Abstract
Transcription factors bind specifically to their target elements in the genome, eliciting specific gene expression programs. The nuclear factor-κB (NF-κB) system is a family of proteins comprising inducible transcription activators, which play a critical role in inflammation and cancer. The NF-κB members function as dimers with each monomeric unit binding the κB-DNA. Despite the available structures of the various NF-κB dimers in complex with the DNA, the structural features of these dimers in the nucleic acid-free form are not well-characterized. Using solution NMR spectroscopy, we characterize the structural features of 73.1 kDa p50 subunit of the NF-κB homodimer in the DNA-free form and compare it with the κB DNA-bound form of the protein. The study further reveals that in the nucleic acid-free form, the two constituent domains of p50, the N-terminal and the dimerization domains, are structurally independent of each other. However, in a complex with the κB DNA, both the domains of p50 act as a single unit. The study also provides insights into the mechanism of κB DNA recognition by the p50 subunit of NF-κB.
Introduction
Nuclear Factor κB (NF-κB) is a family of inducible transcription factor proteins ubiquitously expressed in the vertebrates.1,2 While the NF-κB system is mainly known for its immune response to the pathogenic signals of the environment, its role is also vital in other cellular processes including cell-growth, cell-adhesion, and cell-proliferation.3−5 The family recognizes a 9–11 base pair cognate DNA known as the κB site on the promoter/enhancer region of its target genes to activate transcription.6 Since the transcription of the NF-κB target genes are tightly regulated, in the resting state of the cell NF-κB is sequestered in the cytoplasm by the inhibitor of κB (IκB) proteins. The detection of the external stimulus leads to the proteasomal degradation of the IκB protein, thereby releasing the NF-κB dimer into the nucleus, where it activates the transcription of its target genes.
This family comprises of five members, namely, RelA (aka p65), RelB, c-rel (aka rel), p50, and p52, all of which contain a ∼300 residue long well-conserved region known as the Rel Homology region (RHR). All the NF-κB members function as dimers and can form dimers in various combinations within the family. The RHR can be divided into two domains, namely, the N-terminal DNA binding domain (NTD) and the dimerization domain (DD). The NTD is primarily responsible for the κB-DNA recognition and the DD for the NF-κB dimerization (Figure 1). The five NF-κB subunits are categorized into two classes, class I and class II, based on the presence and the absence of the transactivation domain (TAD), respectively. p50 and p52 NF-κB subunits belong to the class II category as they do not possess the TAD. The lack of TAD renders p50 and p52 homodimers as transcription repressors which can occupy the NF-κB cognate sites on the promoter/enhancers of their target genes but cannot activate their transcription.7−10
Figure 1.
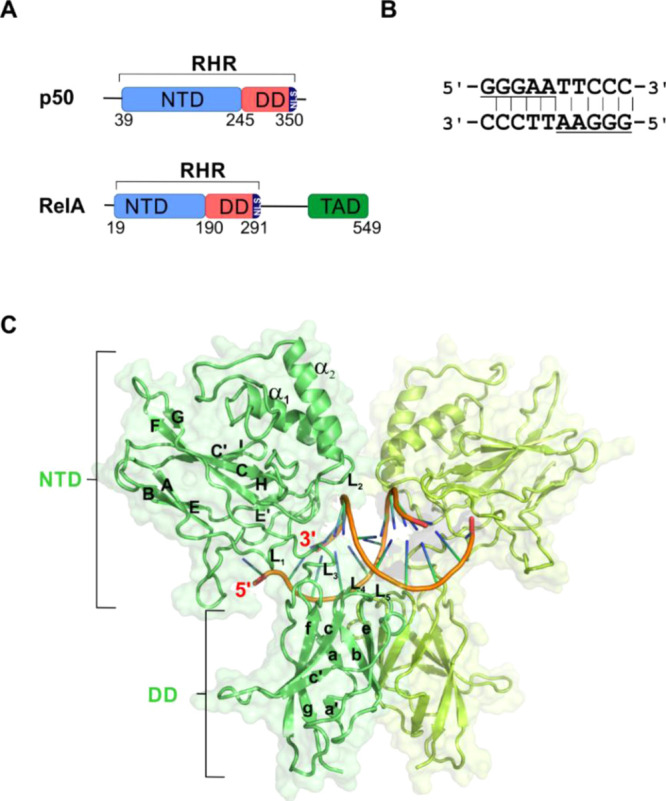
Domain organization of NF-κB subunits. (A) Schematic of the domain organization of p50 and RelA NF-κB subunit. (B) Palindromic full κB DNA sequence. The underlined parts represent each of the two half-κB sites in the sequence. (C) Cartoon depiction of X-ray crystal structure of p50–p50 homodimer bound to κB DNA (orange and blue) (PDB id: 1NFK). Each monomeric unit of p50 is depicted in two different shades of green. The secondary structures are labeled according to the three-dimensional (3D) crystal structure with PDB id: 1NFK (see Figure S1).
The 9–11 base pair κB site with a broad consensus sequence of 5′-GGGNNNNNCC-3′6 (where N = A, T, G, or C) interacts with the different NF-κB dimer forms with variable affinities. The κB sequence consists of two half-κB sites with each half site (5′-GGGNN-3′) binding to one monomeric unit of the NF-κB dimer (Figure 1B). This differential binding affinities of the slightly different κB sites lets a specific NF-κB dimer activate the transcription of a specific set of target genes under a given set of physiological conditions11 which may vary temporally post-stimulation.12 It remains unclear as to how the NTDs of NF-κB discriminate between the closely related κB sites. A number of X-ray crystal structures of NF-κB dimers with various κB sites are available;11,13−17 however, little information about the transcription factor in the nucleic acid-free form is known. Though a report on the NMR characterization of the free-form of RelA-NTD is available, the study focused on the binding of the RelA subunit of NF-κB to its inhibitor protein IκBα.18,19 It is intriguing how the NTD accommodates different κB sequences and whether this differential binding has any implication in the downstream process of the NF-κB signaling pathway. As a first step to understand the mechanism of κB recognition, it is imperative to decipher the structural features of the NF-κB dimers in their DNA-free form.
In this study, we characterized the NF-κB NTD of the p50 subunit in solution in both the DNA-free and κB-bound forms, using NMR spectroscopy. This allowed us to gain residue-level information of the structural elements to further show that each of the two NTDs of the p50 homodimer binds specifically to one-half-κB part of the κB sequence without any influence from the neighboring half of the κB site. We further characterized the full-length p50–p50 homodimer (73 kDa) using NMR spectroscopy in both the DNA-free and the - bound forms.
Materials and Methods
Protein Expression and Purification
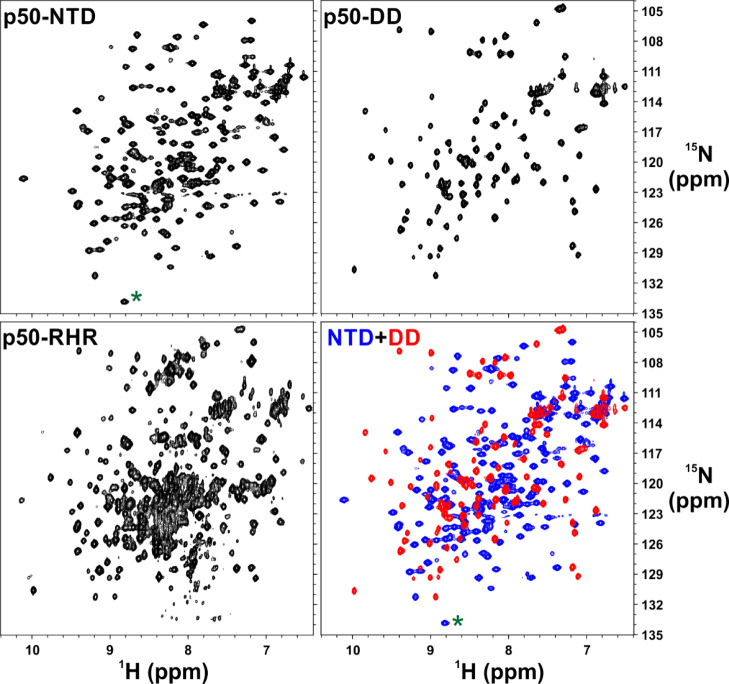
Untagged mouse p50-NTD (39–245; 22.7 kDa) was generated by introducing a stop codon by site-directed mutagenesis in the p50 (39–363) plasmid. p50 (39–363) and p50-DD (245–350) were prepared as described previously.18,20 p50-RHR (39–363) used throughout the study was perdeuterated and isotopically labeled with 15N and/or 13C as described earlier.1915N-Ammonium chloride and 13C-d-glucose were purchased from either Spectra Stable Isotopes or Cambridge Isotope Laboratories. Other required chemicals were from NEB, SRL, and SD Fine-Chemicals unless specifically mentioned. The purification procedure for p50-NTD was similar to that of RelA-NTD mentioned previously.18 All the plasmids used in this study were expressed in Escherichia coli [BL21(DE3)]. For NMR experiments, the NTD (39–245) and the RHR (39–363) fragments were partially/perdeuterated by growing BL21(DE3) cells expressing the fragments in the minimal M9 media made in >98% D2O in the presence of protonated/uniformly-deuterated glucose. The proteins were 15N- and/or 13C-labeled by supplementing the M9 media with 15N-ammonium chloride and 13C-glucose as the sole source of nitrogen and carbon, respectively. For producing p50-NTD specifically unlabeled/labeled amino-acid samples, the above isotopically labeled/unlabeled media were further supplemented with specific unlabeled/labeled amino acids (lysine, arginine, isoleucine, leucine, and/or histidine) to the final concentration of 0.2 g/L.21 The protein overexpression was achieved as described previously.19 For purification of p50-NTD and p50-RHR, the cells were lysed by sonication in a lysis buffer [20 mM Tris (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 0.5 mM ethylenediaminetetraacetic acid, 0.5 mM phenylmethylsulfonyl fluoride, and 10% glycerol] and then centrifuged to remove the insoluble cell debris. The p50-NTD and p50-RHR remained in the supernatant, which was loaded on a tandem Q-Sepharose followed by SP-Sepharose column. The SP-Sepharose column was detached, washed with the lysis buffer, and eluted with batch elution of 150 and 250 mM NaCl concentrations for p50-NTD and p50-RHR, respectively. The elution aliquots containing p50-NTD/p50-RHR, as detected from 12% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE), were pooled, concentrated, and buffer-exchanged with NMR buffer [20 mM Tris (pH 6.8), 50 mM NaCl, and 20 mM β-ME] through repeated washing using 10 kDa cut-off centrifugal filters (Merck Millipore). For purifying the deuterated p50-RHR, an additional step of washing the protein-bound SP-Sepharose column with 1 M urea in the lysis buffer followed by washing with wash buffer without urea was performed before the elution step. p50-DD was purified exactly as mentioned previously.22 In brief, for the purification of the p50-DD, the cells were lysed and then centrifuged as mentioned above. The supernatant containing the p50-DD was loaded onto a Q-Sepharose column (GE Healthcare Life Sciences), washed with lysis buffer, and eluted with batch elution of 200 mM of NaCl concentration. The elution aliquots containing the p50-DD, as detected from 15% SDS–PAGE, were pooled, concentrated, and loaded onto a homemade column filled with Superdex-75 beads (GE-Healthcare) equilibrated with NMR buffer for further purification by size-exclusion chromatography. The purity of the above proteins was checked on SDS–PAGE and [15N, 1H]-HSQC/TROSY spectra (Figures 2 and S2).
Figure 2.
[15N, 1H]-correlation spectra of the isolated domains of p50 NF-κB subunit represent well-folded protein domains that correlate well with that of the full-length p50 homodimer. [15N, 1H]-HSQC spectra of [15N, 2H]-labeled p50-NTD (top left panel) and 15N-labled p50-DD (top right panel). [15N, 1H]-TROSY spectrum of [15N, 2H]-labeled p50 homodimer (bottom left panel) and superimposed [15N–1H] HSQC spectra of p50-NTD (blue) and p50-DD (red). The folded cross-peak is marked with a green asterisk (*). The p50-RHR (39–363) fragment used here has 13 extra residues at the C-terminal end than the p50-DD (245–350) which accounts for the extra peaks for [15N, 1H]-TROSY p50-RHR.
Formation of DNA Duplex
Three commercially purchased 14-mer, 18-mer, and 21-mer κB DNA with sequences 5′-TCGGGAATTCCCGA-3′, 5′-TTCGGGAAATAGAAACTC-3′, and 5′-CTTCGGGAAATAGAAACTCTC-3′, respectively, were purified and annealed as mentioned previously.19 In brief, the oligomers were dissolved in water and loaded onto a priorly equilibrated Q-Sepharose column with 0.1 M NaOH and 0.1 M NaCl. The column was washed with the column equilibration buffer and eluted with a NaCl gradient with a final concentration of 1 M NaCl. The elution was monitored with absorbance at 260 nm and the single-stranded DNA fractions were pooled together and the pH was neutralized with 1 M 2-(N-morpholino)ethanesulfonic acid buffer (pH 5.5). The purified DNA fractions were concentrated and the buffer was exchanged with annealing buffer [5 mM Tris (pH 8.0), 100 mM NaCl]. The 14-mer DNA having a palindromic sequence was self-annealed, whereas for the 18-mer and the 21-mer DNA, the two complimentary strands were mixed in 1:1 stoichiometric ratio followed by annealing by incubating the DNA at 95 °C for 5 min followed by gradual cooling to room temperature to obtain the duplex DNA. Efficient annealing of the oligomers was ensured by checking the ssDNA and the annealed DNA on native PAGE using SyBr Gold (Invitrogen) and ethidium bromide (Bio Basic Canada Inc.) nucleic acid stains.
NMR Spectroscopy
All spectra were acquired at 298 K on Bruker Avance/Ascend NMR spectrometers operating at a 1H resonance frequency of 800 MHz and equipped with a triple resonance cryogenic probe and triple-axis gradients. Partially deuterated uniformly 13C- and 15N-labeled p50-NTD (0.7–0.9 mM) and perdeuterated uniformly 13C- and 15N-labeled p50-RHR (0.28 mM) protein samples were used for triple resonance experiments for backbone resonance assignments. The spectra were processed using NMRPipe23 and analyzed using CARA.24 Backbone resonance assignments for p50-NTD was achieved using multidimensional NMR experiments—HNCA, HNCACB, HNCO, HN(CO)CA, CBCA(CO)NH, and [15N, 1H]-NOESY-HSQC in addition to the specific amino-acid labeling/unlabeling techniques.21,25 The secondary structures of p50-NTD and RelA-NTD were predicted using TALOS-N26 (Figure S3). The RelA-NTD backbone resonance assignments were obtained from the BMRB (BMRB #26647). The backbone resonance assignments of p50-NTD in different DNA-bound complexes were achieved using the standard triple resonance experiments as used for p50-NTD mentioned above. The complete assignments for the p50-DD were recently published by our group.22 The backbone resonance assignments of the p50 homodimer were obtained by the transfer of assignments from the p50-NTD and p50-DD data. These assignments were rechecked by comparing the CA resonances, especially in the overlapping regions of the spectrum, using 3D TROSY-HNCA27 acquired on the [15N, 13C, 2H]-p50 homodimer sample. For obtaining the DNA-bound complexes, the excess DNA was maintained in the sample which was ensured by observing the imino peaks of the free DNA in the 1D 1H spectra in the presence of the protein (Figure S4).
For native hydrogen exchange coupled with NMR (nHX-NMR) experiments, p50-NTD protein (0.7 mM) at pH 6.8 was lyophilized and then dissolved in 100% D2O. This was followed by recording a series of [15N, 1H]-HSQC experiments. The intensities of the cross-peaks in [15N, 1H]-HSQC spectra were plotted with respect to time and fitted to the single exponential decay function to determine the amide protein exchange rate with D2O.
For NMR relaxation parameter measurements, 0.45 mM of partially deuterated 15N-(p50-NTD) concentration was used. 15N R1 and 15N R2 were measured at 298 K using the previously described pulse sequences.28 For R1 measurements, 9 spectra were collected with relaxation delays of 0.2, 0.3, 0.4, 0.5*, 0.6, 0.7, and 0.8* s (the asterisks indicate duplicate measurements). For R2 measurements, 12 spectra were recorded with relaxation delays of 5, 10*, 15, 20, 24, 29*, 39, 49, 59, and 68 ms. The [1H]-15N nuclear Overhauser effect (NOE)29 was measured at 298 K in triplicate in a fully interleaved manner with proton saturation for 4 s and a recycle delay of 6 s. A recycle delay of 10 s was used for the [1H]-15N NOE reference experiments. The relaxation parameters were determined by using relax software.30,31 The global tumbling time (τc) was calculated from the average R2/R1 ratio using the following equation32
| 1 |
where νN is the nitrogen frequency is Hertz.
The average R2/R1 ratio was determined by averaging the R2/R1 ratios for the amide resonances with [1H]-15N NOE values greater than 0.65.
Results
Resonance Assignments of p50-NTD
For obtaining the residue specific backbone resonance assignments of the full length 73.1 kDa p50 homodimer in the DNA-free form, we explored the feasibility of the transfer of assignments from the resonances of the individual domains: p50-NTD and p50-DD. The backbone amide-proton cross-peak resonances of the individual domains, namely, p50-NTD and p50-DD could be readily superimposed on those of the p50 homodimer suggesting the feasibility of the transfer of resonance assignment strategy (Figure 2).
Using the conventional 3D triple resonance experiments, specific labeling and unlabeling amino-acid strategy (Figure S2), we could confirm the amide resonance assignments of p50-NTD in the two-dimensional (2D) [15N, 1H]-HSQC spectrum. We used multiple samples for p50-NTD with differentially 15N-labeled/unlabeled amino-acids, as shown in Table S1. We also performed 2D HN(CO) on the uniformly 13C- and 15N-labeled and selective amino-acid unlabeled samples of p50-NTD (see Table S1) for additional amide resonance assignments.25 Using this strategy, we could obtain amide resonance assignment for p50-NTD to the extent of 84% (Figure 3C). The remaining unassigned residues majorly belonged to the flexible loop regions of the p50-NTD.
Figure 3.
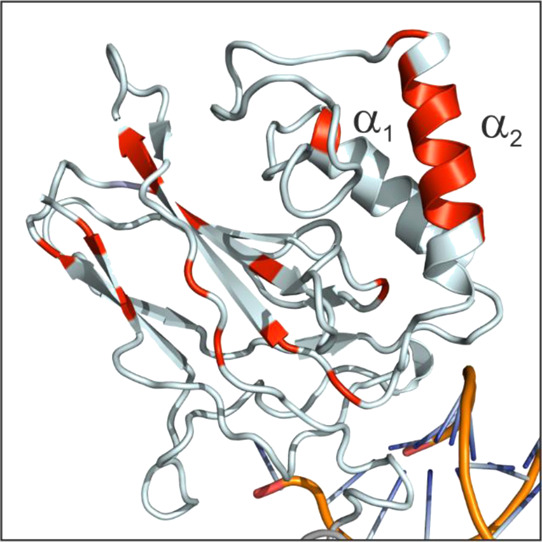
α2 helix is the most stable secondary structure within the p50-NTD. The p50 residues with ΔGHX greater than 5.5 kcal mol-1 is mapped onto the p50 structure (NTD from PDB id: 1NFK)11 and colored red. The two helices are labeled on the structure.
The secondary structure for the DNA-free form of p50-NTD predicted using the TALOS-N software26 closely matched with that of the p50-NTD domain present in the 3D structure of the DNA-bound p50 homodimer11 (Figure S3) except for certain regions. According to the chemical shift-based secondary structure prediction, in the DNA-free p50-NTD, a short helix is predicted at 172–175 which attains a coil structure in the DNA-bound form derived from crystallography. This helix is formed between the α1 and the α2 helices in the region unique to the p50 subunit in the entire NF-κB family. Another helical stretch is predicted at 70–72 that corresponds to the coil in the crystal structure. Apart from the helices, the TALOS-N26 predicted an additional short β-strand at 232–234 which assumes a coiled structure in the available crystal structure. Also, the TALOS-N shows an extended B β-strand when compared with the crystal structure.
α2 Helix is the Most Stable Secondary Structure in p50-NTD
We performed nHX-NMR experiments (see the Materials and Methods section) to study the stability of the H-bonds involved in the formation of the secondary structures of p50-NTD. We observed the complete disappearance of majority of the amide cross-peaks within an hour of the solvent exchange reaction. Among the remaining peaks in the HSQC spectra during the HX reaction, some of the cross-peaks (C85, K92, Q96, K191, F217, L218, V232, and S234) were broad but lasted for a long duration in the reaction. This is in contrast with that observed for the p50-DD,33 where the majority of the amide cross-peaks appeared in the HSQC spectra for more than 24 h under similar experimental conditions. The HX experiments indicate that the H-bonds in the secondary structures of the p50-NTD are weak when compared with that of the p50-DD (Figure 3). Among the secondary structures in the p50-NTD, the α2 helix that is unique to the p50 and p52 subunits of the NF-κB family showed the highest resistance for HX. This suggest that within the NTD, the α2 helix is comparatively the most stable secondary structure with strong H-bonds. Apart from the contiguous residues of the α2 helix, other residues with a relatively high resistance to HX are spread throughout the β-strands.
Backbone Dynamics of the p50-NTD
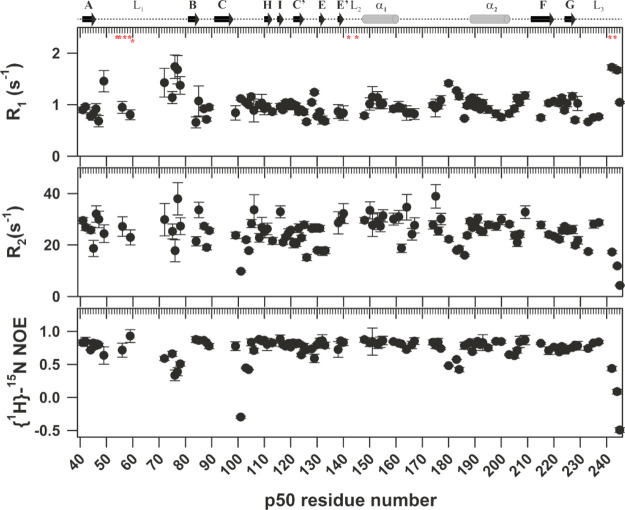
15N-backbone dynamics of the p50-NTD fragment in the DNA-free form was studied using 15N spin relaxation experiments at a static magnetic field strength of 18.8 T. Figure 4 shows the 15N R1, 15N R2, and [1H]-15N heteronuclear NOE spin relaxation constants for the p50-NTD in the DNA-free form. The C-terminal part of the L1 loop in the p50-NTD shows elevated R1 and reduced heteronuclear NOE values suggesting flexibility in this region on the picosecond to nanosecond timescale. However, the R2 values of this region were not used to derive any conclusion because of their high error values. The residues 180–184 belonging to the linker between the α1 and the α2 helices also show elevated R1 values and reduced R2 and heteronuclear NOE values suggesting this region as being more dynamic on the picosecond-to-nanosecond timescale. The α2 helix, consistent with the relatively strong H-bonds as concluded from the nHX experiments, showed reduced R1 and elevated R2 and heteronuclear NOE values suggesting that this helix is less dynamic on the picosecond-to-nanosecond timescale. The global tumbling time (τc) as calculated from R2/R1 ratio of p50-NTD is estimated to be 12.6 ns which corresponds to a molecular weight of 20.8 kDa.32
Figure 4.
15N spin relaxation data for p50-NTD. 15N R1, 15N R2, and [1H]-15N NOE values plotted against the p50 residue numbers. The secondary structural elements of p50 with their labels are shown above the graph for reference. The DNA contacting residues are marked with a red asterisk (*) below the cartoon in the top panel.
DNA-Binding Domain Does Not Discriminate between Half- and Full-κB Sequence
A κB DNA sequence (full-κB site) is comprised of two half-κB parts (half-κB site) (Figure 1B).6 An NF-κB dimer contacts the κB DNA through its loops flanked by its secondary structural elements. Five such loops from each monomer unit contact the κB DNA with three loops from the NTD (L1, L2, and L3) and two from the DD (L4 and L5) (Figures 1C and S1). The κB DNA specificity is attributed to the NTD, also known as the “specificity domain”34 as it contains the four residues (R54, R56, E60, and K241) that make base specific contacts and are indispensable for the κB recognition. The other residues of the DNA-contacting loops interact with the κB DNA mainly through the backbone phosphates; however, these contacts can accommodate subtle changes in the κB sequences by adopting different modes of interaction.11,14−17,34−36 Overall, when a p50 homodimer binds to a complete κB sequence, each of the p50 monomer unit contacts the bases of a half-κB site and interacts with the other half-κB part through the backbone phosphates of the DNA. Thus, the NF-κB dimer complex with a κB DNA assumes a butterfly-like structure with the DDs acting as the body and the two NTDs as the wings grabbing the two half-κB sites of the DNA (Figure 1C).
With the goal to characterize the interaction of the p50-NTD with the κB DNA, we designed a half-κB DNA sequence (5′-TTCGGGAAATAGAAACTC-3′). This was done also to maintain a 1:1 stoichiometric complex of the p50-NTD with its cognate half-κB sites. A full-κB site (5′-TCGGGAATTCCCGA-3′) was also used for comparison with the half-κB DNA to determine any difference that may be observed in the 2D [15N, 1H]-TROSY spectra of p50-NTD upon binding to the two DNA sequences. This also would determine any effect of the second half-κB site (3′ half-κB site) on the interaction of the p50-NTD with the first half-κB (5′ half-κB) sequence. The backbone resonance assignments for all the κB-DNA-bound forms of the p50-NTD were obtained using standard triple resonance experiments.
The 2D [15N, 1H]-TROSY spectrum of the p50-NTD in complex with a full-κB DNA sequence [(p50-NTD)-(full-κB)] (30.8 kDa) yielded well-dispersed cross-peaks (Figure S5), the majority of which showed significant CSPs with respect to its DNA-free form [(p50-NTD)-free] (Figure 5). While the residues that directly interact with the DNA (R54, F55, V57, C59, E60, T142, K144, K145, K241, and P243) could not be assigned in the bound form, the residues adjacent to the DNA-contacting residues in the three-dimensional structure showed significant chemical shift perturbations (CSPs) [Figure 5A (top panel)]. Residues L151, L106, and N172 that are considerably away from the DNA binding site also demonstrated large CSPs upon κB-DNA interaction. Though the p50-NTD physically interacts with κB-DNA through the L1, L2, and L3 loops, the interaction is also sensed by the H, E′, and F β-strands (Figure 5). This shows that the κB interaction with p50-NTD is relayed to the remote areas of the domain through the hydrogen bond network of the secondary structural elements. Notably, the linker connecting the α1 and α2 helices did not undergo any significant CSP upon κB binding.
Figure 5.
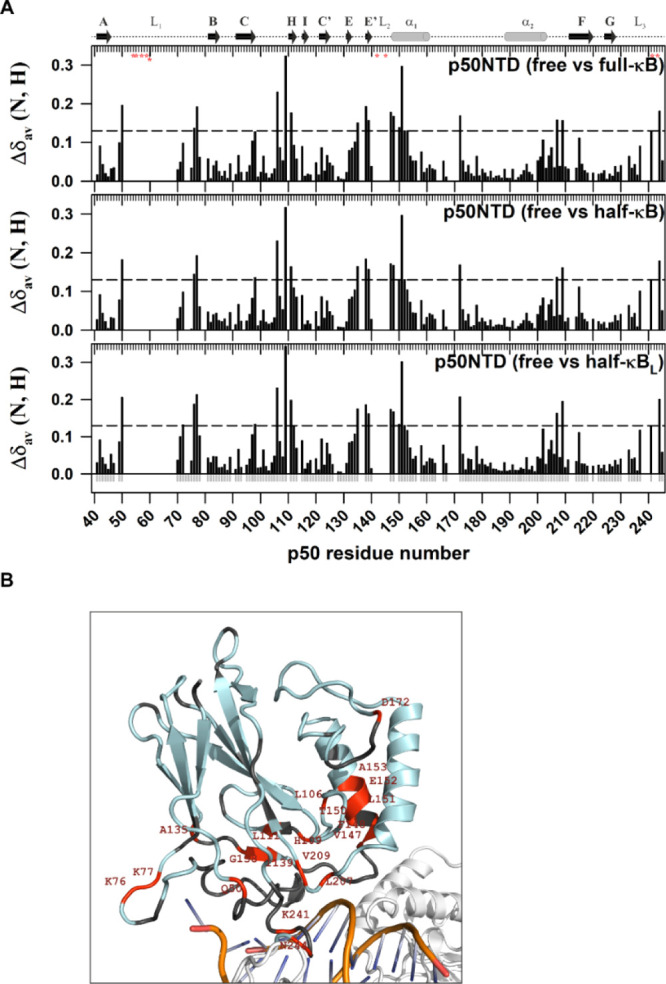
Chemical shift differences between κB DNA-free and -bound forms of the p50-NTD extend beyond the DNA contacting residues. (A) Plot of CSP [Δδav (N, H) = √[(Δδ1H)2 + (Δδ15N/5)2] observed for the p50-NTD in κB DNA-free and -bound forms as a function of the p50 residue number. The secondary structure with the α-helix is shown as a cylinder and β-strand as an arrow corresponding to the p50-NTD sequence is depicted on the top of the plot. The DNA-contacting residues in the protein are shown as red asterisks below the top ruler. The dashed horizontal line in each panel is the reference which is three standard deviations above the average CSP, excluding the outliers. CSPs above the reference in each panel are considered significant. (B) Significant CSPs mapped onto the 3D structure of p50-NTD (PDB id: 1NFK) are depicted in red. The p50-NTD is colored in light-blue; DNA in golden-yellow with bases shown in blue; and the other parts of the p50-RHR structure are shown in white.
No significant differences were observed in the backbone amide chemical shifts of the p50-NTD between the (p50-NTD)-full-κB and the (p50-NTD)-half-κB complexes, thus implying that the κB recognitions in the full-κB and the half-κB sequences are identical [Figure 5A (top and middle panel) and Figure 6 (top panel)]. The half- and the full-κB DNA binding to the p50-NTD show near identical changes in the intensities (Figure 6) and the CSPs suggesting that the p50-NTD conformation is largely indistinguishable when bound to the full- or half-κB DNA sequences. This is because the p50-NTD can bind to the half-κB site through its three DNA binding loops and satisfy all the essential direct base contacts with the DNA.
Figure 6.
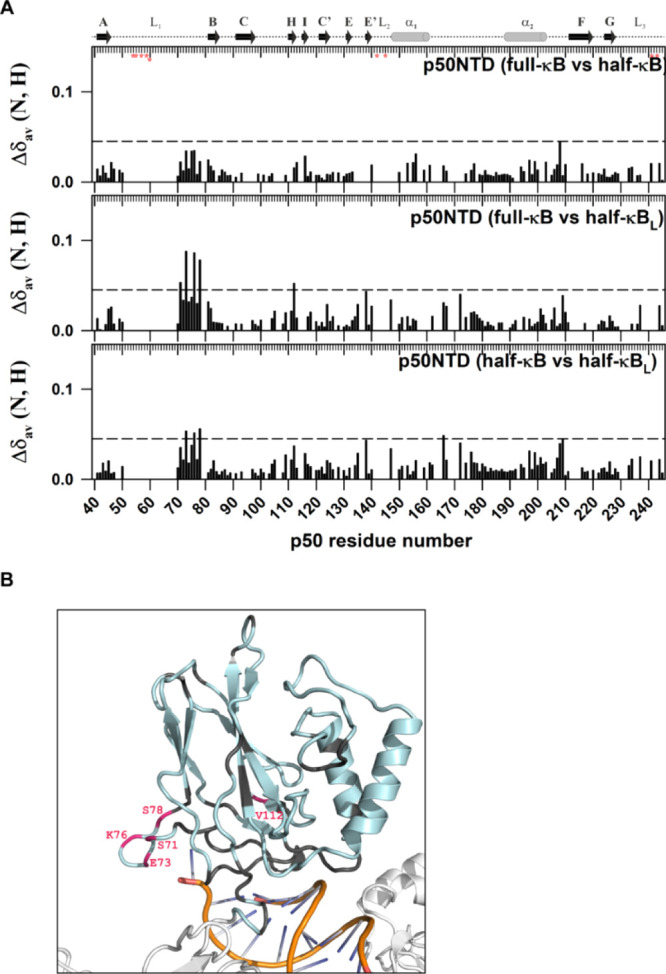
C-terminal part of the L1 DNA-binding loop interacts with the nucleotides beyond the κB sequence. (A) Plot of CSP observed for the p50-NTD in complex with the full-κB with respect to that with the half-κB (top panel); in complex with the full-κB with respect to that with the half-κBL (middle panel); and in complex with the half-κB with respect to that with the half-κBL (lower panel) DNA-bound forms as a function of the p50 residue number. The other details of the figure are the same as mentioned in Figure 5A. (B) Significant CSPs mapped onto the 3D structure of the p50-NTD (PDB id 1NFK) depicted in pink. The p50-NTD is colored in light-blue; DNA in golden-yellow with bases shown in blue; and the other parts of the p50-RHR structure are shown in white.
C-Terminal Part of the L1 Loop of p50-NTD Interacts with Nucleotides Beyond the κB Site
To delineate the effect of the extra base pairs flanking on the 5′ end of the half-κB sequence, two oligomers of different lengths of 18 and 21 bp (5′-CTTCGGGAAATAGAAACTCTC-3′) with the same half-κB sequence in addition to the 14 bp full-κB DNA were used. Based on the (p50–p50)-κB DNA complex structures,11,14 the extra nucleotides in the 5′ end of the κB site are poised to interact with the p50-NTD (Figure 6B). The comparison of the CSPs of the half-κB and the half-κB with extra terminal nucleotides (half-κBL) with respect to the free-p50-NTD showed clear differences only in the C-terminal part of the loop L1 residues of the p50-NTD [Figure 6 (middle and bottom panel)]. The difference was amplified more for the half-κBL sequence compared to the half-κB site by the introduction of an extra bp at the 5′ end of the DNA. This observation implies that the C-terminal part of the L1 loop which undergoes intermediate conformational exchange in the p50-NTD-free form can contact the DNA backbone of nucleotides flanking the κB sequence (Figure 6B).
A comparative analysis of the normalized intensities of the p50-NTD amide resonances in its DNA-free and the DNA-bound forms show nonuniform cross-peak intensities for the (p50-NTD)-κB complexes when compared with the (p50-NTD)-free form (Figure 7). In contrast to the major parts of the NTD, the amide resonances in the C-terminal part of the L1 loop (residues A70-S78) showed enhanced intensities in the complex with κB-DNA, suggesting a decrease in its structural heterogeneity upon binding to the κB DNA. On the other hand, the linker between the two helices, α1 and α2, retains its fast dynamics in the picosecond to nanosecond timescale in both the forms of the NTD.
Figure 7.
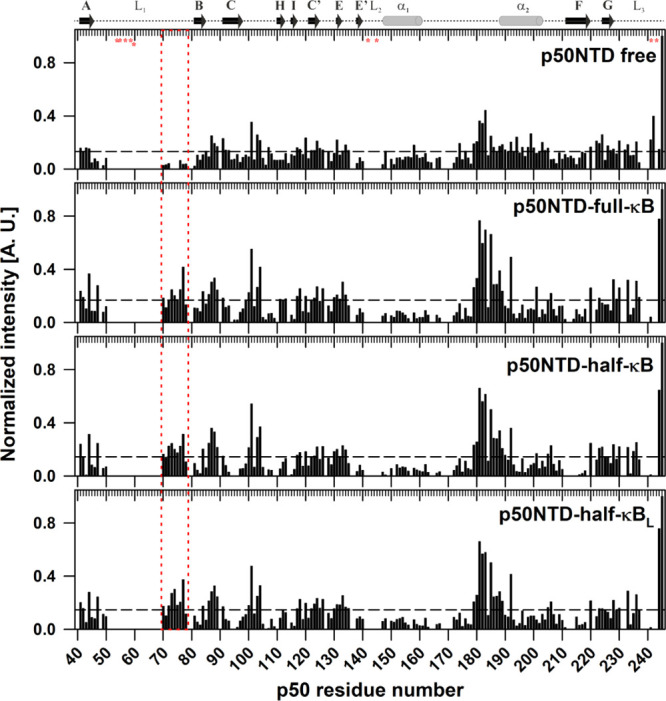
C-terminal part of the L1 DNA-binding loop shows reduced conformational heterogeneity upon binding to κB DNA. Plots of normalized intensity of the p50-NTD in the free (top panel), the full-κB-bound (middle panel), and the half-κB-bound (lower panel) forms as a function of the p50 residue number. The peak height of the respective peaks was normalized with respect to that of the amino-acid residue A245 of the p50-NTD. The dashed horizontal line represents the average normalized intensity in every panel. The inset of the dashed red line highlights the region (residue A70-S78), where the resonances become sharper in the DNA-bound form compared to that in the DNA-free p50-NTD. The fragment of E73-N75 could only be assigned in the (p50-NTD)-κB complex as their corresponding cross-peaks were absent in the [15N–1H]-HSQC spectrum of (p50-NTD)-free form due to excessive broadening. This stretch belongs to the C-terminal part of the L1 loop region of the p50 that contains the DNA contacting residues. A contiguous stretch of amino acid residues from E179-D183 showed augmented intensities in all the κB DNA-bound forms of p50-NTD when compared with the free form of the protein fragment. This stretch belongs to the linker region connecting the two α-helices but adjacent to the N-terminus of the α2 helix which is unique to the p50 and p52 NF-κB subunits only.
Constituent Domains of the p50 are Structurally Independent in Its κB-DNA-Free Form
To characterize the DNA-free form of the p50 homodimer using NMR spectroscopy, we first explored the feasibility to obtain the resonance assignments of the backbone amide-proton cross-peaks of the 73.1 kDa p50 homodimer (p50–p50) using the strategy of transfer of assignments from those of its constitutive individual domains—p50-NTD and p50-DD.
Both the individual domains as well as the full-length p50 yielded well-dispersed 2D [15N, 1H]-HSQC spectra characteristic of a well-folded protein (Figure 2). The amide resonance assignments of p50-DD22 and p50-NTD were then transferred to the [15N, 1H]-TROSY37 spectrum of the 73.1 kDa p50 homodimer (325 amino acids per monomer, excluding the first methionine). While the transfer of the assignment strategy helped in the amide resonance assignments of the well-dispersed regions of p50–p50, assignments of the cross-peaks, especially in the overlapping regions of TROSY spectra remained ambiguous. To unambiguously assign these cross-peaks, TROSY-based 3D HNCA was recorded on the [15N, 13C, 2H]-perdeuterated p50–p50 sample (Figure S6).
A comparison of the chemical shifts of p50 with respect to its constituent isolated domains—p50-NTD and p50-DD showed insignificant differences (Figure 8A). The plot of the intensities of the amide cross-peaks of the [15N, 1H]-TROSY spectra of the perdeuterated p50 homodimer showed differences in the average cross-peak intensities of the residues belonging to the NTD and the DD domains [Figure 8B (top panel); Table 1]. As seen in Table 1, the average intensities of the secondary structural elements of both the domains in the DNA-free p50 homodimer are noticeably different which contrasts with that observed for the full-κB-bound protein. In the p50-full-κB complex, the DD along with the p50-NTD undergoes significant CSPs (Figures 8C and S7). This suggest that the two domains in the DNA-free form of p50 undergo independent motions which upon binding to κB DNA behave as a single unit.
Figure 8.
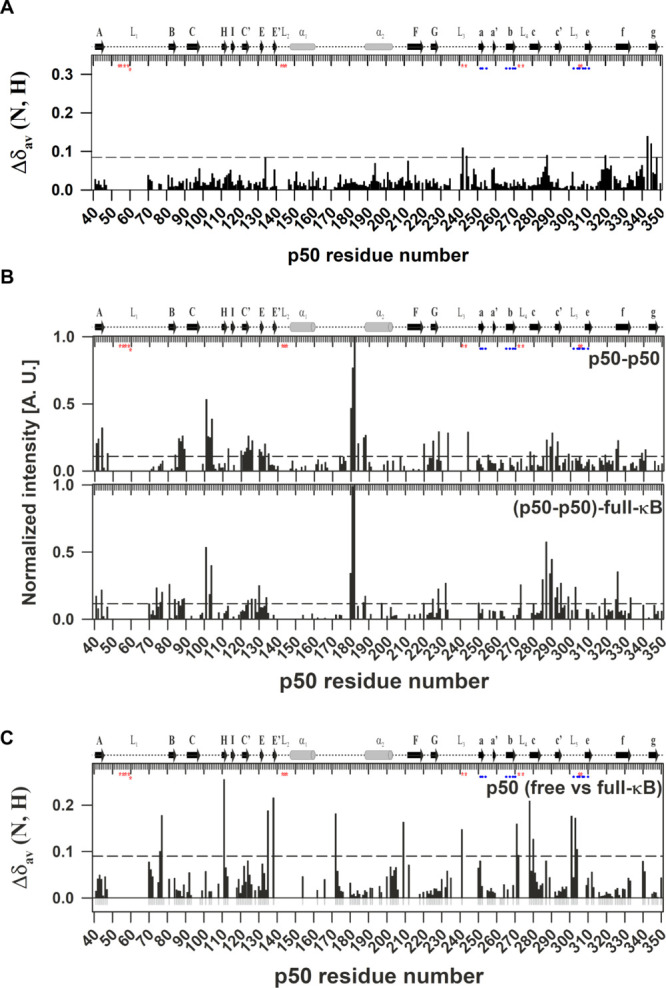
Chemical shift difference between the individual isolated domains and the p50-RHR is insignificant. (A) Plot of CSP observed in full length p50-RHR with respect to its isolated individual domains (p50-NTD and p50-DD) as a function of the p50 residue number. The other details of the figure are the same as mentioned in Figure 5A. Significant chemical shift deviations between the p50-RHR (39–363) and its isolated domains, namely, p50-NTD (39–245) and p50-DD (245–350) were observed in the regions around the termini of the isolated domains, except N288 and N320 that belong to the non-DNA-contacting and non-dimer-interfacial loops in the DD region. (B) Plots of normalized intensity of p50-RHR in the free (top panel) and the full-κB-bound (bottom panel) forms as a function of the p50 residue number. The peak heights of the respective peaks were normalized with respect to that of the residue G182. The dashed horizontal line represents the average normalized intensity in every panel. A contiguous stretch of the amino acid residues from E179-D183 yielded intense peaks in all the forms of p50-RHR. This stretch belongs to the linker region connecting the two α-helices but adjacent to the N-terminus of the α2 helix which is unique to the p50 and p52 NF-κB subunits. Residues in the linker connecting the c and c′β-strands showed augmented normalized intensities in the κB-bound forms when compared with the free protein. This region is remote from the DNA in the 3D structure. The observation implies that this stretch of the protein remains highly flexible in the DNA-bound form of the protein. The resonances from the crowded regions were excluded from the analysis due to the possibility of including ambiguity. The average intensities of the individual domains are given in Table 1. (C) Plot of CSP observed for the p50 in the free with respect to the full-κB-bound form as a function of the p50 residue number. The residues for which unambiguous CSPs were available are shown as gray negative bars. The other details of the figure are same as mentioned in Figure 5A.
Table 1. Average Normalized Intensities of the p50 Domains.
| p50 | free | full-κB bound |
|---|---|---|
| p50-NTD (Δ180–182) | 0.11 ± 0.10 | 0.10 ± 0.09 |
| p50-DD | 0.08 ± 0.05 | 0.11 ± 0.11 |
| p50-NTDa | 0.10 ± 0.10 | 0.09 ± 0.07 |
| p50-DDa | 0.07 ± 0.06 | 0.09 ± 0.08 |
Only secondary structural elements included.
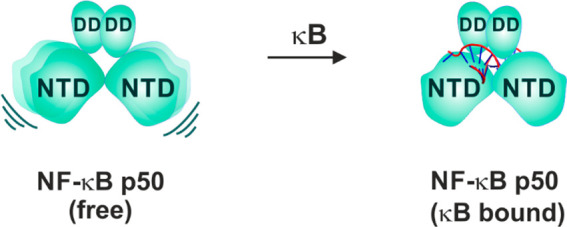
Taking together the near complete correspondence of amide chemical shifts of the individual domains—p50-NTD and p50-DD with those of p50–p50 dimer and distinct average intensity profile of the two domains in p50–p50 suggests the structural independence of the two domains in the DNA-free form of the p50 NF-κB subunit (Figure 9).
Figure 9.
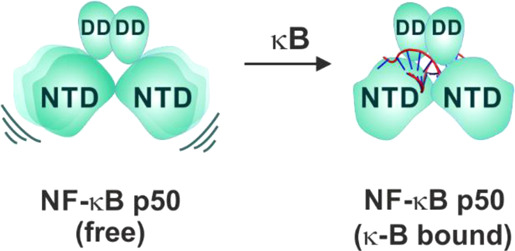
Schematic mechanism of p50 homodimer interaction with a κB DNA. In the DNA-free form, the two NTDs of the p50 homodimer are independent of each other and are anchored by the DD. In the κB-DNA-free form, the NTD is also independent of the DD. Upon binding to κB DNA (full-κB site), both the NTDs stabilize on the DNA as one unit by specifically recognizing one-half-κB site per NTD. The DD further stabilizes the complex through the backbone phosphate interaction. The DNA molecule is represented as a double helix with the full-κB site shown in red.
Discussion
Notwithstanding the structural information available for the NF-κB dimers bound to nucleic acid (DNA as well as RNA), little is known about the DNA-free form of the protein. Based on the experimental secondary structure prediction using the backbone chemical shifts, the p50-NTD in its DNA-free form exhibits certain structural features that are not observed in the available structures of its DNA-bound form. Among the secondary structural elements unique to the DNA-free p50-NTD are a helix in the C-terminal part of Loop L1, a helix in the linker connecting the α1 and α2 helices, extended structures flanking both the ends of the “B” β-strand, and an extended structure in the N-terminal of Loop L3 (Figure S3A). When compared with the similar secondary structure prediction for the DNA-free RelA-NTD (19–191) using the available backbone chemical shifts (BMRB #26647), we did not observe much difference in the secondary structures between the DNA-free and the DNA-bound forms of the protein except for the missing “I” β-strand in the DNA-free form and the formation of a β-strand at the C-terminal end of helix α1 (Figure S3B). The nHX of p50-NTD revealed that the amide resonances of the α2 helix is the most HX-resistant fragment in the domain. This suggests that within the NTD, the α2 helix is the most stable secondary structure with strong hydrogen bonds. The α2 helix along with the linker connecting it with the α1 helix (residues 167–202) does not form a conserved part of the RHR in the NF-κB family and is unique to the p50 subunit. The α2 helix is not affected by the presence or absence of the κB-DNA (Figure 5). We speculate this helix to have a role in the conformational stability of the p50-NTD.
A prominent difference between the DNA-bound and the free forms of the p50-NTD is the dynamics of the C-terminal part of the L1 loop that contains the DNA-contacting residues. This stretch of the L1 loop undergoes a decrease in the conformational heterogeneity upon binding to κB DNA as revealed from the appearance of the amide cross-peaks of the amino acid residues from this region (70–78; Figure 7). This stretch also senses the extra residue on the 5′ end of a κB site (Figure 6). It remains to be determined how this fine-tunes the affinity and specificity of the κB site for a given NF-κB dimer.
Each NTD domain of a NF-κB dimer through its three loops, L1, L2, and L3, make base specific contacts with one-half part of the κB sequence, that is, 4–5 bp of the κB site. The DDs make mainly nonspecific contacts with the backbone phosphates of the κB DNA. This gives the NF-κB-DNA complex a butterfly-like structure with the NTDs acting as the wings and grabbing the two half-κB sites to position itself firmly on the DNA. Given the possibility of the 15 different combinatorial NF-κB dimers, the variability in the sequence of the κB site broadens with the specific sets of target genes activated by a specific NF-κB dimer at the given physiological conditions. It is intriguing if the sequence of one half-κB site influences the binding of the NF-κB NTD bound to the other half-κB site in a full-κB DNA. Our results show that the NTD or the specificity domain of the p50 is not affected by the presence or absence of the second-half of the κB sequence. However, the extra residues in 5′ end of the bound half-κB site is sensed by the C-terminal end of the L1 loop (Figure 6). This implies that each NTD can bind to its cognate half-κB site independent of the partner NTD of the NF-κB dimer. Though the DDs of the NF-κB dimers contact the backbone phosphates of the κB DNA, they potentially can further broaden the sequence space of the κBs by bringing together the NTDs that recognize the distinct half-κB sequences. This also provides reasoning for the observed half-κB sites present in the target genes of other transcription factors like the IRF family which remain bound by the p50 homodimer, thereby acting as repressor of such genes.38
Acknowledgments
The authors thank Gourisankar Ghosh, UCSD, for providing the mouse-p50 (39–363) and mouse-p50-DD (245–350) plasmids and acknowledge the NMR facilities at the NMR Research Centre, IISC Bangalore, India, and the High-Field NMR Facility, TIFR, Mumbai, India. We acknowledge the Heavy Water Board, India. for providing Heavy water required for deuteration. This work was supported by the Young Scientist Scheme of Science and Engineering Research Board, India [grant no YSS/2015/000090] to SPM; Department of Biotechnology, India [grant no BT/PR24430/BRB/10/1630/2017] to S.P.M. and H.S.A.; Ministry of Human Resource Development, India, fellowship to T.R. and N.D.; Council of Scientific and Industrial Research fellowship to D.J.; and Dr. D. S. Kothari Postdoctoral fellowship of University Grants Commission, India [CH/17-18/0123] to V.M.R.K.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01299.
Sequence identity of NF-κB subunits, specific labeling/unlabeling of amino acids of p50-NTD, TALOS-N predicted secondary structure of p50 and RelA NTD, 1D 1H spectrum of κB DNA, [15N, 1H]-HSQC spectra of p50-NTD, [15N–1H]-TROSY spectrum of p50-RHR, p50 residues with high chemical shift changes upon binding to κB DNA, and description of different protein samples (PDF)
Accession Codes
1H, 15N, and 13C backbone chemical shifts assignments and 15N relaxation data for the p50-NTD is deposited in the BMRB (www.bmrb.wisc.edu) under the accession number 28055 and 50819, respectively.
Author Present Address
⊥ Department of NMR-based Structural Biology, Max Planck Institute.
Author Present Address
# Department of Biological Sciences, Columbia University, New York, NY 10027, USA.
The authors declare no competing financial interest.
Author Status
∥ Deceased 30th July 2020.
Supplementary Material
References
- Liou H. C.; Sha W. C.; Scott M. L.; Baltimore D. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Mol. Cell. Biol. 1994, 14, 5349–5359. 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P. A.; Henkel T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994, 12, 141–179. 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- Zhang G.; Ghosh S. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 2001, 107, 13–19. 10.1172/jci11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak P. P.; Firestein G. S. NF-kappaB: a key role in inflammatory diseases. J. Clin. Invest. 2001, 107, 7–11. 10.1172/jci11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J.; Kwon H.; Génin P. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J. Clin. Invest. 2001, 107, 143–151. 10.1172/jci11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero M. C.; Wang V. Y.-F.; Huxford T.; Ghosh G. Genome reading by the NF-kappaB transcription factors. Nucleic Acids Res. 2019, 47, 9967–9989. 10.1093/nar/gkz739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udalova I. A.; Richardson A.; Denys A.; Smith C.; Ackerman H.; Foxwell B.; Kwiatkowski D. Functional consequences of a polymorphism affecting NF-kappaB p50-p50 binding to the TNF promoter region. Mol. Cell. Biol. 2000, 20, 9113–9119. 10.1128/mcb.20.24.9113-9119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohuslav J.; Kravchenko V. V.; Parry G. C.; Erlich J. H.; Gerondakis S.; Mackman N.; Ulevitch R. J. Regulation of an essential innate immune response by the p50 subunit of NF-kappaB. J. Clin. Invest. 1998, 102, 1645–1652. 10.1172/jci3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W. C.; Liou H.-C.; Tuomanen E. I.; Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell 1995, 80, 321–330. 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- Plaksin D.; Baeuerle P. A.; Eisenbach L. KBF1 (p50 NF-kappa B homodimer) acts as a repressor of H-2Kb gene expression in metastatic tumor cells. J. Exp. Med. 1993, 177, 1651–1662. 10.1084/jem.177.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G.; Duyne G. V.; Ghosh S.; Sigler P. B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature 1995, 373, 303–310. 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- Saccani S.; Pantano S.; Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Molecular Cell 2003, 11, 1563–1574. 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Cramer P.; Larson C. J.; Verdine G. L.; Muller C. W. Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution. EMBO J. 1997, 16, 7078–7090. 10.1093/emboj/16.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C. W.; Rey F. A.; Sodeoka M.; Verdine G. L.; Harrison S. C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 1995, 373, 311–317. 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- Chen F. E.; Huang D.-B.; Chen Y.-Q.; Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 1998, 391, 410–413. 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- Berkowitz B.; Huang D.-B.; Chen-Park F. E.; Sigler P. B.; Ghosh G. The x-ray crystal structure of the NF-kappa B p50.p65 heterodimer bound to the interferon beta -kappa B site. Journal of Biological Chemistry 2002, 277, 24694–24700. 10.1074/jbc.m200006200. [DOI] [PubMed] [Google Scholar]
- Escalante C. R.; Shen L.; Thanos D.; Aggarwal A. K. Structure of NF-kappaB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-beta promoter. Structure 2002, 10, 383–391. 10.1016/s0969-2126(02)00723-2. [DOI] [PubMed] [Google Scholar]
- Mukherjee S. P.; Borin B.; Quintas P. O.; Dyson H. J. NMR characterization of a 72 kDa transcription factor using differential isotopic labeling. Protein Science 2016, 25, 597–604. 10.1002/pro.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S. P.; Quintas P. O.; McNulty R.; Komives E. A.; Dyson H. J. Structural characterization of the ternary complex that mediates termination of NF-kappaB signaling by IkappaBalpha. Proc Natl Acad Sci USA 2016, 113, 6212–6217. 10.1073/pnas.1603488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy C. H.; Bergqvist S.; Huxford T.; Ghosh G.; Komives E. A. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004, 13, 1767–1777. 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnarjuna B.; Jaipuria G.; Thakur A.; D’Silva P.; Atreya H. S. Amino acid selective unlabeling for sequence specific resonance assignments in proteins. J. Biomol. NMR 2011, 49, 39–51. 10.1007/s10858-010-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.; Dadhwal P.; Atreya H. S.; Mukherjee S. P. Backbone resonance assignments of the dimeric domain of the p50 NF-kappaB subunit. Biomol NMR Assign 2020, 14, 9–11. 10.1007/s12104-019-09912-y. [DOI] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/bf00197809. [DOI] [PubMed] [Google Scholar]
- Keller R. L. J.Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment; Swiss Federal Institute of Technology Zurich: Zurich, Swizerland, 2004. [Google Scholar]
- Dubey A.; Kadumuri R. V.; Jaipuria G.; Vadrevu R.; Atreya H. S. Rapid NMR Assignments of Proteins by Using Optimized Combinatorial Selective Unlabeling. Chembiochem 2016, 17, 334–340. 10.1002/cbic.201500513. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M.; Pervushin K.; Wider G.; Senn H.; Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proceedings of the National Academy of Sciences 1998, 95, 13585–13590. 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow N. A.; Muhandiram R.; Singer A. U.; Pascal S. M.; Kay C. M.; Gish G.; Shoelson S. E.; Pawson T.; Forman-Kay J. D.; Kay L. E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 1994, 33, 5984–6003. 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Ferrage F.; Piserchio A.; Cowburn D.; Ghose R. On the measurement of 15N-{1H} nuclear Overhauser effects. Journal of Magnetic Resonance 2008, 192, 302–313. 10.1016/j.jmr.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Auvergne E. J.; Gooley P. R. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR 2008, 40, 107–119. 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Auvergne E. J.; Gooley P. R. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR 2008, 40, 121–133. 10.1007/s10858-007-9213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P.; Swapna G. V. T.; Huang Y. J.; Aramini J. M.; Anklin C.; Conover K.; Hamilton K.; Xiao R.; Acton T. B.; Ertekin A.; Everett J. K.; Montelione G. T. A microscale protein NMR sample screening pipeline. J. Biomol. NMR 2010, 46, 11–22. 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.; Dhaka N.; Raza T.; Dadhwal P.; Atreya H. S.; Mukherjee S. P. Domain Stability Regulated through the Dimer Interface Controls the Formation Kinetics of a Specific NF-kappaB Dimer. Biochemistry 2021, 60, 513–523. 10.1021/acs.biochem.0c00837. [DOI] [PubMed] [Google Scholar]
- Müller C. W.; Harrison S. C. The structure of the NF-kappa B p50:DNA-complex: a starting point for analyzing the Rel family. FEBS Lett. 1995, 369, 113–117. 10.1016/0014-5793(95)00541-g. [DOI] [PubMed] [Google Scholar]
- Moorthy A. K.; Huang D.-B.; Wang V. Y.-F.; Vu D.; Ghosh G. X-ray structure of a NF-kappaB p50/RelB/DNA complex reveals assembly of multiple dimers on tandem kappaB sites. Journal of Molecular Biology 2007, 373, 723–734. 10.1016/j.jmb.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.-B.; Vu D.; Cassiday L. A.; Zimmerman J. M.; Maher L. J. 3rd; Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proceedings of the National Academy of Sciences 2003, 100, 9268–9273. 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance M.; Loria J. P.; Palmer A. G. Sensitivity improvement of transverse relaxation-optimized spectroscopy. Journal of Magnetic Resonance 1999, 136, 92–101. 10.1006/jmre.1998.1626. [DOI] [PubMed] [Google Scholar]
- Cheng C. S.; Feldman K. E.; Lee J.; Verma S.; Huang D.-B.; Huynh K.; Chang M.; Ponomarenko J. V.; Sun S.-C.; Benedict C. A.; Ghosh G.; Hoffmann A. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-kappaB p50. Science Signaling 2011, 4, ra11. 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.