SUMMARY
The macroevolutionary transition from terra firma to obligatory inhabitance of the marine hydrosphere has occurred twice in the history of Mammalia: Cetacea and Sirenia. In the case of Cetacea (whales, dolphins, porpoises), molecular phylogenies provide unambiguous evidence that fully aquatic cetaceans and semiaquatic hippopotamids (hippos) are each other’s closest living relatives. Ancestral reconstructions suggest that some adaptations to the aquatic realm evolved in the common ancestor of Cetancodonta (Cetacea+Hippopotamidae). An alternative hypothesis is that these adaptations evolved independently in cetaceans and hippos. Here, we focus on the integumentary system and evaluate these hypotheses by integrating new histological data for cetaceans and hippos, the first genome-scale data for pygmy hippopotamus, and comprehensive genomic screens and molecular evolutionary analyses for protein-coding genes that have been inactivated in hippos and cetaceans. We identified eight skin-related genes that are inactivated in both cetaceans and hippos, including genes that are related to sebaceous glands, hair follicles, and epidermal differentiation. However, none of these genes exhibit inactivating mutations that are shared by cetaceans and hippos. Mean dates for the inactivation of skin genes in these two clades serve as proxies for phenotypic changes and suggest that hair reduction/loss, the loss of sebaceous glands, and changes to the keratinization program occurred ~16 million years earlier in cetaceans (~46.5 Ma) than in hippos (~30.5 Ma). These results, together with histological differences in the integument and prior analyses of oxygen isotopes from stem hippopotamids (“anthracotheres”), support the hypothesis that aquatic skin adaptations evolved independently in hippos and cetaceans.
Keywords: Cetacea, epidermis, gene loss, Hippopotamidae, histology, skin
Graphical Abstract
INTRODUCTION
The evolutionary history of Mammalia is mostly one of deployment and adaptation to different terrestrial habitats. Nevertheless, numerous mammalian clades have returned to aquatic habitats, either on a part-time basis or with full-time commitment to the aquatic realm. Fully aquatic clades include Cetacea and Sirenia, and in both cases there are extinct taxa that document the macroevolutionary transition from land to water [1,2]. Semiaquatic clades are more numerous and examples are found in a wide range of mammalian orders, e.g., Monotremata (platypuses), Rodentia (beavers, capybaras), Carnivora (pinnipeds, otters), and Cetartiodactyla (hippos). Adaptations to aquatic habitats are most extreme in fully aquatic forms where virtually every organ system has been highly modified. Locomotory adaptations in cetaceans include hind limb loss, modification of the front limbs into flippers, and conversion of the tail into a powerful fluke [1]; sensory system modifications include highly modified eyes with reduction or loss of color vision [3], olfactory structures that are highly reduced or absent [4,5], and ultrasonic hearing in odontocetes (toothed whales) [6].
Cetaceans also show changes to their integument. The skin of fully aquatic mammals constantly interacts with dense, viscous, and thermally conductive water, which poses unique physical challenges to its outer surface. The cetacean epidermis is exceptionally thick and undergoes constant cellular renewal [7–13]. Even with its increased thickness there are only three distinct histological layers – a basal layer (stratum basale), an intermediate layer (stratum spinosum), and an outer layer (stratum corneum) [14]. The stratum granulosum, which in land mammals lies beneath the stratum corneum, is ill-defined or absent [10,15]. The thick epidermis provides mechanical and thermal protection, and its fast rate of sloughing and extensive epidermal-dermal interdigitations guard against potential damage from locomotory shear stress [16]. The stratum basale forms deep root-like projections (= rete ridges) that extend into the underlying dermis [17–19]. Extensive cytoplasmic lipid vacuoles are present in the stratum spinosum and stratum corneum keratinocytes [20–22], and may play metabolic and/or thermo-insulating roles [22,23]. Unlike terrestrial mammals, cells of the stratum corneum retain nuclei and do not become fully keratinized in cetaceans [14], possibly because fully aquatic mammals do not need a functional epidermal barrier to a dry environment [24]. The cetacean dermis is thickened and consists of an upper papillary layer that interdigitates with the epidermis and a lower reticular layer that gradually transitions into the underlying blubber [10,11,18,25,26].
Cetaceans have reduced their ectodermal appendages including hair follicles and skin glands [27]. They show no evidence of pelage (i.e., fur) hair follicle formation during embryonic development although vibrissa follicles (i.e., whiskers) form on the head [28–32]. Mysticetes and some odontocetes cyclically grow whiskers as adults, but most adult odontocetes lose their whiskers and convert vibrissa follicles into degenerated pits that may perform sensory functions [30,33,34]. Cetaceans lack oil-secreting sebaceous glands [31,32,35] and sweat glands [1,14,27].
Among semiaquatic forms, hippos are the largest herbivores [36]. Both extant species (Hippopotamus amphibius [river hippo], Choeropsis liberiensis [pygmy hippo]) spend their days in the water and emerge at dusk to feed on grasses and other vegetation. H. amphibius has a thick epidermis and its stratum basale forms distinct projections into the papillary dermis [15]. H. amphibius shows elevated levels of epidermal lipid storage, although unlike cetaceans the most prominent lipid deposits occur in intercellular locations within the stratum corneum [23]. Hippos have bristle-like whiskers on their muzzle, and pelage hairs that are sparsely distributed across most of the body [15]. Sebaceous glands have not been reported, but previous histological studies have only examined limited regions of the body [15,37–39]. Hippo skin contains anatomically complex sweat glands [37,38] that secrete a red-orange pigmented sweat that may have sunscreen and/or antimicrobial properties [40,41].
The traditional view based on morphology is that cetaceans are excluded from a monophyletic Artiodactyla (even-toed hoofed mammals) [42,43] and that hippopotamids are the sister taxon to pigs (Suidae) and/or peccaries (Tayassuidae) [44–48]. However, molecular studies challenged this view and eventually provided conclusive evidence that cetaceans are nested within Artiodactyla [49,50] as the sister to Hippopotamidae [51–54].
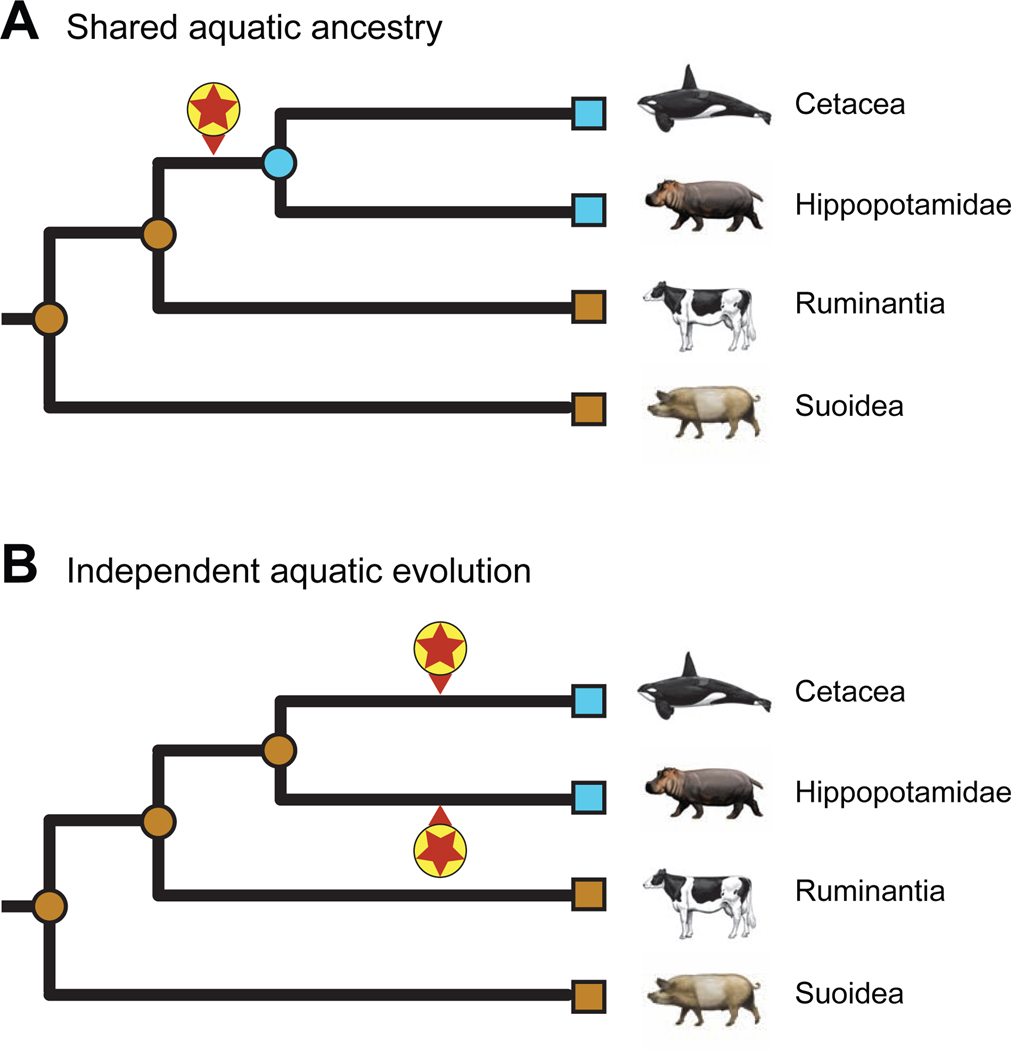
Given that cetaceans and hippopotamids share a variety of morphological and behavioral characters that may be related to aquatic habitats (hairless or nearly hairless body, lack of sebaceous glands, lack of scrotal testes, underwater parturition and nursing, underwater detection of sound directionality), the most parsimonious hypothesis is that these characters evolved in the common ancestor of these two clades (= Cetancodonta) and that this common ancestor was semiaquatic [51,52,55,56] (Figure 1A). O’Leary and Gatesy [57] favored the common origin hypothesis based on ancestral reconstructions of underwater hearing and other aquatic characters. Gatesy et al. [1] favored the aquatic ancestry hypothesis, specifically in freshwater, based on oxygen isotope values in pakicetid cetaceans and the presence of dense, osteosclerotic bones in both hippos and basal stem cetaceans (raoellids, pakicetids). Alternatively, semiaquatic adaptations evolved convergently in hippos and cetaceans [36] (Figure 1B). In part, support for these two competing hypotheses turns on the phylogenetic placement of various ‘Anthracotheriidae’, which collectively are the paraphyletic stem group to Hippopotamidae. Some anthracotheres in the subfamilies Anthrocotheriinae (e.g., Anthracotherium) and Microbunodontinae (e.g., Microbunodon) are inferred to have been terrestrial based on oxygen isotope values, but members of Bothriodontinae (e.g., Bothriogenys, Merycopotamus) have values that are consistent with a semiaquatic lifestyle [58–62]. Cooper et al. [62] performed ancestral reconstructions on bone microanatomy for a data set that included both anthracotheres and stem cetaceans (“archaeocetes”) and concluded that the most recent common ancestor of Cetancodonta was probably semiaquatic. Soe et al. [63] reported skeletal material for the Eocene anthracotheriid Siamotherium, which they recovered as the most basal genus of Anthracotheriidae in their phylogenetic analysis, and concluded there is no evidence of clear-cut aquatic adaptations in Siamotherium.
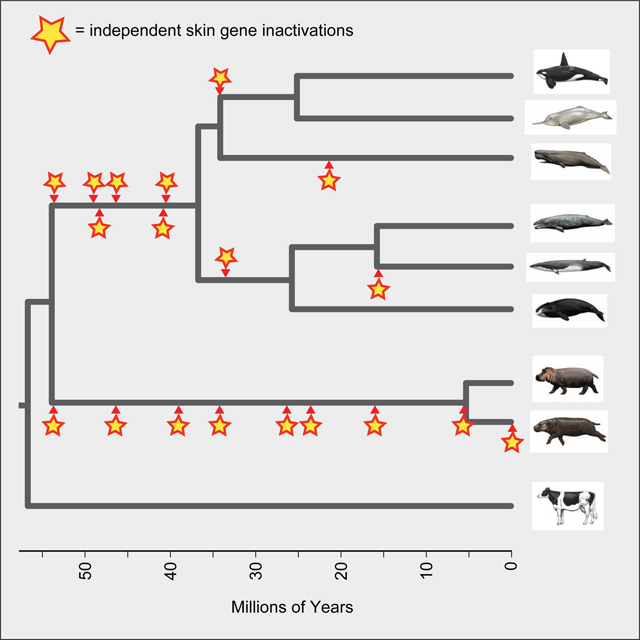
Figure 1. Two hypotheses for the evolution of aquatic adaptations.
(A) Evolution of shared aquatic features in the common ancestor of Hippopotamidae and Cetacea, and (B) independent evolution of aquatic features on the cetacean stem lineage and also on the hippopotamid stem lineage. Encircled red stars mark the initial evolution of behavioral, physiological, and anatomical characteristics associated with adaptation to aquatic environments. Aquatic (blue) and terrestrial (brown) specializations of extant taxa (squares) and ancestral nodes (circles) are indicated in each scenario.
At the molecular level, positive selection analyses provide minimal support for shared aquatic adaptations in the ancestry of Cetancodonta [64]. However, branch-site selection analyses can have weak statistical power. A more promising approach may be to search for skin genes that have been inactivated in the common ancestor of cetaceans and hippos. Previous authors have employed this approach to identify skin genes that were inactivated in the common ancestor of cetaceans [16,24,65–68]. However, only two candidate gene studies [69–70] have searched for skin genes that are inactivated in cetaceans and hippos. It remains to be determined if a comprehensive genomic screen [sensu 67] will reveal additional skin genes that have been inactivated in both cetaceans and hippos.
Here, we present morphological and molecular evidence to evaluate competing hypotheses that aquatic skin adaptations evolved in the common ancestor of cetaceans and hippos versus independently in these two groups (Figure 1). First, we provide histological data for different regions of the integument in two cetacean species and both extant hippos. Next, we perform a comprehensive genomic screen on representative cetaceans and hippopotamids for protein-coding genes that have been inactivated in both of these clades and assess the timing of pseudogenization for these genes. Finally, we discuss our results in the context of the rich fossil record for stem cetaceans (“archaeocetes”) and stem hippos (“anthracotheres”).
RESULTS AND DISCUSSION
Comparative Skin Histology Between Terrestrial and Aquatic Cetartiodactyla
We analyzed skin samples from the face, eyelid, ear, dorsum, ventrum, and tail in both hippopotamid species, and from the facial region of one odontocete (Tursiops truncatus [bottlenose dolphin]) and one mysticete (Eschrichtius robustus [gray whale]). Table 1 summarizes features of the skin in these taxa and also includes data from the literature for terrestrial mammals including humans and two cetartiodactyls (cow, pig) that are close relatives of Cetancodonta [71–79].
Table 1. Anatomical features of the skin.
Anatomical features of the skin in human, cow, pig, hippo, and two cetaceans.
| Cellular/anatomical feature | Human | Pig | Cow | Hippo | Grey whale (facial skin only) | Common bottlenose dolphin (facial skin only) |
|---|---|---|---|---|---|---|
| Epidermal thickness | 29.6 to 96.5 μma | 45.8 to 91.7 μmb | 34.9 to 55.6 μmc | River hippo: 216 to 1,134 μmd (adult trunk); 42 +/− 7 μm (neonatal, facial skin) Pygmy hippo: 44 +/− 12 μm (neonatal, facial skin |
6,334 +/− 872 μm | 945 +/− 267 μm |
| Epidermal rete ridges | Medium-size rete ridges that differ in size across body | Medium-size rete ridges that differ in size across body | Absence of definitive rete ridges | Shallow rete ridges that differ in size across body | Prominent, branched rete ridges | Prominent, branched rete ridges |
| Cytoplasmic lipid vacuoles in epidermal living layers | Absent | N/A | N/A | Extensive cytoplasmic lipid vacuoles in all epidermal layers | Extensive cytoplasmic lipid vacuoles in all epidermal layers | Extensive cytoplasmic lipid vacuoles in all suprabasal epidermal layers |
| Lipid distribution in stratum corneum | Inter-cellular | N/A | N/A | Mostly intercellular | Mostly intracellular | Mostly intracellular |
| Stratum basale | Present | Present | Present | Present | Present | Present |
| Stratum spinosum | Present, thin | Present, medium | Present | Present, medium | Present, very thick | Present, very thick |
| Stratum granulosum | Present | Present | Present | Poorly defined | Absent | Absent |
| Stratum corneum | Present, variable thickness | Present, variable thickness | Present, thin | Present, medium | Present, very thick | Present, very thick |
| Epidermal barrier | Intact | Intact | Intact | Intact | Disrupted | Disrupted |
| Pelage hair follicles | Present, variable hair density across the body | Present, variable hair density across the body | Present, high hair density | Present, variable hair density; sparse over most of the body | Absent | Absent |
| Vibrissa hair follicles | Absent | Present | Present | Present | Present, limited to cranial skin | Present, limited to cranial skin |
| Ability of hair follicles to cycle | Able to cycle | Able to cycle | Able to cycle | Able to cycle | Able to cycle | Hair follicles cycle only once in neonates and degenerate in adults to become pits |
| Hair follicle-associated sebaceous gland | Present, variable in size and often prominent | Present, rudimentary | Present | Absent | Absent | Absent |
| Meibomian glands | Present | Present | Present | Absent | Absent | Absent |
| Sweat glands | Apocrine and eccrine glands | Apocrine glands only | Apocrine glands only | Eccrine glandsd; possibly apocrine glands | Absent | Absent |
| Dermal adipose tissue | Prominent | Prominent | Poorly defined | Absent | Abundant adipocyte clusters throughout the dermis | Abundant adipocyte clusters throughout the dermis |
Cetaceans and hippos have prominent differences in the thickness and organization of the epidermis (Figure 2). Consistent with previous reports [17–19], the facial epidermis in adult Eschrichtius robustus (Figure 2E) and neonatal Tursiops truncatus (Figure 2F-G) is very thick, with a wide stratum spinosum and an undulated stratum basale with deep root-like rete ridges. By contrast, the epidermis in neonatal pygmy hippo is thin, with only shallow rete ridges (Figure 2A-D).
Figure 2. Histological features of the skin in hippos and cetaceans.
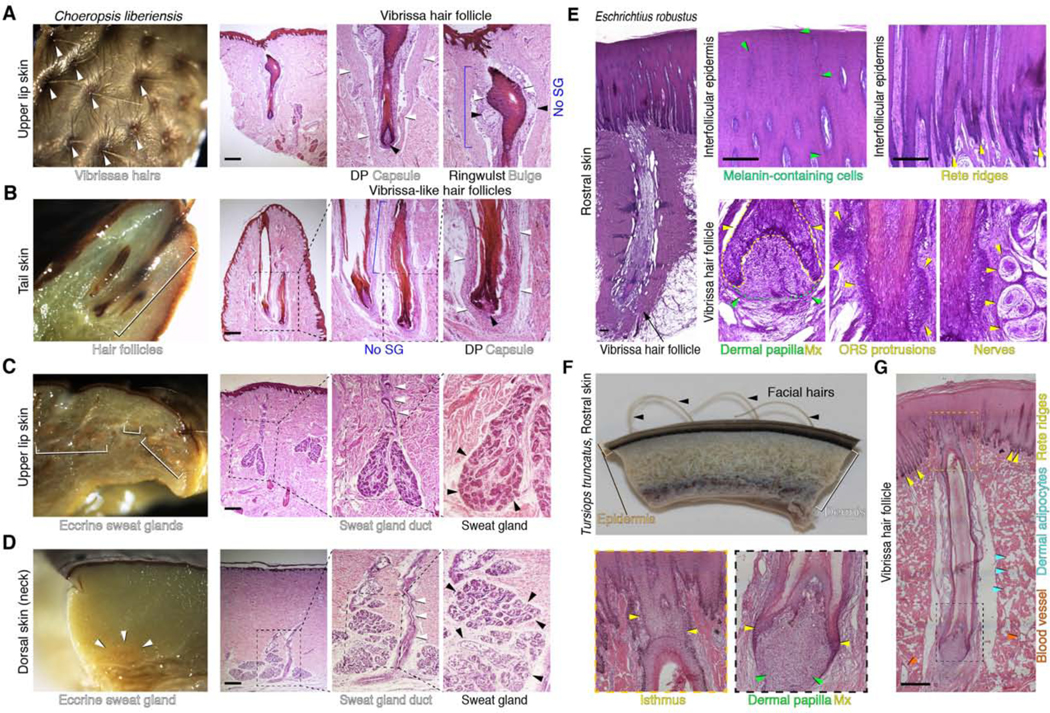
Hippo skin (A-D) and cetacean skin (E-G). (A) Whole mount (left) and histological appearance (middle, right) of the upper lip skin in pigmy hippo. In the lip, vibrissa hairs above the skin surface and anagen (active growth) phase vibrissa hair follicles are prominent. Each lip vibrissa follicle has a prominent mesenchymal dermal papilla (black arrowhead, center), a collagen capsule (white arrowheads, center), an epithelial matrix, a mesenchymal ringwulst (black arrowheads, right), and an epithelial bulge (white arrowheads, right). There is no histological evidence of sebaceous glands. (B) Whole mount of tail skin from pygmy hippo (left) shows large hair follicles (white bracket). Histological analysis (right) suggests that tail hair follicles might be of vibrissa type because they are surrounded by a collagen capsule (white arrowheads). There is no histological evidence of sebaceous glands. (C, D) Whole mount and corresponding histological view of upper lip skin (C) and dorsal skin (D) in the pigmy hippo. At both sites, eccrine sweat glands are present. On histology, secretory coils located deep in the dermis are marked with black arrowheads; associated excretory ducts (where obvious) are marked with white arrowheads. In the dorsal skin (D), secretory coils of the glands reside at the very base of the dermis and come in contact with the underlying skeletal muscle layer. There is no histological evidence of dermal adipocytes. (E) Histology of facial skin and a rostral vibrissa hair follicle in an adult gray whale. The epidermis is thick and its basal layer is heavily undulated. The vibrissa follicle has a typical anagen (active growth) phase morphology with a large mesenchymal dermal papilla (green arrowheads, second panel) surrounded by epithelial matrix (yellow arrowheads, second panel). The epithelial outer root sheath compartment located above hair matrix is uncharacteristically thick and has prominent protrusions (yellow arrowheads, third panel). The vibrissa follicle is associated with distinct nerve bundles on either side (yellow arrowheads, fourth panel). (F) Wholemount side view of rostral skin from neonatal common bottlenose dolphin. Vibrissae hairs (black arrowheads) are clearly visible above the skin surface. (G) Histological view of rostral vibrissa hair follicle from neonatal common bottlenose dolphin. The vibrissa follicle has anagen phase morphology. A large dermal papilla (green arrowheads, bottom middle panel) and an uncharacteristically thin epithelial matrix (yellow arrowheads, bottom middle panel) are obvious. Unlike in gray whale, the outer root sheath lacks undulations, and like in gray whale, the vibrissa follicle lacks sebaceous gland. The overlaying epidermis displays prominent rete ridges (yellow arrowheads, right panel). The surrounding dermis contains numerous adipocyte clusters (blue arrowheads, right panel). Scale bar: A-E, G– 50 μm. See also Figure S1 and Figure S2.
Hippo skin contains hair follicles of both pelage and vibrissa morphology. Prominent vibrissa follicles that contain collagen capsules and ringwulst (ring-like dermal structure that surrounds the follicle) occur in the upper lip skin (Figure 2A; Figure S1C). Some tail-tip hair follicles display collagen capsules (Figure 2B; Figure S1E), suggesting that the tail might contain both pelage and vibrissa hair types. Hair follicles in ear and eyelid skin have typical pelage morphology, and meibomian glands are absent (Figure 2D; Figure S1A-B, S2C). All hippo hair follicles lack sebaceous glands (Figure 2; Figure S1). The facial skin of both cetaceans has prominent vibrissa hair follicles (Figure 2E-G), but their structure differs from hippo vibrissae in lacking collagen capsules and ringwulst. In both cetaceans, mesenchymal dermal papillae and the epithelial hair matrix, two defining structures of actively growing hair follicles, are present. However, the hair matrix in dolphin vibrissae is uncharacteristically thin. There is no evidence of vibrissa-associated sebaceous glands in either cetacean species.
Prominent sweat glands occur in hippo skin in several locations, including the upper lip, dorsum, and ventrum (Figure 2C-D; Figure S2). The structure of the dermis is different in cetaceans and hippos. There are abundant clusters of adipocytes throughout the dermis in dolphin (Figure 2G), but we found no evidence for mature adipocytes in hippos. Hippos exhibit prominent differences in dermal thickness across body sites, with tail and ear dermis being the thinnest.
In conclusion, cetacean skin has a distinctly thick and highly undulated epidermis, a thick and adipocyte-rich dermis, and morphologically-specialized facial vibrissa hair follicles as the only skin appendage. Hippo skin is characterized by a much thinner epidermis, shallow rete ridges, a dermis of variable thickness without adipocytes, highly specialized sweat glands, and both pelage and vibrissa hair follicles. Cetaceans and hippos both lack sebaceous glands (Table 1). Our histological analyses for each species are limited to just one time point. Additional histological features, not captured in our studies, are expected for both hippos and cetaceans. For example, the epidermis undergoes annual molting in some cetaceans including belugas [80] and the Okhotsk population of bowheads [81]. During the spring molt the bowhead epidermis is at its thinnest and contains a highly vacuolated stratum spinosum that is not present in the fall [81,82].
Genomic Screens and Patterns of Gene Inactivation in Cetaceans and Hippos
We evaluated the shared versus independent aquatic ancestry hypotheses using existing and newly-generated genomic data. Changes in both protein-coding sequences and cis-regulatory elements could contribute to the evolution of skin phenotypes, but we focused on the former because the timing of gene inactivation can be estimated from protein-coding sequences [83]. We screened genomic alignments with 63 mammalian taxa for protein-coding genes that are inactivated in Hippopotamidae and Cetacea but not in terrestrial cetartiodactyls. Our screen included Hippopotamus amphibius, one baleen whale (Balaenoptera acutorostrata [common minke whale]), and three toothed whales (Physeter macrocephalus [sperm whale], Orcinus orca [killer whale], Tursiops truncatus). We identified 38 genes that have inactivating mutations (frameshift indels, premature stop codons, splice site mutations, deleted exons) or are completely deleted in these taxa (Table S1) including ten genes that have primary or sole functions related to skin and its ectodermal appendages (ALOX15, AWAT1, KPRP, KRT2, KRT26, KRT77, KRTAP6–2, KRTAP6–3, KRTAP7–1, TCHHL1). Two genes (KRTAP6–2, KRTAP6–3) were excluded because of ambiguous orthology relationships, leaving eight genes for a detailed analysis. ABCC11, which our screen found to be inactivated only in Cetacea, was added to our list because ABCC11 is a candidate gene of interest that is expressed in axillary sweat glands in humans [84,85].
Base errors in genome assemblies can mimic gene-inactivating mutations [86], so we used additional genomic data to confirm the validity of inactivating mutations and thus the loss of gene function. Our investigation of ten additional cetacean genomes revealed that all nine genes have mutations shared between at least two species (as exemplified in Figure 3A-B), which makes base errors highly unlikely. Three of these genes (AWAT1, KRTAP7–1, ABCC11) exhibit inactivating mutations shared between odontocetes and mysticetes, indicating gene loss on the stem cetacean branch (Figure 3C). Three genes exhibit large or entire gene deletions in odontocetes (KRT2, KRT26) or mysticetes (KRT77), and gene loss dating (below) indicates that KRT26 and KRT77 losses happened on the stem cetacean branch. The distribution of mutations in ALOX15 suggests that it was independently inactivated on three different cetacean branches (Figure 3C). Finally, two genes are absent from all cetacean (KPRP and TCHHL1) genomes. However, large deletions and rearrangements in these loci obscure reconstructions of when these genes were lost because there are no obvious breakpoints that are shared by odontocetes and mysticetes.
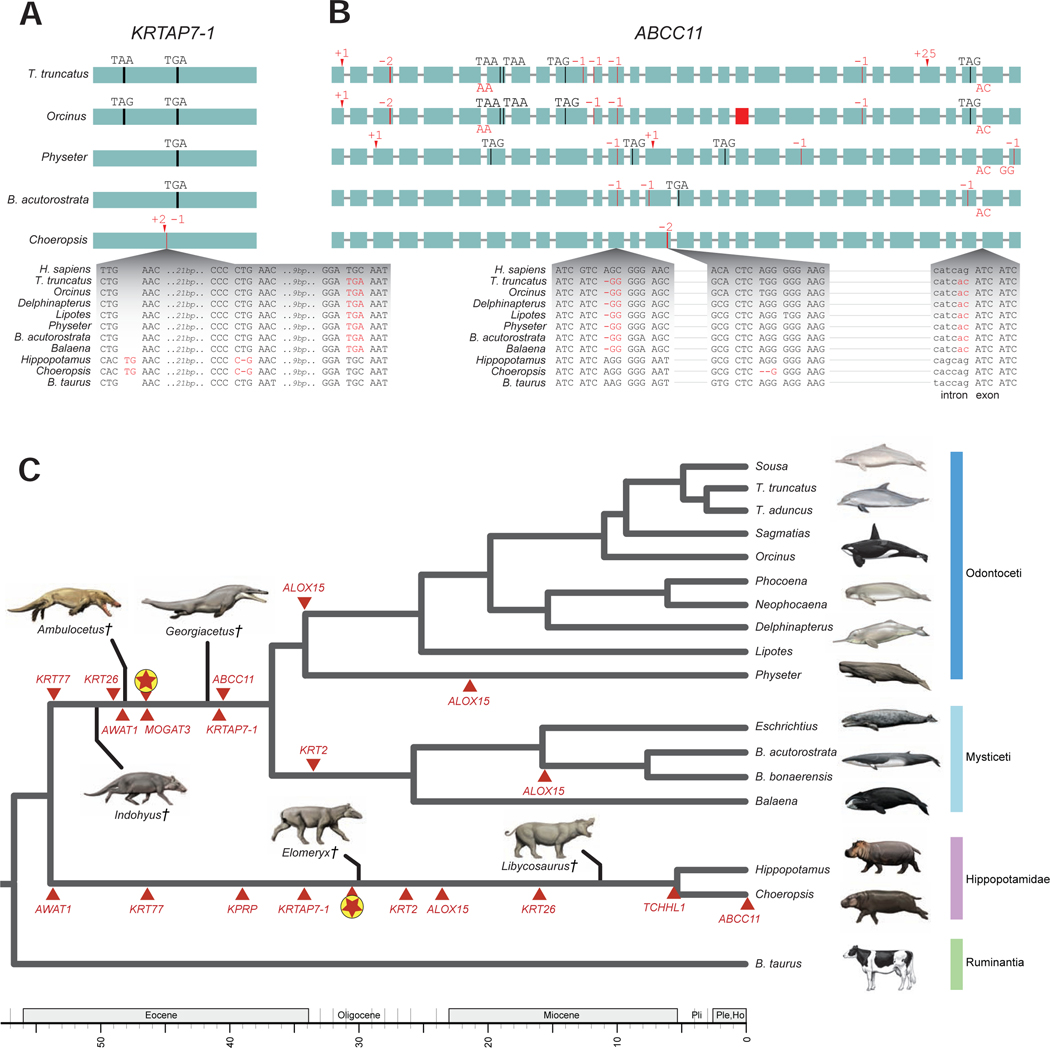
Figure 3. Inactivating mutations in cetaceans and hippos.
Inactivating mutations in KRTAP7–1 (A) and ABCC11 (B). Genes are shown with exons represented by green rectangles proportional to their size and introns represented by horizontal lines. Inactivating mutations are premature stop codons (vertical black line and corresponding triplet), insertions (red arrowhead and corresponding insertion size), deletions (vertical red line and corresponding deletion size, or red rectangle for completely deleted exon), and splice site donor or acceptor mutations (red letters at the end or beginning of an exon, respectively). Insets show the DNA sequence context of representative mutations. Alignment files for inactivating mutations in all genes are available at https://doi.org/10.6084/m9.figshare.13549070.v1. (C) The phylogenetic pattern of skin gene inactivations mapped onto a timetree for Cetancodonta (Hippopotamidae + Cetacea). Independent inactivations of skin genes (red triangles) are marked on branches of the tree; inactivation times are average estimates for each locus based on dN/dS analyses. The mean dates of skin gene knockouts for six loci that were inactivated in the common ancestor of Cetacea and for eight loci that were inactivated in the common ancestor of Hippopotamidae are indicated on the stem lineages to these clades (encircled red stars). The timetree for extant lineages is based on the molecular clock analysis of McGowen et al. [129]. Extinct lineages for stem cetaceans (Indohyus, Ambulocetus, Georgiacetus) and stem hippopotamids (Elomeryx, Libycosaurus) are approximately positioned relative to the geological time scale based on earliest occurrence in the fossil record for each genus and phylogenetic hypotheses based on morphological characters. See also Table S2 and Table S3.
Genomic data for Choeropsis liberiensis are required to investigate whether mutations are shared between both hippopotamid species. Therefore, we generated ~40X coverage of Illumina whole-genome shotgun data for C. liberiensis and mapped these reads to the Hippopotamus amphibius assembly to obtain orthologous sequences. Eight of nine genes (ALOX15, AWAT1, KPRP, KRT2, KRT26, KRT77, KRTAP7–1, TCHHL1) exhibit shared inactivating mutations in both hippopotamid species (Figure 3). ABCC11 is intact in H. amphibius, but a two-bp frameshift deletion in exon 14 of C. liberiensis suggests a recent loss in pygmy hippo (Figure 3B).
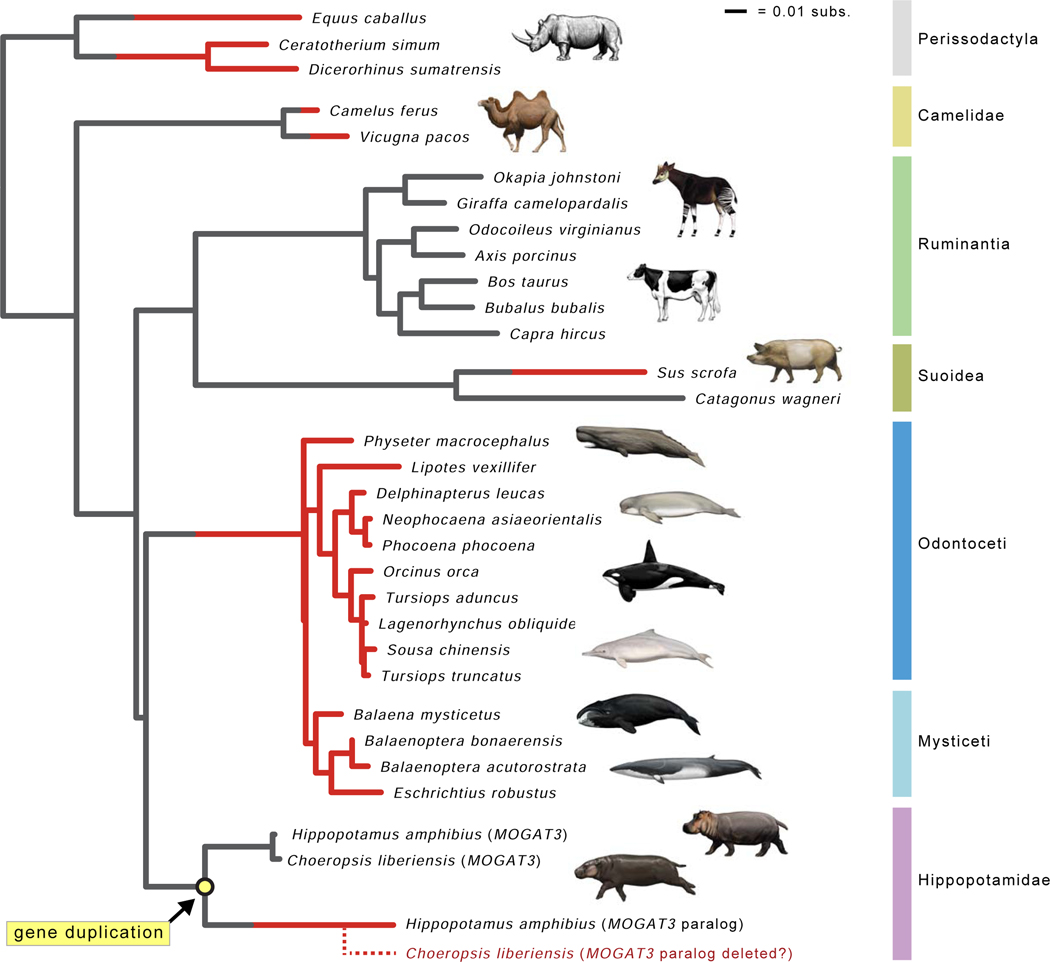
Next we analyzed whether inactivating mutations in any of these nine skin-related genes are shared by cetaceans and hippos, which would indicate gene inactivation in the common ancestor of Cetancodonta. We did not find any shared inactivating mutations. Instead, all nine genes have different inactivating mutations that indicate independent gene loss in cetaceans and hippos (Figure 3, Table S2). Lopes-Marques et al. [70] reported the convergent loss of both AWAT1 and MOGAT3 in Cetacea and Hippopotamus amphibius. Our results confirm the independent inactivation of AWAT1 and further suggest that AWAT1 was inactivated in the common ancestor of H. amphibius and Choeropsis liberiensis. However, our investigation suggests that MOGAT3 underwent a tandem duplication, most likely in the common ancestor of the two extant hippopotamids (Figure 4), and whereas one copy is pseudogenized, a functional copy of MOGAT3 is present ~15.7 kb upstream (NCBI contig PVJP02910399).
Figure 4. Gene tree for MOGAT3.
The gene tree for MOGAT3 shows inferred gene duplication event in Hippopotamidae (yellow circle) and parallel gene inactivations (red lineages). Seven gene knockouts are inferred, including pseudogenization of the MOGAT3 paralog that was derived from a duplication event on the stem hippopotamid branch. Lineages with functional MOGAT3 are gray, and lineages with inactivated MOGAT3 are colored red. Branches where inactivation events (frameshift indels, premature stop codons) were inferred by parsimony optimization of indels are gray and red, indicating the transition from functional to non-functional. The dashed red lineage for Choeropsis liberiensis represents hypothesized deletion of the MOGAT3 paralog in the genome of this species. Branch lengths are in expected numbers of substitutions per site.
Comparison of Inactivated Skin Genes and Epidermal Phenotypes in Cetaceans and Hippos
The cetacean epidermis is very thick and renews rapidly, yet does not fully differentiate into the highly keratinized stratum corneum of terrestrial mammals that confers barrier functions. These modifications are associated with loss of function of numerous genes that belong to the Epidermal Differentiation Complex (EDC) including S100 fused-type protein genes (CRNN, FLG, FLG2, HRNR, RPTN, TCHH, TCHHL1, TCHHL2) [27,65] and suprabasal epidermal keratin genes (KRT1, KRT2, KRT9, KRT10, KRT77, KRT23, KRT24) [16]. Our analyses confirm these findings (Figure 5).
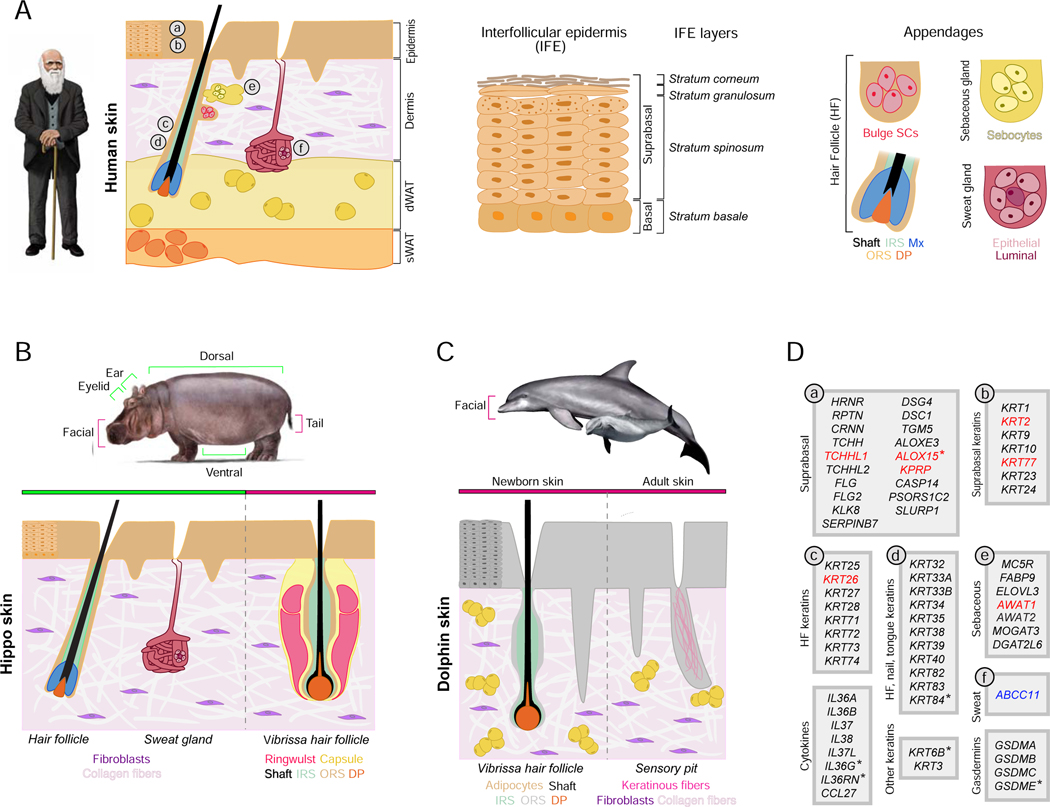
Figure 5. Skin structures and inactivated skin genes.
A comparison of skin structures in human, hippo, and dolphin with skin-associated gene inactivations in hippopotamids and cetaceans. A) Schematic drawing of Homo sapiens skin. Key anatomical structures, including multi-layered epidermis, dermis, dermal white adipose tissue (dWAT), subcutaneous white adipose tissue (sWAT), and ectodermal appendages (hair follicles with sebaceous glands and sweat glands) are shown and color-coded. The epidermis (middle panel) is divided into stratum basale – which houses stem cells, and suprabasal layers of differentiating cells that include stratum spinosum, stratum granulosum, and stratum corneum. dWAT is closely associated with growing hair follicles and secretory coils of sweat glands. Additional abbreviations: IRS – inner root sheath; ORS – outer root sheath; DP – dermal papilla; Mx – hair matrix; SCs – stem cells. (B) Schematic of “average” hippo skin. The epidermis is relatively thin in comparison to cetaceans and displays shallow rete ridges. The dermis lacks identifiable adipocytes. Ventral, dorsal and ear skin contains pelage hair follicles that lack associated sebaceous glands. Facial skin features vibrissae follicles that also lack sebaceous glands. Tail skin contains prominent hair follicles that have vibrissa-like morphology. Several body sites contain distinct sweat glands. (C) Schematic drawing of facial skin in common bottlenose dolphin. The epidermis is very thick and features prominent rete ridges. The dermis contains numerous adipocyte clusters. In newborn dolphin, the facial skin contains actively growing vibrissae hair follicles that lack distinct collagen capsules, ringwulst, and sebaceous glands (left). In adult dolphin, facial vibrissae degrade, forming keratin-filled pits (right). Skin has no sweat glands. (D) Skin-associated genes (excluding KRTAP genes) that are inactivated in some or all cetaceans and sometimes in hippos (see main text). Genes are classified based on their cell type or skin structure-specific expression and ontology as projected on the human skin diagram in A. Genes in black font are inactivated in some (*) or all cetaceans but not in hippos; genes in red font are independently inactivated in some (*) or all cetaceans and both hippos; the single gene in blue font is independently inactivated in cetaceans and pygmy hippo. Inactivated genes are based on the literature [16,24,65–67,69,70,89,117] and new observations reported here. See also Table S1.
Cetaceans and hippos share features such as reduced differentiation of the epidermis, but there are also differences. In particular, the hippo epidermis is much thinner, has shallower rete ridges, and partially preserves epidermal barrier function as hippos spend substantial time on land. These differences may explain why only one EDC gene, TCHHL1 (trichohyalin-like 1), is convergently inactivated in cetaceans and hippos (Figure 5). TCHHL1 is predominantly expressed in the stratum basale and its specific function in keratinocyte differentiation remains unclear [87,88]. Among suprabasal keratins, only KRT2 and KRT77 are independently inactivated in hippos (Figure 5), and the overall degree of suprabasal keratin inactivation in hippos is less than in obligately-aquatic manatees, which are more similar to cetaceans [16]. Other important epidermal genes that were reported as inactivated in cetaceans are intact in hippos. These include terminal keratinocyte differentiation-associated caspase CASP14 [65], PSORS1C2 (psoriasis susceptibility 1 candidate 2) [89], desmosome proteins DSG4 and DSC1, transglutaminase TGM5, and the atypical lipoxygenase ALOXE3 [90]. In summary, cetaceans and hippos both have inactivated copies of EDC and suprabasal keratin genes, but many more genes are knocked out in cetaceans.
Our genome-wide screen identified two new epidermal genes, KPRP and ALOX15, that are independently inactivated in cetaceans and hippos (Figure 5). KPRP (keratinocyte proline-rich protein) is an epidermal terminal differentiation-associated protein, part of the EDC, that is normally expressed in the stratum granulosum [91,92]. This epidermal layer is poorly defined in hippos [15] and absent in cetaceans [10]. A single-nucleotide polymorphism in human KPRP is associated with atopic dermatitis, a condition diagnosed by disrupted epidermal barrier function [93]. ALOX15 (arachidonate 15-lipoxygenase) belongs to the lipoxygenase family of enzymes that catalyze bioactive lipids synthesis, including resolvins that regulate resolution of excessive inflammatory responses [94,95]. ALOX15 is highly expressed in mouse epidermis and Axol15−/− mutant mouse studies suggest a role in epidermal barrier function [96].
We next focused on genes that are associated with hair follicles, sebaceous glands, and sweat glands. We confirm previous findings that some or all cetaceans have inactivated hair inner root sheath keratins (KRT25, KRT26, KRT27, KRT28, KRT71, KRT72, KRT73, KRT74), hair, nail and tongue papillae keratins (KRT32, KRT33A, KRT33B, KRT34, KRT35, KRT38, KRT39, KRT40, KRT82, KRT83, KRT84), as well as keratins KRT3 and KRT6B [16,97]. Of these, hippos have independently inactivated only KRT26. The retention of functional hair and nail keratins in hippos is consistent with the presence of prominent keratinized hoofs [98] and both pelage and vibrissa hairs. Brush-like hairs on the tail aid in spreading feces during defecation, a behavior used by hippos for marking territory [99]. Our analyses validate that cetaceans have inactivating mutations in genes (AWAT1, DGAT2L6, FABP9, ELOVL3, MOGAT3, MC5R) associated with sebaceous gland function [69,70]. AWAT1 is inactivated in both hippos, but in contradiction to Lopes-Marques et al. [70] we show that these species retain an intact copy of MOGAT3 (Figure 4). Finally, we found that ABCC11, which is associated with axillary sweat gland function in humans, is inactivated in all cetaceans and independently in pygmy hippo, which like river hippo has active sweat glands that produce pigmented secretion. Therefore, ABCC11 function does not appear to be critical for pygmy hippos’ sweat gland biology.
The Timing of Gene Inactivations in Hippopotamidae and Cetacea
To understand when gene losses occurred, we performed dN/dS analyses with the coding remnants of inactivated genes and equations from Meredith et al. [83]. To obtain robust inactivation dates, we calculated dates using eight different combinations of codon frequency model (CF1, CF2), fixed versus estimated ω values for the pseudogenic branch category, and one versus two rates for synonymous substitutions (Table S3). Mean inactivation dates based on these estimates are shown in Figure 3 and Table 2. Mean estimates for eight genes (ALOX15, AWAT1, KPRP, KRT2, KRT26, KRT77, KRTAP7–1, TCHHL1) that were inactivated on the stem Hippopotamidae branch range from 53.92 to 5.42 Ma. These inactivation dates suggest that derived changes in hippopotamid skin, including the loss of sebaceous glands, have a long history that encompasses the entirety of the stem hippopotamid branch. The mean date for inactivation of these eight genes is ~30.5 Ma, which is near the midpoint of the stem hippopotamid branch. In addition, ABCC11 appears to have been inactivated very recently in Choeropsis liberiensis (Figure 3; Table 2).
Table 2. Inactivation dates for pseudogenized skin genes.
Mean inactivation dates for skin-related genes in cetaceans and hippos based on eight different combinations of codon frequency model (CF1, CF2), fixed versus estimated dN/dS values for the pseudogenic branch category, and one versus two rates for synonymous substitutions. Estimates based on individual analyses are provided in Table S3.
| Gene | Hippopotamidae | Choeropsis | Cetacea | Mysticeti | Balaenoptera | Delphinida | Physeteroidea |
|---|---|---|---|---|---|---|---|
| ABCC11 | 0 | 40.54 | |||||
| ALOX15 | 23.54 | 15.74 | 34.13 | 21.44 | |||
| AWAT1 | 53.92 | 48.23 | |||||
| KPRP | 39.01 | CDS deleted | |||||
| KRT2 | 26.41 | 33.64a | |||||
| KRT26 | 15.98 | 48.93b | |||||
| KRT77 | 45.75 | 53.92c | |||||
| KRTAP 7–1 | 34.18 | 40.94 | |||||
| MOGAT3 | 46.37 | ||||||
| TCHHL1 | 5.42 | CDS deleted |
CDS (partial) only preset in Mysticeti.
CDS only present in Delphinida.
CDS only present in Mysticti.
For Cetacea, estimated inactivation dates for four genes (ABCC11, AWAT1, KRTAP7–1, MOGAT3) with evidence of pseudogenization on the stem cetacean branch based on shared inactivating mutations range from 48.23 to 40.54 Ma (mean = 44.02 Ma). Three additional genes are inactivated in all cetaceans, but are completely absent from either odontocetes (KRT2, KRT77) or mysticetes (KRT26). As complete gene deletion could have erased smaller mutations that inactivated these genes earlier in evolution, we used the remnants of these genes to estimate whether loss may have occurred on the stem Cetacea branch. Inactivation dates suggest that two of these genes were pseudogenized on the cetacean stem branch (53.92 Ma [KRT77], 48.93 Ma [KRT26]) whereas the date for KRT2 [33.64 Ma] is slightly younger than the most recent common ancestor of crown Cetacea at ~36.72 Ma. The mean date for the six genes (ABCC11, AWAT1, KRT26, KRT77, KRTAP7–1, MOGAT3) inactivated on the stem cetacean branch is ~46.5 Ma, which is near the midpoint of the stem cetacean branch. Overall, the mean inactivation date for these six genes in cetaceans is ~16.0 million years older than the mean inactivation date for eight genes that were inactivated on the stem hippo branch. Among four overlapping genes (AWAT1, KRT26, KRT77, KRTAP7–1), the mean date for inactivation on the stem Cetacea branch (48.00 Ma) is 10.54 Ma older than the mean date for inactivation on the stem hippo branch (37.46 Ma). Two additional genes (KPRP, TCHHL1) are completely absent in all examined cetacean genomes, so the timing of these gene losses on the stem cetacean branch could not be estimated. Finally, ALOX15 was independently inactivated in the common ancestor of Delphinida (34.13 Ma), on the branch leading to Physeter (21.44 Ma), and in the common ancestor of Balaenoptera acutorostrata and B. bonaerensis (15.74 Ma).
Integration of Molecular, Histological, and Paleontological Data
Our analyses shed light on a key question pertaining to the evolution of Cetancodonta: Did shared features associated with a (semi)aquatic lifestyle evolve in the common ancestor of this clade or independently in Cetacea and Hippopotamidae (Figure 1)? If aquatic features of hippos represent an intermediate condition in the transition from land to sea in the ancestry of Cetacea [1], then features of extant hippos might provide insights into the behavior and morphology of the earliest cetaceans from the Eocene [51]. Shared morphological features include the loss of sebaceous glands and the general reduction or complete loss of pelage hairs that cover the body. Previous histological studies that reported the absence of sebaceous glands in river hippo were limited to skin from the trunk, neck, and limbs that has few to no hairs [15]. However, sebaceous glands and hair follicles comprise an anatomically connected pilosebaceous unit. Therefore, we thoroughly investigated several regions of hair-bearing skin and provide more definitive evidence for the absence of sebaceous glands in both extant hippo species. Meibomian glands, which are modified sebaceous glands in the eyelids, are also absent in both hippos (also see [39]). Further, vibrissa follicles in newborn dolphin and adult gray whale lack sebaceous glands. These findings suggest the complete body-wide absence of sebaceous glands in hippopotamids and cetaceans. The most parsimonious explanation is that these glands were lost in the common ancestor of Cetancodonta. Similarly, the most parsimonious explanation for body hair reduction (hippos) or loss (cetaceans) is that pelage density started to decrease in the common ancestor of Cetancodonta. By contrast, the distribution of lipids is intracellular in the stratum corneum of cetaceans whereas hippos have intercellular stratum corneum lipids, which suggests parallel evolution of the highly modified epidermis in these taxa.
If sebaceous gland loss and pelage reduction occurred in the common ancestor of Cetancodonta, then we should find evidence of shared inactivating mutations in one or more skin-specific genes in hippos and cetaceans. By contrast, the independent origins hypothesis predicts convergent gene inactivations in cetaceans and hippos. Our genomic screens identified several skin-specific genes that are inactivated in hippos and cetaceans. Strikingly, while our genomic screens identified several skin-specific genes that are inactivated in hippos and cetaceans, none of these genes have shared inactivating mutations, suggesting that pelage reduction and the loss of sebaceous glands occurred independently in Hippopotamidae and Cetacea.
Mean dates for the inactivation of skin genes in these two clades serve as a proxy for phenotypic changes. These dates suggest that pelage reduction/loss, the loss of sebaceous glands, and changes to the keratinization program occurred 15.95 million years earlier in Cetacea (46.48 Ma) than Hippopotamidae (30.53 Ma) based on all estimates for gene inactivations or 10.54 million years earlier in Cetacea (48.00 Ma) than Hippopotamidae (37.46 Ma) based on an overlapping set of four genes (Figure 3, Table 2). The mean date of ~48.0–46.5 Ma for Cetacea is older than the first obligately aquatic cetaceans in the family Basilosauridae (e.g., Basilosaurus at ~41 Ma) and instead corresponds with the oldest protocetids (e.g., Rodhocetus) from the Lutetian (47.8–41.3 Ma), which may have utilized both land and water as sea lions do today [100]. The mean inactivation date (~37.5–30.5 Ma) for hippopotamid skin genes, in turn, is in the range of early bothriodontine anthracotheres (37.2–33.9 Ma) that are inferred to be the oldest semiaquatic members of Anthracotheriidae [36]. It is also noteworthy that far more skin-related genes have been inactivated in Cetacea than in Hippopotamidae (Figure 5), which is consistent with the more complete reorganization of the epidermis and its derivatives in cetaceans than hippopotamids (Table 1; Figures 2 and 5). Finally, our estimates of individual gene inactivations span tens of millions of years for both stem + crown cetaceans and stem + crown hippos (Figure 3, Table 2), suggesting that macroevolutionary changes to aquatic and semiaquatic habitats in these clades have long, stepwise histories. Indeed, AWAT1 inactivation on the stem hippopotamid branch and ABCC11 inactivation on the Choeropsis liberiensis branch have estimated pseudogenization dates that are separated by ~54 million years.
Paleontological evidence also provides an opportunity to evaluate competing hypotheses that (semi)aquatic features of cetaceans and hippos were acquired independently rather than in the common ancestor of this clade (Figure 1). If the initial shift to a semiaquatic lifestyle occurred in the common ancestor of Cetancodonta, then we might expect to find morphological and/or geochemical evidence for this transition in (1) stem cetancodontans that are close to the crown group, (2) the earliest stem cetaceans, and (3) the earliest stem hippopotamids. For stem cetaceans, the most primitive and earliest branching clade is Raoellidae. Thewissen et al. [101] inferred that the raoellid Indohyus was already semiaquatic based on a thickened medial wall in the auditory bulla (= involucrum) that is associated with underwater hearing. Dense limb bones (for ballast) and oxygen isotopic signatures of its teeth also suggest that Indohyus was semiaquatic [101]. Evidence for semiaquatic specializations in stem cetancodontans and stem hippopotamids is less forthcoming. Definitive stem members of Cetancodonta have been difficult to identify owing to the uncertainty of cladistic analyses. Possible stem cetancodontans have included various anthracotheres (e.g., Anthracotherium, Siamotherium [56]; Elomeryx, Heptacodon, Microbunodon [1]), an entelodontid (Brachyhyops [56], a cebochoerid (Cebochoerus [1,102]), and the enigmatic Andrewsarchus [56]. We are not aware of compelling evidence for aquatic adaptations in any of these taxa. Also, there is an emerging consensus that anthracotheres comprise the paraphyletic stem group that gave rise to Hippopotamidae [36,63,102]. Many anthracotheres (e.g., Anthracotherium, Siamotherium) appear to have been terrestrial based on oxygen isotope values whereas taxa in the subfamily Bothriodontinae have values that are consistent with a semiaquatic lifestyle [58,59,61,62]. However, the phylogenetic placement of presumed terrestrial anthracotheres as basal, stem Hippopotamidae suggests that specializations for an aquatic/semiaquatic lifestyle evolved independently in hippopotamids and cetaceans [36,63]. Thus, available paleontological evidence is largely aligned with distinct inactivating mutations in skin-related genes that favor the independent origins hypothesis. Additional testing of this hypothesis will benefit from more comprehensive taxonomic sampling of anthracotheres in both phylogenetic analyses and oxygen isotope studies.
When sister taxa share the same anatomical, physiological, or behavioral features, as is the case for aquatic features in Hippopotamidae and Cetacea, the simplest hypothesis is that these features evolved in their common ancestor. However, our results suggest otherwise and support the independent evolution of features that are related to the skin in hippos and cetaceans. Given that most synapomorphies for Cetancodonta are aquatic traits [1], morphological and behavioral support is very limited for this group if these traits evolved in parallel. Along the same lines, pinnipeds have lost their sweet and umami taste receptors. However, loss of these receptors occurred independently in the ancestors of the reciprocally monophyletic Phocidae (seals) and Otaroidea (sea lions, fur seals, walruses) rather than in the common ancestor of Pinnipedia [103]. Numerous morphological features related to raptorial feeding and hydrodynamic locomotion also evolved independently within Pinnipedia [104]. In Cetacea, multiple cranial and postcranial specializations for an aquatic lifestyle evolved convergently in odontocetes and mysticetes [1,105,106]. Morphological characters preserved in fossils and pseudogenic remnants of formerly functional genes provide complementary sources of evidence for elucidating such cases of convergent or parallel evolution.
Finally, gene inactivation dates have implications for understanding the physiology, behavior, and appearance of extinct organisms [1,107]. In the case of cetaceans, inactivation dates for AWAT1 and MOGAT3 are older than the inactivation date for ABCC11, which suggests sebaceous glands were lost before sweat glands on the stem cetacean branch. Given the timing of gene inactivations for these three genes, extinct protocetid whales may have retained sweat glands but not sebaceous glands. Similarly, the hair inner root sheath keratin KRT26 was lost relatively early on the stem cetacean branch (~49 Ma), suggesting that the program for generating body pelage hair had already been compromised at this early stage in cetacean evolution. Protocetids, which comprise a paraphyletic grade, were probably the earliest transoceanic cetaceans, but also spent time on land where they may have given birth and nursed their young [100,108,109]. These forms would certainly have looked different than modern whales due to their more prominent hindlimbs and primitive cranial morphology, but in other respects, such as having a largely hairless body, may have been similar to modern cetaceans.
Pelage reduction and other shared morphological features of the skin in Cetancodonta are not obligatory or exclusive features of a (semi)-aquatic lifestyle. For example, pinnipeds and beavers maintain dense hair despite being semi-aquatic. By contrast, sparse hair evolved in some fully terrestrial mammals (e.g., elephants, rhinoceroses) to enhance heat loss in savanna habitats [110]. Selection for efficient thermoregulation is also hypothesized to have contributed to hair reduction in hominins when our ancestors occupied the Africana savanna [110,111].
In summary, the integration of new histological data with comprehensive analyses of inactivated protein-coding genes provides strong support for the hypothesis that aquatic adaptations of the skin evolved independently in cetaceans and hippos. Our study further illustrates the potential of genomic data and in particular remnants of once functional genes as dateable ‘molecular vestiges’ to complement morphological data in providing novel insights into ancestry and timing of key trait changes and macroevolutionary transitions [68,83,86,107,112–117].
STAR*METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mark S. Springer (springer@ucr.edu).
Materials Availability
Gene alignments generated in this study have been deposited to FigShare [https://doi.org/10.6084/m9.figshare.13549070.v1].
Data and Code Availability
The Illumina sequencing data generated for this study are available under NCBI BioProject PRJNA694317.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Genomic DNA from Choeropsis liberiensis (pygmy hippopotamus) was provided by G. Amato (formerly at New York Zoological Society). The sources for skin samples used in histological analyses are as follows: Choeropsis liberiensis (pygmy hippopotamus) skin samples are from Smithsonian National Museum of Natural History – specimen number 395848 (unknown gender, neonatal); Hippopotamus amphibius (river hippopotamus) skin samples are from Smithsonian National Museum of Natural History – specimen number 254870 (male, neonatal); Tursiops truncatus (common bottlenose dolphin) skin samples are from Southwest Fisheries Science Center (NOAA) specimen numbers KKS0032 (unknown gender, neonatal) and KXD0206 (late term fetus); Eschrichtius robustus (gray whale) skin samples are from Southwest Fisheries Science Center (NOAA) Fisheries – specimen number NEB0083 (unknown gender, adult).
METHOD DETAILS
Histology
Formalin-fixed Choeropsis liberiensis and Hippopotamus amphibius skin samples were first hydrated and rinsed in 1X PBS. Samples were then dehydrated through an ethanol gradient (from 25% to 100%), processed through histoclear and embedded in paraffin. Each hydration and dehydration step lasted for 12 hours. Tissues were sectioned at a thickness of 10 μm with a microtome (Leica). Samples were stained with hematoxylin and eosin using standard methods with minor modifications. Tissue sections were mounted with Permount mounting media and visualized with Nikon Ti-E Upright microscope. Tissue whole-mounts were captured with a Nikon dissecting microscope. Individual fields of Eschrichtius robustus rostral skin were visualized and stitched together with Keyence microscope.
Quantification of Epidermal Thickness
Epithelial thickness of Choeropsis liberiensis, Hippopotamus amphibius, Tursiops truncatus and Eschrichtius robustus rostral skin was quantified using ImageJ (NIH). We included measurements ranging from: a) top to start of rete ridge; b) start to end of rete ridge; and c) entire epidermis including rete ridge. Up to 10 individual measurements were included per image field per species. Measurements are reported as average epidermal thickness (μm) ± standard deviation (μm) in Table 1.
Pygmy Hippopotamus Genome
Genomic DNA was sonicated at the University of California, Riverside (UCR), Genomics Core Facility into ~550 bp fragments. We constructed a genomic library using Illumina’s NeoPrep Library Prep System. Paired-end sequencing (150 bp) was performed at UCR. Raw Illumina sequence data have been deposited at NCBI (PRJNA694317).
Genomic Screens for Inactivated Genes
We used a genome alignment of placental mammals with the human hg38 assembly as the reference [118]. Our gene loss detection method [68,119] was used to screen for genes that exhibit inactivating mutations in the genomes of bottlenose dolphin, killer whale, sperm whale, common minke whale, and river hippo. We started with 19,769 genes annotated by Ensembl (http://www.ensembl.org) version 90 [120] in the human genome and considered 18,363 genes that are present in the assemblies of at least 31 of 63 placental mammals. To identify genes that were potentially inactivated on the branch leading to hippos and cetaceans, we further extracted genes that are inactivated in all cetaceans and the river hippo. We excluded genes that are intact in less than three of six terrestrial outgroup artiodactyls included in the screen (Bos taurus [cow], Capra hircus [goat], Camelus ferus [wild Bactrian camel], Pantholops hodgsonii [Tibetan antilope], Bison bison [bison], Vicugna pacos [alpaca]). Finally, we used a more recent assembly of the river hippo genome (GCA_004027065.2) to exclude instances where assembly errors mistakenly led to genes classified as inactivated in the river hippo. This resulted in a final list of 38 genes (Table S1).
BLAST Searches and Alignments
Genomic sequences encoding ten genes of interest (ABCC11, ALOX15, AWAT1, KPRP, KRT2, KRT26, KRT77, KRTAP7–1, MOGAT3, TCHHL1) were downloaded from NCBI for Homo sapiens (human), Bos taurus (cow), and Equus caballus (horse). Sequences for each gene were aligned and exon annotations in Bos and Equus were compared against those in Homo to ensure that orthologous regions were annotated. Protein-coding sequences and flanking introns from Bos and Equus were employed in BLAST searches against other cetartiodactyls and perissodactyls, respectively, in NCBI’s ‘RefSeq Genome’ and ‘Whole-genome shotgun contigs’ databases. Additional perissodactyls included Ceratotherium simum (white rhinoceros) and Dicerorhinus sumatrensis (Sumatran rhinoceros). Additional cetartiodactyls included two camelids (Camelus ferus [wild Bactrian camel], Vicugna pacos [alpaca]), one suid (Sus scrofa [pig]), two bovids (Bubalus bubalis [water buffalo], Capra hircus [goat]), two giraffids (Giraffa camelopardalis [giraffe], Okapia johnstoni [okapi]), two cervids (Axis porcinus [hog deer], Odocoileus virginianus [white-tailed deer]), one hippopotamid (Hippopotamus amphibius [river hippopotamus]), four mysticetes (Balaena mysticetus [bowhead, downloaded from http://www.bowhead-whale.org/], Balaenoptera acutorostrata [common minke whale], B. bonaerensis [Antarctic minke whale], Eschrichtius robustus [gray whale]), and ten odontocetes (Physeter macrocephalus [sperm whale], Lipotes vexillifer [baiji], Delphinapterus leucas [beluga], Phocoena phocoena [harbor porpoise], Neophocaena asiaorientalis [narrow-ridged finless porpoise], Orcinus orca [killer whale], Lagenorhynchus obliquidens [Pacific white-sided dolphin], Sousa chinensis [Indo-Pacific humpback dolphin], Tursiops aduncus [Indo-Pacific bottlenose dolphin], Tursiops truncatus [common bottlenose dolphin]). Additional searches were performed with other perissodactyls or cetartiodactyls when the initial searches with Equus and Bos were unsuccessful in retrieving complete orthologs. Megablast was employed for highly similar sequences and blastn for less similar sequences. Complete protein-coding sequences and intervening introns were imported into Geneious 11.1.5 [121] and aligned against reference sequences with MAFFT [122] with minor adjustments by eye. Aligned sequences were annotated for exons and inspected for splice site mutations. Illumina sequences for Choeropsis liberiensis were imported into Geneious and protein-coding sequences for the above-mentioned genes were obtained using a map to reference approach with probe sequences from the closely related Hippopotamus amphibius. We allowed for a maximum mismatch of 6% per read and required at least two reads for base calling with a consensus threshold of 65%. We also used MAFFT to align complete protein-coding sequences from all taxa for each gene.
Inactivating Mutations
Final alignments for protein-coding sequences for each gene included 15–29 taxa given that some genes are deleted in one or more cetaceans. We inspected the final protein-coding alignment for each gene for inactivating mutations including exon deletions, frameshift insertions and deletions, altered start and stop codons, and premature stop codons (splice site mutations screened above). For each gene, parsimony optimizations with delayed transformation (deltran) were performed with PAUP* 4.0a150 [123] to map inactivating mutations to branches of the species tree (see below).
QUANTIFICATION AND STATISTICAL ANALYSIS
Phylogenetic Analyses
RAxML 8.2.11 [124] was run in Geneious to estimate maximum likelihood gene trees for each protein-coding alignment. Gene trees were inspected for suspicious relationships that conflict with the species tree (e.g., Camelus grouping with Sus instead of Vicugna) but none were found. Instead, gene tree incongruence was confined to conflicts that are readily explained by ILS such as Ruminantia grouping with Suoidea or Physeter grouping with Mysticeti [125]. Rapid bootstrap analysis (100 pseudoreplications) and a search for the best tree were performed in a single run. These analyses were performed with a GTR + Γ model of sequence evolution.
DN/dS Analyses
DN/dS analyses were performed with the codeml program of PAML 4.4 [126]. Analyses for each gene were performed with separate dN/dS categories for functional branches that lack inactivating mutations, fully pseudogenic branches that post-date the occurrence of an inactivating mutation on an earlier branch, and each transitional branch that records the first occurrence of an inactivating mutation (e.g., [83]). Analyses were performed with the codon frequency 1 (CF1) and codon frequency 2 (CF2) models of codeml. We also performed analyses with estimated and fixed (dN/dS =1.00) values for the fully pseudogenic branch category. We employed a species tree with higher level (interordinal, interfamilial) relationships from Meredith et al. [127] and intrafamilial relationships from Hassanin et al. [128] for terrestrial cetartiodactyls and McGowen et al. [129] for cetancodontans.
Estimation of Gene Inactivation Times
Equations from Meredith et al. [83] were used to estimate gene inactivation times in hippopotamids and cetaceans. We performed calculations using eight different combinations of codon model (CF1 or CF2), fixed (1.0) versus estimated values for dN/dS on fully pseudogenic branches, and equations that allow for one versus two synonymous substitution rates [83]. Mean inactivation dates for each gene are averages based on these eight different combinations. Divergence times for relevant nodes in these calculations were taken from McGowen et al. [129].
Supplementary Material
Table S3: Estimated inactivation dates for pseudogenized skin genes, related to Figure 3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Choeropsis liberiensis Illumina whole-genome shotgun sequences | This paper | PRJNA694317 |
| Nexus alignments of inactivated genes | This paper | https://doi.org/10.6084/m9.figshare.13549070.v1 |
| Software and Algorithms | ||
| PAML | [126] | http://abacus.gene.ucl.ac.uk/software/paml.html |
| Geneious 11.1.5 | [121] | https://www.geneious.com |
Springer et al. perform genomic and anatomical comparisons to determine if aquatic adaptations of the skin in hippos and cetaceans are shared derived or convergent features in these two clades. The results of these comparisons support the hypothesis that aquatic adaptions of the skin are convergent characters in hippos and cetaceans.
Cetaceans and hippos have differences in the thickness and organization of the skin
Genomic screens identified 8 skin genes that are inactivated in hippos and cetaceans
None of these 8 genes share inactivating mutations in hippos and cetaceans
Aquatic skin adaptations evolved independently in hippos and cetaceans
ACKNOWLEDGMENTS
This work was supported by NSF grant DEB-1457735 (J.G., M.S.S.), NIH grants U01-AR073159 and P30-AR075047 (M.V.P.), NSF grant DMS-1951144 (M.V.P.), and Pew Charitable Trust and LEO Foundation (M.V.P.). C.F.G-J. was supported by UC Irvine Chancellor’s ADVANCE Postdoctoral Fellowship Program, NSF-Simons Postdoctoral Fellowship, NSF grant (DMS1763272, to Qing Nie), Simons Foundation Grant (594598, to Qing Nie), and by a kind gift from the Howard Hughes Medical Institute Hanna H. Gray Postdoctoral Fellowship Program. M.H. was supported by the Max Planck Society and the LOEWE-Centre for Translational Biodiversity Genomics (TBG) funded by the Hessen State Ministry of Higher Education, Research and the Arts (HMWK). We thank the Smithsonian Institution for skin samples of hippos. C. Buell provided images of mammals. J. Geisler and four anonymous reviewers provided helpful comments on this manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gatesy J, Geisler JH, Chang J, Buell C, Berta A, Meredith RW, Springer MS, and McGowen MR (2013). A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 66(2), 479–506. [DOI] [PubMed] [Google Scholar]
- 2.Springer MS, Signore AV, Paijmans JLA, Vélez-Juarbe J, Domning DP, Bauer CE, He K, Crerar L, Campos PF, Murphy WJ, Meredith RW, Gatesy J, Willerslev E, MacPhee RDE, Hofreiter M, and Campbell KL (2015). Interordinal gene capture, the phylogenetic position of Steller’s sea cow based on molecular and morphological data, and the macroevolutionary history of Sirenia. Mol. Phylogenet. Evol. 91, 178–193. [DOI] [PubMed] [Google Scholar]
- 3.Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS (2013). Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9(4), e1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flower WH, 1883. On whales, past and present, and their probable origin. Proc. R. Inst. Great Brit. 10, 360–376. [Google Scholar]
- 5.Slijper EJ (1962). Whales, second ed. Cornell University Press, Ithaca, NY. [Google Scholar]
- 6.Jensen FH, Johnson M, Ladegaard M, Wisniewska DM,and Madsen PT (2018). Narrow acoustic field of view drives frequency scaling in toothed whale biosonar. Curr. Biol. 28, 3878–3885. [DOI] [PubMed] [Google Scholar]
- 7.Harrison RJ, and Thurley KW (1974). Structure of the epidermis in Tursiops, Delphinus, Orcinus and Phocoena. In: Harrison R. (Ed.), Functional Anatomy of Marine Mammals, Vol. 2. Academic Press, London, pp. 45–72. [Google Scholar]
- 8.Ling J. (1974). The integument of marine mammals. In: Harrison R. (Ed.), Functional Anatomy of Marine Mammals, Vol. 2. Academic Press, London, pp. 1–44. [Google Scholar]
- 9.Durham FE (1980). External morphology of bowhead fetuses and calves. Mar. Fish. Rev. 42, 74–80. [Google Scholar]
- 10.Haldiman JT, Henk WG, Henry RW, Albert TF, Abdelbaki YZ, and Duffield DW (1985). Epidermal and papillary dermal characteristics of the bowhead whale (Balaena mysticetus). Anat. Rec. 211(4), 391–402. [DOI] [PubMed] [Google Scholar]
- 11.Hicks BD, St Aubin DJ, Geraci JR, and Brown WR (1985). Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J. Invest. Dermatol. 85, 60–63. [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer CJ, and Rowntree VJ (1996). Epidermal ultrastructure of the southern right whale calf (Eubalaena australis). J. Submicrosc. Cytol. Pathol. 28, 277–286. [PubMed] [Google Scholar]
- 13.Reeb D, Best PB, and Kidson SH (2007). Structure of the integument of southern right whales, Eubalaena australis. Anat. Rec. 290, 596–613. [DOI] [PubMed] [Google Scholar]
- 14.Sokolov W. (1960). Some similarities and dissimilarities in the structure of the skin among the members of the suborders Odontoceti and Mystacoceti (Cetacea). Nature 185(4715), 745–747. [Google Scholar]
- 15.Luck C, and Wright P. (1964). Aspects of the anatomy and physiology of the skin of the hippopotamus (H. amphibius). Quart. J. Exp. Physiol. 49, 1–14. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich F, Fischer H, Langbein L, Praetzel-Wunder S, Ebner B, Figlak K, Weissenbacher A, Sipos W, Tschachler E, and Eckhart L. (2019). Differential evolution of the epidermal keratin cytoskeleton in terrestrial and aquatic mammals. Mol. Biol. Evol. 36, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokolov W. (1962). Adaptations of the mammalian skin to the aquatic mode of life. Nature 195(4840), 464–466. [Google Scholar]
- 18.Giacometti L. (1967). The skin of the whale (Balaenoptera physalus). Anat. Rec, 159(1), 69–75. [DOI] [PubMed] [Google Scholar]
- 19.Geraci JR, Aubin J, Hicks BD, Bryden MM, and Harrison R. (1986). The epidermis of odontocetes: a view from within. In: Bryden MM, and Harrison R. (Eds.), Research on Dolphins. Clarenden Press, Oxford, pp. 3–21. [Google Scholar]
- 20.Menon GK, Grayson S, Brown BE, and Elias PM (1986). Lipokeratinocytes of the epidermis of a cetacean (Phocena phocena). Cell Tissue Res. 244(2), 385–394. [DOI] [PubMed] [Google Scholar]
- 21.Elias PM, Menon GK, Grayson S, Brown BE, and Rehfeld SJ (1987). Avian sebokeratocytes and marine mammal lipokeratinocytes: structural, lipid biochemical, and functional considerations. Am. J. Anat. 180, 161–177. [DOI] [PubMed] [Google Scholar]
- 22.Pfeiffer CJ, and Jones FM (1993). Epidermal lipid in several cetacean species: ultrastructural observations. Anat. Embryol. 188, 209–218. [DOI] [PubMed] [Google Scholar]
- 23.Meyer W, Schmidt J, Busche R, Jacob R, and Naim HY (2012). Demonstration of free fatty acids in the integument of semi-aquatic and aquatic mammals. Acta Histochemica, 114(2), 145–150. [DOI] [PubMed] [Google Scholar]
- 24.Lachner J, Mlitz V, Tschachler E, and Eckhart L. (2017). Epidermal cornification is preceded by the expression of a keratinocyte-specific set of pyroptosis-related genes. Sci. Rep. 7(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry DA (1949). The structure of whale blubber, and a discussion of its thermal properties. Q. J. Microsc. Sci. 90, 13–25. [PubMed] [Google Scholar]
- 26.Palmer E, and Weddell G. (1964). The relationship between structure, innervation and function of the skin of the bottle nose dolphin (Tursiops truncatus). Proc. Zool. Soc. Lond. 143. 553–568. [Google Scholar]
- 27.Oh JW, Chung O, Cho YS, MacGregor GR, and Plikus MV (2015). Gene loss in keratinization programs accompanies adaptation of cetacean skin to aquatic lifestyle. Exp. Dermatol. 24(8), 572–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai J, and Shida T. (1948). Sinus-hairs of the sei-whale (Balaenoptera borealis). Sci. Rept. Whales Res. Inst. 1, 41–47. [Google Scholar]
- 29.Slijper EJ (1976). Whales and Dolphins. University of Michigan Press, Michigan. [Google Scholar]
- 30.Ling JK (1977). Vibrissae of marine mammals. In: Harrison RJ (Ed.), Functional Anatomy of Marine Mammals 3, Academic Press, London, pp. 387–415. [Google Scholar]
- 31.Berta A, Ekdale EG, Zellmer NT, Demere TA, Kienle SS, and Smallcomb M. (2015). Eye, nose, hair, and throat: external anatomy of the head of a neonate gray whale (Cetacea, Mysticeti, Eschrichtiidae). Anat. Rec. 298, 648–659. [DOI] [PubMed] [Google Scholar]
- 32.Drake SE, Crish SD, George JC, Stimmelmayr R, and Thewissen JG (2015). Sensory hairs in the bowhead whale, Balaena mysticetus (Cetacea, Mammalia). Anat. Rec. 298, 1327–1335. [DOI] [PubMed] [Google Scholar]
- 33.Yablokov AV, and Klevezal GA (1969). Whiskers of whales and seals and their distribution, structure and significance. In: Kleinenberg SE (Ed.), Morphological Characteristics of Aquatic Mammals. Izdatel’stvo Nauka, Moscow, Russia, pp. 48–81. [Google Scholar]
- 34.Czech-Damal NU, Liebschner A, Miersch L, Klauer G, Hanke FD, Marshall C, Dehnhardt G, and Hanke W. (2012). Electroreception in the Guiana dolphin (Sotalia guianensis). Proc. Roy. Soc. B 279, 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huggenberger S, Oelschläger H, and Cozzi B. (2019). Regional anatomy, development, and hydrodynamics including skin anatomy. In: Huggenberger S, Oelschläger H, Cozzi B. (Eds.), Atlas of the Anatomy of Dolphins and Whales. Academic Press, pp. 5–135. [Google Scholar]
- 36.Boisserie JR, Fisher RE, Lihoreau F, and Weston EM (2011). Evolving between land and water: key questions on the emergence and history of the Hippopotamidae (Hippopotamoidea, Cetancodonta, Cetartiodactyla). Biol. Rev. 86(3), 601–625. [DOI] [PubMed] [Google Scholar]
- 37.Allbrook DB, Luck CP, and Wright PG (1959). Histological observations on the cutaneous glands in hippopotamus. J. Anat. 93, 587–588. [Google Scholar]
- 38.Allbrook D. (1962). The morphology of the subdermal glands of Hippopotamus amphibius. Proc. Zool. Soc. Lond. 139, 67–73. [Google Scholar]
- 39.Klećkowska-Nawrot JE, Goździewska-Harłajczuk K, and Paszta W. (2020). Gross anatomy, histological and histochemical analysis of the eyelids and orbital glands of the neonate pygmy hippopotamus (Suina: Choeropsis liberiensis or Hexaprotodon liberiensis, Morton 1849) with reference to its habitat. Anat. Rec. 2020, 1–19. [DOI] [PubMed] [Google Scholar]
- 40.Tomes J. (1850). On the blood-coloured exudation from the skin of the hippopotamus. Proc. Zool. Soc. Lond. 1850, 160–162. [Google Scholar]
- 41.Saikawa Y, Hashimoto K, Nakata M, Yoshihara M, Nagai K, Ida M, and Komiya T. (2004). The red sweat of the hippopotamus. Nature 429, 363. [DOI] [PubMed] [Google Scholar]
- 42.Prothero DR, Manning EM, and Fischer M. (1988). The phylogeny of ungulates. In: Benton MJ (Ed.), The Phylogeny and Classification Classification of the Tetrapods, Vol. 2. Systematics Association Special Volume No. 35B. Clarendon Press, Oxford, pp. 201–234. [Google Scholar]
- 43.Novacek MJ (1992). Fossils, topologies, missing data, and the higher level phylogeny of eutherian mammals. Syst. Biol. 41, 58–73. [Google Scholar]
- 44.Matthew WD (1929). Reclassification of the artiodactyl families. Bull. Soc. Amer. 40, 403–404. [Google Scholar]
- 45.Matthew WD, and Colbert EH (1934). A phylogenetic chart of the Artiodactyla. J. Mammal. 15, 207–209. [Google Scholar]
- 46.Simpson GG (1945). The principles of classification and a classification of mammals. Bull. Amer. Mus. Nat. Hist. 85, 1–350. [Google Scholar]
- 47.Pickford M. (1983). On the origins of Hippopotamidae together with descriptions of two new species, a new genus and a new subfamily from the Miocene of Kenya. Geobios 16(2), 193–217. [Google Scholar]
- 48.Gentry AW, and Hooker JJ (1988). The phylogeny of the Artiodactyla. In: Benton MJ (Ed.), The Phylogeny and Classification of the Tetrapods, Vol. 2: Mammals. Clarendon Press, Oxford, pp. 235–272. [Google Scholar]
- 49.Irwin D, and Arnason U. 1994. Cytochrome b gene of marine mammals: phylogeny and evolution. J. Mammal. Evol. 2, 37–55. [Google Scholar]
- 50.Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, Goto M, Munechika I, and Okada N. (1997). Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature, 388(6643), 666–670. [DOI] [PubMed] [Google Scholar]
- 51.Gatesy J, Hayashi C, Cronin A, Arctander P. (1996). Evidence from milk casein genes that cetaceans are close relatives of hippopotamid artiodactyls. Mol. Biol. Evol. 13, 954–963. [DOI] [PubMed] [Google Scholar]
- 52.Gatesy J. (1997). More DNA support for a Cetacea/Hippopotamidae clade: the bloodclotting protein gene gamma-fibrinogen. Mol. Biol. Evol. 14, 537–543. [DOI] [PubMed] [Google Scholar]
- 53.Gatesy J. (1998). Molecular evidence for the phylogenetic affinities of Cetacea. In: Thewissen JGM (Ed.), The Emergence of Whales. Plenum, New York, pp. 63–111. [Google Scholar]
- 54.Nikaido M, Rooney AP, and Okada N. (1999). Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: hippopotamuses are the closest extant relatives of whales. Proc. Natl. Acad. Sci. USA 96, 10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geisler JH, and Theodor JM (2009). Brief communications arising: hippo and whale phylogeny. Nature 458, E1–E4. [DOI] [PubMed] [Google Scholar]
- 56.Spaulding M, O’Leary MA, and Gatesy J. (2009). Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution. PLoS ONE 4(9), e7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Leary MA, and Gatesy J. (2008). Impact of increased character sampling on the phylogeny of Cetartiodactyla (Mammalia): combined analysis including fossils. Cladistics 24(4), 397–442. [DOI] [PubMed] [Google Scholar]
- 58.Clementz MT, Holroyd PA, and Koch PL (2008). Identifying aquatic habits of herbivorous mammals through stable isotope analysis. Palaios 23, 574–585. [Google Scholar]
- 59.Zin-Maung-Maung-Thein, Takai M, Uno H, Wynn JG, Egi N, Tsubamoto T, Thaung-Htike, Aung-Naing-Soe, Maung-Maung, Nishimura T, and Yoneda M. (2011). Stable isotope analysis of the tooth enamel of Chaingzauk mammalian fauna (late Neogene, Myanmar) and its implication to paleoenvironment and paleogeography. Palaeogeogr. Palaeoclimatol. Palaeoecol. 300, 11–22. [Google Scholar]
- 60.Lihoreau F, Boisserie JR, Blondel C, Jacques L, Likius A, Mackaye HT, Vignaud P, and Brunet M. (2014). Description and palaeobiology of a new species of Libycosaurus (Cetartiodactyla, Anthracotheriidae) from the late Miocene of Toros-Ménalla, northern Chad. J. Syst. Palaeontol. 12(7), 761–798. [Google Scholar]
- 61.Tütken T, and Absolon J. (2015). Late Oligocene ambient temperatures reconstructed by stable isotope analysis of terrestrial and aquatic vertebrate fossils of Enspel, Germany. Paleobiodivers. Paleoenviron. 95(1), 17–31. [Google Scholar]
- 62.Cooper LN, Clementz MT, Usip S, Bajpai S, Hussain ST, and Hieronymus TL (2016). Aquatic habits of cetacean ancestors: integrating bone microanatomy and stable isotopes. Integr. Comp. Biol. 56, 1370–1384. [DOI] [PubMed] [Google Scholar]
- 63.Soe AN, Chavasseau O, Chaimanee Y, Sein C, Jaeger JJ, Valentin X, and Ducrocq S. (2017). New remains of Siamotherium pondaungensis (Cetartiodactyla, Hippopotamoidea) from the Eocene of Pondaung, Myanmar: paleoecologic and phylogenetic implications. J. Vertebr. Paleontol. 37(1), e1270290. [Google Scholar]
- 64.Tsagkogeorga G, McGowen MR, Davies KT, Jarman S, Polanowski A, Bertelsen MF, and Rossiter SJ (2015). A phylogenomic analysis of the role and timing of molecular adaptation in the aquatic transition of cetartiodactyl mammals. Roy. Soc. Open Sci. 2(9), 150156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strasser B, Mlitz V, Fischer H, Tschachler E. and Eckhart L. (2015). Comparative genomics reveals conservation of filaggrin and loss of caspase-14 in dolphins. Exp. Dermatol. 24(5), 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hecker N, Sharma V, and Hiller M. (2017). Transition to an aquatic habitat permitted the repeated loss of the pleiotropic KLK8 gene in mammals. Genome Biol. Evol. 9(11), 3179–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma V, Hecker N, Roscito JG, Foerster L, Langer BE, Hiller M. (2018a). A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Comm. 9(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huelsmann M, Hecker N, Springer MS, Gatesy J, Sharma V, Hiller M. (2019). Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Sci. Adv. 5(9), eaaw6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Springer MS, and Gatesy J. (2018). Evolution of the MC5R gene in placental mammals with evidence for its inactivation in multiple lineages that lack sebaceous glands. Mol. Phylogenet. Evol. 120, 364–374. [DOI] [PubMed] [Google Scholar]
- 70.Lopes-Marques M, Machado AM, Alves LQ, Fonseca MM, Barbosa S, Sinding MHS, Rasmussen MH, Iversen MR, Frost Bertelsen M, Campos PF, and Da Fonseca R. (2019). Complete inactivation of sebum-producing genes parallels the loss of sebaceous glands in Cetacea. Mol. Biol. Evol 36(6), 1270–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montagna W, and Yun JS (1964). The skin of the domestic pig. J. Invest. Dermatol 42, 11–21. [PubMed] [Google Scholar]
- 72.Mawafy M, and Cassens RG (1975). Microscopic structure of pig skin. J. Anim. Sci 41(5), 1281–1290. [DOI] [PubMed] [Google Scholar]
- 73.Debeer S, Le Luduec JB, Kaiserlian D, Laurent P, Nicolas J-F, Dubois D, and Kanitakis J. (2013). Comparative histology and immunohistochemistry of porcine versus human skin. Eur. J. Dermatol 23(4), 456–466. [DOI] [PubMed] [Google Scholar]
- 74.Abu-Samra MT, and Shuaib YA (2014). A study on the nature of association between Demodex mites and bacteria involved in skin and meibomian gland lesions of demodectic mange in cattle. Vet. Med. Int 2014, 413719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jian W, Duangjinda M, Vajrabukka C, and Katawatin S. (2014). Differences of skin morphology in Bos indicus, Bos taurus, and their crossbreds. Int. J. Biometeorol 58(6), 1087–1094. [DOI] [PubMed] [Google Scholar]
- 76.Murgiano L, Shirokova V, Welle MM, Jagannathan V, Plattet P, Oevermann A, Pienkowska-Schelling A, Gallo D, Gentile A, Mikkola M, et al. (2015). Hairless streaks in cattle implicate TSR2 in early hair follicle formation. PLoS Genet. 11(7), e1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turner NJ, Pezzone D, and Badylak SF (2015). Regional variations in the histology of porcine skin. Tissue Eng. Part C: Methods 21(4), 373–384. [DOI] [PubMed] [Google Scholar]
- 78.Wiener DJ, Wiedemar N, Welle MM, and Drogemuller C. (2015). Novel features of the prenatal horn bud development in cattle (Bos taurus). PLoS One 10(5), e0127691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crespo-Moral M, Garcia-Posadas L, Lopez-Garcia A, and Diebold Y. (2020). Histological and immunohistochemical characterization of the porcine ocular surface. PLoS One 15(1), e0227732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aubin DJ St., Smith TG, and Geraci JR (1990). Seasonal epidermal molt in beluga whales, Delphinapterus leucas. Can. J. Zool 68, 359–367. [Google Scholar]
- 81.Chernova OF, Shpak OV, Kiladze AB, Azarova VS, and Rozhnov VV (2016). Summer molting of bowhead whales Balaena mysticetus Linnaeus, 1758, of the Okhotsk Sea population. Doklady Biol. Sci 471, 261–265. [DOI] [PubMed] [Google Scholar]
- 82.Rehorek SJ, Stimmelmayr R, George JC, Suydam R, McBurney DM, and Thewissen JGM (2019). Structure of the external auditory meatus of the bowhead whale (Balaena mysticetus) and its relation to their seasonal migration. J. Anat 234, 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, and Springer MS (2009). Molecular decay of the tooth gene enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 5(9), e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakano M, Miwa N, Hirano A, Yoshiura KI, and Niikawa N. (2009). A strong association of axillary osmidrosis with the wet earwax type determined by genotyping of the ABCC11 gene. BMC Genetics 10(1), 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin A, Saathoff M, Kuhn F, Max H, Terstegen L, and Natsch A. (2010). A functional ABCC11 allele is essential in the biochemical formation of human axillary odor. J. Invest. Dermatol 130(2), 529–540. [DOI] [PubMed] [Google Scholar]
- 86.Sharma V, Hecker N, Walther F, Stuckas H, and Hiller M. (2020). Convergent losses of TLR5 suggest altered extracellular flagellin detection in four mammalian lineages. Mol. Biol. Evol 37, 1847–1854. [DOI] [PubMed] [Google Scholar]
- 87.Yamakoshi T, Makino T, Ur Rehman M, Yoshihisa Y, Sugimori M, and Shimizu T. (2013). Trichohyalin-like 1 protein, a member of fused S100 proteins, is expressed in normal and pathologic human skin. Biochem. Biophys. Res. Comm 432, 66–72. [DOI] [PubMed] [Google Scholar]
- 88.Makino T, Mizawa M, Yoshihisa Y, and Shimizu T. (2019). Ultraviolet B irradiation increases the expression of trichohyalin-like 1 protein in human skin xenotransplants. Clin. Exp. Dermatol 44, 773–776. [DOI] [PubMed] [Google Scholar]
- 89.Abbas Zadeh S, Mlitz V, Lachner J, Golabi B, Mildner M, Pammer J, Tschachler E, and Eckhart L. (2017). Phylogenetic profiling and gene expression studies implicate a primary role of PSORS1C2 in terminal differentiation of keratinocytes. Exp. Dermatol 26, 352–358. [DOI] [PubMed] [Google Scholar]
- 90.Sharma V, Lehmann T, Stuckas H, Funke L, and Hiller M. (2018b). Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals. PLoS Biol. 16(6), e2005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kong W, Longaker MT, and Lorenz HP (2003). Molecular cloning and expression of keratinocyte proline-rich protein, a novel squamous epithelial marker isolated during skin development. J. Biol. Chem 278, 22781–22786. [DOI] [PubMed] [Google Scholar]
- 92.Lee WH, Jang S, Lee JS, Lee Y, Seo EY, You KH, Lee SC, Nam KI, Kim JM, Kee SH, et al. (2005). Molecular cloning and expression of human keratinocyte proline-rich protein (hKPRP), an epidermal marker isolated from calcium-induced differentiating keratinocytes. J. Invest. Dermatol 125, 995–1000. [DOI] [PubMed] [Google Scholar]
- 93.Suga H, Oka T, Sugaya M, Sato Y, Ishii T, Nishida H, Ishikawa S, Fukayama M, and Sato S. (2019). Keratinocyte proline-rich protein deficiency in atopic dermatitis leads to barrier disruption. J. Invest. Dermatol 139, 1867–1875. [DOI] [PubMed] [Google Scholar]
- 94.Serhan CN, Chiang N, and Van Dyke TE (2008). Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 8, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spite M, Norling LV, Summers L, Yang R, Cooper D, Petasis NA, Flower RJ, Perretti M, and Serhan CN (2009). Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature 461, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim SN, Akindehin S, Kwon HJ, Son YH, Saha A, Jung YS, Seong JK, Lim KM, Sung JH, Maddipati KR, et al. (2018). Anti-inflammatory role of 15-lipoxygenase contributes to the maintenance of skin integrity in mice. Sci. Rep 8, 8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nery MF, Arroyo JI, and Opazo JC (2014). Increased rate of hair keratin gene loss in the cetacean lineage. BMC Genomics 15, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Endo H, Yoshida M, Nguyen TS, Akiba Y, Takeda M, and Kudo K. (2019). Three-dimensional CT examination of the forefoot and hindfoot of the hippopotamus and tapir during a semiaquatic walking. Anat. Histol. Embryol 48(1), 3–11. [DOI] [PubMed] [Google Scholar]
- 99.Kranz KR (1982). A note on the structure of tail hairs from a pygmy hippopotamus (Choeropsis liberiensis). Zoo Biol. 1, 237–241. [Google Scholar]
- 100.Thewissen JGM, Cooper LN, George JC, and Bajpai S. (2009). From land to water: the origin of whales, dolphins, and porpoises. Evo. Edu. Outreach, 2(2), 272–288. [Google Scholar]
- 101.Thewissen JGM, Cooper LN, Clementz MT, Bajpai S, and Tiwari BN (2007). Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194. [DOI] [PubMed] [Google Scholar]
- 102.Boisserie JR, Zazzo A, Merceron G, Blondel C, Vignaud P, Likius A, Mackaye HT, and Brunet M. (2005). Diets of modern and late Miocene hippopotamids: evidence from carbon isotope composition and micro-wear of tooth enamel. Palaeogeogr. Palaeoclimatol. Palaeoecol 221(1–2), 153–174. [Google Scholar]
- 103.Wolsan M, and Sato JJ (2020). Parallel loss of sweet and umami taste receptor function from phocids and otarioids suggests multiple colonizations of the marine realm by pinnipeds. J. Biogeogr 47, 235–249. [Google Scholar]
- 104.Paterson RS, Rybczynski N, Kohno N, and Maddin HC (2020). A total evidence phylogenetic analysis of pinniped phylogeny and the possibility of parallel evolution within a monophyletic framework. Front. Ecol. Evol 7, 457. [Google Scholar]
- 105.Geisler JH, and Sanders AE (2003). Molecular evidence for the phylogeny of Cetacea. J. Mamm. Evol 10, 23–27. [Google Scholar]
- 106.Boessenecker RW, Churchill M, Buchholtz EA, Beatty BL, and Geisler JH (2020). Convergent evolution of swimming adaptations in modern whales revealed by a large macrophagous dolphin from the Oligocene of South Carolina. Curr. Biol 30, 1–7. [DOI] [PubMed] [Google Scholar]
- 107.Springer MS, and Gatesy J. (2017). Inactivation of the olfactory marker protein (OMP) gene in river dolphins and other odontocete cetaceans. Mol. Phylogenet. Evol 109, 375–387. [DOI] [PubMed] [Google Scholar]
- 108.Geisler J. (2019). Whale evolution: dispersal by paddle and fluke. Curr. Biol 29, R280–R299. [DOI] [PubMed] [Google Scholar]
- 109.Lambert O, Bianucci G, Salas-Gismondi R, Di Celma C, Steurbaut E, Urbina M, and de Muizon C. (2019). An amphibious whale from the middle Eocene of Peru reveals early South Pacific dispersal of quadrupedal cetaceans. Curr. Biol 29, 1352–1359. [DOI] [PubMed] [Google Scholar]
- 110.Wheeler PE (1985). The loss of functional body hair in man: the influence of thermal environment, body form and bipedality. J. Hum. Evol 14(1), 23–28. [Google Scholar]
- 111.Ruxton GD, and Wilkinson DM (2011). Avoidance of overheating and selection for both hair loss and bipedality in hominins. Proc. Natl. Acad. Sci. USA 108(52), 20965–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meredith RW, Zhang G, Gilbert MTP, Jarvis ED, Springer MS (2014). Evidence for a single loss of mineralized teeth in the common avian ancestor. Science 346, 6215. [DOI] [PubMed] [Google Scholar]
- 113.Springer MS, Emerling CA, Fugate N, Patel R, Starrett J, Morin PA, Hayashi C, and Gatesy J. (2016). Inactivation of cone-specific phototransduction genes in rod monochromatic cetaceans. Front. Ecol. Evol 4, 61. [Google Scholar]
- 114.Springer MS, Starrett J, Morin PA, Lanzetti A, Hayashi C, and Gatesy J. (2016). Inactivation of C4orf26 in toothless placental mammals. Mol. Phylogenet. Evol 95, 34–45. [DOI] [PubMed] [Google Scholar]
- 115.Gaudry MJ, Jastroch M, Treberg JR, Hofreiter M, Paijmans JL, Starrett J, Wales N, Signore AV, Springer MS, and Campbell KL (2017.) Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv 3(7), e1602878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jebb D, and Hiller M. (2018). Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. Elife 7, e38906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Themudo GE, Alves LQ, Machado AM, Lopes-Marques M, da Fonseca RR, Fonseca M, Ruivo R, and Castro LFC (2020). Losing genes: the evolutionary remodeling of Cetacea skin. Front. Mar. Sci 7, 592375. [Google Scholar]
- 118.Sharma V, and Hiller M. (2017). Increased alignment sensitivity improves the usage of genome alignments for comparative gene annotation. Nuc. Acids Res 45(14), 8369–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meyer WK, Jamison J, Richter R, Woods SE, Partha R, Kowalczyk A, Kronk C, Chikina M, Bonde RK, Crocker DE, and Gaspard J. (2018). Ancient convergent losses of Paraoxonase 1 yield potential risks for modern marine mammals. Science, 361(6402), 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. (2018). Ensembl 2018. Nuc. Acids Res 46(D1), D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, and Drummond A. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Swofford DL (2002). PAUP*: phylogenetic analysis using parsimony (*and other methods). 4.0b10 ed Sunderland (Massachusetts): Sinauer Associates, Inc. [Google Scholar]
- 124.Stamatakis A, Ludwig T, and Meier H. (2005). RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21(4), pp.456–463. [DOI] [PubMed] [Google Scholar]
- 125.Schull JK, Turakhia Y, Dally WJ, and Bejerano G. (2019). Champagne: Whole-genome phylogenomic character matrix method places Myomorpha basal in Rodentia. bioRxiv 803957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Z, 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- 127.Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, and Murphy WJ (2011). Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. [DOI] [PubMed] [Google Scholar]
- 128.Hassanin A, Delsuc F, Ropiquet A, Hammer C, Van Vuuren BJ, Matthee C, Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT, and Couloux A. (2012). Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. Comptes Rendus Biologies 335, 32–50. [DOI] [PubMed] [Google Scholar]
- 129.McGowen MR, Tsagkogeorga G, Álvarez-Carretero S, dos Reis M, Struebig M, Deaville R, Jepson PD, Jarman S, Polanowski A, Morin PA, and Rossiter SJ (2020). Phylogenomic resolution of the cetacean tree of life using target sequence capture. Syst. Biol 69, 479–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sandby-Moller J, Poulsen T, and Wulf HC (2003). Epidermal thickness at different body sites: relationship to age, gender, pigmentation, blood content, skin type and smoking habits. Acta Derm. Venereol 83(6), 410–413. [DOI] [PubMed] [Google Scholar]
- 131.Chopra K, Calva D, Sosin M, Tadisina KK, Banda A, De La Cruz C, Chaudhry MR, Legesse T, Drachenberg CB, Manson PN, and Christy MR (2015). A comprehensive examination of topographic thickness of skin in the human face. Aesthet. Surg. J 35(8), 1007–1013. [DOI] [PubMed] [Google Scholar]
- 132.Hole MB, Bhosle NS, and Kapadnis PJ (2008). Study of hair follicles in red kandhari cows. Indian J. Animal Res 42(2), 151–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3: Estimated inactivation dates for pseudogenized skin genes, related to Figure 3.
Data Availability Statement
The Illumina sequencing data generated for this study are available under NCBI BioProject PRJNA694317.