Abstract
Biofilms formed by multidrug-resistant bacteria are a major cause of hospital-acquired infections. Cold atmospheric-pressure plasma (CAP) is attractive for sterilization, especially to disrupt biofilms formed by multidrug-resistant bacteria. However, the underlying molecular mechanism is not clear. In this study, CAP effectively reduced the living cells in the biofilms formed by methicillin-resistant Staphylococcus aureus, and 6 min treatment with CAP reduced the S. aureus cells in biofilms by 3.5 log10. The treatment with CAP caused the polymerization of SaFtsZ and SaClpP proteins in the S. aureus cells of the biofilms. In vitro analysis demonstrated that recombinant SaFtsZ lost its self-assembly capability, and recombinant SaClpP lost its peptidase activity after 2 min of treatment with CAP. Mass spectrometry showed oxidative modifications of a cluster of peaks differing by 16 Da, 31 Da, 32 Da, 47 Da, 48 Da, 62 Da, and 78 Da, induced by reactive species of CAP. It is speculated that the oxidative damage to proteins in S. aureus cells was induced by CAP, which contributed to the reduction of biofilms. This study elucidates the biological effect of CAP on the proteins in bacterial cells of biofilms and provides a basis for the application of CAP in the disinfection of biofilms.
Keywords: cold atmospheric-pressure plasma, biofilm, oxidative damage, reactive species, methicillin-resistant Staphylococcus aureus
1. Introduction
Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), is one of the most frequent causes of infections and generally forms biofilms on infected tissues [1,2,3,4,5]. Biofilms formed by aggregated microbial cells are surrounded by a self-produced extracellular polymeric matrix made of proteins, DNA, and polysaccharides and adhere to surfaces, such as those of living tissues [6,7]. The structure of the biofilm makes the biofilm-associated bacteria more resistant to the host immune defense system, antibiotics, and other antimicrobial agents [8,9]. Methicillin-resistant S. aureus, especially the multidrug-resistant strains, frequently exists in the hospital setting in the form of biofilms [10]. When the multidrug-resistant bacteria infect patients, they also exist in the wounds or other lesions in the form of biofilms [5,6]. The reduction of biofilms is critical for the prevention and treatment of infectious diseases caused by multidrug-resistant bacteria [11]. Therefore, the inactivation of biofilms formed by multidrug-resistant bacteria is a challenging issue in the therapy of infectious diseases.
Cold atmospheric-pressure plasma (CAP), known as the fourth state of matter, contains various reactive species, such as H2O2, 1O2, •OH, and •NO, as well as electrons, ions, and photons. The reactive species of CAP can effectively inactivate microorganisms and cells, which makes it attractive for a range of biomedical applications, such as sterilization and cancer treatment [12,13,14,15,16,17]. Planktonic methicillin-resistant S. aureus was fragile to the surface discharge plasma of CAP, but methicillin-resistant S. aureus usually exists in the form of biofilms in the environment [18]. Therefore, the inactivation effect of CAP on biofilms has been studied further. Xu et al. inactivated approximately 4 log10 S. aureus biofilms after treatment for 10 min with a He/O2 plasma jet, and Wang et al. inactivated approximately 0.5 log10 S. aureus cells in biofilms after 30 min of exposure to a N2 plasma jet [19,20]. Czapka et al. inactivated approximately 2.77 log10 S. aureus biofilms after treatment with dielectric barrier discharge (DBD) for 300 s [21]. However, the molecular mechanism in the bacterial cells in biofilms inactivated by CAP is poorly understood.
The filamentous temperature-sensitive Z (FtsZ) and ATP-dependent caseinolytic protease P subunit (ClpP) are both promising targets for antimicrobial agent development [22,23,24,25,26]. FtsZ is the major cytoskeleton protein of the cell division process in prokaryotes and assembles into short, single-stranded protofilaments with an average length of 120–250 nm in vitro, and into a ring structure in the middle of bacterial cells [22,27,28]. Mutation of Phe138 (F138A) causes a decrease in polymerization activity and guanosine triphosphatase (GTPase) activity [29]. The FtsZ protein is involved in cell division, and the inhibition of FtsZ assembly inhibits cell division [30,31]. FtsZ is essential in S. aureus and cannot be deleted [32]. FtsZ is also involved in biofilm formation, as demonstrated by a previous study showing the upregulation of the FtsZ protein in Aggregatibacter actinomycetemcomitans biofilms versus the planktonic state [33]. The protease ClpP is highly conserved and widely distributed in bacteria and is considered a regulator of the FtsZ protein [30,34]. The ClpP protease is involved in stress responses, the virulence of pathogenic bacteria, and antibiotic resistance [35,36,37,38]. The deletion or inhibition of ClpP proteases resulted in a decrease in persister S. aureus cells and affected virulence and stress response [6,39,40]. The crystal structures of ClpP proteases revealed that the SaClpP tetradecamer, which consisted of two heptameric rings, enclosed a large chamber containing 14 proteolytic active sites (Ser98-His123-Asp172) [41,42,43]. According to the findings of another previous study by our group, CAP can induce oxidative modification of peptides consisting of histidine and phenylalanine; therefore, CAP was proposed to have effects on FtsZ protein and ClpP proteases [44].
In this study, the molecular mechanism of CAP on biofilms was studied. The reactive species of CAP induced aggregation and inactivation of the FtsZ and ClpP proteins in the cells of Staphylococcus aureus biofilm. These results suggest that the reactive species induced protein damage, which contributed to the antibiofilm effects.
2. Materials and Methods
2.1. Biofilm Assay
The culture of S. aureus biofilms was performed as previously described [45]. S. aureus ATCC33591, a methicillin-resistant strain, was purchased from the American Type Culture Collection (ATCC). A single S. aureus ATCC33591 colony was grown in 4 mL of tryptic soy broth (TSB, Oxoid, Hampshire, UK) at 250 rpm at 37 °C overnight. The overnight S. aureus cultures were diluted 1:100 in TSB with 1% glucose. The single-side adhesive silica films (10 × 10 × 0.5 mm, purchased from Taobao) were attached on the bottom of wells of 24-well plates, covered with diluted S. aureus, and cultured at 37 °C for 3 days without medium renewal. The medium was carefully removed, and the silica films with biofilms were gently washed three times with saline solution (0.9% NaCl). Four replicates of biofilms were used for each treatment condition, and untreated biofilms and the biofilms treated with helium and 1% synthetic air without applying a voltage were used as control. After treatment, the biofilms were dispersed in 1 mL of saline in 1.5-mL Eppendorf tubes by sonication for 1 min and vortexing for 5 min. Serial dilutions of each biofilm were performed, and 10 μL of each dilution was spotted onto TSB plates and incubated overnight at 37 °C.
2.2. CAP Device and CAP Treatments
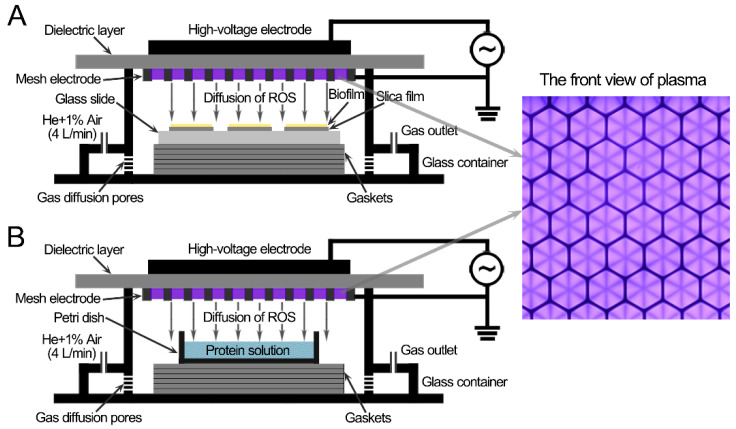
The CAP device was similar to the previous study [45]. The surface discharge structure of the CAP consisted of a high-voltage plane electrode, a liquid-facing grounded mesh electrode with a hexagonal shape, and a dielectric layer (made of polytetrafluoroethylene) sandwiched between the two electrodes (Figure 1). The surface discharge plasma (40 mm × 40 mm) was generated when a sinusoidal high voltage with a frequency of 20 kHz was applied. The applied voltage and discharge current were measured with a high-voltage probe (Tektronix P6015A, Portland, OR, USA) and a current probe (Pearson 2877, Palo Alto, CA, USA), respectively. The discharge power was obtained by integrating the applied voltage and discharge current demonstrated by an oscilloscope (Tektronix, MDO3054, Portland, OR, USA). The discharge power was controlled at approximately 3.2 W (the discharge power density of 0.2 W/cm2) with good mesh-to-mesh homogeneity through regulating the applied voltage. The values of the peak-to-peak voltage and discharge current were approximately 7.44 kV and 40.8 mA, respectively.
Figure 1.
Schematic diagram of the S. aureus biofilms (A) and the protein solutions (B) treated with the surface discharge of cold atmospheric-pressure plasma.
S. aureus biofilms on sterilized glass slides (a diameter of 30 mm) or protein solutions (150 μL) in a Petri dish (a diameter of 15 mm), which was smaller in size than the CAP, were placed under the CAP, and the air gap between the CAP and the surface of the biofilms or liquid surfaces was 8 ± 0.2 mm (Figure 1). The CAP treatment system was housed in a sealed organic glass box, and the gas flow of helium and 1% synthetic air (79% N2 + 21% O2) flowed into the organic glass box at a constant rate of 4 L/min, controlled by the mass flow controller (MFC, Sevenstar, Beijing, China), diffused through pores at the bottom part, and then spread upward to the underneath of the surface plasma. The flow rate at the outlet was less than 1 L/min measured by a rotameter, which indicated that most of the gas leaked through the small gaps that were not completely sealed to prevent the increase in pressure in the chamber (Figure 1). Synthetic air was used as the source of reactive species, while helium was used to enhance the production efficiency of those species as well as their flux on the treated samples via diffusion. The ambient temperature of the gas chamber and the temperature of samples were measured using a thermocouple, and both the temperatures were lower than 35 °C after 6 min of CAP treatment at a room temperature of 28 °C.
2.3. Protein Extraction and Detection
The S. aureus cells from CAP-treated and untreated biofilms were resuspended in solubilization buffer (50 mM imidazole, 50 mM NaCl, 2 mM 6-aminohexanoic acid, 1 mM EDTA, and 1% dodecylmaltoside; pH 7.0), and lysis was performed by bead beating in a MiniBeadbeater-16 (Biospec, Bartlesville, OK, USA). After centrifugation, the supernatants were transferred, and protein concentrations were determined by the BCA Protein Assay Kit (Beyotime, Shanghai, China) using bovine serum albumin (BSA) as the standard. The soluble proteins (10 μg) were mixed with Laemmli buffer (Bio-Rad, Hercules, CA, USA) and subjected to 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue G-250 or transferred to PVDF membranes and subjected to immunoblotting.
2.4. SaFtsZ and SaClpP Protein Expression and Purification
The genes encoding FtsZ (ORF: SACOL1199) and ClpP (ORF: SACOL0833) were amplified from the genomic DNA of S. aureus ATCC33591 by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) (for primer sequences, see Table 1). The PCR product was inserted into the NdeI/XhoI sites of the expression vector pET-28a (for His-tagged SaFtsZ) or pET-32a (for His-tagged and untagged SaClpP) (Novagen, Darmstadt, Germany). Expression plasmids were transformed into E. coli BL21(DE3) cells. The cells were grown and induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG). For His-tagged SaFtsZ and SaClpP, the induced cells were suspended in lysis buffer containing 50 mM Tris-Cl (pH 7.5) and 500 mM NaCl and then lysed at 4 °C by sonication. After centrifugation at 16,000× g for 20 min to remove the insoluble material, the supernatant was loaded onto a Histrap column (1 mL; GE Healthcare, Marlborough, MA, USA), and the protein was eluted with a single gradient of 0.5 M imidazole in lysis buffer. Fractions containing SaFtsZ or SaClpP were pooled and dialyzed against 50 mM HEPES (pH 7.5), 5 mM MgCl2, and 100 mM KCl. For untagged SaClpP, the induced cells were suspended in lysis buffer containing 50 mM Tris-Cl (pH 7.5), 100 mM NaCl, and 1 mM dithiothreitol (DTT) and lysed at 4 °C by sonication. After centrifugation at 16,000× g for 20 min to remove the insoluble material, the supernatant was loaded onto a Hitrap Capto Q column (5 mL; GE Healthcare), and the protein was eluted with a single gradient of 1 M NaCl in lysis buffer. The Q column fractions containing SaClpP were pooled and loaded onto a Superdex G200 column (24 mL; GE Healthcare, Marlborough, MA, USA) equilibrated with 50 mM Tris-Cl (pH 7.5), 100 mM NaCl, and 1 mM DTT. Fractions containing SaClpP were pooled and dialyzed against 10 mM Tris-Cl (pH 7.5), 1 mM EDTA, and 1 mM DTT. Protein concentrations were determined by a BCA Protein Assay Kit (Beyotime, Shanghai, China) using bovine serum albumin (BSA) as the standard.
Table 1.
The primers used in this study.
| Designation | Sequence (5′-3′) |
|---|---|
| ftsZ up | ATCATGCCATGGTAGAATTTGAACAAGGATTTA |
| ftsZ His-tag down | ATACCGCTCGAGACGTCTTGTTCTTCTTGAACGTC |
| clpP up | ATGGGAATTCCATATGAATTTAATTCCTACAGTTATTGAAAC |
| clpP His-tag down | ATACCGCTCGAGTTTTGTTTCAGGTACCATCACTTC |
| clpP down | ATACCGCTCGAGTTATTTTGTTTCAGGTACCATCACTTC |
2.5. Immunoblotting
Anti-SaFtsZ and anti-SaClpP antibodies were raised in rabbits using purified His-tagged SaFtsZ and SaClpP. The blots were blocked in 5% milk/Tris-buffered saline with 0.5% Tween (TBST) at 4 °C overnight with gentle agitation. After incubation with anti-SaFtsZ or anti-SaClpP antibodies for 2 h at room temperature with gentle agitation, the blots were washed four times (10 min per wash) with TBST at room temperature. The blots were incubated with Dylight649 Goat Anti-Rabbit IgG (Abbkine, Wuhan, China) for 1 h at room temperature with gentle agitation and washed again as described above. The blots were detected using a Typhoon FLA9500 biomolecular imager (GE Healthcare, Marlborough, MA, USA).
2.6. Blue Native Polyacrylamide Gel Electrophoresis
The recombinant SaFtsZ and untagged SaClpP proteins (1 mg/mL) in 10 mM Tris-Cl (pH 7.5) and 1 mM EDTA were treated with CAP for 2 min, as shown in Figure 1B. The soluble proteins (20 μg) from the CAP-treated and untreated biofilms were mixed with 4× loading buffer (200 mM imidazole, 200 mM NaCl, 20% glycerol, and 0.02% Ponceau S) and electrophoresed through a 10–20% (for SaFtsZ) or 4–15% (for SaClpP) gradient blue native polyacrylamide gel [46]. The gel was stained with Coomassie Brilliant Blue R-250 or subjected to immunoblotting. The gel for immunoblotting was washed with electroblotting buffer (25 mM Tris-Cl, pH 7.5, 250 mM glycine, and 0.1% sodium dodecyl sulfate (SDS)) for 30 min and transferred to a PVDF membrane using the electroblotting buffer for 3 h at 4 °C with a current of 150 mA. After transfer, methanol was used to destain the background of the PVDF membrane. Then, the PVDF membranes were subjected to immunoblotting.
2.7. In Vitro Assembly Assay of SaFtsZ
The SaFtsZ proteins treated with the CAP, treated with the mixture of H2O2 + NO2− + NO3− (saline with 250 μM H2O2 + 125 μM NO2− + 375 μM NO3−), or left untreated were incubated with 2 mM guanosine triphosphate (GTP) in HMK buffer (50 mM HEPES-OH, pH 7.5, 5 mM MgAc, and 100 mM KCl) for 10 min at 37 °C. Then, the SaFtsZ proteins were diluted five times with HMK buffer. For negative staining, an aliquot (~4 μL) of proteins was dropped onto a glow-discharged thin carbon-coated 200-mesh grid (Carbon Gilder Finder Grids, Cu, F1 UL) and kept for 1 min at 22 °C. Then, the excess solution was removed by touching a piece of filter paper to the back of the grid. The grid was washed by briefly touching the surface of the grid with a drop (~30 μL) of deionized water on Parafilm and then blotted dry with filter paper. The touching and blotting steps were repeated once. Two successive drops (~25 μL/drop) of 1% (w/v) uranyl acetate (UA) solution were applied to Parafilm, and the excess solution was removed by blotting similarly. The grid remained in contact with the last UA drop with the sample side down for 1 min in the dark before the excess stain was removed, and the sample was air-dried at room temperature. The stained samples were examined using an FEI Talos F200C transmission electron microscope operating at 200 kV at 57,000× magnification.
2.8. Enzymatic Activity Assay of SaClpP
Peptidase activity was detected as described previously using the model fluorogenic substrate dipeptide N-succinyl-Leu-Tyr-7-amido-4-methylcoumarin (Suc-LY-AMC) (MP Biomedicals, Santa Ana, CA, USA) [47]. The CAP-treated, H2O2 + NO2− + NO3− treated, or untreated SaClpP (5 μM monomer) was incubated with Suc-LY-AMC (250 μM) in 50 mM HEPES-OH (pH 7.5) and 100 mM NaCl. The hydrolysis of the labeled peptide led to the release of fluorescent AMC. The fluorescence was measured on a microplate reader (Thermo Scientific Varioskan Flash, Waltham, MA, USA) at 37 °C for 30 cycles every 60 s using an excitation wavelength of 340 nm and an emission wavelength of 450 nm, as described previously [48].
2.9. Mass Spectrometry of SaClpP
The CAP-treated and untreated SaClpP (5 μg) were subjected to SDS-PAGE (15%) and visualized by staining with Coomassie Brilliant Blue R-250. The protein bands were excised, and the protein was digested in-gel with trypsin, as described previously [49]. The digested products were dissolved in 0.1% formic acid and purified with ZipTipC18 pipette tips (Millipore, Darmstadt, Germany) according to the manufacturer’s procedure. The tryptic peptides were identified using an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher, Waltham, MA, USA) coupled to an EASY-nLC 1000 UPLC system (ThermoFisher, Waltham, MA, USA).
3. Results
3.1. Inactivation of CAP Treatment on Biofilms
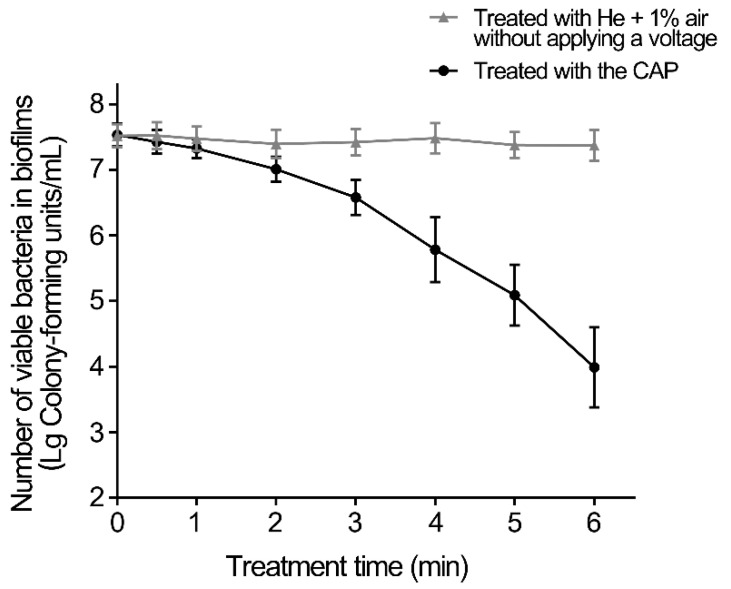
The inactivation effect of CAP on S. aureus biofilms was first determined (Figure 2). The CAP treatment for 2 min decreased the number of living S. aureus cells by approximately 0.5 log10 in biofilms, while that for 4 min decreased them by less than 2 log10. After the biofilms were treated with CAP for 6 min, the living cells in the biofilms were reduced by approximately 3.5 log10 in the biofilms. Therefore, CAP treatment can effectively reduce the living cells in S. aureus biofilms.
Figure 2.
Inactivation effect on S. aureus biofilms of CAP. The biofilms were treated with CAP for the indicated times, and the CAP-treated and untreated biofilms were dispersed in 1 mL saline by sonication and vortexing. Serial dilutions of each biofilm were performed, and 10 μL of each dilution was spotted onto TSB plates and incubated overnight at 37 °C. Error bars represent the standard deviation (SD).
3.2. CAP Treatment Causes Polymerization of the SaFtsZ and SaClpP and Induces Oxidative Modification of Proteins
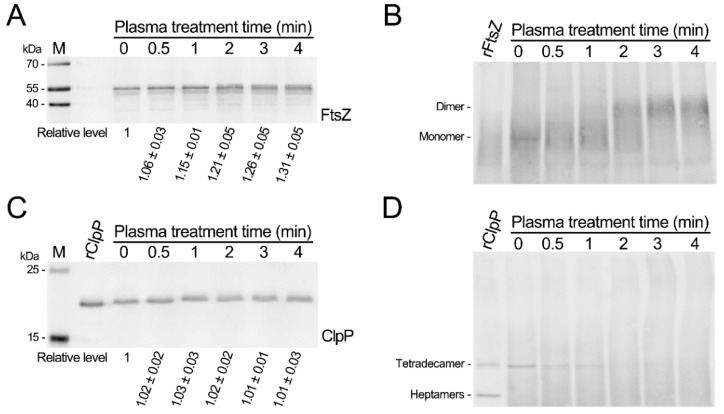
To investigate the molecular mechanisms underlying the inactivation of CAP treatment, the soluble proteins from the CAP-treated and untreated S. aureus biofilms separated by both SDS-PAGE and blue-native gels and the SaFtsZ and SaClpP proteins were detected using immunoblotting. The SaFtsZ protein of the untreated biofilms exhibited a single band, whereas that of the CAP-treated biofilms exhibited two close bands (Figure 3A and Figure S1). The emerging band migrated slightly more slowly than the main band and increased with the time of CAP treatment. Then, blue native gels were employed to identify changes in the SaFtsZ protein. Compared with the SaFtsZ protein of the untreated biofilms, the SaFtsZ protein of the biofilms treated with the CAP for 1 min started to form larger polymers, and the SaFtsZ protein of the biofilms treated with the CAP for 3 min formed larger polymers (Figure 3B). The bands of the SaClpP protein from the biofilms after the CAP treatment for 1 to 4 min migrated slightly more slowly than those from the untreated biofilms (Figure 3C). The bands of the SaClpP protein became dispersive and fuzzy in blue native gels from biofilms treated with the CAP for 1 min compared with untreated biofilms. The SaClpP protein was hardly detectable after the CAP treatment for 2 min to 4 min, and the smear of larger polymers became more intense (Figure 3D). These results indicate that the CAP treatment of S. aureus biofilms caused the polymerization of the SaFtsZ and SaClpP proteins.
Figure 3.
Polymerization of SaFtsZ and SaClpP in S. aureus biofilms induced by plasma treatment. (A) Immunoblotting of SaFtsZ under denaturing conditions. (B) Immunoblotting of SaFtsZ under nondenaturing conditions. (C) Immunoblotting of SaClpP under denaturing conditions. (D) Immunoblotting of SaClpP under nondenaturing conditions. The plasma-treated and untreated biofilms were dispersed and lysed, and then the soluble proteins were extracted and subjected to SDS-PAGE or blue native PAGE. The SaFtsZ and SaClpP proteins were detected by immunoblotting using anti-SaFtsZ and anti-SaClpP antibodies, respectively.
3.3. CAP Treatment Causes Inactivation of the SaFtsZ and SaClpP Proteins
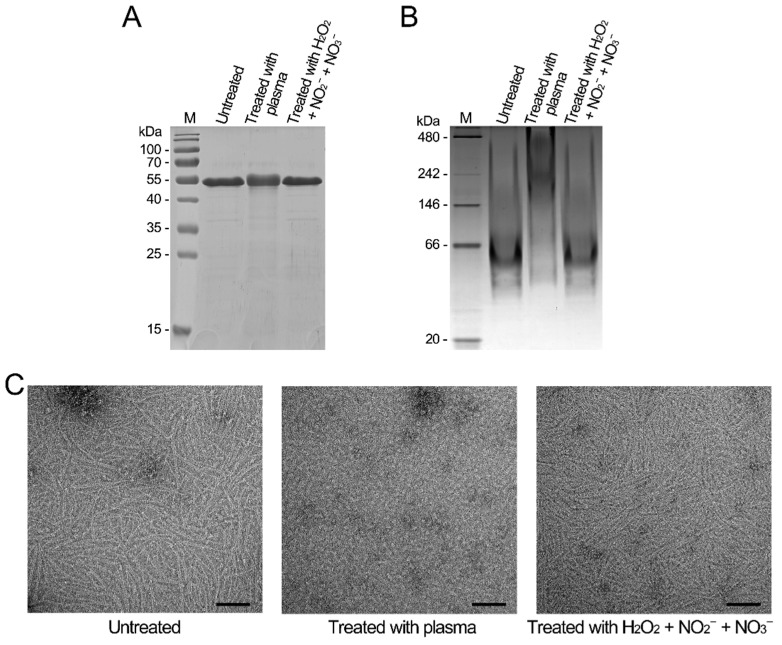
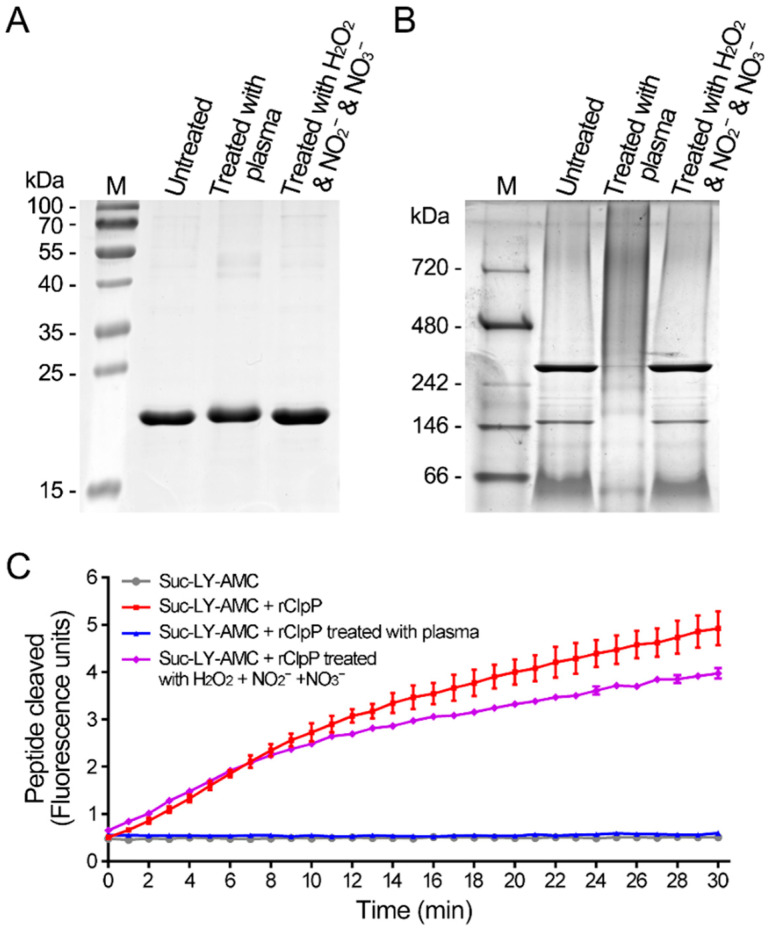
To analyze the biological effects of the CAP treatment on proteins, the recombinant SaFtsZ and SaClpP proteins were treated with the CAP. The SaFtsZ and SaClpP proteins in S. aureus cells were entirely damaged after treatment by CAP for 2 min; therefore, the treatment time of recombinant proteins was 2 min. Based on the previous study, three kinds of long-lived reactive species, namely, H2O2, NO2−, and NO3−, were generated in the same CAP, with concentrations of 53.3 μM, 25.4 μM, and 148.0 μM, respectively [38]. Then, a mixed solution with higher concentrations of H2O2 (100 μM), NO2− (100 μM), and NO3− (500 μM) was also employed for comparison. After the CAP treatment, the denatured SaFtsZ protein migrated slightly more slowly than the untreated SaFtsZ protein (Figure 4A). The untreated SaFtsZ protein was a monomer, and the CAP-treated SaFtsZ protein formed larger polymers (Figure 4B). To further evaluate the effect on the proteins, the self-assembling capacity of SaFtsZ protein was analyzed by electron microscopy (EM). The CAP-treated SaFtsZ protein lost its self-assembly capability, whereas the untreated SaFtsZ protein self-assembled into protofilaments in the presence of GTP (Figure 4C).
Figure 4.
Inactivation of SaFtsZ by plasma treatment in vitro. (A) The plasma-treated, H2O2 + NO2− + NO3−-treated, and untreated SaFtsZ proteins separated by SDS-PAGE. (B) The plasma-treated, H2O2 + NO2− + NO3−-treated, and untreated SaFtsZ proteins separated by blue native PAGE. The SDS-PAGE or blue native PAGE was stained with Coomassie Brilliant Blue R-250. M, molecular weight standards with molecular weights indicated. (C) Self-assembly of SaFtsZ proteins. The plasma-treated, H2O2 + NO2− + NO3−-treated, and untreated SaFtsZ proteins were self-assembled in the presence of GTP. Then, the samples were negatively stained and examined using transmission electron microscopy (TEM).
The denatured CAP-treated SaClpP protein migrated more slowly than the untreated SaClpP protein (Figure 5A). The untreated SaClpP protein exhibited tetradecamers and heptamers, and the CAP-treated SaClpP protein became a larger smear probably formed by large polymers (Figure 5B). To further evaluate the effect on proteins, peptidase activity against peptide substrates was tested. The cleavage of the dipeptide (N-succinyl-Leu-Tyr-7-amido-4-methylcoumarin) by CAP-treated SaClpP was hardly detectable, suggesting that the peptidase activity of the SaClpP protein was inactivated by the CAP treatment (Figure 5C). These results indicated that the CAP treatment caused polymerization and inactivated the activity of the SaFtsZ and SaClpP proteins in vitro.
Figure 5.
Inactivation of SaClpP by plasma treatment in vitro. (A) The plasma-treated, H2O2 + NO2− + NO3−-treated, and untreated SaClpP proteins separated by SDS-PAGE. (B) The plasma-treated, H2O2 + NO2− + NO3−-treated, and untreated SaClpP proteins separated by blue native PAGE. The SDS-PAGE or blue native PAGE was stained with Coomassie Brilliant Blue R-250. M, molecular weight standards with molecular weights indicated. (C) Peptidase activity of SaClpP proteins. The time courses of hydrolysis of the fluorogenic peptide N-succinyl-Leu-Tyr-7-amido-4-methylcoumarin by the plasma-treated, H2O2 + NO2− + NO3− treated, and untreated SaClpP proteins at 37 °C were measured. Error bars represent the standard deviation (SD).
3.4. CAP Induces Oxidative Modification in Proteins
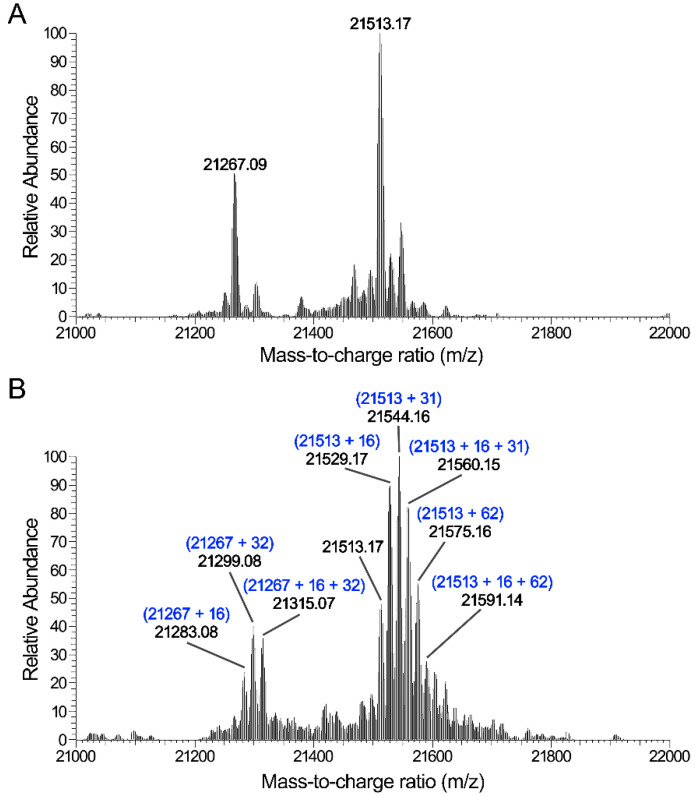
To elucidate the mechanism underlying the effect of the CAP treatment on proteins, the SaClpP proteins were analyzed by mass spectrometry. The mass weights of untreated SaClpP proteins were 21,513.17 (theoretical Mr: 21,513.57) and 21,267.09 with two amino acid deletions of the N-terminus (Figure 6A). The CAP-treated SaClpP appeared to be modified at multiple sites: a cluster of peaks differing by 16 Da, 31 Da, 32 Da, 47 Da, 48 Da, 62 Da, and 78 Da were detected (Figure 6B). The shifts of 16 Da and 32 Da were consistent with the hydroxy group (ROH) and the hydroperoxide group (ROOH) or two hydroxy groups, respectively. The shift of 31 Da corresponded to ROO- or a combination of one RO- and one hydroxy group, and the shift of 62 Da corresponded to the combination of two ROO- groups, one RO-OR (+30 Da) group and one hydroperoxide group, or two hydroxy groups. Mass fingerprinting showed that the SaClpP protein was oxidatively modified at multiple sites (Table S1 and Figure S2). These results demonstrate that the reactive oxygen species from the CAP induced oxidative modification of the SaClpP protein.
Figure 6.
Identification of the modifications of SaClpP induced by plasma treatment. (A) Mass spectrum of untreated SaClpP. (B) Mass spectrum of the plasma-treated SaClpP. A cluster of peaks with mass differences of 16, 31, 32, or 62 Da are shown.
4. Discussion
CAP based on the electrical method can efficiently inactivate bacteria and reduce biofilms. The SaFtsZ and SaClpP proteins could be modified and inactivated by the CAP treatment in both in vitro and S. aureus cells, which contributed to the reduction of biofilms by CAP. The understanding of the molecular mechanism of this process is helpful in optimizing the control of CAP and guiding the application of CAP.
CAP contains various reactive species, such as long-lived H2O2, NO2−, and NO3− and short-lived •OH, 1O2, •NO, and ONOO−, as previously described [18,44]. The mixture of three long-lived species, namely, H2O2, NO2−, and NO3−, exhibited little effect on SaFtsZ and SaClpP proteins, which demonstrated that the short-lived species were critical in the process. The reactive species penetrated bacterial cells in biofilms and induced the polymerization of the SaFtsZ and SaClpP proteins. A previous study also found that CAP-induced polymerization of GFP-tagged proteins caused protein denaturation and insolubility in Saccharomyces cerevisiae [50]. In this study, the polymerized proteins were in the soluble fraction, and this effect could be reversed by the reducing agent. These effects seemed to be similar to the effects of disulfide bond formation, and the reactive oxygen species could also cause cysteine to form disulfide bonds. However, the SaFtsZ protein did not contain cysteine, and further mass spectrometry analysis demonstrated that the oxidative modifications of the SaClpP protein were mainly induced by the CAP treatment. Therefore, oxidative modifications, such as ROOR, occurred between proteins and caused protein polymerization. 1O2 and O2•− are both generated by CAP and can rapidly and selectively react with amino acids, efficiently yielding hydroperoxide groups [44,51,52,53,54]. Therefore, the reactive species of CAP reacted with proteins and caused the polymerization and function loss of proteins through oxidative modifications.
The treatment of CAP exhibited similar effects on the recombinant SaFtsZ and SaClpP proteins and the SaFtsZ and SaClpP proteins from S. aureus biofilms. However, the in vitro analysis could not fully reflect the effects in bacterial cells for two reasons. The first reason is that the reactive species need to cross the cell walls and membranes into the cells and then react with the proteins in the cells [55,56]. Not all of the reactive species of CAP could enter into the cells. The second reason is that the reactive species entering into the cells were affected by the enzymes in the cell or reacted with the reactive species produced by the cells [57]. The types and amounts of reactive species in the cells would change. In the in vitro treatment, the reactive species directly reacted with the recombinant proteins without the obstacle of the cell wall and membrane and the effects of the intracellular enzymes.
It is inevitable that the reactive species of CAP will react with other proteins in S. aureus cells and cause damage due to the non-selectivity and high reactivity of radical species [52]. Previous studies demonstrated that CAP induced double-stranded and single-stranded breaks in DNA and crosslinks between DNA and proteins in Escherichia coli cells [44,58]. Therefore, the reactive species of CAP caused oxidative damage on DNA and various proteins, including SaFtsZ and SaClpP, and the damages synergistically caused the death of S. aureus cells in the biofilm.
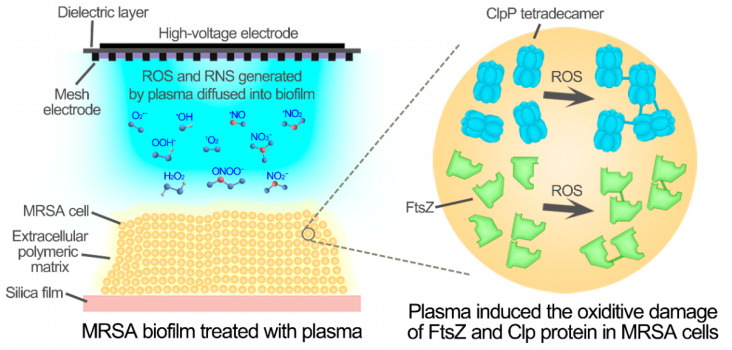
Based on these results, a model of CAP treatment for biofilm eradication was proposed (Figure 7). Reactive oxygen species generated by plasma, such as O2•−, diffused into biofilms and entered into S. aureus cells. The reactive oxygen species reacted with proteins, such as SaFtsZ and SaClpP, and induced oxidative modifications that caused polymerization and functional loss of proteins. Inactivation of SaFtsZ and SaClpP blocked the growth and division of S. aureus cells, resulting in the death and eradication of biofilms. Therefore, cold atmospheric-pressure plasma could be developed and used in strategies for both the disinfection of biofilms and therapy for wound infections.
Figure 7.
The aggregation and inactivation of SaFtsZ and SaClpP induced by the reactive species of CAP.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9051072/s1.
Author Contributions
L.G. (Li Guo), L.Y., Y.Q., G.N., L.H. and Z.W. designed and executed most of the experiments and analyzed the data; L.G. (Lu Gou) and L.Z. assisted in the execution of some experiments; L.G. (Li Guo), L.Y., Y.Q. and D.L. wrote the manuscript; M.G.K., D.L., L.G. (Li Guo), X.W. and H.C. reviewed and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (51977174 and 51722705), the State Key Laboratory of Electrical Insulation and Power Equipment (EIPE19310), the Doctoral Fund of Ministry of Education of China (2017M613106), and the State Key Laboratory of Agricultural Microbiology (AMLKF201705).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [59] partner repository with the dataset identifier PXD023352 and 10.6019/PXD023352.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jain S., Sengupta M., Sarkar S., Ghosh S., Mitra A.N., Sinha A., Chakravorty S. Can EDTA Change MRSA into MSSA? A Future Prospective! J. Clin. Diagn. Res. 2016;10:DC22–DC25. doi: 10.7860/JCDR/2016/17944.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zetola N., Francis J.S., Nuermberger E.L., Bishai W.R. Community-acquired meticillin-resistant Staphylococcus aureus: An emerging threat. Lancet Infect. Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 3.Guillamet C.V., Kollef M.H. How to stratify patients at risk for resistant bugs in skin and soft tissue infections? Curr. Opin. Infect. Dis. 2016;29:116–123. doi: 10.1097/QCO.0000000000000244. [DOI] [PubMed] [Google Scholar]
- 4.Bahamondez-Canas T.F., Heersema L.A., Smyth H.D.C. Current status of in vitro models and assays for susceptibility testing for wound biofilm infections. Biomedicines. 2019;7:34. doi: 10.3390/biomedicines7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyama H., Huh W.K., Yamasaki O., Oono T., Iwatsuki K. Confocal laser scanning microscopic observation of glycocalyx production by Staphylococcus aureus in mouse skin: Does S. aureus generally produce a biofilm on damaged skin? Br. J. Dermatol. 2002;147:879–885. doi: 10.1046/j.1365-2133.2002.04962.x. [DOI] [PubMed] [Google Scholar]
- 6.Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 7.Okuda K., Zendo T., Sugimoto S., Iwase T., Tajima A., Yamada S., Sonomoto K., Mizunoe Y. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013;57:5572–5579. doi: 10.1128/AAC.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Mah T.F.C., O’Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 10.Gao P., Ho P.L., Yan B., Sze K.H., Davies J., Kao R.Y.T. Suppression of Staphylococcus aureus virulence by a small-molecule compound. Proc. Natl. Acad. Sci. USA. 2018;115:8003–8008. doi: 10.1073/pnas.1720520115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Pozo J.L. Biofilm-related disease. Expert Rev. Anti Infect. Ther. 2018;16:51–65. doi: 10.1080/14787210.2018.1417036. [DOI] [PubMed] [Google Scholar]
- 12.Fridman G., Friedman G., Gutsol A., Shekhter A.B., Vasilets V.N., Fridman A. Applied Plasma Medicine. Plasma Process. Polym. 2008;5:503–533. doi: 10.1002/ppap.200700154. [DOI] [Google Scholar]
- 13.Kang M.H., Hong Y.J., Attri P., Sim G.B., Lee G.J., Panngom K., Kwon G.C., Choi E.H., Uhm H.S., Park G. Analysis of the antimicrobial effects of nonthermal plasma on fungal spores in ionic solutions. Free Radic. Biol. Med. 2014;72:191–199. doi: 10.1016/j.freeradbiomed.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Kong M.G., Kroesen G., Morfill G., Nosenko T., Shimizu T., Van Dijk J., Zimmermann J.L. Plasma medicine: An introductory review. New J. Phys. 2009;11:115012. doi: 10.1088/1367-2630/11/11/115012. [DOI] [Google Scholar]
- 15.Moreau M., Orange N., Feuilloley M.G. Non-thermal plasma technologies: New tools for bio-decontamination. Biotechnol. Adv. 2008;26:610–617. doi: 10.1016/j.biotechadv.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Bekeschus S., Clemen R., Niessner F., Sagwal S.K., Freund E., Schmidt A. Medical gas plasma jet technology targets murine melanoma in an immunogenic fashion. Adv. Sci. 2020;7:1903438. doi: 10.1002/advs.201903438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin A., Gorbanev Y., De Backer J., Van Loenhout J., Van Boxem W., Lemiere F., Cos P., Dewilde S., Smits E., Bogaerts A. Non-thermal plasma as a unique delivery system of short-lived reactive oxygen and nitrogen species for immunogenic cell death in melanoma cells. Adv. Sci. 2019;6:1802062. doi: 10.1002/advs.201802062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L., Xu R.B., Zhao Y.M., Liu D.X., Liu Z.J., Wang X.H., Chen H.L., Kong M.G. Gas Plasma Pre-treatment Increases Antibiotic Sensitivity and Persister Eradication in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 2018;9:537. doi: 10.3389/fmicb.2018.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Yu Z., Xu Z., Hu S., Li Y., Xue X., Cai Q., Zhou X., Shen J., Lan Y., et al. Antimicrobial mechanism and the effect of atmospheric pressure N2 plasma jet on the regeneration capacity of Staphylococcus aureus biofilm. Biofouling. 2018;34:935–949. doi: 10.1080/08927014.2018.1530350. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z., Shen J., Cheng C., Hu S., Lan Y., Chu P.K. In Vitro antimicrobial effects and mechanism of atmospheric-pressure He/O2 plasma jet on Staphylococcus aureus biofilm. J. Phys. D Appl. Phys. 2017;50:105201. doi: 10.1088/1361-6463/aa593f. [DOI] [Google Scholar]
- 21.Czapka T., Maliszewska I., Olesiak-Banska J. Influence of atmospheric pressure non-thermal plasma on inactivation of biofilm cells. Plasma Chem. Plasma Process. 2018;38:1181–1197. doi: 10.1007/s11090-018-9925-z. [DOI] [Google Scholar]
- 22.Milam S.L., Osawa M., Erickson H.P. Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 2012;103:59–68. doi: 10.1016/j.bpj.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y., Du R., Cai S., Liu Z., Fang Z., Liu T., So L., Lu Y., Sun N., Wong K. Study of benzofuroquinolinium derivatives as a new class of potent antibacterial agent and the mode of inhibition targeting FtsZ. Front. Microbiol. 2018;9:1937. doi: 10.3389/fmicb.2018.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haydon D.J., Stokes N.R., Ure R., Galbraith G., Bennett J.M., Brown D.R., Baker P.J., Barynin V.V., Rice D.W., Sedelnikova S.E., et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 25.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 26.Culp E., Wright G.D. Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 2017;70:366–377. doi: 10.1038/ja.2016.138. [DOI] [PubMed] [Google Scholar]
- 27.Ma X., Ehrhardt D.W., Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romberg L., Simon M., Erickson H.P. Polymerization of Ftsz, a bacterial homolog of tubulin: Is assembly cooperative? J. Biol. Chem. 2001;276:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- 29.Wagstaff J.M., Tsim M., Oliva M.A., Garcia-Sanchez A., Kureisaite-Ciziene D., Andreu J.M., Lowe J. A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio. 2017;8:e002541-17. doi: 10.1128/mBio.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortiz C., Natale P., Cueto L., Vicente M. The keepers of the ring: Regulators of FtsZ assembly. FEMS Microbiol. Rev. 2016;40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 31.McCool J.D., Long E., Petrosino J.F., Sandler H.A., Rosenberg S.M., Sandler S.J. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- 32.Hu J., Zhang X., Liu X., Chen C., Sun B. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob. Agents Chemother. 2015;59:1352–1355. doi: 10.1128/AAC.04290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llama-Palacios A., Potupa O., Sanchez M.C., Figuero E., Herrera D., Sanz M. Aggregatibacter actinomycetemcomitans growth in biofilm versus planktonic state: Differential expression of proteins. J. Proteome Res. 2017;16:3158–3167. doi: 10.1021/acs.jproteome.7b00127. [DOI] [PubMed] [Google Scholar]
- 34.Feng J., Michalik S., Varming A.N., Andersen J.H., Albrecht D., Jelsbak L., Krieger S., Ohlsen K., Hecker M., Gerth U., et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 2013;12:547–558. doi: 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- 35.Conlon B.P., Nakayasu E.S., Fleck L.E., LaFleur M.D., Isabella V.M., Coleman K., Leonard S.N., Smith R.D., Adkins J.N., Lewis K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frees D., Gerth U., Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int. J. Med. Microbiol. 2014;304:142–149. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Gersch M., Kolb R., Alte F., Groll M., Sieber S.A. Disruption of oligomerization and dehydroalanine formation as mechanisms for ClpP protease inhibition. J. Am. Chem. Soc. 2014;136:1360–1366. doi: 10.1021/ja4082793. [DOI] [PubMed] [Google Scholar]
- 38.McGillivray S.M., Tran D.N., Ramadoss N.S., Alumasa J.N., Okumura C.Y., Sakoulas G., Vaughn M.M., Zhang D.X., Keiler K.C., Nizet V. Pharmacological inhibition of the ClpXP protease increases bacterial susceptibility to host cathelicidin antimicrobial peptides and cell envelope-active antibiotics. Antimicrob. Agents Chemother. 2012;56:1854–1861. doi: 10.1128/AAC.05131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springer M.T., Singh V.K., Cheung A.L., Donegan N.P., Chamberlain N.R. Effect of clpP and clpC deletion on persister cell number in Staphylococcus aureus. J. Med. Microbiol. 2016;65:848–857. doi: 10.1099/jmm.0.000304. [DOI] [PubMed] [Google Scholar]
- 40.Michel A., Agerer F., Hauck C.R., Herrmann M., Ullrich J., Hacker J., Ohlsen K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006;188:5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Ye F., Lan L., Jiang H., Luo C., Yang C.G. Structural switching of Staphylococcus aureus Clp protease: A key to understanding protease dynamics. J. Biol. Chem. 2011;286:37590–37601. doi: 10.1074/jbc.M111.277848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Hartling J.A., Flanagan J.M. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/S0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 43.Ye F., Li J.H., Yang C.G. The development of small-molecule modulators for ClpP protease activity. Mol. Biosyst. 2017;13:23–31. doi: 10.1039/C6MB00644B. [DOI] [PubMed] [Google Scholar]
- 44.Guo L., Zhao Y., Liu D., Liu Z., Chen C., Xu R., Tian M., Wang X., Chen H., Kong M.G. Cold atmospheric-pressure plasma induces DNA-protein crosslinks through protein oxidation. Free Radic. Res. 2018;52:783–798. doi: 10.1080/10715762.2018.1471476. [DOI] [PubMed] [Google Scholar]
- 45.Guo L., Xu R.B., Liu D.X., Qi Y., Guo Y.H., Wang W.T., Zhang J., Liu Z.J., Kong M.G. Eradication of methicillin-resistant Staphylococcus aureus biofilms by surface discharge plasmas with various working gases. J. Phys. D Appl. Phys. 2019;52:425202. doi: 10.1088/1361-6463/ab32c9. [DOI] [Google Scholar]
- 46.Wittig I., Braun H.P., Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 47.Thompson M.W., Singh S.K., Maurizi M.R. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 1994;269:18209–18215. doi: 10.1016/S0021-9258(17)32436-5. [DOI] [PubMed] [Google Scholar]
- 48.Li D.H., Chung Y.S., Gloyd M., Joseph E., Ghirlando R., Wright G.D., Cheng Y.Q., Maurizi M.R., Guarne A., Ortega J. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 2010;17:959–969. doi: 10.1016/j.chembiol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beranova-Giorgianni S., Desiderio D.M. Mass spectrometry of the human pituitary proteome: Identification of selected proteins. Rapid Commun. Mass Spectrom. 2000;14:161–167. doi: 10.1002/(SICI)1097-0231(20000215)14:3<161::AID-RCM859>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Itooka K., Takahashi K., Kimata Y., Izawa S. Cold atmospheric pressure plasma causes protein denaturation and endoplasmic reticulum stress in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2018;102:2279–2288. doi: 10.1007/s00253-018-8758-2. [DOI] [PubMed] [Google Scholar]
- 51.Simpson J.A., Narita S., Gieseg S., Gebicki S., Gebicki J.M., Dean R.T. Long-lived reactive species on free-radical-damaged proteins. Biochem. J. 1992;282:621–624. doi: 10.1042/bj2820621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies M.J. Protein oxidation and peroxidation. Biochem. J. 2016;473:805–825. doi: 10.1042/BJ20151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies M.J. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 2004;3:17–25. doi: 10.1039/b307576c. [DOI] [PubMed] [Google Scholar]
- 54.Davies M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophy. Res. Commun. 2003;305:761–770. doi: 10.1016/S0006-291X(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 55.Huang M., Zhuang H., Zhao J., Wang J., Yan W., Zhang J. Differences in cellular damage induced by dielectric barrier discharge plasma between Salmonella Typhimurium and Staphylococcus aureus. Bioelectrochemistry. 2020;132:107445. doi: 10.1016/j.bioelechem.2019.107445. [DOI] [PubMed] [Google Scholar]
- 56.Kawano A., Yamasaki R., Sakakura T., Takatsuji Y., Haruyama T., Yoshioka Y., Ariyoshi W. Reactive oxygen species penetrate persister cell membranes of Escherichia coli for effective cell killing. Front. Cell Infect. Microbiol. 2020;10:496. doi: 10.3389/fcimb.2020.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vatansever F., de Melo W.C., Avci P., Vecchio D., Sadasivam M., Gupta A., Chandran R., Karimi M., Parizotto N.A., Yin R., et al. Antimicrobial strategies centered around reactive oxygen species--bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 2013;37:955–989. doi: 10.1111/1574-6976.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arjunan K.P., Sharma V.K., Ptasinska S. Effects of atmospheric pressure plasmas on isolated and cellular DNA—A review. Int. J. Mol. Sci. 2015;16:2971–3016. doi: 10.3390/ijms16022971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [59] partner repository with the dataset identifier PXD023352 and 10.6019/PXD023352.