Abstract
The COVID-19 pandemic has placed unprecedented strain on the healthcare system, particularly hospital bed capacity in the setting of large variations in patient length of stay (LOS). Using electronic health record data from 966 COVID-19 patients at a large academic medical center, we developed three machine learning algorithms to predict the likelihood of prolonged LOS, defined as >8 days. The models included 353 variables and were trained on 80% of the cohort, with 20% used for model validation. The three models were created on hospital days 1, 2 and 3, each including information available at or before that point in time. The models’ predictive capabilities improved sequentially over time, reaching an accuracy of 0.765, with an AUC of 0.819 by day 3. These models, developed using readily available data, may help hospital systems prepare for bed capacity needs, and help clinicians counsel patients on their likelihood of prolonged hospitalization.
Keywords: COVID-19, Machine learning
Introduction
The rapid development of a global pandemic following the emergence of SARS-CoV-2 has placed unprecedented strain on the healthcare system. As of April 2021, an estimated 30 million Americans had been infected, with over a million requiring hospitalization and more than 500,000 dying from the resultant illness known as Coronavirus Disease 2019 (COVID-19) [1,2]. Unfortunately, despite public health efforts, the rate of infection has remained high, leaving hospitals struggling to meet the surging demand for beds. This crisis is primed to be compounded by influenza season, which has traditionally strained the healthcare system on its own, as well as the potential for new SARS-CoV-2 variants. The confluence of both of these viral pathogens leaves healthcare providers, administrators, and systems in need of a method to predict the availability of hospital beds so as to appropriately plan for expected capacity.
COVID-19 severity of illness varies greatly, with many patients experiencing few to no symptoms, some needing only short hospitalizations, and others spending weeks to months in the hospital. This variability in length of stay (LOS) makes predicting hospital bed availability difficult. Further, the novel nature of COVID-19 leaves clinicians often ill-equipped to predict which patients will have long lengths of stay and which will be able to quickly return home. We sought to leverage clinically generated data in the electronic health record (EHR) of a large academic medical center to develop a machine learning algorithm to predict prolonged LOS, defined as >8 days, for patients admitted with COVID-19.
Methods
Study population
We examined all patients admitted to Cedars-Sinai Medical Center between April 1st, 2020 and September 6th, 2020 with a diagnosis of COVID-19, based on RT-PCR testing. Cedars-Sinai is a large academic hospital located in Los Angeles, California serving a diverse patient population. Patients who died within 8 days of being admitted to the hospital were excluded from the cohort, as they were not eligible to meet the primary endpoint of prolonged LOS. Eight days was selected as the threshold of prolonged length of stay based on magnitude of deviation from mean and median length of stays during the same time period.
Model structure
LOS prediction models were created using high-dimensional, patient level EHR data. Models were validated on three similar tasks: prediction of LOS with i) data from only day 1, ii) data from the first 2 days of hospitalization, and iii) data from the first 3 days of hospitalization. Automated machine learning through iterative selection of model parameters and model architecture was performed using a structured environment, with selection based on area under the curve (AUC) on a held-out validation cohort. Models evaluated include variations on Elastic-net, gradient boosted trees, random forest, support vector machines, logistic regression, a Eureqa classifier, generalized additive models, a Vowpal Wabbit classifier, K-nearest neighbors classifiers, residual neural network, a Rulefit classifier, and ensemble models, which were a combination of other models listed above, to avoid overfitting of single models on their own. Models were developed using DataRobot (Boston, MA), an automated machine learning method that facilitates parallel algorithms while also supporting ensemble models; the DataRobot method chooses models appropriate to a given data set and prediction target, training those models at different hyperparameter tunings with different groups of features and constraints and then ranking them based on a selected evaluation metric. Models varied in the features that they used for prediction, some using all data fields, while others search over only features that were most highly correlated with the target value.
Data acquisition and preprocessing
All patient information was harvested from the EHR. In order to make predictions at the individual patient level, data sets that contained multiple values for a patient were aggregated. For repeated measures, separate features stored the first value during the applicable time period, the last value during the applicable time period and, if the variable was numeric, the difference between the two. A total of 353 features were used to make the predictions (Supplemental Table 1). Race and ethnicity were explored as potential model features but showed no difference in modeling accuracy and were thus excluded to reduce the risk of model bias. Breakdown of racial and ethnicity data is shown in descriptive tables for completeness (Entire population: 36.7% Non-Hispanic White, 29.2% Hispanic, 17.9% Non-Hispanic Black, 5.1% Asian and 11.1% Other/Unknown) (Supplemental Figure 1). To generate comorbidity related features, we used Charlson and Elixhauser Scores [3], calculated from ICD-10 codes. Additionally, patient day to day location was included to classify patients into intensive care unit (ICU) and non-ICU rooms and also to count the number of days a patient had spent in the ICU. Date of admission was included as a feature. Missing values were imputed to the median if the column exceeded a given model's minimum threshold for number of existing values. If a given model's minimum threshold was not met, the missing values were set to null.
Model validation
Models were evaluated based on the AUC for predicting short LOS, when applied to a set of holdout data, selected based on random stratification of eligible patients. For each model, 80% of the patient data was used for training and 20% was set aside in the holdout portion for model evaluation. Because elements of previous models were being passed forward, the holdout cohort of patients was maintained throughout all 3 versions of the model to reduce the possibility of target leakage between models. The train and test methodology was selected over k-fold cross validation given concerns over target leakage given use of stacked models. All protocols were approved by the Cedars-Sinai Institutional Review Board and the manuscript prepared in accordance with the TRIPOD guidelines [4].
Methodologic process
In summary, models were created to predict short LOS on days 1, 2 and 3 of hospitalization. A total of 42 models were trained on data from 80% of patient population, with all models representing variations on the 12 base models listed above, with ensemble models developed as meta-algorithms of other models. Following training, all models were then tested on the remaining 20% of the population and ranked based on AUC for predicting short LOS. Training and testing were performed for each model on each of the first 3 days of hospitalization. The models with the highest AUC for predicting short LOS using the test data on each of these days were selected as the best model at each timepoint.
Results
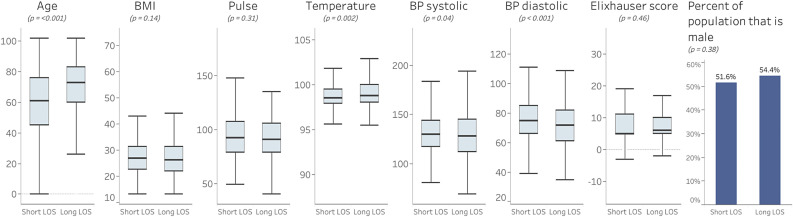
A total of 966 patients were included in this study: 525 of whom had a LOS of ≤8 days, while 441 patients had an LOS of >8 days. The characteristics of those patients are shown in Fig. 1 .
Fig. 1.
Initial patient characteristics for short stay and long stay patients with COVID-19.
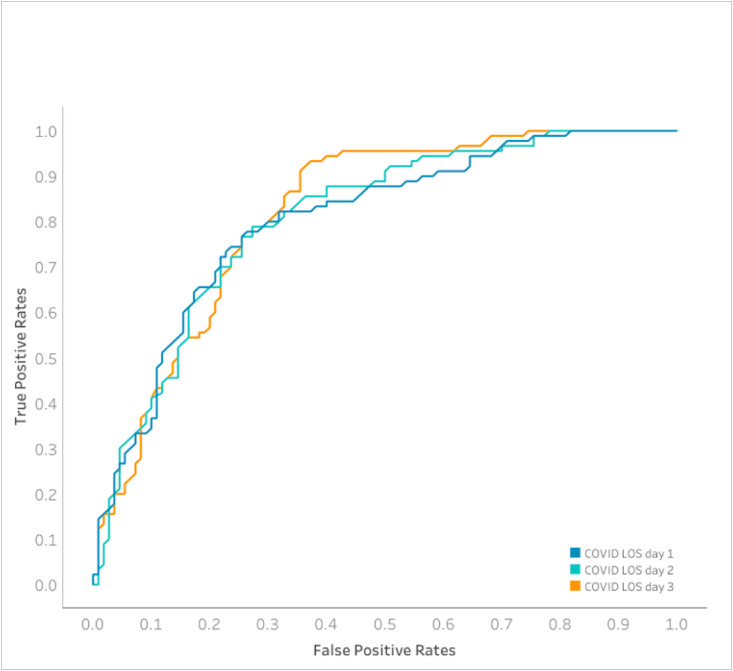
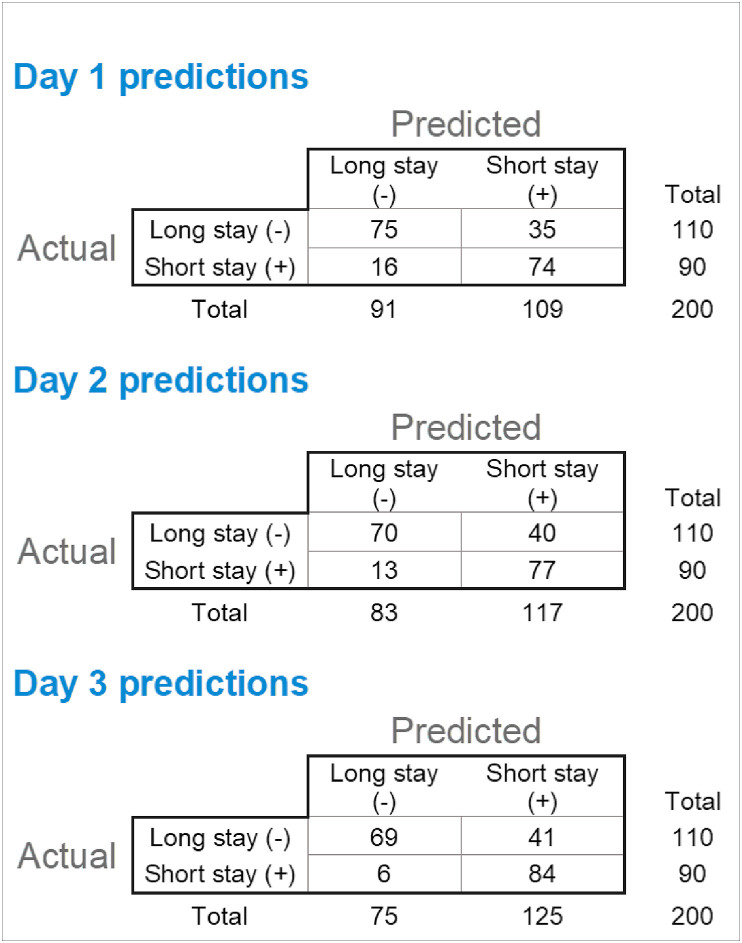
A total of 42 separate models were trained on the data and ranked based on their performance on the AUC metric for predicting short LOS (Supplemental Table 2). For all 3 prediction tasks, ensemble-based models performed best (ENET Blender for days 1 and 2 models and Advanced AVG Blender for day 3 model). Model performance improved with increasing data, with the models trained on culminative day 3 data demonstrated the highest sensitivity (0.93), accuracy (0.765) and AUC (0.819). The sensitivity, specificity and accuracy for the DataRobot ensemble model for each of the first 3 days of hospitalization are shown in Table 1 and AUC for each plotted in Fig. 2 . Fig. 3 shows the comparison of the 2 X 2 confusion matrix for each of these models on the validation data set (n = 200). Model performance was similar on both training and validation data sets for the majority of models, indicating that the model did not overfit to the training data set. Calibration was similar for the top performing model at each time point. In all cases, accuracy was best when the model was predicting values between 0.0 and 0.015 (predicting a long stay for a given patient) (Supplemental Figure 2).
Table 1.
Model statistics comparison.
| Model | AUC | Sensitivity | Specificity | Accuracy | Precision | F1 |
|---|---|---|---|---|---|---|
| 1 day of stay model | 0.803 | 0.82 | 0.68 | 0.745 | 0.68 | 0.74 |
| 2 days of stay model | 0.807 | 0.86 | 0.64 | 0.735 | 0.66 | 0.74 |
| 3 days of stay model | 0.819 | 0.93 | 0.63 | 0.765 | 0.67 | 0.78 |
Fig. 2.
Area under the curve comparison for COVID LOS models created on different of a patient’s LOS
Fig. 3.
Model outcome comparison
Feature importance
For all models developed as part of this study, feature importance was calculated based on scaled (0–1) model accuracy degradation after permutation of feature values (Supplemental Table 3). The top 5 features for the day 1 model were: age at time of admission (feature importance of 1.0), Interleukin 6 values (0.35), the patient's most recent blood urea nitrogen level (0.31), the patient's first temperature measurement (0.31), and whether or not the patient indicated alcohol use (0.24). For the day 2 models, the top 5 features were prediction from the patient's first day of stay (1.0), age at time of admission (0.65), difference in oxygen flow rate from beginning to end of measurement period (0.41), the date of admission (0.32) and the most recent blood urea nitrogen level (0.25). Finally, for the day 3 model, the top 5 features were prediction from the patient's second day of stay (1.0), age at time of admission (0.59), average respiratory rate over the last 12 h (0.28), most recent oxygen flow rate measurement (0.27), and difference in oxygen flow rate from beginning to end of measurement period (0.26).
Discussion
Our results demonstrate that machine learning algorithms, particularly ensemble algorithms, may be useful new tools in predicting hospital LOS, even for novel disease states, such as COVID-19. Physicians have been attempting to predict hospital LOS for over 50 years [5], with varying levels of success. Understandably, LOS is often easier to predict, both clinically and using machine learning algorithms, for well-defined conditions and scheduled admissions such as orthopedic surgeries [6]. As a novel viral pathogen with unknown disease course, COVID-19 represents a unique challenge, leaving clinicians without the experiential knowledge upon which to base their LOS estimations. We present the development of 3 machine learning models (ENET Blender for days 1 and 2 models and Advanced AVG Blender for day 3 model) capable of identifying prolonged LOS among patients admitted with COVID-19, offering physicians and healthcare systems a new tool for predicting outcomes and to plan for hospital capacity needs during the ongoing global pandemic. Model accuracy increased steadily with additional hospitalization data, reaching an AUC of 0.819 by day 3 of hospitalization.
The global focus on battling COVID-19 has provided some insights into important clinical variables that may predict more severe illness. A systematic review demonstrated that, with the exception of China, COVID-19 patients experienced a median hospital LOS of 5 days, but this time period frequently exceeded 3 weeks [7]. In fact, studies in the US indicate that among hospitalized COVID-19 patients, over 41% spent greater than 9 days in the hospital [8]. Patient age [9], presenting temperature [10] and inflammatory marker levels, such as IL-6 [11], have been previously shown to be markers of severity of illness. As expected, our results were also consistent with age as a feature associated with COVID-19 illness severity. Importantly, clinical experience has not yet elucidated what levels of these variables will aid in differentiation between patients who are likely to suffer a prolonged hospital stay and those who will not. Our data indicate that even small perturbations in these factors, for example a presenting temperature difference of only 0.3° Fahrenheit, which may be ignored clinically, may prove important in identifying patients with prolonged LOS.
Prior models have been developed to predict hospital demand across a geographic region via susceptible, infected, removed modeling [12]. Our approach, however, allows for institution-specific estimates using clinical data, allowing for the development of accurate and actionable models at the hospital level. For example, an early awareness of a high number of prolonged LOS COVID-19 patients would allow a hospital to cancel elective procedures, reduce non-urgent transfers from other facilities and expand bed capacity in advance of a potential surge in hospital census. Conversely, a large number of patients predicted to have short LOSs could provide valuable insights for planning care of non-COVID patients. Another benefit of the developed models includes the ease of access of the input variables, which are extracted directly from the institutional EHR.
Machine learning models developed prior to the COVID-19 pandemic have demonstrated the ability to predict prolonged LOS, reaching AUCs as high at 0.84 [13]. Importantly, these results were obtained when examining patients admitted with a multitude of know medical conditions, not including COVID-19. Further, among the most important features in this model was the primary diagnosis at the time of admission, indicating that the reason for hospitalization greatly affects the model's accuracy. As such, our models' ability to identify prolonged LOS with an AUC of 0.819 for a novel disease represents an early and robust result.
We found a rapid decrease in feature importance following patient age and prior model outputs, with other features individually contributing relatively less to overall predictive power. In the context of the models’ robust AUC, this trend in feature importance supports the use of machine learning algorithms that incorporate numerous variables to provide the best predictive output.
There are several limitations of our study that should be considered. The patient population and clinical data were drawn from a single center which may limit generalizability. Given its geographic location, however, the patient population of Cedars-Sinai Medical Center represents one of the most diverse cohorts in the country. The single-center nature of the study also limits the number of patients meeting inclusion criteria. Despite this, the developed algorithms were able to accurately differentiate patients predicated LOS. Future prospective studies, particularly using external datasets from a different geographic location, would provide further validation of these findings. Further, given the rapid development of new therapeutic options for the treatment of COVID-19, such as steroid therapy and convalescent plasma, over time our algorithms may be affected by the introduction of these interventions. A benefit of such model development, however, includes the ability to adapt as new factors, including treatment modalities, are captured in the EHR. Finally, the testing of multiple ML models raises potential limitations around model tuning and multiple testing. Selection of inappropriate tuning parameters for a model may result in selection of a less effective model than may otherwise be found under other parameters. DataRobot addresses this issue by training the same model repeatedly using different standard hyperparameters and selecting the model that provides the highest AUC, minimizing, but not fully eliminating the risk of model parameter mismatch for the goal of a given model. Our results must be interpreted in the context of the recognized limitations of using clinically generated data from the EHR to develop multiple machine learning algorithms. Specifically, unlike in clinical trials, EHR data may contain unrecognized errors which may skew results. The testing of multiple models may compound this issue by introducing error through multiple testing. The decision to pursue testing of multiple models was borne from the lack of clinical information available on the novel SARS-CoV-2 pathogen and what variables may be most predictive for prolonged LOS. Without this prespecified clinical knowledge base, the use of multiple models allowed for inclusion of a greater number of parameters and model fits, with the goal of finding the highest AUC.
In conclusion, the development of machine learning algorithms offer a novel approach to tackling the pressing concern of hospital capacity during the ongoing global pandemic. This work demonstrates that these algorithms are accurate and can be developed for novel disease states for which clinical knowledge is yet unavailable, enhancing clinicians’ ability to make early determinations. Such hospital-level predictions may provide actionable information for healthcare systems and providers in order to maximize capacity to care for a large and critically ill patient population. Lessons learned from these methodologies may be used in the future, if or when we are faced with similar crises.
Funding
This work was supported by NIH/NCI grant U54-CA260591 and NIH/NHLBI grant K23HL153888. The work was additionally supported in part by Cedars Sinai Medical Center and the Erika J. Glazer Family Foundation. Dr Chugh holds the Pauline and Harold Price Endowed Chair at Cedars-Sinai.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ibmed.2021.100035.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Centers for Disease Control and Prevention. CDC COVID data tracker. CDC; Access Date: October 2020.
- 2.The COVID Tracking Project. National: hospitalization. The atlantic monthly group; Access Date: October 2020.
- 3.Gasparini A. Comorbidity Scores. The Comprehensive R Archive Network. Institute for Statistics and mathematics of WU; Access Date: September 2020.
- 4.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) Ann Intern Med. 2015;162:735–736. doi: 10.7326/L15-5093-2. [DOI] [PubMed] [Google Scholar]
- 5.Robinson G.H., Davis L.E., Leifer R.P. Prediction of hospital length of stay. Health Serv Res. 1966;1:287–300. [PMC free article] [PubMed] [Google Scholar]
- 6.Carter E.M., Potts H.W. Predicting length of stay from an electronic patient record system: a primary total knee replacement example. BMC Med Inf Decis Making. 2014;14:26. doi: 10.1186/1472-6947-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees E.M., Nightingale E.S., Jafari Y., Waterlow N.R., Clifford S., Pearson CA B., et al. COVID-19 length of hospital stay: a systematic review and data synthesis. BMC Med. 2020;18:270. doi: 10.1186/s12916-020-01726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson M., Bach P., Baldwin M.R. Hospital length of stay for severe COVID-19: implications for Remdesivir’s value. medRxiv. 2020 doi: 10.1101/2020.08.10.20171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrilli C.M., Jones S.A., Yang J., Rajagopalan H., O'Donnell L., Chernyak Y., et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1966. m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman G.E., Crane-Droesch A., Chivers C., Luong T., Hanish A., Levy M.Z., et al. Locally informed simulation to predict hospital capacity needs during the COVID-19 pandemic. Ann Intern Med. 2020;173:21–28. doi: 10.7326/M20-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton C.B., Milinovich A., Felix C., Vakharia N., Crone T., Donovan C., et al. Personalized predictions of patient outcomes during and after hospitalization using artificial intelligence. NPJ Digit Med. 2020;3:51. doi: 10.1038/s41746-020-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.