We have determined that the results reported in “On the Importance of Well Calibrated Scores for Identifying Shotgun Proteomics Spectra” are problematic due to the way that precursor charge state was handled. Correcting the error leads to systematic changes in most of our results; however, the overall trends that we observe and the main conclusions of our study remain unchanged.
In addition, we made one other change to the analysis. The corrected data in Table 2 are now based on using the XCorr score as implemented in the Crux tool Tide, whereas the original, erroneous table, was generated using an older Crux search engine called “search-for-matches.” We made this change so that all of the results in the paper would be generated by the same search engine.
Table 2. Variability in PSM Discoveries Reported by Different Applications of TDC Using Calibrated and Raw XCorr Scores.
| |
% only in one T-TDC (raw score) |
% only in one T-TDC (calibrated score) |
|||||
|---|---|---|---|---|---|---|---|
| set | FDR | 0.01 | 0.05 | 0.10 | 0.01 | 0.05 | 0.10 |
| yeast | 0.05 quantile | 0.03 | 0.4 | 1.1 | 0.0 | 0.1 | 0.4 |
| median | 0.2 | 0.6 | 1.5 | 0.05 | 0.3 | 0.6 | |
| 0.95 quantile | 5.8 | 3.0 | 3.3 | 3.6 | 1.9 | 2.2 | |
| worm | 0.05 quantile | 0.0 | 0.2 | 1.1 | 0.0 | 0.0 | 0.07 |
| median | 0.2 | 0.6 | 1.2 | 0.09 | 0.2 | 0.4 | |
| 0.95 quantile | 7.8 | 5.2 | 4.6 | 5.2 | 3.5 | 3.3 | |
| Plasmodium | 0.05 quantile | 0.0 | 0.3 | 1.0 | 0.0 | 0.0 | 0.1 |
| median | 0.2 | 0.7 | 1.5 | 0.08 | 0.2 | 0.4 | |
| 0.95 quantile | 7.0 | 3.9 | 4.0 | 2.9 | 1.7 | 2.4 | |
Note that in this correction the method that the paper refers to as “Käll et al.” is now called “STDS-PIT” for “separate target–decoy search with percentage of incorrect targets.” This change was made to be consistent with our subsequent work and also because the new name is more descriptive.
We would also like to take this opportunity to clarify that we implemented TDC by comparing two competing separate searches. This approach coincides with our model and, in the case of Tide and the MS-GF+ raw score, is the same as searching the concatenated DB, but the order of the PSMs might differ in the MS-GF+ E-value case.
We also clarify that when STDS-PIT estimates π0, the proportion of incorrect target hits, we used the R package qvalue with the option bootstrap for this estimation.
Detailed List of Changes
Table 2 and Figures 3–8, 11, and 12 have been updated to reflect the new results. In addition, the following three sections of text from the original paper contain numeric values that have changed due to the reanalysis. In the quoted text, the new numbers are followed by their old values in parentheses.
“At FDR 1% and using Xcorr, we observe an increase in the number of discoveries of 31 (22), 12 (8.0), and 30 (31)% for the yeast, worm, and Plasmodium data sets, respectively. Using the MS-GF+ raw score, the corresponding improvements at the same 1% FDR level are 26 (37), 71 (61), and 27 (27)%. Presumably MS-GF+’s raw score is even less calibrated than XCorr.”
“The MS-GF+ E-value score is designed to be calibrated; thus, it is not surprising that at 1% FDR level there is little difference between using the E-value score and its 10K-calibrated version: 0.2 (1.2) and 1.4(3.3)% more calibrated TDC discoveries in the yeast and worm data sets and 0.5 (0.5)% fewer discoveries in the Plasmodium data set. Similarly, at 5% FDR level, the calibrated version of the MS-GF+ E-value identifies 0.6 (1.5)% more discoveries in the yeast data set and 0.2% more (0.2% fewer) discoveries in the Plasmodium data set. It is, however, surprising that at the same 5% FDR level the calibrated version yields 8.4 (12)% more discoveries in the worm data and that number increases to 9.7 (16)% at 10% FDR level. We suspect that some of the assumptions that go into computing the MS-GF+ E-value are violated for the worm data set, but these do not affect our robust albeit costly calibration procedure.”
“For example, at FDR 1%, STDS-PIT (Käll’s method) suggests that when using XCorr calibration increases the number of discoveries by 39 (43), 21 (20), and 49 (53)% for the yeast, worm, and Plasmodium data sets (again, these are median improvements using 1000 independently drawn decoy sets). Notably, this increase in the number of discoveries is substantially larger than we observed previously for TDC using XCorr, which yielded corresponding percentages of 31 (22), 12 (8.0), and 30 (31)%. The corresponding increases at 1% FDR when using MS-GF+’s raw score are 54 (70), 100 (79), and 46(49)% for the 10-K calibrated STDS-PIT over using the raw score (compared with 26 (37), 71 (61), and 27(27)% increases when using TDC).”
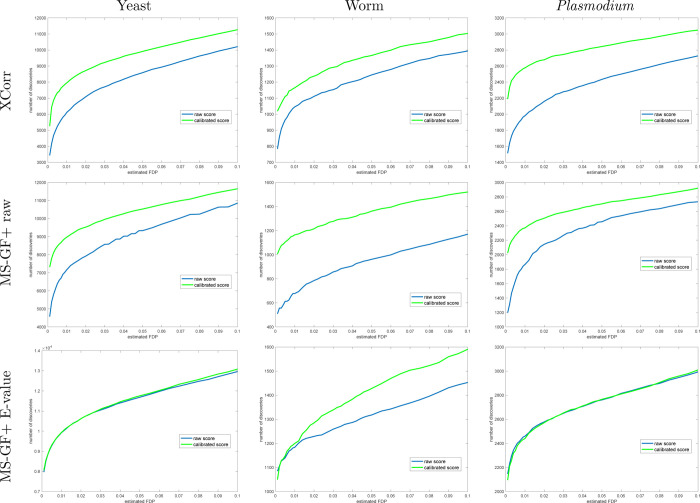
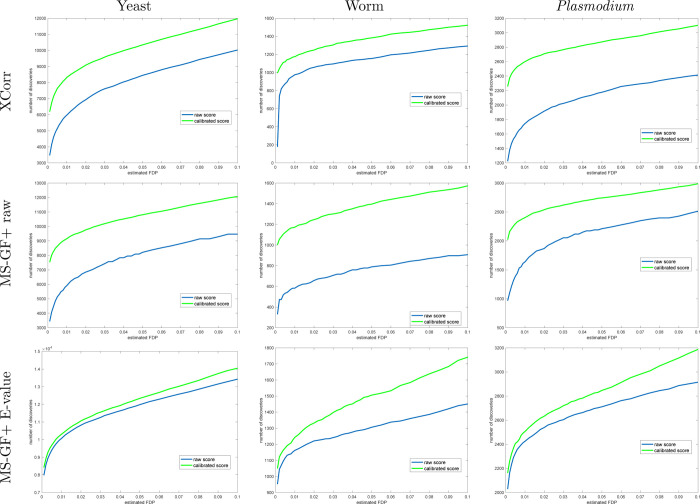
Figure 3.
Calibrating a noncalibrated score on average yields more discoveries (TDC). Each panel plots the median number of discoveries as a function of FDR threshold using TDC applied to the raw and calibrated scores (the median is with respect to 1000 applications, each using a single independently drawn decoy set). Calibration substantially increases the number of TDC discoveries when using the noncalibrated MS-GF+ and Xcorr scores. MS-GF+ E-value is designed as a calibrated score; hence, calibration adds little to the yeast and Plasmodium data sets. Surprisingly, though, calibration makes a substantial impact even on the E-value in the worm data set.
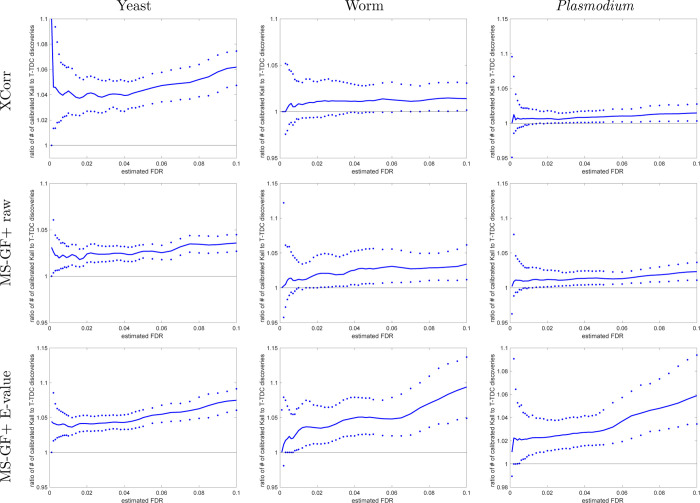
Figure 8.
With calibrated scores, STDS-PIT gives more discoveries than TDC. The ratio of the number of STDS-PIT (Käll) discoveries to TDC discoveries at FDR ≤ 0.1 when using the calibrated score. The median ratio (with respect to 1000 independently drawn decoys) in solid line is flanked by the 0.95 and 0.05 quantiles of the ratios. After calibrating, all three scores typically yield more STDS-PIT than TDC discoveries at any given FDR.
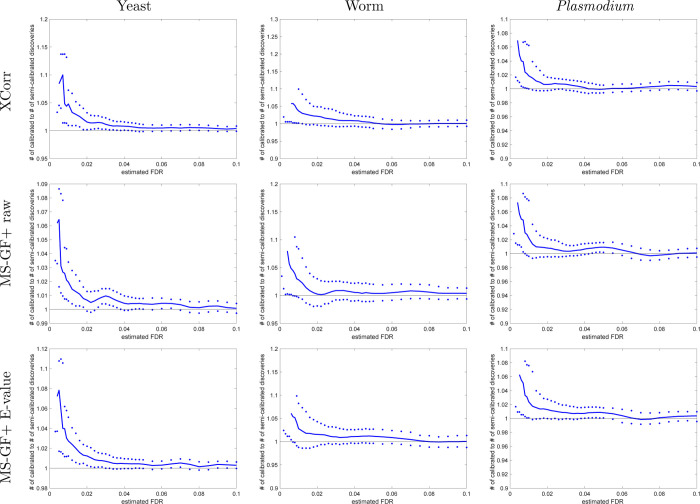
Figure 11.
Semicalibrated scores mostly yield more discoveries than using raw scores (TDC, FDR levels > 2%). Each panel plots, as a function of FDR threshold, the ratio of the number of TDC discoveries at a given FDR threshold when using the semicalibrated score (numerator) versus the number of discoveries at the same FDR when using the raw score (denominator). The solid line represents the median ratio with respect to 1000 independently drawn decoy sets, while the 0.95 and 0.05 quantiles of the ratios are represented as dots.
Figure 12.
1K Semicalibrated scores yield similar improvements to 10K-calibrated scores (TDC, FDR levels > 2%). Same as Figure 11 but now the ratio of the number of TDC discoveries when using the 10K calibrated scores to the number of discoveries when using the 1K (semicalibrated) scores is analyzed. Note that the medians do not extend all the way down to an FDR value of 0.001 because for reasons explained in Section 2.4 of the published paper the number of discoveries using TDC with 1K-calibrated scores is often 0 at the smaller FDR threshold and the ratio is therefore infinity.
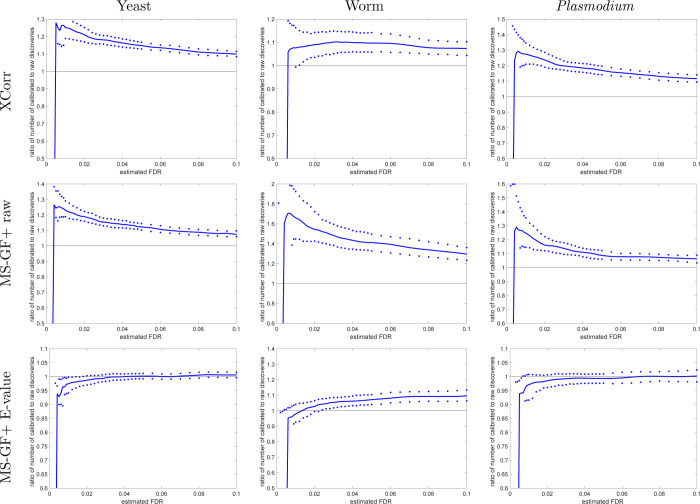
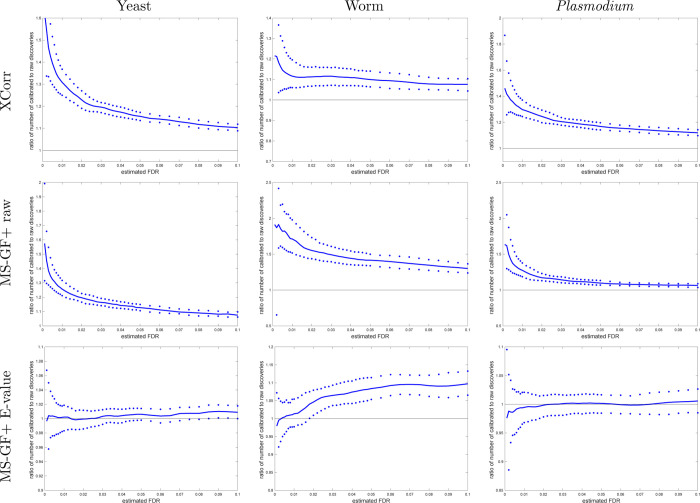
Figure 4.
Calibrating a noncalibrated score mostly yields more discoveries (TDC). Each panel plots, as a function of estimated FDR, the ratio of the number of TDC discoveries at FDR ≤ 0.1 when using the calibrated score (numerator) versus the number of discoveries at the same FDR when using the raw score (denominator). The solid line represents the median ratio (with respect to 1000 ratios, each comparing the raw vs calibrated TDC discoveries using a single independently drawn decoy set), while the 0.95 and 0.05 quantiles of the ratios are represented as dots. For small FDR values, the calibrated score yields considerably more discoveries than the uncalibrated score (MS-GF+ and Xcorr).
Figure 5.
A calibrated score on average yields more discoveries (STDS-PIT Käll) Each panel plots the median number of discoveries as a function of FDR threshold using STDS-PIT (the Käll method) applied to the raw and calibrated scores (the median is with respect 1000 applications, each using a single independently drawn decoy set). For small FDR values, the calibrated score yields considerably more discoveries than the uncalibrated score. Note that even for MS-GF+ E-value our calibration procedure typically increases the number of STDS-PIT discoveries.
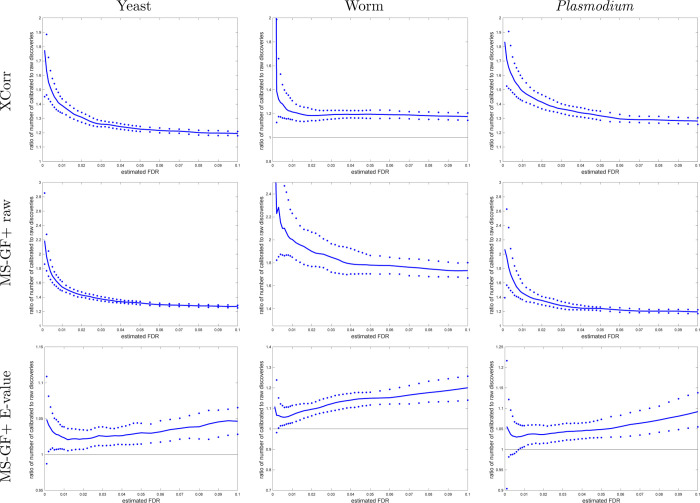
Figure 6.
A calibrated score mostly yields more discoveries (STDS-PIT). Each panel plots, as a function of estimated FDR, the ratio of the number of STDS-PIT (Käll method) discoveries at FDR ≤ 0.1 when using the calibrated score (numerator) versus the number of discoveries at the same FDR when using the raw score (denominator). The solid line represents the median ratio with respect to 1000 independently drawn decoy sets, while the 0.95 and 0.05 quantiles of the ratios are represented as dots.
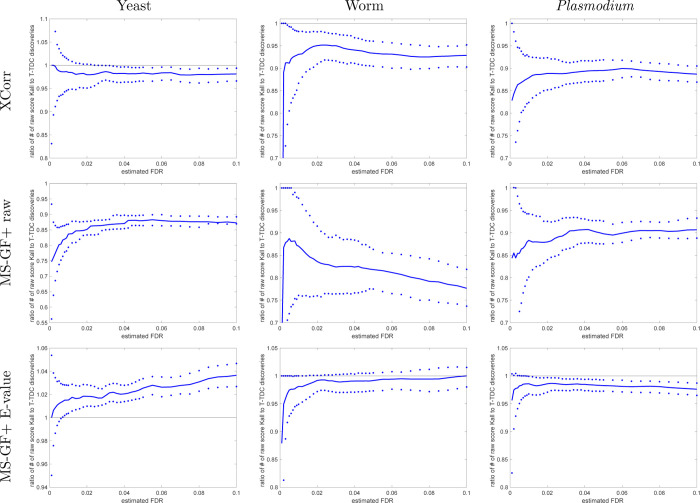
Figure 7.
With noncalibrated scores, STDS-PIT is more conservative than TDC. The ratio of the number of STDS-PIT (Käll) discoveries to TDC discoveries at FDR ≤ 0.1 when using the raw score. The median ratio (with respect to 1000 independently drawn decoys) in solid line is flanked by the 0.95 and 0.05 quantiles of the ratios. Results are for raw scores, but keep in mind that MS-GF+ E-value is partially calibrated even in its “raw” form, so the ratio of STDS-PIT to TDC discoveries fluctuates above and below 1.