confidence intervals.
Inference of gene colinearity
First, the homologous genes were detected using the all-to-all search in Blastp within a species or between two species (E-value <1e-5). Based on the Blast results and general feature format files of the genomes, the colinear genes were identified using MCScanX according to the manual (Wang et al., 2012). The main parameters were set as follows: MATCH_SCORE: 50; MATCH_SIZE: 5; GAP_PENALTY: −1; OVERLAP_WINDOW: 5; and MAX GAPS: 25. The tandem genes were also detected using the detect_collinear_tandem_arrays program of MCScanX software (Wang et al., 2012).
Positive selection analysis
The probability of positive selection was estimated by calculating nonsynonymous mutation rate (Ka)/synonymous mutation rate (Ks), which is the ratio of the Ka to the Ks (Hurst, 2002). When Ka/Ks > 1, the gene was considered as under positive selection during the evolutionary process, and when Ka/Ks <1, the gene was considered as under negative selection. First, the CDS of colinear genes or tandem genes were aligned using Mafft v7.427 (Nakamura et al., 2018). Then, the alignment result was converted to an AXT file using the AXTConvertor program of KaKs_Calculator v2.0 (Wang et al., 2010). Finally, Ka, Ks, and Ka/Ks between colinear genes or tandem genes were calculated using the KaKs_Calculator program with the NG model (Wang et al., 2010).
Gene conversion detection
The gene conversion analyses were conducted according to a previous report (Zhuang et al., 2019). First, a quartet table was constructed using the direct homology between the tetraploid genomes and their corresponding ancestral species. For example, in B. carinata, the BcaB and BcaC subgenomes corresponded to the ancestors B. nigra and B. oleracea, respectively. Then, the quartet table “BcaB_BniB_BcaC_BolC” was constructed according to the colinear relationships (Supplemental Table S28). The quartet tables “BjuA_BraA_BjuB_BniB” and “BnaA_BraA_BnaC_BolC” for B. juncea and B. napus, respectively, were constructed similarly (Supplemental Tables S29 and S30).
By comparing the similarity of homologous genes in the quartet table, the gene conversion in the three tetraploid species was studied. In this study, the sequence similarity of the homologous genes between the tetraploid two subgenomes was assessed. In addition, the sequence similarity of the directly homologous genes between each of the two subgenomes and their corresponding ancestral species was scored. Then, whether gene conversion occurred was judged by comparing the similarity of the two groups. For example, in the study of the B. carinata genome gene conversion model “BcaB_BniB_BcaC_BolC”, BcaB and BcaC were paralogous, whereas BcaB and BniB and BcaC and BolC were orthologous. In theory, orthologues are more similar than paralogues (Gabaldon and Koonin, 2013). However, the opposite can also occur, which was considered as gene conversion between paralogous genes. According to the differences in comparisons, gene conversion was divided into four types: BcaB to BcaC, BcaC to BcaB, no known, and reciprocal (Note S6). The conversion of two or more consecutive genes (gene gap = 0) was defined as segmental gene conversion. The same method was used for gene conversion analysis in the other two tetraploids.
Availability of data and materials
All materials and related data in this study are available upon request.
Accession numbers
The genome sequence and RNA-seq datasets of B. carinata have been deposited in the Genome Sequence Archive (Wang et al., 2017) in BIG Data Center (Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA002151, CRA002152, CRA002162, and CRA002177 that are publicly accessible at http://bigd.big.ac.cn/gsa. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAAMPC000000000. The version described in this article is version JAAMPC010000000. The assembled B. carinata genome and annotated datasets can be downloaded from our Brassica genomics database (http://brassicadb.bio2db.com).
Supplemental data
Supplemental Figure S1. 17-Kmer depth distribution curve of B. carinata used for genome size estimation.
Supplemental Figure S2. The genome and subgenome size of B. carinata and other five U’s triangle species.
Supplemental Figure S3. The (a) number, (b) length, and (c) percentage of Copia and Gypsy repeat sequences in genome of B. carinata and other five U’s triangle species.
Supplemental Figure S4. The long terminal repeat (LTR) analyses in Arabidopsis, B. carinata and other five U’s triangle species.
Supplemental Figure S5. The LTR analyses in B. carinata (Bca), B. nigra (Bni), and B. oleracea (Bol). (a) The density plot of LTR 3′ or 5′ end sequences divergence rate. b, The density plot of age of LTR 3′ or 5′ end sequences.
Supplemental Figure S6. The LTR analyses in B. juncea (Bju), B. rapa (Bra), and B. nigra (Bni). (a) The density plot of LTR 3′ or 5′ end sequences divergence rate. (b) The density plot of age of LTR 3′ or 5′ end sequences.
Supplemental Figure S7. The LTR analyses in B. napus (Bna), B. rapa (Bra), and B. oleracea (Bol). (a) The density plot of LTR 3′ or 5′ end sequences divergence rate. (b) The density plot of age of LTR 3′ or 5′ end sequences.
Supplemental Figure S8. The comparative analyses of gene information in the genome of B. carinata, and other eight related species.
Supplemental Figure S9. The statistics of gene distribution on chromosome of B. carinata, and other six selected species.
Supplemental Figure S10. Comparative analysis of gene family in three tetraploid species, B. carinata B. juncea, and B. napus or pan-genome of B. napus.
Supplemental Figure S11. The density plot of 4dtv sites.
Supplemental Figure S12. The divergence time estimation between B. carinata, and other eight representative species.
Supplemental Figure S13. Ks density plot analyses.
Supplemental Figure S14. The bar chart of the retain and lost syntenic gene pairs among three diploid species, three tetraploid species, and B. carinata.
Supplemental Figure S15. The number of tandem gene pairs on each chromosome of two subgenomes in B. carinata.
Supplemental Figure S16. The expression analyses of tandem genes under drought treatment.
Supplemental Figure S17. The scatter plot of DEGs.
Supplemental Figure S18. The scatter plot of DEGs.
Supplemental Figure S19. The Ks density plot for the tandem genes of two subgenomes of B. carinata.
Supplemental Figure S20. The dot plot of comparative analyses of collinear genes and tandem genes in B. carinata.
Supplemental Figure S21. The positive selection and expression analyses for the tandem genes of two subgenomes of B. carinata.
Supplemental Figure S22. Tandem genes analysis in the two subgenomes of Brassica carinata.
Supplemental Figure S23. The expression analyses of converted genes under drought treatment in B. carinata and its two subgenomes.
Supplemental Figure S24. The common and specific genes among four datasets, including converted genes, colinear genes, BcaB and BcaC tandem genes in B. carinata.
Supplemental Figure S25. The expression analyses of collinear genes under drought treatment.
Supplemental Figure S26. The scatter plot of DEGs.
Supplemental Figure S27. The venn diagram of homoeolog expression dominance genes between two subgenomes.
Supplemental Figure S28. The GO annotation of homoeolog expression dominance genes between two subgenomes in six tissues.
Supplemental Figure S29. The Ks density plot for the collinear genes of two subgenomes of B. carinata.
Supplemental Figure S30. The positive selection and expression analyses for the collinear genes of two subgenomes of B. carinata.
Supplemental Figure S31. Phylogenetic relationship analyses of the NBS gene family.
Supplemental Figure S32. Phylogenetic relationship analyses of the NBS gene family.
Supplemental Figure S33. Phylogenetic relationship analyses of the NBS gene family.
Supplemental Figure S34. Phylogenetic relationship and expression analyses of the NBS gene family in two subgenomes (BcaB and BcaC) of B. carinata.
Supplemental Figure S35. The duplication or losses analyses of NBS family genes.
Supplemental Figure S36. The connection lines of homologous NBS family gene pairs in the (a) Arabidopsis, (b) B. rapa, (c) B. juncea, and (d) B. napus.
Supplemental Figure S37. The common and specific genes among GSLs genes, tandem genes, and collinear genes in B. carinata.
Supplemental Figure S38. The hierarchical clustering heatmap of GSLs biosynthesis genes using FPKM values of six tissues (root, seed, flower, silique, leaf, and stem) by RNA-seq.
Supplemental Figure S39. The hierarchical clustering heatmap of GSLs breakdown genes using FPKM values of six tissues (root, seed, flower, silique, leaf, and stem) by RNA-seq.
Supplemental Table S1. The summary of second-generation sequencing data using for B. carinata genome survey, and the k-mer analyses were used for genome size estimation.
Supplemental Table S2. Summary of B. carinata genome by Nanopore sequencing.
Supplemental Table S3. Statistics of initial assembled B. carinata genome.
Supplemental Table S4. The statistics of assembled B. carinata genome after twice corrected by Pilon with NGS data.
Supplemental Table S5. The summary of the Hi-C sequencing data of B. carinata.
Supplemental Table S6. The Statistic of valid paired-end reads of Hi-C sequencing data.
Supplemental Table S7. The statistics of the scaffold clustering of B. carinata using LACHESIS software.
Supplemental Table S8. The statistics of the 17 chromosomes length and contained scaffold number of the assembled B. carinata genome.
Supplemental Table S9. The statistics of the different type repeat sequences of B. carinata genome.
Supplemental Table S10. Summary of Copia and Gypsy content in six U’ triangle species of Brassica.
Supplemental Table S11. The statistics of full-length RNA sequencing of B. carinata. The sequencing sample includes root, stem, leaf, flower, and silique of B. carinata.
Supplemental Table S12. The genome information of B. carinata and other five U’ triangle species of Brassica.
Supplemental Table S13. The genome information of B. carinata and 10 species used in this study.
Supplemental Table S14. Annotation statistics of protein-encoded genes in B. carinata, and compared with other 8 related species.
Supplemental Table S15. The statistics of the gene structure of other eight representative species for comparative analyses with B. carinata.
Supplemental Table S16. The statistics of gene distribution on each chromosome of B. carinata, and other 6 selected species.
Supplemental Table S17. The summary of gene distribution on all chromosome of B. carinata, and other 6 selected species.
Supplemental Table S18. Statistics of gene functional annotations using six databases for B. carinata genome.
Supplemental Table S19. The statistics of non-coding RNA in B. carinata genome.
Supplemental Table S20. The assessment of B. carinata and other 5 U’s triangle whole-genome genes by BUSCO software (v4.1.4) comparing with embryophyta_odb10 database.
Supplemental Table S21. The statistics of gene family in B. carinata and other two tetraploid species.
Supplemental Table S22. The statistics of gene family in B. carinata, B. juncea, and the pan-genome of B. napus.
Supplemental Table S23. The divergence time estimation of B. carinata and other representative species.
Supplemental Table S24. Protein quartet table listing the homologous gene sets among BniB (B. nigra), BolC (B. oleracea), BcaB and BcaC subgenomes of B. carinata and their putative orthologs in A. thaliana.
Supplemental Table S25. The statistics of the retain and lost syntenic gene pairs among three diploid species B. rapa (BraA), B. nigra (BniB), B. oleracea (BolC), three tetraploid species B. napus (BnaA and BnaC subgenomes), B. juncea (BjuA and BjuB subgenomes), and B. carinata (BcaB and BcaC subgenomes).
Supplemental Table S26. Protein nonet table listing the homologous gene sets among three diploid species B. rapa (BraA), B. nigra (BniB), B. oleracea (BolC), three tetraploid species B. napus (BnaA and BnaC subgenomes), B. juncea (BjuA and BjuB subgenomes), and B. carinata (BcaB and BcaC subgenomes).
Supplemental Table S27. Ka/Ks calculation and divergent time of the tandem gene pairs between subgenome B (BcaB) and subgenome C (BcaC) of B. carinata.
Supplemental Table S28. Protein quartet table used for the gene conversion analyses, which list the homologous gene sets among two diploid species B. nigra (BniB), B. oleracea (BolC), and tetraploid species B. carinata (BcaB and BcaC subgenomes).
Supplemental Table S29. Protein quartet table used for the gene conversion analyses, which list the homologous gene sets among two diploid species B. nigra (BniB), B. rapa (BraA), and tetraploid species B. juncea (BjuA and BjuB subgenomes).
Supplemental Table S30. Protein quartet table used for the gene conversion analyses, which list the homologous gene sets among two diploid species B. oleracea (BolC), B. rapa (BraA), and tetraploid species B. napus (BnaA and BnaC subgenomes).
Supplemental Table S31. The statistics of gene conversion between two subgenomes of B. carinata, B. juncea and B. napus.
Supplemental Table S32. The identification of gene conversion (GCV) between subgenomes BcaB and BcaC of B. carinata genome, between BjuA and BjuB of B. juncea, and between BnaA and BnaC of B. napus genome.
Supplemental Table S33. Segmental gene conversion (GCV) within three Brassica genomes.
Supplemental Table S34. The summary of data quality for the three replicates of six tissues and drought treatment of B. carinata using RNA-Seq.
Supplemental Table S35. The mapping statistic of reads to reference genome for the three replicates of six tissues and drought treatment in B. carinata using RNA-Seq.
Supplemental Table S36. The whole genome gene expression values (FPKM) of six tissues in B. carinata by RNA-seq.
Supplemental Table S37. The basic information and expression of homologous genes between BcaB and BcaC subgenomes of B. carinata.
Supplemental Table S38. Ka/Ks calculation and divergent time of the homologous gene pairs between BcaB and BcaC subgenomes of B. carinata.
Supplemental Table S39. Homoeolog expression dominance analyses in different tissues of B. carinata.
Supplemental Table S40. The common homoeolog expression dominance genes in six tissues of two subgenomes of B. carinata.
Supplemental Table S41. The GO enrichment analyses of common homoeolog expression dominance genes in six tissues of BcaB subgenomes of B. carinata.
Supplemental Table S42. The GO enrichment analyses of common homoeolog expression dominance genes in six tissues of BcaC subgenomes of B. carinata.
Supplemental Table S43. The number of NBS, AP2, and Hsf gene family in A. thaliana and six Brassica U' triangle species.
Supplemental Table S44. The differentially expressed analyses of NBS gene family in B. carinata under drought treatment and control (CK).
Supplemental Table S45. The classification of NBS gene family in B. carinata and other Brassica U' triangle species.
Supplemental Table S46. The number of genes involved in glucosinolate metabolism pathways in B. carinata and other U's triangle Brassica species.
Supplemental Table S47. The expression (FPKM) of genes involved in glucosinolate metabolism pathways in B. carinata.
Supplemental Note S1. Genome size estimation.
Supplemental Note S2. Nanopore sequencing.
Supplemental Note S3. Hi-C sequencing.
Supplemental Note S4. Comparative genomic analyses.
Supplemental Note S5. RNA-Seq.
Supplemental Note S6. Functional genomic analyses.
Funding
This work was supported by the State Key Special Program “Seven Main Crops Breeding” (2016YFD0101701 to C.Z.), the Priority Academic Program Development of Jiangsu Higher Education Institutions to C.Z, the National Natural Science Foundation of China (31801856 to X.S.), the Hebei Province Higher Education Youth Talents Program (BJ2018016 to X.S.), and the China Postdoctoral Science Foundation (2020M673188 to X.S.). The genome sequencing, Hi-C sequencing, and RNA-seq were performed with the help of Nextomics Biosciences.
Consent for publication
All authors are aware of the content and agree with the submission.
Conflict of interest statement. The authors declare no competing interests.
Supplementary Material
X.S., C.Z., and J.Y. conceived the project and were responsible for the project initiation. X.S., C.Z., J.Y., T.L., and X.W supervised and managed the project and research. Data generation was performed by C.Z., X.S., Y.W., D.X., Y.R., and N.L. Bioinformatics analyses were conducted by X.S., X.W., K.G., P.S., X.L., T.W., Q.Y., F.N., S.F., Q.P., and T.Y. The article was organized, written, and revised by X.S., C.Z., J.Y., T.L., and X.W. All authors read and revised the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plphys/pages/general-instructions) is: Changwei Zhang (changweizh@njau.edu.cn).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ban Y, Khan NA, Yu P (2017) Nutritional and metabolic characteristics of Brassica carinata Co-products from biofuel processing in dairy cows. J Agric Food Chem 65: 5994–6001 [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S, Thiel T, Munch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33: 2583–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderoth M, Pfalz M, Kroymann J (2008) Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochemistry Reviews 8: 255 [Google Scholar]
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Durbin R (2000) Using GeneWise in the Drosophila annotation experiment. Genome Research 10: 547–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Cai C, Wang X, Liu B, Wu J, Liang J, Cui Y, Cheng F, Wang X (2017) Brassica rapa Genome 2.0: a reference upgrade through sequence re-assembly and gene re-annotation. Mol Plant 10: 649–651 [DOI] [PubMed] [Google Scholar]
- Cardone M, Prati MV, Rocco V, Seggiani M, Senatore A, Vitoloi S (2002) Brassica carinata as an alternative oil crop for the production of biodiesel in Italy: engine performance and regulated and unregulated exhaust emissions. Environ Sci Technol 36: 4656–4662 [DOI] [PubMed] [Google Scholar]
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953 [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM (2019) tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 1962: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D, Banga S, Gupta M, Bharti S,, Salisbury PA, Banga SS (2016) Resynthesis of Brassica napus through hybridization between B. juncea and B. carinata. Theor Appl Genet 129: 977–990 [DOI] [PubMed] [Google Scholar]
- Cheng F, Mandakova T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Sun R, Hou X, Zheng H, Zhang F, Zhang Y, et al. (2016) Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet 48: 1218–1224 [DOI] [PubMed] [Google Scholar]
- Cheng F, Wu J, Wang X (2014) Genome triplication drove the diversification of Brassica plants. Hortic Res 1: 14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredua-Agyeman R, Coriton O, Huteau V, Parkin IA, Chevre AM, Rahman H (2014) Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus x B. carinata interspecific hybrids. Theor Appl Genet 127: 1305–1318 [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Koonin EV (2013) Functional and evolutionary implications of gene orthology. Nat Rev Genet 14: 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner LJ, Wingen LU, Bailey P, Joynson R, Brabbs T, Wright J, et al. (2019) Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol 20: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Phillippy AM, Koren S (2019) Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Comput Biol 15: e1007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoulatou E, Park SH, Humphreys DT, Ho JW (2014) Verification and validation of bioinformatics software without a gold standard: a case study of BWA and Bowtie. BMC Bioinformatics 15(Suppl 16): S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR (2008) Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpak A, Lan X, Gao Z, Pritchard JK (2017) Frequent nonallelic gene conversion on the human lineage and its effect on the divergence of gene duplicates. Proc Natl Acad Sci USA 114: 12779–12784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Zhang W, Zhang Y, Chang S, Chen L, Chen Y, et al. (2019) Reconstituting the genome of a young allopolyploid crop, Brassica napus, with its related species. Plant Biotechnol J 17: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18: 486. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Jain C, Koren S, Dilthey A, Phillippy AM, Aluru S (2018a) A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics 34: i748–i756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. (2018b) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46: D335–D342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Hartung F, Grau J (2019) GeMoMa: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol 1962: 161–177 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I (2004) Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthauer K, Kimes PK, Duvallet C, Reyes A, Subramanian A, Teng M, Shukla C, Alm EJ, Hicks SC (2019) A practical guide to methods controlling false discoveries in computational biology. Genome Biol 20: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Singh P, Singh DP, Singh H, Sharma HC (1984) Differences in osmoregulation in Brassica species. Ann Bot 54: 537–542 [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Wei L, Li X, Wang Y, Wu J, Liu M, et al. (2019) Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat Commun 10: 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Members BIGDC (2019) Database resources of the BIG Data Center in 2019. Nucleic Acids Res 47: D8–D14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilication. Jpn J Bot 7: 389–452 [Google Scholar]
- Nakamura T, Yamada KD, Tomii K, Katoh K (2018) Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34: 2490–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odongo GA, Schlotz N, Herz C, Hanschen FS, Baldermann S, Neugart S, et al. (2017) The role of plant processing for the cancer preventive potential of Ethiopian kale (Brassica carinata). Food Nutr Res 61: 1271527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojiewo C, Ebert A, Oluoch MO (2014) Brassica carinata (Brassicaceae). A versatile crop for tomorrow. In Nono-Womdim R, Achigan-Dako E, Pichop GN, Maundu P, Baudoin W, Lutaladio NB, Aphane J, Noorani A, Ghosh K, Hodder A, eds. Indigenous Fruit and Vegetables of Tropical Africa. A Guide to a Sustainable Production of Selected Underutilized Crops, Vol 1. Food and Agriculture Organization of the United Nations, Rome, pp 123–136 [Google Scholar]
- Paritosh K, Yadava SK, Singh P, Bhayana L, Mukhopadhyay A, Gupta V, et al. (2020) A chromosome-scale assembly of allotetraploid Brassica juncea (AABB) elucidates comparative architecture of the A and B genomes. Plant Biotechnol J 10.1111/pbi.13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IA, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, et al. (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC,, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Qiu Y, Coombes N, Song J, Kilian A, Raman H (2017) Molecular diversity analysis and genetic mapping of pod shatter resistance loci in Brassica carinata L. Front Plant Sci 8: 1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM (2019) BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962: 227–245 [DOI] [PubMed] [Google Scholar]
- Sharma BB, Kalia P, Yadava DK, Singh D, Sharma TR (2016) Genetics and molecular mapping of black rot resistance locus Xca1bc on chromosome B-7 in Ethiopian mustard (Brassica carinata A. Braun). PLoS One 11: e0152290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS (2016) Polyploidy: pitfalls and paths to a paradigm. Am J Bot 103: 1146–1166 [DOI] [PubMed] [Google Scholar]
- Song JM, Guan Z, Hu J, Guo C, Yang Z, Wang S, et al. (2020) Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannagl M, Nussbaumer T, Bader K, Gundlach H, Mayer KF (2017) PGSB/MIPS PlantsDB database framework for the integration and analysis of plant genome data. Methods Mol Biol 1533: 33–44 [DOI] [PubMed] [Google Scholar]
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33: W465–W467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Wang W, Li P, Zhang B, Li P, Xin X, et al. (2018) A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol Plant 11: 1360–1376 [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N (2009) Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Ch 4: Unit 4.10 [DOI] [PubMed] [Google Scholar]
- Taylor DC, Falk KC, Palmer CD, Hammerlindl J, Babic V, Mietkiewska E, et al. (2010) Brassica carinata—a new molecular farming platform for delivering bio-industrial oil feedstocks: case studies of genetic modifications to improve very long-chain fatty acid and oil content in seeds. Biofuels Bioprod Biorefin 4: 538–561 [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P (2012) The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109: 19–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, de Kraker J-W, Hause B, Gershenzon J, Tokuhisa JG (2007) MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol 144: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonguç M, Griffiths PD (2004) Transfer of powdery mildew resistance from Brassica carinata to Brassica oleracea through embryo rescue. Plant Breed 123: 587–589 [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J (2010) KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8: 77–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu J, Sun S, Liu B, Cheng F, Sun R, Wang X (2011) Glucosinolate biosynthetic genes in Brassica rapa. Gene 487: 135–142 [DOI] [PubMed] [Google Scholar]
- Wang N, Li F, Chen B, Xu K, Yan G, Qiao J, et al. (2014) Genome-wide investigation of genetic changes during modern breeding of Brassica napus. Theor Appl Genet 127: 1–13 [DOI] [PubMed] [Google Scholar]
- Wang X, Tang H, Bowers JE, Feltus FA, Paterson AH (2007) Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wang XY, Paterson AH (2011) Gene conversion in angiosperm genomes with an emphasis on genes duplicated by polyploidization. Genes (Basel) 2: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, et al. (2017) GSA: genome sequence archive. Genomics Proteomics Bioinformatics 15: 14–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E, Velikkakam James G, Ding J, Becker F, Klasen JR, Rawat V, et al. (2013) The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X (2014) Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A 111: 5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci U S A 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35: W265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, et al. (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48: 1225–1232 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hu Y, Yue Y, Pu Y, Yin X, Duan Y, Huang A, Yang Y, Yang Y (2020) Expression profiles of glucosinolate biosynthetic genes in turnip (Brassica rapa var. rapa) at different developmental stages and effect of transformed flavin-containing monooxygenase genes on hairy root glucosinolate content. J Sci Food Agric 100: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yi GE, Robin AH, Yang K, Park JI, Kang JG, Yang TJ, Nou IS (2015) Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies. Molecules 20: 13089–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HM, Srivastava P, Paret ML, Dankers H, Wright DL, Marois JJ, Dufault NS (2012) First report of sclerotinia stem rot caused by Sclerotinia sclerotiorum on Brassica carinata in Florida. Plant Dis 96: 1581. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu Z, Liang J, Wu J, Cheng F, Wang X (2015) Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J Exp Bot 66: 6205–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, et al. (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L (2012) ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419: 779–781 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhao Z, Zhang Z, Fu M, Wu Y, Wang W (2019) Isoform sequencing provides insight into natural genetic diversity in maize. Plant Biotechnol J 17: 1473–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang W, Chen H, Yang M, Wang J, Pandey MK, Zhang C, et al. (2019) The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat Genet 51: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Hu D, Mason AS, Shen X, Wang X, Wang N, et al. (2018) Genetic changes in a novel breeding population of Brassica napus synthesized from hundreds of crosses between B. rapa and B. carinata. Plant Biotechnol J 16: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Mao L, Qiu J, Wang M, Jia L, Wu D, et al. (2019) Genome-wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed. Plant Biotechnol J 17: 1998–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials and related data in this study are available upon request.