Abstract
Trichomes are specialized epidermal cells that act as barriers against biotic and abiotic stresses. Although the formation of trichomes on hairy organs is well studied, the molecular mechanisms of trichome inhibition on smooth organs are still largely unknown. Here, we demonstrate that the CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors inhibit the formation of trichomes on cotyledons in Arabidopsis (Arabidopsis thaliana). The tcp2/3/4/5/10/13/17 septuple mutant produces cotyledons with ectopic trichomes on the adaxial sides. The expression patterns of TCP genes are developmentally regulated during cotyledon development. TCP proteins directly interact with GLABRA3 (GL3), a key component of the MYB transcription factor/basic helix–loop–helix domain protein/WD40-repeat proteins (MYB–bHLH–WD40, MBW) complex essential for trichome formation, to interfere with the transactivation activity of the MBW complex in cotyledons. TCPs also disrupt the MBW complex–R3 MYB negative feedback loop by directly promoting the expression of R3 MYB genes, which enhance the repression of the MBW complex. Our findings reveal a molecular framework in which TCPs suppress trichome formation on adaxial sides of cotyledons by repressing the activity of the MBW complex at the protein level and the transcripts of R3 MYB genes at the transcriptional level.
TEOSINTE BRANCHED1/CYCLOIDEA/PCF transcription factors repress trichome formation on cotyledons by disrupting a trimeric transcriptional activation complex.
Introduction
Trichomes are specialized epidermis cells that act as physical barriers against insect herbivores, photoinhibition, ultraviolet light, and water loss (Hülskamp et al., 1994; Hauser, 2014). Trichomes are varied in morphologies in highly organ-specific and tissue-specific manners. As unicellular structures, Arabidopsis (Arabidopsis thaliana) trichomes have been used as ideal models to answer fundamental biological questions, including those regarding cell fate determination (Ishida et al., 2008). The presence and density of trichomes on different organs varies considerably in Arabidopsis. The adaxial surfaces of true leaves, cauline leaves, stems, and sepals are densely covered with trichomes in general. However, the abaxial surfaces of the first few true leaves, cotyledons, hypocotyls, petals, stamens, and pistils always lack trichomes. The organ-specific developmental schedules of trichomes suggest that the regulation of trichome cell fate determination and differentiation in plants is complicated. The Arabidopsis trichome cells are first specified from an epidermal cell precursor and then undergo rounds of endoreduplication to produce an elongated stalk and trichome branches (Hülskamp et al., 1994). Genetic analysis has identified many factors participating in leaf trichome initiation and patterning. Among them, the key factors form a MYB transcription factor/basic helix–loop–helix domain protein/WD40-repeat proteins (MYB–bHLH–WD40, MBW) complex consisting of the R2R3 MYB-related transcription factor GLABRA1 (GL1; Oppenheimer et al., 1991), the basic helix–loop–helix transcription factor GL3 (Payne et al., 2000) or ENHANCER OF GL3 (EGL3; Zhang et al., 2003), and the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1; Walker et al., 1999). The MBW complex directly activates the expression of GL2 (Rerie et al., 1994), which encodes a homodomain leucine zipper transcription factor essential for trichome development. The gl1 and ttg1 single mutants each exhibit severe deficiency in trichome initiation on leaf surfaces, stems, and sepals (Oppenheimer et al., 1991; Walker et al., 1999; Payne et al., 2000). The gl3 mutant only produces a modest reduction in trichome number on leaf surface (Payne et al., 2000) and the single egl3 mutant has normal trichomes (Zhang et al., 2003). However, the double mutant gl3 egl3 is completely hairless, demonstrating that the two bHLH transcription factors function redundantly in control of trichome formation in true leaves (Zhang et al., 2003). However, because the cotyledons of wild-type Arabidopsis have no trichomes, the glabrous phenotypes of these mutants could not indicate the functions of these genes in cotyledons. In addition to having or not having trichomes, cotyledons have other differences in comparison to the true leaves. For examples, cotyledons are initiated in embryo during seed development, while the leaves are from shoot apical meristem after seed germination. Cotyledons connect plant embryogenesis and juvenile vegetative growth, and function as storage organs of nutrients to support the early seedling growth, while leaves are the major photosynthetic organs at the later growth stage (Chandler, 2008). Also, cotyledons produce simpler vasculatures than the leaves (Meinke, 1992). Although the regulation of trichome formation on the leaves has been thoroughly illustrated, the molecular mechanisms causing glabrous cotyledons are neglected.
The MBW complex is negatively regulated by repressors, such as single-repeat R3 MYB transcription factors (Ishida et al., 2008), DELLAs, and JASMONATE-ZIM DOMAINs (JAZs; Qi et al., 2011, 2014). Single-repeat R3 MYB transcription factors include CAPRICE (CPC; Wada et al., 1997); TRIPTYCHON (TRY; Schellmann et al., 2002); ENHANCER OF TRY AND CPC1, 2, 3 (ETC1, ETC2, and ETC3; Esch et al., 2004; Kirik et al., 2004a, b; Tominaga et al., 2008); and TRICHOMELESS1, 2 (TCL1 and TCL2; Wang et al., 2007; Gan et al., 2011). R3-MYB repressors lack activation domains, and have redundant functions in an organ-specific manner. The single cyc mutant produces denser trichomes on true leaves. The triple etc1 try cpc and etc3 try cpc mutants have clustered trichomes on both true leaves and hypocotyls, while cpc etc1 etc3 produces trichomes on pedicels. The quadruple try cpc etc1 tcl1 mutant displays trichomes on siliques, and the quintuple try cpc etc1 etc3 tcl1 mutant produces ectopic trichomes on cotyledons (Wang et al., 2008). These findings suggest that the R3 MYB proteins are required for suppressing trichome formation in different glabrous organs. However, the molecular mechanisms by which R3 MYB proteins are regulated in glabrous organs to repress trichome formation are not well understood.
TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors play central roles in many aspects of plant development (Martín-Trillo and Cubas, 2010). TCP proteins are conserved in the plant kingdom and are classified into Classes I and II based on the conserved TCP domain (Cubas et al., 1999; Supplemental Figure S1). Class II TCP transcription factors are further divided into CINCINNATA (CIN)-like TCPs and TB1/CYC-like TCPs. Arabidopsis CIN-like TCPs include TCP2, TCP3, TCP4, TCP5, TCP10, TCP13, TCP17, and TCP24, and play essential roles in leaf development (Nath et al., 2003; Koyama et al., 2007, 2010, 2017; Efroni et al., 2008, 2013; Guo et al., 2010; Martín-Trillo and Cubas, 2010), thermomorphogenesis (Han et al., 2019; Zhou et al., 2019), photomorphogenesis (Dong et al., 2019), and other processes (Sarvepalli and Nath, 2011; Vadde et al., 2018; Challa et al., 2019; Lan and Qin, 2020). Overexpression of TCP4 causes a reduced number of trichomes and trichome branches in Arabidopsis (Efroni et al., 2008, 2013; Vadde et al., 2018, 2019). Recently, TCP4 has been reported to directly bind to the promoter regions of TCL1 and TCL2 and activate their expression in repression of the trichome formation on early true leaves (Vadde et al., 2019). Ectopic cotyledon trichomes have been observed in Pro35S:miR319A tcp3/4/5/10/13 in which a microRNA gene MIR319A targeting TCP2, TCP3, TCP4, TCP10, and TCP24 was overexpressed in the tcp3/4/5/10/13 mutant (Koyama et al., 2010). However, the molecular mechanisms of TCPs in control of trichome formation on cotyledons are still elusive.
In this article, by generating high-order tcp mutants, we discover that CIN-like TCP transcription factors play important roles in inhibiting Arabidopsis cotyledon trichome formation. The cotyledons of a septuple tcp2/3/4/5/10/13/17 mutant have ectopic trichomes on the adaxial sides of cotyledons. Biochemical and genetic analysis demonstrate that TCPs negatively regulate the transactivation activity of the MBW complex to repress GL2 expression by interacting with GL3 at the protein level. We further show that TCPs directly activate the expression of a group of trichome-negative regulators including CPC, ETC1, ETC3, and TCL2 in cotyledons at the transcriptional level. We suggest that TCPs may use different molecular mechanisms in suppressing trichome formation on cotyledons.
Results
CIN-like TCP transcription factors inhibit cotyledon trichomes
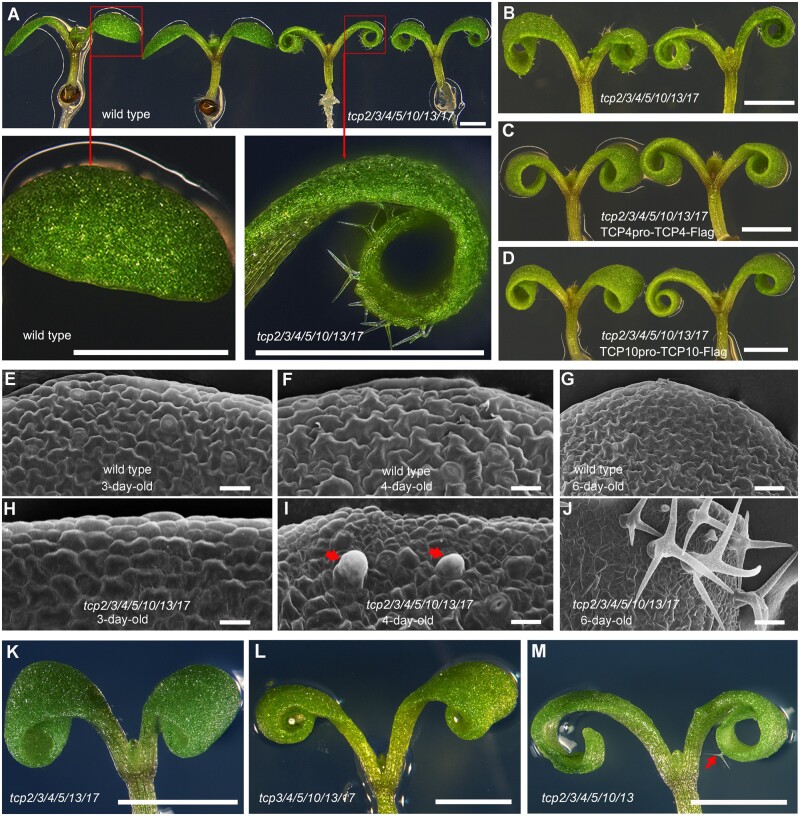
TCP transcription factors have dosage-dependent and highly redundant functions in regulating many aspects of plant development (Nath et al., 2003; Koyama et al., 2007, 2010, 2017; Efroni et al., 2008, 2013; Guo et al., 2010; Martín-Trillo and Cubas, 2010). To further reveal the important roles of CIN-like TCPs, we generated high-order tcp mutants and obtained a septuple tcp2/3/4/5/10/13/17 mutant (Figure 1A; Supplemental Figure S1). We observed ectopic trichomes produced on the adaxial sides of cotyledons in the tcp2/3/4/5/10/13/17 septuple mutant (Figure 1A). To test whether the loss of TCP functions could be responsible for the production of cotyledon trichomes in tcp2/3/4/5/10/13/17, we performed genetic complementation by generating TCP4pro-TCP4-Flag and TCP10pro-TCP10-Flag constructs, in which TCP4 or TCP10 genomic DNA was fused with a flag tag and was driven by a 2,821-bp TCP4 or a 986-bp TCP10 promoter, respectively. We transformed either TCP4pro-TCP4-Flag or TCP10pro-TCP10-Flag into tcp2/3/4/5/10/13/17. The ectopic trichomes disappeared on the cotyledons of tcp2/3/4/5/10/13/17 transformants with either TCP4pro-TCP4-Flag (Figure 1, B and C) or TCP10pro-TCP10-Flag (Figure 1, B and D), indicating that TCPs are very important for suppressing the trichome formation on cotyledons.
Figure 1.
CIN-like TCP transcription factors participate in repressing cotyledon trichome formation. A, Six-day-old seedlings of wild-type and tcp2/3/4/5/10/13/17 mutant plants. The red boxes indicate close-up views of the cotyledons. The ectopic trichomes are clearly produced on cotyledons of tcp2/3/4/5/10/13/17 mutants. Bars = 1 mm. B–D, Images of the 6-d-old seedlings of the TCP4pro-TCP4-Flag (C) or TCP10pro-TCP10-Flag (D) lines showing that TCP4 or TCP10 complements the ectopic trichomes on cotyledons of tcp2/3/4/5/10/13/17 (B). Bars = 1 mm. E–J, Scanning electronic micrographs of the cotyledon trichome cells of 3-, 4-, and 6-d-old wild-type plants (E–G) and tcp2/3/4/5/10/13/17 mutants (H–J). The red arrows indicate the initiated trichome stalks on the 4-d-old tcp2/3/4/5/10/13/17 cotyledons I, Bars = 50 μm. K–M, CIN-like TCP transcription factors have redundant and different contributions to inhibiting trichome formation on cotyledons. The tcp2/3/4/5/10/13 sextuple mutant (M) produced fewer trichomes (red arrow) on cotyledons than the tcp2/3/4/5/10/13/17 septuple mutant, while the tcp2/3/4/5/13/17 (K) and tcp3/4/5/10/13/17 (L) sextuple mutants displayed no trichomes on cotyledons. Bars = 1 mm.
To investigate the developmental stage at which the ectopic trichomes could be initiated on the adaxial sides of cotyledons in tcp2/3/4/5/10/13/17, we first observed embryonic development of the septuple mutant. The results showed that no trichomes were formed on the cotyledons of tcp2/3/4/5/10/13/17 mutants during embryogenesis, and the embryonic development of tcp2/3/4/5/10/13/17 displayed no obvious differences from that of the wild-type control (Supplemental Figure S2). We then observed the cotyledons after germination using scanning electronic microscopy. No trichome initiation was found on the cotyledons of 3-d-old tcp2/3/4/5/10/13/17 mutants (Figure 1, E and H). However, the trichome stalks emerged on the cotyledons of 4-d-old tcp2/3/4/5/10/13/17 mutants (Figure 1, F and I). Mature trichomes with branches were formed on the cotyledons of 6-d-old tcp2/3/4/5/10/13/17 mutants (Figure 1, G and J). No trichomes were observed on the cotyledons of wild-type controls (Figure 1, E–G). These ectopic trichomes on the cotyledons of tcp2/3/4/5/10/13/17 mutants were identical in appearance to those on true leaves (Figure 1J). The mature trichomes carried three branches and exhibited papillae on their surfaces (Figure 1J). These results indicate that tcp2/3/4/5/10/13/17 mutants produce cotyledon trichomes during cotyledon growth after germination.
To examine the possible different contributions of the seven TCPs in the suppression of ectopic cotyledon trichomes in tcp2/3/4/5/10/13/17 mutants, we crossed tcp2/3/4/5/10/13/17 mutants with wild-type plants to segregate different combinations of tcp multiple mutants. The tcp4 single mutant (Supplemental Figure S3B), tcp4/10 double mutant (Supplemental Figure S3C), tcp3/4/10 triple mutant (Figure S3D), and tcp3/4/5/10/13 quintuple mutant (Figure S3E) did not produce ectopic trichomes on cotyledons, indicating that the TCPs had highly redundant functions in inhibiting trichomes on cotyledons. We further obtained different combinations of tcp sextuple mutants. The tcp2/3/4/10/13/17 (Supplemental Figure S3F), tcp2/4/5/10/13/17 (Supplemental Figure S3G), tcp2/3/4/5/13/17 (Figure 1K), and tcp3/4/5/10/13/17 (Figure 1L) mutants did not generate ectopic trichomes on cotyledons, but the cotyledons of the tcp2/3/4/5/10/13 mutant (Figure 1M) exhibited trichomes, but the number was less than that of the septuple tcp2/3/4/5/10/13/17 mutant (Figure 1, A and B; Supplemental Figure S3H). This is consistent with the observation of the ectopic cotyledon trichomes of the tcp3/4/5/10/13 mutant transformed with Pro35S-miR319A (Koyama et al., 2010). These results indicate that the seven CIN-like TCPs contribute to the repression of trichome formation on cotyledons and that the contributions may be variant.
TCP4 interacts with transcription factor GL3 in vitro and in vivo
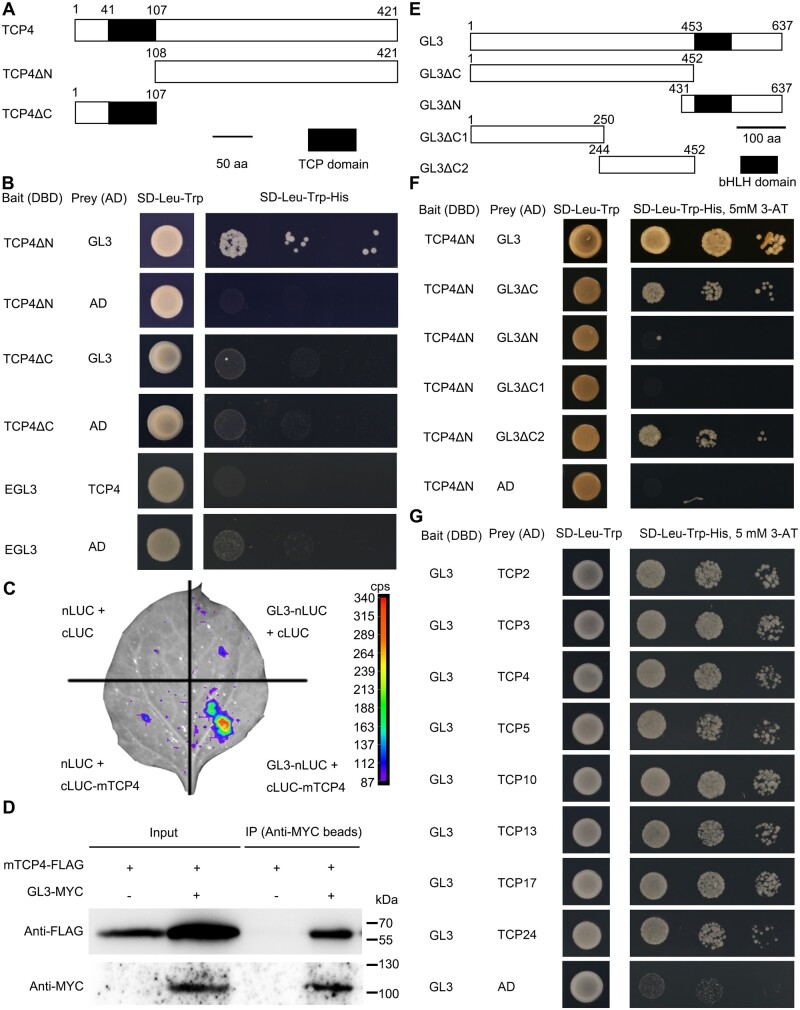
To decipher the molecular mechanisms by which TCPs repress trichome formation in cotyledons after seed germination, we used TCP4 as a bait to screen an Arabidopsis transcription factor library by yeast-two-hybrid assays (Ou et al., 2011). As the full-length TCP4 self-activated the reporter genes, we split TCP4 near the TCP domain into TCP4ΔC and TCP4ΔN. TCP4ΔC contains the TCP domain, and was made by deleting the C-terminal 313 amino acid (aa) residues. TCP4ΔN was obtained by deleting the N-terminal 107 aa residues (Figure 2A). We found that GL3, a key the basic helix-loop-helix (bHLH) transcription factor in the MBW complex, interacted with TCP4ΔN in yeasts (Figure 2B). As the EGL3 is a homolog of GL3 and the two proteins share >75% similarity in aa sequence (Zhang et al., 2003), and EGL3 showed no self-activation activity in yeasts, we tested whether EGL3 could also interact with TCP4 using the full-length EGL3 protein as a bait. The results showed that EGL3 did not interact with TCP4 in yeasts (Figure 2B), suggesting that GL3 and EGL3 may be divergent in functions due to the dissimilarity in their sequences.
Figure 2.
TCPs interacted with the bHLH transcription factor GL3. A, Schematic diagrams of TCP4 protein and truncated forms. The black rectangles represent the TCP domain. B, The yeast-two-hybrid assays of GL3, EGL3, and TCP4 proteins. The transformed yeasts were spotted on selection medium (SD–Leu–Trp–His) containing 5 mM 3-AT at dilutions of 10-, 100-, and 1,000-fold. The TCP4ΔN had weak self-activation activity in yeast, so 5 mM 3-AT (3-amino-1, 2, 4-triazole) was used to inhibit the self-activation for the cotransformed yeasts using TCP4ΔN as the bait. The selection medium used for TCP4ΔC and EGL3 as baits did not contain 3-AT. AD, activation domain; DBD, DNA-binding domain. C, Firefly luciferase complementation assay to test the interaction between TCP4 and GL3 in N. benthamiana. The miR319-resistant TCP4 protein was fused with nLUC at its C-terminus and GL3 protein was fused with cLUC at its N-terminus. nLUC and cLUC fused with GAL4BD were used as negative controls. D, Co-IP assay to confirm the interaction between TCP4 and GL3 in vivo. E, Schematic diagrams of GL3 protein and truncated forms. The black rectangles in the schematic diagrams of GL3 represent the bHLH domain. F, Yeast-two-hybrid assays of truncated GL3 and TCP4. The selection medium contained 5 mM 3-AT. G, Yeast-two-hybrid assays of GL3 and CIN-like TCPs (including TCP2, TCP3, TCP4, TCP5, TCP10, TCP13, TCP17, and TCP24). GL3 was fused with the DNA-binding domain (DBD) as bait. The transformed yeasts were spotted on selection medium (SD–Leu–Trp–His) containing 5 mM 3-AT at dilutions of 10-, 100-, and 1,000-fold.
To determine whether TCP4 can interact with GL3 in planta, we used the firefly luciferase complementation imaging assays using the 35S promoter to drive both the C-terminus of luciferase (cluc) fused with the miR319-resistant mTCP4, or the GL3 protein fused with the N-terminus of luciferase (nluc; Figure 2C). The fluorescence was obviously detected in the co-transformation of mTCP4 and GL3, but not in the controls, suggesting that TCP4 interacted with GL3 in vivo (Figure 2C). To further confirm the interaction, we preformed the co-immunoprecipitation (Co-IP) assays by co-expressing the flag-tagged mTCP4 and myc-tagged GL3 proteins using the transient expression system in Nicotiana benthamiana. The results showed that TCP4 was clearly pulled down by GL3 protein (Figure 2D). These results indicate that TCP4 interacted with GL3 in vivo.
To determine which region in GL3 could be required for the interaction with TCP4, we first divided GL3 into two parts near the bHLH domain as previously described (Wang and Chen, 2008). GL3ΔC was made by deleting the C-terminal region from 451 aa to the stop codon. GL3ΔN containing the bHLH domain was obtained by deleting the N-terminal region from 1 to 430 aa (Figure 2E). The yeast-two-hybrid assays suggested that GL3ΔC interacted with TCP4ΔN, while GL3ΔN containing the bHLH domain could not (Figure 2F). We further truncated GL3ΔC into GL3ΔC1 (from 1 to 250 aa) and GL3ΔC2 (from 244 to 452 aa). The results showed that GL3ΔC2, but not GL3ΔC1, interacted with TCP4ΔN (Figure 2F). These data demonstrate that TCP4 interacts with GL3 and that the TCP domain of TCP4 and the bHLH domain of GL3 are dispensable for their interaction.
To test whether the other members of CIN-like TCPs could interact with GL3, we performed yeast-two-hybrid assays using GL3 as the bait and CIN-like TCPs as the prey. The results showed that all the eight CIN-like TCPs interacted with GL3 (Figure 2G). These results are consistent with the highly redundant function of CIN-like TCP in repressing ectopic trichomes on cotyledons.
TCP4 and GL3 genes are developmentally regulated and have overlapping expression patterns in cotyledons
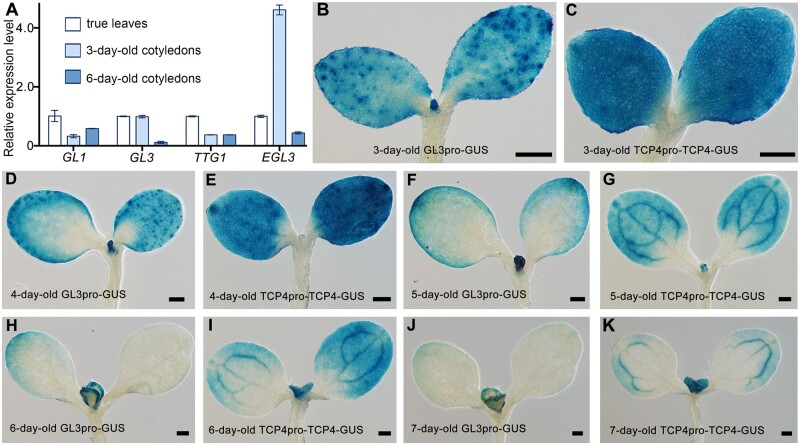
The phenotype of tcp2/3/4/5/10/13/17 and the interaction between TCP4 and GL3 implied that the genes encoding components of the MBW complex may also be expressed in cotyledons, but that the activity of the complex could be repressed by TCPs, causing glabrous cotyledons in wild-type Arabidopsis. To test this hypothesis, we first investigated the expression of GL1, GL3, TTG1, and EGL3 in cotyledons and true leaves. The results showed that all four genes were expressed in the cotyledons of 3-d-old and 6-d-old wild-type plants (Figure 3A). We then examined the expression pattern of GL3 in cotyledons by generating a GL3pro-GUS construct, in which the GUS reporter gene was driven by a 2,135-bp-long GL3 promoter. Similar GUS staining patterns were observed in eight independent transgenic GL3pro-GUS lines. Detailed GUS analysis showed that GL3 was expressed in whole cotyledons in the 3-d-old plants (Figure 3B). Interestingly, as cotyledons grew, the GUS staining faded away from the proximal to distal ends and from the central to marginal areas of cotyledons (Figure 3, D, F, H, and J). The expression of GL3 was mainly found in the marginal areas of cotyledons in 6- and 7-d-old plants (Figure 3, H and J).
Figure 3.
TCP4 and the members of the MBW complex are expressed ubiquitously in cotyledons after germination. A, Relative expression levels of GL1, GL3, TTG1, and EGL3 in 3- and 6-d-old wild-type cotyledons. The expression levels of corresponding genes in the fifth true leaves of 21-d-old wild-type plants were set as 1. ACTIN8 was used as the reference gene. The data are the mean (±SE) of three biological replicates. B–K, Expression patterns of GL3 and TCP4 in 3- to 6-d-old wild-type cotyledons with GUS histochemical staining. Bars = 200 μm. B and C, Expression pattern of GL3 (B) and TCP4 (C) in 3-d-old cotyledons. D and E, Expression pattern of GL3 (D) and of TCP4 (E) in 4-d-old cotyledons. F and G, Expression pattern of GL3 (F) and TCP4 (G) in 5-d-old cotyledons. H and I, Expression pattern of GL3 (H) and TCP4 (I) in 6-d-old cotyledons. J and K, Expression pattern of GL3 (J) and TCP4 (K) in 7-d-old cotyledons.
We next investigated the expression pattern of TCP4 in cotyledons. We generated TCP4pro-TCP4-GUS using a 2,821-bp-long TCP4 promoter to drive TCP4 genomic DNA fused with the GUS reporter gene. Five TCP4pro-TCP4-GUS transgenic lines displayed similar expression patterns overlapping with that of GL3 (Figure 3, B–K). Obvious GUS activity was detected in the whole cotyledons of young seedlings (Figure 3, C and E). Similar to the expression pattern of GL3, the accumulation of TCP4 proteins was also gradually restricted to the marginal areas of cotyledons as cotyledons grew (Figure 3, B–K). The overlapping expression patterns of TCP4 and GL3 in cotyledons further support the interaction of TCP4 and GL3 and are consistent with the development of ectopic trichomes at the marginal areas of cotyledons in the tcp2/3/4/5/10/13/17 septuple mutants.
TCP4 disrupts the transactivation activity of the GL1–GL3–TTG1 complex
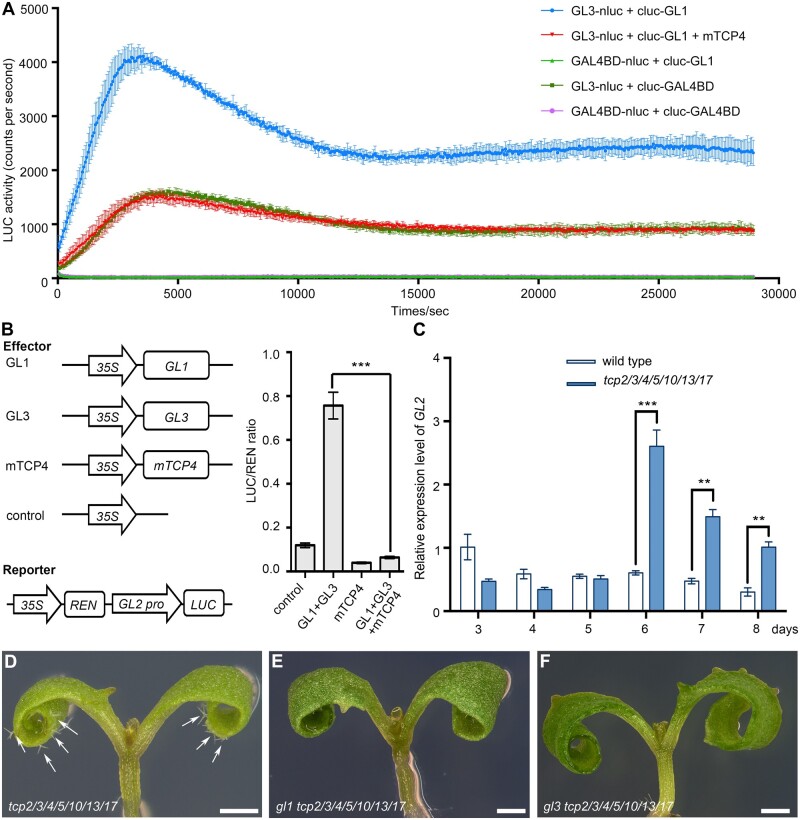
To explore the significance of the interaction between TCP4 and GL3, we tested whether TCP4 could disrupt the integrity of the GL1–GL3–TTG1 complex. We used the Arabidopsis protoplast transient expression system to detect the interaction kinetics between GL1 and GL3 based on firefly luciferase complementation assays. The counts of fluorescence detected per second were used to indicate the strength of the interaction. We first generated GL3-nluc and cluc-GL1 constructs in which GL3 or GL1 was fused with the nluc or cluc and driven by CaMV 35S promoter, respectively. The results showed that high levels of fluorescence were detected in the co-transformation of GL3-nluc and cluc-GL1, but not in the controls (Figure 4A). Interestingly, when the construct 35Spro-mTCP4, in which the microRNA319-resistant TCP4 gene was driven by the CaMV 35S promoter, was co-expressed with GL3-nluc and cluc-GL1, the levels of fluorescence were substantially decreased (Figure 4A), suggesting that TCP4 disrupted the interaction between GL1 and GL3.
Figure 4.
TCP proteins disrupt the function of the MBW complex in repressing trichome formation on cotyledons. A, Firefly luciferase complementation assay showed that TCP4 interfered with the interaction between GL1 and GL3 in Arabidopsis protoplasts. The combinations are listed at the upper right corner. The combinations including GAL4BD-nluc and cluc-GL1, GL3-nluc and cluc-GAL4BD, and GAL4BD-nluc and cluc-GAL4BD were used as negative controls. The data are the means (±se) of three biological replicates. B, The transient expression assay showed that the activation of GL2 expression by GL1/GL3 was repressed by TCP4. The schematic diagram shows the constructs used in the transient expression assays. The GL2pro-LUC reporter was cotransformed with corresponding constructs. The data are the means (±se) of four biological replicates. The asterisks represent Student’s t test significance compared with GL1+GL3. ***P <0.001. C, The relative expression levels of GL2 in the 3- to 8-d-old cotyledons of wild-type and tcp2/3/4/5/10/13/17 plants. The expression level of GL2 in 3-d-old wild-type cotyledons was set as one. ACTIN8 was used as the reference gene. The data are the means (±se) of three biological replicates. The asterisks represent Student’s t test significance. **P < 0.01; ***P < 0.001. D–F, The genetic complementation of ectopic trichome on cotyledons of tcp2/3/4/5/10/13/17 mutants by loss-of-function of GL1 or GL3. Cotyledons of 6-d-old gl1 tcp2/3/4/5/10/13/17 (E) and gl3 tcp2/3/4/5/10/13/17 (F) mutants were glabrous. White arrows indicate the trichomes on tcp2/3/4/510/13/17 cotyledons (D). Bars = 500 μm.
To test whether the interference of the GL1 and GL3 interaction by TCP4 affected the transactivation activity of GL1–GL3–TTG1 complex, we used Arabidopsis protoplasts to transiently express the reporter GL2pro-LUC, in which the Luciferase (LUC) gene was driven by a promoter of the GL2 gene, because GL2 is a direct target of GL1 and GL3 in control of trichome formation. The results showed that co-expression of 35Spro-GL3 and 35Spro-GL1 with GL2pro-LUC could obviously activate LUC activity as previously described (Qi et al., 2014; Figure 4B). Co-expression of the blank construct or 35Spro-mTCP4 with the GL2pro-LUC reporter could not activate LUC activity. However, when the 35Spro-mTCP4 was co-expressed with 35Spro-GL3, 35Spro-GL1, and GL2pro-LUC, the LUC activity was significantly decreased (Figure 4B). These results suggested that the transcription factor TCP4 might not directly regulate the expression of GL2, but possibly disrupted the transactivation activity of the MBW complex to repress the expression of GL2.
We then tested the expression level of GL2 in the cotyledons of tcp2/3/4/5/10/13/17 mutants at different cotyledon growth stages. The GL2 expression levels in the cotyledons of tcp2/3/4/5/10/13/17 mutants were significantly higher than those in wild-type controls after growing 6 d, consistent with the observation that ectopic mature trichomes started to appear in the cotyledons of the 6-d-old tcp2/3/4/5/10/13/17 mutants (Figure 4C). These data demonstrate that the transactivation activity of the MBW complex is negatively regulated to suppress the fate of trichome cells in Arabidopsis cotyledons via the interaction of TCP proteins with GL3.
Because the cotyledons of wild-type Arabidopsis are glabrous, it is difficult to determine by mutant analysis whether the signaling pathway of trichome formation exists in cotyledons. The ectopic trichomes on the cotyledons of tcp2/3/4/5/10/13/17 provide a good material for elucidating the trichome formation pathway in cotyledons using genetic analysis. We disrupted the key components of the signaling pathways including GL1, GL3, and GL2 in the background of tcp2/3/4/5/10/13/17, respectively. The results showed that the deletions of large fragments of GL1 and GL3 genomic regions caused early translation terminations of each protein (Supplemental Figure S4). The octuple mutants gl1 tcp2/3/4/5/10/13/17 (Figure 4E) and gl3 tcp2/3/4/5/10/13/17 (Figure 4F) produced no trichomes on cotyledons (Figure 4, D–F). We also observed that gl2 tcp2/3/4/5/10/13/17 still produced trichomes with no branches and in lower densities when GL2 gene was mutated in different alleles (Supplemental Figure S5). The abnormal and fewer trichomes in gl2 tcp2/3/4/5/10/13/17 were consistent with those observed in the true leaves of gl2 single mutant (Rerie et al., 1994). These data indicate that the signaling pathway of trichome formation exists in cotyledon and is required for the ectopic trichomes on tcp2/3/4/5/10/13/17 cotyledons.
TCP4 directly promotes the expression of R3 MYB genes
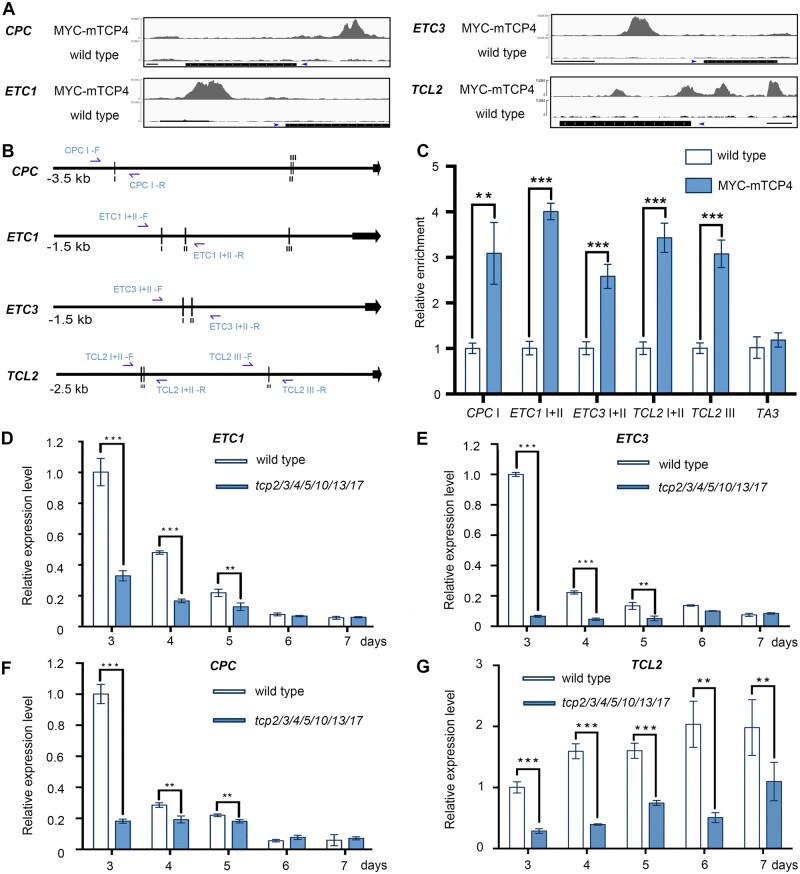
To further reveal the molecular mechanisms by which TCPs repress cotyledon trichomes, we re-analyzed our previous chromatin immunoprecipitation (ChIP)-seq data of TCP4 (Dong et al., 2019). Interestingly, we found that CPC, ETC1, ETC3, and TCL2 were bound by TCP4 (Figure 5A). There are seven R3 MYB proteins in Arabidopsis and they can be classified into two groups according to the similarity of their protein sequences (Gan et al., 2011; Wang and Chen, 2014). TCL1, TCL2, ETC2, and TRY are clustered into one group, ETC1, ETC3, and CPC are classified as the other group (Gan et al., 2011; Wang and Chen, 2014; Supplemental Figure S6A). The ChIP-seq data showed that TRY and ETC2 were not bound by TCP4 in seedlings and there was only a weak enrichment peak at the 5′-untranslated region (UTR) regions of TCL1 (Supplemental Figure S6B). As TCL1 gene is not expressed in cotyledons (Gan et al., 2011), we focused on CPC, ETC1, ETC3, and TCL2 to detect whether they were candidate targets of TCP4 in regulating trichome formation on cotyledons.
Figure 5.
TCPs directly promote the expression levels of R3 MYB genes. A, Peak graphs showing the ChIP-seq raw reads at the potential TCP4-bound gene loci using 4-d-old dark-grown wild-type and 35S-Myc-mTCP4 seedlings. The black bars indicate the genomic regions of the genes. The blue arrows indicate the transcriptional directions. Thin bars = 500 bp. B, Schematic diagrams of the upstream regions of CPC, ETC1, ETC3, and TCL2. The vertical lines indicate the potential TCP4-binding motif GGACCA. The black arrows represent the transcriptional start sites. CPC-I: −2,675 to –2,670 bp; CPC-II: −746 to −741 bp; CPC-III: −736 to −731 bp; ETC1-I: −923 to −918 bp; ETC1-II: −794 to −789 bp; ETC1-III: −232 to −227 bp; ETC3-I: −900 to −895 bp; ETC3-II: −857 to −852 bp; TCL2-I: −1,985 to 1,980 bp; TCL2-II: −1,962 to 1,957 bp; TCL2-III: −892 to −877 bp. The blue arrows indicate the primers used for ChIP-PCR assay. C, ChIP-PCR assays using 5-d-old whole wild-type or 35S-Myc-mTCP4 seedlings grown under normal conditions. The promoter regions containing TCP4-binding motif were amplified with the primer pairs named as in Figure 5B (blue arrows). The relative enrichment of the wild-type group was set to 1.0. The TA3 transposon locus was used as a negative control. The data are the means (±se) of three biological replicates. The asterisks indicate the Student’s t test significance. **P < 0.01; ***P < 0.001. D–G, The relative expression levels of R3 MYB genes (D: ETC1; E: ETC3; F: CPC; G: TCL2) in 3- to 7-d-old cotyledons of wild-type and tcp2/3/4/5/10/13/17 plants. The expression level of each gene in 3-d-old wild-type cotyledons was set as 1.0. ACTIN8 was used as the reference gene. The data are the means (±se) of three biological replicates. The asterisks indicate the Student’s t test significance. **P < 0.01; ***P < 0.001.
We identified the potential TCP4-recognizing motif GGACCA in the upstream regions of these genes (Figure 5B). By performing ChIP-PCR assays using 5-d-old seedlings, we further confirmed that TCP4 was significantly enriched near these cis-elements (Figure 5C). We then tested the expression levels of these genes in the cotyledons of tcp2/3/4/5/10/13/17 septuple mutants and wild-type control. The expression levels of CPC, ETC1, and ETC3 genes were higher in young cotyledons and gradually decreased as cotyledons grew (Figure 5, D–F). Interestingly, the three genes were significantly downregulated in the early developmental stages in the cotyledons of tcp2/3/4/5/10/13/17 (Figure 5, D–F). The other sub-clade TCL2 gene did not show decreasing expression level in cotyledons as cotyledons grew, but the expression level of TCL2 was also significantly downregulated in the cotyledons of tcp2/3/4/5/10/13/17 (Figure 5G). Both TRY and ETC2 were not significantly regulated in cotyledons of tcp2/3/4/5/10/13/17 (Supplemental Figure S6, C and D), consistent with the finding that they were not targeted by TCP4 in cotyledons (Supplemental Figure S6B). These results suggested that TCP4 positively regulates CPC, ETC1, ETC3, and TCL2 by directly binding to the promoters of these genes in cotyledons.
ETC1 and ETC3 are developmentally regulated and have expression patterns similar to that of TCP4 during cotyledon development
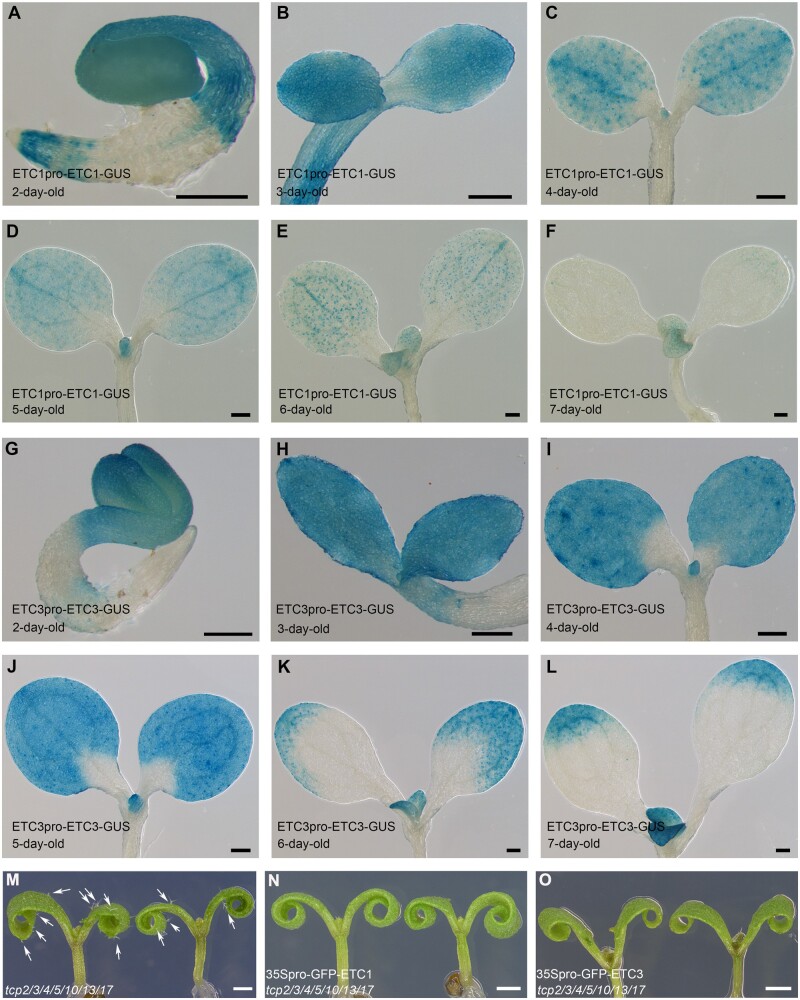
To provide further evidences to support that TCP4 regulates R3 MYB-negative regulators of trichomes on cotyledons, we generated ETC1pro-ETC1-GUS and ETC3pro-ETC3-GUS transgenic lines using a 1,611-bp-long ETC1 and a 2,023-bp-long ETC3 promoter to drive ETC1 or ETC3, respectively, each fused with the GUS reporter gene. GUS staining analysis showed that ETC1 and ETC3 were strongly expressed in young cotyledons (Figure 6, A, B, G, and H). Very interestingly, the expression of ETC1 gradually decreased as cotyledons grew (Figure 6, A–F), while the expression of ETC3 disappeared gradually from the proximal to distal ends and from the central to marginal areas as cotyledons developed (Figure 6, G–L). The developmentally regulated expression patterns of ETC1 and ETC3 were similar to that of TCP4 during cotyledon development (Figure 3, E, G, I, and K) and were consistent with the results of reverse transcription quantitative PCR (RT-qPCR; Figure 5, D and E), further supporting that TCP4 promotes the expression of ETC1 and ETC3.
Figure 6.
The expression pattern of ETC1 or ETC3 overlaps with that of TCP4, and overexpression of ETC1 or ETC3 rescues the ectopic trichome phenotype of tcp2/3/4/5/10/13/17. A–F, The expression patterns of ETC1 in 2- to 7-d-old wild-type cotyledons with GUS histochemical staining. The coding region of ETC1 was driven by its native promoter and fused with GUS gene. Bars = 200 μm. G–L, The expression patterns of ETC3 in 2- to 7-d-old wild-type cotyledons with GUS histochemical staining. The coding region of ETC3 was driven by its native promoter and fused with GUS gene. Bars = 200 μm. M–O, Overexpression of ETC1 or ETC3 rescued the ectopic trichomes on the cotyledons of the tcp2/3/4/510/13/17 mutant. White arrows indicate the trichomes on tcp2/3/4/510/13/17 cotyledons. Bars = 1 mm.
We then generated 35Spro-GFP-ETC1 and 35Spro-GFP-ETC3, in which the CaMV 35S promoter was used to drive GFP fused with ETC1 and ETC3, respectively. We transformed 35Spro-GFP-ETC1 or 35Spro-GFP-ETC3 into the tcp2/3/4/5/10/13/17 septuple mutants. The results showed that overexpression of either ETC1 or ETC3 completely rescued the ectopic trichomes on the cotyledons of tcp2/3/4/5/10/13/17 mutants (Figure 6, M–O). These results provide another layer of TCP repression of trichome formation on cotyledons by directly enhancing the expression of genes encoding R3 MYB transcription factors during cotyledon development.
Discussion
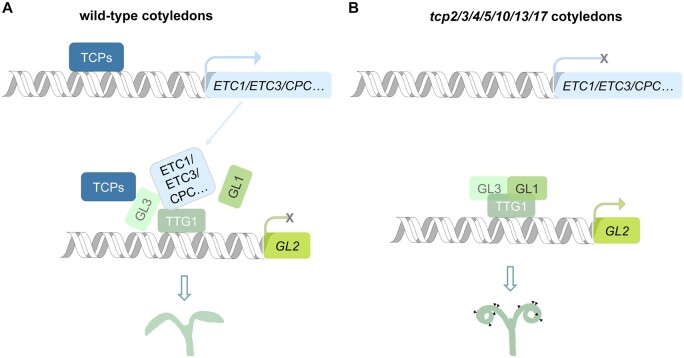
In this study, we demonstrated that CIN-like TCPs repress trichome formation on cotyledons. We showed that the septuple tcp2/3/4/5/10/13/17 mutant produces ectopic trichomes during cotyledon growth after seed germination. We demonstrate that the key components of the MBW complex are all expressed in cotyledons and that TCP genes have overlapping expression patterns with the GL3 gene. TCP4 suppresses the transactivation activity of the MBW complex by directly interacting with GL3 to repress the expression of GL2, which positively regulates trichome formation. We further showed that TCP4 binds to the promoters of genes encoding single R3 MYB proteins, including ETC1, ETC3, CPC, and TCL2, which act as negative regulators in trichome formation, while TCP4 has recently been identified to directly bind to the promoters of TCL1 and TCL2 in control of trichome formation in true leaves (Vadde et al., 2019). We have proposed a working model for the function of TCPs in the determination of trichome formation on cotyledons (Figure 7, A and B). In wild-type cotyledons, on one hand, TCPs directly promote trichome-negative regulators, including CPC, ETC1, ETC3, and TCL2 at the transcriptional level. On the other hand, TCPs directly interact at the protein level with the key component GL3 in the MBW complex. TCPs, CPC, ETC1, ETC3, and TCL2 interfere with the formation of the MBW complex, and the downstream gene GL2 cannot be activated to promote trichome formation on wild-type cotyledons (Figure 7A). In septuple tcp2/3/4/5/10/13/17 cotyledons, disruption of TCPs causes low expression levels of CPC, ETC1, ETC3, and TCL2. The lack of TCPs and these R3 MYB proteins causes the expressed GL3, GL1, and TTG1 to form an active MBW complex to promote the formation of trichomes on cotyledons (Figure 7B). Our findings demonstrate that the machinery of trichome formation exists in cotyledons, and provide molecular mechanisms by which TCPs function redundantly and additively to suppress the activity of the machinery and thus repress trichome formation in Arabidopsis cotyledons. It is worth mentioning that the ectopic formation of trichomes is observed only on the adaxial side but not on abaxial sides of cotyledons in tcp2/3/4/5/10/13/17, suggesting that TCPs may specifically control the trichome formation on the adaxial cotyledons. This implies that the inhibitor mechanisms of ectopic trichomes may be strictly different between the abaxial sides and the adaxial sides of cotyledons.
Figure 7.
The working model of TCP transcription factors controlling trichome formation on cotyledons. A, The schematic representation of the working model of TCPs in repression of trichome formation in wild-type cotyledons. TCPs directly increase the expression levels of R3 MYB genes. R3 MYB proteins and TCPs per se inhibit the transactivation activity of the MBW complex by directly interacting with the components of MBW to suppress the expression of the downstream gene GL2, causing glabrous cotyledons after seed germination. B, The schematic representation of the working model of trichome formation on cotyledons of the tcp2/3/4/510/13/17 mutant. Loss of functions of TCPs result in the inactivation of CPC, ETC1, ETC3, and TCL2. The lack of TCPs and these R3 MYB proteins cause the expressed GL3, GL1, and TTG1 to form an active MBW complex, promoting trichome formation.
Although the cotyledons of Arabidopsis are glabrous, it has been reported that some species in the Brassicaceae family produce trichomes on cotyledons (Beilstein et al., 2006; Chandler, 2008). As trichomes are the specialized epidermis cells that act as physical barriers against insect herbivores, photoinhibition, UV light, and water loss (Hülskamp et al., 1994; Hauser, 2014), the cotyledon trichomes may reflect adaptation strategies to the environment according to the different responsibilities of cotyledons after germination in different species. However, the knowledge about the molecular mechanisms in determining the presence or the absence of trichomes on cotyledons is still limited. Few mutants carrying ectopic trichomes on cotyledons have been identified. As discontinuous-growth organs, cotyledons establish primary morphology during embryogenesis and, after germination, resume development programs interrupted by seed dormancy. The previously reported mutants with cotyledon trichomes including leafy cotyledon1 (lec1; Meinke, 1992; Braybrook and Harada, 2008; Pelletier et al., 2017; Tao et al., 2017, Jo et al., 2019), lec2 (Meinke, 1994; Stone et al., 2001; Tao et al., 2019), fusca3 (Keith et al., 1994; Meinke et al., 1994; Tao et al., 2019), and hydra1 (hyd1; Topping et al., 1997). In this study, we found that the septuple tcp2/3/4/5/10/13/17 mutant produced cotyledon trichomes during growth after germination, suggesting that TCPs and LEC1, LEC2, FUS3, and HYD1 genes repress the trichome formation on cotyledons at different stages of cotyledon development.
The trichome cell fate determination is under delicate regulation involving a negative feedback loop organized by the MBW complex and the single R3 MYB negative regulators. The R3 MYB genes, including ETC1, ETC3, and CPC, are positively regulated by the MBW complex (Digiuni et al., 2008; Zhao et al., 2008). The R3 MYB small proteins in turn repress the transcriptional activity of the MBW complex by competing with GL1 protein (Kurata et al., 2005). The R3 MYB proteins are central for controlling trichome formation at different stages during plant development. The number and density of leaf trichomes on abaxial sides of leaves are used as markers of heteroblasty (Wu et al., 2009). The absence of trichomes on specific glabrous organs, such as cotyledons, and some flower organs, including petals and carpels, is used to represent organ identity (Meinke, 1992; Ó’Maoiléidigh et al., 2018). Spatial and temporal regulations of the conserved MBW-R3 MYB machinery lead to various trichome distribution patterns on different organs. During the leaf development process and the early stage of reproductive growth, the age-dependent SPL/miR156 pathway regulates trichome distribution by directly activating TCL1 and TRY (Yu et al., 2010). TCP4 have been reported to directly promote the expression levels of TCL1 and TCL2 to repress trichome formation at the early developmental stage of true leaves (Vadde et al., 2019). The membrane-associated NAC (No apical meristem, Arabidopsis transcription activation factor, Cup-shape cotyledon) transcription factor NTM1-LIKE8 (NTL8) directly activate TRY and TCL1 to control trichome density in true leaves and inflorescence stems (Tian et al., 2017). In the glabrous organs, the MBW complex and R3 MYB proteins are also meticulously regulated in an organ-specific manner. During flower development, AGAMOUS (AG) activates CPC to inhibit trichome formation on carpels (Ó’Maoiléidigh et al., 2018). The pioneer transcription factor LEC1 controls cotyledon identity by activating a group of R3 MYB genes during embryogenesis (Huang et al., 2015). These reports depict an elegant regulation network in which the functions of the R3 MYB genes are differentially regulated in different organs to control trichome formation. In this article, we demonstrate that TCP transcription factors have a two-pronged function in regulating the activity of MBW complex during cotyledon growth after seed germination. TCP proteins not only repress the activity of the MBW complex by directly interacting with GL3, but also disrupt the negative feedback loop by directly promoting the expression levels of R3 MYB genes to enhance the repression of the MBW complex. However, we find that TCPs directly activate CPC, ETC1, ETC3, and TCL2 (but not TCL1, which has been reported to be a target of TCP4 in control of trichome density of early true leaves; Vadde et al., 2019). These results suggest that even if the same class of TCP proteins controls trichome formation on cotyledons and true leaves, the molecular mechanisms are different in terms of TCL1 regulation. TCP proteins directly activate TCL1 and TCL2 to reduce trichome density in true leaves, while they directly activate CPC, ETC1, ETC3, and TCL2 and interact with MBW complex to completely suppress the trichome formation on cotyledons after seed germination. Interestingly, though CPC, ETC1, ETC3, and TCL1 are targeted by TCPs in cotyledons, we did not observe ectopic trichomes in the pedicels of tcp2/3/4/5/10/13/17 as observed in the triple cpc etc1 etc3 mutant (Supplemental Figure S7; Wang et al., 2008), further suggesting that the regulation of R3 MYB genes by TCPs is organ-specific. The organ-specific mechanism ensures glabrous cotyledons in Arabidopsis and makes cotyledons distinct from true leaves. Our findings reveal an organ-specific regulation of trichome formation on cotyledons.
In addition to having ectopic trichomes, the cotyledons of tcp2/3/4/5/10/13/17 were also curled and serrate (Figure 1A). Compared with the simple vasculature of cotyledons and the complex vasculature of leaves in wild-type controls, the vasculature of the marginal areas of tcp2/3/4/5/10/13/17 cotyledons was more complex than that of wild-type cotyledons and similar to that of true leaves (Supplemental Figure S8, A–D; Koyama et al., 2007, 2010). Both TCP4pro-TCP-Flag and TCP10pro-TCP10-Flag rescued the vascular phenotypes of tcp2/3/4/5/10/13/17 (Supplemental Figure S8, E and F). We emphatically demonstrate in this article that TCPs repress the trichome formation on cotyledons by interfering with the trichome regulation machinery. It has been reported that TCP proteins control auxin biosynthesis (Li and Zachgo, 2013; Lucero et al., 2015; Challa et al., 2016), polar transport (Li and Zachgo, 2013), and signal transduction (Koyama et al., 2010). In terms of polar auxin transport, TCP3 promotes flavonoid production to negatively regulate auxin transport and by interacting with the MBW complexes which control flavonoid biosynthesis. It is possible that the enhanced auxin transport could be one of the reasons causing the more complex vasculature in the cotyledons of tcp2/3/4/5/10/13/17. Interestingly, TCP3 interacts with R2R3 MYBs but not with bHLH protein TT8, and the interaction of TCP3 with R2R3 MYBs promotes the stability of the MBW complexes in control of flavonoid biosynthesis. However, in this article, we demonstrate that TCP4 inhibits the activity of the MBW complex by directly interacting with the bHLH protein GL3, and the interaction interferes with the stability of the MBW complex in control of trichome formation. These results suggest that the molecular mechanisms by which TCP transcription factors regulate the different MBW complexes may be specific in various biological processes. Hence, the abnormal homeostasis of auxin and/or other hormones may contribute to defective vascular patterning in cotyledons of the septuple tcp2/3/4/5/10/13/17. In addition, we previously elucidated that the transcriptional repressor TCP INTERACTOR CONTAINING EAR MOTIF PROTEIN1 (TIE1) interacts with TCP proteins to inhibit their transactivation ability by interacting with TOPLESS (TPL)/TOPLESS-RELATED, which recruit histone deacetylases. Consistently, the serrate and curly leaves in the gain-of-function mutant tie1-D are similar to those of tcp2/3/4/5/10/13/17 (Tao et al., 2013).
CIN-like TCPs have been proposed to promote the differentiation of leaf pavement cells (Efroni et al., 2008), the main type of leaf epidermal cells. Moreover, a recent report and our findings indicate that CIN-like TCPs inhibit the formation of trichomes, another type of epidermal cell, in true leaves and cotyledons (Vadde et al., 2019; this study). We previously demonstrated that CIN-like TCPs repress the differentiation of megaspores (Wei et al., 2015). These findings suggest that CIN-like TCPs regulate cell fate and differentiation in a context-dependent and cell-specific manner.
Materials and methods
Plant materials and growth conditions
All the Arabidopsis (A. thaliana) materials used in this study were in the Columbia-0 (Col-0) accession background. The T-DNA insertion mutants include tcp2 (SAIL_562_D05), tcp3 (wiscDSlox441C3), tcp4 (GABI_363h08), tcp5 (SM_3_29636), tcp10 (SALK_137205), tcp13 (GABI_182B12), and tcp17 (SALK_148580). The tcp multiple mutants were generated by genetic crossing. The seeds were placed on half-strength Murashige and Skoog medium (1/2 MS medium) containing 10 g L−1 sucrose and 8 g L−1 agar (BD Bacto, 214010), and then placed at 4°C for 3 d in dark for stratification. The plates were then transferred to a 22°C chamber for germination under long-day conditions (16-h light and 8-h dark) with a light intensity of 170 mmol m−2 s−1 using fluorescent tubes (Philips F17T8/TL841 17W). The seedlings were planted in soil and grown under the same conditions as described above. N. benthamiana was germinated in soil and was grown under the same conditions as Arabidopsis.
Phylogenetic analysis
For phylogenetic analysis of TCP proteins and R3 MYB proteins, the full-length protein sequences of Arabidopsis TCP transcription factors and R3 MYB proteins were downloaded from The Arabidopsis Information Resource (TAIR) website (https://www.arabidopsis.org) and that of CIN from Antirrhinum majus was downloaded from Phytozome website (https://phytozome.jgi.doe.gov). Multiple alignments of the Arabidopsis TCP and A. majus CIN full-length proteins were conducted using MAFFT Version 7 with L-INS-i iterative refinement methods. Phylogenetic trees were generated with the Maximum Likelihood method using MEGA7 software with the all sites used option and a JTT+G model with 500 bootstrap replications. The alignment file is provided in Supplemental Data Set 1. The multiple alignments of Arabidopsis R3 MYB proteins were performed with the same method. The alignment file is provided in Supplemental Data Set 2.
Genotype analysis and gene expression analysis
For genotype analysis of the T-DNA insertion tcp mutants, the wild-type genomic fragments were amplified by TCPx-F, TCPx-R primer pairs, and the T-DNA insertion fragments were amplified by TCPx-TDNA insertion-F and TCPx-R primer pairs. All the primers used in this study are listed in Supplemental Table S1.
For gene expression analysis, 100 mg cotyledons for each sample were dissected from seedlings and quickly frozen in liquid nitrogen. Total RNA was extracted using Plant Total RNA Purification Kit (GeneMark, TR02-150). Then the genomic DNA was removed with DNase I in the kit. The final concentration and purity of the RNA were evaluated by the ThermoScientific NanoDrop 1000 spectrophotometer.
For preparing cDNA, 2 μg RNA, 1 μL of 100 μM oligo dT, and 1 μL M-MLV reverse transcriptase (Promega, M170A) were added in a 20-μL reaction system. The reverse transcription reaction was performed at 42°C for 60 min.
RT-qPCR was performed using SYBR reagent (CWBIO, CW2601M) in a ThermoFisher QuantStudio 5 real-time system. The experimental conditions were 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. ACTIN8 gene was used as the reference gene. The gene expression level was calculated as normalized relative quantities as described previously (Hellemans et al., 2007). Three biological and technical replicates were performed. Student’s t test was conducted for the statistical analysis.
Generation of binary constructs and transformation
The coding regions of TCPs, GL3, ETC1, and ETC3 were amplified from Arabidopsis cotyledon cDNA. The coding regions for TCPs, GL3, ETC1, and ETC3 were cloned into pENTR/D TOPO (Invitrogen) plasmids. The miR319-resistant forms of TCP4 (mTCP4) and TCP10 (mTCP10) were generated by PCR-based mutagenesis using the primer pairs TCP4-mut-F/R and TCP10-mut-F/R.
For complementation assays, a TCP4 2,821-bp promoter region and a TCP10 986-bp promoter region with full-length genomic region without a stop codon and untranslated regions were amplified from Arabidopsis genomic DNA. The amplified TCP4 and TCP10 fragments were cloned into pENTR/D TOPO vectors (to form pENTR-TCP4pg and pENTR-TCP10pg, respectively), and TCP4pro-TCP4-Flag and TCP10pro-TCP10-Flag constructs, were generated by LR reactions (Invitrogen) with pK7FLAGWG0 using Gateway cloning system, respectively,
To generate the overexpression line for ETC1 or ETC3, 35Spro-GFP-ETC1 or 35Spro-GFP-ETC3 was generated by LR reactions between pENTR-ETC1 or pENTR-ETC3 and pK7WGF2 (Ghent University).
To analyze gene expression patterns using a GUS reporter assay, a 2,455-bp promoter of GL3, a 1,611-bp promoter of ETC1 and a 2,023-bp promoter of ETC3 were amplified. Then, the GL3, ETC1, and ETC3 products were cloned into pENTR/D TOPO and used to generate destination vectors by LR reactions between these entry vectors and pB7GUSWG0 (Ge et al., 2017). For detailed analysis of the TCP4 expression pattern, TCP4pro-TCP4-GUS was generated by LR reactions between pENTR-TCP4pg and pB7GUSWG0.
Scanning electronic microscopy
The cotyledons of 3- to 6-d-old seedlings were isolated for scanning electron microscopy. The plant materials were fixed in FAA solution (100% ethanol:37% (w/v) formaldehyde: 100% acetic acid = 10:2:1), vacuum-dried for 12–24 h, and dehydrated in serial ethanol solutions. The samples were dried using liquid carbon dioxide and observed with a Leica EM CPD300 following the user manual.
Venation and immature embryo observation
For venation observation, the cotyledons of seedlings at 10 d after germination were isolated and fixed in fixation solution (70% ethanol:100% acetic acid = 6:1) for 16 h. The samples were treated with 100% ethanol for 30 min twice and then treated with 70% ethanol for 30 min. To clear the plant material, the samples were treated with venation clearing solution (chloral hydrate:glycerol:H2O = 8 g:1 mL:2 mL) for 24 h. The samples were observed with a Leica M205 FCA stereoscope.
For embryo observation, the siliques were dissected, and the embryos were treated with embryo clearing solution (chloral hydrate:glycerol:H2O = 8 g:1 mL:3 mL) for seconds or a few minutes. Then, the samples were observed with a Zeiss AX10 Imager M2 microscope.
β-Glucuronidase staining
A β-glucuronidase (GUS) histochemical staining assay was performed as described previously (Zhang et al., 2017). The materials were soaked in GUS staining solution at 37°C for 2 h or overnight. The plant samples were moved to 75% (v/v) ethanol for decolorizing and observed with a Leica M205 FCA stereoscope.
Yeast-two-hybrid assays
To test the interaction between TCP4 and GL3, the prey constructs were generated by LR reactions between pENTR-GL3 and pDEST22 (Invitrogen). To avoid the auto-activation activity of TCP4 in yeasts, the truncated coding region of TCP4 containing 108–421 aa at the C-terminus (TCP4△N) was amplified using the primer pair TCP4-108-F and TCP4-R. The N-terminus of TCP4 (TCP4△C) was amplified with the primers TCP4-F and TCP4-107-R. The two truncated versions of the TCP4 coding region were used to generate the bait construct into pDEST32 with LR reactions. The bait constructs and the prey constructs were co-transformed into the AH109 yeast strain. For library screening, the strain AH109 yeasts harboring the bait were mated one by one with the strain Y187 yeasts carrying the preys in the Arabidopsis transcription factor library, as described previously (Ou et al., 2011). The blank pDEST22 was co-transformed with bait blank plasmid as a negative control. Selection medium supplemented with SD–Leu–Trp–His with or without 5 mM 3-AT was used.
To test the interaction between GL3 and CIN-like TCPs, all eight TCPs were cloned into pDEST22 as prey. The coding region of GL3 was cloned into pDEST32 as bait. Selective medium supplemented with SD–Leu–Trp–His with 2.5 mM 3-AT was used.
To determine the domains responsible for the interaction between TCP4 and GL3, GL3 was truncated into GL3△C (1–452 aa at the N-terminus, amplified with the primers GL3-F and GL3-452-R) and GL3△N (431–637 aa at the C-terminus, amplified with the primers GL3-431-F and GL3-R). GL3△C was further truncated into GL3△C1 (1–250 aa, amplified with the primers GL3-F and GL3-250-R) and GL3△C2 (244–452 aa, amplified with the primers GL3-244-F and GL3-452-R). All coding regions were cloned into pDEST22 as prey and co-transformed with pDEST32-TCP4△N.
Firefly luciferase complementation imaging assay
To perform a firefly luciferase complementation imaging assay for confirming the interaction between TCP4 and GL3 in planta, miR319-resistant mTCP4 was cloned into pCambia1300-cluc to generate pCambia1300-cluc-mTCP4 using LR reactions (Zhang et al., 2017). GL3 was cloned into pCambia1300-nluc. The constructs were separately transformed into Agrobacterium tumefaciens strain GV3101. The leaves of N. benthamiana were co-infiltrated with pCambia1300-cluc-mTCP4 or pCambia1300-GL3-nluc and pCAM-P19. The N. benthamiana plants were incubated in the dark for 12 h and then transferred to long-day conditions for 48 h. The infiltrated leaves were dissected and treated with 1 mM luciferin at the abaxial surface and then kept in the dark for 10 min. Then, illumination images were obtained using a low-light cooled CCD imaging apparatus (NightOWL II LB983) with indiGO software. The illumination intensity was 10%. Camera gain was set as high mode.
Co-immunoprecipitation assay
For the Co-IP assay, a 35Spro-mTCP4-FLAG construct was generated by LR reactions between pENTR-mTCP4 and pB7FLAGWG2. 35Spro-GL3-MYC was generated by LR reactions between pENTR-GL3 and pK7MYCWG2. The constructs were co-infiltrated into leaves of N. benthamiana. The leaves were harvested 60 h after infiltration, and homogenized in liquid nitrogen. The total proteins were extracted with IP buffer (50 mM Tris [pH 6.8], 50 mM NaCl, 1 mM EDTA (Ethylene Diamine Tetraacetic Acid), 1 mM DTT (dithiothreitol), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× Protease Inhibitor Cocktail Tablets (Roche, 04693132001), 1‰ MG132, and 1‰ NP-40) and incubated with anti-MYC agarose beads (Sigma, E6654) for 3 h. The concentration of polyacrylamide gel electrophoresis (PAGE) gel used in this study was 10%. The proteins were transferred onto Polyvinylidene difluoride (PVDF) membrane (Millipore, ISEQ00010) and were detected with a chemiluminescence detector (Tanon, 5200Multi). Anti-MYC (CWbio, CW0259), goat-anti-mouse HRP (CWbio, CW0102S) and HRP-conjugated anti-FLAG antibodies (Sigma, A8592) were used for Western blot detection.
Transient dual-luciferase reporter assay and luciferase complementation assay in Arabidopsis protoplasts
Arabidopsis protoplasts were prepared as previously described (Li et al., 2015). For plasmid transformation, each plasmid was adjusted to 10 μg. The total amount of the plasmids used for transformation were <30 μL. The transfected protoplasts were collected and resuspended with 1 mL W5 solution and incubated at room temperature for 12–16 h under mild light conditions.
For the transactivation activity detection assay with transient dual-luciferase reporter system, the mTCP4, GL1, and GL3 coding regions were cloned into pGreen II 62-SK using the FastClone method (Li et al., 2011) to generate effector plasmids. The primers used for preparation of the pGreen II 62-SK backbones were 62sk-FC-F and 62sk-FC-R. The reporter GL2pro-LUC has been described previously (Qi et al., 2014). The LUC/REN intensity was detected using a dual-luciferase reporter assay system (Promega, GLO-MAX 20/20 luminometer). Four biological replicates were performed and the data significance was conducted by Student’s t test.
For the luciferase complementation assay, the protoplasts were collected and resuspended with 100 μL W5 solution, then 90 μL protoplast suspension were mixed with 10 μL 10 mM D-luciferin in 96-well plates. LUC activity was continuously detected with Centro XS3 LB 960 microplate Luminometer.
CRISPR/Cas9-induced mutants
To generate gl1 tcp2/3/4/5/10/13/17, gl3 tcp2/3/4/5/10/13/17, the egg cell-specific promoter-controlled CRISPR/Cas9 system was used as previously reported (Wang et al., 2015). GL1-sgRNA1-U6_26ter-29pro-GL1-sgRNA2 and GL3-sgRNA1-U6_26ter-29pro-GL3-sgRNA2 were amplified from pCBC-dT1T2 and cloned into pHEE401E binary vector using Golden Gate Cloning (Engler et al., 2008). The gl2 tcp2/3/4/5/10/13/17 mutant was generated using a CRISPR/Cas9 system for efficiently generating Cas9-free mutant (Wang and Chen, 2019). The GL2-sgRNA was ligated into pAtU6 plasmid and then cloned into pCAMBIA1300-UBQ10-Cas9-P2A-GFP vector. The primers including sgRNA sequences are listed in Supplemental Table S1. The CRISPR/Cas9 constructs were each transformed into tcp2/3/4/5/10/13/17. The mutants were identified by sequencing the PCR products of the target locus of the offspring of transformants. The mutants generated by CRISPR/Cas9 technology for phenotype observation in Figure 4 and Supplemental Figure S5 were from T2 progenies, and the Cas9 protein had segregated out.
Chromatin immunoprecipitation assay
ChIP-Seq raw data were downloaded from the Gene Expression Omnibus under the accession number GSE115589 (Dong et al., 2019). Quality control of the raw reads was performed by fastp (Chen et al., 2018; 0.12.2). The clean sequencing reads were mapped to the Arabidopsis reference genome TAIR10 (http://www.arabidopsis.org/) by bowtie (Langmead and Salzberg, 2012; v1.2.1.1) with an allowance of two mismatches. PCR duplicates were marked by MarkDuplicates in Picard (http://broadinstitute.github.io/picard/; v2.14.0). Only the unique mapped reads were used for downstream analysis by SAMtools (Li et al., 2009; v1.5). The peaks were denoted by the HOMER (Heinz et al., 2010; v4.10) findPeaks tool with the option-style factor. Bigwig files were generated with binSize 1 and the option normalizeUsingRPKM by bamCoverage from deepTools (Ramírez et al., 2016; v3.1.3). Typical screenshots were captured with Integrative Genomics Viewer (Robinson et al., 2011; IGV, V2.6.2).
A ChIP-qPCR assay was performed as described previously (Zhou et al., 2018). DNA associated with Myc-mTCP4 was enriched with MYC beads (Sigma, A7470) after incubation overnight. Protein A-Sepharose (GE healthcare 17-1279-01) was the negative control, and the supernatant after incubation was used as the input sample. DNA was extracted with phenol/chloroform. The primers used for ChIP-qPCR are listed in Table S1.
Accession numbers
The sequence information from this article can be found in the Arabidopsis Genome Initiative with the following accession numbers: TCP2: AT4G18390; TCP3: AT1G53230; TCP4: AT3G15030; TCP5: AT5G60970; TCP10: AT2G31070; TCP13: AT3G02150; TCP17: AT5G08070; TCP24: AT1G30210; GL1: AT3G27920; GL3: AT5G41315; EGL3: AT1G63650; TTG1: AT5G24520; GL2: AT1G79840; CPC: AT2G46410; ETC1: AT1G01380; ETC2: AT2G30420; TCL1: AT2G30432; TCL2: AT2G30424; TRY: AT5G53200; ETC3: AT4G01060.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The phylogeny of TCP transcription factors in Arabidopsis.
Supplemental Figure S2. The tcp2/3/4/5/10/13/17 septuple mutant had no cotyledon trichomes during embryogenesis.
Supplemental Figure S3. CIN-like TCP transcription factors redundantly inhibit trichome formation on cotyledons.
Supplemental Figure S4. The gl1 and gl3 mutations were generated by CRISPR/Cas9.
Supplemental Figure S5. Disruption of GL2 by CRISPR/Cas9 in various alleles leads to abnormal and fewer ectopic trichomes on the adaxial sides of cotyledons in the gl2 tcp2/3/4/5/10/13/17 octuple mutants.
Supplemental Figure S6. The other three R3 MYB genes, TCL1, TRY, and ETC2 were not directly regulated by TCP4 in cotyledons.
Supplemental Figure S7. No trichomes are produced on pedicels of tcp2/3/4/5/10/13/17 mutant.
Supplemental Figure S8. The cotyledons of tcp2/3/4/5/10/13/17 mutant have more complex vascular systems.
Supplemental Table S1. The list of primers used in this study.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure S1.
Supplemental Data Set 2. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure S6.
Supplementary Material
Acknowledgments
We thank Li-Jia Qu (Peking University) and Hongya Gu (Peking University) for discussions and valuable suggestions. We also thank Yuval Eshed (Weizmann Institute of Science, Israel) for kindly providing the tcp5 tcp13 tcp17 triple mutant seeds, Tomotsugu Koyama (Kyoto University) for kindly providing the seeds of tcp3 tcp4 tcp10 and tcp3 tcp4 tcp5 tcp10 tcp13, and Susheng Song (Capital Normal University) for providing the GL2pro-LUC reporter construct.
Funding
This research was supported by the National Key R&D Program of China (2018YFE0204700), the National Science Fund for Distinguished Young Scholars of China (Grant no. 31725005), and the Science Fund for the Creative Research Groups of the National Natural Science Foundation of China (Grant no. 31621001).
Conflict of interest statement. None declared.
J.L. and G.Q. designed all experiments, analyzed the data, and wrote the article. J.L. performed most of the experiments and collected the data. J.Z., R.Y., H.Y., and F.A. participated in the generation of the septuple tcp2/3/4/5/10/13/17 mutant, constructs, and transgenic plants. L.S., H.C., Y.Z. W.Q., and H.H. contributed to the analysis of ChIP-seq and ChIP-PCR data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Genji Qin (qingenji@pku.edu.cn).
References
- Beilstein MA, Al-Shehbaz IA, Kellogg EA (2006) Brassicaceae phylogeny and trichome evolution. Am J Bot 93: 607–619 [DOI] [PubMed] [Google Scholar]
- Braybrook S, Harada J (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa KR, Rath M, Nath U (2019) The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet 15: e1007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW (2008) Cotyledon organogenesis. J Exp Bot 59: 2917–2931 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Digiuni S, Schellmann S, Geier F, Greese B, Pesch M, Wester K, Dartan B, Mach V, Srinivas BP, Timmer J, et al. (2008) A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol Syst Biol 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Sun N, Yang J, Deng Z, Lan J, Qin G, He H, Deng XW, Irish VF, Chen H, et al. (2019) The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31: 1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han S-K, Kim HJ, Wu M-F, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D (2013) Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch JJ, Chen MA, Hillestad M, Marks MD (2004) Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J 40: 860–869 [DOI] [PubMed] [Google Scholar]
- Gan L, Xia K, Chen J-G, Wang S (2011) Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol 11: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu M-C, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, YuH, YuanR, YangY, AnF, QinG ( . 2019) Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M-T (2014) Molecular basis of natural variation and environmental control of trichome patterning. Front Plant Sci 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, Paepe AD, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hu Y, Liu X, Li Y, Hou X (2015) Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front Plant Sci 6: 955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Miséra S, Jürgens G (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jo L, Pelletier JM, Harada JJ (2019) Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J Integr Plant Biol 61: 564–580 [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) Fusca3: a heterochonic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Hülskamp M, Schiefelbein J (2004a) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hülskamp M (2004b) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55: 389–398 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. (2005) Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Lan J, Qin G (2020) The regulation of CIN-like TCP transcription factors. Int J Mol Sci 21: 4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S (2013) TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76: 901–913 [DOI] [PubMed] [Google Scholar]
- Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y (2011) FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol 11: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Stewart D, Zheng B, Coupland G, Wellmer F (2018) Floral homeotic proteins modulate the genetic program for leaf development to suppress trichome formation in flowers. Development 145: dev157784. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, Zhang L, Miao J, Kondou Y, Matusi M, et al. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JM, Kwong RW, Park S, Le BH, Baden R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB, et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc Natl Acad Sci U S A 114: E6710–E6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26: 1118–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD (1994) The Glabra2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation: TCP4 accelerates plant maturation programs. Plant J. 67: 595–607. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci U S A 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu L-J, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551: 124–128 [DOI] [PubMed] [Google Scholar]
- Tao Z, Hu H, Luo X, Jia B, Du J, He Y (2019) Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat Plants 5: 424–435 [DOI] [PubMed] [Google Scholar]
- Tian H, Wang X, Guo H, Cheng Y, Hou C, Chen J-G, Wang S (2017) NTL8 regulates trichome formation in Arabidopsis by directly activating R3 MYB genes TRY and TCL1. Plant Physiol 174: 2363–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Topping JF, May VJ, Muskett PR, Lindsey K (1997) Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development 124: 4415–4424 [DOI] [PubMed] [Google Scholar]
- Vadde BVL, Challa KR, Nath U (2018) The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J 93: 259–269 [DOI] [PubMed] [Google Scholar]
- Vadde BVL, Challa KR, Sunkara P, Hegde AS, Nath U (2019) The TCP4 transcription factor directly activates TRICHOMELESS1 and 2 and suppresses trichome initiation. Plant Physiol 181: 1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Proc Natl Acad Sci U S A 11: 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen H (2019) A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1: 6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen J-G (2008) Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol 49: 1792–1804 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen J-G (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5: 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen J-G (2008) Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Wang Z-P, Xing H-L, Dong L, Zhang H-Y, Han C-Y, Wang X-C, Chen Q-J (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Zhang J, Pang C, Yu H, Guo D, Jiang H, Ding M, Chen Z, Tao Q, Gu H, et al. (2015) The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res 25: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai W-J, Wang S, Shan C-M, Wang L-J, Chen X-Y (2010) Temporal control of trichome distribution by MicroRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, WeiB, YuanR, WangJ, DingM, ChenZ, YuH, QinG ( . 2017) The Arabidopsis RING-type E3 ligase TEAR1 controls leaf development by targeting the TIE1 transcriptional repressor for degradation. Plant Cell 29: 243–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang Y, Krause K, Yang T, Dongus J.A., Zhang Y, Turck F (2018) Telobox motifs recruit CLF/SWN–PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat Genet 50: 638–644 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Xun Q, Zhang D, Lv M, Ou Y, Li J (2019) TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.