Abstract
Peroxisomes are redox nodes playing a diverse range of roles in cell functionality and in the perception of and responses to changes in their environment.
ADVANCES
Peroxisomal H2O2 regulates pathogen associated processes, DNA repair systems, cell cycles and phytohormone-dependent signalling.
Peroxisomes regulate cellular processes in the cytosol and other cell compartments through moonlighting proteins such as CAT3, which is able to transnitrosylate and degrade GSNOR via autophagy.
Peroxisomes are highly dynamic organelles that are capable of changing their number, size, morphology and speed in response to environmental redox changes.
Peroxules are ROS- and NO-induced dynamic structures that are regulated by PEX11a, which connects peroxisomes to chloroplasts, mitochondria, ER and lipid bodies.
Under basal and stress conditions, peroxisomal populations and quality are regulated by selective autophagy (pexophagy) which is controlled by ROS and the peroxisomal protease LON2.
Abstract
Redox compartmentalization in organelles is an effective evolutionary strategy (Box 1; Jones and Go, 2010). From an evolutionary perspective, peroxisomes, originating from the endoplasmic reticulum (ER), were selected to house a range of metabolic pathways involving the production of certain reactive oxygen species (ROS) such as H2O2 to avoid toxicity to other organelles such as mitochondria (Gabaldón, 2018). Peroxisomes play a diverse range of roles in cell functionality and in the perception of and responses to changes in their environment (Sandalio and Romero-Puertas, 2015; Lismont et al., 2019). The range of functions associated with plant peroxisomes has increased considerably over the last two decades (Table 1). As most of these pathways produce ROS and nitric oxide (NO), disturbances in these metabolic processes trigger transitory changes in ROS/reactive nitrogen species (RNS) production. These changes regulate peroxisomal metabolism, leading to peroxisome-dependent signaling and organelle crosstalk, which triggers specific cell responses (Sandalio and Romero-Puertas, 2015). The biosynthesis of phytohormones jasmonic acid (JA), auxin IAA, and salicylic acid (SA) associated with the β-oxidation pathway contributes to the complex role of peroxisomes in development and stress responses (Kao et al., 2018; Figure 2A). Peroxisomes dynamically regulate their number, shape, and protein content in response to changing environmental conditions and remain in close contact with other subcellular compartments such as mitochondria and chloroplasts (Sandalio and Romero-Puertas, 2015; Shai et al., 2016; Sandalio et al., 2020). Peroxisomes play a key role in the evolution of the metabolic networks of photosynthetic organisms by connecting oxidative and biosynthetic pathways operating in different compartments. This review updates our knowledge of peroxisomal redox homeostasis and the role of ROS and NO in the functionality, biogenesis and abundance of these organelles, as well as their role as redox hubs in metabolic regulation, signaling, and organelle crosstalk.
Box 1.
Subcellular redox compartmentalization
As oxygen-dependent redox reactions came to control life after O2 appeared in the atmosphere, cells developed complex mechanisms to detect and regulate these changes to maintain metabolic functionality. Redox compartmentalization in organelles is an effective evolutionary strategy, which regulates physiological and stress conditions through site-specific footprinting (Jones and Go, 2010). This redox circuit flexibility facilitates rapid responses to changes in intracellular redox equilibrium, which, in turn, favors beneficial signaling and detrimental oxidative stress. Photosynthetic organisms have developed efficient redox control systems using redox signals as the most fundamental forms of information (Foyer and Noctor, 2016). The thiol/disulphide couples GSH/GSSG and Cys/CySS, the ASC/DHA couple and a broad range of redox dependent proteins, which are counterparts of ROS such as H2O2 and other oxidants, form the core of the redox state and regulate the cell signaling, structure and activity of proteins and transcription factors. Apart from ROS, RNS are also redox signaling molecules, which include NO and peroxynitrite (ONOO−). Both ROS and RNS regulate covalent, often reversible, modifications, mainly targeting Cys, which regulates metabolic shifts and triggers signaling cell responses (Noctor et al., 2018; Sánchez-Vicente et al., 2019). Although irreversible oxidation products, such as sulfonic acid, carbonylation, and nitration, adversely affect proteins and lipids, they may be also involved in oxidative signaling (Foyer et al., 2017). Analyses of redox potential in plant tissue identified peroxisomes as some of the most oxidized cellular organelles, with a redox potential of approximately −360 mV (Bratt et al., 2016; Smirnoff and Arnaud, 2019).
Table 1.
Plant peroxisome functions
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
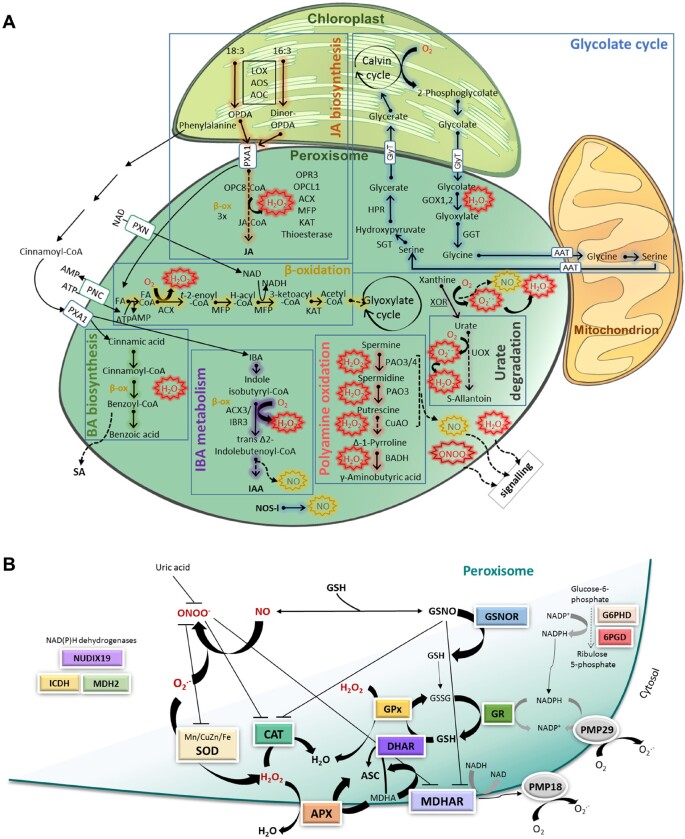
Figure 2.
Oxygen and nitrogen reactive species metabolism in peroxisomes. A, Principal peroxisomal metabolic pathways associated with peroxisomal ROS and NO production. ROS are produced in metabolic pathways such as β-oxidation, photorespiration, ureides metabolism, and polyamine oxidation, and in a small electron transport chain associated with the membrane (peroxisomal membrane proteins, PMP18 and PMP29; Figure 2B). NO is produced in peroxisomes by NOS-like (NOS-l) activity, although other sources, such as XOR, polyamine oxidation, and IBA metabolism, could also be involved. ROS, NO, and other RNS may leak out of the peroxisome (dashed arrows) and act as signal molecules that regulate cell metabolism and gene expression. B, Scheme of peroxisomal antioxidant defenses, RNS scavengers, and NAD(P)H supply. O2.− is regulated by SODs, while H2O2 is controlled by CAT, the ASC-GSH cycle, and GPx. Peroxynitrite (ONOO−) and GSNO are produced in peroxisomes by reaction of NO with O2.− and glutathione (GSH), respectively. GSNO can negatively regulate MDHAR and CAT through S-nitrosylation and nitration, and SOD may be regulated by nitration. SOD may indirectly control ONOO− by regulating O2.− levels. Uric acid acts as an ONOO− scavenger. NAD(P)H is supplied by the oxidative pentose phosphate pathway (G6PD; 6PGD), ICDH, MDH, and NUDIX19. 6PGD, 6 phosphogluconate dehydrogenase; AAT, amino acid translocator; AOC, allene oxide cyclase; AOS, allene oxide synthase; APX, ascorbate peroxidase; BADH, betaine aldehyde dehydrogenase; CAT, catalase; CuAO, copper amine oxidase1; DHAR, dehydroascorbate peroxidase; GOX1,2, glycolate oxidase1,2; G6PD, glucose-6-phosphate dehydrogenase; GGT, glutamate–glyoxylate aminotransferase; GlyT, glycerate–glycolate translocator; GR, glutathione reductase; GPx, glutathione peroxidase; H-Acyl-CoA, 3-hydroxyacyl-CoA; HPR, hydroxypyruvate reductase; IAA, indole-3-acetic acid; IBA, indole-3-butyric acid; IBR3, acyl-coA dehydrogenase/oxidase-like IBR3; ICDH, isocitrate dehydrogenase; KAT, L-3-ketoacyl-CoA-thiolase; LOX, lipoxygenase; MDH2, malate dehydrogenase; MDHAR, monodehydroascorbate peroxidase; MFP, multifunctional protein; OPCL1, OPC-8:0 CoA ligase1; NOS-l, NO synthase-like; NUDIX19, nudix hydrolase homolog 19; OPR3, OPDA reductase3; PAO3, polyamine oxidase3; PAO3/4, polyamine oxidase 3/4; PNC, peroxisomal ATP carrier; PXA1, peroxisomal ABC-transporter1; PXN, peroxisomal NAD carrier; SGT, serin–glyoxylate aminotransferase; UOX, urate oxidase.
Peroxisomes are ROS and NO producers
Peroxisomes produce and scavenge ROS
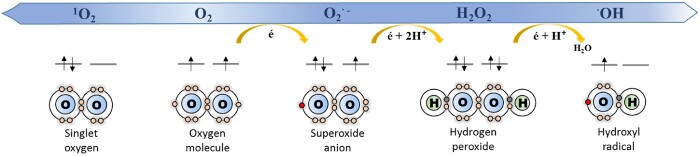
ROS include an array of molecular oxygen derivatives that occur as a normal attribute of aerobic life (Figure 1). Peroxisomes are one of the main sources of cellular ROS production and one of the most oxidized cellular organelles (Smirnoff and Arnaud, 2019). However, peroxisomes have a complex antioxidant system to balance ROS levels, enabling them to strictly regulate organelle functionality, metabolism, and signaling networks. The first step in O2 univale O2.− (Figure 1), is produced in ureide and nucleic acid catabolism by xanthine oxidoreductase (XOR) and urate oxidase or uricase (UO) reaction (Werner and Witte, 2011; Sandalio et al., 2013; Figure 2A); in the sulfite oxidation by sulfite oxidase (SO; Byrne et al., 2009); and in the NADH/NADPH-dependent electron transport chain in the peroxisomal membrane. Superoxide accumulation is regulated by different superoxide dismutases (SOD; reviewed in Sandalio and Romero-Puertas, 2015; Figure 2B).
Figure 1.
Sequential reduction of O2 and ROS production: superoxide (O2.−), hydrogen peroxide (H2O2), and hydroxyl (·OH) radicals.
In photosynthetic tissue, peroxisomes accumulate the highest concentrations of organelle H2O2 (the second step in O2 reduction) with a flux of ∼10,000 nmol−2 m−2 s−1 (Foyer and Noctor, 2003). The use of H2O2 ratiometric reporter HyperAs targeting peroxisomes has facilitated the imaging of changes in peroxisomal H2O2 accumulation in response to Cd treatment (Calero-Muñoz et al., 2019) and the increase in intraperoxisomal Ca2+ levels (Costa et al., 2010). The main source of H2O2 in peroxisomes in green tissue is GOX in the photorespiration cycle (Figure 2A), which contributes up to 70% of total H2O2 production in plant cells (Reumann and Weber, 2006; Foyer et al., 2009). Photorespiration requires coordination of the chloroplast, peroxisome, mitochondrion, and cytosol; and photorespiration-dependent H2O2 production increases considerably under environmental stress conditions such as heat and drought (Talbi et al., 2015; Walker et al., 2016), heavy metal (Gupta et al., 2017), high light (Cui et al., 2016a), and biotic stress (Rojas et al., 2012; Hodges et al., 2016; Yang et al., 2018). Fatty acid β-oxidation, another source of H2O2 in peroxisomes by the Acyl-CoA oxidase (ACX), provides energy during the initial stage of seedling growth by oxidizing fats stored as triacylglycerol (TAG) in oil bodies (Rinaldi et al, 2016; Figure 2A). Other β-oxidation pathways are active in green tissues, including the synthesis of ubiquinone, hormones such as indole acetic acid (IAA) and JA, and secondary metabolites such as benzoic acid (BA) and phenylpropanoids (reviewed in Pan et al., 2020; Figure 2A). Polyamine catabolism and sarcosine oxidase are additional peroxisomal sources of H2O2 (Figure 2A; Goyer et al., 2004; Wang et al., 2019). Peroxisomal H2O2 levels are regulated by balancing H2O2 generation and scavenging rates (Figure 2B) by catalase (CAT), which account for 10%–25% of total peroxisomal proteins (Reumann et al., 2004; Figure 2B). Arabidopsis (Arabidopsis thaliana) plants contain three CAT genes, CAT1, CAT2, and CAT3, with CAT2 being the most important defense against photorespiratory H2O2, accounting for 80% of activity (reviewed in Mhamdi et al., 2012). In fact, physical GOX-CAT interactions regulated by SA occur in rice leaves (Zhang et al., 2016). A protective association between CAT and isocitrate lyase has also been observed in castor bean glyoxysomes (Yanik and Donaldson, 2005) and CAT2 also interacts with ACX2/ACX3 regulating their activity and therefore the SA-mediated regulation of JA biosynthesis, under biotrophic infection (Yuan et al., 2017). Although the extraordinarily low affinity of CAT for H2O2, with a Km of around 43 mM, reduces its efficiency in controlling H2O2 the abundance of CAT compensates for this low affinity (Foyer and Noctor, 2016). The peroxisomal ascorbate–glutathione cycle, which, in Arabidopsis, composed of ascorbate peroxidase (APX3 and APX5), monodehydroascorbate reductase (MDHAR1), dehydroascorbate reductase (DHAR1), and glutathione reductase (GR1; reviewed in Mhamdi et al., 2012; Sandalio and Romero-Puertas 2015; Pan et al., 2020; Figure 2B) also contribute to H2O2 homeostasis. MDHAR and APX are associated with the peroxisomal membrane and the higher affinity for H2O2 of APX (100 μM) as compared to CAT, could regulate H2O2 leakage from peroxisomes to the cytosol (Del Río et al., 2003; Kaur et al., 2009; Eastmond, 2007; Figure 2B). Therefore, CAT and APX are positioned to enable H2O2 to act as a second messenger. Glutathione S-transferases support peroxide regulation in these organelles (Pan and Hu, 2018). The ascorbate–glutathione cycle also facilitates regeneration and maintenance of the peroxisomal redox buffers ASC/DHA and GSH/GSSG. The use of ratiometric glutathione redox potential reporters, such as roGFP2, targeting peroxisomes has facilitated the imaging of peroxisome oxidation under extended dark stress and the application of elicitors (Bratt et al., 2016).
Peroxisomal NO/RNS production and scavenging
Although NO is a well-known signaling molecule in plants, its metabolism has not been fully elucidated (León and Broseta, 2020). Peroxisomal NO production has been associated with a NO synthase-like activity (NOS-l; Figure 2A; Barroso et al., 1999), the conversion of IBA to IAA by β-oxidation (Figure 2A;Schlicht et al., 2013), polyamine catabolism (Figure 2A; Wimalasekera et al., 2011; Agurla et al., 2018), and the XOR reaction (Figure 2A; Antonenkov et al., 2010; Wang et al., 2010). Other nitrogen-derived species, such as peroxynitrite (ONOO–), resulting from the /NO reaction, and nitrosoglutathione (GSNO), resulting from the combination of NO and GSH and considered a cellular NO reservoir, have been detected in peroxisomes (Figure 2A; Ortega-Galisteo et al., 2012; Corpas and Barroso, 2014). Peroxisomal SOD could regulate ONOO– accumulation by controlling O2.− availability and CAT could degrade it, as reported in animal cells (Gebicka and Didik, 2009), and thus play a key modulatory role at the cross-point between H2O2 and NO/ONOO–-mediated signaling pathways (Figure 2B). Urate, a well-known peroxynitrite scavenger (Hooper et al., 2000; Alamillo and García-Olmedo, 2001), may contribute also to regulate ONOO– in peroxisomes (Figure 2B). S-nitrosoglutathione reductase (GSNOR), which balances NO and S-nitrosothiol levels, has been proteomically identified in plant peroxisomes (reviewed in Sandalio et al., 2019), although this requires validation.
NADH/NADPH regeneration in peroxisomes
The concept of redox stress (oxidative and reductive) reflected by changes in NAD(H)/NADP(H) has gained increasing attention. The NAD(P)H cofactor is required to β-oxidation and antioxidative defenses MDHAR and GR. NAD(P)H regeneration in peroxisome take place by the oxidative pentose phosphate pathway (OPPP; Corpas et al., 1999; Reumann et al., 2007; Lansing and Doering, 2020; Figure 2B), NADP-dependent isocitrate dehydrogenase (ICDH; Corpas et al., 1999; Reumann et al., 2007; Figure 2B), NADH phosphorylation by NADH kinase 3 (NADK3; Waller et al., 2010) and possibly betaine aldehyde dehydrogenase (ALDH19; Hou and Bartels, 2015). The peroxisomal NADH pool is supported by malate dehydrogenase MDH2 (Cousins et al., 2008; Figure 2B). Peroxisomes also contain pyrophosphatase Nudix Hydrolase Homolog 19 (NUDT19), which hydrolyzes NADPH to NMNH, as well as 2′,5′-ADP and NADH to NMNH and AMP (Lingner et al., 2011).
ROS- and NO-dependent PTMs in peroxisomal metabolism regulation
Analysis of peroxisomal proteomes shows that a large number of peroxisomal proteins (35%) are targeted by multiple PTMs (Sandalio et al., 2019). Peroxisomal-dependent ROS/RNS can fine-tune post-translational redox changes in proteins, regulating stability, activity, location, and protein–protein interactions (Duan and Walther, 2015; Hashiguchi and Komatsu, 2016; Sandalio et al., 2019; Foyer et al., 2020) supporting peroxisomes capacity to regulate their metabolism and dynamics in response to environmental changes. Hydrogen peroxide leads to rapid and reversible oxidative protein modifications such as sulfenylation, sulfinylation, and intra- and intermolecular disulfide bond formation, which contribute to coordinated regulation of cellular processes, while overoxidation by sulfonylation appears to be an irreversible process (reviewed in Noctor et al., 2018; Young et al., 2019; Sandalio et al., 2019; Sies and Jones, 2020). Given their transient nature, these sulfur modifications are regarded as redox switches (Huang et al, 2018). Peroxisomal antioxidant defenses, fatty acid β-oxidation, and photorespiration are prone to H2O2-dependent redox regulation (reviewed in Sandalio et al., 2019). The glyoxalase 1 (GLX1) homolog is a putative sulfenylated protein involved in protection against carbonyls (Schmitz et al., 2018).
NO, in turn, modifies proteins through covalent PTMs including S-nitrosylation (Martínez-Ruiz et al., 2011; Sánchez-Vicente et al., 2019). Putative peroxisomal S-nitrosylated proteins also include antioxidants and enzymes from the photorespiration cycle (Romero-Puertas and Sandalio, 2016; Sandalio et al., 2019) suggesting that S-nitrosylation plays an important role in regulating peroxisomal H2O2 concentrations under physiological and stress conditions (Ortega-Galisteo et al., 2012). Recently, the noncanonical catalase CAT3, identified as a “repressor of” GSNOR1 (ROG1), was reported to transnitrosylate GSNOR1 to promote its degradation by autophagy, while CAT1 and CAT2 do not do it, thereby CAT3 positively regulates NO signaling and according to Arabidopsis rog1 mutants are more susceptible to NO than WT (Chen et al., 2020). CAT3 is localized in peroxisomes, the cytoplasm, and the plasma membrane (Li et al., 2015; Zou et al., 2015) and is recruited into the nucleus by the cucumber mosaic virus (CMV) 2b protein (Inaba et al., 2011; Murota et al., 2017). Zhan et al. (2018) have reported that S-nitrosylation induces selective autophagy of Arabidopsis GSNOR1 during hypoxia responses. CAT3 also interacts with other proteins in the cytosol and plasma membrane, thus increasing the likelihood that these proteins are also substrates of CAT3 transnitrosylase activity (Chen et al., 2020). These findings suggest NO self-regulation and ROS/NO crosstalk. Zhang et al. (2020) have reported that glutathione denitrosylation is required to maintain the upregulation of GSNOR activity; thus coordinating GSNOR activity with protein S-nitrosylation levels to ensure appropriate signaling involving the SA pathway in response to H2O2.
Some fatty acid β-oxidation enzymes, including ACX2, 3, may be S-nitrosylation targets (Sandalio et al., 2019). OPC-8:0 CoA Ligase1 (OPCL1), involved in activating JA biosynthetic precursors in leaf peroxisomes (Koo et al., 2006), is also a putative target of S-nitrosylation, pointing to NO/JA-crosstalk. Proteomic analyses suggest that BRI1 suppressor 1 (BSU1)-like 3 is targeted by S-nitrosylation (Sandalio et al., 2019) suggesting NO-dependent brassinosteroids signaling. NO-dependent nitration also inhibits peroxisomal antioxidants such CAT (Lozano-Juste et al., 2011; Chaki et al., 2015) and SOD (Holzmeister et al., 2015). Therefore, NO and ROS, apart from self-regulation (Romero-Puertas and Sandalio, 2016), may also regulate specific proteins and/or metabolic pathways and metabolite channeling, depending on the redox environment both inside and outside the peroxisome.
Peroxisome-dependent redox regulation of transcriptional responses
ROS act as secondary messengers that are sensed by specific redox-sensitive proteins, which activate signal transduction pathways and alter gene expression (Suzuki et al., 2012; Mittler, 2017). Different ROS trigger different protein modifications, as shown by different gene expression patterns (Møller and Sweetlove, 2010; Mor et al., 2014). The subcellular site, where the ROS/oxidation state is modified, acts as a specific cellular redox network signal (Foyer and Noctor, 2003; König et al., 2012; Foyer et al., 2017) and leaves a specific imprint on the transcriptome response (Rosenwasser et al., 2011). The selective reactivity, stability, and diffusibility of H2O2 make it an ideal signaling molecule (Sewelam et al., 2014; Sies and Jones, 2020). Mutants lacking peroxisomal CAT2 (cat2) have been extensively studied in Arabidopsis and tobacco (Nicotiana tabacum) plants under control and stress conditions, showing that altering peroxisomal H2O2 induces changes in gene expression profiles (Vandenabeele et al., 2003, 2004; Chaouch et al., 2010; Queval et al., 2012). This profile showed specificity with transcriptional responses that differ from those induced by chloroplast-derived H2O2 (Sewelam et al., 2014). Analyses of WT plants grown at specific atmospheric CO2 levels to boost photorespiration and production of H2O2 (Chaouch et al., 2010; Queval et al., 2012) and of WT plants treated with aminotriazole, a catalase inhibitor (Gechev et al., 2005), have also shown that peroxisomal H2O2 plays a role in signaling, as a transcriptomic footprint have been linked to peroxisomes (Rosenwasser et al., 2013). However, little is known about how peroxisome-derived H2O2 coordinates or relays signaling events. Although peroxisome-dependent gene regulation involves several metabolic categories (reviewed in Sandalio and Romero-Puertas, 2015; Su et al., 2019), those related to protein repair responses under stress conditions are regulated in cat2 mutants (Queval et al., 2007; Sewelam et al., 2014); suggesting that peroxisomes are involved in acclimation and survival processes under changing environmental conditions. The triple mutant cat1 cat2 cat3 shows serious redox disturbance and growth defects under physiological conditions, with differentially expressed genes belonging to plant growth regulation, as well as abiotic and biotic stress response categories. Some of these genes belong to transcription factor and receptor-like protein kinase categories (Su et al., 2019). The ROS signals derived from different cell compartments are proposed to connect in the cytoplasm with MAPK pathway to regulate the expression of nuclear genes (Noctor and Foyer, 2016). Several genes related to MAPK cascade pathways, such as MPK11, MPK13, and serine/threonine kinase oxidative signal inducible 1 (OXI1), are severely altered in the triple cat mutant (Su et al., 2019). Thus, peroxisomal H2O2 appears to participate in retrograde signaling, although little is known about the underlying molecular mechanisms and crosstalk with ROS from other compartments (Sandalio and Romero-Puertas, 2015; Su et al., 2019).
Redox-dependent regulation of peroxisomal plasticity
Peroxisome biogenesis and proliferation
Their high plasticity enables peroxisomes to adapt their number, morphology, movement, and metabolic pathways to changes in their environment (Figure 3). However, why certain signals and molecules trigger these changes, when these changes occur, and how dynamic peroxisomal changes function in relation to tolerance are not well understood. Some evidence shows that peroxisomal proliferation through the division of preexisting peroxisomes is regulated by ROS (López-Huertas et al., 2000; Figure 3; Box 2). Peroxisome proliferation occurs in response to abiotic stresses associated with ROS production: ozone (Oksanen et al., 2004), clofibrate (Nila et al., 2006; Castillo et al., 2008), salinity (Mitsuya et al., 2010; Fahy et al., 2017), cadmium (Romero-Puertas et al., 1999; Rodríguez-Serrano et al., 2016), drought (Ebeed et al., 2018), ABA (Ebeed et al., 2018), and senescence (Pastori and del Río, 1997). Interestingly, NO has recently been reported to be involved in regulating peroxisome proliferation in response to Cd (Terrón-Camero et al., 2020). Proteins involved in peroxisome biogenesis and maintenance are called peroxins (PEXs; Baker et al., 2016; Reumann and Bartel, 2016; Kao et al., 2018; Reumann and Chowdhary, 2018). Peroxisome proliferation under abiotic stress appears to be regulated by specific PEX11 genes which contain five members (PEX11a-PEX11e; Lingard and Trelease, 2006; Orth et al., 2007) and under abiotic stressspecific PEX11 appears to regulate peroxisome proliferation. Thus, salinity upregulated PEX11a and PEX11c in A. thaliana (Fahy et al., 2017), while PEX11a and PEX11e were upregulated in response to Cd exposure in Arabidopsis plants (Rodríguez-Serrano et al., 2016; Terrón-Camero et al., 2020), and PEX11b, PEX11c, and PEX11d were upregulated by hypoxia (Li and Hu, 2015). Gene coexpression analysis in Arabidopsis plants under drought conditions shows clustering of photorespiratory genes and peroxisomal abundance, suggesting that H2O2 plays a role in peroxisomal abundance regulation (Li and Hu, 2015). This is supported by the absence of peroxisome proliferation in gox2 Arabidopsis mutants exposed to Cd (Calero-Muñoz et al., 2019). However, genome analyses of Physcomitrium, A. thaliana, and Triticum aestivum show upregulation of β-oxidation in response to drought, dehydration, and ABA (Ebeed et al., 2018). Interestingly, PEX11 gene family expression differs between drought-sensitive and resistant wheat genotypes, although the significance or not of these differences for drought tolerance has not been established (Ebeed et al., 2018). These findings suggest that peroxisomal H2O2 could be involved in environmental change perception and acclimation through differential PEX11 regulation and peroxisome proliferation. Plant peroxisome proliferation could be a protective response to ROS overflow in cell compartments due to highly efficient peroxisomal antioxidant defenses, as reported during protoplast transition from G0 to G1 (Tiew et al., 2015) and in response to salt stress in Arabidopsis and Oryza sativa by reducing both Na+ accumulation and oxidative stress (Cui et al., 2016b; Fahy et al., 2017). However, in quinoa plants, principal component analyses show a negative correlation of peroxisome abundance with yields in plants exposed to heat, drought or both (Hinojosa et al., 2019). Therefore, the capacity to maintain H2O2 homeostasis and peroxisome quality control/abundance could determine the success of plant adaptation to adverse conditions.
Figure 3.
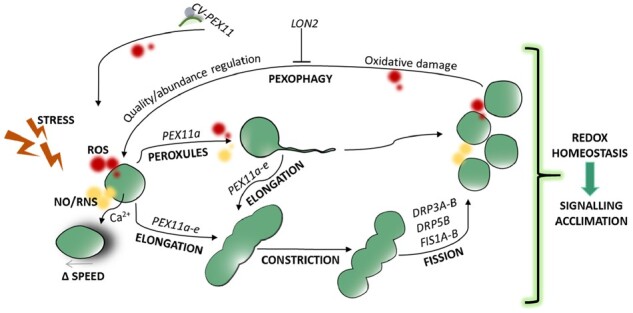
Hypothetical scheme showing changes in peroxisomal dynamics and their regulation, as well as their contribution to cell responses to abiotic stresses such as metal toxicity. Cd stress promotes the generation of ROS and NO, which activate peroxins (PEX11a and PEX11e), probably through ROS-/NO-dependent post-translational modifications (PTMs). PEX11a promotes the formation of peroxules, which may control ROS/NO accumulation and ROS-dependent gene expression. Peroxisomal elongation, constriction, and proliferation, which are regulated by ROS and NO, were later observed. Longer exposure periods increase the speed of peroxisome movement (Δ SPEED), which is also controlled by ROS. The number of peroxisomes, as well as oxidized, damaged peroxisomes, can be regulated by pexophagy or via a process independent of autophagy involving chloroplast vesicle interactions with PEX11 (CV-PEX11), both of which processes are regulated by ROS. All these changes in peroxisomal dynamics may be involved in redox homeostasis and redox-dependent signaling, leading to plant acclimation to the stress. Red color, ROS; yellow color, NO. DRPs, dynamin-related proteins; FIS1A-B, fission protein1A-B; LON2, lon protease homolog 2.
Box 2.
Peroxisome biogenesis and proliferation
Plant peroxisome abundance is governed by (1) biogenesis, associated with physiological processes and division (fission) of a preexisting peroxisome, (2) proliferation, which is related to stress responses, and (3) pexophagy, a selective peroxisome degradation mechanism (Kao et al., 2018; Olmedilla and Sandalio, 2019). Proteins involved in peroxisome biogenesis and maintenance are called peroxins (PEXs; Kao et al., 2018). The import of peroxisomal membrane proteins (PMPs) in Arabidopsis involves peroxins PEX19, acting as the chaperone for PMPs, PEX3 acting as the membrane anchor for PEX19, and PEX16, which recruits PEX3 to the ER before the formation of pre-peroxisomes. Arabidopsis PEX16 also recruits PMPs to the ER in a PEX3/PEX19-independent manner (Pan et al., 2020). The import of peroxisomal matrix proteins containing the C-and N-terminal targeting signals PTS1 and PTS2, respectively, take place by the soluble receptors PEX5 (for PTS1) and PEX7 (for PTS2) in the cytosol (Baker et al., 2016; Reumann and Chowdhary, 2018). PEX5 is recycled from the peroxisomal matrix back to the cytosol by the ubiquitin conjugating enzyme PEX4 and its membrane anchor PEX22, three RING-type ubiquitin ligases, PEX2, PEX10 and PEX12, and two AAA ATPases, PEX1 and PEX6 (reviewed in Reumann and Bartel, 2016; Kao et al., 2018).
Peroxisomes proliferate through the division of preexistent peroxisomes, which involves peroxisome elongation regulated by PEX11, organelle constriction and fission, governed by dynamin-related proteins (DRP3A and DRP3B) and fission proteins (FIS1A and FIS 1B; Pan et al., 2020; Su et al., 2019). The PEX11 gene family in Arabidopsis contain five members: PEX11a, PEX11b, PEX11c, PEX11d and PEX11e, (Lingard and Trelease, 2006; Orth et al., 2007). FIS1A and FIS1B are shared by peroxisomes and mitochondria; DRP3A and DRP3B regulate peroxisomal and mitochondrial fission; and DRP5B is involved in peroxisome and chloroplast fission (Kao et al., 2018), indicating a highly coordinated regulation of organelles populations.
ROS/RNS-dependent formation of peroxules
In vivo observation of plant tissues, with fluorescent proteins targeting peroxisomes, reveals the rapid formation of tubular peroxisome extensions called peroxules, induced in response to changes in ROS levels (Figure 3; Sinclair et al., 2009; Barton et al., 2013; Rodríguez-Serrano et al., 2016). Short periods of Cd exposure (15–30 min) induce peroxule formation, which is considerably reduced by H2O2 scavengers and in rboh mutants, suggesting regulation by external ROS (Rodríguez-Serrano et al., 2016). Confocal images show peroxule contacts with chloroplasts and mitochondria under Cd treatment and high light (Sinclair et al., 2009; Sandalio et al., 2013; Jaipargas et al., 2016; Rodríguez-Serrano et al., 2016). Peroxule formation in response to Cd and As is dependent upon PEX11a, while pex11a Arabidopsis mutants show altered ROS-dependent signaling networks (Rodríguez-Serrano et al., 2016). Peroxule production and peroxisome-dependent signaling are compromised in nia1 nia2 Arabidopsis mutants, which have lower NO levels than wild-type plants in response to Cd treatment, demonstrating the important role of NO in peroxule formation (Terrón-Camero et al., 2020). This could be due to oxidative changes and S-nitrosylation patterns in the antioxidant system (Terrón-Camero et al., 2020), which affect cellular redox balance. PEX11a and peroxule formation therefore play a key role in regulating stress perception and rapid cell responses to environmental cues (Rodríguez-Serrano et al., 2016; Terrón-Camero et al., 2020; Figure 3). Given rapid peroxule induction and no significant changes in PEX11a expression in nox1 mutants (Terrón-Camero et al., 2020), PEX11a can reasonably be assumed to be regulated by specific ROS- and NO-dependent PTMs. Activation of yeast peroxin Pex11p depends on redox changes in its cysteines (Knoblach and Rachubinski, 2010; Schrader et al., 2012).
Although there is no direct evidence, peroxules could participate in the transfer of H2O2 and other metabolites to mitochondria and chloroplast (Figure 4). Stromules, which are dynamic structures similar to peroxules in chloroplasts, transfer H2O2 from chloroplasts to nuclei as part of a retrograde signaling process (Caplan et al., 2015; Kumar et al., 2018); however, to our knowledge, no connection between peroxules and nuclei has been established so far. Peroxules could also be involved in protein transport such as the transfer of the sugar-dependent 1 (SDP1) lipase from the peroxisomal membrane to the lipid body (Thazar-Poulot et al., 2015; Figure 4).
Figure 4.
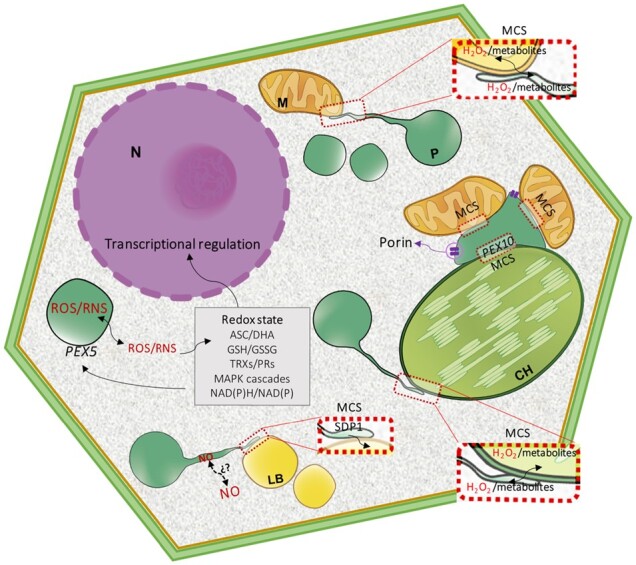
Redox-dependent interorganellar crosstalk. Peroxisomes (P) collaborate and communicate with other cellular organelles, mitochondria (M), and chloroplasts (CH) by exchanging molecules such as H2O2 and redox metabolites, as well as Ca2+ and proteins. These exchanges could take place through porins or MCSs. Peroxule formation contributes to ROS/RNS, metabolite, and protein exchanges such as the transfer of TAG lipase sugar-dependent 1 (SDP1) to lipid bodies (LB). ROS/RNS-dependent posttranslational modifications regulate peroxule formation, MCSs, interorganellar crosstalk, and signaling transduction. Peroxisomal ROS/RNS interferes with cytosolic redox state and signaling processes and vice versa; the cytosolic redox state regulates peroxisomal protein import by affecting the redox state of peroxin 5 (PEX5). The peroxisomal redox state can also regulate redox changes in the nucleus (N), chloroplasts, and mitochondria.
Peroxisomal speed is regulated by ROS
Time course analyses of peroxisomes in response to Cd in Arabidopsis seedlings have highlighted a considerable increase in peroxisomal speed after 24 h of treatment which is regulated by ROS produced by RBOHs and Ca2+ ions (Figure 3; Rodríguez-Serrano et al., 2009, 2016). Increased peroxisomal movement could improve antioxidant defenses where Cd and other factors promote ROS accumulation and/or could aid signaling transduction and metabolite exchanges in different parts of the cell (Rodríguez-Serrano et al., 2009). Information on the role of peroxisomal motility under stress conditions is scarce; however, in myosin loss-of-function Arabidopsis mutants and in Arabidopsis treated with the herbicide 2,4-D, inhibition of organelle movement negatively affects plant growth (Rodríguez-Serrano et al., 2014; Ryan and Nebenführ, 2018). Oikawa et al. (2015) found that light-adapted Arabidopsis peroxisomes are much more mobile than dark-adapted peroxisomes (Oikawa et al., 2015) and, by using photorespiratory mutants shmt1 (defective in serine hydroxymethyltransferase) and ped2 (defective in PEX14), they concluded that both, peroxisome mobility and peroxisome–chloroplast interactions observed under light, are regulated by photosynthesis rather than by photoreceptors or photorespiration (Oikawa et al., 2015).
Pexophagy and peroxisomal homeostasis are regulated by oxidative processes.
Excessive numbers of peroxisomes and those containing obsolete or dysfunctional proteins need to be eliminated to control cellular redox homeostasis. Some evidence shows that ROS and oxidative damage to peroxisomes regulate the degradation of oxidized whole peroxisomes by selective autophagy termed pexophagy (Figure 3; Shibata et al., 2013; Yoshimoto et al., 2014; Olmedilla and Sandalio, 2019). Autophagy-related genes (ATGs) regulate autophagy in all eukaryotic cells including those in plants (Avin-Wittenberg et al., 2018). During photomorphogenesis, several authors have reported pexophagy using Arabidopsis atg mutants (Farmer et al., 2013; Kim et al., 2013; Shibata et al. 2013); however, unlike in mammals and yeast, the mechanism in plants is not fully understood. Shibata et al. (2013) have observed high levels of oxidized CAT and clusters of peroxisomes in atg mutants. Peroxisomal clusters were also observed in H2O2-treated Arabidopsis plants (Yoshimoto et al., 2014) and in atg5 and atg7 Arabidopsis mutants exposed to Cd treatment where phagophore and peroxisome colocalization was observed (Calero-Muñoz et al., 2019). Some evidence shows the important role of oxidative processes in pexophagy induction: (1) ubiquitinated CAT is accumulated in Arabidopsis mutants defective in NBR1 (nbr-1), a pexophagy adaptor (Zhou et al., 2013); (2) ATG8/CAT–CAT/NBR1 interactions have been observed in Arabidopsis plants exposed to Cd (Calero-Muñoz et al., 2019); (3) CAT activity is involved in starvation-induced pexophagy (Tyutereva et al., 2018); and (4) clustered peroxisomes in Arabidopsis atg mutants mainly accumulate in the aerial parts of plants, where oxidative metabolism is higher than in roots (Zhou et al., 2013; Yoshimoto et al., 2014). Glucose-mediated regulation of root meristem activity requires pexophagy to maintain ROS and auxin cellular homeostasis in Arabidopsis plants (Huang et al., 2019). The chaperone activity of peroxisomal protease LON2 negatively regulates pexophagy (Figure 3; Farmer et al., 2013; Young and Bartel, 2016). In plants, specific peroxisomal receptors have not been clearly identified, and the role of adaptors, such as NBR1-like proteins, which specifically interact with ubiquitinilated proteins, is under debate (Olmedilla and Sandalio, 2019; Young et al., 2019). The possibility of both NBR1-dependent and independent pexophagy cannot be ruled out. An alternative process independent of autophagy, induced under high CO2 and increased H2O2 conditions, involves chloroplast vesiculation (CV) proteins which interact with PEX11-1 in rice (Figure 3; Umnajkitikorn et al., 2020).
Peroxisome crosstalk with other organelles
To optimize their multiple functions, peroxisomes collaborate and communicate with other cell organelles by exchanging substrates. Photorespiration is the best example of metabolic cellular interorganelle communication. However, peroxisomes are also cellular redox communication hubs, as well as guardians and modulators of H2O2 levels (Fransen and Lismont, 2019) given the following findings: (1) peroxisomes contain enzymes involved in producing and scavenging H2O2 and NO; (2) they contain proteins regulated by ROS- and NO-dependent PTMs and therefore act as ROS/NO sensors; (3) they regulate NAD(P)+/NAD(P)H, ascorbic (Asc)/dehydroascorbic acid (DHA) and GSH/GSSG pools; and (4) H2O2 and NO act as second messengers in a wide range of developmental, physiological, and stress processes (Fransen and Lismont, 2019; Sandalio et al., 2020).
There is an intimate relationship between the peroxisomal redox state and changes in the redox state of other organelles. In mammalian systems, H2O2 released from peroxisomes into the cytosol diffuses into mitochondria, oxidizing directly or indirectly cysteine residues of mitochondrial proteins (Lismont et al., 2019). Chlamydomonas mutants deficient in peroxisomal NAD+-dependent MDH2 show that MDH2 plays a key role in the reverse coupling of redox/H2O2 signals from peroxisomes to chloroplasts (Kong et al., 2018). Peroxisomal NAD(P)+/NAD(P)H pools in Arabidopsis regulate photosynthesis performance to meet the demand for reducing equivalents under fluctuating light (Li et al., 2019). Peroxisomal basal H2O2 levels greatly affect antioxidative defense regulation in cytosol and chloroplasts, as observed in peroxisomal apx4 knockdown rice plants (Sousa et al., 2018). The inhibition of CAT activity in apx4 Arabidopsis mutants significantly affected networks involved in photosynthetic performance under adverse conditions promoting oxidative stress and favoring antioxidant enzyme accumulation in cytosol and chloroplasts (Sousa et al., 2018).
Despite the central role of H2O2 in peroxisome metabolism and cell functionality, no peroxiporin-like proteins have been identified in the peroxisomal membrane. Although porins are present in plant peroxisomes (Reumann et al., 1997; Corpas et al., 2000; Figure 4), their role in H2O2 permeability remains unclear. In yeast, Pex11A, Pex11B, and Pex11G have been reported to facilitate the permeation of molecules up to 400 Da (Mindthoff et al., 2016) and could be candidates to diffuse H2O2 through the peroxisomal membrane. However, recently, Lismont et al. (2019) provided evidence that neither the porin PXMP2 nor PEX11B is essential for H2O2 permeation across the peroxisomal membrane. Throughout membrane contact sites (MCSs), ROS accumulation could directly facilitate interorganelle signal transmission using as-yet-unknown ROS transporters (Figure 4; Yoboue et al., 2018). Electron microscopy images of leaf cells show physical contact between peroxisome and chloroplasts and, interestingly, H2O2 accumulation inside peroxisomes in the contact site with chloroplasts and vacuoles, suggesting a relationship between ROS accumulation and organelle communication (Romero-Puertas et al., 2004). Using femtosecond laser and optic tweezer techniques, tethering between the chloroplast and peroxisomes has been demonstrated (Oikawa et al., 2015; Gao et al., 2016). The area of peroxisomes interacting with chloroplasts increases under light conditions, whereas, in the dark, peroxisomes lost their connection with chloroplasts (Oikawa et al., 2015). The PEX10 Zn RING finger interacts with the chloroplast envelope’s outer membrane, which is necessary for full photorespiration functionality and could be a candidate for MCSs in plant peroxisomes (Schumann et al., 2007; Figure 4). Although the role of PTMs in regulating protein–protein interactions at interorganelle contact sites remains unexplored, it is reasonable to assume that tethering is regulated by specific PTMs.
The translocation of peroxisomal proteins to other cell compartments is part of interorganelle communication and signaling, although the mechanism(s) by which this occurs is still unknown. Thus, CAT interacts with nonperoxisomal proteins including cytosolic calcium-dependent kinase CDPK8 (Zou et al., 2015), plasma membrane-associated calcium-dependent kinase OsCPK10 (Bundó and Coca, 2017), cytosolic salt overly sensitive 2 (SOS2; Verslues et al., 2007), lesion simulating disease1 (LSD1; Li et al., 2013), receptor-like cytoplasmic kinase STRK1 (Zhou et al., 2018), chloroplast/cytosolic nucleoside diphosphate kinase 2 (NDPK2), no catalase activity 1 (NCA1; Hackenberg et al., 2013; Li et al., 2015), and nucleoredoxin 1 (NRX1; Kneeshaw et al., 2017) in addition to GSNOR (mentioned above), which are all integral stress-signaling proteins. It is unclear whether CAT is translocated from peroxisomes by the ER-associated degradation (ERAD)-like system involved in the export of PEX5 from the peroxisome membrane and export of matrix peroxisomal proteins to be degraded (Lingard et al., 2009) or is retained in the cytosol under oxidative conditions as in the case of mammalian cells (Walton et al., 2017). Walton et al. (2017) have reported that Cys-11 of human PEX5 acts as a redox switch that modulates the import of peroxisomal matrix proteins such as CAT. Under oxidative stress conditions, CAT is retained in the cytosol where it can protect against H2O2-mediated redox changes and reinforce cellular defenses to prevent oxidative damage out of peroxisomes (Walton et al., 2017). Oxidative and nitrosative stress could enable swift control of CAT localization in compartments, thus helping to regulate redox signaling pathways.
In the case of dual-targeted OPPP enzymes, targeting decisions appear to depend on the cytosolic redox state. This has been suggested in relation to Arabidopsis G6PD1/G6PD4 (Meyer et al., 2011), PGL3 (Hölscher et al., 2014), and PGD2 upon interaction with nonperoxisomal isoforms PGD1 or PGD3, which retain heteromeric enzymes in the cytosol (Lutterbey and von Schaewen, 2016). NADPH-oxidase and peroxisomal AtPAO3 cross-talk to balance intracellular O2.−/H2O2, which in turn, affect the cyt-c/AOX pathways in mitochondria and regulates pollen tube elongation (Wu et al., 2010). As catalase (cat2)-deficient Arabidopsis mutants show upregulation of ASC-GSH components in the cytosol (Mhamdi et al., 2010), peroxisomes can interfere with cytosolic redox state. Cytosolic redox changes, in turn, impact the dual targeting of 6-phosphogluconolactonase 3 (PGL3) of chloroplasts and peroxisomes in Arabidopsis leaves, a process requiring thioredoxin m2 (Trxm2) in the cytosol (Hölscher et al., 2014).
Future perspectives
Many challenges in peroxisome redox biology remain to be addressed (Outstanding questions). One of them is to determine the nature of proteins involved in peroxisomal NO production. We need to amplify our limited knowledge of the mechanisms underlying NO regulation of peroxisome dynamics, metabolism, and signaling, together with NO and ROS crosstalk with hormones, such as JA. Additional analyses of the interplay and hierarchy of ROS-, NO-dependent, and other peroxisomal PTMs such as phosphorylation and persulfidation are required. Peroxisome-dependent regulatory components also need to be characterized by analyzing gene network structures and by identifying downstream responses induced by peroxisomal ROS and NO. The components of contact sites and the factors involved in peroxule production, as well as the regulatory role of ROS and NO in both these areas, also need to be determined. Analysis of tethering techniques, specific fluorescence proteins, and ROS mutants, combined with meta-analyses of organelle proteome datasets, should provide a better understanding of peroxisomal dynamics and interorganelle interactions. Finally, identification of pexophagy receptors and adaptors and their regulation by ROS, NO, and S2H should enable us to integrate biochemical processes and organelle dynamics into our understanding of cellular regulatory systems.
OUTSTANDING QUESTIONS
Do carbonylated and nitrated peroxisomal proteins act as signalling messengers?
Does CAT3 transnitrosylate other proteins in addition to GSNOR? Are other transnitrosylases present in peroxisomes?
Is S-nitrosylation involved in retrotranslocation of peroxisomal proteins to the cytosol?
Which pexophagy receptors and adaptors are involved and what role do ROS and NO play in identifying peroxisomes for degradation?
How do cells balance peroxule production, peroxisome proliferation and changes in peroxisomal speed in response to changes in their environment?
Which ROS/redox sensor(s) is (are) present in the peroxisomal membrane?
Acknowledgments
The authors wish to thank Michael O’Shea for proofreading the manuscript and to apologize to colleagues whose work has not been cited due to space limitations.
Funding
Spanish Ministry of Science, Innovation and Universities, State Research Agency, The European Regional Development Fund (MCIU/AEI/ERDF, PGC2018-098372-B-100) and URICI-CSIC.
Conflict of interest statement. The authors declare no conflict of interest.
L.M.S. conceived the original review focus and wrote the article with input and critical discussion from all the authors; M.A.P.-V. and E.M.-M. edited the figures. All authors read and approved the content of the manuscript. L.M.S. has agreed to act as the author responsible for contacts and correspondence.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Luisa M. Sandalio (luisamaria.sandalio@eez.csic.es).
References
- Agurla S, Gayatri G, Raghavendra AS (2018) Polyamines increase nitric oxide and reactive oxygen species in guard cells of Arabidopsis thaliana during stomatal closure. Protoplasma 255: 153–162 [DOI] [PubMed] [Google Scholar]
- Alamillo JM, García-Olmedo F (2001) Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J 25: 529–540 [DOI] [PubMed] [Google Scholar]
- Antonenkov VD, Grunau S, Ohlmeier S, Hiltunen JK (2010) Peroxisomes are oxidative organelles. Antioxid Redox Signal 13: 525–537 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Baluška F, Bozhkov PV, Elander PH, Fernie AR, Galili G, Hassan A, Hofius D, Isono E, Le Bars R, et al. (2018) Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot 69: 1335–1353 [DOI] [PubMed] [Google Scholar]
- Baker A, Hogg TL, Warriner SL (2016) Peroxisome protein import: a complex journey. Biochem Soc Trans 44: 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma M, Lupiáñez JA, del Río LA (1999) Localization of nitric-oxide synthase in plant peroxisomes. Int J Biol Chem 274: 36729–36733 [DOI] [PubMed] [Google Scholar]
- Barton K, Mathur N, Mathur J (2013) Simultaneous live-imaging of peroxisomes and the ER in plant cells suggests contiguity but no luminal continuity between the two organelles. Front Physiol 4: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt A, Rosenwasser S, Meyer A, Fluhr R (2016) Organelle redox autonomy during environmental stress. Plant Cell Environ 39: 1909–1919 [DOI] [PubMed] [Google Scholar]
- Bundó M, Coca M (2017) Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J Exp Bot 68: 2963–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RS, Ha R, Mendel RR, Hille R (2009) Oxidative half-reaction of Arabidopsis thaliana sulfite oxidase generation of superoxide by a peroxisomal enzyme. Int J Bio Chem 284: 35479–35484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Muñoz N, Expósito-Rodríguez M, Collado-Arenal AM, Rodríguez-Serrano M, Laureano-Marín AM, Santamaría ME, Gotor C, Díaz I, Mullineaux PM, Romero-Puertas MC, et al. (2019) Cadmium induces reactive oxygen species-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ 42: 2696–2714 [DOI] [PubMed] [Google Scholar]
- Caplan JL, Kumar AS,, Park E, Padmanabhan MS, Hoban K, Modla S, Czymmek K, Dinesh-Kumar SP (2015) Chloroplast stromules function during innate immunity. Dev Cell 34: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo MC, Sandalio LM, del Río LA, León J (2008) Peroxisome proliferation, wound-activated responses and expression of peroxisome-associated genes are cross-regulated but uncoupled in Arabidopsis thaliana. Plant Cell Environ 31: 492–505 [DOI] [PubMed] [Google Scholar]
- Chaki M, Álvarez de Morales P, Ruiz C, Begara-Morales JC, Barroso JB, Corpas FJ, Palma JM (2015) Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Ann Bot 116: 637–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Van Breusegem F, Saindrenan P, Noctor G (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wu R, Feng J, Feng T, Wang C, Hu J, Zhan N, Li Y, Ma X, Ren B, Zhang J, Song C-P, et al. (2020) Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants. Dev Cell 53: 444–457 [DOI] [PubMed] [Google Scholar]
- Corpas FJ,, Barroso JB (2014) Peroxynitrite () is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann Bot 113: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ,, Barroso JB, Sandalio LM, Palma M, Lupiáñez JA, del Río LA (1999) Characterization and activity regulation during natural senescence. Plant Physiol 121: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Sandalio LM, Brown MJ, del Río LA, Trelease RN (2000) Identification of porin-like polypeptide(s) in the boundary membrane of oilseed glyoxysomes. Plant Cell Physiol 41: 1218–1228 [DOI] [PubMed] [Google Scholar]
- Costa A, Drago I, Behera S, Zottini M, Pizzo P, Schroeder JI, Pozzan T, Lo Schiavo F (2010) H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J 62: 760–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins AB, Pracharoenwattana I, Zhou W, Smith SM, Badger MR (2008) Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol 148: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Lu Y, Li Y, Yang C, Peng X, Lines TR (2016a) Overexpression of glycolate oxidase confers improved photosynthesis under high light and high temperature in rice. Front Plant Sci 7: 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P, Liu H, Islam F, Li L, Farooq MA, Ruan S, Zhou W (2016b) OsPEX11, a peroxisomal biogenesis factor 11, contributes to salt stress tolerance in Oryza sativa. Front Plant Sci 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río LA, Corpas FJ, Sandalio LM, Palma JM, Barroso JB (2003) Plant peroxisomes, reactive oxygen metabolism and nitric oxide. IUBMB Life 55: 71–81 [DOI] [PubMed] [Google Scholar]
- Duan G, Walther D (2015) The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol 11: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ (2007) MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19: 1376–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeed HT, Stevenson SR, Cuming AC, Baker A (2018) Conserved and differential transcriptional responses of peroxisome associated pathways to drought, dehydration and ABA. J Exp Bot 69: 4971–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy D, Sanad MNME, Duscha K, Lyons M, Liu F, Bozhkov P, Kunz HH,, Hu J, Neuhaus HE, Steel PG, et al. (2017) Impact of salt stress, cell death, and autophagy on peroxisomes: quantitative and morphological analyses using small fluorescent probe N-BODIPY. Sci Rep 7: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B (2013) Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Baker A, Wright M, Sparkes IA, Mhamdi A, Schippers JHM, Van Breusegem F (2020) On the move: redox-dependent protein relocation in plants. J Exp Bot 71: 620–631 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G (2009) Photorespiratory metabolism: genes, mutants, energetics and redox signaling. Ann rev Plant Bio 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2003) Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119: 355–364 [Google Scholar]
- Foyer CH, Noctor G (2016) Stress-triggered redox signalling: what’s in pROSpect? Front Plant Sci 39: 951–964 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Ruban A V, Noctor G (2017) Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem J 474: 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M, Lismont C (2019) Redox signaling from and to peroxisomes: Progress, challenges, and prospects. Antioxid Redox Signal 30: 95–112 [DOI] [PubMed] [Google Scholar]
- Gabaldón T (2018) Evolution of the peroxisomal proteome. Subcell Biochem 89: 221–233 [DOI] [PubMed] [Google Scholar]
- Gao H, Metz J, Teanby NA, Ward AD, Botchway SW, Coles B, Pollard MR, Sparkes I (2016) In vivo quantification of peroxisome tethering to chloroplasts in tobacco epidermal cells using optical tweezers. Plant Physiol 170: 263–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebicka L, Didik J (2009) Catalytic scavenging of peroxynitrite by catalase. J Inorg Biochem 103: 1375–1379 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J (2005) Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57: 181–188 [DOI] [PubMed] [Google Scholar]
- Goyer A, Johnson TL, Olsen LJ, Collakova E, Shachar-Hill Y, Rhodes D, Hanson AD (2004) Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from Arabidopsis. J Biol Chem 279: 16947–16953 [DOI] [PubMed] [Google Scholar]
- Gupta DK, Pena LB, Hernández A, Inouhe M, Sandalio LM (2017) NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ 40: 509–526 [DOI] [PubMed] [Google Scholar]
- Hackenberg T, Juul T, Auzina A, Gwizdz S, Małolepszy A, Van Der Kelen K, Dam S, Bressendorff S, Lorentzen A, Roepstorff P, et al. (2013) Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25: 4616–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi A, Komatsu S (2016) Impact of post-translational modifications of crop proteins under abiotic stress. Proteomes 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinojosa L, Sanad MNME, Jarvis DE, Steel P, Murphy K, Smertenko A (2019) Impact of heat and drought stress on peroxisome proliferation in quinoa. Plant J 99: 1144–1158 [DOI] [PubMed] [Google Scholar]
- Hodges M, Dellero Y, Keech O, Betti M, Raghavendra AS, Sage R, Zhu X, Allen DK, Weber APM (2016) Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. J Exp Bot 67: 3015–3026 [DOI] [PubMed] [Google Scholar]
- Hölscher C, Meyer T, Von Schaewen A (2014) Dual-targeting of Arabidopsis 6-phosphogluconolactonase 3 (PGL3) to chloroplasts and peroxisomes involves interaction with Trx m2 in the cytosol. Mol Plant 7: 252–255 [DOI] [PubMed] [Google Scholar]
- Holzmeister C, Gaupels F, Geerlof A, Sarioglu H, Sattler M, Durner J, Lindermayr C (2015) Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J Exp Bot 66: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DC, Scott GS, Zborek A, Mikheeva T, Kean RB, Koprowski H, Spitsin SV (2000) Uric acid, a peroxynitrite scavenger, inhibits CNS inflammation, blood-CNS barrier permeability changes, and tissue damage in a mouse model of multiple sclerosis. FASEB J 14: 691–698 [DOI] [PubMed] [Google Scholar]
- Hou Q, Bartels D (2015) Comparative study of the aldehyde dehydrogenase (ALDH) gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes. Ann Bot 22: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Willems P, Breusegem F, Van Messens J (2018) Pathways crossing mammalian and plant sulfenomic landscapes. Free Radic Biol Med 122: 193–201 [DOI] [PubMed] [Google Scholar]
- Huang L, Yu LJ, Zhang X, Fan B, Wang FZ, Dai YS, Qi H, Zhou Y, Xie LJ, Xiao S (2019) Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 15: 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba J, Kim BM, Shimura H, Masuta C (2011) Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol 156: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaipargas EA, Mathur N, Daher FB, Wasteneys GO, Mathur J (2016) High light intensity leads to increased peroxule-mitochondria interactions in plants. Front Cell Dev Biol 4: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Go YM (2010) Redox compartmentalization and cellular stress. Diabetes Obes Metab 12: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y, González KL, Bartel B (2018) Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol 176: 162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J (2009) Peroxisome biogenesis and function. Arabidopsis Book 7: e0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneeshaw S, Keyani R, Delorme-Hinoux V, Imrie L, Loake GJ, Le Bihan T, Reichheld JP, Spoel SH (2017) Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc Natl Acad Sci USA 114: 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA (2010) Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem 285: 6670–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Burlacot A, Liang Y, Légeret B, Alseekh S, Brotman Y, Fernie AR, Krieger-Liszkay A, Beisson F, Peltier G, et al. (2018) Interorganelle communication: peroxisomal MALATE DEHYDROGENASE2 connects lipid catabolism to photosynthesis through redox coupling in Chlamydomonas. Plant Cell 30: 1824–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Muthuramalingam M, Dietz KJ (2012) Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr Opin Plant Biol 15: 261–268 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Chung HS, Kobayashi Y, Howe GA (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. Int J Biol Chem 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Kumar AS,, Park E, Nedo A, Alqarni A, Ren L, Hoban K, Modla S, McDonald JH, Kambhamettu C, Dinesh-Kumar SP, et al. (2018) Stromule extension along microtubules coordinated with actin-mediated anchoring guides perinuclear chloroplast movement during innate immunity. Elife 7: 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing H, Doering L (2020) Analysis of potential redundancy among Arabidopsis 6-phosphogluconolactonase isoforms in peroxisomes. J Exp Bot 71: 823–836 [DOI] [PubMed] [Google Scholar]
- León J, Broseta AC (2020) Present knowledge and controversies, deficiencies, and misconceptions on nitric oxide synthesis, sensing, and signaling in plants. Plant Cell Environ 43: 1–15 [DOI] [PubMed] [Google Scholar]
- Li J, Hu J (2015) Using co-expression analysis and stress-based screens to uncover Arabidopsis peroxisomal proteins involved in drought response. PLoS One 10: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu J, Wang G, Cha JY, Li G, Chen S, Li Z, Guo J, Zhang C, Yang Y, et al. (2015) A chaperone function of NO CATALASE ACTIVITY1 Is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tietz S, Cruz JA, Strand DD, Xu Y, Chen J, Kramer DM, Hu J (2019) Photometric screens identified Arabidopsis peroxisome proteins that impact photosynthesis under dynamic light conditions. Plant J 97: 460–474 [DOI] [PubMed] [Google Scholar]
- Li Y, Chen L, Mu J, Zuo J (2013) LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol 163: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Trelease RN (2006) Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci 119: 1961–1972 [DOI] [PubMed] [Google Scholar]
- Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen X, Siemsen T, Morgenstern B, Meinicke P, Reumann S (2011) Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23: 1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lismont C, Koster J, Provost S, Baes M, Van Veldhoven PP, Waterham HR, Fransen M (2019) Deciphering the potential involvement of PXMP2 and PEX11B in hydrogen peroxide permeation across the peroxisomal membrane reveals a role for PEX11B in protein sorting. Biochim Biophys Acta Biomembr 1861: 182991. [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-moreno R, Valencia D, Ed CPI, De Vera C (2011) In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot 62: 3501–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbey MC, von Schaewen A (2016) Analysis of homo- and hetero-dimerization among the three 6-phosphogluconate dehydrogenase isoforms of Arabidopsis. Plant Signal Behav 11: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ruiz A, Cadenas S, Lamas S (2011) Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic Biol Med 51: 17–29 [DOI] [PubMed] [Google Scholar]
- Meyer T, Hölscher C, Schwöppe C, Von Schaewen A (2011) Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J 66: 745–758 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Noctor G, Baker A (2012) Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys 525: 181–194 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61: 4197–220 [DOI] [PubMed] [Google Scholar]
- Mindthoff S, Grunau S, Steinfort LL, Girzalsky W, Hiltunen JK, Erdmann R, Antonenkov VD (2016) Peroxisomal Pex11 is a pore-forming protein homologous to TRPM channels. Biochim Biophys Acta 1863: 271–283 [DOI] [PubMed] [Google Scholar]
- Mitsuya S, El-Shami M, Sparkes I, Charlton WL, Lousa CDM, Johnson B, Baker A (2010) Salt stress causes peroxisome proliferation, but inducing peroxisome proliferation does not improve NaCl tolerance in Arabidopsis thaliana. PLoS ONE 5: e9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2017) ROS are good. Trends Plant Sci 22: 11–19 [DOI] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ (2010) ROS signalling-specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Mor A, Koh E, Weiner L, Rosenwasser S, Sibony-benyamini H, Fluhr R (2014) Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiol 165: 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murota K, Shimura H, Takeshita M, Masuta C (2017) Interaction between cucumber mosaic virus 2b protein and plant catalase induces a specific necrosis in association with proteasome activity. Plant Cell Rep 36: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nila AG, Sandalio LM, López MG, Gómez M, del Río LA,, Gómez-Lim MA (2006) Expression of a peroxisome proliferator-activated receptor gene (xPPARα) from Xenopus laevis in tobacco (Nicotiana tabacum) plants. Planta 224: 569–581 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (2016). Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 171: 1581–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Reichheld J, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Matsunaga S, Mano S, Kondo M, Yamada K, Hayashi M, Kagawa T, Kadota A, Sakamoto W, Higashi S, et al. (2015) Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat Plants 1: 15035. [DOI] [PubMed] [Google Scholar]
- Oksanen E, Häikiö E, Sober J, Karnosky DF (2004) Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol 161: 791–799 [DOI] [PubMed] [Google Scholar]
- Olmedilla A, Sandalio LM (2019) Selective autophagy of peroxisomes in plants: from housekeeping to development and stress responses. Front Plant Sci 10: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC (2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J Exp Bot 63: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Hu J (2018) Proteome of plant peroxisomes. Subcell Biochem 89: 3–45 [DOI] [PubMed] [Google Scholar]
- Pan R, Liu J, Wang S, Hu J (2020) Peroxisomes: versatile organelles with diverse roles in plants. New Phytol 225: 1410–1427 [DOI] [PubMed] [Google Scholar]
- Pastori GM, del Río LA (1997) Natural senescence of pea leaves. Plant Physiol 113: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G (2012) Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ 35: 374–387 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Bartel B (2016) Plant peroxisomes: recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr Opin Plant Biol 34: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Bettermann M, Benz R, Heldt HW (1997) Evidence for the presence of a porin in the membrane of glyoxysomes of castor bean. Plant Physiol 115: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Chowdhary G (2018) Prediction of peroxisomal matrix proteins in plants. Subcell Biochem 89: 125–138 [DOI] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L (2004) AraPerox. A database of putative Arabidopsis proteins. Plan Physiol 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Weber APM (2006) Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled-Others remain. Biochim Biophys Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Rinaldi MA, Patel AB, Park J, Lee K, Strader LC, Bartel B (2016) The roles of β-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics 204: 1089–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Pazmiño DM, Sparkes I, Rochetti A, Hawes C, Romero-Puertas MC, Sandalio LM (2014) 2 ,4-Dichlorophenoxyacetic acid promotes S-nitrosylation and oxidation of actin affecting cytoskeleton and peroxisomal dynamics. J Exp Bot 65: 4783–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Sanz-Fernández M, Hu J, Sandalio LM (2016) Peroxisomes extend peroxules in a fast response to stress via a reactive oxygen species-mediated induction of the peroxin PEX11a. Plant Physiol 171: 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Sparkes I, Hawes C, del Río LA, Sandalio LM (2009) Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic Biol Med 48: 979. [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-kumar M, Wang K, Ryu C, Kaundal A, Mysore KS, Division PB, Roberts S, Foundation N (2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plan Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, McCarthy I, Sandalio LM, Palma JM, Corpas FJ, Gómez M, del Río LA (1999) Cadmium toxicity and oxidative metabolism of pea leaf peroxisomes. Free Radic Res 31: 25–31 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Rodríguez-Serrano M,, Corpas FJ, Gómez M, del Río LA, Sandalio LM (2004) Cadmium-induced subcellular accumulation of and H2O2 in pea leaves. Plant Cell Environ 27: 1122–1134 [Google Scholar]
- Romero-Puertas MC, Sandalio LM (2016) Nitric oxide level is self-regulating and also regulates its ROS partners. Front Plant Sci 7: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Fluhr R, Joshi JR, Leviatan N, Sela N, Hetzroni A, Friedman H (2013) ROSMETER: a bioinformatic tool for the identification of transcriptomic imprints related to reactive oxygen species type and origin provides new insights into stress responses. Plant Physiol 163: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Rot I, Sollner E, Meyer AJ, Smith Y, Leviatan N, Fluhr R, Friedman H (2011) Organelles contribute differentially to reactive oxygen species-related events during extended darkness. Plant Cell Environ 156: 185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JM, Nebenführ A (2018) Update on myosin motors: molecular mechanisms and physiological functions. Plant Physiol 176: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vicente I, Fernández-Espinosa MG, Lorenzo O (2019) Nitric oxide molecular targets: reprogramming plant development upon stress. J Exp Bot 70: 4441–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Gotor C, Romero LC, Romero-Puertas MC (2019) Multilevel regulation of peroxisomal proteome by post-translational modifications. Int J Mol Sci 20: 4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Peláez-Vico MA, Romero-Puertas MC (2020) Peroxisomal metabolism and dynamics at the crossroads between stimulus perception and fast cell responses to the environment. Front Cell Dev Biol 8: 2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Rodríguez-Serrano M, Romero-Puertas MC, del Río LA (2013) Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules. Subcell Biochem 69: 231–225 [DOI] [PubMed] [Google Scholar]
- Sandalio LM, Romero-Puertas MC (2015) Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann Bot 116: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht M, Ludwig-m J, Burbach C, Volkmann D, Baluska F (2013) Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytol 200: 473–482 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Rossoni AW, Maurino VG (2018) Dissecting the physiological function of plant glyoxalase I and glyoxalase I-like proteins. Front Plant Sci 9:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Bonekamp NA, Islinger M (2012) Fission and proliferation of peroxisomes. Biochim Biophys Acta Mol Basis Dis 1822: 1343–1357 [DOI] [PubMed] [Google Scholar]
- Schumann U, Prestele J, O’Geen H, Brueggeman R, Wanner G, Gietl C (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA 104: 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N, Jaspert N, Van Der Kelen K, Tognetti VB, Schmitz J (2014) Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol Plant 7: 1191–1210 [DOI] [PubMed] [Google Scholar]
- Shai N, Schuldiner M, Zalckvar E (2016) No peroxisome is an island-Peroxisome contact sites. Biochim Biophys Acta Mol Cell Res 1863: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H, Jones DP (2020) Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol 21: 363–383 [DOI] [PubMed] [Google Scholar]
- Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J (2009) Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J 59: 231–242 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D (2019) Hydrogen peroxide metabolism and functions in plants. New Phytol 221: 1197–1214 [DOI] [PubMed] [Google Scholar]
- Sousa RHV, Carvalho FEL, Lima-Melo Y, Alencar VTCB, Daloso DM, Margis-Pinheiro M, Komatsu S, Silveira JAG (2018) Impairment of peroxisomal APX and CAT activities increases protection of photosynthesis under oxidative stress. J Exp Bot 35: 627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Li W, Wang P, Ma C (2019) Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front Plant Sci 10: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler RON, Miller GAD (2012) ROS and redox signalling in the response of plants to abiotic stress. Plan Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Talbi S, Romero-Puertas MC, Hernández A, Terrón L, Ferchichi A, Sandalio LM (2015) Drought tolerance in a Saharian plant Oudneya africana: role of antioxidant defences. Environ Exp Bot 111: 114–126 [Google Scholar]
- Terrón-Camero LC, Rodríguez-Serrano M, Sandalio LM, Romero-Puertas MC (2020) Nitric oxide is essential for cadmium-induced peroxule formation and peroxisome proliferation. Plant Cell Environ 43: 2492–2507 [DOI] [PubMed] [Google Scholar]
- Thazar-Poulot N, Miquel M, Fobis-Loisy I, Gaude T (2015) Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc Natl Acad Sci USA 112: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiew TW, Sheahan MB, Rose RJ (2015) Peroxisomes contribute to reactive oxygen species homeostasis and cell division induction in Arabidopsis protoplasts. Front Plant Sci 6: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyutereva EV, Dobryakova KS, Schiermeyer A, Shishova MF, Pawlowski K, Demidchik V, Reumann S, Voitsekhovskaja OV (2018) The levels of peroxisomal catalase protein and activity modulate the onset of cell death in tobacco BY-2 cells via reactive oxygen species levels and autophagy. Funct Plant Biol 45: 247–258 [DOI] [PubMed] [Google Scholar]
- Umnajkitikorn K, Sade N, Rubio Wilhelmi MM, Gilbert ME, Blumwald E (2020) Silencing of OsCV (chloroplast vesiculation) maintained photorespiration and N assimilation in rice plants grown under elevated CO2. Plant Cell Environ 43: 920–933 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Kelen Van Der K, Dat J, Gadjev I, Boonefaes T, Morsa S, Van Breusegem F, Rottiers P, Slooten L, Van Montagu M, et al. (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Batelli G, Grillo S, Agius F, Kim Y-S, Zhu J, Agarwal M, Katiyar-Agarwal S, Zhu J-K (2007) Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol Cell Biol 27: 7771–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Vanloocke A, Bernacchi CJ, Ort DR (2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Waller JC, Dhanoa PK, Schumann U, Mullen RT, Snedden WA (2010) Subcellular and tissue localization of NAD kinases from Arabidopsis: compartmentalization of de novo NADP biosynthesis. Planta 231: 305–317 [DOI] [PubMed] [Google Scholar]
- Walton PA, Brees C, Lismont C, Apanasets O, Fransen M (2017) The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim Biophys Acta Mol Cell Res 1864: 1833–1843 [DOI] [PubMed] [Google Scholar]
- Wang W, Paschaladis K, Jian-Can F, Jie S, Ji-Hong L (2019) Polyamine catabolism in plants: a universal process with diverse functions. Front Plant Sci 10: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BL, Tang XY, Cheng LY, Zhang AZ, Zhang WH, Zhang FS, Liu JQ, Cao Y, Allan DL, Vance CP, et al. (2010) Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol 187: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Werner AK, Witte C (2011) The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci 16: 381–387 [DOI] [PubMed] [Google Scholar]
- Wimalasekera R, Tebartz F, Scherer GFE (2011) Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci 181: 593–603 [DOI] [PubMed] [Google Scholar]
- Wu J, Shang Z, Wu J, Jiang X, Moschou PN, Sun W, Roubelakis-Angelakis KA, Zhang S (2010) Spermidine oxidase-derived H2O2 regulates pollen plasma membrane hyperpolarization-activated Ca2+ -permeable channels and pollen tube growth. Plant J 63: 1042–1053 [DOI] [PubMed] [Google Scholar]
- Yang M, Li Z, Zhang K, Zhang X, Zhang Y, Wang X, Han C, Yu J, Xu K, Li D (2018) Barley stripe mosaic virus cb interacts with glycolate oxidase and inhibits peroxisomal ROS production to facilitate virus infection. Molecular Plant 11: 338–341 [DOI] [PubMed] [Google Scholar]
- Yanik T, Donaldson RP (2005) A protective association between catalase and isocitrate lyase in peroxisomes. Arch Biochem Biophys 435: 243–252 [DOI] [PubMed] [Google Scholar]
- Yoboue ED, Sitia R, Simmen T (2018) Redox crosstalk at endoplasmic reticulum (ER) membrane contact sites (MCS) uses toxic waste to deliver messages. Cell Death Dis 9: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y (2014) Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 127: 1161–1168 [DOI] [PubMed] [Google Scholar]
- Young PG, Bartel B (2016) Pexophagy and peroxisomal protein turnover in plants. Biochim Biophys Acta 1863: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D, Pedre Ezeriņa D, De Smet B, Lewandowska A, Tossounian MA, Bodra N, Huang J, Astolfi Rosado L, Van Breusegem F, Messens J (2019) Protein promiscuity in H2O2 signaling. Antioxid Redox Signal 30: 1285–1324 [DOI] [PubMed] [Google Scholar]
- Yuan HM, Liu WC, Lu YT (2017) CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, . ( et al. 2018) S-nitrosylation targets gsnoGSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell 71: 142–154 [DOI] [PubMed] [Google Scholar]
- Zhang T, Ma M, Chen T, Zhang L, Fan L, Zhang W, Wei B, Li S, Xuan W, Noctor G, et al. (2020) Glutathione-dependent denitrosation of GSNOR1 promotes oxidative signalling downstream of H2O2. Plant Cell Environ 43: 1175–1191 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu Y, Xie Z, Li X, He Z, Peng X (2016) Association-dissociation of glycolate oxidase with catalase in rice: a potential switch to modulate intracellular H2O2 levels. Mol Plant 9: 737–748 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Li X, Ratnasekera D, Wang C, Liu W, Song L, Zhang W, Wu W (2015) Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27: 1445–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YB, Liu C, Tang DY, Yan L, Wang D,, Yang YZ, Gui JS, Zhao XY, Li LG, Tang XD, et al. (2018) The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30: 1100–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]