Abstract
Protein phosphorylation is an abundant post-translational modification (PTM) and an essential modulator of protein functionality in living cells. Intrinsically disordered proteins (IDPs) are particular targets of PTM protein kinases due to their involvement in fundamental protein interaction networks. Despite their dynamic nature, IDPs are far from having random-coil conformations but exhibit significant structural heterogeneity. Changes in the molecular environment, most prominently in the form of PTM via phosphorylation, can modulate these structural features. Therefore, how phosphorylation events can alter conformational ensembles of IDPs and their interactions with binding partners is of great interest. Here we study the effects of hyperphosphorylation on the IDP osteopontin (OPN), an extracellular target of the Fam20C kinase. We report a full characterization of the phosphorylation sites of OPN using a combined nuclear magnetic resonance/mass spectrometry approach and provide evidence for an increase in the local flexibility of highly phosphorylated regions and the ensuing overall structural elongation. Our study emphasizes the simultaneous importance of electrostatic and hydrophobic interactions in the formation of compact substates in IDPs and their relevance for molecular recognition events.
Protein phosphorylation is an abundant post-translational modification that adds an extra layer of complexity to the regulation of cellular fate, particularly in intrinsically disordered proteins because of their inherent accessibility.1 Regulation of cellular signaling by phosphorylation is associated with conformational changes2−7 and modulation of binding events8,9 and, recently, has been linked to the formation of membraneless organelles.10
The extracellular matrix (ECM) contains a large fraction of phosphorylated proteins, and many of them have been observed in breast and lung cancer samples.11−13 Among these ECM proteins, osteopontin (OPN) and caseins have the highest fractions of potential phosphorylation sites.12 OPN, also known as secreted phosphoprotein 1 (SPP1), is a secreted extracellular protein that exerts its functionality by binding to integrin and CD44 receptors and is reported to be implicated in apoptosis, wound healing, inflammation, tumor growth, tumor progression, and tumor metastasis.14−16 It is tightly regulated by glycosylation, phosphorylation and cleavage,17,18 and is secreted in its unphosphorylated19 or phosphorylated20 form. Human OPN is mainly phosphorylated by Fam20C (67% of the reported phosphorylated sites).21,22 Fam20C kinase is located in the Golgi lumen and responsible for most of the phosphorylation in the ECM. It recognizes primarily a S-x-E/pS motif but also shows a certain promiscuity with respect to other amino acid motifs (e.g., T-x-E or S-x-D).21 OPN contains 28 potential Fam20C specific motifs, causing ≤14% of the residues being phosphorylated. The degree of OPN phosphorylation has been associated with Raine syndrome, a rare disease characterized by generalized osteosclerosis with periosteal bone formation, characteristic facial dysmorphism, brain abnormalities, including intracerebral calcifications, and in some cases neonatal death.13,23,24 Its abnormal phosphorylation patterns are directly connected to Fam20C mutations. Furthermore, the phosphorylation of OPN regulates its binding interaction with hydroxyapatite and hence the formation and growth of the mineral phase in bone material,25−31 as well as bone remodeling and calcification.13,28,32,33 On top of that, ECM phosphoproteome homeostatis, in particular OPN phosphorylation, has been associated with tumor cell progression,34 macrophage migration,35 and host–cell interactions.36
Nuclear magnetic resonance (NMR) spectroscopy has matured into an exquisite tool to tackle PTMs and to study the structural dynamics of the protein of interest under nativelike conditions.37−44 Although denaturing conditions have been particularly useful for characterizing modified sites,43,44 it is important to take into account the fact that the conformational ensembles of IDPs are drastically affected by the presence of denaturing agents, as IDPs are far from being merely unfolded.45 With respect to OPN, several important features that account for the modulation of compaction, binding and function of the unphosphorylated form have been identified.46−48 Here, we present an NMR-based strategy for structurally characterizing the fully phosphorylated protein and the dynamics of the hyperphosphorylated OPN. For this purpose, a stable HEK293T cell line expressing Fam20C was used to obtain the pure functional kinase.21 The degree of phosphorylation and the homogeneity of the modified phospho-residue patterns were optimized in a controlled in vitro reaction. NMR signal assignment experiments reveal a downfield shift of a majority of the serine and individual threonine 1HN NMR signals due to intraresidue hydrogen bonding between the phosphate and backbone amide groups in unstructured regions.49 The experimental NMR data set is complemented by a mass spectrometry (MS) analysis. The putative biological relevance of these findings is outlined with studies of the interaction with heparin and hyaluronic acid, which are present in proteoglycans and the ECM, and the comparison of our results to reported affinities for integrin receptors, natural binders of OPN.
Results
NMR/MS-Based Phosphoprofiling of OPN
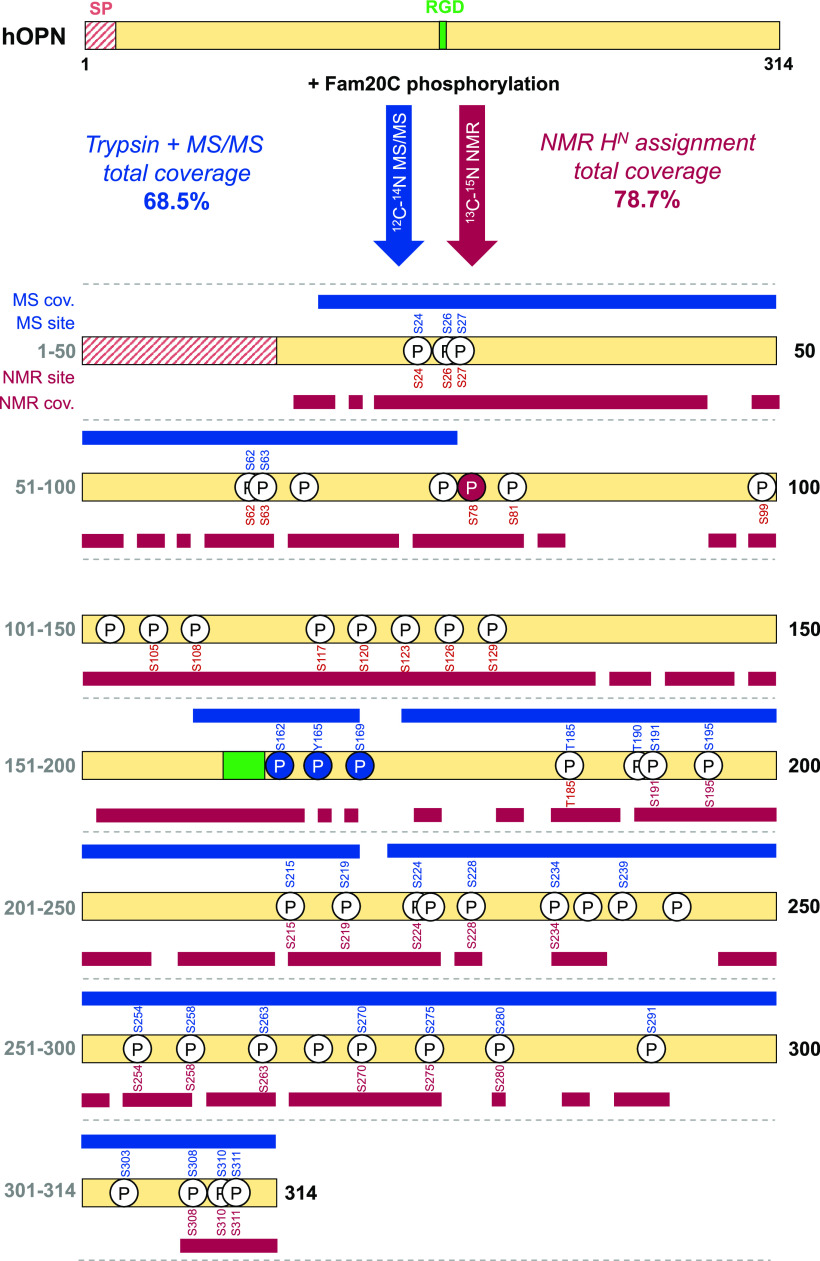
A highly pure unphosphorylated 13C/15N Homo sapiens OPN was expressed recombinantly in Escherichia coli (Figures S1 and S2). The functional wild type (wt) and D478A (kinase-dead mutant) Fam20C kinases were expressed in HEK293T stable expression cells (Figure S3). The in vitro phosphorylation reaction of OPN was optimized from previously reported conditions13 (see the Supporting Information for detailed method protocols). A combined approach using MS and NMR spectroscopy was carried out for the identification of the phosphorylation sites. The results are summarized in Figure 1. The total sequence coverage of the MS/MS experiments is 68.5%, and 28 phosphorylation events are identified (see Figure S4). Among them, 17 of the 22 canonical motifs are found to be phosphorylated (S24, S26, S27, S62, S63, S195, S224, S234, S254, S263, S270, S275, S280, S291, S303, S308, and S310). A plausible alternative motif (T-x-E) is also found to be phosphorylated in position T185. Other phosphorylated residues do not follow the mentioned motif, although some of them were found to be phosphorylated in mammalian cells21 or bodily fluids (milk)22 (Figure S5), suggesting a certain promiscuity of the kinase and/or the activity of other unreported kinases.21
Figure 1.
Scheme of OPN residues phosphorylated in vitro by Fam20C, identified by MS and NMR spectroscopy. White circles represent previously identified phosphorylation sites.21,22 Blue and red circles indicate the phosphorylation sites newly identified by MS and NMR spectroscopy, respectively. The blue and red bars indicate the coverage of MS and HN NMR assignments, respectively.
Among those (e.g., S162, Y165, and S169), previously unreported sites are reliably identified by MS on peptides GDSVVYGLR and GDSVVYGLRSK. The fragment pattern is continuous and shows the properties of phospho spectra. Resonance assignment by NMR spectroscopy was achieved for 78.7% of all HN signals (deposited in the BMRB50 as entry 50447 and Table S1). Twenty-eight phosphorylation events are identified on the basis of the 1HN downfield shifts (Figure 2). Twenty of the 22 canonical phosphorylation motifs are found to be phosphorylated (S24, S26, S27, S62, S63, S78, S81, S120, S126, S129, S195, S224, S234, S254, S263, S270, S275, S280, S308, and S310), and four noncanonical but plausible phosphorylated motifs (T185, T-x-E; and S99, S105, and S108, S-x-D), previously also found in phosphorylated OPN extracted from milk.22 The four remaining phosphorylated residues display a noncanonical phosphorylation motif. Some of these noncanonical phosphorylations are identified both by MS phosphomapping and NMR assignment [S191, S215, S228, and S258 (see Figure 1 and Figure S5)].
Figure 2.
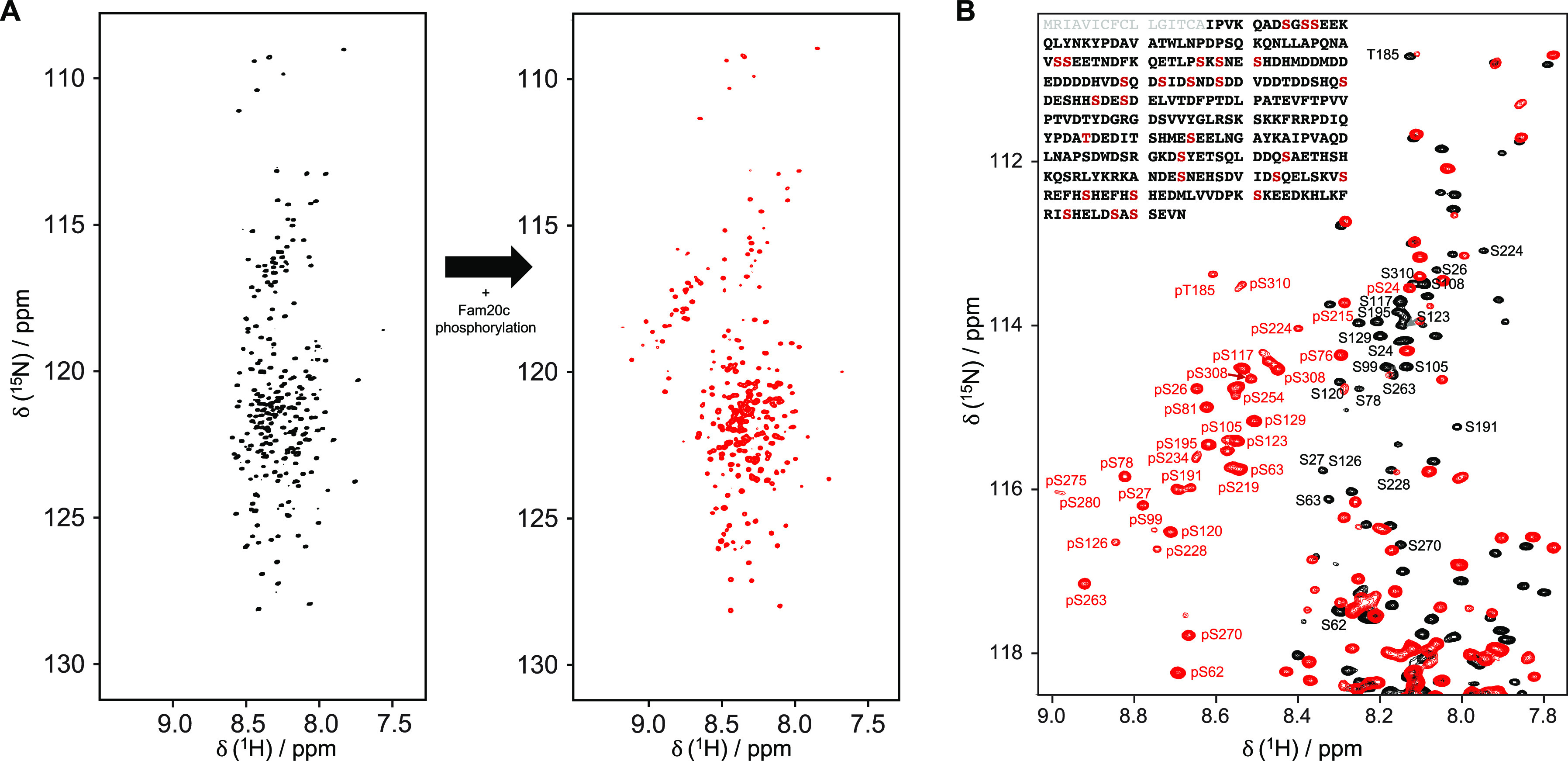
NMR fingerprint of OPN hyperphosphorylation. (A) 1H–15N HSQC NMR spectra of OPN before (black) and after (red) phosphorylation by Fam20C. Note how many serine residues [δ (15N) ≈116 ppm] experience a downfield shift in the 1H dimension. (B) Close-up of the serine region of 1H–15N HSQC NMR spectra of OPN before (black) and after (red) phosphorylation by Fam20C. The protein sequence with the S-x-E/pS sites colored red is shown in the top left corner. The signal peptide, which is not present in our construct, is colored gray.
Phosphorylation Increases Local Flexibility in OPN
NMR observables such as chemical shifts or 15N relaxation rates are very informative for IDP structural dynamics.51,52 Possible changes in the structural dynamics of the protein were studied by a series of 15N relaxation experiments (Figure 3).
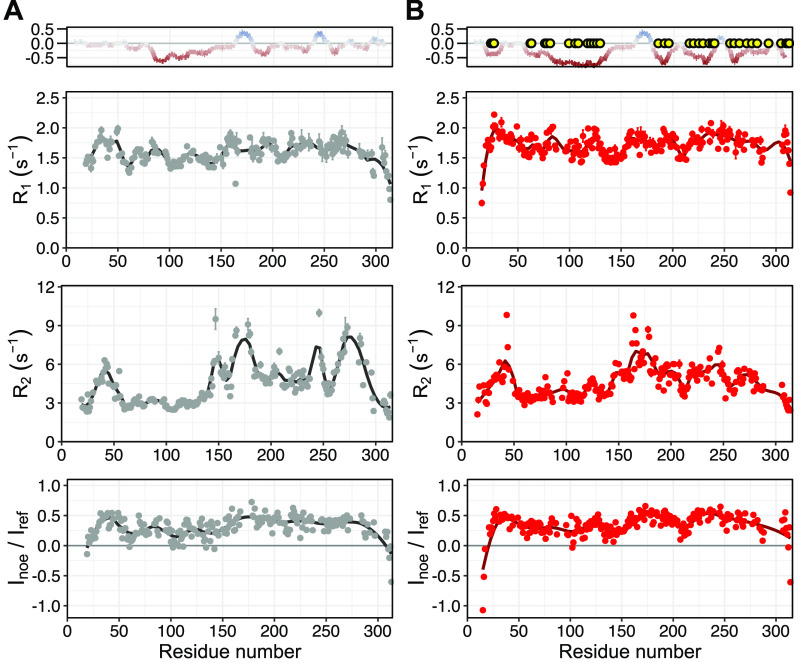
Figure 3.
15N NMR relaxation data of OPN (A) before and (B) after phosphorylation, measured at 18.8 T. A charge plot of the protein sequence is shown at the top. Yellow circles indicate the identified phosphorylated residues. 15N R1, 15N R2, and 15N–{1H} NOE relaxation parameters, from top to bottom, respectively, of OPN measured at 293 K. Error bars indicate the fitting errors (15N R1 and 15N R2) and the error propagation of intensity ratios based on the noise level (hetNOE).
The 15N R1 patterns of both unphosphorylated and phosphorylated OPN show similar features, however with systematically larger R1 values for the modified protein. Interestingly, 15N R2 values of the residues in the second half of the protein (residues 200–314) decrease for the phosphorylated form, while fast NH vector motions are retained, as measured with 15N–{1H} NOE relaxation experiments. Overall, the region of residues 200–314 experiences an increase in backbone dynamics on the nanosecond time scale while faster picosecond time scale motions are nearly unaffected. In summary, the experimental data suggest enhanced dynamics in protein segments that comprise the majority of the phosphorylation sites. Further analysis, e.g., by applying the model free approach, was not pursued because connections between the measured phenomenological relaxation rates and the motions of a protein are far from trivial, especially for IDPs, where experimental rates are a mixture of polymer-like properties and non-uniform chain behaviors caused by secondary structure propensities, residue-dependent motions, and long-range correlated segments.53
Phosphorylation Induces Structural Elongation of the Main Compact State in OPN
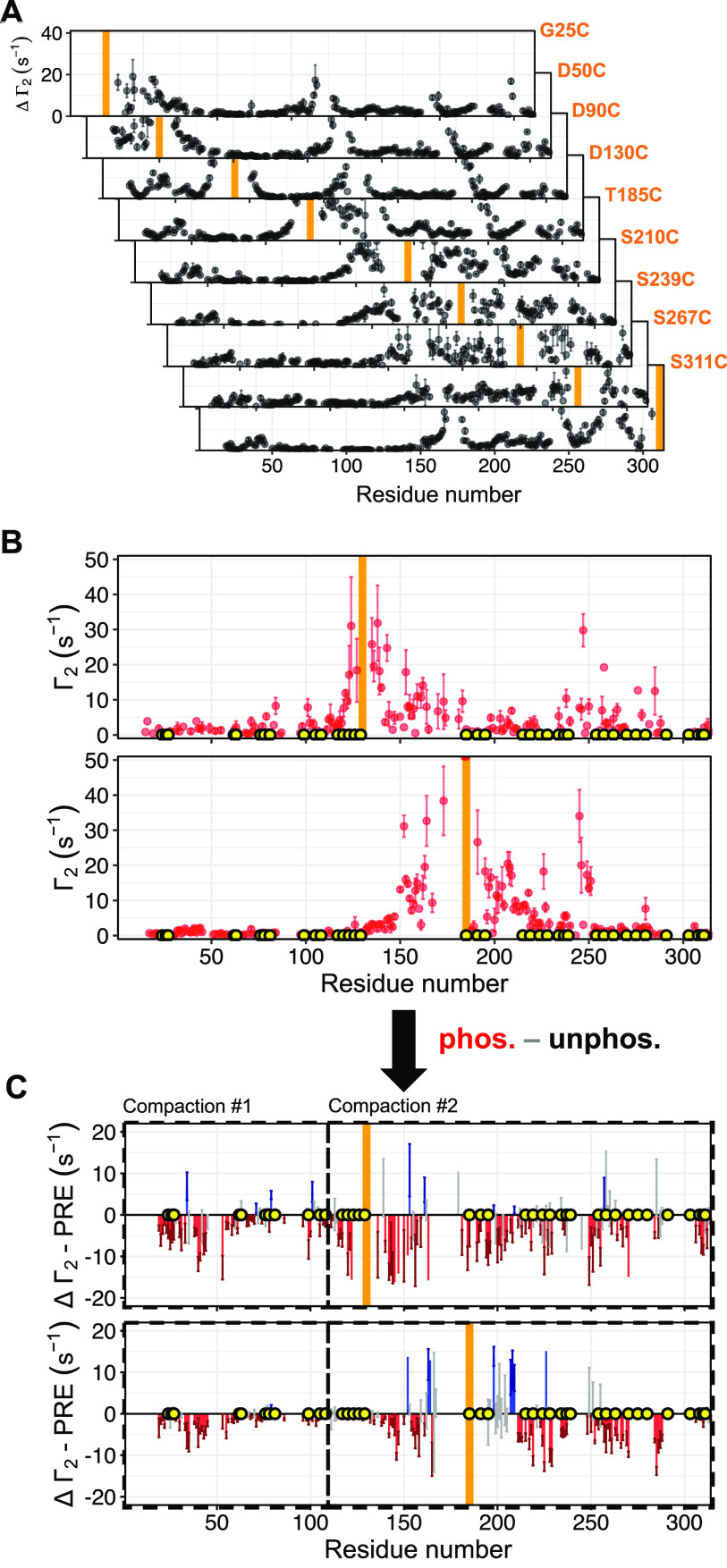
Long-range structural contacts in unphosphorylated (Figure 4A) and hyperphosphorylated OPN (Figure 4B) were probed by measurements of PRE profiles for several cysteine mutants for unphosphorylated OPN, while the hyperphosphorylated state of OPN was probed using the two representative cysteine mutants D130C and T185C. In total, nine cysteine mutants were studied for the unphosphorylated OPN, which is necessary to overcome the intrinsic limitations of PRE measurements due to the r–6 averaging and to achieve a proper modeling of the long-range contacts, as shown elsewhere.54 Importantly, 1H T2 rates were measured instead of intensities, which adds a certain robustness to the experimental PRE data of IDPs. It is important to note that in the case of IDPs the measured R2 rates are the weighted population average, and therefore, conformations with greater R2 enhancements will be heavily weighted even if they are scarcely populated.55 A comparison of the PRE profiles obtained for cysteine mutants D130C and T185C clearly shows a striking reduction of long-range contacts within the central compact core region (residues 120–250), at the N-terminal region around residues 25–30, and within the whole C-terminus of the protein, while most of the negatively charged regions remain unaffected (Figure 4C). To conclude, our data suggest a significant structural elongation of OPN due to hyperphosphorylation, accompanied by an increase in local flexibility in the C-terminal region (residues 200–314) that is particularly rich in phosphorylation sites.
Figure 4.
Effect of phosphorylation on long-range interactions measured by PRE experiments. (A) 1HN Γ2 PRE profiles of different OPN cysteine mutants obtained from 1HNT2 NMR experiments. (B) 1HN Γ2 PRE rates of the phosphorylated OPN mutants D130C (top) and T185C (bottom) determined from 1HNT2 NMR experiments. (C) Plot of the PRE rate difference of OPN and phosphorylated OPN mutants D130C (top) and T185C (bottom). Orange bars indicate the respective mutated cysteine residue with the attached spin-label.
Decrease in the Binding Affinity for Heparin Due to Phosphorylation while Retaining Striking Carbohydrate Specificity
The binding of OPN to heparin, a chemical mimic of the natural glycosaminoglycan heparan sulfate (HS) in the ECM, was investigated by a series of titration experiments employing 1H–15N HSQC NMR spectroscopy (Figure 5A,B). In the unmodified form of OPN, binding to heparin mainly induces chemical shift changes in the positively charged regions (residues 180–190 and 240–260), while modest chemical shift perturbations are found for residues located in the region of residues 140–160 (Figure 5C). A quantitative fit analysis reveals a binding affinity in the micromolar range (48 ± 8 and 52 ± 20 μM) for both positively charged regions (Figure 5C middle panel, blue; Figure S7) in accordance with ITC data from previous work on a protein homologue.46 The observed affinity is very similar to that of the quail homologue form.46 As previously reported, the chemical shifts observed in the region of residues 140–160 may arise from a local “unfolding-upon-binding” process that occurs when OPN binds to this polyanionic carbohydrate.46
Figure 5.
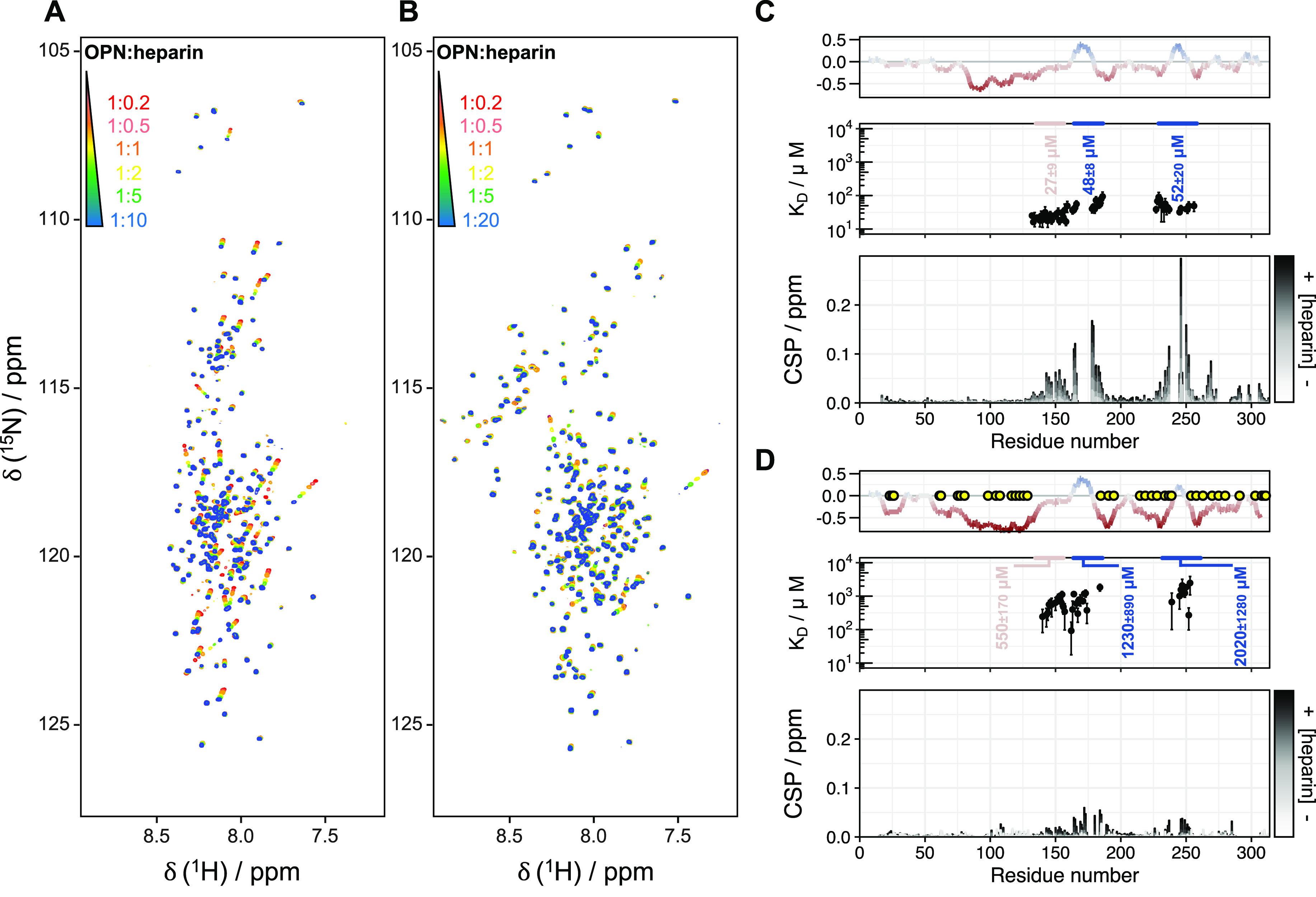
Binding of (phosphorylated) OPN to heparin, monitored by NMR titrations. 1H–15N HSQC NMR spectra in the presence of increasing amounts of heparin (red to blue) for the (A) unphosphorylated and (B) hyperphosphorylated forms. Chemical shift perturbations (bottom panel) and fitted KD of binding regions (middle panel in blue) and the uncompacted region (middle panel in pink) plotted against the residue numbers of (C) OPN and (D) phosphorylated OPN. The corresponding charge plots are shown at the top. Yellow circles indicate the identified phosphorylated residues. The grayscale (from white to black) represents the increasing OPN:hep molar ratio from 1:0.2 to 1:10 (unphosphorylated) and 1:20 (phosphorylated).
This phenomenon, also known as “cryptic disorder”, is a widespread mechanism of folded proteins and IDPs in response to environmental changes (such as binding or protein modifications).56,57 Upon binding, the compensation of entropic loss (from a large conformational ensemble in the free form to a restricted set of conformations in the bound form) can be established in different mechanisms.58 Among them, IDPs may maximize the entropic gain by increasing the flexibility in regions distant from the binding sites, as it was reported for the mechanism of binding of OPN to heparin: a local rigidification in the heparin binding cleft (central core region) leads to a conformational entropy penalty that is reduced by a compensatory increase in the conformational flexibility of the negatively charged regions.46 Upon phosphorylation of OPN, the binding affinity is clearly reduced and consequently more heparin was needed to reach saturation (Figure 5B), presumably due to stronger electrostatic repulsions involving the numerous phosphorylation sites in the region of residues 240–260. Here, the entropic penalty of retaining a partially structured central region in phosphorylated OPN is accommodated by increasing the dynamics of charged regions. A similar mechanism was described for the mode of binding of Sic1 to Cdc4, where entropic compensatory events are also present.59,60 Quantitative analysis of the observed chemical shift changes for both positively charged regions reveals an approximately 20/40-fold decrease in affinity [1230 ± 890 and 2020 ± 1280 μM (Figure 5D)]. Besides that of heparin, the binding of OPN to hyaluronic acid (HA), another ubiquitous extracellular matrix glycosaminoglycan,61,62 was tested (Figure S8). HA is composed of N-acetylglucosamine and glucuronic acid and binds to the abundant extracellular receptor CD44 through a conserved HA binding domain (CD44HABD).63,64 OPN was also described as a binding partner of CD44 (with and without HA).65 Given the polyanionic nature of HA and heparin, a similar mode of binding to OPN can be anticipated. Surprisingly, however, NMR titration experiments show no binding between HA and OPN, in the unphosphorylated or in the phosphorylated form of the protein. Additionally, no binding to CD44HABD is identified in the presence or absence of HA [HA forms a tight complex with CD44HABD (Figure S9)]. This points to an interesting and unexpected (charge-independent) differential specificity of OPN toward the various glycosaminoglycans of the ECM and clearly questions the notion of OPN being a disordered protein lacking structural preformation. Moreover, it restricts the CD44 binding site of OPN to the disordered CD44 region, where the HS modification is present.
Decompaction of OPN Due to Hyperphosphorylation Modulates the Binding Affinity for Integrins
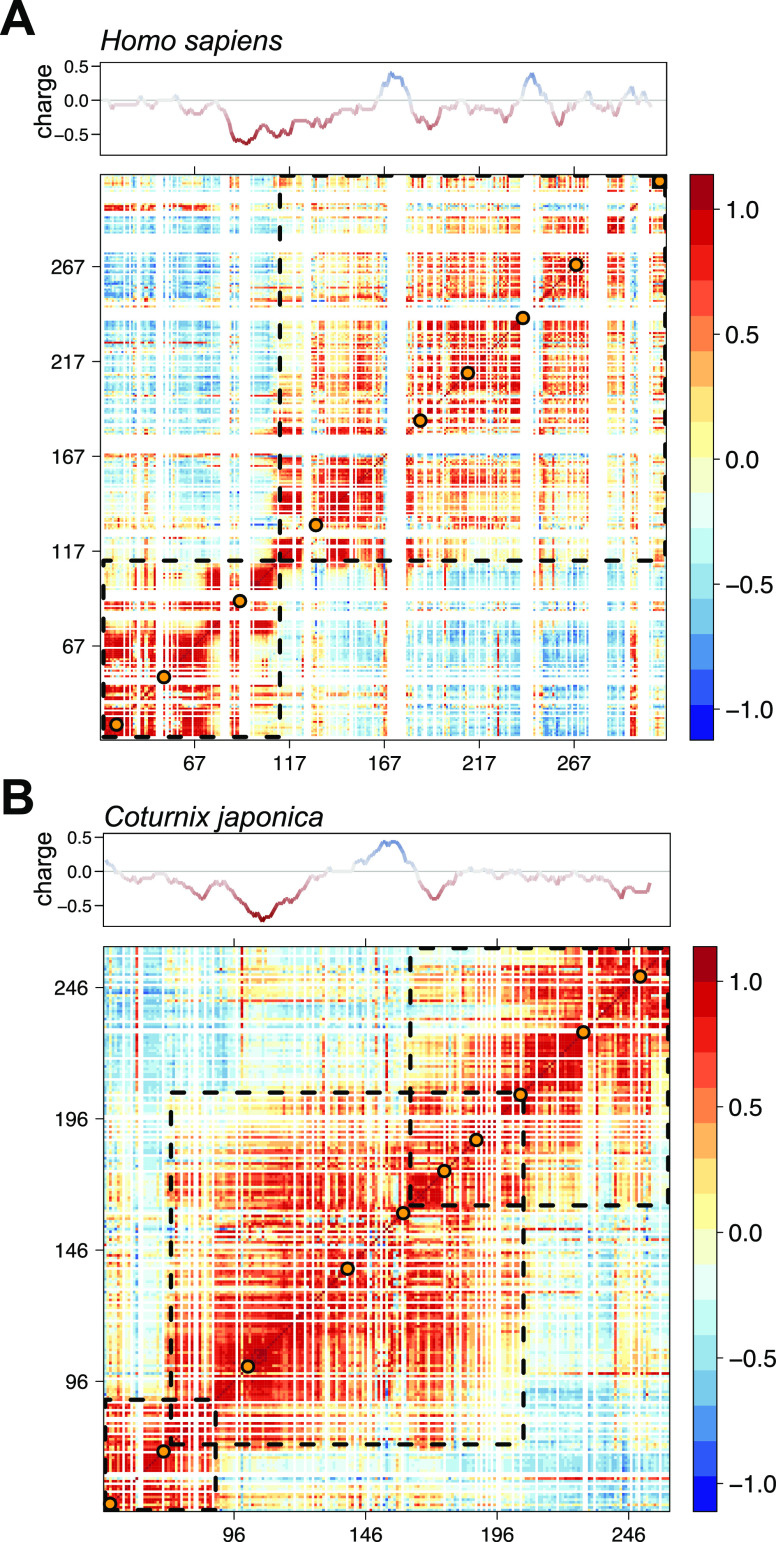
The seminal work of Tagliabbracci et al. on the characterization of Fam20C and its extracellular substrates reported the unexpected observation that Fam20C knockout MDA-MB-231 cells (i.e., no Fam20C-mediated phosphorylations) have superior adhesion properties.21 Moreover, Schytte et al. recently reported that the phosphorylation of an OPN construct, which covers the integrin binding motif, and full-length OPN, co-expressed with Fam20C, strongly hampers the interaction with αvβ3 integrin.66 OPN is not only the most phosphorylated substrate of Fam20C but also a natural binder to integrin receptors.48 Thus, its phosphorylation may have a major impact on the mediation of cell–ECM adhesion properties through integrin binding. Recent studies of Coturnix japonica OPN showed that an expansion of the compact states due to rational mutations of the hydrophobic residues of the central core region leads to higher affinities for heparin47 and lower affinities for integrins. Both C. japonica OPN and H. sapiens OPN form compact central states exploiting electrostatic attractions between differently charged regions as well as backbone hydrophobic interactions.67 The existence of compact substates in OPN has been demonstrated.46,47 Correlated conformational fluctuations within the structure of both H. sapiens OPN and C. japonica OPN are visualized in a Pearson correlation map (Figure 6),54,68 derived from multiple PRE rates. H. sapiens OPN (Figure 6A) reveals two compacted regions (residues 14–115 and 116-314), whereas C. japonica OPN (Figure 6B) reveals three compacted regions (residues 46–90, 80–200, and 160–247). Interestingly, the residue segments of H. sapiens OPN, where the phosphorylation sites are located, show significant correlations. Therefore, we conclude that (hyper)phosphorylation of OPN releases long-range correlations (by weakening stabilizing/attractive electrostatic interactions) and leads to the observed decompaction. Thereby, it abolishes energetically favorable interactions between OPN sites that are distant from the canonical RGD motif and integrin receptors.48,66 However, the central part that contains the (integrin binding) RGD motif (residues 159–161) retains its local rigidity (15N R2 rates) and maintains a preformed template for receptor recognition.
Figure 6.
Pearson correlation maps of (A) H. sapiens OPN determined from nine PRE profiles and (B) C. japonica OPN determined from 10 PRE profiles. The maps show correlated (red to orange), uncorrelated (light yellow to light blue), and anticorrelated (light blue to dark blue) structural fluctuations. The dashed squares enclose regions of distinct structural compaction. The orange dots represent the spin-label sites. The data for the C. japonica OPN correlation matrix were previously published.47 Corresponding charge plots are shown at the top.
Concluding Remarks
In conclusion, we investigated the effect of phosphorylation of OPN by Fam20C. To this end, an optimized protocol for in vitro phosphorylation has been developed using Fam20C expressed in mammalian HEK293T cells. Furthermore, almost complete assignment of phosphorylated S-x-E/pS motifs has been achieved by a combination of MS and NMR spectroscopy. NMR studies of the hyperphosphorylated OPN reveal an increase in flexibility in regions, which comprise the Fam20C phosphorylation sites, and weakened long-range interactions. The role of electrostatics and side chain–backbone interactions has emerged recently as a potential mechanism for modulating the formation of rigid segments and overall compaction.47,69 Moreover, weak side chain–backbone interactions involving proline residues are important for stabilizing OPN’s central compact state. Most importantly, the observed decompaction is also in accordance with the reported biological behavior of OPN (a decreased binding affinity for integrins) and illustrates the importance of compact states for molecular recognition events in which IDPs are involved. However, it is challenging to address the degree of decompaction in a quantitative manner because it is extremely difficult to know the extent to which those conformations are populated. At the same time, the existence of functional minor populations (or excited stats) in IDPs may play a key role in binding events. Low-resolution techniques such as SAXS may not fully grasp the subtleties of IDP ensembles. On the contrary, PRE data accentuate the minor populations that seem to be relevant for understanding OPN function. Furthermore, the unexpected proteoglycan binding preference (heparan sulfate vs hyaluronic acid) of OPN suggests an interaction specificity of IDPs and questions the notion of IDPs being fully disordered and exhibiting random-coil type behavior. To conclude, post-translational modifications, in our case phosphorylation, are effective mechanisms for modifying conformational ensembles of IDPs and populating suitable substates for molecular recognition events. Structural disorder is clearly not adequate for grasping the subtlety of these processes, and more sophisticated concepts have to be involved to fully appreciate how IDPs can respond to changing molecular environments and how they can engage in permanently varying protein interaction networks.
Acknowledgments
The authors thank Vincent Tagliabracci for providing the HEK293T-Fam20C-expressing cell line and useful discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.1c00050.
Materials and Methods, including protein/kinase production, protein phosphorylation, labeling, NMR spectroscopy, and MS experimental details; affinity column chromatogram of OPN; anion exchange chromatogram of (phosphorylated) OPN; sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western plot of Fam20C; phosphopeptide identification by MS; phosphoresidue identification by MS and NMR; secondary shift propensity of phosphorylated OPN; selectred NMR titration curves of OPN:heparin; series of 1H–15N HSQC NMR spectra of OPN:HA; 1H NMR spectra of OPN[+CD44HABD(+HA)]; and tables of backbone chemical shifts of phosphorylated OPN (BMRB ID: 50447)(PDF)
Tables of MS results for unphosphorylated OPN (XLSX)
Tables ofMS results for phosphorylated OPN (XLSX)
Accession Codes
Human OPN, P10451 (Uniprot); quail OPN, Q9I832 (Uniprot); human Fam20C, Q8IXL6 (Uniprot).
Author Contributions
⊥ B.M. and J.H. contributed equally to this work.
J.H. acknowledges Austrian Science Fund Grant (Lise Meitner Program) M2651. G.P. was funded by the Christian Doppler Laboratory for High-Content Structural Biology and Biotechnology, Austria. W.K. and S.Z. thank the Polish National Science Centre for Grant MAESTRO-2015/18/A/ST4/00270. R.K. gratefully acknowledges the financial support by the Austrian Federal Ministry for Digital and Economic Affairs, the National Foundation for Research, Technology and Development, and the Christian Doppler Research Association. This work was supported by the Austrian Science Foundation (FWF, Grants W1258 and P28937-B21).
The authors declare no competing financial interest.
Supplementary Material
References
- Iakoucheva L. M.; Radivojac P.; Brown C. J.; O’Connor T. R.; Sikes J. G.; Obradovic Z.; Dunker A. K. (2004) The Importance of Intrinsic Disorder for Protein Phosphorylation. Nucleic Acids Res. 32, 1037–1049. 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antz C.; Bauer T.; Kalbacher H.; Frank R.; Covarrubias M.; Kalbitzer H. R.; Ruppersberg J. P.; Baukrowitz T.; Fakler B. (1999) Control of K+ Channel Gating by Protein Phosphorylation: Structural Switches of the Inactivation Gate. Nat. Struct. Biol. 6, 146–150. 10.1038/5833. [DOI] [PubMed] [Google Scholar]
- Schwalbe M.; Kadavath H.; Biernat J.; Ozenne V.; Blackledge M.; Mandelkow E.; Zweckstetter M. (2015) Structural Impact of Tau Phosphorylation at Threonine 231. Structure 23, 1448–1458. 10.1016/j.str.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Martin E. W.; Holehouse A. S.; Grace C. R.; Hughes A.; Pappu R. V.; Mittag T. (2016) Sequence Determinants of the Conformational Properties of an Intrinsically Disordered Protein Prior to and upon Multisite Phosphorylation. J. Am. Chem. Soc. 138, 15323–15335. 10.1021/jacs.6b10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs E. B.; Lu F.; Portz B.; Fisher M. J.; Medellin B. P.; Laremore T. N.; Zhang Y. J.; Gilmour D. S.; Showalter S. A. (2017) Phosphorylation Induces Sequence-Specific Conformational Switches in the RNA Polymerase II C-Terminal Domain. Nat. Commun. 8, 15233. 10.1038/ncomms15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska A. A.; Zondlo N. J. (2006) Hyperphosphorylation of Tau Induces Local Polyproline II Helix. Biochemistry 45, 5527–5537. 10.1021/bi052662c. [DOI] [PubMed] [Google Scholar]
- Bah A.; Vernon R. M.; Siddiqui Z.; Krzeminski M.; Muhandiram R.; Zhao C.; Sonenberg N.; Kay L. E.; Forman-Kay J. D. (2015) Folding of an Intrinsically Disordered Protein by Phosphorylation as a Regulatory Switch. Nature 519 (7541), 106–109. 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- Bah A.; Forman-Kay J. D. (2016) Modulation of Intrinsically Disordered Protein Function by Post-Translational Modifications. J. Biol. Chem. 291, 6696–6705. 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona A.; Theillet F. X.; Foster C.; Cheng T. M.; Miralles F.; Bates P. A.; Selenko P.; Treisman R. (2016) Opposing Effects of Elk-1 Multisite Phosphorylation Shape Its Response to ERK Activation. Science 354, 233–237. 10.1126/science.aad1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang B.; Arsenault J.; Vernon R. M.; Lin H.; Sonenberg N.; Wang L. Y.; Bah A.; Forman-Kay J. D. (2019) Phosphoregulated FMRP Phase Separation Models Activity-Dependent Translation through Bidirectional Control of MRNA Granule Formation. Proc. Natl. Acad. Sci. U. S. A. 116, 4218–4227. 10.1073/pnas.1814385116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalak G.; Vogel V. (2012) Extracellular Phosphorylation and Phosphorylated Proteins: Not Just Curiosities but Physiologically Important. Sci. Signaling 5, re7. 10.1126/scisignal.2003273. [DOI] [PubMed] [Google Scholar]
- Tagliabracci V. S.; Pinna L. A.; Dixon J. E. (2013) Secreted Protein Kinases. Trends Biochem. Sci. 38, 121–130. 10.1016/j.tibs.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci V. S.; Engel J. L.; Wen J.; Wiley S. E.; Worby C. a; Kinch L. N.; Xiao J.; Grishin N. V.; Dixon J. E. (2012) Secreted Kinase Phosphorylates Extracellular Proteins That Regulate Biomineralization. Science 336, 1150–1153. 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek J.; Ganss B.; McKee M. D. (2000) Osteopontin. Crit. Rev. Oral Biol. Med. 11, 279–303. 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- Wai P. Y.; Kuo P. C. (2008) Osteopontin: Regulation in Tumor Metastasis. Cancer Metastasis Rev. 27, 103–118. 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- Shevde L. A.; Samant R. S. (2014) Role of Osteopontin in the Pathophysiology of Cancer. Matrix Biol. 37, 131–141. 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanecki C. C.; Uzwiak D. J.; Denhardt D. T. (2007) Control of Osteopontin Signaling and Function by Post-Translational Phosphorylation and Protein Folding. J. Cell. Biochem. 102, 912–924. 10.1002/jcb.21558. [DOI] [PubMed] [Google Scholar]
- Kariya Y.; Kanno M.; Matsumoto-Morita K.; Konno M.; Yamaguchi Y.; Hashimoto Y. (2014) Osteopontin O-Glycosylation Contributes to Its Phosphorylation and Cell Adhesion Properties. Biochem. J. 463, 93–102. 10.1042/BJ20140060. [DOI] [PubMed] [Google Scholar]
- Kubota T.; Zhang Q.; Wrana J. L.; Ber R.; Aubin J. E.; Butler W. T.; Sodek J. (1989) Multiple Forms of SppI (Secreted Phosphoprotein, Osteopontin) Synthesized by Normal and Transformed Rat Bone Cell Populations: Regulation by TGF-β. Biochem. Biophys. Res. Commun. 162, 1453–1459. 10.1016/0006-291X(89)90837-1. [DOI] [PubMed] [Google Scholar]
- Salih E.; Ashkar S.; Gerstenfeld L. C.; Glimcher M. J. (1997) Identification of the Phosphorylated Sites of Metabolically 32P- Labeled Osteopontin from Cultured Chicken Osteoblasts. J. Biol. Chem. 272, 13966–13973. 10.1074/jbc.272.21.13966. [DOI] [PubMed] [Google Scholar]
- Tagliabracci V. S.; Wiley S. E.; Guo X.; Kinch L. N.; Durrant E.; Wen J.; Xiao J.; Cui J.; Nguyen K. B.; Engel J. L.; Coon J. J.; Grishin N.; Pinna L. A.; Pagliarini D. J.; Dixon J. E. (2015) A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell 161, 1619–1632. 10.1016/j.cell.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B.; Nielsen M. S.; Haselmann K. F.; Petersen T. E.; Sorensen E. S. (2005) Post-Translationally Modified Residues of Native Human Osteopontin Are Located in Clusters: Identification of 36 Phosphorylation and Five O-Glycosylation Sites and Their Biological Implications. Biochem. J. 390, 285–292. 10.1042/BJ20050341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine J.; Winter R. M.; Davey A.; Tucker S. M. (1989) Unknown Syndrome: Microcephaly, Hypoplastic Nose, Exophthalmos, Gum Hyperplasia, Cleft Palate, Low Set Ears, and Osteosclerosis. J. Med. Genet. 26, 786–788. 10.1136/jmg.26.12.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. O.; Xu A.; Ogura E.; Manning G.; Irvine K. D. (2012) The Raine Syndrome Protein FAM20C Is a Golgi Kinase That Phosphorylates Bio-Mineralization Proteins. PLoS One 7, e42988. 10.1371/journal.pone.0042988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter G. K.; Kyle C. L.; Goldberg H. A. (1994) Modulation of Crystal Formation by Bone Phosphoproteins: Structural Specificity of the Osteopontin-Mediated Inhibition of Hydroxyapatite Formation. Biochem. J. 300, 723–728. 10.1042/bj3000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke A.; Qin C.; Spevak L.; Fujimoto Y.; Butler W. T.; Sørensen E. S.; Boskey A. L. (2005) Importance of Phosphorylation for Osteopontin Regulation of Biomineralization. Calcif. Tissue Int. 77, 45–54. 10.1007/s00223-004-1288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addison W. N.; Miller S. J.; Ramaswamy J.; Mansouri A.; Kohn D. H.; McKee M. D. (2010) Phosphorylation-Dependent Mineral-Type Specificity for Apatite-Binding Peptide Sequences. Biomaterials 31, 9422–9430. 10.1016/j.biomaterials.2010.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskey A. L.; Christensen B.; Taleb H.; Sørensen E. S. (2012) Post-Translational Modification of Osteopontin: Effects on in Vitro Hydroxyapatite Formation and Growth. Biochem. Biophys. Res. Commun. 419, 333–338. 10.1016/j.bbrc.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter G. K. (2013) Role of Osteopontin in Modulation of Hydroxyapatite Formation. Calcif. Tissue Int. 93, 348–354. 10.1007/s00223-013-9698-6. [DOI] [PubMed] [Google Scholar]
- de Bruyn J. R.; Goiko M.; Mozaffari M.; Bator D.; Dauphinee R. L.; Liao Y.; Flemming R. L.; Bramble M. S.; Hunter G. K.; Goldberg H. A. (2013) Dynamic Light Scattering Study of Inhibition of Nucleation and Growth of Hydroxyapatite Crystals by Osteopontin. PLoS One 8, e56764. 10.1371/journal.pone.0056764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iline-Vul T.; Nanda R.; Mateos B.; Hazan S.; Matlahov I.; Perelshtein I.; Keinan-Adamsky K.; Althoff-Ospelt G.; Konrat R.; Goobes G. (2020) Osteopontin Regulates Biomimetic Calcium Phosphate Crystallization from Disordered Mineral Layers Covering Apatite Crystallites. Sci. Rep. 10, 15722. 10.1038/s41598-020-72786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton S.; Seydel T.; Nylander T.; Holt C.; Härtlein M.; Teixeira S.; Zaccai G. (2015) Dynamic Footprint of Sequestration in the Molecular Fluctuations of Osteopontin. J. R. Soc., Interface 12, 20150506. 10.1098/rsif.2015.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton S.; Grimaldo M.; Roosen-Runge F.; Schreiber F.; Nylander T.; Clegg R.; Holt C.; Härtlein M.; García Sakai V.; Seydel T.; Marujo Teixeira S. C. (2017) Effect of Phosphorylation on a Human-like Osteopontin Peptide. Biophys. J. 112, 1586–1596. 10.1016/j.bpj.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellahcène A.; Castronovo V.; Ogbureke K. U. E.; Fisher L. W.; Fedarko N. S. (2008) Small Integrin-Binding Ligand N-Linked Glycoproteins (SIBLINGs): Multifunctional Proteins in Cancer. Nat. Rev. Cancer 8, 212–226. 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. F.; Zawaideh S.; Hikita S.; Kumar V. A.; Cantor H.; Ashkar S. (2002) Phosphorylation-Dependent Interaction of Osteopontin with Its Receptors Regulates Macrophage Migration and Activation. J. Leukocyte Biol. 72, 752–761. [PubMed] [Google Scholar]
- Nau G. J.; Liaw L.; Chupp G. L.; Berman J. S.; Hogan B. L. M.; Young R. A. (1999) Attenuated Host Resistance against Mycobacterium Bovis BCG Infection in Mice Lacking Osteopontin. Infect. Immun. 67, 4223–4230. 10.1128/IAI.67.8.4223-4230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amata I.; Maffei M.; Igea A.; Gay M.; Vilaseca M.; Nebreda A. R.; Pons M. (2013) Multi-Phosphorylation of the Intrinsically Disordered Unique Domain of c-Src Studied by In-Cell and Real-Time NMR Spectroscopy. ChemBioChem 14, 1820–1827. 10.1002/cbic.201300139. [DOI] [PubMed] [Google Scholar]
- Smith M. J.; Marshall C. B.; Theillet F. X.; Binolfi A.; Selenko P.; Ikura M. (2015) Real-Time NMR Monitoring of Biological Activities in Complex Physiological Environments. Curr. Opin. Struct. Biol. 32, 39–47. 10.1016/j.sbi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Theillet F. X.; Liokatis S.; Jost J. O.; Bekei B.; Rose H. M.; Binolfi A.; Schwarzer D.; Selenko P. (2012) Site-Specific Mapping and Time-Resolved Monitoring of Lysine Methylation by High-Resolution NMR Spectroscopy. J. Am. Chem. Soc. 134, 7616–7619. 10.1021/ja301895f. [DOI] [PubMed] [Google Scholar]
- Theillet F.; Rose H. M.; Liokatis S.; Binolfi A.; Thongwichian R.; Stuiver M.; Selenko P. (2013) Site-Specific NMR Mapping and Time-Resolved Monitoring of Serine and Threonine Phosphorylation in Reconstituted Kinase Reactions and Mammalian Cell Extracts. Nat. Protoc. 8, 1416–1432. 10.1038/nprot.2013.083. [DOI] [PubMed] [Google Scholar]
- Liokatis S.; Dose A.; Schwarzer D.; Selenko P. (2010) Simultaneous Detection of Protein Phosphorylation and Acetylation by High-Resolution NMR Spectroscopy. J. Am. Chem. Soc. 132, 14704–14705. 10.1021/ja106764y. [DOI] [PubMed] [Google Scholar]
- Theillet F. X.; Smet-Nocca C.; Liokatis S.; Thongwichian R.; Kosten J.; Yoon M. K.; Kriwacki R. W.; Landrieu I.; Lippens G.; Selenko P. (2012) Cell Signaling, Post-Translational Protein Modifications and NMR Spectroscopy. J. Biomol. NMR 54, 217–236. 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unione L.; Lenza M. P.; Ardá A.; Urquiza P.; Laín A.; Falcón-Pérez J. M.; Jiménez-Barbero J.; Millet O. (2019) Glycoprofile Analysis of an Intact Glycoprotein As Inferred by NMR Spectroscopy. ACS Cent. Sci. 5, 1554–1561. 10.1021/acscentsci.9b00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M.; Walczak M. J.; Aebi M.; Wider G. (2015) Posttranslational Modifications of Intact Proteins Detected by NMR Spectroscopy: Application to Glycosylation. Angew. Chem., Int. Ed. 54, 7096–7100. 10.1002/anie.201502093. [DOI] [PubMed] [Google Scholar]
- Konrat R. (2015) IDPs: Less Disordered and More Ordered than Expected. Biophys. J. 109, 1309–1311. 10.1016/j.bpj.2015.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbach D.; Schwarz T. C.; Platzer G.; Höfler S.; Hinderberger D.; Konrat R. (2014) Compensatory Adaptations of Structural Dynamics in an Intrinsically Disordered Protein Complex. Angew. Chem., Int. Ed. 53, 3840–3843. 10.1002/anie.201308389. [DOI] [PubMed] [Google Scholar]
- Mateos B.; Conrad-Billroth C.; Schiavina M.; Beier A.; Kontaxis G.; Konrat R.; Felli I. C.; Pierattelli R. (2020) The Ambivalent Role of Proline Residues in an Intrinsically Disordered Protein: From Disorder Promoters to Compaction Facilitators. J. Mol. Biol. 432, 3093–3111. 10.1016/j.jmb.2019.11.015. [DOI] [PubMed] [Google Scholar]
- Mateos B.; Sealey-Cardona M.; Balazs K.; Konrat J.; Staffler G.; Konrat R. (2020) NMR Characterization of Surface Receptor Protein Interactions in Live Cells Using Methylcellulose Hydrogels. Angew. Chem., Int. Ed. 59, 3886–3890. 10.1002/anie.201913585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkiewicz E. A.; Lumb K. J. (1999) Random-Coil Chemical Shifts of Phosphorylated Amino Acids. J. Biomol. NMR 15, 203–206. 10.1023/A:1008375029746. [DOI] [PubMed] [Google Scholar]
- Ulrich E. L.; Akutsu H.; Doreleijers J. F.; Harano Y.; Ioannidis Y. E.; Lin J.; Livny M.; Mading S.; Maziuk D.; Miller Z.; Nakatani E.; Schulte C. F.; Tolmie D. E.; Kent Wenger R.; Yao H.; Markley J. L. (2007) BioMagResBank. Nucleic Acids Res. 36, D402–D408. 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei-Ghaleh N.; Parigi G.; Soranno A.; Holla A.; Becker S.; Schuler B.; Luchinat C.; Zweckstetter M. (2018) Local and Global Dynamics in Intrinsically Disordered Synuclein. Angew. Chem., Int. Ed. 57, 15262–15266. 10.1002/anie.201808172. [DOI] [PubMed] [Google Scholar]
- Camilloni C.; De Simone A.; Vranken W. F.; Vendruscolo M. (2012) Determination of Secondary Structure Populations in Disordered States of Proteins Using Nuclear Magnetic Resonance Chemical Shifts. Biochemistry 51, 2224–2231. 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- Parigi G.; Rezaei-Ghaleh N.; Giachetti A.; Becker S.; Fernandez C.; Blackledge M.; Griesinger C.; Zweckstetter M.; Luchinat C. (2014) Long-Range Correlated Dynamics in Intrinsically Disordered Proteins. J. Am. Chem. Soc. 136, 16201–16209. 10.1021/ja506820r. [DOI] [PubMed] [Google Scholar]
- Silvestre-Ryan J.; Bertoncini C. W.; Fenwick R. B.; Esteban-Martin S.; Salvatella X. (2013) Average Conformations Determined from PRE Data Provide High-Resolution Maps of Transient Tertiary Interactions in Disordered Proteins. Biophys. J. 104, 1740–1751. 10.1016/j.bpj.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D.; Chen J. (2009) Structural Interpretation of Paramagnetic Relaxation Enhancement-Derived Distances for Disordered Protein States. J. Mol. Biol. 390, 467–477. 10.1016/j.jmb.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Jakob U.; Kriwacki R.; Uversky V. N. (2014) Conditionally and Transiently Disordered Proteins: Awakening Cryptic Disorder to Regulate Protein Function. Chem. Rev. 114, 6779–6805. 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M. M.; Kriwacki R. W.; Pappu R. V. (2012) Versatility from Protein Disorder. Science 337, 1460–1461. 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- Flock T.; Weatheritt R. J.; Latysheva N. S.; Babu M. M. (2014) Controlling Entropy to Tune the Functions of Intrinsically Disordered Regions. Curr. Opin. Struct. Biol. 26, 62–72. 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Mittag T.; Orlicky S.; Choy W.-Y.; Tang X.; Lin H.; Sicheri F.; Kay L. E.; Tyers M.; Forman-Kay J. D. (2008) Dynamic Equilibrium Engagement of a Polyvalent Ligand with a Single-Site Receptor. Proc. Natl. Acad. Sci. U. S. A. 105, 17772–17777. 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M.; Mittag T.; Pawson T.; Tyers M.; Forman-Kay J. D.; Chan H. S. (2007) Polyelectrostatic Interactions of Disordered Ligands Suggest a Physical Basis for Ultrasensitive. Proc. Natl. Acad. Sci. U. S. A. 104, 9650–9655. 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Azpurua J.; Hine C.; Vaidya A.; Myakishev-Rempel M.; Ablaeva J.; Mao Z.; Nevo E.; Gorbunova V.; Seluanov A. (2013) High-Molecular-Mass Hyaluronan Mediates the Cancer Resistance of the Naked Mole Rat. Nature 499, 346–349. 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G. J. (2015) Cancer Resistance, High Molecular Weight Hyaluronic Acid, and Longevity. J. Cell Commun. Signal. 9, 91–92. 10.1007/s12079-015-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio C.; Barile E.; Di Sorbo G.; Kipps T. J.; Pellecchia M. (2016) The Cell Surface Receptor CD44: NMR-Based Characterization of Putative Ligands. ChemMedChem 11, 1097–1106. 10.1002/cmdc.201600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M.; Terasawa H.; Sakakura M.; Yamaguchi Y.; Kajiwara M.; Kawashima H.; Miyasaka M.; Shimada I. (2003) Hyaluronan Recognition Mode of CD44 Revealed by Cross-Saturation and Chemical Shift Perturbation Experiments. J. Biol. Chem. 278, 43550–43555. 10.1074/jbc.M308199200. [DOI] [PubMed] [Google Scholar]
- Weber G. F.; Ashkar S.; Glimcher M. J.; Cantor H. (1996) Receptor-Ligand Interaction Between CD44 and Osteopontin (Eta-1). Science 271, 509–512. 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- Schytte G. N.; Christensen B.; Bregenov I.; Kjøge K.; Scavenius C.; Petersen S. V.; Enghild J. J.; Sørensen E. S. (2020) FAM20C Phosphorylation of the RGDSVVYGLR Motif in Osteopontin Inhibits Interaction with the αvβ3 Integrin. J. Cell. Biochem. 121, 4809–4818. 10.1002/jcb.29708. [DOI] [PubMed] [Google Scholar]
- Kurzbach D.; Platzer G.; Schwarz T. C.; Henen M. a; Konrat R.; Hinderberger D. (2013) Cooperative Unfolding of Compact Conformations of the Intrinsically Disordered Protein Osteopontin. Biochemistry 52, 5167–5175. 10.1021/bi400502c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbach D.; Beier A.; Vanas A.; Flamm A. G.; Platzer G.; Schwarz C.; Konrat R. (2017) NMR Probing and Visualization of Correlated Proteins. Phys. Chem. Chem. Phys. 19, 10651–10656. 10.1039/C7CP00430C. [DOI] [PubMed] [Google Scholar]
- Escobedo A.; Topal B.; Kunze M. B. A.; Aranda J.; Chiesa G.; Mungianu D.; Bernardo-Seisdedos G.; Eftekharzadeh B.; Gairí M.; Pierattelli R.; Felli I. C.; Diercks T.; Millet O.; García J.; Orozco M.; Crehuet R.; Lindorff-Larsen K.; Salvatella X. (2019) Side Chain to Main Chain Hydrogen Bonds Stabilize a Polyglutamine Helix in a Transcription Factor. Nat. Commun. 10, 2034. 10.1038/s41467-019-09923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.