Abstract
Hepatocellular carcinoma (HCC) is one of the most commonly cancers with poor prognosis and drug response. Identifying accurate therapeutic targets would facilitate precision treatment and prolong survival for HCC. In this study, we analyzed liver hepatocellular carcinoma (LIHC) RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA), and identified PARD3 as one of the most significantly differentially expressed genes (DEGs). Then, we investigated the relationship between PARD3 and outcomes of HCC, and assessed predictive capacity. Moreover, we performed functional enrichment and immune infiltration analysis to evaluate functional networks related to PARD3 in HCC and explore its role in tumor immunity. PARD3 expression levels in 371 HCC tissues were dramatically higher than those in 50 paired adjacent liver tissues (p < 0.001). High PARD3 expression was associated with poor clinicopathologic feathers, such as advanced pathologic stage (p = 0.002), vascular invasion (p = 0.012) and TP53 mutation (p = 0.009). Elevated PARD3 expression also correlated with lower overall survival (OS, HR = 2.08, 95% CI = 1.45–2.98, p < 0.001) and disease-specific survival (DSS, HR = 2.00, 95% CI = 1.27–3.16, p = 0.003). 242 up-regulated and 71 down-regulated genes showed significant association with PARD3 expression, which were involved in genomic instability, response to metal ions, and metabolisms. PARD3 is involved in diverse immune infiltration levels in HCC, especially negatively related to dendritic cells (DCs), cytotoxic cells, and plasmacytoid dendritic cells (pDCs). Altogether, PARD3 could be a potential prognostic biomarker and therapeutic target of HCC.
Subject terms: Cancer, Computational biology and bioinformatics, Biomarkers, Gastroenterology, Oncology
Introduction
Hepatocellular carcinoma (HCC) is the fifth most commonly diagnosed cancer, accounting for 7% of all cancers worldwide1, and furthermore, the incidence of HCC continues increasing by 2–3% annually2,3. With a 5-year overall survival of 12–18%, HCC ranks second in terms of cancer-related death1,2,4,5. In the past decades, substantial progress has been made in prevention, diagnosis, and treatment for HCC6–8. However, due to insidious onset, rapid progression and a lack of effective screening strategies, less than 30% of HCC patients can be diagnosed at an early stage, and have the opportunity to undergo radical treatments4. Transarterial chemoembolization or systemic therapies are widely recommended for patients with advanced disease4,5, but unfortunately, the improvement in prognosis is not satisfactory enough even with the latest targeted drugs or immune-based therapies9–13. Therefore, identifying more accurate prognostic biomarkers and therapeutic targets would facilitate precision treatment and prolong survival for HCC patients.
Cell polarity is essential for epithelial cells to maintain normal morphology and perform physiological functions14. Aberrant cell polarity, a hallmark of cancers, is implicated in tumor formation, growth, invasion, and metastasis15. Cell polarity is regulated by sets of evolutionarily conserved polarity proteins including the partitioning-defective (Par) complex, Scribble complexes, and Crumbs complexes16. The Par complex, which has the most ubiquitous function among these proteins, consists of Par3, Par6, and atypical protein kinase C (aPKC)17. Par3 serves as an adaptor protein for the assembly of Par complex and multiple proteins, such as the Rac-GEF, Tiam1 or Rho GTPases, thereby activating polarity signaling16,18. PARD3, encoding Par3 protein, is a single-copy gene with 26 exons, and located on chromosome 10p11.22-p11.2117. Deleterious variants of PARD3 were first detected in neural tube defects, coeliac disease and ulcerative colitis19–21, and subsequently, a series of studies identified the dual function of PARD3 in different malignant tumors of epithelial origin14–18,22–27. However, the specific role and detailed mechanism of PARD3 in HCC has not been fully elucidated17.
To screen a biomarker closely related to the formation and progression of HCC, we analyzed the differential expression of PARD3 and its clinicopathological relevance, using liver hepatocellular carcinoma (LIHC) RNA sequencing (RNA-seq) data of HCC patients from The Cancer Genome Atlas (TCGA). Then, we put PARD3 into a prognosis analysis in order to evaluate predictive capacity. Moreover, we performed comparative transcriptome analysis, functional enrichment analysis and correlation analysis between PARD3 and immune cell infiltration to evaluate functional networks related to PARD3 in HCC and explore its role in tumor immunity.
Results
Overexpression of PARD3 in HCC
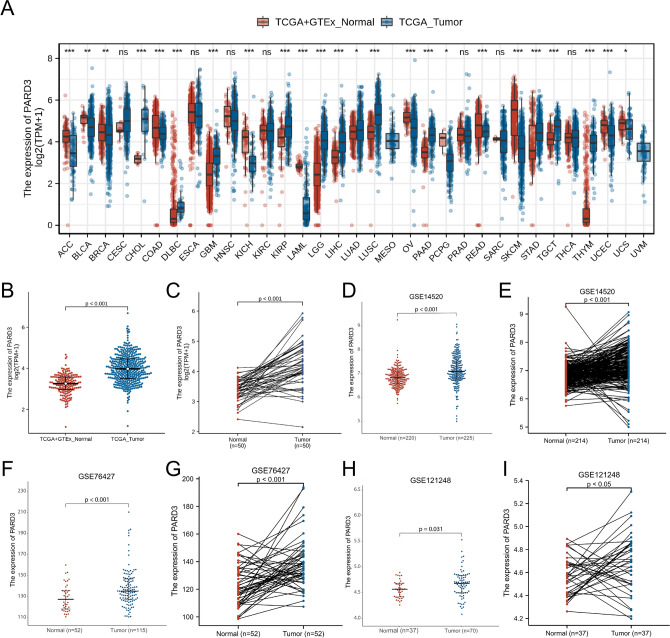
We initially compared PARD3 expression between tumor and normal tissues in multiple cancer types using the UCSC Xena database. As shown in Fig. 1A, PARD3 expression was significantly higher in cholangiocarcinoma (CHOL), Diffuse large B-cell lymphoma (DLBC), glioblastoma (GBM), kidney renal papillary cell carcinoma (KIRP), low-grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), testicular germ cell tumor (TGCT) and thymic carcinoma (THYM). In contrast, PARD3 expression was significantly lower in adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), acute myeloid leukemia (LAML), ovarian serous cystadenocarcinoma (OV), pheochromocytoma and paraganglioma (PCPG), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), uterine corpus endometrial carcinoma (UCEC) and uterine carcinosarcoma (UCS).
Figure 1.
Differential expression of PARD3. (A) Differential expression of PARD3 in 33 types of human cancers and normal tissues from TCGA and GTEx (*p < 0.05, **p < 0.01, ***p < 0.001, ns: p ≥ 0.05). (B) Differential expression of PARD3 in HCC and normal liver tissues from TCGA and GTEx. (C) Differential expression of PARD3 in paired HCC and their adjacent liver tissues from TCGA. (D,F,H) Differential expression of PARD3 in HCC and normal liver tissues from GSE14520, GSE76427 and GSE121248. (E,G,I) Differential expression of PARD3 in paired HCC and their adjacent liver tissues from GSE14520, GSE76427 and GSE121248.
Then, we focused on PARD3 expression in HCC patients. The data from TCGA and GTEx revealed that PARD3 expression levels in 371 HCC tissues were dramatically higher than those in normal liver tissues, which was validated in 50 paired HCC and their adjacent liver tissues (Fig. 1B,C). The data from other three independent cohorts (GSE14520, GSE76427 and GSE121248) confirmed the above results that PARD3 was overexpressed in HCC patients (Fig. 1D–I).
Correlation of PARD3 overexpression with poor clinicopathologic features
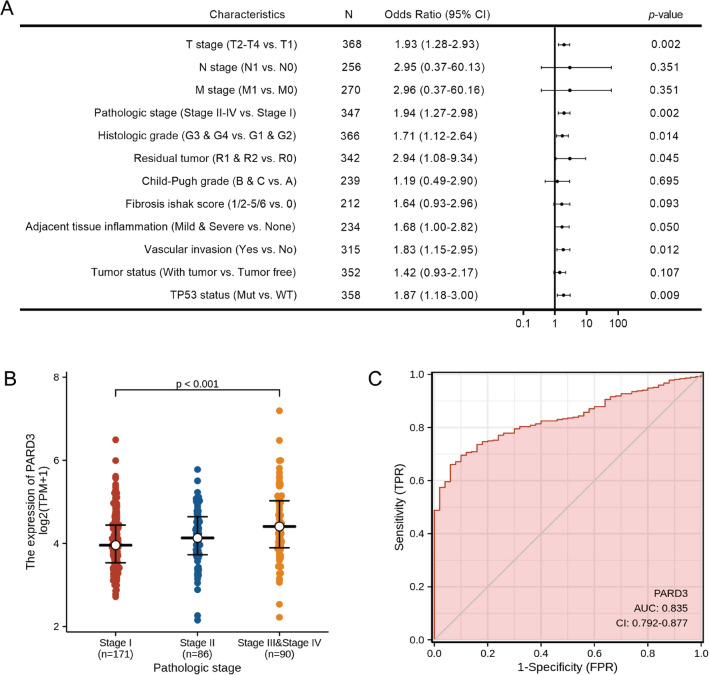
To investigate overexpression of PARD3 and its clinicopathological relevance, 371 HCC samples with detailed patient information (retrieved from TCGA in June 2020) were divided into two groups by the median value of PARD3 expression. As shown in Table 1, high PARD3 expression was associated with advanced T stage, pathologic stage, residual tumor, histologic grade, vascular invasion and higher alpha fetoprotein (AFP). Otherwise, high PARD3 expression group also carried more TP53 mutation (Mut) than low PARD3 expression group. Whereas, the distributions of other clinicopathologic features showed no difference between high and low PARD3 expression group. Univariate logistic regression further confirmed the association between high PARD3 expression and poor clinicopathologic characteristics in HCC patients (Fig. 2A,B). In addition, the area under receiver operation characteristic (ROC) curve (AUC, AUC = 0.835, 95% CI = 0.792–0.877) indicated that PARD3 had a good diagnostic power, and was expected to be a potential biomarker for HCC (Fig. 2C).
Table 1.
Clinicopathologic characteristics in LIHC cohort according to PARD3 expression.
| Characteristics | level | Low expression of PARD3 | High expression of PARD3 | p-value |
|---|---|---|---|---|
| n | 186 | 185 | ||
| Gender (%) | Female | 56 (30.1%) | 65 (35.1%) | 0.356 |
| Male | 130 (69.9%) | 120 (64.9%) | ||
| Race (%) | Asian | 78 (44.3%) | 80 (43.7%) | 0.983 |
| Black or African | 8 (4.5%) | 9 (4.9%) | ||
| White | 90 (51.1%) | 94 (51.4%) | ||
| T stage (%) | T1 | 105 (57.4%) | 76 (41.1%) | 0.006† |
| T2 | 43 (23.5%) | 51 (27.6%) | ||
| T3 | 32 (17.5%) | 48 (25.9%) | ||
| T4 | 3 (1.6%) | 10 (5.4%) | ||
| N stage (%) | N0 | 125 (99.2%) | 127 (97.7%) | 0.622‡ |
| N1 | 1 (0.8%) | 3 (2.3%) | ||
| M stage (%) | M0 | 132 (99.2%) | 134 (97.8%) | 0.622‡ |
| M1 | 1 (0.8%) | 3 (2.2%) | ||
| Pathologic stage (%) | Stage I | 100 (57.5%) | 71 (41.0%) | 0.006†,‡ |
| Stage II | 41 (23.6%) | 45 (26.0%) | ||
| Stage III | 31 (17.8%) | 54 (31.2%) | ||
| Stage IV | 2 (1.1%) | 3 (1.7%) | ||
| Tumor status (%) | Tumor free | 108 (61.4%) | 93 (52.8%) | 0.132 |
| With tumor | 68 (38.6%) | 83 (47.2%) | ||
| Residual tumor (%) | R0 | 172 (97.2%) | 152 (92.1%) | 0.023†,‡ |
| R1 | 4 (2.3%) | 13 (7.9%) | ||
| R2 | 1 (0.6%) | 0 (0.0%) | ||
| Histologic grade (%) | G1 | 36 (19.6%) | 19 (10.4%) | 0.023† |
| G2 | 92 (50.0%) | 85 (46.7%) | ||
| G3 | 52 (28.3%) | 70 (38.5%) | ||
| G4 | 4 (2.2%) | 8 (4.4%) | ||
| Adjacent hepatic tissue inflammation (%) | Mild | 46 (37.4%) | 53 (47.7%) | 0.144 |
| None | 69 (56.1%) | 48 (43.2%) | ||
| Severe | 8 (6.5%) | 10 (9.0%) | ||
| Child–Pugh grade (%) | A | 118 (91.5%) | 99 (90.0%) | 0.810‡ |
| B | 10 (7.8%) | 11 (10.0%) | ||
| C | 1 (0.8%) | 0 (0.0%) | ||
| Fibrosis Ishak score (%) | 0 | 47 (39.8%) | 27 (28.7%) | 0.405 |
| 1–2 | 16 (13.6%) | 15 (16.0%) | ||
| 3–4 | 15 (12.7%) | 13 (13.8%) | ||
| 5–6 | 40 (33.9%) | 39 (41.5%) | ||
| Vascular invasion (%) | No | 116 (72.0%) | 90 (58.4%) | 0.016† |
| Yes | 45 (28.0%) | 64 (41.6%) | ||
| TP53 status (%) | Mut | 40 (22.2%) | 62 (34.8%) | 0.012† |
| WT | 140 (77.8%) | 116 (65.2%) | ||
| Age (median [IQR]) | 61.50 [53.00, 69.00] | 60.50 [51.00, 69.00] | 0.329 | |
| Height (median [IQR]) | 168.00 [161.50, 174.00] | 168.00 [161.00, 173.00] | 0.590 | |
| Weight (median [IQR]) | 72.00 [60.00, 84.00] | 68.00 [59.00, 81.00] | 0.210 | |
| BMI (median [IQR]) | 24.84 [22.14, 28.88] | 24.19 [21.24, 28.43] | 0.256 | |
| AFP (ng/ml) (median [IQR]) | 10.00 [3.00, 52.00] | 28.00 [5.00, 1456.00] | 0.001† | |
| Alb (g/dl) (median [IQR]) | 4.00 [3.42, 4.40] | 4.00 [3.55, 4.30] | 0.520 | |
| PT (s) (median [IQR]) | 1.10 [1.00, 8.93] | 1.10 [1.00, 9.57] | 0.538 |
Mut mutant, WT wild type, BMI body mass index, AFP alpha fetoprotein, Alp albumin, TB total bilirubin, PT prothrombin time.
†Statistically significant.
‡Fisher exact test.
Figure 2.
Correlation between PARD3 expression and clinicopathologic characteristics in HCC. (A) Correlation between PARD3 expression (categorical dependent variable) and clinicopathologic characteristics (logistic regression; Odds Ratio equals to the ratio of oddshigh expression to oddslow expression). (B) High PARD3 expression (categorical dependent variable) was associated with advanced pathologic stages. (C) Time-dependent receiver operation characteristic (ROC) curve of PARD3. False positive rate (FPR) is indicated on the abscissa; and true positive rate (TPR) is indicated on the ordinate.
Correlation of PARD3 overexpression with adverse Outcomes in HCC
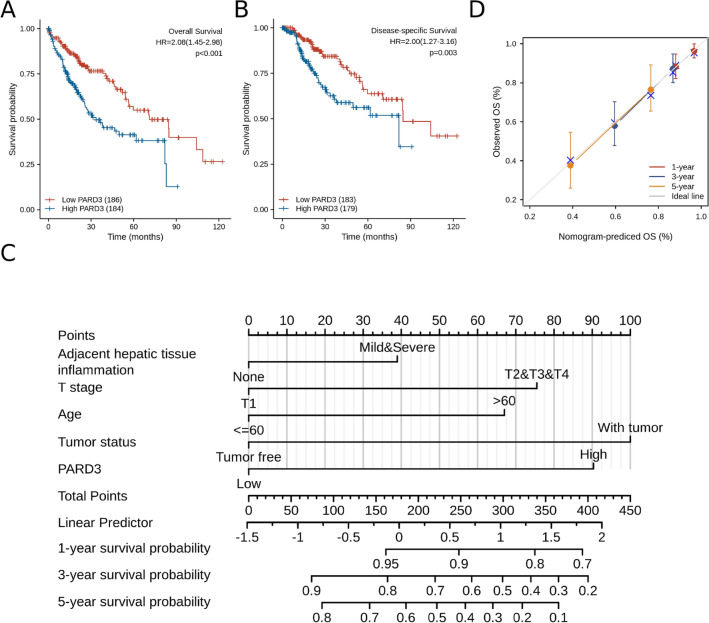
As PARD3 overexpression was correlated with poor clinicopathologic features, we then explored the prognostic value of PARD3 in HCC. Kaplan–Meier survival curve demonstrated that elevated PARD3 expression led to decreased overall survival (OS, HR = 2.08, 95% CI = 1.45–2.98, p < 0.001) and disease-specific survival (DSS, HR = 2.00, 95% CI = 1.27–3.16, p = 0.003) (Fig. 3A,B), which was validated through TISIDB database and other two independent cohorts in GEO database (GSE76427 and GSE14520) (Fig. S1). Further univariate and multivariate Cox analysis showed that PARD3 was an independent risk factor for HCC patients leading to adverse outcomes, and tumor status was the other negative factor (Table S1 and S2)28.
Figure 3.
Impact of PARD3 expression on survival in HCC patients. (A) Overall survival (OS) was significantly higher in low PARD3 expression group (HR = 2.08, 95% CI = 1.45–2.98, p < 0.001); (B) Disease-specific survival (DSS) was significantly higher in low PARD3 expression group (HR = 2.00, 95% CI = 1.27–3.16, p = 0.003); (C) Nomogram for prediction of OS in HCC patients (existing adjacent hepatic tissue inflammation, T2–T4 stage, age > 60 years, with tumor, and high PARD3 expression were converted into 39, 75.5, 67, 100 and 90.25 points, respectively. The total points accumulated by the above covariates correspond to the predicted probability for an HCC patient); (D) Calibration curve of the nomogram.
Based on multivariate COX regression, we constructed a nomogram to provide a visual, intuitive and quantitative description of those risk factors and their weights in HCC, and predict the probability of 1-year, 3-year and 5-year survival. Potential covariates included adjacent hepatic tissue inflammation (39 points), T stage (75.5 points), age (67 points), tumor status (100 points) and PARD3 expression (90.25 points), and higher total points indicated worse prognosis (Fig. 3C).
Then, we made calibration curve of the nomogram, and calculated concordance index (C-index) to assess the predictive ability of PARD3 as a biomarker for HCC. As shown in Fig. 3D, the bias-corrected line in the calibration curve was close to the ideal line, and the C-index was 0.702 (95% CI = 0.668–0.736). In all, the nomogram was available to predict the prognosis of HCC patients, and PARD3 exhibited stable predictive ability.
Co-expression genes and biological functions related to PARD3 in HCC
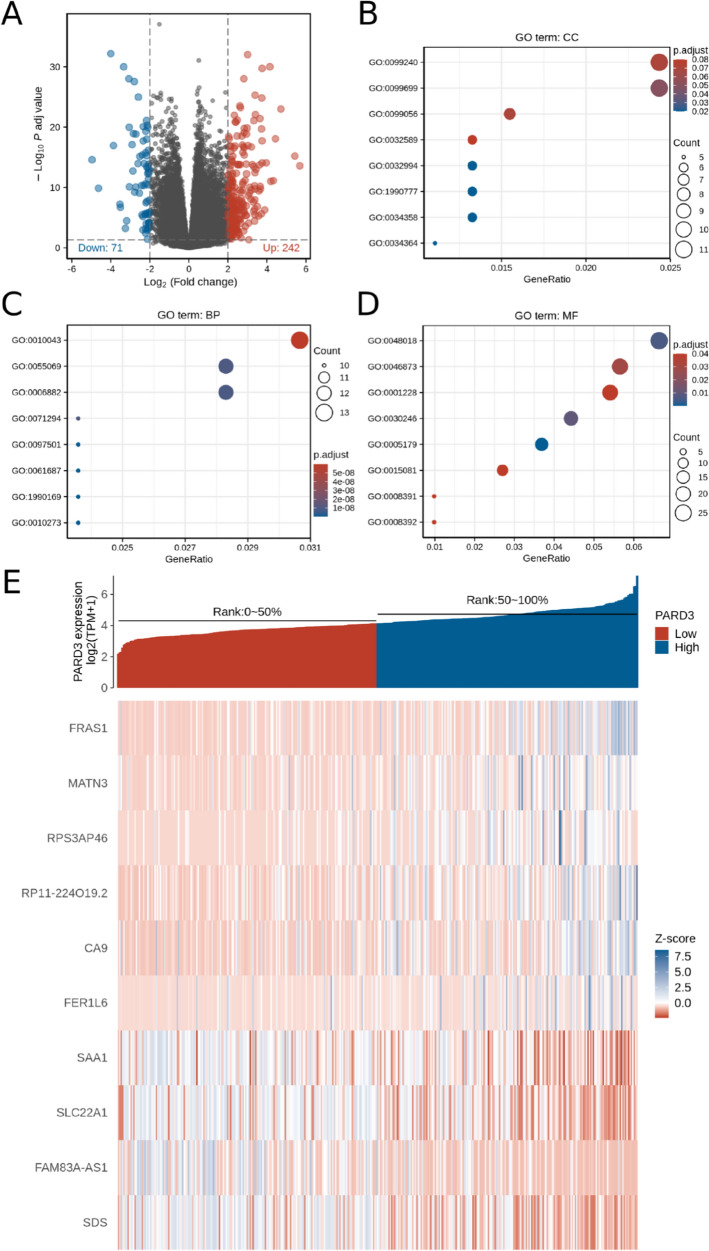
To overview the biological roles of PARD3 in HCC, we conducted gene ontology (GO) enrichment analysis of PARD3 and its associated identify differentially expressed genes (DEGs), including biological processes (BPs), molecular functions (MFs) and cellular components (CCs). As illustrated in the volcano plot (Fig. 4A), 242 up-regulated and 71 down-regulated genes were significantly related with PARD3 expression, top 10 of which were presented in the heatmap (Fig. 4E). As the Par complex consists of Par3, Par6, and aPKC, we specifically analyzed the differentially expressing of PARD6A/B/G, encoding Par6, and PRKCI/Z, encoding aPKC using Spearman correlation test. The correlation values of PARD6A, PARD6B, PARD6G, PRKCI and PRKCIZ were − 0.01 (p > 0.05), 0.51 (p < 0.001), 0.54 (p < 0.001), 0.64 (p < 0.001) and 0.23 (p < 0.001), respectively (Table S3).
Figure 4.
Co-expression genes and biological functions related to PARD3 in HCC. (A) Volcano plot of differential gene profiles between high and low PARD3 expression groups (|logFC| > 2 and adjusted p-value < 0.05); (B) Cellular components (CCs) enrichment; (C) Biological processes (BPs) enrichment; (D) Molecular functions (MFs) enrichment; (E) Expression heatmap of top 10 PARD3-associated genes. Description of GO identifiers: GO:0034358: plasma lipoprotein particle; GO:1990777: lipoprotein particle; GO:0032994: protein-lipid complex; GO:0034364: high-density lipoprotein particle; GO:0099699: integral component of synaptic membrane; GO:0099056: integral component of presynaptic membrane; GO:0099240: intrinsic component of synaptic membrane; GO:0032589: neuron projection membrane. GO:0010273: detoxification of copper ion; GO:1990169: stress response to copper ion; GO:0061687: detoxification of inorganic compound; GO:0097501: stress response to metal ion; GO:0006882: cellular zinc ion homeostasis; GO:0055069: zinc ion homeostasis; GO:0071294: cellular response to zinc ion; GO:0010043: response to zinc ion. GO:0005179: hormone activity; GO:0048018: receptor ligand activity; GO:0030246: carbohydrate binding; GO:0046873: metal ion transmembrane transporter activity; GO:0008392: arachidonic acid epoxygenase activity; GO:0008391: arachidonic acid monooxygenase activity; GO:0001228: DNA-binding transcription activator activity, RNA polymerase II-specific; GO:0015081: sodium ion transmembrane transporter activity.
The result of CCs indicated that PARD3 and its associated DEGs were primarily located in lipoprotein particles, synaptic membranes and neuron projection membranes (Fig. 4B). In terms of BPs and MFs, these genes mainly participated in detoxification of copper ion (Cu), stress response to metal ions, and zinc ion (Zn) homeostasis, which associated with receptor ligand activity, metal ion transmembrane transporter activity, DNA-binding transcription activator activity, hormone activity, carbohydrate binding, and arachidonic acid oxygenase activity (Fig. 4C,D).
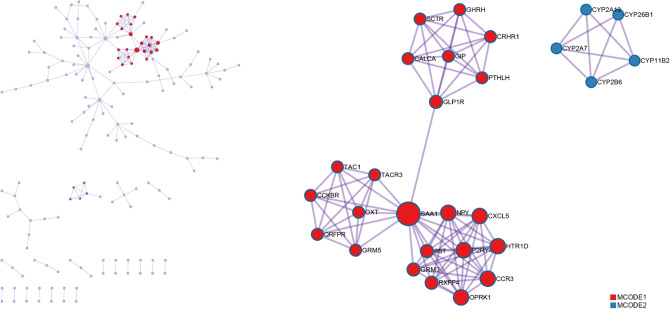
Signaling pathways, working as functional units of gene groups, play an important role in cell biological effects. Thus, we identified significantly enriched signaling pathways between low and high PARD3 expression groups by Gene set enrichment analysis (GSEA). According to normalized enrichment scores (NSEs), 431 pathways were significantly associated with high expression of PARD3, including cell cycle and mitosis, DNA double strand break repair, cell motility, Rho, MAPK, TP53, response to metal ions, and pathways related to metabolism (Table 2). In addition, we made a protein–protein interaction (PPI) to highlight the most important protein functional groups interacting with each other. The two most crucial MCODE subnetworks were SAA1-related cluster and CYP-related cluster, both of which were involved in metabolism and homeostasis (Fig. 5). The above result suggested a negative impact of PARD3 on tumorigenesis and progression of HCC.
Table 2.
Pathways enriched in high expression groups using GSEA.
| Name | NES | Adjusted p-value | FDR q-value |
|---|---|---|---|
| REACTOME_CELL_CYCLE_MITOTIC | 2.718 | 0.019 | 0.013 |
| REACTOME_RHO_GTPASES_ACTIVATE_FORMINS | 2.653 | 0.019 | 0.013 |
| REACTOME_FCERI_MEDIATED_MAPK_ACTIVATION | 2.468 | 0.019 | 0.013 |
| REACTOME_DNA_DOUBLE_STRAND_BREAK_REPAIR | 2.399 | 0.019 | 0.013 |
| REACTOME_MET_PROMOTES_CELL_MOTILITY | 2.314 | 0.019 | 0.013 |
| REACTOME_REGULATION_OF_TP53_ACTIVITY | 2.111 | 0.019 | 0.013 |
| REACTOME_PYRIMIDINE_CATABOLISM | − 2.090 | 0.019 | 0.013 |
| REACTOME_RESPONSE_TO_METAL_IONS | − 2.608 | 0.019 | 0.013 |
| KEGG_FATTY_ACID_METABOLISM | − 2.575 | 0.019 | 0.013 |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM | − 2.727 | 0.019 | 0.013 |
FDR false discovery rate, NES normalized enrichment score.
Figure 5.
Protein–protein interaction (PPI) network of PARD3-associated pathways.
Association of PARD3 with immune infiltration in HCC
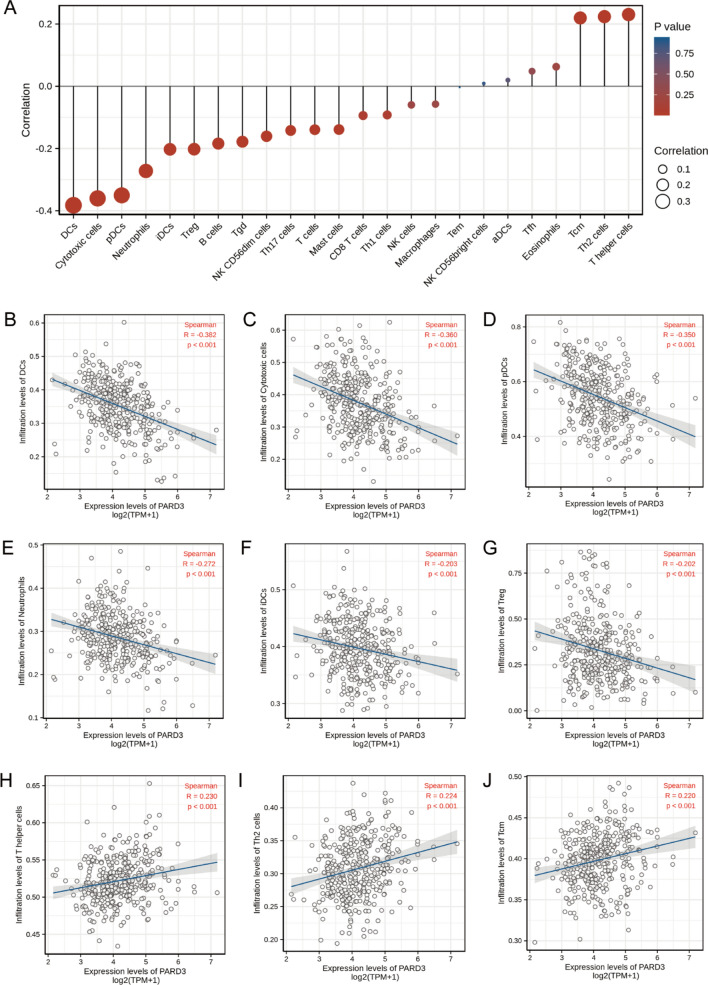
Immune infiltration, which influences tumor purity, is one of the major risk factors in cancers29–31, hence we quantified the enrichment scores (ECs) of 24 types of tumor-infiltrating immune cells (TIICs), in order to evaluate the association between PARD3 and immune infiltration levels in HCC. As illustrated in Fig. 6, PARD3 was involved in infiltration of T helper cells, Th2 cells, and T central memory (Tcm); but negatively related to infiltration of dendritic cells (DCs), cytotoxic cells, plasmacytoid dendritic cells (pDCs), neutrophils, immature DCs (iDCs), and regulatory T cells (Treg). Furthermore, we replicated immune infiltration analysis using another tumor-immune system interaction database (TISIDB), and obtained consistent results (Fig. S2).
Figure 6.
Association of PARD3 with immune infiltration. (A) Association of PARD3 with immune infiltration (DCs dendritic cells, pDCs plasmacytoid DCs, iDCs immature DCs, aDCs activated DCs, Treg regulatory T cells, Tgd T gamma delta, Th helper T cells, Tfh T follicular helper, NK natural killer, Tem T effector memory, Tcm T central memory). (B–G) PARD3 was negatively related to infiltration of DCs, cytotoxic cells, pDCs, neutrophils, iDCs, and Treg (p < 0.01). (H–J) PARD3 was positively related to in infiltration of T helper cells, Th2 cells, and Tcm (p < 0.01).
Discussion
PARD3 plays a crucial role in establishment and maintenance of epithelial cell polarity23. So far, at least five PARD3 variants have been identified in human liver cDNA library32. PARD3 largely engages in cancer cell proliferation, apoptosis, invasion, migration and epithelial-mesenchymal transition (EMT)18,24,33,34. The modulation of PARD3 in tumorigenesis and progression among different cancers seems to be controversial. For instance, PARD3 acts as a tumor suppressor in lung, bladder, breast, cervical, esophageal and pancreatic cancers and malignant melanoma15,22,24,27,35–38, but it is found to be activated in ovarian cancer and clear-cell renal carcinoma18,25,39. In skin cancers, PARD3 shows dual effects depending on the tumor type16. In terms of HCC, a study reported the association between overexpression of PARD3 and extrahepatic metastasis, and suggested one of its possible mechanisms. However, the specific role and detailed mechanism of PARD3 in HCC has not been fully elucidated17. Hence, we performed the bioinformatic analysis using several independent databases to explore the potential functions of PARD3 in HCC, including pathway interactions, immune infiltration, and long-term survival.
Based on TCGA database, PARD3 expressed differentially in 24 types of cancers (Fig. 1A). Thereinto, the expression levels in two types of cancers were inconsistent with previous studies, including lung cancer and pancreatic cancer22,27, which may derive in part from data collection approaches and patients’ biological properties. More specifically, our study provided direct evidences that PARD3 was an independent risk factor for HCC development. Firstly, PARD3 expression was dramatically higher in HCC than in normal liver tissues (Fig. 1B–I). Secondly, high PARD3 expression group contained more patients with advanced pathologic stages, vascular invasion, and TP53 Mut, suggesting that overexpression of PARD3 was significantly associated with poor clinicopathologic features with a good predictive power (AUC = 0.835, Fig. 2 and Table 1). Thirdly, elevated PARD3 expression led to shorter OS and DSS in both whites and Asians regardless of gender, age and Child–Pugh grade (Fig. 3A,B). All these results proved PARD3 as a potential prognostic biomarker of HCC.
Thus, we further explored the possible mechanism by which high PARD3 expression worsens the outcomes of HCC patients. As an adverse prognostic indicator of HCC, PARD3 was involved in many pivotal mechanisms in cancer, including cell cycle40, DNA damage and repair41, and cell motility42 (Table 2). It is well known that genomic instability and mutagenesis, which caused by erroneous DNA repair, are closely correlated with poor prognosis and drug resistance in HCC41. TP53 is universally recognized as a hub gene in responding to DNA damage and guarding the genome, and its mutation is observed in about half of all solid tumors, including HCC43,44. Furthermore, p38MAPK is able to control p53 activation via direct phosphorylation. Based on our result of enrichment analysis that PARD3 was associated with MAPK pathway and TP53 regulation (Table 2), we hypothesized PARD3 may affect the formation and progression of HCC by regulating TP53 via MAPK pathway. However, the hypothesis requires further investigation. Rho Family GTPases, which is closely interact with PARD3, were widely reported to regulate cell cycle and cell motility across human cancer of different origins42. PARD3 directly activates Rac1, promoting proliferation and motility of cancer cells, and leads to tumorigenesis, angiogenesis, invasion and metastasis45–47. Likewise, Par complex also links Rho small GTPases to regulate asymmetrical cell division and cell polarization48, which manipulate EMT and mesenchymal-epithelial transition (MET)49. Our findings that PARD3 was significantly implicated in Rho pathway also provided evidences to confirm this theory (Table 2). In addition, some lncRNAs showed significant correlations with PARD3 expression, such as FAM83A-AS1, which is involved in HCC (Table 2)50.
Remarkably, we found that PARD3 and its associated DEGs mainly participated in cellular response to metal ions (Fig. 4C,D, and Table 2). Previous studies have provided a possible relationship between metal ion homeostasis and vascular invasion in HCC, which may be mediated by p5351–53. Disturbance in Cu and Zn homeostasis has been reported as a significant factor associated with tumor proliferation, angiogenesis and invasion in HCC, and furthermore, cellular response to Cu and Zn is probably involved in mitochondrial accumulation and stability of p53, so as to influence proliferation and apoptosis of hepatoma cells54–56.
In the past decades, reprogramming of energy metabolism was added into the list of cancer hallmarks29. Interestingly, we found many co-expression genes and pathways related to PARD3 were involved in deregulation of metabolisms, covering types of metabolic processes like fat acids, amino acids and pyrimidines (Table 2). In order to sustain prodigious proliferation, tumors exert a specialized metabolism that differs from normal tissues. During the period, tumors recruit abundant nucleotides to maintain unlimited replicative potential, and uptake more nutrients to support unchecked cell growth. In particular, alterations in metabolism fatty acid and glycine, serine and threonine have been investigated as a promoter of HCC initiation and progression57,58. Moreover, SLC22A1, a DEG inversely related to PARD3 (Fig. 4E), is a key regulator of metabolism, which is extensively considered as a suppressor of HCC development59–61.
Besides, PPI enrichment analysis screened SAA1-related cluster and CYP-related cluster as the two most crucial MCODE subnetworks, both of which were involved in metabolism and homeostasis (Fig. 5). Recent study showed that downregulated SAA1 was closely associated with progression of HCC and low anti-tumor immune infiltrating62; and CYP families might impact HCC cell viability via modulating biotransformation63. The results provided supporting evidence that PARD3 might promote HCC via regulating metabolism and homeostasis.
Recently, cellular metabolism has emerged as a determinant of the viability and function of both tumor cells and immune cells. Meanwhile, tumor metabolism is reported as an immune checkpoint58,64. As discussed above, PARD3 was linked to some important metabolic processes, and meanwhile, several enriched pathways were also associated with immune response (Table S4). Thus, we hypothesized that there may be an association between PARD3 and immune infiltration. As expected, PARD3 is correlated with diverse immune infiltration levels in HCC, especially DCs, cytotoxic cells and pDCs (Fig. 6). DCs are a heterogeneous population of professional antigen-presenting cells central to the induction and maintenance of adaptive immunity within tumor microenvironment58,65. In particular, two subsets of DCs exert the most potent antitumor functions, including conventional DCs type 1 (cDC1s) that stimulate T cell proliferation, and pDCs that produce interferon-α (IFN-α)65,66. cDC1s not only take up and cross-present tumor antigens via major histocompatibility complex (MHC) class I to activate naive CD8+ T cells; but also support the cytotoxicity of CD8+ T cells by secreting large amounts of interleukin-12 (IL-12). Then, activated cytotoxic CD8+ T cells migrate to tumors and kill them65. pCDs play two opposite roles in tumor immunity depending on their subsets via inducing Treg or activating cytotoxic T cells respectively65,66. Based on our result that PARD3 negatively correlated with DCs and cytotoxic T cells, we speculate that immune infiltration related to PARD3 may contribute to the unfavorable outcomes for HCC, yet the specific regulation mechanism needed to further elucidate.
In summary, our study reveals that overexpression of PARD3 correlates with poor clinicopathologic features and adverse outcomes in HCC. Moreover, the crosstalk of cellular response of metal ion, metabolism and immune infiltration within tumor microenvironment may partly explain the function of PARD3 in HCC development. However, bioinformatic analysis based on TCGA also has some limitations. First, the sample sizes of blacks and stage IV in LIHC may be too small to show a significant difference between groups. Additionally, transcriptome sequencing cannot directly reflect the protein activity and expression level. Therefore, our results should be verified by further research using sufficient HCC clinical samples, and detailed mechanisms of PARD3 need investigating more intensively. Despite the limitations, our findings provide multilevel evidence for the value of PARD3 as a potential prognostic biomarker and therapeutic target of HCC.
Materials and methods
RNA sequencing data and processing
RNA-Seq data (Workflow Type: HTSeq-FPKM) and corresponding clinical information were retrieved from TCGA-LIHC project, among which 371 HCC patients with complete survival information were retained. Then, level 3 HTseq-FPKM data were transformed to transcripts per million reads (TPM) for further analysis. Unavailable or unknown clinical data were treated as missing values67. RNA-Seq data of multiple cancer types were downloaded from the online database UCSC Xena (https://xenabrowser.net/datapages/), and analyzed using Toil68. This study complied with the publication guidelines provided by TCGA.
Differentially expressed gene analysis
DESeq2 package was used to identify DEGs69. The cut-off value of PARD3 expression was determined by its median value, and the thresholds were defined as |log fold change (log FC)| > 2 and adjusted p-value < 0.05.
The differential expression of PARD3 was simultaneously validated using Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo), including three independent cohorts (GSE14520, GSE76427 and GSE121248)70–72.
Functional enrichment analysis
Metascape analysis
Metascape (http://metascape.org) was used as a gene list analysis tool to conduct GO enrichment analysis of DEGs73, including BPs, MFs and CCs. P-value < 0.05, minimum count > 3 and enrichment factor > 1.5 were considered to be significant. The Cytoscape plug-in MCODE was used to screen crucial clustering subnetworks in PPI network.
Gene set enrichment analysis
GSEA was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, which was performed with 1000 permutations for each analysis using curated gene sets (C2.cp.v7.0.symbols.gmt) as the reference gene set74. Visualization and statistics were carried out by R package clusterProfiler75. Adjusted p-value < 0.05, false discovery rate (FDR) q-value < 0.25 and |normalized enrichment score (NES)| > 1 were considered to be significant.
Immune infiltration analysis
The relative abundance of each immunocyte type was described with EC in single-sample Gene Set Enrichment Analysis (ssGSEA). ECs for 24 types of TIICs were quantified using GSVA package in R as reported previously76.
The immune infiltration analysis of PARD3 was replicated using TISIDB database (http://cis.hku.hk/TISIDB)77.
Prognostic model generation and statistical analysis
All statistics were performed using R (v.3.6.2). The PARD3 expression levels between tumor and normal tissues in paired or non-paired samples were compared using Wilcoxon signed-rank test and Wilcoxon rank sum test, respectively. The discrimination ability of PARD3 in HCC was evaluated using the AUC in ROC78. The correlation between PARD3 expression and screened DEGs were analyzed using Spearman’s correlation. The correlation between PARD3 expression and immune infiltration were analyzed using Spearman’s correlation, while ECs of the two groups with different expression level were compared using Wilcoxon rank test. The relationship between clinicopathological features and PARD3 expression in HCC patients was assessed using Kruskal–Wallis test, Wilcoxon rank test or Spearman’s correlation, while prognostic relevance of the two groups with different expression level were compared using Pearson χ2 test, Fisher exact test or univariate logistic regression. A survival curve was plotted using Kaplan–Meier method, and analyzed by Cox regression. Baseline variables with a p-value < 0.1 on univariate analysis were included in multivariate Cox regression model79,80. A nomogram was generated to predict the prognosis of HCC based on the result of multivariate Cox regression analysis, including significant clinical characteristics and PARD3 expression. C-index was used to validate the predictive power of the model81. Statistical results were displayed with p-value, and hazard ratio (HR) at a 95% confidence interval (95% CI). p-value < 0.05 were considered to be statistically significant.
Supplementary Information
Author contributions
F.Y. and H.Z. and S.L. analysis; J.H. and K.L. data curation; J.H. and Y.T. visualization; K.L. design of the work and original draft revision; S.L. original draft preparation. All authors reviewed the manuscript.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90507-w.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, et al. Guidelines for diagnosis and treatment of primary liver cancer in China (2017 edition) Liver Cancer. 2018;7:235–260. doi: 10.1159/000488035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galle PR, et al. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Kang D, Shim S, Cho J, Lim HK. Systematic review of studies assessing the health-related quality of life of hepatocellular carcinoma patients from 2009 to 2018. Korean J. Radiol. 2020;21:633–646. doi: 10.3348/kjr.2019.0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rau KM, et al. Sequential circulating tumor cell counts in patients with locally advanced or metastatic hepatocellular carcinoma: Monitoring the treatment response. J. Clin. Med. 2020 doi: 10.3390/jcm9010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia F, Zhang D. Expert consensus on multidisciplinary diagnosis and treatment of precancerous lesions of hepatocellular carcinoma (2020 edition) Chin. J. Hepatol. 2020;28:14–20. doi: 10.3760/cma.j.issn.1007-3418.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Cainap C, et al. Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: Results of a randomized phase III trial. J. Clin. Oncol. 2015;33:172–179. doi: 10.1200/jco.2013.54.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu AX, et al. SEARCH: A phase III, randomized, double-blind, placebo-controlled trial of sorafenib plus erlotinib in patients with advanced hepatocellular carcinoma. J. Clin. Oncol. 2015;33:559–566. doi: 10.1200/jco.2013.53.7746. [DOI] [PubMed] [Google Scholar]
- 11.Finn RS, et al. Outcomes of sequential treatment with sorafenib followed by regorafenib for HCC: Additional analyses from the phase III RESORCE trial. J. Hepatol. 2018;69:353–358. doi: 10.1016/j.jhep.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 12.El-Khoueiry AB, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet (London, England) 2017;389:2492–2502. doi: 10.1016/s0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feun LG, et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer. 2019;125:3603–3614. doi: 10.1002/cncr.32339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou PJ, et al. Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway. J. Exp. Clin. Cancer Res. 2017;36:139. doi: 10.1186/s13046-017-0609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mescher M, et al. The epidermal polarity protein Par3 is a non-cell autonomous suppressor of malignant melanoma. J. Exp. Med. 2017;214:339–358. doi: 10.1084/jem.20160596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iden S, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Jan YJ, et al. Expression of partitioning defective 3 (Par-3) for predicting extrahepatic metastasis and survival with hepatocellular carcinoma. Int. J. Mol. Sci. 2013;14:1684–1697. doi: 10.3390/ijms14011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura H, et al. Expression of Par3 polarity protein correlates with poor prognosis in ovarian cancer. BMC Cancer. 2016;16:897. doi: 10.1186/s12885-016-2929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, et al. Quantitative measurement of PARD3 copy number variations in human neural tube defects. Cell. Mol. Neurobiol. 2018;38:605–614. doi: 10.1007/s10571-017-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, et al. Rare deleterious PARD3 variants in the aPKC-binding region are implicated in the pathogenesis of human cranial neural tube defects via disrupting apical tight junction formation. Hum. Mutat. 2017;38:378–389. doi: 10.1002/humu.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wapenaar MC, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463–467. doi: 10.1136/gut.2007.133132. [DOI] [PubMed] [Google Scholar]
- 22.Bonastre E, et al. PARD3 inactivation in lung squamous cell carcinomas impairs STAT3 and promotes malignant invasion. Cancer Res. 2015;75:1287–1297. doi: 10.1158/0008-5472.can-14-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jha HC, et al. KSHV-mediated regulation of Par3 and SNAIL contributes to B-cell proliferation. PLoS Pathog. 2016;12:e1005801. doi: 10.1371/journal.ppat.1005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dugay F, et al. Overexpression of the polarity protein PAR-3 in clear cell renal cell carcinoma is associated with poor prognosis. Int. J. Cancer. 2014;134:2051–2060. doi: 10.1002/ijc.28548. [DOI] [PubMed] [Google Scholar]
- 26.Zen K, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–2918. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 27.Guo X, et al. Par3 regulates invasion of pancreatic cancer cells via interaction with Tiam1. Clin. Exp. Med. 2016;16:357–365. doi: 10.1007/s10238-015-0365-2. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Tumor purity as an underlying key factor in glioma. Clin. Cancer Res. 2017;23:6279–6291. doi: 10.1158/1078-0432.ccr-16-2598. [DOI] [PubMed] [Google Scholar]
- 31.Mao Y, et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag. Res. 2018;10:3569–3577. doi: 10.2147/cmar.s171855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang CM, Xu YH. Down-regulated expression of atypical PKC-binding domain deleted asip isoforms in human hepatocellular carcinomas. Cell Res. 2001;11:223–229. doi: 10.1038/sj.cr.7290090. [DOI] [PubMed] [Google Scholar]
- 33.Song T, et al. Loss of Par3 promotes lung adenocarcinoma metastasis through 14-3-3ζ protein. Oncotarget. 2016;7:64260–64273. doi: 10.18632/oncotarget.11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat. Cell Biol. 2013;15:189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, et al. Downregulation of PKCζ/Pard3/Pard6b is responsible for lung adenocarcinoma cell EMT and invasion. Cell. Signal. 2017;38:49–59. doi: 10.1016/j.cellsig.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimatsu Y, et al. Roles of the PDZ-binding motif of HPV 16 E6 protein in oncogenic transformation of human cervical keratinocytes. Cancer Sci. 2017;108:1303–1309. doi: 10.1111/cas.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namdarian B, et al. Loss of APKC expression independently predicts tumor recurrence in superficial bladder cancers. Urol. Oncol. 2013;31:649–655. doi: 10.1016/j.urolonc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Kitaichi T, et al. Loss of PAR-3 protein expression is associated with invasion, lymph node metastasis, and poor survival in esophageal squamous cell carcinoma. Hum. Pathol. 2017;62:134–140. doi: 10.1016/j.humpath.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Dagher J, et al. Cytoplasmic PAR-3 protein expression is associated with adverse prognostic factors in clear cell renal cell carcinoma and independently impacts survival. Hum. Pathol. 2014;45:1639–1646. doi: 10.1016/j.humpath.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sy SM, Guo Y, Lan Y, Ng H, Huen MS. Preemptive homology-directed DNA repair fosters complex genomic rearrangements in hepatocellular carcinoma. Transl. Oncol. 2020;13:100796. doi: 10.1016/j.tranon.2020.100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oxford G, Theodorescu D. Ras superfamily monomeric G proteins in carcinoma cell motility. Cancer Lett. 2003;189:117–128. doi: 10.1016/s0304-3835(02)00510-4. [DOI] [PubMed] [Google Scholar]
- 43.Akula SM, et al. RAS/RAF/MEK/ERK, PI3K/PTEN/AKT/mTORC1 and TP53 pathways and regulatory miRs as therapeutic targets in hepatocellular carcinoma. Expert Opin. Ther. Targets. 2019;23:915–929. doi: 10.1080/14728222.2019.1685501. [DOI] [PubMed] [Google Scholar]
- 44.Stramucci L, Pranteda A, Bossi G. Insights of Crosstalk between p53 protein and the MKK3/MKK6/p38 MAPK signaling pathway in cancer. Cancers (Basel) 2018 doi: 10.3390/cancers10050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueyama T. Rho-family small GTPases: From highly polarized sensory neurons to cancer cells. Cells. 2019 doi: 10.3390/cells8020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David M, Petit D, Bertoglio J. Cell cycle regulation of Rho signaling pathways. Cell Cycle (Georgetown, Tex.) 2012;11:3003–3010. doi: 10.4161/cc.21088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama M, et al. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev. Cell. 2008;14:205–215. doi: 10.1016/j.devcel.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 49.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 50.He J, Yu J. 2019. Long noncoding RNA FAM83A-AS1 facilitates hepatocellular carcinoma progression by binding with NOP58 to enhance the mRNA stability of FAM83A. Biosci. Rep. [DOI] [PMC free article] [PubMed]
- 51.Imai K, et al. Clinicopathological and prognostic significance of preoperative serum zinc status in patients with hepatocellular carcinoma after initial hepatectomy. Ann. Surg. Oncol. 2014;21:3817–3826. doi: 10.1245/s10434-014-3786-3. [DOI] [PubMed] [Google Scholar]
- 52.Shao J, et al. TPP-related mitochondrial targeting copper (II) complex induces p53-dependent apoptosis in hepatoma cells through ROS-mediated activation of Drp1. Cell Commun. Signal. 2019;17:149. doi: 10.1186/s12964-019-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alshatwi AA, Han CT, Schoene NW, Lei KY. Nuclear accumulations of p53 and Mdm2 are accompanied by reductions in c-Abl and p300 in zinc-depleted human hepatoblastoma cells. Exp. Biol. Med. (Maywood, N.J.) 2006;231:611–618. doi: 10.1177/153537020623100516. [DOI] [PubMed] [Google Scholar]
- 54.Jin C, et al. Novel copper complex CTB regulates methionine cycle induced TERT hypomethylation to promote HCC cells senescence via mitochondrial SLC25A26. Cell Death Dis. 2020;11:844. doi: 10.1038/s41419-020-03048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozeki I, et al. Predictors of hepatocellular carcinoma after hepatitis C virus eradication following direct-acting antiviral treatment: Relationship with serum zinc. J. Clin. Biochem. Nutr. 2020;66:245–252. doi: 10.3164/jcbn.19-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamai Y, et al. Serum copper, zinc and metallothionein serve as potential biomarkers for hepatocellular carcinoma. PLoS One. 2020;15:e0237370. doi: 10.1371/journal.pone.0237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sangineto M, et al. Lipid metabolism in development and progression of hepatocellular carcinoma. Cancers. 2020 doi: 10.3390/cancers12061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leone RD, Powell JD. Metabolism of immune cells in cancer. Nat. Rev. Cancer. 2020;20:516–531. doi: 10.1038/s41568-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heise M, et al. Downregulation of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) in human hepatocellular carcinoma and their prognostic significance. BMC Cancer. 2012;12:109. doi: 10.1186/1471-2407-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaeffeler E, et al. DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. Genome Med. 2011;3:82. doi: 10.1186/gm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herraez E, et al. Expression of SLC22A1 variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology (Baltimore, MD) 2013;58:1065–1073. doi: 10.1002/hep.26425. [DOI] [PubMed] [Google Scholar]
- 62.Zhang W, et al. Immune infiltration-associated serum amyloid A1 predicts favorable prognosis for hepatocellular carcinoma. World J. Gastroenterol. 2020;26:5287–5301. doi: 10.3748/wjg.v26.i35.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lepri SR, et al. Genistein affects expression of cytochrome P450 (CYP450) genes in hepatocellular carcinoma (HEPG2/C3A) cell line. Drug Metab. Lett. 2018;12:138–144. doi: 10.2174/1872312812666180709150440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 65.de Winde CM, Munday C, Acton SE. Molecular mechanisms of dendritic cell migration in immunity and cancer. Med. Microbiol. Immunol. 2020;209:515–529. doi: 10.1007/s00430-020-00680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lurje I, Hammerich L, Tacke F. Dendritic cell and T cell crosstalk in liver fibrogenesis and hepatocarcinogenesis: Implications for prevention and therapy of liver cancer. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21197378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H, Zhang J. Decreased expression of TFAP2B in endometrial cancer predicts poor prognosis: A study based on TCGA data. Gynecol. Oncol. 2018;149:592–597. doi: 10.1016/j.ygyno.2018.03.057. [DOI] [PubMed] [Google Scholar]
- 68.Vivian J, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin. Cancer Res. 2007;13:6275–6283. doi: 10.1158/1078-0432.Ccr-06-2236. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, et al. Identification of a two-layer regulatory network of proliferation-related microRNAs in hepatoma cells. Nucleic Acids Res. 2012;40:10478–10493. doi: 10.1093/nar/gks789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 77.Ru B, et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics (Oxford, England) 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 78.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stone GW, et al. A prospective natural-history study of coronary atherosclerosis. N. Engl. J. Med. 2011;364:226–235. doi: 10.1056/NEJMoa1002358. [DOI] [PubMed] [Google Scholar]
- 80.Kang SJ, et al. Predictors for functionally significant in-stent restenosis: An integrated analysis using coronary angiography, IVUS, and myocardial perfusion imaging. JACC Cardiovasc. Imaging. 2013;6:1183–1190. doi: 10.1016/j.jcmg.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 81.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008;26:1364–1370. doi: 10.1200/jco.2007.12.9791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- He J, Yu J. 2019. Long noncoding RNA FAM83A-AS1 facilitates hepatocellular carcinoma progression by binding with NOP58 to enhance the mRNA stability of FAM83A. Biosci. Rep. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.