Abstract
Patients with ulcerative colitis (UC) are at increased risk for developing colorectal cancer (CRC). In contrast to sporadic colorectal tumorigenesis, TP53 mutations occur early in the progression from inflamed colonic epithelium to dysplasia to CRC, and are sometimes readily detectable in inflamed, (yet) non-dysplastic mucosa. Here, we analyzed formalin-fixed paraffin-embedded tissue samples from 19 patients with long-standing UC (median 18 years, range 3 to 34) who had developed CRC as a consequence of chronic inflammation of the large bowel. We performed microsatellite instability testing, copy number analysis by array-based comparative genomic hybridization, mutation analysis by targeted next generation sequencing (48-gene panel) and TP53 immunostaining. The results were compared to The Cancer Genome Atlas (TCGA) data on sporadic CRC. All UC-CRC lesions in our cohort were microsatellite stable. Overall, genomic imbalances of UC-CRCs showed patterns of chromosomal aneuploidies characteristic for sporadic CRC with the exception of gains of chromosome arm 5p (12 of 23 UC-CRC, 52%), which are rare in sporadic CRCs from TCGA (21 of 144, 15%; FDR adjusted P = 0.006). UC-CRCs showed a predilection for TP53 alterations, which was the most frequently mutated gene in our cohort (20 of 23, 87%). Interestingly, spatially separated tumor lesions from individual patients tended to harbor distinct TP53 mutations. Similar to CRCs arising in a background of Crohn’s colitis, the genetic landscape of UC-CRCs was characterized by TP53 mutations and chromosomal aneuploidies including gains of chromosome arm 5p. Both alterations harbor the potential for early detection in precursor lesions, thus complementing morphologic diagnosis.
Patients with ulcerative colitis (UC) are at an increased risk of developing and dying from colorectal cancer (CRC), which increases as a function of the extent, duration and severity of inflammation of the colorectum (1). The intensity of microscopic inflammation in colonic biopsy specimens has been implicated as another independent risk factor for the development of CRC (2, 3). Furthermore, male sex, young age at UC onset and concomitant primary sclerosing cholangitis have been reported to increase CRC risk (4, 5).
Key phenotypic characteristics of UC-CRC, as opposed to sporadic CRC, include younger patient age at diagnosis, a higher fraction of carcinomas with mucinous and/or signet ring cell histology, and a higher incidence of synchronous or metachronous (precursor) lesions (6). Although colonoscopic surveillance and more effective treatment options have reduced incidence rates of advanced UC-CRC, current numbers still claim a 38% increased risk for developing a CRC with a 25% higher stage-corrected mortality for UC patients compared to non-UC patients (7).
Despite advanced endoscopic imaging techniques, not all dysplastic lesions are endoscopically detectable and histologic identification of dysplasia is still a major factor for cancer surveillance in UC patients (8, 9). However, microscopic appearance of dysplasia can be heterogeneous ranging from the “conventional” intestinal-type histology to so-called “non-conventional” morphological patterns such as hypermucinous and serrated lesions (see (10) for review) and stringent criteria for the identification and grading of dysplasia are lacking (11).
The multistep histologic progression from UC to dysplasia and ultimately invasive carcinoma is driven by the accumulation of genomic alterations, in analogy to the sporadic adenoma-carcinoma sequence, though differences in frequency and timing of genomic alterations exist (12, 13). In silico modeling suggests that half or more of the somatic mutations present in a tumor may originate prior to tumor initiation, i.e., may occur in a histologically occult manner before the development of morphologically recognizable lesions (14). In other words, a significant proportion of the genomic evolution of a tumor may take place in histologically normal appearing tissue without histologic evidence of dysplasia. Compared to sporadic CRC, UC-CRCs are characterized by higher frequencies of TP53 mutations (70% vs. 61%) and lower frequencies of KRAS (34% vs. 43%) and APC mutations (12% vs. 81%) (15–19). TP53 mutations appear to be early events in UC-related colorectal carcinogenesis, in contrast to KRAS mutations, which have rarely been identified as founder events (20, 21). In UC, the process of CRC development might also occur simultaneously and independently in multiple locations in the colon of UC patients as inferred by distinct mutational landscapes (22). Another prevalent genomic characteristic of UC-CRC is aneuploidy, i.e., chromosomal copy number alterations (CNAs) (23, 24). Aneuploidy can already be present in non-dysplastic, inflamed colonic epithelium, and has the potential to trigger dysplasia and/or cancer development (25, 26). This indicates a role for chromosomal instability in UC-CRC development and a possibility to utilize the presence of CNAs as marker for early detection of histologically occult neoplastic clones. Despite genomic instability in cancer genomes (27), cancer cell populations as a whole faithfully maintain tumor-specific patterns of genomic imbalances, which are strongly influenced by the tissue of origin (28–31). Also, etiology-driven distinctive genomic alterations have been reported such as MYC amplification in radiation-induced angiosarcomas (32). Therefore, the landscape of CNAs in UC-CRC warrants investigation. However, unlike analyses of mutations by targeted (or exome) sequencing, only few studies have investigated genome-wide CNA patterns in UC-CRC (24, 33, 34). In particular, studies using high resolution cytogenetic techniques such as array-based comparative genomic hybridization or low-pass whole genome sequencing are sparse (35).
Here, we analyzed formalin-fixed paraffin-embedded tissue samples from 19 patients with long-standing UC (median 18 years, range 3 to 34) who had developed CRC as a consequence of ongoing chronic inflammation of the large intestine. We performed microsatellite instability testing, copy number analysis by array-based comparative genomic hybridization (aCGH), mutation analysis by targeted next generation sequencing (48-gene panel) and TP53 immunostaining. The results suggest an etiology-driven entity-specific landscape of mutations and CNAs in UC-CRC, which is distinct from sporadic CRC.
MATERIALS AND METHODS
Patients and tissue samples
We collected formalin-fixed paraffin-embedded (FFPE) tissue samples from 19 patients with UC-CRC diagnosed between 2003 and 2016 from the archive of the Institute of Pathology in Mannheim. All samples were bowel resection specimen, either partial colectomy or proctocolectomy. UC-related etiology of a CRC was assumed if (i) the patient had long-standing UC at the time of CRC diagnosis, (ii) inflammation in the colon involved the large bowel segment, in which the CRC was located, and (iii) if inflammation and/or chronic inflammatory changes in adjacent mucosa were visible. All intestinal tissue samples were screened for inflammation, regenerative, i.e., inflammatory, changes, and dysplasia according to the histopathologic criteria defined by the Inflammatory Bowel Disease-Dysplasia Morphology Study Group (36). In addition to the initial diagnosis, all samples were re-evaluated by two pathologists (DH, TG). Tumor staging was performed according to the current American Joint Committee on Cancer (AJCC) / Union for International Cancer Control (UICC) staging system (8th edition) (37). The study was approved by the local ethics committee of the Medical Faculty Mannheim of Heidelberg University (2016-819R-MA) and the National Institutes of Health (OHSRP#13229/MTA#41436).
DNA isolation
One to three representative blocks were selected per tumor, depending on tumor size and tumor block availability. DNA was isolated from 20 μm sections from the FFPE blocks after determining the tumor areas on H&E sections, avoiding foci of inflammation as well as foci of high mucin and low cellular content. Samples were deparaffinized in 300 μl of mineral oil (cat # 69794, Sigma-Aldrich, St. Louis, MO, USA) in a thermomixer at 800 rpm at 65 °C overnight before 100 μl of ATL lysis buffer (Qiagen, Hilden, Germany) and 20 μl Proteinase K was added followed by agitation at 450 rpm at 56 °C. After 8 hours, another 20 μl of Proteinase K was added followed by incubation overnight. When digestion of the tissue pieces was complete, 120 μl of the lysate, i.e., the aequous phase, was transferred to a fresh 1.5 ml tube, treated with 2 μl RNAse A (100 mg/ml; Qiagen) for five minutes at room temperature and mixed with 600 μl PM buffer (Qiagen) and 10 μl of 3M sodium acetate (pH 5.2; cat # R1181, Thermo Fisher Scientific, Waltham, MA, USA). Subsequent spin column purification was done using the QIAquick PCR Purification kit (Qiagen) according to the manufacturer’s protocol with minor modifications. DNA concentration and purity were measured by spectrophotometry (NanoDrop 1000 Spectrophotometer, NanoDrop products, Wilmington, DE). In addition, double-stranded DNA was quantified by a Qubit 3.0 fluorometer (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) using the Qubit dsDNA BR (Broad Range) Assay Kit (Life Technologies). Biomed2 multiplex-PCR assay of differently sized amplicons (targeting 400, 300, 200 and 100 bp fragments of AFF1, PLZF, RAG1 and TBXAS1, respectively) was performed to visualize DNA fragment size distribution (38). In addition, amplifiability of the isolated FFPE DNA was assessed by a qPCR assay (Illumina FFPE QC Kit for TruSeqAmplicon – Cancer Panel, Illumina Inc., San Diego, CA, USA) and delta Cq values were determined according to the manufacturer’s instructions.
Microsatellite PCR
Microsatellite PCR was done using a panel of five mononucleotide markers (BAT25, BAT26, NR-21, NR-24, MONO-27; cf. MSI Analysis System, Promega, Madison, WI, USA), and a panel of two mononucleotide (BAT25, BAT26) and three dinucleotide markers (D5S346, D2S123, D17S250; so-called Bethesda panel) as described previously (39).
Array-based comparative genomic hybridization
Array-based comparative genomic hybridization (aCGH) was performed as previously described using ULS labeling (Agilent) and SurePrint G3 CGH 4X180K microarrays (Agilent) (40). Data were visualized and analyzed using Nexus Copy Number software version 9.0 (BioDiscovery, Inc., El Segundo, CA, USA). Arm-level somatic copy number alterations (SCNAs) were defined as single alteration or an aggregate of alterations encompassing half or more (≥50%) of a chromosome arm (29).
The Cancer Genome Atlas (TCGA) copy number data retrieval and processing
SNP array-based copy number data from The Cancer Genome Atlas (TCGA) colorectal adenocarcinoma cohort (41) were retrieved for cases, for which the histotype was known (intestinal-type versus mucinous) and which were microsatellite stable/non-hypermutated (n = 144; n = 129 intestinal-type and n = 15 mucinous). Detailed selection criteria have been described previously (39). Copy number data was subsequently analyzed and visualized using Nexus Copy Number software version 9.0 (BioDiscovery, Inc., El Segundo, CA, USA).
Targeted next generation sequencing
Libraries of FFPE isolated DNA were prepared using the TruSeq Amplicon – Cancer Panel (Illumina). TruSeq Amplicon – Cancer Panel spans mutational hotspots in >35 kilobases (kb) of target genomic sequence in the following genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL. The PhiX control library (Illumina) was spiked in each run at a final concentration of 10 pM to estimate the sequencing error rate. The pooled libraries were paired-end (2×151) sequenced on a MiSeq instrument (Illumina). The mean read depth for targeted regions (mean coverage) was 642X. Alignment and variant calling were done using the BaseSpace TruSeq Amplicon App, Version 3.0.0 (Illumina) using the somatic variant caller. Annotation was done based on RefSeqGene (https://www.ncbi.nlm.nih.gov/refseq/rsg/) and variants were further evaluated based on dbSNP 147 (NCBI; https://www.ncbi.nlm.nih.gov/projects/SNP/), COSMIC database (Catalogue of Somatic Mutations in Cancer; http://cancer.sanger.ac.uk/cosmic) and ClinVar database (42–44). Only non-synonymous variants were considered. Specific filtering criteria included 1) passed variant caller filters, 2) read depth ≥10 with a fraction of alternative reads ≥0.1, 3) ExAC frequency <0.001%. Reads were visualized using the Integrative Genomics Viewer (IGV, Broad Institute) (45, 46). Mutation data was used to determine the phylogenetic relationship between different tumor samples from individual patients as described previously (39, 47). All phylogenetic trees were drawn with a common trunk, representing the normal, i.e., diploid, genome.
Immunohistochemistry
Immunohistochemical staining of tumors and adjacent mucosa was performed as described previously (39). Briefly, the following primary antibodies were used: MLH1 (1:25; clone ES05, cat # M3640, Dako, Agilent Pathology Solutions, Agilent), MSH2 (ready-to-use; clone FE11, cat # IR085, Dako), MSH6 (ready-to-use; clone EP49, cat # IR086, Dako), PMS2 (1:50; clone EP51, cat # M3647, Dako) and TP53 (1:50; clone DO-7, cat # M7001, Dako). Detection was done using the EnVision Detection System, Peroxidase/DAB, Rabbit/Mouse (cat # K5007, Dako). Immunohistochemical stainings were evaluated by two pathologists (DH, TG). Tumor samples lacking nuclear staining for one or more of the following four DNA mismatch repair proteins (MLH1, MSH2, MSH6, PMS2) were considered microsatellite instable while tumor samples with retained expression of all four markers in the tumor cells were considered microsatellite stable. In carcinomas and dysplastic lesions, strong or completely absent nuclear positivity of tumor cells was considered mutant while weak and heterogeneous positivity of tumor cell nuclei was considered a wild-type pattern. In non-dysplastic mucosa, TP53 staining was classified according to nuclear staining intensity and distribution of positive cells as published by Sato and colleagues (48), and Noffsinger and colleagues (49), respectively. Microscopy images were acquired with a digital slide scanner (MoticEasyScan One, Motic, Hongkong).
Data availability
Genomic data have been deposited in Gene Expression Omnibus (GEO) database (accession number GSE156076).
RESULTS
Clinical and pathologic characteristics of UC-CRCs
Our cohort comprised 19 patients with UC who had developed CRC (Table 1). UC patients were diagnosed with CRC at a relatively young age (median 45 years) and had a long history of UC (median 18 years). Four of the 19 patients had synchronous CRCs, resulting in a total of 24 UC-CRCs (Supplementary Table S1). The UC-CRCs were distributed relatively evenly throughout the colorectum (42% right hemicolon, 25% left hemicolon, 33% rectum). Mucinous and/or signet ring cell histology was observed in 7 of 24 (29%) of the UC-CRCs (Supplementary Figure S1). All UC-CRCs in our cohort were microsatellite stable as assessed by immunohistochemistry and PCR, respectively.
Table 1.
Clinico-pathologic characteristics of cases of UC-associated CRCs.
| Variable | UC-associated CRCs n = 19 patients |
|---|---|
| Age at CRC diagnosis, years | |
| Mean ± SD | 47 ± 12 |
| Median (range) | 45 (29 – 64) |
| Duration of UC at CRC diagnosis1 | |
| Mean ± SD | 18 ± 9 |
| Median (range) | 18 (3 – 34) |
| Sex | |
| Male | 10 |
| Female | 9 |
| AJCC stage at diagnosis | |
| 0 | 2 |
| I | 6 |
| II | 4 |
| III | 6 |
| IV | 1 |
| Site of (primary) carcinoma2 | |
| Right hemicolon | 10 |
| Left hemicolon | 6 |
| Rectum | 8 |
| Synchronous CRC(s) | |
| Present | 4 |
| Absent | 15 |
| Histology | |
| Mucinous with signet ring cells | 2 |
| Mucinous without signet ring cells | 5 |
| Intestinal-type | 17 |
| Microsatellite status | |
| Microsatellite stable (MSS) | 24 |
| High-level microsatellite instable (MSI-H) | 0 |
Based on n = 15 patients; four patients without data on precise duration of UC.
Right hemicolon was defined as cecum, ascending and transverse colon; left colon as descending and sigmoid colon.
AJCC, American Joint Committee on Cancer; CRC, colorectal cancer; N/A, not applicable; SD, standard deviation; UC, ulcerative colitis.
Landscape of chromosomal aneuploidies in UC-CRCs
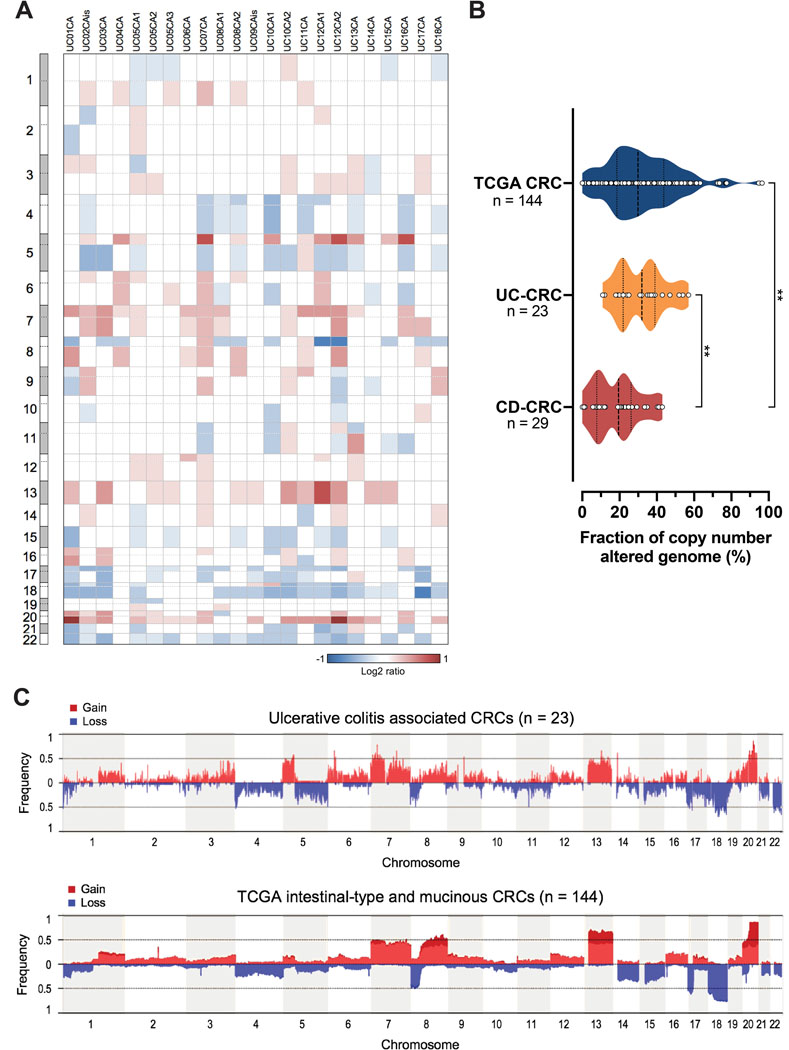
To determine genome alterations that characterize UC-CRCs, we performed aCGH and sequence analysis of a panel of 48 cancer-related genes. All 23 UC-CRC samples that could be successfully analyzed by aCGH showed genomic imbalances (Figure 1A). The fraction of copy number altered genome in UC-CRC was comparable to sporadic CRC based on microsatellite stable/non-hypermutated 144 cases with comparable histology, i.e., intestinal or mucinous type (P = 0.51; Mann-Whitney U test), but however, significantly higher compared to CD-CRC (P = 0.0012; Mann Whitney U test) (Figure 1B) (39). The most common arm-level copy number gains mapped to chromosome arms 20q (17 of 23, 74%), 7p (14 of 23, 61%), 13q (13 of 23, 57%), 5p (12 of 23, 52%), 7q (10 of 23, 43%) and 8q (8 of 23, 35%) (Figure 1C, Supplementary Figure S2). Recurrent arm-level losses were observed on chromosome arms 18q (17 of 23, 74%), 22q (17 of 23, 74%), 17p (14 of 23, 61%), 5q (12 of 23, 52%), 8p (11 of 23, 48%), 4p (9 of 23, 39%), 4q (9 of 23, 39%) and 17q (9 of 23, 39%). Independent from histology (intestinal-type or mucinous), the gain of 5p (harbors e.g., TERT) was usually not observed in sporadic CRC (21 of 144, 15%), making it a distinctive feature of inflammation-related CRC (FDR adjusted P = 0.006) (Supplementary Figure S2). Also, losses of 5q (harbors e.g., APC) were less frequent in sporadic CRC (27 of 144, 19%) than in UC-CRC (FDR adjusted P = 0.04; Fisher’s exact test).
Figure 1. Extent and pattern of genome-wide copy number alterations in ulcerative colitis–associated colorectal carcinomas.
(A) Per sample analysis of CNAs in UC-CRCs, represented as heatmap showing arm-level gains (red) and losses (blue). All UC-CRCs were microsatellite stable. (B) Fraction of copy number altered genome of UC-CRC compared to CRCs (both intestinal-type and mucinous) from the TCGA cohort (41) and CD-CRCs from our previous study (39). All samples included in this comparison were microsatellite stable/non-hypermutated. **, P ≤ 0.01; Mann-Whitney U test. (C) Cumulative copy number frequencies for UC-CRCs compared to CRCs from the TCGA cohort. All samples shown are microsatellite stable/non-hypermutated. Numbers below the graph (x-axis) denote chromosomes, frequency (y-axis) denotes the proportion of samples with a gain or a loss at the respective chromosomal position. CD, Crohn’s disease; CNA, copy number alteration; CRC, colorectal carcinoma; UC, ulcerative colitis.
Landscape of somatic mutations in UC-CRCs
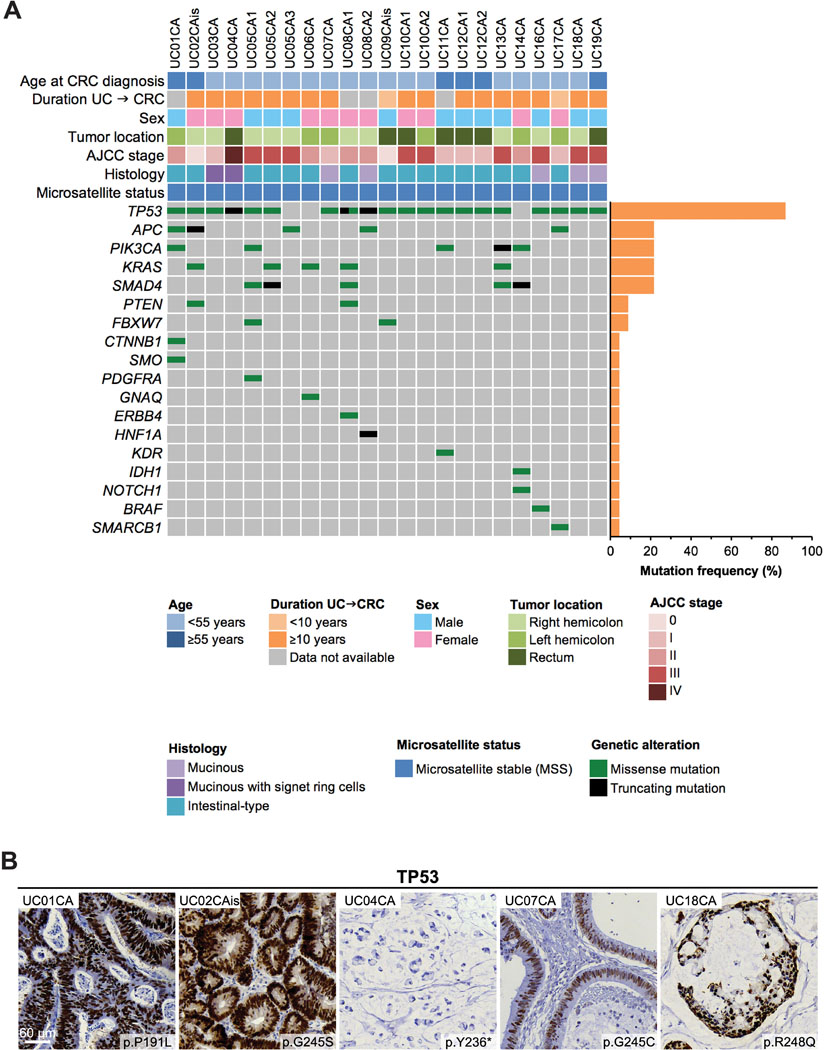
Our panel-based sequencing analysis revealed TP53 as the most frequently mutated gene in 87% (20 of 23) of UC-CRCs (Figure 2A). APC, PIK3CA, KRAS and SMAD4 mutations were detected in 22% (5 of 23) of UC-CRCs, respectively. Mutations in other genes were detected in less than 10% of samples. Of note, one sample harbored an IDH1 mutation (p.R132C). The number of mutations per tumor varied from 1 up to 6 (median: 2 mutations per tumor). The majority of TP53 mutations occurred as missense mutations, which were located predominantly in the DNA binding domain of the protein (Supplementary Figure S3). In line with the sequencing results, TP53 missense mutations showed a very strong nuclear positivity for TP53 while a TP53 truncating mutation presented with a complete absence of staining (Figure 2B). Taken together, the mutational landscape of UC-CRC was dominated by TP53 mutations while mutations in other genes were rare.
Figure 2. Mutational landscape of ulcerative colitis–associated colorectal carcinomas (UC-CRCs).
(A) “Oncoprint” showing all genes, in which mutations could be detected. Each row represents a clinicopathologic feature or gene, each column denotes an individual carcinoma sample. (B) Immunohistochemical staining of TP53 confirms TP53 mutation status. Abnormal overexpression, i.e., strongly intense staining in tumor cell nuclei (UC01CA, UC02CAis, UC07CA, UC18CA), or abnormal complete absence of expression within tumor cell nuclei (UC04CA) indicates TP53 mutation.
Tumor evolution in ulcerative colitis
Using the four patients with synchronous UC-CRC from our cohort as an example, we aimed to gain further insight into the evolution of synchronous UC-CRCs and potential underlying field defects. In addition to genome-wide copy number profiling and targeted sequencing, we performed TP53 immunostaining of the transition zone from carcinoma to adjacent colorectal mucosa and mucosa samples from other colonic sites if available (Supplementary Figure S4). This allowed us to confirm the carcinomas’ TP53 mutational status and to evaluate the presence of TP53 aberrant fields in adjacent and spatially separated colonic mucosa.
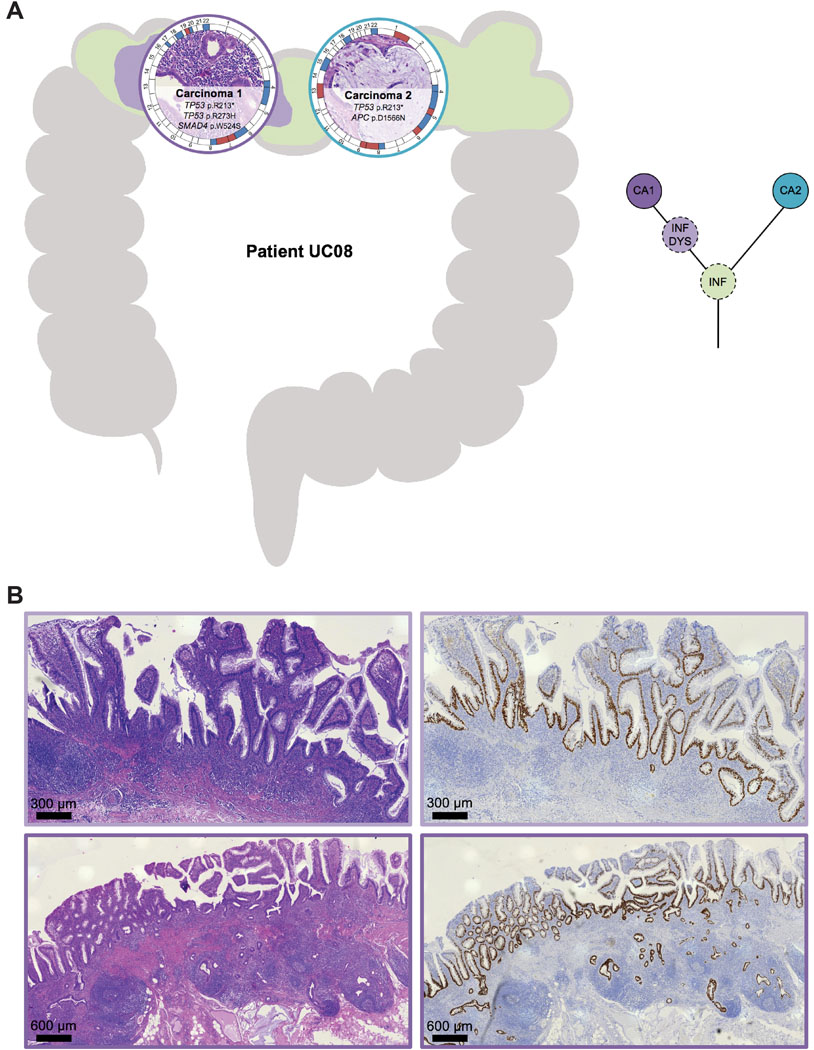
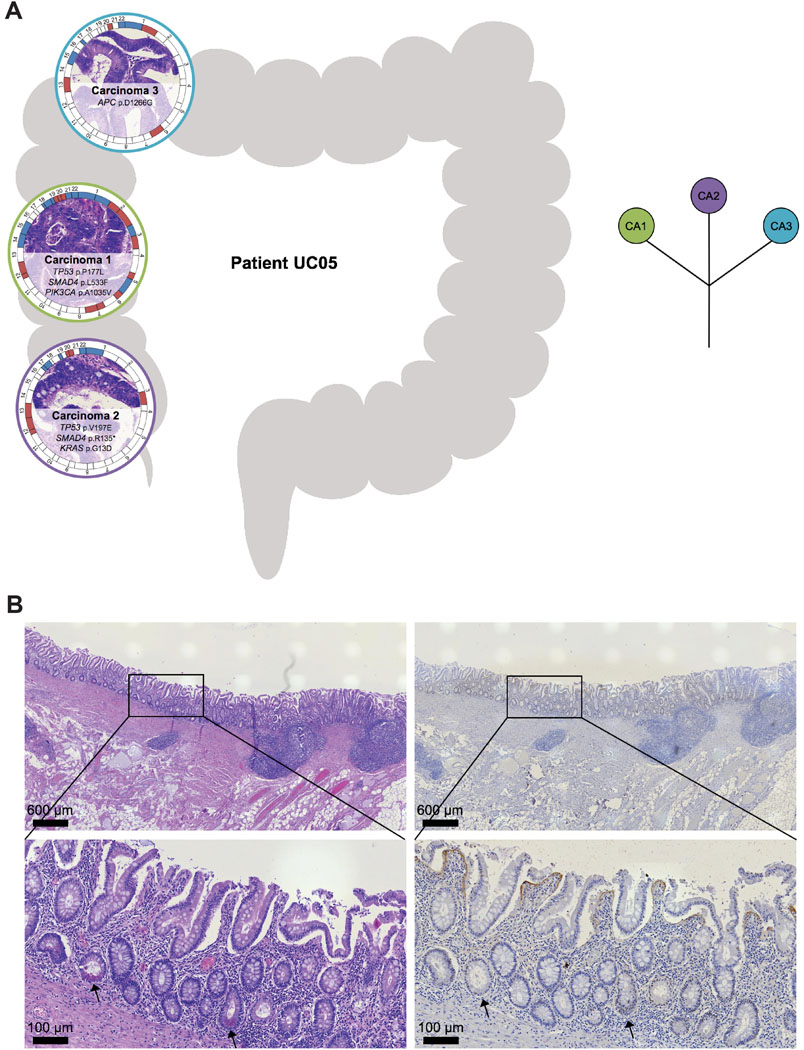
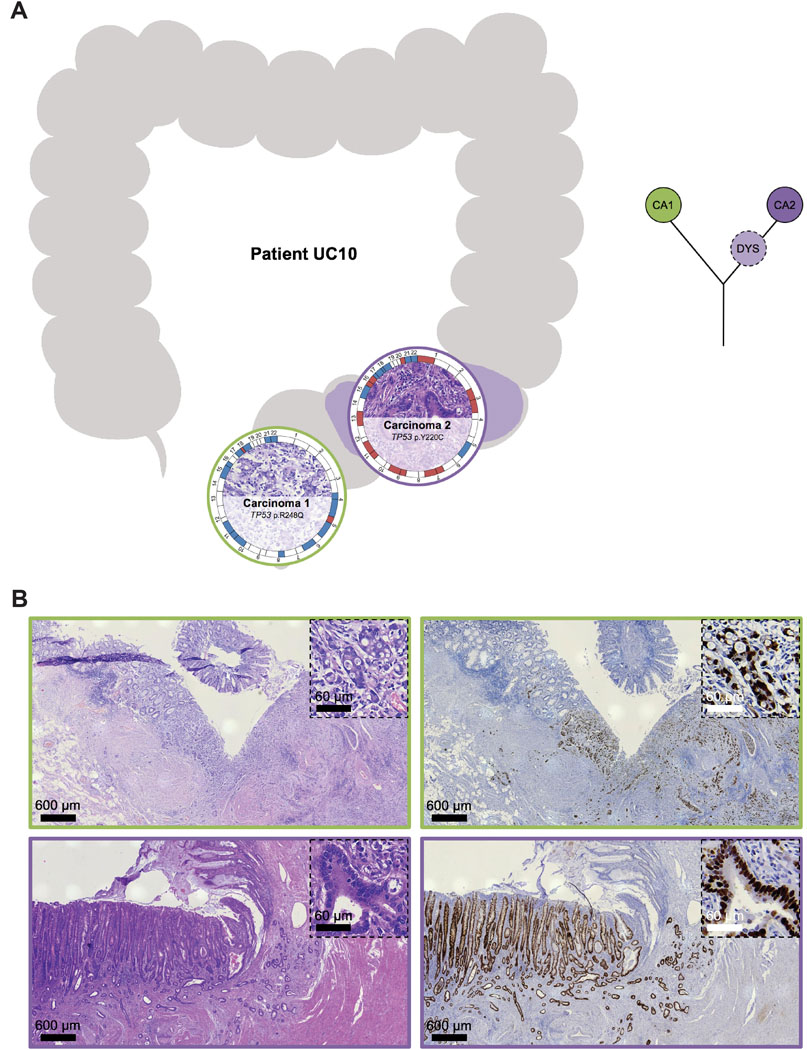
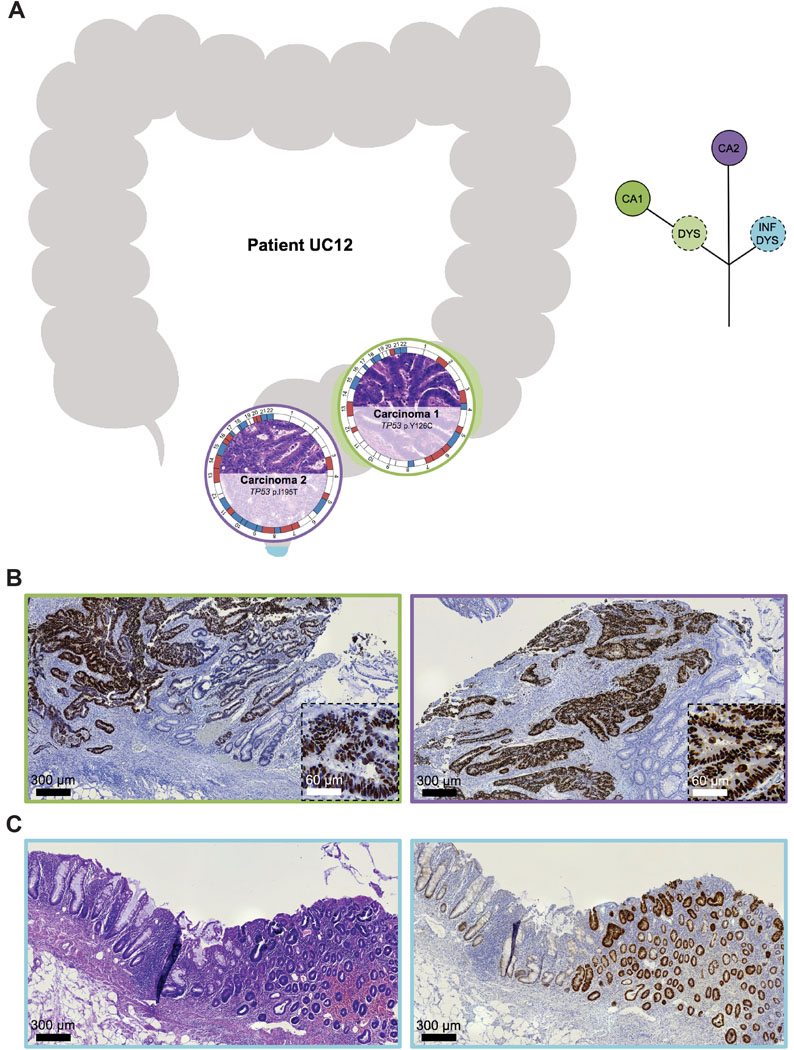
Interestingly, eight of nine synchronous UC-CRCs from the four individual patients had a mutation in TP53 though the specific TP53 mutations of the respective tumors were distinct with the exception of patient UC08 (Supplementary Figure S5). This patient (44-year-old female, exact UC duration not known) had two carcinomas located in the transverse colon (Figure 3). Both carcinomas shared an identical truncating mutation in TP53 (p.R213*), indicating that the two carcinomas emerged from the same mutant field. Carcinoma 1 (UC08CA1) subsequently acquired another TP53 mutation, this time as a missense mutation (p.R273H), which led to a nuclear accumulation of TP53. Nuclear accumulation could be visualized not only in the carcinoma itself but also in adjacent dysplasia (with villous transformation) and inflamed mucosa, again indicating tumor progression through field cancerization. Carcinoma 2 (UC08CA2) did not acquire another TP53 mutation but instead a mutation in APC. In contrast to patient UC08, carcinomas from the other patients with synchronous UC-CRCs (UC05, UC10, UC12) did not share any mutation, indicating multifocal disease and independent tumor progression, despite prevalent convergent evolution for mutations in TP53 (Supplementary Figure S5). For instance, patient UC05 (41-year-old male, UC duration 21 years) harbored three carcinomas in the right hemicolon (Figure 4). Two of the three carcinomas displayed a TP53 mutation (UC05CA1: p.P177L, UC05CA2: p.V197E) along with other mutations involving the TGF-beta, PI3K and RTK/RAS signaling pathways. In contrast, carcinoma 3 (UC05CA3) was solely characterized by an APC mutation targeting the WNT signaling pathway. Patient UC10 (45-year-old female, UC duration 26 years) had two carcinomas located in the rectum and sigmoid colon which both were TP53-mutated though the mutations were distinct (Figure 5). While carcinoma 2 (UC10CA2) apparently emerged from a preexisting inflamed and dysplastic TP53 mutant field as revealed by histology and TP53 immunostaining, carcinoma 1 (UC10CA1) appeared to have developed de novo (i.e., without preceding dysplasia) or alternatively, might be fast growing and rapidly displaced its original localized dysplastic field. The latter would be consistent with the aggressive histologic phenotype of a primarily solid growth pattern and an invasive margin of the infiltrative type with tumor budding. Patient UC12 (58-year-old male, UC for 33 years) also harbored two carcinomas with distinct TP53 mutations in the rectum and rectosigmoid junction 1.2 cm apart from each other (Figure 6). In carcinoma 2 (UC12CA2), small remnants of the original localized TP53 mutant field could be identified at the transition zone from carcinoma to adjacent mucosa, while in carcinoma 2 no abnormal TP53 staining was seen in the adjacent mucosa. Of note, 3.5 cm distal from carcinoma 2 (UC12CA2) another TP53 altered field could be revealed by TP53 immunostaining which has no direct contact with either carcinoma 1 or 2. This highlights the prevalence of multifocal tumor progression in UC patients.
Figure 3. Tumor heterogeneity: spatial and inferred phylogenetic relationship of lesions of patient UC08.
(A) (Left) The anatomic location of the analyzed samples is depicted along the colon frame. Selected somatic mutations and genome-wide copy number alterations are displayed with the respective histology. The outer circle indicates arm-level chromosomal gains (red) and losses (blue); chromosomes are labeled at the centromere. (Right) Corresponding inferred phylogenetic relationship of analyzed lesions. The different colors in the circles correspond to the color of the lesions on the left part. (B) Representative images of histology (H&E, left) and TP53 immunohistochemistry (right) of carcinoma 1 with adjacent dysplasia with villous architecture and inflamed mucosa, showing a ‘nested/diffuse pattern’ characterized by strongly positive nuclei aggregated in most areas of the crypts, sometimes confined to the basal half of the crypts, indicating TP53 mutation. Colors correspond to panel A. CA, carcinoma; DYS, dysplasia; INF, inflamed colonic mucosa.
Figure 4. Tumor heterogeneity: spatial and inferred phylogenetic relationship of lesions of patient UC05.
(A) (Left) The anatomic location of the analyzed samples is depicted along the colon frame. Selected somatic mutations and genome-wide copy number alterations are displayed with the respective histology. The outer circle indicates arm-level chromosomal gains (red) and losses (blue); chromosomes are labeled at the centromere. (Right) Corresponding inferred phylogenetic relationship of analyzed lesions. The different colors in the circles correspond to the color of the lesions on the left part. (B) Representative images of histology (H&E, left) and TP53 immunohistochemistry (right) of inflamed, non-dysplastic colonic mucosa from the left hemicolon, showing a ‘sporadic/scattered pattern’ with only a few positive nuclei dispersed/focused in the crypts, indicating TP53 wild-type status. Arrows (→) indicate Paneth cells. CA, carcinoma.
Figure 5. Tumor heterogeneity: spatial and inferred phylogenetic relationship of lesions of patient UC10.
(A) (Left) The anatomic location of the analyzed samples is depicted along the colon frame. Selected somatic mutations and genome-wide copy number alterations are displayed with the respective histology. The outer circle indicates arm-level chromosomal gains (red) and losses (blue); chromosomes are labeled at the centromere. (Right) Corresponding inferred phylogenetic relationship of analyzed lesions. The different colors in the circles correspond to the color of the lesions on the left part. (B) Representative images of histology (H&E, left) and TP53 immunohistochemistry (right) of carcinoma 1 and 2 with adjacent mucosa, showing a ‘diffuse pattern’ characterized by strongly positive nuclei aggregated in most areas of the crypts, indicating TP53 mutation and emergence of invasive carcinoma 2 from an associated TP53 aberrant field. Colors correspond to panel A. CA, carcinoma; DYS, dysplasia.
Figure 6. Tumor heterogeneity: spatial and inferred phylogenetic relationship of lesions of patient UC12.
(A) (Left) The anatomic location of the analyzed samples is depicted along the colon frame. Selected somatic mutations and genome-wide copy number alterations are displayed with the respective histology. The outer circle indicates arm-level chromosomal gains (red) and losses (blue); chromosomes are labeled at the centromere. (Right) Corresponding inferred phylogenetic relationship of analyzed lesions. The different colors in the circles correspond to the color of the lesions on the left part. (B) Representative images of TP53 immunohistochemistry of carcinoma 1 and 2 with adjacent mucosa. While small remnants of a TP53 altered field can be identified at the transition zone from colonic mucosa to carcinoma 1, in carcinoma 2 no abnormal TP53 staining is visible in the surrounding mucosa. (C) Rectal mucosa with aberrant TP53 staining characterized by strongly positive nuclei aggregated in most areas of the crypts, however, without direct contact to carcinoma 1 or 2. CA, carcinoma; DYS, dysplasia; INF, inflamed mucosa.
Similarly, we looked for field defects based on TP53 immunostaining in patients with single UC-CRCs (UC02, UC03, UC06, UC07, UC09, UC18). In all of these patients except for patient UC09, we could also observe gland formations with aberrant TP53 staining patterns in the colonic mucosa in spatial proximity to the invasive carcinoma (Supplementary Figures S6 to S11). Aberrant TP53 staining pattern presented as either ‘nested’ pattern, i.e., an aggregation of moderately to strongly positive cells confined to the basal half of the glands, or as ‘diffuse’ pattern, i.e., the presence of strongly positive cells in most areas of the glands, as described by Sato (48) and Noffsinger (49). Taken together, we detected an aberrant TP53 staining in tumor surrounding or adjacent colonic mucosa with or without dysplasia in eight of ten (80%) UC-CRC patients from our cohort. Unfortunately, for the other nine patients no tissue samples from non-dysplastic, inflamed mucosa was available for immunohistochemical analysis.
DISCUSSION
While genomic alterations during carcinogenesis have been thoroughly studied in sporadic CRC, less is known about genomic alterations in UC-CRC development. In contrast to sporadic CRC, which typically arises from a well-circumscribed polypoid adenoma, UC-CRC usually develops from flat dysplastic lesions with ill-defined margins in a background of inflammation accompanied by epithelial regeneration, scarring and pseudopolyposis (12). The macroscopic and microscopic heterogeneity of inflammatory and reactive changes complicates endoscopic detection and histological diagnosis of potential UC-CRC precursors or early malignant lesions (8, 9). Several molecular (bio)markers have been proposed for early detection of neoplastic processes in UC including TP53 mutations (50), nuclear aneuploidy (25), chromosomal copy number gains and losses (i.e., chromosomal aneuploidies) (26), telomere shortening (51), epigenetic alterations (52) and microRNA alterations (53). However, as of now, none of these markers is routinely used in the clinic and a deeper knowledge of the molecular alterations underlying UC-CRC development is needed to refine potentially useful molecular diagnostic markers.
Here, we applied high-resolution aCGH and targeted sequencing to analyze the genomic landscape of UC-CRCs. In addition, TP53 immunostaining was used to visualize the extent of morphologically occult tumor evolution.
While the presence of nuclear aneuploidy in non-dysplastic colonic mucosa from UC patients has been associated with an increased risk of progression to UC-CRC (25), specific CNAs or CNA burden might allow an improved risk assessment. The advent of molecular cytogenetics such as CGH (54, 55), spectral karyotyping (56) and multiplex fluorescence in situ hybridization (57) unraveled that most solid tumors do not only display nuclear aneuploidy, but moreover, are characterized by tumor-type specific non-random distributions of chromosomal aneuploidies, despite ongoing genomic instability (28, 30). Chromosomal aneuploidies, i.e. losses and gains of chromosomes or chromosome arms, were present in all UC-CRC samples of our cohort. Overall, the distribution of arm-level CNAs resembled the spectrum in sporadic CRC and chromosomal aneuploidies mainly clustered on the same chromosomes as described for sporadic CRC (41, 58), although some differences in the frequency of individual CNAs were observed. Specifically, the gain of chromosome arm 5p was more prevalent in UC-CRC (12 of 23, 52%), often combined with a loss of 5q. In contrast, a gain of chromosome arm 5p was rarely observed in sporadic CRC (21 of 144, 15%) and the gain of 5p appears to be a distinctive feature of inflammation-related colorectal carcinogenesis. The gain of 5p is prevalent in both UC- and CD-related carcinogenesis and can already be detected in dysplastic lesions with reported frequencies of up to about 50% (15, 35, 59) and sometimes even be observed in non-dysplastic mucosa from IBD patients (39). Sporadic adenomas, on the other hand, do not only have a lower CNA burden compared to IBD-related dysplastic lesions but in particular do virtually not display gains of chromosome arm 5p (<5%) (35, 60, 61). Therefore, the tumor type specific non-random chromosomal aneuploidy patterns in tumors appear not only to be influenced by the tissue of origin (30, 31, 62) but also by the mode of tumor induction, i.e., disease etiology, such as chronic inflammation or radiation, and metabolic stress (63). In line with older data on nuclear aneuploidy (25), preliminary low-pass whole genome sequencing data indicate that CNAs appear to be a potent predictor of UC progression risk from low-grade dysplasia to invasive carcinoma (64).
In addition to chromosomal aneuploidies, another frequently encountered genetic alteration in UC progression to UC-CRC is mutation of TP53. Our genetic profiling of UC-CRC development confirmed TP53 mutations as a key event of chronic inflammation-driven colorectal carcinogenesis occurring in 87% (20 of 23) of UC-CRC samples. This is in concordance with previous next generation sequencing based publications reporting TP53 mutation frequencies in UC-CRC of 57% (n = 7) (15), 63% (n = 16) (16), 53% (n = 15) (17), and 86% (n = 29) (18), respectively. Comparable TP53 mutation frequencies have also been observed in CD-CRC with 100% (n = 3) (15), 88% (n = 8) (16), 76% (n = 29) (39), 69% (n = 13) (17) and 94% (n = 18) (18), respectively. Mutations in other genes were infrequent in both UC-CRC and CD-CRC. Therefore, mutational alterations in UC and CD centered on the TP53 pathway while other pathways such as the WNT (e.g., APC, FBXW7, CTNNB1), TGFß (e.g., SMAD4), and RTK/RAS (e.g., KRAS, BRAF) were rarely affected.
Aneuploidy is a widespread phenomenon in IBD-related carcinogenesis and can sometimes be observed in colonic mucosa before histologically evident dysplasia (25). Furthermore, it is closely correlated with TP53 mutation (50) and we presume that the need for TP53 alterations may be driven by the disruption of the G1 checkpoint by TP53 inactivation which permits survival of cells with chromosomal aneuploidies (65, 66).
Multifocal neoplasia is a common phenomenon in UC-CRC with approximately 38% of patients having synchronous CRCs and 16% having a spatially distinct dysplastic lesion in the colon resection specimen (67). In our cohort, four of 19 patients (21%) had synchronous UC-CRCs. Although we only provide a limited picture of the genetic make-up based on panel sequencing, our data indicate that synchronous UC-CRCs from individual patients display a considerable genetic heterogeneity. In only one of the four patients, synchronous UC-CRCs shared a common ancestral mutation even though in all four patients the synchronous tumor lesions were located within the same or within neighboring colon segments, i.e., in relatively close spatial proximity. Despite the lack of a common ancestral mutation in three of the four patients, synchronous UC-CRCs showed a predilection for mutations in TP53, which were detected in eight of nine synchronous UC-CRCs. To summarize, we largely observed two different evolutionary patterns of genetic CRC progression in UC, which resemble our previous results from CD (39). While some patients develop large precancerous fields with a common ancestral mutation (field cancerization) from which tumors can emerge, other patients develop multifocal tumors without shared mutations (tumor progression through clonal mosaicism). Despite multifocality, we observed a strong convergent evolution for TP53 mutations shaping the genomic alterations in UC-CRC in addition to chromosomal aneuploidies, underlining the critical role of TP53 in UC-related colorectal carcinogenesis. Interestingly, synchronous CRCs not associated with IBD show more variation in terms of affected driver genes, most frequently affecting APC, KRAS, TP53 and PIK3CA, and molecular alterations of lesions from the same patient are usually distinct, indicating independent development of synchronous non-IBD-related CRCs (68–70).
Immunostaining for TP53 confirmed TP53 mutation status of UC-CRCs. In addition, TP53 immunostaining allowed to screen for the presence of potential histologically occult tumor clones in non-dysplastic mucosa. Aberrant TP53 staining pattern according to Sato/Noffsinger (48, 49) was observed in colonic mucosa without dysplasia or with low-grade dysplasia in eight of ten UC-CRC patients from whom tissue samples were available for immunostaining. Among UC patients who underwent immediate colectomy due to dysplasia, the chance for identification of endoscopically undetected invasive UC-CRC is about 20% to 25% for low-grade and 40% to 50% for high-grade dysplasia (67, 71). This highlights the multifocality of CRC development in UC, presumably as a consequence of field cancerization and increased genomic instability in colonic mucosa of these patients.
In conclusion, our data show that inflammatory bowel disease (IBD)-related CRC, whether in a background of UC or CD, follow similar oncogenetic pathways. The major genomic alterations represent TP53 mutations along with cancer-type specific chromosomal aneuploidies including the gain of chromosome arm 5p, which is prevalent in inflammation-related colorectal carcinogenesis. As described above, mounting evidence demonstrates that in IBD progression to cancer molecular changes precede the morphological changes associated with dysplasia. This offers the chance for improving current diagnostic algorithms and may better assist planning colonoscopic surveillance intervals than conventional histology alone (72). However, how these molecular field effects may best be detected in UC patients remains to be established. In addition to UC-CRC-typical early genomic alterations (e.g., TP53) (73), more general / neutral mutant markers identifying clonal expansions such as polyguanine tract genotyping have been proposed (74). In the foreseeable future, whole genome sequencing will provide a comprehensive picture of both random and selected genomic alterations, and their clonal expansion underlying UC progression to cancer. For now, though not perfect, TP53 immunohistochemistry can be a useful, broadly available, easy to implement tool in routine diagnostics to support histopathological discrimination between UC-associated neoplasia and inflammatory regenerative colonic epithelium and to determine the extent of potential histologically occult neoplastic fields.
Supplementary Material
Acknowledgments
The authors would like to thank Yonca Ceribas, Alexandra Eichhorn, Katrin Wolk, Helene Dukal and Dr. Maja Weber for technical assistance. This study was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute. D. Hirsch received an intramural research scholarship (Translational Physician Scientist Program) from the Medical Faculty Mannheim, Heidelberg University, followed by a Mildred Scheel postdoctoral fellowship of the German Cancer Aid (Deutsche Krebshilfe). The results published here are, in part, based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Disclosure of potential conflicts of interest:
The authors declare no potential conflicts of interest.
Supplementary information is available at Modern Pathology’s website
REFERENCES
- 1.Lutgens MW, van Oijen MG, van der Heijden GJ, Vleggaar FP, Siersema PD, Oldenburg B. Declining risk of colorectal cancer in inflammatory bowel disease: an updated meta-analysis of population-based cohort studies. Inflammatory bowel diseases. 2013;19(4):789–99. [DOI] [PubMed] [Google Scholar]
- 2.Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126(2):451–9. [DOI] [PubMed] [Google Scholar]
- 3.Gupta RB, Harpaz N, Itzkowitz S, Hossain S, Matula S, Kornbluth A, et al. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: a cohort study. Gastroenterology. 2007;133(4):1099–105; quiz 340–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10(6):639–45. [DOI] [PubMed] [Google Scholar]
- 5.Kornfeld D, Ekbom A, Ihre T. Is there an excess risk for colorectal cancer in patients with ulcerative colitis and concomitant primary sclerosing cholangitis? A population based study. Gut. 1997;41(4):522–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi PM, Zelig MP. Similarity of colorectal cancer in Crohn’s disease and ulcerative colitis: implications for carcinogenesis and prevention. Gut. 1994;35(7):950–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olen O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, et al. Colorectal cancer in ulcerative colitis: a Scandinavian population-based cohort study. Lancet. 2020;395(10218):123–31. [DOI] [PubMed] [Google Scholar]
- 8.Harpaz N, Talbot IC. Colorectal cancer in idiopathic inflammatory bowel disease. Semin Diagn Pathol. 1996;13(4):339–57. [PubMed] [Google Scholar]
- 9.Itzkowitz SH, Harpaz N. Diagnosis and management of dysplasia in patients with inflammatory bowel diseases. Gastroenterology. 2004;126(6):1634–48. [DOI] [PubMed] [Google Scholar]
- 10.Choi WT, Yozu M, Miller GC, Shih AR, Kumarasinghe P, Misdraji J, et al. Nonconventional dysplasia in patients with inflammatory bowel disease and colorectal carcinoma: a multicenter clinicopathologic study. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2020;33(5):933–43. [DOI] [PubMed] [Google Scholar]
- 11.Harpaz N, Polydorides AD. Colorectal dysplasia in chronic inflammatory bowel disease: pathology, clinical implications, and pathogenesis. Arch Pathol Lab Med. 2010;134(6):876–95. [DOI] [PubMed] [Google Scholar]
- 12.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140(6):1807–16. [DOI] [PubMed] [Google Scholar]
- 13.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67. [DOI] [PubMed] [Google Scholar]
- 14.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(6):1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker AM, Cross W, Curtius K, Al Bakir I, Choi CR, Davis HL, et al. Evolutionary history of human colitis-associated colorectal cancer. Gut. 2019;68(6):985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Din S, Wong K, Mueller MF, Oniscu A, Hewinson J, Black CJ, et al. Mutational Analysis Identifies Therapeutic Biomarkers in Inflammatory Bowel Disease-Associated Colorectal Cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24(20):5133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150(4):931–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaeger R, Shah MA, Miller VA, Kelsen JR, Wang K, Heins ZJ, et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology. 2016;151(2):278–87 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch D. [Colorectal tumor evolution in inflammatory bowel disease : Dynamics of genomic alterations and potential molecular markers to predict malignant progression]. Der Pathologe. 2019;40(Suppl 3):232–8. [DOI] [PubMed] [Google Scholar]
- 20.Holzmann K, Klump B, Borchard F, Hsieh CJ, Kuhn A, Gaco V, et al. Comparative analysis of histology, DNA content, p53 and Ki-ras mutations in colectomy specimens with long-standing ulcerative colitis. International journal of cancer Journal international du cancer. 1998;76(1):1–6. [DOI] [PubMed] [Google Scholar]
- 21.Leedham SJ, Graham TA, Oukrif D, McDonald SA, Rodriguez-Justo M, Harrison RF, et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136(2):542–50 e6. [DOI] [PubMed] [Google Scholar]
- 22.Choi CR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol. 2017;14(4):218–29. [DOI] [PubMed] [Google Scholar]
- 23.Clausen OP, Andersen SN, Stroomkjaer H, Nielsen V, Rognum TO, Bolund L, et al. A strategy combining flow sorting and comparative genomic hybridization for studying genetic aberrations at different stages of colorectal tumorigenesis in ulcerative colitis. Cytometry. 2001;43(1):46–54. [DOI] [PubMed] [Google Scholar]
- 24.Habermann JK, Upender MB, Roblick UJ, Kruger S, Freitag S, Blegen H, et al. Pronounced chromosomal instability and multiple gene amplifications characterize ulcerative colitis-associated colorectal carcinomas. Cancer genetics and cytogenetics. 2003;147(1):9–17. [DOI] [PubMed] [Google Scholar]
- 25.Rubin CE, Haggitt RC, Burmer GC, Brentnall TA, Stevens AC, Levine DS, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103(5):1611–20. [DOI] [PubMed] [Google Scholar]
- 26.Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, Crispin DA, Haggitt RC, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer research. 1999;59(20):5148–53. [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 28.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes, chromosomes & cancer. 1999;25(3):195–204. [DOI] [PubMed] [Google Scholar]
- 29.Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer research. 2012;72(17):4383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer cell. 2018;33(4):676–89 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manner J, Radlwimmer B, Hohenberger P, Mossinger K, Kuffer S, Sauer C, et al. MYC high level gene amplification is a distinctive feature of angiosarcomas after irradiation or chronic lymphedema. The American journal of pathology. 2010;176(1):34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aust DE, Willenbucher RF, Terdiman JP, Ferrell LD, Chang CG, Moore DH 2nd, et al. Chromosomal alterations in ulcerative colitis-related and sporadic colorectal cancers by comparative genomic hybridization. Human pathology. 2000;31(1):109–14. [DOI] [PubMed] [Google Scholar]
- 34.Willenbucher RF, Zelman SJ, Ferrell LD, Moore DH 2nd, Waldman FM. Chromosomal alterations in ulcerative colitis-related neoplastic progression. Gastroenterology. 1997;113(3):791–801. [DOI] [PubMed] [Google Scholar]
- 35.Wanders LK, Cordes M, Voorham Q, Sie D, de Vries SD, d’Haens G, et al. IBD-Associated Dysplastic Lesions Show More Chromosomal Instability Than Sporadic Adenomas. Inflammatory bowel diseases. 2020;26(2):167–80. [DOI] [PubMed] [Google Scholar]
- 36.Riddell RH, Goldman H, Ransohoff DF, Appelman HD, Fenoglio CM, Haggitt RC, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Human pathology. 1983;14(11):931–68. [DOI] [PubMed] [Google Scholar]
- 37.Brierley J, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. Eighth edition. ed. Chichester, West Sussex, UK; Hoboken, NJ: John Wiley & Sons, Inc.; 2017. p. p. [Google Scholar]
- 38.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia. 2003;17(12):2257–317. [DOI] [PubMed] [Google Scholar]
- 39.Hirsch D, Wangsa D, Zhu YJ, Hu Y, Edelman DC, Meltzer PS, et al. Dynamics of Genome Alterations in Crohn’s Disease-Associated Colorectal Carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24(20):4997–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, et al. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer research. 2013;73(5):1454–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, et al. RefSeq: an update on mammalian reference sequences. Nucleic acids research. 2014;42(Database issue):D756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic acids research. 2017;45(D1):D777–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic acids research. 2014;42(Database issue):D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology. 2011;29(1):24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nature communications. 2015;6:8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato A, MacHinami R. p53 immunohistochemistry of ulcerative colitis-associated with dysplasia and carcinoma. Pathol Int. 1999;49(10):858–68. [DOI] [PubMed] [Google Scholar]
- 49.Noffsinger AE, Belli JM, Miller MA, Fenoglio-Preiser CM. A unique basal pattern of p53 expression in ulcerative colitis is associated with mutation in the p53 gene. Histopathology. 2001;39(5):482–92. [DOI] [PubMed] [Google Scholar]
- 50.Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, Rubin CE, Stevens AC, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107(2):369–78. [DOI] [PubMed] [Google Scholar]
- 51.O’Sullivan JN, Bronner MP, Brentnall TA, Finley JC, Shen WT, Emerson S, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nature genetics. 2002;32(2):280–4. [DOI] [PubMed] [Google Scholar]
- 52.Garrity-Park MM, Loftus EV Jr., Bryant SC, Smyrk TC. A Biomarker Panel to Detect Synchronous Neoplasm in Non-neoplastic Surveillance Biopsies from Patients with Ulcerative Colitis. Inflammatory bowel diseases. 2016;22(7):1568–74. [DOI] [PubMed] [Google Scholar]
- 53.Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, et al. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology. 2015;149(4):981–92 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. The American journal of pathology. 1998;152(5):1107–23. [PMC free article] [PubMed] [Google Scholar]
- 55.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends in genetics : TIG. 1997;13(10):405–9. [DOI] [PubMed] [Google Scholar]
- 56.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273(5274):494–7. [DOI] [PubMed] [Google Scholar]
- 57.Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nature genetics. 1996;12(4):368–75. [DOI] [PubMed] [Google Scholar]
- 58.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes, chromosomes & cancer. 1996;15(4):234–45. [DOI] [PubMed] [Google Scholar]
- 59.van Dieren JM, Wink JC, Vissers KJ, van Marion R, Hoogmans MM, Dinjens WN, et al. Chromosomal and microsatellite instability of adenocarcinomas and dysplastic lesions (DALM) in ulcerative colitis. Diagn Mol Pathol. 2006;15(4):216–22. [DOI] [PubMed] [Google Scholar]
- 60.Hirsch D, Camps J, Varma S, Kemmerling R, Stapleton M, Ried T, et al. A new whole genome amplification method for studying clonal evolution patterns in malignant colorectal polyps. Genes, chromosomes & cancer. 2012;51(5):490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter H, Slezak P, Walch A, Werner M, Braselmann H, Jaramillo E, et al. Distinct chromosomal imbalances in nonpolypoid and polypoid colorectal adenomas indicate different genetic pathways in the development of colorectal neoplasms. The American journal of pathology. 2003;163(1):287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auslander N, Heselmeyer-Haddad K, Patkar S, Hirsch D, Camps J, Brown M, et al. Cancer-type specific aneuploidies hard-wire chromosome-wide gene expression patterns of their tissue of origin. bioRxiv 2019;Cancer-type specific aneuploidies hard-wire chromosome-wide gene expression patterns of their tissue of origin(doi: 10.1101/563858). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graham NA, Minasyan A, Lomova A, Cass A, Balanis NG, Friedman M, et al. Recurrent patterns of DNA copy number alterations in tumors reflect metabolic selection pressures. Mol Syst Biol. 2017;13(2):914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Curtius K, Bakir IA, Baker AM, Clarke T, Nasreddin N, Kopczynska MA, et al. Abstract 781: Quantifying evolution of early dysplastic lesions in ulcerative colitis predicts future colorectal cancer risk. Gastroenterology. 2019;156(6):S162–S3. [Google Scholar]
- 65.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188(3):369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soto M, Raaijmakers JA, Bakker B, Spierings DCJ, Lansdorp PM, Foijer F, et al. p53 Prohibits Propagation of Chromosome Segregation Errors that Produce Structural Aneuploidies. Cell Rep. 2017;19(12):2423–31. [DOI] [PubMed] [Google Scholar]
- 67.Choi CH, Rutter MD, Askari A, Lee GH, Warusavitarne J, Moorghen M, et al. Forty-Year Analysis of Colonoscopic Surveillance Program for Neoplasia in Ulcerative Colitis: An Updated Overview. Am J Gastroenterol. 2015;110(7):1022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas V, Cotter MB, Tosetto M, Khaw YL, Geraghty R, Winter DC, et al. Personalised mapping of tumour development in synchronous colorectal cancer patients. NPJ Genom Med. 2020;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Di J, Yang H, Jiang B, Wang Z, Ji J, Su X. Whole exome sequencing reveals intertumor heterogeneity and distinct genetic origins of sporadic synchronous colorectal cancer. International journal of cancer Journal international du cancer. 2018;142(5):927–39. [DOI] [PubMed] [Google Scholar]
- 70.Jesinghaus M, Pfarr N, Kloor M, Endris V, Tavernar L, Muckenhuber A, et al. Genetic heterogeneity in synchronous colorectal cancers impacts genotyping approaches and therapeutic strategies. Genes, chromosomes & cancer. 2016;55(3):268–77. [DOI] [PubMed] [Google Scholar]
- 71.Bernstein CN, Shanahan F, Weinstein WM. Are we telling patients the truth about surveillance colonoscopy in ulcerative colitis? Lancet. 1994;343(8889):71–4. [DOI] [PubMed] [Google Scholar]
- 72.Baker KT, Salk JJ, Brentnall TA, Risques RA. Precancer in ulcerative colitis: the role of the field effect and its clinical implications. Carcinogenesis. 2018;39(1):11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lashner BA, Shapiro BD, Husain A, Goldblum JR. Evaluation of the usefulness of testing for p53 mutations in colorectal cancer surveillance for ulcerative colitis. Am J Gastroenterol. 1999;94(2):456–62. [DOI] [PubMed] [Google Scholar]
- 74.Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(49):20871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data have been deposited in Gene Expression Omnibus (GEO) database (accession number GSE156076).