Brassinosteroid plant hormones are involved in repressing the seed maturation program during the seed-to-seedling transition in Arabidopsis.
Abstract
In flowering plants, repression of the seed maturation program is essential for the transition from the seed to the vegetative phase, but the underlying mechanisms remain poorly understood. The B3-domain protein VIVIPAROUS1/ABSCISIC ACID-INSENSITIVE3-LIKE 1 (VAL1) is involved in repressing the seed maturation program. Here we uncovered a molecular network triggered by the plant hormone brassinosteroid (BR) that inhibits the seed maturation program during the seed-to-seedling transition in Arabidopsis (Arabidopsis thaliana). val1-2 mutant seedlings treated with a BR biosynthesis inhibitor form embryonic structures, whereas BR signaling gain-of-function mutations rescue the embryonic structure trait. Furthermore, the BR-activated transcription factors BRI1-EMS-SUPPRESSOR 1 and BRASSINAZOLE-RESISTANT 1 bind directly to the promoter of AGAMOUS-LIKE15 (AGL15), which encodes a transcription factor involved in activating the seed maturation program, and suppress its expression. Genetic analysis indicated that BR signaling is epistatic to AGL15 and represses the seed maturation program by downregulating AGL15. Finally, we showed that the BR-mediated pathway functions synergistically with the VAL1/2-mediated pathway to ensure the full repression of the seed maturation program. Together, our work uncovered a mechanism underlying the suppression of the seed maturation program, shedding light on how BR promotes seedling growth.
Introduction
During seed maturation, the embryo accumulates storage proteins and oils, and the seed becomes dormant and tolerant to desiccation; for the seed to successfully germinate and grow vegetatively, the seed maturation program (or embryonic program) must be deactivated to prevent the expression of embryonic genes. In Arabidopsis (Arabidopsis thaliana), the seed maturation program is specifically activated during seed development by a network of transcription factors including LEAFY COTYLEDON1 (LEC1), ABSCISIC ACID-INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2), which are collectively known as LAFL proteins, as well as AGAMOUS-LIKE15 (AGL15; Suzuki and McCarty, 2008). LEC1 is a CCAAT-box binding protein, and ABI3, FUS3, and LEC2 are plant-specific B3-DNA binding domain proteins, which recognize and bind to the Sph/RY cis-elements (CATGCA) in their target genes (Wang and Perry, 2013; Jia et al., 2014).
These LAFL transcription factors form complex regulatory networks to promote seed maturation by directly activating the transcription of downstream genes, including seed storage protein genes, late-embryogenesis-abundant genes, and genes that function in various hormone metabolism and signaling pathways (Wang and Perry, 2013). The ectopic expression of individual LAFL genes in seedlings induces the accumulation of embryonic traits (Stone et al., 2001). AGL15 encodes a MCM1, AGAMOUS, DEFICIENS, and SRF [serum response factor]-domain transcriptional regulator (MADS) that is thought to function upstream of the LAFL network (Zheng et al., 2009; Chen et al., 2018). AGL15 directly binds to LEC2, FUS3, and ABI3 during seed development (Zheng et al., 2009). Ectopically constitutive expression of AGL15 in seedlings leads to the upregulation of LAFL genes and somatic embryo formation (Zheng et al., 2009).
The transition from the seed to the vegetative phase is a pivotal stage for flowering plants. During this transition, the seed maturation program must be suppressed. In Arabidopsis, the B3-domain transcription factors VIVIPAROUS1/ABI3-LIKE 1 (VAL1) and VAL2 are essential repressors of the seed maturation program during vegetative growth (Suzuki et al., 2007; Tsukagoshi et al., 2007; Suzuki and McCarty, 2008; Jia et al., 2014). Although they exhibit no embryonic properties, val1-2 mutants show moderate de-repression of seed maturation genes, including LAFL genes and AGL15 (Zhou et al., 2013; Chen et al., 2018). VAL1 and VAL2 are functionally redundant, and both are required to restrain the activation of the seed maturation program during early seedling growth. The simultaneous disruption of VAL1 and VAL2 in seedlings leads to further increases in the expression of seed maturation genes and the formation of embryonic seedlings, which accumulate large amounts of seed storage proteins and oils (Tsukagoshi et al., 2007; Zhou et al., 2013; Chen et al., 2018; Yuan et al., 2021). Recently, it was reported that VAL1 directly binds to the RY elements in the promoter of AGL15 and represses AGL15 expression, leading to the repression of the seed maturation pathway (Chen et al., 2018).
Brassinosteroid (BR) is a major growth-promoting steroid hormone that regulates a wide range of growth and developmental processes (Kim and Wang, 2010; Wang et al., 2012). BR-deficient and BR-insensitive mutants display severe developmental defects, including extreme dwarfism due to reduced cell elongation, dark green leaves, male sterility, reduced seed germination, and photomorphogenesis in the dark (Kim and Wang, 2010; Wang et al., 2012). BR is perceived at the cell surface and activates a signal transduction cascade to control the activities of BRASSINAZOLE-RESISTANT 1 (BZR1) family transcription factors (Kim and Wang, 2010; Wang et al., 2012). The binding of BR to the membrane-anchored receptor kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1) activates its co-receptor kinase BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1; Li et al., 2002), leading to the sequential phosphorylation and activation of BR-SIGNALING KINASEs (BSKs) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) kinases (Tang et al., 2008; Kim et al., 2011). BSKs and CDG1 activate the PP1-type phosphatase BRI-SUPPRESSOR 1 (BSU1; Kim et al., 2011), which in turn inactivates BRASSINAZOLE-INSENSITIVE 2 (BIN2) through dephosphorylation (Kim et al., 2009), allowing rapid dephosphorylation of BZR1 and BRI1-EMS-SUPPRESSOR 1 (BES1) by protein phosphatase 2A (PP2A; Tang et al., 2011). Dephosphorylated BZR1 and BES1 are then translocated into the nucleus and bind to their target genes to regulate gene expression (He et al., 2005; Yin et al., 2005; Sun et al., 2010).
Recent studies have suggested that BR promotes seed germination by counteracting abscisic acid (ABA) activity. The BR biosynthetic mutant det2 (de-etiolated-2) and the BR-insensitive mutant bri1 (brassinosteroid insensitive 1) are more sensitive to ABA during germination than the wild-type (WT; Steber and McCourt, 2001; Xue et al., 2009; Kim et al., 2019). The level of biologically active BR and the expression of BR biosynthetic genes increase in pea (Pisum sativum) seeds during germination (Nomura et al., 2007). Furthermore, ABA signaling pathways negatively regulate BR signaling outputs by activating BIN2 via dephosphorylation by ABI1 and ABI2 (Zhang et al., 2009; Wang et al., 2018). Moreover, BR attenuates ABA sensitivity during early seedling growth via BES1-TOPLESS-HISTONE DEACETYLASE19 co-repressor-mediated repression of ABI3 (Ryu et al., 2014). In addition, BIN2, a critical repressor of BR signaling, interacts with and stabilizes ABI5, whereas BR represses the BIN2–ABI5 pathway to antagonize ABA activity during seed germination (Hu and Yu, 2014). BES1 directly interacts with ABI5 and suppresses ABI5 binding to its downstream genes, leading to their reduced expression and consequently facilitating seed germination (Zhao et al., 2019). Gibberellins (GAs) are other growth-promoting hormones that play important roles in promoting seed germination (Bewley, 1997). BR-activated BZR1/BES1 bind to the promoters of several GA biosynthesis genes in a BR-induced manner to induce their expression and thus to increase the production of bioactive GA (Unterholzner et al., 2015). Although extensively studied for several decades, whether BR plays a role in repressing the seed maturation program during the seed-to-seedling transition is unclear.
In the current study, we demonstrate that BR is involved in repressing the seed maturation program during the seed-to-seedling transition. val1-2 seedlings treated with the BR biosynthesis inhibitors exhibited ectopic formation of embryonic sturctures on seedlings. Loss of BR signaling leads to ectopic induction of the seed maturation program in val1-2 seedlings, whereas the gain-of-function mutants bes1-D and bzr1-1D rescued these embryonic properties, demonstrating that BR is essential and BZR1/BES1 activity is sufficient for preventing the ectopic accumulation of embryonic characteristics in val1 seedlings. Furthermore, the genetic disruption of the BR pathway in WT seedlings de-repressed the expression of the seed maturation gene AGAMOUS-LIKE 15 (AGL15). We further demonstrate that BR-activated BZR1/BES1 directly repressed AGL15 expression and functioned upstream of LAFL genes to suppress the seed maturation program. Together, these findings provide important insights into BR activity and increase our understanding of the mechanism underlying the suppression of the seed maturation program.
Results
Inhibition of BR biosynthesis induces the development of embryonic structures in val1-2 seedlings
BR is essential for promoting the growth of young seedlings. Since a successful seed-to-seedling transition requires the transcriptional repression of seed maturation genes, we reasoned that BR might promote seedling growth by inhibiting the seed maturation program. To test this hypothesis, we treated WT Columbia (Col-0) and val1-2 seedlings with the BR biosynthesis inhibitor BRZ (brassinazole; Asami et al., 2000). Consistent with previous observations, BRZ-treated Col seedlings exhibited dwarfism and altered leaf morphology, including the downward leaf curling and dark green color typically observed in BR-deficient mutants (Asami et al., 2000). However, the seedlings did not show any visible embryonic phenotypes (Figure 1A).
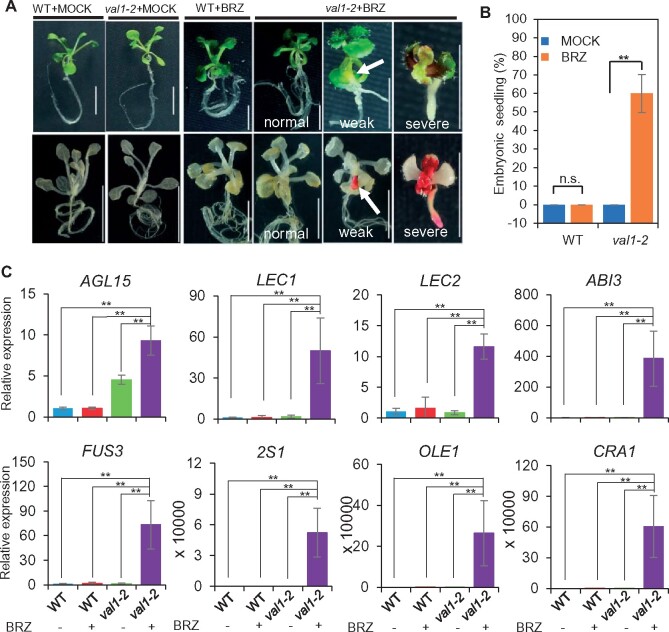
Figure 1.
BRZ treatment on val1 seedlings induces embryonic phenotypes. A, The phenotypes of 14-d-old WT and val1-2 seedlings on 1/2 MS medium containing 1 μM BRZ or 0 μM BRZ (the MOCK control). Lower: seedlings stained with Sudan Red 7B. Normal: seedlings that did not display embryonic properties; Weak: weak embryonic seedlings that developed swollen hypocotyls (white arrow); Severe: severe embryonic seedlings that did not develop true leaves or normal roots and produced embryonic callus in their aerial parts. The white arrow in the upper panel indicates the swollen hypocotyl. The white arrow in the lower panel indicates the swollen hypocotyl stained by Sudan Red. Scale bars = 5 mm. B, The percentage of embryonic phenotypes observed in 14-d-old WT and val1-2 seedlings on 1/2 MS medium containing 1 μM BRZ or the MOCK control. Seedlings with weak and severe embryonic properties were counted as embryonic seedlings. The data represent mean ± SD (n = 5 biological replicates, at least 100 seeds per replicate). n.s. not significant, **P <0.01 (Student’s t test). C, AGL15, LEC1, LEC2, ABI3, FUS3, 2S1, OLE1, and CRA1 expression in 14-d-old Col and val1-2 seedlings treated with (+) or without (−) 1 μM BRZ. The relative expression levels were normalized to the internal control ACT2. The data represent mean ± SD (n = 3 biological replicates). **P <0.01 (Student’s t test).
When val1-2 single mutant seedlings were grown on BRZ-free medium, they did not show visible phenotypes (Figure 1, A and B). In contrast, after BRZ treatment, approximately 54.6% and 4.6% of val1-2 seedlings exhibited weak and strong embryonic phenotypes, respectively (Figure 1, A and B). The BRZ-treated val1-2 seedlings with weak embryonic phenotypes developed true leaves and roots, as did BRZ-treated WT seedlings, but displayed swollen hypocotyls, which were not observed in BRZ-treated WT seedlings (Figure 1A). The swollen hypocotyls were stained by Sudan Red 7B, which stains neutral lipids, whereas WT seedlings were not stained (Figure 1A). On the other hand, the BRZ-treated val1-2 seedlings with strong embryonic phenotypes failed to develop true leaves and normal roots, instead forming stunted primary roots and embryonic calli, which were also stained by Sudan Red (Figure 1A). Real Time Quantitative PCR (RT-qPCR) analysis showed that genes involved in promoting seed maturation, including AGL15, LEC1, LEC2, FUS3, and ABI3, as well as seed storage protein genes SEED STORAGE ALBUMIN 1 (2S1), OLEOSIN 1 (OLE1), and CRUCIFERINA 1 (CRA1), were significantly upregulated in BRZ-treated val1-2 seedlings compared to mock-treated val1-2 seedlings (Figure 1C). In addition, we tested the effects of another BR synthesis inhibitor, propiconazole (PPZ; Hartwig et al., 2012). The val1-2 seedlings treated with PPZ also displayed swollen hypocotyls that were stained by Sudan Red, albeit at a lower rate compared to BRZ-treated val1-2 seedlings (Supplemental Figure S1, A and B). Together, these results indicate that inhibiting BR synthesis leads to the ectopic development of embryonic structures in val1-2 seedlings but not in WT seedlings.
Genetic blocking of BR signaling leads to ectopic induction of the seed maturation program in val1-2 seedlings
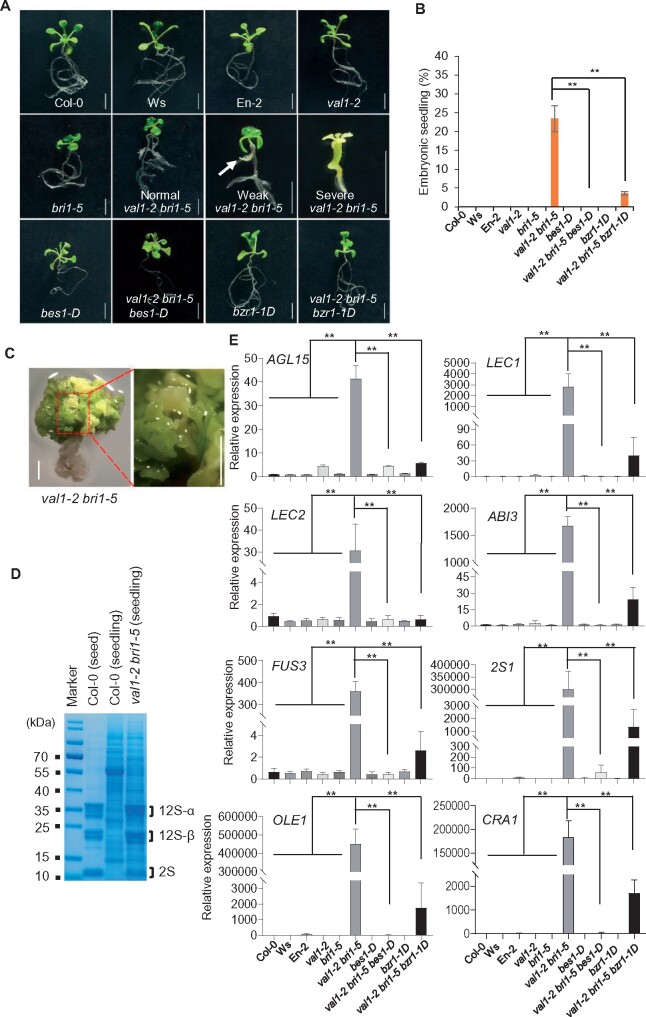
To further confirm that the formation of embryonic structures in BRZ- or PPZ-treated val1-2 seedlings was due to the lack of BR rather than the potential side effects of the two inhibitors, we crossed the BR-insensitive mutant bri1-5 with val1-2 plants. Because the val1-2 and bri1-5 mutants have different genetic backgrounds, we used a pooled collection of multiple independent F3 val1-2 bri1-5 homozygous mutants. Approximately 22% of val1-2 bri1-5 double mutant seedlings produced embryonic structures (including both weak and severe embryonic seedlings; Figure 2, A and B; Supplemental Figure S2). The val1-2 bri1-5 severe embryonic seedlings had white cotyledons, and developed stunted primary roots (Figure 2A), which were completely stained with Sudan Red (Supplemental Figure S2). After one month of growth, the val1-2 bri1-5 severe embryonic seedlings generated somatic embryos (Figure 2C) and accumulated large amounts of 12S globulins and 2S albumins, with a protein profile similar to that of mature seeds (Figure 2D). Moreover, seed maturation genes were highly expressed in val1-2 bri1-5 double mutant seedlings (Figure 2E).
Figure 2.
Ectopic expression of seed maturation genes in the val1-2 bri1-5 mutant. A, The phenotypes of 14-d-old WT (Col-0, Ws, and En-2), val1-2, bri1-5, val1-2 bri1-5, bes1-D, val1-2 bri1-5 bes1-D, bzr1-1D, and val1-2 bri1-5 bzr1-1D seedlings on 1/2 MS medium. Scale bars = 5 mm. Normal: seedlings that did not display embryonic properties; Weak: weak embryonic seedlings that developed stunted roots (white arrow); Severe: severe embryonic seedlings that did not develop true leaves after germination and produced stunted primary roots. B, The percentage of embryonic 14-d-old WT (Col-0, Ws and En-2), val1-2, bri1-5, val1-2 bri1-5, bes1-D, val1-2 bri1-5 bes1-D, bzr1-1D, and val1-2 bri1-5 bzr1-1D seedlings grown on 1/2 MS medium. Seedlings with weak and severe embryonic properties were counted as embryonic seedlings. The data represent mean ± SD (n = 3 biological replicates, at least 100 mature seeds per genotype were used). **P <0.01 (Student’s t test). C, val1-2 bri1-5 produced somatic embryos-like structures after one month of growth on 1/2 MS medium. On the right is a magnified view of somatic embryos. Scale bars = 5 mm. D, Accumulation of seed storage proteins in severe val1-2 bri1-5 seedlings after one month of growth. Proteins extracted from Col-0 seeds and seedlings served as positive and negative controls, respectively. The sizes of the protein standards are indicated on the left. E, AGL15, LEC1, LEC2, ABI3, FUS3, 2S1, OLE1, and CRA1 expression in 14-d-old WT (Col-0, Ws, and En-2), val1-2, bri1-5, val1-2 bri1-5, bes1-D, val1-2 bri1-5 bes1-D, bzr1-1D, and val1-2 bri1-5 bzr1-1D seedlings. The expression levels were normalized to the internal control ACT2. The data represent mean ± SD (n = 3 biological replicates). **P <0.01 (Student’s t test).
DE-ETIOLATED-2 (DET2) is a 5-α-reductase in the BR biosynthesis pathway, and DET2 loss of function leads to typical BR-deficient phenotypes (Li et al., 1996). We crossed BR-deficient det2-1 with val1-2 mutants and found that val1-2 det2-1 seedlings also displayed embryonic structures (Supplemental Figure S3, A–D). Moreover, seed maturation genes were highly expressed in val1-2 det2-1 double mutant seedlings (Supplemental Figure S3E). Together, these results support the notion that blocking the BR pathway in val1 seedlings leads to ectopic induction of the seed maturation program.
BZR1 and BES1 are key transcription factors that are translocated to the nucleus upon receiving BR signals. bes1-D and bzr1-1D are gain-of-function mutants of BES1 and BZR1, respectively (Wang et al., 2002; Yin et al., 2002). Both mutants are insensitive to the BR biosynthesis inhibitor BRZ and can fully rescue BR-deficient mutants. To further confirm the notion that the lack of BR is responsible for the ectopic activation of embryonic properties in val1-2 bri1-5 seedlings, we generated val1-2 bri1-5 bes1-D and val1-2 bri1-5 bzr1-1D mutants. Since the bes1-D mutant is in the En-2 background, the data for val1-2 bri1-5 bes1-D were obtained from multiple independent F3 homozygous lines. The percentage of val1-2 bri1-5 bes1-D and val1-2 bri1-5 bzr1-1D seedlings that displayed embryonic structures was significantly lower than that of val1-2 bri1-5 double mutants (Figure 2, A and B; Supplemental Figure S2), indicating that the gain-of-function mutants bes1-D and bzr1-1D rescued the embryonic phenotypes of val1-2 bri1-5 seedlings. Consistently, the RT-qPCR analysis showed that genes involved in regulating seed maturation, including AGL15, LEC1, LEC2, FUS3, and ABI3, as well as seed storage protein genes 2S1, OLE1, and CRA1, were significantly downregulated in val1-2 bri1-5 bes1-D and val1-2 bri1-5 bzr1-1D compared with val1-2 bri1-5 double mutants (Figure 2E). Additionally, we crossed bes1-D and bzr1-1D with val1-2 to generate val1-2 bes1-D and val1-2 bzr1-1D double mutants. When treated with BRZ, neither val1-2 bes1-D nor val1-2 bzr1-1D seedlings displayed embryonic structures (Supplemental Figure S4). Together, these results demonstrate that BR is essential and BZR1/BES1 activity is sufficient for preventing the ectopic accumulation of embryonic characteristics in val1 seedlings.
BES1 and BZR1 directly bind to the AGL15 promoter
LEC1, LEC2, FUS3, and ABI3 are four master regulators (termed as LAFL regulators) that play critical roles in promoting the seed maturation program (Jia et al., 2014). The MADS family transcription factor AGL15 directly induces the transcription of LEC2, FUS3, and ABI3 to promote the expression of seed maturation genes. As a result, the loss-of-function of AGL15 leads to the ectopic expression of LAFL genes in seedlings (Chen et al., 2018). To determine if BR-activated transcription factors BES1 and BZR1 directly bind to LEC1, LEC2, FUS3, ABI3, and AGL15, we generated Arabidopsis lines expressing Green Fluorescent Protein (GFP)-tagged BES1-D or BZR1-1D driven by their native promoters (Supplemental Figure S5A). Immunoblotting confirmed the accumulation of the corresponding BES1-D-GFP or BZR1-1D-GFP proteins, and, when treated with brassinolide (BL, the most active brassinosteroid), both proteins are induced to generate the nonphosphorylated forms (Supplemental Figure S5B). Both fusion proteins localized to the nucleus (Supplemental Figure S5C), which is consistent with previous reports (Wang et al., 2002; Yin et al., 2002). The transgenic plants were insensitive to the BR biosynthesis inhibitor BRZ (Supplemental Figure S5, D and E). The above results suggest that the two fusion proteins are functional in vivo.
We then performed chromatin immunoprecipitation (ChIP) analysis using the two lines to investigate the enrichment of BES1 or BZR1 on five genes that regulate seed maturation: LEC1, LEC2, FUS3, ABI3, and AGL15. BES1 and BZR1 recognize the E-box (CANNTG), G-box (CACGTG), BRRE (CGTGT/CG), and non-E-box (AAT/ACAAnnnCC/TT) motifs in their target genes to repress or induce their transcription (Yin et al., 2005; Sun et al., 2010; Unterholzner et al., 2015). We designed different primer pairs based on these motifs in the promoter (P) or coding regions (C) of candidate target genes. The enrichment of BZR1 or BES1 was not detected in the tested sites of the promoters or coding regions of LEC1, LEC2, FUS3, or ABI3 (Supplemental Figures S6 and S7). However, both BZR1 and BES1 bound to the proximal promoter region of AGL15 (P1 region) but not to distal upstream regions or coding regions (Figure 3A and Supplemental Figure S7). The P1 region contained a E-box (Figure 3A). DWF4, a known target gene of BES1 and BZR1 (Sun et al., 2010), was significantly enriched by BZR1-1D-GFP or BES1-D-GFP (Supplemental Figure S5, F–G). Furthermore, the enrichment of BZR1-1D-GFP at the P1 site of AGL15 was significantly enhanced by BL treatment (Figure 3B). Therefore, among these master regulators of seed maturation, AGL15 appears to be the direct target of BZR1 and BES1, at least under the conditions used in our analysis. To further verify the ChIP-qPCR data, we conducted a dual-luciferase (LUC) reporter assay in Nicotiana benthamiana leaves. The promoter of AGL15 was fused with LUC to generate the pAGL15::LUC reporter, while the BZR1 or BES1 coding sequence was fused under the 35S promoter to produce effectors (Figure 3C). The results showed that overexpression of BZR1 or BES1 repressed the activity of the AGL15 promoter (Figure 3D).
Figure 3.
BZR1-1D binds to the AGL15 locus. A, ChIP-qPCR analysis of the enrichment of BZR1 at the AGL15 loci. Schematic representations of genes and assayed genomic regions are indicated, with P indicating promoter regions and C indicating coding regions. The black arrow indicates the transcriptional start site. These regions were examined by ChIP-qPCR analysis of 14-d-old transgenic plants harboring pBZR1::bzr1-1D-GFP using an anti-GFP antibody. The putative BES1/BZR1 binding motifs, including the E-box (CANNTG) and G-box (CACGTG) are shown in the examined regions. The VAL1/2 binding motif RY (CATGCA) is also shown. WT plants are used as negative control samples. The amount of DNA after ChIP was normalized using TUB2 as an internal standard. The data represent mean ± SD (n = 3 biological replicates). **P <0.01 (Student’s t test). B, ChIP-qPCR analysis of the enrichment of BZR1 at the AGL15 in WT and pBZR1::bzr1-1D-GFP Arabidopsis plants with or without 250 nM BL. The P1 region of the AGL15 promoter was examined. The amount of DNA after ChIP was normalized using TUB2 as an internal standard. The data represent mean ± SD (n = 3 biological replicates). Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukkey’s HSD test. C–D, Dual-LUC reporter assays showing that BZR1 and BES1 represses the transcription activity of the AGL15 promoter. The AGL15 promoter was fused to the LUC reporter gene and was transformed into N. benthamiana leaves in combination with various effector constructs. LUC activity was normalized to REN activity (as an internal control). The data represent mean ± SD (n = 5 biological replicates). **P <0.01 (Student’s t test).
BR is required for repression of AGL15 expression in seedlings
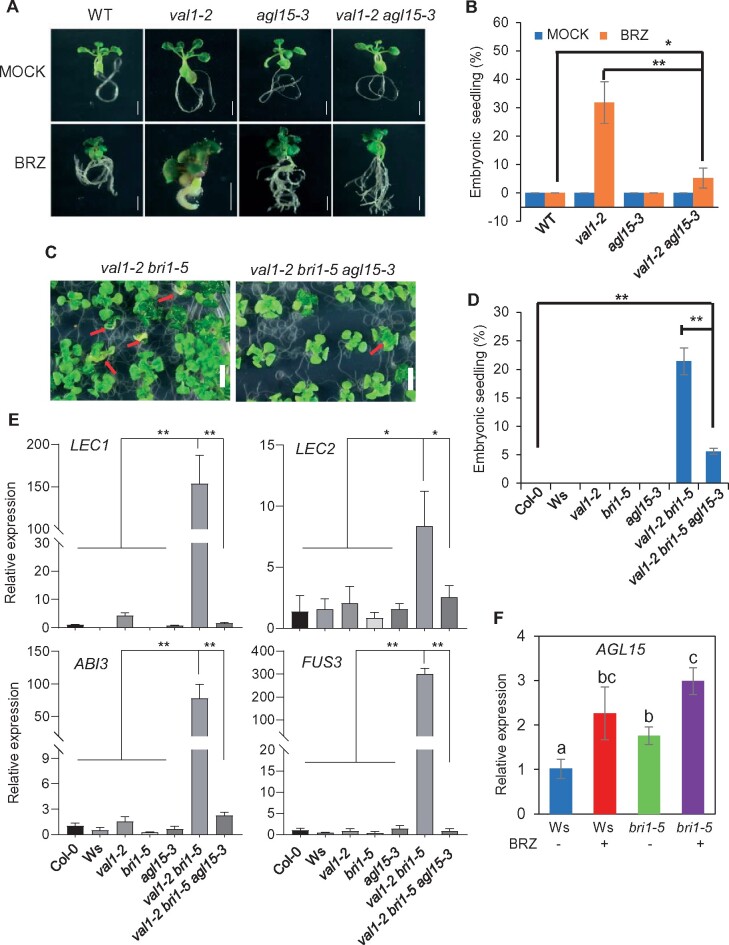
To genetically test if AGL15 is the downstream target of the BR-mediated pathway that represses the embryonic program, we generated val1-2 agl15 double mutants and treated val1-2 and val1-2 agl15 seedlings with BRZ. Compared to BRZ-treated val1-2, the ratio of embryonic structures in BRZ-treated val1-2 agl15 seedlings was significantly reduced (Figure 4, A and B). We also constructed the val1-2 bri1-5 agl15 mutant and observed that the percentage of embryonic structures was significantly lower in this triple mutant than in val1-2 bri1-5 (Figure 4, C and D). The expression levels of the LAFL genes were substantially lower in val1-2 bri1-5 agl15 than those in val1-2 bri1-5 seedlings (Figure 4E). Together, these results demonstrate that the BR pathway acts upstream of AGL15 to inhibit the seed maturation program in seedlings.
Figure 4.
AGL15 functions downstream of BR signaling to repress the expression of seed maturation genes in seedlings. A, The phenotypes of 14-d-old WT, val1-2, agl15-3, and val1-2 agl15-3 seedlings grown on 1/2 MS medium with 1 μM BRZ or the MOCK control. Scale bars = 2 mm. B, The percentage of embryonic WT, val1-2, agl15-3, and val1-2 agl15-3 seedlings grown on 1/2 MS medium containing BRZ or the MOCK control. Seedlings with weak and severe embryonic properties were counted as embryonic seedlings. The data represent mean ± SD (n = 3 biological replicates, at least 120 mature seeds per genotype were used). *P <0.05; **P <0.01 (Student’s t test). C, The phenotypes of 14-d-old val1-2 bri1-5 and val1-2 bri1-5 agl15-3 seedlings. Red arrows point to embryonic seedlings. Scale bars = 5 mm. D, The percentage of embryonic Col-0, Ws, val1-2, bri1-5, agl15-3, val1-2 bri1-5, and val1-2 bri1-5 agl15-3 seedlings. Seedlings with weak and severe embryonic properties were counted as embryonic seedlings. The data represent mean ± SD (n = 3 biological replicates, at least 100 mature seeds per genotype were used). **P <0.01 (Student’s t test). E, LEC1, LEC2, ABI3, and FUS3 expression in 14-d-old Col-0, Ws, val-2, bri1-5, agl15-3, val1-2 bri1-5, and val1-2 bri1-5 agl15-3 seedlings grown on 1/2 MS medium. The expression levels were normalized to the internal control ACT2. The data represent mean ± SD (n = 3 biological replicates). *P <0.05; **P <0.01 (Student’s t test). F, AGL15 expression in 14-d-old WT (Ws) and bri1-5 seedlings with (+) or without (−) 1 μM BRZ. The expression levels were normalized to the internal control ACT2. The data are mean ± SD (n = 3 biological replicates). Different lowercase letters indicate significant differences, as determined by the post hoc Tukey’s HSD test.
We wondered if the lack of the BR pathway alone in WT seedlings would lead to the de-repression of AGL15. RT-qPCR indicated that inhibiting BR biosynthesis by BRZ treatment significantly enhanced the transcription of AGL15 in WT Wassilewskija seedlings (Ws; Figure 4F). Furthermore, AGL15 was expressed at significantly higher levels in bri1-5 seedlings than in the WT (Ws) control (Figure 4F). Additionally, we found that, although the expression of AGL15 in the bzr1-1D and bes1-D mutants was similar to that in the corresponding WT, BL treatment in WT (Col and En-2) significantly repressed the expression of AGL15 (Supplemental Figure S8). Together, these results further support the notion that the BR pathway is involved in repressing AGL15 expression in seedlings.
The genetic interaction between the BR-mediated pathway and the VAL pathway
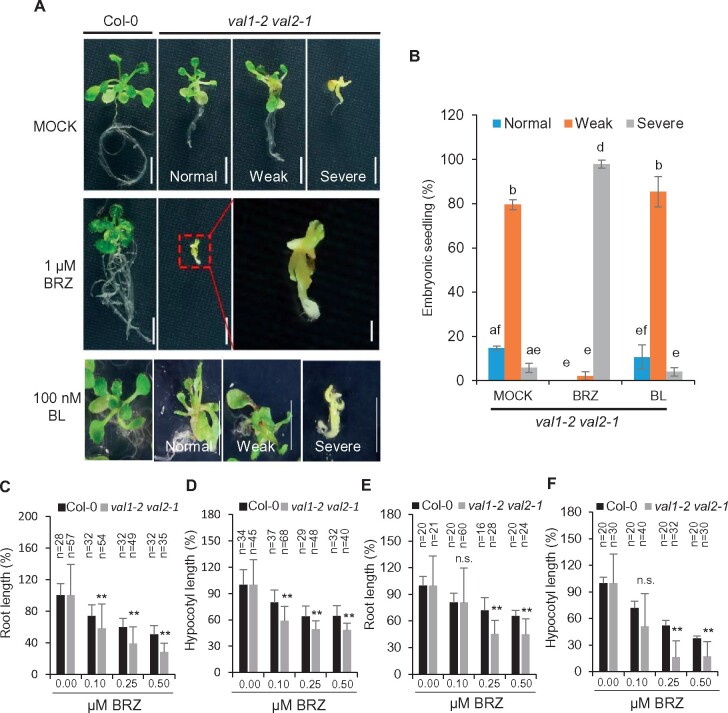
To further explore the genetic relationship between the BR-mediated pathway and the VAL pathway in the repression of the seed maturation program, we treated Col-0 and val1-2 val2-1 seedlings with BRZ or BL. Without treatment, approximately 79.5% and 5.7% of val1-2 val2-1 seedlings exhibited weak and severe embryonic phenotypes, respectively, similar to previous report (Jia et al., 2013; Figure 5, A and B). The 14.8% remaining val1-2 val2-1 seedlings did not show visible embryonic structures at early seedling stages, but exhibited a dwarf phenotype later in development (Supplemental Figure S9). In contrast, when treated by 1 μM BRZ, almost all val1-2 val2-1 double mutant seedlings exhibited severe embryonic phenotypes (Figure 5, A and B). Thus, inhibiting BR biosynthesis strongly enhanced the embryonic phenotypes of val1-2 val2-1 double mutant seedlings. To further investigate the BR responses of the val1-2 val2-1 double mutant, we treated the double mutants using low concentrations of BRZ (0.1, 0.25, and 0.5 μM) under constant light or dark conditions and measured the length of roots and hypocotyls. Both the light- and dark-grown val1-2 val2-1 seedlings exhibited enhanced sensitivity to BRZ treatment in terms of both root growth and hypocotyl elongation compared with Col-0 (Figure 5, C–F). On the other hand, BL treatment did not significantly decrease the ratio of embryonic structures in val1-2 val2-1 double mutant seedlings (Figure 5, A and B).
Figure 5.
Phenotypic analysis of val1-2 val2-1 seedlings treated with BL and BRZ. A, The phenotypes of 14-d-old WT and val1-2 val2-1 seedlings grown on 1/2 MS medium containing 1 μM BRZ, 100 nM BL or the MOCK control. Scale bars = 5 mm. The red dotted line represents the enlarged region. Scale bar = 1 mm. Regarding the severity of the embryonic phenotypes: some val1-2 val2-1 seedlings displayed normal phenotypes and did not develop any type of embryonic phenotypes, seedlings with weak embryonic phenotypes developed embryonic shoot apical meristems, and seedlings with severe embryonic phenotypes did not develop true leaves or roots and displayed embryonic callus. B, The percentage of 14-d-old embryonic val1-2 val2-1 seedlings grown on 1/2 MS medium containing 1 μM BRZ, 100 nM BL, or the MOCK control. Three biological replicates of at least 100 mature seeds per genotype were germinated for each treatment. Error bars, sd from three biological replicates. Different lowercase letters indicate significant differences, as determined by the post hoc Tukey’s HSD test. C–F, The val1-2 val2-1 mutant is hypersensitive to BRZ. Seedlings were grown on various concentrations of BRZ under constant light (C and D) or dark (E and F) for six days before the lengths of root and hypocotyl were measured. Error bars indicate sd. NS, not significant. **P <0.01. The number of ‘n=’ indicates the number of plants that were used.
Discussion
In the current study, we identified a core network by which the growth-promoting hormone BR mediates the repression of the seed maturation program during the seed-to-seedling transition. We demonstrated that BR signaling inhibits the expression of AGL15 in the seedlings to repress the embryonic traits, thus promoting vegetative growth (Figure 6).
Figure 6.
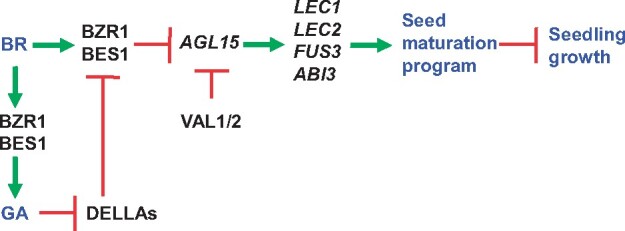
Proposed model of the BR–BZR1–AGL15 pathway, which acts together with VAL1/2–AGL15 pathway to repress the seed maturation program in Arabidopsis seedlings. According to this model, BR-activated BZR1/BES1 directly targets the AGL15 promoter to repress its expression. Because AGL15 activates the expression of LAFL genes including LEC1, LEC2, FUS3, and LEC2, the repression of AGL15 by BZR1/BES1 results in the reduced expression of the LAFL genes. In addition, BZR1/BES1 could also directly induce the expression of GA biosynthesis genes to enhance the GA level. GA promotes the degradation of DELLAs resulting in the release of DELLA from BZR1, forming positive feedback regulation to repress AGL15. Meanwhile, VAL1/2 also directly binds to the proximal promoter region of the AGL15 locus to repress its expression. Together, VAL1/2 and BZR1/BES1 collaborate to directly repress AGL15 transcription, thereby maintaining the appropriate silencing of the seed maturation program during the seed-to-vegetative transition.
Although BR has been extensively studied for several decades, the role of BR in repressing the seed maturation program during seedling growth has been overlooked. Most studies on the biological processes controlled by BR have focused on characterizing the morphological phenotypes of BR-deficient or BR-insensitive mutants. The seedlings of these mutants exhibit multiple morphological deficits but do not display embryonic phenotypes , which has hindered the discovery of the role of BR in repressing embryonic traits the during seed-to-seedling transition. In the current study, we used the val1-2 mutant for analysis. VAL1 is a B3-domain transcriptional repressor required for silencing seed maturation genes in seedlings. The val1-2 seedlings show substantial de-repression of seed maturation genes but do not display visible embryonic structures (Tsukagoshi et al., 2007). Here we found that inhibiting BR synthesis, the loss of the BR receptor, or the knockout of a key BR synthesis enzyme led to ectopic formation of embryonic structures in val1-2 seedlings (Figures 1 and 2, A–D; Supplemental Figure S3), allowing us to uncover the role of BR in suppressing the seed maturation program. The observation that the expression of the seed maturation gene AGL15 was de-repressed in BR-insensitive mutant seedlings further supported the notion that BR signaling is required for repressing the embryonic program during vegetative growth (Figure 4F).
BZR1 and BES1 are key transcription factors in the BR signaling pathway. We demonstrated that BR signaling represses the seed maturation program in seedlings through the BZR1/BES1-mediated direct repression of AGL15 (Figure 3; Supplemental Figure S7). Both bzr1-1D-GFP and bes1-D-GFP were substantially enriched in the promoter region of AGL15 (Figure 3A; Supplemental Figure S7), and the enrichment could be enhanced by BL treatment (Figure 3B). Furthermore, the activity of the AGL15 promoter was repressed by the overexpression of BZR1/BES1, as evidenced by the LUC reporter assays (Figure 3, C and D). Based on these results, we suggest that BZR1/BES1 directly repress AGL15 expression in Arabidopsis seedlings. AGL15 was previously reported to function upstream of the LAFL genes to induce their expression (Chen et al., 2018). AGL15 binds to the FUS3 promoter upstream of the transcription start site and to the second exon–intron boundary of LEC2 (Zheng et al., 2009). Therefore, a decrease in AGL15 levels might result in the downregulation of LEC1, LEC2, ABI3, and FUS3. Indeed, we observed that LAFL gene expression was significantly reduced when agl15 was introduced into the val1-2 bri1-5 double mutants (Figure 4E). Moreover, the lack of AGL15 significantly reduced the formation of embryonic structures in val1-2 bri1-5 or BRZ-treated val1-2 seedlings (Figure 4, A–D). However, agl15 did not completely rescue the embryonic structures in val1-2 bri1-5 or BRZ-treated val1-2 seedlings (Figure 4, B and D). Together, these results indicate that BR acts largely through AGL15 to repress the seed maturation program in seedlings, but additional BR target genes appear to be required for the full repression of this program.
Our observation that ABI3 is not directly targeted by BES1 is in conflict with a previous study (Ryu et al., 2014), which reported that BES1, but not BZR1, is enriched at the 5′ untranslated region (5′ UTR) of ABI3. One possible reason for these conflicting results is that BES1 might bind dynamically to ABI3. Ryu et al. used 5-d-old seedlings in their ChIP assays, whereas we used 14-d-old seedlings in the current study. Perhaps BES1 binds to ABI3 locus only during the early stage after germination and disassociates upon further plant growth. This could explain why we detected BES1 and BZR1 accumulation at the AGL15 locus but not at the LAFL loci in 14-d-old Arabidopsis seedlings. Thus, it remains possible that BES1/BZR1 is enriched at LAFL loci during other developmental stages.
Interestingly, VAL1 directly binds to the proximal promoter region of the AGL15 locus, which contains two RY-elements, to repress its expression (Chen et al., 2018). The RY binding motifs of VAL1 in the AGL15 promoter were close to the E-box in the BZR1/BES1 binding site (Figure 3A). Thus, VAL1 and BZR1/BES1 bind to the same region in the AGL15 promoter, where they may collaborate to repress the seed maturation program. The loss of BR enhanced the embryonic phenotypes of val1-2 and val1-2 val2-1 seedlings, and BL treatment did not decrease the ratio of val1-2 val2-1 seedlings with embryonic properties (Figure 5, A and B). These genetic data further support the notion that the BR-mediated pathway represses seed maturation functions synergistically with the VAL1/2-mediated pathway to ensure the full repression of the seed maturation program. It would be interesting to further explore the molecular crosstalk between the BR- and VAL1/2-mediated pathways in controlling the repression of the seed maturation program.
Both BR and GA are growth-promoting hormones that have similar effects on a wide range of growth and developmental processes (Depuydt and Hardtke, 2011). Previous studies have shown that BR and GA have strong cross-talk and act interdependently to promote cell elongation through a direct interaction between the BZR1 and the DELLA transcription factors (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012). BR-activated BZR1/BES1 directly bind to the non-E-box motif in promoters of GA biosynthesis genes and promote GA biosynthesis (Unterholzner et al., 2015). DELLAs interact with BZR1 to directly inhibit the DNA binding ability of BZR1. GA, through promoting the degradation of DELLAs, could enhance the BZR1 binding activity on target genes (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012). Interestingly, similar to BR, GA was previously shown to enhance the repression of the seed maturation program by VAL1 (Suzuki et al., 2007), suggesting that GA is also involved in the repression of embryonic traits. Based on these results, we propose that BR-activated BZR1/BES1 could directly induce the expression of GA biosynthesis genes to enhance the GA level during seed germination. Increased GA in turn promotes the degradation of DELLAs, resulting in the release of DELLAs from BZR1. The released BZR1/BES1 physically bind to AGL15 promoter and GA biosynthesis genes, forming a positive feedback loop to repress AGL15 expression during the seed-to-vegetative transition (Figure 6). Because AGL15 activates the expression of LAFL genes including LEC1, LEC2, FUS3, and LEC2, the repression of AGL15 by BZR1/BES1 results in the reduced expression of the LAFL genes. Meanwhile, VAL1/2 also directly bind to the proximal promoter region of the AGL15 locus to repress its expression (Chen et al., 2018). Together, BZR1/BES1 and VAL1/2 act synergistically to directly repress AGL15 transcription, thereby maintaining the silencing of the seed maturation program (Figure 6).
Previous studies have identified multiple epigenetic regulators involved in suppressing the seed maturation program. These regulators include the histone deacetylases HDA19 and HDA6 (Tanaka et al., 2008), the Switch defective/sucrose non-fermentable (SWI/SNF) chromatin remodeler ATPase BRAHMA (BRM; Tang et al., 2008), the Chromodomain-Helicase-DNA binding 3-type chromatin remodeler ATPase PICKLE (PKL; Ogas et al., 1999), and Polycomb group (PcG) proteins (Chanvivattana et al., 2004; Lafos et al., 2011). The loss of HDA19 and HDA6 leads to the formation of somatic embryos (Tanaka et al., 2008). We previously showed that HDA19 interacts with VAL2 to mediate the repression of seed maturation genes (Zhou et al., 2013). BRM directly associates with several seed storage protein genes to inhibit their expression during seedling growth (Tang et al., 2008). pkl seedlings exhibit embryonic characteristics and the de-repression of LAFL genes in roots (Ogas et al., 1999; Zhang et al., 2012). PKL-induced repression of LAFL genes is partially mediated by its interaction with PcG proteins (Zhang et al., 2012). Finally, PcG proteins catalyze the formation of repressive H3K27me3 marks on seed maturation genes, and seedlings lacking PcG undergo somatic embryogenesis accompanied by the overexpression of LAFL and AGL15 genes (Chanvivattana et al., 2004; Lafos et al., 2011). Although the functional interplay among these repressors of the seed maturation program is currently unknown, these studies suggest that plants have evolved a complex regulatory network to repress the seed maturation program during germination. Our discovery of the role of BR signaling in the repression of the seed maturation program adds another layer to this complex process. Interestingly, BES1 forms a complex with HDA19 to inhibit ABA-mediated arrest of early seedling development (Oh et al., 2014; Ryu et al., 2014). In addition, PKL physically interacts with BZR1 at cell elongation-related genes to regulate skotomorphogenesis (Zhang et al., 2014). These findings indicate that cross-talk occurs between BR signaling and epigenetic regulators during seedling growth. It would be interesting to investigate the possible links between BR signaling-activated BZR1/BES1 and these epigenetic repressors in the suppression of the seed maturation program.
Plant cells have remarkable developmental plasticity, allowing an entire plant to be regenerated from fully differentiated somatic cells (Ikeuchi et al., 2013). However, intact plants rarely develop somatic embryos under favorable conditions. Therefore, plants are equipped with robust mechanisms to prevent unscheduled dedifferentiation, but little is known about how this process is controlled. Our findings suggest that BR signaling during vegetative development may pose a barrier to somatic embryogenesis from seedlings. Interestingly, mutation of VAL1 does not cause obvious developmental defects. Therefore, our finding that inhibiting BR activity induces the embryonic characteristics in the val1-2 mutant background may facilitate the development of approaches to enhance the efficiency of somatic embryogenesis.
Materials and methods
Plant materials and growth conditions
The val1-2 (salk_088606C; Schneider et al., 2016), val2-1 (salk_058569; Tsukagoshi et al., 2007), and agl15-3 (CS16479; Adamczyk et al., 2007) mutants were obtained from Arabidopsis Biological Resource Center (ABRC). The det2-1 (Li et al., 1996) and bri1-5 (Noguchi et al., 1999) mutants were kindly provided by Prof. Hongwei Xue. The bes1-D (Yin et al., 2002) and bzr1-1D (Wang et al., 2002) mutants were kindly provided by Prof. Junxian He. All plants were in the Columbia-0 (Col-0) background except for bri1-5 and bes1-D, which are in the Ws and Enkheim-2 (En-2) ecotypes, respectively. Homozygous mutants were identified by PCR-based genotyping or sequencing.
Sterilized Arabidopsis seeds were stratified for 4 d at 4°C in darkness. The seeds were then sown on the soil or agar plates containing 2.2 g/L Murashige and Skoog (MS) nutrient mix (Duchefa), 1% (w/v) sucrose (pH 5.8), and 0.45% (w/v) agar. For BRZ, PPZ, or BL treatment, 1 μM BRZ (Sigma, Cat. No. SML1406), 2 μM PPZ (Sigma, Cat. 45642), or 100 nM BL (Sigma, Cat. E1641) was added into the MS plates. Plants were grown in growth rooms with 16-h light/8-h dark cycles at 22°C. For hypocotyl elongation experiments, seeds were placed on 1/2-strength MS medium (1/2 MS) supplied with PPZ or mock solution. After germination was induced by white light for 6 h, the plates were placed vertically under continuous darkness for another 5 d. Hypocotyl lengths were measured using Image J (Schneider et al., 2012).
Microscopy
To detect green fluorescence signal, root tips were cut from 4-d-old seedlings and transferred onto glass slides with 50-μL propidium iodide solution (1 μg/mL). Red fluorescent signals were detected by confocal microscopy (Leica) with excitation at 555 nm and emission at 655 nm. The green fluorescence signals were detected with excitation at 488 nm and emission at 505 nm to 525 nm.
Sudan Red 7B (Fat Red 7B) Staining
Sudan red staining was performed as described (Brundrett et al., 1991). Briefly, a 0.1% (w/v) solution of Sudan red 7B (Sigma) was prepared by dissolving the dye in polyethylene glycol (average molecular mass = 400 D, Sigma) and heating for 1 h at 90°C. An equal volume of 90% (v/v) glycerol was added after cooling down. Whole seedlings grown on 1/2 MS medium were supplemented with 0.1% (w/v) solution of Sudan red 7B for 1 h at room temperature. Samples were then rinsed with 50% ethanol overnight to remove chlorophyll.
Extraction of seed storage proteins
One hundred milligrams of seedlings or dry seeds were used as material for total protein extraction. Materials were ground in liquid nitrogen and then incubated in 200 μL protein extraction buffer ((20 mM Tris–HCl, pH7.5, 150 mM NaCl, 0.5% (v/v) Tween-20, 1 mM Ethylene Diamine Tetraacetic Acid (EDTA), 1 mM DTT, 1 mM Phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail)) for 30 min at 4°C. Supernatant was collected after two cycles of centrifugation (10 min, 16,000 g, 4°C). Proteins were resolved in 4%–20% protein gels (GenScript, SurePAGE, Cat. 4%–20%) and stained with InstaBlue Protein Stain Solution (APExBIO, Cat. B8226).
Generation of transgenic plants
Genomic regions corresponding to full-length bes1-D and bzr1-1D, including a 2-kb promoter region and the coding sequences without the stop codon, were amplified and subcloned into the pDONR221 vector (Invitrogen) by BP reaction. The resulting entry vectors were sequenced to ensure that no error was introduced during PCR amplification. The inserts were then transferred into the pMDC107 vector (Curtis and Grossniklaus, 2003) by LR reaction (to generate pBES1::bes1-D-GFP and pBZR1::bzr1-1D-GFP). The constructs were introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform Col-0 plants using the floral dip method (Clough and Bent, 1998). Sequences for the primers used are listed in Supplemental Table S1.
Chromatin immunoprecipitation
ChIP was performed as described (Gendrel et al., 2005; Yu et al., 2020) with minor modifications. Briefly, for each biological replicate, 2 g of 14-d-old seedlings grown on 1/2 MS medium were collected and cross-linked with 1% (v/v) formaldehyde for 15 min under vacuum. Chromatin was isolated and sonicated to generate DNA fragments with an average size of 500 bp using the Bioruptor sonicator with a 30 s ON and a 30 s OFF cycle (total ON cycles: 27) at HIGH setting. The sonicated chromatin was incubated with 2 μL of antibody to GFP (Abcam, ab290) overnight at 4°C. Precipitated DNA was then recovered and analyzed by quantitative PCR with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711-02) on a Step One Plus Real-Time PCR machine (ABI). ChIP-qPCR was performed with three technical replicates, and relative fold enrichment was calculated by normalizing the amount of a target DNA fragment immunoprecipitated against that of the respective input DNA samples and then against TUB2 promoter fragment. ChIP experiments were performed at least three times. Sequences for the primers used for ChIP-qPCR are listed in Supplemental Table S1.
Immunoblotting
Nuclei were extracted from 14-d-old seedlings according to the ChIP protocol but without the tissue fixation step. Nuclear proteins were released by incubating the nuclei preparation in 300 μL of lysis buffer ((50 mM Tris–HCL, 10 mM EDTA, 1% (w/v) Sodium dodecyl sulfate (SDS), 1× protease inhibitor)) for 3 h at 4°C. The extract was then diluted with 1 volume of ChIP dilution buffer that was centrifuged at 15,000 g for 10 min at 4°C to remove debris. Proteins were resolved in 4%–12% protein gels (GenScript, SurePAGE, Cat. M00653) by electrophoresis and immunoblotted using anti-GFP (Abcam, ab290; 1:5,000 dilution) or histone H3 (Millipore, 07-108; 1:10,000 dilution). Histone H3 was used as the loading control.
Analysis of transcript levels
Total RNA was extracted with RNAprep Pure Plant Plus Kit (TIANGEN; DP441) according to the manufacture’s protocol. The first-strand cDNA was synthesized using HiScript II Q Select RT SuperMix (Vazyme, R233-01). The cDNA samples were diluted 10-fold and then used as PCR templates. RT-qPCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711-02) on a Step One Plus Real-Time PCR machine (ABI). Gene transcript levels were calculated from three replicates using the ΔΔCt method after being normalized to ACT2. All the RT-qPCR analyses were performed for three biological replicates. Data shown are means and SD from three biological replicates. The primers used are listed in Supplemental Table S1.
Dual-LUC assay
The AGL15 promoter was amplified and cloned into BamH I linearized pGreenII-0800-LUC reporter vector (Hellens et al., 2005) to generate pAGL15::LUC reporter construct. The coding sequences of BZR1 and BES1 were amplified and inserted into BamH I linearized pGreenII 62-SK vector (Hellens et al., 2005) using ClonExpress II One Step Cloning Kit (Vazyme) to produce p35S:BZR1 and p35S:BES1 effectors, respectively. The constructed vectors were transformed into A. tumefaciens strain GV3101 along with the pSoup vector (Hellens et al., 2005). An empty pGreenII 62-SK vector was used as a control. After three days, the Firefly LUC and Renilla (REN) activities were quantified using Dual-Luciferase Reporter Gene Assay Kit (Yeasen). Relative LUC activity was expressed as the ratio of LUC to REN. Five biological replicates per combination were performed. Sequences for the primers used for producing reporter constructs and effector constructs are listed in Supplemental Table S1.
Accession numbers
Genes referenced in this article can be found in The Arabidopsis Information Resource (TAIR) under the following accession numbers: AGL15 (AT5G13790); VAL1 (AT2G30470); VAL2 (AT1G60860); BZR1 (AT1G75080); BES1 (AT1G19350); LEC1 (AT1G21970); ABI3 (AT3G24650); FUS3 (AT3G26790); LEC2 (AT1G28300); 2S1 (AT2G05380); OLE1 (AT4G25140); CRA1 (AT5G44120); CNX5, (At5g55130) and DWF4 (AT3G50660).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. PPZ treatment induces embryonic traits in the val1-2 mutant.
Supplemental Figure S2. Sudan red staining of val1-2 bri1-5 mutants.
Supplemental Figure S3. Activation of the embryonic program in val1-2 det2-1 seedlings.
Supplemental Figure S4. Both bes1-D and bzr1-1D rescue the embryonic traits of BRZ-treated val1-2 seedlings.
Supplemental Figure S5. Transgenic plants expressing bes1-D-GFP and bzr1-1D-GFP.
Supplemental Figure S6. BZR1-1D does not associate with the LEC1, LEC2, ABI3, FUS3 loci.
Supplemental Figure S7. BES1-D associates with the AGL15 locus.
Supplemental Figure S8. BL treatment reduces the expression of AGL15 in WT.
Supplemental Figure S9. The phenotype of val1-2 val2-1 plants grown in soil.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the ABRC for seeds of T-DNA insertion lines, Prof. Hongwei Xue for providing det2-1 and bri1-5 seeds, and Prof. Junxian He for providing bes1-D and bzr1-1D seeds.
Funding
This work was supported by the National Natural Science Foundation of China to CL (31870289 and 32070212), LY (32000380), YL (32000249), SH (31470287) and JL (31871716), the Fundamental Research Funds for the Central Universities, Sun Yat-sen University to CL (18lgzd12).
Conflict of interest statement. The authors declare no conflict of interest.
J.R. and C.L. conceived the project. J.R., H.C., T.Z., Y.Y., Y.L., and L.Y., performed experiments. J.R., H.C., T.Z., Y.L., J-F. K., W-J. L., Z-Y.W., J.L., and S.H. analyzed data. C.L. wrote the manuscript.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Chenlong Li (lichlong3@mail.sysu.edu.cn).
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC, Kendrick B, Peterson CA (1991) Efficient lipid staining in plant material with sudan red 7B or fluorol yellow 088 in polyethylene glycol-glycerol. Biotech Histochem 66: 111–116 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen N, Veerappan V, Abdelmageed H, Kang M, Allen RD (2018) HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 30: 600–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–373 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A-V, Lippman Z, Martienssen R, Colot V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Hartwig T, Corvalan C, Best NB, Budka JS, Zhu JY, Choe S, Schulz B (2012) Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yu D (2014) BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A (2013) Plant callus: mechanisms of induction and repression. Plant Cell 25: 3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, McCarty DR, Suzuki M (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Suzuki M, McCarty DR (2014) Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip Rev Dev Biol 3: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Warpeha KM, Huber SC (2019) The brassinosteroid receptor kinase, BRI1, plays a role in seed germination and the release of dormancy by cold stratification. J Plant Physiol 241: 153031. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li QF, Wang C, Jiang L, Li S,, Sun SS, He JX (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal 5: ra72. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Ueno M, Yamada Y, Takatsuto S, Takeuchi Y, Yokota T (2007) Roles of brassinosteroids and related mRNAs in pea seed growth and germination. Plant Physiol 143: 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014) TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5: 4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun 5: 4138. [DOI] [PubMed] [Google Scholar]
- Schneider A, Aghamirzaie D, Elmarakeby H, Poudel AN, Koo AJ, Heath LS, Grene R, Collakova E (2016) Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J 85: 305–319 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W,, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, Reyes JC, Wang A, Keller WA, Harada JJ, Tsang EW, Cui Y (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Morikami A, Nakamura K (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104: 2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B (2015) Brassinosteroids Are Master Regulators of Gibberellin Biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang J, Liu J, Hu J, Liu J, Chen Y, Cai Z, Wang X (2018) Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol Plant 11: 315–325 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Xue LW, Du JB, Yang H, Xu F, Yuan S, Lin HH (2009) Brassinosteroids counteract abscisic acid in germination and growth of Arabidopsis. Z Naturforsch C J Biosci 64: 225–230 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu Y, Liang Z, Song X, Fu W, Xu J, Lei Y, Yuan L, Ruan J, Chen C, Fu W, Cui Y, Huang S, Li C (2020) BRAHMA-interacting proteins BRIP1 and BRIP2 are core subunits of Arabidopsis SWI/SNF complexes. Nat Plants 6: 996–1007 [DOI] [PubMed] [Google Scholar]
- Yuan L, Song X, Zhang L, Yu Y, Liang Z, Lei Y, Ruan J, Tan B, Liu J, Li C (2021) The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res 49: 98–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R (2014) The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W,, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA 106: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Dou L, Gong Z, Wang X (2019) BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol 221: 908–918 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, et al. (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.