Abstract
Objective
To evaluate the agreement, accuracy, and longitudinal reproducibility of quantitative cartilage morphometry from 2D U-Net-based automated segmentations for 3T coronal fast low angle shot (corFLASH) and sagittal double echo at steady-state (sagDESS) MRI.
Methods
2D U-Nets were trained using manual, quality-controlled femorotibial cartilage segmentations available for 92 Osteoarthritis Initiative healthy reference cohort participants from both corFLASH and sagDESS (n = 50/21/21 training/validation/test-set). Cartilage morphometry was computed from automated and manual segmentations for knees from the test-set. Agreement and accuracy were evaluated from baseline visits (dice similarity coefficient: DSC, correlation analysis, systematic offset). The longitudinal reproducibility was assessed from year-1 and -2 follow-up visits (root-mean-squared coefficient of variation, RMSCV%).
Results
Automated segmentations showed high agreement (DSC 0.89–0.92) and high correlations (r ≥ 0.92) with manual ground truth for both corFLASH and sagDESS and only small systematic offsets (≤ 10.1%). The automated measurements showed a similar test–retest reproducibility over 1 year (RMSCV% 1.0–4.5%) as manual measurements (RMSCV% 0.5–2.5%).
Discussion
The 2D U-Net-based automated segmentation method yielded high agreement compared with manual segmentation and also demonstrated high accuracy and longitudinal test–retest reproducibility for morphometric analysis of articular cartilage derived from it, using both corFLASH and sagDESS.
Keywords: Cartilage, Automated segmentation, Knee osteoarthritis, Magnetic resonance imaging, Convolutional neural network
Introduction
Osteoarthritis (OA) is a highly prevalent, chronic disease that affects more than 300 million people world-wide [1, 2]. OA patients experience pain and functional limitations, and the knee is by far the most commonly affected joint [2]. Amongst other structural pathologies of this whole-joint-disease, articular cartilage loss is a hallmark of knee OA. While radiography was previously used to assess the structural progression of OA, quantitative measurement of articular cartilage based on serial magnetic resonance images (MRI) is now the method of choice and provides the high test–retest precision and sensitivity to longitudinal change required for clinical trials [3–5]. The use of quantitative MRI also has revolutionized the conduct of clinical trials on structure or disease modifying OA drugs (S/DMOADs) [5–8], by having recently been scaled up from exploratory to secondary or even primary endpoints for submission for potential regulatory approval [6, 9].
Several groups have proposed semi- or fully automated approaches for reducing the time required for the segmentation of articular cartilage from MRI, including model-, atlas-, graph-, voxel classification-, or active-contour-based methods [10–12]. More recently, convolutional neural networks (CNNs), primarily based on the U-Net architecture [13], have been employed for automated cartilage segmentations and have demonstrated a good segmentation agreement between automated vs. ground-truth approaches [14–23]. Yet, only few of these CNN-based studies examined the accuracy of quantitative cartilage measures (e.g. thickness, volume, and surface area) derived from CNN-based segmentations [14, 16, 23]. Particularly, none of these reported the longitudinal stability or test–retest precision of quantitative cartilage measures derived from CNN-based cartilage segmentation, which is an important prerequisite before a segmentation methodology can be applied to data from a clinical trial, or compared the segmentation and analysis performance between different MRI sequences typically used in osteoarthritis studies [24].
The objective of the current study was, therefore, to evaluate the segmentation agreement as well as the accuracy and longitudinal test–retest reproducibility of quantitative cartilage measures obtained from a 2D U-Net-based methodology for automated femorotibial cartilage segmentation using two different MRI sequences for the same subject. To that end, we used data from the publicly accessible Osteoarthritis Initiative (OAI) cohort, specifically the subcohort of reference knees that were free of symptoms, signs and risk factors of knee OA, and for which cartilage thickness values (and their stability over time) have been reported previously [25–27]. Specifically, this work encompasses:
Evaluating the agreement between automated and quality-controlled, manual segmentation of articular cartilage as “ground truth”.
Testing the accuracy (correlations and systematic offsets) of quantitative cartilage morphometry measures (thickness, volume, surface areas) derived from automated segmentations compared to manual segmentation.
Analysis of the longitudinal test–retest reproducibility of quantitative cartilage measures derived from automated vs. manual segmentation over a 1-year period (using year-1 and -2 follow-up data).
Comparison of the agreement, accuracy, and longitudinal test–retest reproducibility of the automated segmentation (and quantitative cartilage measures derived therefrom) between two different MRI sequences with different contrasts and orientations.
Materials and methods
Participants and MR imaging
This study used data from the OAI (clinicaltrials.gov: NCT00080171) [28]. The OAI was approved by the Committee on Human Research, the Institutional Review Board for the University of California, San Francisco (UCSF). All OAI participants provided written informed consent, and this study was carried out in accordance with the OAI data user agreement. The OAI enrolled participants aged 45–79 years with established knee OA (progression cohort, n = 1390), with risk of developing OA (incidence cohort, n = 3284), and participants without signs, symptoms, or risk factors for developing OA (reference cohort, n = 122, based on the initial clinical site readings). Demographic, clinical and radiographic data, as well as MRIs were collected by four clinical sites at the baseline visit and each of the annual follow-up visits (https://data-archive.nimh.nih.gov/oai/). MRIs were acquired by the OAI using 3T Magnetom Trio scanners (Siemens Medical Solutions, Erlangen, Germany) and quadrature transmit/receive knee coils (USA Instruments, Aurora, OH) [28, 29]. The OAI imaging protocol included coronal fast low angle shot (FLASH) acquisitions with water excitation (in-plane resolution 0.3125 × 0.3125 mm, slice thickness 1.5 mm, flip angle 12°, echo time 7.6 ms, repetition time 20 ms) of the right knees, and sagittal double echo steady state (DESS) with water excitation of both knees (in-plane resolution 0.37 × 0.46 mm, interpolated to 0.37 × 0.37 mm, slice thickness 0.7 mm, flip angle 25°, echo time 4.7 ms, repetition time 16.3 ms) [29].
The current study included all 92 participants from the OAI reference cohort that were confirmed to be free from radiographic signs of OA in both of their knees during post hoc central readings by experienced readers [28], and that had at least the year-1 follow-up MRI available.
Manual segmentation
Manual segmentations of the weight-bearing part of the femorotibial cartilages were available from previous projects for the right knees of the 92 OAI reference cohort participants [25–27]. Segmentations of baseline and year-1 follow-up MRIs from coronal FLASH (corFLASH) MRI were performed for all 92 right knees after the year-1 follow-up data from the OAI became available [25] and were later repeated together with year-2 and -4 follow-up MRIs for 81 of the 92 knees that also had year-4 follow-up MRIs available [26]. Segmentations of baseline, year-1, -2, and -4 follow-up sagittal DESS (sagDESS) MRIs were performed for the same 92 knees, and year-2/year-4 follow-up MRIs were available for 88/82 of the knees, respectively [26, 27].
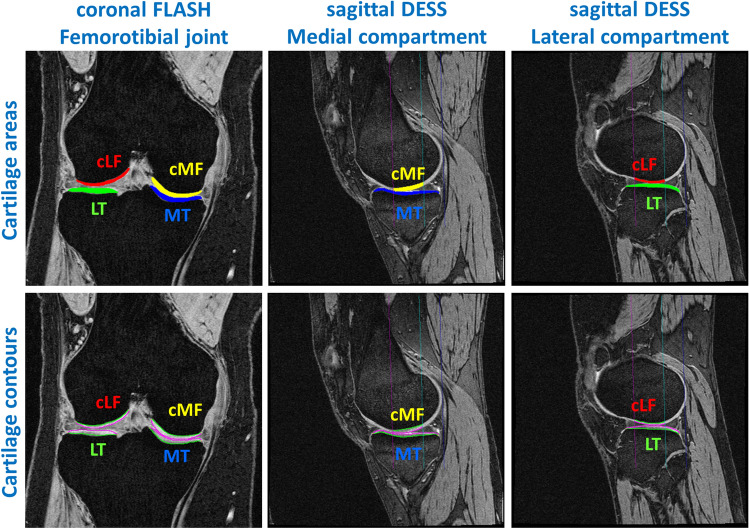
Segmentation comprised the entire medial and lateral tibia (MT/LT), and the central (weight-bearing) part of the medial and lateral femoral condyles (cMF/cLF), defined as 60% of the distance between the inter-condylar notch and the posterior end of the condyles (Fig. 1) [30, 31]. This 60% femoral region of interest (ROI) was necessary to avoid the inclusion of posterior parts of the cartilages in the segmentation, which are affected from partial volume effects in coronal MRIs and display a lesser amount of longitudinal change than the weight-bearing part in knee OA [30]. Manual segmentation was performed by a team of experienced readers using custom software (Chondrometrics GmbH, Ainring, Germany) by tracing the subchondral bone (tAB) and articular cartilage surface area (AC) of all four femorotibial cartilages (Fig. 1) [32]. All visits of each knee were segmented by the same reader, using one of the visits as a reference, but with blinding to the image dates, visit identifiers, and acquisition order. All manual segmentations were quality-controlled by an expert reader.
Fig. 1.
Manual segmentation of the femorotibial cartilages [MT/LT medial/lateral tibia, cMF/cLF central (weight-bearing) part of the medial and lateral femoral condyles] from coronal FLASH and sagittal DESS MRI. The figure shows the cartilage areas (top row) and the cartilage contours (bottom; green: total area of subchondral bone; magenta: cartilage surface area). The sagittal MRIs also show the 60% femoral region of interest (magenta line: anterior margin; blue line: posterior end of the condyles; turquoise line: 60% margin)
Automated, U-Net-based segmentation
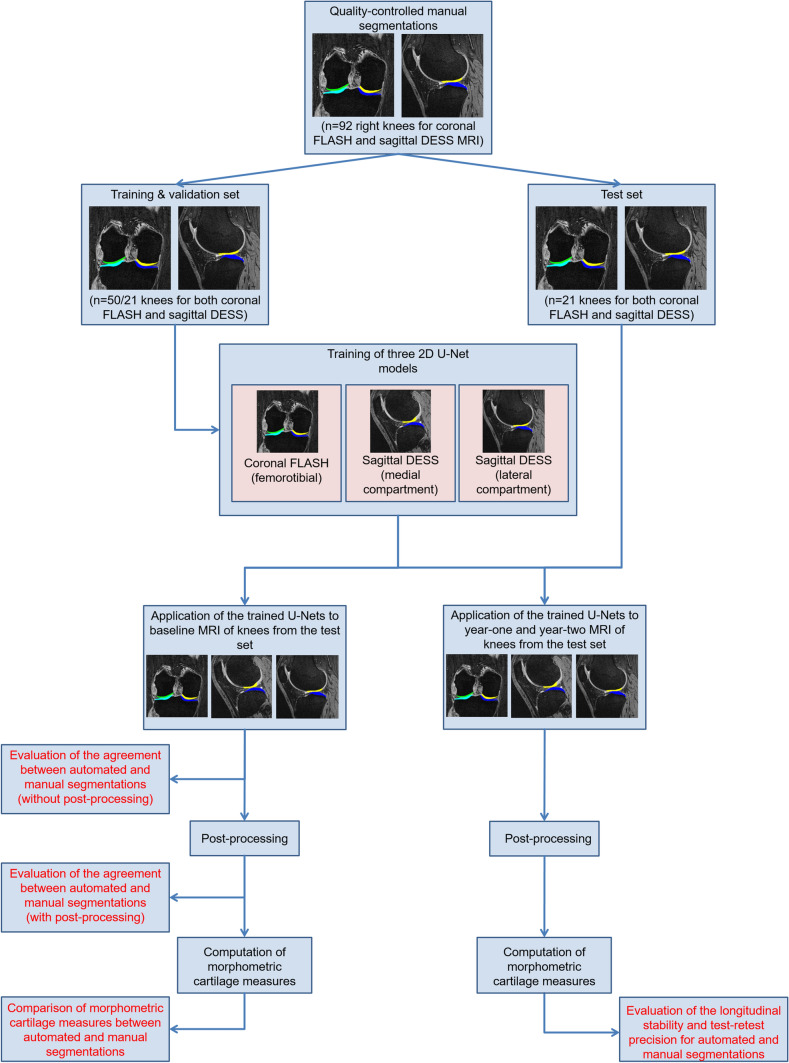
The 92 OAI reference cohort participants were divided into a training (n = 50), validation (n = 21) and test set (n = 21, Fig. 2). The division was controlled to ensure a similar distribution of sex and body height between the sets. Participants for which no manual year-2 segmentations from corFLASH MRI were available, were only considered for inclusion into the training and validation set, to ensure that manual segmentations from year-1 and -2 follow-up MRIs were available for all participants from the test set to evaluate the longitudinal test–retest reproducibility (see below).
Fig. 2.
Overview over the workflow and analysis steps used for the current study
The automated segmentation method was based on the 2D encoder-decoder U-Net architecture proposed by Ronneberger et al. [13] with the number of feature maps in the transpose convolutions of the up-sampling path set to the number of feature classes [33]. This implementation of the 2D U-Net architecture has been previously applied to the segmentation of MRIs of cardiac tissue [33] and thigh muscle cross-sectional areas [34]. In the current study, the U-Net was trained using a weighted cross-entropy loss function (background weight 1/[1 + 2 × number of feature classes]; foreground weight 2/[1 + 2 × number of feature classes]) that was minimized using the adaptive moment estimation (ADAM) optimizer (initial learning rate 0.01, decay rate 0.1, beta1 = 0.9, beta2 = 0.999) [35]. All network weights were randomly initialized using the tensorflow variance scaling initializer. The software was implemented in Python (Python Software Foundation, DE, USA) using the Tensorflow framework (Google LLC, CA, USA).
The training was performed on the training set using full-resolution (512 × 512 pixel for corFLASH, 384× 384 pixel for sagDESS), full-sized MRI slices on a NVIDIA RTX 2080TI GPU. The signal intensity was normalized in each slice by subtracting the mean intensity, and dividing by the standard deviation of the signal intensity. Bright voxels in the image corners (15 × 15 pixels) were set to zero intensity to avoid a negative impact of these imaging artefacts on the signal intensity normalization.
For corFLASH MRI, one network was trained comprising all four femorotibial cartilage plates. For sagDESS MRI, two networks were trained in parallel, one for the medial femorotibial compartment (MFTC) and one for the lateral femorotibial compartment (LFTC). Each cartilage plate was treated as an individual feature class, with the training including only the segmented slices. The three network weights (corFLASH, sagDESS MFTC, sagDESS LFTC) that achieved the best segmentation agreement with the validation set during 50 epochs were eventually applied for automatic segmentation of the test set (Fig. 2). The automated segmentations were not quality-controlled and not manually corrected.
Automated post-processing
Because the predictions made by the U-Net may extend into anatomically implausible locations and because the automated segmentations required adaption before the computation of morphometric cartilage measures, the following, automated rule-based post-processing steps were implemented as a non-interactive command line program using C++:
Filling of small gaps by detecting enclosed, unsegmented areas
Removal of segmentations in slices not connected to the segmentation in the same or other slices (i.e., segmentations at implausible locations)
Removal of spikes (smoothing)
Removal of femoral cartilage segmentations outside the (60%) femoral ROI
Separation of segmentations into the subchondral bone area (tAB), articular cartilage surface area (AC), and inner cartilage (IC), which was required for the computation of morphological parameters (see below).
The separation of segmentations into tAB, AC, and IC was performed for each structure and each slice separately by identifying the two points with the greatest distance from each other and by subsequently assigning the border pixel of the structures’ segmentation to the tAB and AC. When segmentation of the cMF and cLF bordered the femoral ROI (sagDESS only), the intersection between the segmentation and the femoral ROI was used instead. Non-border voxels were assigned to IC.
Statistical analysis
The agreement between automated and quality-controlled manual segmentations was evaluated using the 3D Dice similarity coefficient (DSC), the 3D volume overlap error (VOE), the 3D Hausdorff distance (HD), and the 3D average symmetric surface distance (ASSD) for the knees from the test set, before and after applying the post-processing steps.
To evaluate the repeatability of the training process, training was repeated from scratch using the same sets with a different random initialization of network weights (“repeated run”). To evaluate, whether the agreement between automated vs. manual segmentations is dependent on the assignment of knees to each of the sets, training was repeated using the knees from the validation and test set, together with 8 knees from the original training set as training set, and by assigning the remaining 42 knees from the training set into validation and test sets (each n = 21, “reversed run”). For both these runs, the agreement of automated vs. manual segmentations was again evaluated using the DSC, VOE, HD, and ASSD, after automatically segmenting the cartilages from the respective test set (with and without post-processing).
Cartilage thickness, cartilage volume, and the total area of subchondral bone (tAB) were calculated from both manual and post-processed automated segmentations of the knees in the test set using custom software (Chondrometrics GmbH, Ainring, Germany). Measures for the medial and lateral femorotibial compartment (MFTC/LFTC) were calculated as sums of MT + cMF and LT + cLF, respectively. The accuracy of baseline cartilage measures computed from the automated segmentations vs. measures computed from manual segmentations was evaluated by examining the Pearson correlation. In addition, paired t tests were used to assess differences between cartilage measures computed from automated vs manual segmentations. Bland and Altman plots were used to evaluate potential systematic offsets between both segmentation methods, and between corFLASH and sagDESS. Furthermore, Pearson correlation analyses were conducted to study the association of cartilage thickness differences between automated and manual segmentations vs. DSC, VOE, HD, and ASSD values.
Cartilage thickness has been previously observed to remain stable over periods of 1 year and longer in knees from the OAI reference cohort [25, 26]. Consequently, the year-1 and -2 follow-up visits were used to assess the longitudinal test–retest reproducibility of the automated cartilage analysis over such an observation period typical of interventional clinical trials. The longitudinal stability was assessed using a paired t test and the test–retest reproducibility using the root-mean-square standard deviation (RMSSD) and coefficient of variation (RMSCV%) of repeated measurements. To quantitatively evaluate whether the trained CNNs were overfitting to the data used during the training (training and validation sets), the longitudinal test–retest reproducibility was additionally computed for the knees from the validation and training set that had year-1 and -2 follow-up visits available (n = 19/41 pairs). The standard error of the measurement (SEM) and the smallest detectable change (SDC) threshold were calculated from year-1 and -2 data for the knees in the test set as described previously [36].
Demographic variables were compared between groups using unpaired t tests. The significance level for all statistical testing was set to α = 0.05. Descriptive statistics and t tests were computed using Excel 2010 (Microsoft Corporation, WA, USA).
Results
The 55 female and 37 male OAI reference cohort participants were on average 54.7 ± 7.5 years old, had a BMI of 24.4 ± 3.1 kg/m2 and a body height of 1.68 ± 0.09 m (Table 1). These demographic data did not differ statistically significantly between training, validation, and test set (p ≥ 0.15).
Table 1.
Demographic data
| Training set (n = 50) | Validation set (n = 21) | Test set (n = 21) | ||||
|---|---|---|---|---|---|---|
| N/Mean | %/SD | N/Mean | %/SD | N/Mean | %/SD | |
| Sex | ||||||
| Women | 29 | 58 | 13 | 62 | 13 | 62 |
| Men | 21 | 42 | 8 | 38 | 8 | 38 |
| Age (years) | 54.1 | 7.3 | 53.9 | 8.0 | 56.9 | 7.5 |
| BMI (kg/m2) | 24.1 | 3.0 | 24.5 | 3.2 | 25.1 | 3.3 |
| Height (m) | 1.68 | 0.09 | 1.67 | 0.09 | 1.67 | 0.09 |
SD standard deviation
During the training of the networks, the best segmentation agreement with data from the validation set was achieved for corFLASH/sagDESS LFTC/sagDESS MFTC after 14/33/34 epochs (99/167/159 min of training), and these U-Net weights were subsequently chosen for the automated segmentations on the hold-out test set.
Agreement of the automated U-Net segmentation with manual segmentation
A high agreement was observed between automated and manual cartilage segmentations for both corFLASH and sagDESS MRI already before the post-processing (Table 2). The DSC ranged from 0.88 ± 0.03 to 0.92 ± 0.02, the VOE from 14.9 ± 3.3 to 21.9 ± 4.8%, the HD from 2.8 ± 1.1 to 8.3 ± 13.3 mm, and the ASSD from 0.13 ± 0.03 to 0.28 ± 0.13 mm (Table 2). Post-processing only had a small effect on the DSC (range 0.89 ± 0.03–0.92 ± 0.02) and the VOE (range 14.9 ± 3.3–20.1 ± 4.4%), but notably reduced the HD (range 2.1 ± 0.6–3.2 ± 0.9 mm) and the ASSD (range 0.13 ± 0.03–0.17 ± 0.06 mm, Table 2, Fig. 3).
Table 2.
Agreement between manual and U-Net-based automated segmentations determined from n = 21 knees in the test set in the primary run (top) and the repeated run (bottom)
| DSC | VOE (%) | HD (mm) | ASSD (mm) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| corFLASH | sagDESS | corFLASH | sagDESS | corFLASH | sagDESS | corFLASH | sagDESS | |||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Main run: before post-processing | ||||||||||||||||
| MT | 0.92 | 0.02 | 0.91 | 0.02 | 15.5 | 3.0 | 17.1 | 2.7 | 8.27 | 13.32 | 2.79 | 1.08 | 0.15 | 0.04 | 0.13 | 0.03 |
| cMF | 0.88 | 0.03 | 0.89 | 0.03 | 21.9 | 4.8 | 20.0 | 4.3 | 5.72 | 11.02 | 6.14 | 11.24 | 0.28 | 0.13 | 0.18 | 0.07 |
| LT | 0.92 | 0.02 | 0.92 | 0.02 | 14.9 | 3.3 | 15.4 | 2.9 | 8.06 | 8.80 | 5.31 | 8.46 | 0.17 | 0.05 | 0.17 | 0.04 |
| cLF | 0.88 | 0.02 | 0.90 | 0.02 | 20.8 | 3.8 | 17.8 | 2.8 | 3.66 | 2.33 | 3.86 | 5.54 | 0.26 | 0.09 | 0.14 | 0.03 |
| Main run: after post-processing | ||||||||||||||||
| MT | 0.92 | 0.02 | 0.91 | 0.02 | 15.5 | 3.0 | 17.0 | 2.7 | 2.28 | 0.67 | 2.39 | 0.47 | 0.14 | 0.03 | 0.13 | 0.03 |
| cMF | 0.91 | 0.03 | 0.89 | 0.03 | 16.4 | 4.6 | 20.1 | 4.4 | 2.60 | 1.21 | 3.36 | 0.72 | 0.13 | 0.08 | 0.17 | 0.06 |
| LT | 0.92 | 0.02 | 0.92 | 0.02 | 14.9 | 3.3 | 15.4 | 2.9 | 3.00 | 1.09 | 3.21 | 0.92 | 0.16 | 0.04 | 0.17 | 0.04 |
| cLF | 0.91 | 0.02 | 0.90 | 0.02 | 15.8 | 4.0 | 17.9 | 2.8 | 2.08 | 0.56 | 2.65 | 0.55 | 0.13 | 0.06 | 0.14 | 0.03 |
| Repeated run: before post-processing | ||||||||||||||||
| MT | 0.92 | 0.02 | 0.91 | 0.02 | 15.3 | 2.7 | 16.4 | 2.8 | 6.38 | 8.39 | 2.52 | 0.78 | 0.14 | 0.03 | 0.13 | 0.03 |
| cMF | 0.84 | 0.03 | 0.89 | 0.03 | 26.8 | 5.2 | 20.0 | 4.5 | 8.70 | 10.12 | 8.21 | 11.86 | 0.44 | 0.21 | 0.18 | 0.09 |
| LT | 0.92 | 0.02 | 0.92 | 0.02 | 14.8 | 3.4 | 15.0 | 2.8 | 5.64 | 7.24 | 3.00 | 1.00 | 0.15 | 0.04 | 0.16 | 0.03 |
| cLF | 0.88 | 0.03 | 0.91 | 0.02 | 21.2 | 4.7 | 17.0 | 2.6 | 5.63 | 8.64 | 7.15 | 12.77 | 0.25 | 0.12 | 0.14 | 0.05 |
| Repeated run: after post-processing | ||||||||||||||||
| MT | 0.92 | 0.02 | 0.91 | 0.02 | 15.2 | 2.7 | 16.4 | 2.8 | 2.21 | 0.68 | 2.30 | 0.62 | 0.14 | 0.03 | 0.13 | 0.03 |
| cMF | 0.91 | 0.02 | 0.89 | 0.03 | 17.0 | 3.9 | 20.2 | 4.6 | 2.50 | 1.00 | 3.40 | 0.75 | 0.14 | 0.06 | 0.18 | 0.07 |
| LT | 0.92 | 0.02 | 0.92 | 0.02 | 14.7 | 3.4 | 15.0 | 2.8 | 3.02 | 0.93 | 3.00 | 1.00 | 0.15 | 0.04 | 0.17 | 0.03 |
| cLF | 0.90 | 0.03 | 0.91 | 0.02 | 17.5 | 4.8 | 17.1 | 2.5 | 2.18 | 0.48 | 2.42 | 0.49 | 0.16 | 0.08 | 0.13 | 0.02 |
Agreement was assessed from coronal FLASH (corFLASH) and sagittal DESS (sagDESS) MRI acquired at the OAI baseline visit
DSC dice similarity coefficient, VOE volume overlap error, HD Hausdorff distance, ASSD average symmetric surface distance, MT/LT medial/lateral tibia, cMF/cLF central medial/lateral femoral condyle
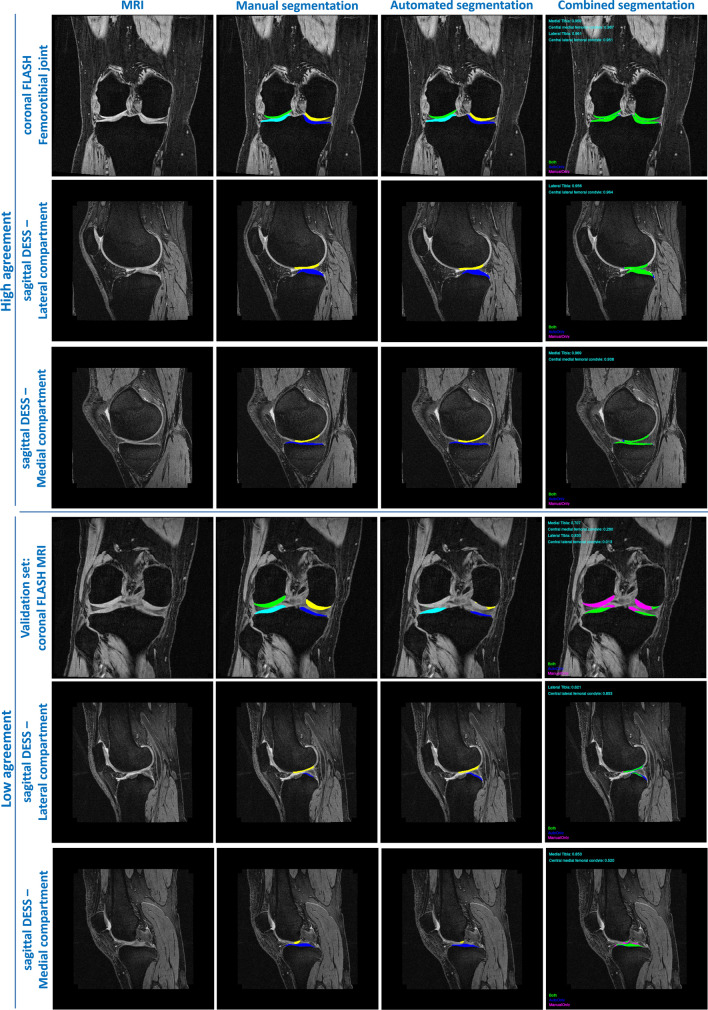
Fig. 3.
Examples of manual and U-Net-based automated segmentations from coronal FLASH and sagittal DESS illustrating the range of agreement observed in this study. Rows 1–3: Examples with high agreement, rows 4–6: examples with low agreement or segmentation errors taken from the training and validation set. Cartilage plates are shown in blue (medial tibia), yellow (central medial femur), turquoise (lateral tibia), and green (central lateral femur) in the two middle columns. The right column shows pixel contained in both manual and automated segmentations in green, pixel only contained in manual segmentations in purple, and pixel only contained in automated segmentations in blue
The agreement between automated vs. manual segmentation obtained in both the repeated run (Table 2) and the reversed run (data now shown) was largely consistent with the results from the main run: A somewhat lower agreement was observed for the cMF in the repeated run for corFLASH MRI (Table 2). A similar observation was made for the cMF (DSC 0.86 ± 0.04, VOE 23.9 ± 5.6%, HD 16.5 ± 16.3 mm, ASSD 0.37 ± 0.20 mm) and cLF (DSC 0.84 ± 0.05, VOE 26.8 ± 7.0%, HD 4.1 ± 13 mm, ASSD 0.40 ± 0.19 mm) with corFLASH MRI in the reversed run (data not shown). These differences were only evident prior to the post-processing.
Accuracy of cartilage morphometry using automated, U-Net vs. manual segmentation
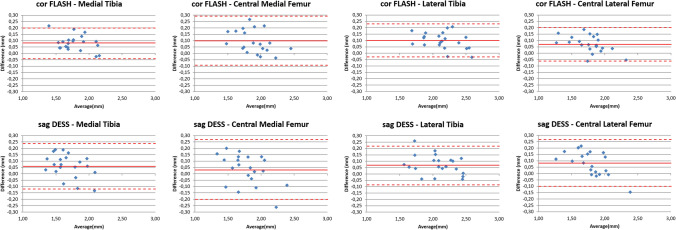
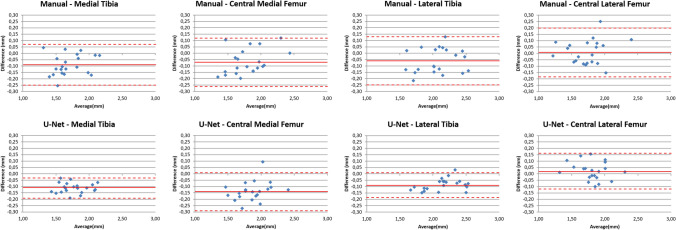
All morphometric cartilage measures computed from the automated segmentations of the baseline MRIs in the test set displayed high correlations with those obtained from manual segmentations (range r = 0.92–0.99, Table 3). Cartilage thickness from the automated segmentation had a slight, but consistent overestimation when compared to the measures derived from manual segmentation for both corFLASH and sagDESS (range 1.9–5.5%, Table 3). This difference was statistically significant in all cartilage plates, except for the cMF with sagDESS (Table 3). Bland and Altman plots comparing cartilage thickness measures computed from automated vs. manual segmentations are shown in Fig. 4, Bland and Altman plots comparing cartilage thickness between corFLASH and sagDESS MRI in Fig. 5. In brief, cartilage thickness computations were highly consistent between corFLASH and sagDESS using both methodological approaches. The mean difference tended to be closer to zero for the manual than for the automated segmentations, whereas the limits of agreement tended to be narrower for the automated than the manual segmentations.
Table 3.
Comparison of quantitative cartilage measures between manual and U-Net-based automated segmentations determined from n = 21 knees from the test set
| Manual | U-Net | Manual vs. U-Net | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Diff (%) | P | r | ||
| Cartilage thickness (mm) | ||||||||
| MFTC | corFLASH | 3.6 | 0.4 | 3.8 | 0.4 | 5.1 | < 0.001 | 0.96 |
| sagDESS | 3.4 | 0.5 | 3.5 | 0.4 | 2.8 | 0.019 | 0.95 | |
| MT | corFLASH | 1.7 | 0.2 | 1.8 | 0.2 | 4.7 | < 0.001 | 0.97 |
| sagDESS | 1.6 | 0.2 | 1.7 | 0.2 | 3.7 | 0.006 | 0.93 | |
| cMF | corFLASH | 1.8 | 0.3 | 1.9 | 0.2 | 5.5 | < 0.001 | 0.93 |
| sagDESS | 1.8 | 0.3 | 1.8 | 0.2 | 1.9 | 0.224 | 0.92 | |
| LFTC | corFLASH | 3.8 | 0.5 | 4.0 | 0.5 | 4.4 | < 0.001 | 0.98 |
| sagDESS | 3.8 | 0.5 | 3.9 | 0.5 | 4.1 | < 0.001 | 0.97 | |
| LT | corFLASH | 2.1 | 0.3 | 2.2 | 0.3 | 4.6 | < 0.001 | 0.97 |
| sagDESS | 2.0 | 0.3 | 2.1 | 0.3 | 3.5 | < 0.001 | 0.97 | |
| cLF | corFLASH | 1.7 | 0.3 | 1.8 | 0.2 | 4.2 | < 0.001 | 0.97 |
| sagDESS | 1.7 | 0.3 | 1.8 | 0.2 | 4.8 | 0.001 | 0.96 | |
| Cartilage volume (mm3) | ||||||||
| MFTC | corFLASH | 2947 | 829 | 3218 | 797 | 9.2 | < 0.001 | 0.99 |
| sagDESS | 2801 | 794 | 2912 | 737 | 4.0 | 0.005 | 0.98 | |
| MT | corFLASH | 1945 | 570 | 2120 | 534 | 9.0 | < 0.001 | 0.99 |
| sagDESS | 1703 | 532 | 1848 | 496 | 8.5 | < 0.001 | 0.98 | |
| cMF | corFLASH | 1003 | 279 | 1099 | 277 | 9.6 | < 0.001 | 0.96 |
| sagDESS | 1098 | 285 | 1064 | 256 | -3.1 | 0.154 | 0.93 | |
| LFTC | corFLASH | 3123 | 859 | 3421 | 904 | 9.6 | < 0.001 | 0.99 |
| sagDESS | 3291 | 928 | 3433 | 872 | 4.3 | < 0.001 | 0.99 | |
| LT | corFLASH | 2083 | 589 | 2294 | 622 | 10.1 | < 0.001 | 0.99 |
| sagDESS | 2144 | 646 | 2295 | 618 | 7.0 | < 0.001 | 0.99 | |
| cLF | corFLASH | 1039 | 298 | 1128 | 302 | 8.5 | < 0.001 | 0.98 |
| sagDESS | 1147 | 311 | 1138 | 264 | -0.8 | 0.623 | 0.97 | |
| Total area of subchondral bone (cm2) | ||||||||
| MFTC | corFLASH | 16.1 | 2.8 | 16.3 | 2.8 | 1.5 | 0.040 | 0.98 |
| sagDESS | 15.3 | 2.7 | 15.7 | 2.7 | 2.5 | < 0.001 | 0.99 | |
| MT | corFLASH | 11.0 | 1.9 | 11.2 | 1.8 | 1.7 | 0.027 | 0.98 |
| sagDESS | 10.0 | 1.7 | 10.4 | 1.8 | 3.8 | < 0.001 | 0.99 | |
| cMF | corFLASH | 5.1 | 1.0 | 5.2 | 1.0 | 1.1 | 0.395 | 0.96 |
| sagDESS | 5.3 | 1.0 | 5.3 | 1.0 | 0.0 | 0.999 | 0.95 | |
| LFTC | corFLASH | 15.2 | 2.6 | 15.5 | 2.7 | 1.9 | 0.015 | 0.98 |
| sagDESS | 15.6 | 2.8 | 15.8 | 2.7 | 1.6 | 0.024 | 0.99 | |
| LT | corFLASH | 9.4 | 1.6 | 9.7 | 1.7 | 2.9 | 0.010 | 0.96 |
| sagDESS | 9.9 | 1.7 | 10.1 | 1.7 | 2.2 | 0.018 | 0.98 | |
| cLF | corFLASH | 5.8 | 1.1 | 5.8 | 1.1 | 0.1 | 0.904 | 0.98 |
| sagDESS | 5.7 | 1.1 | 5.7 | 1.0 | 0.5 | 0.485 | 0.99 | |
Quantitative measures were calculated from coronal FLASH (corFLASH) and sagittal DESS (sagDESS) MRI acquired at the OAI baseline visit
MFTC/LFTC medial/lateral femorotibial compartment, MT/LT medial/lateral tibia, cMF/cLF central medial/lateral femoral condyle, P p value from paired t-tests, r Pearson correlation coefficient
Fig. 4.
Bland and Altman plots relating the cartilage thickness difference between U-Net-based automated vs. manual segmentations to the cartilage thickness averaged over these two segmentation methods for coronal FLASH MRI (corFLASH, top row) and sagittal DESS MRI (sagDESS, bottom row). The mean difference (continuous line) and the 95% limits of agreement (dotted lines) are shown in red for each of the four femorotibial cartilages
Fig. 5.
Bland and Altman plots relating the cartilage thickness difference between coronal FLASH vs sagittal DESS MRI to the cartilage thickness averaged over these two imaging protocols for both manual (top row) and U-Net-based automated segmentations (bottom row). The mean difference (continuous line) and the 95% limits of agreement (dotted lines) are shown in red for each of the four femorotibial cartilages
Cartilage volume also was statistically significantly greater when determined from automated vs. manual segmentation (range − 3.1–10.1%), except for the cMF and cLF in the sagDESS (Table 3). The total area of subchondral bone (tAB) was significantly greater when determined from automated vs. manual segmentation for the tibial cartilages (range 1.7–3.8%), whereas no significant differences were observed for the femoral condyles (range 0.0–1.1%, Table 3).
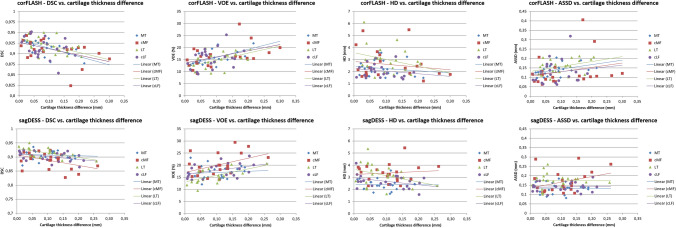
The DSC and VOE were significantly correlated with absolute cartilage thickness differences in the MT, cMF, and LT with corFLASH and for the LT with sagDESS (Table 4, Fig. 6). No statistically significant correlation was observed for the HD, but the ASSD was significantly correlated with thickness differences in the MT with corFLASH MRI (Table 4, Fig. 6).
Table 4.
Correlation between absolute differences in cartilage thickness and measures of agreement between U-Net and manual segmentations
| DSC | VOE | HD | ASSD | |
|---|---|---|---|---|
| corFLASH | ||||
| MT | − 0.61 | 0.61 | − 0.15 | 0.44 |
| cMF | − 0.49 | 0.50 | − 0.17 | 0.26 |
| LT | − 0.50 | 0.50 | − 0.29 | 0.38 |
| cLF | − 0.32 | 0.32 | − 0.19 | 0.14 |
| sagDESS | ||||
| MT | − 0.09 | 0.10 | − 0.02 | − 0.01 |
| cMF | − 0.42 | 0.43 | 0.10 | 0.33 |
| LT | − 0.58 | 0.58 | − 0.35 | 0.00 |
| cLF | − 0.42 | 0.42 | − 0.38 | 0.05 |
Absolute cartilage thickness differences between U-Net-based, automated and manual segmentations calculated from coronal FLASH (corFLASH) and sagittal DESS (sagDESS) MRI acquired at the OAI baseline visit
MT/LT medial/lateral tibia, cMF/cLF central medial/lateral femoral condyle, DSC dice similarity coefficient, HD Hausdorff distance, ASSD average symmetric surface distance
Bold face indicates significant correlation coefficients (p < 0.05)
Fig. 6.
Scatter plots relating the agreement to the absolute difference in cartilage thickness between U-Net-based automated vs. manual segmentations for coronal FLASH MRI (corFLASH, top row) and sagittal DESS MRI. sagDESS bottom row, DSC dice similarity coefficient, VOE volume overlap error, HD Hausdorff distance, ASSD average symmetric surface distance, MT/LT medial/lateral tibia, cMF/cLF central medial/lateral femoral condyle
Longitudinal test–retest reproducibility
The longitudinal change between year-1 and -2 follow-up observed in the 21 test set knees was between − 2.0 and 1.1% for automated and between − 0.9 and 1.6% for manual segmentations, with some of these changes reaching statistical significance, in particular with sagDESS MRI (Table 5).With corFLASH, the RMS SD for cartilage thickness ranged from 0.03 to 0.05 mm for manual, and from 0.02 to 0.06 mm for automated segmentations, with an RMS CV of 1.2–1.9% for manual and an RMS CV of 1.0–2.1% for automated segmentations (Table 5). With sagDESS, the RMS SD ranged from 0.03 to 0.05 mm for manual and automated segmentations, with an RMS CV of 1.2–2.0% for manual, and an RMS CV of 1.3–2.2% for automated segmentations (Table 5). Precision errors for cartilage volume and the total area of subchondral bone are also shown in Table 5.
Table 5.
Longitudinal test–retest reproducibility for manual and U-Net-based segmentations determined from year-1 and -2 follow-up MRIs of n = 21 knees from the test set
| Manual | U-Net | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MC ± SD | MC (%) | P | RMS SD | RMS CV% | MC ± SD | MC (%) | P | RMS SD | RMS CV% | |
| corFLASH | ||||||||||
| Cartilage thickness (mm) | ||||||||||
| MFTC | 0.00 ± 0.06 | 0.1 | 0.76 | 0.04 | 1.2 | 0.00 ± 0.05 | 0.0 | 0.93 | 0.04 | 1.0 |
| MT | 0.00 ± 0.04 | 0.0 | 0.96 | 0.02 | 1.5 | 0.01 ± 0.03 | 0.3 | 0.41 | 0.02 | 1.1 |
| cMF | 0.00 ± 0.04 | 0.2 | 0.67 | 0.03 | 1.5 | 0.00 ± 0.04 | − 0.2 | 0.62 | 0.03 | 1.4 |
| LFTC | 0.01 ± 0.07 | 0.2 | 0.71 | 0.05 | 1.3 | 0.01 ± 0.09 | 0.2 | 0.64 | 0.06 | 1.6 |
| LT | 0.00 ± 0.04 | − 0.2 | 0.72 | 0.03 | 1.3 | 0.01 ± 0.07 | 0.3 | 0.65 | 0.05 | 2.1 |
| cLF | 0.01 ± 0.04 | 0.5 | 0.38 | 0.03 | 1.9 | 0.00 ± 0.05 | 0.2 | 0.81 | 0.04 | 2.1 |
| Cartilage volume (mm3) | ||||||||||
| MFTC | 2.6 ± 63.4 | 0.1 | 0.85 | 43.8 | 1.5 | − 0.7 ± 79.3 | 0.0 | 0.97 | 54.7 | 1.7 |
| MT | − 2.3 ± 51.6 | − 0.1 | 0.84 | 35.6 | 1.9 | 5.5 ± 66.7 | 0.3 | 0.71 | 46.2 | 2.2 |
| cMF | 4.9 ± 30.2 | 0.5 | 0.47 | 21.1 | 2.2 | − 6.3 ± 39.5 | − 0.6 | 0.47 | 27.6 | 2.6 |
| LFTC | − 9.2 ± 69.5 | − 0.3 | 0.55 | 48.4 | 1.6 | − 41.8 ± 108.1 | − 1.2 | 0.09 | 80.2 | 2.4 |
| LT | − 13.5 ± 58.9 | − 0.6 | 0.31 | 41.8 | 2.0 | − 33.1 ± 80.2 | − 1.5 | 0.07 | 60.1 | 2.7 |
| cLF | 4.3 ± 30.4 | 0.4 | 0.52 | 21.2 | 2.1 | − 8.6 ± 55.9 | − 0.8 | 0.49 | 39.1 | 3.5 |
| Total area of subchondral bone (cm2) | ||||||||||
| MFTC | − 0.01 ± 0.12 | − 0.1 | 0.66 | 0.08 | 0.5 | − 0.03 ± 0.31 | − 0.2 | 0.66 | 0.22 | 1.3 |
| MT | − 0.03 ± 0.10 | − 0.3 | 0.20 | 0.07 | 0.6 | 0.00 ± 0.23 | 0.0 | 1.00 | 0.16 | 1.4 |
| cMF | 0.02 ± 0.08 | 0.3 | 0.37 | 0.06 | 1.1 | − 0.03 ± 0.17 | − 0.6 | 0.43 | 0.12 | 2.4 |
| LFTC | − 0.03 ± 0.12 | − 0.2 | 0.33 | 0.09 | 0.6 | − 0.17 ± 0.45 | − 1.1 | 0.09 | 0.33 | 2.2 |
| LT | − 0.02 ± 0.10 | − 0.3 | 0.30 | 0.07 | 0.8 | − 0.10 ± 0.38 | − 1.0 | 0.25 | 0.27 | 2.8 |
| cLF | 0.00 ± 0.07 | − 0.1 | 0.86 | 0.05 | 0.9 | − 0.08 ± 0.24 | − 1.3 | 0.16 | 0.17 | 3.0 |
| sagDESS | ||||||||||
| Cartilage thickness (mm) | ||||||||||
| MFTC | 0.03 ± 0.05 | 0.8 | 0.01 | 0.04 | 1.2 | 0.01 ± 0.07 | 0.3 | 0.47 | 0.05 | 1.4 |
| MT | 0.02 ± 0.03 | 1.1 | 0.02 | 0.03 | 1.5 | 0.02 ± 0.04 | 1.0 | 0.05 | 0.03 | 1.7 |
| cMF | 0.01 ± 0.04 | 0.6 | 0.22 | 0.03 | 1.7 | − 0.01 ± 0.05 | − 0.3 | 0.63 | 0.04 | 2.1 |
| LFTC | 0.01 ± 0.07 | 0.4 | 0.39 | 0.05 | 1.4 | − 0.02 ± 0.07 | − 0.4 | 0.33 | 0.05 | 1.3 |
| LT | 0.02 ± 0.06 | 0.8 | 0.24 | 0.04 | 2.0 | − 0.01 ± 0.05 | − 0.4 | 0.38 | 0.03 | 1.5 |
| cLF | 0.00 ± 0.04 | − 0.1 | 0.90 | 0.03 | 1.6 | − 0.01 ± 0.06 | − 0.4 | 0.61 | 0.04 | 2.2 |
| Cartilage volume (mm3) | ||||||||||
| MFTC | 23.9 ± 63.8 | 0.9 | 0.10 | 47.1 | 1.7 | − 4.0 ± 79.5 | − 0.1 | 0.82 | 54.9 | 1.9 |
| MT | 22.9 ± 44.5 | 1.3 | 0.03 | 34.7 | 2.0 | 3.3 ± 55.6 | 0.2 | 0.79 | 38.4 | 2.1 |
| cMF | 1.1 ± 34.3 | 0.1 | 0.89 | 23.7 | 2.2 | − 7.3 ± 47.3 | − 0.7 | 0.49 | 33.0 | 3.3 |
| LFTC | 34.3 ± 83.2 | 1.0 | 0.07 | 62.3 | 1.9 | − 29.3 ± 123.5 | − 0.9 | 0.29 | 87.7 | 2.7 |
| LT | 29.0 ± 70.6 | 1.4 | 0.07 | 52.9 | 2.5 | − 5.4 ± 84.8 | − 0.2 | 0.77 | 58.7 | 2.6 |
| cLF | 5.3 ± 28.8 | 0.5 | 0.41 | 20.2 | 1.7 | − 23.9 ± 63.0 | − 2.3 | 0.10 | 46.6 | 4.5 |
| Total area of subchondral bone (cm2) | ||||||||||
| MFTC | − 0.02 ± 0.24 | − 0.1 | 0.77 | 0.16 | 1.1 | − 0.13 ± 0.41 | − 0.8 | 0.17 | 0.30 | 1.9 |
| MT | 0.04 ± 0.17 | 0.4 | 0.36 | 0.12 | 1.2 | − 0.09 ± 0.30 | − 0.8 | 0.21 | 0.22 | 2.1 |
| cMF | − 0.05 ± 0.10 | − 1.0 | 0.03 | 0.08 | 1.4 | − 0.04 ± 0.27 | − 0.8 | 0.48 | 0.19 | 3.6 |
| LFTC | 0.06 ± 0.17 | 0.4 | 0.10 | 0.13 | 0.8 | − 0.12 ± 0.45 | − 0.8 | 0.24 | 0.32 | 2.0 |
| LT | 0.05 ± 0.13 | 0.5 | 0.08 | 0.09 | 0.9 | 0.00 ± 0.27 | 0.0 | 0.96 | 0.19 | 1.9 |
| cLF | 0.02 ± 0.12 | 0.3 | 0.56 | 0.08 | 1.4 | − 0.12 ± 0.25 | − 2.1 | 0.04 | 0.19 | 3.4 |
Longitudinal stability and test–retest precision assessed from coronal FLASH (corFLASH) and sagittal DESS (sagDESS) MRI acquired at the OAI year-1 and -2 follow-up visits
MC mean change; SD standard deviation; P p-value from paired t-tests; RMS SD Root mean square standard deviation in mm (cartilage thickness), mm3 (cartilage volume), cm2 (total area of subchondral bone); RMS CV root mean square coefficient of variation (in %); MFTC/LFTC medial/lateral femorotibial compartment; MT/LT medial/lateral tibia; cMF/cLF central medial/lateral femoral condyle
Test–retest precision errors for evaluating the potential effect of network overfitting between year-1 and -2 follow-up MRIs were computed for 39 of the 41 knees from the training set, and for all 19 knees from the validation set that had manual year-1 and -2 MRI segmentations. In two of the knees from the training set, the computation of morphometric cartilage measures failed because of invalid segmentations that could not be corrected by the post-processing steps (Fig. 3). The test–retest precision errors observed in the training and validation sets were similar to those observed in knees from the test set, but tended to be greater for some of the parameters when computed from automated segmentations (data not shown), in particular for knees from the validation set with corFLASH. This can be attributed to three of the knees in the validation set, in which the automated segmentation from corFLASH differed notably between year-1 and -2 follow-up due to obvious segmentation errors (Fig. 3).
The standard error of the measurement (SEM) and the smallest detectable change (SDC) for cartilage thickness were comparable between automated (range SEM 0.04–0.13 mm; range SDC 0.11–0.36 mm) and manual (range SEM 0.05–0.10 mm; range SDC 0.13–0.29 mm) segmentations and between corFLASH (range SEM 0.04–0.13 mm; range SDC 0.11–0.36 mm) and sagDESS MRI (range SEM 0.05–0.10 mm; range SDC 0.13–0.29 mm; Table 6). SEM and SDC for cartilage volume and total area of subchondral bone are shown in Table 6.
Table 6.
Standard error of measurement (SEM) and smallest detectable change (SDC) thresholds computed from year-1 and -2 follow-up MRIs of n = 21 knees from the test set
| corFLASH | sagDESS | |||||||
|---|---|---|---|---|---|---|---|---|
| Manual | U-Net | Manual | U-Net | |||||
| SEM | SDC | SEM | SDC | SEM | SDC | SEM | SDC | |
| Cartilage thickness (mm) | ||||||||
| MFTC | 0.09 | 0.24 | 0.07 | 0.21 | 0.07 | 0.20 | 0.10 | 0.28 |
| LFTC | 0.10 | 0.27 | 0.13 | 0.36 | 0.10 | 0.29 | 0.10 | 0.28 |
| MT | 0.05 | 0.14 | 0.04 | 0.11 | 0.05 | 0.13 | 0.05 | 0.15 |
| cMF | 0.06 | 0.16 | 0.05 | 0.15 | 0.06 | 0.16 | 0.08 | 0.21 |
| LT | 0.06 | 0.15 | 0.09 | 0.26 | 0.08 | 0.23 | 0.06 | 0.18 |
| cLF | 0.06 | 0.17 | 0.08 | 0.21 | 0.06 | 0.16 | 0.08 | 0.22 |
| Cartilage volume (mm3) | ||||||||
| MFTC | 90 | 249 | 112 | 311 | 90 | 250 | 112 | 312 |
| LFTC | 98 | 273 | 153 | 424 | 118 | 326 | 175 | 484 |
| MT | 73 | 202 | 94 | 261 | 63 | 175 | 79 | 218 |
| cMF | 43 | 118 | 56 | 155 | 49 | 135 | 67 | 185 |
| LT | 83 | 231 | 113 | 314 | 100 | 277 | 120 | 332 |
| cLF | 43 | 119 | 79 | 219 | 41 | 113 | 89 | 247 |
| Total area of subchondral bone (cm2) | ||||||||
| MFTC | 0.17 | 0.46 | 0.44 | 1.22 | 0.34 | 0.93 | 0.59 | 1.63 |
| LFTC | 0.18 | 0.49 | 0.64 | 1.77 | 0.25 | 0.68 | 0.63 | 1.75 |
| MT | 0.13 | 0.37 | 0.32 | 0.89 | 0.25 | 0.68 | 0.42 | 1.17 |
| cMF | 0.12 | 0.32 | 0.25 | 0.68 | 0.14 | 0.38 | 0.38 | 1.06 |
| LT | 0.15 | 0.41 | 0.53 | 1.47 | 0.18 | 0.49 | 0.39 | 1.08 |
| cLF | 0.10 | 0.29 | 0.33 | 0.93 | 0.16 | 0.45 | 0.36 | 0.99 |
SEM and SDC were computed from coronal FLASH (corFLASH) and sagittal DESS (sagDESS) MRI acquired at the OAI year-1 and -2 follow-up visits
MFTC/LFTC medial/lateral femorotibial compartment, MT/LT medial/lateral tibia, cMF/cLF central medial/lateral femoral condyle
Discussion
In this study, we have evaluated the segmentation agreement, accuracy, and the longitudinal test–retest reproducibility of an automated, 2D U-Net-based method for the segmentation and quantitative morphometric analysis of articular cartilage, using two MRI acquisition contrasts and orientations frequently used in clinical trials. The results demonstrate not only a high level of agreement of the segmentations, but also a high level of accuracy, and longitudinal test–retest reproducibility of morphometric analyses derived from the automated method, relative to those obtained from quality-controlled, manual segmentations as ground truth.
The U-Net architecture was chosen for automated segmentation, because it was designed to provide precise segmentations even when trained with relatively few examples [13]. The U-Net was originally intended for segmentation of neuronal structures in electron microscopic stacks, but previous studies have successfully applied it to segmentation of various musculoskeletal structures including cartilage [14–21, 23, 34]. The current study extends previous work on the relatively good agreement and accuracy demonstrated by U-Net-based cartilage segmentation methods [14–21, 23] by evaluating the accuracy, and particularly the longitudinal test–retest reproducibility of a U-Net-based segmentation pipeline for cartilage morphometry from two different MRI contrasts and orientations. This is a prerequisite before a segmentation technique can be applied to longitudinal MRI acquisitions from observational or clinical trials, with the main purpose of this technique being to detect small longitudinal changes in clinical trials and to measure the potential impact of disease-modifying treatment on these longitudinal changes. Recent studies also reported that the performance of the U-Net architecture for the segmentation of knee cartilages is on par with that observed for other current network architectures such as the V-Net, SegNet, and DeepLabV3+ [23, 37]. In contrast to the technique proposed by Ronneberger et al. [13], the current study did not employ data augmentation to artificially increase the number of examples for the training. This decision was based on the observation that data augmentation did not improve the agreement with manual segmentation results, when initially evaluating the impact of various parameters before the conduct of this study (data not shown). This observation was likely, because simple data augmentation techniques may not fully capture the heterogeneity of real-world data to improve the internal representations learned by the network. The same was observed when evaluating different loss functions (dice vs. weighted cross-entropy) or different weights for the weighted cross-entropy loss function, which were found to have a negligible impact (data not shown).
Similarly, we observed consistent results when repeating the training of the model using the same data, demonstrating the repeatability of the model training. Similar metrics were also observed when reverting the assignment of data to training, validation, and test set. The combination of features used for training the networks, in contrast, had an important impact on segmentation agreement: some combinations, such as including both the medial and the lateral femoral condyle in one model for sagDESS, did not lead to high segmentation agreement, most likely because of the similarity of the medial and lateral femoral cartilages. We, therefore, trained two separate networks for the segmentation of medial and lateral femorotibial compartment cartilages from sagDESS and this combination showed a similar performance as the one network trained for the cartilage segmentation from corFLASH MRI, despite the differences in orientation, resolution, and contrast. A combined network trained for the segmentation of both sagDESS and corFLASH MRI was also evaluated but showed a worse performance than the chosen combination of separate networks. It remains unknown, whether this is due to the different orientation or contrast (or a combination of both), but we conclude that sequence- and contrast-specific models may be superior to more general models that take greater variability of the features into account.
Most previous studies using CNNs for automated femorotibial cartilage segmentation reported DSCs between 0.78 and 0.92, and VOEs between 17 and 34% [14–16, 18–21, 23] and only one study using a combined bone and cartilage segmentation pipeline reported a higher DSC of 0.98 for femoral cartilage [17]. The agreement observed between automated and manual segmentations in the current study, therefore, compared favorably to that reported previously, both for corFLASH and sagDESS MRI. However, it should be noted that DSC comparisons across studies should be made with caution due to differences in which subjects the automated approaches are tested on.
The main purpose of the post-processing was not to improve agreement between both segmentation methods, but to correct implausible segmentations that precluded the computation of quantitative parameters of cartilage morphology. The post-processing step hence only had a small impact on overlap-based measures of agreement (DSC and VOE), whereas the distance-based measures (HD and ASSD) were improved substantially. This can be attributed to the higher sensitivity of distance-based metrics to implausible segmentations where the real boundaries of the cartilage are missed.
The automated segmentation produced consistently greater cartilage thickness of up to 5% than manual segmentation, with this systematic offset being more pronounced in corFLASH than sagDESS. Similar offsets were observed for cartilage volume, but not for the total area of subchondral bone, indicating that the overestimation is not caused at the edges of the cartilage plates but at the bone–cartilage interface or the articular cartilage surface. A similar overestimation of cartilage thickness and volume has also been observed previously for U-Net-based segmentations [14]. Yet, because these were consistent longitudinally, and because correlations with manual segmentations were high, this does not preclude that longitudinal changes in cartilage thickness (the main focus in clinical trials investigating the efficacy of therapeutic intervention) can be measured with the same sensitivity to change as by manual segmentation.
The current study relied on knees from the healthy reference cohort that were additionally confirmed to be free from radiographic OA. Some of these knees already had joint abnormalities visible on MRI [38] that did, however, not translate into a significant, disease-related change in medial or lateral femorotibial cartilage thickness over the first 2 years after enrollment, the period also included in this study [26, 39]. The statistically significant changes observed in the knees from the test set between year-1 and -2 follow-up for some of the measures can therefore most likely be attributed to statistical artifacts induced by measurement error. In addition, the changes were mostly comparable between cartilage measures computed from automated and manual segmentations, indicating a similar longitudinal reproducibility for both segmentation methods. This was also confirmed by the SEM, which was comparable for cartilage thickness computed from manual and automated segmentations. The precision errors observed in the current study were in the same range as those reported by Brem et al. and Tamez-Pena et al. for sagDESS [12, 40] and lower than the test–retest precision errors previously reported for corFLASH and sagDESS from unpaired, manual segmentations [24]. The low precision errors observed with the automated segmentation method is encouraging and advocates further application to longitudinal image acquisitions of osteoarthritic knees to evaluate its sensitivity to longitudinal change in cartilage thickness (cartilage loss). The test–retest precision errors in the test set were also not observed to be greater than those in the training or validation set. Rather, validation set test–retest errors of the automated segmentations were greater with corFLASH, because of implausible segmentations in a small number of knees. These findings suggest that the U-Net was not affected by overfitting to data used for the training process. At the same time, these findings highlight the importance of expert quality control, to ensure correct and accurate cartilage segmentations, and they highlight the challenge of applying fully automated segmentation blindly, without thoroughly checking segmentation results.
A limitation of the current study is that it only included radiographically normal knees from asymptomatic patients. However, approximately 50% of these knees demonstrated femorotibial cartilage lesions, along with other structural pathologies such as osteophytes, bone marrow lesions, meniscus damage and extrusion, effusion-synovitis and Hoffa-synovitis that affect either the cartilage appearance or that of surrounding tissues [38]. Still, these lesions did not translate into a detectable pathological change in medial or lateral femorotibial cartilage thickness over the first 2 years as previously reported [26, 39]. The OAI healthy reference cohort was, therefore, not only selected as a starting point in testing the U-Net-based segmentation approach, but also because the longitudinal reproducibility of the measurement can only be evaluated in the absence of pathological cartilage change. Hence, the OAI healthy reference cohort was ideally suited for that purpose, and has been previously used to establish progressor thresholds of cartilage loss [26]. Given that test–retest errors using the U-Net segmentation approach were similar to those from manual, quality-controlled segmentations, it can be assumed that the progressor thresholds for cartilage thickness change also apply for automated segmentations. Another limitation of the method is that, although the U-Net provided accurate segmentation for many of the knees, it failed to provide complete cartilage segmentations in some of the slices, and produced implausible segmentations in others. We were able to overcome some of these errors using the post-processing steps, but a simple, rule-based approach cannot compensate for incomplete segmentations. Such incomplete segmentations are most likely explained by the fact that the U-Net has no “real” knowledge about the context of the cartilages, and none about valid shapes. We, therefore, strongly recommend thorough quality control of all segmentations by an expert reader, and to perform manual corrections of automated segmentations where needed. Another limitation of the current study is that the femoral ROI marked by the readers in the manually segmented data sets was applied to both the manual and the automated segmentations to ensure comparability between manual and automated measures. This femoral ROI was, however, necessary to exclude posterior parts of the femoral cartilages from the segmentation, which are affected from partial volume effects in coronal MRIs and display a lesser amount of longitudinal change than the weight-bearing part in knee OA [30]. A strength of the current study is that it did not only confine itself to the analysis of DSCs and other measures of segmentation similarity, but also directly evaluated the accuracy and longitudinal test–retest reproducibility of morphometric cartilage measures, such as thickness, volume, and surface area derived from the automated segmentations. Another strength is that the approach was tested in the same knees for two different MRI contrasts and orientations, which are both frequently applied in clinical trials. Finally, the current study provided progression thresholds based on the SDC methodology [36], which can be used for classifying knees into those showing progression vs those who do not show progression.
In conclusion, this is the first study to test the accuracy and longitudinal test–retest reproducibility of quantitative cartilage morphometry using an automated, U-Net-based segmentation approach, using the two image contrasts and orientations that are most frequently used in clinical trials. We not only demonstrate a high level of agreement between automated vs. manual “ground truth” segmentation, but also a high level of accuracy, and longitudinal test–retest reproducibility for morphometric analysis of articular cartilage derived from the automated method. Yet, post-processing steps and expert quality control are highly recommended. Future research will establish with which level of sensitivity the method is able to detect longitudinal change over time in diseased knees, and the efficacy of therapeutic intervention on stopping or reverting articular cartilage loss in osteoarthritis.
Acknowledgements
The study was supported by a grant from the Paracelsus Medical University research fund (PMU-FFF; E-18/27/146-WIK). The OAI, a public–private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262), was funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners of the OAI include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. The sponsors were not involved in the design and conduct of this particular study, in the analysis and interpretation of the data, and in the preparation, review, or approval of the manuscript.
Author contributions
Study conception and design: WW, JK, FE, AC. Acquisition of data: all authors. Analysis and interpretation of data: all authors. Drafting of manuscript: WW, FE, AC. Critical revision: all authors.
Funding
Open access funding provided by Paracelsus Medical University.
Compliance with ethical standards
Conflict of interest
Wolfgang Wirth: Part-time employee and share-holder of Chondrometrics GmbH and received consulting fees from Galapagos N.V. Felix Eckstein: CEO/CMO and co-owner of Chondrometrics GmbH, and has provided consulting services to Merck KGaA, Samumed, Kolon-Tissuegene, Servier, Galapagos, Roche, Novartis, and ICM. Jana Kemnitz: Nothing to disclose. Christian F. Baumgartner: Nothing to disclose. Ender Konukoglu: Nothing to disclose. David Fuerst: Part-time employee of Chondrometrics GmbH. Akshay Chaudhari: Has provided consulting services to SkopeMR, Inc., Subtle Medical, Chondrometrics GmbH, Image Analysis Group, Edge Analytics, and Culvert Engineering; and is a shareholder of Subtle Medical, LVIS Corporation, and Brain Key.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. The Osteoarthritis Initiative (OAI) was approved by the Committee on Human Research, Institutional Review Board for the University of California, San Francisco. All OAI participants provided written informed consent, and this study was carried out in accordance with the OAI data user agreement.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vos T, Abajobir AA, Abbafati C, Abbas KM, Abate KH, Abd-Allah F, Abdulle AM, Abebo TA, Abera SF, Aboyans V, Abu-Raddad LJ, Ackerman IN, Adamu AA, Adetokunboh O, Afarideh M, Afshin A, Agarwal SK, Aggarwal R, Agrawal A, Agrawal S, Ahmad Kiadaliri A, Ahmadieh H, Ahmed MB, Aichour AN, Aichour I, Aichour MTE, Aiyar S, Akinyemi RO, Akseer N, Al Lami FH, Alahdab F, Al-Aly Z, Alam K, Alam N, Alam T, Alasfoor D, Alene KA, Ali R, Alizadeh-Navaei R, Alkerwi A, Alla F, Allebeck P, Allen C, Al-Maskari F, Al-Raddadi R, Alsharif U, Alsowaidi S, Altirkawi KA, Amare AT, Amini E, Ammar W, Amoako YA, Andersen HH, Antonio CAT, Anwari P, Ärnlöv J, Artaman A, Aryal KK, Asayesh H, Asgedom SW, Assadi R, Atey TM, Atnafu NT, Atre SR, Avila-Burgos L, Avokpaho EFGA, Awasthi A, Ayala Quintanilla BP, Ba Saleem HO, Bacha U, Badawi A, Balakrishnan K, Banerjee A, Bannick MS, Barac A, Barber RM, Barker-Collo SL, Bärnighausen T, Barquera S, Barregard L, Barrero LH, Basu S, Battista B, Battle KE, Baune BT, Bazargan-Hejazi S, Beardsley J, Bedi N, Beghi E, Béjot Y, Bekele BB, Bell ML, Bennett DA, Bensenor IM, Benson J, Berhane A, Berhe DF, Bernabé E, Betsu BD, Beuran M, Beyene AS, Bhala N, Bhansali A, Bhatt S, Bhutta ZA, Biadgilign S, Bienhoff K, Bikbov B, Birungi C, Biryukov S, Bisanzio D, Bizuayehu HM, Boneya DJ, Boufous S, Bourne RRA, Brazinova A, Brugha TS, Buchbinder R, Bulto LNB, Bumgarner BR, Butt ZA, Cahuana-Hurtado L, Cameron E, Car M, Carabin H, Carapetis JR, Cárdenas R, Carpenter DO, Carrero JJ, Carter A, Carvalho F, Casey DC, Caso V, Castañeda-Orjuela CA, Castle CD, Catalá-López F, Chang HY, Chang JC, Charlson FJ, Chen H, Chibalabala M, Chibueze CE, Chisumpa VH, Chitheer AA, Christopher DJ, Ciobanu LG, Cirillo M, Colombara D, Cooper C, Cortesi PA, Criqui MH, Crump JA, Dadi AF, Dalal K, Dandona L, Dandona R, Das Neves J, Davitoiu DV, De Courten B, De Leo D, Degenhardt L, Deiparine S, Dellavalle RP, Deribe K, Des Jarlais DC, Dey S, Dharmaratne SD, Dhillon PK, Dicker D, Ding EL, Djalalinia S, Do HP, Dorsey ER, Dos Santos KPB, Douwes-Schultz D, Doyle KE, Driscoll TR, Dubey M, Duncan BB, El-Khatib ZZ, Ellerstrand J, Enayati A, Endries AY, Ermakov SP, Erskine HE, Eshrati B, Eskandarieh S, Esteghamati A, Estep K, Fanuel FBB, Farinha CSES, Faro A, Farzadfar F, Fazeli MS, Feigin VL, Fereshtehnejad SM, Fernandes JC, Ferrari AJ, Feyissa TR, Filip I, Fischer F, Fitzmaurice C, Flaxman AD, Flor LS, Foigt N, Foreman KJ, Franklin RC, Fullman N, Fürst T, Furtado JM, Futran ND, Gakidou E, Ganji M, Garcia-Basteiro AL, Gebre T, Gebrehiwot TT, Geleto A, Gemechu BL, Gesesew HA, Gething PW, Ghajar A, Gibney KB, Gill PS, Gillum RF, Ginawi IAM, Giref AZ, Gishu MD, Giussani G, Godwin WW, Gold AL, Goldberg EM, Gona PN, Goodridge A, Gopalani SV, Goto A, Goulart AC, Griswold M, Gugnani HC, Gupta R, Gupta R, Gupta T, Gupta V, Hafezi-Nejad N, Hailu AD, Hailu GB, Hamadeh RR, Hamidi S, Handal AJ, Hankey GJ, Hao Y, Harb HL, Hareri HA, Haro JM, Harvey J, Hassanvand MS, Havmoeller R, Hawley C, Hay RJ, Hay SI, Henry NJ, Heredia-Pi IB, Heydarpour P, Hoek HW, Hoffman HJ, Horita N, Hosgood HD, Hostiuc S, Hotez PJ, Hoy DG, Htet AS, Hu G, Huang H, Huynh C, Iburg KM, Igumbor EU, Ikeda C, Irvine CMS, Jacobsen KH, Jahanmehr N, Jakovljevic MB, Jassal SK, Javanbakht M, Jayaraman SP, Jeemon P, Jensen PN, Jha V, Jiang G, John D, Johnson CO, Johnson SC, Jonas JB, Jürisson M, Kabir Z, Kadel R, Kahsay A, Kamal R, Kan H, Karam NE, Karch A, Karema CK, Kasaeian A, Kassa GM, Kassaw NA, Kassebaum NJ, Kastor A, Katikireddi SV, Kaul A, Kawakami N, Keiyoro PN, Kengne AP, Keren A, Khader YS, Khalil IA, Khan EA, Khang YH, Khosravi A, Khubchandani J, Kieling C, Kim D, Kim P, Kim YJ, Kimokoti RW, Kinfu Y, Kisa A, Kissimova-Skarbek KA, Kivimaki M, Knudsen AK, Kokubo Y, Kolte D, Kopec JA, Kosen S, Koul PA, Koyanagi A, Kravchenko M, Krishnaswami S, Krohn KJ, Kuate Defo B, Kucuk Bicer B, Kumar GA, Kumar P, Kumar S, Kyu HH, Lal DK, Lalloo R, Lambert N, Lan Q, Larsson A, Lavados PM, Leasher JL, Lee JT, Lee PH, Leigh J, Leshargie CT, Leung J, Leung R, Levi M, Li Y, Li Y, Li Kappe D, Liang X, Liben ML, Lim SS, Linn S, Liu A, Liu PY, Liu S, Liu Y, Lodha R, Logroscino G, London SJ, Looker KJ, Lopez AD, Lorkowski S, Lotufo PA, Low N, Lozano R, Lucas TCD, Macarayan ERK, Magdy Abd El Razek H, Magdy Abd El Razek M, Mahdavi M, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Malhotra R, Malta DC, Mamun AA, Manguerra H, Manhertz T, Mantilla A, Mantovani LG, Mapoma CC, Marczak LB, Martinez-Raga J, Martins-Melo FR, Martopullo I, März W, Mathur MR, Mazidi M, McAlinden C, McGaughey M, McGrath JJ, McKee M, McNellan C, Mehata S, Mehndiratta MM, Mekonnen TC, Memiah P, Memish ZA, Mendoza W, Mengistie MA, Mengistu DT, Mensah GA, Meretoja A, Meretoja TJ, Mezgebe HB, Micha R, Millear A, Miller TR, Mills EJ, Mirarefin M, Mirrakhimov EM, Misganaw A, Mishra SR, Mitchell PB, Mohammad KA, Mohammadi A, Mohammed KE, Mohammed S, Mohanty SK, Mokdad AH, Mollenkopf SK, Monasta L, Hernandez JM, Montico M, Moradi-Lakeh M, Moraga P, Mori R, Morozoff C, Morrison SD, Moses M, Mountjoy-Venning C, Mruts KB, Mueller UO, Muller K, Murdoch ME, Murthy GVS, Musa KI, Nachega JB, Nagel G, Naghavi M, Naheed A, Naidoo KS, Naldi L, Nangia V, Natarajan G, Negasa DE, Negoi I, Negoi RI, Newton CR, Ngunjiri JW, Nguyen CT, Nguyen G, Nguyen M, Le Nguyen Q, Nguyen TH, Nichols E, Ningrum DNA, Nolte S, Nong VM, Norrving B, Noubiap JJN, O’Donnell MJ, Ogbo FA, Oh IH, Okoro A, Oladimeji O, Olagunju AT, Olagunju TO, Olsen HE, Olusanya BO, Olusanya JO, Ong K, Opio JN, Oren E, Ortiz A, Osgood-Zimmerman A, Osman M, Owolabi MO, Pa M, Pacella RE, Pana A, Panda BK, Papachristou C, Park EK, Parry CD, Parsaeian M, Patten SB, Patton GC, Paulson K, Pearce N, Pereira DM, Perico N, Pesudovs K, Peterson CB, Petzold M, Phillips MR, Pigott DM, Pillay JD, Pinho C, Plass D, Pletcher MA, Popova S, Poulton RG, Pourmalek F, Prabhakaran D, Prasad N, Prasad NM, Purcell C, Qorbani M, Quansah R, Rabiee RHS, Radfar A, Rafay A, Rahimi K, Rahimi-Movaghar A, Rahimi-Movaghar V, Rahman M, Rahman MHU, Rai RK, Rajsic S, Ram U, Ranabhat CL, Rankin Z, Rao PV, Rao PC, Rawaf S, Ray SE, Reiner RC, Reinig N, Reitsma MB, Remuzzi G, Renzaho AMN, Resnikoff S, Rezaei S, Ribeiro AL, Ronfani L, Roshandel G, Roth GA, Roy A, Rubagotti E, Ruhago GM, Saadat S, Sadat N, Safdarian M, Safi S, Safiri S, Sagar R, Sahathevan R, Salama J, Salomon JA, Salvi SS, Samy AM, Sanabria JR, Santomauro D, Santos IS, Santos JV, Santric Milicevic MM, Sartorius B, Satpathy M, Sawhney M, Saxena S, Schmidt MI, Schneider IJC, Schöttker B, Schwebel DC, Schwendicke F, Seedat S, Sepanlou SG, Servan-Mori EE, Setegn T, Shackelford KA, Shaheen A, Shaikh MA, Shamsipour M, Shariful Islam SM, Sharma J, Sharma R, She J, Shi P, Shields C, Shigematsu M, Shinohara Y, Shiri R, Shirkoohi R, Shirude S, Shishani K, Shrime MG, Sibai AM, Sigfusdottir ID, Silva DAS, Silva JP, Silveira DGA, Singh JA, Singh NP, Sinha DN, Skiadaresi E, Skirbekk V, Slepak EL, Sligar A, Smith DL, Smith M, Sobaih BHA, Sobngwi E, Sorensen RJD, Sousa TCM, Sposato LA, Sreeramareddy CT, Srinivasan V, Stanaway JD, Stathopoulou V, Steel N, Stein DJ, Stein MB, Steiner C, Steiner TJ, Steinke S, Stokes MA, Stovner LJ, Strub B, Subart M, Sufiyan MB, Suliankatchi Abdulkader R, Sunguya BF, Sur PJ, Swaminathan S, Sykes BL, Sylte DO, Tabarés-Seisdedos R, Taffere GR, Takala JS, Tandon N, Tavakkoli M, Taveira N, Taylor HR, Tehrani-Banihashemi A, Tekelab T, Temam Shifa G, Terkawi AS, Tesfaye DJ, Tesssema B, Thamsuwan O, Thomas KE, Thrift AG, Tiruye TY, Tobe-Gai R, Tollanes MC, Tonelli M, Topor-Madry R, Tortajada M, Touvier M, Tran BX, Tripathi S, Troeger C, Truelsen T, Tsoi D, Tuem KB, Tuzcu EM, Tyrovolas S, Ukwaja KN, Undurraga EA, Uneke CJ, Updike R, Uthman OA, Uzochukwu BSC, Van Boven JFM, Varughese S, Vasankari T, Venkatesh S, Venketasubramanian N, Vidavalur R, Violante FS, Vladimirov SK, Vlassov VV, Vollset SE, Wadilo F, Wakayo T, Wang YP, Weaver M, Weichenthal S, Weiderpass E, Weintraub RG, Werdecker A, Westerman R, Whiteford HA, Wijeratne T, Wiysonge CS, Wolfe CDA, Woodbrook R, Woolf AD, Workicho A, Wulf Hanson S, Xavier D, Xu G, Yadgir S, Yaghoubi M, Yakob B, Yan LL, Yano Y, Ye P, Yimam HH, Yip P, Yonemoto N, Yoon SJ, Yotebieng M, Younis MZ, Zaidi Z, Zaki MES, Zegeye EA, Zenebe ZM, Zhang X, Zhou M, Zipkin B, Zodpey S, Zuhlke LJ, Murray CJL. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393:1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 3.Eckstein F, Le Graverand MH. Plain radiography or magnetic resonance imaging (MRI): which is better in assessing outcome in clinical trials of disease-modifying osteoarthritis drugs? Summary of a debate held at the World Congress of Osteoarthritis 2014. Semin Arthritis Rheum. 2015;45(3):251–256. doi: 10.1016/j.semarthrit.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Hunter DJ, Altman RD, Cicuttini F, Crema MD, Duryea J, Eckstein F, Guermazi A, Kijowski R, Link TM, Martel-Pelletier J, Miller CG, Mosher TJ, Ochoa-Albiztegui RE, Pelletier JP, Peterfy C, Raynauld JP, Roemer FW, Totterman SM, Gold GE. OARSI clinical trials recommendations: knee imaging in clinical trials in osteoarthritis. Osteoarthr Cartil. 2015;23:698–715. doi: 10.1016/j.joca.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Eckstein F, Guermazi A, Gold G, Duryea J, Hellio Le Graverand M-P, Wirth W, Miller CGG. Imaging of cartilage and bone: promises and pitfalls in clinical trials of osteoarthritis. Osteoarthr Cartil. 2014;22:1516–1532. doi: 10.1016/j.joca.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochberg MC, Guermazi A, Guehring H, Aydemir A, Wax S, Fleuranceau-Morel P, Reinstrup Bihlet A, Byrjalsen I, Ragnar Andersen J, Eckstein F. Effect of intra-articular sprifermin vs placebo on femorotibial joint cartilage thickness in patients with osteoarthritis. JAMA. 2019;322:1360. doi: 10.1001/jama.2019.14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAlindon T, LaValley M, Schneider E, Nuite M, Lee JY, Price LL, Lo G, Dawson-Hughes B. Effect of vitamin D supplementation on progression of knee pain and cartilage volume loss in patients with symptomatic osteoarthritis: a randomized controlled trial. JAMA. 2013;309:155–162. doi: 10.1001/jama.2012.164487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conaghan PG, Bowes MA, Kingsbury SR, Brett A, Guillard G, Tunblad K, Rizoska B, Larsson T, Holmgren Å, Manninen A, Göhlin K, Heber W, Graham P, Jansson Å, Wadell C, Bethell R, Öhd J. Six months’ treatment with MIV-711, a novel Cathepsin K inhibitor induces osteoarthritis structure modification: results from a randomized double-blind placebo-controlled phase IIA trial. Osteoarthr Cartil. 2018;26:S25–S26. doi: 10.1016/j.joca.2018.02.066. [DOI] [Google Scholar]
- 9.Deckx H, Van Der Stoep M, Wooning M, Bernard K, Grankov S, Imbert O, Pueyo M, Eckstein F. Study design of a phase 2 clinical trial with a disease-modifying Osteoarthritis drug candidate GLPG1972/S201086: the roccella trial. Osteoarthr Cartil. 2020;28:S499–S500. [Google Scholar]
- 10.Pedoia V, Majumdar S, Link TM. Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phy. 2016;29:207–221. doi: 10.1007/s10334-016-0532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stammberger T, Eckstein F, Michaelis M, Englmeier KH, Reiser M. Interobserver reproducibility of quantitative cartilage measurements: comparison of b-spline snakes and manual segmentation. Magn Reson Imaging. 1999;17:1033–1042. doi: 10.1016/S0730-725X(99)00040-5. [DOI] [PubMed] [Google Scholar]
- 12.Brem MH, Lang PK, Neumann G, Schlechtweg PM, Schneider E, Jackson R, Yu J, Eaton CB, Hennig FF, Yoshioka H, Pappas G, Duryea J. Magnetic resonance image segmentation using semi-automated software for quantification of knee articular cartilage—initial evaluation of a technique for paired scans. Skelet Radiol. 2009;38:505–511. doi: 10.1007/s00256-009-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical image computing and computer-assisted intervention – MICCAI. Cham: Springer; 2015. pp. 234–241. [Google Scholar]
- 14.Norman B, Pedoia BSV, Majumdar S. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177–185. doi: 10.1148/radiol.2018172322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal. 2019;52:109–118. doi: 10.1016/j.media.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Tack A, Zachow S (2019) Accurate automated volumetry of cartilage of the knee using convolutional neural networks: data from the osteoarthritis initiative. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), Venice, Italy, pp 40–43
- 17.Lee H, Hong H, Kim J (2018) BCD-NET: a novel method for cartilage segmentation of knee MRI via deep segmentation networks with bone-cartilage-complex modeling. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), Washington, DC, pp 1538–1541
- 18.Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H (2018) Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. In: 2018 IEEE 15th international symposium biomedical imaging (ISBI 2018), pp 851–854
- 19.Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med. 2018;80:2759–2770. doi: 10.1002/mrm.27229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2018;79:2379–2391. doi: 10.1002/mrm.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical image computing and computer-assisted intervention – MICCAI. Heidelberg: Berlin; 2013. pp. 246–253. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhari AS, Stevens KJ, Wood JP, Chakraborty AK, Gibbons EK, Fang Z, Desai AD, Lee JH, Gold GE, Hargreaves BA. Utility of deep learning super-resolution in the context of osteoarthritis MRI biomarkers. J Magn Reson Imaging. 2020;51:768–779. doi: 10.1002/jmri.26872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai AD, Caliva F, Iriondo C, Khosravan N, Mortazi A, Jambawalikar S, Torigian D, Ellerman J, Akcakaya M, Bagci U, Tibrewala R, Flament I, O`Brien M, Majumdar S, Perslev M, Pai A, Igel C, Dam EB, Gaj S, Yang M, Nakamura K, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari AS (2020) The international workshop on osteoarthritis imaging knee MRI segmentation challenge: a multi-institute evaluation and analysis framework on a standardized dataset. Archiv (preprint). arxiv:2004.14003 [DOI] [PMC free article] [PubMed]
- 24.Eckstein F, Hudelmaier M, Wirth W, Kiefer B, Jackson R, Yu J, Eaton CB, Schneider E. Double echo steady state magnetic resonance imaging of knee articular cartilage at 3 Tesla: a pilot study for the Osteoarthritis Initiative. Ann Rheum Dis. 2006;65:433–441. doi: 10.1136/ard.2005.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckstein F, Yang M, Guermazi A, Roemer FW, Hudelmaier M, Picha K, Baribaud F, Wirth W, Felson DT. Reference values and Z-scores for subregional femorotibial cartilage thickness—results from a large population-based sample (Framingham) and comparison with the non-exposed Osteoarthritis Initiative reference cohort. Osteoarthr Cartil. 2010;18:1275–1283. doi: 10.1016/j.joca.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirth W, Maschek S, Ladel C, Guehring H, Michaelis M, Eckstein F. Structural progressor thresholds of femorotibial cartilage change over 1 to 4-years for different MRI orientations and contrasts—data from the osteoarthritis initiative. Osteoarthr Cartil. 2019;27:S315–S316. doi: 10.1016/j.joca.2019.02.715. [DOI] [Google Scholar]
- 27.Eckstein F, Mascheck S, Sharma L, Kwoh KC, Wirth W. Quantitative cartilage thickness change in radiographically normal knees with and without OA risk factors—data from the OAI. Osteoarthr Cartil. 2018;26:S426–S427. doi: 10.1016/j.joca.2018.02.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckstein F, Wirth W, Nevitt MC. Recent advances in osteoarthritis imaging-the Osteoarthritis Initiative. Nat Rev Rheumatol. 2012;8:622–630. doi: 10.1038/nrrheum.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthr Cartil. 2008;16:1433–1441. doi: 10.1016/j.joca.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckstein F, Benichou O, Wirth W, Nelson DR, Maschek S, Hudelmaier M, Kwoh CK, Guermazi A, Hunter D. Magnetic resonance imaging-based cartilage loss in painful contralateral knees with and without radiographic joint space narrowing: data from the Osteoarthritis Initiative. Arthritis Rheum. 2009;61:1218–1225. doi: 10.1002/art.24791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckstein F, Ateshian G, Burgkart R, Burstein D, Cicuttini F, Dardzinski B, Gray M, Link TM, Majumdar S, Mosher T, Peterfy C, Totterman S, Waterton J, Winalski CS, Felson D. Proposal for a nomenclature for magnetic resonance imaging based measures of articular cartilage in osteoarthritis. Osteoarthr Cartil. 2006;14:974–983. doi: 10.1016/j.joca.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Wirth W, Eckstein F. A technique for regional analysis of femorotibial cartilage thickness based on quantitative magnetic resonance imaging. IEEE Trans Med Imaging. 2008;27:737–744. doi: 10.1109/TMI.2007.907323. [DOI] [PubMed] [Google Scholar]
- 33.Baumgartner CF, Koch LM, Pollefeys M, Konukoglu E. An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation. In: Pop M, Sermesant M, Jodoin P-M, Lalande A, Zhuang X, Yang G, Young A, Bernard O, editors. Statistical atlases and computational models of the heart. ACDC MMWHS challenges. Berlin: Springer; 2018. pp. 111–119. [Google Scholar]
- 34.Kemnitz J, Baumgartner CF, Eckstein F, Chaudhari A, Ruhdorfer A, Wirth W, Eder SK, Konukoglu E. Clinical evaluation of fully automated thigh muscle and adipose tissue segmentation using a U-Net deep learning architecture in context of osteoarthritic knee pain. Magn Reson Mater Phy. 2020;33(4):483–493. doi: 10.1007/s10334-019-00816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingma DP, Ba JL (2015) ADAM: a method for stochastic optimization. In: Conference paper ICLR 2015, pp 1–15
- 36.de Vet HCW, Terwee CB, Knol DL, Bouter LM. When to use agreement versus reliability measures. J Clin Epidemiol. 2006;59:1033–1039. doi: 10.1016/j.jclinepi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Desai AD, Gold GE, Hargreaves BA, Chaudhari AS (2019) Technical considerations for semantic segmentation in MRI using convolutional neural networks. Archiv (preprint). arxiv:1902.01977
- 38.Roemer FW, Eckstein F, Duda GN, Guermazi A, Maschek S, Wirth W. Baseline structural tissue pathology is not strongly associated with longitudinal change in transverse relaxation time (T2) in knees without osteoarthritis. Eur J Radiol. 2019;118:161–168. doi: 10.1016/j.ejrad.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Eckstein F, Nevitt M, Gimona A, Picha K, Lee JH, Davies RY, Dreher D, Benichou O, Graverand MHLE, Hudelmaier M, Maschek S, Le Graverand MP, Wirth W. Rates of change and sensitivity to change in cartilage morphology in healthy knees and in knees with mild, moderate, and end-stage radiographic osteoarthritis: results from 831 participants from the osteoarthritis initiative. Arthritis Care Res (Hoboken) 2011;63:311–319. doi: 10.1002/art.30191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamez-Pena JG, Farber J, Gonzalez PC, Schreyer E, Schneider E, Totterman S. Unsupervised segmentation and quantification of anatomical knee features: data from the Osteoarthritis Initiative. IEEE Trans Biomed Eng. 2012;59:1177–1186. doi: 10.1109/TBME.2012.2186612. [DOI] [PubMed] [Google Scholar]