Abstract
Emerging evidence in clinical and preclinical studies indicates that success of immunotherapies can be impacted by the state of the microbiome. Understanding the role of the microbiome during immune-targeted interventions could help us understand heterogeneity of treatment success, predict outcomes, and develop additional strategies to improve efficacy. In this review, we discuss key studies that reveal reciprocal interactions between the microbiome, the immune system, and the outcome of immune interventions. We focus on cancer immune checkpoint inhibitor treatment and vaccination as two crucial therapeutic areas with strong potential for immunomodulation by the microbiota. By juxtaposing studies across both therapeutic areas, we highlight three factors prominently involved in microbial immunomodulation: short-chain fatty acids, microbe-associate molecular patterns (MAMPs), and inflammatory cytokines. Continued interrogation of these models and pathways may reveal critical mechanistic synergies between the microbiome and the immune system, resulting in novel approaches designed to influence the efficacy of immune-targeted interventions.
Keywords: microbiome, immune checkpoint inhibitors, vaccines, innate immunity, immuno-oncology, adaptive immunity
Introduction: The Microbiome and the Immune System
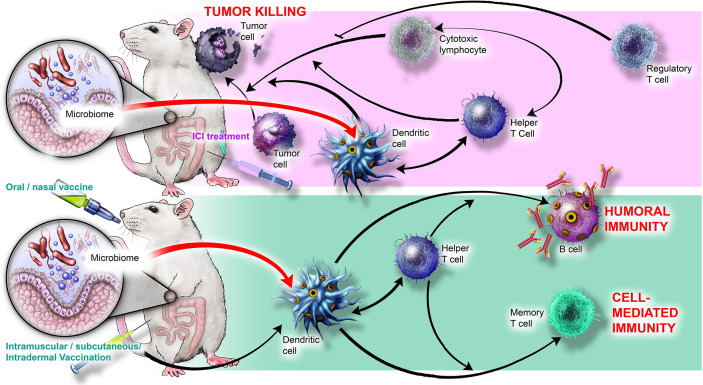
Humans are colonized by trillions of microbes collectively termed the microbiome, consisting of bacteria, archaea, viruses, fungi, and protists (1). Together, these interrelated microbial communities represent a rich source of metabolites and ligands that broadly influence human biology, including nutrient digestion, tissue homeostasis, neuroendocrine signaling, and the development and maintenance of the immune system (reviewed in 2–4). It is becoming clear that the microbiome modulates diverse immune processes, from defense against infection (5–7) to antibody production (8, 9), and from inflammation (10–12) to autoimmunity and allergy (13–15). Immune-microbe interactions also directly regulate homeostasis and development of immune cells such as antigen-presenting cells (16–19) and T cells (20–24). Importantly, the microbiome significantly influences the host response to immune-targeted interventions (25–31), which harness the immune system to treat or prevent diseases including infections, allergy, autoimmunity, inflammatory disorders, and cancer. Two of the most prominent immune interventions currently employed in the clinic are cancer immune checkpoint inhibitors and vaccines, which together are the focus of this review; key cellular players and interactions involved in immune checkpoint inhibitor-mediated tumor killing and vaccine-induced immunity are presented in Figure 1 .
Figure 1.
Cellular players and interactions involved in immune checkpoint inhibitor-mediated tumor killing and vaccine-induced immunity. (Top) ICIs stimulate cytotoxic lymphocytes, Th1 helper T cells, and DCs to kill tumor cells; Tregs inhibit killing. MAMPs produced by bacteria such as B. thetaiotaomicron and B. fragilis may interact with TLR2 and TLR4 on DCs and stimulate Th1 polarization and synergize with ICI activity. Microbial metabolites such as inosine (produced by B. pseudolongum) may also contribute to ICI efficacy by stimulating T cell proliferation. (Bottom) Live, inactivated, or molecular vaccination ultimately activates DCs and helper T cells to yield humoral immunity (B cell antibodies) and cell-mediated immunity (memory T cells). MAMPs, such as flagellin and peptidoglycan, interact with PRRs to stimulate B cells and Tfh cells, thereby augmenting vaccine response. Microbially produced SCFAs may also stimulate DCs.
In the past decade, immune checkpoint inhibitors (ICIs) have become an essential pillar of treatment for numerous cancers (32–34). Currently employed ICIs are monoclonal antibodies that block specific immune checkpoint receptors (CTLA-4, PD-1) or ligands (PD-L1) on the surface of lymphocytes or tumor cells, respectively (32, 34). Normally, immune checkpoint signaling prevents development of hyperactive immune responses and thus damage to healthy tissues (32). However, these checkpoints are exploited by tumor cells to evade immunosurveillance, block anti-tumor cytotoxic T lymphocytes (CTLs), and induce immunosuppressive regulatory T cells (Tregs) (35, 36). By blocking immune checkpoint receptor-ligand interactions, ICIs can restore endogenous anti-tumor immune responses and disrupt cancer progression. However, not all individuals respond to ICIs, and recent evidence suggests the microbiota may play a role in ICI responsiveness by modulating the immune system, particularly the abundances and functions of NK cells, CTLs, and Tregs (37–43).
Equally consequential, vaccines have revolutionized our ability to prevent a myriad of infectious diseases and have had a long-standing impact on global human health (44, 45). During vaccination, individuals are exposed to a foreign antigen, sometimes in the presence of an exogenous adjuvant, to activate the immune system. Immunization leads to development of immune memory: molecules and cells that are able to recognize and eliminate the corresponding pathogen before infection can be established (46). Protection is often mediated by humoral production of antigen-specific antibodies produced by B cells, though cell-mediated protection by T cells is also important for protection elicited by certain vaccines (47). Orchestration of vaccine-specific humoral or cell-mediated immunity requires finely-tuned interactions between antigen-presenting cells (APCs), B cells, and T cells (48–51). However, these cellular interactions may be predisposed to particular responses, which subsequently influence vaccine outcome, depending on the underlying immune state. This “immune tone” comprises the cytokine milieu, basal expression of surface proteins that mediate cell-cell interactions, and antigen presentation by APCs, all of which can be influenced by the microbiome (4, 52–54).
In this review, we summarize key findings in the literature that demonstrate the effect of the microbiome on outcomes of immune interventions, with a focus on ICI treatment and vaccination as the most studied examples. We discuss evidence that immunotherapies can influence the microbiota and that age plays a role in the effects of the microbiota on the immune system, as well as discuss the effects of live biotherapeutic products and prebiotics. We highlight studies that identify cellular and molecular mechanisms by which the microbiome modulates the immune system during immune interventions. Taken together, these studies reveal common microbial and immune elements across both ICI treatment and vaccination that have the potential to shape immune responses across diverse therapeutic spheres.
Linking the Microbiome and Response to Immune Checkpoint Inhibitor Treatment
Immune checkpoint inhibitor (ICI) therapy can improve long-term outcomes in a number of different cancer types, such as melanoma, non-small cell lung cancer (NSCLC), and urothelial cancer. However, the majority of patients experience cancer recurrence or do not respond to treatment (55). A number of factors that are partially predictive of ICI responsiveness have been identified (reviewed in 56, 57). Recently, the gut microbiome, which is known to play a role in the development and function of the immune system, has also been suggested as a determinant of ICI efficacy (58–63). As a result, these studies have sparked interest in the gut microbiome as both a diagnostic and therapeutic target in the context of cancer immunotherapy. In the following sections, we review the body of clinical and preclinical studies that demonstrate a role for the gut microbiome in ICI responsiveness. We highlight potential microbial, molecular, and immune mechanisms by which the microbiome may influence response to ICI treatment, and discuss whether ICI treatment reciprocally modulates the microbiome.
Clinical Findings
Given the role of the gut microbiota in the development and function of the immune system, it is unsurprising that the microbiome may influence ICI outcome measurements in the clinic. Several studies demonstrate that improved patient responses to ICI are associated with a “healthy” gut microbiome, as measured by higher diversity (61, 64). Similarly, antibiotic treatment either shortly before or during ICI therapy has been shown to influence outcomes (65–69). Conversely, other studies have found little to no impact of antibiotics on treatment outcome (70). It is likely that these discrepancies are due in large part to the wide range of antibiotic classes, cancer types, and treatment regimens captured by these clinical studies. Thus, additional work is required to validate these observations and better understand the impact of antibiotics and microbiome diversity on ICI efficacy.
A number of recent studies have identified unique microbial signatures that are associated with ICI treatment outcomes ( Table 1 ). In particular, metastatic melanoma patients that respond to ICI therapy tend to have high abundances of Faecalibacterium prausnitzii (59–61, 64), which comprises a large proportion of the human gut microbiota and is known to influence immune function via production of short-chain fatty acids (SCFAs) (72–74). In addition to F. prausnitzii, other bacteria have been associated with ICI responses in melanoma patients, including Gemmiger formicilis (63), Dorea formicigenerans (60), and Ruminococcus bromii (61), all within the order Clostridia, as well as Bacteroides thetaiotaomicron (60), Holdemania filiformis (60), and the Actinobacteria Bifidobacterium longum and Collinsella aerofaciens (62), which have been shown to promote CD4+ T helper cell 1 (Th1) polarization (75). In contrast to these studies in melanoma patients, a study in a cohort of patients with non-small cell lung carcinoma, renal cell carcinoma, and urothelial carcinoma found that ICI treatment outcome was influenced by abundance of Akkermansia muciniphilia and Enterococcus hirae, suggesting that beneficial microbial signatures could be cancer specific (58). It was recently demonstrated that E. hirae harbors a bacteriophage encoding a MHC class I-binding protein that induces a CD8+ T cell response and cross-reacts with cancer antigens (76), which could explain the influence of E. hirae on ICI treatment. The presence or absence of key microbial taxa may enable the stratification of patient populations and predict potential ICI treatment outcomes based on microbiome composition. Additionally, these findings suggest that the identification of key bacterial species may facilitate development of adjunct therapies to improve ICI outcomes, such as fecal matter transplants (FMT) or probiotics (77–81). Together, these approaches may accelerate development of treatment regimens to improve ICI therapeutic outcomes.
Table 1.
Gut microbial composition is associated with the efficacy of immune checkpoint inhibitor therapy in patients.
| Cancer Type (number of patients) | Immune checkpoint inhibitor | Identified factor | Key associations | Reference |
|---|---|---|---|---|
| Metastatic non-small-cell lung carcinoma (74) | anti-PD-1 | Antibiotic prescription (within 3 months prior) | No association with progression-free survival | 70 |
| Metastatic renal cell (121) and non-small-cell lung (239) carcinomas | anti-PD-L1 | Antibiotic usage (within 30 days prior) | Reduced ICI response a | 65 |
| Metastatic renal cell (67), non-small-cell lung (140), and urothelial carcinoma (42) | anti-PD-1 or anti-PD-L1 anti-PD-1 or anti-PD-L1 |
Bacteria: Akkermansia muciniphilia | Enhanced ICI response a | 58 |
| Metastatic melanoma (26) | anti-CTLA-4 | Bacteria: Faecalibacterium spp. and Firmicutes | Longer progression-free survival | 59 |
| Metastatic melanoma (39) | anti-CTLA-4 | Bacteria: Faecalibacterium prausnitzii, Bacteroides thetaioamicron, Holdemania filiformis | Enhanced ICI response a | 60 |
| anti-PD-1 | Bacteria: Dorea formicigenerans | Enhanced ICI response a | ||
| anti-CTLA-4 or anti-PD-1 | Xenobiotic: anacardic acid | Enhanced ICI response a | ||
| Metastatic melanoma (43) | anti-PD-1 | Bacteria: Faecalibacterium prausnitzii; Ruminococcus bromii; microbial diversity baseline | Enhanced ICI response a | 61 |
| Metastatic melanoma (42) | anti-PD-1; anti-CTLA-4 | Bacteria: Bifidobacterium longum; Collinsella aerofaciens; Enterococcus faecium | Enhanced ICI response a | 62 |
| Metastatic melanoma (27) | anti-PD-1; anti-CTLA-4 | Bacteria: Microbial community richness | Longer progression-free survival | 64 |
| Metastatic non-small-cell lung carcinoma (142) | anti-PD-1; anti-PD-L1 | Antibiotic treatment (concomitant) | Shorter progression-free survival and overall survival | 67 |
| Metastatic renal cell carcinoma (69) | anti-PD-1 | Antibiotic usage (within 2 months prior) | Shorter progression-free survival | 66 |
| Metastatic melanoma (568) | anti-PD-1; anti-CTLA-4 | Antibiotic usage (within 3 months prior) | Shorter overall survival | 68 |
| Metastatic non-small-cell lung carcinoma (2208) | anti-PD-1; anti-CTLA-4 | Antibiotic usage (within 3 months prior or concomitant) | Shorter median overall survival | 69 |
| Metastatic melanoma (50) | anti-CTLA-4 | Bacteria: Faecalibacterium spp.; Gemminger spp | Longer progression-free survival | 63 |
| Metabolite: fecal SCFA butyrate | Shorter progression-free survival | |||
| Metastatic non-small cell lung carcinoma | anti-PD1; anti-PD-1 | Bacterial delivery: Clostridium butyricus | Longer progression-free survival | 71 |
Response determined by biomarker-based disease progression criteria.
Pre-Clinical Findings
The clinical observation that the microbiome can influence ICI treatment response is generally recapitulated in pre-clinical studies of microbiome disruption or supplementation with fecal material, individual microbes, or microbial metabolites ( Table 2 ). Mice with different microbial communities, obtained from different vendors, have been shown to respond differently to ICI treatment (82). Microbiota depletion via administration of a cocktail of broad-spectrum of antibiotics containing ampicillin, streptomycin, and colistin reduces the efficacy of anti-PD-1 and anti-CTLA-4 mAbs, both alone and in combination (58, 83, 85). Furthermore, this effect was observed in multiple tumor types and corresponding models including melanoma (RET), renal cell carcinoma (RENCA), colon cancer (CT26, MC38), and fibrosarcoma (MCA-205), suggesting that the microbiota may contribute to treatment responsiveness in a broad array of cancers (58, 83, 85).
Table 2.
Impact of gut microbiome on immune checkpoint inhibitor therapy: selected preclinical studies.
| Tumor Cell Model (Cancer Type) a | Treatment | Immune checkpoint inhibitor | Key findings | Reference |
|---|---|---|---|---|
| B16.SIY (M) | Comparison of mice from different vendors and different microbial communities | anti-PD-L1 | Differential tumor growth in mice from different vendors; Bifidobacterium promotes antitumor immunity and anti-PD-L1 efficacy | 82 |
| MCA-205 (FS) RET (M) MC38 (CRC) |
Antibiotic cocktail: ampicillin, streptomycin, & colistin | anti-CTLA-4 | Bacteroides fragilis promotes anti-CTLA-4 efficacy via TL2/TLR4 host signaling | 83 |
| RET (M) RENCA (RCC) MCA-205 (FS) |
Antibiotic cocktail: ampicillin, streptomycin, & colistin | anti-PD-1 ± anti-CTLA-4 | Antibiotic exposure decreased ICI efficacy; oral supplementation with Akkermansia muciniphila restored the efficacy of ICI | 58 |
| MC38 (CRC) Braf
V600E
Pten
−/−
(mouse-derived melanoma) |
Colonization with a consortium of 11 fecal strains | anti-PD-1 ± anti-CTLA-4 | Colonization with an 11-strain consortium induces IFN-γ producing CD8+ T cells and increases ICI efficacy | 84 |
| CT26 (CRC) | Antibiotic: ampicillin, streptomycin, & colistin (cocktail); vancomycin; colistin | anti-PD-1 | Antibiotics decreased efficacy of anti-PD-1 therapy and altered glycerophosphlipid metabolism | 85 |
| MC38 (CRC) CT26 (CRC) |
Prebiotic supplementation: butyrate | anti-CTLA-4 | SCFA butyrate supplementation reduces efficacy of anti-CTLA-4 | 63 |
| MC38 (CRC) MB49 (RCC) B16-F10 (M) Genetic CRC (Msh2) |
Colonization with Bifidobacterium pseudolongum; delivery of inosine and microbial ligands | anti-CTLA-4 | Microbial-derived inosine activates anti-tumor T cell via the adenosine A2A receptor in combination with T cell costimulation by MAMPs | 86 |
M, melanoma; FS, fibrosarcoma; CRC, colorectal carcinoma; RCC, renal cell carcinoma.
Fecal microbiome transplant (FMT) from patients into mice has revealed differential effects of microbiota from ICI responders and non-responders, demonstrating microbiome-mediated immunomodulation and impact on ICI outcome. For example, germ-free (GF) or antibiotic-treated mice receiving responder FMT accumulated Th1-polarized cells in the tumor microenvironment following ICI treatment (58). Another study found that GF mice receiving responder FMT had higher frequencies of CTLs, while those receiving non-responder FMT had higher frequencies of immunosuppressive Tregs and Th17 cells (61). Similarly, a third study found that that mice receiving responder FMT displayed elevated levels of tumor-specific CTLs, but not Tregs (62). Together, these studies suggest that the microbiome from patient ICI-responders elicits anti-tumor immunity upon FMT, which in turn promote ICI response. FMT, as a critical research tool, not only demonstrated the direct modulatory effect of gut microbiome, but also opens the door to identify key microbes and/or their metabolites that stimulates the immune response.
Individual microbes have also been identified that promote ICI response by inducing Th1 polarization. Oral supplementation with Bacteroides fragilis, B. thetaiotaomicron, or Burkholderia cepacia restored ICI efficacy in antibiotic treated mice (83). In particular, B. fragilis potentiated ICI treatment by stimulating a Th1 response within the tumor-draining lymph nodes (83). This effect was mediated by stimulation of TLR2 and TLR4, which recognize microbe-associated molecular patterns (MAMPs), leading to the maturation of intratumoral dendritic cells (DCs). In a separate study, oral delivery of A. muciniphila and E. hirae to antibiotic-treated mice increased the incidence of central memory CD4+ T cells within the tumor bed, mesenteric lymph nodes, and draining lymph nodes and also induced production of IL-12 by DCs, a Th1 cytokine that plays a role in ICI response (58). Another study using mice with low baseline ICI response due to a unique microbial community found that administration of Bifidobacterium restored ICI response by stimulating antigen presentation by DCs and CTL activation (82). Additionally, a recent study found that Bifidobacterium pseudolongum was enriched in colon tumors of mice that responded to ICI treatment, and that colonization of germ-free mice with B. pseudolongum promoted ICI response (86). The authors found that this effect was mediated by stimulation of the adenosine A2A receptor on T cells via microbially-produced inosine. Oral delivery of inosine promoted clearance of colon, bladder, and melanoma tumor models in combination with anti-CTLA-4 and CpG treatment.
In addition to inosine, other microbial metabolites have also been shown to mediate the effect of microbes on ICI outcomes. In one study, delivery of a consortium of 11 bacterial species isolated from healthy human fecal material improved ICI response and induced a robust expansion of IFN-γ producing CTLs (84). However, administration of the heat-killed consortium failed to recapitulate the effects, suggesting that active colonization is required. The authors found that mice receiving the live consortium displayed increased levels of mevalonate and dimethylglycine in both cecal contents and sera, which may increase CTL activation or expansion (87, 88). Interestingly, the effect of the consortium was independent of a number of key innate signaling pathways, but loss of CD103+ DCs or MHC class Ia was sufficient to abrogate expansion of CTLs, suggesting that the consortium may interact with tissue-resident DCs in an MHC class Ia-dependent manner to promote ICI response (84).
Elevated levels of microbiota-derived short-chain fatty acids, which are known to induce anti-inflammatory Tregs (40), have also been associated with reduced survival in ICI treatment (63). DCs isolated from butyrate-supplemented mice receiving ICI therapy displayed reduced surface expression of costimulatory molecules on APCs, suggesting that SCFAs may interfere with ICI therapy by inhibiting DC maturation within the tumor-draining lymph nodes (63). Another study found that antibiotics reduced the efficacy of ICI therapy in mice and decreased plasma levels of microbiota-derived metabolites known to be involved in glycerophospholipid metabolism and glycosylphosphatidylinositol (GPI)-anchor biosynthesis (85). The authors also observed reduced levels of the inflammatory cytokines IL-2 and IFN-γ within the tumor microenvironment, leading to the hypothesis that impaired glycerophospholipid metabolism by the microbiota could dampen anticancer immune responses by muting production of proinflammatory cytokines.
Thus far, multiple studies have implicated the microbiome in modulating response to ICI treatment. Clinically, several studies have suggested that the overall diversity of the microbiome is a key determinant of ICI responsiveness (61, 89). However, other studies have identified unique microbial signatures of responders and non-responders, suggesting that the presence or absence of key taxa may be more predictive of response (58–61, 64, 66, 83). Several bacteria identified in ICI responders have been shown to promote ICI efficacy in preclinical studies, indicating that supplementation with these strains could improve outcomes for patients undergoing ICI treatment. Recent molecular and cellular investigations have implicated several pathways by which the microbiota influence ICI treatment outcome, including SCFAs (63), inflammatory cytokines (58, 85), antigen presentation cell function (63, 82–84), and T cell polarization (58, 61, 83).
In addition to the established role of the gut microbiome in regulating immune function, there is also a growing body of evidence that bacteria present within the tumor microenvironment (collectively termed the tumor microbiome) may also impact anti-cancer responses. For example, F. nucleatum is known to form biofilm-like structures within tumor spheroids in vitro and has been shown to directly inhibit NK cells via engaging TIGIT (90–92). Furthermore, a recent analysis of human tumors found that different tumor types displayed unique microbial signatures and that responders to ICI had tumors containing elevated abundances of Clostridia (93). However, it is unclear what role the tumor microbiome plays as a determinant of ICI efficacy. Further investigations of the roles of these microbial communities and their impact on immune function may facilitate target-specific therapeutic approaches to promote ICI response in patients.
Influence of Immune Checkpoint Inhibitors on the Microbiome
Immunomodulatory agents have been shown to change microbial composition, likely by modulating immune-mediated control of the microbiota (27, 94–98). Thus, an open question is whether cancer ICIs influence the microbial community. Emerging evidence suggests that ICIs targeting different immune checkpoint proteins may differentially impact the microbiome depending on their propensity to cause gastrointestinal adverse events such as diarrhea or colitis. However, the immune-microbe interactions that mediate these distinct effects are largely unknown.
Anti-PD-1/PD-L1 drugs appear to have minimal impact on the gut microbiome, in line with a lower incidence of gastrointestinal adverse events compared to other immunotherapy drugs (99). A recent study examining the gut microbiota of melanoma patients receiving anti-PD-1 therapy found no significant differences in microbial diversity or composition (61). Similarly, anti-PD-1 therapy for either renal cell carcinoma or non-small cell lung cancer had no impact on microbiome diversity or gene content after one month treatment, despite of elevation after two months (58). In contrast, anti-CTLA-4 treatment, which is known to be associated with elevated incidence of gastrointestinal adverse events, has been found to increase the abundance of Bacteroides genus in mice and a cohort of metastatic melanoma patients (83). Another study also found that anti-CTLA-4 treatment was associated with a reduction of multiple Firmicute species and an enrichment of Bacteroides, but only in patients were experiencing acute immune-related colitis (59). Lastly, combinatorial therapy with anti-PD-1 and anti-CTLA-4 has been shown to induce limited changes in the gut microbiome (60). However, the changes observed (an increase in Bacteroides stercoris and a reduction in Clostridium boltae) are largely consistent with those seen in other studies examining anti-CTLA-4 alone (59, 60, 83).
Based on these findings, it appears that major changes to the microbial community after ICI treatment are likely the secondary result of immune-related colitis, and it is unlikely that ICIs have a direct impact on microbiome composition. As a result, regimens targeting CTLA-4 are more likely to induce microbiome shifts than those targeting PD-1 due to their greater incidence of gastrointestinal adverse events. However, there are a number of limitations to the existing studies that make it difficult to draw definitive conclusions. Several of the studies have very small sample sizes (n <10), making it challenging to detect potentially small changes in microbial composition. Additionally, these studies differ considerably in methodology of microbiome sequencing and analysis, which limits our capacity to compare microbial signatures across cohorts. Further complicating comparisons between studies are considerable differences between treatment regimens, ICI dosages, and cancer types. Therefore, these limitations highlight a need for additional well-controlled and thorough studies to understand the impact of ICIs on the gut microbiome and whether ICI-induced microbiome changes subsequently influence ICI treatment outcomes.
Microbial Manipulation During ICI Treatment
Clinical and preclinical efforts suggest a link between the state of the microbiome and ICI treatment outcomes. As described above, preclinical studies in mice have found that fecal microbiota transplant, delivery of single or consortia of microbes, or supplementation with microbial molecules can modulate ICI treatment outcomes. Whether microbiome manipulation improves ICI treatment outcomes in clinical patients is a crucial question for ongoing and future studies. In one study, fecal matter transplants (FMT) from donors that have achieved a complete response with anti-PD-1 therapy were shown to improve responsiveness to ICI in some metastatic melanoma patients (100). Additionally, FMT recipients were found to have increased expression of genes involved in the presentation of peptides on MHC-I molecules in APCs along with elevated IL-1-mediated signaling, suggesting a mechanistic link between FMT and improved ICI responsiveness in a clinical setting (100). Another recent study found that supplementation with the probiotic Clostridium butyricum prior to or during ICI therapy improved patient outcomes (71). Although C. butyricum was selectively given to patients based on symptoms of gastrointestinal upset, the effect was observed in patients with or without prior antibiotic treatment. This result suggests that promoting a normobiotic microbiome could not only relieve gastrointestinal side effects of ICI treatment as has been described (101), but may also feedback to enhance ICI treatment outcomes (102).
Linking the Microbiome and Response to Vaccines
Vaccination confers protection against pathogens. However, the response to vaccination varies widely across individuals, which could greatly compromise individual and community protection (reviewed in 103, 104). Several factors that contribute to vaccine non-responsiveness in humans have been identified, including genetics (105, 106), advanced age (reviewed in 107–109), smoking (110), and comorbidities such as infection (111–113), obesity (114), malnutrition (115), kidney disease (116), and autoimmune disorders or allergy (117, 118).
Recently, clinical and preclinical studies have suggested microbial modulation of the immune system is directly responsible for the effects of the microbiome on vaccine response (summarized in Table 3 ). It is also possible the microbiome is the mediator of other factors associated with vaccine nonresponse, for example obesity or celiac disease, which are known to induce changes in the microbiome (147, 148). Thus, manipulation of the microbiota or direct targeting of microbially-regulated immune pathways could represent attractive strategies for promoting vaccine response in the broad healthy population or specific sub-populations with characteristically poor response (4, 147, 149–153).
Table 3.
Summary of preclinical studies linking microbiome and vaccine outcome.
| Vaccine (routea) | Model/Treatment | Key Findings | Reference |
|---|---|---|---|
| Studies in Germ-Free Models | |||
| Bovine gamma-globulin (SC) | Germ-free mice | Reduced serum IgG antibody response | 119 |
| E coli O antigen (PO) | Germ-free pigs | Reduced IgA-positive cells in lamina propria | 120 |
| Sheep red blood cells (IP); Bovine serum albumin (IP) |
Germ-free mice | Reduced serum IgG antibody response | 121 |
| Heat-inactivated E. coli (PO) | Germ-free chickens | Reduced intestinal and serum antibody (IgG, IgA) production | 122 |
| Sheep red blood cells (SC) | Germ-free mice | Reduced delayed-type hypersensitivity response; microbiota restoration restored response | 123 |
| Live attenuated Bacille Calmette–Guerin (IV) | Germ-free mice | Enhanced resistance to Mycobacterium tuberculosis challenge after immunization | 124 |
| Ovalbumin + complete Freund’s adjuvant (SC) | Germ-free mice | Reduced ova-specific antibody response | 125 |
| Trivalent inactivated influenza (SC) | Germ-free mice | Reduced antigen-specific serum IgG | 126 |
| Attenuated human rotavirus (PO) | Germ-free mice | Enhanced antigen-specific antibody response | 127 |
| Human serum albumin + cholera toxin (PO or IN) | Germ-free mice | Reduced ova-specific plasma IgG | 128 |
| Human serum albumin + cholera toxin (PO) | Germ-free mice | Reduced ova-specific plasma IgG | 129 |
| Studies of Antibiotic Treatment | |||
| Tetanus toxoid + alum (SC) Pneumococcal polysaccharides (SC) Hepatitis B surface antigen + alum (IP) Live-attenuated S. typhi Ty21A (IP) |
Antibiotics in mice: Clarithromycin or doxycycline (4 weeks) | Reduced vaccine-specific serum IgM antibody levels | 130 |
| Ovalbumin + complete Freund’s adjuvant (SC) | Antibiotic cocktail in mice: clindamycin, ampicillin, & streptomycin (maternal 5 days) | Reduced ova-specific antibody response in pups from antibiotic-treated dams | 125 |
| Live attenuated human rotavirus (PO) | Antibiotic cocktail in mice: Ampicillin & Neomycin (2 weeks) | Enhanced antigen-specific antibody response | 127 |
| Trivalent inactivated influenza (SC)b | Antibiotics in mice (4 weeks): cocktail of neomycin, ampicillin, Vancomycin, & metronidazole; vancomycin; polymixin B | Reduced antigen-specific serum IgG | 126 |
| Tetanus toxoid + diphtheria toxoid + acellular pertussis antigens + alum (SC) HIV-gp140 + alum (SC) Live attenuated yellow fever YF-17D (SC) |
Antibiotics in mice (4 weeks): cocktail of neomycin, ampicillin, Vancomycin, & metronidazole | No effect on antigen-specific IgG | 126 |
| Human serum albumin + cholera toxin (PO; IN) | Antibiotic cocktail in mice: ampicillin, vancomycin, metronidazole, neomycin (3-4 weeks) | Reduced ova-specific plasma IgG | 128 |
| Live attenuated Bacille Calmette-Guerin (SC) Pneumococcal polysaccharide-diphtheria toxoid conjugate + alum (IP) Meningococcal B surface proteins + outer membrane vesicles (IP) Meningococcal C polysaccharide-tetanus toxoid conjugate + alum (IP) Diphtheria toxoid + tetanus toxoid + pertussis toxoid + pertussis proteins + hepatitis B surface antigen + inactivated polioviruses + Haemophilus influenzae type b-polysaccharide + alum (IP) Trivalent inactivated influenza (SC)b |
Antibiotic cocktail in mice: Ampicillin & neomycin (maternal 2-5 weeks) | Reduced vaccine-specific IgG titer | 131 |
| Live attenuated Bacille Calmette-Guerin (SC) Pneumococcal polysaccharide-diphtheria toxoid conjugate + alum (IP) |
Antibiotic cocktail in mice: ampicillin & neomycin (3 weeks) | No effect on vaccine-specific IgG titer | 131 |
| Ovalbumin + cholera toxin (PO) | Antibiotic cocktail in mice: Metronidazole, vancomycin, ampicillin, kanamycin (10 days) | Reduced ova-specific fecal IgA and serum IgG | 132 |
| Rabies vaccine iLBNSE (IM) | Antibiotic cocktail in mice: metronidazole, vancomycin, ampicillin, neomycin (4 weeks) | Reduced rabies-specific IgG, IgM, neutralizing antibodies; reduced Tfh cells, germinal center B cells, memory response | 133 |
| Studies of Prebiotic Supplementation | |||
| Trivalent inactivated influenza (SC) | Prebiotic cocktail in mice: galacto- and fructo-oligosaccharides | Enhanced delayed-type hypersensitivity response; increased levels of Bifidobacteria and Lactobacilli | 134 |
| Trivalent inactivated influenza (SC) | Prebiotic cocktail in mice: fructo-oligosaccharides and inulin | No effect on delayed-type hypersensitivity response; increased levels of Bifidobacteria and Lactobacilli | 134 |
| Live attenuated Salmonella typhimurium (oral) | Prebiotic cocktail in mice: fructo-oligosaccharides and inulin | Enhanced antigen-specific antibody titer, inflammatory cytokines, and survival after pathogen challenge | 135 |
| Trivalent inactivated influenza (SC) | Prebiotic cocktail in mice: Galacto- and fructo-oligosaccharides | Enhanced delayed-type hypersensitivity response; increased levels of Bifidobacteria and Lactobacilli | 136 |
| Ovalbumin + cholera toxin (oral) | Prebiotic cocktail: acetate and butyrate | Enhanced vaccine response and production of B-cell-activating factors in dendritic cells; effect was dependent on SCFA-receptor GPR43 | 132 |
| Trivalent inactivated influenza (SC) | Prebiotic cocktail in mice: Galacto- and fructo-oligosaccharides 2’FL | Enhanced antigen-specific antibody titer and IL-6 production in male mice, increased levels of Actinobacteria | 137 |
| Cholera toxin + ovalbumin (oral) | Prebiotic cocktail: spirulina, amaranth, flaxseed, micronutrients | Enhanced antigen-specific antibody titer and germinal B cell frequency in mesenteric lymph nodes; effect was dependent on presence of particular microbes | 138 |
| Studies of Microbial Delivery | |||
| Ovalbumin + complete Freund’s adjuvant (SC) | Conventionalization of germ-free mice | Enhanced ova-specific antibody response | 125 |
| Live attenuated human rotavirus (PO) | Probiotic in gnotobiotic neonatal pigs: Lactobacillus acidophilus | Modulated balance of antigen-specific Th1 cells and Tregs in a dose-specific manner | 139 |
| Live attenuated human rotavirus (PO) | Probiotic cocktail in gnotobiotic neonatal pigs: Lactobacillus rhamnosus GG and Bifidobacterium animalis lactis Bb12 |
Enhanced antigen-specific Th1 response and protection from rotavirus challenge | 140 |
| Live attenuated human rotavirus (PO) | Probiotic cocktail in gnotobiotic neonatal pigs: Lactobacillus rhamnosus GG and Bifidobacterium animalis lactis Bb12 |
Enhanced intestinal antigen-specific antibody titers, cell responses, and protection from rotavirus challenge | 141 |
| Live attenuated human rotavirus (PO) | Probiotic in neonatal gnotobiotic pigs pre-colonized with human fecal material: Lactobacillus rhamnosus GG | No effect on protection from rotavirus challenge; modulated production of antigen-specific Th1 cells in a dose- and colonization-dependent manner | 142 |
| Trivalent inactivated influenza (SC) | Single strain in germ-free mice: flagellated or aflagellated E. coli
Conventionalization of germ-free mice |
Enhanced antigen-specific antibody response after conventionalization or colonization with flagellated, but not aflagellated, bacteria MAMP flagellin functions as an endogenous adjuvant |
126 |
| Live attenuated Mycobacterium tuberculosis Ad85A (IM) | Pathobiont in mice: Helicobacter hepaticus colonization | Reduced protection from Mycobacterium tuberculosis
IL10 production reduces vaccine response |
143 |
| Live attenuated yellow fever virus YFV-17D (SC) | Pathobiont in mice: MHV86, MCMV, influenza WSN, & Heligmosomoides polygyrus | Reduced pre-immunization production of inflammatory cytokines correlates with reduced anti-YFV antibody response | 144 |
| Pneumococcal polysaccharide-diphtheria toxoid conjugate + alum (IP) | FMT in antibiotic-treated mice: fecal material from untreated mice | Enhanced vaccine-specific antibody titer | 131 |
| Keyhole limpet hemocyanin + alum (SC); NP conjugated to cholera toxin (PO) | Co-housing of aged mice with young mice FMT in aged mice: fecal material from young mice |
No effect on antibody responses; improved germinal center reactions independent of vaccination | 145 |
| Cholera toxin + ovalbumin (PO) | FMT in germ-free mice: fecal material from undernourished children Co-housing of mice that received different FMT Consortium of five fecal strains: Bacteroides acidifaciens, Bacteroides fragilis, Clostridioides difficile, Costridium innocuum, Fusobacterium mortiferum |
Specific microbes mediate the pro-vaccine effects of prebiotics | 138 |
| Studies in Genetically Engineered Host Models | |||
| Trivalent influenza (SC)b
Inactivated poliovirus (SC) |
TLR5−/− mice | Reduced antigen-specific serum IgG for nonadjuvanted vaccines TLR5 mediates endogenous adjuvant response to vaccine |
126 |
| Live attenuated yellow fever YF-17D (SC) Hepatitis B sAg + alum (SC) |
TLR5−/− mice | No effect on antigen-specific serum IgG | 126 |
| Human serum albumin + cholera toxin (PO; IN) | Nod2−/− mice Nod2-DC(CD11c) specific deletion |
Reduced ova-specific plasma IgG Nod2 in DCs mediates adjuvant activity of cholera toxin |
128 |
| Human serum albumin + cholera toxin (PO; IN) | Myd88−/−; Ripk2−/−; Nod1−/− | No effect on ova-specific plasma IgG | 128 |
| Lactobacillus acidophilus vaccine strain expressing HIV proteins (PO) | Nod2−/− mice | Reduced antigen-specific IgG and IgA Nod2 mediates response to vaccine strain |
146 |
| Trivalent influenza (SC)b | TLR5−/− mice | No effect on antigen-specific serum IgG | 131 |
| Human serum albumin + cholera toxin (PO) | Nod2−/− mice IL1b−/− mice |
Reduced ova-specific plasma IgG IL-1β production via Nod2 is required for cholera toxin adjuvanticity |
129 |
| Ovalbumin + cholera toxin (PO) | GPR43−/− mice | Reduced ova-specific fecal IgA and serum IgG; reduced pro-vaccine effect of SCFA prebiotics
GPR43 mediates pro-vaccine effect of SCFAs |
132 |
aSC, subcutaneous; IP, intraperitoneal; PO, oral; IV, intravenous; IN, intranasal; IM, intramuscular; bNoted discrepancies with regard to effect of antibiotics and TLR5−/− on response to influenza vaccine could be attributed to different underlying microbial communities across study location; Key mechanistic insights are noted in bold text.
In the following sections, we review key clinical and preclinical studies that link the microbiota and vaccine outcome, and we highlight putative immunomodulatory mechanisms by which the microbiome may influence vaccine responsiveness. We also discuss how the connections between the microbiome and vaccine response evolve over the course of a lifetime, and describe current approaches to harness the microbiota to promote vaccine response.
Clinical Findings
Within the last two decades, observational clinical studies have revealed associations between microbiome community composition and host vaccine responses (recently reviewed in 152, 153). One common observation is that decreased vaccine response occurs in individuals with a disrupted microbiota (1). For example, several studies suggest that a normobiotic infant gut microbiome, replete with Bifidobacterium, promotes vaccine response, whereas a more dysbiotic microbiome harboring excess Proteobacteria interferes with vaccination outcome (154–159). Other studies have correspondingly shown that healthy human fecal material transplanted into neonatal gnotobiotic pigs or mice promotes strong responses to vaccines, while transplantation of dysbiotic human samples (harvested from individuals with intestinal enteropathy) dampens immune responses after vaccination (138, 160). Given the important role of the microbiota to educate the immune system during early development (161), it is possible that the dysbiotic infant microbiome could lead to immune deficiencies and reduced vaccine response. However, reducing the frequency of bacterial enteropathogens with azithromycin treatment prior to vaccination did not improve poliovirus vaccine response (162), suggesting that microbial dysbiosis may interfere with vaccine response via complex community changes or long-lasting immune effects.
Specific immunomodulatory bacteria have also been associated with vaccine response. In infants, two parallel studies found that a poor response to rotavirus vaccine was associated with increased Bacteroides and Prevotella spp., whereas a strong response was associated with higher levels of Proteobacteria and Firmicutes, particularly Streptococcus bovis (163, 164). Two additional studies have found that responders tend to have higher levels of Proteobacteria, though these observations were not statistically significant (165, 166). Nonetheless, a clinical study in adults found that vancomycin treatment, which recreates a similar microbial community comprising decreased Bacteroides and increased Proteobacteria, temporarily increased antibody levels in response to rotavirus (167). The authors speculate that in responders, highly immunostimulatory ligands such as LPS, peptidoglycan, or flagellin could promote viral infection or act as endogenous adjuvants to promote vaccine response.
Other immunomodulatory bacteria have been identified in studies of vaccine response in adults. A study with oral typhoid vaccine found that the gut abundance of the Firmicutes Lachnospiraceae and Ruminococcaceae was associated with early cell-mediated response to antigen after vaccination (168). Presence of the Firmicutes Lactobacillus and Streptococcus, as well the Bacteroidetes Bacteroides and Prevotella, in the nasal microbiome were associated with positive response to a nasal influenza vaccine (169). Furthermore, a recent meta-study analysis associated Actinobacteria and Firmicutes with positive vaccine responses, whereas Proteobacteria and Bacteroidetes were associated with poor vaccine outcomes (170). Though differences in study design, vaccine strategy, patient age, as well as other confounding variables complicates the interpretation of cross-study microbial associations, published studies together point toward an association of Firmicutes with successful vaccine outcome.
Interventional clinical studies including extensive immune characterization may facilitate more mechanistic explorations of the role of the microbiome during vaccination. A recent study in humans found that disruption of the microbiota by antibiotic treatment decreased response to influenza vaccination, specifically in individuals with low pre-existing immunity (171). Characterization of the metagenome and immune tone after antibiotic treatment implicated microbial bile acid metabolism, inflammasome signaling, and the underlying inflammatory state as key players that are influenced by the microbiota in the context of vaccination. We anticipate that similar investigational clinical studies may reveal crucial microbial and immune mechanisms that interact to either increase or decrease vaccine response in humans.
Pre-Clinical Findings
Since the 1960s, germ-free mouse, chicken, and pig models have demonstrated that the microbiome influences vaccine response (119–124). More recently, microbiota disruption via antibiotic treatment in mice has corroborated these early observations (125–127, 130, 131, 133). Multiple antibiotics from different classes and with different breadths of activity have been shown to decrease vaccine response ( Table 3 ). This observation suggests that diverse species may play a role in vaccine response. It also appears that the microbiome influences response to multiple classes of vaccines, including live, inactivated, and molecular vaccines ( Table 3 ). Nearly all studies employing molecular or inactivated vaccines have found that microbiome disruption decreases response; in contrast, studies of live vaccines have found the microbiome can either promote, reduce, or have no effect on response (124, 126, 127, 130). These differences raise the possibility that complex interactions between the microbiome and the immune system may influence the ability of a live vaccine to colonize the host (colonization resistance), and thus have clear implications for considering the impact of the microbiome during development of live-attenuated (155, 168, 169) and viral- or bacterial-vectored vaccines (172, 173) and could also influence cellular transfection by mRNA-based vaccines.
Notably, several studies have identified specific microbial products or host factors that mediate the effects of the microbiome on vaccine response. Broadly expressed microbe-associated molecular patterns (MAMPs) may mediate key effects of the microbiota on vaccine response. Indeed, a recent study demonstrated that signaling by the pattern recognition receptor (PRR) TLR5, stimulated by the MAMP flagellin, is important for vaccine adjuvanticity (126). Depletion of the microbiota compromised parenteral vaccine response, and this defect could be reversed by colonization with flagellated bacteria but not by colonization with an aflagellated isogenic strain. Further, loss of TLR5 specifically in B cells compromised response to several unadjuvanted vaccines but did not affect response to adjuvanted vaccines. Since deletion of TLR5 did not alter baseline plasma cell phenotypes (126), the authors hypothesized that gut-derived flagellin spreads systemically and functions as an endogenous adjuvant at the site of immunization (174). However, another group was unable to reproduce the requirement for TLR5 for vaccine response (131), suggesting that differences in the baseline microbial community likely dictate the mechanisms by which the microbiome affects vaccine response. Nonetheless, the data suggest that particular microbial communities may function as an endogenous adjuvant, or that steady-state intestinal MAMP-PRR signaling can alter the systemic immunophenotype and alter response upon subsequent vaccination.
Indeed, altered homeostatic cytokine production in response to microbes has been implicated in vaccine response and protection from infection. One study found that induction of the immunosuppressive cytokine IL-10 by oral introduction of Helicobacter hepaticus (Hh) into immunocompetent mice disrupted response to intramuscular vaccination with live-attenuated Mycobacterium tuberculosis (143). The authors found that, compared to animals with a normal microbiota, Hh-colonized animals exhibited lower antigen-specific cell-mediated responses and higher infection when nasally challenged with M. tuberculosis. Infusion of IL-10-receptor-blocking antibodies restored vaccine response in Hh-colonized animals, demonstrating that the effect was mediated by increased IL-10 signaling. However, Hh colonization had no effect on IL-10 expression or Treg abundances in the lung but did increase IL-10 expression in the intestines, suggesting that intestinal IL-10 and other intermediate factors likely influence systemic immunity. Whether the effects of Hh on intestinal IL-10 production are direct as observed for Hh and T cells (175), or are due to Hh-mediated microbiome disruption (176) remains unknown. A second study also found that colonization of mice with viruses and a helminth disrupted response to vaccination with live-attenuated yellow fever virus, and observed that at the time of vaccination, colonized animals displayed altered expression of several inflammatory cytokines which could compromise vaccine response (144). Thus, microbially-induced changes in the steady-state cytokine milieu likely affect subsequent systemic immunity to vaccines.
Other studies suggest the microbiota are required to supply immune receptor ligands for the activity of mucosal adjuvants. Two independent groups have employed immunization with albumin and cholera toxin (CTx) to demonstrate that germ-free or antibiotic-treated mice have decreased antibody responses, which are rescued by supplementation with microbial molecules (128, 129, 132). Both groups demonstrated that CTx potentiates immune receptor signaling, which require the presence of microbial molecules for full signaling activation in response to CTx. One study found that microbial peptidoglycan signaled through NOD2 during vaccination with CTx, leading to increased IL-1β production and generation of T-follicular helper cells and plasma cells (128, 129). A second group found that microbe-derived short-chain fatty acids (SCFAs) synergized with CT to promote vaccine response, by driving DC-mediated production of B cell activators BAFF and retinoic acid (132). Notably, the authors also found that SCFAs increased vaccine response even in the absence of CTx through the same immune pathway, demonstrating that SCFAs can independently promote pro-vaccine immune tone, in addition to mediating the adjuvant effect of CTx. It is important to note that the two groups employed different antibiotic cocktails, which likely differentially affect the microbiome, which may account for the distinct mechanisms identified.
Taken together, these preclinical studies reveal several possible mechanisms by which the microbiota can promote immune response in the context of vaccine response: as an endogenous adjuvant, as ligands for immune receptors that are potentiated by exogenous adjuvants, and as regulators of systemic immune tone. Germ-free and antibiotic-treated mice represent crucial models to parse the mechanisms by which the microbiome affects vaccine response. Another relevant model for future studies is “dirty” mice, which receive microbes from pet store mice via cohousing or bedding transfer, and may prove a valuable system for understanding vaccine response (177). Given that there are multiple mechanisms by which the microbiome can affect vaccine response and that these effects can vary across different vaccines, additional preclinical studies are critically important to define the microbes and immune pathways involved.
The Microbiome Modulates Vaccine Response in Infants and in the Elderly
Though infants and the elderly are profoundly at risk for severe infection, these populations also consistently fail to respond to vaccines (reviewed in 178–180). Lack of vaccine response can be attributed to maternal antibody interference (reviewed in 181, 182), early life immune immaturity (183–185), and immunosenescence in the elderly (186–192). However, emerging evidence suggests that microbiota may also influence vaccine response at the extremes of life, likely by modulating immune development and senescence (153).
The microbiome plays a key role in the proper maturation of the immune system (reviewed in 161), which is necessary for optimal vaccine response. Alterations in the neonatal microbiome are associated with defects in vaccine response in clinical studies (154, 163, 164) and in preclinical studies (125, 131). Although the mechanisms influencing vaccine response in neonates have not been clearly defined, related studies suggest that early-life microbial stimulation could promote vaccine response indirectly by modulating immune development and function (193–195), or directly by regulating adjuvanticity during vaccination (126). The possibility also exists that microbial dysbiosis during early life could interfere with vaccine response later in life, due to defects in microbial-immune imprinting (reviewed in 196), as has been observed for other immune-related disorders (9, 14, 15, 197). Although this idea has not yet been examined in clinical studies of vaccine response, a study in mice found that early life microbial dysfunction did not affect later vaccine response, provided the microbiome was repaired prior to vaccination (131). Thus, further studies are warranted to explore the immediate and long-term ramifications of early-life microbial dysbiosis on vaccine responses and relevant immune pathways; such discoveries could reveal novel therapeutic strategies to improve vaccine response in neonates by harnessing the microbiota.
In the elderly, emerging evidence suggests that age-induced changes in the microbiome contribute to immunosenescence (145, 198–200) and could thereby mediate changes in vaccine response (201). Indeed, defective intestinal germinal center reactions in aged mice were rescued by transplantation with fecal material from young mice (145), which could suggest an improved capacity for vaccine response after fecal transplantation. In humans, immunophenotypic similarities have been observed between elderly subjects and adults with antibiotic-induced microbial dysbiosis, consistent with the idea that age-mediated effects and antibiotic-mediated effects on vaccine response are both effected by the microbiota (171, 192). Others have speculated that immunosenescence caused by changes in the microbiota could drive other pathological immune conditions such as asthma (202). Thus, in both elderly and neonatal individuals, the microbiome may mediate changes in vaccine response, and microbiome rehabilitation represents a promising approach to promote vaccine response in these populations.
Microbial Manipulation to Improve Vaccine Response
Given the role of the microbiome in vaccine response in clinical and preclinical studies, a key question is whether manipulating the microbiome can improve vaccine outcome, especially in populations at risk for poor vaccine response (182). Many clinical studies have evaluated vaccine response after dietary supplementation with probiotics, primarily species within Lactobacillus or Bifidobacterium. Recent reviews and meta-analyses of these studies highlight that there is significant heterogeneity in the ability of probiotic supplementation to increase antibody titers after vaccination (203–206). Effects are likely to be specific to particular bacterial strains, and may vary between different vaccines and adjuvants (149, 207). Several of these probiotic studies have also found correlates between probiotic supplementation and diverse measures of immune function, including serum levels of pro-inflammatory cytokines (208, 209), T cell responsiveness (210, 211), and innate immune cell activity (212, 213), although the mechanistic implications of these observations are unclear.
The effect of probiotics on vaccine response has also been evaluated in neonatal pigs, and recent studies have begun to elucidate the mechanisms by which Lactobacillus and Bifidobacterium strains promote vaccine response in this model. Early studies using gnotobiotic pigs revealed that colonization by probiotics prior to oral rotavirus immunization enhanced Th1 cellular immunity (139–141). However, prior colonization by human fecal material prevented subsequent Lactobacillus colonization and Lactobacillus-mediated effects on vaccine response (142), suggesting that an intact microbiota can obstruct the effects of probiotics and contribute to the heterogeneity of their effects across individuals.
Efforts are underway to use probiotic strains as vaccine vectors to capitalize on the vaccine-promoting and immunomodulatory effects of certain microbes, by engineering the expression of antigens to induce the desired immune responses including mucosal IgA (reviewed in 214, 215). In particular, Lactobacilli are known to activate the immune cell receptors NOD2, TLR2, TLR6, C-type lectin receptors, and the caspase-1 dependent inflammasome (reviewed in 216). Recently, a study demonstrated that activation of NOD2 was required for Th2 skewing and humoral immune responses to a Lactobacillus vaccine vector strain (146). Further work to elucidate the cell types and functional pathways modulated by Lactobacillus and other probiotics will contribute to understanding of key interactions between the microbiota and the immune system in the context of vaccines.
Several groups have also explored whether prebiotic supplementation improves vaccine outcome. Prebiotics are expected to promote a diverse microbiota and prevent expansion of dysbiosis-inducing microbes (149). Though animal studies suggest prebiotics can stabilize “normobiotic” microbes including Lactobacillus and Bifidobacterium and promote vaccine response (134–138), prebiotics in clinical studies have predominantly had no effect on vaccine outcome (217–222). Why prebiotics do not affect vaccine response remains unclear, as they have been shown to influence other immune-related conditions (reviewed in 149).
Taken together, these and previously discussed studies illustrate the potential of microbiome-modulating interventions to promote vaccine response, but suggest that effective realization of such strategies is challenging. We note that the majority of studies have sought to enhance the abundance of Lactobacilli or Bifidobacteria, which are known to modulate the immune system (216, 223). These families belong respectively to the Firmicutes and Actinobacteria, which have been associated with vaccine response in a meta-analysis of clinical studies (170). It is possible that vaccine response could also be augmented by probiotic supplementation with other families within the Firmicutes, though this possibility has not been widely investigated. An outstanding question for probiotic approaches is whether it is necessary for probiotic strains to colonize within the context of an intact microbiome in order to promote vaccine response. Finally, an alternative strategy may be to leverage the pathways modulated by microbiome, either by delivery of live engineered organisms (146) or of bioactive molecules (132).
Concluding Remarks: Interactions Between Microbiome and the Immune System have the Potential to Shape Response to Immune Interventions
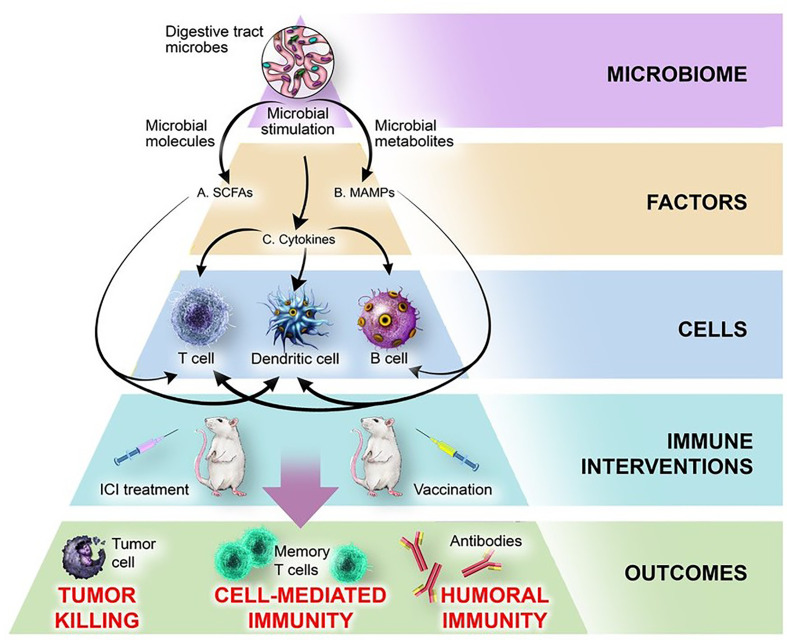
Recent literature reveals that assorted microbes, metabolites, and immune factors interact to influence the patient response during cancer immunotherapy and immunization (summarized in Tables 1 – 3 ). To gain greater perspective into the most meaningful molecular and cellular mechanisms by which the microbiome modulates the immune system, we have juxtaposed two therapeutic spheres: ICIs and vaccines. Evidence from both therapeutic spheres highlights three elements of the microbiome that consistently play an immunomodulatory role: microbially-derived metabolites including short-chain fatty acids, microbe-associated molecular patterns (MAMPs), and inflammatory cytokines ( Figure 2 ).
Figure 2.
Mechanisms by which the microbiome influences response to ICI treatment and vaccination. The microbiome produces microbial factors and influences host factors and cells, thereby influencing the outcomes of immune interventions. (A) Microbiome-derived short-chain fatty acids (SCFAs) bind receptors including GPR43 on DCs and T cells, leading to changes in cytokine production, antigen presentation, cellular polarization, and interactions with other cell types. (B) Microbe-associated molecular patterns (MAMPs) including flagellin, polysaccharide A, fucosylated antigens, unmethylated CpG DNA, and peptidoglycan bind pattern recognition receptors on DCs (NOD2, TLR2, TLR3, TLR9, DC-SIGN) or B cells (TLR5) and modulate activation, cytokine production, and immune cell function. (C) Microbiome-dependent changes in production of cytokines (IL-1β, IL-12, IL-18, IFN-γ, IL-10) produced by intestinal DCs or epithelial cells broadly affect immune cell function.
The microbiome is a rich source of diverse bioactive metabolites that can affect host biology and the immune system (reviewed in 150, 224, 225). One class of bacterial metabolites that influence response to both vaccination and ICI therapy are short-chain fatty acids (SCFAs). SCFAs are immunomodulatory metabolites and are produced by a subset of intestinal microbes (reviewed in 224, 226), some of which are implicated in clinical studies of immunotherapy and vaccine response. SCFAs such as butyrate and propionate appear to alter immunotherapy outcome by modulating CTL activation directly or indirectly by influencing DC maturation (37, 63, 227), whereas butyrate, acetate and some branched-chain fatty acids may alter vaccine response by modulating DC-mediated B cell activation (132, 228, 229). SCFAs are known to broadly influence the immune system as well as other host pathways, and have also been implicated in regulation of Tregs, allergic disease, resistance to infectious disease, obesity, intestinal barrier function, carcinogenesis, and microglia development in the central nervous system (40, 224, 226, 230–235). However, future investigations must continue to define the biology of SCFAs, as well as other immunomodulatory microbial metabolites associated with outcomes of immune-targeted interventions (84, 85), including whether immunotherapies themselves influence the gut metabolome. Determining the unique effects of specific metabolites, including SCFAs, on immune pathways critical for ICI and vaccine response may identify key microbes and signaling pathways that could be harnessed to promote therapeutic success.
The outcomes of ICI and immunization are also influenced by MAMPs, including flagellin, unmethylated CpG DNA, peptidoglycan, muramyl dipeptide, and polysaccharides (6, 83, 126, 129). MAMPs directly and indirectly modulate activation and differentiation of immune cells (236–238), and synthetic MAMPs are already used as exogenous adjuvants during immunization to activate the immune system. In the context of microbial modulation of immunotherapy and vaccination outcomes, the cellular and molecular mechanisms by which commensal MAMPs affect B and T cells are incompletely defined. Commensal MAMPs can bind to host receptors and directly influence B cells during vaccination (126), possibly by promoting antibody production and regulating class-switching (239, 240). Similarly, endogenous MAMPs may also promote T cell function as costimulatory molecules (241–245). B and T cell functions are also indirectly influenced by MAMP-mediated modulation of APC maturation, leading to altered interactions and production of cytokines and chemokines (5, 6, 83, 129, 246, 247). By understanding the role endogenous MAMPs play in outcomes of ICI and vaccination, we may be able to identify key MAMPs and host pathways that promote immunotherapy response, which could lead to development of novel vaccine adjuvants or ICI co-therapies.
Inflammatory cytokines are emerging as a key host mediator of microbial immunomodulation (54). Both ICI and vaccine outcomes have been associated with microbial modulation of inflammatory cytokines, including IL-12, IL-1β, and IFN-γ (6, 58, 83, 85, 129). These effects appear to be primarily mediated by MAMP stimulation of host receptors on APCs, but stimulation of epithelial cells with MAMPs or microbial metabolites can also alter epithelial cytokine production and impact immunity (143, 173, 248, 249). As the core communication toolbox of the immune system, cytokines represent a likely mediator through which the microbiome and intestinal immunity could influence systemic responses during ICI treatment and vaccination (58, 143, 247). Thus, microbiome-derived products could represent an indirect but powerful approach to control cytokine levels and broadly affect the immune system in diverse therapeutic contexts.
The effect of the microbiome on the outcome of immunotherapies certainly extends beyond ICI therapy and vaccination, as has been observed for non-ICI immunostimulatory treatments to control tumor growth (25, 26), anti-inflammatory treatment for irritable bowel syndrome and rheumatoid arthritis (27–30), and antigen desensitization for allergy (31). It is also clear that the microbiome broadly affects the immune system, as has been well-documented in studies of infectious disease and immune function (5, 6, 230, 232, 235, 246, 247). Taken as a whole, the current literature argues that the microbiome likely plays a key role in the heterogeneity of the immune response across individuals. Though the majority of mechanistic studies to date have employed preclinical models, technological developments for ex vivo microbiome studies (58, 61, 138, 160), and microbiome manipulation of human subjects in the clinical setting (162, 167, 171) will facilitate our understanding of the role of the microbiota on the human immune system. As crucial immunomodulatory interactions continue to be identified, novel strategies also need to be developed to manipulate the microbiota and microbial regulation of immunity. Patient microbial profiles could be surveyed to anticipate therapeutic outcomes. Probiotics and prebiotics may play a role in repairing or supporting the microbiota (84, 138). Supplementation with immunomodulatory microbial metabolites or ligands (132), or direct targeting of microbially-regulated immune pathways (6, 247) could also bypass the effects of the microbiota on cytokine production and cellular function. As we develop novel approaches to understand and influence the microbiome, we will also expand our ability to harness the immune system to treat or prevent disease.
Author Contributions
ALR, DJC, XL, JEN, and GS designed the work, collected and interpreted data, and approved the final work. ALR, DJC, and XL drafted the manuscript. GS, AGT, and DJH critically reviewed and approved the final work. All authors contributed to the article and approved the submitted version.
Conflict of Interest
All authors are employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and may hold stock in Merck & Co., Inc. Kenilworth, NJ, USA.
Acknowledgments
The authors wish to acknowledge Laura Maaske for her help with the illustrations. We also thank Michael Citron for his thoughtful comments on the manuscript.
References
- 1. Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature (2012) 486(7402):207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hollister EB, Gao C, Versalovic J. Compositional and Functional Features of the Gastrointestinal Microbiome and Their Effects on Human Health. Gastroenterology (2014) 146(6):1449–58. 10.1053/j.gastro.2014.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, et al. The Intestinal Metabolome: An Intersection Between Microbiota and Host. Gastroenterology (2014) 146(6):1470–6. 10.1053/j.gastro.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng D, Liwinski T, Elinav E. Interaction Between Microbiota and Immunity in Health and Disease. Cell Res (2020) 30(6):492–506. 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramakrishna C, Kujawski M, Chu H, Li L, Mazmanian SK, Cantin EM. Bacteroides Fragilis Polysaccharide A Induces IL-10 Secreting B and T Cells That Prevent Viral Encephalitis. Nat Commun (2019) 10(1):2153. 10.1038/s41467-019-09884-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota Regulates Immune Defense Against Respiratory Tract Influenza A Virus Infection. Proc Natl Acad Sci USA (2011) 108(13):5354–9. 10.1073/pnas.1019378108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseno CG, et al. Oral Antibiotic Treatment of Mice Exacerbates the Disease Severity of Multiple Flavivirus Infections. Cell Rep (2018) 22(13):3440–53.e6. 10.1016/j.celrep.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible Microbial Colonization of Germ-Free Mice Reveals the Dynamics of IgA Immune Responses. Science (2010) 328(5986):1705–9. 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal Microbial Diversity During Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe (2013) 14(5):559–70. 10.1016/j.chom.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seo SU, Kamada N, Munoz-Planillo R, Kim YG, Kim D, Koizumi Y, et al. Distinct Commensals Induce Interleukin-1beta Via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity (2015) 42(4):744–55. 10.1016/j.immuni.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mazmanian SK, Round JL, Kasper DL. A Microbial Symbiosis Factor Prevents Intestinal Inflammatory Disease. Nature (2008) 453(7195):620–5. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- 12. Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, et al. Anti-Commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity (2019) 50(4):1099–114.e10. 10.1016/j.immuni.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity (2016) 44(4):875–88. 10.1016/j.immuni.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome Lps Immunogenicity Contributes to Autoimmunity in Humans. Cell (2016) 165(6):1551. 10.1016/j.cell.2016.05.056 [DOI] [PubMed] [Google Scholar]
- 15. Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced Diversity in the Early Fecal Microbiota of Infants With Atopic Eczema. J Allergy Clin Immunol (2008) 121(1):129–34. 10.1016/j.jaci.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 16. Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The Maternal Microbiota Drives Early Postnatal Innate Immune Development. Science (2016) 351(6279):1296–302. 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- 17. Wu K, Yuan Y, Yu H, Dai X, Wang S, Sun Z, et al. Gut Microbial Metabolite Trimethylamine N-oxide Aggravates GVHD by Inducing M1 Macrophage Polarization in Mice. Blood (2020) 136(4):501–15. 10.1182/blood.2019003990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity (2019) 50(2):432–45.e7. 10.1016/j.immuni.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez-Lopez M, Iborra S, Conde-Garrosa R, Mastrangelo A, Danne C, Mann ER, et al. Microbiota Sensing by Mincle-Syk Axis in Dendritic Cells Regulates Interleukin-17 and -22 Production and Promotes Intestinal Barrier Integrity. Immunity (2019) 50(2):446–61.e9. 10.1016/j.immuni.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of Alpha Beta T-cell Receptor-Bearing Intestinal Intraepithelial Lymphocytes After Microbial Colonization in Germ-Free Mice and its Independence From Thymus. Immunology (1993) 79(1):32–7. [PMC free article] [PubMed] [Google Scholar]
- 21. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific Microbiota Direct the Differentiation of IL-17-producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe (2008) 4(4):337–49. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell (2005) 122(1):107–18. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 23. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated Cd8(+) T Cells. Immunity (2019) 51(2):285–97.e5. 10.1016/j.immuni.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 24. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial Bile Acid Metabolites Modulate Gut RORgamma(+) Regulatory T Cell Homeostasis. Nature (2020) 577(7790):410–5. 10.1038/s41586-019-1865-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science (2013) 342(6161):967–70. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science (2013) 342(6161):971–6. 10.1126/science.1240537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradley SM, Neumann VC, Barr K, Troughton PR, Astbury C, Bird HA, et al. Sequential Study of Bacterial Antibody Levels and Faecal Flora in Rheumatoid Arthritis Patients Taking Sulphasalazine. Br J Rheumatol (1993) 32(8):683–8. 10.1093/rheumatology/32.8.683 [DOI] [PubMed] [Google Scholar]
- 28. Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, et al. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe (2017) 21(5):603–10.e3. 10.1016/j.chom.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, et al. Gut Microbiota Offers Universal Biomarkers Across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. mSystems (2018) 3(1):e00188-17. 10.1128/mSystems.00188-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dewint P, Hansen BE, Verhey E, Oldenburg B, Hommes DW, Pierik M, et al. Adalimumab Combined With Ciprofloxacin is Superior to Adalimumab Monotherapy in Perianal Fistula Closure in Crohn’s Disease: A Randomised, Double-Blind, Placebo Controlled Trial (ADAFI). Gut (2014) 63(2):292–9. 10.1136/gutjnl-2013-304488 [DOI] [PubMed] [Google Scholar]
- 31. Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a Probiotic With Peanut Oral Immunotherapy: A Randomized Trial. J Allergy Clin Immunol (2015) 135(3):737–44.e8. 10.1016/j.jaci.2014.11.034 [DOI] [PubMed] [Google Scholar]
- 32. Wei SC, Duffy CR, Allison JP. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discovery (2018) 8(9):1069–86. 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- 33. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune Checkpoint Inhibitors: Recent Progress and Potential Biomarkers. Exp Mol Med (2018) 50(12):1–11. 10.1038/s12276-018-0191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilky BA. Immune Checkpoint Inhibitors: The Linchpins of Modern Immunotherapy. Immunol Rev (2019) 290(1):6–23. 10.1111/imr.12766 [DOI] [PubMed] [Google Scholar]
- 35. Cai J, Wang D, Zhang G, Guo X. The Role of PD-1/PD-L1 Axis In Treg Development and Function: Implications For Cancer Immunotherapy. Onco Targets Ther (2019) 12:8437–45. 10.2147/OTT.S221340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen DS, Mellman I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Elsevier (2013) p:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 37. Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the Effector Function of CD8(+) T Cells by Gut Microbiota-Derived Metabolite Butyrate. Sci Rep (2018) 8(1):14430. 10.1038/s41598-018-32860-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, et al. Gut Microbiota Modulate T Cell Trafficking Into Human Colorectal Cancer. Gut (2018) 67(11):1984–94. 10.1136/gutjnl-2016-313498 [DOI] [PubMed] [Google Scholar]
- 39. Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G. Anticancer Effects of the Microbiome and its Products. Nat Rev Microbiol (2017) 15(8):465–78. 10.1038/nrmicro.2017.44 [DOI] [PubMed] [Google Scholar]
- 40. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science (2013) 341(6145):569–73. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoeppli RE, Wu D, Cook L, Levings MK. The Environment of Regulatory T Cell Biology: Cytokines, Metabolites, and the Microbiome. Front Immunol (2015) 6:61. 10.3389/fimmu.2015.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poggi A, Benelli R, Venè R, Costa D, Ferrari N, Tosetti F, et al. Human Gut-Associated Natural Killer Cells in Health and Disease. Front Immunol (2019) 10:961. 10.3389/fimmu.2019.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and Anticancer Immunosurveillance. Cell (2016) 165(2):276–87. 10.1016/j.cell.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 44. Andre FE, Booy R, Bock HL, Clemens J, Datta SK, John TJ, et al. Vaccination Greatly Reduces Disease, Disability, Death and Inequity Worldwide. Bull World Health Organ (2008) 86(2):140–6. 10.2471/BLT.07.040089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pezzotti P, Bellino S, Prestinaci F, Iacchini S, Lucaroni F, Camoni L, et al. The Impact of Immunization Programs on 10 Vaccine Preventable Diseases in Italy: 1900-2015. Vaccine (2018) 36(11):1435–43. 10.1016/j.vaccine.2018.01.065 [DOI] [PubMed] [Google Scholar]
- 46. Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From Vaccines to Memory and Back. Immunity (2010) 33(4):451–63. 10.1016/j.immuni.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McNeela EA, Mills KH. Manipulating the Immune System: Humoral Versus Cell-Mediated Immunity. Adv Drug Delivery Rev (2001) 51(1-3):43–54. 10.1016/S0169-409X(01)00169-7 [DOI] [PubMed] [Google Scholar]
- 48. Kleindienst P, Brocker T. Endogenous Dendritic Cells are Required for Amplification of T Cell Responses Induced by Dendritic Cell Vaccines In Vivo. J Immunol (2003) 170(6):2817–23. 10.4049/jimmunol.170.6.2817 [DOI] [PubMed] [Google Scholar]
- 49. Serre K, Cunningham AF, Coughlan RE, Lino AC, Rot A, Hub E, et al. Cd8 T Cells Induce T-bet-dependent Migration Toward CXCR3 Ligands by Differentiated B Cells Produced During Responses to Alum-Protein Vaccines. Blood (2012) 120(23):4552–9. 10.1182/blood-2012-03-417733 [DOI] [PubMed] [Google Scholar]
- 50. Steinman RM. Dendritic Cells In Vivo: A Key Target for a New Vaccine Science. Immunity (2008) 29(3):319–24. 10.1016/j.immuni.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 51. Havenar-Daughton C, Abbott RK, Schief WR, Crotty S. When Designing Vaccines, Consider the Starting Material: The Human B Cell Repertoire. Curr Opin Immunol (2018) 53:209–16. 10.1016/j.coi.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ubeda C, Pamer EG. Antibiotics, Microbiota, and Immune Defense. Trends Immunol (2012) 33(9):459–66. 10.1016/j.it.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pamer EG. Microbial Tuning of the Mammalian Immune System. Trends Mol Med (2017) 23(5):379–80. 10.1016/j.molmed.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell (2016) 167(7):1897. 10.1016/j.cell.2016.11.046 [DOI] [PubMed] [Google Scholar]
- 55. Li B, Chan HL, Chen P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr Med Chem (2019) 26(17):3009–25. 10.2174/0929867324666170804143706 [DOI] [PubMed] [Google Scholar]
- 56. Havel JJ, Chowell D, Chan TA. The Evolving Landscape of Biomarkers for Checkpoint Inhibitor Immunotherapy. Nat Rev Cancer (2019) 19(3):133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gibney GT, Weiner LM, Atkins MB. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy. Lancet Oncol (2016) 17(12):e542–e51. 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut Microbiome Influences Efficacy of PD-1–based Immunotherapy Against Epithelial Tumors. Science (2018) 359(6371):91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 59. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated With Ipilimumab. Ann Oncol (2017) 28(6):1368–79. 10.1093/annonc/mdx108 [DOI] [PubMed] [Google Scholar]
- 60. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated With Immune Checkpoint Therapy Efficacy in Melanoma Patients 1. Neoplasia (2017) 19:848–55. 10.1016/j.neo.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti–PD-1 Immunotherapy in Melanoma Patients. Science (2018) 359(6371):97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The Commensal Microbiome is Associated With Anti–PD-1 Efficacy in Metastatic Melanoma Patients. Science (2018) 359(6371):104–8. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Coutzac C, Jouniaux J-M, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts With Cancer. Nat Commun (2020) 11(1):2168–. 10.1038/s41467-020-16079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, et al. Relating the Gut Metagenome and Metatranscriptome to Immunotherapy Responses in Melanoma Patients. Genome Med (2019) 11(1). 10.1186/s13073-019-0672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative Association of Antibiotics on Clinical Activity of Immune Checkpoint Inhibitors in Patients With Advanced Renal Cell and non-Small-Cell Lung Cancer. Ann Oncol (2018) 29(6):1437–44. 10.1093/annonc/mdy103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur Urol (2020) 78(2):195–206. 10.1016/j.eururo.2020.04.044 [DOI] [PubMed] [Google Scholar]
- 67. Zhao S, Gao G, Li W, Li X, Zhao C, Jiang T, et al. Antibiotics are Associated With Attenuated Efficacy of anti-PD-1/PD-L1 Therapies in Chinese Patients With Advanced non-Small Cell Lung Cancer. Lung Cancer (2019) 130:10–7. 10.1016/j.lungcan.2019.01.017 [DOI] [PubMed] [Google Scholar]
- 68. Mohiuddin JJ, Chu B, Facciabene A, Poirier K, Wang X, Doucette A, et al. Association of Antibiotic Exposure With Survival and Toxicity in Patients With Melanoma Receiving Immunotherapy. J Natl Cancer Inst (2020) 113(2):162–70. 10.1093/jnci/djaa057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lurienne L, Cervesi J, Duhalde L, de Gunzburg J, Andremont A, Zalcman G, et al. Non-Small-Cell Lung Cancer Immunotherapy Efficacy and Antibiotic Use: A Systematic Review and Meta-Analysis. J Thorac Oncol (2020) 15(7):1147–59. 10.1016/j.jtho.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 70. Kaderbhai C, Richard C, Fumet JD, Aarnink A, Foucher P, Coudert B, et al. Antibiotic Use Does Not Appear to Influence Response to Nivolumab. Anticancer Res (2017) 37(6):3195–200. 10.21873/anticanres.11680 [DOI] [PubMed] [Google Scholar]
- 71. Tomita Y, Ikeda T, Sakata S, Saruwatari K, Sato R, Iyama S, et al. Association of Probiotic Clostridium Butyricum Therapy With Survival and Response to Immune Checkpoint Blockade in Patients With Lung Cancer. Cancer Immunol Res (2020) 8(10):1236–42. 10.1158/2326-6066.CIR-20-0051 [DOI] [PubMed] [Google Scholar]
- 72. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium Prausnitzii is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc Natl Acad Sci USA (2008) 105(43):16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Galecka M, Szachta P, Bartnicka A, Lykowska-Szuber L, Eder P, Schwiertz A. Faecalibacterium Prausnitzii and Crohn’s Disease - is There Any Connection? Pol J Microbiol (2013) 62(1):91–5. 10.33073/pjm-2013-013 [DOI] [PubMed] [Google Scholar]
- 74. Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium Prausnitzii Subspecies-Level Dysbiosis in the Human Gut Microbiome Underlying Atopic Dermatitis. J Allergy Clin Immunol (2016) 137(3):852–60. 10.1016/j.jaci.2015.08.021 [DOI] [PubMed] [Google Scholar]
- 75. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell (2017) 168(5):928–43. 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fluckiger A, Daillere R, Sassi M, Sixt BS, Liu P, Loos F, et al. Cross-Reactivity Between Tumor MHC Class I-restricted Antigens and an Enterococcal Bacteriophage. Science (2020) 369(6506):936–42. 10.1126/science.aax0701 [DOI] [PubMed] [Google Scholar]
- 77. Shen H, Yang ESH, Conry M, Fiveash J, Contreras C, Bonner JA, et al. Predictive Biomarkers for Immune Checkpoint Blockade and Opportunities for Combination Therapies. Genes & Diseases (2019) 6(3):232–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lérias JR, Paraschoudi G, de Sousa E, Martins J, Condeço C, Figueiredo N, et al. Microbes as Master Immunomodulators: Immunopathology, Cancer and Personalized Immunotherapies. Front Cell Dev Biol (2020) 7:362–. 10.3389/fcell.2019.00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome Analyses of Blood and Tissues Suggest Cancer Diagnostic Approach. Nature (2020) 579(7800):567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Gavrielatou N, Doumas S, Economopoulou P, Foukas PG, Psyrri A. Biomarkers for Immunotherapy Response in Head and Neck Cancer. Cancer Treat Rev (2020) 84:101977. 10.1016/j.ctrv.2020.101977 [DOI] [PubMed] [Google Scholar]
- 81. Adlung L, Elinav E, Greten TF, Korangy F. Microbiome Genomics for Cancer Prediction. Nat Cancer (2020) 1(4):379–81. 10.1038/s43018-020-0059-x [DOI] [PubMed] [Google Scholar]
- 82. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates anti-PD-L1 Efficacy. Science (2015) 350(6264):1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science (2015) 350(6264):1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tanoue T, Morita S, Plichta D, Skelly AN, Suda W, Sugiura Y, et al. A Defined Commensal Consortium Elicits CD8 T Cells and Anti-Cancer Immunity. Nature (2019) 565(7741):600–5. 10.1038/s41586-019-0878-z [DOI] [PubMed] [Google Scholar]
- 85. Xu X, Lv J, Guo F, Li J, Jia Y, Jiang D, et al. Gut Microbiome Influences the Efficacy of PD-1 Antibody Immunotherapy on MSS-Type Colorectal Cancer Via Metabolic Pathway. Front Microbiol (2020) 11:814. 10.3389/fmicb.2020.00814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-Derived Inosine Modulates Response to Checkpoint Inhibitor Immunotherapy. Science (2020) 369(6510):eabc3421. 10.1126/science.abc3421 [DOI] [PubMed] [Google Scholar]
- 87. Graber CD, Goust JM, Glassman AD, Kendall R, Loadholt CB. Immunomodulating Properties of Dimethylglycine in Humans. J Infect Dis (1981) 143(1):101–5. 10.1093/infdis/143.1.101 [DOI] [PubMed] [Google Scholar]
- 88. Thurnher M, Gruenbacher G. T Lymphocyte Regulation by Mevalonate Metabolism. Sci Signal (2015) 8(370):re4. 10.1126/scisignal.2005970 [DOI] [PubMed] [Google Scholar]
- 89. Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, et al. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti–Programmed Death 1 Immunotherapy in Chinese Patients With Nsclc. J Thorac Oncol (2019) 14(8):1378–89. 10.1016/j.jtho.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 90. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor Tigit Protects Tumors From Immune Cell Attack. Immunity (2015) 42(2):344–55. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium Nucleatum Supresses Anti-Tumor Immunity by Activating CEACAM1. OncoImmunology (2019) 8(6):e1581531. 10.1080/2162402X.2019.1581531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kasper SH, Morell-Perez C, Wyche TP, Sana TR, Lieberman LA, Hett EC. Colorectal Cancer-Associated Anaerobic Bacteria Proliferate in Tumor Spheroids and Alter the Microenvironment. Sci Rep (2020) 10(1):1–13. 10.1038/s41598-020-62139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The Human Tumor Microbiome is Composed of Tumor Type–Specific Intracellular Bacteria. Science (2020) 368(6494):973–80. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Busquets D, Mas-de-Xaxars T, Lopez-Siles M, Martinez-Medina M, Bahi A, Sabat M, et al. Anti-Tumour Necrosis Factor Treatment With Adalimumab Induces Changes in the Microbiota of Crohn’s Disease. J Crohns Colitis (2015) 9(10):899–906. 10.1093/ecco-jcc/jjv119 [DOI] [PubMed] [Google Scholar]
- 95. Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y, et al. Characteristics of Faecal Microbiota in Paediatric Crohn’s Disease and Their Dynamic Changes During Infliximab Therapy. J Crohns Colitis (2018) 12(3):337–46. 10.1093/ecco-jcc/jjx153 [DOI] [PubMed] [Google Scholar]
- 96. Angelakis E, Million M, Kankoe S, Lagier JC, Armougom F, Giorgi R, et al. Abnormal Weight Gain and Gut Microbiota Modifications are Side Effects of Long-Term Doxycycline and Hydroxychloroquine Treatment. Antimicrob Agents Chemother (2014) 58(6):3342–7. 10.1128/AAC.02437-14 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97. Zaza G, Dalla Gassa A, Felis G, Granata S, Torriani S, Lupo A. Impact of Maintenance Immunosuppressive Therapy on the Fecal Microbiome of Renal Transplant Recipients: Comparison Between an Everolimus- and a Standard Tacrolimus-Based Regimen. PloS One (2017) 12(5):e0178228. 10.1371/journal.pone.0178228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ostrov BE, Amsterdam D. Immunomodulatory Interplay of the Microbiome and Therapy of Rheumatic Diseases. Immunol Invest (2017) 46(8):769–92. 10.1080/08820139.2017.1373828 [DOI] [PubMed] [Google Scholar]
- 99. Som A, Mandaliya R, Alsaadi D, Farshidpour M, Charabaty A, Malhotra N. Immune Checkpoint Inhibitor-Induced Colitis: A Comprehensive Review. World J Clin cases (2019) 7(4):405–18. 10.12998/wjcc.v7.i4.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal Microbiota Transplant Promotes Response in Immunotherapy-Refractory Melanoma Patients. Science (2020) 371(6529):602–9. 10.1126/science.abb5920 [DOI] [PubMed] [Google Scholar]
- 101. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal Microbiota Transplantation for Refractory Immune Checkpoint Inhibitor-Associated Colitis. Nat Med (2018) 24(12):1804–8. 10.1038/s41591-018-0238-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Marinelli L, Tenore GC, Novellino E. Probiotic Species in the Modulation of the Anticancer Immune Response. Semin Cancer Biol (2017) 46:182–90. 10.1016/j.semcancer.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 103. Pulendran B. Systems Vaccinology: Probing Humanity’s Diverse Immune Systems With Vaccines. Proc Natl Acad Sci USA (2014) 111(34):12300–6. 10.1073/pnas.1400476111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wiedermann U, Garner-Spitzer E, Wagner A. Primary Vaccine Failure to Routine Vaccines: Why and What to do? Hum Vaccin Immunother (2016) 12(1):239–43. 10.1080/21645515.2015.1093263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. McDermott AB, Cohen SB, Zuckerman JN, Madrigal JA. Hepatitis B Third-Generation Vaccines: Improved Response and Conventional Vaccine non-Response–Evidence for Genetic Basis in Humans. J Viral Hepat (1998) 5 Suppl 2:9–11. 10.1046/j.1365-2893.1998.0050s2009.x [DOI] [PubMed] [Google Scholar]
- 106. Gelder CM, Lambkin R, Hart KW, Fleming D, Williams OM, Bunce M, et al. Associations Between Human Leukocyte Antigens and Nonresponsiveness to Influenza Vaccine. J Infect Dis (2002) 185(1):114–7. 10.1086/338014 [DOI] [PubMed] [Google Scholar]
- 107. Fisman DN, Agrawal D, Leder K. The Effect of Age on Immunologic Response to Recombinant Hepatitis B Vaccine: A Meta-Analysis. Clin Infect Dis (2002) 35(11):1368–75. 10.1086/344271 [DOI] [PubMed] [Google Scholar]
- 108. Chen WH, Kozlovsky BF, Effros RB, Grubeck-Loebenstein B, Edelman R, Sztein MB. Vaccination in the Elderly: An Immunological Perspective. Trends Immunol (2009) 30(7):351–9. 10.1016/j.it.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. McElhaney JE, Zhou X, Talbot HK, Soethout E, Bleackley RC, Granville DJ, et al. The Unmet Need in the Elderly: How Immunosenescence, CMV Infection, Co-Morbidities and Frailty are a Challenge for the Development of More Effective Influenza Vaccines. Vaccine (2012) 30(12):2060–7. 10.1016/j.vaccine.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang S, Tian G, Cui Y, Ding C, Deng M, Yu C, et al. Factors Influencing Immunologic Response to Hepatitis B Vaccine in Adults. Sci Rep (2016) 6:27251. 10.1038/srep27251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Trzonkowski P, Mysliwska J, Szmit E, Wieckiewicz J, Lukaszuk K, Brydak LB, et al. Association Between Cytomegalovirus Infection, Enhanced Proinflammatory Response and Low Level of Anti-Hemagglutinins During the Anti-Influenza Vaccination–an Impact of Immunosenescence. Vaccine (2003) 21(25-26):3826–36. 10.1016/S0264-410X(03)00309-8 [DOI] [PubMed] [Google Scholar]
- 112. Pasricha N, Datta U, Chawla Y, Singh S, Arora SK, Sud A, et al. Immune Responses in Patients With HIV Infection After Vaccination With Recombinant Hepatitis B Virus Vaccine. BMC Infect Dis (2006) 6:65. 10.1186/1471-2334-6-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Akmatov MK, Riese P, Trittel S, May M, Prokein J, Illig T, et al. Self-Reported Diabetes and Herpes Zoster are Associated With a Weak Humoral Response to the Seasonal Influenza A H1N1 Vaccine Antigen Among the Elderly. BMC Infect Dis (2019) 19(1):656. 10.1186/s12879-019-4214-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sheridan PA, Paich HA, Handy J, Karlsson EA, Hudgens MG, Sammon AB, et al. Obesity is Associated With Impaired Immune Response to Influenza Vaccination in Humans. Int J Obes (Lond) (2012) 36(8):1072–7. 10.1038/ijo.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Haque R, Snider C, Liu Y, Ma JZ, Liu L, Nayak U, et al. Oral Polio Vaccine Response in Breast Fed Infants With Malnutrition and Diarrhea. Vaccine (2014) 32(4):478–82. 10.1016/j.vaccine.2013.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Eleftheriadis T, Antoniadi G, Liakopoulos V, Kartsios C, Stefanidis I. Disturbances of Acquired Immunity in Hemodialysis Patients. Semin Dial (2007) 20(5):440–51. 10.1111/j.1525-139X.2007.00283.x [DOI] [PubMed] [Google Scholar]
- 117. Noh KW, Poland GA, Murray JA. Hepatitis B Vaccine Nonresponse and Celiac Disease. Am J Gastroenterol (2003) 98(10):2289–92. 10.1111/j.1572-0241.2003.07701.x [DOI] [PubMed] [Google Scholar]
- 118. Patel DP, Treat JR, Castelo-Socio L. Decreased Hepatitis B Vaccine Response in Pediatric Patients With Atopic Dermatitis, Psoriasis, and Morphea. Vaccine (2017) 35(35 Pt B):4499–500. 10.1016/j.vaccine.2017.07.025 [DOI] [PubMed] [Google Scholar]
- 119. Sell S. Mercaptoethanol-Sensitive Antibody Production in Germ-Free Mice and Guinea Pigs. J Immunol (1965) 95(2):300–5. [PubMed] [Google Scholar]
- 120. Porter P, Kenworthy R, Noakes DE, Allen WD. Intestinal Antibody Secretion in the Young Pig in Response to Oral Immunization With Escherichia Coli. Immunology (1974) 27(5):841–53. [PMC free article] [PubMed] [Google Scholar]
- 121. Ohwaki M, Yasutake N, Yasui H, Ogura R. A Comparative Study on the Humoral Immune Responses in Germ-Free and Conventional Mice. Immunology (1977) 32(1):43–8. [PMC free article] [PubMed] [Google Scholar]
- 122. Parry SH, Allen WD, Porter P. Intestinal Immune Response to E. coli antigens in the germ-free chicken. Immunology (1977) 32(5):731–41. [PMC free article] [PubMed] [Google Scholar]
- 123. MacDonald TT, Carter PB. Requirement for a Bacterial Flora Before Mice Generate Cells Capable of Mediating the Delayed Hypersensitivity Reaction to Sheep Red Blood Cells. J Immunol (1979) 122(6):2624–9. [PubMed] [Google Scholar]
- 124. Hobby GL, Lenert TF, Maier-Engallena J, Wakely-Oliver C, Manty A, Cicenia E, et al. Further Observations on the Immunogenic Effect of BCG in Germfree Mice. II. Am Rev Respir Dis (1968) 97(6):1095–103. 10.1164/arrd.1968.97.6P1.1095 [DOI] [PubMed] [Google Scholar]
- 125. Lamouse-Smith ES, Tzeng A, Starnbach MN. The Intestinal Flora is Required to Support Antibody Responses to Systemic Immunization in Infant and Germ Free Mice. PloS One (2011) 6(11):e27662. 10.1371/journal.pone.0027662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-Mediated Sensing of Gut Microbiota is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity (2014) 41(3):478–92. 10.1016/j.immuni.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. Antibiotic Treatment Suppresses Rotavirus Infection and Enhances Specific Humoral Immunity. J Infect Dis (2014) 210(2):171–82. 10.1093/infdis/jiu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kim D, Kim YG, Seo SU, Kim DJ, Kamada N, Prescott D, et al. Nod2-mediated Recognition of the Microbiota is Critical for Mucosal Adjuvant Activity of Cholera Toxin. Nat Med (2016) 22(5):524–30. 10.1038/nm.4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kim D, Kim YM, Kim WU, Park JH, Nunez G, Seo SU. Recognition of the Microbiota by Nod2 Contributes to the Oral Adjuvant Activity of Cholera Toxin Through the Induction of Interleukin-1beta. Immunology (2019) 158(3):219–29. 10.1111/imm.13105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Woo PC, Tsoi HW, Wong LP, Leung HC, Yuen KY. Antibiotics Modulate Vaccine-Induced Humoral Immune Response. Clin Diagn Lab Immunol (1999) 6(6):832–7. 10.1128/CDLI.6.6.832-837.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe (2018) 23(5):653–60.e5. 10.1016/j.chom.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 132. Yang W, Xiao Y, Huang X, Chen F, Sun M, Bilotta AJ, et al. Microbiota Metabolite Short-Chain Fatty Acids Facilitate Mucosal Adjuvant Activity of Cholera Toxin Through GPR43. J Immunol (2019) 203(1):282–92. 10.4049/jimmunol.1801068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhang Y, Wu Q, Zhou M, Luo Z, Lv L, Pei J, et al. Composition of the Murine Gut Microbiome Impacts Humoral Immunity Induced by Rabies Vaccines. Clin Transl Med (2020) 10(4):e161. 10.1002/ctm2.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vos AP, Haarman M, Buco A, Govers M, Knol J, Garssen J, et al. A Specific Prebiotic Oligosaccharide Mixture Stimulates Delayed-Type Hypersensitivity in a Murine Influenza Vaccination Model. Int Immunopharmacol (2006) 6(8):1277–86. 10.1016/j.intimp.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 135. Benyacoub J, Rochat F, Saudan KY, Rochat I, Antille N, Cherbut C, et al. Feeding a Diet Containing a Fructooligosaccharide Mix can Enhance Salmonella Vaccine Efficacy in Mice. J Nutr (2008) 138(1):123–9. 10.1093/jn/138.1.123 [DOI] [PubMed] [Google Scholar]
- 136. Vos AP, Knol J, Stahl B, M’Rabet L, Garssen J. Specific Prebiotic Oligosaccharides Modulate the Early Phase of a Murine Vaccination Response. Int Immunopharmacol (2010) 10(5):619–25. 10.1016/j.intimp.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 137. van den Elsen LWJ, Tims S, Jones AM, Stewart A, Stahl B, Garssen J, et al. Prebiotic Oligosaccharides in Early Life Alter Gut Microbiome Development in Male Mice While Supporting Influenza Vaccination Responses. Benef Microbes (2019) 10(3):279–91. 10.3920/BM2018.0098 [DOI] [PubMed] [Google Scholar]
- 138. Di Luccia B, Ahern PP, Griffin NW, Cheng J, Guruge JL, Byrne AE, et al. Combined Prebiotic and Microbial Intervention Improves Oral Cholera Vaccination Responses in a Mouse Model of Childhood Undernutrition. Cell Host Microbe (2020) 27(6):899–908 e5. 10.1016/j.chom.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wen K, Li G, Bui T, Liu F, Li Y, Kocher J, et al. High Dose and Low Dose Lactobacillus Acidophilus Exerted Differential Immune Modulating Effects on T Cell Immune Responses Induced by an Oral Human Rotavirus Vaccine in Gnotobiotic Pigs. Vaccine (2012) 30(6):1198–207. 10.1016/j.vaccine.2011.11.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chattha KS, Vlasova AN, Kandasamy S, Rajashekara G, Saif LJ. Divergent Immunomodulating Effects of Probiotics on T Cell Responses to Oral Attenuated Human Rotavirus Vaccine and Virulent Human Rotavirus Infection in a Neonatal Gnotobiotic Piglet Disease Model. J Immunol (2013) 191(5):2446–56. 10.4049/jimmunol.1300678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kandasamy S, Chattha KS, Vlasova AN, Rajashekara G, Saif LJ. Lactobacilli and Bifidobacteria Enhance Mucosal B Cell Responses and Differentially Modulate Systemic Antibody Responses to an Oral Human Rotavirus Vaccine in a Neonatal Gnotobiotic Pig Disease Model. Gut Microbes (2014) 5(5):639–51. 10.4161/19490976.2014.969972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wen K, Tin C, Wang H, Yang X, Li G, Giri-Rachman E, et al. Probiotic Lactobacillus Rhamnosus GG Enhanced Th1 Cellular Immunity But did Not Affect Antibody Responses in a Human Gut Microbiota Transplanted Neonatal Gnotobiotic Pig Model. PloS One (2014) 9(4):e94504. 10.1371/journal.pone.0094504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Arnold IC, Hutchings C, Kondova I, Hey A, Powrie F, Beverley P, et al. Helicobacter Hepaticus Infection in BALB/c Mice Abolishes Subunit-Vaccine-Induced Protection Against M. Tuberculosis. Vaccine (2015) 33(15):1808–14. 10.1016/j.vaccine.2015.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Burger MC, et al. Sequential Infection With Common Pathogens Promotes Human-Like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe (2016) 19(5):713–9. 10.1016/j.chom.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Stebegg M, Silva-Cayetano A, Innocentin S, Jenkins TP, Cantacessi C, Gilbert C, et al. Heterochronic Faecal Transplantation Boosts Gut Germinal Centres in Aged Mice. Nat Commun (2019) 10(1):2443. 10.1038/s41467-019-10430-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bumgardner SA, Zhang L, LaVoy AS, Andre B, Frank CB, Kajikawa A, et al. Nod2 is Required for Antigen-Specific Humoral Responses Against Antigens Orally Delivered Using a Recombinant Lactobacillus Vaccine Platform. PloS One (2018) 13(5):e0196950. 10.1371/journal.pone.0196950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Parker EP, Ramani S, Lopman BA, Church JA, Iturriza-Gomara M, Prendergast AJ, et al. Causes of Impaired Oral Vaccine Efficacy in Developing Countries. Future Microbiol (2018) 13:97–118. 10.2217/fmb-2017-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Jamieson AM. Influence of the Microbiome on Response to Vaccination. Hum Vaccin Immunother (2015) 11(9):2329–31. 10.1080/21645515.2015.1022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Valdez Y, Brown EM, Finlay BB. Influence of the Microbiota on Vaccine Effectiveness. Trends Immunol (2014) 35(11):526–37. 10.1016/j.it.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 150. Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-Modulated Metabolites At the Interface of Host Immunity. J Immunol (2017) 198(2):572–80. 10.4049/jimmunol.1601247 [DOI] [PubMed] [Google Scholar]
- 151. Lynn DJ, Pulendran B. The Potential of the Microbiota to Influence Vaccine Responses. J Leukoc Biol (2018) 103(2):225–31. 10.1189/jlb.5MR0617-216R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ciabattini A, Olivieri R, Lazzeri E, Medaglini D. Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front Microbiol (2019) 10:1305. 10.3389/fmicb.2019.01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. de Jong SE, Olin A, Pulendran B. The Impact of the Microbiome on Immunity to Vaccination in Humans. Cell Host Microbe (2020) 28(2):169–79. 10.1016/j.chom.2020.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool Microbiota and Vaccine Responses of Infants. Pediatrics (2014) 134(2):e362–72. 10.1542/peds.2013-3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Levine MM. Immunogenicity and Efficacy of Oral Vaccines in Developing Countries: Lessons From a Live Cholera Vaccine. BMC Biol (2010) 8:129. 10.1186/1741-7007-8-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Becker-Dreps S, Vilchez S, Bucardo F, Twitchell E, Choi WS, Hudgens MG, et al. The Association Between Fecal Biomarkers of Environmental Enteropathy and Rotavirus Vaccine Response in Nicaraguan Infants. Pediatr Infect Dis J (2017) 36(4):412–6. 10.1097/INF.0000000000001457 [DOI] [PubMed] [Google Scholar]
- 157. Praharaj I, Parker EPK, Giri S, Allen DJ, Silas S, Revathi R, et al. Influence of Nonpolio Enteroviruses and the Bacterial Gut Microbiota on Oral Poliovirus Vaccine Response: A Study From South India. J Infect Dis (2019) 219(8):1178–86. 10.1093/infdis/jiy568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhao T, Li J, Fu Y, Ye H, Liu X, Li G, et al. Influence of Gut Microbiota on Mucosal IgA Antibody Response to the Polio Vaccine. NPJ Vaccines (2020) 5:47. 10.1038/s41541-020-0194-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, et al. Bifidobacterium Abundance in Early Infancy and Vaccine Response At 2 Years of Age. Pediatrics (2019) 143(2):e20181489. 10.1542/peds.2018-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Twitchell EL, Tin C, Wen K, Zhang H, Becker-Dreps S, Azcarate-Peril MA, et al. Modeling Human Enteric Dysbiosis and Rotavirus Immunity in Gnotobiotic Pigs. Gut Pathog (2016) 8:51. 10.1186/s13099-016-0136-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Goenka A, Kollmann TR. Development of Immunity in Early Life. J Infect (2015) 71 Suppl 1:S112–20. 10.1016/j.jinf.2015.04.027 [DOI] [PubMed] [Google Scholar]
- 162. Grassly NC, Praharaj I, Babji S, Kaliappan SP, Giri S, Venugopal S, et al. The Effect of Azithromycin on the Immunogenicity of Oral Poliovirus Vaccine: A Double-Blind Randomised Placebo-Controlled Trial in Seronegative Indian Infants. Lancet Infect Dis (2016) 16(8):905–14. 10.1016/S1473-3099(16)30023-8 [DOI] [PubMed] [Google Scholar]
- 163. Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J Infect Dis (2017) 215(1):34–41. 10.1093/infdis/jiw518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Harris V, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, et al. Rotavirus Vaccine Response Correlates With the Infant Gut Microbiota Composition in Pakistan. Gut Microbes (2018) 9(2):93–101. 10.1080/19490976.2017.1376162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Parker EPK, Praharaj I, Zekavati A, Lazarus RP, Giri S, Operario DJ, et al. Influence of the Intestinal Microbiota on the Immunogenicity of Oral Rotavirus Vaccine Given to Infants in South India. Vaccine (2018) 36(2):264–72. 10.1016/j.vaccine.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Fix J, Chandrashekhar K, Perez J, Bucardo F, Hudgens MG, Yuan L, et al. Association Between Gut Microbiome Composition and Rotavirus Vaccine Response Among Nicaraguan Infants. Am J Trop Med Hyg (2020) 102(1):213–9. 10.4269/ajtmh.19-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL, Jr, et al. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe (2018) 24(2):197–207.e4. 10.1016/j.chom.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Eloe-Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, et al. Impact of Oral Typhoid Vaccination on the Human Gut Microbiota and Correlations With s. Typhi-specific immunological responses. PloS One (2013) 8(4):e62026. 10.1371/journal.pone.0062026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Salk HM, Simon WL, Lambert ND, Kennedy RB, Grill DE, Kabat BF, et al. Taxa of the Nasal Microbiome Are Associated With Influenza-Specific Iga Response to Live Attenuated Influenza Vaccine. PloS One (2016) 11(9):e0162803. 10.1371/journal.pone.0162803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zimmermann P, Curtis N. The Influence of the Intestinal Microbiome on Vaccine Responses. Vaccine (2018) 36(30):4433–9. 10.1016/j.vaccine.2018.04.066 [DOI] [PubMed] [Google Scholar]
- 171. Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell (2019) 178(6):1313–28.e13. 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated With Unique Host and Microbiome Features. Cell (2018) 174(6):1388–405:e21. 10.1016/j.cell.2018.08.041 [DOI] [PubMed] [Google Scholar]
- 173. Stefan KL, Kim MV, Iwasaki A, Kasper DL. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell (2020) 183(5):1312–24:e10. 10.1016/j.cell.2020.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pabst O, Hornef M. Gut Microbiota: A Natural Adjuvant for Vaccination. Immunity (2014) 41(3):349–51. 10.1016/j.immuni.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 175. Kullberg MC, Jankovic D, Gorelick PL, Caspar P, Letterio JJ, Cheever AW, et al. Bacteria-Triggered CD4(+) T Regulatory Cells Suppress Helicobacter Hepaticus-Induced Colitis. J Exp Med (2002) 196(4):505–15. 10.1084/jem.20020556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Kuehl CJ, Wood HD, Marsh TL, Schmidt TM, Young VB. Colonization of the Cecal Mucosa by Helicobacter Hepaticus Impacts the Diversity of the Indigenous Microbiota. Infect Immun (2005) 73(10):6952–61. 10.1128/IAI.73.10.6852-6961.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the Environment Recapitulates Adult Human Immune Traits in Laboratory Mice. Nature (2016) 532(7600):512–6. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Siegrist CA, Aspinall R. B-Cell Responses to Vaccination At the Extremes of Age. Nat Rev Immunol (2009) 9(3):185–94. 10.1038/nri2508 [DOI] [PubMed] [Google Scholar]
- 179. Goronzy JJ, Weyand CM. Understanding Immunosenescence to Improve Responses to Vaccines. Nat Immunol (2013) 14(5):428–36. 10.1038/ni.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Raymond SL, Rincon JC, Wynn JL, Moldawer LL, Larson SD. Impact of Early-Life Exposures to Infections, Antibiotics, and Vaccines on Perinatal and Long-term Health and Disease. Front Immunol (2017) 8:729. 10.3389/fimmu.2017.00729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Niewiesk S. Maternal Antibodies: Clinical Significance, Mechanism of Interference With Immune Responses, and Possible Vaccination Strategies. Front Immunol (2014) 5:446. 10.3389/fimmu.2014.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Church JA, Parker EP, Kirkpatrick BD, Grassly NC, Prendergast AJ. Interventions to Improve Oral Vaccine Performance: A Systematic Review and Meta-Analysis. Lancet Infect Dis (2019) 19(2):203–14. 10.1016/S1473-3099(18)30602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Timens W, Boes A, Rozeboom-Uiterwijk T, Poppema S. Immaturity of the Human Splenic Marginal Zone in Infancy. Possible contribution to the deficient infant immune response. J Immunol (1989) 143(10):3200–6. [PubMed] [Google Scholar]
- 184. Pihlgren M, Tougne C, Bozzotti P, Fulurija A, Duchosal MA, Lambert PH, et al. Unresponsiveness to Lymphoid-Mediated Signals At the Neonatal Follicular Dendritic Cell Precursor Level Contributes to Delayed Germinal Center Induction and Limitations of Neonatal Antibody Responses to T-dependent Antigens. J Immunol (2003) 170(6):2824–32. 10.4049/jimmunol.170.6.2824 [DOI] [PubMed] [Google Scholar]
- 185. Kanswal S, Katsenelson N, Selvapandiyan A, Bram RJ, Akkoyunlu M. Deficient TACI Expression on B Lymphocytes of Newborn Mice Leads to Defective Ig Secretion in Response to BAFF or APRIL. J Immunol (2008) 181(2):976–90. 10.4049/jimmunol.181.2.976 [DOI] [PubMed] [Google Scholar]
- 186. Garg M, Luo W, Kaplan AM, Bondada S. Cellular Basis of Decreased Immune Responses to Pneumococcal Vaccines in Aged Mice. Infect Immun (1996) 64(11):4456–62. 10.1128/IAI.64.11.4456-4462.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Asanuma H, Hirokawa K, Uchiyama M, Suzuki Y, Aizawa C, Kurata T, et al. Immune Responses and Protection in Different Strains of Aged Mice Immunized Intranasally With an Adjuvant-Combined Influenza Vaccine. Vaccine (2001) 19(28-29):3981–9. 10.1016/S0264-410X(01)00129-3 [DOI] [PubMed] [Google Scholar]
- 188. Kang I, Hong MS, Nolasco H, Park SH, Dan JM, Choi JY, et al. Age-Associated Change in the Frequency of Memory CD4+ T Cells Impairs Long Term CD4+ T Cell Responses to Influenza Vaccine. J Immunol (2004) 173(1):673–81. 10.4049/jimmunol.173.1.673 [DOI] [PubMed] [Google Scholar]
- 189. Wagar LE, Gentleman B, Pircher H, McElhaney JE, Watts TH. Influenza-Specific T Cells From Older People are Enriched in the Late Effector Subset and Their Presence Inversely Correlates With Vaccine Response. PloS One (2011) 6(8):e23698. 10.1371/journal.pone.0023698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Cummins NW, Weaver EA, May SM, Croatt AJ, Foreman O, Kennedy RB, et al. Heme Oxygenase-1 Regulates the Immune Response to Influenza Virus Infection and Vaccination in Aged Mice. FASEB J (2012) 26(7):2911–8. 10.1096/fj.11-190017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Haq K, McElhaney JE. Immunosenescence: Influenza Vaccination and the Elderly. Curr Opin Immunol (2014) 29:38–42. 10.1016/j.coi.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 192. Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, et al. Systems Analysis of Immunity to Influenza Vaccination Across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity (2015) 43(6):1186–98. 10.1016/j.immuni.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The Intestinal Microbiome in Early Life: Health and Disease. Front Immunol (2014) 5:427. 10.3389/fimmu.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Nguyen QN, Himes JE, Martinez DR, Permar SR. The Impact of the Gut Microbiota on Humoral Immunity to Pathogens and Vaccination in Early Infancy. PloS Pathog (2016) 12(12):e1005997. 10.1371/journal.ppat.1005997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, Lamouse-Smith ES. Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J Immunol (2016) 196(9):3768–79. 10.4049/jimmunol.1502322 [DOI] [PubMed] [Google Scholar]
- 196. Al Nabhani Z, Eberl G. Imprinting of the Immune System by the Microbiota Early in Life. Mucosal Immunol (2020) 13(2):183–9. 10.1038/s41385-020-0257-y [DOI] [PubMed] [Google Scholar]
- 197. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early Infancy Microbial and Metabolic Alterations Affect Risk of Childhood Asthma. Sci Transl Med. (2015) 7(307):307ra152. 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- 198. Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, et al. Functional Metagenomic Profiling of Intestinal Microbiome in Extreme Ageing. Aging (Albany NY) (2013) 5(12):902–12. 10.18632/aging.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Salazar N, Valdes-Varela L, Gonzalez S, Gueimonde M, de Los Reyes-Gavilan CG. Nutrition and the Gut Microbiome in the Elderly. Gut Microbes (2017) 8(2):82–97. 10.1080/19490976.2016.1256525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Amsterdam D, Ostrov BE. The Impact of the Microbiome on Immunosenescence. Immunol Invest (2018) 47(8):801–11. 10.1080/08820139.2018.1537570 [DOI] [PubMed] [Google Scholar]
- 201. Poland GA, Ovsyannikova IG, Kennedy RB, Lambert ND, Kirkland JL. A Systems Biology Approach to the Effect of Aging, Immunosenescence and Vaccine Response. Curr Opin Immunol (2014) 29:62–8. 10.1016/j.coi.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Murray MA, Chotirmall SH. The Impact of Immunosenescence on Pulmonary Disease. Mediators Inflammation (2015) 2015:692546. 10.1155/2015/692546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Maidens C, Childs C, Przemska A, Dayel IB, Yaqoob P. Modulation of Vaccine Response by Concomitant Probiotic Administration. Br J Clin Pharmacol (2013) 75(3):663–70. 10.1111/j.1365-2125.2012.04404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Lei WT, Shih PC, Liu SJ, Lin CY, Yeh TL. Effect of Probiotics and Prebiotics on Immune Response to Influenza Vaccination in Adults: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients (2017) 9(11):1175. 10.3390/nu9111175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Zimmermann P, Curtis N. The Influence of Probiotics on Vaccine Responses - A Systematic Review. Vaccine (2018) 36(2):207–13. 10.1016/j.vaccine.2017.08.069 [DOI] [PubMed] [Google Scholar]
- 206. Yeh TL, Shih PC, Liu SJ, Lin CH, Liu JM, Lei WT, et al. The Influence of Prebiotic or Probiotic Supplementation on Antibody Titers After Influenza Vaccination: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Drug Des Devel Ther (2018) 12:217–30. 10.2147/DDDT.S155110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Praharaj I, John SM, Bandyopadhyay R, Kang G. Probiotics, Antibiotics and the Immune Responses to Vaccines. Philos Trans R Soc Lond B Biol Sci (2015) 370(1671). 10.1098/rstb.2014.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Olivares M, Diaz-Ropero MP, Sierra S, Lara-Villoslada F, Fonolla J, Navas M, et al. Oral Intake of Lactobacillus Fermentum CECT5716 Enhances the Effects of Influenza Vaccination. Nutrition (2007) 23(3):254–60. 10.1016/j.nut.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 209. Bianchini S, Orabona C, Camilloni B, Berioli MG, Argentiero A, Matino D, et al. Effects of Probiotic Administration on Immune Responses of Children and Adolescents With Type 1 Diabetes to a Quadrivalent Inactivated Influenza Vaccine. Hum Vaccin Immunother (2020) 16(1):86–94. 10.1080/21645515.2019.1633877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Taylor AL, Hale J, Wiltschut J, Lehmann H, Dunstan JA, Prescott SL. Effects of Probiotic Supplementation for the First 6 Months of Life on Allergen- and Vaccine-Specific Immune Responses. Clin Exp Allergy (2006) 36(10):1227–35. 10.1111/j.1365-2222.2006.02553.x [DOI] [PubMed] [Google Scholar]
- 211. Redondo N, Nova E, Gheorghe A, Diaz LE, Hernandez A, Marcos A. Evaluation of Lactobacillus Coryniformis CECT5711 Strain as a Coadjuvant in a Vaccination Process: A Randomised Clinical Trial in Healthy Adults. Nutr Metab (Lond) (2017) 14:2. 10.1186/s12986-017-0193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Bunout D, Barrera G, Hirsch S, Gattas V, de la Maza MP, Haschke F, et al. Effects of a Nutritional Supplement on the Immune Response and Cytokine Production in Free-Living Chilean Elderly. JPEN J Parenter Enteral Nutr (2004) 28(5):348–54. 10.1177/0148607104028005348 [DOI] [PubMed] [Google Scholar]
- 213. Rizzardini G, Eskesen D, Calder PC, Capetti A, Jespersen L, Clerici M. Evaluation of the Immune Benefits of Two Probiotic Strains Bifidobacterium Animalis Ssp. Lactis, BB-12(R) and Lactobacillus Paracasei Ssp. Paracasei, L. Casei 431(R) in an Influenza Vaccination Model: A Randomised, Double-Blind, Placebo-Controlled Study. Br J Nutr (2012) 107(6):876–84. 10.1017/S000711451100420X [DOI] [PubMed] [Google Scholar]
- 214. Mercenier A, Muller-Alouf H, Grangette C. Lactic Acid Bacteria as Live Vaccines. Curr Issues Mol Biol (2000) 2(1):17–25. [PubMed] [Google Scholar]
- 215. Szatraj K, Szczepankowska AK, Chmielewska-Jeznach M. Lactic Acid Bacteria - Promising Vaccine Vectors: Possibilities, Limitations, Doubts. J Appl Microbiol (2017) 123(2):325–39. 10.1111/jam.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Wells JM. Immunomodulatory Mechanisms of Lactobacilli. Microb Cell Fact (2011) 10 Suppl 1:S17. 10.1186/1475-2859-10-S1-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Bunout D, Hirsch S, Pia de la Maza M, Munoz C, Haschke F, Steenhout P, et al. Effects of Prebiotics on the Immune Response to Vaccination in the Elderly. JPEN J Parenter Enteral Nutr (2002) 26(6):372–6. 10.1177/0148607102026006372 [DOI] [PubMed] [Google Scholar]
- 218. Duggan C, Penny ME, Hibberd P, Gil A, Huapaya A, Cooper A, et al. Oligofructose-Supplemented Infant Cereal: 2 Randomized, Blinded, Community-Based Trials in Peruvian Infants. Am J Clin Nutr (2003) 77(4):937–42. 10.1093/ajcn/77.4.937 [DOI] [PubMed] [Google Scholar]
- 219. Salvini F, Riva E, Salvatici E, Boehm G, Jelinek J, Banderali G, et al. A Specific Prebiotic Mixture Added to Starting Infant Formula has Long-Lasting Bifidogenic Effects. J Nutr (2011) 141(7):1335–9. 10.3945/jn.110.136747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Stam J, van Stuijvenberg M, Garssen J, Knipping K, Sauer PJ. A Mixture of Three Prebiotics Does Not Affect Vaccine Specific Antibody Responses in Healthy Term Infants in the First Year of Life. Vaccine (2011) 29(44):7766–72. 10.1016/j.vaccine.2011.07.110 [DOI] [PubMed] [Google Scholar]
- 221. van den Berg JP, Westerbeek EA, van der Klis FR, Berbers GA, Lafeber HN, van Elburg RM. Neutral and Acidic Oligosaccharides Supplementation Does Not Increase the Vaccine Antibody Response in Preterm Infants in a Randomized Clinical Trial. PloS One (2013) 8(8):e70904. 10.1371/journal.pone.0070904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Akatsu H, Nagafuchi S, Kurihara R, Okuda K, Kanesaka T, Ogawa N, et al. Enhanced Vaccination Effect Against Influenza by Prebiotics in Elderly Patients Receiving Enteral Nutrition. Geriatr Gerontol Int (2016) 16(2):205–13. 10.1111/ggi.12454 [DOI] [PubMed] [Google Scholar]
- 223. Medina M, Izquierdo E, Ennahar S, Sanz Y. Differential Immunomodulatory Properties of Bifidobacterium Logum Strains: Relevance to Probiotic Selection and Clinical Applications. Clin Exp Immunol (2007) 150(3):531–8. 10.1111/j.1365-2249.2007.03522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Rooks MG, Garrett WS. Gut Microbiota, Metabolites and Host Immunity. Nat Rev Immunol (2016) 16(6):341–52. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Shibata N, Kunisawa J, Kiyono H. Dietary and Microbial Metabolites in the Regulation of Host Immunity. Front Microbiol (2017) 8:2171. 10.3389/fmicb.2017.02171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Ratajczak W, Ryl A, Mizerski A, Walczakiewicz K, Sipak O, Laszczynska M. Immunomodulatory Potential of Gut Microbiome-Derived Short-Chain Fatty Acids (Scfas). Acta Biochim Pol (2019) 66(1):1–12. 10.18388/abp.2018_2648 [DOI] [PubMed] [Google Scholar]
- 227. Nomura M, Nagatomo R, Inoue K, Doi K, Shimizu J, Baba K, et al. 1249p Association of SCFA in Gut Microbiome and Clinical Response in Solid Cancer Patients Treated With andi-PD-1 Antibody. Ann Oncol (2019) 30(Supplement_5):mdz253.074. 10.1093/annonc/mdz253.074 [DOI] [Google Scholar]
- 228. Sim JR, Kang SS, Lee D, Yun CH, Han SH. Killed Whole-Cell Oral Cholera Vaccine Induces Ccl20 Secretion by Human Intestinal Epithelial Cells in the Presence of the Short-Chain Fatty Acid, Butyrate. Front Immunol (2018) 9:55. 10.3389/fimmu.2018.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Guo CJ, Allen BM, Hiam KJ, Dodd D, Van Treuren W, Higginbottom S, et al. Depletion of Microbiome-Derived Molecules in the Host Using Clostridium Genetics. Science (2019) 366(6471):eaav1282. 10.1126/science.aav1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-cell Generation. Nature (2013) 504(7480):451–5. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence That Asthma is a Developmental Origin Disease Influenced by Maternal Diet and Bacterial Metabolites. Nat Commun (2015) 6:7320. 10.1038/ncomms8320 [DOI] [PubMed] [Google Scholar]
- 232. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of Inflammatory Responses by Gut Microbiota and Chemoattractant Receptor GPR43. Nature (2009) 461(7268):1282–6. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity (2014) 40(1):128–39. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-Sensing Receptors GPR43 and GPR109A Facilitate Dietary Fibre-Induced Gut Homeostasis Through Regulation of the Inflammasome. Nat Commun (2015) 6:6734. 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- 235. Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host Microbiota Constantly Control Maturation and Function of Microglia in the CNS. Nat Neurosci (2015) 18(7):965–77. 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Parker LC, Prince LR, Sabroe I. Translational Mini-Review Series on Toll-like Receptors: Networks Regulated by Toll-like Receptors Mediate Innate and Adaptive Immunity. Clin Exp Immunol (2007) 147(2):199–207. 10.1111/j.1365-2249.2006.03203.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Hua Z, Hou B. TLR Signaling in B-cell Development and Activation. Cell Mol Immunol (2013) 10(2):103–6. 10.1038/cmi.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Jin B, Sun T, Yu XH, Yang YX, Yeo AE. The Effects of TLR Activation on T-cell Development and Differentiation. Clin Dev Immunol (2012) 2012:836485. 10.1155/2012/836485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Heer AK, Shamshiev A, Donda A, Uematsu S, Akira S, Kopf M, et al. TLR Signaling Fine-Tunes Anti-Influenza B Cell Responses Without Regulating Effector T Cell Responses. J Immunol (2007) 178(4):2182–91. 10.4049/jimmunol.178.4.2182 [DOI] [PubMed] [Google Scholar]
- 240. Meyer-Bahlburg A, Khim S, Rawlings DJ. B Cell Intrinsic TLR Signals Amplify But are Not Required for Humoral Immunity. J Exp Med (2007) 204(13):3095–101. 10.1084/jem.20071250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Salerno F, Freen-van Heeren JJ, Guislain A, Nicolet BP, Wolkers MC. Costimulation Through TLR2 Drives Polyfunctional Cd8(+) T Cell Responses. J Immunol (2019) 202(3):714–23. 10.4049/jimmunol.1801026 [DOI] [PubMed] [Google Scholar]
- 242. McCarron M, Reen DJ. Activated Human Neonatal CD8+ T Cells are Subject to Immunomodulation by Direct TLR2 or TLR5 Stimulation. J Immunol (2009) 182(1):55–62. 10.4049/jimmunol.182.1.55 [DOI] [PubMed] [Google Scholar]
- 243. Jun JC, Jones MB, Oswald DM, Sim ES, Jonnalagadda AR, Kreisman LSC, et al. T Cell-Intrinsic TLR2 Stimulation Promotes IL-10 Expression and Suppressive Activity by CD45RbHi T Cells. PloS One (2017) 12(7):e0180688. 10.1371/journal.pone.0180688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Li Q, Yan Y, Liu J, Huang X, Zhang X, Kirschning C, et al. Toll-Like Receptor 7 Activation Enhances Cd8+ T Cell Effector Functions by Promoting Cellular Glycolysis. Front Immunol (2019) 10:2191. 10.3389/fimmu.2019.02191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. TLR2 is Expressed on Activated T Cells as a Costimulatory Receptor. Proc Natl Acad Sci USA (2004) 101(9):3029–34. 10.1073/pnas.0400171101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Gringhuis SI, Kaptein TM, Wevers BA, van der Vlist M, Klaver EJ, van Die I, et al. Fucose-Based PAMPs Prime Dendritic Cells for Follicular T Helper Cell Polarization Via DC-SIGN-dependent Il-27 Production. Nat Commun (2014) 5:5074. 10.1038/ncomms6074 [DOI] [PubMed] [Google Scholar]
- 247. Gringhuis SI, Kaptein TM, Wevers BA, Mesman AW, Geijtenbeek TB. Fucose-Specific DC-SIGN Signalling Directs T Helper Cell Type-2 Responses Via IKKepsilon- and CYLD-dependent Bcl3 Activation. Nat Commun (2014) 5:3898. 10.1038/ncomms4898 [DOI] [PubMed] [Google Scholar]
- 248. Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating Nlrp6 Inflammasome Signaling. Cell (2015) 163(6):1428–43. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Ishiguro K, Ando T, Maeda O, Watanabe O, Goto H. Suppressive Action of Acetate on Interleukin-8 Production Via Tubulin-Alpha Acetylation. Immunol Cell Biol (2014) 92(7):624–30. 10.1038/icb.2014.31 [DOI] [PubMed] [Google Scholar]