Abstract
Among several animals, Rattus rattus (rat) lives in polluted environments and feeds on organic waste/small invertebrates, suggesting the presence of inherent mechanisms to thwart infections. In this study, we isolated gut bacteria of rats for their antibacterial activities. Using antibacterial assays, the findings showed that the conditioned media from selected bacteria exhibited bactericidal activities against Gram-negative (Escherichia coli K1, Klebsiella pneumoniae, Pseudomonas aeruginosa, Serratia marcescens, and Salmonella enterica) and Gram-positive (Bacillus cereus, methicillin-resistant Staphylococcus aureus, and Streptococcus pyogenes) pathogenic bacteria. The conditioned media retained their antibacterial properties upon heat treatment at boiling temperature for 10 min. Using MTT assays, the conditioned media showed minimal cytotoxic effects against human keratinocyte cells. Active conditioned media were subjected to tandem mass spectrometry, and the results showed that conditioned media from Bacillus subtilis produced a large repertoire of surfactin and iturin A (lipopeptides) molecules. To our knowledge, this is the first report of isolation of lipopeptides from bacteria isolated from the rat gut. In short, these findings are important and provide a platform to develop effective antibacterial drugs.
Introduction
Increasing multidrug resistance in pathogenic bacteria highlights an urgent need to discover alternative antibacterials.1−3 Although bacteria isolated from soil have been a tremendous source of antibiotics, bacteria inhabiting unique niches such as the gastrointestinal tract of animals (rats) living in polluted environments may also prove to be a source of potentially novel antimicrobials. In addition to conventional antibiotics, antimicrobial peptides are one of the most promising options that can reduce the risk of antibiotic-resistant bacteria.4−6 In recent years, antimicrobial biosurfactants (lipopeptides) have been extensively studied as a novel class of antimicrobial drugs. These peptides are shown to possess both narrow- and broad-spectrum antibacterial activities against Gram-negative and Gram-positive bacteria and fungi.1,6 Lipopeptides are mostly produced by Gram-positive bacteria including Bacillus species, while a few Gram-negative bacteria especially Pseudomonas have been reported to produce these antimicrobial peptides.1,7 In addition, Bacillus sp. produce several metabolites/molecules including antibacterial polyketides, bacillaenes, isobutanols, polyhydroxyalkanoates, difficidins, and dozens of structurally distinct antibacterial molecules. B. subtilis has key genes in its genome involved in the production of several antibacterial molecules.7 The most valuable and active among them are the cyclic lipopeptides such as surfactin, iturin A, bacillibactin, and fengycin.7−9 Lipopeptides produced by Gram-positive bacteria have been classified into different types based on the amino acids and the length of fatty acid chains.10 The presence of a lipid moiety in the lipopeptides increases their biological activity.1 For example, the chain length of carbon atoms (C10–C12) possesses bactericidal activity, while lipopeptides with 14 or 16 carbons in their lipid chain show higher antifungal activity in addition to antibacterial activity.1,5 These peptides have unique properties such as being easily biodegradable, eco-friendly, highly stable, and nonpolluting biomolecules and nontoxic in nature.5,11 Due to these properties, lipopeptides are gaining importance in several applications such as pharmaceuticals, bioremediation, and food preservation.5,12,13
We have hypothesized previously that gut bacteria of animals/pests are a potential source of novel antibacterials.3Rattus rattus (rat) inhabits polluted environments. They are omnivores eating a wide range of foods including organic wastes, seeds, fruit, stems, leaves, fungi, and several invertebrates and vertebrates.14,15 Rats are among the world’s most invasive species living in close contact with humans and share ecological niches.16 Rats act as a reservoir for many pathogenic species that are transmitted to mammals including humans.17,18 Although rats are known to play a key role in the transmission of several human and domestic animal diseases, it is not clear how they counter infections. A possible explanation is that their gut bacteria produce antibacterials to thwart bacterial infections. Thus, the aim of this study was to mine gut bacteria of a rat for potential antibacterials. We have isolated 30 different bacteria, and among these bacteria, three Bacillus strains showed significant antibacterial properties. Finally, B. subtilis (CM2) exhibited the highest bactericidal activities with minimal cytotoxicity; hence, CM2 was subjected to detailed characterization by tandem mass spectrometry.
Results
Bacterial Species Isolated from the Gut of Rattus rattus (Rat)
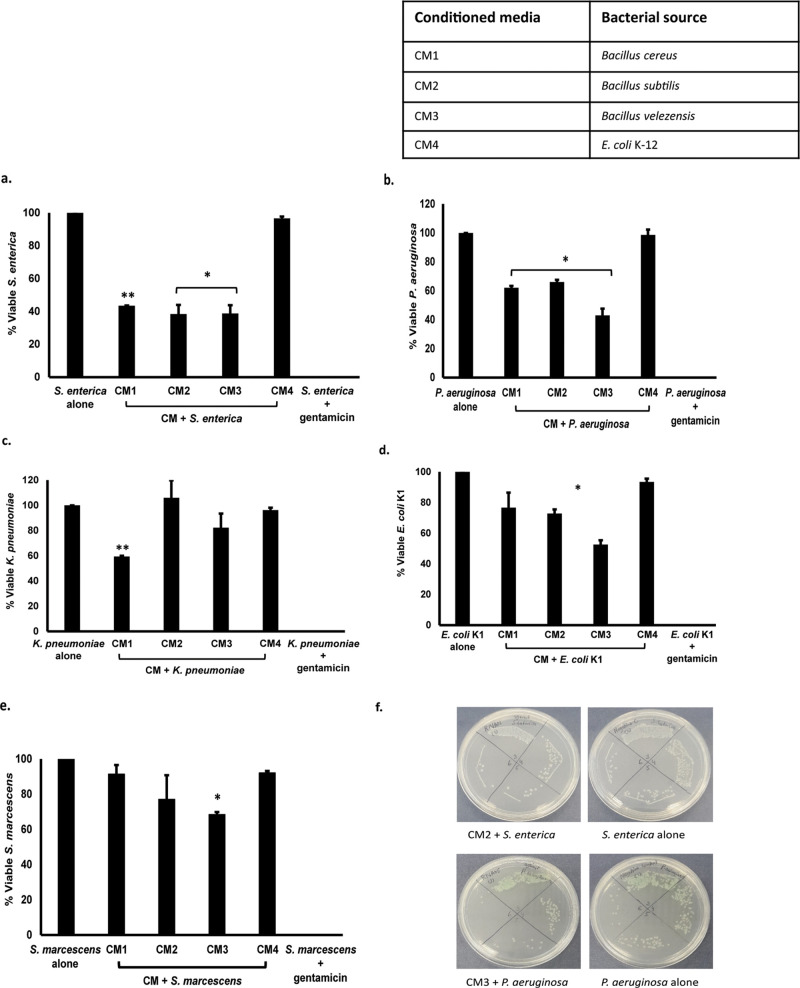
Several bacterial species were isolated from the gut of Rattus rattus (rat). Bacteria isolated were screened against Gram-positive and Gram-negative pathogenic bacteria. Bacteria with profound antibacterial activities were selected, and their identification was done using 16S rRNA gene amplification and sequencing. The results revealed B. velezensis (accession no. MN882653), B. cereus (accession no. MN882654), and B. subtilis (accession no. MN882652) (Figure 1). B. subtilis showed 99 bootstrap values with Bacillus subtilis subsp. natto strain NT-2 sharing the same clade. Next, conditioned media of Bacillus species isolated were prepared as discussed in the materials and methods (Table 1), and codes were given to all CM as shown in Table 2. The CM were then tested against a range of selected Gram-positive and Gram-negative pathogenic bacteria (Table 1).
Figure 1.
Phylogenetic tree representing 15 Bacillus strains with an E. coli strain U 5/41 (purple bold) as the outgroup and the B. subtilis (CM2) strain used in this study (red bold) based on the phylogenetic analysis of 16S rRNA genes. The phylogenetic tree was reconstructed using the maximum likelihood (ML) method (MEGA 7.) based on a GTR+G (8) model with concatenated 16S rRNA sequences. Percentage bootstrap values that were higher than 50% of 1000 replicates are indicated at branching nodes.
Table 1. Bacteria Used in This Study.
| bacteria | strain |
|---|---|
| Bacillus cereus | MTCC 131621 (clinical isolate) |
| methicillin-resistant Staphylococcus aureus | MTCC 381123 (clinical isolate) |
| Streptococcus pyogenes | ATCC 49399 (clinical isolate) |
| Escherichia coli K1 | MTCC 710859 (clinical isolate) |
| Escherichia coli K-12 | MTCC 817356 (nonclinical isolate) |
| Klebsiella pneumoniae | ATCC 13883 (clinical isolate) |
| Pseudomonas aeruginosa | ATCC 10145 (clinical isolate) |
| Salmonella enterica | ATTC 14028 (clinical isolate) |
| Serratia marcescens | MTTC 13880 (clinical isolate) |
Table 2. Bacterial Species Isolated from a Rat’s Gut and Their Conditioned Media.
| conditioned media | bacterial source |
|---|---|
| CM1 | Bacillus cereus |
| CM2 | Bacillus subtilis |
| CM3 | Bacillus velezensis |
| CM4 | E. coli K-12 |
Conditioned Media of Rattus rattus Gut Bacteria Exhibited Notable Antibacterial Activities against Gram-Negative and Gram-Positive Bacteria
Conditioned media were tested for their bactericidal effects against pathogenic bacteria, and the results revealed that all CM except CM4 showed significant antibacterial activities against S. enterica and P. aeruginosa (P < 0.05 using Student’s t-test, two-tailed distribution) (Figure 2a,b and Table 3). When tested against K. pneumoniae, only CM1 showed promising bactericidal properties (P < 0.05) (Figure 2c). CM2 and CM3 presented important antibacterial effects when tested against E. coli K1 (P < 0.05) (Figure 2d), while in the case of S. marcescens, only CM3 exhibited significant bactericidal activities (P < 0.05) (Figure 2e).
Figure 2.
Conditioned media of rat gut bacteria possessed significant antibacterial activities against Gram-negative bacteria. CM were incubated with 1 million bacteria for 2 h at 37 °C. Next, cultures were serially diluted and plated onto nutrient agar plates, and plates were incubated overnight at 37 °C. Viable bacterial cfu were calculated, and results were recorded. (a) CM tested against S. enterica, (b) against P. aeruginosa, (c) against K. pneumoniae, (d) against E. coli K1, and (e) against S. marcescens and (f) representative effects of CM against S. enterica and P. aeruginosa. Data are expressed as the mean ± standard error of several independent experiments performed in duplicate. P values were determined using Student’s t-test. An asterisk (*) denotes P ≤ 0.05.
Table 3. Overall Representation of Antibacterial Activities of Conditioned Media against Gram-Positive and Gram-Negative Bacteria.
| antibacterial
activities against Gram-positive bacteria |
antibacterial
activities against Gram-negative bacteria |
|||||||
|---|---|---|---|---|---|---|---|---|
| conditioned media | MRSA | B. cereus | S. pyogenes | E. coli K1 | P. aeruginosa | S. enterica | S. marcescens | K. pneumoniae |
| CM1 | – | + | – | – | + | + | – | + |
| CM2 | + | + | + | + | + | + | – | – |
| CM3 | – | + | + | + | + | + | + | – |
| CM4 | – | – | – | – | – | – | – | – |
Similarly, when CM were evaluated against Gram-positive bacteria, all CM except CM4 possessed antibacterial properties against B. cereus (P < 0.05) (Figure 3a and Table 3). CM2 showed the highest activity among all CM tested (Figure 3a). When CM were tested against S. pyogenes, CM2 and CM3 revealed notable bactericidal activities (P < 0.05) (Figure 3b), whereas only CM2 showed remarkable bacterial killing properties against MRSA (P < 0.05) (Figure 3c).
Figure 3.
Conditioned media exhibited important bactericidal effects against Gram-positive bacteria. Approximately 1 × 106 bacterial cells were exposed to CM from rat gut bacteria and incubated at 37 °C for 2 h. After this incubation, cultures were serially diluted, plated on nutrient agar, and incubated at 37 °C for 24 h. Bacterial colonies were enumerated on the following day. Data are expressed as the mean ± standard error of several independent experiments performed in duplicate. P values were determined using two sample t-tests. An asterisk (*) denotes P ≤ 0.05 (a) when CM were tested against B. cereus, (b) against S. pyogenes, and (c) against MRSA and (d) demonstrative effects of CM against MRSA and S. pyogenes.
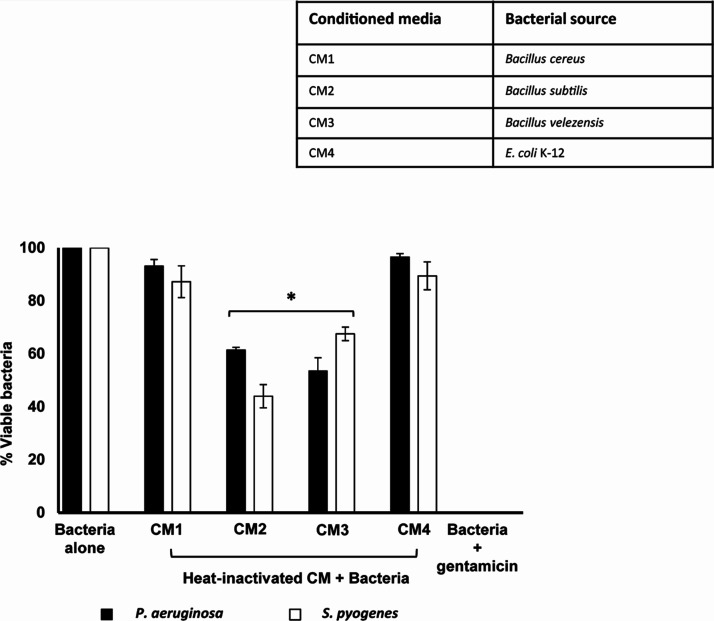
In some experiments, CM were incubated at 95 °C for 10 min, and then, their antibacterial properties were tested. Results showed that heat treatment did not abolish CM2 and CM3 and exhibited significant bactericidal effects against P. aeruginosa and S. pyogenes (P < 0.05) (Figure 4).
Figure 4.
Heat-treated CM-mediated antibacterial properties against MRSA and S. pyogenes. Heat-treated CM were incubated with MRSA and S. pyogenes for 2 h at 37 °C. Cultures were serially diluted and subsequently plated on nutrient agar plates, and plates were incubated at 37 °C overnight. The next day, bacterial cfu were enumerated, and results were recorded. Data are expressed as the mean ± standard error of three independent experiments performed in duplicate.
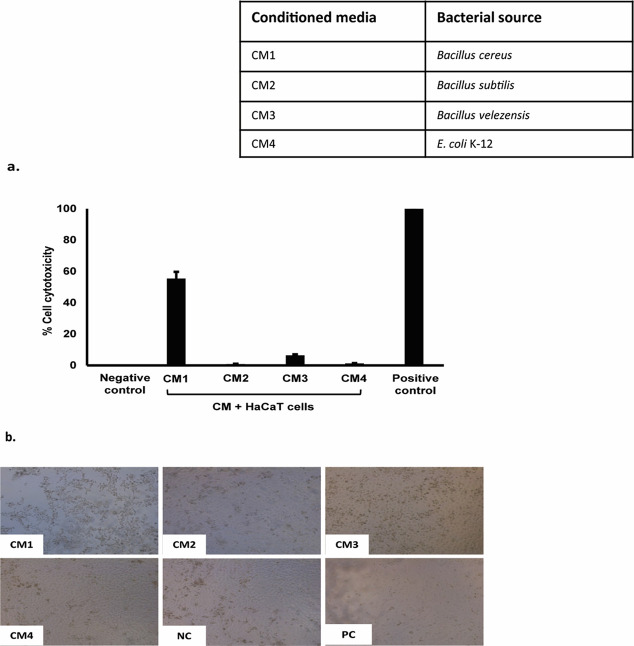
Conditioned Media Showed Limited Cytotoxicity
To determine the cytotoxic effects of CM, lactate dehydrogenase assays were performed against HaCaT cell lines. Results from cytotoxicity assays revealed that CM from Rattus rattus gut bacteria showed minimal effects against human cells except CM1 (B. cereus) with 55% cytotoxicity (Figure 5a,b).
Figure 5.
Conditioned media showed limited cytotoxicity properties against HaCaT cell lines. Briefly, HaCaT cells were challenged with CM from the rat gut bacteria for 24 h at 37 °C in the presence of 95% humidity and 5% CO2. The next day, LDH released from HaCaT cells was measured as described in the materials and methods. (a) CM tested were nontoxic against HaCaT cells except CM1 (55%) and (b) representation of CM cytotoxic effects incubated with a human cell monolayer.
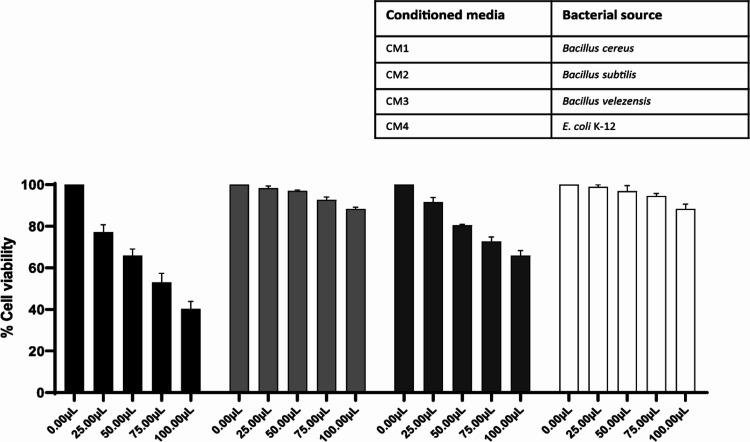
For MTT assays, CM were tested against human cell lines in a graduated concentration as discussed in the materials and methods. Results showed parallel results with LDH assays where CM1 showed moderate to higher cytotoxic effects, while other CM (CM2–CM4) showed negligible cytotoxic effects against human cell lines (Figure 6). Values for CC50 and MNTD90 highlighting CM2 (B. subtilis) as a safe antibacterial drug candidate used for its medicinal therapeutic use are shown in Table 4.
Figure 6.
Conditioned media of rat gut bacteria produced negligible cytotoxicity against human cell lines. CM from all three Bacillus species were tested against human cells at their graduated volume/concentration overnight at 37 °C with 5% CO2 and 95% humidity. Only CM1 (B. cereus) showed moderate to high cytotoxic effects, and other CM showed limited effects. Data are presented as the mean ± SE of three independent experiments performed in duplicate.
Table 4. Cytotoxic Concentration 50 (CC50) and Maximum Nontoxic Dose (MNTD90) Values of CM of Rat Gut Bacteriaa.
| sample/conditioned media | CC50 | MNTD90 |
|---|---|---|
| CM1 | 78.51 | 12.54 |
| CM2 | 295.3 | 90.76 |
| CM3 | 171.5 | 27.04 |
| CM4 | 281.9 | 97.51 |
CC50 = cytotoxic concentration. MNTD90 = maximum nontoxic dose. CC50 and MNTD90 are the concentrations at which 50 and 90% cells survive.
Mass Spectrometry Revealed Lipopeptides (Surfactin and Iturin)
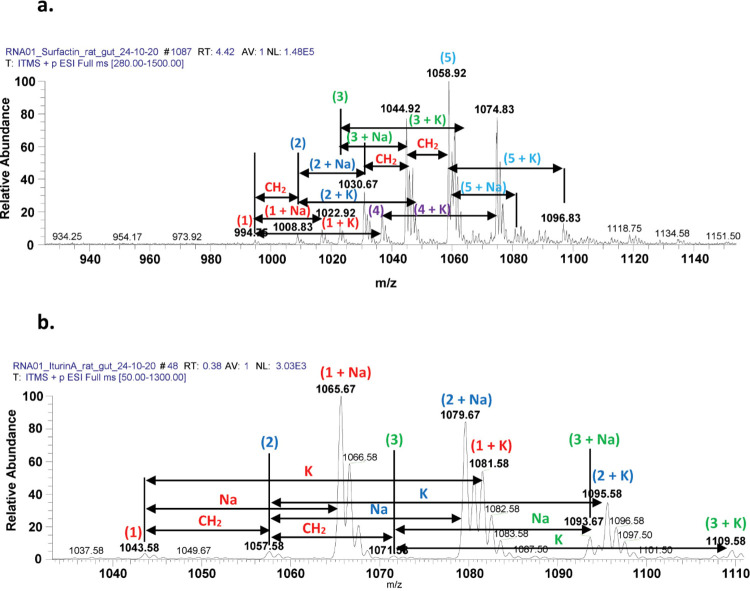
Among all CM tested, CM2 (B. subtilis with accession no. MN882652) showed the highest bactericidal effects, and it was further subjected to electrospray ionization (ESI)-MS/MS analysis. The direct syringe pump method was employed, and CM were injected. Tandem mass spectrometry was performed at positive total ion full scan mode. Results showed that CM2 (B. subtilis) revealed numerous lipopeptide molecules (Figure 7 and Table 5).
Figure 7.
LCMS spectrum of B. subtilis isolated from the rat gut revealed the presence of several homologues of lipopeptides labeled as (a) surfactin molecules and (b) spectrum illustrating iturin A molecular ion species at positive ion mode.
Table 5. Identification of Lipopeptides Using ESI MS/MS Analysis Produced by B. subtilis Isolated from the Rat Gut.
| metabolites/homologues | M | [M+H]+ | [M+Na]+ | [M+K]+ | references |
|---|---|---|---|---|---|
| C12 surfactin | 993.92 | 994.75 | 1016.83 | 1033.75 | (28) |
| C13 surfactin | 1007.75 | 1008.83 | 1030.92 | 1047.83 | (7, 28) |
| C14 surfactin | 1021.83 | 1022.92 | 1044.92 | 1061.75 | (7, 27, 28) |
| C15 surfactin | 1035.50 | 1036.83 | 1058.75 | 1074.75 | (7, 27, 28) |
| C16 surfactin | 1049.83 | 1050.92 | 1072.83 | 1089.83 | (7, 27) |
| C14 iturin A | 1042.67 | 1043.58 | 1065.67 | 1081.58 | (7, 27) |
| C15 iturin A | 1056.50 | 1057.58 | 1079.67 | 1095.58 | (7, 27) |
| C16 iturin A | 1070.67 | 1071.58 | 1093.67 | 1109.58 | (29) |
In this study, we identified and characterized lipopeptide (surfactin and Iturin A) molecules ranging from m/z 1000 to 1100 (Figure 7a,b). Lipopeptides produced by B. subtilis isolated from the rat gut were first analyzed through ESI-MS in positive full scan mode. The initial set of peaks observed belonged to the surfactin family with strong signals at m/z 1030.67, 1044.92, and 1058.75 [M+Na]+ corresponding to sodiated peaks; moderate signals of 994.75, 1008.83, 1022.92, 1036.92 [M+H]+; and finally 1046.92, 1060.67, 1074.83 and 1096.83 corresponding to [M+K]+ (Figure 7a, Supplementary Figure S1a–e). Surfactins are believed to be a potent alternative to antibiotic compounds.19 The presence of these metabolites was confirmed by tandem mass spectrometry as well as through comparing the values with literature data.
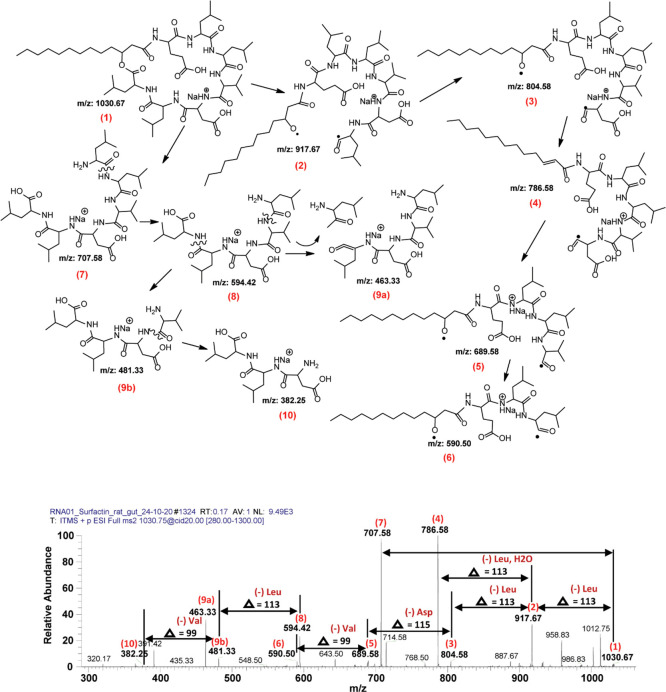
Among surfactin molecular peaks, sodiated adduct at m/z 1030.67 [M+Na]+ was further analyzed by tandem mass spectrometry MS/MS analysis due to its higher ion intensity, and the fragmentation pattern revealed two different series of daughter ion peaks with the first series ranging from m/z 590.50 to 1012.75 (Figure 8).
Figure 8.
LCMS spectrum representing surfactin molecular ion species. The fragmentation pattern and product ion spectra of sodiated molecules of surfactin at m/z 1030.67 [M+Na]+at positive ion mode.
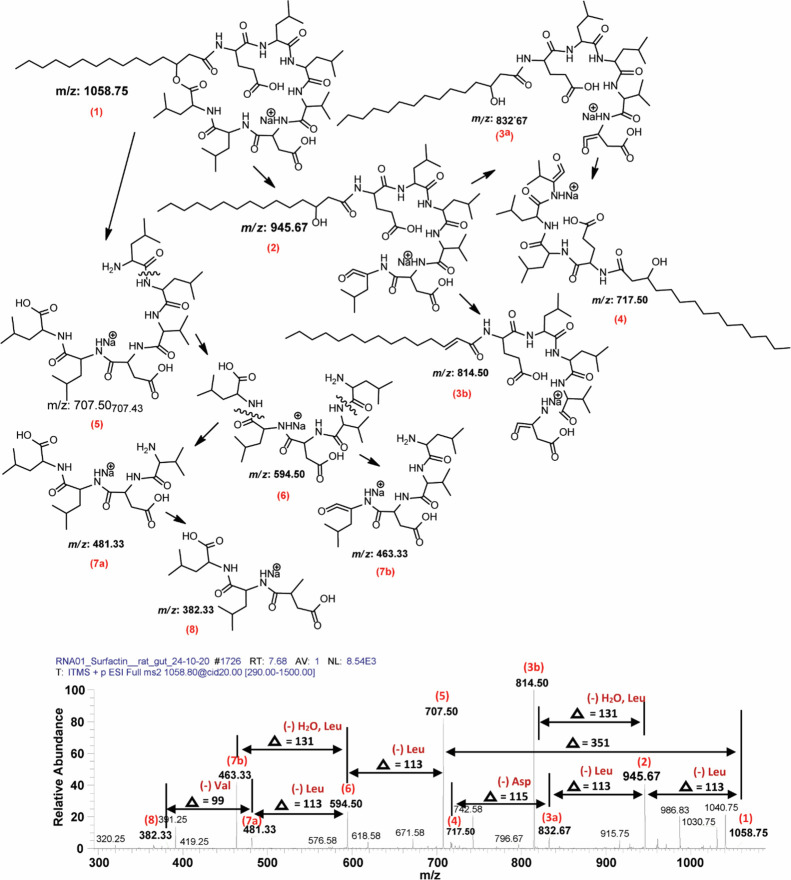
Fragmentation of these surfactin molecules resulted in two different series. Series 1 represents loss of amino acid residues (Figure 8), while Series 2 shows loss of fatty acid chain length (CH2) (Figure 8). Upon CID@20, product ions of m/z 1030.67 yielded both patterns (m/z 707.58–1012.75, Series 1, and m/z 382.25 to 707.58, Series 2). In detail, the peak at m/z 1030.67 [M+Na]+ (1) loses two consecutive leucine (Δ = 113) molecules with one mole of H2O (Δ = 18) leaving 917.67 (2) and 804.58 (3) and 786.58 (4). Further dissociation yielded 689.58 (5) (−asparagine), 590.50 (6) (−valine), 481.33 (9b), 463.33 (9a), and 382.25 (10) with loss of leucine, H2O, and finally valine (Figure 8). On the other hand, when CID was applied, molecule (1) generated 707.58 (7) that further dissociated into (8), (9a), (9b), and (10) as shown in Figure 8. Correspondingly, another sodiated peak at m/z 1058.75 [M+Na]+ (1) resulted in a similar fragmentation pattern and produced its fingerprint daughter ion peaks such as 707.50 (5), 594.50 (6), 481.33 (7a), 463.33 (7b), and 382.33 (8) (Figure 9). In both cases, both variants (Leu and Val) are present. For example, in the case of m/z at 1030.67 (Figure 8), the difference between 707.58 (7) and 594.42 (8) highlighted the loss of leucine molecules (−113), while at the same time, the difference between 689.58 (5) and 590.50 (6) signifies the loss of a valine molecule (−99). A similar pattern has been observed for m/z at 1058.75 where due to the loss of leucine (−113) from 707.50 (5) resulted in 594.50 (6), and the alternate loss of valine (−99) from 717.50 (4) produced 618.58 (not labeled). All the other surfactin homologues showed almost the same pattern of fragmentation upon tandem mass spectrometry analysis (Supplementary Figure S1a–e).
Figure 9.
Profiling of fragmentation data obtained from tandem mass spectrometry of a surfactin homologue at m/z 1058.75 [M+Na]+ at positive ion mode.
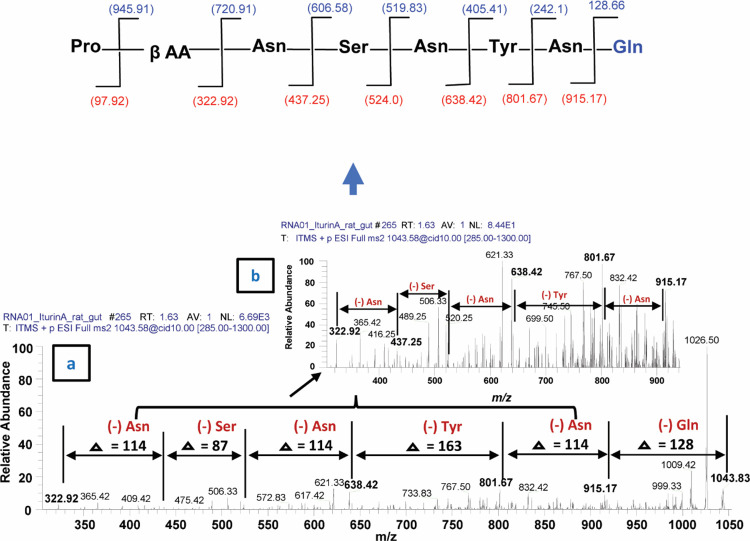
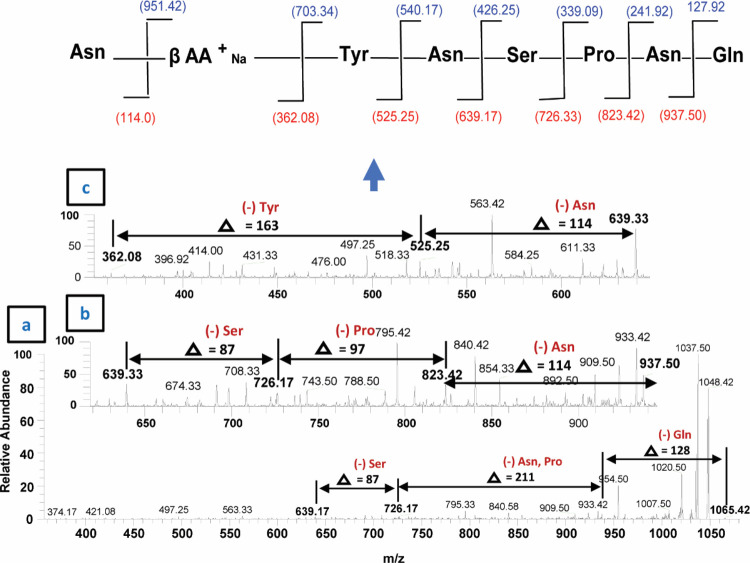
The second class of lipopeptides produced by B. subtilis corresponds to prominent peaks of iturin A molecules (Figure 7b, Supplementary Figure S2a–c). Among them, when 1043.83 [M+H]+ was subjected to tandem mass spectrometry analysis (MS2), upon CID @10, the iturin A precursor ion produced 915.17 after losing one glutamine molecule (−128) (Figure 10). Further fragmentation (MS3 and MSn) upon CID produced several daughter ion peaks, i.e., 801.67 (loss of asparagine −114), 638.42 (loss of tyrosine −163), 524.58 (loss of asparagine, −144), 437.25 (loss of serine, −87) and 322.92 (loss of asparagine, −114), leaving the proline (−98) moiety with a fatty acid chain (Figure 10). Next, the sodiated peak 1065.42 [M+Na]+ was characterized by MS/MS analysis (Figure 11). The fragmentation pattern revealed its daughter ion peaks that confirmed its identity as an iturin A molecule as previously characterized.20,21 When CID @11.0 was applied, the sodiated peak yielded 937.50 after losing one mole of glutamine residues (−128). Likewise, further fragmentation generated 823.42 (shed off asparagine, −114), 726.17 (loss of proline, −97), 639.33 (loss of serine, −87), 525.25 (loss of asparagine), and finally 362.08 (loss of tyrosine, −163) while leaving asparagine (114) with a sodiated fatty acid chain. A similar fragmentation pattern has been observed for the rest of iturin A homologues by ESI-MS/MS (Supplementary Figure S2a–c).
Figure 10.
Fragmentation pattern and product ion spectra of sodiated molecules of the iturin A homologue having m/z at 1043.83 [M+H]+.
Figure 11.
ESI–MS/MS spectrum of the iturin A precursor ion [M+Na]+ at m/z 1065.42 and their daughter ion peaks upon CID.
Discussion and Conclusions
Increasing antibiotic resistance and a decline in antibacterial discovery are of major concerns to human and animal health.22,23 Therefore, there is an urgent need to search for safe and effective drugs to combat antibiotic-resistant bacteria. Natural products isolated from microbes are considered as the most promising source of future antibacterials.24 For example, bacteria from panda feces were shown to produce bioactive molecules with strong antibacterial activities against enterotoxigenic E. coli, Salmonella, and S. aureus.25 Similarly, gut bacteria of fish have been exploited for their antibacterial activities against multidrug-resistant pathogenic bacteria including Aeromonas hydrophila, Salmonella enteritidis, Campylobacter jejuni, Vibrio cholera, and Enterobacter cloacae.26 Sponge-associated bacteria showed antimicrobial activities against Candida albicans, S. aureus, and Vibrio parahaemolyticus.27 A recent study showed that actinomycetes isolated from insects produced a potent antimicrobial agent “cyphomycin”. This antibacterial showed robust bioactivity against multidrug-resistant fungal species and Gram-negative bacteria.28 A recent study reported lipopeptides from Gram-negative P. aeruginosa with potent antibacterial and antifungal activities.29 Several other microbial species also produced structurally diverse lipopeptides having antagonistic effects against other pathogens.30 Similarly, B. subtilis R1 significantly reduced surface tension and enhanced oil recovery.31
In this study, aerobic and culturable bacteria were isolated from the gut of Rattus rattus. Isolated bacteria were identified and cultured in RPMI (minimal medium) to prepare CM. CM were evaluated for their antibacterial activities before and after heat treatment at 95 °C for 10 min. CM exhibited significant antibacterial activities against selected pathogenic bacteria. CM2 and CM3 retained the activity post heat treatment and showed notable bactericidal effects against P. aeruginosa and S. pyogenes. CM were tested for their cytotoxicity against human cell lines, and only CM1 had 55% cytotoxic effects, while other CM showed no cytotoxicity against HaCaT cells. CM2 showed consistent and broad-spectrum activity and showed no human cell cytotoxicity. Furthermore, CM2 were subjected to liquid chromatography–mass spectrometry (LCMS) analysis. Tandem mass spectrometry analysis revealed that CM2 (B. subtilis) produced abundant isoforms of surfactant molecules during cultivation of potent antibacterial lipopeptides, i.e., “surfactin”32,33 and pore-forming iturin A molecules.34,35
Molecular ion peaks with high intensity were subjected to MS/MS and MSn analysis, and results showed that B. subtilis produced several lipopeptides and their derivatives. Previously, Mandal et al., (2013) isolated and identified lipopeptides from Enterobacter and Citrobacter species that showed significant antibacterial activities against S. aureus (MTCC1430) (1).1 Likewise, in another study, lipopeptides were isolated and characterized for their antifungal activities against plant fungal pathogens.36 Ali et al., (2014) isolated and identified Bacillus sp. from a plant rhizosphere. The bacterium was characterized for isolation of lipopeptides surfactin and iturin A. The lipopeptides exhibited broad-spectrum antifungal activities.7Bacillus safensis isolated from olive oil (contaminated with soil) produced several lipopeptide molecules with antibacterial activities against several bacterial pathogens and antibiofilm activity of Staphylococcus epidermidis.37 A novel lipopeptide, paenibacterin, was identified from Paenibacillus sp. isolated from soil. The lipopeptide showed broad-spectrum antibacterial activities against Gram-positive and Gram-negative pathogenic bacteria.38 Similarly, here we identified lipopeptides surfactin and Iturin A from bacteria isolated from the rat gut with broad-spectrum antibacterial activities against selected bacterial pathogens, and our results are in agreement with the abovementioned literature. Lipopetides (fengycin) were identified from B. subtilis, a Banyan endophyte with strong antifungal activities,39 while surfactin molecules were identified from B. subtilis. Molecules showed significant antibacterial activities against both bacteriocin-sensitive and bacteriocin-resistant Listeria monocytogenes at lower concentrations. Antibacterial activity was retained after heat treatment for 10 min and 121 °C as well as proteolytic incubation.40 In this study, rat gut bacteria produced several lipopeptides, and further characterization and functional studies of these molecules could be a basis for the development of novel antibacterial(s).
In summary, bacteria here were isolated from a unique source, i.e., rats, that inhibit polluted environments. The rat-derived gut bacteria showed broad-spectrum antibacterial activities against several Gram-positive and Gram-negative pathogenic bacteria. Mass spectrometry analysis revealed several lipopeptides from B. subtilis that could be the basis for rational development of antibacterial molecules.
Experimental Section
Isolation and Identification of Rat Gut Bacteria
The use of animals was approved by the Sunway University Research Ethics Committee, SUNREC 2017/042. We confirm that all experiments including dissection were done according to appropriate regulations and guidelines as described earlier.3,41 Prior to dissection, all instruments were sterilized as well as surface sterilized using 70% alcohol throughout the dissection. The whole gut was removed aseptically. Following this, bacteria were isolated from the gut with sterile cotton swabs followed by plating on blood agar plates and incubated for 24 h at 37 °C. A number of bacterial species were isolated from the gut. These bacteria were differentiated from each other based on the appearance, color, shape, and texture on blood agar plates. Bacterial colonies selected were cultured on nutrient agar plates at 37 °C overnight. Subsequently, bacterial identification was performed using molecular identification by 16S rRNA amplification and sequencing.42 Single pure bacterial colonies were cultured in RPMI (minimal medium) at 37 °C for 24 h aerobically to prepare their conditioned media (CM). Next, overnight cultures were centrifuged at 10,000g at 4 °C for 1 h. Finally, culture supernatants were collected, filter-sterilized using a 0.22 μm pore size filter, and CM were stored at −80 °C until further use.
Bacterial Cultures
Several bacteria used in this study including Gram-positive (B. cereus, methicillin-resistant S. aureus (MRSA), and S. pyogenes) and Gram-negative neuropathogenic E. coli K1, E. coli K-12, K. pneumoniae, P. aeruginosa, S. enterica, and S. marcescens) (Table 1). MRSA was isolated from the blood culture of a patient with sepsis, while E. coli K1 (018: K1:H7), strain E44, was originally isolated from the cerebrospinal fluid (CSF) of a meningitis patient (obtained from the Luton & Dunstable NHS Foundation Trust, Luton, England, UK). All other bacteria were isolated from clinical samples including B. cereus, K. pneumoniae, P. aeruginosa, S. enterica, S. marcescens, and S. pyogenes (Table 1). Bacteria were grown in nutrient broth at 37 °C overnight aerobically prior to experiments as previously described.3,43−45
Evaluation of Bacterial Supernatants for Antibacterial Assays
To determine bactericidal properties of CM, antibacterial assays were performed as previously described.3,44,46 Briefly, 1 × 106 bacteria were challenged with 100 μL of CM at 37 °C for 2 h. Following this, bacterial cultures were 10-fold serially diluted, plated on nutrient agar plates, and incubated overnight at 37 °C. Bacterial colonies were counted the following day. Bacteria incubated in PBS and E. coli K-12 CM were used as the negative control, while bacteria incubated with gentamicin (100 μg/mL) were used as the positive control. Additionally, in some experiments, CM were heat-treated at 95 °C for 10 min and then tested for their antibacterial activities.3,47
In Vitro Cytotoxicity Assays
To determine host cell cytotoxic effects of CM, cytotoxicity assays were performed using the human cell line (HaCaT).48,49 Briefly, HaCaT cells (P14) were obtained from a cell tissue culture laboratory (Department of Biological Sciences, Sunway University). HaCaT monolayers were exposed to CM (100 μL) at 37 °C with 5% CO2 and 95% humidity for 24 h. Next, cell supernatants were collected, and cytotoxicity was evaluated using the lactate dehydrogenase (LDH) assay kit (cytotoxicity detection kit). To estimate LDH released by human cells, percent cytotoxicity was determined as follows
cytotoxicity (%) = (sample value – negative control value)/(positive control value – negative control value) × 100.
For the negative control, cells were grown in RPMI alone and incubated with E. coli K-12 CM, while cells incubated with Triton X-100 (0.1%) were taken as the positive control. In addition (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or MTT assay was performed by growing a HaCaT cell up to 80–90% confluency in a 96-well plate for 24 h at 37 °C in the presence of 95% humidity and 5% CO2.44 Subsequently, cells were exposed to CM at graduated concentrations, i.e., 25, 50, 75, and 100 μL and incubated at 37 °C for 24 h with 5% CO2 and humidified conditions. After this, freshly prepared MTT dye solution (10 μL) was added and incubated for 3–4 h. DMSO (100 μL) was added to each well to dissolve formazan crystals formed by live cells. HaCaT monolayer cells incubated with DMSO alone were taken as the negative control, and absorbance was measured at 540 nm, and percent viability was determined using the given formula, % viability = mean OD of the test sample/mean OD of the negative control × 100.
Calculations for 50% cytotoxic concentration (CC50) and maximum nontoxic dose (MNTD90) were performed using GraphPad Prism 8.0.2 software.
Extraction and Purification of Lipopeptides
B. subtilis was grown in 1 L of the minimal salt medium (MSM) at 30 ± 2 °C for 24 h as described earlier.7 After this incubation, the culture was centrifuged at 10000g for 45 min at 4 °C, and the culture supernatant was extracted using 1:3 of n-butanol in three rounds, and bacterial extracts were precipitated by pH adjustment of the medium to 2.0 with 6 N hydrochloric acid (HCL). The mixture was incubated at 4 °C for 2 h. Next, the final precipitate was achieved through centrifugation at 4 °C for 30 min, and resultant residues were dissolved in 2 mL of LCMS grade methanol (MeOH) and type 1 water at 2:1. It was further filtered through a 0.22 μm syringe filter to avoid contamination. Finally, the extract was evaporated under reduced pressure by rotary evaporation and resuspended in 2 mL of LCMS grade MeOH and stored at −20 °C until further use.
Metabolic Profiling of Bacillus subtilis (CM2) Supernatant Extracts Using Mass Spectrometric Analysis
For characterization, the extract (lipopeptides) was subjected to tandem mass spectrometry analysis using a mass spectrometer (LTQ XL Linear Ion Trap mass spectrophotometer, Thermo Scientific, USA) equipped with an ESI source as described previously.44 Samples were filter-sterilized first and then injected through a direct syringe pump with a flow rate of 5 μL min–1. Samples were scanned at positive total ion full scan mode (mass scan range m/z 50–2000) with a source voltage and a capillary voltage of 4.8 kV and 23 V, respectively. Sheath gas flow (N2) and capillary temperature were 30 arbitrary units and 350 °C, respectively, in both scan modes. Selected analytes were further fragmented at positive ion modes by employing a collision-induced dissociation (CID) energy of 35 (percentage of 5 V). Mass spectra for molecule(s) present in CM2 were compared against the NIST Mass Spectral Search Program for identification of their analogues. Compounds were identified after correlation with already published data.
Statistical Analysis
Data analysis was done using Student’s t-test to determine statistical significance. Data are presented as the mean ± standard error of several independent experiments performed in duplicate. P values ≤ 0.05 were considered statistically significant. Cytotoxic concentration (CC50) and maximum nontoxic dose (MNTD), i.e., CC90 values, were determined with GraphPad Prism version 8.0.2 (GraphPad Software, San Diego, CA, USA) software.
Acknowledgments
The authors acknowledge the Sunway University, Malaysia, the National Institute for Biotechnology and Genetic Engineering Faisalabad, Pakistan (NRPU nos. 2056 and 3786), the American University of Sharjah, and the University of Sharjah.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01137.
ESI–MS spectrum of the B. subtilis CM2 extract demonstrating the presence of surfactin analogues analyzed at positive ion mode (Figure S1); putative structures of the fragment ion generated through CID of m/z at 1057.58, 1079.58, and 1081.50 [M+H]+ (Figure S2); ESI–MS spectrum of the surfactin standard (Figure S3); and ESI–MS spectrum of the iturin A standard (Figure S4) (PDF)
Author Contributions
K.S. and N.A. sourced the animals and carried out dissections. N.A. performed all the experiments under the supervision of R.S., M.I., K.S.K., and N.A.K. and wrote the first draft of the manuscript. N.A. carried out LC/MS data analyses under the supervision of M.I. F.H. helped N.A. in LCMS sample analysis. R.S. and N.A.K. conceived the idea, obtained funding, supervised all work, and corrected the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Mandal S. M.; Sharma S.; Pinnaka A.; Kumari A.; Korpole S. Isolation and characterization of diverse antimicrobial lipopeptides produced by Citrobacter and Enterobacter. BMC Microbiol. 2013, 13, 152. 10.1186/1471-2180-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R.; Duse A.; Wattal C.; Zaidi A. K. M.; Wertheim H. F. L.; Sumpradit N.; Vlieghe E.; Hara G. L.; Gould I. M.; Goossens H.; Greko C.; So A. D.; Bigdeli M.; Tomson G.; Woodhouse W.; Ombaka E.; Peralta A. Q.; Qamar F. N.; Mir F.; Kariuki S.; Bhutta Z. A.; Coates A.; Bergstrom R.; Wright G. D.; BrowN E. D.; Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Akbar N.; Siddiqui R.; Iqbal M.; Sagathevan K.; Khan N. A. Gut bacteria of cockroaches are a potential source of antibacterial compound (s). Lett. Appl. Microbiol. 2018, 66, 416–426. 10.1111/lam.12867. [DOI] [PubMed] [Google Scholar]
- Gill E. E.; Franco O. L.; Hancock R. E. W. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 2015, 85, 56–78. 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena K. R.; Kanwar S. S. Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed Res. Int. 2015, 1–9. 10.1155/2015/473050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduszynska M.; Maciejewska M.; Greber K.; Sawicki W.; Kamysz W. Antibacterial activities of lipopeptide (C10) 2-KKKK-NH2 applied alone and in combination with lens liquids to fight biofilms formed on polystyrene surfaces and contact lenses. Int. J. Mol. Sci. 2019, 20, 393. 10.3390/ijms20020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S.; Hameed S.; Imran A.; Iqbal M.; Lazarovits G. Genetic, physiological and biochemical characterization of Bacillus sp. strain RMB7 exhibiting plant growth promoting and broad-spectrum antifungal activities. Microb. Cell Fact. 2014, 13, 144. 10.1186/s12934-014-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.; Patel S. K.; Kalia V. C. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb. Cell Factories. 2009, 8, 38. 10.1186/1475-2859-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Huang D.; Li Y.; Wen J.; Jia X. Rational improvement of the engineered isobutanol-producing Bacillus subtilis by elementary mode analysis. Microb. Cell Fact. 2012, 11, 101. 10.1186/1475-2859-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M.; Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Rodrigues L.; Banat I. M.; Teixeira J.; Oliveira R. Biosurfactants: potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. 10.1093/jac/dkl024. [DOI] [PubMed] [Google Scholar]
- Shaligram N. S.; Singhal R. S. Surfactin–a review on biosynthesis, fermentation, purification and applications. Food Technol. Biotechnol. 2010, 48, 119–134. [Google Scholar]
- Chen W. C.; Juang R. S.; Wei Y. H. Applications of a lipopeptide biosurfactant, surfactin, produced by microorganisms. Biochem. Eng. J. 2015, 103, 158–169. 10.1016/j.bej.2015.07.009. [DOI] [Google Scholar]
- Mushtaq-Ul-Hassan M.; Hussain I.; Shehzadi B.; Shaheen M.; Mahmood M.; Rafique A.; Mahmood-Ul-Hassan M. Occurrence of some zoonotic microorganisms in faecal matter of house rat (Rattus rattus) and house mouse (Musmusculus) trapped from various structures. Pak. Vet. J. 2008, 28, 171–174. [Google Scholar]
- Masi E.; Pino F. A.; Santos M. D. G. S.; Genehr L.; Albuquerque J. O. M.; Bancher A. M.; Alves J. C. M. Socioeconomic and environmental risk factors for urban rodent infestation in Sao Paulo, Brazil. J Pest Sci. 2010, 83, 231–241. 10.1007/s10340-010-0290-9. [DOI] [Google Scholar]
- Banks P. B.; Hughes N. K. A review of the evidence for potential impacts of black rats (Rattus rattus) on wildlife and humans in Australia. Wildl. Res. 2012, 39, 78–88. 10.1071/WR11086. [DOI] [Google Scholar]
- Ellis B. A.; Regnery R. L.; Beati L.; Bacellar F.; Rood M.; Glass G. G.; Marston E.; Ksiazek T. G.; Jones D.; Childs J. E. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease?. J Infect Dis 1999, 180, 220–224. 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- Umali D. V.; Lapuz R. R. S. P.; Suzuki T.; Shirota K.; Katoh H. Transmission and shedding patterns of Salmonella in naturally infected captive wild roof rats (Rattus rattus) from a Salmonella-contaminated layer farm. Avian Dis 2012, 56, 288–294. 10.1637/9911-090411-Reg.1. [DOI] [PubMed] [Google Scholar]
- Horng Y. B.; Yu Y. H.; Dybus A.; Hsiao F. S. H.; Cheng Y. H. Antibacterial activity of Bacillus species-derived surfactin on Brachyspira hyodysenteriae and Clostridium perfringens. AMB Express 2019, 9, 188. 10.1186/s13568-019-0914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Wang L.; Su C. X.; Gong G. H.; Wang P.; Yu Z. L. Isolation and characterization of lipopeptide antibiotics produced by Bacillus subtilis. Lett. Appl. Microbiol. 2008, 47, 180–186. 10.1111/j.1472-765X.2008.02412.x. [DOI] [PubMed] [Google Scholar]
- Gong A. D.; Li H. P.; Yuan Q. S.; Song X. S.; Yao W.; He W. J.; Zhang J. B.; Liao Y. C. Antagonistic mechanism of Iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One 2015, 10, e0116871 10.1371/journal.pone.0116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk T. U.; Manaia C. M.; Merlin C.; Fatta-Kassinos D.; Cytryn E.; Walsh F.; Bürgmann H.; Sørum H.; Norström M.; Pons M. N.; Kreuzinger N.; Huovinen P.; Stefani S.; Schwartz T.; Kisand V.; Baquero F.; Martinez J. L. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 2015, 13, 310. 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- Santajit S.; Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res. Int. 2016, 1–8. 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gislin D.; Sudarsanam D.; Raj G. A.; Baskar K. Antibacterial activity of soil bacteria isolated from Kochi, India and their molecular identification. J Genet Eng Biotechnol. 2018, 16, 287–294. 10.1016/j.jgeb.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Ni X.; Wang Q.; Peng Z.; Niu L.; Xie M.; Lin Y.; Zhou Y.; Sun H.; Pan K.; Jing B.; Zeng D. Investigation of lactic acid bacteria isolated from giant panda feces for potential probiotics in vitro. Probiotics Antimicrob. Proteins 2018, 85. 10.1007/s12602-017-9381-8. [DOI] [PubMed] [Google Scholar]
- Hatha A.; Shubhankar Ghosh A.; Selvam D.; Neethu C.; Saramma A. Diversity and antimicrobial activity of lactic acid bacteria from the gut of marine fish Rastrelliger kanagurta against fish, shrimp and human pathogens. J. Mar. Biol. Ass. India 2014, 55, 22–27. 10.6024/jmbai.2013.55.2.01778-04. [DOI] [Google Scholar]
- Kuo J.; Yang Y. T.; Lu M. C.; Wong T. Y.; Sung P. J.; Huang Y. S. Antimicrobial activity and diversity of bacteria associated with Taiwanese marine sponge Theonella swinhoei. Ann. Microbiol. 2019, 69, 253–265. 10.1007/s13213-018-1414-3. [DOI] [Google Scholar]
- Chevrette M. G.; Carlson C. M.; Ortega H. E.; Thomas C.; Ananiev G. E.; Barns K. J.; Book A. J.; Cagnazzo J.; Carlos C.; Flanigan W.; Grubbs K. J.; Horn H. A.; Hoffmann F. M.; Klassen J. L.; Knack J. J.; Lewin G. R.; McDonald B. R.; Muller L.; Melo W. G. P.; Pinto-Tomás A. A.; Schmitz A.; Wendt-Pienkowski E.; Wildman S.; Zhao M.; Zhang F.; Bugni T. S.; Andes D. R.; Pupo M. T.; Currie C. R. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran-Gracia E.; Macedo-Raygoza G.; Villafaña-Rojas J.; Martinez-Rodriguez A.; Chavez-Castrillon Y. Y.; Espinosa-Escalante F. M.; Di Mascio P.; Ogura T.; Beltran-Garcia M. J. Production of lipopeptides by fermentation processes: Endophytic bacteria, fermentation strategies and easy methods for bacterial selection. Ferment Pro 2017, 199–222. 10.5772/64236. [DOI] [Google Scholar]
- Raaijmakers J. M.; De Bruijn I.; Nybroe O.; Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- Jha S. S.; Joshi S. J.; Geetha S. J. Lipopeptide production by Bacillus subtilis R1 and its possible applications. Braz. J. Microbiol 2016, 47, 955–964. 10.1016/j.bjm.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A. Peptide-based antibiotics: A potential answer to raging antimicrobial resistance. Drug Dev. Res. 2000, 50, 440–447. 10.1002/1098-2299(200007/08)50. [DOI] [Google Scholar]
- Vazquez L.; Teixeira Da Silva Ferreira A.; Cavalcante F. S.; Garcia I. J. P.; Dos Santos K. R. N.; Barbosa L. A. D. O.; Almeida M. D. S.; Mignaco J. A.; Fontes C. F. L. Properties of novel surfactin-derived biosurfactants obtained through solid-phase synthesis. J. Pept. Sci. 2018, 24, e3129. 10.1002/psc.3129. [DOI] [PubMed] [Google Scholar]
- Maget-Dana R.; Ptak M.; Peypoux F.; Michel G. Pore-forming properties of iturin A, a lipopeptide antibiotic. Biochim. Biophys. Acta, Biomembr. 1985, 815, 405–409. 10.1016/0005-2736(85)90367-0. [DOI] [PubMed] [Google Scholar]
- Inès M.; Dhouha G. Lipopeptide surfactants: production, recovery and pore forming capacity. Peptides 2015, 71, 100–112. 10.1016/j.peptides.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Kim P. I.; Ryu J.; Kim Y. H.; Chi Y. T. Production of biosurfactant lipopeptides Iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 2010, 20, 138–145. 10.4014/jmb.0905.05007. [DOI] [PubMed] [Google Scholar]
- Abdelli F.; Jardak M.; Elloumi J.; Stien D.; Cherif S.; Mnif S.; Aifa S. Antibacterial, anti-adherent and cytotoxic activities of surfactin (s) from a lipolytic strain Bacillus safensis F4. Biodegradation 2019, 30, 287. 10.1007/s10532-018-09865-4. [DOI] [PubMed] [Google Scholar]
- Huang E.; Guo Y.; Yousef A. E. Draft genome sequence of Paenibacillus sp. strain OSY-SE, a bacterium producing the novel broad-spectrum lipopeptide antibiotic paenibacterin. J. Bacteriol. 2012, 194, 6306–6306. 10.1128/JB.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak K. V.; Keharia H.; Gupta K.; Thakur S. S.; Balaram P. Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J. Am. Soc. Mass Spectrom. 2012, 23, 1716–1728. 10.1007/s13361-012-0437-4. [DOI] [PubMed] [Google Scholar]
- Sabaté D. C.; Audisio M. C. Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against Listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol. Res. 2013, 168, 125–129. 10.1016/j.micres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Siddiqui R.; Jeyamogan S.; Ali S. M.; Abbas F.; Sagathevan K. A.; Khan N. A. Crocodiles and alligators: Antiamoebic and antitumor compounds of crocodiles. Exp. Parasitol. 2017, 183, 194–200. 10.1016/j.exppara.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Ferris R. A.; Palmer B. A.; Borlee B. R.; Mccue P. M. Ability of chromogenic agar, MALDI-TOF, API 20E and 20 strep strips, and BBL crystal enteric and gram-positive identification kits to precisely identify common equine uterine pathogens. J. Equine Vet. Sci. 2017, 57, 35–40. 10.1016/j.jevs.2017.06.009. [DOI] [Google Scholar]
- Anwar A.; Masri A.; Rao K.; Rajendran K.; Khan N. A.; Shah M. R.; Siddiqui R. Antimicrobial activities of green synthesized gums-stabilized nanoparticles loaded with flavonoids. Sci. Rep. 2019, 9, 3122. 10.1038/s41598-019-39528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar N.; Khan N. A.; Sagathevan K.; Iqbal M.; Tawab A.; Siddiqui R. Gut bacteria of Cuora amboinensis (turtle) produce broad-spectrum antibacterial molecules. Sci. Rep. 2019, 9, 17012. 10.1038/s41598-019-52738-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar N.; Siddiqui R.; Iqbal M.; Khan N. A. Antibacterial activities of selected pure compounds isolated from gut bacteria of animals living in polluted environments. Antibiotics 2020, 9, 190. 10.3390/antibiotics9040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri A.; Anwar A.; Ahmed D.; Siddiqui R.; Raza Shah M.; Khan N. Silver Nanoparticle Conjugation-Enhanced Antibacterial Efficacy of Clinically Approved Drugs Cephradine and Vildagliptin. Antibiotics 2018, 7, 100. 10.3390/antibiotics7040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J.; Siddiqui R.; Khan N. Acanthamoeba and bacteria produce antimicrobials to target their counterpart. Parasit Vectors 2014, 7, 56. 10.1186/1756-3305-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glady A.; Tanaka M.; Moniaga C. S.; Yasui M.; Hara-Chikuma M. Involvement of NADPH oxidase 1 in UVB-induced cell signaling and cytotoxicity in human keratinocytes. Biochem. Biophys. Rep. 2018, 14, 7–15. 10.1016/j.bbrep.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas K.; Shang L. Silver Nanoparticle-Mediated Cellular Responses in Human Keratinocyte Cell Line HaCaT in Vitro. Nanoscale Rep. 2019, 2, 1–9. 10.26524/nr1921. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.