Abstract
We hypothesized that the gut microbiome in patients with diabetes secondary to chronic pancreatitis (Type 3c) is different from those with Type 1 and Type 2 diabetes. This was a cross-sectional preliminary study that included 8 patients with Type 1, 10 with Type 2, 17 with Type 3c diabetes and 9 healthy controls. Demographic, clinical, biochemical, imaging and treatment data were recorded and sequencing of the V3–V4 region of the bacterial 16SrRNA was done on fecal samples. Bioinformatics and statistical analyses was performed to evaluate the differences in the diversity indices, distance matrices, relative abundances and uniqueness of organisms between the types of diabetes. There was significant difference in the species richness. Beta diversity was significantly different between patients with Type 3c diabetes and the other groups. 31 genera were common to all the three types of diabetes. There was significant differences in the species level taxa between Type 3c diabetes and the other groups. The unique bacterial species signature in Type 3c diabetes compared to Type 1 and Type 2 diabetes included Nesterenkonia sp. AN1, Clostridium magnum, Acinetobacter lwoffii, Clostridium septicum, Porphyromonas somerae, Terrabacter tumescens, and Synechococus sp.
Subject terms: Chronic pancreatitis, Diabetes
Introduction
India harbours 77 million diabetics, which amounts to a national prevalence of 8.9%1. While Type 2 diabetes is the most common form of diabetes, the prevalence of Type 1 DM is also on the rise in India at a rate of 3–5% every year2. Likewise, there is also a substantially high frequency (~ 38%) of Type 3c diabetes in India3. Type 3c diabetes is defined as diabetes secondary to disease of the exocrine pancreas4. The frequency of Type 3c diabetes among patients with CP in India ranges from around 17% in early onset CP to over 60% in late onset CP5–8. Moreover, the age of onset of this condition is generally lower in India compared to that in the west6,7.
It is now clear that the normal gut microbial ecology has a substantial role in maintaining health through optimal nutrient digestion, maintenance of gastrointestinal function, synthesis of vitamins and pro-health metabolites, metabolism of drugs and xenobiotics, and glycemic and lipidomic control9–12. There is now substantial evidence from the west and a handful from India that there is significant gut microbial dysbiosis in Type 1 and Type 2 diabetes13–19. In our recent study on gut microbial dysbiosis in CP, we observed alteration of the microbiome even in patients with Type 3c diabetes20. From a mechanistic perspective, the pathogenesis of the three types of diabetes differs profoundly. While Type 1 and Type 2 diabetes are associated with insulin deficiency resulting from autoimmunity and insulin resistance respectively, Type 3c diabetes shows a combination of insulin deficiency and hepatic insulin resistance4,21–25. Further, current evidence suggests infiltration of Th17 cells into the islets in CP that secretes IFN-γ resulting in a functional defect in the beta-cells26–28. There is also a loss of pancreatic polypeptide response that results in hepatic insulin resistance in Type 3c diabetes3.
On the premise that the pathophysiology of Type 3c diabetes differ from that of Type 1 and Type 2 diabetes, we hypothesized that the gut microbiome in Type 3c diabetes will also differ from Type 1 and Type 2 diabetes. Even though gut microbial dysbiosis has been reported in all these three types of diabetes separately, no head to head comparison have yet been evaluated. We therefore embarked on the current study wherein the objective was to evaluate any difference in the gut microbiome between the three types of diabetes.
Results
Patients and fecal metagenomic characteristics
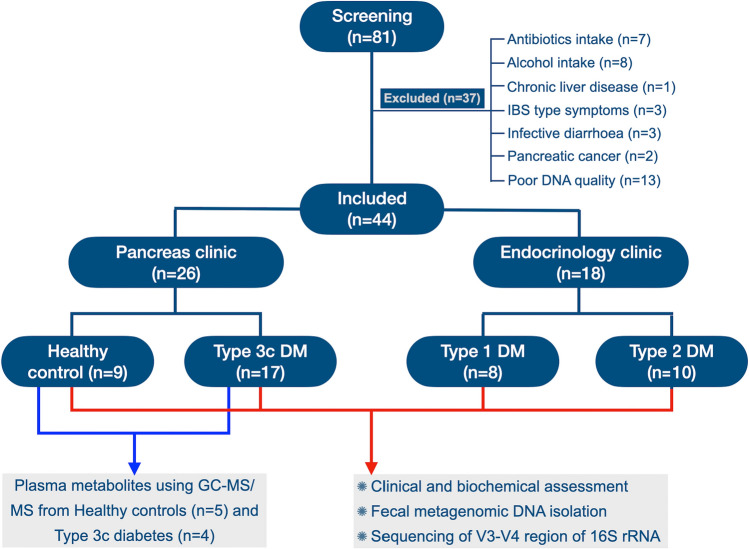
A total of 81 individuals were screened out of which 9 healthy volunteers, 8 patients with Type 1, 10 with Type 2 and 17 with Type 3c diabetes were finally included for analyses. The patient flow and clinical characteristics have been depicted in Fig. 1 and Supplementary Table 1 respectively. Supplementary Table 2 represents the metagenomic quality, number of sequences and singletons, and Good’s coverage of the fecal metagenome of individual control and patients.
Figure 1.
Study groups and patient distribution.
Fecal microbial diversity among study participants
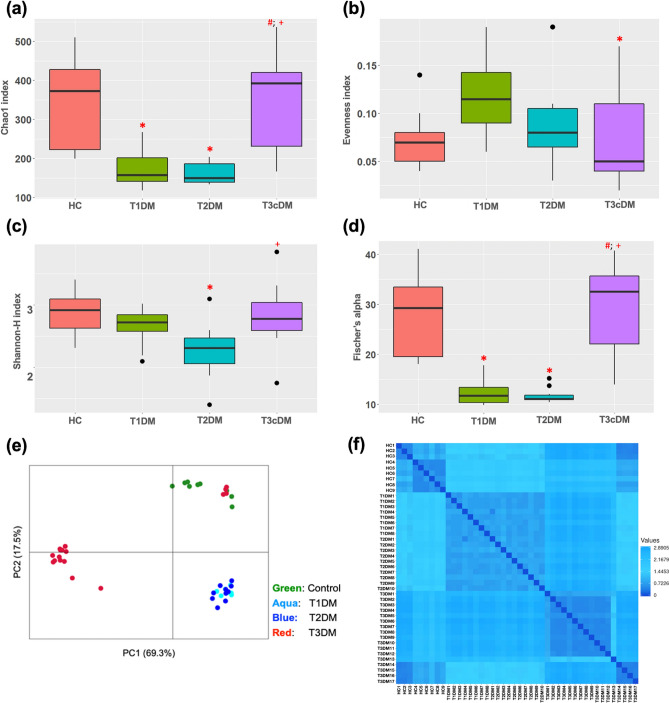
The microbial alpha diversity indices and beta diversity are depicted in Fig. 2a–f and Supplementary Fig. 1. There were significant difference in the species richness (Chao 1) (p < 0.0001) as shown by a reduction among patients with Type 1 and Type 2 diabetes compared to healthy controls and those with Type 3c diabetes (adjusted p values for HC vs. T1DM = 0.03, HC vs. T2DM = 0.003, T1DM vs. T3cDM = 0.004 and T2DM vs. T3cDM < 0.0001, based on Tukey’s post hoc test) (Supplementary Table 3). Shannon index was found to be lowest in the patients with Type 2 diabetes compared to that of healthy controls and Type 3c diabetes (adjusted p values for HC vs. T2DM = 0.01, T2DM vs. T3cDM = 0.002, based on Tukey’s post hoc test) (Supplementary Table 3).
Figure 2.
Fecal microbial diversity among the study patients. Box and whisker plots in panels a-d represent the alpha diversity indices. (a) Chao I, (b) Evenness index, (c) Shannon index, (d) Fischer’s alpha. Panels (e) Principal coordinate analyses [PCoA] plot using Euclidean distance and (f) Heatmap representing the intergroup and intragroup beta diversity, based on Euclidean distance, among the study groups. The p values were derived using the Kruskal–Wallis test with Tukey’s post-hoc pairwise analyses. * indicates statistically significant difference compared to healthy control; # compared to Type 1 diabetes and + compared to Type 2 diabetes. Refer to Supplementary Table 3 for exact p values.
Figure 2e,f shows the beta diversity between the groups. The beta diversity was similar between the patients with Type 1 and Type 2 diabetes, while it was significantly different between patients with Type 3c diabetes and the other groups including healthy controls (Bonferroni corrected p values for HC vs. T1D = 0.001; HC vs. T2D = 0.001; HC vs. T3cD = 0.002; T1D vs. T3cD = 0.001, T2D vs. T3cD = 0.001).
Differences in bacterial taxa between study groups
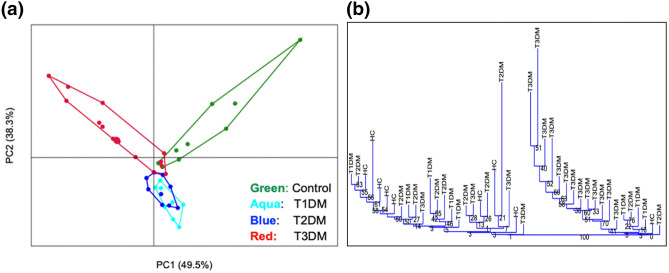
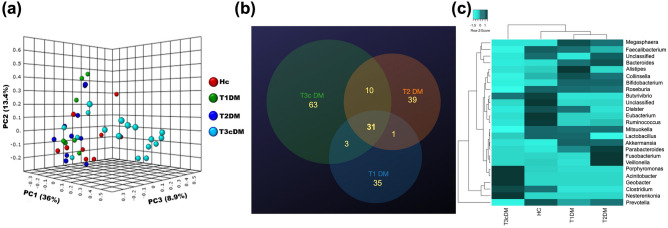
As shown in the PCA plot in Fig. 3a, there was significant differences in the species level taxa between Type 3c diabetes, healthy controls and, Type 1 and Type 2 diabetes combined (Bonferroni corrected p values for HC vs. T1D = 0.01; HC vs. T2D = 0.004; HC vs. T3cD = 0.002; T1D vs. T3cD = 0.009, T2D vs. T3cD = 0.002). This was confirmed in a hierarchical dendrogram (Fig. 3b). The scree plot for the PCA analyses have been depicted in Supplementary Fig. 2. The clustering of the different species between healthy controls and all diabetic patients, and between the three diabetic groups are shown in Supplementary Fig. 3.
Figure 3.
Clustering of gut microbiota at species level taxa among the study groups. (a) Principal component analysis (PCA) of gut microbial species abundance showing distinct clustering in the four study groups. The relation between the microbiota and disease status was assessed with PERMANOVA using 10,000 permutation [Bonferroni corrected p < 0.0001] followed by post hoc pair wise comparison. (b) Agglomerative clustering dendrogram showing hierarchical cluster analysis based on Euclidean similarity matrix (with boot strapping to 1000) to measure closeness between individual samples.
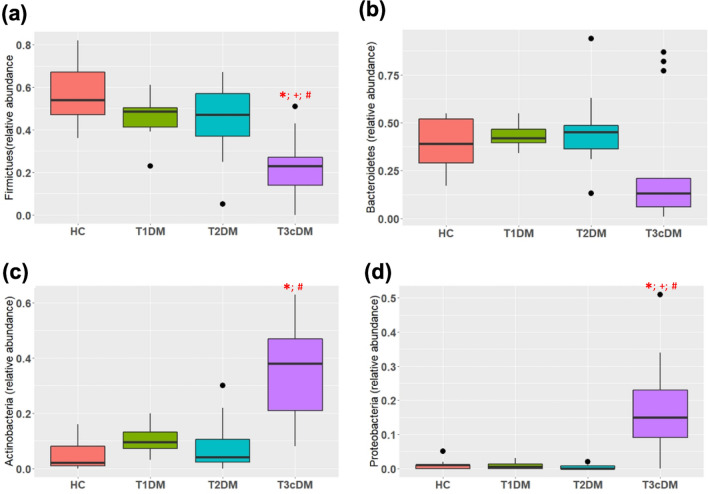
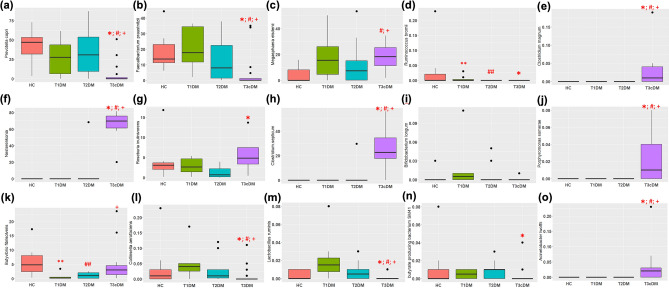
As shown in Fig. 4a–d, there were significant differences in the abundances in the phylum Firmicutes (p < 0.0001), Bacteroidetes (p = 0.04), Actinobacteria (p < 0.0001) and Proteobacteria (p < 0.0001), which together constituted the majority of the phyla. The Firmicutes to Bacteroidetes ratio was least in Type 3c diabetes compared to healthy controls (Supplementary Fig. 4). On post-hoc analyses (Supplementary Table 4), significant differences were observed in Firmicutes between Type 3c diabetes and healthy control (adjusted p < 0.0001), Type 1 diabetes (adjusted p = 0.03) and Type 2 diabetes (adjusted p = 0.01). Significant differences between these groups were also observed with Actinobacteria and Proteobacteria.
Figure 4.
Relative abundances of the four most common phylum. Box and whisker plots depicting relative abundance of (a) Firmicutes; (b) Bacteroidetes; (c) Actinobacteria; (d) Proteobacteria. * indicates statistically significant difference between healthy control and T3c diabetes; + between Type 1 and Type 3c diabetes, # between Type 2 and Type 3c diabetes. Refer to Supplementary Table 4 for exact p values.
The relative abundances of organisms for each participant at the phylum, class, order and family level taxa has been depicted in Supplementary Fig. 5a–d respectively. Supplementary Fig. 6a–c shows the clustering of the study groups at the order, class and family level taxa while Supplementary Fig. 6d–e depicts abundance of these taxa according to the groups.
As shown in Fig. 5a, there was significant clustering of genus between the study groups with maximum difference for patients with Type 3c diabetes (p < 0.0001). The Venn diagram in Fig. 5b shows that there were 63 unique genera in the group of Type 3c diabetes while there were 35 and 39 in the Type 1 and Type 2 diabetes respectively. There were 31 genera common to all the three types of diabetes. Figure 5c and Supplementary Table 5 represents the abundances and intergroup differences of the top 25 genera. The core bacterial genera in the healthy control cohort included Prevotella, Faecalibacterium, Roseburia, Eubacterium, Clostridium, Collinsella, Lactobacillus, Bacteroides, Ruminococcus, Parabacteroides, Dialister, Blautia, Butyricicoccus, Butyrivibrio, and Acetivibrio. While, Alistipes was found uniquely in patients with Type 1 diabetes, both Type 1 and 2 group contained a core of Prevotella, Faecalibacterium, Bacteroides, Collinsella, Ruminococcus, Clostridium, Eubacterium, Megasphaera, Bifidobacterium, and unclassified Clostridiales. Patients with Type3c diabetes had a distinctive pattern of fecal bacteriome, containing Nesterenkonia, Geobacter, Acinetobacter, Porphyromonas and Arthrobacter as the unique core.
Figure 5.
Bacterial characteristics at the genus level taxa. (a) PCoA plot, based on Bray Curtis distance matrix, showing significantly different clustering of Type 3c diabetes (Light blue dots) compared to the other groups (Red dots- healthy control, Green dots- Type 1 diabetes, Deep blue dots- Type 2 diabetes). (b) Venn diagram showing the numbers of unique and common genera between the three diabetes groups. (c) Heatmap with clustering dendrograms depicting the group wise difference in abundance of the organisms at the genus level taxa. The colour intensity representing the median relative abundances is based on the row z scores.
Supplementary Fig. 7 shows the top 25 species in the study groups while Fig. 6 and Supplementary Table 6 shows the abundances of the species with most significant difference. There was significant reduction of Prevotella copri, Faecalibacterium praustnitzii, Collinsella aerofaciens and Lactobacillus ruminis while significant increase uniquely in Nesterenkonia sp. AN1, Clostridium magnum, Acinetobacter lwoffii, Clostridium septicum, Porphyromonas somerae, Terrabacter tumescens, and Synechococus sp. in Type 3c diabetes compared to Type 1 and Type 2 diabetes. Butyrivibrio fibrisolvens was significantly low in abundance in Types 1 and 2 diabetes compared to healthy controls.
Figure 6.
Relative abundances of top 15 species showing the most significant differences. Box and whisker plots depicting relative abundance of (a) Prevotella copri; (b) Faecalibacterium praustnitzii; (c) Megasphaera elsdenii; (d) Ruminococus bromii; (e) Clostridium magnum; (f) Nesterenkonia.sp AN1; (g) Roseburia inulivorans; (h) Clostridium septicum; (i) Bifidobacterium longum; (j) Porphyromonas somerae; (k) Butyrivibrio fibrisolvens; (l) Collinsella aerofaciens; (m) Lactobacillus ruminis; (n) Butyrate producing bacteria SM41 (o) Acinetobacter lwoffi. * indicates statistically significant difference between healthy control and T3c diabetes; ** between healthy controls and Type 1 diabetes, # between Type 1 and Type 3c diabetes, ## between healthy control and Type 2 diabetes, and + between Type 2 and Type 3c diabetes. Refer to Supplementary Table 6 for exact p values.
Correlations between different genera in the study groups
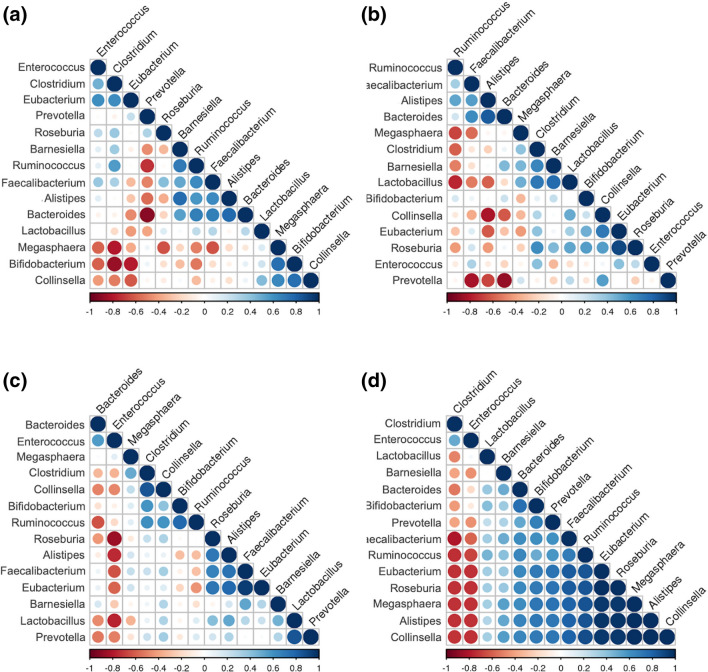
As shown in Fig. 7, while there was a strong negative correlation between Prevotella and Bacteroides within the healthy control (r = − 0.90; p = 0.001) and Type 1 diabetes (r = − 0.88; p = 0.004) groups, the correlation was lost among patients with Types 2 and 3c diabetes. Furthermore, there was a strong negative correlation between Bifidobacterium and Clostridium among healthy controls (r = − 0.87; p = 002), which was not observed in the diabetes groups. In addition, among patients with Type 2 diabetes, there was a strong negative correlation of Enterococcus with Lactobacillus (r = − 0.77; p = 0.009) and Roseburia (r = − 0.86; p = 0.001). Furthermore, among patients with Type 3c diabetes, there were strong negative correlations of Clostridium with Fecalibacterium (r = − 0.80; p < 0.0001), Ruminococcus (r = − 0.74, p = 0.001), Eubacterium (r = − 0.73, p = 0.001), Roseburia (r = − 0.74; p = 0.001) and Collinsella (r = − 0.72; p = 0.001). There were also strong negative correlations of Enterococcus with Eubacterium (r = − 0.73, p = 0.001), Roseburia (r = − 0.73, p = 0.001) and Collinsella (r = − 0.73, p = 0.001) among the patients with Type 3c diabetes.
Figure 7.
Corrogram depicting correlation of abundance of different genus. (a) Healthy control; (b) Type 1 diabetes; (c) Type 2 diabetes; (d) Type 3c diabetes.
Analysis of plasma metabolites
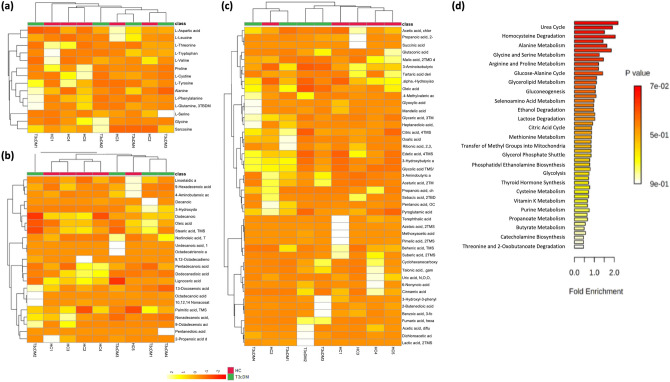
We conducted preliminary evaluation for plasma metabolites and pathway enrichment analyses (Fig. 8a–d; Supplementary Fig. 8) in 5 healthy controls and 4 patients with Type 3c diabetes. Although we did not find any significant unique metabolite signature based on the type of diabetes, we did observe a trend towards reduction of l-threonine, l-cystine, l-phenylalanine in Type 3c diabetes compared to healthy controls. Moreover, patients with Type 3c diabetes showed an increasing trend for fatty acids and sphingolipids (eg. myoinositol) compared to the healthy controls. As shown in Supplementary Fig. 9, we could not demonstrate statistically significant differences in the interactions between the microbiome and the metabolome.
Figure 8.
Metabolomic parameters of controls (n = 5) and patients with Type 3c diabetes (n = 4). Heatmaps with dendrograms depicting (a) Amino acid profile; (b) Fatty acid profile; (c) Non-fatty organic acid profile; Enrichment plot depicting (d) Metabolic pathways.
Discussion
In this preliminary cross-sectional study, we have shown that patients with Type 3c diabetes has a unique gut bacterial signature that differs from Type 1 and Type 2 diabetes. Since the gut microbiota is shaped by a variety of physiological and pathological factors, and the pathophysiology of the three types of diabetes are different, we hypothesized that gut microbial dysbiosis in these patients could also be variable. This speculation was triggered by the results from our earlier studies on CP which suggested that Type 3c diabetes was associated with islet inflammation and infiltration with Th17 cells26–28. It was also suggested that islet inflammation could be contributed by endotoxemia that resulted from gut microbial dysbiosis and altered gut mucosal permeability in these patients20. Even though there are earlier studies that had evaluated the gut microbiome in diabetes, to the best of our knowledge this the first study to have compared the gut microbiome head to head in the three types of diabetes. The other factor that makes the current study relevant is the recent evidence that confirms the Indian gut microbiota to be unique from that seen in the west29. Dubey et al. reported 993 unique organisms in a pan Indian study of which 390 were common to all geographic locations and the most abundant species were Prevotella copri and Faecalibacterium praustnitzii30.
In the current study, the predominant organisms in the healthy controls included Prevotella, Fecalibacterium, Ruminococcus, Roseburia, which were in alignment with the earlier reports of normal Indian gut microbiota29,31,32. In the Type 2 diabetic group, we observed a reduction in abundance in Fecalibacterium, Eubacterium, and Ruminococcus compared to controls, which was similar to that observed in earlier studies on Indian Type 2 diabetes patients17,18. Our current results were also similar to our earlier report in which we demonstrated significant reduction in Fecalibacterium praustnitzii and Ruminococcus bromii in patients with CP with diabetes20. These data had ensured the reliability of the findings on altered microbiome in the diabetic patients in this study. There are so far no Indian studies that reported on gut microbiome in Type 1 diabetes. In the current study, we observed reduced abundance of Prevotella and Faecalibacterium while increase in Bacteroides, Alistepes, Bifidobacterium and Parabacteroides that was similar to studies from the west13,21.
Although we observed significant difference in the dysbiosis between the three type of diabetes, it would be difficult to comment on the cause-effect relationship sans experimental evidence. Nevertheless, the dysbiosis that we observed is likely to have functional implications in the pathophysiology of diabetes. An increased abundance of Bacteroides sp. have been shown to result in altered mucosal integrity and gut permeability by affecting zonulin level, that further modulates the intercellular junctions and macromolecular passages in the cell15. Additionally, a higher abundance of Vellionella sp. could impact host health by producing lactate which eventually could weakens cellular tight junctions. Further, the reduction of butyrate producing genera Faecalibacterium, Ruminococcus, and Eubacterium is known to result in a proinflammatory environment within the gut as butyrate induces colonic T-reg cells, decreases pro-inflammatory macrophage production, thereby enhancing the gut barrier integrity. It also enhances mucin production which is also helpful in maintaining the gut permeability33. Additionally R. bromii has been considered as a keystone species which metabolizes starch and the reduction of the species may lead to the poor starch degrading capacity of the host, which is often observed in the Type 3c diabetics34. All these genera were significantly altered in the patients with Type 3c diabetes compared Type 1 and Type 2 diabetes.
Recent studies have shown that Prevotella copri is among the most abundant core organism in the heathy Indian gut29–31. We observed a significant reduction in the abundance of this organism in patients with Type 3c diabetes compared to healthy control, Type 1 and Type 2 diabetes. While we did not observe any difference of Prevotella copri between healthy controls and Type 1 diabetics, it was earlier reported to be increased in Type 1 diabetics15. Prevotella has been linked to produce propionate, succinate and acetate which are responsible for gut mucosal and tight junction integrity, and T-reg differentiation. Prevotella has been linked with a diet rich in plant derived carbohydrates and fiber and is in inverse relationship with Bacteroides35. The high abundance of Prevotella in our healthy controls can be explained by a vegetarian predominant diet in India, and the loss of correlation of Prevotella and Bacteroides in the Type 3c diabetes group (Fig. 3) suggests its potential implication in Type 3c diabetes.
We observed significantly lower abundance of Megasphaera in Type 3c diabetes compared to the patients with Type 1 and Type 2 diabetes. It has now been shown that Megasphaera elsdenii, Megasphaera sp. NM10 and Megasphaera sp. BL7 renders a positive effect to the host health by utilizing lactate to produce SCFAs and synthesizing riboflavin36. This points towards a potential implication of reduced Megasphaera in Type 3c diabetes. We also observed unique bacterial profiles in Type 3c diabetes group namely, Nesterenkonia sp., Geobacter sp., Clostridium sp. Acinetobacter sp., and Porphyromonas sp. Porphyromonas somarae has been linked with an increase in pro-inflammatory cytokines. This may have an implication in Type 3c diabetes since it has been associated with chronic inflammation of the pancreas. Nesterenkonia sp. has also been considered as pathogenic and has been shown to be associated with the inflammatory condition in the host37.
CP is characterized by chronic, progressive inflammation involving the exocrine and endocrine pancreas. It is characterized by exocrine insufficiency that results in fat malabsorption. Excess fat in the intestine results in dysbiosis, which could eventually lead to intestinal inflammation, gut barrier dysfunction, and endotoxemia38. Circulating endotoxin could eventually cause islet inflammation via TLR4 involving the NFkB pathway39. The resulting hyperglycemia could further lead to gut microbial dysbiosis, resulting in a vicious cycle. This hypothesis appears to explain the significant differences in the gut microbiome in patients with Type 3c diabetes compared to Type 1 and 2 diabetes Furthermore, patients with CP are associated with intestinal dysmotility and small intestinal bacterial overgrowth, irrespective of glycemic status, and the morphological and biochemical characteristics of CP are heterogenous40,41. This could further explain our observation that the dysbiosis was particularly high in some of the Type 3c diabetics compared to others within the group, and the higher abundance of the phylum Proteobacteria was particularly high in this group. Further support to our observation comes from the fact that these patients had a poorer glycemic control. However, this hypothesis needs to be confirmed by experimental approaches using established models of CP.
A drawback of our study is the small sample size. Since there were no prior comparative studies, we decided to initially conduct a preliminary study. Since the results are encouraging, a better structured study with a large sample size is now mandated. Moreover, we did not observe significant differences in the metabolome profile in spite of differences in the bacteriome profile. This could also be explained by the very small sample size. Since there were some trend in significance in a few of the metabolites, it is likely that studies with larger sample sizes would yield meaningful differences. Furthermore, use of other techniques such as LC–MS/MS could also provide plausible results.
In conclusion, in this preliminary cross-sectional study, we report significant differences in gut microbiome in patients with Type 3c or pancreatogenic diabetes compared to patients with Type 1 and 2 diabetes. Our data needs to be validated in larger multicentre cohorts of patients and the cause-effect relationship needs to be evaluated in experimental studies in order to make the results generalizable.
Methods
Patient recruitment and clinical data acquisition
This preliminary cross-sectional study was conducted at a tertiary care academic centre in accordance with principles of the Declaration of Helsinki as revised in 2008. Ethical approval was obtained from the AIG Institutional Review Board prior to initiation of the study, and written informed consent was taken from all the participants prior to recruitment. All experimental procedures were performed according to standard guidelines and procedures that were approved by the Asian Institute of Gastroenterology Institutional Review Board. Patients with CP of at least 3 yrs duration and having poorly controlled Type 3c diabetes were screened for enrolment criteria in the Pancreas Clinic over a 9-month period. Patients with poorly controlled Type 1 and Type 2 diabetes were recruited from the Endocrinology outpatient clinic over a 3-month period. Individuals with history of inflammatory bowel disease, irritable bowel syndrome, chronic liver disease, recent critical illness, ongoing pregnancy, constipation and diarrhoea in the past 3 months, and antibiotics and probiotics intake in the past 3 months were excluded. We recruited healthy family members between 18 and 60yrs age who lived with the patients for at least 10 years as controls. Patients and healthy controls who were obese, smoked cigarettes, consumed alcohol and had poor quantity/quality metagenomic DNA were also excluded.
Data pertaining to demographic characteristics of patients and controls, diabetic status, clinical and imaging parameters of the patients with CP, details of treatment of diabetes and CP were captured in a structured proforma.
Definitions
CP was defined as per the MANNHEIM classification and confirmed with contrast enhanced computed tomography (CECT) scan42. Severity of CP was defined using magnetic resonance cholangiopancreatography (MRCP) using the Cambridge criteria for endoscopic retrograde cholangiopancreatography (ERCP) or endoscopic ultrasonography (EUS) using the Rosemont criteria43. The different types of diabetes were defined according to the ADA 2014 criteria44. We adopted the term Type 3c diabetes for diabetes secondary to exocrine pancreas as per recent literature. We considered the diagnosis of Type 3c diabetes in the patients with CP if it was diagnosed after a consistent lag period following the onset of CP and had a low to low-normal plasma C-peptide levels. Poorly controlled diabetes was defined as HbA1c of 7% and/or preprandial capillary plasma glucose between 80 and 130 mg/dL and/or peak postprandial capillary plasma glucose of 180 mg/dL.
Sample collection, biochemical evaluation, imaging and treatment
After recording clinical data, fecal samples were collected in a sterile fecal container, containing 3 ml RNA Later (Qiagen, Germany cat.no/ID: 76,106). Blood samples were collected after 8 h of fasting and 2 h post meal in K3-EDTA tubes for estimation of blood glucose, glycosylated hemoglobin (HbA1c) and C-peptide. Blood samples were also collected for evaluation of plasma metabolites in a proportion of patients and healthy controls. Both fecal and plasma samples were stored at − 80 °C immediately after collection until further analysis.
We estimated blood glucose using the Glucose Oxidase–Peroxidase (GOD-POD) method with ERBA Glucose kit (Transasia Bio-Medicals Ltd, HP, India). High performance liquid chromatography (HPLC) using a National Glycohemoglobin Standardised Program (NGSP) certified automated analyser from Bio-Rad was used to measure HbA1c. Plasma C-peptide assay was estimated using the sandwiched Electrochemical Immunoassay (ECLIA) technique in the Roche Cobas ‘e’ 601 Immunoassay Analyzer. Briefly, double incubations were done with 20 μl biotinylated monoclonal antibodies (anti C-peptide) followed by streptavidin coated micro particles. The bound micro particles were then magnetically captured onto the surface of the electrode that resulted in induction of chemiluminescent emission upon current application. The emission was measured on a photomultiplier and quantified by 2-point calibration with a master curve.
Disease morphology in the patients with CP was evaluated by cross sectional imaging [contrast enhanced computed tomography (CECT) or magnetic resonance cholangiopancreatography (MRCP)] and endoscopic retrograde cholangiography (ERCP). All controls and patients with Type 1 and Type 2 diabetes underwent transabdominal ultrasonography to evaluate for the pancreas and the liver.
Patients with CP were treated with antioxidant cocktails, pancreatic enzyme supplementation and analgesics on demand. Patients who had recurrent intractable pain that did not respond to medical therapy were treated with endoscopic treatment. Patient with Type 1 diabetes were treated with insulin, while patients with Type 2 and Type 3c diabetes were initiated on oral hypoglycemic agents (OHAs). If glycemic control was suboptimal with OHAs, then insulin treatment was initiated.
Fecal DNA extraction
The metagenomic DNA isolation was performed (200 mg stool/sample) using Qiagen mini stool DNA isolation kit (Germany, Cat. No 81504) as per the manufacturer’s instruction. DNA quantity and quality were assessed using the Nano drop 2000 spectrophotometer (Thermo Scientific, IL, USA) based on A260/A280 absorbance and agarose gel electrophoresis (1% wt vol−1 agarose in Tris acetate EDTA). Reagents from the same lot was used for all pre-sequencing sample processing.
Next generation sequencing (NGS)
NGS of the V3–V4 region of 16SrDNA in the fecal metagenome was conducted on the Illumina MiSeq platform at Xcelris Genomics (Ahmedabad, India) with 2*250 (i.e. paired end) chemistry. The paired end sequence data were assembled into contigs following V3–V4 region amplification using the universal primers (V3-F: 5′ CCTACGGGNGGCWGCAG3′ and V4-R: 5′GACTACHVGGGTATCTAATCC3′). The subsequent steps such as ligation with the adaptors, library preparation, barcode additions were processed as described previously20. The R1 and R2 reads from each sample were stitched or assembled and then proceeded for quality check i.e. phred (Q20) quality score, and sequences with less than Q20 were removed. Chimeric sequences, mismatched sequences, barcodes and the sequences with length less than 100 bp were also removed from the final sequences, following which the rest of the reads were uploaded to the MG-RAST server45. The metadata and raw sequences have also been submitted to the NCBI SRA portal with BioProject accession PRJNA723868 (SRR1430686-SRR1430729). Taxonomic assignment was performed with 95% similarity cut off with the RDP (Ribosomal Database Project) database in the MG-RAST server. We used positive and negative controls in the sequence runs to evaluate for batch effect and inadvertent contamination respectively.
Sample preparation for metabolite analysis
Plasma was separated within 30 min of blood withdrawal from the study participants. Plasma metabolite profiles were screened using the Gas Chromatography Mass Spectrometry (GCMS). Briefly, 300 µl of plasma samples were extracted in n-methanol (1:1, vv−1) followed by 15 h incubation on a shaker incubator at room temperature. Samples were centrifuged at 12,000 rpm for 5 min at 4 °C. A 200 µl extract was preserved for drying in a desiccator for 24 h. Derivatization was performed using 50 µl of Pyridine (20 mg/ml, Sigma-Aldrich, (CAS nos. 110-86-1) followed by incubation at 30 °C for 90 min and 30 µl N-trimethylsilyl-N-methyl trifluoroacetamide (MSTFA, Sigma-Aldrich, CAS no. 24589-78-4) followed by re-incubation at 37 °C for 30 min. We did not use any internal standard since the study was focused on untargeted metabolite profiling of the samples.
Gas Chromatography mass spectrometry (GCMS)
1 µl of the derivatized samples were prepared for GCMS analysis under the Splitless injection condition as per the program described earlier. We used a triple quadrupole Shimadzu GC 2010 Plus-TP-8030 system (equipped with EB5MS column), and helium was used as the carrier gas. The peak area for each metabolite which was based on the m/z ratio and peak intensity was considered for further analysis. Background peaks and column bleed peaks consisting of silanes and siloxanes were not considered. The peak area of a single compound showing multiple derivatized forms were pooled for the downstream analysis. National Institute of Standards and Technology (NIST) library was used to identify the peaks. Further functional analysis of the metabolite was carried out using the human metabolite database (HMDB) (http://www.hmdb.ca)46 and the Kyoto Encyclopedia of Genes and Genomes database (KEGG, http://www.genome.jp/kegg/)47.
Statistical analyses
Since there are no prior parallel comparison of the gut microbiota between the three types of diabetes, we decided to undertake the current comparison as a preliminary study. Therefore, we did not perform any formal sample size calculation. A database was generated in Excel for Mac and statistical analyses were conducted using the Statistical Package of Social Scientists (SPSS) (IBM SPSS 20, SPSS Inc, Chicago, IL), Paleontological Statistics Software (PAST) (Version 3.11 for Mac)48 and R studio (Version 1.3.959)49 statistical platforms. Continuous clinical data were expressed as mean (± SD) while categorical data were expressed as proportions.
Microbial richness and alpha diversity were expressed as Chao 1, and Shannon-H index, Evenness index, Fischer’s alpha respectively based on the species level abundance. The data were represented as box and whisker plots generated in ggplot2 package for R. Beta diversity was assessed by principal coordinate analysis calculating the Euclidean distance matrix. Per sample rarefaction curves were constructed using the MicrobiomeAnalyst online platform based on minimum library depth after normalization to rule out artifacts, as per the method described by Weiss et al.50. The initial comparison among the microflora in all the groups were made using the absolute abundance /counts of the taxa in each group. Principal Component analysis (PCA) with convex hulls was created to evaluate the intergroup differences and the groups were compared by one-way permutational multivariate analysis of variance (PERMANOVA) using 10,000 permutations with species abundance values in PAST (PAleontological STatistics software, Mac version 3.11). Significant differences in the microbiome profiles were tested using the non-parametric Kruskal–Wallis test (KW) followed by Tukey’s post-hoc pairwise analyses in SPSS with Bonferroni corrections for multiple hypothesis testing. Relative abundances of the different taxonomic levels, (i.e. phylum, class, order, family, genus and species) were expressed by box and whisker plots. Heatmaps were constructed based on the abundance value for microbes and area value of the peaks detected by GCMS for metabolites in R script based Metaboanalyst 4.0 and Heatmapper data expression tools51,52. The correlation matrices were generated using Spearman correlation in SPSS and the matrices were visualized as corrograms with the R script based STHDA online platform53. Venn diagram was used to express common genera among the different groups using the Venn diagram maker54. Microbiome-metabolome correlation networks were generated using Cytoscape v 3.855. The pathway enrichment plots were generated using the MetaboAnalyst v.4.0.
Supplementary information
Acknowledgements
We thank Xcelris Laboratories, Ahmedabad for conducting the bacterial metagenomic sequencing and Vimta Laboratories, Hyderabad for conducting the GC-MS/MS. We also thank Asian Healthcare Foundation for providing unconditional intramural research grant to conduct this study.
Abbreviations
- CECT
Contrast enhanced computed tomography
- CP
Chronic pancreatitis
- ECLIA
Electrochemical immunoassay
- ERCP
Endoscopic retrograde cholangiopancreatography
- EUS
Endoscopic ultrasound
- GOD-POD
Glucose oxidase–peroxidase
- KEGG
Kyoto encyclopedia of genes and genomes database
- MG-RAST
Metagenomic rapid annotations using subsystems technology
- MRCP
Magnetic resonance cholangiopancreatography
- NGS
Next generation sequencing
- NGSP
National glycohemoglobin standardised program
- OHA
Oral hypoglycemic agents
- PAST
Paleontological statistics software
- PCA
Principal component analysis
- PERMANOVA
Permutational multivariate analysis of variance
- SPSS
Statistical package for social scientists
- Th
T-helper
Author contributions
Study design was done by R.T.; Patient screening, recruitment and clinical data recording were done by R.T., M.A.; D.N.R.; Fecal metagenomic DNA isolation was conducted by A.J., M.J.; Biochemical analyses were conducted by D.G.; Metabolomic profiling and analysis was performed by P.S.; Statistical and bioinformatics analyses along with visualizations were done by R.T., P.S., S.S.; Manuscript drafting and editing was done by P.S., R.T., M.U.; Manuscript review and intellectual inputs were provided by D.N.R. All the authors approved the final version of the manuscript.
Funding
The study was supported partially by a fellowship grant from the Wellcome DBT India Alliance (IA/CPHS/17/1/503358) and Institutional Research Grant from the Asian Healthcare Foundation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90024-w.
References
- 1.https://idf.org/our-network/regions-members/south-east-asia/members/94-india.html.
- 2.Das A, et al. Type 1 diabetes in India: overall insights. Indian J. Endocr. Metab. 2015;19:31. doi: 10.4103/2230-8210.155372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslam M, Vijayasarathy K, Talukdar R, Sasikala M, Nageshwar Reddy D. Reduced pancreatic polypeptide response is associated with early alteration of glycemic control in chronic pancreatitis. Diabetes Res. Clin. Pract. 2020;160:107993. doi: 10.1016/j.diabres.2019.107993. [DOI] [PubMed] [Google Scholar]
- 4.Hart PA, et al. Type 3c (pancreatogenic) diabetes mellitus secondary to chronic pancreatitis and pancreatic cancer. Lancet Gastroenterol. Hepatol. 2016;1:226–237. doi: 10.1016/S2468-1253(16)30106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha AK, Goenka MK, Goenka U. Chronic pancreatitis in Eastern India: Experience from a tertiary care center. Indian J. Gastroenterol. 2017;36:131–136. doi: 10.1007/s12664-017-0733-9. [DOI] [PubMed] [Google Scholar]
- 6.Rajesh G, Veena AB, Menon S, Balakrishnan V. Clinical profile of early-onset and late-onset idiopathic chronic pancreatitis in South India. Indian J. Gastroenterol. 2014;33:231–236. doi: 10.1007/s12664-013-0421-3. [DOI] [PubMed] [Google Scholar]
- 7.Bhasin DK, et al. Clinical profile of idiopathic chronic pancreatitis in North India. Clin. Gastroenterol. Hepatol. 2009;7:594–599. doi: 10.1016/j.cgh.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharjee PK, Mukerjee A, Adhikary C. Demographic and clinicopathological profile of patients with chronic pancreatitis in a tertiary referral teaching hospital of West Bengal: Personal experience. Indian J. Gastroenterol. 2015;34:365–371. doi: 10.1007/s12664-015-0594-z. [DOI] [PubMed] [Google Scholar]
- 9.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Mitnala S, Reddy DN. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppel N, Maini RV, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356:6344. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, He J, Jia W. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin. Drug Metab. Toxicol. 2016;12:31–40. doi: 10.1517/17425255.2016.1121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamshidi P, et al. Is there any association between gut microbiota and type 1 diabetes? A systematic review. Gut Pathog. 2019;11:49. doi: 10.1186/s13099-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harsch IA, Konturek PC. The Role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into “old” diseases. Med. Sci. 2018;17(6):32. doi: 10.3390/medsci6020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leiva-Gea I, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. 2018;41:2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 16.Muñoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol. Nutr. 2016;63:560–568. doi: 10.1016/j.endonu.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Pushpanathan P, et al. Gut microbiota in type 2 diabetes individuals and correlation with monocyte chemoattractant protein1 and interferon gamma from patients attending a tertiary care centre in Chennai, India. Indian J. Endocr Metab. 2016;20:523–530. doi: 10.4103/2230-8210.183474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaike, A.H., et. al. The Gut Microbial Diversity of Newly Diagnosed Diabetics but Not of Prediabetics Is Significantly Different from That of Healthy Nondiabetics. mSystems.5, e00578–19 (2020). [DOI] [PMC free article] [PubMed]
- 19.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 20.Jandhyala SM, et al. Altered intestinal microbiota in patients with chronic pancreatitis: implications in diabetes and metabolic abnormalities. Sci. Rep. 2017;7:43640. doi: 10.1038/srep43640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibbò S, Dore MP, Pes GM, Delitala G, Delitala AP. Is there a role for gut microbiota in type 1 diabetes pathogenesis? Ann. Med. 2017;49:11–22. doi: 10.1080/07853890.2016.1222449. [DOI] [PubMed] [Google Scholar]
- 22.DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med. Clin. North Am. 2004;88:787–835. doi: 10.1016/j.mcna.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2019;15:635–650. doi: 10.1038/s41574-019-0254-y. [DOI] [PubMed] [Google Scholar]
- 24.Conwell DL, et al. American pancreatic association practice guidelines in chronic pancreatitis: evidence-based report on diagnostic guidelines. Pancreas. 2014;43:1143–1162. doi: 10.1097/MPA.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rickels MR, et al. Detection, evaluation and treatment of diabetes mellitus in chronic pancreatitis: recommendations from PancreasFest 2012. Pancreatology. 2013;13:336–342. doi: 10.1016/j.pan.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasikala M, et al. Bach2 repression mediates Th17 cell induced inflammation and associates with clinical features of advanced disease in chronic pancreatitis. United Eur. Gastroenterol. J. 2018;6:272–282. doi: 10.1177/2050640617716596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talukdar R, et al. T-Helper cell-mediated islet inflammation contributes to β-cell dysfunction in chronic pancreatitis. Pancreas. 2016;45:434–442. doi: 10.1097/MPA.0000000000000479. [DOI] [PubMed] [Google Scholar]
- 28.Sasikala M, et al. β-Cell dysfunction in chronic pancreatitis. Dig. Dis. Sci. 2012;57:1764–1772. doi: 10.1007/s10620-012-2086-7. [DOI] [PubMed] [Google Scholar]
- 29.Pulipati P, et al. The Indian gut microbiota-Is it unique? Indian J. Gastroenterol. 2020;39:133–140. doi: 10.1007/s12664-020-01037-8. [DOI] [PubMed] [Google Scholar]
- 30.Dubey AK, et al. LogMPIE, pan-India profiling of the human gut microbiome using 16S rRNA sequencing. Sci. Data. 2018;5:180232. doi: 10.1038/sdata.2018.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dehingia M, et al. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 2015;5:18563. doi: 10.1038/srep18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhakan DB, et al. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience. 2019;8:giz004. doi: 10.1093/gigascience/giz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamer HM, et al. Review article: the role of butyrate on colonic function: review: role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2007;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 34.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shetty, S. A., Marathe, N. P., Lanjekar, V., Ranade, D., & Shouche, Y. S. Comparative Genome Analysis of Megasphaera sp. Reveals Niche Specialization and Its Potential Role in the Human Gut. PLoS One.8, e79353 (2013). [DOI] [PMC free article] [PubMed]
- 37.Chander AM, et al. Genome insight and comparative pathogenomic analysis of nesterenkonia jeotgali strain CD087 isolated from duodenal mucosa of celiac disease patient. Front. Microbiol. 2017;8:1. doi: 10.3389/fmicb.2017.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care. 2015;18:515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amyot J, Semache M, Ferdaoussi M, Fontés G, Poitout V. Lipopolysaccharides impair insulin gene expression in isolated islets of langerhans via toll-like receptor-4 and nf-κb signalling. PLoS ONE. 2012;7:36200. doi: 10.1371/journal.pone.0036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura T, et al. Pancreatic dysfunction and treatment options. Pancreas. 1998;29:329–336. doi: 10.1097/00006676-199804000-00020. [DOI] [PubMed] [Google Scholar]
- 41.El Kurdi B, et al. Factors that affect prevalence of small intestinal bacterial overgrowth in chronic pancreatitis: a systematic review and meta-regression. Clin. Tansl. Gastroenterol. 2019;10:e00072. doi: 10.14309/ctg.0000000000000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider A, Löhr JM, Singer MV. The M-ANNHEIM classification of chronic pancreatitis: introduction of a unifying classification system based on a review of previous classifications of the disease. J. Gastroenterol. 2007;42:101–119. doi: 10.1007/s00535-006-1945-4. [DOI] [PubMed] [Google Scholar]
- 43.Catalano MF, et al. EUS-based criteria for the diagnosis of chronic pancreatitis: the Rosemont classification. Gastrointest. Endosc. 2009;69:1251–1261. doi: 10.1016/j.gie.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 44.American Diabetes Association. Standards of Medical Care in Diabetes. Diabetes Care1(37)(Supplement_1), S14–80 (2014). [DOI] [PubMed]
- 45.Keegan KP, Glass EM, Meyer F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. In: Martin F, Uroz S, editors. Microbial Environmental Genomics (MEG) Springer; 2020. pp. 207–233. [DOI] [PubMed] [Google Scholar]
- 46.Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucl. Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucl. Acids Res. 2016;44(D1):D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammer O, Harper DAT, Ryan P. Past: paleontological statistics software package for education and: data analysis. Palaeontol. Electron. 2001;4(1):1–9. [Google Scholar]
- 49.R Studio: Integrated Development for R. RStudio, PBC, Boston, MA. http://www.rstudio.com (2020).
- 50.Weiss S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucl. Acids Res. 2009;37:W652–660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babicki S, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.http://www.sthda.com/english/wiki/correlation-analyses-in-r.
- 54.https://www.meta-chart.com/venn#/display.
- 55.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.