Abstract

The abnormally high expression of glutathione transferases is closely associated with cancer incidence and drug resistance. By introducing a hydrophobic moiety to the inhibitor structure, we organized a series of degraders of glutathione transferases and demonstrated them potently inducing apoptosis in cancer cells, presenting their pharmacological potential in cancer therapy.
Keywords: glutathione transferase pi, protein degradation, hydrophobic tagging
The glutathione transferases (GSTs) are a group of typical phase II detoxification enzymes found mostly in the cytosol. These enzymes catalyze conjugations of glutathione with a wide variety of exogenous and endogenous chemicals with electrophilic functional groups, forming water-soluble GSH conjugates that are readily transported out of the cell. Thus, GSTs play a pivotal role in cellular protection from environmental and oxidative stress, as well as in cancer cell resistance to drugs.1 Up to now, seven classes of the mammalian cytosolic GSTs are recognized, namely, alpha, mu, pi, theta, sigma, zeta, and omega.2 Many of them also catalyze isomerase, glutathione peroxidase, or thiol transferase reactions in the synthesis of leukotrienes, prostaglandins, and steroid hormones as well as in the degradation of tyrosine and inactivation or reduction of oxidative stress byproducts. In addition to their enzymatic activities, several GST family members also exhibit nonenzymatic functions in the regulation of cell signaling pathways implicated in stress responses, cell survival, and apoptosis.1−3 Specifically, GST pi (GSTP) has been known as an endogenous inhibitor of c-Jun N-terminal kinase (JNK)4 and tumor necrosis factor receptor associated factor 2 (TRAF2).5 GSTA1 has been shown to physically interact with JNK,6 and GSTM1 has inhibitory interaction directly with the apoptosis signal-regulated kinase 1 (ASK1).7 Of these enzymes, GSTP is of particular interest with regard to cancer because many tumors and cancer cell lines are characterized by high GSTP expression which can be linked with cancer incidence and drug resistance.8 It is anticipated that GST, especially the pi class, can be developed as a biomarker for early cancer detection or diagnosis as well as for targeted preventive and therapeutic interventions.9 Indeed, inhibition of GSTP expression through antisense cDNA was able to increase the cancer cell sensitivity to adriamycin, cisplatin, melphalan, and etoposide.10 Meanwhile, a significant number of GST inhibitors has been synthesized to modulate drug resistance as sensitizers to chemotherapy due to the predominant overexpression of GSTs in tumor cells.11,12
In recent years, proteolysis-targeting chimera (PROTAC) has emerged as a useful tool for the chemical knockdown of a protein of interest (POI). It employs E3 ligase ligands, fused via a flexible chemical linker to a targeting element for the POI, to elicit ectopic ubiquitination, resulting in proteasomal protein degradation.13 Alternatively, replacement of the E3 ligase ligand with a strong hydrophobic molecule like adamantyl, which is known as hydrophobic tagging (HyT), has been demonstrated as an efficient approach to downregulate the target protein in many cases.14,15 Considering the promising pharmacological utility of GSTs, especially GSTP, regarding tumor genesis and chemoresistance, we wonder whether chemical downregulation of GSTs could generate a better therapeutic effect compared with functional inhibition.
In 2012, Hedstrom et al. reported that a kind of chimera molecule connecting ethacrynic acid (EA), a covalent GST inhibitor, to Boc3Arg by a (CH2)6 linker (Figure 1), was capable to degrade cytosolic GST up to 80% at 80 μM.16 The mechanism was later demonstrated through direct localization of GSTP to 20S proteasome that subsequently executed degradation.17
Figure 1.
Reaction of EA or EA-Boc3Arg with GST.
In this study, we synthesized a series of HyT molecules that connect EA with the adamantyl moiety through either multiple ethylene or diethoxyl chains (Figure 2), aiming at increasing the hydrophobicity of the GSTP surface and thus triggering the cell’s protein quality control machinery and ultimately proteasomal degradation. Synthesis of these compounds was achieved from commercially available EA in 2 or 3 steps. For compounds with carbon linker ADC0EA–ADC4EA, EA reacted with different N-Boc protected diamines to form amides (3a–3c). Then, the −NH2 groups were released by removal of protecting groups to react with 1-adamantaneacetic acid to afford corresponding amides (5a–5c). For compounds ADE1EA–ADE4EA, 1-bromoadamantane was converted to ether with different polyethylene glycols with the terminal −OH, finally reacting with EA to give the targeted compounds 8a–8d (Figure 2).
Figure 2.
Structure and synthesis of EA-based HyTs. Reagents and conditions: (i) 1-adamantanamine hydrochloride, EDCI, HOBT, NMM, DMF, rt, overnight (92%); (ii) N-tert-butoxycarbonyl-1,6-hexanediamine (N-tert-butoxy carbonyl-1,2-ethylenediamine/N-tert-butoxycarbonyl-1,4-butanediamine), EDCI, HOBT, NMM, DMF, rt, overnight (73–93%); (iii) TFA, DCM, rt; (iv) 1-adamantaneacetic acid, EDCI, HOBT, NMM, DMF, rt, overnight (69–87%, over 2 steps); (v) ethylene glycol (diethylene/triethylene/tetraethylene glycol), Et3N, 180 °C, 18 h; (vi) ethacrynic acid, EDCI, DMAP, DMF, rt, overnight (57–72%).
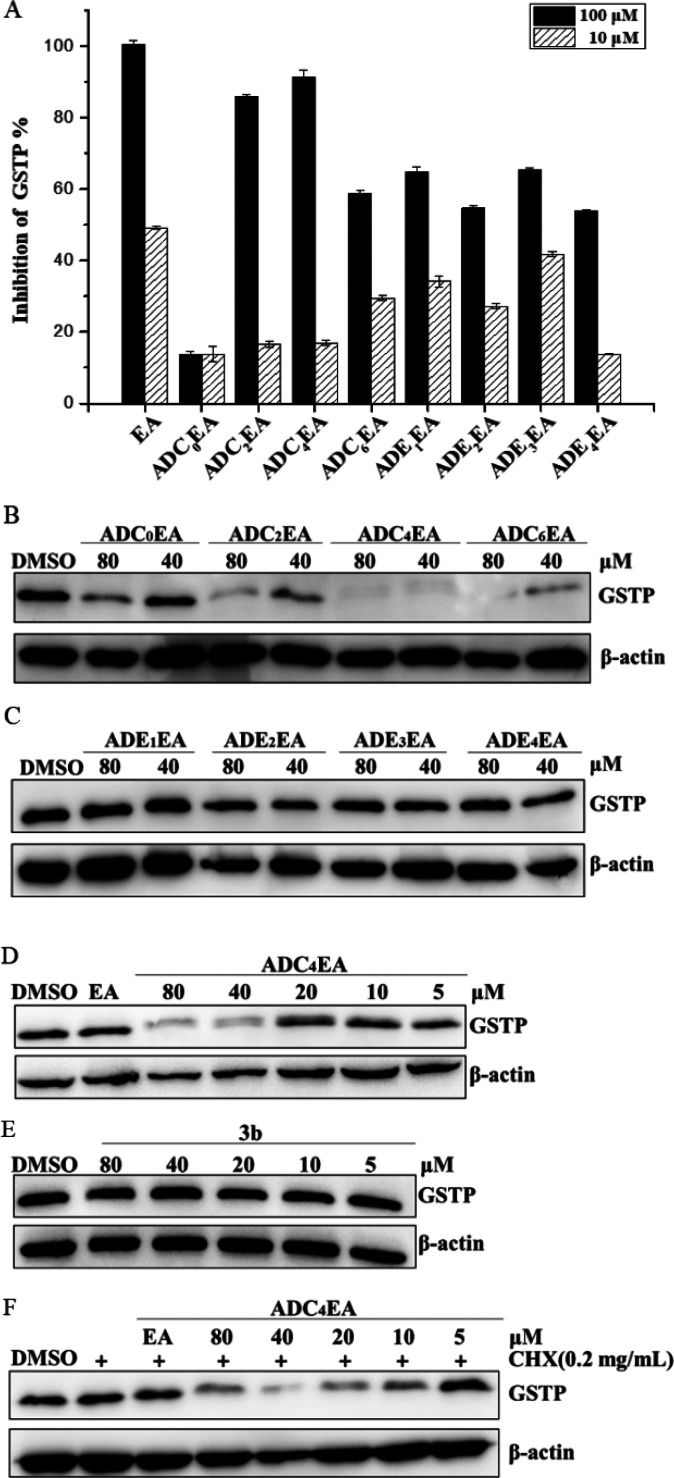
To ensure that attaching of the adamantyl moiety does not interfere with EA binding to GSTP, we measured the enzyme inhibitory activity of these bivalent compounds against GSTP using 1-chloro-2,4-dinitrobenzene (CDNB) assay in which the inhibition of GSTP catalyzed a conjugation reaction between GSH and CDNB, measured by the decrease in absorbance at 340 nm specific to the conjugate.18 Results showed that except for ADC0EA, which suffered a loss of GSTP inhibition potency, all other compounds were able to inhibit GSTP enzymatic activity, especially at high concentration of 100 μM (Figure 3A). Exposure of Hela (human cervical carcinoma) cells in which GSTP is expressed in high degree to either 40 or 80 μM of the −CH2– linked compounds for 3 h induced obvious degradation of GSTP (Figure 3B). However, this was not the case for diethoxy linked compounds (Figure 3C). The most potent compound was ADC4EA, which generated over 80% clearance of GSTP protein at 40 μM. Consistent with our expectation, this compound was observed to dose-dependently downregulate the GSTP protein level at concentrations higher than 20 μM (Figure 3D and S1). In contrast, the synthetic intermediate 3b which carries the C4 linker but no adamantyl tag exhibited no degradation at all tested concentrations (Figure 3E). This result emphasized the importance of the adamantyl tag for degradation. Because the intracellular protein homeostasis is maintained by a balance of both protein synthesis and degradation, and to reduce the impact of GSTP expression on protein level, we further pretreated the cells with cycloheximide (CHX) to inhibit protein synthesis before ADC4EA treatment. Indeed, ADC4EA dose-dependently downregulated the GSTP protein level (Figures 3F and S2), suggesting this downregulation was a result of improved degradation instead of decreased expression.
Figure 3.
(A) Inhibitory activities of the HyTs against GSTP. The values are an average of three experiments in parallel. GSTP protein levels in Hela cells after (B) ADC0EA to ADC4EA and (C) ADE0EA to ADE4EA treatment for 3 h. Dose–response of GSTP protein levels in Hela cells treated with (D) ADC4EA or (E) 3b for 3 h. (F) Dose–response of GSTP protein levels in Hela cells pretreated with CHX followed ADC4EA treatment for 3 h. The protein levels were measured in Western blot assay with specific antibodies using total cell lysates. Data are representative of three independent experiments.
We also compared the potencies of ADC4EA and 3b in GSTP degradation in two more cell lines, namely A549 lung adenocarcinoma and HT1080 fibrosarcoma cell lines, in which the GSTP are expressed in high or medium levels.19 Likewise, ADC4EA was found to degrade GSTP in these two cell lines, while 3b was not measured at high concentrations (Figure 4B)
Figure 4.
(A) The GSTP protein levels in A549 cells after treatment with ADC4EA or 3b for 3 h. (B) The GSTP protein levels in HT1080 cells after treatment with ADC4EA or 3b for 3 h.
To further validate that this degradation was processed through the ubiquitin-proteasome pathway (UPP), we used bortezomib, a 20S proteasome inhibitor, to cotreat the cells, and as expected, the degradation induced by ADC4EA was significantly reversed. In addition, the degradation can also be reversed by PYR-41, which is an inhibitor of ubiquitin-activating enzyme E1, suggesting the degradation relies on the ubiquitination of the target protein (Figure 5). Interestingly, when the cells were cotreated with bafilomycin A1, a specific inhibitor of vacuolar type H (+)-ATPase that prevents autophagy at late stage by inhibiting fusion between autophagosomes and lysosomes, the ADC4EA induced GSTP degradation was also greatly reversed. This suggested that in addition to UPP, ADC4EA can also induce GSTP protein degradation through autophagy pathway.
Figure 5.
(A) The GSTP levels in Hela cells after treatment of ADC4EA or the combination of ADC4EA with indicated agents for 3 h. (B) Quantitation of panel A by ImageJ and Origin 9.0 software. The protein levels were measured via Western blot assay with specific antibodies using total cell lysates. Data are representative of three independent experiments.
As an endogenous inhibitor of JNK, GSTP degradation is speculated to release JNK from the GSTP–JNK complex. Through activation by cytosol MAPKKs,20 the released JNK is going to induce chain events, starting from the phosphorylation of c-Jun, resulting in alteration in cell cycle, DNArepair, and apoptosis.21 Clearly, we have observed increased phosphorylation of JNK and c-Jun in ADC4EA treated cells, whereas no obvious alteration was observed in the EA treated cells (Figure 6A and S3). Interestingly, the total JNK levels seemed also downregulated by a certain degree in the 20, 40, or 80 μM treated groups (Figures 6A and B and S4), indicating the degradation of the entire GSTP–JNK complex induced by ADC4EA. Considering that reduction of oxidative stress byproducts is one of the major biofunctions of GST family members, the GSTP degradation is supposed to result in an accumulation of the reactive oxygen species (ROS) in cells. We thus measured the ROS level using DCFH-DA ROS florescent probe.22 As shown in Figure 6C, there was a significant ROS accumulation in the ADC4EA (20 μM) treated cells, which was comparable to the positive control (50 μg/mL of Rosup), while almost no ROS can be detected in the EA group even at a concentration as high as 80 μM.
Figure 6.

ADC4EA promoted phosphorylation of JNK and c-Jun and dose-dependently induced increase in ROS level. (A) Hela cells were treated with ADC4EA for 3 h, followed by Western blot assay with specific antibodies using total cell lysates. Data are representative of three independent experiments. (B) Quantitation of total JNK of panel A by ImageJ and Origin 9.0 software. (C) Hela cells were treated with the indicated concentrations of ADC4EA before the ROS level was quantitatively detected as a measure of fluorescence at Ex/Em: 488/525 with a ultrahigh-resolution laser confocal microscope (Nikon, Japan).
Considering JNK phosphorylation initiates multiple pro-apoptotic signaling and accumulation of ROS is a strong stimulus of cellular apoptosis, we further measured the apoptotic population in the treated cells using flow cytometry. Dose-dependently, ADC4EA induced apoptosis in cells after treatment for 3 h (Figure 7A). Treatment with 80 μM of ADC4EA has generated 61% apoptotic population. In contrast, only a marginal portion (∼15%) of apoptotic cells was observed in the EA treated cells (Figure 7B). The above observations indicated that compared with functional inhibition of GSTP, the HyT induced GSTP degradation should be more lethal to cancer cells due to its strong potency in triggering apoptotic cell death. Therefore, HyT-based GSTP degraders might have pharmacological potentials not limited to sensitizing chemotherapy but also as a single therapeutic agent.
Figure 7.
ADC4EA dose-dependently induced apoptosis in Hela cells. (A) The percentage of cells in different apoptotic states was measured by flow cytometric analysis using Annexin V-FITC and PI double-staining. (B) Quantitation of panel A by Origin 9.0.
In this study, EA was utilized as a ligand to bind with GSTP. However, it is known that EA also reacts with other GST isoenzymes and shows a preference toward GSTP and GSTA1. The observed degradation is also supposed to happen on other GST isoenzymes, for example, GSTM (Figure S5). The induced apoptosis should be the total effect resulting from degradation of all potential targets, although only GSTP was emphasized in this study. Because there are also some selective GSTP inhibitors available, for example the GSH analogue TLK199, it will be interesting and applicable to develop an HyT molecule using a selective GSTP inhibitor, by which a clearer contribution of GSTP to the apoptosis can be observed. Unlike the specific E3 ligase-based PROTACs, the structure of the hydrophobic moiety of HyT-based degraders can be very diverse and simple, facilitating the fast design and synthesis of the degrader. As reported by Mignani et al.23 that the poor antiproliferative capacity of EA could be modified by introduction of relevant lateral chain leading to the activation of the apoptotic cascade, although the degradation of the GSTs was not measured in their study. Indeed, we have observed that addition of a simple hexyl chain to EA had generated a compound potent enough to induce GSTP degradation at 80 μM (Figure S6). This study also showed that the hydrophobicity of the linker between EA and AD played an important role in the degradation effect. The hydrophobic carbon chain was more preferential, and the degradation potency could be modified by shifting the length of the linker.
In conclusion, by linking an adamantyl moiety to a GST inhibitor, we developed a series of HyT molecules that can potently degrade GSTP in cancer cells. Upon GST degradation, significant activation of JNK and accumulation of ROS was observed, which finally led to apoptotic cell death. The degradation was found through both proteasomal and autophagic pathways that updated the current knowledge about HyT technique. The work also provided preliminary structure–activity relationship regarding the HyT linker to facilitate the design of more degraders with pharmacological potential.
Acknowledgments
This work was generously supported by the National Natural Science Foundation of China (Grants 81872731 and 21977006). The authors also appreciate Dr. Paola Ripani from the Biochemistry Division, University of Konstanz for advices on GST subtypes.
Glossary
Abbreviations
- GST
glutathione transferase
- JNK
c-Jun N-terminal kinase
- TRAF2
tumor necrosis factor receptor associated factor 2
- ASK1
apoptosis signal-regulated kinase 1
- PROTAC
proteolysis-targeting chimera
- POI
protein of interest
- HyT
hydrophobic tagging
- EA
ethacrynic acid
- CDNB
1-chloro-2,4-dinitrobenzene
- CHX
cycloheximide
- UPP
ubiquitin-proteasome pathway
- ROS
reactive oxygen species
- EDCI
1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide hydrochloride
- HOBT
1-hydroxybenzotriazole
- NMM
N-methyl morpholine
- DMF
N,N-dimethylformamide
- TFA
trifluoroacetic acid
- DCM
dichloromethane
- Et3N
triethylamine
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00627.
Typical experimental procedures; supporting figures (S1–S5); synthesis and characterization of target compounds with NMR and mass spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hayes J. D.; Flanagan J. U.; Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Sheehan D.; Meade G.; Foley V. M.; Dowd C. A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. 10.1042/bj3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board P. G.; Menon D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim. Biophys. Acta, Gen. Subj. 2013, 1830 (5), 3267–88. 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Adler V.; Yin Z. M.; Fuchs S. Y.; Benezra M.; Rosario L.; Tew K. D.; Pincus M. R.; Sardana M.; Henderson C. J.; Wolf C. R.; Davis R. J.; Ronai Z. Regulation of JNK signaling by GSTp. EMBO J. 1999, 18 (5), 1321–1334. 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Fan Y.; Xue B.; Luo L.; Shen J.; Zhang S.; Jiang Y.; Yin Z. Human glutathione S-transferase P1–1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene 2006, 25 (42), 5787–800. 10.1038/sj.onc.1209576. [DOI] [PubMed] [Google Scholar]
- Romero L.; Andrews K.; Ng L.; O’Rourke K.; Maslen A.; Kirby G. Human GSTA1–1 reduces c-Jun N-terminal kinase signalling and apoptosis in Caco-2 cells. Biochem. J. 2006, 400 (1), 135–41. 10.1042/BJ20060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. G.; Lee Y. H.; Park H. S.; Ryoo K.; Kang K. W.; Park J.; Eom S. J.; Kim M. J.; Chang T. S.; Choi S. Y.; Shim J.; Kim Y.; Dong M. S.; Lee M. J.; Kim S. G.; Ichijo H.; Choi E. J. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 2001, 276 (16), 12749–55. 10.1074/jbc.M005561200. [DOI] [PubMed] [Google Scholar]
- Tew K. D.; Townsend D. M. Regulatory functions of glutathione S-transferase P1–1 unrelated to detoxification. Drug Metab. Rev. 2011, 43 (2), 179–93. 10.3109/03602532.2011.552912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Gupta S. The multifaceted role of glutathione S-transferases in cancer. Cancer Lett. 2018, 433, 33–42. 10.1016/j.canlet.2018.06.028. [DOI] [PubMed] [Google Scholar]
- Ban N. T. Y.; Takayama T.; Kura T.; Katahira T.; Sakamaki S.; Niitsu Y. Transfection of Glutathione S-Transferase (GST)-Π Antisense Complementary DNA Increases the Sensitivity of a Colon Cancer Cell Line to Adriamycin, Cisplatin, Melphalan, and Etoposide. Cancer Res. 1996, 56, 3577–3582. [PubMed] [Google Scholar]
- Mahajan S.; Atkins W. M. The chemistry and biology of inhibitors and pro-drugs targeted to glutathione S-transferases. Cell. Mol. Life Sci. 2005, 62 (11), 1221–33. 10.1007/s00018-005-4524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pljesa-Ercegovac M.; Savic-Radojevic A.; Matic M.; Coric V.; Djukic T.; Radic T.; Simic T. Glutathione Transferases: Potential Targets to Overcome Chemoresistance in Solid Tumors. Int. J. Mol. Sci. 2018, 19 (12), 3785. 10.3390/ijms19123785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G. M.; Crews C. M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181 (1), 102–114. 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T. K.; Crews C. M. Greasy tags for protein removal. Nature 2012, 487 (7407), 308–309. 10.1038/487308a. [DOI] [PubMed] [Google Scholar]
- Cromm P. M.; Crews C. M. Targeted Protein Degradation: from Chemical Biology to Drug Discovery. Cell Chem. Biol. 2017, 24 (9), 1181–1190. 10.1016/j.chembiol.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. J.; Gollapalli D. R.; Hedstrom L. Inhibitor mediated protein degradation. Chem. Biol. 2012, 19 (5), 629–37. 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Long M. J.; Rosenberg M. M.; Li S.; Kobjack A.; Lessans P.; Coffey R. T.; Hedstrom L. Boc3Arg-Linked Ligands Induce Degradation by Localizing Target Proteins to the 20S Proteasome. ACS Chem. Biol. 2016, 11 (12), 3328–3337. 10.1021/acschembio.6b00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido Y.; Tomoike F.; Kuwata K.; Fujikawa H.; Sekido Y.; Murakami-Tonami Y.; Kameda T.; Abe N.; Kimura Y.; Shuto S.; Abe H. A Covalent Inhibitor for Glutathione S-Transferase Pi (GSTP1–1) in Human Cells. ChemBioChem 2019, 20 (7), 900–905. 10.1002/cbic.201800671. [DOI] [PubMed] [Google Scholar]
- Di Pietro G.; Magno L. A. V; Rios-Santos F. Glutathione S-transferases: an overview in cancer research. Expert Opin. Drug Metab. Toxicol. 2010, 6 (2), 153–170. 10.1517/17425250903427980. [DOI] [PubMed] [Google Scholar]
- Davis R. J. Signal transduction by the JNK group of MAP kinases. Cell 2000, 103 (2), 239–52. 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Karin M.; Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 2005, 57 (4–5), 283–95. 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- Lyublinskaya O. G.; Ivanova J. S.; Pugovkina N. A.; Kozhukharova I. V.; Kovaleva Z. V.; Shatrova A. N.; Aksenov N. D.; Zenin V. V.; Kaulin Y. A.; Gamaley I. A.; Nikolsky N. N. Redox environment in stem and differentiated cells: A quantitative approach. Redox Biol. 2017, 12, 758–769. 10.1016/j.redox.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignani S.; El Brahmi N.; El Kazzouli S.; Eloy L.; Courilleau D.; Caron J.; Bousmina M. M.; Caminade A. M.; Cresteil T.; Majoral J. P. A novel class of ethacrynic acid derivatives as promising drug-like potent generation of anticancer agents with established mechanism of action. Eur. J. Med. Chem. 2016, 122, 656–673. 10.1016/j.ejmech.2016.05.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.