Abstract
Shiga toxin is an AB5 toxin produced by Shigella species, while related toxins are produced by Shiga toxin-producing Escherichia coli (STEC). Infection by Shigella can lead to bloody diarrhea followed by the often fatal hemolytic uremic syndrome (HUS). In the present paper, we aimed for a simple and effective toxin inhibitor by comparing three classes of carbohydrate-based inhibitors: glycodendrimers, glycopolymers, and oligosaccharides. We observed a clear enhancement in potency for multivalent inhibitors, with the divalent and tetravalent compounds inhibiting in the millimolar and micromolar range, respectively. However, the polymeric inhibitor based on galabiose was the most potent in the series exhibiting nanomolar inhibition. Alginate and chitosan oligosaccharides also inhibit Shiga toxin and may be used as a prophylactic drug during shigella outbreaks.
Introduction
Bacterial dysentery or shigellosis has been identified as one of the major causes of mortality in children under 5 years of age.1 Shigellosis is caused by gram-negative bacteria from the following four species of Shigella: Shigella dysenteriae, Shigella flexneri,Shigella boydii, and Shigella sonnei through the fecal–oral route. The pathology can include bloody diarrhea (hemorrhagic colitis) followed by the often fatal hemolytic uremic syndrome (HUS). HUS can occur if the pathogen also produces the Shiga toxin (Stx). The toxin is produced by S. dysenteriae serotype 1, but closely related toxins Stx1 and Stx2 are produced by Shiga toxin-producing Escherichia coli (STEC) or enterohemorrhagic E. coli (EHEC), where Stx2 has been reported to cause more severe infections.2 STEC outbreaks are mostly food-borne with the largest ever reported in Germany (2011) linked to sprout consumption.3
The Shiga toxin is an AB5 toxin composed of the toxic A subunit and a pentameric B subunit that is responsible for the binding of the toxin to its cell surface receptor globotriaosylceramide (Gb3; Galα1-4Galβ1-4Glcβ1-ceramide, also known as CD77 or the Pk blood group antigen).4 Each of the five B subunits can bind three Gb3 molecules simultaneously.5,6 After the initial bloody diarrhea, the toxin enters the bloodstream by poorly understood mechanisms.7 The ample presence of Gb3 molecules in the kidney targets the toxin to this location. Once endocytosed, the toxin induces multiple signaling pathways leading to blockage of protein synthesis and induction of apoptosis8 and HUS. STEC infections are treated with antibiotics, although their use is controversial with respect to their ability to increase the risk of HUS.9 The recent emergence of toxin-producing strains of S. flexneri and S. sonnei points toward increased future morbidity and mortality.10−19 As an alternative to antibiotics, synthetic molecules based on Gb3 have been explored as potential prophylactic treatment for STEC.20 Synsorb Pk, silicon dioxide coupled to synthetic Pk, showed promising results in the trapping of toxins and preventing toxic effects on renal cells.21 However, a subsequent clinical trial was unsuccessful in diminishing diarrhea-associated HUS possibly due to late administration of the drug to the GI tract, while the toxin was already active systemically.22 Recommendations were made for intervention in the circulation. This approach was explored in several cases with antibodies and nanobodies, as summarized recently.23 Smaller dendritic molecules were also explored in this respect. The soluble STARFISH inhibitor with a decavalent display of the Gb3 trisaccharide, reported by Bundle et al., exhibited subnanomolar inhibition of Stx1, with large potency gains over the divalent analogue and the Pk trisaccharide itself.24 A modification of the STARFISH named DAISY was observed to be effective against both Stx1 and Stx2 with nanomolar inhibition and in vivo activity in EHEC orally infected mice by subcutaneous injection 24 h after infection.25 Several related SUPERTWIG structures (based on Gb3 conjugated to carbosilane dendrimers), developed by Nishikawa et al., were also identified as effective neutralizers of Stx with a dependency on their valency and structure.26 A hexavalent structure provided protection after intravenous injections starting 3 days after oral infection.
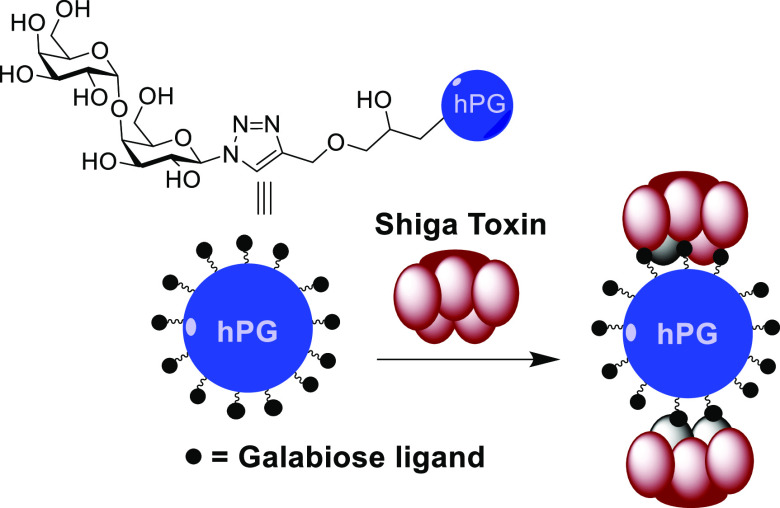
In the present paper, three classes of carbohydrate-containing structures were investigated: dendritic-synthesized multivalent inhibitors, glycopolymers, and natural oligosaccharides. As the ligand, we chose to explore the potential of the disaccharide (Galα1-4Galβ; galabiose) as a possible monovalent alternative to Gb3-based inhibitors. The intention here was to explore what the minimal structural requirements for potent toxin inhibition would be by minimizing the ligand and the multivalent scaffold. For the dendrimers, ease of preparation was central to the selection of di- and tetravalent dendrimers utilized. Polymeric scaffolds were selected for potency comparison. For the polymer scaffold, hyperbranched polyglycerol (hPG) was used for their easy synthesis, high functionalization, biocompatibility, and low in vivo toxicity.27 A polyalkyne and a polyazide variant of hPG were prepared for their conjugation by employing the copper-catalyzed alkyne azide cycloaddition (CuAAC) conjugations. In addition to the synthesized compounds, commercially available oligosaccharides such as nontoxic food grade alginate, chitosan, and fructo- and galacto-oligosaccharides [alginate oligosaccharide (AOS), chitosan oligosaccharide (COS), galactose oligosaccharide (GOS), fructose oligosaccharide (FOS)]28 were tested for Stx inhibition. These could serve as an even more viable practical alternative that could be a part of a preventative food-based approach during outbreaks with a focus on the gastrointestinal phase of the toxin-producing Shigella pathogenicity.
Results
For the synthesis of the monovalent galabiose reagent, galactose pentaacetate was used as the common precursor for the synthesis of the glycosyl donor and acceptor (Scheme 1). Glycosyl donor 1a was synthesized in three steps by thioglycoside preparation as the first step followed by silyl protection of the sugar. Glycosyl donor 1b was synthesized by azidation using trimethylsilyl azide and benzoyl protection. Trifluoromethanesulfonic anhydride-mediated glycosylation afforded the disaccharide 1c in moderate yields. Deprotection was performed over two steps without purification followed by acetylation to obtain 1d which was used for conjugation with various dendrimers.
Scheme 1. Synthesis of Galabiose Azide.

Reagents and conditions: (i) HSPh, BF3·Et2O, DCM, r.t., 16 h, 90%; NaOMe, MeOH, r.t. 90% (ii) tBu2Si(OTf)2, pyridine, DMF, −40°C, >90% (iii) TBDMSOTf, DMAP, pyridine, r.t. 70% (iv) TMSN3, SnCl4, DCM, 95%; NaOMe, MeOH, r.t., 16 h, 100% (v) BzCl, pyridine, DCM, −80°C, 2 h, 50% (vi) Tf2O, Ph2SO, TTBP, DCM, −60°C, 1 h, 72% (vii) NaOMe, MeOH; HF, pyridine; Ac2O, pyridine, 63% (viii) NaOH, MeOH, 90%.
Building block 3,5-dihydroxybenzoic acid was used as the starting material for the synthesis of all four dendrimers (Figure 1). 2a and 2c were synthesized using previously reported procedures.29,30 Divalent 2b was conveniently prepared by coupling methyl 3,5-bis(2-aminoethoxy)benzoate to propargyl chloroformate and was obtained in 88% yield. Amide coupling of 2a to dodecane-1,12-diamine using BOP gave tetravalent dendrimer 2d in 60% yield. Dendrimers (2a, 2b, 2c, and 2d) were conjugated to 1d by CuAAC and deprotected to obtain final compounds 3, 4, 5, and 6 (Figure 3) in good yields.
Figure 1.

Di- and tetravalent dendrimers.
Figure 3.

Dendrimeric- and polymeric-galabiose conjugates.
Glycidol, a reactive hydroxy-epoxide, was used as an AB2 monomer, and polymerization was initiated using tris(hydroxymethyl)propane (TMP). TMP was partially deprotonated and used as an initiator for the anionic polymerization carried out by slow monomer addition and yielding hPG-OH of ca. 9.4 kDa with 125 OH end groups, calculated using inverse-gated carbon and proton NMR.31 Azidation of the hPG was performed in two steps by first substituting the hydroxy groups of the hPG with the more reactive mesyl groups followed by azide substitution using sodium azide.32 Mesyl substitution of the hPG was calculated at 8% (ca. 10 mesyl end groups per molecule) using proton NMR, and complete substitution with azide groups was confirmed by the absence of the mesyl protons (1H NMR) and the appearance of the azide stretching in the infrared spectra (IR) at 2110 cm–1. Propargylation of hPG was performed in a single step using propargyl bromide in 72% yield.33 The polymer was calculated to be 16% functionalized, which means ca. 20 propargyl end groups per molecule based on proton NMR, and the IR spectra further confirmed this via the 2110 cm–1 peak (see the Supporting Information).
hPG azide was conjugated by CuAAC to globotriose-NAc-propargyl (7, Scheme 2) to obtain 8 (Figure 3) in 80% yield. Similarly, conjugation of 1d to hPG-propargyl following deprotection yielded final compound 9 (Figure 3) in 75% yield over two steps. Final polymers 8 and 9 were characterized by 1H NMR and also by IR to check for the absence of the azide and alkyne stretching peaks, respectively.
Scheme 2. Synthesis of Hyperbranched Polymers.
Reagents and conditions: (i) MsCl, TEA, DMF, 0 °C–r.t., 16 h, 88%; NaN3, DMF, 60 °C, quant. (ii) NaH, KI, propargyl bromide, DMF, 0 °C–r.t., 72%.
Previously, inhibitors were tested for inhibition in ELISA assays using immobilization of the B subunit of Stx1 (Stx1B).24 In contrast, we used an assay in which FSL-Gb3 was immobilized instead of the toxin (Figure S2), as this was deemed more realistic since in vivo the toxin is also free to move. FSL-GB3 is comprised of a functional component (F) which is GB3, conjugated via an O(CH2)3NH spacer (S) to an activated adipate derivative of dioleoylphosphatidylethanolamine (L). Monovalent 1e was used as the reference in the ELISA and, as expected, showed millimolar inhibition of the toxin with an IC50 of approx. 5 mM. Divalent 3 and 4 also inhibited the toxin in the millimolar range (1 and 1.2 mM respectively) (Table 1). Clearly, the small variation in the spacer length between dendrimer 2a and 2b did not cause any significant variation in potency. It was anticipated that if the divalent ligands bridge between sites 1 and 2 on a single toxin subunit,6 this would be more easily possible with the longer spacer of 4. A stronger enhancement of the inhibition was observed with the tetravalent compounds 5 and 6 as both showed micromolar inhibition (20 and 13 μM respectively). Here again, the toxin did not discriminate between the elongated and more flexible dendrimer 2d backbone with respect to 2c (Figure 2).
Table 1. Results of Inhibition in the Stx1B ELISA Assaya.
| entry | Construct | ligand | valency (% functionalization of polymer) | IC50 (μM) | rel.pot.b | rel. pot. per sugarc |
|---|---|---|---|---|---|---|
| 1 | 1e | galabiose | 1 | 4968 ± 1232 | 4968 | 1 |
| 2 | 3 | galabiose | 2 | 1070 ± 283 | 4.6 | 2.3 |
| 3 | 4 | galabiose | 2 | 1245 ± 169 | 4 | 2 |
| 4 | 5 | galabiose | 4 | 19.9 ± 2.4 | 250 | 62.5 |
| 5 | 6 | galabiose | 4 | 13.5 ± 2.6 | 367 | 92 |
| 6 | 8 | globotriose | 10 (8%) | 2.8 ± 0.2 | 1,774 | 187 |
| 7 | 9 | galabiose | 20 (16%) | 0.0083 ± 0.0006 | 598,554 | 29,928 |
Determined in an ELISA-like assay with Stx1B (0.1 μg/mL) and wells coated with Gb3.
Relative to the potency of galabiose for 3, 4, 5, 6, and 9.
Relative potency divided by the valency.
Figure 2.
Inhibition of STx1B (0.1 μg/mL) binding to a GB3 covered surface by compounds from left to right, 9 (blue), 8 (red), 1e (black), and 5 (white).
We expected both compounds to bridge between the strongest of the three binding sites per subunit, the so-called site 2,34 of the same pentamer separated by ca. 30 Å.6 For the decavalent hPG-Gb3 polymeric inhibitor 8, low micromolar inhibition was seen (IC50 = 3 μM). Indeed, the compound was more potent than the tetravalent 5 and 6 but not much and the inherently stronger trisaccharide ligand it contains could easily be responsible for this difference. Gratifyingly, the more highly substituted hPG-galabiose conjugate 9 was much more potent with an IC50 of 8 nM and a relative potency per sugar of ca. 30,000. These data make it the first nanomolar Stx inhibitor based on the disaccharide galabiose to the best of our knowledge.
A number of natural or synthesized oligosaccharides were subsequently tested for activity at a maximal nontoxic concentration of 2% (Figure 4).35,36 Chitosan oligosaccharide (COS) is a cationic polymer obtained from crustaceans and consists of glucosamine repeating units and has several promising applications.37 COS (degree of acetylation: ≥95%) showed a 71% inhibition of the Stx1B with inhibitory effects seen as low as a 0.5% COS concentration (Figure 5). AOS, another naturally occurring polyuronic saccharide, is composed of β-d-mannuronic acid and α-l-guluronic acid.38 AOS have, among others, antitumor, antioxidative, immunoregulatory, and anti-inflammatory activity.39 AOS did show 51% inhibition at a concentration of 0.5%. Curiously, higher AOS concentrations reduced inhibition. FOS and GOS did not inhibit the toxin. Lactose was used as a negative control and did not show any activity.
Figure 4.
Structures of natural and synthetic inhibitors of Stx.
Figure 5.

Stx1B binding inhibition by COS.
In order to evaluate the potential toxicity of the most effective Stx inhibitor, polymer 9, toxicity tests were undertaken. Different concentrations of glycopolymer 9 (1, 10, and 100 nM) did not impair T84 cell viability after 24 h exposure as indicated by the MTT assay, while 10% ethanol (positive control) significantly reduced the cell viability (Figure 6a). Furthermore, as depicted in Figure 6b, 1, 10, and 100 nM of glycopolymer 9 did not significantly alter the transepithelial electrical resistance (TEER) values compared to untreated cells after 24 h, whereas 10% ethanol strongly decreased the TEER values. TEER values (transepithelial electrical resistance) indicate the barrier integrity of epithelial cells.
Figure 6.
Effect of glycopolymer 9 on intestinal cell viability and integrity. (a) T84 cells grown on 96-well plates were exposed to 1, 10, and 100 nM glycopolymer 9 or 10% ethanol (positive control) for 24 h, and cell viability was measured by the MTT reduction assay. The MTT values were presented as the percentage MTT released by nontreated T84 cells as mean ± SEM of three independent experiments each performed in triplicate. (b) T84 cells grown on transwell inserts exposed to 1, 10, and 100 nM glycopolymer 9 or 10% ethanol (positive control) for 24 h, and TEER was measured as described in the Experimental Section. The TEER values are presented as mean (Ω·cm2) ± SEM of three independent experiments each performed in triplicate. ** = P < 0.001 compared to control. **** = P < 0.0001 compared to control).
Discussion and Conclusions
A growing number of Shigella infections contain the deadly Shiga toxin, and the related STEC is also still a major threat without a proper therapeutic approach. In this study, we aimed for a simple and effective toxin inhibitor by comparing three classes of carbohydrate-based inhibitors: glycodendrimers, glycopolymers, and oligosaccharides. The glycodendrimers needed at least a tetravalent ligand to reach significant inhibition. One reason could be that it requires bridging40 between the two highest affinity sites (sites 2) of neighboring toxin subunits for a significant inhibitory effect. The smaller divalent compounds were too short to bridge ca. 30 Å. It is likely that in addition to the chelation binding mode, aggregation of the toxin also takes place, as previously noted,41 and also for the related cholera toxin.42,43 Of the two glycopolymers, it is striking that the more highly functionalized 9 was much more potent than 8, despite having the weaker galabiose ligand. Clearly, the high density of binding sites helps in inhibition as seen for the related cholera toxin inhibition with similar polymers;44,45 however, with three binding sites per subunit, that is, 15 in total, the effects are more dramatic than those for the cholera toxin with one binding site per subunit. Prior works, both theoretical and practical involving the Shiga-like toxin, have clearly indicated that the avidity effects as seen here are caused by intrinsic inter- and intramolecular recognition events, but on top of that, there is an important combinatorial factor that describes the probabilities of binding events. This factor is very important and favorable and was shown to increase rapidly for higher valency systems, provided that the geometry of the multivalent ligand is appropriate for the target. In the case at hand, the particle-like nature of the polymer is particularly suitable for toxins in comparison with other polymers.44 Furthermore, the polymers were both ca. 10 kDa, but the ligand density is vastly different (valencies of 10 vs 20 for 8 and 9). Clearly, the statistical possibilities for higher ligand density 9 are far greater and can overcome the lower intrinsic binding potency of the disaccharide versus trisaccharide ligand.52
Shigella spp. are highly infective bacteria. 10–100 microbes are enough to cause infection that could become fatal, especially when it produces the toxin, which is also the case for STEC. The initial diarrhea followed by the toxin moving into circulation provides a challenge for therapy. It takes ca. 5–9 days between the initial gastroenteritis until HUS occurs.46 In this time window, a GI-based agent, for example, a food-grade polysaccharide, such as, COS, can be beneficial. This is true also as a preventative, in case of an outbreak which can happen with Shigella. In order to prevent systemic diseases, that is, HUS, a soluble nontoxic multivalent glycan with sufficient potency will likely be helpful. As such, a further optimized dendrimer or the glycopolymer 9 based on hPG can be used. hPGs can be prepared on a large scale in an economical manner and have also been used in circulation.47 The utility of hPG is also well established in terms of safety and biocompatibility. We have also previously used a hPG backbone to target the cholera toxin and the flu virus with good results.44,48
Experimental Section
Plasmid Construction, Protein Expression, and Protein Purification
The Stx1B expression plasmid was constructed by using the Gateway recombinant cloning kit (Thermo Fisher, Spain). Briefly, a synthetic DNA cassette (Invitrogen, Spain) that encodes residues 21–89 of Stx1B (GenBank: AAA98348.1) with a C-terminal 6× His tag was first inserted into an entry plasmid pENTR1A through restriction sites Dra I and Xho I. The resultant plasmid Stx1B-His-pENTR1A together with a destination vector pDEST 14 was further subjected to the Gateway LR Clonase cloning reaction to achieve the final protein expression construct Stx1B-His-pDEST14 following the manufacturer’s instructions.
Protein expression was carried out using E. coli BL21 cells transformed with the plasmid Stx1B-His-pDEST14. The above E. coli cells were grown in LB broth media containing 100 mg/mL ampicillin at 37 °C until OD450 reached 0.6, and then, IPTG was added to the culture at a final concentration of 1 mM to induce the expression of recombinant proteins at room temperature for 16 h. At the end of IPTG induction, E. coli cells were immediately lysed in the culture using a B-PER direct bacterial protein extraction kit (Thermo Fisher, Spain) with the supplementation of the protease inhibitor (EDTA-free), DNase I, and lysozyme following the manufacturer’s instructions. After centrifugation for 10 min at 12,000 rpm at 4 °C, the supernatant was collected for further protein purification.
Protein purification was performed using a home-made column packed with HisPur Ni-NTA resin (Thermo Fisher, Spain) according to the manufacturer’s instructions. After the column was washed with an equilibration buffer (20 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole, pH 7.4), it was then loaded with the above supernatant containing 6× His tagged Stx1B at 4 °C for 1 h. Unbound proteins were removed from the column by using a washing buffer (20 mM sodium phosphate, 300 mM sodium chloride, 20 mM imidazole, pH 7.4). Finally, 6× His tagged Stx1B was eluted from the column using an elution buffer (20 mM sodium phosphate, 500 mM sodium chloride, and 300 mM imidazole, pH 7.4) and later confirmed by SDS-PAGE (Figure S1). Before applying for the later binding assay, imidazole residues in the eluted proteins were removed using 10 kDa molecular weight cut off protein concentrators (Thermo Fisher, Spain) and a buffer containing 20 mM sodium phosphate and 500 mM sodium chloride.
Shiga Toxin Inhibition Assay
A (Stx1B ELISA) 96-well plate (Nunc PolySorp) was coated with a solution of FSL-GB3 (50 μL, 2 μg/mL) in phosphate-buffered saline (PBS) for 3 h at room temperature. Unattached GB3 was removed by washing with PBS (0.2% BSA), and the remaining binding sites of the surface were blocked with BSA (1%) for 1 h, followed by washing with PBS (0.2% BSA). Samples of Stx1B (50 μL, 0.1 μg/mL) and inhibitors were transferred to the GB3-coated plate and incubated at r.t. for 1 h followed by washing with PBS (0.2% BSA). HisProbe-HRP (4 mg/mL, 1:2000 dilution, 100 μL/well) was incubated for 0.5 h followed by washing with PBS (0.2% BSA).49 The HRP activity was measured using the 1-Step Ultra TMB-ELISA substrate solution (100 μL/well) for a maximum of 10 min. After quenching with H2SO4, the absorbance in each well was measured at 450 nm. Compounds 3, 4, 5, 6, 8, and 9 were tested at least twice in duplicate or triplicate, whereas compound 1e was tested once. Inhibition data from the experiments were averaged and fitted in GraphPad Prism 8.3.0 with a nonfixed Hill-slope.
Cell Culture
Human colonic epithelial T84 cells (ATCC CCL-248) were cultured in Dulbecco’s modified Eagle’s medium: nutrient mixture F-12 (DMEM/F-12; Gibco, Invitrogen, Carlsbad, CA, USA) (1:1) supplemented with 10% fetal calf serum (FCS; Gibco), penicillin (100 U/mL), and streptomycin (100 g/mL) (Biocambrex) and maintained at 37 °C in a humidified incubator with 5% CO2. Τ84 cells were grown on plastic culture flasks (75 cm2) at a density of 3 × 106 cells/mL. After 7 days, T84 cells were seeded on 0.3 cm2 high pore density polyethylene terephthalate membrane transwell inserts with 0.4 μm pores (Falcon, BD Biosciences, Franklin Lakes, NJ, USA) placed in a 24-well plate (density of 3 × 105 cells/insert) or in 96-well microtiter plates (Costar 3614, Corning, NY, USA) at a density of 3 × 104 cells/well. Cells were passaged by addition of trypsin ethylenediaminetetraacetic acid (EDTA) at 100% confluency every week. The experiments were performed at passage number 51–55 on fully confluent monolayers with TEER values >1000 Ω·cm2.
Oligosaccharides
Fructo-oligosaccharides (FOS) isolated from chicory were obtained from Orafti (Wijchen, The Netherlands) (purity >97%). Galacto-oligosaccharides (GOS) (Vivinal GOS Powder, purity >70%) produced from lactose were provided by FrieslandCampina (Amersfoort, The Netherlands). AOS prepared by degradation of algin (purity >85%) and chitosan oligosaccharides (COS) derived from rich marine biological sources (shrimp & crab shells) (purity >90%) were both purchased from BZ Oligo Biotech Co., Ltd. (Qingdao, Shandong, China). All oligosaccharide solutions were freshly prepared through dissolution in DMEM/F12, and their pH was adjusted to pH = 7.2–7.4.
TEER Measurement
For evaluating the epithelial integrity of the T84 monolayer, TEER values were measured using a Millicell-ERS Volt-Ohm-meter (Millipore, Temecular, CA, USA). As described above, T84 cells were seeded at a density of 3 × 105 cells/insert and cultured for 3 weeks. The inserts were placed in a 24-well plate with 300 μL of medium at the apical compartment and 700 μL of medium at the basolateral compartment. Different concentrations of the Glycopolymer 9 (1, 10 and 100 nM) were added to the apical compartment of the transwell inserts. Transwell inserts of T84 cells, treated with medium, were considered as the control group. The TEER values were measured before and 24 h after exposure to different concentrations of Glycopolymer 9 incubated at 37 °C in 5% CO2. The TEER was expressed as Ω·cm2.
Viability—MTT Assay
Cell viability was measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reduction assay (Sigma-Aldrich, St. Louis, Mo, USA). T84 cells were seeded on a flat-bottomed 96-well plate at a density of 3 × 104 cells/well and grown for 7 days until they reached 100% confluency. Thereafter, cells were exposed to three concentrations of glycopolymer 9 (1, 10, and 100 nM), and 10% ethanol was used as positive control. The wells with medium (no treatment) were considered as the control group. After a 24 h incubation at 37 °C in 5% CO2, the medium was removed, and 120 μL of MTT working solution [20 μL MTT (5 mg/mL) and 100 μL medium] was added to each well and incubated for 2 h under the same conditions. Finally, DMSO was added to lyse the cells and dissolve the purple-blue sediment. After 5 min of mild shaking, the absorbance value of each well was measured at 595 nm using a Glomax Discover microplate reader. The viability of the T84 cells was calculated based on the following equation: (mean absorbance of treatment cells/mean absorbance of control cells) × 100.
Statistical Analysis
Data were reported as mean values ± SEM of three independent experiments (n = 3) routinely performed in triplicate (3 wells/condition). Results were analyzed using Prism 8.0 GraphPad software (GraphPad, San Diego, CA, USA). Statistical significance was determined using one-way ANOVA followed by Bonferroni post-hoc test. Differences were considered statistically significant when P < 0.05.
Chemistry
Chemicals were obtained from commercial sources and were used without further purification unless noted otherwise. The solvents were obtained as synthesis grade and stored on molecular sieves (4 Å). TLC was performed on Merck-precoated silica plates. Spots were visualized by UV light and 10% H2SO4 in MeOH. Microwave reactions were carried out in a Biotage microwave initiator (300W, Uppsala, Sweden). The microwave power was limited by temperature control once the desired temperature was reached. Sealed vessels of 2–5 and 10–20 mL were used. 1H NMR, HSQC, COSY (600 MHz), and 13C (151 MHz) were performed on a Bruker 600 spectrometer. Infrared (IR) spectroscopy was performed using a universal attenuated total reflectance (UATR) accessory of a PerkinElmer Spectrum Two FT-IR spectrometer. High-resolution mass spectrometry analysis was recorded using an Agilent 6560 Ion Mobility Q-TOF LC/MS instrument. Analytical HPLC and Preparative HPLC runs were performed on a Shimadzu 20A HPLC system. Analytical HPLC was performed using a Dr Maisch GmBh C18-AQ column (5 μm) at a flow rate of 0.5 mL/min. The used buffers were H2O (buffer A) and CH3CN (buffer B). Runs were performed using a standard protocol: 2–100% gradient buffer B in 35 min; UV-absorption was measured at 254 nm. For Preparative HPLC, a Waters XBridge BEH Prep Amide column (5 μm, 250 × 10 mm) at a flow rate of 2.4 mL/min was used. Runs were performed using a standard protocol: 95–50% gradient buffer B in 60 min. UV-absorption was measured at 254 and 210 nm. For commercial oligosaccharides, AOS (food grade) was purchased from Qingdao Bz Oligo Biotech Co. Ltd. COS (9012-76-4) with a degree of deacetylation ≥95%. GOS was purchased from Friesland Campina (Vivinal GOS powder, 69%). Bis-alkyne 2a(29) and Tetra-alkyne 2c(30) were synthesized according to the reported procedures, with the spectral data in agreement with the reported values.
General CuAAC Procedure for the Synthesis of Multivalent Galabiose Compounds 3, 4, 5, and 6
All tested compounds were >95% pure by HPLC. The alkyne (2a, 2b, 2c, 2d, and 1 equiv) was dissolved in DMF followed by the addition of the ligand 1d (1.2 equiv). Copper sulfate pentahydrate (0.1 equiv) was dissolved in water separately and added to the reaction mixture. Sodium ascorbate (0.3 equiv) was also dissolved in water separately and added to the reaction mixture. The reaction was performed at 80 °C in the microwave for 1 h. The reaction mixture was extracted using EtOAc and water, followed by column purification (6% MeOH in DCM) to obtain the purified compound which was further subjected to deacetylation as described below.
General Procedure for the Deacetylation Reaction
The peracetylated compound was dissolved in anhydrous methanol, followed by addition of a catalytic amount of an aqueous NaOH solution (1 M) and stirred at room temperature. The reaction was monitored by TLC. Upon completion of the reaction, the mixture was neutralized by the addition of Dowex marathon resin. The solvent was evaporated, and the crude mixture was purified by preparative HPLC to obtain the pure product (1e, 3, 4, 5, and 6) in >80% yields.
Azide 1c
Compound 1a (2.00 g, 3.13 mmol, 1 equiv), diphenyl sulfoxide (1.2 g, 6.0 mmol, 2.6 equiv), and 2,4,6-tris-tert-butylpyrimidine (2.232 g, 9.0 mmol, 3.0 equiv) were dissolved in anhydrous DCM (45 mL) under an atmosphere of argon. Activated molecular sieves (3 Å) were added. The solution was then cooled to −40 °C, and trifluoromethanesulfonic anhydride (500 μL, 3 mmol, 1.3 equiv) was added. The mixture was stirred for 10 min, and galactose acceptor 1b (2.261 g, 4.373 mmol, 1.4 equiv) was added as a solution in anhydrous DCM (40 mL). The reaction was stirred for ca. 1.5 h at −40 °C and then quenched by addition of triethylamine (5 mL, excess). The mixture was diluted with DCM (100 mL) and washed with 1 M HCl and saturated aqueous sodium bicarbonate. The organic layer was dried with NaSO4, filtered, and concentrated in vacuo. Purification was done by column chromatography (0–20% EtOAc in petroleum ether) to yield the product (1.655 g, 1.58 mmol, 50%). 1H NMR (600 MHz, CDCl3): δ 8.09 (dd, J = 8.1, 1.4 Hz, 2H, 2 × CH Bz ortho), 8.04 (dd, J = 8.3, 1.3 Hz, 2H, 2× CH Bz ortho), 7.93 (dd, J = 8.2, 1.4 Hz, 2H, 2× CH Bz ortho), 7.62–7.56 (m, 1H, CH Bz para), 7.50 (dddd, J = 8.8, 6.1, 3.0, 1.3 Hz, 2H, 2× CH Bz para), 7.46 (t, J = 7.8 Hz, 2H, 2× CH Bz meta), 7.36 (td, J = 7.8, 3.7 Hz, 4H, 4× CH Bz meta), 5.69 (dd, J = 10.5, 8.6 Hz, 1H, H-2), 5.46 (dd, J = 10.5, 2.4 Hz, 1H, H-3), 5.02 (d, J = 3.1 Hz, 1H, H′-1), 4.83–4.76 (m, 2H, H-1, H-6a), 4.68 (dd, J = 12.1, 7.3 Hz, 1H, H-6b), 4.41 (d, J = 2.5 Hz, 1H, H-4), 4.38 (d, J = 2.4 Hz, 1H, H′-4), 4.24–4.16 (m, 3H, H′-2, H-5, H′-6a), 4.12–4.07 (m, 2H, H′-3, H′-5), 4.05 (dd, J = 12.7, 1.9 Hz, 1H, H′-6b), 1.03 (s, 9H, SiC(CH3)3), 1.00 (s, 9H, SiC(CH3)3), 0.97 (s, 9H, SiC(CH3)3), 0.86 (s, 9H, SiC(CH3)3), 0.19 (s, 3H, SiCH3), 0.17 (s, 3H, SiCH3), 0.08 (s, 3H, SiCH3), 0.04 (s, 3H, SiCH3). 13C NMR (151 MHz, CDCl3): δ 166.15 (C=O, Bz), 165.82 (C=O, Bz), 165.12 (C=O, Bz), 133.60 (CH, Bz para), 133.36 (CH, Bz para), 133.21 (CH, Bz para), 130.01 (2× CH, Bz ortho), 129.83 (2× CH, Bz ortho), 129.80 (2× CH, Bz ortho), 129.78 (C Bz), 128.94 (C Bz), 128.67 (C Bz), 128.57 (2× CH, Bz meta), 128.43 (2× CH, Bz meta), 128.38 (2× CH, Bz meta), 101.45 (C′-1), 88.38 (C-1), 75.69 (C′-5), 75.05 (C′-4), 75.04 (C-4), 73.17 (C-3), 70.94 (C′-3), 70.04 (C′-2), 69.17 (C′-5), 68.68 (C-2), 67.02 (C′-6), 64.12 (C-6), 27.46 (SiC(CH3)3), 27.36 (SiC(CH3)3), 26.23 (SiC(CH3)3), 26.21 (SiC(CH3)3), 23.41 (2× SiC(CH3)3), 18.36 (SiC(CH3)3), 18.22 (SiC(CH3)3), −3.91 (SiCH3), −4.23 (SiCH3), −4.35 (SiCH3), −4.66 (SiCH3).
Azide 1d
Compound 1c (1655 mg, 1.58 mmol 1 equiv) was solved in MeOH (30 mL), and an excess of K2CO3 was added. The reaction was stirred at r.t. for 16 h and then filtered and concentrated in vacuo. The crude product was dissolved in EtOAc and extracted with aqueous NaHCO3, and the organic layer was separated, dried with NaSO4, filtered, and concentrated again in vacuo. The product was purified using column chromatography (10–50% EtOAc in petroleum ether) to yield the product (1034 mg, 1.41 mmol, 89%).The debenzoylated product (332 mg, 0.452 mmol, 1 equiv) was dissolved in DCM (5 mL), and HF-Pyridine (70%, 0.4 mL) was added dropwise at r.t. under continuous argon flow. The reaction was stirred for 2 h and quenched by addition of solid CaCl2 (99 mg, 0.904 mmol, 2 equiv), pyridine (10 mL, 124 mmol, 272 equiv), Ac2O (5 mL, 53 mmol, 117 equiv), and DMAP (3 mg, 0.02 mmol, 0.05 equiv). The reaction was stirred for 16 h at r.t. and then diluted with EtOAc and washed with saturated, aqueous K2CO3. The organic phase was dried with NaSO4, filtered, and concentrated in vacuo. The product was purified using silica gel flash chromatography using a gradient of 40–100% EtOAc in petroleum ether (154 mg, 0.232 mmol, 51% (over 3 steps)). The spectral data was in accordance with published data.501H NMR (600 MHz, CDCl3): δ 5.53 (dd, J = 3.4, 1.3 Hz, 1H, H′-4), 5.33 (dd, J = 11.0, 3.4 Hz, 1H, H′-3), 5.16 (dd, J = 11.0, 3.7 Hz, 1H, H′-2), 5.12 (dd, J = 10.7, 8.6 Hz, 1H, H-2), 4.99 (d, J = 3.7 Hz, 1H, H′-1), 4.83 (dd, J = 10.7, 2.7 Hz, 1H, H-3), 4.63 (d, J = 8.6 Hz, 1H, H-1), 4.45 (ddd, J = 7.6, 5.8, 1.3 Hz, 1H, H′-5), 4.40 (dd, J = 11.4, 6.9 Hz, 1H, H-6a), 4.12 (dd, J = 11.3, 6.1 Hz, 1H, H-6b), 4.10–4.04 (m, 3H, H′6ab; H-4), 3.87 (t, J = 6.5 Hz, 1H, H-5), 2.09 (s, 3H, CH3 Ac), 2.07 (s, 3H, CH3 Ac), 2.05 (s, 3H, CH3 Ac), 2.04 (s, 3H, CH3 Ac), 2.04 (s, 3H, CH3 Ac), 2.00 (s, 3H, CH3 Ac), 1.95 (s, 3H, CH3 Ac). 13C NMR (151 MHz, CDCl3): δ 170.45 (C=O, Ac), 170.37 (C=O, Ac), 170.32 (C=O, Ac), 170.27 (C=O, Ac), 169.98 (C=O, Ac), 169.66 (C=O, Ac), 168.90 (C=O, Ac), 98.97 (C′-1), 88.11 (C-1), 76.27 (C-4), 74.03 (C-5), 72.40 (C-3), 68.30 (C′-2), 67.88 (C-2), 67.67 (C′-4), 67.18 (C′-5), 67.07 (C′-3), 61.81 (C-6), 60.42 (C′-6), 20.76 (CH3, Ac), 20.62 (CH3, Ac), 20.58 (CH3, Ac), 20.54 (CH3, Ac), 20.51 (CH3, Ac), 20.47 (CH3, Ac).
Azide 1e
Compound 1d was deprotected using the general procedure described above to obtain the final compound 1e in 90% yield. The spectral data was in accordance with published data.511H NMR (600 MHz, MeOD): δ 4.94 (d, J = 3.7 Hz, 1H, H′-1), 4.52 (d, J = 8.4 Hz, 1H, H-1), 4.18 (t, J = 6.2 Hz, 1H, H-5), 3.99 (d, J = 3.0 Hz, 1H, H-4), 3.88 (d, J = 3.2 Hz, 1H, H′-4), 3.85–3.71 (m, 6H, H′-2, H′-3, H′-5, H′-6ab, H-6a), 3.65 (dd, J = 11.2, 5.1 Hz, 1H, H-6b), 3.52 (dd, J = 10.0, 2.9 Hz, 1H, H-3), 3.41 (dd, J = 10.3, 1.9 Hz, 1H, H-2). 13C NMR (151 MHz, MeOD): δ 101.43 (C′-1), 91.39 (C-1), 78.37 (C-5), 76.70 (C-4), 73.43 (C-3), 71.62 (C′-5), 71.02 (C-2), 69.83 (C′-4), 69.66 (C′-2), 69.20 (C′-3), 61.30 (C-6), 59.70 (C′-6).
Compound 2b
Methyl 3,5-bis(2-(boc-amino)ethoxy)benzoate (110 mg, 0.25 mmol, 1 equiv) was prepared as reported30 and dissolved in 1:1 TFA/DCM and stirred at r.t. for 2 h before concentrating in vacuo. The residue was dissolved in DCM (10 mL), and TEA (139 μL, 1 mmol, 4 equiv) was added, and the mixture was left stirring for 5 min at r.t. before cooling to 0 °C. Propargyl chloroformate (55 μL, 0.55 mmol, 2.2 equiv) was added dropwise, and the reaction was allowed to slowly warm up to r.t. and was left stirring for 16 h. The reaction was diluted with an excess of DCM and 1 M aq HCl, and the organic layer was collected, dried with NaSO4, filtrated, and concentrated in vacuo. The compound was purified by column chromatography using a gradient of 0–50% EtOAc in petroleum ether and with 1% TFA yielding the free acid (89 mg, 0.22 mmol, 88%). 1H NMR (600 MHz, methanol-d4): δ 7.20 (d, J = 2.4 Hz, 2H, 2× CHarom-2,6), 6.79 (d, J = 2.5 Hz, 1× CHarom-4), 4.68 (d, J = 2.4 Hz, 4H, 2× CH2, propargyl), 4.07 (t, J = 5.5 Hz, 4H, 2× OCH2), 3.41 (t, J = 5.5 Hz, 4H, 2× NCH2), 2.89 (t, J = 2.5 Hz, 2H, 2× C≡CH). 13C NMR (151 MHz, Methanol-d4): δ 169.61 (C=O, acid), 161.30 (CO, aromatic), 158.13 (2× C=O, carbamate), 134.17 (Carom-COOH), 109.32 (2× CHarom-2,6), 107.26 (CHarom-4), 79.44 (2× C≡CH), 75.80 (2× C≡CH), 68.11 (2× OCH2), 53.20 (2× CH2, propargyl), 41.43 (2× NCH2). HR-ESI-TOF/MS (m/z): [M + Na]+ calcd for C19H20N2O8, 427.1117; found, 427.1116.
Compound 2d
To a solution of 2a (16.4 mg, 82 μmol) in DCM (1 mL) was added (COCl)2 (31 μL, 246 μL, 3 equiv) and DMF (10 μL). After stirring at r.t. for 1.5 h, the mixture was concentrated. The resulting residue was coevaporated with 10 mL of anhydrous toluene and then redissolved in DCM (1 mL) and cooled to 0 °C. A solution of pyridine (1 mL), DCM (1 mg), and dodecane-1,12-diamine (10 mg) was added slowly to the reaction flask. The resulting mixture was stirred at r.t. overnight. Solvents were removed, and the residue was partitioned between EtOAc and water. The organic layer was separated, washed with brine (1×), dried with anhydrous MgSO4, filtered, and concentrated. The residue was purified by silica gel flash chromatography and yielded a product (20 mg, 80%). 1H NMR (600 MHz, acetone-d6): δ 7.57 (t, J = 5.9 Hz, 2H, NH), 7.02 (s, 4H, 4× CH arom), 6.64 (s, 2H, 2× CH arom), 4.71–4.67 (m, 8H, (4× OCH2), 3.24 (q, J = 6.7 Hz, 4H, 4× CH propargyl), 2.97 (s, 4H, 2× NHCH2), 1.92 (s, 4H, 2× CH2), 1.46 (t, J = 7.1 Hz, 4H, 2× CH2), 1.28–1.02 (m, 12H, 6× CH2). 13C NMR (151 MHz, acetone-d6): δ 165.64 (2× C=O), 158.77 (4× CO, arom), 137.57 (2× C-CONH), 106.68 (4× CH), 104.54 (2× CH), 78.54 (4× CCH, alkyne), 76.38 (4× CCH, alkyne), 55.70 (4× OCH2), 39.64 (2× N-CH2), 29.32 (CH2), 29.30 (CH2), 29.17 (CH2), 29.15 (CH2), 29.04 (CH2), 29.01 (CH2), 28.90 (CH2), 28.77(CH2), 28.64 (CH2). HR-ESI-TOF/MS (m/z): [M + H]+ calcd for C38H44N2O6, 625.3277; found, 625.3302.
Compound 3
1H NMR (600 MHz, Deuterium Oxide): δ 8.23 (s, 2H, triazole), 7.07 (dd, J = 2.3, 0.9 Hz, 2H, 2× CHarom-2,6)), 6.68 (td, J = 2.3, 0.8 Hz, 1H, 1× CHarom-4), 5.58 (dd, J = 9.0, 0.8 Hz, 2H, 2× H-1), 5.18 (s, 4H, 2× OCH2), 4.90 (d, J = 4.0 Hz, 2H, 2× H′-1), 4.29 (t, J = 6.4 Hz, 2H, 2× H-5), 4.20 (t, J = 9.5 Hz, 2H, 2× H-2), 4.07 (d, J = 3.2 Hz, 2H, 2× H′-5), 3.96–3.68 (m, 14H, 2× H′-4, 2× H′-2, 2× H′-6, 2× H-4, 2× H′-3, 2× H-3), 3.64–3.55 (m, 4H, 2× H-6).
13C NMR (151 MHz, deuterium oxide): δ 174.24 (-COOH), 158.33 (2× Carom-3), 143.27 (2× −OCH2-C), 139.24 (COOH–C), 125.28 (2× N–CH), 108.97 (2× CHarom-2,6)), 105.72 (1× CHarom-4), 100.48 (2× C′-1), 88.10 (2× C-1), 78.28 (2× C′-5), 77.24 (2× C-4), 72.69 (2× C′-3), 70.90 (2× C-5), 69.61 (2× C-3), 69.05 (2× C′-2), 68.98 (2× C′-4), 68.63 (2× C-2), 61.39 (2× OCH2), 60.54 (2× C′-6), 60.06 (2× C-6). HR-ESI-TOF/MS (m/z): [M + Na]+ calcd for C37H52N6O24, 987.2930; found, 987.2934.
Compound 4
1H NMR (600 MHz, deuterium oxide): δ 8.23 (s, 2H, triazole), 7.07 (s, 2H, 2× CHarom-2,6)), 6.66 (s, 1H, 1× CHarom-4), 5.66 (d, J = 9.0 Hz, 2H, 2× H-1), 5.20 (d, J = 2.4 Hz, 4H, 2× NH–CO–CH2), 5.03 (d, J = 4.0 Hz, 2H, 2× H′-1), 4.41 (t, J = 6.5 Hz, 2H, 2× H-5), 4.31 (t, J = 9.6 Hz, 2H 2× H-2), 4.20 (d, J = 3.0 Hz, 2H, 2× H-3), 4.13 (t, J = 5.1 Hz, 4H, 2× CONH–CH2–CH2), 4.07–3.84 (m, 14H, 2× H′-4, 2× H′-3, 2× H′-2, 2× H′-5, 2× H-4, 2× H-6), 3.78–3.68 (m, 4H, 2× H′-6), 3.52 (t, J = 5.2 Hz, 4H, 2× CONH-CH2-CH2). 13C NMR (151 MHz, deuterium oxide): δ 157.89 (2× Carom3,5), 125.11 (2× N–CH), 108.29 (2× CHarom-2,6)), 100.47 (2× C′-1), 88.08 (2× C-1), 78.23 (2× C′-5), 77.20 (2× C-4), 72.68 (2× C′-3), 70.89 (2× C-5), 69.59 (2× C-3), 69.06 (2× C′-2), 68.97 (2× C′-4), 68.62 (2× C-2), 67.18 (2× OCH2), 60.53 (2× C′-6), 60.00 (2× C-6), 57.63 (2× NHCO–O–CH2), 40.09 (2× OCH2–CH2).
HR-ESI-TOF/MS (m/z): [M + Na]+ calcd for C43H62N8O28, 1161.3571; found, 1161.3574.
Compound 5
1H NMR (600 MHz, deuterium oxide): δ 8.26 (s, 4H, triazole), 7.06 (s, J = 1.9 Hz, 2H, 2× CHarom-2,6), 6.70 (s, 1H, 1× CHarom-4), 6.62 (d, J = 2.2 Hz, 4H, 4× CHarom-2′,6′), 6.44 (s, 2H, 2× CHarom-4′), 5.64 (d, J = 9.0 Hz, 4H, 4× H-1), 5.00 (d, J = 4.0 Hz, 4H, 4× H′-1), 4.95 (s, 8H, 4× triazole-CH2–), 4.35 (t, J = 6.4 Hz, 4H, 4× H-5), 4.29 (t, J = 9.6 Hz, 4H, 4× H-2), 4.17 (d, J = 3.2 Hz, 8H, 4× H-4, 2× CONH–CH2–CH2−), 4.00 (t, J = 6.3 Hz, 4H, 4× H′-5), 3.97–3.88 (m, 12H, 4× H-3, 4× H′-3, 4× H′4), 3.88–3.78 (m, 12H, 4× H′-6, 4× H′-2), 3.68 (d, J = 6.4 Hz, 8H, 4× H-6), 3.64 (s, 4H, 2× CONH–CH2–CH2−). 13C NMR (151 MHz, deuterium oxide, extracted from HSQC): δ 125.03 (triazole), 108.89 (2× CHarom-2,6, 104.45 1× CHarom-4), 106.57 (4× CHarom-2′,6′), 105.32 (2× CHarom-4′), 88.11(C-1), 100.53 (C′-1), 61.07 (triazole-CH2-), 70.93 (C-5), 69.65 (C-2), 77.35, 66.73, 78.13 (C′-5), 68.94, 72.72, 59.97, 68.70, 60.52 (C-6), 39.84 (CONH–CH2–CH2−). HR-ESI-TOF/MS (m/z): [M – H]− calcd for C85H116N14O50, 2132.6964; found, 2132.6869.
Compound 6
1H NMR (600 MHz, deuterium oxide): δ 8.18 (s, 4H, 4H, triazole), 6.92 (s, 4H, 4× CHarom-2′,6′), 6.56 (s, 2H, 2× C CHarom-4′), 5.54 (s, 4H, 4× H-1), 4.98 (s, 6H, 4× H′-1), 4.76 (s, 8H, 4× triazole-CH2−), 4.32 (s, 4H, 4× H-5), 4.25 (s, 4H, 4× H-2), 4.12 (s, 4H, 4× H-4), 4.01–3.49 (m, 32H, 4× H′-2, 4× H′-3, 4× H′-4, 4× H′-5, 4× H′-6, 4× H-3, 4× H-6), 3.19 (s, 4H, 2× CONH–CH2−), 1.42 (s, 4H, 2× CONH–CH2–CH2−), 1.05 (s, 16H, CONH-(CH2)12). HR-ESI-TOF/MS (m/z): [M – H]− calcd for C86H128N14O46, 2092.8107; found, 2092.8028.
Compound 8
hPG-azide (2.5 mg, 0.002 mmol of azide groups) was dissolved in water followed by the addition of ligand 7 (1.8 mg, 0.0032 mmol, 1.6 equiv). Copper sulfate pentahydrate (0.1 equiv) was dissolved in water separately and added to the reaction mixture. 0.3 equiv of sodium ascorbate was also dissolved in water separately and added to the reaction mixture. The reaction was carried out at 100 °C in the microwave for 1 h. Cuprisorb resin was added to the reaction mixture and stirred to adsorb excess copper. The solvent was evaporated, and the crude reaction mixture was purified by dialysis using a cellulose based dialysis cassette (MWCO: 2k) against deionized water for 3–4 days and freeze dried to get 8 in 80% yield as an off-white solid. The disappearance of the azide stretching peak in the IR spectra of the final compound confirmed that all of the azido polymer were consumed. 1H NMR (600 MHz, deuterium oxide): δ 8.03 (s, triazole), 5.18–3.16 (m, CH2 and CH, hPG-OH backbone; GB3), 2.31–2.18 (m, GB3; CH3), 0.85 (s, hPG core, CH2).
Compound 9
hPG-propargyl (5 mg, 0.0095 mmol of propargyl groups) was dissolved in water followed by the addition of 1d (8.2 mg, 0.012 mmol, 1.3 equiv) which was dissolved in DMF. Copper sulfate pentahydrate (0.1 equiv) was dissolved in water separately and added to the reaction mixture. 0.3 equiv of sodium ascorbate was also dissolved in water separately and added to the reaction mixture. The reaction was carried out at 80 °C in the microwave for 60 min. Cuprisorb resin was added to the reaction mixture and stirred to adsorb excess copper. The crude mixture was extracted using ethyl acetate and water. The protected polymer conjugate was then subjected to deacetylation using the standard procedure described above. The solvent was evaporated, and the crude reaction mixture was purified by dialysis using a cellulose-based dialysis cassette (MWCO: 2k) against deionized water for 3–4 days and freeze-dried. The final product 9 was obtained in 75% yield as a white solid. The disappearance of the C≡CH stretching peak in the IR spectra of the final compound confirmed that all the polymer was consumed. 1H NMR (600 MHz, deuterium oxide): δ 8.30 (s, triazole), 5.69 (d, J = 9.2 Hz, galabiose; H-1), 5.00 (d, J = 3.9 Hz, galabiose; H′-1), 4.51–3.30 (m, CH2 and CH, hPG-OH backbone; galabiose; H-2, H-3, H-4, H-5, H-6, H′-2, H′-3, H′-4, H′-5, H′-6), 1.25 (s, CH2 core), 0.84 (s, CH3 core).
Acknowledgments
We are grateful to the Tres Cantos Open Lab Foundation for funding this project (TC-232), as well as for the participation of GSK and making the Open Lab available.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c00152.
General information; Stx1B inhibition curves; synthesis; NMR spectra; IR spectra; and analytical HPLC traces (PDF)
Author Present Address
∥ Present address: Key Laboratory of Carbohydrate Chemistry & Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, 214122, Wuxi, China.
Author Contributions
The manuscript was written through contributions of all authors. All authors approved of the final version of the manuscript
The authors declare no competing financial interest.
Supplementary Material
References
- Liu J.; Platts-Mills J. A.; Juma J.; Kabir F.; Nkeze J.; Okoi C.; Operario D. J.; Uddin J.; Ahmed S.; Alonso P. L.; Antonio M.; Becker S. M.; Blackwelder W. C.; Breiman R. F.; Faruque A. S. G.; Fields B.; Gratz J.; Haque R.; Hossain A.; Hossain M. J.; Qamar F.; Iqbal N. T.; Kwambana B.; Mandomando I.; McMurry T. L.; Ochieng C.; Ochieng J. B.; Ochieng M.; Onyango C.; Panchalingam S.; Kalam A.; Aziz F.; Qureshi S.; Ramamurthy T.; Roberts J. H.; Saha D.; Sow S. O.; Stroup S. E.; Sur D.; Tamboura B.; Taniuchi M.; Tennant S. M.; Toema D.; Wu Y.; Zaidi A.; Nataro J. P.; Kotloff K. L.; Levine M. M.; Houpt E. R. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS Case-Control Study. Lancet 2016, 388, 1291–1301. 10.1016/s0140-6736(16)31529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh V. L.; Burris J. A.; Owens J. W.; Gordon V. M.; Wadolkowski E. A.; O’Brien A. D.; Samuel J. E. Comparison of the relative toxicities of shiga-like toxins type I and type II for mice. Infect. Immun. 1993, 61, 3392–3402. 10.1128/iai.61.8.3392-3402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M.; Hammerl J. A.; Hertwig S.; Appel B.; Brüssow H. Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl. Environ. Microbiol. 2012, 78, 4065–4073. 10.1128/aem.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-S.; Koo S.; Jeong D. G.; Tesh V. L. Shiga toxins as multi-functional proteins: induction of host cellular stress responses, role in pathogenesis and therapeutic applications. Toxins 2016, 8, 77. 10.3390/toxins8030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergan J.; Dyve Lingelem A. B.; Simm R.; Skotland T.; Sandvig K. Shiga toxins. Toxicon 2012, 60, 1085–1107. 10.1016/j.toxicon.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Ling H.; Boodhoo A.; Hazes B.; Cummings M. D.; Armstrong G. D.; Brunton J. L.; Read R. J. Structure of the Shiga-like Toxin I B-Pentamer Complexed with an Analogue of Its Receptor Gb3,. Biochemistry 1998, 37, 1777–1788. 10.1021/bi971806n. [DOI] [PubMed] [Google Scholar]
- Lingwood C. Verotoxin receptor-based pathology and therapies. Front. Cell. Infect. Microbiol. 2020, 10, 123. 10.3389/fcimb.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paletta A. C. C.; Castro V. S.; Conte-Junior C. A. Shiga toxin-producing and enteroaggregative Escherichia coli in animal, foods, and humans: pathogenicity mechanisms, detection methods, and epidemiology. Curr. Microbiol. 2020, 77, 612–620. 10.1007/s00284-019-01842-1. [DOI] [PubMed] [Google Scholar]
- Wong C. S.; Jelacic S.; Habeeb R. L.; Watkins S. L.; Tarr P. I. The Risk of the Hemolytic-Uremic Syndrome after Antibiotic Treatment ofEscherichia coliO157:H7 Infections. N. Engl. J. Med. 2000, 342, 1930–1936. 10.1056/nejm200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogolari M.; Mavian C.; Angeletti S.; Salemi M.; Lampel K. A.; Maurelli A. T. Distribution and characterization of Shiga toxin converting temperate phages carried by Shigella flexneri in Hispaniola. Infect., Genet. Evol. 2018, 65, 321–328. 10.1016/j.meegid.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. C.; Fierer J.; Chiu W. W.; Looney D. J.; Strain M.; Mehta S. R. A novel Shiga toxin 1a-converting bacteriophage of Shigella sonnei with close relationship to Shiga toxin 2-converting pages of Escherichia coli. Open Forum Infect. Dis. 2016, 3, ofw079. 10.1093/ofid/ofw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm O.; Lienemann T.; Halkilahti J.; Mero S.; Rimhanen-Finne R.; Lehtinen V.; Salmenlinna S.; Siitonen A. Characterization ofShigella sonneiIsolate Carrying Shiga Toxin 2-Producing Gene. Emerg. Infect. Dis. 2015, 21, 891–892. 10.3201/eid2105.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth I.; Sváb D.; Bálint B.; Brown-Jaque M.; Maróti G. Comparative analysis of the Shiga toxin converting bacteriophage first detected in Shigella sonnei. Infect., Genet. Evol. 2016, 37, 150–157. 10.1016/j.meegid.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Gray M. D.; Lampel K. A.; Strockbine N. A.; Fernandez R. E.; Melton-Celsa A. R.; Maurelli A. T. Clinical Isolates of Shiga Toxin 1a-ProducingShigella flexneriwith an Epidemiological Link to Recent Travel to Hispañiola. Emerg. Infect. Dis. 2014, 20, 1669–1677. 10.3201/eid2010.140292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K.; Fair M. A.; Strockbine N.; Hise K.; Mintz E.; Omondi M. Emergence of Shiga Toxin 1 Genes within Shigella Dysenteriae Type 4 Isolates from Travelers Returning from the Island of Hispañola. Am. J. Trop. Med. Hyg. 2007, 76, 1163–1165. 10.4269/ajtmh.2007.76.1163. [DOI] [PubMed] [Google Scholar]
- Sváb D.; Bálint B.; Vásárhelyi B.; Maróti G.; Tóth I. Comparative genomic and phylogenetic analysis of a Shiga toxin producing Shigella sonnei (STSS) strain. Front. Cell. Infect. Microbiol. 2017, 7, 229. 10.3389/fcimb.2017.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S.; Pilon P. A.; Cloutier N.; Doualla-bell F.; Longtin J. Identification of Shigella flexneri isolates carrying the Shiga Toxin 1-producing gene in Quebec, Canada, linked to travel to Haiti. Can. J. Microbiol. 2015, 61, 995–996. 10.1139/cjm-2015-0538. [DOI] [PubMed] [Google Scholar]
- Beutin L.; Strauch E.; Fischer I. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 1999, 353, 1498. 10.1016/s0140-6736(99)00961-7. [DOI] [PubMed] [Google Scholar]
- Lamba K.; Nelson J. A.; Kimura A. C.; Poe A.; Collins J.; Kao A. S.; Cruz L.; Inami G.; Vaishampayan J.; Garza A.; Chaturvedi V.; Vugia D. J. Shiga Toxin 1-ProducingShigella sonneiInfections, California, United States, 2014-2015. Emerg. Infect. Dis. 2016, 22, 679–686. 10.3201/eid2204.151825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M.; Igai K.; Matsuoka K.; Miyagawa A.; Watanabe T.; Yanoshita R.; Samejima Y.; Terunuma D.; Natori Y.; Nishikawa K. Structural Analysis of the Interaction between Shiga Toxin B Subunits and Linear Polymers Bearing Clustered Globotriose Residues. Infect. Immun. 2006, 74, 1984–1988. 10.1128/iai.74.3.1984-1988.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T.; Yoshino K.-i.; Adachi E.; Sato Y.; Yamagata K. In VitroAssessment of a Chemically Synthesized Shiga Toxin Receptor Analog Attached to Chromosorb P (Synsorb Pk) as a Specific Absorbing Agent of Shiga Toxin 1 and 2. Microbiol. Immunol. 1999, 43, 331–337. 10.1111/j.1348-0421.1999.tb02413.x. [DOI] [PubMed] [Google Scholar]
- Trachtman H.; Cnaan A.; Christen E.; Gibbs K.; Zhao S.; DWK A.; Weiss R.; FJ K.; Spitzer A.; GH H. Effect of an Oral Shiga Toxin-Binding Agent on Diarrhea-Associated Hemolytic Uremic Syndrome in ChildrenA Randomized Controlled Trial. JAMA, J. Am. Med. Assoc. 2003, 290, 1337–1344. 10.1001/jama.290.10.1337. [DOI] [PubMed] [Google Scholar]
- Ganji A.; Islami M.; Ejtehadifar M.; Zarei-Mehrvarz E.; Darvish M. Nanobody and aptamer as targeting moiety against bacterial toxins: therapeutic and diagnostic applications. Rev. Med. Microbiol. 2019, 30, 183–190. 10.1097/mrm.0000000000000175. [DOI] [Google Scholar]
- Kitov P. I.; Sadowska J. M.; Mulvey G.; Armstrong G. D.; Ling H.; Pannu N. S.; Read R. J.; Bundle D. R. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature 2000, 403, 669–672. 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- Mulvey G. L.; Marcato P.; Kitov P. I.; Sadowska J.; Bundle D. R.; Armstrong G. D. Assessment in mice of the therapeutic potential of tailored, multivalent Shiga toxin carbohydrate ligands. J. Infect. Dis. 2003, 187, 640–649. 10.1086/373996. [DOI] [PubMed] [Google Scholar]
- Nishikawa K.; Matsuoka K.; Kita E.; Okabe N.; Mizuguchi M.; Hino K.; Miyazawa S.; Yamasaki C.; Aoki J.; Takashima S.; Yamakawa Y.; Nishijima M.; Terunuma D.; Kuzuhara H.; Natori Y. A therapeutic agent with oriented carbohydrates for treatment of infections by Shiga toxin-producing Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 7669–7674. 10.1073/pnas.112058999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón M.; Quadir M. A.; Sharma S. K.; Haag R. Dendritic polyglycerols for biomedical applications. Adv. Mater. 2010, 22, 190–218. 10.1002/adma.200902144. [DOI] [PubMed] [Google Scholar]
- Asadpoor M.; Peeters C.; Henricks P. A. J.; Varasteh S.; Pieters R. J.; Folkerts G.; Braber S. Anti-pathogenic functions of non-digestible oligosaccharides in vitro. Nutrients 2020, 12, 1789. 10.3390/nu12061789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers D. T. S.; van Esse G. W.; Merkx R.; Brouwer A. J.; Jacobs H. J. F.; Pieters R. J.; Liskamp R. M. J.; van Esse G. W. Efficient microwave-assisted synthesis of multivalent dendrimeric peptides using cycloaddition reaction (click) chemistry. Chem. Commun. 2005, 13, 4581–4583. 10.1039/b507975f. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf I.; Rijnders A. Y.; Soede A.; Dechesne A. C.; Van Esse G. W.; Brouwer A. J.; Corstens F. H. M.; Boerman O. C.; Rijkers D. T. S.; Liskamp R. M. J. Synthesis of DOTA-conjugated multivalent cyclic-RGD peptide dendrimers via 1,3-dipolar cycloaddition and their biological evaluation: implications for tumor targeting and tumor imaging purposes. Org. Biomol. Chem. 2007, 5, 935–944. 10.1039/b615940k. [DOI] [PubMed] [Google Scholar]
- Sunder A.; Hanselmann R.; Frey H.; Mülhaupt R. Controlled Synthesis of Hyperbranched Polyglycerols by Ring-Opening Multibranching Polymerization. Macromolecules 1999, 32, 4240–4246. 10.1021/ma990090w. [DOI] [Google Scholar]
- Bhatia S.; Lauster D.; Bardua M.; Ludwig K.; Angioletti-Uberti S.; Popp N.; Hoffmann U.; Paulus F.; Budt M.; Stadtmüller M.; Wolff T.; Hamann A.; Böttcher C.; Herrmann A.; Haag R. Linear polysialoside outperforms dendritic analogs for inhibition of influenza virus infection in vitro and in vivo. Biomaterials 2017, 138, 22–34. 10.1016/j.biomaterials.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Kurniasih I. N.; Liang H.; Möschwitzer V. D.; Quadir M. A.; Radowski M.; Rabe J. P.; Haag R. Synthesis and transport properties of new dendritic core-shell architectures based on hyperbranched polyglycerol with biphenyl-PEG shells. New J. Chem. 2012, 36, 371–379. 10.1039/c1nj20466a. [DOI] [Google Scholar]
- Shimizu H.; Field R. A.; Homans S. W.; Donohue-Rolfe A. Solution Structure of the Complex between the B-Subunit Homopentamer of Verotoxin VT-1 fromEscherichia coliand the Trisaccharide Moiety of Globotriaosylceramide†. Biochemistry 1998, 37, 11078–11082. 10.1021/bi980946+. [DOI] [PubMed] [Google Scholar]
- Akbari P.; Braber S.; Alizadeh A.; Verheijden K. A.; Schoterman M. H.; Kraneveld A. D.; Garssen J.; Fink-Gremmels J. Galacto-oligosaccharides protect the intestinal barrier by maintaining the tight junction network and modulating the inflammatory responses after a challenge with the mycotoxin deoxynivalenol in human caco-2 cell monolayers and B6C3F1 mice. J. Nutr. 2015, 145, 1604–1613. 10.3945/jn.114.209486. [DOI] [PubMed] [Google Scholar]
- Varasteh S.; Braber S.; Garssen J.; Fink-Gremmels J. Galacto-oligosaccharides exert a protective effect against heat stress in a caco-2 cell model. J. Funct. Foods 2015, 16, 265–277. 10.1016/j.jff.2015.04.045. [DOI] [Google Scholar]
- Naveed M.; Phil L.; Sohail M.; Hasnat M.; Baig M. M. F. A.; Ihsan A. U.; Shumzaid M.; Kakar M. U.; Mehmood Khan T.; Akabar M.; Hussain M. I.; Zhou Q.-G. Chitosan oligosaccharide (COS): an overview. Int. J. Biol. Macromol. 2019, 129, 827–843. 10.1016/j.ijbiomac.2019.01.192. [DOI] [PubMed] [Google Scholar]
- Liu J.; Yang S.; Li X.; Yan Q.; Reaney M. J. T.; Jiang Z. Alginate oligosaccharides: production, biological activities, and potential applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1859–1881. 10.1111/1541-4337.12494. [DOI] [PubMed] [Google Scholar]
- Xing M.; Cao Q.; Wang Y.; Xiao H.; Zhao J.; Zhang Q.; Ji A.; Song S. Advances in research on the bioactivity of alginate oligosaccharides. Mar. Drugs 2020, 18, 144. 10.3390/md18030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann V.; Pieters R. J. Bridging lectin binding sites by multivalent carbohydrates. Chem. Soc. Rev. 2013, 42, 4492–4503. 10.1039/c3cs60089k. [DOI] [PubMed] [Google Scholar]
- Hilaire P. M. S.; Boyd M. K.; Toone E. J. Interaction of the Shiga-like toxin type 1 B-subunit with its carbohydrate receptor. Biochemistry 1994, 33, 14452–14463. [DOI] [PubMed] [Google Scholar]
- Fu O.; Pukin A. V.; van Ufford H. C. Q.; Branson T. R.; Thies-Weesie D. M. E.; Turnbull W. B.; Visser G. M.; Pieters R. J. Tetra- versus pentavalent inhibitors of cholera toxin. ChemistryOpen 2015, 4, 471–477. 10.1002/open.201500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer-van Ommen D. D.; Pukin A. V.; Fu O.; Quarles van Ufford L. H. C.; Janssens H. M.; Beekman J. M.; Pieters R. J. Functional characterization of cholera toxin inhibitors using human intestinal organoids. J. Med. Chem. 2016, 59, 6968–6972. 10.1021/acs.jmedchem.6b00770. [DOI] [PubMed] [Google Scholar]
- Haksar D.; de Poel E.; van Ufford L. Q.; Bhatia S.; Haag R.; Beekman J.; Pieters R. J. Strong inhibition of cholera toxin B subunit by affordable, polymer-based multivalent inhibitors. Bioconjugate Chem. 2019, 30, 785–792. 10.1021/acs.bioconjchem.8b00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haksar D.; Quarles van Ufford L.; Pieters R. J. A Hybrid polymer to target blood group dependence of cholera toxin. Org. Biomol. Chem. 2020, 18, 52–55. 10.1039/c9ob02369k. [DOI] [PubMed] [Google Scholar]
- Armstrong G. D.; Rowe P. C.; Goodyer P.; Orrbine E.; Klassen T. P.; Wells G.; Mackenzie A.; Lior H.; Blanchard C.; Auclair F.; Thompson B.; Rafter D. J.; McLaine P. N.; Rowe P. C.; Goodyer P.; Orrbine E.; Klassen T. P.; Wells G.; Mackenzie A.; Lior H.; Blanchard C.; Auclair F.; Thompson B.; Rafter D. J.; McLaine P. N. A phase I study of chemically synthesized verotoxin (Shiga-like toxin) Pk-trisaccharide receptors attached to chromosorb for preventing hemolytic-uremic syndrome. J. Infect. Dis. 1995, 171, 1042–1045. 10.1093/infdis/171.4.1042. [DOI] [PubMed] [Google Scholar]
- Chapanian R.; Constantinescu I.; Brooks D. E.; Scott M. D.; Kizhakkedathu J. N. In vivo circulation, clearance, and biodistribution of polyglycerol grafted functional red blood cells. Biomaterials 2012, 33, 3047–3057. 10.1016/j.biomaterials.2011.12.053. [DOI] [PubMed] [Google Scholar]
- Lu W.; Du W.; Somovilla V. J.; Yu G.; Haksar D.; de Vries E.; Boons G.-J.; de Vries R. P.; de Haan C. A. M.; Pieters R. J. Enhanced inhibition of influenza A virus adhesion by di- and trivalent hemagglutinin inhibitors. J. Med. Chem. 2019, 62, 6398–6404. 10.1021/acs.jmedchem.9b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin I.-S.; Nishikawa K.; Maruyama H.; Ishii S. Histidine-tagged Shiga toxin B subunit binding assay: simple and specific determination of Gb3 content in mammalian cells. Chem. Pharm. Bull. 2006, 54, 522–527. 10.1248/cpb.54.522. [DOI] [PubMed] [Google Scholar]
- Novoa A.; Machida T.; Barluenga S.; Imberty A.; Winssinger N. PNA-Encoded Synthesis (PES) of a 10 000-Member Hetero-Glycoconjugate Library and Microarray Analysis of Diverse Lectins. ChemBioChem 2014, 15, 2058–2065. 10.1002/cbic.201402280. [DOI] [PubMed] [Google Scholar]
- Ohlsson J.; Larsson A.; Haataja S.; Alajääski J.; Stenlund P.; Pinkner J. S.; Hultgren S. J.; Finne J.; Kihlberg J.; Nilsson U. J. Structure–activity relationships of galabioside derivatives as inhibitors of E. coli and S. suis adhesins: nanomolar inhibitors of S. suis adhesins. Org. Biomol. Chem. 2005, 3, 886–900. 10.1039/b416878j. [DOI] [PubMed] [Google Scholar]
- Kitov P. I.; Bundle D. R. On the nature of the multivalency effect: a thermodynamic model. J. Am. Chem. Soc. 2002, 125, 16271–16284. 10.1021/ja038223n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.