Abstract
Legumes form a symbiosis with rhizobia that convert atmospheric nitrogen (N2) to ammonia and provide it to the plant in return for a carbon and nutrient supply. Nodules, developed as part of the symbiosis, harbor rhizobia that are enclosed in a plant-derived symbiosome membrane (SM) to form an organelle-like structure called the symbiosome. In mature nodules exchanges between the symbionts occur across the SM. Here we characterize Yellow Stripe-like 7 (GmYSL7), a Yellow stripe-like family member localized on the SM in soybean (Glycine max) nodules. It is expressed specifically in infected cells with expression peaking soon after nitrogenase becomes active. Unlike most YSL family members, GmYSL7 does not transport metals complexed with phytosiderophores. Rather, it transports oligopeptides of between four and 12 amino acids. Silencing GmYSL7 reduces nitrogenase activity and blocks infected cell development so that symbiosomes contain only a single bacteroid. This indicates the substrate of YSL7 is required for proper nodule development, either by promoting symbiosome development directly or by preventing inhibition of development by the plant. RNAseq of nodules where GmYSL7 was silenced suggests that the plant initiates a defense response against rhizobia with genes encoding proteins involved in amino acid export downregulated and some transcripts associated with metal homeostasis altered. These changes may result from the decrease in nitrogen fixation upon GmYSL7 silencing and suggest that the peptide(s) transported by GmYSL7 monitor the functional state of the bacteroids and regulate nodule metabolism and transport processes accordingly. Further work to identify the physiological substrate for GmYSL7 will allow clarification of this role.
Soybean Yellow Stripe-like 7 encodes an oligopeptide transporter that localizes to the symbiosome membrane in nodules and contributes to nitrogen fixation and symbiosome development.
Introduction
Legumes form a symbiosis with soil bacteria, rhizobia, that allows them to access N2 from the atmosphere. This symbiosis is an important contributor to the biological nitrogen cycle. The rhizobia fix N2 via the enzyme nitrogenase to produce ammonia and provide it to the plant in return for reduced carbon generated via photosynthesis. This biological N2-fixation provides a large proportion of the nitrogen in the natural environment (Fowler et al., 2013) and is an important component of sustainable agricultural systems, reducing the requirement for expensive nitrogen fertilizers and the pollution that can arise from their overuse (Vance, 2001).
The establishment of this symbiosis involves signaling between the two partners and results in rhizobia moving through an infection thread derived from an invaginated root cell wall into the root cortex where a new organ, the nodule is initiated. The cell wall of the root cells is degraded and the rhizobia released into the cell. Within the nodule infected cells, the rhizobia are enclosed in a plant-derived membrane to form an organelle-like compartment called the symbiosome. Within this symbiosome, the rhizobia differentiate into their symbiotic form, the bacteroid. The symbiosome membrane (SM), initially derived from the plasma membrane (PM), becomes specialized as an interface between the bacteroid and its plant host, segregating the bacteroids from the plant cytoplasm and “protecting” them from any plant defense response (Mohd-Noor et al., 2015).
The major metabolite exchange across the SM is fixed nitrogen (principally ammonia) to the plant and a carbon source, most likely malate, to the bacteroids. However, transport of many other compounds into the symbiosome across the SM must occur as the enclosed bacteroids depend on the plant for all of their nutrients, including iron, zinc, calcium, and cobalt amongst others (Brear et al. 2013; Udvardi and Poole, 2013; Clarke et al., 2014). The SM effectively controls the symbiosis via a suite of transport proteins synthesized by the plant. The plant can control what moves into the symbiosome and, presumably, can withhold sustenance if required. It has been suggested that the plant can impose sanctions on nonfixing rhizobia (Kiers et al., 2003), and controlling transport across the SM could regulate this. It is also probable that compounds other than ammonia/ammonium move from the bacteroids to the plant (Udvardi and Poole 2013).
Transport studies with isolated symbiosomes have demonstrated the presence of a malate transporter and an ammonium channel on the SM, as well as metal ion transporters, but their molecular identity remains elusive (Udvardi and Day, 1997; Udvardi and Poole 2013; González-Guerrero et al. 2016). A number of proteomic analyses of the SM have been reported (Wienkoop and Saalbach, 2003; Catalano et al. 2004; Clarke et al. 2015), and although the earlier studies were limited by the lack of genome sequences for the legumes studied, an array of putative transport proteins have been identified. An example is LjSST1, a sulfate transporter later shown to be essential for nitrogen fixation in the Lotus japonicus–rhizobia interaction (Krusell et al. 2005; Schneider et al. 2019).
The most recent analysis of the soybean SM proteome (Clarke et al. 2015) identified a protein from the Yellow Stripe-like (YSL) family, Glyma.11G203400 (known then as Glyma11g31870). YSL proteins are members of the wider oligopeptide transporter (OPT) family, generally considered to transport metals chelated to phytosiderophores (PS), such as deoxymugineic acid and nicotianamine (NA; Curie et al., 2009). In monocots, PS is excreted to the rhizosphere chelate ferric iron, and the complexes are transported into the plant cytoplasm by YSL transporters. Maize mutants for the first characterized member of this family show a phenotype of interveinal chlorosis that is characteristic of iron deficiency, and it is this phenotype that gave rise to the name Yellow-stripe 1 (YS1). YSL proteins, often localized in xylem parenchyma, can also transport other metal chelates (Dai et al. 2018; Chu et al., 2010; Sasaki et al., 2011; Zheng et al., 2012) and are important for intracellular iron transport and iron homeostasis, with both ferric and ferrous-PS complexes transported (Lubkowitz, 2011). YSL proteins are also involved in mobilization of intracellular stores of metals (Divol et al., 2013; Conte et al., 2013). YSL transporters operate through proton cotransport driven by the membrane potential (Schaaf et al., 2004), and whether localized to the PM or internal membranes, transport is always into the cell cytosol (Lubkowitz, 2011).
Despite the biochemical characterization of some members of the YSL family, the functional role of other members is less clear. In particular, members of one phylogenetic clade of the YSL family, including YSL5, 7, and 8 (Group III) are not well characterized. A recent study showed that Arabidopsis YSL7 and 8 are responsible for the import of a Pseudomonas syringae virulence factor, syringolin A (Syl A), into the plant cytoplasm (Hofstetter et al., 2013). Syl A is a peptide derivative that acts by inhibiting the plant proteasome, and peptides of 4–8 amino acids in length inhibited its transport in plants and in yeast expressing AtYSL7. Consequently, it was suggested that AtYSL7 and AtYSL8 act as OPTs, although direct evidence of oligopeptide transport was not shown (Hofstetter et al., 2013).
In this study, we show that both AtYSL7 and soybean YSL7 (GmYSL7, encoded by Glyma.11G203400) can transport oligopeptides and that the soybean protein, which is localized to the SM in nodule infected cells, is important for nitrogen fixation.
Results
GmYSL7 is a transporter of the YSL family
In our proteomic study (Clarke et al., 2015), we identified Glyma.11G203400 on the SM of soybean nodules. The protein is a member of the OPT superfamily (Saier, 2000; Yen et al., 2001; Stacey et al., 2008) and has significant homology with members of the YSL family (Curie et al., 2009). We named it YSL7, as its closest Arabidopsis homolog is AtYSL7 (74% amino acid identity and 85% similarity; Yordem et al., 2011).
GmYSL7 belongs to a family consisting of 15 members in soybean (Supplemental Figure S1; Schmutz et al., 2010) which in phylogenetic analysis fall into the three clades with both monocots and dicots members (Groups I–III, Supplemental Figure S1). Group IV has only monocot members. In GenBank, six proteins are annotated as “probable metal-nicotianamine transporter YSL7” but in the phylogenetic analysis only GmYSL7, Glyma.11G203400, associates closely with AtYSL7 in Group III, also clustering with the chickpea protein CaYSL7 (Ca08876) and three Medicago truncatula proteins [Medtr3g063490 (MtYSL7), Medtr3g063520 (MtYSL9), and Medtr5g091600 (MtYSL8); Supplemental Figure S1]. Of the other soybean proteins annotated as YSL7, Glyma.09G164500 and Glyma.16G212900 are more closely related to AtYSL5 and AtYSL8, while Glyma.09G281500, Glyma.20G004200, and Glyma.20G004300, although part of Group III, form a subclade not associated with any YSL proteins from other plants included in the phylogeny (Supplemental Figure S1).
GmYSL7 is expressed in infected cells of soybean root nodules
Publicly available transcriptomic data for soybean suggests nodule-specific expression of GmYSL7 (Severin et al., 2010; Supplemental Figure S2). We confirmed this by measuring GmYSL7 transcript abundance in leaves, roots of 8-d-old seedlings, nodules, and denodulated roots of 32-d-old plants using reverse transcription–quantitative PCR (RT-qPCR). GmYSL7 transcript was abundant in nodules but almost undetectable in other plant organs examined (Figure 1A). We investigated the expression patterns of other YSL genes and all had lower nodule expression than YSL7 and transcripts present in other tissues (Supplemental Figure 2B).
Figure 1.
GmYSL7 is expressed in infected cells of soybean root nodules. A, Transcript level of GmYSL7 in tissue samples from different organs. DNR, denodulated roots; nd, not detected. B, Transcript level of GmYSL7 during nodule development. 6R, roots 6 d after inoculation (DAI); 10RN, roots and nodules 10 DAI; 13N-26N, nodules the indicated DAI. Data shown are from three independent time courses. Error bars are SE (n = 3). Nitrogenase activity was first detected at 18 DAI. The relative expression was quantified by RT-qPCR and normalized to a soybean ubiquitin gene (GmUBI3, Glyma20g27950.1). C, Transgenic root expressing pYSL7:GFP-GUS. GUS staining was not detectable in the very early stages of nodule development. Arrowhead indicates a nodule initiation. D, Transgenic pYSL7:GFP-GUS 10-d-old nodule primordia. IR, infected region. E, Transgenic pYSL7:GFP-GUS mature nodule. GUS staining is restricted to infected cells. OC, outer cortex. F, Magnification of E. Scale bars, 150 µm.
Expression of GmYSL7 during nodule development was examined. Transcripts were almost undetectable in young (6- to 10-d-old) inoculated roots, but abundance increased sharply before nitrogenase activity was first detected (Day 18). Expression peaked in nodules of 18-d-old plants, and steadily decreased in older nodules (Figure 1B; Supplemental Figure 3).
As some characterized YSL proteins are involved in transport of iron complexes we examined expression of GmYSL7 in nodules grown in hydroponics with a range of iron concentrations (0–100 µM). GmYSL7 expression was largely insensitive to iron concentration (Supplemental Figure 3A). This was in contrast to two AtYSL3 homologs with clear upregulation in high iron conditions (Supplemental Figire 3B and C).
We investigated GmYSL7 cellular expression pattern in nitrogen-fixing nodules. The 2 kb genomic fragment immediately upstream of the coding region of GmYSL7 was inserted upstream of a promoter-less green fluorescent protein-β-glucuronidase (GFP-GUS) coding sequence to give pGmYSL7:GFP-GUS. GUS staining of pGmYSL7:GFP-GUS transformed roots and nodules agreed well with our RT-qPCR data, with no staining detectable in roots or in early nodule initials (Figure 1C). GUS staining became evident as nodules developed (Figure 1D) and was strongest in maturing nodules. In mature nodules, GUS staining was detected in the infected region and appeared to be confined to rhizobia-infected cells (Figure 1E and F). No GUS staining was detected in the outer cortex of the nodule (Figure 1E).
Localization of GmYSL7 on the SM in rhizobia-infected cells was confirmed using transgenic nodules expressing pGmLBc3:GFP-GmYSL7 and analyzed by confocal microscopy. FM4-64, a lipophilic dye that fluoresces when bound to membrane (Vida and Emr, 1995), was used to counterstain the SM (Limpens et al., 2009, Gavrin et al. 2014). GFP-GmYSL7 signal was on internal membranes within infected cells but not on the PM (Figure 2A and B). Colocalization of GFP-GmYSL7 and FM4-64 signals was analyzed by JACoP and Coloc 2 ImageJ plugins and quantified as Overlap coefficient with standard error (0.87 ± 0.05; Figure 2C and D). Discrete localization of GFP-GmYSL7 on the SM can be seen also in Figure 2E, with GFP-YSL forming a clear “halo” around the perimeter of symbiosomes. The GFP-YSL7 fluorescence pattern in infected cells was distinct from free GFP, detected in the cytoplasm (Figure 2F), and from that of a construct targeted to the symbiosome space (Figure 2G).
Figure 2.
Localization of GmYSL7 in soybean nodule cells infected with rhizobia. A, Localization of GFP-GmYSL7 on SMs in an infected cell counterstained by membrane lipophilic dye FM4-64; IC, infected cell; NI, noninfected cell; ROI, region of interest. B, Fluorescent intensity plot of ROI from (A). C, Colocalization analysis of GFP-YSL7 with FM4-64. The colocalized points are highlighted by white in colocalization map. D, Scatterplot between the intensities of the GFP-YSL7 and FM4-64 in (C). E, Superimposed confocal z-stack sections of GFP-GmYSL7 signal in rhizobia-infected nodule cells. F, Free GFP localizes to the cytoplasmic spaces surrounding symbiosomes in infected cells. G, MtNOD25-GFP (Hohnjec et al., 2009) localizes to the peribacteroid space inside the symbiosomes. Scale bars, 5µm.
Further confirmation that GmYSL7 was localized on the SM and not the PM was obtained by proteomic analysis of isolated SM and microsomal extract enriched in PM and endoplasmic reticulum. Approximately 6 times more peptides from the well-characterized, SM-localized GmNOD26 were in the SM sample compared to the microsomal membrane sample, indicating enrichment of the SM in the purified sample. GmYSL7 peptides were only in the purified SM sample (Supplemental Table S1).
Silencing of GmYSL7 interrupts development of the symbiosis
Since GmYSL7 is localized to the SM, we investigated whether it is important for development of the symbiosis and nitrogen fixation by rhizobia using RNA interference (RNAi). Nodules from transgenic roots silenced for GmYSL7 have analyzed 24 d post-inoculation (24 dpi; Figure 3). Expression of GmYSL7 in RNAi nodules was approximately 40% of the control but expression of the closest homologs, Glyma.16G212900 (GmYSL8) and Glyma.09G164500 (GmYSL5), was not affected (Figure 3A). Acetylene-reduction analyses showed that nitrogenase activity was reduced in silenced nodules to only 25% of the activity of control nodules (Figure 3B). GmYSL7 silenced nodules were smaller (Figure 3C), paler (Supplemental Figure S4A–C), and displayed a delay in development (Figure 3G–I) in comparison to empty vector control nodules (Figure 3D–F).
Figure 3.
RNAi silencing of GmYSL7 affects nodule development. A, Transcript level of GmYSL7 and its closest homologs in 24-d-old nodules of empty vector control and GmYSL7-RNAi plants (error bars represent SD; n = 4; t test: *P < 0.05). B, Nitrogenase enzyme activity in 24-d-old nodules of empty vector control and GmYSL7-RNAi plants (error bars represent SD; n = 8; t test: **P < 0.01). C, Fresh weight of 24-d-old nodules of empty vector control and RNAi-GmYSL7 (error bars represent SD; t test: *P < 0.05). D. Longitudinal section of a 24-d-old nodule from an empty vector control plant. Scale bar, 200 µm. E, Magnification of (D) showing developed (Stage IV) infected cells. Scale bar, 10µm. F, EM of infected cells of a 24-d-old nodule from empty vector control containing developed multibacteroid symbiosomes. Scale bar, 2µm. G, Longitudinal section of a GmYSL7-RNAi 24-d-old nodule. Scale bar, 200µm. H, Magnification of (G) showing undeveloped (Stage II) infected cells. Scale bar, 10 µm. I, EM of infected cells of a GmYSL7-RNAi 24-d-old nodule containing undeveloped single-bacteroid symbiosomes. Scale bar, 2µm. Yellow arrowheads indicate vacuoles. Orange arrowheads indicate symbiosomes.
Silencing of GmYSL7 did not affect bacteria release, but infected cells remained small and, unlike the control nodules, contained small, single-bacteroid symbiosomes (Figure 3I). Numerous small vacuoles were localized around the nucleus (Figure 3H) whereas control nodules had no vacuoles (Figure 3E). To pinpoint the developmental stage in wild-type nodules that matches the RNAi nodules we completed an analysis of nodules from soybean infected with Bradyrhizobium diazoefficiens strain 1042-45 carrying the lacZ fusion driven by the nifD promoter (Acuña et al., 1987). Four stages of development were identified and images can be seen in Supplemental Figure S5. The morphology of infected cells of GmYSL7-silenced nodules (Figure 3G–I) appeared to be arrested at Stage II of normal nodule development (Figure 3D–F, Supplemental Figure 5E–H) where numerous small vacuoles were present in infected cells and most symbiosomes contained single elongated bacteroids. Electron microscopy (EM) of the silenced nodules confirmed that the infected cells were small and under-developed, packed with symbiosomes containing only a single bacteroid (Figure 3I). Bacteroids appeared elongated as seen in control nodules during Stage II (Supplemental Figure S5F and G). Infected cells also contained numerous vacuoles of different sizes and apparent endosomes fusing with symbiosomes, reminiscent of the formation of a lytic compartment, which usually occurs during nodule senescence (Figure 3I). Symbiosomes were isolated from silenced and control nodules, and this showed that in the silenced nodules, symbiosomes contained only single bacteroids compared to the control symbiosomes, which had multiple bacteroids (Supplemental Figure S4D), confirming the phenotype seen by EM analysis. The results suggest that silencing of GmYSL7 arrests development of soybean nodules at Stage II.
RNAseq of nodules in which GmYSL7 is silenced
We used RNAseq to compare the transcriptome in 22-d-old nodules from GmYSL7-RNAi plants and empty vector controls. Before library construction, the level of GmYSL7 transcript in each RNAseq sample was measured by RT-qPCR, and silencing of GmYSL7 was confirmed in each of the GmYSL7 RNAi samples (Supplemental Figure S6). Principal component analysis (Figure 4A) and a heatmap of gene expression of all differentially expressed genes shows clear differences between the RNAi and control samples (Figure 4B). There were no significant changes in expression of other YSL genes.
Figure 4.
RNAseq analysis of GmYSL7-RNAi nodules. A, Principal component analysis of RNAseq samples. B, Heatmap of clustering of differentially regulated genes in each nodule sample. Gene-expression values are normalized by using a z-score transformation on TPM. E1-5 indicates the RNA libraries derived from nodules transformed with empty vector and Y1-5 indicates those transformed with the GmYSL7 RNAi construct.
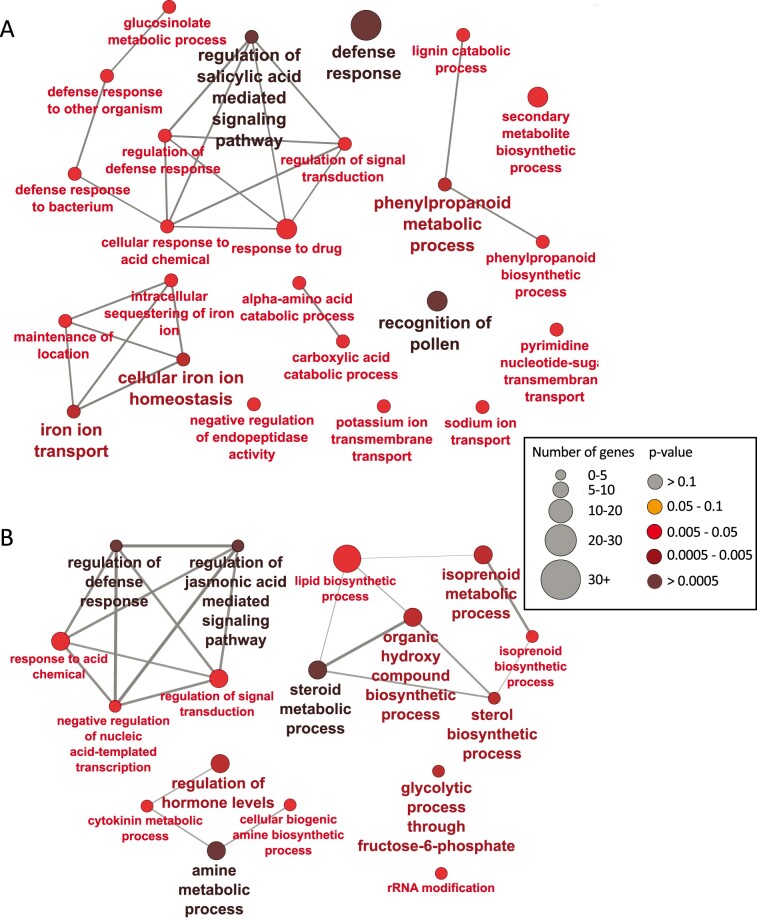
There were 924 genes with log2fold change of 1 or greater in nodules in which YSL7 was silenced, while 1,180 genes had log2fold change of −1 or greater (with adjusted P < 0.05). Gene ontology (GO) enrichment analysis showed that genes involved in defense responses (e.g., defense response to bacterium, defense response to other organism), “negative regulation of endopeptidase activity” and a network associated with iron homeostasis, sequestration, and transport, are overrepresented in the upregulated genes (Figure 5A). A network of genes with GO terms including regulation of defense response, regulation of jasmonic acid (JA)-mediated signaling pathway, and regulation of signal transduction, and another including those associated with lipid biosynthesis, are overrepresented in the downregulated transcripts (Figure5B).
Figure 5.
Gene ontology enrichment analysis of biological processes in up and downregulated genes from GmYSL7-RNAi nodules. GO term enrichment analyses were performed using the ClueGO v2.5.5 plugin (Bindea et al., 2009) in Cytoscape v3.5.1 (Shannon et al., 2003). Circles represent an enriched group of genes based on their GO terms. Circle size and color indicate the number of mapped genes and associated Term P-value corrected with Bonferroni step down.
Details of expression of all genes in the RNAi and control nodules are available in Supplemental Table S2. Among the genes with significantly higher expression in the silenced nodules were those encoding homologs of a senescence-associated gene 13 (Glyma.12G059200), NRT1.5 (nitrate transporter 1.5, Glyma.18G260000), organic cation/carnitine transporter4 (Glyma.12G216400), ferritin (Glyma.01G124500, Glyma.11G232600), vacuolar iron transporter (VIT)-like proteins (Glyma.05G121200, Glyma.08G076000), plantacyanin (Glyma.08G128100), a copper transport protein (Glyma.09G179800), cation efflux family protein (Glyma.08G164800), a cationic amino acid transporter 2 (Glyma.19G116500), nitrate transporter 2.4 (Glyma.11G195200), and a number of protease inhibitors (Supplemental Table S2).
Genes with significantly lower expression in the GmYSL7 silenced nodules were those encoding homologues of sucrose-proton symporter 2 (Glyma.16G156900), glutamine dumper 2 (Glyma.18G277600), a number of carboxyesterases (alpha/beta-Hydrolases or carboxyesterase 17, Glyma.12G096400.1, Glyma.18G298000, Glyma.08G364100, Glyma.04G081600), Protein TIFY 5A-Related (Glyma.20G065500), NA synthase (MtNAS2 homologue, Glyma.15G251300), nodulin MtN21/EamA-like transporter family protein (Glyma.06G246000), cytochrome P450, family 94, subfamily C, polypeptide 1 (Glyma.11G212000), and GmNIC1a (Glyma.12G208900) a Clavata3/ESR (CLE)-related homologue (Supplemental Table S2).
GmYSL7 transports oligopeptides and Syl A but not Fe(II)-NA
Yeast complementation was used to try to identify a substrate for GmYSL7. Initially, we tested for transport of Fe(II)-NA by complementation of the fet3/fet4/ftr1 mutant; however, although the positive control ZmYS1 (Curie et al., 2001, Schaaf et al., 2004) complemented the mutant, GmYSL7 and AtYSL7 did not (Figure 6).
Figure 6.
GmYSL7 does not transport iron or Fe(II)-NA. DEY1530 yeast (fet3/fet4/ftr1) was transformed with the empty vector plasmid pDR196, AtYSL7, GmYSL7, or ZmYS1 in pDR196GW. Serial dilutions of each yeast transformant were applied to SD plates (that include 1.6 µM FeCl3) with 10 µM Fe-citrate, Fe(II)-NA or no added iron (no iron), and the plates were grown for 3–5 d.
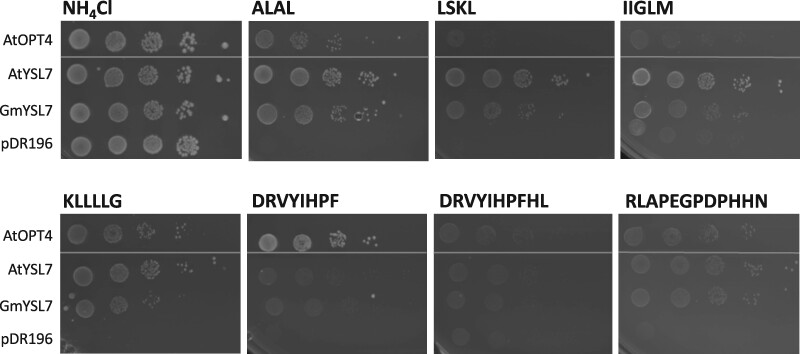
Since the YSL family is part of the wider OPT family, we next tested whether GmYSL7 could complement the yeast oligopeptide transport opt1 mutant, using different oligopeptides as the sole source of nitrogen for growth. The peptides chosen included peptides used in other yeast assays for transport of oligopeptides (Osawa et al., 2006), peptides used in Hofstetter et al. (2013), and a peptide associated with a biological role in nodules (CLE peptide, GmRIC1a encoded by Glyma.13G292300; Hastwell et al., 2015). When the transformants were grown with four (ALAL, LSKL), five (IIGLM), and six (KLLLLG) amino acid peptides as the only N source, cells expressing AtYSL7, GmYSL7, or AtOPT4, but not with the empty vector pDR196, grew (Figure 7). On media containing larger peptides (8, 10, or 12 amino acid), the growth of the transformants was more varied. AtOPT4 supported growth on the eight amino acid peptide DRVYIHPF, while growth was weak for AtYSL7 and GmYSL7. Growth on the 10 amino acid peptides DRVYIHPFHL was close to background for all transformants, but all grew better than vector control on the 12 amino acid peptide RLAPEGPDPHHN (GmRIC1a, Figure 7).
Figure 7.
GmYSL7 and AtYSL7 transport oligopeptides. AtOPT4, AtYSL7, GmYSL7 in pDR196GW, and the empty vector (pDR196) were introduced into the yeast opt1 mutant, Y11213. Serial dilutions of each transformant were grown as above on minimal medium containing either 10 mM NH4Cl (positive control) or 100 µM peptide (with sequence as indicated) as the sole source of nitrogen.
AtYSL7 is involved in SylA uptake and, when expressed in yeast, exposure to SylA inhibited growth (Hofstetter et al., 2013, Figure 8) suggesting the transporter-mediated uptake of this toxic peptide derivative. We used this assay to test for transport of SylA by GmYSL7. GmYSL7, AtYSL7, and the empty vector pDR195 were expressed in the yeast pdr5 mutant that lacks the ABC transporter PDR5, and plated as a lawn (Hofstetter et al., 2013). When a disk containing SylA was placed on the plate, growth of yeast expressing GmYSL7 and AtYSL7, but not the empty vector pDR195, was inhibited. Inhibition of growth caused by SylA on the AtYSL7 plate showed as clear patches on the plate for all concentrations of SylA tested, while inhibition of yeast expressing GmYSL7 was weaker (Figure 8).
Figure 8.
GmYSL7 transports Syl A BY4742 yeast Δpdr5:KanMX6 transformed with the empty vector (pDR196) or the vector expressing AtYSL7 or GmYSL7 were plated as a lawn on solid synthetic defined (SD) media. Filter disks with the indicated SylA solutions were placed onto the plates and inhibition of growth examined after 2 d.
GmYSL7 can replace MtYSL7
MtYSL7 (Medtr3g063490) is the closest homolog of GmYSL7 in M. truncatula, although it is localized on the PM in the vasculature and nodule cortex (Castro-Rodríguez et al., 2020). To determine whether GmYSL7 and MtYSL7 proteins play similar roles in the different cell types in which they are located, we expressed GmYSL7 in the Mtysl7-1 mutant (Castro-Rodríguez et al., 2020). Expression was driven by the MtYSL7 promoter to ensure expression in the cells in which MtYSL7 is present (vasculature and nodule cortex but not infected cells). Although GmYSL7 localizes to the SM in soybean, it was able to complement the Mtysl7-1 transposon insertion mutant to restore nitrogenase activity to wild type levels and increase the dry weight of the transformed plants compared to the mutant (Figure 9). Based on this result, we assume that in the Mtysl7-1 mutant, when expressed in the cells where MtYSL7 is normally active, GmYSL7 at least partially localizes to the PM.
Figure 9.
GmYSL7 is functionally equivalent to MtYSL7. A, Growth of representative wild type (WT), ysl7-1, and ysl7-1 transformed with GmYSL7 controlled by the MtYSL7 promoter (ysl7-1 MtYSL7prom:: GmYSL7). Bar = 1 cm. B, Dry weight of 28 dpi WT, ysl7-1, and ysl7-1 MtYSL7prom:: GmYSL7 plants. Data are the mean ± SE of five transformed plants. C, Nitrogenase activity of 28 dpi WT, ysl7-1, and ysl7-1 MtYSL7prom:: GmYSL7 plants. Acetylene reduction was measured in duplicate from two sets of three to four pooled plants. Data are the mean ± SE. * indicates statistically significant differences in Students t test (P < 0.05)
Discussion
We have characterized a member of the YSL family, GmYSL7, in soybean. The protein is part of a clade of YSL proteins (Group III) that includes AtYSL5, AtYSL7, and AtYSL8. AtYSL7 and 8 are involved in transport of the P. syringae virulence factor into Arabidopsis cells across the PM (Hofstetter et al., 2013), but their physiological role in plants has not been determined. GmYSL7, AtYSL7, CaYSL7, and three M. truncatula proteins (MtYSL7, 8, and 9) form a cluster in phylogenetic analyses, but the soybean protein’s expression profile is distinct from that of AtYSL7 and MtYSL7. AtYSL7 is expressed mainly in flowers but also in siliques and roots. MtYSL7, has highest expression in nodules, but is also expressed in roots. Soybean, on the contrary, appears to lack a YSL7 paralog with expression similar to AtYSL7 and MtYSL7. Rather, GmYSL7 expression is linked specifically to symbiotic nitrogen fixation, occurring only in infected nodule cells where the protein is present on the SM, but not the PM, in contrast to AtYSL7 and MtYSL7. Furthermore, its expression is only marginally affected by the iron concentration of the growth medium (Supplemental Figure S3). This seems a clear example of neofunctionalization with the loss of the paralog (Xu et al., 2018). Either there is no requirement in soybean for the role played by AtYSL7 in other organs or another soybean gene with functional redundancy fulfills that role. The closest homologs of GmYSL7, Glyma.16G054200 (GmYSL8), and Glyma.19G094800 (GmYSL5) are expressed in almost all tissues (Supplemental Figure S2), but we know nothing about their function at this stage.
An important role for GmYSL7 in nitrogen-fixing nodules is shown by knockdown of its expression, which resulted in smaller nodules, with reduced leghemoglobin and a decrease in nitrogenase activity. The GmYSL7-RNAi nodules appear to have been developmentally arrested, with infected cells having small single bacteroid symbiosomes, in contrast to the large symbiosomes containing multiple bacteroids in infected cells of control nodules. The GmYSL7-RNAi infected cell ultrastructure is similar to control nodules in the early stages of development. This suggests that the activity of GmYSL7, and the substrate(s) it transports across the SM, is important for the continued development of the symbiosis and maturation of infected cells.
Our results show clearly that GmYSL7 transports a range of small peptides. When considering the activity of SM transporters, it is important to bear in mind the orientation and energization of the SM (Udvardi and Day, 1997), as this influences the direction that any given substrate is transported. A P-type ATPase on the SM together with the rhizobial electron transport chain pump protons into the symbiosome space, creating an electrochemical gradient across the SM, with the membrane potential positive on the inside and acidifying the interior of the symbiosome (symbiosome space or peribacteroid space; Udvardi and Day, 1997). All YSL proteins characterized to date transport compounds across cell membranes into the cytoplasm (Lubkowitz, 2011), with proton symport the most likely mechanism (Schaaf et al., 2004). Assuming that GmYSL7 has a similar mechanism, it is consequently likely to transport its peptide substrate out of the symbiosome and into the plant cell cytosol. The phenotype seen in GmYSL7-RNAi nodules is, therefore, related to the lack of provision of this substrate to the plant cell.
A M. truncatula homolog of GmYSL7, MtYSL7, has been characterized and also transports oligopeptides. It is not localized on the SM but the Mtysl7-mutant has a phenotype that also affects the symbiosis and nitrogen fixation (Castro-Rodríguez et al., 2020). The difference in cellular localization might be explained by the fact that M. truncatula produces indeterminant nodules, where the meristem continues to be active throughout development, and soybean determinant nodules, in which mature nodules have no meristem. As well as structural differences the different nodule types have a number of metabolic differences including in the mechanism for nitrogen assimilation and the compounds transported from the nodules (Smith and Atkins, 2002). However, GmYSL7 was able to complement the Mtysl7-1 mutant, restoring nitrogenase activity and growth in low N conditions. This suggests that the two YSL7 proteins are able to transport the same substrate/s and that while MtYSL7 brings this substrate into the cell across the PM, GmYSL7 moves its substrate out of the symbiosome and into the cytosol.
In contrast to many other YSL proteins, GmYSL7, AtYSL7, and MtYSL7 were not able to transport Fe(II)-NA (this study, Castro-Rodríguez et al., 2020). Additionally, MtYSL7 could not transport Fe(III), zinc, or copper complexed with NA (Castro-Rodríguez et al., 2020). On the contrary, complementation of the yeast opt1 mutant showed that the three proteins could transport oligopeptides of various sizes and amino acid sequences. Like AtYSL7, when expressed in yeast, GmYSL7 could also transport Syl A, a peptide derivative that is the virulence factor for P. syringae (Hofstetter et al., 2013). Inhibition of yeast growth caused by the transported SylA was not as strong as for AtYSL7 suggesting that GmYSL7 may not transport it as effectively or have the same specificity for the compound. In our assays for direct uptake of oligopeptides in yeast, both AtYSL7 and GmYSL7 supported growth on media with oligopeptides of 4–6 and 12 amino acids as their sole N source, but there was little growth when the 8 amino acid peptide, DRVYIHPF was used, despite its ability to reduce the effect of SylA on Arabidopsis roots (Hofstetter et al., 2013).
While it is clear that YSL7 proteins are peptide transporters, their physiological role in legumes is not clear. Glutathione is a three amino acid peptide derivative found in nodules and bacteroids but as MtYSL7 cannot transport GSH (Castro-Rodríguez et al., 2020) and GmYSL7 is able to replace the function of MtYSL7, it is unlikely that transport of GSH out of the symbiosome is the physiological role of GmYSL7. The symbiosome contains a number of proteases on the SM and in the symbiosome space (Clarke et al., 2015) and appears to act like a vacuole containing large amounts of free peptides (Clarke et al., 2015). Some of these could be substrates for GmYSL7, but why blocking their exit from the symbiosome would inhibit N-fixation and symbiosome development to such an extent is not obvious. While it is possible that GmYSL7 acts to scavenge N by transporting peptides from the symbiosome space into the plant cytosol, it is unlikely that this would have such a profound effect on nodule development.
It is tempting to speculate that the release of peptides from the symbiosomes has a more direct role in manipulating plant gene expression and organogenesis. Cyclic peptides act as signaling molecules in some symbioses (Abbamondi et al., 2014) and it is possible that GmYSL7 transports an oligopeptide derivative produced in the bacteroids. In this scenario, release of the oligopeptide signal could be required to relieve plant inhibition of bacteroid division or as a positive signal for symbiosome development. Supporting this idea is the fact that a protein annotated as an OPT was specifically induced in symbiotic B. japonicum (Pessi et al., 2007) and a number of transcription factors are up- or downregulated in nodules of GmYSL7-RNAi plants.
We used RNAseq of GmYSL7-RNAi nodules to investigate further the effects of inhibiting transport by GmYSL7. Overrepresented GO terms among the downregulated genes included regulation of defense response and regulation of JA signaling pathway and also a range of terms associated with lipid metabolic processes (lipid biosynthetic process, isoprenoid metabolic process). The lipid-associated terms likely relate to the failure of the symbiosome to develop with multiple bacteroids. The change from a single bacteroid symbiosome to one with multiple bacteroids is likely to require synthesis of large amounts of lipid. With the development of the infected cell blocked at an early stage, this synthesis would not be required.
Overrepresented terms in the upregulated genes included “defense response,” “defense response to bacteria,” “defense response to other organism,” and “regulation of defense response,” suggesting that blocking transport by GmYSL7 causes a general defense response against the rhizobia. This links with the downregulation of genes associated with the regulation of defense response GO term which may indicate that the inhibition of the defense response that allows the symbiotic association to develop, has been lifted in the RNAi nodules. The changes in genes associated with a defense response may be an indirect effect of a decrease in nitrogen fixation, with the plant sanctioning the bacteria as ineffective (Kiers et al., 2003). Upregulation of Glyma.11G195200, a soybean homolog of AtNRT2.4, a nitrate transporter that is upregulated in response to nitrogen starvation, suggests that as nitrogenase activity was reduced, the nodules in GmYSL7-RNAi plants may indeed be nitrogen starved. A laser ablation electrospray ionization-mass spectrometry (LAESI-MS) study comparing ineffective nifH nodules with wild type (Agtuca et al., 2020) found JA-associated metabolites were increased in the nitrogen deprived nifH nodules. JA is known to mediate defense responses with an associated increase in secondary metabolites (Pauwels et al., 2009). Two of the transcripts with the strongest downregulation, Glyma.20G065500.1 and .2, and a third, Glyma.11G212000, are likely to encode proteins that repress JA defense responses, which would likely lead to an increase in JA and initiation of a defense response to rhizobia.
Zeatin is another metabolite associated with functional nodules that has reduced abundance in nonfixing nifH nodules (Agtuca et al., 2020). In the GmYSL7-RNAi nodules, that also have reduced nitrogen fixation, a gene encoding isopentyl transferase 5 (GmIPT5, Mens et al., 2018) is downregulated. GmNIC1, a peptide responsible for nitrate regulation of nodulation, is also downregulated in the RNAi nodules indicating that there is no requirement for control of nodule numbers in the RNAi plants.
A link to peptide signaling in the RNAi nodules is the downregulation of three genes encoding glutamine dumpers (GDU). Glyma.18G277600 is one of the most downregulated genes in RNAi nodules with expression reduced 29-fold, while Glyma.14G105200 and Glyma.17G083000 are also reduced significantly. In Arabidopsis, most GDUs are regulated via a peptide receptor PEPR that interacts with AtPep1 (Ma et al., 2014). There is no orthologue of AtPep1 in soybean but homologs of the PEPR gene (Glyma.20G194400 and Glyma.10G195700) are highly expressed in soybean nodules (this study) and could be involved in perception of the peptide transported by YSL7. In Arabidopsis, glutamine dumpers are involved in regulating amino acid exporters via an interaction with a ubiquitin ligase (LOG2; Guerra et al., 2017), homologs of which are expressed in nodules.
Although soybean exports most nitrogen from nodules as ureides, it also exports some amides (Smith and Atkins, 2002), so it is possible that the GDU downregulated in GmYSL7-RNAi nodules is involved in export of fixed nitrogen. A purine permease, likely required for uptake of these purines in the uninfected cells, is also downregulated in the RNAi nodules. It is possible, therefore, that the peptide transported via GmYSL7 from the symbiosome provides a link between nitrogen fixation in the bacteroid and fixed-N transport from the nodule. Export of a peptide from the bacteroid to the plant that increases fixed-N transport out of the nodule, would prevent feedback inhibition of nitrogen fixation by the plant. Disruption of nitrogen fixation in the bacteroid, on the contrary, may inhibit peptide transport, decreasing fixed-N efflux from the nodule to maintain C/N balance in the nodule.
Another possibility is that GDU regulates export of amino acids across the SM. A number of studies suggest that the majority of the fixed nitrogen is exported and not assimilated by the bacteroids (reviewed by Patriarca et al., 2002); bacteroids downregulate glutamine synthetase and symbiotic auxotrophy for branched-chain amino acids has been demonstrated in pea (Prell et al., 2009) and Phaseolus vulgaris (Prell, 2010). In bacteroids of soybean nodules, genes encoding proteins associated with amino acid biosynthesis are downregulated when compared to free-living bacteria (Pessi et al., 2007), suggesting that plants may supply amino acids to support nitrogen fixation. As import into the symbiosome is equivalent to export from the cell this would require the action of an amino acid exporter. In this scenario, export of the peptide that is the substrate for YSL7 would regulate GDU expression and the supply of amino acids to the symbiosome and bacteroid allowing continuation of the symbiotic interaction.
Another group of GO terms that are overrepresented in the upregulated genes in GmYSL7-RNAi nodules is “intracellular sequestering of iron ion,” “iron ion transport,” and “cellular iron ion homeostasis.” Some of the genes associated with these terms include ferritins and VIT gene homologs. This is accompanied by upregulation of a number proteins potentially associated with metal transport (Cu transport protein, cation efflux family protein, MATE efflux family protein) and a transcription factor, WRKY9, associated with a GO term “cellular response to iron ion starvation,” and downregulation of a gene encoding NAS, responsible for synthesis of NA, a PS involved in metal transport by YSLs. MtNAS2 is essential for iron supply to the nodules in M. truncatula and in the nas2-1 mutant nitrogenase activity is reduced (Escudero et al., 2020). These results indicate that metal homeostasis is dysregulated in the GmYSL7-RNAi nodules. There are two possible explanations for this. GmYSL7 may play a direct role in metal ion homeostasis with the peptide transported acting as a signal to regulate metal homeostasis. However, since the genes with modified expression are not those most highly expressed in nodules, we favor the second explanation, namely that when symbiosome development is stalled and nitrogen fixation blocked, then metals being supplied to the symbiosome by the plant may accumulate in the nodules and need to be sequestered to avoid cellular damage. This may result in storage of metals, particularly iron, in uninfected cells with ferritin, or transport into the vacuole via VIT proteins.
Further study is required to decide between these options. Identification of the peptide substrate for GmYSL7 will be key to determining its role.
Conclusion
We have identified a member of the OPT family, GmYSL7, which is localized to the SM in nitrogen-fixing soybean nodules. It transports an array of small oligopeptides out of the symbiosome and into the plant cell cytosol, and its disruption arrests infected cell development and symbiosome maturation, inhibiting nitrogen fixation. GmYSL7’s disruption affects expression of a number of genes involved in plant defense responses, nitrogen metabolism, and in metal homeostasis suggesting a role monitoring the functional state of the bacteroids and regulating nodule metabolism and transport processes. The SM localization has been ideal to illustrate the effects of oligopeptide transport by GmYSL7 but the ability to transport oligopeptides is shared by AtYSL7 and presumably other YSL7 proteins. It will be interesting in future to determine whether the same substrates are physiologically important in different tissues and other plants and the roles this oligopeptide transport plays in plant growth and development.
Methods
Plant growth conditions
Soybean (Glycine max L.) cv Stevens seeds were inoculated at planting and 1 w after planting with B. diazoefficiens (Soybean group H, New Edge Microbials). Plants were grown as described in Clarke et al. (2015) and fertilized once a week with a nitrogen-free B&D nutrient solution (Broughton and Dilworth, 1971). Nitrogenase activity in nodules was assessed using an acetylene reduction assay as described by Unkovich and Baldock (2008).
For limited and excess iron conditions plants were grown in B&D solution with 0, 1, 10 (control concentration), or 100 µM Fe-citrate, which was renewed every 2 d to maintain pH and stable nutrient supply. Two biological replicates were done. Fe status was determined by elemental analysis (Lee M, School of Land and Environment, University of Melbourne) using the Perchloric Nitric Acid Method. Fifteen plants per treatment were analyzed to determine shoot iron content using an Inductively Coupled Plasma Optical Emission Spectrometer (Varian Medical Systems, Palo Alto, CA, USA).
Cloning and constructs
Genomic DNA for cloning the GmYSL7 promoter was extracted from mature soybean leaves using DNeasy Plant Minikit (Qiagen). RNA was extracted from plant tissues using an RNeasy Plant mini kit (Qiagen) and cDNA synthesized using an iScript cDNA synthesis kit (Invitrogen). All constructs were PCR amplified from Arabidopsis thaliana seedling or soybean nodule cDNA, gDNA, or available plasmids using either Platinum Pfx50 (Invitrogen) or Phusion (Thermo Fisher Scientific) high fidelity polymerases and cloned using the Gateway cloning system (Invitrogen) to produce entry clones (Brear et al., 2020) for GmYSL7, AtYSL7, ZmYS1, and AtIRT1. A list of primers used can be found in Supplemental Table S3. The entry clones were used as a basis for LR recombination to produce most expression constructs.
For GmYSL7 promoter, GUS fusion constructs, a 2 kb genomic fragment immediately upstream of the GmYSL7 coding region was recombined into either pKGW-GGRR (Gavrin et al., 2016) or pKGWFS7 (Karimi et al., 2002). The full-length coding sequence of GmYSL7 was recombined into pGmLBC3-pK7GWIWG2 Gateway vector (Gavrin et al., 2016) to create a hairpin RNAi vector for silencing the gene. N-terminal GFP fusion constructs for GmYSL7 were constructed from the full-length coding sequence recombined into either pGmLBC3-pK7WGF2-R or a modified pK7WGF2 (pGmLBC3-pK7WGF2) where the 35S promoter is replaced by the GmLBC3 promoter (Gavrin et al., 2016). The free GFP construct was made by EcoRV digestion and religation of the pGmLBC3-pK7WGF2 vector to remove the intervening Gateway cassette. The symbiosome space MtNOD25-GFP construct is from Hohnjec et al. (2009) and encodes the first 24 amino acids of MtNOD25 fused to GFP. For yeast expression, full-length open reading frames of GmYSL7, AtYSL7, and AtIRT1 were recombined into the pDR196GW vector (Brear et al., 2020). AtOPT4 (Osawa et al., 2006) and ZmYS1 (Curie et al., 2001) in pDR196 (Rentsch et al., 1995) were provided by Doris Rentsch and Catherine Curie, respectively.
The GmYSL7 coding sequence was synthesized with the MtYSL7 promoter and flanked by attL recombination sites inserted in the pUC57 (Synbio). The construct was recombined into pGBW13 using Gateway Cloning technology.
Transformation of soybean and medicago
Hairy root transformation of soybeans (cv Stevens) used Agrobacterium rhizogenes K599 and was as described by Mohammadi-Dehcheshmeh et al. (2014). Transformed roots were inoculated with B. diazoefficiens CB1809 (Becker Underwood, Somersby, NSW, Australia). Plants were grown under controlled temperature and lighting conditions (26°C d, 24°C night; 16 h d; 120–150 µmol m−2 s−1). Transformed nodules were examined 2–4 w post-inoculation.
Transformation of M. truncatula was as described by Boisson-Dernier et al. (2001) using A. rhizogenes ARqua1.
Microscopy
Confocal imaging of GFP-fusion proteins was done on transgenic nodules either hand sectioned or sectioned in low melt agarose using a vibratome (752M Vibroslice, Campden Instruments, Loughborough, Leics., UK). In some instances, nodules were counterstained by FM4-64 (30 μg/ml). Nodule sections were immediately imaged as described previously (Limpens et al., 2009) using either an LSM Pascal 410 (Zeiss) or an SP5 II (Leica) confocal laser-scanning microscope equipped with an argon laser with a 488-nm laser line and helium-neon laser with a 543-nm laser line. Confocal settings: excitation at 488 nm for GFP and 543 nm FM4-64; GFP emission was selectively detected using a 505–530 nm range; FM4-64 emission was detected in another channel using a 560–615 nm range.
Imaging of GUS expression was done as described in Clarke et al. (2015). Sections were either counterstained with ruthenium red or mounted directly in Milli-Q water, and imaged using an Axiophot epifluorescence microscope with a set of Achroplan objective lens (Zeiss).
The protocol for tissue preparation for light and EM has been described previously (Limpens et al., 2009). Semithin sections (0.6 μm) for light microscopy and thin sections (60 nm) for EM of transgenic nodules were cut using a Leica Ultracut ultramicrotome UC7 (Leica). Sections were collected on 400 mesh nickel grids and examined using a Jeol JEM 1400 transmission electron microscope (Jeol Ltd, Tokyo, Japan).
Reverse transcription quantitative-PCR
RT-qPCR assays were used to measure transcript abundance in soybean tissues of control and YSL7 RNAi plants grown in sand or hydroponics. cDNA was synthesized from 500 ng total RNA using Iscript reverse transcriptase (Bio-Rad, Hercules, CA, USA), according to manufacturer’s instructions. RT-qPCR assays were done in a volume of 5 µl in triplicate and contained 1 µl of cDNA diluted 1/5, 1 X LightCycler® 480 SYBR Green I mix (Roche Applied Science, Castle Hill, Australia) and 0.5 µM of each primer (GmYSL7 and GmUBI3 QRT primers; Supplemental Table S3). Assays were done using a LightCycler® 480 (Roche Applied Science) and the following conditions: 95°C 10 min, 45 cycles of 95°C 10 s, 56°C 10 s, 72°C 20 s, followed by ramping the temperature from 55°C to 95°C for melt curve analysis. PCR efficiency for each primer pair was determined using the LinRegPCR software (Ramakers et al., 2003) and data analyzed using the LightCycler® 480 software package (Roche Applied Science). Data were normalized using GmUBI3 (Glyma20g27950; Trevaskis et al., 2002) or cons6 expression (Libault et al., 2008). Stable GmUBI3 expression in the tissues examined in this study was confirmed through comparison of its expression with five characterized soybean reference genes (cons4, 6, 7, and 15; Libault et al., 2008) using geNorm software (Vandesompele et al., 2002). The amplified product from the real-time reaction was cloned and sequenced to confirm the specificity of the amplification product.
Yeast complementation
To test for the transport of Fe(II)NA AtYSL7, GmYSL7, ZmYS1 in pDR196GW and the empty vector were introduced into the Saccharomyces cerevisiae (yeast) fet3/fet4/ftr1 mutant (Spizzo et al., 1997; DEY1530: MATa ade2 his3 leu2 lys2 trp1 ura3 fet3-2::HIS3 fet4-1::LEU2 ftr1D1::TRP1) using the method described by Dohmen et al. (1991). Fe(II)-NA plates were prepared by mixing 15 µl 10 mM FeSO4 in 200 mM MES/Tris pH 7.4 with 250 µl 200 mM Na-ascorbate and 8 µl of 50 mM NA and heating at 65°C for 10 min to produce a clear solution that was added to 25 ml of SD media to produce the Fe(II)-NA plate. Transformants were grown in liquid media to an OD600 of 1 and then serially spotted in 10-fold dilutions on either SD plates with no added iron, Fe(II)-NA plates or SD-plates with 10 µM Fe-citrate.
For the peptide transport assay AtOPT4, AtYSL7, GmYSL7 in pDR196GW and the empty vector were introduced into the yeast opt1 mutant (Y11213: BY4742; MATα; ura3Δ0; leu2Δ0; his3Δ1; lys2Δ0; YJL212c::kanMX4, Euroscarf). Transformants were grown as above on minimal medium (0.17% YNB without amino acids and [NH4]2SO4, supplemented with amino acids as required, and containing either 10 mM NH4Cl (positive control) or 100 µM of the following peptides, ALAL, LSKL, IIGLM, KLLLLG, DRVYIHPF, DRVYIHPFHL, or RLAPEGPDPHHN, as the sole source of nitrogen).
Syl A transport assay
An assay for transport of Syl A by AtYSL7 and GmYSL7 in the yeast strain Δpdr5 (Y12409: BY4742; MATα; ura3Δ0; leu2Δ0; his3Δ1; lys2Δ0; YOR153w::kanMX4, Euroscarf) was done as described in Hofstetter et al. (2013). Syl A was kindly provided by Robert Dudler, University of Zurich.
Statistical analyses
A one-way ANOVA with Tukey’s HSD (SAS Enterprise Guide Version 4.3; SAS Institute Inc., Cary, NC, USA) was used to analyze differences in plant organ dry mass after growth in varying Fe concentrations. Differences are reported as significant where P < 0.05.
RNAseq analysis of transcriptome in GmYSL7-RNAi nodules
The transcriptome for GmYSL7-RNAi nodules was compared to those transformed with an empty vector control using RNAseq. Hairy root transformation with pGmLBC3-pK7GWIWG2 vector or the vector containing GmYSL7 coding sequence was used to produce transformed nodules. RNA was isolated from nodules 21 or 22 d after inoculation using an RNeasy kit (Qiagen). Five replicates for each construct were done, each with nodules from 5 to 7 transformed plants. RNA integrity number (RIN) was determined on a 2100 bioanalyzer (Agilent) and was between 7 and 8.3 for all samples. RNAseq library construction and analysis were completed at Institute for Molecular Bioscience Sequencing Facility, The University of Queensland. A combination of the Ribo Zero rRNA removal bacteria (Illumina) and Ribo Zero rRNA removal plant (seed and root; Illumina) was used to eliminate the rRNA from the sample. The library was constructed using a TruSeq® Stranded mRNA LT—SetA and SetB (Illumina). Sequencing was performed using the Illumina NextSeq500 (NextSeq control software v1.4/Real Time Analysis v2.1). The library pool was diluted and denatured according to the standard NextSeq protocol, and sequenced to generate single-end 76 bp reads using a 75 cycle NextSeq500/550 High Output reagent Kit (Illumina).
Raw sequence reads were aligned to the JGI Wm82.a2 soybean assembly. DESeq2 (Love et al., 2014) was used to test for differential expression between control and YSL7-RNAi samples. Genes with log2 fold change (log2 FC) >1 and adjusted P < 0.05 were considered differentially expressed. Overrepresented biological terms were identified from the list of differentially expressed genes. GO term enrichment analysis was based on the information in SoyBase (https://soybase.org/). Enriched biological terms and their linkage were analyzed and visualized using ClueGO v2.5.5 (Bindea et al., 2009), implemented in the Cytoscape v3.5.1 environment (Shannon et al., 2003; https://cytoscape.org/cy3.html). ClueGO parameters were as follow: Analysis Mode, Functional Analysis; Load Markers List, G. max (3847); Visual Style: Significance Shape, ellipse; ClueGO settings, Ontology/Pathway; GO, Biological Process/KEGG: downloaded the 12/12/2017; Evidence type, All Evidences; Statistical Test Used = Enrichment/Depletion (Two-sided hypergeometric test); Correction Method Used = Bonferroni step down; Min GO Level = 3, Max GO Level = 8, Min Percentage = 4.0, GO Fusion = true, GO Group = true, Kappa Score Threshold = 0.4; Over View Term = SmallestPValue; Group By Kappa Statistics = true; Initial Group Size = 1;Sharing Group Percentage = 50.0.
Symbiosome isolation, SM and microsomal membrane isolation, and proteomic analysis
Symbiosomes were isolated as described by Clarke et al. (2015). SM was collected after pelleting of the membrane and resuspended in 1M Urea for proteomic analysis. Microsomal membrane was isolated from nodules ground and filtered through miracloth as described in Clarke et al. (2015). Symbiosomes and other intact organelles were pelleted by centrifugation at 20,000 g. The membrane in the supernatant (enriched in PM and endoplasmic reticulum) was collected by centrifugation at 100,000 g for 1 h at 4°C and the pellet resuspended in 8M urea. Proteomic analysis was completed at the La Trobe Comprehensive Proteomics Platform (La Trobe University). Data were collected on a Q Exactive HF (Thermo-Fisher Scientific) in Data Dependent Acquisition mode using m/z 350–1,500 as MS scan range at 60 000 resolution. HCD MS/MS spectra were collected for the 7 most intense ions per MS scan at 60 000 resolution with a normalized collision energy of 28% and an isolation window of 1.4 m/z. Dynamic exclusion parameters were set as follows: exclude isotope on, duration 30 s, and peptide match preferred. Other instrument parameters for the Orbitrap were MS maximum injection time 30 ms with AGC target 3 × 106, MSMS for a maximum injection time of 110 ms with AGT target of 1 × 105.
Raw files consisting of high-resolution MS/MS spectra were processed with MaxQuant version 1.5.5.1 to detect features and identify proteins using the search engine Andromeda. Sequence data for soybean from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Gmax) was used as the database for the search engine.
Accession numbers
The accession number for GmYSL7 is NM_001289202.2.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. Unique GmYSL7 and GmNOD26 peptides1 identified in purified SM, a microsomal membrane fraction, and a symbiosome-enriched membrane sample from soybean nodule homogenate.
Supplemental Table S2. Genes upregulated or downregulated in GmYSL7-RNAi nodules and data for all genes expressed in the nodules.
Supplemental Table S3. Primers used in this study.
Supplemental Figure S1. Phylogenetic analysis of YSL proteins.
Supplemental Figure S2. Expression analysis of the YSL genes in soybean.
Supplemental Figure S3. Expression of GmYSL7 and two YSL3 homologs in response to Fe status during nodule development.
Supplemental Figure S4. Physical characteristics of nodules and FM4-64 stained symbiosomes extracted from empty vector control and GmYSL7-RNAi nodules.
Supplemental Figure S5. Detailed morphological analysis of determinate nodule development.
Supplemental Figure S6. Expression level of GmYSL7 in RNAseq samples measured by RT-qPCR.
Supplementary Material
Acknowledgments
We thank Catherine Curie for providing the plasmid containing ZmYS1 and useful discussions about YSL transporters, Sarah Conte and Elsbeth Walker for providing advice about the methods for yeast assays for transport of Fe(II)-NA and Robert Dudler for providing Syl A.
Funding
This research was funded by the Australian Research Council Discovery Projects DP0772452, DP120102780, and DP150102264 and Industrial Transformation Research HUB IH140100013.
A.G. completed the gene silencing, RNAseq analysis, and contributed to the localization. P.C.L. cloned the gene and contributed to the localization, promoter GUS analysis, and real-time analysis. E.B., S.N.M.-N., M.S.G., D.R., and P.M.C.S. completed the analysis of YSL7 in yeast. O.W.G., E.B., M.B.O., F.B., and P.M.C.S. analyzed the RNAseq results. V.E., M.R., and M.G.-G. completed the complementation of MtYSL7. Y.Q. completed the real-time analysis. C.C. completed the promoter GUS analysis. P.M.C.S., F.B., E.B., and D.A.D. isolated symbiosome and microsomal membrane and did proteomic analysis. P.M.C.S. and D.A.D. conceived the project and were involved in experimental design and analysis. P.M.C.S. and A.G. wrote the manuscript. All authors contributed to editing of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Penelope Mary Collina Smith (p.smith3@latrobe.edu.au).
References
- Abbamondi GR, De Rosa S, Iodice C, Tommonaro G (2014) Cyclic dipeptides produced by marine sponge-associated bacteria as quorum sensing signals. Nat Prod Commun 9: 229–232 [PubMed] [Google Scholar]
- Acuña G, Alvarez-Morales A, Hahn M, Hennecke H (1987) A vector for the site-directed, genomic integration of foreign DNA into soybean root-nodule bacteria. Plant Mol Biol 9: 41–50 [DOI] [PubMed] [Google Scholar]
- Agtuca BJ, Stopka SA, Evans S, Samarah L, Liu Y, Xu D, Stacey MG, Koppenaal DW, Paša-Tolić L, et al. (2020) Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J 103: 1937–1958. doi: 10.1111/tpj.14815 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant-Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Brear EM, Day DA, Smith PMC (2013) Iron: an essential micronutrient for the legume–rhizobium symbiosis. Front Plant Sci. 4: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brear EM, Bedon F, Gavrin A, Kryvoruchko IS, Torres-Jerez I, Udvardi MK, Day DA, Smith PMC (2020) GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. New Phytol 228: 667–681. doi: 10.1111/nph.16734 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Rodríguez R, Reguera M, Escudero V, Gil-Díez P, Quintana J, Prieto RI, Kumar RK, Brear EM, Grillet L, Wen J, et al. (2020) Medicago truncatula Yellow Stripe-Like7 encodes a peptide transporter required for symbiotic nitrogen fixation. BioRiv doi: 10.1101/2020.03.26.009159 [DOI] [PubMed] [Google Scholar]
- Catalano CM, Lane WS, Sherrier DJ (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25: 519–531 [DOI] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL (2010) Successful Reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke VC, Loughlin PC, Day DA, Smith PMC (2014) Transport processes of the legume symbiosome membrane. Front in Plant Sci 5: 699. doi:10.3389/fpls.2014.00699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke VC, Loughlin PC, Gavrin A, Chen C, Brear EM, Day DA, Smith PMC (2015) Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol Cell Proteomics 14: 1301–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte SS, Chu HH, Chan-Rodriguez D, Punshon T, Vasques KA, Salt DE, Walker EL (2013) Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis. Front Plant Sci 4: 283. doi:10.3389/fpls.2013.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349. doi: 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, Divol F, Higuchi K, Jean M, Misson J, Schikora A, Czernic P, Mari S (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wang NQ, Xiong HC, Qiu W, Nakanishi H, Kobayashi T, Nishizawa NK, Zuo YM (2018) The Yellow Stripe-Like (YSL) gene functions in internal copper transport in peanut. Genes 9: 635. doi:10.3390/genes9120635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divol F, Couch D, Conéjéro G, Roschzttardtz H, Mari S, Curie C (2013) The Arabidopsis YELLOW STRIPE LIKE4 and 6 transporters control iron release from the chloroplast. Plant Cell 25: 1040–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AWM, Honer CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7: 691–692 [DOI] [PubMed] [Google Scholar]
- Escudero V Abreu I del Sastre E Tejada-Jiménez M Larue C Novoa-Aponte L Castillo-González J Wen J Mysore KS Abadía J et al. (2020) Nicotianamine Synthase 2 Is Required for Symbiotic Nitrogen Fixation in Medicago truncatula Nodules. Front Plant Sci 10. doi: 10.3389/fpls.2019.01780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D, Coyle M, Skiba U, Sutton MA, Cape JN, Reis S, Sheppard LJ, Jenkins A, Grizzetti B, Galloway JN, et al. (2013) The global nitrogen cycle in the twenty-first century. Philos Trans R Soc B Biol Sci 368: 20130164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrin A, Chiasson D, Ovchinnikova E, Kaiser BN, Bisseling T, Fedorova EE (2016) VAMP721a and VAMP721d are important for pectin dynamics and release of bacteria in soybean nodules. New Phytol 210: 1011–1021 [DOI] [PubMed] [Google Scholar]
- Gavrin A, Kaiser BN, Geiger D, Tyerman SD, Wen Z, Bisseling T, Fedorova EE (2014) Adjustment of host cells for accommodation of symbiotic bacteria: vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell 26: 3809–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guerrero M, Escudero V, Sáez Á, Tejada-Jiménez M (2016) Transition metal transport in plants and associated endosymbionts. Arbuscular mycorrhizal fungi and rhizobia. Front Plant Sci 7: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D, Chapiro SM, Pratelli R, Yu S, Jia W, Leary J, Pilot G, Callis J (2017) Control of amino acid homeostasis by a ubiquitin ligase-coactivator protein complex. J Biol Chem 292: 3827–3840. doi: 10.1074/jbc.M116.766469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell AH, Gresshoff PM, Ferguson BJ (2015) Genome-wide annotation and characterization of CLAVATA/ESR (CLE) peptide hormones of soybean (Glycine max) and common bean (Phaseolus vulgaris), and their orthologues of Arabidopsis thaliana. J Exp Bot 66: 5271–5287. doi: 10.1093/jxb/erv351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter SS, Dudnik A, Widmer H, Dudler R (2013) Arabidopsis YELLOW STRIPE-LIKE7 (YSL7) and YSL8 transporters mediate uptake of pseudomonas virulence factor syringolin A into plant cells. Mol Plant Microbe Interact 26: 1302–1311 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Lenz F, Fehlberg V, Vieweg MF, Baier MC, Hause B, Küster H (2009) The signal peptide of the Medicago truncatula modular nodulin MtNOD25 operates as an address label for the specific targeting of proteins to nitrogen-fixing symbiosomes. Mol Plant Microbe Interact 22: 63–72 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY((TM)) vectors for agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume-rhizobium mutualism. Nature 425: 78–81 [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Kramer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1: 44–54 [Google Scholar]
- Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T (2009) Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21: 2811–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowitz M (2011) The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol Plant 4: 407–415 [DOI] [PubMed] [Google Scholar]
- Ma C, Guo J, Kang Y, Doman K, Bryan AC, Tax FE, Yamaguchi Y, Qi Z (2014) AtPEPTIDE RECEPTOR2 mediates the AtPEPTIDE1-induced cytosolic Ca2+ rise, which is required for the suppression of Glutamine Dumper gene expression in Arabidopsis roots. J Integr Plant Biol 56: 684–94. doi: 10.1111/jipb.12171 [DOI] [PubMed] [Google Scholar]
- Mens C Li D Haaima LE Gresshoff PM Ferguson BJ (2018) Local and Systemic Effect of Cytokinins on Soybean Nodulation and Regulation of Their Isopentenyl Transferase (IPT) Biosynthesis Genes Following Rhizobia Inoculation. Front Plant Sci 9. 10.3389/fpls.2018.01150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi-Dehcheshmeh M, Ebrahimie E, Tyerman SD, Kaiser BN (2014) A novel method based on combination of semi-in vitro and in vivo conditions in Agrobacterium rhizogenes-mediated hairy root transformation of Glycine species. In Vitro Cell Dev 50: 282–291 [Google Scholar]
- Mohd-Noor SN, Day DA, Smith PMC (2015) Chapter 68: The symbiosome membrane. In F de Bruijn, ed, Biological Nitrogen Fixation. Vol 2. Wiley-Blackwell, HOBOKEN, NJ 07030 USA, pp 683–694. ISBN: 978-1-118-63704-3
- Osawa H, Stacey G, Gassmann W (2006) ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem J 393: 267–75. doi: 10.1042/BJ20050920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarca EJ, Tatè R,, Iaccarino M (2002) Key role of bacterial metabolism in Rhizobium-plant symbiosis. Microbiol Mol Biol Rev 66: 203–22. doi: 10.1128/mmbr.66.2.203-222.2002. PMID: 12040124; PMCID: PMC120787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Inze D, Goossens A (2009) Jasmonate-inducible gene: what does it mean? Trends Plant Sci 14: 87–91 [DOI] [PubMed] [Google Scholar]
- Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer HM, Hennecke H (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20: 1353–1363 [DOI] [PubMed] [Google Scholar]
- Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ,, Poole PS (2009) Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc Natl Acad Sci USA 106: 12477–12482. doi: 10.1073/pnas.0903653106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, Bourdès A, Kumar S, Lodwig E, Hosie A, Kinghorn S, White J, Poole P (2010) Role of symbiotic auxotrophy in the Rhizobium-legume symbioses. PLoS One 5: e13933. doi: 10.1371/journal.pone.0013933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Saier MH (2000) Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146: 1775–1795 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Xia JX, Ma JF (2011) OsYSL6 is involved in the detoxification of excess manganese in rice. Plant Physiol 157: 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279: 9091–9096 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schneider S, Schintlmeister A, Becana M, Wagner M, Woebken D, Wienkoop S (2019) Sulfate is transported at significant rates through the symbiosome membrane and is crucial for nitrogenase biosynthesis. Plant Cell Environ 42: 1180–1189. doi: 10.1111/pce.13481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. doi: 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N,, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Atkins CA (2002) Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol 128: 793–802. doi: 10.1104/pp. 010912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo T, Byersdorfer C, Duesterhoeft S, Eide D (1997) The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol Gen Genet 256: 547–556. doi: 10.1007/pl00008615 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Wandrey M, Colebatch G, Udvardi MK (2002) The soybean GmN6L gene encodes a late nodulin expressed in the infected zone of nitrogen-fixing nodules. Mol Plant Microbe Interact 15: 630–636 [DOI] [PubMed] [Google Scholar]
- Udvardi M, Poole PS (2013) Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64: 781–805 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Unkovich M, Baldock J (2008) Measurement of a symbiotic N2 fixation in Australian agriculture. Soil Biol Biochem 40: 2915–2921 [Google Scholar]
- Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127: 390–397 [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkoop S, Saalbach G (2003) Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Nadon BD, Kim KD, Jackson SA (2018) Genetic and epigenetic divergence of duplicate genes in two legume species. Plant Cell Environ 41: 2033–2044 [DOI] [PubMed] [Google Scholar]
- Yen MR, Tseng YH, Saier MH (2001) Maize yellow Stripe1, an iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family. Microbiology 147: 2881–2883 [DOI] [PubMed] [Google Scholar]
- Yordem BK Conte SS Ma Jian F Yokosho K Vasques KA Gopalsamy SN Walker EL (2011) Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Annals of Botany 108(5): 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L Yamaji N Yokosho K Ma Jian F (2012) YSL16 Is a Phloem-Localized Transporter of the Copper-Nicotianamine Complex That Is Responsible for Copper Distribution in Rice. The Plant Cell 24(9): 3767–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.