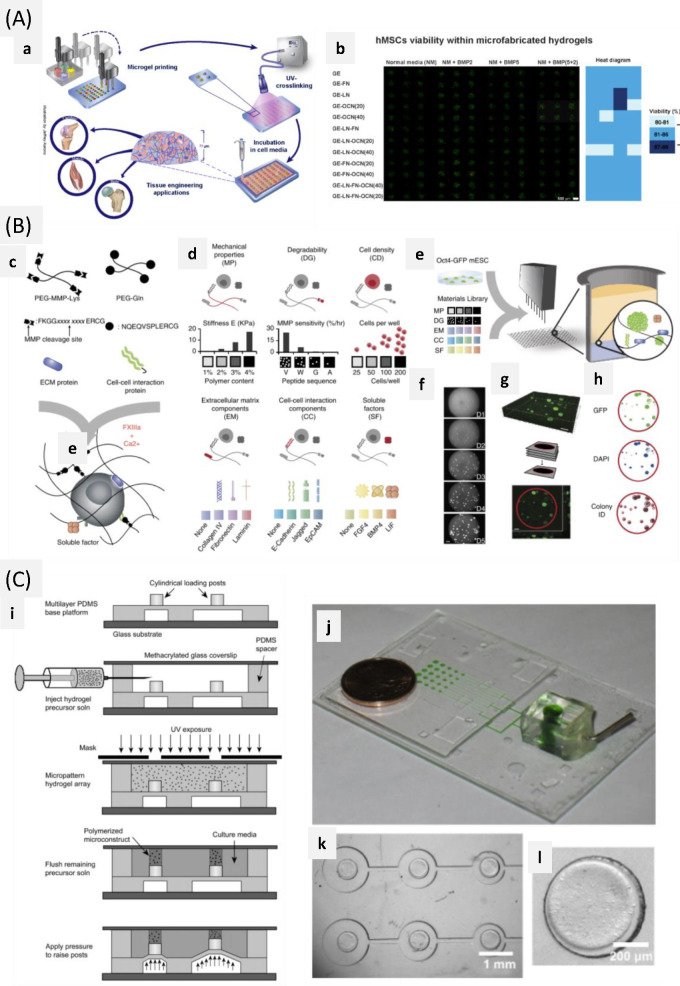
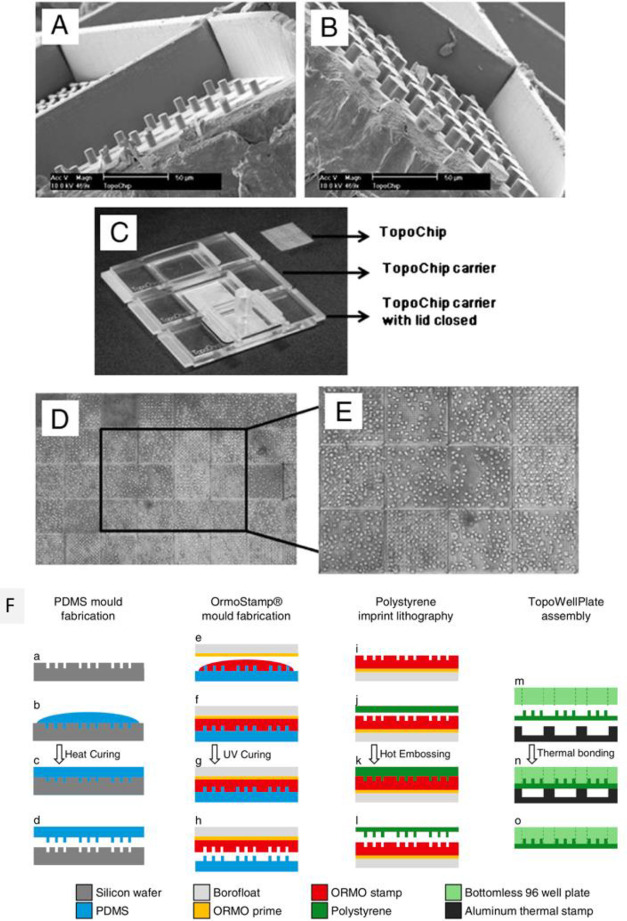
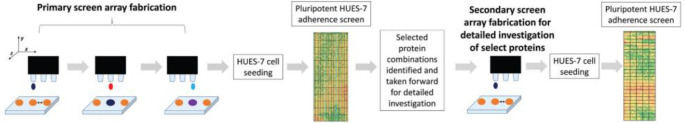
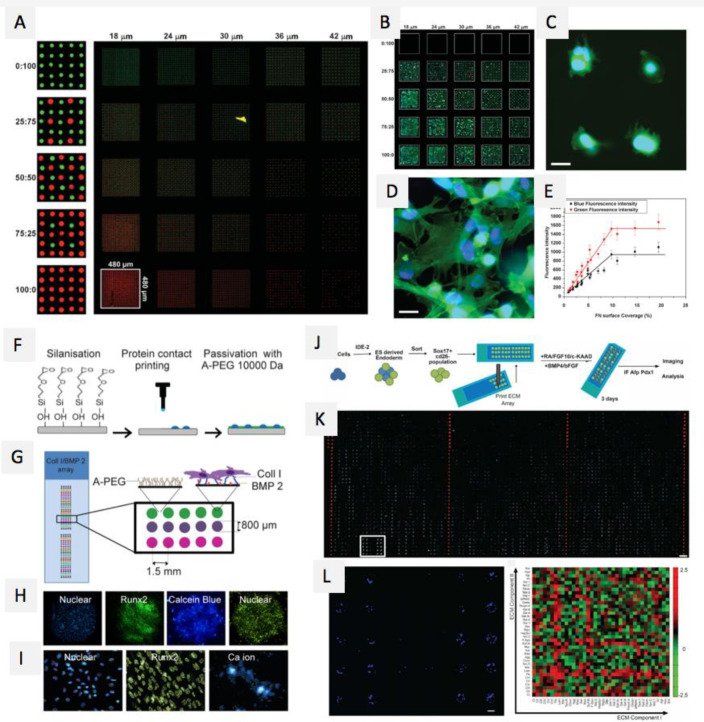
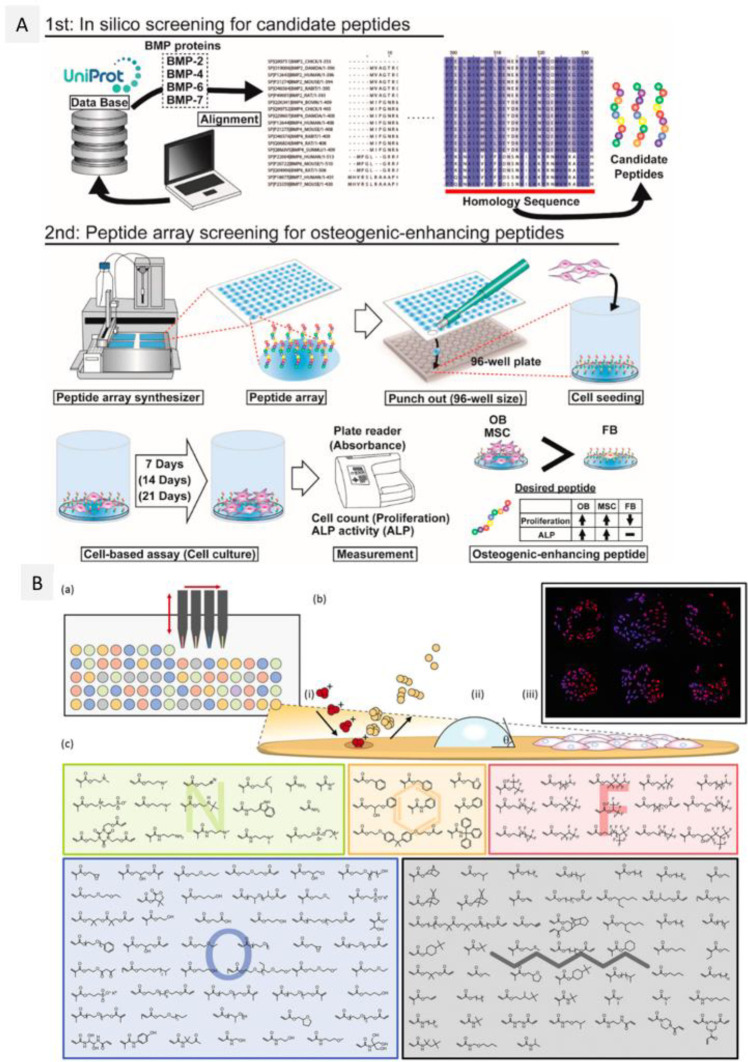
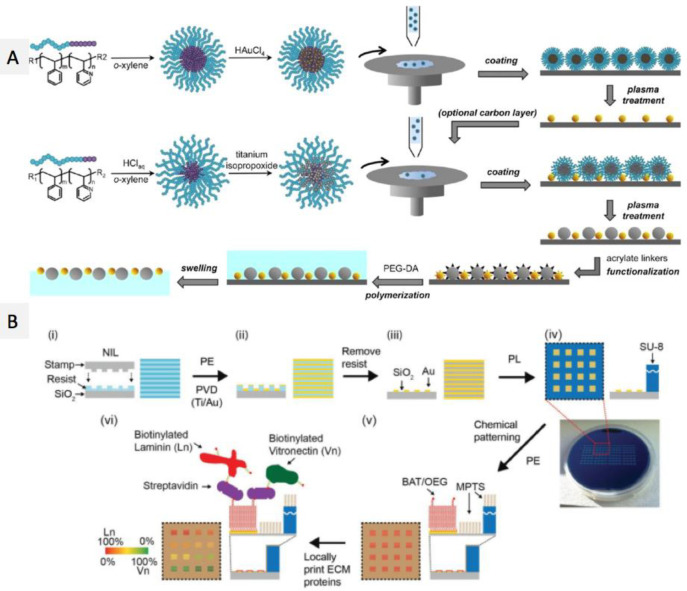
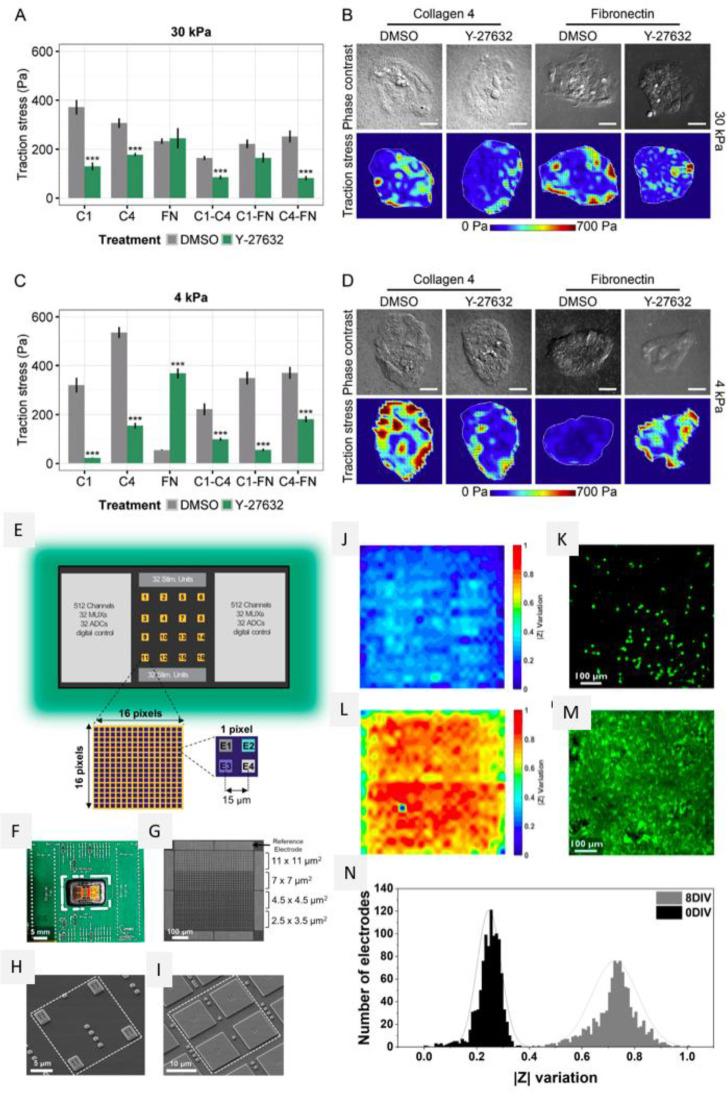
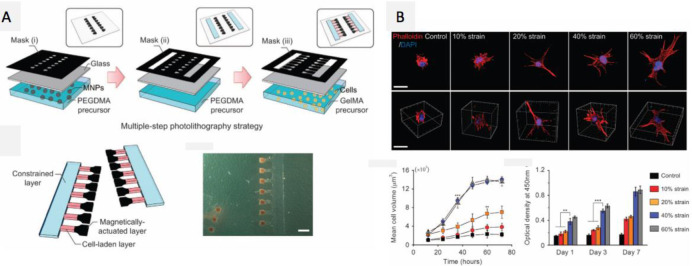
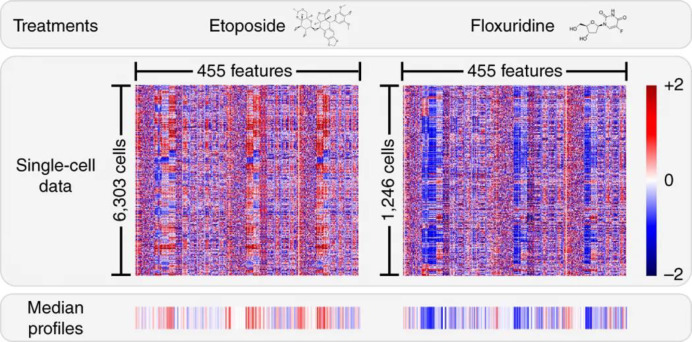
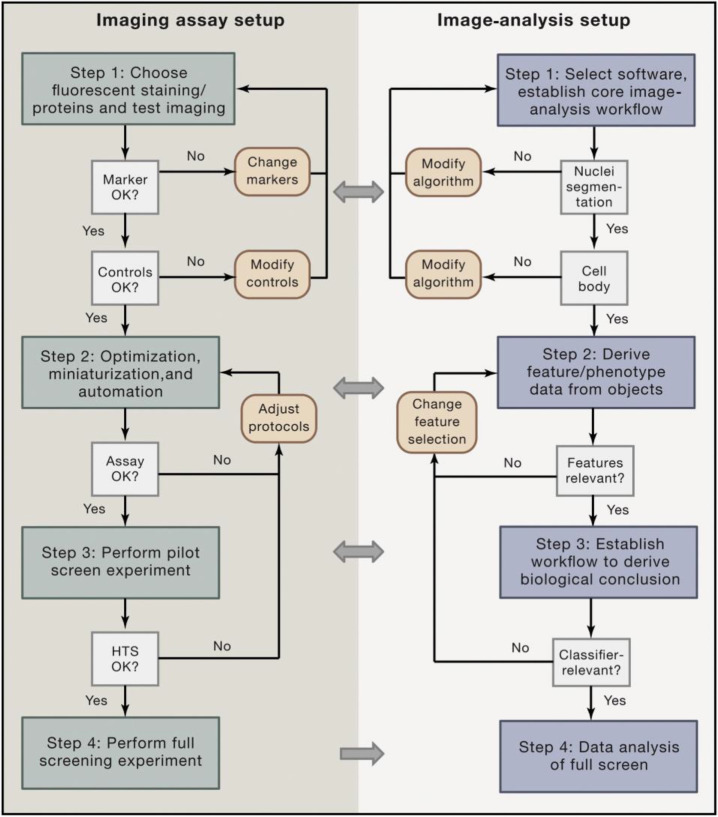
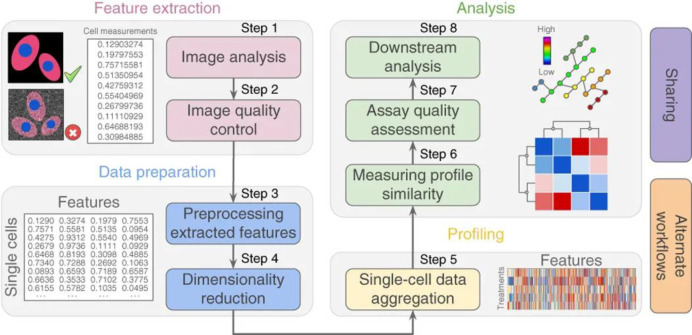
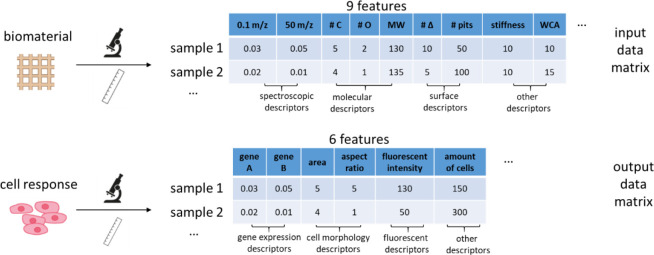
Abstract
The complex interaction of cells with biomaterials (i.e., materiobiology) plays an increasingly pivotal role in the development of novel implants, biomedical devices, and tissue engineering scaffolds to treat diseases, aid in the restoration of bodily functions, construct healthy tissues, or regenerate diseased ones. However, the conventional approaches are incapable of screening the huge amount of potential material parameter combinations to identify the optimal cell responses and involve a combination of serendipity and many series of trial-and-error experiments. For advanced tissue engineering and regenerative medicine, highly efficient and complex bioanalysis platforms are expected to explore the complex interaction of cells with biomaterials using combinatorial approaches that offer desired complex microenvironments during healing, development, and homeostasis. In this review, we first introduce materiobiology and its high-throughput screening (HTS). Then we present an in-depth of the recent progress of 2D/3D HTS platforms (i.e., gradient and microarray) in the principle, preparation, screening for materiobiology, and combination with other advanced technologies. The Compendium for Biomaterial Transcriptomics and high content imaging, computational simulations, and their translation toward commercial and clinical uses are highlighted. In the final section, current challenges and future perspectives are discussed. High-throughput experimentation within the field of materiobiology enables the elucidation of the relationships between biomaterial properties and biological behavior and thereby serves as a potential tool for accelerating the development of high-performance biomaterials.
1. Introduction
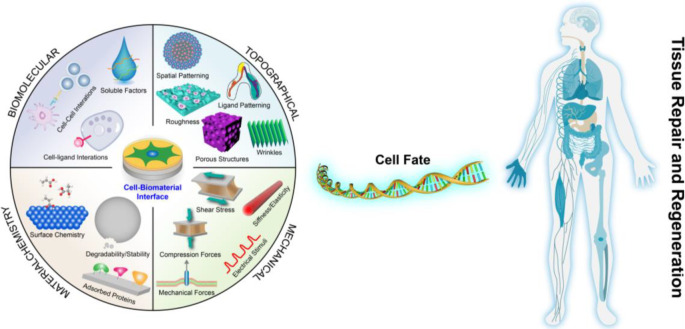
Faced with an ever-increasing burden of disease, congenital abnormalities, and accidents each year, tissue engineering and regenerative medicine (TERM) hold great promise to repair or replace tissues or even entire organs on demand for a better quality of life, which has been gaining widespread attention.1−7 In human tissues and organs, every cell is exposed to an intricate 3D network of nanofibers and specific molecules or components named the extracellular matrix (ECM), which is composed of proteins and glycosaminoglycans.8,9 For that reason, numerous research groups have been trying to design and develop biomaterials that include features (Figure 1) of the ECM (biochemical or physicochemical) or reconstruct its interface based on recent discoveries about the macro-/micro-/nano-/molecular-level architecture of the natural ECM and its interaction with cells to treat, repair, or regenerate tissue and its functions for basic biological studies and clinical applications.10−13
Figure 1.
Variables within the cell–microenvironment interface can invoke a biological response and decide cell fate in the process of tissue repair and regeneration.
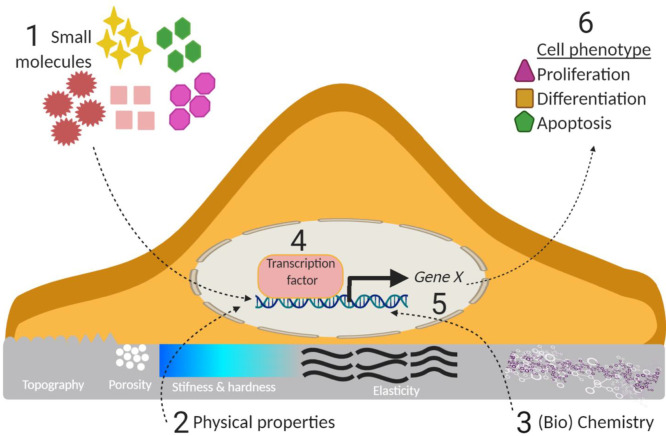
“Biomaterials are those materials—be it natural or synthetic, alive or lifeless, and usually made of multiple components—that has been engineered to interact with biological systems for a medical purpose—either a therapeutic (treat, augment, repair, or replace a tissue function of the body) or a diagnostic one.”.14,15 Previously, biomaterials have been considered as a passive supporting substrate in which the resident cells were regarded as the major actors. Biomaterials are undoubtedly increasingly recognized as a bioactive structure, which offers structural, mechanical, and compositional signals that can direct cell activities and functions in the natural process of tissue regeneration.16,17 Biomaterials play a pivotal role in the development of implants, biomedical devices, and tissue engineering scaffolds to treat diseases, aid in the restoration of bodily functions, or construct healthy tissues or regenerate diseased ones.18,19 While not too long ago, inert biomaterials were thought to be the best approach not to intervene with biological processes, in fact, there is no such thing as inert. It was found that cells are inherently sensitive to their surrounding microenvironment including (bio)materials, any material for that matter, and cells respond to the properties of these materials (Figure 1) such as the physical cues (e.g., mechanical properties, wettability, 2D topography, and 3D geometry), (bio)chemical signals (e.g., material component and ECM proteins), or other stimuli.5,20 It is well documented that these biomaterial properties can regulate cell behaviors (e.g., cell adhesion, migration, proliferation, and the self-maintenance or differentiation of stem cells) and tissue/organ functions. Initially, most of these studies used independent substrates with different and randomly selected biomaterial properties, which provided interesting yet limited information. The conventional approaches are incapable of screening the huge amount of potential material parameter combinations to identify the optimal cell responses, and involves a combination of serendipity and many series of trial-and-error experiments. For TERM, highly efficient and complex bioanalysis platforms are expected to explore smart and biomimetic biomaterials that offer desired complex microenvironments during healing, development, and homeostasis.21
With the advent of new fabrication technologies and advanced analytic equipment, high-throughput screening (HTS) platforms (e.g., gradient and microarray) provide an ideal strategy to analyze thousands of combinations of interactions among biomaterials, biomolecules, and cells for cell–material interaction and drug screening.21−30 Numerous research groups developed gradient surfaces with different material features (e.g., topography, stiffness, wettability, or chemical/biochemical composition) to successfully identify the optimal cell responses (such as adhesion, spreading, proliferation, and differentiation)31−39 and study cell-directed migration (e.g., topotaxis, chemotaxis, and durotaxis).40−49 The microarray also offers a powerful platform for the high-throughput assessment of material-cell interactions or biomolecular influences (e.g., DNA, growth factor, etc.) by presenting a single substrate at specific addressed locations.21,50 From the point of dimension, conventional 2D HTS platforms provided valuable information and great insight into key cell–material interaction mechanisms responsible for cellular events. As compared to 2D platforms, 3D HTS platforms can mimic certain physiological properties of the in vivo 3D microenvironment. This HTS approach is time and cost-efficient, expedites analysis procedures, combines multiple factors on a single platform, elicits more accurately the optimum material conditions for promoting cellular processes, and minimizes systematic or methodological errors. Revolutionary advances provide a better understanding of the structure–function relationships between surface properties and biological performance and identify the optimal condition for cell response. These approaches enable researchers to precisely adjust cell responses by optimizing the features of biomaterials, accelerate the development of high-quality biomaterials and then propel the clinical translation of biomaterials.
Although 2D and 3D HTS platform technologies have been developing rapidly, it remains a huge challenge to collect and analyze big data efficiently and accurately. Recently, HTS platforms based on microfluidics offer important advantages over conventional analysis systems, for example, integration of sensors for direct readout, higher reliability, and the possibility to enhance the throughput of screening by utilizing parallelization, multiplexing, and automation.24 Microfluidics-based HTS platforms may result in more breakthrough discoveries in both fundamental research and clinical application. In addition, array/grad-wellplates (array or gradient surfaces integrated into commercial well plate technology) were developed, which consists of a standard well plate with bottoms containing arrays or gradients.51−54 The array/grad-wellplate offers several key merits compared to the prior technologies, including preventing cross-communication of cells and cross-contamination of soluble factors, incorporate high throughput assays such as ELISA, as well as allowing for robotic liquid handling and implementation of multiwell plate-based instrumentation. Importantly, the array/grad-wellplate enables straightforward studying of synergetic effects of drugs/biomolecular (gene, protein, enzyme) and material properties, to identify the optimal conditions. Therefore, combinatorial HTS platforms with real-time detection and high-content analysis systems are being developed for deeper exploration of the interactions between cells and biomaterials, which significantly advances the field of biomaterials for specific target applications. It is expected that the true success of HTS platforms will rely on their translation toward commercial uses and the clinic, for example, drug discovery, toxicology, pharmaceutical science, and cellular therapies.
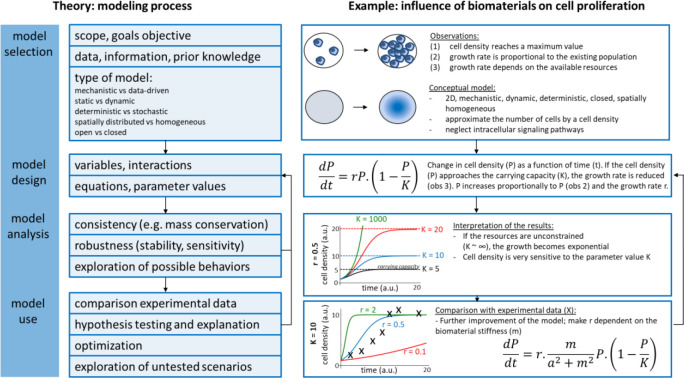
Clearly, high-throughput platforms enable measuring hundreds of parameters and conduct thousands of experiments at relatively low cost, which offer valuable and powerful tools for deciphering cell–material interactions. However, subsequent investigations have raised more questions than they answered, especially facing with massive amounts of information and data.55 Recently, computational simulations based on machine learning algorithms and mathematical modeling were used to deal with these data that relate the biomaterial properties as input to the cell behaviors as output.56−58 More holistic biological knowledge can be generated when biological-omics approaches are performed on cells that are in contact with biomaterial. With the advent of next-generation sequencing and the ready availability of commercial options to produce transcriptomics data, more and more researchers perform transcriptomics analysis. New biological mechanisms have thus been uncovered59 and give mechanistic insight into the molecular biology behind material-induced responses. One of the strengths of -omics experiments is that a high degree of standardization in the generation of the data has been achieved by the industry, and a lot of open-access software is available for data analysis. The next challenge for the field of material engineering is to achieve an equally high level of standardization in material characterization, or in other words, the metadata describing the biomaterials analyzed in transcriptomics experiments should be well registered. To this end, a publically available database was created and named The Compendium for Biomaterial Transcriptomics (cBiT).60 It allows researchers to search, select, and download materiobiology data as well as invite scientists to contribute their data to cBiT. Cloud platform, even materiobiology genome, could be built to help scientists to pose explicit hypotheses or make predictions of biomaterial parameters as a function of cell response.
In this review, we first introduce materiobiology and its HTS. Then we present an in-depth of the recent progress of 2D/3D HTS platforms (i.e., gradient and microarray) in the principle, preparation, screening for materiobiology, and combination with other advanced technologies. cBIT and high content imaging (HCI), computational simulations, and their translation toward commercial and clinical uses are highlighted. In the final section, current challenges and future perspectives are discussed.
2. Materiobiology
Until now, it is well-demonstrated that there is a close correlation between material features and biological responses.5−7 Increasing evidence has indicated that the physicochemical properties of biomaterials can decide cell survival, adhesion, morphology (e.g., cell shape, spreading, elongation, and alignment), proliferation, stiffness, migration, function, the pluripotency or differentiation of stem cells as well as even tissue repair and regeneration. Recent advances have reached a consensus view that manipulating the properties of implantable materials, such as biophysical and biochemical cues, can precisely regulate cell responses ranging from the macroscopic to the molecular level.12,61−63 On the basis of the information above, Li and co-workers proposed the concept of “materiobiology”: “Materiobiology is a scientific discipline that studies the biological effects of the properties of biomaterials on biological functions at different levels (e.g., cells, tissues, organs, and the whole organism)”.7 The presentation of this new concept further highlights and motivates the synergism between cell biology and biomaterial technologies that jointly accelerate the clinical and commercial translation of biomaterials.
2.1. Biophysical Cues Eliciting Cell Responses
The biophysical cues of biomaterials, including topography and stiffness, serve as an important indirect signal (Figure 1), which can be sensed at the cell membrane receptors and initiate intracellular signaling cascades through mechanosensing and mechanotransduction.64−67 These cues are increasingly considered as key mechanical regulators of cell fate and tissue regeneration. These cues can be transduced into intracellular biochemical information through the integrin-focal adhesion cytoskeleton actin transduction pathway, and vice versa the intercellular signals can be transformed back to dynamic mechanical signals (e.g., traction forces). Therefore, the degree of biophysical cues from biomaterials independently or synergistically can mediate cell behavior, function, and fate.
2.1.1. Stiffness Eliciting Cell Response
Tissues in the human body have various mechanical characteristics ranging from soft (brain, ∼0.1 kPa), to moderately stiff (skin and muscles, ∼10 kPa) and stiff (precalcified bone, >1 GPa).68,69 The mechanical features of the ECM in which cells reside were found to be correlated with cell adhesion, morphology, stiffness, proliferation, migration, and differentiation.70,71 Biomaterial stiffness has been regarded as an important regulator for cell fate.63 It was found that cell spreading and stiffness increased with increasing substrate stiffness, because cells on a stiffer substrate induce the maturation of focal adhesions and reorganize their actin cytoskeleton into stress fibers.64 Also, it was found that cells can recruit nearby fibers on lower stiffness substrates via cellular forces, which increased ligand density at the cell surface and thereby facilitated the generation of FAs and cell spreading.70,72 Interestingly, the effects of biomaterial stiffness on cell behaviors were dependent on cell types.73 For instance, neutrophils are not sensitive to the substrate stiffness at all.70,74 For the proliferation, fibroblasts, endothelial cells, and epithelial cells displayed better proliferation on stiffer surfaces, whereas neural stem cells showed enhanced proliferation on softer substrates, likely due to the softer brain tissue.75 Engler et al. reported that MSCs displayed different morphologies and phenotypes when cultured on polyacrylamide (PAAm) substrates with stiffness ranging from 1 to 40 kPa.76 MSCs cultured on a soft substrate (1 kPa) induced more specific neuronal cytoskeleton markers (ß-3-tubulin), whereas cells on stiffer samples (11 and 34 kPa) showed expression of early myogenic and osteogenic transcription factors, such as MyoD and CBFα-1, respectively. In addition, changing the substrate stiffness could influence noninvasive gene delivery, regulating a cell’s ability to uptake exogenous molecules.77 Taken together, engineered topographical and mechanical cues on substrates are powerful tools for directing cell interactions with the ECM.
2.1.2. Topography Eliciting Cell Response
For biomaterial topographical cues, there are different scales (i.e., macroscale, microscale, and nanoscale), patterns (i.e., isotropic and anisotropic), etc.65,78−80 It is well-known that in the body, tissues and organs have specific macro-shapes/sizes, such as ear, nose, skin, bone, and heart. Proper macroscale design for tissue engineering scaffolds is highly important because the shape and size can influence the integration with adjacent tissues, the efficiency of tissue repair and regeneration, which can be conveniently fabricated by custom-designed molds or computer-aided additive manufacturing technologies (e.g., 3D/4D printing). In the natural ECM, there is a highly porous microstructure with water and soluble factors filling.8,9,16 Biomimetic materials with a microporous structure can not only provide a large surface area for cell adhesion, but also enable efficient transport of nutrition and waste as well as allowing cells to grow into the scaffold for generating real tissues. In detail, some key porous features, such as porosity, pore size, and interconnectivity, have been demonstrated to have significant effects on cell responses.81 Generally, the improvement in pore size, porosity, or interconnectivity is able to enhance cell ingrowth, ECM secretion, stem cell differentiation, tissue formation, soluble factor diffusion, and vascularization. The optimal range of pore size varies for specific cells and tissues. For instance, the range of pore size with 200–400 μm was demonstrated to be effective for the repair and regeneration of bone tissue.82 The pore size of 50–200 μm was appropriate for the growth of smooth muscle cells.83In vivo, macrophages tended to the pro-inflammatory type M1 macrophage polarization induced by the scaffolds with a pore size above 80 μm.84 Nanoscale pores with the high specific surface area could facilitate cell attachment.85 In addition, it was reported that 90% porosity in the interconnected porous structure is ideal for the flow of nutrients, cell penetration, ECM deposition, and thereby promoting the growth of peripheral nerve and neovascularization.86 The 3D printed scaffolds and hydrogels are often used as microporous materials. Further, ultrastructural analysis of the native ECM in tissues indicates that the framework is composed of fibers with different diameters and structures. Fibrils varying in diameter are from a few nanometers to ∼150 nm and actual fiber bundles from several hundred nanometers to ∼400 μm in diameter.87−89 Nanoscale structural features have been widely applied in the biomedical fields owing to their ECM like structure, high surface area, structural diversity, favorable biological properties, and sufficient mechanical strength, which can significantly affect cell adhesion, morphology, proliferation, migration, and differentiation, as well as subcellular structures. The diameter of nanofibers can significantly affect cell behaviors. However, the ideal size of nanofiber for different types of cells and conditions varies substantially. For instance, it was found that the adhesion of human mesenchymal stem cells on the fibers with 1 μm diameter was better than that of 500 nm fibers,90 while the fibroblast 3T3 adhesion on the fibers with 425 nm diameter was more as compared to the fiber samples with 641 and 900 nm.91 The elongation of human endothelial cells on the fibers with 1–5 μm was more than that of nanofiber films with 10–200 nm.92 The formation of calciumphosphate in osteoblasts cultured on 60 and 100 nm carbon fibers was greatly enhanced as compared to those on 125 and 200 nm fiber.93 Strategies for preparing nanostructures include phase separation, lithography, electrospinning, and self-assembly.94
In addition to the material dimension, topographical patterns are critical to cell fate. There are two kinds of tissues depending on the ECM organization, that is, isotropic and anisotropic tissues.88,95 Anisotropic ECM structures and their interaction with cells have induced great interest among investigators. As demonstrated, anisotropic micro/nanotopographies (e.g., groove, grating, wrinkle, aligned fibers, etc.) that replicate the natural ECM in vivo of anisotropic tissues (e.g., blood vessels, nerve, cardiac muscle, tendon, and ligament), can regulate cell morphology (alignment and elongation), migration, ECM reorganization, and differentiation through contact guidance.96,97 For instance, aligned nanofibers offered specific contact guidance for tendon fibroblasts, oriented the cells, and promoted the secretion of ECM into an ordered structure.98 Also, smooth muscle cells cultured on aligned fibers expressed more contractile-related genes compared to those on the random fibers.99 Also, the size of aligned topography can greatly affect cell behaviors.34,100
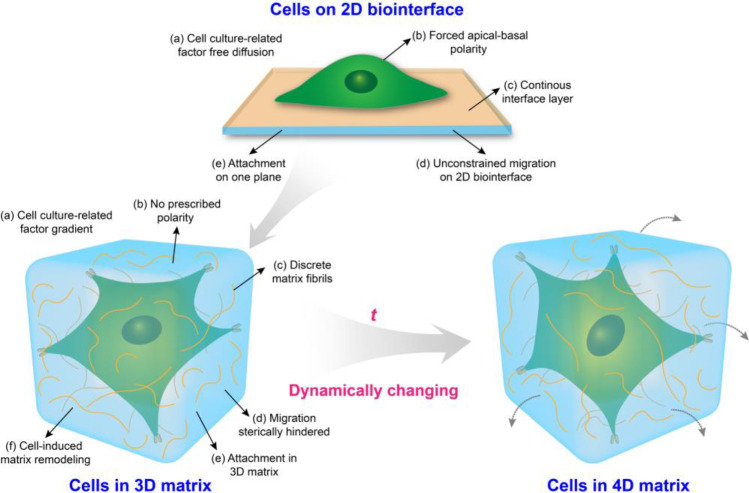
In the past few decades, conventional two-dimensional (2D) cell culture systems as a simplified approach have provided fundamental insights in decoding the effects of biochemical and biophysical cues on cell fate due to the relatively high accessibility and reproducibility (Figure 2). Till now, the most frequently used substrate culturing cells is rigid polystyrene tissue-culture plastic (TCP). However, in vitro 2D culture conditions do not fully capture the physicochemical features experienced by cells in the body.101 Increasing evidence suggests that cells cultured on 2D surfaces differ significantly from those grown in vivo. As compared to 2D substrates, bioinspired 3D geometry that recapitulates as many aspects of the natural ECM as possible, provides combinative biochemical/physical cues to mediate cell responses (Figure 2).102 For instance, the speed and direction of cell migration on 2D substrates are mediated by the actin cytoskeleton, integrin-related attachment, and myosin-associated contraction. However, cell migration in a 3D geometry is complicated, involving surrounding stiffness, the excitation of the nuclear piston, microtubule dynamics, etc. In addition, cell culture-related factors (such as medium, oxygen, growth factors, and differentiation inducer) on 2D substrates can undergo free diffusion, while these factors can be limited in a 3D geometry, generating gradients (Figure 2).62 Also, dynamic mechanostructural changes play a critical role in mediate cell responses in embryonic development as well as tissue repair and regeneration. The development of 4D biomimetic matrices respond to external stimuli, for example, heat, light, and magnetic fields, gained much attention in biomedical engineering.103
Figure 2.
Schematic diagram of the interactions between cells and biointerface/matrix.
2.2. Biochemical Cues Eliciting Cell Response
In addition to (bio)physical parameters, the (bio)chemical cues on biomaterials can significantly affect cell responses (Figure 1), such as cell adhesion, morphology, migration, proliferation, and differentiation.5,7,12,104 Therefore, the chemical modification of biomaterials provides the potential to govern cell behaviors.
Natural ECM contains many bio(macro)molecules, such as glycoproteins, proteoglycans, glycosaminoglycans, collagens, laminins, and growth factors, which have the ability to enhance cell–ECM adhesions via integrin binding.9 They can be used to improve most synthetic materials without adhesion ligands. Particularly, as compared to the use of diffusible growth factors, immobilized growth factors in biomaterials may enable prolonged delivery of growth factors, resist enzymatic degradation, and modulate specific growth factor bioactivity and signaling.105,106 Therefore, optimizing the chemical properties of biomaterials via growth factor or peptide immobilization is a key design consideration. Cell adhesion can also be controlled by the direct decoration from small molecules, such as peptides, and oligosaccharides.107,108 In addition, some biopolymers, such as collagen and its derivative (gelatin), alginate, chitosan, hyaluronic acid (HA), etc., can be fabricated into hydrogels as a 3D scaffold.109
The chemical functional groups on biomaterials can significantly influence the mass and conformation of the adsorbing proteins and then regulate cell response, which plays a critical role in the subsequent cellular behaviors.110,111 When biomaterials are placed into a biological environment, cells will not directly respond to the material surface but always via a protein conditioning film that originates from either the culture medium supplemented with fetal bovine serum (FBS) or proteins from biological fluids such as blood, saliva, etc. This protein adsorption is generally much faster than the cell adhesion events and hence any alterations in this film will directly affect cellular behavior.112
Generally, different chemical functional groups on biomaterials have various performances, such as wettability, solubility, reactivity, charge, and so on. Surface wettability, indicating interface energies of the biomaterial surface (quantified as water contact angle, WCA), has previously been correlated with protein adsorption and cell behavior.110,111 Wei and co-workers investigated the effect of surface wettability on competitive protein adsorption (albumin, Alb; fibronectin, Fn) and found that Fn adsorbed more on hydrophilic surfaces, whereas Alb predominantly adsorbed on hydrophobic surfaces. The initial attachment of osteoblastic cells increased with increasing surface wettability, which correlated well with Fn adsorption in the competitive mode.113 In addition, surface wettability is critical for cell spreading and differentiation. Generally, cells have good spreading, proliferation, and differentiation on hydrophilic surfaces. Mouse osteoblasts on hydrophilic surfaces ranging from 24 to 31° WCA, showed higher metabolic activity and expressed more osteogenic proteins (alkaline phosphatase (ALP) and osteocalcin (OCN)) than those on unmodified counterparts (WCA 72°).114
Surface wettability originating from material chemical functionalities (e.g., positive, negative, or neutral) can affect protein adsorption and then mediate cell response. For instance, Lee and co-workers described how alkylsilane self-assembled monolayers (SAM) with different functional groups (OH, COOH, NH2, and CH3) affected 125I-labeled fibronectin adsorption where at pH 7, COOH is actually COO- and NH2 becomes NH3+ while OH remains unaffected. They found the adhesion constant and binding efficiency of the adsorbed Fn for the α5β1 integrins (CH3 ≈ NH2 < COOH ≈ OH). Fibronectin interacted more strongly with α5β1 integrins when adsorbed on COOH versus OH surfaces suggesting that negative charge may be a critical component of inducing efficient cellular adhesion.115 The above studies indicate that the specific biomaterial chemistry plays a major role in protein/cell–material interaction and this chemistry is reflected by the numerous different synthetic polymers and inorganic materials used as biomaterials. In addition, it illustrates that relatively simple concepts such as wettability, are far more complex with large consequences for cell–material interactions. Therefore, solely relying on wettability measured using WCA and indicating it as the main contributing factor to cell response is therefore debated and said not to be the most reliable approach.116 In fact, this increased complexity for seemingly simple attributes also holds for mechanical properties, topographical cues, and many others.
2.3. Mechanism of Materiobiology
Specific biological response induced by robust spatiotemporal control of biophysical and biochemical cues by biomaterials through mechanisms that are not yet fully understood, remains an open challenge. To date, it has been reported that in the mechanotransduction key processes are focal adhesions (bundles of integrin receptors), cell–cell interactions (such as E-cadherins), mechanosensors (such as talin), and nuclear signaling elements (such as the YAP, the YAP-transcriptional coactivator (TAZ) and lamin A/C).62 Cells sense physical stimuli on biomaterials mainly via the integrin-based signaling pathways. The basic process is that the interactions between cells and (bio)materials activate the integrin-focal adhesion cytoskeleton actin transduction pathway, stimulating cytoskeletal tension exerted by myosin motors, driving actin filament polymerization, inducing cell morphology deformation and associated signaling cascades that thereby alter gene expression to regulate cell functions and promote tissue regeneration.63,71,117 Initially, integrin clusters directly in contact with biomaterials, collect the biointerface information, activate focal adhesion kinase (FAK), induce the maturation of focal adhesions, and stimulate actin filament growth and actin–myosin assembly. In spite of various features on biomaterials, a common contractility-based effect that mediates (stem) cells toward specific responses depends on the degree of activation. In addition, integrin-adhesion-ligand bonds as morphology-independent sensors are pivotal to govern cell fate by designing materials for medical applications. As demonstrated, inhibition of the generation of cytoskeleton actin by blebbistatin renders cells unable to react to biomaterial effects.118 In addition, the cells can adhere to specific substrates through an integrin-independent mechanism because these substrates inhibit protein adsorption.119,120 In other words, these substrate surfaces were exposed to cells, allowing direct cell–interface interaction. The adhesion force between integrin-independent and integrin-dependent mechanisms would likely be different.120 Generally, the anchoring strength of cells following an integrin-independent mechanism was lower than that of cells following an integrin-dependent mechanism. It was reported that the expression of key transcription factors is related to the properties of biomaterials. For instance, cell contraction against substrates with various stiffness or topographies induce different nuclear localization of the transcription factor Yes-associated protein (YAP)/Tafazzin (TAZ),121−123 guiding cells into an osteogenic differentiation. In addition, the biomaterial stimulus mediates integrins and then influences the expression of the associated signaling proteins, for example, Ras homologue gene family member A (RhoA)), which regulates myosin contraction through myosin light chain kinase (MLC) and rho kinase (ROCK) and ultimately modulates cell responses.118,124,125
2.4. Design of Experiments (DOE)
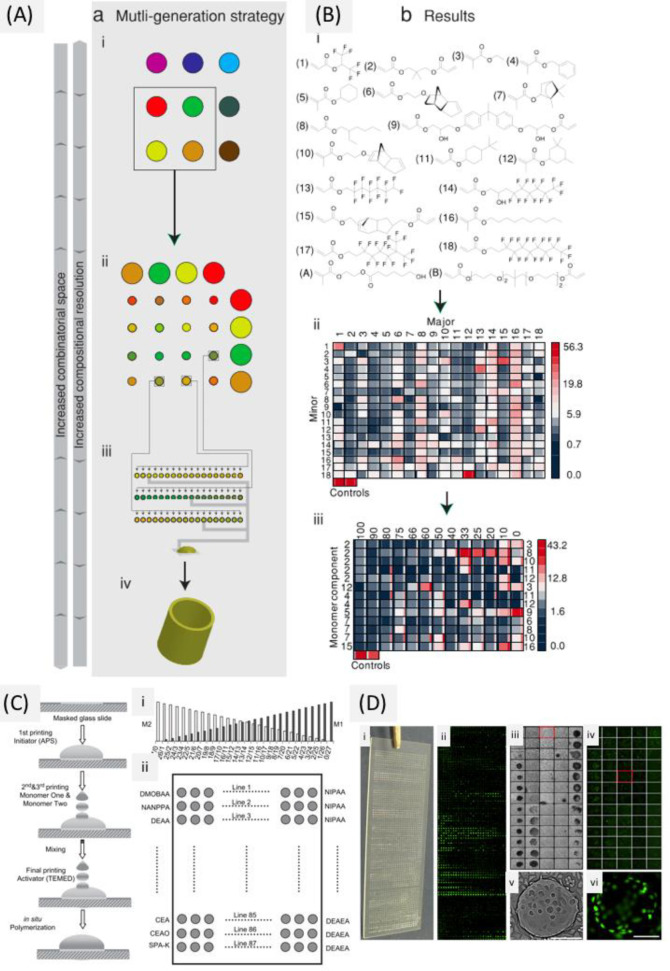
The biomaterial design space is very vast and continues to increase as new discoveries are made, including material stiffness, topography, chemistry, degradation rates, etc., which can be arranged in a combinatorial way. Even though recent progress in efficient 2D/3D high throughput screening platforms allow us to screen thousands of conditions in parallel and are generating more data both on the material (e.g., polymer and material gradient arrays) as well as the biological (e.g., high content imaging and gene expression microarrays) side of the cell–biomaterial interface, it is essential to properly design these advanced bioanalysis platforms to maximize the resulting information.126,127
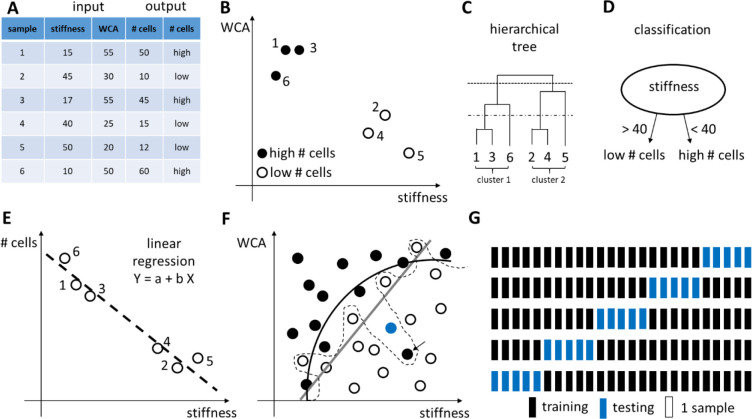
Design of Experiments (DOE) is a methodology to investigate the relationship between experimental variables (also called “factors”) and outcomes.128 The purpose of DOE is to find a suitable experimental design, meaning a number of factor combinations to be tested experimentally, that will maximize the amount of information about the influence of the factors on the outcome while minimizing the costs (i.e., the number of experimental conditions). Importantly, in some cases, it is not known which experimental variables influence the outcome. Under those circumstances, DOE can help design the first experiment with putative factors, from which the most influential ones are determined, followed by a second experiment to analyze those influential experimental variables and their potential interactions in more detail.
The DOE method consists of three steps: (1) determine a suitable experimental design, (2) perform the experiment with the proposed factor combinations, and (3) analyze the results and draw conclusions.129 A number of experimental designs are available; here we will focus on one-at-a-time analysis, full and fractional factorial designs to illustrate the concepts of DOE. We refer the interested reader to more dedicated and advanced resources for more details.128,130−133
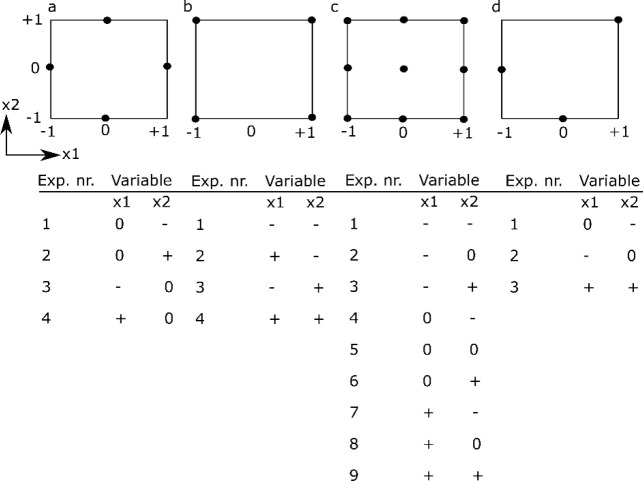
The simplest design is a one-at-a-time (OAT) analysis, which uses a reference condition and then changes each factor individually to higher and lower values, while keeping the other factors constant at the reference value (see Figure 3a). The importance of a factor is determined from the difference between the outcome for the high and low factor value. The main advantage of the OAT design is its simplicity and the fact that it requires a limited amount of experiments (i.e., twice the amount of factors studied). The main disadvantage of the OAT design, however, is that it is impossible to investigate the interactions between factors.
Figure 3.
Schematic overview of different experimental designs for two factors x1 and x2, including a table of the experimental runs to be performed. (a) OAT design, (b) two-level full factorial design, (c) three-level full factorial design, (d) three-level fractional factorial design. Note that for the OAT-design it is assumed that the intermediate level (0) is the standard level. −1 refers to the low level and +1 refers to the high level of a particular factor.
To determine the experimental variables (and their interactions) that have a significant influence on the outcome, a screening experiment can be performed where one or several outcomes are measured. In screening studies, full factorial or fractional factorial designs are commonly used.128 Factorial designs are classified depending on the number of levels that are chosen for each factor: for example, in two-level designs, two different levels, a high (+) and a low (−) level, are chosen; in three-level designs, a high (+), low (−), and an intermediate level (0) are chosen. In full factorial designs all possible combinations are examined (see Figure 3b,c). The advantage of full factorial designs is that the effect of each factor can be studied, as well as all potential interactions between factors. The main disadvantage is the high amount of experiments and the associated costs. For example, if one wants to investigate seven factors, each at three levels, this would require 37 or 2187 experiments.
However, in most investigations, it is reasonable to assume that the higher-order interactions between factors are very small and negligible. As such, if one only wants to estimate the mean value, the influence of the main factors and the second-order interactions, fewer experiments are needed if a suitable set of experimental combinations are chosen.128 A fractional factorial design is thus a trade-off between experimental cost and accuracy and is often used to identify a subset of most influential factors that require more extensive investigation (see Figure 3d).
Once a suitable design has been set up and all the required experimental combinations have been performed, the results need to be analyzed. Analysis of Variance (ANOVA) is suited for analyzing the outcome of a full or fractional factorial design, although other analysis methods are available (see chapter 8). Also, there exist various computer software programs to help create as well as analyze experimental designs, such as JMP,134 Minitab,135 Design-Expert,136 MODDE,137 or open-source, free packages in R138 and Python.139
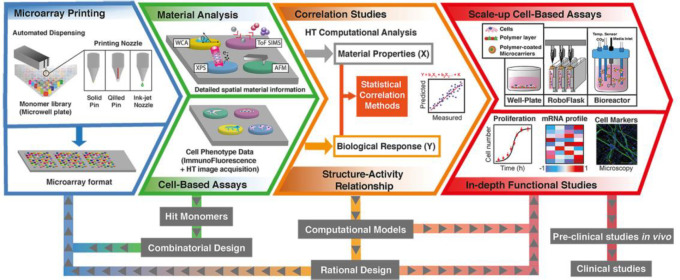
3. HT Screening in Materiobiology
Biomaterials possess many physicochemical parameters that are being studied independently as well as in a combinatorial fashion to better understand the interaction with biological systems. These studies, together with the emergence of completely new biomaterials, and the complexity that arises from the many biological and biochemical possibilities poses as a tremendous challenge and may not be addressed using conventional strategies. Particularly as conventional scientific approaches one start with a hypothesis on whether a specific property has a particular influence on a biological behavior. However, when there is a tremendous amount of variations with many codependent parameters, starting with a clear focused hypothesis is useless due to the lack of specific knowledge to make an educated initial assessment. At such a stage, HT screening comes in, as there is no assumption specific knowledge because that knowledge is obtained by generating large amounts of data in an unbiased fashion from which one can derive a clear understanding in what area refinement of research focus is required. This refinement with lead to a more focused research question. With the advent of new fabrication technologies and advanced analysis strategies, high-throughput (HT) screening systems, including HT screening substrates (gradients and microarrays), HT data collection, HT analysis software, and in silico assessment, provide an ideal strategy to determine thousands of combinations of interactions among biomaterials, biomolecules, and cells for cell–material interaction and drug screening (Figure 4). Particularly, when there is a poor theoretical basis with which to predict the response to a certain material based on structure/chemical composition.21−30 This approach is particularly relevant to biomaterial engineering for cell-based strategies, as there is currently no broad theoretical relationship between material composition and cellular response. In a time when it is becoming increasingly apparent that reproducibility in science is not as high as it ought to be, automated strategies have the potential to bridge the reproducibility gap.140 Equally, assembling multicomponent cellular microenvironments generates such a large design space that only high-throughput methods allow for their efficient exploration. Finally, the discovery process generates large databases of material structure-cell performance data, allowing insight into be gained into the controlling relationships between material physicochemical properties and cell phenotype. Discovery and mechanistic insight are not mutually exclusive pursuits, but rather highly complementary strategies currently being combined to generate in silico models and, ultimately, further increase the design space available to biomaterial engineering.116
Figure 4.
Schematic diagram of the combinations of HT screening, materiobiology genome, as well as tissue repair and regeneration.
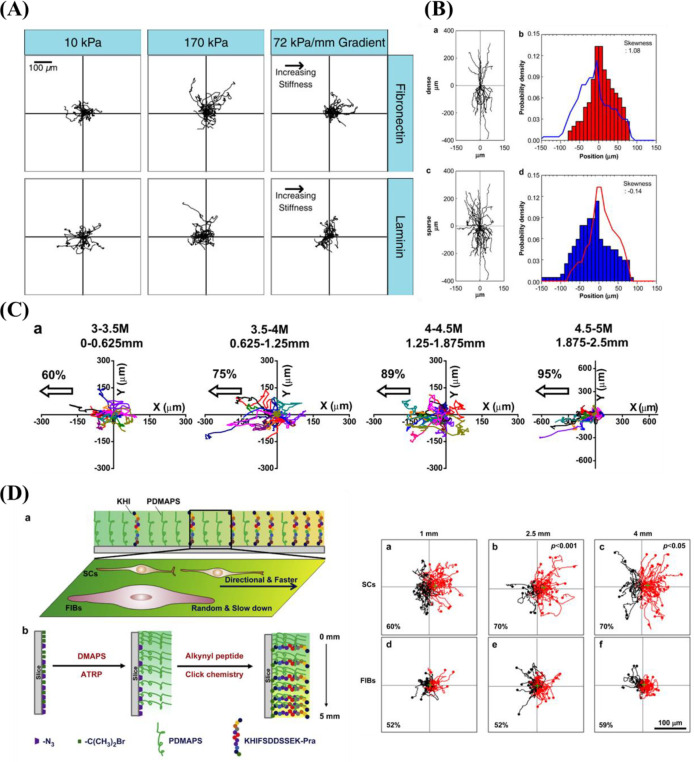
4. Gradient-Based HTS Approaches for Biomaterials Discovery and Materiobiology
As indicated before, physicochemical properties at the biointerface can influence cellular behavior such as adhesion, spreading, migration, proliferation and differentiation.70,141−143 The proper control over physical properties and the testing of cell behavior is much easier on 2D samples than in a 3D environment. Therefore, HTS approaches for in vitro testing of cell responses toward certain parameters have been developed. Using systematical changes in material composition111 or topography144 is time-consuming and costly and hence alternative methods are developed as the preparation of numerous single samples is highly labor-intensive. One of the alternatives used approaches to study cell–material interactions more efficiently is the use of surface gradients. A surface gradient is a surface, on which one parameter changes gradually from one side to the other. This gradual change between a minimum and a maximum value allows for systematical testing of a parameter using only one sample making it more cost and time-efficient than individual samples34,100,145,146 (Figure 5a). The working principle of gradients is simple as one can image and analyze cells in a regular fashion without complicated procedures. However, there are a few working principles that need to be followed. Cell seeding requires to be homogeneous. Cells migrate and attach in response to surface properties. Therefore, deviations in cell density should arise from either of them and not be artificially induced by cell seeding inconsistencies. Another key feature is awareness of the location on the surface. While the location on a surface is conventionally trivial, on a linear gradient changing the location will alter the magnitude of a parameter when going in the direction of the gradient. Alternatively, no change in magnitude is occurring when moving perpendicular to the gradient direction. To always keep track of the surface location is pertinent, as minor changes will result possibly in a different cell–material interaction. It goes without saying that on the forehand, the parameter development along the gradient needs to be very carefully determined. To avoid any potential mismatches between location and cell response, one can choose to image the whole surface and all cells on it, which will provide the possibility of postdetermination of location when a reference point is known. This approach is particularly useful when systems as orthogonal double gradients are being used where the magnitude of two surface parameters change in all direction. Hence, gradients are powerful in generating many insights but require delicate and highly systematic approaches in their use.
Figure 5.
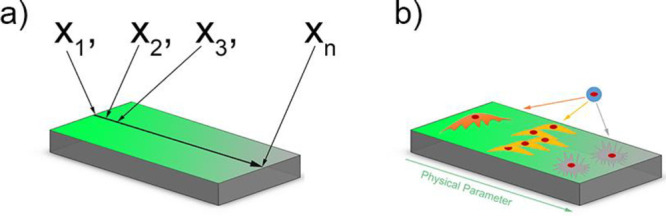
(a) Schematic illustration of a 2D surface gradient and the advantages over single sample measuring. (b) Illustration of cellular responses to a physicochemical parameter on a surface gradient.
These surface gradients can be produced by changing different physicochemical properties over this surface. This distinct parameter can then be tested toward cell response using only a single sample (Figure 5b). Where changing one parameter is already a huge improvement over separate samples, a two-parameter gradient can give exponentially more information. In a double orthogonal gradient approach, the influence of parameter combinations can be determined, which is not possible using a single surface gradient approach. Where the influence of one physicochemical parameter was already shown, the combinatorial effect of two or more at the same time is still relatively less reported. This section focuses on the preparation and use of gradients for studying cell–material interactions in a high-throughput fashion.
4.1. Preparation Approaches of Gradients for Studying Biointerfaces
Researchers have used high-throughput approaches to quantify cell response to many material properties in a single experiment. These efforts have led to the development of a wide range of combinatorial methods including libraries on surfaces (2D), and in 3D scaffolds. In this part, we focus on the preparation methods and developments of gradient surfaces with different single and double parameters used for high-throughput approaches.
4.1.1. Gradients as 2D Biointerfaces
Generally, cellular microenvironmental signals can be categorized into physical and (bio)chemical cues. Physical signals include substrate mechanical properties and topography. Material chemical composition, soluble growth factors, the biochemical composition of ECM, and cell–secreted proteins are examples of (bio)chemical cues.
4.1.1.1. Stiffness Gradients
A variety of materials, including polyacrylamide (PAA), polydimethylsiloxane (PDMS), polyethylene glycol (PEG), and HA, have been adopted to prepare stiffness gradients for studying cell behaviors.38Table 1 summarizes the various materials and methods for preparing the stiffness gradient, and representative schematic diagrams are shown in Figure 6.
Table 1. Various Materials and Methods Used to Engineer Varied Stiffness Gradients.
| materials | preparation methods | stiffness range | ref |
|---|---|---|---|
| PVA hydrogel | LN2-contacting gradual freezing and thawing method | 1–24 kPa | (38) |
| PVA/HA hydrogel | 20–200 kPa | (147) | |
| poly(allylamine hydrochloride) and poly (acrylic acid) | controlled dipping into a cross-linking solution | 0.5–110 MPa | (39) |
| PEG diacrylate hydrogels | photopolymerization using a sliding mask | 2–100 kPa | (150) |
| polyacrylamide | 2.5–11 kPa | (151) | |
| HA hydrogel system | 3–100 kPa | (152) | |
| acrylamide/bis-acrylamide | 1–240 kPa | (153) | |
| PDMS | temperature gradients during cross-linking | 190 kPa–3.1 MPa | (148) |
| PEG hydrogels | graded exposure to UV | 7–32 kPa | (159) |
| PDMS | shielded plasma oxidation with a mask | 6–89 MPa | (146) |
Figure 6.
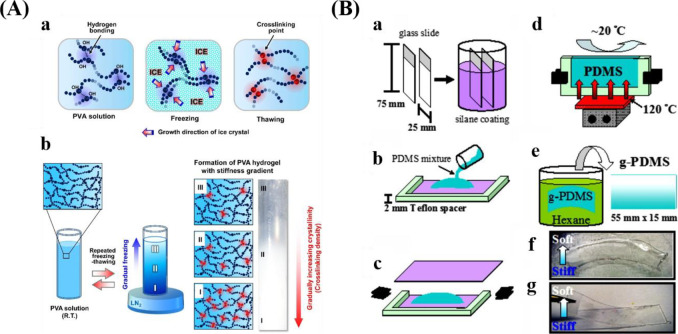
Different methods for preparing stiffness gradients. (A) Schematic representations displaying the formation of PVA hydrogel with stiffness gradient by the freeze–thaw method. Reprinted with permission from ref (38). Copyright 2015 Elsevier, Ltd. (B) Schematic preparation process for PDMS stiffness gradients via a temperature gradient during curing. Reprinted with permission from ref (148). Copyright 2012 Elsevier, Ltd.
To prepare stiffness gradients, researchers have proposed various methods, for example, gradual freeze–thaw by liquid nitrogen (LN2)38,147 (Figure 6A), heat gradients within polymerizing PDMS148 (Figure 6B), the incorporation of particles within a hydrogel,40 and controlled dipping into a cross-linking solution.39 Lee and co-workers reported poly(vinyl alcohol) (PVA)38 and PVA/HA147 stiffness gradients via gradual freeze–thaw by LN2. Liquid PVA can be converted into hydrogels by the generation of crystallites.149 During the process of freezing, ice facilitates the increase of crystallinities, which is beneficial for the cross-linking of polymer chains. Gradual freezing (freezing time and temperature) produced a stiffness gradient within the hydrogel. This method is simple without the addition of harmful agents, complex machines and procedures, but allows for the generation of a wide stiffness gradient with similar chemical property.38
Other techniques for preparing stiffness gradient involve photoinitiators and a patterned150,151 or a moving photomask.152,153 UV will give rise to radical polymerization within polymers. Therefore, the degree of cross-linking relies on light density, which could be adjusted by utilizing a photomask.151 Sunyer et al. fabricated a PAA hydrogel containing a linear gradient (115 kPa/mm, 1–240 kPa) involving the photopolymerization of films covered with a mask.153 Although this method is easy to implement, the low resolution of the mask is not good for the accurate regulation of stiffness.153 In fact, investigations based on photomasks have shown poor repeatability.154 In addition, the toxicity from prepolymer and cross-linker are also considered as the potential limitations.155−158
4.1.1.2. Topography Gradients
Soft lithography (microcontact printing), capillary force lithography (CFL), photolithography (hard lithography), photopolymerization, and shielded plasma oxidation methods are typical methods used to create anisotropic topography gradients such as grating, pillar, and wrinkle gradients.
Electron beam lithography (EBL) is one of the top-down approaches. An electron beam is focused on the surface filled with photoresist. The solid part will become a liquid after exposure to the beam. While small dimensions could be achieved, the disadvantage of this method is very expensive and a slow speed. Combined with other techniques, for example, ultraviolet-assisted capillary force lithography (UV-CFL) or plasma etching, grating and pillar gradients can be fabricated. For instance, Kim et al.160 described using EBL combined with UV-CFL to prepare parallel micro/nanogratings (Figure 7A). The substrate was composed of a fixed ridge diameter (1 μm) and depth (400 nm) but varied in groove diameter (1–9.1 μm). With EBL and plasma etching, M. Reynolds et al.161 prepared nanopillar arrays and the height ranges between flat and 250 nm. For pillar gradients, except for the method introduced above, the combination of photolithography and soft-lithography could also be employed.
Figure 7.
Schematic diagrams for preparing anisotropic gradient. (A) Arrays with varied intervals prepared by UV-assisted capillary force lithography. (a) Schematic of the preparation process for pattern arrays. (b) 3D AFM picture. (c) SEM image. Reprinted with permission from ref (160). Copyright 2009 Elsevier, Ltd. (B) (a) Procedure for the fabrication of wrinkle gradient. (b) AFM pictures along the gradient. Reprinted with permission from ref (100). Copyright 2017 American Chemical Society.
Photolithography was initially used in the semiconductor industry.162 The lowest resolution for photolithography is about 1 μm and the pattern can be created onto large substrates. This technique requires a clean environment and costly equipment. Photolithographic methods are suitable for many surfaces, for example, metal oxides,163 polymers,164 glass,165 and hydrogels.166
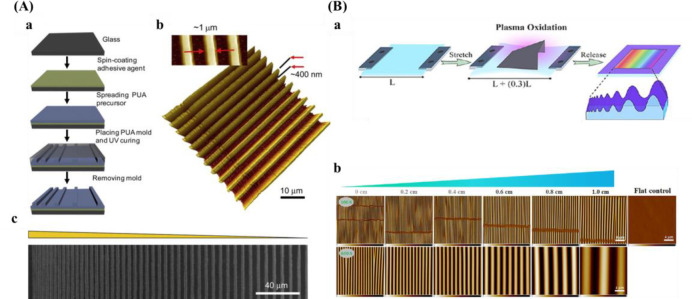
Soft lithography is another popular method for surface patterning.167 The size is restricted by the photomask resolution and the spread of inks, so the degree of complication is relatively low. Microcontact printing (μCP) is a commonly used soft lithographic method. Although the process is simple and relatively inexpensive, this method does not allow the simultaneous printing of multiple inks. In addition, there is a problem with the diffusion of ink. Wang et al.168 fabricated pillar arrays consisting of a height gradient by the combination of photolithography and soft-lithography. The parameters of PDMS pillars are 2 μm in diameter, 5.5 μm pitch size, and varied heights increasing from 3.8 to 10.1 μm. The methods introduced above always require complicated equipment and procedures. In addition to gratings and pillars, another common anisotropic topography gradient is wrinkles, which are fabricated by plasma oxidation and can even be combined with soft imprint lithography to transfer these topographies to soft hydrogels.169 A PDMS substrate is stretched, partially shielded with a mask, and the surface oxidized by plasma. After that, releasing the strain induces the generation of a wave-like topography gradient. Wrinkle amplitude and wavelength increase from the closed to the open side of the mask, resulted from increased thickness of the silica-like layer (Figure 7B). With this method, the van Rijn group prepared PDMS-based wrinkled topography gradients with varied wavelengths (W) and amplitudes (A).34,100,170−173Table 2 presents an overview of varied fabrication techniques for the creation of topography gradients.
Table 2. Preparation Methods of Gradient Surfaces with Anisotropic Topographies.
| gradient patterns | preparation methods | topographical size | ref |
|---|---|---|---|
| grating | EBL and UV-assisted capillary force lithography | constant ridge width (1 μm) and depth (400 nm) and variable groove widths (1–9.1 μm) | (160) |
| combination of nanoimprint lithography and photopolymerization | gradient in pattern height (0–350 nm) | (174) | |
| pillar | EBL and plasma etching | pillar height changes from planar to 250 nm over 9 mm | (161) |
| photolithography and soft-lithography | pillar diameter 2 μm, pitch size 5.5 μm and variable heights between 3.8 and 10.1 μm | (168) | |
| wrinkle | unidirectional strain during surface oxidation using shielded plasma oxidation by applying a mask | W: 464–7121 nm | (100) |
| A: 49–2561 nm | |||
| W: 200–1087 nm | (34) | ||
| A: 0.1 nm–260 nm | |||
| W: 0.8–14 μm | (172) | ||
| A: 144–3000 nm | |||
| W: 4–30 μm | (173) | ||
| A: 144–3000 nm | |||
| W: 464–10990 nm | (171) | ||
| A: 49–3425 nm | |||
| W: 1520–9934 nm | (170) | ||
| A: 176–2168 nm |
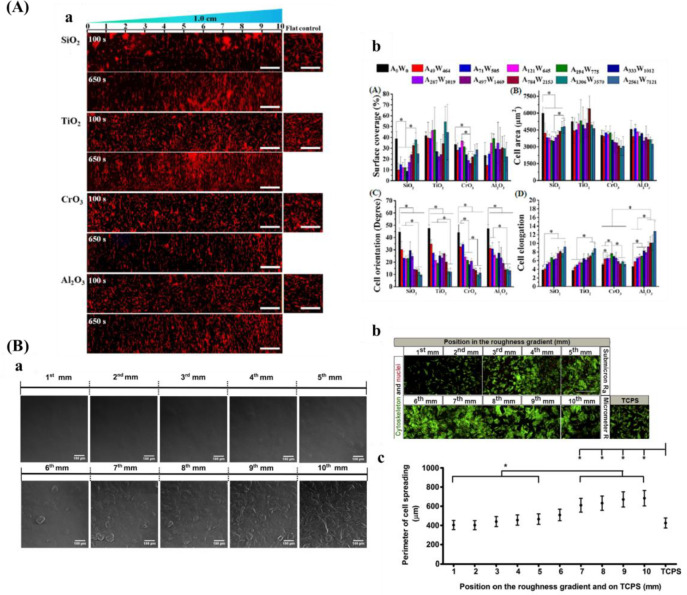
In addition to anisotropic topography gradients, isotropic topography gradients (e.g., roughness, particles, and pores) are commonly used to study the interaction between cells and topographical structures. Different fabrication methods for isotropic gradients are summarized in Table 3.
Table 3. Isotropic Gradients with Various Structures and Preparation Methods.
| isotropic gradient types | preparation methods | gradient parameters | ref |
|---|---|---|---|
| pores | centrifugation | pore size: 88–405 μm | (175) |
| pore size: 90–400 μm | (176) | ||
| anodic etching and chemical etching | pore size: 50 nm–3 μm | (177) | |
| electrochemical etching of silicon wafers in electrolytes containing hydrofluoric acid (HF) | pore size: 29–226 nm | (36) | |
| pore size: 5–3000 nm | (178) | ||
| pore size: 10–500 nm | (179) | ||
| roughness | annealing temperature | roughness: 0.5 to 13 nm | (180) |
| combination of sand-blasting and chemical-polishing technique | roughness: 0.5–4.7 μm | (181) | |
| roughness: 1.12–5.7 μm | (182) | ||
| roughness: 0.87–4.41 μm | (183) | ||
| sputter deposition | roughness: 1–16 nm | (184) | |
| particle | dip-coating | maximum particle coverage is 21% corresponding to a mean particle spacing of 190 nm | (185) |
| dip-coating | particle coverage range from 35% to 0, and particle diameter was 73 nm | (186) | |
| controlled immersion into the solution of gold nanoparticles in a time-controlled manner | root–mean–square roughness change from 0 to 15 nm, and diameters of nanoparticles are 16, 38, or 68 nm | (187) | |
| roughness: ∼2.5 to 5, and nanoparticles of diameters of 16, 68 nm | (188) | ||
| electrospray | surface roughness (root–mean–square value) range from 80–900 nm, and the average size of the deposited particles was about 3 μm | (189) |
It has been demonstrated that pore size and porosity play a critical role in guiding cell behavior. Porosity is described as the proportion of pore interspace in a solid.190 Pore size influences cell migration, spreading, and transportation of nutrients.175 Generally, larger pore size or higher porosity can afford enough supplements, and is beneficial for the removal of waste, but not favorable for cell attachment, while the smaller pore size or lower porosity has a reverse effect.81,191,192 In addition, porosity improves the physical connection between bone and the implanted material, thus providing stronger mechanical stability at the interface.193 However, it has to be noted that higher porosity is harmful to the mechanical properties of the substrate.194
For the fabrication of substrates, the optimum pore size relies on a specific purpose, for example, different tissues for tissue engineering.81,175,191 Therefore, substrates consisting of pore-size gradients afford an accelerated screening platform, which contributes to the identification/validation of the optimum pore size.
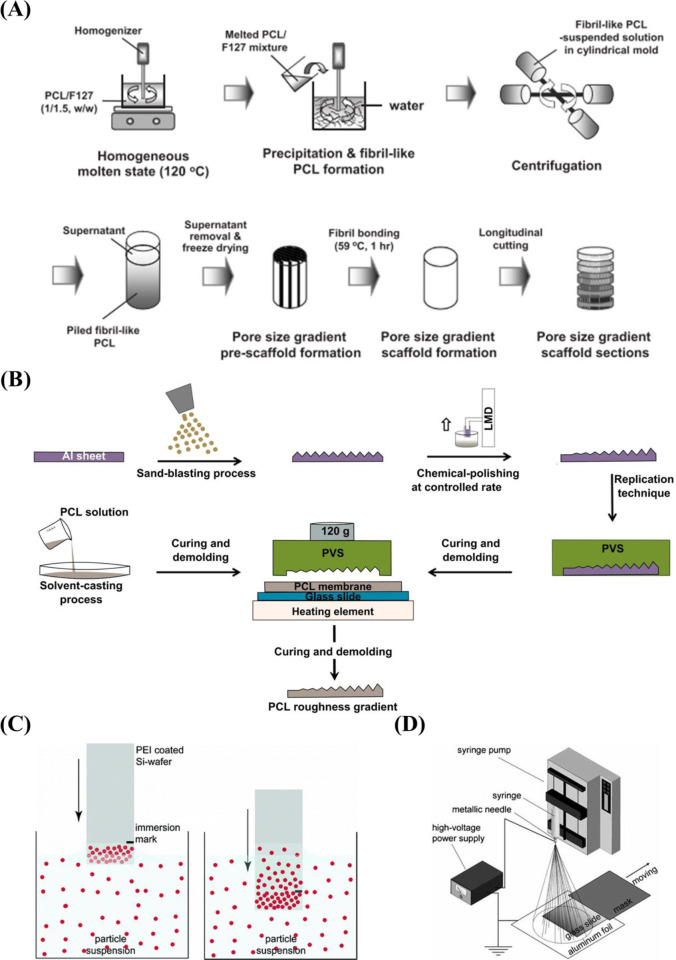
A popular candidate for the preparation of surfaces with gradient pore size is porous silicon (pSi). The advantages of pSi are biocompatible and biodegradable,195 adjustable porosity,196 and chemical property.197 pSi is fabricated by electrochemical anodic etching of silicon wafers in electrolytes containing HF.198 The pore size of pSi is easily controlled by varying the etching conditions (e.g., current density, the ratio of HF-to-surfactant) to fabricate surfaces with pore size increasing from nanometers to micrometers.178 The pSi gradients with varied pore sizes36,178,179 prepared by electrochemical etching are listed in Table 3. While it is easy for preparation and further functionalization, stability is a problem for long-term cell culture. Therefore, a stable substrate with a pore gradient is important. Wang et al.177 fabricated porous alumina (pAl) with pore sizes between 50 nm and 3 μm by anodic etching and chemical etching methods. For long-term cell culture, compared with pSi, the main advantage of pAl is higher stability in water.199 Besides, there are many advantages for aluminum-based materials, for example, suitable for implanting in vivo,199 easy for further surface functionalization.200,201 Therefore, pAl has drawn much attention in recent years.202,203 Since the pore size ranging from several nanometers to hundreds of nanometer with the methods mentioned above, therefore, Oh et al. prepared a larger pore size gradient (∼90 to ∼400 μm) by a centrifugation method175,176 (Figure 8A).
Figure 8.
(A) Schematic representation displaying the preparation process of pore size gradient by a centrifugation technique. Reprinted with permission from ref (175). Copyright 2007 American Chemical Society. (B) Schematic diagrams illustrating the fabrication of PCL roughness-gradient. Reprinted with permission from ref (181). Copyright 2014 Elsevier, Ltd. (C) Schematic drawing of the dip-coating process to prepare particle gradient. Reprinted with permission from ref (186). Copyright 2007 American Chemical Society. (D) Schematic diagram for producing a density gradient of microparticles by electrospray method. Reprinted with permission from ref (189). Copyright 2010 WILEY-VCH.
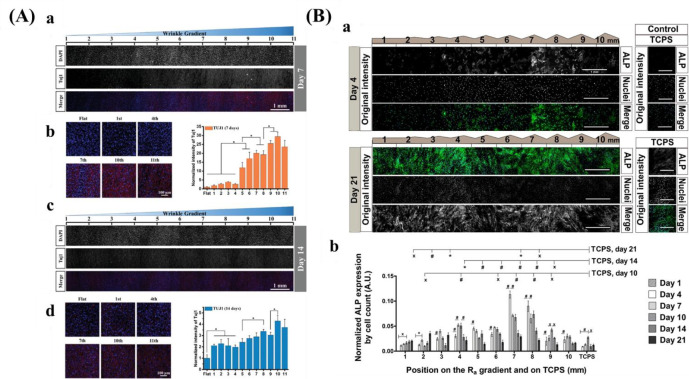
Another popular isotropic topography gradient is based on surface roughness, which is denoted as the surface texture where deviation from a normal vector determined and given generally as Ra, the arithmetic average of the roughness profile. It is evidenced that roughness influences various cell behaviors, including cell adhesion, migration, viability, and differentiation.204,205 It has been suggested that adjusting surface roughness plays a crucial role for osteointegration.206 In that regard, roughness could mimic the topography cues that were found after bone resorption by osteoclasts.207 Furthermore, the roughness increases the surface area of the biomaterial and increases the amount of adsorbed proteins, which allows for more deposition of ECM and earlier bone ingrowth.208
There are several techniques for the preparation of roughness gradients, for example, the gradient in annealing temperature, and sand-blasting combined with chemical-polishing techniques. With the former method, Washburn et al.180 prepared a roughness gradient in the range of nanometer size. Crystallinity gradients of polymer were engineered by using a gradient in annealing temperature. A poly(L-lactic acid) (PLLA) membrane was annealed on a temperature gradient stage. This range produced varied crystallinity, giving rise to a roughness gradient (root-mean-square roughness between 0.5 and 13 nm). However, this technique has several drawbacks, for example, lower roughness range, and restriction to specific biomaterials. For the latter method, it produces roughness gradients with nano/micrometer over a centimeter-scaled substrate.181−183 Kunzler et al.182 fabricated micrometer-scale roughness gradients (1.12 to 5.70 μm) by sandblasting followed by chemical etching. Faia-Torres et al.181 fabricated a roughness gradient via the same process but added a material transfer step by imprinting the topography ultimately into polycaprolactone (Figure 8B). Via this method, a roughness was obtained that increased from 0.5 to 4.7 μm. Nevertheless, this multistep procedure includes some translational issues as it is not straightforward transferable to real components and the coating step may change the chemical property of the substrate surface. The production of the topography in metallic implants, a possible good translation could be expected as sandblasting and chemical etching are commonly used treatments and transferable to larger objects. It has to be noted that transference to clinically relevant products is the main problem for many screening approaches. Previous work demonstrated that HF etching of zirconia implants with excellent performances could improve bone attachment.209,210 Flamant et al. produced a roughness gradient at the surface of zirconia using HF etching.35
Alternatively, topography gradients can be fabricated by the manipulation of nano/microscale particles. It has been demonstrated that a density gradient of particles can be produced by controlling the adsorption of particles.185−187,211,212 Specifically, the gradient of −NH2 was formed by vapor deposition, followed by attaching nanoparticles to the surface through immersing the substrate into a gold solution.212 Alternatively, Huwiler et al.186 fabricated nanoparticle density gradient by immersing a positively charged poly(ethylene imine) (PEI)-coated silicon wafer into the solution filled with negatively charged silica nanoparticles (Figure 8C). Although those techniques are successful, there are still some disadvantages, for instance, complicated processes, specific reagents or substrates. Therefore, a simple and effective technique still needs to be identified for the preparation of particle gradients and particularly together with the possibility of transferring the topographies to biomedical products.189
Electrospray is a technique with tremendous potential for the preparation of particle gradients. During the process, a liquid is forced through a capillary onto a collector while kilovolts are applied between the capillary and the collector.213 It is a simple approach for producing homogeneous particles with sizes between nano- to micrometers, which is suitable for serving as carriers for chemotherapeutics, proteins, and biomacromolecules.213,214 Moreover, different than previous techniques, this method allows a precise adjustment of both microparticle density and size. Li et al.189 created density gradients of microparticles (Figure 8D) by changing the deposition time of electrosprayed microparticles.
4.1.1.3. (Bio)Chemistry Gradients
A number of techniques have been developed for the preparation of chemical gradients, for example, plasma-assisted approaches,215 corona discharge,216 SAM-based techniques,217,218 UV,219 plasma polymerization,220−226 and click reactions based methods.227
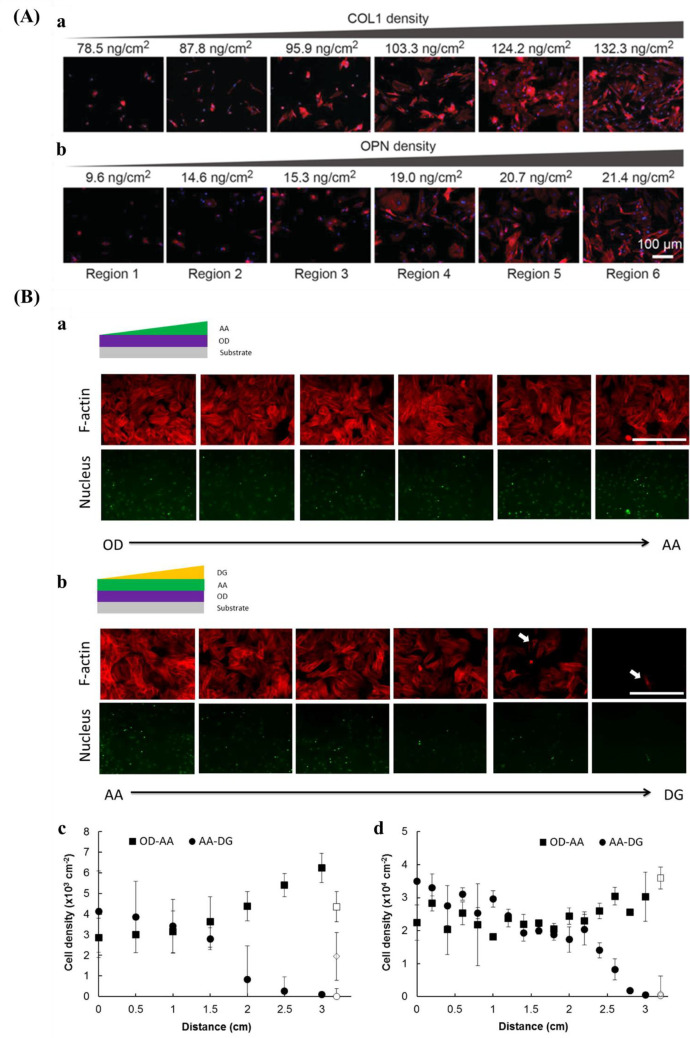
Plasma polymerization is a facile technique for surface decoration of biomaterials as it deposits a very thin functional layer on any kind of material without the requirement of premodifying the surface. Not needing complex modification is a critical aspect for medical devices based on polymers, ceramics, metals, and composites.228 The plasma modifies the surface of a material by bombarding the substrate with high-energy particulates. The polymerization degree is based on the intensity and number of particulates.47 Various functional groups, for instance, amine, carboxyl, hydroxyl, and sulfonic acid could be coated onto the surface via nitrogen, ammonia, oxygen, and sulfur dioxide plasma, respectively.229,230 Furthermore, surface gradients could be formed via a shielded approach using masks during plasma polymerization. For example, Wang et al.231 prepared two surface chemical gradients: 1,7-octadiene (OD)–acrylic acid (AA) gradient, and AA–diethylene glycol dimethyl ether (DG) gradient.
Another popular method for surface modification is the use of SAM-based techniques. SAMs of alkanethiols are popular since they could generate a steady organic coating on gold surfaces. Importantly, varied chemical properties could be simply fabricated by generating alkanethiols with diverse end-groups.232,233 For example, Morgenthaler et al.217 prepared density gradients of thiolates by dipping a gold-deposited surface into a thiol solution, then followed by dipping the surface into the complementary thiol solution. In addition, chemical gradients have also been fabricated by UV irradiation. Peroxides could be formed by a radical-based photo-oxidation mechanism.234 With this method, Li et al.219,235 fabricated a density gradient of carboxylates.
Bioactive molecules can significantly improve the interaction between cells and biomaterials.143,236−238 Various biomolecules, including proteins, peptides, and growth factors, have been successfully grafted on the surfaces of biomaterials in a gradient manner. Proteins of ECM will enhance the interactions between synthetic biomaterials and tissues in vivo.239
Protein gradients have been fabricated by the combination of using SAMs and polymerization techniques for fibronectin,240 prepared by adsorption onto a density gradient of PEG.241 Additionally, protein gradients have also been prepared using nanofiber systems. Nanofibers mimic the fibrous ECM in vivo.242 With a controlled filling method, Shi et al.243 fabricated the Fn gradient within the nanofibers.
For peptide gradients, different kinds of peptides have been used and implemented via various methods. For example, RGD peptide gradients mediated by atomic transfer radical polymerization (ATRP) and carbodiimide chemistry,244 using a “universal gradient substrate for click biofunctionalization” methodology,245 immobilized osteogenic growth peptide,246 density gradient of Val-Ala-Pro-Gly (VAPG) peptides,247 and Arg-Glu-Asp-Val peptide gradient248 synthesized via click chemistry have been used to produce peptide-based gradients. Also, the growth factor gradient has been prepared based on plasma oxidation,249 surface electrochemistry,250 and injection methods.251
4.1.1.4. Multiparameter Gradients
The development of double gradients allows for studying the interaction between cells and two or more surface parameters. For linear double gradients, the two parameters are arranged in the same or opposite direction. For orthogonal gradients, the two parameters are perpendicular to each other.252
The linear/orthogonal double gradient method significantly decreases the sample number for investigating potential combinations of different gradients, which is beneficial for HTS cell response.253 In this part, we mainly focus on the linear double gradient and orthogonal double gradients. Table 4 summarizes the multiparameter gradients and corresponding parameters.
Table 4. Multiparameter Gradients and Corresponding Parameters.
| multiparameter gradient | parameters | ref |
|---|---|---|
| linear double gradient | composition: the gradients range from 25–100% PLLA | (254) |
| roughness: starting from the PDLLA-rich end, surface roughness increased as the fraction of PLLA increased; roughness then reached a plateau between 60 and 80% PLLA before becoming somewhat smoother from 80–100% PLLA | ||
| wettability: 30–94° | (145) | |
| stiffness: 85 MPa to ∼7 MPa | ||
| orthogonal gradient | pore size: ranging from hundreds to tens of nanometers | (255) |
| peptide density: not shown | ||
| dual topography gradient: on one axis varied from 8–100 μm space between the 8 μm grooves and on the other axis from ∼5 nm to ∼1 μm in depth | (256) | |
| topography: the groove width ranging from 5–95 μm with a constant ridge depth of 3.4 μm | (257) | |
| chemistry: varying from a hydrocarbon to a nitrogen-containing polymer | ||
| topography: parallel grooves with widths varying from 5–95 μm, separated by 5 μm wide ridges | (253) | |
| wettability: WCA ranging from 55–96° | ||
| roughness: Ra value ranging from 0.8–4.1 μm | (31) | |
| nanoparticle density: particle number decreased linearly from around 74 particles per μm2 to 0 | ||
| constant groove width of 8 μm and with ridge width increasing from 8 μm in 0.5 μm steps across 10 mm | (33) | |
| gradient of groove depth spanning more than 2 orders of magnitude (less than 10 nm to over 1000 nm) | ||
| peptide concentration gradient: GRGDS concentrations ranging from ∼15–90 pmol/cm2 and BMP-2 concentration range from ∼0–25 pmol/cm2 | (258) | |
| stiffness: 6–89 MPa | (146) | |
| WCA: 29–90° | ||
| stiffness: 0.5–1.3 kPa | (259) | |
| Fn: a roughly 5-fold difference between the highest and lowest densities |
C.G. Simon Jr. and co-workers254 created a gradient using a polymer blend composed of PLLA and poly(d,l-lactic acid) (PDLLA). Surface roughness varied with composition where the areas were smooth for the regions rich in PDLLA, it was rougher for the regions rich in PLLA. Kühn et al.146 developed a double linear wettability-stiffness gradient by a single-step shielded air plasma treatment.
There are also some strategies for designing and preparing orthogonal double gradients. Huethorst et al.256 described using UV and dry etching to produce a dual microgradient substrate. For the microgrooves, the deepness (Y-axis) and dimension (X-axis) increased from 5 nm to 1 μm and 8–100 μm, respectively. Roach et al.257 produced an orthogonal gradient of topography and chemistry by hot embossing and plasma polymer deposition. The grooves of the substrate varied between 5 and 95 μm and chemistry changed from hydrocarbon to nitrogen. With a similar method, Yang and co-workers253 prepared orthogonal chemical and topographical gradients on a poly(methyl methacrylate) substrate. In one direction, the topographical gradient consisted of constant width (5 μm) and depth (3 μm), but the size of the groove gradually increased from 5 to 95 μm. In the other direction (vertical), a wettability gradient was introduced ranging from WCA 95–55°, which was fabricated by diffusion-controlled plasma deposition. Zink et al.31 used the combination of sand-blasting and nanoparticle adsorption to produce roughness gradients from two directions by introducing particle density gradient onto a roughness gradient.
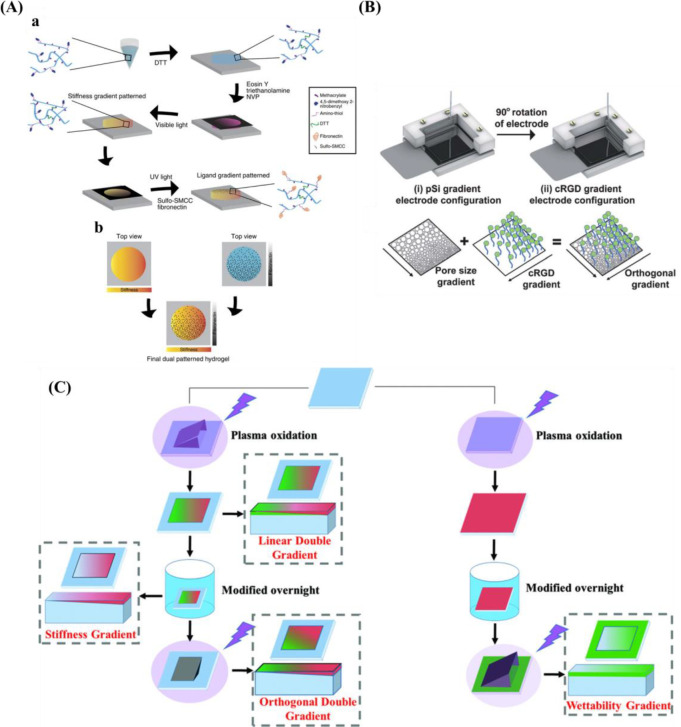
In addition to orthogonal gradient with topography and topography or chemistry, gradients with physical cues, for example, stiffness and protein have also been fabricated. Rape et al.259 fabricated a simple, high-throughput platform with ligand density (Fn) and substrate stiffness based on light-modulated HA hydrogels (Figure 9A). Clements et al.255 prepared an orthogonal gradient composed of pore size gradient and peptide density gradient via an electrochemical approach (Figure 9B).
Figure 9.
(A) Schematic diagram for the design double orthogonal gradient composed of stiffness gradient and Fn density gradient. Reprinted with permission from ref (259). Copyright 2015 Nature Publishing Group. (B) Schematic presentation for the fabrication of an orthogonal gradient comprising pore size gradient and RGD gradient. Reprinted with permission from ref (260). Copyright 2012 Royal Society of Chemistry. (C) Formation of unidirectional single and orthogonal double surface gradients (stiffness and wettability). Reprinted with permission from ref (146). Copyright 2018 WILEY-VCH.
In light of the plasma-generated double linear gradient of stiffness and wettability, also the orthogonal double gradient was developed via a similar approach. The stiffness and wettability were decoupled by first generating a stable stiffness gradient using a harsh plasma oxidation treatment using air plasma with subsequent silanization to recover the hydrophobic properties. Then a short plasma treatment of 20 s at 500 mTorr again via a shielding approach provided the wettability gradient (Figure 9C). One drawback is that samples needed to be used directly as the substrates based on PDMS display hydrophobic recovery and care needed to be taken not to alter the added properties.
4.1.2. Gradients in 3D Culture Systems
For many applications, 2D culture platforms are able to provide proper insights into how materials interact with biological systems such as cells with medical implants. However, 2D systems cannot fully mimic the complicated 3D microenvironment found in vivo, which is important to gain insights into cell behavior in their native environment or in specific tissue pathologies.21 Until now, only a few techniques are used for fabricating 3D matrices with gradients, for example, scaffolds with porosity and hydrogels. Several techniques have been reported for fabricating physicochemical gradients within scaffolds, especially gradients in pore size or porosity. Roy et al.261 and Woodfield et al.262 fabricated scaffolds with porosity gradients and pore size gradients by 3D printing.
So far, most 3D gradients are dependent on hydrogels since it mimics the hydrated state in soft tissue. Several methods have been applied to fabricate gradients in hydrogels, including gradient maker and diffusion. For the gradient maker, various gradient markers are used to fabricate linear-gradient hydrogels. PEG hydrogel gradients with a stiffness gradient between 10 and 300 kPa were prepared via a gradient maker.263
Molecular diffusion is another technique for fabricating 3D gradients. In this method, hydrogels are exposed to molecules, which will spread within the hydrogel, forming a density gradient in the direction of the molecular diffusion. Vepari and co-workers264 prepared immobilized enzyme gradients with a 3D porous material using the principles of diffusion. In addition, growth factor gradients in the 3D porous matrix could also be prepared by the combination of centrifugation and surface immobilization.265
4.2. Interaction between Biological Species and Gradients
4.2.1. Interactions of Proteins with Gradient Substrates
It is well-demonstrated that the interactions between cell and biomaterial are regulated by the type and conformation of the adsorbed proteins that can interplay with specific integrins present on the cell surface. Importantly, the surface physicochemical properties of the biomaterials can significantly affect the amount, orientation, and conformation of adsorbed proteins.184 Interfacial interactions are key for implanted devices.266 On one hand, proteins adsorbed onto the surface of biomaterials facilitate the activation of inflammatory cells.267 On the other hand, the ECM proteins are also important signaling factors known to control cell attachment, and adjust subsequent cell activity (proliferation, migration, differentiation).268 Consequently, tailoring cell interactions at the interface of biomaterials is pivotal for the designing and eventual success of implantable medical devices and engineered tissues.269
To date, many studies have been focusing on the understanding and adjustment of protein adsorption on gradient substrates. The surface physicochemical properties of the biomaterials modulate the adsorption behavior of proteins, and for studying the protein adsorption behavior wettability gradients,270,271 charge gradients,272 and roughness gradients184 have been used.
Fn facilitates cell adhesion and has been previously reported to adsorb in greater amounts onto hydrophobic rather than hydrophilic surfaces.273,274 For instance, Mohan et al.218 fabricated the monotonically varying surface chemistry gradients (WCA ranging from 10–100°) on PDMS substrates to study their influence on Fn adsorption. The results displayed that the adsorption of fibronectin enhanced monotonically with increasing hydrophobicity. Cantini and co-workers270 also found the amount of Fn decreased monotonically with increasing wettability. However, other researchers have contradictory findings. Liu et al.228 also prepared a wettability gradient (WCA from 70–90°) with an increasing N/C ratio. When BSA was adsorbed onto the gradient, the amount of adsorbed protein decreased from the hydrophobic part to the hydrophilic part, and the adsorption of Fn from an Fn solution showed no significant changes along the gradient. However, when exposed to the mixture of BSA and Fn, a significant increase of Fn was found from hydrophobic to the hydrophilic part.
In addition to material wettability, several groups also prepared PEG density gradients.223,241,275 PEG can prevent protein adsorption.276,277 Pei et al.266 fabricated density gradients of PEG to carry out a systematic study on protein adsorption. The results demonstrated that the adsorption of single proteins (fibrinogen and Alb) increased as the density of PEG decreased, and competitive adsorption including both proteins indicated more fibrinogen adsorption than Alb along the PEG gradient.
In addition to chemical cues, topography cues also play an important role in protein adsorption. For roughness gradients, previous investigations have shown that adsorption of protein is sensitive to nanoscale roughness.278,279 Rockwell et al.184 studied protein adsorption via the Ti roughness gradient (roughness varying from 1–16 nm). The results showed that the adsorption behaviors of fibrinogen and Alb along the gradients were similar.
4.2.2. Macroscopic Cell Behaviors
Cell adhesion is the first step and a critical requirement for anchorage-dependent cells to survive, proliferate, and consequently functionalize or differentiate on a substrate. Poor adherence of these cells to substrates causes cell quiescence or even apoptosis.280 Therefore, cell adhesion is regarded as the initial indicator of cell interplay with its surrounding microenvironments, which precedes all other cellular behaviors. Following cell adhesion, they start to conform to the microenvironments, which could result in a transformation in cell morphology, spreading, orientation, migration, and eventually stem cell differentiation.
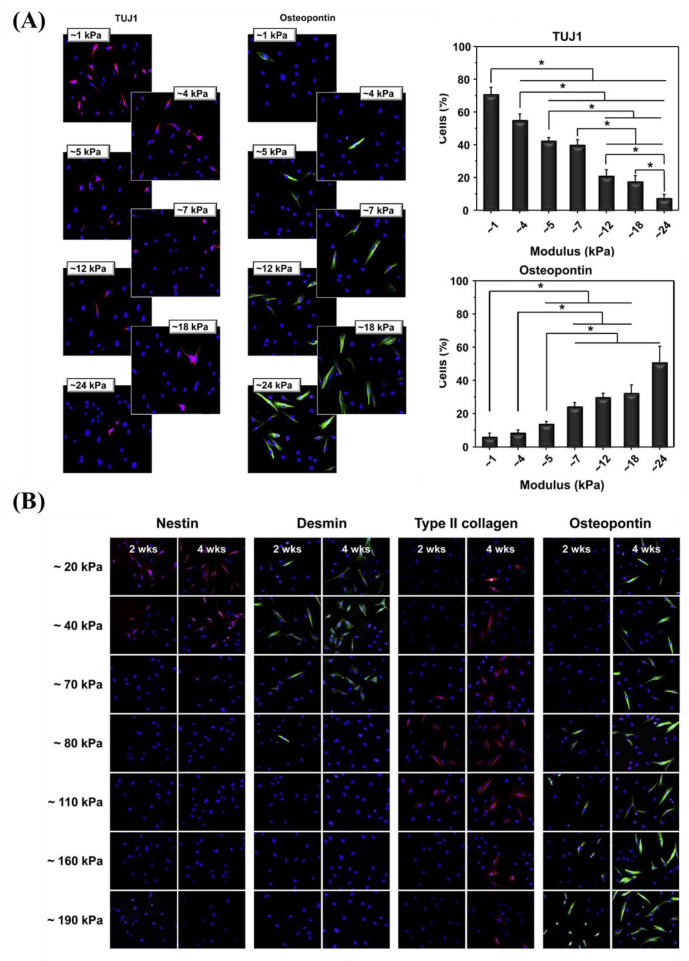
4.2.2.1. Stiffness Gradient
The mechanical properties of biomaterials play a substantial role in influencing cell adhesion and proliferation.
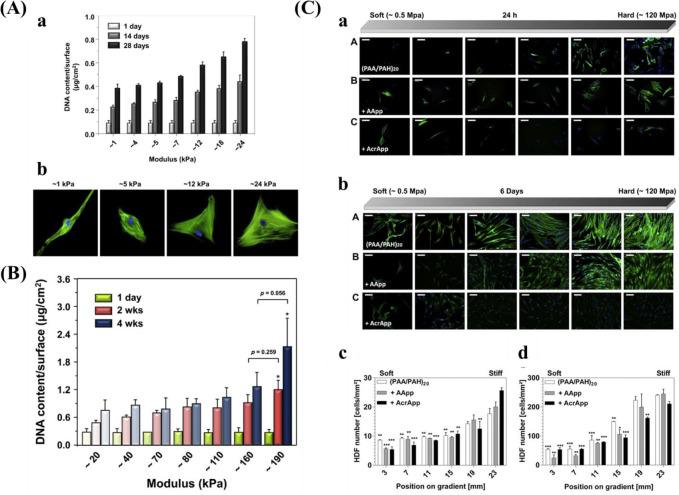
Kim et al.38 prepared a mechanical gradient varying from 1 to 24 kPa composed of poly(vinyl alcohol) (PVA) hydrogels to explore adhesion and proliferation of hMSCs. The results in Figure 10A show that there is no significant difference for original cell adhesion, and that stiffer hydrogel sections allow for better cell proliferation compared to the softer parts. After 4 weeks, cells exhibited better adhesion and more spreading when grown on the stiffer region, while cells showed elongated shape on the softer part. While this study used a gradient spanning 1 order of magnitude, other researchers prepared stiffness gradients with spanning several orders more. For example, Oh et al. found a similar cell response of MSCs with stiffness gradient ranging between 20–200 kPa, indicating that there was no obvious change for cell adhesion after 1 day, but that cells on the stiffer regions displayed increased cell proliferation compared to those on the softer parts (Figure 10B). In addition to MSCs, fibroblasts showed a similar response to stiffness. Sunyer et al.153 prepared a hydrogel with a stiffness gradient ranging from 1–240 kPa and found that material stiffness has a significant effect on cell spreading.
Figure 10.
(A) (a) DNA contents of hBM-MSC grown on the stiffness gradient for different days and (b) immunofluorescence staining of hBM-MSCs after culturing for 28 days. Reprinted with permission from ref (38). Copyright 2015 Elsevier, Ltd. (B) DNA contents of hBM-MSC cultured on the stiffness gradient for different time. Reprinted with permission from ref (147). Copyright 2016 Elsevier, Ltd. (C) Fluorescent images of stained human dermal fibroblasts (HDFs) across the gradient after (a) 24 h attachment, and 6 days proliferation. (III) Cell density of HDF after (c) 1 day and (d) 6 days. Blue for cell nucleus and green for cytoskeleton. Reprinted with permission from ref (39). Copyright 2013 Elsevier, Ltd.
For the softer region, cells displayed a circular shape and low spreading. Conversely, cells showed better attachment and spreading in the stiffer region. In addition to stiffness in the range of kPa, researchers also studied the cell response on substrates with stiffness in the MPa range. Hopp and co-workers39 showed that the adhesion of fibroblasts also depended on the stiffness of the substrate. Cells attached much better and proliferated quicker when grown on the stiffer parts (110 MPa). Cells grown on the softer parts (0.5 MPa) displayed a weaker actin structure compared to the more organized cytoskeleton fibers for cells grown on stiffer regions (Figure 10C).
4.2.2.2. Topography Gradients
It has been demonstrated that the surface topography of biomaterials has a crucial influence on various cell behaviors such as morphology,281−283 adhesion,284 and proliferation.285,286 In particular, gratings are a common and popular model to study the influence of nanotopography on cellular behavior and function.160 Kim et al.160 prepared micro- and nanotopographic arrays comprising of different densities (fixed width for ridge and depth, and varied widths for groove (1–9.1 μm)), and the elongation and orientation of fibroblast were susceptive to them. For example, fibroblasts showed a higher degree of orientation on the denser parts compared to cells grown on parts with a lower density of surface structures. In addition to density, height also plays an important role in cellular behaviors. Lin-Gibson et al.174 cultured MC3T3-E1 murine preosteoblasts onto nanograting gradients (height (H) ranging from 0 to 350 nm), and found that cells grown on lower height (H < 30 nm) showed a lower degree of orientation compared with those on higher height.
Compared to the grating patterned surface consisting of sharpened corners, which is not supportive for cell attachment, wavy-like architectures (e.g., wrinkle) may more appropriately mimic the natural structure of ECM.287,288 Zhou et al.34 prepared directional wrinkle gradients (amplitudes increasing from 0.1 to 260 nm and wavelengths changing from 200 to 1087 nm), and found that topography with lower amplitude and smaller wavelength is better for osteoblast attachment, and higher amplitude and wavelength are beneficial for cell orientation. Furthermore, the authors proposed a new method translating PDMS-based wrinkle gradients to the inorganic surface (SiO2, TiO2, CrO3, and Al2O3).100 Results showed that the optimum parameter for improving hBM-MSC behaviors, for example, orientation, focal adhesion assembly, was different for the different materials used (Figure 11A). From the evidence mentioned above, anisotropic structure, for example, grating or wrinkle, has an important influence on cell behaviors.
Figure 11.
(A) (a) Fluorescent staining of the gradients. Red color is for cytoskeleton. (b) Cell macroscopic behaviors. Reprinted with permission from ref (100). Copyright 2017 American Chemical Society. (B) (a) SEM of the polycaprolactone roughness gradient. (b) F-actin staining at day 4. (c) Quantified cell perimeter along the gradient after 4 days. Reprinted with permission from ref (181). Copyright 2014 Elsevier, Ltd.
In addition to topography with a specific orientation, more random-like topography without specific orientations such as roughness are often used, also because they are more easily applied because of the use of etching techniques. However, cell responses found throughout many studies were different or even opposite with similar topography but using different cell types. For example, Faia-Torres et al.181 explored the morphology of MSCs on a roughness gradient. They found that the cell perimeter was enhanced with increasing roughness (Figure 11B). Kunzler et al.182 also studied the response of osteoblast and fibroblast using a similar roughness gradient. However, the response for the two cell types is different. For osteoblasts, cell proliferation rate markedly increased with increasing surface roughness, and the cell area increased significantly when the extent of roughness became lower. Interestingly, fibroblasts displayed the inverse trend for proliferation, and a higher level of roughness reduced cell proliferation. For similar roughness gradients, different groups obtained contradictory results. Washburn et al.180 showed that the smoother parts of the roughness gradient increased the proliferation rate of MC3T3-E1 cells compared to the rough parts. Furthermore, there was no distinct difference between cell spreading and density of focal adhesions.
In addition, pore size also has a crucial influence on cell macroscopic behaviors. Wang et al.36 prepared a pore size gradient ranging from 29–226 nm in diameter, and found that rat MSCs showed a weaker attachment on areas with a smaller pore size and attachment increased with increasing pore size. However, cell proliferation was not affected by the surface topography. In another study, they also prepared a pore size gradient increasing from 50 nm to 3 μm,177 and found the density and spreading area of MSCs decreased with increasing pore size. These results demonstrate that pore size with different orders of magnitude could result in different cell responses. In addition to the small pore size, other research groups also engineered large pore sizes and investigated the response of different cell types. Oh et al.175 fabricated a pore gradient with larger pore sizes (88–405 μm). The substrate with 380–405 μm facilitated cell expansion for chondrocytes and osteoblasts, while 186–200 μm was beneficial for fibroblasts. However, using a similar pore size gradient (90–400 μm),176 the same group found larger pore size down-regulated the proliferation of stem cells. These pieces of evidence show that the pore size gradient is a good platform for studying cell–biomaterial interactions.
In addition, several groups investigated cell response mediated by nanoparticle-density gradients. Kunzler et al.185 seeded osteoblasts onto particle-density gradients (73 nm in diameter) and found a significantly diminished cell quantity on regions with a higher density of particles, and lower density of particles improved cell spreading and formation of cytoskeleton structure. However, inverse cell response was found by Bachhuka and co-workers.187 In their study, they fabricated gold nanoparticles with diameters of 16, 38, or 68 nm. The results showed that regions with a higher density of nanoparticles enhanced the attachment and proliferation rate of human dental pulp derived stem cells. In addition to nanoparticles, Xia et al.189 prepared a density gradient of microparticles (3 μm in diameter) and studied the effect on neurite growth. The moderate roughness increased the length of neurite compared to a low and high degree of roughness. The optimum roughness with a Ra value of 594 ± 89 nm was identified for maximized cell adhesion and neurite extension. These results show that cell adhesion and proliferation are significantly dependent on particle size and cell type.
4.2.2.3. (Bio)Chemical Gradients
Surfaces with different kinds of (bio)chemistry have been prepared, for example, density of poly(ethylene glycol) brushes,266 Fn,289,239 laminin,235 gelatin,290 collagen type I (COL1), and osteopontin (OPN),291 osteogenic growth peptide (OGP),246 RGD,244 −CH3,218 −COOH,219 plasma polymerized hexane,225 Acrylic acid/diethylene glycol dimethyl ether,231 PLLA/PDLLA,254 poly(caprolactone) and poly(d,l-lactide),292 poly(2-hydroxyethyl methacrylate).240 Mohan et al.218 fabricated chemical gradients on the surface of PDMS, and water contact angle (WCA) increased from ∼10° to ∼100°. Their results showed that the spreading of fibroblasts was weaker when grown on the hydrophilic region and better on the hydrophobic part. However, other researchers obtained different results. Zelzer et al.225 also fabricated a chemical gradient with varied WCA. WCA ∼60° facilitated the attachment and proliferation of fibroblasts compared with WCA ∼93°. Similar results were achieved by the same research group by preparing a wettability gradient between <10° and 98°,222 and found that cell density decreased from the hydrophilic side to the hydrophobic side. A critical but often overlooked result from the work of Zelzer et al.,225 is that the cell densities observed on the gradients were not the same as those observed on uniform samples of the same chemistry. This was rationalized as differences in the cell–cell signaling processes or protein production from surrounding cells on the gradient compared to the uniform sample format. This is a note of caution indicating that cell response studies on gradients require validation before extrapolation to predict the cell behavior on homogeneous samples.
In addition to chemical cues, biochemistry signals, for example, proteins or protein composition could also modulate various cell behaviors. Ghemei et al.291 prepared density gradients of OPN and COL1, and the results (Figure 12A) showed that the COL1 gradient from 78.5 to 124.2 ng/cm2 facilitated the attachment and proliferation of MSCs and there was no prominent difference detected when cells were seeded onto region with a higher density of COL1. For the OPN gradient, the trend was similar as for COL1, and the best density for prompting cell attachment was 20.7 ng/cm2. These results demonstrate that cell adhesion and proliferation are appreciably dependent on protein concentration.
Figure 12.
(A) Attachment of MSC on (a) COL1 and (b) OPN gradient after 5 h. Reprinted with permission from ref (291). Copyright 2019 WILEY-VCH. (B) Cell morphology on (a) OD–AA and (b) AA–DG gradient. Quantification for cell number after (c) 1 day and (d) 6 days. Reprinted with permission from ref (231). Copyright 2015 Elsevier, Ltd.
Several groups used plasma polymer deposition, for example, allylamine (AA), octadiene (OD), and diethylene glycol (DG) to fabricate chemical gradients. Vasilev et al.228 prepared OD–AA gradient (ratio of N/C between 0.04 and 0.16), and the regions riched in AA enhanced the attachment and cell spreading of stem cells under normal culture medium with serum. However, the differences were significant when a serum-containing medium was used and all differences were lost under serum-free conditions, which indicates that preferred adsorption of serum proteins plays a determining role here. A similar trend was found by Short and co-workers,220 that is the attachment of mouse embryonic stem cell increased from the OD to the AA region. However, Voelcker et al.293 found the attachment of embryonic stem cells prompted with increasing N/C ratio to 0.1, and higher ratios negatively influenced cell attachment. Furthermore, the group of Voelcker also developed an AA–DG gradient221 and found that the attachment of embryonic stem cells depended on the gradient. Cell attachment decreased from the region rich in AA to the region rich in DG. Similar conclusions were obtained when using different cell types. For instance, in another study, Voelcker et al.231 developed OD–AA and AA–DG gradients simultaneously. MSCs density decreased from AA to OD, and from AA to DG end (Figure 12B). Taken together, these results demonstrate that (bio)chemical cues have a significant influence on cell macroscopic behaviors.
4.2.2.4. Multiparameter Gradients
In addition to the single parameter gradients, researchers also prepared multiparameter gradient platforms, for example, orthogonal nanometer-micrometer roughness gradients,31 orthogonal chemistry-topography gradient,257 orthogonal topography and surface chemistry gradients.253,260 Huethorst et al.256 investigated the influence of orthogonal topography on the morphology of cardiomyocytes. The width of the grating (horizontally) increased from 8 to 100 μm and the deepness (vertically) from ∼5 nm to ∼1 μm. Compared to a flat surface, deeper and wider gratings facilitated the alignment and stretching of cardiomyocyte. Yang and co-workers253 reported a versatile combinatorial method to explore cell–surface interactions, and the system consists of orthogonal chemical (WCA: 60–90°) and topographical gradients (the widths of the grooves increasing from 5 to 95 μm, and 5 μm for ridge width). They used 3T3 dermal fibroblast as a model cell type and found that after 2 and 3 days cell culture, WCA of 68–76° and groove widths of 40–60 μm had the highest cell coverage and greatest proliferation rate. Furthermore, the van Rijn group prepared an orthogonal double gradient (stiffness, 6–89 MPa; WCA, 29–90°).146 The optimum parameter for cell attachment was WCA: 35–39° and stiffness: 8.2–9.3 MPa, stiffness: 8–50 MPa and WCA: 79–85° for largest cell area, and stiffness: 40–89 MPa and WCA: 29–34° for maximum expression of vinculin. These pieces of evidence highlight that the orthogonal double gradient approach is a powerful tool for identifying optimum combined parameters for specific cell behaviors, therefore accelerating the development of high-performance biomaterials.
4.2.3. Stem Cell Differentiation
The microenvironment in which cells reside is complicated and dynamic. It is extremely important for stem cells to controllably differentiate into different cell types in tissue engineering and regenerative medicine. The microenvironment of the stem cell niche is important for regulating and controlling the fate of stem cells. The microenvironment is composed of stem cells, growth factors, and ECM.143
It was shown that physical properties (for instance, substrate stiffness, and topography) and (bio)chemistry of biomaterials significantly affect stem cell behaviors. So far, many studies examined the influence of these parameters on stem cell differentiation via an HTS approach.
4.2.3.1. Stiffness Gradient
In vivo, tissues have a broad range of mechanical properties, for instance, ∼1 kPa for brain294 and ∼10 GPa for bone.295,296 The stiffness of biomaterials is a crucial factor for regulating stem cell fate.70,151,297,298 Many researchers prepared different kinds of materials and stiffness gradients of different ranges to study the influence on stem cell differentiation.38,147,148,299 Polyacrylamide (PAAm) hydrogels have been widely used as a mechanically tunable substrate. For example, Engler et al.300 prepared a PAAm stiffness gradient which ranged from 1–14 kPa, and the stiffer region enhanced myogenic differentiation of MSCs. However, this kind of material may induce cytotoxicity301,302 and the surface always requires a protein coating process.
Freeze–thaw cycles facilitate liquid poly(vinyl alcohol) (PVA) to form a hydrogel.149 Kim et al.38 prepared PVA hydrogel with stiffness gradients (from 1 to 24 kPa). The softer region enhanced neuron differentiation while the stiffer region improved osteogenic differentiation of MSCs (Figure 13A). Furthermore, the same research group also prepared PVA/HA stiffness gradients with a broad stiffness range of 20–200 kPa with the same method.147 The results (Figure 13B) showed that the optimum mechanical value for neurogenesis, myogenesis, chondrogenesis, and osteogenesis of MSCs was ∼20 kPa, ∼40 kPa, ∼80 kPa, and ∼190 kPa, respectively. The mechanical property of hydrogels mentioned above is always in the kPa-range and it is very soft compared to the stiffness of some tissues in vivo.296 As stem cells probably become a particular cell type on the surface of a biomaterial with a similar value as a tissue in vivo,6,71 enlarging the range of stiffness gradient is important and necessary.
Figure 13.
(A) Immunofluorescent staining of hBM-MSC grown on 1–24 kPa gradient after 28 days and the results for quantitation. Reprinted with permission from ref (38). Copyright 2015 Elsevier, Ltd. (B) Immunofluorescent staining of hBM-MSC grown on 20–200 kPa gradient after 2 and 4 weeks. Reprinted with permission from ref (147). Copyright 2016 Elsevier, Ltd.
PDMS is a common and popular biomaterial suitable for many applications.303−306 Compared to hydrogels, PDMS is stable in an aqueous environment, helpful for preparing stiffness gradient.307 In addition, the stiffness value of this material is always about kPa or MPa,308 much higher than the mechanical properties of the hydrogel. Wang et al.148 fabricated stiffness gradients with PDMS (190 kPa–3.1 MPa) to investigate osteogenesis of MSCs. After 1 week of culturing cells in differentiation medium, stiffer regions enhanced osteogenic differentiation compared to the softer regions, as evaluated by Alizarin Red staining that indicates mineral production. Taken together, the stiffness gradient is a convenient platform for screening the best mechanical value for fate commitment of stem cells, therefore helpful for designing biomaterials in vitro.
4.2.3.2. Topography Gradient
It is well-known that topography can adjust various cell behaviors and stem cell fate.309 In this part, we will focus on the stem cell differentiation stimulated by anisotropic and isotropic topography, which has been studied using gradients.
For anisotropic gradients, ECM architecture of some tissues, for example, nerve, bone, vessel, muscle, comprise complicated and parallel structure.12 Wave-like topography gradients with a height between 541 and 3073 nm and wavelengths increasing from 4 to 30 μm were used to study the fate commitment of MSCs toward neuron lineage.173 They found that the substrate with wavelength 26 μm/height 2.9 μm was optimum for neuron differentiation (Figure 14A). These results demonstrate that an anisotropic gradient platform can serve as an effective system to obtain the optimum parameter for specific cellular behaviors, which could improve regenerative medicine of stem cell therapies.
Figure 14.
(A) Immunostaining of Tuj1 of MSCs grown on wrinkle gradients for (a) 1 week and (c) 2 weeks. (b, d) Zoomed picture of the typical position and quantitative results of Tuj1. Reprinted with permission from ref (173). Copyright 2020 Wiley-VCH. (B) (a) Immunostaining of ALP for cells cultured on roughness gradient after 4 and 21 days. (b) Quantitative results of ALP. Reprinted with permission from ref (181). Copyright 2014 Elsevier, Ltd.
For the isotropic gradient, structures such as roughness,181 nanoparticle,187 nanotube,310 and pore size36,177 are the commonly used models to study their effect on the stem cell fate. The roughness of the biomaterial surface is one of the most important parameters for successful osteointegration.206 Faia-Torres et al.181 investigated the effect of roughness gradients (Ra: ∼0.5–4.7 μm) on the osteogenesis of MSCs under differentiation medium. They found that a specific roughness of Ra: ∼2.1–3.1 μm was the optimum for improving osteogenesis of MSCs (Figure 14B). Soluble factors also play an important role in modulating stem cell fate.311 For instance, dexamethasone (Dex) is always added into the differentiation medium,311 and has been shown to support osteogenesis.312 However, Dex negatively influences the migration of MSCs.313,314 For this reason, the same research group engineered the same parameters of roughness gradient (Ra: 0.5–4.7 μm) to investigate the influence on osteogenesis of MSCs cultured in differentiation medium without the addition of Dex and in growth medium.315 The results showed that Ra ∼1.53 μm in differentiation medium without Dex, and Ra ∼0.93 μm in growth medium exhibited the highest capacity for improving osteogenesis. Alternatively, roughness gradients can be fabricated through particles.189 For example, Bachhuka et al.187 prepared density gradients of gold nanoparticles (diameter: 16, 38, and 68 nm, respectively), and the results showed that higher roughness was beneficial for osteogenesis.
During the process of bone regeneration, architectures with pores are beneficial for the exchange of nutrients, cell movement, growth, and vascularization. In addition, a porous surface of a biomaterial promotes connection with the natural bone.7 It has been demonstrated that different pore sizes have a significant effect on osteogenesis. For instance, scaffolds with smaller pore sizes are beneficial for chondrogenic differentiation, while larger pore sizes enhance osteogenic differentiation.7 Wang et al.36 produced gradients with pore sizes between 29 and 226 nm in diameter and roughness decreasing from 22 to 3 nm. They found that roughness smaller than 10 nm promoted osteogenic differentiation of MSCs, while adipogenic differentiation is not dependent on pore size. In addition to osteogenic and adipogenic differentiation mediated by pore size, the influence of pore size on chondrogenesis has also been studied. Oh et al. fabricated PCL scaffolds with pore sizes ranging from 90 to 400 μm and porosity between 80 and 97%. The results showed that pore size 370–400 μm was the optimum for chondrogenesis of stem cells.
However, the stability of biomaterials is a common problem. Therefore, a stable gradient substrate composed of varied pore sizes is desired.177 Compared to pSi, porous alumina (pAl) has attracted much attention199,203,316 because of high stability in aqueous environments199 and has served as implant material.202 Wang and co-workers177 reported the influence of pAl comprising pore size gradients between 50 nm and 3 μm on the osteogenesis of MSCs. The optimum parameter for osteogenic differentiation was 120–230 nm. These results show that isotropic gradients, for example, roughness, nanoparticle, pore size, allow for screening of the optimum parameter for varied stem cell differentiation, which improves the advancement of biomaterials for tissue engineering.
4.2.3.3. (Bio)Chemical Gradients
The MSC differentiation depends not only on the mechanical and topographical properties of the biomaterial but also on the chemical environment. Several groups have shown that chemistry has an important effect on stem cell fate and used gradients to study this.219,228,231,317 Voelcker et al.291 prepared density gradients of COL1 (ranging from 78.5 to 132.3 ng/cm2) and OPN (increasing from 9.6 to 21.4 ng/cm2) (Figure 15A). The osteogenesis increased with increasing COL1 density, and the optimum density was 124.2 ng/cm2, and 19.0 ng/cm2 for the density of OPN. Moreover, Vasilev et al.228 fabricated OD–AA gradient (N/C ratio between 0.04 and 0.16) and found that the AA side improved the osteogenesis of stem cells, while the OD side enhanced adipogenesis. With a similar gradient platform, the authors also investigated the differentiation into endoderm, mesoderm, and ectoderm in the absence of bioactive differentiation factors.317 They found that high ratios of AA/OD were beneficial for mesoderm and ectoderm. Furthermore, OD–AA and AA–DG gradients were fabricated to systematically screen MSCs differentiation.231 Using osteogenic induction medium, osteogenic differentiation increased along with the OD–AA gradient, and decreased along the AA–DG gradient. In adipogenic induction medium, there was no obvious difference for adipogenesis for cells cultured on the OD–AA gradient, but declined from the AA side to DG side. Furthermore, osteogenesis was significantly enhanced by a thick surface of AA (>40 nm) under a mixed differentiation medium on an OD–AA gradient. However, adipogenic differentiation did not show remarkable differences (Figure 15). These results show that stem cell fate can be influenced by (bio)chemical cues and (bio)chemical gradients are a powerful approach for screening the optimum condition for specific differentiation behavior.
Figure 15.

(A) Influence of COL1 and OPN gradient on Runx2 expression after 7 day under (a, c) osteogenic medium (OSM) and (b, d) DMEM. (e) Quantitative results of Runx2 on the COL1 and OPN gradients. Reprinted with permission from ref (291). Copyright 2019 Wiley-VCH. (B) Adipogenesis/osteogenesis on OD–AA gradient under mixed medium after 14 days. (a) Nile red staining and (b) Calcein Blue staining, respectively. (c) Quantification results along the gradient. Reprinted with permission from ref (231). Copyright 2015 Elsevier, Ltd.
While these approaches focused on engineering gradients with individual parameters (e.g., mechanical property, topography, and (bio)chemistry), it is meaningful to prepare various parameters on one substrate, as those may interact with each other to affect cell behaviors in a synergistic manner.318 Several groups prepared multiparameter gradient platforms to study stem cell differentiation.256,258,259 Rape et al. created hydrogel composed of stiffness and Fn density combined in an orthogonal way. After 1 week under mixed differentiation medium conditions, differentiation of cells was assessed. A higher density of Fn and higher stiffness was better for osteogenesis, while adipogenesis depended on stiffness but not on Fn density. For stem cell differentiation, 3D structures better mimic the environment in vivo.319 Vega et al.320 fabricated a hydrogel substrate with RGD and HAV peptide gradients to investigate the influence on MSC chondrogenesis. The results demonstrated that higher HAV but lower RGD density was beneficial for chondrogenesis.
4.3. Cell Migration on Gradients
For regenerative medicine, tissue reconstruction requires the (stem) cell to be recruited to the injured site.321 To elucidate the mechanism for this process and to provide more information for preparing biomaterials, it is important to investigate cell migration under physical, and (bio)chemical cues.47 Cell migration always occurs in many physiological and pathological activities, for instance, morphogenesis,322 angiogenesis,323 immune responses,324 the renewal of skin and intestinal cells,325 and tumor metastasis.326,327 Cell migration includes various complex procedures, including the formation of filopodia, change of cell contractility, the development of focal adhesions.328,329 Similar to other cell activities, cell migration is also controlled by physical parameters (such as stiffness and topography) and the (bio)chemistry of the microenvironments of cells.330,331 The type of migration could be divided into several types, including dissolved chemoattractants (chemotaxis), immobilized molecules (haptotaxis), and biophysical contact cues (topotaxis, durotaxis).332,333
4.3.1. Durotaxis on Stiffness Gradients
The stiffness of biomaterials is able to not only adjust cell attachment, morphology, proliferation, and differentiation but also modulate cell migration.334 The procedure to guide cell migration by stiffness is termed “durotaxis”.335 Durotaxis modulates cell migration by mechanical communication or mechano-biochemical transduction.336,337 In addition, durotaxis is related to many biological procedures, including epithelial-to-mesenchymal transition,338,339 nerve tissue development,340,341 innate immunity,342 and cancer metastasis.343,344 Durotaxis always triggers cells to move toward stiffer parts.154 Wong et al.345 further indicated that durotaxis of vascular smooth muscle cells increased with increasing magnitude of the gradient, but there was no relationship with the absolute modulus.
Until now, there is no conclusion about MSC differentiation to be prior to migration or postmigration. To answer this question, Engler et al. fabricated a hydrogel containing a physiological gradient of 1.0 ± 0.1 kPa/mm, and found that MSCs migrate to the stiffer region and then start to differentiate into the specific cell type. Furthermore, Hadden et al.48 developed polyacrylamide hydrogels with varying stiffness gradients to study the migration of human adipose-derived stem cells (hASCs). They found that the average speed was similar for cells cultured on a shallow gradient (2.9 kPa/mm) compared with that on a steep gradient (8.2 kPa/mm). However, cells displayed obviously higher speed toward y-direction, suggesting a stiffer region is beneficial for durotaxis.
In vivo it has been evidenced that mechanical gradients always accompany changes in the composition of ECM for many diseases. For example, for lung fibrosis, mechanical properties increasing in the lung parenchymal tissue are associated with the enhancement of COL1,346 and in breast cancer, an increase of mechanical property from the tumor core to the periphery is accompanied by elevated COL1 and laminin.347 For this reason, Hartman and co-workers348 compared the durotaxis of vascular smooth muscle cells (VSMCs) on stiffness gradients decorated with Fn or laminin. The results (Figure 16A) demonstrated that VSMCs grown on Fn-coated surface showed durotactic behavior. Nonetheless, cells grown on laminin-coated surfaces did not exhibit durotaxis.
Figure 16.
(A) Representative migration behaviors for VSMCs cultured on the stiffness gradient and uniform stiffness gels decorated with Fn or laminin. Reprinted with permission from ref (348). Copyright 2016 National Academy of Sciences. (B) Migration trajectories of cells on an (a) densely and (c) sparsely spaced ridged arrays. The skewness of the distributions is shown in each panel (b, d). Reprinted with permission from ref (160). Copyright 2009 Elsevier, Ltd. (C) Cell migration trajectories on the swelling gradient. The arrows mean the ratios of cells moving to the direction of the lower degree of hydration. Reprinted with permission from ref (357). Copyright 2013 Elsevier, Ltd. (D) Left: Schematic representation for the structure of a complementary density gradient of PDMAPS and KHI. Right: The effect on the migration of SCs and FIBs. Reprinted with permission from ref (368). Copyright 2015 Elsevier, Ltd.
Although most studies have focused on the durotaxis in 2D, it is important to explore the influence of mechanical property on cell migration in a 3D microenvironment as it could better mimic the structure in vivo. Several groups studied the influence of the stiffness gradient on cell migration in 3D.44,349 For instance, Joaquin et al.44 demonstrated that cell migration speed was dependent on the development of the stiffness gradient and not on the absolute stiffness value.
4.3.2. Topotaxis on Topography Gradients
Living cells in vivo can sense the structure of ECM to modulate their morphology, movement, and fate.350,351 It is well-known that surface geometry of biomaterial can profoundly affect cell migration.41,352,353 Cells are inclined to migrate along the substrate grooves, while random movement happens on flat surface.352 NIH3T3 fibroblasts grown on a grooved surface with 550–1100 nm spacing displayed faster migration speed than those on 2750 nm spacing.354 Kim et al.160 found that fibroblasts adhering to the areas with higher density patterns showed higher alignment and elongation along the ridges; while cells on the areas with the lower density of pattern displayed a biphasic dependence of the migration speed on the pattern density (Figure 16B). Recently, Ge et al.172 developed a topographical gradient with wavelike features that gradually change in wavelength and amplitude to explore the migration behavior of fibroblasts. The wound coverage speed was quantified on the regions with wavelength of 2, 5, and 8 μm both in the perpendicular direction and parallel direction. The results showed that smaller wavelength (2 μm) facilitated migration compared to the larger wavelength, and the lower amplitude was beneficial for cell migration. However, cells showed quicker speed on a larger wavelength under the same amplitude. These results provide important insights into topography-induced cell migration.
4.3.3. Chemotaxis on Chemical Gradients
Various biological and chemical cues are involved in cell migration. Introducing these signals onto the surface of biomaterials will affect cell migration behaviors.355 Many chemical cues have been decorated onto biomaterials to study the movement of cells, for example, chemical density,356 swelling,324,357 molecular weight (MW),358,359 ECM proteins (fibronectin,250,360,361 collagen,362,363 lamin,364 and gelatin290) and their derived peptides,247,248,365−370 and growth factors.250,251,370−372
Collagen is the main component within ECM and it can significantly enhance cell attachment and spreading.47 Gelatin composed of various peptides and proteins resulted from collagen hydrolysis can also improve cell attachment and proliferation.373 Fn is another important protein within the ECM and it is beneficial for promoting angiogenesis.374 Smith et al.360 investigated the movement of bovine aortic endothelial cells (BAECs) on Fn gradients bound on the gold surface. They used two different methods (nonconfluent cells versus confluent cells) to explore the migration behavior of BAECs on the Fn gradients. The results displayed that the cell migration speed increased with increasing the Fn gradient compared with the uniform control substrate. The Fn slope of 0.15 ng/mm2 increased the migration speed of BAECs from −0.15 and 4.45 μm/h to 1.71 and 7.82 μm/h for nonconfluent and confluent cells, respectively. Furthermore, migration speed is also dependent on the density of the protein. For example, Cai and co-workers362 prepared collagen density gradients onto PDLLA film to investigate the migration of endothelial cells (ECs). ECs grown on the gradient surface with low or moderate densities of collagen showed a strong motility tendency. However, there was a reverse trend for cells cultured on the gradient area with low collagen density. These pieces of evidence demonstrate that cell migration adjusted by the collagen gradient depends on the protein density. However, it should be noted that most gradients are relatively oversimplified compared to degradable biomaterials. Therefore, Yu et al.290 prepared a gelatin density gradient (from 0.49 to 1.57 μg/cm2) on poly(ε-caprolactone) membrane, and investigated the influence on the migration behavior of ECs. Cells showed directional migration toward the higher concentration of gelatin. In contrast, the ECs showed random movement on the PCL membrane and the uniform protein surfaces. Peptides with amino acid sequences mimicking functional features of ECM proteins have been used as an alternative for the above-mentioned protein-density gradients because of their higher stability and low MW. Most cells prefer to move toward the higher concentration of peptide and migration behavior is based on the gradient slope of the peptide. For instance, Guarnieri et al.366 used a decorated RGD gradient on a hydrogel surface and studied the influence on cell migration. Results suggest that cells migrated to the higher density of RGD compared to a uniform density of RGD, and increased their migration by increasing the gradient slope (0.7, 1, and 2 mM/cm).
Except for ECM proteins and peptides, different kinds of growth factors (basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF)) have also been prepared into gradients to explore the influence on cell migration. Wu et al.251 reported the influence of the density gradient of bFGF (density increased with a slope of 17 ng/cm2/mm) on the directional migration of VSMCs. They found that up to 70% of the VSMCs migrated toward the part with a higher density of bFGF. However, the bFGF gradient did not influence the cell migration rate. A similar conclusion was also drawn by Cai and co-workers372 with a different cell type. They fabricated a gradient density of VEGF from 54 to 132 ng/cm2 with a slope of 7.8 ng/cm2/mm. About 72% of ECs migrated toward the part with a higher density of VEGF. However, the cell migration rate was not influenced.
Although various gradients of biological molecules such as proteins, peptides, and growth factors have been proved effective in adjusting cell migration, it remains challenging to design biomaterials for precisely modulating cell migration because of the complexity of natural macromolecules in the ECM where cells reside. In addition, these biological molecules are costly and easy to diminish bioactivity or even denature, limiting their applications in vitro. For this reason, Han et al.357 prepared the swelling gradient and found that the VSMCs migrated to the low hydration region under a suitable cell density (1.5 × 104/cm2) (Figure 16C). To avoid direct contact between cells and only study the cell–substrate interactions, the migration of cells at low density (5 × 103/cm2) was also investigated. In this case, cell migration was not influenced. Ren et al.358 prepared an MW gradient of poly(2-hydroxyethyl methacrylate) (PHEMA) brush with a thickness between 3 and 30 nm and slopes of 0.8–3.2 nm/mm, and then studied the directional migration of VSMCs. The VSMCs migrated toward the region with reduced thickness.
The investigations mentioned above are mainly focused on nonselective cell migration, however, selective cell migration is extremely crucial in many physiological procedures, for example, cancer metastasis, and inflammation.328 Undesired cell migration can lead to serious problems.375,376 For example, after vascular injury, the appropriate directional migration of SMCs is important. However, compared to SMCs, fibroblasts (FIBs) may have a quicker response to inflammatory signals,377,378 leading to the generation of scars. This could cause the reduction in contractile function of SMCs and final angiosclerosis and adventitia fibrosis.379 Therefore, it is important to design biomaterials that could facilitate the directional migration of SMCs over FIBs.380
However, most kinds of biomaterials are not suitable for selective cell migration and only some studies have reported this.247,248,356,368,369 Yu and co-workers247 prepared density gradients of Val-Ala-Pro-Gly (VAPG) peptides on a PEG surface. This peptide can specifically bind to SMCs through cell surface receptors.381 The results demonstrated that the migration of SMCs was prompted toward the higher density of VAPG. In contrast, the movement ability of FIBs was markedly decreased. In addition, endothelium composed of endotheliocytes (ECs), is damaged during atherosclerosis. The migration speed of SMCs is faster than ECs, leading to further damage to the vasculature.382 Therefore, it is important to develop and design a material that improves the migration of ECs over SMCs. For this purpose, Yu et al.248 studied the density gradient of Arg-Glu-Asp-Val (REDV) peptide and the influence on the directional migration of ECs rather than SMCs. REDV is the smallest active sequence of Fn and can be recognized by the integrin α4β1 receptor on ECs.383 The REDV gradient selectively improved directional migration of ECs toward the higher density of REDV, while it markedly decreased the attachment of SMCs without influencing the migration speed and directionality. Furthermore, selective improvement of directional migration of Schwann cells (SCs) rather than FIBs is extremely important in peripheral nerve regeneration, which is beneficial for neuron regeneration and prevents fibrosis. Therefore, a complementary density gradient of poly(3-dimethyl-methacryloyloxyethylammonium propane sulfonate) (PDMAPS) and KHIFSDDSSE peptide (KHI) was fabricated.368 The SCs displayed improved migration toward the lower density of PDMAPS and higher density of KHI, while the migration behavior of FIBs was not influenced by the surface gradient (Figure 16D). Taken together, successful adjustment of the selective directional migration plays an important role for guided tissue regeneration.
4.4. Bacterial Behaviors on Gradients
Bacteria, as one of the smallest and most tremendous organisms on Earth, use various mechanisms to guarantee their survival and flourish.384 Bacteria are capable of adhering to biomaterial implants, and the subsequent colonization and formation of biofilms are responsible for the failure of implantable biomedical devices.385 Furthermore, about 60% of nosocomial infections are associated with biomaterial-associated infections.386 Physicochemical properties of biomaterial surface play a crucial role in the process of bacterial adhesion, and several methods have been proposed to develop different coatings, for instance, polymer brush-coatings,387 poly(ethylene glycol) (PEG) derivatives or zwitterionic polymers,388,389 polycationic coatings (e.g., N,N-dodecyl, methyl-PEI),390 polyurethanes,391 microgel.392 Our group has demonstrated that microgel coatings based on poly-N-isopropylmethacrylamide prevent bacterial adhesion, and cross-linking density/mechanical property, the thickness of the microgel coating have a significant influence on the adhesion of bacteria. In addition, Vliet et al.393 prepared weak polyelectrolyte multilayered substrata, ranging from 1 to 100 MPa, and found the adhesion of S. epidermidis and E. coli correlates positively with the mechanical property of these substrates, independently of other surface properties, for example, roughness, charge density, and interaction energy. Studies of bacterial adhesion on different coatings have made excellent progress in understanding bacterial-surface interactions, and several distinguished relevant reviews already exist.394−397
However, there are relatively few studies devoted to investigating bacterial behaviors on biomaterial surfaces via gradient-based methods. Indeed, concentration/chemical gradients in solution generated by, for example, microfluidic device is a common method to study bacterial behavior, that is, chemotaxis.398,399,408,400−407 These types of gradients bare little connection to physicochemical properties of biomaterials let alone any relationship with surface gradients. Although gradients are already known and produced for decades, still it shows that with these approaches many unresolved questions are there for the solving. Future endeavors with physicochemical gradients will surely enter the field of implant-associated infections and be used to resolve fundamental questions between surfaces and bacteria.
4.5. Limitations of Gradient-Based High-Throughput Systems
Although a lot of progress has been achieved for preparing gradient-based HTS platforms and study the physicochemical influence of materials on various cell behaviors, it is challenging to establish an HTS platform that possesses physical and (bio)chemical properties. Until now, most studies about the HTS platform are performed in 2D, only a few are in 3D. While 2D would represent a tissue-implant interface, a 3D environment would better mimic physiological conditions and recreate pathologies. Therefore, 3D HTS platforms would generate more insights that could also apply to 2D interfaces. For determining the behavior of cells on implants, medical device surfaces, or tissue engineering scaffolds, 2D platforms still offer good insights into the behavior of cells. Although a note of caution needs to be given as the initial seeding density may already be a cell biological parameter that is influenced that makes further studies more complex as cell–cell interaction and paracrine cell–cell communication may affect further development. Therefore, postverification is required to draw unambiguous conclusions. Furthermore, the HTS platform combined with dynamic environments will be helpful for better understanding cell behaviors. Another limitation for HTS is analyzing images and processing data. At last, the final goal of HTS should be a focus on the application in vivo, and so far no HTS in vivo has been investigated, which would be an important step for regenerative medicine and tissue engineering.
5. Microarray Strategies Applied in Biomaterial Screening and Used to Model the Cellular Microenvironment
5.1. Introduction
While the previous section focused on gradient-based technologies, here array-based systems are addressed for studying the biological-biomaterial interactions. The merits of HTS microarray technologies have already been discussed in section 1, and include cost-efficiency, reproducibility, and parallel experimental conditions for up to hundreds to thousands of discrete conditions. Gradients are ideal for investigating the effect of a gradual change of up to 2–3 parameters upon cell response; for example, the concentration of amine surface groups mixed with a hydrocarbon functionality,225 or the relative influence of topography versus chemistry for a range of dimensions of the former and concentrations of the latter.257 Micorarrays on the other hand are well suited to look at many unique and spatially defined identities in parallel, for example, a library of 141 monomers, polymerized alone and mixed to form 909 unique polymer spots on a microarray.409 As such, microarray technology provides discrete representations of unique materials, or material-topography combinations in the case of the ChemoTopoChip presented by Burroughs et al.410 with 1008 unique environments, allowing large parameter space to be explored, unrestricted by the need for them to be related as in the case of gradients.
Within this part, section 5.2 outlines the methods used in microarray preparation for cellular applications, while section 5.3 will review the HTS methodologies developed to investigate how defined chemical and physical properties within man-made biomaterials can invoke particular cellular responses. Section 5.4 will consider the biological models and assays used in high-throughput technologies, and their impact in biology and biomedicine.
5.2. Methods for HTS of Cell–Biomaterial Interactions
Brocchini et al.411 published one of the first cell-based screenings of a library of synthetic polymers in 1997. The concept of combinatorial design in monomer systems was used to create libraries of structurally related polymers that would vary in a predictable and systemic fashion, which were then tested to assess biological performance.411 In particular, 14 tyrosine-derived diphenols and eight aliphatic diacids were prepolymerized in glass chromatography vials combinatorially to produce 112 polyacrylates, characterized by using gel permeation chromatography, WCA measurement, differential scanning calorimetry, and thermogravimetric analysis. The polymers were then coated onto glass coverslips in quadruplicate and tested for their ability to promote cell attachment and growth of rat lung fibroblasts in 24-well nontissue culture-treated polystyrene plates; with tissue culture-treated polystyrene as the positive control. The authors performed correlation studies between chemical descriptors and cellular performance, showing that WCA and the level of polymer backbone substitution with oxygen correlated with higher cell growth.412 This study illustrated well that combinatorial approaches were indeed a valuable tool to (1) increase the number of polymers tested for any given application and (2) facilitate the systematic study of the relationship between material properties and biological response.412
To increase the number of materials that may be investigated, polymer microarrays were developed where small polymer spots in the μm-range afford high density on a substrate. A large variety of techniques, such as but not limited to, photolithography,413 soft-lithography,414 microfluidics,415 nanolithography,416 contact pin,417 and inkjet418 automated printing, and on-chip synthesis,419 have been used to fabricate microarrays. These approaches vary in their flexibility; from the type and range of defined biomaterials, they are able to support, to their solvent compatibility, their freedom to control object shape, and their polymerization strategies.
The microarray format is an ideal platform for the rapid assessment of material-cell responses, utilizing robotics to facilitate near parallel processing and chemical and cell analysis at a rate that increased exponentially from traditional methods. However, for microarray technologies to yield reliable biological results, careful preparation and quality control methods are required. In this section, the basis for the fabrication of microarrays for cellular studies will be outlined, along with the surface characterization required to elucidate relationships between the cellular response achieved and the properties of the biomaterial. A schematic of a typical workflow for cell-based studies using microarrays is presented in Figure 17.
Figure 17.
Illustrative workflow for high-throughput microarray studies in cellular applications (representations not to scale). First, microarray fabrication can be done through automated liquid dispensing systems (contact printing with a solid or quilled pin, or inkjet printing). Second, high-throughput is carried out for (1) material analysis technologies (e.g., time-of-flight mass spectrometry (ToF SIMS), WCA, X-ray photoelectron spectroscopy (XPS), atomic force microscopy (AFM)), and (2) biological performance. Third, biological and structural data are correlated, and used to generate structure–activity relationship models. The material library can be further mined through combinatorial microarrays of the highest performing biomaterials, and computational models can be generated from the available data sets. After extensive mining of the microarray-generated data, scale-up studies take the best-performing polymers onto bigger platforms, such as multiwell tissue culture plates, robot-assisted automated cell culture platforms using RoboFlasks, or 3D culture by generating microparticles from the hit polymers and using them in Bioreactor-based strategies. In-depth cell-based studies (e.g., proliferation assessment, transcriptional analysis, and specific cell marker immunostaining) can then be performed in parallel to investigate the long-term effect of “hit” polymers on cellular behavior. Ultimately, preclinical studies on animals, followed by clinical trials are undertaken to ensure the biomaterial’s safety and effectiveness in biomedical applications.
5.2.1. Microarray Preparation
In general, three key features are shared among most high-throughput biomaterial screening strategies for cell-based assays: (1) a patterning technique that allows for uniform spot distribution and fast preparation; (2) a cell resistant background polymer that can enable ready spot location and maximize the signal-to-noise ratio during the cell-analysis; and (3) fluorescent cell labeling or transformation to allow for automated high throughput end-point biological data collection and analysis.420
5.2.1.1. 2D Microarrays
Patterning Strategies. The techniques that aim to fabricate flat polymer spots, or 2D microarrays where cells are supported by the material of interest, have already been reviewed in a number of articles.26,421 Typically, they have been produced by using high-throughput patterning technologies, which can be divided into two main groups: liquid dispensing systems and microfabrication technologies.422 The liquid dispensing systems, which are generally robotic, include both contact printing (normally pins) and noncontact printing strategies419,423 (i.e., inkjet printing) and can require a polymerization strategy (e.g., UV-based photopolymerization424 or 2-part reactions418). Other techniques, such as microfabrication-based strategies, have mainly been used for controlling biomaterial physical properties, such as topography or elasticity, and will be discussed in section 5.2.1.2. Contact and inkjet printing is the most common and widely used methods for the preparation of 2D microarrays, as they can pattern thousands of different materials in discrete spots and in a rapid manner;423,425 by changing the compositions of the source solutions, different chemistries are easily placed into spatially defined spots, typically 100–500 μm in diameter.
Contact printing technologies use a rigid pin (solid or split/quilled) to transfer the prepolymerized polymer or monomer for on-slide polymerization from a source plate (e.g., polypropylene 384-well plate426) to the destination (e.g., glass slide) (Figure 17). The process involves dipping the pin into a small volume of sample, and then placing the pin in contact with the desired substrate for polymer drop transference, typically in the picoliter-nanoliter range. Contact printing is regarded as a simple and robust technology capable of printing high-density microarrays (e.g., Hook et al. in 2012427 used it to print 576 unique polymer solutions in triplicate), but with the limitation of having a fixed dispensing volume. It has also been reported to have higher spot size spreading, when compared to inkjet printing, in one particular sytem.423 Sequential overprinting has also been reported to increase spot area and polymer deposition.426
Drop-on-demand inkjet printing systems, such as piezoelectric-inkjet423 and thermal-inkjet,428 have also been reported in microarray fabrication for cellular applications.418,423,429,430 Some of the advantages of inkjet printing over contact printing are the ability to tune drop volume, lack of contact with the substrate (so overprinting can be performed without fear of cross-contamination), and fast and robust cleaning by flushing solvent between solutions.423 Piezoelectric dispensing uses a piezoelectric crystal that is in contact with a capillary tube to apply force to the fluid and eject the polymer droplet when a voltage is applied,423 while thermal-inkjet, also known as bubble-jet, utilizes rapid heating of the samples to create a pocket of gas (or bubble), which then pushes or “ejects” the polymer out as a droplet.428 Piezoelectric dispensers can create drops in the picoliter range and operate at 1000–5000 drops per second.429 However, given the small size of the orifice and its dependence on surface tension, it is highly sensitive to precipitation and solvent viscosity, so there are limitations to the polymers that can be printed.429 Thermal-inkjet technology can generate small volume droplets (150–200 picoliters), however, the solvent chemistry is limited to vaporizable and thermally stable inks,431 which is potentially why it has not been commonly used in combination with wide range of biomaterial libraries.
Celiz et al.423 compared the spot formation of a wide combinatorial library of acrylates and methacrylates (mixing monomers pairwise) on microarrays produced by contact printing with quilled pins and piezoelectric inkjet printing. TOF-SIMS surface analysis on the differentially printed microarrays showed that inkjet printing generated discrete spots of 250–400 μm with reduced chemical spreading when compared to contact printing. Monomer combinations that had spread (in the order of mm) and cross-contaminated other spots in the contact printed microarray, had been contained to their spatially defined spots by using inkjet printing. It is worth noting that not all monomers could successfully be printed using the piezoelectric-inkjet printer; this was attributed to air bubble formation within the nozzles, which can disrupt polymer droplet formation. Solution viscosity and polymer solubility are also limiting parameters to consider when using inkjet printing strategies.429,432
Background Polymer. The chemistry of the underlying substrate material plays an important role in the formation and subsequent performance of microarrays in biomaterial discovery for cellular applications. This background must be both resistant to the polymer printing process (e.g., not be overly disrupted by the printing solvent), contain the spreading of the printed polymer solutions to reduce cross-contamination of the separate spots, and repel cell attachment and protein adsorption to optimize signal-to-noise ratio in the subsequent biological assay.26 The polymer spots also need to withstand subsequent sterilization protocols and biological assays for data collection, and thus, they need to be immobilized within the background with sufficient strength and durability, with physical entanglement within a background polymer being commonly used (e.g., poly(2-hydroxyethyl methacrylate) (pHEMA)).26
Several substrates have been reported, with a preference for cheap and robust coating methodologies. For example, dip-coating epoxy functionalized glass slides into a pHEMA solution creates a nontoxic layer that repels most cell attachment as well as provide a stable matrix in which the polymer can physically entangle, retaining stability and spot size during printing.433 A 30–40% humidity atmosphere can induce swelling of the pHEMA and facilitate polymer penetration, subsequently reducing spot “spreading” or cross-contamination, and further immobilizing the resulting polymer and decreasing spot-delamination postprinting.433 Consequently, pHEMA thickness is surprisingly important in controlling spot spreading, where polymer concentration and parameters such as the number of dips and withdrawal speed control the resulting pHEMA thickness.423 The epoxy groups are intended to form covalent linkages with the pHEMA, although they may just act as a compatible surface, and dip-coating is readily achieved by immersion and removal of the slide into a 4% (w/v) pHEMA solution in ethanol, preferably under automation.433 Agarose dip-coating on aminoalkylsilanated slides is also a widely used strategy, showing negligible background cell attachment and retaining stability throughout the polymer printing and UV sterilization processes.425,434 Another reported polymer is PEG, synthesized by incorporating a small proportion of functionalized PEG molecules into the underlying silane formulation, it generates a substrate that can be used for the covalent binding of peptides, natural polymers, hydrogels or synthetic polymer biomaterials.434,435 Preparations of the nonionic surfactant Pluronic436 have also been used, typically in combination with biomolecule-based microarrays, as a commercially available and cell-compatible formulation that effectively repels cell binding.437
Masking. The incorporation of a mask to pattern the polymers into a defined space has been used to increase spot density by confining the liquid monomers prior to curing. For example, Hansen et al.438 used such an approach to print a high-density microarray (7,316 distinct polymer features on a standard glass slide).438 First, a 20% w/w sucrose solution was inkjet-printed at 400 μm distance onto acrylate functionalized glass slides, followed by passivation with (tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-dimethylchlorosilane (FDS) and removal of the sucrose mask using water and acetone. The exposed glass surface was then functionalized with 3-(trimethoxysilyl)propyl methacrylate, allowing spatially defined anchoring and in situ polymerization of the different monomer/initiator solutions.438 The fluorinated-mask defined the sizes of the subsequently printed polymers while also reducing nonspecific cellular attachment. The microarray printing required a two-step production process, first printing half of the polymer spots, with 20 min polymerization under UV light and cooling conditions on ice, followed by a second printing batch of the rest of the spots, increasing printing time significantly. This masking-based approach allowed for the fabrication of a high-density polymer microarray, with polymer spots of 200 ± 20 μm diameter. X-ray spectroscopy analysis showed uniform intensity profiles across the features and spot confinement of the chemical identities.438
Polymerization Strategies. Polymerization of each of the printed spots can be achieved before, during, or after the printing process; for example, the polymers can be spotted as prepolymerized solutions412,426 or monomer solutions can be dispensed and subsequently polymerized on-slide.419,420 Monomer solutions can also be printed over each other to achieve unique, automatically generated monomer mixtures and polymerized in situ.419 While prepolymerization and deposition from solution allow the use of standard polymer characterization techniques (e.g., gel permeation chromatography), on-slide polymerization translates to higher flexibility in monomer combination, increasing the high throughput nature of these studies.111,409,427,439−441 In 2004, Anderson et al.424 presented the first on-slide polymerization of acrylate monomers.424 Using a contact printer, 25 different acrylate, diacrylate, dimethacrylate and triacrylate monomers were mixed pairwise in a 384-well black polypropylene plate at a ratio of 70:30 (v/v) and printed in triplicate on pHEMA coated, epoxy-functionalized glass slides. Some required adjustments over the traditional prepolymerized polymer printing were: (1) incorporation of 25% DMF to reduce viscosity; (2) argon atmosphere with less than 0.1% oxygen composition, as oxygen was found to inhibit radical polymerization at such small volumes; and (3) high humidity to minimize failed printing, presumably reducing static effects.424 The final printing setup consisted of 75% v/v monomer in DMF, with 1% w/v of the initiator 2,2-dimethoxy-2-phenyl acetophenone.424 This on-slide polymerization approach readily allowed for multiple generational screening by using off-the-shelf commercial monomer libraries. As proof of principle, human ECSs were used to investigate the effect of the polymer microarray on cell phenotype, with cell attachment showing confinement to the defined spots.424
In 2012, Hook et al.427 used the same approach to produce 576 unique copolymers in a two-generation microarray setup. A library of 22 commercially available monomers of wide chemical diversity was used to generate a large combinatorial space; 16 monomers were premixed as the major monomer to the other 6 minor monomers in ratios of 100:0 (homopolymer), 90:10, 85:15, 80:20, 75:25, and 70:30, totaling 576 combinations.427 They contact printed each solution on a pHEMA coated epoxy glass slide using the same modified conditions (<1% oxygen, argon atmosphere, high humidity) as Anderson et al.424 Three different GFP-transfected bacterial species (S. aureus, P. aeruginosa, and uropathogenic E. coli (UPEC) were incubated on the arrays for 24 h or 72 h at 37 °C. Polymers that were resistant to bacterial attachment were identified, and correlation studies showed that some polymers had superior biological behavior as copolymers than as homopolymers, indicating a synergistic effect (found in 11 of the 96 monomer pairs).427 Five high-performing monomers were selected and mixed pairwise at 14 different ratios, creating a “second-generation” array of reduced combinatorial space but increased combinatorial resolution.427 Subsequently, an even more diverse array was published by Hook et al.441 in 2013 combining 116 unique off-the-shelf meth(acrylate) monomers to generate 1273 distinct polymers tested against multiple pathogens and environments in a combinatorial manner (Figure 18A/B), totaling 19 870 separate measurements in three generations of combinatorial arrays of increased compositional resolution.441
Figure 18.
(A, B) Combinatorial strategy applied to microarray formation Reprinted with permission from ref (441). Copyright 2013 Wiley-VCH. (A) Schematic of the combinatorial design; (i) first generation array consists of 116 homopolymers, (ii) second generation array consisted of 324 copolymers formed by mixing 18 “hit” monomers pairwise, (iii) third generation array explored 13 “hit” compositions from the second generation array via incremental compositional variations, (iv) lead compositions from the third generation were selected for scale-up and additional testing. (B) Results from applying the microarray strategy from Hook et al.;441 (i) chemical structures of hit monomers selected from the 1st generation array; (ii) intensity scale image representing bacterial attachment averaged value (iota) for each of the materials in the 2nd generation array, the scale on the right is nonlinear to highlight the range of the array, the central square is the iota value, while the narrow columns to the left indicate standard deviation (n = 3), the major or minor monomer is indicated across the row or column, respectively; (iii) intensity scale image of the iota value for each of the materials in the third generation array, the monomers used are indicated to the left and right of the intensity scale, and refer to the monomers shown in (i), the content (%) of each of the monomers listed on the left is indicated in the top row. (C, D) Microarray fabrication. Reprinted with permission from ref (418). Copyright 2019 Elsevier, Ltd. (C) Representation of the spot-in-spot fabrication strategy, (i) ratio of monomer drops printed on each spot, (ii) schematic pattern of the printed microarray. (D) Results by Zhang et al.:418 (i) image of a microarray printed on a microscope slide with 28 × 87 polymer spots, (ii) corresponding fluorescent image of the spots, (iii) mosaic made with brightfield images of representative spots on the microarray, (iv) corresponding fluorescent images for the spots represented in (iii), (v) enlarged brightfield image of a feature with cells (red box in (iii)), (vi) enlarged fluorescent image of cells on a spot (red box in (iv)), scale bar = 200 μm.
Bradley and co-workers in 2007419 presented the concept of spot-to-spot fabrication using in situ pico-nano liter-scale polymerization to form hydrogels on glass slides. They used inkjet printing to first print initiator drops, followed by drops containing a series of monomers with a reductant which stimulates polymerization in combination with the initiator; combining 7 different monomers to create 36 different polymers in situ. To show that the polymers prepared in situ were analogous to those prepared in conventional conditions, the same polymers were synthesized on glass slides (printing 200 features) and under identical conditions in glass vials. The polymers were dissolved and characterized via gel permeation chromatography. The average MW of the polymers ranged from 3.7–8.2 × 105 Da (on the slide) to 3.3–12 × 105 Da (in the glass vial), and the polydispersity index range for the in situ generated polymers (6.1–8.8) was similar to the range on the polymers polymerized in the glass vials (3.3–9.8). This approach allowed access to a broad range of new polymers in a highly miniaturized manner.
In 2009, Bradley and co-workers418 used the same spot-to-spot fabrication strategy in combination with masking of areas of the slide to generate a high-density microarray, with 2280 unique polymers on a standard microscope slide (2436 total spots) (Figure 18B). A sucrose solution was inkjet-printed to pattern 28 × 87 spots/slide, with an interspot distance of 0.8 × 0.8 mm2, followed by masking. On-slide polymer synthesis was achieved as follows for each line of spots: (i) printing of 28 spots with 10 drops APS solution per spot, (ii) overprinting monomer 1 with the number of drops descending from 27 to 0, (iii) overprinting of monomer 2 with drop numbers ascending from 0 to 27, and (iv) activation of the polymerization by overprinting 10 drops of TEMED per spot (Figure 18C,D). The microarray was then used to assess cell attachment and release 37, 20, and 10 °C, and temperature-related performance was correlated to the polymer composition. Hit polymers were identified that allowed cell attachment and growth at 37 °C, and enzyme-free release by incubating the cells at 10 °C for 20 min, reporting 92% detachment, and 88% viability of the recovered cells.418 Hansen et al.438 used a similar approach to produce a high-density microarray with 7316 features and used it to identify a polymer (1:1 ratio of 4-tert-butylcyclohexyl acrylate and n-butyl methacrylate) capable of supporting attachment, expansion, and differentiation of human pluripotent stem cells (PSCs).
Solvent. The solvent used for liquid dispensing of solid polymers/monomers is typically related to the library of polymers chosen. Early studies used very small amounts of solvent, but later work increased the solvent:monomer ratio to facilitate spotting strategies, achieving stable spot diameters when using more diverse chemical libraries.442 Indeed, Hook et al.443 investigated variable features within spot printing, and showed that when using diverse polymer libraries chemical incompatibilities can arise, and thus, quality control strategies are required to monitor spot spreading, noncircularity, chemical heterogeneity and roughness (Figure 19).443 Studies with acrylate/acrylamide-based libraries have typically used DMF (up to 50% v/v), printed at 30–40% humidity, and low oxygen (<2000 ppm).424,433 After the printing and polymerization process, the microarray is typically dried in a vacuum oven (<50 mTorr) or in atmospheric conditions, to remove solvent and unpolymerized volatile monomers.433 The choice of solvent influences the system and can affect other processes, such as the background polymer. Some groups have used 1-methyl-2-pyrrolidinone (NMP) as a printing solvent, which was selected on the basis that the majority (>95%) of their polyurethane-based library is soluble in it.425 However, because NMP can disrupt pHEMA due to dissolution, agarose was used as the background polymer on the basis of its resistance to the solvent and stability through the UV-sterilization process.425
Figure 19.
Microarrays assessed for spot defects. Reprinted with permission from ref (443). Copyright 2013 Royal Society of Chemistry. (a) Overview of a combinatorial microarray prepared using premixed combinations of commercially available monomers and on-slide polymerization. (b) Representative images of defective copolymer spots showing noncircularity, spreading, and roughness (light microscopy), and chemical heterogeneity (ToF SIMS images of the C3H3– ion); images of the corresponding nondefective homopolymers (left images); (c) AFM images of polymer spots assigned with a roughness defect. Images are 5 × 5 μm2.
The coffee-ring effect, in which the inner and outer regions of the spot display different surface chemistry profiles, can be a big problem in patterned printing of polymer solutions.443 Solvent evaporation during automated dispensing can alter the spot chemical patterning; thus drop spacing, number of drops per spot, and printing atmosphere (temperature and humidity) need to be carefully evaluated to generate uniform spots.444 The use of high humidity (73%) conditions has been reported to prevent solvent evaporation during the printing process.419 Another strategy by Bradley et al.445 used a mineral paraffin oil layer to control solvent evaporation. Monomer droplets (50% w/v in NMP) were inkjet-printed onto an agarose-coated slide that had been pretreated with a thin oil layer. The monomer solution would sink and settle into the agarose layer, and the oil would prevent any solvent evaporation before the UV-based polymerization step spatially contained the polymers. The thickness and density of the oil layer were shown to be crucial for accurate fabrication (15 μm and 0.89 g/mL); a thick enough layer was necessary for uniform slide coverage, while too thick of a layer allowed drops to slide and loose spatial definition. Sequential weighing of the microarray after printing (every 1 min for 4 h) was performed to show the lack of evaporation. Gel permeation chromatography was used to investigate the MW of the resulting polymers, with most polymers showing the same MW as obtained with traditional polymerization methods (identical conditions in a glass vial).445 The microarrays were then validated for cell-based assays using mouse ECSs, with several spots showing successful survival and growth of these sensitive cells.
Postprocessing. Before cell assays can be performed, a sterilization step is required to remove any ambient pathogens. Polymer microarrays are typically sterilized by UV-exposure, ranging from 10 to 30 min depending on polymer stability.433,438 In cases where UV-exposure could damage the microarray patterning (i.e., protein-based patterned formulations), incubation in a 70% ethanol solution,446 or a high-concentration antibiotic and antimycotic solution317,447 have been reported as sterilization methods. The microarrays are then thoroughly washed with ultrapure water, phosphate buffer saline or cell culture medium to prepare them for cell culture conditions.
5.2.1.2. 2.5/3D Microarrays
HTS technologies have also been used to study the effect of varying mechanical/physical cues on cell behavior, and to gain insight into the 3D cellular microenvironment. In these types of microarrays, the cells are typically immersed within the biomaterial or sitting on a topographically patterned surface. Fabrication strategies for 2.5/3D microarrays include direct printing, embossing, and lithography methodologies.
Direct Printing. Direct printing of 3D features is typically done through automated contact or inkjet printing. To achieve direct cell encapsulation within the biomaterial, nanoliter volumes of a solution containing the polymer precursor are premixed together with the cells and potentially relevant proteins. An example of this technology is the work published in 2015 by Dolatshahi-Pirouz et al.,448 which used in situ silane adhesion to 3-(trimethoxysilyl)propyl methacrylate-treated slides to pattern droplets via contact printing of methacrylated gelatin premixed with human MSCs (mesenchymal stem or stromal cells) and ECM proteins (fibronectin, OCN, and laminin), with a very quick UV treatment step to polymerize the gels without affecting cell viability (Figure 20).448 Prior to printing, combinations of ECM proteins, cells, and hydrogel solutions were premixed and injected into 384-well plates, with a total of 96 different combinations. During the printing process, the temperature was kept above 22 °C and the humidity above 90% to avoid polymer evaporation and cell death. A 15 s UV light treatment (320–500 nm, 800 mW power, 10 cm height) was used to cross-link the cell-laden hydrogel. The gels showed uniform cell distribution and high viability for up to 7 days, after which cell viability slightly decreased (Figure 20A(b)).448 The authors attributed this lower viability after long-term culture to the high cross-linking density of the hydrogels. However, a more in-depth analysis was needed to investigate the cell response to the UV treatment. Elastic modulus, protein content, and soluble growth factors were probed against viability and osteogenic differentiation of the MSC; via ALP, OPN, and alizarin red staining.448 The results of this screening showed that combinations of ECM proteins (gelatin, laminin, fibronectin, and OCN) induced significantly higher ALP expression, and thus osteogenic differentiation, than individual components.
Figure 20.
Microarray fabrication strategies for 2.5/3D microenvironment investigation. (A) Microarray production. Reprinted with permission from ref (448). Copyright 2015 Nature Publishing Group. (a) Schematic of the microarray production process, (b) human MSC viability after 7 days in culture along with a color-diagram (right) displaying the quantified cell viability per system. (B) Microarray. Reprinted with permission from ref (450). Copyright 2014 Nature Publishing Group. (c) Enzymatically mediated cross-linking scheme represents a specific peptide sequence, (d) biologically relevant factors are used to generate a combinatorial toolbox in a categorized form, (e) schematic of the experimental process: combining the components library with reporter cells (Oct4-GFP mouse ECSs) using robotic mixing and dispensing technology into 1,536 well plates, (f) representative images of automated microscopy, used to determine colony growth in every single well over a 5-day experiment, (g) 3D confocal reconstruction and (h) image segmentation using automated microscopy and computational methods; scale bar = 200 μm. (C) Microfabrication strategy. Reprinted with permission from ref (452). Copyright 2010 Elsevier, Ltd., (i) schematic representation of the fabrication process for mechanically active 3D cell culture arrays, (j) photo of the entire array connected to the solenoid valve, the green dye in the pressurized actuation channels), (k) increasing actuation cavity size across the array enables a range of mechanical conditions to be created simultaneously, (l) cylindrical hydrogel polymerized on a loading post of the active culture array.
To generate 3D cell-laden biomaterials through direct printing, the cells need to be embedded within the biomaterial. This embedding is ideally performed during the printing process. However, the robotic handling of living cells has its own unique challenges; as the precursor solution, cross-linking strategy and handling technique must not be cytotoxic or carcinogenic. As such, a range of “bio-inks” (biocompatible printing solutions) have been developed, based on advances in the field of cell culture using autogelation, photoinitiation, enzymatic-mediated cross-linking or thermal gelation.449 For example, Lutolf and co-workers450 have developed a multimodal assay platform by using the coagulation enzyme-activated transglutaminase factor XIIIa (FXIIIa) sequence to cross-link PEG-based macromers into 3D hydrogel networks (Figure 20B).450 The insertion of a short peptidic substrate from FXIIIa allows for site-specific enzymatically mediated amide bond formation between the PEG chains under physiological conditions, thus increasing cell viability. A programmable nanoliter-range liquid handling robot generates 1:l of each unique condition in triplicate and in a completely automated manner, and subsequently prints them onto a glass slide or a standard 1536-well plate (Figure 20B(e)). The microwell plate format is used as an ideal surface-to-volume ratio for the hydrogel drops, physically isolating each well from its neighbors (in contrast to the glass slide format) and rendering each well as a truly independent experimental condition free of soluble factors cross-contamination.450 This microarray platform allowed for the modulation and study of five key parameters: (1) hydrogel modulus, (2) proteolytic degradability through the insertion of a peptide sequence susceptible to cell-mediated cleavage, (3) ECM protein composition, (4) cell–cell interactions, and (5) soluble cues.450 By using an Oct4-GFP reporter mouse embryonic stem cell (ESC) line in conjunction with automated imaging, the platform worked as a multimodal assay platform allowing multiple readouts in parallel (Figure 20B(f)). The cell-laden hydrogels were imaged the day after printing to obtain initial fluorescent levels per well, and subsequently, every day to assess changes in cell number and colony formation. In the most permissive conditions, the cells formed spherical colonies within 3 days, and kept proliferating until fixation and staining at day 5. High-resolution confocal microscopy confirmed that colonies grew in a 3D space of approximately 500 μm thickness (Figure 20B(g)). An automated analysis script developed in CellProfiler (Broad Institute) was used to segment and analyze colony area, the measure of mouse ESC proliferation, and GFP intensity, as a marker of undifferentiated ESCs (Figure B20(h)).
In 2017, Sharma et al.451 used contact printing together with electrospinning to generate highly tunable fibrous PEG-based microarrays amenable to the spatially controlled presentation of peptides.451 Their technology used the selective reactivity and chemical robustness of the thiol and norbornene click reaction to generate cross-linked fibrous PEG-based scaffolds via electrospinning. By reacting off-stoichiometry, free norbornenes could be preserved along the fibers, and subsequently functionalized via a second-step photo “click” reaction for the introduction of cysteine/thiol-containing peptides into the fibrous matrix, which was contact printed for defined spot generation. As proof of concept, the authors showed cell attachment of several mice and human cell lines, and their morphological changes to variable peptide residue presentation in the 3D fibrous tissue-mimicking environment.451
Direct Embossing. Lithography-based printing techniques have also been used to generate microarrays for cellular assays. For example, Moraes et al.452 used mask-based soft lithography to investigate the effect of mechanical compressive strains (ranging from 6% to 26%) on cells encapsulated in PEG-hydrogels. The device pitch was based on a 1536-well plate and consisted of 25 vertically actuated loading posts arranged in a 5 × 5 array, with 5 replicates per mechanical condition (Figure 20C(j)). For the device fabrication, PDMS was cured and bonded in different steps to generate a spatially defined actuation cavity chamber network. A solution of PEG monomers mixed with cell medium, a photoinitiator, and live mouse MSCs, was then injected into the PDMS chamber using a long needle, and masking was used to selectively photopolymerize the cell-laden hydrogel pillars with UV light (Figure 20C(i)). The array of pillars was suspended over actuation cavities of varying diameters, allowing the formation of a range of vertical displacements by applying a 55 kPa pressure via a solenoid valve (Figure 20C(i,j)). During the hydrogel polymerization, cell viability was found to decrease significantly for UV-polymerization times greater than 300s. Therefore, a 1 μm thick layer of Parylene-C was used to protect the cell-laden hydrogel from UV overexposure, also reducing oxygen permeation onto the system and formation of cell-toxic free radicals. A UV exposure time of 195s at 17.5 mW/cm2 was found to obtain initial cell viability of 69.4 ± 3.4%, comparable to the viabilities achieved in similar PEG hydrogels.453
Indirect Patterning. Lithography techniques have also been used to create a reverse mold to indirectly pattern structures onto polymers and create microarrays of varying micro- and nanotopographies. Such as the technology developed by de Boer and co-workers,454 which increased the number of topographies available in a single microarray to 4000+, allowing libraries of randomized surface topographies generated in silico to be explored in vitro and moving beyond simple geometric shapes such as pits and grooves. Microsized “primitive” shapes (circles, rectangles, and triangles) were combined to form a range of 10 μm high “topofeatures”, and patterned into 300 μm × 300 μm TopoUnits455 (Figure 21A,B). Photolithography is used to etch the micropatterns onto a master mold made from a silicon wafer, followed by hot embossing of PLA films (250 μm thick) creating the final microarray, called a TopoChip.455 The TopoChip is divided into four quadrants, where quadrant A has identical TopoUnits to that of Ai and quadrant B is identical to quadrant Bi, to ensure that the different TopoUnits have duplicates in different spatial areas (in the middle of the array or the periphery), and reduce localization-induced variation on cell responses. The TopoChips can also be fitted to a variety of materials used in tissue engineering, such as on microscope slides (Figure 21C),455 at the bottom of tissue-culture plates for a TopoWellPlate (Figure 21F),51,456 and nanosized to generate the so-called NanoTopoChip.457 Cells are shown to distribute evenly through gravity over the entirety of the array (Figure 21D,E).455 After cell seeding and incubation, automated microscopy is used to evaluate a large number of varying cell parameters number, such as total and nuclear size, spreading, orientation, morphology, proliferation, and expression of cell-type-specific markers.
Figure 21.
Representative microarrays to investigate the effect of topographical features on cell phenotype. (A–E) TopoChip characterization. Reprinted with permission from ref (455). Copyright 2011 National Academy of Sciences. (A, B) SEM images of a section of TopoChips, displaying accurate feature replication (scale bar = 50 μm), (C) image of the TopoChip carrier, lid and chip assembly, (D, E) light microscopy images of cells seeded onto the TopoChip displaying homogeneity of cell distribution within and between TopoUnits. (F) TopoWellPlate fabrication scheme. Reprinted with permission from ref (51). Copyright 2011 Wiley-VCH. representing the following steps (a) silicon master mold containing the inverse structures of the selected topographies is used to (b) cast a layer of PDMS, followed by (c) curing of the PDMS layer and (d) peeling off from the silicon master. (e) OrmoPrime is applied to a Borofloat wafer, followed by application of OrmoStamp onto the PDMS mold, (f) which is spread by capillary forces, (g) UV curated and (h) peeled off from the mold. The hot embossing process involves (i) the inverse OrmoStamp mold is aligned (j) to the polystyrene film followed by (k) hot embossing and (l) gentle peeling off. The patterned polystyrene film is then (m) aligned to the bottomless 96-well plate and an aluminum thermal stamp, followed by (n) thermal bonding, resulting in (o) a leakage-free well plate containing defined surface substrates.
For the fabrication of the TopoWellPlate, De Boer and co-workers51 used polystyrene instead of PLA, as it is the golden standard for tissue-culture substrates. Polystyrene is regarded as a stiff and brittle polymer, so Zhao et al.458 optimized their hot embossing methodology using OrmoStamp, a UV-curable hybrid polymer that was developed as a tool for UV nanoimprint lithography. The fabrication method used (1) a TopoChip silicon mold fabricated through micromachining technologies, to (2) emboss an intermediate PDMS mold, which was then used to (3) pattern an OrmoStamp solution on a Borofloat wafer, using UV polymerization and nanoimprint lithography; (4) the OrmoStamp mold was then baked at 130 °C, cooled down and treated with a gentle plasma treatment; (5) this mold was then used for the hot embossing of a polystyrene film, followed by gentle O2 plasma treatment of the free-standing polystyrene chip.458 For the fitting into a well plate, Beijer et al.51 used thermal bonding; the polystyrene chips were aligned with the chimneys of a bottomless 96-well plate and an aluminum heat-transmitting stamp, and placed in a temperature-controlled press; effectively generating a leakage-free polystyrene 96-well plate with 87 topographically enhanced and 9 control unpatterned wells (Figure 21F). As proof of principle, primary bone marrow-derived MSCs were seeded onto the TopoWellPlates, and their metabolic activity was measured using a Presto Blue assay. A clear 2.5-fold difference between the lowest and highest scoring conditions demonstrated the effect of the different topographies on cell metabolism. The TopoWellPlate was presented as a platform for transcriptomic, proteomic, and metabolomic analysis, as the well-plate format allowed for completely isolated conditions.
5.2.2. Microarray Analysis for Characterization, Quality Control, and the Construction of Structure–Performance Relationships
5.2.2.1. Array Material Characterization
Array preparation is a complex process, and many aspects can go wrong. To name a few, monomers can carry over due to insufficient washing of pins or piezo pipettes, contaminants can be introduced by dirty solvents, spots can spread unexpectedly, contaminants in the supporting hydrogel can alter the spots’ surface chemistry, the background chemistry itself can contribute to the spotted biomaterial, spots can fail to print, and there are many other occurrences virtually invisible to the human eye or even microscopy, which can affect the cellular response to a biomaterial.423 Consequently, chemical surface analysis is essential in the quality control of polymer microarrays.51 Presynthesized polymers can be characterized using analysis of their MW, wettability, and purity before spotting onto the array.425 However, for microarray strategies that use on-slide polymerization, bulk characterization techniques can only be performed if many spots of the same chemistry are printed, polymerized, and then removed in solution to be analyzed using gel permeation chromatography and nuclear magnetic resonance instruments.419 Moreover, traditional bulk-polymer characterization techniques miss surface segregation and contaminants, which when analyzed using solution-based strategies can represent subdetectable levels.
Surface chemical analysis of printed arrays is therefore a required step; initially, as a development tool to control and optimize the fabrication process, and subsequently as quality control, detecting differences between manufactured microarrays and generating accurate in situ biomaterial characterization data for correlation studies and computational modeling.51,459 To achieve full exploitation of the data generated from polymer microarrays, the same analysis given to the “hit” polymers has to be performed for the nonfunctioning ones, to truly be able to generate in silico models for structure-polymer interaction.
WCA was the gold standard at the beginning of polymer microarray characterization.425 However, studies using diverse polymer libraries have shown no correlation between cell attachment and wettability, and other characterization techniques have taken relevance in polymer characterization (reviewed in116). Furthermore, wettability is a very unspecific measurement, that is, contamination may not be detected by small differences in expected WCA values (if known) and are even less likely to be assigned from such measurement. Consequently, time-of-flight secondary ion mass spectrometry (ToF SIMS) and XPS, which are able to detect small contaminant levels, have become prevalent in microarray analysis and characterization, and are carried out as standard in some laboratories. In particular, ToF SIMS has proven exceptionally well suited to achieve structure-performance relationships between the materials surface mass spectrum and cellular outputs in eukaryotic and prokaryotic cells.111,427,460
5.2.2.2. Adsorbed Layer Screening and Characterization
Synthetic materials have traditionally required the adsorption of proteins onto the surface to achieve robust cell attachment.461−463 These proteins serve as anchors/translators for cell surface engagement and subsequent cell adhesion, spreading and migration processes, with eventual ECM deposition by the adhered cells themselves.461 Immediately after submerging a synthetic material in a protein-containing liquid, protein adsorption begins, as it happens within a second of exposure and is mediated by an entropic gain as the binding releases the otherwise low-entropy water surrounding the protein and material surface.464 The composition, distribution, and spatial conformation of these absorbed proteins have been proven to play a role in cell behavior.465 Material features such as surface charge, wettability, curvature, chemisorbed functional groups and the conditions during protein adsorption all have been shown to influence protein presentation and function, and characterization of these adsorbed proteins is essential.466,467
Analysis of the propensity of the different polymers to adsorb and present ECM proteins gives useful data, and can be correlated to their ability to support cell attachment and proliferation in high throughput.462,468−470 Therefore, high throughput methods for protein–polymer interaction have been developed to predict cell attachment. In 2008, Taylor et al.471 correlated the measurement of protein adsorption using fluorescent labeling and protein adhesion strength through AFM.471 In 2009, Hook et al.468 showed that surface plasmon resonance imaging could measure real-time adsorption and desorption of Alb, collagen, and fibronectin to an array of polymer spots in parallel and in a label-free manner.468 Hook et al.469 also used fluorescently labeled Alb to measure protein adsorption onto a microarray containing 70 different poly(meth)acrylate spots, and correlate its fluorescence intensity to surface chemistry (obtained through ToF SIMS analysis), using multivariate curve resolution image analysis to investigate the relationship between surface chemistry and protein adsorption in an unbiased manner.469
However, to accurately determine protein composition in complex settings, scale up or combinatorial protein spotting is required.463,472,473 Rao et al.473 used desorption electrospray ionization and liquid extraction surface analysis mass spectrometry coupled with in situ surface tryptic digestion to identify protein species on polymer surfaces. For the first time, in situ automated tandem mass spectrometry was able to separate and identify protein entities on a biomaterial surface.473 Hammad et al.474 took a different approach, using extraction by washing and mass spectrometry to identify proteins absorbed onto biomaterials that supported stem cell expansion.474 They first used gel electrophoresis and liquid chromatography–mass spectrometry (LC–MS) to identify 14 proteins (out of 71) that uniquely deposited onto oxygen etched over untreated polystyrene when incubated with a complex media known to support human embryonic stem cell expansion. These identified proteins were then commercially sourced and used to generate a combinatorial protein microarray to investigate protein adsorption-related performance of a polymer known to support human embryonic stem cell expansion, N-(4-hydroxyphenyl)-methacrylamide (polyHPhMA) (Figure 22).474 Inkjet printing was used to mix and spot 13 proteins pairwise (30/70) resulting in 169 protein combinations onto a contact-printed array of defined polyHPhMA spots. Human PSCs were cultured on the protein/polymer arrays using a serum-free medium (StemPro) and PSC number was assessed through fluorescent microscopy (total nuclei using DAPI and stem cells through Oct4 staining). Five protein combinations were selected for further investigation in a secondary array (Heat shock protein-1 (HSP):Heat shock protein 90 (HSP90), Platelet factor 4 (PF4):HSP, PF4:glyceraldehyde-3-phosphate dehydrogenase (GAPDH), HSP:Fibronectin (FN), and GAPDH:serum amyloid P (SAP)) at a range of dosing compositions (30%, 50%, and 70%) and concentrations (0.1, 0.5, 1, 2, and 4 fmol). In some instances, protein concentration did not affect cell adhesion (HSP:FN and HSP:HSP90 displayed high cell adherence across all concentrations), and protein ratios were also not linearly correlated to cell number (as HSP increased from 0 to 70% in combination with HSP90, cell numbers increase, but decreased for 100% HSP coating). This suggested a synergistic effect of HSP and HSP90 at the cell–polymer interface. Interestingly, HSP:HSP90 adsorbed polyHPhMA showed levels of cell adherence comparable to that seen for FN, a well-established integrin-binding substrate for stem cells, and scale-up studies showed expansion and maintenance of a human PSC line for 3 days. This was the first report of heat-shock proteins mediating stem cell attachment.472 As our knowledge of the ECM composition of biological systems improves, chemical structure-protein adsorption correlation studies may eventually support the rational design of bioinstructive materials.
Figure 22.
Strategy used by Hammad et al. From left to right: Sequential inkjet printing of biomolecules on the same coordinates as the polymer spots (in orange) in the primary screen: proteins were mixed pairwise at a 70/30% ratio and at 0.1, 0.5, and 1 fmol. Table showing results of the primary screen, displaying color coding for human PSC line (HUES7) cell number (OCT-4-positive cell quantification) per spot. A secondary microarray screen was generated from “hit” protein combinations, mixed pairwise at 30, 50, and 70% ratios and at 0.1, 0.5, 1, 2, and 4 fmol. The table shows representative results of OCT4-positive cells per spot in the second generation array. Reprinted with permission from ref (474). Copyright 2010 Royal Society of Chemistry.
5.2.2.3. Computational Models in High-Throughput Cell Phenotyping
Data processing and computer simulation have the potential to evolve the biomaterial discovery field toward modeling/predictive strategies. The dimensionality of materials property space is too large to be explored even by high-throughput methods, and so computational models are the only means for covering the entire spectrum of biomaterials. Nowadays, microarray screening is often accompanied by computational data analysis and modeling for maximum data extraction from the experiment, for example, by cellular image analysis, and then subsequently to mine the data for structure–activity/performance relationships.144,417,475 Many different approaches have been developed for cellular data mining to increase data extraction from single experiments. A recent example is the Cell Painting high-content image-based technology,476 a system that uses automated high throughput microscopy and image software analysis to measure ∼1500 morphological features (variables in size, shape, texture, intensity, and more) using only 6 chromophores that, when combined, provide a powerful phenotypic assignment tool.476 The overall time for imaging acquisition and analysis is 3–4 weeks, evidencing the complexity of such methods. With numerous strategies for cellular data mining having been reported, researchers are also putting efforts into unifying the different strategies for image-based cell profiling and data mining. As it is becoming evident that each laboratory has developed and optimized its own in-house method, there is a need for a generalized approach to facilitate data comparison between studies (reviewed in477).
Many papers have correlated the cell-assay data with polymer composition to model polymer-cell interactions and identify functional groups or polymer characteristics that improve biomaterial performance in specific biological conditions.462,469,478 For example, Epa et al.479 derived a computational model from microarray data based on the bacterial attachment of three separate clinical pathogens to the first-generation microarray of varying polymers, and accurately and quantitatively predicted attachment to a second-generation combinatorial microarray.479 More recently, three predictive models based on three separate screening studies have been combined to generate a single model capable of quantitative predictions for the attachment of three different pathogens together to a large library of polymers.475 High-throughput technologies generate the data necessary to create models and predict the responses of future biomaterials, which will be addressed in more depth in section 8. However, with the increasing amount of data generated using different experimental strategies, biomaterial properties and cell models; the challenge lies in the quality control and processing strategies for these big data sets.55 For that reason, an initiative is being started to offer a publicly accessible repository called the The cBiT, which will be further discussed in section 7.
5.3. Cell–Biomaterial Interactions
As has been already been discussed in section 2.3, the cellular microenvironment consists of both biochemical and physical cues.480 The cell-surface receptors and molecules that mediate cell adhesion have the capacity not only to bind to chemical cues, but also to sense their physical presence in a wide variety of ways (e.g., shape, distribution, size, binding strength, elasticity, and many more).481−483 Thus, the study of the cell–biomaterial interface has expanded to include all biochemical and biophysical properties of the biomaterial.
5.3.1. Biomolecules
The presence and density of certain biomolecules in the ECM have a major impact on cell behavior. Changes in ECM composition have been directly linked to disease development; for example, solid cancer development includes a desmoplastic response (the growth of fibrous tissue in pathology) in which its surrounding environment goes through an incremental increase in ECM molecules such as fibronectin, and displays higher levels of reorganized and cross-linked type COL1.484 These changes in ECM composition stiffen the tissue stroma and correlate to a worse patient prognosis.484 From large macromolecules such as hyaluronan,485 to small peptides,486 a large variety of cell–ligand molecules have been investigated for their ability to induce cell attachment and evoke a cellular phenotype.
5.3.1.1. ECM Molecules
Though more expensive than synthetic polymers and carrying a risk of animal-derived immunogens or pathogens, ECM proteins are a powerful tool to evoke specific cell responses, and as such, they have been studied using microarray strategies. For these microarrays, the key features are stable protein conformation and sterile printing techniques. One of the first cell-based protein microarrays as described in 2005 by Flaim et al.,463 who used a commercial DNA spotter to contact-print small drops with combinations of ECM proteins onto polyacrylamide-coated glass slides, which resisted unspecific protein adsorption and cell attachment in serum-containing medium, and confined the different protein spots while retaining hydration and native protein conformation.463 Flaim et al.487 further developed this technique to study the effect of growth factors in combination with ECM proteins by printing 20 combinations of 5 proteins (fibronectin, laminin, COL1, collagen III and collagen IV) into a multiwell system. Each microwell was used to array 12 different growth factors, allowing for the simultaneous generation of 1200 experiments in 240 unique signaling environments.487 The microarray was used to examine the differentiation of human stem cells along the cardiac lineage, using a confocal microarray scanner to monitor myosin heavy chain expression (a marker of cardiac cells) as compared to cell growth (nuclear DNA).487 The comparison of the single and combinatorial protein effects showed interesting results. For example, the attachment molecules COL1 and III were found to individually decrease cardiac differentiation, but increase it when combined. The growth factors Wnt3a and Activin A also affected differentiation negatively on their own, whereas they induced cardiac commitment if added together. Attachment molecules and growth factor cross-talk also showed unique combinatorial effects; for example, fibronectin was found to have an antagonistic effect on cardiac cell differentiation when combined with Wnt3a and Activin A.487
Mei et al.437 focused on achieving spot resolution at a subcellular scale. An automated printing technique based on AFM allowed the programmable generation of surfaces containing multiple ECM proteins at the subcellular size (6–9 μm, slightly bigger than a cell’s focal-adhesion site); with differential spacing and protein surface covering, modeling the in vivo spatially defined cell microenvironment.437 The generated microarray contained 16 800 patterned spots on a glass slide (Figure 23A). Effective, error-free printing of this many spots and different protein formulations required method optimization, with humidity and background surface (hydrophobic octyltrichlorosilane-treated glass) playing a key role in defined spot generation. Mouse myoblasts were incubated on the microarrays and stained with DAPI (nucleus) and actin (cytoskeleton filaments) to determine the cell number, size and shape (Figure 23A–C). Results showed that cells had a more spread-out morphology on arrays with smaller space between spots (i.e., 18 and 24 μm) (Figure 23C), while on arrays with the larger interspot distance the cells showed a circular shape with a smaller size (less spreading) (Figure 23B). The authors concluded that 10% or greater protein (Fn) covering of a surface is necessary for maxim cell adhesion and spreading (Figure 23D).
Figure 23.
Microarray technologies based on animal-derived biomolecules. Reprinted with permission from ref (437). Copyright 2008 Wiley-VCH. (A) Fluorescent image of a two-component protein array, composed of 25 different combinations of mini-arrays (white square marks one mini-array), red spots are Alexa-546-labeled fibrinogen and green spots are FITC-labeled BSA, the numbers at the top of representing the distance between spots and the numbers on the left are the ratio of red: green spots. (B) Collage of fluorescence images of C2C12 myoblast cell adhesion on fibronectin arrays after a 3h incubation on protein arrays with different spot spacing, (C, D) enlarged images of the red squares gown in panel B, scale bar = 20 μm. (E) Correlation between fibronectin surface coverage and cell density (black line) or cell spreading (red line). (F–I) Microarray fabrication. Reprinted with permission from ref (417). Copyright 2016 Elsevier, Ltd. (F) Schematic of the cell microarray formation, (G) Typical cell microarray platform with various protein ratios. (H) Fluorescent images of the slide acquired using automated fluorescence microscopy for nuclear localization (Hoechst 33342) and osteogenic markers (Runx2, Calcein Blue). (I) Automated analysis for the detection and measurement of nuclear count and osteogenic marker presence (yellow line in Runx2 and blue line in Calcein Blue, Ca ion). (J–L) Microarray strategy. Reprinted with permission from ref (489). Copyright 2016 Elsevier, Ltd. (J) Schematic of the experimental approach, with 4000 features (including positive and negative controls) on a single glass slide, (K) representative image of complete ECM microarray 12 h post cell-seeding with definitive endoderm cells, image represents the 4000 features in the array, nucleus and cytoplasm are labeled with fluorescent dyes (blue and green, respectively) and red spots are rhodamine-dextran spots utilized as negative controls and for alignment of image acquisition, scale bar = 1 mm. (L) Inset from panel B (white box), including cell nuclei label only and illustrating a representative set of ECM islands in quintuplicate, scale bar = 100 μm. Right graph shows a heatmap quantification of endoderm cell adhesion 12 h postseeding. Each feature represents the combination formed by 2 different ECM molecules on the x and y axes.
In 2013, Ghemei et al.435 further modified these techniques to achieve long-term culture stability of the immobilized biomolecules.435 The protein solutions were printed and covalently bound onto epoxy-silane-coated slides, after which the surface was passivated using the covalent coupling of PEG-bis-amine to mitigate protein degradation and avoid unspecific cell attachment over long-term cell culture (Figure 23B). The covalent PEG immobilization was carried out under cloud-point conditions (meaning marginal solvation conditions as compared to good solubility of PEG488), achieved using K2SO4 concentrations of 0.5–0.8 M at 37 °C (temperature required to preserve protein functionality). Under these conditions, the authors assumed interchain repulsion due to the hydration shell surrounding the PEG chains being decreased, resulting in higher PEG-bis-amine immobilization density, PEG chain alignment, and protein adsorption repulsion. Using this method, they successfully cultured human MSCs on the microarrays over a three-week span, investigating the effect of cell–ligand proteins alone435 or combinations of cell–ligands and growth factors417 on the cells’ osteogenic differentiation.
In 2015, Beachley et al.447 generated microarrays representative of in vivo microenvironments using complete animal tissue samples. A physiologically broad tissue ECM library set was generated by harvesting 11 different porcine tissues and organs (spleen, small, intestine, bladder, bone, brain, cartilage, heart, kidney, liver, lung, and adipose tissue). The tissue samples were chemically treated using a combination of acid, detergent, and DNase, followed by lyophilization and mechanical breakdown into small microparticles using a cryomill.447 For the microarray production, silicon gaskets with arrays of 3 mm-diameter wells were placed on dry acrylamide-coated slides, creating 40 wells per array. The wells were collagen-coated and single or combinations of tissue particle suspension were spotted on each well.447 The microparticle solution was dried overnight, after which the gaskets were removed and the microarray was UV-sterilized. Histochemical staining (Masson’s trichrome) and proteomic analysis (SDS-PAGE and LC–MS) were used to validate the reproducibility of the spotting technique. A variety of cell types were seeded onto the arrays and assayed for cell binding, proliferation, and shape, correlating different cell type adhesion and phenotype to ECM tissue formulation.447
Other reports have focused on expanding the library of extracellular proteins used and extracting more mechanistic data from the arrays. For example, in 2016, Malta et al.489 combined 38 different ECM molecules (proteins, proteoglycans, and glycosaminoglycans) to create 741 unique pairwise combinations in quintuplicate on a polyacrylamide-coated microarray (Figure 23K). Mouse endoderm progenitor cells were seeded onto the microarrays and treated with either hepatocyte or pancreatic priming medium. Cell phenotype was assessed through immunofluorimetric analysis of the hepatic marker alpha-fetoprotein (AFP), the pancreatic marker Pdx1, and total cell number (DAPI, nuclear staining) (Figure 23L).489 Quantified adherent cell numbers at 24, 48, and 72 h after differentiation induction showed that cell numbers per condition were similar at 24 h, but diverged at 48–72 h. Cell phenotype was further investigated by quantifying apoptosis (activated (cleaved) caspase-3 staining), activated endoderm-related signaling (phosphorylated SMAD1/5/8), and epigenetic modulators (Histone Acetylation marks H3K9ac and H3K14ac),489 and correlated with differentiation efficiency. Their results found a previously unreported synergistic effect of Fibronectin + Merosin in hepatic differentiation, showing increased H3K14 acetylation, SMAD1/5/8 phosphorylation, and AFP marker expression.489
5.3.1.2. Synthetic Peptides
Since the introduction of solid-phase peptide synthesis in 1963,490 the technique has been implemented, improved, and modified for the synthesis of long and complex peptides and small organic molecule libraries, which has given way to high throughput screening technologies using synthetic biomolecules.491,492 While animal-derived proteins have shown great potential in cell-phenotype control, their application is limited due to their high cost, batch-to-batch variability, and risk of immunogenic or pathogenic contaminants. In contrast, short peptides can be fabricated at a relatively low cost and with high purity. Since the description in 1987 of the precise sequence in fibronectin that induced cell attachment, there has been a wide interest in designing short peptides capable of replicating the functionality of full proteins for cellular control.493 One of the first reports of peptide microarrays in 2001 by Falsey et al.107 described a chemical microchip on glass slides where the small molecule ligands and peptides were bound through site-specific oxime bonds or thiazolidine ring ligation reactions.107 The biological performance was measured using three biological assays: (1) fluorescent detection of protein-binding, (2) functional phosphorylation to label peptide spots, and (3) a live cell adhesion measurement.107
Since then, the patterning of short peptides and organic molecules on microarrays for cell-interaction studies has become well established.494 For example, Kiessling and co-workers495,496 developed a method for the generation of self-assembled monolayer (SAM) thiolated-peptide arrays on gold surfaces, forming reproducible and well-ordered surfaces that could be patterned into the size and shape-specific elements and present variable epitope density for cellular studies. In 2016, Derda and co-workers497 used this technique to investigate the epithelial-to-mesenchymal cell transition. To fabricate the SAM-based peptide microarrays, alkanethiol-terminated peptides were contact printed onto glass slides coated with pure gold using thermal evaporation. A mammary gland cell line that undergoes epithelial to mesenchymal transition in response to soluble transforming growth factor (TGF) was used to investigate the effect of a surface-immobilized library of TGF-receptor, and integrin targeting peptides. The cells were grown for 4 days, after which cell phenotype was assessed through nuclear (DAPI) and cytoskeleton (smooth muscle actin) and cell–cell adhesions (E-cadherin). Results showed that only one of their TGF-receptor targeting peptides reduced E-cadherin expression (a marker of epithelial phenotype) and an increase in cell area (a marker of mesenchymal phenotype), while integrin-binding peptides did not induce a mesenchymal phenotype over a 4-day incubation.
In 2014, Kilian and Zhang498 presented a peptide-based microarray fabricated using a single-step azide–alkyne “click” chemistry approach. They manually synthesized a peptide library using standard F-moc solid-phase methodology, purified them via HPLC, and capped them with an alkyne-containing reagent to yield a terminal domain amenable to bioconjugation to an azide-terminated surface.498 This strategy allowed for the investigation of combinations of peptides by simple premixing of stock peptide solutions in the desired ratio. Alkanethiols in the spotted solutions adsorbed immediately to the gold forming SAMs, retaining spatial resolution. The array was used to investigate the adhesion characteristics of mouse embryonic fibroblasts and adipose-derived stem cells, as well as the differentiation of the stem cells to the osteogenic fate.498 The use of the “click” chemistry approach allowed for the peptide-functionalized surfaces to be used in cell culture for up to 21 days; however, spot fidelity was found to decrease over time, even when postspotting incubation was done in low vacuum desiccator conditions, so the microarrays had to be used within a short time of fabrication.498
Another strategy developed by Gupta et al.499 in 2010 reported a microarray strategy where biomolecules were covalently linked on a PEG-based hydrogel. Two strategies were presented; (1) a PEG matrix containing a functional group reactive to thiol–ene chemistry to covalently attach peptides through allyl ester protective groups or cysteines; and (2) orthogonal functionalization via the printing of a heterobifunctional linker (such as N-hydroxysuccinimide ester) that could interact with both the PEG matrix and the biomolecules.499 The microarrays were fabricated by first printing the biomolecules on a glass slide, after which a liquid PEG prepolymer mixture was cured onto the array using a short UV treatment. The viscosity of the prepolymer PEG solution and the rapid cross-linking ensured the printed spots remained unperturbed, also the mild conditions allowed for the effective incorporation of a wide range of biomolecules. The spots showed uniform size, with at least 100 mg/mL peptide composition and 50 wt % PEG groups. As proof of concept, cell adhesion was assayed using four different peptides on PEG hydrogel using system (i), with fibroblasts showing specific cell attachment onto RGD-based functionalized peptides, as compared to control nonadhesive peptide (RGRES) spots. In 2016, Jia et al.500 used solid-phase synthesis and isocyanation chemistry to generate methacrylate-peptides that would be soluble in DMF and have high miscibility with low MW-PEGDA, allowing ratiometric incorporation in a homogeneous manner within a PEG-hydrogel microarray. The technology was used to print 12 peptide sequences, and was tested on human PSC-derived cardiomyocytes. The cells were cultured on the microarrays for 3 days, after which cell number and sarcomere structure (a marker of cardiomyocyte maturity) were examined using fluorescence microscopy. The peptides performed differently even though all of them contained an RGD region, with the sequenced peptide from the laminin β4 subunit (PMQKMRGDVFSP) showing the highest number of cell adhesion and sarcomere formation.
In 2016, Kanie et al.501 combined peptide microarrays with in silico screening to define candidate peptides that could induce osteogenic-selective proliferation and differentiation of human MSCs. The sequences of 19 types of natural BMPs (known to be bone regenerative molecules) were obtained from UniProt, and aligned with a peptide database (graphical image of the alignment, Figure 24A). Twenty-five candidate 9-mer peptides, a positive control RGD, and negative control blanks were synthesized on cellulose membranes using F-moc methodology, cut, and deposited at the bottom of a 96-well plate for cell-based experiments (Figure 24B). Normal human osteoblasts, human MSCs from umbilical cord matrix, and normal human dermal fibroblasts were used to investigate selective osteoprogenitor cell proliferation and osteogenic differentiation of MSCs, with the final determination of peptide sequences that could selectively induce osteogenic cell phenotype in detriment of other cell types.501 Kanie et al.486 later increased the scope of their in silico assessment by using a clustering-assisted focused screening method, which minimized the risk of redundantly including highly similar peptides by first clustering a large library of peptides sequences according to physicochemical properties, and then selecting ten peptides from each category cluster.486 The study screened a total of 500 peptides for their ability to selectively bind collagen IV and enhance endothelialization. Endothelialization of a biomedical device surface is known to reduce the risk of complications, especially in long-term implantation of medical devices used to treat cardiovascular diseases,486 and is therefore a desirable bioinstructive trait in a biomaterial.
Figure 24.
Microarray strategies for 2D/surface printing of biomolecules and synthetic polymers. (A) Schematic of the strategy. Reprinted with permission from ref (501). Copyright 2016 MDPI. First, in silico screening of candidate peptides is performed, and candidate 9-mer peptides are selected from the homologous sequences. Second, the microarray is fabricated using a peptide array synthesizer on a cellulose membrane, which is cut and deposited in a 96-well plate. Cells are seeded, cultured, and assayed for cell proliferation and osteogenic differentiation. (B) Polymer microarray. Reprinted with permission from ref (442). Copyright 2014 Royal Society of Chemistry. (a) Microarray fabrication using contact printing of monomer solution followed by UV polymerization, (b) high-throughput screening and characterization of polymer spots for (i) surface chemistry by ToF-SIMS, (ii) surface wettability using WCA, (iii) human stem cell adhesion and hit polymers with high cell attachment (inset); (c) structures of the 141 monomers used in the microarray, grouped by side-chain chemistry.
5.3.1.3. Polysaccharides
Polysaccharides or glycans participate in almost every aspect of biology, from cell-surface recognition markers,502 to modulating cell differentiation,503 to being an intrinsic part of the ECM.503,504 The diversity within naturally occurring carbohydrate chains and in composition has only just begun being explored, with synthetic and natural glycan libraries being compiled.504−506 The high binding specificity of glycans has been widely recognized as a powerful tool for diagnostic and drug development strategies of clinical impact.504,507−509
Since the first reports in 2002,510,511 carbohydrate microarrays have been successfully employed for the rapid screening of carbohydrate-binding proteins, such as antibodies or lectins,512,513 profiling enzyme–substrate specificity,514 and detecting pathogens.511 These arrays provide a critical initial screen for binding specificity, and are free of the bias shown by other technologies (e.g., surface plasmon resonance, frontal affinity chromatography) that require preliminary knowledge of the binding affinity.515 Typically, these microarrays display a variety of saccharide chains immobilized on epoxy-activated glass slides using contact printing at high humidity, and require a postprinting blocking step to remove any exposed hydroxyl groups that could promote protein deposition and cell adhesion during the out-put assays.505 Covalently bound glycan microarrays have also been described, where maleimide-activated surfaces are used to bind a thiol-terminated linker on the carbohydrate chain.516 The most challenging feature of these arrays has been the source glycans, with large libraries being assembled on the basis of their biological source and potential binding-specificity.517 For example, Toonstra et al.505 described a chemoenzymatic approach to synthesize a library of high-mannose N-glycans and related neoglycoproteins through sequential enzymatic trimming of two readily available natural N-glycans; the Man9GlcNAc2Asn from soybean flour and the sialoglycopeptide from chicken egg yolk. The resulting glycans were separated through chromatographic techniques, obtaining a full-range of natural high-mannose N-glycans.505
Chemoenzymatic approaches have also been reported for the generation of synthetic oligosaccharides libraries.518 For example, Seeberger et al.519 used a modular strategy to generate a library of chemically synthesized heparan sulfate glycans ranging from di- to hexamers of different sequences and sulfate-group patterns. An amine-terminated thiol was installed at the terminal pentenyl glycoside end of the oligosaccharide and used to immobilize the glycans onto N-hydroxysuccinimide-activated slides via automated printing, creating synthetic glycan microarrays.520 Key features of this modular synthesis were the influence of the amine linker on the glycosidation efficiency and the compatibility of the protecting group strategy with the sulfate-group decorating step.519 Recent reports have combined the specificity of carbohydrate libraries with computational modeling. Such as Amon et al.,521 who used a comprehensive sialoglycan microarray to challenge a monoclonal antibody (mAb) developed against the tumor-associated carbohydrate antigen sialyl-Tn; the mAb specificity, key residues in the binding site and antibody–glycan contact surface were identified, and used as source data for in silico 3D-modeling.521
5.3.2. Material Surface Chemistry
Over the last 20 years, synthetic chemical libraries composed of polymeric acrylates/acrylamides,111,409 polyurethanes,425 polyesters,420 and others,415 have been probed for cellular applications using a range of cell types, phenotypic readouts, and media formulations. Protein adsorption onto the biomaterials has been clearly related to cell-performance,442,472 with synthetic polymers exhibiting cell-type418 and species-specific418,427,522 effects. Even though there have been numerous studies searching for correlations between chemical moieties displayed on the biomaterial and cell performance, a unified model for cell–biomaterial performance remains elusive.116 Therefore, current efforts have been focused on increasing our understanding of the relationship between polymer-surface chemical profiles and cell performance.
5.3.2.1. Polymer Surface Chemistry
Yang et al.478 reported the first quantitative structure–functionality correlation between surface chemistry and cell-adhesion. Microarrays were fabricated using 16 major and 6 minor monomers combinations (ratios: 100:0, 90:10, 85:15, 80:20, 75:25, 70:30), and cell attachment of randomly differentiated human ECSs was recorded.478,523 Surface structural characterization was undertaken using high throughput measurements of wettability, topography, surface chemistry, protein adsorption, and cell adhesion, termed high-throughput surface characterization.478,523 Acquisition of elastic modulus data from high throughput AFM was added to this approach later.524 Multivariate analysis technique partial least-squares (PLS) regression was then used to identify chemical structure–function relationships. In their study, only ToF SIMS spectra had information to correlate with the cell adhesion, indicating the importance of molecular-level chemical identity in controlling cell response.478 The cell attachment numbers predicted showed an R2 value of 0.62 with the real cell attachment data, validating the PLS model. The ions with high regression coefficients in the training sample PLS model were also consistent with those pulled from the larger library (576) microarray data, supporting the notion that surface-specific chemical composition data is crucial for cell–biomaterial performance correlation models. The same approach was then applied to identify materials that promoted undifferentiated human PSCs attachment and proliferation,106 and to discover polymers that deterred bacterial pathogen adhesion,419 providing a general paradigm for the combinatorial development of synthetic materials. In 2012, human PSC attachment data was used to present the first report of a computational model, where the experimental adhesion of differentiated stem cells onto a library of untested polymers was accurately predicted in silico using only computational descriptors.525
5.3.2.2. Chemical Diversity
Another important parameter of synthetic polymer high throughput screening has been the number of unique polymers that can be probed in parallel using a single microarray. Since the first microarray combined 22 unique monomers to generate 112 polyacrylates,411 researchers have worked on improving the patterning methods, spot size, and combinatorial strategies to create microarrays with higher chemical diversity. Celiz et al.442 increased the chemical space featured on the arrays by successfully printing 141 chemically diverse, unique homopolymers on a single microarray to investigate human PSC attachment.442 The monomers were clustered according to side-chain chemistry (oxygen, nitrogen, fluorine, aromatic and aliphatic) and main chemistry (acrylate or acrylamide). To reduce polymer spreading during printing, a diluted monomer/initiator solution was used (50% v/v monomer, 1% w/v initiator), allowing the generation of ∼400 μm spots, and high throughput characterization techniques (i.e., ToF SIMS) were used to monitor cross-contamination442 (Figure 24B). The authors also investigated the correlation between biological performance and chemical properties, probing WCA and ToF SIMS ion profile against stem cell attachment (total cell number and stem cell marker Oct4-positive cells). While WCA was found to not accurately correlate to cell performance, ToF-SIMS analysis paired with PLS regression analysis identified certain chemical moieties displayed recurrently in high or low performing homopolymers.442 Among these moieties pHEMA fragments were found, unintentionally leaking to the spot surface from the support material, which was intentionally copolymerized in the scaled-up “hit” copolymers to replicate their optimal cell performance.
5.3.2.3. Combinatorial Polymers
In a similar manner that combinations of different biomolecules have been shown to induce cell phenotypes that they did not support on their own, the notion that copolymers could perform better than their separate homopolymers has been considered and addressed by combinatorial methods. To this end, Bradley et al.418,438 designed a spot-to-spot polymerization strategy to generate unique polymers in situ during the automated process by an overprinting varying numbers of drops from different monomer solutions into defined spots. Alexander et al.409,441,526 have used multigeneration arrays starting from large monomer libraries. Using this strategy, a library of 100+ monomers comprises the “first-generation” microarray, where the best performing homopolymers are identified. A “second-generation” microarray is then fabricated, where the first-generation “hits” are arrayed in all multiple combinations at a set ratio (e.g., 2:1), generating a library of copolymers. The best performing copolymers are further combined using multiple ratios (i.e., 1:0, 9:1, 4:1, 3:1, 2:1, 1:1, 1:2, 1:3, 1:4, 1:9, and 0:1.) on a “third-generation” microarray (Figure 18A,B). This strategy is aimed at the identification of high-performing copolymers and synergistic effects, starting from a large library of monomers and subsequently combining the highest performing features and testing them against one biological setting.442
5.3.2.4. Chemical Surface Patterning
Cells communicate with each other through both physical and biochemical cues, and using both close contacts and secreted factors that carry through the porous matrix in vivo, or the liquid media in vitro settings.527 To investigate this effect in microarray format, chemical surface patterning strategies have been developed to isolate and measure cell–cell communication. One such technology is the droplet microarray (Patented technology by Aquarray, www.aquarray.com) developed by Levkin and co-workers,528,529 which is composed of hydrophilic spots surrounded by a superhydrophobic polymer. The fabrication is based on a combination of photomasking and UV-initiated surface grafting, effectively generating geometrically defined microreservoirs for cell separation. The resulting microarray generates unique cellular microenvironments for high-throughput testing, as a thin layer of liquid avoids the superhydrophobic regions effectively collecting as small droplets into the patterned hydrophilic spots. This technology was tested for long-term cultivation of several (more than three) cell types in separate but adjacent compartments, allowing limited cell-to-cell communication via media contact and secreted factor exchange and effectively investigating the impact of coculture between different cell types in a high-throughput manner.528
The droplet-microarray has been used by Levkin and collaborators in several studies23,530−533 as a miniaturized platform for cell-based HTS. For example, in 2016, Neto et al.531 used these patterned arrays to generate alginate hydrogels of defined sizes and shapes, by using the effect of discontinuous dewetting on the patterned microarrays. In 2017, Tronser et al.534 used the droplet-microarray to generate a microarray of mouse ECSs. The cell solution was easily confined to the patterned spots, generating microenvironments for the mouse embryonic stem cell growth. Results showed the cells viable and proliferating over 72 h, and maintained pluripotency (through Oct4-GFP markers assessment). This study presented the droplet microarray as a simple and translatable platform for pharmacological cell-based high throughput assays.534
5.3.2.5. Chemical Surface Nanopatterning
Significant research has also been devoted to investigating the mechanisms behind the phenotypical cell behavior changes induced by chemical nanopatterning at the subcellular level. Pioneer work by Spartz and co-workers535 in 2004 presented highly defined nanopatterned rigid ligand arrays to study the molecular arrangements of single integrins in cell adhesion. They designed a hexagonally close-packed rigid array of cell-adhesive gold nanodots coated with cyclic RGDfK peptide using block-copolymer micelle lithography.535 The diameter of the adhesive dots (<8 nm), allowing for the binding of just one integrin, and the dots were positioned with high precision at 28, 58, 73, and 85 nm spacing.535 Three cell lines were used to investigate the effect of ligand spacing on integrin dynamics, assayed through immunofluorescent staining of integrin, vinculin, FAK, and actin filaments followed by microscopy analysis. Data analysis showed that adhesive dots separated by 73 nm or more impaired or reduced cell adhesion, cell spreading, and focal adhesion formation, while 58 nm or less allowed for effective adhesion.535 This feature was attributable to the restriction of integrin clustering by the covalent binding of the RGDfK-peptide, giving insight into the dynamics of integrin cluster formation and cell spreading.535
In 2016, Spatz and co-workers536 improved upon their early work by designing a quasi-hexagonal array of Au- and TiO2-nanoparticles (AuNPs, TiO2NPs). To prepare the arrays, reverse micelles of a block copolymer consisting of polystyrene (PS) and poly(2-vinylpyridine) (P2VP) (PS(x)-b-P2VP(y)) were formed, with the length of the block copolymer ((PS)/(P2VP)) determining the adjustable interparticle distance. The addition of Au (HAuCl4) or titanium isopropoxide generated the different AuNPs and TiO2NPs, respectively. A functionalized glass slide was dip-coated onto the micelle-containing solution, followed by plasma-etching to remove the polystyrene coating and expose the nanoparticles (Figure 25A). Once both NPs were functionalized onto the slide, a PEG layer was UV-polymerized in situ, creating PEG hydrogels with nanostructured AuNPs and TiO2NPs on the surface, which could be peeled off and used for cell assays. Peptide functionalization could be performed through a phosphonic acid ligand onto the TiO2NPs, or thiol groups onto the AuNPs. As proof of concept, the authors created a binary nanostructure, orthogonally functionalizing the AuNPs and TiO2NPs with either cyclic RGD for cell attachment, or cyclic RAD as a noncell binding control, in varying combinations (2:0, 1:1, 0:2). Rat embryonic fibroblasts showed good spreading on the microarrays, with larger cell area and focal adhesion formation in the areas functionalized with 2 cell-binding peptides, and the smallest cell area on the noncell binding peptides. As per the nanopattern, the interparticle distance of the peptide-functionalized AuNPs showed the greatest effect in cell adhesion.536
Figure 25.
Microarray strategies using chemical nanopatterning. (A) Schematic representing the key steps involved in the generation of binary nanostructured hydrogels consisting of PEG substrates decorated with AuNPs and TiO2NPs. Reprinted with permission from ref (536). Copyright 2016 American Chemical Society. (B) Microarray fabrication workflow steps: (i) parallel nanoimprint lithograph of a silicon wafer is performed using a stamp containing 16 replicates of 8 different line patterns, (ii) followed by surface treatment with plasma etching (PE) and physical vapor deposition (PVD) of Ti and Au, which leads to (iii) fabrication of multiple Si/Au nanoscale patterns within every 9 mm2 areas; (iv) microwell generation around each patterned area via SU-8 photolithography, (v) chemical activation and modification of the SU-8 surfaces, and (vi) finally dispensing of a streptavidin solution on all patterned areas, followed by protein solutions (ratios of LN and VN) locally printed in duplicates for each pattern size, creating 64 combinations of nanopatterned ECM protein patterns. Representations not to scale. Reprinted with permission from ref (537). Copyright 2016 Wiley-VCH.
In 2016, Amin et al.537 presented a combinatorial biomolecular nanopatterning method, in which multiple biomolecular ligands were patterned into multiple nanoscale dimensions on a single surface via chemical moiety patterning (Figure 25B). Parallel nanoimprint lithography was used to transfer patterns from a stamp, containing 16 replicates of 8 different line patterns with equal width to pitch sizes (100–1500 nm, 100 nm depth), to a silicon wafer (Figure 25B(i)). Physical vapor deposition was used to add silica and gold, generating multiple Si/Au nanoscale patterns (Figure 25B(ii, iii)). Wells (300 μm height) around the patterned regions (9 mm2) were spin-coated with a photonegative resist polymer (SU-8) (Figure 25B(iv)), activated through O2-plasma etching, and chemically and sequentially modified to enable selective chemical patterning of SU-8/Si and Au/thiol regions (Figure 25B(v)). This rendered SU-8 and silica regions nonfouling, with Au regions binding biotinylated proteins (Figure 25B(vi)). Biotinylated-proteins (laminin (LN) and vitronectin (VN)) were premixed at ratios (100:0, 90:10, 80:20, 60:40, 40:60, 20:80, 10:90, and 0:100 w/v) and patterned in duplicate; generating 128 combinatorial dual protein nanopatterns on a single surface. Human-derived dental pulp stem cells were seeded onto the arrays, incubated for 4 h, and immunostained for data collection. Results showed that the cell protrusions interacted only with the Au/thiol regions. Cell number and alignment to the nanopattern were also assessed, and compared with a flat control surface. An increase in VN protein content and pattern size correlated with increased cell number, alignment, and size. 500 nm was shown to be a pivotal size for the patterns; line widths ≤500 nm showed increased cell area due to focal adhesion bridging, which allowed cells to spread over the gaps between the lines, while ≥700 nm line patterns displayed cell spreading mainly along the line, thus increasing cell alignment.
5.3.3. Physical Properties
As presented in section 2.2, even when the biomolecules of biological systems adsorb to the surface of nonporous materials, the physical properties of such materials can play a key role in their final biological performance. In recent years, many efforts have been directed toward the development of microarray strategies to investigate the cellular effect of physical parameters in a robust and high-throughput manner.
5.3.3.1. Bulk Stiffness
Biomaterial stiffness, typically characterized by the elastic or Young’s modulus, or surface compliance, has emerged as one of the most relevant mechanical features of biomaterials influencing cell behavior. Traditional 2D microarrays have the shortcoming of being built on surfaces supports with nonphysiological stiffness (such as tissue culture plastic, ∼106 kPa, or glass in the range of GPa538), while natural extracellular environments can present a rather wide range of Young’s Modulus. For example, Gilbert et al.538 used hydrogels with tunable Young’s Modulus to show the effect of surface stiffness on stem cell preservation. They fabricated PEG with varying starting monomer concentrations, and tested their effect on skeletal muscle stem cell self-renewal. Their results showed that when these cells were grown on soft substrates (12 kPa) they were able to self-renew in vitro, while cells grown on rigid plastic surfaces (∼106 kPa) lost their self-renewing capacity quickly after seeding.538
In 2015, Gobaa et al.539 combined biomolecular microarrays with substrate elasticity by contact printing a peptide-laden stamp onto PEG hydrogels, simultaneously probing biochemical and biophysical factors in confined niches.540 To generate the confined microenvironments, 11 different biomolecules were robotically spotted, in single and two-way combinations (totaling 67 features) directly onto a topographically structured stamp.539 The protein-laden stamp was then used to emboss a partially cross-linked hydrogel, transferring the protein combinations and generating a microwell protein array. The hydrogel contained different ratios of thiol and vinyl terminated PEG macromers, resulting in three different hydrogel modulus along with the array (10, 30, and 50 kPa). Human MSCs were trapped within the patterned microwells by using gravitational sedimentation, and assayed for adipogenic differentiation. Results showed that increased stiffness correlated with impaired adipogenic differentiation, with protein effect on adipogenesis being decreased in the higher stiffness hydrogels. Overall, the data suggested that the elastic modulus had a bigger effect on adipogenic differentiation of MSCs than protein composition.
Kourouklis et al.541,542 correlated modulus, ECM composition, and cell-generated traction stress on a microarray-based strategy. Polyacrylamide hydrogels of 4, 13, and 30 kPa elastic moduli were polymerized onto glass slides,542 and contact printed with all two-factor combinations of COL1, collagen III, collagen IV, FN and VN proteins541 (Figure 26). Liver progenitor cell differentiation was induced on the microarray via soluble cues to either hepatocyte or cholangiocyte fate, and cell phenotype was assessed through total nuclei (DAPI), hepatocyte, and cholangiocyte marker (OPN) staining. Results showed that in hepatocyte priming medium, 4 kPa microarrays had a modest overall reduction in total cell number and hepatocyte differentiation across all protein conditions, while collagen IV showed a discrete boosting effect in hepatocyte differentiation regardless of stiffness.541 In cholangiocyte conditions, data analysis showed that at 30 and 13 kPa, the protein composition did not have a highly pronounced effect in cholangiocyte differentiation, while at 4 kPa collagen IV achieved the greatest number of OPN positive cells.541 To gain more insight into the signaling pathways involved, the cells were incubated on the microarrays in cholangiocyte priming medium supplemented with a control (DMSO) or a Rho-associated protein kinase (ROCK) inhibitor. ROCK is a key component of the mammalian cell’s mechano-sensing machinery.543 Regression analysis showed that ROCK inhibition increased OPN positive cells on both 30 and 4 kPa substrates (P > 0.001). The increase in cholangiocyte differentiation using ROCK-inhibitor prompted the authors to investigate cell-generated traction stress using traction force microscopy (Figure 26C–F). ROCK inhibition was found to reduce cell traction stress on both 4 and 30 kPa substrates in all protein conditions except fibronectin (Figure 26C–F). ROCK has been shown to promote cell contractility,544 so the authors hypothesized that ROCK-inhibition on the microarray could increase cell–cell interactions, promoting cholangiocyte differentiation in a manner that is independent of cell-matrix traction.
Figure 26.
Microarray strategies to investigate cell responses to mechanical stimuli. (A–D) Evaluation of cell traction stress. (A, C) Cell traction stress (Pa) measured on the microarrays on 30 kPa (A) and 4 kPa (C) substrates on the different protein compositions, in control (DMSO) and ROCK inhibiting (Y-27623, 10 μM) media conditions. (B, D) Representative phase contrast micrographs and heat maps of traction stress for 30 kPa (B) and 4 kPa (D) substrates. Student’s t tests were performed against DMSO within each ECM combination, and data is presented as mean ± SEM with n = 3 and ≈ 20 total islands per condition (P < 0.05 (∗), P < 0.01 (∗∗), P < 0.001 (∗∗∗)). The scale bar is 50 μm. Reprinted with permission from ref (541). Copyright 2016 Elsevier, Ltd. (E–N) Microarray strategy. (E) Schematic of the overall microarray chip with zoom on a single active area and on a single pixel, (F) picture of the chip bonded on the carrier, (G) SEM image of one active area featuring the 4 different electrode sizes and the 8 reference electrodes. (H, I) SEM images of a single pixel, containing electrodes of sizes 2.5 × 3.5 μm2 (H) or 11 × 11 μm2 (I). (J–N) Electrical and confocal imaging of primary hippocampal neuron culture at 0 and 8 days in vitro. (J, K) Electrical impedance map and confocal image of the cells stained with Calcein AM 4.5 h after seeding. (L, M) Electrical impedance map and confocal images corresponding to the same chip surface area (2500 μm2). (N) Histogram illustrating the distribution of the relative impedance variation recorded by the electrodes for cells after 0 (black) and 8 (gray) days in vitro. Reprinted with permission from ref (551). Copyright 2019 Frontiers.
5.3.3.2. Electrical Stimuli
Other factors such as electrical stimuli can also affect cell behavior, for example, in 2017 Wang et al.545 showed that stem cell-derived cardiomyocytes display better myofibril ultrastructural organization when cultured on superconductive graphene surfaces, proving the importance of electrically active substrates.545 In 2019, Vaithilingam et al.546 developed a conductive composite material for 3D biomimetic structure fabrication.546 The base polymer (pentaerythritol triacrylate, PETrA) was identified using high-throughput microarray screening of a large library of acrylates/acrylamides as a suitable biomaterial for adhesion of mature human stem cell-derived cardiomyocytes. The electrical properties of the polymer were modified by using a dispersion of conductive multiwalled carbon nanotubes, turning the material electroactive, and two-photon polymerization was used to fabricate a biomimetic topography spanning the macro-nanoscale onto the electroconductive polymer. Cardiomyocytes were plated onto the polymer-containing conductive nanotubes, which were coated on top of an in-house 6-well plate setup that provided a low electric potential (2 V and 3 Hz). On all topographies, the cardiomyocytes showed higher sarcomere length, a sign of cytoskeletal maturity, in the presence of a low current over the nonelectrically stimulated counterparts. Different chemical substrates and microgrooved topographies were also tested on their system, showing that all three characteristics (chemistry, topography and electrical stimuli) affected cardiomyocyte shape, morphology and maturity.546
To investigate electrophysiological events in biological systems, high-density microelectrode arrays have been developed with the capacity to record and stimulate single cells within a cellular network. In 2015, Müller et al.547 presented a complementary metal-oxide semiconductor (CMOS) high-density microelectrode array capable of recording and stimulating with bidirectional microelectrodes at high spatial resolution and signal-to-noise ratio. This work, based on earlier work by Frey et al.,548 was capable of circumventing the trade-off between electrode pitch and readout noise performance, using the available area below the electrodes (3.85 × 2.10 mm2) to implement programmable routings of electrodes (26 400 platinum microelectrodes) as readout channels. This proof of concept study achieved the recording of a neural network composed of primary rat cortical neuronal and glial cells at different levels of spatial resolution. Single neuronal stimulation could also be achieved within the microarray with electrical pulses, while simultaneously tracking the affected neuron’s activity over multiple recording sites of its axonal branches at a high spatial resolution over a distance exceeding 1.5 mm.547
Lopez et al.549−551 presented another strategy for high-density electrical recording based on CMOS and Electrochemical impedance spectroscopy (EIS) technology. EIS technology uses a small alternating current applied across a cell-laden surface to measure changes via voltage drop.552 EIS has been presented as a real-time cell monitoring technology; it can be used to measure cell adhesion and growth, does not require labels, is noninvasive for cells and can be performed at several frequencies to assess different physiological phenomena.553 The microarray setup consisted of 16 independent active areas patterned onto a chip, with 1024 electrodes per area grouped into 256 pixels (Figure 26E–I)551 and enabling 6 different cell-interfacing modalities: (1) extracellular and (2) intracellular recording, (3) constant-voltage stimulation for controlled electrode potential, (4) constant-current stimulation for controlled charge delivery, (5) fast impedance monitoring, and (6) impedance spectroscopy.550 Lopez et al.549,550 used this strategy for high-throughput electrical activity monitoring of cardiomyocytes, measuring cardiac cell contractility in real-time. During the contraction phase, the interface impedance dropped, indicating a separation between the cell and the electrode, while during the relaxation phase the impedance increased again. This work also demonstrated the importance of multimodal recording by treating the cells with Nifedipine (calcium channel blocker) and recording electrical output using the extracellular and intracellular recording modalities. Only the intracellular recording showed that Nifedipine significantly decreased the cell’s action potential duration.550 Miccoli et al.551 presented the use of this technology to measure cell adhesion and growth of primary rat hippocampal neurons on the microarray at seeding and after 8 days growth (Figure 26J–N), and to record spontaneous electrical activity under the form of a single unit and synchronized network activity.551
5.3.3.3. Compression, Stretching, and Shear Stress
In their natural microenvironment, cells are also subjected to compression, stretching and shear stress. These forces are part of their natural process and can trigger important cellular events such as YAP-mediated increase of cell motility.554 For example, a study in organogenesis showed that oscillating waves of stretching and compaction of entire fields of cells can pattern intracellular YAP/TAZ signaling, resulting in the orchestration of defined tissue boundaries and therefore, achieving controlled organ development.555
High throughput technologies have also been used to study the effect of varying mechanical stress on cell behavior. One of the first reports of such designs was by Moraes et al.,452 who developed a microarray capable of applying cyclic compressive strain onto 3D PEG hydrogels, with a compressive strain range of 6 to 26%, stimulating individual cell deformations under compression. Confocal microscopy was used to assess the hydrogel deformation by mixing FITC-labeled fluorescent beads into the hydrogel solution, and cell viability by using a fluorescent-based LIFE/DEAD cell assay, informing on cell viability and shape.452 Results showed that cell deformation was significantly increased on the largest force sample, but not for the rest of the compression range, while nuclear deformation was not significantly different under any of the mechanical conditions. The authors investigated this lack of linear correlation by using parametric finite element simulations, and suggested that the relative stiffness difference between the cells and the matrix (E*matrix) affected cell deformation due to local “shielding” effects. The cell deformation was not linearly correlated to mechanical strain in the lower end of E*matrix ratios showed by PEG hydrogels (0.1–1 kPa) because the local stiffness inside the hydrogel was much lower than within the cytoplasm, and therefore it “shielded” it from deformation. As the modulus of the matrix equaled that of the cells, a large increase in cell deformation occurs. In a similar way, as the nucleus is three to four times stiffer than the cytoplasm, it was “shielded” by its soft environment and thus its deformation response to a linear increase in compression force was not actually linear.
More recently, Li et al.556 fabricated microscale magnetically actuated cell-laden hydrogels to investigate the cellular response induced by microenvironment strain.556 These microarrays could undergo reversible, relatively homogeneous deformation through magnetic fields, enabling the application of strains of up to 60%. Using a multistep photolithography model and sequential masking steps, they polymerized a compressive layer, a magnetically actuated layer, and a cell layer; all fused together in one device (Figure 27A).556 Cell experiments started 6 h after encapsulation, and the mechanical strain was applied for 10 h per day. After the different treatments, the cells were fixed and stained using phalloidin and DAPI (for cytoskeleton and nuclear staining, respectively), and imaged using confocal microscopy. 3D reconstructions of confocal Z-stack images generated data on cell spreading over the hydrogels (Figure 27A).556 Results showed enhanced spreading of the fibroblast cells in response to the mechanical forces (Figure 27B),556 while the magnetic field showed no statistically significant changes in cell morphology. Data also showed that a mechanical strain up to 40% could significantly enhance cell proliferation after 1 day of culture, and that in all experiments strain activation was required for substantial cell division, indicating a strong correlation between mechanical strains and fibroblast proliferation.556
Figure 27.
Magnetically actuated, cell-laden hydrogels (μMACs). Three types of masks are designed, one for each of the layers: (i) a “magnetically actuated” PEGDMA layer encapsulating magnetic Fe3O4 nanoparticles, (ii) a stiff, constrained layer, and (iii) a gelatin methacrylate layer encapsulating the cells. The bottom right shows a photo of the hydrogel and cells setup, scale bar = 2 mm. (B) Fluorescent confocal images (top panel) and 3D reconstructions (bottom panel) of encapsulated fibroblasts in μMACs (modulus = 6 kPa) under different strain conditions. Cellular F-actin fibers stained using phalloidin (red), and nuclei (DAPI, blue). Graphs show (left) mean cell spreading volume increase over culture time and increased strain levels, rising quickly to an asymptotic saturation level at a critical strain in the range of 40–60%, and (right) cell proliferation increases with time and strain levels, saturating at the ∼40% strain condition. Scale bars = 500 μm. Error bars, SD (n = 10 μMACs for each strain level, ∗∗P < 0.01, ∗∗∗P < 0.001). Reprinted with permission from ref (556). Copyright 2016 Nature Publishing Group.
In 2018, Horner et al.446 used electrospun 3D scaffold discs for the assessment of varying dynamic compressive strain on hMSCs under osteogenic conditions. The scaffold fabrication was performed through the electrospinning of poly(ε-caprolactone) into a fiber mat, and a 6 mm diameter biopsy punch was used to generate cylindrical scaffolds of 3 mm thickness. The scaffolds were plasma-treated, followed by COL1 conjugation using a cross-linking agent, sterilized in 70% ethanol, and transferred to 24-well plates for cell culture assays using human MSCs. The cells were allowed to infiltrate the scaffold for 5 days before starting the osteogenic induction conditions through soluble cues and mechanical stimulation. The mechanical strain was applied using a custom-made compression system in four separate magnitudes: 5, 10, 15, 20% of the scaffold thicknesses; statically cultured samples (0%) served as a control. The strain was applied for 2 h each day for up to 28 days of differentiation, after which histology and transcriptional analysis were used to evaluate osteogenic and chondrogenic markers. The authors found transcriptional upregulation of chondrogenic markers and glycosaminoglycan synthesis in response to the increased magnitudes of compressive strain, whereas most osteogenic markers and calcium deposition were decreased by compressive loading in a magnitude-dependent manner. The dynamic mechanical analysis showed enhanced viscoelastic modulus of the scaffolds in response to increased dynamic strain, peaking at 15% (≈ 45 kPa), which coincided with the maximal GAG synthesis. This study suggested that the differentiation of human MSCs toward osteogenic or chondrogenic lineage was related and dependent on the magnitude of dynamic compressive strain, and therefore that mechanical strain is a powerful feature for cartilage and osteogenic cell regeneration strategies.
5.3.4. Topographical Cues
There is plenty of evidence showing that cells are able to respond to the topographical features of biomaterials by integrin receptor-binding and development of focal adhesion points.557,558 In their natural setting, cells experience a complex 3D microenvironment, and studies have shown that cells behave closer to in vivo models when cultured in 3D conditions.559 Roughness has been traditionally regarded as a random topography and an effective surface modification strategy to improve cell adhesion and infiltration of biomaterials, especially in osteointegration.560 As such, it has been an important feature of microarray-based biomaterial discovery, especially in combination with surface chemistry and wettability. Early studies combined topography and surface roughness studies in a reduced number of biomaterials to investigate their effect on cell adhesion.561 Subsequent high throughput studies have assessed the chemical structure and roughness measurement in their microarray characterization and data mining strategies. For example, in 2010 Yang et al.478 used data correlation analysis to investigate the relationship between roughness and cell performance. They fabricated microarrays using a library of 496 copolymers and recorded data on human embryoid body–cell attachment, WCA, elastic modulus, protein adsorption, and surface roughness. Multivariate analysis technique PLS regression was then used to identify material properties–functional relationships. In that study, only ToF SIMS spectra were found to correlate to cell adhesion with nanometer-level roughness observed not to have a significant influence.478
In more recent approaches, advanced photolithography and soft-lithography techniques have been combined to fabricate micro- and nanopatterns to study cell phenotype. For example, Dalby et al.562 used electron-beam lithography to fabricate defined nanopits of 120 nm diameter and 100 nm depth arranged in 5 different patterns to compare varying degrees of disorder, recreating the levels of disarray found in the collagen fibers of natural ECM.562 Results showed that one particular disordered topography, with nanopits 300 nm apart in a square array and randomly displaced by up to 50 nm, was able to induce osteogenic differentiation comparable to cells that were chemically induced by dexamethasone on tissue-culture plastic. Analysis using ingenuity pathways and canonical analysis (i.e., related to a specific signaling pathway), showed activation of certain signaling events, in particular fibroblast growth factor and epithelial growth factor signaling, that have been associated with bone development.562 The authors followed up by altering integrin clustering at the nanoscale level to investigate the effect of small nanotopographical changes, and showed that organized nanoscale patterning of cell-adhesive features affects cytoskeletal contraction and intracellular tension, subsequently affecting cell differentiation.562 Indeed, other groups have shown a direct relationship between cell shape through surface-induced topography and cell epigenetic state, which in turn alters the cell phenotype through gene expression (reviewed in ref (563)).
De Boer and co-workers51,144,455−457,564 have developed their own strategy for high-throughput topography studies. In 2011, Unadkat et al.455 presented a mathematical algorithm that can design over 150 million (154 320 600) unique topographies by combinatorial mixing of three primitive shapes (triangles, circles, and rectangles), generating a very large number of relevant geometrical features such as smooth areas, angles and stretched elements. The topographical design is generated in silico by randomly selecting parameter values such as total size, number of primitive shapes to be used, their distribution, and the degree of alignment between the shapes. Next, each primitive is placed at a random position within the feature, with the overlapping of primitives being allowed. The different shapes range from 3 to 10 μm and are arranged into imaginary squares of the variable area and 5 μm height. From the in silico library, 2176 unique TopoUnits were randomly selected to create the first “TopoChip”455 (information on fabrication in section 5.2.1.2). Human MSCs were used to investigate the bioactivity of the topographies, with cell number (TOTO-3 staining) and proliferation (EdU staining) measured after 8 h of culture using fluorescent microscopy. A classifier algorithm was trained from a subset of the data set (the highest and lowest 10% of the data), and used to identify specific topographical features with high cell phenotype modulation. Feature size and the Fourier-based parameters WN1 and WN1.5 were found to display the best predictive power on cell behavior, suggesting that cell proliferation could be triggered by concentrating on designs composed of certain spatial distributions.455 The authors also investigated the osteogenic differentiation of MSCs (ALP staining), and correlated positive cell numbers to cell morphology features on the different topologies, showing that smaller area and higher major axis cell length identified with increased osteogenic cell phenotype.455
In 2015, Hulsman et al. presented a materiomics-based computation analysis for the exploration of the cell morphology responses to the different TopoChip features. First, data from one human MSC cell line on 8 identical TopoChips was gathered for the nucleus (DAPI) and cytoskeleton (phalloidin), generating 16 replicate measurements in parallel. The fluorescent images were analyzed using CellProfiler software for cell number, cell localization, and cell morphology data acquisition; followed by extensive computational analysis based on 33 cell morphological properties and 30+ topographical parameters. Interestingly nucleus form factor (i.e., “roundness” of the nucleus, defined as 4π × area/perimeter2) showed the most significant response to topography (Figure 28A), with cell orientation and maximum DAPI staining intensity also showing highly significant variation. On the other hand, the cell area was shown to only moderately responsive to the micropatterned substrates. The authors followed up by ranking the features’ effect on cell morphology, thus finding spectra of surfaces that could modulate cell properties and identifying relevant surface parameter combinations that could predict cell responses on untested surfaces. Since de Boer’s topographical library is too large to study even in high-throughput, the use of computational models allowed for a first in silico test to identify features that can most likely evoke the desired cell response, thus improving microarray design.565
Figure 28.
Features of cell-based topographical microarrays. (A) Fluorescent images ranking surfaces by nucleus roundness of human MSCs growing on patterned PLA surfaces; the three lowest and three highest-ranked surfaces are represented. The topographies are represented and colored according to a Mann–Whitney U-test ranking. Cells show nuclear (red) and cytoskeleton (green) staining. Reprinted with permission from ref (144). Copyright 2015 Elsevier, Ltd. (B) High fidelity of PDMS patterns generated from the MARC master molds by soft lithography. SEM images of (i) unpatterned PDMS control, and (ii–viii) patterned topographies, with the description of patterned size (line width or pillar and well diameter, μm), pattern spacing (μm) and pattern height (μm), in that order. The perpendicular pattern is also indicated with the abbreviation (pr). Scale bar = 5 μm. Reprinted with permission from ref (571). Copyright 2013 Elsevier, Ltd.
De Boer and co-workers have since optimized the TopoChip technology and generated variant formats, such as the TopoWellPlate51 and the Nano-TopoChip,457 and investigated the effect of topography on a variety of different cell models and assays; such as secretory capacity456 and osteogenic differentiation566 of human MSCs, tenocyte cell phenotype,567 human epidermal stem cell differentiation,568 and “hit” topographies have even been tested in vivo in animal studies of osteointegration.566 For example, Leuning et al.456 used the TopoWellPlateto study the effect of surface topology on the cell secretion profile of primary bone-marrow derived MSCs and kidney-derived perivascular stromal cells. Stromal cells have immunomodulatory and regenerative potential in vivo, and these properties have been tightly linked to their secretory profile.569,570 The authors cultured the cell types in a TopoWellPlate containing 76 unique topographies plus unpatterned controls. The cytokine secretion profile after 48 h was assayed using a custom-made multiplex ELISA system, and total cell number and morphology were measured via fluorescent microscopy.456 Secretion levels were adjusted for cell number, and comparison between topographies showed a high range of variability. Most importantly, the coefficient of variation between technical replicates was low for the majority of factors, showing a robust response to the topographical features. Data analysis also showed noticeable cell type-specific responses, with differential secretory responses to the same defined topographies among the different stromal cell populations.456
The Nano-TopoChip was presented by Hulsholf et al.457 in 2017, with high-quality designed nanometer-scale features fabricated on polystyrene, on a large area (4 cm2), using deep UV projection lithography together with conventional lithography. The study used osteosarcoma cells to compare cell number and morphology changes on the microsized and nanosized TopoChips. Cell nuclear morphology showed the biggest difference, with cells showing marked nuclear changes when cultured on the microsized topographies, as compared to almost no effect on the Nano-TopoChip. Cytoskeleton morphology was markedly affected by both the nano- and the microtopographies. Machine learning methods were used for data analysis of the cellular effects of nanotopography; identifying the size and spacing of the nanotopographies, as well as the ratio of patterned versus nonpatterned area, to have significant and reproducible effects on cell spreading. Since nanotopographies do not restrict the shape of the cells, the authors suggested an effect on the availability and localization of cell attachment sites through spatial constraints (e.g., alignment of attachment proteins and availability of recognition sites such as RGD). This work translated the large topographical design space of the algorithm by de Boer et al.455 from micro- to nanosize scale, generating data on topographical effects at the subcellular level.
In 2012, Yim and co-workers571,572 developed another microarray strategy to investigate the influence of topography in cell behavior. The microarray included topographies ranging from micro- to nanodimensions, with not only varying lateral dimensions but also height and complexity. Gratings, pillars, wells, and complex 3D hierarchical structures were included in the so-called multiarchitecture (MARC) chip (Figure 28B), which was composed of patterned PDMS substrates featuring 18 different topographies in duplicate.571,572 Ankam et al.571 used the MARC chip to investigate the neural commitment of human ECSs. Human ECSs were cultured on the topographical arrays for 7 days in neural induction media, and subsequently stained for glia cell markers (glial fibrillary acidic protein, GFAP) and neural cell markers (tubulin III, Tuj1). Results showed a significant effect of topography on neuronal differentiation, with planar surfaces generating an even mixture of neuronal and glial cells, while anisotropic micro and nanogratings displayed higher neuronal markers (Tuj-1 positive cells), and isotropic pillar patterns resulting in greater glial cell number (GFAP-positive cells).571
In 2018, Kukumberg et al.573 further developed this technology to create the MARC chamber and the MARC plate. The MARC chamber contained 42 distinct patterns (2 × 2 mm2) mimicking the topographies on the MARC chip, with the added feature that it could robustly fit into conventional cell culture plates or dishes. The authors also generated a MARC plate, with one pattern in each well of a multiwell plate, providing a scale-up format with a larger area for microarray data validation. The MARC chamber was used to investigate the effect of topography, COL1 coating, and cell density seeding in umbilical vein endothelial cells phenotype. ICAM-1 expression and monocyte adhesion were assayed as markers of functional endothelial cell layers. Pillar structure topographies showed improved performance as compared to lens topographies, resulting in significantly enhanced endothelial cell monolayer phenotype on the study.573 In 2020, Arora et al.574 followed up on this work by culturing primary arterial endothelial cells, primary venous endothelial cells, and human PSC-derived endothelial cells on MARC chips containing 16 different topologies. Only 2 μm wide simple grating and hierarchical gratings produced significant alignment of the primary cells, while the PSC-derived cells showed alignment to a wider range of topographies. Results also showed that when the primary endothelial cells were cultured on gratings topography, there was an induction of the venous over the arterial phenotype.574
5.3.5. Next-Generation Multiparameter Microarrays
As individual characteristics of biomaterials have been thoroughly investigated in different cellular models, the next logical step has been to combine the screening of set characteristics to achieve a basic understanding of the correlation between the different properties of biomaterials and their biological performance in a systematic manner. One such combination has been presented in 2020 by Burroughs et al.,410 where the TopoChip technology from de Boer and co-workers51,455,458 has been combined with the large chemical libraries studied by Alexander and co-workers409,441,575 to generate a microarray containing 980 unique chemo-topo-features, herein called ChemoTopoChips. The microarray design comprised 36 TopoUnits (including a flat control), patterned onto tri(3-mercaptopropionate):tetra(ethylene glycol) diacrylate (1:2 TMPMP:TEGDA), arranged in a 3 × 3 mm2 grid and repeated 28 times for variable chemical functionalization via UV-polymerization of monomer/DMF solutions to react with thiols on the patterned substrate.410 The ChemoTopoChips were used to assay both human MSCs and primary monocytes isolated from human blood, as cross-talk between macrophages and MSCs has been shown to be essential for successful biomaterial osteointegration. First, the human MSCs were cultured on the microarrays using the basal (nonosteoinductive) medium for 5 days, followed by immunostaining. CellProfiler software was used to extract data on total cell number (nucleus), cell morphology (ß-3-tubulin), and osteogenic phenotype (ALP staining intensity); which was normalized to the TMPMP-co-TEGDA flat control. Analysis of the mean integrated ALP intensity per cell revealed 113 features with a significant ALP upregulation and 103 features with a higher cell number (p < 0.05). Interestingly, all combinations containing microtopographies displayed higher cell numbers than the counterpart chemistry-functionalized flat control.410
The 3D microarray system developed in 2016 by Lutolf and co-workers450 is another example of highly combinatorial systems for cell niche bioengineering. The platform was fabricated on 1536-well plates, and the combination of a library of molecular building blocks allowed the generation of a highly tunable system to investigate five distinct ECM parameters.450 The peptide derived from FXIIIa was used to cross-link PEG-based macromers into 3D hydrogel networks of tunable stiffness in physiological conditions. The addition of lysine-containing peptides with various MMP-sensitive sequences onto the PEG-macromers allowed the hydrogel to be susceptible to cell-mediated degradation, an important feature of tissue homeostasis (Figure 20B). Tunable cell number and ECM protein addition to the prepolymerization solution generated an even larger combinatorial space. Finally, the well-plate format allowed for multiple media formulations in closed environments. The combination of all these variables generated a platform for the independent control of five key cell microenvironment parameters: mechanical properties, proteolytic degradability, ECM protein composition, cell–cell interactions, and soluble cues.450 In the study by Ranga et al.450 an Oct4-GFP reporter mouse embryonic stem cell line was grown in the multiparameter 3D hydrogel microarray, and real-time data acquisition for colony formation and cell proliferation was achieved using confocal microscopy and GFP-fluorescence monitoring (Figure 20B).
Cells in vivo are exposed to a complex, spatially organized 3D microenvironment, generating polarization, gradients, and spatially defined cues that direct cell migration, proliferation, differentiation, and metabolic responses over time.576 Matrix degradation and remodeling are also an intrinsic part of tissue homeostasis, thus spatiotemporal cues have been recognized as useful parameters in biomaterial engineering, managing to evoke specific biological responses over time. Recently, Brehm et al.530 have described the miniaturization and parallelization of combinatorial organic synthesis, through the development of an on-chip solid-phase combinatorial library synthesis method. The microarray platform used was the Droplet-Microarray developed by Levkin and co-workers,528,533 and a hydroxyethyl linker was used for the solid-phase synthesis, as it can be cleaved using a UV light of 365 nm (shown to be nontoxic to cells577). The photolinker allowed for compound release at any point during the assay, adding temporal flexibility to the platform. The Ugi four-component reaction,578 which can yield a large number of different products with a small input of variation, was used for compound synthesis; using five different amino acids and surface-bound amines, three isocyanides, seven carboxylic acids, and eight carbonyls; and generating of a total of 840 unique products. The nanomolar library was then retrieved from the surface through photorelease, and analyzed using mass spectrometry. As a proof of concept, the drug chlorambucil was covalently bound through the photosensitive linkers onto the microarray and Chinese hamster ovary (CHO) cells were used to assess response to culture on a drug-laden microarray with and without a UV-treatment for drug release, and culture on “naked” nonfunctionalized microarrays with or without a UV treatment. CalceinAM and Propidium iodide staining were used to assess cell viability. Non-UV treated drug-laden arrays had similar cell viability to naked microarray controls (97%), while UV-triggered drug release reduced cell viability (<5%). UV treatment on the naked microarray did not have a significant effect on cell viability (96%).530 These results showed how the surface-bound compounds were only active on the cells after UV-triggered release, thus evidencing the flexibility of their platform’s screening conditions in a spatial, temporal, and quantitative manner.530
Another drawback of microarray technologies is that they typically rely on “end-point” cell assays, with data collection requiring the termination of the experiment. Informative data on how the cells attach, grow, and react over time to the biomaterial can greatly increase our understanding of the cell–biomaterial interface. For example, natural heart development starts from a soft, disorganized matrix, and naturally evolves through embryo development toward the adult heart, which presents a highly organized and stiff matrix capable of enduring cardiomyocyte contraction and conductive of electrical stimuli.579 This is a timely and spatially defined process, with changes occurring not only within the cells, but with cells modifying their surroundings over time. Real-time data collection methods, so-called 4D tracking (including time as the fourth dimension) are gaining momentum, with a number of new methodologies being developed for live-cell tracking.
An example of real-time data collection on cell behavior within a biomaterial was reported by Blakely et al.,580 who developed a system using molecular tension probes based on DNA hairpins conjugated to fluorophore-quenchers to reveal cellular traction forces applied within biomolecule-laden hydrogels, giving spatiotemporal information on cell “pulling” within their matrix.580 Another system for real-time cell assessment is the so-called 4D cell culture imaging systems,581 which are able to track cell migration, matrix remodeling, proliferation, and phenotypical changes in a spatial and temporal manner. Samal et al.582 have recently presented a microengineered high-content imaging platform for 4D tracking of single cells in an in vitro morphogenesis model. The platform fabrication used a free-forming variant of negative microscale pressure thermoforming, where a transparent polycarbonate film was patterned into microcavities arranged in an array fashion, with a ∼500 μm width and a ∼500–1250 μm length.582 The thin-walled cavities allowed for multidimensional time-lapse imaging using confocal microscopy, translatable to many common laboratories. The authors created a data analysis pipeline using free software that could be used to evaluate and quality-control single-cell migration and aggregate morphogenesis, making this time/spatial single-cell tracking method an open-access resource.582 These novel 4D technologies have the potential to widely increasing our understanding of cell–biomaterial interactions over long-term exposure, and as such, better predict their performance in vivo applications.581
Mass spectrometry techniques and elemental analysis will also be key to the success of cell-based microarrays strategies, improving our ability to detect individual protein species on and within biomaterials in a high-throughput manner.473 Moreover, biomaterials that enable reversible biophysical and biochemical properties will also be great additions to high throughput assays.530 For example, the use of photoresponsive and photocleavable reactions on hydrogels, which permits user-defined quantities of proteins to be anchored within distinct subvolumes of a 3D matrix, and tailored subsequent removal using ortho-nitrobenzyl ester photoscission.583 In this manner, hydrogels can be designed with spatially defined ligand availability, and modified over time of cell culture to induce a certain cellular response.
5.4. Biological Models in Microarray Studies
Cells in vivo interact with a highly complex and specific ECM, responding to its cues, gathering information and generating signaling events that ultimately control cell behavior.584 The lack of complete and accurate information on natural niches complicates rational engineering. To bridge the gap, high throughput technologies have the ability to present one particular biological system with a large number of varying biomaterials in parallel, therefore reducing biological variability while increasing sample size and reducing experimental cost and time. However, because the biological response is ultimately the key to the success of these arrays, the biological assay design and data analysis are determining characteristics, and they need to be tailored to the specific biological system investigated, and to the desired cell response. This section will review the variety of biological models used in high throughput technologies, with emphasis on the output/functional assays used in relation to the desired cellular behavior.
5.4.1. Mammalian Cells
Most of our current understanding of mammalian cell biology comes from artificial, 2D culture methods on glass or tissue culture plastic. These cultural methods dominate biomedical research because they are easy to operate and highly reproducible. However, 3D biomaterials are the ones that most accurately recapitulate the native microenvironment of mammalian cells. It has been long established that cells in 2D conditions behave differently than their 3D counterparts, and that in sensitive applications such as drug development or organ formation, a native microenvironment is paramount to collect accurate data that can be readily translated to clinical applications.559,585,586 As we learn more about the biological response to artificial niches, we are better able to understand the different ways that mammalian cells communicate with and respond to their immediate surrounding microenvironment.
5.4.1.1. Stem Cells: Differentiation and Self-Renewal
Regenerative medicine holds great promise for treating dysfunctional or damaged tissue resulting from trauma, disease, or aging. In particular, cell therapies and tissue engineering strategies have focused on two types of cells: (1) human ESCs or PSCs, which are derived or mirror the postimplantation epiblast embryonic state; and (2) multipotent stem or stromal cells, such as MSCs, which are tissue-specific cells with limited differentiation capacity.586−589 Stem cells are defined by two key characteristics: the ability to self-renew, that is, to proliferate and replenish their population, and the potential to differentiate into other cell types. While these cells offer the possibility to create unlimited amounts of any cell type, there are still major biological roadblocks to overcome before they become an easily accessible biomedical tool. In particular, one of the major problems for cell therapies is that current in vitro differentiation protocols still fail to produce mature and functional organs, typically generating disarrayed cell mixtures with embryonic or immature features.526,590 For example, cardiomyocyte cell replacement therapies have met two key issues that are still being resolved: (1) foreign cardiomyocyte cells do not easily integrate within adult hearts, showing low engraftment efficiency and potentially creating ventricular arrhythmias591 and (2) human PSC-derived cardiomyocytes are phenotypically immature, with features closer to prenatal heart.526,592 With soluble cues having been amply optimized, the focus is now on biomaterial engineering as the key to improved cell differentiation.526
The field of biomaterials has focused greatly on the development of biomaterials to influence, direct, and determine the fate of differentiating stem cells (reviewed in refs (22 and 422)). Stem cells respond to both (bio)chemical and (bio)physical cues in their immediate microenvironment, and these changes influence both their differentiation potential, efficiency, and maturation into tissue-specific functioning cells.593−595 In particular, MSCs have shown to be greatly influenced by surface chemistry, topography, and stiffness, with studies showing that biomaterials resembling the mechanical features of the desired tissue can induce directed differentiation as effectively as soluble cues.76,410,456,596,597 Since biomolecular cues used in stem cell culture can be expensive to produce and quality control,598 the induction of stem cell differentiation through interaction with synthetic chemical groups tethered to a spatially defined structure opens the door to more powerful, simple and cheap strategies for therapeutic treatments.596
Universal substrates for stem cell culture in vitro have been investigated. Such as the study by Celiz et al.,409 which used a large library of chemical monomers to find an improved synthetic polymer that could support both expansion and multilineage differentiation of human ECSs.409 During the microarray discovery step, only one stem cell marker (Oct4) was used to determine stem cell survival and maintenance onto the different polymers. It was during the scale-up of the hit combinatorial polymer Poly(HphMA-co-HEMA) that multilineage differentiation potential was investigated. In this instance, a polymer that could be easily adapted to several differentiation protocols was the goal, as a defined and economical alternative to the widespread Matrigel formulation (an ECM formulation derived from a mouse sarcoma599). The mechanism of initial cell adhesion was also investigated through integrin blocking experiments, reporting for the first time β1 and αv integrins as the main mediators of cell attachment to a synthetic polymer.409
Improved stem cell phenotype and generation have also been investigated using high throughput strategies. Caiazzo et al.586 showed that an engineered biomimetic 3D microenvironment not only promoted mouse embryonic stem cell features (even in the absence of supporting biochemical factor LIF), but also increased the efficiency of cell reprogramming to PSCs 3-fold as compared to 2D culture.586 Their data showed that early events during reprogramming of adult cells toward a pluripotent stem cell state are accompanied by profound morphological changes, which may facilitate the mesenchymal-to-epithelial transition and chromatin remodeling required for the reprogramming toward PSCs.586 Higher histone acetylation and trimethylation in lysine 4 (H3K4me3) were detected in 3D conditions, showing that biophysical cues can modulate the epigenetic state and improve the efficiency of cell reprogramming in 3D hydrogel microarrays.586
Other reports have focused on differentiation efficiency in closely related cell types. Malta et al.489 investigated the differentiation of mouse ECSs primed to definitive endoderm. 741 unique protein combinations together with soluble factors (media for pancreatic or hepatic differentiation) were combined. The authors found that, as the differentiation progressed, cell survival and proliferation changed, and that the ECM composition had a marked and differential effect in the differentiation efficiency of these very close cell types.489 In 2016, Delalat et al.600 combined 7 ECM proteins and 4 soluble growth factors to generate 201 unique microenvironments to investigate cell survival of insulin-producing cells. The results obtained using a mouse insulinoma cell line were compared with cells harvested from mouse and human pancreatic islets. Total cell adhesion number of mouse insulinoma cells on the microarray showed that triple protein combinations dominated the top 10% of candidates identified, with FGF-2 (fibroblast growth factor II) being present as the major components in 7/20 top-performing combinations.600 The authors then used the same microarray to assess mouse and human primary islet cell attachment and growth at 48h. Interestingly, the results did not correlate with the ones obtained on the insulinoma cell line, but did have some correlation between the two primary cell populations. Collagen IV recurred as the major components of the highest-ranking protein combinations for both primary cell samples. This study showed the importance of the biological system in microarray technologies, as the biomaterial hits shown by an immortalized cell line did not accurately model the ones shown by cells sourced from adult tissue.
Hou et al.601 used a microarray platform to investigate the effect of random protein combinations in embryonic stem cell-derived-endothelial cell functionality, specifically investigating a potential cell therapy treatment for critical limb ischemia. Their platform combined 6 ECM components, generating 63 single or multicomponent mixtures, which were printed onto glass slides in 378 distinct spots, including Matrigel599 as a positive control. Human PSC-derived endothelial cells were incubated onto the microarrays for overnight cell attachment in normoxic conditions at 37 °C, after which the cells were subjected to conditions frequently found at sites of tissue ischemia, namely hypoxia and reduced nutrients, replicated by media exchange to basal (no added growth factors) and then transfer to a hypoxia chamber set to 1% O2, 5% CO2, 94% N2.601 Cell viability (Calcein-AM), cell number (Hoechst 33342), CD31 expression (endothelial cell marker), and nitric oxide production (via incubation with a fluorescent NO probe) were assessed using high-throughput microscopy. The authors correlated the large data sets, and identified several protein combinations that performed better than the positive control (Matrigel), showing the importance of nonintuitive protein combinations in specific biological processes.601
5.4.1.2. Immunology
An important feature of a biomaterial to be used in cell therapies is its immunological response, which needs to be thoroughly investigated beforehand to avoid complications such as host-rejection, excessive inflammatory response, or fibrosis. The term “immune-instructive” material has been used to characterize biomaterials that can elicit a desired immunological response in the target host.602 In 2020, Rostam et al.603 presented the high-throughput combinatorial study on macrophage phenotype activation using a large library of chemistries (141 distinct monomers),427 followed by a second-generation array of “hit” monomer combinations, and scale-up of the best performing chemistries for in vitro and in vivo studies. Human blood-derived monocytes were cultured on the primary arrays, after which they were fixed and stained for calprotectin (M1 pro-inflammatory phenotype), CD206 (M2 anti-inflammatory phenotype) and DAPI for total cell number. The average M2/M1 ratio was calculated for each polymer to identify “hit” materials that could evoke an M2, M1, or M0 (representing either naïve or evenly distributed) phenotype. Data mining showed that cell attachment did not correlate with high polarizing scores, and thus the authors generated a secondary copolymer microarray of 400 features composed of the combination of the 10 highest macrophage polarizing monomers with the 10 monomers with the highest cell attachment. This second-generation array showed a high level of macrophage polarization and cell attachment. Four high-performing polymers were then selected and scaled up to 24-well plate format for the culture of monocytes for 6 days, followed by ELISA-based analysis of cytokine secretion. The scaled-up polymers showed high cell viability (>95%), with two polymers found to induce highly specified cytokine secretion profiles. Finally, the authors dip-coated 6 “hit” chemistries (inducing all three phenotypes) onto clinical-grade silicone rubber tube segments and implanted them subcutaneously into mice for a period of 28 days. Hematoxylin and eosin and Masson’s trichrome stains were used to assess the tissue inflammatory response, and the results obtained correlated with the in vitro data as compared to the uncoated silicone control, which showed the thickest collagen layer, a sign of fibrosis. The correlation of the microarray and the in vivo data suggested that unbiased and combinatorial in vitro screening of large polymer libraries can successfully translate to bioinstructive biomaterials for clinical applications.
Recent work by Vassey et al.602 investigated the effect of topography on macrophage polarization. Plasma etched polystyrene TopoChips458 were used to culture blood-derived monocytes from five different donors in serum-containing medium. Rank order analysis of the cell attachment (total nuclei, DAPI) and morphology (CellMask), was compared across the different donors showing consistency for the high and low attachment topographies. Overall, monocyte attachment was significantly higher in the presence of topography when compared to flat surfaces. Computational regression analysis was used to determine topography parameters that highly influenced monocyte adhesion. Descriptors that highlighted the size of the individual TopoUnit components, specifically the presence of micro pillars with a small surface pattern area, dominated the model and were able to capture the differences between high and low performing features (Figure 29A).602 The ability of topography to modulate macrophage attachment was challenged by using protein coatings (Fn or COL1), with results showing the same cell attachment changes as compared to flat controls independent of the preadsorbed protein composition.602 Interestingly, a differential monocyte–biomaterial interaction was observed, with the small micropillar topographies being fully engulfed by the monocytes, in contrast with selective attachment on larger topographies (Figure 29B–D).602 Polarization of naïve macrophages was also investigated, via immunostaining for calprotectin (M1-phenotype) and mannose receptor (M2-phenotype). M2:M1 ratio was calculated (per cell) and normalized to the flat TopoUnit on each chip. Overall, the proportion of the three potential phenotypes across the TopoChip indicated there was a range of phenotypic responses and no one predominant macrophage polarization state. Trained machine learning models were used to investigate the relationship between a range of topographical parameters and macrophage phenotype. Interestingly, the micropillar pattern area was found again to have a strong predictive quality on M2 polarization.602
Figure 29.
Macrophage cell attachment is mediated by small circular pillar topography. (A) Macrophage cell attachment represented as high (blue), medium (green), or low (orange) plotted against total pattern area (μm2). Categories of macrophage attachment were determined through cluster analysis using Euclidian distance. Composite confocal images showing cell membrane (green) and nucleus (blue), representative of (B) low attachment, or (C) high attachment TopoUnits with inset (D) with orthogonal views of Z-stack images showing cellular engulfment of the cylindrical pillar feature. Scale bar = 10 μm. Reprinted with permission from ref (602). Copyright 2020 Wiley-VCH.
Acharya et al.604 used a so-called “immunoarray” for the high-throughput investigation of the effect of adjuvants and toll-like receptor (TLR) ligands on dendritic cell response. Adjuvants are critical components of vaccines, as they potentiate the specific type and magnitude of the immune response to the coformulated antigens; the mechanism underlying adjuvant activity is key to the optimization and generation of vaccine formulations. In the immune response, the recognition of pathogen-associated molecular patterns by the immune system is typically achieved through TLRs. To address the development of polymeric, particle-based approaches in antigen/adjuvant combinations, a cell-based microarray screen was developed where the adjuvants and TLR-ligands monophosphoryl lipid A (MPLA), CpG, and polyinosinic:polycytidylic acid (poly I:C) were encapsulated in PLGA microparticles and contact-printed onto glass coverslips, generating 216 distinct dosing combinations in triplicate. Dendritic cell number and marker expression were assessed through immunostaining and fluorescent microscopy, showing the cells were able to attach, spread, and survive on the arrays for 24 h. The platform was then used to investigate the interplay between TLR signaling on cells and the magnitude of the antigen-specific immune response, with cell activation assessed via several pro-inflammatory markers. All adjuvant combinations showed significant marker expression variation as compared to negative (no-particles) conditions, confirming a measurable effect of the particles on the cells in the microarray format. A ratio consisting of 1:2:2 CpG:MPLA:poly I:C generated the highest pro-inflammatory cell phenotype on the microarray.
5.4.1.3. Soluble Factors and Paracrine Signaling
Another important feature of the cell niche is the paracrine cues, which are soluble signals secreted by the cells themselves onto the media, which then can bind to specific cell-receptors in neighboring cells and drive key processes such as proliferation, survival or differentiation.576 Microarray technologies for cell–biomaterial assays have typically used glass slides as printing support, due to their ready availability and easy handling during the microarray fabrication process. This glass slide would then be exposed to one cell solution, with the cell culture done in one continuous liquid form. While this approach reduces biological variability between biomaterial samples, it also implies that cell-secreted factors diffuse freely between the different biomaterial conditions, as well as any toxic molecules derived from any particular biomaterial over time of culture, or from biological processes such as apoptosis. Very little work has been done to investigate the effect of these paracrine signals on individual polymer data, with scale-up and subsequent isolated testing being the ultimate proof of biological performance. To partially mitigate these effects, several strategies have been presented. For example, randomized printing has been used to reduce the effect of localized events on polymer efficiency.605 Other groups, such as Lutolf and co-workers,450 have used microwell plates to create truly isolated biological systems, and including soluble cues as a microarray variable.450
Another approach by Levkin et al.528 has used chemical patterning on 2D microarrays to generate superhydrophobic borders in between hydrophilic polymer islands of defined geometry,528 called droplet-microarray.529 The superhydrophobic area repels protein adsorption, cell attachment, and even liquid deposition, with individual media drops spontaneously generating on top of each polymer island, physically separated by the hydrophobic strip but close enough to yield a high number of spots tested per slide.531 In 2020, Levkin and co-workers532 used the droplet-microarray529 for the HTS of the cell-transfection inducing capacity of 774 different FDA-approved drugs, using different concentrations and totaling 42 000 individual experiments. The amounts of drug required were 20 nL droplets and only 200 pmoles of drug, about 2500-times smaller than if the experiment had been performed in 384-well plates. A GFP-encoding plasmid was used as a reporter gene to monitor transfection efficiency. An FDA-approved compound library was diluted and arrayed into 96-well plates, followed by their contact printing onto the droplet microarrays. Complexes of GFP plasmid DNA and a transfection reagent were prepared and printed into each spot, and incubated with drugs at three concentrations along with a drug-free control (DMSO). Cells were added onto the arrays in one solution, which rapidly formed uniform microdroplets for each spot due to the superhydrophobic edges, and incubated onto the array for 24 h. The relative transfection enhancement was calculated as a ratio of the mean number of GFP positive cells for each drug over the mean number in GFP positive cells of drug-free controls. The 425 compounds showed transfection enhancement, with some compounds showing up to a 5-fold increase of cell transfection efficiency. This study highlighted the importance of cell-isolated experiments, as it presented a highly miniaturized soluble compound screening platform on a single 2D substrate.
5.4.1.4. Chemically Defined Conditions
As described in section 5.2.2, protein adsorption onto biomaterials has been defined as the key mediator of cell attachment.462,467 These proteins are typically part of complex media formulations used during the biological assay, and contain variable amounts of cell–ligand molecules (e.g., cell-binding proteins such as Vitronectin or Interalpha-inhibitor are found in serum, a common cell media culture additive606,607). While great efforts have been directed into predicting the protein deposition potential of biomaterials,471−473 highly complex media formulations have made the investigation of the exact features mediating cell attachment complicated and, ultimately, unsuccessful in creating a unified theory for rational polymer design. Only fully controlled environments can elucidate the direct impact biomaterial characteristics have on biological entities.63
In 2016, Reimer et al.564 used polystyrene TopoChips458 to investigate human pluripotent stem cell attachment (number of cells), proliferation (EdU staining), and maintenance of pluripotency (Oct4 expression) in vitro in completely defined media (E8, based on the study from Chen et al.589) without the addition of any cell-attachment proteins. Human PSCs are traditionally difficult to grow in vitro, with the typical use of complex protein solutions (such as Matrigel599) required for their undifferentiated expansion.608 The cell data showed that at 4 h, the 100 top and 100 bottom topographies did not differ significantly from the control flat topographies in the number of cells. At 24 h postseeding, the top 100 topographies showed approximately 3-fold more Oct4 and EdU-positive cells as compared to the control (flat polystyrene). Cell behavior revealed that the top 100 topographies supported cell-cluster formation with a higher cell number per cluster. These observations correlated with the literature, as human PSCs are dependent on cell–cell contact for survival and proliferation.609 After 4 days of culture, the top 100 topographies had supported the formation of large colonies of undifferentiated human PSCs, expressing the pluripotency markers Oct4 and Sox2. Computational tools were used to predict the topographical features most supportive of human PSC proliferation, and feature size (less than 60 μm2) and wavenumber (WN0.2), smaller than 0.065 au (i.e., small features more frequent) showed high prevalence among the top 100 topographies. Their results showed the possibility of topography-based substrate design for short-term studies of human PSCs in chemically defined and extracellular-matrix free conditions.
In 2020, Ireland et al.610 presented another matrix microarray screening strategy to study the effect of ECM composition and substrate stiffness on stem cell fate. They used O2-plasma treated PDMS to generate soft (1–2 kPa) or stiff (61.5 ± 2.37 kPa) substrates on glass slides, which were then covalently functionalized with single or combinations of fibronectin (FN), Vitronectin (VN), Laminin-521 (LN521), and Collagen IV (COLIV) to generate 32 unique matrix islands. The proteins were chosen on the basis of previous use in human pluripotent stem cell culture. A chemically defined and “lean” medium (E8589), which contains no ECM proteins, was used to create a highly defined setup. Human PSCs were incubated on the arrays for 4 days before fixing and staining for the pluripotent marker (Oct4) and nuclear detection (DAPI). Only LN521 could support hPSCs for 4 days as a single protein, and the combinations FN+VN+LN521 and FN+LN521 on stiff substrates were the ones that performed the best in PSC attachment and growth, with no loss of pluripotency (Oct4 expression) in any condition tested. FN and LN521 showed the best cell performance, and the stiffer PDMS substrate performed best in all protein conditions over the soft substrate. When investigating the combinatorial feature effect, the authors found that fibronectin+laminin-521 conditions have a strong synergistic effect (as compared to single protein conditions) in the soft array, but not in the stiff conditions. The authors suggested that the synergy of these proteins was masked in stiff conditions due to the high performance of the stiff hydrogel. Four combinations were scaled-up and used in hPSC culture for 15 passages to investigate long-term effects alongside VN- and Matrigel-coated positive controls. Not all conditions performed, such as FN+VN+LN521+COLIV on stiff PDMS, which produced a loss of pluripotency of hPSCs at passage 3. This highlighted the importance of scale-up studies to validate microarray data. The other three conditions supported the expansion of pluripotent hPSCs for over 10 passages, with increased proliferation and pluripotency marker expression when compared to traditional coatings Matrigel and Vitronectin.610
5.4.1.5. Cancer
Cancer diagnosis and treatment strategies continue to be one of the most important medical challenges to date. It has been described that changes in cellular glycosylation, aberrant expression of glycosyltransferases, glycosidases, and transporters render cancer cells with altered carbohydrate profiles on the cellular surface. These changes can result in unique antigenic glycans called tumor-associated carbohydrate antigens (TACAs). TACAs are useful clinical disease markers, with important potential for early disease diagnosis and for engineering targeted drug-delivery.611 One such antigen is sialyl Lewis a (sLeA), which is used in pancreatic cancer diagnosis as a cancer-stage indicator.521 High-throughput strategies also hold great potential for cancer diagnostic and drug-discovery purposes. For example, recent work by Amon et al.521 developed a generalizable approach using high throughput strategies for the characterization of structure and specificity of monoclonal antibodies against the tumor-associated carbohydrate antigen sialyl-Tn. Site-specific mutagenesis was employed to fabricate a library of possible antibodies and probed against the cancer-related antigen, determining quantitative glycan deposition on each spot. They further characterized the binding site using nuclear magnetic resonance, and used the experimental features collected to generate a 3D-model of the antibody-glycan complex, which was then further challenged through computational screening against a human sialyl-Tn-glycome library for specificity.521
5.4.2. Bacterial Cells
Controlling the colonization of materials by microorganisms has become important for a wide range of settings, especially in the clinical and pharmacological industries. In particular, medical devices (e.g., venous catheters) can become colonized by bacteria, which can grow to form what is called a biofilm.612 Biofilms are surface-associated microbial communities that support robust growth and improved survival of the individual bacteria, and can increase their antibiotic resistance up to a thousand-fold in comparison to their planktonic form.613 Since there is currently no universal theory as to how bacteria interact with material surfaces, high throughput technologies have been employed to investigate the mechanisms behind the ability of certain biomaterials to resist biofilm formation (reviewed in detail in ref (612)). The unified knowledge of bacterial-repellent and cell-adhesive properties in biomaterials is the key to engineering safe medical devices.
One of the first high throughput studies of bacterial attachment was reported by Pernagallo et al.522 in 2011, where they investigated attachment of the food-borne pathogens Salmonella enterica and Escherichia coli onto a microarray of 370 polyurethane and polyacrylate-based polymers.522 GFP-expressing plasmids were transformed onto the bacteria prior to the attachment study to streamline data collection, and rich LB broth was used as the medium for the overnight microarray-bacteria incubation. The bacterial binding to the polymers was determined on the array, and separated into: High binding polymers (with the high-density attachment of both strains), polymers for selective binding (high-density attachment of one strain but negligible binding for the other) and polymers repellent for both strains (nonbinding).522 Results showed that polymers containing methyl methacrylate (MMA), methacrylic acid (MA-H), and 2-(diethylamino)ethyl methacrylate (DEAEMA) were highly successful in preventing adhesion of both bacteria strains.522 The highest-binding and the lowest-binding polymer were further explored by coating on a coverslip, incubating with both strains overnight, and analyzed the next day for bacterial attachment and biofilm formation. The scale-up biomaterial performance accurately replicated the microarray results.522
Hook et al.427 focused on polymers for medical applications, specifically aiming at urinary catheter biomaterial design. 576 unique copolymers were tested in a two-generation microarray setup against three different bacterial species; representing Gram-positive (S. aureus) and Gram-negative (P. aeruginosa and uro-pathogenic E. coli (UPEC)) pathogens frequently found in medical device-associated infections; in conditions replicating urinary catheter infection conditions (using artificial urine).427 To assess the resistance of any given material to attachment by diverse bacterial species and exposure conditions, the authors developed a composite bacterial attachment parameter called iota; the fluorescent intensity per unit area from all three strains on each polymer and in each condition was normalized to the maximum value on the slide, and averaged over all three replicate to produce an estimate of bacterial attachment.427 A high iota value meant high bacterial attachment and therefore a poor biomaterial performance. Polymers that were resistant to bacterial attachment were identified, and correlation studies showed synergistic effects of some monomers (found in 11 of the 96 copolymers).427 To investigate the effectiveness of the polymer in vivo, control (commercial catheter) and “hit” polymer-coated silicone catheters were inserted subcutaneously into mice. After 24 h the catheter lumen was inoculated with bioluminescent Staphylococcus aureus. Immediately after inoculation, similar bioluminescence was found in both samples. However, 24 h later there was already a 10-fold reduction between control and polymer-coated catheters, and after 4 days bacterial numbers were shown to be nearly 2 orders of magnitude reduced in the polymer-coated catheter surrounding area, including kidneys and spleen.427
Follow-up work from Hook and co-workers441,614−616 has expanded on this study to cover all facets of high-throughput microarray technologies. Hook et al.441 in 2013 further increased the combinatorial space accessible with available off-the-shelf (meth)acrylate monomers, with 116 monomers combined to create 1273 unique polymers tested in 3 generations of microarray designs.441 Hook and co-workers475,614,617 also extensively mined the large microarray data sets generated to investigate the correlation between polymer chemical structure and biological response,617 and present computational models for biomaterial-performance prediction.475,614 Dundas et al.616 focused on optimizing the biomaterial hits to increase their coating potential and adapt them to the desired biophysical properties of the biomaterial for a biomedical coating, such as the optimal flexing fatigue required for catheters. A recent study by Hook and co-workers615 presented a clinical trial report to evaluate bacterial attachment and biofilm formation on a urinary tract medical catheter coated with a synthetic polymer identified by microarray high-throughput methodologies. This first in-man pilot study evaluated 10 polymer-coated catheters against 12 commercially available silicone catheters, allocated nonrandomly for bladder management after urethral reconstitution. Biomass and biomineralization were quantified, showing a reduction in biomass on polymer-coated catheters compared to uncoated conventional catheters, replicating in vitro data on bacterial attachment and biofilm formation.615
5.4.3. In Vivo Studies
Preclinical trials are a crucial step to investigate the foreign-body response and toxicity generated by newly developed biomaterials. High throughput preclinical testing can be useful to advance biomaterial development, greatly reducing the number of animals needed for testing in a cost-effective and ethically superior manner. Oliveira et al.605 developed this technique by implanting a microarray chip containing 36 different scaffolds on Wistar rats.605 The microarrays were used to study the immune response by analyzing lymphocyte recruitment at 24 h and 7 days after implantation. The microscaffolds showed different inflammatory responses, depending on their physical and chemical properties. Moreover, histology analysis of the tissue surrounding the microchip showed evidence of unique inflammatory responses for each independent material. This study, therefore, proved that microarray technology could be used in vivo with sufficient resolution to distinguish biological response differences between the different spots. However, large scaffold areas were used within the microarray, reducing the number of biomaterials that could be tested on one animal to 36 combinations.
Recently, Vegas et al.618 developed a combinatorial biomaterial library of alginate-based hydrogels that was tested in vivo by using a rapid subcutaneous mouse model to measure inflammation. From their library, 634 alginates were evaluated as bulk hydrogels using an in vivo assay where multiple implantations were performed per mouse on the dorsal surface in an eight-array format, 0.8 cm paramedian to the midline and 1 cm between adjacent sites. The assay monitored subcutaneous events through an imaging agent that yielded increased fluorescence in response to increased cathepsin activity, a marker of immune cell activation. Each mouse was implanted with a control unmodified alginate to serve as an internal control for fluorescence normalization during the 7-day imaging. About 200 hydrogels displayed lower fluorescent levels than unmodified alginate. As an end-point assay, Masson’s trichrome (MT) staining of tissue sections was performed, and three modified alginates (Z2-Y12, Z1-Y15, Z1-Y19) produced microcapsules with lower fibrotic growth over the implant. The microcapsules were then implanted in the intraperitoneal space of C57BL/6J mice to investigate if the same lower inflammation was achieved in other tissue areas. The same three lead materials showed almost no fibrous deposition, all three containing a triazole function, which was associated with the improved performance overall. The performance of the rest of the chemistries varied significantly, so it was hypothesized that the triazole modification may be a versatile chemical space for biomaterial design in applications that require reduced foreign body response. In line with other studies,410 material chemistry strongly correlated with reduced inflammation in this study. The materials were then separately implanted intraperitoneally into nonhuman primates (n = 3 each) using a minimally invasive laparoscopic procedure to investigate species-related effects and potential in human clinical applications.618 The three “hit” materials showed substantially reduced fibrotic response after 4 weeks compared to the controls. This study showed a novel approach to perform preclinical studies on large libraries of polymers. The most promising alginate derivate (Z1-Y15) was later used to generate an implant for the protection of glucose-responsive allogeneic cell islets implanted in a nonhuman primate model for 4 months. The hit materials maintained viable, glucose-responsive cells for the entire time of the implant and showed low foreign body response without the need for immunosuppression treatment.619
5.5. Future Perspectives
Cell culture in vitro inevitably involves the removal of the cells from their natural environment and culturing them in artificial conditions. Most used are 2D tissue culture plates or stripped-down hydrogels with disproportionate volumes of media, which necessarily dilutes secreted factors and alters cell behavior. Scientists have been arguing for integrative technologies that can control cellular environments at multiple length scales, and help to replicate the natural extracellular environment to generate cell populations more similar to their natural counterparts for some years.620 Most of the studies described in this chapter have used strategies with limited automation, and focused on controlling mainly one specific aspect of the microenvironment-cell interphase. However, recent work has leaned toward multiparameter microarray technologies and computer algorithms, to increase the output value of these high throughput technologies. We speculate three areas that will become of increasing importance in coming microarray work.
First, multiparameter microarrays will improve to include not only material chemistry or topography in the screens, but the combination of these to exercise greater control over cellular phenotype on solid biomaterials. Moving into the 3D biomaterial strategies to control architecture and the screening of these features with the full design freedom afforded by additive manufacturing 3D printing approaches. 3D control is already happening in hydrogel screens to a degree, with the help of biomolecular cues. However, the incorporation of the extensive current knowledge on the influence of chemical structure in cell behavior has yet to be explored in combination with topographical design, spatial distribution, and mechanical forces.
Second, the incorporation of increased automation in microarray technologies, ultimately with artificial intelligence control over literature surveys, library selection, hypothesis generation, and experimental control. It has been postulated that machine learning algorithms and robotics are now advanced enough to be efficiently incorporated in biomaterial discovery.621 Vasilevich and de Boer621 recently presented a theoretical robot called Toby and described the biomaterial discovery flow that would be used in this robotic approach. Toby would perform from the exhaustive initial literature review to the final biomaterial optimization, if a well-defined question or task was presented, and that all the required materials and equipment was available to it. Although it may seem far-fetched, the robot-assisted material discovery could become a reality, with highly improved and unbiased data generation, collection, interpretation, and optimization. Indeed, the use of robotics has already reached chemical discovery. Recently, the Cronin group has developed an organic synthesis robot that can perform chemical reactions and real-time analysis, as well as use machine learning algorithms for subsequent decision making.622,623 The robot Eve has already been used to discover drugs using only 10% of the compound collection in closed-loop automation.624 This platform is constantly testing new chemical combinations to generate real-time data that is immediately analyzed and used for more chemical reaction design and experimental testing. Moreover, the data generated by the robot can be accessed in real-time, and subsequently replicated by human scientists. This successful automation of chemical processes opens the door to robot-assisted biomaterial discovery, greatly increasing the available material space for cellular applications.143
Finally, knowledge will be accumulated on the biointerface by building on the library of understanding of cell–material interactions and the resultant theory will be used to generate universally translatable relationships across different material classes in controlling cell responses that will include chemistry, mechanics, topography, 3D architecture and perhaps most importantly biomolecular adsorbates. Ultimately, the success of biomaterials research lies in the number of translatable technologies that become adopted in the clinic. Therefore, it is important that we keep striving to accurately investigate biologically relevant models, and focus on their translation potential. In section 9, the clinical translation from the high throughput biomaterial discovery platforms will be reviewed in more detail.
6. Microfluidic Technologies for Biomaterials Discovery and Biointerface Understanding
Microfluidics, a technology that manipulates fluids at small scales (typically from 10–8 to 10–18 liters) in narrow (10–6 to 10–8 meters) channels,625 provides engineered and integrated platforms for chemical, biotechnological, and biological analysis applications.626 The development of fabrication techniques to produce polymeric devices in the late 1990s627 opened a new paradigm in micrototal analysis systems (μTAS) and lab-on-a-chip (LOC) technology, especially for biological applications including chemotaxis,628−630 cell biophysics,631,632 DNA sequencing,633−635 and cell culture assay.636−638 Besides benefits from a small volume, for example, low sample consumption, fast analysis, and low cost, microfluidics offers possibilities for high-throughput and combinatorial analysis by parallelization, automation, and integration with gradient generators,639,640,649,650,641−648 sensors,651,652 and actuators.597,653,662,663,654−661 Also, microfluidic devices coupled with various micro/nano-patterning658,661 and surface modification techniques414 enabled to mimic and study fundamental cellular processes, including cell adhesion, proliferation, and differentiation. Recent technical development in multilayer device fabrication to construct 3D models offered new tools that resemble complex microenvironments in nature for studying physiological processes.664−670 In this chapter, section 6.1 will review microfluidic HTS approaches that have been developed to improve efficiency and accuracy of data collection and analysis, and section 6.2 will outline microfluidic systems to recreate physicochemical stimuli and physiological/biomimetic microenvironments for screening cell–material interactions.
6.1. Microfluidics-Based Screening Platforms Improving the Throughput, Efficiency, and Accuracy of Data Collection and Analysis
HTS, an experimental tool for the principal evaluation of a target system with a large set of processes,671 provides an enormous amount of information to address the requested screening on material and parameter libraries.672,673 By automation of the operations, miniaturization of testing, and computational data processing, HTS enables fast analysis of a massive number of samples not only for basic research in biology and biochemistry but also in industrial, biopharmaceutical, and clinical applications.674,675 Indeed, miniaturization of an assay platform is an easy and practical way to increase the efficiency of HTS because it decreases the cost of reagents and materials as well as decreases sample read-out time. The most common HTS platform is a microtiter plate, which comprises a series of wells with microscale volumes. However, the surface to volume ratio increases according to the decreasing reactor (i.e., well) size, which affects not only the adsorption characteristics and stability of a target but also the sample handling processes. Therefore, a better understanding of the fluidic phenomena in tiny volumes is required to implement miniaturization successfully. Microfluidics has been able to support HTS by exploiting the fundamental study of the physical properties of fluids in a small volume and engineering of essential components to control the fluids. This section of the review will survey microfluidics-based HTS approaches by classifying them into four categories based on the integration concept of the technology used: (1) standard multiwell plate based-microfluidics, (2) microstructure-based cell culture array, (3) valve-assisted large-scale integration, and (4) droplet-based microfluidics (Figure 30). Within each category, we highlight recent microfluidic technologies that combined multiple functions on the same platform toward a fully integrated cell culture system.
Figure 30.
High-throughput microfluidic platforms. (A) Standard multiwell plate based-microfluidics. (a) Sliced tissue culture system for drug screening. Reprinted with permission from ref (679). Copyright 2014 Royal Society of Chemistry. (b) Multiwell plate-based 3D cell culture platform with 40 culture chambers. Reprinted with permission from ref (677). Copyright 2013 Royal Society of Chemistry. (c) A 384 hanging drop spheroid culture array plate. Reprinted with permission from ref (680). Copyright 2011 Royal Society of Chemistry. (B) Microstructure-based cell culture array. (a) Microfluidic cell culture unit for long-term cellular monitoring. Reprinted with permission from ref (636). Copyright 2005 Wiley-VCH. (b) Microfluidic device containing thousands of cell traps for cell capture and pairing. Scale bar, 20 μm. Reprinted with permission from ref (705). Copyright 2009 Nature Publishing Group. (C) Valve-assisted microfluidic platforms. (a) Integration of 96 parallel cell culture chambers on a chip. Reprinted and modified with permission from ref (638). Copyright 2007 American Chemical Society. (b) Microfluidic living cell array. Reprinted with permission from ref (728). Copyright 2007 Royal Society of Chemistry. (D) Droplet-based microfluidics. (a) Microfluidic channel array for droplet immobilization. Scale, 40 μm. Reprinted with permission from ref (734). Copyright 2009 Royal Society of Chemistry. (b) Microfluidic droplet printer for dispensing programmed combinations of cells and reagents on the substrate. Reprinted with permission from ref (737). Copyright 2017 National Academy of Sciences.
6.1.1. Microfluidics Integrated Multiwell Plate for HTS with High-Content Screening Equipment
Microfluidic platforms combined with standard multiwell plates are one of the promising approaches to improve the functionality and efficiency of HTS.676 Also, the systems can address practical issues in the commercialization of microfluidic-based HTS, including user-friendly operation, full compatibility to standard equipment, relevance to industry, and cost reduction.677 In microtiter plate-based microfluidics, a microfluidic channel network replaces tubing and connectors thereby significantly decreasing the consumption of reagents with minimal dead volume. The accurate and reliable control of fluids, for example, cell loading and continuous medium perfusion, is achieved by the applied pressure678,679 and gravity-driven flow.677,679 Lee et al. developed a microfluidic cell-based assay compatible with a commercial 96-well plate for cytotoxicity screening of HeLa cells.678 HeLa cells were loaded by a modularized cell-loading manifold and long-term cultured under gravity-driven perfusion. The process was semiautomated by standardized instruments such as an incubation chamber and a plate reader. Chang et al. further advanced the concept and design to culture organotypic tissue slices for screening chemotherapeutic drug activity (Figure 30A(a)).679 The system consisted of a microfluidic channel layer, hydrophilic polytetrafluoroethylene (PTFE) porous membrane, and multiwell plate. Fluids were delivered into open wells via microfluidic channels and solutes were transported to tissue slices placed on a PTFE membrane. After culture and exposure to drugs, the tissues were collected for further analysis with the membranes, and the platform was cleaned and reused with new membranes. Besides 2D culture, Sebastiaan et al. proposed an array of 3D cell culture chambers incorporated in a microtiter plate format (Figure 30A(b)).677 The microfluidic titer plate included 40 culture chambers, and three lanes of gels and liquids were patterned by phase guides in each chamber. They demonstrated the 3D culture of HepG2 hepatocytes under perfusion, coculture with fibroblasts, and invasion model using 4T1 breast cancer cells. Tung et al. developed another concept of high-throughput 3D cell culture array by adapting a hanging drop technique (Figure 30A(c)).680 The authors fabricated a hydrophilic plate with 384 access holes, which could create cells-containing hanging drops, and integrated with a 96-well plate based humidity chamber for the formation and culture of A431.H9 spheroids and combinatorial drug treatment.
6.1.2. Microfluidic Cell Culture Arrays
Microfluidic cell culture arrays have shown the potential to create more compact and comprehensive screening platforms with tiny sample volumes, few orders of magnitude smaller than conventional well-plates, by parallelizing microsized bioreactors.625 Components and processes required for cell culture and screening were constructed on a single microfluidic chip as the concept of lab-on-a-chip. Microstructures were designed to isolate cells from the cell suspension, and microchannels were connected to the reactors for the continuous perfusion of medium or delivery of other solutes. Because of the small size of the microfluidic devices, it was not practical to make a direct interface with conventional instruments; however, the progress in microfabrication techniques allowed to manufacture and integrate essential parts for cell culture, including heaters,681−685 gas controllers,686−693 and optical monitoring,694−697 with the devices. Overviews of internal/external components to build up complete microfluidic platforms for complex sequential chemical and biological processes were given by Nguyen et al.,698 Oh et al.,699 and Boyd-Moss et al.700 The first microfluidic cell culture array for long-term cell culture and monitoring was presented by Lee’s group (Figure 30B(a)).636 They combined 100 microfluidic culture units in which multiple narrow side channels for perfusion surrounded a culture chamber. In addition, a concentration gradient generator, which will be discussed later, allowed for long-term cell culture under 100 different conditions in parallel. Following this development, the concept of microfluidic cell culture array has been exploited to study drug-dose response,637,701,702 cell growth dynamics,703,704 and cell–cell interactions.705 Furthermore, microfluidic-based cell arrays have been employed for single-cell analysis due to the capability for the isolation of single-cells by microfabricated cell capture geometries.705−707 Over hundreds of microfluidic single-cell capture units were easily parallelized to increase the throughput capabilities in a reactor with relatively low fabrication cost compared to other techniques, such as a single cell micromanipulator708−710 or optical tweezer.711,712 An integrated channel network allowed further manipulation and analysis on isolated single cells such as cell culture,713,714 drug dose–response,649,715,716 and DNA sequencing.717−720 Skelley et al. developed a device to trap and pair thousands of cells by adapting microfluidic capture structures (Figure 30B(b)) and showed reprogramming in a hybrid between mouse ECSs and mouse embryonic fibroblasts (mEFs) through on-chip electrofusion.705 Interesting reviews on single-cell analysis on microfluidic platforms were published by Wyatt Shields Iv et al.721 and Hosic et al.722
6.1.3. Valve-Assisted Platforms
A microvalve is one of the essential microfluidic components for the accurate control of fluid flows, especially in a sophisticated design where multiple reagents need to be manipulated sequentially.700 The Quake group developed a monolithic valve, an elastic membrane that enables fluid flow control in a channel under applying pressure in an adjacent channel, by PDMS multilayer soft lithography.723 The “Quake valve” allowed high-order parallel integration of cell culture and stimuli control. Also, the material properties of PDMS, being transparent, biocompatible, and gas permeable,627 makes it applicable for various studies ranging from cytotoxicity analysis724 and single-cell imaging725,726 to stem cell differentiation638 and mammalian cell transfection.727Figure 30C(a) shows a fully automated microfluidic cell culture array for quantitative measurements of the influence of transient stimulation on cell motility presented by Gomez-Sjoberg et al.638 The integration of a peristaltic pump, chaotic mixers, and 96 culture chambers enabled the monitoring of human mesenchymal stem cell (hMSC) culture and osteogenic differentiation under unattended stimulation for a week. King et al. developed a microfluidic real-time gene expression imaging platform using another type of microvalve arrays to control row-seeding and column-stimulation (Figure 30C(b)).728 Their microfluidic platform consisted of a 16 × 16 array of cell visualization chambers and pneumatic valves to control cell seeding and chemical stimulation configurations. An excellent overview on large-scale microfluidic integration was published by Melin et al.729
6.1.4. Droplet-Based Microfluidics
Droplet-based microfluidics has great potential in the HTS of soluble contents, including biomarkers and DNA/RNA.730,731 Droplet reactors offer dimensional scaling benefits that enable to reduce sample volume resulting in a significant reduction in cost. Advances in technologies for droplet microfluidics such as monodispersed droplet formation, high-speed manipulation, and compartmentalization enabled an increase in assay sensitivity and throughput.732 By introducing the controlled encapsulation of cells within droplets, Edd et al. demonstrated the potential of droplet microfluidics in a cell-based screening assay.733 An example of cell culture in droplets was given by Schmitz et al.,734 who developed a microfluidic channel array to immobilize thousands of drops containing cells for monitoring growth rates of single cells (Figure 30D(a)). Beside on-chip incubation, the concept of modulized microfluidics which stored droplets in commercially available flexible tubing was proposed to provide easy accessible quantitative statistical cell assay by Trivedi et al.,735 and Baraban et al.736 The droplet-based microfluidic platform was further advanced by Brouzes et al., who adapted an optically coded droplet library for high-throughput cytotoxicity screening. Recently, Cole et al. developed a fully integrated droplet printing system by coupling single-cell encapsulation, fiber-optic detection, electric sorting, and printing an oil moat substrate (Figure 30D(b)).737 They presented on-demand printing of cells and reagents in deterministic combinations to construct a large number of single-cellular and multicellular libraries. Programmed combinations of cells and reagents were printed on the substrate by additive droplet manipulation with a laser-guided printer head. The technical developments in droplet microfluidics were reviewed by The et al.738 and Solvas et al.,732 and overviews of recent applications of droplet-based microfluidics were given by Guo et al.730 and Mashaghi et al.731
6.2. Microfluidics Recreating Physicochemical Stimuli and Physiological/Biomimetic Microenvironments for Screening Cell–Material Interactions
Besides the highlighted advantages for HTS based on its small scale, microfluidics offers beneficial opportunities to design sophisticated microenvironments for multifunctional screening by applying various physicochemical cues in a spatiotemporally controlled manner. Deep insights into the fundamental physics of microfluidics and advances in microfabrication techniques enabled a robust chemical gradient generation and dynamic physical stimulation, alone, or even in combination, to study cellular responses to direct or indirect stimuli. Moreover, three-dimensional compartmentalization allowed creating microenvironments that resemble the natural systems for studying physiological processes. This section will contemplate the potential of microfluidic systems to generate physical and chemical stimulations and recreate biomimetic microenvironments for cell screening applications within four categories: (1) chemical concentration gradients formed by microfluidics, (2) mechanical stimulation generated by microfluidics, (3) microfluidic 3D cell culture models, and (4) organ-like microfluidic models.
6.2.1. Chemical Concentration Gradients Formed by Microfluidics
One of the unique and most valuable capabilities of microfluidic technology for screening processes is handling and producing a stable concentration gradient of soluble species. In most HTS applications such as cell culture-based screening assays, significant numbers of molecules at varying concentrations need to be tested to identify the dynamic responses of target cells. In conventional microfluidics, when two fluid streams flow together in a channel, fluid flows are laminar and mix by diffusion only, without turbulent mixing or thermal convection.739,740 Hence, additional techniques, such as chaotic mixers,741 rotary pumps,648 or acoustic mixers,742 are necessary to enhance the mixing efficiency in microchannels. Nevertheless, solely diffusive mixing in microfluidics enables the formation of well-defined concentration gradients along with the interfaces between multiple flow streams.
Figure 31A(a) shows a microfluidic chemical measurement device designed by Kamholz et al.743 It combines two fluid streams at a T-junction to flow alongside each other in a microchannel. Since the two input fluids flow in a laminar fashion, the width and length of the interdiffusive region depended on the diffusion coefficients of two input molecules in a steady state. Later, Ismagilov et al. visualized the solely diffusive mixing behavior in the microfluidic channel in 3D using confocal microscopy.744 Lucchetta et al. used the design to control the temperature of two halves of an embryo for the spatial and temporal regulation of embryonic development by introducing two fluids, one heated and the other cooled down, into channel.745 Somaweera et al. introduced 256 cell culture chambers on the sides of the main channel in a microfluidic Y-mixer to apply a concentration gradient in each cell culture chamber (Figure 31A(b)).746 Two laminar streams flowed along the main channel and mixed by diffusion, exposing cell culture chambers, located orthogonal to the flow axis, to different concentrations of reagents. The gradient range was determined by the initial concentrations and flow rates of the two input solutions.
Figure 31.
Concentration gradient generation in microfluidic devices. (A) Chemical concentration gradient generation on microfluidic platforms. (a) Microfluidic T-channel with two input fluids in a steady state. Reprinted with permission from ref (743). Copyright 1999 American Chemical Society. (b) Gradient culture device. Reprinted with permission from ref (746). Copyright 2015 Royal Society of Chemistry. (c) Microfluidic channel network to create the gradients of two dyes in solution. Reprinted with permission from ref (640). Copyright 2001 American Chemical Society. (d) Microfluidic high-throughput cell-based assay. Reprinted with permission from ref (636). Copyright 2005 Wiley-VCH. (e) Multilayer microfluidic device to create 3 replicates of 10 independent concentration gradients. Reprinted with permission from ref (650). Copyright 2013 Royal Society of Chemistry. (B) Oxygen concentration gradient control in microfluidic devices. (a) Microfluidic device to formulate oxygen gradients by diffusing oxygen through PDMS side walls. Reprinted with permission from ref (689). Copyright 2011 Royal Society of Chemistry. (b) Microfluidic oxygen tension generator integrated with a sandwiched gas permeable membrane. Reprinted with permission from ref (691). Copyright 2013 Royal Society of Chemistry. (C) Multiple gradients formation. (a) Microfluidic cell culture platform capable of generating chemical and oxygen gradients. Reprinted with permission from ref (749). Copyright 2014 Royal Society of Chemistry. (b) High-throughput photodynamic therapy (PDT) screening chip. Scale bar 5 mm (right). Reprinted with permission from ref (751). Copyright 2014 Royal Society of Chemistry.
Jeon et al. expanded the concept of T-mixer to generate gradients of soluble species in solution by interconnecting multiple T-junctions within a network of microchannels (Figure 31A(c)).639,640 The microfluidic gradient generation network was composed of multiple T-junction branches where fluid streams were combined, mixed, and split. Solutions containing different soluble species were loaded from ports at the top of the device and flowed through the network to yield distinct mixtures in each branch channel. The branches were recombined to establish a concentration gradient in a single cell culture channel to study various processes such as chemotaxis642,643 and haptotaxis.644,645 The range and profile of a concentration gradient were determined by the initial concentration of solutions and the flow rates of fluid streams, while the resolution of gradients could be adjusted by increasing the number of branches.640
Hung et al. integrated the concentration gradient generator with an array of cell culture chambers for high-throughput cell-based assays (Figure 31A(d)).636 They split the gradient generator channel into 10 branches. The branches were connected to columns in the 10 × 10 cell culture array for 100 different cell-based screens under 10 various conditions. The microfluidic HTS device enabled the real-time optical analysis of long-term perfusion cell culture, including repetitive cycles of cell proliferation and passaging. In recent studies, gradient generators were applied in microfluidic 3D cell spheroid culture platforms. This enabled high-throughput drug screens on 3D tumor models.646,647
Microfluidic valves based on an elastomeric membrane deflection have shown to be useful for controlling fluid flows in a microchannel network with fast switching of flow directions.638,723,729 Also, the valve system enabled to design a microfluidic peristaltic mixer for enhanced mixing, and highly integrated parallel mixers allowed to create a stepwise concentration gradient.648,649 Kim et al. described the integration of flow-switching valves with a gradient-generating network to form 64 individual pairwise concentration gradients in an array of 8 × 8 cell culture chambers.641Figure 31A(e) shows a microfluidic cell culture device for combinatorial cell screening developed by Frank et al.650 Diffusion-based gradients of platelet-derived growth factor, tumor necrosis factor alpha, and lipopolysaccharide were controlled by programmable flow-switching with pneumatic microfluidic valves to generate 30 parallel gradients with 10 different formulations, and 3 replicates for mammalian fibroblast and macrophage cell screening. While the reagents and cells were loaded and flowed through the culture chambers, pneumatic valves allowed the control of fluid flows for the formation of independent chemical conditions in the gradient chambers.
Oxygen plays an essential role in cell behavior and influences a variety of vital biological functions such as metabolism, homeostasis, and differentiation.747,748 Hence, oxygen concentration control in in vitro cell culture is important to reconstruct physiologically realistic microenvironments. Various microfluidic devices have been developed to enable real-time monitoring of cell response to the dynamic regulation of gaseous conditions.687−693 To control oxygen concentration, gases were delivered from pressurized gas cylinders into gas channels and diffused to cell culture channels through a gas-permeable membrane.687,688,690 Furthermore, chemical reactions such as between sodium hydroxide and pyrogallol (for oxygen scavenging) and between hydrogen peroxide and sodium hypochlorite (for oxygen generation), were adapted to control oxygen concentration without bubble generation and medium drying caused by direct injection of gases.689,691,692 The oxygen gradient devices were fabricated in a single layer design with a centered cell culture channel with neighboring gas channels (Figure 31B(a)), or in multilayer architectures by sandwiching a membrane between cell culture and gas supply layers (Figure 31B(b)). Such microfluidic platforms enabled high-throughput analysis of cellular responses to various oxygen concentrations to investigate how cells adapt to heterogeneous oxygen conditions found in physiological processes.
Further advancements of microfluidic systems for combinatorial and high-throughput cell-based screening were achieved by integrating gradients of multiple microenvironmental factors in a single device. For example, Chang et al. developed a platform for anticancer-drug screening under various oxygen conditions (Figure 31C(a)).749 They sandwiched a PDMS membrane between a top layer for oxygen gradients formation and a bottom layer for chemical gradients generation and embedded a polycarbonate film in the top layer as a gas diffusion barrier. Using the platform, they investigated the effect of hypoxia-activated cytotoxicity of tirapazamine on human lung adenocarcinoma epithelial cell (A549) apoptosis. Liu et al. introduced a liquid–air dual-gradient device composed of an array of the agarose liquid–air interfaces for the biological evaluation of air pollutant effects.750 They quantified the synergistic effects of cigarette smoke dose and the inflammatory level of A549 lung cancer cells on cell viability. Lou et al. developed a high-throughput photodynamic therapy (PDT) screening platform by integrating individual controls of photosensitizer concentration, oxygen level, and light influence for evaluating the anticancer efficacy of PDT drugs (Figure 31C(b)).751 Using the device, they demonstrated PDT efficiency assays on C6 glioma cells for over 324 various conditions within an hour.
6.2.2. Mechanical Stimulation Generated by Microfluidics
Microfluidic HTS platforms have become an attractive tool for the study of mechanotransduction by providing the precise control of mechanical stimuli and the real-time monitoring of cell response in parallelized cell cultures.752 Mechanotransduction is the process by which cells sense mechanical stimuli either exerted by the mechanical properties of a material or via motion or pressure, and convert them into intracellular biochemical signals inducing responses in cell behavior. The mechanical inputs are not originating from material properties, for example, shear stress, compression, and stretching, start the process, which results in the biological output, including protein secretion, cell migration, and cell differentiation.753,754
In the past decade, various microfluidic devices were designed to study the effect of continuous shear stress755−759 and shear stress gradients760 on adherent cell culture. Some of these devices included parallel culture channels to generate multiple shear stress levels for HTS.755−757,759 For example, Kim et al. designed a channel network to form logarithmic shear stress for monitoring qualitative perfusion-dependent proliferation (Figure 32A(a)).756 They designed various channel outlets to change the flow rates in the chambers, and ABJ1 mouse ESCs were cultured under continuous perfusion with logarithmically scaled flow rates. Moreover, the integration of ECM molecule-patterned PDMS microposts with a microfluidic shear device was engineered by Lam et al. (Figure 32A(b)).414 The elastomeric micropost array functionalized with fibronectin enabled to measure subcellular contractile forces in the morphological realignment process of human umbilical vein endothelial cells (HUVECs) under a sustained directional fluid shear.
Figure 32.
Mechanochemical stimulation generated by microfluidic components. (A) Shear stress. (a) Cell culture array with a channel network to apply a logarithmic range of shear stress. Reprinted with permission from ref (756). Copyright 2006 Royal Society of Chemistry. (b) Microfluidic shear device with the channel embedded with ECM molecule-coated PDMS posts. Scale bars, 6 μm (right) and 20 μm (bottom left). Reprinted with permission from ref (414). Copyright 2012 Royal Society of Chemistry. (B) Compression. (a) Compression device to evaluate differentiation of hMSCs and hASCs toward osteogenesis under cyclic pneumatic force. Scale bar 100 μm. Reprinted with permission from ref (654). Copyright 2012 PLOS. (b) Microfluidic axon injury microcompression device. Scale bar 25 μm. Reprinted with permission from ref (762). Copyright 2011 Royal Society of Chemistry. (C) Stretching. (a) High-throughput cell stretcher chip combined with 9 × 12 arrays of mechanically active culture units. Reprinted and modified with permission from ref (597). Copyright 2010 Royal Society of Chemistry. (b) Microfluidic stretcher consisting of eight stretching units with nanosized posts patterned actuation cavity layer. Reprinted with permission from ref (661). Copyright 2015 Nature Publishing Group.
Recent developments in multilayer device fabrication have allowed high-throughput and effective investigation on the effect of planar loads on cell migration and differentiation.653−655 The microfluidic designs included an elastomeric membrane to apply compression forces on a layer of cells by pneumatically actuating the membrane. Dynamic and cyclic membrane deflection was obtained by coupling fast-switching valves. Sim et al. developed a microfluidic platform that can generate various amplitudes of compressive pressures in cell culture chambers to induce the proliferation and osteogenic differentiation of hMSCs.653 Park et al. advanced the microfluidic model of mechanical stimuli-induced osteogenic differentiation by expanding the number of cell culture units (Figure 32B(a)).654 They designed an array of cell chambers to culture adipose tissue-derived stem cells (ASCs) and hMSCs on the same device concurrently for the comparison of their osteogenic differentiation under dynamic compressive stimulation. Kim et al. presented a microfluidic biomechanical chip to monitor the viability of mammary gland epithelial (MCF7) cells under controlled compressive loads. They showed a potential use of the device for the mechanical lysis of cells by evaluating the gradual rupture of cells according to elevated compression pressures.761 Hosmane et al. used the membrane-based microfluidic compression architecture to modulate various pressure levels on single central nervous system (CNS) axons for the quantitative characterization of axon injury, degeneration, and regrowth (Figure 32B(b)).762 In this system, an elastomeric injury pad was deflected by applying pressure into a top microchannel and the contact pressure of the deflected injury pad according to input pressure and membrane thickness was calibrated by finite element modeling.
Recently, Ho et al. exerted cyclic compression at the single cell level by patterning fibronectin islands to trap single cells in compression chambers.655 Furthermore, the systems incorporating an elastomeric membrane have facilitated stretching of cells cultured on the deformable substrate with a linear strain profile.597,656−663 Membrane deflection was operated by pneumatic,597,657−662 piezoelectric,656 and electromagnetic663 actuations to regulate cell adhesion and function such as the change in morphology and orientation,656−660 nuclear protein accumulation,597,661 and stem cell differentiation.763 Kamotani et al. showed the alignment and elongation of human dermal microvascular endothelial cells (HDMECs) and mouse myogenic C2C12 cells in response to cyclic stretch, while A549 human lung adenocarcinoma epithelial cells did not respond to the stretch with a Braille display-actuated microfluidic device.656 An excellent example of a high-throughput microfluidic cell stretcher was presented by Moraes et al. (Figure 32C(a)).597 They combined one hundred eight stretching units on a device for applying cyclic equibiaxial substrate strains with various magnitudes to small populations of adherent cells and demonstrated the mechanical strain-induced activation of the canonical Wnt/β-catenin signaling pathway in cardiac valve mesenchymal progenitor cells. They cultured primary porcine aortic valvular interstitial cells in a static condition and under 3-h stimulation with cyclic substrate strains of 3%, 8%, and 15% and showed the influence of mechanical stimulation on the β-catenin accumulation in the cell nuclei.
Furthermore, an advanced soft lithography technique allowed to pattern the arrays of micro-658 or nanopillars661 on stretchable elastomeric substrates. Mann et al. microfabricated a micropost array membrane device to measure the live-cell subcellular contractile response of VSMCs to static equibiaxial cell stretch,658 and Cui et al. used a nanopillar patterned stretching membrane to separate mechanical force effects from substrate rigidity effects in stretching-induced cell spreading and proliferation (Figure 32C(b)).661 Also, integrated microfluidic devices were proposed to investigate the combined effects of flow-induced shear and solid mechanical stresses on cell death and detachment in a microfluidic alveolar model657 and in vivo-like endothelial cell alignment.662
6.2.3. Microfluidic 3D Cell Culture Models
In the past decade, 3D cell culture techniques have been highlighted in cell biology, tissue engineering, and regenerative medicine research, because cell function and tissue evolution, including cell–cell interaction, proliferation, differentiation, and ECM formation vastly differ between conventional in vitro two-dimensional (2D) culture and the in vivo cellular microenvironment.764−766 Recent advances in microfluidic technology, together with developments in the field of natural and synthetic 3D biomaterial matrices allow for the generation of physiologically more representative in vitro 3D microenvironments to study fundamental biological processes and cell/tissue responses to different cues/treatments/materials. Biocompatible, ECM-mimicking hydrogels are most commonly used as matrices in the microfluidic 3D culture systems. They allow easy patterning with high resolution, and they are amenable for cell encapsulation as well as an effective transport of soluble cues throughout the matrix.767 Various hydrogels, for example, collagen,664,665,670,768−770 gelatin,768,771,772 fibrin,668 Matrigel,669,670,773,774 agarose,767,775 HA,670 PEG,667,776 and poly(ethylene glycol) diacrylate (PEGDA),777,778 were processed in microfluidic platforms to design 3D microenvironments recapitulating natural cell–cell, and cell–matrix interactions as well as to study responses to soluble and insoluble, surface-bound factors. Largely, these approaches are complementary with the ECM–hydrogel systems as discussed in the HTS approach using array systems. However, the microfluidic approach has the potential of introducing other parameters as well be it in a less-high throughput fashion with respect to material-cell interaction. The 3D boundaries between the cell-laden hydrogel and soluble species were constructed by compartmentalization,664−670 patterning using sacrificial elements,768,771,773 and molding.769,770,772,777
One of the most popular designs for the compartmentalization of microchannels to create a 3D hydrogel-based cellular microenvironment employs arrays of microstructures.664−668 For example, Toh et al. developed a microfluidic device containing an array of micropillars to trap hepatocytes and form a localized 3D matrix for in vitro dose-dependent 3D drug screening to predict in vivo hepatotoxicity664 (Figure 33A(a)). They separated a channel into three compartments by an array of micropillars for trapping and culturing cells inside the pillar array under continuous perfusion through the two side compartments. 3D cocultures in compartmentalized microchannel networks generated using periodic microstructures were also used for the formation of neuronal networks by Dinh et al.667 and for the recreation of the perivascular niche by Carrion et al.668 Recently, techniques to compartmentalize microchannel networks without microstructures were also developed to increase the contact surface area between ECM and cells.669,670,677,779−781 Bang et al.669 described a microfluidic approach to align neural networks to construct a neural circuit in 3D, by using Matrigel aligned by a continuous fluid as a guide (Figure 33A(b)). They aligned Matrigel in the middle of the 3D cell culture system and demonstrated the network formation between presynaptic and postsynaptic neuron groups following the direction of Matrigel alignment. Vulto et al.781 developed a phaseGuide technology that can control the filling and emptying of microstructures using the meniscus pinning effect. Trietsch et al.677 and Moreno et al.779 integrated the phaseGuide within cell culture channels to pattern 3D ECM-like gels with a large cell–matrix interaction area. Han et al.670 used butyl methacrylate-ethylene dimethacrylate (BMA-EDMA) micropatterns as phaseGuide to introduce 3D ECM-like gels into a microchannel and evaluated their technique with various hydrogels including COL1, Matrigel, HA, and cell-laden Matrigel (Figure 33A(c)).
Figure 33.
Cell culture in 3D matrices in microfluidic chips. (A) Compartmentalization. (a) Microfluidic chip for hepatocyte 3D culture and multiple drug testing. Reprinted with permission from ref (664). Copyright 2009 Royal Society of Chemistry. (b) Microfluidic device for constructing a neural circuit in 3D. Scale bar 200 μm. Reprinted with permission from ref (669). Copyright 2016 Wiley-VCH. (c) Microfluidic 3D blood capillary model using BMA-EDMA-supported hydrogel patterning. Scale bar 50 μm. Reprinted with permission from ref (670). Copyright 2018 Wiley-VCH. (B) Patterning using sacrificial elements. Fabrication process to create microfluidic channels within collagen gels (top). Photos show a device design, a fabricated device, and DAPI stained-HUVECs culture in the channel (bottom). Reprinted with permission from ref (768). Copyright 2013 Royal Society of Chemistry. (C) Molding. (a) Fabrication of PDMS/PEG-hydrogel microfluidic networks. Reprinted with permission from ref (777). Copyright 2010 Elsevier, Ltd. (b) Schematic fabrication process of hydrogel microfluidic device by a twice-cross-linking technique. Reprinted with permission from ref (772). Copyright 2018 Wiley-VCH.
The other attractive technique to construct hydrogel-based 3D microenvironments is patterning using sacrificial elements such as gelatin structures768,771 and chitosan membranes.773 In combination with soft lithography, the process enabled patterning of various microfluidic channel designs inside hydrogels (Figure 33B). Using this method, Baker et al.768 demonstrated the invasion of human umbilical vascular endothelial cells into the 3D matrix in the presence of a chemokine and growth factor gradient. Furthermore, advances in photo-cross-linking techniques allowed the direct casting of hydrogel on structured molds as well as bonding of two patterned hydrogel parts to develop hydrogel microfluidic devices.772,777 Cuchiara et al. presented a multilayer replica molding technique to fabricate microfluidic PEGDA hydrogel networks embedded within PDMS housing777 (Figure 33C(a)). Nie et al.772 described a method to produce hydrogel-based microfluidic chips using a twice-cross-linking strategy (Figure 33C(b)). To evaluate the technique, they investigated the bonding strength and surface morphology of various combinations of hydrogels, for example, gelatin, gelatin methacrylate, and alginate and demonstrated the formation of endothelialized channels with the vascular function.
6.2.4. Organ-like Microfluidic Models
By exploiting 3D cell culture systems, in the past few years, microfluidics-based technologies have developed organ-like 3D models, called organ-on-a-chip, to recapitulate particular biological architectures and functions including the interface between different tissues,782−784 regulation of presence/supply of biochemical species,785−788 and physiological or pathological conditions.789−792 The microengineered human in vitro models allowed the characterization of the fundamental mechanisms of disease etiology,790,793−796 cell differentiation,797−799 and organogenesis798 in tissue- or organ-level contexts, benefiting drug safety and efficacy screening and target identification and promising to replace animal testing.782−784,793 The key aspects for the construction of such biomimetic platforms, supported by recent innovations in microfluidic technologies, include coculture systems separated by ECM800−802 or a porous membrane,782−784,791,792,797−799,803,804 well-defined dynamic mechanical stimulations,782,783,798,799 and microcirculation networks for accurate mass and nutrient transport.786,805,806
Advances in 3D gel-based microenvironments compartmentalized by microstructure arrays, as discussed in the previous section, were expanded to construct interdependent organ models such as blood-brain barrier,800,801 and neuron-on-a-chip.802 Bang et al. presented a microfluidic network to construct a blood-brain barrier by coculturing HUVECs with cortical neural cells under independent perfusion of different types of media (Figure 34A).801 The vascular network had a permeability comparable to in vivo values and a high degree of neurovascular interfacing between astrocytic endfeet and the adjacent capillaries with the presence of synapses. Jang et al. identified the electrophysiological properties of neurons under repeatable activation in a microfluidic chip integrated with microelectrode array measurements.802 The neurons were arranged in a microgroove region within a cell culture unit that was activated by a neurophysiology setup capable of optical stimulation and electrical recording.
Figure 34.
Microfluidic organ-on-a-chip models. (A) Microfluidics-based 3D neurovascular unit platform. (a) Design of the platform to create a blood–brain barrier. (b) Fabrication steps to form the microfluidic blood–brain barrier. Reprinted with permission from ref (801). Copyright 2017 Nature Publishing Group. (B) Gut-on-a-chip microfluidic platform. (a) Device design with a porous stretchable membrane. (b) Comparison of cell shape and polarity in a static Transwell model and the recreated gut microenvironment. Scale bars, 20 μm. Reprinted with permission from ref (798). Copyright 2012 Royal Society of Chemistry. (C) 3D printed oviduct-on-a-chip platform. (a) Schematics of the device. (b) 3D cultured bovine oviduct epithelial cells (BOECs) in the device. Scale bars, 25 μm. (c) Ciliated cells at the air–liquid interface during culture. (d) On-chip monospermic oocyte penetrations. Scale bar, 50 μm. Reprinted with permission from ref (797). Copyright 2017 Royal Society of Chemistry. (D) Multiorgan-on-a-plate system. (a) Multiorgan models and microfluidic circulatory networks. (b) Assembly of the device. (c) Evaluation of three anticancer drugs in the four-organ system composed of the intestine, liver, cancer, and connective tissue models. Reprinted with permission from ref (787). Copyright 2017 Royal Society of Chemistry.
Moreover, porous membrane-based coculture systems have been leveraged to incorporate endothelium–epithelium782−784 or air–liquid797 interfaces in sophisticated organ-on-a-chip models. To mimic human biological barriers in more in vivo-like architectures, epithelial cells from various organs, for example, brain,803 oviduct,797,804 lung,782−784,807 intestine,798,799 and kidney,791,792 were cultured and polarized in a microchannel compartmentalized by a perforated membrane while endothelial interconnection, chemical gradients, and nutrients were provided from the other side of the membrane. A PDMS membrane cast on micropillar arrays is one of the most commonly used membranes in the organ-on-a-chip platforms due to easy fabrication and deformability for cyclic stretching;782,783,798,799,803,807 however, other types of membranes, for example, polycarbonate,797,804 polyester,791,792 and poly(lactic-co-glycolic acid) nanofiber784 membranes were also adapted to provide smaller pore size and scaffold for 3D cell culture.
Kilic et al. developed a brain-on-a-chip platform by the fabrication of a mixed neuronal-glial cell population layer interacting with a human brain microvascular endothelial cells monolayer through a porous membrane.803 The microfabricated model allowed the analysis of human neuronal differentiation and cellular interactions between migrating human neural progenitor cells and the adjacent brain tissues. Huh et al. reproduced a functional interface between the layers of human alveolar epithelial and microvascular endothelial cells under air and fluid flow, as well as controlled cyclic mechanical strain resembling human breathing motions.782,783 The microsystem was explored to investigate nanomaterial toxicology under mechanical strain-induced oxidation782 and drug toxicity-induced pulmonary edema.783 The integrated microarchitecture with a porous flexible membrane was adapted to develop a microfluidic human intestine model by Kim et al. (Figure 34B).798 They recreated a gut microenvironment with intestinal villi structures in the microchannel by applying shear stress and exerting cyclic strain on a human intestinal epithelial cell monolayer. Recently Ferraz et al. presented a 3D-printed oviduct culture device integrated with a polycarbonate membrane for improving bovine embryo production (Figure 34C).797 Bovine oviduct epithelial cell culture at the air–liquid interface enhanced epithelial polarization and differentiation, and in vitro fertilization (IVF) in the reconstituted oviduct allowed monospermic oocyte penetrations without the parthenogenic activation of oocytes. Although this first oviduct-on-a-chip was a big improvement in the field, the resins used for device 3D-printing were found to release components toxic to developing embryos but not the epithelial cells.808 Recently, an improved nontoxic oviduct-on-a-chip fabricated out of PDMS has been created.804 The device was successfully applied to perform an entire IVF process with embryos cultured up to the blastocyst stage. Moreover, the zygotes resulting from the oviduct-on-a-chip culture were more similar to their in vivo counterparts than to conventional in vitro zygotes, in terms of their global DNA methylation level and transcriptome.
Engineering of functional microvascular systems has been highlighted in organ-on-a-chip technologies not only for creating in vivo-like microenvironments with biological barriers between epithelial and endothelial cells782,783,803 but also for probing the (blood) transport function of the cardiovascular system809,810 and cardiovascular diseases such as atherosclerosis793 and thrombosis.790,794−796 Zheng et al. described a microfluidic early stage atherosclerosis model recapitulating atherosclerosis-prone hemodynamic conditions.793 The atherogenic responses of endothelial cells were recaptured in the biomimetic system obtaining the antiatherosclerosis efficiency of platinum-nanoparticles comparable to the values seen with testing in animals. Zhang et al. developed a 3D bioprinted thrombosis-on-a-chip model for thrombus formation by infusing whole blood into a channel network coated with HUVECs.790 Recently, microfluidic vascular networks were proposed for integrating organ-on-a-chip modules into a single circulatory system.786,805,806
Furthermore, the concept of multiorgan-on-a-chip has been established by interconnecting various organ modules in a physiologically relevant scale to reproduce organ–organ interaction in the human body for mimicking the whole-body response to drugs.788,811,812 Kamei et al. combined two individual microfluidic cell cultures with an artificial blood circulatory loop to reproduce the exchange of drug metabolites between liver cancer cells and human heart cells for modeling the cardiotoxic side effect of an anticancer drug.788 The multiorgan drug screening system was expanded in a plate-formatted microfluidic device by Satoh et al.787 who demonstrated the evaluation of three anticancer drugs in four-organ systems (Figure 34D).
6.3. Opportunities and Challenges
From the overview provided in section 6, it is evident that microfluidic technologies offer promising tools for increasing throughput, efficiency, and accuracy of screening of interactions between biomaterials and cells, for the recreation of physiological/biomimetic microenvironments including physical, chemical and mechanical stimuli, and complementary to some of the other discussed techniques, for the formation of complex 3D cellular models. Especially these 3D models will be valuable for getting deeper insights into property-function relationships of biomaterials in physiological-like microenvironments.
In this context, an opportunity lies in combining microfluidics with other HTS (material) platforms, such as gradient- or array-based ones. This can be achieved by integrating microchannels into the platforms,677−679 which enables accurate fluid flow control for supply and removal of solutions to HTS platforms, including seeding of cells, medium perfusion and generation of gradients of soluble compounds. The feasibility and success of integration are dependent on advancements in microengineering and microfabrication technologies. Methods like plasma-activated bonding,813 anodic bonding,814 thin glue layer deposition by microstamping,815 and microchannel fabrication in a double-sided tape816 are some of the recent developments that enabled the robust assembly of microfluidic compartments onto various substrates with 2D and 3D gradients.
While such integrated platforms hold the promise of delivering synergistic models where the complexity of a physiological-like microenvironment is combined with HTS, they also present challenges regarding fast and reliable data collection. For this, highly parallelized microfluidic chips can be combined with various optical, mechanical, and electrical sensing techniques. Data acquisition can be automated by synchronizing, for example, imaging-based assays with the device location. An alternative to sensor development and integration is the control over temperature and gas environment on the platform, which enables (real-time) assessment of cell behavior even using a general microscope. Such systems can be built in-house; however, nowadays, increasingly more combined operating and monitoring systems for microfluidic devices are also commercially available. For example, Olympus Co. (Tokyo, Japan), ibidi GmbH (Gräfelfing, Germany), OKOLAB S.R.L. (Pozzuoli, Italy), and Tokai Hit Co. (Fujinomiya, Japan) provide stage-top incubation systems for live-cell imaging with temperature and gas control, which allow monitoring of cell migration, proliferation, and phenotypical changes. Besides, other high-resolution analytical methods such as surface-enhanced Raman spectroscopy,817 mass spectrometry,818 and nuclear magnetic resonance spectroscopy819 have been combined with microfluidic platforms as alternatives to imaging techniques to quantitatively assess changes in chemical or biomolecular compounds occurring during cell–material interactions.
Despite the fact that microfluidics has emerged into a powerful technology to study interactions between biomaterials and the biological system, and, more specifically, to use a biomaterial (microenvironment) for the exogenous control of cell fate decisions, it remains challenging to make microfluidic models applicable for screening a variety of functional biomaterials of all types including metals, ceramics, polymers and composites thereof. Such materials do not meet the criteria of transparency, gas-permeability, and processability that are common to materials used for the fabrication of microfluidic platforms such as PDMS. Therefore, their integration into a microfluidic system and evaluation of their interaction with cells and extracellular matrices poses challenges and requires further development of microfabrication techniques and adequate assays. Moreover, these functional materials dynamically interact with the biological systems through protein adsorption, degradation, etc., which makes it important to study the level of miniaturization required to have them match the on-chip microenvironment. The latter is a part of a greater challenge, which is the validation of the predictive value of on-chip cell–material interactions for the actual interactions that take place in the body. While this challenge is not unique to the systems discussed here and is applicable to all in vitro models, the aspect of functional materials integration makes it more complex and deserves particular attention in the years to come.
7. cBIT and High-Content Imaging
7.1. cBIT
A major factor in terms of studying cell–material interactions is the variance between different laboratories using not only different material preparations and characterizations as a biomaterial analogue, also the variations in cell types, culture conditions, the specific type of cell behavior, and the analysis approach. To truly come to the core parameters that drive cell behavior, which is induced by physicochemical properties of biomaterials or used biochemical stimulation, a more unified approach and general comprehension needs to be created concerning the complete development route and analysis phase. This general comprehension can only be achieved when data from various sources with defined analysis data and biological approaches is shared and available for reuse. For this purpose, cBiT60 has been created to facilitate the sharing of results from the biointerface field ranging from various methodologies for material preparation and characterization as described in the previous sections as well as performing generalized biological assessments, outcomes, and high-content imaging in a standardized fashion, which is the focus of this section.
7.1.1. Measuring Biological Outcome: Gene Expression
Measuring gene expression is one of the widely used ways to evaluate how cells respond to biomaterials, because it gives a snapshot of cellular activity at the molecular scale. There are several methods by which gene expression can be measured, and each has its advantages and challenges. In this section, we will focus on the basic mechanism of gene expression and the relevance of the different measurement techniques. Furthermore, we will summarize gene expression measurement tools, and how they have been implemented to understand biomateria–cell interactions.
7.1.2. Relevance of Gene Expression in Research
Genes are the basic unit of the genome sequence, which encodes a set of functional products, proteins, and ultimately dictates cell phenotype and function. For this reason, gene expression levels are measured in many areas of biological research such as oncology820−823 and developmental biology.824−827 In addition, gene expression levels have been measured in biomaterials research to determine whether the engineered biomaterial is capable of inducing stem cell differentiation,828−830 or can support the phenotype of differentiated cells for prospective tissue engineering applications.831−833 To make conclusions regarding the compatibility of the biomaterial, researchers select specific “marker” genes based on their scientific questions. Marker genes can be used to identify cell types or cell phenotypes. For instance, ALP has been used in bone tissue engineering to assess early osteogenic differentiation.834 When the expression of ALP upregulated in vitro, it indicates that the biomaterial has potential osteoinductive properties in vivo. For some tissues, there is more than one marker gene. For instance, in tendon tissue engineering, the combination of the expression of three or four genes, including scleraxis gene (SCX) and tenomodulin (TNMD), are used to determine the tenogenic capacity of the engineered biomaterial.835,836 Furthermore, marker genes can be used to show the activity of a signaling pathway that regulates the response of a cell to the biomaterial; and can provide valuable insights into the fundamentals of cell–biomaterial interactions.837,838 Hence, information obtained from gene expression data can then be used to modify and specifically tailor the biomaterial.
7.1.3. Regulation of Gene Expression
When a cell receives signals from its environment, it can respond to it by changing its shape and proliferation rate, differentiating into another cell type, maintaining its phenotype, or dying. These signals can be small molecules (Figure 35(1)), or the physical properties of the material such as surface topography (Figure 35(2)), and surface (bio)chemistry (Figure 35(3)). They are then transmitted to the nucleus through certain biological pathways. In the nucleus, there are intermediary elements such as transcription factors and enzymes, both of which regulate gene expression (Figure 35(4)). Transcription factors are transcriptional activator proteins that bind to the DNA sequence, and stimulate the expression of target genes (Figure 35(5)). Enzymes, such as RNA polymerase, are involved in the production of mRNA (mRNA) that can be considered as a copy of the gene. Afterward, genes are converted into proteins, and ultimately result in cellular response, such as proliferation, apoptosis, or differentiation (Figure 35(6)). In the gene expression regulatory machine, there are also repressors that regulate the gene expression process by binding to a specific DNA sequence or other transcription factors, and inhibit gene expression.
Figure 35.
Cells receive stimuli from their surroundings, such as small molecules (1), the physical (i.e., surface topography, porosity, stiffness, elasticity etc.) (2) and (bio)chemical (2) and surface (bio)chemical (3) properties of a material, and produce a response via signaling pathways. The pathways can, in turn, lead to the production of transcription enhancers, such as transcription factors (4) that enable the expression of certain genes (5). This gene expression can lead to different cell phenotypes (6) such as proliferation, differentiation, or cell death.
Gene expression can also be controlled through epigenetic regulation mechanisms, such as DNA methylation and histone modifications. Measurement of DNA methylation (the process of gene methylation) provides information on the epigenetic makeup of a cell, which can indicate how the particular biomaterial affects gene expression. For instance, Schellenberg et al. investigated the DNA methylation profiles of mesenchymal stem cells cultured on TCP or on PDMS.839 They showed that matrix elasticity does not have a major impact on DNA methylation profiles, but that it influences differentiation toward adipogenic and osteogenic lineage. This information is vital, as it emphasizes the relevance of matrix elasticity when designing a biomaterial with which cells will directly interact. Similar to DNA methylation, histone (the proteins in cell nuclei that DNA is rolled over) acetylation and deacetylation are involved in gene expression regulation by changing the accessibility of DNA to transcription factors. Downing et al. reported that biophysical cues provided by parallel microgrooves decreased histone deacetylase activity, and therefore, lead to increased histone H3 acetylation and methylation.840 They concluded that a change in cell morphology is responsible for the modulation of the epigenetic state of cells. Hence, understanding the regulation of gene expression can help to manipulate cellular states or cellular differentiation.
7.1.4. Methods to Measure Gene Expression
Since the discovery of DNA, scientists have been developing different methods to measure gene expression (Box 1, History of the Measurement of Gene Expression Technologies). Each method has advantages and challenges, but the selection of the appropriate tool can provide valuable information. In this section, we describe the commonly used gene expression measurement tools, their advantages, and limitations, and finally, give examples from biomaterials research.
Box 1. History of the Measurement of Gene Expression Technologies.
History of the measurement of gene expression technologies Ever since Rosalind Franklin, Francis Crick, James Watson and Maurice Wilkins discovered the structure of DNA, scientists have put their efforts to further understanding the–secret– language of DNA. In 1961, Sydney Brenner and Fran–ois Jacob discovered mRNA (mRNA). The same year, Sydney Brenner and Francis Crick cracked the secret language of the genetic code. In the following years, Phil Sharp and Rich Roberts discovered the basis of introns and alternative splicing. The same year, James Alwine, David Kemp, and George Stark at Stanford University developed a technique called Northern blot to detect the levels of gene expression by using electrophoresis to separate RNA by its size, then detected the signal with a hybridization probe that is complementary to a target sequence. In 1980, Frederick Sanger was awarded the Nobel Prize due to his discovery of a method DNA sequencing. Also in the early 1980s, another method called fluorescence in situ hybridization (FISH) was developed. The method was based on using fluorescent DNA probes to detect and target the specific locations on chromosome or mRNA in the cell. Our understanding on the complexity of DNA and gene regulatory networks continued to expand dramatically when Kary B. Mullis and co-workers invented the method called polymerase chain reaction (PCR). Their invention was awarded with a Nobel Prize for chemistry in 1993. Later, Russell Higuchi added a fluorescent label that binds to the accumulating PCR product. He reasoned that the intensity of the fluorescence signal has a linear relation with the concentration of the PCR product. This method is called qPCR and it is one of the most frequently used techniques in the gene expression today. Later, more established transcriptomics technologies have been invented to measure the sum of all of the RNA transcripts in cell. There are two key techniques that have been used under transcriptome technologies: DNA microarray and RNA sequencing. In principle, microarray determines transcript abundance by measuring the hybridization of fluorescently labeled transcripts. RNA sequencing (RNA-Seq) sequences the whole transcriptome of a cell. The advanced gene expression measurement technologies have been used in molecular biology for many years to understand how cells change at a disease state or how they respond to certain chemicals and biomaterials. Understanding the phenotype and function of a cell provides information about the tissue, organ and organism itself.
7.1.4.1. Fluorescence In Situ Hybridization (FISH)
The presence of a biomaterial not only affects the gene expression of the cells that are in direct contact with it, but also that of the cells in the vicinity. It is, therefore, important to understand gene expression in the context of the spatial organization of cells within the tissue. FISH allows quantification of both in vitro and in vivo spatial gene expression, which makes it a very useful technique. For instance, Zreiqat et al. investigated the expression of osteoblastic genes in hBM-MSCs that were cultured on pure titanium and titanium alloy.841 They used FISH to quantify the gene expression, and found that there was an increase in the expression of osteoblastic genes on the cells cultured on pure titanium and titanium alloy compared to TCP plates. To confirm the validity of their results, they performed quantitative immunohistochemistry to measure the expression of corresponding proteins. Their results provided initial information on how small differences in the surface chemistry and structure of biomaterials can induce osteogenic differentiation. Similarly, Knabe et al. showed the effect of bioactive glass-ceramics on the expression of osteogenic genes and corresponding proteins by performing Q-FISH and immunohistochemistry, respectively.842 They reported that glass-ceramics could be regarded as potential bone substitutes. Maeda and colleagues employed FISH to evaluate the effect of micropillar substrate, an important parameter in the materiobiology research, with varying height on the gene expression of MMP-1 in tendon fibroblast and reported that 8-μm-height micropillar is the most optimal compared to flat, 2 and 4 μm height.843
Although it is a useful technique to study materiobiology, FISH does have several limitations. First, the optimization of samples, multiplexing, and the number of cells and materials to be used in the detection all require a lot of time and effort. Second, to detect the gene expression, histological slides are prepared out of the material or tissue, and this requires extra handling of the samples. Therefore, FISH is not as straightforward as other gene expression methods. However, this technique is very useful for identifying spatial gene expression in particular.
7.1.4.2. Quantitative Polymerase Chain Reaction (qPCR)
In materiobiology, qPCR is used to answer various biological questions. In our recent study, we employed qPCR to measure the expression of ALP, OCN, and OPN genes to identify the surface topographies that can induce osteogenic differentiation.566 Here, we reported that certain surface topographies induce osteogenic differentiation more than others in vitro; and these selected surfaces are capable of osteogenic differentiation in vivo. In addition, qPCR can be performed to understand and evaluate the initial biological response of a cell to a material. For instance, Kato et al. investigated the gene expression of Heat-Shock Protein (HSP) 70, 90, and 47 in HeLa S3 cells cultured on different materials, such as TCP, silicone, tetrafluoroethylene-hexafluoropropylene copolymer, and cellulose.844 Their results showed that HSP 70mRNA can be used as a marker for the investigation of cell–polymer interactions. In another study, the same group investigated the expression of the oncogenes c-myc, c-fos, and p53 (the tumor suppressor gene) in human fibroblasts cultured on polymeric biomaterials. They evaluated the carcinogenic activity of cells, and showed that p53 is a very sensitive marker in the evaluation of the carcinogenic potential of a biomaterial.845 Substrate stiffness is another important parameter in the materiobology. To elaborate its relevance, Forte et.al. executed qPCR to evaluate neo-angiogenesis capacity of neonatal murine cardiomyocytes on poly-ε-caprolactone planar layers with Young’s modulus ranging from 1 to 133 MPa; and showed that softer substrates induce an increase in the gene expression of alpha actinin and myosin heavy chain.846
For practical reasons, typically 1–10 genes can be investigated in qPCR. For this reason, marker genes are predefined when performing qPCR. Another limitation of qPCR is the fact that it does not provide gene expression data with single-cell resolution. Therefore, when the marker genes are not known, and gene expression in single-cell resolution is desired, other gene expression methods are used.
7.1.4.3. Transcriptomics Technologies
Although using the aforementioned tools to measure gene expression provides useful data when marker genes are defined, using such methods when marker genes are not defined is more challenging. Transcriptomics has been developed over the last three decades, and is more advanced in that it enables assessment of the global gene expression. An advantage of implementing transcriptomics technologies in biomaterial research is that it enables the discovery of novel genes, and the identification of target genes and signaling pathways. This information can then be used to enhance the performance of biomaterials. In our recent work, we investigated the influence of material properties, such as microporosity and ion composition, of a set of synthesized osteoinductive and noninductive calcium phosphate ceramic materials on the transcriptomics profile of osteogenic cells.59 Transcriptomics revealed the crucial role of ECM deposition in osteoinduction, which is controlled by surface topography and calcium phosphate ions.
7.1.4.4. Microarray and RNA Sequencing
DNA microarray and RNA sequencing (RNA-seq) are the two methods used in transcriptomics-based techniques. They provide mapping and quantification of the whole transcriptome of cells. Microarray relies on the hybridization of fluorescently labeled complementary DNA (cDNA) obtained from cells to complementary DNA sequences on probes. In RNA-seq, adaptors are added to cDNA fragments to allow recognition for each cDNA fragment, and fragments with adapters are amplified. The gene expression is determined based on the fluorescence intensity. Microarray differs from RNA-seq in that it relies on predefined DNA sequences, whereas RNA-seq sequences the whole transcriptome of cells. Therefore, RNA-seq provides a complete picture of the cell transcriptome, and increases the likelihood of discovering novel genes. Given these reasons, a microarray is being replaced by RNA-seq in transcriptomics-based research.
Both microarray and RNA-seq provide transcriptomics data belonging to a cell population; however, in some cases, it is important to understand the gene expression profile of individual cells. For such cases, single-cell RNA sequencing (scRNA-seq) is used. It works in a manner similar to RNA-seq, but instead provides the whole transcriptome of each individual cell. This technology has been used in cancer research847−849 and in developmental biology;850,851 however, its use has not yet expanded to biomaterials research. scRNA-seq can be extremely useful for understanding the influence of biomaterials that possess heterogeneous structures, such as gradient surfaces or surfaces with various surface chemistry or topographical structures.
7.1.5. Transcriptomics in Biomaterial Research
As mentioned above, the use of transcriptomics-based technology in research has been growing. In 2000, there was only one publication that included transcriptomics-based methods.852 In 2019, there are more than 50 publications. We divided the most abundant topics into three categories: (1) 2D/3D culture systems, (2) surface topography, and (3) titanium, which is one of the most frequently used materials in the clinic and biomaterials research.
7.1.5.1. Impact of 2D/3D Culture Systems on Cell Differentiation
Herlofsen et al. investigated the chondrogenic differentiation of hMSCs cultured on 2D and 3D environments at different time points.853 They employed the Illumina microarray platform as a transcriptomics tool and identified 1969 differentially expressed genes between monolayer hMSCs and chondrogenic cells. With further bioinformatics analysis, they highlighted the role of genes involved in the ECM and transcription factors involved in ECM synthesis. Similarly, Kumar et al. investigated stem cell response to a library of scaffolds with varied 3D structures.854 Their transcriptome analysis revealed that each type of scaffold induced a unique gene expression signature. Furthermore, of each structure, only the nanofibrous morphology induced osteogenic differentiation of hBMSC in the absence of osteogenic supplements. Baker et al. also adopted transcriptomics-based research to investigate the influence of 2D and 3D culture of hBMSC.855 They reported that 3D scaffolds induce the activity of TGF-β and cell-adhesion/ECM-receptor pathways that can be used to control hBMSC differentiation. In addition, Zhang et al. performed RNA-seq, and reported that gene expression profiles of endothelial cells and pericytes cocultured in 3D were similar. However, in 2D culture, each cell type displayed different vascular signatures.856
7.1.5.2. Influence of Surface Topography on Cell Fate
This subject has been the focus of many research groups;857−860 however, only a few studies have made use of transcriptomics to obtain a broader picture of cellular response to a biomaterial. For example, Dalby et al. investigated the response of fibroblasts to microgrooved topographies to understand the events taking place at initial cell contact with the scaffold.861 In their study, they demonstrated that the genes involved in cell signaling, cytoskeleton, ECM remodeling, and DNA transcription are differentially expressed between microgrooves and flat surfaces. The same group also employed RNA-seq technology to understand the mechanisms through which skeletal stem cells (SSCs) preserve their multipotency or differentiate toward fibroblastic lineage.862 Their results indicated that activation of mitogen-activated protein kinases (MAPKs) is involved in the control of SSC self-renewal and differentiation. With a similar approach, Abagnale et al. showed that different surface topographies induce differentiation of stem cells toward osteogenic and adipogenic differentiation.863 Furthermore, Darnell et al. examined the effect of surface stiffness on the global gene expression of mouse mesenchymal stem cells.864 They found that stiffer hydrogels induce the expression of genes that are involved in bone remodeling, cell-matrix adhesion, proteolysis, and IL- 1 and androgen receptor signaling.
7.1.5.3. Effect of Titanium on Cell Phenotype
Titanium is one of the most frequently used materials in the clinic. Therefore, many researchers have focused on understanding the interaction between cells and titanium surfaces. Ku et al. investigated the effect of different titanium surface treatments on the global gene expression of osteoblasts.865 They reported that the expression of the genes involved in the regulation of FAK and apoptosis depending on the metal ion release from the surface. Similarly, Carinci et al. investigated the titanium-cell interaction based on global gene expression profiling by using a custom-made microarray called Human 19.2 K DNA microarray.866 They reported that the titanium surfaces induced a broad range of functional activities, including apoptosis, vesicular transport, and structural function. Similarly, in our recent work, we made use of microarray technology to explore cell–biomaterial interactions to assess and improve material properties.867 For this, we tested the response of MG-63 cells to 23 different materials relevant for bone regeneration. Our results showed the role of TGF-β and WNT signaling in the cellular response to osteoinductive materials and activity of FAK signaling in differential cell adhesion kinetics.
7.1.6. Other Omics Technologies
Alongside transcriptomics, proteomics and metabolomics are the two other omics technologies through which cellular responses to a biomaterial can be investigated. Proteomics allows protein expression profiling, and enables identification of post-translational modification events and subcellular localization, and the relevant functional aspects of the proteome of cells. Metabolomics is another omics technology, which provides chemical fingerprints–metabolites–that specific cellular processes leave behind. Alakpa et al. used supramolecular hydrogels to target the range of stem cell phenotypes by measuring the metabolites that they leave behind.868 On the basis of their results, they identified the bioactive metabolites that can target bone and cartilage formation. Proteomics and metabolomics can also be used to generate complementary information to transcriptomics data. Guerette et al. showed how the integration of high-throughput RNA-seq with proteomics accelerated biomimetic engineering.869
7.1.7. Current Standing and Future of Transcriptomics in Biomaterials Research
Understanding the biology of cells, their healthy and diseased states, and their response to external signals, are the goals in both fundamental and applied biology. Advancements in transcriptomics technology have enabled the collection of the whole transcriptome of all cells in a tissue or organ, which has significantly contributed to our understating of cell biology. Given this, there are attempts to gather transcriptome data in a consortium, and make it useful for public research. One of the consortiums founded with this purpose is The Human Cell Atlas.870 The Human Cell Atlas is a collection of maps that describes and defines the cellular basis of health and disease. This consortium benefits from the technological power of single-cell profiling. For instance, MacParland et al. created a map of cells in the human liver by using scRNA-seq, and provided transcriptional profiles of 8,444 parenchymal and nonparenchymal cells.871 They further identified 20 discrete cell populations by using gene expression patterns, flow cytometry, and immunohistochemical examinations. A similar approach was taken by Ledergor et al.,872 Vento-Tormo et al.,873 and Reyfman et al.,874 where they revealed the transcriptome of different organs in healthy and diseased donors. The Connectivity Map (CMap) database875,876 creates a genome-scale library of cellular signatures, and catalogs transcriptional responses to chemical, genetic, and disease perturbation to accelerate the discovery of novel therapeutics. Both CMap and The Human Cell Atlas benefit from the collection of large data sets, which in turn informs the research of its users.
Currently, there are several repositories that are used to contain, curate, and maintain metadata, and raw and processed transcriptomics data. The Gene Expression Omnibus (GEO) database877 and ArrayExpress878 provide up-to-date raw and processed transcriptomics data of various species. Once downloaded, data is ready for further processing or analysis. The cBiT is an open access online web tool that provides data storage.60 In fact, it is the only repository that offers biomaterial-based transcriptomics along with metadata of materials, including the biological and technical properties. Therefore, cBiT encourages both material scientists and cell biologists all around the world to submit their transcriptomics data to make it publically available. The fact that transcriptome data and metadata of material properties is publically available enables its application in the research of the wider scientific community. Furthermore, comparing and combining the transcriptomics data of different data sets opens the door for the discovery of new materials. A paradigm shift from traditional experimental sciences to omics-based sciences can be achieved by combining the information from all the omics data into one repository. When all these data are gathered with the information coming from high content imaging, we will have a better understanding of cellular response to materials, enhance existing materials, and engineer new materials. Therefore, the future of cell-based biomaterials research lies in the collaboration, sharing, and interpretation of existing data. When the biomaterials field follows the slipstream of other transcriptome-based research present in other fields, the discovery of new materials will be inevitable.
7.2. What Is the High Content Imaging?
High content imaging (HCI) refers to the microscopy-based acquisition of a large number of images of single cells or whole organisms following treatment with various perturbations, chemical or genetic. Biomaterials are a special class of perturbance that affects cells simultaneously by chemical and physical cues, as was reviewed above. HCI is also interchangeably called High-Content Screening (HCS) with more focus on throughput and typically includes a screening of more than 100 000 conditions; and High-Content Analysis (HCA) sometimes refers to the analysis part of HCI.879 The “High-Content”, part of the definition, implies that thousands of properties are typically measured. This is in contrast to the HTS, where usually only one parameter is assessed. The exception is Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS) technology, which can also be considered as HCI type approach applied by a different instrument. ToF-SIMS, for example, is used for imaging lipids.880 The ability to produce a significant amount of data is both the power and the challenge of HCI.881
7.2.1. Applications of the HCI
HCI is applied when conventional methods are not able to screen for a vast number of parameter combinations and thus extensively used in both drug discovery and cell biology. This allows screening, for example, a library of small molecules that can consist of hundreds of thousands of compounds.882 One of the key advantages of HCI is that it produces rich data sets at a low cost. What makes HCI different from any other HTS approach is the ability to generate per cell data with high spatial resolution. This advantage is widely used for assessment of the functional gap junctions,883 neurite outgrowth,884 translocation of proteins between cytoplasm and nucleus,885 expression of differentiation markers by single cells,886 cell shape during epithelial to mesenchymal transition,887 internalization of the proteins or vesicles882 or collective cells behaviors such as wound healing.888 Another advantage of HCI is the ability to visually inspect any tested condition and thus increase the confidence of the outcome (Figure 36).
Figure 36.
Comparison of the high-throughput and high content assay readouts as reviewed by Mercola et al.889
A typical readout of HCI is the expression of specific markers, nuclei, and cell shape.890 Such setup allows single-cell segmentation by using three fluorescent dyes for capturing. Automatic microscopes used in HCI typically can image up to five fluorescent channels, which allow assessing several parameters simultaneously. For example, cytotoxicity and expression of the protein of interest. Also, the predictive power of a single measurement for some assays is not sufficient to judge the statistical significance of the test. Therefore, multiple parameters can be measured to increase confidence in the results. Another application of HCI is screening cell lines against a library of compounds that enable RNA knockdown or protein overexpression.891 Such an approach is used in studying pathways, or mechanisms of action of the disruptive factors, and therefore can be used for studying the biological activity of biomaterials.892
The examples reviewed so far belong to the screening type of experiments. However, there is another type of HCI application that is called profiling.893 The goal of the profiling assays is to extract signatures of the cell state from the imaging data. The idea is that compounds with a similar mechanism of action also induce the same changes in cell morphology, which can be captured by imaging. Cell painting assay was explicitly designed for profiling purposes.476 It allows for imaging of eight broadly relevant cellular components by using five fluorescent channels. This approach was successfully applied to group compounds based on their mechanism of action and was proven to be more accurate than classification based on the chemical structure.894 Profiling experiments can be used in biomaterial research to find biomaterials with a unique effect on the cells, or predict cell response to the new biomaterial without thorough biological assays. For example, Figure 37 demonstrates how distinct single-cell profiles treated by two different chemical compounds, as demonstrated by Caicedo et al.895 Another example is a research done by P. Moghe and colleagues, who were able to predict cell commitment to osteogenesis based on the images of stem cells cytoskeleton cultured on different coatings.896
Figure 37.
Single-cell, treated with either floxuridine or etoposide, show distinct image-based signatures. Adopted from Caicedo et al.895
Above, we have reviewed the general application of the HCI to provide the reader with more insights in the type of studies that can be done, below we will discuss HCI application in the Biomaterial field that was done so far.
7.2.2. Overview of HCI Applications in the Biomaterials Field
Several biomaterial research groups already recognized the power of HCI. What is typical for these laboratories is that they used miniaturization as a means to make their standard assays compatible with automated imaging, which enables a high-throughput format.22
HCI enabled testing an unprecedented chemical space. For example, Celiz et al. have developed a microarray that allowed to identify materials that can support the culturing of the human (PSC). As a result, screening was performed with 909 unique polymers, tested in 4356 individual assays, and generated in three cycles of contact printed arrays. The first generation consisted of the large library of the 141 (meth)acrylate and (meth)acrylamide monomers used to identify homopolymers that could support hPSC. The second and third generations were used to optimize copolymers by varying homopolymers ratios and combinations. As a result, Ceiliz et al. have identified the first synthetic polymeric substrate that achieves both pluripotent hPSC expansion and subsequent multilineage differentiation.409
Another example of the successful application of the HCI approach is the TopoChip platform. It enables exploration of the cell response to an extensive library of microtopographical surfaces. In total, it contains 2176 unique topographical structures; each of them is formed by arrays of pillars with a fixed height of 10 μm. The shape of the pillars in XY projection was generated randomly by the combinatorial mixing of three primitive shapes: circles, lines, and triangles.455 Cells seeded on the TopoChip and screened with HCI methods enabled discovery of the topographies that, for example, promote osteogenesis,566 form biomechanical niche,567 or control ICAM-1 expression.897
Lutolf and colleagues used a liquid dispensing robot to prepare hydrogel microarrays compatible with HCI format. The approach allowed to systematically investigate the combined effect of elasticity, proteolytic degradation, and signaling proteins on the self-renewal of ECSs. In total, they were able to simultaneously explore the effect of 1024 unique hydrogel microenvironments, which allowed to broaden understanding of multifactorial 3D cell–matrix interactions.450
Another example of the HCI screening platform that allows systematic exploration of the cell–biomaterial interaction is gradient surfaces. Such platforms allow assessing continuous material properties on cell behavior and are less prone to variations as it allows us to control for systematic error. For example, Zhou et al. developed a nanotopography gradient on a PDMS substrate by applying a unidirectional strain during surface oxidation by air plasma. The obtained platform was used to investigate how topography aspect ratio affected osteoblasts adhesion. This experiment allowed to discover a nonlinear relationship between cell adhesion and aspect ratio.34 In another study, Zhou et al. were able to combine orthogonal double gradient, effectively combining surface stiffness and wettability. They discovered the nonlinear effect of stiffness and wettability combination on a broad range of the mesenchymal stromal cells properties such as adhesion, spreading, nucleus size, and vinculin expression.146
Below we will outline typical steps that should be considered during HCI setup with biomaterials. The general HCI outline was proposed by M. Boutros et al.890 (Figure 38).
Figure 38.
Workflow for assay development and image analysis for HCI, proposed by Boutros et al.890
On the basis of the type of the assay, biomaterial screening platforms can be classified into three groups:
2D platforms. Examples of such platforms can be polymer microarrays, reviewed above, and in Section 5. One of the challenges with imaging such arrays is the autofluorescence of the polymers. What complicates the matter is that different polymers can have autofluorescence in different fluorescent channels. We will discuss a solution for this later in this section.
2.5D platforms. 2.5D substrates typically refer to platforms that have repeating isotropic architecture in the third dimension, in the micrometer range.898 One of the examples of the 2.5D screening platform is the TopoChip. The challenge of such platforms is that cells can either be confined between pillars or located on top of them, depending on the density of the posts, affecting the shape and size of the cells. Another challenge is the different density of the pillars that can distort light differently and affect background intensity.
3D platforms. Here, cells are cultured either in the hydrogels or as spheroid cultures. Such platforms offer several challenges for the HCI, such as the homogeneous distribution of the immunofluorescent antibodies inside the 3D material, autofluorescence, transparency, thickness, and density. A recent review covers the basic setup of organoids in HCI imaging.899
Some biomaterial platforms are not compatible with imaging technology, for instance, 3D printed scaffolds from nontransparent polymers.
7.3. Experimental Design of HCI Screening
7.3.1. Assay Optimization and Development
During the development of the biological assay for HCI, it is important to remember the GIGO principle, which stands for “garbage in—garbage out.” It emphasizes the fact that the quality of the obtained results will not be better than the quality of the data.900 It is easier to improve the quality of the data during the assay development step than during the analysis. Therefore, an adequately developed assay is an essential part of any successful HCI experiment. Typical assay optimization for the HCI includes optimizing cell culturing, staining protocols, and testing them in high throughput format by running a screening pilot.901 Here we will focus on challenges specific to the biomaterial field. We want to emphasize again that the quality of the assay will affect imaging and data analysis; therefore, it requires appropriate expertise.
HCI is based on converting light emitted by excited fluorescent tags/compounds to a digital image. These fluorescent tags coupled to antibodies or fluorescent chemicals can specifically bind to the molecules proportionally to their amount.902 Different fluorophores can be excited and emit the light of a specific wavelength, the property that allows decoupling different cell measurements in the same experiment.
The light coming from the target is called signal, and off-target noise contributes to the background. During assay development, one of the goals should be to increase the signal-background ratio.903 The application of HCI to biomaterial research is complicated by the autofluorescence of many materials. This is a phenomenon when a substance or material emits light upon excitation without the addition of a specific fluorescent label or marker. The first rule is to avoid using fluorescent dyes that emit the same wavelength as the material of choice. It is not possible when screening includes a library of different materials that have different emission profiles. One of the promising solutions to overcome material autofluorescence was proposed by Berezin and Achilefu, who suggested using special fluorophores with an emitting lifetime longer than of the materials.904 Another solution can be adopted from histology, where treatment with various chemical agents such as ammonia-ethanol, sodium borohydride, and Sudan Black B, are commonly used to reduce tissue autofluorescence.905 Viegas et al. further improved autofluorescence removal by combining treatment with Sudan Black B and short-duration, high-intensity UV irradiation in paraffin sections.906
Low-intensity, uniform autofluorescence can be eliminated digitally by subtracting or spectral unmixing postacquisition.907 Besides, autofluorescence of the materials can be reduced by applying optical techniques. For example, multispectral imaging (MSI),908 or total internal reflection fluorescence TIRF,909 but due to complexity, they are not very compatible with high throughput format. Adopting these imaging techniques to the HCI is under active development.910
Other sources of off-target fluorescence are cell culturing medium and not properly washed out unbound fluorophores. Some sources of background intensity are equipment-specific and cannot be controlled in the lab, such as the offset of the detector and light leakage from the excitation source.911 The last two examples represent the systematic type of error and will be discussed later.
Besides the problem of background intensity, critical parameters to pay attention to are homogeneous cell density, and staining quality. Typically, before starting the full screening a pilot experiment should be performed, to ensure optimum performance.890
7.3.2. Controls
Controls serve multiple purposes in the HCI experiment. First, controls are used to judge the quality of the assay. For example, if is not possible to distinguish positive and negative controls, the chosen readout should be reconsidered. Usually, positive and negative controls are selected from the same compound classes as the screening one, which is often not possible. For example, we cannot choose topography as a positive control for a screening on topographies if such studies were not performed before. In this case, it is acceptable to select another type of compound, for example, treatment with a chemical compound. However, the intensity of the effect should be comparable with the anticipated magnitude of the screening compounds. Positive control with a strong effect will cause numerous problems during data analysis, for example, during the normalization step. When the screening is done in several batches, controls can be used to normalize the data.
For excluding any effects with respect to the sample position, the position of the controls should be random. However, this is hard to implement practically.
7.3.3. Replicates
The number of replicates in the HCI experiment depends on many factors. One of the elements is the cost of the screening. The majority of the HCI experiments are performed in duplicates, by introducing the third replica the cost of the whole trial will increase by 50%. However, adding more replicates is an efficient way to reduce the number of false negatives without increasing the number of false positives. The presence of sufficient amounts of replicates will enable proper statistical analysis. If the phenomenon is complex and differences are subtle, the number of replicas should be enough to be able to test statistical significance. Tools that allow taking all the variation into account and estimate the number of replicas are proposed by Malo et al.912
7.4. Imaging and Image Analysis
7.4.1. Imaging of the Samples
7.4.1.1. Automation
To be able to image a large number of conditions, automatized microscopy systems are used. Such systems are often equipped with a motorized stage that allows precise placement of the sample under the objective and rapid autofocusing but also supports automatic handling of the plates. This allows for producing images with maximum efficiency. Companies like PerkinElmer and ThermoFisher are offering commercial setups designed explicitly for high-content screening such as “Opera Phenix” and “CellInsight”, respectively. At the same time, many microscope brands, for example, Nikon, Zeiss, and Leica, produce highly customizable instruments that can be constructed to meet specific demands.
7.4.1.2. Programming
Microscopes used for HCI are fully programmable. Customizable microscopes mentioned in the previous section are enabled for high-throughput systems by using software such as “Jobs”, “Zen 2”, “LAS X”. The image acquisition software is highly customizable and allows us to set up different options such as the number and location of images per sample, number of channels, autofocusing algorithm, and many others.913 Even though the microscopes are programmable, it is possible to set them up without any knowledge of the programming language because often they are utilizing a so-called visual programming concept, where every function is represented as a block, and for setting the microscope instructions, it is sufficient to drag and drop all these blocks together in the right sequence. Microscopes already have presetup settings for most common tasks. The time required to customize those settings for untrained cell biologists depends on the complexity of the task. Depending on the microscope and brand, companies might provide training, support, or service for creating custom scripts. In the end, efforts spend on customization will be paid off by time saved during unattended automated imaging. Open-source software, such as uManager, allows for programming the microscope directly from the data analysis tools such as Matlab or Python. This allows unifying image acquisition and data analysis and provides additional possibilities for automation.
7.4.1.3. Autofocusing
Autofocusing is a critical part of imaging as it directly influences the quality of the data. There are different technical solutions to allow autofocusing.914 Some of them are algorithmic, for example, using mathematical transformations to identify in-focus images from a set of images, acquired along the Z-axis;915 others are based on laser focusing. The last is fastest, but at the same time, it fails to work in multiple reflection interfaces, for example, between the objective and the specimen, which is a case for a biomaterial screening.916 It is critical to find the right balance between image quality and speed. Therefore, the approach that permits the fastest autofocusing should be preferred. Contemporary microscopes allow customizing the imaging pipeline, which permits finding this balance. For example, instead of autofocusing for every image, one might choose to autofocus every second image and use the focus plane from the previous image as a focusing point. Such an approach can help to reduce imaging time significantly without losing quality.
7.4.1.4. Type of Imaging
There are two types of imaging that can be used with HCI. Most widespread is epi-fluorescent imaging; it is based on exciting (shining light) the whole specimen visible in the field of view. In parallel, a camera is detecting all the emitted light. Confocal imaging was explicitly designed to overcome this limitation, by using a specifically configured optical path and excitation source, it can only capture emission close to the focal point, a phenomenon called optical sectioning.917 This allows capturing an image with higher optical resolution and contrast. However, this comes with a cost. Confocal imaging is slower; it requires a brighter signal and can damage the sample.
The disadvantages of both methods can be overcome by light-sheet fluorescence microscopy (LSFM). The fundamental difference from traditional microscopes is that illumination and detection of optical pathways are decoupled, allowing novel illumination strategies.918 Imaging systems that are utilizing LSFM for HCI applications in mind were recently developed.899
7.4.1.5. Image Resolution
Image resolution directly affects many parameters such as details in the image, speed of acquisition, or the total size of the photos. It is crucial to determine the right image resolution that suits the purpose of the screening upfront. For example, if one is interested in the analysis of the actin fibers, the resolution of the image should be high enough to capture these finite elements, on the other hand, if the goal is to see the only cell shape the resolution can be lower.919
7.4.1.6. Bit Depth and File Type
Bit depth is a significant parameter; it reflects the number of bits that span an intensity range. Most of the microscope cameras are acquiring images with 12-bit depth, allowing 4096 levels of intensity (shades of gray). Note that most of the screens are only capable of displaying 8 bits (256 shades of gray). As a result, not all information is visible, and the 12-bit image is black upon viewing in standard software. Some programs such as ImageJ and IrfanView allows shrinking intensities range to 8-bit images, allowing its viewing. During image acquisition, it is the rule of thumb to use all the intensity range to allow better separation between signal and background. However, it is vital to avoid saturation of the signal; otherwise, best-performing conditions will be indistinguishable.920
To avoid any image distortion, data should be saved in a lossless format, which does not compress the data and allows saving all the data without any loss such as tiff or png. Jpeg is not a lossless file format. Microscopy brands often have proprietary lossless imaging types, which may not be natively supported by image processing software. In this case, images can be converted to broadly supported tiff files.
7.4.2. Image Preprocessing
It is important to realize that raw images from the microscope, regardless of the quality of the assay will always contain some errors, which can be systematic or random. That is why before image analysis these errors should be corrected during a preprocessing step.
The systematic error is represented by the unevenness of the intensity across the image. It is not uncommon to have more than a 1.5-fold difference in intensity level within the same image, caused by this type of error.921 It can result from illumination field unevenness, variation in the output of the light source, optical aberrations, and artifact presence in the well or other reasons. The methods that allow correcting for this type of error are called illumination, the background, or flat field corrections. They can be fixed, either by subtraction or normalization procedures. There are three types of algorithms that allows background correction:
-
a.
Prospective based on correction from the reference images, which can be obtained by imaging a material without cells. However, this approach contains several disadvantages and rarely applied in HCI practice.
-
b.
Retrospective single image method based on the computational estimation of the background separately for every image. Nevertheless, the result can vary between images, causing alterations in relative intensity level.
-
c.
The retrospective multi-image method estimates correction function based on all images in the screening or batch. It is the most robust method and allows identifying the systematic error across all images.
Retrospective methods use a computational model to calculate correction function that can be estimated using smoothing, surface fitting, or energy minimization approaches. The systematic error should be determined on per channel level as different channels have different distortion patterns. In the case when different materials have different autofluorescence levels, the background should be defined and subtracted in the additional step. There are multiple computational tools available for this task, for example, Horvath and colleagues developed CIDRE application: an illumination-correction method for optical microscopy software.922 Singh et al. proposed a method for illumination correction for HCI implemented in CellProfiler software.923
7.4.2.1. Artifacts Removal
Artifacts are usually unspecific fluorescent objects such as dust particles and can severely disturb cell segmentation or can be analyzed as cells during the analysis. Depending on the amount of the artifact, the image can be excluded entirely, for example, based on the percentage of the saturated pixels or using the computational approach excluded during image analysis or right after.
7.4.3. Image Analysis Pipeline
The goal of the image analysis is to extract information from digital images that represent the state of the cells in every condition. Typically, this kind of analysis is performed on per cell level. The group of pixels represents every cell. The task of segmentation is to identify single cells based on this information. Figure 39 sketches the main steps of extracting quantitative information from the images.
Figure 39.
Main steps to extract quantitative information from images.895
7.4.3.1. Segmentation
Segmentation can be performed using two standard methods.
Model-based: The researcher needs to choose an appropriate algorithm and manually optimize parameters based on the visual inspection of the segmentation results. Typically, the first step is to identify the nucleus based on DNA staining and use the results as seeds to identify the cytoplasm of the cell. The model requires some parameters to be known upfront, such as the typical size and shape of the nucleus. This type of segmentation can be performed in software such as CellProfiler921 or ImageJ.924
Machine learning-based: The researcher is training a model that can find single cells and elements of the cells in the image. The training is done by manually labeling parts of the cells on the image and assigning them to specific classes such as the nucleus and cytoplasm. Once the model is trained, it can be applied to all the images. This type of approach can be performed in free software such as Ilastik925 and Cells Cognition.926 Such a method can be more straightforward than model-based, but it requires a significant amount of manual labeling.
Even though segmentation of cytoplasm and nucleus are commonly performed, it is possible to segment any other subcellular structures such as focal adhesion, actin fibers, nucleoli, mitochondria, and others.
7.4.3.2. Feature Extraction
Once the separate objects are identified, the next step is to extract measurements that are often called features. Typically, there are four groups of features:
Shape-based: computed based on the outline of the objects and consists of typical shape measurements such as area, which is a number of pixels that can be fitted within the object outline, the perimeter is the total length of the outline, eccentricity is the elongation of the objects and so on, see Figure 40 for an example.927
Intensity-based: measured directly from pixel intensity values and can be helpful, for example, for identification of total protein amount per cell. They also include simple statistics such as min, max, median, and mean intensities per cell. The intensity that reflects the total amount of marker is Integrated Intensity measurement, which is calculated by summation of all the pixel intensities values and thus indicates not only average intensity but also the entire area. By using Integrated Intensity, it is possible to compare expression values of cells that have different sizes, which is commonly the case on topographical-based screenings.
Texture-based: this metric quantifies spatial arrangements of pixel intensities in the image. The periodic patterns in the image can be found by applying the statistical approach and mathematical transformations. Typically, texture features can create a unique signature, but the interpretability of these measurements can be very poor.
Microenvironment or context-based: this type of feature allows quantification of a spatial relationship between objects. For example, the number of neighbors for every cell, the location of the nuclei inside the cytoplasm to name a few. Such features are useful in analyzing the collective behavior of the cells.
Figure 40.

Scatterplot of cell silhouettes according to compactness and elongation values. The cells indicated using black arrowheads share similar elongation values and differ markedly in their circularity and border complexity. As quantified by Bitar et al.927
Although there are only four most commonly used groups of features, the total amount of elements in these groups can be more than 200. Given that per condition, it is possible to acquire a separate image per subcellular structure, in the case of Cell Painting, the total number of features per treatment can exceed a thousand.
7.4.4. Free, Open-Source Image Analysis Software
There is plenty of free imaging analysis software available such as ImageJ,924 CellCognition,926 Icy,928 and others. However, the CellProfiler921 was initially designed to meet the image analysis needs of the HCI screenings. The significant advantage of CellProfiler is that it was intended for biologists and does not assume any prior knowledge. Also, it supports plugins from the popular image analysis software ImageJ. Therefore, it is possible to use the advantages of both applications.
By means of programming, it is also possible to create an image analysis pipeline by using specially designed libraries directly in Python,929 R,930 Matlab,931 or Mathematica.932
7.4.5. Alternative Approaches for Analysis of Imaging Data
Classical Image analysis pipelines based on object segmentation and feature extraction is currently the most popular method. However, alternative approaches exist.
One of the examples is PhenoRipper,933 which positions itself as easy to use the HCI image analysis tool. It is utilizing a segmentation free approach by splitting the original multiplex image into blocks of pixels and perform feature extraction from these blocks rather than individual cells. Mainly it analyzes characteristic patterns of neighboring blocks and describes the picture regarding occurrence frequencies of these patterns.
Another approach is based on convolution neural networks (CNN). CNN recently gained popularity and became a state-of-the-art approach in the image recognition field.934 Initially, it was developed for recognition of handwritten text and proved to be superior to all other methods.935 As was reported by Caicedo et al., CNN is able to improve computation times by at least a factor of 16× in comparison to CellProfiler.936
CNN is also a segmentation free approach. The CNN algorithm has a multilayer structure that consists of nodes. Nodes are the schematic representation of the nonlinear transformation step and only nodes that are in adjacent layers are interconnected And these connections depict data flow. Layers can have an arbitrary number of nodes. The first layer of nodes uses the intensity values of individual pixels as input. In simple architecture, the last node outputs a probability that the image contains the specific object. CNN can adjust itself during training by modifying nodes.937
Moreover, Pawlowski et al. have shown that it is possible to use generic deep convolutional networks to extract features for cell morphological profiling. This approach is more accurate and faster in comparison with currently used methods and enables fully automated image processing without single-cell identification.938 Besides, Lafarge et al. have demonstrated that neural networks can be used for learning representations and generating realistic single cell reconstructions, which can be useful to gain better insight into cellular structure variations that are driving differences between populations of cells.939
7.5. Data Mining and Quality Control
7.5.1. Image Quality Control
In HCI, it is impossible to check the quality of all the images visually. Therefore, automatic methods should be performed. The most common problem is out of focus images. Assessing the log–log slope of the power spectrum of pixel intensities has been shown to perform well in measuring image blurriness in HCI. Its value is always negative. Images that typically have values in the range from −3 to −2 are in focus.940 However, it is advisable to check images that represent the whole range to decide on the threshold value to use. There is a potentially different source of Artifact in the images. The advice is to perform different metrics that can capture artifacts, and by using data, analytical approaches find the best threshold value that will be able to separate data with the acceptable quality. The rule of thumb, always checks what kind of images are eliminated by filtering. There is still a risk of removing unique phenotypes that are different from the rest of the data but are not artifacts.
One of the most accessible approaches that can be utilized is to use supervised machine learning, for example, implemented in CellProfiler Analyst as was proposed by Bray et al.941 It is based on training the model to capture a specific type of. The disadvantage of this approach is obtaining enough examples to teach the model.
7.5.2. Data Preparation for Downstream Analysis
When the data are sifted through a quality control filter, they should be prepared for the downstream analysis. Usually, in the result data, features are represented as columns and single cells as rows. First, it should be checked if any missing values are present. Many analytical tools are not able to use data that has missing values, in such cases, values can be imputed, or the whole row or column can be removed from further analysis. If data was collected in batches, it should be normalized by using the batch normalization technique as reviewed by Caicedo et al.895 The data also should be checked if any spatial biases exist in the data. For example, it is well-known that edge effects should be taken into account in HCI screening. After the data are normalized, they should be scaled. Features usually are represented by a different range of numbers. For example, the cell area can range from tens to a couple thousand while eccentricity from 0 to 1. If not scaled, some analytical techniques can weigh the “Area” feature as more important just because of the higher magnitude of numbers. Many analysis procedures assume a normal distribution of the data, which may not be the case. Data transformation can be used to make it normally distributed. Not all features in the HCI results are potentially useful for the downstream analysis. For example, features that have near-zero variance and cannot be used for distinguishing between conditions and can be excluded from further analysis. The dimensions can be reduced even further by excluding features that are highly redundant and correlate with each other. Examples of such features are Area and Perimeter. There are more complicated dimension reduction approaches that exist which have been reviewed by Li.942 It is not uncommon to aggregate data by averaging per cell data to per treatment. Finding medians of the feature per condition was found to be a robust approach. Alternatively, the median can be found per image, and then the average of those images per treatment. Once the general data preparation pipeline is executed, data can be used for downstream analysis. For example, for finding unique phenotypes or correlations between biomaterial properties and cell response as was reviewed by Vasilevich et al.55
7.5.3. Challenges in the Analysis of the Imaging Data
To be able to derive the right conclusions from the HCI data, it should be thoroughly understood. There are known problems that are inherent to all HCI screenings, such as plate layout effects, interbatch variation, out of focus images, materials autofluorescence, and if not addressed, can lead to false conclusions.943 However, issues can be screening-dependent. To identify the issues, it is advisible to assess the raw images carefully. The next step is to come up with a strategy to correct for them or take them into account in the downstream analysis. Sometimes raw images can be in complete disagreement with the data; this is possible if, for example, segmentation went wrong. In such cases, saving segmentation results and observing them in parallel to the raw images will be very beneficial.
7.5.4. Visualizations and Computational Tools for Data Exploration
It is not possible to spot all the issues by investigation of the raw images. In this case, data visualization techniques can be convenient, allowing understanding the data better. For example, visualization of one of the features in the form of a heat map that mimics the layout of the plate can immediately show a plate layout effect. Boutros and colleagues have developed HTSvis, a platform for interactive visualization of the raw data, to perform quality control and assess screening results.944
Commonly used dimension reduction techniques such as principal component analysis (PCA) can quickly indicate the batch effect or severe outliers, which can be artifacts or unique phenotypes.945 To distinguish them, the raw image should be observed.
Another handy tool in data exploration is interactive plots, which are available in R and Python as reviewed by Perkel.946 By using this kind of visualization, it is possible to explore data quickly and focus on data understanding, not the scripting.
Data exploration can be done in any spreadsheet program, for example, Excel. The most significant disadvantage is that it does not provide an opportunity to record all the data manipulations and therefore, does not guarantee reproducibility. Scripting based approaches, for example, by using most popular tools such as R, Python, and Matlab, allow for reproducing precisely the same results by rerunning code.947 Tools like R Markdown, IPython, or Jupyter notebook enable sharing and discussing results with the noncomputational colleagues by keeping scripts, detailed annotation, results, and visualizations in one place. In the next section, we will discuss in detail other computational approaches that are useful during biomaterial discovery.
8. Computational Tools for Materiobiology and Biomaterials Discovery
The amount of data created by HTS methods can be overwhelming and biomaterial scientists might be faced with questions like “Which are the best performing biomaterials?” or “How do I analyze such a large set of data and what is required for such analysis?”.55 The first part of this section will give an overview of the computational methods that allow finding patterns in cellular responses and make statistical predictions on cell–material interactions. Importantly, the biomaterial design space is very vast and continues to increase as new discoveries are made, including material stiffness, topography, chemistry, degradation rates, etc., which can also be arranged in a combinatorial way. As such, even though HTS methods allow us to screen thousands of materials in parallel, these methods alone will not be sufficient to navigate the enormous biomaterial design space.948 Recent developments in computational power do allow, however, developing computational simulations that can test hypotheses and perform experiments in silico, many orders of magnitude faster than real experiments. The second part of this section will provide insight into the powerful field of computational modeling and how it can be used to advance the field of materiobiology and biomaterial discovery.
8.1. Basic Principles
8.1.1. Basic Principles of Data Analysis
8.1.1.1. Parameterization of Biomaterials
To use computational and statistical tools for HTS data analysis, both the biomaterial as well as the cell response (see section 8.1.1.2) need to be characterized in a quantitative way. Defining and quantitatively measuring parameters is also called parametrization where parameters or features are defined as “a set of physical properties whose values determine the characteristics of an entity”.55 More specifically, possible parameters for biomaterials are (a quantitative description of) the chemistry, adhesivity, wettability, mechanical properties such as stiffness, elasticity, and hardness, electrical properties, degradability, topography at the macro-, micro-, and nanoscale, etc. (see refs (63, 523, and 563) for an extensive list of biomaterial properties). Note that many biomaterial parameters influence each other and cannot easily be separated experimentally. Moreover, even though many biomaterial properties are known to highly affect cell behavior, describing the biomaterial properties in a quantitative way is challenging and an increasing number of advanced descriptors are continuously being developed. Here, we give three examples of material descriptors: spectroscopy descriptors, molecular descriptors, and surface topography descriptors (Figure 41).
Figure 41.
Basic principles of data analysis: parametrization. The biomaterial is characterized by 9 features or descriptors whereas the cell response is described by 6 features or descriptors in this toy example. m/z, mass to charge ratio; #C–O, number of C or O atoms; #Δ, number of triangles present in a surface topography; #pits, number of pits present in a surface topography; gene A-B, relative gene expression versus a control; area, cell area; aspect ratio, cell aspect ratio (length/width); fluorescent intensity, the fluorescent intensity of a marker of interest.
Spectroscopy Descriptors. Spectroscopy descriptors, that is, features that represent the complete mass spectrum such as the position and height of the ion peaks, are one of the ways materials can be parametrized. Perez-Luna et al. used spectroscopic measurements (from static secondary ion mass spectroscopy and electron spectroscopy for chemical analysis) and WCA measurements of 21 polymers to predict fibrinogen adsorption and retention.949 Similarly, Urquhart et al. have used time-of-flight secondary ion mass spectrometry (ToF-SIMS) data, automated XPS, and WCA measurements of 576 copolymers to identify surface structure–property relationships from large polymer libraries.523 Using principle component analysis (PCA–see section 8.1.1.3) on the ToF-SIMS data, polymer groups could be distinguished based on their monomers. In another study, Taylor et al. used partial least-squares regression (PLS) to relate ToF-SIMS data to surface energy and investigate how the prediction accuracy varies with the number of samples used to build the PLS model.950
Molecular Descriptors. Molecular descriptors, which represent a molecule in a mathematical form, have been first developed in the chemistry field to derive quantitative structure–activity relationships (QSAR) or quantitative structure–property relationships (QSPR), that is, to predict the properties of chemical compound based on their molecular structure.951,952 Several software packages such as the Molecular Operating Environment (MOE),953 the CODESSA package,954 PaDEL,955 GRIND,956 and DRAGON957 can calculate the molecular descriptors based on theoretical considerations. Lewis et al. used 1D, 2D and 3D molecular descriptors to describe a new library of amphiphilic macromolecules and construct QSAR models for their antiatherogenic activity.958 They identified five key descriptors that were then used to correctly predict the efficacy of three newly designed amphiphilic macromolecules. Similarly, Bygd et al. used 36 molecular descriptors to relate in vivo macrophage polarization to particular biomaterial surface chemistries.959 Using the DRAGON and ADRIANA software, Epa et al. generated 68 chemically interpretable molecular descriptors and related those to the attachment data of three clinically important pathogens (i.e., Pseudomonas aeruginosa, Staphylococcus aureus, and uropathogenic Escherichia coli).614 Interestingly, the use of molecular descriptors rather than spectroscopic ToF-SIMS data generated QSPR models of higher predictive power and robustness.
Surface Topography Descriptors. In comparison with the development of accurate molecular descriptors, far less work has been performed on describing the surface structure or topography of biomaterials. Anselme et al. have used fractal dimension, in addition to classical roughness descriptors, to describe the surface organization.960 Their work showed, based on 180 samples, a lower proliferation of human osteoblasts on less organized surfaces (i.e., the sand-blasted orthopedic Ti6Al4 V alloys). Similarly, Wickens et al. used multifractal spectra to investigate the effects of surface topography on bacterial distribution.961 Interestingly, they showed that both bacteria were influenced by surface topographical and chemical effects, although Staphylococcus aureus appeared to be more influenced by surface topography and Staphylococcus epidermidis by surface chemistry. Another approach to characterize the surface structure is to directly use the surface design parameters. For example, Robotti et al. created 12 different surface structures consisting of hexagonal pits, aligned in hexagonal and squared patterns.962 As parameters they used the diameter of the pits, the interpit distance, and the shape of the pit pattern, although no further modeling was performed. A more diverse set of surface topographies was developed by Unadkat et al., who used primitives (circles, triangles, and rectangles) to design 2178 different surface topographies combined on a HTS platform called the “TopoChip” (see section 5.2.1.2).455 Several predictors have been identified that are able to reveal the effect of surface topography on cell morphology,144 Oct4 expression in human induced PSCs,564 osteogenic differentiation,566 and epidermal stem cell differentiation.568
Once all the parameters describing the biomaterial have been selected, each biomaterial sample can be represented by a feature vector, that is, the set of parameter values for that sample. The size of the feature vector represents the dimensionality of the feature space (9 in the toy example of Figure 41). The set of all feature vectors, corresponding to all samples in an experiment represents the input data matrix.
8.1.1.2. Parameterization of Cellular Responses
Similar to the biomaterial, also the cellular response, the other side of the cell–biomaterial interface, needs to be parametrized (Figure 41). Here, possible parameters include cell shape, gene expression, cell differentiation potential, etc. As the measurement methods need to be high-throughput compatible and ideally produce a rich description of the cellular state by producing multiple distinct measurements at the same time, high-content imaging and gene expression analysis are two widely used techniques. A detailed description of these techniques can be found in Section 7. The set of parameter values that describes the cellular state of a biological sample, that is, gene expression values or fluorescent intensities, represents the biological feature vector. The set of all biological feature vectors, corresponding to all biological samples in an experiment is the output data matrix, assuming that the cellular response will typically be the output that needs to be predicted. For example, Dhaliwal et al. used machine learning to link the 3D spatial organization of splicing factor SC-35, captured by high content image analysis, to hMSC cell phenotype in three different 3D scaffolds (collagen hydrogels, salt leached scaffolds and electrospun mats).892 More specifically, the 22 textural descriptors for the 3D SC-35 organization were linearly reduced to a minimum number of eigenvectors that accounted for 95% of the data variance by principal component analysis. The predictive classification model was made using J48 decision tree analysis in the Weka software.
8.1.1.3. Data Exploration
In some cases, HTS data will be used to answer well-defined questions, such as finding which biomaterial performs best and which biomaterial properties relate to biomaterial performance (see section 8.1.1.4 below).963 It can also be interesting to explore the HTS data (see Figure 42A for an instructive example) to find novel, maybe nonintuitive, patterns in an unbiased fashion. For example, which biomaterial groups induce similar cell responses or, which cell morphologies are related to a particular cell differentiation status? The challenge of finding groups of biomaterials (or cell morphologies) can be translated into finding clusters of data points that are close together. To find these clusters of data points, the measurement results are typically visualized in, for example, scatter plots (Figure 42B). A scatterplot visualization is relatively straightforward for two parameters (note that it is not known a priori which parameters will yield informative plots), but plotting three or more parameters simultaneously, which is common for biomaterial and cell response descriptors, is not trivial. A solution to this problem is to decrease the number of parameters (dimensions) in such a way that the data can be visualized but that the distribution of the data in the original space is still maintained. In other words, plotting “transformed” data points in 2D but making sure that the data points that are close to each other in the original space are also close in the transformed 2D space. The transformation from multiple dimensions to two can be performed by various dimension scaling methods such as principle component analysis (PCA964), independent component analysis (ICA), multidimensional scaling (MDS), and many others. The main differences between these methods lie in how they decide which parameters are important and how the multidimensional set of parameters is distributed over a reduced set of (combined) parameters.963 However, for all scaling methods, the interpretation of the transformed, 2D results in terms of the original parameters is difficult as the newly transformed parameters (plotted along the x and y-axis) are complex combinations of the original set of parameters. A detailed description of these methods is beyond the scope of this chapter, but we refer the reader to the following reviews.965−967
Figure 42.
Basic principles of data analysis: data exploration and statistical modeling. (A) Input and output data matrix. (B) Scatterplot of the number of cells as a function of stiffness and WCA. Every sample is one data point. (C) Hierarchical clustering of the 6 data points into a dendrogram. The dendrogram or tree can be cut at any height to determine data clusters. Cutting the tree at the highest level (dashed line) results in 2 clusters (samples 1–3–6 and 2–4–5). Cutting the tree at a lower level (broken line) results in 4 clusters (samples 1–3, 6, 2–4, and 5). (D) On the basis of stiffness, the data points can be classified into two groups: high # cells and low # cells. (E) Simple linear regression where the dependent variable Y (here: the amount of cells) is predicted using the independent variable X (here: stiffness). (F) Classification of data points into two classes (open and solid circles) with an underfitted (gray line), reasonable (black curve), and overfitted (dashed curve) decision boundaries. The overfit is influenced by one black data point (arrow) and would classify a new point (blue dot) as a solid circle, which would probably be an error. (G) Example of K-fold cross-validation with K = 5. Reprinted with permission from refs (55) and (972). Copyright 2017 Elsevier, Ltd. and 2016 Nature Publishing Group.
Another way to explore HTS data is to group the data using clustering techniques. K-means clustering and hierarchical clustering are two common methods to find data points that are close together. Briefly, first, the number of clusters (k) to group the data points into is specified to initialize the k-means clustering algorithm. Then the algorithm will randomly pick k points as starting centers for the k clusters and assigns each data point to the closest center. Next, the algorithm will update the centers of each cluster to a new center point (based on the assigned data points). This process is repeated until the cluster centers, or assigned data points, do not change significantly from one iteration to the next. Note that the k-means clustering algorithm requires some a priori knowledge on the number of clusters, although this can be accommodated by trying several values for k and quantifying cluster quality using, for example, the silhouette coefficient (see refs (968 and 969) for more information).
Hierarchical clustering uses a different approach to group data points, that is, it iteratively groups the most similar data points together into larger clusters, resulting in a tree or dendrogram (Figure 42C). The clusters of data points can be obtained by (arbitrarily) cutting the tree at a particular height. Importantly, when interpreting the result of a clustering algorithm, one should remember that the above clustering algorithms will always generate a clustering result, even if there is no real evidence for clusters in the data. As such, the clustering results should be further inspected by visualizing them with complementary methods such as a heat map or a scatter plot.963 Besides the k-means and hierarchical clusters, several other techniques have been comprehensively reviewed by Jain et al.970 and Xu et al.971
8.1.1.4. Statistically Modeling the Relation between Biomaterial Performance and Biomaterial Descriptors
Ideally, the biomaterial performance can be predicted from first-principles, that is, from known “physical laws” such as quantum mechanics laws that allow to predict the molecular structure from the molecular composition. However, there are two main limitations to this modeling approach: (1) quantum mechanical or molecular dynamics simulations quickly become computationally impossible for large-scale systems, (2) “physical laws” describing the relationship between biomaterial properties and cell behavior are currently inexistent.963 To solve this problem, statistical models are being developed that explain the experimental results starting from sufficient available data (top-down), in contrast to computational models that start from known principles (bottom-up; see also section 8.1.2).
Model Choice. Finding a plausible model that explains the experimental results, comes down to finding the function f in y = f(x) with y the outcome measurement (e.g., cell response) and x the input biomaterial parameters (features) using training examples. Machine learning algorithms will use the training examples to find a model that accurately predicts the outcome measurement given the biomaterial features. This phase of model development is also called model training. The outcome measurement can be broadly specified into two different categories: the outcome measurement is a continuous value (e.g., the amount of bone, the fluorescent intensity of a biomarker, the fold change of a gene) or it is a discrete value (e.g., bone formation/no bone formation, differentially expressed gene or not). The machine learning algorithms to train the first type of models are called regression algorithms whereas the latter is called classification algorithms. The name classification comes from the machine learning field where a set of data points with a common label is called a class, for example, the classes of high and low number of cells (Figure 42D) or low, medium, and high bone forming materials. In other words, classification algorithms require labeled data, that is, data points that belong to particular discrete classes, which may be known from the experimental design or can be found through clustering techniques (see section 8.1.1.3).
For both classification as well as regression, a wide variety of algorithms is available depending on the required complexity of the data. For example, the simpler algorithms such as linear regression fit a linear regression function y = β0 + β1x1 + β2x2 + . . . + βnxn to the data. More specifically, the algorithm will estimate the βi values from the training examples that best predict the outcome y (e.g., cell response) from the inputs x1, x2, . . .xn (e.g., the biomaterial parameters) (Figure 42E). Note that finding the βi values not only allows predicting the outcome of novel biomaterials but immediately also sheds light on the most important biomaterial properties contributing to that response (i.e., the biomaterial parameters xi with high βi). If necessary, more complex algorithms are available to model nonlinear responses such as neural networks, decision trees, random forests, and support vector machines,973−978 all available in versions for classification (to predict a class) and regression (to predict a continuous value).
Choosing the adequate algorithm and associated complexity is difficult as potential nonlinearities or other relations between input and output are not known a priori. Moreover, the model choice is a balancing act between overfitting and generalization (Figure 42F); a good model should be complex enough to capture the underlying relations but simple enough so that it has sufficient generalization power. In other words, a complex algorithm with many degrees of freedom will perfectly reproduce the training data responses but will fail to properly predict novel data as it does not capture the true underlying relations (overfitting). This can be compared to a student who learned all the answers of a list of exam questions by heart (overfitting on the training examples) but cannot correctly answer to variations of these questions (generalization) as the student does not grasp the underlying concepts or theory.963 To avoid overfitting one can increase the training set so that the model is not able to capture the response of every individual data point, forcing it to generalize. A large training set, however, comes at a cost as it requires many experimental data that are not necessarily available. Another solution it to reduce the model complexity (less degrees of freedom) so that the model cannot fit every parameter perfectly, again forcing the model to generalize.
Model Testing. To test whether a model is sufficiently generalizable, the predicted response of a model f(x) will be compared with the real measured value y, for a set of data points y and x that were not used during the training phase. To maximize the use of the data points, cross-validation can be used (Figure 42G). In this scheme, one splits the data into K equally sized partitions. Then one partition is held out (test set), the model is trained on the other partitions and the model performance is calculated on the test set. This process is repeated for each of the partitions and the overall model performance is calculated by averaging the performances of all K iterations. Other model testing techniques are discussed in other works.979 To quantify the difference between predicted and measured biomaterial performance, various measures exist. For classification models, one typically uses accuracy, which measures the number of correct class predictions, known from the comparison between the predicted and the real class labels.
For regression models, where a continuous output instead of a discrete class label is predicted, one can use the Pearson correlation coefficient between the known and predicted performances as a measure of performance.
Model Use. Once a (statistical) model has been build and tested, a mathematical input-output relation has been found and can be used for various purposes. First, the model components can be investigated in more detail to improve the understanding of the underlying system. For example, a (linear) regression function y = β0 + β1x1 + β2x2 + ... + βnxn identifies which biomaterial parameters (xi) influence the biological output (y). Moreover, the coefficients corresponding to these biomaterial parameters (βi) highlight which biomaterial parameters contribute most to the biological output (i.e., the biomaterial parameters xi with high βi). Second, the statistical model can be used to predict the biological output of new biomaterials, given their biomaterial parameters. For example, assume a model was built to predict the amount of cell proliferation based on the following calcium phosphate biomaterial parameters: type of calcium phosphate, dissolution rate, crystallinity and water contact angle. The model was trained and tested on various calcium phosphate materials. Next we can use the model to predict the amount of cell proliferation from the four above biomaterial characteristics of other materials, for example, calcium phosphate materials with new (untested) ratios of HA and TCP. Third, the above example model can also be used to determine the optimal dissolution rate that maximizes or minimizes cell proliferation. Note that for the last two applications, prediction and optimization, it is important to critically assess the model results. More specifically, a (statistical) model is trained on a particular data set, and thus its predictions are valid within a particular parameter space. In other words, the predictions of the above example model might hold well for all types of calcium phosphate materials but might not extrapolate well to polymers, which might require another data set or are characterized by a different set of predictive biomaterial parameters.
8.1.1.5. Statistical Analysis
Does a Biomaterial Property Influence the Cell Response? One question that can be asked when analyzing an HTS data set is whether the biomaterial design significantly affects the biological performance, that is, induces the desired cell response. For example, can we show, based on the experimental data, that the biomaterial stiffness has a significant influence on cell differentiation? Such questions can be answered through a “one-way analysis of variance” ANOVA test.980 More specifically, given the assumption that the actual material performances of k materials are equal (i.e., y1 = y2 = . . . = yk), the ANOVA test will calculate a p-value representing the likelihood of measuring material performance differences exceeding those that have been observed.963 In other words, one tests whether the variance in measurements can explain the observed performance differences. If the p-value is below a user-defined threshold (typically 0.05 or 0.01), the equality assumption is rejected resulting in the conclusion that there is a significant material effect on the performance of (one of the) materials. Importantly, a p-value above the user-defined threshold does not imply that there is no material effect. The effect might be hidden by other sources of (technical) variation. When the material performance is expressed by multiple values, a multivariate version of ANOVA, MANOVA, can be used.981
Does Material A Perform Better than Material B? To answer this question, which determines whether one of the tested biomaterials performs better than a reference or a control condition, a Student’s t test can be used.982 This statistical test determines for a particular material mi whether the hypothesis that it performs equally well or worse than a reference material can be rejected. Importantly, in a high-throughput setting this test is performed for every material, leading to the multiple testing effect. More specifically, for a single test, there is a chance of finding a false positive result once in every 20 tests (given a p-value threshold of α = 0.05). For single tests, this error is typically accepted but when this test is applied to, for example, 1000 materials, 50 materials will significantly outperform the reference material only by chance (false positives). Many corrections have been developed to account for the multiple testing effect, including the Bonferroni correction, the false discovery rate, the Benjamini-Hochberg procedure etc.983,984 Importantly, the higher amount of tested materials in HTS platforms does not reduce the required number of replicates per material. On the contrary, due to the multiple testing corrections, the requirements for significance are more stringent, resulting in more statistical power (more replicates) required for a significant result.963
Which Materials Are “Hits”? How to Rank Materials Based on Their Performance? A straightforward way to find the most promising biomaterials in terms of performance would be to order them according to their observed performance (averaged across replicates). A problem with this approach lies in the potential influence of outliers: materials could be ranked highly only because of one highly performing replicate, skewing the average performance of that material. A better measure is the median value across replicates as this is less affected by outliers. Another alternative is ranking materials based on their p-value obtained from a performance comparison with a reference material. This method is more robust to outlier effects as the variance of the replicates as well as the number of replicates is taken into account.
8.1.1.6. Software and Tools
There are many software tools for machine learning available, a nonexhaustive summary is given in Table 5. Most software tools require some programming knowledge. Fortunately, excellent online courses and tutorials985−987 exist such as Codecademy, Udacity, and Software Carpentry.
Table 5. Overview of Popular Machine Learning Tools.
| tool | license | programming language | data set size | development mode | website |
|---|---|---|---|---|---|
| Weka | open source | Java | small data sets | GUI-based; many algorithms are already implemented | https://www.cs.waikato.ac.nz/ml/weka/ |
| DL4J | open source | Java | larger data sets | code-based | https://deeplearning4j.org/ |
| TensorFlow | open source | Python | big data sets | code-based | https://www.tensorflow.org/ |
| PyTorch | open source | Python | big data sets | code-based; many inbuilt algorithms | https://pytorch.org/ |
| R | open source | R | big data sets | code-based; many inbuilt algorithms | https://www.r-project.org/ |
| RapidMiner (similar to Weka) | open source | Java | big data sets | GUI-based | https://rapidminer.com/ |
| MATLAB | closed source | MATLAB; can interface with programs written in C++, Java, R, and Python | medium sized data sets | code-based; many inbuilt algorithms | https://nl.mathworks.com/?s_tid=gn_logo |
| GNU Octave (alternative for MATLAB) | open source | Octave | medium sized data sets | code-based | https://www.gnu.org/software/octave/ |
8.1.2. Basic Principles of Computer Simulations
8.1.2.1. Motivations for Mathematical Modeling
As mentioned in the sections of this review, the biomaterial design space is so enormous that even HTS methods will not allow us to explore all possible biomaterial combinations. To further speed up development, however, one can make use of mathematical models and computational simulations to test hypotheses and perform experiments in silico, many orders of magnitude faster than real experiments and make extrapolations to patterns that are not present in the original data (see Table 2).948 Importantly, the biomaterial design space is not only very vast; the cell–biomaterial interface is also a very complex biological system due to four main reasons. First, the cell–biomaterial interface consists of a very large number of components, that is, different biomaterials and various types of tissues and cells, on their own consisting of thousands of genes and proteins. Second, there is a multitude of spatiotemporal (unknown) interaction processes among this very large number of components. Third, many biological processes are nonlinear, implying that if input m1 leads to output y1 and input m2 leads to output y2, input m1+m2 does not lead to output y1+y2. In other words, we cannot analyze nonlinear biological systems by analyzing the system piece by piece and in the end add all input-output relations as one runs the risk of destroying some system properties emerging from the interactions between them.988 Fourth, many biological systems have invisible thresholds, that is, as long as a parameter is below a threshold, the system is in a stable state (e.g., inactive) but as soon as the parameter exceeds the threshold the system responds in a different manner, for example, by activating other downstream components.988 Because of this complexity, one cannot make reliable predictions (or extrapolations) about biological systems with intuition alone. Interestingly, mathematics naturally deals with thousands of variables and nonlinearities, providing us with an excellent tool to explore the cell–biomaterial interface.
8.1.2.2. What Is a Mathematical Model?
A mathematical model is defined as an abstract representation of a complex system in terms of equations or rules and a description of the region (spatially or temporally) on which they are valid.990 Modeling is the process of creating such an abstract representation and generating new insights from it. The task of creating a mathematical model for the cell–biomaterial interface is not trivial, as we cannot resort to fundamental laws and principles as in physics (e.g., Newton’s law to describe falling objects). Considering the astonishing complexity and diversity of biological phenomena, it is logical that mathematical models also come in many types and forms, which all have their strengths and weaknesses. Indeed, the art of modeling consists, among others, in defining the correct framework suitable for the investigation of the research question at hand. Therefore, the next paragraph will focus on the common aspects of the modeling process itself, without providing an extensive survey of all possible approaches and numerical implementations for which we refer the reader to the following excellent reviews.988,991−994
8.1.2.3. Steps of Building Mathematical Models
The generic modeling process consists of four phases: model selection, model design, model analysis, and diagnosis, and model use (see Figure 43).988 To find the right balance between capturing the underlying mechanisms without adding unnecessary complexities, some iterations might be necessary. Selecting which simplifications to make requires detailed knowledge of the system that is studied as well as mathematical skills. As such, a close collaboration between modelers, material scientists, and biologists can be very fruitful, especially when they understand each other’s jargon and work in a tight feedback loop going from in silico prediction and hypothesis generation to in vitro or in vivo validation. To illustrate all the steps of the modeling process, a simple example will be used throughout: “how do biomaterials influence cell proliferation?”.
Figure 43.
Flowchart of the modeling process, illustrated by a toy example of the influence of biomaterials on cell proliferation. a.u.: arbitrary units. Adapted from ref (988) and reprinted with permission from ref (55). Copyright 2008 Georg Thieme Verlag KG and 2017 Elsevier, Ltd.
Step 1: Model Selection. Goals and Data Availability. Before selecting a proper mathematical model type, one needs to define the purpose and goals of the model as well as make an overview of the data available to calibrate and validate the model. As such, the research question defined above - “how do biomaterials influence cell proliferation?” - needs to be much more specific. Is one interested in a particular cell type or should the model be generalized to all cell types? Is one interested in one biomaterial type, and which particular properties thereof? Are these properties the same throughout the material or is there local variation that needs to be included? Is one interested in the in vitro response or the in vivo one (if so, which tissue, animal?)? Is one interested in capturing the 3D dynamics or is a 2D approximation sufficient? Which scale does one want to capture, the intracellular gene and protein dynamics occurring during cell proliferation or cell or tissue level proliferation? Is one interested in an end-point or the dynamics over time? etc. Importantly, this wish list of details to be included in the model will depend on the a priori available knowledge of the system and the available data (e.g., if one wants to include temporal dynamics, experimental measurements at different time-points will be essential to calibrate the model).
Model Selection. Once the goals of the model are clear, decisions need to be made regarding the model type. Note that these decisions do not relate to the model’s usefulness or quality, they are solely based on the available data, a priori knowledge and scope of the model.988
Mechanistic versus Data-Driven. A mechanistic model will try to link the inputs and outputs of a biological system by describing the actual underlying (biological) mechanisms (see Table 6). For the toy model, this implies having a basic understanding of cell proliferation (e.g., functioning of the cell cycle, essential proteins and genes involved therein, the dependence of the proliferation rate on the cell density and available resources, etc.) and a corresponding data set. In contrast, a data-driven model will correlate the inputs and outputs in a direct (statistical) way, often called a black box. In the context of the toy model, a data-driven model might be a linear regression between stiffness values and proliferation rates, based on an extensive HTS data set (see section 8.1.1.4 and Figure 42). Note that these data-driven models can find correlations, but do not explicitly explain the causes and mechanistic relations between input and response.
Table 6. Advantages and Disadvantages of Mathematical Modeling and Data-Driven Modelinga.
| mathematical modeling (section 8.2.2) | data-driven modeling (section 8.2.1) |
|---|---|
| provides a mechanistic relation between inputs and outputs | provides a statistical relationship between inputs and outputs |
| capable of handling small (dedicated) data sets | requires large (HTS) data sets |
| upon validation, the model can be used as a predictive tool for difficult or costly to perform experiments | upon validation, the model can only make predictions related to existing patterns in the supplied data |
| deductive | inductive |
Adapted from ref (989).
Static versus Dynamic. If the system changes appreciably over time, the temporal dynamics might be important to capture. In the toy example, one would like to understand cell proliferation, implying that the amount of cells changes over time, requiring a dynamic model.
Deterministic versus Stochastic. In a deterministic model, all information is known at the beginning and once all settings are defined, all future predictions are fully determined (always the same for the same parameter value settings). In other words, a deterministic model neglects the influence of noise or fluctuations on the biological process. Depending on the importance of noise in the modeled process, this simplification is justified, as probabilistic modeling is mathematically much more complicated and may not be worth the effort. In the toy example, given that, the available data focuses on recording average cell densities over time (neglecting single cell proliferation states); a deterministic model would be a good starting point.
Spatially Distributed versus Spatially Homogeneous. If the biological process under study varies greatly with space, it might be warranted to account for the spatial effects instead of using a spatial average. Assuming that cell proliferation is spatially homogeneous, one can opt for a nonspatial model. Note that in Figure 43, the model design captures the change of P with respect to time with a simple derivative (i.e., ordinary differential equation); the changes of P with respect to space are neglected.
Open versus Closed. A model can be open or closed, depending on whether components can enter or leave the system. In the toy model, one can decide to start from an initial population of cells (e.g., the initial seeding density in the culture flask) and follow this density as a function of time without the addition of new cells (closed system). In vivo, on the other hand, migration of cells toward the region of interest might occur, obscuring the proliferation effect of the biomaterial and requiring an open model formulation.
Step 2: Model Design. Model Components. Most models will contain several or all of the following components: variables, parameters, and processes that link the variables.988 The variables represent the biological and biomaterial entities of interest, such as genes, proteins, cells, biomaterial properties, etc. In the toy example, the variables are the cell density (P), time (t), and biomaterial stiffness (m). The parameters are the numerical values that characterize particular features of a variable or process. For example, the carrying capacity (K), the growth rate (r) or a stiffness threshold (a). The processes describe how the variables are linked, for example, the change in cell density is proportional to the existing population or the higher the stiffness m of a biomaterial, the lower the cell proliferation.
Model Implementation. Once the mathematical structure is clear and the variables and parameter values are known, the model can be implemented. Typically, one will first formulate the corresponding set of equations and then convert them into computer code to solve the set of equations numerically. Note that, depending on the complexity of the mathematical model, the equations can also be solved by hand or analytically. To be able to run the simulations, actual values need to be specified for the parameters and initial settings of the variables. Parameter values can be determined in many ways (e.g., literature, other mathematical models, dedicated experimental data) but very often parameter determination is the most challenging bottleneck in the modeling process. For detailed approaches to determine parameter values, we refer the reader to other literature.995−997
Step 3: Model Analysis and Diagnosis. Before using the model for its intended goals, one typically first tests the model as thoroughly as possible to (1) ensure there are no obvious implementation errors, (2) delineate which parts of the biological process under study are reasonably modeled and which fall beyond the scope of the in silico model, (3) explore the robustness of the predictions and the sensitivity of these predictions to the parameter values, (4) explore nonintuitive dynamics.988 Some of these tests can be done with mathematical methods or through computer simulations where one repeatedly introduces deviations (e.g., changes a parameter value 1–50%) and studies how the system responds.998,999 In the toy example, cell proliferation becomes exponential if the resources are unconstrained (see Figure 43), which might be a reasonable approximation or warrant a more complex description to capture, for example, the process of contact inhibition. From Figure 43, it is also clear that the model predictions are very sensitive to the exact value of K (the carrying capacity), implying that this parameter value should be accurately determined. If the simulations need to represent experiments that were executed in a lab, one will question whether the simulations generate responses that are consistent with the experimental observations (see Figure 43).
Step 4: Model Use. Once the model has been sufficiently tested, the model can be used to answer “what if?” scenarios. For example, one can vary the growth rate (r) at any magnitude, in combination with variations in the carrying capacity (K) (which is difficult to modify experimentally) or on its own, to explore the influence of r on the cell density. This set of in silico experiments can be complemented with an explanation of how the input (in the computational model) leads to the observed response. In particular, with an in silico model, the data can be nondestructively recorded at a higher frequency and spatial resolution than in an experimental setting, limited only by the temporal and spatial resolution of the numerical simulation. This allows capturing all interesting dynamics versus the static and user-defined timing of data collection, potentially overlooking interesting phenomena.55 Another use of the computational model is to find optimal settings for a particular goal, for example, an optimal culture refreshment regime (amount and timing in between) that maximizes in vitro cell proliferation. Finally, the computational model can be used to test and explain hypotheses, for example, why is the first step of the cell cycle influenced by the stiffness of the biomaterial and not the second or third? This type of exploration could be done by making two types of models, one with stiffness feedback to the first step and one to the second step and comparing the resulting system behavior. Importantly, although valid model parameters (determined by physical laws) and model structures are required for realistic predictions, running a model with “unrealistic” values or processes might yield interesting observations (e.g., what kind of growth dynamics would be predicted for an infinite amount of resources? See Figure 43).
8.1.2.4. Software and Tools
An overview of some popular simulation tools can be found in Table 7. Note that there is a wide variety of tools depending on the type of simulation (e.g., solving for fluid flow using Comsol Multiphysics versus intracellular signaling dynamics using VCell).
Table 7. Overview of Popular Simulation Tools.
| tool | description | programming language | license | development mode | website |
|---|---|---|---|---|---|
| COPASI | software application for simulation and analysis of biochemical networks and their dynamics | C, SMBL | open source | GUI + code-based | http://copasi.org/ |
| VCell | VCell, (Virtual Cell) is a comprehensive platform for modeling cell biological systems that is built on a central database and disseminated as a web application. | VCell Math description language, SBML | open source | GUI + code-based | https://vcell.org/ |
| COMPUCELL3D | COMPUCELL3D is a software framework for three-dimensional (3D) simulation of morphogenesis in different organisms that uses a stochastic cellular potts model (CPM) and PDE solvers. | C++ | open source | GUI-based | http://www.compucell3d.org/ |
| FLAME | FLAME is a generic agent-based modeling system | XML | open source | GUI-based | http://flame.ac.uk/ |
| FreeFEM | FreeFEM is a partial differential equation solver for nonlinear multiphysics systems in 2D and 3D | C++ | open source | code-based; many algorithms implemented | https://freefem.org/ |
| FEBIO | FEBio software suite is a set of software tools for nonlinear finite element analysis in biomechanics and biophysics | C++ | open source | GUI-based | https://febio.org/ |
| Comsol Multiphysics | a cross-platform finite element analysis, solver, and multiphysics simulation software. | Comsol language | closed source | GUI-based | https://www.comsol.com/ |
| MATLAB | is a multiparadigm numerical computing environment and proprietary programming language developed by MathWorks. | MATLAB; can interface with programs written in C++, Java, R, and Python | closed source | code-based; many inbuilt algorithms | https://nl.mathworks.com/?s_tid=gn_logo |
| GNU Octave (alternative for MATLAB) | GNU Octave is a high-level language, primarily intended for numerical computations | Octave | open source | code-based | https://www.gnu.org/software/octave/ |
8.2. Data-Driven Models in Biomaterials Research
As 90% of the world’s data has been generated in the last five years,1000 the use of data-driven models has vastly increased in the biomedical and clinical sciences to keep up with the rate of data generation, find novel correlations and define new hypotheses. Here we will focus on data-driven models related to cell–biomaterial interactions, and particularly those that use data derived from HTS platforms.
8.2.1. Surface Chemistry
To relate the microscopic or molecular properties of polymers to their biological effects, and in particular human embryoid body cell adhesion,525 bacterial attachment,475,614 and human dental pulp-derived stem cell behavior,1001 polymer microarrays and machine learning methods have been developed previously (see also sections 5 and 7). In these studies, they have explored various polymer descriptors including computational (molecular) descriptors,475,1001 WCA,116 ToF-SIMS spectra,475,1001 or mechanical properties.525 Ideally, purely computational descriptors are used as this will accelerate new materials discovery by reducing the need for additional experimental measurements to characterize the material properties of large materials libraries.525 For computational modeling, among others multiple linear regression,475 partial least-squares regression,1001 and three-layer neural networks,475,525 have been explored.
8.2.2. Surface Topography
Hulsman et al. explored various statistical tests to rank the biomaterial surfaces.144 They found that the Mann–Whitney U-test significantly outperforms the mean or Welch t test as it takes into account the distribution of the replicate measurements and can better handle outliers. To relate cell behavior to morphological descriptors or surface topographical descriptors, random forest algorithms,897 logistic regression,564 ordinary linear regression,144 Lasso regression,144 M-estimation regression,144 and support vector regression144 were explored.
8.2.3. Cellular Profiling
Next to other forms of cellular parametrization, cell morphology has also been identified as a potential indicator of cell–biomaterial response. Chen et al. used support vector machines to quantify the morphological response of hBMSCs to different microenvironments.1002 Their analysis was conducted at a multicell level, where a “supercell” represented the average shape measurements of small groups of single cells to account for heterogeneous populations and microenvironments. They found that smaller, more elongated shapes are strong indicators of hBMSCs response to fibrous microenvironments whereas osteogenic supplements triggered morphological changes in terms of cell boundary concavity and roughness.1002 Similarly, Treiser et al. used 43 morphometric descriptors, with a particular focus on the spatial and structural organization of actin to forecast the lineage commitment of hMSCs.896 They used MDS to reduce the number of descriptors such that the observed similarities and dissimilarities between cells are maximized while being able to plot each cell in 3D. They could discern hMSCs cultured under osteogenic versus fat-inductive conditions as well as predict the degree of bone predisposition across a series of synthetic polymeric materials.896 Rostam et al. compared various machine learning methods (i.e., logistic regression, random forest, naïve Bayes, support vector machines and k-nearest neighbors) to identify M1 and M2 macrophage phenotypes.1003 Logistic regression and random forest showed the highest classification accuracy while support vector machines and k-nearest neighbors regularly misclassified cells. As such, high-content imaging and analysis in combination with data-driven modeling can be used to phenotype diverse cell populations without the need for multiple markers.
8.3. Hypothesis-Driven Models in Biomaterials Research
Many hypothesis-driven models are being developed to improve our fundamental biological understanding as well as to develop optimized treatments and devices. Here, we will focus specifically on the modeling of cell–biomaterial and cell–ECM interactions.
8.3.1. Hypothesis-Driven Models for a Fundamental Understanding
A recent review of Cheng et al.,1004 including commentaries thereon,1005−1008 provides an excellent overview of current mathematical models describing the responses of a cell to various biophysical cues in their microenvironment, such as dynamic strain, fluid shear stress, mechanical force, matrix rigidity, and matrix porosity. Here, we provide some highlights, particularly focusing on cell–biomaterial and cell–ECM interactions.
When cells interact with the ECM, they will deform and reorganize their actin fibrils, which feeds back on cell contractility and signaling. Abhilash et al. developed a computational model to capture cell-mediated ECM remodeling using finite element-based discrete fiber network simulations.1009 Interestingly, in agreement with experimental observations, the sensing distance of a cell is ten times the cell size due to long-range force transmission in the ECM through tension-driven fiber alignment. Similarly, van Oers et al. used a hybrid Cellular Potts and finite element computational model to show that a combination of contractile forces exerted by endothelial cells on the ECM, the resulting strains, and cellular response to these strains is sufficient to reproduce vascular network formation and sprouting.1010 Focusing more on the intracellular aspects of ECM sensing, Sun. et al. have developed a novel computational model of YAP/TAZ signaling and cytoskeleton dynamics in which the ECM stiffness is converted into biochemical signals.1011 They show that the synergistic effect of YAP/TAZ activity between the mechanosensing and Hippo pathways might be explained by the interaction of LIM-kinase and large tumor suppressor (LATS).
Besides shedding light on cell–ECM interactions, computational models can also play an important role in understanding the response to engineered biomaterial surfaces and implanted biomedical devices. Kang et al. have developed a mathematical model to understand biomaterial-mediated inflammatory responses.1012 For the parameter estimation, they used a global optimization algorithm called discrete selection Levenberg–Marquardt (DSLM). Albert and Schwarz have developed mathematical models, based on a cellular Potts model formalism, to predict the dynamical forces and cell shapes on micropatterned substrates.1013−1016 The in silico models are able to describe how the cell contour adapts to the pattern’s geometry as well as predict the traction forces in agreement with experimental data. In the next step the model was combined with genetic algorithms to optimize the migration of cells on ratchet micropatterns.1016 As was demonstrated experimentally, a triangular shape was predicted to guide cell migration in the direction of the tip of the triangle. However, to achieve unidirectional migration the most effective pattern consisted of connected, asymmetric and rotated triangles. An overview of computational models of substrate-based cell motility is given by Ziebert et al.1017
8.3.2. Hypothesis-Driven Models for Biomaterial Optimization
To improve cell viability after implantation of tissue engineering constructs, Pedraza et al. have developed and optimized hydrolytically activated oxygen-generating biomaterials.1018 More specifically, COMSOL Multiphysics was used to predict the oxygen gradients within cell-loaded agarose constructs with varying total cell loads, external oxygen tension and the presence or absence of the oxygen-generating biomaterial (modeled with first-order reaction rate kinetics). From these calculations, they could optimize the dose, geometry and surface/volume ratio of the oxygen-generating biomaterial. Similarly, others have focused on optimizing the release of ions from biodegradable scaffolds. For example, calcium phosphate materials are widely used for dental and orthopedic applications and their degradation products, Ca2+ and Pi ions, are believed to affect bone cell chemotaxis, proliferation and differentiation. However, the ion release rate from these scaffolds depends on a number of biophysicohemical phenomena such as dissolution and diffusion, which in turn depend on the scaffold composition and geometry. Manhas et al. have developed a finite element method to investigate the local Ca2+ ion release from CaP-based scaffolds.1019 Combining this work with a computational model of Ca2+ influence on osteogenic cell behavior and bone formation,999 allowed for CaP scaffold optimization.1020 Besides optimizing tissue engineering constructs for oxygen transport and release rate, others have optimized the macroscopic pore shapes of scaffolds to control the kinetics of tissue deposition, for example, bone formation. For example, Bidan et al. developed a model to predict the behavior of osteoblasts on curved surfaces.1021 They show that in cross-shaped pores the initial overall tissue deposition is twice as fast as in square-shaped pores, opening new avenues to improve the speed of bone ingrowth into porous scaffolds.
9. Clinical and Commercial Translation
Biomaterials with appropriate chemical, physical, and biological properties have the ability to revolutionize the diagnosis and treatment of challenging diseases such as cancer, tissue injury, and defects.5,1022,1023 Millions of patients worldwide are benefiting from biomaterial-based products, such as surgical meshes, dermal fillers, pacemakers, stents, joint replacement devices, controlled drug delivery devices, dental and cosmetic implants, surgical adhesives, antiadhesive devices, hemostats, etc. Particularly, in the field of tissue engineering, recent decades have been dedicated to the discovery and translation of biomaterials-inspired therapies which has led to a rapid increase in technologies and methods with an incredible capability on assisting patients.1024 For instance, Atala et al. reported urothelial and muscle cells were seeded on a bladder-shaped scaffold consist of collagen, or collagen and polyglycolic acid composite.1025 The autologous engineered bladder constructs cultured 7 days in vitro were implanted into seven patients with myelomeningocele and poorly compliant bladders. After a mean 46 months, the mean bladder leak point pressure decreased at capacity, and the volume and compliance increase were greatest in the composite engineered bladders with an omental wrap (56%, 1.58-fold, and 2.79-fold, respectively). The engineered bladder biopsies displayed an appropriate structure and phenotype. In addition, a 30-year-old woman was the first person to receive a tissue-engineering tracheal segment, a procedure that saved her left lung.1026 Significant strides have been made in terms of engineering materials with cells and biochemical factors to promote tissue repair and regeneration in patients, which facilitate the development of the next generation of medicine. Although remarkable examples have been reported in the clinical translation of biomaterials-based strategies, it remains uncertain whether they can fulfill the requirements in the clinical translation and commercialization process. Unitl 2018, a small number of cellular-based strategies have been approved by the FDA, in which only a few contain a combination of cells with materials.1024,1027 Often, advanced biomaterials that show great promise in the laboratory setting fail on the road to clinical and commercial translation.
Translating biomaterial-based products from the bench to the clinic takes a lot of effort, determination, and funds. Tissue-engineering (TE) products based on biomaterials represent relatively complex product details combined with cells and active molecules, compared to biomaterials alone. It is well-known that the more complex the medical product the more challenges there will be to overcome to meet the guidelines of regulatory agencies for host compatibility, sterility, and functionality before allowing them to be used for clinical and commercial settings. Similar to drugs, TE products need a very long time for approval, because of the complex mechanisms between biomaterials and cells as well as funding challenges. Also, the costs associated with the technical hurdles, timelines, and regulatory may be difficult to motivate and opportunities for clinical and commercial translation may be lost. Importantly, the familiarity and predictability for the interaction between biomaterials and cells is beneficial for the long-term development of TE-based diagnosis.
Efforts to develop more efficient platforms rather than a persistent attachment to previously used and well-established models will accelerate and improve the process of bringing TE products from the bench to the clinic. Also, it is very important to have the ability to accurately predict, from preclinical data, the human health risks associated with a new product. According to the challenges described above, HT analysis platforms of materiobiology and biomaterial discovery provide an ideal method to measure the complicated interactions among biomaterials, biomolecules, and cells for cell–material interaction and drug screening.21 The output results provide a better understanding of the structure–function relationships between surface properties and biological performance and identify the optimal condition for cell response, which would accelerate the development of high-quality biomaterials and then achieve the clinical translation of biomaterials. In addition, HTS platforms combined with other advanced analysis technologies (i.e., microfluidics or arrays) provide critical strengths over conventional systems,24,51,52 for example, integration of sensors for direct readout, higher reliability, and the possibility to enhance the throughput of screening by utilizing parallelization, multiplexing, and automation, preventing cross-communication of cells and cross-contamination of soluble factors, incorporate high throughput assays such as ELISA, as well as allow for robotic liquid handling and implementation of multiwell plate-based instrumentation. The true success of HTS platforms is to alter the way of doing R&D and then assist their translation toward commercial uses and the clinic. Recently, computational simulations based on machine learning algorithms and mathematical modeling were used to deal with these data that relate the biomaterial properties as input to the cell behaviors as output.55,56 A publically available database was created and named The cBiT.60 It allows researchers to search, select, and download materiobiology data as well as invite scientists to contribute their data to cBiT. Cloud platform, even materiobiology genome, could be built to help scientists to pose explicit hypotheses or make predictions of biomaterial parameters as a function of cell response.
10. Conclusion and Outlook
For the effective development of high-performance biomaterials for various biomedical applications, a comprehensive understanding of the combinatorial effects of biomaterials’ properties on cellular behaviors is pivotal. Gradient and microarray platforms can be used to investigate and mechanisms the interactions between cells and biomaterials and identify optimal formulations of biomaterials in a high-throughput manner for specific target applications. Further, microfluidics and array technologies offer important advantages over conventional analysis systems, to collect the data from HTS of materiobiology. Also, high content imaging and computational tools are extremely valuable to efficiently obtain, analyze, and predict big data, illuminate the relationship between cells and biomaterials, and help scientists pose explicit hypotheses or make predictions of biomaterial parameters as a function of cell response. The materiobiology genome is expected to be built and used to help scientists to pose explicit hypotheses or make predictions of biomaterial parameters as a function of cell response in the development of high-performance biomaterials.
However, to translate the output from HT analysis of materiobiology for efficient biomaterial discovery and development of these findings into commercial clinical uses, some bottlenecks need to be overcome and extensive works are needed to be carried out.
HTS platforms could have exciting applications in animal studies and clinical testing. Multiple materials and material properties could be screened in a single specimen, which would reduce the number of animals needed for experimentation. This approach would also enable more systematic in vivo screenings since the same individual animal could be used for all the materials in a library.
Our ability to engineer and control biomaterial properties such as surface topography, stiffness, chemistry, mechanical properties, etc. has progressed significantly in the last decades but our understanding of how to control and stimulate the desired cell–biomaterial responses with complex combinations of physicochemical biomaterial properties is still insufficient. To optimize these cell–biomaterial systems, high-throughput assays have been developed, resulting in overwhelming amounts of data. It is essential that the biomaterials field taps into the advances in the field of machine learning and computational simulations to (1) make full use of the available data, (2) reduce the experimental trial-and-error through in silico design, (3) speed-up the discovery process. However, to reach the full potential of the above-mentioned computational tools, several open questions need to be tackled. First, the biomaterials field needs to create improved descriptors for parametrizing biomaterials as well as standardize them. Such a parametrization standard would not only allow a better comparison between different studies and increase the reproducibility of the results, it would also facilitate the creation of data repositories of cell–biomaterial interactions, which are essential for data-driven tools. Second, the experimental design of HTS platforms should be optimized to ensure that most are learned from a (small) number of experiments. Instead of random sampling, “design of experiments” (DOE) can be used to decide which combinations of materials or material properties yield the best sampling of the entire parameter space. Moreover, the knowledge of previous experiments should be used to determine the next set of samples to test, employing, for example, genetic algorithms. Third, the biological response at the interface between a biomaterial and the human body is dynamic, with changing cellular players, evolving requirements and controlled at different length scales. Capturing the dynamic and multiscale nature of the processes occurring at the cell–biomaterial interface is not easy. Mechanistic models are more suited to approach these kinds of questions, but further developments are needed to link different (single-scale, separately developed) models into complex multiscale models and numerically handle such computationally challenging coupled equations. Finally, the biomaterials and computational modeling community need to further build bridges and train the next generation of scientists to develop interdisciplinary competencies in both computing as well as biomaterial sciences. By understanding each other’s jargon and working in a tight feedback loop going from in silico predictions to in vitro or in vivo experiments, the field of materiobiology can take the step toward making scientific breakthroughs.
Acknowledgments
The authors are very grateful for financial support of by the National Natural Science Foundation of China (Grant No. 31900957), Shandong Provincial Natural Science Foundation (Grant No. ZR2019QC007), China Postdoctoral Science Foundation (Grant No. 2019M652326), Innovation and technology program for the excellent youth scholars of higher education of Shandong province (Grant No. 2019KJE015), Key Laboratory of Tianjin Hyaluronic Acid Application Research Enterprise (Grant No. KTRDHA-Y201902), the Scientific Research Foundation of Qingdao University (Grant No. DC1900009689), the China Scholarship Council (Grant Nos. 201608310113, 201707720058, and 201406630003), the Wellcome Trust (Sir Henry Wellcome Postdoctoral Fellowship, 201457/Z/16/Z), the European Union’s Horizon 2020 Programme (H2020-MSCA-ITN-2015; Grant Agreement No. 676338), The Netherlands Organisation for Scientific Research Vidi grant (15604) and the Dutch Province of Limburg (LINK project), the Gravitation Program ‘Materials-Driven Regeneration’ funded by The Netherlands Organisation for Scientific Research (024.003.013), the Dutch province of Limburg in the LINK (FCL67723) (“Limburg INvesteert in haar Kenniseconomie”) knowledge economy project, and a VENI grant (No. 15075) from the Dutch Science Foundation (NWO).
Biographies
Liangliang Yang received his M.Sc. degree in biochemistry and molecular biology from Donghua University, China, in 2015. He is currently pursuing his Ph.D. degree under the supervision of Prof. Patrick van Rijn at University of Groningen via a personal grant offered by the China Scholarship Council. His research focuses on the design biomaterials to mimic the native ECM and further guide stem cell differentiation by physical cues.
Sara Pijuan-Galitó is currently a Sir Henry Wellcome postdoctoral fellow in the School of Pharmacy at the University of Nottingham. She received her Ph.D. from Uppsala University in 2015, and her thesis focused on pluripotent stem cell signaling pathways and culture optimization through adhesion biomolecules. In 2015, she was awarded the International Postdoctoral Fellowship from the Swedish Research Council and moved to the University of Nottingham to further investigate the stem cell niche. In 2016, she was awarded the Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust. Her research interests center around the biomaterial–cell interface and its implication in stem cell expansion and differentiation for clinical applications.
Hoon Suk Rho received his Ph.D. (2016) degree in chemical engineering from University of Twente (The Netherlands). From 2016 to 2018, he worked as a postdoctoral researcher in Cancer-ID program to develop novel technologies for monitoring cancer therapy through revealing the extracellular vesicle identity. In March 2018, he joined the Department of Instructive Biomaterials Engineering at MERLN institute, Maastricht University, The Netherlands, as a postdoctoral researcher. His research interests include microfabrication, microfluidic, single cell analysis, and biomaterials.
Aliaksei S. Vasilevich studied biomedical sciences and ecology in the International Sakharov Environmental University, Minsk, Belarus. During the Master thesis, he developed a method to rapidly measure the sensitivity of cells to drugs using image analysis. He obtained his Ph.D. under the supervision of Prof. Jan de Boer, where he applied machine learning algorithms to study the biomaterials design parameters that affect cell fate. He is keen on the application of data science solutions for biomaterials discovery.
Aysegul Dede Eren received her M.Sc. degree in Neuroscience department from Bilkent University, Turkey in 2017. The same year she joined the group of Prof. Jan de Boer on a Marie-Curie ITN “Tendon Therapy Train” scholarship to pursue her Ph.D. studies in The Netherlands. Her research focuses on the understanding the role of surface topology on the regulation of cell shape and cell phenotype. She combines material science with cell biology and uses transcriptomics tools to contribute to biomaterial discovery.
Lu Ge received her M.Sc. degree in cell biology from Kunming University of Science and Technology, China, in 2016. Currently, she is a Ph.D. candidate in the group of Dr. Patrick van Rijn at the University Medical Center from Groningen University via a personal grant offered by the China Scholarship Council. She is working on understanding cell–materials interaction and tissue engineering based on physical cues.
Pamela Habibović received her Ph.D. (2005) degree in biomaterials science from University of Twente (The Netherlands). She did her postdoctoral research at Children’s Hospital Boston and at McGill University. From 2008 until 2014, she led a research group at the University of Twente, first as assistant and later as associate professor. In 2014, she moved to Maastricht University, where she became full professor of Inorganic Biomaterials and where she cofounded MERLN Institute for Technology-Inspired Regenerative Medicine. Currently, she is the Scientific Director of the institute and she chairs the Department of Instructive Biomaterials Engineering. Her research group focuses on synthetic biomaterials for regenerative medicine. Different research lines include ceramic bone graft substitutes, bioinorganics, nanomaterials for targeted delivery, and microfluidics-based (high-throughput) screening approaches in biomaterials research. She serves on editorial boards of several journals in the field of biomaterials and regenerative medicine, and she is currently the president of the European Society for Biomaterials.
Morgan Alexander is Professor of Biomedical Surfaces in the School of Pharmacy at the University of Nottingham. He is the Director of the EPSRC Programme Grant in Next Generation Biomaterials Discovery and a Wellcome Trust Senior Investigator. He received his B.Sc. in Materials (1988) and his Ph.D. from the same department at The University of Sheffield in 1992. His work involves developing materials for application in human healthcare and characterizing relationships between surface structure and biological properties. Understanding these relationships is critical to the development of next generation biomaterials, and it is the theme running through his group’s work across a variety of areas spanning control of bacterial adhesion to engineering cell response with applications in medical devices and cell manufacture.
Jan de Boer is chair of the BioInterface Science lab at the Institute for Complex Molecular Systems and the department of Biomedical Engineering at Eindhoven University of Technology. His research is focused on understanding and implementing molecular biology in the field of tissue engineering, regenerative medicine, and medical devices interface biology. His research is characterized by a holistic approach to both discovery and application, aiming at combining high throughput technologies such as the TopoChip platform, computational modeling, and experimental cell biology to streamline the wealth of biological knowledge to clinical applications.
Aurélie Carlier is tenure track assistant professor at the MERLN Institute for Technology-Inspired Regenerative Medicine in Maastricht, The Netherlands. She received her M.Sc. (2010) and Ph.D. (2014) in Biomedical Engineering at the KU Leuven, Belgium. Her research interests encompass the multiscale computational modelling of biological processes, with a particular focus on bone tissue engineering applications and cell–biomaterial interactions and using a range of data-driven to mechanistic modelling approaches. Her research achievements have been awarded with a number of distinctions including the Reinhart Heinrich Doctoral Thesis Award (ESMTB, 2015) and the Best Doctoral Thesis Award (ESB, 2015). She has also been elected as member of the Tissue Engineering Young Investigator Council (2016) and received a prestigious VENI career development grant (0.25 M€) from the Dutch Science Foundation.
Patrick van Rijn received his Ph.D. in Supramolecular Chemistry from Delft University of Technology in 2010 after he moved as an Alexander von Humboldt-fellow to the Leibniz Institute for Interactive Materials in Aachen. In 2013, he started his independent research group at the University Medical Center Groningen where his current research interests are on developing new biointerfaces to understand the relationship between material properties and cellular behavior and therefore develop the BiomACS platform. The research that is conducted in his group is both of fundamental and applied character with strong connections to the clinic and industry. Patrick is chair of The Netherlands Society for Biomaterials and Tissue Engineering (NBTE), topical editor biomedical polymer for the open access journal Polymers (MPDI).
Qihui Zhou received his Ph.D. in Biomedical Engineering from the University of Groningen in 2018. Upon graduation, immediately he was employed as an Associate Professor and Director at the Center for Precision Medicine at Qingdao University (QDU). Also, he is the Scientific Director of the Stomatology Center at the Affiliated Hospital of QDU. Currently, as the Principal Investigator at QDU, he directs the Laboratory of Biomaterials for Translational Medicine at the University. His research interests focus on the design and development of advanced biomaterials with unique chemical, physical, and biological properties to assist the diagnosis and treatment of challenging diseases such as cancer, sepsis, tissue injury, and defects. He is a member of the youth editorial board for the journal Military Medical Research.
Author Contributions
Δ L.Y., S.P.-G., H.S.R., A.S.V., A.D.E., and L.G. are cofirst authors.
The authors declare the following competing financial interest(s): P.v.R is cofounder/scientific advisor/shareholder of BiomACS BV. There are no other conflicts to declare.
References
- Chen F.-M.; Liu X. Advancing Biomaterials of Human Origin for Tissue Engineering. Prog. Polym. Sci. 2016, 53, 86–168. 10.1016/j.progpolymsci.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza R.; Langer R.; Vacanti J. P.. Principles of Tissue Engineering; Academic Press, 2011. [Google Scholar]
- Griffith L. G.; Naughton G. Tissue Engineering--Current Challenges and Expanding Opportunities. Science (Washington, DC, U. S.) 2002, 295, 1009–1014. 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- Vacanti J. P.; Langer R. Tissue Engineering: The Design and Fabrication of Living Replacement Devices for Surgical Reconstruction and Transplantation. Lancet 1999, 354, S32–S34. 10.1016/S0140-6736(99)90247-7. [DOI] [PubMed] [Google Scholar]
- Huang G.; Li F.; Zhao X.; Ma Y.; Li Y.; Lin M.; Jin G.; Lu T. J.; Genin G. M.; Xu F. Functional and Biomimetic Materials for Engineering of the Three-Dimensional Cell Microenvironment. Chem. Rev. 2017, 117, 12764–12850. 10.1021/acs.chemrev.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi A.; Ling Q. D.; Chang Y.; Hsu S. T.; Umezawa A. Physical Cues of Biomaterials Guide Stem Cell Differentiation Fate. Chem. Rev. 2013, 113, 3297–3328. 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- Li Y.; Xiao Y.; Liu C. The Horizon of Materiobiology: A Perspective on Material-Guided Cell Behaviors and Tissue Engineering. Chem. Rev. 2017, 117, 4376–4421. 10.1021/acs.chemrev.6b00654. [DOI] [PubMed] [Google Scholar]
- Theocharis A. D.; Skandalis S. S.; Gialeli C.; Karamanos N. K. Extracellular Matrix Structure. Adv. Drug Delivery Rev. 2016, 97, 4–27. 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Frantz C.; Stewart K. M.; Weaver V. M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N.; Mooney D. J. Inspiration and Application in the Evolution of Biomaterials. Nature 2009, 462, 426–432. 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegst U. G. K.; Bai H.; Saiz E.; Tomsia A. P.; Ritchie R. O. Bioinspired Structural Materials. Nat. Mater. 2015, 14, 23–36. 10.1038/nmat4089. [DOI] [PubMed] [Google Scholar]
- Lutolf M. P.; Gilbert P. M.; Blau H. M. Designing Materials to Direct Stem-Cell Fate. Nature 2009, 462, 433–441. 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q.; Chen J.; Luan Y.; Vainikka P. A.; Thallmair S.; Marrink S. J.; Feringa B. L.; Van Rijn P. Unidirectional Rotating Molecular Motors Dynamically Interact with Adsorbed Proteins to Direct the Fate of Mesenchymal Stem Cells. Sci. Adv. 2020, 6, eaay2756 10.1126/sciadv.aay2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biomaterials. Nature Research; Springer, 2020. https://www.nature.com/subjects/biomaterials (accessed 05-01-2020).
- Biomaterials; Elsevier, 2020. https://www.journals.elsevier.com/biomaterials (accessed 05-01-2020).
- Hynes R. O. The Extracellular Matrix: Not Just Pretty Fibrils. Science (Washington, DC, U. S.) 2009, 326, 1216–1219. 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F.; Cohen D. M.; Estes B. T.; Gimble J. M.; Liedtke W.; Chen C. S. Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix. Cell Stem Cell 2009, 5, 17–26. 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R.; Tirrell D. a. Designing Materials for Biology and Medicine. Nature 2004, 428, 487–492. 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- Hao Y.; Zhao W.; Zhang L.; Zeng X.; Sun Z.; Zhang D.; Shen P.; Li Z.; Han Y.; Li P.; et al. Bio-Multifunctional Alginate/Chitosan/Fucoidan Sponges with Enhanced Angiogenesis and Hair Follicle Regeneration for Promoting Full-Thickness Wound Healing. Mater. Des. 2020, 193, 108863. 10.1016/j.matdes.2020.108863. [DOI] [Google Scholar]
- Bao M.; Lou X.; Zhou Q.; Dong W.; Yuan H.; Zhang Y. Electrospun Biomimetic Fibrous Scaffold from Shape Memory Polymer of PDLLA- Co -TMC for Bone Tissue Engineering. ACS Appl. Mater. Interfaces 2014, 6, 2611–2621. 10.1021/am405101k. [DOI] [PubMed] [Google Scholar]
- Seo J.; Shin J. Y.; Leijten J.; Jeon O.; Camci-Unal G.; Dikina A. D.; Brinegar K.; Ghaemmaghami A. M.; Alsberg E.; Khademhosseini A. High-Throughput Approaches for Screening and Analysis of Cell Behaviors. Biomaterials 2018, 153, 85–101. 10.1016/j.biomaterials.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. K.; Tibbitt M. W.; Celiz A. D.; Davies M. C.; Langer R.; Denning C.; Alexander M. R.; Anderson D. G. High Throughput Screening for Discovery of Materials That Control Stem Cell Fate. Curr. Opin. Solid State Mater. Sci. 2016, 20, 202–211. 10.1016/j.cossms.2016.02.002. [DOI] [Google Scholar]
- Tronser T.; Popova A. A.; Levkin P. A. Miniaturized Platform for High-Throughput Screening of Stem Cells. Curr. Opin. Biotechnol. 2017, 46, 141–149. 10.1016/j.copbio.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Barata D.; Van Blitterswijk C.; Habibovic P. High-Throughput Screening Approaches and Combinatorial Development of Biomaterials Using Microfluidics. Acta Biomater. 2016, 34, 1–20. 10.1016/j.actbio.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Fernandes T. G.; Diogo M. M.; Clark D. S.; Dordick J. S.; Cabral J. M. S. High-Throughput Cellular Microarray Platforms: Applications in Drug Discovery, Toxicology and Stem Cell Research. Trends Biotechnol. 2009, 27, 342–349. 10.1016/j.tibtech.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L.; Anderson D. G.; Langer R.; Williams P.; Davies M. C.; Alexander M. R. High Throughput Methods Applied in Biomaterial Development and Discovery. Biomaterials 2010, 31, 187–198. 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- Kim H. D.; Lee E. A.; Choi Y. H.; An Y. H.; Koh R. H.; Kim S. L.; Hwang N. S. High Throughput Approaches for Controlled Stem Cell Differentiation. Acta Biomater. 2016, 34, 21–29. 10.1016/j.actbio.2016.02.022. [DOI] [PubMed] [Google Scholar]
- Kim M. S.; Khang G.; Lee H. B. Gradient Polymer Surfaces for Biomedical Applications. Prog. Polym. Sci. 2008, 33, 138–164. 10.1016/j.progpolymsci.2007.06.001. [DOI] [Google Scholar]
- Oliveira M. B.; Mano J. F. High-Throughput Screening for Integrative Biomaterials Design: Exploring Advances and New Trends. Trends Biotechnol. 2014, 32, 627–636. 10.1016/j.tibtech.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Simon C. G.; Sheng L. G. Combinatorial and High-Throughput Screening of Biomaterials. Adv. Mater. 2011, 23, 369–387. 10.1002/adma.201001763. [DOI] [PubMed] [Google Scholar]
- Zink C.; Hall H.; Brunette D. M.; Spencer N. D. Orthogonal Nanometer-Micrometer Roughness Gradients Probe Morphological Influences on Cell Behavior. Biomaterials 2012, 33, 8055–8061. 10.1016/j.biomaterials.2012.07.037. [DOI] [PubMed] [Google Scholar]
- Hulander M.; Lundgren A.; Faxälv L.; Lindahl T. L.; Palmquist A.; Berglin M.; Elwing H. Gradients in Surface Nanotopography Used to Study Platelet Adhesion and Activation. Colloids Surf., B 2013, 110, 261–269. 10.1016/j.colsurfb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Reynolds P. M.; Pedersen R. H.; Riehle M. O.; Gadegaard N. A Dual Gradient Assay for the Parametric Analysis of Cell-Surface Interactions. Small 2012, 8, 2541–2547. 10.1002/smll.201200235. [DOI] [PubMed] [Google Scholar]
- Zhou Q.; Kühn P. T.; Huisman T.; Nieboer E.; Van Zwol C.; Van Kooten T. G.; Van Rijn P. Directional Nanotopographic Gradients: A High-Throughput Screening Platform for Cell Contact Guidance. Sci. Rep. 2015, 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamant Q.; Stanciuc A. M.; Pavailler H.; Sprecher C. M.; Alini M.; Peroglio M.; Anglada M. Roughness Gradients on Zirconia for Rapid Screening of Cell-Surface Interactions: Fabrication, Characterization and Application. J. Biomed. Mater. Res., Part A 2016, 104, 2502–2514. 10.1002/jbm.a.35791. [DOI] [PubMed] [Google Scholar]
- Wang P. Y.; Clements L. R.; Thissen H.; Jane A.; Tsai W. B.; Voelcker N. H. Screening Mesenchymal Stem Cell Attachment and Differentiation on Porous Silicon Gradients. Adv. Funct. Mater. 2012, 22, 3414–3423. 10.1002/adfm.201200447. [DOI] [Google Scholar]
- Hu B.; Shi W.; Wu Y. L.; Leow W. R.; Cai P.; Li S.; Chen X. Orthogonally Engineering Matrix Topography and Rigidity to Regulate Multicellular Morphology. Adv. Mater. 2014, 26, 5786–5793. 10.1002/adma.201402489. [DOI] [PubMed] [Google Scholar]
- Kim T. H.; An D. B.; Oh S. H.; Kang M. K.; Song H. H.; Lee J. H. Creating Stiffness Gradient Polyvinyl Alcohol Hydrogel Using a Simple Gradual Freezing-Thawing Method to Investigate Stem Cell Differentiation Behaviors. Biomaterials 2015, 40, 51–60. 10.1016/j.biomaterials.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Hopp I.; Michelmore A.; Smith L. E.; Robinson D. E.; Bachhuka A.; Mierczynska A.; Vasilev K. The Influence of Substrate Stiffness Gradients on Primary Human Dermal Fibroblasts. Biomaterials 2013, 34, 5070–5077. 10.1016/j.biomaterials.2013.03.075. [DOI] [PubMed] [Google Scholar]
- Kuo C. H. R.; Xian J.; Brenton J. D.; Franze K.; Sivaniah E. Complex Stiffness Gradient Substrates for Studying Mechanotactic Cell Migration. Adv. Mater. 2012, 24, 6059–6064. 10.1002/adma.201202520. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Seo C. H.; Han K.; Kwon K. W.; Levchenko A.; Suh K. Y. Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy. Adv. Funct. Mater. 2009, 19, 1579–1586. 10.1002/adfm.200801174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Mao Z.; Tan H.; Han L.; Ren T.; Gao C. Gradient Biomaterials and Their Influences on Cell Migration. Interface Focus 2012, 2, 337–355. 10.1098/rsfs.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale N. A.; Yang Y.; Rajagopalan P. Cell Migration at the Interface of a Dual Chemical-Mechanical Gradient. ACS Appl. Mater. Interfaces 2010, 2, 2317–2324. 10.1021/am100346k. [DOI] [PubMed] [Google Scholar]
- Joaquin D.; Grigola M.; Kwon G.; Blasius C.; Han Y.; Perlitz D.; Jiang J.; Ziegler Y.; Nardulli A.; Hsia K. J. Cell Migration and Organization in Three-Dimensional in Vitro Culture Driven by Stiffness Gradient. Biotechnol. Bioeng. 2016, 113, 2496–2506. 10.1002/bit.26010. [DOI] [PubMed] [Google Scholar]
- Wu J.; Mao Z.; Han L.; Xi J.; Zhao Y.; Gao C. Directional Migration of Vascular Smooth Muscle Cells Guided by Synergetic Surface Gradient and Chemical Pattern of Poly(Ethylene Glycol) Brushes. J. Bioact. Compat. Polym. 2013, 28, 605–620. 10.1177/0883911513506665. [DOI] [Google Scholar]
- Park J. S.; Kim D. H.; Levchenko A. Topotaxis: A New Mechanism of Directed Cell Migration in Topographic ECM Gradients. Biophys. J. 2018, 114, 1257–1263. 10.1016/j.bpj.2017.11.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Mao Z.; Tan H.; Han L.; et al. Gradient Biomaterials and Their Influences on Cell Migration. Interface Focus 2012, 2, 337–355. 10.1098/rsfs.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden W. J.; Young J. L.; Holle A. W.; McFetridge M. L.; Kim D. Y.; Wijesinghe P.; Taylor-Weiner H.; Wen J. H.; Lee A. R.; Bieback K.; et al. Stem Cell Migration and Mechanotransduction on Linear Stiffness Gradient Hydrogels. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 5647–5652. 10.1073/pnas.1618239114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S.; Kim D. H.; Kim H. N.; Wang C. J.; Kwak M. K.; Hur E.; Suh K. Y.; An S. S.; Levchenko A. Directed Migration of Cancer Cells Guided by the Graded Texture of the Underlying Matrix. Nat. Mater. 2016, 15, 792–801. 10.1038/nmat4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W.; Ueda E.; Levkin P. A. Droplet Microarrays: From Surface Patterning to High-Throughput Applications. Adv. Mater. 2018, 30, 1706111. 10.1002/adma.201706111. [DOI] [PubMed] [Google Scholar]
- Beijer N. R. M.; Vasilevich A. S.; Pilavci B.; Truckenmüller R. K.; Zhao Y.; Singh S.; Papenburg B. J.; de Boer J. TopoWellPlate: A Well-Plate-Based Screening Platform to Study Cell-Surface Topography Interactions. Adv. Biosyst. 2017, 1, 1700002. 10.1002/adbi.201700002. [DOI] [PubMed] [Google Scholar]
- Hu J.; Gondarenko A. A.; Dang A. P.; Bashour K. T.; O’Connor R. S.; Lee S.; Liapis A.; Ghassemi S.; Milone M. C.; Sheetz M. P.; et al. High-Throughput Mechanobiology Screening Platform Using Micro-and Nanotopography. Nano Lett. 2016, 16, 2198–2204. 10.1021/acs.nanolett.5b04364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Ge L.; Zhou Q.; Jurczak K. M.; van Rijn P. Decoupling the Amplitude and Wavelength of Anisotropic Topography and the Influence on Osteogenic Differentiation of Mesenchymal Stem Cells Using a High-Throughput Screening Approach. ACS Appl. Bio Mater. 2020, 3, 3690–3697. 10.1021/acsabm.0c00330. [DOI] [PubMed] [Google Scholar]
- An S.; Lim J.; Choi D.; Hong H.; Kim H. W.; Park S. M.; Rhie J. W.; Kim D. S. Fabrication of Polystyrene-Based Multi-Well Screening Platform for Micrometer-Scale Surface Topographies Promoting Stem Cell Functions. Microelectron. Eng. 2017, 174, 28–34. 10.1016/j.mee.2016.12.020. [DOI] [Google Scholar]
- Vasilevich A. S.; Carlier A.; de Boer J.; Singh S. How Not to Drown in Data: A Guide for Biomaterial Engineers. Trends Biotechnol. 2017, 35, 743–755. 10.1016/j.tibtech.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Geris L.; Lambrechts T.; Carlier A.; Papantoniou I. The Future Is Digital: In Silico Tissue Engineering. Curr. Opin. Biomed. Eng. 2018, 6, 92–98. 10.1016/j.cobme.2018.04.001. [DOI] [Google Scholar]
- Semple J. L.; Woolridge N.; Lumsden C. J. In Vitro, in Vivo, in Silico: Computational Systems in Tissue Engineering and Regenerative Medicine. Tissue Eng. 2005, 11, 341–356. 10.1089/ten.2005.11.341. [DOI] [PubMed] [Google Scholar]
- Giannitelli S. M.; Accoto D.; Trombetta M.; Rainer A. Current Trends in the Design of Scaffolds for Computer-Aided Tissue Engineering. Acta Biomater. 2014, 10, 580–594. 10.1016/j.actbio.2013.10.024. [DOI] [PubMed] [Google Scholar]
- Groen N.; Yuan H.; Hebels D. G. A. J.; Koçer G.; Mbuyi F.; LaPointe V.; Truckenmüller R.; van Blitterswijk C. A.; Habibović P.; de Boer J. Linking the Transcriptional Landscape of Bone Induction to Biomaterial Design Parameters. Adv. Mater. 2017, 29, 1603259. 10.1002/adma.201603259. [DOI] [PubMed] [Google Scholar]
- Hebels D. G. A. J.; Carlier A.; Coonen M. L. J.; Theunissen D. H.; de Boer J. CBiT: A Transcriptomics Database for Innovative Biomaterial Engineering. Biomaterials 2017, 149, 88–97. 10.1016/j.biomaterials.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Schaap-Oziemlak A. M.; Kühn P. T.; van Kooten T. G.; van Rijn P. Biomaterial-Stem Cell Interactions and Their Impact on Stem Cell Response. RSC Adv. 2014, 4, 53307–53320. 10.1039/C4RA07915A. [DOI] [Google Scholar]
- Bao M.; Xie J.; Huck W. T. S. Recent Advances in Engineering the Stem Cell Microniche in 3D. Adv. Sci. 2018, 5, 1800448. 10.1002/advs.201800448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. L.; McDevitt T. C.; Engler A. J. Materials as Stem Cell Regulators. Nat. Mater. 2014, 13, 547–557. 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V.; Sheetz M. Local Force and Geometry Sensing Regulate Cell Functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Prager-Khoutorsky M.; Lichtenstein A.; Krishnan R.; Rajendran K.; Mayo A.; Kam Z.; Geiger B.; Bershadsky A. D. Fibroblast Polarization Is a Matrix-Rigidity-Dependent Process Controlled by Focal Adhesion Mechanosensing. Nat. Cell Biol. 2011, 13, 1457–1465. 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Zhou Q.; Xie J.; Bao M.; Yuan H.; Ye Z.; Lou X.; Zhang Y. Engineering Aligned Electrospun PLLA Microfibers with Nano-Porous Surface Nanotopography for Modulating the Responses of Vascular Smooth Muscle Cells. J. Mater. Chem. B 2015, 3, 4439–4450. 10.1039/C5TB00051C. [DOI] [PubMed] [Google Scholar]
- Mokabber T.; Zhou Q.; Vakis A. I.; van Rijn P.; Pei Y. T. Mechanical and Biological Properties of Electrodeposited Calcium Phosphate Coatings. Mater. Sci. Eng., C 2019, 100, 475–484. 10.1016/j.msec.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Swift J.; Ivanovska I. L.; Buxboim A.; Harada T.; Dingal P. C. D. P.; Pinter J.; Pajerowski J. D.; Spinler K. R.; Shin J.-W.; Tewari M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. L. K.; Simmons C. A. Lessons from (Patho) Physiological Tissue Stiffness and Their Implications for Drug Screening, Drug Delivery and Regenerative Medicine. Adv. Drug Delivery Rev. 2011, 63, 269–276. 10.1016/j.addr.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Discher D. E.; Janmey P.; Wang Y.-L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Huebsch N.; Arany P. R.; Mao A. S.; Shvartsman D.; Ali O. A.; Bencherif S. A.; Rivera-Feliciano J.; Mooney D. J. Harnessing Traction-Mediated Manipulation of the Cell/Matrix Interface to Control Stem-Cell Fate. Nat. Mater. 2010, 9, 518–526. 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. M.; Trappmann B.; Wang W. Y.; Sakar M. S.; Kim I. L.; Shenoy V. B.; Burdick J. A.; Chen C. S. Cell-Mediated Fibre Recruitment Drives Extracellular Matrix Mechanosensing in Engineered Fibrillar Microenvironments. Nat. Mater. 2015, 14, 1262–1268. 10.1038/nmat4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T.; Georges P. C.; Flanagan L. A.; Marg B.; Ortiz M.; Funaki M.; Zahir N.; Ming W.; Weaver V.; Janmey P. A. Effects of Substrate Stiffness on Cell Morphology, Cytoskeletal Structure, and Adhesion. Cell Motil. Cytoskeleton 2005, 60, 24–34. 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Evans E.; Leung A.; Zhelev D. Synchrony of Cell Spreading and Contraction Force as Phagocytes Engulf Large Pathogens. J. Cell Biol. 1993, 122, 1295–1300. 10.1083/jcb.122.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K.; Keung A. J.; Irwin E. F.; Li Y.; Little L.; Schaffer D. V.; Healy K. E. Substrate Modulus Directs Neural Stem Cell Behavior. Biophys. J. 2008, 95, 4426–4438. 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J.; Sen S.; Sweeney H. L.; Discher D. E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Kong H. J.; Liu J.; Riddle K.; Matsumoto T.; Leach K.; Mooney D. J. Non-Viral Gene Delivery Regulated by Stiffness of Cell Adhesion Substrates. Nat. Mater. 2005, 4, 460–464. 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- Yang G.; Li X.; He Y.; Ma J.; Ni G.; Zhou S. From Nano to Micro to Macro: Electrospun Hierarchically Structured Polymeric Fibers for Biomedical Applications. Prog. Polym. Sci. 2018, 81, 80–113. 10.1016/j.progpolymsci.2017.12.003. [DOI] [Google Scholar]
- Zhou Q.; Zhang H.; Zhou Y.; Yu Z.; Yuan H.; Feng B.; van Rijn P.; Zhang Y. Alkali-Mediated Miscibility of Gelatin/Polycaprolactone for Electrospinning Homogeneous Composite Nanofibers for Tissue Scaffolding. Macromol. Biosci. 2017, 17, 1–10. 10.1002/mabi.201700268. [DOI] [PubMed] [Google Scholar]
- Yuan H.; Zhou Q.; Zhang Y.. 6 - Improving Fiber Alignment during Electrospinning. In Woodhead Publishing Series in Textiles; Afshari M. B. T.-E. N., Ed.; Woodhead Publishing, 2017; pp 125–147. [Google Scholar]
- Zeltinger J.; Sherwood J. K.; Graham D. A.; Müeller R.; Griffith L. G. Effect of Pore Size and Void Fraction on Cellular Adhesion, Proliferation, and Matrix Deposition. Tissue Eng. 2001, 7, 557–572. 10.1089/107632701753213183. [DOI] [PubMed] [Google Scholar]
- Boyan B. D.; Hummert T. W.; Dean D. D.; Schwartz Z. Role of Material Surfaces in Regulating Bone and Cartilage Cell Response. Biomaterials 1996, 17, 137–146. 10.1016/0142-9612(96)85758-9. [DOI] [PubMed] [Google Scholar]
- Lee M.; Wu B. M.; Dunn J. C. Y. Effect of Scaffold Architecture and Pore Size on Smooth Muscle Cell Growth. J. Biomed. Mater. Res., Part A 2008, 87, 1010–1016. 10.1002/jbm.a.31816. [DOI] [PubMed] [Google Scholar]
- Madden L. R.; Mortisen D. J.; Sussman E. M.; Dupras S. K.; Fugate J. A.; Cuy J. L.; Hauch K. D.; Laflamme M. A.; Murry C. E.; Ratner B. D. Proangiogenic Scaffolds as Functional Templates for Cardiac Tissue Engineering. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 15211–15216. 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S.; Wendorff J. H.; Greiner A. Use of Electrospinning Technique for Biomedical Applications. Polymer 2008, 49, 5603–5621. 10.1016/j.polymer.2008.09.014. [DOI] [Google Scholar]
- Shruti S.; Salinas A. J.; Lusvardi G.; Malavasi G.; Menabue L.; Vallet-Regi M. Mesoporous Bioactive Scaffolds Prepared with Cerium-, Gallium- and Zinc-Containing Glasses. Acta Biomater. 2013, 9, 4836–4844. 10.1016/j.actbio.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Chaterji S.; Kim P.; Choe S. H.; Tsui J. H.; Lam C. H.; Ho D. S.; Baker A. B.; Kim D.-H. Synergistic Effects of Matrix Nanotopography and Stiffness on Vascular Smooth Muscle Cell Function. Tissue Eng., Part A 2014, 20, 2115–2126. 10.1089/ten.tea.2013.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester-Beltrán J.; Biggs M. J. P.; Dalby M. J.; Salmeron-Sanchez M.; Leal-Egaña A. Sensing the Difference: The Influence of Anisotropic Cues on Cell Behavior. Front. Mater. 2015, 2, 1–12. 10.3389/fmats.2015.00039. [DOI] [Google Scholar]
- Carson D.; Hnilova M.; Yang X.; Nemeth C. L.; Tsui J. H.; Smith A. S. T.; Jiao A.; Regnier M.; Murry C. E.; Tamerler C.; et al. Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS Appl. Mater. Interfaces 2016, 8, 21923–21932. 10.1021/acsami.5b11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finne-Wistrand A.; Albertsson A.-C.; Kwon O. H.; Kawazoe N.; Chen G.; Kang I.-K.; Hasuda H.; Gong J.; Ito Y. Resorbable Scaffolds from Three Different Techniques: Electrospun Fabrics, Salt-Leaching Porous Films, and Smooth Flat Surfaces. Macromol. Biosci. 2008, 8, 951–959. 10.1002/mabi.200700328. [DOI] [PubMed] [Google Scholar]
- Chen M.; Patra P. K.; Warner S. B.; Bhowmick S. Role of Fiber Diameter in Adhesion and Proliferation of NIH 3T3 Fibroblast on Electrospun Polycaprolactone Scaffolds. Tissue Eng. 2007, 13, 579–587. 10.1089/ten.2006.0205. [DOI] [PubMed] [Google Scholar]
- Rubenstein D.; Han D.; Goldgraben S.; El-Gendi H.; Gouma P.-I.; Frame M. D. Bioassay Chamber for Angiogenesis with Perfused Explanted Arteries and Electrospun Scaffolding. Microcirculation 2007, 14, 723–737. 10.1080/10739680701410173. [DOI] [PubMed] [Google Scholar]
- Elias K. L.; Price R. L.; Webster T. J. Enhanced Functions of Osteoblasts on Nanometer Diameter Carbon Fibers. Biomaterials 2002, 23, 3279–3287. 10.1016/S0142-9612(02)00087-X. [DOI] [PubMed] [Google Scholar]
- Wei G.; Ma P. X. Nanostructured Biomaterials for Regeneration. Adv. Funct. Mater. 2008, 18, 3568–3582. 10.1002/adfm.200800662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S.; Levengood S. K. L.; Zhang M. Anisotropic Materials for Skeletal-muscle-tissue Engineering. Adv. Mater. 2016, 28, 10588–10612. 10.1002/adma.201600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H.; Provenzano P. P.; Smith C. L.; Levchenko A. Matrix Nanotopography as a Regulator of Cell Function. J. Cell Biol. 2012, 197, 351–360. 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. T.; Sathe S. R.; Yim E. K. F. From Nano to Micro: Topographical Scale and Its Impact on Cell Adhesion, Morphology and Contact Guidance. J. Phys.: Condens. Matter 2016, 28, 183001. 10.1088/0953-8984/28/18/183001. [DOI] [PubMed] [Google Scholar]
- Xie J.; Li X.; Lipner J.; Manning C. N.; Schwartz A. G.; Thomopoulos S.; Xia Y. Aligned-to-Random” Nanofiber Scaffolds for Mimicking the Structure of the Tendon-to-Bone Insertion Site. Nanoscale 2010, 2, 923–926. 10.1039/c0nr00192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayatpisheh S.; Heath D. E.; Shakouri A.; Rujitanaroj P.-O.; Chew S. Y.; Chan-Park M. B. Combining Cell Sheet Technology and Electrospun Scaffolding for Engineered Tubular, Aligned, and Contractile Blood Vessels. Biomaterials 2014, 35, 2713–2719. 10.1016/j.biomaterials.2013.12.035. [DOI] [PubMed] [Google Scholar]
- Zhou Q.; Castañeda Ocampo O.; Guimarães C. F.; Kühn P. T.; van Kooten T. G.; van Rijn P. Screening Platform for Cell Contact Guidance Based on Inorganic Biomaterial Micro/Nanotopographical Gradients. ACS Appl. Mater. Interfaces 2017, 9, 31433–31445. 10.1021/acsami.7b08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbagheri M.; Adibnia V.; Hughes B. R.; Waldman S. D.; Banquy X.; Hwang D. K. Advanced Cell Culture Platforms: A Growing Quest for Emulating Natural Tissues. Mater. Horiz. 2019, 6, 45–71. 10.1039/C8MH00803E. [DOI] [Google Scholar]
- Baker B. M.; Chen C. S. Deconstructing the Third Dimension-How 3D Culture Microenvironments Alter Cellular Cues. J. Cell Sci. 2012, 125, 3015–3024. 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto K.; Tsui J. H.; DeForest C. A.; Kim D.-H. Dynamically Tunable Cell Culture Platforms for Tissue Engineering and Mechanobiology. Prog. Polym. Sci. 2017, 65, 53–82. 10.1016/j.progpolymsci.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q.; Zhao Z.; Zhou Z.; Zhang G.; Chiechi R. C.; van Rijn P. Directing Mesenchymal Stem Cells with Gold Nanowire Arrays. Adv. Mater. Interfaces 2018, 5, 1–8. 10.1002/admi.201800334. [DOI] [Google Scholar]
- Belair D. G.; Le N. N.; Murphy W. L. Design of Growth Factor Sequestering Biomaterials. Chem. Commun. 2014, 50, 15651–15668. 10.1039/C4CC04317K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Wang Z.; Lu W. W.; Zhen W.; Yang D.; Peng S. Novel Biomaterial Strategies for Controlled Growth Factor Delivery for Biomedical Applications. NPG Asia Mater. 2017, 9, e435 10.1038/am.2017.171. [DOI] [Google Scholar]
- Falsey J. R.; Renil M.; Park S.; Li S.; Lam K. S. Peptide and Small Molecule Microarray for High Throughput Cell Adhesion and Functional Assays. Bioconjugate Chem. 2001, 12, 346–353. 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- Gehlen D. B.; De Lencastre Novaes L. C.; Long W.; Ruff A. J.; Jakob F.; Haraszti T.; Chandorkar Y.; Yang L.; van Rijn P.; Schwaneberg U.; et al. Rapid and Robust Coating Method to Render Polydimethylsiloxane Surfaces Cell-Adhesive. ACS Appl. Mater. Interfaces 2019, 11, 41091–41099. 10.1021/acsami.9b16025. [DOI] [PubMed] [Google Scholar]
- Lee K. Y.; Mooney D. J. 193 Hydrogels for Tissue Engineering. Chem. Rev. 2001, 101, 1869–1880. 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- Tamada Y.; Ikada Y. Effect of Preadsorbed Proteins on Cell Adhesion to Polymer Surfaces. J. Colloid Interface Sci. 1993, 155, 334–339. 10.1006/jcis.1993.1044. [DOI] [Google Scholar]
- Mei Y.; Saha K.; Bogatyrev S. R.; Yang J.; Hook A. L.; Kalcioglu Z. I.; Cho S.-W.; Mitalipova M.; Pyzocha N.; Rojas F.; et al. Combinatorial Development of Biomaterials for Clonal Growth of Human Pluripotent Stem Cells. Nat. Mater. 2010, 9, 768–778. 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroman L. Effect of Adsorbed Proteins on the Wettability of Hydrophilic and Hydrophobic Solids. Nature 1962, 196, 476–477. 10.1038/196476a0. [DOI] [PubMed] [Google Scholar]
- Wei J.; Igarashi T.; Okumori N.; Igarashi T.; Maetani T.; Liu B.; Yoshinari M. Influence of Surface Wettability on Competitive Protein Adsorption and Initial Attachment of Osteoblasts. Biomed. Mater. 2009, 4, 45002. 10.1088/1748-6041/4/4/045002. [DOI] [PubMed] [Google Scholar]
- Yildirim E. D.; Besunder R.; Pappas D.; Allen F.; Güçeri S.; Sun W. Accelerated Differentiation of Osteoblast Cells on Polycaprolactone Scaffolds Driven by a Combined Effect of Protein Coating and Plasma Modification. Biofabrication 2010, 2, 14109. 10.1088/1758-5082/2/1/014109. [DOI] [PubMed] [Google Scholar]
- Lee M. H.; Ducheyne P.; Lynch L.; Boettiger D.; Composto R. J. Effect of Biomaterial Surface Properties on Fibronectin-β5β1 Integrin Interaction and Cellular Attachment. Biomaterials 2006, 27, 1907–1916. 10.1016/j.biomaterials.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Alexander M. R.; Williams P. Water Contact Angle Is Not a Good Predictor of Biological Responses to Materials. Biointerphases 2017, 12, 02C201 10.1116/1.4989843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P.; Iskratsch T.; Sheetz M. P. Finding the Weakest Link - Exploring Integrin-Mediated Mechanical Molecular Pathways. J. Cell Sci. 2012, 125, 3025–3038. 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R.; Pirone D. M.; Nelson C. M.; Bhadriraju K.; Chen C. S. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 2004, 6, 483–495. 10.1016/S1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Hoshiba T.; Tanaka M. Integrin-Independent Cell Adhesion Substrates: Possibility of Applications for Mechanobiology Research. Anal. Sci. 2016, 32, 1151–1158. 10.2116/analsci.32.1151. [DOI] [PubMed] [Google Scholar]
- Hoshiba T.; Nemoto E.; Sato K.; Orui T.; Otaki T.; Yoshihiro A.; Tanaka M. Regulation of the Contribution of Integrin to Cell Attachment on Poly(2-Methoxyethyl Acrylate) (PMEA) Analogous Polymers for Attachment-Based Cell Enrichment. PLoS One 2015, 10, e0136066 10.1371/journal.pone.0136066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Rowe R. G.; Botvinick E. L.; Kurup A.; Putnam A. J.; Seiki M.; Weaver V. M.; Keller E. T.; Goldstein S.; Dai J.; et al. MT1-MMP-Dependent Control of Skeletal Stem Cell Commitment via a B1-Integrin/YAP/TAZ Signaling Axis. Dev. Cell 2013, 25, 402–416. 10.1016/j.devcel.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S.; Morsut L.; Aragona M.; Enzo E.; Giulitti S.; Cordenonsi M.; Zanconato F.; Le Digabel J.; Forcato M.; Bicciato S.; et al. Role of YAP/TAZ in Mechanotransduction. Nature 2011, 474, 179–183. 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Abagnale G.; Sechi A.; Steger M.; Zhou Q.; Kuo C. C.; Aydin G.; Schalla C.; Müller-Newen G.; Zenke M.; Costa I. G.; et al. Surface Topography Guides Morphology and Spatial Patterning of Induced Pluripotent Stem Cell Colonies. Stem Cell Rep. 2017, 9, 654–666. 10.1016/j.stemcr.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton S. R.; Kim P. D.; Ghajar C. M.; Seliktar D.; Putnam A. J. The Effects of Matrix Stiffness and RhoA on the Phenotypic Plasticity of Smooth Muscle Cells in a 3-D Biosynthetic Hydrogel System. Biomaterials 2008, 29, 2597–2607. 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo C. H.; Furukawa K.; Montagne K.; Jeong H.; Ushida T. The Effect of Substrate Microtopography on Focal Adhesion Maturation and Actin Organization via the RhoA/ROCK Pathway. Biomaterials 2011, 32, 9568–9575. 10.1016/j.biomaterials.2011.08.077. [DOI] [PubMed] [Google Scholar]
- Lipsitz Y. Y.; Timmins N. E.; Zandstra P. W. Quality Cell Therapy Manufacturing by Design. Nat. Biotechnol. 2016, 34, 393–400. 10.1038/nbt.3525. [DOI] [PubMed] [Google Scholar]
- Archer R.; Williams D. J. Why Tissue Engineering Needs Process Engineering. Nat. Biotechnol. 2005, 23, 1353–1355. 10.1038/nbt1105-1353. [DOI] [PubMed] [Google Scholar]
- Lundstedt T.; Seifert E.; Abramo L.; Thelin B.; Nyström Å.; Pettersen J.; Bergman R. Experimental Design and Optimization. Chemom. Intell. Lab. Syst. 1998, 42, 3–40. 10.1016/S0169-7439(98)00065-3. [DOI] [Google Scholar]
- Van Schepdael A.; Carlier A.; Geris L.. Sensitivity Analysis by Design of Experiments BT - Uncertainty in Biology: A Computational Modeling Approach; Geris L., Gomez-Cabrero D., Eds.; Springer International Publishing: Cham, 2016; pp 327–366. [Google Scholar]
- George E. P.; Hunter J. S.; Hunter W. G.; Bins R.; Kirlin K. IV; Carroll D.. Statistics for Experimenters: Design, Innovation, and Discovery; Wiley: New York, 2005. [Google Scholar]
- Montgomery D. C.Montgomery Design and Analysis of Experiments; John Wiley, 1997. [Google Scholar]
- Myers R. H.; Montgomery D. C.; Anderson-Cook C. M.. Response Surface Methodology: Process and Product Optimization Using Designed Experiments; John Wiley & Sons, 2016. [Google Scholar]
- Saltelli A.; Chan K.; Scott E. M.. Sensitivity Analysis; John Wiley & Sons, Ltd., 2000. [Google Scholar]
- Design of Experiments Software; JMP Statistical Discovery, 2020. https://www.jmp.com/en_gb/offers/design-of-experiments.html?utm_campaign (accessed 10-29-2020).
- Designing an Experiment; Minitab LLC, 2019. https://support.minitab.com/en-us/minitab/18/getting-started/designing-an-experiment/ (accessed 10-29-2020).
- Design of Experiments (DOE) Made Easy. Design Expert; Statease, 2020. https://www.statease.com/software/design-expert/ (accessed 10-29-2020).
- Design of Experiments Software That Accelerates Progress. MODDE; Sartorius, 2020. https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software/doe-software/modde (accessed 10-29-2020).
- DoE. Base: Full Factorials, Orthogonal Arrays and Base Utilities for DoE Packages; Cran-R, 2020. https://cran.r-project.org/web/packages/DoE.base/index.html (accessed 10-29-2020).
- PyDOE: The Experimental Design Package for Python; Abraham Lee, 2014. https://pythonhosted.org/pyDOE/ (accessed 10-29-2020).
- Baker M. Is There a Reproducibility Crisis? A Nature Survey Lifts the Lid on How Researchers View the ‘crisis Rocking Science and What They Think Will Help. Nature 2016, 533, 452–455. 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- Ayala R.; Zhang C.; Yang D.; Hwang Y.; Aung A.; Shroff S. S.; Arce F. T.; Lal R.; Arya G.; Varghese S. Engineering the Cell-Material Interface for Controlling Stem Cell Adhesion, Migration, and Differentiation. Biomaterials 2011, 32, 3700–3711. 10.1016/j.biomaterials.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Stevens M. M.; George J. H. Exploring and Engineering the Cell Surface Interface. Science 2005, 310, 1135–1138. 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- Discher D. E.; Mooney D. J.; Zandstra P. W. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science 2009, 324, 1673–1677. 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsman M.; Hulshof F.; Unadkat H.; Papenburg B.; Stamatialis D.; Truckenmüller R.; van Blitterswijk C.; de Boer J.; Reinders M. J. T. Analysis of High-Throughput Screening Reveals the Effect of Surface Topographies on Cellular Morphology. Acta Biomater. 2015, 15, 29–38. 10.1016/j.actbio.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Kuhn P. T.; Zhou Q.; van der Boon T. A. B.; Schaap-Oziemlak A. M.; van Kooten T. G.; van Rijn P. Double Linear Gradient Biointerfaces for Determining Two- Parameter Dependent Stem Cell Behavior. ChemNanoMat 2016, 2, 407–413. 10.1002/cnma.201600028. [DOI] [Google Scholar]
- Zhou Q.; Ge L.; Guimarães C. F.; Kühn P. T.; Yang L.; van Rijn P. Development of a Novel Orthogonal Double Gradient for High-Throughput Screening of Mesenchymal Stem Cells-Materials Interaction. Adv. Mater. Interfaces 2018, 5, 4–11. 10.1002/admi.201800504. [DOI] [Google Scholar]
- Oh S. H.; An D. B.; Kim T. H.; Lee J. H. Wide-Range Stiffness Gradient PVA/HA Hydrogel to Investigate Stem Cell Differentiation Behavior. Acta Biomater. 2016, 35, 23–31. 10.1016/j.actbio.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Wang P. Y.; Tsai W. B.; Voelcker N. H. Screening of Rat Mesenchymal Stem Cell Behaviour on Polydimethylsiloxane Stiffness Gradients. Acta Biomater. 2012, 8, 519–530. 10.1016/j.actbio.2011.09.030. [DOI] [PubMed] [Google Scholar]
- Stauffer S. R.; Peppast N. A. Poly(Vinyl Alcohol) Hydrogels Prepared by Freezing-Thawing Cyclic Processing. Polymer 1992, 33, 3932–3936. 10.1016/0032-3861(92)90385-A. [DOI] [Google Scholar]
- Nemir S.; Hayenga H. N.; West J. L. PEGDA Hydrogels with Patterned Elasticity: Novel Tools for the Study of Cell Response to Substrate Rigidity. Biotechnol. Bioeng. 2010, 105, 636–644. 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- Wong J. Y.; Velasco A.; Rajagopalan P.; Pham Q. Directed Movement of Vascular Smooth Muscle Cells on Gradient-Compliant Hydrogels. Langmuir 2003, 19, 1908–1913. 10.1021/la026403p. [DOI] [Google Scholar]
- Marklein R. A.; Burdick J. A. Spatially Controlled Hydrogel Mechanics to Modulate Stem Cell Interactions. Soft Matter 2010, 6, 136–143. 10.1039/B916933D. [DOI] [Google Scholar]
- Sunyer R.; Jin A. J.; Nossal R.; Sackett D. L. Fabrication of Hydrogels with Steep Stiffness Gradients for Studying Cell Mechanical Response. PLoS One 2012, 7, 1–9. 10.1371/journal.pone.0046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent L. G.; Choi Y. S.; Alonso-Latorre B.; Del Álamo J. C.; Engler A. J. Mesenchymal Stem Cell Durotaxis Depends on Substrate Stiffness Gradient Strength. Biotechnol. J. 2013, 8, 472–484. 10.1002/biot.201200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.; Kim M.; Kim S.; Park M.; Kong K.; Letters H. K.-T.; et al. Acrylamide Induces Cell Death in Neural Progenitor Cells and Impairs Hippocampal Neurogenesis. Toxicol. Lett. 2010, 193, 86–93. 10.1016/j.toxlet.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Regehr K.; Domenech M.; Koepsel J.; Chip K. C.-L.; et al. Biological Implications of Polydimethylsiloxane-Based Microfluidic Cell Culture. Lab Chip 2009, 9, 2132–2139. 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.; Malik A.; Kim T.; Biomaterials P. M.; et al. Variable Cytocompatibility of Six Cell Lines with Photoinitiators Used for Polymerizing Hydrogels and Cell Encapsulation. Biomaterials 2005, 26, 1211–1218. 10.1016/j.biomaterials.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Hao Y.; Xu P.; He C.; Yang X.; Huang M.; Xing J.; Chen J. Impact of Carbondiimide Crosslinker Used for Magnetic Carbon Nanotube Mediated GFP Plasmid Delivery. Nanotechnology 2011, 22, 285103. 10.1088/0957-4484/22/28/285103. [DOI] [PubMed] [Google Scholar]
- Kloxin A. M.; Benton J. A.; Anseth K. S. In Situ Elasticity Modulation with Dynamic Substrates to Direct Cell Phenotype. Biomaterials 2010, 31, 1–8. 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H.; Han K.; Gupta K.; Kwon K. W.; Suh K. Y.; Levchenko A. Mechanosensitivity of Fibroblast Cell Shape and Movement to Anisotropic Substratum Topography Gradients. Biomaterials 2009, 30, 5433–5444. 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. M.; Pedersen R. H.; Stormonth-Darling J.; Dalby M. J.; Riehle M. O.; Gadegaard N. Label-Free Segmentation of Co-Cultured Cells on a Nanotopographical Gradient. Nano Lett. 2013, 13, 570–576. 10.1021/nl304097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconnet D.; Csucs G.; Grandin H.; Biomaterials M. T. Surface Engineering Approaches to Micropattern Surfaces for Cell-Based Assays. Biomaterials 2006, 27, 3044–3063. 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Michel R.; Lussi J. W.; Csucs G.; Reviakine I.; Danuser G.; Ketterer B.; Hubbell J. A.; Textor M.; Spencer N. D. Selective Molecular Assembly Patterning: A New Approach to Micro- and Nanochemical Patterning of Surfaces for Biological Applications. Langmuir 2002, 18, 3281–3287. 10.1021/la011715y. [DOI] [Google Scholar]
- Falconnet D.; Koenig A.; Assi F.; Textor M. A Combined Photolithographic and Molecular-Assembly Approach to Produce Functional Micropatterns for Applications in the Biosciences. Adv. Funct. Mater. 2004, 14, 749–756. 10.1002/adfm.200305182. [DOI] [Google Scholar]
- Jang K.; Xu Y.; Sato K.; Tanaka Y.; Mawatari K.; Kitamori T. Micropatterning of Biomolecules on a Glass Substrate in Fused Silica Microchannels by Using Photolabile Linker-Based Surface Activation. Microchim. Acta 2012, 179, 49–55. 10.1007/s00604-012-0856-8. [DOI] [Google Scholar]
- Hahn M.; Taite L.; Moon J.; Rowland M.; Ruffino K.; West J. Photolithographic Patterning of Polyethylene Glycol Hydrogels. Biomaterials 2006, 27, 2519–2524. 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Whitesides G. M.; Ostuni E.; Takayama S.; Jiang X.; Ingber D. E. Soft Lithography in Biology and Biochemistry. Annu. Rev. Biomed. Eng. 2001, 3, 335–373. 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- Wang B.; Shi J.; Wei J.; Wang L.; Tu X.; Tang Y.; Chen Y. Fabrication of Elastomer Pillar Arrays with Height Gradient for Cell Culture Studies. Microelectron. Eng. 2017, 175, 50–55. 10.1016/j.mee.2017.01.014. [DOI] [Google Scholar]
- Zhou Q.; Wünnemann P.; Kühn P. T.; de Vries J.; Helmin M.; Böker A.; van Kooten T. G.; van Rijn P. Mechanical Properties of Aligned Nanotopologies for Directing Cellular Behavior. Adv. Mater. Interfaces 2016, 3, 1600275. 10.1002/admi.201600275. [DOI] [Google Scholar]
- Almonacid Suarez A. M.; Zhou Q.; van Rijn P.; Harmsen M. C. Directional Topography Gradients Drive Optimum Alignment and Differentiation of Human Myoblasts. J. Tissue Eng. Regener. Med. 2019, 13, 2234–2245. 10.1002/term.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori G. R.; Zhou Q.; Liguori T. T. A.; Barros G. G.; Kühn P. T.; Moreira L. F. P.; Van Rijn P.; Harmsen M. C. Directional Topography Influences Adipose Mesenchymal Stromal Cell Plasticity: Prospects for Tissue Engineering and Fibrosis. Stem Cells Int. 2019, 2019, 5387850. 10.1155/2019/5387850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L.; Yang L.; Bron R.; Burgess J. K.; van Rijn P. Topography-Mediated Fibroblast Cell Migration Is Influenced by Direction, Wavelength, and Amplitude. ACS Appl. Bio Mater. 2020, 3, 2104–2116. 10.1021/acsabm.0c00001. [DOI] [PubMed] [Google Scholar]
- Yang L.; Jurczak K. M.; Ge L.; van Rijn P. High-Throughput Screening and Hierarchical Topography-Mediated Neural Differentiation of Mesenchymal Stem Cells. Adv. Healthcare Mater. 2020, 9, 2000117. 10.1002/adhm.202000117. [DOI] [PubMed] [Google Scholar]
- Sun J.; Ding Y.; Lin N. J.; Zhou J.; Ro H.; Soles C. L.; Cicerone M. T.; Lin-Gibson S. Exploring cellular contact guidance using gradient nanogratings. Biomacromolecules 2010, 11, 3067–3072. 10.1021/bm100883m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. H.; Park I. K.; Kim J. M.; Lee J. H. In Vitro and in Vivo Characteristics of PCL Scaffolds with Pore Size Gradient Fabricated by a Centrifugation Method. Biomaterials 2007, 28, 1664–1671. 10.1016/j.biomaterials.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Oh S. H.; Kim T. H.; Im G. Il; Lee J. H. Investigation of Pore Size Effect on Chondrogenic Differentiation of Adipose Stem Cells Using a Pore Size Gradient Scaffold. Biomacromolecules 2010, 11, 1948–1955. 10.1021/bm100199m. [DOI] [PubMed] [Google Scholar]
- Wang P.-Y.; Clements L. R.; Thissen H.; Tsai W.-B.; Voelcker N. H. High-Throughput Characterisation of Osteogenic Differentiation of Human Mesenchymal Stem Cells Using Pore Size Gradients on Porous Alumina. Biomater. Sci. 2013, 1, 924. 10.1039/c3bm60026b. [DOI] [PubMed] [Google Scholar]
- Khung Y. L.; Barritt G.; Voelcker N. H. Using Continuous Porous Silicon Gradients to Study the Influence of Surface Topography on the Behaviour of Neuroblastoma Cells. Exp. Cell Res. 2008, 314, 789–800. 10.1016/j.yexcr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Collins B. E.; Dancil K. P. S.; Abbi G.; Sailor M. J. Determining Protein Size Using an Electrochemically Machined Pore Gradient in Silicon. Adv. Funct. Mater. 2002, 12, 187–191. . [DOI] [Google Scholar]
- Washburn N. R.; Yamada K. M.; Simon C. G.; Kennedy S. B.; Amis E. J. High-Throughput Investigation of Osteoblast Response to Polymer Crystallinity: Influence of Nanometer-Scale Roughness on Proliferation. Biomaterials 2004, 25, 1215–1224. 10.1016/j.biomaterials.2003.08.043. [DOI] [PubMed] [Google Scholar]
- Faia-Torres A. B.; Guimond-Lischer S.; Rottmar M.; Charnley M.; Goren T.; Maniura-Weber K.; Spencer N. D.; Reis R. L.; Textor M.; Neves N. M. Differential Regulation of Osteogenic Differentiation of Stem Cells on Surface Roughness Gradients. Biomaterials 2014, 35, 9023–9032. 10.1016/j.biomaterials.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Kunzler T. P.; Drobek T.; Schuler M.; Spencer N. D. Systematic Study of Osteoblast and Fibroblast Response to Roughness by Means of Surface-Morphology Gradients. Biomaterials 2007, 28, 2175–2182. 10.1016/j.biomaterials.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Kunzler T. P.; Drobek T.; Sprecher C. M.; Schuler M.; Spencer N. D. Fabrication of Material-Independent Morphology Gradients for High-Throughput Applications. Appl. Surf. Sci. 2006, 253, 2148–2153. 10.1016/j.apsusc.2006.04.014. [DOI] [Google Scholar]
- Rockwell G. P.; Lohstreter L. B.; Dahn J. R. Fibrinogen and Albumin Adsorption on Titanium Nanoroughness Gradients. Colloids Surf., B 2012, 91, 90–96. 10.1016/j.colsurfb.2011.10.045. [DOI] [PubMed] [Google Scholar]
- Kunzler T. P.; Huwiler C.; Drobek T.; Vörös J.; Spencer N. D. Systematic Study of Osteoblast Response to Nanotopography by Means of Nanoparticle-Density Gradients. Biomaterials 2007, 28, 5000–5006. 10.1016/j.biomaterials.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Huwiler C.; Kunzler T. P.; Textor M.; Vörös J.; Spencer N. D. Functionalizable Nanomorphology Gradients via Colloidal Self-Assembly. Langmuir 2007, 23, 5929–5935. 10.1021/la0700422. [DOI] [PubMed] [Google Scholar]
- Bachhuka A.; Delalat B.; Ghaemi S. R.; Gronthos S.; Voelcker N. H.; Vasilev K. Nanotopography Mediated Osteogenic Differentiation of Human Dental Pulp Derived Stem Cells. Nanoscale 2017, 9, 14248–14258. 10.1039/C7NR03131A. [DOI] [PubMed] [Google Scholar]
- MacGregor-Ramiasa M.; Hopp I.; Bachhuka A.; Murray P.; Vasilev K. Surface Nanotopography Guides Kidney-Derived Stem Cell Differentiation into Podocytes. Acta Biomater. 2017, 56, 171–180. 10.1016/j.actbio.2017.02.036. [DOI] [PubMed] [Google Scholar]
- Li X.; MacEwan M. R.; Xie J.; Siewe D.; Yuan X.; Xia Y. Fabrication of Density Gradients of Biodegradable Polymer Microparticles and Their Use in Guiding Neurite Outgrowth. Adv. Funct. Mater. 2010, 20, 1632–1637. 10.1002/adfm.201000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon y Leon C. A New Perspectives in Mercury Porosimetry. Adv. Colloid Interface Sci. 1998, 76–77, 341–372. 10.1016/S0001-8686(98)00052-9. [DOI] [Google Scholar]
- Yang S.; Leong K.-F.; Du Z.; Chua C.-K. The Design of Scaffolds for Use in Tissue Engineering. Part I. Traditional Factors. Tissue Eng. 2001, 7, 679–689. 10.1089/107632701753337645. [DOI] [PubMed] [Google Scholar]
- van Tienen T. G.; Heijkants R. G. J. C.; Buma P.; de Groot J. H.; Pennings A. J.; Veth R. P. H. Tissue Ingrowth and Degradation of Two Biodegradable Porous Polymers with Different Porosities and Pore Sizes. Biomaterials 2002, 23, 1731–1738. 10.1016/S0142-9612(01)00280-0. [DOI] [PubMed] [Google Scholar]
- Story B. J.; Wagner W. R.; Gaisser D. M.; Cook S. D.; Rust-Dawicki A. M. In Vivo Performance of a Modified CSTi Dental Implant Coating. Int. J. Oral Maxillofac. Implants 1998, 13, 749–757. [PubMed] [Google Scholar]
- Karageorgiou V.; Kaplan D. Porosity of 3D Biomaterial Scaffolds and Osteogenesis. Biomaterials 2005, 26, 5474–5491. 10.1016/j.biomaterials.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Canham L. T. Bioactive Silicon Structure Fabrication through Nanoetching Techniques. Adv. Mater. 1995, 7, 1033–1037. 10.1002/adma.19950071215. [DOI] [Google Scholar]
- Janshoff A.; Dancil K.-P. S.; Steinem C.; Greiner D. P.; Lin V. S.-Y.; Gurtner C.; Motesharei K.; Sailor M. J.; Ghadiri M. R. Macroporous P-Type Silicon Fabry-Perot Layers. Fabrication, Characterization, and Applications in Biosensing. J. Am. Chem. Soc. 1998, 120, 12108–12116. 10.1021/ja9826237. [DOI] [Google Scholar]
- Stewart M. P.; Buriak J. M. Chemical and Biological Applications of Porous Silicon Technology. Adv. Mater. 2000, 12, 859–869. . [DOI] [Google Scholar]
- Sapelkin A.; Bayliss S.; Unal B.; Charalambou A. Interaction of B50 Rat Hippocampal Cells with Stain-Etched Porous Silicon. Biomaterials 2006, 27, 842–846. 10.1016/j.biomaterials.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Dronov R.; Jane A.; Shapter J. G.; Hodges A.; Voelcker N. H. Nanoporous Alumina-Based Interferometric Transducers Ennobled. Nanoscale 2011, 3, 3109–3114. 10.1039/c0nr00897d. [DOI] [PubMed] [Google Scholar]
- Jani A. M. M.; Kempson I. M.; Losic D.; Voelcker N. H. Dressing in Layers: Layering Surface Functionalities in Nanoporous Aluminum Oxide Membranes. Angew. Chem., Int. Ed. 2010, 49, 7933–7937. 10.1002/anie.201002504. [DOI] [PubMed] [Google Scholar]
- Mutalib Md Jani A.; Anglin E. J.; McInnes S. J. P.; Losic D.; Shapter J. G.; Voelcker N. H. Nanoporous Anodic Aluminium Oxide Membranes with Layered Surface Chemistry. Chem. Commun. 2009, 3062–3064. 10.1039/b901745c. [DOI] [PubMed] [Google Scholar]
- Gultepe E.; Nagesha D.; Sridhar S.; Amiji M. Nanoporous Inorganic Membranes or Coatings for Sustained Drug Delivery in Implantable Devices. Adv. Drug Delivery Rev. 2010, 62, 305–315. 10.1016/j.addr.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Ingham C. J.; ter Maat J.; de Vos W. M. Where Bio Meets Nano: The Many Uses for Nanoporous Aluminum Oxide in Biotechnology. Biotechnol. Adv. 2012, 30, 1089–1099. 10.1016/j.biotechadv.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Flemming R.G.; Murphy C.J.; Abrams G.A.; Goodman S.L.; Nealey P.F. Effects of Synthetic Micro-and Nano-Structured Surfaces on Cell Behavior. Biomaterials 1999, 20, 573–588. 10.1016/S0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- Brunette D. M.; Chehroudi B. The Effects of the Surface Topography of Micromachined Titanium Substrata on Cell Behavior in Vitro and in Vivo. J. Biomech. Eng. 1999, 121, 49–57. 10.1115/1.2798042. [DOI] [PubMed] [Google Scholar]
- Elias C. N.; Oshida Y.; Lima J. H. C.; Muller C. A. Relationship between Surface Properties (Roughness, Wettability and Morphology) of Titanium and Dental Implant Removal Torque. J. Mech. Behav. Biomed. Mater. 2008, 1, 234–242. 10.1016/j.jmbbm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Hefti T.; Frischherz M.; Spencer N. D.; Hall H.; Schlottig F. A Comparison of Osteoclast Resorption Pits on Bone with Titanium and Zirconia Surfaces. Biomaterials 2010, 31, 7321–7331. 10.1016/j.biomaterials.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Yeo A.; Wong W. J.; Khoo H. H.; Teoh S. H. Surface Modification of PCL-TCP Scaffolds Improve Interfacial Mechanical Interlock and Enhance Early Bone Formation: An in Vitro and in Vivo Characterization. J. Biomed. Mater. Res., Part A 2010, 92A, 311–321. 10.1002/jbm.a.32366. [DOI] [PubMed] [Google Scholar]
- Gahlert M.; Röhling S.; Wieland M.; Sprecher C. M.; Kniha H.; Milz S. Osseointegration of Zirconia and Titanium Dental Implants: A Histological and Histomorphometrical Study in the Maxilla of Pigs. Clin. Oral Implants Res. 2009, 20, 1247–1253. 10.1111/j.1600-0501.2009.01734.x. [DOI] [PubMed] [Google Scholar]
- Gahlert M.; Röhling S.; Wieland M.; Eichhorn S.; Küchenhoff H.; Kniha H. A Comparison Study of the Osseointegration of Zirconia and Titanium Dental Implants. A Biomechanical Evaluation in the Maxilla of Pigs. Clin. Implant Dent. Relat. Res. 2010, 12, 297–305. 10.1111/j.1708-8208.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Krämer S.; Xie H.; Gaff J.; Williamson J. R.; Tkachenko A. G.; Nouri N.; Feldheim D. A.; Feldheim D. L. Preparation of Protein Gradients through the Controlled Deposition of Protein-Nanoparticle Conjugates onto Functionalized Surfaces. J. Am. Chem. Soc. 2004, 126, 5388–5395. 10.1021/ja031674n. [DOI] [PubMed] [Google Scholar]
- Bhat R. R.; Fischer D. A.; Genzer J. Fabricating Planar Nanoparticle Assemblies with Number Density Gradients. Langmuir 2002, 18, 5640–5643. 10.1021/la025524m. [DOI] [Google Scholar]
- Xie J.; Marijnissen J. C. M.; Wang C. H. Microparticles Developed by Electrohydrodynamic Atomization for the Local Delivery of Anticancer Drug to Treat C6 Glioma in Vitro. Biomaterials 2006, 27, 3321–3332. 10.1016/j.biomaterials.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Hanna M. A. Electrospray Encapsulation of Water-Soluble Protein with Polylactide. Effects of Formulations on Morphology, Encapsulation Efficiency and Release Profile of Particles. Int. J. Pharm. 2006, 320, 30–36. 10.1016/j.ijpharm.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Sardella E.; Salama R. A.; Waly G. H.; Habib A. N.; Favia P.; Gristina R. Improving Internal Cell Colonization of Porous Scaffolds with Chemical Gradients Produced by Plasma Assisted Approaches. ACS Appl. Mater. Interfaces 2017, 9, 4966–4975. 10.1021/acsami.6b14170. [DOI] [PubMed] [Google Scholar]
- Choee J. H.; Lee S. J.; Lee Y. M.; Rhee J. M.; Lee H. B.; Khang G. Proliferation Rate of Fibroblast Cells on Polyethylene Surfaces with Wettability Gradient. J. Appl. Polym. Sci. 2004, 92, 599–606. 10.1002/app.20048. [DOI] [Google Scholar]
- Morgenthaler S.; Lee S.; Zürcher S.; Spencer N. D. A Simple, Reproducible Approach to the Preparation of Surface-Chemical Gradients. Langmuir 2003, 19, 10459–10462. 10.1021/la034707l. [DOI] [PubMed] [Google Scholar]
- Mohan G.; Gallant N. D. Surface Chemistry Gradients on Silicone Elastomers for High-Throughput Modulation of Cell-Adhesive Interfaces. J. Biomed. Mater. Res., Part A 2015, 103, 2066–2076. 10.1002/jbm.a.35349. [DOI] [PubMed] [Google Scholar]
- Li B.; Ma Y.; Wang S.; Moran P. M. Influence of Carboxyl Group Density on Neuron Cell Attachment and Differentiation Behavior: Gradient-Guided Neurite Outgrowth. Biomaterials 2005, 26, 4956–4963. 10.1016/j.biomaterials.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Wells N.; Baxter M. A.; Turnbull J. E.; Murray P.; Edgar D.; Parry K. L.; Steele D. A.; Short R. D. The Geometric Control of E14 and R1Mouse Embryonic Stem Cell Pluripotency by Plasma Polymer Surface Chemical Gradients. Biomaterials 2009, 30, 1066–1070. 10.1016/j.biomaterials.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Harding F. J.; Clements L. R.; Short R. D.; Thissen H.; Voelcker N. H. Assessing Embryonic Stem Cell Response to Surface Chemistry Using Plasma Polymer Gradients. Acta Biomater. 2012, 8, 1739–1748. 10.1016/j.actbio.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Zelzer M.; Alexander M. R.; Russell N. A. Hippocampal Cell Response to Substrates with Surface Chemistry Gradients. Acta Biomater. 2011, 7, 4120–4130. 10.1016/j.actbio.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Menzies D. J.; Cowie B.; Fong C.; Forsythe J. S.; Gengenbach T. R.; McLean K. M.; Puskar L.; Textor M.; Thomsen L.; Tobin M.; et al. One-Step Method for Generating PEG-like Plasma Polymer Gradients: Chemical Characterization and Analysis of Protein Interactions. Langmuir 2010, 26, 13987–13994. 10.1021/la102033d. [DOI] [PubMed] [Google Scholar]
- Balamurugan A.; Balossier G.; Michel J.; Kannan S.; Benhayoune H.; Rebelo A. H. S.; Ferreira J. M. F. System — Preparation and In Vitro Characterization. J. Biomed. Mater. Res., Part B 2007, 5, 546–553. 10.1002/jbm.b.30827. [DOI] [PubMed] [Google Scholar]
- Zelzer M.; Majani R.; Bradley J. W.; Rose F. R. A. J.; Davies M. C.; Alexander M. R. Investigation of Cell-Surface Interactions Using Chemical Gradients Formed from Plasma Polymers. Biomaterials 2008, 29, 172–184. 10.1016/j.biomaterials.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Whittle J. D.; Barton D.; Alexander M. R.; Short R. D. A Method for the Deposition of Controllable Chemical Gradients. Chem. Commun. 2003, 9, 1766–1767. 10.1039/b305445b. [DOI] [Google Scholar]
- Guan Z. Y.; Wu C. Y.; Wu J. T.; Tai C. H.; Yu J.; Chen H. Y. Multifunctional and Continuous Gradients of Biointerfaces Based on Dual Reverse Click Reactions. ACS Appl. Mater. Interfaces 2016, 8, 13812–13818. 10.1021/acsami.6b03908. [DOI] [PubMed] [Google Scholar]
- Liu X.; Shi S.; Feng Q.; Bachhuka A.; He W.; Huang Q.; Zhang R.; Yang X.; Vasilev K. Surface Chemical Gradient Affects the Differentiation of Human Adipose-Derived Stem Cells via ERK1/2 Signaling Pathway. ACS Appl. Mater. Interfaces 2015, 7, 18473–18482. 10.1021/acsami.5b04635. [DOI] [PubMed] [Google Scholar]
- Pitt W. G. Fabrication of a Continuous Wettability Gradient by Radio Frequency Plasma Discharge. J. Colloid Interface Sci. 1989, 133, 223–227. 10.1016/0021-9797(89)90295-6. [DOI] [Google Scholar]
- Golander C.-G.; Pitt W. G. Characterization of Hydrophobicity Gradients Prepared by Means of Radio Frequency Plasma Discharge. Biomaterials 1990, 11, 32–35. 10.1016/0142-9612(90)90048-U. [DOI] [PubMed] [Google Scholar]
- Wang P. Y.; Clements L. R.; Thissen H.; Tsai W. B.; Voelcker N. H. Screening Rat Mesenchymal Stem Cell Attachment and Differentiation on Surface Chemistries Using Plasma Polymer Gradients. Acta Biomater. 2015, 11, 58–67. 10.1016/j.actbio.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Ulman A. Formation and Structure of Self-Assembled Monolayers. Chem. Rev. 1996, 96, 1533–1554. 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- Ostuni E.; Yan L.; Whitesides G. M. The Interaction of Proteins and Cells with Self-Assembled Monolayers of Alkanethiolates on Gold and Silver. Colloids Surf., B 1999, 15, 3–30. 10.1016/S0927-7765(99)00004-1. [DOI] [Google Scholar]
- Gijsman P.; Dozeman A. Comparison of the UV-Degradation Chemistry of Polypropylene, Polyethylene, Polyamide 6 and Polybutylene Terephthalate. Polym. Degrad. Stab. 1996, 53, 45–50. 10.1016/0141-3910(96)00027-4. [DOI] [Google Scholar]
- Li B.; Ma Y.; Wang S.; Moran P. M. A Technique for Preparing Protein Gradients on Polymeric Surfaces: Effects on PC12 Pheochromocytoma Cells. Biomaterials 2005, 26, 1487–1495. 10.1016/j.biomaterials.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Gomez N.; Lu Y.; Chen S.; Schmidt C. E. Immobilized Nerve Growth Factor and Microtopography Have Distinct Effects on Polarization versus Axon Elongation in Hippocampal Cells in Culture. Biomaterials 2007, 28, 271–284. 10.1016/j.biomaterials.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Venugopal J.; Low S.; Choon A. T.; Ramakrishna S. Interaction of Cells and Nanofiber Scaffolds in Tissue Engineering. J. Biomed. Mater. Res., Part B 2008, 84B, 34–48. 10.1002/jbm.b.30841. [DOI] [PubMed] [Google Scholar]
- Rønning S. B.; Pedersen M. E.; Andersen P. V.; Hollung K. The Combination of Glycosaminoglycans and Fibrous Proteins Improves Cell Proliferation and Early Differentiation of Bovine Primary Skeletal Muscle Cells. Differentiation 2013, 86, 13–22. 10.1016/j.diff.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Faia-Torres A. B.; Goren T.; Ihalainen T. O.; Guimond-Lischer S.; Charnley M.; Rottmar M.; Maniura-Weber K.; Spencer N. D.; Reis R. L.; Textor M.; et al. Regulation of Human Mesenchymal Stem Cell Osteogenesis by Specific Surface Density of Fibronectin: A Gradient Study. ACS Appl. Mater. Interfaces 2015, 7, 2367–2375. 10.1021/am506951c. [DOI] [PubMed] [Google Scholar]
- Wu T.; Xu C.; Beers K. L.; Vogt B. D.; Elliott J. T.; Amis E. J.; Langenbach K. J.; Mei Y.; Washburn N. R. Tuning Cell Adhesion on Gradient Poly(2-Hydroxyethyl Methacrylate)-Grafted Surfaces. Langmuir 2005, 21, 12309–12314. 10.1021/la050668x. [DOI] [PubMed] [Google Scholar]
- Vasilev K.; Hook A. L.; Mierczynska A.; Voelcker N. H.; Short R. D.; Chan J. Creating Gradients of Two Proteins by Differential Passive Adsorption onto a PEG-Density Gradient. Biomaterials 2010, 31, 392–397. 10.1016/j.biomaterials.2009.09.056. [DOI] [PubMed] [Google Scholar]
- Seidi A.; Ramalingam M.; Elloumi-Hannachi I.; Ostrovidov S.; Khademhosseini A. Gradient Biomaterials for Soft-to-Hard Interface Tissue Engineering. Acta Biomater. 2011, 7, 1441–1451. 10.1016/j.actbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Shi J.; Wang L.; Zhang F.; Li H.; Lei L.; Liu L.; Chen Y. Incorporating Protein Gradient into Electrospun Nanofibers as Scaffolds for Tissue Engineering. ACS Appl. Mater. Interfaces 2010, 2, 1025–1030. 10.1021/am9007962. [DOI] [PubMed] [Google Scholar]
- Li L.; Wu J.; Gao C. Gradient Immobilization of a Cell Adhesion RGD Peptide on Thermal Responsive Surface for Regulating Cell Adhesion and Detachment. Colloids Surf., B 2011, 85, 12–18. 10.1016/j.colsurfb.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Acharya A. P.; Dolgova N. V.; Moore N. M.; Xia C. Q.; Clare-Salzler M. J.; Becker M. L.; Gallant N. D.; Keselowsky B. G. The Modulation of Dendritic Cell Integrin Binding and Activation by RGD-Peptide Density Gradient Substrates. Biomaterials 2010, 31, 7444–7454. 10.1016/j.biomaterials.2010.06.025. [DOI] [PubMed] [Google Scholar]
- Moore N. M.; Lin N. J.; Gallant N. D.; Becker M. L. The Use of Immobilized Osteogenic Growth Peptide on Gradient Substrates Synthesized via Click Chemistry to Enhance MC3T3-E1 Osteoblast Proliferation. Biomaterials 2010, 31, 1604–1611. 10.1016/j.biomaterials.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Yu S.; Zuo X.; Shen T.; Duan Y.; Mao Z.; Gao C. A Density Gradient of VAPG Peptides on a Cell-Resisting Surface Achieves Selective Adhesion and Directional Migration of Smooth Muscle Cells over Fibroblasts. Acta Biomater. 2018, 72, 70–81. 10.1016/j.actbio.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Yu S.; Gao Y.; Mei X.; Ren T.; Liang S.; Mao Z.; Gao C. Preparation of an Arg-Glu-Asp-Val Peptide Density Gradient on Hyaluronic Acid-Coated Poly (ε-Caprolactone) Film and Its Influence on the Selective Adhesion and Directional Migration of Endothelial Cells. ACS Appl. Mater. Interfaces 2016, 8, 29280–29288. 10.1021/acsami.6b09375. [DOI] [PubMed] [Google Scholar]
- Delalat B.; Mierczynska A.; Ghaemi S. R.; Cavallaro A.; Harding F. J.; Vasilev K.; Voelcker N. H. Materials Displaying Neural Growth Factor Gradients and Applications in Neural Differentiation of Embryoid Body Cells. Adv. Funct. Mater. 2015, 25, 2737–2744. 10.1002/adfm.201500595. [DOI] [Google Scholar]
- Liu L.; Ratner B. D.; Sage E. H.; Jiang S. Endothelial Cell Migration on Surface-Density Gradients of Fibronectin, VEGF, or Both Proteins. Langmuir 2007, 23, 11168–11173. 10.1021/la701435x. [DOI] [PubMed] [Google Scholar]
- Wu J.; Mao Z.; Han L.; Zhao Y.; Xi J.; Gao C. A Density Gradient of Basic Fibroblast Growth Factor Guides Directional Migration of Vascular Smooth Muscle Cells. Colloids Surf., B 2014, 117, 290–295. 10.1016/j.colsurfb.2014.02.043. [DOI] [PubMed] [Google Scholar]
- Rasi Ghaemi S.; Harding F. J.; Delalat B.; Gronthos S.; Voelcker N. H. Exploring the Mesenchymal Stem Cell Niche Using High Throughput Screening. Biomaterials 2013, 34, 7601–7615. 10.1016/j.biomaterials.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Yang J.; Rose F. R. A. J.; Gadegaard N.; Alexander M. R. A High-Throughput Assay of Cell-Surface Interactions Using Topographical and Chemical Gradients. Adv. Mater. 2009, 21, 300–304. 10.1002/adma.200801942. [DOI] [Google Scholar]
- Simon C. G.; Eidelman N.; Kennedy S. B.; Sehgal A.; Khatri C. A.; Washburn N. R. Combinatorial Screening of Cell Proliferation on Poly(l-Lactic Acid)/Poly(d, l-Lactic Acid) Blends. Biomaterials 2005, 26, 6906–6915. 10.1016/j.biomaterials.2005.04.050. [DOI] [PubMed] [Google Scholar]
- Clements L. R.; Wang P. Y.; Tsai W. B.; Thissen H.; Voelcker N. H. Electrochemistry-Enabled Fabrication of Orthogonal Nanotopography and Surface Chemistry Gradients for High-Throughput Screening. Lab Chip 2012, 12, 1480–1486. 10.1039/c2lc20732j. [DOI] [PubMed] [Google Scholar]
- Huethorst E.; Hortigon M.; Zamora-Rodriguez V.; Reynolds P. M.; Burton F.; Smith G.; Gadegaard N. Enhanced Human-Induced Pluripotent Stem Cell Derived Cardiomyocyte Maturation Using a Dual Microgradient Substrate. ACS Biomater. Sci. Eng. 2016, 2, 2231–2239. 10.1021/acsbiomaterials.6b00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P.; Parker T.; Gadegaard N.; Alexander M. R. A Bio-Inspired Neural Environment to Control Neurons Comprising Radial Glia, Substrate Chemistry and Topography. Biomater. Sci. 2013, 1, 83–93. 10.1039/C2BM00060A. [DOI] [PubMed] [Google Scholar]
- Ma Y.; Policastro G. M.; Li Q.; Zheng J.; Jacquet R.; Landis W. J.; Becker M. L. Concentration-Dependent HMSC Differentiation on Orthogonal Concentration Gradients of GRGDS and BMP-2 Peptides. Biomacromolecules 2016, 17, 1486–1495. 10.1021/acs.biomac.6b00088. [DOI] [PubMed] [Google Scholar]
- Rape A. D.; Zibinsky M.; Murthy N.; Kumar S. A Synthetic Hydrogel for the High-Throughput Study of Cell-ECM Interactions. Nat. Commun. 2015, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements L. R.; Wang P.-Y.; Tsai W.-B.; Thissen H.; Voelcker N. H. Electrochemistry-Enabled Fabrication of Orthogonal Nanotopography and Surface Chemistry Gradients for High-Throughput Screening. Lab Chip 2012, 12, 1480. 10.1039/c2lc20732j. [DOI] [PubMed] [Google Scholar]
- Dutta Roy T.; Simon J. L.; Ricci J. L.; Rekow E. D.; Thompson V. P.; Parsons J. R. Performance of Hydroxyapatite Bone Repair Scaffolds Created via Three-Dimensional Fabrication Techniques. J. Biomed. Mater. Res., Part A 2003, 67A, 1228–1237. 10.1002/jbm.a.20034. [DOI] [PubMed] [Google Scholar]
- Woodfield T. B. F.; Blitterswijk C. A.; Wijn J. De; Sims T. J.; Hollander A. P.; Riesle J. Polymer Scaffolds Fabricated with Pore-Size Gradients as a Model for Studying the Zonal Organization within Tissue-Engineered Cartilage Constructs. Tissue Eng. 2005, 11, 1297–1311. 10.1089/ten.2005.11.1297. [DOI] [PubMed] [Google Scholar]
- Chatterjee K.; Lin-Gibson S.; Wallace W. E.; Parekh S. H.; Lee Y. J.; Cicerone M. T.; Young M. F.; Simon C. G. The Effect of 3D Hydrogel Scaffold Modulus on Osteoblast Differentiation and Mineralization Revealed by Combinatorial Screening. Biomaterials 2010, 31, 5051–5062. 10.1016/j.biomaterials.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. D.; Tan H. P.; Li L. H.; Gao C. Y. Covalently Immobilized Gelatin Gradients within Three-Dimensional Porous Scaffolds. Chin. Sci. Bull. 2009, 54, 3174–3180. 10.1007/s11434-009-0215-2. [DOI] [Google Scholar]
- Oh S. H.; Kim T. H.; Lee J. H. Creating Growth Factor Gradients in Three Dimensional Porous Matrix by Centrifugation and Surface Immobilization. Biomaterials 2011, 32, 8254–8260. 10.1016/j.biomaterials.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Pei J.; Hall H.; Spencer N. D. The Role of Plasma Proteins in Cell Adhesion to PEG Surface-Density-Gradient-Modified Titanium Oxide. Biomaterials 2011, 32, 8968–8978. 10.1016/j.biomaterials.2011.08.034. [DOI] [PubMed] [Google Scholar]
- Ratner B. D. The Blood Compatibility Catastrophe. J. Biomed. Mater. Res. 1993, 27, 283–287. 10.1002/jbm.820270302. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli J.-P.; Klein-Soyer C.; Beretz A.; Brisson C.; Archipoff G.; Cazenave J.-P. Endothelial Cell Seeding: Coating Dacron and Expanded Polytetrafluoroethylene Vascular Grafts with a Biological Glue Allows Adhesion and Growth of Human Saphenous Vein Endothelial Cells. Int. J. Artif. Organs 1991, 14, 482–490. 10.1177/039139889101400806. [DOI] [PubMed] [Google Scholar]
- Bhat R. R.; Chaney B. N.; Rowley J.; Liebmann-Vinson A.; Genzer J. Tailoring Cell Adhesion Using Surface-Grafted Polymer Gradient Assemblies. Adv. Mater. 2005, 17, 2802–2807. 10.1002/adma.200500858. [DOI] [Google Scholar]
- Cantini M.; Sousa M.; Moratal D.; Mano J. F.; Salmerón-Sánchez M. Non-Monotonic Cell Differentiation Pattern on Extreme Wettability Gradients. Biomater. Sci. 2013, 1, 202–212. 10.1039/C2BM00063F. [DOI] [PubMed] [Google Scholar]
- Lee J. H.; Lee H. B. A Wettability Gradient as a Tool to Study Protein Adsorption and Cell Adhesion on Polymer Surfaces. J. Biomater. Sci., Polym. Ed. 1993, 4, 467–481. 10.1163/156856293X00717. [DOI] [PubMed] [Google Scholar]
- Ekblad T.; Andersson O.; Tai F. I.; Edeith T.; Liedberg B. Lateral Control of Protein Adsorption on Charged Polymer Gradients. Langmuir 2009, 25, 3755–3762. 10.1021/la803443d. [DOI] [PubMed] [Google Scholar]
- Altankov G.; Thom V.; Groth T.; Jankova K.; Jonsson G.; Ulbricht M. Modulating the Biocompatibility of Polymer Surfaces with Poly(Ethylene Glycol): Effect of Fibronectin. J. Biomed. Mater. Res. 2000, 52, 219–230. . [DOI] [PubMed] [Google Scholar]
- Iuliano D. J.; Saavedra S. S.; Truskey G. A. Effect of the Conformation and Orientation of Adsorbed Fibronectin on Endothelial Cell Spreading and the Strength of Adhesion. J. Biomed. Mater. Res. 1993, 27, 1103–1113. 10.1002/jbm.820270816. [DOI] [PubMed] [Google Scholar]
- Lin Y. S.; Hlady V.; Gölander C. G. The Surface Density Gradient of Grafted Poly(Ethylene Glycol): Preparation, Characterization and Protein Adsorption. Colloids Surf., B 1994, 3, 49–62. 10.1016/0927-7765(93)01114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingshott P.; McArthur S.; Thissen H.; Castner D. G.; Griesser H. J. Ultrasensitive Probing of the Protein Resistance of PEG Surfaces by Secondary Ion Mass Spectrometry. Biomaterials 2002, 23, 4775–4785. 10.1016/S0142-9612(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Pasche S.; De Paul S. M.; Vörös J.; Spencer N. D.; Textor M. Poly(l-Lysine)-Graft-Poly(Ethylene Glycol) Assembled Monolayers on Niobium Oxide Surfaces: A Quantitative Study of the Influence of Polymer Interfacial Architecture on Resistance to Protein Adsorption by ToF-SIMS and in Situ OWLS. Langmuir 2003, 19, 9216–9225. 10.1021/la034111y. [DOI] [Google Scholar]
- Rechendorff K.; Hovgaard M. B.; Foss M.; Zhdanov V. P.; Besenbacher F. Enhancement of Protein Adsorption Induced by Surface Roughness. Langmuir 2006, 22, 10885–10888. 10.1021/la0621923. [DOI] [PubMed] [Google Scholar]
- Dolatshahi-Pirouz A.; Rechendorff K.; Hovgaard M. B.; Foss M.; Chevallier J.; Besenbacher F. Bovine Serum Albumin Adsorption on Nano-Rough Platinum Surfaces Studied by QCM-D. Colloids Surf., B 2008, 66, 53–59. 10.1016/j.colsurfb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Schwarz U. S.; Bischofs I. B. Physical Determinants of Cell Organization in Soft Media. Med. Eng. Phys. 2005, 27, 763–772. 10.1016/j.medengphy.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Teixeira A. I.; Abrams G. A.; Bertics P. J.; Murphy C. J.; Nealey P. F. Epithelial Contact Guidance on Well-Defined Micro- and Nanostructured Substrates. J. Cell Sci. 2003, 116, 1881–1892. 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch A. S.; Miller D.; Luebke K. J.; Hu W. Correlation of Anisotropic Cell Behaviors with Topographic Aspect Ratio. Biomaterials 2009, 30, 1560–1567. 10.1016/j.biomaterials.2008.11.041. [DOI] [PubMed] [Google Scholar]
- Huang N. F.; Patel S.; Thakar R. G.; Wu J.; Hsiao B. S.; Chu B.; Lee R. J.; Li S. Myotube Assembly on Nanofibrous and Micropatterned Polymers. Nano Lett. 2006, 6, 537–542. 10.1021/nl060060o. [DOI] [PubMed] [Google Scholar]
- Karuri N. W.; Liliensiek S.; Teixeira A. I.; Abrams G.; Campbell S.; Nealey P. F.; Murphy C. J. Bilogical Length Scale Topography Enhances Cell-Substratum Adhesion of Human Corneal Epithelial Cells. J. Cell Sci. 2004, 117, 3153–3164. 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liliensiek S. J.; Campbell S.; Nealey P. F.; Murphy C. J. The Scale of Substratum Topographic Features Modulates Proliferation of Corneal Epithelial Cells and Corneal Fibroblasts. J. Biomed. Mater. Res., Part A 2006, 79A, 185–192. 10.1002/jbm.a.30744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson G. T.; Song H.; Mao H. Q. The Influence of Fiber Diameter of Electrospun Substrates on Neural Stem Cell Differentiation and Proliferation. Biomaterials 2009, 30, 556–564. 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Choi Y. J.; Park S. J.; Yi H. G.; Lee H.; Kim D. S.; Cho D. W. Muscle-Derived Extracellular Matrix on Sinusoidal Wavy Surfaces Synergistically Promotes Myogenic Differentiation and Maturation. J. Mater. Chem. B 2018, 6, 5530–5539. 10.1039/C8TB01475B. [DOI] [PubMed] [Google Scholar]
- Song K. H.; Park S. J.; Kim D. S.; Doh J. Sinusoidal Wavy Surfaces for Curvature-Guided Migration of Tlymphocytes. Biomaterials 2015, 51, 151–160. 10.1016/j.biomaterials.2015.01.071. [DOI] [PubMed] [Google Scholar]
- Comelles J.; Hortigüela V.; Samitier J.; Martínez E. Versatile Gradients of Covalently Bound Proteins on Microstructured Substrates. Langmuir 2012, 28, 13688–13697. 10.1021/la3025638. [DOI] [PubMed] [Google Scholar]
- Yu S.; Mao Z.; Gao C. Preparation of Gelatin Density Gradient on Poly(ε-Caprolactone) Membrane and Its Influence on Adhesion and Migration of Endothelial Cells. J. Colloid Interface Sci. 2015, 451, 177–183. 10.1016/j.jcis.2015.03.056. [DOI] [PubMed] [Google Scholar]
- Rasi Ghaemi S.; Delalat B.; Cavallaro A.; Mierczynska-Vasilev A.; Vasilev K.; Voelcker N. H. Differentiation of Rat Mesenchymal Stem Cells toward Osteogenic Lineage on Extracellular Matrix Protein Gradients. Adv. Healthc. Mater. 2019, 8, 1–11. 10.1002/adhm.201900595. [DOI] [PubMed] [Google Scholar]
- Zapata P.; Su J.; García A. J.; Meredith J. C. Quantitative High-Throughput Screening of Osteoblast Attachment, Spreading, and Proliferation on Demixed Polymer Blend Micropatterns. Biomacromolecules 2007, 8, 1907–1917. 10.1021/bm061134t. [DOI] [PubMed] [Google Scholar]
- Harding F.; Goreham R.; Short R.; Vasilev K.; Voelcker N. H. Surface Bound Amine Functional Group Density Influences Embryonic Stem Cell Maintenance. Adv. Healthcare Mater. 2013, 2, 585–590. 10.1002/adhm.201200119. [DOI] [PubMed] [Google Scholar]
- van Dommelen J. A. W.; van der Sande T. P. J.; Hrapko M.; Peters G. W. M. Mechanical Properties of Brain Tissue by Indentation: Interregional Variation. J. Mech. Behav. Biomed. Mater. 2010, 3, 158–166. 10.1016/j.jmbbm.2009.09.001. [DOI] [PubMed] [Google Scholar]
- van Eijden T. M. G. J.; van Ruijven L. J.; Giesen E. B. W. Bone Tissue Stiffness in the Mandibular Condyle Is Dependent on the Direction and Density of the Cancellous Structure. Calcif. Tissue Int. 2004, 75, 502–508. 10.1007/s00223-004-0295-6. [DOI] [PubMed] [Google Scholar]
- Data Book on Mechanical Properties of Living Cells, Tissues, and Organs; Abé H., Hayashi K., Sato M., Eds.; Springer, 1996. [Google Scholar]
- Engler A. J.; Griffin M. A.; Sen S.; Bönnemann C. G.; Sweeney H. L.; Discher D. E. Myotubes Differentiate Optimally on Substrates with Tissue-like Stiffness: Pathological Implications for Soft or Stiff Microenvironments. J. Cell Biol. 2004, 166, 877–887. 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.-Y.; Yu J.; Lin J.-H.; Tsai W.-B. Modulation of Alignment, Elongation and Contraction of Cardiomyocytes through a Combination of Nanotopography and Rigidity of Substrates. Acta Biomater. 2011, 7, 3285–3293. 10.1016/j.actbio.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Mosley M. C.; Lim H. J.; Chen J.; Yang Y.-H.; Li S.; Liu Y.; Smith Callahan L. A. Neurite Extension and Neuronal Differentiation of Human Induced Pluripotent Stem Cell Derived Neural Stem Cells on Polyethylene Glycol Hydrogels Containing a Continuous Young’s Modulus Gradient. J. Biomed. Mater. Res., Part A 2017, 105, 824–833. 10.1002/jbm.a.35955. [DOI] [PubMed] [Google Scholar]
- Tse J. R.; Engler A. J. Stiffness Gradients Mimicking in Vivo Tissue Variation Regulate Mesenchymal Stem Cell Fate. PLoS One 2011, 6, e15978. 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz R. D.; Andrawes F. F.; Barvenik F. W.; Koehn A. C. Acrylamide Monomer Leaching from Polyacrylamide-Treated Irrigation Furrows. J. Environ. Qual. 2008, 37, 2293–2298. 10.2134/jeq2007.0574. [DOI] [PubMed] [Google Scholar]
- Park H. R.; Kim M. S.; Kim S. J.; Park M.; Kong K. H.; Kim H. S.; Kwack S. J.; Kang T. S.; Kim S. H.; Kim H. S.; et al. Acrylamide Induces Cell Death in Neural Progenitor Cells and Impairs Hippocampal Neurogenesis. Toxicol. Lett. 2010, 193, 86–93. 10.1016/j.toxlet.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Lötters J. C.; Olthuis W.; Veltink P. H.; Bergveld P. The Mechanical Properties of the Rubber Elastic Polymer Polydimethylsiloxane for Sensor Applications. J. Micromech. Microeng. 1997, 7, 145–147. 10.1088/0960-1317/7/3/017. [DOI] [Google Scholar]
- Kumar A.; Biebuyck H. A.; Whitesides G. M. Patterning Self-Assembled Monolayers: Applications in Materials Science. Langmuir 1994, 10, 1498–1511. 10.1021/la00017a030. [DOI] [Google Scholar]
- Duffy D. C.; McDonald J. C.; Schueller O. J. A.; Whitesides G. M. Rapid Prototyping of Microfluidic Systems in Poly(Dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Pino C. J.; Haselton F. R.; Chang M. S. Seeding of Corneal Wounds by Epithelial Cell Transfer from Micropatterned PDMS Contact Lenses. Cell Transplant. 2005, 14, 565–571. 10.3727/000000005783982783. [DOI] [PubMed] [Google Scholar]
- Lee J. N.; Park C.; Whitesides G. M. Solvent Compatibility of Poly(Dimethylsiloxane)-Based Microfluidic Devices. Anal. Chem. 2003, 75, 6544–6554. 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- Mata A.; Fleischman A. J.; Roy S. Characterization of Polydimethylsiloxane (PDMS) Properties for Biomedical Micro/Nanosystems. Biomed. Microdevices 2005, 7, 281–293. 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- Yim E. K. F.; Darling E. M.; Kulangara K.; Guilak F.; Leong K. W. Nanotopography-Induced Changes in Focal Adhesions, Cytoskeletal Organization, and Mechanical Properties of Human Mesenchymal Stem Cells. Biomaterials 2010, 31, 1299–1306. 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P.; Li Y.; Zhang Y.; Yang Y.; Hu R.; Zhao X.; Huang A.; Zhang R.; Liu X.; Huang Q.; et al. High-Throughput Screening of Rat Mesenchymal Stem Cell Behavior on Gradient TiO2 Nanotubes. ACS Biomater. Sci. Eng. 2018, 4, 2804–2814. 10.1021/acsbiomaterials.8b00488. [DOI] [PubMed] [Google Scholar]
- Vater C.; Kasten P.; Stiehler M. Culture Media for the Differentiation of Mesenchymal Stromal Cells. Acta Biomater. 2011, 7, 463–477. 10.1016/j.actbio.2010.07.037. [DOI] [PubMed] [Google Scholar]
- Martins A.; Duarte A. R. C.; Faria S.; Marques A. P.; Reis R. L.; Neves N. M. Osteogenic Induction of HBMSCs by Electrospun Scaffolds with Dexamethasone Release Functionality. Biomaterials 2010, 31, 5875–5885. 10.1016/j.biomaterials.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Nair A.; Shen J.; Lotfi P.; Ko C. Y.; Zhang C. C.; Tang L. Biomaterial Implants Mediate Autologous Stem Cell Recruitment in Mice. Acta Biomater. 2011, 7, 3887–3895. 10.1016/j.actbio.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasso R.; Fais F.; Reverberi D.; Tortelli F.; Cancedda R. The Recruitment of Two Consecutive and Different Waves of Host Stem/Progenitor Cells during the Development of Tissue-Engineered Bone in a Murine Model. Biomaterials 2010, 31, 2121–2129. 10.1016/j.biomaterials.2009.11.064. [DOI] [PubMed] [Google Scholar]
- Faia-Torres A. B.; Charnley M.; Goren T.; Guimond-Lischer S.; Rottmar M.; Maniura-Weber K.; Spencer N. D.; Reis R. L.; Textor M.; Neves N. M. Osteogenic Differentiation of Human Mesenchymal Stem Cells in the Absence of Osteogenic Supplements: A Surface-Roughness Gradient Study. Acta Biomater. 2015, 28, 64–75. 10.1016/j.actbio.2015.09.028. [DOI] [PubMed] [Google Scholar]
- Chung S. H.; Son S. J.; Min J. The Nanostructure Effect on the Adhesion and Growth Rates of Epithelial Cells with Well-Defined Nanoporous Alumina Substrates. Nanotechnology 2010, 21, 125104. 10.1088/0957-4484/21/12/125104. [DOI] [PubMed] [Google Scholar]
- Delalat B.; Goreham R. V.; Vasilev K.; Harding F. J.; Voelcker N. H. Subtle Changes in Surface Chemistry Affect Embryoid Body Cell Differentiation: Lessons Learnt from Surface-Bound Amine Density Gradients. Tissue Eng., Part A 2014, 20, 1715–1725. 10.1089/ten.tea.2013.0350. [DOI] [PubMed] [Google Scholar]
- Engler A.; Sheehan M.; Sweeney H. L.; Discher D. E. Substrate Compliance vs Ligand Density in Cell on Gel Responses. Biophys. J. 2003, 6, 7–8. 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubashkin M. G.; Ou G.; Weaver V. M. Deconstructing Signaling in Three Dimensions. Biochemistry 2014, 53, 2078–2090. 10.1021/bi401710d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega S. L.; Kwon M. Y.; Song K. H.; Wang C.; Mauck R. L.; Han L.; Burdick J. A. Combinatorial Hydrogels with Biochemical Gradients for Screening 3D Cellular Microenvironments. Nat. Commun. 2018, 9, 614. 10.1038/s41467-018-03021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin J. W.; Brockes J. P. Regeneration, Tissue Injury and the Immune Response. J. Anat. 2006, 209, 423–432. 10.1111/j.1469-7580.2006.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano R.; Biology S. H.-T. J. Signal Transduction from the Extracellular Matrix. J. Cell Biol. 1993, 120, 5775–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D.; Lie M.; Gao C. Preparation and Applications of Ordered and Gradient Scaffolds in Regenerative Medicine. Prog. Chem. 2011, 23, 2550. [Google Scholar]
- Han L.; Mao Z.; Wu J.; Guo Y.; Ren T.; Gao C. Unidirectional Migration of Single Smooth Muscle Cells under the Synergetic Effects of Gradient Swelling Cue and Parallel Groove Patterns. Colloids Surf., B 2013, 111, 1–6. 10.1016/j.colsurfb.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Jin T.; Xu X.; Hereld D. Chemotaxis, Chemokine Receptors and Human Disease. Cytokine+ 2008, 44, 1–8. 10.1016/j.cyto.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kempen L. C. L. T.; van den Oord J. J.; van Muijen G. N. P.; Weidle U. H.; Bloemers H. P.J.; Swart G. W. M. Activated Leukocyte Cell Adhesion Molecule/CD166, a Marker of Tumor Progression in Primary Malignant Melanoma of the Skin. Am. J. Pathol. 2000, 156, 769–774. 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A. J.; Brenot A.; Duong M.; Chan B. S.; Werb Z. Collective Epithelial Migration and Cell Rearrangements Drive Mammary Branching Morphogenesis. Dev. Cell 2008, 14, 570–581. 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D. A.; Horwitz A. F. Cell Migration: A Physically Integrated Molecular Process. Cell 1996, 84, 359–369. 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lo C.-M.; Wang H.-B.; Dembo M.; Wang Y.-L. Cell Movement Is Guided by the Rigidity of the Substrate. Biophys. J. 2000, 79, 144–152. 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J. E; Haugh J. M Directed Migration of Mesenchymal Cells: Where Signaling and the Cytoskeleton Meet. Curr. Opin. Cell Biol. 2014, 30, 74–82. 10.1016/j.ceb.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C.-H.; Cheng Y.-C.; Chao P.-H. G. The Influence and Interactions of Substrate Thickness, Organization and Dimensionality on Cell Morphology and Migration. Acta Biomater. 2013, 9, 5502–5510. 10.1016/j.actbio.2012.11.024. [DOI] [PubMed] [Google Scholar]
- Redd M. J.; Kelly G.; Dunn G.; Way M.; Martin P. Imaging Macrophage Chemotaxis in Vivo: Studies of Microtubule Function in Zebrafish Wound Inflammation. Cell Motil. Cytoskeleton 2006, 63, 415–422. 10.1002/cm.20133. [DOI] [PubMed] [Google Scholar]
- Cara D. C.; Kaur J.; Forster M.; McCafferty D.-M.; Kubes P. Role of P38 Mitogen-Activated Protein Kinase in Chemokine-Induced Emigration and Chemotaxis in Vivo. J. Immunol. 2001, 167, 6552–6558. 10.4049/jimmunol.167.11.6552. [DOI] [PubMed] [Google Scholar]
- Dubin-Thaler B. J.; Giannone G.; Dobereiner H.-G.; Sheetz M. P. Nanometer Analysis of Cell Spreading on Matrix-Coated Surfaces Reveals Two Distinct Cell States and STEPs. Biophys. J. 2004, 86, 1794–1806. 10.1016/S0006-3495(04)74246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.; Berkland C.; Detamore M. S. Strategies and Applications for Incorporating Physical and Chemical Signal Gradients in Tissue Engineering. Tissue Eng., Part B 2008, 14, 341–366. 10.1089/ten.teb.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P.; Sunyer R.; Trepat X. Mechanical Guidance of Cell Migration: Lessons from Chemotaxis. Curr. Opin. Cell Biol. 2013, 25, 543–549. 10.1016/j.ceb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Plotnikov S. V.; Pasapera A. M.; Sabass B.; Waterman C. M. Force Fluctuations within Focal Adhesions Mediate ECM-Rigidity Sensing to Guide Directed Cell Migration. Cell 2012, 151, 1513–1527. 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.-h.; Frey M. T.; Burnham N. A.; Wang Y.-L. Substrate Rigidity Regulates the Formation and Maintenance of Tissues. Biophys. J. 2006, 90, 2213–2220. 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J.; Kerstens A.; Danuser G.; Schwartz M. A.; Waterman-Storer C. M. Integrin-Dependent Actomyosin Contraction Regulates Epithelial Cell Scattering. J. Cell Biol. 2005, 171, 153–164. 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L. A.; Ju Y.-E.; Marg B.; Osterfield M.; Janmey P. A. Neurite Branching on Deformable Substrates. NeuroReport 2002, 13, 2411. 10.1097/00001756-200212200-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch D.; Rosoff W. J.; Jiang J.; Geller H. M.; Urbach J. S. Strength in the Periphery: Growth Cone Biomechanics and Substrate Rigidity Response in Peripheral and Central Nervous System Neurons. Biophys. J. 2012, 102, 452–460. 10.1016/j.bpj.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville J. T. H.; Lawson M. A.; Maxfield F. R. Dynamic Imaging of Neutrophil Migration in Three Dimensions: Mechanical Interactions between Cells and Matrix. J. Leukocyte Biol. 1997, 61, 188–200. 10.1002/jlb.61.2.188. [DOI] [PubMed] [Google Scholar]
- Ulrich T. A.; de Juan Pardo E. M.; Kumar S. The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells. Cancer Res. 2009, 69, 4167–4174. 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M. A.; Desai R.; Solski P. A.; Der C. J.; Keely P. J. ROCK-Generated Contractility Regulates Breast Epithelial Cell Differentiation in Response to the Physical Properties of a Three-Dimensional Collagen Matrix. J. Cell Biol. 2003, 163, 583–595. 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg B. C.; DiMilla P. A.; Walker M.; Kim S.; Wong J. Y. Vascular Smooth Muscle Cell Durotaxis Depends on Substrate Stiffness Gradient Strength. Biophys. J. 2009, 97, 1313–1322. 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Mih J. D.; Shea B. S.; Kho A. T.; Sharif A. S.; Tager A. M.; Tschumperlin D. J. Feedback Amplification of Fibrosis through Matrix Stiffening and COX-2 Suppression. J. Cell Biol. 2010, 190, 693–706. 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plodinec M.; Loparic M.; Monnier C. A.; Obermann E. C.; Zanetti-Dallenbach R.; Oertle P.; Hyotyla J. T.; Aebi U.; Bentires-Alj M.; Lim R. Y. H.; et al. The Nanomechanical Signature of Breast Cancer. Nat. Nanotechnol. 2012, 7, 757–765. 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- Hartman C. D.; Isenberg B. C.; Chua S. G.; Wong J. Y. Vascular Smooth Muscle Cell Durotaxis Depends on Extracellular Matrix Composition. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11190–11195. 10.1073/pnas.1611324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjipanayi E.; Mudera V.; Brown R. A. Guiding Cell Migration in 3D: A Collagen Matrix with Graded Directional Stiffness. Cell Motil. Cytoskeleton 2009, 66, 121–128. 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- Kim D.-H.; Wong P. K.; Park J.; Levchenko A.; Sun Y. Microengineered Platforms for Cell Mechanobiology. Annu. Rev. Biomed. Eng. 2009, 11, 203–233. 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- Patel S.; Kurpinski K.; Quigley R.; Gao H.; Hsiao B. S.; Poo M.-M.; Li S. Bioactive Nanofibers: Synergistic Effects of Nanotopography and Chemical Signaling on Cell Guidance. Nano Lett. 2007, 7, 2122–2128. 10.1021/nl071182z. [DOI] [PubMed] [Google Scholar]
- Diehl K. A.; Foley J. D.; Nealey P. F.; Murphy C. J. Nanoscale Topography Modulates Corneal Epithelial Cell Migration. J. Biomed. Mater. Res., Part A 2005, 75, 603–611. 10.1002/jbm.a.30467. [DOI] [PubMed] [Google Scholar]
- Kaiser J. P.; Reinmann A.; Bruinink A. The Effect of Topographic Characteristics on Cell Migration Velocity. Biomaterials 2006, 27, 5230–5241. 10.1016/j.biomaterials.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kim H. N.; Hong Y.; Kim M. S.; Kim S. M.; Suh K.-Y. Effect of Orientation and Density of Nanotopography in Dermal Wound Healing. Biomaterials 2012, 33, 8782–8792. 10.1016/j.biomaterials.2012.08.038. [DOI] [PubMed] [Google Scholar]
- Genzer J. Surface-Bound Gradients for Studies of Soft Materials Behavior. Annu. Rev. Mater. Res. 2012, 42, 435–468. 10.1146/annurev-matsci-070511-155050. [DOI] [Google Scholar]
- Liang S.; Yu S.; Zhou N.; Deng J.; Gao C. Controlling the Selective and Directional Migration of Hepatocytes by a Complementary Density Gradient of Glycosylated Hyperbranched Polymers and Poly(Ethylene Glycol) Molecules. Acta Biomater. 2017, 56, 161–170. 10.1016/j.actbio.2016.12.032. [DOI] [PubMed] [Google Scholar]
- Han L.; Mao Z.; Wu J.; Guo Y.; Ren T.; Gao C. Directional Cell Migration through Cell-Cell Interaction on Polyelectrolyte Multilayers with Swelling Gradients. Biomaterials 2013, 34, 975–984. 10.1016/j.biomaterials.2012.10.041. [DOI] [PubMed] [Google Scholar]
- Ren T.; Mao Z.; Guo J.; Gao C. Directional Migration of Vascular Smooth Muscle Cells Guided by a Molecule Weight Gradient of Poly(2-Hydroxyethyl Methacrylate) Brushes. Langmuir 2013, 29, 6386–6395. 10.1021/la4004609. [DOI] [PubMed] [Google Scholar]
- Wu J.; Mao Z.; Gao C. Controlling the Migration Behaviors of Vascular Smooth Muscle Cells by Methoxy Poly(Ethylene Glycol) Brushes of Different Molecular Weight and Density. Biomaterials 2012, 33, 810–820. 10.1016/j.biomaterials.2011.10.022. [DOI] [PubMed] [Google Scholar]
- Smith J. T.; Tomfohr J. K.; Wells M. C.; Beebe T. P.; Kepler T. B.; Reichert W. M. Measurement of Cell Migration on Surface-Bound Fibronectin Gradients. Langmuir 2004, 20, 8279–8286. 10.1021/la0489763. [DOI] [PubMed] [Google Scholar]
- Smith J. T.; Elkin J. T.; Reichert W. M. Directed Cell Migration on Fibronectin Gradients: Effect of Gradient Slope. Exp. Cell Res. 2006, 312, 2424–2432. 10.1016/j.yexcr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Cai K.; Kong T.; Wang L.; Liu P.; Yang W.; Chen C. Regulation of Endothelial Cells Migration on Poly(d, l-Lactic Acid) Films Immobilized with Collagen Gradients. Colloids Surf., B 2010, 79, 291–297. 10.1016/j.colsurfb.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Tan H.; Wan L.; Wu J.; Gao C. Microscale Control over Collagen Gradient on Poly(l-Lactide) Membrane Surface for Manipulating Chondrocyte Distribution. Colloids Surf., B 2008, 67, 210–215. 10.1016/j.colsurfb.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Gunawan R. C.; Silvestre J.; Gaskins H. R.; Kenis P. J. A.; Leckband D. E. Cell Migration and Polarity on Microfabricated Gradients of Extracellular Matrix Proteins. Langmuir 2006, 22, 4250–4258. 10.1021/la0531493. [DOI] [PubMed] [Google Scholar]
- Adams D. N.; Kao E. Y. C.; Hypolite C. L.; Distefano M. D.; Hu W. S.; Letourneau P. C. Growth Cones Turn and Migrate up an Immobilized Gradient of the Laminin IKVAV Peptide. J. Neurobiol. 2005, 62, 134–147. 10.1002/neu.20075. [DOI] [PubMed] [Google Scholar]
- Guarnieri D.; De Capua A.; Ventre M.; Borzacchiello A.; Pedone C.; Marasco D.; Ruvo M.; Netti P. A. Covalently Immobilized RGD Gradient on PEG Hydrogel Scaffold Influences Cell Migration Parameters. Acta Biomater. 2010, 6, 2532–2539. 10.1016/j.actbio.2009.12.050. [DOI] [PubMed] [Google Scholar]
- DeLong S. A.; Gobin A. S.; West J. L. Covalent Immobilization of RGDS on Hydrogel Surfaces to Direct Cell Alignment and Migration. J. Controlled Release 2005, 109, 139–148. 10.1016/j.jconrel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Ren T.; Yu S.; Mao Z.; Gao C. A Complementary Density Gradient of Zwitterionic Polymer Brushes and NCAM Peptides for Selectively Controlling Directional Migration of Schwann Cells. Biomaterials 2015, 56, 58–67. 10.1016/j.biomaterials.2015.03.052. [DOI] [PubMed] [Google Scholar]
- Ren T.; Yu S.; Mao Z.; Moya S. E.; Han L.; Gao C. Complementary Density Gradient of Poly(Hydroxyethyl Methacrylate) and YIGSR Selectively Guides Migration of Endotheliocytes. Biomacromolecules 2014, 15, 2256–2264. 10.1021/bm500385n. [DOI] [PubMed] [Google Scholar]
- Miller E. D.; Li K.; Kanade T.; Weiss L. E.; Walker L. M.; Campbell P. G. Spatially Directed Guidance of Stem Cell Population Migration by Immobilized Patterns of Growth Factors. Biomaterials 2011, 32, 2775–2785. 10.1016/j.biomaterials.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong S. A.; Moon J. J.; West J. L. Covalently Immobilized Gradients of BFGF on Hydrogel Scaffolds for Directed Cell Migration. Biomaterials 2005, 26, 3227–3234. 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Wu P.; Fu Y.; Cai K. Regulation of the Migration of Endothelial Cells by a Gradient Density of Vascular Endothelial Growth Factor. Colloids Surf., B 2014, 123, 181–190. 10.1016/j.colsurfb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Au J. L.-S.; Li D.; Gan Y.; Gao X.; Johnson A. L.; Johnston J.; Millenbaugh N. J.; Jang S. H.; Kuh H.-J.; Chen C.-T. Pharmacodynamics of Immediate and Delayed Effects of Paclitaxel: Role of Slow Apoptosis and Intracellular Drug Retention. Cancer Res. 1998, 58, 2141–8. [PubMed] [Google Scholar]
- Nicosia R. F.; Bonanno E.; Smith M. Fibronectin Promotes the Elongation of Microvessels during Angiogenesis in Vitro. J. Cell. Physiol. 1993, 154, 654–661. 10.1002/jcp.1041540325. [DOI] [PubMed] [Google Scholar]
- Ceradini D. J.; Kulkarni A. R.; Callaghan M. J.; Tepper O. M.; Bastidas N.; Kleinman M. E.; Capla J. M.; Galiano R. D.; Levine J. P.; Gurtner G. C. Progenitor Cell Trafficking Is Regulated by Hypoxic Gradients through HIF-1 Induction of SDF-1. Nat. Med. 2004, 10, 858–864. 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- Kunkel E. J.; Butcher E. C. Chemokines and the Tissue-Specific Migration of Lymphocytes. Immunity 2002, 16, 1–4. 10.1016/S1074-7613(01)00261-8. [DOI] [PubMed] [Google Scholar]
- Solouk A.; Cousins B.; Mirahmadi F.; et al. Biomimetic Modified Clinical-Grade POSS-PCU Nanocomposite Polymer for Bypass Graft Applications: A Preliminary Assessment of Endothelial Cell Adhesion. Mater. Sci. Eng., C 2015, 46, 400–408. 10.1016/j.msec.2014.10.065. [DOI] [PubMed] [Google Scholar]
- Sartore S.; Chiavegato A.; Faggin E.; Franch R.; Puato M.; Ausoni S.; Pauletto P. Contribution of Adventitial Fibroblasts to Neointima Formation and Vascular Remodeling. Circ. Res. 2001, 89, 1111–1121. 10.1161/hh2401.100844. [DOI] [PubMed] [Google Scholar]
- Wilcox J. N.; Waksman R.; King S. B.; Scott N. A. The Role of the Adventitia in the Arterial Response to Angioplasty: The Effect of Intravascular Radiation. Int. J. Radiat. Oncol., Biol., Phys. 1996, 36, 789–796. 10.1016/S0360-3016(96)00299-4. [DOI] [PubMed] [Google Scholar]
- Huebner K. D.; Jassal D. S.; Halevy O.; Pines M.; Anderson J. E. Functional Resolution of Fibrosis in Mdx Mouse Dystrophic Heart and Skeletal Muscle by Halofuginone. Am. J. Physiol. Circ. Physiol. 2008, 294, H1550–H1561. 10.1152/ajpheart.01253.2007. [DOI] [PubMed] [Google Scholar]
- Mann B. K.; West J. L. Cell Adhesion Peptides Alter Smooth Muscle Cell Adhesion, Proliferation, Migration, and Matrix Protein Synthesis on Modified Surfaces and in Polymer Scaffolds. J. Biomed. Mater. Res. 2002, 60, 86–93. 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- Tedgui A.; Mallat Z. Cytokines in Atherosclerosis: Pathogenic and Regulatory Pathways. Physiol. Rev. 2006, 86, 515–581. 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- Massia S. P.; Hubbell J. A. Vascular Endothelial Cell Adhesion and Spreading Promoted by the Peptide REDV of the IIICS Region of Plasma Fibronectin Is Mediated by Integrin Alpha 4 Beta 1. J. Biol. Chem. 1992, 267, 14019–14026. 10.1016/S0021-9258(19)49672-5. [DOI] [PubMed] [Google Scholar]
- Wolfram C. J.; Rubloff G. W.; Luo X. Perspectives in Flow-Based Microfluidic Gradient Generators for Characterizing Bacterial Chemotaxis. Biomicrofluidics 2016, 10, 061301. 10.1063/1.4967777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G.; Xue H.; Li G.; Jiang S. Integrated Antimicrobial and Nonfouling Hydrogels to Inhibit the Growth of Planktonic Bacterial Cells and Keep the Surface Clean. Langmuir 2010, 26, 10425–10428. 10.1021/la101542m. [DOI] [PubMed] [Google Scholar]
- Bartlett J. G. Treatment of Infections Associated with Surgical Implants. Infect. Dis. Clin. Pract. 2004, 12, 258–259. 10.1097/01.idc.0000130890.12611.f3. [DOI] [Google Scholar]
- Senaratne W.; Andruzzi L.; Ober C. K. Self-Assembled Monolayers and Polymer Brushes in Biotechnology: Current Applications and Future Perspectives. Biomacromolecules 2005, 6, 2427–2448. 10.1021/bm050180a. [DOI] [PubMed] [Google Scholar]
- Chang Y.; Yandi W.; Chen W. Y.; Shih Y. J.; Yang C. C.; Chang Y.; Ling Q. D.; Higuchi A. Tunable Bioadhesive Copolymer Hydrogels of Thermoresponsive Poly(N -Isopropyl Acrylamide) Containing Zwitterionic Polysulfobetaine. Biomacromolecules 2010, 11, 1101–1110. 10.1021/bm100093g. [DOI] [PubMed] [Google Scholar]
- Cheng G.; Li G.; Xue H.; Chen S.; Bryers J. D.; Jiang S. Zwitterionic Carboxybetaine Polymer Surfaces and Their Resistance to Long-Term Biofilm Formation. Biomaterials 2009, 30, 5234–5240. 10.1016/j.biomaterials.2009.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer T. P.; Stewart S.; Hsu B. B.; Klibanov A. M. Hydrophobic Polycationic Coatings That Inhibit Biofilms and Support Bone Healing during Infection. Biomaterials 2012, 33, 1245–1254. 10.1016/j.biomaterials.2011.10.038. [DOI] [PubMed] [Google Scholar]
- Li M.; Mondrinos M. J.; Chen X.; Gandhi M. R.; Ko F. K.; Lelkes P. I. S. Epidermidis Biofilm Formation: Effects of Biomaterial Surface Chemistry and Serum Proteins. J. Biomed. Mater. Res. Part A 2006, 79, 963–973. 10.1002/jbm.a.31103. [DOI] [Google Scholar]
- Keskin D.; Mergel O.; Van Der Mei H. C.; Busscher H. J.; Van Rijn P. Inhibiting Bacterial Adhesion by Mechanically Modulated Microgel Coatings. Biomacromolecules 2019, 20, 243–253. 10.1021/acs.biomac.8b01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter J. A.; Thompson M. T.; Delgadillo M.; Nishikawa T.; Rubner M. F.; Van Vliet K. J. Substrata Mechanical Stiffness Can Regulate Adhesion of Viable Bacteria. Biomacromolecules 2008, 9, 1571–1578. 10.1021/bm701430y. [DOI] [PubMed] [Google Scholar]
- Hucknall A.; Rangarajan S.; Chilkoti A. In Pursuit of Zero: Polymer Brushes That Resist the Adsorption of Proteins. Adv. Mater. 2009, 21, 2441–2446. 10.1002/adma.200900383. [DOI] [Google Scholar]
- Banerjee I.; Pangule R. C.; Kane R. S. Antifouling Coatings: Recent Developments in the Design of Surfaces That Prevent Fouling by Proteins, Bacteria, and Marine Organisms. Adv. Mater. 2011, 23, 690–718. 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- Tuson H. H.; Weibel D. B. Bacteria-Surface Interactions. Soft Matter 2013, 9, 4368–4380. 10.1039/c3sm27705d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H.; Ren D. Materials and Surface Engineering to Control Bacterial Adhesion and Biofilm Formation: A Review of Recent Advances. Front. Chem. Sci. Eng. 2014, 8, 20–33. 10.1007/s11705-014-1412-3. [DOI] [Google Scholar]
- Park D.; Park S. J.; Cho S.; Lee Y.; Lee Y. K.; Min J. J.; Park B. J.; Ko S. Y.; Park J. O.; Park S. Motility Analysis of Bacteria-Based Microrobot (Bacteriobot) Using Chemical Gradient Microchamber. Biotechnol. Bioeng. 2014, 111, 134–143. 10.1002/bit.25007. [DOI] [PubMed] [Google Scholar]
- Ahmed T.; Shimizu T. S.; Stocker R. Bacterial Chemotaxis in Linear and Nonlinear Steady Microfluidic Gradients. Nano Lett. 2010, 10, 3379–3385. 10.1021/nl101204e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. H.; Lee S. H.; Kim J. M.; Kim H. E.; Kim Y. G.; Yoo J. Y.; Chang W. S.; Lee C. S. Microfluidic Monitoring of Pseudomonas Aeruginosa Chemotaxis under the Continuous Chemical Gradient. Biosens. Bioelectron. 2010, 26, 351–356. 10.1016/j.bios.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Kim M.; Kim T. Diffusion-Based and Long-Range Concentration Gradients of Multiple Chemicals for Bacterial Chemotaxis Assays. Anal. Chem. 2010, 82, 9401–9409. 10.1021/ac102022q. [DOI] [PubMed] [Google Scholar]
- Diao J.; Young L.; Kim S.; Fogarty E. A.; Heilman S. M.; Zhou P.; Shuler M. L.; Wu M.; DeLisa M. P. A Three-Channel Microfluidic Device for Generating Static Linear Gradients and Its Application to the Quantitative Analysis of Bacterial Chemotaxis. Lab Chip 2006, 6, 381–388. 10.1039/B511958H. [DOI] [PubMed] [Google Scholar]
- Jeon H.; Lee Y.; Jin S.; Koo S.; Lee C. S.; Yoo J. Y. Quantitative Analysis of Single Bacterial Chemotaxis Using a Linear Concentration Gradient Microchannel. Biomed. Microdevices 2009, 11, 1135–1143. 10.1007/s10544-009-9330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert D. L.; Manson M. D.; Jayaraman A. Flow-Based Microfluidic Device for Quantifying Bacterial Chemotaxis in Stable, Competing Gradients. Appl. Environ. Microbiol. 2009, 75, 4557–4564. 10.1128/AEM.02952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert D. L.; Manson M. D.; Jayaraman A. A Microfluidic Device for Quantifying Bacterial Chemotaxis in Stable Concentration Gradients. J. Vis. Exp. 2010, 2–7. 10.3791/1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavifar A.; Xu J.; Hovaizi M.; Hesketh P.; Daley W.; Britton D. A Nitrocellulose-Based Microfluidic Device for Generation of Concentration Gradients and Study of Bacterial Chemotaxis. J. Electrochem. Soc. 2014, 161, B3064–B3070. 10.1149/2.009402jes. [DOI] [Google Scholar]
- Murugesan N.; Singha S.; Panda T.; Das S. K. A Diffusion Based Long-Range and Steady Chemical Gradient Generator on a Microfluidic Device for Studying Bacterial Chemotaxis. J. Micromech. Microeng. 2016, 26, 35011. 10.1088/0960-1317/26/3/035011. [DOI] [Google Scholar]
- Roggo C.; Picioreanu C.; Richard X.; Mazza C.; van Lintel H.; van der Meer J. R. Quantitative Chemical Biosensing by Bacterial Chemotaxis in Microfluidic Chips. Environ. Microbiol. 2018, 20, 241–258. 10.1111/1462-2920.13982. [DOI] [PubMed] [Google Scholar]
- Celiz A. D.; Smith J. G. W.; Patel A. K.; Hook A. L.; Rajamohan D.; George V. T.; Flatt L.; Patel M. J.; Epa V. C.; Singh T.; et al. Discovery of a Novel Polymer for Human Pluripotent Stem Cell Expansion and Multilineage Differentiation. Adv. Mater. 2015, 27, 4006–4012. 10.1002/adma.201501351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs L.; Amer M. H.; Vassey M.; Koch B.; Figueredo G. P.; Mukonoweshuro B.; Mikulskis P.; Vasilevich A.; Vermeulen S.; Dryden I. L.; et al. Synergistic Material-Topography Combinations to Achieve Immunomodulatory Osteogenic Biomaterials. bioRxiv 2020. 10.1101/2020.04.29.067421. [DOI] [PubMed] [Google Scholar]
- Brocchini S.; James K.; Tangpasuthadol V.; Kohn J. A Combinatorial Approach for Polymer Design. J. Am. Chem. Soc. 1997, 119, 4553–4554. 10.1021/ja970389z. [DOI] [Google Scholar]
- Brocchini S.; James K.; Tangpasuthadol V.; Kohn J. Structure-Property Correlations in a Combinatorial Library of Degradable Biomaterials. J. Biomed. Mater. Res. 1998, 42, 66–75. . [DOI] [PubMed] [Google Scholar]
- Rao C.; Prodromakis T.; Kolker L.; Chaudhry U. A. R.; Trantidou T.; Sridhar A.; Weekes C.; Camelliti P.; Harding S. E.; Darzi A.; et al. The Effect of Microgrooved Culture Substrates on Calcium Cycling of Cardiac Myocytes Derived from Human Induced Pluripotent Stem Cells. Biomaterials 2013, 34, 2399–2411. 10.1016/j.biomaterials.2012.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam R. H. W.; Sun Y.; Chen W.; Fu J. Elastomeric Microposts Integrated into Microfluidics for Flow-Mediated Endothelial Mechanotransduction Analysis. Lab Chip 2012, 12, 1865–1873. 10.1039/c2lc21146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry O. Y. F.; Piletsky S. A.; Cullen D. C. Fabrication of Molecularly Imprinted Polymer Microarray on a Chip by Mid-Infrared Laser Pulse Initiated Polymerisation. Biosens. Bioelectron. 2008, 23, 1769–1775. 10.1016/j.bios.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Laing S.; Suriano R.; Lamprou D. A.; Smith C.-A.; Dalby M. J.; Mabbott S.; Faulds K.; Graham D. Thermoresponsive Polymer Micropatterns Fabricated by Dip-Pen Nanolithography for a Highly Controllable Substrate with Potential Cellular Applications. ACS Appl. Mater. Interfaces 2016, 8, 24844–24852. 10.1021/acsami.6b03860. [DOI] [PubMed] [Google Scholar]
- Rasi Ghaemi S.; Delalat B.; Cetó X.; Harding F. J.; Tuke J.; Voelcker N. H. Synergistic Influence of Collagen i and BMP 2 Drives Osteogenic Differentiation of Mesenchymal Stem Cells: A Cell Microarray Analysis. Acta Biomater. 2016, 34, 41–52. 10.1016/j.actbio.2015.07.027. [DOI] [PubMed] [Google Scholar]
- Zhang R.; Liberski A.; Sanchez-Martin R.; Bradley M. Microarrays of over 2000 Hydrogels - Identification of Substrates for Cellular Trapping and Thermally Triggered Release. Biomaterials 2009, 30, 6193–6201. 10.1016/j.biomaterials.2009.07.055. [DOI] [PubMed] [Google Scholar]
- Zhang R.; Liberski A.; Khan F.; Diaz-Mochon J. J.; Bradley M. Inkjet Fabrication of Hydrogel Microarrays Using in Situ Nanolitre-Scale Polymerisation. Chem. Commun. 2008, 1317–1319. 10.1039/b717932d. [DOI] [PubMed] [Google Scholar]
- Anderson D. G.; Putnam D.; Lavik E. B.; Mahmood T. A.; Langer R. Biomaterial Microarrays: Rapid, Microscale Screening of Polymer-Cell Interaction. Biomaterials 2005, 26, 4892–4897. 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Algahtani M. S.; Scurr D. J.; Hook A. L.; Anderson D. G.; Langer R. S.; Burley J. C.; Alexander M. R.; Davies M. C. High Throughput Screening for Biomaterials Discovery. J. Controlled Release 2014, 190, 115–126. 10.1016/j.jconrel.2014.06.045. [DOI] [PubMed] [Google Scholar]
- Coyle R.; Jia J.; Mei Y. Polymer Microarray Technology for Stem Cell Engineering. Acta Biomater. 2016, 34, 60–72. 10.1016/j.actbio.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiz A. D.; Hook A. L.; Scurr D. J.; Anderson D. G.; Langer R.; Davies M. C.; Alexander M. R. ToF-SIMS Imaging of a Polymer Microarray Prepared Using Ink-Jet Printing of Acrylate Monomers. Surf. Interface Anal. 2013, 45, 202–205. 10.1002/sia.5042. [DOI] [Google Scholar]
- Anderson D. G.; Levenberg S.; Langer R. Nanoliter-Scale Synthesis of Arrayed Biomaterials and Application to Human Embryonic Stem Cells. Nat. Biotechnol. 2004, 22, 863–866. 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- Tourniaire G.; Collins J.; Campbell S.; Mizomoto H.; Ogawa S.; Thaburet J. F.; Bradley M. Polymer Microarrays for Cellular Adhesion. Chem. Commun. 2006, 2118–2120. 10.1039/b602009g. [DOI] [PubMed] [Google Scholar]
- Pernagallo S.; Unciti-Broceta A.; Díaz-Mochón J. J.; Bradley M. Deciphering Cellular Morphology and Biocompatibility Using Polymer Microarrays. Biomed. Mater. 2008, 3, 034112. 10.1088/1748-6041/3/3/034112. [DOI] [PubMed] [Google Scholar]
- Hook A. L.; Chang C. Y.; Yang J.; Luckett J.; Cockayne A.; Atkinson S.; Mei Y.; Bayston R.; Irvine D. J.; Langer R.; et al. Combinatorial Discovery of Polymers Resistant to Bacterial Attachment. Nat. Biotechnol. 2012, 30, 868–875. 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth E. A.; Xu T.; Das M.; Gregory C.; Hickman J. J.; Boland T. Inkjet Printing for High-Throughput Cell Patterning. Biomaterials 2004, 25, 3707–3715. 10.1016/j.biomaterials.2003.10.052. [DOI] [PubMed] [Google Scholar]
- Begines B.; Hook A. L.; Alexander M. R.; Tuck C. J.; Wildman R. D. Development, Printability and Post-Curing Studies of Formulations of Materials Resistant to Microbial Attachment for Use in Inkjet Based 3D Printing. Rapid Prototyp. J. 2016, 22, 835–841. 10.1108/RPJ-11-2015-0175. [DOI] [Google Scholar]
- Hansen A.; Zhang R.; Bradley M. Fabrication of Arrays of Polymer Gradients Using Inkjet Printing. Macromol. Rapid Commun. 2012, 33, 1114–1118. 10.1002/marc.201200193. [DOI] [PubMed] [Google Scholar]
- Li J.; Rossignol F.; Macdonald J. Inkjet Printing for Biosensor Fabrication: Combining Chemistry and Technology for Advanced Manufacturing. Lab Chip 2015, 15, 2538–2558. 10.1039/C5LC00235D. [DOI] [PubMed] [Google Scholar]
- Yun Y. H.; Kim J. D.; Lee B. K.; Yoo B.; Lee J. H.; Cho Y. W. Construction of Micro-Patterned Polymer Structures by Piezoelectric Inkjet Printing. Polym.-Plast. Technol. Eng. 2009, 48, 1318–1323. 10.1080/03602550903204196. [DOI] [Google Scholar]
- Hook A. L.; Chang C.-Y.; Yang J.; Scurr D. J.; Langer R.; Anderson D. G.; Atkinson S.; Williams P.; Davies M. C.; Alexander M. R.; et al. Polymer Microarrays for High Throughput Discovery of Biomaterials. J. Vis. Exp 2012, 59, 3636. 10.3791/3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel E. A.; Larson B. L.; Luly K. M.; Kim J. D.; Langer R. Non-Cell-Adhesive Substrates for Printing of Arrayed Biomaterials. Adv. Healthcare Mater. 2015, 4, 501–505. 10.1002/adhm.201400594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasi Ghaemi S.; Harding F.; Delalat B.; Vasani R.; Voelcker N. H. Surface Engineering for Long-Term Culturing of Mesenchymal Stem Cell Microarrays. Biomacromolecules 2013, 14, 2675–2683. 10.1021/bm400531n. [DOI] [PubMed] [Google Scholar]
- Tharmalingam T.; Ghebeh H.; Wuerz T.; Butler M.. Pluronic Enhances the Robustness and Reduces the Cell Attachment of Mammalian Cells. In Molecular Biotechnology; Springer, 2008; Vol. 39, pp 167–177. [DOI] [PubMed] [Google Scholar]
- Mei Y.; Cannizzaro C.; Park H.; Xu Q.; Bogatyrev S. R.; Yi K.; Goldman N.; Langer R.; Anderson D. G. Cell-Compatible, Multicomponent Protein Arrays with Subcellular Feature Resolution. Small 2008, 4, 1600–1604. 10.1002/smll.200800363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A.; Mjoseng H. K.; Zhang R.; Kalloudis M.; Koutsos V.; de Sousa P. A.; Bradley M. High-Density Polymer Microarrays: Identifying Synthetic Polymers That Control Human Embryonic Stem Cell Growth. Adv. Healthcare Mater. 2014, 3, 848–853. 10.1002/adhm.201300489. [DOI] [PubMed] [Google Scholar]
- Hook A. L.; Scurr D. J.; Anderson D. G.; Langer R.; Williams P.; Davies M.; Alexander M.. High Throughput Discovery of Thermo-Responsive Materials Using Water Contact Angle Measurements and Time-of-Flight Secondary Ion Mass Spectrometry. In Surf. Interface Anal.; Wiley-Blackwell, 2013; Vol. 45, pp 181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal J. B.Microarrays Vol. 1: Synthesis Methods, 2nd ed.; Humana Press, 2007. [Google Scholar]
- Hook A. L.; Chang C. Y.; Yang J.; Atkinson S.; Langer R.; Anderson D. G.; Davies M. C.; Williams P.; Alexander M. R. Discovery of Novel Materials with Broad Resistance to Bacterial Attachment Using Combinatorial Polymer Microarrays. Adv. Mater. 2013, 25, 2542–2547. 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiz A. D.; Smith J. G. W.; Patel A. K.; Langer R.; Anderson D. G.; Barrett D. A.; Young L. E.; Davies M. C.; Denning C.; Alexander M. R. Chemically Diverse Polymer Microarrays and High Throughput Surface Characterisation: A Method for Discovery of Materials for Stem Cell Culture. Biomater. Sci. 2014, 2, 1604–1611. 10.1039/C4BM00054D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L.; Scurr D. J.; Burley J. C.; Langer R.; Anderson D. G.; Davies M. C.; Alexander M. R. Analysis and Prediction of Defects in UV Photo-Initiated Polymer Microarrays. J. Mater. Chem. B 2013, 1, 1035–1043. 10.1039/C2TB00379A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltman D.; Subramanian V. Inkjet-Printed Line Morphologies and Temperature Control of the Coffee Ring Effect. Langmuir 2008, 24, 2224–2231. 10.1021/la7026847. [DOI] [PubMed] [Google Scholar]
- Liberski A.; Zhang R.; Bradley M. Inkjet Fabrication of Polymer Microarrays and Grids - Solving the Evaporation Problem. Chem. Commun. 2009, 334–336. 10.1039/B816920A. [DOI] [PubMed] [Google Scholar]
- Horner C. B.; Hirota K.; Liu J.; Maldonado M.; Hyle Park B.; Nam J. Magnitude-Dependent and Inversely-Related Osteogenic/Chondrogenic Differentiation of Human Mesenchymal Stem Cells under Dynamic Compressive Strain. J. Tissue Eng. Regen. Med. 2018, 12, e637 10.1002/term.2332. [DOI] [PubMed] [Google Scholar]
- Beachley V. Z.; Wolf M. T.; Sadtler K.; Manda S. S.; Jacobs H.; Blatchley M. R.; Bader J. S.; Pandey A.; Pardoll D.; Elisseeff J. H. Tissue Matrix Arrays for High-Throughput Screening and Systems Analysis of Cell Function. Nat. Methods 2015, 12, 1197–1204. 10.1038/nmeth.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatshahi-Pirouz A.; Nikkhah M.; Gaharwar A. K.; Hashmi B.; Guermani E.; Aliabadi H.; Camci-Unal G.; Ferrante T.; Foss M.; Ingber D. E.; et al. A Combinatorial Cell-Laden Gel Microarray for Inducing Osteogenic Differentiation of Human Mesenchymal Stem Cells. Sci. Rep. 2015, 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donderwinkel I.; Van Hest J. C. M.; Cameron N. R. Bio-Inks for 3D Bioprinting: Recent Advances and Future Prospects. Polym. Chem. 2017, 8, 4451–4471. 10.1039/C7PY00826K. [DOI] [Google Scholar]
- Ranga A.; Gobaa S.; Okawa Y.; Mosiewicz K.; Negro A.; Lutolf M. P. 3D Niche Microarrays for Systems-Level Analyses of Cell Fate. Nat. Commun. 2014, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Floren M.; Ding Y.; Stenmark K. R.; Tan W.; Bryant S. J. A Photoclickable Peptide Microarray Platform for Facile and Rapid Screening of 3-D Tissue Microenvironments. Biomaterials 2017, 143, 17–28. 10.1016/j.biomaterials.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Moraes C.; Wang G. H.; Sun Y.; Simmons C. A. A Microfabricated Platform for High-Throughput Unconfined Compression of Micropatterned Biomaterial Arrays. Biomaterials 2010, 31, 577–584. 10.1016/j.biomaterials.2009.09.068. [DOI] [PubMed] [Google Scholar]
- Panda P.; Ali S.; Lo E.; Chung B. G.; Hatton T. A.; Khademhosseini A.; Doyle P. S. Stop-Flow Lithography to Generate Cell-Laden Microgel Particles. Lab Chip 2008, 8, 1056–1061. 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S.; Vermeulen S.; Van Der Veer B.; Carlier A.; De Boer J.; Subramanyam D. Shaping Cell Fate: Influence of Topographical Substratum Properties on Embryonic Stem Cells. Tissue Eng., Part B 2018, 24, 255–266. 10.1089/ten.teb.2017.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unadkat H. V.; Hulsman M.; Cornelissen K.; Papenburg B. J.; Truckenmüller R. K.; Carpenter A. E.; Wessling M.; Post G. F.; Uetz M.; Reinders M. J. T.; et al. An Algorithm-Based Topographical Biomaterials Library to Instruct Cell Fate. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16565–16570. 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuning D. G.; Beijer N. R. M.; Du Fossé N. A.; Vermeulen S.; Lievers E.; Van Kooten C.; Rabelink T. J.; De Boer J. The Cytokine Secretion Profile of Mesenchymal Stromal Cells Is Determined by Surface Structure of the Microenvironment. Sci. Rep. 2018, 8, 7716. 10.1038/s41598-018-25700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshof F. F. B.; Zhao Y.; Vasilevich A.; Beijer N. R. M.; de Boer M.; Papenburg B. J.; van Blitterswijk C.; Stamatialis D.; de Boer J. NanoTopoChip: High-Throughput Nanotopographical Cell Instruction. Acta Biomater. 2017, 62, 188–198. 10.1016/j.actbio.2017.08.023. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Truckenmuller R.; Levers M.; Hua W. S.; de Boer J.; Papenburg B. High-Definition Micropatterning Method for Hard, Stiff and Brittle Polymers. Mater. Sci. Eng., C 2017, 71, 558–564. 10.1016/j.msec.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Sanni O.; Chang C. Y.; Anderson D. G.; Langer R.; Davies M. C.; Williams P. M.; Williams P.; Alexander M. R.; Hook A. L. Bacterial Attachment to Polymeric Materials Correlates with Molecular Flexibility and Hydrophilicity. Adv. Healthcare Mater. 2015, 4, 695–701. 10.1002/adhm.201400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart A. J.; Taylor M.; Anderson D. G.; Langer R.; Davies M. C.; Alexander M. R. TOF-SIMS Analysis of a 576 Micropatterned Copolymer Array to Reveal Surface Moieties That Control Wettability. Anal. Chem. 2008, 80, 135–142. 10.1021/ac071560k. [DOI] [PubMed] [Google Scholar]
- Koenig A. L.; Gambillara V.; Grainger D. W. Correlating Fibronectin Adsorption with Endothelial Cell Adhesion and Signaling in Polymer Substrates. J. Biomed Mater. Res. A 2002, 64, 20–37. 10.1002/jbm.a.10316. [DOI] [PubMed] [Google Scholar]
- Mei Y.; Gerecht S.; Taylor M.; Urquhart A. J.; Bogatyrev S. R.; Cho S. W.; Davies M. C.; Alexander M. R.; Langer R. S.; Anderson D. G. Mapping the Interactions among Biomaterials, Adsorbed Proteins, and Human Embryonic Stem Cells. Adv. Mater. 2009, 21, 2781–2786. 10.1002/adma.200803184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaim C. J.; Chien S.; Bhatia S. N. An Extracellular Matrix Microarray for Probing Cellular Differentiation. Nat. Methods 2005, 2, 119–125. 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- Norde W. Protein Adsorption at Solid Surfaces: A Thermodynamic Approach. Pure Appl. Chem. 1994, 66, 1–8. 10.1351/pac199466030491. [DOI] [Google Scholar]
- Llopis-Hernández V.; Cantini M.; González-García C.; Cheng Z. A.; Yang J.; Tsimbouri P. M.; García A. J.; Dalby M. J.; Salmerón-Sánchez M. Material-Driven Fibronectin Assembly for High-Efficiency Presentation of Growth Factors. Sci. Adv. 2016, 2, 1–10. 10.1126/sciadv.1600188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L.; Cheng K.; Zhou Y.; Yu M.; Gong J.; Lin Y.; Luo Q.; Wang Q.; Weng W.; Wang H. Surface Atomic Structure Directs the Fate of Human Mesenchymal Stem Cells. ACS Appl. Mater. Interfaces 2017, 9, 15274–15285. 10.1021/acsami.7b02411. [DOI] [PubMed] [Google Scholar]
- Ngandu Mpoyi E.; Cantini M.; Reynolds P. M.; Gadegaard N.; Dalby M. J.; Salmerón-Sánchez M. Protein Adsorption as a Key Mediator in the Nanotopographical Control of Cell Behavior. ACS Nano 2016, 10, 6638–6647. 10.1021/acsnano.6b01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L.; Thissen H.; Voelcker N. H. Surface Plasmon Resonance Imaging of Polymer Microarrays to Study Protein-Polymer Interactions in High Throughput. Langmuir 2009, 25, 9173–9181. 10.1021/la900735n. [DOI] [PubMed] [Google Scholar]
- Hook A. L.; Williams P. M.; Alexander M. R.; Scurr D. J. Multivariate ToF-SIMS Image Analysis of Polymer Microarrays and Protein Adsorption. Biointerphases 2015, 10, 019005. 10.1116/1.4906484. [DOI] [PubMed] [Google Scholar]
- Celiz A. D.; Harrington H. C.; Hook A. L. High Throughput Assessment and Chemometric Analysis of the Interaction of Epithelial and Fibroblast Cells with a Polymer Library. Appl. Surf. Sci. 2014, 313, 926–935. 10.1016/j.apsusc.2014.06.111. [DOI] [Google Scholar]
- Taylor M.; Urquhart A. J.; Anderson D. G.; Williams P. M.; Langer R.; Alexander M. R.; Davies M. C. A Methodology for Investigating Protein Adhesion and Adsorption to Microarrayed Combinatorial Polymers. Macromol. Rapid Commun. 2008, 29, 1298–1302. 10.1002/marc.200800171. [DOI] [Google Scholar]
- Hammad M.; Rao W.; Smith J. G. W.; Anderson D. G.; Langer R.; Young L. E.; Barrett D. A.; Davies M. C.; Denning C.; Alexander M. R. Identification of Polymer Surface Adsorbed Proteins Implicated in Pluripotent Human Embryonic Stem Cell Expansion. Biomater. Sci. 2016, 4, 1381–1391. 10.1039/C6BM00214E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao W.; Celiz A. D.; Scurr D. J.; Alexander M. R.; Barrett D. A. Ambient DESI and LESA-MS Analysis of Proteins Adsorbed to a Biomaterial Surface Using in-Situ Surface Tryptic Digestion. J. Am. Soc. Mass Spectrom. 2013, 24, 1927–1936. 10.1007/s13361-013-0737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlstedt M. M.; Anderson D.; Sharp J. S.; McGilvray R.; Barbadillo MuÃtoz M. D.; Buttery L. D.; Alexander M. R.; Rose F. R. A. J.; Denning C. Maintenance of Pluripotency in Human Embryonic Stem Cells Cultured on a Synthetic Substrate in Conditioned Medium. Biotechnol. Bioeng. 2010, 105, 130–140. 10.1002/bit.22520. [DOI] [PubMed] [Google Scholar]
- Mikulskis P.; Hook A.; Dundas A. A.; Irvine D.; Sanni O.; Anderson D.; Langer R.; Alexander M. R.; Williams P.; Winkler D. A. Prediction of Broad-Spectrum Pathogen Attachment to Coating Materials for Biomedical Devices. ACS Appl. Mater. Interfaces 2018, 10, 139–149. 10.1021/acsami.7b14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M.-A.; Singh S.; Han H.; Davis C. T.; Borgeson B.; Hartland C.; Kost-Alimova M.; Gustafsdottir S. M.; Gibson C. C.; Carpenter A. E. Cell Painting, a High-Content Image-Based Assay for Morphological Profiling Using Multiplexed Fluorescent Dyes. Nat. Protoc. 2016, 11, 1757–1774. 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo J. C.; Cooper S.; Heigwer F.; Warchal S.; Qiu P.; Molnar C.; Vasilevich A. S.; Barry J. D.; Bansal H. S.; Kraus O.; et al. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14, 849–863. 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Mei Y.; Hook A. L.; Taylor M.; Urquhart A. J.; Bogatyrev S. R.; Langer R.; Anderson D. G.; Davies M. C.; Alexander M. R. Polymer Surface Functionalities That Control Human Embryoid Body Cell Adhesion Revealed by High Throughput Surface Characterization of Combinatorial Material Microarrays. Biomaterials 2010, 31, 8827–8838. 10.1016/j.biomaterials.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epa V. C.; Hook A. L.; Chang C.; Yang J.; Langer R.; Anderson D. G.; Williams P.; Davies M. C.; Alexander M. R.; Winkler D. A. Modelling and Prediction of Bacterial Attachment to Polymers. Adv. Funct. Mater. 2014, 24, 2085–2093. 10.1002/adfm.201302877. [DOI] [Google Scholar]
- Alsberg E.; von Recum H. A.; Mahoney M. J. Environmental Cues to Guide Stem Cell Fate Decision for Tissue Engineering Applications. Expert Opin. Biol. Ther. 2006, 6, 847–866. 10.1517/14712598.6.9.847. [DOI] [PubMed] [Google Scholar]
- Ngandu Mpoyi E.; Cantini M.; Reynolds P. M.; Gadegaard N.; Dalby M. J.; Salmerón-Sánchez M.; Mpoyi E. N.; Cantini M.; Reynolds P. M.; Gadegaard N.; et al. Protein Adsorption as a Key Mediator in the Nanotopographical Control of Cell Behavior. ACS Nano 2016, 10, 6638–6647. 10.1021/acsnano.6b01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining K. H.; Mooney D. J.. Mechanical Forces Direct Stem Cell Behaviour in Development and Regeneration. Nature Reviews Molecular Cell Biology; Nature Publishing Group, 2017; pp 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. L.; Hua X.; Somsel H.; Reichart F.; Kessler H.; Spatz J. P. Integrin Subtypes and Nanoscale Ligand Presentation Influence Drug Sensitivity in Cancer Cells. Nano Lett. 2020, 20, 1183–1191. 10.1021/acs.nanolett.9b04607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acerbi I.; Cassereau L.; Dean I.; Shi Q.; Au A.; Park C.; Chen Y. Y.; Liphardt J.; Hwang E. S.; Weaver V. M. Human Breast Cancer Invasion and Aggression Correlates with ECM Stiffening and Immune Cell Infiltration. Integr. Biol. 2015, 7, 1120–1134. 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier Leach J.; Bivens K. A.; Patrick Jr C. W.; Schmidt C. E. Photocrosslinked Hyaluronic Acid Hydrogels: Natural, Biodegradable Tissue Engineering Scaffolds. Biotechnol. Bioeng. 2003, 82, 578–589. 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- Kanie K.; Kondo Y.; Owaki J.; Ikeda Y.; Narita Y.; Kato R.; Honda H. Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays. Bioengineering 2016, 3, 31. 10.3390/bioengineering3040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaim C. J.; Teng D.; Chien S.; Bhatia S. N. Combinatorial Signaling Microenvironments for Studying Stem Cell Fate. Stem Cells Dev. 2008, 17, 29–40. 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- Kingshott P.; Thissen H.; Griesser H. J. Effects of Cloud-Point Grafting, Chain Length, and Density of PEG Layers on Competitive Adsorption of Ocular Proteins. Biomaterials 2002, 23, 2043–2056. 10.1016/S0142-9612(01)00334-9. [DOI] [PubMed] [Google Scholar]
- Malta D. F. B.; Reticker-Flynn N. E.; da Silva C. L.; Cabral J. M. S.; Fleming H. E.; Zaret K. S.; Bhatia S. N.; Underhill G. H. Extracellular Matrix Microarrays to Study Inductive Signaling for Endoderm Specification. Acta Biomater. 2016, 34, 30–40. 10.1016/j.actbio.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield R. B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. 10.1021/ja00897a025. [DOI] [Google Scholar]
- Renil M.; Ferreras M.; Delaisse J. M.; Foged N. T.; Meldal M. PEGA Supports for Combinatorial Peptide Synthesis and Solid-Phase Enzymatic Library Assays. J. Pept. Sci. 1998, 4, 195–210. . [DOI] [PubMed] [Google Scholar]
- Lam K. S.; Salmon S. E.; Hersh E. M.; Hruby V. J.; Kazmierski W. M.; Knapp R. J. A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature 1991, 354, 82–84. 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Singer I. I.; Kawka D. W.; Scott S.; Mumford R. A.; Lark M. W. The Fibronectin Cell Attachment Sequence Arg-Gly-Asp-Ser Promotes Focal Contact Formation during Early Fibroblast Attachment and Spreading. J. Cell Biol. 1987, 104, 573–584. 10.1083/jcb.104.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner N.; Dargaville T. R.; Forget A. Discovering Cell-Adhesion Peptides in Tissue Engineering: Beyond RGD. Trends Biotechnol. 2018, 36, 372–383. 10.1016/j.tibtech.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Orner B. P.; Derda R.; Lewis R. L.; Thomson J. A.; Kiessling L. L. Arrays for the Combinatorial Exploration of Cell Adhesion. J. Am. Chem. Soc. 2004, 126, 10808–10809. 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- Derda R.; Li L.; Orner B. P.; Lewis R. L.; Thomson J. A.; Kiessling L. L. Defined Substrates for Human Embryonic Stem Cell Growth Identified from Surface Arrays. ACS Chem. Biol. 2007, 2, 347–355. 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- Lin E.; Sikand A.; Wickware J.; Hao Y.; Derda R. Peptide Microarray Patterning for Controlling and Monitoring Cell Growth. Acta Biomater. 2016, 34, 53–59. 10.1016/j.actbio.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Zhang D.; Kilian K. A. Peptide Microarrays for the Discovery of Bioactive Surfaces That Guide Cellular Processes: A Single Step Azide-Alkyne “Click” Chemistry Approach. J. Mater. Chem. B 2014, 2, 4280–4288. 10.1039/C4TB00375F. [DOI] [PubMed] [Google Scholar]
- Gupta N.; Lin B. F.; Campos L. M.; Dimitriou M. D.; Hikita S. T.; Treat N. D.; Tirrell M. V.; Clegg D. O.; Kramer E. J.; Hawker C. J. A Versatile Approach to High-Throughput Microarrays Using Thiol-Ene Chemistry. Nat. Chem. 2010, 2, 138–145. 10.1038/nchem.478. [DOI] [PubMed] [Google Scholar]
- Jia J.; Coyle R. C.; Richards D. J.; Berry C. L.; Barrs R. W.; Biggs J.; James Chou C.; Trusk T. C.; Mei Y. Development of Peptide-Functionalized Synthetic Hydrogel Microarrays for Stem Cell and Tissue Engineering Applications. Acta Biomater. 2016, 45, 110–120. 10.1016/j.actbio.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie K.; Kurimoto R.; Tian J.; Ebisawa K.; Narita Y.; Honda H.; Kato R. Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications. Materials 2016, 9, 730. 10.3390/ma9090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H.; Uchiyama N.; Kuno A.; Togayachi A.; Sato T.; Narimatsu H.; Hirabayashi J. A Novel Strategy for Mammalian Cell Surface Glycome Profiling Using Lectin Microarray. Glycobiology 2007, 17, 1138–1146. 10.1093/glycob/cwm084. [DOI] [PubMed] [Google Scholar]
- Smith R. A. A.; Meade K.; Pickford C. E.; Holley R. J.; Merry C. L. R. Glycosaminoglycans as Regulators of Stem Cell Differentiation. Biochem. Soc. Trans. 2011, 39, 383–387. 10.1042/BST0390383. [DOI] [PubMed] [Google Scholar]
- Chen Y. H.; Narimatsu Y.; Clausen T. M.; Gomes C.; Karlsson R.; Steentoft C.; Spliid C. B.; Gustavsson T.; Salanti A.; Persson A.; et al. The GAGOme: A Cell-Based Library of Displayed Glycosaminoglycans. Nat. Methods 2018, 15, 881–888. 10.1038/s41592-018-0086-z. [DOI] [PubMed] [Google Scholar]
- Toonstra C.; Wu L.; Li C.; Wang D.; Wang L. X. Top-Down Chemoenzymatic Approach to Synthesizing Diverse High-Mannose N-Glycans and Related Neoglycoproteins for Carbohydrate Microarray Analysis. Bioconjugate Chem. 2018, 29, 1911–1921. 10.1021/acs.bioconjchem.8b00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Sheng J.; Liu Y.; Li J.; Liu J.; Wang F. Heparosan-Derived Heparan Sulfate/Heparin-Like Compounds: One Kind of Potential Therapeutic Agents. Med. Res. Rev. 2013, 33, 665–692. 10.1002/med.21263. [DOI] [PubMed] [Google Scholar]
- Syed P.; Gidwani K.; Kekki H.; Leivo J.; Pettersson K.; Lamminmäki U. Role of Lectin Microarrays in Cancer Diagnosis. Proteomics 2016, 16, 1257–1265. 10.1002/pmic.201500404. [DOI] [PubMed] [Google Scholar]
- Kamena F.; Tamborrini M.; Liu X.; Kwon Y. U.; Thompson F.; Pluschke G.; Seeberger P. H. Synthetic GPI Array to Study Antitoxic Malaria Response. Nat. Chem. Biol. 2008, 4, 238–240. 10.1038/nchembio.75. [DOI] [PubMed] [Google Scholar]
- Mietzsch M.; Broecker F.; Reinhardt A.; Seeberger P. H.; Heilbronn R.; Imperiale M. J. Differential Adeno-Associated Virus Serotype-Specific Interaction Patterns with Synthetic Heparins and Other Glycans. J. Virol. 2014, 88, 2991–3003. 10.1128/JVI.03371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui S.; Feizi T.; Galustian C.; Lawson A. M.; Chai W. Oligosaccharide Microarrays for High-Throughput Detection and Specificity Assignments of Carbohydrate-Protein Interactions. Nat. Biotechnol. 2002, 20, 1011–1017. 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- Wang D.; Liu S.; Trummer B. J.; Deng C.; Wang A. Carbohydrate Microarrays for the Recognition of Cross-Reactive Molecular Markers of Microbes and Host Cells. Nat. Biotechnol. 2002, 20, 275–281. 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- Padler-Karavani V.; Song X.; Yu H.; Hurtado-Ziola N.; Huang S.; Muthana S.; Chokhawala H. A.; Cheng J.; Verhagen A.; Langereis M. A.; et al. Cross-Comparison of Protein Recognition of Sialic Acid Diversity on Two Novel Sialoglycan Microarrays. J. Biol. Chem. 2012, 287, 22593–22608. 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer A. R.; Hokke C. H.; Deelder A. M.; Wuhrer M. General Microarray Technique for Immobilization and Screening of Natural Glycans. Anal. Chem. 2007, 79, 8107–8113. 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- Hyun J. Y.; Kang N. R.; Shin I. Carbohydrate Microarrays Containing Glycosylated Fluorescent Probes for Assessment of Glycosidase Activities. Org. Lett. 2018, 20, 1240–1243. 10.1021/acs.orglett.8b00180. [DOI] [PubMed] [Google Scholar]
- Wang L.; Cummings R. D.; Smith D. F.; Huflejt M.; Campbell C. T.; Gildersleeve J. C.; Gerlach J. Q.; Kilcoyne M.; Joshi L.; Serna S.; et al. Cross-Platform Comparison of Glycan Microarray Formats. Glycobiology 2014, 24, 507–517. 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngundi M. M.; Taitt C. R.; McMurry S. A.; Kahne D.; Ligler F. S. Detection of Bacterial Toxins with Monosaccharide Arrays. Biosens. Bioelectron. 2006, 21, 1195–1201. 10.1016/j.bios.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J. Y.; Pai J.; Shin I. The Glycan Microarray Story from Construction to Applications. Acc. Chem. Res. 2017, 50, 1069–1078. 10.1021/acs.accounts.7b00043. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Cai C.; Chandarajoti K.; Hsieh P.-H.; Li L.; Pham T. Q.; Sparkenbaugh E. M.; Sheng J.; Key N. S.; Pawlinski R.; et al. Homogeneous Low-Molecular-Weight Heparins with Reversible Anticoagulant Activity. Nat. Chem. Biol. 2014, 10, 248–250. 10.1038/nchembio.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paz J. L.; Noti C.; Seeberger P. H. Microarrays of Synthetic Heparin Oligosaccharides. J. Am. Chem. Soc. 2006, 128, 2766–2767. 10.1021/ja057584v. [DOI] [PubMed] [Google Scholar]
- Scurr D. J.; Horlacher T.; Oberli M. A.; Werz D. B.; Kroeck L.; Bufali S.; Seeberger P. H.; Shard A. G.; Alexander M. R. Surface Characterization of Carbohydrate Microarrays. Langmuir 2010, 26, 17143–17155. 10.1021/la1029933. [DOI] [PubMed] [Google Scholar]
- Amon R.; Grant O. C.; Leviatan Ben-Arye S.; Makeneni S.; Nivedha A. K.; Marshanski T.; Norn C.; Yu H.; Glushka J. N.; Fleishman S. J.; et al. A Combined Computational-Experimental Approach to Define the Structural Origin of Antibody Recognition of Sialyl-Tn, a Tumor-Associated Carbohydrate Antigen. Sci. Rep. 2018, 8, 10786. 10.1038/s41598-018-29209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernagallo S.; Wu M.; Gallagher M. P.; Bradley M. Colonising New Frontiers - Microarrays Reveal Biofilm Modulating Polymers. J. Mater. Chem. 2011, 21, 96–101. 10.1039/C0JM01987A. [DOI] [Google Scholar]
- Urquhart A. J.; Anderson D. G.; Taylor M.; Alexander M. R.; Langer R.; Davies M. C. High Throughput Surface Characterisation of a Combinatorial Material Library. Adv. Mater. 2007, 19, 2486–2491. 10.1002/adma.200700949. [DOI] [Google Scholar]
- Vining K. H.; Scherba J. C.; Bever A. M.; Alexander M. R.; Celiz A. D.; Mooney D. J. Synthetic Light-Curable Polymeric Materials Provide a Supportive Niche for Dental Pulp Stem Cells. Adv. Mater. 2018, 30, 1704486. 10.1002/adma.201704486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epa V. C.; Yang J.; Mei Y.; Hook A. L.; Langer R.; Anderson D. G.; Davies M. C.; Alexander M. R.; Winkler D. A. Modelling Human Embryoid Body Cell Adhesion to a Combinatorial Library of Polymer Surfaces. J. Mater. Chem. 2012, 22, 20902–20906. 10.1039/c2jm34782b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. K.; Celiz A. D.; Rajamohan D.; Anderson D. G.; Langer R.; Davies M. C.; Alexander M. R.; Denning C. A Defined Synthetic Substrate for Serum-Free Culture of Human Stem Cell Derived Cardiomyocytes with Improved Functional Maturity Identified Using Combinatorial Materials Microarrays. Biomaterials 2015, 61, 257–265. 10.1016/j.biomaterials.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N.; Ohta H.; Nakayama Y.; Konishi M. Roles of FGF Signals in Heart Development, Health, and Disease. Front. Cell Dev. Biol. 2016, 4, 1–11. 10.3389/fcell.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A. N.; Stanganello E.; Welle A.; Scholpp S.; Levkin P. A. Micropatterned Superhydrophobic Structures for the Simultaneous Culture of Multiple Cell Types and the Study of Cell-Cell Communication. Biomaterials 2013, 34, 1757–1763. 10.1016/j.biomaterials.2012.11.034. [DOI] [PubMed] [Google Scholar]
- Popova A. A.; Demir K.; Hartanto T. G.; Schmitt E.; Levkin P. A. Droplet-Microarray on Superhydrophobic-Superhydrophilic Patterns for High-Throughput Live Cell Screenings. RSC Adv. 2016, 6, 38263–38276. 10.1039/C6RA06011K. [DOI] [Google Scholar]
- Brehm M.; Heissler S.; Afonin S.; Levkin P. A. Nanomolar Synthesis in Droplet Microarrays with UV-Triggered On-Chip Cell Screening. Small 2020, 16, 1905971. 10.1002/smll.201905971. [DOI] [PubMed] [Google Scholar]
- Neto A. I.; Demir K.; Popova A. A.; Oliveira M. B.; Mano J. F.; Levkin P. A. Fabrication of Hydrogel Particles of Defined Shapes Using Superhydrophobic-Hydrophilic Micropatterns. Adv. Mater. 2016, 28, 7613–7619. 10.1002/adma.201602350. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Tronser T.; Peravali R.; Reischl M.; Levkin P. A. High-Throughput Screening of Cell Transfection Enhancers Using Miniaturized Droplet Microarrays. Adv. Biosyst. 2020, 4, 1900257. 10.1002/adbi.201900257. [DOI] [PubMed] [Google Scholar]
- Popova A. A.; Depew C.; Permana K. M.; Trubitsyn A.; Peravali R.; Ordiano J. Á. G.; Reischl M.; Levkin P. A. Evaluation of the Droplet-Microarray Platform for High-Throughput Screening of Suspension Cells. SLAS Technol. 2017, 22, 163–175. 10.1177/2211068216677204. [DOI] [PubMed] [Google Scholar]
- Tronser T.; Popova A. A.; Jaggy M.; Bastmeyer M.; Levkin P. A. Droplet Microarray Based on Patterned Superhydrophobic Surfaces Prevents Stem Cell Differentiation and Enables High-Throughput Stem Cell Screening. Adv. Healthcare Mater. 2017, 6, 1700622. 10.1002/adhm.201700622. [DOI] [PubMed] [Google Scholar]
- Arnold M.; Cavalcanti-Adam E. A.; Glass R.; Blümmel J.; Eck W.; Kantlehner M.; Kessler H.; Spatz J. P. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. ChemPhysChem 2004, 5, 383–388. 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Guasch J.; Diemer J.; Riahinezhad H.; Neubauer S.; Kessler H.; Spatz J. P. Synthesis of Binary Nanopatterns on Hydrogels for Initiating Cellular Responses. Chem. Mater. 2016, 28, 1806–1815. 10.1021/acs.chemmater.5b04910. [DOI] [Google Scholar]
- Amin Y. Y. I.; Runager K.; Simoes F.; Celiz A.; Taresco V.; Rossi R.; Enghild J. J.; Abildtrup L. A.; Kraft D. C. E.; Sutherland D. S.; et al. Combinatorial Biomolecular Nanopatterning for High-Throughput Screening of Stem-Cell Behavior. Adv. Mater. 2016, 28, 1472–1476. 10.1002/adma.201504995. [DOI] [PubMed] [Google Scholar]
- Gilbert P. M.; Havenstrite K. L.; Magnusson K. E. G.; Sacco A.; Leonardi N. A.; Kraft P.; Nguyen N. K.; Thrun S.; Lutolf M. P.; Blau H. M. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science (Washington, DC, U. S.) 2010, 329, 1078–1081. 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobaa S.; Hoehnel S.; Lutolf M. P. Substrate Elasticity Modulates the Responsiveness of Mesenchymal Stem Cells to Commitment Cues. Integr. Biol. (United Kingdom) 2015, 7, 1135–1142. 10.1039/C4IB00176A. [DOI] [PubMed] [Google Scholar]
- Gobaa S.; Hoehnel S.; Roccio M.; Negro A.; Kobel S.; Lutolf M. P. Artificial Niche Microarrays for Probing Single Stem Cell Fate in High Throughput. Nat. Methods 2011, 8, 949–955. 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- Kourouklis A. P.; Kaylan K. B.; Underhill G. H. Substrate Stiffness and Matrix Composition Coordinately Control the Differentiation of Liver Progenitor Cells. Biomaterials 2016, 99, 82–94. 10.1016/j.biomaterials.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Kaylan K. B.; Kourouklis A. P.; Underhill G. H. A High-Throughput Cell Microarray Platform for Correlative Analysis of Cell Differentiation and Traction Forces. J. Vis. Exp. 2017, 1–11. 10.3791/55362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Erlach T. C.; Bertazzo S.; Wozniak M. A.; Horejs C. M.; Maynard S. A.; Attwood S.; Robinson B. K.; Autefage H.; Kallepitis C.; Del Río Hernández A.; et al. Cell-Geometry-Dependent Changes in Plasma Membrane Order Direct Stem Cell Signalling and Fate. Nat. Mater. 2018, 17, 237–242. 10.1038/s41563-017-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E.; Marshall C. J. ROCK and Dia Have Opposing Effects on Adherens Junctions Downstream of Rho. Nat. Cell Biol. 2002, 4, 408–415. 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- Wang J.; Cui C.; Nan H.; Yu Y.; Xiao Y.; Poon E.; Yang G.; Wang X.; Wang C.; Li L.; et al. Graphene Sheet-Induced Global Maturation of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells. ACS Appl. Mater. Interfaces 2017, 9, 25929–25940. 10.1021/acsami.7b08777. [DOI] [PubMed] [Google Scholar]
- Vaithilingam J.; Sanjuan-Alberte P.; Campora S.; Rance G. A.; Jiang L.; Thorpe J.; Burroughs L.; Tuck C. J.; Denning C.; Wildman R. D.; et al. Multifunctional Bioinstructive 3D Architectures to Modulate Cellular Behavior. Adv. Funct. Mater. 2019, 29, 1902016. 10.1002/adfm.201902016. [DOI] [Google Scholar]
- Müller J.; Ballini M.; Livi P.; Chen Y.; Radivojevic M.; Shadmani A.; Viswam V.; Jones I. L.; Fiscella M.; Diggelmann R.; et al. High-Resolution CMOS MEA Platform to Study Neurons at Subcellular, Cellular, and Network Levels. Lab Chip 2015, 15, 2767–2780. 10.1039/C5LC00133A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U.; Sedivy J.; Heer F.; Pedron R.; Ballini M.; Mueller J.; Bakkum D.; Hafizovic S.; Faraci F. D.; Greve F.; et al. Switch-Matrix-Based High-Density Microelectrode Array in CMOS Technology. IEEE J. Solid-State Circuits 2010, 45, 467–482. 10.1109/JSSC.2009.2035196. [DOI] [Google Scholar]
- Lopez C. M.; Chun H. S.; Berti L.; Wang S.; Putzeys J.; Van Den Bulcke C.; Weijers J. W.; Firrincieli A.; Reumers V.; Braeken D.; et al. A 16384-Electrode 1024-Channel Multimodal CMOS MEA for High-Throughput Intracellular Action Potential Measurements and Impedance Spectroscopy in Drug-Screening Applications. In Digest of Technical Papers - IEEE International Solid-State Circuits Conference; Institute of Electrical and Electronics Engineers Inc., 2018; Vol. 61, pp 464–466.
- Lopez C. M.; Chun H. S.; Wang S.; Berti L.; Putzeys J.; Van Den Bulcke C.; Weijers J. W.; Firrincieli A.; Reumers V.; Braeken D.; et al. A Multimodal CMOS MEA for High-Throughput Intracellular Action Potential Measurements and Impedance Spectroscopy in Drug-Screening Applications. IEEE J. Solid-State Circuits 2018, 53, 3076–3086. 10.1109/JSSC.2018.2863952. [DOI] [Google Scholar]
- Miccoli B.; Lopez C. M.; Goikoetxea E.; Putzeys J.; Sekeri M.; Krylychkina O.; Chang S. W.; Firrincieli A.; Andrei A.; Reumers V.; et al. High-Density Electrical Recording and Impedance Imaging with a Multi-Modal CMOS Multi-Electrode Array Chip. Front. Neurosci. 2019, 13, 641. 10.3389/fnins.2019.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J. S.; Pourmand N.. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis. John Wiley & Sons, Ltd, 2007; pp 1239–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer M.; Jahnke H. G.; Krinke D.; Zitzmann F. D.; Hill K.; Schaefer M.; Robitzki A. A. Quantitative Characterization of Capsaicin-Induced TRPV1 Ion Channel Activation in HEK293 Cells by Impedance Spectroscopy. Anal. Bioanal. Chem. 2016, 408, 8529–8538. 10.1007/s00216-016-9978-x. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Diaz M. F.; Price K. M.; Ozuna J. A.; Zhang S.; Sevick-Muraca E. M.; Hagan J. P.; Wenzel P. L. Fluid Shear Stress Activates YAP1 to Promote Cancer Cell Motility. Nat. Commun. 2017, 8, 1–14. 10.1038/ncomms14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaud A.; Regev I.; Mahadevan L.; Pourquié O. Excitable Dynamics and Yap-Dependent Mechanical Cues Drive the Segmentation Clock. Cell 2017, 171, 668–682. 10.1016/j.cell.2017.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Huang G.; Gao B.; Li M.; Genin G. M.; Lu T. J.; Xu F. Magnetically Actuated Cell-Laden Microscale Hydrogels for Probing Strain-Induced Cell Responses in Three Dimensions. NPG Asia Mater. 2016, 8, e238 10.1038/am.2015.148. [DOI] [Google Scholar]
- Lehnert D.; Wehrle-Haller B.; David C.; Weiland U.; Ballestrem C.; Imhof B. A.; Bastmeyer M. Cell Behaviour on Micropatterned Substrata: Limits of Extracellular Matrix Geometry for Spreading and Adhesion. J. Cell Sci. 2004, 117, 41–52. 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- Dalby M. J.; Gadegaard N.; Oreffo R. O. C. Harnessing Nanotopography and Integrin-Matrix Interactions to Influence Stem Cell Fate. Nat. Mater. 2014, 13, 558–569. 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- Fischbach C.; Chen R.; Matsumoto T.; Schmelzle T.; Brugge J. S.; Polverini P. J.; Mooney D. J. Engineering Tumors with 3D Scaffolds. Nat. Methods 2007, 4, 855–860. 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- Lossdörfer S.; Schwartz Z.; Wang L.; Lohmann C. H.; Turner J. D.; Wieland M.; Cochran D. L.; Boyan B. D. Microrough Implant Surface Topographies Increase Osteogenesis by Reducing Osteoclast Formation and Activity. J. Biomed. Mater. Res., Part A 2004, 70A, 361–369. 10.1002/jbm.a.30025. [DOI] [PubMed] [Google Scholar]
- Anselme K.; Bigerelle M. Statistical Demonstration of the Relative Effect of Surface Chemistry and Roughness on Human Osteoblast Short-Term Adhesion. J. Mater. Sci.: Mater. Med. 2006, 17, 471–479. 10.1007/s10856-006-8475-8. [DOI] [PubMed] [Google Scholar]
- Dalby M. J.; Gadegaard N.; Tare R.; Andar A.; Riehle M. O.; Herzyk P.; Wilkinson C. D. W.; Oreffo R. O. C. The Control of Human Mesenchymal Cell Differentiation Using Nanoscale Symmetry and Disorder. Nat. Mater. 2007, 6, 997–1003. 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Crowder S. W.; Leonardo V.; Whittaker T.; Papathanasiou P.; Stevens M. M. Material Cues as Potent Regulators of Epigenetics and Stem Cell Function. Cell Stem Cell 2016, 18, 39–52. 10.1016/j.stem.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer A.; Vasilevich A.; Hulshof F.; Viswanathan P.; Van Blitterswijk C. A.; De Boer J.; Watt F. M. Scalable Topographies to Support Proliferation and Oct4 Expression by Human Induced Pluripotent Stem Cells. Sci. Rep. 2016, 6, 18948. 10.1038/srep18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J. Shifting Gears in Biomaterials Discovery. Matter 2020, 2, 1358–1360. 10.1016/j.matt.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshof F. F. B.; Papenburg B.; Vasilevich A.; Hulsman M.; Zhao Y.; Levers M.; Fekete N.; de Boer M.; Yuan H.; Singh S.; et al. Mining for Osteogenic Surface Topographies: In Silico Design to in Vivo Osseo-Integration. Biomaterials 2017, 137, 49–60. 10.1016/j.biomaterials.2017.05.020. [DOI] [PubMed] [Google Scholar]
- Vermeulen S.; Vasilevich A.; Tsiapalis D.; Roumans N.; Vroemen P.; Beijer N. R. M.; Dede Eren A.; Zeugolis D.; de Boer J. Identification of Topographical Architectures Supporting the Phenotype of Rat Tenocytes. Acta Biomaterialia 2018, 83, 277–290. 10.1016/j.actbio.2018.10.041. [DOI] [PubMed] [Google Scholar]
- Zijl S.; Vasilevich A. S.; Viswanathan P.; Helling A. L.; Beijer N. R. M.; Walko G.; Chiappini C.; de Boer J.; Watt F. M. Micro-Scaled Topographies Direct Differentiation of Human Epidermal Stem Cells. Acta Biomater. 2019, 84, 133–145. 10.1016/j.actbio.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuning D. G.; Reinders M. E. J.; Li J.; Peired A. J.; Lievers E.; de Boer H. C.; Fibbe W. E.; Romagnani P.; van Kooten C.; Little M. H.; et al. Clinical-Grade Isolated Human Kidney Perivascular Stromal Cells as an Organotypic Cell Source for Kidney Regenerative Medicine. Stem Cells Transl. Med. 2017, 6, 405–418. 10.5966/sctm.2016-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K.; Fibbe W. E. Unraveling Mechanisms of Mesenchymal Stromal Cell-Mediated Immunomodulation through Patient Monitoring and Product Characterization. Ann. N. Y. Acad. Sci. 2016, 1370, 15–23. 10.1111/nyas.12984. [DOI] [PubMed] [Google Scholar]
- Ankam S.; Suryana M.; Chan L. Y.; Moe A. A. K.; Teo B. K. K.; Law J. B. K.; Sheetz M. P.; Low H. Y.; Yim E. K. F. Substrate Topography and Size Determine the Fate of Human Embryonic Stem Cells to Neuronal or Glial Lineage. Acta Biomater. 2013, 9, 4535–4545. 10.1016/j.actbio.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Moe A. A. K.; Suryana M.; Marcy G.; Lim S. K.; Ankam S.; Goh J. Z. W.; Jin J.; Teo B. K. K.; Law J. B. K.; Low H. Y.; et al. Microarray with Micro- and Nano-Topographies Enables Identification of the Optimal Topography for Directing the Differentiation of Primary Murine Neural Progenitor Cells. Small 2012, 8, 3050–3061. 10.1002/smll.201200490. [DOI] [PubMed] [Google Scholar]
- Kukumberg M.; Yao Y.; Goh S. H.; Neo D. J. H.; Yao J. Y.; Yim E. K. F. Evaluation of the Topographical Influence on the Cellular Behavior of Human Umbilical Vein Endothelial Cells. Adv. Biosyst. 2018, 2, 1700217. 10.1002/adbi.201700217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S.; Lin S.; Cheung C.; Yim E. K. F.; Toh Y. C. Topography Elicits Distinct Phenotypes and Functions in Human Primary and Stem Cell Derived Endothelial Cells. Biomaterials 2020, 234, 119747. 10.1016/j.biomaterials.2019.119747. [DOI] [PubMed] [Google Scholar]
- Rostam H. M.; Singh S.; Salazar F.; Magennis P.; Hook A.; Singh T.; Vrana N. E.; Alexander M. R.; Ghaemmaghami A. M. The Impact of Surface Chemistry Modification on Macrophage Polarisation. Immunobiology 2016, 221, 1237–1246. 10.1016/j.imbio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Scadden D. T. The Stem-Cell Niche as an Entity of Action. Nature 2006, 441, 1075–1079. 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Usui K.; Kikuchi T.; Tomizaki K. Y.; Kakiyama T.; Mihara H. A Novel Array Format for Monitoring Cellular Uptake Using a Photo-Cleavable Linker for Peptide Release. Chem. Commun. 2013, 49, 6394–6396. 10.1039/c3cc41632a. [DOI] [PubMed] [Google Scholar]
- Ugi I. The A□Addition of Immonium Ions and Anions to Isonitriles Accompanied by Secondary Reactions. Angew. Chem., Int. Ed. Engl. 1962, 1, 8–21. 10.1002/anie.196200081. [DOI] [Google Scholar]
- Majkut S.; Idema T.; Swift J.; Krieger C.; Liu A.; Discher D. E. Heart-Specific Stiffening in Early Embryos Parallels Matrix and Myosin Expression to Optimize Beating. Curr. Biol. 2013, 23, 2434–2439. 10.1016/j.cub.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely B. L.; Dumelin C. E.; Trappmann B.; McGregor L. M.; Choi C. K.; Anthony P. C.; Duesterberg V. K.; Baker B. M.; Block S. M.; Liu D. R.; et al. A DNA-Based Molecular Probe for Optically Reporting Cellular Traction Forces. Nat. Methods 2014, 11, 1229–1232. 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C. F.; Reichardt I.; Berger C.; Lendl T.; Knoblich J. A. Long-Term Live Cell Imaging and Automated 4D Analysis of Drosophila Neuroblast Lineages. PLoS One 2013, 8, e79588 10.1371/journal.pone.0079588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal P.; Maurer P.; van Blitterswijk C.; Truckenmüller R.; Giselbrecht S. A New Microengineered Platform for 4D Tracking of Single Cells in a Stem-Cell-Based In Vitro Morphogenesis Model. Adv. Mater. 2020, 32, 1907966. 10.1002/adma.201907966. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Tirrell D. A. A Photoreversible Protein-Patterning Approach for Guiding Stem Cell Fate in Three-Dimensional Gels. Nat. Mater. 2015, 14, 523–531. 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- Chen L.; Yan C.; Zheng Z. Functional Polymer Surfaces for Controlling Cell Behaviors. Mater. Today 2018, 21, 38–59. 10.1016/j.mattod.2017.07.002. [DOI] [Google Scholar]
- Gjorevski N.; Sachs N.; Manfrin A.; Giger S.; Bragina M. E.; Ordóñez-Morán P.; Clevers H.; Lutolf M. P. Designer Matrices for Intestinal Stem Cell and Organoid Culture. Nature 2016, 539, 560–564. 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- Caiazzo M.; Okawa Y.; Ranga A.; Piersigilli A.; Tabata Y.; Lutolf M. P. Defined Three-Dimensional Microenvironments Boost Induction of Pluripotency. Nat. Mater. 2016, 15, 344–352. 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- Dolatshahi-Pirouz A.; Nikkhah M.; Gaharwar A. K.; Hashmi B.; Guermani E.; Aliabadi H.; Camci-Unal G.; Ferrante T.; Foss M.; Ingber D. E.; et al. A Combinatorial Cell-Laden Gel Microarray for Inducing Osteogenic Differentiation of Human Mesenchymal Stem Cells. Sci. Rep. 2015, 4, 3896. 10.1038/srep03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury Y.; Scadden D. T. Mesenchymal Cell Contributions to the Stem Cell Niche. Cell Stem Cell 2015, 16, 239–253. 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Chen G.; Gulbranson D. R.; Hou Z.; Bolin J. M.; Ruotti V.; Probasco M. D.; Smuga-Otto K.; Howden S. E.; Diol N. R.; Propson N. E.; et al. Chemically Defined Conditions for Human IPSC Derivation and Culture. Nat. Methods 2011, 8, 424–429. 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G.; Manfrin A.; Lutolf M. P. Progress and Potential in Organoid Research. Nat. Rev. Genet. 2018, 19, 671–687. 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- Chong J. J. H.; Yang X.; Don C. W.; Minami E.; Liu Y. W.; Weyers J. J.; Mahoney W. M.; Van Biber B.; Cook S. M.; Palpant N. J.; et al. Human Embryonic-Stem-Cell-Derived Cardiomyocytes Regenerate Non-Human Primate Hearts. Nature 2014, 510, 273–277. 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach V.; Passier R.. Native Cardiac Environment and Its Impact on Engineering Cardiac Tissue. Biomaterials Science; Royal Society of Chemistry, 2019; pp 3566–3580. [DOI] [PubMed] [Google Scholar]
- Alakpa E. V.; Burgess K. E. V.; Chung P.; Riehle M. O.; Gadegaard N.; Dalby M. J.; Cusack M. Nacre Topography Produces Higher Crystallinity in Bone than Chemically Induced Osteogenesis. ACS Nano 2017, 11, 6717–6727. 10.1021/acsnano.7b01044. [DOI] [PubMed] [Google Scholar]
- Wang X.; Maruotti J.; Majumdar S.; Roman J.; Mao H. Q.; Zack D. J.; Elisseeff J. H. Collagen Vitrigels with Low-Fibril Density Enhance Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cell Maturation. J. Tissue Eng. Regener. Med. 2018, 12, 821–829. 10.1002/term.2598. [DOI] [PubMed] [Google Scholar]
- Lanniel M.; Huq E.; Allen S.; Buttery L.; Williams P. M.; Alexander M. R. Substrate Induced Differentiation of Human Mesenchymal Stem Cells on Hydrogels with Modified Surface Chemistry and Controlled Modulus. Soft Matter 2011, 7, 6501–6514. 10.1039/c1sm05167a. [DOI] [Google Scholar]
- Benoit D. S. W.; Schwartz M. P.; Durney A. R.; Anseth K. S. Small Functional Groups for Controlled Differentiation of Hydrogel-Encapsulated Human Mesenchymal Stem Cells. Nat. Mater. 2008, 7, 816–823. 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes C.; Chen J. H.; Sun Y.; Simmons C. A. Microfabricated Arrays for High-Throughput Screening of Cellular Response to Cyclic Substrate Deformation. Lab Chip 2010, 10, 227–234. 10.1039/B914460A. [DOI] [PubMed] [Google Scholar]
- Celiz A. D.; Smith J. G. W.; Langer R.; Anderson D. G.; Winkler D. A.; Barrett D. A.; Davies M. C.; Young L. E.; Denning C.; Alexander M. R. Materials for Stem Cell Factories of the Future. Nat. Mater. 2014, 13, 570–579. 10.1038/nmat3972. [DOI] [PubMed] [Google Scholar]
- Orkin R. W.; Gehron P.; McGoodwin E. B.; Martin G. R.; Valentine T.; Swarm R. A Murine Tumor Producing a Matrix of Basement Membrane. J. Exp. Med. 1977, 145, 204–220. 10.1084/jem.145.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delalat B.; Rojas-Canales D.; Rasi Ghaemi S.; Waibel M.; Harding F.; Penko D.; Drogemuller C.; Loudovaris T.; Coates P.; Voelcker N. A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations. Microarrays 2016, 5, 21. 10.3390/microarrays5030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L.; Coller J.; Natu V.; Hastie T. J.; Huang N. F. Combinatorial Extracellular Matrix Microenvironments Promote Survival and Phenotype of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells in Hypoxia. Acta Biomater. 2016, 44, 188–199. 10.1016/j.actbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassey M. J.; Figueredo G. P.; Scurr D. J.; Vasilevich A. S.; Vermeulen S.; Carlier A.; Luckett J.; Beijer N. R. M.; Williams P.; Winkler D. A.; et al. Immune Modulation by Design: Using Topography to Control Human Monocyte Attachment and Macrophage Differentiation. Adv. Sci. 2020, 7, 1903392. 10.1002/advs.201903392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostam H. M.; Fisher L. E.; Hook A. L.; Burroughs L.; Luckett J. C.; Figueredo G. P.; Mbadugha C.; Teo A. C. K.; Latif A.; Kämmerling L.; et al. Immune-Instructive Polymers Control Macrophage Phenotype and Modulate the Foreign Body Response In Vivo. Matter 2020, 2, 1564–1581. 10.1016/j.matt.2020.03.018. [DOI] [Google Scholar]
- Acharya A. P.; Carstens M. R.; Lewis J. S.; Dolgova N.; Xia C. Q.; Clare-Salzler M. J.; Keselowsky B. G. A Cell-Based Microarray to Investigate Combinatorial Effects of Microparticle-Encapsulated Adjuvants on Dendritic Cell Activation. J. Mater. Chem. B 2016, 4, 1672–1685. 10.1039/C5TB01754H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. B.; Ribeiro M. P.; Miguel S. P.; Neto A. I.; Coutinho P.; Correia I. J.; Mano J. F. In Vivo High-Content Evaluation of Three-Dimensional Scaffolds Biocompatibility. Tissue Eng., Part C 2014, 20, 851–864. 10.1089/ten.tec.2013.0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Galitó S.; Tamm C.; Schuster J.; Sobol M.; Forsberg L.; Merry C. L. R.; Annerén C. Human Serum-Derived Protein Removes the Need for Coating in Defined Human Pluripotent Stem Cell Culture. Nat. Commun. 2016, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman E. G.; Pierschbacher M. D.; Suzuki S.; Ruoslahti E. Vitronectin-A Major Cell Attachment-Promoting Protein in Fetal Bovine Serum. Exp. Cell Res. 1985, 160, 245–258. 10.1016/0014-4827(85)90173-9. [DOI] [PubMed] [Google Scholar]
- Dakhore S.; Nayer B.; Hasegawa K. Human Pluripotent Stem Cell Culture: Current Status, Challenges, and Advancement. Stem Cells Int. 2018, 7396905. 10.1155/2018/7396905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K.; Ueno M.; Kamiya D.; Nishiyama A.; Matsumura M.; Wataya T.; Takahashi J. B.; Nishikawa S.; Nishikawa S. I.; Muguruma K.; et al. A ROCK Inhibitor Permits Survival of Dissociated Human Embryonic Stem Cells. Nat. Biotechnol. 2007, 25, 681–686. 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Ireland R. G.; Kibschull M.; Audet J.; Ezzo M.; Hinz B.; Lye S. J.; Simmons C. A. Combinatorial Extracellular Matrix Microarray Identifies Novel Bioengineered Substrates for Xeno-Free Culture of Human Pluripotent Stem Cells. Biomaterials 2020, 248, 120017. 10.1016/j.biomaterials.2020.120017. [DOI] [PubMed] [Google Scholar]
- Soliman C.; Yuriev E.; Ramsland P. A. Antibody Recognition of Aberrant Glycosylation on the Surface of Cancer Cells. Curr. Opin. Struct. Biol. 2017, 44, 1–8. 10.1016/j.sbi.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Magennis E. P.; Hook A. L.; Davies M. C.; Alexander C.; Williams P.; Alexander M. R. Engineering Serendipity: High-Throughput Discovery of Materials That Resist Bacterial Attachment. Acta Biomater. 2016, 34, 84–92. 10.1016/j.actbio.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. P.; Kim Y.-G.; Choi C.-H.; Kim H.-E.; Lee S.-H.; Chang W.-S.; Lee C.-S. In Situ Monitoring of Antibiotic Susceptibility of Bacterial Biofilms in a Microfluidic Device. Lab Chip 2010, 10, 3296. 10.1039/c0lc00154f. [DOI] [PubMed] [Google Scholar]
- Epa V. C.; Hook A. L.; Chang C.; Yang J.; Langer R.; Anderson D. G.; Williams P.; Davies M. C.; Alexander M. R.; Winkler D. A. Modelling and Prediction of Bacterial Attachment to Polymers. Adv. Funct. Mater. 2014, 24, 2085–2093. 10.1002/adfm.201302877. [DOI] [Google Scholar]
- Jeffery N.; Kalenderski K.; Dubern J.; Lomiteng A.; Dragova M.; Frost A.; Macrae B.; Mundy A.; Alexander M.; Williams P.; et al. A New Bacterial Resistant Polymer Catheter Coating to Reduce Catheter Associated Urinary Tract Infection (CAUTI): A First-in-Man Pilot Study. Eur. Urol., Suppl. 2019, 18, e377 10.1016/S1569-9056(19)30282-9. [DOI] [Google Scholar]
- Dundas A. A.; Sanni O.; Dubern J. F.; Dimitrakis G.; Hook A. L.; Irvine D. J.; Williams P.; Alexander M. R. Validating a Predictive Structure-Property Relationship by Discovery of Novel Polymers Which Reduce Bacterial Biofilm Formation. Adv. Mater. 2019, 31, 1903513. 10.1002/adma.201903513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanni O.; Chang C. Y.; Anderson D. G.; Langer R.; Davies M. C.; Williams P. M. P.; Williams P. M. P.; Alexander M. R.; Hook A. L. Bacterial Attachment to Polymeric Materials Correlates with Molecular Flexibility and Hydrophilicity. Adv. Healthcare Mater. 2015, 4, 695–701. 10.1002/adhm.201400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegas A. J.; Veiseh O.; Doloff J. C.; Ma M.; Tam H. H.; Bratlie K.; Li J.; Bader A. R.; Langan E.; Olejnik K.; et al. Combinatorial Hydrogel Library Enables Identification of Materials That Mitigate the Foreign Body Response in Primates. Nat. Biotechnol. 2016, 34, 345–352. 10.1038/nbt.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochenek M. A.; Veiseh O.; Vegas A. J.; McGarrigle J. J.; Qi M.; Marchese E.; Omami M.; Doloff J. C.; Mendoza-Elias J.; Nourmohammadzadeh M.; et al. Alginate Encapsulation as Long-Term Immune Protection of Allogeneic Pancreatic Islet Cells Transplanted into the Omental Bursa of Macaques. Nat. Biomed. Eng. 2018, 2, 810–821. 10.1038/s41551-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijten J.; Khademhosseini A. From Nano to Macro: Multiscale Materials for Improved Stem Cell Culturing and Analysis. Cell Stem Cell 2016, 18, 20–24. 10.1016/j.stem.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Vasilevich A.; de Boer J. Robot-Scientists Will Lead Tomorrow’s Biomaterials Discovery. Curr. Opin. Biomed. Eng. 2018, 6, 74–80. 10.1016/j.cobme.2018.03.005. [DOI] [Google Scholar]
- Kitson P. J.; Marie G.; Francoia J.-P.; Zalesskiy S. S.; Sigerson R. C.; Mathieson J. S.; Cronin L. Digitization of Multistep Organic Synthesis in Reactionware for On-Demand Pharmaceuticals. Science 2018, 359, 314–319. 10.1126/science.aao3466. [DOI] [PubMed] [Google Scholar]
- Granda J. M.; Donina L.; Dragone V.; Long D.-L.; Cronin L. Controlling an Organic Synthesis Robot with Machine Learning to Search for New Reactivity. Nature 2018, 559, 377–381. 10.1038/s41586-018-0307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland E., Williams K., Sparkes A., King R. D., Oliver S.. Yeast-Based Drug Discovery and Development by the Robot Scientist “Eve”; Wiley: Hoboken, NJ, 2015; Vol. 32. [Google Scholar]
- Whitesides G. M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Sackmann E. K.; Fulton A. L.; Beebe D. J. The Present and Future Role of Microfluidics in Biomedical Research. Nature 2014, 507, 181–189. 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Whitesides G. M. SOFT LITHOGRAPHY. Annu. Rev. Mater. Sci. 1998, 28, 153–184. 10.1146/annurev.matsci.28.1.153. [DOI] [Google Scholar]
- Mao H.; Cremer P. S.; Manson M. D. A Sensitive, Versatile Microfluidic Assay for Bacterial Chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 5449–5454. 10.1073/pnas.0931258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C.; Allen S. G.; Ingram P. N.; Buckanovich R.; Merajver S. D.; Yoon E. Single-Cell Migration Chip for Chemotaxis-Based Microfluidic Selection of Heterogeneous Cell Populations. Sci. Rep. 2015, 5, 9980. 10.1038/srep09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Kumar-Kanojia A.; Hombach-Klonisch S.; Klonisch T.; Lin F. A Radial Microfluidic Platform for Higher Throughput Chemotaxis Studies with Individual Gradient Control. Lab Chip 2018, 18, 3855–3864. 10.1039/C8LC00981C. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Shojaei-Baghini E.; Azad A.; Wang C.; Sun Y. High-Throughput Biophysical Measurement of Human Red Blood Cells. Lab Chip 2012, 12, 2560–2567. 10.1039/c2lc21210b. [DOI] [PubMed] [Google Scholar]
- Huang Y. L.; Segall J. E.; Wu M. Microfluidic Modeling of the Biophysical Microenvironment in Tumor Cell Invasion. Lab Chip 2017, 17, 3221–3233. 10.1039/C7LC00623C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartalov E. P.; Quake S. R. Microfluidic Device Reads up to Four Consecutive Base Pairs in DNA Sequencing-by-Synthesis. Nucleic Acids Res. 2004, 32, 2873–2879. 10.1093/nar/gkh613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J. S.; Anderson W. F.; Quake S. R. Parallel Picoliter RT-PCR Assays Using Microfluidics. Anal. Chem. 2006, 78, 956–958. 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Rho H. S.; Stevens M.; Tibbe A. G. J.; Gardeniers H.; Terstappen L. W. M. M. Microfluidic Device for DNA Amplification of Single Cancer Cells Isolated from Whole Blood by Self-Seeding Microwells. Lab Chip 2015, 15, 4331–4337. 10.1039/C5LC00816F. [DOI] [PubMed] [Google Scholar]
- Hung P. J.; Lee P. J.; Sabounchi P.; Lin R.; Lee L. P. Continuous Perfusion Microfluidic Cell Culture Array for High-Throughput Cell-Based Assays. Biotechnol. Bioeng. 2005, 89, 1–8. 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- Lee P. J.; Hung P. J.; Rao V. M.; Lee L. P. Nanoliter Scale Microbioreactor Array for Quantitative Cell Biology. Biotechnol. Bioeng. 2006, 94, 5–14. 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- Gómez-Sjöberg R.; Leyrat A. A.; Pirone D. M.; Chen C. S.; Quake S. R. Versatile, Fully Automated, Microfluidic Cell Culture System. Anal. Chem. 2007, 79, 8557–8563. 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- Jeon N. L.; Dertinger S. K. W.; Chiu D. T.; Choi I. S.; Stroock A. D.; Whitesides G. M. Generation of Solution and Surface Gradients Using Microfluidic Systems. Langmuir 2000, 16, 8311–8316. 10.1021/la000600b. [DOI] [Google Scholar]
- Dertinger S. K. W.; Chiu D. T.; Jeon N. L.; Whitesides G. M. Generation of Gradients Having Complex Shapes Using Microfluidic Networks. Anal. Chem. 2001, 73, 1240–1246. 10.1021/ac001132d. [DOI] [Google Scholar]
- Kim J.; Taylor D.; Agrawal N.; Wang H.; Kim H.; Han A.; Rege K.; Jayaraman A. A Programmable Microfluidic Cell Array for Combinatorial Drug Screening. Lab Chip 2012, 12, 1813–1822. 10.1039/c2lc21202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jeon N.; Baskaran H.; Dertinger S. K. W.; Whitesides G. M.; Van De Water L.; Toner M. Neutrophil Chemotaxis in Linear and Complex Gradients of Interleukin-8 Formed in a Microfabricated Device. Nat. Biotechnol. 2002, 20, 826–830. 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- Saadi W.; Wang S. J.; Lin F.; Jeon N. L. A Parallel-Gradient Microfluidic Chamber for Quantitative Analysis of Breast Cancer Cell Chemotaxis. Biomed. Microdevices 2006, 8, 109–118. 10.1007/s10544-006-7706-6. [DOI] [PubMed] [Google Scholar]
- Dertinger S. K. W.; Jiang X.; Li Z.; Murthy V. N.; Whitesides G. M. Gradients of Substrate-Bound Laminin Orient Axonal Specification of Neurons. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 12542–12547. 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick J. A.; Khademhosseini A.; Langer R. Fabrication of Gradient Hydrogels Using a Microfluidics/Photopolymerization Process. Langmuir 2004, 20, 5153–5156. 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- Fan Y.; Nguyen D. T.; Akay Y.; Xu F.; Akay M. Engineering a Brain Cancer Chip for High-Throughput Drug Screening. Sci. Rep. 2016, 6, 25062. 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W.; Park S. A Microfluidic Spheroid Culture Device with a Concentration Gradient Generator for High-Throughput Screening of Drug Efficacy. Molecules 2018, 23, 3355. 10.3390/molecules23123355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. P.; Unger M. A.; Quake S. R. A Microfabricated Rotary Pump. Biomed. Microdevices 2001, 3, 323–330. 10.1023/A:1012412916446. [DOI] [Google Scholar]
- Yang Y.; Le Gac S.; Terstappen L. W. M. M.; Rho H. S. Parallel Probing of Drug Uptake of Single Cancer Cells on a Microfluidic Device. Electrophoresis 2018, 39, 548–556. 10.1002/elps.201700351. [DOI] [PubMed] [Google Scholar]
- Frank T.; Tay S. Flow-Switching Allows Independently Programmable, Extremely Stable, High-Throughput Diffusion-Based Gradients. Lab Chip 2013, 13, 1273–1281. 10.1039/c3lc41076e. [DOI] [PubMed] [Google Scholar]
- Ymeti A.; Kanger J. S.; Greve J.; Besselink G. A. J.; Lambeck P. V.; Wijn R.; Heideman R. G. Integration of Microfluidics with a Four-Channel Integrated Optical Young Interferometer Immunosensor. Biosens. Bioelectron. 2005, 20, 1417–1421. 10.1016/j.bios.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Ryu S.; Yoo I.; Song S.; Yoon B.; Kim J. M. A Thermoresponsive Fluorogenic Conjugated Polymer for a Temperature Sensor in Microfluidic Devices. J. Am. Chem. Soc. 2009, 131, 3800–3801. 10.1021/ja808077d. [DOI] [PubMed] [Google Scholar]
- Sim W. Y.; Park S. W.; Park S. H.; Min B. H.; Park S. R.; Yang S. S. A Pneumatic Micro Cell Chip for the Differentiation of Human Mesenchymal Stem Cells under Mechanical Stimulation. Lab Chip 2007, 7, 1775–1782. 10.1039/b712361m. [DOI] [PubMed] [Google Scholar]
- Park S. H.; Sim W. Y.; Min B. H.; Yang S. S.; Khademhosseini A.; Kaplan D. L. Chip-Based Comparison of the Osteogenesis of Human Bone Marrow- and Adipose Tissue-Derived Mesenchymal Stem Cells under Mechanical Stimulation. PLoS One 2012, 7, e46689 10.1371/journal.pone.0046689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho K. K. Y.; Wang Y. L.; Wu J.; Liu A. P. Advanced Microfluidic Device Designed for Cyclic Compression of Single Adherent Cells. Front. Bioeng. Biotechnol. 2018, 6, 148. 10.3389/fbioe.2018.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamotani Y.; Bersano-Begey T.; Kato N.; Tung Y. C.; Huh D.; Song J. W.; Takayama S. Individually Programmable Cell Stretching Microwell Arrays Actuated by a Braille Display. Biomaterials 2008, 29, 2646–2655. 10.1016/j.biomaterials.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville N. J.; Zamankhan P.; Tung Y. C.; Li R.; Vaughan B. L.; Tai C. F.; White J.; Christensen P. J.; Grotberg J. B.; Takayama S. Combination of Fluid and Solid Mechanical Stresses Contribute to Cell Death and Detachment in a Microfluidic Alveolar Model. Lab Chip 2011, 11, 609–619. 10.1039/C0LC00251H. [DOI] [PubMed] [Google Scholar]
- Mann J. M.; Lam R. H. W.; Weng S.; Sun Y.; Fu J. A Silicone-Based Stretchable Micropost Array Membrane for Monitoring Live-Cell Subcellular Cytoskeletal Response. Lab Chip 2012, 12, 731–740. 10.1039/C2LC20896B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.; Shunori A.; Morimoto K.; Hashida M.; Konishi S. Development of a Biochip with Serially Connected Pneumatic Balloons for Cell-Stretching Culture. Sens. Actuators, B 2011, 156, 486–493. 10.1016/j.snb.2011.04.048. [DOI] [Google Scholar]
- Tremblay D.; Chagnon-Lessard S.; Mirzaei M.; Pelling A. E.; Godin M. A Microscale Anisotropic Biaxial Cell Stretching Device for Applications in Mechanobiology. Biotechnol. Lett. 2014, 36, 657–665. 10.1007/s10529-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y.; Hameed F. M.; Yang B.; Lee K.; Pan C. Q.; Park S.; Sheetz M. Cyclic Stretching of Soft Substrates Induces Spreading and Growth. Nat. Commun. 2015, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R.; Le Gac S.; Verdonschot N.; Van Den Berg A.; Koopman B.; Rouwkema J. Endothelial Cell Alignment as a Result of Anisotropic Strain and Flow Induced Shear Stress Combinations. Sci. Rep. 2016, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. Y.; Kim J.; Song H. J.; Kim K.; Choi K. C.; Park S.; Sung G. Y. Development of Wrinkled Skin-on-a-Chip (WSOC) by Cyclic Uniaxial Stretching. J. Ind. Eng. Chem. 2018, 68, 238–245. 10.1016/j.jiec.2018.07.050. [DOI] [Google Scholar]
- Toh Y. C.; Lim T. C.; Tai D.; Xiao G.; Van Noort D.; Yu H. A Microfluidic 3D Hepatocyte Chip for Drug Toxicity Testing. Lab Chip 2009, 9, 2026–2035. 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- Shin Y.; Han S.; Jeon J. S.; Yamamoto K.; Zervantonakis I. K.; Sudo R.; Kamm R. D.; Chung S. Microfluidic Assay for Simultaneous Culture of Multiple Cell Types on Surfaces or within Hydrogels. Nat. Protoc. 2012, 7, 1247–1259. 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecka E.; Zukowski K.; Jastrzebska E.; Chudy M.; Brzozka Z. Microsystem with Micropillar Array for Three- (Gel-Embaded) and Two-Dimensional Cardiac Cell Culture. Sens. Actuators, B 2018, 254, 973–983. 10.1016/j.snb.2017.07.186. [DOI] [Google Scholar]
- Dinh N.-D.; Chiang Y.-Y.; Hardelauf H.; Baumann J.; Jackson E.; Waide S.; Sisnaiske J.; Frimat J.-P.; Thriel C. v.; Janasek D.; Peyrin J.-M.; West J. Microfluidic Construction of Minimalistic Neuronal Co-Cultures. Lab Chip 2013, 13, 1402–1412. 10.1039/c3lc41224e. [DOI] [PubMed] [Google Scholar]
- Carrion B.; Huang C. P.; Ghajar C. M.; Kachgal S.; Kniazeva E.; Jeon N. L.; Putnam A. J. Recreating the Perivascular Niche Ex Vivo Using a Microfluidic Approach. Biotechnol. Bioeng. 2010, 107, 1020–1028. 10.1002/bit.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang S.; Na S.; Jang J. M.; Kim J.; Jeon N. L. Engineering-Aligned 3D Neural Circuit in Microfluidic Device. Adv. Healthcare Mater. 2016, 5, 159–166. 10.1002/adhm.201500397. [DOI] [PubMed] [Google Scholar]
- Han S.; Kim J.; Li R.; Ma A.; Kwan V.; Luong K.; Sohn L. L. Hydrophobic Patterning-Based 3D Microfluidic Cell Culture Assay. Adv. Healthcare Mater. 2018, 7, 1800122. 10.1002/adhm.201800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persidis A. High-Throughput Screening. Bio/Technology 1998, 16, 488. [DOI] [PubMed] [Google Scholar]
- Zhang J. H.; Chung T. D. Y.; Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screening 1999, 4, 67–73. 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Liu B.; Li S.; Hu J. Technological Advances in High-Throughput Screening. Am. J. PharmacoGenomics 2004, 4, 263–276. 10.2165/00129785-200404040-00006. [DOI] [PubMed] [Google Scholar]
- Broach J. R.; Thorner J. High-Throughput Screening for Drug Discovery. Nature 1996, 384, 14–16. 10.1038/384014a0. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P.; Pope A. J. High-Throughput Screening: New Technology for the 21st Century. Curr. Opin. Chem. Biol. 2000, 4, 445–451. 10.1016/S1367-5931(00)00110-1. [DOI] [PubMed] [Google Scholar]
- Lee P.; Gaige T.; Hung P. Microfluidic Systems for Live Cell Imaging. Methods Cell Biol. 2011, 102, 77. 10.1016/B978-0-12-374912-3.00004-3. [DOI] [PubMed] [Google Scholar]
- Trietsch S. J.; Israëls G. D.; Joore J.; Hankemeier T.; Vulto P. Microfluidic Titer Plate for Stratified 3D Cell Culture. Lab Chip 2013, 13, 3548–3554. 10.1039/c3lc50210d. [DOI] [PubMed] [Google Scholar]
- Lee P. J.; Ghorashian N.; Gaige T. A.; Hung P. J. Microfluidic System for Automated Cell-Based Assays. JALA 2007, 12, 363–367. 10.1016/j.jala.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C.; Mikheev A. M.; Huynh W.; Monnat R. J.; Rostomily R. C.; Folch A. Parallel Microfluidic Chemosensitivity Testing on Individual Slice Cultures. Lab Chip 2014, 14, 4540–4551. 10.1039/C4LC00642A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Y. C.; Hsiao A. Y.; Allen S. G.; Torisawa Y. S.; Ho M.; Takayama S. High-Throughput 3D Spheroid Culture and Drug Testing Using a 384 Hanging Drop Array. Analyst 2011, 136, 473–478. 10.1039/C0AN00609B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandurina J.; McKnight T. E.; Jacobson S. C.; Waters L. C.; Foote R. S.; Ramsey J. M. Integrated System for Rapid PCR-Based DNA Analysis in Microfluidic Devices. Anal. Chem. 2000, 72, 2995–3000. 10.1021/ac991471a. [DOI] [PubMed] [Google Scholar]
- Xiang Q.; Xu B.; Fu R.; Li D. Real Time PCR on Disposable PDMS Chip with a Miniaturized Thermal Cycler. Biomed. Microdevices 2005, 7, 273–279. 10.1007/s10544-005-6069-8. [DOI] [PubMed] [Google Scholar]
- Roper M. G.; Easley C. J.; Legendre L. A.; Humphrey J. A. C.; Landers J. P. Infrared Temperature Control System for a Completely Noncontact Polymerase Chain Reaction in Microfluidic Chips. Anal. Chem. 2007, 79, 1294–1300. 10.1021/ac0613277. [DOI] [PubMed] [Google Scholar]
- Heo Y. S.; Cabrera L. M.; Song J. W.; Futai N.; Tung Y.-C.; Smith G. D.; Takayama S. Characterization and Resolution of Evaporation-Mediated Osmolality Shifts That Constrain Microfluidic Cell Culture in Poly(Dimethylsiloxane) Devices. Anal. Chem. 2007, 79, 1126–1134. 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Cao W.; Wen W.; Chang D. C.; Sheng P. Polydimethylsiloxane Microfluidic Chip with Integrated Microheater and Thermal Sensor. Biomicrofluidics 2009, 3, 012005. 10.1063/1.3058587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. C.; Raghavan S. R.; Forry S. P. Regulating Oxygen Levels in a Microfluidic Device. Anal. Chem. 2011, 83, 8821–8824. 10.1021/ac202300g. [DOI] [PubMed] [Google Scholar]
- Polinkovsky M.; Gutierrez E.; Levchenko A.; Groisman A. Fine Temporal Control of the Medium Gas Content and Acidity and On-Chip Generation of Series of Oxygen Concentrations for Cell Cultures. Lab Chip 2009, 9, 1073–1084. 10.1039/b816191g. [DOI] [PubMed] [Google Scholar]
- Lo J. F.; Sinkala E.; Eddington D. T. Oxygen Gradients for Open Well Cellular Cultures via Microfluidic Substrates. Lab Chip 2010, 10, 2394–2401. 10.1039/c004660d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. A.; King A. D.; Shih H. C.; Peng C. C.; Wu C. Y.; Liao W. H.; Tung Y. C. Generation of Oxygen Gradients in Microfluidic Devices for Cell Culture Using Spatially Confined Chemical Reactions. Lab Chip 2011, 11, 3626–3633. 10.1039/c1lc20325h. [DOI] [PubMed] [Google Scholar]
- Rexius-Hall M. L.; Mauleon G.; Malik A. B.; Rehman J.; Eddington D. T. Microfluidic Platform Generates Oxygen Landscapes for Localized Hypoxic Activation. Lab Chip 2014, 14, 4688–4695. 10.1039/C4LC01168F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C. C.; Liao W. H.; Chen Y. H.; Wu C. Y.; Tung Y. C. A Microfluidic Cell Culture Array with Various Oxygen Tensions. Lab Chip 2013, 13, 3239–3245. 10.1039/c3lc50388g. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Liu Z.; Li L.; Liang Q. Investigation into the Hypoxia-Dependent Cytotoxicity of Anticancer Drugs under Oxygen Gradient in a Microfluidic Device. Microfluid. Nanofluid. 2015, 19, 1271–1279. 10.1007/s10404-015-1637-6. [DOI] [Google Scholar]
- Khan D. H.; Roberts S. A.; Cressman J. R.; Agrawal N. Rapid Generation and Detection of Biomimetic Oxygen Concentration Gradients in Vitro. Sci. Rep. 2017, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas G.; Martins J. S.; Ribeiro J. C.; Wolffenbuttel R. F.; Correia J. H. Biological Microsystem for Measuring Uric Acid in Biological Fluids. Sens. Actuators, A 2004, 110, 33–38. 10.1016/j.sna.2003.10.049. [DOI] [Google Scholar]
- Kim S. B.; Bae H.; Cha J. M.; Moon S. J.; Dokmeci M. R.; Cropek D. M.; Khademhosseini A. A Cell-Based Biosensor for Real-Time Detection of Cardiotoxicity Using Lensfree Imaging. Lab Chip 2011, 11, 1801–1807. 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. B.; Koo K. in; Bae H.; Dokmeci M. R.; Hamilton G. A.; Bahinski A.; Kim S. M.; Ingber D. E.; Khademhosseini A. A Mini-Microscope for in Situ Monitoring of Cells. Lab Chip 2012, 12, 3976–3982. 10.1039/c2lc40345e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. S.; Ribas J.; Nadhman A.; Aleman J.; Selimović Š.; Lesher-Perez S. C.; Wang T.; Manoharan V.; Shin S. R.; Damilano A.; et al. A Cost-Effective Fluorescence Mini-Microscope for Biomedical Applications. Lab Chip 2015, 15, 3661–3669. 10.1039/C5LC00666J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.-T.; Wu Z. Micromixers—a Review. J. Micromech. Microeng. 2005, 15, R1. 10.1088/0960-1317/15/2/R01. [DOI] [Google Scholar]
- Oh K. W.; Ahn C. H. A Review of Microvalves. J. Micromech. Microeng. 2006, 16, R13. 10.1088/0960-1317/16/5/R01. [DOI] [Google Scholar]
- Boyd-Moss M.; Baratchi S.; Di Venere M.; Khoshmanesh K. Self-Contained Microfluidic Systems: A Review. Lab Chip 2016, 16, 3177–3192. 10.1039/C6LC00712K. [DOI] [PubMed] [Google Scholar]
- Lee P. J.; Hung P. J.; Lee L. P. An Artificial Liver Sinusoid with a Microfluidic Endothelial-like Barrier for Primary Hepatocyte Culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- Wang H. Y.; Bao N.; Lu C. A Microfluidic Cell Array with Individually Addressable Culture Chambers. Biosens. Bioelectron. 2008, 24, 613–617. 10.1016/j.bios.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Albrecht D. R.; Underhill G. H.; Resnikoff J.; Mendelson A.; Bhatia S. N.; Shah J. V. Microfluidics-Integrated Time-Lapse Imaging for Analysis of Cellular Dynamics. Integr. Biol. 2010, 2, 278–287. 10.1039/b923699f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimov I. K.; Kijanka G.; Park Y.; Ducrée J.; Kang T.; Lee L. P. Integrated Microfluidic Array Plate (IMAP) for Cellular and Molecular Analysis. Lab Chip 2011, 11, 2701–2710. 10.1039/c1lc20105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelley A. M.; Kirak O.; Suh H.; Jaenisch R.; Voldman J. Microfluidic Control of Cell Pairing and Fusion. Nat. Methods 2009, 6, 147–152. 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel S.; Valero A.; Latt J.; Renaud P.; Lutolf M. Optimization of Microfluidic Single Cell Trapping for Long-Term on-Chip Culture. Lab Chip 2010, 10, 857–863. 10.1039/b918055a. [DOI] [PubMed] [Google Scholar]
- Frimat J. P.; Becker M.; Chiang Y. Y.; Marggraf U.; Janasek D.; Hengstler J. G.; Franzke J.; West J. A Microfluidic Array with Cellular Valving for Single Cell Co-Culture. Lab Chip 2011, 11, 231–237. 10.1039/C0LC00172D. [DOI] [PubMed] [Google Scholar]
- Buican T. N.; Smyth M. J.; Crissman H. A.; Salzman G. C.; Stewart C. C.; Martin J. C. Automated Single-Cell Manipulation and Sorting by Light Trapping. Appl. Opt. 1987, 26, 5311–5316. 10.1364/AO.26.005311. [DOI] [PubMed] [Google Scholar]
- Hagiwara M.; Kawahara T.; Yamanishi Y.; Masuda T.; Feng L.; Arai F. On-Chip Magnetically Actuated Robot with Ultrasonic Vibration for Single Cell Manipulations. Lab Chip 2011, 8, 134–142. 10.1039/c1lc20164f. [DOI] [PubMed] [Google Scholar]
- Ino K.; Okochi M.; Konishi N.; Nakatochi M.; Imai R.; Shikida M.; Ito A.; Honda H. Cell Culture Arrays Using Magnetic Force-Based Cell Patterning for Dynamic Single Cell Analysis. Lab Chip 2008, 8, 134–142. 10.1039/B712330B. [DOI] [PubMed] [Google Scholar]
- Chiou P. Y.; Ohta A. T.; Wu M. C. Massively Parallel Manipulation of Single Cells and Microparticles Using Optical Images. Nature 2005, 436, 370–372. 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Liu K. K. Optical Tweezers for Single Cells. J. R. Soc., Interface 2008, 5, 671–690. 10.1098/rsif.2008.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Sun J.; Wolvetang E.; Cooper-White J. High-Throughput, Deterministic Single Cell Trapping and Long-Term Clonal Cell Culture in Microfluidic Devices. Lab Chip 2015, 15, 1072–1083. 10.1039/C4LC01176G. [DOI] [PubMed] [Google Scholar]
- Lin C. H.; Hsiao Y. H.; Chang H. C.; Yeh C. F.; He C. K.; Salm E. M.; Chen C.; Chiu I. M.; Hsu C. H. A Microfluidic Dual-Well Device for High-Throughput Single-Cell Capture and Culture. Lab Chip 2015, 15, 2928–2938. 10.1039/C5LC00541H. [DOI] [PubMed] [Google Scholar]
- Bithi S. S.; Vanapalli S. A. Microfluidic Cell Isolation Technology for Drug Testing of Single Tumor Cells and Their Clusters. Sci. Rep. 2017, 7, 41707. 10.1038/srep41707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstetter A.; Stellamanns E.; Deshpande S.; Uppaluri S.; Engstler M.; Pfohl T. Microfluidics-Based Single Cell Analysis Reveals Drug-Dependent Motility Changes in Trypanosomes. Lab Chip 2015, 15, 1961–1968. 10.1039/C5LC00124B. [DOI] [PubMed] [Google Scholar]
- Navin N.; Kendall J.; Troge J.; Andrews P.; Rodgers L.; McIndoo J.; Cook K.; Stepansky A.; Levy D.; Esposito D.; et al. Tumour Evolution Inferred by Single-Cell Sequencing. Nature 2011, 472, 90–94. 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. R.; Neff N. F.; Kalisky T.; Dalerba P.; Treutlein B.; Rothenberg M. E.; Mburu F. M.; Mantalas G. L.; Sim S.; Clarke M. F.; et al. Quantitative Assessment of Single-Cell RNA-Sequencing Methods. Nat. Methods 2014, 11, 41–46. 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Swennenhuis J. F.; Rho H. S.; Le Gac S.; Terstappen L. W. M. M. Parallel Single Cancer Cell Whole Genome Amplification Using Button-Valve Assisted Mixing in Nanoliter Chambers. PLoS One 2014, 9, e107958 10.1371/journal.pone.0107958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerling R. J.; Lee Szeto G.; Li J. W.; Genshaft A. S.; Kazer S. W.; Payer K. R.; De Riba Borrajo J.; Blainey P. C.; Irvine D. J.; Shalek A. K.; et al. A Microfluidic Platform Enabling Single-Cell RNA-Seq of Multigenerational Lineages. Nat. Commun. 2016, 7, 1–7. 10.1038/ncomms10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt Shields C. IV; Reyes C. D.; López G. P. Microfluidic Cell Sorting: A Review of the Advances in the Separation of Cells from Debulking to Rare Cell Isolation. Lab Chip 2015, 15, 1230–1249. 10.1039/C4LC01246A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosic S.; Murthy S. K.; Koppes A. N. Microfluidic Sample Preparation for Single Cell Analysis. Anal. Chem. 2016, 88, 354–380. 10.1021/acs.analchem.5b04077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger M. A.; Chou H. P.; Thorsen T.; Scherer A.; Quake S. R. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science 2000, 288, 113–116. 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Kim M. C.; Marquez M.; Thorsen T. High-Density Microfluidic Arrays for Cell Cytotoxicity Analysis. Lab Chip 2007, 7, 740–745. 10.1039/b618734j. [DOI] [PubMed] [Google Scholar]
- Taylor R. J.; Falconnet D.; Niemisto A.; Ramsey S. A.; Prinz S.; Shmulevich I.; Galitski T.; Hansen C. L. Dynamic Analysis of MAPK Signaling Using a High-Throughput Microfluidic Single-Cell Imaging Platform. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 3758–3763. 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconnet D.; Niemistö A.; Taylor R. J.; Ricicova M.; Galitski T.; Shmulevich I.; Hansen C. L. High-Throughput Tracking of Single Yeast Cells in a Microfluidic Imaging Matrix. Lab Chip 2011, 11, 466–473. 10.1039/C0LC00228C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff K.; Maerkl S. J. A High-Throughput Microfluidic Platform for Mammalian Cell Transfection and Culturing. Sci. Rep. 2016, 6, 1–12. 10.1038/srep23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. R.; Wang S.; Irimia D.; Jayaraman A.; Toner M.; Yarmush M. L. A High-Throughput Microfluidic Real-Time Gene Expression Living Cell Array. Lab Chip 2007, 7, 77–85. 10.1039/B612516F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin J.; Quake S. R. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 213–231. 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- Guo M. T.; Rotem A.; Heyman J. A.; Weitz D. A. Droplet Microfluidics for High-Throughput Biological Assays. Lab Chip 2012, 12, 2146–2155. 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- Mashaghi S.; Abbaspourrad A.; Weitz D. A.; van Oijen A. M. Droplet Microfluidics: A Tool for Biology, Chemistry and Nanotechnology. TrAC, Trends Anal. Chem. 2016, 82, 118–125. 10.1016/j.trac.2016.05.019. [DOI] [Google Scholar]
- Casadevall I Solvas X.; Demello A. Droplet Microfluidics: Recent Developments and Future Applications. Chem. Commun. 2011, 47, 1936–1942. 10.1039/C0CC02474K. [DOI] [PubMed] [Google Scholar]
- Edd J. F.; Di Carlo D.; Humphry K. J.; Köster S.; Irimia D.; Weitz D. A.; Toner M. Controlled Encapsulation of Single-Cells into Monodisperse Picolitre Drops. Lab Chip 2008, 8, 1262–1264. 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C. H. J.; Rowat A. C.; Köster S.; Weitz D. A. Dropspots: A Picoliter Array in a Microfluidic Device. Lab Chip 2009, 9, 44–49. 10.1039/B809670H. [DOI] [PubMed] [Google Scholar]
- Trivedi V.; Doshi A.; Kurup G. K.; Ereifej E.; Vandevord P. J.; Basu A. S. A Modular Approach for the Generation, Storage, Mixing, and Detection of Droplet Libraries for High Throughput Screening. Lab Chip 2010, 10, 2433–2442. 10.1039/c004768f. [DOI] [PubMed] [Google Scholar]
- Baraban L.; Bertholle F.; Salverda M. L. M.; Bremond N.; Panizza P.; Baudry J.; De Visser J. A. G. M.; Bibette J. Millifluidic Droplet Analyser for Microbiology. Lab Chip 2011, 11, 4057–4062. 10.1039/c1lc20545e. [DOI] [PubMed] [Google Scholar]
- Cole R. H.; Tang S.-Y.; Siltanen C. A.; Shahi P.; Zhang J. Q.; Poust S.; Gartner Z. J.; Abate A. R. Printed Droplet Microfluidics for on Demand Dispensing of Picoliter Droplets and Cells. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 8728–8733. 10.1073/pnas.1704020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh S. Y.; Lin R.; Hung L. H.; Lee A. P. Droplet Microfluidics. Lab Chip 2008, 8, 198–220. 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Kenis P. J. A.; Ismagilov R. F.; Whitesides G. M. Microfabrication inside Capillaries Using Multiphase Laminar Flow Patterning. Science (Washington, DC, U. S.) 1999, 285, 83–85. 10.1126/science.285.5424.83. [DOI] [PubMed] [Google Scholar]
- Squires T. M.; Quake S. R. Microfluidics: Fluid Physics at the Nanoliter Scale. Rev. Mod. Phys. 2005, 77, 977. 10.1103/RevModPhys.77.977. [DOI] [Google Scholar]
- Stroock A. D.; Dertinger S. K. W.; Ajdari A.; Mezić I.; Stone H. A.; Whitesides G. M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- Sritharan K.; Strobl C. J.; Schneider M. F.; Wixforth A.; Guttenberg Z. Acoustic Mixing at Low Reynold’s Numbers. Appl. Phys. Lett. 2006, 88, 054102. 10.1063/1.2171482. [DOI] [Google Scholar]
- Kamholz A. E.; Weigl B. H.; Finlayson B. A.; Yager P. Quantitative Analysis of Molecular Interaction in a Microfluidic Channel: The T-Sensor. Anal. Chem. 1999, 71, 5340–5347. 10.1021/ac990504j. [DOI] [PubMed] [Google Scholar]
- Ismagilov R. F.; Stroock A. D.; Kenis P. J. A.; Whitesides G.; Stone H. A. Experimental and Theoretical Scaling Laws for Transverse Diffusive Broadening in Two-Phase Laminar Flows in Microchannels. Appl. Phys. Lett. 2000, 76, 2376–2378. 10.1063/1.126351. [DOI] [Google Scholar]
- Lucchetta E. M.; Lee J. H.; Fu L. A.; Patel N. H.; Ismagilov R. F. Dynamics of Drosophila Embryonic Patterning Network Perturbed in Space and Time Using Microfluidics. Nature 2005, 434, 1134–1138. 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somaweera H.; Haputhanthri S. O.; Ibraguimov A.; Pappas D. On-Chip Gradient Generation in 256 Microfluidic Cell Cultures: Simulation and Experimental Validation. Analyst 2015, 140, 5029–5038. 10.1039/C5AN00481K. [DOI] [PubMed] [Google Scholar]
- Boonstra J.; Post J. A. Molecular Events Associated with Reactive Oxygen Species and Cell Cycle Progression in Mammalian Cells. Gene 2004, 337, 1–13. 10.1016/j.gene.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Mohyeldin A.; Garzón-Muvdi T.; Quiñones-Hinojosa A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 2010, 7, 150–161. 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Chang C. W.; Cheng Y. J.; Tu M.; Chen Y. H.; Peng C. C.; Liao W. H.; Tung Y. C. A Polydimethylsiloxane-Polycarbonate Hybrid Microfluidic Device Capable of Generating Perpendicular Chemical and Oxygen Gradients for Cell Culture Studies. Lab Chip 2014, 14, 3762–3772. 10.1039/C4LC00732H. [DOI] [PubMed] [Google Scholar]
- Liu X. J.; Hu S. W.; Xu B. Y.; Zhao G.; Li X.; Xie F. W.; Xu J. J.; Chen H. Y. Microfluidic Liquid-Air Dual-Gradient Chip for Synergic Effect Bio-Evaluation of Air Pollutant. Talanta 2018, 182, 202–209. 10.1016/j.talanta.2018.01.087. [DOI] [PubMed] [Google Scholar]
- Lou X.; Kim G.; Yoon H. K.; Lee Y. E. K.; Kopelman R.; Yoon E. A High-Throughput Photodynamic Therapy Screening Platform with on-Chip Control of Multiple Microenvironmental Factors. Lab Chip 2014, 14, 892–901. 10.1039/c3lc51077h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck W. J.; Li R.; Uzel S. G. M.; Kamm R. D. Microfluidic Platforms for Mechanobiology. Lab Chip 2013, 13, 2252–2267. 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe C. L.; Engler A. J. Mechanical Forces Reshape Differentiation Cues That Guide Cardiomyogenesis. Circ. Res. 2016, 118, 296–310. 10.1161/CIRCRESAHA.115.305139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M.; Panciera T.; Manfrin A.; Giulitti S.; Michielin F.; Elvassore N.; Dupont S.; Piccolo S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 2013, 154, 1047–1059. 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Gray B. L.; Lieu D. K.; Collins S. D.; Smith R. L.; Barakat A. I. Microchannel Platform for the Study of Endothelial Cell Shape and Function. Biomed. Microdevices 2002, 4, 9–16. 10.1023/A:1014211627166. [DOI] [Google Scholar]
- Kim L.; Vahey M. D.; Lee H. Y.; Voldman J. Microfluidic Arrays for Logarithmically Perfused Embryonic Stem Cell Culture. Lab Chip 2006, 6, 394–406. 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- Chau L.; Doran M.; Cooper-White J. A Novel Multishear Microdevice for Studying Cell Mechanics. Lab Chip 2009, 9, 1897–1902. 10.1039/b823180j. [DOI] [PubMed] [Google Scholar]
- Wan J.; Ristenpart W. D.; Stone H. A. Dynamics of Shear-Induced ATP Release from Red Blood Cells. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 16432–16437. 10.1073/pnas.0805779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou S.; Pan L.; van Noort D.; Meng G.; Wu X.; Sun H.; Xu J.; Lee I. A Multishear Microfluidic Device for Quantitative Analysis of Calcium Dynamics in Osteoblasts. Biochem. Biophys. Res. Commun. 2011, 408, 350–355. 10.1016/j.bbrc.2011.04.044. [DOI] [PubMed] [Google Scholar]
- Rossi M.; Lindken R.; Hierck B. P.; Westerweel J. Tapered Microfluidic Chip for the Study of Biochemical and Mechanical Response at Subcellular Level of Endothelial Cells to Shear Flow. Lab Chip 2009, 9, 1403–1411. 10.1039/b822270n. [DOI] [PubMed] [Google Scholar]
- Kim Y. C.; Kang J. H.; Park S. J.; Yoon E. S.; Park J. K. Microfluidic Biomechanical Device for Compressive Cell Stimulation and Lysis. Sens. Actuators, B 2007, 128, 108–116. 10.1016/j.snb.2007.05.050. [DOI] [Google Scholar]
- Hosmane S.; Fournier A.; Wright R.; Rajbhandari L.; Siddique R.; Yang I. H.; Ramesh K. T.; Venkatesan A.; Thakor N. Valve-Based Microfluidic Compression Platform: Single Axon Injury and Regrowth. Lab Chip 2011, 11, 3888–3895. 10.1039/c1lc20549h. [DOI] [PubMed] [Google Scholar]
- Wan C. R.; Chung S.; Kamm R. D. Differentiation of Embryonic Stem Cells into Cardiomyocytes in a Compliant Microfluidic System. Ann. Biomed. Eng. 2011, 39, 1840–1847. 10.1007/s10439-011-0275-8. [DOI] [PubMed] [Google Scholar]
- Petersen O. W.; Ronnov-Jessen L.; Howlett A. R.; Bissell M. J. Interaction with Basement Membrane Serves to Rapidly Distinguish Growth and Differentiation Pattern of Normal and Malignant Human Breast Epithelial Cells. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 9064–9068. 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H.; Murphy C. L.; Murphy C.; Kimura M.; Kawai S.; Polak J. M. Chondrogenic Differentiation of Murine Embryonic Stem Cells: Effects of Culture Conditions and Dexamethasone. J. Cell. Biochem. 2004, 93, 454–462. 10.1002/jcb.20171. [DOI] [PubMed] [Google Scholar]
- Birgersdotter A.; Sandberg R.; Ernberg I. Gene Expression Perturbation in Vitro - A Growing Case for Three-Dimensional (3D) Culture Systems. Semin. Cancer Biol. 2005, 15, 405–412. 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ling Y.; Rubin J.; Deng Y.; Huang C.; Demirci U.; Karp J. M.; Khademhosseini A. A Cell-Laden Microfluidic Hydrogel. Lab Chip 2007, 7, 756–762. 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- Baker B. M.; Trappmann B.; Stapleton S. C.; Toro E.; Chen C. S. Microfluidics Embedded within Extracellular Matrix to Define Vascular Architectures and Pattern Diffusive Gradients. Lab Chip 2013, 13, 3246–3252. 10.1039/c3lc50493j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M. D.; Golden A. P.; Tien J. Molding of Three-Dimensional Microstructures of Gels. J. Am. Chem. Soc. 2003, 125, 12988–12989. 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- Price G. M.; Chu K. K.; Truslow J. G.; Tang-Schomer M. D.; Golden A. P.; Mertz J.; Tien J. Bonding of Macromolecular Hydrogels Using Perturbants. J. Am. Chem. Soc. 2008, 130, 6664–6665. 10.1021/ja711340d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A. P.; Tien J. Fabrication of Microfluidic Hydrogels Using Molded Gelatin as a Sacrificial Element. Lab Chip 2007, 7, 720–725. 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- Nie J.; Gao Q.; Wang Y.; Zeng J.; Zhao H.; Sun Y.; Shen J.; Ramezani H.; Fu Z.; Liu Z.; et al. Vessel-on-a-chip with Hydrogel-based Microfluidics. Small 2018, 14, 1802368. 10.1002/smll.201802368. [DOI] [PubMed] [Google Scholar]
- Tibbe M. P.; Leferink A. M.; van den Berg A.; Eijkel J. C. T.; Segerink L. I. Microfluidic Gel Patterning Method by Use of a Temporary Membrane for Organ-On-Chip Applications. Adv. Mater. Technol. 2018, 3, 1700200. 10.1002/admt.201700200. [DOI] [Google Scholar]
- Lii J.; Hsu W. J.; Parsa H.; Das A.; Rouse R.; Sia S. K. Real-Time Microfluidic System for Studying Mammalian Cells in 3D Microenvironments. Anal. Chem. 2008, 80, 3640–3647. 10.1021/ac8000034. [DOI] [PubMed] [Google Scholar]
- Si G.; Yang W.; Bi S.; Luo C.; Ouyang Q. A Parallel Diffusion-Based Microfluidic Device for Bacterial Chemotaxis Analysis. Lab Chip 2012, 12, 1389–1394. 10.1039/c2lc21219f. [DOI] [PubMed] [Google Scholar]
- Cosson S.; Kobel S. A.; Lutolf M. P. Capturing Complex Protein Gradients on Biomimetic Hydrogels for Cell-Based Assays. Adv. Funct. Mater. 2009, 19, 3411–3419. 10.1002/adfm.200900968. [DOI] [Google Scholar]
- Cuchiara M. P.; Allen A. C. B.; Chen T. M.; Miller J. S.; West J. L. Multilayer Microfluidic PEGDA Hydrogels. Biomaterials 2010, 31, 5491–5497. 10.1016/j.biomaterials.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Ostrovidov S.; Annabi N.; Seidi A.; Ramalingam M.; Dehghani F.; Kaji H.; Khademhosseini A. Controlled Release of Drugs from Gradient Hydrogels for High-Throughput Analysis of Cell-Drug Interactions. Anal. Chem. 2012, 84, 1302–1309. 10.1021/ac202256c. [DOI] [PubMed] [Google Scholar]
- Moreno E. L.; Hachi S.; Hemmer K.; Trietsch S. J.; Baumuratov A. S.; Hankemeier T.; Vulto P.; Schwamborn J. C.; Fleming R. M. T. Differentiation of Neuroepithelial Stem Cells into Functional Dopaminergic Neurons in 3D Microfluidic Cell Culture. Lab Chip 2015, 15, 2419–2428. 10.1039/C5LC00180C. [DOI] [PubMed] [Google Scholar]
- Trietsch S. J.; Naumovska E.; Kurek D.; Setyawati M. C.; Vormann M. K.; Wilschut K. J.; Lanz H. L.; Nicolas A.; Ng C. P.; Joore J.; et al. Membrane-Free Culture and Real-Time Barrier Integrity Assessment of Perfused Intestinal Epithelium Tubes. Nat. Commun. 2017, 8, 1–8. 10.1038/s41467-017-00259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulto P.; Podszun S.; Meyer P.; Hermann C.; Manz A.; Urban G. A. Phaseguides: A Paradigm Shift in Microfluidic Priming and Emptying. Lab Chip 2011, 11, 1596–1602. 10.1039/c0lc00643b. [DOI] [PubMed] [Google Scholar]
- Huh D.; Matthews B. D.; Mammoto A.; Montoya-Zavala M.; Yuan Hsin H.; Ingber D. E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D.; Leslie D. C.; Matthews B. D.; Fraser J. P.; Jurek S.; Hamilton G. A.; Thorneloe K. S.; McAlexander M. A.; Ingber D. E. A Human Disease Model of Drug Toxicity-Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice. Sci. Transl. Med. 2012, 4, 159ra147. 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Li K.; Zhang X.; Liu C.; Guo B.; Wen W.; Gao X. Nanofiber Membrane Supported Lung-on-a-Chip Microdevice for Anti-Cancer Drug Testing. Lab Chip 2018, 18, 486–495. 10.1039/C7LC01224A. [DOI] [PubMed] [Google Scholar]
- Domansky K.; Inman W.; Serdy J.; Dash A.; Lim M. H. M.; Griffith L. G. Perfused Multiwell Plate for 3D Liver Tissue Engineering. Lab Chip 2010, 10, 51–58. 10.1039/B913221J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan D. T. T.; Wang X.; Craver B. M.; Sobrino A.; Zhao D.; Chen J. C.; Lee L. Y. N.; George S. C.; Lee A. P.; Hughes C. C. W. A Vascularized and Perfused Organ-on-a-Chip Platform for Large-Scale Drug Screening Applications. Lab Chip 2017, 17, 511–520. 10.1039/C6LC01422D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T.; Sugiura S.; Shin K.; Onuki-Nagasaki R.; Ishida S.; Kikuchi K.; Kakiki M.; Kanamori T. A Multi-Throughput Multi-Organ-on-a-Chip System on a Plate Formatted Pneumatic Pressure-Driven Medium Circulation Platform. Lab Chip 2018, 18, 115–125. 10.1039/C7LC00952F. [DOI] [PubMed] [Google Scholar]
- Kamei K. I.; Kato Y.; Hirai Y.; Ito S.; Satoh J.; Oka A.; Tsuchiya T.; Chen Y.; Tabata O. Integrated Heart/Cancer on a Chip to Reproduce the Side Effects of Anti-Cancer Drugs: In Vitro. RSC Adv. 2017, 7, 36777–36786. 10.1039/C7RA07716E. [DOI] [Google Scholar]
- Grosberg A.; Alford P. W.; McCain M. L.; Parker K. K. Ensembles of Engineered Cardiac Tissues for Physiological and Pharmacological Study: Heart on a Chip. Lab Chip 2011, 11, 4165–4173. 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. S.; Davoudi F.; Walch P.; Manbachi A.; Luo X.; Dell’Erba V.; Miri A. K.; Albadawi H.; Arneri A.; Li X.; et al. Bioprinted Thrombosis-on-a-Chip. Lab Chip 2016, 16, 4097–4105. 10.1039/C6LC00380J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K. J.; Suh K. Y. A Multi-Layer Microfluidic Device for Efficient Culture and Analysis of Renal Tubular Cells. Lab Chip 2010, 10, 36–42. 10.1039/B907515A. [DOI] [PubMed] [Google Scholar]
- Jang K. J.; Mehr A. P.; Hamilton G. A.; McPartlin L. A.; Chung S.; Suh K. Y.; Ingber D. E. Human Kidney Proximal Tubule-on-a-Chip for Drug Transport and Nephrotoxicity Assessment. Integr. Biol. (United Kingdom) 2013, 5, 1119–1129. 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Huang R.; Jiang B.; Zhao Y.; Zhang W.; Jiang X. An Early-Stage Atherosclerosis Research Model Based on Microfluidics. Small 2016, 12, 2022–2034. 10.1002/smll.201503241. [DOI] [PubMed] [Google Scholar]
- Stroock A. D.; Kermani P.; Choi N. W.; Diaz-Santana A.; Zheng Y.; Fischbach-Teschl C.; Hempstead B.; Chen J.; Totorica S.; Craven M.; et al. In Vitro Microvessels for the Study of Angiogenesis and Thrombosis. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9342–9347. 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.; Kita A.; Leach J.; Rounsevell R.; Huang J. N.; Moake J.; Ware R. E.; Fletcher D. A.; Lam W. A. In Vitro Modeling of the Microvascular Occlusion and Thrombosis That Occur in Hematologic Diseases Using Microfluidic Technology. J. Clin. Invest. 2012, 122, 408–418. 10.1172/JCI58753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Hotaling N. A.; Ku D. N.; Forest C. R. Microfluidic Thrombosis under Multiple Shear Rates and Antiplatelet Therapy Doses. PLoS One 2014, 9, e82493 10.1371/journal.pone.0082493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraz M. A. M. M.; Henning H. H. W.; Costa P. F.; Malda J.; Melchels F. P.; Wubbolts R.; Stout T. A. E.; Vos P. L. A. M.; Gadella B. M. Improved Bovine Embryo Production in an Oviduct-on-a-Chip System: Prevention of Poly-Spermic Fertilization and Parthenogenic Activation. Lab Chip 2017, 17, 905–916. 10.1039/C6LC01566B. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Huh D.; Hamilton G.; Ingber D. E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow. Lab Chip 2012, 12, 2165–2174. 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Pocock K.; Delon L.; Bala V.; Rao S.; Priest C.; Prestidge C.; Thierry B. Intestine-on-A-Chip Microfluidic Model for Efficient in Vitro Screening of Oral Chemotherapeutic Uptake. ACS Biomater. Sci. Eng. 2017, 3, 951–959. 10.1021/acsbiomaterials.7b00023. [DOI] [PubMed] [Google Scholar]
- Adriani G.; Ma D.; Pavesi A.; Kamm R. D.; Goh E. L. K. A 3D Neurovascular Microfluidic Model Consisting of Neurons, Astrocytes and Cerebral Endothelial Cells as a Blood-Brain Barrier. Lab Chip 2017, 17, 448–459. 10.1039/C6LC00638H. [DOI] [PubMed] [Google Scholar]
- Bang S.; Lee S. R.; Ko J.; Son K.; Tahk D.; Ahn J.; Im C.; Jeon N. L. A Low Permeability Microfluidic Blood-Brain Barrier Platform with Direct Contact between Perfusable Vascular Network and Astrocytes. Sci. Rep. 2017, 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. M.; Lee J.; Kim H.; Jeon N. L.; Jung W. One-Photon and Two-Photon Stimulation of Neurons in a Microfluidic Culture System. Lab Chip 2016, 16, 1684–1690. 10.1039/C6LC00065G. [DOI] [PubMed] [Google Scholar]
- Kilic O.; Pamies D.; Lavell E.; Schiapparelli P.; Feng Y.; Hartung T.; Bal-Price A.; Hogberg H. T.; Quinones-Hinojosa A.; Guerrero-Cazares H.; et al. Brain-on-a-Chip Model Enables Analysis of Human Neuronal Differentiation and Chemotaxis. Lab Chip 2016, 16, 4152–4162. 10.1039/C6LC00946H. [DOI] [PubMed] [Google Scholar]
- Ferraz M. A. M. M.; Rho H. S.; Hemerich D.; Henning H. H. W.; van Tol H. T. A.; Hölker M.; Besenfelder U.; Mokry M.; Vos P. L. A. M.; Stout T. A. E.; et al. An Oviduct-on-a-Chip Provides an Enhanced in Vitro Environment for Zygote Genome Reprogramming. Nat. Commun. 2018, 9, 1–14. 10.1038/s41467-018-07119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimek K.; Busek M.; Brincker S.; Groth B.; Hoffmann S.; Lauster R.; Lindner G.; Lorenz A.; Menzel U.; Sonntag F.; et al. Integrating Biological Vasculature into a Multi-Organ-Chip Microsystem. Lab Chip 2013, 13, 3588–3598. 10.1039/c3lc50217a. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Zhang Y. S.; Bakht S. M.; Aleman J.; Shin S. R.; Yue K.; Sica M.; Ribas J.; Duchamp M.; Ju J.; et al. Elastomeric Free-Form Blood Vessels for Interconnecting Organs on Chip Systems. Lab Chip 2016, 16, 1579–1586. 10.1039/C6LC00001K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva T.; Hsiao J. C.; Charest J. L.; Borenstein J. T. A Microfluidic Respiratory Assist Device with High Gas Permeance for Artificial Lung Applications. Biomed. Microdevices 2011, 13, 315–323. 10.1007/s10544-010-9495-1. [DOI] [PubMed] [Google Scholar]
- de Almeida Monteiro Melo Ferraz M.; Henning H. H. W.; Ferreira da Costa P.; Malda J.; Le Gac S.; Bray F.; van Duursen M. B. M.; Brouwers J. F.; van de Lest C. H. A.; Bertijn I.; et al. Potential Health and Environmental Risks of Three-Dimensional Engineered Polymers. Environ. Sci. Technol. Lett. 2018, 5, 80–85. 10.1021/acs.estlett.7b00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther A.; Yasotharan S.; Vagaon A.; Lochovsky C.; Pinto S.; Yang J.; Lau C.; Voigtlaender-Bolz J.; Bolz S. S. A Microfluidic Platform for Probing Small Artery Structure and Function. Lab Chip 2010, 10, 2341–2349. 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeon J. H.; Ryu H. R.; Chung M.; Hu Q. P.; Jeon N. L. In Vitro Formation and Characterization of a Perfusable Three-Dimensional Tubular Capillary Network in Microfluidic Devices. Lab Chip 2012, 12, 2815–2822. 10.1039/c2lc40131b. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Zhao Z.; Abdul Rahim N. A.; Van Noort D.; Yu H. Towards a Human-on-Chip: Culturing Multiple Cell Types on a Chip with Compartmentalized Microenvironments. Lab Chip 2009, 9, 3185–3192. 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- Materne E. M.; Ramme A. P.; Terrasso A. P.; Serra M.; Alves P. M.; Brito C.; Sakharov D. A.; Tonevitsky A. G.; Lauster R.; Marx U. A Multi-Organ Chip Co-Culture of Neurospheres and Liver Equivalents for Long-Term Substance Testing. J. Biotechnol. 2015, 205, 36–46. 10.1016/j.jbiotec.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S.; Datta A.; Berg J. M.; Gangopadhyay S. Studies on Surface Wettability of Poly(Dimethyl) Siloxane (PDMS) and Glass under Oxygen-Plasma Treatment and Correlation with Bond Strength. J. Microelectromech. Syst. 2005, 14, 590–597. 10.1109/JMEMS.2005.844746. [DOI] [Google Scholar]
- Schasfoort R. B. M.; Schlautmann S.; Hendrikse J.; van den Berg A. Field-Effect Flow Control for Microfabricated Fluidic Networks. Science 1999, 286, 942–945. 10.1126/science.286.5441.942. [DOI] [PubMed] [Google Scholar]
- Chueh B.; Huh D.; Kyrtsos C. R.; Houssin T.; Futai N.; Takayama S. Leakage-Free Bonding of Porous Membranes into Layered Microfluidic Array Systems. Anal. Chem. 2007, 79, 3504–3508. 10.1021/ac062118p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler T. E.; Feil M.; Stronkman E. F. G. J.; Matthiesen I.; Herland A. Low-Cost Microphysiological Systems: Feasibility Study of a Tape-Based Barrier-on-Chip for Small Intestine Modeling. Lab Chip 2020, 20, 1212–1226. 10.1039/D0LC00009D. [DOI] [PubMed] [Google Scholar]
- Jahn I. J.; Žukovskaja O.; Zheng X.-S.; Weber K.; Bocklitz T. W.; Cialla-May D.; Popp J. Surface-Enhanced Raman Spectroscopy and Microfluidic Platforms: Challenges, Solutions and Potential Applications. Analyst 2017, 142, 1022–1047. 10.1039/C7AN00118E. [DOI] [PubMed] [Google Scholar]
- Liszka B. M.; Rho H. S.; Yang Y.; Lenferink A. T. M.; Terstappen L. W. M. M.; Otto C. A Microfluidic Chip for High Resolution Raman Imaging of Biological Cells. RSC Adv. 2015, 5, 49350–49355. 10.1039/C5RA05185A. [DOI] [Google Scholar]
- Bart J.; Kolkman A. J.; Oosthoek-de Vries A. J.; Koch K.; Nieuwland P. J.; Janssen H. J. W. G.; van Bentum J. P. J. M.; Ampt K. A. M.; Rutjes F. P. J. T.; Wijmenga S. S.; et al. A Microfluidic High-Resolution NMR Flow Probe. J. Am. Chem. Soc. 2009, 131, 5014–5015. 10.1021/ja900389x. [DOI] [PubMed] [Google Scholar]
- Vinagre J.; Almeida A.; Pópulo H.; Batista R.; Lyra J.; Pinto V.; Coelho R.; Celestino R.; Prazeres H.; Lima L.; et al. Frequency of TERT Promoter Mutations in Human Cancers. Nat. Commun. 2013, 4, 1–6. 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- Herranz D.; Muñoz-Martin M.; Cañamero M.; Mulero F.; Martinez-Pastor B.; Fernandez-Capetillo O.; Serrano M. Sirt1 Improves Healthy Ageing and Protects from Metabolic Syndrome-Associated Cancer. Nat. Commun. 2010, 1, 1–8. 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Getz G.; Miska E. A.; Alvarez-Saavedra E.; Lamb J.; Peck D.; Sweet-Cordero A.; Ebert B. L.; Mak R. H.; Ferrando A. A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Nilsson J.; Skog J.; Nordstrand A.; Baranov V.; Mincheva-Nilsson L.; Breakefield X. O.; Widmark A. Prostate Cancer-Derived Urine Exosomes: A Novel Approach to Biomarkers for Prostate Cancer. Br. J. Cancer 2009, 100, 1603–1607. 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragazzini R.; Pérez-Palacios R.; Baymaz I. H.; Diop S.; Ancelin K.; Zielinski D.; Michaud A.; Givelet M.; Borsos M.; Aflaki S.; et al. EZHIP Constrains Polycomb Repressive Complex 2 Activity in Germ Cells. Nat. Commun. 2019, 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella C. M.; Ruiz-Ruano F. J.; Dion-Côté A.-M.; Charles A. J.; Gossmann T. I.; Cabrero J.; Kappei D.; Hemmings N.; Simons M. J. P.; Camacho J. P. M.; et al. Programmed DNA Elimination of Germline Development Genes in Songbirds. bioRxiv 2018, 444364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seishima R.; Leung C.; Yada S.; Bte K.; Murad A.; Tan L. T.; Hajamohideen A.; Tan S. H.; Itoh H.; Murakami K.; et al. Neonatal Wnt-Dependent Lgr5 Positive Stem Cells Are Essential for Uterine Gland Development. Nat. Commun. 2019, 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster K. A.; Schach U.; Ordaz A.; Steinfeld J. S.; Draper B. W.; Siegfried K. R. Dmrt1 Is Necessary for Male Sexual Development in Zebrafish. Dev. Biol. 2017, 422, 33–46. 10.1016/j.ydbio.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt-Leal A. M.; Carpenedo R. L.; Ungrin M. D.; Zandstra P. W.; McDevitt T. C. Incorporation of Biomaterials in Multicellular Aggregates Modulates Pluripotent Stem Cell Differentiation. Biomaterials 2011, 32, 48–56. 10.1016/j.biomaterials.2010.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. A.; Liu X.; Hu J.; Ma P. X. The Enhancement of Human Embryonic Stem Cell Osteogenic Differentiation with Nano-Fibrous Scaffolding. Biomaterials 2010, 31, 5526–5535. 10.1016/j.biomaterials.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Itaka K.; Ohba S.; Nishiyama N.; Chung U. il; Yamasaki Y.; Kataoka K. 3D Spheroid Culture System on Micropatterned Substrates for Improved Differentiation Efficiency of Multipotent Mesenchymal Stem Cells. Biomaterials 2009, 30, 2705–2715. 10.1016/j.biomaterials.2009.01.030. [DOI] [PubMed] [Google Scholar]
- Schuh E.; Kramer J.; Rohwedel J.; Notbohm H.; Müller R.; Gutsmann T.; Rotter N. Effect of Matrix Elasticity on the Maintenance of the Chondrogenic Phenotype. Tissue Eng., Part A 2010, 16, 1281–1290. 10.1089/ten.tea.2009.0614. [DOI] [PubMed] [Google Scholar]
- Prasad C. K.; Krishnan L. K. Regulation of Endothelial Cell Phenotype by Biomimetic Matrix Coated on Biomaterials for Cardiovascular Tissue Engineering. Acta Biomater. 2008, 4, 182–191. 10.1016/j.actbio.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Brodkin K. R.; García A. J.; Levenston M. E. Chondrocyte Phenotypes on Different Extracellular Matrix Monolayers. Biomaterials 2004, 25, 5929–5938. 10.1016/j.biomaterials.2004.01.044. [DOI] [PubMed] [Google Scholar]
- Yuan H.; Fernandes H.; Habibovic P.; de Boer J.; Barradas A. M. C.; de Ruiter A.; Walsh W. R.; van Blitterswijk C. A.; de Bruijn J. D. Osteoinductive Ceramics as a Synthetic Alternative to Autologous Bone Grafting. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 13614–13619. 10.1073/pnas.1003600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Luo Q.; Deng B.; Morita Y.; Ju Y.; Song G. Construction of Tendon Replacement Tissue Based on Collagen Sponge and Mesenchymal Stem Cells by Coupled Mechano-Chemical Induction and Evaluation of Its Tendon Repair Abilities. Acta Biomater. 2018, 74, 247–259. 10.1016/j.actbio.2018.04.047. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Li B.; Wang J. H. C. The Role of Engineered Tendon Matrix in the Stemness of Tendon Stem Cells in Vitro and the Promotion of Tendon-like Tissue Formation in Vivo. Biomaterials 2011, 32, 6972–6981. 10.1016/j.biomaterials.2011.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddappa R.; Martens A.; Doorn J.; Leusink A.; Olivo C.; Licht R.; van Rijn L.; Gaspar C.; Fodde R.; Janssen F.; et al. CAMP/PKA Pathway Activation in Human Mesenchymal Stem Cells in Vitro Results in Robust Bone Formation in Vivo. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 7281–7286. 10.1073/pnas.0711190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B.; Song G.; Ju Y.; Li X.; Song Y.; Watanabe S. RhoA/ROCK, Cytoskeletal Dynamics, and Focal Adhesion Kinase Are Required for Mechanical Stretch-Induced Tenogenic Differentiation of Human Mesenchymal Stem Cells. J. Cell. Physiol. 2012, 227, 2722–2729. 10.1002/jcp.23016. [DOI] [PubMed] [Google Scholar]
- Schellenberg A.; Joussen S.; Moser K.; Hampe N.; Hersch N.; Hemeda H.; Schnitker J.; Denecke B.; Lin Q.; Pallua N.; et al. Matrix Elasticity, Replicative Senescence and DNA Methylation Patterns of Mesenchymal Stem Cells. Biomaterials 2014, 35, 6351–6358. 10.1016/j.biomaterials.2014.04.079. [DOI] [PubMed] [Google Scholar]
- Downing T. L.; Soto J.; Morez C.; Houssin T.; Fritz A.; Yuan F.; Chu J.; Patel S.; Schaffer D. V.; Li S. Biophysical Regulation of Epigenetic State and Cell Reprogramming. Nat. Mater. 2013, 12, 1154–1162. 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zreiqat H.; Howlett C. R. Titanium Substrata Composition Influences Osteoblastic Phenotype: In Vitro Study. J. Biomed. Mater. Res. 1999, 47, 360–366. . [DOI] [PubMed] [Google Scholar]
- Knabe C.; Stiller M.; Berger G.; Reif D.; Gildenhaar R.; Howlett C. R.; Zreiqat H. The Effect of Bioactive Glass Ceramics on the Expression of Bone-Related Genes and Proteins in Vitro. Clin. Oral Implants Res. 2005, 16, 119–127. 10.1111/j.1600-0501.2004.01066.x. [DOI] [PubMed] [Google Scholar]
- Maeda E.; Kuroyanagi K.; Ando Y.; Matsumoto T. Effects of Substrate Stiffness on Morphology and MMP-1 Gene Expression in Tenocytes Stimulated With Interleukin-1β. J. Orthop. Res. 2020, 38, 150–159. 10.1002/jor.24403. [DOI] [PubMed] [Google Scholar]
- Kato S.; Akagi T.; Kishida A.; Sugimura K.; Akashi M. Evaluation of Biological Responses to Polymeric Biomaterials by RT-PCR Analysis III: Study of HSP 70, 90 and 47 MRNA Expression. J. Biomater. Sci., Polym. Ed. 1997, 8, 809–814. 10.1163/156856297X00335. [DOI] [PubMed] [Google Scholar]
- Kato S.; Akagi T.; Kishida A.; Sugimura K.; Akashi M. Evaluation of Biological Responses to Polymeric Biomaterials by RT-PCR Analysis IV: Study of c-Myc, c-Fos and P53 MRNA Expression Shinya. Biomaterials 2000, 21, 521–527. 10.1016/S0142-9612(99)00214-8. [DOI] [PubMed] [Google Scholar]
- Forte G.; Pagliari S.; Ebara M.; Uto K.; Van Tam J. K.; Romanazzo S.; Escobedo-Lucea C.; Romano E.; Di Nardo P.; Traversa E.; et al. Substrate Stiffness Modulates Gene Expression and Phenotype in Neonatal Cardiomyocytes In Vitro. Tissue Eng., Part A 2012, 18, 1837–1848. 10.1089/ten.tea.2011.0707. [DOI] [PubMed] [Google Scholar]
- Chung W.; Eum H. H.; Lee H.-O.; Lee K.-M.; Lee H.-B.; Kim K.-T.; Ryu H. S.; Kim S.; Lee J. E.; Park Y. H.; et al. Single-Cell RNA-Seq Enables Comprehensive Tumour and Immune Cell Profiling in Primary Breast Cancer. Nat. Commun. 2017, 8, 15081. 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D.; Luo S.; Wang Y.-C.; Li R.; Deng Q.; Faridani O. R.; Daniels G. A.; Khrebtukova I.; Loring J. F.; Laurent L. C.; et al. Full-Length MRNA-Seq from Single-Cell Levels of RNA and Individual Circulating Tumor Cells. Nat. Biotechnol. 2012, 30, 777. 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C. W.; Lopez-Diaz F. J.; Khan S. Y.; Tariq M. A.; Dayn Y.; Vaske C. J.; Radenbaugh A. J.; Kim H. J.; Emerson B. M.; Pourmand N. Single-Cell Analyses of Transcriptional Heterogeneity during Drug Tolerance Transition in Cancer Cells by RNA Sequencing. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E4726 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A. A.; Nowakowski T. J.; Shuga J.; Wang X.; Leyrat A. A.; Lui J. H.; Li N.; Szpankowski L.; Fowler B.; Chen P.; et al. Low-Coverage Single-Cell MRNA Sequencing Reveals Cellular Heterogeneity and Activated Signaling Pathways in Developing Cerebral Cortex. Nat. Biotechnol. 2014, 32, 1053–1058. 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z.; Huang K.; Cai C.; Cai L.; Jiang C.; Feng Y.; Liu Z.; Zeng Q.; Cheng L.; Sun Y. E.; et al. Genetic Programs in Human and Mouse Early Embryos Revealed by Single-Cell RNA Sequencing. Nature 2013, 500, 593. 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xynos I. D.; Edgar A. J.; Buttery L. D. K.; Hench L. L.; Polak J. M. Ionic Products of Bioactive Glass Dissolution Increase Proliferation of Human Osteoblasts and Induce Insulin-like Growth Factor II MRNA Expression and Protein Synthesis. Biochem. Biophys. Res. Commun. 2000, 276, 461–465. 10.1006/bbrc.2000.3503. [DOI] [PubMed] [Google Scholar]
- Herlofsen S. R.; Küchler A. M.; Melvik J. E.; Brinchmann J. E. Chondrogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells in Self-Gelling Alginate Discs Reveals Novel Chondrogenic Signature Gene Clusters. Tissue Eng., Part A 2011, 17, 1003–1013. 10.1089/ten.tea.2010.0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G.; Tison C. K.; Chatterjee K.; Pine P. S.; McDaniel J. H.; Salit M. L.; Young M. F.; Simon C. G. The Determination of Stem Cell Fate by 3D Scaffold Structures through the Control of Cell Shape. Biomaterials 2011, 32, 9188–9196. 10.1016/j.biomaterials.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. A.; Pine P. S.; Chatterjee K.; Kumar G.; Lin N. J.; McDaniel J. H.; Salit M. L.; Simon C. G. Ontology Analysis of Global Gene Expression Differences of Human Bone Marrow Stromal Cells Cultured on 3D Scaffolds or 2D Films. Biomaterials 2014, 35, 6716–6726. 10.1016/j.biomaterials.2014.04.075. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Schwartz M. P.; Hou Z.; Bai Y.; Ardalani H.; Swanson S.; Steill J.; Ruotti V.; Elwell A.; Nguyen B. K.; et al. A Genome-Wide Analysis of Human Pluripotent Stem Cell-Derived Endothelial Cells in 2D or 3D Culture. Stem Cell Rep. 2017, 8, 907–918. 10.1016/j.stemcr.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieuchot L.; Marteau J.; Guignandon A.; Dos Santos T.; Brigaud I.; Chauvy P. F.; Cloatre T.; Ponche A.; Petithory T.; Rougerie P.; et al. Curvotaxis Directs Cell Migration through Cell-Scale Curvature Landscapes. Nat. Commun. 2018, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa D. O.; Prowse P. D. H.; Chrones T.; Sims S. M.; Hamilton D. W.; Rizkalla A. S.; Dixon S. J. The Differential Regulation of Osteoblast and Osteoclast Activity Bysurface Topography of Hydroxyapatite Coatings. Biomaterials 2013, 34, 7215–7226. 10.1016/j.biomaterials.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Khang D.; Choi J.; Im Y. M.; Kim Y. J.; Jang J. H.; Kang S. S.; Nam T. H.; Song J.; Park J. W. Role of Subnano-, Nano- and Submicron-Surface Features on Osteoblast Differentiation of Bone Marrow Mesenchymal Stem Cells. Biomaterials 2012, 33, 5997–6007. 10.1016/j.biomaterials.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Viswanathan P.; Ondeck M. G.; Chirasatitsin S.; Ngamkham K.; Reilly G. C.; Engler A. J.; Battaglia G. 3D Surface Topology Guides Stem Cell Adhesion and Differentiation. Biomaterials 2015, 52, 140–147. 10.1016/j.biomaterials.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M. J.; Riehle M. O.; Yarwood S. J.; Wilkinson C. D. W.; Curtis A. S. G. Nucleus Alignment and Cell Signaling in Fibroblasts: Response to a Micro-Grooved Topography. Exp. Cell Res. 2003, 284, 274–282. 10.1016/S0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- Lee L. C. Y.; Gadegaard N.; de Andrés M. C.; Turner L. A.; Burgess K. V.; Yarwood S. J.; Wells J.; Salmeron-Sanchez M.; Meek D.; Oreffo R. O. C.; et al. Nanotopography Controls Cell Cycle Changes Involved with Skeletal Stem Cell Self-Renewal and Multipotency. Biomaterials 2017, 116, 10–20. 10.1016/j.biomaterials.2016.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abagnale G.; Steger M.; Nguyen V. H.; Hersch N.; Sechi A.; Joussen S.; Denecke B.; Merkel R.; Hoffmann B.; Dreser A.; et al. Surface Topography Enhances Differentiation of Mesenchymal Stem Cells towards Osteogenic and Adipogenic Lineages. Biomaterials 2015, 61, 316–326. 10.1016/j.biomaterials.2015.05.030. [DOI] [PubMed] [Google Scholar]
- Darnell M.; Gu L.; Mooney D. RNA-Seq Reveals Diverse Effects of Substrate Stiffness on Mesenchymal Stem Cells. Biomaterials 2018, 181, 182–188. 10.1016/j.biomaterials.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C. H.; Browne M.; Gregson P. J.; Corbeil J.; Pioletti D. P. Large-Scale Gene Expression Analysis of Osteoblasts Cultured on Three Different Ti-6Al-4V Surface Treatments. Biomaterials 2002, 23, 4193–4202. 10.1016/S0142-9612(02)00161-8. [DOI] [PubMed] [Google Scholar]
- Carinci F.; Volinia S.; Pezzetti F.; Francioso F.; Tosi L.; Piattelli A. Titanium-Cell Interaction: Analysis of Gene Expression Profiling. J. Biomed. Mater. Res. 2003, 66, 341–346. 10.1002/jbm.b.10021. [DOI] [PubMed] [Google Scholar]
- Groen N.; Tahmasebi N.; Shimizu F.; Sano Y.; Kanda T.; Barbieri D.; Yuan H.; Habibovic P.; van Blitterswijk C. A.; de Boer J. Exploring the Material-Induced Transcriptional Landscape of Osteoblasts on Bone Graft Materials. Adv. Healthcare Mater. 2015, 4, 1691–1700. 10.1002/adhm.201500171. [DOI] [PubMed] [Google Scholar]
- Alakpa E. V.; Jayawarna V.; Lampel A.; Burgess K. V.; West C. C.; Bakker S. C. J.; Roy S.; Javid N.; Fleming S.; Lamprou D. A.; et al. Tunable Supramolecular Hydrogels for Selection of Lineage-Guiding Metabolites in Stem Cell Cultures. Chem. 2016, 1, 298–319. 10.1016/j.chempr.2016.07.001. [DOI] [Google Scholar]
- Guerette P. A.; Hoon S.; Seow Y.; Raida M.; Masic A.; Wong F. T.; Ho V. H. B.; Kong K. W.; Demirel M. C.; Pena-Francesch A.; et al. Accelerating the Design of Biomimetic Materials by Integrating RNA-Seq with Proteomics and Materials Science. Nat. Biotechnol. 2013, 31, 908–915. 10.1038/nbt.2671. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O.; Stubbington M. J. T.; Regev A.; Teichmann S. A. The Human Cell Atlas: From Vision to Reality. Nature 2017, 550, 451–453. 10.1038/550451a. [DOI] [PubMed] [Google Scholar]
- MacParland S. A.; Liu J. C.; Ma X. Z.; Innes B. T.; Bartczak A. M.; Gage B. K.; Manuel J.; Khuu N.; Echeverri J.; Linares I.; et al. Single Cell RNA Sequencing of Human Liver Reveals Distinct Intrahepatic Macrophage Populations. Nat. Commun. 2018, 9, 1–21. 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledergor G.; Weiner A.; Zada M.; Wang S. Y.; Cohen Y. C.; Gatt M. E.; Snir N.; Magen H.; Koren-Michowitz M.; Herzog-Tzarfati K.; et al. Single Cell Dissection of Plasma Cell Heterogeneity in Symptomatic and Asymptomatic Myeloma. Nat. Med. 2018, 24, 1867–1876. 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R.; Efremova M.; Botting R. A.; Turco M. Y.; Payne R. P.; Goncalves A.; Zou A.; Henriksson J. Reconstructing the Human First Trimester Fetal-Maternal Interface Using Single Cell Transcriptomics. bioRxiv 2018, 429589. [Google Scholar]
- Reyfman P. A.; Walter J. M.; Joshi N.; Anekalla K. R.; McQuattie-Pimentel A. C.; Chiu S.; Fernandez R.; Akbarpour M.; Chen C. I.; Ren Z.; et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1517–1536. 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.; Narayan R.; Corsello S. M.; Peck D. D.; Natoli T. E.; Lu X.; Gould J.; Davis J. F.; Tubelli A. A.; Asiedu J. K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molecules S.; Lamb J.; Crawford E. D.; Peck D.; Modell J. W.; Blat I. C.; Wrobel M. J.; Lerner J.; Brunet J.; Subramanian A. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Clough E.; Barrett T.. The Gene Expression Omnibus Database. In Statistical Genomics: Methods and Protocols; Mathé E., Davis S., Eds.; Springer New York: New York, 2016; pp 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H.; Kapushesky M.; Shojatalab M.; Abeygunawardena N.; Coulson R.; Farne A.; Holloway E.; Kolesnykov N.; Lilja P.; Lukk M.; et al. ArrayExpress - A Public Database of Microarray Experiments and Gene Expression Profiles. Nucleic Acids Res. 2007, 35, 747–750. 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchser W.; Ph D.. Assay Development Guidelines for Image-Based High Content Screening, High Content Analysis and High Content Imaging. Assay Guid. Man. [Internet]. Eli Lilly Co. Natl. Cent. Adv. Transl. Sci. 2014. [PubMed] [Google Scholar]
- Passarelli M. K.; Winograd N. Lipid Imaging with Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811, 976–990. 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickischer D.; Elkin L.; Cloutier N.; O’Connell J.; Banks M.; Weston A.. Challenges and Opportunities in Enabling High-Throughput, Miniaturized High Content Screening. In Methods Mol. Biol.; Humana Press Inc., 2018; Vol. 1683, pp 165–191. [DOI] [PubMed] [Google Scholar]
- Korn K.; Krausz E. Cell-Based High-Content Screening of Small-Molecule Libraries. Curr. Opin. Chem. Biol. 2007, 11, 503–510. 10.1016/j.cbpa.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Li Z.; Yan Y.; Powers E. A.; Ying X.; Janjua K.; Garyantes T.; Baron B. Identification of Gap Junction Blockers Using Automated Fluorescence Microscopy Imaging. J. Biomol. Screening 2003, 8, 489–499. 10.1177/1087057103257309. [DOI] [PubMed] [Google Scholar]
- Al-Ali H.; Blackmore M.; Bixby J. L.; Lemmon V. P.. High Content Screening with Primary Neurons Assay Guidance Manual [Internet]. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences 2014. [PubMed] [Google Scholar]
- Trask O. J.Nuclear Factor Kappa B (NF-KB) Translocation Assay Development and Validation for High Content Screening Assay Guidance Manual [Internet]. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences 2012. [PubMed] [Google Scholar]
- Reimer A.; Vasilevich A.; Hulshof F.; Viswanathan P.; Van Blitterswijk C. A.; De Boer J.; Watt F. M. Scalable Topographies to Support Proliferation and Oct4 Expression by Human Induced Pluripotent Stem Cells. Sci. Rep. 2016, 6, 18948. 10.1038/srep18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett S. E.; Sim J. Y.; Rubins J. E.; Neronha Z. J.; Williams E. K.; Wong I. Y. Morphological Single Cell Profiling of the Epithelial-Mesenchymal Transition. Integr. Biol. (United Kingdom) 2016, 8, 1133–1144. 10.1039/C6IB00139D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang P.; Yeow K.; Nichols A.; Scheer A. Cellular Imaging in Drug Discovery. Nat. Rev. Drug Discovery 2006, 5, 343–356. 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- Mercola M.; Colas A.; Willems E. Induced Pluripotent Stem Cells in Cardiovascular Drug Discovery. Circ. Res. 2013, 112, 534–548. 10.1161/CIRCRESAHA.111.250266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M.; Heigwer F.; Laufer C.. Microscopy-Based High-Content Screening. Cell; Cell Press, 2015; pp 1314–1325. [DOI] [PubMed] [Google Scholar]
- Conrad C.; Gerlich D. W. Automated Microscopy for High-Content RNAi Screening. J. Cell Biol. 2010, 188, 453–461. 10.1083/jcb.200910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal A.; Brenner M.; Wolujewicz P.; Zhang Z.; Mao Y.; Batish M.; Kohn J.; Moghe P. V. Profiling Stem Cell States in Three-Dimensional Biomaterial Niches Using High Content Image Informatics. Acta Biomater. 2016, 45, 98–109. 10.1016/j.actbio.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo J. C.; Singh S.; Carpenter A. E. Applications in Image-Based Profiling of Perturbations. Curr. Opin. Biotechnol. 2016, 39, 134–142. 10.1016/j.copbio.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Wawer M. J.; Li K.; Gustafsdottir S. M.; Ljosa V.; Bodycombe N. E.; Marton M. A.; Sokolnicki K. L.; Bray M. A.; Kemp M. M.; Winchester E.; et al. Toward Performance-Diverse Small-Molecule Libraries for Cell-Based Phenotypic Screening Using Multiplexed High-Dimensional Profiling. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 10911–10916. 10.1073/pnas.1410933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo J. C.; Cooper S.; Heigwer F.; Warchal S.; Qiu P.; Molnar C.; Vasilevich A. S.; Barry J. D.; Bansal H. S.; Kraus O.; et al. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14, 849–863. 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiser M. D.; Yang E. H.; Gordonov S.; Cohen D. M.; Androulakis I. P.; Kohn J.; Chen C. S.; Moghe P. V. Cytoskeleton-Based Forecasting of Stem Cell Lineage Fates. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 610–615. 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilevich A. S.; Mourcin F.; Mentink A.; Hulshof F.; Beijer N.; Zhao Y.; Levers M.; Papenburg B.; Singh S.; Carpenter A. E.; et al. Designed Surface Topographies Control ICAM-1 Expression in Tonsil-Derived Human Stromal Cells. Front. Bioeng. Biotechnol. 2018, 6, 87. 10.3389/fbioe.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner A. M.; Richter B.; Bastmeyer M. Micro-Engineered 3D Scaffolds for Cell Culture Studies. Macromol. Biosci. 2012, 12, 1301–1314. 10.1002/mabi.201200132. [DOI] [PubMed] [Google Scholar]
- Li L.; Zhou Q.; Voss T. C.; Quick K. L.; LaBarbera D. V.. High-Throughput Imaging: Focusing in on Drug Discovery in 3D. Methods; Academic Press Inc, 2016; pp 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hurley G.; Sjöstedt E.; Rahman A.; Li B.; Kampf C.; Pontén F.; Gallagher W. M.; Lindskog C.. Garbage in, Garbage out: A Critical Evaluation of Strategies Used for Validation of Immunohistochemical Biomarkers. Molecular Oncology; John Wiley and Sons Ltd, 2014; pp 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M.; Ph D.. Advanced Assay Development Guidelines for Image-Based High Content Screening and Analysis. Assay Guid. Man. [Internet] Eli Lilly Co. Natl. Cent. Adv. Transl. Sci. 2017. [PubMed] [Google Scholar]
- Zanella F.; Lorens J. B.; Link W. High Content Screening: Seeing Is Believing. Trends Biotechnol. 2010, 28, 237–245. 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Walters W. P.; Namchuk M.. Designing Screens: How to Make Your Hits a Hit. Nature Reviews Drug Discovery; European Association for Cardio-Thoracic Surgery, 2003; pp 259–266. [DOI] [PubMed] [Google Scholar]
- Berezin M. Y.; Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschong W.; Suetterlin R.; Hubert Laeng R. Control of Autofluorescence of Archival Formaldehyde-Fixed, Paraffin-Embedded Tissue in Confocal Laser Scanning Microscopy (CLSM). J. Histochem. Cytochem. 2001, 49, 1565–1571. 10.1177/002215540104901210. [DOI] [PubMed] [Google Scholar]
- Viegas M.; Martins T.; Seco F.; Carmo A. do. An Improved and Cost-Effective Methodology for the Reduction of Autofluorescence in Direct Immunofluorescence Studies on Formalin-Fixed Paraffin-Embedded Tissues. Eur. J. Histochem. 2007, 59–66. [PubMed] [Google Scholar]
- Noller C. M.; Boulina M.; McNamara G.; Szeto A.; McCabe P. M.; Mendez A. J. A Practical Approach to Quantitative Processing and Analysis of Small Biological Structures by Fluorescent Imaging. J. Biomol. Technol. 2016, 27, 90–97. 10.7171/jbt.16-2703-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson R. M.; Mansfield J. R. Multispectral Imaging in Biology and Medicine: Slices of Life. Cytometry, Part A 2006, 69, 748–758. 10.1002/cyto.a.20319. [DOI] [PubMed] [Google Scholar]
- Johnson D. S.; Jaiswal J. K.; Simon S. Total Internal Reflection Fluorescence (TIRF) Microscopy Illuminator for Improved Imaging of Cell Surface Events. Curr. Protoc. Cytom. 2012, 61, 12–29. 10.1002/0471142956.cy1229s61. [DOI] [PubMed] [Google Scholar]
- Fang Y.Total Internal Reflection Fluorescence Quantification of Receptor Pharmacology. Biosensors; MDPI AG, 2015; pp 223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal P. P.; Diaspro A.. Fundamentals of Fluorescence Microscopy: Exploring Life with Light; Springer Science & Business Media, 2013. [Google Scholar]
- Malo N.; Hanley J. a; Cerquozzi S.; Pelletier J.; Nadon R. Statistical Practice in High-Throughput Screening Data Analysis. Nat. Biotechnol. 2006, 24, 167–175. 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- Eliceiri K. W.; Berthold M. R.; Goldberg I. G.; Ibáñez L.; Manjunath B. S.; Martone M. E.; Murphy R. F.; Peng H.; Plant A. L.; Roysam B.; et al. Biological Imaging Software Tools. Nat. Methods 2012, 9, 697. 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder M.; Pernthaler J. Multispot Live-Image Autofocusing for High-Throughput Microscopy of Fluorescently Stained Bacteria. Cytometry, Part A 2009, 75, 781–788. 10.1002/cyto.a.20770. [DOI] [PubMed] [Google Scholar]
- Alvarez-Borrego J. Fast Autofocus Algorithm for Automated Microscopes. Opt. Eng. 2005, 44, 063601. 10.1117/1.1925119. [DOI] [Google Scholar]
- Liron Y.; Paran Y.; Zatorsky N. G.; Geiger B.; Kam Z. Laser Autofocusing System for High-Resolution Cell Biological Imaging. J. Microsc. 2006, 221, 145–151. 10.1111/j.1365-2818.2006.01550.x. [DOI] [PubMed] [Google Scholar]
- Rost F. W. D.Fluorescence Microscopy Practical. Quantitative Fluorescence Microscopy; Cambridge University Press, 1991; Vol. 1, p 267. [Google Scholar]
- Verveer P. J.; Swoger J.; Pampaloni F.; Greger K.; Marcello M.; Stelzer E. H. K. High-Resolution Three-Dimensional Imaging of Large Specimens with Light Sheet-Based Microscopy. Nat. Methods 2007, 4, 311–313. 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- Wolf D. E.; Samarasekera C.; Swedlow J. R. Quantitative Analysis of Digital Microscope Images. Methods Cell Biol. 2007, 81, 365–396. 10.1016/S0091-679X(06)81017-4. [DOI] [PubMed] [Google Scholar]
- North A. J. Seeing Is Believing? A Beginners’ Guide to Practical Pitfalls in Image Acquisition. J. Cell Biol. 2006, 172, 9–18. 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E.; Jones T. R.; Lamprecht M. R.; Clarke C.; Kang I. H.; Friman O.; Guertin D. a; Chang J. H.; Lindquist R. a; Moffat J.; et al. CellProfiler: Image Analysis Software for Identifying and Quantifying Cell Phenotypes. Genome Biol. 2006, 7, R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.; Li Y.; Piccinini F.; Csucs G.; Balazs C.; Bevilacqua A.; Horvath P. CIDRE: An Illumination-Correction Method for Optical Microscopy. Nat. Methods 2015, 12, 404–406. 10.1038/nmeth.3323. [DOI] [PubMed] [Google Scholar]
- Singh S.; Bray M. A.; Jones T. R.; Carpenter A. E. Pipeline for Illumination Correction of Images for High-Throughput Microscopy. J. Microsc. 2014, 256, 231–236. 10.1111/jmi.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T. J. ImageJ for Microscopy. BioTechniques 2007, 43, S25–S30. 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Sommer C.; Straehle C.; Koethe U.; Hamprecht F. A.. Ilastik: Interactive Learning and Segmentation Toolkit. In 2011 IEEE international symposium on biomedical imaging: From nano to macro; IEEE, 2011; pp 230–233.
- Held M.; Schmitz M. H. a; Fischer B.; Walter T.; Neumann B.; Olma M. H.; Peter M.; Ellenberg J.; Gerlich D. W. CellCognition: Time-Resolved Phenotype Annotation in High-Throughput Live Cell Imaging. Nat. Methods 2010, 7, 747–754. 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- Bitar M.; Friederici V.; Imgrund P.; Brose C.; Bruinink A. In Vitro Bioactivity of Micro Metal Injection Moulded Stainless Steel with Defined Surface Features. Eur. Cell Mater. 2012, 23, 333–347. 10.22203/eCM.v023a26. [DOI] [PubMed] [Google Scholar]
- De Chaumont F.; Dallongeville S.; Chenouard N.; Hervé N.; Pop S.; Provoost T.; Meas-Yedid V.; Pankajakshan P.; Lecomte T.; Le Montagner Y.; et al. Icy: An Open Bioimage Informatics Platform for Extended Reproducible Research. Nat. Methods 2012, 9, 690. 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- Chityala R.; Pudipeddi S.. Image Processing and Acquisition Using Python; Chapman and Hall/CRC, 2014. [Google Scholar]
- Pau G.; Fuchs F.; Sklyar O.; Boutros M.; Huber W. EBImage—an R Package for Image Processing with Applications to Cellular Phenotypes. Bioinformatics 2010, 26, 979–981. 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon C.; Breckon T.. Fundamentals of Digital Image Processing: A Practical Approach with Examples in Matlab; John Wiley & Sons, 2011. [Google Scholar]
- Sarid D.Exploring Scanning Probe Microscopy with Mathematica; Wiley Online Library, 2007. [Google Scholar]
- Rajaram S.; Pavie B.; Wu L. F.; Altschuler S. J. PhenoRipper: Software for Rapidly Profiling Microscopy Images. Nat. Methods 2012, 9, 635. 10.1038/nmeth.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G.; Kooi T.; Bejnordi B. E.; Setio A. A. A.; Ciompi F.; Ghafoorian M.; van der Laak J. A. W. M.; van Ginneken B.; Sánchez C. I.. A Survey on Deep Learning in Medical Image Analysis. Medical Image Analysis; Elsevier B.V., 2017; pp 60–88. [DOI] [PubMed] [Google Scholar]
- Lecun Y.; Bengio Y.; Hinton G.. Deep Learning. Nature; Nature Publishing Group, 2015; pp 436–444. [DOI] [PubMed] [Google Scholar]
- Caicedo J. C.; Roth J.; Goodman A.; Becker T.; Karhohs K. W.; Broisin M.; Molnar C.; McQuin C.; Singh S.; Theis F. J. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. Cytometry, Part A 2019, 95, 952. 10.1002/cyto.a.23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.; Lee B.; Yoon S. Deep Learning in Bioinformatics. Briefings in bioinformatics 2017, 18, 851–869. 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- Pawlowski N.; Caicedo J. C.; Singh S.; Carpenter A. E.; Storkey A. Automating Morphological Profiling with Generic Deep Convolutional Networks. bioRxiv 2016, 4–8. [Google Scholar]
- Lafarge M. W.; Caicedo J. C.; Carpenter A. E.; Pluim J. P. W.; Singh S.; Veta M. Capturing Single-Cell Phenotypic Variation via Unsupervised Representation Learning. Int. Conf. Med. Imaging with Deep Learn. 2018, 315–325. [PMC free article] [PubMed] [Google Scholar]
- Bray M. A.; Fraser A. N.; Hasaka T. P.; Carpenter A. E. Workflow and Metrics for Image Quality Control in Large-Scale High-Content Screens. J. Biomol. Screening 2012, 17, 266–274. 10.1177/1087057111420292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. A.; Carpenter A. E.. Quality Control for High-Throughput Imaging Experiments Using Machine Learning in Cellprofiler. In Methods Mol. Biol.; Humana Press Inc., 2018; Vol. 1683, pp 89–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Dimension Reduction for High-Dimensional Data. In Statistical methods in molecular biology; Springer, 2010; pp 417–434. [DOI] [PubMed] [Google Scholar]
- Leek J. T.; Scharpf R. B.; Bravo H. C.; Simcha D.; Langmead B.; Johnson W. E.; Geman D.; Baggerly K.; Irizarry R. A. Tackling the Widespread and Critical Impact of Batch Effects in High-Throughput Data. Nat. Rev. Genet. 2010, 11, 733. 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeder C.; Heigwer F.; Boutros M. HTSvis: A Web App for Exploratory Data Analysis and Visualization of Arrayed High-Throughput Screens. Bioinformatics 2017, 33, 2960–2962. 10.1093/bioinformatics/btx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi A. S.; Imon A. H. M. R.; Werner M. Detection of Outliers. Wiley Interdiscip. Rev. Comput. Stat. 2009, 1, 57–70. 10.1002/wics.6. [DOI] [Google Scholar]
- Perkel J. M. Data Visualization Tools Drive Interactivity and Reproducibility in Online Publishing. Nature 2018, 554, 133–134. 10.1038/d41586-018-01322-9. [DOI] [PubMed] [Google Scholar]
- Peng R. D. Reproducible Research in Computational Science. Science (Washington, DC, U. S.) 2011, 334, 1226–1227. 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L.; Alexander M. R.; Winkler D. A.. Materiomics: A Toolkit for Developing New Biomaterials. In Tissue Engineering; Elsevier, 2014; pp 253–281. [Google Scholar]
- Pérez-Luna V. H.; Horbett T. A.; Ratner B. D. Developing Correlations between Fibrinogen Adsorption and Surface Properties Using Multivariate Statistics. J. Biomed. Mater. Res. 1994, 28, 1111–1126. 10.1002/jbm.820281002. [DOI] [PubMed] [Google Scholar]
- Taylor M.; Urquhart A. J.; Anderson D. G.; Langer R.; Davies M. C.; Alexander M. R. Partial Least Squares Regression as a Powerful Tool for Investigating Large Combinatorial Polymer Libraries. Surf. Interface Anal. 2009, 41, 127–135. 10.1002/sia.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A.; Consonni V.; Todeschini R. Molecular Descriptors. Handb. Comput. Chem. 2017, 2065–2093. 10.1007/978-3-319-27282-5_51. [DOI] [Google Scholar]
- Winkler D. A. Sparse QSAR Modelling Methods for Therapeutic and Regenerative Medicine. J. Comput.-Aided Mol. Des. 2018, 32, 497–509. 10.1007/s10822-018-0106-1. [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment Integrated Computer-Aided Molecular Design Platform; Chemical Computing Group, 2020. https://www.chemcomp.com/Products.htm (accessed 05-01-2020).
- Karelson M.; Maran U.; Wang Y.; Katritzky A. R. QSPR and QSAR Models Derived Using Large Molecular Descriptor Spaces. A Review of Codessa Applications. Collect. Czech. Chem. Commun. 1999, 64, 1551–1571. 10.1135/cccc19991551. [DOI] [Google Scholar]
- Yap C. W. PaDEL-descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Pastor M.; Cruciani G.; McLay I.; Pickett S.; Clementi S. GRid-INdependent Descriptors (GRIND): A Novel Class of Alignment-Independent Three-Dimensional Molecular Descriptors. J. Med. Chem. 2000, 43, 3233–3243. 10.1021/jm000941m. [DOI] [PubMed] [Google Scholar]
- Mauri A.; Consonni V.; Pavan M.; Todeschini R. Dragon Software: An Easy Approach to Molecular Descriptor Calculations. MATCH Communications in Mathematical and in Computer Chemistry 2006, 56, 237–248. [Google Scholar]
- Lewis D. R.; Kholodovych V.; Tomasini M. D.; Abdelhamid D.; Petersen L. K.; Welsh W. J.; Uhrich K. E.; Moghe P. V. In Silico Design of Anti-Atherogenic Biomaterials. Biomaterials 2013, 34, 7950–7959. 10.1016/j.biomaterials.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygd H. C.; Forsmark K. D.; Bratlie K. M. Altering in Vivo Macrophage Responses with Modified Polymer Properties. Biomaterials 2015, 56, 187–197. 10.1016/j.biomaterials.2015.03.042. [DOI] [PubMed] [Google Scholar]
- Anselme K.; Bigerelle M.; Noel B.; Dufresne E.; Judas D.; Iost A.; Hardouin P. Qualitative and Quantitative Study of Human Osteoblast Adhesion on Materials with Various Surface Roughnesses. J. Biomed. Mater. Res. 2000, 49, 155–166. . [DOI] [PubMed] [Google Scholar]
- Wickens D.; Lynch S.; West G.; Kelly P.; Verran J.; Whitehead K. A. Quantifying the Pattern of Microbial Cell Dispersion, Density and Clustering on Surfaces of Differing Chemistries and Topographies Using Multifractal Analysis. J. Microbiol. Methods 2014, 104, 101–108. 10.1016/j.mimet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Robotti F.; Bottan S.; Fraschetti F.; Mallone A.; Pellegrini G.; Lindenblatt N.; Starck C.; Falk V.; Poulikakos D.; Ferrari A. A Micron-Scale Surface Topography Design Reducing Cell Adhesion to Implanted Materials. Sci. Rep. 2018, 8, 10887. 10.1038/s41598-018-29167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer J.; Van Blitterswijk C. A.. Materiomics: High-Throughput Screening of Biomaterial Properties; Cambridge University Press, 2013. [Google Scholar]
- Lever J.; Krzywinski M.; Altman N. Points of Significance: Principal Component Analysis. Nat. Methods 2017, 14, 641–642. 10.1038/nmeth.4346. [DOI] [Google Scholar]
- Burges C. J. C. Dimension Reduction: A Guided Tour. FNT in Machine Learning 2010, 2, 275–365. 10.1561/2200000002. [DOI] [Google Scholar]
- Sorzano C. O. S.; Vargas J.; Montano A. P.. A Survey of Dimensionality Reduction Techniques. arXiv Prepr. arXiv1403.2877 2014. [Google Scholar]
- Van Der Maaten L.; Postma E.; Van den Herik J. Dimensionality Reduction: A Comparative. J. Mach Learn Res. 2009, 10, 66–71. [Google Scholar]
- Altman N.; Krzywinski M. Points of Significance: Clustering. Nat. Methods 2017, 14, 545–546. 10.1038/nmeth.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P. J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- Jain A. K.; Murty M. N.; Flynn P. J. Data Clustering: A Review. ACM Comput. Surv. 1999, 31, 264–323. 10.1145/331499.331504. [DOI] [Google Scholar]
- Xu R.; Wunsch D. C. Clustering Algorithms in Biomedical Research: A Review. IEEE Rev. Biomed. Eng. 2010, 3, 120–154. 10.1109/RBME.2010.2083647. [DOI] [PubMed] [Google Scholar]
- Lever J.; Krzywinski M.; Altman N. Points of Significance: Model Selection and Overfitting. Nat. Methods 2016, 13, 703–704. 10.1038/nmeth.3968. [DOI] [Google Scholar]
- Krzywinski M.; Altman N. Points of Significance: Classification and Regression Trees. Nat. Methods 2017, 14, 757–758. 10.1038/nmeth.4370. [DOI] [Google Scholar]
- Altman N.; Krzywinski M. Points of Significance: Ensemble Methods: Bagging and Random Forests. Nat. Methods 2017, 14, 933–934. 10.1038/nmeth.4438. [DOI] [Google Scholar]
- Cios K. J.; Pedrycz W.; Swiniarski R. W.. Data Mining and Knowledge Discovery. In Data mining methods for knowledge discovery; Springer, 1998; pp 1–26. [Google Scholar]
- Burges C. J. C. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discovery 1998, 2, 121–167. 10.1023/A:1009715923555. [DOI] [Google Scholar]
- Afanador N. L.; Smolinska A.; Tran T. N.; Blanchet L. Unsupervised Random Forest: A Tutorial with Case Studies. J. Chemom. 2016, 30, 232–241. 10.1002/cem.2790. [DOI] [Google Scholar]
- Jain A. K.; Mao J.; Mohiuddin K. M. Artificial Neural Networks: A Tutorial. Computer (Long. Beach. Calif). 1996, 31–44. [Google Scholar]
- James G.; Witten D.; Hastie T.; Tibshirani R.. An Introduction to Statistical Learning; Springer, 2013; Vol. 112. [Google Scholar]
- Krzywinski M.; Altman N.. Points of Significance: Analysis of Variance and Blocking; Nature Publishing Group, 2014; pp 699–700. [DOI] [PubMed] [Google Scholar]
- Stahle L.; Wold S. Multivariate Analysis of Variance (MANOVA). Chemom. Intell. Lab. Syst. 1990, 9, 127–141. 10.1016/0169-7439(90)80094-M. [DOI] [Google Scholar]
- Krzywinski M.; Altman N. Points of Significance: Significance, P Values and t-Tests. Nat. Methods 2013, 10, 1041–1042. 10.1038/nmeth.2698. [DOI] [PubMed] [Google Scholar]
- Noble W. S. How Does Multiple Testing Correction Work?. Nat. Biotechnol. 2009, 27, 1135. 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Lutz M.; Ascher D.. Learning Python; O’Reilly Media, 1999. [Google Scholar]
- McKinney W.Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython; O’Reilly Media, Inc., 2012. [Google Scholar]
- Wickham H.; Grolemund G. R.. R for Data Science; O’Reilly Media, 2016. [Google Scholar]
- Voit E. O.; Qi Z.; Miller G. W. Steps of Modeling Complex Biological Systems. Pharmacopsychiatry 2008, 41, S78–S84. 10.1055/s-2008-1080911. [DOI] [PubMed] [Google Scholar]
- Baker R. E.; Peña J.-M.; Jayamohan J.; Jérusalem A. Mechanistic Models versus Machine Learning, a Fight Worth Fighting for the Biological Community?. Biol. Lett. 2018, 14, 20170660. 10.1098/rsbl.2017.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern J.; Pitt-Francis J.; Whiteley J.; Stokeley D.; Kobashi H.; Nobes R.; Kadooka Y.; Gavaghan D. Multi-Scale Computational Modelling in Biology and Physiology. Prog. Biophys. Mol. Biol. 2008, 96, 60–89. 10.1016/j.pbiomolbio.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F.; Megason S. G. Abstracting the Principles of Development Using Imaging and Modeling. Integr. Biol. 2015, 7, 633–642. 10.1039/c5ib00025d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholma J.; Schivo S.; Camacho R. A. U.; van de Pol J.; Karperien M.; Post J. N. Biological Networks 101: Computational Modeling for Molecular Biologists. Gene 2014, 533, 379–384. 10.1016/j.gene.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Brodland G. W.How Computational Models Can Help Unlock Biological Systems. In Seminars in cell & developmental biology; Elsevier, 2015; Vol. 47, pp 62–73. [DOI] [PubMed] [Google Scholar]
- Mogilner A.; Wollman R.; Marshall W. F. Quantitative Modeling in Cell Biology: What Is It Good For?. Dev. Cell 2006, 11, 279–287. 10.1016/j.devcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Geris L.; Gomez-Cabrero D.. Uncertainty in Biology; Springer, 2016. [Google Scholar]
- Ashyraliyev M.; Fomekong-Nanfack Y.; Kaandorp J. A.; Blom J. G. Systems Biology: Parameter Estimation for Biochemical Models. FEBS J. 2009, 276, 886–902. 10.1111/j.1742-4658.2008.06844.x. [DOI] [PubMed] [Google Scholar]
- Chou I.-C.; Voit E. O. Recent Developments in Parameter Estimation and Structure Identification of Biochemical and Genomic Systems. Math. Biosci. 2009, 219, 57–83. 10.1016/j.mbs.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schepdael A.; Carlier A.; Geris L.. Sensitivity Analysis by Design of Experiments. In Uncertainty in Biology; Springer, 2016; pp 327–366. [Google Scholar]
- Carlier A.; Chai Y. C.; Moesen M.; Theys T.; Schrooten J.; Van Oosterwyck H.; Geris L. Designing Optimal Calcium Phosphate Scaffold-Cell Combinations Using an Integrative Model-Based Approach. Acta Biomater. 2011, 7, 3573–3585. 10.1016/j.actbio.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Society R.Machine Learning: The Power and Promise of Computers That Learn by Example: An Introduction; Royal Society, 2017.
- Rasi Ghaemi S.; Delalat B.; Gronthos S.; Alexander M. R.; Winkler D. A.; Hook A. L.; Voelcker N. H. High-Throughput Assessment and Modeling of a Polymer Library Regulating Human Dental Pulp-Derived Stem Cell Behavior. ACS Appl. Mater. Interfaces 2018, 10, 38739–38748. 10.1021/acsami.8b12473. [DOI] [PubMed] [Google Scholar]
- Chen D.; Sarkar S.; Candia J.; Florczyk S. J.; Bodhak S.; Driscoll M. K.; Simon Jr C. G.; Dunkers J. P.; Losert W. Machine Learning Based Methodology to Identify Cell Shape Phenotypes Associated with Microenvironmental Cues. Biomaterials 2016, 104, 104–118. 10.1016/j.biomaterials.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostam H. M.; Reynolds P. M.; Alexander M. R.; Gadegaard N.; Ghaemmaghami A. M. Image Based Machine Learning for Identification of Macrophage Subsets. Sci. Rep. 2017, 7, 3521. 10.1038/s41598-017-03780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B.; Lin M.; Huang G.; Li Y.; Ji B.; Genin G. M.; Deshpande V. S.; Lu T. J.; Xu F. Cellular Mechanosensing of the Biophysical Microenvironment: A Review of Mathematical Models of Biophysical Regulation of Cell Responses. Phys. Life Rev. 2017, 22, 88–119. 10.1016/j.plrev.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A. Cell Adhesion Mechanosensitivity, an Active Biological Process: Comment on” Cellular Mechanosensing of the Biophysical Microenvironment: A Review of the Mathematical Models of Biophysical Regulation of Cell Responses” by Bo Cheng et Al. Phys. Life Rev. 2017, 22, 123. 10.1016/j.plrev.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Wu J.; LeDuc P.; Steward Jr R. How Can We Predict Cellular Mechanosensation?: Comment on “Cellular Mechanosensing of the Biophysical Microenvironment: A Review of Mathematical Models of Biophysical Regulation of Cell Responses” by Bo Cheng et. Al. Phys. Life Rev. 2017, 22, 120. 10.1016/j.plrev.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spill F.; Zaman M. H. Multiscale Dynamics of the Biophysical and Biochemical Microenvironment: Comment on” Cellular Mechanosensing of the Biophysical Microenvironment: A Review of Mathematical Models of Biophysical Regulation of Cell Responses” by Bo Cheng et Al. Phys. Life Rev. 2017, 22, 127. 10.1016/j.plrev.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Cheng B.; Lin M.; Huang G.; Li Y.; Ji B.; Genin G. M.; Deshpande V. S.; Lu T. J.; Xu F. Energetics: An Emerging Frontier in Cellular Mechanosensing: Reply to Comments on “Cellular Mechanosensing of the Biophysical Microenvironment: A Review of Mathematical Models of Biophysical Regulation of Cell Responses. Phys. Life Rev. 2017, 22, 130–135. 10.1016/j.plrev.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Abhilash A. S.; Baker B. M.; Trappmann B.; Chen C. S.; Shenoy V. B. Remodeling of Fibrous Extracellular Matrices by Contractile Cells: Predictions from Discrete Fiber Network Simulations. Biophys. J. 2014, 107, 1829–1840. 10.1016/j.bpj.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers R. F. M.; Rens E. G.; LaValley D. J.; Reinhart-King C. A.; Merks R. M. H. Mechanical Cell-Matrix Feedback Explains Pairwise and Collective Endothelial Cell Behavior in Vitro. PLoS Comput. Biol. 2014, 10, e1003774 10.1371/journal.pcbi.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.; Spill F.; Zaman M. H. A Computational Model of YAP/TAZ Mechanosensing. Biophys. J. 2016, 110, 2540–2550. 10.1016/j.bpj.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.; Tang L.; Gao J. Computational Modeling of Phagocyte Transmigration for Foreign Body Responses to Subcutaneous Biomaterial Implants in Mice. BMC Bioinf. 2016, 17, 111. 10.1186/s12859-016-0947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. J.; Schwarz U. S. Dynamics of Cell Shape and Forces on Micropatterned Substrates Predicted by a Cellular Potts Model. Biophys. J. 2014, 106, 2340–2352. 10.1016/j.bpj.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. J.; Schwarz U. S. Modeling Cell Shape and Dynamics on Micropatterns. Cell Adh. Migr. 2016, 10, 516–528. 10.1080/19336918.2016.1148864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. J.; Schwarz U. S. Dynamics of Cell Ensembles on Adhesive Micropatterns: Bridging the Gap between Single Cell Spreading and Collective Cell Migration. PLoS Comput. Biol. 2016, 12, e1004863 10.1371/journal.pcbi.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert P. J.; Schwarz U. S. Optimizing Micropattern Geometries for Cell Shape and Migration with Genetic Algorithms. Integr. Biol. 2016, 8, 741–750. 10.1039/C6IB00061D. [DOI] [PubMed] [Google Scholar]
- Ziebert F.; Aranson I. S. Computational Approaches to Substrate-Based Cell Motility. npj Comput. Mater. 2016, 2, 16019. 10.1038/npjcompumats.2016.19. [DOI] [Google Scholar]
- Pedraza E.; Coronel M. M.; Fraker C. A.; Ricordi C.; Stabler C. L. Preventing Hypoxia-Induced Cell Death in Beta Cells and Islets via Hydrolytically Activated, Oxygen-Generating Biomaterials. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 4245–4250. 10.1073/pnas.1113560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhas V.; Guyot Y.; Kerckhofs G.; Chai Y. C.; Geris L. Computational Modelling of Local Calcium Ions Release from Calcium Phosphate-Based Scaffolds. Biomech. Model. Mechanobiol. 2017, 16, 425–438. 10.1007/s10237-016-0827-9. [DOI] [PubMed] [Google Scholar]
- Manhas V. In Silico Modeling of Bone Formation under the Influence of Calcium Phosphate-Based Biomaterials and Osteochondrogenic Growth Factors; Université de Liège, Liège: Belgique, 2017. [Google Scholar]
- Bidan C. M.; Kommareddy K. P.; Rumpler M.; Kollmannsberger P.; Fratzl P.; Dunlop J. W. C. Geometry as a Factor for Tissue Growth: Towards Shape Optimization of Tissue Engineering Scaffolds. Adv. Healthcare Mater. 2013, 2, 186–194. 10.1002/adhm.201200159. [DOI] [PubMed] [Google Scholar]
- Bhat S.; Kumar A. Biomaterials and Bioengineering Tomorrow’s Healthcare. Biomatter 2013, 3, e24717 10.4161/biom.24717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban M. A. Translational Biomaterials—the Journey from the Bench to the Market—Think ‘Product’. Curr. Opin. Biotechnol. 2016, 40, 31–34. 10.1016/j.copbio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Hoffman T.; Khademhosseini A.; Langer R. S. Chasing the Paradigm: Clinical Translation of 25 Years of Tissue Engineering. Tissue Eng., Part A 2019, 25, 679–687. 10.1089/ten.tea.2019.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orabi H.; Bouhout S.; Morissette A.; Rousseau A.; Chabaud S.; Bolduc S. Tissue Engineering of Urinary Bladder and Urethra: Advances from Bench to Patients. Sci. World J. 2013, 2013, 154564. 10.1155/2013/154564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place E. S.; Evans N. D.; Stevens M. M. Complexity in Biomaterials for Tissue Engineering. Nat. Mater. 2009, 8, 457. 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- Mao A. S.; Mooney D. J. Regenerative Medicine: Current Therapies and Future Directions. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 14452–14459. 10.1073/pnas.1508520112. [DOI] [PMC free article] [PubMed] [Google Scholar]