Abstract
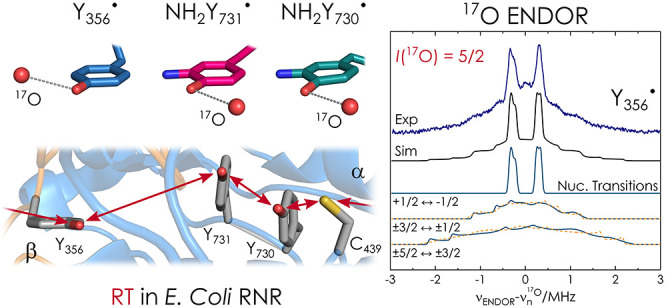
The role of water in biological proton-coupled electron transfer (PCET) is emerging as a key for understanding mechanistic details at atomic resolution. Here we demonstrate 17O high-frequency electron–nuclear double resonance (ENDOR) in conjunction with H217O-labeled protein buffer to establish the presence of ordered water molecules at three radical intermediates in an active enzyme complex, the α2β2E. coli ribonucleotide reductase. Our data give unambiguous evidence that all three, individually trapped, intermediates are hyperfine coupled to one water molecule with Tyr-O···17O distances in the range 2.8–3.1 Å. The availability of this structural information will allow for quantitative models of PCET in this prototype enzyme. The results also provide a spectroscopic signature for water H-bonded to a tyrosyl radical.
Water is no longer known as just the solvent in which biochemical reactions take place but has been recognized as an essential player in these reactions.1 Of particular interest is water involvement in electron transfer processes,2−5 its action as a proton wire6−8 or its role in proton-coupled electron transfer (PCET).9−12 The identification of internal water in proteins can be achieved by X-ray diffraction.13−15 However, the crystallization of transient protein complexes is difficult. One key approach for detection of water in biological systems has been the use of 17O-enriched water in conjunction with magnetic resonance spectroscopy.16−21 Among these methods, electron paramagnetic resonance (EPR) can take advantage of high selectivity, as it detects nuclei only in the ligand sphere (r ≲ 1.5 nm)22 of paramagnetic centers.
EPR-based 17O hyperfine (hf) spectroscopy has been established for the detection of water binding to transition-metal ions, where the oxygen usually coordinates to the ion and large hyperfine couplings (several MHz) can be observed.23−25 However, the most common water coordination motif to biological radicals occurs via H-bond interactions. The hf coupling to 17O is diminished in comparison to the metal ion coordination, due to a longer interspin distance. In addition, the small 17O gyromagnetic ratio (γH/γ17O ≈ 7.4)26 and high nuclear spin (I = 5/2) have rendered the 17O hf coupling difficult to resolve.
Here we illustrate that high-frequency (94 and 263 GHz) electron–nuclear double resonance (ENDOR) spectroscopy can detect the 17O signal of ordered water molecules at an H-bond distance to radical intermediates in E. coli ribonucleotide reductase (RNR). The enzyme uses a long-range (32 Å) radical transfer (RT) to initiate nucleotide reduction (Scheme 1).27 Three tyrosines (Y356, Y731, and Y730) are essential pathway residues, which form transient intermediates in the active complex α2β2, consisting of the two homodimeric subunits α2 and β2.13,27 Water has been observed only crystallographically in inactive α2s without β2.13,14,28,29 Using site-selectively inserted tyrosine analogues to trap Y intermediates,30 our previous 1H/2H ENDOR and DFT studies10,11,31 revealed H-bonds attributed to water molecules and proposed a role of water in RT. However, all active sites of proteins have exchangeable protons, and thus alternative interpretations to our water proposal were possible. Recently, a cryo-EM structure of α2β2 was reported but the resolution was insufficient for water observation.32 Since our original proposals, studies using photo-RNRs and MD simulations implying waters in α2β2 have appeared.33,34 However, water has never been directly detected.
Scheme 1. Current PCET Model of the 32 Å (Y122 to C439) RT in E. coli RNR27,32,
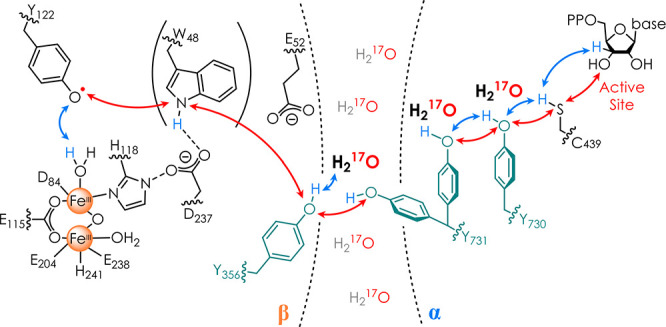
Redox-active tyrosines 356, 731, and 730 are shown in cyan, electron transfer steps as red arrows, and proton transfer steps as blue arrows. Water molecules revealed in this study in respective site-selective mutants are shown in boldface.
Therefore, we explored the capability of H217O ENDOR spectroscopy by exchanging the RNR buffer with H217O. α2β2-Y356• was generated by a 2,3,5-F3Y122• mutation in β2,35 whereas radicals at Y731 and Y730 were trapped by replacing the respective residue with 3-aminotyrosine (NH2Y),36 leading to α2β2-NH2Y731• and α2β2-NH2Y730•. The individual variants were mixed with the complementary α2 or β2 protein, CDP as a substrate, and ATP as an effector. The reaction was then quenched after a few seconds inside EPR tubes. Details on the sample preparation are given in sections SI1 and SI2.
Figure 1 displays representative 94 GHz 17O Mims37 ENDOR spectra of the radical intermediates.
Figure 1.
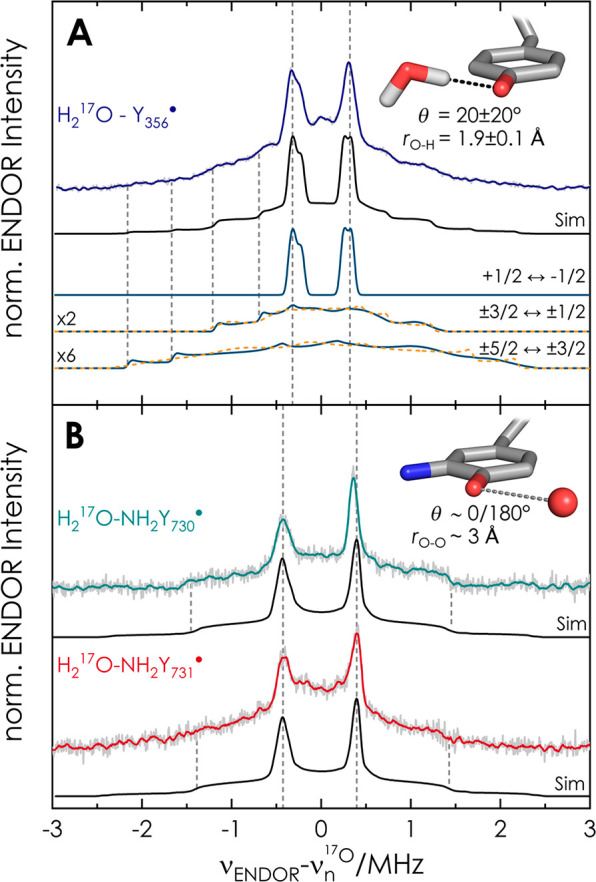
94 GHz 17O Mims37 ENDOR spectra of (A) the intermediate Y356• and (B) NH2Y731• and NH2Y730• at B0∥gy in the EPR line (T = 50 K, τMims = 390 ns). Acquisition time: 46 h (Y356•), 40 h (NH2Y731•), and 18 h (NH2Y730•). Y356• is from β2-F3Y122•/α2-Y730F, which gives the highest radical yield (section SI2). Experimental spectra are shown in gray, with a Savitzky–Golay filter (fourth-order polynomial, 20-point window) shown in color. Simulations used Easyspin38 (section SI1.6) with parameters given in Table 1 and section S3. Solid lines (teal) represent transitions among mI > 0 manifolds and dashed lines (orange) those among mI < 0 manifolds. The simulation does not distinguish between dihedral θ = 0°or θ = 180°.
Each spectrum shows a sharp doublet centered on the 17O Larmor frequency (19.3 MHz at 3.4 T), which can be assigned to the central spin transition (mI(17O) = +1/2 → −1/2) of one coupled 17O nucleus. As 17O is contained only in the water of the protein buffer, these sharp signals must arise from water molecules coupled to the radicals. Control experiments with only β2 protein confirmed that the signal is associated to the radicals generated in α2β2 (section SI3). The broad resonances at ±2.5 MHz are attributed to other nuclear transitions of the I = 5/2 spin system, broadened by nuclear quadrupole coupling (Figure 1A). Additionally, we note asymmetry of the doublet, which arises from second-order effects of the quadrupole coupling (section SI4). A comparison of the ENDOR spectra at the low (B0∥gx) and high-field (B0∥gz) edges of the EPR line (section SI5) indicates an almost isotropic hf coupling, with the dipolar contribution dominating the line width of the central doublet. The 17O ENDOR spectra could be simulated with one 17O nucleus, from which the asymmetry of the central peaks resulted using full diagonalization of the spin Hamiltonian (Figure 1 and section SI8). Parameters are given in Table 1 and section SI3. The spectra of Y356• and NH2Y731• additionally contain signals close to the Larmor frequency not reproduced in the simulations, which likely originate from second-sphere water molecules at the subunit interface. Additional broadening is also observed, particularly at NH2Y731•. It might be caused by conformational distribution of this residue, which was found to have flexibility.39,40,33
Table 1. Simulation and DFT Parameters for 17O and 1H hf Couplings of Water in RNR Intermediatesa.
| Y356• sim/DFTsmall | NH2Y731• sim | NH2Y730• sim/DFTlargeb | |
|---|---|---|---|
| Ax (17O) | 0.43/0.19 | 0.70 | 0.65/0.24 |
| Ay (17O) | 0.66/0.59 | 0.84 | 0.80/0.6 |
| Az (17O) | 0.70/0.65 | 0.89 | 0.89/0.6 |
| A(H1) | 6.231/7.4 | ≲2.5b | 2.7b/4.2 |
| ρ(17O)c (%) | 0.05 | 0.03 | |
| rO···17O (Å) | 2.9 ± 0.1 | ∼3.0 | ∼3.0 |
To rationalize the coupling, we began with a DFT-optimized small model (25 atoms, details in section SI1) of Y356•, as previous ENDOR spectra revealed 1H couplings consistent with one water at the H-bond distance rO–H ≈ 1.8 Å.31 The 17O coupling from this model was Amax(17O) ≈ 1 MHz, slightly exceeding the present experimental value of 0.6 ± 0.05 MHz. To optimize the model, we computed dihedral θ (C3–C4–O···H) and distance scans for 17O couplings, including the quadrupole tensor and the relative energies (section SI6). The DFT equilibrium distance always resulted in rO–H ≈ 1.8 Å. We found that hf couplings and energies vary significantly with θ, while the quadrupole coupling is less affected (Figure S9A–C). Axyz values of ≲1 MHz are found for θ in the range ≲±30° (or equivalently 150° ≲ θ ≲ 240°): i.e., close to the ring plane. Water coordination in the ring plane also results in minimal relative energies (Figure S9B). Importantly, predicted spin densities on 17O are <0.1% but are sufficient for producing a marked 17O isotropic splitting. The spin density transfer or spin polarization is likely related to the H-bond nature. A distance scan for the optimized dihedral of +20° predicts Amax(17O) in the range 0.75–0.56 MHz (Figure S10A) for rO–H ≈ 1.8–2.0 Å. Consideration of the DFT-predicted 1H couplings (Figure S10B) and comparison with the experimental values31 of ∼6.2 (H1) and ∼1.6 MHz (H2) indicates that the water is located at rTyr-O···17O = 2.9 ± 0.1 Å, corresponding to an rO–H value of 1.9 ± 0.1 Å. Notably, the DFT-predicted dipolar coupling (T|| ≈ 0.3 MHz, section SI6) is consistent with the point-dipole model and the aforementioned broadening of the sharp peaks.
Analogous DFT calculations were performed on the isolated amino tyrosyl NH2Y•.10,11,36 We observed a trend for the 17O hf coupling in the dihedral and distance scans (section SI7) very similar to the Y• model. The calculation predicts that Aiso(17O) of NH2Y• is slightly larger (10–15%) than that of Y• at similar Tyr-O···17O distances and orientations, which could explain the experimental observation. The amino group introduces an asymmetry in the radical, and the energetically most favored water orientation is found at the opposite side of the amino group (Figure S11B). Nevertheless, this small model could not account simultaneously for the 17O and 1H couplings observed for these two intermediates (Figure S12). As noted in a previous gx calculation,10 the coordination of the water molecule to NH2Y•s is influenced by the surrounding second-sphere residues, as these two intermediates are buried in α2β2 (Scheme 1).
Having established that at least one water molecule is hf-coupled to each of the three intermediates, we examine their current molecular models in light of this finding. First, we consider the radical site Y356• (Scheme 1). To explain the unprecedented gx value of Y356• (gx = 2.0062), we previously proposed that two almost equivalent waters might be simultaneously bonded to Y356•.31 While the present results are most consistent with the distance and orientation proposed for one water, the 94 GHz 17O ENDOR spectra (Figure 1A) cannot resolve a second water. We note that the spectral line shape and 17O hf coupling in Figure 1A are conserved in other RNR constructs that generate Y356• (section SI8), including the F3Y122•/E52Q-β2 double mutant used to solve a recent cryo-EM structure.43
To gain spectral resolution, we recorded 17O ENDOR spectra of Y356• at 263 GHz/9.4 T (Figure 2).44−46
Figure 2.
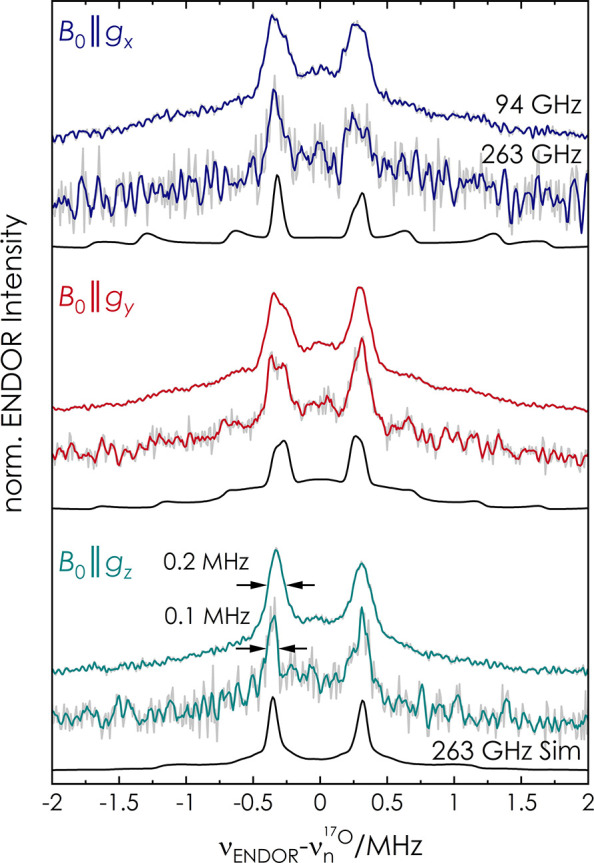
Comparison of 94 and 263 GHz Mims ENDOR of Y356• at the three canonical positions in the EPR line. Total acquisition time for 263 GHz (T = 20 K): 18 h (B0∥gx), 10 h (B0∥gy), and 11 h (B0∥gz). Experimental spectra are shown in gray, with a Savitzky–Golay filter (fourth-order polynomial, 10 point window) in color. Simulations of 263 GHz spectra are in black with parameters as for 94 GHz (see Table 1 and Table S4).
The results illustrate that the line width of the central doublet substantially narrows, particularly at B0∥gz (Figure 2). Despite the narrowing, a factor of approximately 2 from 94 to 263 GHz, we cannot discern two distinct 17O contributions. Simulations of the 263 GHz spectra with the same parameters used at 94 GHz reproduce the line narrowing and support the analysis at 94 GHz. The lack of evidence for a second, almost equivalent water H-bonded to Y356• strongly suggests that the two-water model has become very unlikely and alternative explanations for the shifted gx value of Y356• will have to be examined. The precise location of second-sphere residues might play a role,12 which will require further experimental and computational investigation.
For the radical intermediates in the subunit α, a previous combined ENDOR/DFT model of NH2Y730• proposed a water molecule coordinated in plane at a distance rNH2Y730-O···17O ≈ 3.0 Å.10 The present results are consistent with this model and provide direct evidence for this postulated water in the enzyme complex α2β2-NH2Y730•. The DFT-predicted hf parameters (DFTlarge) for this large model (140 atoms) are reported in Table 1, and the model is displayed in section SI9.
Finally, for α2β2-NH2Y731•, large-scale (215 atoms) DFT calculations previously proposed three models of the trapped intermediate (section SI10). Among these models, only one (model 3, Figure S15) contained a water molecule at an H-bond distance. The DFT-predicted 17O hf couplings of model 3 (∼2.5 MHz), however, largely exceed the present experimental values (Table S5). However, this DFT model did not include residues from the β subunit, which we now know are close to this residue in the active complex.43 Therefore, the model will require further refinement. Nevertheless, the present results give evidence for a water molecule coordinated almost in the plane of NH2Y731•.
In conclusion, we have reported the capability of 17O high-frequency ENDOR to detect water H-bonded to tyrosyl radicals. The spectroscopic approach led to the first detection of ordered water molecules at three trapped radicals proposed to be representative of Y• intermediates in the PCET of E. coli RNR. These results verify previous hypotheses on the presence and role of water in the RNR mechanism and provide a new starting point for computational studies. Knowledge of this 17O signature will also be generally useful for many other biological systems, in which tyrosyl radicals are involved.
Acknowledgments
We thank Brandon Greene (UCSB) for his help in producing protein samples in 17O-labeled buffer. We also thank Markus Hiller (MPIBPC) for discussions about DFT modeling, Igor Tkach (MPIBPC) for assistance with instrumentation, and Andreas Meyer (MPIBPC) for discussions and suggestions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c01359.
Experimental procedure, radical yield determination, ENDOR spectra of Y122• and F3Y122•, ENDOR spectra of I = 5/2 nuclei, orientation-selective ENDOR spectra, DFT models of Y• and NH2Y•, 17O Y356• spectra of different mutants, previous large DFT models of α2β2-NH2Y730• and α2β2-NH2Y731• (PDF)
This work was funded by the Max Planck Society and the DFG priority program SPP1601 to M.B. as well as an NIH grant 29595 to J.S.
The authors declare no competing financial interest.
Supplementary Material
References
- Ball P. Water as an active constituent in cell biology. Chem. Rev. 2008, 108 (1), 74–108. 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- Tezcan F. A.; Crane B. R.; Winkler J. R.; Gray H. B. Electron tunneling in protein crystals. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (9), 5002–6. 10.1073/pnas.081072898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amsterdam I. M.; Ubbink M.; Einsle O.; Messerschmidt A.; Merli A.; Cavazzini D.; Rossi G. L.; Canters G. W. Dramatic modulation of electron transfer in protein complexes by crosslinking. Nat. Struct. Biol. 2002, 9 (1), 48–52. 10.1038/nsb736. [DOI] [PubMed] [Google Scholar]
- Lin J.; Balabin I. A.; Beratan D. N. The nature of aqueous tunneling pathways between electron-transfer proteins. Science 2005, 310 (5752), 1311–1313. 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Lande A.; Marti S.; Parisel O.; Moliner V. Long distance electron-transfer mechanism in peptidylglycine alpha-hydroxylating monooxygenase: a perfect fitting for a water bridge. J. Am. Chem. Soc. 2007, 129 (38), 11700–7. 10.1021/ja070329l. [DOI] [PubMed] [Google Scholar]
- Luecke H.; Schobert B.; Richter H. T.; Cartailler J. P.; Lanyi J. K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999, 291 (4), 899–911. 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Sass H. J.; Buldt G.; Gessenich R.; Hehn D.; Neff D.; Schlesinger R.; Berendzen J.; Ormos P. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature 2000, 406 (6796), 649–53. 10.1038/35020607. [DOI] [PubMed] [Google Scholar]
- Linke K.; Ho F. M. Water in Photosystem II: structural, functional and mechanistic considerations. Biochim. Biophys. Acta, Bioenerg. 2014, 1837 (1), 14–32. 10.1016/j.bbabio.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Saito K.; Shen J. R.; Ishida T.; Ishikita H. Short hydrogen bond between redox-active tyrosine Y(Z) and D1-His190 in the photosystem II crystal structure. Biochemistry 2011, 50 (45), 9836–44. 10.1021/bi201366j. [DOI] [PubMed] [Google Scholar]
- Argirevic T.; Riplinger C.; Stubbe J.; Neese F.; Bennati M. ENDOR spectroscopy and DFT calculations: evidence for the hydrogen-bond network within α2 in the PCET of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2012, 134 (42), 17661–70. 10.1021/ja3071682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick T. U.; Lee W.; Kossmann S.; Neese F.; Stubbe J.; Bennati M. Hydrogen bond network between amino acid radical intermediates on the proton-coupled electron transfer pathway of E. coli α2 ribonucleotide reductase. J. Am. Chem. Soc. 2015, 137 (1), 289–98. 10.1021/ja510513z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohiwal A.; Neese F.; Pantazis D. A. Microsolvation of the Redox-Active Tyrosine-D in Photosystem II: Correlation of Energetics with EPR Spectroscopy and Oxidation-Induced Proton Transfer. J. Am. Chem. Soc. 2019, 141 (7), 3217–3231. 10.1021/jacs.8b13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin U.; Eklund H. Structure of ribonucleotide reductase protein R1. Nature 1994, 370 (6490), 533–9. 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- Eriksson M.; Uhlin U.; Ramaswamy S.; Ekberg M.; Regnström K.; Sjöberg B.-M.; Eklund H. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure 1997, 5 (8), 1077–1092. 10.1016/S0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- Umena Y.; Kawakami K.; Shen J. R.; Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 2011, 473 (7345), 55–60. 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- Schmidt B.; McCracken J.; Ferguson-Miller S. A discrete water exit pathway in the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (26), 15539–42. 10.1073/pnas.2633243100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennati M.; Hertel M. M.; Fritscher J.; Prisner T. F.; Weiden N.; Hofweber R.; Sporner M.; Horn G.; Kalbitzer H. R. High-frequency 94 GHz ENDOR characterization of the metal binding site in wild-type Ras x GDP and its oncogenic mutant G12V in frozen solution. Biochemistry 2006, 45 (1), 42–50. 10.1021/bi051156k. [DOI] [PubMed] [Google Scholar]
- Potapov A.; Goldfarb D. The Mn(2+)-bicarbonate complex in a frozen solution revisited by pulse W-band ENDOR. Inorg. Chem. 2008, 47 (22), 10491–8. 10.1021/ic8011316. [DOI] [PubMed] [Google Scholar]
- McConnell I. L.; Grigoryants V. M.; Scholes C. P.; Myers W. K.; Chen P. Y.; Whittaker J. W.; Brudvig G. W. EPR-ENDOR characterization of (17O, 1H, 2H) water in manganese catalase and its relevance to the oxygen-evolving complex of photosystem II. J. Am. Chem. Soc. 2012, 134 (3), 1504–12. 10.1021/ja203465y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa A.; Malferrari M.; Lubitz W.; Venturoli G.; Mobius K.; Savitsky A. Local water sensing: water exchange in bacterial photosynthetic reaction centers embedded in a trehalose glass studied using multiresonance EPR. Phys. Chem. Chem. Phys. 2017, 19 (41), 28388–28400. 10.1039/C7CP03942E. [DOI] [PubMed] [Google Scholar]
- Rowlands L. J.; Marks A.; Sanderson J. M.; Law R. V. 17O NMR spectroscopy as a tool to study hydrogen bonding of cholesterol in lipid bilayers. Chem. Commun. 2020, 56 (92), 14499–14502. 10.1039/D0CC05466F. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Dechert S.; Dey S.; Hobartner C.; Bennati M. Measurement of Angstrom to Nanometer Molecular Distances with 19F Nuclear Spins by EPR/ENDOR Spectroscopy. Angew. Chem., Int. Ed. 2020, 59 (1), 373–379. 10.1002/anie.201908584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitsimring A. M.; Astashkin A. V.; Baute D.; Goldfarb D.; Caravan P. W-Band 17O Pulsed Electron-Nuclear Double Resonance Study of Gadolinium Complexes with Water. J. Phys. Chem. A 2004, 108 (35), 7318–7323. 10.1021/jp040306i. [DOI] [Google Scholar]
- Baute D.; Goldfarb D. The 17O hyperfine interaction in V17O(H217O)52+ and Mn(H217O)62+ determined by high field ENDOR aided by DFT calculations. J. Phys. Chem. A 2005, 109 (35), 7865–71. 10.1021/jp052132q. [DOI] [PubMed] [Google Scholar]
- Rapatskiy L.; Cox N.; Savitsky A.; Ames W. M.; Sander J.; Nowaczyk M. M.; Rogner M.; Boussac A.; Neese F.; Messinger J.; Lubitz W. Detection of the water-binding sites of the oxygen-evolving complex of Photosystem II using W-band 17O electron-electron double resonance-detected NMR spectroscopy. J. Am. Chem. Soc. 2012, 134 (40), 16619–34. 10.1021/ja3053267. [DOI] [PubMed] [Google Scholar]
- Tiesinga E.; Mohr P. J.; Newell D. B.; Taylor B. N.. CODATA Recommended Values of the Fundamental Physical Constants: 2018; https://physics.nist.gov/cuu/Constants/index.html (accessed 2021-01-15). [DOI] [PMC free article] [PubMed]
- Greene B. L.; Kang G.; Cui C.; Bennati M.; Nocera D. G.; Drennan C. L.; Stubbe J. Ribonucleotide Reductases: Structure, Chemistry, and Metabolism Suggest New Therapeutic Targets. Annu. Rev. Biochem. 2020, 89, 45–75. 10.1146/annurev-biochem-013118-111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K.; Uhlin U.; Stubbe J. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J. Am. Chem. Soc. 2010, 132 (24), 8385–97. 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan E. C.; Seyedsayamdost M. R.; Uhlin U.; Stubbe J. Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases. J. Am. Chem. Soc. 2011, 133 (24), 9430–40. 10.1021/ja201640n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan E. C.; Nocera D. G.; Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc. Chem. Res. 2013, 46 (11), 2524–35. 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick T. U.; Ravichandran K. R.; Stubbe J.; Kasanmascheff M.; Bennati M. Spectroscopic Evidence for a H Bond Network at Y356 Located at the Subunit Interface of Active E. coli Ribonucleotide Reductase. Biochemistry 2017, 56 (28), 3647–3656. 10.1021/acs.biochem.7b00462. [DOI] [PubMed] [Google Scholar]
- Kang G.; Taguchi A. T.; Stubbe J.; Drennan C. L. Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex. Science 2020, 368 (6489), 424–427. 10.1126/science.aba6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt C. R.; Li P.; Kang G.; Stubbe J.; Drennan C. L.; Hammes-Schiffer S. Conformational Motions and Water Networks at the alpha/beta Interface in E. coli Ribonucleotide Reductase. J. Am. Chem. Soc. 2020, 142 (32), 13768–13778. 10.1021/jacs.0c04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C.; Greene B. L.; Kang G.; Drennan C. L.; Stubbe J.; Nocera D. G. Gated Proton Release during Radical Transfer at the Subunit Interface of Ribonucleotide Reductase. J. Am. Chem. Soc. 2021, 143 (1), 176–183. 10.1021/jacs.0c07879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan E. C.; Young D. D.; Schultz P. G.; Stubbe J. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J. Am. Chem. Soc. 2011, 133 (40), 15942–5. 10.1021/ja207719f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.; Kasanmascheff M.; Huynh M.; Quartararo A.; Costentin C.; Bejenke I.; Nocera D. G.; Bennati M.; Tommos C.; Stubbe J. Properties of Site-Specifically Incorporated 3-Aminotyrosine in Proteins To Study Redox-Active Tyrosines: Escherichia coli Ribonucleotide Reductase as a Paradigm. Biochemistry 2018, 57 (24), 3402–3415. 10.1021/acs.biochem.8b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mims W. B. Pulsed endor experiments. Proc. R. Soc. London, Ser. A 1965, 283 (1395), 452–457. 10.1098/rspa.1965.0034. [DOI] [Google Scholar]
- Stoll S.; Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178 (1), 42–55. 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Kasanmascheff M.; Lee W.; Nick T. U.; Stubbe J.; Bennati M. Radical transfer in E. coli ribonucleotide reductase: a NH2Y731/R411A-α mutant unmasks a new conformation of the pathway residue 731. Chem. Sci. 2016, 7 (3), 2170–2178. 10.1039/C5SC03460D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B. L.; Taguchi A. T.; Stubbe J.; Nocera D. G. Conformationally Dynamic Radical Transfer within Ribonucleotide Reductase. J. Am. Chem. Soc. 2017, 139 (46), 16657–16665. 10.1021/jacs.7b08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds D. T.; Zussman A. Pure quadrupole resonance of 17O in ice. Phys. Lett. A 1972, 41 (2), 167–169. 10.1016/0375-9601(72)91097-3. [DOI] [Google Scholar]
- Löwdin P. O. On the Non-Orthogonality Problem Connected with the Use of Atomic Wave Functions in the Theory of Molecules and Crystals. J. Chem. Phys. 1950, 18 (3), 365–375. 10.1063/1.1747632. [DOI] [Google Scholar]
- Lin Q.; Parker M. J.; Taguchi A. T.; Ravichandran K.; Kim A.; Kang G.; Shao J.; Drennan C. L.; Stubbe J. Glutamate 52-β at the α/β subunit interface of Escherichia coli class Ia ribonucleotide reductase is essential for conformational gating of radical transfer. J. Biol. Chem. 2017, 292 (22), 9229–9239. 10.1074/jbc.M117.783092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen H. J.; Bildsøe H.; Brorson M.; Wu G.; Gor’kov P. L.; Gan Z.; Hung I. High-Field 17O MAS NMR Reveals 1J(17O-127I) with its Sign and the NMR Crystallography of the Scheelite Structures for NaIO4 and KIO4. J. Phys. Chem. C 2015, 119 (25), 14434–14442. 10.1021/acs.jpcc.5b03721. [DOI] [Google Scholar]
- Keeler E. G.; Michaelis V. K.; Colvin M. T.; Hung I.; Gor’kov P. L.; Cross T. A.; Gan Z.; Griffin R. G. 17O MAS NMR Correlation Spectroscopy at High Magnetic Fields. J. Am. Chem. Soc. 2017, 139 (49), 17953–17963. 10.1021/jacs.7b08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach I.; Bejenke I.; Hecker F.; Kehl A.; Kasanmascheff M.; Gromov I.; Prisecaru I.; Hofer P.; Hiller M.; Bennati M. 1H high field electron-nuclear double resonance spectroscopy at 263 GHz/9.4T. J. Magn. Reson. 2019, 303, 17–27. 10.1016/j.jmr.2019.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


