Abstract
When a dark-germinated seedling reaches the soil surface and perceives sunlight for the first time, light signaling is activated to adapt the plant’s development and transition to autotrophism. During this process, functional chloroplasts assemble in the cotyledons and the seedling’s cell expansion pattern is rearranged to enhance light perception. Hypocotyl cells expand rapidly in the dark, while cotyledon cell expansion is suppressed. However, light reverses this pattern by activating cell expansion in cotyledons and repressing it in hypocotyls. The fact that light-regulated developmental responses, as well as the transcriptional mechanisms controlling them, are organ-specific has been largely overlooked in previous studies of seedling de-etiolation. To analyze the expansion pattern of the hypocotyl and cotyledons separately in a given Arabidopsis (Arabidopsis thaliana) seedling, we define an organ ratio, the morphogenic index (MI), which integrates either phenotypic or transcriptomic data for each tissue and provides an important resource for functional analyses. Moreover, based on this index, we identified organ-specific molecular markers to independently quantify cotyledon and hypocotyl growth dynamics in whole-seedling samples. The combination of these marker genes with those of other developmental processes occurring during de-etiolation will allow improved molecular dissection of photomorphogenesis. Along with organ growth markers, this MI contributes a key toolset to unveil and accurately characterize the molecular mechanisms controlling seedling growth.
Organ-specific molecular markers provide an important toolset to assess seedling growth in Arabidopsis.
Introduction
The ability to transition from heterotrophic to autotrophic growth is crucial for plant survival. When a seedling germinating under darkness perceives light for the first time, it must undergo a myriad of developmental adaptations to succeed in this transition and develop photomorphogenically, a process known as de-etiolation (Arsovski et al., 2012). Most of these light-regulated biological processes are organ-specific (Montgomery, 2016), including chloroplast development and photosynthesis, which are strongly induced upon light exposure in cotyledons while only minimally increased in the hypocotyl. Among the organ-specific responses, cell expansion, which drives seedling morphogenesis, is remarkable in that it is oppositely affected in these two organs by both light and dark. In the dark, cell expansion is active in the hypocotyl and suppressed in cotyledons (skotomorphogenesis), but light reverses this pattern, inducing cell expansion in cotyledons and repressing it in the hypocotyl (photomorphogenesis). This growth switch allows plants to rapidly expose their photosynthetic tissues (cotyledons) to sunlight.
Although the different physiological programs occurring in hypocotyls and cotyledons during skoto and photomorphogenic development are well known, the molecular pathways accounting for their specificity are not fully understood (Gommers and Monte, 2018). Both phytochrome interacting factors (PIFs) and elongated hypocotyl 5 (HY5) have been found to play key roles in the control of light-regulated morphogenesis (Shi et al., 2018). PIFs are basic helix-loop-helix (bHLH) transcription factors negatively regulated by photoactivated phytochromes (PHYs), the red (R) and far-red (FR) light plant photoreceptors (Al-Sady et al., 2006; Leivar and Monte, 2014), and are, therefore, active in the dark to repress photomorphogenesis. Consequently, higher-order mutants for four of the seven PIFs (pif1pif3pif4pif5; pifq) exhibit a constitutive photomorphogenic phenotype in the dark: open, expanded cotyledons and short hypocotyls (Leivar et al., 2008; Shin et al., 2009). By contrast, the bZIP transcription factor HY5 remains inactive in the dark due to the repressive action of the ubiquitin E3 ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and SUPRESSOR of PHYA (SPA) proteins (Osterlund et al., 2000; Saijo et al., 2003; Gangappa and Botto, 2016). Once dark-germinated seedlings detect the presence of light, HY5 action becomes crucial to control the expression of light-regulated genes. In line with its molecular function, mutants for the corresponding gene display hyposensitivity to light (Oyama et al., 1997; Lee et al., 2007).
Molecular markers used to globally quantify photomorphogenesis often encode chloroplast proteins markedly induced by light (Tobin and Silverthorne, 1985; Ma et al., 2001; Leivar et al., 2009). Because of their transcriptional misregulation in Arabidopsis (Arabidopsis thaliana) mutants displaying constitutive photomorphogenic phenotypes under darkness (Ma et al., 2002; Alabadí et al., 2004; Leivar et al., 2009), it has always been assumed that misregulation of these genes is associated with alterations in the seedling’s photomorphogenic status. However, as photomorphogenesis comprises multiple physiological processes, this link between photomorphogenesis and the expression of chloroplast-related genes may not stand for the quantification of other physiological processes with a more complex light and/or organ regulation, as we demonstrate here for growth responses. To circumvent these conceptual and experimental limitations of classical molecular markers for morphological changes associated with de-etiolation, we identify here marker genes for early growth dynamics that take the distinct cell-expansion pattern of hypocotyl and cotyledons into account. To integrate both these growth patterns in a given seedling, we define a ratio between the two organs called morphogenic index (MI), which can be applied to either phenotypic or molecular data (molMI) and provides a key tool to assess precisely seedling growth responses.
Results
Classical markers of photomorphogenesis fail to quantify accurately seedling growth dynamics
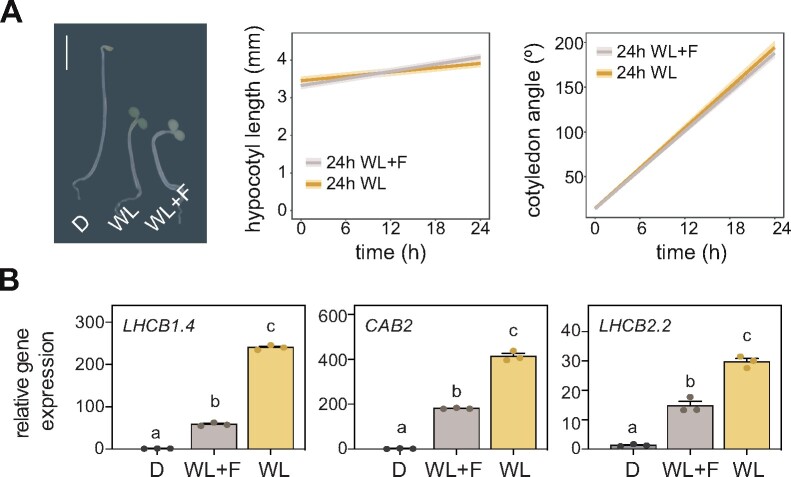
To evaluate the need for defining photomorphogenic markers focused on growth responses, we tested the association between the expression levels of classical photomorphogenic markers and morphogenic responses under particular growth conditions. Given that classical markers encode for chloroplast proteins, we challenged plants with the carotene biosynthesis inhibitor fluridone (Bartels and Watson, 1978), which affects chloroplast function without impacting growth responses (Figure 1A). Interestingly, under these conditions, classical photomorphogenic markers were downregulated (Figure 1B) in line with chloroplast function repression, but this did not correlate with the seedling’s photomorphogenic phenotype, which was unaffected by the drug (Figure 1A). This experiment demonstrated the experimental limitations of classical molecular markers of photomorphogenesis to quantify accurately growth responses during de-etiolation.
Figure 1.
Expression levels of classical molecular markers of photomorphogenesis under specific de-etiolation conditions. (A) Representative image of seedlings grown for 2 d in the dark and then treated for 24 h with dark (D), WL or white light and fluridone (WL + F; left), as well as quantification of hypocotyl elongation (center) and cotyledon aperture (right) in seedlings subjected to the L or L + F treatments. Bar, 2.5 mm. Measurements from the initial and final time points are shown. Thick lines and shaded areas represent respectively the median and the interquartile range of at least 45 seedlings. (B) RT-qPCR analysis of the expression of the classical photomorphogenic marker genes LHCB1.4, CAB2, and LHCB2.2 in seedlings grown as in A. The expression of each gene was normalized to that of PP2A and transcript levels expressed relative to the value of darkness, set to one. Data represent mean ± SE of three biological replicates. Different letters denote statistically significant differences between means (Tukey test; P < 0.05).
MI allows identification of organ-specific genes
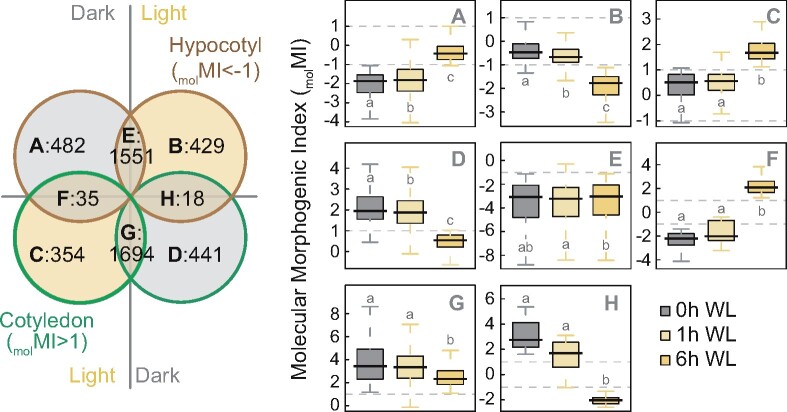
For an adequate assessment of seedling growth responses, we needed to take into account the different expansion dynamics of hypocotyl and cotyledon cells. Seedling dissection and collection of isolated organs, the most direct approach to study tissue-specific responses at the molecular level, is technically challenging particularly in the dark. We, therefore, aimed at defining organ-specific marker genes to score cell expansion in whole-seedling samples. To identify these markers, we first analyzed available data from an RNA-seq experiment where cotyledon and hypocotyls were independently collected from seedlings grown under darkness and during the first 6 h of light exposure (Sun et al., 2016). Given the opposite cell-expansion pattern in cotyledons and hypocotyls under dark and light (Supplemental Figure S1), we assumed that bona fide organ-expansion markers cannot be similarly expressed in both organs and defined tissue-specific genes at time 0 (dark) and after 6 h of light exposure (light). We performed a differential expression analysis between cotyledons and hypocotyls (fold change [FC] = cotyledon/hypocotyl) and called the logarithm of this value, molecular MI (molMI). We named genes with molMI > 1 cotyledon genes, while hypocotyl genes were those with molMI < −1. Based on the molMI value under the two light conditions, we then classified genes into eight categories, A–H (Figure 2)—hypocotyl or cotyledon genes only in the dark (A or D), only in the light (B or C), in both light and dark (E or G), or opposite patterns in dark and light (F and H; Supplemental Data set 1).
Figure 2.
Gene classification based on organ-specific expression patterns. Venn diagram of genes differentially expressed in cotyledon and hypocotyl samples in the dark and WL using RNA-seq data from Sun et al. (2016) (left), and boxplot representation of the molecular MI (molMI, log2 of cotyledon expression/hypocotyl expression) for genes belonging to the eight (A–H) defined categories (right). Boxplots indicate the median (center line), interquartile range (box limits), and minimum and maximum values (whiskers). Different letters denote statistically significant differences between medians (Kruskal–Wallis test; P < 0.05).
To address the consistency of the groups defined based on the data of Sun et al. (2016), we quantified the expression of genes in each group using two time-course experiments where cotyledons and hypocotyls were collected from seedlings exposed to light for 12 h (Burko et al., 2020; Supplemental Figure S2B), or treated with reduced R/FR ratio light (Kohnen et al., 2016; Supplemental Figure S2C), which mimics darkness in the regulation of seedling growth dynamics (Casal, 2013). Supplemental Figure S2 shows very similar molMI patterns observed in the three experiments for all gene groups, with changes in response to low R/FR ratio mirroring those observed in dark-grown seedlings.
Identification of organ-specific genes expressed under growth-promoting conditions
Once genes were grouped according to their tissue specificity during de-etiolation, we searched for expression patterns correlating with cell expansion. Group F comprises 35 genes expressed in expanding tissues, i.e. hypocotyls in the dark and cotyledons in the light (Figure 2; Supplemental Figure S3A), while the 18 genes in Group H exhibit the opposite pattern, being expressed in non-expanding tissues, i.e. cotyledon genes in the dark that become hypocotyl genes upon light exposure (Figure 2; Supplemental Figure S3B). Although in principle the most interesting, genes from these two groups were discarded because their contrasting organ expression pattern may fail to reflect light responsiveness in the whole seedling (Supplemental Figure S4A). In fact, even if the whole seedling’s expression changes in response to light, this will merely reflect the dynamics of the organ displaying the most marked expression changes or with the highest mRNA abundance (Supplemental Figure S4B). We also discarded genes similarly expressed in the same organ under both dark and light—groups E and G containing genes always expressed in hypocotyls or cotyledons, respectively (Figure 2; Supplemental Figure S5)—because the growth pattern of each tissue changes after light exposure.
We thus focused on the 1,706 genes of the remaining four categories (A, B, C, and D), which are tissue-specific under only one condition, dark or light (Figure 2; Supplemental Figure S6). Groups B and D genes are expressed in non-expanding tissues, i.e. hypocotyls in the light and cotyledons in the dark, respectively. Contrastingly, Groups A and C contain genes expressed specifically in expanding tissues. The 482 genes from Group A are expressed in hypocotyls in the dark, with mRNA levels dropping during the first hours of light exposure (Supplemental Figure S6A). This group of genes expressed in growing hypocotyls is enriched in cytoskeletal and cell-wall related functions (Supplemental Figure S7A), both closely linked to hypocotyl elongation. Inversely, Group C contains 354 genes lowly expressed in the dark and induced upon light exposure only in cotyledons (Supplemental Figure S6C), hence correlating with induction of cell expansion in this tissue. Functions of these genes expressed in expanding cotyledons include those related to translation (Supplemental Figure S7B), consistent with a recent report that this process is crucial for proper light-regulated cotyledon development (Chen et al., 2018).
Identification of organ-expansion genes misregulated in light-signaling mutants
To narrow down our list of organ-expansion genes, we next searched for genes expressed in expanding hypocotyl and cotyledon cells that are misregulated in seedlings with extensive growth defects. To this end, we performed an RNA-seq experiment comparing wild-type (WT) seedlings with pifq, hy5, and phyab mutants grown for 3 d in darkness or light. The importance of harboring functional copies of these genes for a proper skotomorphogenic or photomorphogenic development has been extensively reported (Whitelam et al., 1998; Gommers and Monte, 2018). Thus, in accordance with a myriad of previous studies, pifq mutants exhibited short hypocotyls and expanded cotyledons when growing in the dark, while hy5 and phyab showed the most dramatic morphogenic defects when exposed to light, namely longer hypocotyls and/or more closed cotyledons (Supplemental Figure S8).
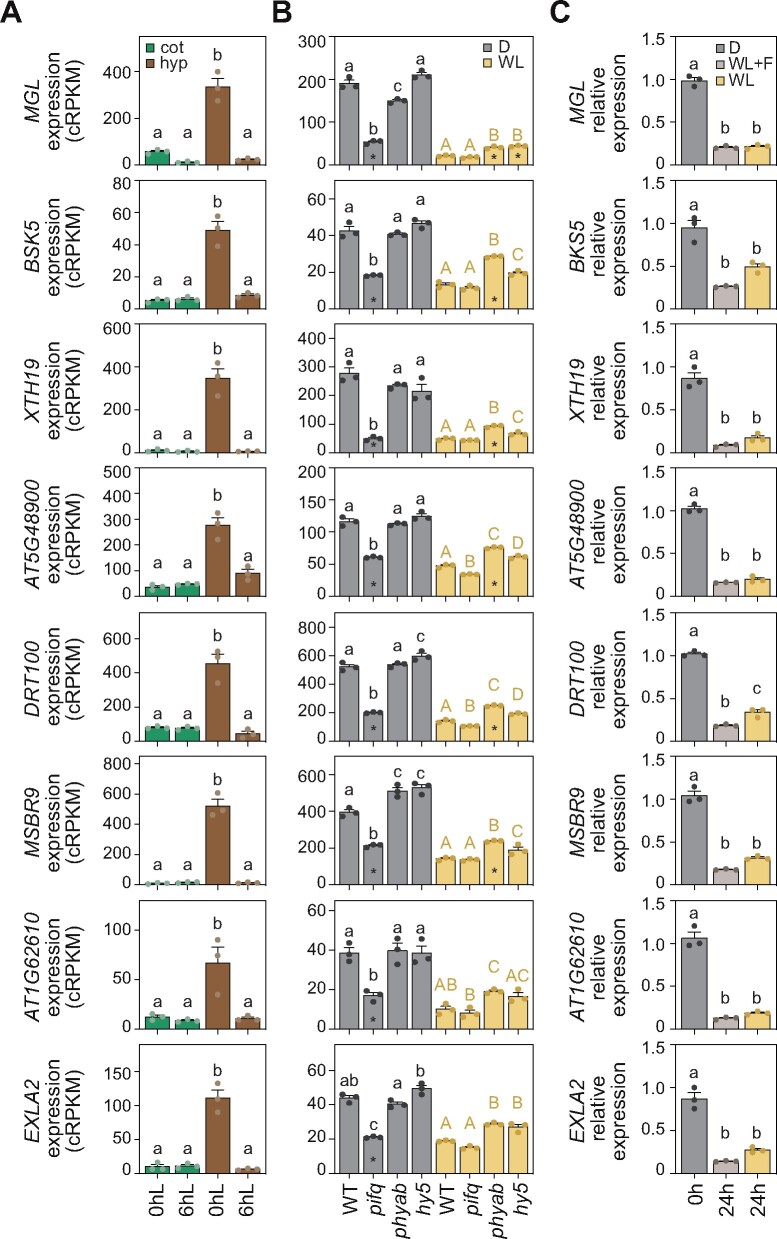
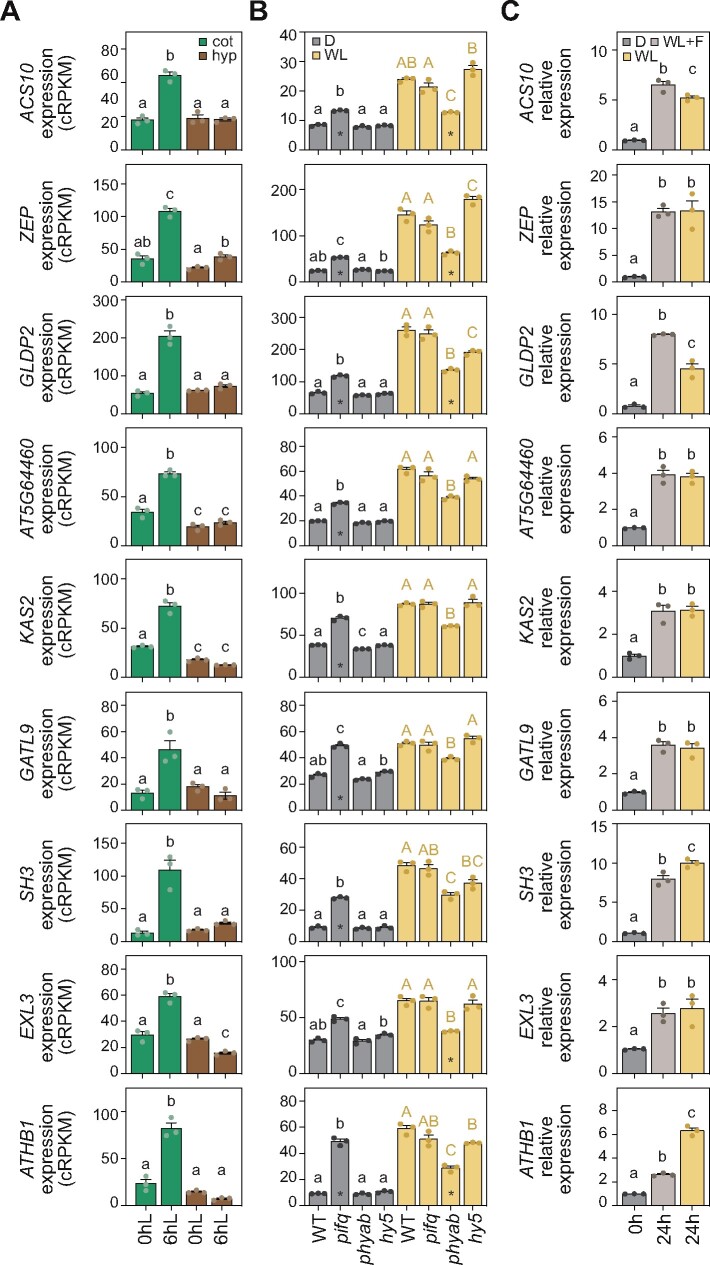
Based on statistical and fold-change criteria (see “Material and methods”), we selected genes expressed in growing hypocotyls (Group A, Figure 3A), that are strongly downregulated in dark-grown pifq mutants and/or upregulated in phyab and hy5 light-grown seedlings (Figure 3B). These expression patterns clearly match the hypocotyl phenotypes observed in these seedlings (Supplemental Figure S8). We then followed the same rationale for genes expressed in expanding cotyledons agreeing with mutant cotyledon phenotypes (Supplemental Figure S8). We thus selected genes from Group C (Figure 4A) differentially upregulated only in dark-grown pifq seedlings and/or downregulated only in phyab seedlings grown in the light (Figure 4B). These criteria defined 18 putative organ-expansion markers, for which expression analysis in hypocotyl and cotyledon tissues isolated from WT and phyab light-grown seedlings indicated that the differences found between genotypes were largely caused by differences in the relevant organ (Supplemental Figure S9). Noteworthily, for ATHB1 and MSBR9, we could not reproduce the differences in expression observed when whole-seedling WT and phyab samples were compared (Figures 3B, 4B; Supplemental Data set 2). We, therefore, discarded these two genes from our list of putative organ-expansion molecular markers. Next, to ensure misexpression of the remaining genes, we used available RNA-seq data of two mutants that like pifq present a constitutive photomorphogenic phenotype in the dark: the COP1 (cop1) mutant and the quadruple mutant of SPA (spaQ) proteins (Pham et al., 2018). In general, selected hypocotyl-expansion genes were downregulated in cop1 and spaQ mutants (Supplemental Figure S10A), correlating with these plants’ shorter hypocotyls, while cotyledon-expansion genes were highly expressed in these mutants (Supplemental Figure S10B), which exhibit expanded cotyledons. However, among all genes analyzed, we discarded the putative hypocotyl-expansion marker BEE1 (Figure 3), as its expression is unaffected in dark-grown spaQ mutants (Supplemental Figure S10A), indicating disassociation between downregulation of this gene and shortened hypocotyls.
Figure 3.
Gene expression analysis of putative hypocotyl-expansion molecular markers. (A) Expression values of selected hypocotyl-expansion marker genes in cotyledon and hypocotyl samples from Sun et al. (2016) collected in the dark and after 6 h of light. (B) Expression values from RNA-seq data of the set of genes analyzed in A in WT, pifq, hy5-2, and phyab seedlings grown for 3 d in darkness or light. Asterisks indicate a change of at least 1.5-fold. (C) RT-qPCR analysis of the set of genes analyzed in A and B in WT seedlings grown for 2 d in the dark and then treated for 24 h with dark (D), WL, or white light and fluridone (WL + F). Expression of each gene was normalized to that of PP2A and transcript levels were expressed relative to the value of dark set to one. In A–C, data represent mean ± SE of three biological replicates, and different letters denote statistically significant differences between means (Tukey test; P < 0.05).
Figure 4.
Gene expression analysis of putative cotyledon-expansion molecular markers. (A) Expression values of selected cotyledon-expansion marker genes in cotyledon and hypocotyl samples from Sun et al. (2016) collected in the dark and after 6 h of light. (B) Expression values from RNA-seq data of the set of genes analyzed in A in WT, pifq, hy5-2, and phyab seedlings grown for 3 d in darkness or light. Asterisks indicate a change of at least 1.5-fold. (C) RT-qPCR analysis of the set of genes analyzed in A and B in WT seedlings grown for 2 d in the dark and then treated for 24 h with dark (D), WL, or white light and fluridone (WL + F). Expression of each gene was normalized to that of PP2A and transcript expressed relative to the value of dark set to one. In A–C, data represent the mean ± SE of three biological replicates, and different letters denote statistically significant differences between means (Tukey test; P < 0.05).
Validation of organ-specific growth molecular markers
Finally, to experimentally validate the selected genes as hypocotyl and cotyledon growth markers, we tested their expression in samples where classical markers had failed to quantify light-regulated morphological patterns (Figure 1). Remarkably, the expression of hypocotyl genes was downregulated, while cotyledon genes were upregulated in light-grown seedlings (Figures 3C, 4C). Also, for the vast majority of these genes, no significant differences were observed between light samples, with or without fluridone, clearly matching the observed phenotypes (Figure 1A). Among the 18 genes analyzed (Figures 3, 4), only two were not confirmed: the cotyledon genes GLDP2 and ATHB1, the latter of which had already been discarded (Supplemental Figure S9B). Although the expected expression pattern trend was observed for both genes, statistical analysis revealed important differences between light-grown samples treated with or without fluridone (P-value < 0.001; Figure 4C). Having previously also eliminated MSBR9 and BEE1 (Supplemental Figures S9A, S10A), we thus defined the 14 genes that fulfilled all the criteria as organ-specific molecular markers for hypocotyl and cotyledon growth during early plant development (Table 1). For at least two of these molecular markers, XTH19 and EXLA2, seedling organ growth defects have been previously reported (Miedes et al., 2013; Boron et al., 2015).
Table 1.
List of organ-specific growth molecular markers
| Locus ID | Name | Short description | Classification |
|---|---|---|---|
| AT1G64660 | MGL | Methionine gamma-lyase | Hypocotyl marker |
| AT5G59010 | BSK5 | Protein kinase protein with tetratricopeptide repeat domain | Hypocotyl marker |
| AT4G30290 | XTH19 | Xyloglucan endotransglucosylase/hydrolase 19 | Hypocotyl marker |
| AT5G48900 | – | Pectin lyase-like superfamily protein | Hypocotyl marker |
| AT3G12610 | DRT100 | Leucine-rich repeat family protein | Hypocotyl marker |
| AT1G62610 | – | NAD(P)-binding Rossmann-fold superfamily protein | Hypocotyl marker |
| AT4G38400 | EXLA2 | Expansin-like A2 | Hypocotyl marker |
| AT1G62960 | ACS10 | ACC synthase 10 | Cotyledon marker |
| AT5G67030 | ZEP | Zeaxanthin epoxidase | Cotyledon marker |
| AT5G64460 | – | Phosphoglycerate mutase family protein | Cotyledon marker |
| AT1G74960 | KAS2 | Fatty acid biosynthesis 1 | Cotyledon marker |
| AT1G70090 | GATL9 | Glucosyl transferase family 8 | Cotyledon marker |
| AT5G64850 | SH3 | NA | Cotyledon marker |
| AT5G51550 | EXL3 | EXORDIUM LIKE 3 | Cotyledon marker |
MI applied to phenotyping for a direct assessment of the plant’s growth pattern
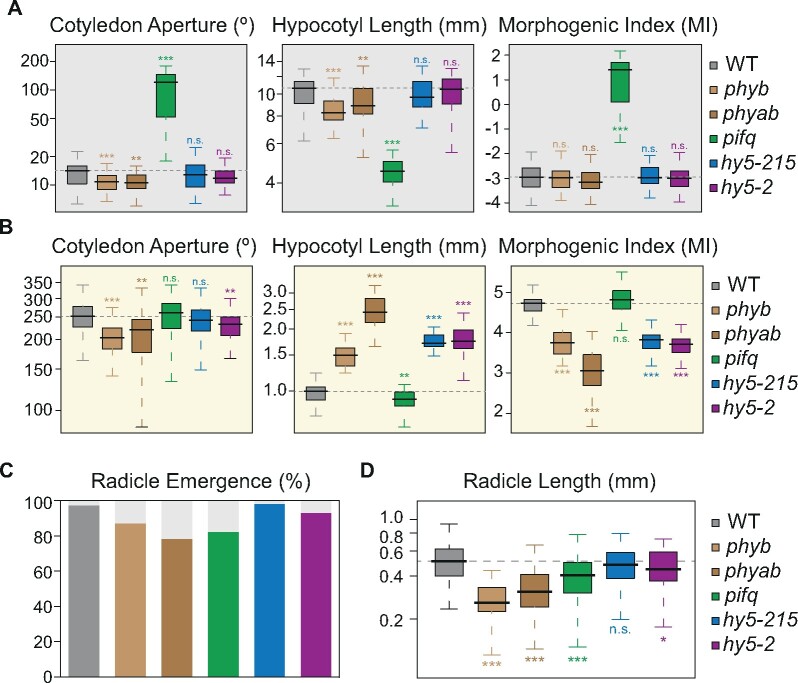
Importantly, the proposed MI, which integrates data from two organs with opposite cell-expansion patterns, can prove useful in the phenotypical analysis of dark- and light-grown seedlings by reducing measurements from different traits (cotyledon angle and hypocotyl length) to a single value (MI = log2 of the ratio between cotyledon angle and 10 times the hypocotyl length). It is also an evident asset when comparing morphogenesis of different seedling genotypes. As seen in Figure 5, A and B, the MI allows not only identifying seedlings with cell-expansion defects in either hypocotyl or cotyledons, or simultaneously in both tissues but with opposite trends, but also discarding mutants with converging defects in the two tissues, e.g. dark-grown phytochrome mutants with shorter hypocotyls and more closed cotyledons, which could be due to a delay in seed germination when compared to the WT (Figure 5, C and D). The suitability of the MI for the analysis of phenotypic data is also evident when assessing skoto and photomorphogenic growth of WT seedlings from different seed batches (Supplemental Figure S11), where no differences in skoto and photomorphogenesis are expected. Distinct hypocotyl elongation rates (Supplemental Figure S11, A and B), likely arising from developmental differences associated with the speed of germination (Supplemental Figure S11, C and D), are annulled by the MI ratio, which is similar for all seedling batches.
Figure 5.
MI assessment of seedling development in light-signaling mutants. (A–B) Boxplot representation of the cotyledon aperture (left), hypocotyl length (center), and MI (right) of seedlings of different genotypes grown in continuous dark (A) or light (B) for 3 d (n ≥ 44). (MI, log2 of cotyledon angle/10*hypocotyl length). (C–D) Percentage of emerged radicles (C) and boxplot representation of the radicle length (D), both measured upon 24 h in darkness after stratification of WT and different light-signaling mutant seeds (n ≥ 45). Boxplots indicate the median (center line), interquartile range (box limits), and minimum and maximum values (whiskers). In A–B and D, asterisks indicate statistically significant differences between each mutant and WT seedlings (Mann–Whitney test; *P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
Plants respond to a given stimulus by modulating a range of physiological processes, with these responses varying significantly among plant organs. A good illustration of this heterogeneity is the response of dark-germinated seedlings to light, which includes the regulation of different plant developmental processes such as growth, chloroplast biogenesis, or gravitropism. Importantly, these processes do not occur equally in the different seedling organs and neither do the molecular mechanisms controlling them (Li et al., 2011; Sun et al., 2016; Bellstaedt et al., 2019; Dong et al., 2019; Hamasaki et al., 2020; Yang et al., 2020). Although the tissue complexity of photomorphogenesis is widely recognized, molecular studies are only recently beginning to center on the particular dynamics of each organ. Hypocotyls and cotyledons display opposite growth regulation in darkness and light, being active in one organ when inactive in the other (Supplemental Figure S1). Focusing on this particular case, we show here that organ-centered studies are not only advantageous, but also essential to avoid erroneous conclusions. As illustrated in Supplemental Figure S4, genes that do not change their expression in response to light in whole-seedling samples would lead to the conclusion that their expression is not influenced by light. However, in some cases, their expression may be changing in opposite directions in each organ, which could represent an efficient light-regulation mechanism.
With the purpose of defining organ-specific molecular markers that allow an accurate quantification of growth responses during early seedling development, we introduce a measure combining hypocotyl and cotyledon information, the MI. Applying this ratio to transcriptomic data (molMI) successfully identified hypocotyl- and cotyledon-specific molecular markers that independently quantify the growth dynamics of each organ in whole-seedling samples. The possibility of quantifying organ-specific responses using the whole seedling avoids isolating the tissues and synthesizing cDNA from their RNA, which is not only technically difficult, especially for non-expanding tissues, but also implies processing twice the number of samples. The identified marker genes could prove useful in molecular screenings to detect growth regulators during early seedling development or in rapidly inferring phenotypic patterns from transcriptomic studies of specific genotypes. Moreover, complementing the use of these organ-specific growth markers with classical photomorphogenic marker genes focused on chloroplast-related functions will allow for an improved molecular dissection of the different developmental processes comprising photomorphogenesis. We have also shown that applying the MI to phenotypic data provides an important resource for functional analyses. In reducing different measures to a single value, this index allows discarding phenotypical differences resulting from developmental processes not related to photomorphogenesis such as germination defects. In sum, both the MI and the organ-expansion markers defined here are bound to provide key tools to unveil and characterize the molecular mechanisms controlling seedling growth responses.
Conclusion
This study underscores the need for more specific molecular markers to study light-regulated morphological changes and introduces a set of hypocotyl- and cotyledon-specific markers based on a proposed ratio between measurements in the two organs, the MI. This index also enables a direct assessment of the plant’s growth pattern—a positive value indicates that plants are developing photomorphogenically (cotyledon cell expansion), whereas a negative MI points to skotomorphogenic development (hypocotyl cell expansion). We expect this toolset to substantially improve the molecular dissection of the different developmental processes occurring during photomorphogenesis and the mechanisms controlling seedling growth responses.
Materials and methods
Seedling growth and phenotypic measurements
Previously described Arabidopsis (A. thaliana) mutants were used in this study, namely phyb (Leivar et al., 2012), phyab (Cerdán and Chory, 2003), pifq (Leivar et al., 2008), hy5-215 (Oyama et al., 1997), and hy5-2 (van Gelderen et al., 2018), together with the Columbia (Col-0) ecotype used as the WT. Seeds were surface sterilized and sown on MS medium containing 1× Murashige and Skoog (MS) salts (Duchefa Biochemie), 2.5 mM MES (pH 5.7), 0.5 mM myo-inositol, and 0.8% agar (w/v). After stratification for 4 d at 4°C in darkness, seeds were submitted to a pulse of 3 h of light to induce germination and then transferred to continuous darkness or white light (WL; 45 µmol·m−2·s−1) for 3 d (Figure 5, A and B). For the phenotypical analyses shown in Figure 1A and Supplemental Figure S1, after germination induction with the 3-h light pulse, seeds were placed for 2 d in continuous darkness and then kept in continuous WL or darkness for 24 h. In Figures 1, A and B, 3C, 4C, fluridone (Fluka) was added to 2-d-old dark-grown seedlings at 1 µM for 24 h in the light and compared with non-treated plants. Radicle length and percentage of emergence shown in Figure 5, C and D and Supplemental Figure S11, C and D were measured 24 h after stratification in darkness. Phenotypic measurements (hypocotyl length, cotyledon aperture, and radicle length) were performed using the National Institutes of Health ImageJ software and at least 25 seedlings were measured to calculate the mean and SE in at least two biological replicates. Statistical differences between median values for the different morphogenic traits were determined by non-parametric tests using GraphPad Prism8.
MI
The MI assesses the molecular or morphological phenotype of a seedling and represents the log2 ratio between either gene expression values in cotyledons and the hypocotyl, or the cotyledon angle and 10 times the hypocotyl length, respectively. In both cases, a positive value indicates photomorphogenic development (cell expansion preferentially in cotyledons), while a negative index reflects skotomorphogenic development (cell expansion preferentially in the hypocotyl).
RNA extraction and gene expression analyses
Total RNA (1 µg) was extracted from whole Arabidopsis seedlings using the innuPREP Plant RNA kit (Analytik Jena BioSolutions) and treated with DNase I (Promega) according to the manufacturer’s instructions. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and oligo dT as a primer in the presence of RNase Out (Invitrogen). Reverse transcription-quantitative PCR (RT-qPCR) was performed using a QuantStudio™ 7 Flex Real-Time PCR System 384-well format and the Absolute SYBR Green ROX Mix (Thermo Scientific) on 2.5 μL of cDNA (diluted 1:10) per 10 μL of reaction volume, containing 300 nM of each gene-specific primer (Supplemental Table S1). For tissue-collected samples (Supplemental Figure S9), cDNA was synthesized from 150 ng of total RNA and 3.5 µL of cDNA (diluted 1:7) was used to perform RT-qPCR. For each condition tested, three independent biological replicates were performed, and the PP2A gene was used for normalization (Shin et al., 2007). Statistical differences between mean values obtained from RT-qPCR experiments (Figures 1B, 3C, 4C; Supplemental Figure S9) were determined by Tukey-b post hoc multiple comparison test using GraphPad Prism8. Statistically significant differences were defined as those with a P-value < 0.05.
RNA-seq datasets
Data from five independent available experiments were used in this study: GSE79576 (Sun et al., 2016), GSE132861 (Burko et al., 2020), GSE81202 (Kohnen et al., 2016), GSE112662 (Pham et al., 2018), and GSE70575 (Hartmann et al., 2016). To complement these datasets, we extracted RNA from WT, pifq, phyab, and hy5-2 seedlings grown for 3 d in continuous dark or light (13 µmol·m−2·s−1). OligodT, strand-specific libraries from triplicate biological replicates were built and sequenced using Illumina HiSeq 2500 at the Centre for Genomic Regulation Genomics Unit (Barcelona). An average of 50 million 125-nucleotide paired-end reads was generated per sample.
Gene expression analyses using RNA-seq experiments
To quantify gene expression levels from these RNA-seq data, we used the cRPKM metric (“corrected for mappability Reads Per Kilobase of uniquely mappable positions per Million mapped reads”), calculated as described by Labbé et al. (2012). Briefly, for each gene, a reference transcript was selected —the *.1 transcript according to the TAIR 10 nomenclature or else that with the lowest suffix (33,602 genes in total). Uniquely mappable positions for each of these transcripts were then calculated as previously described (Labbé et al., 2012), after which RNA-seq data were mapped to the library of reference transcripts using Bowtie version 1, with –m 1 and –v 2 parameters (uniquely mapping with two or fewer mismatches). In all cases, only the first 50 nucleotides of the forward read mate (if the RNA-seq was paired-end) were mapped to reduce batch effects. The cRPKMs were then calculated for each transcript as the number of mapped reads per million mapped reads divided by the number of uniquely mappable positions of the transcript (see Labbé et al., 2012 for further details). The biological seedling replicates shown in Supplemental Figure S4 were obtained by merging values from the three individual hypocotyl samples with those of the three individual cotyledon samples. To validate the calculated seedling expression values, we compared them with samples of 6-d-old seedlings exposed to 6 h of WL (Hartmann et al., 2016; Supplemental Figure S12). The results indicated that the light-responsiveness of our calculated seedling “samples” is representative of that of a true seedling.
To identify differentially expressed genes, quantile normalization of cRPKM values was first performed with “normalizeBetweenArrays” within the “limma” package. Next, for each pairwise comparison, genes that were not expressed at cRPKM >5 and read counts >50 across all the replicates of at least one of the groups being compared were filtered out. Only expression and MI values from genes that passed these cut-offs are shown in Figure 2 and Supplemental Figures S2, S3, S5, S6, S12. Finally, differentially-expressed genes were defined as those with a FC of at least 2 between each of the individual replicates from each group.
Gene Ontology analysis was performed with the online tool DAVID (https://david.ncifcrf.gov/version 6.8), using all genes that passed the coverage filters (minimum expression and read count) as a background.
Selection of organ-expansion genes misregulated in light-signaling mutants
To select genes from Groups A and C (expressed in growing organs) with interesting expression patterns in light-signaling mutants, we performed pairwise comparisons between the expression values of WT seedlings and the three mutants sequenced (pifq, hy5-2, and phyab), in either dark or light conditions. Genes with a FC of at least 1.5 were defined as differentially expressed. Of the genes that passed this criterion under the conditions in which the most relevant phenotypes were observed (Supplemental Figure S8), dark-grown pifq (Group A: upregulated and Group C: downregulated) or light-grown phyab seedlings (Group A: downregulated and Group C: upregulated) centered our attention. Among them, we then discarded those that were differentially expressed, or displayed an FC well >1, in phyab and hy5-2 dark-grown and pifq light-grown mutants, because no relevant phenotypes were observed in these mutants under these specific conditions. Finally, we focused on genes that though being only differentially expressed in one context of interest, dark-grown pifq or light-grown phyab seedlings, their FC in the other was close to 1.5. We also centered our attention on genes that, in addition to the previous criteria, exhibited expression levels in hy5-2 light-grown seedlings correlating with their phenotypes, i.e. cotyledon-expansion markers not regulated in the hy5-2 mutant and hypocotyl-expansion markers upregulated only in the light. This selection resulted in the final list of 18 candidates for organ-specific growth molecular markers.
Accession numbers
Raw sequencing data were submitted to the Sequence Read Archive (accession number GSE164122).
Supplemental Data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Cell expansion is oppositely regulated in hypocotyls and cotyledons.
Supplemental Figure S2. The MI accurately reflects similar growth patterns under different light conditions.
Supplemental Figure S3. Expression pattern of genes from groups F and H in hypocotyls and cotyledons.
Supplemental Figure S4. Whole-seedling expression levels can mask organ-specific light regulation.
Supplemental Figure S5. Expression pattern of genes from groups E and G in hypocotyls and cotyledons.
Supplemental Figure S6. Expression pattern of genes from groups A to D in hypocotyls and cotyledons.
Supplemental Figure S7. Functional analysis of genes from groups A and C.
Supplemental Figure S8. Phenotypes of light-signaling mutants grown under the experimental conditions used for RNA-seq.
Supplemental Figure S9. Expression of putative organ-expansion molecular markers in hypocotyls and cotyledons.
Supplemental Figure S10. Expression of selected genes from groups A and C in constitutive photomorphogenic mutants in the dark.
Supplemental Figure S11. MI assessment of seedling development in WT plants from different seed batches.
Supplemental Figure S12. Whole-seedling expression of light-regulated genes in an independent RNA-seq experiment.
Supplemental Table S1. List of primers used in this study.
Supplemental References. List of references cited in Supplemental Data.
Supplemental Data set S1. Gene classification based on the molecular MI (molMI) under dark and light conditions.
Supplemental Data set S2. Expression values of genes from Groups A and C in light-signaling mutants.
Supplementary Material
Acknowledgments
We thank E. Monte and C.M.M. Gommers for sharing seed resources.
Funding
G.M. was supported by an EMBO Long-Term Fellowship (ALTF 1576-2016) and a Marie Skłodowska-Curie Individual Postdoctoral Fellowship (EU project 750469). Work in our lab is funded by Fundação para a Ciência e a Tecnologia (FCT) through grants PTDC/BIA-FBT/31018/2017 and PTDC/BIA-BID/30608/2017. Funding from the research unit GREEN-it “Bioresources for Sustainability” (UIDB/04551/2020) is also acknowledged.
Conflict of interest statement. None declared.
G.M. designed the study, performed the experiments, and analyzed the data. P.D. helped analyze the data, and both authors discussed the results and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is (https://academic.oup.com/plphys/pages/general-instructions): Guiomar Martín (guiomarm@igc.gulbenkian.pt).
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Gil J, Blázquez MA, García-Martínez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL (2012) Photomorphogenesis. Arab B 10: e0147–e0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels PG, Watson CW (1978) Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci 26: 198–203 [Google Scholar]
- Bellstaedt J, Trenner J, Lippmann R, Poeschl Y, Zhang X, Friml J, Quint M, Delker C (2019) A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol 180: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron AK, Van Loock B, Suslov D, Markakis MN, Verbelen J-P, Vissenberg K (2015) Over-expression of AtEXLA2 alters etiolated arabidopsis hypocotyl growth. Ann Bot 115: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale U V, Ecker JR, Chory J (2020) Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32: 967–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881. [DOI] [PubMed] [Google Scholar]
- Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH (2018) TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc Natl Acad Sci 115: 12823–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Sun N, Yang J, Deng Z, Lan J, Qin G, He H, Deng XW, Irish VF, Chen H et al. (2019) The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31: 1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Paalman R, Keuskamp D, Hayes S, Pierik R (2018) Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30: 101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CMM, Monte E (2018) Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol 176: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki H, Ayano M, Nakamura A, Fujioka S, Asami T, Takatsuto S, Yoshida S, Oka Y, Matsui M, Shimada Y (2020) Light activates brassinosteroid biosynthesis to promote hook opening and petiole development in Arabidopsis thaliana. Plant Cell Physiol 61: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Hartmann L, Drewe-Boß P, Wießner T, Wagner G, Geue S, Lee HC, Obermüller DM, Kahles A, Behr J, Sinz FH et al. (2016) Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28: 2715–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen M V, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DDR, Ross EJ, Voisin V, Bader GD, Blencowe BJ et al. (2012) A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30: 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH (2012) Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Swaminathan K, Hudson ME (2011) Rapid, organ-specific transcriptional responses to light regulate photomorphogenic development in dicot seedlings. Plant Physiol 156: 2124–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW (2002) Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Suslov D, Vandenbussche F, Kenobi K, Ivakov A, Van Der Straeten D, Lorences EP, Mellerowicz EJ, Verbelen JP, Vissenberg K (2013) Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J Exp Bot 64: 2481–2497 [DOI] [PubMed] [Google Scholar]
- Montgomery BL (2016) Spatiotemporal phytochrome signaling during photomorphogenesis: from physiology to molecular mechanisms and back. Front Plant Sci 7: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Xu X, Huq E (2018) Molecular bases for the constitutive photomorphogenic phenotypes in Arabidopsis. Development 145: dev169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lyu M, Luo Y, Liu S, Li Y, He H, Wei N, Deng XW, Zhong S (2018) Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc Natl Acad Sci 115: 6482–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin EM, Silverthorne J (1985) Light regulation of gene expression in higher plants. Annu Rev Plant Physiol 36: 569–593 [Google Scholar]
- Whitelam GC, Patel S, Devlin PF (1998) Phytochromes and photomorphogenesis in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 353: 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wen Q, Yu R, Han X, Deng XW, Chen H (2020) Light modulates the gravitropic responses through organ-specific PIFs and HY5 regulation of LAZY4 expression in Arabidopsis. Proc Natl Acad Sci 117: 18840–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.