Abstract
Immune checkpoint inhibitors (ICIs) have become a promising area of research for cancer treatment. In addition to the well-known ICIs targeting PD-1/PD-L1, HLA-G/ILT-2/-4 is relatively new immune checkpoint that has been evaluated in early clinical trials in patients with advanced solid tumors. In this study, the expression of HLA-G (n=157), ILT-2/4 (n=82), and PD-L1 (n=70) in epithelial cell adhesion molecule (EpCAM)-positive colorectal cancer (CRC) cells was analyzed by multicolor flow cytometry, and the prognostic significance of these molecules was evaluated. In EpCAM+ CRC cells, the median percentages of HLA-G, ILT-2, ILT-4, and PD-L1 were 14.90%, 67.70%, 8.55% and 80.30%, respectively. In addition, a positive correlation was observed between them (all p<0.001). Higher levels of these immune checkpoint proteins are associated with lymph node metastasis. In addition to the AJCC stage (p=0.001), Kaplan-Meier survival analysis showed that higher levels of HLA-G (p=0.041), ILT-2 (p=0.060), ILT-4 (p<0.001), PD-L1 (p=0.012), HLA-GILT4 (p<0.001) and ILT-2ILT-4 (p<0.001) were significantly associated with shorter survival of CRC patients. When CRC patients were stratified by early and advanced AJCC stages, HLA-G levels were only related to the survival among CRC patients with early disease stage (p=0.024), while ILT-4 levels were significant for both CRC patients with early (p=0.001) and advanced (p=0.020) disease stages. Multivariate cox regression analysis revealed that advanced AJCC stage (HR=2.435; p=0.005) and higher ILT-4 levels (HR=2.198; p=0.063) were independent risk factors for poor outcomes in patients with CRC. In summary, among the immune checkpoints, HLA-G/ILT-2/4 and PD-L1, ILT-4 is the most significant prognostic indicator of CRC. This finding indicated that a combination of immunotherapy strategies, such as ILT-4 blockade, could improve the clinical outcomes in patients with cancer. Moreover, multicolor flow cytometry can be employed as a reliable and efficient, alternative to immunohistochemistry, for evaluating the immune checkpoint proteins expressed in tumor lesions.
Keywords: HLA-G, ILT-2, ILT-4, PD-L1, colorectal cancer, prognosis
Introduction
Immune checkpoint protein expression is one of the dominant mechanisms by which tumor cells evade the innate and adaptive anti-tumor immune responses and promote disease progression (1). Fortunately, blocking the immune checkpoint signaling pathway with immune checkpoint inhibitors (ICIs) has become a promising strategy in cancer treatment (2), which has shown remarkable outcomes in considerable proportion of patients with advanced solid tumors. The ICIs targeting the programmed death-1 (PD-1)/programmed death ligand-1 (PD-L1) axis have been developed and have marked a milestone in cancer immunotherapy (3). However, resistance to ICI treatment is common among many patients suffering from cancer. Either therapy with multiple combined ICIs or exploration of novel ICIs is necessary to overcome such resistance and ultimately improve the long-term clinical benefit (2).
HLA-G, a non-classical HLA class I antigen, is predominantly expressed in extravillous cytotrophoblasts during pregnancy, and under physiological conditions, its expression is restricted to a few immune-privileged tissues (4). However, an aberrant induction of HLA-G expression is observed in most cancer histological types, which is closely related to tumor metastasis and poor prognosis in clinical settings as well as in murine laboratory models (5). The expression of HLA-G, an immune tolerant and tumor promoting factor, has been extensively investigated, and its role as a novel immune checkpoint has been established (6, 7).
The immunosuppressive function of HLA-G is mediated by its interaction with immune inhibitory receptors, immunoglobulin-like transcript (ILT)-2 and/or ILT-4 (8). These receptors are expressed in tumor cells, as well as in a wide range of immune cells, such as B cells, T cells, natural killer cells (NK), dendritic cells (DCs), and myeloid-derived suppressor cells (MDSCs) (5). Interaction between tumor HLA-G and ILT-2/-4 expressed on the surface of immune cells can induce a broad spectrum of immune suppressive responses by (i) inhibiting T-cell proliferation and B-cell immunoglobulin production (9–11), (ii) impairing T-cell and NK-cell cytotoxicity, as well as maturation of DCs (12–14), (iii) promoting the proliferation of regulatory MDSCs and accumulation of M2-type macrophages, as well as inducing anergy by the generation of regulatory T cells (Treg), tolerant DC-10 cells, and invariant natural killer T cells (iNKTs) (15–19), and (iv) dampening the expression of chemokine receptors on B cells, T cells, and NK cells, thereby hampering their infiltration into the tumor microenvironment (20). In this context, the cytotoxic functions of tumor-infiltrating CD8+PD-L1-ILT-2+ cells could be selectively inhibited by tumor cell-expressing HLA-G (21). Moreover, ILT-2/-4 has been observed to be expressed in both solid tumors, such as stomach, lung, and liver cancers, as well as in hematological malignancies (22). Higher levels of ILT-2 expression have been found to be related to a shorter overall survival in patients with colon mucinous adenocarcinoma and gastric mixed adenocarcinoma. However, the level of ILT-2 expression was comparable among rectal mucinous adenocarcinomas and markedly lower in hepatocellular cancers than in that normal controls (23, 24). Additionally, previous studies have shown that the expression of ILT-4 is associated with tumor metastasis, advanced disease, and poor prognosis (25).
Currently, although immunohistochemistry is widely used in the evaluation of immune checkpoint protein expression in tumor lesions, many challenges, such as high inter-assay variability in routine performance remain to be addressed (26). In this study, a suspension of CRC cells was prepared from the frozen lesions and immune checkpoints, HLA-G, ILT-2, ILT-4, and PD-L1, were analyzed by multicolor flow cytometry gated using epithelial cell adhesion molecule (EpCAM); subsequently, the prognostic significance of the checkpoint proteins was evaluated.
Materials and methods
Patients With Colorectal Cancer
A total of 157 frozen primary CRC lesions from the Biological Resource Center, Taizhou Hospital of Zhejiang Province (National Human Genetic Resources Platform of China YCZYPT [2017]02) were included in the study. All patients with CRC were diagnosed and treated between June 2006 and May 2012 at Taizhou Hospital of Zhejiang Province, China. No immunotherapy was applied for these CRC patients. Written informed consent was obtained from each participant prior to surgery, and the study protocol was approved by the Ethics Review Board of Taizhou Hospital of Zhejiang Province (K20190766).
Detailed clinicopathological information was retrieved from the electronic medical records in the hospital. Among 157 patients with CRC (colon=90, rectal =67), 89 were male and 68 were female, with a median age of 69.5 years (range, 30 ~ 89 years). With respect to TNM staging, 10, 144, and 3 patients had pT2, T3 and T4, respectively, and 75, 45, and 37 patients had pN0, N1, and N2, respectively. Similarly, 152 and 5 patients had pM0 and M1, respectively. AJCC disease stages I, II, III, and IV were found in 6, 68, 78, and 5 patients, respectively (AJCC 7th TNM staging system) (27).
Follow-up information was available for 82 of the 157 patients with CRC. The last follow-up was carried out in November 2016, with a median follow-up of 31 months, and 60 cancer-related events were recorded. Overall survival (OS) was determined from the date of surgery to the date of the event. The detailed clinicopathological characteristics of CRC patients are shown in Table 1 .
Table 1.
Association between high and low levels of immune checkpoint expression with clinical parameters in CRC patients.
| Variables | No. | HLA-G (n=157; median=14.9%) | No. | ILT-2 (n=82; median=67.7%) | ILT-4 (n=82; median=8.55%) | No. | PD-L1 (n=70; median=80.30%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | p | Low | High | p | Low | High | p | Low | High | p | ||||
| Type | |||||||||||||||
| colon | 90 | 46 (51.1%) | 44 (48.9%) | 0.377 | 60 | 32 (53.3%) | 28 (46.7%) | 0.527 | 29 (48.3%) | 31 (51.7%) | 0.894 | 51 | 27 (52.9%) | 24 (47.1%) | 0.420 |
| rectal | 67 | 39 (58.2%) | 28 (41.8%) | 22 | 10 (45.5%) | 12 (54.5%) | 11 (50.0%) | 11 (50.0%) | 19 | 8 (42.1%) | 11 (57.9%) | ||||
| Gender | |||||||||||||||
| male | 89 | 43 (48.3%) | 46 (51.7%) | 0.094 | 40 | 22 (55.0%) | 18 (45.0%) | 0.504 | 19 (47.5%) | 21 (52.5%) | 0.821 | 35 | 18 (51.4%) | 17 (48.6%) | 0.811 |
| female | 68 | 42 (61.8%) | 26 (38.2%) | 42 | 20 (47.6%) | 22 (52.4%) | 21 (50.0%) | 21 (50.0%) | 35 | 17 (48.6%) | 18 (51.4%) | ||||
| Age | |||||||||||||||
| <69.5yrs | 82 | 43 (52.4%) | 39 (47.6%) | 0.655 | 41 | 17 (41.5%) | 24 (58.5%) | 0.077 | 19 (46.3%) | 22 (53.7%) | 0.579 | 35 | 15 (42.9%) | 20 (57.1%) | 0.232 |
| >69.5 yrs | 75 | 42 (56.0%) | 33 (44.0%) | 41 | 25 (61.0%) | 16 (39.0%) | 21 (51.2%) | 19 (46.3%) | 35 | 20 (57.1%) | 15 (42.9%) | ||||
| pT stage | |||||||||||||||
| T2 | 10 | 7 (70.0%) | 3 (30.0%) | 0.923 | 6 | 1 (16.7%) | 5 (83.3%) | 0.115 | 4 (66.7%) | 2 (33.3%) | 0.363 | 4 | 2 (50.0%) | 2 (50.0%) | 1.000 |
| T3+T4 | 147 | 105 (71.4%) | 42 (28.6%) | 76 | 38 (50.0%) | 38 (50.0%) | 36 (47.4%) | 40 (52.6%) | 66 | 33 (50.0%) | 33 (50.0%) | ||||
| pN stage | |||||||||||||||
| N0 | 75 | 46 (61.3%) | 29 (38.7%) | 0.028 | 32 | 20 (62.5%) | 12 (37.5%) | 0.055 | 21 (65.4%) | 11 (32.4%) | 0.005 | 27 | 16 (59.3%) | 11 (40.7%) | 0.064 |
| N1 | 45 | 26 (57.8%) | 19 (42.2%) | 27 | 15 (55.6%) | 12 (44.4%) | 13 (48.1%) | 14 (51.9%) | 22 | 13 (59.1%) | 9 (40.9%) | ||||
| N2 | 37 | 13 (35.17%) | 24 (64.9%) | 23 | 7 (30.4%) | 16 (69.4%) | 6 (26.1%) | 17 (73.9%) | 21 | 6 (28.6%) | 15 (71.4%) | ||||
| pM stage | |||||||||||||||
| M0 | 152 | 39 (25.7%) | 113 (74.3%) | 0.088 | 78 | 40 (51.3%) | 38 (48.7%) | 0.960 | 39 (50.0%) | 39 (50.0%) | 0.329 | 66 | 33 (50.0%) | 33 (50.0%) | 1.000 |
| M1 | 5 | 3 (60.0%) | 2 (40.0%) | 4 | 2 (50.0%) | 2 (50.0%) | 1 (25.0%) | 3 (75.0%) | 4 | 2 (50.0%) | 2 (50.0%) | ||||
| AJCC stage | |||||||||||||||
| I+II | 74 | 45 (60.8%) | 29 (39.2%) | 0.113 | 32 | 20 (62.5%) | 12 (37.5%) | 0.102 | 21 (65.6%) | 11 (34.4%) | 0.015 | 27 | 16 (59.3%) | 11 (40.7%) | 0.220 |
| III+IV | 83 | 40 (48.2%) | 43 (51.8%) | 50 | 22 (44.0%) | 28 (56.0%) | 19 (38.0%) | 31 (62.0%) | 43 | 19 (44.2%) | 24 (55.8%) | ||||
Sample Preparation
Single-cell suspensions from frozen CRC lesions were prepared by mechanical dissociation and enzymatic digestion. These lesions were kept at -80°C in an ultra-low temperature freezer at the Biological Resource Center, Taizhou Hospital of Zhejiang Province (National Human Genetic Resources Platform of China).
After taking them out from the freezer, the lesions were balanced at room temperature for 5 ~ 10 min, washed three times with HBSS buffer (Thermo Fisher Scientific, Grand Island, NY, USA), sectioned into small pieces (5 ~ 6 mm) and washed with HBSS buffer again. After dissociation, small lesion pieces were digested in HBSS buffer containing 2% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA), collagenase type IV (1 mg/ml, Thermo Fisher Scientific, Grand Island, NY, USA), and hyaluronidase (10 ng/mL, Merck, St. Louis, MO, USA) at 37°C for 4 h (28). After digestion, the cells were filtered with a 70 µm cell strainer (Corning, Durham, NC, USA) to obtain single cell suspension that was used for flow cytometry analysis.
Flow Cytometry Analysis
To analyze HLA-G, ILT-2, ILT-4, and PD-L1 expression in CRC cells, monoclonal antibodies (mAbs), such as anti-EpCAM-FITC (CD326, Thermo Fisher Scientific, Carlsbad, CA, USA), anti-HLA-G-PE (MEM-G/09, Exbio, Vestec, Czech Republic), anti-ILT4-APC (CD85d, Thermo Fisher Scientific, Carlsbad, CA, USA), anti-ILT2-PE/Cy7 (CD85j, Biolegend, San Diego, CA, USA), and anti-PD-L1-PerCP/Cyanine 5.5 (Biolegend, San Diego, CA, USA), were used.
Flow cytometry analysis was performed using BD FACS Canto Plus (Becton Dickinson, San Jose, CA). Single cell suspensions (1×106 ~ 5×106) were resuspended in 100 µL PBS containing 2% FBS, and 5 µL of each of the aforementioned multicolor mAbs were added and incubated in the dark at 4°C for 30 min. For each mAb, a negative control, that is, an appropriate fluorescent-labeled isotype control, was used. Data were analyzed using FlowJo Jo™ software (Becton-Dickinson, San Jose, CA). CRC cells were identified as EpCAM (CD326)-positive cells.
Statistical Analyses
Statistical analyses were performed using the statistical software package SPSS (version 13.0, SPSS, Inc. Chicago, IL, USA). Bivariate correlation analysis between HLA-G, ILT-2, ILT-4, and PD-L1 expression was performed using the Pearson test. The relation between the expression of HLA-G, ILT-2, ILT-4, and PD-L1 alone or combination in CRC cells and the clinicopathological status of patients with CRC was analyzed using the chi-square test. Survival analysis was performed using the Kaplan-Meier method (log-rank test). The significance of the appropriate variables for survival was evaluated using univariate and/or covariate Cox regression methods. Statistical significance was set at p<0.05 (Flowchart of the study protocol was shown in Supplementary Figure 2 ).
Results
Expression of HLA-G, ILT-2, ILT-4 and PD-L1 in CRC Lesions
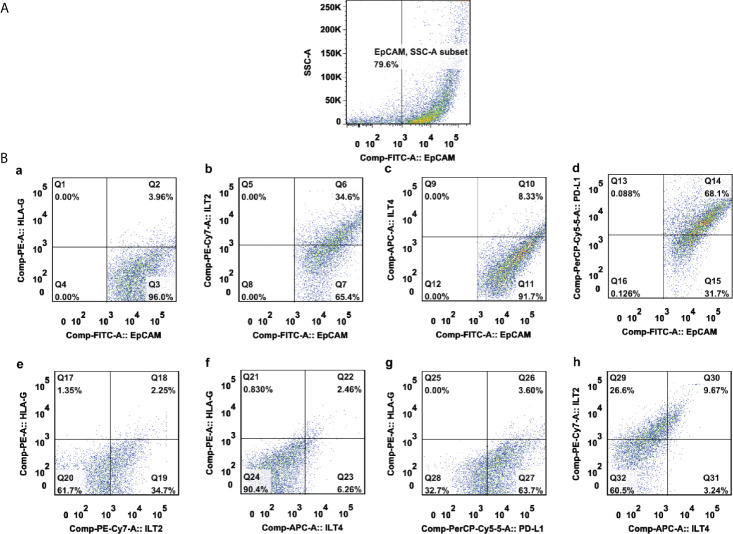
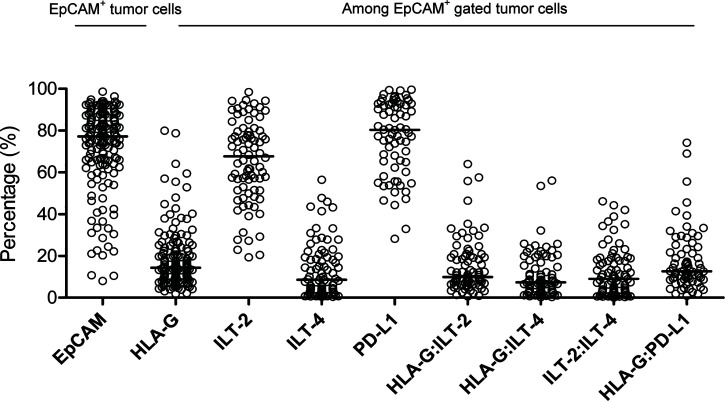
Representative flow cytometry analysis is shown in Figure 1 . Most samples (86.0%, 135/157) had more than 50% of EpCAM+ CRC cells. Among these samples, the median percentage of EpCAM (CD326)+ CRC cells was 77.2% (range: 7.6% ~ 98.60%), out of which the median percentage of the expression of HLA-G, ILT-2, ILT-4 and PD-L1 was 14.90% (n=157; range: 1.81% ~ 79.90%), 67.70% (n=82; range: 19.30% ~ 98.40%), 8.55% (n=82; range: 0.41% ~ 56.40%), and 80.30% (n=70; range: 28% ~ 99.60%), respectively. Moreover, the median percentage of the expression of HLA-G:ILT-2, HLA-G:ILT-4, ILT-2:ILT4 and HLA-G:PD-L1 was 9.90% (n=82; range: 0.61% ~ 64.0%), 7.35% (n=82; range: 0.26% ~ 56.0%), 8.99% (n=82; range: 0.01% ~ 46.10%), and 12.60% (n=70; range: 0.85% ~ 74.20%), respectively ( Figure 2 ).
Figure 1.
A representative multicolor flow cytometry analysis of either HLA-G, ILT-2, ILT-4, and PD-L1 alone or combination in a CRC lesion. (A) EpCAM+-gated CRC tumor cells; (B) expression percentage of (a) HLA-G, (b) ILT-2, (c) ILT-4, (d) PD-L1, (e) HLA-G:ILT-2, (f) HLA-G:ILT-4, (g) HLA-G:PD-L1, and (h) ILT-2:ILT-4 among EpCAM+-gated CRC tumor cells.
Figure 2.
Percentage distribution of EpCAM+ CRC cells, as well as HLA-G, ILT-2, ILT-4, PD-L1, HLA-G:ILT-2, HLA-G:ILT-4, ILT2:ILT-4 and HLA-G:PD-L1 expression in EpCAM+ gated CRC cells.
Bivariate correlation analysis revealed a significant positive correlation between HLA-G, ILT-2, ILT-4, and PD-L1 expression in EpCAM+ CRC cells. HLA-G and ILT-2 (r=0.463; p<0.001), HLA-G and ILT-4 (r=0.744; p<0.001), HLA-G and PD-L1 (r=0.573; p<0.001), ILT-2 and ILT-4 (r=0.408; p<0.001), ILT-2 and PD-L1 (r=0.603; p<0.001), and ILT-4 and PD-L1 (r=0.703; p<0.001) ( Supplementary Figure 1 ).
Relationship Between HLA-G, ILT-2, ILT-4 and PD-L1 Expression and CRC Clinical Features
Based on the median levels of HLA-G, ILT-2, ILT-4, and PD-L1 expression, each of them was divided into high-and low-level expression groups. HLA-Ghigh and HLA-Glow groups had 72 and 85 cases, respectively, whereas ILT-2high and ILT-2low had 40 and 42 cases, respectively; ILT-4high and ILT-4low had 42 and 40 cases, respectively, and PD-L1high and PD-L1low had 35 and 35 cases, respectively ( Table 1 ).
Data showed that patients with CRC having distant lymph node metastasis (pT stage) were significantly associated with high levels of HLA-G (p=0.028), ILT-2 (p=0.055), ILT-4 (p=0.005), and PD-L1 (p=0.064). Moreover, high levels of HLA-G, ILT-2, ILT-4, and PD-L1 were more evident among patients in advanced disease stages. However, no marked difference was found between the HLA-G, ILT-2, ILT-4 and PD-L1 expression status and the clinicopathological parameters, such as patient age, sex, and colon or rectal cancer types ( Table 1 ).
Levels of HLA-G, ILT-2, ILT-4 and PD-L1 Expression and CRC Survival
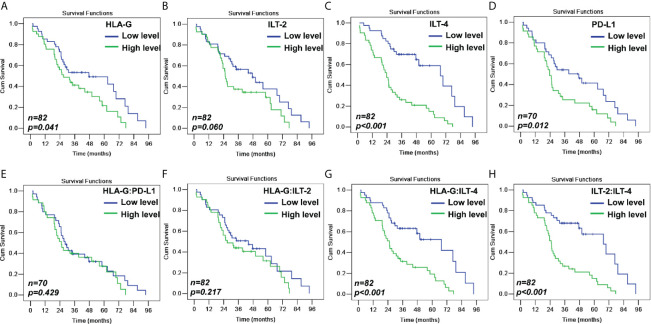
In addition to the traditional pN (p<0.001), pM (p=0.004), and AJCC disease stage (p=0.001), Kaplan-Meier log-rank test results showed that higher expression of HLA-G, ILT-2, ILT-4, and PD-L1 was significantly associated with poor survival. A remarkably shorter overall survival (OS) for patients with CRC was observed between HLA-Ghigh and HLA-Glow (35.5 months vs. 48.7 months; p=0.041), ILT-2high and ILT-2low (36.1 months vs. 47.1 months; p=0.060), ILT-4high and ILT-4low (28.6 months vs. 58.1 months; p<0.001), and PD-L1high and PD-L1low (29.9 months vs. 45.5 months; p=0.012). Furthermore, patients with higher percentages of HLA-GILT-4 and ILT-2ILT-4 had significantly worse survival that those with lower HLA-GILT-4 and ILT-2ILT-4 expression (all p<0.001). No notable differences were found in CRC survival in terms of patient age, sex, pT stage, colon or rectal cancer types, and patients with low or high levels of HLA-G:ILT-2, and HLA-G:PD-L1 expression ( Table 2 and Figure 3 ).
Table 2.
Kaplan-Meier log-rank analysis of variables for CRC patient survival.
| Variables | No. Total | No. Events | Survival (months) Mean (95% CI) | p | |
|---|---|---|---|---|---|
| Subtypes | colon | 60 | 42 | 42.4 (34.4 – 50.4) | 0.528 |
| rectal | 22 | 18 | 40.2 (29.0 – 51.4) | ||
| Gender | male | 40 | 32 | 43.0 (33.6-52.3) | 0.444 |
| female | 42 | 28 | 39.3 (37.5-47.1) | ||
| Age | <69-year | 41 | 30 | 43.3 (34.3-52.3) | 0.563 |
| >69-year | 41 | 30 | 39.6 (30.6-48.6) | ||
| pT stage | T2 | 6 | 4 | 58.1 (37.1-79.2) | 0.341 |
| T3+4 | 76 | 56 | 40.3 (33.7-46.9) | ||
| pN stage | N0 | 32 | 18 | 56.6 (45.5-67.7) | <0.001 |
| N1 | 27 | 22 | 40.4 (30.8-49.9) | ||
| N2 | 23 | 20 | 23.7 (15.5-31.8) | ||
| pM stage | M0 | 78 | 56 | 42.9 (36.4-49.4) | 0.004 |
| M1 | 4 | 4 | 16.5 (6.30-26.7) | ||
| AJCC stage | I+II | 32 | 18 | 56.7 (45.5-67.7) | 0.001 |
| III+IV | 50 | 42 | 32.7 (25.9-35.6) | ||
| Percentage (%)* | |||||
| HLA-G | Low vs. high | 41 | 27 | 48.7 (38.7-58.6) | 0.041 |
| 41 | 33 | 35.5 (27.8-43.2) | |||
| HLA-G:PD-L1 | 35 | 30 | 39.0 (29.4- 48.6) | 0.429 | |
| 35 | 29 | 35.5 (26.9- 44.2) | |||
| ILT-2 | 42 | 28 | 47.1 (37.3-56.9) | 0.060 | |
| 40 | 32 | 36.1 (28.1-44.0) | |||
| ILT-4 | 40 | 20 | 58.1 (48.0-68.2) | <0.001 | |
| 42 | 40 | 28.6 (21.9-35.4) | |||
| PD-L1 | 35 | 26 | 45.5 (35.1-55.9) | 0.012 | |
| 35 | 33 | 29.9 (22.3-37.4) | |||
| HLA-G:ILT-2 | 41 | 28 | 45.1 (35.1-55.0) | 0.217 | |
| 41 | 32 | 38.5 (30.2-46.8) | |||
| HLA-G:ILT-4 | 41 | 22 | 54.7 (44.0-65.4) | <0.001 | |
| 41 | 38 | 31.6 (24.4-38.8) | |||
| ILT-2:ILT-4 | 41 | 21 | 56.0 (45.5-66.5) | <0.001 | |
| 41 | 39 | 30.0 (23.4-36.6) |
*Percentage among EpCAM+ gated CRC tumor cells.
Figure 3.
Kaplan-Meier survival analysis for patients with CRC between (A) HLA-Ghigh and HLA-Glow (p=0.041); (B) ILT-2high and ILT-2low (p=0.060); (C) ILT-4high and ILT-4low (p<0.001); (D) PD-L1high and PD-L1low (p=0.012); (E) HLA-G:PD-L1high and HLA-G:PD-L1low (p=0.429); (F) HLA-G:ILT-2high and HLA-GILT-2low (p=0.217); (G) HLA-GILT-4high and HLA-G:ILT-4low (p<0.001); and (H) ILT-2:ILT-4high and ILT-2ILT-4low (p<0.001).
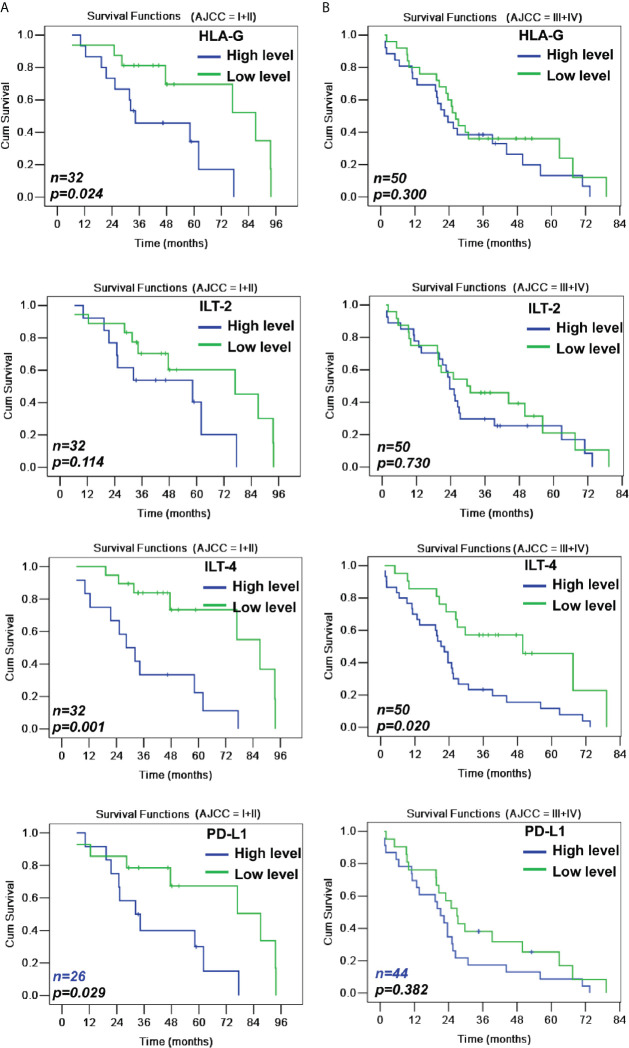
When CRC patients were stratified by early and advanced AJCC stages, HLA-G levels were only related to the survival among CRC patients with early disease stage (43.4 months vs. 70.4 months; p=0.024), while ILT-4 levels were significant for both CRC patients with early (36.2 months vs. 73.6 months; p=0.001) and advanced (25.5 months vs. 47.4 months; p=0.020) disease stages ( Table 3 and Figure 4 ).
Table 3.
Log-rank Mantel-Cox analysis of stratified variables in survival by tumor AJCC status in CRC patients.
| Variables | Stratified variables | AJCC I+II | p | AJCC III+IV | p | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. Total | No. Events | Survival Mean (95% CI) | No. Total | No. Events | Survival Mean (95% CI) | ||||
| HLA-G | low | 16 | 8 | 70.4 (53.9 – 87.0) | 0.024 | 25 | 19 | 37.2 (26.5 – 47.9) | 0.300 |
| high | 15 | 11 | 43.4 (30.5 – 56.2) | 26 | 22 | 30.4 (21.1 – 39.7) | |||
| ILT-2 | low | 18 | 10 | 64.7 (48.3 – 81.1) | 0.114 | 24 | 18 | 36.7 (25.8 – 47.5) | 0.730 |
| high | 13 | 9 | 46.3 (32.1 – 60.4) | 27 | 23 | 31.3 (22.0 – 40.5) | |||
| ILT-4 | low | 19 | 8 | 73.6 (59.3 – 87.8) | 0.001 | 21 | 12 | 47.4 (34.5 – 60.3) | 0.020 |
| high | 12 | 11 | 36.2 (22.4 – 50.0) | 30 | 29 | 25.5 (18.0 – 33.0) | |||
| PD-L1 | low | 14 | 8 | 67.9 (49.4 – 86.3) | 0.029 | 21 | 18 | 34.2 (23.4 – 45.0) | 0.382 |
| high | 12 | 10 | 41.3 (27.5 – 55.0) | 23 | 23 | 23.8 (15.6 – 32.1) | |||
Figure 4.
Kaplan-Meier survival analysis of low and high levels of HLA-G, ILT-2, ILT-4 and PD-L1 for survival either in AJCC stage I+II (A) or in AJCC stage III+IV (B) CRC patients.
Prognostic Value of HLA-G, ILT-2, ILT-4 and PD-L1 Expression in Patients With CRC
Finally, using univariate and covariate Cox regression analyses, we analyzed the prognostic value of AJCC disease stage and HLA-G, ILT-2, ILT-4 and PD-L1 expression, respectively and evaluated how these variables were significantly related to the survival of patients with CRC. Although AJCC disease stage, HLA-G, ILT-2, ILT-4, and PD-L1 expression were closely related to the survival of patients with CRC, covariate Cox regression data revealed that only AJCC disease stage (HR=2.435, 95% CI: 1.300 ~ 4.562; p=0.005) and ILT-4 (HR=2.198, 0.985 ~ 5.042; p=0.063) were independent prognostic factors for those patients ( Table 4 ).
Table 4.
Cox regression analysis for prognostic value of variables for CRC patients.
| Variables | Categories | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| AJCC stage | III/IV vs. I/II | 2.694 (1.502-4.831) | 0.001 | 2.435 (1.300-4.562) | 0.005 |
| HLA-G | high vs. low | 1.727 (1.016-2.936) | 0.043 | 0.859 (0.428-1.723) | 0.668 |
| ILT-2 | 1.652 (0.974-2.801) | 0.062 | 0.933 (0.488-1.783) | 0.834 | |
| ILT-4 | 3.537 (1.955-6.272) | <0.001 | 2.198 (0.985-5.042) | 0.063 | |
| PD-L1 | 1.962 (1.146-3.360) | 0.014 | 1.224 (0.569-2.631) | 0.605 | |
HR, hazard ratio; 95% CI, 95% confidence interval.
Discussion
Considerable therapeutic success has been achieved by using ICIs, such as clinically validated antibodies targeting the PD-1/PD-L1 and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)/B7 signaling pathways. Exploration of novel immune checkpoints and development of new ICIs has gained momentum in the field of cancer immunotherapy (29). Among them, the HLA-G/ILTs signaling pathway has been recently recognized as a new immune checkpoint, and ICIs targeting this interaction are in early clinical trials to treat different types of advanced solid tumors (30).
EpCAM-based cell surface markers for identification of epithelial cancers and as a target for circulating tumor cell (CTC) isolation and enrichment have long been established (31). In this study, the percentage of EpCAM+ CRC cells varied, but most samples (86.0%, 135/137) had more than 50% of these cells. In a study with 21 fresh CRC lesions that were analyzed using a protocol similar to ours, Satio et al. (32) reported that both low and high counts of EpCAM+ cells detected by flow cytometry in cell suspensions isolated from the lesions were tumor cells of a proportion of 64.0%±15.8% as compared to 72.1±20.0% observed in our study. This marginal difference might be attributed to the different compositions of the CRC cohorts analyzed.
We then analyzed the expression of immune checkpoint proteins, HLA-G, ILT-2, ILT-4, and PD-L1, using multicolor flow cytometry in EpCAM+-gated tumor cells from frozen CRC samples. Our results revealed that in samples with higher counts of EpCAM+ tumor cells, the expression of HLA-G, ILT-2, ILT-4, and PD-L1 was significantly related to distant lymph node metastasis, advanced disease stage, and shorter patient survival. To be noted, when CRC patients were stratified by early and advanced AJCC stages, our data showed that HLA-G levels were only related to the survival in CRC patients with early disease stage, and ILT-4 levels were significant for both CRC patients with early and advanced disease stages. In addition to well-known and clinically proven ICIs targeting the PD-1/PD-L1 signaling pathway, it has been demonstrated using the Cox regression data that a higher ILT-4 expression could be an independent prognostic risk factor for patients with CRC. Phase I clinical trials based on the HLA-G/ILTs interaction blockade for the treatment of different types of advanced solid tumors have been performed, and the clinical benefits are yet to be evaluated (30).
The expression of ILT-2 and ILT-4 has been observed in various immune and tumor cells (22). Several studies have demonstrated that the binding of HLA-G expressed on tumor cells to immune cells bearing ILT-2 and ILT-4 aids the evasion of the anti-tumor immune responses of the host. This interaction can suppress both the peripheral and intra-tumor functions of immune-competent cells by (i) inhibiting the proliferation and cytolytic functions of T cells and NK cells, thereby hampering the maturation and antigen-presentation ability of DCs, and (ii) inducing the generation of immune regulatory cells, such as Tregs, MDSCs, DC-10 and M2 type macrophages (5). In this context, our findings in this study support the conclusion drawn from previous studies that tumor HLA-G expression is closely related to tumor progression and poor clinical outcomes in patients with cancer. A large-scale genome-wide association (GWAS) study of East Asians (22,775 CRC patients and 47,731 controls) revealed that HLA-G is one of the leading loci associated with the risk of CRC (33). At the protein level, enhanced HLA-G expression has been found to be greatly correlated with weak anti-tumor immune response, disease progression, and poor survival, and HLA-G is an independent prognostic predictor of CRC (34, 35).
The association of ILT-2 and ILT-4 expression with tumor malignancy and poor prognosis has been addressed in many previous reports (25). Pre-clinical data have shown that ILT-2/4 blockade can remarkably restore the cytotoxic capacity of CD8+ T cells or NK cells against tumor cells, thereby promoting antitumor immune responses (36–38). We found that the expression of ILT-2 and ILT-4 in CRC lesions was significantly correlated with HLA-G and PD-L1 expression. It has been previously reported that the counts of antigen-presenting cells and NK cells, and the expression of ILT-2/4 on T cell surface were increased by HLA-G (39). Moreover, there has been evidence that HLA-G upregulates ILT-4 expression in CRC cell lines in an autocrine manner, and HLA-G/ILT-4 engagement enhances CRC cell proliferation, migration, and invasion by the activation of protein kinase B (AKT)/extracellular signal regulated kinase (ERK) signaling (40). HLA-G/ILT-4 engagement via ERK signaling may be involved in the upregulation of vascular endothelial growth factor C (VEGF-C) expression in non-small cell lung cancer (NSCLC) cells (41). García et al. (42) reported more evidence regarding the mechanism by which HLA-G/ILT-4 interaction increased VEGF-C expression in clear cell renal cell carcinoma. It indicated the significance of HLA-G/ILT-4 signaling in the promotion of angiogenesis during tumor development, which is the rationale behind the findings of Carosella et al. (43) who proposed that a combination of anti-angiogenic therapies can enhance the synergistic antitumor activity of ICI monotherapy.
In summary, our findings revealed that ILT-4 is the most significant prognostic indicator among HLA-G/ILT-2/4 and PD-L1 immune checkpoint proteins in patients with CRC. Combinatorial immunotherapy with different ICIs could improve the clinical outcomes in patients with cancer. To be noted, ICIs such as PD1 antibody are only approved for solid cancer patients with microsatellite instability-high (MSI-H), mismatch-repair deficiency (dMMR) or high tumor mutational burden (TMB-high). Though MSI-H, dMMR or TMB-high molecular phenotype varies among cancer patients, and not all patients with the MSI-H, dMMR or TMB-high phenotype respond to the ICIs therapy. In this context, exploration of mechanisms underlying resistance to ICIs is necessary (44, 45). Moreover, due to many challenges, such as high variation in inter-assay precision and cut-off values associated with routine immunohistochemistry, multicolor flow cytometry can serve as a reliable and efficient alternative method. However, due to these CRC samples were collected and frozen between 2006 and 2012, no case-matched fresh samples were analyzed for the expression of HLA-G, ILT-2, ILT-4 and PD-L1 with flow cytometry method. Whether cryopreservation with different time could affect the expression of these molecules remains to be evaluated in our future prospective studies.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Review Board of Taizhou Hospital of Zhejiang Province (K20190766). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Concept and design, manuscript drafting and statistical analysis: W-HY and AL. Experiments and methods: Q-YC and Y-XC. Data collection and interpretation of data: Q-YH, J-GZ, W-JZ, XZ, and Y-HY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Science and Technology Bureau of Taizhou (1901ky01; 1901ky04; 20ywa01; 20ywa03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.679090/full#supplementary-material
| Bivariate correlation analysis between the expression of HLA-G, ILT-2, ILT-4, and PD-L1 performed using Pearson test.
Flowchart of the study protocol.
References
- 1. Mortezaee K. Immune Escape: A Critical Hallmark in Solid Tumors. Life Sci (2020) 258:118110. 10.1016/j.lfs.2020.118110 [DOI] [PubMed] [Google Scholar]
- 2. Morganti S, Curigliano G. Combinations Using Checkpoint Blockade to Overcome Resistance. Ecancermedicalscience (2020) 14:1148. 10.3332/ecancer.2020.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. Pd-L1 as a Biomarker of Response to Immune-Checkpoint Inhibitors. Nat Rev Clin Oncol (2021). 10.1038/s41571-021-00473-5 [DOI] [PubMed] [Google Scholar]
- 4. Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent Advances in Our Understanding of HLA-G Biology: Lessons From a Wide Spectrum of Human Diseases. J Immunol Res (2016) 2016:4326495. 10.1155/2016/4326495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. 10.3389/fimmu.2018.02164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loustau M, Anna F, Dréan R, Lecomte M, Langlade-Demoyen P, Caumartin J. Hla-G Neo-Expression on Tumors. Front Immunol (2020) 11:1685. 10.3389/fimmu.2020.01685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. Hla-G: An Immune Checkpoint Molecule. Adv Immunol (2015) 127:33–144. 10.1016/bs.ai.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 8. Attia JVD, Dessens CE, van de Water R, Houvast RD, Kuppen PJK, Krijgsman D. The Molecular and Functional Characteristics of HLA-G and the Interaction With Its Receptors: Where to Intervene for Cancer Immunotherapy? Int J Mol Sci (2020) 21(22):8678. 10.3390/ijms21228678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naji A, Menier C, Maki G, Carosella ED, Rouas-Freiss N. Neoplastic B-cell Growth is Impaired by HLA-G/ILT2 Interaction. Leukemia (2012) 26(8):1889–92. 10.1038/leu.2012.62 [DOI] [PubMed] [Google Scholar]
- 10. Morandi F, Marimpietri D, Görgens A, Gallo A, Srinivasan RC, El-Andaloussi S, et al. Human Amnion Epithelial Cells Impair T Cell Proliferation: The Role of HLA-G and. Cells (2020) 9(9):2123. 10.3390/cells9092123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-Like Transcript 2 Receptor Suppresses B Cell Responses. J Immunol (2014) 192(4):1536–46. 10.4049/jimmunol.1300438 [DOI] [PubMed] [Google Scholar]
- 12. Naji A, Durrbach A, Carosella ED, Rouas-Freiss N. Soluble HLA-G and HLA-G1 Expressing Antigen-Presenting Cells Inhibit T-cell Alloproliferation Through ILT-2/ILT-4/FasL-mediated Pathways. Hum Immunol (2007) 68(4):233–9. 10.1016/j.humimm.2006.10.017 [DOI] [PubMed] [Google Scholar]
- 13. Riteau B, Menier C, Khalil-Daher I, Martinozzi S, Pla M, Dausset J, et al. Hla-G1 Co-Expression Boosts the HLA Class I-mediated NK Lysis Inhibition. Int Immunol (2001) 13(2):193–201. 10.1093/intimm/13.2.193 [DOI] [PubMed] [Google Scholar]
- 14. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6–STAT3 signaling pathwa. Proc Natl Acad Sci USA (2008) 105(24):8357–62. 10.1073/pnas.0803341105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Köstlin N, Ostermeir AL, Spring B, Schwarz J, Marmé A, Walter CB, et al. HLA-G Promotes Myeloid-Derived Suppressor Cell Accumulation and Suppressive Activity During Human Pregnancy Through Engagement of the Receptor ILT4. Eur J Immunol (2017) 47(2):374–84. 10.1002/eji.201646564 [DOI] [PubMed] [Google Scholar]
- 16. Zhang YH, He M, Wang Y, Liao AH. Modulators of the Balance Between M1 and M2 Macrophages During Pregnancy. Front Immunol (2017) 8:120. 10.3389/fimmu.2017.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melsted WN, Matzen SH, Andersen MH, Hviid TVF. The Choriocarcinoma Cell Line JEG-3 Upregulates Regulatory T Cell Phenotypes and Modulates Pro-Inflammatory Cytokines Through HLA-G. Cell Immunol (2018) 324:14–23. 10.1016/j.cellimm.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 18. Amodio G, Comi M, Tomasoni D, Gianolini ME, Rizzo R, LeMaoult J, et al. HLA-G Expression Levels Influence the Tolerogenic Activity of Human DC-10. Haematologica (2015) 100(4):548–57. 10.3324/haematol.2014.113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu CL, Caumartin J, Amodio G, Anna F, Loustau M, Gregori S, et al. Inhibition of Inkt Cells by the HLA-G-ILT2 Checkpoint and Poor Stimulation by HLA-G-Expressing Tolerogenic Dc. Front Immunol (2021) 11:608614. 10.3389/fimmu.2020.608614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morandi F, Rouas-Freiss N, Pistoia V. The Emerging Role of Soluble HLA-G in the Control of Chemotaxis. Cytokine Growth Factor Rev (2014) 25(3):327–35. 10.1016/j.cytogfr.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 21. Dumont C, Jacquier A, Verine J, Noel F, Goujon A, Wu CL, et al. Cd8+Pd-1-Ilt2+ T Cells are an Intratumoral Cytotoxic Population Selectively Inhibited by the Immune-Checkpoint Hla-G. Cancer Immunol Res (2019) 7(10):1619–32. 10.1158/2326-6066.CIR-18-0764 [DOI] [PubMed] [Google Scholar]
- 22. Kang X, Kim J, Deng M, John S, Chen H, Wu G, et al. Inhibitory Leukocyte Immunoglobulin-Like Receptors: Immune Checkpoint Proteins and Tumor Sustaining Factors. Cell Cycle (2016) 15(1):25–40. 10.1080/15384101.2015.1121324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng J, Gao X, Zhang X, Guo H, Chen S, Gou X. Leukocyte Immunoglobulin-Like Receptor Subfamily B Member 1 Potentially Acts as a Diagnostic and Prognostic Target in Certain Subtypes of Adenocarcinoma. Med Hypotheses (2020) 144:109863. 10.1016/j.mehy.2020.109863 [DOI] [PubMed] [Google Scholar]
- 24. Cheng J, Luan J, Chen P, Kuang X, Jiang P, Zhang R, et al. Immunosuppressive Receptor LILRB1 Acts as a Potential Regulator in Hepatocellular Carcinoma by Integrating With SHP1. Cancer Biomark (2020) 28(3):309–19. 10.3233/CBM-190940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao A, Sun Y, Peng G. ILT4 Functions as a Potential Checkpoint Molecule for Tumor Immunotherapy. Biochim Biophys Acta Rev Cancer (2018) 1869(2):278–85. 10.1016/j.bbcan.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 26. Kerr KM, Hirsch FR. Programmed Death Ligand-1 Immunohistochemistry: Friend or Foe? Arch Pathol Lab Med (2016) 140(4):326–31. 10.5858/arpa.2015-0522-SA [DOI] [PubMed] [Google Scholar]
- 27. Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann Surg Oncol (2010) 17(6):1471–4. 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 28. Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, et al. γδt17 Cells Promote the Accumulation and Expansion of Myeloid-Derived Suppressor Cells in Human Colorectal Cancer. Immunity (2014) 40(5):785–800. 10.1016/j.immuni.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hahn AW, Gill DM, Pal SK, Agarwal N. The Future of Immune Checkpoint Cancer Therapy After PD-1 and CTLA-4. Immunotherapy (2017) 9(8):681–92. 10.2217/imt-2017-0024 [DOI] [PubMed] [Google Scholar]
- 30. Gilead Buys into Tizona’s Anti-HLA-G Strategy. Cancer Discov (2020) 10(10):1433. 10.1158/2159-8290.CD-NB2020-077 [DOI] [PubMed] [Google Scholar]
- 31. Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells (2020) 9(8):1836. 10.3390/cells9081836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saito A, Tojo M, Kumagai Y, Ohzawa H, Yamaguchi H, Miyato H, et al. Flow Cytometry Detection of Cell Type-Specific Expression of Programmed Death Receptor Ligand-1 (PD-L1) in Colorectal Cancer Specimens. Heliyon (2020) 7(1):e05880. 10.1016/j.heliyon.2020.e05880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai Q, et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated With Risk for Colorectal Cancer. Gastroenterology (2019) 156(5):1455–66. 10.1053/j.gastro.2018.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human Leukocyte Antigen G Expression: As a Significant Prognostic Indicator for Patients With Colorectal Cancer. Mod Pathol (2007) 20(3):375–83. 10.1038/modpathol.3800751 [DOI] [PubMed] [Google Scholar]
- 35. Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive Value of Different Proportion of Lesion HLA-G Expression in Colorectal Cancer. Oncotarget (2017) 8(64):107441–51. 10.18632/oncotarget.22487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen H, Chen Y, Deng M, John S, Gui X, Kansagra A, et al. Antagonistic anti-LILRB1 Monoclonal Antibody Regulates Antitumor Functions of Natural Killer Cells. J Immunother Cancer (2020) 8(2):e000515. 10.1136/jitc-2019-000515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim A, Han CJ, Driver I, Olow A, Sewell AK, Zhang Z, et al. Lilrb1 Blockade Enhances Bispecific T Cell Engager Antibody-Induced Tumor Cell Killing by Effector Cd8+ T Cells. J Immunol (2019) 203(4):1076–87. 10.4049/jimmunol.1801472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen HM, van der Touw W, Wang YS, Kang K, Mai S, Zhang J, et al. Blocking Immunoinhibitory Receptor LILRB2 Reprograms Tumor-Associated Myeloid Cells and Promotes Antitumor Immunity. J Clin Invest (2018) 128(12):5647–62. 10.1172/JCI97570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J (2005) 19(6):662–4. 10.1096/fj.04-1617fje [DOI] [PubMed] [Google Scholar]
- 40. Cai Z, Wang L, Han Y, Gao W, Wei X, Gong R, et al. Immunoglobulin-Like Transcript 4 and Human Leukocyte Antigen-G Interaction Promotes the Progression of Human Colorectal Cancer. Int J Oncol (2019) 54(6):1943–54. 10.3892/ijo.2019.4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang P, Guo X, Li J, Yu S, Wang L, Jiang G, et al. Immunoglobulin-Like Transcript 4 Promotes Tumor Progression and Metastasis and Up-Regulates VEGF-C Expression Via ERK Signaling Pathway in non-Small Cell Lung Cancer. Oncotarget (2015) 6(15):13550–63. 10.18632/oncotarget.3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. García M, Palma MB, Verine J, Miriuka S, Inda AM, Errecalde AL, et al. The Immune-Checkpoint HLA-G/ILT4 is Involved in the Regulation of VEGF Expression in Clear Cell Renal Cell Carcinoma. BMC Cancer (2020) 20(1):624. 10.1186/s12885-020-07113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carosella ED, Gregori S, Tronik-Le Roux D. HLA-G/Lilrbs: A Cancer Immunotherapy Challenge. Trends Cancer (2021) 7(5):389–92. 10.1016/j.trecan.2021.01.004 [DOI] [PubMed] [Google Scholar]
- 44. Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. ESMO Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and its Relationship With PD-1/PD-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann Oncol (2019) 30(8):1232–43. 10.1093/annonc/mdz116 [DOI] [PubMed] [Google Scholar]
- 45. Kwon M, An M, Klempner SJ, Lee H, Kim KM, Sa JK, et al. Determinants of Response and Intrinsic Resistance to PD-1 Blockade in Microsatellite Instability-High Gastric Cancer. Cancer Discov (2021). 10.1158/2159-8290.CD-21-0219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| Bivariate correlation analysis between the expression of HLA-G, ILT-2, ILT-4, and PD-L1 performed using Pearson test.
Flowchart of the study protocol.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.