Abstract
Tip60, a pan-acetyltransferase encoded by the Kat5 gene, is enriched in the myocardium; however, its function in the heart is unknown. In cancer cells, Tip60 acetylates Atm (Ataxia-telangiectasia mutated), enabling its auto-phosphorylation (pAtm), which activates the DNA damage response (DDR). It was recently reported that activation of pAtm at the time of birth induces the DDR in cardiomyocytes (CMs), resulting in proliferative senescence. We therefore hypothesized that Tip60 initiates this process, and that depletion of Tip60 accordingly diminishes the DDR while extending the duration of CM cell-cycle activation. To test this hypothesis, an experimental model was used wherein a Myh6-driven Cre-recombinase transgene was activated on postnatal day 0 (P0) to recombine floxed Kat5 alleles and induce Tip60 depletion in neonatal CMs, without causing pathogenesis. Depletion of Tip60 resulted in reduced numbers of pAtm-positive CMs during the neonatal period, which correlated with reduced numbers of pH2A.X-positive CMs and decreased expression of genes encoding markers of the DDR as well as inflammation. This was accompanied by decreased expression of the cell-cycle inhibitors Meis1 and p27, activation of the cell-cycle in CMs, reduced CM size, and increased numbers of mononuclear/diploid CMs. Increased expression of fetal markers suggested that Tip60 depletion promotes a fetal-like proliferative state. Finally, infarction of Tip60-depleted hearts at P7 revealed improved cardiac function at P39 accompanied by reduced fibrosis, increased CM cell-cycle activation, and reduced apoptosis in the remote zone. These findings indicate that, among its pleiotropic functions, Tip60 induces the DDR in CMs, contributing to proliferative senescence.
Keywords: Tip60, Atm, DNA Damage Response, Neonatal Cardiomyocytes, Cell-Cycle, Myocardial Infarction
1. Introduction
During the past two decades, evidence indicating that the adult heart is regenerable has ignited various approaches designed to optimize this process. As recently reviewed (1), these approaches include transplantation of adult cardiac stem cells or cardiomyocytes (CMs) derived from induced pluripotent stem cells (iPSCs), and efforts to re-initiate the proliferation of pre-existing CMs in vivo. Efforts directed toward the latter have intensified in recent years, pursuant to findings indicating that stimulation of pro-proliferative pathways, or inhibition of anti-proliferative pathways, can activate the cell-cycle in adult CMs, resulting in improved function (1); however, the issue concerning whether these interventions result in bona fide proliferation of CMs resulting in the generation of daughter cells remains controversial. Considering the profound senescence of adult CMs, it has been speculated that the attainment of bona fide CM proliferation sufficient to re-muscularize the myocardium may require the removal of multiple layers of cell-cycle inhibition (2).
Previous results in this laboratory suggested that Tip60 (Tat-interactive protein, 60 kD), a protein containing a chromodomain as well as an acetyltransferase domain, inhibits the CM cell-cycle (3). Although discovered as a viral protein, Tip60, which is encoded by the Kat5 gene, is widely expressed in mammalian cells, wherein it has been shown to be indispensable for the vitality of mouse blastocysts (4), hematopoietic stem cells (5), hippocampal neurons (6), as well as CMs (7). Tip60 is a pan-acetyltransferase, with targets ranging from histones wherein it functions within the NuA4 complex to promote chromatin remodeling (8), to non-histone proteins that regulate diverse cellular functions. Most information regarding its function has been derived from experiments performed on cell-lines, which have shown that Tip60 possesses at least three mutually non-exclusive tumor-suppressive functions: pro-apoptosis (9, 10), anti-proliferation (8), and pro-DNA damage response (DDR; (11–13)). Although the in vivo function of Tip60 remains unclear, these in vitro findings indicate that Tip60, via acetylation of multiple targets, maintains organ function by limiting cellular growth.
It was recently reported that exposure to the aerobic environment at the time of birth causes DNA damage in CMs, resulting in the phosphorylation of Atm (serine-threonine kinase ataxia-telangiectasia mutated); in turn, pAtm induces DDR signaling, which culminates in the replicative senescence of CMs by the end of the neonatal period (14, 15). Because the in vitro studies cited above have documented that Tip60 acetylates Atm (11–13), resulting in its auto-phosphorylation and activation of the DDR, we hypothesized that Tip60 initiates these events in the neonatal heart, in which instance genetic depletion of Tip60 from CMs should down-regulate the DDR, extending the duration of cell-cycle activity in neonatal CMs and the ability to regenerate following cardiac injury. The experiments reported here, which were designed to test these possibilities, indicate that among its pleiotropic functions, Tip60 induces the DDR in CMs, contributing to cell-cycle senescence, and that Tip60 depletion extends the period during which the neonatal heart can regenerate.
2. Materials and Methods
2.1. Animal Care & Use:
This investigation adhered to the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (NIH Pub. Nos. 85–23, Revised 1996). All protocols described in the authors’ Animal Use Application (AUA #000225), which were approved by the Medical College of Wisconsin’s Institutional Animal Care and Use Committee (IACUC), were adhered to in this study. The IACUC has an Animal Welfare Assurance status from the Office of Laboratory Welfare (A3102–01).
We previously described the preparation of mice containing floxed Kat5 alleles, wherein Cre-recombinase removes exons 3–11 consisting of 71% of the exon structure including the chromo and acetyltransferase domains (7). For the experiments described here, these mice were mated with a line obtained from the Jackson Laboratory (Jax #005650) that expresses an α-myosin heavy chain (Myh6)-driven merCremer-recombinase transgene that, upon administration of tamoxifen, enters the nucleus to recombine floxed alleles (16).
In these experiments, neonatal mice were administered a single injection of 250 μg tamoxifen (Sigma #T5648) suspended in 6.25% ethanol/sunflower oil into the scruff of the neck on the day of birth (P0). On the day before harvest, mice were injected with 0.7 mg 5’-bromodeoxyuridine (BrdU). On the day of harvest, hearts were perfused with ice-cold ~3 ml cardioplegic solution (25 mM KCl/5% dextrose in PBS) followed by apportionment of transverse sections for histology, RNA isolation, and western blotting, for which these portions were respectively placed in ice-cold 4% paraformaldehyde (PFA), TRIzol Reagent (Thermo-Fisher #15591626), and RIPA buffer (Thermo-Fisher #89901) containing Halt™ anti-protease/antiphosphatase cocktail (Thermo-Fisher #78440). Tissue for histology remained in ice-cold 4% PFA overnight, transferred to 70% EtOH for a minimum of one overnight, followed by embedding in paraffin. Samples for RNA were homogenized with a teflon pestle, and, samples for protein were minced, homogenized, and sonicated; both were stored at −80°C until final processing.
2.2. Genotyping
was performed by PCR, using 20 μl reactions including 2x GoTaq Green Mastermix (Promega #M7123), 1.1 mM MgCl2, 0.5 μM each primer, 0.5 μM internal control primers, and 4.0 μl template. Templates consisted of 1,200g supernatants of ear tissue punches or tail tip samples that were boiled for 10 minutes in 0.3 ml 10 mM NaOH/1 mM EDTA. Sequences of primer pairs for PCR are listed in Table 1. PCR products were amplified in an AB Applied Biosystems GeneAmp PCR System 9700 using the following programs: for LoxP, one 5-minute cycle at 95°C, thirty-five cycles at 94°C 30 sec/61°C 45 sec/72°C 45 sec, followed by one 10-minute cycle at 72°C; for Myh6-merCremer, one 5-minute cycle at 95 °C, thirty-five cycles at 94°C 30 sec/54°C 45 sec/72°C 45 sec, followed by one 10-minute cycle at 72°C. Amplicons were separated at 80–95 V for two hours in 2% agarose and imaged after ethidium bromide staining.
Table 1.
Primers & Probes
| Primers for PCR Genotyping | ||||
|---|---|---|---|---|
| Allele | Sequence (5’-3’) and Working Conc. | Amplicon (bp) | Annealing °C | |
| LoxP in intron 2 | FWD GGAGGGAGTCAACGATCGCA 0.5 μM | 687 LoxP 586 WT | 61 | |
| REV AATGGGGGACCTACTCACCA 0.5 μM | ||||
| Cycling Details: 94 °C 5 min, then 35 cycles of 94 °C 30sec/61 °C 45sec/72 °C 45sec, then 72 °C 10 mi | ||||
| LoxP in intron 11 | FWD GCACTCATCCAGGCTGTCC 0.5 μM | 655 LoxP 554 WT | 61 | |
| REV TCGGTTCTCAGAGACTAGC 0.5 μM | ||||
| Cycling Details: 94 °C 5 min, then 35 cycles of 94 °C 30sec/61 °C 45sec/72 °C 45sec, then 72 °C 10 min | ||||
| Myh6-Cre transgene | FWD ATACCGGAGATCATGCAAGC 0.5 μM | 440 | 54 | |
| REV AGGTGGACCTGATCATGGAG 0.5 μM | ||||
| Cycling Details: 94 °C 5 min, then 35 cycles of 94 °C 30sec/54 °C 45sec/72 °C 45sec, then 72 °C 10 min | ||||
| Primers/Probes for Taqman qRT-PCR Gene Expression Analysis | ||||
| Gene Target | Taqman Probe Kit (Thermo-Fisher catalog #) | |||
| Acta1 | Mm00808218_g1 | |||
| Atm | Mm01177457_m1 | |||
| Bax | Mm00432051_m1 | |||
| Ccna2 (Cyclin A2) | Mm00438063_m1 | |||
| Ccnb1 (Cyclin B1) | Mm03053893_gH | |||
| Ccnd1 (Cyclin D1) | Mm00432359_m1 | |||
| Ccnd2 (Cyclin D2) | Mm00438070_m1 | |||
| Cdk1 | Mm00772472_m1 | |||
| Cdk4 | Mm00726334_s1 | |||
| Cdkn1a (p21) | Mm00432448_m1 (4930567H1+) | |||
| Cdkn1b (p27) | Mm00438168_m1 | |||
| Gapdh | Mm99999915_g1 | |||
| Kat5 (Tip60) | Mm01231512_m1 | |||
| Meis1 | Mm00487664_m1 | |||
| Myh6 | Mm00440359_m1 | |||
| Myh7 | Mm00600555_m1 | |||
| NFκB1 | Mm00476361_m1 | |||
| Nppa | Mm01255747_g1 | |||
| Nppb | Mm01255770_g1 | |||
| p53 | Mm01731290_g1 | |||
| Rpl37α | Mm01546394_s1 | |||
| Vcam1 | Mm01320970_m1 | |||
| Wee1 | Mm00494175_m1 | |||
2.3. Western Blotting:
For the blot in Figure 1C, protein was extracted from the basal third of hearts from which atria had been removed. After harvesting in ice-cold RIPA buffer (Thermo-Fisher #89900) containing Halt™ Protease Inhibitor (Thermo-Fisher #78440), samples were minced and homogenized, followed by sonicating (Misonix Sonicator 3000) for 10 seconds at an output setting of 2.5 and centrifugation at 1,000 g for 5 minutes. Total protein concentration in supernatants was determined using the standard Bradford Assay (Bio-Rad #500–0006), followed by dilution in Laemmli Sample Buffer (Bio-Rad #161–0747) to a loading concentration of 1 μg/μl. For the blot in Supplemental Figure 3, protein was extracted from hearts at the indicated embryonic and neonatal stages as previously described; at the earliest stages, groups of 5–10 hearts were pooled to obtain sufficient protein (3). For electrophoresis, 10 μg of each sample were loaded into lanes of pre-cast Bio-Rad 10% or 4–15% acrylamide gels, and, separated proteins were electroblotted for 60 minutes at 100V onto 0.45 μm nitrocellulose membranes (Bio-Rad #162–0146). Blots were blocked with NFDM/TBST (5% non-fat dry milk/10mM Tris-HCl (pH 7.6)/150mM NaCl/0.05% Tween-20) or 5% BSA in TBST. Primary and secondary antibodies, and dilutions for western blotting, are listed in Table 2. Blots were reacted with primary antibody in 5% NFDM or 5% BSA blocking buffer overnight at 4°C. Secondary antibodies were diluted in 5% NFDM/TBST or 5% bovine serum albumin (BSA) and applied for 60 minutes at RT. Reacted blots were covered with HRP-substrate (Thermo-Fisher #34580) for 5 minutes at RT, followed by chemiluminescent imaging and densitometry using Bio-Rad ChemiDoc and ImageJ software, respectively.
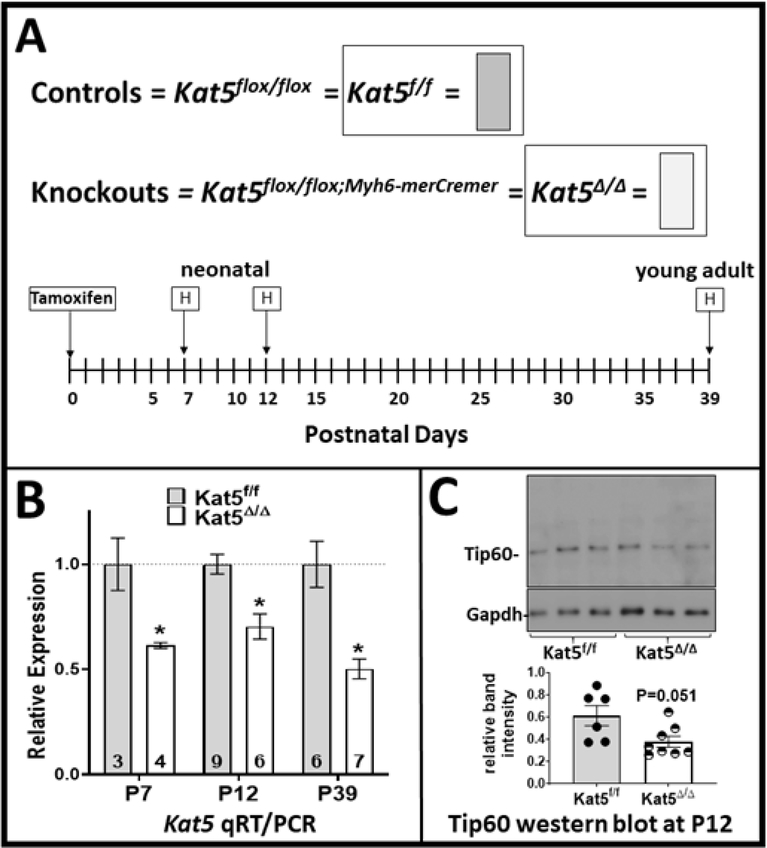
Figure 1. Tamoxifen-induced activation of Cre-recombinase at P0 causes Tip60 depletion in the heart at later postnatal stages.
Control (Kat5f/f) and experimental (Kat5f/f;Myh6-merCremer, denoted Kat5Δ/Δ) pups were injected with 250 μg tamoxifen on the day of birth (P0) to initiate recombination of the floxed Kat5 gene. Panel A schematically displays how the control and knockout genotypes are designated in this paper, and the postnatal days when hearts were harvested (H) for analysis. Panel B shows results from qRT-PCR analyses revealing the extent of Kat5 mRNA knockdown on each neonatal day. Panel C is a western blot showing Tip60 protein depletion at P12. Error bars denote ±SEM. Statistical significance was determined using an unpaired two-tailed t-test. *P<0.05 vs Kat5f/f.
Table 2.
Antibodies for Western Blotting
| Antigen | Manufacturer | Catalog# | Made in | Dilution |
|---|---|---|---|---|
| 1° Tip60 (N1) | Bethyl | custom | rabbit | 1:1000 |
| 2° goat anti-rabbit IgG HRP | Thermo Fisher | 32460 | goat | 1:5000 |
| 1° GAPDH | Adv ImmunoChem | 2-RGM2 | mouse | 1:1000 |
| 2° goat anti-mouse IgG HRP | Thermo Fisher | 32430 | goat | 1:5000 |
| 1° phosphorylated Atm (pATM) (S1981 human, S1987 mouse) |
Santa Cruz | sc-47739 | mouse | 1:1000 |
| 2° goat anti-mouse IgG HRP | Thermo Fisher | 32430 | goat | 1:5000 |
| 1° Atm | Santa Cruz | sc-23921 | mouse | 1:750 |
| 2° goat anti-mouse IgG HRP | Thermo Fisher | 32430 | goat | 1:5000 |
2.4. Quantitative RT-PCR (qPCR):
Heart tissue in Trizol was purified using PureLink RNA Mini-Kits (Invitrogen #12183018A), including a genomic DNA removal step (PureLink DNase; Thermo-Fisher #12185–010) according to the manufacturer’s instructions. RNA Yield & Quality were determined using an Eppendorf Biophotometer Plus Instrument.
cDNA was synthesized as follows. After diluting totalRNA from each heart so that precisely 1.0 μg was suspended in 14 μl nuclease-free distilled water (NFDW), 4.0 μl 5x VILO reaction mixture (Invitrogen #100002277) were added. To start the reverse-transcription reaction, 2.0 μl 10x SuperScript Enzyme Mix (Invitrogen #100002279) were added, followed by incubation for 10 minutes at 25°C, 60 minutes at 42°C, and 5 minutes at 85°C. cDNA templates were diluted with NFDW to a concentration of 6.25 ng/μl and stored at −20°C.
qPCR was carried-out by subjecting each biological replicate (i.e. sample from each individual heart) to triplicate determinations. Each reaction was performed in a total volume of 20 μl using 96-well arrays, each well containing 1x Taqman Fast-Advanced Master Mix (Thermo-Fisher #4444557), 1x Taqman Probe Kit (Table 1), and 25 ng cDNA as template. The arrayed samples were amplified in a Bio-Rad CFX96 Real Time System (C1000 Touch) programmed as follows: 2 minutes at 50°C → 20 seconds at 95°C → 3 seconds at 95°C → 30 seconds at 60°C; the last two steps were repeated 39 times. Results were processed using Bio-Rad CFX Manager 3.1 software.
2.5. Immunostaining & Cell Counting:
On the day before harvest, mice were injected with 0.7 mg 5’-bromo-2’-deoxyuridine (BrdU; Sigma #B9285). Following removal, hearts were perfused with ice-cold cardioplegic solution and atria were removed. Ventricles were fixed overnight in fresh ice-cold 4% paraformaldehyde/PBS, processed through ethanol series, and embedded in paraffin. Sections (4 μm thick) mounted on microscope slides were de-waxed, subjected to antigen retrieval (100°C in 10 mM trisodium citrate pH6.0/0.05% Tween-20 for 20 minutes) followed by 30 minutes’ cooling at RT, and blocked with 2% goat serum/0.1% Triton-X-100 in PBS. Primary antibodies were diluted in blocking buffer and applied overnight at 4°C; secondary antibodies were applied for one hour in the dark. Combinations and dilutions of primary and secondary antibodies employed to immunostain each target antigen are provided in Table 3.
Table 3.
Antibodies for Immunofluorescent Staining
| Antigen | Manufacturer | Catalog # | Made in | Dilution |
|---|---|---|---|---|
| 1° 5’-Bromodeoxyuridine (BrdU) | Abcam | ab6326 | rat | 1:200 |
| 2° goat anti-rat 488 | Invitrogen | A-11006 | goat | 1:500 |
| 1° phospho-Histone H3 (pH3) | EMD Millipore | 05–806 | mouse | 1:200 |
| 2° goat anti-mouse 488 | Invitrogen | A-11029 | goat | 1:500 |
| 1° phospho-Atm (pAtm) | Novus | NB100–306 | mouse | 1:200 |
| 2° goat anti-mouse 488 | Invitrogen | A-11029 | goat | 1:500 |
| 1° Ki67 | Invitrogen | 14–5698-82 | rat | 1:250 |
| 2° goat anti-rat 488 | Invitrogen | A-11006 | goat | 1:500 |
| 1° cardiac-Troponin-T (cTnT) | Abcam | ab8295 | mouse | 1:200 |
| 2° goat anti-mouse 488 | Invitrogen | A-11029 | goat | 1:500 |
| 1° GATA-4 | Cell Signaling Technology | D3A3M 36966S | rabbit | 1:100 |
| 2° goat anti-rabbit 594 | Invitrogen | A-11037 | goat | 1:500 |
| 1° phospho-Histone H2A.X S139 | EMD Millipore | 05–636 | mouse | 1:400 |
| 2° goat anti-mouse 488 | Invitrogen | A-11029 | goat | 1:500 |
| 1° 8-oxoG | Abcam | Ab62623 | mouse | 1:100 |
| 2° goat anti-mouse IgG 568 | Invitrogen | A-11031 | goat | 1:500 |
| 1° alpha-Actinin | Abcam | ab68167 | rabbit | 1:500 |
| 2° goat anti-rabbit 594 | Invitrogen | A-11037 | goat | 1:500 |
| 1° Caspase-3 | Cell Signaling | 9661S | rabbit | 1:50 |
| 2° goat anti-rabbit 594 | Invitrogen | A-11037 | goat | 1:500 |
| Wheat Germ Agglutinin-488 | Thermo-Fisher | W11261 |
Microscopy was performed on a Nikon Eclipse 50i microscope equipped with a Nikon DSU3 digital camera. For most determinations, it was strongly preferred to identify CMs based on expression of the nuclear marker GATA4, for which an antibody having remarkable specificity and sensitivity was employed (Cell Signaling #36966); CM identity was verified by immunostaining the cytoplasmic marker cardiac troponin-T (cTnT), or α-actinin.
In non-injured Tip60-depleted hearts, percentages of CMs expressing Ki67, BrdU, phosphorylated histone H3 (pH3), or phosphorylated Atm (pATM), were assessed by monitoring fluorescent signal in the FITC channel, after confirming the identity of each GATA4-stained CM in the Texas Red channel according to the following criteria: (i) GATA4 staining was verified as nuclear by co-staining with DAPI, (ii) counting was restricted to ovoid/spherical nuclei that were relatively large (≥1.5 μm diam.), (iii) only nuclei residing in the myocardium, as revealed by auto-fluorescence in the FITC channel, were counted, (iv) nuclei located in interstitial spaces, epicardium, or vasculature were ignored. At least 500 GATA4-positive cells were evaluated in each section during scanning at 1,000x magnification. To verify pAtm staining, only nuclei that were at least half-filled with FITC fluorescence were counted; pAtm-positive nuclei were also confirmed to be DAPI-positive. To enumerate CMs exhibiting phosphorylated histone H2A.X signal, both α-actinin and GATA4 were used to identify CMs, and, the total number of double-positive cells in each section was manually counted; pH2A.X signals were counted only if confined to DAPI-positive nuclei.
In infarcted hearts, CM cell-cycle activation was assessed in six randomly selected 200x microscopic fields representing (i) the remote zone, specifically the area of myocardium ~2 mm distal to the boundary of the infarct, and (ii) the border zone, specifically the area immediately adjacent to the infarct zone. For these determinations, cardiac troponin-T (cTnT) was employed to identify CMs, and, cells that were double-positive for cTnT and Ki67/BrdU/pH3 were counted and presented as the average number per microscopic field.
2.6. Apoptosis:
Apoptosis was assessed using the DeadEnd Fluorometric TUNEL System (Promega #G3250) per the manufacturer’s instructions. The total number of TUNEL-positive nuclei present in each section was manually counted at 400x magnification. TUNEL signal was counted only if confined to a DAPI-positive nucleus. Nuclei were scored as TUNEL-positive only if at least 50% of the nucleus contained fluorescent signal. Because the use of proteinase-K in the TUNEL method disabled ability to co-stain antigens to determine CM identity, apoptosis was also determined by immunostaining cleaved caspase-3; in these determinations, WGA was employed as described below to identify CMs based on cellular outlines, and, the total number of cleaved caspase-3 positive CMs present in each section was manually enumerated at 400x magnification.
2.7. Wheat Germ Agglutinin (WGA) Staining
was performed using Thermo-Fisher #W11261 Alexa Fluor 488 conjugate. Sections on microscope slides were stained with 50 μg/ml WGA in PBS for 10 minutes at room temperature, followed by washing. In each heart, six images of CMs in transverse orientation were photographed at 400x magnification and processed using ImageJ software to calculate average pixel numbers/CM, as indicative of CM size. Briefly, the green (488) channel displaying CMs was isolated, followed by thresholding to fill-in spaces occupied by CM cytoplasm, then adjusting settings to acquire particle sizes in the 600−∞ range having a circularity of 0.25–1.0. After results (which were set to “include holes”) were obtained, particles representing incorrectly oriented CMs, and blood vessels, were removed. To assess CM density, the total number of CMs in each 400x microphotograph was manually counted by blinded observers and presented as the average number per 400x field.
2.8. Cardiomyocyte (CM) Nucleation & Ploidy:
Cells in the myocardium were separated by perfusing collagenase II (1 mg/ml/Calcium-free Tyrodes) retro-aortically using a Langendorff apparatus, as previously described (17). Digested ventricles were isolated, triturated in KB buffer (20 mM KCl/10 mM KH2PO4/70 mM K-glutamate/1 mM MgCl2/25 mM glucose/10 mM β-hydroxybutyric acid/20 mM taurine/0.5 mM EGTA/0.1% albumin/10 mM HEPES [pH 7.4]), and filtered through 250 μm mesh. While homogeneously suspended, the cells were fixed by adding PFA to 2%, followed by immunofluorescent identification of CMs using primary cardiac troponin-T (cTnT) antibody (diluted 1:1,000, overnight at 4 °C) and a goat anti-mouse secondary (ThermoFisher A11001, 1:500), plus DAPI staining to evaluate nucleation and ploidy. Aliquots of stained cell suspension were spread across microscope slides, followed by mounting coverslips with Prolong Gold Mounting Media (Thermo-Fisher P36930) and microscopic examination at 200x magnification. CM nucleation (mono-, bi, tri-, tetra-) was determined by evaluating approximately 400 CMs per heart. Nuclear ploidy within the mononuclear fraction was estimated from photomicrographs subjected to ImageJ analysis to determine the relative concentration of DNA in each nucleus inferred from the summation of pixel intensity within each DAPI-stained nuclear area (18). Approximately 20 mononuclear cells were analyzed per heart and normalized to diploid nuclei identified in tri- and tetra-nucleated CMs.
2.9. Myocardial Infarction:
P7 mice were anesthetized by induction of hypothermia for ~3–5 minutes. A lateral thoracotomy was performed at the fourth intercostal space to expose the heart, followed by opening of the pericardium and placement of an 8.0 prolene suture under the main coronary artery at the mid-ventricular level, with the aid of a microscope. Ischemia was induced by tying the suture, after which successful occlusion was verified by blanching of the myocardium below the ligature. The incision was closed using two stitches of 8–0 non-absorbable prolene to enclose ribs, muscle, and skin. Pups were placed into a warming chamber until fully recovered, then returned to the mother. On the day before harvest, mice were injected intraperitoneally with BrdU (0.7 mg). On the day of harvest, mice were subjected to echocardiography to assess heart function, then were euthanized with CO2. Hearts were perfused with ice-cold cardioplegic solution (25 mM KCl/5% dextrose in PBS) followed by 4% PFA. After overnight fixation in PFA, hearts were transferred to 70% EtOH, followed by embedding in paraffin. Paraffinized hearts were transversely sectioned, in entirety from apex to base, after which eight 4 μm thick sections from equidistant (~0.5 mm) intervals were placed on microscope slides. The slides were stained with Masson trichrome, examined with a Nikon SMZ800 microscope, and photographed at 10x magnification using a SPOT Insight camera (Nikon Instruments). MIQuant software was used to quantitate infarct size in sections between the apex and the site of ligation; results were expressed as percentage area of the left ventricle.
2.10. Echocardiography:
Echocardiography was performed on mice lightly anesthetized with isoflurane delivered via a nose cone. Parasternal long-axis, short-axis, and apical 4-chamber views were obtained using a VisualSonics high-frequency ultrasound imaging system (Vevo 3100) equipped with a transducer (MX550D) operating at 30–40 mHz. Short-axis views in M-mode were used to measure the left ventricular (LV) anteroposterior internal diameter (LVID) at the mid-ventricular level, at end-diastole (d) and end-systole (s). Long-axis views in B-mode were used to measure LV internal area (LVA) and length (L) at end-diastole and end-systole. LV systolic function was assessed by (i) fractional shortening (FS%) = ([LVIDd − LVIDs] / LVIDd) * 100, and (ii) ejection fraction (EF%) = (end-diastolic volume - end-systolic volume)/end-diastolic volume, whereby volumes were estimated by 4π/3 * L/2 * (LVA ÷ π(L/2))2. In addition, global LV function was assessed by calculating the myocardial performance index: MPI = (isovolumic contraction time + isovolumic relaxation time) / ejection time. Time intervals were obtained from pulsed Doppler waveforms of mitral valve inflow and aortic valve outflow from apical 4-chamber views.
2.11. Data Analysis and Statistics:
All determinations were performed by blinded observers. Data are reported as means ± SEM. Data encompassing the P7 thru P39 timepoints were analyzed using two-way ANOVA to determine global effects of time and of genotype; see Table 4. If a global test indicated an effect of genotype, post hoc contrasts at each timepoint were compared using Bonferroni multiple comparison tests correction. For the qPCR studies, ΔCq values of genes expressed in individual hearts (normalized to Rpl37a or Gapdh) were plotted versus individual Kat5 values and subjected to linear regression analysis to test for correlation. All other data were compared using an unpaired, two-tailed Student’s t test.
Table 4.
Two-Way ANOVA Comparison of Gene Expression in Neonatal Hearts Containing Tip60-depleted (Kat5Δ/Δ) Cardiomyocytes Data presented as expression relative to control (Kat5f/f = 1.0)
| Gene | Postnatal Day | Genotype P-value | ||
|---|---|---|---|---|
| 7 | 12 | 39 | ||
| Cell-Cycle Activators | ||||
| Ccnd1 (cyclin D1) | 0.742 ± 0.053 | 0.829 ± 0.081 | 1.037 ± 0.050 | 0.053 |
| Ccnd2 (cyclin D2) | 0.810 ± 0.031 | 0.873 ± 0.081 | 0.990 ± 0.049 | 0.093 |
| Cdk4 | 0.815 ± 0.083 | 0.867 ± 0.079 | 0.884 ±0.050 | 0.033 |
| Ccna2 (cyclin A2) | 0.935 ±0.113 | 1.269 ± 0.155 | 1.120 ± 0.153 | 0.308 |
| Ccnb1 (cyclin B1) | 0.986 ± 0.098 | 1.190 ± 0.177 | 1.501 ± 0.205 | 0.142 |
| Cdk1 | 0.886 ± 0.133 | 1.016 ± 0.110 | 1.302 ± 0.135 | 0.534 |
| Cell-Cycle Inhibitors | ||||
| Cdkn1a (p21) | 1.116 ± 0.177 | 0.971 ± 0.106 | 1.330 ± 0.114 | 0.162 |
| Cdkn1b (p27) | 0.837 ± 0.034 | 0.758 ± 0.062* | 0.725 ± 0.048* | 0.001 |
| Meis1 | 0.917 ± 0.092 | 0.939 ± 0.090 | 0.694 ± 0.050** | 0.033 |
| DDR Markers | ||||
| Atm | 1.219 ± 0.130 | 0.711 ± 0.065 | 0.756 ± 0.412 | 0.155 |
| Wee1 | 1.092 ± 0.079 | 0.933 ± 0.089 | 0.738 ± 0.045 | 0.325 |
| P53 | 1.514 ± 0.100 | 1.007 ± 0.095 | 0.746 ± 0.051 | 0.297 |
| Bax | 0.986 ± 0.082 | 0.906 ± 0.115 | 0.575 ± 0.040 | 0.027 |
| SASP Markers | ||||
| Vcam1 | 0.595 ± 0.045 | 0.605 ± 0.101** | 0.679 ± 0.038** | <0.001 |
| NFκb1 | 0.1.122 ± 0.079 | 0.866 ± 0.079 | 0.611 ± 0.036** | 0.042 |
| Fetal Gene Markers | ||||
| Nppa | 0.976 ± 0.088 | 2.107 ± 0.580* | 2.202 ± 0.290* | 0.010 |
| Nppb | 2.269 ± 0.246** | 0.753 ± 0.175 | 1.674 ± 0.115* | 0.022 |
| Acta1 | 1.036 ± 0.138 | 1.116 ± 0.207 | 2.655 ± 0.362*** | 0.010 |
| Myh6 | 1.073 ± 0.116 | 0.906 ± 0.160 | 0.746 ± 0.057 | 0.435 |
| Myh7 | 1.430 ± 0.189 | 1. 404 ± 0.193 | 4.244 ± 0.916*** | 0.003 |
P<0.05
P<0.02
P<0.001 vs Kat5f/f
3. Results
3.1. Experimental Model & Timeline.
The objective of this study was to assess the effects of depleting Tip60 via disruption of the Kat5 gene in postnatal CMs, beginning at the time of birth (P0). According to our hypothesis, Tip60 depletion should prevent activation of the DDR, as indicated by reduced levels of DDR markers, resulting in extension of the period of CM cell-cycle activity in the neonatal heart, in accord with recent findings (14). Importantly, because long-term constitutive depletion of Tip60 is lethal to cells (4–6) including CMs (7), conditional activation of the Myh6-cre transgene was employed by administering only a single injection of tamoxifen to control (Kat5flox/flox, hereafter Kat5f/f) and experimental (Kat5flox/flox;Myh6-merCremer, hereafter Kat5Δ/Δ) mice on the day of birth (P0). Figure 1A depicts the experimental timeline wherein hearts in mice that were injected with tamoxifen on P0 were evaluated on postnatal days P7, P12, and P39, the latter to assess the effect of neonatal Tip60 depletion at an early adult stage. Although this regimen produced a Kat5 phenotype that is less pronounced than the effect we observe after administering multiple tamoxifen injections to adult mice, the single-dose regimen used here significantly, but not exhaustively, depleted Kat5 mRNA in Kat5Δ/Δ hearts (Fig. 1B) at each stage of the timeline. (The developmental pattern of Kat5 expression in the heart at the RNA level in control mice (Kat5flox/flox) is shown in Supplemental Figure 1.) Western blotting at P12 indicated that the level of Tip60 protein was accordingly reduced (Fig. 1C). The absence of effects on cardiac mass or apoptosis (Supp. Fig. 2), as well as fibrosis (not shown), in Tip60-depleted hearts indicated that this regimen did not elicit untoward effects during neonatal heart development.
3.2. Diminished DNA Damage Response (DDR) in Tip60-depleted neonatal hearts.
Based on the hypothesis, it was reasoned that depletion of Tip60 would diminish activation of Atm, and consequently the DDR, in postnatal CMs. Prior to assessing effects of Tip60 depletion on Atm, it was of interest to examine the pattern of Atm phosphorylation (pAtm), as well as levels of bulk Atm, in wild-type (i.e. non-floxed) hearts at successive stages of heart development. Western blotting of protein from embryonic and adult hearts at successive stages of development indicated that pAtm levels increased beginning at P2, the stage when the Tip60α isoform curiously begins to be replaced by alternatively-spliced Tip60β (7), and that this was accompanied by increased levels of non-phosphorylated (bulk) Atm (Supp. Fig. 3).
To assess pAtm levels in Tip60-depleted CMs, immunofluorescent microscopy was employed because low levels of this large phosphorylated protein (~350 kD), combined with the presence of non-CMs in heart tissue samples, precluded definitive assessment by western blotting. As described in Materials and Methods, quantitation was restricted to enumerating CM nuclei that exhibited pAtm signal in at least 50% of the nuclear area (Fig. 2A). In comparison with percentages of pAtm-positive CM nuclei in Kat5f/f control hearts (which declined during P7-P39), percentages in Tip60-depleted CMs were significantly lower during the timeline (Fig. 2A).
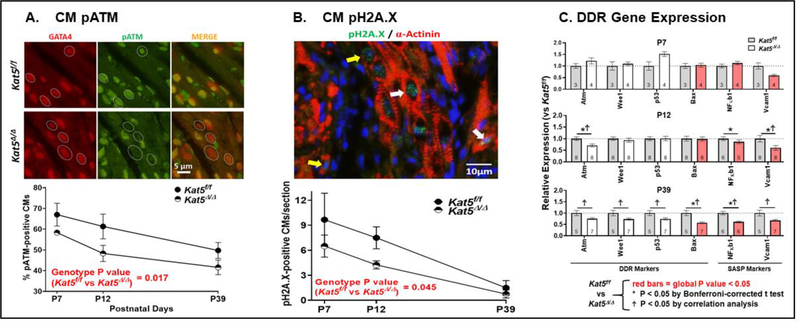
Figure 2. DDR mitigation in Tip60-depleted neonatal hearts.
Hearts in control (Kat5f/f) and experimental (Kat5Δ/Δ) neonates that were administered tamoxifen at P0 were harvested on the indicated postnatal days. Panel A shows sections that were double-immunostained to detect GATA4 (red) and pAtm (green), with quantitation expressed as percentages of pAtm-positive CMs enumerated by manually scanning entire sections at 1,000x magnification. Panel B shows numbers of pH2A.X CMs in sections double-immunostained to detect pH2A.X and α-actinin. White arrows point to examples of nuclei that are over half-filled with FITC fluorescence and were counted as pH2A.X-positive CMs whereas yellow arrows highlight examples of nuclei that were not counted as pH2A.X-positive. Panel C shows the expression of genes in the DDR pathway (DDR markers), and genes associated with the senescence-associated secretory phenotype (SASP markers), determined using qRT-PCR. Data in panels A-C are presented as mean ±SEM. In panels A-B, numbers (N) of individual hearts evaluated in each group were as follows: P7, 3 vs 4; P12, 7 vs 8; P39, 7 vs 7. Two-way ANOVA revealed that Tip60 depletion was associated with statistically significant reduced levels of pAtm (panel A), pH2A.X (panel B), and the genes denoted by red bars in panel C, across the neonatal timeline. In panel C, symbols * and Ϯ adjacent to individual data points respectively denote P<0.05 vs. Kat5f/f by post-hoc analysis using the Bonferroni multiple comparison test, and by correlation analysis.
[Note: This figure requires color reproduction.]
To verify that depletion of Tip60 diminished the DDR in CMs, the presence of phosphorylated histone H2A.X, a widely employed marker of definitive DNA damage (19) that is phosphorylated by pAtm (20, 21), was also assessed in CMs, identified by counterstaining with α-actinin (Fig 2B; (22, 23)) as well as GATA4 (Supp. Fig 4). Although the incidence of pH2A.X, like pAtm, became diminished during the neonatal period, pH2A.X levels were further reduced (30–50%) in Tip60-depleted CMs. However, Tip60 depletion did not alter the extent of DNA damage, which increases during neonatal heart development (14), as revealed by 8-oxoG (8-oxo-7,8-dihydroguanine) immunostaining (Supp. Fig. 5); we speculate that this reflects Tip60’s position downstream of DNA damage (11, 24).
In addition, we employed qPCR to interrogate the expression of genes that serve as markers of the DDR pathway (Atm, Bax, p53, Wee1), as well as markers indicative of the senescence-associated secretory phenotype (SASP (25); NFκB1, Vcam1), which was recently shown to accompany myocardial DNA damage (23). Figure 2C shows mRNA levels as fold-changes in Tip60-depleted (Kat5Δ/Δ) hearts relative to Kat5f/f controls at each developmental stage. To assess statistically significance changes in these and in subsequent qPCR datasets, results were subjected to analysis using two-way ANOVA to assess global effects of time and genotype (Table 4); also, ΔCq values of target genes in each heart were plotted against the normalized extent of Kat5 and subjected to regression analysis to identify significant correlations within gene pairs. This revealed that Tip60 depletion was associated with reduced levels of mRNAs encoding all of the DDR and SASP markers, which became highly significant at P39 (Fig. 2C; Table 4).
3.3. Increased cell-cycle activation in cardiomyocytes of Tip60-depleted neonatal hearts.
In accord with our hypothesis, reduction of the DDR consequent to Tip60 depletion should delay the onset of replicative senescence in CMs. Findings presented in Figures 3 and 4 are consistent with this possibility.
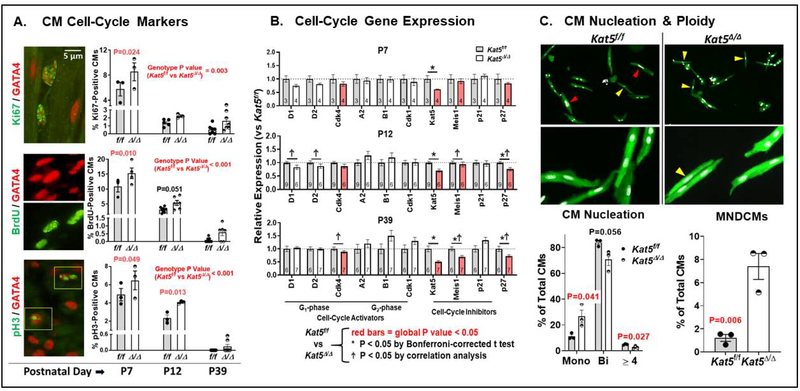
Figure 3. Cell-cycle activation in Tip60-depleted neonatal hearts.
Following injection of tamoxifen at P0, hearts harvested on the indicated postnatal days were subjected to the following determinations. Panel A shows results of immunostaining to detect Ki67, BrdU, and pH3, plus GATA4 to identify CMs. Percentages of cell-cycle-activated CMs were enumerated by blinded observers; the entirety of each section was scanned at 1,000x magnification. Panel B shows results of qPCR determinations to detect the expression of genes that activate and inhibit the cell-cycle. Panel C shows assessment of percentages of CMs isolated from P12 hearts that were mononuclear, and mononuclear/diploid (MNDCM). CMs immunostained with cTnT (green) are shown at two magnifications (CM nuclei are pseudo-colored white). Yellow and red arrowheads denote CMs that are mononuclear/diploid and mononuclear/polyploid, respectively. All data are presented as means ±SEM. For Panels A and B, two-way ANOVA revealed that the changes in Ki67, BrdU, pH3, and the genes labeled with red bars (Panel B) in Tip60-depleted hearts across neonatal time-points were statistically significant (P<0.05). P values and asterisks (*P<0.05) next to individual data points report results of post-hoc analysis using a Bonferroni-corrected multiple comparison test. Ϯ next to individual data points denotes P<0.05 vs. Kat5f/f by correlation analysis. For Panel C, data were analyzed by an unpaired, two-tailed Student’s t test.
[Note: This figure requires color reproduction.]
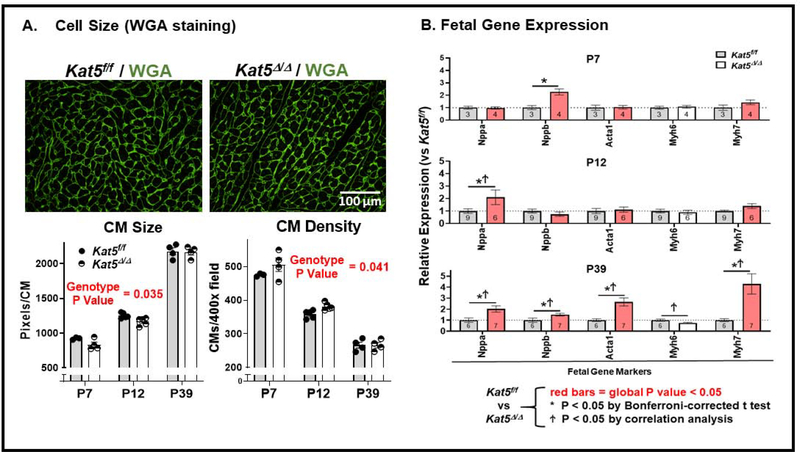
Figure 4. Increased CM density and expression of fetal gene markers in Tip60-depleted hearts.
Following injection of tamoxifen at P0, hearts were harvested on the indicated postnatal days and subjected to the following determinations. Panel A: WGA staining of Tip60-depleted CMs in transverse sections reveals reduced size and increased density. Panel B: qPCR shows increased expression of fetal gene markers in Tip60-depleted hearts at P39. All data are presented as means ±SEM. Two-way ANOVA revealed that the changes in CM size/density (Panel A) and expression of the genes labeled with red bars (Panel B) in Tip60-depleted hearts across neonatal time-points were statistically significant. Asterisks (*) next to individual data points indicate P<0.05 vs Kat5f/f by post-hoc comparison using a Bonferroni multiple comparison test. Ϯ next to individual data points denotes P<0.05 vs. Kat5f/f by correlation analysis.
[Note: This figure requires color reproduction.]
First, to assess whether Tip60 depletion increased the percentages of CMs exhibiting markers of cell-cycle activation, sections of ventricular myocardium at developmental stages P7, P12, and P39 were immunostained for Ki67, BrdU, and pH3 (Fig. 3A). Because it was our preference to identify CMs by probing nuclear antigens, GATA4 staining was employed, imaged upon the background of auto-fluorescent CM cytoplasm. Enumeration of Ki67-, BrdU-, and pH3-/GATA4-positive CMs revealed significantly increased percentages of each in Tip60-depleted hearts. To verify CM identity, sections immunostained for BrdU were also counter-stained using a CM-specific cytoplasmic marker, cardiac troponin-T (cTnT), which yielded similar results (Supp. Fig. 6).
Also, we performed qPCR to assess whether Tip60 depletion altered the expression of genes that inhibit or activate the cell-cycle. Most remarkably, decreased expression of genes encoding the cell-cycle inhibitors Meis1 and p27 was observed in Tip60-depleted hearts, as revealed by decreasing trends at P7 and P12 that became highly significant at P39 (Fig. 3B; Table 4); curiously, this was accompanied by a trend toward increased expression of the gene encoding the cell-cycle inhibitor p21 (Fig. 3B). qPCR also revealed trends toward increased expression of genes that activate late (G2) cell-cycle phases (Cyclins A2, B1, Cdk1) in Tip60-depleted hearts, despite trends toward reduced expression of genes that activate early (G1) cell-cycle phases (cyclins D1, D2 and Cdk4; Fig. 3B & Table 4).
Based on findings in Figures 3A–B, it was of interest to assess whether Tip60-depleted hearts contained increased numbers of CMs that were mononuclear and diploid (i.e. MNDCMs), as evidence of delayed CM senescence (17). To address this possibility, the frequency of MNDCMs was quantified in CMs isolated from hearts at P12, as by this time the majority of CMs have entered senescence and become polyploid (26). As shown in Figure 3C, single-cell suspensions of ventricular CMs from Kat5f/f and Kat5Δ/Δ hearts that were analyzed for multinuclearity and ploidy revealed that the percentage of mononucleated CMs, which was ~11% in control hearts at this timepoint, was increased ~2.5-fold in Tip60-depleted (Kat5Δ/Δ) hearts, at the expense of bi-nucleated and tetra-nucleated CMs. Of perhaps greater significance, the population of CMs that were both mononuclear and diploid (MNDCMs) was increased ~7-fold in Tip60-depleted hearts (Fig. 3C). The possibility that Tip60 depletion increased CM numbers was further supported by staining hearts with wheat germ agglutinin (WGA; Fig. 4A) to outline the perimeter of transversely-sectioned CMs, which indicated that CM size and density were respectively decreased and increased, suggestive of increased proliferation.
At the time of birth, CMs transition from a glycolytic environment wherein CMs undergo proliferation, to an oxidative environment in which CM endomitosis and proliferative senescence occur. Because this glycolytic-oxidative transition is characterized by the repression of fetal gene markers (27), a gene set that includes Acta1, Nppa, Nppb, and Myh7 (28), it was of interest to assess whether expression of these genes was affected in Tip60-depleted CMs. As shown in Figure 4B, trends toward increased expression of these genes were observed at the early timepoints, culminating in significantly increased expression of all fetal gene markers at P39, when expression of the adult gene marker Myh6 was depressed (Fig. 4B & Table 4).
The above experiments were performed by administering a single dose of tamoxifen to control (Kat5f/f) and experimental (Kat5flox/flox;Myh6-merCremer i.e. Kat5Δ/Δ) neonatal mice containing a Myh6-cre transgene at P0, followed by assessing effects of Tip60 depletion no earlier than P7. To ensure that these results were not affected by transient off-target effects of Cre-recombinase on CM cell-cycle activation that we recently reported in adult mice (29), control determinations were performed comparing tamoxifen-injected wild-type (Kat5+/+) mice with wild-type mice containing the Cre transgene (Kat5+/+;Myh6-merCremer). This revealed no effects on cardiac morphology, CM cell-cycle activation, or expression of cell-cycle regulatory genes (Supp. Fig. 7). Moreover, our observations that DNA damage markers were never increased, but in most instances decreased in Kat5Δ/Δ hearts, indicated the absence of off-target effects.
To summarize, the findings shown in Figures 3 and 4 indicate that depletion of Tip60 is associated with activation of the CM cell-cycle during neonatal stages of heart development, maintaining a fetal environment that is amenable to CM proliferation (30).
3.4. Improved regeneration in Tip60-depleted neonatal hearts after MI.
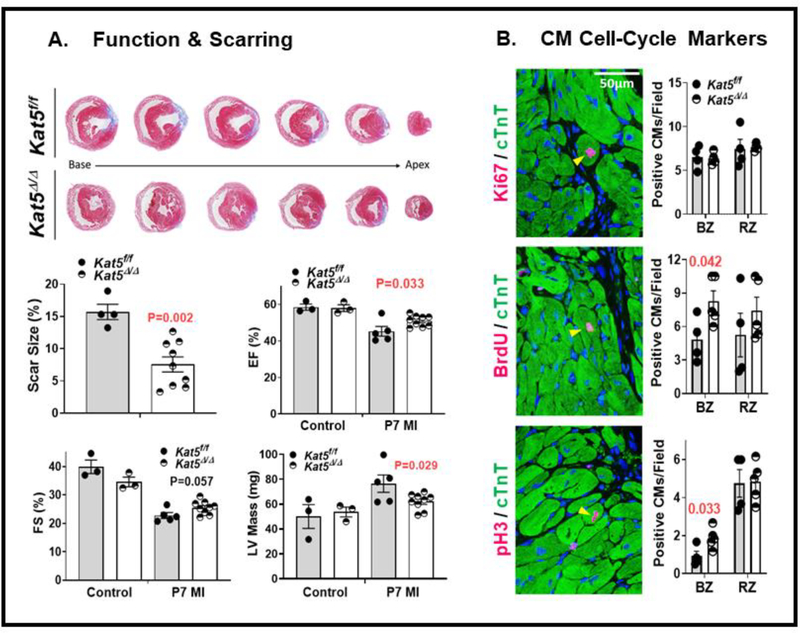
Because the above results correlating Tip60 depletion with increased CM cell-cycle activation and numbers of MNDCMs suggested that hearts containing reduced levels of Tip60 might exhibit improved regeneration after myocardial infarction (MI), the assessments described in Figures 5 and 6 were performed. In these experiments, Kat5f/f and Kat5Δ/Δ hearts that had been treated with tamoxifen on P0 were infarcted on neonatal day P7, when regenerative potential has normally abated (31), by permanently ligating the left main coronary artery, followed by echocardiographic and histologic assessments at P39. The extent of scarring in transverse segments of the LV below the site of ligation was assessed by Masson trichrome staining (Fig. 5A), revealing that levels were reduced by nearly 50% in Tip60-depleted hearts. Echocardiography revealed subtle but significant improvement of LV function in Tip60-depleted hearts, including increased ejection fraction (EF)) and fractional shortening (FS), which was accompanied by reduced LV mass (Fig. 5A); Tip60 depletion did not improve function in hearts that were not infarcted (see Controls in Fig. 5A).
Figure 5. Reduced scarring, improved cardiac function, and increased cell-cycle activation after MI in Tip60-depleted hearts.
Kat5f/f and Kat5Δ/Δ hearts treated with tamoxifen on P0 were infarcted on P7 via permanent ligation of the left main coronary artery. On P39, cardiac function was assessed by echocardiography, after which hearts were processed for histology, and transverse sections were removed at equal intervals below the site of ligation. Panel A (upper) shows effects on scarring; quantification of scar size and echocardiographic function are shown below. ‘Control’ denotes hearts that were not infarcted. Panel B shows increased cell-cycle activity in Tip60-depleted CMs; BZ and RZ respectively denote the border and remote zones. In both panels, statistical significance was determined using unpaired, two-tailed Student t tests. Yellow arrowheads denote Ki67/BrdU/pH3-positive CMs.
[Note: This figure requires color reproduction.]
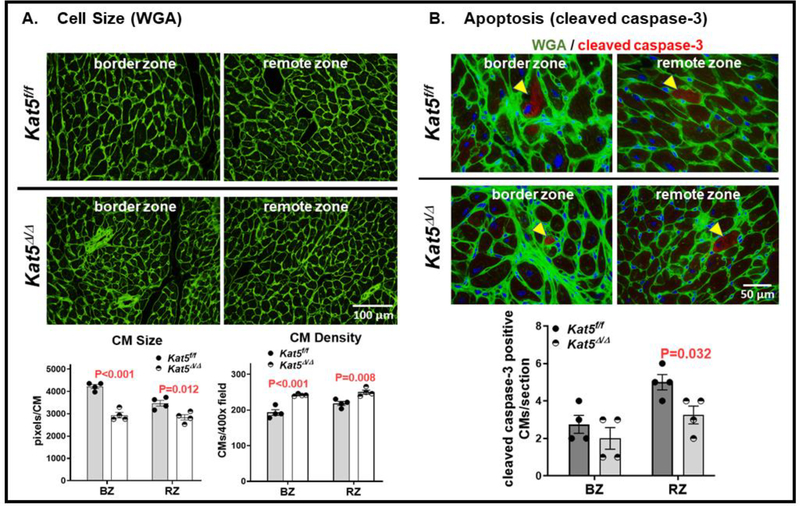
Figure 6. Increased density, reduced size, and diminished apoptosis of CMs in Tip60-depleted/infarcted hearts at P39.
Kat5f/f and Kat5Δ/Δ hearts were treated with tamoxifen on P0, infarcted on P7, and harvested on P39 followed by histological processing. Panel A: Sections were stained with fluorescently-labeled wheat germ agglutinin (WGA), and areas containing CMs in transverse orientation were photographed and processed with ImageJ to estimate CM size and density within both the infarct border (BZ) and remote zones (RZ). Panel B: Sections were double-stained with fluorescent WGA to identify CM outlines and with anti-caspase-3 to identify apoptotic CMs, followed by enumerating the total number of caspase-positive CMs (examples are denoted by yellow arrowheads) in each section. In both panels, statistical significance was determined using unpaired, two-tailed Student t tests.
[Note: This figure requires color reproduction.]
To assess whether reduced scarring and functional improvement were accompanied by increased activation of the CM cell-cycle, the immunostains depicted in Figure 5B were performed. Although numbers of Ki67-positive nuclei in CMs identified by cTnT staining were not altered in Tip60-depleted/infarcted hearts, nuclei exhibiting pH3 staining and BrdU incorporation were significantly increased in the border zone (Fig. 5B). Increased cell-cycle activity was accompanied by a reduction in CM size and increased CM density in both the border and remote zones of Tip60-depleted/infarcted hearts as revealed by WGA staining (Fig. 6A). And, consistent with Tip60’s pro-apoptotic activity (9, 32), its depletion correlated with reduced numbers of CMs in the remote zone of infarcted hearts that exhibited cleaved caspase-3 staining (Fig. 6B).
To address the question of whether depletion of Tip60 prior to MI conferred an immediate cardioprotective effect, hearts were infarcted at P7, followed by echocardiography and removal for histological analysis at P10. As shown in Supplemental Figure 8, pre-depletion of Tip60 did not improve cardiac function (Supp. Fig. 8A), or confer protection from cardiac damage (Supp. Fig. 8B) or apoptosis (Supp. Fig. 8C).
To summarize, the findings described in Figures 5–6 suggest that pre-depletion of Tip60 from the neonatal heart diminishes the long-term effects of MI by permitting CM regeneration and reducing apoptosis.
4. Discussion
Based on findings in cancer cells that Tip60 activates the DDR by enabling Atm phosphorylation (33), and on subsequent findings in neonatal CMs that activation of the DDR by pAtm at the time of birth promotes proliferative senescence by the end of the neonatal period (14), we examined the possibility that Tip60 regulates these events, and accordingly that genetic depletion of Tip60 mitigates the DDR, extending the duration of CM cell-cycle activity in the neonatal heart. Despite the requirement to employ an experimental model that avoids exhaustive depletion of this vital protein, findings shown in Figures 2–4 are supportive of this hypothesis. And, findings described in Figures 5–6 indicate that Tip60-depletion, perhaps by reducing proliferative senescence and/or apoptosis, may enhance cardiac regeneration and/or confer protection from the effects of myocardial ischemia.
Increased CM cell-cycle activation in Tip60-depleted neonatal hearts, which implies but does not prove bona fide proliferation (34), is consistent with our previous findings in the adult heart (3). However, the extent of cell-cycle activation observed at P7 and P12 was not sustained at P39 (Fig. 3A). Although this may be explained in part by the modest tamoxifen regimen, it more likely reflects the intransigence of endogenous mechanisms that cause CM senescence, similar to previous findings showing that although pharmacological prevention of oxidative damage in CMs at early neonatal stages reduces the DDR, proliferative senescence of CMs nonetheless ensues (14).
Among possible molecular responses to Tip60 depletion that permit cell-cycle activation, the most remarkable were decreased expression of Meis1 and p27 (Fig. 3B & Table 4), both of which promote proliferative senescence in CMs at neonatal stages (35, 36). To our knowledge, whether Tip60 mechanistically regulates these cell-cycle inhibitors has not been addressed. By contrast, the trend toward increased expression of the gene encoding p21 at P39 (Fig. 3B & Table 4) was interesting because, similar to the paradoxical finding that forced activation of the CM cell-cycle increases levels of p21 protein (37), this suggests a cellular response designed to maintain this inhibitor at levels sufficient to block CM cell-cycle activity. In addition, it was recently reported that intracellular localization of p21 modulates cell-cycle activation in CMs (38). These findings, in addition to those cited below, should compel further investigation into how Tip60 regulates levels of p21 protein.
An emerging principle in the field of cardiac regeneration holds that the hypoxic environment, present in the embryo as well as in ischemic myocardium, is accompanied by glycolytic metabolism that is permissive for CM proliferation, whereas the aerobic environment encountered at birth induces a shift toward oxidative metabolism wherein CMs undergo proliferative senescence to accommodate increased contractile function (14, 15, 39, 40). In this regard it was recently reported that impairment of glycolysis inhibits regeneration of the zebrafish heart (41). The transition from hypoxia to normoxia is accompanied by reduced expression of fetal marker genes (27, 28, 30), including those interrogated in Figure 4B, nearly all of which are increased in Tip60-depleted hearts by P39. Despite the caveat necessitated by our finding indicating that depletion of Tip60 did not alter 8-oxoG labeling (Supp. Fig. 5), this result suggests that Tip60 suppresses the glycolytic pro-proliferative environment.
In the adult heart, it has been speculated that the virtual absence of CM proliferation after MI is due to multiple layers of inhibition that become established during mid to late neonatal stages (2, 42). To date, several proteins capable of inhibiting CM proliferation have been identified, including retinoblastoma (43, 44), Meis1 (35) and Meis2 (44), components of the Hippo pathway (45), and glycogen synthase kinase (Gsk). Interestingly, Gsk, depletion of which in CMs causes mitotic catastrophe (46, 47), directly activates Tip60 via phosphorylation (48–50). Ongoing work in this laboratory designed to assess the effect of genetically depleting Tip60 from the adult heart three days after MI has revealed preservation of cardiac function four weeks later, accompanied by reduced scarring and activation of the CM cell-cycle. These results are consistent with the findings in Figures 5–6 showing that depletion of Tip60 from the neonatal heart prior to MI on P7 results in beneficial effects at P39; this also suggests that Tip60 depletion extends the window of competence to regenerate after cardiac injury (31). Because these effects were not observed three days post-MI (i.e. at P10; Supp. Fig. 8), immediate cardioprotection was not conferred by reduced Tip60 levels.
As recently reviewed (51), despite wide interest in the management cardiac dysfunction by pharmaceutically targeting deacetylase proteins, the targeting of acetyltransferases has received relatively little attention. We believe that our findings justify further investigation into the role of Tip60 in the myocardium. For example, definitive evidence that Tip60 activates the DDR to initiate CM proliferative senescence could be obtained using mice engineered to enable a conditionally-induced lysine to arginine mutation at Atm residue 3026 in CMs (13). It is also possible, and perhaps likely due to its multiple targets, that Tip60 drives proliferative senescence outside the direct pAtm→pH2A.X signaling axis. Hence, experiments to ascertain whether Tip60 acetylates other proteins that regulate CM cell-cycle activation must be considered. For example, as mentioned above, this includes assessment of whether Tip60 site-specifically acetylates p21 in CMs, since it has been reported that Tip60 acetylates p21 in colon cancer cells (52), at the same lysine residues recently shown to be required to maintain inhibitory levels of p21 in CMs (53). Moreover, it was recently reported that Tip60-mediated acetylation of the transcription factor specificity protein 1 (Sp1) prevents expression of the gene encoding Tert polymerase (Tert, (54)), down-regulation of which accompanies attrition of CM telomeres (55) and the onset of CM proliferative senescence (56) during neonatal stages of heart development. And, although we did not observe reduced levels of apoptosis in Tip60-depleted neonatal hearts three days after infarction (Supp. Fig. 8), the reduced incidence of cleaved caspase-3-positive CMs four weeks later (Fig. 6) warrants further investigation into whether depletion of Tip60, in accord with its ability to activate p53 with pro-apoptotic consequences (9, 32), prevents CM death in the injured myocardium. Finally, questions remain at the cellular level regarding the extent to which depletion of Tip60 promotes cytokinesis and bona fide CM proliferation, as indicated by results in Figure 3C. Although this is a matter of crucial importance in the heart regeneration field (1, 34, 57–59), recently reported findings showing that genetic ablation of cell-cycle activated CMs from the infarcted heart diminishes the regenerative response indicate that CM cell-cycle activation per se may be beneficial (60).
Supplementary Material
Highlights.
It is known that activation of Atm induces the DDR and proliferative senescence in neonatal cardiomyocytes (CMs).
It is unknown whether Tip60, which activates Atm→DDR signaling in cancer cells, activates this process in CMs.
Here, genetic depletion of Tip60 in neonatal CMs de-activates Atm, reduces DDR markers, and increases activation of the cell-cycle in CMs.
Also, after MI, function is preserved and scarring is reduced in Tip60-depleted hearts.
Acknowledgments
Grants
Supported by NIH 5R01HL131788 (JA & JL), S10 OD025038, and Grant FP00012308 from the Medical College of Wisconsin Cardiovascular Center
Abbreviations:
- Atm
Ataxia–telangiectasia mutated
- BrdU
5’-bromo-deoxyuridine
- CM
cardiomyocyte
- cTnT
cardiac troponin T
- DDR
DNA Damage Response
- MI
myocardial infarction
- pH2A.X
phosphorylated histone H2A.X
- pH3
phosphorylated histone H3
- Tip60
Tat-Interactive Protein U60U kD
Footnotes
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sadek H, Olson EN. Toward the Goal of Human Heart Regeneration. Cell Stem Cell. 2020;26(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galdos FX, Guo Y, Paige SL, VanDusen NJ, Wu SM, Pu WT. Cardiac Regeneration: Lessons From Development. Circ Res. 2017;120(6):941–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher JB, Kim MS, Blinka S, Ge ZD, Wan T, Duris C, et al. Stress-induced cell-cycle activation in Tip60 haploinsufficient adult cardiomyocytes. PLoS One. 2012;7(2):e31569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238(11):2912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Numata A, Kwok HS, Zhou QL, Li J, Tirado-Magallanes R, Espinosa Angarcia V, et al. Lysine acetyltransferase Tip60 is required for hematopoietic stem cell maintenance. Blood. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urban I, Kerimoglu C, Sakib MS, Wang H, Benito E, Thaller C, et al. TIP60/KAT5 is required for neuronal viability in hippocampal CA1. Sci Rep. 2019;9(1):16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher JB, Horst A, Wan T, Kim MS, Auchampach J, Lough J. Depletion of Tip60 from In Vivo Cardiomyocytes Increases Myocyte Density, Followed by Cardiac Dysfunction, Myocyte Fallout and Lethality. PLoS One. 2016;11(10):e0164855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16(9):433–42. [DOI] [PubMed] [Google Scholar]
- 9.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9(5):930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11(11):1376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27(24):8502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157(3):565–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541(7636):222–7. [DOI] [PubMed] [Google Scholar]
- 16.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89(1):20–5. [DOI] [PubMed] [Google Scholar]
- 17.Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet. 2017;49(9):1346–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan P, Patterson M, Velasquez A, Wang K, Tian D, Windle JJ, et al. Tnni3k alleles influence ventricular mononuclear diploid cardiomyocyte frequency. PLoS Genet. 2019;15(10):e1008354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Zglinicki T, Saretzki G, Ladhoff J, d’Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126(1):111–7. [DOI] [PubMed] [Google Scholar]
- 20.Sulli G, Di Micco R, d’Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer. 2012;12(10):709–20. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26(56):7741–8. [DOI] [PubMed] [Google Scholar]
- 22.Nakada Y, Nhi Nguyen NU, Xiao F, Savla JJ, Lam NT, Abdisalaam S, et al. DNA Damage Response Mediates Pressure Overload-Induced Cardiomyocyte Hypertrophy. Circulation. 2019;139(9):1237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higo T, Naito AT, Sumida T, Shibamoto M, Okada K, Nomura S, et al. DNA single-strand break-induced DNA damage response causes heart failure. Nature communications. 2017;8:15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10(2):261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271(5 Pt 2):H2183–9. [DOI] [PubMed] [Google Scholar]
- 27.Wei Y, Peng S, Wu M, Sachidanandam R, Tu Z, Zhang S, et al. Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 2014;24(3):278–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox EJ, Marsh SA. A systematic review of fetal genes as biomarkers of cardiac hypertrophy in rodent models of diabetes. PLoS One. 2014;9(3):e92903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Lauth A, Wan TC, Lough JW, Auchampach JA. Myh6-driven Cre-recombinase activates the DNA damage response and the cell-cycle in the myocardium in the absence of loxP sites. Dis Model Mech. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monroe TO, Hill MC, Morikawa Y, Leach JP, Heallen T, Cao S, et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev Cell. 2019;48(6):765–79 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24(6):827–39. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102(37):13182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan P, Patterson M, Sucov HM. Cardiomyocyte Polyploidy and Implications for Heart Regeneration. Annu Rev Physiol. 2020;82:45–61. [DOI] [PubMed] [Google Scholar]
- 35.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497(7448):249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tane S, Ikenishi A, Okayama H, Iwamoto N, Nakayama KI, Takeuchi T. CDK inhibitors, p21(Cip1) and p27(Kip1), participate in cell cycle exit of mammalian cardiomyocytes. Biochem Biophys Res Commun. 2014;443(3):1105–9. [DOI] [PubMed] [Google Scholar]
- 37.Tane S, Kubota M, Okayama H, Ikenishi A, Yoshitome S, Iwamoto N, et al. Repression of cyclin D1 expression is necessary for the maintenance of cell cycle exit in adult mammalian cardiomyocytes. J Biol Chem. 2014;289(26):18033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volland C, Schott P, Didie M, Manner J, Unsold B, Toischer K, et al. Control of p21Cip by BRCA1-associated protein is critical for cardiomyocyte cell cycle progression and survival. Cardiovasc Res. 2020;116(3):592–604. [DOI] [PubMed] [Google Scholar]
- 39.Medley TL, Furtado M, Lam NT, Idrizi R, Williams D, Verma PJ, et al. Effect of oxygen on cardiac differentiation in mouse iPS cells: role of hypoxia inducible factor-1 and Wnt/beta-catenin signaling. PLoS One. 2013;8(11):e80280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guimaraes-Camboa N, Stowe J, Aneas I, Sakabe N, Cattaneo P, Henderson L, et al. HIF1alpha Represses Cell Stress Pathways to Allow Proliferation of Hypoxic Fetal Cardiomyocytes. Dev Cell. 2015;33(5):507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda R, Marin-Juez R, El-Sammak H, Beisaw A, Ramadass R, Kuenne C, et al. Stimulation of glycolysis promotes cardiomyocyte proliferation after injury in adult zebrafish. EMBO Rep. 2020:e49752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Berlo JH, Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med. 2014;20(12):1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sdek P, Zhao P, Wang Y, Huang CJ, Ko CY, Butler PC, et al. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J Cell Biol. 2011;194(3):407–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alam P, Haile B, Arif M, Pandey R, Rokvic M, Nieman M, et al. Inhibition of Senescence-Associated Genes Rb1 and Meis2 in Adult Cardiomyocytes Results in Cell Cycle Reentry and Cardiac Repair Post-Myocardial Infarction. J Am Heart Assoc. 2019;8(15):e012089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Ahmad F, Parikh S, Hoffman NE, Rajan S, Verma VK, et al. Loss of Adult Cardiac Myocyte GSK-3 Leads to Mitotic Catastrophe Resulting in Fatal Dilated Cardiomyopathy. Circ Res. 2016;118(8):1208–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerkela R, Kockeritz L, Macaulay K, Zhou J, Doble BW, Beahm C, et al. Deletion of GSK-3beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J Clin Invest. 2008;118(11):3609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, et al. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol Cell. 2011;42(5):584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336(6080):477–81. [DOI] [PubMed] [Google Scholar]
- 50.Cheng X, Ma X, Zhu Q, Song D, Ding X, Li L, et al. Pacer Is a Mediator of mTORC1 and GSK3-TIP60 Signaling in Regulation of Autophagosome Maturation and Lipid Metabolism. Mol Cell. 2019;73(4):788–802 e7. [DOI] [PubMed] [Google Scholar]
- 51.Li P, Ge J, Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nature reviews Cardiology. 2020;17(2):96–115. [DOI] [PubMed] [Google Scholar]
- 52.Lee MS, Seo J, Choi DY, Lee EW, Ko A, Ha NC, et al. Stabilization of p21 (Cip1/WAF1) following Tip60-dependent acetylation is required for p21-mediated DNA damage response. Cell death and differentiation. 2013;20(4):620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Li M, Li X, Li H, Lai Y, Huang S, et al. Sirt1-inducible deacetylation of p21 promotes cardiomyocyte proliferation. Aging (Albany NY). 2019;11(24):12546–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajagopalan D, Pandey AK, Xiuzhen MC, Lee KK, Hora S, Zhang Y, et al. TIP60 represses telomerase expression by inhibiting Sp1 binding to the TERT promoter. PLoS pathogens. 2017;13(10):e1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aix E, Gutierrez-Gutierrez O, Sanchez-Ferrer C, Aguado T, Flores I. Postnatal telomere dysfunction induces cardiomyocyte cell-cycle arrest through p21 activation. J Cell Biol. 2016;213(5):571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh H, Taffet GE, Youker KA, Entman ML, Overbeek PA, Michael LH, et al. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc Natl Acad Sci U S A. 2001;98(18):10308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vujic A, Natarajan N, Lee RT. Molecular mechanisms of heart regeneration. Semin Cell Dev Biol. 2020;100:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derks W, Bergmann O. Polyploidy in Cardiomyocytes: Roadblock to Heart Regeneration? Circ Res. 2020;126(4):552–65. [DOI] [PubMed] [Google Scholar]
- 59.Patterson M, Swift SK. Residual Diploidy in Polyploid Tissues: A Cellular State with Enhanced Proliferative Capacity for Tissue Regeneration? Stem Cells Dev. 2019;28(23):1527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradley LA, Young A, Li H, Billcheck HO, Wolf MJ. Loss of Endogenously Cycling Adult Cardiomyocytes Worsens Myocardial Function. Circ Res. 2021;128(2):155–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.