Abstract
Ultrasensitivity is a ubiquitous emergent property of biochemical reaction networks. The design and construction of synthetic reaction networks exhibiting ultrasensitivity has been challenging, but would greatly expand the potential properties of life-like materials. Herein, we exploit a general and modular strategy to reversibly regulate the activity of enzymes using light and show how ultrasensitivity arises in simple out-of-equilibrium enzymatic systems upon incorporation of reversible photoswitchable inhibitors (PIs). Utilizing a chromophore/warhead strategy, PIs of the protease α-chymotrypsin were synthesized, which led to the discovery of inhibitors with large differences in inhibition constants (Ki) for the different photoisomers. A microfluidic flow setup was used to study enzymatic reactions under out-of-equilibrium conditions by continuous addition and removal of reagents. Upon irradiation of the continuously stirred tank reactor with different light pulse sequences, i.e., varying the pulse duration or frequency of UV and blue light irradiation, reversible switching between photoisomers resulted in ultrasensitive responses in enzymatic activity as well as frequency filtering of input signals. This general and modular strategy enables reversible and tunable control over the kinetic rates of individual enzyme-catalyzed reactions and makes a programmable linkage of enzymes to a wide range of network topologies feasible.
Introduction
Living systems display unique capabilities, e.g., adaptation to the environment, self-healing, homeostasis, or converting chemical energy into directed motion, growth, and division. These processes are governed by complex chemical reaction networks that operate far from equilibrium and allow a precise regulation of a wide range of cellular mechanisms, e.g., signaling or metabolism.1−3 A characteristic feature found in many biochemical networks is ultrasensitivity, which means that (in contrast to a standard hyperbolic Michaelis–Menten response) the response to a stimulus yields a sharp, switch-like sigmoidal function (see Figure 1A for a general schematic of the phenomenon).4−7 This property enables signaling systems to filter out noise and be readily activated once a certain required threshold stimuli is present. Different mechanisms have been identified that can generate this nonlinear input–output relationship, e.g., multisite phosphorylations,6 molecular titrations (buffering),8 substrate competition,9,10 or zero-order kinetics.4
Figure 1.
(A) Schematic of Michaelis–Menten (black) and ultrasensitive response (blue). (B) Incorporation of competitive photoswitchable inhibitors into out-of-equilibrium enzymatic systems enables reversible and tunable activity control using light. Enz = enzyme, W = warhead.
A central goal of systems chemistry is to investigate and translate the common design principles of nature into a practical and modular approach, ultimately enabling a programmable and rational design of life-inspired systems exhibiting tunable properties.11−18 While (light-induced) sigmoidal responses in enzymatic logic gate systems have been reported,19−22 the bottom-up construction of ultrasensitive, life-inspired enzymatic systems remains challenging due to the lack of a general strategy enabling the reversible and tunable regulation of enzymes under out-of-equilibrium conditions.
Although a plethora of different external stimuli to reversibly and spatiotemporally control the activity of enzymes have been applied in the last decades,23 light is an ideal external control element: it is bioorthogonal (λ > 360 nm),24−27 offers high spatiotemporal resolution, can be precisely tuned in terms of photon flux, and introduces the opportunity to control chemical reaction networks using optoelectronic devices.28−30 A prominent approach to gain photocontrol over diverse biological processes is the (mostly) covalent installation of photoswitchable chromophores into the biomolecule of interest.31−35 These so-called “molecular photoswitches” undergo a reversible change in their three-dimensional structure between two or more isomeric forms upon irradiation with light of suitable wavelengths. A number of biological processes, such as protein folding,36 membrane transport,37 or transcription and translation,38 have successfully been photoregulated by the incorporation of photosensitizable units. Thus, an attractive option for obtaining optical control over enzyme function is by installation of molecular photoswitches into the scaffold of enzyme inhibitors.23,34,39
Herein, we exploit a general and modular strategy to reversibly regulate the activity of enzymes using light and show how ultrasensitivity arises in simple out-of-equilibrium enzymatic systems upon incorporation of reversible photoswitchable inhibitors (PIs). We studied enzymatic activity of a α-chymotrpysin/PI-based system under out-of-equilibrium conditions (continuous feeding of substrates and waste removal) using a continuously stirred tank reactor (CSTR) and investigated the response to a series of light pulses (UV (365 nm) and blue (460 nm) light) as input stimuli (Figure 1B). Surprisingly, we discovered a very strong ultrasensitive response to combinations of UV and blue light pulses under out-of-equilibrium conditions. This nonlinear response was also observed in a bottom-up constructed enzymatic cascade based on the proteolytic activity of α-chymotrypsin (Cr) and aminopeptidase M (Ap). Furthermore, we observed that a combination of flow and reversible activity regulation yields a simple enzymatic system with frequency filtering properties upon gradually increasing the UV pulse irradiation frequency and switching back the PI using blue light.
Results and Discussion
Synthesis and Characterization of Competitive, Photoswitchable Inhibitors of α-Chymotrypsin (PIs)
Due to our long-standing expertise in the bottom-up construction of enzymatic reaction networks using proteases,29,40−42 we started by synthesizing reversible photoswitchable inhibitors of α-chymotrypsin (PIs) in order to externally control its enzymatic activity using light. To sufficiently regulate the output of out-of-equilibrium enzymatic systems, the photoswitchable inhibitors need to fulfill several criteria:
-
(1)
The switching factor (SF), i.e., the ratio between the inhibition constants of the cis and trans isomer (SF = Ki,cis/Ki,trans), should be bigger than 10. This ensures that the activity of the respective enzyme can be sufficiently externally modulated using light.
-
(2)
The PIs should exhibit high photostationary states (PSS, defined as the percentage of cis isomer present at equilibrium under irradiation, PSS = ([cis-PI]/([cis-PI ] + [trans-PI]) × 100) under UV (365 nm) irradiation. A high PSS (>90%) implies superior concentrations of the cis isomer relative to its trans isomer, thereby enabling decent levels of photocontrol over the activity of the respective enzyme.
-
(3)
In order to reversibly regulate the activity of enzymes using light, the PI needs to exhibit robust photoreversibility. This implies that no observable decomposition of the PI should take place upon several consecutive UV/vis switching cycles (>10).
-
(4)
Ideally, the inhibition constants (Ki) of both the trans and cis isomer of the PI should be in the μM range or lower. Thereby, only small quantities of PIs, enzymes, and substrates have to be used throughout the performance of flow experiments.
-
(5)
Additionally, the PIs have to be sufficiently water-soluble, should exhibit high photostability under irradiation, and should show slow thermal cis to trans isomerization, as complete photocontrol over the state of the PIs using light is preferred.
Diverse photoswitchable inhibitors for the photoregulation of the activity of proteases have been described in the literature.43−47 These effectors were mostly rationally designed by incorporating a molecular photoswitch (usually an azobenzene) into a previously identified pharmacophore (a reversible inhibitor functionality; “warhead”), thus applying a so-called chromophore/warhead strategy.34 However, the reported PIs exhibit suboptimal properties for successful application in out-of-equilibrium enzymatic systems due to several reasons, e.g., the best Cr PI, bearing a trifluoromethyl ketone warhead, had a SF of only 4.7.47
Therefore, we set out to design and synthesize improved photoswitchable Cr inhibitors to fulfill these criteria and render them suitable for application in out-of-equilibrium enzymatic systems. The synthesis of PIs was performed by applying a chromophore/warhead strategy,34 aiming to maximize the difference in Ki between both isomeric states (switching factor), while keeping the other (photo-)physical requirements for later flow applications in mind. Due to their excellent photophysical properties and straightforward synthetic construction, azobenzenes were chosen for the chromophore.48 Based on initial experiments, phenylalanine-, tyrosine-, valine-, tryptophan-, boronate-, or boronic-acid-based warheads were identified as unsuitable when installed into the azobenzene scaffold. Weak inhibitory properties, negligible differences in Ki between both isomeric states, and insufficient water solubility or (photochemical) degradation of the respective PIs over time were observed. Inspired by a report from Smoum et al. on the competitive inhibition of α-chymotrypsin by trifluoro(organo)borates,49 we aimed to synthesize azobenzene-based photoswitchable inhibitors bearing this warhead. A systematic structure–activity-relationship (SAR) analysis was performed, in which the position of the BF3K warhead on the azobenzene chromophore was altered and diverse substituents with different steric and electronic properties were introduced (see 2.1–2.5 of the Supporting Information (SI) for details). In total, 21 photoswitchable α-chymotrypsin inhibitors have been synthesized throughout the performed SAR study. The synthesized PIs have been carefully characterized with respect to (photo-)physical and inhibitory properties (see 3.1–4.6 of the SI for detailed results). Among them, we identified α-chymotrypsin inhibitor PI, bearing a meta-trifluoroborate functionality, as an ideal candidate for our envisaged applications (Figure 2A, see 2.1–2.5 of the SI for details).
Figure 2.
(A) Structure and enzymological data of the photoswitchable α-chymotrypsin inhibitor PI identified throughout structure–activity-relationship analysis. Inhibition constants were determined using a fluorogenic assay. For details on other synthesized photoswitchable α-chymotrypsin inhibitors, experimental procedures, and results, see 2.1–4.6 of the SI. SF = switching factor; PSS = photostationary state. (B) Batch photoswitching using PI illustrating reversible photoswitching and competitive mode of inhibition. [Cr] = 10 nM, [PI] = 400 μM, [AAPF-AMC] = 80 μM (see 4.5 of the SI for details).
PI exhibits a good solubility in commonly used aqueous protease buffers (TRIS (200 mM), Ca2+ (20 mM), pH = 7.81), and no substantial hydrolysis of the trifluoroborate was observed within 24 h using 19F NMR analysis. In addition, the photophysical characterization revealed both a high photostability and photoreversibility of PI upon 365/460 nm excitation and a very slow thermal cis to trans relaxation (t1/2 = 274 h), thereby enabling full photocontrol over the isomeric state of PI using 365/460 nm light. Importantly, the content of cis isomer obtained upon 365 nm irradiation of PI in the protease buffer was sufficiently high (PSS = 93%, see 3.1–3.4 of the SI for details regarding photophysical characterization of PI).
Using PI, the protease α-chymotrypsin, N-succinyl-Ala-Ala-Pro-Phe-7-amido-4-methylcoumarin (AAPF-AMC) as substrate and TRIS/Ca2+/BSA as buffer (200 mM, 20 mM, 0.1 wt %, pH = 7.81), we determined the inhibition constants (Ki) for the enzyme/inhibitor interaction of trans/cis-PI and Cr using a fluorogenic assay.42 To our delight, trans-PI was identified to be a (moderately) strong competitive inhibitor of α-chymotrypsin (Ki,trans = 12.6 ± 0.6 μM), whereas cis-PI exhibited significantly weaker inhibitory properties (Ki,cis = 264.2 ± 50.6 μM). The three-dimensional structural change caused by photoinduced trans to cis isomerization thus leads to weaker interactions of cis-PI and Cr compared to trans-PI. Combined, this leads to a good switching factor of 21.0 and enables excellent reversible photoregulation of Cr activity.
Having identified PI as a suitable Cr inhibitor, we experimentally probed and verified its reversible photoswitching properties upon consecutive 365/460 nm irradiation cycles as well as its competitive mode of inhibition. We performed a batch photoswitching experiment, in which a mixture of PI and α-chymotrypsin in a TRIS/Ca2+/BSA (200 mM, 20 mM, 0.1 wt %, pH = 7.81) buffer was successively irradiated several times for 5 min with light of a 365 or 460 nm LED, followed by the addition of AAPF-AMC as substrate to an aliquot of the mixture (see 4.5 of the SI for detailed procedure and results). As depicted in Figure 2B, the degree of AMC production over time was reproducible throughout multiple 365/460 nm photoswitching cycles, thereby demonstrating the reversible switching properties of PI as well as its competitive mode of α-chymotrypsin inhibition. In addition, these results point toward high photostability of Cr with respect to irradiation, as its performance is still consistent and reproducible after a total of 40 min of each 365 and 460 nm irradiation.
Multi-State Activity Control over Cr in Enzymatic Systems
With suitable PIs in hand, we aimed to regulate the activity of the protease Cr in a reversible and tunable manner under out-of-equilibrium conditions (continuous addition and removal of PI, enzyme, and buffer to/from CSTR) using 365/460 nm irradiation pulses. To this end, a microfluidic flow setup with a CSTR was constructed. The latter could be selectively irradiated with light from surrounding 365 and 460 nm LEDs, which in turn were externally controlled by connecting both to an Arduino device. The construction of this setup enabled a programmable timing of the irradiation source and its intensity (photon flux), the precise control over the flow rates of the respective stock solutions, and on-line fluorescence monitoring (Figure 3A; see 5.1 and 6 of the SI for experimental details of the flow setup and computer code to control irradiation patterns).
Figure 3.
(A) Schematic of the experimental setup developed for the performance of flow experiments. (B) Network motif of the Cr-catalyzed cleavage of AAPF-AMC to give AMC. (C) Precise reversible photocontrol over the activity of α-chymotrypsin under out-of-equilibrium conditions: generation of an electrocardiogram heartbeat AMC-output pattern. See 5.2.1 of the SI for details regarding timing, duration, and intensity of 365 and 460 nm irradiation. Concentrations in the flow cuvette: [Cr] = 40 nM, [PI] = 200 μM, [AAPF-AMC] = 80 μM. Flow rates: Cr, PI = 400 μL/h; AAPF-AMC, buffer = 200 μL/h. Reactor volume = 196 μL.
To demonstrate multi-state activity control over Cr in this setup, we used PI (Ki,trans = 12.6 ± 0.6 μM; Ki,cis = 264.2 ± 50.6 μM) and AAPF-AMC as fluorogenic substrate (Figure 3B). Irradiation with 365 nm light converts the thermally adapted trans-PI into the weaker inhibitory cis isomer, which induces an increase in Cr activity. By consecutive irradiation with 460 nm light, the cis isomer of PI is converted back to the stronger inhibitory trans isomer, leading to a decrease in Cr activity. The amount of cis-PI present at the 365 nm photostationary state depends strongly on the applied photon flux,48 and variation in light intensity therefore introduces an easy route to modulate the effective inhibitor strength. Thus, similar to the recently described light engineering of the PSS for wavelength-gated adaptation of hydrogel properties,50 a wide range of enzymatic activity can be accessed by utilizing PIs, which enables fine-tuning the kinetics of enzyme-catalyzed reactions using light. Figure 3C shows how irradiation with 365 and 460 nm light is translated into a well-controlled temporal pattern of Cr activity. By applying the depicted light pulse sequence, which is characterized by changes in the photon flux of the 365 nm light source and differently long pulses of 365 and 460 nm irradiation with 100% power, a characteristic pattern could be introduced (see 5.2.1 of the SI for details regarding timing, duration, and intensity of 365 and 460 nm irradiation). Interestingly, it appears that irradiation with blue light (460 nm) can sharpen the peak generated by irradiation with 365 nm (see large peak in the center). The photoinduced trans to cis isomerization of PI is rapidly reversed upon 460 nm irradiation, leading to the formation of a stronger inhibitor and thus to a sharper decrease in Cr activity than observed when the inhibitor concentration decreases only due to efflux from the reactor (see smaller peaks).
Ultrasensitivity in Out-of-Equilibrium Enzymatic Systems by Incorporation of PIs
Encouraged by the initial results shown above, we decided to study the impact of varying the dynamics of the UV input. In cellular signaling pathways, information is encoded not only in the amplitude of a certain signal, but also in its dynamics.51,52 Hence, cellular output decisions can be altered by differences in signal duration and frequency of the input.53 It has been reported that changes to dynamic signal transduction properties, e.g., by cancer mutations or drugs targeting the respective pathway, can lead to improper cell decisions and dysfunction.54−56
Inspired by these biological processes, we first systematically altered the duration of irradiation with 365 nm light as input stimuli, both with and without irradiation by blue light immediately afterwards, in order to optionally switch the chromophore back (Figure 4A). Due to the very slow thermal cis to trans isomerization of PI (t1/2 = 274 h), the isomeric state of the photoswitchable inhibitor is controlled exclusively by 365 and 460 nm irradiation. Thus, PI remains in its cis state after 365 nm irradiation and can only be converted back to the corresponding trans state by 460 nm irradiation. In both pulse sequences, the duration of UV irradiation (365 nm) was gradually increased from 1 s to 30 min. The CSTR was irradiated with the next UV pulse only after the AMC concentration had stabilized to the initial concentration prior to UV irradiation. In the absence of blue light irradiation, PI cannot undergo photoinduced cis to trans isomerization. Hence, the AMC output of the system to values prior to irradiation can only be recovered “passively” by relying on flow to flush the cis isomer out of the CSTR over time. In the pulse sequence with blue light irradiation, however, a 15 s long pulse of blue light directly after UV irradiation induces complete cis to trans photoisomerization, thereby directly resetting the system to the Cr activity state prior to UV irradiation. Figure 4A shows the enzymatic systems response to the two pulse sequences, where the UV pulse duration is logarithmically plotted against the corresponding normalized AMC output recorded on-line using our flow setup.
Figure 4.
(A) Impact of different UV pulse durations and pulse sequences (with and without 15 s of 460 nm CSTR irradiation following the UV pulse) on the enzymatic systems AMC output (see 5.2.2 of the SI for details). [Cr] = 40 nM, [PI] = 200 μM, [AAPF-AMC] = 80 μM. Flow rates: Cr, PI = 400 μL/h; AAPF-AMC, buffer = 200 μL/h. Reactor volume = 196 μL. (B) Modeling of the behavior observed in A, using a system of rate equations with experimentally determined rate constants and roughly assumed photoswitching rates (see 7 of the SI). (C) Impact of different UV pulse durations and pulse sequences (with and without 10 s of 460 nm CSTR irradiation following a UV pulse) on the Cr/Ap-based enzymatic systems AMC output (see 5.2.3 of the SI). [Cr] = 20 nM, [PI] = 200 μM, [Z-Phe-Arg-AMC] = 100 μM, [Ap] = 0.1 U/mL. Flow rates: Cr, Ap = 200 μL/h; PI, Z-Phe-Arg-AMC = 400 μL/h; buffer = 100 μL/h. Reactor volume = 196 μL.
In the absence of a blue light pulse, a sigmoidal response curve was observed in which higher concentrations of AMC are formed with increasing UV irradiation times. Just 5 s of UV irradiation led to an increase in AMC output. The maximum Cr activity of the enzymatic system was reached at UV irradiation times of 5 min and longer. By irradiating the CSTR with a 15 s long pulse of blue light immediately after each UV irradiation step, a significantly different AMC response curve was obtained. Only UV pulses of 3 min or longer led to an increase of AMC production. Shorter UV pulses are filtered out, as the combination of flow and the 460 nm induced cis to trans isomerization results in a quick recovery of the system to the Cr activity prior to UV irradiation. In both pulse sequences, we observed an ultrasensitive input–output relationship, but especially pulsing with blue light resulted in sharp, switch-like behavior.5 This ultrasensitive response is caused by the interplay of residence time of the Cr/PI mixture within the CSTR and the duration and intensity of UV/blue light pulses. We wish to remind the reader that an ultrasensitive response formally refers to an increase from 10% to 90% of the maximum response upon a less than 81-fold change in input stimulus. We can quantify the degree of ultrasensitivity by fitting our experimental data to the Hill equation (eq 1):
| 1 |
where 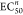 is the stimulus value needed to produce
50% of the maximum response. This plotting allowed us to determine
the Hill coefficient (n), which is a quantitative
measure of an ultrasensitive response as defined according to eq 2:57
is the stimulus value needed to produce
50% of the maximum response. This plotting allowed us to determine
the Hill coefficient (n), which is a quantitative
measure of an ultrasensitive response as defined according to eq 2:57
| 2 |
Herein, EC90 and EC10 are the stimulus values needed to produce 90% and 10% of the maximum response, respectively. In this equation, a Michaelis–Menten response is characterized by a Hill coefficient of n = 1. In signal transduction, the Hill coefficients of ultrasensitive signaling systems are typically in the range n = 1.3–20,5 for example with n = 3.5 for the Cdk1/Wee1A stimulus–response relationship or n = 11 for Cdk1/Cdc25C.9,58
In the absence of blue light pulses, the ultrasensitivity of the enzymatic system is weak, with n = 1.25. In contrast, a highly ultrasensitive response is obtained when UV and blue light pulses are combined (n = 5.71), due to 460 nm induced cis to trans isomerization causing a quick recovery of the system to the Cr activity prior to UV irradiation.
To further rationalize and support our experimental findings, we simulated the system’s response to the different pulse sequences and UV pulse durations using mass-action kinetics (Figure 4B). The model qualitatively reproduces the sigmoidal, switch-like behavior observed experimentally very well. We note that there is a global shift to shorter UV pulse durations leading to a faster response in the simulations compared to the experimental data. This discrepancy is caused by the fact that we only have rough estimates of the rates of photoswitching (for details, see 7 of the SI). The exact position of the plateau is determined by both trans to cis and cis to trans photoisomerization rates as well as by the applied UV pulse durations.
To illustrate how our regulatory strategy can be applied to control more complex systems, we designed an enzymatic cascade that consists of two elemental steps. The first one is the Cr-catalyzed proteolytic cleavage of the phenylalanine/arginine peptide bond (KM = 180 μM) of the substrate Z-Phe-Arg-AMC, which produces H-Arg-AMC (Figure 4C).41 The peptide bond of the latter molecule is then cleaved by the action of aminopeptidase M (Ap),59 thereby producing the AMC reporter molecule (KM = 5 mM). As the AMC production of the cascadic network depends on the Cr-catalyzed formation of H-Arg-AMC, the whole output of the network can be controlled via photoinduced isomerization of PI. Similar to the experiments performed before, we systematically altered the duration of irradiation with 365 nm and optionally switched the chromophore back with blue light irradiation immediately afterwards. Regardless of the applied pulse sequence, both curves show an ultrasensitive response (Figure 4C). Interestingly, in the absence of blue light pulses, we determined a Hill coefficient of n = 1.46, which correlates with stronger ultrasensitivity for the Cr/Ap-based system compared to the single enzyme Cr/PI-based system discussed above (n = 1.25). In the two-step cascade, short UV pulses do not lead to the formation of significant amounts of H-Arg-AMC. As also the Ap-catalyzed cleavage is slow due to weak binding of the substrate (KM = 5 mM), no substantially increased amounts of AMC can be formed before the enzyme/substrate mixture reaches the fluorescence detector unit. Thus, even in the absence of a blue light pulse, the enzymatic system is able to filter short UV pulses due to the kinetics of the two underlying elemental steps. By irradiating the CSTR with a 10 s long pulse of blue light after each UV-irradiation step, the system became more ultrasensitive compared to the other pulse sequence (n = 2.78); however, this ultrasensitivity is significantly less pronounced compared to that of the simpler Cr/PI-based system (n = 5.71). The lowered ultrasensitivity is a result of the irreversible production of H-Arg-AMC, which will proceed to AMC production, even when Cr is inhibited again after the blue light pulse. These results provide a proof-of-concept demonstration of our strategy to induce and control the ultrasensitivity in enzymatic cascades and to selectively modulate the activity of enzymes within multi-enzymatic systems.
Stimulus Frequency Filtering in Out-of-Equilibrium Enzymatic Systems by Incorporation of PIs
Finally, we sought to characterize the temporal resolution of our strategy to regulate the activity of enzymes under out-of-equilibrium conditions (continuous addition and removal of PI, enzyme, and buffer to/from CSTR) by applying different stimuli frequencies to our system (Figure 5A). In two applied pulse sequences, the offset between four UV pulses of 30 s duration was gradually increased, ranging from as short as 1 s to 60 min. If no blue light pulses are installed after UV irradiation, the PI within the CSTR remains in its weaker inhibitory cis state due to the very slow thermal cis to trans isomerization (t1/2 = 274 h). Consecutively, the depicted exemplary [AMC]–t plot of the pulse sequence without blue irradiation shows that the temporal resolution of the system is insufficient to separate the four UV pulses at higher pulse frequency, and they fuse into a single peak. As a result, the maximum AMC concentration observed after four UV pulses increases. Figure 5B shows the normalized change in AMC output of the system as a function of the time offset between multiple UV pulses (see 5.2.4 of the SI). In the absence of blue light irradiation, a pulse offset of 30 min or shorter is not sufficient to completely flush out all formed cis-PI of the reactor, and the residual cis-PI leads to a higher AMC output upon the next consecutive UV pulse. Thus, in the absence of a blue light pulse, the temporal resolution of our regulatory strategy is strictly limited by the underlying flow properties. In contrast, a similar sequence with 5 s long blue pulses after UV irradiation shows strong “filtering” of the UV input pulses. The blue light immediately switches PI back to its strongly inhibiting trans state, and infrequent pulses will thus not lead to an appreciable buildup of active enzyme. Only when the pulse offset is decreased to 2 s or shorter do we observe an increase in AMC response. This is a first promising demonstration that a combination of flow and reversible enzyme activation yields a system with frequency filtering properties.
Figure 5.
(A) Impact of different UV pulse periodicity (pulse duration = 30 s) and pulse sequences (with and without 5 s of 460 nm CSTR irradiation following UV pulse) on the enzymatic systems AMC output over time. [Cr] = 40 nM, [PI] = 200 μM, [AAPF-AMC] = 80 μM. Flow rates: Cr, PI = 400 μL/h; AAPF-AMC, buffer = 200 μL/h. Reactor volume = 196 μL. (B) Plot of the normalized change in AMC output of the system as a function of the time offset between multiple UV pulses (see 5.2.4 of the SI for details).
Conclusions
Ultrasensitivity is an emergent property of many biochemical reaction networks. It enables these networks to generate switch-like responses upon small changes in input parameters. Introducing ultrasensitivity in synthetic systems is a desirable goal in systems chemistry, as it would open up new routes to control life-inspired, complex systems. However, ultrasensitivity in biological systems is often the result of rather complex motifs, which are challenging to recapitulate in synthetic systems. Here, we have shown that enzymatic reactions operating under out-of-equilibrium conditions generate an ultrasensitive response in a surprisingly simple topology upon incorporation of photoswitchable inhibitors. Introduction of a photoswitchable inhibitor with sufficiently large differences in inhibitory properties between the two photoisomers yielded switch-like responses in enzymatic activity. Furthermore, we discovered that by reversibly switching the photoisomers, even single enzymatic reactions can filter out short pulses under flow conditions. In the absence of photoinduced cis to trans irradiation (blue light pulse), a period of over 30 min between two UV light pulses was required to ensure an “on–off” switching of the system, whereas short (5 s) blue light irradiation immediately after UV pulses allowed us to detect pulses that were only 5 s apart.
The simple building blocks introduced here set the scene for more complex motifs that are now becoming feasible. We can introduce photoswitchable inhibitors with different chromophores, couple multiple enzymes into reaction networks, and explore more complex motifs. The ability to modulate the activity of individual enzymes in such networks using light will provide an exceedingly versatile tool for creating ever more advanced complex molecular systems.
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 862081 (CLASSY). Generous financial support by the NWO (Spinoza prize to W.T.S.H.) and by the German National Academy of Sciences Leopoldina (Postdoctoral fellowship LPDS2020-04 to M.T.) is gratefully acknowledged. We thank C. L. Fernández Regueiro, Dr. J. Taylor, Dr. O. R. Maguire, and Dr. W. A. Velema (all Radboud University Nijmegen) for fruitful discussions and experimental advice. Furthermore, we would like to thank Prof. F. Glorius and F. Sandfort (WWU Münster) for their support with mass spectrometric analysis of PIs.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c12956.
Experimental details, photophysical and enzymological characterization data, copies of NMR spectra of new compounds, simulation details, and assay procedures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Bhalla U. S.; Iyengar R. Emergent Properties of Networks of Biological Signaling Pathways. Science 1999, 283, 381–387. 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- Kholodenko B. N. Cell-Signalling Dynamics in Time and Space. Nat. Rev. Mol. Cell Biol. 2006, 7, 165–176. 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J.; Fernie A. R. The Role of Dynamic Enzyme Assemblies and Substrate Channelling in Metabolic Regulation. Nat. Commun. 2018, 9, 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A.; Koshland D. E. An Amplified Sensitivity Arising from Covalent Modification in Biological Systems. Proc. Natl. Acad. Sci. U. S. A. 1981, 78, 6840–6844. 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E.; Ha S. H. Ultrasensitivity Part I: Michaelian Responses and Zero-Order Ultrasensitivity. Trends Biochem. Sci. 2014, 39, 496–503. 10.1016/j.tibs.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E.; Ha S. H. Ultrasensitivity Part II: Multisite Phosphorylation, Stoichiometric Inhibitors, and Positive Feedback. Trends Biochem. Sci. 2014, 39, 556–569. 10.1016/j.tibs.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E.; Ha S. H. Ultrasensitivity Part III: Cascades, Bistable Switches, and Oscillators. Trends Biochem. Sci. 2014, 39, 612–618. 10.1016/j.tibs.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler N. E.; Louis M. Molecular Titration and Ultrasensitivity in Regulatory Networks. J. Mol. Biol. 2008, 384, 1106–1119. 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- Kim S. Y.; Ferrell J. E. Substrate Competition as a Source of Ultrasensitivity in the Inactivation of Wee1. Cell 2007, 128, 1133–1145. 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Rafael S. P.; Vallée-Bélisle A.; Fabregas E.; Plaxco K.; Palleschi G.; Ricci F. Employing the Metabolic “Branch Point Effect” to Generate an All-or-None, Digital-like Response in Enzymatic Outputs and Enzyme-Based Sensors. Anal. Chem. 2012, 84, 1076–1082. 10.1021/ac202701c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow R. F.; Otto S. Systems Chemistry. Chem. Soc. Rev. 2008, 37, 101–108. 10.1039/B611921M. [DOI] [PubMed] [Google Scholar]
- Ashkenasy G.; Hermans T. M.; Otto S.; Taylor A. F. Systems Chemistry. Chem. Soc. Rev. 2017, 46, 2543–2554. 10.1039/C7CS00117G. [DOI] [PubMed] [Google Scholar]
- Grzybowski B. A.; Huck W. T. S. The Nanotechnology of Life-Inspired Systems. Nat. Nanotechnol. 2016, 11, 585–592. 10.1038/nnano.2016.116. [DOI] [PubMed] [Google Scholar]
- Singh N.; Lainer B.; Formon G. J. M.; De Piccoli S.; Hermans T. M. Re-Programming Hydrogel Properties Using a Fuel-Driven Reaction Cycle. J. Am. Chem. Soc. 2020, 142, 4083–4087. 10.1021/jacs.9b11503. [DOI] [PubMed] [Google Scholar]
- Maity I.; Dev D.; Basu K.; Wagner N.; Ashkenasy G. Signaling in Systems Chemistry: Programing Gold Nanoparticles Formation and Assembly Using a Dynamic Bistable Network. Angew. Chem., Int. Ed. 2021, 60, 4512–4517. 10.1002/anie.202012837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. C.; Kosikova T.; Philp D. Encoding Multiple Reactivity Modes within a Single Synthetic-Replicator. J. Am. Chem. Soc. 2020, 142, 11139–11152. 10.1021/jacs.0c03527. [DOI] [PubMed] [Google Scholar]
- Afrose S. P.; Bal S.; Chatterjee A.; Das K.; Das D. Designed Negative Feedback from Transiently Formed Catalytic Nanostructures. Angew. Chem., Int. Ed. 2019, 58, 15783–15787. 10.1002/anie.201910280. [DOI] [PubMed] [Google Scholar]
- Kriebisch B. A. K.; Jussupow A.; Bergmann A. M.; Kohler F.; Dietz H.; Kaila V. R. I.; Boekhoven J. Reciprocal Coupling in Chemically Fueled Assembly: A Reaction Cycle Regulates Self-Assembly and Vice Versa. J. Am. Chem. Soc. 2020, 142, 20837–20844. 10.1021/jacs.0c10486. [DOI] [PubMed] [Google Scholar]
- Privman V.; Fratto B. E.; Zavalov O.; Halámek J.; Katz E. Enzymatic AND Logic Gate with Sigmoid Response Induced by Photochemically Controlled Oxidation of the Output. J. Phys. Chem. B 2013, 117, 7559–7568. 10.1021/jp404054f. [DOI] [PubMed] [Google Scholar]
- Katz E.; Privman V. Enzyme-Based Logic Systems for Information Processing. Chem. Soc. Rev. 2010, 39, 1835–1857. 10.1039/b806038j. [DOI] [PubMed] [Google Scholar]
- Domanskyi S.; Privman V. Design of Digital Response in Enzyme-Based Bioanalytical Systems for Information Processing Applications. J. Phys. Chem. B 2012, 116, 13690–13695. 10.1021/jp309001j. [DOI] [PubMed] [Google Scholar]
- Bakshi S.; Zavalov O.; Halámek J.; Privman V.; Katz E. Modularity of Biochemical Filtering for Inducing Sigmoid Response in Both Inputs in an Enzymatic AND Gate. J. Phys. Chem. B 2013, 117, 9857–9865. 10.1021/jp4058675. [DOI] [PubMed] [Google Scholar]
- Claaßen C.; Gerlach T.; Rother D.. Stimulus-Responsive Regulation of Enzyme Activity for One-Step and Multi-Step Syntheses. Adv. Synth. Catal. 2019, 361, 2387–−2401. 10.1002/adsc.201900169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K.; Chin J. W. Bioorthogonal Reactions for Labeling Proteins. ACS Chem. Biol. 2014, 9, 16–20. 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- Song W.; Wang Y.; Qu J.; Lin Q. Selective Functionalization of a Genetically Encoded Alkene-Containing Protein via “Photoclick Chemistry” in Bacterial Cells. J. Am. Chem. Soc. 2008, 130, 9654–9655. 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- Yu Z.; Ohulchanskyy T. Y.; An P.; Prasad P. N.; Lin Q. Fluorogenic, Two-Photon-Triggered Photoclick Chemistry in Live Mammalian Cells. J. Am. Chem. Soc. 2013, 135, 16766–16769. 10.1021/ja407867a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Wang Y.; Qu J.; Madden M. M.; Lin Q. A Photoinducible 1,3-Dipolar Cycloaddition Reaction for Rapid, Selective Modification of Tetrazole-Containing Proteins. Angew. Chem., Int. Ed. 2008, 47, 2832–2835. 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]
- Willner I.; Willner B. Photoswitchable Biomaterials as Grounds for Optobioelectronic Devices. Bioelectrochem. Bioenerg. 1997, 42, 43–57. 10.1016/S0302-4598(96)05152-5. [DOI] [Google Scholar]
- Pogodaev A. A.; Wong A. S. Y.; Huck W. T. S. Photochemical Control over Oscillations in Chemical Reaction Networks. J. Am. Chem. Soc. 2017, 139, 15296–15299. 10.1021/jacs.7b08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.; Bezold D.; Jessen H. J.; Walther A. Multiple Light Control Mechanisms in ATP-Fueled Non-equilibrium DNA Systems. Angew. Chem., Int. Ed. 2020, 59, 12084–12092. 10.1002/anie.202003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner I.; Rubin S. Control of the Structure and Functions of Biomaterials by Light. Angew. Chem., Int. Ed. 1996, 35, 367–385. 10.1002/anie.199603671. [DOI] [Google Scholar]
- Beharry A. A.; Woolley G. A. Azobenzene Photoswitches for Biomolecules. Chem. Soc. Rev. 2011, 40, 4422–4437. 10.1039/c1cs15023e. [DOI] [PubMed] [Google Scholar]
- Mayer G.; Heckel A. Biologically Active Molecules with a “Light Switch”.. Angew. Chem., Int. Ed. 2006, 45, 4900–4921. 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- Szymański W.; Beierle J. M.; Kistemaker H. A. V.; Velema W. A.; Feringa B. L. Reversible Photocontrol of Biological Systems by the Incorporation of Molecular Photoswitches. Chem. Rev. 2013, 113, 6114–6178. 10.1021/cr300179f. [DOI] [PubMed] [Google Scholar]
- Hüll K.; Morstein J.; Trauner D. In Vivo Photopharmacology. Chem. Rev. 2018, 118, 10710–10747. 10.1021/acs.chemrev.8b00037. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Zarrine-Afsar A.; Al-Abdul-Wahid M. S.; Prosser R. S.; Davidson A. R.; Woolley G. A. Structure-Based Approach to the Photocontrol of Protein Folding. J. Am. Chem. Soc. 2009, 131, 2283–2289. 10.1021/ja807938v. [DOI] [PubMed] [Google Scholar]
- Van Bergeijk P.; Adrian M.; Hoogenraad C. C.; Kapitein L. C. Optogenetic Control of Organelle Transport and Positioning. Nature 2015, 518, 111–114. 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Asanuma H.; Komiyama M. Azobenzene-Tethered T7 Promoter for Efficient Photoregulation of Transcription. J. Am. Chem. Soc. 2006, 128, 1009–1015. 10.1021/ja055983k. [DOI] [PubMed] [Google Scholar]
- Hoorens M. W. H.; Szymanski W. Reversible, Spatial and Temporal Control over Protein Activity Using Light. Trends Biochem. Sci. 2018, 43, 567–575. 10.1016/j.tibs.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Semenov S. N.; Wong A. S. Y.; van der Made R. M.; Postma S. G. J.; Groen J.; van Roekel H. W. H.; de Greef T. F. A.; Huck W. T. S. Rational Design of Functional and Tunable Oscillating Enzymatic Networks. Nat. Chem. 2015, 7, 160–165. 10.1038/nchem.2142. [DOI] [PubMed] [Google Scholar]
- Helwig B.; van Sluijs B.; Pogodaev A. A.; Postma S. G. J.; Huck W. T. S. Bottom-Up Construction of an Adaptive Enzymatic Reaction Network. Angew. Chem., Int. Ed. 2018, 57, 14065–14069. 10.1002/anie.201806944. [DOI] [PubMed] [Google Scholar]
- Pogodaev A. A.; Fernández Regueiro C. L.; Jakštaitė M.; Hollander M. J.; Huck W. T. S. Modular Design of Small Enzymatic Reaction Networks Based on Reversible and Cleavable Inhibitors. Angew. Chem., Int. Ed. 2019, 58, 14539–14543. 10.1002/anie.201907995. [DOI] [PubMed] [Google Scholar]
- Westmark P. R.; Kelly J. P.; Smith B. D. Photoregulation of Enzyme Activity. Photochromic, Transition-State-Analog Inhibitors of Cysteine and Serine Proteases. J. Am. Chem. Soc. 1993, 115, 3416–3419. 10.1021/ja00062a003. [DOI] [Google Scholar]
- Harvey A. J.; Abell A. D. Azobenzene-Containing, Peptidyl α-Ketoesters as Photobiological Switches of α-Chymotrypsin. Tetrahedron 2000, 56, 9763–9771. 10.1016/S0040-4020(00)00883-8. [DOI] [Google Scholar]
- Pearson D.; Abell A. D. Photoswitch Inhibitors of α-Chymotrypsin—Increased Substitution and Peptidic Character in Peptidomimetic Boronate Esters. Org. Biomol. Chem. 2006, 4, 3618–3625. 10.1039/B609320P. [DOI] [PubMed] [Google Scholar]
- Abell A. D.; Jones M. A.; Neffe A. T.; Aitken S. G.; Cain T. P.; Payne R. J.; McNabb S. B.; Coxon J. M.; Stuart B. G.; Pearson D.; Lee H. Y. -Y.; Morton J. D. Investigation into the P3 Binding Domain of M-Calpain Using Photoswitchable Diazo- and Triazene-Dipeptide Aldehydes: New Anticataract Agents. J. Med. Chem. 2007, 50, 2916–2920. 10.1021/jm061455n. [DOI] [PubMed] [Google Scholar]
- Pearson D.; Alexander N.; Abell A. D. Improved Photocontrol of α-Chymotrypsin Activity: Peptidomimetic Trifluoromethylketone Photoswitch Enzyme Inhibitors. Chem. - Eur. J. 2008, 14, 7358–7365. 10.1002/chem.200800082. [DOI] [PubMed] [Google Scholar]
- Bandara H. M. D.; Burdette S. C. Photoisomerization in Different Classes of Azobenzene. Chem. Soc. Rev. 2012, 41, 1809–1825. 10.1039/C1CS15179G. [DOI] [PubMed] [Google Scholar]
- Smoum R.; Rubinstein A.; Srebnik M. Noncovalent Inhibition of the Serine Proteases, α-Chymotrypsin and Trypsin by Trifluoro(Organo)Borates. Org. Biomol. Chem. 2005, 3, 941–944. 10.1039/B415957H. [DOI] [PubMed] [Google Scholar]
- Ludwanowski S.; Skarsetz O.; Creusen G.; Hoenders D.; Straub P.; Walther A. Wavelength-Gated Adaptation of Hydrogel Properties via Photo-Dynamic Multivalency in Associative Star Polymers. Angew. Chem., Int. Ed. 2021, 60, 4358–4367. 10.1002/anie.202011592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch N.; Rukhlenko O. S.; Kolch W.; Kholodenko B. N. MAPK Kinase Signalling Dynamics Regulate Cell Fate Decisions and Drug Resistance. Curr. Opin. Struct. Biol. 2016, 41, 151–158. 10.1016/j.sbi.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Purvis J. E.; Karhohs K. W.; Mock C.; Batchelor E.; Loewer A.; Lahav G. P53 Dynamics Control Cell Fate. Science 2012, 336, 1440–1444. 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis J. E.; Lahav G. Encoding and Decoding Cellular Information through Signaling Dynamics. Cell 2013, 152, 945–956. 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj L. J.; Sabnis A. J.; Mitchell A.; Garbarino J. E.; Toettcher J. E.; Bivona T. G.; Lim W. A.. Cancer Mutations and Targeted Drugs Can Disrupt Dynamic Signal Encoding by the Ras-Erk Pathway. Science 2018, 361, eaao3048. 10.1126/science.aao3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P. T. C.; Garnett M. J.; Roe S. M.; Lee S.; Niculescu-Duvaz D.; Good V. M.; Project C. G.; Jones C. M.; Marshall C. J.; Springer C. J.; Barford D.; Marais R. Mechanism of Activation of the RAF-ERK Signaling Pathway by Oncogenic Mutations of B-RAF. Cell 2004, 116, 855–867. 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Behar M.; Barken D.; Werner S. L.; Hoffmann A. The Dynamics of Signaling as a Pharmacological Target. Cell 2013, 155, 448–461. 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. N. The Hill Equation Revisited: Uses and Misuses. FASEB J. 1997, 11, 835–841. 10.1096/fasebj.11.11.9285481. [DOI] [PubMed] [Google Scholar]
- Trunnell N. B.; Poon A. C.; Kim S. Y.; Ferrell J. E. Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Mol. Cell 2011, 41, 263–274. 10.1016/j.molcel.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales T.; Robert-Baudouy J. Bacterial Aminopeptidases: Properties and Functions. FEMS Microbiol. Rev. 1996, 18, 319–344. 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








