Abstract
Osteoarthritis (OA) is a multifactorial pathology and comprises a wide range of distinct phenotypes. In this context, the characterization of the different molecular profiles associated with each phenotype can improve the classification of OA. In particular, OA can coexist with type 2 diabetes mellitus (T2DM). This study investigates lipidomic and proteomic differences between human OA/T2DM– and OA/T2DM+ cartilage through a multimodal mass spectrometry approach. Human cartilage samples were obtained after total knee replacement from OA/T2DM– and OA/T2DM+ patients. Label-free proteomics was employed to study differences in protein abundance and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) for spatially resolved-lipid analysis. Label-free proteomic analysis showed differences between OA/T2DM– and OA/T2DM+ phenotypes in several metabolic pathways such as lipid regulation. Interestingly, phospholipase A2 protein was found increased within the OA/T2DM+ cohort. In addition, MALDI-MSI experiments revealed that phosphatidylcholine and sphingomyelin species were characteristic of the OA/T2DM– group, whereas lysolipids were more characteristic of the OA/T2DM+ phenotype. The data also pointed out differences in phospholipid content between superficial and deep layers of the cartilage. Our study shows distinctively different lipid and protein profiles between OA/T2DM– and OA/T2DM+ human cartilage, demonstrating the importance of subclassification of the OA disease for better personalized treatments.
Keywords: label-free proteomics, MALDI-MSI, spatially resolved-lipid analysis, cartilage, osteoarthritis, diabetes
Introduction
Osteoarthritis (OA) is mainly characterized by the progressive deterioration of articular cartilage, and it is the most common form of arthritis worldwide, affecting over 10% of the population above 60 years.1−3 OA is recognized as an age-related joint disease,4,5 but a variety of other risk factors have been associated with the development of the pathology.6 This observation suggests that OA could be subclassified into distinct phenotypes rather than being a single disease.7,8 The classification of OA patients into different groups can significantly transform OA clinical trials and enhance their efficiency toward a personalized medicine approach.9 In this sense, different models for patient classification have been proposed. For instance, the study carried out by Herrero-Beaumont et al. proposed four clinical phenotypes (biomechanical, osteoporotic, metabolic, and inflammatory), whereas another study suggested the existence of six clinical phenotypes (chronic pain, inflammatory, metabolic syndrome (MetS), bone and cartilage metabolism, mechanical overload, and minimal joint disease) to stratify patients with OA knee.10,11 However, the molecular mechanisms underlying all these OA phenotypes are still poorly described with only two metabolomic studies performed on synovial fluid (SF), revealing the existence of OA subgroups and showing differences from early versus late OA patients, respectively.12,13 Deciphering the biomolecular profiles of each OA phenotype is the first step toward the development and design of more specific and efficient drugs.14,15
From the phenotypes proposed in the literature, the MetS phenotype is defined by chronic low-grade inflammation, which leads to systemic and local molecular changes.16 MetS is a health disorder affecting 10–30% of the worldwide population with a prevalence of 59% in OA patients.17 MetS is diagnosed when a patient has at least three of the following conditions: (i) high blood pressure (≥130/ ≥ 85 mmHg), (ii) high blood glucose levels (≥110 mg/dL at fasting glucose), (iii) high triglyceride levels (≥150 mg/dL), (iv) low HDL-cholesterol (<40 mg/dL in men and <50 mg/dL in women), and (v) a large waist circumference (>102 cm in men and >88 cm in women).18 Moreover, MetS also increases the risk of type 2 diabetes mellitus (T2DM), hypertension, or dyslipidemia.19
Interestingly, different studies have shown a correlation between T2DM and OA20,21 and a higher prevalence of OA in T2DM patients.22,23 Zhang et al. used a targeted approach to study metabolic differences in plasma from OA, T2DM, and OA/T2DM+ patients.24 In this work, different phosphatidylcholine (PC) species were detected with significantly lower plasma concentrations in OA patients with diabetes than OA patients without diabetes suggesting that T2DM might alter PC metabolism leading to further joint deterioration. Other authors have studied the inflammatory response in chondrocytes obtained from OA/T2DM– and OA/T2DM+ patients.25 These results showed that T2DM chondrocytes promote cartilage degradation via interleukin 6 (IL-6), prostaglandin E2 (PGE2) release, reactive oxygen species (ROS), and nitric oxide (NO) production. However, the molecular mechanisms underlying the coexistence of the diseases is not clearly defined yet.
In this work, we hypothesized that OA/T2DM– and OA/T2DM+ human knee cartilage present differential molecular profiles. Here, an untargeted multimodal mass spectrometry (MS) approach was employed for the first time on human knee cartilage to compare the differential protein and lipid profiles of OA/T2DM– and OA/T2DM+ patients. To this end, we combined both label-free proteomics and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) methodologies.
Materials and Methods
Experimental Design and Statistical Rationale
The experimental design and statistical rationale for both the proteomics and MALDI-MSI experiments conducted in this work will be described in more detail in each subsection. Proteomic experiments were performed on cartilage samples obtained from ten OA/T2DM– and ten OA/T2DM+ patients. MALDI-MSI experiments were performed on six OA/T2DM– and six OA/T2DM+ patients due to the amount of human cartilage material obtained after the total knee joint replacement surgery, which differs from patient to patient.
Sample Collection
The Maastricht University Medical Centre’s institutional policy on the use of residual human surgical material states that no informed consent is needed in the case of residual surgical material (METC number of the WMO approval waste material of total knee arthroplasty (TKA): 2017–0183). Human cartilage from OA/T2DM– and OA/T2DM+ patients was obtained from donors undergoing total knee joint replacement. Additional information such as age, gender, affected knee, body mass index (BMI), and Kellgren-Lawrence (KL) scores can be found in Supporting Table S1. Unwounded cartilage areas were selected macroscopically, cut into punches of 8 mm and heat stabilized (Denator, Uppsala, Sweden) to avoid molecular degradation before being snap-frozen in liquid nitrogen and stored at −80 °C.26−28 The punches were finally sectioned at 12 μm thickness with a cryostat (Leica Microsystems, Wetzlar, Germany) at −20 °C (Supporting Figure S1).
Label-Free Proteomics: Protein Extraction
For label-free proteomics experiments, 30 consecutive tissue sections per patient were collected in Eppendorf tubes. 100 μL of 50 mM ammonium bicarbonate (ABC) buffer (Sigma-Aldrich, Zwijndrecht, The Netherlands) containing 5 M Urea (GE Healthcare, Eindhoven, The Netherlands) and 0.2% Rapigest (Waters Corporation, Milford, MA, USA) were added. Proteins were then extracted by freeze–thaw cycles (3×), with 1 min of sonication step between each cycle. After 30 min of centrifugation at 14 000 rpm at 4 °C, the supernatant containing proteins was finally transferred into new tubes and stored at −80° until further analysis.
Label-Free Proteomics: Gel Electrophoresis
Bradford assay (Biorad, Lunteren, The Netherlands) was performed to assess the protein concentration. Ten μg of proteins of each patient were loaded on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Biorad) and run shortly for 4 min at 180 V. The gel was stained with Coomassie blue (Sigma-Aldrich) for protein visualization. The protein bands were excised from the gel and processed on a MassPREP digestion robot (Waters, Manchester, UK). The destaining of the Coomassie was performed using 50 mM of ABC buffer mixed with 50% acetonitrile (Biosolve, Valkenswaard, The Netherlands). Then, cysteines were reduced with 10 mM dithiothreitol (Sigma-Aldrich) in 100 mM ABC for 30 min followed by alkylation with 55 mM iodoacetamide (Sigma-Aldrich) in 100 mM ABC for 20 min in the dark. Samples were washed with 100 mM ABC and dehydrated with 100% acetonitrile. Trypsin (Promega, Leiden, The Netherlands) at 6 ng/μL in 50 mM ABC was added to the gel plugs and incubated at 37 °C for 5 h. The peptides were finally extracted three times with 50 μL of 1% formic acid (Biosolve) and 2% acetonitrile. The volume of the peptide extract was reduced in a speedvac (Eppendorf, Nijmegen, The Netherlands) until a final volume of 50 μL was reached.
Label-Free Proteomics: Data Acquisition and Processing
400 ng of proteins were injected for liquid-chromatography mass spectrometry (LC-MS/MS) analysis. The samples were randomized and blanks between runs were injected to minimize the carry over. In addition, the samples were run the same day to avoid batch effects. Peptide separation was performed on a Thermo Fisher Scientific Dionex Ultimate 3000 Rapid Separation ultrahigh-performance liquid-chromatography (HPLC) system (Thermo Scientific, Waltham, MA, USA) equipped with an Acclaim PepMap C18 analytical column (2 μm, 75 μm × 500 mm, 100 Å). The samples were first trapped on an online C18 column for desalting. The peptides were then separated on the analytical column with a 180 min linear gradient from 5% to 45% acetonitrile/0.1% formic acid and a flow rate set at 300 nL/min. The HPLC system was coupled online to a high-mass resolution Orbitrap MS Q-Exactive instrument (Thermo Scientific) with a nanoelectrospray Flex ion source (Proxeon, Thermo Scientific). The mass spectrometer was operated in positive ion polarity in data-dependent acquisition (DDA) mode with the following settings: Full MS scan of the mass range m/z 250–1250 at a resolution of 70 000 at m/z 200 with a maximum injection time of 120 ms, followed by tandem mass spectrometry (MS/MS) scans for the fragmentation of the 10 most intense ions at a resolution of 17 500. Isolation of precursors was performed with a 1.8 m/z window and a maximum injection time of 200 ms. The ions already selected for fragmentation were dynamically excluded for 30 s. External calibration of the instrument was performed using a standard calibration solution for positive ion mode (Pierce LTQ Velos ESI positive ion calibration solution, Thermo Scientific).
For protein identification, raw files were processed within the Proteome Discoverer software version 2.2 (Thermo Scientific) using the search engine Sequest version 2.2 with the Swiss-Prot human database Homo sapiens version 2020–02–10 (TaxID 9606). The following parameters were used for the database search: Carbamidomethylation of C for fixed modifications; oxidation of M and acetylation of protein N-term for variable modifications; trypsin for enzyme with a maximum of two missed cleavages; precursor mass tolerance was set at 10 ppm and fragment tolerance at 0.02 Da; minimum and maximum peptide length of 6 and 144 amino acids, respectively. Normalization of the data was performed on the total peptide amount. Percolator was used for the decoy database search and the false discovery rate (FDR) was fixed at 1% maximum. For protein quantitation, the Minora Feature Detector node in the processing step and the Feature Mapper node combined with the Precursor Ions Quantifier node in the consensus step were used with default settings.
Label-Free Proteomics: Data Analysis
ANOVA test was performed within the Proteome Discoverer software to analyze the statistical significance of variation observed in protein abundances between the OA/T2DM– and OA/T2DM+ patients. Proteins were considered modulated with an adjusted p-value ≤0.05 and a fold change (FC) cutoff set at 1.5-fold.29−31 The modulated proteins were then imported within the EnrichR software to display the top 10 down or up-regulated pathways ranked by the combined score, as it has been reported to be the best ranking (https://maayanlab.cloud/Enrichr/help#background&q=4).32 WikiPathways and KEGG were used as databases (version 2019, Human).
MALDI-MSI: Matrix Application
Twelve μm tissue sections were thaw mounted over cleaned indium tin oxide (ITO) glass slides (Delta Technologies, Loveland, CO, USA) coated with double-sided conductive copper tape 1182 (3 M Science, St. Paul, MN, USA). Samples were randomized to correct for any potential batch effect. Norharmane (Sigma-Aldrich) was used as a matrix for lipid analysis. Eight layers of matrix at 7 mg/mL in chloroform/methanol (2:1, (v/v)) were sprayed at 30 °C with a flow rate of 0.12 mL/min using the HTX TM-Sprayer system (HTX Imaging, Chapel Hill, NC, USA). A drying time of 30 s between each layer and a velocity set at 1200 mm/min were used to enable homogeneous matrix application.
MALDI-MSI: Data Acquisition and Processing
MALDI-MSI experiments were performed in positive ion polarity at 50 μm of lateral resolution using a RapifleX MALDI Tissue-typer instrument operating in reflectron mode (Bruker, Bremen, Germany) to compare the lipidomic profiles of OA/T2DM– and OA/T2DM+ patients. Lipids were detected over the mass range m/z 300–1600.
A Waters Synapt G2Si coupled to a μMALDI source (Waters, Manchester, UK) with a lateral resolution of 15 μm in combination with the Waters Research Enabled Software (WREnS) was employed for high spatial resolution MALDI-MSI experiments.33 The following settings were used for data acquisition: mass range m/z 400–1200, automatic quadrupole mode profile and laser pulse energy of 300 (a.u.). External calibration on both instruments was performed using the standard calibration mixture of red phosphorus (Sigma-Aldrich) with a mass error below 10 ppm.
MALDI-MSI: Data Analysis
For the MALDI-MSI comparative approach, raw data were processed by SCiLS lab software version 2019b (SCiLS, Bremen, Germany). Principal component analysis (PCA) and linear discriminant analysis (LDA) were performed after peak picking using an in-house-built ChemomeTricks toolbox for MATLAB version 2012B (The MathWorks, Natick, MA, USA).34
For the μMALDI experiments, data were processed using the High-Definition Imaging software version 1.4 (Waters, Manchester, UK) by extracting the 1000 most abundant ions with the following settings: m/z window set at 0.02 Da and MS resolution at 15 000. Processed data were then exported to imzML format before being imported into SCiLS lab MultiVendor Support (MVS) software version 2019b. Within the software, the standard segmentation pipeline using bisecting k-means algorithm and a probabilistic latent semantic analysis (pLSA) with random initialization were performed on the data normalized by total ion current (TIC) with weak denoising and with an m/z interval width set to ±0.1 Da.
MALDI: Lipid Identification
Lipids of interest from the PCA-LDA analysis were identified using targeted MS/MS analysis performed in the ion trap of an Orbitrap Elite hybrid ion trap mass spectrometer (Thermo Scientific). Collision-induced dissociation (CID) was used to fragment the precursors with an isolation window of 1 Da and with a collision energy (CE) set up at 30 eV. Each precursor from the inclusion list was measured for 1 min in a continuous acquisition mode. To obtain the accurate mass, 25 full MS scans over the mass range m/z 340–1300 were acquired with the Orbitrap (resolution of 240 000 at m/z 400). For low abundant, odd-fatty acyl containing species, a lipid extraction was performed in chloroform/methanol (2:1, (v/v)) with 50 μM ammonium formate (Biosolve) to enhance their detection. The extract was then analyzed using a high-mass resolution Orbitrap MS Q-Exactive instrument (resolution of 240 000 at m/z 200) (Thermo Scientific) with electrospray ionization (ESI). These species were detected in negative ion mode as formate adducts.35 MS/MS was performed in the higher-energy collisional dissociation (HCD) cell with a normalized collision energy (NCE) of 30 and 1 Da isolation window. External calibration of the instrument was performed as described in the Label-Free Proteomics: Data Acquisition and Processing section. Lipids were assigned manually using Xcalibur software version 4.2 (Thermo Scientific) and the online ALEX123 library.36,37
Histological Staining
Matrix was removed by immersion in 70% ethanol. The sections were then stained using hematoxylin (Sigma-Aldrich) for 8 min, Safranin O 0,1% (Sigma-Aldrich) dissolved in ultrapure H2O for 2 min and Fast Green FCF 0.1% (Sigma-Aldrich) dissolved in ultrapure H2O for 4 min. Finally, digital images were acquired with the M8 scanner (Precipoint, Freising, Germany) after dehydrating steps.
Results
Protein Pathway Differences between OA Phenotypes
A label-free proteomic analysis has been performed to assess the differences in protein abundances between OA/T2DM– and OA/T2DM+ cartilage samples. After data processing, a total of 1619 proteins were identified. Among them, 115 displayed statistically significant differences (adjusted p-value ≤0.05; FC cutoff set at 1.5-fold) between the two conditions (Supporting Table S2). Among those, 75 were more abundant in OA/T2DM– samples, whereas 40 were more abundant in OA/T2DM+ patients. Enrichment analysis performed on these proteins revealed specific pathways of interest (Table 1 and Supporting Table S3).
Table 1. Top 10 Enriched Pathways Ranked by the Combined Scorea.
| (A) WikiPathways database (Human) – OA/T2DM– group | ||
|---|---|---|
| pathway ID | pathway description | combined score |
| WP3601 | composition of lipid particles* | 451.49 |
| WP1533 | vitamin B12 metabolism* | 217.78 |
| WP4522 | metabolic pathway of LDL, HDL and TG* | 214.40 |
| WP12 | osteoclast signaling | 214.40 |
| WP408 | oxidative stress | 200.60 |
| WP3967 | miR-509-3p alteration of YAP1/ECM axis | 197.94 |
| WP176 | folate metabolism* | 147.28 |
| WP2911 | miRNA targets in ECM and membrane receptors | 140.40 |
| WP100 | glutathione metabolism | 132.24 |
| WP111 | electron transport chain (OXPHOS system in mitochondria) | 130.27 |
| (B) KEGG database (Human) – OA/T2DM– group | |
|---|---|
| name | combined score |
| protein digestion and absorption | 158.74 |
| riboflavin metabolism* | 117.32 |
| ECM-receptor interaction | 107.60 |
| glutathione metabolism | 95.87 |
| oxidative phosphorylation | 88.73 |
| arachidonic acid metabolism* | 80.90 |
| taurine and hypotaurine metabolism | 77.74 |
| Huntington disease | 77.55 |
| focal adhesion | 73.86 |
| fatty acid biosynthesis* | 62.42 |
| (C) WikiPathways database (Human) – OA/T2DM+ group | ||
|---|---|---|
| pathway ID | pathway description | combined score |
| WP1589 | folate-alcohol and cancer pathway hypotheses | 223.62 |
| WP4225 | pyrimidine metabolism and related diseases | 223.62 |
| WP4146 | macrophage markers | 223.62 |
| WP206 | fatty acid omega oxidation* | 117.34 |
| WP497 | urea cycle and metabolism of amino groups | 80.93 |
| WP2533 | glycerophospholipid biosynthetic pathway* | 75.94 |
| WP3580 | methionine de novo and salvage pathway | 71.45 |
| WP1539 | angiogenesis | 63.73 |
| WP2878 | PPAR alpha pathway* | 57.32 |
| WP167 | eicosanoid synthesis* | 54.52 |
| (D) KEGG database (Human) – OA/T2DM+ group | |
|---|---|
| name | combined score |
| fat digestion and absorption* | 347.52 |
| pantothenate and CoA biosynthesis* | 86.52 |
| renin-angiotensin system | 67.40 |
| vitamin digestion and absorption* | 63.73 |
| alpha-linolenic acid metabolism* | 60.38 |
| linoleic acid metabolism* | 49.56 |
| beta-alanine metabolism | 45.32 |
| pancreatic secretion | 41.93 |
| drug metabolism | 36.35 |
| glycine, serine, and threonine metabolism | 32.04 |
(A) Top 10 enriched pathways in the OA/T2DM– group from WikiPathways database. (B) Top 10 enriched pathways in the OA/T2DM– group from KEGG database. (C) Top 10 enriched pathways in the OA/T2DM+ group from WikiPathways database. (D) Top 10 enriched pathways in the OA/T2DM+ group from KEGG database. The (*) displays the pathways related to lipid regulation.
Interestingly, a high number of lipid-related pathways were found in both OA/T2DM– and OA/T2DM+ phenotypes (Table 1A–D). In particular, composition of lipid particles, vitamin B12 metabolism, and metabolic pathway of low-density lipoprotein (LDL), high-density lipoprotein (HDL), and triglyceride (TG) were considered significantly characteristic of the OA/T2DM– group (Table 1A).
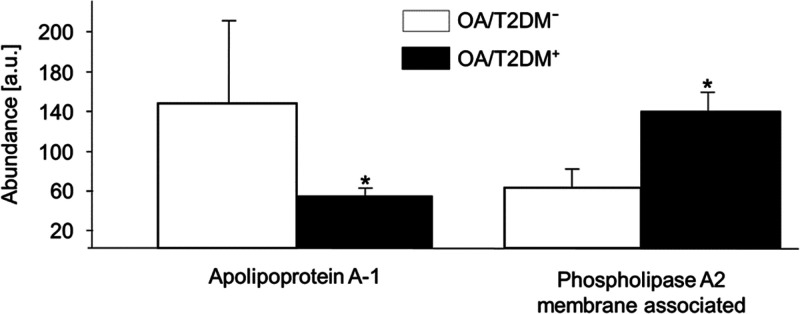
Using the KEGG database, we also identified three lipid metabolic pathways upregulated in the OA/T2DM– group (Table 1B). In addition, fat digestion and absorption, pantothenate and CoA biosynthesis, vitamin digestion and absorption, alpha-linolenic acid metabolism and linoleic acid metabolism were upregulated in the OA/T2DM+ group (Table 1D). In particular, the apolipoprotein A-1 (APOA1) has been found in 4/7 of the lipid metabolic pathways in the OA/T2DM– group (Supporting Table S3A,B). The phospholipase A2 protein, membrane associated (PLA2G2A) has been found linked to 5/9 lipid metabolic pathways in the OA/T2DM+ group (Supporting Table S3C,D). The relative intensities of these proteins are displayed in Figure 1.
Figure 1.
Relative intensities of apolipoprotein A-1 and phospholipase A2 proteins in OA/T2DM– and OA/T2DM+ cohorts. Apolipoprotein A1 protein has been found increased in the OA/T2DM– group, whereas phospholipase A2 protein has been found increased in the OA/T2DM+ group (*adjusted p-value ≤0.05).
In addition to the lipid-related pathways, oxidative stress-related pathways (oxidative stress; glutathione metabolism; electron transport chain [OXPHOS system in mitochondria]; oxidative phosphorylation) and extracellular matrix (ECM) related pathways (osteoclast signaling; miR-509-3p alteration of YAP1/ECM axis; miRNA targets in ECM and membrane receptors; ECM-receptor interaction; focal adhesion) are found enriched in OA/T2DM– patients (Table 1A,B). Moreover, pathways related to organic compounds metabolism (pyrimidine metabolism and related diseases; urea cycle and metabolism of amino groups; methionine de novo and salvage pathway; beta-Alanine metabolism; glycine, serine, and threonine metabolism) have been found enriched in OA/T2DM+ patients (Table 1C,D).
Lipid Profile Differences between OA Phenotypes and Intratissue Heterogeneity
Since several lipid metabolism-related proteins were altered in both OA groups, spatially resolved MALDI-MSI experiments were performed to study lipid differences between both phenotypes. These experiments followed by PCA-LDA analysis on 1354 spectra, showed overall differences between OA/T2DM– and OA/T2DM+ patients based on their specific lipidomic profiles, with a PCA and DA variances of 38.1% and 1.15%, respectively. The projection of the discriminant function 1 (DF1) scores confirmed the lipid content differences between OA/T2DM– and OA/T2DM+ patients (Figure 2A and Supporting Figure S2). Interestingly, more lysolipid species were found in OA/T2DM+, whereas sphingomyelin (SM) and PC species were more characteristic of OA/T2DM– patients (Figure 2B).
Figure 2.
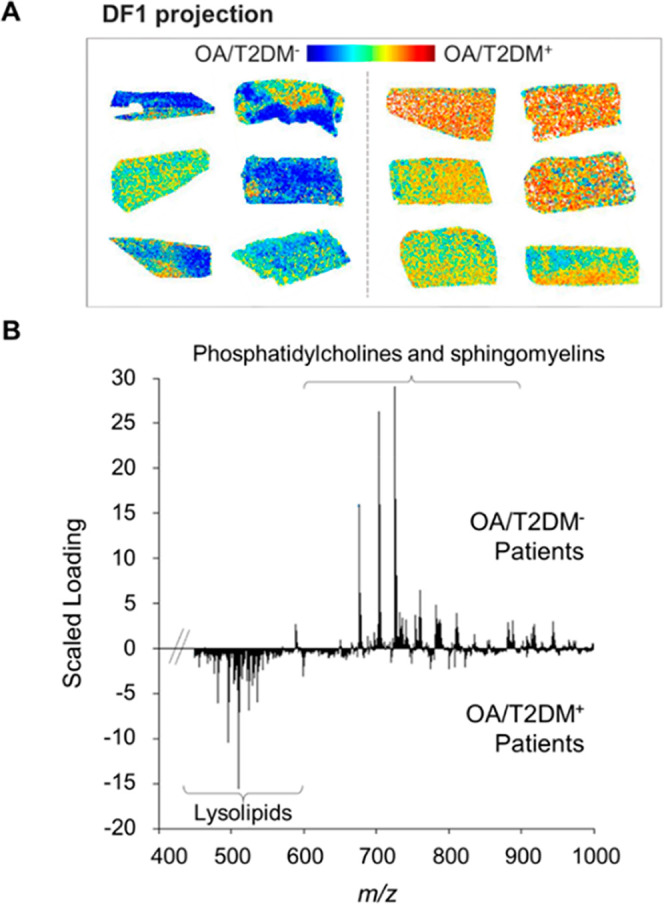
Linear discriminant analysis of OA/T2DM– and OA/T2DM+ patients based on their lipid signature. (A) Discriminant function 1 (DF1) score projection. (B) DF1 scaled loading spectrum.
MALDI-MSI experiments also revealed overall differences between deep and superficial layers of the cartilage tissues based on their specific lipidomic profiles. PCA followed by LDA was performed, resulting in variances of 38.2% and 8.2%, respectively. The projection of the DF1 scores (Figure 3A) and the associated loading spectrum (Figure 3B) confirmed the differences between superficial and deep layers.
Figure 3.
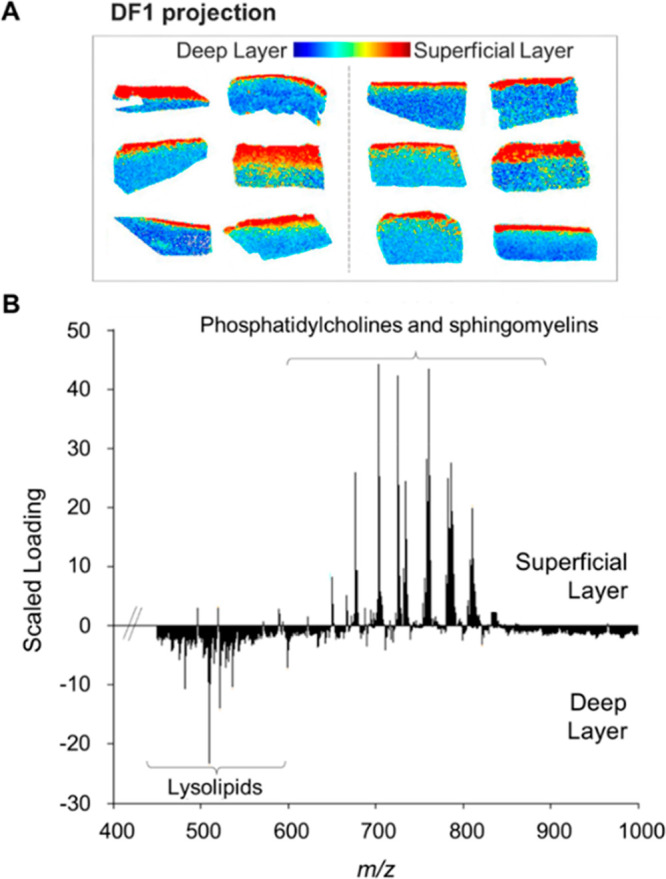
Linear discriminant analysis of superficial and deep cartilage layers based on their specific lipid signature. (A) DF1 score projection. (B) DF1 scaled loading spectrum of the superficial and deep layers of human cartilage. The LDA analysis was performed on both groups.
Since MALDI-MSI preserves the spatial molecular information, a second PCA-LDA analysis was performed after manual annotation to compare the superficial and deep layers of both groups independently. These results showed that PC and SM species were characteristic of the superficial layers and more abundant in OA/T2DM– patients, whereas lysolipid species were more characteristic of the deep layers and more present in the OA/T2DM+ cohort (Supporting Figure S3). Additional high spatial resolution MALDI-MSI was performed to confirm this differential lipid distribution through the cartilage. Experiments at 15 μm of lateral resolution followed by segmentation and pLSA analysis, are in line with the previous data (Figure 4) with additional spatial information: transitional layers (represented by yellow and pink colors), lacunae (represented in light blue color), and chondrocyte cells (represented in blue color). Figure 4B shows that the lipid differences between superficial and deep areas are mainly caused by the specific lipid profiles of chondrocyte cells from these layers.
Figure 4.
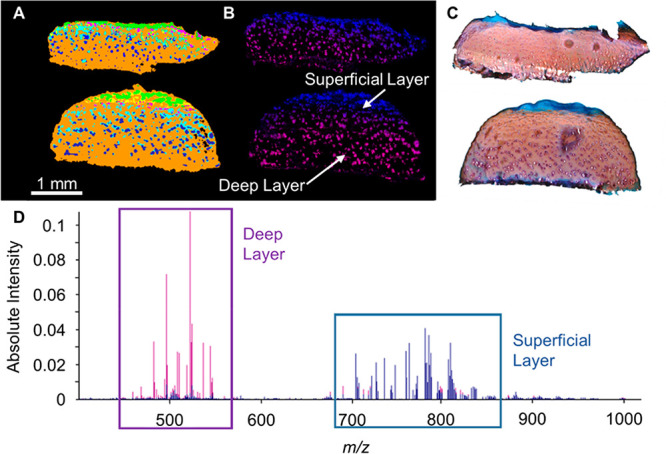
μMALDI experiment at 15 μm raster size. (A) Segmentation of OA/T2DM– tissue (top) and OA/T2DM+ tissue (bottom). The transitional layers are represented by yellow and pink colors, the lacunae in light blue color and the chondrocyte cells in blue color. (B) pLSA analysis with random initialization. The superficial layer (blue) is represented by component 1 and the deep layer (purple) is represented by component 4. (C) Safranin O staining. (D) pLSA loading spectra of selected components 1 and 4.
Finally, targeted MS/MS experiments were performed to identify the lipids of interest from the PCA-LDA analysis (Supporting Table S4). Low abundances prohibited the complete identification of some of the lipids and resulted in tentative assignments.
Discussion
Studies that highlight proteomic and lipidomic differences between healthy and OA tissues or fluids are common in the literature.38−40 However, OA is a multifactorial disease and could be subclassified in different phenotypes. To the best of our knowledge, this is the first study that compares the molecular profiles of OA/T2DM– and OA/T2DM+ human knee cartilage.
In this study, 7 pathways involved in lipid metabolism have been found enriched in the OA/T2DM– group. Interestingly, the APOA1 protein has been found in 4/7 of these lipid metabolic pathways. APOA1 protein is the major constituent of HDL, and therefore plays a key role in the lipoprotein metabolism.41 De Seny et al. demonstrated the potential pro-inflammatory properties of the protein in the context of OA disease.42 A higher level of APOA1 was observed in the serum of healthy control individuals compared to OA patients.43 Moreover, T2DM has been recently correlated with low levels of APOA1 in serum.44
Second, a total of 9 lipid metabolic pathways have been found enriched in the OA/T2DM+ group. Here, the PLA2G2A protein has been found linked to 5/9 of these pathways. PLA2G2A is an enzyme that hydrolyzes phospholipid species, liberating free fatty acids (most importantly arachidonic acid) and lysophospholipids. Arachidonic acid is the common precursor of the eicosanoids, a family of compounds with multiple roles in the inflammation process.45 Interestingly, the PLA2G2A protein has been found to be linked to T2DM46,47 and OA pathologies,48,49 but no comparison between OA/T2DM– and OA/T2DM+ groups has been performed so far.
From all of the lipid metabolic pathways, many were related to the fatty acid metabolism. Fatty acids are important components of membrane lipids and are considered as an attractive target to regulate diseases such as obesity, diabetes, cancer, and cardiovascular complications.50 Interestingly, the fatty acid omega oxidation pathway has been found enriched in the OA/T2DM+ group. The omega oxidation of fatty acids is an alternative pathway when the beta oxidation is blocked and increases with diabetes.51
In addition to lipid metabolic pathways, the proteome analysis revealed other important biological processes. For instance, four pathways associated with cellular oxidative stress have been found increased in the OA/T2DM– group. This biological process is related to OA disease in the literature.52−54 The oxidative stress reflects the imbalance between free radicals such as ROS and antioxidants in the body, which can induce inflammatory responses and contribute to the pathophysiology of diseases.55 Interestingly, mitochondrial superoxide dismutase (SOD2) and extracellular superoxide dismutase (SOD3) proteins are associated with these pathways. SOD2 and SOD3 represent two out of three members of the SOD family and are the most important line of antioxidant defense systems against ROS.56 In our case, we hypothesized that more ROS could be generated by the coexistence of both OA and T2DM diseases when compared to OA/T2DM– patients, which might be caused by a lower expression of both SOD2 and SOD3 in OA/T2DM+ patients. In addition to cellular oxidative stress pathways, ECM-related pathways have been found enriched in the OA/T2DM– group. The degradation of ECM is known to be linked to the cartilage loss over the progression of OA pathology. In comparison, site-specific advanced glycation end-product (AGEs) modifications in the ECM proteins have been shown in T2DM disease.57 Future studies applied on cartilage comparing both phenotypes and targeting specifically the ECM structure should be considered in order to investigate the key molecules associated with each disease.
Five other pathways have been found associated with the metabolism of organic compounds within the OA/T2DM+ cohort. From these pathways, the organic compounds displayed have been associated with T2DM,58−60 but so far, no studies investigated the complete role of these metabolites in the context of the disease.
Proteins that play an important role in OA and/or T2DM can also be pointed out in this study. In fact, most of the 75 up-regulated proteins found in the OA/T2DM– samples have been identified and linked to cartilage tissues and/or OA disease in other studies (Supporting Table S2).61,62 Moreover, other proteins such as the apolipoprotein A-IV (APOA4) and the serine protease HTRA1 have been found increased in the OA/T2DM+ group (FC: 1.59 and 1.63, respectively). APOA4 protein is involved in protection against atherosclerosis and diabetes, which renders it as a potential target for therapeutic purposes.63 The serine protease HTRA1 protein might affect the chondrocyte metabolism and therefore play a role in the progression of OA.64
Since many proteins related to lipid signaling were differently modulated in both groups, we then focused on the investigation of lipid patterns between both patient groups. MALDI-MSI has been described as a powerful method to investigate the distribution of lipids through cartilage tissue sections. Especially, MALDI-MSI applied on cartilage enables the differentiation of the superficial layer and the deep layer of the cartilage, and provides spatial molecular information.26,65 Lipid content differences between healthy and OA cartilage have been studied by time-of-flight secondary ion mass spectrometry (TOF-SIMS).66 However, this technology causes extensive molecular fragmentation and needs long acquisition times.
In our study, a low lipid signal was detected using MALDI-MSI in negative ion mode. This problem has been already shown with the same matrix on cartilage tissue.67 SM, PC, and lysolipid (mostly lysophosphatidylcholine (LPC)) species have been detected using MALDI-MSI in positive ion mode. SM species are structural components of the cell membrane and play a role in cell growth, cell differentiation, and programmed cell death.68−70 On the other hand, PC species are one of the most abundant phospholipids in all mammalian cell membranes,71 whereas LPC species are lipids with pro-inflammatory properties produced under pathological conditions.72,73
PC and SM have been mainly detected in the superficial layer of the cartilage. Interestingly, a study conducted by Sarma et al. showed that these species were the major components of the lipid layer present at the surface of articular cartilage.74 This specific localization of PC and SM species could also be linked to a different chondrocyte type/role in the superficial and the deep layers.75 Interestingly, SM 34:1 is the most abundant SM species in SF and its level is elevated in OA and rheumatoid arthritis.76 This lipid has been also found in chondrocyte cell pellets cultured in 2.5% of oxygen tension.77
According to Zhang et al., the ratio between LPC and PC in the serum could be used to predict advanced knee OA.78 Indeed, the activation of the PC to LPC pathway seems to be associated with OA knee cartilage volume loss over time.48 Here, the higher level of LPC in OA/T2DM+ compared to OA/T2DM– condition could be linked to the coexistence of the two diseases, accentuating the inflammatory process and the view of the OA pathology as a systemic rather than a local pathology. This hypothesis is supported by studies showing an increased level of LPC in the plasma of diabetic patients79 and the link between saturated fatty acids to insulin resistance.80 This insulin resistance can also be associated with the pancreatic secretion pathway found within the OA/T2DM+ group (Table 1D).
The combination of the proteomics data with the spatially resolved lipidomics data also revealed interesting findings. For instance, we observed a higher abundance of PLA2G2A protein in the OA/T2DM+ group, which could be in line with the higher LPC content also associated with this group and the low PC levels found in plasma by other authors.24 Indeed, studies have found that PLA2G2A protein was enriched in the deep layer of the cartilage, which supports this hypothesis as LPC species have been mainly localized in the same area.61,81 We therefore postulated that the overexpression of PLA2G2A protein within the OA/T2DM+ cohort could explain an increased production of LPC in the deep layers of the cartilage tissue. In addition, we previously hypothesized that more ROS could be generated by the coexistence of both OA and T2DM diseases. Interestingly, the production of more ROS can be linked in the literature to a higher LPC content.82−84 Altogether, the data demonstrated the need to consider the LPC species as potential new targets to distinguish the two OA phenotypes described in this study.
In our study, we also demonstrated new spatial information using high spatial resolution MALDI-MSI. For instance, we showed that the chondrocyte cells, the lacunae, and the transitional layer displayed their own lipid content. In addition, hypertrophic chondrocyte cells seem to have a lower phospholipid content compared to chondrocytes from the superficial areas. These data show the need to study the molecular complexity of cartilage tissue at cellular resolution in the future.
One limitation of our study is the sample size, and future efforts should hence focus on the validation of these findings in a larger cohort. A small sample size restricts indeed the possibility to correct for other biological parameters such as gender differences (four males and two females in the OA/T2DM– group, and five males and one female in the OA/T2DM+). In addition, drug treatments that may affect lipid levels have not been taken into account in this study but should also be considered in the future.85,86
In this work, we showcased specific proteins and lipids associated with each phenotype as well as differences in phospholipid content between superficial and deep layers of the cartilage. Future efforts should focus on the biological validation of the data presented with functional studies, which will ultimately improve the existing classification models.
Acknowledgments
The work was performed as part of the M4i research program that was financially supported by the Dutch Province of Limburg through the “LINK” program and by the MUMC institutional grant for clinical collaborative research. The authors thank Ronny Mohren for his support on proteomics method optimization and data interpretation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00186.
Table S1: Patient information; Table S2: Significantly up- and down-regulated proteins found in OA/T2DM– compared to OA/T2DM+; Table S3: Top 10 enriched pathways associated with the significant up- and down-regulated proteins found in OA/T2DM– compared to OA/T2DM+; Table S4: Assignments of 15 lipid species based on targeted MS/MS experiments performed at high-mass resolution; Figure S1: Sample collection and processing; Figure S2: Example of the spatial distribution of SM and LPC species in OA/T2DM– and OA/T2DM+ patients; and Figure S3: Linear discriminant analysis of superficial and deep layers of both groups independently (PDF)
Author Contributions
M.R.E.-M., P.J.E., A.B., R.M.A.H., and B.C.-P. designed the experiments. M.R.E.-M., B.S.R.C., and F.G.B. performed experiments. M.R.E.-M., B.S.R.C., F.G.B., R.M.A.H., and B.C.-P. analyzed the data. M.R.E.-M., P.J.E., A.B., B.S.R.C., F.G.B., R.M.A.H., and B.C.-P. wrote the paper. P.J.E. provided cartilage tissues.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identified PXD021487.
Supplementary Material
References
- Gonzalez Gil A. B.; Llombart Blanco R.; Diaz de Rada P. Validity of magnetic resonance arthrography as a diagnostic tool in femoroacetabular impingement syndrome. Rev. Esp. Cir. Ortop. Traumatol. 2015, 59, 281–286. 10.1016/j.recote.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Helmick C. G.; Felson D. T.; Lawrence R. C.; Gabriel S.; Hirsch R.; Kwoh C. K.; Liang M. H.; Kremers H. M.; Mayes M. D.; Merkel P. A.; Pillemer S. R.; Reveille J. D.; Stone J. H. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Arthritis Rheum. 2008, 58, 15–25. 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q.; Jordan J. M. Epidemiology of Osteoarthritis. Clinics in Geriatric Medicine 2010, 26, 355–369. 10.1016/j.cger.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeser R. F. Aging and osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 492–496. 10.1097/BOR.0b013e3283494005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacitharan P. K. Ageing and Osteoarthritis. Subcell. Biochem. 2019, 91, 123–159. 10.1007/978-981-13-3681-2_6. [DOI] [PubMed] [Google Scholar]
- Haq I.; Murphy E.; Dacre J. Osteoarthritis. Postgrad. Med. J. 2003, 79, 377–83. 10.1136/pmj.79.933.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruyere O.; Cooper C.; Arden N.; Branco J.; Brandi M. L.; Herrero-Beaumont G.; Berenbaum F.; Dennison E.; Devogelaer J. P.; Hochberg M.; Kanis J.; Laslop A.; McAlindon T.; Reiter S.; Richette P.; Rizzoli R.; Reginster J. Y. Can We Identify Patients with High Risk of Osteoarthritis Progression Who Will Respond to Treatment? A Focus on Epidemiology and Phenotype of Osteoarthritis (vol 32, pg 179, 2015). Drugs Aging 2017, 34, 411–411. 10.1007/s40266-017-0456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Spil W. E.; Kubassova O.; Boesen M.; Bay-Jensen A. C.; Mobasheri A. Osteoarthritis phenotypes and novel therapeutic targets. Biochem. Pharmacol. 2019, 165, 41–48. 10.1016/j.bcp.2019.02.037. [DOI] [PubMed] [Google Scholar]
- Dell’Isola A.; Steultjens M. Classification of patients with knee osteoarthritis in clinical phenotypes: Data from the osteoarthritis initiative. PLoS One 2018, 13, e0191045. 10.1371/journal.pone.0191045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Beaumont G.; Roman-Blas J. A.; Bruyere O.; Cooper C.; Kanis J.; Maggi S.; Rizzoli R.; Reginster J. Y. Clinical settings in knee osteoarthritis: Pathophysiology guides treatment. Maturitas 2017, 96, 54–57. 10.1016/j.maturitas.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Dell’Isola A.; Allan R.; Smith S. L.; Marreiros S. S. P.; Steultjens M. Identification of clinical phenotypes in knee osteoarthritis: a systematic review of the literature. BMC Musculoskeletal Disord. 2016, 17, 425. 10.1186/s12891-016-1286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Likhodii S.; Zhang Y.; Aref-Eshghi E.; Harper P. E.; Randell E.; Green R.; Martin G.; Furey A.; Sun G.; Rahman P.; Zhai G. Classification of osteoarthritis phenotypes by metabolomics analysis. BMJ. Open 2014, 4, e006286. 10.1136/bmjopen-2014-006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson A. K.; Rawle R. A.; Wallace C. W.; Brooks E. G.; Adams E.; Greenwood M. C.; Olmer M.; Lotz M. K.; Bothner B.; June R. K. Characterization of synovial fluid metabolomic phenotypes of cartilage morphological changes associated with osteoarthritis. Osteoarthritis Cartilage 2019, 27, 1174–1184. 10.1016/j.joca.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveza L. A.; Nelson A. E.; Loeser R. F. Phenotypes of osteoarthritis: current state and future implications. Clin. Exp. Rheumatol. 2019, 37, 64–72. [PMC free article] [PubMed] [Google Scholar]
- Blanco F.; Ruiz-Romero C. Metabolomic characterization of metabolic phenotypes in OA. Nat. Rev. Rheumatol. 2012, 8, 130–132. 10.1038/nrrheum.2012.11. [DOI] [PubMed] [Google Scholar]
- Mobasheri A.; Saarakkala S.; Finnila M.; Karsdal M. A.; Bay-Jensen A. C.; van Spil W. E. Recent advances in understanding the phenotypes of osteoarthritis. F1000Research 2019, 8, 2091. 10.12688/f1000research.20575.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clanche S.; Bonnefont-Rousselot D.; Sari-Ali E.; Rannou F.; Borderie D. Inter-relations between osteoarthritis and metabolic syndrome: A common link?. Biochimie 2016, 121, 238–252. 10.1016/j.biochi.2015.12.008. [DOI] [PubMed] [Google Scholar]
- Sherling D. H.; Perumareddi P.; Hennekens C. H. Metabolic Syndrome: Clinical and Policy Implications of the New Silent Killer. J. Cardiovasc. Pharmacol. Ther. 2017, 22, 365–367. 10.1177/1074248416686187. [DOI] [PubMed] [Google Scholar]
- Mendrick D. L.; Diehl A. M.; Topor L. S.; Dietert R. R.; Will Y.; La Merrill M. A.; Bouret S.; Varma V.; Hastings K. L.; Schug T. T.; Emeigh Hart S. G.; Burleson F. G. Metabolic Syndrome and Associated Diseases: From the Bench to the Clinic. Toxicol. Sci. 2018, 162, 36–42. 10.1093/toxsci/kfx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louati K.; Vidal C.; Berenbaum F.; Sellam J. Association between diabetes mellitus and osteoarthritis: systematic literature review and meta-analysis. Rmd Open 2015, 1, e000077. 10.1136/rmdopen-2015-000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. F.; London D. A.; Husni E. M.; Navaneethan S.; Kashyap S. R. Type 2 diabetes and osteoarthritis: a systematic review and meta-analysis. Journal of Diabetes and Its Complications 2016, 30, 944–950. 10.1016/j.jdiacomp.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Veronese N.; Cooper C.; Reginster J. Y.; Hochberg M.; Branco J.; Bruyere O.; Chapurlat R.; Al-Daghri N.; Dennison E.; Herrero-Beaumont G.; Kaux J. F.; Maheu E.; Rizzoli R.; Roth R.; Rovati L. C.; Uebelhart D.; Vlaskovska M.; Scheen A. Type 2 diabetes mellitus and osteoarthritis. Semin. Arthritis Rheum. 2019, 49, 9–19. 10.1016/j.semarthrit.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courties A.; Sellam J. Osteoarthritis and type 2 diabetes mellitus: What are the links?. Diabetes Res. Clin. Pract. 2016, 122, 198–206. 10.1016/j.diabres.2016.10.021. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Sun G.; Likhodii S.; Aref-Eshghi E.; Harper P. E.; Randell E.; Green R.; Martin G.; Furey A.; Rahman P.; Zhai G. Metabolomic analysis of human synovial fluid and plasma reveals that phosphatidylcholine metabolism is associated with both osteoarthritis and diabetes mellitus. Metabolomics 2016, 12, 24. 10.1007/s11306-015-0937-x. [DOI] [PubMed] [Google Scholar]
- Laiguillon M. C.; Courties A.; Houard X.; Auclair M.; Sautet A.; Capeau J.; Fève B.; Berenbaum F.; Sellam J. Characterization of diabetic osteoarthritic cartilage and role of high glucose environment on chondrocyte activation: toward pathophysiological delineation of diabetes mellitus-related osteoarthritis. Osteoarthritis Cartilage 2015, 23, 1513–22. 10.1016/j.joca.2015.04.026. [DOI] [PubMed] [Google Scholar]
- Eveque-Mourroux M. R.; Emans P. J.; Zautsen R. R. M.; Boonen A.; Heeren R. M. A.; Cillero-Pastor B. Spatially resolved endogenous improved metabolite detection in human osteoarthritis cartilage by matrix assisted laser desorption ionization mass spectrometry imaging. Analyst 2019, 144, 5953–5958. 10.1039/C9AN00944B. [DOI] [PubMed] [Google Scholar]
- Kultima K.; Skold K.; Boren M. Biomarkers of disease and post-mortem changes - Heat stabilization, a necessary tool for measurement of protein regulation. J. Proteomics 2011, 75, 145–59. 10.1016/j.jprot.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Jerneren F.; Soderquist M.; Karlsson O. Post-sampling release of free fatty acids - effects of heat stabilization and methods of euthanasia. J. Pharmacol. Toxicol. Methods 2015, 71, 13–20. 10.1016/j.vascn.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Massafra V.; Milona A.; Vos H. R.; Burgering B. M.; van Mil S. W. Quantitative liver proteomics identifies FGF19 targets that couple metabolism and proliferation. PLoS One 2017, 12, e0171185. 10.1371/journal.pone.0171185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J.; Ferluga S.; Sharma V.; Futschik M.; Hilton D. A.; Adams C. L.; Lasonder E.; Hanemann C. O. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 2019, 40, 77–91. 10.1016/j.ebiom.2018.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.; Belugali Nataraj N.; Perry G.; Loayza Puch F.; Nagel R.; Marin I.; Balint N.; Bossel N.; Pavlovsky A.; Barshack I.; Kaufman B.; Agami R.; Yarden Y.; Dadiani M.; Geiger T. Proteomic patterns associated with response to breast cancer neoadjuvant treatment. Mol. Syst. Biol. 2020, 16, e9443. 10.15252/msb.20209443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y.; Tan C. M.; Kou Y.; Duan Q.; Wang Z.; Meirelles G. V.; Clark N. R.; Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013, 14, 128. 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre F.; Rocha B.; Dewez F.; Towers M.; Murray P.; Claude E.; Cillero-Pastor B.; Heeren R.; Siegel T. P. Faster raster matrix-assisted laser desorption/ionization mass spectrometry imaging of lipids at high lateral resolution. Int. J. Mass Spectrom. 2019, 437, 38–48. 10.1016/j.ijms.2018.09.015. [DOI] [Google Scholar]
- Eijkel G. B.; Kaletas B. K.; van der Wiel I. M.; Kros J. M.; Luider T. M.; Heeren R. M. A. Correlating MALDI and SIMS imaging mass spectrometric datasets of biological tissue surfaces. Surf. Interface Anal. 2009, 41, 675–685. 10.1002/sia.3088. [DOI] [Google Scholar]
- Calvano C. D.; Bianco M.; Ventura G.; Losito I.; Palmisano F.; Cataldi T. R. I. Analysis of Phospholipids, Lysophospholipids, and Their Linked Fatty Acyl Chains in Yellow Lupin Seeds (Lupinus luteus L.) by Liquid Chromatography and Tandem Mass Spectrometry. Molecules 2020, 25, 805. 10.3390/molecules25040805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling J. K.; Hermansson M.; Hartler J.; Christiansen K.; Gallego S. F.; Peng B.; Ahrends R.; Ejsing C. S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS One 2017, 12, e0188394. 10.1371/journal.pone.0188394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Fahy E.; Aoki J.; Dennis E. A.; Durant T.; Ejsing C. S.; Fedorova M.; Feussner I.; Griffiths W. J.; Köfeler H.; Merrill A. H. Jr; Murphy R. C.; O’Donnell V. B.; Oskolkova O.; Subramaniam S.; Wakelam M. J. O.; Spener F. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobezie R.; Kho A.; Krastins B.; Sarracino D. A.; Thornhill T. S.; Chase M.; Millett P. J.; Lee D. M. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res. Ther. 2007, 9, R36. 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi M.; Deberg M.; Henrotin Y. Application for Proteomic Techniques in Studying Osteoarthritis: A Review. Front. Physiol. 2011, 2, 90. 10.3389/fphys.2011.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pousinis P.; Gowler P. R. W.; Burston J. J.; Ortori C. A.; Chapman V.; Barrett D. A. Lipidomic identification of plasma lipids associated with pain behaviour and pathology in a mouse model of osteoarthritis. Metabolomics 2020, 16, 32. 10.1007/s11306-020-01652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attie A. D.; Kastelein J. P.; Hayden M. R. Pivotal role of ABCA1 in reverse cholesterol transport influencing HDL levels and susceptibility to atherosclerosis. J. Lipid Res. 2001, 42, 1717–1726. 10.1016/S0022-2275(20)31498-X. [DOI] [PubMed] [Google Scholar]
- de Seny D.; Cobraiville G.; Charlier E.; Neuville S.; Lutteri L.; Le Goff C.; Malaise D.; Malaise O.; Chapelle J. P.; Relic B.; Malaise M. G. Apolipoprotein-A1 as a Damage-Associated Molecular Patterns Protein in Osteoarthritis: Ex Vivo and In Vitro Pro-Inflammatory Properties. PLoS One 2015, 10, e0122904. 10.1371/journal.pone.0122904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido L.; Ayoglu B.; Fernandez-Tajes J.; Henjes F.; Schwenk J. M.; Ruiz-Romero C.; Nilsson P.; Blanco F. J. Identification of a Serum Protein Biomarker Panel for the Diagnosis of Knee Osteoarthritis. Osteoarthritis and Cartilage 2016, 24, S23–S23. 10.1016/j.joca.2016.01.070. [DOI] [Google Scholar]
- Wu X.; Yu Z.; Su W.; Isquith D. A.; Neradilek M. B.; Lu N.; Gu F.; Li H.; Zhao X. Q. Low levels of ApoA1 improve risk prediction of type 2 diabetes mellitus. J. Clin. Lipidol. 2017, 11, 362–368. 10.1016/j.jacl.2017.01.009. [DOI] [PubMed] [Google Scholar]
- Higgins A. J.; Lees P. The acute inflammatory process, arachidonic acid metabolism and the mode of action of anti-inflammatory drugs. Equine Vet. J. 1984, 16, 163–75. 10.1111/j.2042-3306.1984.tb01893.x. [DOI] [PubMed] [Google Scholar]
- Monroy-Munoz I. E.; Angeles-Martinez J.; Posadas-Sanchez R.; Villarreal-Molina T.; Alvarez-Leon E.; Flores-Dominguez C.; Cardoso-Saldana G.; Medina-Urrutia A.; Juarez-Rojas J. G.; Posadas-Romero C.; Alarcon G. V. PLA2G2A polymorphisms are associated with metabolic syndrome and type 2 diabetes mellitus. Results from the genetics of atherosclerotic disease Mexican study. Immunobiology 2017, 222, 967–972. 10.1016/j.imbio.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Lopes M. B.; Freitas R. C.; Hirata M. H.; Hirata R.; Rezende A. A.; Silbiger V. N.; Bortolin R. H.; Luchessi A. D. mRNA-miRNA integrative analysis of diabetes-induced cardiomyopathy in rats. Front. Biosci. (School Ed.) 2017, 9, 194–229. 10.2741/s483. [DOI] [PubMed] [Google Scholar]
- Zhai G. J.; Pelletier J. P.; Liu M.; Aitken D.; Randell E.; Rahman P.; Jones G.; Martel-Pelletier J. Activation of The Phosphatidylcholine to Lysophosphatidylcholine Pathway Is Associated with Osteoarthritis Knee Cartilage Volume Loss Over Time. Sci. Rep. 2019, 9, 9648. 10.1038/s41598-019-46185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido L.; Calamia V.; Fernandez-Puente P.; Mateos J.; Rocha B.; Fernandez-Costa C.; Blanco F. J.; Ruiz-Romero C. Proteomics Analysis Of Cartilage Secretome: A Powerful Tool For The Discovery Of OA Biomarkers. Arthritis Rheum. 2013, 65, S800–S800. [Google Scholar]
- Wakil S. J.; Abu-Elheiga L. A. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 2009, 50, S138–43. 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y. The biological significance of ω-oxidation of fatty acids. Proc. Jpn. Acad., Ser. B 2013, 89, 370–82. 10.2183/pjab.89.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet B.; Beier F. Targeting oxidative stress to reduce osteoarthritis. Arthritis Res. Ther. 2016, 18, 32. 10.1186/s13075-015-0908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetsos P.; Papavassiliou A. G. ROS/oxidative stress signaling in osteoarthritis. Biochim. Biophys. Acta, Mol. Basis Dis. 2016, 1862, 576–591. 10.1016/j.bbadis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Zhu S. A.; Makosa D.; Miller B.; Griffin T. M. Glutathione as a mediator of cartilage oxidative stress resistance and resilience during aging and osteoarthritis. Connect. Tissue Res. 2020, 61, 34–47. 10.1080/03008207.2019.1665035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J.; Rosenblatt-Velin N.; Parapanov R.; Liaudet L. The role of oxidative stress during inflammatory processes. Biol. Chem. 2014, 395, 203–30. 10.1515/hsz-2013-0241. [DOI] [PubMed] [Google Scholar]
- Fukai T.; Folz R. J.; Landmesser U.; Harrison D. G. Extracellular superoxide dismutase and cardiovascular disease. Cardiovasc. Res. 2002, 55, 239–249. 10.1016/S0008-6363(02)00328-0. [DOI] [PubMed] [Google Scholar]
- Voziyan P.; Brown K. L.; Chetyrkin S.; Hudson B. Site-specific AGE modifications in the extracellular matrix: a role for glyoxal in protein damage in diabetes. Clin. Chem. Lab. Med. 2014, 52, 39–45. 10.1515/cclm-2012-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjara S.; Sochor M.; Ali M.; Drake A.; Greenbaum A. L.; McLean P. Pyrimidine nucleotide synthesis in the rat kidney in early diabetes. Biochem. Med. Metab. Biol. 1991, 46, 215–25. 10.1016/0885-4505(91)90069-W. [DOI] [PubMed] [Google Scholar]
- Bauer J. A. The urea cycle as a source of nitric oxide implicated in the pathogenesis of insulin-dependent diabetes mellitus. Med. Hypotheses 1998, 51, 71–3. 10.1016/S0306-9877(98)90258-4. [DOI] [PubMed] [Google Scholar]
- Yin J.; Ren W.; Chen S.; Li Y.; Han H.; Gao J.; Liu G.; Wu X.; Li T.; Woo Kim S.; Yin Y. Metabolic Regulation of Methionine Restriction in Diabetes. Mol. Nutr. Food Res. 2018, 62, e1700951. 10.1002/mnfr.201700951. [DOI] [PubMed] [Google Scholar]
- Hsueh M. F.; Khabut A.; Kjellstrom S.; Onnefjord P.; Kraus V. B. Elucidating the Molecular Composition of Cartilage by Proteomics. J. Proteome Res. 2016, 15, 374–388. 10.1021/acs.jproteome.5b00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J.; Amhare A. F.; Wang L.; Lv Y.; Deng H.; Gao H.; Guo X.; Han J.; Lammi M. J. Proteomic analysis of knee cartilage reveals potential signaling pathways in pathological mechanism of Kashin-Beck disease compared with osteoarthritis. Sci. Rep. 2020, 10, 6824. 10.1038/s41598-020-63932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J.; Ko C. W.; Tso P.; Bhargava A. Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells 2019, 8, 319. 10.3390/cells8040319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polur I.; Lee P. L.; Servais J. M.; Xu L.; Li Y. Role of HTRA1, a serine protease, in the progression of articular cartilage degeneration. Histol. Histopathol. 2010, 25, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillero-Pastor B.; Eijkel G. B.; Kiss A.; Blanco F. J.; Heeren R. M. Matrix-assisted laser desorption ionization-imaging mass spectrometry: a new methodology to study human osteoarthritic cartilage. Arthritis Rheum. 2013, 65, 710–20. 10.1002/art.37799. [DOI] [PubMed] [Google Scholar]
- Cillero-Pastor B.; Eijkel G.; Kiss A.; Blanco F. J.; Heeren R. M. Time-of-flight secondary ion mass spectrometry-based molecular distribution distinguishing healthy and osteoarthritic human cartilage. Anal. Chem. 2012, 84, 8909–16. 10.1021/ac301853q. [DOI] [PubMed] [Google Scholar]
- Barre F.; Flinders B.; Garcia J. P.; Jansen I.; Huizing L. R.; Porta T.; Creemers L. B.; Heeren R. M.; Cillero-Pastor B. Derivatization Strategies for the Detection of Triamcinolone Acetonide in Cartilage by Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2016, 88, 12051–12059. 10.1021/acs.analchem.6b02491. [DOI] [PubMed] [Google Scholar]
- Merrill A. H. Jr.; Schmelz E. M.; Dillehay D. L.; Spiegel S.; Shayman J. A.; Schroeder J. J.; Riley R. T.; Voss K. A.; Wang E. Sphingolipids--the enigmatic lipid class: biochemistry, physiology, and pathophysiology. Toxicol. Appl. Pharmacol. 1997, 142, 208–25. 10.1006/taap.1996.8029. [DOI] [PubMed] [Google Scholar]
- Harrell C. R.; Markovic B. S.; Fellabaum C.; Arsenijevic A.; Volarevic V. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomed. Pharmacother. 2019, 109, 2318–2326. 10.1016/j.biopha.2018.11.099. [DOI] [PubMed] [Google Scholar]
- Rocha B.; Cillero-Pastor B.; Eijkel G.; Bruinen A. L.; Ruiz-Romero C.; Heeren R. M. A.; Blanco F. J. Characterization of lipidic markers of chondrogenic differentiation using mass spectrometry imaging. Proteomics 2015, 15, 702–713. 10.1002/pmic.201400260. [DOI] [PubMed] [Google Scholar]
- van der Veen J. N.; Kennelly J. P.; Wan S.; Vance J. E.; Vance D. E.; Jacobs R. L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 1558–1572. 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Matsumoto T.; Kobayashi T.; Kamata K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr. Med. Chem. 2007, 14, 3209–20. 10.2174/092986707782793899. [DOI] [PubMed] [Google Scholar]
- Barre F.; Claes B. S. R.; Dewez F.; Peutz-Kootstra C.; Munch-Petersen H. F.; Grønbæk K.; Lund A. H.; Heeren R. M. A.; Côme C.; Cillero-Pastor B. Specific Lipid and Metabolic Profiles of R-CHOP-Resistant Diffuse Large B-Cell Lymphoma Elucidated by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging and in Vivo Imaging. Anal. Chem. 2018, 90, 14198–14206. 10.1021/acs.analchem.8b02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma A. V.; Powell G. L.; LaBerge M. Phospholipid composition of articular cartilage boundary lubricant. J. Orthop. Res. 2001, 19, 671–6. 10.1016/S0736-0266(00)00064-4. [DOI] [PubMed] [Google Scholar]
- Stockwell R. A. Chondrocytes. J. Clin. Pathol. 1978, 31, 7–13. 10.1136/jcp.31.Suppl_12.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinska M. K.; Liebisch G.; Lochnit G.; Wilhelm J.; Klein H.; Kaesser U.; Lasczkowski G.; Rickert M.; Schmitz G.; Steinmeyer J. Sphingolipids in human synovial fluid--a lipidomic study. PLoS One 2014, 9, e91769. 10.1371/journal.pone.0091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B.; Eijkel G. B.; Heeren R. M. A.; Karperien M.; Post J. N.; Cillero-Pastor B. Oxygen-Dependent Lipid Profiles of Three-Dimensional Cultured Human Chondrocytes Revealed by MALDI-MSI. Anal. Chem. 2017, 89, 9438–9444. 10.1021/acs.analchem.7b02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Sun G.; Aitken D.; Likhodii S.; Liu M.; Martin G.; Furey A.; Randell E.; Rahman P.; Jones G.; Zhai G. Lysophosphatidylcholines to phosphatidylcholines ratio predicts advanced knee osteoarthritis. Rheumatology (Oxford, U. K.) 2016, 55, 1566–74. 10.1093/rheumatology/kew207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabini R. A.; Galassi R.; Fumelli P.; Dousset N.; Solera M. L.; Valdiguie P.; Curatola G.; Ferretti G.; Taus M.; Mazzanti L. Reduced Na(+)-K(+)-ATPase activity and plasma lysophosphatidylcholine concentrations in diabetic patients. Diabetes 1994, 43, 915–919. 10.2337/diabetes.43.7.915. [DOI] [PubMed] [Google Scholar]
- Han M. S.; Lim Y. M.; Quan W.; Kim J. R.; Chung K. W.; Kang M.; Kim S.; Park S. Y.; Han J. S.; Park S. Y.; Cheon H. G.; Rhee S. D.; Park T. S.; Lee M. S. Lysophosphatidylcholine as an effector of fatty acid-induced insulin resistance. J. Lipid Res. 2011, 52, 1234–1246. 10.1194/jlr.M014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzanski W.; Bogoch E.; Wloch M.; Vadas P. The role of phospholipase A2 in the physiopathology of osteoarthritis. J. Rheumatol. Suppl. 1991, 27, 117–119. [PubMed] [Google Scholar]
- Takeshita S.; Inoue N.; Gao D.; Rikitake Y.; Kawashima S.; Tawa R.; Sakurai H.; Yokoyama M. Lysophosphatidylcholine enhances superoxide anions production via endothelial NADH/NADPH oxidase. J. Atheroscler. Thromb. 2000, 7, 238–46. 10.5551/jat1994.7.238. [DOI] [PubMed] [Google Scholar]
- Li X.; Fang P.; Li Y.; Kuo Y. M.; Andrews A. J.; Nanayakkara G.; Johnson C.; Fu H.; Shan H.; Du F.; Hoffman N. E.; Yu D.; Eguchi S.; Madesh M.; Koch W. J.; Sun J.; Jiang X.; Wang H.; Yang X. Mitochondrial Reactive Oxygen Species Mediate Lysophosphatidylcholine-Induced Endothelial Cell Activation. Arterioscler., Thromb., Vasc. Biol. 2016, 36, 1090–100. 10.1161/ATVBAHA.115.306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi S. J.; Seo S. Y.; Cho K.; Nam M. H.; Park K. Y. Lysophosphatidylcholine enhances susceptibility in signaling pathway against pathogen infection through biphasic production of reactive oxygen species and ethylene in tobacco plants. Phytochemistry 2014, 104, 48–59. 10.1016/j.phytochem.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Pahan K. Lipid-lowering drugs. Cell. Mol. Life Sci. 2006, 63, 1165–1178. 10.1007/s00018-005-5406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. X.; Wu Y. J. Lipid-Modifying Drugs: Pharmacology and Perspectives. Adv. Exp. Med. Biol. 2020, 1177, 133–148. 10.1007/978-981-15-2517-9_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.