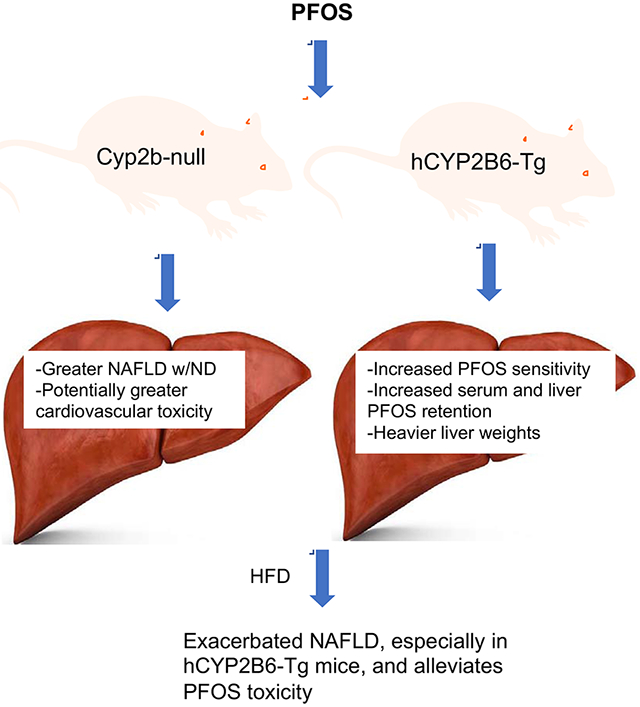
Abstract
PFOS is a persistent, fluorosurfactant used in multiple products. Murine Cyp2b’s are induced by PFOS and high-fat diets (HFD) and therefore we hypothesized that human CYP2B6 may alleviate PFOS-induced steatosis. Cyp2b-null and hCYP2B6-Tg mice were treated with 0, 1, or 10mg/kg/day PFOS by oral gavage for 21-days while provided a chow diet (ND) or HFD. Similar to murine Cyp2b10, CYP2B6 is inducible by PFOS. Furthermore, three ND-fed hCYP2B6-Tg females treated with 10mg/kg/day PFOS died during the exposure period; neither Cyp2b-null nor HFD-fed mice died. hCYP2B6-Tg mice retained more PFOS in serum and liver than Cyp2b-null mice presumably causing the observed toxicity. In contrast, serum PFOS retention was reduced in the HFD-fed hCYP2B6-Tg mice; the opposite trend observed in HFD-fed Cyp2b-null mice. Hepatotoxicity biomarkers, ALT and ALP, were higher in PFOS-treated mice and repressed by a HFD. However, PFOS combined with a HFD exacerbated steatosis in all mice, especially in the hCYP2B6-Tg mice with significant disruption of key lipid metabolism genes such as Srebp1, Pparg, and Hmgcr. In conclusion, CYP2B6 is induced by PFOS but does not alleviate PFOS toxicity presumably due to increased retention. CYP2B6 protects from PFOS-mediated steatosis in ND-fed mice, but increases steatosis when co-treated with a HFD.
Keywords: Perflourooctane sulfonate (PFOS), Non-alcoholic fatty liver disease (NAFLD), Cytochrome P450, CYP2B6, hepatotoxicity, polyunsaturated fatty acids (PUFAs)
Graphical Abstract
1. Introduction
Per and Polyfluoroalkylsubstances (PFAS) such as perflurooctanesulfonic acid (PFOS) are used in stain repellents, varnishes, cleaning products, semiconductors, and firefighting foams (Development, 2006). PFOS was phased out of production in the United States (US) and Europe for most uses; however, it is still manufactured in China. PFOS is persistent and bioaccumulates in the environment (Yamashita et al., 2005), and therefore is found in surface, ground, and drinking water (Chang et al., 2016; Development, 2006). PFOS is also persistent in humans with a half-life of approximately 5.4 years (Olsen et al., 2007), and in turn is present in the majority of US citizens where obesity related illnesses are the leading causes of death (Gleason et al., 2015; Heron, 2019). It is detected in the blood of 98% of Americans with serum PFOS concentrations of approximately 1,000 ng/mL in occupationally exposed personnel and 9-30 ng/mL in the general US population (Frisbee et al., 2009; Gleason, et al., 2015; Kato et al., 2011; Olsen et al., 2012; Olsen, et al., 2007).
Exposure to PFOS and other PFAS chemicals is associated with elevated serum markers of hepatotoxicity in rodents and humans (Das et al., 2017; Xu et al., 2016). PFOS administration is known to decrease body weight, increase liver weight, and increase hepatic steatosis in mice (Fletcher et al., 2013; Lau et al., 2007; Wang et al., 2014). This is accompanied by decreased serum glucose, triglycerides, and cholesterol in mice (Bijland et al., 2011; Wang, et al., 2014). Some of these effects may be caused by PFOS activation of peroxisome proliferator activated receptors (PPARs) that play vital roles in the regulation of lipid metabolism, distribution, energy homeostasis, and adipocyte maturation (Corton et al., 2014; Huck et al., 2018; Klaunig et al., 2012; Xu, et al., 2016). PFOS has also been shown to induce hepatic steatosis by interfering with key processes such as lipid synthesis, lipid hepatic export process, and β-oxidation (Cheng et al., 2016). Steatosis, which is also associated with cardiovascular disease (Berlanga et al., 2014), can develop into non-alcoholic steatohepatitis (NASH) after a “second hit”. PFOS and other PFAS chemicals may be part of that second hit as they are associated with an increase in NASH in rodents and humans, including children (Jin et al., 2020).
In addition to modulating the expression of genes that encode for lipid metabolism and transporters, Cyp2b10 is one of the most highly induced transcripts in PFOS treated mice and hepatocytes (Abe et al., 2017; Pfohl et al., 2021; Rosen et al., 2010). PFOS moderately activates CAR and PXR in mice (Rosen, et al., 2010) and induces murine Cyp2b9 and Cyp2b10 (Das, et al., 2017). PFOS also induces human CYP2B6 in vitro but this has not been verified in vivo in humans or in a humanized model (Rosen et al., 2013).
Perturbations in several CYPs have been shown to increase NAFLD, including Cyp3a members, Cyp2b members, Cyp2j members, and Cyp2e1 (Heintz et al., 2019; Kumar et al., 2018; Olona et al., 2018; Seth et al., 2014). CYP2B6 is the only CYP2B member in humans. CYP2B6 metabolizes a large array of pharmaceuticals, environmental chemicals, steroids, and polyunsaturated fatty acids (PUFA’s) (Desta et al., 2017; Hodgson et al., 2007; Wang et al., 2008), including pesticides, polyaromatic hydrocarbons, and industrial chemicals such as chlorpyrifos, benzo(a)pyrene, and styrene (Hodgson, et al., 2007; Kim et al., 1997; Tang et al., 2001). CYP2B6 is regulated by the Pregnane X-receptor (PXR), Constitutive androstane receptor (CAR), and Forkhead box A2 (FoxA2); all crucial xenobiotic and/or metabolic transcription factors (Hashita et al., 2008; Kretschmer et al., 2005).
Our laboratory has recently developed a Cyp2b-null mouse lacking the primarily hepatic Cyp2b9, 10, and 13 genes found in tandem repeat (Heintz, et al., 2019; Kumar et al., 2017). In addition, we recently developed a humanized CYP2B6-Tg (hCYP2B6-Tg) mouse on the Cyp2b-null background to reduce some of the uncertainty when extrapolating from mice to humans. The hCYP2B6-Tg mouse will allow the human CYP2B6 gene to be studied in vivo and provide a better understanding of the role it plays in PFOS toxicity and the co-treatment of PFOS and a high-fat diet (HFD). Cyp2b9/10/13-null mice are diet-induced obese in males (Heintz, et al., 2019); however, these effects are not observed in females and in fact Cyp2b-null females are protected from NASH (Heintz et al., 2020). Cyp2b9 and Cyp2b10 are highly inducible in the liver by HFD, and in several cases Cyp2b9 was the most highly induced gene following a HFD (Heintz, et al., 2019; Hoek-van den Hil et al., 2015; Leung et al., 2016). Whether CYP2B6 is induced by a HFD is not known.
Because murine Cyp2b members are highly induced by PFOS, the purpose of this study was to determine if human CYP2B6 is induced by PFOS and/or a HFD, to test whether CYP2B6 alleviates the development of PFOS-mediated NAFLD, and determine whether the combination of a HFD and PFOS (similar to the condition of many global and US citizens) increases the hepatotoxicity of PFOS and escalates PFOS-induced NAFLD. To determine these aims, we compared male and female mice treated with 0, 1, or 10 mg/kg/day PFOS in both in Cyp2b9/10/13-null (Cyp2b-null) and hCYP2B6-Tg mice. This study will help identify CYP2B6’s and a HFD’s role in PFOS-mediated hepatotoxicity and NAFLD.
2. Materials and Methods
2.1. Animals and Diet:
All studies were reviewed and approved by Clemson University’s Institution Animal Care and Use Committee (IACUC). Housing and studies were performed in a controlled environment with a temperature of 24 ± 2°C, 60-70% relative humidity, and a 12:12 h light:dark cycle. C57BL/6J-Cyp2b10em1(Del(7Cyp2b10-Cyp2b9))Fatso/Mmnc mice, often called Cyp2b9/10/13-null or abbreviated as Cyp2b-null mice, lack the murine genes Cyp2b9, Cyp2b10, and Cyp2b13 on a C57Bl/6J (B6) background. Cyp2b-null mice were developed using Crispr/Cas9 to delete a 287 kb region of chromosome 7 (Kumar, et al., 2017). Humanized CYP2B6/2A13/2F1-transgenic mice on the Cyp2b-null background were generated by breeding CYP2B6/2A13/2F1-transgenic mice generated previously (Wei et al., 2012) to our Cyp2b-null mice in order to generate Cyp2b-null mice containing the human BAC clone of CYP2B6/2A13/2F1 to what we abbreviate as hCYP2B6-Tg mice (Heintz, 2020).
Mice were genotyped from extracted genomic DNA isolated from tails using the QuantaBio (Beverly, MA USA) AccuStart II Mouse Genotyping Kit according to the manufacturer’s instructions. Each mouse was genotyped to confirm that the Cyp2b9/10/13 cluster on murine chromosome 7 was deleted using the F2/R2 primer set: (F2: 5’-gccagggtcagcatattcaccaa-3’/ R2: 5’-gcacagacatcatgaggttctggtg-3’; 59°C) to confirm the deletion followed by a Cyp2b13 specific primer set (F1: 5’-cagactcttgttagaccggaccat-3’ /R1: 5’-ccccaaggaataaaattctacatg-3’; 59°C) to ensure the mice were not heterozygous (Kumar, et al., 2017). hCYP2B6-2A13-Tg primer set (F1: 5’-cctggacagatgcctttaactccg-3’ / R1: 5’-tggctttgcacctgcctgact-3’; 63°C) then confirmed the presence of the human BAC clone containing CYP2B6 and the CYP2B6/2A13/2F1 P450 cluster on human chromosome 19 (Wei, et al., 2012).
Mice, 10-12 weeks old, were separated into 19 different groups based on their genotype, PFOS (Sigma-Aldrich, St. Louis, MO, USA) dose, gender, and diet (Table 1). PFOS, dissolved in a solution of water and 0.5% Tween 20, was administered by gavage each morning for three weeks. Mice were either fed a normal diet (ND; 2018S-Envigo Teklad Diet, Madison, WI) consisting of 18% kcal of fat, or a high-fat diet (HFD; Envigo, TD.06414 Adjusted calorie diet) consisting of 60.3% kcal of fat. Water was provided ad libitum. Male Cyp2b-null male mice were not provided a HFD in conjunction with PFOS because fewer Cyp2b-null male pups were born than female pups during this round of breeding. The hCYP2B6-Tg mice provided a HFD and 1 mg/kg/day PFOS were also eliminated because of a lack of available humanized mice after breeding (Table 1).
Table 1:
Description of the different treatment groups by gender, diet and genotype with replicate numbers in parenthesis.
| Males | Females | ||||||
|---|---|---|---|---|---|---|---|
| Normal Diet | High Fat Diet | Normal Diet | High Fat Diet | ||||
| Cyp2b-null | hCYP2B6 | Cyp2b-null | hCYP2B6 | Cyp2b-null | hCYP2B6 | Cyp2b-null | hCYP2B6 |
| 0 mg/kg PFOS (n = 6) | 0 mg/kg PFOS (n = 7) | 0 mg/kg PFOS (n = 5) | 0 mg/kg PFOS (n = 5) | 0 mg/kg PFOS (n = 7) | 0 mg/kg PFOS (n = 5) | 0 mg/kg PFOS (n = 6) | |
| 1 mg/kg PFOS (n = 6) | 1 mg/kg PFOS (n = 7) | 1 mg/kg PFOS (n = 5) | 1 mg/kg PFOS (n = 7) | 1 mg/kg PFOS (n = 5) | |||
| 10 mg/kg PFOS (n = 5) | 10 mg/kg PFOS (n = 7) | 10 mg/kg PFOS (n = 5) | 10 mg/kg PFOS (n = 5) | 10 mg/kg PFOS (n = 7) | 10 mg/kg PFOS (n = 5) | 10 mg/kg PFOS (n = 5) | |
2.2. Necropsy:
Mice were weighed prior to euthanasia. Euthanasia was performed by heart puncture to collect blood while under 3% isoflurane anesthesia followed by CO2 asphyxiation. Whole blood was kept at room temperature for 30 minutes to allow for clotting and then centrifuged at 6,000 rpm for 10 minutes to collect serum. Liver and gonadal and perirenal visceral white adipose tissue (WAT) were harvested and weighed. Liver was diced into several pieces for lipid sampling, RNA extraction, protein preparation, and histology preparation. Samples were snap frozen in liquid nitrogen prior to storage at −80°C.
2.3. Serum Lipids and Liver Cholesterol Screening:
Serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), total bilirubin, direct bilirubin, blood urea nitrogen (BUN), cholesterol, creatine kinase (CK), creatinine, gamma-glutamyl transferase (GGT), glucose, lactate dehydrogenase (LDH), calcium, magnesium, phosphorous, albumin, total protein, triglycerides, and total carbon dioxide (CO2) were measured using a Beckmann-Coulter AU480 analyzer with the appropriate Beckmann-Coulter kits according to the manufacturer’s instructions (Heintz, et al., 2019). Serum HDL and LDL/VLDL cholesterol was determined using the HDL and LDL/VLDL Colorimetric Quantitation Assay from Sigma Aldrich (St. Louis, MO) according to the manufacturer’s instructions. Total liver cholesterol was determined using the Cholesterol/ Cholesteryl Ester Quantitation Assay Kit from Abcam (Cambridge, MA USA) according to the manufacturer’s instructions. Each measured serum and liver parameters were statistically compared across groups using GraphPad Prism 7.0 (La Jolla, CA USA). Principle component analysis (PCA) was performed using RStudio to associate serum and liver parameters with PFOS dose, diet, or genotype.
2.4. Principal Component Analysis (PCA) biplot:
Two PCA biplots were generated, male and female, to compare differences in groups based on diet, PFOS, and genotype using 19 different parameters. The PCA biplots were generated using the data visualization package, ggplot2, in RStudio. Treatment groups were color coded and overlaid on each of the plots, respectively. A normal data ellipse was drawn for each group, a scale factor of 2 was applied to observations, and a scale factor of 2 was applied to variables. An aspect ratio of 1 was used to make the plots square. The x-axis (PC1) and y-axis (PC2) show the percent of explained variability amongst the data.
2.5. Oil Red O Staining:
Liver samples (n=3) from each group were sectioned and stained with Oil Red O according to standard protocol (Dong et al., 2009). Stained slides were imaged (400X magnification) on a Leica Acquire Light Microscope and analyzed using ImageJ Fiji Particle Analysis to quantify the lipids and triglycerides in the liver (Schindelin et al., 2012). Briefly, the scale bar was set to 0.05 mm and the threshold color was set to red, with red pass selected and green/blue pass deselected. The wavelength was set to 120-255 and the range of the size (mm2) was set to 0.00001-infinity for all images as described previously (Heintz, et al., 2020).
2.6. RNA Quantification and quantitative Real-time Polymerase Chain Reaction (qPCR):
RNA was isolated in TRIzol (Invitrogen, Carlsbad, CA USA) according to the manufacturer’s instructions, initially quantified on a NanoDrop, and stored at −80°C. DNA contamination was eliminated with the Turbo DNA-Free Kit (Invitrogen, Carlsbad, CA). RNA was then re-quantified with the Qubit RNA BR Assay (Invitrogen, Carlsbad, CA) using the Qubit 4 Fluorometer (Invitrogen, USA) for improved accuracy. qPCR was performed using primers specific for Srebp1, Cpt1a, CYP2B6, Pparγ, Cd68, Cyp2a5, Cyp4a14, Hmgcr, CYP2A13, 18S and Gapdh (Supplemental Table 1). 1 μL of cDNA was mixed and incubated with 12.5 μL RT2 SYBR Green (Qiagen Frederick, MD USA), 9.5 μL Millipore water, 1 μL of the forward and reverse primers (25 μL per well). All plates were heated to 95°C for 1 minute followed by 50 cycles of denaturation at 95°C for 30 seconds, annealing for 30 seconds (temperature varied for each primer; Supplemental Table 1), and elongation for 45 seconds at 72°C. To determine the melt curve, samples were heated to 76.5°C for 5 seconds and then 95°C for 5 minutes. Efficiency of the reaction was determined with a standard curve performed in triplicate with a mix of samples diluted from 1:1 - 1:1024. Samples were diluted 1:20 and fluorescence measured using a Bio-Rad CFX96 Real-Time System. Gene expression was quantified and normalized to the geometric means of 18S and Gapdh as the housekeepers using the inverted Muller’s equation (Muller et al., 2002; Roling et al., 2004).
2.7. Western Blot:
Livers were individually homogenized with a Dounce Homogenizer and microsomes isolated by differential centrifugation (Van der Hoeven et al., 1974). Protein concentrations were determined using the Bradford Protein Assay according to the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). Western blots were performed to determine CYP2B6 and CYP2A protein expression with 25 μg of microsomal protein. Briefly, samples were separated via gel electrophoresis, transferred to a Bio-Rad PVDF (Hercules, CA USA), and quantified using antibodies (diluted 1:1000) specific to CYP2B6 (Chemicon International, Temecula, CA USA), CYP2A (Thermo-Fisher Scientific, Rockford, IL USA), with β-actin (Sigma Aldrich, St. Louis, MO) as the reference protein (Hernandez et al., 2006). An alkaline phosphatase conjugated goat anti-rabbit IgG (Bio-Rad) secondary antibody (diluted 1:500) was used to identify the CYP2B6 and CYP2A primary antibodies. Goat anti-mouse IgG (Bio-Rad) secondary antibody (diluted 1:500) was used to recognize β-actin primary antibody. Individual proteins were detected with the chemiluminescent Immun-Star AP detection kits from Bio-Rad, and band intensities were quantified on a Bio-Rad ChemiDoc Imaging System with iLab.
2.8. Vivid (Blue) CYP2B6 Enzyme Inhibition Assay:
PFOS inhibition of CYP2B6 activity was determined and compared to nonylphenol using the Vivid CYP2B6 Blue Screening Kit from Life Technologies (Carlsbad, CA USA).
2.9. Liver PFOS Extractions:
Liver PFOS was extracted using a slightly modified previously published method (Chang et al., 2017). Frozen liver tissues (~50 mg) was homogenized in 2 mL Omni Hard Tissue Homogenizing tubes containing 1.4 mm ceramic beads, with 400 μL cold, deionized water spiked with a fixed amount of a stable isotope-labeled internal standard (13C4-PFOS, Wellington Laboratories, Ontario, Canada, Product code: MPFOS). Using an Omni Bead Ruptor Elite (Omni International, Kennesaw, GA), the mixture was homogenized for 30 seconds at 4 m/s. 250 μL of homogenate was digested overnight at room temperature in 10% 1N KOH. 100 μL of digested homogenate was further treated with 100 μL of 2N HC1, 500 μL 1N formic acid, 500 μL of saturated ammonium sulfate, and 5 mL methyl tert-butyl ether (MTBE). The solution was mixed on a shaker (20-30 min at room temperature). The organic and aqueous layers were separated by centrifugation (2500 × g, 5 min), and an exact volume of MTBE (4.5 mL) was removed from the solution. The top organic layer was subsequently transferred to a new tube and evaporated. The resulting sample was reconstituted with 20 mL of acetonitrile and water (1:1) prior to LC-MS/MS analysis.
2.10. Serum PFOS Extraction:
Serum collected at necropsy was prepared according to methods described by Hansen (Hansen et al., 2001). Briefly, 20 μL of sera, 2 μL of isotope-labeled internal standard (13C4-PFOS, Wellington Laboratories, Ontario, Canada, Product code: MPFOS), 200 μL of 0.5M tetrabutylammonium bisulfate (TBA; adjusted to pH 10), and 400 μL of 0.25M sodium carbonate were added to a 15-mL polypropylene tube, and thoroughly mixed. 5 mL of MTBE was added to the solution, and the mixture was placed on a shaker for 20-30 min at room temperature. The organic and aqueous layers were separated by centrifugation (2500 × g, 5 min), and an exact volume of MTBE (4.5 mL) was removed from the solution. The top organic layer was subsequently transferred to a new tube and evaporated overnight. The resulting sample was reconstituted with 20 mL of acetonitrile and water (1:1) prior to LC-MS/MS analysis
2.11. PFOS Quantification by LC-MS MS:
Liver and serum samples were vortexed for 30 s and passed through a 0.2 μm polyethersulfone membrane syringe filter (MDI Membrane Technologies, Harrisburg, PA) into an autosampler vial. Ultra-fast liquid chromatography (UFLC) was performed on a SHIMADZU Prominence UFLC system consisting of three LC-20AD pumps, a DGU-20A degassing unit, SIL-20AC autosampler, CTO-20AC column oven and CBM-20A communication bus module (Columbia, MD). Chromatographic separation was performed on a Waters XBridge® C18 column (100 mm X 4.6 mm i.d., 5 μm, Milford, MA). The mobile phase consisted of 0.1% (v/v) formic acid/water (A) and 0.1% (v/v) formic acid/acetonitrile (B) with a gradient elution of 70% of B increased to 90% of B over 8 min; at 8 min the gradient was revered to original conditions. The column temperature was 40°C, flow rate was 0.6000 mL/min, and the injection volume was 10.00 μL. Mass spectrometry was performed on the QTRAP 4500 system coupled with an electrospray ionization (ESI) interface (AB Sciex, Framingham, MA). Nitrogen was used in all cases. The parameters were optimized as follows: negative ionization, IonSpray voltage, −4500; nebulizer gas, 40; auxiliary heater gas, 45; curtain gas, 20; turbo gas temperature, 400; declustering potential, −60; entrance potential, −10; collision energy, −122; collision cell exit potential, −15. The samples were analyzed in MRM (Multiple Reaction Monitoring) mode. The MRM ion pair used for PFOS quantification was 498.9/79.8 and compared to a standard curve prepared in either liver or serum matrix to account for matrix effects. The data were acquired using Analyst 1.6.3 software and processed using MultiQuant 3.0.1 software.
2.12. Tests of Significance:
Tests of significance were performed using GraphPad Prism software 7.0 & 8.0 (LaJolla, CA USA). Student’s t-tests were performed when comparing two groups, and one-way ANOVAs followed by Fisher’s LSD as the post-hoc test were performed when comparing more than two groups. A p-value < 0.05 was considered significantly different.
3. Results
3.1. PFOS toxicity and effects on tissue, and body mass:
Unexpectedly, greater toxicity was observed in the ND-fed hCYP2B6-Tg female mice given 10 mg/kg PFOS (10-PFOS) than other groups. Of the 110 mice in the study, only three mice, all female treated with 10-PFOS and fed a ND, died during the 21-day exposure. Two died on day 20 and the third died on day 21 at the onset of the necropsies. Neither males nor HFD-fed females provided 10-PFOS died indicating increased sensitivity in females compared to males and HFD-mediated protection from lethality (Fig. 1).
Fig. 1: Survival curve of female mice treated with different concentrations of PFOS via oral gavage.
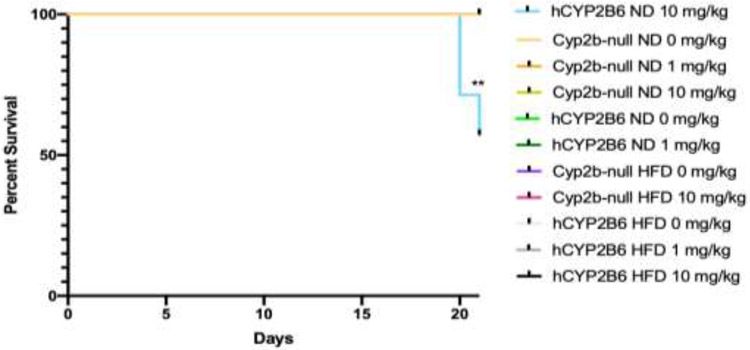
This survival curve was generated using GraphPad Prism 8. Statistical differences in survival were determined by Gehan-Breslow-Wilcoxon test where ** indicates p < 0.001 (n = 5-7).
PFOS increased liver mass and decreased body mass in a dose-dependent manner, especially at 10 mg/kg PFOS. Weight loss can be in part attributed to the significantly reduced white adipose tissue in these mice (Table 2 & 3). The addition of a HFD increased liver, body, and WAT weights. PFOS negated the gains in HFD-mediated body and WAT weight, but had little effect on liver weight with the exception of the 1-PFOS dose in females. Furthermore, the mean liver weight of hCYP2B6-Tg male mice fed a ND and treated with 10-PFOS was 10% heavier than their Cyp2b-null counterparts (p < 0.0001) (Table 2), suggesting increased hepatic sensitivity to PFOS in the humanized mice.
Table 2:
Body weights, Liver weights, and WAT weights of Male mice.
| Diet | Genotype | PFOS (mg/kg) |
Body Weight (g) | Liver Weight (g) | WAT Weight (g) |
|---|---|---|---|---|---|
| Normal Diet (ND) | Cyp2b-null | 0 | 22.58±0.54p**g** | 0.958±0.024p*** | 0.335±0.014p |
| 1 | 23.77±0.70g*** | 1.372±0.038g*** | 0.388±0.045 | ||
| 10 | 18.11±0.37p**g | 1.820±0.12p***b | 0.0382±0.025p | ||
| hCYP2B6 | 0 | 23.20±0.55p**g**d** | 0.927±0.022p***g | 0.426±0.058pd*** | |
| 1 | 23.52±0.50g*** | 1.350±0.027g*** | 0.310±0.025 | ||
| 10 | 18.51±0.81p**g* | 2.004±0.067p***gd*b | 0.0714±0.022pd* | ||
| High Fat Diet (HFD) | 0 | 27.55±1.87p***gd** | 1.066±0.10p*** | 1.358±0.290p***gd*** | |
| 10 | 20.29±0.70p***g** | 2.250±0.070p***g**d* | 0.260±0.051p***d* |
Data represented as mean +/− SEM. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test.
‘p’ indicates difference between PFOS concentration
‘g’ indicates difference between gender (see Table 3)
‘d’ indicates difference between diet
‘b’ indicated difference between genotype
Letter with no asterisk indicates p < 0.05
indicates p < 0.01
indicates p < 0.001
indicates p < 0.0001.
Table 3:
Body weights, Liver weights, and WAT weights of Female mice
| Diet | Genotype | PFOS (mg/kg) |
Body Weight (g) | Liver Weight (g) | WAT Weight (g) |
|---|---|---|---|---|---|
| Normal Diet (ND) | Cyp2b-null | 0 | 18.37±0.34p*g**d** | 0.79±0.042p*** | 0.212±0.036p*d** |
| 1 | 18.15±0.65g*** | 0.956±0.035g*** | 0.156±0.037 | ||
| 10 | 15.15±0.78p*g | 1.69±0.039p***d* | 0.024±0.016p*d* | ||
| hCYP2B6 | 0 | 18.42±0.23p*g**d*** | 0.730±0.028p***gd** | 0.199±0.022p*d*** | |
| 1 | 17.38±0.23g***d*** | 0.854±0.060g***d*** | 0.1471±0.014d*** | ||
| 10 | 14.68±0.29p*g*d* | 1.763±0.20p***g | 0.025±0.025p*d* | ||
| High Fat Diet (HFD) | Cyp2b-null | 0 | 22.76±0.93p***d** | 0.902±0.036p*** | 0.868±0.15p***d* |
| 10 | 16.09±0.69p*** | 1.95±0.105p***d* | 0.178±0.030p***d* | ||
| hCYP2B6 | 0 | 24.87±1.12p***gd*** | 1.028±0.033p***d** | 1.00±0.240p***gd*** | |
| 1 | 23.32±1.63d*** | 1.23±0.074d*** | 0.988±0.290d*** | ||
| 10 | 16.15±0.27p***g**d* | 1.92±0.030p***g** | 0.150±0.021p***d* |
Data represented as mean +/− SEM. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test.
‘p’ indicates difference between PFOS concentration
‘g’ indicates difference between gender
‘d’ indicates difference between diet
‘b’ indicated difference between genotype
Letter with no asterisk indicates p < 0.05
indicates p < 0.01
indicates p < 0.001
indicates p < 0.0001.
3.2. CYP2B6 inhibition and induction:
PFOS is a strong inhibitor of CYP2B6 in vitro with an IC50 of 165 nM (95% CI: 104 - 255 nM) (Fig. 2). PFOS shows similar, but slightly lower, potency than nonylphenol (IC50 = 77.3 nM; 95% CI: 57.5 – 103 nM), which was used as a positive control (Acevedo et al., 2005). Thus, PFOS is a CYP2B6 inhibitor in vitro and may act as an inhibitor in vivo in humans because its IC50 is within measured serum and liver concentrations of some exposed individuals and these concentrations are associated with adverse effects (Baldwin et al., 2020; Tanner et al., 2018).
Fig. 2: Concentration-response curve for the inhibition of CYP2B6 by PFOS in comparison to the positive control, nonylphenol.
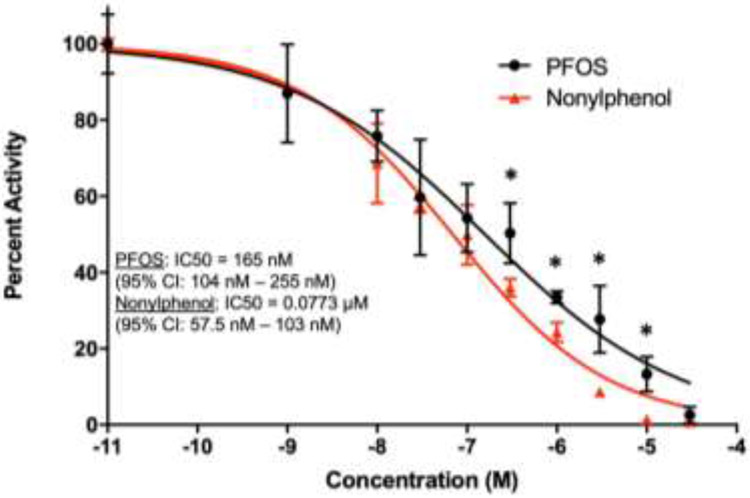
IC50 and 95% CI values were determined using a sigmoidal dose response least squares fit test performed on GraphPad Prism 7 (n = 3). Multiple Student t-test’s were performed to determine significance between PFOS and Nonylphenol. * indicates a significant difference of p < 0.05.
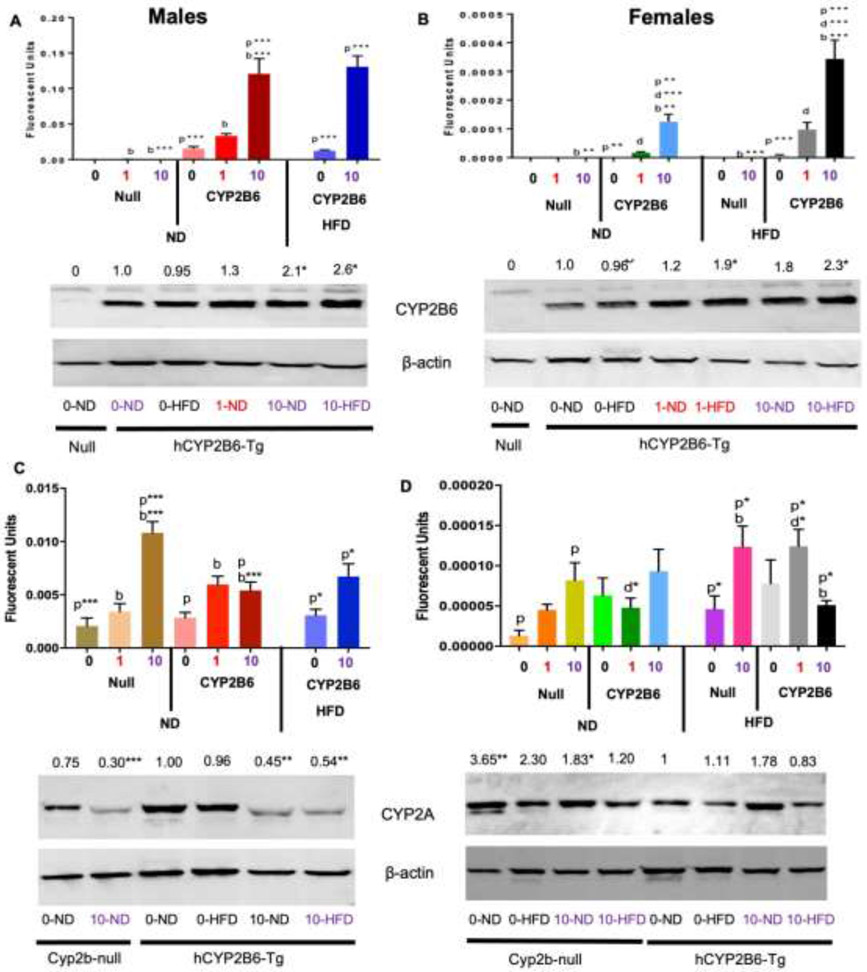
Previous research has indicated that PFOS is a potent murine Cyp2b inducer (Cheng et al., 2012; Das, et al., 2017; Rosen et al., 2017). PFOS has been shown to induce CYP2B6 in vitro (Rosen, et al., 2013); however whether it is an inducer of human CYP2B6 in vivo is in question. This transgenic model contains the CAR and FoxA2 upstream promoter and enhancer regions of CYP2B6 (Wei, et al., 2012). CYP2B6 gene expression was significantly induced by PFOS in both male (7.8X) and female (163X) mice fed a ND as determined by qPCR (Fig. 3ab). Western blots confirmed dose-dependent CYP2B6 induction at the protein level in males and females, including HFD-mediated CYP2B6 induction in the females (Fig. 3ab) although induction was much lower than measured by qPCR. In addition, HFD did exacerbate PFOS-mediated CYP2B6 gene expression in female mice (2.7X) when compared to their ND counterparts (Fig. 3b). However, HFD alone did not induce CYP2B6 gene expression although previous work has demonstrated incredibly high murine Cyp2b9/10 induction by a HFD in mice (Heintz, et al., 2019). In summary, PFOS induces CYP2B6 in a hCYP2B6-Tg model that contains the CYP2B6 human promoter and enhancer regions (Wei, et al., 2012).
Fig. 3: CYP2B6 (A & B) and Cyp2a5 (C & D) hepatic gene and protein expression in male and female mice.
Mice were treated as described in the Materials and Methods and CYP2B6 and Cyp2a5 expression measured by qPCR (n = 4-5) and Western blotting (n = 2-3). ). The values above the Western blots reflect relative change in protein expression compared to 0-PFOS ND. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD when comparing 3 or more groups (qPCR) and Student’s t-test when comparing two groups (Western blots). For qPCR, ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets, ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001. * indicates a significant difference of p < 0.05 in Western blots.
CYP2A13 along with CYP2F1 were both a part of the BAC clone used to generate the original CYP2B6/2A13/2F1-Tg mouse. Both CYP2A13 and CYP2F1 are predominantly expressed in the respiratory tract and previous research indicates they are not expressed in the liver (Wei, et al., 2012). qPCR was performed to confirm human CYP2A13 is not expressed in liver because members of the CYP2A subfamily are known to metabolize xenobiotics and endogenous lipids (Lu et al., 2006). Human CYP2A13 expression was not detected in the liver by qPCR (data not shown), but genomic CYP2A13 is detected from tail snips demonstrating that our assay worked and there was no CYP2A13 hepatic expression. Murine Cyp2a5 was also measured because it is the predominant CYP2A enzyme in adult murine liver, Cyp2a5 and several other Cyp2a members were induced in Cyp2b-null mice or a HFD in comparison to WT mice, and it is orthologous to the human CYP2A6 hepatic enzyme (Abu-Bakar et al., 2012; Heintz, et al., 2019). Cyp2a5 gene expression was increased by 5.3x and 6.3x in ND-fed Cyp2b-null male and female mice, respectively, treated with 10-PFOS compared to Cyp2b-null controls (0-PFOS), with a 2.7X increase in HFD-fed Cyp2b-null mice provided 10-PFOS compared to HFD-fed 0-PFOS Cyp2b-null mice as well. Cyp2a5 gene expression increased 1.9x and 2.2x in ND- and HFD-fed hCYP2B6-Tg male mice treated with 10-PFOS compared to their 0-PFOS counterparts (Fig. 3cd). Cyp2a5 gene expression was not induced by 10-PFOS in hCYP2B6-Tg female mice. In contrast, PFOS repressed male protein expression of CYP2A with slight induction in the Cyp2b-null females (Fig. 3cd). In summary, human CYP2A13 is not expressed in murine livers. Murine Cyp2a5 compensates for a lack of Cyp2b expression in null mice, is responsive to PFOS by qPCR, strongly repressed by PFOS in males, and repressed by a HFD in females.
3.3. Retention of PFOS in hCYP2B6 mice:
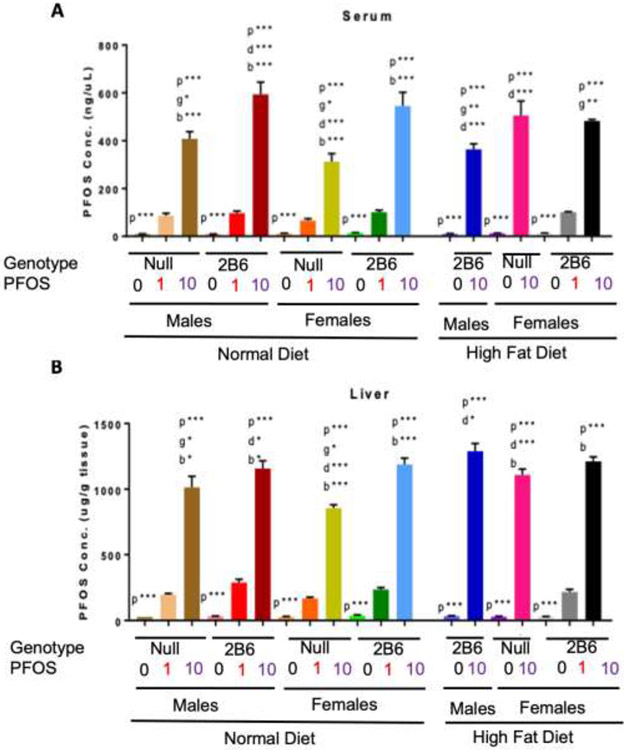
Serum and liver PFOS concentrations were determined by LC-MS/MS analysis and demonstrated concentration-dependent increases. hCYP2B6-Tg mice showed significantly increased PFOS retention compared to Cyp2b-null mice in males and females treated with 10-PFOS and fed a ND (Fig. 4ab). Average serum PFOS was 1.46X greater in ND-fed hCYP2B6-Tg male mice than ND-fed Cyp2b-null male mice, and 1.74X greater in ND-fed hCYP2B6-Tg female mice than identically treated Cyp2b-null female mice (Fig. 4a). hCYP2B6-Tg mice fed a HFD showed a significant decrease in PFOS retention, unlike the Cyp2b-null mice. In humanized mice, the average serum PFOS level was 1.63X lower in HFD-fed male mice than their ND-fed counterparts and 1.13X lower in HFD-fed female mice than their ND-fed counterparts. In contrast, in Cyp2b-null female mice the average serum PFOS level was 1.62X greater in the HFD-fed mice than ND-fed mice at 10 mg/kg/day (Fig. 4a).
Fig. 4: LC-MS/MS analysis of PFOS concentrations in the serum (A) and liver (B) of hCYP2B6 and Cyp2b-null mice.
Data are presented as mean ± SEM. Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test (n=4-5). ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets, ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001.
Liver PFOS retention followed a similar pattern to serum PFOS retention with greater PFOS retention in ND-fed hCYP2B6-Tg male and female mice compared to their Cyp2b-null counterparts (1.14X and 1.39X, respectively) (Fig. 4b). HFD increased hepatic PFOS retention within all groups with the most significant increase in the Cyp2b-null female mice compared to their ND-fed counterparts (1.3X) (Fig. 4b). This data suggests that a HFD may play a protective role in PFOS toxicity in hCYP2B6-Tg mice but not Cyp2b-null mice. The only groups of mice that showed lower serum PFOS coupled with higher liver PFOS were 10-PFOS treated mice provided a HFD. Typically, hCYP2B6-Tg ND-fed mice treated with 10-PFOS showed higher serum and liver PFOS. The greater PFOS retention measured in humanized mice was unexpected and may explain the decreased survival in the hCYP2B6-Tg female mice fed a ND (Fig. 1).
3.4. Serum markers of liver toxicity are increased by PFOS and reduced by HFD cotreatment:
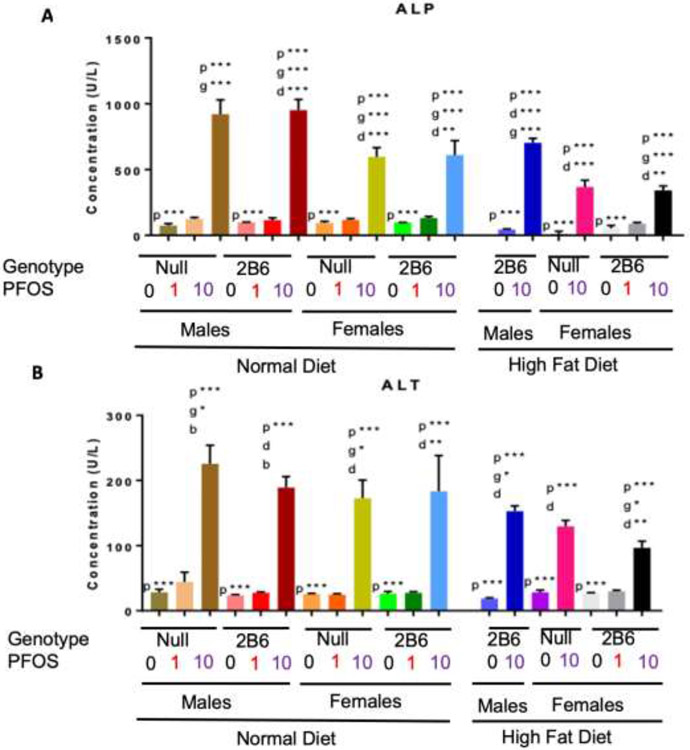
Alkaline phosphatase (ALP) and alanine aminotransferase (ALT), which are indicative of liver damage, were both significantly increased by PFOS in a dose-dependent manner (Fig. 5ab). Genotype only had an effect on ALT levels in 10-PFOS treated hCYP2B6-Tg males compared to Cyp2b-null males where the presence of CYP2B6 was protective from liver damage (Fig. 5b) despite greater measured PFOS concentrations (Fig. 4). Interestingly, ALP and ALT levels were decreased in HFD-fed mice treated with 10-PFOS compared to their ND-fed counterparts (Fig. 5). These data indicate that a HFD decreases PFOS-mediated hepatotoxicity in hCYP2B6-Tg and Cyp2b-null mice despite greater PFOS retention in HFD-fed hCYP2B6-Tg male and Cyp2b-null females.
Fig. 5: Serum concentrations of Alkaline Phosphatase (ALP) (A) and Alanine Aminotransferase (ALT) (B) from PFOS-treated mice.
Data are presented as mean ± SEM. Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test (n = 5). ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001.
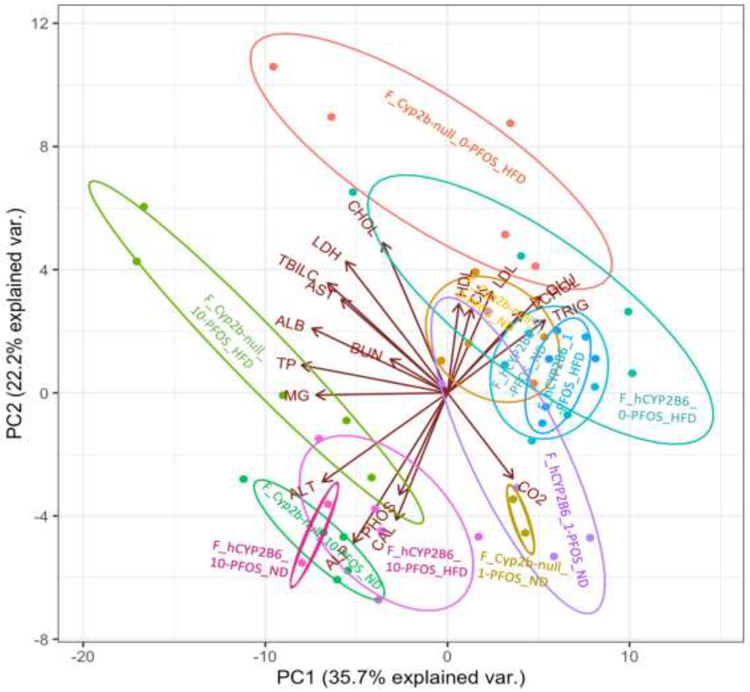
3.5. PCA biplot investigating associations between key serum parameters in different treatment groups:
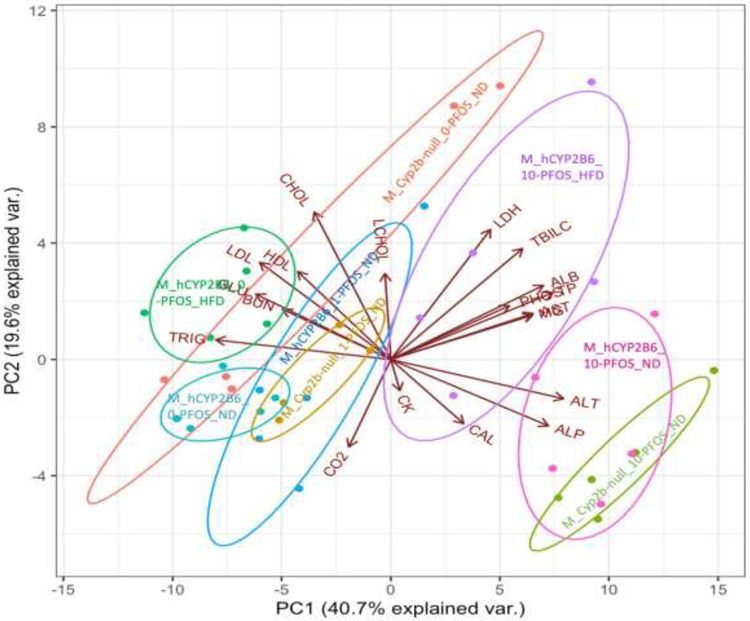
Other serum parameters are also perturbed by PFOS, diet or genotype, including glucose, triglycerides, total cholesterol, LDL/VLDL and HDL, and LDH (Supplementary Figures 1-5). Therefore, PCA biplots examining associations between 19 different parameters were performed to compare relationships among treatment groups in males and females. In both males and females, the biplots account for approximately 60% of the variation amongst the data. The male mice treated with 0-PFOS or 1-PFOS grouped together skewing towards triglycerides (TAG) and glucose (GLU) (Fig. 6). The ND-fed male mice treated with 10-PFOS grouped towards biomarkers indicative of hepatotoxicity, ALT and ALP (bottom right of plot). The HFD-fed hCYP2B6-Tg male mice treated with 10-PFOS grouped more towards the 0-PFOS or 1-PFOS treated groups when compared to their 10-PFOS counterparts, consistent with data that a HFD is playing a protective role against PFOS toxicity in hCYP2B6-Tg male mice (Fig. 6).
Fig. 6: Principal component analysis (PCA) biplot composed of 19 different parameters compared across the 8 male groups.
Graphical representation of multivariate data. Treatment groups are color coded and the group descriptions are overlaid on the plot.
The female mice treated with 0-PFOS or 1-PFOS also grouped together with TAG and GLU. HFD-fed Cyp2b-null female mice, regardless of PFOS concentration, showed lots of variation with several mice weighted toward cardiovascular toxicity biomarkers, cholesterol and lactate dehydrogenase (top left of plot), suggesting that Cyp2b-null female mice may be at an increased risk of cardiovascular disease when fed a HFD (Fig. 7). All of the female mice treated with 10-PFOS, regardless of diet, grouped separately from the female mice treated with 0-PFOS or 1-PFOS and towards biomarkers indicative of liver damage such as ALT and ALP (bottom left of plot) (Fig. 7) with surprisingly little variation between genotypes given the greater PFOS concentrations and toxicity observed in the hCYP2B-Tg females treated with 10-PFOS. Of course, serum was not collected from the mice that died and therefore their data is not reflected in the PCA biplots. However, parameters associated with toxicity are reduced in female HFD-fed Cyp2b-null mice compared to ND-fed Cyp2b-null female mice (Fig. 7).
Fig. 7: Principal component analysis (PCA) biplot composed of 19 different parameters compared across the 11 female groups.
Graphical representation of multivariate data. Treatment groups are color coded and the group descriptions are overlaid on the plot.
3.6. Diet effects PFOS and CYP2B6-mediated increases in hepatic triglycerides:
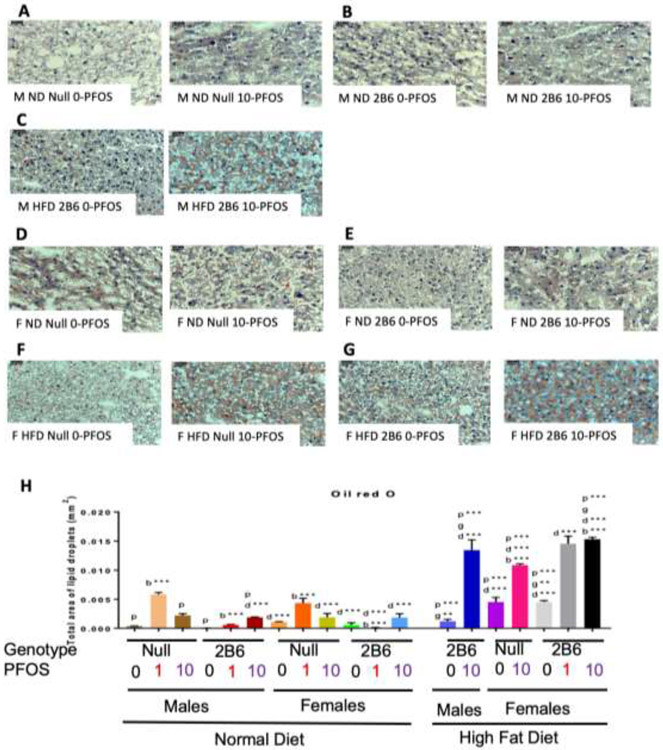
Oil Red O staining demonstrates an increase in liver triglyceride concentrations in the ND-fed male and female Cyp2b-null mice treated with 1-PFOS compared to their hCYP2B6-Tg counterparts (10.33X and 50.4X, respectively) (Fig. 8). This indicates a prominent role for CYP2B6 in protection from PFOS-mediated fatty liver disease. HFD increased liver triglyceride concentrations in every group except hCYP2B6-Tg male mice treated with 0-PFOS, indicating CYP2B6’s role in protection from NAFLD. PFOS addition exacerbated NAFLD progression significantly in all exposed groups (Fig. 8). In contrast to a ND, hCYP2B6-Tg female mice fed a HFD and treated with 10-PFOS had 1.41X greater triglyceride accumulation in the liver compared to their Cyp2b-null counterparts (Fig. 8h). Greater toxicity observed in the hCYP2B6-Tg mice may play a role. In summary, ND-fed Cyp2b-null mice had a greater accumulation of hepatic triglycerides than their hCYP2B6-Tg counterparts treated with 1-PFOS; however, HFD-fed Cyp2b-null mice had significantly less liver triglycerides than their hCYP2B6-Tg counterparts treated with 10-PFOS (Fig. 8), indicating the progression of NAFLD is slowed by the presence of CYP2B6 in a ND but exacerbated by a HFD in this group.
Fig. 8: Triglyceride content in livers as measured by Oil Red O.
Male ND Cyp2b-null 0-PFOS (A), Male ND hCYP2B6 0-PFOS (B), Male HFD hCYP2B6 0-PFOS (C), Female ND Cyp2b-null 0-PFOS (D), Female ND hCYP2B6 0-PFOS (E), Female HFD Cyp2b-null 0-PFOS (F), Female HFD hCYP2B6 0-PFOS (G), and quantified using Image J Fiji Particle Analysis (H). Images were taken at 400x (0.05mm) magnification. Data are presented as mean ± SEM (n = 3). Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test. ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets, ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001.
3.7. qPCR:
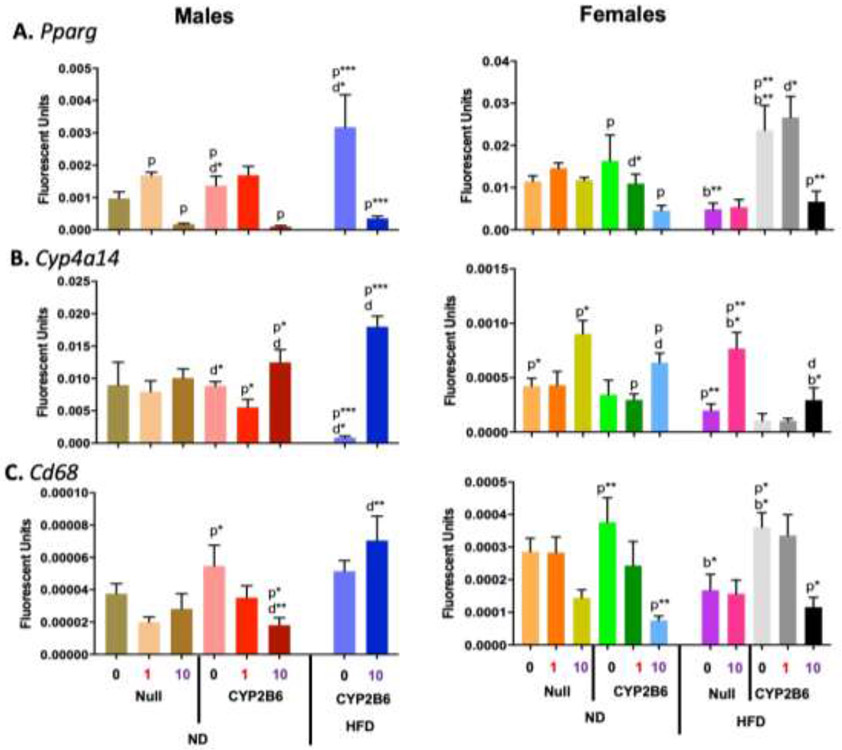
qPCR was performed to identify expression changes in genes associated with peroxisome proliferation (Pparγ, Cyp4a14) (Wafer et al., 2017; Zhang et al., 2017), inflammation (Cd68) (Chistiakov et al., 2017), and lipid metabolism (Hmgcr; Cpt1a; Srebp1) (Jiang et al., 2018; Ruiz et al., 2014; Schlaepfer et al., 2020). Pparγ expression was significantly repressed in all PFOS-treated male mice and hCYP2B6-Tg female mice (Fig. 9a), suggesting that PFOS is interfering with adipogenesis. Interestingly, Pparγ was significantly increased in females by a HFD, but only in hCYP2B6-Tg mice; not Cyp2b-null mice. Cyp4a14 gene expression was increased in all groups treated with 10-PFOS except ND-fed Cyp2b-null male mice and HFD-fed hCYP2B6-Tg female mice. Cyp4a14 expression was significantly induced by the combination of PFOS and a HFD in hCYP2B6-Tg male mice (21.5X) and Cyp2b-null female mice (3.9X). However, PFOS-treated hCYP2B6-Tg female mice showed significantly less induction than correspondingly treated Cyp2b-null mice suggesting less PPARα activation in the transgenic mice (Fig. 9b).
Fig. 9: Changes in hepatic gene expression caused by genotype, PFOS, HFD, or a combination of PFOS and a HFD in male and female mice.
Gene expression of Pparg (A), Cyp4a14 (B), and Cd68 (C) in male and female mice was determined by qPCR as described in Material and Methods. Data are presented as mean ± SEM (n = 4-5). Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test. ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets, ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001.
Because Cyp4a14 is regulated by Pparα and has been associated with NAFLD (Zhang, et al., 2017), this data indicates that PFOS activates Pparα and promotes the progression of NAFLD, which is corroborated by the Oil Red O stains. Cd68 gene expression was repressed in all of the humanized mice treated with 10-PFOS except for the HFD-fed hCYP2B6-Tg male mice (Fig. 9c). PFOS had no significant effect on Cd68 gene expression in Cyp2b-null mice; suggesting that although PFOS is hepatotoxic, there may be less inflammatory signaling in the liver when exposed to PFOS in the presence of the human CYP2B6 gene or untreated hCYP2B6-Tg mice are more prone to diet-induced inflammation.
Several lipid metabolism genes were also investigated by qPCR. Hmgcr gene expression in 10-PFOS-treated hCYP2B6-Tg mice was down-regulated 60% in HFD-fed hCYP2B6-Tg male mice, 93% in ND-fed hCYP2B6-Tg female mice, and 88% in HFD-fed hCYP2B6-Tg female mice (Fig. 10a). These results suggest that PFOS may inhibit the metabolism of cholesterol in the liver of humanized mice potentially leading to greater NAFLD (Jiang, et al., 2018) consistent with HFD-PFOS cotreatments. Cpt1a gene expression was lower in all groups treated with 10-PFOS except for the HFD-fed Cyp2b-null female mice (Fig. 10b). Cpt1a gene expression was significantly repressed in all humanized groups except HFD-fed male mice potentially because they show greater constitutive expression. Because Cpt1a is essential for fatty acid mitochondrial transport and oxidation (Schlaepfer, et al., 2020), this data suggests that PFOS inhibits lipid metabolism and the humanized mice are more affected PFOS although they may also contain higher constitutive expression. Humanized male and female Srebp1 gene expression was induced by a HFD (2.3X and 2.1X, respectively) and negated by 10-PFOS (1.8X and 1.6X, respectively) (Fig. 10c). Srebp1 was also down-regulated by PFOS, but in a different manner depending on the genotype in females. This data is consistent with Srebp1’s role in lipogenesis, TAG synthesis, and NAFLD (Ruiz, et al., 2014), especially following a HFD and in the HFD-fed hCYP2B6-Tg mice whom are the most sensitive to NAFLD. Srebp1 was also associated with loss of NAFLD in ND-fed PFOS treated mice.
Fig. 10: Changes in hepatic gene expression caused by genotype, PFOS, HFD, or a combination of PFOS and a HFD in male and female mice.
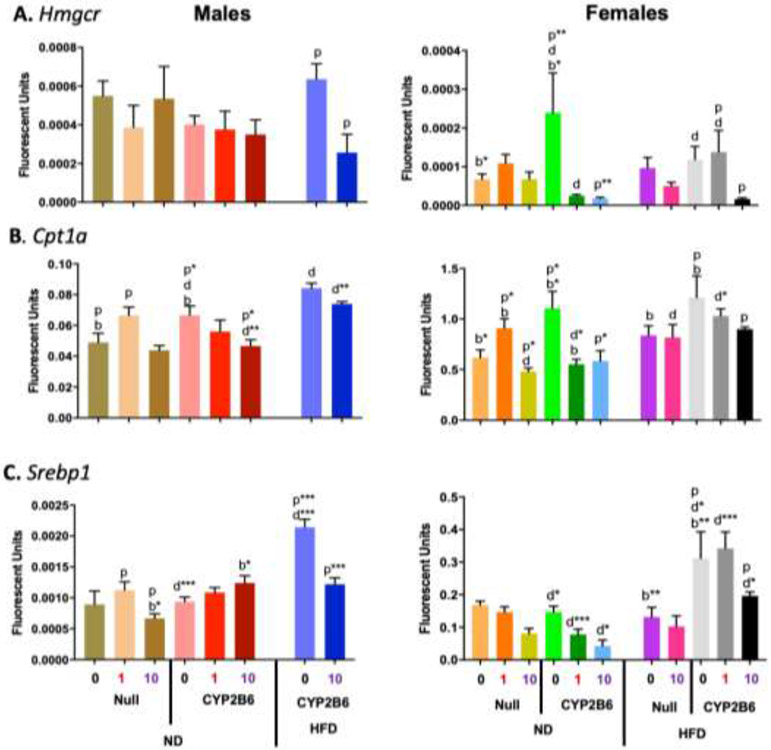
Gene expression of Hmgcr (A), Cpt1a (B), and Srebp1 (C) in male and female mice was determined by qPCR as described in Material and Methods. Data are presented as mean ± SEM (n = 4-5). Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test. ‘p’ indicates difference between PFOS concentrations, ‘g’ indicates difference between gender, ‘d’ indicates difference between diets, ‘b’ indicated difference between genotypes. A letter indicates a p < 0.05, letter w/ * indicates p < 0.01, letter w/ ** indicates p < 0.001 and a letter w/ *** indicates p < 0.0001.
4. Discussion
The purposes of this study were to determine if PFOS induces human CYP2B6, and test whether CYP2B6 protects from PFOS toxicity and NAFLD. We were able to address these questions and examine the adverse effects of co-treatment of PFOS and a HFD. First, CYP2B6 is inducible by PFOS. Surprisingly, female humanized mice are significantly more sensitive to PFOS toxicity than their Cyp2b-null counterparts, probably due to increased PFOS retention in hCYP2B6-Tg mice. However, sublethal measurements, such as NAFLD, were more sensitive in Cyp2b-null mice than hCYP2B6-Tg mice unless the mice were treated with a HFD in which case hCYP2B6-Tg female mice were more sensitive to NAFLD as demonstrated by Oil Red O and several biomarkers of NAFLD.
Toxicity was significantly greater in hCYP2B6-Tg females than Cyp2b-null females. Three ND-fed hCYP2B6-Tg female mice exposed to 10-PFOS died and these were the only mice to die during the 3 week exposure period (Fig. 1) clearly demonstrating the sensitivity of ND-fed hCYP2B6-Tg mice to PFOS. The increased toxicity in the hCYP2B6-Tg mice is most likely due to increased PFOS retention and the subsequent effects caused by greater PFOS. LC-MS/MS analysis showed that serum and liver PFOS was significantly greater in the hCYP2B6-Tg mice compared to their counterparts when fed a ND; more so in female mice (174% and 139%, respectively) than male mice (146% and 114%, respectively).
Interestingly, their HFD-fed counterparts survived (Fig. 1), indicating that a HFD was protective from lethality probably due to the increased adiposity. PFOS causes wasting and loss of adipose tissue potentially by activating uncoupling proteins in the mitochondria (Shabalina et al., 2015), mimicking fatty acids, or antagonizing PPARγ and in turn inhibiting fat accumulation in white adipose tissue (Wen et al., 2016). We observed decreased HFD and CYP2B6-mediated PPARγ expression by qPCR suggestive of decreased activity. Excess fatty acids could delay the lethal effects of the lipid depleting PFOS. However, differences in transport expression and decreased uptake may also play a role in hCYP2B6-Tg mice (Pfohl, et al., 2021). HFD significantly decreased serum PFOS concentrations in all hCYP2B6-Tg mice. In contrast, a HFD led to a significant increase in serum and liver PFOS retention in female Cyp2b-null mice compared to their ND-fed counterparts, nevertheless hCYP2B6-Tg female mice still had greater liver PFOS retention than their Cyp2b-null counterparts.
PFOS induced human CYP2B6 in male and female mice (Fig. 3ab). These results confirm that CYP2B6 is inducible by PFOS in a dose dependent manner similar to murine Cyp2b10 (Rosen, et al., 2010). HFD exacerbated human CYP2B6 induction in females (Fig. 3b); however, a HFD alone was not a CYP2B6 inducer. Murine Cyp2b9 and Cyp2b10 induction by a HFD has been observed previously (Heintz, et al., 2019; Hoek-van den Hil, et al., 2015; Leung, et al., 2016), potentially due to activation of CAR by PUFAs (Finn et al., 2009), or activation of FoxA2 due to insulin resistance (Wolfrum et al., 2004), glucagon or glucocorticoid signaling (Zhang et al., 2005). CYP2B6 protein induction was moderate in comparison to changes in RNA levels. Overall, it is unlikely that CYP2B6 directly regulates PFOS metabolism, retention, or elimination as most PFOS is not metabolized but retained and then eliminated slowly as parent compound through the gastrointestinal tract and kidney (Alexander et al., 2008; Cui et al., 2010). Therefore, increased PFOS retention in hCYP2B6-Tg mice is most likely due to repression of the expression of transporters, but this has not been confirmed.
PFOS also inhibited CYP2B6 with an IC50 of 165 nM (95% CI of 104 – 255 nM), which is approximately 82.5 ng/ml (95% CI: 52-127 ng/ml). Most PFOS is found in the serum and retained in the liver (Cui, et al., 2010; Kim et al., 2016). In this study, serum concentration typically averaged about 400-600 μg/ml and liver concentrations typically averaged about 1000-1250 μg/g in the 10-PFOS group and about 6X lower in the 1-PFOS group. These concentrations are much higher than the IC50 and would likely inhibit most CYP2B6 activity. Liver PFOS concentrations from autopsy specimens of random human samples vary from 4.5 to 57 ng/g in the USA (Olsen et al., 2003), and from below detection to 402 ng/g in southern Spain with a mean of 102 ng/g and a median of 42 ng/g PFOS (Perez et al., 2013). Fishing subsistent Gullah populations on the South Carolina coast have serum PFOS concentrations that vary from 5 – 155 ng/ml with a mean of 53 ng/ml. Serum PFOS concentrations are dropping in the Gullah population and estimated half-life of PFOS was 6.0 years (Gribble et al., 2015). Last, the Hudson River and NHANES studies showed serum PFOS concentrations from below detection to 217 and 425 ng/g, respectively, with geometric means of 34.3 and 27.5 ng/g (Tanner, et al., 2018). This indicates that many people are harboring PFOS concentrations that can significantly inhibit CYP2B6 metabolism based on our data although for most individuals this inhibtion would be less than 50%. However, it should be considered that other PFAS chemicals are also present (Baldwin, et al., 2020; Gribble, et al., 2015; Tanner, et al., 2018) and they most likely contribute to CYP2B6 inhibition and the adverse effects of PFAS.
Hepatic CYP2A gene and protein expression was determined to ensure that one of the human genes from the original construct, CYP2A13 (Wei, et al., 2012), was not being expressed in the liver. qPCR confirmed that CYP2A13 is not expressed in the liver in hCYP2B6-Tg mice confirming previous results (Wei, et al., 2012). Cyp2a5 is the predominant hepatic CYP2A member in adult mice and has been shown to be female predominant (Lu, et al., 2006; Poça et al., 2017). Cyp2a5 gene expression increased significantly in all of the Cyp2b-null groups (Fig. 3cd), potentially as a compensatory mechanism (Kumar, et al., 2017). However, CYP2A protein expression was repressed in male mice exposed to PFOS regardless of genotype and in Cyp2b-null female mice (Fig. 3cd). Based on these results, neither CYP2A13 or other CYP2A members are playing roles in the increased PFOS retention and toxicity observed in hCYP2B6-Tg mice.
PFOS exhibits severe toxicity on different tissues and metabolic pathways, which was evident by biomarkers from the serum panel. Hepatotoxicity was observed in the groups exposed to 10-PFOS based on significantly elevated ALP and ALT levels (Fig. 5). However, increased toxicity caused by higher PFOS serum and liver concentrations did not always manifest themselves as higher serum markers of tissue toxicity. ALT and ALP levels indicate greater liver toxicity in ND-fed mice than HFD-fed mice even though hepatic PFOS concentrations were higher in the HFD mice and these mice showed much greater NAFLD (Fig. 8). PCA indicates a clearly protective effect of a HFD from PFOS toxicity in male mice, but not female mice (Fig. 6-7). This protective effect was highly pronounced in the hCYP2B6-Tg male mice as this group was found between the 0- to 1-PFOS groups and the other 10-PFOS groups.
PCA also indicates an increase in markers indicative of poor cardiovascular health such as cholesterol, triglycerides, LDH, and LDL in Cyp2b-null mice compared to hCYP2B6 mice. This may be due to differences in fatty acid metabolism as CYP2B6 and other CYPs are involved in PUFA metabolism (Bishop-Bailey et al., 2014; Heintz, et al., 2019; Nelson et al., 2004). There is overlap between the genotypes in the PCA plot; however, lack of Cyp2b in both males and females pushes the groups towards these adverse cardiovascular parameters and in some cases it is clear that the combination of a Cyp2b-null genotype and a HFD are deleterious to some parameters such as LDH and cholesterol (SM 3-4). Last, serum glucose and triglyceride levels were significantly lower in groups treated with 10-PFOS compared to 0-PFOS suggesting PFOS interferes with gluconeogenesis and fatty acid esterification as published previously (Das, et al., 2017; Hagenaars et al., 2008) (SM 1-2).
PFOS increases NAFLD. Previous work has demonstrated that male Cyp2b-null mice are more susceptible to NAFLD than wildtype mice (Heintz, et al., 2019). Similarly, ND-fed Cyp2b-null male and female mice were more susceptible to 1-PFOS mediated NAFLD than hCYP2B6-Tg mice. However, the combination of PFOS and a HFD potentiated NAFLD, especially in hCYP2B6-Tg mice in comparison to Cyp2b-null mice (Fig. 8), which is somewhat surprising. Recent results investigating the effects of a HFD provided for 16 weeks on hCYP2B6-Tg mice show that CYP2B6 is an anti-obesity CYP; however, the presence of CYP2B6 increased NAFLD while still protecting male mice from diabetes (Heintz, et al., 2020). Female hCYP2B6-Tg mice were also more susceptible to the HFD-PFOS co-treatment than Cyp2b-null mice. New data suggests that CYP2B6 is not protective from NAFLD and possibly increases NAFLD susceptibility through increased production of oxylipins (Deol et al., 2017; Heintz, et al., 2020). These results suggest a complicated role of CYP2B6 in PUFA metabolism, but only during a HFD in which hepatic PUFA concentrations are high and PFOS exacerbates this effect.
NAFLD progression is associated with decreases in Cpt1a (Fig. 10), a key gene involved in mitochondrial fatty acid uptake prior to β-oxidation in the liver (Schlaepfer, et al., 2020). There was a significant decrease of relative Cpt1a gene expression in HFD-fed hCYP2B6-Tg mice treated with 10-PFOS compared to their 0-PFOS counterparts. LDL/VLDL levels in the serum were also significantly lower in PFOS treated groups (SM 5; Fig 6-7). A decrease in Cpt1a gene expression and oxidation coupled with lower LDL production and release may explain the exacerbated increase in hepatic triglyceride content in the liver. These findings support other studies that have suggested PFOS induced steatosis by inhibiting mitochondrial β-oxidation and decreasing LDL content that, normally, would help transport triglycerides out of the liver (Cheng, et al., 2016).
Srebp1, Pparγ, and to a lesser extent Cd68 all followed similar gene expression profiles following HFD or PFOS treatments with increased expression in HFD-fed hCYP2B6-Tg mice in comparison to HFD-fed Cyp2b-null mice (Fig. 9-10). Srebp1 is a key regulator of lipogenesis and therefore it is not surprising that it is associated with NAFLD (Moslehi et al., 2018). Srebp1 also regulates Pparγ (Fajas et al., 1999) and Cd68 expression (Jump et al., 2013). Expression of Hmgcr, the rate determining step in cholesterol biosynthesis (Jiang, et al., 2018), was slightly higher in hCYP2B6-Tg mice and repressed by PFOS which is consistent with the liver and serum cholesterol data (Supplementary Figures 3,5) (Supplementary Material 6). The greater expression of these genes in the hCYP2B6-Tg mice when fed a HFD may help explain their sensitivity to NAFLD.
PPARα and PPARγ are important in maintaining energy homeostasis and lipid metabolism (Corton, et al., 2014; Klaunig, et al., 2012; Peraza et al., 2006; Rosen, et al., 2010). PFOS is a PPARα activator, and several studies have indicated that PFOS has no activity towards PPARγ (Behr et al., 2020; Takacs et al., 2007). However, antagonism was not examined in these studies and one recent study indicates that PFOS is a PPARγ antagonist (Wen, et al., 2016), which is consistent with our observed down-regulation of Pparγ gene expression (Fig. 9). Pparγ, found in most tissues, plays a role in lipid metabolism and is responsible for adipocyte differentiation and lipid uptake into white adipose tissues (Wafer, et al., 2017; Zhang et al., 2014). This antagonism of PPARγ may help explain PFOS’s ability to cause WAT wasting and in response NAFLD.
Pparα regulates a wide variety of genes including certain CYPs such as Cyp4a14 and Cpt1a that are associated with NAFLD but overall Pparα activation is associated with preventing NAFLD (Patsouris et al., 2006; Zhang, et al., 2017). Cyp4a14 levels were increased in groups treated with 10-PFOS and exacerbated by a HFD suggesting the activation of Pparα by PFOS and diet. Interestingly, Cpt1a expression increased with diet but decreased with PFOS; Cyp4a14 was repressed by the presence of CYP2B6 in HFD-fed mice and induced by PFOS with the exception of Cyp2b-null males (Fig. 9-10). The down regulation of Pparγ activation of Cyp4a14, and down regulation of Cpt1a is consistent with data that PFOS alters lipid distribution and promotes NAFLD and in turn the liver is responding. Further, these results support the Oil Red O analysis that the HFD-fed hCYP2B6-Tg mice exposed to 10-PFOS were further along in NAFLD progression.
Human CYP2B6, is inducible in vivo by PFOS, associated with increased PFOS toxicity in ND-fed mice, and increased NAFLD when provided in combination with a HFD. Many of the effects of CYP2B6 were not protective with the exception of decreased hepatic toxicity as measured by ALT following combined PFOS – HFD exposure in hCYP2B6-Tg female mice. A HFD was protective of acute PFOS liver toxicity as measured by ALT and ALP in male and female hCYP2B6-Tg mice and the female Cyp2b-null mice. Male Cyp2b-null mice were not part of this study. In contrast, Cyp2b-null mice had increased levels of biomarkers indicative of cardiovascular disease. The increased toxicity observed in the hCYP2B6-Tg mice fed a ND is most likely a result of the increased serum and liver PFOS retention in these mice. It is unknown why there was an increase in PFOS retention in humanized mice but it is clearly a result of the presence of the CYP2B6 gene and resultant compensatory effects (Heintz, et al., 2019; Kumar, et al., 2017). Further research is needed in this area and to tease out the combined effects of a HFD and CYP2B6 on toxicity and lipid markers. In conclusion, both CYP2B6 and a HFD effect PFOS toxicity and NAFLD with CYP2B6 enhancing toxicity and enhancing NAFLD when co-treated with a HFD. Interestingly, the lack of Cyp2b (Cyp2b-null mice) enhanced NAFLD following a ND, protected female mice from overt toxicity, but not from hepatotoxicity.
Supplementary Material
Highlights.
Human CYP2B6 is inducible by PFOS in an in vivo model
hCYP2B6-Tg mice retain more PFOS than Cyp2b-nulls and in turn show greater toxicity
A high-fat diet (HFD) reduced PFOS-mediated hepatotoxicity, but increased steatosis
HFD has opposing effects on PFOS retention in Cyp2b-null and hCYP2B6 mice
CYP2B6 is protective from PFOS-mediated steatosis but not HFD-mediated steatosis
Acknowledgements:
Funding was provided by NIH grant R15ES017321 and P20GM121342 to WSB and P42ES027706 to ALS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- Abe T, Takahashi M, Kano M, Amaike Y, Ishii C, Maeda K, Kudoh Y, Morishita T, Hosaka T, Sasaki T, et al. (2017). Activation of nuclear receptor CAR by an environmental pollutant perfluorooctanoic acid. Arch Toxicol 91, 2365–2374. [DOI] [PubMed] [Google Scholar]
- Abu-Bakar A. e., Hakkola J, Juvonen R, Rahnasto-Rilla M, Raunio H, and A. Lang M (2012). Function and Regulation of the Cyp2a5/CYP2A6 Genes in Response to Toxic Insults in the Liver. Current Drug Metabolism 14(1), 137–150. [PubMed] [Google Scholar]
- Acevedo R, Villanueva H, Parnell PG, Chapman LM, Gimenez T, Gray SL, and Baldwin WS (2005). The contribution of hepatic steroid metabolism to serum estradiol and estriol concentrations in nonylphenol treated MMTVneu mice and its potential effects on breast cancer incidence and latency. J Appl Toxicol 25, 339–353. [DOI] [PubMed] [Google Scholar]
- Alexander J, Auðunsson GA, Benford D, Cockburn A, Cravedi J-P, Dogliotti E, Di Domenico A, Fernández-Cruz ML, Fink-Gremmels J, Fürst P, et al. (2008). Perfluorooctane sulfonate (PFOS), perfluorooctanoic acid (PFOA) and their salts: Scientific Opinion of the Panel on Contaminants in the Food chain1 (Question No EFSA-Q-2004-163). The EFSA Journal 653, 1–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, Bain LJ, Di Giulio R, Kullman S, Rice CD, Ringwoo AH, and van den Hurk P (2020). 20th pollutant responses in marine organisms (PRIMO): Global issues and fundamental mechanisms caused by pollutant stress in marine and freshwater organisms. Aquat Toxicol 227, 105620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr A-C, Plinsch C, Braeuning A, and Buhrke T (2020). Activation of human nuclear receptors by perfluoroalkylated substances (PFAS). Toxicol In Vitro 62, 104700. [DOI] [PubMed] [Google Scholar]
- Berlanga A, Guiu-Jurado E, Porras JA, and Auguet T (2014). Molecular pathways in non-alcoholic fatty liver disease. Clin Exp Gastroenterol 7, 221–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijland S, Rensen PCN, Pieterman EJ, Maas ACE, Van Der Hoorn JW, Van Erk MJ, Havekes LM, Willems Van Dijk K, Chang S-C, Ehresman DJ, et al. (2011). Perfluoroalkyl Sulfonates Cause Alkyl Chain Length-Dependent Hepatic Steatosis and Hypolipidemia Mainly by Impairing Lipoprotein Production in APOE*3-Leiden CETP Mice. Toxicol Sci 123(1), 290–303. [DOI] [PubMed] [Google Scholar]
- Bishop-Bailey D, Thomson S, Askari A, Faulkner A, and Wheeler-Jones C (2014). Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism. Annu Rev Nutr 34, 261–279. [DOI] [PubMed] [Google Scholar]
- Chang ET, Adami HO, Boffetta P, Wedner HJ, and Mandel JS (2016). A critical review of perfluorooctanoate and perfluorooctanesulfonate exposure and immunological health conditions in humans Crit Rev Toxicol 46, 279–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Allen BC, Andres KL, Ehresman DJ, Falvo R, Provencher A, Olsen GW, and Butenhoff JL (2017). Evaluation of Serum Lipid, Thyroid, and Hepatic Clinical Chemistries in Association With Serum Perfluorooctanesulfonate (PFOS) in Cynomolgus Monkeys After Oral Dosing With Potassium PFOS. Toxicol Sci 156(2), 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Lv S, Nie S, Liu J, Tong S, Kang N, Xiao Y, Dong Q, Huang C, and Yang D (2016). Chronic perfluorooctane sulfonate (PFOS) exposure induces hepatic steatosis in zebrafish. Aquat Toxicol 176, 45–52. [DOI] [PubMed] [Google Scholar]
- Cheng X, and Klaassen CD (2012). Perfluorocarboxylic Acids Induce Cytochrome P450 Enzymes in Mouse Liver through Activation of PPAR-α and CAR Transcription Factors. Toxicol Sci 125(1), 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, and Bobryshev YV (2017). CD68/macrosialin: not just a histochemical marker. Laboratory Investigation 97, 4–13. [DOI] [PubMed] [Google Scholar]
- Corton JC, Cunningham ML, Hummer BT, Lau C, Meek B, Peters JM, Popp JA, Rhomberg L, Seed J, and Klaunig JE (2014). Mode of action framework analysis for receptor-mediated toxicity: The peroxisome proliferator-activated receptor alpha (PPARα) as a case study. Crit Rev Toxicol 44(1), 1–49. [DOI] [PubMed] [Google Scholar]
- Cui L, Liao C-Y, Zhou Q-F, Xia T-M, Yun Z-J, and Jiang G-B (2010). Excretion of PFOA and PFOS in male rats during a subchronic exposure. Arch Environ Contam Toxicol 58(1), 205–213. [DOI] [PubMed] [Google Scholar]
- Das KP, Wood CR, Lin MT, Starkov AA, Lau C, Wallace KB, Corton JC, and Abbott BD (2017). Perfluoroalkyl acids-induced liver steatosis: Effects on genes controlling lipid homeostasis. Toxicology 378, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol P, Fahrmann J, Yang J, Evans JR, Rizo A, Grapov D, Salemi M, Wanichthanarak K, Fiehn O, PHinney B, et al. (2017). Omega-6 and omega-3 oxylipins are implicated in soybean oil induced obesity in mice. Sci Rep 7, 12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z, and Flockhart DA (2017). Pharmacogenetics of Drug Metabolism. In Clinical and Translational Science: Principles of Human Research (Robertson D, and Williams G, Eds.) doi: 10.1016/B978-0-12-802101-9.00018-1, pp. 327–345. Elsevier Inc., Elsevier. [DOI] [Google Scholar]
- Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, et al. (2009). Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A 106(44), 18831–18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, Martin G, Fruchart J-C, Briggs M, Spiegelman BM, and Auwerx J (1999). Regulation of Peroxisome Proliferator-Activated Receptor γ Expression by Adipocyte Differentiation and Determination Factor 1/Sterol Regulatory Element Binding Protein 1: Implications for Adipocyte Differentiation and Metabolism. Molecular and Cellular Biology 19(8), 5495–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Henderson CJ, Scott CL, and Wolf CR (2009). Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J 417, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher T, Galloway TS, Melzer D, Holcroft P, Cipelli R, Pilling LC, Mondal D, Luster M, and Harries LW (2013). Associations between PFOA, PFOS and changes in the expression of genes involved in cholesterol metabolism in humans. Environment International 57-58, 2–10. [DOI] [PubMed] [Google Scholar]
- Frisbee SJ, Brooks AP, Maher A, Flensborg P, Arnold S, Fletcher T, Steenland K, Shankar A, Knox SS, Pollard C, et al. (2009). The C8 health project: Design, methods, and participants. Environmental Health Perspectives 117(12), 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JA, Post GB, and Fagliano JA (2015). Associations of perfluorinated chemical serum concentrations and biomarkers of liver function and uric acid in the US population (NHANES), 2007-2010. Envirol Res 136, 8–14. [DOI] [PubMed] [Google Scholar]
- Gribble MO, Bartell SM, Kannan K, Wu Q, Fair PA, and Kamen DL (2015). Longitudinal measures of perfluoroalkyl substances (PFAS) in serum of Gullah African Americans in South Carolina: 2003–2013. Environ Res 143, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars A, Knapen D, Meyer IJ, van der Ven K, Hoff P, and De Coen W (2008). Toxicity evaluation of perfluorooctane sulfonate (PFOS) in the liver of common carp (Cyprinus carpio). Aquatic Toxicology 88(3), 155–163. [DOI] [PubMed] [Google Scholar]
- Hansen KJ, Clemen LA, Ellefson ME, and Johnson HO (2001). Compound-specific, quantitative characterization of organic fluorochemicals in biological matrices. Environmental Science and Technology 35(4), 766–770. [DOI] [PubMed] [Google Scholar]
- Hashita T, Sakuma T, Akada M, Nakajima A, Yamahara H, Ito S, Takesako H, and Nemoto N (2008). Forkhead box A2-mediated regulation of female-predominant expression of the mouse Cyp2b9 gene. Drug Metab Dispos 36, 1080–1087. [DOI] [PubMed] [Google Scholar]
- Heintz M (2020). Role of CYP2B in Unsaturated Fatty Acid Metabolism, Obesity, and Non-Alcoholic Fatty Liver Disease. doi: https://tigerprints.clemson.edu/all_dissertations/2661. [Google Scholar]
- Heintz MM, Kumar R, Rutledge MM, and Baldwin WS (2019). Cyp2b-null male mice are susceptible to diet–induced obesity and perturbations in lipid homeostasis J Nutr Biochem 70, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz MM, McRee R, Kumar R, and Baldwin WS (2020). Gender differences in diet-induced steatotic disease in Cyp2b-null mice. PLoS ONE 15(3), e0229896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Chapman LM, Kretschmer XC, and Baldwin WS (2006). Gender Specific Induction of Cytochrome P450s in Nonylphenol-Treated FVB/NJ Mice. Toxicol Appl Pharmacol 216, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron M (2019). National Vital Statistics Reports Volume 68, Number 6, June 24, 2019, Deaths: Leading Causes for 2017. [PubMed] [Google Scholar]
- Hodgson E, and Rose RL (2007). The importance of cytochrome P450 2B6 in the human metabolism of environmental chemicals. Pharmacol Ther 113, 420–428. [DOI] [PubMed] [Google Scholar]
- Hoek-van den Hil EF, van Schothorst EM, van der Stelt I, Swarts HJ, van Vliet M, Amolo T, Vervoort JJ, Venema D, Hollman PC, Rietjens IM, et al. (2015). Direct comparison of metabolic health effects of the flavonoids quercetin, hesperetin, epicatechin, apigenin and anthocyanins in high-fat-diet-fed mice. Genes Nutr 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck I, Beggs K, and Apte U (2018). Paradoxical Protective Effect of Perfluorooctanesulfonic Acid Against High-Fat Diet-Induced Hepatic Steatosis in Mice. International Journal of Toxicology 37(5), 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Li H, Tang JJ, Wang J, Luo J, Liu B, Wang JK, Shi XJ, Cui HW, Tang J, et al. (2018). Discovery of a potent HMG-CoA reductase degrader that eliminates statin-induced reductase accumulation and lowers cholesterol. Nature Communications 9(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, McConnell R, Catherine C, Xu S, Walker DI, Stratakis N, Jones DP, Miller GW, Peng C, Conti DV, et al. (2020). Perfluoroalkyl Substances and Severity of Nonalcoholic Fatty Liver in Children: An Untargeted Metabolomics Approach. Environ Int 134, 105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump DB, Tripathy S, and Depner CM (2013). Fatty Acid–Regulated Transcription Factors in the Liver. Annual Review of Nutrition 33(1), 249–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Wong LY, Jia LT, Kuklenyik Z, and Calafat AM (2011). Trends in exposure to polyfluoroalkyl chemicals in the U.S. population: 1999-2008. Environmental Science and Technology 45(19), 8037–8045. [DOI] [PubMed] [Google Scholar]
- Kim H, Wang RS, Elovaara E, Raunio H, Pelkonen O, Aoyama T, Vainio H, and Nakajima T (1997). Cytochrome P450 isozymes responsible for the metabolism of toluene and styrene in human liver microsomes. Xenobiotica 27(7), 657–665. [DOI] [PubMed] [Google Scholar]
- Kim S-J, Heo S-H, Lee D-S, Hwang IG, Lee Y-B, and Cho H-Y (2016). Gender differences in pharmacokinetics and tissue distribution of 3 perfluoroalkyl and polyfluoroalkyl substances in rats. Food Chem Toxicol 97, 243–255. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Hocevar BA, and Kamendulis LM (2012). Mode of Action analysis of perfluorooctanoic acid (PFOA) tumorigenicity and Human Relevance. Reproductive Toxicology 33(4), 410–418. [DOI] [PubMed] [Google Scholar]
- Kretschmer XC, and Baldwin WS (2005). CAR and PXR: Xenosensors of Endocrine Disrupters? Chem-Biol. Interac 155, 111–128. [DOI] [PubMed] [Google Scholar]
- Kumar R, Litoff EJ, Boswell WT, and Baldwin WS (2018). High fat diet induced obesity is mitigated in Cyp3a-null female mice. Chem-Biol Interact 289, 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Mota LC, Litoff EJ, Rooney JP, Boswell WT, Courter E, Henderson CM, Hernandez JP, Corton JC, Moore DD, et al. (2017). Compensatory changes in CYP expression in three different toxicology mouse models: CAR-null, Cyp3a-null, and Cyp2b9/10/13-null mice. PLOS ONE 12(3), e0174355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, and Seed J (2007). REVIEW Perfluoroalkyl Acids: A Review of Monitoring and Toxicological Findings. TOXICOLOGICAL SCIENCES 99(2), 366–394. [DOI] [PubMed] [Google Scholar]
- Leung A, Trac C, Du J, Natarajan R, and Schones DE (2016). Persistent chromatin modifications induced by a high fat diet. J Biol Chem 291(20), 10446–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, and Cederbaum A (2006). Alcohol Upregulation of CYP2A5: Role of Reactive Oxygen Species. Reactive Oxygen Species 1(2), 117–130. [PMC free article] [PubMed] [Google Scholar]
- Moslehi A, and Hamidi-Zad Z (2018). Role of SREBPs in liver diseases: A mini-review. J Clin Transl Hepatol 6(3), 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, and Dobbie Z (2002). Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32, 1372–1379. [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, and Nebert DW (2004). Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative splice variants. Pharmacogenetics 14, 1–18. [DOI] [PubMed] [Google Scholar]
- Olona A, Terra X, Ko J-H, Grau-Bove C, Pinent M, Ardevol A, Diaz AG, Moreno-Moral A, Edin M, Bishop-Bailey D, et al. (2018). Epoxygenase inactivation exacerbates diet and aging-associated metabolic dysfunction resulting from impaired adipogenesis. Mol Metab 11, 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GW, Hansen KJ, Stevenson LA, Burris JM, and Mandel JH (2003). PFOS : random people. Human Donor Liver and Serum Concentrations of Perfluorooctanesulfonate and Other Perfluorochemicals. Environ Sci Technol 37(5), 888–891. [DOI] [PubMed] [Google Scholar]
- Olsen GW, Lange CC, Ellefson ME, Mair DC, Church TR, Goldberg CL, Herron RM, Medhdizadehkashi Z, Nobiletti JB, Rios JA, et al. (2012). Temporal trends of perfluoroalkyl concentrations in American Red Cross adult blood donors, 2000-2010. Environmental Science and Technology 46(11), 6330–6338. [DOI] [PubMed] [Google Scholar]
- Olsen GW, and Zobel LR (2007). Assessment of lipid, hepatic, and thyroid parameters with serum perfluorooctanoate (PFOA) concentrations in fluorochemical production workers. International Archives of Occupational and Environmental Health 81(2), 231–246. [DOI] [PubMed] [Google Scholar]
- Organization for Economic and Community Development (OECD) (2006). Results of the 2006 survey on the production and use of PFOS, PFAS, PFOA, PFCA, their related substances and products/mixtures containing these substances OECD Environment, Health and Safety Publications Series on Risk Management. [Google Scholar]
- Patsouris D, Reddy JK, Müller M, and Kersten S (2006). Peroxisome proliferator-activated receptor α mediates the effects of high-fat diet on hepatic gene expression. Endocrinology 147(3), 1508–1516. [DOI] [PubMed] [Google Scholar]
- Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, and Peters JM (2006). The Toxicology of Ligands for Peroxisome Proliferator-Activated Receptors (PPAR). Toxicological Sciences 90(2), 269–295. [DOI] [PubMed] [Google Scholar]
- Perez F, Nadal M, Navarro-Ortega A, Fabrega F, Domingo JL, Barcelo D, and Farre M (2013). Accumulation of perfluoroalkyl substances in human tissues. Environ Int 59, 354–562. [DOI] [PubMed] [Google Scholar]
- Pfohl M, Marques E, Auclair A, Barlock B, Jamwal R, Goedken M, Akhlaghi F, and Slitt AL (2021). An ‘Omics Approach to Unraveling the Paradoxical Effect of Diet on Perfluorooctanesulfonic Acid (PFOS) and Perfluorononanoic Acid (PFNA)-Induced Hepatic Steatosis. Toxicol Sci doi: doi: 10.1093/toxsci/kfaa172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poça KS, Parente TEM, Chagas LF, Leal BS, Leal HS, Paumgartten FJR, and De-Oliveira ACAX (2017). Interstrain differences in the expression and activity of Cyp2a5 in the mouse liver. BMC Research Notes 10(1), 125–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roling JA, Bain LJ, and Baldwin WS (2004). Differential gene expression in mummichogs (Fundulus heteroclitus) following treatment with pyrene: comparison to a creosote contaminated site. Mar Environ Res 57, 377–395. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Das KP, Rooney J, Abbott B, Lau C, and Corton JC (2017). PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology 387, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MB, Das KP, Wood CR, Wolf CJ, Abbott BD, and Lau C (2013). Evaluation of perfluoroalkyl acid activity using primary mouse and human hepatocytes. Toxicology 308, 129–137. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Schmid JR, Corton JC, Zehr RD, Das KP, Abbott BD, and Lau C (2010). Gene expression profiling in wild-type and PPARa-null mice exposed perfluorooctane sulfonate reveals PPARa-independent effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz R, Jideonwo V, Ahn MY, Surendran S, Tagliabracci VS, Hou Y, Gamble A, Kerner J, Irimia-Dominguez JM, Puchowicz MA, et al. (2014). Sterol regulatory elementbinding protein-1 (SREBP-1) is required to regulate glycogen synthesis and gluconeogenic gene expression in mouse liver. J Biol Chem 289(9), 5510–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, and Joshi M (2020). CPT1A-mediated Fat Oxidation, Mechanisms, and Therapeutic Potential. Endocrinology 161(3), bqz046. [DOI] [PubMed] [Google Scholar]
- Seth RK, Das S, Kumar A, Chanda A, Kadiiska MB, Michelotti G, Manautou J, Diehl AM, and Chatterjee S (2014). CYP2E1-dependent and leptin-mediated hepatic CD57 expression on CD8+ T cells aid progression of environment-linked nonalcoholic steatohepatitis. Toxicol Appl Pharmacol 274(1), 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina IG, Kramarova TV, Mattsson CL, Petrovic N, Qazi MR, Csikasz RI, Chang S-C, Butenhoff J, DePierre JW, Cannon B, et al. (2015). The Environmental Pollutants Perfluorooctane Sulfonate and Perfluorooctanoic Acid Upregulate Uncoupling Protein 1 (UCP1) in Brown-Fat Mitochondria Through a UCP1-Dependent Reduction in Food Intake Toxicol Sci 146(2), 334–343. [DOI] [PubMed] [Google Scholar]
- Takacs ML, and Abbott BD (2007). Activation of mouse and human peroxisome proliferator-activated receptor (a,b/d,g) perfluorooctanoic acid and perfluorooctane sulfonate. Toxicol Sci 95, 108–117. [DOI] [PubMed] [Google Scholar]
- Tang J, Cao Y, Rose RL, Brimfield AA, Dai D, Goldstein JA, and Hodgson E (2001). Metabolism of chlorpyrifos by human cytochrome P450 isoforms and human, mouse and rat liver microsomes. Drug Metab Dispos 29, 1201–1204. [PubMed] [Google Scholar]
- Tanner EM, Bloom MS, Wu Q, Kannan K, Yucel RM, Shrestha S, and Fitzgerald EF (2018). Occupational exposure to perfluoroalkyl substances and serum levels of perfluorooctanesulfonic acid (PFOS) and perfluorooctanoic acid (PFOA) in an aging population from upstate New York: a retrospective cohort study. Int Arch Occup Environ Health 91(12), 145–154. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven TA, and Coon MJ (1974). Preparation and properties of partially purified cytochrome P450 and NADPH-cytochrome P450 reductase from rabbit liver microsomes. J Biol Chem 249, 6302–6310. [PubMed] [Google Scholar]
- Wafer R, Tandon P, and Minchin JEN (2017). The role of peroxisome proliferator-activated receptor gamma (PPARG) in adipogenesis: Applying knowledge from the fish aquaculture industry to biomedical research. Front Endocrinol 8, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, and Tompkins LM (2008). CYP2B6: New insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9, 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Y, Liang Y, Li J, Liu Y, Zhang J, Zhang A, Fu J, and Jiang G (2014). PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci Rep 4, 4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wu H, Li L, Liu Z, Zhou X, Zhang Q-Y, Weng Y, D’Agostino J, Ling G, Zhang X, et al. (2012). Generation and Characterization of a CYP2A13/2B6/2F1-Transgenic Mouse Model. Drug Metab Dispos 40, 1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L-L, Lin C-Y, Chou H-C, Chang C-C, Lo H-Y, and Juan S-H (2016). Perfluorooctanesulfonate Mediates Renal Tubular Cell Apoptosis through PPARgamma Inactivation. PLoS One 11(5), e0155190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Asilmaz E, Luca E, Friedman JM, and Stoffel M (2004). Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature 432, 1027–1032. [DOI] [PubMed] [Google Scholar]
- Xu J, Shimpi P, Armstrong L, Salter D, and Slitt AL (2016). PFOS induces adipogenesis and glucose uptake in association with activation of Nrf2 signaling pathway. Toxicol Appl Pharmacol 290, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Kannan K, Taniyasu S, Horii Y, Petrick G, and Gamo T (2005). A global survey of perfluorinated acids in oceans Mar Pollut Bull 51, 658–668. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren XM, Wan B, and Guo LH (2014). Structure-dependent binding and activation of perfluorinated compounds on human peroxisome proliferator-activated receptor γ. Toxicology and Applied Pharmacology 279(3), 275–283. [DOI] [PubMed] [Google Scholar]
- Zhang L, Rubins NE, Ahima RS, Greenbaum LE, and Kaestner KH (2005). Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab 2(2), 141–148. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li S, Zhou Y, Su W, Ruan X, Wang B, Zheng F, Warner M, Gustafsson JÅ, and Guan Y (2017). Ablation of cytochrome P450 omega-hydroxylase 4A14 gene attenuates hepatic steatosis and fibrosis. Proc Natl Acad Sci USA 114(12), 3181–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.