Abstract
Haloferax sp strain NRS1 (MT967913) was isolated from a solar saltern on the southern coast of the Red Sea, Jeddah, Saudi Arabia. The present study was designed for estimate the potential capacity of the Haloferax sp strain NRS1 to synthesize (silver nanoparticles) AgNPs. Biological activities such as thrombolysis and cytotoxicity of biosynthesized AgNPs were evaluated. The characterization of silver nanoparticles biosynthesized by Haloferax sp (Hfx-AgNPs) was analyzed using UV–vis spectroscopy, transmission electron microscopy (TEM), X-ray diffraction (XRD), and Fourier-transform infrared spectroscopy (FTIR). The dark brown color of the Hfx-AgNPs colloidal showed maximum absorbance at 458 nm. TEM image analysis revealed that the shape of the Hfx-AgNPs was spherical and a size range was 5.77- 73.14 nm. The XRD spectra showed a crystallographic plane of silver nanoparticles, with a crystalline size of 29.28 nm. The prominent FTIR peaks obtained at 3281, 1644 and 1250 cm− 1 identified the Functional groups involved in the reduction of silver ion reduction to AgNPs. Zeta potential results revealed a negative surface charge and stability of Hfx-AgNPs. Colloidal solution of Hfx-AgNPs with concentrations ranging from 3.125 to 100 μg/mL was used to determine its hemolytic activity. Less than 12.5 μg/mL of tested agent showed no hemolysis with high significant decrease compared with positive control, which confirms that Hfx-AgNPs are considered non-hemolytic (non-toxic) agents according to the ISO/TR 7405-1984(f) protocol. Thrombolysis activity of Hfx-AgNPs was observed in a concentration-dependent manner. Further, Hfx-AgNPs may be considered a promising lead compound for the pharmacological industry.
Keywords: Biosynthesis, Silver-nanoparticles, Haloferax sp, Thrombolysis, Cytotoxicity
Key points
The haloarchaeon Haloferax sp. strain NRS1 (MT967913), isolated from solar saltern in Jeddah, Saudi Arabia, has promising ability when synthesized with silver nanoparticles.
Characterization of silver nanoparticles (AgNPs) synthesized by haloarchaeon Haloferax sp. strain NRS1 was performed by UV–vis spectroscopy, transmission electron microscope (TEM), X-ray diffraction (XRD), Fourier-transform infrared spectroscopy (FTIR).
The low hemolytic activity exhibited by Hfx-AgNPs, confirmed their promising role as nano-drug delivery agents. Hfx-AgNPs displayed clot lysis’s properties which may be attributed to the activation of the cascade reaction of clot lysis.
Introduction
Haloarchaea may be the best candidates for synthesizing nanoparticles because to S-layer glycoproteins present in their membrane. The unique property of haloarchaeal cells to quickly lyse at low concentrations of salt, leading to intracellular and membrane components being released, allowing S-proteins recovery efficient in cost. S-layer proteins of haloarchaea serve as building blocks for the synthesis of main biomolecules (proteins, lipids, glycans, nucleic acids, or their combinations) which are appropriate for nanobiotechnology (Sleytr et al. 2014). Hence, the intracellular biosynthesis of nanoparticles in haloarchaea, suggests these extremophilic microorganisms are promising in the field of nanobiotechnology (Beeler and Singh 2016).
Nanoparticles are promising in different fields such as biomedicine, drug delivery, bio-imaging, bio-labeling, photovoltaics, photocatalysis, solar cells, and data storage (Bera et al. 2010; Garcia 2011; Issa et al. 2013; Javed et al. 2020; Aisida et al. 2019a, b, c, 2021a, b). Biosynthesis of nanoparticles by microorganisms is a non-toxic and eco-friendly method (Srivastava et al. 2014). Biosynthesized nanoparticles by prokaryotes have attracted significant attention in the biomedical field because of their biocompatibility, nontoxicity (Abdullaeva 2017), and antimicrobial nature (Aisida et al. 2021c, d, e). Depending on microorganisms' metabolic activity and nature, the nanoparticle's biosynthesis can either be an intra- or extracellular process. Haloarchaea possess different metal resistance mechanisms and biosynthesis of nanoparticles, however, most are still unknown (Srivastava and Kowshik 2013). Few haloarchaeal strains Halococcus salifodinae BK3, Halococcus salifodinae BK6, have been reported for intracellular synthesis of silver nanoparticles (AgNPs) with antibacterial activity against gram-positive and gram-negative bacteria (Srivastava et al. 2013, 2014). The same action has been reported by the cells of Haloferax alexandrinus (Patil et al. 2014). Also, Costa et al. (2020) reported the ability of Haloferax volcanii to synthesize silver and gold nanoparticles biosynthesis with distinct properties for nanobiotechnological applications. Haloferax sp playing a potential role as an in-vitro antioxidant and antimicrobial agent (Zalazar et al. 2019). Recently, De Castro et al. (2020) reported that haloarchaea have a biotechnological potential that still needs to be explored for its significance in pharmaceutical applications. Further studies are therefore required to explore more biosynthesized nanoparticles from microorganisms with unique properties for future applications. In this study, we report the biosynthesis of silver nanoparticles intracellularly by unclassified species of Haloferax, Haloferax sp strain NRS1, and bioactivities for AgNPs. As well, characterization of biosynthesized AgNPs by UV- vis spectroscopy, TEM, XRD, and FTIR. The main objective of this work is finding a novel application for non-toxic biosynthesized AgNPs by Haloferax sp strain NRS1 as a significant fibrinolytic agent.
Materials and methods
Site description and sampling
Sediment and brine samples were collected from a solar saltern located at the southern coast of Jeddah, Saudi Arabia (21o10′16.04''N, 39o11′5.94''E) in April 2019 (Fig. 1) into 1000 ml bottles of sterile Pyrex. All samples have been stored at 4 °C and upon arrival, the samples have been subjected to microbiological examination within 24 h.
Fig. 1.
Map and A satellite image showing the location of the study area. B The studied solar saltern in the southern coast of Jeddah; C. The salt crystals were collected from the saltern
Isolation and growth conditions
Isolation of haloarchaea from sediment and brine described by Srivastava et al. (2013) which involves NTYE (25% NaCl, 0.5% tryptone, 0.3% yeast extract, 2% MgSO4-7H2O and 0.5% KCl, 2% Agar) was used. This was also in accordance to the isolation procedures described by Dyall-Smith (2008). Pure isolates were obtained by successive cultivation on NTYE and was also maintained on the same medium.
Screening for silver-resistant haloarchaeal isolates
Five different haloarchaeal isolates, obtained after two weeks of incubation at 40 °C, were tested for the growth in the presence of AgNO3 in a concentration range of 0.05–1 mM according to the method described by Srivastava et al. (2014). 1 ml aliquot of cell culture was measured at 600 nm on a UV–Visible spectrophotometer (UV-2600 Series, SHIMADZU) at intervals of 24 h, culture medium without AgNO3, and non-inoculated medium with AgNO3 considered as positive and negative control respectively.
Silver nitrate biosynthesis
The selected haloarchaeal isolate (NRS1), kept in the dark for 10 days was inoculated in NTYE-medium supplemented with 0.5 mM AgNO3, incubated at 37 °C and stirred at 110 rpm. The synthesis of nanoparticles was observed visually by the change of biomass color from orange-red to brown-black. Then the cells were harvested by centrifugation (10,000 rpm for 20 min). The obtained pellet was washed several times with distilled water, then freeze dried at 60 °C over-night then the nanoparticles were obtained as a powder.
Identification of the isolates
Genomic DNA was extracted from the selected isolate using a modified method from Experimental Techniques in Bacterial Genetics, described by Maloy (1990). The 16S rRNA gene was amplified with a set of Archaea-universal primers (Invitrogen, USA), 5'-ATTCCGGTTGATCCTGCCGG-3' primers (positions 6–25 in Escherichia coli numbering) and 5'AGGAGGTGATCCAGCCGCAG-3' primers (positions 1540–1521) reported by Ventosa et al. (2004). The PCR conditions were as follows: 50 μl of reaction system, reaction cycles 30 times, 95 °C pre-denaturation 5 min, 94 °C denaturation 1 min, 60 °C annealing 1 min, 72 °C extension 1 min 30 s, 72 °C final extension 10 min, 4 °C hold. A total of 50 ng/μl of each PCR product was used to prepare the samples delivered to MacroGen Company in Korea following their specifications. The sequences were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) to get a preliminary identify of the strain. The cluster analysis was performed using the MEGA 7software package.
Hemolytic activity
Cytotoxicity of Hfx-AgNPs was evaluated through hemolytic assay according to the method described by Powell et al. (2000). To prepare erythrocyte suspension, 1 ml of fresh blood was centrifuged for five minutes at 10,000 rpm. 200 μl of the erythrocytes precipitate was added to 9.8 ml of Phosphate Buffered Saline (PBS; pH 7.4). Then the mixture was centrifuged for 15 min at 4000 rpm. The supernatant was discarded and the process repeated three times. Finally, the washed erythrocytes were diluted with PBS to make the cell suspension (2%). Hfx-AgNPs were prepared in six different concentrations (3.125, 6.25, 12.5, 25, 50, 100 μg/ml). Each concentration of Hfx-AgNPs was tested in three replicates. Aliquots of 20 µl of Hfx-AgNPs have aseptically placed into a microcentrifuge tube; then, RBC suspension (200 µl) was added and mixed gently. The samples were incubated at 35 °C for 60 min, then centrifuged at 3000 rpm for fifteen minutes. The absorbance (Abs) of the supernatant was read at 545 nm. PBS and Deionized water were added to erythrocyte suspension represents; the negative control (no hemolysis) and the positive control (100% hemolysis), respectively. Percentage hemolysis (%) was calculated according to the following formula (Taniyama et al. 2003).
Clot lysis
Clot lysis experiments have been conducted as previously mentioned by Prasad et al. (2006). Concisely, venous blood was drawn from the healthy volunteers, then 500 μl of blood was transferred to ten pre-weighed sterile Eppendorf. The blood was incubated for 45 min at 37 °C for allowing blood to coagulate. Then the samples were centrifuged for 15 min, at 5000 rpm. Serum has been entirely removed without disrupting the clot. Every Eppendorf has been weighed again to assess the clot's weight; 200 μl of different concentrations of synthesized NPs colloidal solution (100, 50, 25, 12.5, 6.25 and 3.125 μg/ml) were added separately. As a positive control, 200 μl of streptokinase and as a negative non-thrombolytic control, 200 μl of saline solution were separately added to the clot. All the tubes were then incubated at 37 °C for 90 min and observed for clot lysis. After incubation, the supernatant was removed, and tubes were again weighed to monitor the weight difference after clot disruption. The experiment was repeated with the blood samples of the 5 volunteers.
Characterization of synthetized silver nanoparticles
The extinction coefficient of the synthesized Hfx-AgNPs colloidal solution was performed to monitor the bioreduction of silver by Haloferax sp strain NRS1 using a UV–Visible spectrophotometer (UV-2600 Series, SHIMADZU). Transmission electron microscopy (TEM HF-3300- Hitachi High-Tech Canada, Inc.) was used to determine the shape and size of Hfx-AgNPs. The size distributions from the TEM images for Hfx-AgNPs were further analyzed using image j Freeware Version 1.53d downloaded from the NIH website (http://rsb.info.nih.gov/ij). Zeta potential of the synthesized Hfx-AgNPs was determined by nanoparticle analyzer (Zetasizer Ver. 7.13, Malvern Instrument Ltd, UK). The structural characterization was determined using X-ray diffractometer (XRD Malvern Panalytical, UK). Finally, the structural units were identified with the use of Fourier Transmission Infra-Red spectroscopic (FTIR) (Perkin Elmer Spectrum GX Range Spectrometer).
Statistical analysis
Statistical analysis was performed using the software SPSS for Windows (IBM Corporation, New York, USA). One-way ANONA test was applied for the statistical analysis for the present data according to the mathematical principles described by Kutner et al. (2005) followed by Post hoc comparisons using the Duncan's test. The result was considered to be significant when P is less than 0.05.
Results
Biosynthesis of silver nanoparticles
Out of five tested isolates, one isolate (NRS1) exhibited a wide range of growth in presence of different concentrations of AgNO3 (0.05 up to 1 mM) and showed the highest growth rate at 0.5 mM AgNO3 Biosynthesis of silver nanoparticles change in the color from orange-red to brown-black was observed after one week of incubation at 37 °C, in the presence of 0.5 mM AgNO3. No color change was observed in control (culture medium without AgNO3). The appearance of brown-black color indicated the formation of AgNPs by the potential strain (Fig. 2). A peak at 458 nm in the UV–Vis spectrum further confirmed the formation of AgNPs (Fig. 3).
Fig. 2.

Growth of Haloferax sp strain NRS1 on NTYE medium in the absence and presence of 0.5 mM AgNO3. A, B Haloferax sp strain NRS1 on NTYE medium appears orange-red on agar-plate and broth respectively; C, D Haloferax sp strain NRS1 turned dark brown in the presence of 0.5 mM AgNO3 on NTYE agar and liquid media, respectively
Fig. 3.
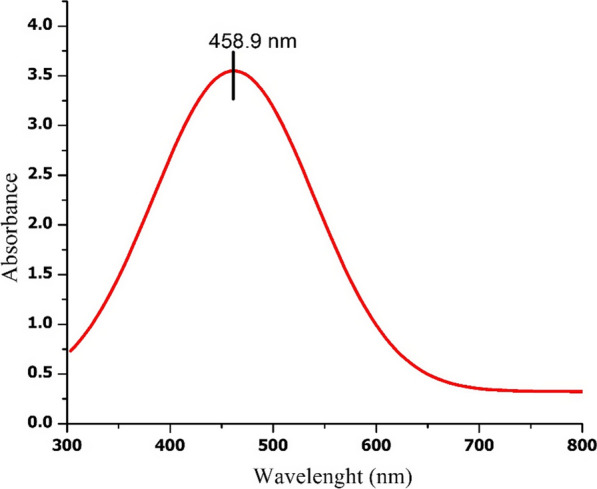
UV–Vis-Spectrum of biosynthesized AgNPs by Haloferax sp strain NRS1
Identification of the potential strain
Analysis of 16S rRNA gene sequencing indicated that the potential strain is a member of Haloferax (unidentified species) with 90% sequence similarity. The 16S rRNA gene data of the haloarchaeal strain reported in this study have been deposited in the NCBI and GenBank nucleotide sequence databases under the accession number MT967913 (Fig. 4). The strain has been deposited in the Egyptian Microbial Culture Collection (EMCC) under the code Hfx-NRS1 EMCC 23999.
Fig. 4.
Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences showing the relationship between Haloferax sp strain NRS1 and closely related taxa. Scale bar indicates 0.005 substitutions per nucleotide position
Characterization of silver nanoparticles
TEM investigation was conducted to determine Hfx-AgNPs size, as shown in Fig. 5. The shape of Ag nanoparticles synthesized by Haloferax sp strain NRS1 is mostly spherical; and the average particle size is 27.7 nm. The diameter range is 5.77- 73.14 nm. Geometric mean equal value of 24.96 ± 1.65 nm. The stability of Hfx-AgNPs was evaluated in terms of zeta potential (Fig. 6A). It was found that the zeta potential value was negative, i.e., − 25.5 ± 3.15 mV, which implies the good colloidal nature of synthesized nanoparticles.
Fig. 5.
A Histogram of the particle diameter size distribution of the Hfx-AgNPs based on TEM image analysis. Red line: Gaussian distribution fit. B Transmission electron microscopy (TEM) images of (AgNPs) (scale bar = 100 nm). C High-resolution TEM image of Ag nanoparticle (scale bar = 20 nm)
Fig. 6.
Physical characterization. A Zeta Potential of biosynthesized AgNPs by Haloferax sp strain NRS1. B X-ray diffraction pattern of biosynthesized AgNPs by Haloferax sp strain NRS1. C Williamson–Hall analysis of Hfx-Ag nanoparticles. The slope and crystalline size from the intercept of the fit were used to determine strain lattice
Figure 6B shows the powder XRD pattern of the Hfx-AgNPs. The diffraction peaks appeared at four 2θ angles equal to 38.21°, 44.63°, 64.39° and 77.61° which correspond to (hkl) planes (111), (200), (220) and (311), respectively. The crystallite of Hfx-AgNPs sizes was 29.28 nm as calculated using Debye Scherrer's equation. The full width at half maximum (FWHM) values of the nanoparticles was consistent with their size and was ranged from 0.190 to 0.511 (Table 1). Lattice strain in the biosynthesized AgNPs by Haloferax sp strain is determined from the slope W–H plot (Fig. 6C), which equal to − 0.259 ± 0.114.
Table 1.
The crystalline size of Silver Nano-powder synthesized by Haloferax sp strain NRS1
| Peak position (2θ°) | Hkl | FWHM Radians (β) |
Amplitude | Crystalline Size of the particles (D, nm) |
β cos θ | 4 sin θ |
|---|---|---|---|---|---|---|
| 38.21 | 111 | 0.511 | 16.788 | 17.197 | 0.446 | 1.956 |
| 44.63 | 200 | 0.475 | 9.655 | 18.897 | 0.379 | 2.413 |
| 64.39 | 220 | 0.393 | 9.391 | 24.968 | 0.0050 | 3.999 |
| 77.61 | 311 | 0.190 | − 2.097 | 56.082 | − 0.114 | 3.206 |
| (D) Average | 29.28 |
The FTIR spectrum showed characteristic absorption frequencies of the O–H stretching group at 3281 cm−1 wavelengths, which probably present either in phenol or alcohol. The C=O stretch of carbonyl groups was observed at 1644 cm−1, which was related to amino acid residues and peptides and had a strong capability to bind to metal nanoparticles. Furthermore, C–O stretching vibrations were determined at 1250 cm−1, being related to aromatic esters, where Ag2O has a stretching vibration; Ag2O may form at Ag nanoparticles' surface (Fig. 7).
Fig. 7.

FTIR spectra of Hfx-AgNPs
Bioactivity of silver nanoparticles
All materials that enter the blood get in contact with red blood cells (RBC). A hemolysis test was accomplished to assess Hfx-AgNPs impact on erythrocyte by measuring hemoglobin release after exposure to various concentrations of Hfx-AgNPs colloidal solution. The hemolysis assay performance was tested by the negative control PBS and positive control deionized distilled water (Fig. 8). It was observed that the hemolytic percentage of Hfx-AgNPs was concentration-dependent. Hemolysis was not detected at concentrations below 12.5 µg/ml, while 2.01% hemolysis was seen at the concentration of 100 µg/ml. According to the ISO/TR 7405-1984(f), the samples were considered as hemolytic if the hemolytic percentage was above 5%.
Fig. 8.
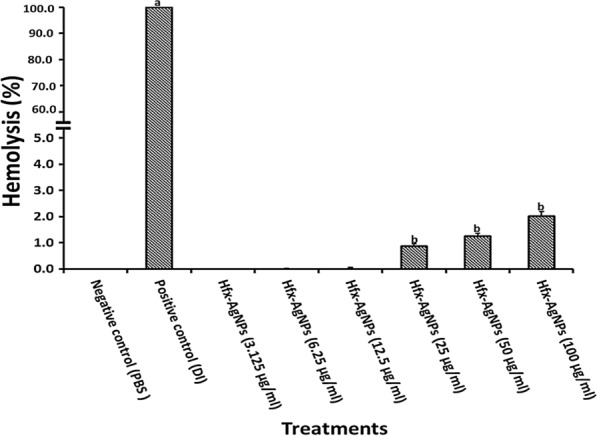
Hemolysis assay for Hfx-AgNPs colloidal solution against human erythrocytes, using DI as a positive control (+) and PBS as a negative control (−). Different concentration of Hfx-AgNPs were incubated with RBCs for 60 min for determining hemolytic activity. Data is represented as mean ± SD (n = 3). The mean difference between treatments considered significant at p < 0.05 using a one-way analysis of variance followed by Duncan's post hoc test. a statistically significant difference when compared with negative control; b statistically significant difference when compared with positive control
The effectiveness clot lysis by Hfx-AgNPs, positive thrombolytic control (streptokinase), and negative control is represented in Fig. 9. The addition of 200 µl SK, a positive control to the clots at 37 °C, showed 76.7% clot lysis. When treated with a saline solution (negative control), clots showed only negligible clot lysis (6.0%). The mean difference in clot lysis percentage between positive and negative control was significant (p value < 0.05). After treating clots with different concentrations of Hfx-AgNPs colloidal solution, the highest percentage of clot lysis was displayed at 100 μg/ml of Hfx-AgNPs with 50.218%, which means the percentage of the clot lysis compared with the negative control was significant (p-value < 0.05). Also, it has been observed that the lysis activity of tested agent was dose-dependent.
Fig. 9.

In vitro thrombolysis activity of Hfx-AgNPs. Positive control- streptokinase (SK), Negative control (saline solution). Data are expressed as mean ± SD (n = 3). The mean difference between different Hfx-AgNPs concentrations was considered significant at p < 0.05 using a one-way analysis of variance followed by Duncan's post hoc test for multiple comparisons. Lowercase letters compare the positive and negative control with different concentrations of Hfx-AgNPs colloidal solution. Same lowercase letters mean no statistically significant difference. However, Different lowercase letters mean a statistically significant difference
Discussion
The results of the present study demonstrated the biosynthesis of silver nanoparticles mediated Haloferax sp strain NRS1. The brown-black color of the culture confirmed the reduction of silver ions (Ag+) with the formation of AgNPs, according to Abdollahnia et al. (2020). Silver nanoparticles synthesis by Hfx. sp strain NRS1 was potential occurred in the presence of 0.5 mM AgNO3, 25% NaCl (w/v), at 37 °C. The nanostructures' size, shape, and distribution are related to the intensity and frequency of the plasmon surface's absorption bands (Petryayeva and Krull, 2011). Sharp peaks in silver nanoparticles' spectral extinction at visible and near-infrared frequencies rely on the resonances of localized surface plasmon (Mayer and Hafner, 2011). In the wavelength range of 300–800 nm at a resolution of 1 nm (Peiris et al. 2017). Owing to the elevated optical density (OD) of the colloidal suspension, a 1 ml colloidal aliquot was diluted with 15 mL of distilled water. Distilled water was used as blank. The analysis of Hfx-AgNPs colloidal solution by UV–Vis spectroscopy showed a characteristic absorbance peak at 458 nm, mostly similar to those for AgNPs synthesized by Halococcus salifodinae with maximum absorption peaks at 440 nm (Srivastava et al. 2013). Also, a peak equal to 446 nm was demonstrated for colloidal nanoparticles synthesized by Haloferax denitrificans (Abdollahnia et al. 2020).
Transmission electron microscopy (TEM)
The size and morphology of Hfx-AgNPs were done using TEM. Typically, the TEM pictures of 10 replicates of tested samples were taken at different 3 magnifications (30,000, 80,000, and 100,000 X). A drop of aqueous AgNPs sample was placed on a carbon-coated copper grid. The samples were allowed to dry under an infrared lamp before the examination; the micrographs were obtained using TEM operating at an accelerating voltage of 200 kV (Schrand et al. 2010). Dimension and morphology of Hfx-AgNPs were performed using the threshold method after set scale than the "Analyze Particle "feature to measure each particle area and diameter (Woehrle et al. 2006; Schneider et al. 2012). The particle size has been measured using image analysis. The present results confirmed the spherical-oblate shape of biosynthesized nanoparticles by Haloferax sp strain NRS1, in size ranges from 5.77 to 73.14 nm. The average particle size for AgNPs synthesized by Halococcus salifodinae was found to be around 50.3 nm (Srivastava et al. 2013).
Zeta potential analysis
Zeta potential determination is an essential and easiest means of predicting the stability and understanding of nanoparticle surface quality (Gregory et al. 2016); which displays the magnitude of repulsions between adjacent, equally charged dispersed particles whose values are correlated to colloidal dispersion stability (Larsson et al. 2012). the aqueous suspension of Hfx-AgNPs was placed in a DTS0112-low volume disposable cuvette. The potential of zeta is determined by the electrophoretic mobility principle, induced by applying an electric field through dispersion media. The silver nanoparticles have carried the charge of capping agents (Singh et al. 2014). The current investigation results revealed that; zeta potential shows negative value (− 25.5 ± 3.15 mV) for the Hfx-AgNPs reaction mixture. The negative values of zeta potential analysis confirmed the stability of Hfx-AgNPs. As formulated by Meléndrez et al. (2010), the magnitude of zeta potential gives an implication of colloid's possible stability. Similar observation revealed that; zeta potential with a negative value equal to − 18.9 mV is considered stable (Salvioni et al. 2016). The zeta potentials of silver nanoparticles produced by other halophiles such as Haloferax sp. and Halomonas sp isolated from solar saltern were equal to − 33.12, − 35.9, respectively (Gregory et al. 2016). These data confirmed that the haloarchaeon Haloferax sp strain NRS1 used in the present study produced the nanoparticles which are comparably stable as previously synthesized silver nanoparticles.
X-ray diffraction (XRD)
Dried Hfx-AgNPs were coated on an XRD grid to determine their structural characteristics using an XRD analysis. X-ray diffractometer was recorded by monochromatic Cu kα radiation (λ = 1.78 Å) with TD-2500 XRD which ran at 40 kV. The scanning was measured in the region of 30°–80°. For 2θ (Bragg diffraction angle) at 0.02°/min, and the time constant was 2 s (Klung and Alexander 1962). FWHM (full width at half maximum) of peaks was determined by Origin pro using the "Quick fit" command. Then the crystallite size was calculated using the Debey-Scherrer equation (Eq. 1) (Holzwarth and Gibson 2011):
| 1 |
L is particle size, θ is peak position (2θ/2) in radian. λ is the wavelength of the X-ray diffraction. K = 0.89 is the Debye–Scherrer constant. The lattice strain, ε, were estimated using the Williamson–Hall (WH) plot, determined according to Singaravelan and Alwar (2015) using the following equation (Eq. 2).
| 2 |
where β is FWHM in radian, t is the grain size in nm, ε is the strain, and C is a correction factor taken as 0.94.
The XRD peaks of Hfx-AgNPs exhibited a strong and narrow pattern, implying that nanomaterials synthesized by Haloferax sp strain NRS1 displayed a high degree of crystallinity. Peaks from other impurities were not detected, which confirming the high purity of the synthesized AgNPs. XRD provided significant evidence for the complete reduction of silver ions resulting in nanomaterials' production (Bhainsa and D'souza 2006). The XRD patterns of silver nanoparticles produced by archaeal strain Haloferax sp strain NRS1 showed an unassigned peak at 38.21°, 44.63°, 64.39°, and 77.61°. Thus, the XRD pattern clearly illustrated that the Ag-NPs formed in this study were crystalline in nature. According to the previous study, the peaks corresponding to our findings confirm that the main crystalline phase was silver (Sadhasivam et al. 2010; Shameli et al. 2012; Gurunathan et al. 2013). Concerning the FWHM data, the silver nanocrystalline structure obtained by haloarchaeal strain Haloferax sp. NRS1 (29.28 nm), which is similar to a previous report for NPs by the halophilic strain, Halococcus salifodinae BK 3 (22 nm) (Srivastava et al. 2013). In comparison with our previous work at Sayed et al. (2018) who investigate the characterization of nano silver powder (Sigma Aldrich, 576832) and nano silver ink (Metalon, JS-B25HV) X-ray diffraction pattern revealed four peaks 111, 200, 220, 311, these results were in accordance with our findings. Approving the formation of silver nanoparticle by the studied strain NRS1.
Fourier Transmission Infra-Red (FT-IR) spectroscopic
For analysis of FTIR of Hfx-AgNPs, the colloidal solution was centrifuged at 10,000 rpm for 30 min. The pellet was washed three times using deionized water to eliminate the free proteins and enzymes. Then a small quantity of dried powder was grinded with potassium bromide (KBr). The sample's FTIR spectrum was recorded at 400–4000 cm−1 (Taran et al. 2016). The results of FTIR confirmed the capping agents of some functional groups responsible for the biosynthesized nanoparticles' stability (Ajitha et al. 2018). The bands were observed at 3281 cm−1, 1644 cm−1, 1250 cm−1 and 611 cm−1. The current result confirmed that the amide group from amino acid residues and peptides of proteins has a stronger ability to bind to metal. The amino group could form a coat covering the metal nanoparticles to stabilize silver nanoparticles formed intracellularly. This evidence suggests that the biological molecules could perform the function of stabilizing the intracellular biosynthesized nanoparticles (Soenen et al. 2010). According to Makarov et al. (2014), the silver nanoparticle surface proteins act as capping agent amino acid residues. Peptides have a strong ability to bind to silver ions. Comparing the present findings with our previous work, Sayed et al. (2018), FTIR of nano silver powder (Sigma Aldrich, 576832) and nano silver ink (Metalon, JS-B25HV) revealed presence of O–H stretch, C=C stretch and N–H groups which are almost similar to Hfx-AgNPs composition.
Bioactivity of silver nanoparticles
Previous studies reported the potential antimicrobial activity of synthesized silver nanoparticles by Haloarchaea (Srivastava et al. 2014; Patil et al. 2014; Abdollahnia et al. 2020). The current study reports new bioactivities for green synthesized silver nanoparticles by Haloferax strain. Hemolytic activity against erythrocytes determines the tested material's potential membrane-stabilizing capability (Seeman and Weinstein 1966). The results of the present study revealed significant membrane-stabilizing potential associated with low toxicity of Hfx-AgNPs. This finding is supported by the negative value recorded for Hfx-AgNPs zeta potential. The nanoparticles' negative charge may prevent RBCs from interacting with the nanoparticles, consequently (Rothen-Rutishauser et al. 2006). Thus, Hfx-AgNPs may display their favorable hemolysis rate due to carboxylate group presence, leading to repulsion with RBC. Our results are in accordance with Singh et al. (2020). They stated that Fe3 O4 -Au Cys and Fe3 O4 -Au Cyt nanoparticles show low hemolytic activity due to their negative charges related to the presence of functional groups with a negative charge, which may lead to repulsion with RBC. According to Guidance for Industry and Food and Drug Administration Staff (FDA)-2013-D-0350 (ISO 10993-1) protocol, when the hemolysis level is above 5%, the tested material is considered a hemolytic agent.
At the present time there is an urgent need to discover drugs to break up or remove blood clots, as the formation of clots is the main cause of heart disease and stroke (Almalk and Zhang 2020). Regarding clot lysis assay, negative control revealed that clot dissolution did not occur when saline was added to the clot. However, significant thrombolytic activity was observed after treating the clots with Hfx-AgNPs colloidal solution as compared with the negative control. The Hfx-AgNPs thrombolysis activity was compared with streptokinase. The current result identified the potent thrombolytic activity of the tested material. According to Marder et al. (2001), thrombolytic agents dissolve blood clots by activating plasminogen that converted to a proteolytic enzyme called plasmin. Plasmin is capable of breaking cross-links between fibrin molecules, which provide the structural integrity of blood clots. Recently Colasuonno et al. (2018) summarize that some nanotherapeutic agent exerts its action through tissue plasminogen activation, maximizing thrombolytic activity. A variety of nanoparticles with clot-specific targeting capability release some thrombolytic agents (Landowski et al. 2020). Moreover Deng et al. (2018) stated that fibrin-targeted nanoparticle considered as promising drug delivery system which improve systemic hemostasis in vivo.
To sum up, the haloarchaeon Haloferax sp strain NRS1 (MT967913), isolated from solar saltern in Jeddah, Saudi Arabia, can accumulate silver in a nanoparticle form. AgNPs biosynthesized in NTYE medium with an average particle size equal to 27.7 nm. The biosynthesis of silver nanoparticles was confirmed using UV–visible spectroscopy and TEM, Moreover, FTIR showed that Hfx-AgNPs considered as both reducing and stabilizing agent. X-ray diffraction analysis revealed that the AgNPs were crystalline in nature. The relatively stable nature of Hfx-AgNPs could be attributed to their zeta-potential value, which displayed a realistic surface charge magnitude. Hfx-AgNPs exhibited low hemolytic activity, which may be related to negative zeta-potential, confirming their promising role as nano-drug delivery agents. However, increased clot lysis properties may be attributed to the activation of the clot lysis's cascade reaction.
Acknowledgements
This work was funded by the University of Jeddah, Jeddah, under Grant No. (UJ-02-061-DR). The authors, therefore, acknowledge with thanks the University of Jeddah for its technical and financial support.
Abbreviations
- AgNPs
Silver nanoparticles
- Hfx-AgNPs
Silver nanoparticles biosynthesized by Haloferax sp
- TEM
Transmission Electron Microscopy
- XRD
X-ray Diffraction
- FT-IR
Fourier Transmission Infra-Red
- RBC
Red blood cells
- FWHM
Full width at half maximum
Authors' contributions
AS, HT, MA and NH conceived and designed research. HT and NH conducted experiments and analyzed data. MA analyzed data. HT and NH wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by the University of Jeddah, Jeddah, under Grant No. (UJ-02–061-DR).
Data availability
Available upon request.
Declarations
Institutional Review Board Statement
Ethical review and approval were waived for this study due to the study was applied on the blood from volunteer donors. An informed blood donors' consent form was submitted to the volunteer donors, which informed them of the title, the name, and details of the researchers and the research purpose. This individual study sample was limited and not presented in the consent form for future research projects. In this study, the potential donor harm, wounds, discomfort, or discomfort were added as a statement of informed consent.
Consent for publication
All authors have read and approved the manuscript for submission and publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hend M. Tag, Email: hmtaha@uj.edu.sa
Nashwa Hagagy, Email: niibrahem@uj.edu.sa.
References
- Abdollahnia M, Makhdoumi A, Mashreghi M, Eshghi H. Exploring the potentials of halophilic prokaryotes from a solar saltern for synthesizing nanoparticles: the case of silver and selenium. PLoS ONE. 2020;15(3):e0229886. doi: 10.1371/journal.pone.0229886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullaeva Z. Synthesis of nanoparticles and nanomaterials. Cham: Springer; 2017. Synthesis of nanomaterials by prokaryotes; pp. 25–54. [Google Scholar]
- Aisida SO, Ugwu K, Akpa PA, Nwanya AC, Ejikeme PM, Botha S, Ahmad I, Maaza M, Ezema FI. Biogenic synthesis and antibacterial activity of controlled silver nanoparticles using an extract of Gongronema Latifolium. Mater Chem Phys. 2019;237:121859. doi: 10.1016/j.matchemphys.2019.121859. [DOI] [Google Scholar]
- Aisida SO, Ugwoke E, Iroegbu UA, Botha S, Ahmad I, Maaza M, Ezema FI. Incubation period induced biogenic synthesis of PEG enhanced Moringa oleifera silver nanocapsules and its antibacterial activity. J Polym Res. 2019;26:225. doi: 10.1007/s10965-019-1897-z. [DOI] [Google Scholar]
- Aisida SO, Ugwu K, Akpa PA, Nwanya AC, Nwankwo U, Botha S, Ejikeme PM, Ahmad I, Maaza M, Ezema FI. Biosynthesis of silver nanoparticles using bitter leave (Veronica amygdalina) forantibacterial activities. Surf Interf. 2019;17:100359. doi: 10.1016/j.surfin.2019.100359. [DOI] [Google Scholar]
- Aisida SO, Ugwu K, Nwanya AC, Bashir AKH, Nwankwo NU, Ahmed I, Ezema FI. Biosynthesis of silver oxide nanoparticles using leave extract of Telfairia Occidentalis and its antibacterial activity. Mater Today Proc. 2021;36(2):208–213. doi: 10.1016/j.matpr.2020.03.005. [DOI] [Google Scholar]
- Aisida SO, Ugwu K, Akpa PA, Nwanya AC, Nwankwo NU, Bashir AKH, Madiba IG, Ahmad I, Ezema FI. Synthesis and characterization of iron oxide nanoparticles capped with Moringa Oleifera: the mechanisms of formation effects on the optical, structural, magnetic and morphological properties. Mater Today Proc. 2021;36(2):214–218. doi: 10.1016/j.matpr.2020.03.167. [DOI] [Google Scholar]
- Aisida SO, Ali A, Oyewande OE, Ahmad I, Ul-Hamid A, Zhao T-Z, Maaza M, Ezema FI. Biogenic synthesis enhanced structural, morphological, magnetic and optical properties of zinc ferrite nanoparticles for moderate hyperthermia applications. J Nanopart Res. 2021;23:47. doi: 10.1007/s11051-021-05149-w. [DOI] [Google Scholar]
- Aisida SO, Ugwu K, Nwanya AC, Akpa PA, Madiba IG, Bashir AKH, Botha S, Ejikeme PM, Zhao T-Z, Ahmad I, Maaza M, Ezema FI. Dry Gongronema latifolium aqueous extract mediated silver nanoparticles by one-step in-situ biosynthesis for antibacterial activities. Surf Interf. 2021;24:101116. doi: 10.1016/j.surfin.2021.101116. [DOI] [Google Scholar]
- Aisida SO, Ugwu K, Akpa PA, Nwanya AC, Ejikeme PM, Botha S, Ahmad I, Ezema FI. Morphological, optical and antibacterial study of green synthesized silver nanoparticles via Vernonia amygdalina. Mater Today Proc. 2021;36(2):199–203. doi: 10.1016/j.matpr.2020.03.167. [DOI] [Google Scholar]
- Ajitha B, Reddy YAK, Jeon HJ, Ahn CW. Synthesis of silver nanoparticles in an eco-friendly way using Phyllanthus amarus leaf extract: antimicrobial and catalytic activity. Adv Pow Technol. 2018;29(1):86–93. doi: 10.1016/j.apt.2017.10.015. [DOI] [Google Scholar]
- Almalki WH, Zhang W. Emerging paradigms in treating cerebral infarction with nanotheranostics: opportunities and clinical challenges. Drug Discov Today. 2020;26(3):826–835. doi: 10.1016/j.drudis.2020.12.018. [DOI] [PubMed] [Google Scholar]
- Beeler E, Singh OV. Extremophiles as sources of inorganic bio-nanoparticles. W J Microbiol Biotech. 2016;32(9):156. doi: 10.1007/s11274-016-2111-7. [DOI] [PubMed] [Google Scholar]
- Bera RK, Mandal SM, Raj CR. Antimicrobial activity of fluorescent Ag nanoparticles. Lett App Microbiol. 2010;58:520–526. doi: 10.1111/lam.12222. [DOI] [PubMed] [Google Scholar]
- Bhainsa KC, D'souza SF. Extracellular biosynthesis of silver nanoparticles using the fungus Aspergillus fumigatus. Colloids Surf B. 2006;47(2):160–164. doi: 10.1016/j.colsurfb.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Costa MI, Álvarez-Cerimedo MS, Urquiza D, Ayude MA, Hoppe CE, Hoppe CE, Fasce DP, De Castro RE, Giménez MI. Synthesis, characterization and kinetic study of silver and gold nanoparticles produced by the archaeon Haloferax volcanii. J App Microbiol. 2020;129:1297–1308. doi: 10.1111/jam.14726. [DOI] [PubMed] [Google Scholar]
- De Castro I, Mendo S, Caetano T. Antibiotics from Haloarchaea: what can we learn from comparative genomics? Marine Biotechnol. 2020;22(2):308–316. doi: 10.1007/s10126-020-09952-9. [DOI] [PubMed] [Google Scholar]
- Deng J, Mei H, Shi W, Pang ZQ, Zhang B, Guo T, Hu Y. Recombinant tissue plasminogen activator-conjugated nanoparticles effectively targets thrombolysis in a rat model of middle cerebral artery occlusion. Curr Med Sci. 2018;38(3):427–435. doi: 10.1007/s11596-018-1896-z. [DOI] [PubMed] [Google Scholar]
- Dyall-Smith M (2008) The Halohandbook: protocols for halobacterial genetics, Mark Dyall-Smith, Martinsried, Germany
- Garcia MA. Surface plasmons in metallic nanoparticles: fundamentals and applications. J Phys D App Phys. 2011;44(28):283001. doi: 10.1088/0022-3727/44/28/283001. [DOI] [Google Scholar]
- Gregory L, Reghan V, Hill J, Harper S, Rawle FA, Hendren OC, Klaessig F, Nobbmann UIF, Sayre P, Rumble J. "Guidance to improve the scientific value of zeta-potential measurements in nanoEHS. Environ Sci Nano. 2016;3(5):953–965. doi: 10.1039/c6en00136j. [DOI] [Google Scholar]
- Gurunathan S, Raman J, Abd Malek SN, John PA, Vikineswary S. Green synthesis of silver nanoparticles using Ganoderma neo-japonicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int J Nanomed. 2013;8:4399. doi: 10.2147/IJN.S51881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzwarth U, Gibson N. The Scherrer equation versus the 'Debye-Scherrer equation'. Nat Nanotech. 2011;6(9):534–534. doi: 10.1038/nnano.2011.145. [DOI] [PubMed] [Google Scholar]
- Issa B, Ihab M, Obaidat MI, Albiss AB, Haik Y. Magnetic nanoparticles: surface effects and properties related to biomedicine applications. Int J Mol Sci. 2013;14:21266–21305. doi: 10.3390/ijms141121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed R, Zia M, Naz S, Aisida SO, Ul-Ain N, Ao Q. Role of capping agents in the application of nanoparticles in biomedicine and environmental remediation: recent trends and future prospects. J Nanobiotechnol. 2020;18:172. doi: 10.1186/s12951-020-00704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klung HP, Alexander LE. X-ray diffraction procedures. New York: Wiley; 1962. p. 974. [Google Scholar]
- Kutner MH, Nachtsheim CJ, Neter J, Li W. Applied linear statistical models. New York: McGraw-Hill, Irwin; 2005. [Google Scholar]
- Landowski LM, Niego BE, Sutherland BA, Hagemeyer CE, Howells DW. Applications of nanotechnology in the diagnosis and therapy of stroke. Seminars Thromb Hemostasis. 2020;46(05):592–605. doi: 10.1055/s-0039-3399568. [DOI] [PubMed] [Google Scholar]
- Larsson M, Hill A, Duffy J (2012) Suspension stability; why particle size, zeta potential and rheology are important. Ann Trans Nord Rheol Soc 20:209–214. https://nrs.blob.core.windows.net/pdfs/nrspdf-6f0ecb18-3077-4226-af5c-166c2340e756.pdf
- Makarov VV, Love AJ, Sinitsyna OV, Makarova SS, Yaminsky IV, Taliansky ME, Kalinina NO. “Green” nanotechnologies: synthesis of metal nanoparticles using plants. Acta Naturae. 2014;6(1):20. doi: 10.32607/20758251-2014-6-1-35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR. Experimental techniques in bacterial genetics. Burlington: Jones and Bartlet Publisher Inc; 1990. pp. 125–139. [Google Scholar]
- Marder JV, Landskroner K, Novokhatny V, Zimmerman PT, Kong M, Kanouse J, Jesmok G. Plasmin induces local thrombolysis without causing hemorrhage: a comparison with tissue plasminogen activator in the rabbit. Throm Haemo. 2001;86(09):739–745. doi: 10.1055/s-0037-1616127. [DOI] [PubMed] [Google Scholar]
- Mayer KM, Hafner JH. Localized surface plasmon resonance sensors. Chem Rev. 2011;111(6):3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- Meléndrez MF, Cárdenas G, Arbiol J. Synthesis and characterization of gallium colloidal nanoparticles. J Coll Interf Sci. 2010;346(2):279–287. doi: 10.1016/j.jcis.2009.11.069. [DOI] [PubMed] [Google Scholar]
- Patil S, Fernandes J, Tangasali R, Furtado I. Exploitation of Haloferax alexandrinus for biogenic synthesis of silver nanoparticles antagonistic to human and lower mammalian pathogens. J Clust Sci. 2014;25:423–433. doi: 10.1007/s10876-013-0621-0. [DOI] [Google Scholar]
- Peiris MK, Gunasekara CP, Jayaweera PM, Arachchi ND, Fernando N. Biosynthesized silver nanoparticles: are they effective antimicrobials? Mem Inst Oswaldo Cruz. 2017;112(8):537–543. doi: 10.1590/0074-02760170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryayeva E, Krull UJ. Localized surface plasmon resonance: nanostructures, bioassays and biosensing—a review. Anal Chim Acta. 2011;706(1):8–24. doi: 10.1016/j.aca.2011.08.020. [DOI] [PubMed] [Google Scholar]
- Powell WA, Catranis CM, Maynard CA. Design of self-processing antimicrobial peptides for plant protection. Lett Appl Microbiol. 2000;31:163–168. doi: 10.1046/j.1365-2672.2000.00782.x. [DOI] [PubMed] [Google Scholar]
- Rothen-Rutishauser BM, Schürch S, Haenni B, Kapp N, Gehr P. Interaction of fine particles and nanoparticles with red blood cells visualized with advanced microscopic techniques. Envir Sci Technol. 2006;40(14):4353–4359. doi: 10.1021/es0522635. [DOI] [PubMed] [Google Scholar]
- Sadhasivam S, Shanmugam P, Yun K. Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms. Colloids Surf, B. 2010;81(1):358–362. doi: 10.1016/j.colsurfb.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Salvioni L, Galbiati E, Collico V, Alessio G, Avvakumova S, Corsi F, Tortora P, Prosperi D, Colombo M. Negatively charged silver nanoparticles with potent antibacterial activity and reduced toxicity for pharmaceutical prep-arations. Int J Nanomed. 2016;12:2517. doi: 10.2147/IJN.S127799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed R, Saad H, Hagagy N. Silver nanoparticles: characterization and antibacterial properties. Rendiconti Lincei Scienze Fisiche e Naturali. 2018;29(1):81–86. doi: 10.1007/s12210-017-0663-6. [DOI] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrand AM, Schlager JJ, Dai L, Hussain SM. Preparation of cells for assessing ultrastructural local-ization of nanoparticles with transmission electron microscopy. Nat Protoc. 2010;5(4):744. doi: 10.1038/nprot.2010.2. [DOI] [PubMed] [Google Scholar]
- Seeman P, Weinstein JI. Erythrocyte membrane stabilization by tranquilizers and antihistamines. Biochem Pharma. 1966;15(11):1737–1752. doi: 10.1016/0006-2952(66)90081-5. [DOI] [Google Scholar]
- Shameli K, Ahmad MB, Zamanian A, Sangpour P, Shabanzadeh P, Abdollahi Y, Zargar M. Green biosynthesis of silver nanoparticles using Curcuma longa tuber powder. Int j Nanomed. 2012;7:5603. doi: 10.2147/ijn.s36786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelan R, Alwar SBS. Electrochemical synthesis, characterization and phytogenic properties of silver nanoparticles. Appl Nanosci. 2015;5:983–991. doi: 10.1007/s13204-014-0396-0. [DOI] [Google Scholar]
- Singh S, Bharti A, Meena VK. Structural, thermal, zeta potential and electrical properties of disaccharide reduced silver nanoparticles. J Mater Sci Mater Electro. 2014;25(9):3747–3752. doi: 10.1007/s10854-014-2085-x. [DOI] [Google Scholar]
- Singh N, Sahoo SK, Kumar R. Hemolysis tendency of anticancer nanoparticles changes with type of blood group antigen: An insight into blood nanoparticle interactions. Mat Sci Engin. 2020;109:110645. doi: 10.1016/j.msec.2020.110645. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Schuster B, Egelseer EM, Pum D. S-layers: principles and applications. FEMS Microbiol Rev. 2014;38(5):823–864. doi: 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soenen SJ, Himmelreich U, Nuytten N, Pisanic TR, Ferrari A, De Cuyper M. Intracellular nanoparticle coating stability determines nanoparticle diagnostics efficacy and cell functionality. Small. 2010;6(19):2136–2145. doi: 10.1002/smll.201000763. [DOI] [PubMed] [Google Scholar]
- Srivastava P, Kowshik M. Mechanisms of metal resistance and homeostasis in haloarchaea. Archaea. 2013;2013:1–16. doi: 10.1155/2013/732864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Bragança J, Ramanan SR, Kowshik M. Synthesis of silver nanoparticles using haloarchaeal isolate Halococcus salifodinae BK 3. Extremophiles. 2013;17(5):821–831. doi: 10.1007/s00792-013-0563-3. [DOI] [PubMed] [Google Scholar]
- Srivastava P, Braganca J, Ramanan SR, Kowshik M. Green synthesis of silver nanoparticles by Haloarchaeon Halococcus salifodinae BK6. Adv Mat Res. 2014;938:236–241. [Google Scholar]
- Taniyama S, Arakawa O, Terada M, Nishio S, Takatani T, Mahmud Y, Noguchi T. Ostreopsis sp., a possible origin of palytoxin (PTX) in parrotfish Scarus ovifrons. Toxicon. 2003;42(1):29–33. doi: 10.1016/s0041-0101(03)00097-7. [DOI] [PubMed] [Google Scholar]
- Taran M, Rad M, Alavi M. Characterization of Ag nanoparticles biosynthesized by Bacillus sp. HAI4 in different conditions and their antibacterial effects. J Appl Pharma Sci. 2016;6(11):094–99. doi: 10.7324/japs.2016.601115. [DOI] [Google Scholar]
- Ventosa A, Gutierrez MC, Kamekura M, Zvyagintseva IS, Oren A. Taxonomic study of Halorubrum distributum and proposal of Halorubrum terrestre sp. nov. Int J Syst Evol Microbiol. 2004;54:389–392. doi: 10.1099/ijs.0.02621-0. [DOI] [PubMed] [Google Scholar]
- Woehrle GH, Hutchison JE, Özkar S, FINKE RG (2006) Analysis of nanoparticle transmission electron microscopy data using a public-domain image-processing program, image. Turk J Chem 30(1):1–13. https://journals.tubitak.gov.tr/chem/issues/kim-06-30-1/kim-30-1-1-0508-1
- Zalazar L, Pagola P, Miró MV, Churio MS, Cerletti M, Martínez C, De Castro R. Bacterioruberin extracts from a genetically modified hyperpigmented Haloferax volcanii strain: antioxidant activity and bioactive properties on sperm cells. J App Microbiol. 2019;126(3):796–810. doi: 10.1111/jam.14160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available upon request.






