We show that the Sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) family proteins SERCA2 and SERCA3 have redundant functions that are required for RAG gene expression and DNA breaks during V(D)J recombination. Loss of SERCA activity leads to B lymphopenia in mice and humans.
Abstract
A whole-genome CRISPR/Cas9 screen identified ATP2A2, the gene encoding the Sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) 2 protein, as being important for V(D)J recombination. SERCAs are ER transmembrane proteins that pump Ca2+ from the cytosol into the ER lumen to maintain the ER Ca2+ reservoir and regulate cytosolic Ca2+-dependent processes. In preB cells, loss of SERCA2 leads to reduced V(D)J recombination kinetics due to diminished RAG-mediated DNA cleavage. SERCA2 deficiency in B cells leads to increased expression of SERCA3, and combined loss of SERCA2 and SERCA3 results in decreased ER Ca2+ levels, increased cytosolic Ca2+ levels, reduction in RAG1 and RAG2 gene expression, and a profound block in V(D)J recombination. Mice with B cells deficient in SERCA2 and humans with Darier disease, caused by heterozygous ATP2A2 mutations, have reduced numbers of mature B cells. We conclude that SERCA proteins modulate intracellular Ca2+ levels to regulate RAG1 and RAG2 gene expression and V(D)J recombination and that defects in SERCA functions cause lymphopenia.
Introduction
All developing lymphocytes must assemble the second exon of genes encoding antigen receptor chains (Bassing et al., 2002). This occurs through V(D)J recombination, a reaction where the RAG endonuclease, composed of the RAG1 and RAG2 proteins, introduces DNA double-strand breaks (DSBs) at the border of RAG recognition sequences (RSs) and variable (V), diversity (D), or joining (J) gene segments undergoing recombination (Fugmann et al., 2000). RAG-mediated DNA cleavage results in a signal end (SE) and coding end (CE) at each DSB, and the nonhomologous end-joining pathway of DNA DSB repair joins the two SEs to form a signal join (SJ) and the two CEs to form a coding join (CJ; Helmink and Sleckman, 2012).
The V(D)J recombination reaction can be studied in mouse preB cells transformed by the viral abl kinase, hereafter referred to as abl preB cells (Bredemeyer et al., 2006; Helmink and Sleckman, 2012). Treating these cells with the abl kinase inhibitor imatinib leads to cell cycle arrest in G0/G1, induction of RAG1 and RAG2 gene expression, and V(D)J recombination at the endogenous immunoglobulin light chain κ (Igk) locus and chromosomally integrated retroviral V(D)J recombination substrates (Bredemeyer et al., 2006; Helmink and Sleckman, 2012; Hung et al., 2018; Muljo and Schlissel, 2003). Here we used abl preB cells for a CRISPR/Cas9 whole-genome guide RNA (gRNA) screening approach to identify novel genes encoding proteins required for V(D)J recombination. This approach revealed the Sarco/ER Ca2+-ATPase 2 (SERCA2) protein, encoded by the ATP2A2 gene, as a potential regulator of V(D)J recombination.
SERCA proteins are ATP-dependent Ca2+ transporters that move Ca2+ from the cytosol into the ER in order to maintain low cytosolic, and high ER, Ca2+ concentrations (Stammers et al., 2015; Vandecaetsbeek et al., 2011). There are two additional SERCA family members, SERCA1 and SERCA3, encoded by distinct genes, ATP2A1 and ATP2A3, respectively (Stammers et al., 2015; Vandecaetsbeek et al., 2011). SERCA1 is expressed primarily in skeletal muscle, and SERCA3 is expressed in nonmuscle cells, including lymphocytes (Vandecaetsbeek et al., 2011; Wu et al., 1995; Wuytack et al., 1994). Mutations in ATP2A2 result in a rare autosomal dominant skin disorder, Darier disease (Ahn et al., 2003; Burge and Wilkinson, 1992; Leong et al., 2017; Tavadia et al., 2002).
Here we show that in developing B cells, SERCA2 and SERCA3 have redundant functions required to promote RAG1 and RAG2 expression and V(D)J recombination. Moreover, SERCA2 deficiency leads to B lymphopenia in mice and humans. We propose that SERCA proteins regulate intracellular Ca2+ homeostasis in a way that is required for normal RAG1 and RAG2 expression and efficient V(D)J recombination.
Results and discussion
SERCA2 is required for efficient V(D)J recombination
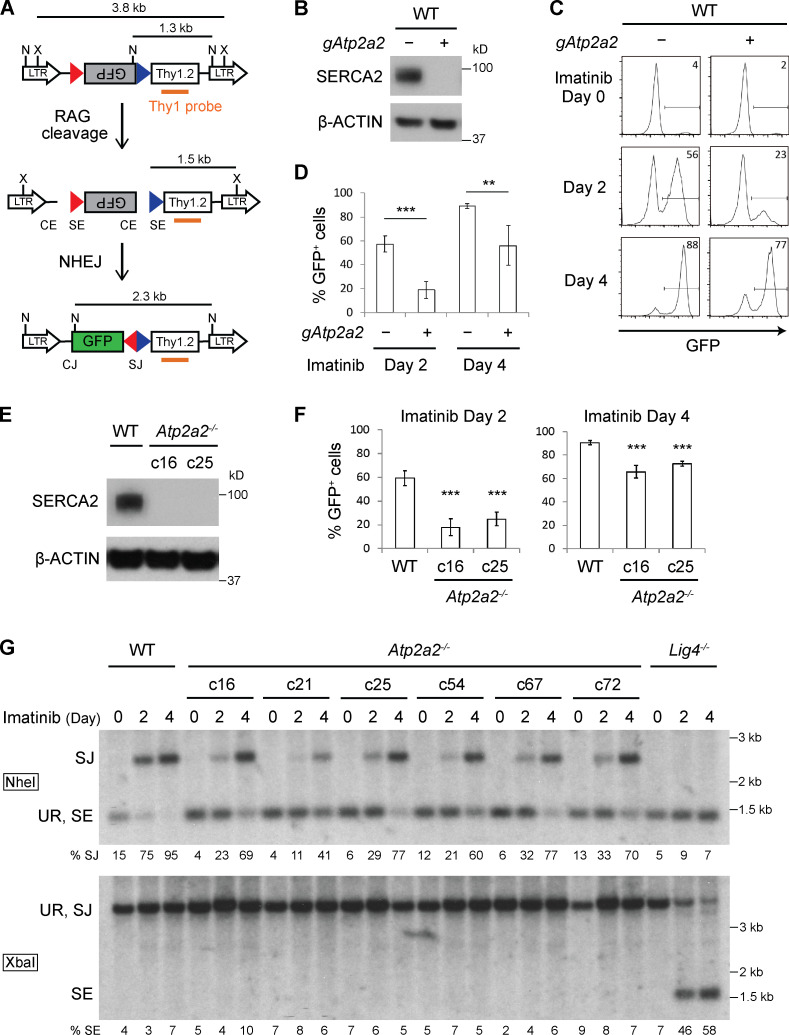
We generated WT abl preB cells containing a doxycycline-inducible Cas9 cDNA and pMGINV retroviral V(D)J recombination substrate (Fig. 1 A; Hung et al., 2018). pMGINV has an anti-sense GFP cDNA flanked by two RSs that mediate V(D)J recombination by inversion placing the GFP cDNA in the sense orientation leading to GFP expression (Hung et al., 2018). A lentiviral murine whole-genome gRNA library was introduced into these cells followed by doxycycline treatment for 7 d to promote Cas9 expression and gene inactivation (Tzelepis et al., 2016). These cells were treated with imatinib for 4 d to induce RAG expression and V(D)J recombination of pMGINV (Fig. 1 A). Abl preB cells that had not (GFP−) or had (GFP+) completed V(D)J recombination were isolated by flow-cytometric cell sorting, and gRNAs were sequenced from both populations.
Figure 1.
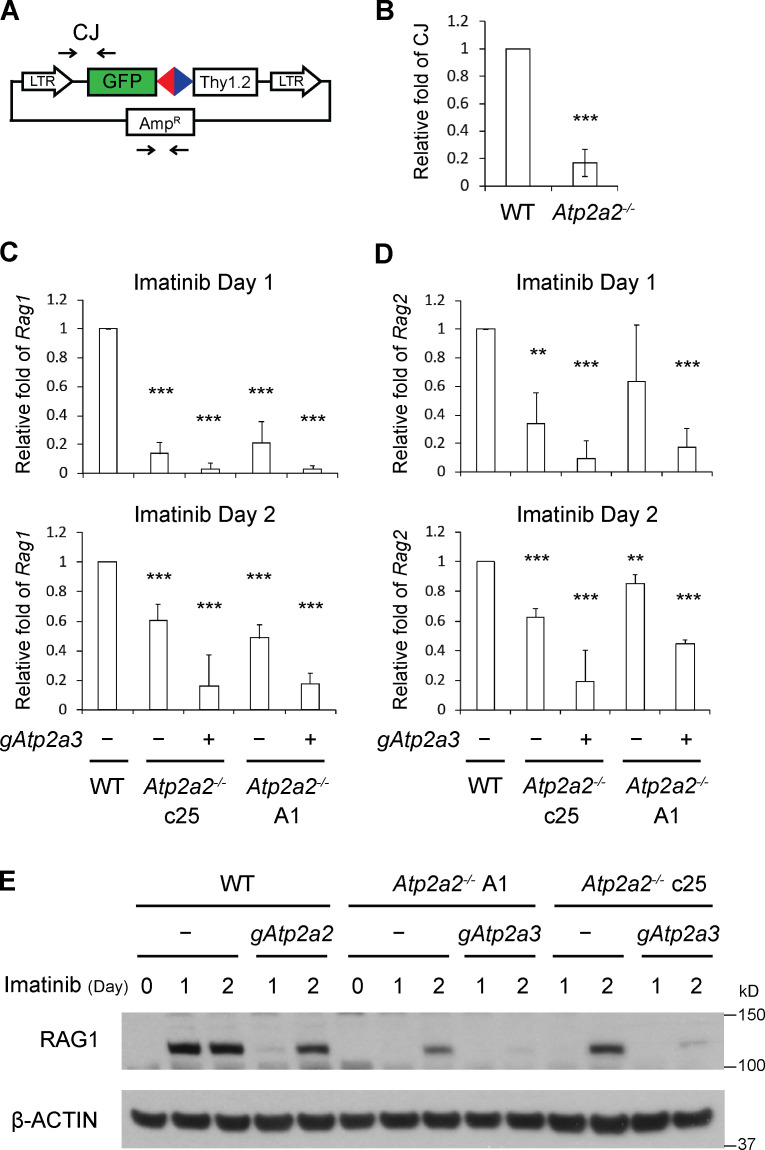
Loss of SERCA2 impairs RAG cleavage. (A) Schematic of pMGINV showing the antisense (gray rectangle) and sense (green rectangle) GFP cDNA, RSs (red and blue triangles), CE, SE, CJ, and SJ. The Thy1.2 cDNA (open rectangle), Thy1 probe (orange line), and NheI (N) and XbaI (X) sites are indicated. (B) Western blot for SERCA2 and β-actin in WT abl preB cells (Tet-On Cas9 clone 302) without (−) an Atp2a2 gRNA (gAtp2a2) and bulk Atp2a2 inactivated with (+) gAtp2a2 (n = 5). (C) Flow-cytometric analysis for GFP expression in WT (−) or bulk Atp2a2 inactivated (+) abl preB cells with pMGINV that were treated with imatinib for 0, 2, or 4 d. Percentage of GFP-expressing cells is indicated. (D) Mean ± SD of five experiments performed as in C. P values were calculated using Student’s t test throughout the figures. **, P < 0.01; ***, P < 0.001. (E) Western blot analysis of SERCA2 in two Atp2a2−/− abl preB clones (c16 and c25) and the parent WT abl preB cell line (c302) as described in B (n = 3). (F) Analysis of pMGINV rearrangement in Atp2a2−/− abl preB clones and WT abl preB cells as described in D (mean ± SD; n = 4). ***, P < 0.001 compared with WT. (G) Southern blot analyses of NheI (top) or XbaI (bottom) digested genomic DNA probed with the Thy1 probe as shown in A. Shown are Lig4−/−, WT, and Atp2a2−/− abl preB clones treated with imatinib for 0, 2, or 4 d. Bands reflecting unrearranged (UR) pMGINV and pMGINV SEs and SJs are indicated. The percentages of pMGINV that have SJs (%SJ) or SEs (%SE) are indicated and quantified as described in Materials and methods (n = 4 for SJ analysis in Atp2a2−/− c25; n = 1 for other Atp2a2−/− clones).
GFP− cells exhibited significant enrichment in gRNAs to Rag1 and Rag2, and Lig4 and Xrcc6, which encode the DNA ligase 4 and Ku70 proteins, respectively, that are required for the repair of RAG-mediated DSBs, demonstrating the veracity of our approach (Table S1; Helmink and Sleckman, 2012). Several distinct gRNAs to Atp2a2 were significantly enriched (approximately sixfold) in the GFP− population, suggesting that the SERCA2 may regulate V(D)J recombination (Table S1).
SERCA2 promotes RAG cleavage
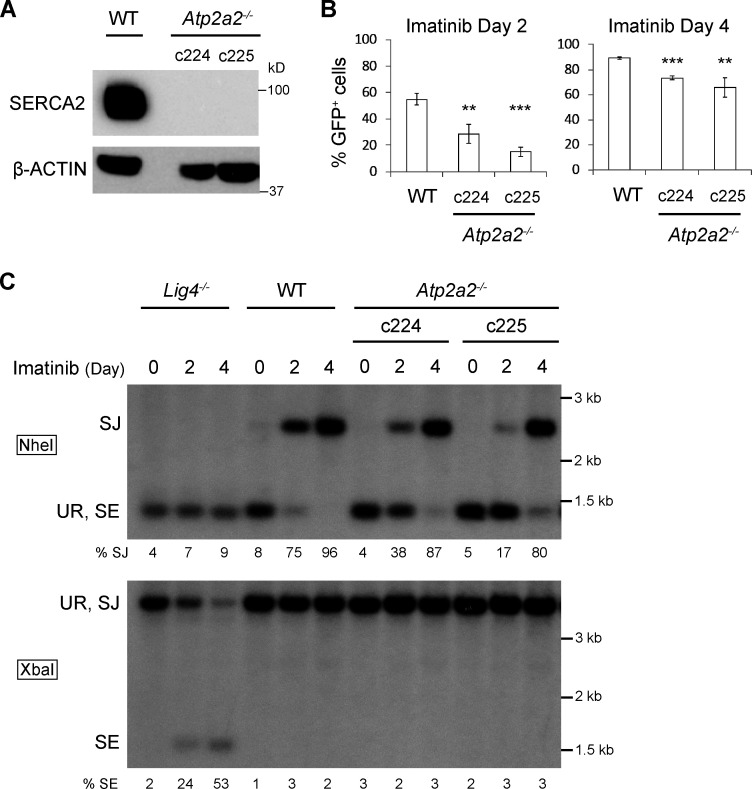
To validate our finding, we generated Atp2a2 “bulk inactivated” abl preB cells by introducing a gRNA to the 5′ region of Atp2a2 into WT abl preB cells containing a doxycycline-inducible Cas9 cDNA and pMGINV. These cells were treated with doxycycline for 6 d, leading to inactivation of most Atp2a2 alleles in the bulk population, as evidenced by the loss of SERCA2 protein (Fig. 1 B). Atp2a2 mRNA splice variants leading to different SERCA2 isoforms are generated by alternative splicing in the 3′ region of Atp2a2 and can be detected by the antibody we use for Western blotting (Lipskaia et al., 2014; Vandecaetsbeek et al., 2011). Thus, we conclude that SERCA2 isoforms are not expressed in the bulk Atp2a2 inactivated abl preB cells. Bulk Atp2a2 inactivated abl preB cells exhibited diminished pMGINV V(D)J recombination kinetics, as evidenced by the reduced fraction of GFP+ cells at 2 and 4 d after imatinib treatment (23% and 77%, respectively) compared with WT cells (56% and 88%, respectively; Fig. 1, C and D). Several Atp2a2−/− abl preB cell clones (c16, c21, c25, c54, c67, c72, c224, and c225) were generated from two distinct lines of WT abl preB cells, and SERCA2 loss was confirmed by Western blotting (Fig. 1 E and Fig. S1 A). As assessed by flow cytometry, Atp2a2−/− abl preB cell clones also exhibited impaired V(D)J recombination kinetics compared with WT abl preB cells (Fig. 1 F and Fig. S1 B).
Figure S1.
SERCA2 deficiency impairs V(D)J recombination. (A) Western blot analysis preformed as described in Fig. 1 B in Atp2a2−/− abl preB clones (c224 and c225) derived from a distinct WT abl preB cell line (BA600; n = 1). (B) Analysis of pMGINV rearrangement preformed as described in Fig. 1 D (mean ± SD; n = 3). **, P < 0.01; ***, P < 0.001 compared with WT. (C) Southern blot analyses of pMGINV in abl preB cells described in A performed as described in Fig. 1 G (n = 1).
To determine whether the requirement for SERCA2 is at the RAG DNA cleavage or DNA DSB repair step of V(D)J recombination, we assayed pMGINV by Southern blotting, which permits the detection of unrepaired SEs and normally repaired SJs (Fig. 1 A). In Atp2a2−/− abl preB cells treated with imatinib, the hybridizing band reflecting pMGINV SJs in NheI-digested DNA formed with slower kinetics than that in WT cells (Fig. 1 G and Fig. S1 C). Moreover, while hybridizing bands reflecting unrepaired pMGINV SEs were readily detected in XbaI-digested DNA from imatinib-treated Lig4−/− abl preB cells, they were not detected in Atp2a2−/− abl preB cells (Fig. 1 G and Fig. S1 C ). We conclude that loss of SERCA2 leads to diminished V(D)J recombination due to impaired RAG cleavage.
SERCA2 and SERCA3 function redundantly during V(D)J recombination
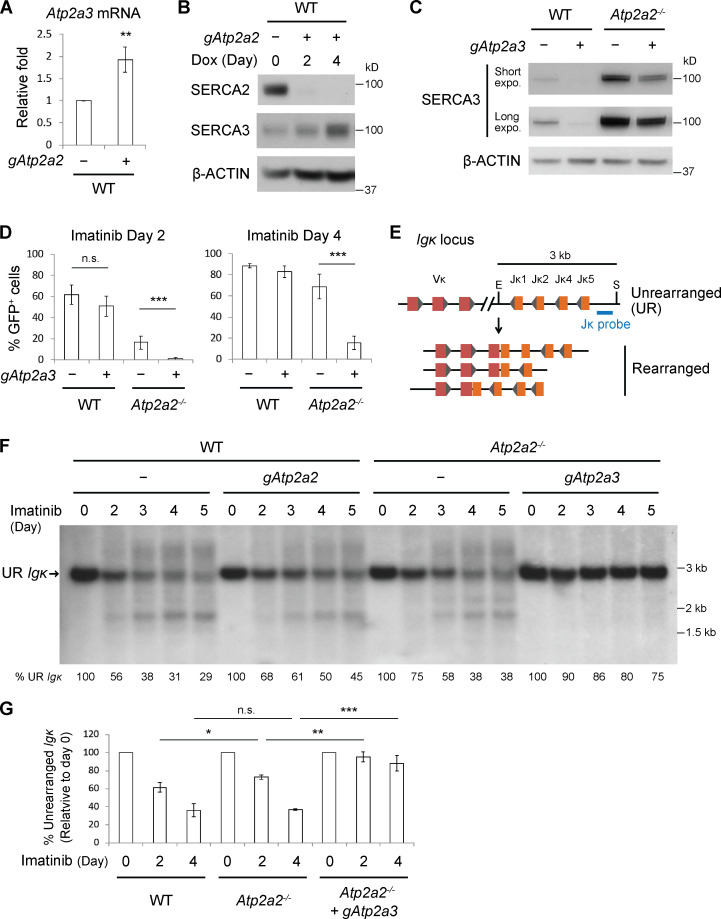
SERCA3, a less potent ER Ca2+ transporter, is often coexpressed with SERCA2, including in lymphoid cells (Chandrasekera et al., 2009; Vandecaetsbeek et al., 2011; Wu et al., 1995; Wuytack et al., 1994). Abl preB cells could compensate for loss of SERCA2 by up-regulating SERCA3 (Lipskaia et al., 2014). Indeed, bulk Atp2a2 inactivated abl preB cells had increased levels of Atp2a3 transcripts (Fig. 2 A) and SERCA3 protein (Fig. 2 B). While bulk Atp2a3 inactivation led to a near complete loss of SERCA3 protein in WT abl preB cells, it caused only partial loss of SERCA3 in Atp2a2−/− abl preB cells, suggesting that these cells cannot tolerate the loss of SERCA2 and SERCA3 (Fig. 2 C). Bulk Atp2a3 inactivation in WT abl preB cells did not lead to significant defects in V(D)J recombination of pMGINV (Fig. 2 D). However, bulk Atp2a3 inactivation in Atp2a2−/− abl preB cells, which led to a reduction in SERCA3 protein, resulted in a near complete block in pMGINV V(D)J recombination (Fig. 2 D).
Figure 2.
SERCA3 supports V(D)J recombination in SERCA2-deficient cells. (A) Quantitative RT-PCR analysis of Atp2a3 mRNA in WT abl preB cells without (−) gAtp2a2 and bulk Atp2a2 inactivated with (+) gAtp2a2 normalized to β-actin mRNA (mean ± SD; n = 3). **, P < 0.01. (B) Western blot analysis of SERCA2, SERCA3, and β-actin in WT and bulk Atp2a2 inactivated abl preB cells performed as in Fig. 1 B (n = 1 for Dox 2 d; n = 4 for Dox 4 d or more). (C) Western blot analysis of SERCA3 and β-actin in WT and Atp2a2−/− abl preB cells without (−) gAtp2a3 and bulk Atp2a3 inactivated with (+) gAtp2a3 (n = 3). (D) Flow-cytometric analysis of pMGINV rearrangement in WT and Atp2a2−/− abl preB cells (−) and these cells after bulk Atp2a3 inactivation with (+) gAtp2a3 (mean ± SD; n = 3). ***, P < 0.001. (E) Mouse Igk locus schematic showing multiple Vk and Jk gene segments, relative positions of SacI (S) and EcoRI (E) restriction sites, and Jk probe. The 3-kb Jk probe-hybridizing SacI–EcoRI fragment is shown, as are several different Vk to Jk rearrangements that will give rise to Jk probe-hybridizing SacI–EcoRI fragments of different sizes. (F) Southern blot of SacI and EcoRI digested genomic DNA probed with the Jk probe from WT and Atp2a2−/− abl preB cells (−) and the same cells bulk Atp2a2 (gAtp2a2) or Atp2a3 (gAtp2a3) inactivated and treated with imatinib for the indicated number of days. The fragment generated by the unrearranged (UR) Igk locus is indicated. The percentage of lane hybridization that corresponds to the UR Igk locus is shown with 0-d imatinib set at 100%. Quantification described in Materials and methods. (G) Mean ± SD of the percentage of UR Igk locus quantified as described in F in imatinib-treated WT, Atp2a2−/−, and bulk Atp2a3 inactivated Atp2a2−/− abl preB cells (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Dox, doxycycline; expo., exposure; n.s., not significant.
Southern blot analysis of V(D)J recombination at the Igk locus yielded similar results (Fig. 2, E and F). Treatment of WT abl preB cells with imatinib led to a progressive reduction in the intensity of the hybridizing band corresponding to the unrearranged Igk locus and the appearance of heterogeneously sized bands reflecting diverse Vκ to Jκ rearrangements (Fig. 2, E and F). Southern blot analyses of cells with bulk Atp2a2 inactivation or clonal Atp2a2−/−abl preB cells revealed a mild reduction in the kinetics of Igk locus rearrangement (Fig. 2, F and G). Strikingly, bulk Atp2a3 inactivation in Atp2a2−/− abl preB cells led to a near complete block of Igk rearrangements with persistence of the band reflecting the unrearranged Igk locus and undetectable Vk to Jk rearrangements after imatinib treatment for up to 5 d (Figs. 2, F and G). We conclude that SERCA2 and SERCA3 have redundant functions that are critical for the RAG cleavage step of the V(D)J recombination reaction.
Loss of SERCA2 and SERCA3 disrupts Ca2+ homeostasis
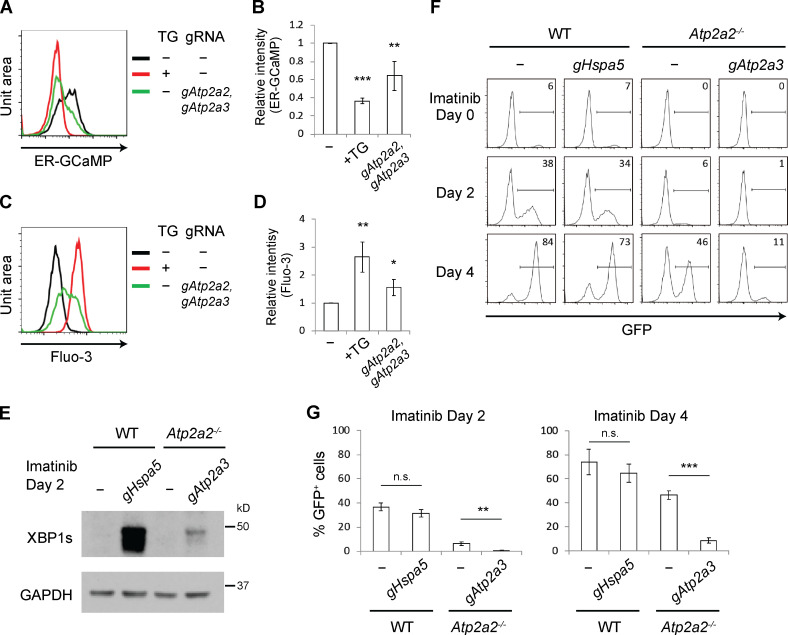
We assessed ER Ca2+ levels in abl preB cells using ER-GCaMP6-150, a low-affinity calcium indicator designed to detect µM Ca2+ concentrations present in the ER lumen (de Juan-Sanz et al., 2017). This indicator contains ER-targeting and retention sequences to ensure high-fidelity measurement of ER Ca2+. Upon Ca2+ binding, ER-GCaMP6-150 fluorescence changes ∼45-fold, similar to the original GCaMP6 reporter (de Juan-Sanz et al., 2017). ER-GCaMP6-150 was expressed in WT abl preB cells, and GFP fluorescence indicative of ER Ca2+ could be readily detected by flow cytometry (Fig. 3 A). Treating these cells with the SERCA inhibitor thapsigargin led to a significant reduction in ER-GCaMP6-150 fluorescence, indicative of loss of ER Ca2+ (Fig. 3, A and B) and concomitant increase in cytosolic Ca2+, as evidenced by Fluo-3 fluorescence (Fig. 3, C and D). Bulk Atp2a2 and Atp2a3 inactivation also led to reduced ER-GCaMP6-150 fluorescence (Fig. 3, A and B) and increased Fluo-3 fluorescence (Fig. 3, C and D). We conclude that in abl preB cells, loss of SERCA2 and SERCA3 leads to decreased ER Ca2+ and increased cytosolic Ca2+.
Figure 3.
Loss of SERCA2 and SERCA3 disrupts Ca2+ homeostasis. (A–D) Flow-cytometric analysis of WT abl preB cells expressing ER-GCaMP6-150 (A) or incubated with Fluo-3 AM Ca2+ indicator (C) without further treatment (black histogram), treated with thapsigargin (TG; red histogram), or after bulk inactivation of Atp2a2 and Atp2a3 genes (green histogram). Median fluorescence intensity was calculated and normalized to that of the black histogram in each experiment to derive the relative intensity of ER-GCaMP6-150 (B; mean ± SD; n = 4) and Fluo-3 dye (D; mean ± SD; n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the untreated sample. (E) Western blot analysis of XBP1s in imatinib-treated WT abl preB cells bulk Hspa5 (gHspa5) inactivated or Atp2a2−/− cells bulk Atp2a3 (gAtp2a3) inactivated (n = 3). (F) Flow-cytometric analysis of GFP expression from pMGINV rearrangement in cells from E treated with imatinib for 0, 2, or 4 d. (G) Mean ± SD of three pMGINV experiments performed as in F. **, P < 0.01; ***, P < 0.001. n.s., not significant.
Reduced ER Ca2+ promotes IRE1α activation, which leads to expression of the spliced form of XBP1, XBP1s, and activation of the unfolded protein response (UPR; Hetz and Glimcher, 2009). As expected, loss of SERCA2 and SERCA3 led to activation of the UPR, as evidenced by XBP1s expression in Atp2a2−/− abl preB cells after bulk Atp2a3 inactivation (Fig. 3 E). BiP, encoded by Hspa5, normally binds IRE1α, preventing its activation until the accumulation of unfolded proteins prompts its dissociation from IRE1α and activation of the UPR (Hetz and Glimcher, 2009). Indeed, bulk Hspa5 inactivation in abl preB cells led to high levels of XBP1s expression (Fig. 3 E). However, unlike loss of SERCA2 and SERCA3, loss of BiP had minimal effects on V(D)J recombination (Fig. 3, F and G). We conclude that pathways other than the UPR are regulated by SERCA2 and SERCA3 to promote efficient V(D)J recombination.
SERCAs are required for RAG gene expression
V(D)J recombination relies on pathways that induce RAG expression and promote accessibility of antigen receptor genes to the RAG proteins (Sleckman et al., 1998). We assayed V(D)J recombination on episomal pMGINV plasmid substrates transiently introduced into abl preB cells, but not integrated into the chromosome (Fig. 4 A). pMGINV plasmids were transfected into WT and Atp2a2−/− abl preB cells treated with imatinib and then recovered 2 d later, and CJ formation was assayed by PCR. These analyses revealed a decrease in pMGINV CJ formation in Atp2a2−/− compared with WT abl preB cells, suggesting that SERCA proteins have functions regulating the expression or basic biochemical activities of the RAG proteins (Fig. 4 B). Indeed, Atp2a2−/− abl preB clones had lower levels of Rag1 and Rag2 transcripts at 1 and 2 d after imatinib treatment compared with WT cells (Fig. 4, C and D). Strikingly, bulk Atp2a3 inactivation in Atp2a2−/− abl preB cell clones led to a profound reduction in Rag1 and Rag2 transcripts after imatinib treatment (Fig. 4, C and D). In agreement, RAG1 protein levels were lower in bulk Atp2a2 inactivated abl preB cells, Atp2a2−/− abl preB cell clones, and after bulk Atp2a3 inactivation in Atp2a2−/− abl preB cell clones (Fig. 4 E). We conclude that SERCA2 and SERCA3 function redundantly to promote Rag1 and Rag2 expression.
Figure 4.
Loss of SERCA2 and SERCA3 leads to diminished Rag expression. (A) Schematic of pMGINV with locations of PCR primers (arrows) used to amplify CJ and the plasmid backbone (AmpR). (B) Quantitative PCR analysis of pMGINV CJ formation in plasmids recovered from WT and Atp2a2−/− abl preB cells treated with imatinib (mean ± SD; n = 3). ***, P < 0.001. (C and D) Levels of Rag1 (C) and Rag2 (D) mRNA assayed by quantitative RT-PCR in abl preB cells treated with imatinib for 1 or 2 d. WT and Atp2a2−/− abl preB clones (−) were assayed, as were Atp2a2−/− abl preB clones with bulk Atp2a3 inactivation (+). Rag1 and Rag2 mRNA levels were normalized to β-actin mRNA and relative to the WT value, which was set at 1. Rag1 and Rag2 mRNAs were not detected at significant levels in cells not treated with imatinib (mean ± SD; n = 3). **, P < 0.01; ***, P < 0.001 compared with WT. (E) Western blot analysis for RAG1 and actin in WT and Atp2a2−/− abl preB cell clones (−) and cells bulk Atp2a2 (gAtp2a2) or Atp2a3 (gAtp2a3) inactivated and treated with imatinib for 1 or 2 d (n = 1 for 1 d, n = 2 for 2 d).
SERCA2 deficiency leads to B lymphopenia in mice and humans
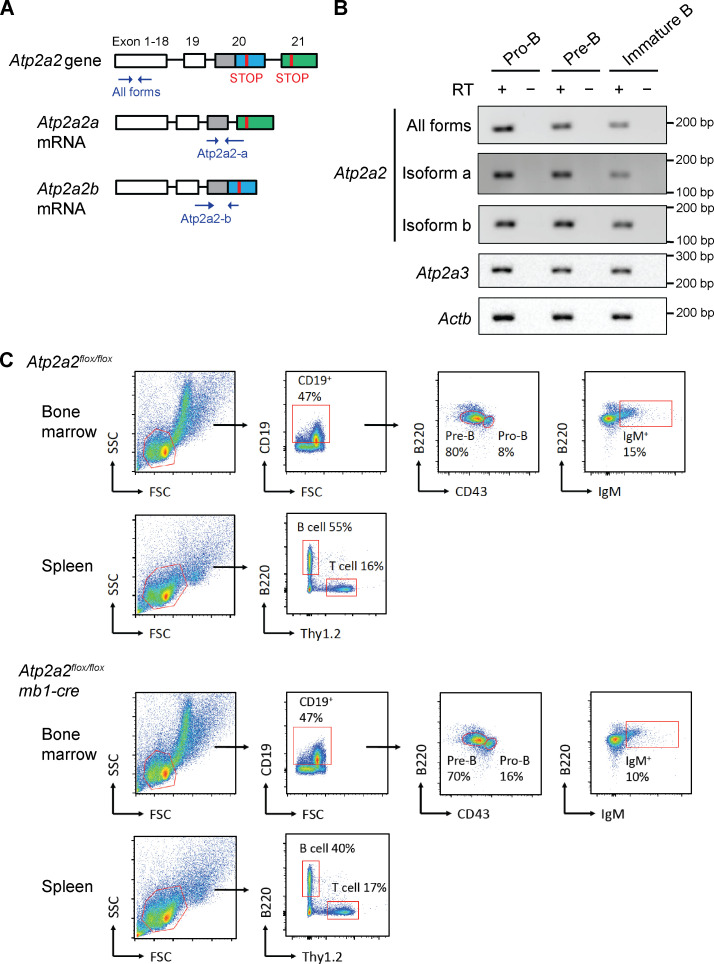
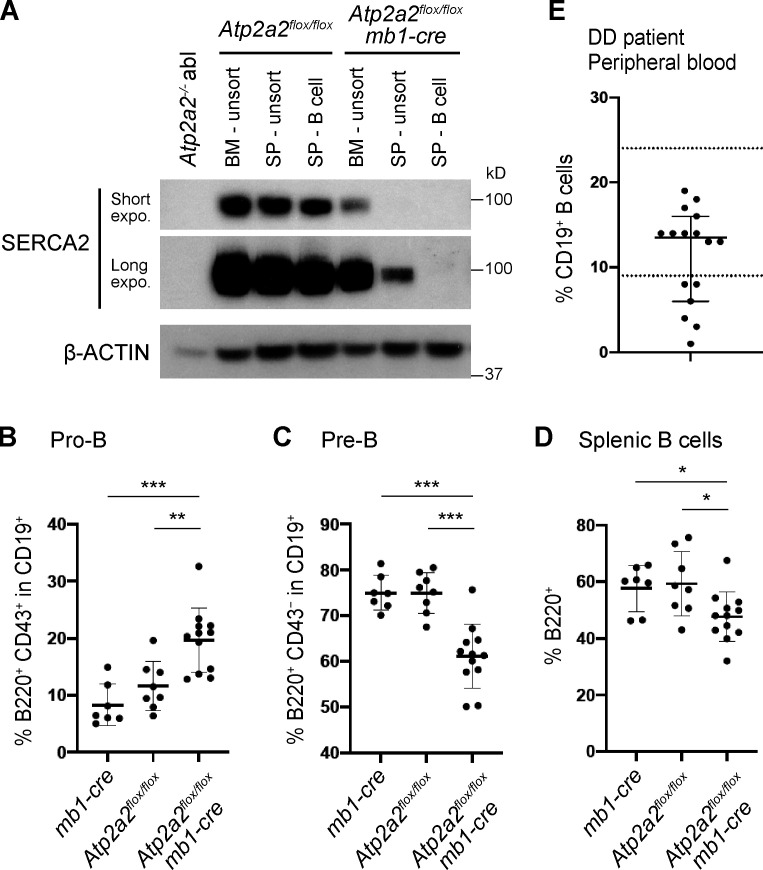
There are at least four splice variants of SERCA2, with SERCA2a and SERCA2b being the main variants (Fig. S2 A; Gélébart et al., 2003; Stammers et al., 2015; Vandecaetsbeek et al., 2011). Analyses of mRNA from proB, preB, and immature B cells purified from the bone marrow of WT mice revealed SERCA2a, SERCA2b, and SERCA3 mRNA in all of these cells (Fig. S2 B). As Atp2a2−/− mice exhibit embryonic lethality, we generated mice carrying a conditional Atp2a2 floxed allele (Atp2a2flox) and the Cd79a/mb1-cre allele (mb1-cre), which leads to Cre expression in the early proB cells (Hobeika et al., 2006; Ikeda et al., 2017; Periasamy et al., 1999). SERCA2 protein expression was effectively abolished in B cells from Atp2a2flox/flox:mb1-cre mice (Fig. 5 A). Atp2a2flox/flox:mb1-cre mice exhibited an increase in the percentage of bone marrow proB cells compared with Atp2a2flox/flox or mb1-cre mice (Fig. 5 B and Fig. S2 C). Moreover, Atp2a2flox/flox:mb1-cre mice exhibited a decreased percentage of bone marrow preB cells (Fig. 5 C and Fig. S2 C) and a decrease in the percentage of mature splenic B cells compared with Atp2a2flox/flox or mb1-cre mice (Fig. 5 D and Fig. S2 C). These findings are consistent with potential defects in antigen receptor gene assembly in developing B cells.
Figure S2.
Analysis of SERCA2-deficient primary B cells. (A) PCR strategy used to distinguish the Atp2a2a and Atp2a2b mRNA isoforms. (B) RT-PCR analysis of mRNA representing all Atp2a2 isoforms and Atp2a2b or Atp2a2a isoforms in addition to Atp2a3 transcripts in proB (B220+CD43+), preB (B220+CD43−), and immature B (B220+IgM+) cells purified from bone marrow of WT B6 mice (n = 2). (C) Gating strategies for the flow-cytometric analysis of B cell development in mouse bone marrow and spleen shown in Fig. 5, B–D. The analyses shown are of 4-wk-old littermates of Atp2a2flox/flox and Atp2a2flox/flox:mb1-cre mice. SSC, side scatter; FSC, forward scatter.
Figure 5.
SERCA2 deficiency impairs B cell development in vivo. (A) Western blot analysis of SERCA2 and actin in cells isolated from Atp2a2flox/flox and Atp2a2flox/flox:mb1-cre mice. Bone marrow (BM), whole spleen (SP - unsort), and purified splenic B cells (SP - B cell) were analyzed (n = 1). (B–D) Mean ± SD of the percentages of proB (B), preB (C), and mature splenic B (D) cells assayed by flow-cytometric analysis of BM (B and C) and spleen (D) from mb1-cre (n = 7), Atp2a2flox/flox (n = 8), and Atp2a2flox/flox:mb1-cre (n = 12) mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) Percentage of CD19+ B cells in the peripheral blood of Darier disease (DD) individuals (median with 95% CI; mean, 11.4%; n = 16). The normal laboratory range (9–24%) for the percentage of B cells in peripheral blood is indicated by the dotted lines. expo., exposure.
Darier disease is an autosomal-dominant disease caused by heterozygous ATP2A2 mutations that span the entire gene, consistent with the notion that the disease is caused by ATP2A2 haploinsufficiency (Korosec et al., 2006; Leong et al., 2017). We assayed the peripheral blood of 16 Darier disease patients. Remarkably, six patients exhibited a reduction in the percentage of B cells in peripheral blood compared with the normal reference ranges for healthy individuals (Fig. 5 E). The remaining 10 patients could have SERCA2 levels that can cause the disease but are adequate for normal B cell development. Darier disease patients are prone to skin infections, likely due to disrupted skin integrity, but could also be impacted by concomitant immune dysfunction (Tayabali et al., 2019; von Eiff et al., 2001; Wheeland et al., 1985).
We have shown that loss of SERCA functions in abl preB cells leads to decreased ER Ca2+ and increased cytosolic Ca2+, which are accompanied by diminished RAG expression and a block in V(D)J recombination. This is not due to activation of the UPR. Rather, it likely reflects the perturbation in signaling pathways required for RAG gene expression (Kuo and Schlissel, 2009). In this regard, the binding of Ca2+ to calmodulin leads to the activation of calcineurin, a phosphatase that can activate NFAT, which can inhibit RAG expression (Amin and Schlissel, 2008; Clapham, 2007; Patra et al., 2006). Thus, we speculate that the increased cytosolic Ca2+ in SERCA-deficient B cells could negatively regulate RAG expression through the activation of NFAT or other transcriptional pathways. Changes in intracellular Ca2+ levels could also modulate RAG activity as Ca2+ inhibits RAG enzymatic activity in vitro (Hiom and Gellert, 1997). Increased Ca2+ can also promote chromatin condensation, which would reduce RAG accessibility to antigen receptor genes (Phengchat et al., 2016; Schatz and Ji, 2011).
Materials and methods
Mice
All mice were maintained on B6 background, and all animal protocols were approved by the Weill Cornell Institutional Animal Care and Use Committee or the University of Alabama at Birmingham Animal Resources Program. Atp2a2flox (tm1c allele) mice were obtained from S. Kajimura (Ikeda et al., 2017). Genotyping was performed by PCR using Platinum Taq Polymerase (Invitrogen; 11304011) with the following primers: Atp2a2 Fwd: 5′-CACCTTGTTTAGCCTAGCTTTTTAC-3′; and Atp2a2 Rev: 5′-GTTGCACACTCTTTCTGTCCTG-3′. The PCR program is as follows: 94°C/2 min → [94°C/15 s → 58°C/30 s → 68°C/40 s] ×34 cycles → 68°C/3 min. The product size is 589 bp for WT and 700 bp for flox allele. To detect the knockout allele after crossing with Cre mice, a third primer (Atp2a2 3′LOXP.R1: 5′-ACTGATGGCGAGCTCAGACC-3′) was added together with the Fwd/Rev primer set. The product size for Cre-deleted allele is 200 bp.
Mb1-cre allele genotyping was performed using the above PCR conditions with the following primers: Mb1 Fwd: 5′-CTGCGGGTAGAAGGGGGTC-3′; Mb1 Rev: 5′-CCTTGCGAGGTCAGGGAGCC-3′; Mb1-Cre Fwd: 5′-ACCTCTGATGAAGTCAGGAAGAAC-3′; and Mb1-Cre Rev: 5′-GGAGATGTCCTTCACTCTGCTTCT-3′ (Yen et al., 2019). The expected product size is 400 bp for WT and 500 bp for the Cre allele.
Cell culture
abl preB cells were cultured in DMEM-HG (Gibco; 11960044) supplemented with 10% FBS (Gemini; 100106), nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin–streptomycin, and 60 µM β-mercaptoethanol. For viral infection with gRNA expression virus, 1 × 106 abl preB cells were mixed with viral supernatant and Polybrene (Santa Cruz; sc134220; 10 µg/ml final concentration) in 6-well plates and centrifuged at 1,800 rpm for 1.5 h at room temperature. Cells were purified by flow-cytometric cell sorting on day 3 after infection based on the expression of blue fluorescent protein (BFP) or human CD2 from the pKLV-U6gRNA lentiviral vector (Addgene; 50946) using a BD FACSAria Fusion Sorter. gRNAs used were as follows (protospacer adjacent motif, or PAM, sequence underlined): gAtp2a2: ATGGGCAAAGTGTATCGACAGG; gAtp2a3: TGATCCGGTCTGACCGCAAGGG; gHspa5: CTCCGGCGTGAGGTAGAAAAGG; and gRosa26 control: CTCCAGTCTTTCTAGAAGATGG. Abl preB cell lines containing pMGINV were generated as previously described (Hung et al., 2017). Cells were treated with 3 µM imatinib (Sigma-Aldrich; CDS022173) at the density of 1 × 106 cells/ml to induce G1 arrest. GFP signals arising from rearranged pMGINV were assayed on a BD LSRII flow cytometer, and the data were analyzed using FlowJo.
CRISPR/Cas9 genetic screening
The WT abl preB cell line carrying a Tet-On Cas9 transgene (clone 302) was infected with a lentiviral gRNA library containing 90,230 gRNAs targeting 18,424 mouse genes (Addgene; 67988; Tzelepis et al., 2016). 140 × 106 cells were infected to achieve >500-fold coverage using the infection described above at low multiplicity of infection. 3 d after infection, cells expressing BFP indicative of positive infection were isolated by flow-cytometric cell sorting and treated with 3 µg/ml doxycycline for 1 wk to induce Cas9 expression. Cells were then treated with 3 µM imatinib for 4 d as described above to induce V(D)J recombination of pMGINV. Cells that had (GFP+) and had not (GFP−) undergone pMGINV recombination were purified by flow-cytometric cell sorting, and genomic DNA was isolated from these cells. The gRNAs from each population were amplified using nested PCR and sequenced on an Illumina HiSeq 2500 platform performed in the Weill Cornell Epigenomics Core. The sequencing primer for the pKLV2 gRNA plasmid used in the library was 5′-GGCTTTATATATCTTGTGGAAAGGACGAAACACCG-3′.
The gRNA sequence regions were retrieved from FASTQ files using Seqkit (Shen et al., 2016). The derived sequences were mapped to the original gRNA sequences in the library (Tzelepis et al., 2016). The number of reads of each gRNA was normalized as follows:
(Shalem et al., 2014). The enrichment score of a gRNA is calculated as a ratio of normalized reads of the gRNA between two samples (GFP− versus GFP+).
Southern blotting
Southern blot analyses of pMGINV were performed as previously described (Hung et al., 2018). To quantify the percentage of SJs and SEs in pMGINV, the background of the scanned blot was subtracted in ImageJ, and the measured intensity of SJ and SE bands was divided by the sum of SJ plus unrearranged (UR)/SE bands and SE plus UR/SJ bands, respectively, in the same lane. To quantify Igk rearrangement, the intensity of the germline UR Igk band was measured after background subtraction and then divided by the intensity of whole lane (1–5 kb) that represents different Igk rearrangement products hybridizing to Jk probe. The derived values were normalized to that of day 0 of imatinib treatment in each genotype to indicate the remaining levels of UR Igk locus.
Western blotting
3–5 × 106 abl preB cells were lysed in 50 µl buffer (10 mM Tris, pH 8, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 400 mM NaCl, 1 mM dithiothreitol, and 1× protease/phosphatase inhibitor cocktails) on ice for 30 min and centrifuged at 14,000 rpm for 15 min to collect supernatants. 20 µg of the lysates were boiled in 1× lithium dodecyl sulfate sample buffer (Invitrogen) for 5 min and separated on NuPAGE 4–12% Bis-Tris mini gels (Invitrogen). Proteins were transferred to a nitrocellulose membrane in transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol) at 100 V for 90 min at room temperature using a wet transfer apparatus (Bio-Rad). The membrane was blocked in 5% milk/Tris buffered saline–Tween20 (TBST) at room temperature for 1 h. Primary antibody was diluted in 2.5% milk/TBST and probed overnight at 4°C. The membrane was then washed in TBST and probed with HRP-conjugated secondary antibodies (Promega) at room temperature for 1 h followed by TBST wash and visualization with enhanced chemiluminescence reagents (Thermo Fisher Scientific; 32109). Primary antibodies used were anti-SERCA2, rabbit polyclonal, 1:2,000 (Cell Signaling Technology; 4388); anti-SERCA3, rabbit polyclonal, 1:5,000 (GeneTex; GTX102381); anti-RAG1, rabbit monoclonal, 1:1,000 (Abcam; ab172637); anti-XBP1s, rabbit monoclonal, 1:1,000 (Cell Signaling Technology; 12782); anti–β-actin, mouse monoclonal, 1:20,000 (Sigma-Aldrich; A2228); and anti-GAPDH, mouse monoclonal, 1:5,000 (Sigma-Aldrich; G8795).
RNA analysis
RNA was extracted from 5 × 106 abl preB cells using the RNeasy Mini Kit (Qiagen; 74104). 1 µg RNA was used for cDNA synthesis using SuperScript III Reverse transcription (Invitrogen; 18080044) with oligo-dT primer. The quantitative PCR reaction was set up with 0.5 µl cDNA, 2 µl 10 mM forward/reverse primer mix, and 5 µl LightCycler 480 SYBR Green I Master Mix (Roche; 04707516001) to a total volume of 10 µl and analyzed on a Roche LightCycler 480. The Ct value of a target transcript was normalized to that of the β-actin transcript of the same sample to calculate ΔCt, which was then normalized to the ΔCt of control sample to analyze fold changes (2−ΔΔCt). The forward/reverse primers were designed to bind different exons to minimize cross-amplification from genomic DNA. Primer sequences for mouse mRNAs were as follows: Rag1 Fwd: 5′-AGGCCTGTGGAGCAAGGTA-3′; Rag1 Rev: 5′-AGGATCTCACCCTAAACAGC-3′; Rag2 Fwd: 5′-AGGATTCAGAGAGGGATAAGC-3′; Rag2 Rev: 5′-CCATCTGCAGGGACATTTTTG-3′; β-actin Fwd2: 5′-CATGGCATTGTTACCAACTGG-3′; β-actin Rev2: 5′-ATGGCTACGTACATGGCTGG-3′; Atp2a3 Fwd: 5′-TTGGAGTGTATGTAGGCCTG-3′; and Atp2a3 Rev: 5′-TCAGAGACGCTGTTGAGGG-3′. The following primers were used to amplify Atp2a2 transcripts: all forms Fwd: 5′-TCGACAGGACAGAAAGAGTGTG-3′; all forms Rev: 5′-GTATGCTTGATGACGGAGACAG-3′; isoform-a Fwd: 5′-GCTCATTTTCCAGATCACACCG-3′; isoform-a Rev: 5′-GTTACTCCAGTATTGCGGGTTG-3′; isoform-b Fwd: 5′-ACCTTTGCCGCTCATTTTCCAG-3′; and isoform-b Rev: 5′-AGGCTGCACACACTCTTTACC-3′.
Calcium indicator assay
The ER-GCaMP6-150 plasmid, provided by T.A. Ryan (de Juan-Sanz et al., 2017), is a variant of GCaMP6 Ca2+ indicator containing the signal peptide of calreticulin and KDEL ER retention motif suitable for detecting Ca2+ in the ER. WT abl preB cells carrying Tet-On Cas9 transgene were infected with lentivirus carrying ER-GCaMP6-150. Cells emitting green fluorescence were isolated by flow-cytometric cell sorting and single-cloned. The responsiveness of ER-GCaMP6-150 to Ca2+ fluctuation in the ER was confirmed by flow-cytometric analysis following treatments with 50 nM SERCA inhibitor thapsigargin for 30 min. The levels of ER-GCaMP6-150 fluorescence in the cycling abl preB cells expressing gRNA were analyzed on day 3 or 4 of doxycycline treatment. For cytosolic Ca2+ analysis, abl preB cells (without ER-GCaMP6-150) were treated with 2 µM Fluo 3-AM (Sigma-Aldrich; 39294) at 37°C for 30 min in growth medium. The stained cells were washed once in prewarmed growth medium and then aliquoted for thapsigargin treatment before flow cytometry or analyzed directly.
Episomal V(D)J recombination assay
Abl preB cells were pretreated with 3 µM imatinib overnight to induce G1 arrest. 15 × 106 cells were transfected with 8 µg pMGINV plasmid using Amaxa Nucleofector (Program X-001; Lonza). The cells were incubated in media containing imatinib for 2 d, and the plasmid was isolated from these cells. pMGINV CJ formation was quantified by quantitative PCR using the primers 23RS-Fwd2: 5′-TCTCCCCCTTGAACCTCCTC-3′ and CJ-GFP-Strat: 5′-GAACAGCTCCTCGCCCTTG-3′. The CJ products were normalized to the levels of AmpR gene on the vector backbone to control for transfection efficiency (primers: AmpR Fwd1: 5′-ATAAACCAGCCAGCCGGAAG-3′; and AmpR Rev1: 5′-AACCGGAGCTGAATGAAGC-3′).
Lymphocyte development
Bone marrow and spleen cells from 4- to 5-wk-old mice were incubated with mouse Fc Block (BD; 553141; 1:100 dilution) on ice for 15 min. Cells were incubated with fluorophore-conjugated primary antibody (1:500 dilution) at 4°C for 30 min, then analyzed on a BD LSRII flow cytometer. For bone marrow, viable CD19+ cells were analyzed for B220 and CD43 expression. For spleen samples, viable cells were analyzed for B220 and Thy1.2 expression. Antibodies used were PacificBlue-anti-h/mB220 (BioLegend; 103227), APC-anti-mCD43 (BioLegend; 143208), FITC-anti-mCD19 (BioLegend; 115506), APC-anti-mCD90.2/Thy1.2 (BioLegend; 105312), and PE/Cy7-anti-mIgM (BD; 552867).
Human peripheral blood analysis
Ethical permit for blood sampling of Darier disease patients was granted by the regional ethical review board in Stockholm, Sweden (dnr 2017/1098-32). Prior to sampling, patients gave informed written consent. Patients were treated according to the principles of the Helsinki declaration. CD19+ B cells were analyzed in peripheral blood by flow cytometry (https://www.karolinska.se/KUL/Alla-anvisningar/Anvisning/10106). Reference data are based on healthy adult individuals at the Karolinska University Hospital clinical laboratory (Stockholm, Sweden) and comprise values between the 5th and 95th percentiles.
Statistical analysis
Statistical comparisons between two samples were analyzed by unpaired Student’s t test. P values are indicated in the figure legends.
Online supplemental material
Fig. S1 shows supporting information for Fig. 1: loss of SERCA2 impairs RAG cleavage. Fig. S2 showssupporting information for Fig. 5: SERCA2 deficiency impairs B cell development in vivo.Table S1 shows CRISPR/Cas9 screening results of candidate genes regulating V(D)J recombination.
Supplementary Material
shows CRISPR/Cas9 screening results of candidate genes regulating V(D)J recombination.
Acknowledgments
This work was supported by National Institutes of Health grants R01 AI047829 (B.P. Sleckman, University of Alabama at Birmingham), R01 AI074953 (B.P. Sleckman, University of Alabama at Birmingham), R01 DK097441 (S. Kajimura, Beth Israel Deaconess Medical Center), R01 CA95641 (J.K. Tyler, Weill Cornell Medicine), R01 AI 032524 (D.G. Schatz, Yale University), R37 NS036942 (T.A. Ryan, Weill Cornell Medicine), a Leukemia & Lymphoma Society Career Development Program Fellowship (C.-C. Chen), and Vetenskapsrådet, Hudfonden, Svenska Sällskapet förr Medicinsk Forskning, ALF Medicin Stockholm, Jeanssons Stiftelser, and Tore Nilssons Stiftelse (J.D. Wikstrom).
Author contributions: C.-C. Chen and B.P. Sleckman designed research. C.-C. Chen, B.-R. Chen, Y. Wang, P. Curman, H.A. Beilinson, R.M. Brecht, and C.C. Liu performed research. R.J. Farrell, J. de Juan-Sanz, S. Kajimura, and T.A. Ryan provided reagents and resources. C.-C. Chen, B.-R. Chen, L.-M. Charbonnier, T.A. Ryan, D.G. Schatz, T. Chatila, J.D. Wikstrom, J.K. Tyler, and B.P. Sleckman interpreted data. C.-C. Chen and B.P. Sleckman wrote the paper.
References
- Ahn, W., Lee M.G., Kim K.H., and Muallem S.. 2003. Multiple effects of SERCA2b mutations associated with Darier’s disease. J. Biol. Chem. 278:20795–20801. 10.1074/jbc.M301638200 [DOI] [PubMed] [Google Scholar]
- Amin, R.H., and Schlissel M.S.. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622. 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing, C.H., Swat W., and Alt F.W.. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109(2suppl):S45–S55. 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- Bredemeyer, A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., and Sleckman B.P.. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470. 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Burge, S.M., and Wilkinson J.D.. 1992. Darier-White disease: a review of the clinical features in 163 patients. J. Am. Acad. Dermatol. 27:40–50. 10.1016/0190-9622(92)70154-8 [DOI] [PubMed] [Google Scholar]
- Chandrasekera, P.C., Kargacin M.E., Deans J.P., and Lytton J.. 2009. Determination of apparent calcium affinity for endogenously expressed human sarco(endo)plasmic reticulum calcium-ATPase isoform SERCA3. Am. J. Physiol. Cell Physiol. 296:C1105–C1114. 10.1152/ajpcell.00650.2008 [DOI] [PubMed] [Google Scholar]
- Clapham, D.E.2007. Calcium signaling. Cell. 131:1047–1058. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- de Juan-Sanz, J., Holt G.T., Schreiter E.R., de Juan F., Kim D.S., and Ryan T.A.. 2017. Axonal Endoplasmic Reticulum Ca2+ Content Controls Release Probability in CNS Nerve Terminals. Neuron. 93:867–881.e6. 10.1016/j.neuron.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann, S.D., Lee A.I., Shockett P.E., Villey I.J., and Schatz D.G.. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527. 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Gélébart, P., Martin V., Enouf J., and Papp B.. 2003. Identification of a new SERCA2 splice variant regulated during monocytic differentiation. Biochem. Biophys. Res. Commun. 303:676–684. 10.1016/S0006-291X(03)00405-4 [DOI] [PubMed] [Google Scholar]
- Helmink, B.A., and Sleckman B.P.. 2012. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30:175–202. 10.1146/annurev-immunol-030409-101320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz, C., and Glimcher L.H.. 2009. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell. 35:551–561. 10.1016/j.molcel.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom, K., and Gellert M.. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 88:65–72. 10.1016/S0092-8674(00)81859-0 [DOI] [PubMed] [Google Scholar]
- Hobeika, E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, P.J., Chen B.R., George R., Liberman C., Morales A.J., Colon-Ortiz P., Tyler J.K., Sleckman B.P., and Bredemeyer A.L.. 2017. Deficiency of XLF and PAXX prevents DNA double-strand break repair by non-homologous end joining in lymphocytes. Cell Cycle. 16:286–295. 10.1080/15384101.2016.1253640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, P.J., Johnson B., Chen B.R., Byrum A.K., Bredemeyer A.L., Yewdell W.T., Johnson T.E., Lee B.J., Deivasigamani S., Hindi I., et al. 2018. MRI Is a DNA Damage Response Adaptor during Classical Non-homologous End Joining. Mol. Cell. 71:332–342.e8. 10.1016/j.molcel.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., Kang Q., Yoneshiro T., Camporez J.P., Maki H., Homma M., Shinoda K., Chen Y., Lu X., Maretich P., et al. 2017. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 23:1454–1465. 10.1038/nm.4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosec, B., Glavac D., Rott T., and Ravnik-Glavac M.. 2006. Alterations in the ATP2A2 gene in correlation with colon and lung cancer. Cancer Genet. Cytogenet. 171:105–111. 10.1016/j.cancergencyto.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Kuo, T.C., and Schlissel M.S.. 2009. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr. Opin. Immunol. 21:173–178. 10.1016/j.coi.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, I.U.S., Stuckey A., Ahanian T., Cederlöf M., and Wikstrom J.D.. 2017. Novel mutations in Darier disease and association to self-reported disease severity. PLoS One. 12:e0186356. 10.1371/journal.pone.0186356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipskaia, L., Keuylian Z., Blirando K., Mougenot N., Jacquet A., Rouxel C., Sghairi H., Elaib Z., Blaise R., Adnot S., et al. 2014. Expression of sarco (endo) plasmic reticulum calcium ATPase (SERCA) system in normal mouse cardiovascular tissues, heart failure and atherosclerosis. Biochim. Biophys. Acta. 1843:2705–2718. 10.1016/j.bbamcr.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo, S.A., and Schlissel M.S.. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37. 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Patra, A.K., Drewes T., Engelmann S., Chuvpilo S., Kishi H., Hünig T., Serfling E., and Bommhardt U.H.. 2006. PKB rescues calcineurin/NFAT-induced arrest of Rag expression and pre-T cell differentiation. J. Immunol. 177:4567–4576. 10.4049/jimmunol.177.7.4567 [DOI] [PubMed] [Google Scholar]
- Periasamy, M., Reed T.D., Liu L.H., Ji Y., Loukianov E., Paul R.J., Nieman M.L., Riddle T., Duffy J.J., Doetschman T., et al. 1999. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J. Biol. Chem. 274:2556–2562. 10.1074/jbc.274.4.2556 [DOI] [PubMed] [Google Scholar]
- Phengchat, R., Takata H., Morii K., Inada N., Murakoshi H., Uchiyama S., and Fukui K.. 2016. Calcium ions function as a booster of chromosome condensation. Sci. Rep. 6:38281. 10.1038/srep38281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz, D.G., and Ji Y.. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263. 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- Shalem, O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., Le S., Li Y., and Hu F.. 2016. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One. 11:e0163962. 10.1371/journal.pone.0163962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman, B.P., Bassing C.H., Bardon C.G., Okada A., Khor B., Bories J.C., Monroe R., and Alt F.W.. 1998. Accessibility control of variable region gene assembly during T-cell development. Immunol. Rev. 165:121–130. 10.1111/j.1600-065X.1998.tb01235.x [DOI] [PubMed] [Google Scholar]
- Stammers, A.N., Susser S.E., Hamm N.C., Hlynsky M.W., Kimber D.E., Kehler D.S., and Duhamel T.A.. 2015. The regulation of sarco(endo)plasmic reticulum calcium-ATPases (SERCA). Can. J. Physiol. Pharmacol. 93:843–854. 10.1139/cjpp-2014-0463 [DOI] [PubMed] [Google Scholar]
- Tavadia, S., Mortimer E., and Munro C.S.. 2002. Genetic epidemiology of Darier’s disease: a population study in the west of Scotland. Br. J. Dermatol. 146:107–109. 10.1046/j.1365-2133.2002.04559.x [DOI] [PubMed] [Google Scholar]
- Tayabali, K., Pothiwalla H., and Lowitt M.. 2019. Eczema herpeticum in Darier’s disease: a topical storm. J. Community Hosp. Intern. Med. Perspect. 9:347–350. 10.1080/20009666.2019.1650590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis, K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O.M., Mupo A., Grinkevich V., Li M., Mazan M., et al. 2016. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 17:1193–1205. 10.1016/j.celrep.2016.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecaetsbeek, I., Vangheluwe P., Raeymaekers L., Wuytack F., and Vanoevelen J.. 2011. The Ca2+ pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harb. Perspect. Biol. 3:a004184. 10.1101/cshperspect.a004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff, C., Becker K., Metze D., Lubritz G., Hockmann J., Schwarz T., and Peters G.. 2001. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with darier’s disease. Clin. Infect. Dis. 32:1643–1647. 10.1086/320519 [DOI] [PubMed] [Google Scholar]
- Wheeland, R.G., Donaldson M.L., and Bulmer G.S.. 1985. Localized Darier’s disease of the scalp complicated by Trichophyton tonsurans infection. Arch. Dermatol. 121:905–907. 10.1001/archderm.1985.01660070095025 [DOI] [PubMed] [Google Scholar]
- Wu, K.D., Lee W.S., Wey J., Bungard D., and Lytton J.. 1995. Localization and quantification of endoplasmic reticulum Ca(2+)-ATPase isoform transcripts. Am. J. Physiol. 269:C775–C784. 10.1152/ajpcell.1995.269.3.C775 [DOI] [PubMed] [Google Scholar]
- Wuytack, F., Papp B., Verboomen H., Raeymaekers L., Dode L., Bobe R., Enouf J., Bokkala S., Authi K.S., and Casteels R.. 1994. A sarco/endoplasmic reticulum Ca(2+)-ATPase 3-type Ca2+ pump is expressed in platelets, in lymphoid cells, and in mast cells. J. Biol. Chem. 269:1410–1416. 10.1016/S0021-9258(17)42273-3 [DOI] [PubMed] [Google Scholar]
- Yen, W.F., Sharma R., Cols M., Lau C.M., Chaudhry A., Chowdhury P., Yewdell W.T., Vaidyanathan B., Sun A., Coffre M., et al. 2019. Distinct Requirements of CHD4 during B Cell Development and Antibody Response. Cell Rep. 27:1472–1486.e5. 10.1016/j.celrep.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows CRISPR/Cas9 screening results of candidate genes regulating V(D)J recombination.